
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol., 24 November 2023
Sec. Molecular and Cellular Pathology
Volume 11 - 2023 | https://doi.org/10.3389/fcell.2023.1267792
Circular RNAs (circRNAs) belong to a unique class of endogenously expressed non-protein-coding RNAs with a distinct circularized structure, characterized by the absence of 5′-cap and 3′-polyadenylate ends. They are generally formed through back-splicing from pre-mRNAs. They serve as regulators of transcription and splicing, and act as sponges for microRNAs (miRNAs) and RNA-binding proteins, thereby modulating the expression of target genes. As a result, they exert a substantial impact on a diverse array of cellular and biological processes, including cell proliferation, migration, inflammation, and oxidative stress. Asthma and COPD are chronic airway conditions that currently have no cure. In recent years, emerging evidence suggests that altered expression of circRNAs in airway, bronchial and immune cells is involved in asthma and COPD pathogenesis. Studies exploring circRNA dysregulation in asthma have showcased their involvement in regulating the proliferation, migration, and inflammation of airway smooth muscle and bronchial epithelial cells, as well as impacting goblet cell metaplasia, Th2 cell differentiation, and macrophage activation, primarily through interactions with miRNAs. Similarly, in COPD, circRNAs have shown altered expression patterns in the blood and lungs of patients, and these changes have been linked to modulating inflammation, oxidative stress, and airway remodeling in preclinical models. Furthermore, certain circRNAs have demonstrated promising potential as diagnostic and prognostic biomarkers for both asthma and COPD. This review delves into the current understanding of the function and molecular mechanisms of circRNAs in asthma and COPD, along with exploring their potential as biomarkers in these respiratory conditions.
Chronic respiratory diseases (CRDs) represent a group of lung conditions that affects the respiratory system, particularly the airways and lungs. These diseases are characterized by reduced lung function, and, in some cases, irreversible damage to respiratory tissues. In 2019, CRDs ranked as the third leading cause of global mortality associated with a significant social and economic burden, thereby posing a significant healthcare challenge (Diseases and Injuries, 2020; Syamlal et al., 2020). The most common CRDs include asthma and chronic obstructive pulmonary disease (COPD).
Asthma is a multifaceted and heterogeneous airway condition, affecting an estimated 260 million people globally (Ali et al., 2020; Liu et al., 2022; The Global Asthma Report, 2022). Its cardinal clinical manifestations encompass airflow obstruction, breathing difficulties, coughing, chest tightness, and wheezing (Habib et al., 2022). While asthma can develop at any age, it is most common in children. The exact cause of asthma remains unknown; however, it is believed to arise from a combination of genetic factors (individuals with a family history of asthma) and environmental influences (such as allergies, air pollution, cold air, and exercise). These contributing factors influence the pathogenic changes within the airways, including inflammation, mucous hypersecretion, airway remodeling, airway hyper-responsiveness (AHR), and airway obstruction (Gysens et al., 2022). Airway remodeling involves significant structural alterations in the airways, which include hypertrophy/hyperplasia of airway smooth muscle cells (ASMCs), alterations in airway epithelial cells (AECs), sub-epithelial fibrosis, goblet cell metaplasia, and increased expression of angiogenic factors (Curran and Cohn, 2010). While available medications can effectively manage the symptoms, a cure for the disease is currently lacking. Therefore, there is a pressing need to develop an effective therapy that can target the underlying causes of the disease.
Chronic Obstructive Pulmonary Disease (COPD) comprises a spectrum of lung conditions, including pulmonary emphysema and chronic bronchitis, which lead to breathing difficulties and airflow obstruction (Guo et al., 2022). COPD is a major global contributor to mortality and morbidity, placing a substantial economic and social burden worldwide (Safiri et al., 2022). While COPD can affect individuals of all ages, it is more commonly observed in older populations (Tudorache et al., 2017; Matera et al., 2023). Although smokers are at a higher risk of developing COPD, non-smokers can also be affected by the disease due to exposure to environmental pollutants, occupational dust and chemicals, respiratory infections, and genetic factors (Feizi et al., 2022; He et al., 2023). Despite extensive efforts to identify effective therapies for COPD, the disease remains incurable, and the existing treatment options only aim to manage clinical symptoms, slow down disease progression, and enhance the quality of life (Bollmeier and Hartmann, 2020). Consequently, the development of efficient treatments for this debilitating condition is urgently needed.
Non-coding RNAs (ncRNAs) are a class of RNA molecules that do not encode proteins. NcRNAs play essential regulatory roles in various cellular and biological functions, including cellular proliferation, migration, apoptosis, angiogenesis, inflammation, and metabolism in both health and disease (Bhatti et al., 2021). Based on their size, ncRNAs can be categorized into different subtypes, which include microRNAs (miRNAs, approximately 21–25 nucleotides in length), long non-coding RNAs (lncRNAs, over 200 nucleotides), circular RNAs (circRNAs, roughly 100 nucleotides to 4 kilobases, but in a circularized form), and more. They are involved in the regulation of gene expression at various levels (Bhatti et al., 2021). NcRNAs have gained significant attention in recent years due to their roles in various diseases, including cancer, neurodegenerative disorders, and lung diseases (Bhatti et al., 2021). Their study has opened new avenues for understanding disease mechanisms and exploring potential therapeutic interventions. While miRNAs and lncRNAs have been extensively investigated in both healthy and diseased states, circRNAs have received comparatively less attention in research.
CircRNAs represent a distinct class of endogenously expressed circularized RNA molecules lacking 5′end caps and 3′poly(A) ends. Generally, circRNAs do not have the capacity to encode proteins. Nevertheless, recent studies have unveiled some exceptions, where certain circRNAs have been found to translate into proteins. An example of this is circ-FBXW7, which has been identified as a circRNA capable of translating into the protein FBXW7-185aa (Yang et al., 2018), although, its functions remain largely unknown. CircRNAs are characterized by their distinct covalently closed ring structure, mainly generated via head-to-tail splicing. This structure of circRNAs confers protection against degradation by RNA endonucleases, resulting in their remarkable stability and high abundance in blood and tissues (Wang et al., 2017; Li X. et al., 2018). Additionally, this structural characteristic helps them to be highly conserved across different species. Most circRNAs display prominent expression patterns that are specific to particular cells, tissues, and stages of development.
Most circRNAs are generated through back-splicing, which involves the circularization of pre-mRNA derived from protein-coding genes, antisense transcripts, the intergenic spacer in tRNA, or lncRNAs (Ali et al., 2022). CircRNAs can originate from both exons and intron regions. Currently, circRNAs are categorized into four groups based on their synthesis mechanisms and the genomic splice junctions where they are formed: intronic, exonic, exon-intron, and tRNA intronic circRNAs, the latter being generated by tRNA introns. Exonic circRNAs, being the most prevalent type among circRNAs, are primarily located in the cytoplasm. In contrast, other types of circRNAs are mostly detected in the nucleus, suggesting their potential role in regulating gene transcription. There are three well-established models for circRNA formation: 1) Intron pairing-driven circularization, where inverted repeat elements like ALU repeats, facilitate splicing by bringing splice donor and acceptor sites together; 2) intron lariat generation and exon skipping and, where exonic circRNAs are formed by joining exon ends; and 3) RBP-associated mechanism, where RBPs bridge flanking introns to facilitate loop formation (Figure 1). The biogenesis of the circRNAs can be modulated by several factors, including the cis-elements (intronic complementary sequences), canonical spliceosomal machinery, and transfactors (RBPs) (Ali et al., 2022).
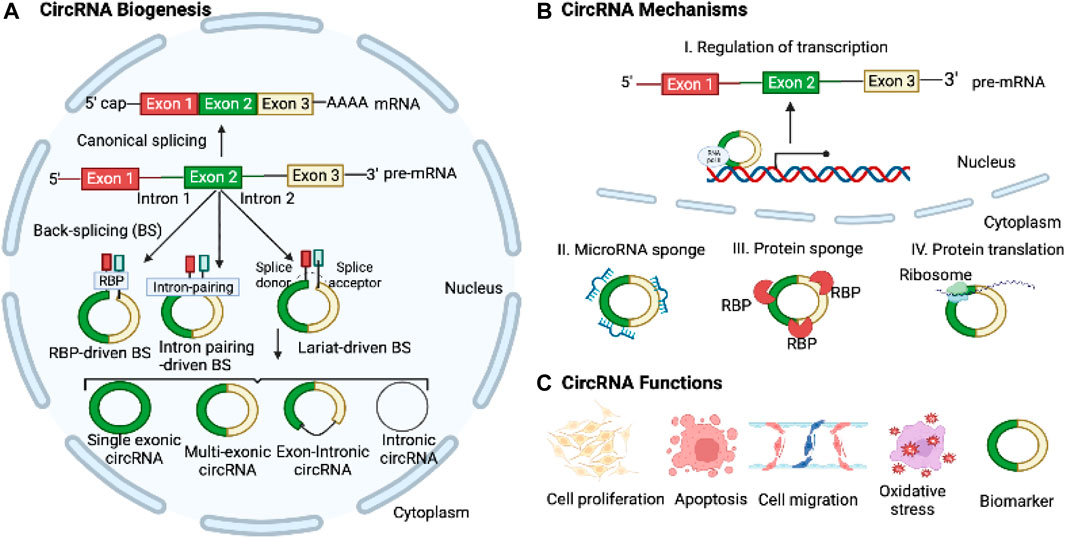
FIGURE 1. Biogenesis, Mechanisms, and Functions of CircRNAs. (A) CircRNAs are derived from pre-mRNAs through three widely accepted mechanisms: intron-pairing-driven, RBP-mediated, and lariat-driven back-splicing processes. (B) CircRNAs play regulatory roles in transcription and translation and act as microRNA sponges to control target gene expressions. (C) CircRNAs can modulate cell proliferation, apoptosis, migration, EMT, and oxidative stress. Moreover, they hold potential as diagnostic and prognostic biomarkers for various diseases.
CircRNAs have emerged as key regulators in a diverse array of physiological, biological, and cellular processes, encompassing cellular proliferation, migration, and apoptosis through their interactions with other RNAs and proteins (Salzman, 2016; Li X. et al., 2018; Mao and Xu, 2020; Ali et al., 2022). MicroRNA sponging is a well-established mechanism of circRNAs, wherein they modulate the expression of target mRNA genes by competitively binding to specific microRNAs (Salzman, 2016). CircRNAs also bind to RBPs and other proteins, acting as translational regulators and serving as protein scaffolds to regulate target gene expression (Salzman, 2016). Recently, it has been discovered that circRNAs can also be translated into polypeptides (Zhou et al., 2020). The covalently closed structure of circRNAs gives them a longer half-life and greater resistance to RNase R compared to linear RNAs. Moreover, circRNAs exhibit differential expression patterns in various diseases, encompassing diverse conditions such as cancer and cardiovascular diseases (Altesha et al., 2019; Li et al., 2021; Kristensen et al., 2022). Given these features, circRNAs hold great promise as potential diagnostic and therapeutic biomarkers for a wide range of diseases.
Recent findings indicate dysregulation of circRNAs in both the blood and lungs of patients with asthma and COPD (Tables 1–4). These circRNAs have demonstrated significant regulatory effects on the pathophysiology of these respiratory conditions (Figures 2, 3). This review presents a comprehensive overview of circRNAs and their relevance to the pathophysiology of asthma and COPD. We specifically concentrate on asthma and COPD among the various CRDs, as they are the most prevalent, and currently there is no comprehensive reviews solely regarding the involvement of circRNAs in these conditions. Our focus lies primarily on the function and mechanisms of circRNAs in the pathophysiology of these respiratory conditions. Moreover, we delve into the potential of circRNAs as biomarkers and therapeutic targets for asthma and COPD.
Although mild asthma is effectively manageable, severe asthma remains a substantial source of economic and social burden. It often involves substantial changes in ASMCs, such as hyperplasia, hypertrophy, and alterations in inflammation and fibrosis-related proteins (Camoretti-Mercado and Lockey, 2021). These aberrant ASMCs greatly influenced airway remodeling in severe asthma (Hershenson et al., 2008). Unfortunately, there are limited interventions to address these ASMC abnormalities. Thus, a solid understating the pathogenesis of asthma is crucial for developing effective therapeutic strategies to alleviate the impact of severe asthma on ASMCs and improve patient outcomes.
Recent advancements in next-generation sequencing technology have facilitated the identification of numerous circRNAs across a wide range of cell types and species (Kristensen et al., 2019). However, limited research has explored the involvement of circRNAs in ASMCs dysfunction in asthma. Over the past few years, a growing body of research studies has uncovered circRNAs as regulators of various cellular and biological processes in asthmatic ASMCs, including proliferation, apoptosis, migration, ECM production, and secretion of inflammatory mediators, primarily through acting as ceRNAs of miRNAs to regulate target gene mRNAs.
One such circRNA is circERBB2, derived from the ERBB2 gene locus, associated with tumorigenesis. Elevated circERBB2 expression was observed in bronchial biopsy tissues of asthmatic individuals and in Platelet-Derived Growth Factor BB (PDGF-BB)-stimulated ASMCs, an in vitro model of asthma (Huang J. Q. et al., 2021). PDGF-BB is a well-known factor that stimulates airway remodeling and inflammation in asthma (Kardas et al., 2020). It promotes proliferation and migration of ASMCs (Ito et al., 2009; Spinelli et al., 2012). It is also crucial to highlight that the upregulation of PDGF-BB expression is closely associated with the severity of asthma, and increased proliferation and migration of ASMCs are commonly observed in individuals with asthma (Ohno et al., 1995). Knockdown of circERBB2 using small hairpin RNA (shRNA) led to significant amelioration of inflammation, proliferation, and migration in ASMCs induced by PDGF-BB (Huang J. Q. et al., 2021). Mechanistically, circERBB2 functions as a sponge for miR-98-5p, modulating IGF1R expression and accelerating ASMC proliferation and migration upon PDGF-BB treatment (Huang J. Q. et al., 2021).
Another circRNA, circHIPK3, also known as circ_0000284, was upregulated in ASMCs when stimulated with PDGF-BB (Lin et al., 2020; Jiang et al., 2021). CircHIPK3 was found to promote migration and proliferation while suppressing apoptosis in PDGF-BB-stimulated ASMCs. It achieved this through interacting with miR-326 to regulate stromal interaction molecule 1 (STIM1) (Lin et al., 2020), an endoplasmic reticulum membrane protein known that acts as a calcium sensor and stimulator of ASMCs remodeling and AHR in asthma (Zou et al., 1985; Johnson et al., 2022). Moreover, circHIPK3 was shown to boost ASMC proliferation, invasion, and migration upon PDGF treatment, potentially through the regulation of the miR-375/matrix metalloproteinase-16 (MMP-16) axis (Jiang et al., 2021).
PDGF-treated ASMCs also exhibited upregulation of circ_CSNK1E and circ_0002594, while the expression of circ_0000029 was downregulated (Ding et al., 2022; Quan et al., 2022; Wang R. et al., 2023). Furthermore, the expression of circ_0002594 and circ_CSNK1E was significantly increased in samples obtained from patients with asthma (Ding et al., 2022; Quan et al., 2022). Functional studies demonstrated that the shRNA-mediated knockdown of circ_CSNK1E significantly reduced the proliferative and migratory responses of ASMCs to PDGF (Ding et al., 2022). Computation analysis and experimental validation studies revealed that circ_CSNK1E binds to miR-34a-5p to modulate vesicle-associated membrane protein 2, thereby accelerating PDGF-induced ASMCs proliferation and migration (Ding et al., 2022). Moreover, circ_0002594 was identified as a facilitator of inflammation, migration, and proliferation in ASMCs in response to PDGF treatment. The authors further demonstrated that circ_0002594 acts as a sponge for miR-139-5p to modulate the expression of tripartite motif 8 (TRIM8), which in turn stimulates cellular damage (Quan et al., 2022). Studies have also shown that miR-139-5p treatment and TRIM8 inhibition decreases ASMCs proliferation through inhibiting a chromatin remodeling factor BRG1, and inactivating NF-kb signaling pathway, respectively (Zhang et al., 2017; Dang et al., 2020). Another circRNA, circ_0000029 was found to regulate ASMC migration and proliferation by modulating miR-576-5p and KCNA1 expression (Wang R. et al., 2023). These findings underscore the substantial role of circRNAs in modulating critical functions in asthmatic ASMCs through their interactions with microRNAs and target genes. However, the full extent of circRNA functions with other mechanisms in asthmatic ASMCs remains largely unexplored. Further research is required to gain a comprehensive understanding of the role played by circRNAs in asthmatic ASMCs.
Airway epithelial cells serve as the initial protective barrier, guarding the airway and lungs against inflammatory stimuli and antigens. Dysfunction of airway and bronchial epithelial cells is commonly found in all types of asthma. Emerging evidence indicates that aberrant expression of circRNAs may exert a substantial influence on the functionality of airway and bronchial epithelial cells (Table 1).
Jia et al. conducted a comprehensive profiling study of circRNAs using micro-arrays in human bronchial epithelial cells (BEAS-2B) exposed to PM2.5, a known respiratory hazard associated with lung diseases, including asthma (Jia et al., 2020). Among the dysregulated circRNAs, circ_406961 showed a remarkable fold change difference. Experimental investigations revealed a dose-dependent decrease in circ_406961 expression upon PM2.5 exposure in BEAS-2B cells. Silencing circ_406961 using siRNA reduced cell viability and increased inflammation, while its overexpression reversed these effects. Mechanistic analysis highlighted the role of circ_406961 in modulating inflammation by interacting with ILF2 and activating the STAT3/JNK pathways.
Another circRNA circVPS33A was shown to be upregulated in the plasma of asthmatics and BEAS-2B cells treated with house dust mites (HDM) protein (Der p1) (Su et al., 2021). Der p1 treatment resulted in decreased cell viability, proliferation, migration, invasion, and induced apoptosis, autophagy, and inflammation in BEAS-2B cells. Silencing circVPS33A using siRNA ameliorated these cellular damages induced by Der p1. Mechanistically, circVPS33A was shown to promote these cellular damages by acting as a sponge for miR-192-5p, leading to modulating the expression of HMGB1. Furthermore, circARRDC3 was found to enhance inflammation and mucus production in nasal epithelial cells stimulated with IL-13 in allergic rhinitis, a substantial risk factor for the development of asthma (Wang T. et al., 2021). This regulatory effect was mediated through the miR-375/KLF4 signaling axis. Together, these findings suggest that dysregulated circRNAs may contribute to bronchial epithelial cell dysfunction associated with asthma. However, these findings are needed to further confirm in in vivo animal models and clinical studies.
Airway goblet cells play a vital role in maintaining airway homeostasis through the production and secretion of mucins (Whitsett, 2018). In response to genetic factors, growth factors, inflammation or environmental insults, goblet cells differentiate from club (Clara) cells, a type of airway epithelial cells (Whitsett, 2018; Kim et al., 2019). Uncontrolled differentiation of the goblet cells leads to increased production and secretion of mucins within the airways (Fahy and Dickey, 2010). Excessive accumulation of mucins is a common manifestation of many airway diseases, such as asthma, COPD, and cystic fibrosis (Fahy and Dickey, 2010). In asthma, goblet cell hyperplasia (increased number of goblet cells) is one of the major features of airway remodeling and is associated with chronic mucus hypersecretion and resultant airway obstruction (Rogers, 2002). Recently, circRNAs have been shown to induce goblet cell hyperplasia. For example, Wang et al., discovered that circZNF652, also known as circ_0000782, can promote goblet cell hyperplasia in allergic epithelial (Wang X. et al., 2022). CircZNF652 exhibited significant and specific expression in the airway epithelium of both children with asthma and ova-induced mice models of experimental asthma (Wang X. et al., 2022). The increased expression of circZNF652 was found to contribute to the promotion of bronchial goblet cell metaplasia and the excessive secretion of mucus. It was discovered that circZNF652 accelerated goblet cell metaplasia through sponging miR-452-5p and activating JAK2/STAT6 signaling axis (Wang X. et al., 2022). Furthermore, the splicing factor ESRP1 was identified as a facilitator of circZNF652 biogenesis, thereby accelerating goblet cell metaplasia (Wang X. et al., 2022). These findings highlight the significant role of the ESRP1/circZNF652/miR-452-5p/JAK2/STAT6 axis in the modulation of goblet cell metaplasia, mucus hypersecretion, and AHR in mice models of experimental asthma. Therefore, targeting circZNF652 or activating miR-452-5p could offer a potential therapeutic strategy for intervening in epithelial remodeling in experimental asthma. Nevertheless, the exact underlying mechanism responsible for the elevation of ESRP1 expression in allergic airway epitheliums is yet unclear. Furthermore, circZNF652 has been identified as a regulator of EIF5, RhoA, JAK2, and SRSF3. While the involvement of JAK2 in goblet cell metaplasia in asthma has been established, the roles of other factors in this process are still unclear and require further investigation.
Asthma involves the infiltration and activation of various immune cells in the airways. These immune cells encompass dendritic cells (DCs), T cells (Th1, Th2, Th9, Th17, Th22 cells), innate lymphoid cells (ILCs), eosinophils, neutrophils, B cells, and mast cells (Lommatzsch, 2020). Asthma can be categorized into two primary types: type 2 (the most common) and non-type 2 asthma. Type 2 asthma is typically triggered by allergens but can also be provoked by non-allergic factors. In contrast, non-type 2 asthma is primarily induced by non-allergic factors such as pollutants and infections. Upon recognizing allergens, Th2 cells or ILCs, especially ILC2, release cytokines like IL-4, IL-5, and IL-13, leading to the recruitment of eosinophils, which cause airway inflammation (Lommatzsch, 2020). Mast cells release histamine, contributing to bronchoconstriction. Neutrophils may also be involved in severe asthma. Dendritic cells present antigens to T cells, initiating immune responses. Macrophages can promote either inflammation or resolution depending on the types of macrophages (M1 or M2). B cells produce IgE antibodies, which trigger allergic responses. These immune mechanisms collectively lead to the characteristic symptoms of asthma, such as bronchoconstriction and AHR.
Emerging studies have provided substantial evidence supporting the pivotal role of circRNAs as competing endogenous RNAs (ceRNAs) for microRNAs in T cell development and function. An example of circRNA involvement in immune regulation is the LPS-induced circRNA, m_circRasGEF1B, which regulates the stability of ICAM-1 mRNA, contributing to immune regulation in the body (Ng et al., 2016). ICAM1, a cell surface glycoprotein, is expressed in endothelial cells and specific immune cells and is well-recognized as a biomarker for inflammation. Analysis of circRNA expression profiles in different cell subpopulations has been demonstrated to possess distinct expression profiles, indicating their regulatory roles in specific cellular processes. For example, studies have revealed that the decreased hsa_circ_0012919 expression leads to CD11a and CD70 DNA methylation in CD4+ T cells (Wang et al., 2015; Li L. J. et al., 2018; Zhang et al., 2018; Gaffo et al., 2019). Furthermore, hsa_circ_0045272 has been demonstrated to potentially act as a negative regulator of T cell apoptosis promotion and IL2 release by interacting with hsa-miR-6127 (Li L. J. et al., 2018). Moreover, circTNIK, circIKZF1, circFBXW7, and circTXK are circRNAs specifically expressed in T cells (Gaffo et al., 2019).
In recent years, a wealth of evidence has emphasized the substantial role of circRNAs in the pathophysiology of asthma through modulating immune responses (Ghafouri-Fard et al., 2020; Chen et al., 2021). Substantial evidence underscores the essential role of CD4+ T cells in driving the disease by regulating IgE production in response to allergic conditions, secreting pro-inflammatory mediators, and activating and recruiting inflammatory cells like eosinophils, macrophages, and neutrophils (DiCosmo et al., 1994; Erpenbeck et al., 2003; Ferreira, 2003; Fujiwara et al., 2007; Holgate, 2008; Lloyd and Hessel, 2010; Fujimoto et al., 2011; Lambrecht and Hammad, 2015; Chen et al., 2016; Cheng et al., 2016; Hammad and Lambrecht, 2021). CD4+ T cells exhibit notable cytokine production abilities, with Th1 cells producing IFN-γ, Th2 cells secreting IL-4, IL-5, IL-6, IL-9, and IL-13, and Th17 cells releasing IL-17A and IL-17F (Lambrecht and Hammad, 2015). The complex immune response observed in asthma is regulated by this intricate network of diverse cytokines (Erpenbeck et al., 2003; Zhu and Paul, 2010; Fujimoto et al., 2011; Chen et al., 2016; Cheng et al., 2016). To explore the critical involvement of circRNAs in CD4+ T cells in asthma, Huan et al. profiled the expression of circRNAs using micro-array analysis in CD4+ T cells isolated from 5 asthmatics and 5 healthy subjects (Huang et al., 2019). This study revealed 597 dysregulated circRNAs in asthmatic patients compared to the control group. Among the dysregulated circRNAs, hsa_circ_0005519 exhibited one of the highest fold of up-regulations, which was further validated through subsequent qRT-PCR analysis in the existing cohort samples as well as in a separate cohort comprising 65 asthma patients and 30 healthy subjects. The authors employed bioinformatic analysis, reporter assays, and literature search to uncover that hsa_circ_0005519 functions as a regulator of the expression of IL13 and IL6, possibly through interacting with let-7a-5p. Notably, IL-13 and IL-6 have been widely recognized for their significant involvement in asthma pathogenesis (DiCosmo et al., 1994; Zhu et al., 1999; Pope et al., 2001; Kimura and Kishimoto, 2010). Furthermore, the expression of hsa_circ_0005519 in CD4+ T cells was found to exhibit a negative correlation with let-7a-5p, and a positive correlation with IL13 and IL6 mRNA, as well as FeNO and peripheral blood eosinophil ratio. These results indicate that hsa_circ_0005519 may be a promising biomarker for asthma.
In a separate study, the authors identified another circRNA, hsa_circ_0002594, which exhibited upregulation in CD4+ T cells using the existing micro-array dataset consisting of 5 patients with asthma and 5 healthy controls (Huang Z. et al., 2021). To validate the findings, a second cohort consisting of 83 asthmatics and 54 healthy subjects was examined, and the result revealed a significant increase in hsa_circ_0002594 levels in CD4+ T cells of asthmatics (Huang Z. et al., 2021). In asthmatics, the expression of hsa_circ_0002594 in CD4+ T cells showed a positive correlation with FeNO and a negative association with PD20 (the methacholine dose required to induce a 20% decrease in FEV1). The authors performed a correlation analysis of hsa_circ_0002594 expression in the subgroups of asthmatic patients with clinical features. The researchers observed that the subgroup with high expression of hsa_circ_0002594 exhibited elevated FeNO levels and lower PD20 values compared to the subgroup with low expression of hsa_circ_0002594. Furthermore, the subgroup with high expression of hsa_circ_0002594 had higher Th2-high and SPT-positive individuals, when compared to non-Th2 inflammation and SPT-negative cases. These findings suggest that the subgroup of asthmatics with high expression of hsa_circ_0002594 also exhibits clinical indicators consistent with Th2-driven allergic response. The treatment with inhaled ICS led to a significant reduction in hsa_circ_0002594 expression, highlighting its potential as a therapeutic target. Moreover, in exploring the diagnostic potential of hsa_circ_0002594 for asthma, the authors observed that its expression demonstrated high sensitivity and specificity values in diagnosing asthma, regardless of the presence or absence of ICS treatment. Furthermore, the authors investigated the potential molecular mechanisms of hsa_circ_0002594 in the development of asthma through computational analysis and identified potential target miRNAs of the circRNA. They proposed the hsa_circ_0002594 could competitively sequester the activity of hsa-let-7e-5p, hsa-miR-503-5p, hsa-miR-16-5p, hsa-miR-587, and hsa-miR-514a-3p. Although this study had several limitations: 1) Small sample size necessitates larger, diverse studies. 2) MicroRNA interactions with hsa_circ_0002594 lack experimental validation. 3) Circulating hsa_circ_0002594 levels were not assessed for biomarker potential.
In a recent study, it was discovered that mmu_circ_0001359 exhibited downregulation in the lungs of an OVA-induced murine asthma model (Shang et al., 2020). Moreover, the delivery of exosomes derived from ADSCs, which were modified with mmu_circ_0001359, was observed to mitigate airway remodeling by facilitating the activation of M2-like macrophages through the regulation of the miR-183-5p/FoxO1 signaling pathway (Shang et al., 2020).
In another recent study, circRNAs were profiled in the lung of mice with experimental asthma developed by HDM (Bao et al., 2020). Through the analysis of the circRNA-miRNA signaling axis, two upregulated circRNAs, circ_0000629 and circ_0000455, were identified to target miR-29b and miR-15a, respectively. Earlier investigations have demonstrated an inverse association between these miRNAs and the occurrence of allergic reactions (Nakano et al., 2013; Yan et al., 2019). In contrast, two downregulated circRNAs, circ_0000723 and circ_0001454 were found to target miR-214 and miR-146b respectively. MiR-214 and miR-146b have previously been positively associated with asthma (Feng et al., 2012; Qiu et al., 2017). Thus, these four circRNAs represent promising candidates for further investigation concerning their association with asthma.
Increasing evidence suggests that circRNAs are implicated in the pathophysiology of COPD, as shown in Tables 3, 4 and Figure 3. To identify circRNAs involved in COPD, several circRNAs profile studies have been profiled using NGS technology. As an example, Duan et al., profiled circRNA expression in PBMCs of 21 patients with COPD and 21 healthy individuals (Duan et al., 2020). The analysis revealed 2,132 differentially expressed circRNAs and 2,734 differentially expressed mRNAs in COPD patients. The further in-depth analysis identified an association between circ_0008672 and key COPD pathways, including Th17 cell differentiation, NK cell-driven cytotoxicity, and NOD-like receptor pathway. The authors also constructed a circRNA-microRNA-mRNA network to investigate circRNAs’ role as microRNA sponges in COPD. Their findings showed that circRNAs can regulate mRNAs by acting as sponges for one or multiple miRNAs. For instance, circ_0008672 was discovered to interact with miR-1265 and thereby regulate MAPK1 expression. Notably, MAPK signaling activation plays a crucial role in the development of various pathophysiological features of COPD, including, but not limited to, lung inflammation, airway mucus hypersecretion, airway fibrosis, and T cell activation (Mercer and D'Armiento, 2006). These findings underscore the ceRNA network’s importance in COPD, but limitations exist. Validation in a larger COPD cohort is essential. Experimental studies on circRNA functions are needed for precise roles and also subsets of PBMCs warrant assessment for comprehensive insights.
Circ-HACE1, also known as circ_0077520, was over-expressed in the serum of 21 smokers and 24 smokers with COPD compared to 17 non-smokers (Zhou F. et al., 2021). Additionally, in human bronchial epithelial cells (16HBE cells) exposed to cigarette smoke extract (CSE), a well-established in vitro model of COPD, a significant increase in the expression of circ-HACE1 was observed (Zeng et al., 2019; Zhou F. et al., 2021). Targeting circ-HACE1 expression using siRNA demonstrated an improvement in CSE-induced cellular damage in 16HBE cells, as evidenced by increased cell viability and reduced inflammation, oxidative stress, and apoptosis. This beneficial effect was achieved through circ-HACE1’s interaction with miR-485-3p, which modulated the expression level of TLR4. TLR4 plays a crucial role in innate immune activation and is closely linked to inflammatory responses in several conditions, including diabetes. These findings highlight circ-HACE1 as a promising target to enhance the diagnostic accuracy of COPD and suggest that inhibiting its expression could hold potential benefits in the therapeutic management of COPD.
Furthermore, circ_0040929 showed significant upregulation in both serum of patients with COPD and 16HBE cells exposed to CSE (Miao et al., 2022). Circ_0040929 silencing protected from CSE-treated 16HBE cell injuries via the regulation of the miR-515-5p/IGFBP3 signaling axis. Circ-RBMS1, also known as has_circ_0002136, originates from the RBMS1 gene and was shown to be over-expressed in the PBMCs of patients with COPD (Duan et al., 2020). However, the specific role and molecular mechanisms of circ-RBMS1 were not further investigated in this circRNA profiling and bioinformatic analysis study. Recently, Qiao et al. also found upregulation of circ-RBMS1 in both patients with COPD (Qiao et al., 2021). Furthermore, circ-RBMS1 expression was shown to increase in a dose-dependent manner in CSE-treated 16HBE cells (Qiao et al., 2021). Functional analysis demonstrated that inhibiting circ-RBMS1 with siRNA resulted in the attenuation of CSE-induced 16HBE cell injuries. Mechanistically, circ-RBMS1 was found to interact with miR-197-3p to modulate FBXO11, a member of the F Box family of proteins known for its role in ubiquitination and degradation of substrates, as well as genome stability regulation. A previous study also associated FBXO11 with airway remodeling, apoptosis, and inflammation induced by CSE (Mei et al., 2020). However, it is important to consider that these findings were obtained from in vitro analysis. To validate and strengthen these results, further investigation using in vivo assays and a larger COPD patient cohort will be necessary.
CircFOXO3 was increased in mouse alveolar epithelial cells exposed to CSE and in the lung of mice with experimental COPD induced by CS (Zhou L. et al., 2021). However, silencing CircFOXO3 using lentivirus attenuated CS-induced lung inflammation in mice. This effect was mediated through the CircFOXO3/miR-214-3p/NF-κB signaling pathway, targeting the inflammatory response induced by CS in the lungs of mice. Thus, targeting circFOXO3 could serve as a novel preventive approach to counter CS-induced inflammation.
The Circ-OSBPL2 was found to regulate apoptosis, oxidative stress, and inflammation in HBECs exposed to CSE by modulating the miR-193a-5p/BRD4 axis (Zheng et al., 2021). Circ_0061052 was upregulated in CSE-exposed HBECs and modulated airway remodeling and EMT induced by CS through sponging miR-515-5p, thereby regulating downstream targets FoxC1 and Snail (Ma et al., 2020). Circ_0026466 exhibited upregulation in the blood of smokers with COPD and CSE-exposed HBECs (Wang C. et al., 2023). Knockdown of circ_0026466 mitigated CSE-induced inhibition of viability and proliferation, induced apoptosis, oxidative stress, and inflammation in HBECs through the circ_0026466/miR-153-3p/TRAF6/NF-κB pathway (Wang C. et al., 2023). Similarly, circXPO1 displayed upregulation in the lung of mice with experimental COPD induced by CS and CSE-treated AT2 cell lines (Du et al., 2022). Inhibition of circXPO1 was found to suppress inflammatory response and cellular senescence induced by CSE. Mechanistic experiments revealed that circXPO1 regulates TAB3 mRNA through sponging miR-23b-3p to modulate the inflammatory response and cellular senescence processes. The findings suggest that circXPO1 contributes to COPD pathogenesis potentially through sponging miR-23b-3p, thereby modulating TAB3 expression.
In contrast, the downregulation of circRNAs can also contribute to COPD pathogenesis. For instance, circ_0006892 was found to be downregulated in the lungs of COPD patients and CSE- 16HBE and BEAS-2B cells exposed to CSE, as measured by qRT-PCR (Zhang et al., 2022). Furthermore, the expression of circ_0006892 positively correlated with FEV1% in COPD patients. To explore the impact of circ_0006892, gain-of-function experiments were conducted on 16HBE and BEAS-2B cells exposed to CSE, assessing cell survival, apoptosis, and inflammation. The results indicated that over-expression of circ_0006892 through plasmid transfection mitigated cell apoptosis and the inflammatory response induced by CSE, indicating its protective function in CSE-associated bronchial epithelial injury. Conversely, inhibition of circ_0006892 led to increased apoptosis and inflammation in HBECs exposed to CSE, and this effect was regulated through the modulation of the miR-24/PHLPP2 axis. However, it is crucial to consider that because this study only examined the in vitro performance of circ_0006892, its conclusions might not accurately reflect the complicated environment experienced by COPD patients. To gain a comprehensive understanding of COPD pathogenesis, further investigations using preclinical animal models are warranted.
CircRNAs have also been shown to play a regulatory role in pulmonary endothelial and smooth muscle cells in the context of COPD in vitro models. For example, circANKRD11 demonstrated upregulation in the lungs of smokers with or without COPD and CSE-treated human pulmonary microvascular endothelial cells (HPMECs) (Wang Z. et al., 2021). Knocking down circANKRD11 in HPMECs resulted in a reduction of cellular apoptosis, inflammatory response, and oxidative stress induced by CSE, achieved through the modulation of the miR-145-5p/BRD4 axis. Elevated expression of Circ-BPTF was found to be over-expressed in the pulmonary arteries of patients with COPD, and it facilitated the proliferation of hypoxic pulmonary artery smooth muscle cells (PASMCs). Circ-BPTF/miR-486-5p/CEMIP signaling axis promoted the proliferation of the PASMCs (Wang C. et al., 2022). While dysfunction of pulmonary vascular smooth muscle cells and endothelial cells is well-established in pulmonary arterial hypertension, their involvement as major contributors to chronic vascular inflammation and lung damage in COPD is increasingly recognized. Future studies are warranted to explore the causal function of circRNAs in these cells through in vivo and clinical investigations.
Overall, dysregulation of circRNAs is evident in COPD cells, and they play significant roles in disease development and progression. However, it is important to note that these findings primarily originate from in vitro experiments and may not fully represent the complex environment of COPD patients. To gain a better understanding of COPD pathogenesis and further investigate the role of circRNAs, future studies utilizing pre-clinical animal models are warranted.
As mentioned above, circRNAs exhibit stability due to their closed-loop structures, conservation across species, and display tissue- and developmental stage-specific expression manners. Moreover, circRNAs are abundantly present in blood, saliva, exosomes, and urine, enabling their detection with remarkable sensitivity and specificity. These distinctive features make circRNAs promising candidates for biomarker discovery.
CircRNAs have emerged as potential biomarkers in asthma and COPD. For instance, receiver operating characteristic (ROC) curve analysis of circ_0002594 expression in the CD4+ T cells of 83 asthmatics who were not using inhaled corticosteroids (ICS) revealed a significantly high area under the curve (AUC) value of 0.727 (Huang Z. et al., 2021). Similarly, an AUC value of 0.767 were found when assessing the diagnostic utility of circ_0002594 in the context of ICS treatment in 48 asthmatic patients (Huang Z. et al., 2021). Furthermore, the expression of circ_0002594 showed a positive correlation with exhaled nitric oxide levels, the patient’s family history, positive skin prick test (SPT) results, and Th2 cytokine expression levels (Huang Z. et al., 2021). Additionally, elevated expression of circSORT1 and circSERPINB1 was observed in the induced sputum of 68 asthmatic patients compared to 20 healthy individuals, demonstrating higher specificity and sensitivity in predicting asthma (Xu et al., 2023). CircSORT1 expression correlated with various asthma clinical parameters, including FeNO, EOS%, IL-17A, IFN-γ, and PC20 (Xu et al., 2023). On the other hand, CircSERPINB1 expression showed correlations with IL-6, IL-17A, IFN-γ, FEV1%, and FVC% (Xu et al., 2023). Furthermore, a significantly increased expression of circ_0005519 was observed in PBMCs and CD4+ T cells of 30 asthmatics compared to 24 healthy individuals, and this increased expression was associated with exhaled nitric oxide levels (Huang et al., 2019). The serum levels of circ_0001859 were observed to be reduced in 38 patients with COPD and 24 patients with acute exacerbation of COPD (AE-COPD) compared to 28 healthy controls. This decreased expression exhibited higher specificity and sensitivity in predicting both COPD and AE-COPD and correlated with the FEV1% predicted. Furthermore, another circRNA, circ_004092 expression was found to be increased in the blood of smokers with COPD (n = 22) or smokers without COPD patients (n = 22) in comparison to non-smokers (n = 22). Collectively, these findings indicate that circRNAs hold promise as potential diagnostic and prognostic biomarkers for asthma and COPD. However, the potential of circRNAs as biomarkers in asthma and COPD requires validation in larger sample cohorts.
Circular RNAs (circRNAs) show promise in asthma and COPD research, offering potential as therapeutic targets and biomarkers. Despite valuable insights gained, significant challenges remain. Limited circRNAs have been linked to asthma and COPD, underscoring the need for a more comprehensive circRNA profile in these conditions. These molecules play crucial roles in various airway and immune cell functions, including AECs, 16HBE, ASMCs, CD4+ T cells, and macrophages in asthma, and 16HBE and AECs in COPD. However, no research has delved into circRNA roles in ASMCs dysfunction in COPD, despite its pivotal role in airway remodeling. Further investigations are necessary to enhance our understanding of COPD. Moreover, the primary focus of asthma and COPD studies has been on circRNAs’ capacity to sponge microRNAs, leaving other regulatory mechanisms such as protein sponges and scaffolds. Additionally, while most circRNA studies have been confined to in vitro settings, their in vivo roles remain mostly unexplored. Only a select few, such as circZNF652 in asthma, and circFOXO3, circFOXOP1, and circBbs9 in COPD, have been studied in animal models, with no assessment of their impact on lung function changes in asthma and COPD. Exploring the clinical applicability of circRNAs as diagnostic tools, prognostic markers, and predictive biomarkers requires studies in extensive patient cohorts. Finally, circRNAs offer potential for targeted therapeutic approaches, demanding identification of circRNAs with therapeutic value and the development of strategies to modulate their expression or activity, potentially leading to innovative asthma and COPD treatments.
XL: Conceptualization, Writing–original draft. MA: Conceptualization, Writing–original draft. KD: Conceptualization, Writing–review and editing. YM: Conceptualization, Writing–review and editing. JL: Conceptualization, Writing–original draft, Writing–review and editing.
The authors declare financial support was received for the research, authorship, and/or publication of this article. This work is funded by the People’s Livelihood Science and Technology Program Joint Plan of Liaoning Province, China (Grant No. 2021JH2/10300131).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Ali, M. K., Kim, R. Y., Brown, A. C., Mayall, J. R., Karim, R., Pinkerton, J. W., et al. (2020). Crucial role for lung iron level and regulation in the pathogenesis and severity of asthma. Eur. Respir. J. 55 (4), 1901340. doi:10.1183/13993003.01340-2019
Ali, M. K., Schimmel, K., Zhao, L., Chen, C. K., Dua, K., Nicolls, M. R., et al. (2022). The role of circular RNAs in pulmonary hypertension. Eur. Respir. J. 60 (6), 2200012. doi:10.1183/13993003.00012-2022
Altesha, M. A., Ni, T., Khan, A., Liu, K., and Zheng, X. (2019). Circular RNA in cardiovascular disease. J. Cell. Physiol. 234 (5), 5588–5600. doi:10.1002/jcp.27384
Bao, H., Zhou, Q., Li, Q., Niu, M., Chen, S., Yang, P., et al. (2020). Differentially expressed circular RNAs in a murine asthma model. Mol. Med. Rep. 22 (6), 5412–5422. doi:10.3892/mmr.2020.11617
Bhatti, G. K., Khullar, N., Sidhu, I. S., Navik, U. S., Reddy, A. P., Reddy, P. H., et al. (2021). Emerging role of non-coding RNA in health and disease. Metab. Brain Dis. 36 (6), 1119–1134. doi:10.1007/s11011-021-00739-y
Bollmeier, S. G., and Hartmann, A. P. (2020). Management of chronic obstructive pulmonary disease: a review focusing on exacerbations. Am. J. Health Syst. Pharm. 77 (4), 259–268. doi:10.1093/ajhp/zxz306
Camoretti-Mercado, B., and Lockey, R. F. (2021). Airway smooth muscle pathophysiology in asthma. J. Allergy Clin. Immunol. 147 (6), 1983–1995. doi:10.1016/j.jaci.2021.03.035
Chen, D., Wu, W., Yi, L., Feng, Y., Chang, C., Chen, S., et al. (2021). A potential circRNA-miRNA-mRNA regulatory network in asthmatic airway epithelial cells identified by integrated analysis of microarray datasets. Front. Mol. Biosci. 8, 703307. doi:10.3389/fmolb.2021.703307
Chen, H., Xu, X., Teng, J., Cheng, S., Bunjhoo, H., Cao, Y., et al. (2016). CXCR4 inhibitor attenuates ovalbumin-induced airway inflammation and hyperresponsiveness by inhibiting Th17 and Tc17 cell immune response. Exp. Ther. Med. 11 (5), 1865–1870. doi:10.3892/etm.2016.3141
Chen, S., Yao, Y., Lu, S., Chen, J., Yang, G., Tu, L., et al. (2020). CircRNA0001859, a new diagnostic and prognostic biomarkers for COPD and AECOPD. BMC Pulm. Med. 20 (1), 311. doi:10.1186/s12890-020-01333-1
Cheng, S., Chen, H., Wang, A., Bunjhoo, H., Cao, Y., Xie, J., et al. (2016). Blockade of IL-23 ameliorates allergic lung inflammation via decreasing the infiltration of Tc17 cells. Arch. Med. Sci. 12 (6), 1362–1369. doi:10.5114/aoms.2016.62923
Curran, D. R., and Cohn, L. (2010). Advances in mucous cell metaplasia: a plug for mucus as a therapeutic focus in chronic airway disease. Am. J. Respir. Cell. Mol. Biol. 42 (3), 268–275. doi:10.1165/rcmb.2009-0151TR
Dang, X., He, B., Ning, Q., Liu, Y., Chang, Y., and Chen, M. (2020). Suppression of TRIM8 by microRNA-182-5p restricts tumor necrosis factor-α-induced proliferation and migration of airway smooth muscle cells through inactivation of NF-Κb. Int. Immunopharmacol. 83, 106475. doi:10.1016/j.intimp.2020.106475
DiCosmo, B. F., Geba, G. P., Picarella, D., Elias, J. A., Rankin, J. A., Stripp, B. R., et al. (1994). Airway epithelial cell expression of interleukin-6 in transgenic mice. Uncoupling of airway inflammation and bronchial hyperreactivity. J. Clin. Investig. 94 (5), 2028–2035. doi:10.1172/JCI117556
Ding, L., Liu, G. L., Lu, L., Ge, L., and Wang, J. Y. (2022). circ_CSNK1E modulates airway smooth muscle cells proliferation and migration via miR-34a-5p/VAMP2 axis in asthma. Cell. Signal 95, 110340. doi:10.1016/j.cellsig.2022.110340
Diseases, G. B. D., and Injuries, C. (2020). Global burden of 369 diseases and injuries in 204 countries and territories, 1990-2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet 396 (10258), 1204–1222. doi:10.1016/S0140-6736(20)30925-9
Du, Y., Ding, Y., Shi, T., He, W., Mei, Z., Feng, X., et al. (2022). Suppression of circXPO1 attenuates cigarette smoke-induced inflammation and cellular senescence of alveolar epithelial cells in chronic obstructive pulmonary disease. Int. Immunopharmacol. 111, 109086. doi:10.1016/j.intimp.2022.109086
Duan, R., Niu, H., Yu, T., Cui, H., Yang, T., Hao, K., et al. (2020). Identification and bioinformatic analysis of circular RNA expression in peripheral blood mononuclear cells from patients with chronic obstructive pulmonary disease. Int. J. Chron. Obstruct Pulmon Dis. 15, 1391–1401. doi:10.2147/COPD.S252896
Erpenbeck, V. J., Hohlfeld, J. M., Volkmann, B., Hagenberg, A., Geldmacher, H., Braun, A., et al. (2003). Segmental allergen challenge in patients with atopic asthma leads to increased IL-9 expression in bronchoalveolar lavage fluid lymphocytes. J. Allergy Clin. Immunol. 111 (6), 1319–1327. doi:10.1067/mai.2003.1485
Fahy, J. V., and Dickey, B. F. (2010). Airway mucus function and dysfunction. N. Engl. J. Med. 363 (23), 2233–2247. doi:10.1056/NEJMra0910061
Feizi, H., Alizadeh, M., Nejadghaderi, S. A., Noori, M., Sullman, M. J. M., Ahmadian Heris, J., et al. (2022). The burden of chronic obstructive pulmonary disease and its attributable risk factors in the Middle East and North Africa region, 1990-2019. Respir. Res. 23 (1), 319. doi:10.1186/s12931-022-02242-z
Feng, M. J., Shi, F., Qiu, C., and Peng, W. K. (2012). MicroRNA-181a, -146a and -146b in spleen CD4+ T lymphocytes play proinflammatory roles in a murine model of asthma. Int. Immunopharmacol. 13 (3), 347–353. doi:10.1016/j.intimp.2012.05.001
Ferreira, M. A. (2003). Cytokine expression in allergic inflammation: systematic review of in vivo challenge studies. Mediat. Inflamm. 12 (5), 259–267. doi:10.1080/09629350310001619717
Fujimoto, M., Nakano, M., Terabe, F., Kawahata, H., Ohkawara, T., Han, Y., et al. (2011). The influence of excessive IL-6 production in vivo on the development and function of Foxp3+ regulatory T cells. J. Immunol. 186 (1), 32–40. doi:10.4049/jimmunol.0903314
Fujiwara, M., Hirose, K., Kagami, S. i., Takatori, H., Wakashin, H., Tamachi, T., et al. (2007). T-bet inhibits both TH2 cell-mediated eosinophil recruitment and TH17 cell-mediated neutrophil recruitment into the airways. J. Allergy Clin. Immunol. 119 (3), 662–670. doi:10.1016/j.jaci.2006.12.643
Gaffo, E., Boldrin, E., Dal Molin, A., Bresolin, S., Bonizzato, A., Trentin, L., et al. (2019). Circular RNA differential expression in blood cell populations and exploration of circRNA deregulation in pediatric acute lymphoblastic leukemia. Sci. Rep. 9 (1), 14670. doi:10.1038/s41598-019-50864-z
Ghafouri-Fard, S., Shoorei, H., Taheri, M., and Sanak, M. (2020). Emerging role of non-coding RNAs in allergic disorders. Biomed. Pharmacother. 130, 110615. doi:10.1016/j.biopha.2020.110615
Guo, P., Li, R., Piao, T. H., Wang, C. L., Wu, X. L., and Cai, H. Y. (2022). Pathological mechanism and targeted drugs of COPD. Int. J. Chron. Obstruct Pulmon Dis. 17, 1565–1575. doi:10.2147/COPD.S366126
Gysens, F., Mestdagh, P., de Bony de Lavergne, E., and Maes, T. (2022). Unlocking the secrets of long non-coding RNAs in asthma. Thorax 77 (5), 514–522. doi:10.1136/thoraxjnl-2021-218359
Habib, N., Pasha, M. A., and Tang, D. D. (2022). Current understanding of asthma pathogenesis and biomarkers. Cells 11 (17), 2764. doi:10.3390/cells11172764
Hammad, H., and Lambrecht, B. N. (2021). The basic immunology of asthma. Cell. 184 (6), 1469–1485. doi:10.1016/j.cell.2021.02.016
He, Y., Qian, D. C., Diao, J. A., Cho, M. H., Silverman, E. K., Gusev, A., et al. (2023). Prediction and stratification of longitudinal risk for chronic obstructive pulmonary disease across smoking behaviors. medRxiv. doi:10.1101/2023.04.04.23288086
Hershenson, M. B., Brown, M., Camoretti-Mercado, B., and Solway, J. (2008). Airway smooth muscle in asthma. Annu. Rev. Pathol. 3, 523–555. doi:10.1146/annurev.pathmechdis.1.110304.100213
Holgate, S. T. (2008). Pathogenesis of asthma. Clin. Exp. Allergy 38 (6), 872–897. doi:10.1111/j.1365-2222.2008.02971.x
Huang, J. Q., Wang, F., Wang, L. T., Li, Y. M., Lu, J. L., and Chen, J. Y. (2021a). Circular RNA ERBB2 contributes to proliferation and migration of airway smooth muscle cells via miR-98-5p/igf1r signaling in asthma. J. Asthma Allergy 14, 1197–1207. doi:10.2147/JAA.S326058
Huang, Z., Cao, Y., Zhou, M., Qi, X., Fu, B., Mou, Y., et al. (2019). Hsa_circ_0005519 increases IL-13/IL-6 by regulating hsa-let-7a-5p in CD4(+) T cells to affect asthma. Clin. Exp. Allergy 49 (8), 1116–1127. doi:10.1111/cea.13445
Huang, Z., Fu, B., Qi, X., Xu, Y., Mou, Y., Zhou, M., et al. (2021b). Diagnostic and therapeutic value of Hsa_circ_0002594 for T helper 2-mediated allergic asthma. Int. Arch. Allergy Immunol. 182 (5), 388–398. doi:10.1159/000511612
Ito, I., Fixman, E. D., Asai, K., Yoshida, M., Gounni, A. S., Martin, J. G., et al. (2009). Platelet-derived growth factor and transforming growth factor-beta modulate the expression of matrix metalloproteinases and migratory function of human airway smooth muscle cells. Clin. Exp. Allergy 39 (9), 1370–1380. doi:10.1111/j.1365-2222.2009.03293.x
Jia, Y., Li, X., Nan, A., Zhang, N., Chen, L., Zhou, H., et al. (2020). Circular RNA 406961 interacts with ILF2 to regulate PM(2.5)-induced inflammatory responses in human bronchial epithelial cells via activation of STAT3/JNK pathways. Environ. Int. 141, 105755. doi:10.1016/j.envint.2020.105755
Jiang, Y., Guo, X., and Qin, J. (2021). Silencing of circHIPK3 hampers platelet-derived growth factor-induced proliferation and migration in airway smooth muscle cells through the miR-375/MMP-16 axis. Cytotechnology 73 (4), 629–642. doi:10.1007/s10616-021-00483-2
Johnson, M. T., Xin, P., Benson, J. C., Pathak, T., Walter, V., Emrich, S. M., et al. (2022). STIM1 is a core trigger of airway smooth muscle remodeling and hyperresponsiveness in asthma. Proc. Natl. Acad. Sci. U. S. A. 119 (1), e2114557118. doi:10.1073/pnas.2114557118
Kardas, G., Daszyńska-Kardas, A., Marynowski, M., Brząkalska, O., Kuna, P., and Panek, M. (2020). Role of platelet-derived growth factor (PDGF) in asthma as an immunoregulatory factor mediating airway remodeling and possible pharmacological target. Front. Pharmacol. 11, 47. doi:10.3389/fphar.2020.00047
Kim, H. T., Yin, W., Nakamichi, Y., Panza, P., Grohmann, B., Buettner, C., et al. (2019). WNT/RYK signaling restricts goblet cell differentiation during lung development and repair. Proc. Natl. Acad. Sci. U. S. A. 116 (51), 25697–25706. doi:10.1073/pnas.1911071116
Kimura, A., and Kishimoto, T. (2010). IL-6: regulator of Treg/Th17 balance. Eur. J. Immunol. 40 (7), 1830–1835. doi:10.1002/eji.201040391
Kristensen, L. S., Andersen, M. S., Stagsted, L. V. W., Ebbesen, K. K., Hansen, T. B., and Kjems, J. (2019). The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 20 (11), 675–691. doi:10.1038/s41576-019-0158-7
Kristensen, L. S., Jakobsen, T., Hager, H., and Kjems, J. (2022). The emerging roles of circRNAs in cancer and oncology. Nat. Rev. Clin. Oncol. 19 (3), 188–206. doi:10.1038/s41571-021-00585-y
Lambrecht, B. N., and Hammad, H. (2015). The immunology of asthma. Nat. Immunol. 16 (1), 45–56. doi:10.1038/ni.3049
Li, J., Sun, C., Cui, H., Sun, J., and Zhou, P. (2021). Role of circRNAs in neurodevelopment and neurodegenerative diseases. J. Mol. Neurosci. 71 (9), 1743–1751. doi:10.1007/s12031-021-01882-y
Li, L. J., Zhu, Z. W., Zhao, W., Tao, S. S., Li, B. Z., Xu, S. Z., et al. (2018b). Circular RNA expression profile and potential function of hsa_circ_0045272 in systemic lupus erythematosus. Immunology 155 (1), 137–149. doi:10.1111/imm.12940
Li, M., Hua, Q., Shao, Y., Zeng, H., Liu, Y., Diao, Q., et al. (2020). Circular RNA circBbs9 promotes PM(2.5)-induced lung inflammation in mice via NLRP3 inflammasome activation. Environ. Int. 143, 105976. doi:10.1016/j.envint.2020.105976
Li, X., Yang, L., and Chen, L. L. (2018a). The biogenesis, functions, and challenges of circular RNAs. Mol. Cell. 71 (3), 428–442. doi:10.1016/j.molcel.2018.06.034
Lin, H., and Wan, N. (2022). Circular RNA has circ 001372-reduced inflammation in ovalbumin-induced asthma through sirt1/NFAT5 signaling pathway by miRNA-128-3p. Mol. Biotechnol. 64 (9), 1034–1044. doi:10.1007/s12033-022-00480-6
Lin, J., Feng, X., and Zhang, J. (2020). Circular RNA circHIPK3 modulates the proliferation of airway smooth muscle cells by miR-326/STIM1 axis. Life Sci. 255, 117835. doi:10.1016/j.lfs.2020.117835
Liu, X., Ali, M. K., Dua, K., and Xu, R. (2022). The role of zinc in the pathogenesis of lung disease. Nutrients 14 (10), 2115. doi:10.3390/nu14102115
Lloyd, C. M., and Hessel, E. M. (2010). Functions of T cells in asthma: more than just T(H)2 cells. Nat. Rev. Immunol. 10 (12), 838–848. doi:10.1038/nri2870
Lommatzsch, M. (2020). Immune modulation in asthma: current concepts and future strategies. Respiration 99 (7), 566–576. doi:10.1159/000506651
Ma, H., Lu, L., Xia, H., Xiang, Q., Sun, J., Xue, J., et al. (2020). Circ0061052 regulation of FoxC1/Snail pathway via miR-515-5p is involved in the epithelial-mesenchymal transition of epithelial cells during cigarette smoke-induced airway remodeling. Sci. Total Environ. 746, 141181. doi:10.1016/j.scitotenv.2020.141181
Mao, Y., and Xu, R. (2020). Circular RNA CDR1-AS contributes to pemetrexed and cisplatin chemoresistance through EGFR/PI3K signaling pathway in lung adenocarcinoma. Biomed. Pharmacother. 123, 109771. doi:10.1016/j.biopha.2019.109771
Matera, M. G., Hanania, N. A., Maniscalco, M., and Cazzola, M. (2023). Pharmacotherapies in older adults with COPD: challenges and opportunities. Drugs Aging 40 (7), 605–619. doi:10.1007/s40266-023-01038-0
Mei, J., Zhang, Y., Lu, S., and Wang, J. (2020). Long non-coding RNA NNT-AS1 regulates proliferation, apoptosis, inflammation and airway remodeling of chronic obstructive pulmonary disease via targeting miR-582-5p/FBXO11 axis. Biomed. Pharmacother. 129, 110326. doi:10.1016/j.biopha.2020.110326
Mercer, B. A., and DArmiento, J. M. (2006). Emerging role of MAP kinase pathways as therapeutic targets in COPD. Int. J. Chron. Obstruct Pulmon Dis. 1 (2), 137–150. doi:10.2147/copd.2006.1.2.137
Miao, Y., Wu, J., Wu, R., Wang, E., and Wang, J. (2022). Circ_0040929 serves as promising biomarker and potential target for chronic obstructive pulmonary disease. Int. J. Chron. Obstruct Pulmon Dis. 17, 2079–2092. doi:10.2147/COPD.S364553
Nakano, T., Inoue, Y., Shimojo, N., Yamaide, F., Morita, Y., Arima, T., et al. (2013). Lower levels of hsa-mir-15a, which decreases VEGFA, in the CD4+ T cells of pediatric patients with asthma. J. Allergy Clin. Immunol. 132 (5), 1224–1227. doi:10.1016/j.jaci.2013.06.041
Ng, W. L., Marinov, G. K., Liau, E. S., Lam, Y. L., Lim, Y. Y., and Ea, C. K. (2016). Inducible RasGEF1B circular RNA is a positive regulator of ICAM-1 in the TLR4/LPS pathway. RNA Biol. 13 (9), 861–871. doi:10.1080/15476286.2016.1207036
Ohno, I., Nitta, Y., Yamauchi, K., Hoshi, H., Honma, M., Woolley, K., et al. (1995). Eosinophils as a potential source of platelet-derived growth factor B-chain (PDGF-B) in nasal polyposis and bronchial asthma. Am. J. Respir. Cell. Mol. Biol. 13 (6), 639–647. doi:10.1165/ajrcmb.13.6.7576701
Pope, S. M., Brandt, E. B., Mishra, A., Hogan, S. P., Zimmermann, N., Matthaei, K. I., et al. (2001). IL-13 induces eosinophil recruitment into the lung by an IL-5- and eotaxin-dependent mechanism. J. Allergy Clin. Immunol. 108 (4), 594–601. doi:10.1067/mai.2001.118600
Qiao, D., Hu, C., Li, Q., and Fan, J. (2021). Circ-RBMS1 knockdown alleviates CSE-induced apoptosis, inflammation and oxidative stress via up-regulating FBXO11 through miR-197-3p in 16HBE cells. Int. J. Chron. Obstruct Pulmon Dis. 16, 2105–2118. doi:10.2147/COPD.S311222
Qiu, Y. Y., Zhang, Y. W., Qian, X. F., and Bian, T. (2017). miR-371, miR-138, miR-544, miR-145, and miR-214 could modulate Th1/Th2 balance in asthma through the combinatorial regulation of Runx3. Am. J. Transl. Res. 9 (7), 3184–3199.
Quan, L., Ren, G., Liu, L., Huang, W., and Li, M. (2022). Circular RNA circ_0002594 regulates PDGF-BB-induced proliferation and migration of human airway smooth muscle cells via sponging miR-139-5p/TRIM8 in asthma. Autoimmunity 55 (5), 339–350. doi:10.1080/08916934.2022.2062596
Rogers, D. F. (2002). Airway goblet cell hyperplasia in asthma: hypersecretory and anti-inflammatory? Clin. Exp. Allergy 32 (8), 1124–1127. doi:10.1046/j.1365-2745.2002.01474.x
Safiri, S., Carson-Chahhoud, K., Noori, M., Nejadghaderi, S. A., Sullman, M. J. M., Ahmadian Heris, J., et al. (2022). Burden of chronic obstructive pulmonary disease and its attributable risk factors in 204 countries and territories, 1990-2019: results from the Global Burden of Disease Study 2019. BMJ 378, e069679. doi:10.1136/bmj-2021-069679
Salzman, J. (2016). Circular RNA expression: its potential regulation and function. Trends Genet. 32 (5), 309–316. doi:10.1016/j.tig.2016.03.002
Shang, Y., Sun, Y., Xu, J., Ge, X., Hu, Z., Xiao, J., et al. (2020). Exosomes from mmu_circ_0001359-modified ADSCs attenuate airway remodeling by enhancing FoxO1 signaling-mediated M2-like macrophage activation. Mol. Ther. Nucleic Acids 19, 951–960. doi:10.1016/j.omtn.2019.10.049
Spinelli, A. M., González-Cobos, J. C., Zhang, X., Motiani, R. K., Rowan, S., Zhang, W., et al. (2012). Airway smooth muscle STIM1 and Orai1 are upregulated in asthmatic mice and mediate PDGF-activated SOCE, CRAC currents, proliferation, and migration. Pflugers Arch. 464 (5), 481–492. doi:10.1007/s00424-012-1160-5
Su, Y., Geng, L., Ma, Y., Yu, X., Kang, Z., and Kang, Z. (2021). Identification of circular RNA circVPS33A as a modulator in house dust mite-induced injury in human bronchial epithelial cells. Exp. Lung Res. 47 (8), 368–381. doi:10.1080/01902148.2021.1974125
Syamlal, G., Bhattacharya, A., and Dodd, K. E. (2020). Medical expenditures attributed to asthma and chronic obstructive pulmonary disease among workers - United States, 2011-2015. MMWR Morb. Mortal. Wkly. Rep. 69 (26), 809–814. doi:10.15585/mmwr.mm6926a1
The Global Asthma Report 2022 (2022). The global asthma Report 2022. Int. J. Tuberc. Lung Dis. 26 (1), 1–104. doi:10.5588/ijtld.22.1010
Tudorache, E., Fildan, A. P., Frandes, M., Dantes, E., and Tofolean, D. E. (2017). Aging and extrapulmonary effects of chronic obstructive pulmonary disease. Clin. Interv. Aging 12, 1281–1287. doi:10.2147/CIA.S145002
Wang, C., Liu, Y., Zhang, W., Huang, J., Jiang, J., Wang, R., et al. (2022b). circ-BPTF serves as a miR-486-5p sponge to regulate CEMIP and promotes hypoxic pulmonary arterial smooth muscle cell proliferation in COPD. Acta Biochim. Biophys. Sin. (Shanghai) 55 (3), 438–448. doi:10.3724/abbs.2022178
Wang, C., Tang, Y., Hou, H., Su, C., Gao, Y., and Yang, X. (2023b). CIRC_0026466 KNOCKDOWN PROTECTS HUMAN BRONCHIAL EPITHELIAL CELLS FROM CIGARETTE SMOKE EXTRACT-INDUCED INJURY BY PROMOTING THE MIR-153-3P/TRAF6/NF-ΚB PATHWAY. Shock 60, 121–129. doi:10.1097/SHK.0000000000002141
Wang, J., and Cheng, Y. (2022). The interaction of hsa_circ_0002594 and eIF4A3 promotes T-helper 2 cell differentiation by the regulation of PTEN. Clin. Exp. Med. 23, 887–895. doi:10.1007/s10238-022-00862-9
Wang, M., Yu, F., Wu, W., Zhang, Y., Chang, W., Ponnusamy, M., et al. (2017). Circular RNAs: a novel type of non-coding RNA and their potential implications in antiviral immunity. Int. J. Biol. Sci. 13 (12), 1497–1506. doi:10.7150/ijbs.22531
Wang, R., Tan, Y., Bao, X., Xiong, S., Liang, R., Cai, M., et al. (2023a). Circ_0000029 interacts with the miR-576-5p/KCNA1 Axis to hamper the development of pediatric asthma in an asthma-like in vitro assessment. Ann. Clin. Lab. Sci. 53 (2), 200–211.
Wang, T., Wang, P., Chen, D., Xu, Z., and Yang, L. (2021a). circARRDC3 contributes to interleukin-13-induced inflammatory cytokine and mucus production in nasal epithelial cells via the miR-375/KLF4 axis. Mol. Med. Rep. 23 (2), 141. doi:10.3892/mmr.2020.11780
Wang, X., Xu, C., Cai, Y., Zou, X., Chao, Y., Yan, Z., et al. (2022a). CircZNF652 promotes the goblet cell metaplasia by targeting the miR-452-5p/JAK2 signaling pathway in allergic airway epithelia. J. Allergy Clin. Immunol. 150 (1), 192–203. doi:10.1016/j.jaci.2021.10.041
Wang, Y. H., Yu, X. H., Luo, S. S., and Han, H. (2015). Comprehensive circular RNA profiling reveals that circular RNA100783 is involved in chronic CD28-associated CD8(+)T cell ageing. Immun. Ageing 12, 17. doi:10.1186/s12979-015-0042-z
Wang, Z., Zuo, Y., and Gao, Z. (2021b). CircANKRD11 knockdown protects HPMECs from cigarette smoke extract-induced injury by regulating miR-145-5p/BRD4 Axis. Int. J. Chron. Obstruct Pulmon Dis. 16, 887–899. doi:10.2147/COPD.S300332
Whitsett, J. A. (2018). Airway epithelial differentiation and mucociliary clearance. Ann. Am. Thorac. Soc. 15 (Suppl. 3), S143-S148–S148. doi:10.1513/AnnalsATS.201802-128AW
Xu, C., Du, L., Guo, Y., and Liang, Y. (2023). Differential circular RNA expression profiles in induced sputum of patients with asthma. Int. Arch. Allergy Immunol. 184 (5), 460–470. doi:10.1159/000528129
Xue, M., Peng, N., Zhu, X., and Zhang, H. (2021). Hsa_circ_0006872 promotes cigarette smoke-induced apoptosis, inflammation and oxidative stress in HPMECs and BEAS-2B cells through the miR-145-5p/NF-κB axis. Biochem. Biophys. Res. Commun. 534, 553–560. doi:10.1016/j.bbrc.2020.11.044
Yan, J., Zhang, X., Sun, S., Yang, T., Yang, J., Wu, G., et al. (2019). miR-29b reverses T helper 1 cells/T helper 2 cells imbalance and alleviates airway eosinophils recruitment in OVA-induced murine asthma by targeting inducible Co-stimulator. Int. Arch. Allergy Immunol. 180 (3), 182–194. doi:10.1159/000501686
Yang, Y., Gao, X., Zhang, M., Yan, S., Sun, C., Xiao, F., et al. (2018). Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J. Natl. Cancer Inst. 110 (3), 304–315. doi:10.1093/jnci/djx166
Zeng, H., Gao, H., Zhang, M., Wang, J., Gu, Y., Wang, Y., et al. (2021). Atractylon treatment attenuates pulmonary fibrosis via regulation of the mmu_circ_0000981/miR-211-5p/TGFBR2 Axis in an ovalbumin-induced asthma mouse model. Inflammation 44 (5), 1856–1864. doi:10.1007/s10753-021-01463-6
Zeng, N., Wang, T., Chen, M., Yuan, Z., Qin, J., Wu, Y., et al. (2019). Cigarette smoke extract alters genome-wide profiles of circular RNAs and mRNAs in primary human small airway epithelial cells. J. Cell. Mol. Med. 23 (8), 5532–5541. doi:10.1111/jcmm.14436
Zhang, C., Gu, S., and Kang, X. (2022). CircRNA circ_0006892 regulates miR-24/PHLPP2 axis to mitigate cigarette smoke extract-induced bronchial epithelial cell injury. Biotechnol. Appl. Biochem. 69 (2), 735–748. doi:10.1002/bab.2148
Zhang, C., Wang, X., Chen, Y., Wu, Z., and Shi, W. (2018). The down-regulation of hsa_circ_0012919, the sponge for miR-125a-3p, contributes to DNA methylation of CD11a and CD70 in CD4(+) T cells of systemic lupus erythematous. Clin. Sci. (Lond) 132 (21), 2285–2298. doi:10.1042/CS20180403
Zhang, H., Sun, Z., Yu, L., and Sun, J. (2017). MiR-139-5p inhibits proliferation and promoted apoptosis of human airway smooth muscle cells by downregulating the Brg1 gene. Respir. Physiol. Neurobiol. 246, 9–16. doi:10.1016/j.resp.2017.07.004
Zheng, C., Zhang, Y., Zhao, Y., Duan, Y., Mu, Q., and Wang, X. (2021). Circ-OSBPL2 contributes to smoke-related chronic obstructive pulmonary disease by targeting miR-193a-5p/BRD4 Axis. Int. J. Chron. Obstruct Pulmon Dis. 16, 919–931. doi:10.2147/COPD.S298465
Zhou, F., Cao, C., Chai, H., Hong, J., and Zhu, M. (2021a). Circ-HACE1 aggravates cigarette smoke extract-induced injury in human bronchial epithelial cells via regulating toll-like receptor 4 by sponging miR-485-3p. Int. J. Chron. Obstruct Pulmon Dis. 16, 1535–1547. doi:10.2147/COPD.S304859
Zhou, L., Wu, B., Yang, J., Wang, B., Pan, J., Xu, D., et al. (2021b). Knockdown of circFOXO3 ameliorates cigarette smoke-induced lung injury in mice. Respir. Res. 22 (1), 294. doi:10.1186/s12931-021-01883-w
Zhou, W. Y., Cai, Z. R., Liu, J., Wang, D. S., Ju, H. Q., and Xu, R. H. (2020). Circular RNA: metabolism, functions and interactions with proteins. Mol. Cancer 19 (1), 172. doi:10.1186/s12943-020-01286-3
Zhu, J., and Paul, W. E. (2010). Heterogeneity and plasticity of T helper cells. Cell. Res. 20 (1), 4–12. doi:10.1038/cr.2009.138
Zhu, Z., Homer, R. J., Wang, Z., Chen, Q., Geba, G. P., Wang, J., et al. (1999). Pulmonary expression of interleukin-13 causes inflammation, mucus hypersecretion, subepithelial fibrosis, physiologic abnormalities, and eotaxin production. J. Clin. Investig. 103 (6), 779–788. doi:10.1172/JCI5909
Keywords: CircRNAs, miRNAs, lung disease, asthma, COPD
Citation: Liu X, Ali MK, Dua K, Mao Y and Liu J (2023) Circular RNAs: emerging players in asthma and COPD. Front. Cell Dev. Biol. 11:1267792. doi: 10.3389/fcell.2023.1267792
Received: 27 July 2023; Accepted: 14 November 2023;
Published: 24 November 2023.
Edited by:
David Wei Chang, University of Texas MD Anderson Cancer Center, United StatesReviewed by:
Dharmendra Kumar Soni, Uniformed Services University of the Health Sciences, United StatesCopyright © 2023 Liu, Ali, Dua, Mao and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun Liu, bGl1ajVAc2otaG9zcGl0YWwub3Jn
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.