
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell Dev. Biol. , 24 August 2023
Sec. Cellular Biochemistry
Volume 11 - 2023 | https://doi.org/10.3389/fcell.2023.1225628
Obesity has become a global pandemic. WDTC1 is a WD40-containing protein that functions as an anti-obesity factor. WDTC1 inhibits adipogenesis by working as an adaptor of the CUL4-DDB1 E3 ligase complex. It remains unclear about how WDTC1 is regulated. Here, we show that the TRiC/CCT functions as a chaperone to facilitate the protein folding of WDTC1 and proper function in adipogenesis. Through tandem purification, we identified the molecular chaperone TRiC/CCT as WDTC1-interacting proteins. WDTC1 bound the TRiC/CCT through its ADP domain, and the TRiC/CCT recognized WDTC1 through the CCT5 subunit. Disruption of the TRiC/CCT by knocking down CCT1 or CCT5 led to misfolding and lysosomal degradation of WDTC1. Furthermore, the knockdown of CCT1 or CCT5 eliminated the inhibitory effect of WDTC1 on adipogenesis. Our studies uncovered a critical role of the TRiC/CCT in the folding of WDTC1 and expanded our knowledge on the regulation of adipogenesis.
WD40 and tetratricopeptide repeats 1 (WDTC1) is an anti-obesity factor conserved from worms to mammals (Suh et al., 2007). Mutation in Adipose, the Drosophila homolog of WDTC1, caused obesity in Drosophila (Doane, 1960a; Doane, 1960b; Hader et al., 2003). Wdtc1 heterozygous mice are obese with enlarged adipocytes, whereas fat-specific Wdtc1 transgenic mice are lean with dramatically decreased fat mass (Suh et al., 2007). In humans, the expression level of WDTC1 is negatively correlated with obesity (Lai et al., 2009; Galgani et al., 2013). WDTC1 is identified as an adaptor of the CRL4 E3 ligase complex (Groh et al., 2016). We have previously shown that the CRL4-WDTC1 complex targets MED20, a subunit of the mediator complex, for degradation to inhibit adipogenesis (Tang et al., 2021). MED20 directly binds C/EBPβ and is required for the transcription of C/EBPα and PPARγ, two central players in adipogenesis (Tang et al., 2021). However, it remains unclear about how WDTC1 is regulated.
The tailless complex protein-1 (TCP-1) ring complex (TRiC), also known as complex containing TCP-1 (CCT), is the primary eukaryotic cytosolic chaperonin that is essential for cytosolic protein folding and assembly (Lopez et al., 2015; Skjaerven et al., 2015). The TRiC/CCT consists of two stacked rings, which was composed of eight homologous but distinct subunits (CCT1-8) (Cong et al., 2010). The TRiC/CCT facilitates the folding of approximately 10% of the newly synthesized cytosolic polypeptides (Yam et al., 2008), including WD40 domain-containing proteins (Miyata et al., 2014; Lopez et al., 2015). Dysfunction of the TRiC/CCT has been associated with a number of diseases (Lopez et al., 2015), and its function is absolutely essential for viability (Yam et al., 2008). It remains unclear whether the TRiC/CCT regulates adipogenesis.
Here, in an effort to purify WDTC1-interacting proteins, we identified the TRiC/CCT as a major binding partner of WDTC1. The TRiC/CCT binds WDTC1 primarily through the CCT5 subunit and facilitates the folding of WDTC1. We demonstrate that the TRiC/CCT is required for the inhibitory effect of WDTC1 on adipogenesis. Our findings uncover a role of the TRiC/CCT in regulating adipogenesis.
We obtained Strep-Tactin Superflow (2-1206-002) and desthiobiotin (2-1000-001) from IBA life sciences; dexamethasone, isobutylmethylxanthine (IBMX), biotin, pioglitazone, bovine insulin, urea, SDS, DTT, DMSO, and Triton X-100 from Sigma-Aldrich; Dulbecco’s modified Eagle’s medium (DMEM) with low (1 g/L) or high (4.5 g/L) glucose, fetal and neonatal bovine serum, blasticidin, and puromycin from Thermo Fisher Scientific; donkey anti-rabbit IgG or anti-mouse IgG conjugated to horseradish peroxidase from Jackson Immuno Research; protease inhibitor cocktail from Roche Applied Science; and all other chemicals from local suppliers unless otherwise specified.
Full-length cDNAs of human CCT1-8 subunits of the CCT were generous gifts from Dr. Jiahuai Han at Xiamen University, China. The coding regions of these genes were cloned into pcDNA3.3 (Thermo Fisher Scientific) with an N-terminal Flag tag. Mouse WDTC1 were cloned from a cDNA library prepared from testis mRNA of a C57BL6 mouse. A pIRESpuro-GLUE vector (Addgene, #15100) (Angers et al., 2006) was used to generate a stable cell line for the purification of WDTC1-interacting proteins. For lentiviral overexpression, genes were cloned into either pCDH-EF1-MCS-IRES-Puro (System Biosciences) or pCDH-EF1-MCS-IRES-BSD (modified from the former one by replacing the puromycin resistance gene to blasticidin resistant gene) with or without the N-terminal Flag tag. For knockdown of genes, a pLKO.1 vector (Addgene, 10878) (Moffat et al., 2006) was used. The primer sequences are listed in Supplementary Table S1.
HEK293T cells were cultured as previously described (Wang et al., 2019; Hao et al., 2020). In brief, cells were cultured in Medium A (DMEM high glucose, 10% (v/v) FCS, 100 U/ml penicillin, and 100 mg/ml streptomycin) at 37°C in an atmosphere of 5% CO2. To generate stable cell lines, HEK293T cells were set up at 5×104 cells per 10-cm dish on day 0. On day 1, cells were transfected with either 0.5 μg pIRESpuro-GLUE (pGlue) or WDTC1/pGlue using polyetherimide (PEI, Polysciences, 239660) at 2.4:1 (wt/wt) of PEI to plasmids. Starting from day 2, cells were selected with Medium A containing 1 μg/ml puromycin until colonies were formed, which were then screened by Western blot using the anti-HA antibody (1:2000, Roche, 11867423001). A positive clone of either construct was subcloned and used for the current studies.
3T3-L1 preadipocytes were obtained from the American Type Culture Collection and cultured in Medium B (DMEM low glucose, 10% (v/v) NCS, 100 U/ml penicillin, and 100 mg/ml streptomycin) at 37°C in an atmosphere of 8.8% CO2. Cells were maintained at no more than 50% confluence.
To differentiate into mature adipocytes, 3T3-L1 cells were subjected to a standard cocktail hormone as previously described (Wang et al., 2021). In brief, cells were cultured to 100% confluence and maintained in Medium B for another 2 days. On day 0 of differentiation, cells were treated with Medium C (DMEM low glucose, 10% (v/v) FCS, 100 U/ml penicillin, and 100 mg/ml streptomycin) containing 5 μg/ml insulin, 0.5 mM IBMX, 1 μM dexamethasone, and 1 μM pioglitazone. On days 2 and 4, the medium was changed to Medium C containing 5 μg/ml insulin. On day 6, cells were fed with Medium C. On day 8, complete differentiation was achieved.
For the cell growth curve, control and CCT1/5 knockdown 3T3-L1 cells were set up at 1 × 104 cells/35-mm dish. Cells were counted every day for 4 days.
For Oil Red O staining, fully differentiated 3T3-L1 cells were washed with PBS and fixed in 2.5% glutaraldehyde at room temperature for 1 h. Cells were then treated with 60% isopropanol for 5 min and stained with freshly prepared Oil Red O (1.8 mg/ml) for 5 min. For quantification of triglycerides in 3T3-L1 adipocytes, cells were harvested in PBS containing 0.5% SDS, gradually heated up to 95°C, and kept at 95°C for another 5 min. The cooled samples were subjected to triglyceride measurement using a commercial kit from Wako Chemicals. The content of triglycerides was normalized to protein content, which was quantified by using a Pierce BCA kit (Thermo Fisher Scientific).
To produce lentivirus in HEK293T cells, on day 0, cells were set up at 2.5×105 cells per 60-mm dish. On day 2, cells were transfected with 1 μg lentiviral vector and two packaging plasmids, 0.75 μg psPAX2 (Addgene, 12260) and 0.25 μg pMD2.G (Addgene, 12259), using polyetherimide (PEI, Polysciences, 239660). On day 3, cells were refed with Medium A. On days 4 and 5, lentiviral particles were collected from the media after centrifugation at 1,000 g for 5 min, concentrated at 70,000 g for 2 h or directly aliquoted, and stored at −80°C until further use.
For lentiviral infections, 3T3-L1 preadipocytes were set up, cultured to 50%–70% confluence, and infected with lentivirus in the medium containing 8–10 μg/ml polybrene. Cells, 24 h after infection, were selected against 5 μg/ml puromycin and/or 5 μg/ml blasticidin for at least 48 h and then used for the described experiments.
To purify WDTC1-interacting proteins, nuclear extracts were prepared from 10 of 10-cm dishes of stable cell lines of pGLUE or WDTC1/pGLUE as previously described (Nilsen, 2013). The nuclear extracts were incubated with 0.1 ml Strep-Tactin beads at 4°C for 1 h, washed with 10 ml Buffer A (10 mM HEPES, pH 8.0, and 150 mM NaCl), and eluted six times in 0.1 ml Buffer B (10 mM HEPES, pH 8.0, 150 mM NaCl, 10 mM β-mercaptoethanol, 1 mM MgOAc, 1 mM imidazole, 2 mM CaCl2, and 2.5 mM desthiobiotin). The eluted fractions were combined and incubated with 0.1 ml calmodulin beads at 4°C for 3 h, washed with 5 ml Buffer C (50 mM ammonium bicarbonate, pH 8.3, 75 mM NaCl, 1 mM MgOAc, 1 mM imidazole, and 2 mM CaCl2), and eluted in 0.1 ml Buffer B. The extracts were subjected to silver staining and protein ID analysis by AB5600+ mass spectrometry.
To express recombinant WDTC1, the coding region of WDTC1 was cloned into a pET28a vector and transformed into BL21(DE3)pLys bacterial cells. On day 0, a colony was inoculated in the LB medium and cultured at 37°C overnight. On day 1, the bacteria were scaled up at 1:100, cultured to an OD600 around 0.8, and induced with 1 mM IPTG for 3 h. Cells were then harvested and lysed in PBS containing 1 mg/ml lysozyme supplemented with protease inhibitors. As WDTC1 was expressed in the inclusion bodies, the pellet from the lysis was collected and thoroughly washed with PBS containing 1% Triton X-100 five times. The final product shows about 95% purity on SDS-PAGE.
To refold WDTC1 in vitro, purified WDTC1 proteins were denatured in 6 M urea in 10 mM HEPES pH 8.0 at 25°C overnight. On the next day, cytosolic fractions were prepared from cells infected with control shRNA, shCC1, or shCCT5. In brief, cells were harvested by spinning at 1,000 g for 5 min and resuspended in hypotonic buffer (10 mM HEPES, pH 7.9, 1.5 mM MgCl2, and 10 mM KCl) to swell for 10 min on ice. Cells were homogenized in a Dounce homogenizer with 10 strokes using a loose-fitting pestle and spun at 1,000 g for 15 min. The resultant was supplemented with 0.11 volume of 10× cytoplasmic extract buffer (0.3 M HEPES, pH 7.9, 1.4 M KCl, and 0.03 M MgCl2) and spun at 100,000 g at 4°C for 1 h. The supernatant is the cytosolic fraction. The denatured WDTC1 proteins (100 ng) were diluted into cytosolic fractions and incubated at 25°C for 1 h. The samples were then centrifuged at 20,000 g for 10 min, and the supernatant and pellet fractions were subjected to SDS-PAGE and Western blot.
Western blot was carried out as previously described (Zhang et al., 2015). The following antibodies were used: anti-CCT1 (1:1,000, Protein Tech, 10320-1-AP), anti-CCT5 (1:1,000, Protein Tech, 11603-1-AP), anti-WDTC1 (1:1,000, Abgent, AP4944b), anti-CD36 (1:1,000, Sino Biological Inc., 80263-T48), anti-PPARγ (1:1,000, Santa Cruz, sc-7273), anti-C/EBPα (1:1,000, CST, 8178s), anti-perilipin (1;1,000, CST, 9349s), anti-GAPDH (1:5,000, CST, 5174s), and anti-Flag M2 (1:1,000, Sigma, F1804). Membranes were developed in a ChemStudio imaging system (Analytik Jena AG).
All the statistical analyses were performed using Student’s two-tailed paired t-test. The value represents mean ± SEM. Statistical details of all experiments can be found in the figure legends, including the exact number of cell samples. Asterisks (*) indicate levels of statistical significance. *, p < 0.05; **, p < 0.01; and ***, p < 0.001. No data were excluded from any of the experiments.
To search for potential interacting proteins of WDTC1, we generated a stable HEK293T cell line expressing Glue (SBP-HA-CBP)-tagged WDTC1 and performed tandem purification (Figure 1A). When subjected to silver staining, several bands were specifically identified in the Glue-WDTC1-expressing cells (Figure 1B). Mass spectrometry analysis showed that these bands contained DDB1, all eight subunits of the TRiC/CCT, Hsp70, and Hsp90, which was confirmed by two separate experiments (Figure 1C).
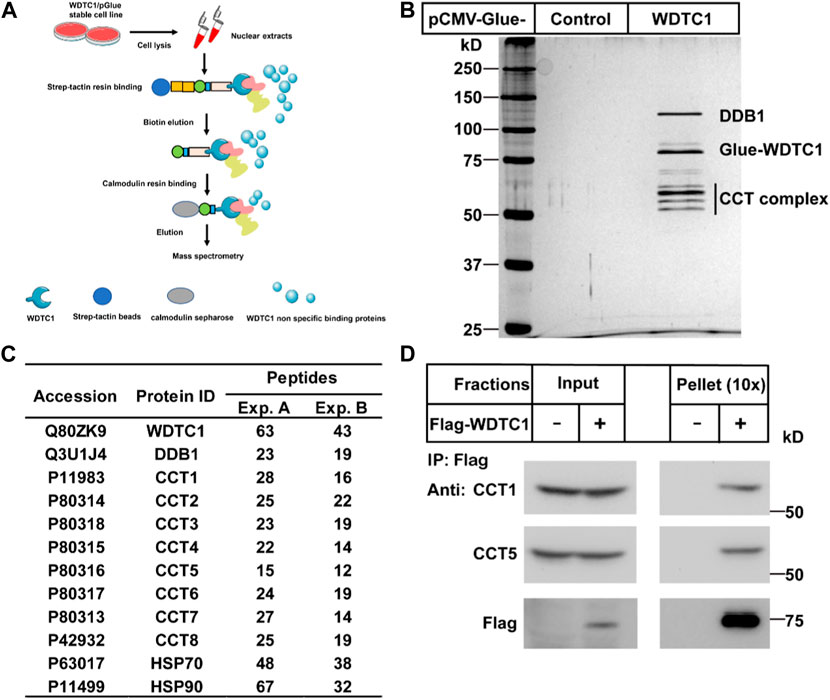
FIGURE 1. Identification of the CCT complex as WDTC1-interacting proteins. (A) Strategy to purify WDTC1-interacting proteins in HEK293T cells using Glue-WDTC1 as a bait. (B,C) On day 0, nuclear fractions were prepared from stable HEK293T cells expressing WDTC1/pGlue or vector alone and subjected to purification with Strep-Tactin beads as described. The eluted fractions were subjected to silver staining (B) and mass spectrometry for protein ID analysis. Potential candidates are listed in (C). (D) On day 0, HEK293T cells were set up at 250,000 cells/60-mm dish. On day 2, cells were transfected with pEF-Flag-WDTC1 or vector alone. On day 3, immunoprecipitation was performed using anti-Flag M2 beads and endogenous CCT1 and CCT5 were detected.
The chaperonin TRiC/CCT has been shown to facilitate the folding of WD40-domain containing proteins (Miyata et al., 2014; Lopez et al., 2015). As WDTC1 is also a WD40-domain containing protein, we hypothesized that the TRiC/CCT might be required for the folding and function of WDTC1. We first overexpressed Flag-tagged WDTC1 in HEK293T cells and performed co-immunoprecipitation (co-IP) assay. As shown in Figure 1D, WDTC1 pulled down endogenous CCT1 and CCT5, indicating that the TRiC/CCT indeed binds WDTC1.
We next examined which part of WDTC1 binds the TRiC/CCT. Previous studies have shown that WDTC1 has six WD40 domains, one ADP domain, and three tetratricopeptide domains (TPR) (Hader et al., 2003). To map the interaction domain of WDTC1 with the CCT, we generated different truncates of WDTC1 (Figure 2A) and transfected them into HEK293T cells for co-IP assay. As shown in Figure 2B, the M1 truncate (aa1-241) of WDTC1 pulled down a small fraction of endogenous CCT1 and CCT5, but M2 truncate (aa 1–343) pulled down comparable amounts of CCT1 and CCT5 with full-length WDTC1, suggesting that the ADP domain is the primary domain that interacts with the TRiC/CCT. Consistently, the M3 and M4 truncates, which lacked the ADP domain, showed no interaction with the TRiC/CCT.
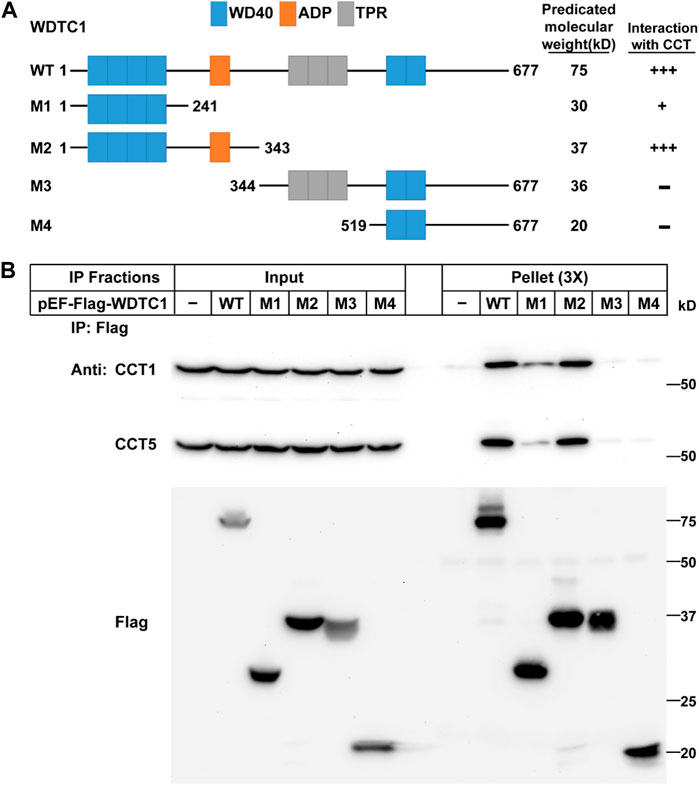
FIGURE 2. WDTC1 interacts with the CCT through its ADP domain. (A) Illustration of the domain structures of WT and different truncates of WDTC1. (B) HEK293T cells were set up, transfected with indicated plasmids, and subjected to immunoprecipitation as in Figure 1D. Input and pellet fractions were immunoblotted with indicated antibodies.
We then determined which subunit of the TRiC/CCT interacts with WDTC1. We co-transfected WDTC1 with each of the Flag-tagged subunit of the TRiC/CCT into HEK293T cells and performed a Co-IP experiment. As shown in Figure 3, the immunoprecipitation of Flag-CCT5 pulled down comparable amounts of WDTC1 than other subunits, indicating that the TRiC/CCT binds WDTC1 primarily through CCT5.
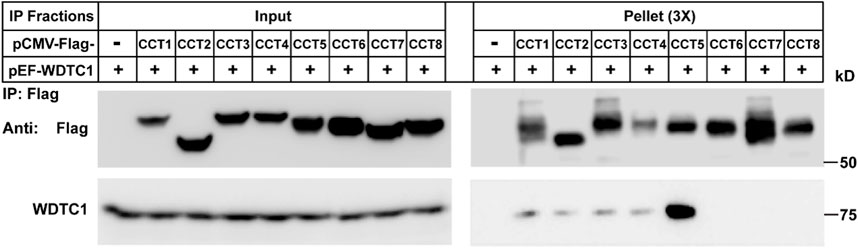
FIGURE 3. TRiC/CCT binds WDTC1 through CCT5. On day 0, HEK293T cells were set up at 250,000 cells/60-mm dish. On day 2, cells were transfected with pEF-Flag-CCT1-8 or vector alone. On day 3, immunoprecipitation was performed using anti-Flag M2 beads and endogenous WDTC1 were detected.
We then went on to check the effect of the TRiC/CCT on WDTC1. We generated a stable cell line in 3T3-L1 cells that overexpress WDTC1 and knocked down CCT1 or CCT5 to check their effect on the protein and RNA levels of WDTC1. Consistent with the work of Trinidad et al. (2013), knockdown of either CCT1 or CCT5 resulted in a decreased protein level of the other (Figures 4A, C), suggesting that all subunits are required for the integrity of the TRiC/CCT. Depletion of either CCT1 or CCT5 dramatically reduced the protein, but not the mRNA, level of WDTC1 (Figures 4A–C), indicating that the TRiC/CCT is required for the stability of WDTC1. Moreover, knockdown of CCT1 or CCT5 showed no difference on cell viability with control (Figure 4D).
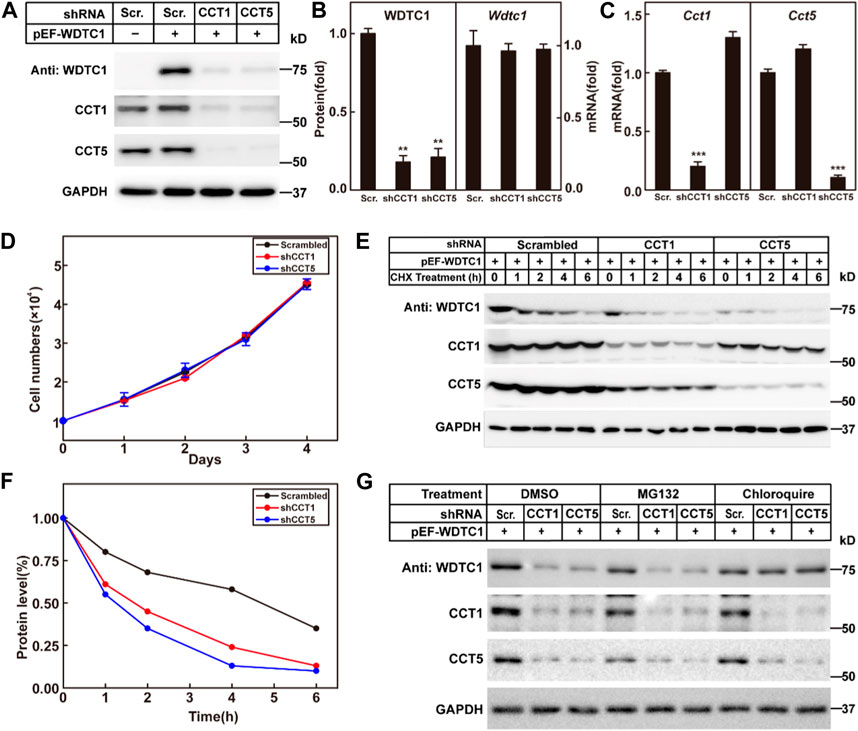
FIGURE 4. TRiC/CCT is required for the stability of WDTC1. (A–C) On day 0, 3T3-L1 preadipocytes infected with lentivirus expressing WDTC1 and the indicated shRNAs were set up at 1 × 105 cells per 35-mm dish. On day 2, cells were harvested to detect the protein (A) and RNA (B, C) levels of WDTC1, CCT1, and CCT5. Each value represents mean ± SEM of three replicates. (D) Control, CCT1, and CCT5 knockdown 3T3-L1 cells were set up at 1 × 104 cells/35-mm dish. Cells were counted every day for 4 days. Cell numbers are plotted in (D). (E,F) On day 0, 3T3-L1 preadipocytes infected with lentivirus expressing the indicated shRNAs and WDTC1 were set up at 1 × 105 cells per 35-mm dish. On day 2, cells were treated with CHX (100 μg/ml) for 0, 1, 2, 4, and 6 h, then cells were harvested, and total cell lysate was immunoblotted with indicated antibodies (E); band intensities of WDTC1 are quantified and plotted in (F). (G) On day 0, 3T3-L1 preadipocytes infected with lentivirus expressing WDTC1 and the indicated shRNAs were set up at 1 × 105 cells per 35-mm dish. On day 2, cells were treated with MG132(10 μg/ml) or chloroquine (50 μM) for 6 h, then cells were harvested, and total cell lysate was immunoblotted with indicated antibodies. For all panels, asterisks (*) denote the level of statistical significance (Student’s t-test). **p < 0.01 and ***p < 0.001.
To further confirm the results, we overexpressed WDTC1 in control and CCT1 or CCT5 knockdown 3T3-L1 cells, treated with cycloheximide, and examined the half-life of WDTC1. As shown in Figures 4E, F, the half-life of WDTC1 in the control cells was about 4 h, but it was shortened to about 2 h in CCT1 or CCT5 knockdown cells.
To examine how WDTC1 was degraded in cells lack of the TRiC/CCT, we treated CCT1 or CCT5 knockdown 3T3-L1 cells with MG132 or chloroquine and found that chloroquine, but not MG132, largely restored the level of WDTC1, suggesting a lysosomal degradation of WDTC1 in TRiC/CCT-depleted cells (Figure 4G).
To directly test whether the TRiC/CCT was required for the folding of WDTC1, we carried out an in vitro refolding assay. We expressed WDTC1 in E. coli and purified it from the inclusion bodies (Figure 5A). The purified WDTC1 was then denatured in urea and diluted to allow refolding in the cytosol prepared from control (scrambled shRNA), CCT1, or CCT5 knockdown 3T3-L1 cells. As shown in Figure 5B, WDTC1 was undetectable in the supernatant when refolded in buffer alone, whereas a significant amount of WDTC1 was detected in the cytosol prepared from the control cells. However, in the cytosol of either CCT1 or CCT5 knockdown cells, the amount of WDTC1 in the supernatant was almost undetectable, indicating that the TRiC/CCT was required for the refolding of WDTC1.
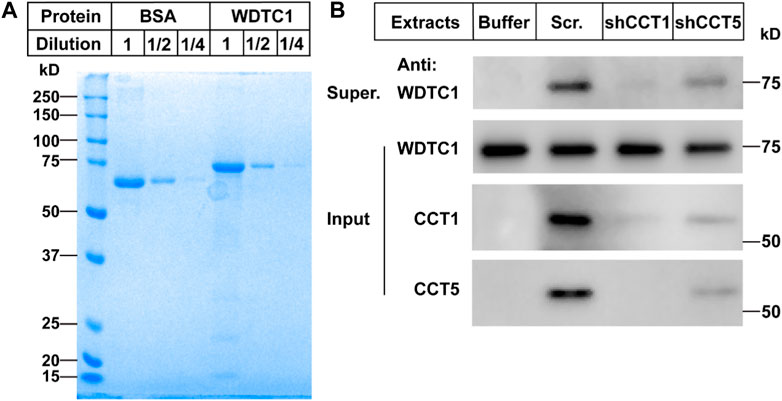
FIGURE 5. TRiC/CCT is required for the protein folding of WDTC1. (A) On day 0, the WDTC1 colony was incubated in the LB medium overnight. On day 1, the bacteria were induced with 1 mM IPTG for 3 h. Cells were then harvested and lysed in PBS containing 1 mg/ml lysozyme supplemented with protease inhibitors. The final product was subjected to SDS-PAGE and Coomassie Blue Staining. (B) On day 0, the purified WDTC1 was then denatured in 6 M urea in 10 mM HEPES pH 8.0. On day 1, 100 ng denatured WDTC1 protein was diluted into cytosolic fractions prepared from 3T3-L1 cell medium buffer or cells infected with scrambled shRNA, shCC1, or shCCT5 and incubated at 25°C for 1 h. The supernatant and pellet fractions of samples were immunoblotted with indicated antibodies.
We then went on to test the effect of the TRiC/CCT on the inhibitory effect of WDTC1 on adipogenesis. We overexpressed WDTC1 in 3T3-L1 cells, knocked down either CCT1 or CCT5, and then, subjected the cells to differentiation. After 8 days of differentiation, while the overexpression of WDTC1 suppressed differentiation, knockdown of either CCT1 or CCT5 largely restored the differentiation, as illustrated by lighter Oil Red O staining, intracellular triglyceride content, and the expression of the adipocyte markers (Figures 6A–C). These results indicate that the TRiC/CCT is required for the inhibitory effect of WDTC1.
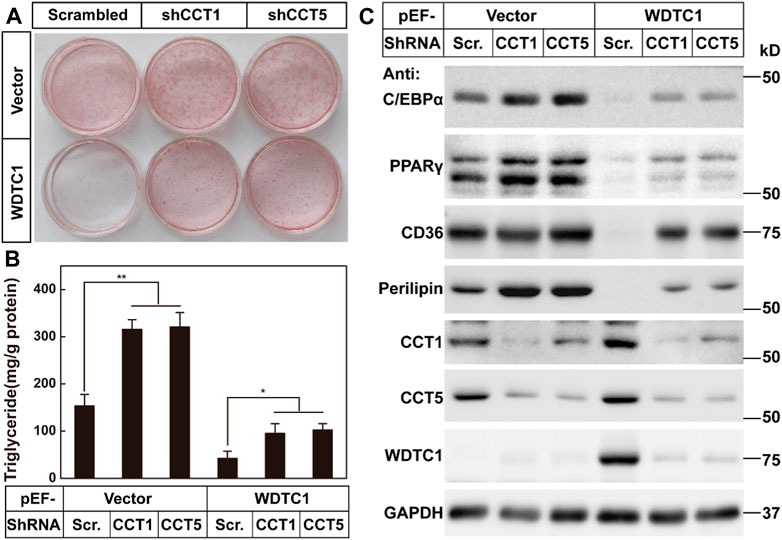
FIGURE 6. TRiC/CCT is required for the inhibitory effect of WDTC1 on adipogenesis. (A–C) 3T3-L1 preadipocytes infected with lentivirus expressing WDTC1 and the indicated shRNAs on pLKO.1 vector were set up at 1 × 105 cells per 35-mm dish and subjected to differentiation. On day 8 of differentiation, cells were harvested for Oil Red O staining (A), measurement of intracellular triglyceride content (B), and Western blot tests (C). Each value represents means ± SEM of three samples. Asterisks (*) denote the level of statistical significance (Student’s t-test) between scrambled and shCCT1 or shCCT5 cells; *p < 0.05 and **p < 0.01.
Adipogenesis is closely associated with obesity and related metabolic disorders (Rosen and MacDougald, 2006). Over the decades, numerous factors have been identified as key regulators of adipogenesis (Siersbaek et al., 2012). Among them, WDTC1 is very intriguing, as it is an evolutionally conserved anti-obesity factor (Hader et al., 2003; Suh et al., 2007). Functioning as an adaptor of the CRL4 E3 complex, WDTC1 binds MED20, which is essential for the transcriptional activation of C/EBPβ, and leads to its ubiquitination and degradation to inhibit adipogenesis (Tang et al., 2021). Here, we identify the TRiC/CCT as a key regulator of WDTC1. The TRiC/CCT binds WDTC1 and facilitates its protein folding and function. Disruption of the complex leads to the degradation of WDTC1 and eliminates the inhibitory effect of WDTC1 on adipogenesis. Our findings help better understand the regulatory mechanism of WDTC1 and adipogenesis.
In addition, we demonstrated a clear role of the TRiC/CCT in regulating adipogenesis. The TRiC/CCT is a common molecular chaperone found in all eukaryotes, and it is involved in many cellular activities by helping the folding of its substrates (Sternlicht et al., 1993). WD40-containing proteins have been shown to be substrates of the TRiC/CCT. Here, we clearly proved that WDTC1 is a substrate of the TRiC/CCT and, thus, expanded the role of the complex in adipogenesis. Furthermore, knockdown of either CCT1 or CCT5 largely promotes differentiation and lipid accumulation, suggesting that the endogenous WDTC1 might be regulated by the TRiC/CCT. However, we cannot rule out whether there are other substrates of the TRiC/CCT that might also be involved in adipogenesis. In the future, it will be interesting to find whether there are clinical mutants of the TRiC/CCT that are closely associated with obesity and related metabolic diseases.
Moreover, there are also some limitations of this study. In the adipogenesis study, we only choose 3T3-L1 preadipocytes, which, although widely applied in adipogenesis (Kuri-Harcuch et al., 2019), cannot really reflect human adipogenesis. Some other types of adipocytes need to be tested in the future study. Additionally, the roles of WDTC1 and the TRiC/CCT in other cell types also remain elusive.
All data generated or analyzed during this study are included in this published article and its Supplementary Information Files.
W-ST, XC, S-SY, S-TY, and LW performed the experiments. T-JZ, XW, and W-ST designed the experiments, analyzed the data, and wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (32101046 and 32201063) and the Excellent Youth Project of Natural Science Research of Anhui Education Department (No. 2022AH030079).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2023.1225628/full#supplementary-material
Angers, S., Li, T., Yi, X., MacCoss, M. J., Moon, R. T., and Zheng, N. (2006). Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature 443 (7111), 590–593. doi:10.1038/nature05175
Cong, Y., Baker, M. L., Jakana, J., Woolford, D., Miller, E. J., Reissmann, S., et al. (2010). 4.0-A resolution cryo-EM structure of the mammalian chaperonin TRiC/CCT reveals its unique subunit arrangement. Proc. Natl. Acad. Sci. U. S. A. 107 (11), 4967–4972. doi:10.1073/pnas.0913774107
Doane, W. W. (1960a). Developmental physiology of the mutant female sterile(2)adipose of Drosophila melanogaster. I. Adult morphology, longevity, egg production, and egg lethality. J. Exp. Zool. 145, 1–21. doi:10.1002/jez.1401450102
Doane, W. W. (1960b). Developmental physiology of the mutant female sterile(2)adipose of Drosophila melanogaster. II. Effects of altered environment and residual genome on its expression. J. Exp. Zool. 145, 23–41. doi:10.1002/jez.1401450103
Galgani, J. E., Kelley, D. E., Albu, J. B., Krakoff, J., Smith, S. R., Bray, G. A., et al. (2013). Adipose tissue expression of adipose (WDTC1) gene is associated with lower fat mass and enhanced insulin sensitivity in humans. Obes. (Silver Spring) 21 (11), 2244–2248. doi:10.1002/oby.20371
Groh, B. S., Yan, F., Smith, M. D., Yu, Y., Chen, X., and Xiong, Y. (2016). The antiobesity factor WDTC1 suppresses adipogenesis via the CRL4WDTC1 E3 ligase. EMBO Rep. 17 (5), 638–647. doi:10.15252/embr.201540500
Hader, T., Muller, S., Aguilera, M., Eulenberg, K. G., Steuernagel, A., Ciossek, T., et al. (2003). Control of triglyceride storage by a WD40/TPR-domain protein. EMBO Rep. 4 (5), 511–516. doi:10.1038/sj.embor.embor837
Hao, J. W., Wang, J., Guo, H., Zhao, Y. Y., Sun, H. H., Li, Y. F., et al. (2020). CD36 facilitates fatty acid uptake by dynamic palmitoylation-regulated endocytosis. Nat. Commun. 11 (1), 4765. doi:10.1038/s41467-020-18565-8
Kuri-Harcuch, W., Velez-delValle, C., Vazquez-Sandoval, A., Hernandez-Mosqueira, C., and Fernandez-Sanchez, V. (2019). A cellular perspective of adipogenesis transcriptional regulation. J. Cell. Physiol. 234 (2), 1111–1129. doi:10.1002/jcp.27060
Lai, C. Q., Parnell, L. D., Arnett, D. K., Garcia-Bailo, B., Tsai, M. Y., Kabagambe, E. K., et al. (2009). WDTC1, the ortholog of Drosophila adipose gene, associates with human obesity, modulated by MUFA intake. Obes. (Silver Spring) 17 (3), 593–600. doi:10.1038/oby.2008.561
Lopez, T., Dalton, K., and Frydman, J. (2015). The mechanism and function of group II chaperonins. J. Mol. Biol. 427 (18), 2919–2930. doi:10.1016/j.jmb.2015.04.013
Miyata, Y., Shibata, T., Aoshima, M., Tsubata, T., and Nishida, E. (2014). The molecular chaperone TRiC/CCT binds to the Trp-Asp 40 (WD40) repeat protein WDR68 and promotes its folding, protein kinase DYRK1A binding, and nuclear accumulation. J. Biol. Chem. 289 (48), 33320–33332. doi:10.1074/jbc.M114.586115
Moffat, J., Grueneberg, D. A., Yang, X., Kim, S. Y., Kloepfer, A. M., Hinkle, G., et al. (2006). A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 124 (6), 1283–1298. doi:10.1016/j.cell.2006.01.040
Nilsen, T. W. (2013). Preparation of nuclear extracts from HeLa cells. Cold Spring Harb. Protoc. 2013 (6), 579–583. doi:10.1101/pdb.prot075176
Rosen, E. D., and MacDougald, O. A. (2006). Adipocyte differentiation from the inside out. Nat. Rev. Mol. Cell. Biol. 7 (12), 885–896. doi:10.1038/nrm2066
Siersbaek, R., Nielsen, R., and Mandrup, S. (2012). Transcriptional networks and chromatin remodeling controlling adipogenesis. Trends Endocrinol. Metab. 23 (2), 56–64. doi:10.1016/j.tem.2011.10.001
Skjaerven, L., Cuellar, J., Martinez, A., and Valpuesta, J. M. (2015). Dynamics, flexibility, and allostery in molecular chaperonins. FEBS Lett. 589 (1), 2522–2532. doi:10.1016/j.febslet.2015.06.019
Sternlicht, H., Farr, G. W., Sternlicht, M. L., Driscoll, J. K., Willison, K., and Yaffe, M. B. (1993). The t-complex polypeptide 1 complex is a chaperonin for tubulin and actin in vivo. Proc. Natl. Acad. Sci. U. S. A. 90 (20), 9422–9426. doi:10.1073/pnas.90.20.9422
Suh, J. M., Zeve, D., McKay, R., Seo, J., Salo, Z., Li, R., et al. (2007). Adipose is a conserved dosage-sensitive antiobesity gene. Cell. Metab. 6 (3), 195–207. doi:10.1016/j.cmet.2007.08.001
Tang, W. S., Weng, L., Wang, X., Liu, C. Q., Hu, G. S., Yin, S. T., et al. (2021). The Mediator subunit MED20 organizes the early adipogenic complex to promote development of adipose tissues and diet-induced obesity. Cell. Rep. 36 (1), 109314. doi:10.1016/j.celrep.2021.109314
Trinidad, A. G., Muller, P. A., Cuellar, J., Klejnot, M., Nobis, M., Valpuesta, J. M., et al. (2013). Interaction of p53 with the CCT complex promotes protein folding and wild-type p53 activity. Mol. Cell. 50 (6), 805–817. doi:10.1016/j.molcel.2013.05.002
Wang, J., Hao, J. W., Wang, X., Guo, H., Sun, H. H., Lai, X. Y., et al. (2019). DHHC4 and DHHC5 facilitate fatty acid uptake by palmitoylating and targeting CD36 to the plasma membrane. Cell. Rep. 26 (1), 209–221. doi:10.1016/j.celrep.2018.12.022
Wang, X., Wang, H. Y., Hu, G. S., Tang, W. S., Weng, L., Zhang, Y., et al. (2021). DDB1 binds histone reader BRWD3 to activate the transcriptional cascade in adipogenesis and promote onset of obesity. Cell. Rep. 35 (12), 109281. doi:10.1016/j.celrep.2021.109281
Yam, A. Y., Xia, Y., Lin, H. T., Burlingame, A., Gerstein, M., and Frydman, J. (2008). Defining the TRiC/CCT interactome links chaperonin function to stabilization of newly made proteins with complex topologies. Nat. Struct. Mol. Biol. 15 (12), 1255–1262. doi:10.1038/nsmb.1515
Keywords: TRiC/CCT, WDTC1, adipogenesis, protein folding, obesity
Citation: Tang W-S, Cen X, Yao S-S, Yin S-T, Weng L, Zhao T-J and Wang X (2023) TRiC/CCT chaperonin is required for the folding and inhibitory effect of WDTC1 on adipogenesis. Front. Cell Dev. Biol. 11:1225628. doi: 10.3389/fcell.2023.1225628
Received: 19 May 2023; Accepted: 08 August 2023;
Published: 24 August 2023.
Edited by:
Marie-Pierre Golinelli, UPR2301 Institut de Chimie des Substances Naturelles (ICSN CNRS), FranceReviewed by:
Hengbin Wang, Virginia Commonwealth University, United StatesCopyright © 2023 Tang, Cen, Yao, Yin, Weng, Zhao and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xu Wang, d2FuZ3h1QGFobXUuZWR1LmNu; Tong-Jin Zhao, emhhb3RqQGZ1ZGFuLmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.