- MDI Biological Laboratory, Bar Harbor, ME, United States
The highly conserved integrated stress response (ISR) reduces and redirects mRNA translation in response to certain forms of stress and nutrient limitation. It is activated when kinases phosphorylate a key residue in the alpha subunit of eukaryotic translation initiation factor 2 (eIF2). General Control Nonderepressible-2 (GCN2) is activated to phosphorylate eIF2α by the presence of uncharged tRNA associated with nutrient scarcity, while protein kinase R-like ER kinase-1 (PERK) is activated during the ER unfolded protein response (UPRER). Here, we investigated the role of the ISR during nutrient limitation and ER stress with respect to changes in protein synthesis, translationally driven mRNA turnover, and survival in Caenorhabditis elegans. We found that, while GCN2 phosphorylates eIF2α when nutrients are restricted, the ability to phosphorylate eIF2α is not required for changes in translation, nonsense-mediated decay, or lifespan associated with dietary restriction (DR). Interestingly, loss of both GCN2 and PERK abolishes increased lifespan associated with dietary restriction, indicating the possibility of other substrates for these kinases. The ISR was not dispensable under ER stress conditions, as demonstrated by the requirement for PERK and eIF2α phosphorylation for decreased translation and wild type-like survival. Taken together, results indicate that the ISR is critical for ER stress and that other translation regulatory mechanisms are sufficient for increased lifespan under dietary restriction.
Introduction
Aging is associated with a decline in mechanisms governing cellular resilience. This decline results in increased susceptibility to the negative effects of nutrient imbalances and stress, disrupting essential processes such as proteostasis and energy metabolism, and contributing to the accumulation of misfolded proteins and cellular dysfunction (Butcher and Lord, 2004; Haigis and Yankner, 2010; Kyriakakis et al., 2015). In particular, impaired proteostasis is closely linked to the development of neurodegenerative disorders, such as Alzheimer’s, Parkinson’s, and Huntington’s diseases (Kim et al., 2022). Interventions that help maintain cellular homeostasis hold the potential to help treat such disorders.
The ISR is a conserved signaling pathway for translationally-mediated adaptation to various forms of stress that perturb cellular homeostasis (Pakos-Zebrucka et al., 2016). In recent years, the ISR has gained notoriety due to its role in neurodegenerative disease and possible role in aging (Bond et al., 2020; Derisbourg et al., 2021a; Derisbourg et al., 2021b). The key event in the ISR is phosphorylation of the eukaryotic translation initiation factor subunit (eIF) 2α, which inhibits the exchange of eIF2 GDP-to-GTP by eIF2B and thereby suppresses the translation-initiating ternary complex (TC). This complex is a crucial molecular component involved in the initiation of protein synthesis comprising eIF2, GTP and Methionyl-tRNA (Jackson et al., 2010). Inhibiting GDP-to-GTP recharging leads to global attenuation of most translation except for certain genes, notably those containing upstream open reading frames (uORFs) in the 5’ untranslated region, such as the activating transcription factor 4 (ATF4) (Costa-Mattioli and Walter, 2020). Under normal conditions, such transcripts are inefficiently translated and destabilized due to the presence of premature termination codons (PTC) associated with the uORFs.
In mammals, eIF2α is phosphorylated at S51 by one of four kinases depending on the stress encountered. These include GCN2 (general amino acid control nonderepressible 2), PKR (double stranded RNA-dependent protein kinase), HRI (heme-regulated inhibitor), and PERK (PKR-like ER kinase). When tRNAs are uncharged, as happens during amino acid deprivation, GCN2 activates the ISR, reducing and redirecting protein synthesis to conserve energy and resources (Lee et al., 2008; Bunpo et al., 2010; Jonsson et al., 2019). PERK activates the ISR during ER stress when proteins are misfolded or when there are perturbations in calcium as a part of the unfolded protein response (UPR)ER (Korennykh and Walter, 2012; Wang and Kaufman, 2016). PKR is involved in antiviral defense activated by double-stranded RNA (Harding et al., 2000; Barber, 2001), whereas HRI is a heme sensor that reduces globin synthesis during heme deficiency (Chen, 2014). In each case, phosphorylation of eIF2α results in rapid attenuation of mRNA translation and redeployment of remaining translation to stress responsive genes (Harding et al., 2000; Teske et al., 2011).
The Caenorhabditis elegans genome only encodes two ISR activators. These are GCN-2 (GCN2) and PEK-1 (PERK), which target eIF-2α at S49. Research on the ISR in C. elegans has provided valuable insights into the regulatory mechanisms and functional significance of the ISR. The amino acid sequence of eIF-2α shares nearly 50% identity with the human ortholog and the key phosphorylation site is conserved (Derisbourg et al., 2021a). ER stress triggered by overexpression of the glutamine-fructose 6-phosphate aminotransferase homolog gfat-1 increases the ISR-dependent expression of ATF-4 (Horn et al., 2020), indicating that the key ISR components are conserved. The kinase PEK-1 is essential to protect larvae against bacterial infection (Richardson et al., 2011) and transition into dauer state under ER stress (Kulalert and Kim, 2013). In addition, GCN-2 mediates mitochondrial stress in C. elegans, whereas HRI is responsible for ISR activation under mitochondrial stress in mammals (Baker et al., 2012).
The ISR may play a role in the effects of dietary restriction (DR), which increases healthy lifespan by reducing and redirecting mRNA translation. Many types of DR exist, including caloric restriction, restriction of specific macronutrients or micronutrients, and intermittent fasting (Greer and Brunet, 2009). In C. elegans, dilution of their bacterial food source is a frequently used method of DR, increasing lifespan by 40% (Rollins et al., 2019). Research on understanding how changes in mRNA translation mediate beneficial effects on health and longevity has focused mostly on anabolic machinery controlled by the nutrient-sensing mechanistic Target of Rapamycin (mTOR). Little is known about how mTOR-independent ISR contributes to mRNA translation changes important for increasing healthy lifespan under DR.
Healthy lifespan is also governed by the proteostasis-regulating UPRER, which governs translation changes through PERK/PEK-1 and the ISR. A variety of internal and external stimuli can cause ER stress that overlap with the ISR, including hypoxia, nutrient scarcity, and pathogen infection (Wang and Kaufman, 2016). When misfolded proteins accumulate in the ER lumen, it creates proteotoxic stress that triggers the UPRER, which is controlled by three transmembrane proteins comprising transcription factor 6 (ATF6), inositol-requiring enzyme 1 (IRE1), and PERK. Once activated, these sensors initiate distinct signaling pathways to regulate gene expression both transcriptionally and translationally and restore protein-folding capacity (Ron and Walter, 2007; Wek and Cavener, 2007). Loss of PERK is associated with disturbed ER morphology and calcium signaling (Verfaillie et al., 2012). Pathologies like cell death, progressive diabetes mellitus, and exocrine pancreatic insufficiency are observed in PERK-deficient mice (Harding et al., 2001). Loss of the C. elegans ortholog, pek-1, leaves animals relatively healthy under standard conditions and partial knockdown of pek-1 with RNAi does not reduce survival of wild-type C. elegans during ER stress (Howard et al., 2016). However, a study using a pek-1 mutant shows that this gene may be important for survival under ER stress in C. elegans (Derisbourg et al., 2021b).
Translation initiation regulates longevity and is controlled by two complexes. One comprises the cap-binding complex downstream of mTOR, which circularizes mRNA and helps recruit additional translation factors (Kapahi et al., 2010). mTOR is a nutrient sensor that can act independently or in parallel to the ISR and other initiation-regulating complex, the TC (Sonenberg and Hinnebusch, 2009). Studies showed that amino acid depletion extends the lifespan in yeast and activates the ISR, but there is no direct evidence proving that the ISR is essential for the longevity benefit associated with this form of DR (Jiang et al., 2000; Ecker et al., 2010; Kaya et al., 2015). Interestingly, it was found that abrogating the ISR in phosphorylation-defective eIF-2α mutants increases the lifespan of C. elegans, suggesting that chronic inhibition of the ISR favors protein homeostasis in the absence of stress (Derisbourg et al., 2021b).
The ISR and nonsense-mediated RNA decay (NMD) pathways are connected via translation. Concurrent with, and dependent on ribosome translocation during protein synthesis, NMD degrades aberrant mRNA containing PTCs. The aberrant mRNA includes mRNA bearing retained introns or those containing uORFs in the 5’ UTR of genes translationally activated by the ISR. Thus, it serves as an mRNA quality control mechanism but also regulates a significant proportion of regular (i.e., non-PTC bearing) mRNAs through mechanisms that are not well characterized (Hug et al., 2016). Under hypoxic stress, NMD is inhibited in an ISR dependent manner in mammalian tissue culture. Interestingly, this was found to favor stabilization of certain stress-induced “normal” (i.e., not bearing PTCs) mRNAs normally targeted by NMD under non-stressed conditions (Gardner, 2008). Besides hypoxia, ER stress and amino acid starvation also trigger the escape of stress-associated mRNA that are usually targeted by NMD, including the ISR effector ATF4 (Karam et al., 2015; Martin and Gardner, 2015; Li et al., 2017). Under nutrient deprivation stress associated with DR, overall NMD activity is diminished, but factors mediating its regulation are required for lifespan extension (Rollins et al., 2019). Whether the ISR is essential to alter NMD and promote longevity associated with DR remains unclear.
Here, we investigated the role of the ISR in translation and lifespan extension under DR in C. elegans. We validated that DR activates the ISR by GCN-2 but found that eIF-2α phosphorylation is not required for lifespan extension and downregulation of protein synthesis. Additionally, changes in the prevalence of NMD targets are not ISR dependent under nutrient-limited conditions. Furthermore, while the ISR is important for ER stress adaptation, it is not necessary to respond to starvation and heat stress. Interestingly, while neither pek-1 or gcn-2 genes are individually required for increased lifespan under DR, double mutants show a complete loss of the lifespan phenotype under this condition.
Materials and methods
Caenorhabditis elegans strains and culture
All strains used are listed in Supplementary Table S1. The worms were cultured at 20°C and maintained on nematode growth medium (NGM) plates seeded with Escherichia coli OP50 unless indicated otherwise. All worms grew at least three generations under normal, unstressed conditions prior to use in experiments. Mutant strains were obtained from the CGC and backcrossed with the wild type Bristol N2 strain at least three times.
Lifespan analysis
Caenorhabditis elegans were developmentally synchronized from a 4 h egg lay and transferred to 10 cm peptone-free nematode growth media (NMG) plates spotted with 1011 CFU/mL (AL) or 109 CFU/mL (DR) OP50 upon reaching day 1 of adulthood. Plates had 25 mg/mL of carbenicillin to prevent bacterial growth. The worms were transferred daily to fresh plates until they reached the post-reproductive stage and then transferred every 4 days. The animals were scored for survival every other day by monitoring the movement after tapping the worm gently with a platinum wire.
Heat stress recovery
Synchronized worms were transferred to fresh plates daily for 1 week and then shifted to 35°C for 4 h. After the heat shock, the nematodes were monitored daily for survival.
ER stress induction
NGM plates were spotted with OP50. After 1 day, either DMSO or 25 μg/mL tunicamycin dissolved in DMSO was added to each plate. Another day was allowed to pass before animals were exposed to treated agar plates. Due to the large number of animals required for experiments involving polysome profiling, animals were synchronized by bleaching gravid adults. In translation and eIF-2α phosphorylation experiments, synchronized adults were transferred to tunicamycin or DMSO plates for 3 h and washed 3 times in M9 prior to analysis. To minimize vulval ruptures in survival assays, adult worms were treated with 50 μg/mL 5-fluoro-2-deoxyuridine (FUDR) for 2 days prior to exposure to tunicamycin or DMSO.
Polysome profiling
150 µL of pelleted day 1 adult worms were lysed on ice in 350 µL of solubilization buffer (300 mM NaCl, 50 mM Tris–HCl [pH 8.0], 10mM MgCl2, 1 mM EGTA, 400 U RNasin/mL, 3.55 µM cycloheximide, 1% Triton X-100, 1 mM PMSF, 1 mini tablet of protease inhibitor [Thermo Sicentific] and 0.1% sodium deoxycholate). An additional 200 µL of solubilization buffer was added after homogenization and the samples were incubated on ice for 1 h. Lysates were centrifuged at 14,000 × g at 4°C for 5 min and the supernatants were collected. 300 μL of each sample was loaded to the top of a 5%–50% sucrose gradient in high salt resolving buffer (140 mM NaCl, 25 mM Tris–HCl [pH 8.0], and 10 mM MgCl2) and centrifuged in a Beckman SW41Ti rotor at 38,000 × g at 4°C for 2 h. Gradient fractionation was conducted with a Piston Gradient Station equipped with a Triax flow cell (BioComp Instruments) and the absorbance was monitored at 260 nm continuously. Polysome profile quantification was carried out using R with codes adapted from Shaffer and Rollins (2020).
SUnSET puromycin assay
Bacteria were spotted into 12-well plates containing 2 mL of peptone-free NGM agar. Day 1 adults were assigned to AL (ad libitum, i.e., full fed), fasting (no bacteria, FT), or AL supplemented with 10 mM cycloheximide and incubated for 3 h. 300 μL of 1 mM puromycin was added to each well and incubated for 1 h. The worms were washed three times with M9 and 50 µL of worm slurry was frozen immediately in liquid nitrogen. To extract protein, worms were lysed on ice in 100 µL of solubilization buffer (300 mM NaCl, 50 mM Tris–HCl [pH 8.0], 10mM MgCl2, 1 mM EGTA, 1% Triton X-100, 1 mini tablet of protease inhibitor, and 1 mM PMSF) for 1 h and centrifuged at 14,000 × g at 4°C for 5 min. The Western blotting was conducted as described in the next section.
Western blotting
Animals used in experiments were Day 1 adults. Worm collection, lysis, and protein extraction were performed as described in the “SUnSET puromycin assay” section. For SDS-PAGE, 20 µg of total protein as determined by Bradford protein assay was loaded on 4%–20% mini-Protean TGX stain-free gels (Bio-Rad) and separated at 100 V for 5 min followed by 150 V for 1 h. The SDS-PAGE gels were exposed to UV for 2.5 min for total protein quantity detection and then transferred to PVDF membranes. The membranes were incubated with primary antibody in EveryBlot blocking buffer (Bio-Rad) at 4°C overnight and further incubated with secondary antibody for 1 h after washing. The protein density was quantified using ImageJ 1.47V and normalized by the total quantity of protein. For puromycin assays, anti-puromycin Ab (clone 12D10, Sigma; 1:5000) was used followed by anti-mouse secondary Ab. For eIF2α S49 phosphorylation detection, the antibodies were anti-phospho-eIF2α rabbit polyclonal Ab (1:3000 dilution; Cell Signaling) and anti-rabbit secondary Ab. Three replicates were performed.
NMD reporter and worm imaging
The NMD reporter strain PTCXi was described previously (Longman et al., 2007). Larval 4 stage worms were added to AL, DR or smg-2 RNAi (Y48G8AL.6; Vidal library) (Rual et al., 2004) plates and transferred daily in the same conditions for 3 days. Fluorescence microscopy was performed with individual C. elegans using a Leica M165FC stereo microscope in the GFP channel (narrow band filter set, excitation ET470/40 nm, emission ET510/10 nm). Each worm was imaged on a 1% agarose pad on a glass slide and immobilized with a drop of 20 mM levamisole. Quantification of fluorescence was conducted using ImageJ for mean pixel intensity after correcting for background fluorescence. At least 15 worms were included in each replicate. Three replicates were performed.
qPCR analysis
Adult worms were treated as described and then collected in TRIzol reagent (Invitrogen). RNA extraction and purification were performed using RNA Clean and Concentrator kit (Zymo Research) according to the manufacturer’s protocol followed by cDNA synthesis (QuantiTect Reverse Transcription kit [Qiagen]). qPCR analysis was conducted in technical triplicate using KAPA SYBR FAST qPCR Master Mix on a LightCycler 480 (Roche Applied Science, Indianapolis, IN, USA). Primers used are listed in Supplementary Table S2. The housekeeping gene cdc-42 was used for target gene mRNA normalization. Gene expression changes were analyzed by the 2−ΔΔCT method. Three replicates were conducted.
Development and fecundity
Worms were synchronized by a 1-h timed egg lay. Time to reproductive development was measured from the time an egg was laid until it became an egg-laying adult. The fecundity of nematodes was determined by counting hatched progeny. Both development and fecundity assays were conducted at 20°C. At least 12 worms were included in each replicate. Three replicates were performed.
Results
Fasting upregulates eIF-2α phosphorylation that depends on GCN-2
In order to understand the role of ISR in response to nutrient deprivation, we started by assessing changes in translation. Polysome profiling provides an assessment of translation based on ribosome association with mRNA. A redistribution of ribosomes from high translation polysomes (i.e., mRNA bound by 2 or more ribosomes) to low translation monosomes was observed on young adult N2 worms after fasting (FT) for 3 h compared to full fed (ad libitum, AL) (Figure 1A). Quantitatively, the relative abundance of polysomes decreased by 20.3% ± 1.7% in this time (p = 0.0009). However, more signal was lost in the highest translation polysomes (mRNA bound by 4 or more ribosomes), indicating that absolute levels of translation were likely decreased by more than 20%. As a separate measure of translation rate, surface sensing of translation (SUnSET) was optimized for use under DR conditions in C. elegans. Previous application of SUnSET in C. elegans relied on ingestion of bacteria mixed with puromycin (Arnold et al., 2014; Heissenberger et al., 2020). To allow puromycin incorporation in worms without food (i.e., from fasted animals), the mutant strain bus-5(br19) was used due to its enhanced cuticle permeability (Xiong et al., 2017), allowing this strain to incorporate puromycin at a higher rate compared to N2 (Somers et al., 2022). In this assay, bus-5(br19) animals were incubated with puromycin for 1 h followed by Western blot to detect puromycin signal. N2 animals were included for comparison. As expected, since the 1 h incubation was much shorter than the traditional puromycin assay, almost no puromycin signal could be detected in N2 (Figure 1B). In contrast, the bus-5(br19) mutant showed clear puromycin incorporation in both AL and fasting conditions. However, almost no signal was observed in worms treated with cycloheximide, an inhibitor of mRNA translation. A reduction in translation by 37.3% was detected in worms fasted for 1 h and 48.9% after 3 h (Figure 1B, lower panel). Taken together, both types of translation assay demonstrate translation reduction under short-term FT. Furthermore, the modified SUnSET method utilizing bus-5(br19) mutants provides an effective approach for assessing translational changes in C. elegans that is not affected by differences in food availability or pharyngeal pumping rate of worms.
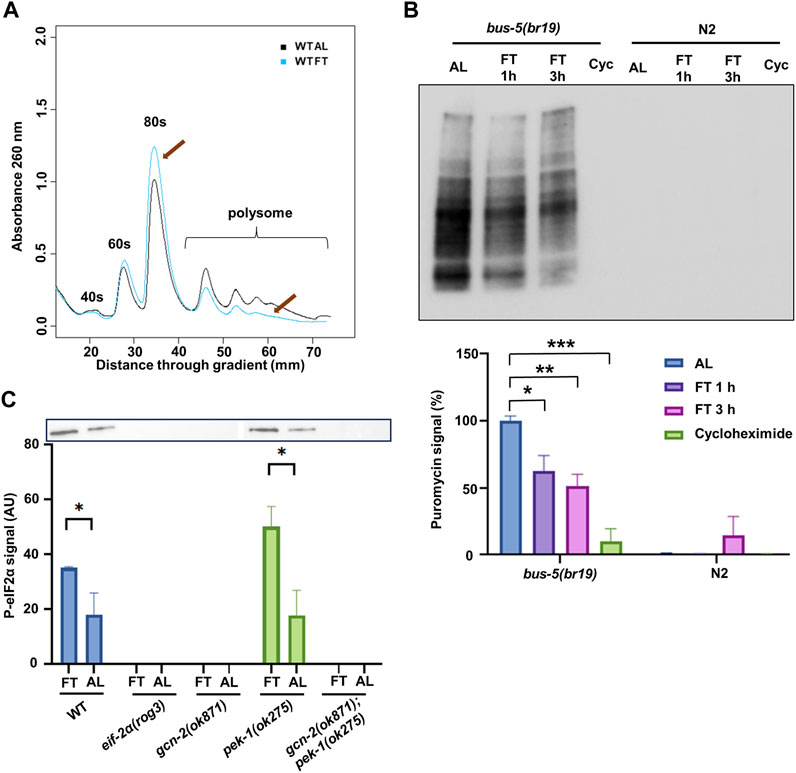
FIGURE 1. DR reduces translation and activates the ISR via GCN-2. (A) Polysome profiling of day 1 adult N2 with ad libitum feeding (AL) or fasting (FT) for 3 h. Representative of 3 replicates is shown. Arrows indicate the change of monosome and polysome. (B) SUnSET puromycin assay using Western blot with anti-puromycin antibodies to detect translation change in bus-5(br19) and N2 under AL, 1 h FT, 3 h FT and 10 mM cycloheximide treatments. Representative membrane is shown. The lower panel shows analysis from 3 experiments. (C) Western blot with anti-eIF2α phosphorylation antibody performed on day 1 adult under AL or FT for 3 h. Representative membrane is shown. Quantification was conducted with 3 independent experiments. Error bars represent means ± SEM, t-test was performed within each strain (*p < 0.05; **p < 0.01; ***p < 0.001).
It is not clear what role ISR plays in the attenuated translation and extended longevity under DR conditions. We performed Western blot to detect the activation of the ISR by eIF-2α phosphorylation. We included the knockout mutants gcn-2(ok871) and pek-1(ok275), which were crossed to generate the double mutant gcn-2(ok871);pek-1(ok275). We also included a mutant eif-2α(rog3), hitherto referred to simply as eif-2α, with serine 49 replaced by alanine, which cannot be phosphorylated. All these mutants and N2 were treated with AL or FT conditions for 3 h (Figure 1C). In N2, fasted animals showed higher eIF-2α phosphorylation compared to AL, indicating that the ISR was activated under the fasting condition. In the mutants, only pek-1(ok275) showed a similar pattern to wild type, whereas the signal could not be detected in other mutants. Results indicate that GCN-2, but not PEK-1, is responsible for eIF-2α phosphorylation under FT conditions as expected.
Early changes in translation upon fasting are similar in WT and ISR mutants
Next, we wanted to see how translation is affected in phosphorylation and kinase mutants under short-term fasting conditions. Therefore, we used polysome profiling and SUnSET to determine how translation is influenced in ISR mutants after 3 h FT. Polysome profiling showed that, regardless of the mutations, translational repression was observed in the eif-2α phospho-null mutant as well as gcn-2(ok871) and pek-1(ok275) (Figure 2A). In each instance, polysomes were reduced about 20% (Figure 2B), indicating that phosphorylation of eIF-2α is not required for these early changes in translation when food is removed.
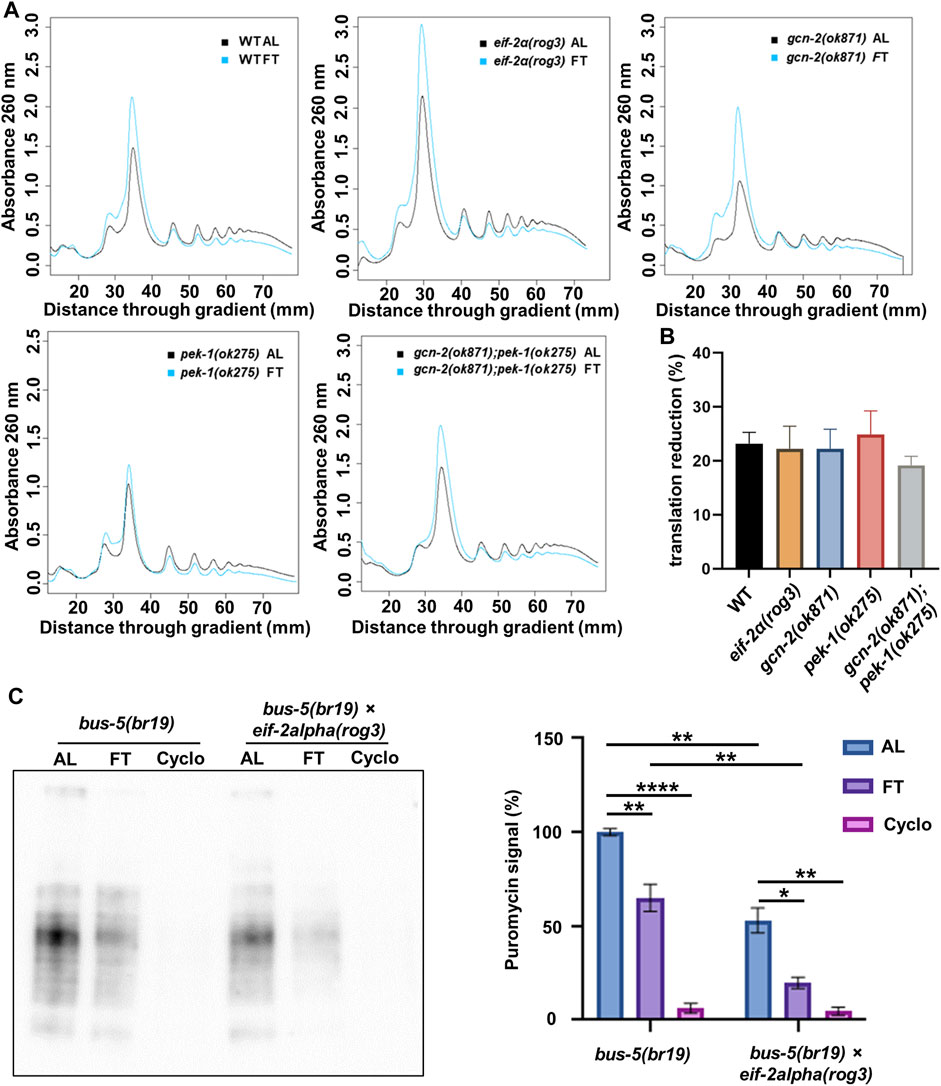
FIGURE 2. eIF-2α phosphorylation is not required for downregulation of translation upon withdrawal of food. (A) polysome profiling of day 1 adult N2, eif-2α(rog3), gcn-2(ok871), pek-1(ok275), and gcn-2(ok871);pek-1(ok275) after ad libitum feeding (AL) or fasting (FT) for 3 h. Each profile is representative of 3 independent experiments. (B) Quantification of translation reduction (%). Error bars represent means ± SEM. One-way ANOVA was performed and no statistical difference was detected. (C) SUnSET puromycin assay using Western blot with anti-puromycin antibodies to detect translation change in bus-5(br19) and bus-5(br19);eif-2α(rog3) day 1 adult worms under 3 h AL, 3 h FT and 10 mM cycloheximide treatments. Representative membrane is shown. Quantification was conducted with 3 replicates. Error bars represent means ± SEM. One-way ANOVA with Dunnett’s post hoc test was performed (*p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001).
In order to compare total changes in translation between control and the eif-2α phosphorylation mutant using SUnSET, bus-5(br19) was crossed with eif-2α mutants and animals were fasted for 3 h (Figure 2C). Both bus-5(br19) and bus-5(br19);eif-2α mutants showed reduced translation under FT conditions. Interestingly, translation is constitutively low in eif-2α phospho-null mutants compared to WT animals in both AL and fasting conditions (Figure 2C). Constitutively low translation is interesting from the point of view of NMD, which requires translation for its activity and which is, itself required for longevity associated with DR in C. elegans (Rollins et al., 2019; Kim et al., 2020). Importantly, results indicate that translation is downregulated in eif-2α phospho-mutants under FT compared to AL conditions (Figure 2C). Thus, the ISR is not required for translation attenuation under DR conditions.
Translation-dependent changes in NMD driven by fasting do not require the ISR
The activation of NMD relies on translation, thus we sought to confirm whether the ISR downregulates NMD when food is removed. To test its activity, we used the NMD reporter strain PTCXi harboring a PTC that normally abrogates expression of a GFP reporter (Longman et al., 2007). In environments without stress where NMD is highly active, the PTC containing transcript is degraded by NMD, precluding expression of GFP. We tested wild type and the eif-2α phospho-deficient mutant in response to DR. smg-2 RNAi was used as a control, since this gene is required for NMD. As expected, the worms on smg-2 RNAi showed the highest GFP signal. The animals under DR also increased their fluorescence significantly compared to AL, but not as strong as the one in smg-2 RNAi, indicating NMD decreased under DR, but did not turn off completely (Figures 3A, B). The fluorescence in the NMD reporter crossed with eif-2α presented a very similar pattern. The worms under DR increased GFP signal significantly, suggesting that NMD was also downregulated in the phospho-null mutant and that the ISR was not required for suppressed NMD activity under DR.
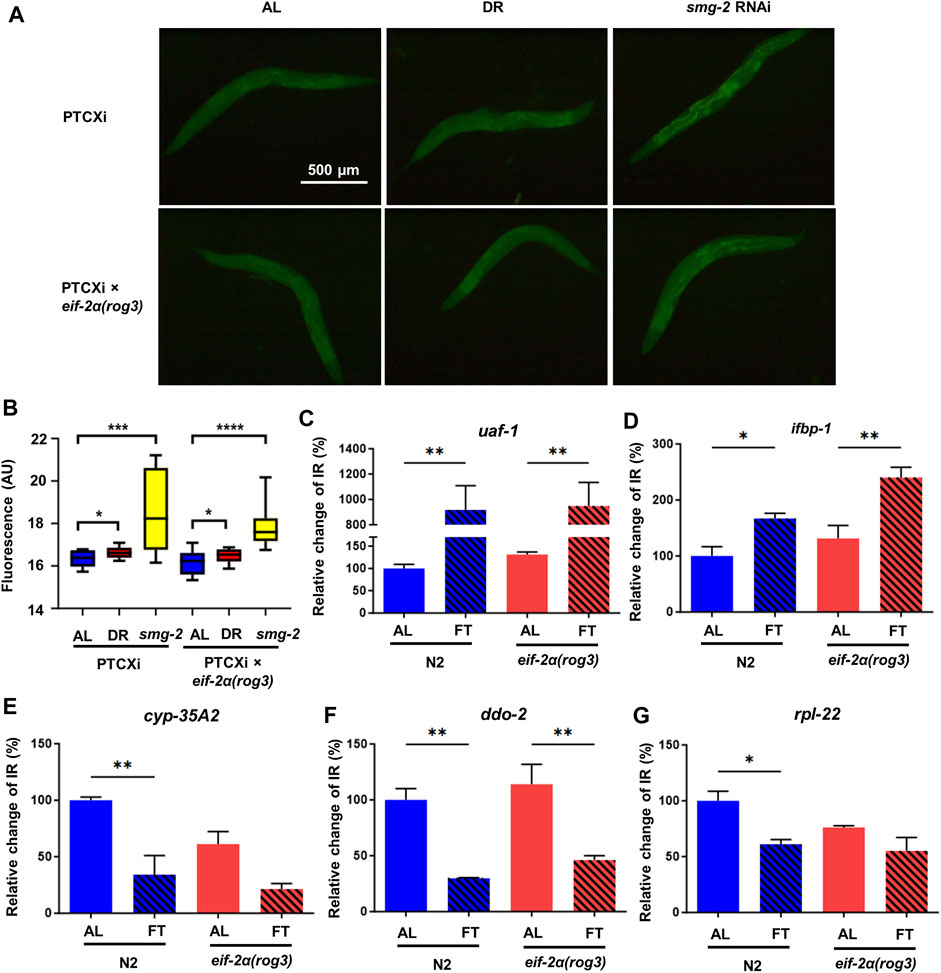
FIGURE 3. eIF-2α phosphorylation is dispensable for NMD under DR conditions. (A) Representative microscopy images of NMD reporter PTCXi and PTCXi crossed with eif-2α(rog3). (B) GFP signal quantification shown in (A) from 3 experiments performed. For (A) and (B), day 1 adult worms were placed under ad libitum feeding (AL), dietary restriction (DR) or smg-2 RNAi for 3 days prior to imaging. (C–G) Intron retained (IR) isoform expression of uaf-1 (C), ifbp-1 (D), cyp-35A2 (E), ddo-2 (F), and rpl-22 (G) in N2 and eif-2α(rog3) after 3 days of AL or fasting (FT) in young adult worms. IR expression was normalized by expression of all isoforms of each gene. Error bars represent means ± SEM (n = 3). One-way ANOVA with Dunnett’s post hoc test was performed. *p < 0.05; **p < 0.01; ***p < 0.001; and ****p < 0.0001.
To better understand the relationship between ISR and NMD, we measured expression of certain transcripts with intron retention (IR), which causes out-of-frame translation and generates at least one PTC. We tested the isoform expression relative to productively spliced transcripts in N2 and eif-2α phospho-mutants under AL and FT conditions for 3 days. The first set of genes tested were the alternatively spliced ribosomal protein mRNA rpl-1, rpl-3, rpl-7A, and rpl-12 that are known NMD targets (Mitrovich and Anderson, 2000). Except rpl-7A that did not show expression change, the IR containing isoforms of rpl-1, rpl-3, and rpl-12 decreased in FT compared to AL significantly (Supplementary Figure S1). Results suggest that alternative splicing decreased and/or NMD increased for these factors. We also analyzed the expression of atf-4, which is the main ISR effector (Supplementary Figure S1E) but did not observe any statistical change in the expression level (p = 0.09 for N2 AL-DR comparison). Furthermore, the expression pattern was very consistent in wild type and eif-2α in all these genes, indicating that these large ribosomal subunit transcripts are not dependent on the ISR for changes in NMD. We further tested the expression of the same IR transcripts in gcn-2(ok871);pek-1(ok275) to confirm the relationship between ISR and NMD. Similarly, under FT only uaf-1 and ifbp-1 increased their expression, but not any other transcripts tested (Supplementary Figure S2 and Supplementary Figure S3), The same pattern was observed in the wild type, confirming again that ISR does not regulate NMD under FT.
We then analyzed the expression of IR containing mRNA with increased abundance under conditions of a food dilution form of DR according to RNAseq data from a previous study (Rollins et al., 2019). We selected the isoforms with IR of uaf-1, ifbp-1, cyp-35A2, ddo-2, and rpl-22. UAF-1 is a large subunit of splicing factor U2AF (U2 auxiliary factor) involved in the recognition of 3’ splice sites (Zorio and Blumenthal, 1999; Ma and Horvitz, 2009). IFBP-1 is an ortholog of human IRF2BP2 and IRF2BPL, which are members of the IRF2BP (interferon regulatory factor 2 binding protein) family of transcriptional repressors (Childs, 2003), while CYP-35A2 is a protein in Cytochrome P450 family (Lim et al., 2022). DDO-2 encodes D-aspartate oxidase that degrades D-amino acids (Saitoh et al., 2012). RPL-22 is a large subunit of ribosomal protein. In these mRNAs, uaf-1 showed the highest increase of expression from 100% to 934.6% in N2 and 134.3% of the N2 AL value to 963.9% in eif-2α respectively (Figure 3C). The fact that UAF-1 is a splicing factor highlights the self-regulation of splicing factors through Alternative splicing and NMD (AS-NMD) (Lejeune, 2022) The alternatively spliced isoform of ifbp-1 also went up 81.9% in N2 and 72.55% in eif-2α respectively (Figure 3D). The higher levels of uaf-1 and ifbp-1 with IR under FT conditions is suggestive of downregulation of NMD for these genes. Interestingly, cyp-35A2, ddo-2, and rpl-22 that increased IR events under DR as previously reported did not increase their abundance in fasted animals (Figures 3E–G), which could be due to experimental condition variation (i.e., food dilution versus FT). However, the gene expression pattern in all these genes were very similar between N2 and eif-2α, suggesting again that the ISR is not a key player to regulate NMD when nutrients are limited.
The ISR is dispensable for increased lifespan under DR
To address whether inactivation of the ISR displays detrimental effects on longevity under DR, we compared lifespan of eif-2α phosphorylation and ISR kinase mutants under AL and DR. Similar to N2, DR promoted lifespan in eif-2α, gcn-2(ok871), and pek-1(ok275), indicating that neither phosphorylation of eIF-2α, nor the individual kinases targeting this translation factor, are needed for lifespan extension under DR (Figure 4; Supplementary Table S3). Interestingly, the lifespan of double mutant gcn-2(ok871);pek-1(ok275) under DR was not significantly different from AL (Figure 4E). This observation suggests more than one phosphorylation target is regulated by these kinases. As with eIF-2α, the target may be shared between GCN-2 and PEK-1. Taken together with earlier results, the ISR is not required for changes in translation, NMD, or lifespan associated with DR. Furthermore, eif-2α lived longer than wild type under AL feeding (p = 0.0053; Supplementary Figure S4 and Supplementary Table S3), indicating that the inability to activate the ISR positively influences longevity under the conditions tested. This finding is consistent with previously reported results (Derisbourg et al., 2021b).
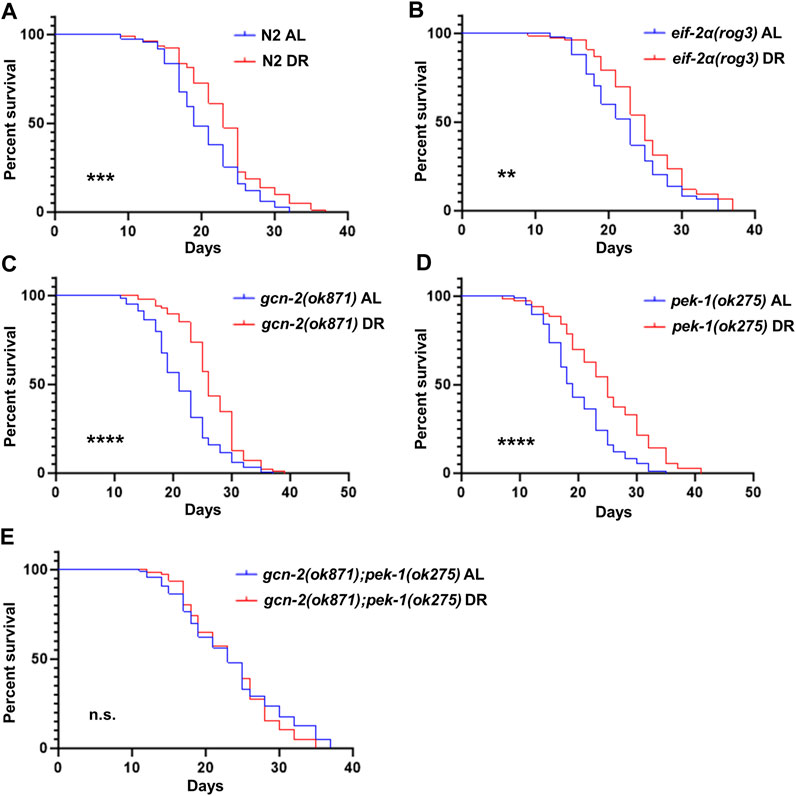
FIGURE 4. eIF-2α phosphorylation is not required for increased survival under DR. Worms were fed AL or DR diets starting from day 1 of adulthood. Percent survival was plotted for (A) N2, (B) eif-2α(rog3), (C) gcn-2(ok871), (D) pek-1(ok275), and (E) gcn-2(ok871);pek-1(ok275). Kaplan–Meier survival curves were compared using the Mantel–Cox log-rank test. *p < 0.05; **p < 0.01; ***p < 0.001; and ****p < 0.0001. See Supplementary Table S3 for additional details and replicates.
The ISR is required for protection from ER stress, but not for fasting or thermal stress
Considering that the ISR is dispensable under DR involving food dilution, we wanted to see if it is required for other forms of stress. To induce ER stress, C. elegans were treated continuously with tunicamycin starting from young adulthood. Compared to N2, the survival of eif-2α was reduced significantly under ER stress (Figure 5A; Supplementary Table S6). Thus, the ISR is required for a wild-type level of survival under conditions of ER stress.
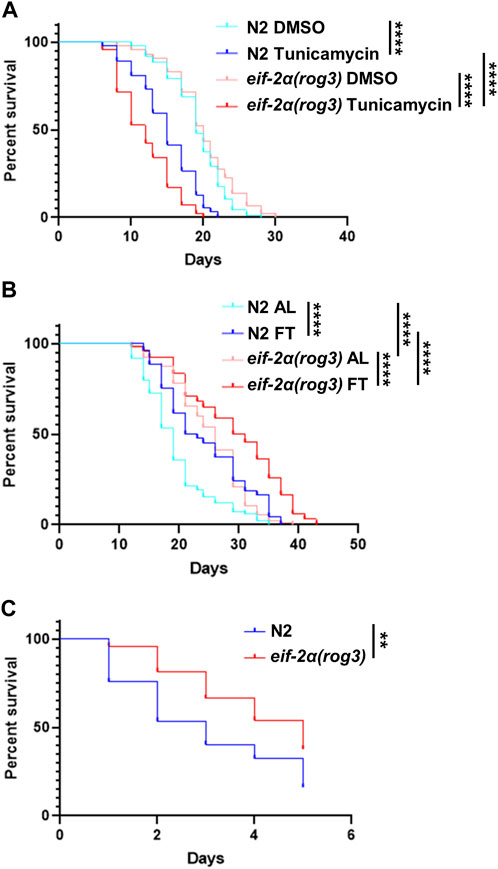
FIGURE 5. eIF-2α phosphorylation is required for normal survival under ER stress, but not for thermal stress. (A) Survival of worms in DMSO (control) or 25 μg/mL tunicamycin starting from day 1 adulthood. (B) Lifespan of worms under AL or fasting (FT) treatments starting from day 1 adulthood. (C) Heat stress recovery assay. Worms were incubated at 35°C for 4 h at day 7 of adulthood then shifted to 20°C. The survival was scored for 5 days continuously. Kaplan–Meier survival curves were compared using the Mantel–Cox log-rank test. *p < 0.05; **p < 0.01; ***p < 0.001; and ****p < 0.0001. See Supplementary Tables S4–S6 for additional details and replicates.
Given the lack of requirement for the ISR in lifespan extension under DR (Figure 4), we wondered if imposing a stronger form of DR requires the ISR for enhanced survival, since this may lead to a greater level of uncharged tRNA associated with its activation. Under fasting conditions starting from young adulthood, both N2 and eif-2α exhibited an extension in lifespan characteristic of this form of DR (Figure 5B; Supplementary Table S5). Notably, under both AL and fasting conditions, the mutant demonstrated a longer lifespan compared to wild-type animals. We also tested the lifespan of gcn-2(ok871);pek-1(ok275) under fasting due to its non-extended lifespan under DR (Supplementary Figure S5). The results were similar to the one under DR. The survival rate of the double mutant did not differ from AL, also suggesting an unidentified shared target exists between GCN-2 and PEK-1 essential for lifespan extension in nutrient deprivation.
Although heat stress is not typically associated with the ISR, we included it since increased resistance to heat stress is frequently associated with gene changes that increase lifespan as observed for the eif-2α mutant under food dilution DR (Figure 4B and Supplementary Table S3) and fasting (Figure 5B). To investigate whether the ISR is involved in the response to heat stress, day 7 adult worms were incubated at 35°C for 4 h and their survival was scored daily thereafter. The inhibition of EIF-2α phosphorylation did not decrease the survival rate after heat stress (Figure 5C; Supplementary Table S4). As observed with survival under standard conditions, the eif-2α phospho-null mutant showed increased survival compared to N2.
Extended lifespan in eIF-2α phospho-null mutants is associated with decreased fecundity
Since eif-2α lived longer than N2 animals, we investigated if ISR inhibition affects development and reproduction of animals. We measured the development time of eif-2α and found that it was not significantly different between the mutant and N2, with both taking between 78 and 80 h to go from egg to egg-laying adult (Figure 6A). However, the total number of progeny was significantly lower in eif-2α compared to N2 (Figure 6B). Therefore, inhibition of the ISR leads to an extension of lifespan in the absence of stress, but it also showed diminished reproductive ability. This is consistent with trade-offs frequently observed in long-lived mutants.
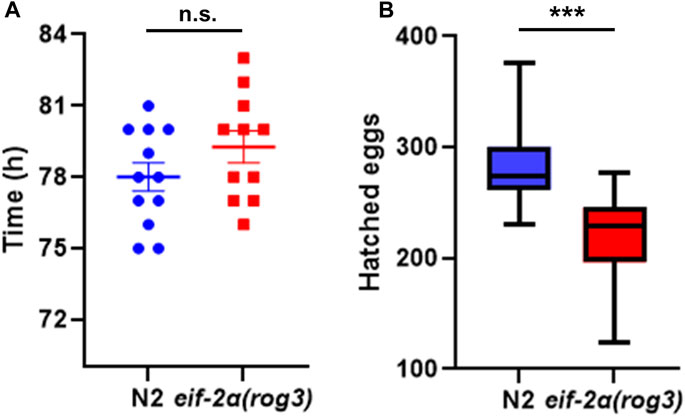
FIGURE 6. eIF-2α phospho-null mutant has smaller brood size. (A) Development time and (B) hatched egg numbers of N2 and eif-2α(rog3). Representative results of 3 independent experiments are shown. In each experiment, at least 12 worms of each strain were included. Error bars represent means ± SEM. Student’s t-test was performed to determine significance. *p < 0.05; **p < 0.01; ***p < 0.001; and ****p < 0.0001.
The ISR is only activated by PEK-1 in response to ER stress
To gain further insights into the response of the ISR to ER stress, we tested the eIF-2α phosphorylation status of kinase mutants upon exposure to tunicamycin. After 3 h of tunicamycin exposure, only N2 and the gcn-2(ok871) mutant were able to phosphorylate eIF2α, as expected. Conversely, pek-1(ok275) and gcn-2(ok871);pek-1(ok275) double mutants could not (Figure 7A), showing that only PEK-1 is responsible for ISR activation under this condition.
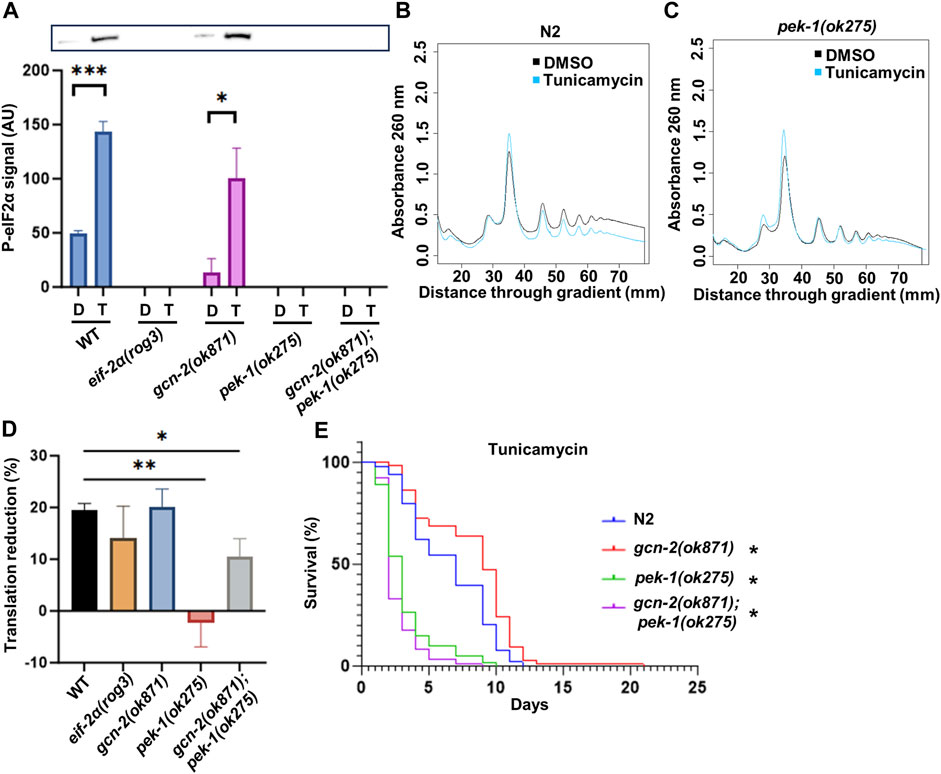
FIGURE 7. The importance of eIF-2α phosphorylation under ER stress. (A) Western blot with anti-eIF2α phosphorylation antibody. Representative membrane is shown. (B,C) Polysome profiling of day 1 adult N2 (B) and pek-1(ok871) (C). Day 1 adult worms were treated with DMSO or 25 μg/mL tunicamycin for 3 h prior to cell lysis. (D) Quantification of translation reduction. Error bars represent means ± SEM (n = 3). One-way ANOVA with Dunnett’s post hoc test was performed. See Supplementary Figure S6 for the polysome profiling of the other strains. (E) Survival of worms in 25 μg/mL tunicamycin starting from day 1 adulthood. See Supplementary Table S7 for additional details and replicates. Kaplan–Meier survival curves were compared using the Mantel–Cox log-rank test. *p < 0.05; **p < 0.01; ***p < 0.001; and ****p < 0.0001.
Since PEK-1 is required for normal survival and eIF-2α phosphorylation in response to tunicamycin, we wondered whether translational downregulation associated with ER stress and ISR activation was impaired in pek-1 mutants. Polysome profiling was conducted after the worms were challenged with tunicamycin for 3 h. In N2, the polysome fraction was reduced by 19.5% (Figures 7B, D). In the pek-1 mutant, translation remained near DMSO control levels in the presence of tunicamycin (Figures 7C, D), which is consistent with the fact that pek-1 is essential to activate ISR under ER stress. The double mutant gcn-2(ok871);pek-1(ok275) also exhibited a muted response compared to N2 (Figure 7D and Supplementary Figure S6C). There was no statistical difference between pek-1(ok275) and the double mutant (p = 0.093). Surprisingly, the eif-2α phospho-mutant showed similar reduction in translation to N2 (Figure 7D and Supplementary Figure S6A), despite the importance of this residue for survival under ER stress (Figure 5A). To confirm the importance of PEK-1 for survival under ER stress, we challenged the ISR kinase mutants with long-term tunicamycin exposure. As expected, the survival rate of pek-1(ok275) and gcn-2(ok871);pek-1(ok275) dropped dramatically (Figure 7E) while the DMSO control worms all showed similar lifespans (Supplementary Figure S6D). Taken together, PEK-1 activates the ISR under ER stress and the ISR is necessary for the ER stress response.
Discussion
In this study, we explored the role of the ISR in various stress conditions. We found that the ISR plays a critical role in the ER stress response. Specifically, eIF2α was phosphorylated by PEK-1 under ER stress, and inhibition of ISR activation not only hindered attenuation of total translation but also showed detrimental effects on survival. Conversely, the ISR was not required for lifespan extension under food dilution or fasting conditions. Despite an increase in GCN-2 mediated eIF-2α phosphorylation during fasting, the ISR was not required for translation suppression in gcn-2 or eif-2α phospho-mutants. Furthermore, ISR inhibition did not impact the ability to differentially regulate NMD according to levels of intron-bearing mRNA of several genes tested. Interestingly, inhibiting phosphorylation of eIF2α at S49 via S-to-A mutation extended lifespan in the absence of stress. However, this came at the cost of reduced fecundity, indicating a trade-off between lifespan extension and reproductive ability.
Monitoring translation changes that suppress and redirect protein synthesis is critical to understand stress response mechanisms. Low translation states are frequently associated with increased lifespan. Consequently, the availability of reliable methods to measure translation within specific time frames and under particular treatments is critical to comprehending the underlying genetic mechanisms. By accurately measuring translation changes, researchers can assess the impact of specific stress or treatments, and further unravel the complex interplay between genetic regulation and stress responses. Various approaches have been developed to measure translation quantitatively. One common method is polysome profiling, which allows for isolation of actively translated mRNA and separates ribosomal subunits, monosomes, and polysomes (Merret et al., 2015; Chassé et al., 2017; Rollins et al., 2019). This type of profiling measures not just total translation but can also be used to measure gene-specific translation efficiency according to expression of specific mRNA within isolated polysomes. However, polysomal analysis is time consuming and requires a large quantity of worms to generate profiles. Therefore, it is advantageous to have an alternative method to quantify translation. The SUnSET puromycin assay is a fitting solution in this regard. Puromycin replaces aa-tRNA to enter the ribosome, causing translation termination and subsequent release of puromycin-bound peptidyl-tRNA. The levels of puromycylation directly correlate with the overall rate of translation (Aviner, 2020). Thus, its incorporation enables the quantification of global translation. Quantification can be achieved biochemically via Western blotting or by imaging fluorescently labeled puromycin in individual worms (Somers et al., 2022). In this study, we further refined the assay by optimizing the protocol using the bus-5(br19) mutant and short-period puromycin incubation. In this way, incorporation does not rely on food source, making it ideal to measure translation even under fasting conditions.
The phosphorylation of eIF2α can be uncoupled from downstream ISR effectors (Wanders et al., 2016; Pettit et al., 2017). While activated, the ISR attenuates global protein synthesis and promotes the translation of specific mRNAs. The four kinases GCN2, PERK, PKR, and HRI phosphorylate eIF2α under different stress conditions. Each of these kinases bear conserved kinase and regulatory domains allowing them to respond to diverse conditions (Costa-Mattioli and Walter, 2020). In nutrient restriction stress, studies reported increased levels of eIF2α phosphorylation and ATF4 expression (Lee et al., 2008; Sikalidis and Stipanuk, 2010; Sikalidis et al., 2011). However, eIF2α phosphorylation does not always correlate with ATF4 expression and overall protein changes. Feeding a methionine-restricted diet to wild-type and Gcn2−/− mice had similar effects on many genes regulated by the ISR, including ATF4 (Pettit et al., 2017). Similarly, methionine restricted diet was reported to activate the ISR (Rajanala et al., 2019) and results in phenotypes (Wanders et al., 2016) that are independent from GCN2 and PERK. In our study, the role of C. elegans ATF-4 was not directly assessed, although its expression was not significantly changed under DR in either N2 wild type or eif-2α phospho-null mutants. Indeed, although GCN-2 activates the ISR in C. elegans during fasting (Figure 1C), it is not required to downregulate translation or to increase lifespan under nutrient-limiting conditions (Figures 2, 4).
ISR kinases harbor functions beyond eIF-2α phosphorylation. PERK not only phosphorylates eIF2α to induce global attenuation of protein synthesis (Harding et al., 2000) and stimulate expression of stress response proteins, but is also involved in activation of ATF6, which increases expression of UPR target genes in mice (Teske et al., 2011). Our findings are in line with the function of PERK across various species, particularly in regard to translational downregulation and survival under ER stress. Interestingly, one study found that, while the phosphorylation of eIF2α was reduced in PERK−/− fibroblasts under ER stress, GCN2 was able to compensate partially for the loss of PERK (Hamanaka et al., 2005). In our study, GCN-2 was only active under FT and PEK-1 was only active under ER stress with respect to eIF-2α phosphorylation (Figures 1C, 7A). This does not necessarily mean that GCN-2 and PEK-1 cannot compensate for each other’s activity. For example, the compensation may target a different phosphorylation site on eIF-2α besides S49. The notion that other kinase targets may exist is supported by results from the gcn-2(ok871);pek-1(ok275) double mutant, which did not show lifespan extension under DR whereas the single kinase and eIF-2α phospho-mutants did (Figure 4). Additionally, the translation reduction was completely inhibited in the loss-of-function pek-1 mutant, but not in the eif-2α phospho-mutant (Figure 7D). Both observations suggest that eIF2α may have other important phosphosites related to ISR activation or some other targets of GCN-2 or PEK-1 may exist. For example, Cullinan et al. showed that PERK also phosphorylates Nrf2 transcription factor (SKN-1 in C. elegans) and stimulates dissociation of the cytoplasmic Nrf2/Keap1 complex (Cullinan et al., 2003). Nrf2 deletion decreased cell survival rate compared to wild type after inducing ER stress (Cullinan et al., 2003). To our knowledge, Nrf2 is the only target of PERK besides eIF2α that has been identified.
As previously reported, the ISR and NMD coregulate each other in response to stress. Both hypoxia and ER stress suppress NMD via eIF2α phosphorylation in mammalian cell lines (Gardner, 2008; Usuki et al., 2013) and the ISR factor ATF4 is otherwise subject to degradation by NMD. Conversely, it was reported recently that NMD inhibition via pharmacologic disruption of SMG1 activated the ISR in mice, revealing a bidirectional relationship. (De La Peña et al., 2023). However, in our study, eIF2α phosphorylation was not required for NMD changes during fasting (Figure 3, Supplementary Figure S1). Whether NMD inactivation affects ISR activity was not examined and will need further exploration. Under DR, NMD is downregulated but not completely inactive (Figure 3). In fact, NMD still plays an important role in specific gene expression regulation, since loss of NMD factors negatively affects benefits of increased longevity under DR (Rollins et al., 2019; Kim et al., 2020).
AS-NMD is usually coupled to regulate expression of specific genes to adapt to environmental stress. One study showed that RNP-6 modulates alternative splicing events and regulates longevity by mTORC1 downregulation (Huang et al., 2022). Similarly, under DR, as a result of AS-NMD, IR events increased in both translated and total fraction showing specific gene expression regulation (Rollins et al., 2019). The existence of PTC-containing transcripts among polysomes suggests that a higher proportion of these transcripts evaded the surveillance by NMD. Most of PTC-containing transcripts remain unfunctional, but a few of them can be translated and synthesize different protein variants (Ge and Porse, 2014). In this study, we noticed that IR events increased for specific genes under fasting, specifically for those encoding the splicing factor UAF-1 and transcriptional regulator IFBP-1 (Figures 3C,D). IR favors producing more varieties of protein isoforms, which is critical for restoring transcriptome homeostasis in response to stress. Some of the intron-retained isoforms decreased under FT in our experiments (e.g., cyp-35A2, ddo-2 and rpl-22; Figures 3E–G). This can be due to two reasons. One is that under FT the total translation activity goes down causing the lower transcription activity of these isoforms. Another is that even though NMD is downregulated in nutrient scarcity, it is still critical for specific gene expression regulation. Thus, NMD may further degrade the IR isoforms of these genes.
This study explored the role of ISR, especially eIF-2α phosphorylation, under stress and DR conditions. Further studies are needed to look into the potential for additional targets of GCN-2 and PEK-1, as well as the role AS-NMD plays in changing mRNA processing to differentially regulate gene expression under nutrient limiting conditions.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics statement
The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
ZM: Writing–original draft, Writing–review and editing. JH: Writing–original draft. DM: Writing–original draft. MC: Writing–original draft. MR: Writing–original draft. JR: Writing–original draft. AR: Writing–original draft, Writing–review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the grant from the NIH (R21 AG056743) and the Morris Scientific Discovery Fund. In addition, research reported in this publication was supported by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant numbers P20GM103423 and P20GM104318. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2023.1263344/full#supplementary-material
References
Arnold, A., Rahman, M. M., Lee, M. C., Muehlhaeusser, S., Katic, I., Gaidatzis, D., et al. (2014). Functional characterization of C. elegans Y-box-binding proteins reveals tissue-specific functions and a critical role in the formation of polysomes. Nucleic Acids Res. 42, 13353–13369. doi:10.1093/nar/gku1077
Aviner, R. (2020). The science of puromycin: from studies of ribosome function to applications in biotechnology. Comput. Struct. Biotechnol. J. 18, 1074–1083. doi:10.1016/j.csbj.2020.04.014
Baker, B. M., Nargund, A. M., Sun, T., and Haynes, C. M. (2012). Protective coupling of mitochondrial function and protein synthesis via the eIF2α kinase GCN-2. PLOS Genet. 8, e1002760. doi:10.1371/journal.pgen.1002760
Barber, G. N. (2001). Host defense, viruses and apoptosis. Cell Death Differ. 8, 113–126. doi:10.1038/sj.cdd.4400823
Bond, S., Lopez-Lloreda, C., Gannon, P. J., Akay-Espinoza, C., and Jordan-Sciutto, K. L. (2020). The integrated stress response and phosphorylated eukaryotic initiation factor 2α in neurodegeneration. J. Neuropathology Exp. Neurology 79, 123–143. doi:10.1093/jnen/nlz129
Bunpo, P., Cundiff, J. K., Reinert, R. B., Wek, R. C., Aldrich, C. J., and Anthony, T. G. (2010). The eIF2 kinase GCN2 is essential for the murine immune system to adapt to amino acid deprivation by asparaginase. J. Nutr. 140, 2020–2027. doi:10.3945/jn.110.129197
Butcher, S. K., and Lord, J. M. (2004). Stress responses and innate immunity: aging as a contributory factor. Aging Cell 3, 151–160. doi:10.1111/j.1474-9728.2004.00103.x
Chassé, H., Boulben, S., Costache, V., Cormier, P., and Morales, J. (2017). Analysis of translation using polysome profiling. Nucleic Acids Res. 45, e15. doi:10.1093/nar/gkw907
Chen, J.-J. (2014). Translational control by heme-regulated eIF2α kinase during erythropoiesis. Curr. Opin. Hematol. 21, 172–178. doi:10.1097/MOH.0000000000000030
Childs, K. S., and Goodbourn, S. (2003). Identification of novel co-repressor molecules for interferon regulatory factor-2. Nucleic Acids Res. 31, 3016–3026. doi:10.1093/nar/gkg431
Costa-Mattioli, M., and Walter, P. (2020). The integrated stress response: from mechanism to disease. Science 368, eaat5314. doi:10.1126/science.aat5314
Cullinan, S. B., Zhang, D., Hannink, M., Arvisais, E., Kaufman, R. J., and Diehl, J. A. (2003). Nrf2 is a direct PERK substrate and effector of PERK-dependent cell survival. Mol. Cell. Biol. 23, 7198–7209. doi:10.1128/MCB.23.20.7198-7209.2003
De La Peña, J. B., Chase, R., Kunder, N., Smith, P. R., Lou, T.-F., Stanowick, A., et al. (2023). Inhibition of nonsense-mediated decay induces nociceptive sensitization through activation of the integrated stress response. J. Neurosci. 43, 2921–2933. doi:10.1523/JNEUROSCI.1604-22.2023
Derisbourg, M. J., Hartman, M. D., and Denzel, M. S. (2021a). Perspective: modulating the integrated stress response to slow aging and ameliorate age-related pathology. Nat. Aging 1, 760–768. doi:10.1038/s43587-021-00112-9
Derisbourg, M. J., Wester, L. E., Baddi, R., and Denzel, M. S. (2021b). Mutagenesis screen uncovers lifespan extension through integrated stress response inhibition without reduced mRNA translation. Nat. Commun. 12, 1678. doi:10.1038/s41467-021-21743-x
Ecker, N., Mor, A., Journo, D., and Abeliovich, H. (2010). Induction of autophagic flux by amino acid deprivation is distinct from nitrogen starvation-induced macroautophagy. Autophagy 6, 879–890. doi:10.4161/auto.6.7.12753
Gardner, L. B. (2008). Hypoxic inhibition of nonsense-mediated RNA decay regulates gene expression and the integrated stress response. Mol. Cell. Biol. 28, 3729–3741. doi:10.1128/MCB.02284-07
Ge, Y., and Porse, B. T. (2014). The functional consequences of intron retention: alternative splicing coupled to NMD as a regulator of gene expression. BioEssays 36, 236–243. doi:10.1002/bies.201300156
Greer, E. L., and Brunet, A. (2009). Different dietary restriction regimens extend lifespan by both independent and overlapping genetic pathways in C. elegans. Aging Cell 8, 113–127. doi:10.1111/j.1474-9726.2009.00459.x
Haigis, M. C., and Yankner, B. A. (2010). The aging stress response. Mol. Cell 40, 333–344. doi:10.1016/j.molcel.2010.10.002
Hamanaka, R. B., Bennett, B. S., Cullinan, S. B., and Diehl, J. A. (2005). PERK and GCN2 contribute to eIF2alpha phosphorylation and cell cycle arrest after activation of the unfolded protein response pathway. MBoC 16, 5493–5501. doi:10.1091/mbc.e05-03-0268
Harding, H. P., Novoa, I., Zhang, Y., Zeng, H., Wek, R., Schapira, M., et al. (2000). Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 6, 1099–1108. doi:10.1016/S1097-2765(00)00108-8
Harding, H. P., Zeng, H., Zhang, Y., Jungries, R., Chung, P., Plesken, H., et al. (2001). Diabetes mellitus and exocrine pancreatic dysfunction in Perk−/− mice reveals a role for translational control in secretory cell survival. Mol. Cell 7, 1153–1163. doi:10.1016/S1097-2765(01)00264-7
Heissenberger, C., Rollins, J. A., Krammer, T. L., Nagelreiter, F., Stocker, I., Wacheul, L., et al. (2020). The ribosomal RNA m5C methyltransferase NSUN-1 modulates healthspan and oogenesis in Caenorhabditis elegans. eLife 9, e56205. doi:10.7554/eLife.56205
Horn, M., Denzel, S. I., Srinivasan, B., Allmeroth, K., Schiffer, I., Karthikaisamy, V., et al. (2020). Hexosamine pathway activation improves protein homeostasis through the integrated stress response. iScience 23, 100887. doi:10.1016/j.isci.2020.100887
Howard, A. C., Rollins, J., Snow, S., Castor, S., and Rogers, A. N. (2016). Reducing translation through eIF4G/IFG-1 improves survival under ER stress that depends on heat shock factor HSF-1 in Caenorhabditis elegans. Aging Cell 15, 1027–1038. doi:10.1111/acel.12516
Huang, W., Kew, C., Fernandes, S. de A., Löhrke, A., Han, L., Demetriades, C., et al. (2022). Decreased spliceosome fidelity and egl-8 intron retention inhibit mTORC1 signaling to promote longevity. Nat. Aging 2, 796–808. doi:10.1038/s43587-022-00275-z
Hug, N., Longman, D., and Cáceres, J. F. (2016). Mechanism and regulation of the nonsense-mediated decay pathway. Nucl. Acids Res. 44, 1483–1495. doi:10.1093/nar/gkw010
Jackson, R. J., Hellen, C. U. T., and Pestova, T. V. (2010). The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 11, 113–127. doi:10.1038/nrm2838
Jiang, J. C., Jaruga, E., Repnevskaya, M. V., and Jazwinski, S. M. (2000). An intervention resembling caloric restriction prolongs life span and retards aging in yeast. FASEB J. 14, 2135–2137. doi:10.1096/fj.00-0242fje
Jonsson, W. O., Margolies, N. S., and Anthony, T. G. (2019). Dietary sulfur amino acid restriction and the integrated stress response: mechanistic insights. Nutrients 11, 1349. doi:10.3390/nu11061349
Kapahi, P., Chen, D., Rogers, A. N., Katewa, S. D., Li, P. W.-L., Thomas, E. L., et al. (2010). With tor, less is more: a key role for the conserved nutrient-sensing tor pathway in aging. Cell Metab. 11, 453–465. doi:10.1016/j.cmet.2010.05.001
Karam, R., Lou, C.-H., Kroeger, H., Huang, L., Lin, J. H., and Wilkinson, M. F. (2015). The unfolded protein response is shaped by the NMD pathway. EMBO Rep. 16, 599–609. doi:10.15252/embr.201439696
Kaya, A., Lee, B. C., and Gladyshev, V. N. (2015). Regulation of protein function by reversible methionine oxidation and the role of selenoprotein MsrB1. Antioxidants Redox Signal. 23, 814–822. doi:10.1089/ars.2015.6385
Kim, E. J. E., Son, H. G., Park, H.-E. H., Jung, Y., Kwon, S., and Lee, S.-J. V. (2020). Caenorhabditis elegans algn-2 is critical for longevity conferred by enhanced nonsense-mediated mRNA decay. iScience 23, 101713. doi:10.1016/j.isci.2020.101713
Kim, S., Kim, D. K., Jeong, S., and Lee, J. (2022). The common cellular events in the neurodegenerative diseases and the associated role of endoplasmic reticulum stress. Int. J. Mol. Sci. 23, 5894. doi:10.3390/ijms23115894
Korennykh, A., and Walter, P. (2012). Structural basis of the unfolded protein response. Annu. Rev. Cell Dev. Biol. 28, 251–277. doi:10.1146/annurev-cellbio-101011-155826
Kulalert, W., and Kim, D. H. (2013). The unfolded protein response in a pair of sensory neurons promotes entry of C. elegans into dauer diapause. Curr. Biol. 23, 2540–2545. doi:10.1016/j.cub.2013.10.058
Kyriakakis, E., Princz, A., and Tavernarakis, N. (2015). “Stress responses during ageing: molecular pathways regulating protein homeostasis,” in Stress responses. Editor C. M. Oslowski (New York, NY: Springer New York), 215–234. Available at: http://link.springer.com/10.1007/978-1-4939-2522-3_16 (Accessed March 26, 2015).
Lee, J.-I., Dominy, J. E., Sikalidis, A. K., Hirschberger, L. L., Wang, W., and Stipanuk, M. H. (2008). HepG2/C3A cells respond to cysteine deprivation by induction of the amino acid deprivation/integrated stress response pathway. Physiol. Genomics 33, 218–229. doi:10.1152/physiolgenomics.00263.2007
Lejeune, F. (2022). Nonsense-mediated mRNA decay, a finely regulated mechanism. Biomedicines 10, 141. doi:10.3390/biomedicines10010141
Li, Z., Vuong, J. K., Zhang, M., Stork, C., and Zheng, S. (2017). Inhibition of nonsense-mediated RNA decay by ER stress. RNA 23, 378–394. doi:10.1261/rna.058040.116
Lim, S. Y. M., Alshagga, M., Kong, C., Alshawsh, M. A., Alshehade, S. A., and Pan, Y. (2022). CYP35 family in Caenorhabditis elegans biological processes: fatty acid synthesis, xenobiotic metabolism, and stress responses. Arch. Toxicol. 96, 3163–3174. doi:10.1007/s00204-022-03382-3
Longman, D., Plasterk, R. H. A., Johnstone, I. L., and Caceres, J. F. (2007). Mechanistic insights and identification of two novel factors in the C. elegans NMD pathway. Genes and Dev. 21, 1075–1085. doi:10.1101/gad.417707
Ma, L., and Horvitz, H. R. (2009). Mutations in the Caenorhabditis elegans U2AF large subunit UAF-1 alter the choice of a 3′ splice site in vivo. PLoS Genet. 5, e1000708. doi:10.1371/journal.pgen.1000708
Martin, L., and Gardner, L. B. (2015). Stress-induced inhibition of nonsense-mediated RNA decay regulates intracellular cystine transport and intracellular glutathione through regulation of the cystine/glutamate exchanger SLC7A11. Oncogene 34, 4211–4218. doi:10.1038/onc.2014.352
Merret, R., Nagarajan, V. K., Carpentier, M.-C., Park, S., Favory, J.-J., Descombin, J., et al. (2015). Heat-induced ribosome pausing triggers mRNA co-translational decay in Arabidopsis thaliana. Nucleic Acids Res. 43, 4121–4132. doi:10.1093/nar/gkv234
Mitrovich, Q. M., and Anderson, P. (2000). Unproductively spliced ribosomal protein mRNAs are natural targets of mRNA surveillance in C. elegans. Genes Dev. 14, 2173–2184. doi:10.1101/gad.819900
Pakos-Zebrucka, K., Koryga, I., Mnich, K., Ljujic, M., Samali, A., and Gorman, A. M. (2016). The integrated stress response. EMBO Rep. 17, 1374–1395. doi:10.15252/embr.201642195
Pettit, A. P., Jonsson, W. O., Bargoud, A. R., Mirek, E. T., Peelor, F. F., Wang, Y., et al. (2017). Dietary methionine restriction regulates liver protein synthesis and gene expression independently of eukaryotic initiation factor 2 phosphorylation in mice. J. Nutr. 147, 1031–1040. doi:10.3945/jn.116.246710
Rajanala, S. H., Ringquist, R., and Cryns, V. L. (2019). Methionine restriction activates the integrated stress response in triple-negative breast cancer cells by a GCN2- and PERK-independent mechanism. Am. J. Cancer Res. 9, 1766–1775.
Richardson, C. E., Kinkel, S., and Kim, D. H. (2011). Physiological IRE-1-XBP-1 and PEK-1 signaling in Caenorhabditis elegans larval development and immunity. PLoS Genet. 7, e1002391. doi:10.1371/journal.pgen.1002391
Rollins, J. A., Shaffer, D., Snow, S. S., Kapahi, P., and Rogers, A. N. (2019). Dietary restriction induces posttranscriptional regulation of longevity genes. Life Sci. Alliance 2, e201800281. doi:10.26508/lsa.201800281
Ron, D., and Walter, P. (2007). Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 8, 519–529. doi:10.1038/nrm2199
Rual, J.-F., Ceron, J., Koreth, J., Hao, T., Nicot, A.-S., Hirozane-Kishikawa, T., et al. (2004). Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 14, 2162–2168. doi:10.1101/gr.2505604
Saitoh, Y., Katane, M., Kawata, T., Maeda, K., Sekine, M., Furuchi, T., et al. (2012). Spatiotemporal localization of d -amino acid oxidase and d -aspartate oxidases during development in Caenorhabditis elegans. Mol. Cell. Biol. 32, 1967–1983. doi:10.1128/MCB.06513-11
Shaffer, D., and Rollins, J. (2020). Fluorescent polysome profiling in Caenorhabditis elegans. Bio Protoc. 10, e3742. doi:10.21769/BioProtoc.3742
Sikalidis, A. K., Lee, J.-I., and Stipanuk, M. H. (2011). Gene expression and integrated stress response in HepG2/C3A cells cultured in amino acid deficient medium. Amino Acids 41, 159–171. doi:10.1007/s00726-010-0571-x
Sikalidis, A. K., and Stipanuk, M. H. (2010). Growing rats respond to a sulfur amino acid-deficient diet by phosphorylation of the alpha subunit of eukaryotic initiation factor 2 heterotrimeric complex and induction of adaptive components of the integrated stress response. J. Nutr. 140, 1080–1085. doi:10.3945/jn.109.120428
Somers, H. M., Fuqua, J. H., Bonnet, F. X. A., and Rollins, J. A. (2022). Quantification of tissue-specific protein translation in whole C. elegans using O-propargyl-puromycin labeling and fluorescence microscopy. Cell Rep. Methods 2, 100203. doi:10.1016/j.crmeth.2022.100203
Sonenberg, N., and Hinnebusch, A. G. (2009). Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136, 731–745. doi:10.1016/j.cell.2009.01.042
Teske, B. F., Wek, S. A., Bunpo, P., Cundiff, J. K., McClintick, J. N., Anthony, T. G., et al. (2011). The eIF2 kinase PERK and the integrated stress response facilitate activation of ATF6 during endoplasmic reticulum stress. MBoC 22, 4390–4405. doi:10.1091/mbc.e11-06-0510
Usuki, F., Fujimura, M., and Yamashita, A. (2013). Endoplasmic reticulum stress preconditioning attenuates methylmercury-induced cellular damage by inducing favorable stress responses. Sci. Rep. 3, 2346. doi:10.1038/srep02346
Verfaillie, T., Rubio, N., Garg, A. D., Bultynck, G., Rizzuto, R., Decuypere, J.-P., et al. (2012). PERK is required at the ER-mitochondrial contact sites to convey apoptosis after ROS-based ER stress. Cell Death Differ. 19, 1880–1891. doi:10.1038/cdd.2012.74
Wanders, D., Stone, K. P., Forney, L. A., Cortez, C. C., Dille, K. N., Simon, J., et al. (2016). Role of GCN2-independent signaling through a noncanonical PERK/NRF2 pathway in the physiological responses to dietary methionine restriction. Diabetes 65, 1499–1510. doi:10.2337/db15-1324
Wang, M., and Kaufman, R. J. (2016). Protein misfolding in the endoplasmic reticulum as a conduit to human disease. Nature 529, 326–335. doi:10.1038/nature17041
Wek, R. C., and Cavener, D. R. (2007). Translational control and the unfolded protein response. Antioxidants Redox Signal. 9, 2357–2371. doi:10.1089/ars.2007.1764
Xiong, H., Pears, C., and Woollard, A. (2017). An enhanced C. elegans based platform for toxicity assessment. Sci. Rep. 7, 9839–9911. doi:10.1038/s41598-017-10454-3
Keywords: integrated stress response, nonsense mediated decay, aging, dietary restriction, endoplasmic reticulum stress
Citation: Ma Z, Horrocks J, Mir DA, Cox M, Ruzga M, Rollins J and Rogers AN (2023) The integrated stress response protects against ER stress but is not required for altered translation and lifespan from dietary restriction in Caenorhabditis elegans. Front. Cell Dev. Biol. 11:1263344. doi: 10.3389/fcell.2023.1263344
Received: 20 July 2023; Accepted: 24 November 2023;
Published: 14 December 2023.
Edited by:
Jiang-An Yin, University of Zurich, SwitzerlandReviewed by:
Yunpeng Xu, The State University of New Jersey, United StatesYusuke Sekine, University of Pittsburgh, United States
Copyright © 2023 Ma, Horrocks, Mir, Cox, Ruzga, Rollins and Rogers. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aric N. Rogers, YXJvZ2Vyc0BtZGlibC5vcmc=
 Zhengxin Ma
Zhengxin Ma Jordan Horrocks
Jordan Horrocks Dilawar A. Mir
Dilawar A. Mir Matthew Cox
Matthew Cox Aric N. Rogers
Aric N. Rogers