- 1Department of Stem Cell and Regenerative Biotechnology, Institute of Advanced Regenerative Science, Konkuk University, Seoul, Republic of Korea
- 2Guangdong Provincial Key Laboratory of Large Animal Models for Biomedicine, Wuyi University, Jiangmen, China
- 3Department of Animal Science, Sangji University, Wonju, Republic of Korea
- 4Department of Surgery, School of Medicine, Konkuk University, Seoul, Republic of Korea
The aberrant function of ATP-dependent chromatin remodeler INO80 has been implicated in multiple types of cancers by altering chromatin architecture and gene expression; however, the underlying mechanism of the functional involvement of INO80 mutation in cancer etiology, especially in breast cancer, remains unclear. In the present study, we have performed a weighted gene co-expression network analysis (WCGNA) to investigate links between INO80 expression and breast cancer sub-classification and progression. Our analysis revealed that INO80 repression is associated with differential responsiveness of estrogen receptors (ERs) depending upon breast cancer subtype, ER networks, and increased risk of breast carcinogenesis. To determine whether INO80 loss induces breast tumors, a conditional INO80-knockout (INO80 cKO) mouse model was generated using the Cre-loxP system. Phenotypic characterization revealed that INO80 cKO led to reduced branching and length of the mammary ducts at all stages. However, the INO80 cKO mouse model had unaltered lumen morphology and failed to spontaneously induce tumorigenesis in mammary gland tissue. Therefore, our study suggests that the aberrant function of INO80 is potentially associated with breast cancer by modulating gene expression. INO80 mutation alone is insufficient for breast tumorigenesis.
1 Introduction
Breast cancer is a devastating disease caused by genetic and epigenetic aberrations, which lead to alterations in gene expression and subsequently cellular functions. Numerous genetic mutations in epigenetic factors have been identified so far, and may play a key role in breast cancer development and therapy resistance (Wang et al., 2007; Cancer Genome Atlas Network, 2012; Stephens et al., 2012; Wang et al., 2014b; Helming et al., 2014; Kumar et al., 2016; Chu et al., 2017; Nickerson et al., 2017; Swinstead et al., 2018; Li et al., 2021). Among such epigenetic factors, ATP-dependent chromatin remodelers have emerged as potential biomarkers for breast cancer due to their role in regulating a distinct set of gene expression programs. For example, approximately 11% of breast cancer is related to mutations of the SWI/SNF complex, a member of the ATP-dependent chromatin remodeler family, and their roles are implicated in breast cancer cell plasticity and therapeutic response (Kadoch et al., 2013; Helming et al., 2014; Hohmann and Vakoc, 2014; Chu et al., 2017; Nagarajan et al., 2020; Xu et al., 2020). Mechanistically, dysregulation of such chromatin remodelers in breast cancer alter DNA compaction and accessibility, resulting in changes in 3D epigenomic and transcriptional profiles, particularly leading to aberrant expression of oncogenes (Bochar et al., 2000; Guerrero-Martínez and Reyes, 2018; Nagarajan et al., 2020; Kim et al., 2021). It is still unclear, however, whether the ATP-dependent chromatin remodelers are drivers or mere passengers of tumorigenesis as the factors are also linked to mammary stem cell function and mammary gland development in mice (Smalley and Ashworth, 2003; Cohet et al., 2010; Devinoy and Rijnkels, 2010; Macias and Hinck, 2012; Dravis et al., 2018; Holliday et al., 2018; Hanin and Ferguson-Smith, 2020; Ivanova et al., 2021).
Recent studies have shown that both fetal and adult basal cells share common epigenetic features and multi-lineage differentiation potential, and that the transcription factor SOX10 is critical for lineage determination of mammary epithelial cells and breast cancer metastasis (Dravis et al., 2015; Dravis et al., 2018). These findings suggest that breast cancer cells often acquire epigenetic and transcriptional features similar to those of the developing mammary gland. Understanding these similarities may provide insights into the underlying mechanisms of breast cancer development and help identify potential therapeutic targets. Studies have shown that the ATP-dependent chromatin remodeler families are essential for luminal cell identity and promote cell cycle decisions during mammary gland development (Cohet et al., 2010; Serber et al., 2012; Skibinski et al., 2014; Frey et al., 2017). Depletion of these factors results in attenuation of multiple signaling pathways crucial to regulation of mammary epithelial cell fate decisions and proliferation (Skibinski et al., 2014; Frey et al., 2017).
INO80 complex, a member of the ATP-dependent chromatin remodeler family, is involved in multiple functions related to cancer stem cells and cancer progression, through both canonical and non-canonical INO80 complexes that directly modulate chromatin architecture and gene expression (Min et al., 2013; Wang et al., 2014a; Lafon et al., 2015; Runge et al., 2018). Aberrant INO80 function has been associated with progression of multiple types of cancer through its binding to key enhancer and super-enhancer elements involved in oncogenic gene expression, including CXCL5 and MAP3K1 in non-small-cell lung cancer (Zhang et al., 2017), BMPR1A in live cancer (Wang et al., 2019), and MITF and SOX9 in melanoma (Zhou et al., 2016a). Furthermore, silencing of INO80 appears to have a similar effect to dysfunction of KRAS, MYC, PIK3CA, and ERRB2, inhibiting the migration and metastatic abilities of cancer cells (Zhang et al., 2017). While the association between INO80 and breast cancer has been demonstrated elsewhere (Segala et al., 2016), the specific function of the INO80 subunit and the underlying molecular mechanism involved in mammary development and breast cancer have not been fully elucidated.
In this study, we aimed to explore the functional role of INO80 in both breast cancer progression and mammary gland development through analysis of publicly available TCGA datasets and a conditional knockout (cKO) mouse model. Our findings demonstrate a significant, albeit heterogeneous, correlation between INO80 expression and breast cancer progression. Additionally, our study provides evidence to suggest that INO80 loss alone is not sufficient to induce the development of breast cancer in mice.
2 Materials and methods
2.1 Mouse models
All animal experiments in the present study were performed under the guidelines of the Institute of Animal Care and Use Committee of Konkuk University (IACUC# KU21020). The Ino80 cKO allele (Ino802f/2f) and Tg(MMTV-Cre) animals were obtained from the Institut Clinique de la Souris (ICS; llkirch, France) and the Jackson laboratory. Female Ino802f/2f mice were bred with Ino802f/+; MMTV-Cre males to produce littermate control (Ino802f/2f) and experimental (Ino802f/2f; MMTV-Cre) females. PCR genotyping for the Ino80 cKO allele was carried out with the following primers: 5′-AGGCCTTATTTAGCTCAGGTTGGC-3’ (forward) and 5′- CCACTACACACAGCAGATACACAT -3’ (reverse). The PCR amplicons for wildtype and the conditional alleles were 224 and 382 bp, respectively.
2.2 Tissue collection and whole-mount carmine staining
Mammary gland samples were harvested 4, 8, and 16 weeks after birth. Samples from inguinal mammary glands (#1, #2, and #5) on the right side were quickly frozen in liquid nitrogen (LN2) and then kept at −80°C, allowing RNA and protein extraction at later stages. The inguinal mammary glands (#2 and #4) were harvested and fixed in 4% paraformaldehyde (PFA)/PBS overnight at 4°C. After fixation, the glands were preserved in 70% EtOH/PBS for long-term use if needed. The procedure for whole-mount carmine staining was as described in a previous study (Seong et al., 2018). Briefly, samples were fixed in 4% PFA/PBS for 4–6 h, and then lipids were removed from the fat pad with Clarke’s solution (25% acetic anhydride in 75% EtOH) for 16–18 h at room temperature (RT). The slides were washed with 70% EtOH/PBS and PBS, followed by staining in Carmine Alum (C1022; Sigma-Aldrich, Burlington, MA, United States) solution overnight at RT. Samples were rinsed with PBS and 70% EtOH/PBS before incubation in a de-staining solution (2% HCl in 70% EtOH/PBS) for 3–6 h. Tissues were dehydrated in a series of EtOH (70%, 95%, and 100%) and xylene for at least 3 h at each step and mounted on a cover glass. Images were taken using an Olympus SZX7 microscope (Olympus, Tokyo, Japan).
2.3 H&E staining and immunofluorescence
The histology samples were cut into small pieces, placed into cassettes, and submerged in 4% PFA/PBS overnight at 4°C. Then, the samples were dehydrated with 70%, 95%, 100%, and 100% EtOH for 1 h at each step, incubated in xylene for 4 times for 30 min each time, and finally twice in paraffin for 1 h each time. The paraffin blocks were cut to a 5 μm thickness. For H&E staining, sections were rehydrated and incubated in hematoxylin (#1051750500; Sigma-Aldrich) for 5 min, washed in distilled water before being placed in eosin, and then soaked in EtOH 95% for washing. In addition, slides were dried and washed with an EtOH-xylene mixture before mounting with the mounting solution (#25608-33-7; Sigma-Aldrich). For immunofluorescence (IF) staining, tissue samples were steamed in antigen retrieval buffer (#E-IR-R104; Elabscience, Houston, TX, United States) for 40 min to recover antigen epitopes. Non-specific binding was blocked with blocking solution (2% donkey serum plus 3% BSA and 0.1% Triton-X 100 in PBS) for 1 h at RT in advance. Sections were incubated with primary antibodies [anti-KRT18 (1:250, #ab133263; Abcam, Cambridge, United Kingdom), anti-SMA (1:250, #ab124964; Abcam), anti-Ino80 (1:200, #18810-1-AP; Proteintech, Chicago, IL, United States)] overnight at 4°C, incubated with secondary antibodies [anti-rabbit Alexa Fluor 568 (1:250, #ab175471; Abcam) and anti-mouse Alexa Fluor 488 (1:250, #ab150077; Abcam)] in the dark for 1 h, washed with 1X PBS thrice, and briefly counter-stained with DAPI (1:1000, #ab228549; Abcam). The sections were mounted and imaged using a confocal microscope (LSM800; Carl Zeiss, Oberkochen, Germany).
2.4 TCGA and METABRIC analysis
The analysis of breast cancer patient survival was conducted using the Kaplan-Meier Plotter (https://kmplot.com/analysis/index.php?p=service), as described in Nagy et al. (2018). In this analysis, the expression levels of INO80 were utilized to investigate their relevance to patient survival. The mean expression value of all INO80 probes was calculated, and patients were categorized based on the auto cut-off of INO80 expression, with default parameters applied for the analysis, including the ER subtype, overall survival (OS), distant metastasis-free survival (DMFS), and recurrence-free survival (RFS).
Data on invasive breast carcinoma (BRCA) legacy level 3 was obtained from The Cancer Genome Atlas (TCGA) cohort, and raw RNA-seq counts and clinical data (1108 primary tumors and 114 normal samples) were retrieved using the TCGAbiolinks R package. The data was filtered and normalized using the TCGAanalyze_Normalization function to adjust the GC-content effect on read counts. Normalized transcriptomics and clinical data of METABRIC dataset (1966 tumor and 133 normal samples) were retrieved from MetaGxData package and cBioportal, respectively. Patients with INO80 expression were classified into molecular subtypes, such as normal, basal-like, HER2-enriched, luminal A, luminal B, and normal-like, and by tumor stage in each database. Samples without standards were excluded from the analysis. The INO80 expression sample also groups based on the ER_IHC, PR_IHC, or HER2_IHC subgroups. After statistical analysis with unpaired t-tests, plots were generated using the ggpubr and ggplot2 packages to visualize the relative INO80 expression between breast cancer subtypes, IHC groups, and cancer stage classification.
2.5 Weighted gene co-expression correlation network analysis (WGCNA)
Clinical features, gene copy number variation (CNV) data, and gene expression profiles of patients with breast cancer were obtained from TCGA and METABRIC, and analyzed using cBioportal. Only patients with estrogen immunohistochemistry (IHC) staining and gene expression data were included in the analysis. Weighted gene co-expression network analysis (WGCNA, ver.1.70) was performed on mRNA data to identify modules of co-expressed genes based on GISTIC and IHC traits. Poorly expressed genes and outliers were removed, and data was normalized using the limma package, with the top 5,000 genes selected. A β = 4 threshold power was chosen based on co-expression similarity to the scale-free topology fit index curve (h = 0.9), and the tree cut parameters were set at 0.15 with a minimum module size of 30 genes. The highest correlated module was selected for downstream analysis. Specific gene networks were identified using unsupervised clustering, and heatmaps were generated using the pheatmap package (ver. 1.0.12). Gene ontology analysis was performed using DAVID (ver. 6.8) (Huang et al., 2009a; Huang et al., 2009b), and gene–trait correlations were illustrated using gene significance (GS) scores and VisANT network analysis (Hu et al., 2004).
2.6 Quantification and image analyses
ImageJ (ver. 1.52, https://imagej.nih.gov/ij/) was utilized for the analyses of whole-mount carmine staining, immunofluorescence, and H&E staining in our study. The software enabled the measurement of various parameters, including the length of the mammary gland ductal tree in millimeters from the nipple to the last branch, the number of branching points, and the relative area of the fat pad in the mammary gland.
2.7 Statistical analyses
SigmaPlot (ver. 14; Systat Software, Chicago, IL, United States) and GraphPad Prism (ver. 5; GraphPad Software, La Jolla, CA, United States) were used for statistical analysis and producing graphs. Data are presented as mean and standard error of the mean (±SEM). Student’s t-test and one-way ANOVA with Bonferroni post hoc tests were used to determine statistical significance.
3 Results
3.1 INO80 expression in breast cancer
To investigate the contribution of the INO80 complex to breast cancer, we first examined the frequency of alteration in the expression of INO80 complex subunits in various cancer types using the cBioportal platform. To that end, a large amount of sequencing data from patients with cancer was retrieved from TCGA and METABRIC to investigate the impact of INO80 on breast cancer progression and outcome. As shown in Supplementary Figure S1A, INO80 complex subunits exhibit a high frequency of alteration in multiple cancer types. For instance, alterations in the INO80 complex were found in approximately 80% of non-small-cell lung carcinomas (PanCancer Atlas), and in approximately 60% of breast, lung, and colorectal cancers (PanCancer Atlas). Interestingly, INO80 showed an alteration frequency of 1%–8% in most cancer types and approximately 5% in all patients with breast cancer (Supplementary Figure S1B).
Next, we aimed to determine the expression levels of INO80 in normal tissue and different breast cancer PAM50 subtypes, including basal, luminal A, luminal B, HER2, and normal-like. The PAM50 approach is closely associated with pathological classification and provides a means to identify or characterize cancer subtypes using RNA analysis rather than traditional histological staining methods. PAM50 intrinsic breast cancer subtypes, along with the associated proliferation score and risk of recurrence score (ROR-PT), are independent prognostic factors that enhance the classification of breast cancer patients into prognostic groups (Nielsen et al., 2010; Liu et al., 2016; Ohnstad et al., 2017). A remarkably lower expression of INO80 was found in all breast cancer subtypes, with significantly downregulated INO80 in the basal type compared with that in the normal sample (Figure 1A; Supplementary Figure S1C). Meanwhile, a relatively higher level of mean INO80 expression was found in patients with luminal breast cancer classified as ER and/or progesterone receptor (PR) IHC-positive (Figures 1A, B; Supplementary Figures S1C, D). Furthermore, there was no significant correlation between INO80 expression and tumor stage or HER2 IHC classification (Supplementary Figures S1E, F). On the other hand, lower expression of INO80 affects overall survival (OS) rate, Distant Metastasis-free survival (DMFS), and Recurrence-free survival (RFS) endpoints in breast cancer. The Kaplan-Meier survival analysis of the TCGA dataset revealed that breast cancer patients with lower INO80 expression had reduced survival probabilities over a 150-month follow-up period (Figure 1C).
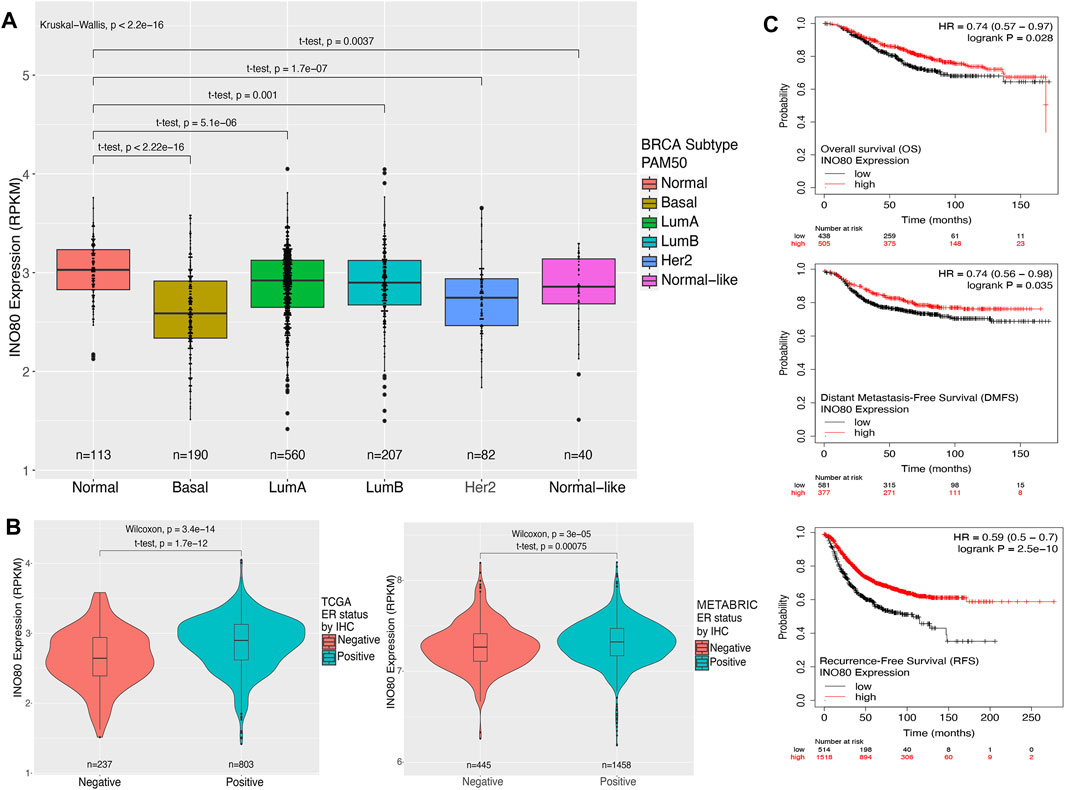
FIGURE 1. Expression and survival analysis of INO80 in breast cancer. (A) The level of INO80 expression in normal tissue and breast cancer subtypes. RNA-seq FPKMs values were retrieved from TCGA and analyzed based on clinical classification. Statistical analysis was performed using unequal t-tests, with significance set at p ≤ 0.05. (B) Comparison of INO80 expression between ER-positive and ER-negative cohorts in TCGA (left) and METABRIC data (right). Two cohorts were classified by immunohistochemistry (IHC) clinical data on ER and INO80 FPKMs between groups and statistically analyzed using the t-test with unequal sample sizes. (C) Kaplan–Meier survival analysis of patients with breast cancer and INO80 mutation and wild-type (WT) groups was conducted using the DriverDBv3 tool. The analysis used default parameters with gene symbols and means on the survival function, with overall survival (OS—top), disease-specific survival (DSS—center), and progression-free survival (PFI—bottom). BRCA, Breast Cancer LumA; Luminal subtype A; LumB, Luminal subtype B; HER2, Human epidermal growth factor receptor 2 subtype; PAM50, Prediction Analysis of Microarray 50; RPKM, reads per kilobase of transcript per million reads mapped.
3.2 INO80 mutations are commonly detected in human breast cancer and correlated with breast cancer subtype
Given the significant increase in the risk of breast cancer and decreased survival rate observed in patients with INO80 complex dysfunction (Figure 1C), as well as the need for further investigation into the association between INO80 deregulation and breast cancer, WGCNA was performed to investigate gene expression, CNV, and clinical IHC annotations of INO80 in patients with breast cancer (Cancer Genome Atlas Network, 2012; Ciriello et al., 2015) followed the workflow in Supplementary Figure S2A. Our analysis primarily focused on the effect of INO80 mutation on breast cancer subtype and changes in gene cohorts associated with breast cancer. The gene expression profile was divided into 13 co-expression modules, each containing 34–687 genes (Figure 2A). Notably, the MEblue module was strongly correlated with INO80 CNV (cor = 0.76), with a significant p-value of 1.3e-128 (Figure 2B). This cohort contained a subset of genes that could be considered potential biomarkers for breast cancer progression, including FOXA1, MLPH, ESR1, AR, GATA3, TFF1, THSD4, and TBC1D9 (Supplementary Figure S2B). Moreover, analysis of the ER-associated co-expressed network uncovered several key genes involved in ER signaling, such as ESR1, AR, GATA3, and TFF1, and showed a strong correlation between differential gene expression in the MEblue cohort and INO80 CNV. Based on gene expression and clinical patient data, our analysis classified three clusters and showed that INO80 deletion, including both shallow and deep deletion, was associated with IHC status (Figures 2A, C).
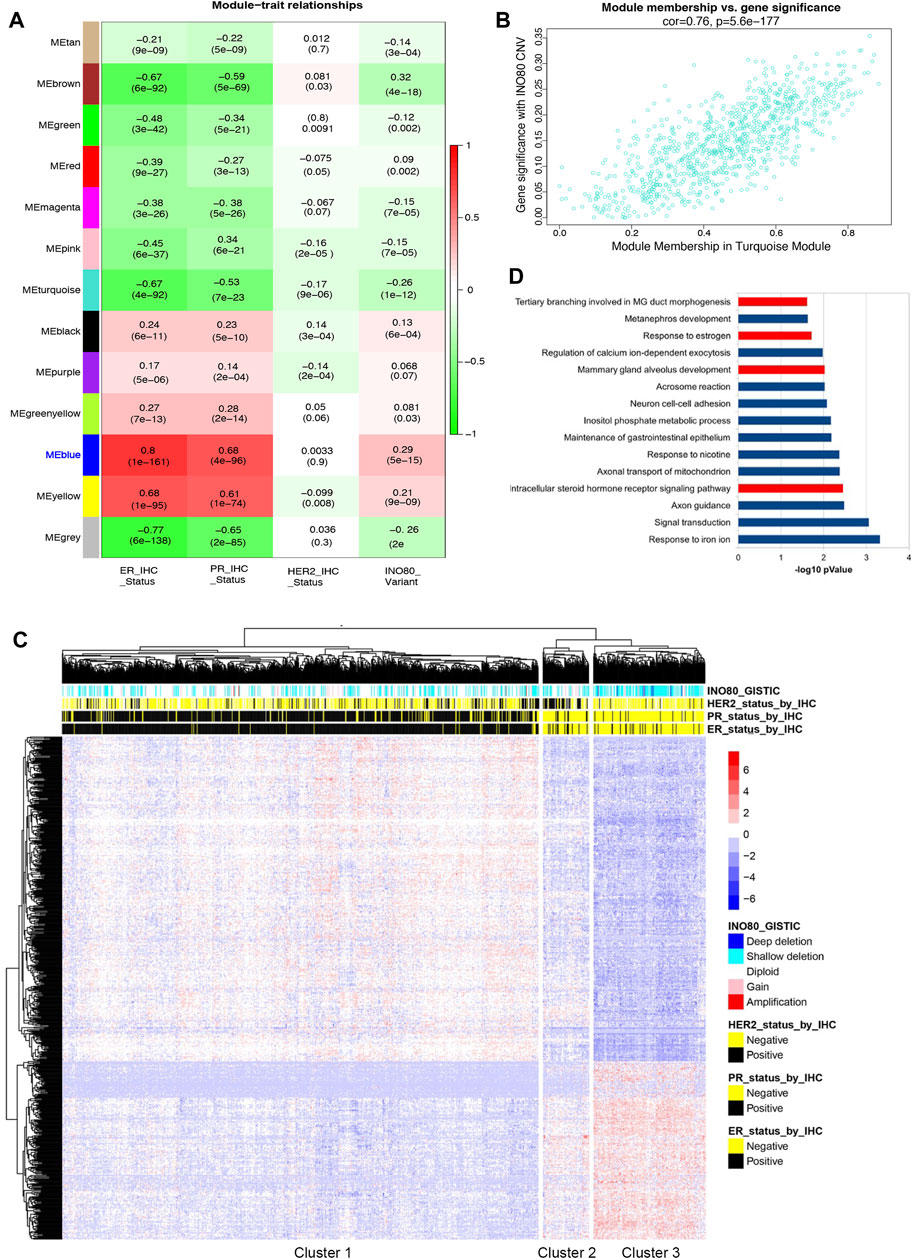
FIGURE 2. The correlation between INO80 expression level and breast cancer. (A) Weighted gene co-expression network analysis (WGCNA) was performed to identify the correlation between INO80 copy number variation (CNV) and gene set modules. The MEblue module was found most associated with INO80 CNV Genomic Identification of Significant Targets in Cancer (GISTIC) among the 13 modules, where red indicates co-expression and green indicates negative co-regulation. (B) The correlation between INO80 copy number variation (CNV) and the significant module, MEblue (793 genes, correlation value 0.76, p = 5.6e-177). (C) A heatmap showing unsupervised clustering of subset genes in the MEblue module of TCGA. The relation between INO80 traits (IHC and CNV) and expression (z-score) of genes is classified into three clusters, with each cluster associated with a certain subtype of breast cancer. (D) The top 15 gene ontology biological processes (GOBPs) from DAVID analysis of gene set data overlapped genes between the MEblue module of TCGA and the MEblue module of METABRIC. GOBPs related to mammary gland development are highlighted in red. ER, estrogen receptor; PR, progesterone receptor; MG, mammary gland; HER2, Human epidermal growth factor receptor 2; CNV, Copy Number Variation.
Based on INO80 CNV and IHC status, the unsupervised clustering identified three clusters: the primary luminal (cluster 1), mainly composed of patients with ER+/PR+/HER2-status; the primary HER2+ (cluster 2), mainly composed of patients with ER-/PR-/HER2+ status; and the primary triple-negative breast cancer (TNBC) (cluster 3), mainly composed of patients with ER-/PR-/HER2-status (Figure 2C). The odds ratio (OR) suggests that the risk of breast cancer is 6.5724 times higher in patients with INO80 deletion in the TNBC cluster than in those with other mutations, which is statistically significant [CI (4.55–9.49) and p < 0.0001]. Similarly, the OR shows that the risk of breast cancer is 5.1429 times higher in cluster 3 than in cluster 2 (HER2+ subtype) [CI (2.75–9.63) and p < 0.0001]. Additionally, the analysis of the METABRIC dataset showed consistency with our TCGA data analysis, demonstrating a significant correlation between INO80 CNV and luminal signatures, as well as an association with the TNBC cluster compared to other breast cancer subtypes (Supplementary Figures S2D, E). The OR for the primary TNBC cluster was 4.1385 times higher than that for the primary HER2 cluster [CI (2.8158–6.0825), p < 0.0001] and 7.1482 times higher than that for the primary luminal cluster [CI (5.3870–9.4853), p < 0.0001]. Therefore, loss of INO80 likely increases the risk of breast carcinogenesis, especially in the TNBC subtype.
Up to 189 genes overlapped between 2 cohorts (MEblue from TCGA analysis and MEblue from METABRIC analysis), accounting for 25%–30% of the total genes in each cohort (Supplementary Figure S2F). Gene ontology (GO) term analysis showed that the MEblue genes were associated with mammary gland epithelial cell differentiation and mammary gland duct branching morphogenesis (Supplementary Figure S2G). The top list of gene ontology biological processes (GOBP) was obtained by overlapping two modules associated with INO80 CNV, including terms related to mammary gland development and estrogen regulation (Figure 2D). Therefore, the findings suggest an underlying mechanism by which INO80 function is involved in mammary gland development and breast cancer.
3.3 INO80 mutation impairs mammary gland development
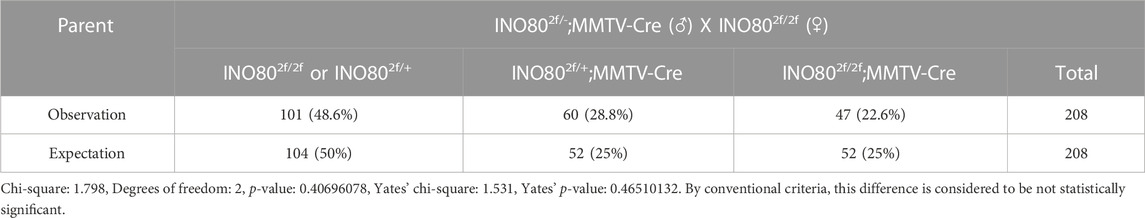
INO80 is highly expressed in the lungs, colon, breasts, and several parts of the brain, such as the nerves, cerebellar hemisphere, and cerebellum (Figure 3A). To investigate whether INO80 plays a role in tumorigenesis, a mouse model was generated in which the INO80 gene was explicitly deleted in the mammary gland. Complete elimination of the INO80 gene in mice results in embryonic lethality (Min et al., 2013; Wang et al., 2014a; Qiu et al., 2016); therefore, we utilized a Cre-loxP system with Tg(MMTV-Cre) mice (line D) (Wagner et al., 1997; Wagner et al., 2001) to generate control (INO80f/f) and cKO [Tg(MMTV-Cre); INO80f/f] females (Supplementary Figure S3A). Genotyping analysis confirmed that the INO80 cKO mice were viable (Supplementary Figure S3B), and their live offspring displayed an expected Mendelian ratio (Table 1). Immunofluorescence confirmed suppression of the INO80 expression in the ductal lumen of the knockout mouse model (Supplementary Figure S3C).
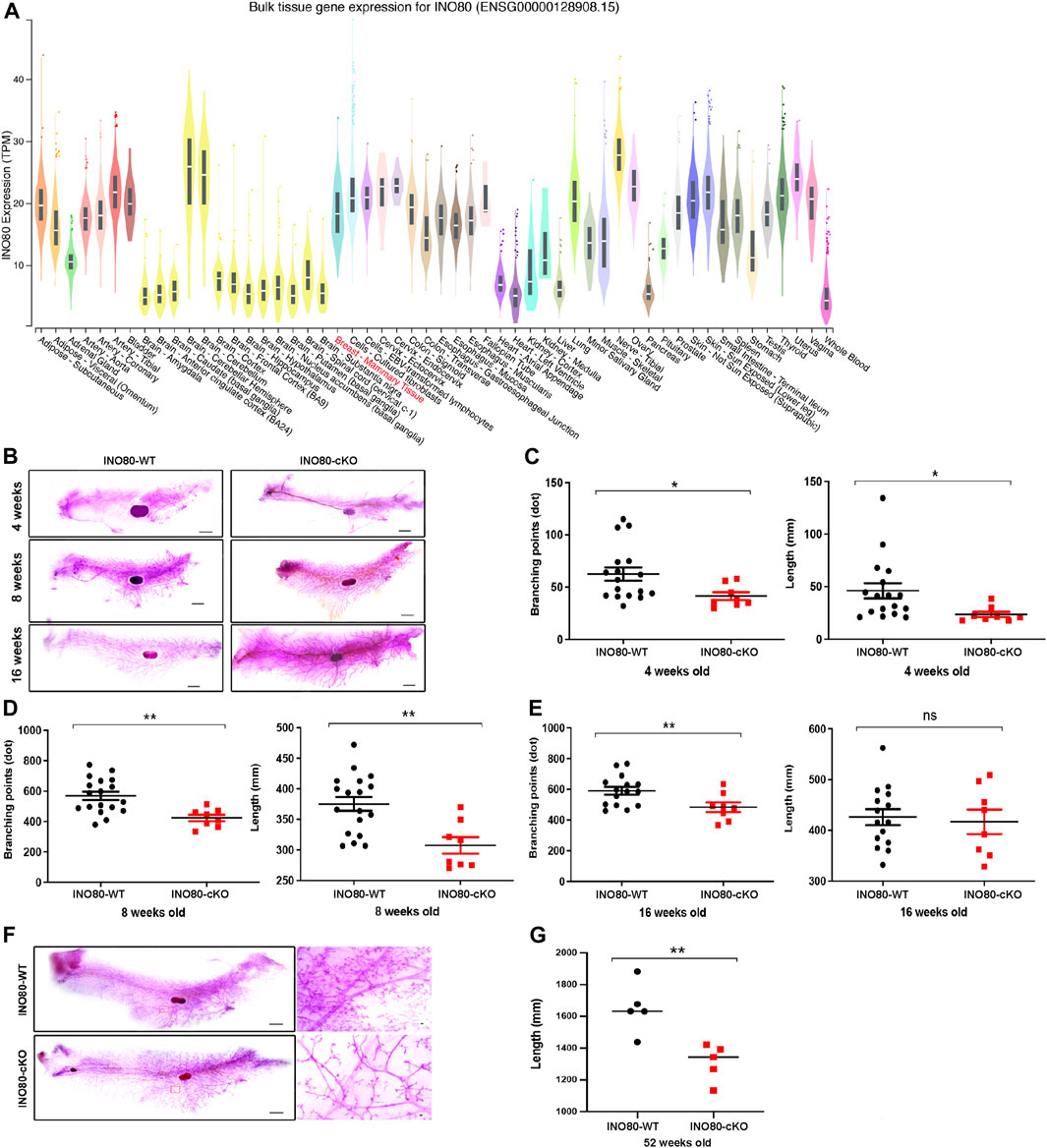
FIGURE 3. INO80 loss impairs mouse mammary gland development. (A) INO80 expression in bulk RNA sequencing of GTEx data. (B) Whole-mount carmine staining of mammary glands from 4-, 8-, and 16-week-old mice. (C–E) Quantification of branching points and length of mammary glands from INO80 cKO mice versus those from wild-type mice at 4-weeks old [in (C)], 8-weeks old [in (D)], and 16 weeks old [in (E)]. (F,G) The mammary gland of 52-week-old mice stained with whole carmine [in (F)] and the length quantification of mammary gland ductal analysis with 5 mice per group. Ductal length was significantly decreased in INO80-cKO compared with that in WT mice, which was consistent with the data on development stages [in (B)]. Significant statistic: *p-value =< 0.05, **p-value =< 0.005; GTEx, Genotype-Tissue Expression; WT, wide type; cKO, Conditional knockout.
Next, to determine whether the loss of INO80 causes phenotypic changes in mammary tissue, whole-mount carmine staining was performed on mammary glands harvested from 4-week-, 8-week-, and 16-week-old INO80 cKO and littermate control females (Figure 3B). The staining was used to examine the ductal growth within the mammary glands in juvenile and adult virgin mice. Quantification analysis of the whole-mount carmine staining revealed that the number of measurable branching points was reduced, and mammary duct length was significantly decreased in the INO80 cKO mice compared to values in control mice (Figures 3C–E). The data suggest that INO80 deficiency resulted in hypoplasia in developing mouse mammary glands. Next, to investigate whether the loss of INO80 is sufficient to induce mammary tumorigenesis, we examined the lifespan and mammary gland morphology of cKO mice up to 2 years of age. Our analysis showed no incidence of spontaneous mammary tumorigenesis in the INO80 cKO mice. Although there were no significant lifespan differences between the INO80 cKO and control mice for up to 2 years, morphometric analysis showed consistent reductions in the length and branching points of mammary glands in the INO80 cKO mice (Figures 3F, G). Notably, histological examination of the mammary glands using CK18(+)/α-SMA(+) ratio and H&E staining revealed the presence of morphologically normal-like ductal structures in the INO80 cKO mammary glands (Supplementary Figures S3D, E). Furthermore, the degree of mammary gland hypoplasia was alleviated in pregnant and lactating INO80 cKO mice (Supplementary Figure S3E), which may account for the comparable weight of newborn pups between the two groups (Supplementary Figure S3F). Therefore, our findings suggest that INO80 loss alone is insufficient to induce mammary tumor formation in mice.
4 Discussion
Understanding the intricate molecular mechanisms underlying normal mammary gland development is crucial for developing effective treatment strategies for breast cancer, which is the most prevalent cancer in women worldwide, accounting for 24.5% of all cancer cases, and is the fifth leading cause of cancer-related death among women, responsible for 15% of all cancer deaths (Sung et al., 2021; Siegel et al., 2022). The mammary gland undergoes numerous phases of development and differentiation, including ductal tree expansion and invasion into the fat pad, pregnancy, lactation, and involution stages (Macias and Hinck, 2012; Biswas et al., 2022), all of which are tightly regulated by a complex interplay between genetic and epigenetic factors (Bae and Hennighausen, 2014; Holliday et al., 2018).
Driver epi-mutations can disrupt normal mammary development and promote breast cancer progression, while passenger epi-mutations are typically neutral and do not provide a growth advantage to cancer cells, but may be associated with functionally altered signaling pathways (McFarland et al., 2017; Fernandez-Moya et al., 2020; Ying and Beronja, 2020). For instance, the absence of certain ATP-dependent chromatin remodeling factors of the SWI/SNF complex affects mammary gland development (Cohet et al., 2010; Skibinski et al., 2014), which has helped clarify the specific roles of these factors in breast cancer development and resistance to therapy (García-Pedrero et al., 2006; Nagarajan et al., 2020; Xu et al., 2020).
INO80 is essential for cell reprogramming, blastocyst development (Wang et al., 2014a; Zhou et al., 2016b), and organ development, including in spermatogenesis (Serber et al., 2016; Chakraborty and Magnuson, 2022), ventricular compaction, and coronary vascularization during heart development (Rhee et al., 2018, 2021). Recent studies have shown that complete abolition of INO80 causes embryonic lethality in mice (Min et al., 2013; Wang et al., 2014a; Lee et al., 2014; Lafon et al., 2015; Qiu et al., 2016) by altering compaction, accessibility of DNA within chromatin, and various molecular processes, including DNA replication, transcription, and DNA damage response (Shen et al., 2000; Gospodinov et al., 2011; Lange et al., 2011; Volokh et al., 2016). Furthermore, this complex has been implicated in both maintenance of stem cell and progression of cancer cell by functioning as a critical regulator of super-enhancers in both contexts (Wang et al., 2014a; Zhou et al., 2016a; Serber et al., 2016; Zhang et al., 2017). Additionally, INO80 is required for H2A.Z dynamics in ER signaling, and silencing of INO80 reduces stimulation of endogenous GREB1 and TFF1 enhancers in breast cancer (García-Pedrero et al., 2006; Papamichos-Chronakis et al., 2011; Segala et al., 2016). Therefore, these findings, along with our INO80 cKO study, suggest that INO80 plays a key role in mammary gland development and breast cancer progression.
The INO80 complex comprises highly conserved principal subunits in humans, mice, flies, and yeast (Shen et al., 2000, 2003; Jin et al., 2005). The canonical INO80 complex comprises several subunits, including INO80, RUVBL1, RUVBL2, MCRS1, and YY1, and is involved in active transcription regulation by physically interacting with P300 and MED1. This complex is also associated with active histone modifications, including H3K4me1, H3K4me3, and H3K27ac (Zhou et al., 2016a; Zhang et al., 2017; Runge et al., 2018). RUVBL1 and YY1 promote tumor growth (Wang et al., 2015; Fan et al., 2017), and inhibiting RUVBL1 expression in metastatic breast cancer cells can reduce both cell proliferation and invasion (Fan et al., 2017). Additionally, YY1 promotes tumor growth by suppressing the expression of p27 and interacting with it (Wang et al., 2015). Conversely, the noncanonical class of the INO80 complex is linked to a repressive histone modification, H3K27me3, suggesting that the INO80 complex can act as a tumor suppressor (Runge et al., 2018; Chakraborty and Magnuson, 2022). INO80 occupancy affects replication forks, and its silencing can activate the replication stress-induced ATR-CHK1 signaling pathway in colon cancer (Lee et al., 2017).
In breast cancer, INO80 expression is generally lower, although it correlates with the ER-positive breast cancer subtype (Figures 1A, B). Higher median INO80 expression was found in ER-positive than in ER-negative breast cancer. Furthermore, the results from our WGCNA analysis indicated the existence of unknown networks between INO80 and a subset of luminal breast cancer biomarkers, including FOXA1, ESR1, GATA3, TFF1, and AR (Supplementary Figure S2B). The FOXA1 transcription factor is a key regulator of breast cancer identity, as it controls ER activity (Hurtado et al., 2011). Furthermore, FOXA1 and MLPH upregulate the expression of luminal-like genes, and both have emerged as prognostic indicators for breast cancer (Thorat et al., 2008; Hurtado et al., 2011; Bernardo et al., 2013; Thakkar et al., 2015). Some genes in ER-associated co-expressed networks alter the gene expression profile of luminal breast cancer cells and are predictive of patient response to breast cancer therapy (Tozlu et al., 2006; Chen et al., 2011; Zhu et al., 2020). THSD4 and TBC1D9 are two factors that drive TNBC, and their expression is also linked to the epithelial–mesenchymal transition (EMT), metastatic dissemination, and the plasticity of breast cancer cells (Cohen et al., 2014; Kothari et al., 2020; Miao et al., 2020; Kothari et al., 2021). Previous studies have also revealed the opposing effects of GATA3 function on the tumor suppressor gene THSD4 (Cohen et al., 2014). These findings suggest a potential role for INO80 in ER signaling, where the presence of INO80 is required for ER activity by forming a physical interaction with the ER-INO80 complex in breast cancer, as reported in previous studies (Segala et al., 2016). Moreover, our analysis suggests that there is an increasing risk of breast cancer and reduced survival rates in patients harboring the INO80 mutation. In addition, unsupervised clustering from the WGCNA analysis revealed an association between patients with INO80 mutation and breast cancer biomarkers (ER/PR/HER2 IHC data). INO80 copy number status is associated with breast cancer subtype, whereas patients with primary breast cancer and TNBC subtypes harbored higher odd ratios than did those with luminal or HER2 subtypes (Figure 2C; Supplementary Figure S2C).
The gene cohort from the WGCNA analysis suggests that INO80 may be involved in mammary gland development (Figure 2D; Supplementary Figure S2F), which is consistent with our in vivo model showing that INO80 affects ductal morphogenesis in the mammary gland (Figures 3C–E, G). Several factors, including steroid hormone, BMP, Wnt, cell cycle, and peptide hormone signaling pathways, are known to play roles in mammary gland development (Bocchinfuso et al., 2000; Feng et al., 2007; Hens et al., 2007; Timmermans-Sprang et al., 2019). Previous studies have demonstrated the involvement of INO80 in stem cell differentiation and mammary tumors via the Wnt pathway (Wang et al., 2014a; Zhou et al., 2016b; Zhang et al., 2017; Timmermans-Sprang et al., 2019) and its regulation of BMP signaling in embryonic and liver cancer stem cells (Hens et al., 2007; Qiu et al., 2016; Wang et al., 2019). Another study demonstrated that the overexpression of INO80 and NANOG could promote cervical cancer cell proliferation and tumorigenesis (Hu et al., 2016). It is possible that the downregulation of these signaling pathways by INO80 KO led to changes in the mammary gland phenotype in our model. INO80 loss in our mouse model failed to cause tumor formation (Figure 3F). We cannot rule out the possibility of insufficient or unexpected CRE activity in the Tg(MMTV-cre) line (De Vos et al., 2008; Diegel et al., 2010). However, based on our breast cancer data analysis and the observed mammary gland phenotype, it is reasonable to hypothesize that INO80 may play a role in enhancing signaling abnormalities associated with breast cancer oncogenesis. Nevertheless, further investigation is required to determine the exact nature of INO80’s involvement in breast cancer development and progression.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics statement
The animal study was approved by the Institute of Animal Care and Use Committee of Konkuk University (IACUC# KU21020). The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
Conceptualization, KH; methodology, NT and DH; validation, NT, DH, CP, HLe, HLa (HLa), SY, HeL (HeL), SU, HS, JD, KP, YC, and KH; formal analysis, NT, DH, CP, HLe, HLa (HLa), SY, HeL (HeL), SU, HS, JD, YC, and KH; investigation, NT, DH, CP, HLe, HLa (HLa), SY, HeL (HeL), SU, HS, JD, YC, and KH; writing–original draft preparation, NT, DH, and KH. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Ministry of Science and ICT (RS-2023-00221200) of the Korean government.
Acknowledgments
The authors are indebted to all the members of the Hong and Han labs for their helpful discussion.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2023.1253274/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | Expression of INO80 components in breast cancer. (A) Copy number variation analysis of subunits in the INO80 complex obtained from TCGA. The cBioportal tool was used for the analysis. Note that up to 60% of INO80 complex alterations are associated with breast cancer. (B) Histogram showing that the alteration frequencies of INO80 in Pan-Cancer Atlas patients differ between cancer types. The average INO80 alteration frequency in breast cancer datasets can be as much as 5%. (C, D) Violin plots showing INO80 FPKMs expression based on PR (C) and HER2 IHC clinical data (D) generated from TCGA (top) and METABRIC (bottom) datasets. A statistical significance between the groups was found only in the PR group. (E) Boxplot assessed the expression of INO80 in each breast cancer subtype using the MetaGxBreast R package. The statistics were analyzed using unpaired t-tests, comparing each breast cancer subtype with the corresponding normal subset. (F) Correlation between INO80 expression and tumor stage (I, II, III, and IV) in TCGA (left) and METABRIC (right) databases. An unpaired t-test was used for statistical analysis, with significance set at p ≤ 0.05. CNV, Copy Number Variation; CNA, Copy Number Alteration; IHC, immunohistochemistry; RPKM, reads per kilobase of transcript per million reads mapped.
SUPPLEMENTARY FIGURE S2 | The correlation between INO80 and breast cancer. (A) The workflow of WGCNA analysis using TCGA and METABRIC breast cancer data. (B) Network of MEblue (TCGA) genes identified in the WGCNA analysis. Each node represents a gene, and lines show the known connections. (C) A correlation plot between INO80 CNV and genes of the MEblue (METABRIC) module. (D) Relationship between modules (subsets of genes) and INO80 traits (IHC status and CNV). Red indicates co-expression, and green presents no correlation in expression. (E) Unsupervised clustering of METABRIC data analyzed by the WGCNA. Heatmap showing the correlation between gene expression and INO80 traits (IHC status and CNV) of the MEblue module. (F) A venn diagram showing overlapping genes between the two most co-expressing modules based on INO80 CNV and METABRIC data. (G) Top 15 GOBPs in the MEblue module of the WGCNA analysis using TCGA data. ER, estrogen receptor; PR, progesterone receptor; MG, mammary gland; HER2, Human epidermal growth factor receptor 2; CNV, Copy Number Variation.
SUPPLEMENTARY FIGURE S3 | Characterization of INO80 cKO mice. (A) A breeding scheme to produce the INO80 conditional knockout (cKO) mouse model. Exon 6 of the INO80 gene is flanked by loxP sites and deleted by the Tg(MMTV-Cre) line. (B) PCR results of offspring genotyping where WT (224 bp band), hemizygous (242 and 382 bp bands), and homozygous (382 bp band) of INO80 gene. (C) Confirmation of INO80 depletion by immunofluorescence with INO80 antibody (red), CK18 antibody (luminal cell—turquoise) or alpha-SMA antibody (basal cell—turquoise), and DAPI (blue). (D) GraphPad Prism illustrates the ratio between luminal cells and basal cells, and an unpaired t-test showed that the difference was insignificant. (E) H&E staining of postnatal (4, 8, and 16 weeks after delivery), pregnant (18 days post coitus), and lactating (lactation day 2, L2) mammary glands. (F) Measurement of body weight of pups delivered from WT or cKO females.
References
Bae, W. K., and Hennighausen, L. (2014). Canonical and non-canonical roles of the histone methyltransferase EZH2 in mammary development and cancer. Mol. Cell Endocrinol. 382, 593–597. doi:10.1016/j.mce.2013.05.002
Bernardo, G. M., Bebek, G., Ginther, C. L., Sizemore, S. T., Lozada, K. L., Miedler, J. D., et al. (2013). FOXA1 represses the molecular phenotype of basal breast cancer cells. Oncogene 32, 554–563. doi:10.1038/onc.2012.62
Biswas, S. K., Banerjee, S., Baker, G. W., Kuo, C.-Y., and Chowdhury, I. (2022). The mammary gland: basic structure and molecular signaling during development. Int. J. Mol. Sci. 23, 3883. doi:10.3390/ijms23073883
Bocchinfuso, W. P., Lindzey, J. K., Hewitt, S. C., Clark, J. A., Myers, P. H., Cooper, R., et al. (2000). Induction of mammary gland development in estrogen receptor-alpha knockout mice. Endocrinology 141, 2982–2994. doi:10.1210/endo.141.8.7609
Bochar, D. A., Wang, L., Beniya, H., Kinev, A., Xue, Y., Lane, W. S., et al. (2000). BRCA1 is associated with a human SWI/SNF-related complex: linking chromatin remodeling to breast cancer. Cell 102, 257–265. doi:10.1016/s0092-8674(00)00030-1
Cancer Genome Atlas Network (2012). Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70. doi:10.1038/nature11412
Chakraborty, P., and Magnuson, T. (2022). INO80 requires a polycomb subunit to regulate the establishment of poised chromatin in murine spermatocytes. Development 149, dev200089. doi:10.1242/dev.200089
Chen, Y., Chen, C., Yang, B., Xu, Q., Wu, F., Liu, F., et al. (2011). Estrogen receptor-related genes as an important panel of predictors for breast cancer response to neoadjuvant chemotherapy. Cancer Lett. 302, 63–68. doi:10.1016/j.canlet.2010.12.014
Chu, X., Guo, X., Jiang, Y., Yu, H., Liu, L., Shan, W., et al. (2017). Genotranscriptomic meta-analysis of the CHD family chromatin remodelers in human cancers - initial evidence of an oncogenic role for CHD7. Mol. Oncol. 11, 1348–1360. doi:10.1002/1878-0261.12104
Ciriello, G., Gatza, M. L., Beck, A. H., Wilkerson, M. D., Rhie, S. K., Pastore, A., et al. (2015). Comprehensive molecular portraits of invasive lobular breast cancer. Cell 163, 506–519. doi:10.1016/j.cell.2015.09.033
Cohen, H., Ben-Hamo, R., Gidoni, M., Yitzhaki, I., Kozol, R., Zilberberg, A., et al. (2014). Shift in GATA3 functions, and GATA3 mutations, control progression and clinical presentation in breast cancer. Breast Cancer Res. 16, 464. doi:10.1186/s13058-014-0464-0
Cohet, N., Stewart, K. M., Mudhasani, R., Asirvatham, A. J., Mallappa, C., Imbalzano, K. M., et al. (2010). SWI/SNF chromatin remodeling enzyme ATPases promote cell proliferation in normal mammary epithelial cells. J. Cell Physiol. 223, 667–678. doi:10.1002/jcp.22072
Devinoy, E., and Rijnkels, M. (2010). Epigenetics in mammary gland biology and cancer. J. Mammary Gland. Biol. Neoplasia 15, 1–4. doi:10.1007/s10911-010-9171-3
De Vos, L., Declercq, J., Rosas, G. G., Van Damme, B., Roebroek, A., Vermorken, F., et al. (2008). MMTV-cre-mediated Fur inactivation concomitant with PLAG1 proto-oncogene activation delays salivary gland tumorigenesis in mice. Int. J. Oncol. 32, 1073–1083. doi:10.3892/ijo.32.5.1073
Diegel, C. R., Cho, K. R., El-Naggar, A. K., Williams, B. O., and Lindvall, C. (2010). Mammalian target of rapamycin-dependent acinar cell neoplasia after inactivation of Apc and Pten in the mouse salivary gland: implications for human acinic cell carcinoma. Cancer Res. 70, 9143–9152. doi:10.1158/0008-5472.CAN-10-1758
Dravis, C., Chung, C.-Y., Lytle, N. K., Herrera-Valdez, J., Luna, G., Trejo, C. L., et al. (2018). Epigenetic and transcriptomic profiling of mammary gland development and tumor models disclose regulators of cell state plasticity. Cancer Cell 34, 466–482. doi:10.1016/j.ccell.2018.08.001
Dravis, C., Spike, B. T., Harrell, J. C., Johns, C., Trejo, C. L., Southard-Smith, E. M., et al. (2015). Sox10 regulates stem/progenitor and mesenchymal cell States in mammary epithelial cells. Cell Rep. 12, 2035–2048. doi:10.1016/j.celrep.2015.08.040
Fan, W., Xie, J., Xia, J., Zhang, Y., Yang, M., Wang, H., et al. (2017). RUVBL1-ITFG1 interaction is required for collective invasion in breast cancer. Biochim. Biophys. Acta Gen. Subj. 1861, 1788–1800. doi:10.1016/j.bbagen.2017.03.016
Feng, Y., Manka, D., Wagner, K.-U., and Khan, S. A. (2007). Estrogen receptor-alpha expression in the mammary epithelium is required for ductal and alveolar morphogenesis in mice. Proc. Natl. Acad. Sci. U. S. A. 104, 14718–14723. doi:10.1073/pnas.0706933104
Fernandez-Moya, A., Morales, S., Arancibia, T., Gonzalez-Hormazabal, P., Tapia, J. C., Godoy-Herrera, R., et al. (2020). Germline variants in driver genes of breast cancer and their association with familial and early-onset breast cancer risk in a Chilean population. Cancers (Basel) 12, 249. doi:10.3390/cancers12010249
Frey, W. D., Chaudhry, A., Slepicka, P. F., Ouellette, A. M., Kirberger, S. E., Pomerantz, W. C. K., et al. (2017). BPTF maintains chromatin accessibility and the self-renewal capacity of mammary gland stem cells. Stem Cell Rep. 9, 23–31. doi:10.1016/j.stemcr.2017.04.031
García-Pedrero, J. M., Kiskinis, E., Parker, M. G., and Belandia, B. (2006). The SWI/SNF chromatin remodeling subunit BAF57 is a critical regulator of estrogen receptor function in breast cancer cells. J. Biol. Chem. 281, 22656–22664. doi:10.1074/jbc.M602561200
Gospodinov, A., Vaissiere, T., Krastev, D. B., Legube, G., Anachkova, B., and Herceg, Z. (2011). Mammalian Ino80 mediates double-strand break repair through its role in DNA end strand resection. Mol. Cell Biol. 31, 4735–4745. doi:10.1128/MCB.06182-11
Guerrero-Martínez, J. A., and Reyes, J. C. (2018). High expression of SMARCA4 or SMARCA2 is frequently associated with an opposite prognosis in cancer. Sci. Rep. 8, 2043. doi:10.1038/s41598-018-20217-3
Hanin, G., and Ferguson-Smith, A. C. (2020). The evolution of genomic imprinting: epigenetic control of mammary gland development and postnatal resource control. Wiley Interdiscip. Rev. Syst. Biol. Med. 12, e1476. doi:10.1002/wsbm.1476
Helming, K. C., Wang, X., and Roberts, C. W. M. (2014). Vulnerabilities of mutant SWI/SNF complexes in cancer. Cancer Cell 26, 309–317. doi:10.1016/j.ccr.2014.07.018
Hens, J. R., Dann, P., Zhang, J.-P., Harris, S., Robinson, G. W., and Wysolmerski, J. (2007). BMP4 and PTHrP interact to stimulate ductal outgrowth during embryonic mammary development and to inhibit hair follicle induction. Development 134, 1221–1230. doi:10.1242/dev.000182
Hohmann, A. F., and Vakoc, C. R. (2014). A rationale to target the SWI/SNF complex for cancer therapy. Trends Genet. 30, 356–363. doi:10.1016/j.tig.2014.05.001
Holliday, H., Baker, L. A., Junankar, S. R., Clark, S. J., and Swarbrick, A. (2018). Epigenomics of mammary gland development. Breast Cancer Res. 20, 100. doi:10.1186/s13058-018-1031-x
Hu, J., Liu, J., Chen, A., Lyu, J., Ai, G., Zeng, Q., et al. (2016). Ino80 promotes cervical cancer tumorigenesis by activating Nanog expression. Oncotarget 7, 72250–72262. doi:10.18632/oncotarget.12667
Hu, Z., Mellor, J., Wu, J., and DeLisi, C. (2004). VisANT: an online visualization and analysis tool for biological interaction data. BMC Bioinforma. 5, 17. doi:10.1186/1471-2105-5-17
Huang, D. W., Sherman, B. T., and Lempicki, R. A. (2009a). Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13. doi:10.1093/nar/gkn923
Huang, D. W., Sherman, B. T., and Lempicki, R. A. (2009b). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57. doi:10.1038/nprot.2008.211
Hurtado, A., Holmes, K. A., Ross-Innes, C. S., Schmidt, D., and Carroll, J. S. (2011). FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat. Genet. 43, 27–33. doi:10.1038/ng.730
Ivanova, E., Le Guillou, S., Hue-Beauvais, C., and Le Provost, F. (2021). Epigenetics: new insights into mammary gland biology. Genes (Basel) 12, 231. doi:10.3390/genes12020231
Jin, J., Cai, Y., Yao, T., Gottschalk, A. J., Florens, L., Swanson, S. K., et al. (2005). A mammalian chromatin remodeling complex with similarities to the yeast INO80 complex. J. Biol. Chem. 280, 41207–41212. doi:10.1074/jbc.M509128200
Kadoch, C., Hargreaves, D. C., Hodges, C., Elias, L., Ho, L., Ranish, J., et al. (2013). Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat. Genet. 45, 592–601. doi:10.1038/ng.2628
Kim, J., Jang, G., Sim, S. H., Park, I. H., Kim, K., and Park, C. (2021). SMARCA4 depletion induces cisplatin resistance by activating YAP1-mediated epithelial-to-mesenchymal transition in triple-negative breast cancer. Cancers (Basel) 13, 5474. doi:10.3390/cancers13215474
Kothari, C., Clemenceau, A., Ouellette, G., Ennour-Idrissi, K., Michaud, A., C-Gaudreault, R., et al. (2021). TBC1D9: an important modulator of tumorigenesis in breast cancer. Cancers (Basel) 13, 3557. doi:10.3390/cancers13143557
Kothari, C., Osseni, M. A., Agbo, L., Ouellette, G., Déraspe, M., Laviolette, F., et al. (2020). Machine learning analysis identifies genes differentiating triple negative breast cancers. Sci. Rep. 10, 10464. doi:10.1038/s41598-020-67525-1
Kumar, R., Li, D.-Q., Müller, S., and Knapp, S. (2016). Epigenomic regulation of oncogenesis by chromatin remodeling. Oncogene 35, 4423–4436. doi:10.1038/onc.2015.513
Lafon, A., Taranum, S., Pietrocola, F., Dingli, F., Loew, D., Brahma, S., et al. (2015). INO80 chromatin remodeler facilitates release of RNA polymerase II from chromatin for ubiquitin-mediated proteasomal degradation. Mol. Cell 60, 784–796. doi:10.1016/j.molcel.2015.10.028
Lange, M., Demajo, S., Jain, P., and Di Croce, L. (2011). Combinatorial assembly and function of chromatin regulatory complexes. Epigenomics 3, 567–580. doi:10.2217/epi.11.83
Lee, H.-S., Lee, S.-A., Hur, S.-K., Seo, J.-W., and Kwon, J. (2014). Stabilization and targeting of INO80 to replication forks by BAP1 during normal DNA synthesis. Nat. Commun. 5, 5128. doi:10.1038/ncomms6128
Lee, S.-A., Lee, H.-S., Hur, S.-K., Kang, S. W., Oh, G. T., Lee, D., et al. (2017). INO80 haploinsufficiency inhibits colon cancer tumorigenesis via replication stress-induced apoptosis. Oncotarget 8, 115041–115053. doi:10.18632/oncotarget.22984
Li, Y., Gong, H., Wang, P., Zhu, Y., Peng, H., Cui, Y., et al. (2021). The emerging role of ISWI chromatin remodeling complexes in cancer. J. Exp. Clin. Cancer Res. 40, 346. doi:10.1186/s13046-021-02151-x
Liu, M. C., Pitcher, B. N., Mardis, E. R., Davies, S. R., Friedman, P. N., Snider, J. E., et al. (2016). PAM50 gene signatures and breast cancer prognosis with adjuvant anthracycline- and taxane-based chemotherapy: correlative analysis of C9741 (Alliance). NPJ Breast Cancer 2, 15023-. doi:10.1038/npjbcancer.2015.23
Macias, H., and Hinck, L. (2012). Mammary gland development. Wiley Interdiscip. Rev. Dev. Biol. 1, 533–557. doi:10.1002/wdev.35
McFarland, C. D., Yaglom, J. A., Wojtkowiak, J. W., Scott, J. G., Morse, D. L., Sherman, M. Y., et al. (2017). The damaging effect of passenger mutations on cancer progression. Cancer Res. 77, 4763–4772. doi:10.1158/0008-5472.CAN-15-3283-T
Miao, K., Lei, J. H., Valecha, M. V., Zhang, A., Xu, J., Wang, L., et al. (2020). NOTCH1 activation compensates BRCA1 deficiency and promotes triple-negative breast cancer formation. Nat. Commun. 11, 3256. doi:10.1038/s41467-020-16936-9
Min, J.-N., Tian, Y., Xiao, Y., Wu, L., Li, L., and Chang, S. (2013). The mINO80 chromatin remodeling complex is required for efficient telomere replication and maintenance of genome stability. Cell Res. 23, 1396–1413. doi:10.1038/cr.2013.113
Nagarajan, S., Rao, S. V., Sutton, J., Cheeseman, D., Dunn, S., Papachristou, E. K., et al. (2020). ARID1A influences HDAC1/BRD4 activity, intrinsic proliferative capacity and breast cancer treatment response. Nat. Genet. 52, 187–197. doi:10.1038/s41588-019-0541-5
Nagy, Á., Lánczky, A., Menyhárt, O., and Győrffy, B. (2018). Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci. Rep. 8, 9227. doi:10.1038/s41598-018-27521-y
Nickerson, J. A., Wu, Q., and Imbalzano, A. N. (2017). Mammalian SWI/SNF enzymes and the epigenetics of tumor cell metabolic reprogramming. Front. Oncol. 7, 49. doi:10.3389/fonc.2017.00049
Nielsen, T. O., Parker, J. S., Leung, S., Voduc, D., Ebbert, M., Vickery, T., et al. (2010). A comparison of PAM50 intrinsic subtyping with immunohistochemistry and clinical prognostic factors in tamoxifen-treated estrogen receptor-positive breast cancer. Clin. Cancer Res. 16, 5222–5232. doi:10.1158/1078-0432.CCR-10-1282
Ohnstad, H. O., Borgen, E., Falk, R. S., Lien, T. G., Aaserud, M., Sveli, M. A. T., et al. (2017). Prognostic value of PAM50 and risk of recurrence score in patients with early-stage breast cancer with long-term follow-up. Breast Cancer Res. 19, 120. doi:10.1186/s13058-017-0911-9
Papamichos-Chronakis, M., Watanabe, S., Rando, O. J., and Peterson, C. L. (2011). Global regulation of H2A.Z localization by the INO80 chromatin-remodeling enzyme is essential for genome integrity. Cell 144, 200–213. doi:10.1016/j.cell.2010.12.021
Qiu, Z., Elsayed, Z., Peterkin, V., Alkatib, S., Bennett, D., and Landry, J. W. (2016). Ino80 is essential for proximal-distal axis asymmetry in part by regulating Bmp4 expression. BMC Biol. 14, 18. doi:10.1186/s12915-016-0238-5
Rhee, S., Chung, J. I., King, D. A., D’amato, G., Paik, D. T., Duan, A., et al. (2018). Endothelial deletion of Ino80 disrupts coronary angiogenesis and causes congenital heart disease. Nat. Commun. 9, 368. doi:10.1038/s41467-017-02796-3
Rhee, S., Paik, D. T., Yang, J. Y., Nagelberg, D., Williams, I., Tian, L., et al. (2021). Endocardial/endothelial angiocrines regulate cardiomyocyte development and maturation and induce features of ventricular non-compaction. Eur. Heart J. 42, 4264–4276. doi:10.1093/eurheartj/ehab298
Runge, J. S., Raab, J. R., and Magnuson, T. (2018). Identification of two distinct classes of the human INO80 complex genome-wide. G3 (Bethesda) 8, 1095–1102. doi:10.1534/g3.117.300504
Segala, G., Bennesch, M. A., Pandey, D. P., Hulo, N., and Picard, D. (2016). Monoubiquitination of histone H2B blocks eviction of histone variant H2A.Z from inducible enhancers. Mol. Cell 64, 334–346. doi:10.1016/j.molcel.2016.08.034
Seong, J., Kim, N.-S., Kim, J.-A., Lee, W., Seo, J.-Y., Yum, M. K., et al. (2018). Side branching and luminal lineage commitment by ID2 in developing mammary glands. Development 145, dev165258. doi:10.1242/dev.165258
Serber, D. W., Rogala, A., Makarem, M., Rosson, G. B., Simin, K., Godfrey, V., et al. (2012). The BRG1 chromatin remodeler protects against ovarian cysts, uterine tumors, and mammary tumors in a lineage-specific manner. PLoS One 7, e31346. doi:10.1371/journal.pone.0031346
Serber, D. W., Runge, J. S., Menon, D. U., and Magnuson, T. (2016). The mouse INO80 chromatin-remodeling complex is an essential meiotic factor for spermatogenesis. Biol. Reprod. 94, 8. doi:10.1095/biolreprod.115.135533
Shen, X., Mizuguchi, G., Hamiche, A., and Wu, C. (2000). A chromatin remodelling complex involved in transcription and DNA processing. Nature 406, 541–544. doi:10.1038/35020123
Shen, X., Ranallo, R., Choi, E., and Wu, C. (2003). Involvement of actin-related proteins in ATP-dependent chromatin remodeling. Mol. Cell 12, 147–155. doi:10.1016/s1097-2765(03)00264-8
Siegel, R. L., Miller, K. D., Fuchs, H. E., and Jemal, A. (2022). Cancer statistics, 2022. CA Cancer J. Clin. 72, 7–33. doi:10.3322/caac.21708
Skibinski, A., Breindel, J. L., Prat, A., Galván, P., Smith, E., Rolfs, A., et al. (2014). The Hippo transducer TAZ interacts with the SWI/SNF complex to regulate breast epithelial lineage commitment. Cell Rep. 6, 1059–1072. doi:10.1016/j.celrep.2014.02.038
Smalley, M., and Ashworth, A. (2003). Stem cells and breast cancer: a field in transit. Nat. Rev. Cancer 3, 832–844. doi:10.1038/nrc1212
Stephens, P. J., Tarpey, P. S., Davies, H., Van Loo, P., Greenman, C., Wedge, D. C., et al. (2012). The landscape of cancer genes and mutational processes in breast cancer. Nature 486, 400–404. doi:10.1038/nature11017
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249. doi:10.3322/caac.21660
Swinstead, E. E., Paakinaho, V., and Hager, G. L. (2018). Chromatin reprogramming in breast cancer. Endocr. Relat. Cancer 25, R385–R404. doi:10.1530/ERC-18-0033
Thakkar, A., Raj, H., Ravishankar, N., Muthuvelan, B., Balakrishnan, A., and Padigaru, M. (2015). High expression of three-gene signature improves prediction of relapse-free survival in estrogen receptor-positive and node-positive breast tumors. Biomark. Insights 10, 103–112. doi:10.4137/BMI.S30559
Thorat, M. A., Marchio, C., Morimiya, A., Savage, K., Nakshatri, H., Reis-Filho, J. S., et al. (2008). Forkhead box A1 expression in breast cancer is associated with luminal subtype and good prognosis. J. Clin. Pathol. 61, 327–332. doi:10.1136/jcp.2007.052431
Timmermans-Sprang, E., Collin, R., Henkes, A., Philipsen, M., and Mol, J. A. (2019). P-cadherin mutations are associated with high basal Wnt activity and stemness in canine mammary tumor cell lines. Oncotarget 10, 2930–2946. doi:10.18632/oncotarget.26873
Tozlu, S., Girault, I., Vacher, S., Vendrell, J., Andrieu, C., Spyratos, F., et al. (2006). Identification of novel genes that co-cluster with estrogen receptor alpha in breast tumor biopsy specimens, using a large-scale real-time reverse transcription-PCR approach. Endocr. Relat. Cancer 13, 1109–1120. doi:10.1677/erc.1.01120
Volokh, O. I., Derkacheva, N. I., Studitsky, V. M., and Sokolova, O. S. (2016). Structural studies of chromatin remodeling factors. Mol. Biol. Mosk. 50, 922–934. doi:10.7868/S0026898416060215
Wagner, K. U., McAllister, K., Ward, T., Davis, B., Wiseman, R., and Hennighausen, L. (2001). Spatial and temporal expression of the Cre gene under the control of the MMTV-LTR in different lines of transgenic mice. Transgenic Res. 10, 545–553. doi:10.1023/a:1013063514007
Wagner, K. U., Wall, R. J., St-Onge, L., Gruss, P., Wynshaw-Boris, A., Garrett, L., et al. (1997). Cre-mediated gene deletion in the mammary gland. Nucleic Acids Res. 25, 4323–4330. doi:10.1093/nar/25.21.4323
Wang, G. G., Allis, C. D., and Chi, P. (2007). Chromatin remodeling and cancer, Part I: covalent histone modifications. Trends Mol. Med. 13, 363–372. doi:10.1016/j.molmed.2007.07.003
Wang, J., Zhou, L., Li, Z., Zhang, T., Liu, W., Liu, Z., et al. (2015). YY1 suppresses FEN1 over-expression and drug resistance in breast cancer. BMC Cancer 15, 50. doi:10.1186/s12885-015-1043-1
Wang, L., Du, Y., Ward, J. M., Shimbo, T., Lackford, B., Zheng, X., et al. (2014a). INO80 facilitates pluripotency gene activation in embryonic stem cell self-renewal, reprogramming, and blastocyst development. Cell Stem Cell 14, 575–591. doi:10.1016/j.stem.2014.02.013
Wang, L., Zhao, Z., Meyer, M. B., Saha, S., Yu, M., Guo, A., et al. (2014b). CARM1 methylates chromatin remodeling factor BAF155 to enhance tumor progression and metastasis. Cancer Cell 25, 21–36. doi:10.1016/j.ccr.2013.12.007
Wang, Y., Zhu, P., Luo, J., Wang, J., Liu, Z., Wu, W., et al. (2019). LncRNA HAND2-AS1 promotes liver cancer stem cell self-renewal via BMP signaling. EMBO J. 38, e101110. doi:10.15252/embj.2018101110
Xu, G., Chhangawala, S., Cocco, E., Razavi, P., Cai, Y., Otto, J. E., et al. (2020). ARID1A determines luminal identity and therapeutic response in estrogen-receptor-positive breast cancer. Nat. Genet. 52, 198–207. doi:10.1038/s41588-019-0554-0
Ying, Z., and Beronja, S. (2020). Embryonic barcoding of equipotent mammary progenitors functionally identifies breast cancer drivers. Cell Stem Cell 26, 403–419. doi:10.1016/j.stem.2020.01.009
Zhang, S., Zhou, B., Wang, L., Li, P., Bennett, B. D., Snyder, R., et al. (2017). INO80 is required for oncogenic transcription and tumor growth in non-small cell lung cancer. Oncogene 36, 1430–1439. doi:10.1038/onc.2016.311
Zhou, B., Wang, L., Zhang, S., Bennett, B. D., He, F., Zhang, Y., et al. (2016a). INO80 governs superenhancer-mediated oncogenic transcription and tumor growth in melanoma. Genes Dev. 30, 1440–1453. doi:10.1101/gad.277178.115
Zhou, B., Zou, J., Zou, S., and Li, X. (2016b). INO80 is required for osteogenic differentiation of human mesenchymal stem cells. Sci. Rep. 6, 35924. doi:10.1038/srep35924
Keywords: mammary gland development, breast cancer, INO80, transcriptional regulation, estrogen
Citation: Thang NX, Han DW, Park C, Lee H, La H, Yoo S, Lee H, Uhm SJ, Song H, Do JT, Park KS, Choi Y and Hong K (2023) INO80 function is required for mouse mammary gland development, but mutation alone may be insufficient for breast cancer. Front. Cell Dev. Biol. 11:1253274. doi: 10.3389/fcell.2023.1253274
Received: 05 July 2023; Accepted: 16 October 2023;
Published: 01 November 2023.
Edited by:
Andrea Stoccoro, University of Pisa, ItalyReviewed by:
Berta N. Vazquez, Josep Carreras Leukaemia Research Institute (IJC), SpainYiran Guo, Duke University, United States
Copyright © 2023 Thang, Han, Park, Lee, La, Yoo, Lee, Uhm, Song, Do, Park, Choi and Hong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kwonho Hong, aG9uZ2tAa29ua3VrLmFjLmty
†These authors have contributed equally to this work
 Nguyen Xuan Thang
Nguyen Xuan Thang Dong Wook Han
Dong Wook Han Chanhyeok Park1
Chanhyeok Park1 Hyeonwoo La
Hyeonwoo La Hyuk Song
Hyuk Song Jeong Tae Do
Jeong Tae Do Kyoung Sik Park
Kyoung Sik Park Kwonho Hong
Kwonho Hong