
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell Dev. Biol. , 07 November 2023
Sec. Cell Adhesion and Migration
Volume 11 - 2023 | https://doi.org/10.3389/fcell.2023.1240920
This article is part of the Research Topic Mechanical Coupling between Extracellular and Intracellular Microenvironment View all 5 articles
 Shin Young Ahn1†
Shin Young Ahn1† Mina Bagheri Varzaneh2†
Mina Bagheri Varzaneh2† Yan Zhao2
Yan Zhao2 Jacob Rozynek2
Jacob Rozynek2 Sriram Ravindran2
Sriram Ravindran2 Jonathan Banks2
Jonathan Banks2 Minahil Chaudhry2
Minahil Chaudhry2 David A. Reed2*
David A. Reed2*The migration of mandibular fibrochondrocytes is important for the development of the mandible, the homeostasis of the mandibular cartilage, and for the capacity of the tissue to respond to injury. Mandibular fibrochondrocytes have to overcome formidable obstacles during migration including a dense and heterogeneous three-dimensional matrix. Guiding the direction of cell migration and commitment to a migratory phenotype in this microenvironment necessitates a multivalent response to chemotactic and extracellular matrix-mediated stimuli. One of the key matrix components in the cartilage of the temporomandibular joint is type VI collagen. Neuron/glial antigen 2 (NG2/CSPG4) is a transmembrane proteoglycan that binds with collagen VI and has been implicated in a wide range of cell behaviors including cell migration, motility, adhesion, and proliferation. While NG2/CSPG4 has been shown to be a key regulator of mandibular cartilage homeostasis, its role in the migration of mandibular fibrochondrocytes during normal and cell stress conditions has yet to be resolved. Here, we address this gap in knowledge by characterizing NG2/CSPG4-dependent migration in mandibular fibrochondrocytes using primary mandibular fibrochondrocytes isolated from control and full length NG2/CSPG4 knockout mice, in primary mandibular fibrochondrocytes isolated from NG2|DsRed reporter mice and in an immortalized mandibular fibrochondrocyte cell line with a mutated NG2/CSPG4 ectodomain. All three cells demonstrate similar results, with loss of the full length or truncated NG2/CSPG4 increasing the rate of cell migration in serum starvation/cell stress conditions. These findings clearly implicate NG2/CSPG4 as a key molecule in the regulation of cell migration in mandibular fibrochondrocytes in normal and cell stress conditions, underscoring the role of NG2/CSPG4 as a mechanosensitive signaling hub in the mandibular cartilage.
The migration of chondrocytes through a dense extracellular matrix is a remarkable and understudied phenomenon in the cell and molecular biology of cartilage. Migration is critical for both development and the tissue’s response to injury. During degenerative arthropathies such as osteoarthritis (OA), the post-traumatic injury response of the cartilage is facilitated through cell migration, enabling progenitor and inflammatory cells to initiate the repair and resolution of the affected area. Migratory chondrocyte progenitor cells synthesize a fibrocartilaginous neo-matrix that improves the overall integrity of the tissue but does not perfectly recapitulate the higher-order structure and material properties of the original tissue (Seol et al., 2012; Jiang and Tuan, 2015). These progenitor cells have stem cell-like properties and can be induced to commit to a chondrogenic fate. A similar process has been reported in the cartilage of the temporomandibular joint (TMJ), with migratory cells within cartilage defects observed shortly after injury (Cledes et al., 2006). It is unknown if these migratory cells in the TMJ are the fibrocartilage stem cells recently identified from the rodent and human condyle (Robinson et al., 2015; Embree et al., 2016; Bi et al., 2020; Fan et al., 2021). Despite the mobilization of these cells, many post-natal injuries are not repaired. This is likely the result of the difficulties associated with migration through a dense and heterogeneous extracellular matrix that impairs the infiltration of repair cells into the affected area following post-traumatic injury (Morales, 2007). There is an important gap in knowledge related to the molecular mechanisms regulating the migratory potential and phenotype of chondrocytes.
When moving through a heterogeneous substrate such as cartilage, cells adaptively modify their cell surface microdomain by modifying the composition and organization of receptors, proteoglycans, cell adhesion molecules, and proteases, switching between “path finding” and “path generating” behaviors (Cattaruzza and Perris, 2005). Cells migrate through a substrate by active polymerization of the leading edge of the cell, generating cytoskeletal protrusions that adhere to the extracellular matrix substratum (Lauffenburger and Horwitz, 1996; Yamada and Sixt, 2019). Adhesion to the substrate is achieved through integrin binding with coordinated mechanosensory integration with intracellular focal adhesions. These substrate interactions potentiate, in part, the capacity of cells to sense differences in the material properties of the extracellular matrix (Yamada and Sixt, 2019). Cells integrate this adhesion information from the substrate with the mechanics of forward actin protrusion and actomyosin contractions to control cell polarity, migration directionality, and coordinated cell movements (Haeger et al., 2015).
Therefore, the direction of cell migration and commitment to a migratory phenotype necessitates a multivalent response to chemotactic and extracellular matrix-mediated stimuli. Growth factors affecting cell migration include platelet-derived growth factor (PDGF), fibroblast growth factor (FGF), insulin-like growth factor (IGF), and transforming growth factor (TGF)-β. PDGF, FGF, and IGF are present in post-traumatic cartilage (Mishima and Lotz, 2008; Hopper et al., 2015). Of these growth factors, PDGF is one of the most potent. In chondrocytes and mesenchymal stem cells, PDGF-BB and -AB induce a robust migratory response from the cells, with a lower response being elicited from PDGF-AA (Mishima and Lotz, 2008). PDGF is of special interest in biomedical studies as a prominent component of the serum used in cell culture media. TGF-β is also involved in the injury response of the cell, regulating the fibrogenic signaling axis. This TGF-β-mediated injury response is associated with the accumulation of cell surface and extracellular matrix proteoglycans (Cattaruzza and Perris, 2005).
The accumulation of membrane-associated proteoglycans after injury can have multiple roles including regulating growth factor-mediated signaling, ectodomain shedding, the synthesis of scaffolding proteins, the composition of the subcellular microdomain, and the localization of glycosaminoglycan chains to the cell. These modifications are associated with both anti-adhesion and anti-migratory effects and impact the regenerative and reparative potential of the tissue (Krekoski et al., 2001; Bradbury et al., 2002; Zuo et al., 2002). Specifically, sulfated GAG side chains exert anti-adhesion and anti-migratory effects on the cell due to the negative charge of the molecule (Davies et al., 2008). The anti-migratory effects of chondroitin sulfate chains in the cartilage is well-documented, particularly in the major proteoglycan of articular cartilage aggrecan (Hunziker and Rosenberg, 1996; Perris et al., 1996; Johnson et al., 2005; Johnson et al., 2006; Davies et al., 2008). Less is known about the role of other chondroitin sulfates containing membrane-associated proteoglycans in the migration of cartilage cells.
One of these proteoglycans is neuron glial antigen 2 (NG2; human homolog, CSPG4; mouse homolog, AN2). NG2/CSPG4 is a single-pass transmembrane proteoglycan with chondroitin sulfate chains present on the ectodomain. NG2/CSPG4 is present in cartilage (Fukushi et al., 2003; Yotsuya et al., 2019) and regulates the homeostasis of the cartilage during health and disease (Midwood and Salter, 1998; Midwood and Salter, 2001; Reed et al., 2022). NG2/CSPG4 has been implicated in cell migration and motility in other cell types (Fukushi et al., 2004; Makagiansar et al., 2007) but has not been studied in temporomandibular joint cartilage. NG2-/CSPG4-mediated migration/motility is achieved through both direct and indirect mechanisms. NG2/CSPG4 directly engages with the cell surface microdomain through beta-1 integrin (Burg et al., 1996; Burg et al., 1997; Goretzki et al., 1999; Fukushi et al., 2004; Makagiansar et al., 2007; Chekenya et al., 2008; Jamil et al., 2016) and is a cell surface receptor for type VI collagen (Stallcup et al., 1990; Nishiyama and Stallcup, 1993; Burg et al., 1996; Burg et al., 1997; Tillet et al., 2002; Huang et al., 2010). NG2/CSPG4 also regulates cell motility through binding with pro-migratory growth factors, PDGF-AA and FGF2 (Nishiyama et al., 1996a; Nishiyama et al., 1996b; Goretzki et al., 1999; Grako et al., 1999). Indirectly, the addition of NG2/CSPG4 enhances cellular motility even in cells that do not contain substantial endogenous cell surface NG2/CSPG4 such as vascular endothelial cells, promoting in vitro endothelial tube formation and in vivo blood vessel development (Fukushi et al., 2004).
The role of NG2/CSPG4 in the migratory potential of a cell is regulated by differential phosphorylation at two sites. Phosphorylation at Thr2256 through PKCα promotes migration by enhancing NG2/CSPG4 colocalization with beta-1 integrin on the leading edge lamellipodia. Phosphorylation at Thr2314 through ERK-1/2 enhances NG2/CSPG4 colocalization with β-1 integrin on the apical surface microprotrusions and promotes a proliferative phenotype (Makagiansar et al., 2007). The ERK-1/2 and PCKα signaling pathways are important mediators of chondrocyte differentiation and mechanotransduction (Zhen et al., 2001; Lee et al., 2002). PKCα activation occurs in an mTOR-dependent manner, with mTOR signaling being a potent upstream regulator of migration (Zhou and Wong, 2006). Chondrocyte migration is associated with the ERK-1/2 pathway. Together, the ERK, PKC, and mTOR pathways represent parallel and complementary signaling pathways regulating cell migration in cartilage (Fujita et al., 2004; Davies et al., 2008; Lu et al., 2013).
NG2/CSPG4 contains large chondroitin sulfate chains, suggesting that it should have anti-migratory effects on the cells. However, multiple studies have demonstrated that the presence of NG2/CSPG4 in cancers is a strong indicator of the metastatic potential of the tumor (Burg et al., 1997; Burg et al., 1998; Chekenya and Pilkington, 2002), with NG2/CSPG4 loss of function experiments resulting in attenuated cell migration (Cattaruzza et al., 2013b; Jamil et al., 2016; Yang et al., 2019a; Wilms et al., 2022). Given the multivalent nature of NG2-/CSPG4-dependent migratory signaling, with cytoskeletal-mediated/growth factor-independent pathways and convergent growth factor/extracellular matrix pathways potentially influencing the phenotype and behavior of the cell (Cattaruzza and Perris, 2005), the role of NG2/CSPG4 as a regulator of cell migration in cartilage health and disease has yet to be resolved.
The interaction of NG2/CSPG with OMI/HTRA2 to regulate oxidative stress implicates the proteoglycan in the transcriptional regulation of the cell stress response (Maus et al., 2015). Oxidative stress is an important factor in the progression of degenerative conditions such as osteoarthritis (Tanaka et al., 2008). Oxidative stress can be modeled in an in vitro environment using serum starvation (Tangtrongsup and Kisiday, 2018), not only elevating reactive oxygen species but also removing a number of key growth factors known to interact with NG2/CSPG4, synchronizing the cell-cycle kinetics of cells, suppressing proliferation, and altering signaling molecules associated with NG2/CSPG4 functionality such as ERK-1/2, PKCα, and mTOR signaling (Hasan et al., 1999; Chen and Long, 2014; Raja et al., 2018). Here, we will leverage this cell culture model together with live cell imaging of migration to determine the role of NG2/CSPG4 as a regulator of cell migration during normal physiological and cell stress conditions.
Control mice from a C57BL/6J background were purchased from Jackson Laboratory. Knockout mice were acquired from the KOMP repository (Cspg4tm1a(KOMP)Wtsi/Bcm) and were generated using the knockout-first allele, promoter-driven section kit. The knockout mice used in this study were generated by cross-breeding with a Cre-expressing line to generate a reporter-tagged deletion allele. Heterogeneous mice with the reporter-tagged deletion allele were backcrossed with a C57BL/6J line and then mated to generate a reporter-tagged deletion allele for NG2/CSPG4. All animals were housed together to minimize confounding conditions. NG2/CSPG4 knockout mice were viable through skeletal maturity with no strong developmental phenotypes aside from those reported, including increased lean body mass (Mousephenotype.org). There is a mild but significant phenotype in the TMJ cartilage (Reed et al., 2022). DSRed-NG2/CSPG4 (DSRed|NG2) reporter mice were acquired from a commercial source (Tg(Cspg4–DsRed.T1)1Akik, Jackson Laboratory) and bred to generate offspring hemizygous for the reporter construct. TMJ osteoarthritis was induced using the unilateral partial discectomy model as previously described (Yotsuya et al., 2019; Reed et al., 2022). The use of all animal tissues followed an approved animal use protocol (UIC ACC #23-042/#20-068).
For immunohistochemistry, the sections were deparaffinized, treated with sodium borohydride (132.2 mM, 452882, Sigma), permeabilized with methanol and 0.5% Triton (v/v), blocked in 5% donkey serum (D9663, Sigma, St. Louis, MO) for 2 h, and incubated with primary antibodies against NG2/CSPG4 (1:200, AB5320, Sigma Millipore, Santa Cruz, CA) or a custom monoclonal antibody raised against the NG2/CSPG4 intracellular domain (1:200, GGQPDPELLQFCRTPNPALRNGQYWV, UIC Protein Core, Chicago, IL, United States). Secondary labeling was done with Alexa Fluor donkey anti-rabbit 568 (1:500, Invitrogen, Carlsbad, CA). Nuclei were labeled with DAPI (D9542-1 MG, 1 μg/μL, Sigma, St. Louis, MO). The sections were imaged using an inverted fluorescence microscope using a ×10 objective (DMI6000B, Leica, Buffalo Grove, IL). Laser intensity, gain, and magnification were standardized for all acquisitions. Brightness and contrast settings were standardized for all images during post-processing. All data were compared to those of a no primary antibody control and isotype control. For immunocytochemistry, the cells were fixed using HistoChoice tissue fixative (VWRVH102, VWR, Radnor, PA). Antigen retrieval is the same as that described for immunohistochemistry. All samples were imaged using a laser scanning confocal microscope with a ×63 oil immersion objective (LSM 710, Zeiss) using identical laser intensity, brightness, and gain standardized for all image acquisitions. The images are representative of four biological replicates for each experimental group.
Primary cell isolation was carried out by following the published methods for chondrocytes (Gosset et al., 2008; Reed et al., 2021; Reed et al., 2022; Bagheri Varzaneh et al., 2023). Mandibular condylar cartilages (MCCs) were collected from wild-type, NG2/CSPG2 knockout, and/or DsRed reporter mice from 10–14-day-old pups. Extracted MCCs were placed in collection medium DMEM (12492–013, Gibco, Gaithersburg, MD) and rinsed twice with sterile phosphate-buffered saline (PBS) solution with 25 mg/mL Plasmocin (ant-mmp, InvivoGen, San Diego, CA), 50 U/mL penicillin, and 0.05 mg/mL streptomycin (P0781, Sigma, St. Louis, MO) under a sterile flow hood. For cell isolation, MCCs were digested in type II collagenase (S004174, Worthington Biochemical, Lakewood, NJ) suspended in 3 mg/mL Dulbecco’s modified Eagle medium (11966–025, Gibco, Gaithersburg, MD) for 45 min in a thermal incubator under 5% CO2 at 37°C. The tissue fragments were agitated using a pipette to detach soft tissues and were then washed with PBS solution. The cartilage pieces were retrieved and then transferred to 1.5 mg/mL type II collagenase digestion medium overnight in 5% CO2 at 37°C. The cell solution was retrieved and placed in a 15-mL tube and then dispersed by using a transfer pipette. The cell suspension was filtered using a sterile 48 uM cell strainer and then centrifuged at 10,000 g for 10 min at room temperature. The pellet was retrieved and washed with PBS and resuspended with 15 mL of the culture medium. The cell density was calculated using a hemocytometer. All cells were cultured, grown to confluence, and used before the third passage.
The bulk RNA-seq analysis follows previously published methods (Reed et al., 2022). In short, RNA was isolated using the Qiagen RNeasy Mini Kit (79216 Qiagen, Germantown, MD). A Poly(A) RNA sequencing library was prepared with Illumina’s TruSeq Stranded mRNA sample preparation protocol, including oligo-(dT) magnetic bead purification, poly(A) RNA fragmentation, DNA library construction, and Agilent Technologies 2100 Bioanalyzer high-sensitivity DNA chip quality control. Sequencing was performed using Illumina’s NovaSeq 6000 sequencing system. HISAT2 was used to carry out mapping of reads for the genome, and the reads were assembled using StringTie. All transcriptomes were merged using perl scripts and gffcompare. StringTie and edgeR were used to estimate the expression levels and perform mRNA expression levels. Three biological replicates were used for the analysis. Differential gene expression was used for a Gene Ontology analysis using the ShinyGO platform (Ge et al., 2020) using the biological processes pathway database (ShinyGO 0.77), with differentially up- and downregulated genes analyzed together (q-value <0.05) against background genes.
For in vitro and in vivo protein isolation of cultured cells, the plates were washed in ice-cold, 1× PBS, lysed using an extraction reagent (M-PER, 78501, Thermo Fisher, Waltham, MA) with protease (cOmplete, 4693116001, Sigma, St. Louis, MO) and phosphatase (PhosSTOP, 4906845001, Sigma, St. Louis, MO) inhibitors. For in vitro protein isolation of the cell–agarose scaffolds, the samples were rinsed in 1× PBS for 20 min, placed in Laemmli buffer, boiled for 5 min, cooled on ice, and spun down for 2 h using a mini-spin column (Pierce Spin Cups, 69700, Thermo Fisher, Waltham, MA). For all samples, insoluble lysates were removed by centrifugation at 14,000 g for 15 min at 4°C. For the monolayer cell and tissue samples tested for NG2/CSPG4, the supernatant was incubated with chondroitinase ABC (100330-1, AMSBio, Cambridge, MA) added at 0.05 units/mL for 3 h at 37°C. For all samples, the lysates were adjusted to a 4x Protein Sample Loading Buffer (928–40004, LI-COR, Lincoln, NE), heated at 100°C for 5 min, run on a 4%–15% sodium dodecyl sulfate polyacrylamide gel (SDS–PAGE), and analyzed using Western blot with antibodies against NG2/CSPG4 (1:500, AB5320, Sigma Millipore, Santa Cruz, CA) and PCNA (1:1000, 2586S, Cell Signaling Technologies, Danvers, Massachusetts). The blots were imaged using a LI-COR fluorescence quantitative western blot. Fluorescence values were normalized to β-actin and standardized to experimental control samples. Four biological replicates were used for all western blot.
Primary cells under three passages were used for the study. Primary cells were trypsinized and seeded in a three-well culture insert for cell migration (ibidi, Gräfelfing) at a concentration of 55 × 103 cells per 100 ul. The cells were incubated in secondary growth conditions overnight to achieve an 80% confluence. To suppress cell proliferation, mitomycin C (5 μg/mL; Sigma-Aldrich, St. Louis, MO) was added to cell culture media, and the cells were incubated for 2 h at 37°C at 5% CO2. To initiate cell migration, the insert was removed and the culture plate was placed in a live-cell microscope stage at 37°C and 5% CO2 (Leica, DMI6000B). The cells were imaged under phase contrast microscopy every 15 min for 24 h. Each experimental group consisted of four biological replicates and four technical replicates per sample. To quantify cell migration, the gap closure rate was analyzed using ImageJ software. The leading edges of the cells on each image were manually traced to create an outline of the target cell-free areas at 0, 4, 12, and 24 h. The ImageJ measure tool was used to calculate the areas in pixels at each time point and were transferred to the data window. The cell-free area at 0 h is used as a baseline to calculate % closure of the area over time: % closure = [(cell-free area at 0 h) − (cell-free area at x h)]/(cell-free area at 0 h). Cell counting was performed by counting the total number of cells in the cell-free zone defined at time 0 for each experimental time point. Four biological replicates and two or four technical replicates were used for each experimental group.
For quantifying gene expression changes, RNA was isolated using the RNeasy Mini Kit (74104, Qiagen, Germantown, MD). All target genes were amplified with the SYBR® Select Master Mix (4385610, Applied Biosystems, Waltham, MA) in a Bio-Rad iQ5 (Bio-Rad, Des Plaines, IL). Primer sequences are reported in Supplementary Table S3. Validation of all primers using negative controls substituting molecular grade water for cDNA was carried out for each primer for standard quality control. Gene expression changes were calculated by the comparative threshold cycle method with data standardized to a sample control and normalized to GAPDH using the ΔΔCq method. Negative controls with no cDNA were run for all primers. Four biological replicates and two technical replicates were used for each experimental group.
Primary cells from the mandibular condyles of DSRed|NG2 reporter mice were plated in a 12-well plate and grown to confluence. For the migration assay, a scratch was made across the length of the plate with the tip of a 10-μL pipette, creating a higher signal intensity of DsRed positive cells at the injury site than using the insert. The cells were cultured in serum-supplemented and low-serum condition during the migration assay. Live-cell microscopy followed the methods previously described with a fluorescent channel (588 excitation/583 emission). Laser intensity, brightness, and the contrast of image acquisition were established before starting the experiment to optimize imaging DsRed-positive cells. Exported data were processed and cell motility performance was quantified using TrackMate (Tinevez et al., 2017). DsRed-positive and -negative cells were tracked using the software and manually correct by the frame. The migration, velocity, and distance of each cell were calculated and output by TrackMate. Four biological replicates and five technical replicates (i.e., cells) were used for each experimental group.
Primary cells from a 10–14-day-old mouse mandibular condylar cartilage were isolated as described in the previous section. Once confluent in a 100-mm dish, the cells were transduced with the supernatant of a recombinant retroviral vector containing cDNAs expressing hTERT containing a GFP-expressing protein. After one round of retroviral transduction, the cells were given serum-supplemented media and grown to confluence. Confluent cells were then trypsinized to a single-cell suspension and sorted for GFP using a flow cytometer (Bio-Plex, Bio-Rad). GFP-positive cells were then plated in a 96-well plate using a single-cell cloning approach. The clone with the highest growth rate was selected and frozen for preparing a stock solution. The resuspended cells from this clone were used for all experiments and for the CRISPR/Cas9 modifications from cells under 10 passages.
The collagen VI binding sequence for NG2/CSPG4 was identified from published sequences (Tamburini et al., 2018). The sgRNA sequences were designed from this target sequence using the ALT-R® CRISPR–Cas9 system from Integrated DNA Technologies (IDT, Coralville, IA, United States). All of the off- and on-targets were designed using IDT software (https://eu.idtdna.com/pages). The ALT-R® CRISPR–Cas9 system was used for transfection and includes the Cas9 protein, trans-activating CRISPR RNA (tracrRNA), and CRISPR RNA (crRNA). TracrRNA (5 nmol; 3 μL) was mixed with target-specific crRNA (2 nmol, 3 μL) in the IDT nuclease-free duplex buffer. The solution was incubated for 5 min at 95°C and slowly annealed at room temperature for 10 min. The sgRNA (400 ng in total) and Cas9 (1 μg) were complexed for 5 min in reduced serum medium (Opti-MEM, Gibco, NY, United States). The solution was combined with 150,000 cells, placed in a cuvette, and electroporated at 225 V for six pulses (Gene Pulser Xcell, Bio-Rad) following the published optimization values for chondrocytes (Schönenberger et al., 2011). The cells were then incubated in 10% FBS/DMEM for a further 48 h and allowed to recover. After the recovery period, dead cells were removed and adherent cells were trypsinized to generate a single-cell suspension. These cells were then plated for single-cell clonal expansion using the dilution method. Clone colonies were expanded into two plates. One plate was screened by PCR with primers designed to span the target deletion (Table 1). The second plate was screened using Sanger sequencing (3730xl Analyzer, Life Technologies). NG2/CSPG4 sequences from the control and CRISPR/Cas9 truncated cells were manually aligned to confirm the truncation of the amino acid sequence corresponding to the collage VI binding region. One clone with the best growth properties and the appropriate genotype was isolated and used for all subsequent studies.
A one-way ANOVA was used for all statistical tests. Post hoc Bonferroni tests were carried out for multi-group comparisons (SPSS, Chicago, IL). A p-value of <0.05 was considered statistically significant for all studies.
NG2/CSPG4 is abundant in a restricted cell population in the mandibular condylar cartilage. The mandibular condylar cartilage of the TMJ is a secondary cartilage that forms from a migratory cell population derived from the periosteum of the developing mandibular mesenchyme (Hinton and Carlson, 2005). In the mandibular condylar cartilage, the superficial perichondrium is continuous with the fibrous bony periosteum and the underlying prechondroblastic layer is continuous with the osteogenic layer of the bony periosteum. Both of these cell layers are superficial to chondroblastic cells evident by the presence of proteoglycans such as aggrecan (Figures 1A, B). During MCC development, NG2/CSPG4 is abundant in the prechondroblastic layer (Figures 1C, D). In the skeletally mature mandibular condylar cartilage, the continuity of the perichondrium with the periosteum is still apparent near the pole of the condyle (Figures 1E, F). There is also an NG2/CSPG4-positive cell population deep in this perichondrium, in the prechondroblastic and chondroblastic cell layer (Figures 1G, H). During the early stages of TMJ OA, the pole of the condyle expands as the condyle flattens and the perichondrium thickens (Figures 1I, J), with a concomitant increase in the NG2/CSPG4-positive layer deep in the perichondrium (Figure 1K, L). These data illustrate that NG2/CSPG4 is not localized to the fibrous perichondrium but is concentrated in cells that have committed to chondrogenic differentiation.
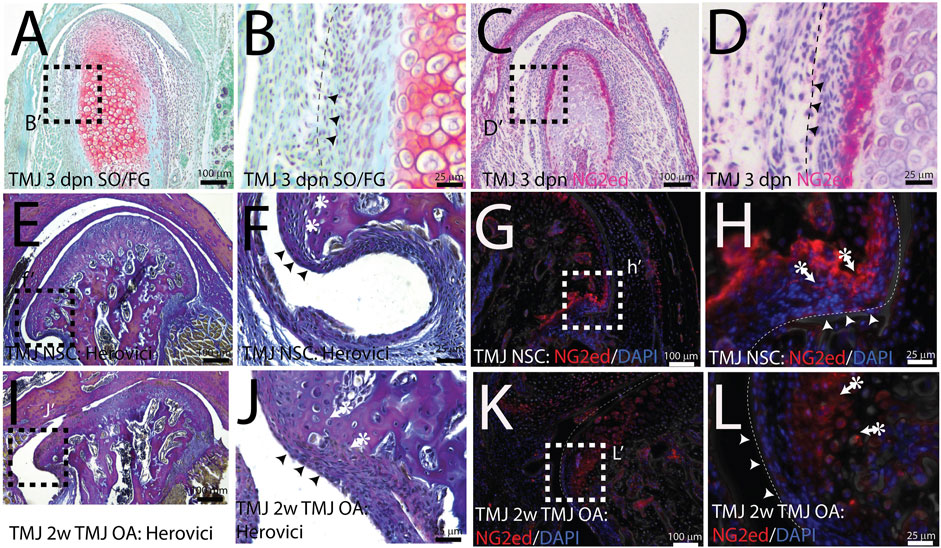
FIGURE 1. NG2/CSPG4 is abundant in a restricted population of cells in the mandibular condyle. (A, B) Safranin O/Fast Green staining of a 3-day post-natal mouse TMJ illustrating that the superficial fibrous layer of the condylar cartilage (arrows) and an underlying prechondroblastic layer. The chondroblastic layer is delineated by the presence of proteoglycans (red). (C, D) Immunohistochemistry illustrating that NG2/CSPG4 is strongly expressed in the prechondroblastic layer. (E, F) Herovici’s polychrome stain of the pole of the mandibular condyle illustrating that the fibrous layer of the bony periosteum is continuous with the articular layer of the mandibular cartilage and that the underlying prechondroblastic layer is continuous with the osteogenic layer of the bony periosteum; (G, H) immunofluorescent staining illustrating that NG2/CSPG4 is localized to the prechondroblastic cells in the mandibular condylar cartilage and on a cell layer adjacent to the bone in the osteogenic layer of the condylar periosteum. (I, J) Herovici’s polychrome stain of early-stage TMJ OA illustrating an increase in the thickness of prechondroblastic/chondroblastic layers at the pole of the condyle and a thickening of the periosteum. (K, L) Immunofluorescence staining of NG2/CSPG4 in early-stage TMJ OA illustrating an increase in immune-positive cells in the prechondroblastic layer of the mandibular condylar cartilage and negligible staining in the fibrous and osteogenic layers of the periosteum.
NG2/CSPG4 regulates the transcriptional control of cell-matrix interactions and migration. The bulk RNA-seq analysis of primary mandibular fibrochondrocytes derived from the mandibular condylar cartilage of the control and NG2/CSPG4 knockout mice cultured in serum-supplemented and serum starvation conditions was analyzed using Gene Ontology (GO) enrichment analysis. Using the biological process ontology set, several significant GO terms associated with cell migration during serum culture conditions were identified, including the “Regulation of cell migration” (-log10 FDR = 16; 200 genes), “Cell migration” (–log10 FDR = 24; 300 genes), and “Cell motility” (–log10 FDR = 16; 300 genes). In serum starvation conditions, significant pathways include “Cell migration” (–log10 FDR = 4; 200 genes) and “Cell motility” (–log10 FDR = 4; 200 genes). In both serum-supplemented and serum starvation conditions, NG2/CSPG4 knockout cells have a transcriptionally distinct profile affecting genes that regulate cell migration (Figure 2).
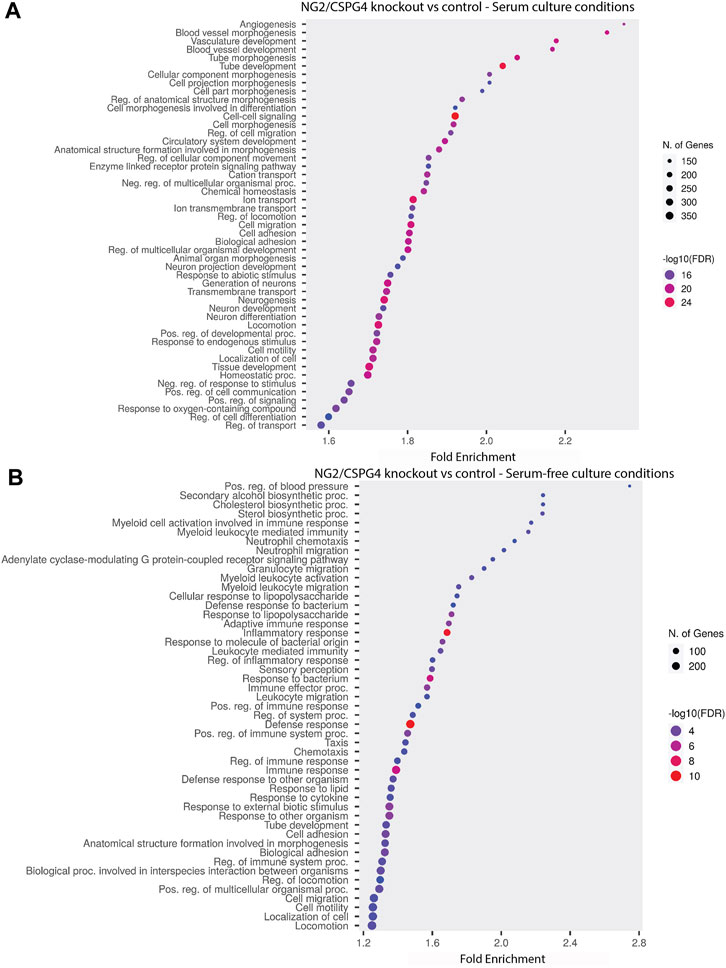
FIGURE 2. NG2/CSPG4 regulates the transcriptional control of cell-matrix interactions and migration in serum-supplemented and serum starvation conditions. (A) Gene Ontology enrichment analysis comparing the differences in the biological process terms from a comparison of NG2/CSPG4 knockout and control c57 primary mandibular condylar cartilage cells in serum (A) and serum-free (B) cell-culture conditions. Data represent sequencing results from n = 3/genotype per treatment group.
NG2/CSPG4 knockout cells migrate more quickly under serum starvation conditions. Control and NG2/CSPG4 knockout cells cultured in serum-supplemented media and in serum starvation conditions illustrate that NG2/CSPG4 is only present on the control cells (Figures 3A, B). Western blot analysis of the control cells illustrates that serum starvation does not affect the amount of full-length NG2/CSPG4 protein (Figures 3C, D). In serum-supplemented cell culture conditions, there was no difference in cell migration between the control and NG2/CSPG4 knockout cells measured by the percent closure or cell-free area after 24 h of migration (Figure 3E). In serum starvation conditions, NG2/CSPG4 knockout cells migrated more than the controls, as measured by both the percent closure (Figure 3E; p <0.05; n = 4/genotype) and the number of cells in the cell-free region after 24 h (Figure 3F; p <0.05; n = 4/genotype). Furthermore, there was no significant difference in the percent closure in NG2/CSPG4 knockout cells in serum-supplemented and serum-free culture conditions (Figure 3E).
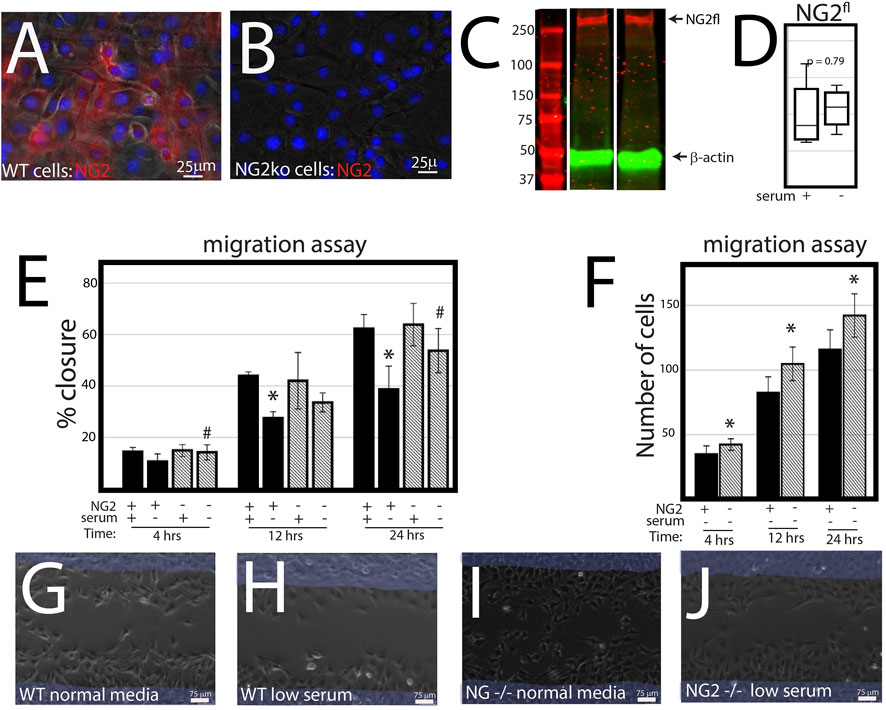
FIGURE 3. NG2/CSPG4 knockout cells migrate more quickly in serum starvation conditions. (A) Immunocytochemistry with immunostaining for NG2/CSPG4 in control primary mandibular fibrochondrocytes. (B) Immunocytochemistry with immunostaining for NG2/CSPG4 in NG2/CSPG4 knockout primary mandibular fibrochondrocytes. (C) Western blot of NG2/CSPG4 from the protein collected from control primary mandibular fibrochondrocytes cultured in serum-supplemented and serum starvation conditions. The full-length NG2/CSPG4 fragment is indicated at 300 kDa. Continuous western blot are provided in Supplementary Figure S1. (D) Quantification of the full length NG2/CSPG4 fragment with values standardized to β-actin and fold-change is calculated relative to the serum supplemented condition. n = 4/experimental group. (E) Quantification of a migration assay from the control and NG2/CSPG4 knockout primary mandibular fibrochondrocytes in serum-supplemented and serum starvation conditions measured at 4, 12, and 24 h. Data represent the percent closures of the cell-free region relative to time 0. (F) Quantification of the number of cells that passed in the cell free from the migration assay from serum starvation conditions at 4, 12, and 24 h. (G–J) Raw data visualization of the migration data with the cell-free range at time 0 indicated in yellow, superimposed over the same region after 24 h of migration. For all graphs, * represents the comparisons of serum-supplemented and serum starvation conditions with * = p < 0.05; # represents comparisons of the control versus NG2/CSPG4 knockout cells with # = p < 0.05.* = p <0.05 and *** = p <0.001; # represents comparisons of serum-supplemented and serum starvation conditions with # = p < 0.05 and *** = p < 0.001; # represents the comparisons of the control versus NG2/CSPG4 knockout cells with # = p < 0.05 and ### = p < 0.001. Representative live-cell videos of migration are provided as Supplementary Videos S1–S4.
NG2/CSPG4 knockout cells migrate more quickly in a cell proliferation-independent manner. NG2/CSPG4 cells have significantly lower levels of cell proliferation during serum-supplemented cell culture conditions (Figures 4A, B; p <0.001; n = 4/experimental group). Serum starvation suppresses cell proliferation in both the control and NG2/CSPG4 knockout cells, equilibrating proliferation to statistically indistinguishable, but non-zero levels. To remove the confounding impact of proliferation, we repeated all migration experiments following 2 h of 5 μg/mL mitomycin C treatment to suppress DNA synthesis. In serum-supplemented cell culture conditions, mitomycin C pretreatment resulted in a decrease in the rate of cell migration in NG2/CSPG4 knockout cells when compared with the control as measured by the percent closure or cell-free area after 24 h of migration (Figure 4C; p < 0.05; n = 4/experimental group). In serum starvation conditions, mitomycin C pretreatment results in an increase in the rate of cell migration in NG2/CSPG4 knockout cells when compared with controls, as measured by both the percent closure (Figure 4C; p <0.05; n = 4/genotype) and an increase in the number of cells in the cell-free region after 4 and 12 h (Figure 4D; p <0.05; n = 4/genotype). The percent closure and number of cells in the NG2/CSPG4 knockout cells decreased at 24 h, likely due to elevated levels of cell death resulting from prolonged exposure to the stress conditions resulting from mitomycin C pretreatment and serum starvation (see Figure 4F).
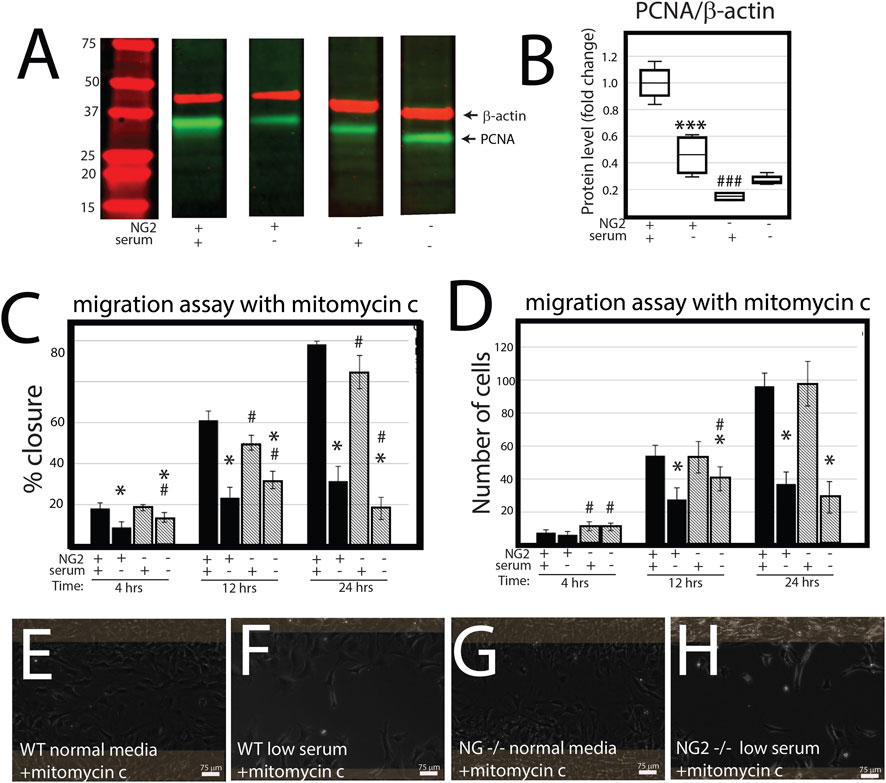
FIGURE 4. NG2/CSPG4 knockout cells have suppressed proliferation, but the rate of migration occurs in a proliferation-independent manner. (A) Western blot of PCNA from protein collected from the control and NG2/CSPG4 knockout primary mandibular fibrochondrocytes cultured in serum-supplemented and serum starvation conditions. The bands are representative of n = 4/experimental group. Continuous Western blots are provided in Supplementary Figure S1. (F) Quantification of PCNA with values standardized to β-actin and fold change is calculated relative to the serum-supplemented serum condition. (C) Quantification of a migration assay from the control and NG2/CSPG4 knockout primary mandibular fibrochondrocytes in serum-supplemented and serum starvation conditions measured at 4, 12, and 24 h pretreated with 5 μg/mL mitomycin C to suppress proliferation. Data represent the percent closures of the cell-free region at time 0. (D) Quantification of the number of cells that passed in the cell free region from the migration assay from serum starvation conditions at 24 h. (E–H) Raw data visualization of the migration data with the cell-free range at time 0 indicated in yellow, superimposed over the same region after 24 h of migration. For all graphs, * represents comparisons of serum-supplemented and serum starvation conditions with * = p < 0.05 and *** = p < 0.001; # represents the comparisons of the control versus NG2/CSPG4 knockout cells with # = p < 0.05 and ### = p < 0.001.
NG2/CSPG4 is diminished during active migration in mandibular fibrochondrocytes. To characterize NG2/CSPG6 levels in actively migrating cells, we conducted a migration assay on a glass slide and fixed the cells after 6 h. Confocal immunofluorescence using an antibody against full-length NG2/CSPG4 illustrates that cell–cell contact is associated with high levels of NG2/CSPG4, while migration is associated with lower levels of NG2/CSPG4 (Figures 5A–C). It has been previously reported that NG2/CSPG4 levels are linked with cell density (Asher et al., 2005). To determine if the heterogeneous distribution of NG2/CSPG4 protein in these cells was related to cell density or the migratory phenotype, we characterized NG2/CSPG4 levels from a confluent and sub-confluent population of primary mandibular fibrochondrocytes (Figures 5D, E). There was no difference in the gene expression levels between the confluent and sub-confluent population (Figure 5F; p = 0.194; n = 4/experimental group). Western blot analysis illustrates that the sub-confluent population of cells had a significantly higher level of both full-length and shed-membrane tethered fragments of NG2/CSPG4 (Figures 5.G–I; p <0.05; n = 4/experimental group). Together, these findings illustrate that the turnover of the NG2/CSPG4 protein in active migration cells is a property of the migratory phenotype and not of the density of the cells.
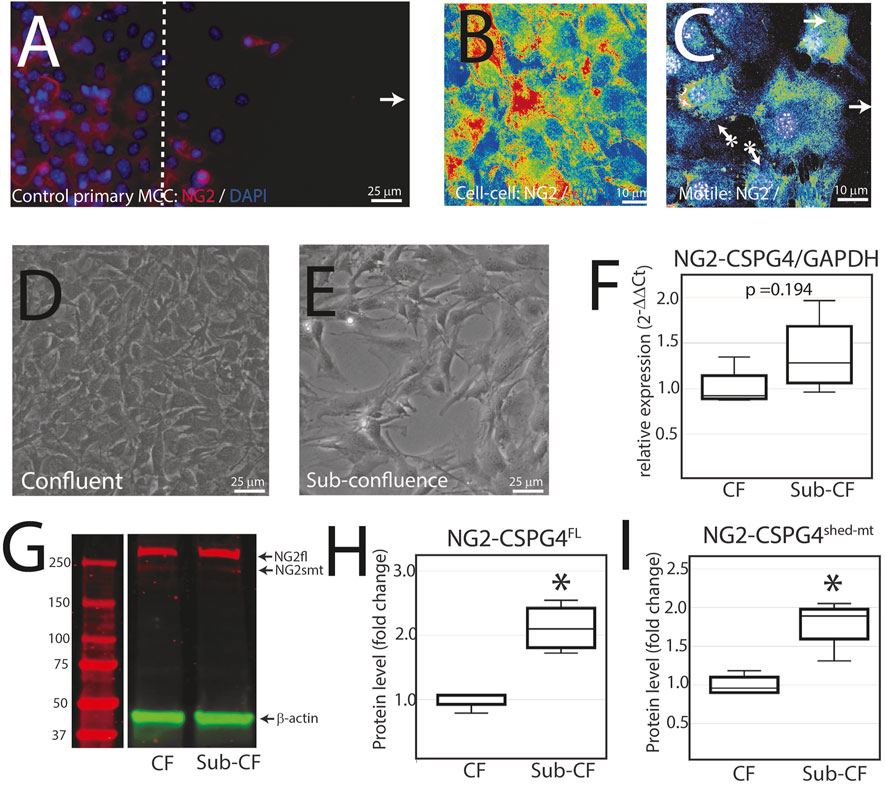
FIGURE 5. NG2/CSPG4 is diminished during active migration in mandibular fibrochondrocytes. (A) Immunocytochemistry of primary control mandibular fibrochondrocytes fixed and immuno-labeled with NG2/CSPG4 6 h after the start of migration. (B) Confocal immunofluorescence of NG2/CSPG4 pseudo-colored to illustrate the signal intensity from a region of the migration assay with high cell–cell contact. (C) Confocal immunofluorescence of NG2/CSPG4 pseudo-colored to illustrate the signal intensity from a region of the migration assay with actively migrating cells. (D) Bright-field microscopy of control primary mandibular fibrochondrocytes from a confluent plate. (E) Bright-field microscopy of control primary mandibular fibrochondrocytes from a sub-confluent plate. (F) RT-qPCR illustrating gene expression changes in NG2/CSPG4 in the confluent (CF) and sub-confluent (sub-CF) samples of control primary mandibular fibrochondrocytes. (G) Western blot of NG2/CSPG4 from the protein collected from confluent and sub-confluent samples of control primary mandibular fibrochondrocytes. The full-length fragment is indicated at 300 kDa and the shed membrane-tethered fragment is indicated at 275/260 kDa. These bands are representative of an n = 4/experimental group. Continuous blots are provided in Supplementary Figure S2. (H–I) Quantification of full-length NG2/CSPG4 with values standardized to β-actin and fold change is calculated relative to the confluent experimental group. * represents the comparisons of confluent and sub-confluent experimental groups with * = p <0.05.
DSRed|NG2-positive cells migrate slower in serum starvation conditions. Since NG2/CSPG4 knockout cells have a distinct transcriptional profile, we conducted a scratch assay on primary mandibular fibrochondrocytes derived from DSRed|NG2 reporter mice. Primary cells extracted from the mandibular condylar cartilage reflect the heterogeneity associated with the cartilage, with only some of the cells expressing NG2/CSPG4 (i.e., DsRed positive). Cell tracing software was used to characterize the distance and velocity of DSRed|NG2-positive and -negative cells during migration. DSRed|NG2-positive cells migrated with a significantly lower distance and velocity than NG2/CSPG4–DsRed-negative cells (Figure 6. D–E; p <0.05; n = 5/reporter status). This finding illustrates that NG2/CSPG4 expressing cells have an attenuated migratory phenotype, consistent with the knockout data.
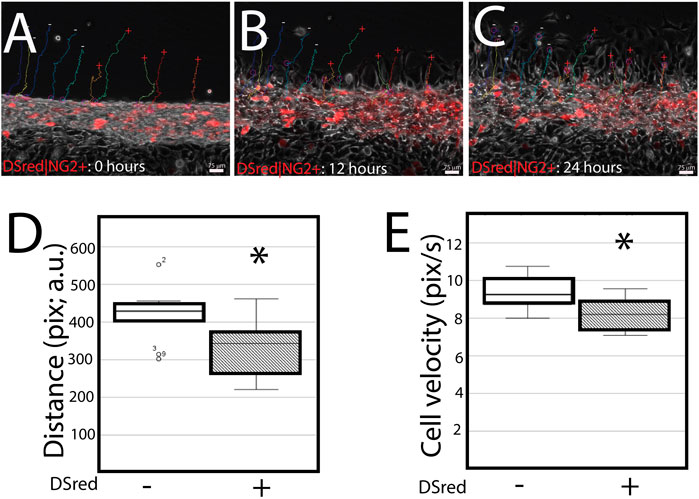
FIGURE 6. DSRed|NG2-positive cell migrate slower in serum starvation conditions. (A–C) Cell migration dynamics after a scratch assay at 0 (A), 12 (B), and 24 (C) hours. DSRed|NG2-positive are red. The migration path of DSRed|NG2-positive (+) and -negative (−) cells is superimposed over each image. (D) Quantification of the distance traveled by each DSRed|NG2-positive and -negative cell reported in pixels, arbitrary units. (E) Quantification of the velocity of each cell in DSRed|NG2-positive and -negative cells reported in pixels/second, arbitrary units. For all experiments, data were collected from two biological replicates, with five DSRed|NG2-positive and -negative cells quantified from each biological replicate. * represents the comparisons of DSRed|NG2-positive and -negative cells with p <0.05. A representative live-cell video of cell migration is provided as Supplementary Video S05.
NG2/CSPG4 ectodomain perturbation impacts cell migration. Since the NG2/CSPG4 ectodomain interacts with the extracellular matrix through type VI collagen, we next probed if the NG2/CSPG4 ectodomain was a select and specific regulator of the migratory phenotype of the cell. Mandibular fibrochondrocytes from C57BL/6J mice were immortalized using hTERT and stabilized for five passages. To mutate the ectodomain, the type VI collagen binding region was truncated from the NG2/CSPG4 ectodomain using CRISPR/Cas9 (NG2|EDmut). Western blot analysis of these cells illustrates that the NG2|EDmut lacks robust full length and shed membrane-tethered bands compared to the control (Figure 7A). Immunocytochemistry using a polyclonal antibody against the NG2/CSPG4 ectodomain shows a similar trend, with the control cells having robust membrane-associated NG2/CSPG4 that is greatly diminished in the NG2|EDmut cells (Figures 7B, C). Immunocytochemistry using a monoclonal antibody against the NG2/CSPG4 intracellular domain illustrates robust intracellular/cytosolic NG2/CSPG4 in the control and NG2|EDmut cells (Figures 7D, E). The NG2/CSPG4 intracellular domain could not be resolved on the Western blot in either sample but has been previously validated (Reed et al., 2022). The gene expression of NG2/CSPG4 is significantly higher in the NG2|EDmut cells than in the control (Figure 7F; p <0.05; n = 4/genotype). There is no statistically significant difference in the rate of proliferation for the transgenic or serum experimental conditions (Figures 7G–H). The rate of migration was significantly different in a genetic and serum-dependent manner. In serum-supplemented culture conditions, the control and NG2|EDmut cells migrate at the same rate. In serum starvation conditions, the NG2|EDmut cells migrate significantly faster than the controls (Figure 7I; p <0.05; n = 4/genotype) and have significantly more cells migrating into the cell-free regions after 24 h (Figure 7J; p <0.05; n = 4/genotype). Together, these data illustrate that the NG2/CSPG4 ectodomain is an important regulator of the migratory phenotype of mandibular fibrochondrocytes. The bulk RNA-seq analysis of the NG2|EDmut and control cells in serum supplemented and serum starvation conditions was carried out using Gene Ontology enrichment analysis. Using the biological process ontology set, several significant GO terms associated with cell migration were identified in serum culture conditions including the “Regulation of cell migration” (–log10 FDR = 12.5; 200 genes), “Cell migration” (–log10 FDR = 15; 300 genes), and “Cell motility” (–log10 FDR = 12.5; 300 genes). In serum starvation conditions, significant pathways include “Positive regulation of cell migration” (–log10 FDR = 10; 200 genes), “Regulation of cell migration” (–log10 FDR = 12.5; 300 genes), “Cell migration” (–log10 FDR = 17.5; 400 genes), and “Cell motility” (–log10 FDR = 12.5; 400 genes). In both serum and serum starvation conditions, NG2|EDmut cells have a transcriptionally distinct profile affecting genes that regulate cell migration (Figure 8).
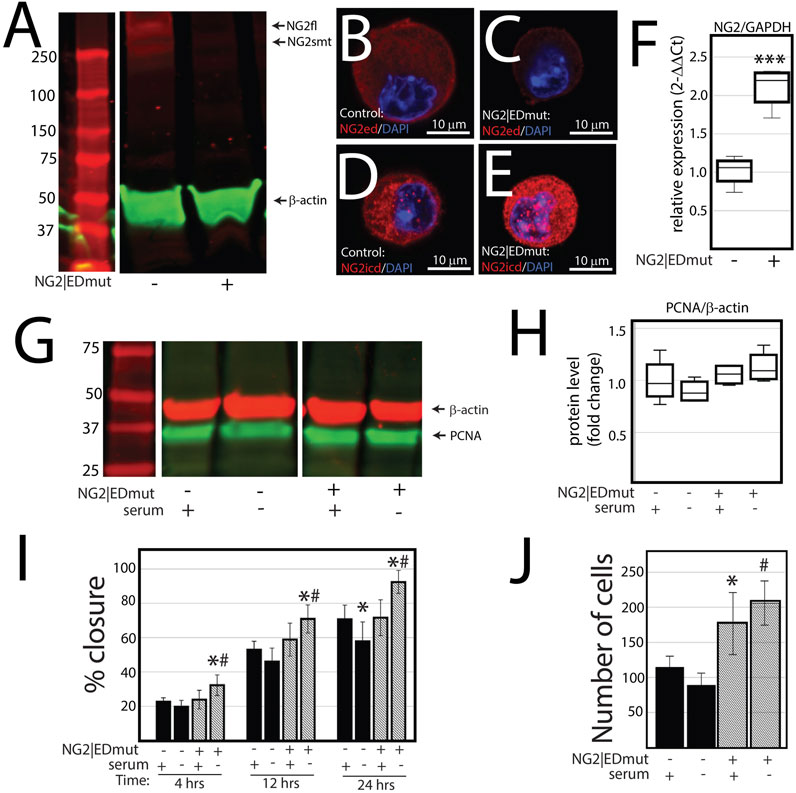
FIGURE 7. NG2/CSPG4 ectodomain perturbation impacts cell migration. (A) Western blot analysis of NG2/CSPG4 of the control and NG2|EDmut cells. These bands are representative of the n = 4/experimental group. Continuous Western blots are provided in Supplementary Figure S3. (B) Immunocytochemistry from control cells using a polyclonal antibody raise the NG2/CSPG4 ectodomain. (C) Immunocytochemistry from NG2|EDmut cells using a polyclonal antibody raise the NG2/CSPG4 ectodomain. (D) Immunocytochemistry from control cells using a monoclonal antibody raised the NG2/CSPG4 intracellular domain. (E) Immunocytochemistry from NG2|EDmut cells using a monoclonal antibody raised the NG2/CSPG4 intracellular domain. (F) RT-qPCR illustrating gene expression changes in NG2/CSPG4 in control and NG2|EDmut cells. (G) Western blot analysis of PCNA of the control and NG2|EDmut cells in serum-supplemented and serum-free culture conditions. The bands are representative of n = 4/experimental group. Continuous Western blots are provided in Supplementary Figure S3. (H) Quantification of PCNA Western blots with values standardized to β-actin and fold change calculated relative to the control–serum-supplemented experimental group. (I) Quantification of a migration assay from the control and NG2|EDmut cells in serum-supplemented and serum starvation conditions measured at 4, 12, and 24 h. Data represent the percent closures of the cell-free region time 0. (J) Quantification of the number of cells that passed in the cell-free region from the migration assay from serum starvation conditions at 24 h. For all graphs, * represents the comparisons of serum-supplemented and serum starvation conditions with * = p <0.05 and *** = p <0.001; # represents the comparisons of the control versus NG2|EDmut cell with # = p < 0.05.
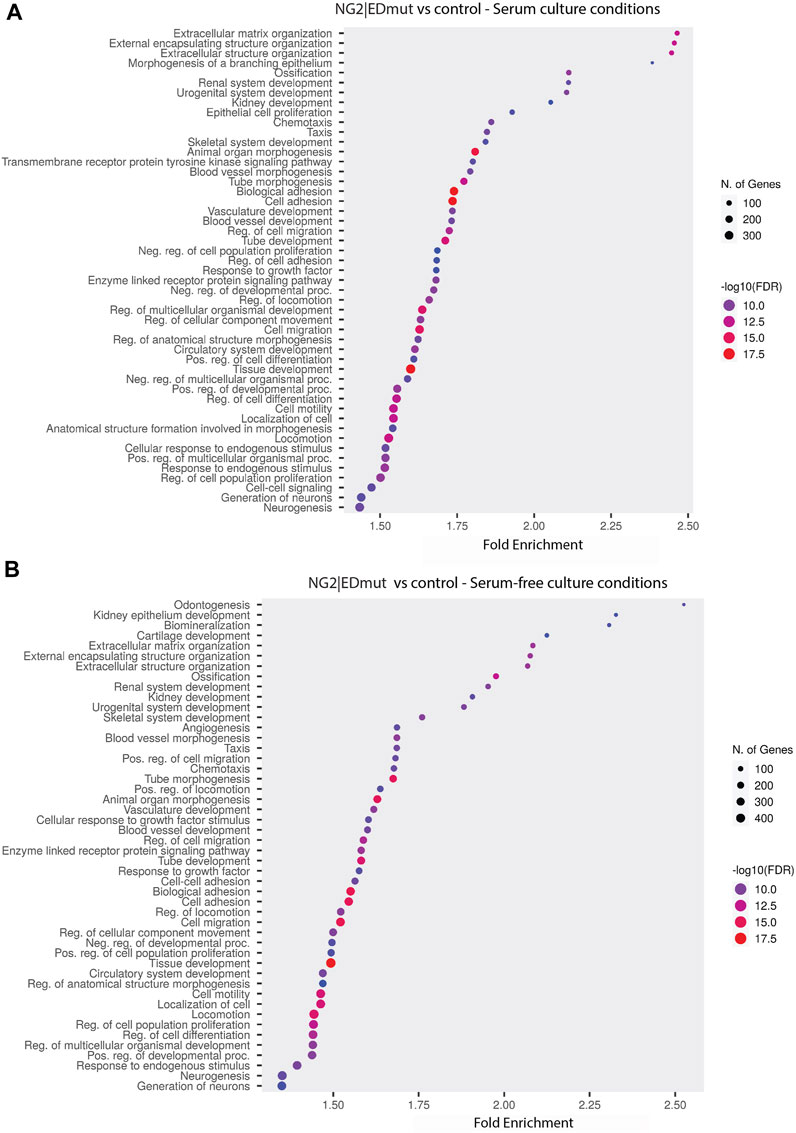
FIGURE 8. NG2/CSPG4 ectodomain perturbation impacts the transcriptional regulation of cell migration in serum starvation conditions. (A, B) Gene Ontology enrichment analysis comparing the differences in biological process terms from a comparison of NG2|EDmut and control immortalized primary mandibular condylar cartilage cells in serum (A) and serum-free (B) cell culture conditions. Data represent sequencing results from an n = 3/genotype per treatment group.
This study defines the role of NG2/CSPG4 in the migration of mandibular fibrochondrocytes during normal physiological and cell stress conditions. NG2/CSPG4 has multivalent properties due to the ability to bind the pro-migratory growth factor PDGF and bind with the extracellular matrix through type VI collagen. In serum-supplemented culture conditions, containing the pro-migratory growth factor PDGF, there is no difference in the migration potential of control and NG2/CSPG4 knockout cells. However, pretreatment with mitomycin C resulted in the suppression of cell migration in NG2/CSPG4 knockout cells. In serum starvation conditions, NG2/CSPG4 knockout cells migrated at a higher rate than control cells, irrespective of being pretreated with and without mitomycin C. Using NG2–DsRed reporter cells and cell tracking software, we find that NG2–DsRed-positive cells migrate a shorter distance and at a slower rate than NG2–DsRed-negative cells in serum starvation conditions. When the NG2/CSPG4 ectodomain is perturbed, a similar pattern of migration potential is observed. NG2|EDmut cells migrate faster in serum starvation conditions. Together, these three model systems illustrate that the presence of full-length NG2/CSPG4 on the cell surface attenuates the migratory potential of mandibular fibrochondrocytes.
Our findings from healthy, primary mandibular chondrocytes cultured in serum-supplemented conditions and pretreated with mitomycin C are in agreement with the consensus view from studies using cancer cells. In these cells, NG2/CSPG4 is hypothesized to be a prognostic indicator of metastasis and promotes migration. In chondrosarcoma, siRNA suppression of NG2/CSPG4 attenuated the rate of migration (Jamil et al., 2016). In glia and glioma cells, SP-1 binding to the enhancer region of NG2/CSPG4, and subsequent increase in NG2/CSPG4 protein, increased the migratory potential of the cells (Wilms et al., 2022). A similar pattern is observed in neural cells, with the addition of NG2/CSPG4 antibodies attenuating migration rates (Stegmüller et al., 2002). However, the stage of the cancer can influence the role of NG2/CSPG4 (Hsu et al., 2018), underscoring that NG2/CSPG4 can have different roles in different contexts due to complex ecto- and intracellular domain-mediated processing. In contrast, carrying out the migration assay in serum starvation conditions illustrates that NG2/CSPG4 knockout cells migrate faster than the controls, at a rate more similar to that of the serum-supplemented conditions. This finding underscores that there is potentially an important interaction effect between the transcriptional regulation of the cell and the exposure to cell stress conditions in a high ROS intracellular environment.
During the cell migration assay, cells that migrate into the cell-free area have low levels of cell surface NG2/CSPG4. This could indicate that the primary cells are heterogeneously fated and/or that only cells with low basal levels of NG2/CSPG4 are migrating. This hypothesis is supported by the findings of bulk RNA-seq analysis, illustrating that NG2/CSPG4 knockout cells are transcriptionally distinct with differential gene expression profiles that favor cell migration in both serum-supplemented and serum starvation conditions. However, results from the NG2 reporter line provide a more opaque explanation, with the NG2|DsRed-positive cells migrating slower than the NG2|DsRed-negative cells, but with both cell types moving into the cell-free region. Lower levels of full-length NG2/CSPG4 protein and/or transcript may not necessarily indicate that the protein is non-functional in the molecular mechanics of migration. These cells adjacent to the cell-free region may commit to a migratory phenotype through local endocytic recycling of cell surface NG2/CSPG4 and/or through proteolytic cleavage of the NG2/CSPG4 ectodomain.
NG2/CSPG4 colocalization of β1-integrin at the leading edge of the cell is important and may indicate turnover or recycling of the protein through the lysosomal machinery or proteolytic processing of the ectodomain, as the cell interacts with the endogenous extracellular matrix of the tissue. The endocytosis of NG2/CSPG4 has been implicated in focal adhesion dynamics, with Stonin1 knockout fibroblasts, an NG2 endocytic adapter, associated with the accumulation of cell-surface NG2/CSPG4 and impaired focal adhesion-mediated motility (Feutlinske et al., 2015). The proposed mechanism of action includes the Arf6-integrin disruption of focal adhesions, impacting the directionality of the cells. The capacity for NG2/CSPG4 to regulate migration in melanoma cells was associated with the localization of activated focal adhesion kinases to lipid rafts through the transmembrane cysteine residue (C2230)-mediated assembly of molecular complexes related to syntenin-1 (Yang et al., 2019b). The shedding of the NG2/CSPG4 ectodomain has been illustrated in primary mandibular fibrochondrocytes; however, the molecular functionality of this shedding has yet to be fully resolved (Bagheri Varzaneh et al., 2023). Thus, subcellular localization, and not abundance, could be the most critical parameter regulating migration.
Conversely, high levels of NG2/CSPG4 at cell–cell contacts could indicate contact inhibition of locomotion, where contact with another cell confers polarity and directionality to the cell. This biophysical property is important for regulating the coordinated collective cell migration behaviors such as those in a cell migration assay. In cultured primary mandibular chondrocytes, these data indicate that the abundance of NG2/CSPG4 is dependent on the density of the cells in the plate, confirming previous reports using other cell types (Asher et al., 2005). This result implicates NG2/CSPG4 in the mechanocoupling of cell–cell/matrix junctions and illustrates that this cell–cell/matrix contact is important for retaining cell surface NG2/CSPG4.
The functionality of cell surface abundance may also indicate the need for the cell to interact with the endogenous extracellular matrix produced when confluences is reached, either acting as a cell surface receptor for type VI collagen or as a binding partner with β1-integrins. Changes in collagen VI are one of the characteristics of end-stage temporomandibular osteoarthritis (Reed et al., 2019; Yotsuya et al., 2019). The loss of full-length NG2/CSPG4 or the ectodomain leads to dysfunctional collagen VI matrix adhesion and migration. When collagen VI binding is dysfunctional, sarcoma cells respond with convergent cell survival and adhesion/migration pathways. In other cell types, the targeted deletion of the C-terminal cytoplasmic region also impacts matrix adhesion and cell motility (Fang et al., 1999; Makagiansar et al., 2004; Cattaruzza et al., 2013a). Thus, the proteolytic and endocytic processing of NG2/CSPG4 integrates multiple external stimuli and intracellular signaling networks when shifting toward a migratory phenotype.
NG2/CSPG4 knockout mandibular fibrochondrocytes show significant changes in several key pathways implicated in cell motility and adhesion, including PI3K and MAPK (Reed et al., 2022). NG2/CSPG4 binding with collagen VI promotes the PI3K pathway, regulating cell spreading and motility through bypassing of the canonical integrin transduction mechanisms (Cattaruzza et al., 2013a). The PI3K pathway is associated with cell survival and mTORC1. This could potentially implicate PI3K cell survival as a common signaling pathway regulating migration in serum starvation conditions. PI3K is further implicated in the migratory phenotype of chondrocytes through Runx2 (Fujita et al., 2004). One of the other key cell signaling pathways for chondrocyte migration includes ERK-1/2 (Morales, 2007; Lu et al., 2013). NG2/CSPG4 is necessary for the sustained activation of the ERK cascade (Reed et al., 2022), with ERK differentially phosphorylating the NG2/CSPG4 intracellular domain to regulate proliferation and migration (Makagiansar et al., 2007).
There are several study limitations that were beyond the current scope of this work. Chemotaxis is an important regulator of cell migration, including growth factors such as FGF2 and PDGF. The NG2/CSPG4 ectodomain contains a binding region with high affinity for PDGF-AA, but not PDGF-BB (Nishiyama et al., 1996a; Grako et al., 1999). Future studies will use recombinant PDGF in serum-free conditions to control for the confounding chemotactic effects of PDGF. The NG2/CSPG4 ectodomain also contains chondroitin sulfate chains that can inhibit migration in the cartilage (Davies et al., 2008). Future studies will focus on treatment of cells with chondroitinase ABC for cleaving chondroitin sulfate chains prior to migration. Technical limitations also prevent us from experimentally testing the role of type VI collagen interactions. We attempted to replicate experiments on murine collagen VI-coated plates, but the cells failed to adequately adhere to the plates for the duration of the experiment, particularly so in NG2/CSPG4 knockouts. This finding is consistent with the reports from the literature that NG2/CSPG4 knockout cells have an impaired ability to adhere to type VI collagen. A second technical limitation in the study concerns the use of only two-dimensional culture assays. We replicated the migration experiments using a Boyden chamber, with the serum as the chemotactic gradient, but the primary mandibular fibrochondrocytes failed to migrate through transwell in sufficient quantity to quantify migration.
Despite these limitations, this study has yielded important insights into the functionality of NG2/CSPG4 in cell migration and motility. Future studies on NG2/CSPG4 will broaden our understanding of the migration of stem cells and chondroprogenitor cells as an important part of skeletogenesis. These pro-migratory developmental programs could be utilized for tissue regeneration strategies or improved wound healing outcomes. Tissue engineering approaches would also benefit by the development of homing molecules for stem and/or progenitor cells into the tissue, improving the integration of the engineered scaffold. In the pathophysiology of degenerative arthropathy such as rheumatoid and osteoarthritis, activated fibroblasts like synoviocytes migrate on the articular cartilage and promote cartilage degeneration. The migration of these cells could be targeted to slow the progression of degenerative arthropathy. Together, these findings illustrate that NG2/CSPG4 represents an underappreciated target since it is abundant on the cell surface and contributes to a diverse set of cellular behaviors.
RNAseq data deposited in the Gene Expression Omnibus: Series GSE245589 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE245589. All other datasets are provided in the manuscript and/or Supplementary Material.
The animal study was approved by the UIC Animal Care and Use Committee, protocol 23-042. The study was conducted in accordance with the local legislation and institutional requirements.
SA: data acquisition and interpretation and drafted and critically revised the manuscript. MB: data acquisition and interpretation and critically revised the manuscript. YZ: data acquisition and interpretation and critically revised the manuscript. JR: data acquisition and interpretation and critically revised the manuscript. SR: data acquisition and interpretation and critically revised the manuscript. JB: data acquisition and interpretation and critically revised the manuscript. MC: data acquisition and interpretation and critically revised the manuscript. DR: Contributed to the conception, design, data acquisition and interpretation; performed all statistical analyses; and drafted and critically revised the manuscript.
This investigation was supported by the National Institute of Dental and Craniofacial Research of the National Institutes of Health under Award 1R56DE029835 (DR) and 1R01DE029835-01 (DR).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2023.1240920/full#supplementary-material
Asher, R. A., Morgenstern, D. A., Properzi, F., Nishiyama, A., Levine, J. M., and Fawcett, J. W. (2005). Two separate metalloproteinase activities are responsible for the shedding and processing of the NG2 proteoglycan in vitro. Mol. Cell. Neurosci. 29, 82–96. doi:10.1016/j.mcn.2005.02.001
Bagheri Varzaneh, M., Zhao, Y., Rozynek, J., Han, M., and Reed, D. (2023). Disrupting mechanical homeostasis promotes matrix metalloproteinase-13 mediated processing of neuron glial antigen 2 in mandibular condylar cartilage. Eur. Cells Mater. 45, 113–130. doi:10.22203/eCM.v045a08
Bi, R., Yin, Q., Mei, J., Chen, K., Luo, X., Fan, Y., et al. (2020). Identification of human temporomandibular joint fibrocartilage stem cells with distinct chondrogenic capacity. Osteoarthr. Cartil. 28, 842–852. doi:10.1016/j.joca.2020.02.835
Bradbury, E. J., Moon, L. D., Popat, R. J., King, V. R., Bennett, G. S., Patel, P. N., et al. (2002). Chondroitinase ABC promotes functional recovery after spinal cord injury. Nature 416, 636–640. doi:10.1038/416636a
Burg, M. A., Grako, K. A., and Stallcup, W. B. (1998). Expression of the NG2 proteoglycan enhances the growth and metastatic properties of melanoma cells. J. Cell. physiology 177, 299–312. doi:10.1002/(SICI)1097-4652(199811)177:2<299::AID-JCP12>3.0.CO;2-5
Burg, M. A., Nishiyama, A., and Stallcup, W. B. (1997). A central segment of the NG2 proteoglycan is critical for the ability of glioma cells to bind and migrate toward type VI collagen. Exp. Cell Res. 235, 254–264. doi:10.1006/excr.1997.3674
Burg, M. A., Tillet, E., Timpl, R., and Stallcup, W. B. (1996). Binding of the NG2 proteoglycan to type VI collagen and other extracellular matrix molecules. J. Biol. Chem. 271, 26110–26116. doi:10.1074/jbc.271.42.26110
Cattaruzza, S., Nicolosi, P. A., Braghetta, P., Pazzaglia, L., Benassi, M. S., Picci, P., et al. (2013a). NG2/CSPG4–collagen type VI interplays putatively involved in the microenvironmental control of tumour engraftment and local expansion. J. Mol. cell Biol. 5, 176–193. doi:10.1093/jmcb/mjt010
Cattaruzza, S., Ozerdem, U., Denzel, M., Ranscht, B., Bulian, P., Cavallaro, U., et al. (2013b). Multivalent proteoglycan modulation of FGF mitogenic responses in perivascular cells. Angiogenesis 16, 309–327. doi:10.1007/s10456-012-9316-7
Cattaruzza, S., and Perris, R. (2005). Proteoglycan control of cell movement during wound healing and cancer spreading. Matrix Biol. 24, 400–417. doi:10.1016/j.matbio.2005.06.005
Chekenya, M., Krakstad, C., Svendsen, A., Netland, I., Staalesen, V., Tysnes, B., et al. (2008). The progenitor cell marker NG2/MPG promotes chemoresistance by activation of integrin-dependent PI3K/Akt signaling. Oncogene 27, 5182–5194. doi:10.1038/onc.2008.157
Chekenya, M., and Pilkington, G. (2002). NG2 precursor cells in neoplasia: functional, histogenesis and therapeutic implications for malignant brain tumours. J. Neurocytol. 31, 507–521. doi:10.1023/a:1025795715377
Chen, J., and Long, F. (2014). mTORC1 signaling controls mammalian skeletal growth through stimulation of protein synthesis. Development 141, 2848–2854. doi:10.1242/dev.108811
Cledes, G., Felizardo, R., Foucart, J.-M., and Carpentier, P. (2006). Validation of a chemical osteoarthritis model in rabbit temporomandibular joint: a compliment to biomechanical models. Int. J. oral Maxillofac. Surg. 35, 1026–1033. doi:10.1016/j.ijom.2006.05.003
Davies, L. C., Blain, E. J., Caterson, B., and Duance, V. C. (2008). Chondroitin sulphate impedes the migration of a sub-population of articular cartilage chondrocytes. Osteoarthr. Cartil. 16, 855–864. doi:10.1016/j.joca.2007.12.005
Embree, M. C., Chen, M., Pylawka, S., Kong, D., Iwaoka, G. M., Kalajzic, I., et al. (2016). Exploiting endogenous fibrocartilage stem cells to regenerate cartilage and repair joint injury. Nat. Commun. 7, 13073. doi:10.1038/ncomms13073
Fan, Y., Cui, C., Li, P., Bi, R., Lyu, P., Li, Y., et al. (2021). Fibrocartilage stem cells in the temporomandibular joint: insights from animal and human studies. Front. Cell Dev. Biol. 9, 665995. doi:10.3389/fcell.2021.665995
Fang, X., Burg, M. A., Barritt, D., Dahlin-Huppe, K., Nishiyama, A., and Stallcup, W. B. (1999). Cytoskeletal reorganization induced by engagement of the NG2 proteoglycan leads to cell spreading and migration. Mol. Biol. cell 10, 3373–3387. doi:10.1091/mbc.10.10.3373
Feutlinske, F., Browarski, M., Ku, M.-C., Trnka, P., Waiczies, S., Niendorf, T., et al. (2015). Stonin1 mediates endocytosis of the proteoglycan NG2 and regulates focal adhesion dynamics and cell motility. Nat. Commun. 6, 8535. doi:10.1038/ncomms9535
Fujita, T., Azuma, Y., Fukuyama, R., Hattori, Y., Yoshida, C., Koida, M., et al. (2004). Runx2 induces osteoblast and chondrocyte differentiation and enhances their migration by coupling with PI3K-Akt signaling. J. cell Biol. 166, 85–95. doi:10.1083/jcb.200401138
Fukushi, J.-I., Makagiansar, I. T., and Stallcup, W. B. (2004). NG2 proteoglycan promotes endothelial cell motility and angiogenesis via engagement of galectin-3 and alpha3beta1 integrin. Mol. Biol. cell 15, 3580–3590. doi:10.1091/mbc.e04-03-0236
Fukushi, J. I., Inatani, M., Yamaguchi, Y., and Stallcup, W. B. (2003). Expression of NG2 proteoglycan during endochondral and intramembranous ossification. Dev. Dyn. 228, 143–148. doi:10.1002/dvdy.10359
Ge, S. X., Jung, D., and Yao, R. (2020). ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics 36, 2628–2629. doi:10.1093/bioinformatics/btz931
Goretzki, L., Burg, M. A., Grako, K. A., and Stallcup, W. B. (1999). High-affinity binding of basic fibroblast growth factor and platelet-derived growth factor-AA to the core protein of the NG2 proteoglycan. J. Biol. Chem. 274, 16831–16837. doi:10.1074/jbc.274.24.16831
Gosset, M., Berenbaum, F., Thirion, S., and Jacques, C. (2008). Primary culture and phenotyping of murine chondrocytes. Nat. Protoc. 3, 1253–1260. doi:10.1038/nprot.2008.95
Grako, K. A., Ochiya, T., Barritt, D., Nishiyama, A., and Stallcup, W. B. (1999). PDGF (alpha)-receptor is unresponsive to PDGF-AA in aortic smooth muscle cells from the NG2 knockout mouse. J. cell Sci. 112, 905–915. doi:10.1242/jcs.112.6.905
Haeger, A., Wolf, K., Zegers, M. M., and Friedl, P. (2015). Collective cell migration: guidance principles and hierarchies. Trends cell Biol. 25, 556–566. doi:10.1016/j.tcb.2015.06.003
Hasan, N. M., Adams, G. E., and Joiner, M. C. (1999). Effect of serum starvation on expression and phosphorylation of PKC-alpha and p53 in V79 cells: implications for cell death. Int. J. cancer 80, 400–405. doi:10.1002/(sici)1097-0215(19990129)80:3<400::aid-ijc11>3.0.co;2-u
Hinton, R. J., and Carlson, D. S. (2005). Regulation of growth in mandibular condylar cartilage. Seminars Orthod. 11, 209–218. doi:10.1053/j.sodo.2005.07.005
Hopper, N., Henson, F., Brooks, R., Ali, E., Rushton, N., and Wardale, J. (2015). Peripheral blood derived mononuclear cells enhance osteoarthritic human chondrocyte migration. Arthritis Res. Ther. 17.1, 1–10. doi:10.1186/s13075-015-0709-z
Hsu, S.-H. C., Nadesan, P., Puviindran, V., Stallcup, W. B., Kirsch, D. G., and Alman, B. A. (2018). Effects of chondroitin sulfate proteoglycan 4 (NG2/CSPG4) on soft-tissue sarcoma growth depend on tumor developmental stage. J. Biol. Chem. 293, 2466–2475. doi:10.1074/jbc.M117.805051
Huang, F.-J., You, W.-K., Bonaldo, P., Seyfried, T. N., Pasquale, E. B., and Stallcup, W. B. (2010). Pericyte deficiencies lead to aberrant tumor vascularizaton in the brain of the NG2 null mouse. Dev. Biol. 344, 1035–1046. doi:10.1016/j.ydbio.2010.06.023
Hunziker, E. B., and Rosenberg, L. C. (1996). Repair of partial-thickness defects in articular cartilage: cell recruitment from the synovial membrane. JBJS 78, 721–733. doi:10.2106/00004623-199605000-00012
Jamil, N. S., Azfer, A., Worrell, H., and Salter, D. M. (2016). Functional roles of CSPG4/NG2 in chondrosarcoma. Int. J. Exp. pathology 97, 178–186. doi:10.1111/iep.12189
Jiang, Y., and Tuan, R. S. (2015). Origin and function of cartilage stem/progenitor cells in osteoarthritis. Nat. Rev. Rheumatol. 11, 206–212. doi:10.1038/nrrheum.2014.200
Johnson, W. E., Caterson, B., Eisenstein, S. M., and Roberts, S. (2005). Human intervertebral disc aggrecan inhibits endothelial cell adhesion and cell migration in vitro. Spine 30, 1139–1147. doi:10.1097/01.brs.0000162624.95262.73
Johnson, W. E., Sivan, S., Wright, K. T., Eisenstein, S. M., Maroudas, A., and Roberts, S. (2006). Human intervertebral disc cells promote nerve growth over substrata of human intervertebral disc aggrecan. Spine 31, 1187–1193. doi:10.1097/01.brs.0000217669.04903.61
Krekoski, C. A., Neubauer, D., Zuo, J., and Muir, D. (2001). Axonal regeneration into acellular nerve grafts is enhanced by degradation of chondroitin sulfate proteoglycan. J. Neurosci. 21, 6206–6213. doi:10.1523/JNEUROSCI.21-16-06206.2001
Lauffenburger, D. A., and Horwitz, A. F. (1996). Cell migration: a physically integrated molecular process. cell 84, 359–369. doi:10.1016/s0092-8674(00)81280-5
Lee, H.-S., Millward-Sadler, S., Wright, M., Nuki, G., Al-Jamal, R., and Salter, D. (2002). Activation of Integrin-RACK1/PKCalpha signalling in human articular chondrocyte mechanotransduction. Osteoarthr. Cartil. 10, 890–897. doi:10.1053/joca.2002.0842
Lu, Y., Xu, Y., Yin, Z., Yang, X., Jiang, Y., and Gui, J. (2013). Chondrocyte migration affects tissue-engineered cartilage integration by activating the signal transduction pathways involving Src, PLCγ1, and ERK1/2. Tissue Eng. Part A 19, 2506–2516. doi:10.1089/ten.TEA.2012.0614
Makagiansar, I. T., Williams, S., Dahlin-Huppe, K., Fukushi, J.-I., Mustelin, T., and Stallcup, W. B. (2004). Phosphorylation of NG2 proteoglycan by protein kinase C-alpha regulates polarized membrane distribution and cell motility. J. Biol. Chem. 279, 55262–55270. doi:10.1074/jbc.M411045200
Makagiansar, I. T., Williams, S., Mustelin, T., and Stallcup, W. B. (2007). Differential phosphorylation of NG2 proteoglycan by ERK and PKCalpha helps balance cell proliferation and migration. J. cell Biol. 178, 155–165. doi:10.1083/jcb.200612084
Maus, F., Sakry, D., Binamé, F., Karram, K., Rajalingam, K., Watts, C., et al. (2015). The NG2 proteoglycan protects oligodendrocyte precursor cells against oxidative stress via interaction with OMI/HtrA2. PloS one 10, e0137311. doi:10.1371/journal.pone.0137311
Midwood, K. S., and Salter, D. M. (1998). Expression of NG2/human melanoma proteoglycan in human adult articular chondrocytes. Osteoarthr. Cartil. 6, 297–305. doi:10.1053/joca.1998.0128
Midwood, K. S., and Salter, D. M. (2001). NG2/HMPG modulation of human articular chondrocyte adhesion to type VI collagen is lost in osteoarthritis. J. pathology 195, 631–635. doi:10.1002/path.985
Mishima, Y., and Lotz, M. (2008). Chemotaxis of human articular chondrocytes and mesenchymal stem cells. J. Orthop. Res. 26, 1407–1412. doi:10.1002/jor.20668
Morales, T. I. (2007). Chondrocyte moves: clever strategies? Osteoarthr. Cartil. 15, 861–871. doi:10.1016/j.joca.2007.02.022
Nishiyama, A., Lin, X. H., Giese, N., Heldin, C. H., and Stallcup, W. (1996a). Co-localization of NG2 proteoglycan and PDGF alpha-receptor on O2A progenitor cells in the developing rat brain. J. Neurosci. Res. 43, 299–314. doi:10.1002/(SICI)1097-4547(19960201)43:3<299::AID-JNR5>3.0.CO;2-E
Nishiyama, A., Lin, X. H., Giese, N., Heldin, C. H., and Stallcup, W. (1996b). Interaction between NG2 proteoglycan and PDGF alpha-receptor on O2A progenitor cells is required for optimal response to PDGF. J. Neurosci. Res. 43, 315–330. doi:10.1002/(SICI)1097-4547(19960201)43:3<315::AID-JNR6>3.0.CO;2-M
Nishiyama, A., and Stallcup, W. B. (1993). Expression of NG2 proteoglycan causes retention of type VI collagen on the cell surface. Mol. Biol. cell 4, 1097–1108. doi:10.1091/mbc.4.11.1097
Perris, R., Perissinotto, D., Pettway, Z., Bronner-Fraser, M., Mörgelin, M., and Kjmata, K. (1996). Inhibitory effects of PG-H/aggrecan and PG-M/versican on avian neural crest cell migration. FASEB J. 10, 293–301. doi:10.1096/fasebj.10.2.8641562
Raja, R., Lata, S., Trivedi, S., and Banerjea, A. C. (2018). Serum deprivation/starvation leads to reactivation of HIV-1 in latently infected monocytes via activating ERK/JNK pathway. Sci. Rep. 8, 14496–14499. doi:10.1038/s41598-018-32316-2
Reed, D. A., Yotsuya, M., Gubareva, P., Toth, P. T., and Bertagna, A. (2019). Two-photon fluorescence and second harmonic generation characterization of extracellular matrix remodeling in post-injury murine temporomandibular joint osteoarthritis. PloS one 14, e0214072. doi:10.1371/journal.pone.0214072
Reed, D. A., Zhao, Y., Bagheri Varzaneh, M., Soo Shin, J., Rozynek, J., Miloro, M., et al. (2022). NG2/CSPG4 regulates cartilage degeneration during TMJ osteoarthritis. Front. Dent. Med. 69. doi:10.3389/fdmed.2022.1004942
Reed, D. A., Zhao, Y., Han, M., Mercuri, L. G., and Miloro, M. (2021). Mechanical loading disrupts focal adhesion kinase activation in mandibular fibrochondrocytes during murine temporomandibular joint osteoarthritis. J. Oral Maxillofac. Surg. 79, 2058.e1–2058.e15. doi:10.1016/j.joms.2021.05.001
Robinson, J., O’Brien, A., Chen, J., and Wadhwa, S. (2015). Progenitor cells of the mandibular condylar cartilage. Curr. Mol. Biol. Rep. 1, 110–114. doi:10.1007/s40610-015-0019-x
Seol, D., Mccabe, D. J., Choe, H., Zheng, H., Yu, Y., Jang, K., et al. (2012). Chondrogenic progenitor cells respond to cartilage injury. Arthritis and Rheumatism 64, 3626–3637. doi:10.1002/art.34613
Stallcup, W. B., Dahlin, K., and Healy, P. (1990). Interaction of the NG2 chondroitin sulfate proteoglycan with type VI collagen. J. cell Biol. 111, 3177–3188. doi:10.1083/jcb.111.6.3177
Stegmüller, J., Schneider, S., Hellwig, A., Garwood, J., and Trotter, J. (2002). AN2, the mouse homologue of NG2, is a surface antigen on glial precursor cells implicated in control of cell migration. J. Neurocytol. 31, 497–505. doi:10.1023/a:1025743731306
Tanaka, E., Detamore, M., and Mercuri, L. (2008). Degenerative disorders of the temporomandibular joint: etiology, diagnosis, and treatment. J. Dent. Res. 87, 296–307. doi:10.1177/154405910808700406
Tangtrongsup, S., and Kisiday, J. D. (2018). Modulating the oxidative environment during mesenchymal stem cells chondrogenesis with serum increases collagen accumulation in agarose culture. J. Orthop. Research® 36, 506–514. doi:10.1002/jor.23618
Tillet, E., Gential, B., Garrone, R., and Stallcup, W. B. (2002). NG2 proteoglycan mediates beta1 integrin-independent cell adhesion and spreading on collagen VI. J. Cell. Biochem. 86, 726–736. doi:10.1002/jcb.10268
Tinevez, J.-Y., Perry, N., Schindelin, J., Hoopes, G. M., Reynolds, G. D., Laplantine, E., et al. (2017). TrackMate: an open and extensible platform for single-particle tracking. Methods 115, 80–90. doi:10.1016/j.ymeth.2016.09.016
Wilms, C., Lepka, K., Häberlein, F., Edwards, S., Felsberg, J., Pudelko, L., et al. (2022). Glutaredoxin 2 promotes SP-1-dependent CSPG4 transcription and migration of wound healing NG2 glia and glioma cells: enzymatic Taoism. Redox Biol. 49, 102221. doi:10.1016/j.redox.2021.102221
Yamada, K. M., and Sixt, M. (2019). Mechanisms of 3D cell migration. Nat. Rev. Mol. Cell Biol. 20, 738–752. doi:10.1038/s41580-019-0172-9
Yang, H., Wen, Y., Zhang, M., Liu, Q., Zhang, H., Zhang, J., et al. (2019a). MTORC1 coordinates the autophagy and apoptosis signaling in articular chondrocytes in osteoarthritic temporomandibular joint. Autophagy 16, 271–288. doi:10.1080/15548627.2019.1606647
Yang, J., Price, M. A., Wanshura, L. E. C., He, J., Yi, M., Welch, D. R., et al. (2019b). Chondroitin sulfate proteoglycan 4 enhanced melanoma motility and growth requires a cysteine in the core protein transmembrane domain. Melanoma Res. 29, 365–375. doi:10.1097/CMR.0000000000000574
Yotsuya, M., Bertagna, A. E., Hasan, N., Bicknell, S., Sato, T., and Reed, D. A. (2019). Neuron/glial antigen 2-type VI collagen interactions during murine temporomandibular joint osteoarthritis. Sci. Rep. 9, 56. doi:10.1038/s41598-018-37028-1
Zhen, X., Wei, L., Wu, Q., Zhang, Y., and Chen, Q. (2001). Mitogen-activated protein kinase p38 mediates regulation of chondrocyte differentiation by parathyroid hormone. J. Biol. Chem. 276, 4879–4885. doi:10.1074/jbc.M004990200
Zhou, H. Y., and Wong, A. S. (2006). Activation of p70S6K induces expression of matrix metalloproteinase 9 associated with hepatocyte growth factor-mediated invasion in human ovarian cancer cells. Endocrinology 147, 2557–2566. doi:10.1210/en.2005-1404
Keywords: cell migration, NG2/CSPG4, chondrocytes, temporomandibular joint, collagen VI, cell adhesion, cell motility
Citation: Ahn SY, Bagheri Varzaneh M, Zhao Y, Rozynek J, Ravindran S, Banks J, Chaudhry M and Reed DA (2023) NG2/CSPG4 attenuates motility in mandibular fibrochondrocytes under serum starvation conditions. Front. Cell Dev. Biol. 11:1240920. doi: 10.3389/fcell.2023.1240920
Received: 15 June 2023; Accepted: 10 October 2023;
Published: 07 November 2023.
Edited by:
Saumendra Bajpai, Indian Institute of Technology Madras, IndiaReviewed by:
Kazuhiro Yamamoto, University of Liverpool, United KingdomCopyright © 2023 Ahn, Bagheri Varzaneh, Zhao, Rozynek, Ravindran, Banks, Chaudhry and Reed. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: David A. Reed, cmVlZGRAdWljLmVkdQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.