- 1School of Basic Medicine, Anhui Medical University, Hefei, Anhui, China
- 2College of Veterinary Medicine, Yunnan Agricultural University, Kunming, Yunnan, China
Background: Neural crest cells constitute a distinct set of multipotent cells that undergo migration along predefined pathways, culmination in the differentiation into a plethora of cell types, including components of the pharyngeal cartilage. The neurocranium is composite structure derived from both cranial neural crest and mesoderm cells, whereas the pharyngeal skeletal elements-including the mandibular and branchial arches-are exclusively formed by craniofacial neural crest cells. Previous studies have elucidated the critical involvement of the chemokine signaling axis Cxcl12b/Cxcr4a in craniofacial development in zebrafish (Danio rerio). Nonetheless, the function contribution of Cxcl12a and Cxcr4b-the homologous counterparts of Cxcl12b and Cxcr4a-remain largely unexplored.
Methods: In the present study, mutant lines for cxcl12a and cxcr4b were generated employing CRISPR/Cas9 system. Temporal and spatial expression patterns of specific genes were assessed using in situ hybridization and dual-color fluorescence in situ hybridization techniques. High-resolution confocal microscopy was utilized for in vivo imaging to detect the pharyngeal arch or pouch patterning. Additionally, cartilage formation within the craniofacial region was analyzed via Alcian blue staining, and the proliferation and apoptosis rates of craniofacial neural crest cells were quantified through BrdU incorporation and TUNEL staining.
Results: Our data reveals that the deletion of the chemokine gene cxcl12a results in a marked diminution of pharyngeal cartilage elements, attributable to compromised proliferation of post-migratory craniofacial neural crest cells. Subsequent experiments confirmed that Cxcl12a and Cxcl12b exhibit a synergistic influence on pharyngeal arch and pouch formation.
Conclusion: Collectively, the present investigation furnishes compelling empirical evidence supporting the indispensable role of Cxcl2a in craniofacial cartilage morphogenesis, albeit cxcr4b mutants exert a minimal impact on this biological process. We delineate that Cxcl12a is essential for chondrogenesis in zebrafish, primarily by promoting the proliferation of craniofacial neural crest cells. Furthermore, we proposed a conceptual framework wherein Cxcl12a and Cxcl12b function synergistically in orchestrating both the pharyngeal arch and pouch morphogenesis.
1 Introduction
The craniofacial skeletal framework constitutes a pivotal element in vertebrate morphogenesis, serving indispensable functions in the encapsulation and protection of the brain, as well as facilitating vital processes such as respiration, alimentary intake, and communicative functions. Predominantly, the craniofacial bones emanate from cranial neural crest cells (CNCCs), a specialized subpopulation of neural crest cells (NCCs) with multipotent characteristics (LaBonne and Bronner-Fraser, 1998). These NCCs exhibit remarkable migratory capabilities and differentiate into a broad spectrum of cellular lineages, including the components of the pharyngeal skeleton. Notably, the developmental dynamics of CNCCs have attracted considerable scholarly attention, largely attributable to the plethora of congenital human maladies, such as Waardenburg’s syndrome (Waardenburg, 1951), Treacher Collins syndrome (Sulik et al., 1987) and Pierre-Robin syndrome (Dennison, 1965). These conditions are manifest as aberrant craniofacial phenotypes and arise from perturbations in CNCC migration and pharyngeal arch development. In orchestrating these intricate developmental processes, CNCCs interact with a complex molecular milieu, involving signaling pathways such as Sonic Hedgehog (Shh) (Wada et al., 2005; Eberhart et al., 2006), Fibroblast Growth Factor (FGF) (Crump et al., 2004), and Retinoic Acid (RA) (Kopinke et al., 2006). Although recent years have witnessed substantial advancements in our understanding of NCC ontogeny and pharyngeal cartilage morphogenesis, the precise molecular mechanisms underlying these processes remain incompletely elucidated.
Chemokines, small proteins with chemoattractant properties, represent another layer of complexity in vertebrate development and are classified into four subgroups based on the spatial arrangement of cysteine residues (C, CC, CXC, and CX3C) (Raz and Mahabaleshwar, 2009). Particularly, the stromal cell-derived factor 1 (SDF-1, also known as CXCL12), a member of the CXC subfamily, and its cognate receptor CXCR4, have garnered interest for their roles in modulating cellular proliferation and adhesion, and migration during embryogenesis (Peled et al., 1999; Raz and Mahabaleshwar, 2009; Boldajipour et al., 2011; Liu et al., 2019). Earlier reports have ascertained that Cxcl12 exhibits specific expression in the pharyngeal endoderm concomitant with CNCC migration in murine and avian models, while its receptor-Cxcr4 is expressed in the pharyngeal NCCs (Escot et al., 2016). Disruptions in the CXCR4 signaling axis impair CNCC migration into the pharyngeal arches, culminating in congenital malformations of the lower jaw and hyoid bone (Escot et al., 2016). Given the genomic duplications in Danio rerio (zebrafish), two homologous CXCL12 ligands (Cxcl12a and Cxcl12b) and two CXCR4 receptors (Cxcr4a and Cxcr4b) are present, each with distinct expression patterns and functional roles in cellular cell adhesion and migration (Peled et al., 1999; Colak-Champollion et al., 2019). The Cxcl12a-Cxcr4b axis has been implicated in a myriad of physiological processes, including but not limited to the primordial germ cell and lateral line primordium migration, the guidance of truck lymphatic network (David et al., 2002; Doitsidou et al., 2002; Knaut et al., 2003). Conversely, the Cxcl12b-Cxcr4a signaling axis has been predominantly studied for its role in endodermal migration and vascular patterning, with particular emphasis on craniofacial development (Nair and Schilling, 2008; Olesnicky Killian et al., 2009; Siekmann et al., 2009; Li et al., 2018).
In light of the demonstrated impact of the Cxcl12b-Cxcr4a signaling axis on craniofacial morphogenesis, the present study aims to elucidate the functional significance of their homologous counterparts, Cxcl12a and Cxcr4b, in this intricate developmental process. Utilizing D. rerio (zebrafish) as a model organism, we meticulously investigated the role of Cxcl12a in orchestrating craniofacial development. Initially, the spatiotemporal expression profiles of cxcl12a and cxcr4b were ascertained within the developing pharyngeal region. Leveraging in vivo gene manipulation techniques, namely, knock-down and knock-out strategies, we discerned that Cxcl12a is indispensable for the genesis of craniofacial cartilage—an effect that could be substantially ameliorated via the exogenous administration of cxcl12a mRNA. Intriguingly, the observed cartilaginous defects in cxcl12a mutant lines emanated from compromised proliferation of cranial neural crest cells (CNCCs) subsequent to their migratory phase. Furthermore, our results unveil a synergistic functionality between cxcl12a and cxcl12b in the formation of pharyngeal arches and pouches. Collectively, our findings delineate the crucial role of Cxcl12a in craniofacial cartilage development and accentuate the integral contributions of chemokine signaling networks to the ontogenesis of the pharyngeal apparatus.
2 Materials and methods
2.1 Zebrafish husbandry and strains
The wild-type embryos employed in the present study were procured through natural mating of the AB strain zebrafish. Post-fertilization, embryos were incubated in a Holtfreter’s solution at an ambient temperature of 28.5°C. Embryonic staging was conducted according to discernible morphological landmarks. For the purpose of visualizing pharyngeal pouch cells, transgenic lines Tg(sox17:GFP) and Tg(nkx2.3-mCherry) were utilized. Tg(sox10:GFP; nkx2.3-mCherry) and Tg(fli1:GFP; nkx2.3-mCherry) lines were employed to delineate both the pharyngeal arch cells and pouch cells.
2.2 Zebrafish cxcl12a and cxcr4b mutant line generation
Utilizing the CRISPR/Cas9 genome-editing technique, single guide RNAs (sgRNAs) were engineered to target exon 1 of cxcl12a and exon 2 of cxcr4b. The sequence corresponding to the sgRNA target in cxcl12a exon 1 was: 5’-GGTCGCCATTCATGCACCGATTTCCAA-3’. For cxcr4b exon 2, the sequence was: 5’-CTTACTGTGCCGGCATCCGG-3’. Subsequent to the CRISPR/Cas9-mediated manipulation, frameshift mutations were observed—specifically, a four-base-pair deletion in cxcl12a and a thirteen-base-pair deletion in cxcr4b. These deletions resulted in premature stop codons in the respective mRNAs. Homozygous mutants of cxcl12a−/− and cxcr4b−/− were isolated through the screening of progeny from heterozygous mutant parents.
2.3 Microinjection
For cxcl12a and cxcl12b, capped messenger RNAs (mRNAs) were synthesized in vitro from the linearized plasmid templates employing the mMessage mMachine kit (Ambion). Control morpholinos, constituting the sequence (5’-CCTCTTACCTCAGTTACAATTTATA-3’), were obtained along with ATG morpholinos (MOs) specifically targeting cxcl12b (5'-CGCTACTACTTTGCTATCCATGCCA-3') and MO targeting the start codon of cxcl12a (5'-ATCACTTTGAGATCCATGTTTGCA-3'). These were procured from Gene Tools (Philomath, OR, United States), were microinjected in accordance with previously established protocols (Dambly-Chaudiere et al., 2007; Hollway et al., 2007; Nair and Schilling, 2008).
2.4 Whole-mount in situ hybridization
Whole-mount in situ hybridization assays were executed in strict adherence to previously published protocols (Ning et al., 2013). In dual-color fluorescence in situ hybridization, anti-Digoxigenin-POD (11633716001, Roche) and anti-Fluorescein-POD (11426346910, Roche) were deployed as the primary antibodies to discern Digoxigenin-labeled mCherry probes and Fluorescein-labeled cxcl12a or cxcr4b probes, respectively. Subsequent analysis was performed utilizing the Perkin Elmer TAS fluorescein system (NEL701A001KT) as delineated in earlier publications (Mao et al., 2019). The composite in situ hybridization images display the following annotations: upper left quadrant designates the genotype, upper right indicates developmental stage, lower left specifies the probe in use, and the lower right reveals the aggregate count of representative samples alongside detected samples.
2.5 Reverse-transcription quantitative polymerase chain reaction (RT-qPCR)
Transcript levels of cxcl12a, cxcr4b, hand2, and sox10 were ascertained from a composite pool of 30 embryos at specified developmental intervals. Total RNA was extracted employing TRIzol reagent (Invitrogen, Beijing, China) and purified to an A260/A280 optical density ratio of 2.0. Subsequently, 2 µg of each sample was subjected to reverse transcription utilizing 5X RT MasterMix (Abcam, Beijing, China, G592). Quantitative PCR assays were conducted employing TB Green Premix Ex Taq II (Takara, RR820A) within an Mx3000P real-time PCR system (Stratagene). Expression levels of the genes under investigation were quantified via the 2−ΔΔCT method and were normalized relative to β-actin transcripts. The fold-change in mRNA expression is presented in relation to wild-type embryo controls.
2.6 Confocal imaging
Zebrafish embryos were anaesthetized and positioned in a 2% low melting point agarose matrix (0815, AMRESCO) immersed in egg water. Temporally specific live imaging was executed employing a Nikon A1R+ confocal microscope. All resultant confocal images were processed and analyzed utilizing Nikon NIS-Elements AR 4.13.00 software.
2.7 Alcian blue staining
Embryos were initially fixed in a 4% paraformaldehyde solution overnight and subsequently rinsed with a 0.1% Tween-20 infused distilled water until transparency was achieved. The specimens were then stained with an Alcian blue buffer solution (comprising 0.015% Alcian Blue, 80% ethanol, and 20% acetic acid) overnight at ambient temperature. A destaining process ensued, utilizing ethanol as the solvent. Subsequently, the embryos were enzymatically treated with a 0.5% trypsin solution (0458, AMRESCO) in a supersaturated borax medium until the specimens attained a softer consistency. Post-trypsin treatment, embryos were incubated in a 1% KOH solution for 30 min, followed by rinsing in 0.1% Tween-20 infused water. Lastly, embryos were subjected to a gradient of glycerol concentrations and subsequently imaged via microscopy.
2.8 Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL)
The In Situ Cell Death Detection Kit, TMR Red (Roche, 12156792910), was employed for the TUNEL assays in accordance with the manufacturer’s guidelines. Embryos harvested at specified stages were fixed in 4% paraformaldehyde at 4°C overnight, followed by a triple wash in PBST and proteinase K-mediated permeabilization. The permeabilized embryos were subsequently re-fixed in 4% paraformaldehyde for 30 min at room temperature, washed thrice in PBST, and incubated overnight at 4°C in a TUNEL-labeling milieu. Following triple PBST washes, the embryos were mounted in 2% low-melting-point agarose and visualized through confocal microscopy.
2.9 Proliferation analyses
For BrdU incorporation assays, embryos were exposed to a 10 mM BrdU solution at designated stages for a duration of 20 min and were subsequently harvested. Incorporated BrdU and GFP were detected using anti-BrdU antibodies (1:1,000; B5002, Sigma) and rabbit anti-GFP antibodies (1:1,000, ab183734, Abcam) via whole-mount immunostaining.
2.10 Statistical analysis
Data were subjected to analytical scrutiny utilizing GraphPad Prism 9 software and are articulated as the mean ± standard deviation (SD). Comparisons between control and experimental cohorts were conducted through an unpaired, two-tailed Student’s t-test. Levels of statistical significance were demarcated as follows: p < 0.05 was considered statistically significant and designated with a single asterisk (*), p < 0.01 was deemed highly significant and marked with double asterisks (**), and p < 0.001 was categorized as exceedingly significant and annotated with triple asterisks (***).
3 Results
3.1 cxcl12a has specific expression in pharyngeal pouch
Previous investigations have elucidated the pivotal role of cxcl12b in cranial neural crest cell (CNCC) migration and neurocranium morphogenesis, originating from the pharyngeal endoderm and arch mesenchyme (Olesnicky Killian et al., 2009). Additionally, dmrt2b has been identified as an upstream regulator of cxcl12b, thereby influencing pharyngeal neural crest cell ontogenesis (Li et al., 2018). In light of these findings, our study aims to interrogate the influence of cxcl12a and its cognate receptor, cxcr4b—homologous counterparts to cxcl12b and cxcr4a in zebrafish—on craniofacial development. An initial examination of the spatiotemporal expression patterns of cxcl12a and cxcr4b was conducted employing whole-mount in situ hybridization (WISH) experiments at distinct developmental stages. Intriguingly, while no expression of cxcl12a was discerned in CNCCs within the timeframe of 14–17 h post-fertilization (hpf) (from ZFIN. org), specific expression was observed in the pharyngeal region commencing at 18 hpf, coinciding with the manifestation of the first pharyngeal pouch (Figure 1A). During subsequent developmental intervals, ranging 24 hpf to 72 hpf, there was a conspicuous enrichment of cxcl12a expression in the pharyngeal sector (Figure 1B). To further delineate the precise anatomical locus of cxcl12a expression within the pharyngeal domain, dual-color fluorescence in situ hybridization assays were deployed to examine the exact expression of cxcl12a in the Tg (nkx2.3: mCherry) transgenic embryos. The outcomes, illustrated in Figure 1C unequivocally established co-localization of cxcl12a transcripts with mCherry-tagged pharyngeal pouch cells at 36 hpf. In contrast, cxcr4b exhibited expression within the pharyngeal region but was conspicuously absent in the pharyngeal pouches (Supplementary Figures S1A–C). Collectively, these data cogently substantiate that cxcl12a is distinctly localized within the pharyngeal pouches.
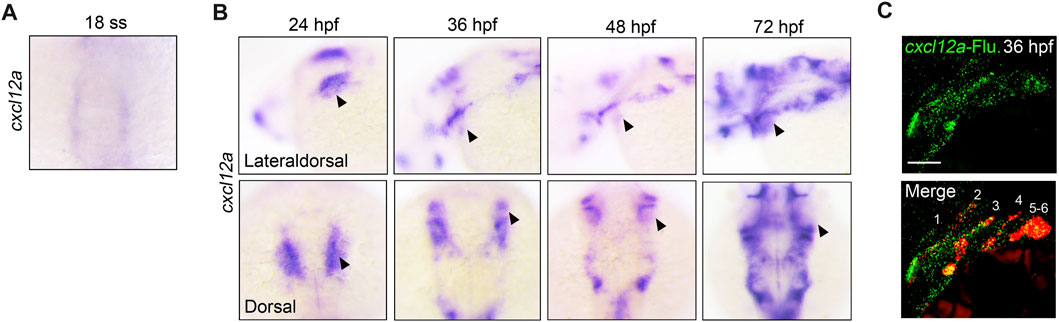
FIGURE 1. Expression of cxcl12a in endodermal pouches. (A) Temporal evaluation of cxcl12a transcriptional activity at 18-somite stage. (B) Spatiotemporal dynamics of cxcl12a expression within the developmental window ranging from 24 hpf to 72 hpf. Triangular markers delineate regions manifesting cxcl12a transcripts in the pharyngeal area. (C) Concomitant fluorescence in situ hybridization of cxcl12a at 36 hpf employing Tg(nkx2.3:mCherry) transgenic embryos. Probes used for staining include cxcl12a-Fluorescence probe (green) and mCherry-DNP (red) probes. Scale bar, 50 μm.
3.2 Loss of cxcl12a leads to pharyngeal cartilage malformation
In pursuit of further delineating the contributory roles of cxcl12a and cxcr4b in craniofacial ontogenesis, targeted mutations in both genes were engendered via the CRISPR/Cas9 genomic editing system (Supplementary Figures S2A, B). Notably, aberrations in nanos3 localization, a marker for the primordial germ cells, were discernibly manifested in homozygous cxcl12a and cxcr4b mutants (Supplementary Figure S2C) (Boldajipour et al., 2008). Subsequent evaluations employing whole-mount in situ hybridization (WISH) and quantitative real-time polymerase chain reaction (qRT-PCR) revealed precipitous decrements in the expression levels of both cxcl12a and cxcr4b within these mutants (Supplementary Figures S2D, E). These observations rigorously authenticate these mutants as bona fide loss-of-function strains.
A comprehensive phenotypic scrutiny revealed that cxcl12a mutants exhibited symptoms of pericardial edema coupled with mandibular hypoplasia at 5 days post-fertilization (dpf), anomalies amenable to amelioration via cxcl12a mRNA microinjection (Figure 2A). In contrast, cxcr4b ablation failed to induce any overt morphological perturbations in the resultant mutants (Figure 2A). Alcian blue histochemical analyses revealed a substantive attenuation in the structural integrity of both Meckel’s cartilage and palatoquadrate, as well as a marked diminution in the number of ceratobranchial cartilages in cxcl12a mutants, thereby implicating compromised chondrogenic differentiation (Figure 2B). Remarkably, these phenotypic aberrations could be substantially rectified through cxcl12a mRNA microinjection (Figure 2B). Concomitantly, cxcr4b mutants exhibited no such craniofacial cartilage deformities (Figure 2B). Furthermore, gene expression assays targeting the extracellular primary cartilage matrix marker, col2a1a, undertaken at 72 hpf, ascertained a notable reduction in its expression levels exclusively in cxcl12a mutants, a condition reversible upon cxcl12a mRNA supplementation (Figure 2B).
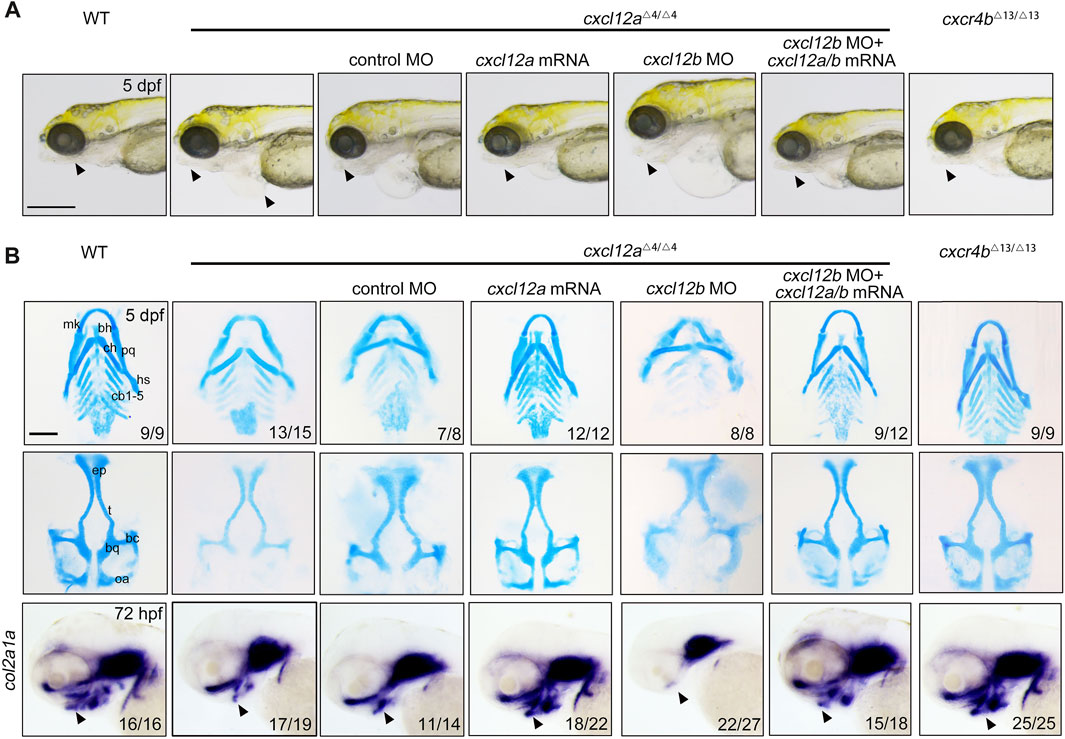
FIGURE 2. Depletion of cxcl12a impairs craniofacial cartilage development. (A) Phenotypic assessment of embryos at designated developmental stages, noting discernible craniofacial aberrations and pericardial edema in cxcl12a mutant embryos. The triangle annotates the zebrafish mandibular region. Scale bars set at 200 mm. (B) Alcian blue histochemical staining of craniofacial cartilage structures at 5 dpf and corresponding in situ hybridization images of col2a1a. Anatomical components denoted are: ep, ethmoid plate; tc, trabeculae cranii; mk, Meckel’s cartilage; bh, basihyal; ch, ceratohyal; pq, palatoquadrate; hs, hyosymplectic; cb, ceratobranchial. The triangle designates the mandibular sector. Scale bars: 100 µm.
To augment our understanding of cxcl12a's involvement in craniofacial cartilaginous morphogenesis, an antisense cxcl12a MO targeting cxcl12a mRNA—a MO previously validated—was employed to attenuate cxcl12a expression (Gamba et al., 2010; Jiang et al., 2019). To assess the efficacy of this MO-mediated silencing, a plasmid harboring both the target sequence of the cxcl12a MO and eGFP was synthesized. MO efficiency was subsequently substantiated through the quantification of fluorescence intensity (Supplementary Figure S3A). Perturbations in nanos3 expression were also evident following cxcl12a knockdown (Supplementary Figure S3B), corroborating both the specificity and efficacy of the cxcl12a MO. Consonant with observations in cxcl12a mutants, morphants displayed manifest aberrations in pharyngeal cartilage formation, defects that were ameliorable by cxcl12a mRNA injection (Supplementary Figure S2D). Collectively, Based on these data, these findings validate that the observed craniofacial cartilage malformations in either cxcl12a mutants and morphants emanate from cxcl12a transcriptomic attenuation, effectively obviating off-target effect of CRISPR/Cas9 effects.
Previously, the Cxcl12b-Cxcr4a axis has been delineated as requisite for CNCC migration and patterning during craniofacial ontogenesis (Olesnicky Killian et al., 2009). Given this, we postulated that cxcl12a and cxcl12b might exert synergistic functions in this developmental process. To interrogate this hypothesis, we deployed a previously characterized cxcl12b MO to suppress cxcl12b expression in cxcl12a mutants (Cha et al., 2012; Nguyen et al., 2014). The efficaciousness of cxcl12b MO was ascertained via fluorescence intensity metrics and in situ hybridization employing a cxcl12b-specific probe (Supplementary Figures S3A, C). Evaluation of cartilaginous elements in cxcl12b morphants revealed a pronounced diminution or outright absence of the ethmoid plate, coupled with trabecular fusion, corroborating prior literature consistent with previous report (Supplementary Figure S3D) (Olesnicky Killian et al., 2009). Remarkably, when cxcl12b MO was introduced into cxcl12a mutants, the resultant phenotypic distortions in pharyngeal cartilage were exacerbated; specifically, both Meckel’s cartilage and palatoquadrate were gravely malformed, and ceratobranchial cartilages were virtually obliterated (Figures 2A, B). Intriguingly, the simultaneous microinjection of cxcl12a and cxcl12b mRNA substantively restored craniofacial cartilage integrity in double-knockdown morphants (Figures 2A, B). Thus, our findings affirm that cxcl12a principally orchestrates craniofacial cartilage formation, whilst cxcl12b downregulation independently induces ethmoid plate atrophy and trabecular fusion.
3.3 cxcl12a has minor effect on CNCC migration
It is universally acknowledged that cranial neural crest cells (CNCCs) originate from the neural plate and subsequently undergo an epithelial-to-mesenchymal (ETM) transition. Following this transformation, CNCCs traverse toward pharyngeal arches and eventually differentiate into the chondrocytes constituting facial cartilage (Dash and Trainor, 2020). In order to elucidate the ramifications of cxcl12a deletion on CNCC activity, we ascertained the expression profiles of specific neural crest cell (NCC) markers across temporal stages. Remarkably, the expression of the neural crest cell specification marker foxd3 remained congruent among control embryos, cxcl12a mutants and cxcl12a cxcl12b MO at the 5-somite stage (Figure 3). This uniformity in expressions trends indicates that cxcl12a is inconsequential to the induction and specification of the early neural crest cells.
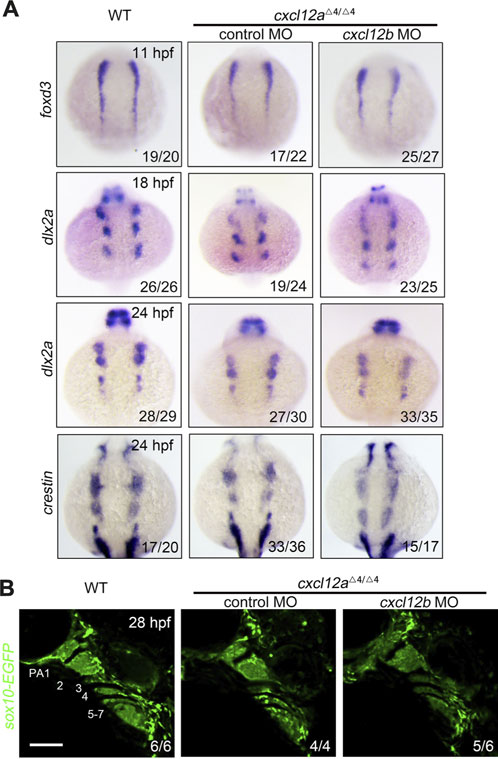
FIGURE 3. NCC specification and migration. (A) Comparative in situ hybridization analysis of wild type and cxcl12a mutant embryos administered either control MO or cxcl12b MO, harvested at indicated time points and probed for foxd3, dlx2a and crestin. (B) In vivo imaging of the pharyngeal compartment of wild type and cxcl12a mutants injected with either control MO or cxcl12b MO, set against a Tg(sox10:EGFP) reporter background. Pharyngeal arches are numerically annotated. PA denotes pharyngeal arch. Scale bars are set at 50 μm.
To further scrutinize the impact on NCC migration, we employed dlx2a and crestin as migration markers. Within the time frame of 18–24 h post-fertilization (hpf) in control embryos, both dlx2a and crestin transcripts were profusely localized within the three neural crest cell groups. Nonetheless, only a marginal decrement in the expression levels of dlx2a and crestin was discernible in cxcl12a mutants at both 18 hpf and 24 hpf (Figure 3A). Conversely, injection of cxcl12b MO into cxcl12a mutants exacerbated the impediments to NCC migration (Figure 3A). In vivo imaging of the Tg(sox10-EGFP) line at 24 hpf corroborated that arch structures in control embryos and cxcl12a mutants were comparable, whereas the number of CNCCs in cxcl12a cxcl12b MO organisms was conspicuously reduced, aligning with prior studies (Figure 3B). Collectively, these findings substantiate the notion that the abrogation of cxcl12a exerts a minimal influence on CNCC migratory patterns.
3.4 cxcl12a is necessary pharyngeal arch patterning through promotes CNCC proliferation
Extant literature delineates that the post-migratory CNCCs in the arches ultimately differentiate into cartilaginous chondrocytes (Hirata and Hall, 2000; Clouthier and Schilling, 2004), a process modulated by pharyngeal pouches serving as pivotal signaling centers (Piotrowski and Nusslein-Volhard, 2000; Couly et al., 2002; Crump et al., 2004). Given the localized expression of cxcl12a in these pharyngeal pouches post-CNCC migration (Figure 1C), we scrutinized the repercussions of cxcl12a depletion on these cells by assessing the expression profiles of hand2 and sox10 at 36 hpf. Notably, a quantitative decline in CNCCs was observed in both cxcl12a mutants and morphants in comparison to these control embryos, a reduction that could be successfully mitigated through the exogenous administration of cxcl12a mRNA (Figure 4A and Supplementary Figure S3E). To corroborate these observations, we employed quantitative reverse-transcription polymerase chain reaction (RT-qPCR) and whole-mount in situ hybridization (WISH) analyses, both of which yielded results in consonance with the initial assessments (Supplementary Figure S4). In line with these findings, comparable results were observed in cxcl12a morphants in Tg(nkx2.3:mCherry;fli1:GFP) or Tg(nkx2.3:mCherry; sox10:GFP) background, characterized by GFP expression in CNCCs at 36 hpf. These morphants exhibited disarrayed and abbreviated pharyngeal arch structures subsequent to cxcl12a knock-down (Figures 4B, C). Moreover, pharyngeal pouch patterning was exacerbated in cxcl12a cxcl12b MO mutants (Figures 4A–C).
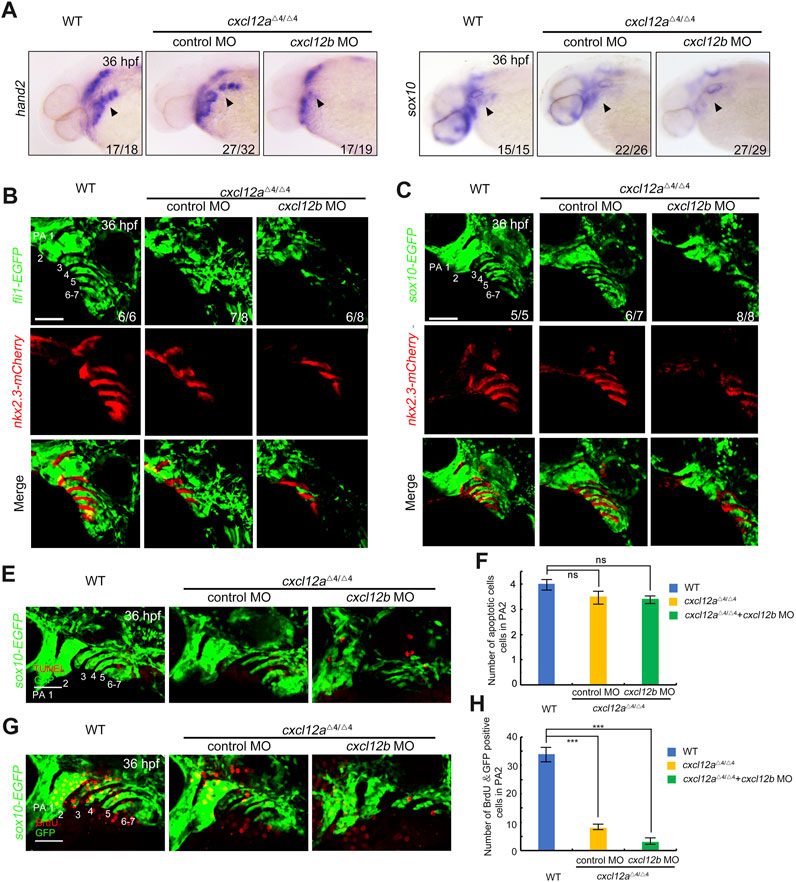
FIGURE 4. Cxcl12a function in CNCC proliferation after migration. (A) WT and cxcl12a mutant embryos subjected to control MO or cxcl12b MO administration were harvested at 36 hpf and interrogated via in situ hybridization employing hand2 and sox10 specific probes. Embryonic lateral views are presented with anterior orientation to the left. (B–C) In vivo live confocal micrographs of Tg(nkx2.3:mCherry;fli1:GFP) or Tg(nkx2.3:mCherry;sox10:EGFP) transgenic embryos injected with control MO, cxcl12a MO, or a combination of cxcl12a and cxcl12b MO at 24 hpf. Scale bar, 50 μm. (E,F) Apoptotic cell assessment in pharyngeal domains via TUNEL assays in cxcl12a mutants and wild-type siblings, injected with either control MO or cxcl12b MO against a Tg(sox10:EGFP) background. Pharyngeal arches (PA) are indicated. Scale bars represent 50 μm. Quantitative analysis of TUNEL-positive and GFP-positive cells within the pharyngeal arch was performed from nine independent embryos (F). Error bars depict standard deviation (SD). NS signifies statistical non-significance as per Student’s t-test. (G,H) Bromodeoxyuridine (BrdU) incorporation assays reveal attenuated proliferative activity within CNCC populations in cxcl12a mutants subjected to control MO or cxcl12b MO treatment. Harvested embryos expressing sox10:EGFP were immunostained with anti-BrdU (red) and anti-GFP (green) antibodies. Pharyngeal regions were visualized through confocal microscopy (G). Scale bars are set at 50 μm. Quantification of BrdU-positive and GFP-positive cells within the pharyngeal arch was achieved from nine independent embryos (H). Error bars represent standard deviation (SD). Statistical significance denoted by ***, p < 0.001 as determined by Student’s t-test.
To discern whether this observed phenotype could be attributed to alterations in CNCC apoptosis or proliferation, TUNEL staining and Bromodeoxyuridine (BrdU) incorporation assays were conducted on the Tg(sox10:EGFP) transgenic reporter line. Intriguingly, apoptosis levels in chondrogenic cells remained invariant between cxcl12a mutants and control embryos (Figures 4E, F). However, a pronounced diminution in the proliferation of BrdU+ CNCCs was detected following cxcl12a depletion (Figures 4G, H). Further compounding this effect, injection of cxcl12b MO into cxcl12a mutants culminated in even more severe pharyngeal arch malformations, attributable to a substantially diminished proliferative capacity of CNCCs (Figure 4 and Supplementary Figure S4).
Collectively, these empirical findings substantiate the indispensable role of cxcl12a in fostering CNCC proliferation subsequent to their migration.
3.5 Cxcl12a promotes pharyngeal pouch formation
Previous empirical evidence presented in Figures 4B, C elucidated that microinjection of cxcl12a MO in transgenic embryos with the Tg(nkx2.3:mCherry;fli1:GFP) or Tg(nkx2.3:mCherry;sox10:EGFP) background engendered a discernible diminution in mCherry-expressing pharyngeal pouches at 36 hpf. Given the co-localization of cxcl12a expression within the pharyngeal pouch, we conducted a meticulous examination of pharyngeal endoderm marker nkx2.3 and cv2 in both control and cxcl12a mutant embryos, either in the presence or absence of cxcl12b MO injections. Corroborating our earlier observations, the patterning of the pharyngeal pouches appeared perturbed in cxcl12a mutants consistent with our observation (Figure 5A).
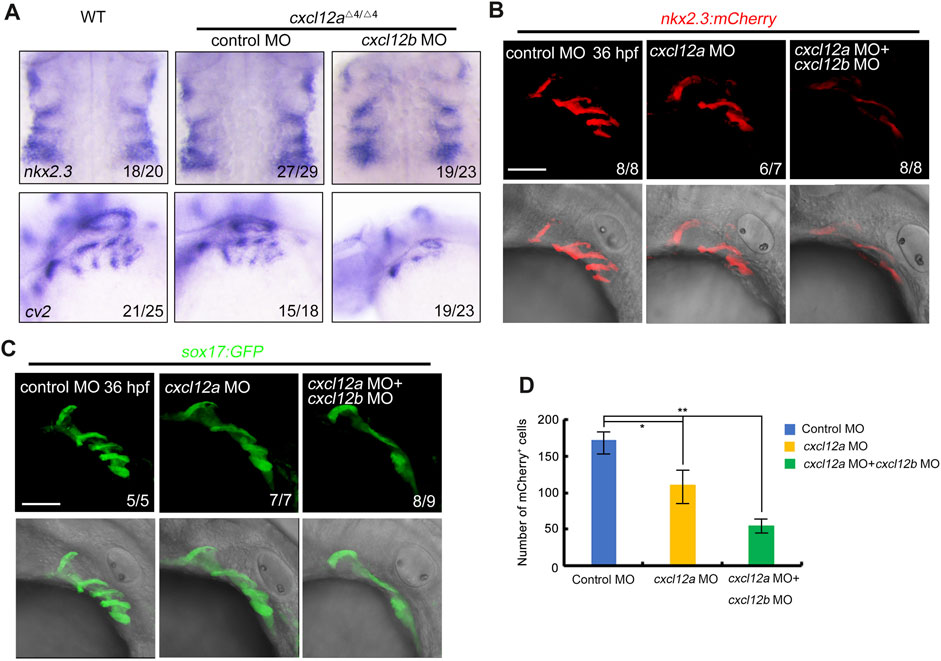
FIGURE 5. cxcl12a mutants exhibit reduced pharyngeal pouch cells. (A) Expression levels of nkx2.3 and cv2 expression were assayed in wild-type or cxcl12a mutants post-injection with either control MO or cxcl12b MO. (B–C) Confocal microscopy elucidates images showing pharyngeal pouch cellular architecture in wild-type and cxcl12a, or combined cxcl12a and cxcl12b knockdown embryos within a Tg(nkx2.3:mCherry) (B) or Tg(sox17:GFP) (C) background at 36 hpf. Scale bar, 50 μm. (D) Quantification of mCherry-positive cells within the Tg(nkx2.3-mCherry) transgenic line was executed from a sample size of nine embryos. Error bars are indicative of standard deviation (SD). Statistical significance denoted by *, p < 0.05; **, p < 0.01, as gauged by Student’s t-test.
To substantiate our preliminary findings, we deployed transgenic embryos featuring the Tg(nkx2.3-mCherry) and Tg(sox17-GFP) genetic backgrounds to serve as indicators of pharyngeal pouch cells. Our analysis revealed that the structural integrity of the pharyngeal pouch was compromised upon cxcl12a MO injection, manifesting as disorganized patterning (Figures 5B–D). Additionally, concomitant microinjection of cxcl12a MO with cxcl12b MO resulted in exacerbated perturbations in pharyngeal pouch morphology (Figures 5B–D). In summation, these empirical observations validate the cooperative functionality of cxcl12a and cxcl12b in orchestrating the formation of the pharyngeal pouch.
4 Discussion
In the present investigation, we delineated that cxcl12a exhibits a spatially restricted expression within the pharyngeal pouches, and its abrogation precipitates anomalies in pharyngeal cartilage formation, attributable to attenuated post-migratory cranial neural crest cell (CNCC) proliferation. Concurrent knockdown cxcl12b in cxcl12a mutations or morpholinos intensifies the severity of defects in both craniofacial cartilage and pharyngeal pouch formation, thereby substantiating the hypothesis that cxcl12a and cxcl12b have synergistic influence on pharyngeal cartilage morphogenesis and pouch development. Collectively, these findings elucidate the pivotal role of cxcl12a in the proliferative dynamics of post-migratory CNCCs.
The specificity of cxcl12a expression to the pouch endoderm (Figure 1) contrasts with cxcl12b's broader expression profile, which encompasses both the pharyngeal pouch and arch mesenchyme (Olesnicky Killian et al., 2009). These differential expression patterns engender the supposition that cxcl12a performs a cell-autonomous function in the formation of the pharyngeal pouch and a cell non-autonomous function in the pharyngeal arch, whereas cxcl12b exhibits a dual role, influencing both arch and pouch development through cell-autonomous mechanisms. The veracity of this conjecture could be elucidated via tissue-specific ablation of cxcl12a/b, necessitating further experimental validation.
In this study, cxcr4b null mutants were generated through the CRISPR/Cas9-mediated gene editing. Notably, these mutants exhibited no conspicuous anomalies, corroborating prior reports (Liu et al., 2022). CXCR7, a recently characterized G-protein coupled receptor, demonstrates affinity for two chemokine ligands-CXCL11 and CXCL12—with a pronounced binding affinity for CXCL12 (Naumann et al., 2010; Sanchez-Martin et al., 2013). Given these antecedents, we postulate that CXCR7 could potentially interact with Cxcl12a to facilitate CNCC proliferation in the absence of Cxcr4b, albeit this hypothesis warrants further empirical scrutiny.
Previous literature has indicated that Cxcl12a augments chondrocyte proliferation via the ERK1/2-cyclin D1 signaling pathway, a finding concordant with our results (Kim et al., 2015). Nonetheless, the downstream signaling cascade implicated in cxcl12a-mediated CNCC proliferation remains elusive and calls for further investigative efforts. Previous reports have demonstrated that Cxcr4 and Sdf1 are genetically downstream of Tbx1 in pharyngeal neural crest cell ontogeny, and a decrement in CXCR4 signaling has been linked to impaired CNCC migration in murine and avian models (Escot et al., 2016; Page et al., 2018). In zebrafish embryos, cxcl12b operates downstream of dmrt2b within endodermal pouches and modulates CNC cell compaction within the pharyngeal arches (Li et al., 2018). Nevertheless, morphants of cxcl12b and cxcr4a exhibit only modest defects in CNCC migration and craniofacial cartilage development (Olesnicky Killian et al., 2009). We posit that while Cxcl12b/Cxcr4a predominantly modulates CNCC migration, cxcl12a exerts a crucial influence on CNCC proliferation post-migration, an assertion that merits comprehensive molecular elucidation in future studies.
5 Conclusion
In summation, the ontogenetic paradigms that govern craniofacial cartilage morphogenesis and NCC development are intricate, encompassing a multifaceted interplay of signaling cascades and cellular interactions across diverse cell types. The current investigation underscores the quintessential function of the zebrafish-specific chemokine cxcl12a in the orchestration of craniofacial cartilage formation and post-migratory CNCC proliferation. Consequently, this study illuminates a heretofore uncharacterized molecular mechanism that modulates craniofacial cartilage development through the promotion of CNCC proliferation in the post-migratory phase, mediated by the pharyngeal pouch-localized chemokine cxcl12a.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The animal study was approved by the Laboratory Animal Ethics Committee of Anhui Medical University. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
JL supervised and designed the project. ZW wrote the manuscript. ZW, QH, and ZD analyzed the data. ZW performed all the experiments. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (32300681), Anhui Province Scientific Research Plan Project (2023AH050657), and Scientific research start-up funds of Anhui Medical University (0101045201).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2023.1243265/full#supplementary-material
References
Boldajipour, B., Doitsidou, M., Tarbashevich, K., Laguri, C., Yu, S. R., Ries, J., et al. (2011). Cxcl12 evolution-subfunctionalization of a ligand through altered interaction with the chemokine receptor. Development 138 (14), 2909–2914. doi:10.1242/dev.068379
Boldajipour, B., Mahabaleshwar, H., Kardash, E., Reichman-Fried, M., Blaser, H., Minina, S., et al. (2008). Control of chemokine-guided cell migration by ligand sequestration. Cell 132 (3), 463–473. doi:10.1016/j.cell.2007.12.034
Cha, Y. R., Fujita, M., Butler, M., Isogai, S., Kochhan, E., Siekmann, A. F., et al. (2012). Chemokine signaling directs trunk lymphatic network formation along the preexisting blood vasculature. Dev. Cell 22 (4), 824–836. doi:10.1016/j.devcel.2012.01.011
Clouthier, D. E., and Schilling, T. F. (2004). Understanding endothelin-1 function during craniofacial development in the mouse and zebrafish. Birth Defects Res. C Embryo Today 72 (2), 190–199. doi:10.1002/bdrc.20007
Colak-Champollion, T., Lan, L., Jadhav, A. R., Yamaguchi, N., Venkiteswaran, G., Patel, H., et al. (2019). Cadherin-mediated cell coupling coordinates chemokine sensing across collectively migrating cells. Curr. Biol. 29 (15), 2570–2579. doi:10.1016/j.cub.2019.06.061
Couly, G., Creuzet, S., Bennaceur, S., Vincent, C., and Le Douarin, N. M. (2002). Interactions between Hox-negative cephalic neural crest cells and the foregut endoderm in patterning the facial skeleton in the vertebrate head. Development 129 (4), 1061–1073. doi:10.1242/dev.129.4.1061
Crump, J. G., Maves, L., Lawson, N. D., Weinstein, B. M., and Kimmel, C. B. (2004). An essential role for Fgfs in endodermal pouch formation influences later craniofacial skeletal patterning. Development 131 (22), 5703–5716. doi:10.1242/dev.01444
Dambly-Chaudiere, C., Cubedo, N., and Ghysen, A. (2007). Control of cell migration in the development of the posterior lateral line: antagonistic interactions between the chemokine receptors CXCR4 and CXCR7/RDC1. BMC Dev. Biol. 7, 23. doi:10.1186/1471-213X-7-23
Dash, S., and Trainor, P. A. (2020). The development, patterning and evolution of neural crest cell differentiation into cartilage and bone. Bone 137, 115409. doi:10.1016/j.bone.2020.115409
David, N. B., Sapede, D., Saint-Etienne, L., Thisse, C., Thisse, B., Dambly-Chaudiere, C., et al. (2002). Molecular basis of cell migration in the fish lateral line: role of the chemokine receptor CXCR4 and of its ligand, SDF1. Proc. Natl. Acad. Sci. U. S. A. 99 (25), 16297–16302. doi:10.1073/pnas.252339399
Dennison, W. M. (1965). The Pierre robin syndrome. Pediatrics 36 (3), 336–341. doi:10.1542/peds.36.3.336
Doitsidou, M., Reichman-Fried, M., Stebler, J., Koprunner, M., Dorries, J., Meyer, D., et al. (2002). Guidance of primordial germ cell migration by the chemokine SDF-1. Cell 111 (5), 647–659. doi:10.1016/s0092-8674(02)01135-2
Eberhart, J. K., Swartz, M. E., Crump, J. G., and Kimmel, C. B. (2006). Early Hedgehog signaling from neural to oral epithelium organizes anterior craniofacial development. Development 133 (6), 1069–1077. doi:10.1242/dev.02281
Escot, S., Blavet, C., Faure, E., Zaffran, S., Duband, J. L., and Fournier-Thibault, C. (2016). Disruption of CXCR4 signaling in pharyngeal neural crest cells causes DiGeorge syndrome-like malformations. Development 143 (4), 582–588. doi:10.1242/dev.126573
Gamba, L., Cubedo, N., Ghysen, A., Lutfalla, G., and Dambly-Chaudiere, C. (2010). Estrogen receptor ESR1 controls cell migration by repressing chemokine receptor CXCR4 in the zebrafish posterior lateral line system. P Natl. Acad. Sci. U. S. A. 107 (14), 6358–6363. doi:10.1073/pnas.0909998107
Hirata, M., and Hall, B. K. (2000). Temporospatial patterns of apoptosis in chick embryos during the morphogenetic period of development. Int. J. Dev. Biol. 44 (7), 757–768.
Hollway, G. E., Bryson-Richardson, R. J., Berger, S., Cole, N. J., Hall, T. E., and Currie, P. D. (2007). Whole-somite rotation generates muscle progenitor cell compartments in the developing zebrafish embryo. Dev. Cell 12 (2), 207–219. doi:10.1016/j.devcel.2007.01.001
Jiang, D., Jiang, Z., Lu, D., Wang, X., Liang, H. S., Zhang, J. F., et al. (2019). Migrasomes provide regional cues for organ morphogenesis during zebrafish gastrulation. Nat. Cell Biol. 21 (8), 966–977. doi:10.1038/s41556-019-0358-6
Kim, G. W., Han, M. S., Park, H. R., Lee, E. J., Jung, Y. K., Usmani, S. E., et al. (2015). CXC chemokine ligand 12a enhances chondrocyte proliferation and maturation during endochondral bone formation. Osteoarthr. Cartil. 23 (6), 966–974. doi:10.1016/j.joca.2015.01.016
Knaut, H., Werz, C., Geisler, R., Nusslein-Volhard, C., and Tubingen Screen, C. (2003). A zebrafish homologue of the chemokine receptor Cxcr4 is a germ-cell guidance receptor. Nature 421 (6920), 279–282. doi:10.1038/nature01338
Kopinke, D., Sasine, J., Swift, J., Stephens, W. Z., and Piotrowski, T. (2006). Retinoic acid is required for endodermal pouch morphogenesis and not for pharyngeal endoderm specification. Dev. Dyn. 235 (10), 2695–2709. doi:10.1002/dvdy.20905
LaBonne, C., and Bronner-Fraser, M. (1998). Neural crest induction in Xenopus: evidence for a two-signal model. Development 125 (13), 2403–2414. doi:10.1242/dev.125.13.2403
Li, L., Mao, A., Wang, P., Ning, G., Cao, Y., and Wang, Q. (2018). Endodermal pouch-expressed dmrt2b is important for pharyngeal cartilage formation. Biol. Open 7 (12), bio035444. doi:10.1242/bio.035444
Liu, J., Zhang, M., Dong, H., Liu, J., Mao, A., Ning, G., et al. (2022). Chemokine signaling synchronizes angioblast proliferation and differentiation during pharyngeal arch artery vasculogenesis. Development 149 (23), dev200754. doi:10.1242/dev.200754
Liu, J., Zhu, C., Ning, G., Yang, L., Cao, Y., Huang, S., et al. (2019). Chemokine signaling links cell-cycle progression and cilia formation for left-right symmetry breaking. PLoS Biol. 17 (8), e3000203. doi:10.1371/journal.pbio.3000203
Mao, A., Zhang, M., Liu, J., Cao, Y., and Wang, Q. (2019). PDGF signaling from pharyngeal pouches promotes arch artery morphogenesis. J. Genet. Genomics 46 (12), 551–559. doi:10.1016/j.jgg.2019.11.004
Nair, S., and Schilling, T. F. (2008). Chemokine signaling controls endodermal migration during zebrafish gastrulation. Science 322 (5898), 89–92. doi:10.1126/science.1160038
Naumann, U., Cameroni, E., Pruenster, M., Mahabaleshwar, H., Raz, E., Zerwes, H. G., et al. (2010). CXCR7 functions as a scavenger for CXCL12 and CXCL11. PLoS One 5 (2), e9175. doi:10.1371/journal.pone.0009175
Nguyen, P. D., Hollway, G. E., Sonntag, C., Miles, L. B., Hall, T. E., Berger, S., et al. (2014). Haematopoietic stem cell induction by somite-derived endothelial cells controlled by meox1. Nature 512 (7514), 314–318. doi:10.1038/nature13678
Ning, G., Liu, X., Dai, M., Meng, A., and Wang, Q. (2013). MicroRNA-92a upholds Bmp signaling by targeting noggin3 during pharyngeal cartilage formation. Dev. Cell 24 (3), 283–295. doi:10.1016/j.devcel.2012.12.016
Olesnicky Killian, E. C., Birkholz, D. A., and Artinger, K. B. (2009). A role for chemokine signaling in neural crest cell migration and craniofacial development. Dev. Biol. 333 (1), 161–172. doi:10.1016/j.ydbio.2009.06.031
Page, M., Ridge, L., Diaz, D. G., Tsogbayar, T., Scambler, P. J., and Ivins, S. (2018). Loss of CXCL12/CXCR4 signalling impacts several aspects of cardiovascular development but does not exacerbate Tbx1 haploinsufficiency. Plos One 13 (11), e0207251. doi:10.1371/journal.pone.0207251
Peled, A., Petit, I., Kollet, O., Magid, M., Ponomaryov, T., Byk, T., et al. (1999). Dependence of human stem cell engraftment and repopulation of NOD/SCID mice on CXCR4. Science 283 (5403), 845–848. doi:10.1126/science.283.5403.845
Piotrowski, T., and Nusslein-Volhard, C. (2000). The endoderm plays an important role in patterning the segmented pharyngeal region in zebrafish (Danio rerio). Dev. Biol. 225 (2), 339–356. doi:10.1006/dbio.2000.9842
Raz, E., and Mahabaleshwar, H. (2009). Chemokine signaling in embryonic cell migration: A fisheye view. Development 136 (8), 1223–1229. doi:10.1242/dev.022418
Sanchez-Martin, L., Sanchez-Mateos, P., and Cabanas, C. (2013). CXCR7 impact on CXCL12 biology and disease. Trends Mol. Med. 19 (1), 12–22. doi:10.1016/j.molmed.2012.10.004
Siekmann, A. F., Standley, C., Fogarty, K. E., Wolfe, S. A., and Lawson, N. D. (2009). Chemokine signaling guides regional patterning of the first embryonic artery. Genes Dev. 23 (19), 2272–2277. doi:10.1101/gad.1813509
Sulik, K. K., Johnston, M. C., Smiley, S. J., Speight, H. S., and Jarvis, B. E. (1987). Mandibulofacial dysostosis (treacher Collins syndrome): A new proposal for its pathogenesis. Am. J. Med. Genet. 27 (2), 359–372. doi:10.1002/ajmg.1320270214
Waardenburg, P. J. (1951). A new syndrome combining developmental anomalies of the eyelids, eyebrows and nose root with pigmentary defects of the iris and head hair and with congenital deafness. Am. J. Hum. Genet. 3 (3), 195–253. doi:10.1111/j.1399-0004.2009.01239.x
Keywords: Cxcl12a, chemokine signal, pharyngeal arches, craniofacial cartilage, neural crest cells
Citation: Wei Z, Hong Q, Ding Z and Liu J (2023) cxcl12a plays an essential role in pharyngeal cartilage development. Front. Cell Dev. Biol. 11:1243265. doi: 10.3389/fcell.2023.1243265
Received: 20 June 2023; Accepted: 25 September 2023;
Published: 04 October 2023.
Edited by:
Walid D. Fakhouri, University of Texas Health Science Center at Houston, United StatesReviewed by:
Andre Luiz Pasqua Tavares, George Washington University, United StatesLindsey Barske, Cincinnati Children’s Hospital Medical Center, United States
Annita Achilleos, University of Nicosia, Cyprus
Copyright © 2023 Wei, Hong, Ding and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jingwen Liu, MTIwNTA5OTgwMkBxcS5jb20=
 Zhaohui Wei
Zhaohui Wei Qiang Hong1
Qiang Hong1 Jingwen Liu
Jingwen Liu