
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell Dev. Biol. , 02 August 2023
Sec. Molecular and Cellular Reproduction
Volume 11 - 2023 | https://doi.org/10.3389/fcell.2023.1177774
This article is part of the Research Topic Mammalian Spermatogenesis: Genetic and Environmental Factors View all 9 articles
 Jiachen Bai1,2,3†
Jiachen Bai1,2,3† Guizhen Zhou2†
Guizhen Zhou2† Shaopeng Hao3,4
Shaopeng Hao3,4 Yucheng Liu3
Yucheng Liu3 Yanhua Guo3
Yanhua Guo3 Jingjing Wang3
Jingjing Wang3 Hongtao Liu3
Hongtao Liu3 Longfei Wang2,3
Longfei Wang2,3 Jun Li5
Jun Li5 Aiju Liu2
Aiju Liu2 Wendell Q. Sun1
Wendell Q. Sun1 Pengcheng Wan3*
Pengcheng Wan3* Xiangwei Fu2,3*
Xiangwei Fu2,3*Semen cryopreservation is a promising technology employed in preserving high-quality varieties in animal husbandry and is also widely applied in the human sperm bank. However, the compromised qualities, such as decreased sperm motility, damaged membrane structure, and reduced fertilization competency, have significantly hampered the efficient application of this technique. Therefore, it is imperative to depict various molecular changes found in cryopreserved sperm and identify the regulatory network in response to the cryopreservation stress. In this study, semen was collected from three Chinese Merino rams and divided into untreated (fresh semen, FS) and programmed freezing (programmed freezing semen, PS) groups. After measuring different quality parameters, the ultra-low RNA-seq and tandem mass tag-based (TMT) proteome were conducted in both the groups. The results indicated that the motility (82.63% ± 3.55% vs. 34.10% ± 2.90%, p < 0.05) and viability (89.46% ± 2.53% vs. 44.78% ± 2.29%, p < 0.05) of the sperm in the FS group were significantly higher compared to those in the PS group. In addition, 45 upregulated and 291 downregulated genes, as well as 30 upregulated and 48 downregulated proteins, were found in transcriptomics and proteomics data separately. Moreover, three integrated methods, namely, functional annotation and enrichment analysis, Pearson’s correlation analysis, and two-way orthogonal partial least squares (O2PLS) analysis, were used for further analysis. The results suggested that various differentially expressed genes and proteins (DEGs and DEPs) were mainly enriched in leishmaniasis and hematopoietic cell lineage, and Fc gamma receptor Ia (FCGR1A) was significantly downregulated in cryopreserved sperm both at mRNA and protein levels in comparison with the fresh counterpart. In addition, top five genes (FCGR1A, HCK, SLX4, ITGA3, and BET1) and 22 proteins could form a distinct network in which genes and proteins were significantly correlated (p < 0.05). Interestingly, FCGR1A also appeared in the top 25 correlation list based on O2PLS analysis. Hence, FCGR1A was selected as the most potential differentially expressed candidate for screening by the three integrated multi-omics analysis methods. In addition, Pearson’s correlation analysis indicated that the expression level of FCGR1A was positively correlated with sperm motility and viability. A subsequent experiment was conducted to identify the biological role of FCGR1A in sperm function. The results showed that both the sperm viability (fresh group: 87.65% ± 4.17% vs. 75.8% ± 1.15%, cryopreserved group: 48.15% ± 0.63% vs. 42.45% ± 2.61%, p < 0.05) and motility (fresh group: 83.27% ± 4.15% vs. 70.41% ± 1.07%, cryopreserved group: 45.31% ± 3.28% vs. 35.13% ± 2.82%, p < 0.05) were significantly reduced in fresh and frozen sperm when FCGR1A was blocked. Moreover, the cleavage rate of embryos fertilized by FCGR1A-blocked sperm was noted to be significantly lower in both fresh (95.28% ± 1.16% vs. 90.44% ± 1.56%, p < 0.05) and frozen groups (89.8% ± 1.50% vs. 82.53% ± 1.53%, p < 0.05). In conclusion, our results revealed that the downregulated membrane protein FCGR1A can potentially contribute to the reduced sperm fertility competency in the cryopreserved sheep sperm.
Cryopreservation of semen has emerged as an important technology for efficient reproduction of livestock and poultry and protection of endangered species. It can aid to propagate animal offspring with excellent production performance, break the barriers to facilitate the exchange of genetic information caused by geographical isolation, and accelerate the promotion as well as application of in vivo and in vitro embryo production technologies (Kumar et al., 2019; Bolton et al., 2022). At present, slow freezing (also called programmed freezing) of semen has been widely used in animal breeding (Oldenhof et al., 2021) as well as in the donation and fertility preservation of human males (Tao et al., 2020; Agarwal et al., 2021).
It has been reported previously that cryoprotectant toxicity, osmotic pressure alteration, and ice crystal formation could inevitably induce distinct functional and structural damage in sperms during cryopreservation (Ezzati et al., 2020; Peris-Frau et al., 2020). Although optimized freezing procedures and the application of low-toxic cryoprotectants can help limit the formation of ice crystals and reduce the potential toxicity to the cell, decreased sperm viability as well as motility, reduced acrosome integrity, and compromised fertilization capability were observed due to the imbalance between the cooling rate and permeability velocity in the slow freezing program (Whaley et al., 2021). Moreover, it was reported that the elevated level of reactive oxygen species in the cryopreserved sperm could result in substantial DNA damage, plasma membrane damage, lipid peroxidation, and acrosomal membrane damage (Khan et al., 2021). Interestingly, a previous study has indicated that decreased DNA integrity induced by freezing could activate the repair pathway and alter the epigenetic reprogramming profile in the fertilized embryo (Wyck et al., 2018; Esteves et al., 2021). In addition, evidence has shown that both intact acrosomes and adequate sperm motility were required for proper fertilization (Yoshida et al., 2008). At the onset of the acrosome reaction, plasma membrane and acrosomal membrane were fused to form pores, which can release acrosome enzymes (Stival et al., 2016), and thus the intactness of the plasma/acrosomal membrane is pivotal for the successful acrosome reaction. Moreover, previous studies have also indicated that the plasma membrane intactness and acrosomal membrane integrity in the bovine cryopreserved sperm were reduced to 65.2% and 34.0%, respectively (Zoca et al., 2021). Furthermore, cryopreservation could also induce decreased sperm motility and viability (Estudillo et al., 2021). Interestingly, it was reported that decreased plasma membrane integrity could effectively lead to reduced sperm motility (Schulz et al., 2020). However, the molecular mechanisms underlying the compromised fertility in the cryopreserved sperm have not been fully identified yet.
Nowadays, significant progress has been made in understanding the sperm cryodamage mechanisms with the aid of high-throughput omics technologies. For instance, transcriptome analysis has revealed that numerous genes could be directly correlated with sperm fertility and freezing tolerance (Khan et al., 2021). Non-coding RNA sequencing of cryopreserved bull semen demonstrated that the differentially expressed miRNA and mRNA fragments were related to fertilization, ATP production, and apoptosis (Shangguan et al., 2020). miRNA sequencing of cryopreserved porcine semen indicated that various differentially expressed miRNAs were associated with energy metabolism, sperm structure, motility, and apoptosis (Zhang et al., 2017). The semen transcriptomes of porcine showed that DEGs, related to inflammation and apoptosis, spermatogenesis, autophagy, protein phosphorylation, and energy metabolism, were significantly upregulated in the semen with low freezability (Fraser et al., 2020). Furthermore, proteome determination of boar sperm and bull seminal plasma identifies cryogenic biomarkers to assess sperm motility (Moura and Memili, 2016; Parrilla et al., 2019). It has been established that compared to a single omics analysis, integrated multi-omics analysis is based on large and multidimensional data, which can substantially reduce bias as well as noise and identify the possible relationship between phenotypes and molecules (DNA, RNA, proteins, and metabolites) with greater precision (Krassowski et al., 2020). In fact, few prior studies have been performed on cryopreserved livestock sperm, especially sheep, using omics techniques. In recent years, integrated multi-omics analysis has emerged as the method of choice to decipher the cellular molecular information flow with spatial–temporal specificity (Subramanian et al., 2020). Thus, in order to comprehensively understand the regulatory network of RNA and protein in response to the cryopreservation stress, integrated ultra-low RNA-seq and TMT proteomics assay was applied in the present study. We expect that our result will definitely aid to provide novel ideas and theoretical basis for improving the cryopreservation technology of the sheep semen.
Three adult Chinese Merino rams with good nutrition, well-proportioned body, and normal libido were selected for the semen collection. All the rams were provided consistent feeding and management conditions with availability of free water. These rams were first trained in two semen collections, and finally the semen was collected for the third time. The collected semen was divided into two different parts: one was directly preprocessed before RNA and protein extraction (fresh semen, FS, n = 3), while the other was preprocessed after the freeze–thawing (programmed freezing semen, PS, n = 3). The RNA and proteins were extracted for ultra-low RNA-seq and TMT proteome determination (Figure 1A). However, only semen samples with motility higher than 80% in the FS group and higher than 30% in the PS group were used for omics analysis in the present study.
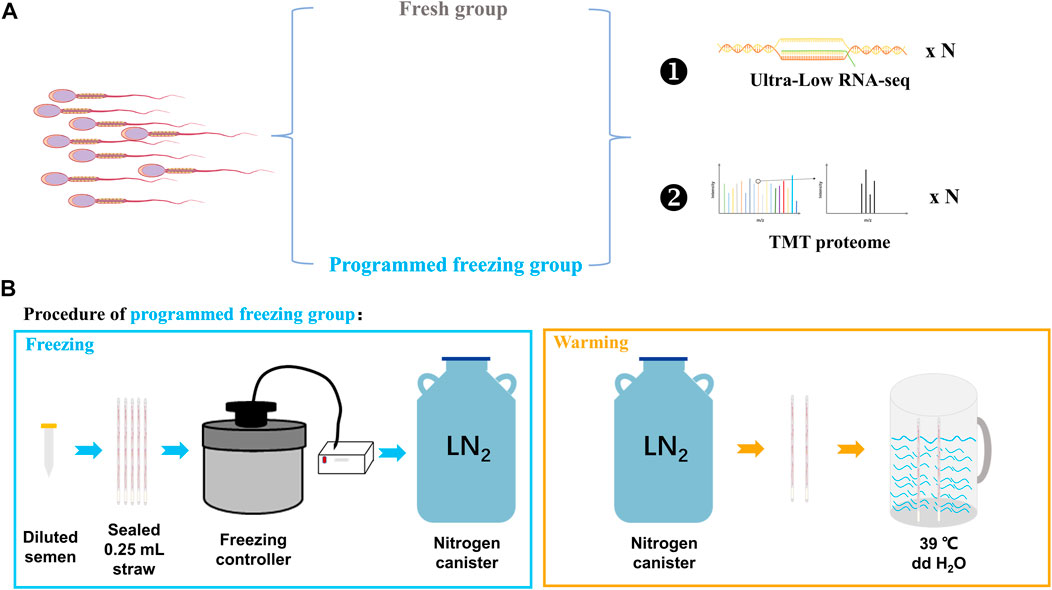
FIGURE 1. Experimental design for integrated multi-omics analysis before and after the programmed freezing semen in sheep. (A) Experimental grouping and omics analysis methods. (B) Schematic diagram depicting programmed freezing and thawing of sheep semen. ddH2O represents distillation–distillation H2O.
The collected fresh semen (sperm concentration: 2–3 × 109 sperm/mL) was diluted at a ratio of 1:2 with the freezing Medium for Semen (IMV Technologies, Aigle, France). Furthermore, 200–220 μL diluted semen was pipetted from the centrifuge tube into 0.25-mL straws (IMV Technologies, Aigle, France) by using a modified 1-mL syringe. The open mouths of the straws were then sealed with tweezers heated using an alcohol lamp. After pre-cooling in a 4°C refrigerator freezer for 20 min, the sealed straws were thereafter inserted into slots of the freezer which have been pre-cooled with liquid nitrogen controlled using a freezing controller (CryoLogic CL5500TC, Australia) in advance. The freezing program of the freezing controller was that the temperature cooled down from 4°C to −30°C at a cooling rate of 3°C/min and from −30°C to −80°C at a cooling rate of 10°C/min (Supplementary Table S1). Finally, the straws were quickly removed from the freezing controller and stored in liquid nitrogen. For thawing, the sealed end of the straw was held with the forceps, removed from the liquid nitrogen, and shaken 2–3 times in the air. The straw was then quickly placed into a rewarming cup at 39°C for 30 s (Figure 1B). Finally, the rewarmed straw was incubated in a 38.5°C incubator for temporary storage.
The semen was diluted with the Freezing Medium for Semen to achieve the condition such that the sperm concentration was 3–5 × 107 sperm/mL. A volume of 3 μL of semen was then pipetted onto an object slide from the upper-middle layers of the diluted semen. The sperm quality (viability, motility, velocity straight line, velocity curve line, average path velocity, and amplitude of lateral head) was measured for both FS and PS groups using the Mailang Animal Sperm Quality Analyzer (Nanning SongJing Tianlun Biotechnology Co., Ltd., Nanning, China).
A volume of 400 μL of the diluted semen (sperm concentration: 6.6–10 × 108 sperm/mL) was first centrifuged at 2,600 g for 3 min at room temperature, and then the supernatant was removed. A volume of 1 mL of physiological saline was added to resuspend the precipitate, and the mixture was centrifuged again at 2,600 g for 3 min, and then the supernatant was removed. The precipitate of the semen was mixed and resuspended in 500 μL of somatic cell lysis buffer (Allwegene, Beijing, China). After incubation for 5 min, the mixture was centrifuged at 10,000 g for 3 min; the supernatant was removed completely and stored in the liquid nitrogen.
The micro-amplification of RNA samples was carried out based on the instructions of the RNALib Single cell WTA Kit (Allwegene, Beijing, China). cDNA library construction was performed based on the instructions of Lifeint Transpose DNA Library Prep Kit for Illumina (Allwegene, Beijing, China). Thereafter, cDNAs, which could meet the quality requirements for the concentration and fragment size, were sequenced using the SE50 read length of the BGISEQ platform (Allwegene, Beijing, China). Fastp version 0.23.0, FastQC version 0.11.9, and MultiQC version 1.11 were used for cutting out the sequencing adapters and low-quality bases. HISAT version 2.2.1 was used to align various quality-controlled reads to the reference genome of Ovis aries (ARS-UI_Ramb_v2.0). After sorting using SAMtools version 1.12, the read numbers of the genes were counted using StringTie version 2.16. The DEGs were tested and compared using Student’s t-test and Benjamini and Hochberg (BH) method.
A volume of 100 μL diluted semen (sperm concentration: 6.6–10 × 108 sperm/mL) was initially directly mixed with 500 μL of 4°C pre-cooled normal saline and centrifuged at 100 g for 5 min, and then the supernatant was discarded. The precipitate of the semen was then mixed again with 300 μL of the pre-cooled normal saline and centrifuged at 1,000 g for 5 min, and thereafter the supernatant was discarded. The precipitate of the semen was mixed with 500 μL of 4% SDS solution in the centrifuge tube which was maintained at 95°C ddH2O for 10 min. After centrifugation at 13,800 g for 20 min, the supernatant was dispensed into cryovials and stored in liquid nitrogen.
Protein extraction and digestion, labeling, TMT labeling, and fractionation were performed by Allwegene Technology Inc., Beijing, China. LC-MS/MS analysis was carried out using a Q Exactive mass spectrometer coupled to the Easy-nLC system by Allwegene Company. The MS raw data for each sample were thereafter searched using Mascot version 2.2 (Matrix Science, London, United Kingdom) embedded into Proteome Discoverer 1.4 software to facilitate both identification and quantitation analysis. Different parameters and instructions of Mascot are summarized in Supplementary Table S2.
PCoA and PCA were performed using the prcomp function of the R stats package. K-means clustering analysis of the samples of each omics was presented as heatmaps with dendrograms using the R package pheatmap. Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) enrichment analyses were performed using KOBAS (http://bioinfo.org/kobas/) for determining DEGs and DEPs. Gene set enrichment analysis (GSEA) was carried out on the identified clusters of the genes to reveal the functions of quantificational genes using OmicStudio tools (https://www.omicstudio.cn/tool). Clusters of Orthologous Groups of proteins and euKaryotic Orthologous Group (COG/KOG) classification analysis and subcellular localization analysis were performed at http://pantherdb.org/webservices/go/overrep.jsp.
First, Co-KEGG analysis of DEGs and DEPs was carried out using KOBAS. Second, Pearson’s correlation analysis was used to analyze the correlation between different genes and proteins enriched for various significant pathways from single KEGG analysis of RNA-seq and TMT proteome. The |rho| > 0.8 and p < 0.05 were used as the potential thresholds for a significant correlation. Third, O2PLS analysis between RNA-seq and TMT proteome was performed at https://www.omicshare.com/tools/Home/Soft/o2pls. The top 25 joint loading genes and proteins were selected as the significant correlation components between the two omics. All the plotting was performed using OmicShare (https://www.omicshare.com/tools) and OmicStudio (https://www.omicstudio.cn/tool).
The blocking was carried out based on a method described in a previous study (Rival et al., 2019) with some modifications. Briefly, 500 µL fresh semen (sperm concentration: 2–3 × 109 spermatozoa per mL) was added to 500 µL of in vitro fertilization (IVF) solution and then centrifuged at 43 g for 3 min. The supernatant was transferred to 500 µL IVF solution and centrifuged at 160 g for 3 min, and thereafter the sediment was collected. For the frozen semen, 130 µL semen (sperm concentration: 6.6–10 × 108 sperm/mL) was added to 1 mL of IVF solution after thawing and centrifuged at 43 g for 5 min, and thereafter the sediment was obtained. A volume of 100 μL of the sediment from fresh and frozen semen was used for the subsequent blocking experiments. For blocking, the anti-FCGR1A antibody (Bioss, Beijing, China) was incubated with semen (sperm concentration: 2–3 × 108 sperm/mL) at a working concentration of 50 μg/mL for 30 min.
The sheep ovaries were collected from the local slaughterhouse. Cumulus–oocyte complexes (COCs) were obtained via aspiration of the follicles with 3–6 mm size. Thereafter, the COCs were subjected to in vitro maturation for 24 h at 38.5°C and 5% CO2. A volume of 3 μL semen (sperm concentration: 2–3 × 108 sperm/mL) was added to 80 μL droplet containing 35–40 oocytes for fertilization. After 18 h of IVF, the embryos were finally washed and transferred to the IVC solution to facilitate embryonic development.
Statistically significant differences for various sperm quality parameters, cleavage rates, and blastocyst rates were determined using Student’s t-test. All statistical analyses were carried out using SPSS version 24.0 (IBM Corporation, Armonk, USA), and the data are presented as mean ± SEMs. p values <0.05 were considered significant, and p values <0.001 were considered extremely significant.
The results indicated that sperm motility (82.63% vs. 34.10%, p < 0.001) and viability (89.46% vs. 44.78%, p < 0.001) of the FS group were significantly higher than those of the PS group. The sperm motion parameters including velocity curve line (87.71 um/s vs. 68.39 um/s, p < 0.05), average path velocity (61.99 um/s vs. 48.36 um/s, p < 0.05), and amplitude of lateral head (25.69 um vs. 20.03 um, p < 0.05) were also significantly higher in the FS group compared to those of the PS group. However, the velocity straight line (43.65 um/s vs. 38.42 um/s, p > 0.05) was not significantly different between the two groups (Table 1).
After the quality control of RNA-seq data, total reads of RNA-seq were aligned and counted. Among the total 310,699,628 reads, 186,669,343 (60.08%) unique mapped reads were obtained (Supplementary Table S3). Variance analysis of fragments per kilobase of the exon model per million mapped fragments (FPKM) for all the genes indicated that a significant difference could be found between FS and PS groups (p < 0.001) (Figure 2A). The six samples of the two groups were distributed in two different triangular areas under PCo1 (47.25%) and PCo2 (30.47%) after projection of the multidimensional gene expression data (Figure 2B). K-means clustering analysis indicated that the six samples were correctly clustered in both FS and PS groups through the centralization and normalization of the read numbers of DEGs (Figure 2C). A total of 7,682 genes were then identified from RNA-seq. Additionally, under the threshold value of Log2|fold change| > 1 and false discovery rate (FDR) < 0.05, 45 upregulated and 291 downregulated genes were obtained (Figure 2D). The top 20 DEGs are listed in Table 2.

FIGURE 2. Quality assessment of RNA-seq data and the subsequent analysis of DEGs. (A) Boxplot of RNA-seq differences between FS and PS groups. (B) PCoA distribution of the samples obtained from FS and PS groups. (C) Heatmap of expression comparison of DEGs and clustering of the samples obtained from FS and PS groups. (D) Volcano plot of various quantificational genes for RNA-seq compared with FS and PS groups. F_SR represents the sample from RNA-seq of the FS group. P_SR represents the sample from RNA-seq of the PS group. Sig_Up represents the upregulated DEGs. Sig_Down represents the downregulated DEGs.
The DEGs were then subjected to GO classification and KEGG enrichment analyses. GO classification analysis revealed that the number of DEGs distributed in the translation biological process (BP), intracellular protein transport BP, cytosol cellular component (CC), nucleus CC, metal ion-binding molecular function (MF), and identical protein-binding MF was 18, 9, 68, 56, 37, and 24, respectively (Figure 3A; Supplementary Table S4). A total of nine different significant pathways were identified from the KEGG enrichment analysis including oxytocin signaling pathway, Fanconi anemia pathway, and selenocompound metabolism. (Figure 3B; Supplementary Table S5). GSEA based on the quantificational genes indicated that the differential gene sets were mainly enriched in leishmaniasis, ferroptosis, cholinergic synapse, B-cell receptor signaling pathway, antifolate resistance, and alcoholism (Figures 4A–F; Supplementary Table S6). In addition, various DEGs, including CYBB, FCGR1A, GABRA3, HDAC10, INPP5D, LOC106991388, PIK3CG, and SLCO1A2, were also involved in these pathways (Figure 4G).
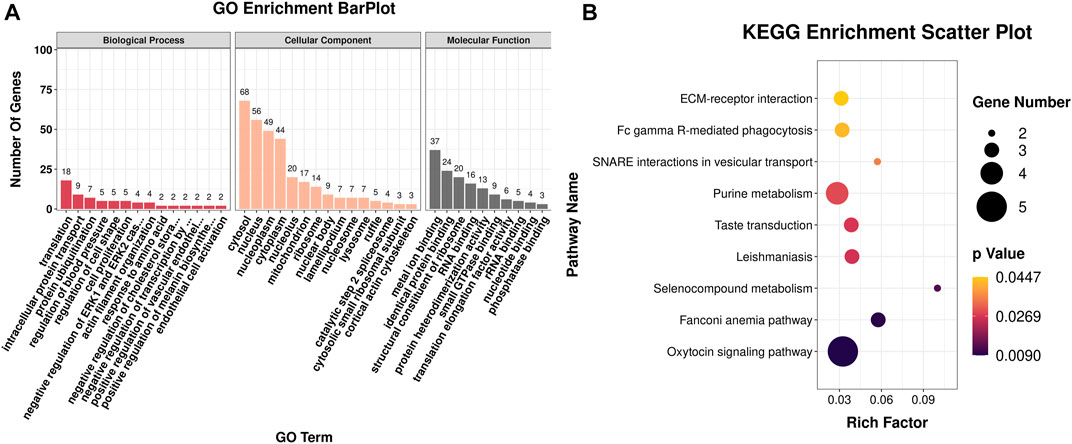
FIGURE 3. GO classification and KEGG enrichment analyses of DEGs obtained from RNA-seq. (A) Bar plot of GO classification of DEGs. (B) Scatter plot of the KEGG enrichment pathways of DEGs.
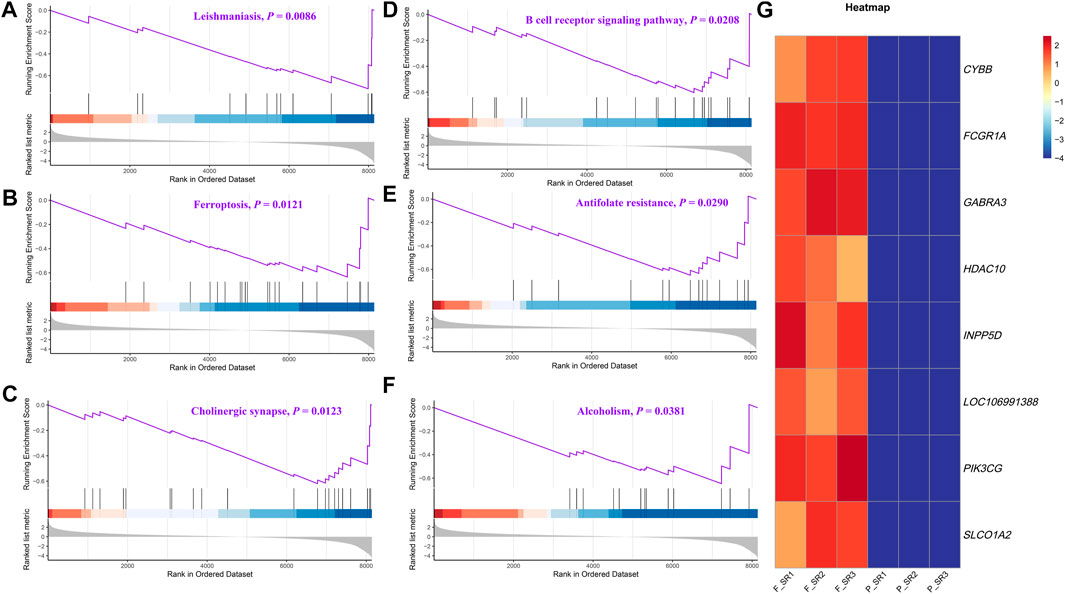
FIGURE 4. GSEA of the quantificational genes from RNA-seq. Representative GSEA significance pathways of gene sets, including those of leishmaniasis (A), ferroptosis (B), cholinergic synapse (C), B-cell receptor signaling pathway (D), antifolate resistance (E), and alcoholism (F). (G) Heatmap of the relative expression of DEGs enriched in significant pathways by GSEA.
After quality control, a total of 926,536 spectrums were acquired from TMT proteome, including 11,357 unique peptides and 2,639 identified proteins. Finally, 2,634 quantifiable proteins were obtained (Figure 5A). PCA demonstrated that six samples from FS and PS groups were aggregated within the group and then separated between the groups under PC1 (39.3%) and PC2 (26.1%) after projection of the multidimensional protein expression data (Figure 5B). K-means clustering analysis of DEPs also confirmed the PCA results (Figure 5C). A total of 2,631 proteins were identified from TMT proteome. Under the threshold value of Log2|fold change|> 0.263 and FDR <0.05, 30 up- and 48 downregulated proteins were obtained (Figure 5D). The top 20 DEPs are listed in Table 3.
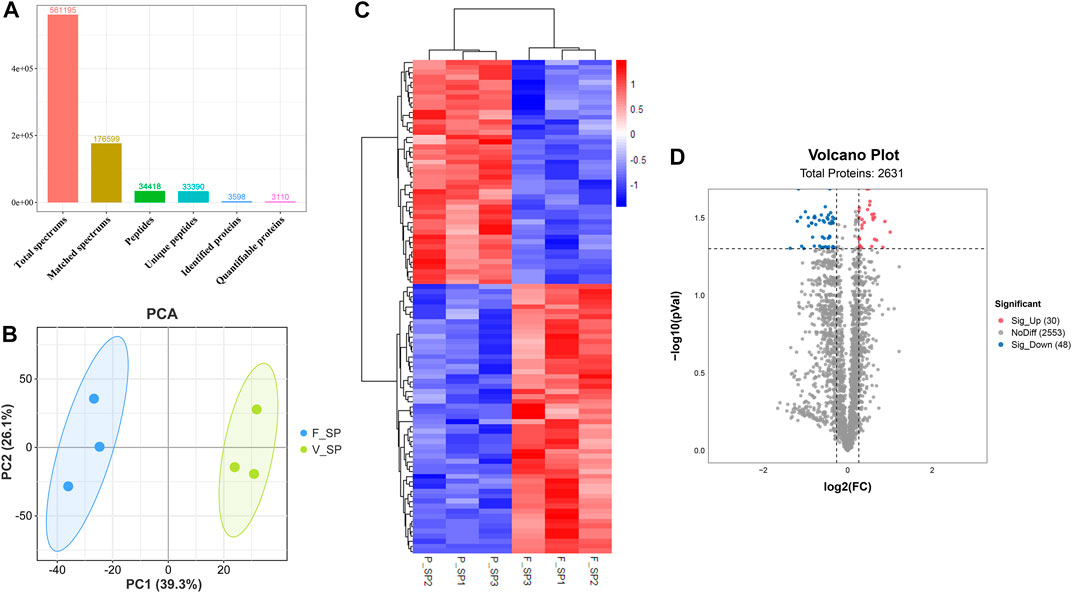
FIGURE 5. Quality assessment of TMT proteome data and analysis of DEPs. (A) Key information for each procedure of the proteome analysis. (B) PCA distribution of the samples from FS and PS groups. (C) Expression comparison of proteins and clustering of the samples from FS and PS groups. (D) Volcano plot of various quantificational proteins for proteome obtained from FS and PS groups. F_SP represents the sample from TMT proteome of the FS group. P_SP represents the sample from TMT proteome of the PS group. Sig_Up represents the upregulated DEPs. Sig_Down represents the downregulated DEPs.
COG/KOG category analysis indicated that 12 and 7 DEPs were mainly involved in signal transduction mechanisms and post-translational modification, respectively (Figure 6A; Supplementary Table S7). The subcellular localization analysis suggested that DEPs mainly belonged to the protein-modifying enzyme (27.08%) and metabolite interconversion enzyme (27.08%) (Figure 6B; Supplementary Table S8). Based on BP, CC, and MF classifications, proteins were mainly found to be involved in the cellular process BP, metabolic process BP, cellular anatomical entity CC, catalytic activity MF, and bind MF with the percentage of 49.3%, 32.4%, 67.6%, 33.8% and 19.7%, respectively (Figure 6C; Supplementary Table S9). KEGG enrichment analysis revealed that various DEPs were enriched in different metabolic pathways, such as amino sugar and nucleotide sugar metabolism, pyrimidine metabolism, fructose and mannose metabolism, and lysosome (Figure 6D; Supplementary Table S10).
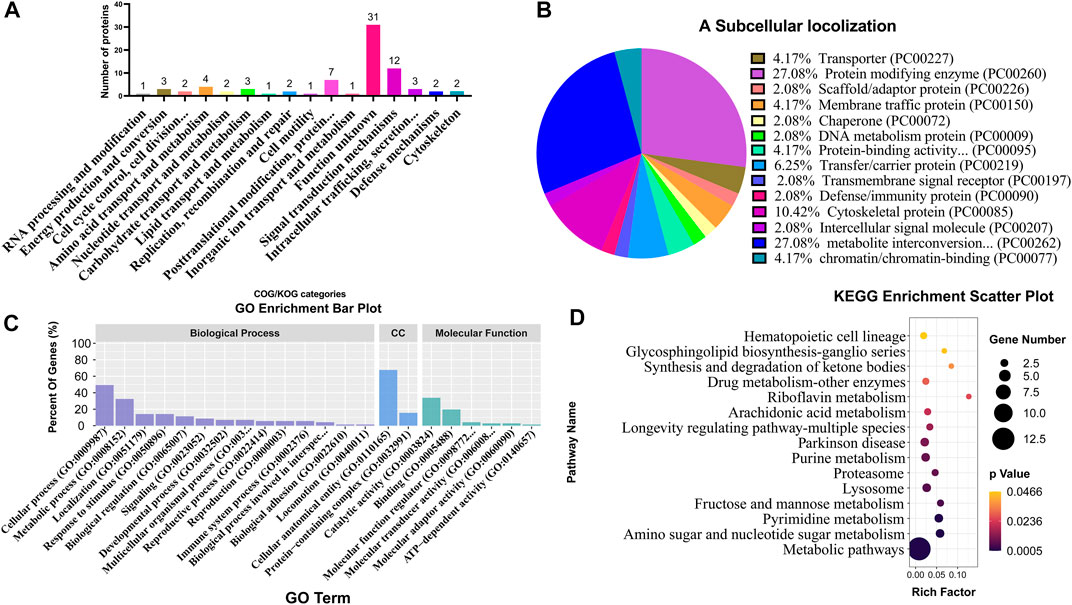
FIGURE 6. Classification and enrichment analyses of different DEPs from TMT proteome. COG/KOG classification analysis (A), subcellular localization analysis (B), GO classification analysis (C), and KEGG enrichment analysis (D) of DEPs. CC represents cellular components.
Co-KEGG enrichment analysis of DEGs and DEPs revealed that the differentially expressed candidates were involved in 14 different pathways (p < 0.05; Figure 7A). Among these 14 pathways, four major pathways were enriched: SNARE interactions involved in the vesicular transport (BET1 and STX6), leishmaniasis (FCGR1A, CYBB, and FOS), hematopoietic cell lineages (FCGR1A, ITGA3, MME, and CD5) and Fanconi anemia pathway (SLX4, ATR, and TELO2) (Figure 7B; Supplementary Table S11).
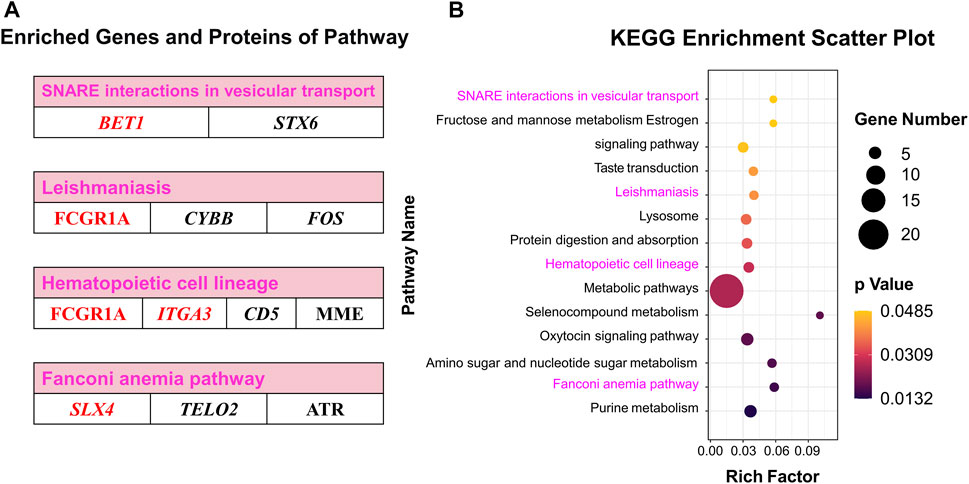
FIGURE 7. Co-KEGG enrichment analysis and specific DEGs and DEPs contained in significant pathways. (A) Enriched genes and proteins in interesting pathway. (B) Scatter plot of the Co-KEGG enrichment pathways of DEGs and DEPs. Words in red represent interesting genes or proteins.
Pearson’s correlation analysis indicated that there were 22 distinct proteins (JEQ12_018837, MME, TKFC, PTGDS, JEQ12_020033, HEXB, FCGR1A, PARK7, DNASE2, PDE10A, UCHL1, GALT, MPI, SOD1, JEQ12_002945, HSPA1L, CMPK1, JEQ12_010623, PSME4, OXCT1, CTSD, and PSMA8) which were significantly correlated with the top five genes: FCGR1A, HCK, SLX4, ITGA3, and BET1. The number of proteins significantly associated with FCGR1A, HCK, SLX4, ITGA3, and BET1 was 22, 19, 15, 14, and 11, respectively (Figures 8A,B; Supplementary Table S12).
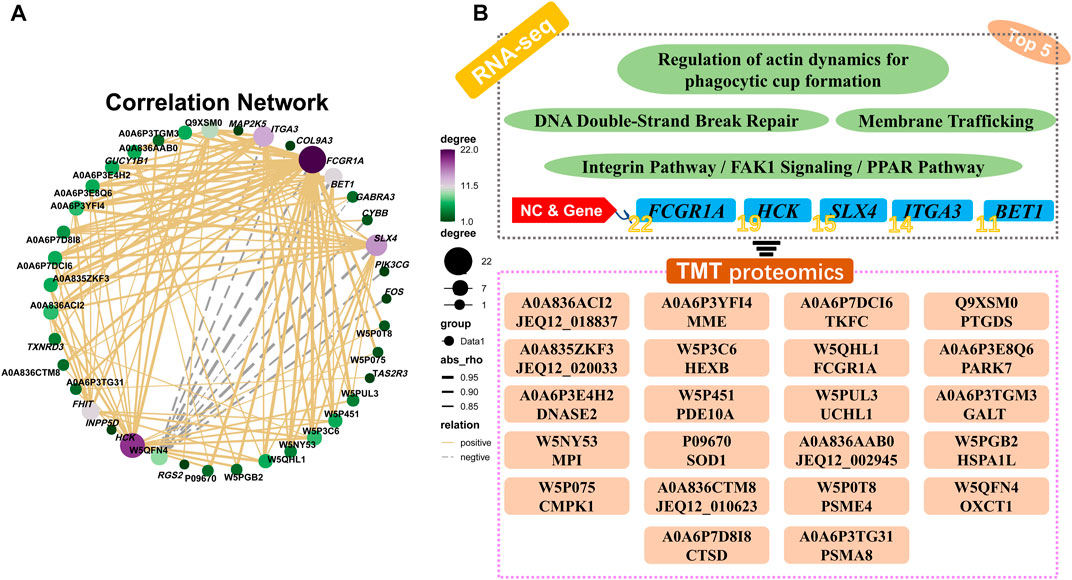
FIGURE 8. Pearson’s correlation analysis and the relational network between pathway-enriched DEGs and DEPs. (A) Pearson’s correlation analysis of DEGs and DEPs in significant pathways obtained by KEGG enrichment analyses of RNA-seq and TMT proteome. (B) Potential relationships and functional pathways of DEGs and DEPs.
O2PLS analysis was performed between all the quantificational genes and proteins obtained from RNA-seq and proteome analyses. Thereafter, the top 25 genes and proteins contributing to the changes after cryopreservation were plotted (Figure 9A; Supplementary Table S13). Interestingly, FCGR1A appeared in both the top 25 gene and protein lists. Notably, FCGR1A was included in the list of 15 DEGs and DEPs (BET1, FCGR1A, ITGA3, SLX4, CMPK1, CTSD, DNASE2, GALT, HEXB, HSPA1L, MME, MPI, PDE10A, PTGDS, and TKFC) based on Co-KEGG enrichment analysis and Pearson’s correlation analysis (Figure 9B). In addition, it was also the only gene screened by three integrated multi-omics analysis methods (Figure 9C).
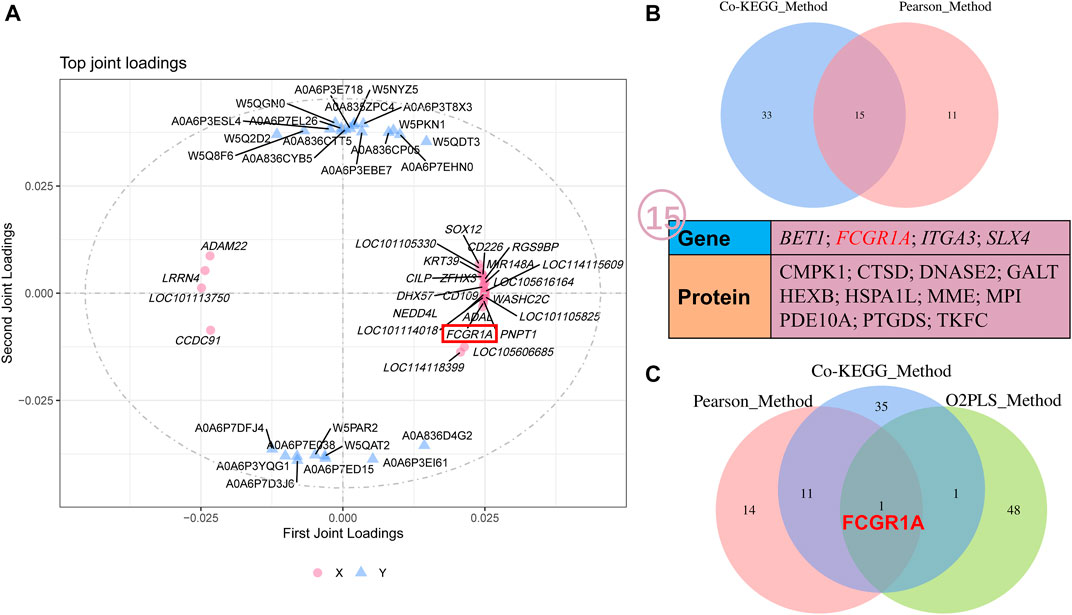
FIGURE 9. O2PLS analyses of all the quantificational genes and proteins derived from RNA-seq and TMT proteome. (A) O2PLS analysis of all the quantificational genes and proteins. (B) Collection of various genes and proteins between Co-KEGG enrichment analysis and Pearson’s correlation analysis. (C) Collection of genes and proteins between Co-KEGG enrichment analysis, Pearson’s correlation analysis, and O2PLS analysis. KEGG_Method represents Co-KEGG enrichment analysis. Pearson_Method represents Pearson’s correlation analysis. O2PLS_Method represents O2PLS analysis.
We first evaluated the viability and motility after sperm preparation, and the results indicated that the sperm viability (87.65% ± 4.17% vs. 48.15% ± 0.63%, p < 0.001) and motility (83.27% ± 4.15% vs. 45.31% ± 3.28%, p < 0.001) of the fresh group was significantly higher than those of the frozen counterpart (Figures 10A,B). Pearson’s correlation analysis showed that both the sperm motility and viability were positively correlated with FCGR1A abundance based on omics data (Table 4; p < 0.05). Furthermore, blocking experiments were performed, and the result suggested that the viability (87.65% ± 4.17% vs. 75.8% ± 1.15%, p < 0.05) as well as motility (83.27% ± 4.15% vs. 70.41% ± 1.07%, p < 0.05) was significantly reduced in the fresh sperm blocked with the FCGR1A antibody. Interestingly, the dramatically decreased viability (48.15% ± 0.63% vs. 42.45% ± 2.61%, p < 0.05) and motility (45.31% ± 3.28% vs. 35.13% ± 2.82%, p < 0.05) were also observed in the frozen sperm blockage (Figures 10A,B). Moreover, when FCGR1A was blocked, the cleavage rate of embryos derived from either the fresh sperm (95.28% ± 1.16% vs. 90.44% ± 1.56%, p < 0.05) or the frozen sperm (89.8% ± 1.50% vs. 82.53% ± 1.53%, p < 0.05) was significantly lower compared to that of the corresponding unblocked counterparts (Figure 10C). However, no significant difference in the blastocyst formation rate was observed (Figure 10D).
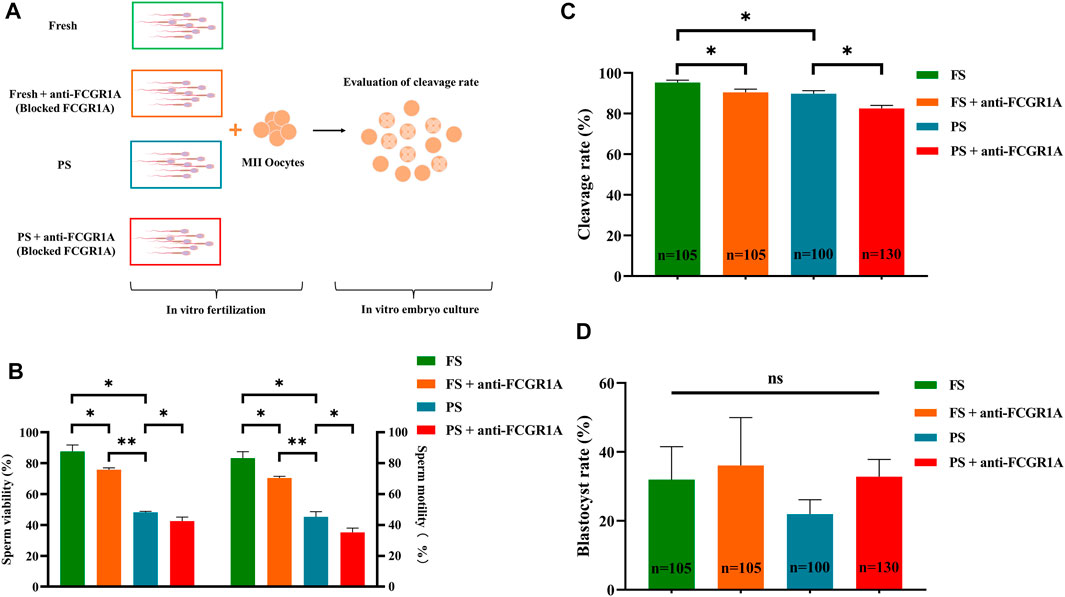
FIGURE 10. Correlation of FCGR1A with sperm fertilization ability. (A) Experimental design for the evaluation of the fertilization ability of FCGR1A-blocked sperm. (B) Difference between the sperm motility and viability after IVF with sperm obtained from different treatments. (C) Difference between the cleavage rate after IVF with sperm obtained from the different treatments. (D) Difference between blastocyst rates after in vitro fertilization with sperm from different treatments. FS represents fresh sperm after floating. FS + anti-FCGR1A represents fresh sperm blocked by anti-FCGR1A. PS represents programmed freezing sperm after floating. PS + anti-FCGR1A represents fresh sperm blocked by anti-FCGR1A. *p < 0.05, **p < 0.001; ns, non-significance.
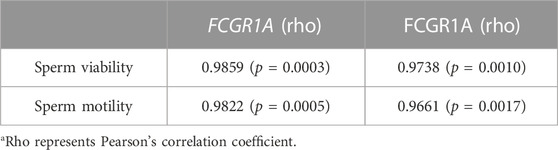
TABLE 4. List of possible correlations between the sperm motility, viability, and FCGR1A expression level.
During programmed freezing, the stresses originating from multiple sources could lead to substantial structural damage and metabolic changes in sperm, which subsequently might impair sperm motility and fertilization ability. However, the molecular mechanisms underlying compromised fertilization competency have not been identified yet. In the present study, we have identified that FCGR1A was primarily responsible for the decreased sperm fertilization capacity using integrated transcriptomics and proteomics assays.
It has been reported previously that compared to the fresh semen, the total motility as well as progressive motility of the semen was significantly reduced after programmed freezing in Slovak dairy, Native Wallachian, and Improved Wallachian sheep rams (Vozaf et al., 2022). In consistent with the previous studies, we also discovered that severe cryodamage could effectively directly lead to reduced viability and motility in sheep. A number of prior studies have also indicated that the plasma and acrosome membranes can play an essential role in the capacitation, acrosome reaction, and sperm–oocyte recognition (Ritagliati et al., 2018; Bianchi and Wright, 2020). In addition, sperm motility is associated with ion channel proteins at the plasma membrane that can regulate potential and pH of the plasma membrane, which are essential for sperm fertilization (Nowicka-Bauer and Szymczak-Cendlak, 2021). However, till date, the specific mechanisms underlying sperm fertilization ability remain unclear.
To thoroughly illustrate the regulatory network involved in reduced sperm fertilization due to cryopreservation stress, integrated transcriptome and proteome analysis was first performed in the current study. A large number of up- and downregulated DEGs and DEPs were identified. It has been established that mature sperm are often in a state of transcriptional and translational silencing due to compacted DNA and absence of organelles (Ren et al., 2017). Without new RNA produced in the mature sperm before and after freezing, only differential degradation amount of RNA that was already synthesized and stored prior to transcriptional arrest was taken into account (Lalancette et al., 2008). For instance, the m6A modification can regulate the stabilization, degradation, and translation of mRNA (Wang et al., 2015; Edupuganti et al., 2017; Qin et al., 2021). Thus, significant changes in methylation modifications on sperm transcripts caused by freezing could result in up- and downregulation of mRNA at the transcriptome level due to possible differences in mRNA degradation rates between the fresh and frozen semen. As expected, numerous studies have also identified several up- and downregulated transcripts in the frozen semen (Dai et al., 2019; Wang et al., 2022), and a previous study has indicated that the number of up- and downregulated genes in bull frozen semen compared with the fresh semen was 241 and 662, respectively (Ebenezer Samuel King et al., 2022). In addition, numerous past studies have also explored potential changes in mRNA and protein levels in the frozen semen of various species, including boar (Chen et al., 2014; Dai et al., 2019; Fraser et al., 2020), human (Wang et al., 2022), and sheep (Jia et al., 2022) by transcriptomics or proteomics. For example, in the transcriptome analysis of the fresh and frozen boar semen, differentially expressed mRNA and miRNA were found to be mainly related to environmental stress, apoptosis, and metabolism (Dai et al., 2019). Comparative transcriptomics analysis has revealed that compared with the fresh sperm, DEGs of human frozen and vitrified sperm were associated with different immune and infectious diseases (Wang et al., 2022). These findings further corroborate with the results of our study that the enriched pathways could be mainly categorized into metabolism, immune response, disease, and signal transduction. Comparative proteomics analysis of the fresh and frozen boar semen has revealed that most of the DEPs primarily participated in the sperm premature capacitation, adhesions, energy supply, and sperm–oocyte binding and fusion (Chen et al., 2014). Isobaric tags for the relative and absolute quantification combined with parallel reaction monitoring (PRM) proteomics analysis of the fresh and frozen sheep semen indicated that DEPs were enriched in metabolic activities, disease, and oxidative phosphorylation pathways (Jia et al., 2022). In this study, DEPs were mainly involved in the heterogeneous metabolic pathways. The variability between the studies could be attributed to the possible differences in experimental species, freezing procedures, and detection methods.
For in-depth analysis, integrated Co-KEGG enrichment analysis and Pearson’s correlation analysis were conducted, and the results revealed that FCGR1A, ITGA3, SLX4, and BET1 gene clusters were correlated with 11 distinct DEPs (Figures 11A,B). Our findings indicated that FCGR1A was more than a member of DEG/DEP as it could also associate with the highest number of proteins in Pearson’s correlation analysis. Interestingly, a prior report has indicated that FCGR1A was involved in the modulation of immune response and mainly expressed on the cell membrane of leukocytes (Barb, 2021). Additionally, in neutrophilic granulocytes, FCGR1A can act as a receptor for signal transduction and induce dynamic changes in kinase expression, which can finally result in cell morphology modification to enhance phagocytosis (Futosi et al., 2013). Furthermore, in the monocytes, the binding of FCGR1A to the ligands can lead to phagocytosis and promote the release of various inflammatory factors and reactive oxygen species (Swisher and Feldman, 2015). Moreover, a previous study has reported that FCGR1A was located on the sperm plasma membrane (Hellstrom et al., 1988). However, the function of FCGR1A in sperm fertilization remains undetermined until now. In the present study, ITGA3 was also associated with DEPs. ITGA3, a member of a huge integrin family, can mainly combine with ITGB1 to form a complete integrin and play a vital role in sperm adhesion and sperm–oocyte fusion (Merc et al., 2021). It has been reported that the integrin could act as an essential transmembrane protein and play an important role in the signal transduction process by interacting with intracellular kinases or acting indirectly with actin (Green and Brown, 2019). Therefore, we hypothesized that the downregulated ITGA3 by freezing might inhibit the binding of the frozen sperm to the oocytes. Interestingly, SLX4 was identified to be correlated with DEPs. Another previous study has shown that the SLX1–SLX4 complex plays a vital role in maintaining the genome stability, including preventing the collapse of the replication forks and cleaving Holliday junctions (Fricke and Brill, 2003; Garner et al., 2013). Xu et al. (2021) discovered that SLX1 could process 5′-flap DNA efficiently and rapidly primarily by cooperating with SLX4 in the presence of the SAP domain. In addition, it has been reported that increased sperm DNA fragmentation was found in the frozen sheep sperm (Palazzese et al., 2018), and the phenomenon might be explained by the reduced SLX4 transcript discovered in the present study since SLX4 could contribute to the DNA instability. In addition, BET1 was identified as another vital gene associated with DEPs. For example, a prior study has indicated that BET1 could be involved in endoplasmic reticulum-secreted vesicle recognition, fusion, and molecule transport to the Golgi apparatus (Zhang et al., 1997; Malsam and Söllner, 2011). It is well known that the acrosome is developed from the Golgi apparatus and contains diverse digestive enzymes required for sperm penetration (Khawar et al., 2019). As a membrane protein, BET1 probably can integrate into the Golgi apparatus, which may eventually evolve into the acrosome membrane, and the reduced BET1 content might impair sperm fertilization capability.
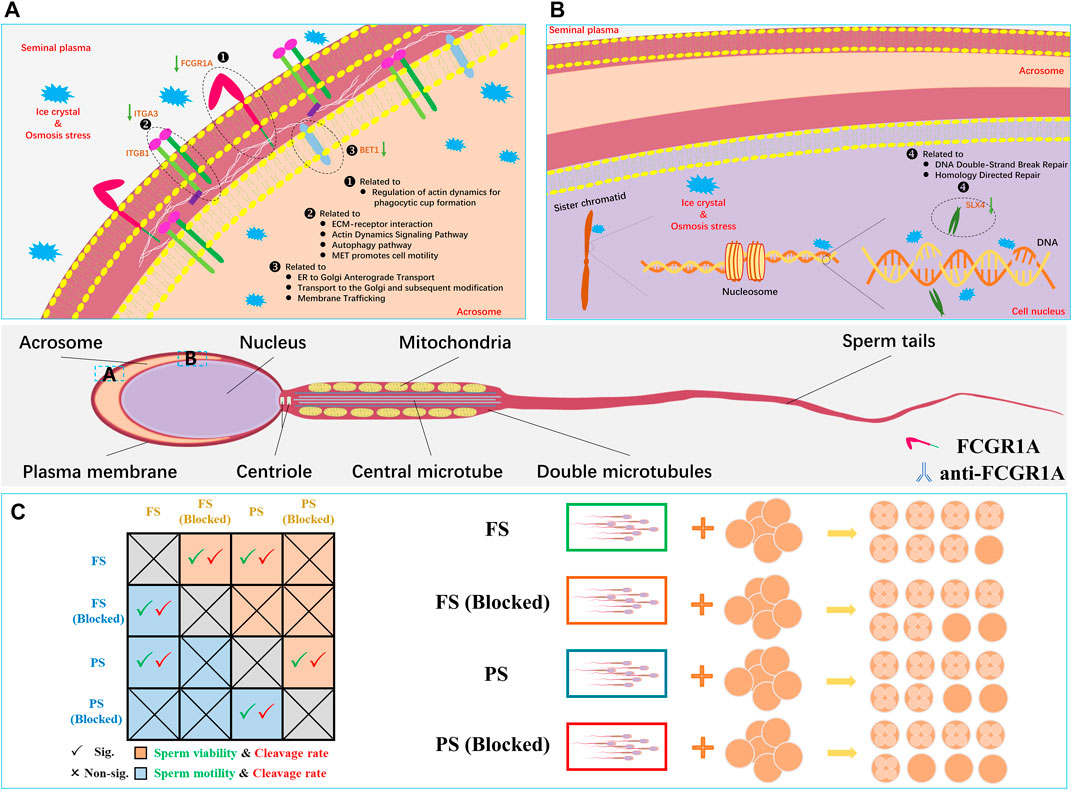
FIGURE 11. Representative sperm mRNA and protein response mechanisms in the frozen semen of sheep. (A) Located in the sperm acrosomal and cytoplasm membrane, FCGR1A, ITGA3, and BET1 proteins are downregulated related to the pathway and function. (B) Located in the sperm nucleus, SLX4 is downregulated related to the pathway and function. (C) Schematic diagram showing the differences in sperm motility, viability, cleavage rate blocked, and unblocked sperm.
Moreover, we found that there were 11 proteins associated with four genes under Pearson’s correlation analysis. Interestingly, most of these 11 proteins were associated with sperm motility and fertilization capacity. In the reproductive system, MME has been associated with sperm formation and development and can regulate follicle maturation and ovulation (Nalivaeva et al., 2020). However, its specific mechanism remains unclear. It has been reported that the addition of the inhibitor (thiorphan) to inhibit CD10 (also called MME) can significantly enhance human sperm motility (Subirán et al., 2010). However, the reduction in sperm motility caused by freezing was observed to be accompanied by the downregulation of MME, which requires further research. L-PGDS (also called PTGDS) can mainly localize in the head of the epididymis and in the apical ridge of the acrosome (Gerena er al., 2000). Analysis by ELISA and flow cytometry indicated that L-PGDS can improve sperm motility (Chen et al., 2007). The bovine sperm or oocytes treated with the L-PGDS antibody could improve the sperm–oocyte binding, thereby reducing the fertilization ability (Gonçalves et al., 2008). In addition, compared with the semen with the highest freezability, L-PGDS with the lowest freezability showed a decreasing trend in boar frozen semen (Valencia et al., 2017). In this study, downregulated PTGDS could potentially induce polyspermism, thereby resulting in decreased oocyte quality and fertilization failure of frozen sperm. Male mice with knocked-out HEXB can exhibit a lower fertilization rate at approximately 109.2 days of age compared to wild-type mice (Juneja, 2002). Thus, we believed that the downregulated HEXB could be the probable reason for the low fertilization rate observed in the frozen semen. A high-throughput phenotypic screening platform has revealed that sperm motility was elevated when PDE10A was inhibited (Gruber, et al., 2022). For instance, in the brain, PDE10A has been reported to be involved in the regulation of cAMP and cGMP synthesis and is commonly used as a therapeutic inhibitor for psychiatric disorder treatment (Wilson and Brandon, 2015). Since cAMP signaling pathway and calcium ions can play crucial roles in sperm capacitation and motility acquisition (Balbach et al., 2018), it was hypothesized that the downregulation of PDE10A in the frozen group could be responsible for reduced mobility in the frozen sperm. During fertilization, the GALT protein present on the sperm membrane can adhere to zona pellucida 3 (ZP3) on the oocyte membrane (Shur et al., 2006). Thus, the downregulated GALT in the frozen sperm might reduce the sperm–oocyte adhesion in the pre-acrosome reaction. It was demonstrated that the expression of HSPA1L was significantly reduced in the low-motility sperm, and the antibody neutralization experiment indicated that blocking HSPA1L could significantly reduce the sperm motility (Liu et al., 2022). Moreover, a previous study has indicated that stress can lead to the activation of HSPA1L (Murphy, 2013). Therefore, we hypothesized that the low HSPA1L levels in the frozen sperm could also attribute to the reduced sperm motility in our study. It has been reported that CTSD could be transported from the epididymis to the surface of sperm during sperm maturation (Asuvapongpatana et al., 2013). A previous study has also indicated that CTSD in the human sperm could be activated during capacitation (Saewu et al., 2012). However, in the present study, reduced CTSD abundance was found in the frozen sperm, which could possibly disturb sperm fertilization through impaired sperm capacitation. Notably, FCGR1A was the only gene identified upon conducting the three integrated multi-omics analyses, and it was found to be downregulated in both RNA-seq and proteome profiles. FCGR1A, also known as CD64 (FCGR1A), is a member of the differentiation (CD) family. CD proteins are mainly localized on the surface of immune cells and play vital roles in signal recognition during immune response. Notably, numerous studies have revealed that CD9 protein belonging to this family is associated with sperm–oocyte binding and fusion (Siu et al., 2021). The results of a binding assay using radio-iodinated immunoglobulin showed that both human and mouse sperm possessed Fc receptors on the surface, which could effectively bind to the Fc γ region (Sethi and Brandis, 1980). Additionally, another study has indicated that human seminal plasma containing Fc γ receptor proteins could significantly protect sperm from immune damage in the female reproductive tract (Chiu and Chamley, 2003). In this report, we have demonstrated that FCGR1A abundance is directly related to sperm motility and viability. Sperm-blocking experiments further revealed that the loss of FCGR1A could effectively reduce sperm viability and motility and also result in the decreased cleavage rate (Figure 11C). In addition, evidence has indicated that FCGR1A could affect diverse biological events, including cytoskeletal changes, ROS production, and cellular survival, through modulating SYK, SYK partners, and other signaling intermediates (Mócsai et al., 2010; van der Poel et al., 2011). Interestingly, a previous finding has identified that FCGR (including FCGR1A) recruitment is required for SYK-mediated cytoskeleton reorganization (Jaumouillé et al., 2014). It was demonstrated that the sperm motility is primarily dependent on sperm tail motility, which is maintained by the normal cytoskeletal axonemal structure and adequate ATP supply in a constant and stable manner (Lehti and Sironen, 2017). Thus, the low-content FCGR1A could possibly cause insufficient recruitment and further disturb the cytoskeleton reorganization, and hence we hypothesized that the downregulated FCGR1A expression in the frozen sperm could lead to reduced motility by interrupting the cytoskeletal organization. Moreover, it has been discovered that Fc γ receptors also existed on the oocyte membrane surface, and the binding of the ligand to the receptor is required for the correct signal transduction during sperm recognition in the fertilization process (Bronson, 1998). Moreover, another recent study has highlighted that Fc receptor-like 3 located on the oocyte membrane could also act as a binding receptor similar to the JUNO and IZUMO1 proteins (Vondrakova et al., 2022). Overall, we speculate that FCGR1A might also play a role in sperm–egg recognition, which needs to be further validated in future studies.
In summary, our results have demonstrated that integrated multi-omics could be effectively used as a potent tool in identifying various candidate genes responsible for sperm cryoinjuries. Moreover, we have discovered that downregulated FCGR1A could result in reduced motility and viability, which can further attribute to the compromised fertility capacity in frozen sheep sperm.
This raw data of this article will be available by the iProX (Project ID: PXD044096) and National Genomics Data Center with identifier (Project ID: PRJCA018587).
The animal study was reviewed and approved by the Ethics Committee of Xinjiang Academy of Agricultural and Reclamation Sciences.
XF, PW, JB, and GZ contributed to the conception and design of the study. JB and GZ organized the database. JB and GZ performed the statistical analysis. SH, YL, YG, JW, HL, and LW contributed to the cell and animal experiments. JB wrote the first draft of the manuscript. GZ and SH wrote sections of the manuscript. JL, AL, WS, PW, and XF revised the manuscript. All authors contributed to the article and approved the submitted version.
The work was supported by grants from the Development and Demonstration of Key Technologies for Cryopreservation of Gametes and Embryos in Cattle and Sheep (S2021BC505), the central government guides local special projects for science and technology development—Development of Cattle and Sheep Oocytes and Embryos Vitrification Freezing System Based on New Type Composite Nanoparticles and 3D Printed Microporous Carriers, Special Program for Training and Guiding Outstanding Young and Middle-aged Talents (SKLSGIHP2021A01), and China Agriculture Research System (CARS-39-07).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling editor KC declared a past co-authorship with the authors JL and XF.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2023.1177774/full#supplementary-material
Agarwal, A., Baskaran, S., Parekh, N., Cho, C. L., Henkel, R., Vij, S., et al. (2021). Male in-fertility. Lancet 397, 319–333. doi:10.1016/S0140-6736(20)32667-2
Asuvapongpatana, S., Saewu, A., Chotwiwatthanakun, C., Vanichviriyakit, R., and Weerachatyanukul, W. (2013). Localization of cathepsin D in mouse reproductive tissues and its acquisition onto sperm surface during epididymal sperm maturation. Acta histochem. 115, 425–433. doi:10.1016/j.acthis.2012.10.005
Balbach, M., Beckert, V., Hansen, J. N., and Wachten, D. (2018). Shedding light on the role of cAMP in mammalian sperm physiology. Mol. Cell. Endocrinol. 468, 111–120. doi:10.1016/j.mce.2017.11.008
Barb, A. W. (2021). Fc γ receptor compositional heterogeneity: Considerations for immunotherapy development. J. Biol. Chem. 296, 100057. doi:10.1074/jbc.REV120.013168
Bianchi, E., and Wright, G. J. (2020). Find and fuse: Unsolved mysteries in sperm-egg recognition. PLoS Biol. 18, e3000953. doi:10.1371/journal.pbio.3000953
Bolton, R. L., Mooney, A., Pettit, M. T., Bolton, A. E., Morgan, L., Drake, G. J., et al. (2022). Resurrecting biodiversity: Advanced assisted reproductive technologies and biobanking. Reprod. Fertil. 3, R121–R146. doi:10.1530/RAF-22-0005
Bronson, R. (1998). Is the oocyte a non-professional phagocyte? Hum. Reprod. Update 4, 763–775. doi:10.1093/humupd/4.6.763
Chen, D. Y., Zhu, M. Y., Cui, Y. D., and Huang, T. H. (2007). Relationship between contents of lipocalin-type prostaglandin D synthase on the surface of infertility sperm and in seminal plasma. Biochem. (Mosc) 72, 215–218. doi:10.1134/s0006297907020125
Chen, X., Zhu, H., Hu, C., Hao, H., Zhang, J., Li, K., et al. (2014). Identification of differentially expressed proteins in fresh and frozen-thawed boar spermatozoa by iTRAQ-coupled 2D LC-MS/MS. Reproduction 147, 321–330. doi:10.1530/REP-13-0313
Chiu, W. W., and Chamley, L. W. (2003). Human seminal plasma antibody-binding proteins. Am. J. Reprod. Immunol. 50, 196–201. doi:10.1034/j.1600-0897.2003.00067.x
Dai, D. H., Qazi, I. H., Ran, M. X., Liang, K., Zhang, Y., Zhang, M., et al. (2019). Exploration of miRNA and mRNA profiles in fresh and frozen-thawed boar sperm by transcriptome and small RNA sequencing. Int. J. Mol. Sci. 20, 802. doi:10.3390/ijms20040802
Ebenezer Samuel King, J. P., Sinha, M. K., Kumaresan, A., Nag, P., Das Gupta, M., Arul Prakash, M., et al. (2022). Cryopreservation process alters the expression of genes involved in pathways associated with the fertility of bull spermatozoa. Front. Genet. 13, 1025004. doi:10.3389/fgene.2022.1025004
Edupuganti, R. R., Geiger, S., Lindeboom, R. G. H., Shi, H., Hsu, P. J., Lu, Z., et al. (2017). N6-methyladenosine (m6A) recruits and repels proteins to regulate mRNA homeostasis. Nat. Struct. Mol. Biol. 24, 870–878. doi:10.1038/nsmb.3462
Esteves, S. C., Zini, A., Coward, R. M., Evenson, D. P., Gosálvez, J., Lewis, S. E. M., et al. (2021). Sperm DNA fragmentation testing: Summary evidence and clinical practice recommendations. Andrologia 53, e13874. doi:10.1111/and.13874
Estudillo, E., Jiménez, A., Bustamante-Nieves, P. E., Palacios-Reyes, C., Velasco, I., and López-Ornelas, A. (2021). Cryopreservation of gametes and embryos and their molecular changes. Int. J. Mol. Sci. 22, 10864. doi:10.3390/ijms221910864
Ezzati, M., Shanehbandi, D., Hamdi, K., Rahbar, S., and Pashaiasl, M. (2020). Influence of cryopreservation on structure and func-tion of mammalian spermatozoa: An overview. Cell. Tissue Bank. 21, 1–15. doi:10.1007/s10561-019-09797-0
Fraser, L., Brym, P., Pareek, C. S., Mogielnicka-Brzozowska, M., Paukszto, Ł., Jastrzębski, J. P., et al. (2020). Transcriptome analysis of boar spermatozoa with different freezability using RNA-Seq. Theriogenology 142, 400–413. doi:10.1016/j.theriogenology.2019.11.001
Fricke, W. M., and Brill, S. J. (2003). Slx1-Slx4 is a second structure-specific endonuclease functionally redundant with Sgs1-Top3. Genes. Dev. 17, 1768–1778. doi:10.1101/gad.1105203
Futosi, K., Fodor, S., and Mócsai, A. (2013). Neutrophil cell surface receptors and their intracellular signal transduction pathways. Int. Immunopharmacol. 17, 638–650. doi:10.1016/j.intimp.2013.06.034
Garner, E., Kim, Y., Lach, F. P., Kottemann, M. C., and Smogorzewska, A. (2013). Human GEN1 and the SLX4-associated nucleases MUS81 and SLX1 are essential for the resolution of replication-induced Holliday junctions. Cell. Rep. 5, 207–215. doi:10.1016/j.celrep.2013.08.041
Gerena, R. L., Irikura, D., Eguchi, N., Urade, Y., and Killian, G. J. (2000). Immunocytochemical localization of lipocalin-type prosta-glandin D synthase in the bull testis and epididymis and on ejaculated sperm. Biol. Reprod. 62, 547–556. doi:10.1095/biolreprod62.3.547
Gonçalves, R. F., Barnabe, V. H., and Killian, G. J. (2008). Pre-treatment of cattle sperm and/or oocyte with antibody to lipocalin type prostaglandin D synthase inhibits in vitro fertilization and increases sperm-oocyte binding. Anim. Reprod. Sci. 106, 188–193. doi:10.1016/j.anireprosci.2007.12.019
Green, H. J., and Brown, N. H. (2019). Integrin intracellular machinery in action. Exp. Cell. Res. 378, 226–231. doi:10.1016/j.yexcr.2019.03.011
Gruber, F. S., Johnston, Z. C., Norcross, N. R., Georgiou, I., Wilson, C., Read, K. D., et al. (2022). Compounds enhancing human sperm motility identified using a high-throughput phenotypic screening platform. Hum. Reprod. 37, 466–475. doi:10.1093/humrep/deac007
Hellstrom, W. J., Overstreet, J. W., Samuels, S. J., and Lewis, E. L. (1988). The relationship of circulating antisperm antibodies to sperm surface antibodies in infertile men. J. Urol. 140, 1039–1044. doi:10.1016/s0022-5347(17)41922-7
Jaumouillé, V., Farkash, Y., Jaqaman, K., Das, R., Lowell, C. A., and Grinstein, S. (2014). Actin cytoskeleton reorganization by Syk regulates Fcγ receptor responsiveness by increasing its lateral mobility and clustering. Dev. Cell. 29, 534–546. doi:10.1016/j.devcel.2014.04.031
Jia, B., Larbi, A., Lv, C., Liang, J., Xiang, D., Zhang, B., et al. (2022). Identification and validation of ram sperm proteins associated with cryoinjuries caused by the cryopreservation process. Theriogenology 184, 191–203. doi:10.1016/j.theriogenology.2022.03.015
Juneja, S. C. (2002). Development of infertility at young adult age in a mouse model of human Sandhoff disease. Reprod. Fertil. Dev. 14, 407–412. doi:10.1071/rd02060
Khan, I. M., Cao, Z., Liu, H., Khan, A., Rahman, S. U., Khan, M. Z., et al. (2021). Impact of cryopreservation on spermatozoa freeze-thawed traits and relevance OMICS to assess sperm cryo-tolerance in farm animals. Front. Vet. Sci. 8, 609180. doi:10.3389/fvets.2021.609180
Khawar, M. B., Gao, H., and Li, W. (2019). Mechanism of acrosome biogenesis in mammals. Front. Cell. Dev. Biol. 7, 195. doi:10.3389/fcell.2019.00195
Krassowski, M., Das, V., Sahu, S. K., and Misra, B. B. (2020). State of the field in Multi-Omics research: From computational needs to data mining and sharing. Front. Genet. 11, 610798. doi:10.3389/fgene.2020.610798
Kumar, A., Prasad, J. K., Srivastava, N., and Ghosh, S. K. (2019). Strategies to minimize various stress-related freeze-thaw damages during conventional cryopreservation of mammalian spermatozoa. Biopreserv. Biobank. 17, 603–612. doi:10.1089/bio.2019.0037
Lalancette, C., Miller, D., Li, Y., and Krawetz, S. A. (2008). Paternal contributions: New functional insights for spermatozoal RNA. J. Cell. Biochem. 104, 1570–1579. doi:10.1002/jcb.21756
Lehti, M. S., and Sironen, A. (2017). Formation and function of sperm tail structures in association with sperm motility defects. Biol. Reprod. 97, 522–536. doi:10.1093/biolre/iox096
Liu, X., Teng, Z., Wang, Z., Zhu, P., Song, Z., and Liu, F. (2022). Expressions of HSPA1L and HSPA9 are associated with poor sperm quality of low-motility spermatozoa in fertile men. Andrologia 54, e14321. doi:10.1111/and.14321
Malsam, J., and Söllner, T. H. (2011). Organization of SNAREs within the Golgi stack. Cold Spring Harb. Perspect. Biol. 3, a005249. doi:10.1101/cshperspect.a005249
Merc, V., Frolikova, M., and Komrskova, K. (2021). Role of integrins in sperm activation and fertilization. Int. J. Mol. Sci. 22, 11809. doi:10.3390/ijms222111809
Mócsai, A., Ruland, J., and Tybulewicz, V. L. (2010). The SYK tyrosine kinase: A crucial player in diverse biological functions. Nat. Rev. Immunol. 10, 387–402. doi:10.1038/nri2765
Moura, A. A., and Memili, E. (2016). Functional aspects of seminal plasma and sperm proteins and their potential as molecular markers of fertility. Anim. Reprod. Sci. 13, 191–199. doi:10.21451/1984-3143-AR884
Murphy, M. E. (2013). The HSP70 family and cancer. Carcinogenesis 34, 1181–1188. doi:10.1093/carcin/bgt111
Nalivaeva, N. N., Zhuravin, I. A., and Turner, A. J. (2020). Neprilysin expression and functions in development, ageing and disease. Mech. Ageing Dev. 192, 111363. doi:10.1016/j.mad.2020.111363
Nowicka-Bauer, K., and Szymczak-Cendlak, M. (2021). Structure and function of ion channels regulating sperm motility-An overview. Int. J. Mol. Sci. 22, 3259. doi:10.3390/ijms22063259
Oldenhof, H., Wolkers, W. F., and Sieme, H. (2021). Cryopreservation of semen from domestic livestock: Bovine, equine, and porcine sperm. Methods Mol. Biol. 2180, 365–377. doi:10.1007/978-1-0716-0783-1_15
Palazzese, L., Gosálvez, J., Anzalone, D. A., Loi, P., and Saragusty, J. (2018). DNA fragmentation in epididymal freeze-dried ram spermatozoa impairs embryo development. J. Reprod. Dev. 64, 393–400. doi:10.1262/jrd.2018-033
Parrilla, I., Perez-Patiño, C., Li, J., Barranco, I., Padilla, L., Rodriguez-Martinez, H., et al. (2019). Boar semen proteomics and sperm preservation. Theriogenology 137, 23–29. doi:10.1016/j.theriogenology.2019.05.033
Peris-Frau, P., Soler, A. J., Iniesta-Cuerda, M., Martín-Maestro, A., Sánchez-Ajofrín, I., Medina-Chávez, D. A., et al. (2020). Sperm cryodamage in ruminants: Understanding the molecular changes induced by the cryopreservation process to optimize sperm quality. Int. J. Mol. Sci. 21, 2781. doi:10.3390/ijms21082781
Qin, Z., Wang, W., Ali, M. A., Wang, Y., Zhang, Y., Zhang, M., et al. (2021). Transcriptome-wide m6A profiling reveals mRNA post-transcriptional modification of boar sperm during cryopreservation. BMC genomics 22, 588. doi:10.1186/s12864-021-07904-8
Ren, X., Chen, X., Wang, Z., and Wang, D. (2017). Is transcription in sperm stationary or dynamic? J. Reprod. Dev. 63, 439–443. doi:10.1262/jrd.2016-093
Ritagliati, C., Baro Graf, C., Stival, C., and Krapf, D. (2018). Regulation mechanisms and implications of sperm membrane hyperpolarization. Mech. Dev. 154, 33–43. doi:10.1016/j.mod.2018.04.004
Rival, C. M., Xu, W., Shankman, L. S., Morioka, S., Arandjelovic, S., Lee, C. S., et al. (2019). Phosphatidylserine on viable sperm and phagocytic machinery in oocytes regulate mammalian fertilization. Nat. Commun. 10, 4456. doi:10.1038/s41467-019-12406-z
Saewu, A., Asuvapongpatana, S., Chotwiwatthanakun, C., Tantiwongse, A., Weerachatyanukul, W., and Thitilertdecha, S. (2012). Cathepsin D in human reproductive tissues: Cellular localization in testis and epididymis and surface distribution in different sperm conditions. J. Androl. 33, 726–734. doi:10.2164/jandrol.111.014639
Schulz, M., Risopatrón, J., Uribe, P., Isachenko, E., Isachenko, V., and Sánchez, R. (2020). Human sperm vitrification: A scientific report. Andrology 8, 1642–1650. doi:10.1111/andr.12847
Sethi, K. K., and Brandis, H. (1980). IgG Fc-binding receptors on spermatozoa. Eur. J. Immunol. 10, 964–965. doi:10.1002/eji.1830101213
Shangguan, A., Zhou, H., Sun, W., Ding, R., Li, X., Liu, J., et al. (2020). Cryopreservation induces alterations of miRNA and mRNA fragment profiles of bull sperm. Front. Genet. 11, 419. doi:10.3389/fgene.2020.00419
Shur, B. D., Rodeheffer, C., Ensslin, M. A., Lyng, R., and Raymond, A. (2006). Identification of novel gamete receptors that mediate sperm adhesion to the egg coat. Mol. Cell. Endocrinol. 250, 137–148. doi:10.1016/j.mce.2005.12.037
Siu, K. K., Serrão, V. H. B., Ziyyat, A., and Lee, J. E. (2021). The cell biology of fertilization: Gamete attachment and fusion. J. Cell. Biol. 220, e202102146. doi:10.1083/jcb.202102146
Stival, C., Puga Molina Ldel, C., Paudel, B., Buffone, M. G., Visconti, P. E., and Krapf, D. (2016). Sperm capacitation and acrosome reaction in mammalian sperm. Adv. Anat. Embryol. Cell. Biol. 220, 93–106. doi:10.1007/978-3-319-30567-7_5
Subirán, N., Pinto, F. M., Agirregoitia, E., Candenas, L., and Irazusta, J. (2010). Control of APN/CD13 and NEP/CD10 on sperm motility. Asian J. Androl. 12, 899–902. doi:10.1038/aja.2010.82
Subramanian, I., Verma, S., Kumar, S., Jere, A., and Anamika, K. (2020). Multi-omics data integration, interpretation, and its application. Bioinform. Biol. Insights 14, 1177932219899051. doi:10.1177/1177932219899051
Swisher, J. F., and Feldman, G. M. (2015). The many faces of FcγRI: Implications for therapeutic antibody function. Immunol. Rev. 268, 160–174. doi:10.1111/imr.12334
Tao, Y., Sanger, E., Saewu, A., and Leveille, M. C. (2020). Human sperm vitrification: The state of the art. Reprod. Biol. Endocrinol. 18, 17. doi:10.1186/s12958-020-00580-5
Valencia, J., Gómez, G., López, W., Mesa, H., and Henao, F. J. (2017). Relationship between HSP90a, NPC2 and L-PGDS proteins to boar semen freezability. J. Anim. Sci. Biotechnol. 8, 21. doi:10.1186/s40104-017-0151-y
van der Poel, C. E., Spaapen, R. M., van de Winkel, J. G., and Leusen, J. H. (2011). Functional characteristics of the high affinity IgG receptor, FcγRI. FcγRI. J. Immunol. 186, 2699–2704. doi:10.4049/jimmunol.1003526
Vondrakova, J., Frolikova, M., Ded, L., Cerny, J., Postlerova, P., Palenikova, V., et al. (2022). MAIA, Fc receptor-like 3, supersedes JUNO as IZUMO1 receptor during human fertilization. Sci. Adv. 8, eabn0047. doi:10.1126/sciadv.abn0047
Vozaf, J., Svoradová, A., Baláži, A., Vašíček, J., Olexiková, L., Dujíčková, L., et al. (2022). The cryopreserved sperm traits of various ram breeds: Towards biodiversity conservation. Anim. (Basel) 12, 1311. doi:10.3390/ani12101311
Wang, M., Todorov, P., Wang, W., Isachenko, E., Rahimi, G., Mallmann, P., et al. (2022). Cryoprotectants-free vitrification and conventional freezing of human spermatozoa: A comparative transcript profiling. Int. J. Mol. Sci. 23, 3047. doi:10.3390/ijms23063047
Wang, X., Zhao, B. S., Roundtree, I. A., Lu, Z., Han, D., Ma, H., et al. (2015). N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 161, 1388–1399. doi:10.1016/j.cell.2015.05.014
Whaley, D., Damyar, K., Witek, R. P., Mendoza, A., Alexander, M., and Lakey, J. R. (2021). Cryopreservation: An overview of principles and cell-specific considerations. Cell. Transpl. 30, 963689721999617. doi:10.1177/0963689721999617
Wilson, L. S., and Brandon, N. J. (2015). Emerging biology of PDE10A. Curr. Pharm. Des. 21, 378–388. doi:10.2174/1381612820666140826114744
Wyck, S., Herrera, C., Requena, C. E., Bittner, L., Hajkova, P., Bollwein, H., et al. (2018). Oxidative stress in sperm affects the epigenetic reprogramming in early embryonic development. Epigenetics Chromatin 11, 60. doi:10.1186/s13072-018-0224-y
Xu, X., Wang, M., Sun, J., Yu, Z., Li, G., Yang, N., et al. (2021). Structure specific DNA recognition by the SLX1-SLX4 en-donuclease complex. Nucleic Acids Res. 49, 7740–7752. doi:10.1093/nar/gkab542
Yoshida, M., Kawano, N., and Yoshida, K. (2008). Control of sperm motility and fertility: Diverse factors and common mechanisms. Cell. Mol. Life Sci. 65, 3446–3457. doi:10.1007/s00018-008-8230-z
Zhang, T., Wong, S. H., Tang, B. L., Xu, Y., Peter, F., Subramaniam, V. N., et al. (1997). The mammalian protein (rbet1) homologous to yeast Bet1p is primarily associated with the pre-Golgi intermediate compartment and is involved in ve-sicular transport from the endoplasmic reticulum to the Golgi apparatus. J. Cell. Biol. 139, 1157–1168. doi:10.1083/jcb.139.5.1157
Zhang, Y., Dai, D., Chang, Y., Li, Y., Zhang, M., Zhou, G., et al. (2017). Cryopreservation of boar sperm induces differential microRNAs expression. Cryobiology 76, 24–33. doi:10.1016/j.cryobiol.2017.04.013
Keywords: sperm, programmed freezing, multi-omics, FCGR1A, fertilization, sheep
Citation: Bai J, Zhou G, Hao S, Liu Y, Guo Y, Wang J, Liu H, Wang L, Li J, Liu A, Sun WQ, Wan P and Fu X (2023) Integrated transcriptomics and proteomics assay identifies the role of FCGR1A in maintaining sperm fertilization capacity during semen cryopreservation in sheep. Front. Cell Dev. Biol. 11:1177774. doi: 10.3389/fcell.2023.1177774
Received: 02 March 2023; Accepted: 17 July 2023;
Published: 02 August 2023.
Edited by:
Keren Cheng, University of Pennsylvania, United StatesReviewed by:
Brendan J. Houston, The University of Melbourne, AustraliaCopyright © 2023 Bai, Zhou, Hao, Liu, Guo, Wang, Liu, Wang, Li, Liu, Sun, Wan and Fu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiangwei Fu, eGlhbmd3ZWlmdUAxMjYuY29t; Pengcheng Wan, d2FucGNAaG90bWFpbC5jb20=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.