
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol., 06 July 2023
Sec. Developmental Epigenetics
Volume 11 - 2023 | https://doi.org/10.3389/fcell.2023.1212199
This article is part of the Research TopicIn Celebration of Women in Developmental EpigeneticsView all 11 articles
DNA methylation is the most commonly studied epigenetic mark in humans, as it is well recognised as a stable, heritable mark that can affect genome function and influence gene expression. Somatic DNA methylation patterns that can persist throughout life are established shortly after fertilisation when the majority of epigenetic marks, including DNA methylation, are erased from the pre-implantation embryo. Therefore, the period around conception is potentially critical for influencing DNA methylation, including methylation at imprinted alleles and metastable epialleles (MEs), loci where methylation varies between individuals but is correlated across tissues. Exposures before and during conception can affect pregnancy outcomes and health throughout life. Retrospective studies of the survivors of famines, such as those exposed to the Dutch Hunger Winter of 1944-45, have linked exposures around conception to later disease outcomes, some of which correlate with DNA methylation changes at certain genes. Animal models have shown more directly that DNA methylation can be affected by dietary supplements that act as cofactors in one-carbon metabolism, and in humans, methylation at birth has been associated with peri-conceptional micronutrient supplementation. However, directly showing a role of micronutrients in shaping the epigenome has proven difficult. Recently, the placenta, a tissue with a unique hypomethylated methylome, has been shown to possess great inter-individual variability, which we highlight as a promising target tissue for studying MEs and mixed environmental exposures. The placenta has a critical role shaping the health of the fetus. Placenta-associated pregnancy complications, such as preeclampsia and intrauterine growth restriction, are all associated with aberrant patterns of DNA methylation and expression which are only now being linked to disease risk later in life.
According to the Developmental Origins of Health and Disease hypothesis, environmental exposures in early life affects later life risk. Part of this connection may come through DNA methylation, patterns of which are known to change under different conditions. The placenta is important for mediating the connection between mother and fetus, both able to respond to the environment itself and controlling the environment of the fetus. Many pregnancy complications are linked to placenta function and birth outcomes can have a large effect on later disease risk. This review will summarise current knowledge on the effect of early environmental exposure on later disease risk, especially where this may be mediated by DNA methylation. We will highlight the unique nature of the placenta epigenome and its potential as a connection between environment and health.
Epigenetic marks are heritable DNA modifications that can influence gene expression without changing the DNA sequence. These include chemical modifications of DNA bases, post-translational histone modifications and chromatin structure, and their configuration can be affected by a variety of environmental exposures.
Histones wrapped with DNA form the nucleosome, that can alter gene accessibility by forming transcriptionally inactive heterochromatin or transcriptionally active euchromatin (Li et al., 2007). Expression can be controlled by reversible post-translational modifications on histone amino acid tails, with complex cross-talk between modifications (Kouzarides, 2007). For example, lysine 9 or 27 acetylation on histone 3 (H3K9ac or H3K27ac) weakens DNA-histone interactions, and so opens chromatin to facilitate transcription, whereas trimethylation of the same lysines is associated with heterochromatin formation (Quina et al., 2006).
5-methylcytosine (5 mC) is well established as a stable, heritable DNA methylation mark that affects gene expression and genome stability. It is created by the addition of a methyl group to the 5-carbon atom of the cytosine ring (Moore et al., 2013). This modification is important for many processes, including tissue-specific gene regulation, imprinting and X-chromosome inactivation, working in combination with histone modifications (Mikkelsen et al., 2007; Moore et al., 2013). In promoters and intergenic regions, it is generally associated with transcriptional silencing through recruiting gene suppressor proteins, promoting heterochromatin and disrupting transcription factor binding (McMahon et al., 2017; Kaluscha et al., 2022). In bodies of highly expressed genes, high levels of 5 mC stop promiscuous transcription initiation (Neri et al., 2017).
Most 5 mC is found on CpG dinucleotides (a cytosine base next to a guanine), up to 80% of which are methylated in mammalian genomes (Lister et al., 2009). These CpG dinucleotides are often clustered into CpG islands, which are associated with 70% of known gene promoters, where methylation can silence gene expression (Mohn et al., 2008; Illingworth et al., 2010). They are also enriched in repetitive elements, satellite DNA and transposable elements to help maintain genome stability (Vera et al., 2008; Lister et al., 2009; Si et al., 2009).
In humans, 5 mC is created and maintained by DNA methyltransferases (DNMTs) using S-adenyl methionine (SAM) as a methyl donor. DNMTs include: DNMT3a and 3b for de novo methylation in embryos and germ cells, with the non-catalytic DNMT3L as a cofactor (Bourc’his and Bestor, 2004; Kaneda et al., 2004), and DNMT1 for maintenance during DNA replication and repair (Hermann et al., 2004; Mortusewicz et al., 2005). Methylation can be removed passively through replication without methylation maintenance, or actively, through several intermediates catalysed by Ten-Eleven-Translocation (TET) proteins (Hackett et al., 2013). One of these intermediates, 5-hydroxymethylcytosine (5hmC), has been proposed to also play a regulatory role as its distribution shows strand and sequence bias. 5hmC is enriched in regulatory elements and promoters in embryonic stem cells and occasionally in placenta (Hernandez Mora et al., 2018), as well as in actively transcribed genes in neuronal cells (Mellén et al., 2012; Yu et al., 2012).
Another suggested regulatory modification is methylation to the adenine base (m6dA), which is a known epigenetic mark in bacteria, protists (Wion and Casadesús, 2006), and eukaryotes such as C. elegans (Greer et al., 2015). In humans, m6dA is a common RNA modification but is also detected on DNA, where it has been associated with actively transcribed genes and risk of tumorigenesis (Xiao et al., 2018). However, suggestions for a human DNA adenine methylase or demethylase have failed to be replicated (Xie et al., 2018; Musheev et al., 2020) and other studies suggest DNA m6dA in humans arises purely through nucleotide salvage from RNA degradation (Liu et al., 2020; Musheev et al., 2020).
The synthesis of both post-translational protein methylation, including histones, and DNA methylation is dependent on one-carbon metabolism, along with many other cellular processes (Ducker and Rabinowitz, 2017). The methyl group is donated from S-adenosylmethionine (SAM), leaving S-adenosyl homocysteine (SAH) (Figure 1). Many components of this pathway must come from the diet, both methyl donors, including folate, choline and betaine, and cofactors, including vitamins B-2, B-6, and B-12 (James et al., 2018). Studies in animal models demonstrate that supplementing these nutrients can alter DNA methylation patterns (Waterland and Jirtle, 2003; Sinclair et al., 2007).
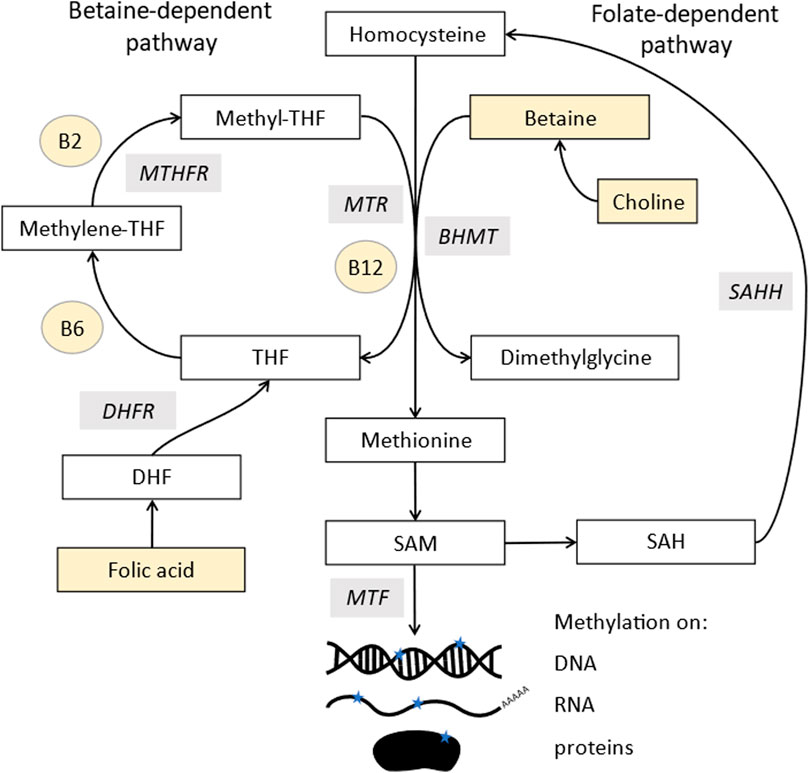
FIGURE 1. Methylation pathway. Homocysteine (top middle) is converted into methionine by two pathways: methionine synthase (MTR), which uses vitamin B12 as a cofactor and acquires a methyl group from the conversion of 5-methyltetrahydrofolate (methyl-THF) into tetrahydrofolate (THF). MethyL-THF is acquired from dietary folates which is converted from dihydrofolate (DHF) by dihydrofolate reductase (DHFR) and 5,10- methylenetetrahydrofolate (5,10MTHF) by methylene tetrahydrofolate reductase (MTHFR). Methionine can also acquire a methyl group from betaine via a reaction involving betaine homocysteine methyl-transferases (BHMT). Methionine is further converted to s-adenosylmethionine (SAM), the major methyl donor for all methyl transferases (MTF), which add methyl groups to DNA, RNA, lipids, and proteins. SAM is recycled via s-adenosylhomocysteine (SAH) which is converted to homocysteine in a reversible reaction by s-adenosylhomocysteine hydrolase (SAHH). Components highlighted in yellow are derived from the diet.
SAM is derived from methionine, which comes from methylated homocysteine. There are two enzymes that can make methionine, most commonly methionine synthase, which requires vitamin B12 as a cofactor. It uses 5-methyltetrahydrofolate as a methyl donor, whose synthesis is folate dependent and also links into purine biosynthesis. The second enzyme, betaine-homocysteine methyltransferases (BHMT), has two isoforms, of which one is expressed only in the liver and kidneys, with the other (BHMT2) expressed more widely (Pajares and Pérez-Sala, 2006). This uses betaine as a methyl donor, which comes either from choline or direct dietary supplementation.
The donation of the methyl group from SAM is catalysed by methyl transferases, which are inhibited by SAH. Therefore, methylation stops if the process of converting SAH back to homocysteine and methionine and SAM is inhibited. Ways to measure methylation potential therefore include the SAM:SAH ratio as well as levels of the pathway components (Mason, 2003).
DNA methylation can change dynamically throughout life but there are many loci where it remains consistent. For these loci, there are two major reprogramming events to consider. The first is in the primordial germ cells that give rise to the sperm and oocyte, where DNA is passively demethylated during initial formation of the germ cells to remove any previous marks (Hackett et al., 2013; Guo et al., 2015). The DNA can then be remethylated to establish parent-of-origin (PoO) specific methylation, which according to studies in mice occurs before birth in sperm but not until maturation for oocytes (Hiura et al., 2006; Guo et al., 2014; 2015; Gahurova et al., 2017).
The next major reprogramming stage occurs after fertilisation, where a wave of demethylation before the blastocyst stage results in the lowest developmental level of genome methylation (Weaver et al., 2009). Parental DNA from sperm begins almost entirely methylated, except in CpG islands, but at this stage most of its methylation is actively removed (Guo et al., 2014; Shen et al., 2014; Smith et al., 2014). In contrast, maternal, oocyte-derived DNA is methylated mostly at active gene bodies and maintains more methylation (Smallwood et al., 2011). It is demethylated passively by dilution through replication (Hirasawa et al., 2008; Shen et al., 2014).
Differential maintenance of methylation at this stage has the potential to create changes in gene expression. This can create metastable epialleles, loci with variable methylation between, but not within, individuals. Regions that maintain PoO specific methylation through this process create germline differentially methylated regions (gDMRs), with the potential to be imprinted.
Where differential methylation is maintained, alleles are protected against demethylation by ZFP57 and ZFP445, which targets DNMT1 (Li et al., 2008; Quenneville et al., 2011; Takahashi et al., 2019). The opposite alleles are kept unmethylated by many different factors, either acting together or on different genes, including CFP1, SP1 or CTCF (CCTC-binding factor) (Macleod et al., 1994; Fedoriw et al., 2004).
Imprinted genes are found in therian (eutherian and marsupial) mammals and are defined by differential expression between maternal and paternal alleles, which can be specific to developmental stage, tissue or isoform (Renfree et al., 2013). Incorrect imprinting can have long term effects, such as imprinting diseases (Peters, 2014; Monk et al., 2019). Imprinting is generally a result of allele-specific repression from gDMRs, with most imprinted genes found within regions containing an imprinting control region (ICR) (Figure 2) (Spahn and Barlow, 2003; Schulz et al., 2010). As well as directly controlling expression, imprinted DMRs are often associated with repressive histone modifications, such as H3K9me3 on methylated alleles (Monk et al., 2006; Court et al., 2014).
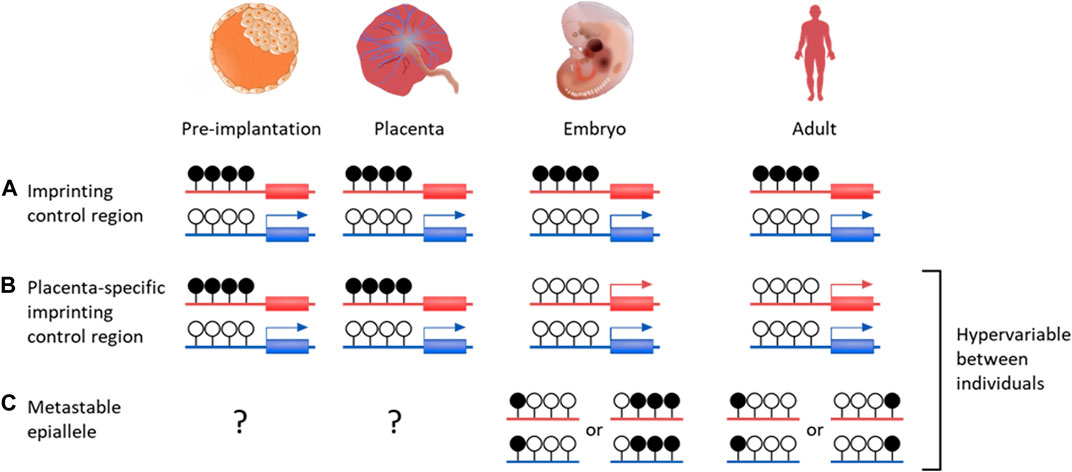
FIGURE 2. DNA methylation profiles of various genetic features that are important for placenta development and growth. Canonical genomic imprinting is associated with regions of allelic methylation inherited from one gamete. (A) Oocyte-derived methylation is faithfully maintained throughout development on the maternal allele, resulting in paternal expression. (B) An example of a germline-derived, placenta-specific imprint. During pre-implantation development these regions are indistinguishable from canonical imprinted DMRs. Upon implantation, embryonic tissues become demethylated through to adulthood, while allelic methylation is maintained solely in the placenta. (C) While establishment of metastable epialleles during early development is currently unknown, these intervals are particularly vulnerable to environmental influences. Once systematically methylated, they are stable over time, despite showing inter-individual variability. Blue genes are paternally expression, red genes are maternally expressed. Black filled lollipops are methylated CpG intervals, while unfilled are unmethylated.
There are over 120 confirmed imprinted genes in humans, of which most are associated with maternally methylated regions (Okae et al., 2014; Humanimprints.net; Geneimprint, 2023). Some gDMRs maintain allelic methylation throughout life but the majority are transient, surviving the pre-implantation demethylation then becoming entirely methylated or unmethylated after implantation (Proudhon et al., 2012). It is possible these have a function as ICRs in the pre-implantation embryo, or in the placenta, where many transient gDMRs are maintained (Figure 2) (Sanchez-Delgado et al., 2016).
Examples of non-canonical imprinting are so far restricted to mice. In these cases, imprinted genes have no detectable methylation differences between parental gametes. Instead, they have the histone modification H3K27me3 in the oocyte, which later leads to maternal allele methylation in extra-embryonic tissues (including placenta) (Inoue et al., 2017).
MEs are loci with variable methylation between individuals, but consistent cross-tissue methylation within individuals, suggesting the methylation is established early in development before separation of the germ layers. They are often associated with transposable elements, which evolve fast and often give rise to new cis-regulatory DNA elements or even new exons and genes (Imbeault et al., 2017; Bourque et al., 2018).
The Agouti viable yellow (Avy) mouse allele is a well characterised example of a mammalian metastable epiallele associated with a transposable element, which causes a yellow coat colour when expressed (Dickies, 1962; Duhl et al., 1994). This allele was created by insertion of an Intracisternal A-particle (IAPs), a Class II endogenous retrovirus (ERV) with protein-coding sequences between long terminal repeats (LTRs) (Cole et al., 1981). The insertion created a cryptic promoter in the LTR which drives expression when unmethylated and crucially the epigenetic states, including DNA methylation and histone profiles, are variable between individuals (Duhl et al., 1994; Morgan et al., 1999; Kazachenka et al., 2018). The phenotype is heritable, with maternal inheritance causing an incomplete shift in yellow coats in the litter (Morgan et al., 1999). The pattern in phenotype can also be shifted by environment, for example, a preconception diet rich in methyl donors and cofactors in utero leads to increased methylation and fewer offspring with yellow coats (Wolff et al., 1998; Cooney et al., 2002; Waterland and Jirtle, 2003; Cropley et al., 2006).
Subsequent studies have found other variably methylation IAPs, which vary between but not within individuals (Kazachenka et al., 2018). Most are not heritable and many are not changed by environmental exposures like bisphenol A or methyl donor supplementation (Bertozzi et al., 2021). Sites also seem to function independently, as methylation is generally not correlated across different sites within individuals but is found at consistent levels across populations at each site (Kazachenka et al., 2018).
The Developmental Origins of Health and Disease (DOHaD) hypothesis states that environmental exposures in early life affect the risk of adverse health outcomes later in life (Barker, 1990). Epidemiological studies have shown many associations between early environment and later disease outcomes in humans.
There are several possible mechanisms that could link early environmental exposures to lifelong health of which DNA methylation changes, causing gene expression changes, are a leading candidate (Fleming et al., 2018). When investigating these cases, the time period around conception may be a critical window because, as previously discussed, shortly after fertilisation patterns of DNA methylation are established that can persist throughout life.
Adverse birth outcomes can be caused by many different factors, including a wide variety of environmental exposures. Both maternal obesity and preconception physical activity are associated with risk of preeclampsia (Aune et al., 2014; Marchi et al., 2015; Poston et al., 2016). Alcohol and caffeine intake have been linked to reduced birthweight (Konje and Cade, 2008; Popova et al., 2021), and tobacco smoke exposure to worse birth outcomes, including reduced fetal growth (Peterson and Hecht, 2017). These adverse birth outcomes can subsequently be linked to later life disease, as discussed previously with placenta-related outcomes.
Important exposures often occur over long time periods in humans, so the critical windows of sensitivity are easier to separate in animal models. These studies show short term effects during pregnancy, such as placental growth being affected by a preconception zinc deficiency in rodents (Tian et al., 2014). Longer term effects include increased risks of offspring hypertension and adiposity from a preimplantation low protein diet in rodents (Kwong et al., 2000; Watkins et al., 2008a; Watkins et al., 2008b). These models suggest the blastocyst is capable of sensing nutrient status and altering the phenotype of placental development (Eckert et al., 2012; Watkins et al., 2015).
In humans, famines occurring over well-defined dates allow the study of exposures in specific developmental windows. One well-studied example is the Dutch Hunger Winter, a severe famine in 1944-45. Prenatal exposure to this famine increased the risk of diabetes, schizophrenia and other diseases in later life (Lumey et al., 2011).
Many examples of studies on the effect of early life nutrition and health outcomes are from a Sub-Saharan population in rural Gambia that experiences two distinct seasons with very different environmental, especially nutritional, exposures. In the rainy/hungry season, food availability is lower and energy status is poorer than in the dry/harvest season (Prentice et al., 1981). Babies born in the rainy season are more likely to have intrauterine growth restriction (Ceesay et al., 1997). They are also 10 times more likely to die prematurely, which may be linked to infection (Moore et al., 1997; 1999), as babies born in the hungry season have altered T cell immunity, with lower CD3+ and CD4+ lymphocyte counts (Ngom et al., 2011).
Micronutrition supplementation studies can test the association between nutritional environment and health more directly, for example, folic acid is known to reduce neural tube defects by up to 70% when taken in the months before and after conception and may also reduce the risk of small for gestational age births (Mastroiacovo and Leoncini, 2011; Hodgetts et al., 2015; Gao et al., 2016). The UK based UPBEAT trial used an intervention during pregnancy that reduced processed and snack food consumption and found a decrease in infant adiposity at 6 months of age (Patel et al., 2017).
An alternative approach to isolating early life environment is the study of assisted reproductive technologies (ART), such as in-vitro fertilisation, which provide a different environment only around conception. These show increased risk of high blood pressure in children and altered heart shape and chamber size in infants (Ceelen et al., 2008; Valenzuela-Alcaraz et al., 2013).
As discussed, methylation could be a mechanism for the link between early environmental exposures and health. In humans, ART has been linked to an increased risk of imprinting disorders (Lazaraviciute et al., 2014) and changes in methylation and histone modifications at imprinted genes (Choux et al., 2020). However, another study found no variability in 25 imprinted DMRs and allelic expression in placenta or cord blood in spontaneously conceived and assisted pregnancies (Camprubí et al., 2013).
Many environmental factors are known to affect methylation, for example, there are changes in methylation at repetitive regions with fetal bisphenol A exposure (Faulk et al., 2016) and changes in methylation at 12 CpGs in blood and 12 in buccal epithelial cells with prenatal exposure to phthalates (England-Mason et al., 2022). Aflatoxin B1 exposure in utero in The Gambia was associated with differential methylation at 71 CpG sites in infant white blood cells, including in growth factor and immune-related genes (Hernandez-Vargas et al., 2015).
Nutrition exposure may be key to investigating methylation variability, due to the direct links of micronutrients into the 1C pathway that provides methyl groups required for DNA methylation. Again, animal studies can provide more detailed exposure timing and insight into molecular mechanisms. In mouse studies of periconceptional maternal obesity, fetal growth rate is directly affected by alterations in methylation at the ribosomal DNA promoter which controls the levels of ribosomal RNA (Denisenko et al., 2016; Holland et al., 2016).
In humans, data from the Dutch Hunger Winter shows adults whose mothers were exposed to the famine only before and during the periconceptional period have modestly decreased DNA methylation in the IGF2 gene (Heijmans et al., 2008). Further studies have shown exposure to this famine in early gestation affects methylation in many regions, mostly annotated as regulatory and with nearby genes expressed in early developmental phases (Tobi et al., 2014).
Identification of MEs in humans is complex, but advances in molecular techniques have suggested their existence in the human genome and identified putative links with environment and disease. They are a useful tool when exploring the link between environment, methylation and health because they are established very early in development and are hypervariable. They can also be studied in easily available samples, such as fetal blood obtained from the umbilical cord.
The levels of micronutrients essential for the 1C cycle in diet can affect methylation levels (Waterland and Jirtle, 2003; Sinclair et al., 2007). In The Gambia, micronutrients related to the 1C cycle vary across the seasons, as measured by maternal blood biomarkers (Dominguez-Salas et al., 2013). Studies have found higher concentration of methyl donors in the rainy season, potentially giving a higher methylation potential (Figure 3).
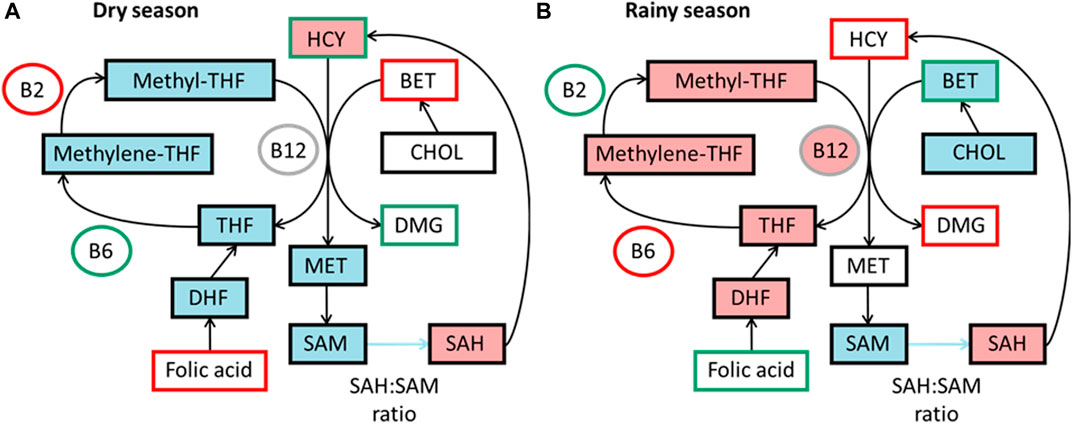
FIGURE 3. Simplified summaries of metabolic pathways highlighting seasonal differences reported from studies in The Gambia. Comparative levels between seasons in diet as measured by blood biomarkers and components predicting DNA methylation in each season, data from James et al. (2019) (A) Components outlined in green are higher in dry season and components outlined in red are lower. Components highlighted in blue are positive predictors of DNA methylation and pink negative predictors in dry season. (B) Components outlined in green are higher in rainy season and components outlined in red are lower. Components highlighted in blue are positive predictors of DNA methylation and pink negative predictors in rainy season. BET, betaine; B12, vitamin B-12; B2, vitamin B-2; B6, vitamin B6; CHOL, choline; DHF, dihydrofolate; DMG, dimethyl glycine; HCY, homocysteine; MET, methionine; methylene-THF, N5,10-methylene tetrahydrofolate; methyl-THF, N5-methyl tetrahydrofolate; SAH, S-adenosyl homocysteine; SAM, S-adenosyl methionine; THF, tetrahydrofolate.
Non-imprinted DMRs in cord blood, placental tissue or peripheral blood from Gambian infants have also been associated with birthweight, and 25 DMRs in infant blood at 12 months are associated with length for age (Quilter et al., 2021). A meta-analysis found DNA methylation in neonatal blood has been linked with many pregnancy exposures and is associated with birthweight through childhood, but not adulthood. There is also an overlap between CpGs related to birthweight and intrauterine exposures (Küpers et al., 2019).
These changes were first associated with significant changes in methylation at seven infant MEs, with increased levels of homocysteine and B6 and decreased levels of B2 correlating with higher methylation (Dominguez-Salas et al., 2014). A model found different biomarkers predicted methylation between seasons, with vitamin B-2 and methionine as positive predictors in the dry season, and choline and vitamin B-6 as positive and folate and vitamin B-12 as negative predictors in the rainy season (Figure 3) (James et al., 2019). The different seasonal effects suggest a switch between the betaine dependent pathway in the rainy season to the folate dependent pathway in the dry season.
In further studies, children conceived in the Gambian rainy season showed consistently increased methylation at multiple MEs (Waterland et al., 2010; Khulan et al., 2012; Silver et al., 2022). Methylation changes extend beyond MEs; Silver et al. (Silver et al., 2022) identified 259 CpGs that showed a robust signature of differential methylation between seasons of conception (SoC-CpGs) across two independent cohorts. In one of these cohorts methylation measured in blood in early and mid-childhood showed that the seasonality effect was attenuated in the older cohort.
Many SoC-CpGs overlapped with MEs identified in previous studies and many have been associated with sex (Kessler et al., 2018). They were also enriched for CpGs previously identified to be methylated on PoO specific alleles (Zink et al., 2018), which were also enriched for placenta-specific oocyte gDMRs (Sanchez-Delgado et al., 2016).
One example of a non-coding RNA gene responsive to SoC is VTRNA2-1, which is also a putative ME (Silver et al., 2015). This tumour suppressor gene has been linked to cancer and there is evidence that it is maternally imprinted (Romanelli et al., 2014; Silver et al., 2015).
The EMPHASIS (Epigenetic Mechanisms linking Preconceptional nutrition and Health Assessed in India and sub-Saharan Africa) study conducted two randomised controlled trials that used different micronutrient interventions taken preconception and during pregnancy. Six differentially methylated CpGs were found in Gambian children, of which four were around the endothelial cell-specific molecule 1 (ESM1) gene (Saffari et al., 2020). Another randomised controlled trial found sex-specific epigenetic changes in Gambian cord and infant blood from periconceptional micronutrient supplementation (Khulan et al., 2012). In these children, girls had reduced methylation at IGFR2 and boys at MEG3/GTL2, but these differences disappeared by 9 months (Cooper et al., 2012).
One of the genes with increased methylation associated with periconceptional micronutrient supplementation and conception in the Gambia rainy seasons is PAX8 (paired-box 8) (Waterland et al., 2010; Silver et al., 2015; Saffari et al., 2020). It is a master regulator of thyroid gene expression, and its methylation has been associated with thyroid function in Gambian children (Candler et al., 2021). Another example is POMC, where methylation has been separately shown to reduce POMC transcription and is associated with obesity in children (Kuehnen et al., 2012; Kühnen et al., 2016).
The placenta is a transient organ that acts as a physical connector and barrier between maternal and fetal blood, mediating nutrient and gas exchange, and waste removal between the embryo and the mother. It also functions as an endocrine organ, producing hormones including human chorionic gonadotropin (hCG) (Lopata et al., 1997). Placenta development and structure is highly species-specific; the human placenta displays deep trophoblast invasion which is unique to great apes (Imakawa et al., 2015). Correct functioning is vital for appropriate fetal development, for example, increased glucose transfer in gestational diabetes increases fetal adiposity and macrosomia, which subsequently is associated with disease in later life (Pettitt et al., 1993; Tint et al., 2020).
Many pregnancy complications are linked with placental development defects, including preeclampsia, fetal growth restriction, recurrent miscarriage and still-birth (Brosens et al., 2011). These conditions contribute to a high proportion of maternal and neonatal morbidity and mortality, especially in sub-Saharan Africa (Graham et al., 2016).
Intrauterine growth retardation (IUGR) affects 10%–15% pregnancies and is a subset of small for gestational age, which is defined as birth weight below the 10th percentile (Harkness and Mari, 2004). Many IUGR cases are caused by placental insufficiency. In these cases, there is deficient remodeling of uterine spiral arteries that supply the placenta creating a lower villous volume and surface area for maternal-fetal exchange, and dysregulation of many genes (Burton and Jauniaux, 2018). Babies with IUGR have a high risk of hypertension, type 2 diabetes and heart disease later in life (Curhan et al., 1996; Rich-Edwards et al., 1997).
Preeclampsia affects 3%–5% of pregnancies in the developed world and forms part of the hypertensive disorders that cause 12% of maternal deaths during and after pregnancy (WHO, 2005). It is characterised by hypertension and proteinuria in the second half of pregnancy (Roberts and Cooper, 2001) and is also associated with defective uterine spiral artery remodeling. Its effects range from mild to multiorgan failure and it is caused by low perfusion in the placenta (Brosens et al., 2011).
Overall placenta morphology has also been associated with later disease risk. For example, in the Helsinki Birth Cohort that experienced prenatal famine exposure, placenta thickness has been associated with risk of sudden cardiac death (Barker et al., 2012). Another study linked longer and more oval placentas in this cohort with colorectal cancer (Barker et al., 2013).
The placenta is hypomethylated overall, with around 3% cytosine bases methylated compared to 4% in somatic tissue (Fuke et al., 2004). Global placenta methylation increases from 2.8% in first trimester to 3.1% in term placentas, unlike global methylation in somatic tissues which decreases with age (Fuke et al., 2004). The location of methylation also changes with gestational age, suggesting that genes act at specific stages (Novakovic et al., 2011; Camprubí et al., 2013). Methylation changes with gestational age are accompanied by a change in gene expression levels in 25% of placenta expressed genes (Sitras et al., 2012; Uusküla et al., 2012). Most imprinted genes show reduced expression with advancing gestational age independent of allelic methylation (Monteagudo-Sánchez et al., 2019; Pilvar et al., 2019).
The hypomethylation in placenta is not confined to specific genomic features but does include repetitive elements, including transposable elements, as well as large (>100 kb), partially methylated domains (PMDs) that are largely stable throughout gestation (Schroeder et al., 2013; Schroeder and Lasalle, 2013). Repetitive elements are highly methylated in somatic tissue but variably methylated in placenta and highly species specific, which may allow them to act as drivers for placenta evolution. There are multiple placenta-specific promotors derived from TEs, which includes endogenous retroviruses, including for the CYP19A1, NOS3 and PTN genes (Dunn-Fletcher et al., 2018). ERV families have been identified with regulatory potential that are close to trophoblast-specific expressed genes (Sun et al., 2021; Frost et al., 2023). In human trophoblast stem cells, genetic editing showed these elements act as enhancers (Frost et al., 2023).
HERV-W is a co-opted human ERV envelope gene which illustrates how these elements can exhibit placenta-specific function. One of its copies produces Syncitin-1, which is important for syncytiotrophoblast fusion, a multinucleated cell layer around placenta villi (Blond et al., 1999; Mi et al., 2000). Its 5′ UTR contains a LTR with a trophoblast-specific enhancer and a promoter region with a CpG island, whose methylation controls syncitin-1 expression (Matoušková et al., 2006; Gao et al., 2012). HERV-W also highlights how these elements can be important for pregnancy outcomes as it is hypomethylated and expressed in placenta, with lower expression in preeclampsia and IUGR placentas (Ruebner et al., 2013).
Placenta methylation at gene promoters is highly variable between individuals, with increased variability in term compared to first trimester placentas (Novakovic et al., 2011). However, there is also variability across a placenta depending on the sampling site (Janssen et al., 2015) which may reflect cellular composition.
Placental methylation at many genes has been associated with pregnancy outcomes. Placentas from pregnancies with preeclampsia have higher global DNA methylation and within this, blood pressure increases correlate with methylation increases (Kulkarni et al., 2011). Studies looking at preterm infants found lower SAM:SAH ratio and global methylation (Khot et al., 2017). Sinclair et al. (2007) found 15 loci where methylation associated with birthweight, of which four had corresponding changes in transcript levels in placenta.
In recent years, genome-wide DNA methylation values have been used to show discrepancies between chronological and biological age. Such “epigenetic clocks” use algorithms to calculate biological age on the basis of hundreds to thousands of CpG sites across the genome (Horvath, 2013). By calculating multi-organ clocks it is possible to predict clinically useful biomarkers associated with age-related disease and mortality (Lu et al., 2019). Whilst a pan-tissue epigenetic clock cannot reliably estimate gestational age, Lee et al. (2019) generated bioinformatic pipelines to generate placenta-derived epigenetic clocks that can track gestational age.
Deviations from normal placenta aging have been reported for several pregnancy complications. Accelerated epigenetic aging has been linked to early onset preeclampsia (Mayne et al., 2017), and lower fetal weight (Tekola-Ayele et al., 2019), while deceleration has been associated with maternal weight gain during pregnancy (Workalemahu et al., 2021). Together this indicates that both epigenetic age acceleration and deceleration are associated with distinct risk and protective factors, with studies from many laboratories are trying to identify distinct, tissue and cell-type specific trajectories to predict pregnancy outcomes.
Although the placenta is hypomethylated overall compared to somatic tissue, it exclusively maintains PoO methylation and expression at many transient gDMRs (Figure 2). In humans, over 150 maternally methylated DMRs have been identified in the placenta, around half of which have confirmed paternal expression (Yuen et al., 2011; Barbaux et al., 2012; Sanchez-Delgado et al., 2015; Hamada et al., 2016; Hanna et al., 2016; Sanchez-Delgado et al., 2016). Placenta-specific imprinting is highly polymorphic between individuals in humans and poorly conserved between humans and mice, though it may be more similar in other primates (Hanna et al., 2016; Sanchez-Delgado et al., 2016).
Hanna et al. (2016) found a monoallelic methylation in 50% of heterozygous samples at selected DMRs, looking just at trophoblast and villi cells to remove differences from cell composition, and there was significantly less variability at the same sites in somatic tissues. Complementary studies have showed that of 104 placenta-specific gDMRs studied, 52% intervals possess low methylation and biallelic expression in multiple term biopsies (Sanchez-Delgado et al., 2016; Monteagudo-Sanchez et al., 2019), although the frequency was variable between loci. It is unknown if this variation arises from the interactions with in cis genetic variants, environmental exposures, or is truly stochastic.
One well characterised transiently imprinted gene in mice is Zdbf2 (zinc finger DBF-type containing 2) (Duffié et al., 2014). Generally, imprinting is poorly conserved between humans and mice, but the methylation pattern is the same at Zdbf2. The locus has a maternal pre-implantation gDMR, causing paternal expression of an alternative long isoform Liz (Duffié et al., 2014). This leads to creation of a paternal secondary DMR, which blocks the repressive H3K27me3 mark allowing paternal expression from the canonical Zdbf2 promoter. In somatic tissue, the maternal gDMR is lost after implantation, leaving the paternal sDMR and Zdbf2 expression. In extraembryonic lineages, including the placenta, the maternal gDMR and Liz expression are maintained instead (Kobayashi et al., 2009; Duffie et al., 2014). In humans, decreases in placental ZDBF2 expression have been associated with intrauterine growth restriction (Monteagudo-Sánchez et al., 2019).
The impact of imprinted genes in placenta has been well studied in mice. Mouse models have demonstrated that generally deletions of genes which are paternally expressed, including Igf2, Peg1 and Peg3, reduce placental size and increase the incidence of IUGR (DeChiara et al., 1991; Lefebvre et al., 1998). The opposite happens with deletions on maternally expressed genes, such as Grb10 and Phlda2, causing fetal over-growth (Charalambous et al., 2003; Salas et al., 2004).
In humans, IUGR has been associated with placental expression of the imprinted GPR1-AS1 and ZDBF2 genes (Monteagudo-Sánchez et al., 2019). It is also associated with increases in HYMA1 expression, and, in girls only, lower PLAGL1 expression (Iglesias-Platas et al., 2014). PLAGL1 is a transcription factor and its expression changes impact a network of genes downstream.
The placenta is particularly interesting in the context of DOHaD because early environment can influence placental function, impacting development and pregnancy outcomes. Further, the placenta has a large impact on fetal development and placenta-related adverse pregnancy outcomes can influence later life disease. Placental function can be influenced by many environmental exposures, such as maternal smoking and obesity (Reijnders et al., 2019; Scott et al., 2022). Increased physical activity before and during early pregnancy can reduce the incidence of preeclampsia (Aune et al., 2014). ART is associated with increased risk of IUGR or premature birth (Camprubí et al., 2013).
Adequate nutrition in the first trimester and periconceptional folic acid supplementation are associated with lower resistance of the uterine and umbilical arteries in second and third trimesters, in which high resistance can be associated with preeclampsia and fetal growth restriction risk (Reijnders et al., 2019). The effect of the previously discussed Dutch Famine has also been studied, with famine exposure reducing placenta weight, birthweight and increasing preterm births, stillbirths and neonatal death (Susser and Stein, 1994; Lumey, 1998). A study in Gambia found that periconceptional multiple micronutrient supplementation improved placenta vascular function (Owens et al., 2015).
A systematic analysis of the effects of micronutrient supplementation looked at six placenta-related outcomes: preeclampsia, small for gestational age, low birthweight, preterm birth, stillbirth and maternal death (Kinshella et al., 2021). No single factor affected all six, but one or more were affected by vitamins C and E, vitamin D, vitamin D and calcium co-supplementation, calcium, iron and/or folic acid, zinc and multiple micronutrient supplementation. In another review, 14 of 25 nutritional factors reviewed were significantly associated with preeclampsia incidence, including vitamin D deficiency and high serum iron (Kinshella et al., 2022).
In rodents, placenta methylation is affected by maternal diet. A gestational high-fat diet leads to placenta global hypomethylation and changes in methylation at enzymes involved in epigenetic machinery and expression (Gallou-Kabani et al., 2010). In mice, folate deficiency leads to lower overall placental methylation (Mahajan et al., 2019). There are also changes at many genes in the transcriptome and methylome, and the genes vary with sex, even being involved in different biological functions (Gabory et al., 2012).
Knowledge on the effect of diet on human placental methylation is limited, but changes in methylation linked to many other maternal exposures have been more widely studied. For example, ART has been linked to methylation changes in placenta, specifically reduced methylation at the H19 and MEST DMRs with an increase in H19 expression (Nelissen et al., 2013). In another study, ART placentas had increased expression of INSIG1 and SREBF1, linked to cholesterol metabolism, with decreased methylation (Lou et al., 2014). A recent review looked at other exposures associated with methylation changes in placentas both globally and at specific sites, including air pollution, maternal smoking, bisphenol A and trace metals (Mortillo and Marsit, 2022). They observed that the effects are generally stronger when the exposure occurs in the first trimester. In other studies, maternal depression and anxiety have also been linked to a decrease in overall placenta methylation levels, with some changes in gene expression (Chen et al., 2014).
There are some studies on the effects of maternal diet on placental methylation in humans, for example, prenatal vitamin intake has been associated with a small reduction in placental methylation, including at sites associated with neuronal developmental pathways (Dou et al., 2022). Looking at specific components, folate has been widely studied during pregnancy, as its deficiency is associated with low birth weight, preterm birth, spontaneous abortion and birth outcomes including neural tube defects (Tamura and Picciano, 2006; Fekete et al., 2010; Greenberg et al., 2016; Hoffbrand et al., 2014). One-carbon components, including folate, have been studied in complicated pregnancies to see how they affect DNA methylation. There are three components important for folate transport through the placenta: Three main components participate in this process: folate receptor α (FRα), reduced folate carrier (RFC) and proton-coupled folate transporter (PCFT) (Solanky et al., 2010; Castano-Morena et al., 2020). FRα mRNA was lower in preterm placentas compared to term, which correlated with increased methylation on the fetal side (chorionic plate). The increased methylation in preterm placentas also correlated with higher folate and lower B12 concentrations in the cord blood (Piñuñuri et al., 2020). The syncytiotrophoblasts in IUGR placentas had lower folate update and reduced levels of RFC but not FR-α (Chen et al., 2018).
In preterm placentas, expression of MAT2A and AHCY, enzymes involved in producing SAM, is higher in preterm placentas, and a SAH:SAM ratio, giving a lower methylation potential (Khot et al., 2017). These studies highlight how maternal nutrition can have an effect on methylation through one-carbon cycle components. However, not much is known still about how the overall methylation changes are acting to alter the function of the placenta.
The placental epigenome is hypomethylated and hypervariable so has the potential for creating large inter-individual differences. This gives it great potential as a tissue to investigate the effects of environmental exposures on DNA methylation. So far, there has been limited study on the effects non-nutritional exposures and no study on the correlation in methylation at MEs between embryonic and extra-embryonic tissues. If MEs in placenta and somatic tissue are matched, this could suggest MEs are established before the trophectoderm lineage separates. Many of the MEs identified as responsive to season of conception in The Gambia overlap placenta DMRs which could suggest a mechanistic link, perhaps an issue with residual DMRs rather than somatically acquired. However, this is unlikely to explain all MEs, those that are not related to gDMRs must have a different mechanism of variability.
MEs are also known to disappear with age, so a readily obtainable early life tissue such as the placenta is an excellent place to look. This would also diversify the tissues used, as MEs are commonly identified only in cord blood. It is possible that more variation may be maintained in placenta as it is only needed for 9 months so is not regulated as highly as somatic tissue, or the unique epigenetic landscape may play an important role. Perhaps if there is a correlation, the placenta epigenome and transcriptome would serve as a good biomarker for later life disease. There is need for more work on MEs in humans to determine the effects of genetic variation and their relationship to environmental exposures and health.
RS wrote the first draft of the manuscript.
This work was funded by the Medical Research Council (MR/T032863/1), UKRI Biotechnology and Biological Sciences Research Council (BB/V016156/1) and the BBSRC Norwich Research Park Biosciences Doctoral Training Partnership (BB/T008717/1).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aune, D., Saugstad, O. D., Henriksen, T., and Tonstad, S. (2014). Physical activity and the risk of preeclampsia: A systematic review and meta-analysis. Epidemiol. Camb. Mass.) 25 (3), 331–343. doi:10.1097/ede.0000000000000036
Barbaux, S., Gascoin-Lachambre, G., Buffat, C., Monnier, P., Mondon, F., Tonanny, M. B., et al. (2012). A genome-wide approach reveals novel imprinted genes expressed in the human placenta. Epigenetics 7 (9), 1079–1090. doi:10.4161/epi.21495
Barker, D. J., Larsen, G., Osmond, C., Thornburg, K. L., Kajantie, E., and Eriksson, J. G. (2012). The placental origins of sudden cardiac death. Int. J. Epidemiol. 41 (5), 1394–1399. doi:10.1093/ije/dys116
Barker, D. J., Osmond, C., Thornburg, K. L., Kajantie, E., and Eriksson, J. G. (2013). The shape of the placental surface at birth and colorectal cancer in later life. Am. J. Hum. Biol. 25 (4), 566–568. doi:10.1002/ajhb.22409
Barker, D. J. P. (1990). The fetal and infant origins of adult disease. BMJ 301 (6761), 1111. doi:10.1136/bmj.301.6761.1111
Bertozzi, T. M., Becker, J. L., Blake, G. E. T., Bansal, A., Nguyen, D. K., Fernandez-Twinn, D. S., et al. (2021). Variably methylated retrotransposons are refractory to a range of environmental perturbations. Nat. Genet. 53 (8), 1233–1242. doi:10.1038/S41588-021-00898-9
Blond, J. L., Besème, F., Duret, L., Bouton, O., Bedin, F., Perron, H., et al. (1999). Molecular characterization and placental expression of HERV-W, a new human endogenous retrovirus family. J. Virology 73 (2), 1175–1185. doi:10.1128/jvi.73.2.1175-1185.1999
Bourc’his, D., and Bestor, T. H. (2004). Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 431, 96–99. doi:10.1038/nature02886
Bourque, G., Burns, K. H., Gehring, M., Gorbunova, V., Seluanov, A., Hammell, M., et al. (2018). Ten things you should know about transposable elements. Genome Biol. 19 (1), 199. doi:10.1186/s13059-018-1577-z
Brosens, I., Pijnenborg, R., Vercruysse, L., and Romero, R. (2011). The “Great Obstetrical Syndromes” are associated with disorders of deep placentation. Am. J. Obstetrics Gynecol. 204 (3), 193–201. doi:10.1016/j.ajog.2010.08.009
Burton, G. J., and Jauniaux, E. (2018). Pathophysiology of placental-derived fetal growth restriction. Am. J. Obstetrics Gynecol. 218 (2S), S745–S761. doi:10.1016/j.ajog.2017.11.577
Camprubí, C., Iglesias-Platas, I., Martin-Trujillo, A., Salvador-Alarcon, C., Rodriguez, M., Barredo, D. R., et al. (2013). Stability of genomic imprinting and gestational-age dynamic methylation in complicated pregnancies conceived following assisted reproductive technologies. Biol. Reproduction 89 (3), 50. doi:10.1095/biolreprod.113.108456
Candler, T., Kessler, N. J., Gunasekara, C. J., Ward, K. A., James, P., Laritsky, E., et al. (2021). DNA methylation at a nutritionally sensitive region of the PAX8 gene is associated with thyroid volume and function in Gambian children. Sci. Adv. 7 (45), 1561. doi:10.1126/sciadv.abj1561
Castaño-Moreno, R., Piñuñuri, E. R., Peñailillo, R., Casanello, P., Llanos, M., and Ronco, A. M. (2020). Folates transport in placentas. Mol. Nutr. Vitam. 2020, 345–365. doi:10.1016/B978-0-12-811907-5.00009-9
Ceelen, M., van Weissenbruch, M. M., Vermeiden, J. P., van Leeuwen, F. E., and Delemarre-van de Waal, H. A. (2008). Cardiometabolic differences in children born after in vitro fertilization: Follow-up study. J. Clin. Endocrinol. Metabolism 93 (5), 1682–1688. doi:10.1210/jc.2007-2432
Ceesay, S. M., Prentice, A. M., Cole, T. J., Foord, F., Weaver, L. T., Poskitt, E. M., et al. (1997). Effects on birth weight and perinatal mortality of maternal dietary supplements in rural Gambia: 5 year randomised controlled trial. BMJ 315 (7111), 786–790. doi:10.1136/bmj.315.7111.786
Charalambous, M., Prentice, A. M., Cole, T. J., Foord, F., Weaver, L. T., Poskitt, E. M., et al. (2003). Disruption of the imprinted Grb10 gene leads to disproportionate overgrowth by an Igf2-independent mechanism. Proc. Natl. Acad. Sci. 100 (14), 8292–8297. doi:10.1073/pnas.1532175100
Chen, J., Li, Q., Rialdi, A., Mystal, E., Ly, J., Finik, J., et al. (2014). Influences of maternal stress during pregnancy on the epigenome: Comparison of placenta and umbilical cord blood. J. Depress. Anxiety 3 (2), 152. doi:10.4172/2167-1044.1000152
Chen, Y. Y., Gupta, M. B., Grattton, R., Powell, T. L., and Jansson, T. (2018). Down-regulation of placental folate transporters in intrauterine growth restriction. J. Nutr. Biochem. 59, 136–141. doi:10.1016/j.jnutbio.2018.06.003
Choux, C., Petazzi, P., Sanchez-Delgado, M., Hernandez Mora, J. R., Monteagudo, A., Sagot, P., et al. (2020). The hypomethylation of imprinted genes in IVF/ICSI placenta samples is associated with concomitant changes in histone modifications. Epigenetics 15 (12), 1386–1395. doi:10.1080/15592294.2020.1783168
Cole, M. D., Ono, M., and Huang, R. C. (1981). Terminally redundant sequences in cellular intracisternal A-particle genes. J. Virology 38 (2), 680–687. doi:10.1128/JVI.38.2.680-687.1981
Cooney, C. A., Dave, A. A., and Wolff, G. L. (2002). Maternal methyl supplements in mice affect epigenetic variation and DNA methylation of offspring. J. Nutr. 132 (8), 2393S–2400S. doi:10.1093/jn/132.8.2393S
Cooper, W. N., Khulan, B., Owens, S., Elks, C. E., Seidel, V., Prentice, A. M., et al. (2012). DNA methylation profiling at imprinted loci after periconceptional micronutrient supplementation in humans: Results of a pilot randomized controlled trial. FASEB J. 26 (5), 1782–1790. doi:10.1096/fj.11-192708
Court, F., Tayama, C., Romanelli, V., Martin-Trujillo, A., Iglesias-Platas, I., Okamura, K., et al. (2014). Genome-wide parent-of-origin DNA methylation analysis reveals the intricacies of human imprinting and suggests a germline methylation-independent mechanism of establishment. Genome Res. 24 (4), 554–569. doi:10.1101/gr.164913.113
Cropley, J. E., Suter, C. M., Beckman, K. B., and Martin, D. I. (2006). Germ-line epigenetic modification of the murine Avy allele by nutritional supplementation. Proc. Natl. Acad. Sci. 103 (46), 17308–17312. doi:10.1073/pnas.0607090103
Curhan, G. C., Chertow, G. M., Willett, W. C., Spiegelman, D., Colditz, G. A., Manson, J. E., et al. (1996). Birth weight and adult hypertension and obesity in women. Circulation 94 (6), 1310–1315. doi:10.1161/01.cir.94.6.1310
DeChiara, T. M., Robertson, E. J., and Efstratiadis, A. (1991). Parental imprinting of the mouse insulin-like growth factor II gene. Cell. 64 (4), 849–859. doi:10.1016/0092-8674(91)90513-X
Denisenko, O., Lucas, E. S., Sun, C., Watkins, A. J., Mar, D., Bomsztyk, K., et al. (2016). Regulation of ribosomal RNA expression across the lifespan is fine-tuned by maternal diet before implantation. Biochimica biophysica acta 1859 (7), 906–913. doi:10.1016/j.bbagrm.2016.04.001
Dickies, M. M. (1962). A new viable yellow mutation in the house mouse. J. Hered. 53 (2), 84–86. doi:10.1093/oxfordjournals.jhered.a107129
Dominguez-Salas, P., Moore, S. E., Cole, D., da Costa, K. A., Cox, S. E., Dyer, R. A., et al. (2013). DNA methylation potential: Dietary intake and blood concentrations of one-carbon metabolites and cofactors in rural african women. Am. J. Clin. Nutr. 97 (6), 1217–1227. doi:10.3945/ajcn.112.048462
Dominguez-Salas, P., Moore, S. E., Cole, D., da Costa, K. A., Cox, S. E., Dyer, R. A., et al. (2014). Maternal nutrition at conception modulates DNA methylation of human metastable epialleles. Nat. Commun. 5, 3746. doi:10.1038/ncomms4746
Dou, J. F., Middleton, L. Y. M., Zhu, Y., Benke, K. S., Feinberg, J. I., Croen, L. A., et al. (2022). Prenatal vitamin intake in first month of pregnancy and DNA methylation in cord blood and placenta in two prospective cohorts. Epigenetics Chromatin 15 (1), 28. doi:10.1186/S13072-022-00460-9
Ducker, G. S., and Rabinowitz, J. D. (2017). One-carbon metabolism in health and disease. Cell. Metab. 25 (1), 27–42. doi:10.1016/j.cmet.2016.08.009
Duffié, R., Ajjan, S., Greenberg, M. V., Zamudio, N., Escamilla del Arenal, M., Iranzo, J., et al. (2014). The Gpr1/Zdbf2 locus provides new paradigms for transient and dynamic genomic imprinting in mammals. Genes. & Dev. 28 (5), 463–478. doi:10.1101/gad.232058.113
Duhl, D. M., Vrieling, H., Miller, K. A., Wolff, G. L., and Barsh, G. S. (1994) ‘Neomorphic agouti mutations in obese yellow mice’, Nat. Genet. 8 (1), 59–65. doi:10.1038/ng0994-59
Dunn-Fletcher, C. E., Muglia, L. M., Pavlicev, M., Wolf, G., Sun, M. A., Hu, Y. C., et al. (2018). Anthropoid primate-specific retroviral element THE1B controls expression of CRH in placenta and alters gestation length. PLoS Biol. 16 (9), e2006337. doi:10.1371/journal.pbio.2006337
Eckert, J. J., Porter, R., Watkins, A. J., Burt, E., Brooks, S., Leese, H. J., et al. (2012). Metabolic induction and early responses of mouse blastocyst developmental programming following maternal low protein diet affecting life-long health. PLoS One 7 (12), e52791. doi:10.1371/journal.pone.0052791
England-Mason, G., G., Merrill, S. M., Gladish, N., Moore, S. R., Giesbrecht, G. F., Letourneau, N., et al. (2022). Prenatal exposure to phthalates and peripheral blood and buccal epithelial DNA methylation in infants: An epigenome-wide association study. Environ. Int. 163, 107183. doi:10.1016/j.envint.2022.107183
Faulk, C., Kim, J. H., Anderson, O. S., Nahar, M. S., Jones, T. R., Sartor, M. A., et al. (2016). Detection of differential DNA methylation in repetitive DNA of mice and humans perinatally exposed to bisphenol A. Epigenetics 11 (7), 489–500. doi:10.1080/15592294.2016.1183856
Fedoriw, A. M., Stein, P., Svoboda, P., Schultz, R. M., and Bartolomei, M. S. (2004). Transgenic RNAi reveals essential function for CTCF in H19 gene imprinting. Science 303 (5655), 238–240. doi:10.1126/science.1090934
Fekete, K., Berti, C., Cetin, I., Hermoso, M., Koletzko, B. V., and Decsi, T. (2010). Perinatal folate supply: Relevance in health outcome parameters. Maternal Child Nutr. 6, 23–38. doi:10.1111/J.1740-8709.2010.00261.X
Fleming, T. P., Watkins, A. J., Velazquez, M. A., Mathers, J. C., Prentice, A. M., Stephenson, J., et al. (2018). Origins of lifetime health around the time of conception: Causes and consequences. Lancet 391, 1842–1852. doi:10.1016/S0140-6736(18)30312-X
Frost, J. M., Amante, S. M., Okae, H., Jones, E. M., and Ashley, B. (2023). Regulation of human trophoblast gene expression by endogenous retroviruses. Nat. Struct. Mol. Biol. 30 (4), 527–538. doi:10.1038/s41594-023-00960-6
Fuke, C., Shimabukuro, M., Petronis, A., Sugimoto, J., Oda, T., Miura, K., et al. (2004). Age related changes in 5-methylcytosine content in human peripheral leukocytes and placentas: An HPLC-based study. Ann. Hum. Genet. 68 (3), 196–204. doi:10.1046/J.1529-8817.2004.00081.X
Gabory, A., Ferry, L., Fajardy, I., Jouneau, L., Gothié, J. D., Vigé, A., et al. (2012). Maternal diets trigger sex-specific divergent trajectories of gene expression and epigenetic systems in mouse placenta. PLoS One 7 (11), 47986. doi:10.1371/journal.pone.0047986
Gahurova, L., Tomizawa, S. I., Smallwood, S. A., Stewart-Morgan, K. R., Saadeh, H., Kim, J., et al. (2017). Transcription and chromatin determinants of de novo DNA methylation timing in oocytes. Epigenetics Chromatin 10 (1), 25. doi:10.1186/s13072-017-0133-5
Gallou-Kabani, C., Gabory, A., Tost, J., Karimi, M., Mayeur, S., Lesage, J., et al. (2010). Sex- and diet-specific changes of imprinted gene expression and DNA methylation in mouse placenta under a high-fat diet. PLoS One 5 (12), 14398. doi:10.1371/journal.pone.0014398
Gao, Y., He, Z., Wang, Z., Luo, Y., Sun, H., Zhou, Y., et al. (2012). Increased expression and altered methylation of HERVWE1 in the human placentas of smaller fetuses from monozygotic, dichorionic, discordant twins. PLoS One 7 (3), 33503. doi:10.1371/journal.pone.0033503
Gao, Y., Sheng, C., Xie, R. H., Sun, W., Asztalos, E., Moddemann, D., et al. (2016). New perspective on impact of folic acid supplementation during pregnancy on neurodevelopment/autism in the offspring children - a systematic review. PloS One 11 (11), e0165626. doi:10.1371/journal.pone.0165626
Geneimprint (2023). Geneimprint. Retrieved from: https://www.geneimprint.com.
Graham, W., Woodd, S., Byass, P., Filippi, V., Gon, G., Virgo, S., et al. (2016). Diversity and divergence: The dynamic burden of poor maternal health. Lancet 388 (10056), 2164–2175. doi:10.1016/S0140-6736(16)31533-1
Greenberg, M. V. C., Glaser, J., Borsos, M., Marjou, F. E., Walter, M., Teissandier, A., et al. (2016). Transient transcription in the early embryo sets an epigenetic state that programs postnatal growth. Nat. Genet. 49 (1), 110–118. doi:10.1038/ng.3718
Greer, E. L., Blanco, M. A., Gu, L., Sendinc, E., Liu, J., Aristizábal-Corrales, D., et al. (2015). DNA methylation on N6-adenine in C. elegans. Cell. 161 (4), 868–878. doi:10.1016/j.cell.2015.04.005
Guo, F., Yan, L., Guo, H., Li, L., Hu, B., Zhao, Y., et al. (2015). The transcriptome and DNA methylome landscapes of human primordial germ cells. Cell. 161 (6), 1437–1452. doi:10.1016/j.cell.2015.05.015
Guo, H., Zhu, P., Yan, L., Li, R., Hu, B., Lian, Y., et al. (2014). The DNA methylation landscape of human early embryos. Nature 511 (7511), 606–610. doi:10.1038/nature13544
Hackett, J. A., Sengupta, R., Zylicz, J. J., Murakami, K., Lee, C., Down, T. A., et al. (2013). Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science 339 (6118), 448–452. doi:10.1126/science.1229277
Hamada, H., Okae, H., Toh, H., Chiba, H., Hiura, H., Shirane, K., et al. (2016). Allele-specific methylome and transcriptome analysis reveals widespread imprinting in the human placenta. Am. J. Hum. Genet. 99 (5), 1045–1058. doi:10.1016/J.AJHG.2016.08.021
Hanna, C. W., Peñaherrera, M. S., Saadeh, H., Andrews, S., McFadden, D. E., Kelsey, G., et al. (2016). Pervasive polymorphic imprinted methylation in the human placenta. Genome Res. 26 (6), 756–767. doi:10.1101/gr.196139.115
Harkness, U. F., and Mari, G. (2004). Diagnosis and management of intrauterine growth restriction. Clin. Perinatology 31 (4), 743–764. doi:10.1016/j.clp.2004.06.006
Heijmans, B. T., Tobi, E. W., Stein, A. D., Putter, H., Blauw, G. J., Susser, E. S., et al. (2008). Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc. Natl. Acad. Sci. U. S. A. 105 (44), 17046–17049. doi:10.1073/pnas.0806560105
Hermann, A., Goyal, R., and Jeltsch, A. (2004). The Dnmt1 DNA-(cytosine-C5)-methyltransferase methylates DNA processively with high preference for hemimethylated target sites. J. Biol. Chem. 279 (46), 48350–48359. doi:10.1074/jbc.M403427200
Hernandez Mora, J. R., Sanchez-Delgado, M., Petazzi, P., Moran, S., Esteller, M., Iglesias-Platas, I., et al. (2018). Profiling of oxBS-450K 5-hydroxymethylcytosine in human placenta and brain reveals enrichment at imprinted loci. Epigenetics 13 (2), 182–191. doi:10.1080/15592294.2017.1344803
Hernandez-Vargas, H., Castelino, J., Silver, M. J., Dominguez-Salas, P., Cros, M. P., Durand, G., et al. (2015). Exposure to aflatoxin B1 in utero is associated with DNA methylation in white blood cells of infants in the Gambia. Int. J. Epidemiol. 44 (4), 1238–1248. doi:10.1093/ije/dyv027
Hirasawa, R., Chiba, H., Kaneda, M., Tajima, S., Li, E., Jaenisch, R., et al. (2008). Maternal and zygotic Dnmt1 are necessary and sufficient for the maintenance of DNA methylation imprints during preimplantation development. Genes. & Dev. 22 (12), 1607–1616. doi:10.1101/gad.1667008
Hiura, H., Obata, Y., Komiyama, J., Shirai, M., and Kono, T. (2006). Oocyte growth-dependent progression of maternal imprinting in mice. Genes. Cells 11 (4), 353–361. doi:10.1111/J.1365-2443.2006.00943.X
Hodgetts, V. A., Morris, R. K., Francis, A., Gardosi, J., and Ismail, K. M. (2015). Effectiveness of folic acid supplementation in pregnancy on reducing the risk of small-for-gestational age neonates: A population study, systematic review and meta-analysis. BJOG 122 (4), 478–490. doi:10.1111/1471-0528.13202
Hoffbrand, A. V., Martin, J., John, S., and Scott, M. (2014). Professor John Scott, folate and neural tube defects. Br. J. Haematol. 164 (4), 496–502. doi:10.1111/bjh.12654
Holland, M. L., Lowe, R., Caton, P. W., Gemma, C., Carbajosa, G., Danson, A. F., et al. (2016). Early-life nutrition modulates the epigenetic state of specific rDNA genetic variants in mice. Science 353 (6298), 495–498. doi:10.1126/science.aaf7040
Horvath, S. (2013). DNA methylation age of human tissues and cell types. Genome Biol. 14 (10), R115–R120. doi:10.1186/gb-2013-14-10-r115
Iglesias-Platas, I., Martin-Trujillo, A., Petazzi, P., Guillaumet-Adkins, A., Esteller, M., and Monk, D. (2014). Altered expression of the imprinted transcription factor PLAGL1 deregulates a network of genes in the human IUGR placenta. Hum. Mol. Genet. 23 (23), 6275–6285. doi:10.1093/hmg/ddu347
Illingworth, R. S., Gruenewald-Schneider, U., Webb, S., Kerr, A. R., James, K. D., Turner, D. J., et al. (2010). Orphan CpG islands identify numerous conserved promoters in the mammalian genome. PLoS Genet. 6 (9), 1001134. doi:10.1371/journal.pgen.1001134
Imakawa, K., Nakagawa, S., and Miyazawa, T. (2015). Baton pass hypothesis: Successive incorporation of unconserved endogenous retroviral genes for placentation during mammalian evolution. Genes. Cells 20 (10), 771–788. doi:10.1111/gtc.12278
Imbeault, M., Helleboid, P. Y., and Trono, D. (2017). KRAB zinc-finger proteins contribute to the evolution of gene regulatory networks. Nature 543 (7646), 550–554. doi:10.1038/nature21683
Inoue, A., Jiang, L., Lu, F., Suzuki, T., and Zhang, Y. (2017). Maternal H3K27me3 controls DNA methylation-independent imprinting. Nature 547 (7664), 419–424. doi:10.1038/nature23262
James, P., Sajjadi, S., Tomar, A. S., Saffari, A., Fall, C. H. D., Prentice, A. M., et al. (2018). Candidate genes linking maternal nutrient exposure to offspring health via DNA methylation: A review of existing evidence in humans with specific focus on one-carbon metabolism. Int. J. Epidemiol. 47 (6), 1910–1937. doi:10.1093/ije/dyy153
James, P. T., Dominguez-Salas, P., Hennig, B. J., Moore, S. E., Prentice, A. M., and Silver, M. J. (2019). Maternal one-carbon metabolism and infant DNA methylation between contrasting seasonal environments: A case study from the Gambia. Curr. Dev. Nutr. 3 (1), nzy082. doi:10.1093/cdn/nzy082
Janssen, A. B., Tunster, S. J., Savory, N., Holmes, A., Beasley, J., Parveen, S. A., et al. (2015). Placental expression of imprinted genes varies with sampling site and mode of delivery. Placenta 36 (8), 790–795. doi:10.1016/j.placenta.2015.06.011
Kaluscha, S., Domcke, S., Wirbelauer, C., Stadler, M. B., Durdu, S., Burger, L., et al. (2022) ‘Evidence that direct inhibition of transcription factor binding is the prevailing mode of gene and repeat repression by DNA methylation’, Nat. Genet. 54 (12), 1895–1906. doi:10.1038/s41588-022-01241-6
Kaneda, M., Okano, M., Hata, K., Sado, T., Tsujimoto, N., Li, E., et al. (2004). Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature 429 (6994), 900–903. doi:10.1038/nature02633
Kazachenka, A., Bertozzi, T. M., Sjoberg-Herrera, M. K., Walker, N., Gardner, J., Gunning, R., et al. (2018). Identification, characterization, and heritability of murine metastable epialleles: Implications for non-genetic inheritance. Cell. 175 (5), 1259–1271. doi:10.1016/j.cell.2018.09.043
Kessler, N. J., Waterland, R. A., Prentice, A. M., and Silver, M. J. (2018). Establishment of environmentally sensitive DNA methylation states in the very early human embryo. Sci. Adv. 4 (7), eaat2624. doi:10.1126/sciadv.aat2624
Khot, V. V., Chavan-Gautam, P., Mehendale, S., and Joshi, S. R. (2017). Variable methylation potential in preterm placenta: Implication for epigenetic programming of the offspring. Reprod. Sci. 24 (6), 891–901. doi:10.1177/1933719116671001
Khulan, B., Cooper, W. N., Skinner, B. M., Bauer, J., Owens, S., Prentice, A. M., et al. (2012). Periconceptional maternal micronutrient supplementation is associated with widespread gender related changes in the epigenome: A study of a unique resource in the Gambia. Hum. Mol. Genet. 21 (9), 2086–2101. doi:10.1093/hmg/dds026
Kinshella, M. L. W., mar, S., Scherbinsky, K., Vidler, M., Magee, L. A., von Dadelszen, P., et al. (2021). Effects of maternal nutritional supplements and dietary interventions on placental complications: An umbrella review, meta-analysis and evidence map. Nutrients 13 (2), 1–29. doi:10.3390/nu13020472
Kinshella, M. L. W., Pickerill, K., Bone, J. N., Prasad, S., Campbell, O., Vidler, M., et al. (2022). An evidence review and nutritional conceptual framework for pre-eclampsia prevention. Br. J. Nutr. 2022, 1–12. doi:10.1017/S0007114522003889
Kobayashi, H., Yamada, K., Morita, S., Hiura, H., Fukuda, A., Kagami, M., et al. (2009). Identification of the mouse paternally expressed imprinted gene Zdbf2 on chromosome 1 and its imprinted human homolog ZDBF2 on chromosome 2. Genomics 93 (5), 461–472. doi:10.1016/j.ygeno.2008.12.012
Konje, J. C., and Cade, J. E. (2008). Maternal caffeine intake during pregnancy and risk of fetal growth restriction: A large prospective observational study. BMJ 337 (7682), 1334–1338. doi:10.1136/bmj.a2332
Kouzarides, T. (2007). Chromatin modifications and their function. Cell. 128, 693–705. doi:10.1016/j.cell.2007.02.005
Kuehnen, P., Mischke, M., Wiegand, S., Sers, C., Horsthemke, B., Lau, S., et al. (2012). An Alu element-associated hypermethylation variant of the POMC gene is associated with childhood obesity. PLoS Genet. 8 (3), e1002543. doi:10.1371/journal.pgen.1002543
Kühnen, P., Handke, D., Waterland, R. A., Hennig, B. J., Silver, M., Fulford, A. J., et al. (2016). Interindividual variation in DNA methylation at a putative POMC metastable epiallele is associated with obesity. Cell. Metab. 24 (3), 502–509. doi:10.1016/j.cmet.2016.08.001
Kulkarni, A., Chavan-Gautam, P., Mehendale, S., Yadav, H., and Joshi, S. (2011). Global DNA methylation patterns in placenta and its association with maternal hypertension in pre-eclampsia. DNA Cell. Biol. 30 (2), 79–84. doi:10.1089/dna.2010.1084
Küpers, L. K., Monnereau, C., Sharp, G. C., Yousefi, P., Salas, L. A., Ghantous, A., et al. (2019). Meta-analysis of epigenome-wide association studies in neonates reveals widespread differential DNA methylation associated with birthweight. Nat. Commun. 10 (1), 1893. doi:10.1038/s41467-019-09671-3
Kwong, W. Y., Wild, A. E., Roberts, P., Willis, A. C., and Fleming, T. P. (2000). Maternal undernutrition during the preimplantation period of rat development causes blastocyst abnormalities and programming of postnatal hypertension. Development 127 (19), 4195–4202. doi:10.1242/dev.127.19.4195
Lazaraviciute, G., Kauser, M., Bhattacharya, S., Haggarty, P., and Bhattacharya, S. (2014). A systematic review and meta-analysis of DNA methylation levels and imprinting disorders in children conceived by IVF/ICSI compared with children conceived spontaneously. Hum. Reprod. Update 20 (6), 840–852. doi:10.1093/humupd/dmu033
Lee, Y., Choufani, S., Weksberg, R., Wilson, S. L., Yuan, V., Burt, A., et al. (2019). Placental epigenetic clocks: estimating gestational age using placental DNA methylation levels. Aging 11 (12), 4238. doi:10.18632/aging.102049
Lefebvre, L., Viville, S., Barton, S. C., Ishino, F., Keverne, E. B., and Surani, M. A. (1998). Abnormal maternal behaviour and growth retardation associated with loss of the imprinted gene Mest. Nat. Genet. 20 (2), 163–169. doi:10.1038/2464
Li, B., Carey, M., and Workman, J. L. (2007). The role of chromatin during transcription. Cell. 128, 707–719. doi:10.1016/j.cell.2007.01.015
Li, X., Ito, M., Zhou, F., Youngson, N., Zuo, X., Leder, P., et al. (2008). A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev. Cell. 15 (4), 547–557. doi:10.1016/j.devcel.2008.08.014
Lister, R., Pelizzola, M., Dowen, R. H., Hawkins, R. D., Hon, G., Tonti-Filippini, J., et al. (2009). Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462 (7271), 315–322. doi:10.1038/nature08514
Liu, X., Ito, M., Zhou, F., Youngson, N., Zuo, X., Leder, P., et al. (2020). N6-methyladenine is incorporated into mammalian genome by DNA polymerase. Cell. Res. 31 (1), 94–97. doi:10.1038/s41422-020-0317-6
Lopata, A., Oliva, K., Stanton, P. G., and Robertson, D. M. (1997). Analysis of chorionic gonadotrophin secreted by cultured human blastocysts. Mol. Hum. Reprod. 3 (6), 517–521. doi:10.1093/molehr/3.6.517
Lou, H., Le, F., Zheng, Y., Li, L., Wang, L., Wang, N., et al. (2014). Assisted reproductive technologies impair the expression and methylation of insulin-induced gene 1 and sterol regulatory element-binding factor 1 in the fetus and placenta. Fertil. Steril. 101 (4), 974–980. doi:10.1016/j.fertnstert.2013.12.034
Lu, A. T., Quach, A., Wilson, J. G., Reiner, A. P., Aviv, A., Raj, K., et al. (2019). DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging 11 (2), 303. doi:10.18632/aging.101684
Lumey, L. H. (1998). Compensatory placental growth after restricted maternal nutrition in early pregnancy. Placenta 19 (1), 105–111. doi:10.1016/S0143-4004(98)90105-9
Lumey, L. H., Stein, A. D., and Susser, E. (2011). Prenatal famine and adult health. Annu. Rev. Public Health 32, 237–262. doi:10.1146/annurev-publhealth-031210-101230
Macleod, D., Charlton, J., Mullins, J., and Bird, A. P. (1994). Sp1 sites in the mouse aprt gene promoter are required to prevent methylation of the CpG island. Genes. & Dev. 8 (19), 2282–2292. doi:10.1101/gad.8.19.2282
Mahajan, A., Sapehia, D., Thakur, S., Mohanraj, P. S., Bagga, R., and Kaur, J. (2019). Effect of imbalance in folate and vitamin B12 in maternal/parental diet on global methylation and regulatory miRNAs. Sci. Rep. 9 (1). doi:10.1038/s41598-019-54070-9
Marchi, J., Berg, M., Dencker, A., Olander, E. K., and Begley, C. (2015). Risks associated with obesity in pregnancy, for the mother and baby: A systematic review of reviews. Obes. Rev. 16 (8), 621–638. doi:10.1111/obr.12288
Mason, J. B. (2003). Biomarkers of nutrient exposure and status in one-carbon (methyl) metabolism. J. Nutr. 133 (3), 941S–947S. doi:10.1093/jn/133.3.941S
Mastroiacovo, P., and Leoncini, E. (2011). More folic acid, the five questions: Why, who, when, how much, and how. BioFactors 37 (4), 272–279. doi:10.1002/biof.172
Matousková, M., Blazková, J., Pajer, P., Pavlícek, A., and Hejnar, J. (2006). CpG methylation suppresses transcriptional activity of human syncytin-1 in non-placental tissues. Exp. Cell. Res. 312 (7), 1011–1020. doi:10.1016/j.yexcr.2005.12.010
Mayne, B. T., Leemaqz, S. Y., Smith, A. K., Breen, J., Roberts, C. T., and Bianco-MiT, T. (2017). Accelerated placental aging in early onset preeclampsia pregnancies identified by DNA methylation. Epigenomics 9 (3), 279–289. doi:10.2217/epi-2016-0103
McMahon, K. W., Karunasena, E., and Ahuja, N. (2017). The roles of DNA methylation in the stages of cancer. Cancer J. 23 (5), 257–261. doi:10.1097/ppo.0000000000000279
Mellén, M., Ayata, P., Dewell, S., Kriaucionis, S., and Heintz, N. (2012). MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 151 (7), 1417–1430. doi:10.1016/j.cell.2012.11.022
Mi, S., Lee, X., Li, X., Veldman, G. M., Finnerty, H., Racie, L., et al. (2000). Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 403 (6771), 785–789. doi:10.1038/35001608
Mikkelsen, T. S., Ku, M., Jaffe, D. B., Issac, B., Lieberman, E., Giannoukos, G., et al. (2007). Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448 (7153), 553–560. doi:10.1038/nature06008
Mohn, F., Weber, M., Rebhan, M., Roloff, T. C., Richter, J., Stadler, M. B., et al. (2008). Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol. Cell. 30 (6), 755–766. doi:10.1016/j.molcel.2008.05.007
Monk, D., Arnaud, P., Apostolidou, S., Hills, F. A., Kelsey, G., Stanier, P., et al. (2006). Limited evolutionary conservation of imprinting in the human placenta. Proc. Natl. Acad. Sci. U. S. A. 103 (17), 6623–6628. doi:10.1073/pnas.0511031103
Monk, D., Mackay, D. J. G., Eggermann, T., Maher, E. R., and Riccio, A. (2019). Genomic imprinting disorders: Lessons on how genome, epigenome and environment interact. Nat. Rev. Genet. 20, 235–248. doi:10.1038/s41576-018-0092-0
Monteagudo-Sánchez, A., Sánchez-Delgado, M., Mora, J. R. H., Santamaría, N. T., Gratacós, E., Esteller, M., et al. (2019). Differences in expression rather than methylation at placenta-specific imprinted loci is associated with intrauterine growth restriction. Clin. Epigenetics 11 (1), 35. doi:10.1186/s13148-019-0630-4
Moore, L. D., Le, T., and Fan, G. (2013). DNA methylation and its basic function. Neuropsychopharmacology 38 (1), 23–38. doi:10.1038/npp.2012.112
Moore, S. E., Cole, T. J., Collinson, A. C., Poskitt, E. M., McGregor, I. A., and Prentice, A. M. (1999). Prenatal or early postnatal events predict infectious deaths in young adulthood in rural Africa. Int. J. Epidemiol. 28 (6), 1088–1095. doi:10.1093/ije/28.6.1088
Moore, S. E., Cole, T. J., Poskitt, E. M., Sonko, B. J., Whitehead, R. G., McGregor, I. A., et al. (1997). Season of birth predicts mortality in rural Gambia. Nature 388 (6641), 434. doi:10.1038/41245
Morgan, H. D., Sutherland, H. G., Martin, D. I., and Whitelaw, E. (1999). Epigenetic inheritance at the agouti locus in the mouse. Nat. Genet. 23 (3), 314–318. doi:10.1038/15490
Mortillo, M., and Marsit, C. J. (2022). Select early-life environmental exposures and DNA methylation in the placenta. Curr. Environ. Health Rep. 10 (1), 22–34. doi:10.1007/S40572-022-00385-1
Mortusewicz, O., Schermelleh, L., Walter, J., Cardoso, M. C., and Leonhardt, H. (2005). Recruitment of DNA methyltransferase I to DNA repair sites. Proc. Natl. Acad. Sci. U. S. A. 102 (25), 8905–8909. doi:10.1073/pnas.0501034102
Musheev, M. U., Baumgärtner, A., Krebs, L., and Niehrs, C. (2020). The origin of genomic N6-methyl-deoxyadenosine in mammalian cells. Nat. Chem. Biol. 16 (6), 630–634. doi:10.1038/s41589-020-0504-2
Nelissen, E. C., Dumoulin, J. C., Daunay, A., Evers, J. L., Tost, J., and van Montfoort, A. P. (2013). Placentas from pregnancies conceived by IVF/ICSI have a reduced DNA methylation level at the H19 and MEST differentially methylated regions. Hum. Reprod. 28 (4), 1117–1126. doi:10.1093/humrep/des459
Neri, F., Rapelli, S., Krepelova, A., Incarnato, D., Parlato, C., Basile, G., et al. (2017). Intragenic DNA methylation prevents spurious transcription initiation. Nature 543 (7643), 72–77. doi:10.1038/nature21373
Ngom, P. T., Solon, J., Moore, S. E., Morgan, G., Prentice, A. M., and Aspinall, R. (2011). Thymic function and T cell parameters in a natural human experimental model of seasonal infectious diseases and nutritional burden. J. Biomed. Sci. 18 (1), 41–11. doi:10.1186/1423-0127-18-41
Novakovic, B., Yuen, R. K., Gordon, L., Penaherrera, M. S., Sharkey, A., Moffett, A., et al. (2011). Evidence for widespread changes in promoter methylation profile in human placenta in response to increasing gestational age and environmental/stochastic factors. BMC Genomics 12 (1), 529–614. doi:10.1186/1471-2164-12-529
Okae, H., Chiba, H., Hiura, H., Hamada, H., Sato, A., Utsunomiya, T., et al. (2014). Genome-wide analysis of DNA methylation dynamics during early human development. PLoS Genet. 10 (12), 1004868. doi:10.1371/journal.pgen.1004868
Owens, S., Gulati, R., Fulford, A. J., Sosseh, F., Denison, F. C., Brabin, B. J., et al. (2015). Periconceptional multiple-micronutrient supplementation and placental function in rural Gambian women: A double-blind, randomized, placebo-controlled trial. Am. J. Clin. Nutr. 102 (6), 1450–1459. doi:10.3945/AJCN.113.072413
Pajares, M. A., and Pérez-Sala, D. (2006). Betaine homocysteine S-methyltransferase: Just a regulator of homocysteine metabolism? ’, Cell. Mol. Life Sci. 63 (23), 2792–2803. doi:10.1007/S00018-006-6249-6
Patel, N., Godfrey, K. M., Pasupathy, D., Levin, J., Flynn, A. C., Hayes, L., et al. (2017). Infant adiposity following a randomised controlled trial of a behavioural intervention in obese pregnancy. Int. J. Obes. 41 (7), 1018–1026. doi:10.1038/ijo.2017.44
Peters, J. (2014). The role of genomic imprinting in biology and disease: An expanding view. Nat. Rev. Genet. 15 (8), 517–530. doi:10.1038/nrg3766
Peterson, L. A., and Hecht, S. S. (2017). Tobacco, e-cigarettes, and child health. Curr. Opin. Pediatr. 29 (2), 225–230. doi:10.1097/MOP.0000000000000456
Pettitt, D. J., Nelson, R. G., Saad, M. F., Bennett, P. H., and Knowler, W. C. (1993). Diabetes and obesity in the offspring of Pima Indian women with diabetes during pregnancy. Diabetes Care 16 (1), 310–314. doi:10.2337/diacare.16.1.310
Pilvar, D., Reiman, M., Pilvar, A., and Laan, M. (2019). Parent-of-origin-specific allelic expression in the human placenta is limited to established imprinted loci and it is stably maintained across pregnancy. Clin. Epigenetics 11 (1), 94–14. doi:10.1186/s13148-019-0692-3
Piñuñuri, R., Castaño-Moreno, E., Llanos, M. N., and Ronco, A. M. (2020). Epigenetic regulation of folate receptor-α (FOLR1) in human placenta of preterm newborns. Placenta 94, 20–25. doi:10.1016/j.placenta.2020.03.009
Popova, S., Dozet, D., Shield, K., Rehm, J., and Burd, L. (2021). Alcohol’s impact on the fetus. Nutrients 13 (10), 3452. doi:10.3390/nu13103452
Poston, L., Caleyachetty, R., Cnattingius, S., Corvalán, C., Uauy, R., Herring, S., et al. (2016). Preconceptional and maternal obesity: Epidemiology and health consequences. Lancet Diabetes & Endocrinol. 4 (12), 1025–1036. doi:10.1016/s2213-8587(16)30217-0
Prentice, A. M., Whitehead, R. G., Roberts, S. B., and Paul, A. A. (1981). Long-term energy balance in child-bearing Gambian women. Am. J. Clin. Nutr. 34 (12), 2790–2799. doi:10.1093/ajcn/34.12.2790
Proudhon, C., Duffié, R., Ajjan, S., Cowley, M., Iranzo, J., Carbajosa, G., et al. (2012). Protection against de novo methylation is instrumental in maintaining parent-of-origin methylation inherited from the gametes. Mol. Cell. 47 (6), 909–920. doi:10.1016/j.molcel.2012.07.010
Quenneville, S., Verde, G., Corsinotti, A., Kapopoulou, A., Jakobsson, J., Offner, S., et al. (2011). In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanucleotide to affect chromatin and DNA methylation of imprinting control regions. Mol. Cell. 44 (3), 361–372. doi:10.1016/j.molcel.2011.08.032
Quilter, C. R., Harvey, K. M., Bauer, J., Skinner, B. M., Gomez, M., Shrivastava, M., et al. (2021). Identification of methylation changes associated with positive and negative growth deviance in Gambian infants using a targeted methyl sequencing approach of genomic DNA. FASEB BioAdvances 3 (4), 205–230. doi:10.1096/fba.2020-00101
Quina, A. S., Buschbeck, M., and di Croce, L. (2006). Chromatin structure and epigenetics. Biochem. Pharmacol. 72 (11), 1563–1569. doi:10.1016/j.bcp.2006.06.016
Reijnders, I. F., Mulders, A. G. M. G. J., van der Windt, M., Steegers, E. A. P., and Steegers-Theunissen, R. P. M. (2019). The impact of periconceptional maternal lifestyle on clinical features and biomarkers of placental development and function: A systematic review. Hum. Reprod. update 25 (1), 72–94. doi:10.1093/humupd/dmy037
Renfree, M. B., Suzuki, S., and Kaneko-Ishino, T. (2013). The origin and evolution of genomic imprinting and viviparity in mammals. Philosophical Trans. R. Soc. B Biol. Sci. 368, 20120151. doi:10.1098/rstb.2012.0151
Rich-Edwards, J. W., Stampfer, M. J., Manson, J. E., Rosner, B., Hankinson, S. E., Colditz, G. A., et al. (1997). Birth weight and risk of cardiovascular disease in a cohort of women followed up since 1976. BMJ 315 (7105), 396–400. doi:10.1136/bmj.315.7105.396
Roberts, J. M., and Cooper, D. W. (2001). Pathogenesis and genetics of pre-eclampsia. Lancet 357 (9249), 53–56. doi:10.1016/S0140-6736(00)03577-7
Romanelli, V., Nakabayashi, K., Vizoso, M., Moran, S., Iglesias-Platas, I., Sugahara, N., et al. (2014). Variable maternal methylation overlapping the nc886/vtRNA2-1 locus is locked between hypermethylated repeats and is frequently altered in cancer. Epigenetics 9 (5), 783–790. doi:10.4161/epi.28323
Ruebner, M., Strissel, P. L., Ekici, A. B., Stiegler, E., Dammer, U., Goecke, T. W., et al. (2013). Reduced syncytin-1 expression levels in placental syndromes correlates with epigenetic hypermethylation of the ERVW-1 promoter region. PloS One 8 (2), 56145. doi:10.1371/journal.pone.0056145
Saffari, A., Shrestha, S., Issarapu, P., Sajjadi, S., Betts, M., Sahariah, S. A., et al. (2020). Effect of maternal preconceptional and pregnancy micronutrient interventions on children’s DNA methylation: Findings from the EMPHASIS study. Am. J. Clin. Nutr. 112 (4), 1099–1113. doi:10.1093/ajcn/nqaa193
Salas, M., John, R., Saxena, A., Barton, S., Frank, D., Fitzpatrick, G., et al. (2004). Placental growth retardation due to loss of imprinting of Phlda2. Mech. Dev. 121 (10), 1199–1210. doi:10.1016/j.mod.2004.05.017
Sanchez-Delgado, M., Court, F., Vidal, E., Medrano, J., Monteagudo-Sánchez, A., Martin-Trujillo, A., et al. (2016). Human oocyte-derived methylation differences persist in the placenta revealing widespread transient imprinting. PLoS Genet. 12 (11), 1006427. doi:10.1371/journal.pgen.1006427
Sanchez-Delgado, M., Martin-Trujillo, A., Tayama, C., Vidal, E., Esteller, M., Iglesias-Platas, I., et al. (2015). Absence of maternal methylation in biparental hydatidiform moles from women with NLRP7 maternal-effect mutations reveals widespread placenta-specific imprinting. PLoS Genet. 11 (11), 1005644. doi:10.1371/journal.pgen.1005644
Schroeder, D. I., Blair, J. D., Lott, P., Yu, H. O., Hong, D., Crary, F., et al. (2013). The human placenta methylome. Proc. Natl. Acad. Sci. U. S. A. 110 (15), 6037–6042. doi:10.1073/pnas.1215145110
Schroeder, D. I., and Lasalle, J. M. (2013). How has the study of the human placenta aided our understanding of partially methylated genes? ’, Epigenomics 5 (6), 645–654. doi:10.2217/epi.13.62
Schulz, R., Proudhon, C., Bestor, T. H., Woodfine, K., Lin, C. S., Lin, S. P., et al. (2010). The parental non-equivalence of imprinting control regions during mammalian development and evolution. PLoS Genet. 6 (11), 1001214. doi:10.1371/journal.pgen.1001214
Scott, H., Grynspan, D., Anderson, L. N., and Connor, K. L. (2022). Maternal underweight and obesity are associated with placental pathologies in human pregnancy. Reprod. Sci. 29 (12), 3425–3448. doi:10.1007/s43032-022-00983-2
Shen, L., Inoue, A., He, J., Liu, Y., Lu, F., and Zhang, Y. (2014). Tet3 and DNA replication mediate demethylation of both the maternal and paternal genomes in mouse zygotes. Cell. Stem Cell. 15 (4), 459–471. doi:10.1016/j.stem.2014.09.002
Si, H. C., Worswick, S., Byun, H. M., Shear, T., Soussa, J. C., Wolff, E. M., et al. (2009). Changes in DNA methylation of tandem DNA repeats are different from interspersed repeats in cancer. Int. J. Cancer. 125 (3), 723–729. doi:10.1002/ijc.24384
Silver, M. J., Kessler, N. J., Hennig, B. J., Dominguez-Salas, P., and Laritsky, E. (2015). Independent genomewide screens identify the tumor suppressor VTRNA2-1 as a human epiallele responsive to periconceptional environment. Genome Biol. 16 (1), 118. doi:10.1186/s13059-015-0660-y
Silver, M. J., Saffari, A., Kessler, N. J., Chandak, G. R., Fall, C. H. D., Issarapu, P., et al. (2022). Environmentally sensitive hotspots in the methylome of the early human embryo. eLife 11, 72031. doi:10.7554/elife.72031
Sinclair, K. D., Allegrucci, C., Singh, R., Gardner, D. S., Sebastian, S., Bispham, J., et al. (2007). DNA methylation, insulin resistance, and blood pressure in offspring determined by maternal periconceptional B vitamin and methionine status. Proc. Natl. Acad. Sci. U. S. A. 104 (49), 19351–19356. doi:10.1073/pnas.0707258104
Sitras, V., Fenton, C., Paulssen, R., Vårtun, Å., and Acharya, G. (2012). Differences in gene expression between first and third trimester human placenta: A microarray study. PLoS One 7 (3), 33294. doi:10.1371/journal.pone.0033294
Smallwood, S. A., Tomizawa, S., Krueger, F., Ruf, N., Carli, N., Segonds-Pichon, A., et al. (2011). Dynamic CpG island methylation landscape in oocytes and preimplantation embryos. Nat. Genet. 43 (8), 811–814. doi:10.1038/ng.864
Smith, Z. D., Chan, M. M., Humm, K. C., Karnik, R., Mekhoubad, S., Regev, A., et al. (2014). DNA methylation dynamics of the human preimplantation embryo. Nature 511 (7511), 611–615. doi:10.1038/nature13581
Solanky, N., Requena Jimenez, A., D’Souza, S. W., Sibley, C. P., and Glazier, J. D. (2010). Expression of folate transporters in human placenta and implications for homocysteine metabolism. Placenta 31 (2), 134–143. doi:10.1016/J.placenta.2009.11.017
Spahn, L., and Barlow, D. P. (2003). An ICE pattern crystallizes. Nat. Genet. 35 (1), 11–12. doi:10.1038/ng0903-11
Sun, M. A., Wolf, G., Wang, Y., Senft, A. D., Ralls, S., Jin, J., et al. (2021). Endogenous retroviruses drive lineage-specific regulatory evolution across primate and rodent placentae. Mol. Biol. Evol. 38 (11), 4992–5004. doi:10.1093/molbev/msab223
Susser, M., and Stein, Z. (1994). Timing in prenatal nutrition: A reprise of the Dutch famine study. Nutr. Rev. 52 (3), 84–94. doi:10.1111/J.1753-4887.1994.TB01395.X
Tamura, T., and Picciano, M. F. (2006). Folate and human reproduction. Am. J. Clin. Nutr. 83 (5), 993–1016. doi:10.1093/ajcn/83.5.993
Takahashi, N., Coluccio, A., Thorball, C. W., Planet, E., Shi, H., Offner, S., et al. (2019). ZNF445 is a primary regulator of genomic imprinting. Genes. & Dev. 33 (1–2), 49–54. doi:10.1101/gad.320069.118
Tekola-Ayele, F., Workalemahu, T., Gorfu, G., Shrestha, D., Tycko, B., Wapner, R., et al. (2019). Sex differences in the associations of placental epigenetic aging with fetal growth. Aging 11 (15), 5412. doi:10.18632/aging.102124
Tian, X., Anthony, K., Neuberger, T., and Diaz, F. J. (2014). Preconception zinc deficiency disrupts postimplantation fetal and placental development in mice. Biol. Reproduction 90 (4), 83–12. doi:10.1095/biolreprod.113.113910
Tint, M. T., Sadananthan, S. A., Soh, S. E., Aris, I. M., Michael, N., Tan, K. H., et al. (2020). Maternal glycemia during pregnancy and offspring abdominal adiposity measured by MRI in the neonatal period and preschool years: The Growing up in Singapore towards healthy Outcomes (GUSTO) prospective mother–offspring birth cohort study. Am. J. Clin. Nutr. 112 (1), 39–47. doi:10.1093/ajcn/nqaa055
Tobi, E. W., Goeman, J. J., Monajemi, R., Gu, H., Putter, H., Zhang, Y., et al. (2014). DNA methylation signatures link prenatal famine exposure to growth and metabolism’, Nat. Commun. 5, 5592–5614. doi:10.1038/ncomms6592
Uusküla, L., Männik, J., Rull, K., Minajeva, A., Kõks, S., Vaas, P., et al. (2012). Mid-gestational gene expression profile in placenta and link to pregnancy complications. PLoS One 7 (11), 49248. doi:10.1371/journal.pone.0049248
Valenzuela-Alcaraz, B., Crispi, F., Bijnens, B., Cruz-Lemini, M., Creus, M., Sitges, M., et al. (2013). Assisted reproductive technologies are associated with cardiovascular remodeling in utero that persists postnatally. Circulation 128 (13), 1442–1450. doi:10.1161/circulationaha.113.002428
Vera, E., Canela, A., Fraga, M. F., Esteller, M., and Blasco, M. A. (2008). Epigenetic regulation of telomeres in human cancer. Oncogene 27 (54), 6817–6833. doi:10.1038/onc.2008.289
Waterland, R. A., and Jirtle, R. L. (2003). Transposable elements: Targets for early nutritional effects on epigenetic gene regulation. Mol. Cell. Biol. 23 (15), 5293–5300. doi:10.1128/mcb.23.15.5293-5300.2003
Waterland, R. A., Kellermayer, R., Laritsky, E., Rayco-Solon, P., Harris, R. A., Travisano, M., et al. (2010). Season of conception in rural Gambia affects DNA methylation at putative human metastable epialleles. PLoS Genet. 6 (12), 10012522–e1001310. doi:10.1371/journal.pgen.1001252
Watkins, A. J., Lucas, E. S., Marfy-Smith, S., Bates, N., Kimber, S. J., and Fleming, T. P. (2015). Maternal nutrition modifies trophoblast giant cell phenotype and fetal growth in mice. Reproduction 149 (6), 563–575. doi:10.1530/rep-14-0667
Watkins, A. J., Ursell, E., Panton, R., Papenbrock, T., Hollis, L., Cunningham, C., et al. (2008a). Adaptive responses by mouse early embryos to maternal diet protect fetal growth but predispose to adult onset disease. Biol. Reproduction 78 (2), 299–306. doi:10.1095/biolreprod.107.064220
Watkins, A. J., Wilkins, A., Cunningham, C., Perry, V. H., Seet, M. J., Osmond, C., et al. (2008b). Low protein diet fed exclusively during mouse oocyte maturation leads to behavioural and cardiovascular abnormalities in offspring. J. Physiology 586 (8), 2231–2244. doi:10.1113/jphysiol.2007.149229
Weaver, J. R., Susiarjo, M., and Bartolomei, M. S. (2009). Imprinting and epigenetic changes in the early embryo. Mamm. Genome 20 (9–10), 532–543. doi:10.1007/S00335-009-9225-2
WHO (2005). The world health report 2005. Make every mother and child count - health and education advice and resource team. Geneva, Switzerland: World Health Organization, 1–243. Avaliable At: http://www.heart-resources.org/doc_lib/the-world-health-report-make-every-mother-and-child-count/ (Accessed: March 13, 2023).
Wion, D., and Casadesús, J. (2006). N6-methyl-adenine: An epigenetic signal for DNA-protein interactions. Nat. Rev. Microbiol. 4 (3), 183–192. doi:10.1038/nrmicro1350
Wolff, G. L., Kodell, R. L., Moore, S. R., and Cooney, C. A. (1998). Maternal epigenetics and methyl supplements affect agouti gene expression in Avy/a mice. FASEB J. 12 (11), 949–957. doi:10.1096/fasebj.12.11.949
Workalemahu, T., Shrestha, D., Tajuddin, S. M., and Tekola-Ayele, F. (2021). Maternal cardiometabolic factors and genetic ancestry influence epigenetic aging of the placenta. J. Dev. Orig. Health Dis. 12 (1), 34–41. doi:10.1017/S2040174419000801
Xiao, C. L., Zhu, S., He, M., Chen, D., Zhang, Q., Chen, Y., et al. (2018). N 6 -methyladenine DNA modification in the human genome. Mol. Cell. 71 (2), 306–318.e7. doi:10.1016/j.molcel.2018.06.015
Xie, Q., Wu, T. P., Gimple, R. C., Li, Z., Prager, B. C., Wu, Q., et al. (2018). N6-methyladenine DNA modification in glioblastoma. Cell. 175 (5), 1228–1243.e20. doi:10.1016/j.cell.2018.10.006
Yu, M., Hon, G. C., Szulwach, K. E., Song, C. X., Zhang, L., Kim, A., et al. (2012). Base-Resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 149 (6), 1368–1380. doi:10.1016/j.cell.2012.04.027
Yuen, R. K., Jiang, R., Peñaherrera, M. S., McFadden, D. E., and Robinson, W. P. (2011). Genome-wide mapping of imprinted differentially methylated regions by DNA methylation profiling of human placentas from triploidies. Epigenetics Chromatin 4 (1), 10–16. doi:10.1186/1756-8935-4-10
Keywords: epigentics, DNA methylation, placenta, imprinting, metastable epialles, DOHaD
Citation: Sainty R, Silver MJ, Prentice AM and Monk D (2023) The influence of early environment and micronutrient availability on developmental epigenetic programming: lessons from the placenta. Front. Cell Dev. Biol. 11:1212199. doi: 10.3389/fcell.2023.1212199
Received: 25 April 2023; Accepted: 27 June 2023;
Published: 06 July 2023.
Edited by:
Masako Suzuki, Texas A and M University, United StatesReviewed by:
Courtney W. Hanna, University of Cambridge, United KingdomCopyright © 2023 Sainty, Silver, Prentice and Monk. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rebecca Sainty, Yi5zYWludHlAdWVhLmFjLnVr
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.