
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell Dev. Biol., 15 May 2023
Sec. Developmental Epigenetics
Volume 11 - 2023 | https://doi.org/10.3389/fcell.2023.1191797
This article is part of the Research TopicEpigenetics of Cell Programming and ReprogrammingView all 5 articles
Dynamic-related protein 1 (DRP1) is a key protein of mitochondrial fission. In this study, we found that inhibition of activity of DRP1 led to increased levels of cleavage embryo genes in mouse embryonic stem cells (mESCs), which might reflect a transient totipotency status derived from pluripotency. This result indicates that DRP1 inhibition in mESCs leads to a tendency to obtain a new expression profile similar to that of the 2C-like state. Meanwhile, we also noticed that the glycolysis/gluconeogenesis pathway and its related enzymes were significantly downregulated, and the key glycolytic enzymes were also downregulated in various 2C-like cells. Moreover, when DRP1 activity was inhibited from the late zygote when cleavage embryo genes started to express, development of early embryos was inhibited, and these cleavage embryo genes failed to be efficiently silenced at the late 2-cell (2C) stage. Taken together, our result shows that DRP1 plays an important role in silencing cleavage embryo genes for totipotency-to-pluripotency transition.
Mitochondria are important and dynamic double membrane-bound organelles, and the balance of fission and fusion maintains their physiological functions (Youle and van der Bliek, 2012; Wai and Langer, 2016; Giacomello et al., 2020; Li et al., 2021). The guanosine triphosphatases (GTPases), which are well conserved in mammals, are responsible for this dynamic process (Youle and van der Bliek, 2012; Wai and Langer, 2016). As for the fission process, the dynamic-related protein 1 (DRP1), a member of the cytosolic dynamin family, is recruited by Mid49, Mid51, and Mff to function in mammalian mitochondria (Schmitt et al., 2018; Kleele et al., 2021; Li et al., 2021). Mitochondrial fission is essential for development, and it is demonstrated that the knockout of Drp1 in mice caused death of early embryos. In addition, the loss of Drp1 seriously affects the maturation of oocytes and the establishment of maternal epigenome in preimplantation embryos (Udagawa et al., 2014; Li et al., 2021; Adhikari et al., 2022). Interestingly, the CRISPR-Cas9 system has been used to generate DRP1-knockout embryonic stem cells (ESCs), which showed significantly impacted mitochondrial metabolism and global gene expression profiles (Seo et al., 2020).
Mitochondria produce ATP for energy through oxidative phosphorylation (OXPHOS), which, in stem cells, is thought to rely mainly on glycolysis. Because of mitochondrial elongation, deletion of mitochondrial fission factors such as Drp1, Mff, or Fis resulted in increased OXPHOS and intracellular ATP concentration and decreased glycolysis (Seo et al., 2020). Recent studies have shown that mitochondria play an important role in the regulation of stem cell activity in terms of metabolism (Matilainen et al., 2017; Zhang et al., 2018; Ludikhuize et al., 2020). The intermediates in the mitochondrial metabolic pathway are closely related to key enzymes, which regulate chromatin and protein modification such as histones, methylation, and acetylation (Gafni et al., 2013; Carbognin et al., 2016; Matilainen et al., 2017; Zhang et al., 2018). Upregulation of mitochondrial transcripts and metabolism with activated STAT3 promotes the proliferation of mouse ESCs and the reprogramming of EpiSCs (Li et al., 2021). The inhibition of histone deacetylase (HDAC), which is dependent on NAD+, drove ESCs to a more naive state in vitro (Giacomello et al., 2020). In addition, sirtuin enzymes and mitochondrial metabolite NAD+ together regulate the post-translational modifications (PTMs) of proteins to orchestrate cell fate (Wai and Langer, 2016; Chini et al., 2021).
During early embryonic development, the developmental program depends on the degradation of maternal mRNAs and zygotic genome activation (ZGA) (Aoki et al., 1997; Srinivasan et al., 2020; Cheng et al., 2022). A great mass of transcripts and widespread changes in chromatin that occurred during the ZGA event complicated the regulatory module at this specific point. ZGA starts at the zygote stage and becomes robust at the 2-cell (2C) stage in mice, and thousands of 2C-specific genes (2C genes, ZGA genes, or cleavage embryo genes) (we refer to 2C gene in the following) are abruptly upregulated, such as Zscan4 and Dux (De Iaco et al., 2017; Srinivasan et al., 2020; Grow et al., 2021). ZGA is controlled by different mechanisms including the expression of specific transcription factors (TFs) and the activity of RNA polymerase (Grow et al., 2021; Wang et al., 2022).
Our previous studies have shown that DRP1 deactivation led to mitochondrial abnormalities and thus affected the normal development of embryos (Li et al., 2021), but its effect on cell fate control remains unclear. In particular, mitochondrial division inhibitor-1 (Mdivi-1), which can inhibit the activity of DRP1, has been proposed to selectively inhibit mitochondrial division in mammals (Cassidy-Stone et al., 2008; Bordt et al., 2017). Herein, we use both mouse early embryos and mouse ESCs to investigate how DRP1 activity is involved in totipotency-to-pluripotency transition.
The mouse ESC line AB2.2 was seeded on mitotically inactivated MEF feeder cells in the N2B27 medium with 2i, 0.4 μM PD0325901 (Stemgent, United States), 3 μM CHIR99021 (Stemgent, United States), and LIF (1000 U/mL) in tissue culture dishes (Buecker et al., 2014). Mdivi-1 was dissolved by DMSO for dilution to the designated concentration. ESCs were treated with 50 μM Mdivi-1 (MedChemExpress, United States) and cultured for a certain amount of time for further examination. The medium was changed every day, and the cells were cultured in humidified conditions with 5% CO2 at 37°C.
Female ICR mice (7–8 weeks) were injected with 10 IU of pregnant mare serum gonadotropin (PMSG, Sansheng, China). After 48 h, female mice were injected with 10 IU of human chorionic gonadotropin (hCG, Sansheng, China) and then let to mate with adult male ICR mice. Zygotes were collected from the oviduct ampulla of female mice with vaginal plugs 16 h later. Zygotes were cultured in the KSOM medium with or without 200 μM Mdivi-1 at 37°C in a humidified atmosphere of 5% CO2.
Total RNA was extracted from 50 oocytes or 5 × 105 cells using the TRIzol reagent (Invitrogen, United States), following the manufacturer’s instructions, and cDNA synthesis was completed using the Hifair 1st Strand cDNA Synthesis Kit (Yeasen Biotechnology Co., Ltd., China). RT-PCR was performed with the SYBR Green Master Mix (Yeasen Biotechnology Co., Ltd., China) with the ABI 7500 Real-Time PCR system (Applied Biosystems, United States). Mouse Zscan4 (Forward: 5′- GAGATTCATGGAGAGTCTGACTGATGAGTG-3′ and Reverse: 5′- GCTGTTGTTTCAAAAGCTTGATGACTTC-3′), Zfp352 (Forward: 5′- ACCACCTCAAAGAACACCAG-3′ and Reverse: 5′- ACAAGGGACAAGCGTAGAAC-3′), were normalized against Gapdh (Forward: 5′-TCTTCCAGGAGCGAGACCC-3′ and Reverse: 5′-CGGAGATGATGACCCTTTT-3′), and the quantification of the fold change was determined by the comparative CT method.
The samples (106 cells/line) were lysed with the 100 μL RIPA buffer (Sangon Biotech, China), and the lysates were separated by SDS–polyacrylamide gel electrophoresis at 110 V for 60 min. Separated proteins were then transferred onto the nitrocellulose filter membrane at 350 mA for 60 min in ice. The membranes were blocked in PBS (Solarbio, China) with 1% (wt/vol) BSA (Sangon Biotech, China) for 1 h at room temperature and then incubated with primary antibodies overnight at 4°C. The primary antibodies against DRP1 (1:1000, 8570, CST, United States) and GAPDH (1:1000, 10494-1-AP, Proteintech Group, Inc., United States) were used. After washing three times, the membrane was incubated with HRP-conjugated goat anti-rabbit IgG (H + L) (1:5000, SA00001-2, Proteintech Group, Inc., United States). Solutions A and B of the ECL chemiluminescent solution (Affinity Biosciences, K002, United States) were mixed in a ratio of 1:1 to cover the membrane for incubation. The membrane was protected from exposure to light. Images were collected using a gel imager (Image Lab Software, Bio-Rad) in the dark.
For mouse ESCs, total RNA was extracted by the TRIzol reagent (Invitrogen, United States) from 5 × 105 cells for each group, followed by standard RNA-seq library preparation (TruSeq RNA Sample Preparation Kit, Illumina). For late 2-cell embryos, 8–10 embryos were collected after 16 h in vitro culture for each group in lysis components with ribonuclease inhibitors, and amplification was further carried out by the Smart-Seq2 method by BGI Genomics Co., Ltd., China. Qualified libraries were loaded onto the Illumina HiSeq platform for PE150 sequencing. Raw reads were processed with cutadapt v1.16 to remove adapters and perform quality trimming with default parameters except for a quality cutoff of 20 and a minimum length of 20. Trimmed reads were mapped to the mouse genome (GENCODE release M23) using STAR with default settings. Reads were counted in exons of the mouse genome, using the STAR-quantMode GeneCounts setting. RSEM was used to calculate the FPKM value. Differentially regulated genes in DESeq2 analysis were defined as those which were more than two-fold increased or decreased with adjusted p < 0.05. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were performed by Metascape (https://metascape.org). Volcano plot, heatmap, bar chart, and bubble chart were generated by R software.
ChIP-seq was performed using a Hyperactive In-Situ ChIP Library Prep Kit for Illumina (pG-Tn5) (Vazyme, Nanjing, China), according to the manufacturer’s procedure with the appropriate antibodies H3K4me3 (9297, CST, United States), H3K27me3 (ab6002, Abcam, United Kingdom), and Pol II (39097, Active motif, United States).
Data were presented as the mean ± SEM. All experiments were replicated more than three times. Statistical comparisons were made with the Mann–Whitney U test for the analysis of two groups. Analyses were conducted by SPSS 20 software (IBM). A p-value <0.05 (*p < 0.05) was considered statistically significant.
We first analyzed the RNA-seq data (GSE148959) on mouse ESCs with knocked-out mitochondrial fission factors (Seo et al., 2020). The results showed that deletion of mitochondrial fission factors, such as Drp1, Mff, or Fis, resulted in the change in gene expression in ESCs compared with the control group (Figure 1A). There were more than 700 differentially expressed genes in Drp1-KO ESCs, and nearly half of them were upregulated or downregulated (Figure 1A). There were about 1000 differentially expressed genes in Mff-KO ESCs and 600 differentially expressed genes in Fis-KO ESCs (Figure 1A). There was no significant difference in the expression of pluripotency factors such as Oct4, Sox2, and Nanog in Drp1-KO, Mff-KO, and Fis-KO ESCs, respectively, compared with the control group (Figure 1B). It was also reported that the pluripotency of the three knockout ESC lines was not affected (Seo et al., 2020). Next, we scrutinized the differentially expressed genes of the three knockout ESC lines (Drp1-KO, Mff-KO, and Fis-KO) and found that deletion of mitochondrial fission factors led to the upregulation of many 2C genes (cleavage embryo genes) (Figure 1C). In the Drp1-KO ESC line, 2C genes were mostly obviously upregulated (Figure 1C). GO analysis showed that DEGs were enriched in mRNA processing and mitochondrial metabolism in Drp1-KO cell lines (Figure 1D). The mitochondrial metabolic pathway is closely related to the pluripotent state of ESCs (Mandal et al., 2011; Rodrigues et al., 2015) and also participates in the regulation of 2C gene expression in ESCs and embryos (Nagaraj et al., 2017; Hu et al., 2020). These results suggest that the mitochondrial fission factor Drp1 is involved in the regulation of 2C gene levels in mouse ESCs.
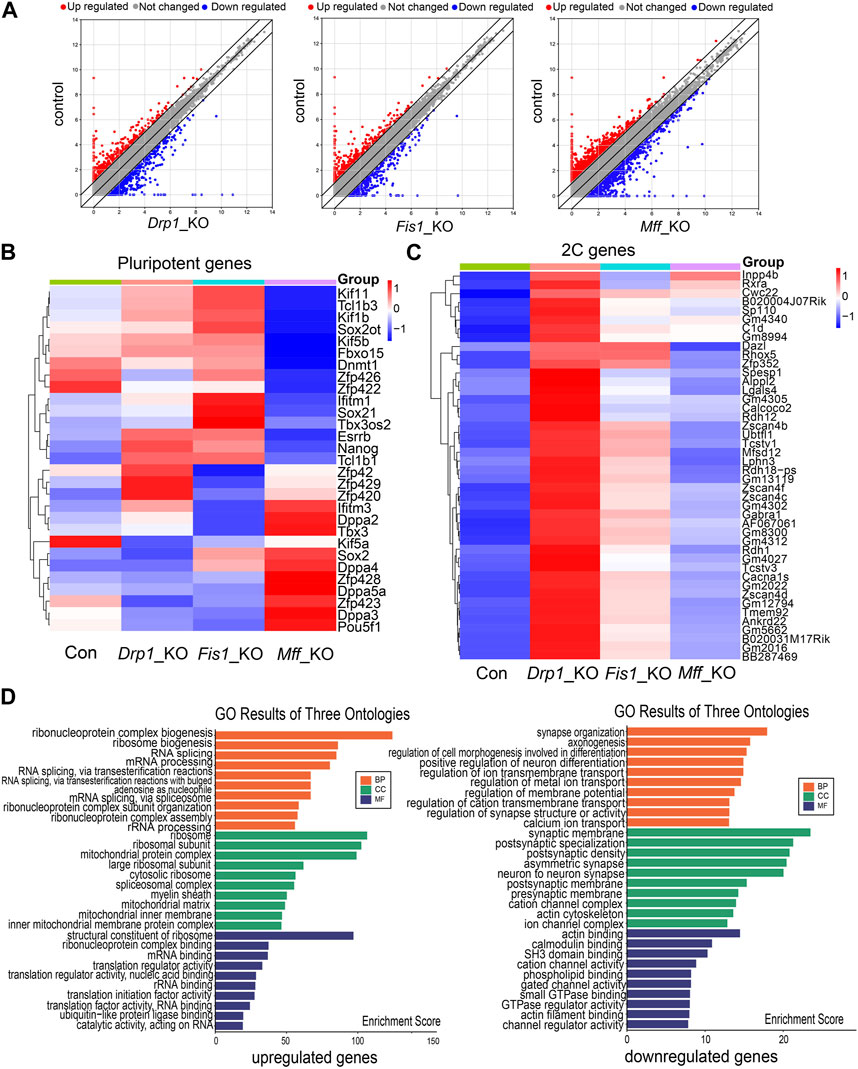
FIGURE 1. RNA-seq data analysis on ESC lines with knockout of various mitochondrial fission factors. (A) Scatter plots of differentially expressed genes in Drp1-KO ESCs (left), Fis-KO ESCs (middle), and Mff-KO ESCs (right) compared with control ESCs. Heatmap of differential expression of pluripotent genes (B) and 2C genes (C) in Drp1-KO ESCs, Fis-KO ESCs, and Mff-KO ESCs compared to the control group. (D) Gene Ontology (GO) enrichment analysis of upregulated (left) and downregulated genes (right) in Drp1-KO ESCs compared to control ESCs.
Mdivi-1 is a cellular permeable selective inhibitor of DRP1. In this study, we inhibited the activity of DRP1 in mouse ESCs by adding Mdivi-1 to the ESC medium. We first treated ESCs with Mdivi-1 at different concentrations for 24 h. However, when the drug concentration was very high, the morphology of ESC clones worsened and the cells died (Supplementary Figure S1). After comparison, we treated mouse ESCs with a concentration of 50 μM. We then examined the effects of drug treatment time on stem cell status. However, ESCs gradually died after treatment with 50 μM Mdivi-1 for more than 24 h (Figure 2A). Therefore, in this study, we treated ESCs with 50 μM Mdivi-1 for 24 h for subsequent detection. Western blot analysis confirmed that the expression of DRP1 was decreased after treatment of ESCs with Mdivi-1 (Figures 2B, C). Next, RNA-seq data on ESCs before (control group) and after (treatment group) Mdivi-1 treatment were analyzed. Compared with the control group, there were 1,129 differentially expressed genes in the treatment group, among which 647 genes were upregulated and 482 genes were downregulated (Figure 2D). Among these significantly upregulated genes, we noticed several typical 2C genes (Figure 2E and Supplementary Figure S2A). Moreover, the results of qRT-PCR proved that 2C genes such as Zscan4 and Zfp352 were indeed significantly upregulated (Figure 2F). In addition, we noticed no significant changes of transposon expression upon Mdivi-1 treatment (Figure 2G). Notably, our results also showed no significant changes in the expression of pluripotent genes in Mdivi-1-treated ESCs (Figure 2H). These results implied that the transition of totipotency-to-pluripotency may be disrupted by the abnormal activity of mitochondria, resulting in failed silencing of 2C genes.
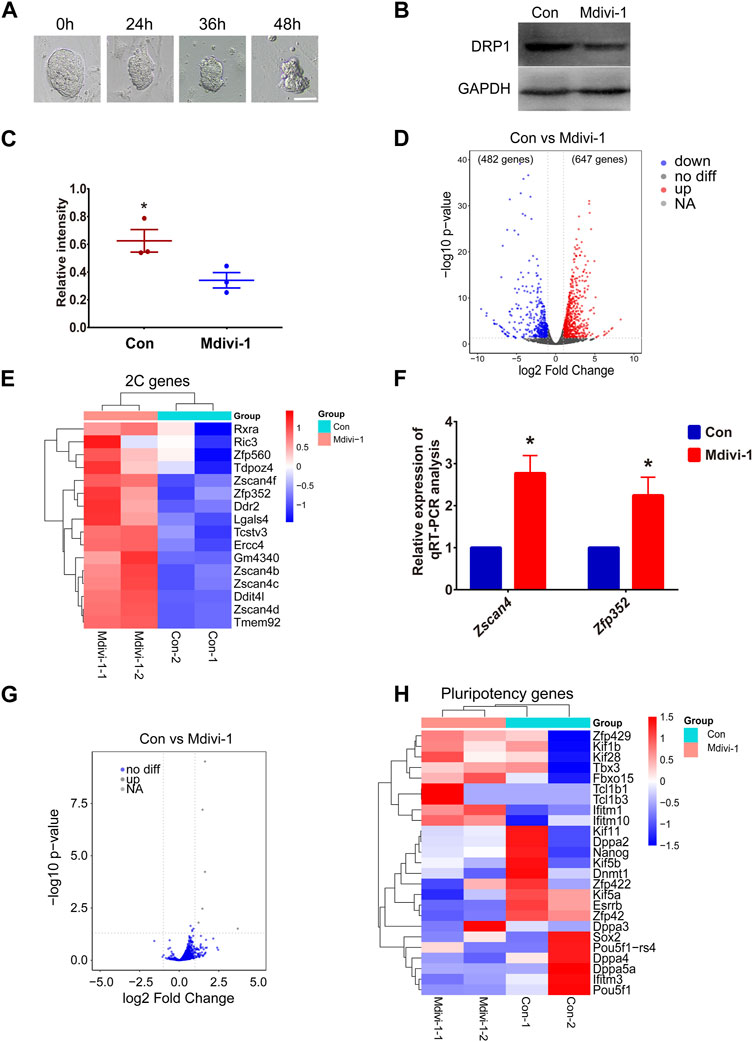
FIGURE 2. The expression level of 2C genes in Mdivi-1-treated ESCs increased compared to control ESCs. (A) DIC images of ESCs in 50 µM Mdivi-1 treatment after culturing for 0, 24, 36, and 48 h, respectively. Scale bar, 50 µm. (B, C) Western blotting of DRP1 and the statistical analysis of DRP1 protein level in control and Mdivi-1-treated ESCs. GAPDH was used as the internal control. *p < 0.05. (D) Volcano plot comparing differentially expressed genes (1129 genes) between Mdivi-1-treated ESCs and the control group. The value of |log2 fold-change| to mark the upregulated (red) (647 genes) and downregulated (blue) (482 genes) genes. (E) Heatmap of differential expression of 2C genes in Mdivi-1-treated mESCs compared to the control group. (F) The relative expression of Zscan4, Zfp352, mRNA in control and Mdivi-1-treated ESCs. *p < 0.05. (G) Volcano plot shows differential expression of transposons in control and Mdivi-1 treatment ESCs. (H) Heatmap of differentially expressed genes of pluripotent genes in control and Mdivi-1-treated ESCs.
In addition, gene enrichment analysis showed that these upregulated genes were enriched in “p53” and other pathways (Figure 3A). Studies have shown that P53 promoted Dux-mediated 2C gene upregulation (Li et al., 2021). Subsequently, we found that the downregulated genes were enriched in the “glycolysis/gluconeogenesis” pathway (Figure 3B). In these differentially expressed genes, multiple gene encoding enzymes in the glycolysis/gluconeogenesis pathway were downregulated (Figure 3C). These results showed that the glycolysis activity of Mdivi-1-treated cells might be decreased. Previous reports showed that glycolysis activity in Drp1-KO ESCs was indeed decreased (Seo et al., 2020), supporting our discovery. Decreased glycolysis pathway activity promoted the activation of the 2C program in ESCs (Hu et al., 2020). In conclusion, inhibition of DRP1 by Mdivi-1 can promote the expression of 2C genes by changing a variety of biological pathways in mouse ESCs.
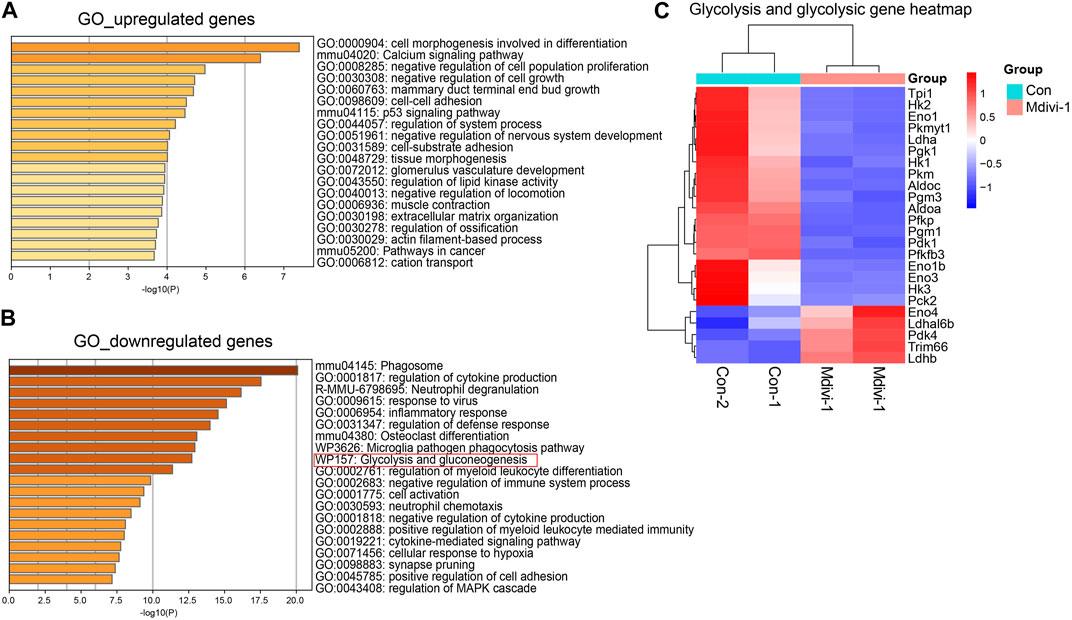
FIGURE 3. Schematic diagram of RNA-seq data analysis. GO enrichment analysis of upregulated (A) and downregulated genes (B) in Mdivi-1-treated ESCs compared to the control group. (C) Heatmap of differentially expressed genes of glycolysis/gluconeogenesis genes in control and Mdivi-1-treated ESCs.
To further understand the mechanism of 2C gene upregulation, we detected the enrichment of H3K4me3, H3K27me3, and RNA polymerase II (Pol II) in mouse ESCs after Mdivi-1 treatment. We found that H3K4me3, a marker of transcription initiation, had significantly increased accumulation at transcription start sites (TSSs) of upregulation genes (Figure 4A). In contrast, H3K27me3 was considered a marker of transcriptional inhibition, and accumulation at TSSs of upregulated genes was significantly reduced (Figure 4B). At the same time, we found that Pol II occupation around the upregulated TSSs was increased after Mdivi-1 treatment (Figure 4C). These results suggest that inhibition of DRP1 expression by Mdivi-1 can enhance transcriptional activity by strengthening H3K4me3 and reducing H3K27me3 at TSSs, thereby promoting 2C gene expression.
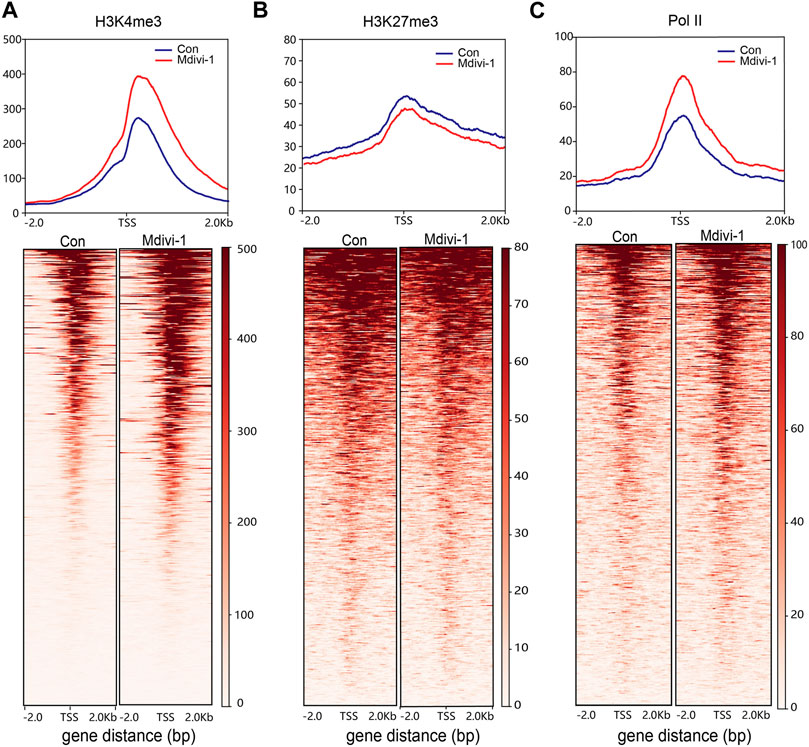
FIGURE 4. Comparison of H3K4me3, H3K27me3, and Pol II occupancy. The signal intensity diagram of H3K4me3 (A), H3K27me3, (B) and Pol II (C) at gene promoters (TSS flanking 2 kb) of upregulated genes in the Mdivi-1-treated group compared to the control group (upper) and the heatmap of upregulated genes at gene promoter regions (lower).
To identify how DRP1 activity is involved in the cell fate control in early mouse embryos, we treated mouse early embryos with Mdivi-1 at the late zygote stage when embryonic transcription started. After culturing the embryos for 96 h, we noticed significant developmental delay upon Mdivi-1 treatment (Figure 5A). The RNA-seq analysis of late 2-cell (2C) embryos showed that 861 genes were differentially expressed in the Mdivi-1-treated group when compared with the control group (Figure 5B). Among the 861 DEGs, 438 genes were overexpressed and 423 genes were downregulated in late 2C embryos. Interestingly, we found the 2C genes, such as Zfp352 and Zscan4d, were upregulated in Mdivi-1-treated embryos (Figure 5C and Supplementary Figure S2B), and this was similar to what happened in mouse ESCs. We also found no general differences in retrotransposon expression at the late 2C stage after DRP1 inhibition, which was also similar to that in mouse ESCs (Figure 2G; Figure 5D). In conclusion, inhibition of DRP1 can effectively promote the expression of 2C genes in late 2C embryos, while 2C genes are supposed to be silenced at this stage to start mid-implantation development and facilitate totipotency-to-pluripotency transition. In addition, the enrichment of Pol II at the differentially expressed genes in the late 2C stage with and without Mdivi-1 treatment was also examined. As expected, Pol II had significantly increased accumulation at TSSs of upregulated genes in late 2C embryos upon Mdivi-1 treatment (Figure 5E). In contrast, the enrichment of Pol II at TSSs of these downregulated genes was reduced obviously (Figure 5E).
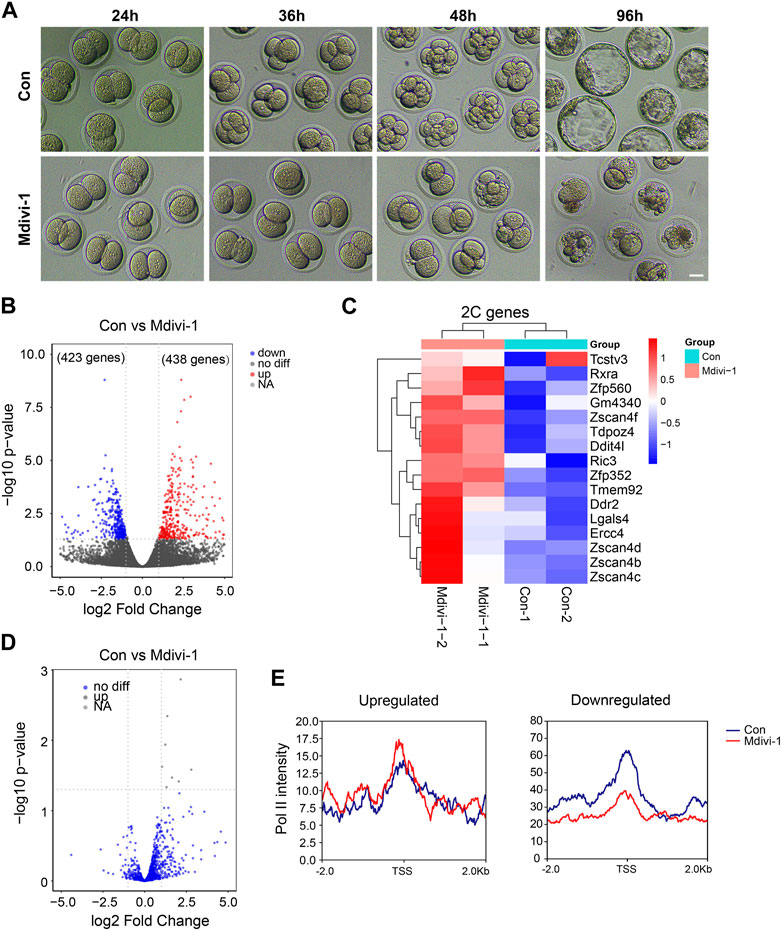
FIGURE 5. Mdivi-1 treatment impacted 2C gene expression in late 2-cell mouse embryos. (A) DIC images of early embryonic development in control and 200 µM Mdivi-1-treated groups after culturing for 24, 36, 48, and 96 h, respectively. Bar = 20 µm. (B) Volcano plot showing differentially expressed genes (861 genes) between Mdivi-1-treated embryos and control embryos. |log2 fold-change| marks the upregulated (red) (438 genes) and downregulated (blue) (423 genes) genes. (C) Heatmap of differential expression of ZGA genes in Mdivi-1-treated embryos compared to control embryos. (D) Volcano plot showing the expression changes of transposons in Mdivi-1-treated mouse embryos compared with control embryos. (E) Signal intensity diagram of Pol II at TSSs of upregulated and downregulated genes in Mdivi-1-treated group (red) compared to the control group (blue).
Taken together, our results showed that DRP1 activity is important in early mouse embryos to silence 2C genes at the late 2C stage to drive developmental programs for the totipotency-to-pluripotency transition.
A transient ESC subpopulation displays totipotent features by expressing a set of genes, which are activated in 2C embryos (Le et al., 2020). In the mouse ESC culture medium, the 2C-like state is transient and can be transformed back into pluripotency, and the process is dynamic and reversible (Fu et al., 2020). Studies have shown that pluripotent stem cells and 2C-like cells have different metabolic pathways (Zhao et al., 2021). Mitochondrial function maintained the proliferation of self-renewing ESCs (Mandal et al., 2011). RNA-seq analysis shows knocked-out mitochondrial fission-related factors, such as Drp1, Fis, or Mff, does not affect the expression of pluripotent genes in ESCs (Seo et al., 2020). However, we noticed that the 2C gene levels were increased in KO-ESCs (Seo et al., 2020). In our study, the mitochondrial fission inhibitor Mdivi-1 was used to effectively inhibit DRP1 activity, even at the protein level. In addition, treatment with Mdivi-1 could effectively increase the expression of 2C genes such as Zscan4 and Zfp352 in ESCs. This is in agreement with the result from Drp1-KO ESCs. It was suggested that the cellular pluripotency transformation disorder appeared due to inhibition of mitochondrial activity, which resulted in failed silencing of totipotency-related genes.
In this study, we identified that Mdivi-1 treatment facilitated the expression of 2C genes through accumulated H3K4me3 modification and reduced H3K27me3 modification. H3K4me3 is associated with gene activation (Liu et al., 2016). It is reported that highly upregulated 2C genes are correlated with the deposition of H3K4me3 (Zhang et al., 2021). The proportion of H3K4me3 tended toward TSSs was significantly increased in 2C-like cells compared with ESCs, which is similar to 2C embryos (Zhang et al., 2021). In our previous study, we noticed disturbed ZGA events in early 2C embryos treated with Mdivi-1 (Li et al., 2021). Interestingly, 2C genes were increased in late 2C embryos upon Mdivi-1 treatment. The role of retrotransposons in the regulation of transcription cannot be ignored, but our analysis results showed that the changes in transposons were not obvious, indicating that the upregulation of 2C genes may not be regulated by transposons. We propose that it was due to important roles of mitochondrial fission not only in totipotency establishment but also in the totipotency-to-pluripotency transition.
GO analysis in this study showed that the activity of the glycolysis/gluconeogenesis pathway decreased upon DRP1 inhibition, and the glycolysis/gluconeogenesis pathway-related enzymes were indeed significantly downregulated. Many stem cells including ESCs appear to be more dependent on glycolysis to produce adenosine-5 ′-triphosphate (ATP) than differentiated cells, and this phenomenon is similar to cancer cells (Rafalski et al., 2012). The highly active glycolysis pathway maintains the expression of pluripotent factors and the pluripotent state of mouse ESCs (Mandal et al., 2011). Notably, glycolysis participation is a key step in the transformation of terminally differentiated cells into induced pluripotent stem cells (iPSCs) (Varum et al., 2009; Rafalski et al., 2012). The expression level of pluripotency markers was increased when the activity of mitochondria was inhibited by chemical conditions, while pluripotency gene expression was not significantly affected in ESCs after Mdivi-1 treatment in our study. In Drp1-KO ESCs, the decreased activity of the glycolysis pathway was found, but there was no significant difference in the expression of pluripotent factors (Seo et al., 2020). Related studies have shown that the inhibition of the glycolysis pathway can promote the expression of 2C genes in ESCs (Hu et al., 2020). Pyruvate produced by the glycolysis pathway will participate in the mitochondrial TCA cycle. Acetyl-CoA is produced by mouse ESCs through the action of threonine dehydrogenase as an important source of energy metabolism (Mandal et al., 2011; Shyh-Chang et al., 2013). In addition to the downregulation of the glycolysis/gluconeogenesis pathway, a decrease in lipid kinase may lead to a decrease in lipid metabolic efficiency. Recent studies indicated that lipid metabolism has gradually become a critical regulator of stem cell differentiation (Mandal et al., 2011; Knobloch et al., 2013). The products of lipid metabolism also eventually participate in the TCA cycle. Although we did not find the significant changes in the TCA cycle in GO analysis, RNA-seq analysis showed that the expression of Pck2 was downregulated. Therefore, we hypothesized that decreased glycolytic pathway activity caused by abnormal mitochondrial fission would indirectly promote the upregulation of 2C genes in ESCs. This may be due to the change in metabolite quantity in the glycolysis/gluconeogenesis pathway, which indirectly affects TCA products and 2C gene expression. This deserves further experimental verification.
Our results showed that inhibition of DRP1 induced the increased levels of 2C genes in both mouse ESCs and mouse late 2C embryos, which may be due to the disturbed transition from totipotency-to-pluripotency status. After Mdivi-1 treatment, the intensity of H3K27me3, which is associated with gene repression, was decreased at the TSSs of upregulated genes in ESCs. In contrast, increased H3K4me3 occupancy and Pol II deposition at TSSs were observed. Therefore, inhibition of DRP1 stimulated H3K4me3 and Pol II accumulation at 2C genes. In addition, these results implied the change in metabolite quantity in the glycolysis/gluconeogenesis pathway may indirectly affect TCA products and the expression levels of 2C genes.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE221989.
The animal study was reviewed and approved by the Institutional Animal Care and Use Committee of Tongji Medical College, Huazhong University of Science and Technology.
YY and L-QZ conceived the idea and revised the manuscript. S-MG and Y-RZ performed the experiments and wrote the initial manuscript. B-XM helped with embryo manipulation. All authors contributed to the article and approved the final manuscript.
This work was supported by the National Natural Science Foundation of China (NSFC 32000488, 32170820, and 31771661), the National Key R&D Program of China (2018YFC1004000), and the program for HUST Academic Frontier Youth Team.
The authors thank all members of L-QZ’s laboratory for help on this project.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2023.1191797/full#supplementary-material
Adhikari, D., Lee, I. W., Al-Zubaidi, U., Liu, J., Zhang, Q. H., Yuen, W. S., et al. (2022). Depletion of oocyte dynamin-related protein 1 shows maternal-effect abnormalities in embryonic development. Sci. Adv. 8 (24), eabl8070. doi:10.1126/sciadv.abl8070
Aoki, F., Worrad, D. M., and Schultz, R. M. (1997). Regulation of transcriptional activity during the first and second cell cycles in the preimplantation mouse embryo. Dev. Biol. 181 (2), 296–307. doi:10.1006/dbio.1996.8466
Bordt, E. A., Clerc, P., Roelofs, B. A., Saladino, A. J., Tretter, L., Adam-Vizi, V., et al. (2017). The putative Drp1 inhibitor mdivi-1 is a reversible mitochondrial complex I inhibitor that modulates reactive oxygen species. Dev. Cell. 40 (6), 583–594. doi:10.1016/j.devcel.2017.02.020
Carbognin, E., Betto, R. M., Soriano, M. E., Smith, A. G., and Martello, G. (2016). Stat3 promotes mitochondrial transcription and oxidative respiration during maintenance and induction of naive pluripotency. Embo J. 35 (6), 618–634. doi:10.15252/embj.201592629
Cassidy-Stone, A., Chipuk, J. E., Ingerman, E., Song, C., Yoo, C., Kuwana, T., et al. (2008). Chemical inhibition of the mitochondrial division dynamin reveals its role in Bax/Bak-dependent mitochondrial outer membrane permeabilization. Dev. Cell. 14 (2), 193–204. doi:10.1016/j.devcel.2007.11.019
Cheng, S., Altmeppen, G., Welp, L. M., Penir, S., Ruhwedel, T., et al. (2022). Mammalian oocytes store mRNAs in a mitochondria-associated membraneless compartment. Sci. (New York, N.Y.) 378 (6617), eabq4835. doi:10.1126/science.abq4835
Chini, C. C. S., Zeidler, J. D., Kashyap, S., Warner, G., and Chini, E. N. (2021). Evolving concepts in NAD+ metabolism. Cell. Metab. 33 (6), 1076–1087. doi:10.1016/j.cmet.2021.04.003
De Iaco, A., Planet, E., Coluccio, A., Verp, S., Duc, J., and Trono, D. (2017). DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nat. Genet. 49 (6), 941–945. doi:10.1038/ng.3858
Fu, X., Djekidel, M. N., and Zhang, Y. (2020). A transcriptional roadmap for 2C-like-to-pluripotent state transition. Sci. Adv. 6 (22), eaay5181. doi:10.1126/sciadv.aay5181
Gafni, O., Weinberger, L., Mansour, A. A., Manor, Y. S., Chomsky, E., Ben-Yosef, D., et al. (2013). Derivation of novel human ground state naive pluripotent stem cells. Nature 504 (7479), 282–286. doi:10.1038/nature12745
Giacomello, M., Pyakurel, A., Glytsou, C., and Scorrano, L. (2020). The cell biology of mitochondrial membrane dynamics. Nat. Rev. Mol. Cell. Biol. 21 (4), 204–224. doi:10.1038/s41580-020-0210-7
Grow, E. J., Weaver, B. D., Smith, C. M., Guo, J., Stein, P., Shadle, S. C., et al. (2021). p53 convergently activates Dux/DUX4 in embryonic stem cells and in facioscapulohumeral muscular dystrophy cell models. Nat. Genet. 53 (8), 1207–1220. doi:10.1038/s41588-021-00893-0
Hu, Z., Tan, D. E. K., Chia, G., Tan, H., Leong, H. F., Chen, B. J., et al. (2020). Maternal factor NELFA drives a 2C-like state in mouse embryonic stem cells. Nat. Cell. Biol. 22 (2), 175–186. doi:10.1038/s41556-019-0453-8
Kleele, T., Rey, T., Winter, J., Zaganelli, S., Mahecic, D., Perreten Lambert, H., et al. (2021). Distinct fission signatures predict mitochondrial degradation or biogenesis. Nature 593 (7859), 435–439. doi:10.1038/s41586-021-03510-6
Knobloch, M., Braun, S. M. G., Zurkirchen, L., von Schoultz, C., Zamboni, N., Araúzo-Bravo, M. J., et al. (2013). Metabolic control of adult neural stem cell activity by Fasn-dependent lipogenesis. Nature 493 (7431), 226–230. doi:10.1038/nature11689
Le, R., Huang, Y., Zhao, A., and Gao, S. (2020). Lessons from expanded potential of embryonic stem cells: Moving toward totipotency. J. Genet. Genomics 47 (3), 123–130. doi:10.1016/j.jgg.2020.02.003
Li, Y., Mei, N. H., Cheng, G. P., Yang, J., and Zhou, L. Q. (2021). Inhibition of DRP1 impedes zygotic genome activation and preimplantation development in mice. Front. Cell. Dev. Biol. 9, 788512. doi:10.3389/fcell.2021.788512
Liu, X., Wang, C., Liu, W., Li, J., Li, C., Kou, X., et al. (2016). Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos. Nature 537 (7621), 558–562. doi:10.1038/nature19362
Ludikhuize, M. C., Meerlo, M., Gallego, M. P., Xanthakis, D., Burgaya Julià, M., Nguyen, N. T. B., et al. (2020). Mitochondria define intestinal stem cell differentiation downstream of a FOXO/notch Axis. Cell. Metab. 32 (5), 889–900. doi:10.1016/j.cmet.2020.10.005
Mandal, S., Lindgren, A. G., Srivastava, A. S., Clark, A. T., and Banerjee, U. (2011). Mitochondrial function controls proliferation and early differentiation potential of embryonic stem cells. Stem Cells 29 (3), 486–495. doi:10.1002/stem.590
Matilainen, O., Quirós, P. M., and Auwerx, J. (2017). Mitochondria and Epigenetics - crosstalk in homeostasis and stress. Trends Cell. Biol. 27 (6), 453–463. doi:10.1016/j.tcb.2017.02.004
Nagaraj, R., Sharpley, M. S., Chi, F., Braas, D., Zhou, Y., Kim, R., et al. (2017). Nuclear localization of mitochondrial TCA cycle enzymes as a critical step in mammalian zygotic genome activation. Cell. 168 (1-2), 210–223. doi:10.1016/j.cell.2016.12.026
Rafalski, V. A., Mancini, E., and Brunet, A. (2012). Energy metabolism and energy-sensing pathways in mammalian embryonic and adult stem cell fate. J. Cell. Sci. 125 (23), 5597–5608. doi:10.1242/jcs.114827
Rodrigues, A. S., Correia, M., Gomes, A., Pereira, S. L., Perestrelo, T., Sousa, M. I., et al. (2015). Dichloroacetate, the pyruvate dehydrogenase complex and the modulation of mESC pluripotency. PLoS One 10 (7), e0131663. doi:10.1371/journal.pone.0131663
Schmitt, K., Grimm, A., Dallmann, R., Oettinghaus, B., Restelli, L. M., Witzig, M., et al. (2018). Circadian control of DRP1 activity regulates mitochondrial dynamics and bioenergetics. Cell. Metab. 27 (3), 657–666. doi:10.1016/j.cmet.2018.01.011
Seo, B. J., Choi, J., La, H., Habib, O., Choi, Y., Hong, K., et al. (2020). Role of mitochondrial fission-related genes in mitochondrial morphology and energy metabolism in mouse embryonic stem cells. Redox Biol. 36, 101599. doi:10.1016/j.redox.2020.101599
Shyh-Chang, N., Locasale, J. W., Lyssiotis, C. A., Zheng, Y., Teo, R. Y., Ratanasirintrawoot, S., et al. (2013). Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science 339 (6116), 222–226. doi:10.1126/science.1226603
Srinivasan, R., Nady, N., Arora, N., Hsieh, L. J., Swigut, T., Narlikar, G. J., et al. (2020). Zscan4 binds nucleosomal microsatellite DNA and protects mouse two-cell embryos from DNA damage. Sci. Adv. 6 (12), eaaz9115. doi:10.1126/sciadv.aaz9115
Udagawa, O., Ishihara, T., Maeda, M., Matsunaga, Y., Tsukamoto, S., Kawano, N., et al. (2014). Mitochondrial fission factor Drp1 maintains oocyte quality via dynamic rearrangement of multiple organelles. Curr. Biol. 24 (20), 2451–2458. doi:10.1016/j.cub.2014.08.060
Varum, S., Momcilović, O., Castro, C., Ben-Yehudah, A., Ramalho-Santos, J., and Navara, C. S. (2009). Enhancement of human embryonic stem cell pluripotency through inhibition of the mitochondrial respiratory chain. Stem Cell. Res. 3 (2-3), 142–156. doi:10.1016/j.scr.2009.07.002
Wai, T., and Langer, T. (2016). Mitochondrial dynamics and metabolic regulation. Trends Endocrinol. Metabolism TEM 27 (2), 105–117. doi:10.1016/j.tem.2015.12.001
Wang, C., Chen, C., Liu, X., Li, C., Wu, Q., Chen, X., et al. (2022). Dynamic nucleosome organization after fertilization reveals regulatory factors for mouse zygotic genome activation. Cell. Res. 32 (9), 801–813. doi:10.1038/s41422-022-00652-8
Youle, R. J., and van der Bliek, A. M. (2012). Mitochondrial fission, fusion, and stress. Science 337 (6098), 1062–1065. doi:10.1126/science.1219855
Zhang, H., Menzies, K. J., and Auwerx, J. (2018). The role of mitochondria in stem cell fate and aging. Development 145 (8), dev143420. doi:10.1242/dev.143420
Zhang, Y., Huang, Y., Dong, Y., Liu, X., Kou, X., Zhao, Y., et al. (2021). Unique patterns of H3K4me3 and H3K27me3 in 2-Cell-like embryonic stem cells. Stem Cell. Rep. 16 (3), 458–469. doi:10.1016/j.stemcr.2021.01.020
Keywords: embryonic stem cell, DRP1, zygotic genome activation, totipotency, embryo
Citation: Guo S-M, Zhang Y-R, Ma B-X, Zhou L-Q and Yin Y (2023) Regulation of cleavage embryo genes upon DRP1 inhibition in mouse embryonic stem cells. Front. Cell Dev. Biol. 11:1191797. doi: 10.3389/fcell.2023.1191797
Received: 22 March 2023; Accepted: 26 April 2023;
Published: 15 May 2023.
Edited by:
Yi-Liang Miao, Huazhong Agricultural University, ChinaReviewed by:
Xiaogang Weng, Northeast Agricultural University, ChinaCopyright © 2023 Guo, Zhang, Ma, Zhou and Yin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li-Quan Zhou, emhvdWxpcXVhbkBodXN0LmVkdS5jbg==; Ying Yin, eWlueWluZ0BodXN0LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.