
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell Dev. Biol. , 30 October 2023
Sec. Embryonic Development
Volume 11 - 2023 | https://doi.org/10.3389/fcell.2023.1238546
 Wen-Jie Jiang1
Wen-Jie Jiang1 Song-Hee Lee1
Song-Hee Lee1 Geun Heo1
Geun Heo1 Hak Jae Chung2
Hak Jae Chung2 Eun Seok Cho2
Eun Seok Cho2 Soo Jin Sa3
Soo Jin Sa3 Shinichi Hochi4
Shinichi Hochi4 Xiang-Shun Cui1*
Xiang-Shun Cui1*Y-box binding protein 1 (YBX1) plays important roles in RNA stabilization, translation, transcriptional regulation, and mitophagy. However, its effects on porcine preimplantation embryos remain unclear. In this study, we knocked down YBX1 in the one-cell (1C) stage embryo via small interfering RNA microinjection to determine its function in porcine embryo development. The mRNA level of YBX1 was found to be highly expressed at the four-cell (4C) stage in porcine embryos compared with one-cell (1C) and two-cell (2C) stages. The number of blastocysts was reduced following YBX1 knockdown. Notably, YBX1 knockdown decreased the phosphatase and tensin homolog-induced kinase 1 (PINK1) and parkin RBR E3 ubiquitin protein ligase (PRKN) mRNA levels. YBX1 knockdown also decreased PINK1, active mitochondria, and sirtuin 1 levels, indicating reduced mitophagy and mitochondrial biogenesis. Furthermore, YBX1 knockdown increased the levels of glucose-regulated protein 78 (GRP78) and calnexin, leading to endoplasmic reticulum (ER) stress. Additionally, YBX1 knockdown increased autophagy and apoptosis. In conclusion, knockdown of YBX1 decreases mitochondrial function, while increasing ER stress and autophagy during embryonic development.
A mitochondrion is an essential organelle that controls energy conversion and ATP production (Matsumoto et al., 2012). Mitochondrial fission and fusion maintain the mitochondrial morphology, homeostasis, and inheritance via mitochondrial biogenesis and mitophagy (Ma et al., 2020). Mitophagy is crucial for mitochondrial quality control (Dlamini et al., 2021). Dysfunctional mitochondria can generate apoptotic signals to induce cell death (Zhang, 2013). Endoplasmic reticulum (ER) is another important organelle which coordinates stress-related signaling pathways critical for maintaining the crosstalk between intracellular and extracellular environments (Lin et al., 2008; Ron and Hubbard, 2008; Kuo et al., 2022). Excess unfolded or misfolded proteins accumulate in the ER lumen leading to ER stress, disrupting ER function, thereby activating the unfolded protein response (UPR) (Senft and Ronai, 2015). ER and mitochondria together regulate various cellular processes, for example, lipid biosynthesis, apoptosis, and mitophagy (Dlamini et al., 2021).
Y-box binding protein 1 (YBX1) is a cytoplasmic messenger ribonucleoprotein that can bind to RNA (Deng et al., 2022). It modulates RNA stability, translation, and transcription (Mordovkina et al., 2020). YBX1 is necessary for cancer cell proliferation and embryonic development. In human cervical cancer HeLa cells, YBX1 affects mitochondrial oxidative phosphorylation proteins expression (Matsumoto et al., 2012). In goat embryos, YBX1 was studied to regulate splicing and maternal mRNA decay (Deng et al., 2022). In mouse, YBX1 is involved in early mouse development, including neural tube closure and cell proliferation (Uchiumi et al., 2006). In zebrafish, YBX1 affects oocyte maturation and maternal-to-zygotic transition process (Sun et al., 2018). Phosphatase and tensin homolog-induced kinase 1 (PINK1) is associated with Parkinsonian disorders (Valente et al., 2004a; Valente et al., 2004b). PINK1 plays a key role in mitochondrial quality control via the PINK1 and Parkin pathways (Niu et al., 2019). YBX1 knockdown decreases the stability of PINK1 and parkin RBR E3 ubiquitin protein ligase (PRKN) to regulate mitophagy for brown adipogenesis and thermogenesis (Wu et al., 2022). PINK1 knockdown impairs embryonic development and induces mitochondrial dysfunction, autophagy, and apoptosis (Niu et al., 2019).
In this study, we hypothesized that YBX1 is crucial for porcine embryonic development. To evaluate this hypothesis, we reduced the expression of YBX1 by microinjection of YBX1 siRNA to explore the role of YBX1 during the early development of porcine embryos. Additionally, we examined mitochondrial function, GRP78, calnexin, LC3, and caspase 3 following YBX1 knockdown. The results suggest that YBX1 affects embryonic development by influencing mitochondrial function, ER stress, autophagy, and apoptosis.
Unless otherwise stated, all chemicals were purchased from Sigma-Aldrich (St. Louis, MO, United States of America).
Pre-pubertal porcine ovaries were collected from a local abattoir (Farm Story Dodarm B&F, Umsung, Chungbuk, South Korea) and transported to the laboratory at 37°C in phosphate-buffered saline (PBS). Porcine cumulus–oocyte complexes (COCs) were aspirated from small antral follicles (diameter: 3–6 mm). Oocytes surrounded by intact cumulus layers were washed thrice with the in vitro maturation (IVM) medium (11150-059; Thermo Fisher Scientific, Waltham, MA, United States) containing TCM199, 0.1 g/L sodium pyruvate, 0.6 mM cysteine, 10 ng/mL epidermal growth factor, 10% (v/v) porcine follicular fluid, 10 IU/mL luteinizing hormone, and 10 IU/mL follicle-stimulating hormone. COCs were randomly divided and cultured in IVM medium (500 μL) for 44 h at 38.5°C under 5% CO2.
Briefly, 1 mg/mL hyaluronidase was used to detach the cumulus cells. Oocyte with a polar body was selected for activation. Denuded oocytes were parthenogenetically activated using two direct-current pulses of 110 V for 60 µs in 280 mM mannitol containing 0.1 mM CaCl2, 0.05 mM MgSO4, 0.01% polyvinyl alcohol (PVA, w/v), and 0.5 mM 4-(2-hydroxyethyl) piperazine-l-ethanesulfonic acid. After that, activated oocytes were cultured with 0.4% bovine serum albumin (BSA) and 7.5 μg/mL cytochalasin B in PZM-5 for 3 h to inhibit pseudo-second polar body extrusion. Activated oocytes were washed 3 times and cultured in PZM-5 with 0.4% BSA in a 4-well plate under the same condition as IVM. Subsequently, embryos were cultured for 7 days, and then examined the blastocyst development rate (blastocyst development rate = number of blastocysts/number of embryos).
For the knockdown groups, the sequences of the small interfering RNAs (siRNAs) was the same with previous study (Jiang et al., 2023b). YBX1 siRNA (50 μM) was microinjected into the cytoplasm of a parthenogenetically activated oocyte via an Eppendorf Femto-Jet (Eppendorf, Hamburg, Germany) and Nikon Diaphot Eclipse TE300 inverted microscope (Nikon, Tokyo, Japan) equipped with the Narishige MM0-202N hydraulic 3-dimensional micromanipulator (Narishige, Amityville, NY, United States). As a control, the company provided negative siRNA (sense: UUCUCCGAACGUGUCACGUTT, antisense: ACGUGACACGUUCGGAGAATT) was microinjected into the cytoplasm of a parthenogenetically activated oocyte under the same conditions. Embryos were cultured in PZM-5 medium for 1 or 2 or 7 d after microinjection.
According to previous article (Niu et al., 2019), 4% paraformaldehyde in PBS was used to fixed embryos for 1 h at room temperature. Then, embryos were washed thrice and incubated with 1% Triton X-100 in PBS for 1 h at room temperature. Embryos were washed thrice and blocked with 3% BSA in PVA-PBS for 1 h. Embryos were then incubated overnight in different primary antibodies: rabbit anti-YBX1 (1:100, 20339-1-AP, Proteintech), rabbit anti-PINK1 (1:100, 23274-1-AP, Proteintech), rabbit anti-GRP78 (1:100, ab21685, Abcam), rabbit anti-calnexin (1:100, ab22595, Abcam), light chain 3 (LC3) (1:100, NB100-2220, Novus biologicals), mouse anti-TOM20 (1:50, SC-17764, Santa Cruz Biotechnology) and rabbit anti-caspase 3 (1:100, 9664S, Cell Signaling Technology) at 4°C. Following three washes (5 min each) with PVA-PBS, embryos were stained with different Alexa Fluor secondary antibodies for 1 h at room temperature. After washing 3 times, embryos were mounted on slides, and examined by confocal microscope (Zeiss LSM 710 Meta). Images were processed using the Zen software (v.8.0; Zeiss).
For the colocalization of mitochondria and TOM20, 4-cell stage embryos were incubated with 500 nM MitoTracker Red CMXRos (M7512; Thermo Fisher Scientific) at 38.5°C for 30 min. After 3 washes with PZM-5, the staining of TOM20 was the same as in the immunofluorescence staining.
60 embryos per group (control group and YBX1 knockdown group) were placed in 20 μL of ice-cold 1 × sodium dodecyl sulfate [SDS] sample buffer and incubated at 98°C for 10 min. Based on the previous article (Sui et al., 2020; Sun et al., 2023), the proteins in each sample were separated using 10% SDS-polyacrylamide gel electrophoresis and transferred to a polyvinylidene fluoride membrane (Millipore, Bedford, MA, United States) via electroblotting. The target protein binding site was blocked with Tris-buffered saline containing Tween-20 (TBST) and 5% skim milk powder for 1 h at room temperature. Subsequently, the membranes were incubated with different antibodies: rabbit anti-YBX1 (1:1,000, 20339-1-AP, Proteintech), rabbit anti-GRP78 (1:1,000, ab21685, Abcam), rabbit anti-calnexin (1:1,000, ab22595, Abcam), light chain 3 (LC3) (1:1,000, NB100-2220, Novus biologicals) and mouse anti-SIRT1 (1:1,000, 60303-1, Proteintech) overnight at 4°C. The membranes were washed three times with TBST for 10 min each. The membranes were then incubated in secondary antibody (1:20,000) for 1 h at room temperature. The membranes were then washed three times with TBST and exposed to the SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific). To quantify the Western blot results, the intensities of the bands were analyzed by the ImageJ software.
RNA was extracted from a pool of 30 embryos per group (control group and YBX1 knockdown group) using the Dynabead mRNA DIRECT kit (61,012; Thermo Fisher Scientific). cDNA was synthesized using a cDNA synthesis kit (Thermo Fisher Scientific), according to the manufacturer’s instructions. Quantitative reverse transcription-PCR was performed using a fast real-time PCR system (ABI StepOnePlus). Real-time quantitative PCR (qPCR) was performed according to previous article (Jiang et al., 2023a). 18S rRNA was used as the reference gene. All primer sequences used in this study are listed in Table 1. Relative genes expression levels were determined using the 2−ΔΔCT method.
Each experiment was repeated at least three times and results were presented as the mean ± standard error of the mean. The GraphPad Prism 5 software (GraphPad, San Diego, CA, United States of America) was used for statistical analysis. A t-test was used to compare the results between groups. p < 0.05 was considered statistically significant.
To investigate its subcellular localization during embryonic development, we performed immunofluorescence staining to determine the location of YBX1 in two-cell (2C; n = 6), four-cell (4C; n = 6), morula (MO; n = 5) and blastocyst (BL; n = 6) stage embryos. As shown in Figures 1A,B, YBX1 localized in the cytoplasm and the immunofluorescence (IF) intensity of YBX1 was gradually increased. Next, we examined the mRNA levels of YBX1 during embryonic development. We observed that the mRNA levels of YBX1 were increased from the 4C to BL stage compared with the 1C and 2C stages (Figure 1C), indicating that YBX1 is a zygotic gene. Moreover, the results of Western blotting were similar to those of IF and real-time quantitative PCR. Taken together, these data indicate the presence of YBX1 in porcine embryos, which may play a key role in embryonic development.
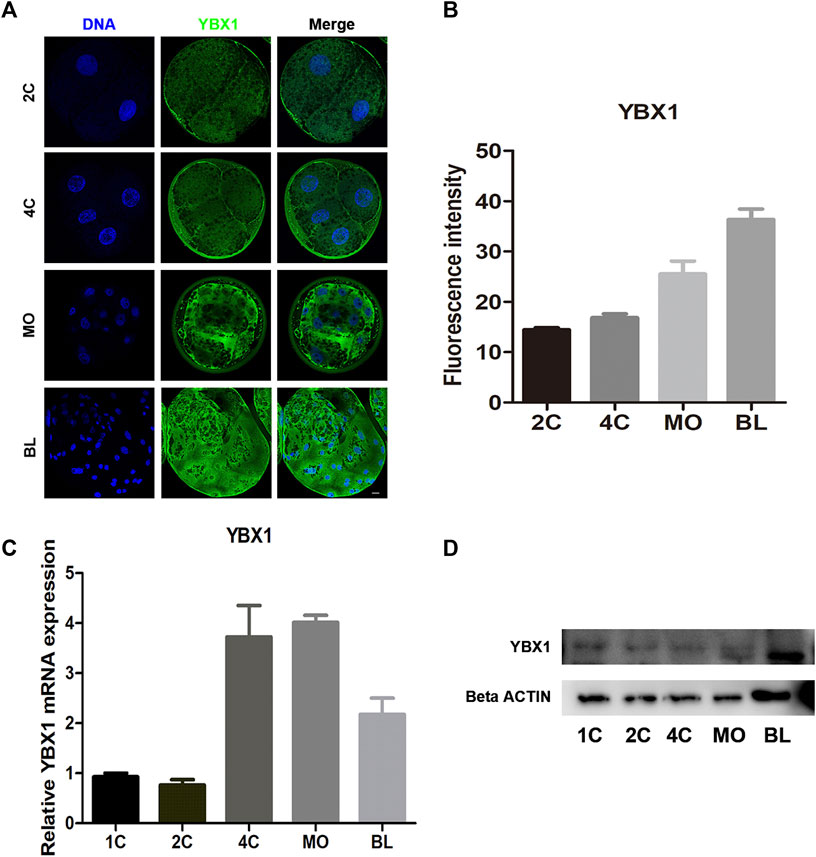
FIGURE 1. Subcellular distribution and expression of YBX1 during embryo development. (A) Immunofluorescence images of YBX1 at two-cell (2C), four-cell (4C), morula (MO) and blastocyst (BL) stages. Blue, DNA; green, YBX1. Scale bar, 40 µm. (B) Fluorescence intensity for YBX1 expression at 2C, 4C, MO, BL stages. (C) Real-time quantitative PCR results of YBX1 mRNA expression levels during early porcine embryonic development. (D) Western blotting results of YBX1 protein expression levels during early porcine embryonic development.
To explore the functional roles of YBX1 during embryonic development in pigs, we knocked down YBX1 at the 1C stage. As shown in Figure 2A, mRNA levels of YBX1 were decreased by approximately 62% in the YBX1 knockdown group compared to those in the control group at the 2C, 4C and blastocyst stage. Knockdown of YBX1 was verified by Western blotting (0.62 ± 0.08 vs 1; p < 0.05; Figures 2B,C) at blastocyst stage and immunofluorescence staining (0.91 ± 0.03, n = 11 vs 1 ± 0.01, n = 12; p < 0.05; Figures 2D,E) at 4C stage. Moreover, blastocyst development rate (25.94 ± 4.42, n = 218 vs 36.66 ± 4.30, n = 217; p < 0.05; Figure 2F) and cell number in each blastocyst were decreased in the YBX1 knockdown group than in the control group (31.86 ± 0.35, n = 31 vs 21.02 ± 0.89, n = 27; p < 0.05; Figure 2G). These results suggest that YBX1 is important for embryonic development.
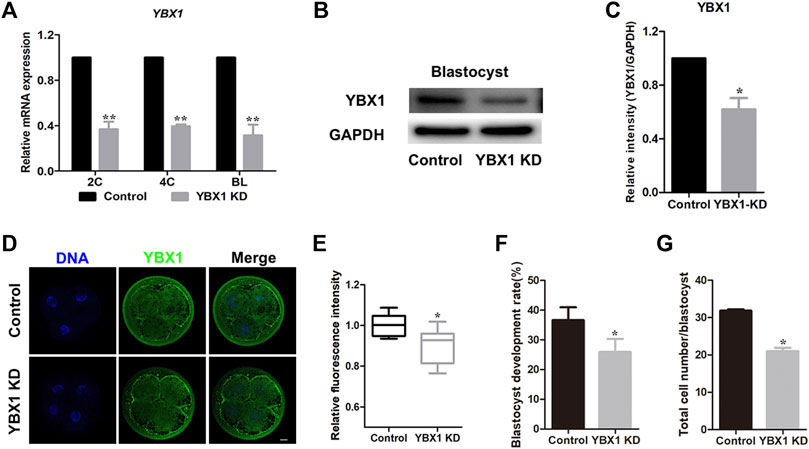
FIGURE 2. YBX1 knockdown impairs embryo development. (A) Real-time quantitative PCR results of YBX1 mRNA expression levels in the control and YBX1 KD groups. Compared with the control group, the expression level of YBX1 mRNA was significantly lower in the YBX1 KD group. (B) YBX1 knockdown was confirmed via Western blotting. (C) Relative YBX1 protein intensity after YBX1 knockdown. (D) Immunofluorescence staining of YBX1 in the control and YBX1 KD groups at the four-cell stage. Blue, DNA; green, YBX1. Scale bar, 20 µm. (E) Relative fluorescence intensity of YBX1 at the four-cell stage. The relative fluorescence intensity of YBX1 was significantly lower in the YBX1 KD group at the 4C stage compared to the control group. (F) Blastocyst development rate after YBX1 knockdown. (G) Total cell number/blastocyst after YBX1 knockdown. *p < 0.05, **p < 0.01 indicate significant differences between groups.
It has been reported that YBX1 knockdown reduces the mRNA stability of two important mitophagy-associated proteins, PINK1 and PRKN (Wu et al., 2022). Therefore, we measured the expression levels of PINK1 and PRKN following YBX1 knockdown in this study. First, we examined PINK1 expression levels in 4C and blastocyst stage embryos via fluorescent staining (Figure 3A). We found that the expression levels of PINK1 were decreased in 4C (1 ± 0.03, n = 29 vs 0.85 ± 0.02, n = 28; p < 0.001; Figure 3B) and blastocyst (1 ± 0.06, n = 13 vs 0.76 ± 0.07, n = 12; p < 0.01; Figure 3C) stage embryos. Moreover, mRNA levels of PINK1 (1 vs 0.39 ± 0.04; p < 0.01; Figure 3D) and PRKN (1 vs 0.51 ± 0.07; p < 0.05; Figure 3E) were decreased after YBX1 knockdown. These results suggest that YBX1 affects the PINK1 and PRKN expression levels. Next, we investigated the influence of YBX1 knockdown on mitochondrial biogenesis. SIRT1 is a marker of mitochondrial biogenesis. Western blotting revealed significantly decreased SIRT1 expression after YBX1 knockdown (1 vs 0.63 ± 0.1; p < 0.05; Figure 3F). Moreover, MitoTracker Red CMXRos was used to detect mitochondrial activity (Figure 3G). Mitochondrial activity was significantly decreased in YBX1 KD group (0.95 ± 0.05, n = 32 vs 0.68 ± 0.11, n = 30; p < 0.001; Figure 3H), these results indicate that YBX1 knockdown decreases mitochondrial function.
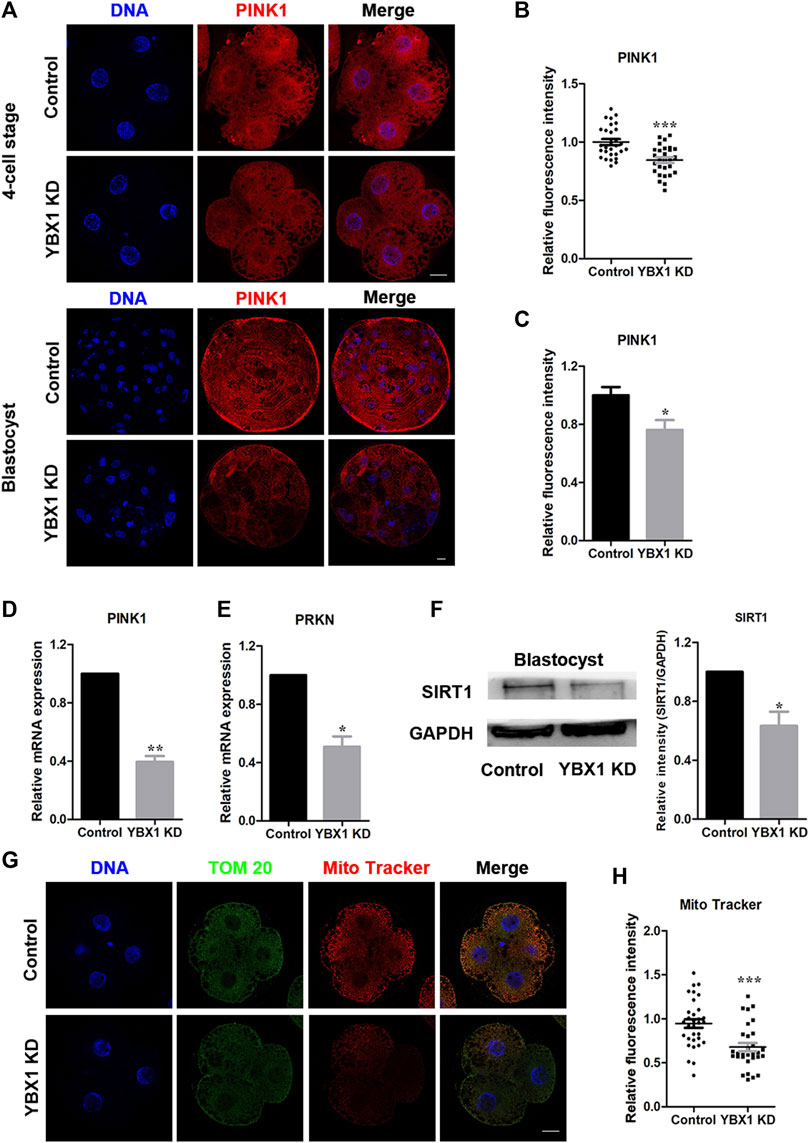
FIGURE 3. YBX1 knockdown impairs mitochondrial function. (A) Immunofluorescence staining of PINK1 in the control and YBX1 KD groups at the four-cell and blastocyst stages. Blue, DNA; red, PINK1. Scale bar, 20 µm. (B) Relative fluorescence intensity of PINK1 at the four-cell stage. The relative fluorescence intensity of PINK1 was significantly lower in the YBX1 KD group at the 4C stage compared to the control group. (C) Relative fluorescence intensity of PINK1 at the blastocyst stage. The relative fluorescence intensity of YBX1 was significantly lower in the YBX1 KD group at the BL stage compared to the control group. (D) Relative mRNA expression of PINK1 at the blastocyst stage. (E) Relative mRNA expression of PRKN at the blastocyst stage. (F) Protein levels of SIRT1 in the control and YBX1 KD groups. (G) Immunofluorescence staining of TOM20, Mito Tracker in the control and YBX1 KD groups at the four-cell stage. Blue, DNA; Green, TOM20; Red, Mito Tracker; Scale bar, 20 µm. (H) Relative fluorescence intensity of Mito Tracker at four-cell stage. Compared with the control group, the relative fluorescence intensity of Mito Tracker in 4C stage in the YBX1 KD group was significantly lower. *p < 0.05, **p < 0.01, ***p < 0.001 indicate significant differences between groups.
Next, we investigated the association of YBX1 knockdown with ER stress and determined the ER stress-related proteins expression, calnexin and GRP78 (Figure 4 A and C). As shown in Figures 4B,D, expression levels of GRP78 (1 ± 0.03, n = 23 vs 1.18 ± 0.04, n = 22; p < 0.001) and calnexin (1 ± 0.03, n = 21 vs 1.16 ± 0.03, n = 21; p < 0.001) were all significantly increased after YBX1 knockdown. Meanwhile, protein expression levels of BiP/GRP78 (1 vs 1.16 ± 0.02; p < 0.05; Figures 4E,F) and calnexin (1 vs 1.07 ± 0.01; p < 0.05; Figures 4E,F) were significantly increased after YBX1 knockdown. Taken together, these results indicate that YBX1 knockdown induces ER stress during embryonic development.
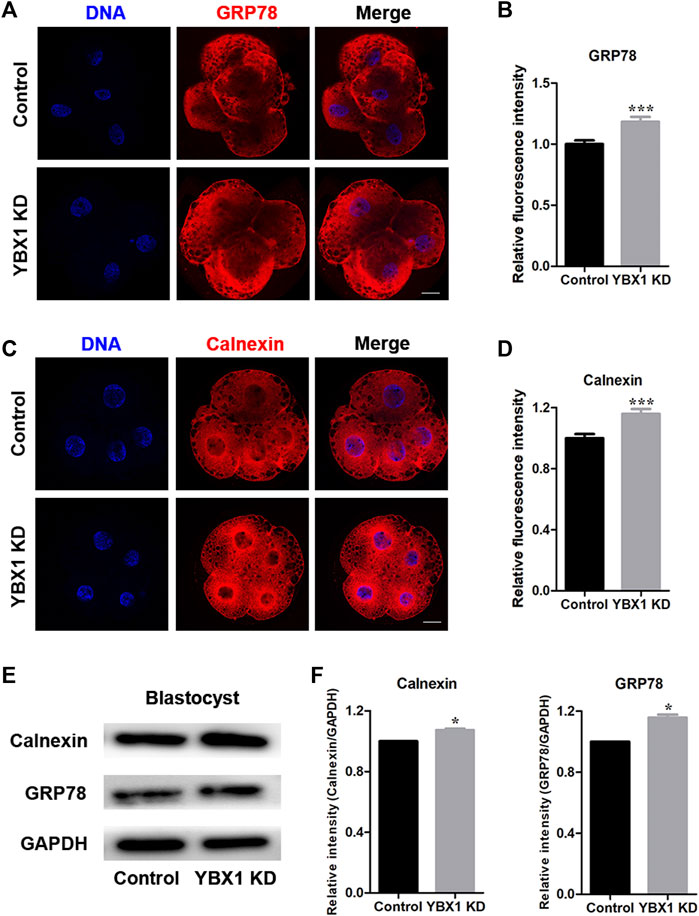
FIGURE 4. YBX1 knockdown induces ER stress. (A) Immunofluorescence staining of GRP78 in the control and YBX1 KD groups at the four-cell stage. Blue, DNA; red, GRP78. Scale bar, 20 µm. (B) Relative fluorescence intensity of GRP78 at the four-cell stage. Compared with the control group, the relative fluorescence intensity of GRP78 in 4C stage in the YBX1 KD group was significantly higher. (C) Immunofluorescence staining of calnexin in the control and YBX1 KD groups at the four-cell stage. Blue, DNA; red, calnexin. Scale bar, 20 µm. (D) Relative fluorescence intensity of calnexin at the four-cell stage. Compared with the control group, the relative fluorescence intensity of calnexin in 4C stage in the YBX1 KD group was significantly higher. (E) Protein levels of calnexin and GRP78 in the control and YBX1 KD groups. (F) Relative protein levels of calnexin and GRP78. *p < 0.05 and ***p < 0.001 indicate significant differences between groups.
Autophagy and apoptosis are important for maintaining organismal and cellular homeostasis (Fan and Zong, 2013). As mitochondrial and ER impairment can induce autophagy and apoptosis, we first evaluated the influence of YBX1 knockdown on autophagy. As shown in Figures 5A–D, LC3 levels were significantly increased in 4C (1 ± 0.02, n = 25 vs 1.29 ± 0.03, n = 19; p < 0.001) and blastocyst (1 ± 0.02, n = 21 vs 1.09 ± 0.04, n = 19; p < 0.05) stage embryos of the YBX1 knockdown group compared to the control group. This result was confirmed by Western blotting (1 vs 1.29 ± 0.03; p < 0.05; Figure 5F), although the mRNA level of LC3 decreased (1 vs 0.70 ± 0.01; p < 0.01; Figure 5E). Therefore, autophagy was observed in YBX1 knocked down porcine embryos. Next, we assessed cell apoptosis via immunofluorescence staining for caspase 3. As shown in Figures 5G,H, staining intensity of caspase 3 was significantly increased in YBX1 knockdown group than that in the control group (1 ± 0.04, n = 18 vs 1.27 ± 0.13, n = 13; p < 0.05). Moreover, the apoptosis related genes Caspase 3 (1 vs 1.62 ± 0.2; p < 0.05; Figure 5I), BAX (1 vs 1.69 ± 0.24; p < 0.05; Figure 5I) and BCL2 (1 vs 1.95 ± 0.17; p < 0.05; Figure 5I) were significantly increased. According to the above results, YBX1 knockdown induces autophagy and apoptosis.
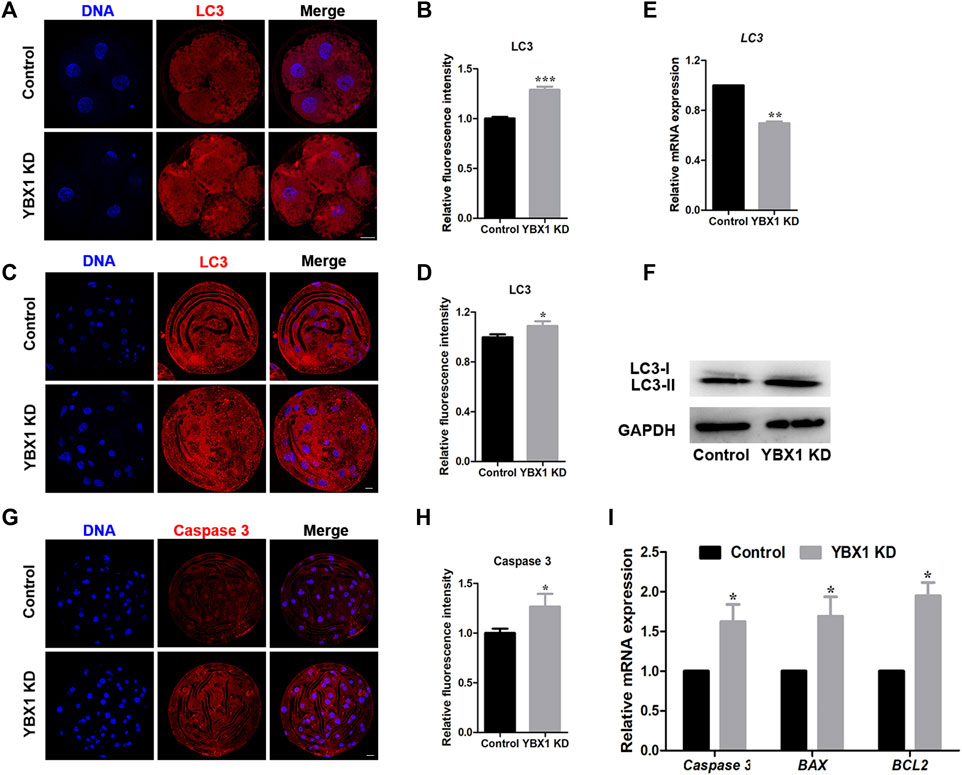
FIGURE 5. YBX1 knockdown induces autophagy and apoptosis. (A) Immunofluorescence staining of LC3 in the control and YBX1 KD groups at the four-cell stage. Blue, DNA; red, LC3. Scale bar, 20 µm. (B) Relative fluorescence intensity of LC3 at the four-cell stage. Compared with the control group, the relative fluorescence intensity of LC3 in 4C stage in the YBX1 KD group was significantly higher. (C) Immunofluorescence staining of LC3 in the control and YBX1 KD groups at the blastocyst stage. Blue, DNA; red, LC3. Scale bar, 20 µm. (D) Relative fluorescence intensity of LC3 at the blastocyst stage. Compared with the control group, the relative fluorescence intensity of LC3 in BL stage in the YBX1 KD group was significantly higher. (E) Relative mRNA expression of LC3 at the four-cell stage. (F) Relative protein levels of LC3 in the control and YBX1 KD groups. (G) Immunofluorescence staining of caspase 3 in the control and YBX1 KD groups at the blastocyst stage. Blue, DNA; red, caspase 3. Scale bar, 20 µm. (H) Relative fluorescence intensity of caspase 3. Compared with the control group, the relative fluorescence intensity of Caspase 3 in blastocyst stage in the YBX1 KD group was significantly higher. (I) Relative mRNA expression of Caspase3, BAX and BCL2 at the four-cell stage. *p < 0.05, **p < 0.01 and ***p < 0.001 indicate significant differences between groups.
In this study, we found that the loss of YBX1 decreased PINK1 and PRKN expression levels, thereby decreasing mitochondrial function and increasing ER stress autophagy as well as apoptosis during embryonic development (Figure 6).
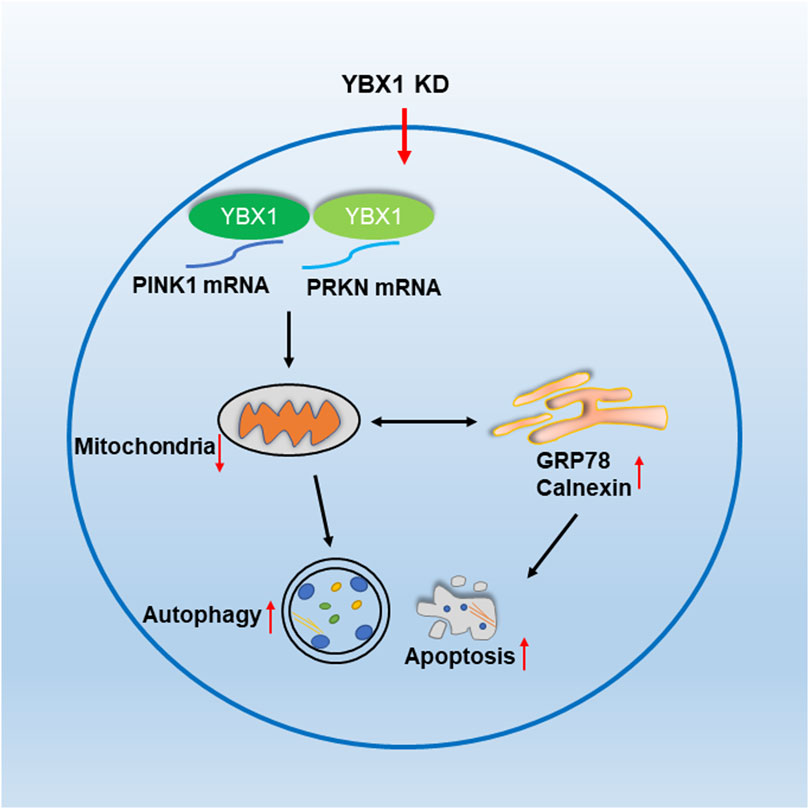
FIGURE 6. YBX1 is important for embryonic development in pigs. YBX1 knockdown decreases PINK1 and PRKN levels and affects mitochondrial function, thereby inducing ER stress, autophagy, and apoptosis.
Mitophagy is the selective degradation of mitochondria via autophagy (Dlamini et al., 2021). It usually occurs in mitochondria that are defective after injury or stress. Mitophagy promotes the renewal of mitochondria and prevents the accumulation of dysfunctional mitochondria, which may lead to cellular degeneration (Wang et al., 2022). Mitophagy is activated in mammalian cells by PINK1/Parkin-mediated mitophagy (Iorio et al., 2021). PINK1 affects the health of mitochondrial (Nguyen et al., 2016). PINK1 activates Parkin via phosphorylation, which is an E3 ubiquitin ligase located in the cytoplasm (Bingol and Sheng, 2016; Wang et al., 2022). YBX1, a positive regulator of mitophagy, by enhancing PINK1/PRKN-mediating mitophagy in brown adipocyte (Wu et al., 2022). In this study, YBX1 knockdown decreased PINK1 and PRKN mRNA levels and reduced PINK1 protein levels, indicating that YBX1 may regulate their mRNA stability and protein expression to influence mitophagy. SIRT1 is a marker of mitochondrial biogenesis (Majeed et al., 2021). Previous studies have shown that SIRT1 has emerged as an important regulator of mitochondrial function (Jian et al., 2012; Li et al., 2017; Niu et al., 2020). MitoTracker Red CMXRos was used to detect mitochondrial activity. It has been shown that decreased mitochondrial activity decreases mitochondrial function (Niu et al., 2019; Niu et al., 2020). Additionally, in aged oocytes, decreased MitoTracker Red were accompanied by decreased PINK1 and PRKN (Niu et al., 2020). Here, knockdown of YBX1 significantly reduced SIRT1 expression and mitochondrial activity, indicating that YBX1 is important for mitochondrial biogenesis. Therefore, YBX1 is crucial for mitophagy and mitochondrial biogenesis.
ER serves as a crucial organelle involved in the biosynthesis of lipids, proteins, and secreted proteins as well as an important site of calcium homeostasis (Lin et al., 2019). ER stress is triggered by the accumulation of unfolded or misfolded proteins in the ER that induce UPR (Feldman et al., 2005). UPR is mainly regulated by three sensors, including BiP (also known as GRP78) (Ghemrawi et al., 2018; Song et al., 2018). ER–mitochondrial play important roles in regulating the mitochondrial dynamics, inflammation, autophagy, and apoptosis. Therefore, we examined ER function in porcine embryos. The results indicated that YBX1 knockdown increased the expression levels of calnexin and GRP78, inducing ER stress. Subsequently, increased ER stress induced UPR. Previous article indicated that YBX1 depletion induces UPR (Sun et al., 2018), which is consistent with our finding.
Autophagy is a crucial cellular response to stress that degrades and removes unfolded proteins and damaged organelles to protect the cells (Yu et al., 2004; Dikic and Elazar, 2018). As mitophagy and ER stress induce autophagy and apoptosis, we determined the expression levels of their markers, LC3 and caspase 3 during embryonic development. We found that YBX1 knockdown increased LC3 protein level. In cancer cells and preadipocytes, YBX1 knockdown reduces LC3 protein expression (Wu et al., 2022; Gong and Zhang, 2023; Wu et al., 2023). In this study, LC3 mRNA levels were decreased but protein expression was increased, contrary to the results, which may be due to the uniqueness of different cells or tissues. Additionally, YBX1 has been identified as an RNA-binding protein and a DNA-binding protein, mainly involved in translational repression, RNA stabilization, and transcriptional regulation. One study showed that overall translation levels were increased in YBX1-depleted embryos (Sun et al., 2018), which may lead to increased LC3 protein levels. GRP78 is important for both ER stress and autophagy (Li et al., 2008). PINK1 knockdown increases autophagy and apoptosis (Niu et al., 2019). Here, YBX1 knockdown increased GRP78 level and decreased PINK1 level, thereby increasing autophagy. In addition, YBX1 knockdown has been reported to upregulate the levels of apoptosis-related genes, such as FAS, tumor necrosis factor, and caspase (Kloetgen et al., 2020). One study reported that oxygen-glucose deprivation/reoxygenation downregulates YBX1 expression, whereas YBX1 overexpression attenuates growth inhibition and apoptosis in PC12 cells (Tuerxun et al., 2021). However, the mRNA levels of BCL2 were elevated after YBX1 knockdown, contrary to the study by Feng et al. (Feng et al., 2021). The elevation of BCL2 may be due to cellular self-protection, which inhibits apoptosis and autophagy. BCL-x1 is a member of the Bcl-2 family of proteins, which are anti-apoptotic proteins. A previous study showed that apoptosis was increased but the mRNA levels of BCL-x1, BAX, and caspase-3 were decreased (Zhang et al., 2020). In addition, it was found that Bcl-2 and Bcl-xL enhance autophagy under certain conditions, such as in response to treatment with etoposide and staurosporine (Fan and Zong, 2013). Moreover, YBX1 is a DNA/RNA-binding protein that affects transcription and translation. The levels of transcription and translation are not exactly the same; therefore, an increase in BCL2 mRNA level does not mean that the protein is also elevated. Therefore, it is possible that BCL2 is increased in YBX1 knockdown embryos. These findings indicate that YBX1 knockdown induces autophagy and apoptosis.
Taken together, our results indicate that YBX1 decreases PINK1 and PRKN expression levels and mitochondrial function and induces ER stress, thereby causing autophagy and apoptosis during embryonic development.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
W-JJ and X-SC designed the research; W-JJ conducted the experiments, analyzed the results, and wrote the manuscript; S-HL, and GH, HC, EC, SS, SH contributed to the materials; and X-SC revised the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by a National Research Foundation (NRF) of Korea grant funded by the Korean government (MSIT) (No. 2022R1A2C300769), Republic of Korea.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Bingol, B., and Sheng, M. (2016). Mechanisms of mitophagy: PINK1, parkin, USP30 and beyond. Free Radic. Biol. Med. 100, 210–222. doi:10.1016/j.freeradbiomed.2016.04.015
Deng, M., Chen, B., Liu, Z., Wan, Y., Li, D., Yang, Y., et al. (2022). YBX1 mediates alternative splicing and maternal mRNA decay during pre-implantation development. Cell Biosci. 12 (1), 12. doi:10.1186/s13578-022-00743-4
Dikic, I., and Elazar, Z. (2018). Mechanism and medical implications of mammalian autophagy. Nat. Rev. Mol. Cell Biol. 19 (6), 349–364. doi:10.1038/s41580-018-0003-4
Dlamini, M. B., Gao, Z., HasenbiligeJiang, L., Geng, C., Li, Q., Shi, X., et al. (2021). The crosstalk between mitochondrial dysfunction and endoplasmic reticulum stress promoted ATF4-mediated mitophagy induced by hexavalent chromium. Environ. Toxicol. 36 (6), 1162–1172. doi:10.1002/tox.23115
Fan, Y. J., and Zong, W. X. (2013). The cellular decision between apoptosis and autophagy. Chin. J. Cancer 32 (3), 121–129. doi:10.5732/cjc.012.10106
Feldman, D. E., Chauhan, V., and Koong, A. C. (2005). The unfolded protein response: a novel component of the hypoxic stress response in tumors. Mol. Cancer Res. 3 (11), 597–605. doi:10.1158/1541-7786.Mcr-05-0221
Feng, M., Xie, X., Han, G., Zhang, T., Li, Y., Li, Y., et al. (2021). YBX1 is required for maintaining myeloid leukemia cell survival by regulating BCL2 stability in an m6A-dependent manner. Blood 138 (1), 71–85. doi:10.1182/blood.2020009676
Ghemrawi, R., Battaglia-Hsu, S. F., and Arnold, C. (2018). Endoplasmic reticulum stress in metabolic disorders. Cells 7 (6). doi:10.3390/cells7060063
Gong, W., and Zhang, S. (2023). YB1 participated in regulating mitochondrial activity through RNA replacement. Front. Oncol. 13, 1145379. doi:10.3389/fonc.2023.1145379
Iorio, R., Celenza, G., and Petricca, S. (2021). Mitophagy: molecular mechanisms, new concepts on parkin activation and the emerging role of AMPK/ULK1 Axis. Cells 11 (1). doi:10.3390/cells11010030
Jian, B., Yang, S., Chaudry, I. H., and Raju, R. (2012). Resveratrol improves cardiac contractility following trauma-hemorrhage by modulating Sirt1. Mol. Med. 18 (1), 209–214. doi:10.2119/molmed.2011.00365
Jiang, W. J., Sun, M. H., Li, X. H., Lee, S. H., Heo, G., Zhou, D., et al. (2023a). E2F4 regulates cell cycle to mediate embryonic development in pigs. Theriogenology 196, 227–235. doi:10.1016/j.theriogenology.2022.10.040
Jiang, W. J., Sun, M. H., Li, X. H., Lee, S. H., Heo, G., Zhou, D., et al. (2023b). Y-box binding protein 1 influences zygotic genome activation by regulating N6-methyladenosine in porcine embryos. J. Cell Physiol. 2023. doi:10.1002/jcp.31040
Kloetgen, A., Duggimpudi, S., Schuschel, K., Hezaveh, K., Picard, D., Schaal, H., et al. (2020). YBX1 indirectly targets heterochromatin-repressed inflammatory response-related apoptosis genes through regulating CBX5 mRNA. Int. J. Mol. Sci. 21 (12). doi:10.3390/ijms21124453
Kuo, T. J., Jean, Y. H., Shih, P. C., Cheng, S. Y., Kuo, H. M., Lee, Y. T., et al. (2022). Stellettin B-induced oral cancer cell death via endoplasmic reticulum stress-mitochondrial apoptotic and autophagic signaling pathway. Int. J. Mol. Sci. 23 (15). doi:10.3390/ijms23158813
Li, J., Ni, M., Lee, B., Barron, E., Hinton, D. R., and Lee, A. S. (2008). The unfolded protein response regulator GRP78/BiP is required for endoplasmic reticulum integrity and stress-induced autophagy in mammalian cells. Cell Death Differ. 15 (9), 1460–1471. doi:10.1038/cdd.2008.81
Li, P., Liu, Y., Burns, N., Zhao, K. S., and Song, R. (2017). SIRT1 is required for mitochondrial biogenesis reprogramming in hypoxic human pulmonary arteriolar smooth muscle cells. Int. J. Mol. Med. 39 (5), 1127–1136. doi:10.3892/ijmm.2017.2932
Lin, T., Lee, J. E., Kang, J. W., Shin, H. Y., Lee, J. B., and Jin, D. I. (2019). Endoplasmic reticulum (ER) stress and unfolded protein response (UPR) in mammalian oocyte maturation and preimplantation embryo development. Int. J. Mol. Sci. 20 (2). doi:10.3390/ijms20020409
Lin, Y. D., Chen, S., Yue, P., Zou, W., Benbrook, D. M., Liu, S., et al. (2008). CAAT/enhancer binding protein homologous protein-dependent death receptor 5 induction is a major component of SHetA2-induced apoptosis in lung cancer cells. Cancer Res. 68 (13), 5335–5344. doi:10.1158/0008-5472.Can-07-6209
Ma, K., Chen, G., Li, W., Kepp, O., Zhu, Y., and Chen, Q. (2020). Mitophagy, mitochondrial homeostasis, and cell fate. Front. Cell Dev. Biol. 8, 467. doi:10.3389/fcell.2020.00467
Majeed, Y., Halabi, N., Madani, A. Y., Engelke, R., Bhagwat, A. M., Abdesselem, H., et al. (2021). SIRT1 promotes lipid metabolism and mitochondrial biogenesis in adipocytes and coordinates adipogenesis by targeting key enzymatic pathways. Sci. Rep. 11 (1), 8177. doi:10.1038/s41598-021-87759-x
Matsumoto, S., Uchiumi, T., Tanamachi, H., Saito, T., Yagi, M., Takazaki, S., et al. (2012). Ribonucleoprotein Y-box-binding protein-1 regulates mitochondrial oxidative phosphorylation (OXPHOS) protein expression after serum stimulation through binding to OXPHOS mRNA. Biochem. J. 443 (2), 573–584. doi:10.1042/bj20111728
Mordovkina, D., Lyabin, D. N., Smolin, E. A., Sogorina, E. M., Ovchinnikov, L. P., and Eliseeva, I. (2020). Y-box binding proteins in mRNP assembly, translation, and stability control. Biomolecules 10 (4). doi:10.3390/biom10040591
Nguyen, T. N., Padman, B. S., and Lazarou, M. (2016). Deciphering the molecular signals of PINK1/parkin mitophagy. Trends Cell Biol. 26 (10), 733–744. doi:10.1016/j.tcb.2016.05.008
Niu, Y. J., Nie, Z. W., Shin, K. T., Zhou, W., and Cui, X. S. (2019). PINK1 regulates mitochondrial morphology via promoting mitochondrial fission in porcine preimplantation embryos. Faseb J. 33 (7), 7882–7895. doi:10.1096/fj.201802473R
Niu, Y. J., Zhou, W., Nie, Z. W., Zhou, D., Xu, Y. N., Ock, S. A., et al. (2020). Ubiquinol-10 delays postovulatory oocyte aging by improving mitochondrial renewal in pigs. Aging (Albany NY) 12 (2), 1256–1271. doi:10.18632/aging.102681
Ron, D., and Hubbard, S. R. (2008). How IRE1 reacts to ER stress. Cell 132 (1), 24–26. doi:10.1016/j.cell.2007.12.017
Senft, D., and Ronai, Z. A. (2015). UPR, autophagy, and mitochondria crosstalk underlies the ER stress response. Trends Biochem. Sci. 40 (3), 141–148. doi:10.1016/j.tibs.2015.01.002
Song, S., Tan, J., Miao, Y., and Zhang, Q. (2018). Crosstalk of ER stress-mediated autophagy and ER-phagy: involvement of UPR and the core autophagy machinery. J. Cell Physiol. 233 (5), 3867–3874. doi:10.1002/jcp.26137
Sui, X., Hu, Y., Ren, C., Cao, Q., Zhou, S., Cao, Y., et al. (2020). METTL3-mediated m(6)A is required for murine oocyte maturation and maternal-to-zygotic transition. Cell Cycle 19 (4), 391–404. doi:10.1080/15384101.2019.1711324
Sun, J., Yan, L., Shen, W., and Meng, A. (2018). Maternal Ybx1 safeguards zebrafish oocyte maturation and maternal-to-zygotic transition by repressing global translation. Development 145 (19). doi:10.1242/dev.166587
Sun, M. H., Jiang, W. J., Li, X. H., Lee, S. H., Heo, G., Zhou, D., et al. (2023). ATF7-dependent epigenetic changes induced by high temperature during early porcine embryonic development. Cell Prolif. 56 (2), e13352. doi:10.1111/cpr.13352
Tuerxun, T., Li, X., Lou, F., Wang, X., and Ma, L. (2021). YBX1 protects against apoptosis induced by oxygen-glucose deprivation/reoxygenation in PC12 cells via activation of the AKT/GSK3β pathway. Folia Biol. (Praha) 67 (4), 150–157.
Uchiumi, T., Fotovati, A., Sasaguri, T., Shibahara, K., Shimada, T., Fukuda, T., et al. (2006). YB-1 is important for an early stage embryonic development: neural tube formation and cell proliferation. J. Biol. Chem. 281 (52), 40440–40449. doi:10.1074/jbc.M605948200
Valente, E. M., Abou-Sleiman, P. M., Caputo, V., Muqit, M. M., Harvey, K., Gispert, S., et al. (2004a). Hereditary early-onset Parkinson's disease caused by mutations in PINK1. Science 304 (5674), 1158–1160. doi:10.1126/science.1096284
Valente, E. M., Salvi, S., Ialongo, T., Marongiu, R., Elia, A. E., Caputo, V., et al. (2004b). PINK1 mutations are associated with sporadic early-onset parkinsonism. Ann. Neurol. 56 (3), 336–341. doi:10.1002/ana.20256
Wang, Q., Xue, H., Yue, Y., Hao, S., Huang, S. H., and Zhang, Z. (2022). Role of mitophagy in the neurodegenerative diseases and its pharmacological advances: a review. Front. Mol. Neurosci. 15, 1014251. doi:10.3389/fnmol.2022.1014251
Wu, R., Cao, S., Li, F., Feng, S., Shu, G., Wang, L., et al. (2022). RNA-binding protein YBX1 promotes brown adipogenesis and thermogenesis via PINK1/PRKN-mediated mitophagy. Faseb J. 36 (3), e22219. doi:10.1096/fj.202101810RR
Wu, R., Feng, S., Li, F., Shu, G., Wang, L., Gao, P., et al. (2023). Transcriptional and post-transcriptional control of autophagy and adipogenesis by YBX1. Cell Death Dis. 14 (1), 29. doi:10.1038/s41419-023-05564-y
Yu, L., Alva, A., Su, H., Dutt, P., Freundt, E., Welsh, S., et al. (2004). Regulation of an ATG7-beclin 1 program of autophagic cell death by caspase-8. Science 304 (5676), 1500–1502. doi:10.1126/science.1096645
Zhang, H., Pan, Z., Ju, J., Xing, C., Li, X., Shan, M., et al. (2020). DRP1 deficiency induces mitochondrial dysfunction and oxidative stress-mediated apoptosis during porcine oocyte maturation. J. Anim. Sci. Biotechnol. 11, 77. doi:10.1186/s40104-020-00489-4
Keywords: YBX1, mitochondria, er stress, autophagy, pig
Citation: Jiang W-J, Lee S-H, Heo G, Chung HJ, Cho ES, Sa SJ, Hochi S and Cui X-S (2023) Knockdown of Y-box binding protein 1 induces autophagy in early porcine embryos. Front. Cell Dev. Biol. 11:1238546. doi: 10.3389/fcell.2023.1238546
Received: 13 June 2023; Accepted: 16 October 2023;
Published: 30 October 2023.
Edited by:
Anne-Sophie Armand, Université Paris Cité, FranceCopyright © 2023 Jiang, Lee, Heo, Chung, Cho, Sa, Hochi and Cui. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiang-Shun Cui, eHNjdWlAY2JudS5hYy5rcg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.