
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol., 16 March 2023
Sec. Developmental Epigenetics
Volume 11 - 2023 | https://doi.org/10.3389/fcell.2023.1164706
As a medium among pregnant women, environment and fetus, placenta owns powerful and delicate epigenetic processes to regulate gene expression and maintain cellular homeostasis. N6-methyladenosine (m6A) is the most prevalent modification that determines the fate of RNA, and its dynamic reversibility indicates that m6A may serve as a sensitive responder to environmental stimuli. Emerging evidence suggests that m6A modifications play an essential role in placental development and maternal-fetal crosstalk, and are closely related to gestational diseases. Herein, we summarized the latest techniques for m6A sequencing and highlighted current advances of m6A modifications in maternal-fetal crosstalk and the underlying mechanisms in gestational diseases. Therefore, proper m6A modifications are important in placental development, but its disturbance mainly caused by various environmental factors can lead to abnormal placentation and function with possible consequences of gestational diseases, fetal growth and disease susceptibility in adulthood.
The placenta acts as a medium for pregnant women and fetuses, enabling the exchange of nutrients, gas and also waste productions, protecting fetuses from maternal immune attack, and also secreting hormones and factors to support fetal growth. The defects of placentation will result in a range of gestational diseases (Jauniaux et al., 2006; Ananth, 2014; Roberts, 2014; Ashraf et al., 2021), such as miscarriage, preterm birth, gestational diabetes mellitus, pre-eclampsia and fetal growth restriction. Placental development during the whole gestation involves a variety of cellular function changes and transformations (Caniggia et al., 2000; Pringle et al., 2010). This rapid development makes the prenatal period particularly vulnerable because of the complicated trajectory of cells and environmental perturbations. To maintain placental homeostasis, several epigenetic mechanisms have been highlighted to drive short- and long-term gene expression changes (Nelissen et al., 2011), including DNA methylation (Koukoura et al., 2012; Lorincz and Schubeler, 2017), histone modifications (Meister et al., 2021), non-coding RNAs (Du et al., 2021; Zhang et al., 2021; Zarkovic et al., 2022), and also RNA methylation (Mu et al., 2022). As one of the most prevalent internal modifications in RNAs, N6-methyladenosine (m6A) extensively regulate RNA metabolism (Jia et al., 2022). Because of its involvement in RNA-related bioprocesses, m6A is essential in decision of RNA fate and plays an irreplaceable role in cell proliferation, differentiation, stress responses and other activities (Wang et al., 2014a; Zhang et al., 2017a; Zhang et al., 2017b). A recent study based on multi-organ m6A sequencing showed that placental tissues are rich in m6A modifications, and tissue specific m6A may be related to unique biofunctions (Xiao et al., 2019; Zhang et al., 2020a). Some other studies also reported the relationship between m6A dysregulation and gestational diseases (Li et al., 2019; Taniguchi et al., 2020; Qiu et al., 2021; Wang et al., 2021; Xu et al., 2021; Zhang et al., 2021). Therefore, in this review, we summarize the role of m6A in placental development and gestational diseases, and provide novel insights for underling mechanisms during these processes.
Over 100 kinds of chemical modifications of RNA have been identified in organisms, including protein-coding RNAs and non-coding RNAs (Saletore et al., 2012). Among them, m6A modification is the most abundant internal methylation of mRNA in eukaryotes, which typically accounts for 0.1%–0.4% of total RNA adenosines (Dominissini et al., 2012). In mammalian cells, m6A is mostly enriched in the 3′-untranslated regions, near stop codons and long exons, and with a consensus sequence of RRACH (R = G or A; H = A, C or U) (Meyer et al., 2012). Transcriptome-wide profile of m6A through m6A antibody-based immunoprecipitation followed by high-throughput sequencing has validated that m6A modification may regulate more than 7000 mRNAs in human transcripts and also lncRNAs, miRNAs and circRNAs (Dominissini et al., 2012; Meyer et al., 2012; Zhao et al., 2014). Similar to DNA and histone modifications, m6A is dynamic and reversible. RNA m6A modifications can be methylated under the action of methyltransferases and demethylated mediated by demethylases (Table 1). Then, binding proteins can recognize this specific modification and regulate RNA metabolism (Bokar et al., 1994; Liu et al., 2014; Ping et al., 2014). High conserved m6A is widely involved in the decision of RNA fate, including alternative splicing, transportation, stability, and translation (Figure 1).
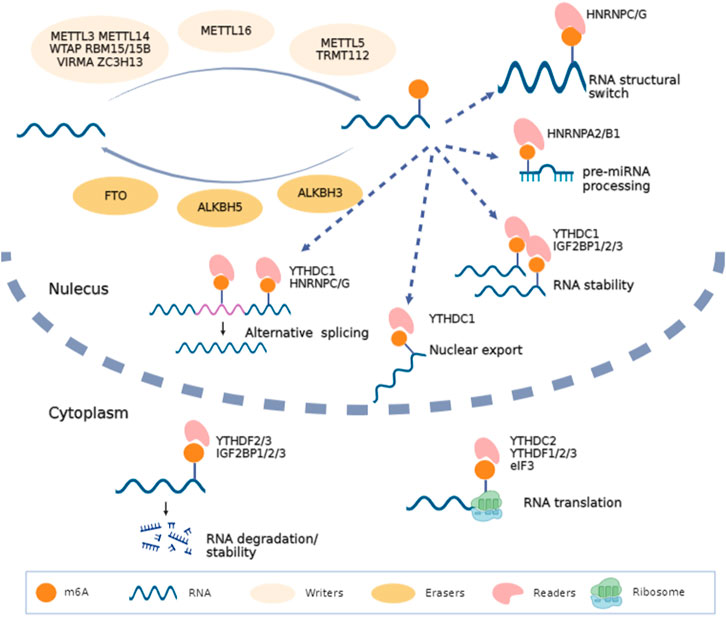
FIGURE 1. Biofunctions of m6A in RNA metabolism. RNA m6A is methylated under the action of methyltransferases (METTL3, WTAP, METTL14, RBM15/15B, VIRMA, ZC3H13, METTL16, METTL5 and TRMT112), demethylated mediated by demethylases (FTO, ALKBH5 and ALKBH3) and recognized by binding proteins (YTHDC1/2, YTHDF1/2/3, IGF2BP1/2/3, HNRNP and eIF3). This figure was drawn using BioRender (https://www.biorender.com).
Installation of m6A can be catalyzed by a methyltransferase complex (MTC) composed of several proteins, or through MTC-independent manners. Methyltransferase-like 3 (METTL3) is the most important component of MTC, with an internal S-adenosyl methionine (SAM)-binding domain and catalyzes methyl group transfer through its highly active methyltransferase domain (Liu et al., 2014; Wang et al., 2016). METTL14, another active component of MTC, is co-localized in nuclear speckles with METTL3 and stabilizes the structure of MTC and identifies specific RNA sequence (Liu et al., 2014; Wang et al., 2016). Other auxiliary proteins are also essential for the stability of MTC, and Wilms tumor 1-associated protein (WTAP) is the first identified protein which recruits METTL3-METTL14 heterodimer into nuclear speckles (Ping et al., 2014). Moreover, RNA-binding motif protein 15 (RAB15/15B) and Vir like m6A methyltransferase associated (VIRMA/KIAA1429) can direct MTC to specific RNA sites for m6A modification (Knuckles et al., 2018; Yue et al., 2018). Additionally, zinc finger CCCH-type containing 13 (ZC3H13) interacts with WTAP to keep MTC in nuclear speckles (Knuckles et al., 2018; Wen et al., 2018). Among these components, except METTL3, all other factors lack methyltransferase activity. In addition, another MTC of METTL5 and tRNA methyltransferase activator subunit 112 (TRMT112) has been identified as a m6A methyltransferase for 18S rRNA (Huang et al., 2022; Sepich-Poore et al., 2022).
METTL16 and Zinc finger CCHC-type Containing 4 (ZCCHC4) are newly identified independent RNA methyltransferases. METTL16 contains two structural domains and catalyzes m6A in the 3′UTR in mRNA and on A43 of U6 snRNA (Shima et al., 2017). METTL16 also plays an important role in RNA splicing, which targets for pre-mRNAs and non-coding RNAs (Warda et al., 2017). In addition, ZCCHC4 contains an N-terminal specific zinc finger domain and a C-terminal CCHC domain for RNA binding and catalyzes m6A in 28S rRNA (Ren et al., 2019; Pinto et al., 2020).
Fat mass and obesity-associated protein (FTO) is the first reported m6A demethylase which was discovered in 2011 (Jia et al., 2011). After that, the second eraser, AlkB homolog 5 (ALKBH5), was discovered (Zheng et al., 2013). Both of them belong to the alpha-ketoglutarate-dependent dioxygenase family and remove the m6A modification labeled in mRNAs and ncRNAs through an Fe (II) and α-ketoglutaric acid-dependent manner. FTO is partially located in nuclei and can be recruited to the spliceosome center by its nuclear speckle partner, and then participates in RNA processing. Similarly, ALKBH5 is verified to co-localize in nuclear speckles with other RNA processing factors and is essential in RNA synthesis, transport and stability. In addition, ALKBH3 is newly reported to demethylate m6A in mammalian tRNA (Ueda et al., 2017; Chen et al., 2019).
Different downstream biofunctions of RNA m6A depend on various RNA binding proteins called m6A readers. These readers discovered to date include YT521-B homology (YTH) domain-containing proteins (YTHDF1/2/3 and YTHDC1/2), heterogeneous nuclear ribonucleoproteins (HNRNPA/B/C/G), insulin-like growth factor 2 mRNA binding proteins (IGF2BP1/2/3) and eukaryotic initiation factor 3 (eIF3). Considering the different cellular localizations during RNA metabolism, readers can be categorized as nuclear readers and cytoplasmic readers. The former is more likely to participate in alternative splicing, RNA structural switch and nuclear export (Xiao et al., 2016; Roundtree et al., 2017). Typically, YTHDC1 can recruit serine- and arginine-rich splicing factor 3 (SRSF3) and SRSF10 to promote exon inclusion and skipping (Xiao et al., 2016). YTHDC2 also promotes the nuclear export of m6A-modified mRNA through interacting with nuclear RNA export factor 1 (NXF1) and the three prime repair exonuclease (TREX) mRNA export complex (Lesbirel et al., 2018). HNRNPC/G act as nuclear RNA binding proteins and are responsible for pre-mRNA processing; m6A modified mRNA alters their local structure and regulate the HNRNPC/G activities to finally affect the abundance as well as alternative splicing (Liu et al., 2015; Liu et al., 2017). In addition, IGF2BPs can identify m6A and further promote RNA stability and translation (Sun et al., 2022). Some evidence shows that m6A in 5′UTR can recruit eIF3 directly and then increase the efficacy of cap-independent translation (Meyer et al., 2015). Hence, the final fate of RNA depends on its m6A sites and the type of readers recognizing the effective m6A motif (Figure 1).
The development of high-throughput m6A sequencing significantly promote the exploration in epitranscriptomics field, especially in the distribution, proportion and function of m6A in transcriptome wide (Table 2).
In 2012, MERIP-seq and m6A-seq were the first high-throughput sequencing methods which were in dependently developed by two laboratories (Dominissini et al., 2012; Meyer et al., 2012). The theory and experimental methods of these two techniques are similar. The RNA fragments containing m6A modified sites are incubated and enriched by m6A antibody, and then they are subjected to the library preparation and sequencing. Because of simple operation and reagents, this technique is successfully applied to reveal the transcriptome methylation across different species (Schwartz et al., 2013; Luo et al., 2014; Ma et al., 2017). However, high-resolution site information is difficult to obtain because of the limited size of fragmented RNA which could only be about 100–200 nt. Antibody-based IP systems also requires a large amount of initial RNA (400 µg mRNA or 2.5 mg total RNA) and cannot identify precise m6A sites (Dominissini et al., 2012). In next year, Dominissini et al. improved the methods and reduced the initial requirement of total RNA (5 µg mRNA or 300 µg total RNA) (Dominissini et al., 2013), which expanded the application of this technology. MeRIP-seq is widely used to uncover the dynamic modifications in physiological and pathological processes (Lence et al., 2016; Zhang et al., 2017a; Zhao et al., 2017).
In 2016, m6A-LAIC-seq (m6A-level and isoform-characterization sequencing) was developed to quantify m6A stoichiometry on the transcriptome level (Molinie et al., 2016). In general, the experimental steps of m6A-LAIC-seq are similar to MERIP-seq and m6A-seq. There are two main differences: excess m6A antibodies are used to enrich full-length transcripts rather than RNA fragments; equal amount of ERCC RNA Spike-In Mix is used for normalization and calculation of m6A levels. This technique also provides new insights into the dynamic range and isoform complexity of the m6A epitranscriptome. However, use this method alone cannot obtain m6A positions and fractions, and the combination with other methods may compensate this drawback.
In 2015, Chuan He et al. developed a photo-crosslinking-assisted m6A sequencing strategy (PA-m6A-seq) (Chen et al., 2015). In this technique, the mixture of m6A antibody and purified RNA incorporated with 4-thiouridine (4SU) is exposed to 365 nm UV light to trigger the covalent crosslinking between antibody and m6A modified RNA. Then, crosslinked RNA is digested to around 30 nt, which greatly improve the resolution of sequencing data. Incorporation of 4SU can induce a T-to-C transition which serves as hallmarks of nearby m6A and further improves the resolution to 23 nt. However, this method only works in cultured cell lines and may get false negative results when 4SU is not involved in m6A sites.
m6A-CLIP (Ke et al., 2015) and miCLIP (m6A individual-nucleotide-resolution cross-linking and immunoprecipitation) (Linder et al., 2015) are similar to PA-m6A-seq. The RNA-antibody mixture is crosslinked by 254 nm UV light, and then RNA is released by proteinase K. Mutations (C-to-T transition) and truncations induced by crosslink are introduced during reverse transcription, and their patterns are predictable which helps elevating single-nucleotide resolution of m6A. However, these techniques still represent an indirect way to analyze m6A positions and suffer from a low crosslinking yield. They are also difficult to precisely identify m6A sites and detect m6A clusters (Shu et al., 2020).
Based on photo-crosslinking, m6ACE (m6A-crosslinking-exonuclease-sequencing) combines exonuclease to improve the sequencing resolution (Koh et al., 2019). In this technique, UV-crosslinked RNA with m6A antibody is exempt from digestion by exonuclease XRN1. After sequencing, XRN1 treated reads start exactly at the m6A location when compared to input reads. Hence, m6ACE-seq can directly map transcriptome-wide m6A at quantitative single-base-resolution. This method is also useful to explore the target m6A modifications which are uniquely mediated by specific m6A regulators. However, the use of splice fragments with specific molecular identification to decrease the deviation may cause a higher cost.
Escherichia coli MazF toxin is an endoribonuclease sensitive to m6A which specifically cleaves unmodified ACA-sequence rather than m6ACA sites (Imanishi et al., 2017). m6A-REF-seq (Zhang et al., 2019) and MAZTER-seq (Garcia-Campos et al., 2019) use MazF enzyme to directly fragment RNA, cleaving the motif sequence ACA from the 5’ side of first A and leaving the m6ACA motif intact. This method can identify single base m6A at the transcriptome level, and also quantify the methylation level of each m6A site by calculating the ratio of reads with internal ACA reverse reads split at the motif. However, this technique can only cover a limited proportion of m6A sites because the ACA motif MazF recognized accounts for 16% of total m6A sites in mammals and 25% in yeast (Garcia-Campos et al., 2019).
In 2019, Kate D Meyer presented DART-seq (deamination adjacent to RNA modification targets) using APOBEC1-YTH recombinant protein (Meyer, 2019). APOBEC1 is a cytidine deaminase which can induce C-to-U transition. Because YTH domain can specifically recognize m6A site, the deaminase activity of APOBEC1-YTH is mainly induced by YTH. Meanwhile, a mutant APOBEC1-YTH without m6A binding function is transfected into cells as a negative control. DART-seq can identify m6A using as little as 10 ng of initial RNA, and facilitate exploration of m6A in limited samples. Even more surprising, DART-seq can be coupled with single cell isolation and achieves single-cell m6A detection (Tegowski et al., 2022). scDART-seq distinguishes cellular subpopulations based on m6A signatures rather than gene expression, revealing abundant m6A features in cellular functions. However, the efficacy of vector transfection during cell culture limits the application of DART-seq on tissues samples. YTH domain only recognize around 60% of total m6A sites, which may miss several modification sites.
In 2020, m6A-SEAL was developed which relies on FTO activity (Wang et al., 2020). In principle, FTO recognizes and binds to m6A sites, and then converts m6A into N6-hydroxymethyladenosine [hm (Pringle et al., 2010) A]. After treatment with DTT, hm6A is converted into a stable chemical N6-dithiolsitolmethyladenosine [dm (Pringle et al., 2010) A] which can be tagged with biotin for streptavidin enrichment and sequencing. Moreover, optimization of the FTO oxidation, m6A-SEAL can also be used to specifically detect cap m6A. When compared to other m6A sequencing methods and specific validation methods, m6A-SEAL shows great sensitivity, specificity and reliability for transcriptome-wide m6A detection. Considering the rich tagging ability, it may expand the application of m6A, especially in m6A enrichment and imaging.
m6A-label-seq is a metabolic labeling method to map transcriptome-wide m6A modifications at single-base resolution through converting stable m6A structure into a reactive one (Shu et al., 2020). In this technique, cells are incubated with a methionine analog, Se-allyl-L-selenohomocysteine, to incorporate allyl-group into m6A sites and introduce allyladenosine (a6A). Then, anti-a (Pringle et al., 2010) A antibody is used to enrich the a6A modified mRNA to achieve high coverage of m6A target. Under mild conditions, iodine can undergo addition reaction with a6A and obtain CycA through spontaneous cyclization. During the reverse transcription, misincorporation at the opposite site in cDNA will occur and m6A sites can be detected after sequencing. However, this method only works in cultured cell lines and may also get false negative results because of the low incorporation efficacy of a6A. Similar to m6A-SEAL (Wang et al., 2020), m6A-label-seq offers a new option for m6A specific mapping, but lacks stoichiometric information.
In 2022, Chuan He et al. developed a new technique based on selective allyl chemical labeling, m6A-SAC-seq (Hu et al., 2022), which can map the transcriptome-wide m6A at single-nucleotide resolution and also achieve stoichiometric information. MjDim1, a member in Dim1/KsgA family, is used to transfer the methyl group from allylic-S-adenosyl-L-methionine (SAM) to adenosines, converting m6A into allyl-modified m6A (N6-allyl, N6-methyladenosine or a6m6A). a6m6A can undergo cyclization following I2 treatment, and then mutations will occur in the process of reverse transcription. The m6A positions can be detected according to the mutation sites, and the accurate proportion can be calculated by converting the mutation rate through the standard curve. This method requires only 30 ng of poly(A) or rRNA-depleted RNA which largely expand the application in limited samples.
Placentation is a physiological process with dynamic and precise regulation in time and space. Epigenetic processes which drive short- and long-term gene expression changes are particularly powerful and delicate in placentas. RNA m6A methylomes across fetus and adult tissues depicted the dynamic m6A methylation across different tissue types, covering both broadly or tissue-specifically m6A sites (Xiao et al., 2019; Zhang et al., 2020a). In agreement with previous studies, the m6A modifications in placental tissues are mostly enriched around stop codons, with a consensus motif of RRACH, indicating the high conservation of m6A in human placentas (Taniguchi et al., 2020; She et al., 2022). However, abundant tissue-differential m6A peaks are identified in placenta which is higher than that in brain, heart, kidney and other tissues (Xiao et al., 2019). More than half of placental specific m6As is located in introns, suggesting the potentially higher activity of RNA splicing in placentas (Xiao et al., 2019). Hence, the detailed mechanism of RNA m6A in placentation and maternal-fetal crosstalk needs to be further understood (Figure 2).

FIGURE 2. RNA m6A act as sensitive effectors and regulators in maternal-fetal crosstalk. Several environmental factors act on the mothers, and affect the m6A modifications in placentas (orange) and fetal tissues (blue). Alterations of m6A modifications regulate the expression of hub genes and disturb the placental and fetal development. This figure was drawn using BioRender (https://www.biorender.com).
The growing prevalence of maternal obesity or overweight shows a higher risk of abnormal fetal growth and failed placentation. In obese pregnant women, global m6A levels are decreased in placental tissues, along with decreased expression of WTAP, RBM15B and VIRMA; while, the obesity also triggers genome-wide DNA hypermethylation, especially in 5-methylcytosine (5 mC) (Shen et al., 2022). In pigs, maternal obesity is associated with low birth weight (LBW) of piglets. In LBW placentas derived from sows with obese pregnancy, the protein level of FTO is decreased, with an elevated m6A level (Song et al., 2018). FTO demethylates m6A of PPARγ, VEGFA, ABHD5, and GPR120, and the expression levels of these genes in both mRNA and protein are decreased in LBW placentas (Song et al., 2018). Therefore, maternal obesity may regulate multifaceted gene network to affect normal placentation and fetal growth (Song et al., 2018; Shen et al., 2022).
External stimuli such as fear stress can cause significant harm to pregnant women and increase the risk of fetal malformations and placental structural alterations (Tsui et al., 2006; Mizrak and Kabakci, 2021). In pregnant rats, fear stress during gestation significantly reduces placental weight and offspring viability (Wang et al., 2022a). Stress also increases the expression levels of METTL3, METTL14 and WTAP, decreases the level of FTO, and leads to a higher overall m6A level in placental tissues (Wang et al., 2022a). While, stress has no obvious change in the distribution of m6A-enriched regions and motif. Based on MeRIP-seq data, fear stress mainly affects m6A-modified genes involved in utero embryonic development, protein stabilization, angiogenesis and embryonic digit morphogenesis (Wang et al., 2022a). Similar changes are also found in brain development and adult neurons (Widagdo et al., 2016; Walters et al., 2017). Fear stress can alter the levels of METTL3 and FTO, thereby affecting m6A modifications and synaptic plasticity (Widagdo et al., 2016; Walters et al., 2017).
In the first trimester, the presence of extravillous trophoblast plugs in the spiral arteries changes the level of oxygen in placental circulation, inducing trophoblast differentiation and invasion, and arterial transformation (Caniggia et al., 2000; Iriyama et al., 2015; Albers et al., 2019). Disturbance of this orderly process may lead to abortion and ischemic placental diseases (Ananth, 2014; Roberts, 2014). Hypoxia treatment can upregulate the expression level of ALKBH5 in trophoblast cells, and it also promotes ALKBH5 to translocate from nuclear to cytoplasm to demethylate m6A-modified SMAD1/5 mRNA. High level of SMAD1/5 further activates TGF-β signaling pathway and increases the expression of MMP9 and ITGA1 to promote the cellular viability of trophoblast (Zheng et al., 2022). Inhibition of ALKBH5 promotes cellular viability and inhibits cell apoptosis, oxidative stress in hypoxia/reoxygenation treated trophoblast cells, and finally alleviates preeclampsia-like symptoms in pregnant mice. ALKBH5 knockdown facilitates the m6A modification of PPARG mRNA and further activates the Wnt/β-catenin pathway (Guo et al., 2022). Consistent with other reports in cancers (Chen et al., 2020a; Dong et al., 2021; Sun et al., 2023), hypoxia-responsive ALKBH5 may be an important regulator in trophoblast activity and placental development.
Oxidized oil or lipid oxidation products, especially maternal oxidative stress damage, have toxicological effects of placenta and fetus. The placental tissues from rat fed with oxidized soybean oil (OSO) had no significant change in the distribution of m6A-modified regions and motif. However, the mRNA expression levels of Mettl3, Mettl14, Alkbh5 and Fto are decreased in placentas after OSO ingestion, with a lower global m6A level. The placental genes with differential m6A levels are related to nutrient metabolic process and hormone activity (Wang et al., 2022b). In addition, exposure to BPDE [benzo(a)pyrene-7,8-dihydrodiol-9,10-epoxide], the metabolite of Benzo(a)pyrene (BaP) which is one representative of PAHs (polycyclic aromatic hydrocarbons), upregulates the expression level of m6A-modified lnc-HZ01 and inhibits the proliferation of trophoblast cells. More interestingly, m6A modified lnc-HZ01 upregulates the expression level of MXD1, and the latter promotes the transcription of METTL14 which in turn catalyzes m6A in lnc-HZ01, forming a positive feedback loop to inhibit trophoblast viability (Xu et al., 2021).
Environmental chemicals can not only disturb the normal progression of placental development and affect intrauterine growth, but also increase the long-term risk of offspring. Epigenetics is a possible link between the environment and fetal growth, and RNA m6A is also inherited across generations. Maternal ingestion of oxidized soybean oil (OSO) during gestation and lactation disturbs the homeostasis of DNA/RNA methylation and negatively affects the placental function and intestinal development in offspring, with a reduced height of villi and a lower level of anti-inflammatory factors (Wang et al., 2022b). In addition, maternal engineered nanomaterial (ENM) exposure increases the m6A level in the 3′UTR region of mitochondria phospholipid hydroperoxide glutathione peroxidase (mPHGPx) which is an antioxidant enzyme that protects fetal cells from oxidative stress. The m6A-modified mPHGPx diminishes antioxidant capacity, damages the mitochondrial function and causes cardiac deficits, which persists into adulthood following ENM exposure through gestation (Kunovac et al., 2021).
Beforehand, RNA m6A modification has been reported to be involved in neurodevelopment and neurotransmitter signaling (Du et al., 2019; Shafik et al., 2020). FTO regulates the activity of the dopaminergic midbrain circuitry (Hess et al., 2013), neuronal growth and plasticity (Yu et al., 2018). Environmental stimuli during pregnancy alter the m6A modification pattern in fetus and induce the neurodevelopmental impairments. Pregnant mice exposed to carbon black nanoparticles (CBNPs) cause obvious alterations on maternal behaviors, and neurobehavioral and muscular developmental impairments of offspring (Zhang et al., 2020b). Moreover, maternal CBNPs exposure significantly decreases the expression levels of Mettl3, Mettl14 and Wtap, increases the expression level of Alkbh5, and further decreases the global m6A level in the cerebral cortex tissues, indicating the potential relation between m6A alteration induced by maternal CBNPs exposure and neurodevelopment of offspring at postnatal time (Zhang et al., 2020b). Anesthetic exposure during gestation, especially repeated sevoflurane exposure, significantly damages the memory and learning ability of the offspring mice through inhibiting the axon growth and branching. After repeated sevoflurane exposure, differential m6A-modified genes are related to neurodevelopment, especially synapse, main axon and postsynaptic density membrane (Chen et al., 2020b). In addition, a recent study reported that maternal treatment with resveratrol, an anti-inflammatory and synaptic plasticity inductor produced from grapes, can decrease the expression levels of Mettl3 and Fto, as well as increase the global m6A levels in adult offspring (Izquierdo et al., 2021). Maternal consumption of resveratrol can prevent cognitive impairment induced by a high-fat diet and this improvement is associated with increased m6A levels (Izquierdo et al., 2021).
GOS involves a serious of pregnancy-related disorders with a placental component as one part of etiology, including spontaneous abortion, preterm birth, preeclampsia, stillbirth and abnormal fetal growth (Brosens et al., 2011; Prins et al., 2022). Most etiologies arise from events during maternal-fetal exchange, such as nutrients, oxygen, waste products and toxins. Gestational diabetes mellitus is a particular example of pregnancy disorders involving environmental exposome which disturbs the maternal-fetal interaction (Gabbay-Benziv and Baschat, 2015; Valero et al., 2022). Whether and how m6A dysregulation contributes to the pathological mechanisms remains to be elucidated (Table 3).
The known pathological factors of SA include chromosomal abnormalities, maternal infections, endocrine disorders, nutrition, occupational and environmental factors, immunological factors, and inherited thrombophilia. Epigenetics also participates in the pathogenesis, while the detailed mechanism has not been fully understood. Based on MeRIP-seq data of villous tissues from SA, m6A peaks are still mainly located in the codding region and near the stop codon, with a consensus sequence of RRACH (She et al., 2022). Differential m6A-modified genes are mainly involved in the Hippo and Wnt signal pathways, phosphatase activity regulation and transcription inhibitor activity (She et al., 2022). At the maternal-fetal interface, FTO, IGF2BP1 and IGF2BP2 are decreased in abortion tissues, with a decreased level of FTO-bound HLA-G, VEGFR and MMP9 mRNA (Qiu et al., 2021). Hence, aberrant FTO level changes the m6A modifications of hub genes involved in immune tolerance, angiogenesis and trophoblast invasion, indicating the potential pathogenesis of m6A in SA. Research focusing on trophoblast function indicates that aberrant m6A regulation inhibits the trophoblast activity and leads to SA. Upregulated ALKBH5 demethylates m6A of CYP61 mRNA and further decreases the half-life of CYR61 mRNA, inhibiting the trophoblast invasion (Li et al., 2019). ALKBH5 is also sensitive to hypoxic condition which has been supposed to be an important regulator in trophoblast activity, especially in the first trimester (Guo et al., 2022; Zheng et al., 2022). ALKBH5 proteins translocate to the cytoplasm under hypoxia and then demethylates m6A-modified SMAD1/SMAD5 mRNA, consequently enhancing the efficacy of translation (Zheng et al., 2022). Trophoblast-specific knockdown of ALKBH5 in mice significantly inhibits the trophoblast invasion and causes abortion (Zheng et al., 2022). In addition, upregulated METTL14 was also reported to inhibit trophoblast proliferation and induce miscarriage through catalyzing m6A on lnc-HZ01 and enhancing its RNA stability (Xu et al., 2021).
The increasing prevalence of type 2 diabetes in general and in younger people in particular, has led to an increasing number of affected pregnancies. Its maternal and fetal complications include abortion, malformations, preterm delivery, preeclampsia, etc. Several researchers performed high throughput sequencing and analyzed the data from GEO to explore the underlying mechanism of m6A in GDM. Du et al. generated a lncRNA-mediated competitive endogenous RNA (ceRNA) network, and found that hub genes were mainly involved in fatty acid metabolism pathway, which play a role in the development and adverse outcomes of GDM (Du et al., 2022a). Other GDM-associated hormones were also enriched, such as thyroid hormone and oxytocin (Du et al., 2022a). In addition, m6A modified genes related to monocyte infiltration were also clinically important in GDM, including CD81, CFH, FABP5, GBP1 etc. (Du et al., 2022b) Decreased level of METTL4 was found in placentas from GDM patients, and the m6A levels of BAMBI, INSR and IRS1 which are GDM-related genes were also significantly decreased, with the same change in mRNA and protein levels (Wang et al., 2021). Hence, m6A modification may regulate placental metabolism, hormone secretion and immune infiltration in GDM.
An imposing number of mechanisms have been proposed to explain the occurrence of preeclampsia, and those currently considered important include abnormal trophoblast invasion, immune intolerance, maternal maladaptation to inflammation, and genetic factors including inherited predisposing genes and epigenetics. Nowadays, abundant studies have identified the role of epigenetics in preeclampsia and FGR (Nelissen et al., 2011; Koukoura et al., 2012; Ashraf et al., 2021; Meister et al., 2021), while the potential function and mechanism of m6A need to be further explored. The MeRIP-seq data of placentas showed a correlation between higher m6A at 5′UTR and small-for-date placentas, and the decreased m6A near stop codon was related to heavy-for-date placentas, revealing the different m6A modified sites may be important for fetal and placenta growth (Taniguchi et al., 2020). Meanwhile, the m6A labeled at 5′UTR promoted the protein translation of SMPD1 mRNA (Taniguchi et al., 2020). Upregulated METTL3 significantly elevated the expression level of HNRNPC1/2, further inducing vitamin D deficiency, trophoblast dysfunction and preeclampsia (Gu et al., 2021). Moreover, METTL14 was upregulated in preeclamptic placentas: its high expression inhibited trophoblast proliferation and invasion, but induced autophagy and apoptosis (Fan et al., 2022). On the mechanism, METTL14 catalyzed m6A on FOXO3a and enhanced RNA stability and translation (Fan et al., 2022). Another MeRIP-seq data showed that METTL14 was upregulated and total m6A levels of circRNAs were increased in preeclampsia. METTL14 modified m6A on circPAPPA2 and the latter was identified by IGF2BP3 to maintain RNA stability (Zhang et al., 2021). The knockdown of circPAPPA2 suppressed trophoblast invasion and the expression level of circPAPPA2 was reduced in preeclampsia. In addition, m6A-related bioinformatic analysis was performed to seek the correlation between m6A modifications and clinical characteristics of preeclampsia (Li et al., 2022a). Higher m6A level was associated with higher maternal age and even a higher rate of FGR.
The rapid developments in m6A sequencing and its relevant methodology have conclusively highlighted the abundant existence and dynamic regulation network of m6A modification in RNA metabolism and fate decision, including alternative splicing, translocation, stability and translation (Wang et al., 2014b; Berulava et al., 2015; Tang et al., 2020; Akhtar et al., 2021). The whole-transcriptome m6A methylomes across major human tissues depicted the dynamic m6A methylation across different tissue types, covering both broadly and tissue-specifically m6A sites (Xiao et al., 2019). Moreover, m6A modifications have been discovered in various biological functions, including self-renewal and transition of stem cells, cell differentiation, cellular response to stress, hypoxia adaptation, metabolism and secretion, and other bioprocesses (Batista et al., 2014; Lin and Gregory, 2014; Zhang et al., 2017a; Zhang et al., 2017b; Cui et al., 2017; Xu et al., 2017; Frye et al., 2018; Tang et al., 2018; Wang et al., 2018). Hence, m6A modification serves as an essential regulator in physiological and pathophysiological conditions. Placental-specific m6A modifications are observed when compared with other human tissues, indicating its potentially unique biofunctions in placental development. In this study, we summarize current advances of m6A modifications during gestational period and obstetric diseases, and also highlight that m6A plays as sensitive effectors and regulators in maternal-fetal interaction.
Considering the existence and regulatory mechanism of epigenetics in placenta, RNA m6A may act as a versatile checkpoint that correlates different layers of gene regulation and forms a more complicated regulatory network for cellular homeostasis (Kan et al., 2022). Similar to mRNA, the metabolic processes of non-coding RNAs are also regulated by m6A modifications, including RNA synthesis, cellular localization, translation and degradation (Jia et al., 2022). What’s more, RNA m6A modifications interact with DNA methylation and histone modifications. ALKBH5 is verified to demethylate m6A in DNMT3B mRNA and inhibit the degradation, inducing the pathogenesis of intervertebral disc disorders (Li et al., 2022b). Maternal and environmental factors may affect multiple checkpoints of epigenetics and further induce the pathogenesis of placenta-related diseases.
Nevertheless, our knowledge of m6A in placenta, especially placental development and gestational diseases is far from complete. We still have limited information about the physiological changes of m6A in developmental placentas which is the basis of placental research. More importantly, m6A target RNAs in maternal serum can be a new direction and strategy for the development of novel biomarkers for prenatal diagnosis. Follow-up studies need to address these key issues more specifically.
SuqW wrote the manuscript, KL draw the figures, BZ and SuwW revised the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Akhtar, J., Lugoboni, M., and Junion, G. (2021). m(6 A RNA modification in transcription regulation. Transcription 12 (5), 266–276. doi:10.1080/21541264.2022.2057177
Alarcon, C. R., Goodarzi, H., Lee, H., Liu, X., Tavazoie, S., and Tavazoie, S. F. (2015). HNRNPA2B1 is a mediator of m(6)a-dependent nuclear RNA processing events. Cell 162 (6), 1299–1308. doi:10.1016/j.cell.2015.08.011
Albers, R. E., Kaufman, M. R., Natale, B. V., Keoni, C., Kulkarni-Datar, K., Min, S., et al. (2019). Trophoblast-specific expression of hif-1α results in preeclampsia-like symptoms and fetal growth restriction. Sci. Rep. 9 (1), 2742. doi:10.1038/s41598-019-39426-5
Ananth, C. V. (2014). Ischemic placental disease: A unifying concept for preeclampsia, intrauterine growth restriction, and placental abruption. Semin. Perinatol. 38 (3), 131–132. doi:10.1053/j.semperi.2014.03.001
Ashraf, U. M., Hall, D. L., Rawls, A. Z., and Alexander, B. T. (2021). Epigenetic processes during preeclampsia and effects on fetal development and chronic health. Clin. Sci. (Lond). 135 (19), 2307–2327. doi:10.1042/CS20190070
Batista, P. J., Molinie, B., Wang, J., Qu, K., Zhang, J., Li, L., et al. (2014). m(6 A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell 15 (6), 707–719. doi:10.1016/j.stem.2014.09.019
Berulava, T., Rahmann, S., Rademacher, K., Klein-Hitpass, L., and Horsthemke, B. (2015). N6-adenosine methylation in MiRNAs. Plos One 10 (2), e0118438. doi:10.1371/journal.pone.0118438
Bokar, J. A., Rath-Shambaugh, M. E., Ludwiczak, R., Narayan, P., and Rottman, F. (1994). Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei. Internal mRNA methylation requires a multisubunit complex. J. Biol. Chem. 269 (26), 17697–17704. doi:10.1016/s0021-9258(17)32497-3
Brosens, I., Pijnenborg, R., Vercruysse, L., and Romero, R. (2011). The "Great Obstetrical Syndromes" are associated with disorders of deep placentation. Am. J. Obstet. Gynecol. 204 (3), 193–201. doi:10.1016/j.ajog.2010.08.009
Caniggia, I., Mostachfi, H., Winter, J., GassMann, M., Lye, S. J., Kuliszewski, M., et al. (2000). Hypoxia-inducible factor-1 mediates the biological effects of oxygen on human trophoblast differentiation through TGFbeta(3). J. Clin. Invest. 105 (5), 577–587. doi:10.1172/JCI8316
Chang, G., Shi, L., Ye, Y., Shi, H., Zeng, L., Tiwary, S., et al. (2020). YTHDF3 induces the translation of m(6)a-enriched gene transcripts to promote breast cancer brain metastasis. Cancer Cell 38 (6), 857–871.e7. doi:10.1016/j.ccell.2020.10.004
Chen, B., Liu, Y., Cai, Y., Tang, D., Xu, S., Gao, P., et al. (2020). Hippocampus is more vulnerable to neural damages induced by repeated sevoflurane exposure in the second trimester than other brain areas. Acta Biochim. Biophys. Sin. (Shanghai) 52 (8), 864–874. doi:10.1093/abbs/gmaa060
Chen, G., Liu, B., Yin, S., Li, S., Guo, Y., Wang, M., et al. (2020). Hypoxia induces an endometrial cancer stem-like cell phenotype via HIF-dependent demethylation of SOX2 mRNA. Oncogenesis 9 (9), 81. doi:10.1038/s41389-020-00265-z
Chen, K., Lu, Z., Wang, X., Fu, Y., Luo, G. Z., Liu, N., et al. (2015). High-resolution N(6) -methyladenosine (m(6) A) map using photo-crosslinking-assisted m(6) A sequencing. Angew. Chem. Int. Ed. Engl. 54 (5), 1587–1590. doi:10.1002/anie.201410647
Chen, Z., Qi, M., Shen, B., Luo, G., Wu, Y., Li, J., et al. (2019). Transfer RNA demethylase ALKBH3 promotes cancer progression via induction of tRNA-derived small RNAs. Nucleic Acids Res. 47 (5), 2533–2545. doi:10.1093/nar/gky1250
Choe, J., Lin, S., Zhang, W., Liu, Q., Wang, L., Ramirez-Moya, J., et al. (2018). mRNA circularization by METTL3-eIF3h enhances translation and promotes oncogenesis. Nature 561 (7724), 556–560. doi:10.1038/s41586-018-0538-8
Cui, Q., Shi, H., Ye, P., Li, L., Qu, Q., Sun, G., et al. (2017). m(6 A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 18 (11), 2622–2634. doi:10.1016/j.celrep.2017.02.059
Dominissini, D., Moshitch-Moshkovitz, S., Salmon-Divon, M., Amariglio, N., and Rechavi, G. (2013). Transcriptome-wide mapping of N(6)-methyladenosine by m(6)A-seq based on immunocapturing and massively parallel sequencing. Nat. Protoc. 8 (1), 176–189. doi:10.1038/nprot.2012.148
Dominissini, D., Moshitch-Moshkovitz, S., Schwartz, S., Salmon-Divon, M., Ungar, L., Osenberg, S., et al. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485 (7397), 201–206. doi:10.1038/nature11112
Dong, F., Qin, X., Wang, B., Li, Q., Hu, J., Cheng, X., et al. (2021). ALKBH5 facilitates hypoxia-induced paraspeckle assembly and IL8 secretion to generate an immunosuppressive tumor microenvironment. Cancer Res. 81 (23), 5876–5888. doi:10.1158/0008-5472.CAN-21-1456
Du, K., Zhang, L., Lee, T., and Sun, T. (2019). m(6 A RNA methylation controls neural development and is involved in human diseases. Mol. Neurobiol. 56 (3), 1596–1606. doi:10.1007/s12035-018-1138-1
Du, R., Bai, Y., and Li, L. (2022). Biological networks in gestational diabetes mellitus: Insights into the mechanism of crosstalk between long non-coding RNA and N(6)-methyladenine modification. BMC Pregnancy Childbirth 22 (1), 384. doi:10.1186/s12884-022-04716-w
Du, R., Li, L., and Wang, Y. (2022). N6-Methyladenosine-Related gene signature associated with monocyte infiltration is clinically significant in gestational diabetes mellitus. Front. Endocrinol. (Lausanne). 13, 853857. doi:10.3389/fendo.2022.853857
Du, R., Wu, N., and Li, L. (2021). Aberrantly expressed non-coding RNAs in the placenta and their role in the pathophysiology of gestational diabetes mellitus. Diabetes Metab. Syndr. Obes. 14, 3719–3732. doi:10.2147/DMSO.S325993
Fan, W., Zhou, W., Yan, Q., Peng, Y., Wang, H., Kong, C., et al. (2022). Upregulation of METTL14 contributes to trophoblast dysfunction by elevating FOXO3a expression in an m(6)A-dependent manner. Placenta 124, 18–27. doi:10.1016/j.placenta.2022.05.008
Frye, M., Harada, B. T., Behm, M., and He, C. (2018). RNA modifications modulate gene expression during development. Science 361 (6409), 1346–1349. doi:10.1126/science.aau1646
Gabbay-Benziv, R., and Baschat, A. A. (2015). Gestational diabetes as one of the "great obstetrical syndromes"--the maternal, placental, and fetal dialog. Best. Pract. Res. Clin. Obstet. Gynaecol. 29 (2), 150–155. doi:10.1016/j.bpobgyn.2014.04.025
Garcia-Campos, M. A., Edelheit, S., Toth, U., Safra, M., Shachar, R., Viukov, S., et al. (2019). Deciphering the "m(6)A code" via antibody-independent quantitative profiling. Cell 178 (3), 731–747. doi:10.1016/j.cell.2019.06.013
Gu, Y., Chu, X., Morgan, J. A., Lewis, D. F., and Wang, Y. (2021). Upregulation of METTL3 expression and m6A RNA methylation in placental trophoblasts in preeclampsia. Placenta 103, 43–49. doi:10.1016/j.placenta.2020.10.016
Guo, Y., Song, W., and Yang, Y. (2022). Inhibition of ALKBH5-mediated m(6) A modification of PPARG mRNA alleviates H/R-induced oxidative stress and apoptosis in placenta trophoblast. Environ. Toxicol. 37 (4), 910–924. doi:10.1002/tox.23454
Hess, M. E., Hess, S., Meyer, K. D., Verhagen, L. A. W., Koch, L., Bronneke, H. S., et al. (2013). The fat mass and obesity associated gene (Fto) regulates activity of the dopaminergic midbrain circuitry. Nat. Neurosci. 16 (8), 1042–1048. doi:10.1038/nn.3449
Hu, L., Liu, S., Peng, Y., Ge, R., Su, R., Senevirathne, C., et al. (2022). m6A RNA modifications are measured at single-base resolution across the mammalian transcriptome. Nat. Biotechnol. 40 (8), 1210–1219. doi:10.1038/s41587-022-01243-z
Huang, H., Li, H., Pan, R., Wang, S., Khan, A. A., Zhao, Y., et al. (2022). Ribosome 18S m(6)A methyltransferase METTL5 promotes pancreatic cancer progression by modulating cMyc translation. Int. J. Oncol. 60 (1), 9. doi:10.3892/ijo.2021.5299
Huang, X. T., Li, J. H., Zhu, X. X., Huang, C. S., Gao, Z. X., Xu, Q. C., et al. (2021). HNRNPC impedes m(6)A-dependent anti-metastatic alternative splicing events in pancreatic ductal adenocarcinoma. Cancer Lett. 518, 196–206. doi:10.1016/j.canlet.2021.07.016
Imanishi, M., Tsuji, S., Suda, A., and Futaki, S. (2017). Detection of N(6)-methyladenosine based on the methyl-sensitivity of MazF RNA endonuclease. Chem. Commun. (Camb). 53 (96), 12930–12933. doi:10.1039/c7cc07699a
Iriyama, T., Wang, W., Parchim, N. F., Song, A., Blackwell, S. C., Sibai, B. M., et al. (2015). Hypoxia-independent upregulation of placental hypoxia inducible factor-1α gene expression contributes to the pathogenesis of preeclampsia. Hypertension 65 (6), 1307–1315. doi:10.1161/HYPERTENSIONAHA.115.05314
Izquierdo, V., Palomera-Avalos, V., Pallas, M., and Grinan-Ferre, C. (2021). Resveratrol supplementation attenuates cognitive and molecular alterations under maternal high-fat diet intake: Epigenetic inheritance over generations. Int. J. Mol. Sci. 22 (3), 1453. doi:10.3390/ijms22031453
Jauniaux, E., Poston, L., and Burton, G. J. (2006). Placental-related diseases of pregnancy: Involvement of oxidative stress and implications in human evolution. Hum. Reprod. Update 12 (6), 747–755. doi:10.1093/humupd/dml016
Jia, G., Fu, Y., Zhao, X., Dai, Q., Zheng, G., Yang, Y., et al. (2011). N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7 (12), 885–887. doi:10.1038/nchembio.687
Jia, J., Wu, S., Jia, Z., Wang, C., Ju, C., Sheng, J., et al. (2022). Novel insights into m(6)A modification of coding and non-coding RNAs in tumor biology: From molecular mechanisms to therapeutic significance. Int. J. Biol. Sci. 18 (11), 4432–4451. doi:10.7150/ijbs.73093
Kan, R. L., Chen, J., and Sallam, T. (2022). Crosstalk between epitranscriptomic and epigenetic mechanisms in gene regulation. Trends Genet. 38 (2), 182–193. doi:10.1016/j.tig.2021.06.014
Ke, S., Alemu, E. A., Mertens, C., Gantman, E. C., Fak, J. J., Mele, A., et al. (2015). A majority of m6A residues are in the last exons, allowing the potential for 3' UTR regulation. Genes Dev. 29 (19), 2037–2053. doi:10.1101/gad.269415.115
Klinge, C. M., Piell, K. M., Tooley, C. S., and Rouchka, E. C. (2019). HNRNPA2/B1 is upregulated in endocrine-resistant LCC9 breast cancer cells and alters the miRNA transcriptome when overexpressed in MCF-7 cells. Sci. Rep. 9 (1), 9430. doi:10.1038/s41598-019-45636-8
Knuckles, P., Lence, T., Haussmann, I. U., Jacob, D., Kreim, N., Carl, S. H., et al. (2018). Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m6 A machinery component Wtap/Fl(2)d. Gene Dev. 32 (5-6), 415–429. doi:10.1101/gad.309146.117
Koh, C., Goh, Y. T., and Goh, W. (2019). Atlas of quantitative single-base-resolution N(6)-methyl-adenine methylomes. Nat. Commun. 10 (1), 5636. doi:10.1038/s41467-019-13561-z
Koukoura, O., Sifakis, S., and Spandidos, D. A. (2012). DNA methylation in the human placenta and fetal growth (review). Mol. Med. Rep. 5 (4), 883–889. doi:10.3892/mmr.2012.763
Kunovac, A., Hathaway, Q. A., Pinti, M. V., Durr, A. J., Taylor, A. D., Goldsmith, W. T., et al. (2021). Enhanced antioxidant capacity prevents epitranscriptomic and cardiac alterations in adult offspring gestationally-exposed to ENM. Nanotoxicology 15 (6), 812–831. doi:10.1080/17435390.2021.1921299
Lence, T., Akhtar, J., Bayer, M., Schmid, K., Spindler, L., Ho, C. H., et al. (2016). m(6 A modulates neuronal functions and sex determination in Drosophila. Nature 540 (7632), 242–247. doi:10.1038/nature20568
Lesbirel, S., Viphakone, N., Parker, M., Parker, J., Heath, C., Sudbery, I., et al. (2018). The m(6)A-methylase complex recruits TREX and regulates mRNA export. Sci. Rep. 8 (1), 13827. doi:10.1038/s41598-018-32310-8
Li, G., Luo, R., Zhang, W., He, S., Wang, B., Liang, H., et al. (2022). m6A hypomethylation of DNMT3B regulated by ALKBH5 promotes intervertebral disc degeneration via E4F1 deficiency. Clin. Transl. Med. 12 (3), e765. doi:10.1002/ctm2.765
Li, X. C., Jin, F., Wang, B. Y., Yin, X. J., Hong, W., and Tian, F. J. (2019). The m6A demethylase ALKBH5 controls trophoblast invasion at the maternal-fetal interface by regulating the stability of CYR61 mRNA. Theranostics 9 (13), 3853–3865. doi:10.7150/thno.31868
Li, Y., Chen, C., Diao, M., Wei, Y., Zhu, Y., and Hu, W. (2022). Gene model-related m6A expression levels predict the risk of preeclampsia. Bmc Med. Genomics 15 (1), 103. doi:10.1186/s12920-022-01254-4
Lin, S., and Gregory, R. I. (2014). Methyltransferases modulate RNA stability in embryonic stem cells. Nat. Cell Biol. 16 (2), 129–131. doi:10.1038/ncb2914
Linder, B., Grozhik, A. V., Olarerin-George, A. O., Meydan, C., Mason, C. E., and Jaffrey, S. R. (2015). Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 12 (8), 767–772. doi:10.1038/nmeth.3453
Liu, J., Yue, Y., Han, D., Wang, X., Fu, Y., Zhang, L., et al. (2014). A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10 (2), 93–95. doi:10.1038/nchembio.1432
Liu, N., Dai, Q., Zheng, G., Parisien, M., and Pan, T. (2015). N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 518 (7540), 560–564. doi:10.1038/nature14234
Liu, N., Zhou, K. I., Parisien, M., Dai, Q., Diatchenko, L., and Pan, T. (2017). N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 45 (10), 6051–6063. doi:10.1093/nar/gkx141
Lorincz, M. C., and Schubeler, D. (2017). Evidence for converging DNA methylation pathways in placenta and cancer. Dev. Cell 43 (3), 257–258. doi:10.1016/j.devcel.2017.10.009
Luo, G. Z., MacQueen, A., Zheng, G., Duan, H., Dore, L. C., Lu, Z., et al. (2014). Unique features of the m6A methylome in Arabidopsis thaliana. Nat. Commun. 5, 5630. doi:10.1038/ncomms6630
Ma, L., Zhao, B., Chen, K., Thomas, A., Tuteja, J. H., He, X., et al. (2017). Evolution of transcript modification by N(6)-methyladenosine in primates. Genome Res. 27 (3), 385–392. doi:10.1101/gr.212563.116
Meister, S., Hahn, L., Beyer, S., Kuhn, C., Jegen, M., von Schonfeldt, V., et al. (2021). Epigenetic modification via H3K4me3 and H3K9ac in human placenta is reduced in preeclampsia. J. Reprod. Immunol. 145, 103287. doi:10.1016/j.jri.2021.103287
Meyer, K. D. (2019). DART-Seq: An antibody-free method for global m(6)A detection. Nat. Methods 16 (12), 1275–1280. doi:10.1038/s41592-019-0570-0
Meyer, K. D., Patil, D. P., Zhou, J., Zinoviev, A., Skabkin, M. A., Elemento, O., et al. (2015). 5' UTR m(6)A promotes cap-independent translation. Cell 163 (4), 999–1010. doi:10.1016/j.cell.2015.10.012
Meyer, K. D., Saletore, Y., Zumbo, P., Elemento, O., Mason, C. E., and Jaffrey, S. R. (2012). Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell 149 (7), 1635–1646. doi:10.1016/j.cell.2012.05.003
Mizrak, S. B., and Kabakci, E. N. (2021). The experiences of pregnant women during the COVID-19 pandemic in Turkey: A qualitative study. Women Birth 34 (2), 162–169. doi:10.1016/j.wombi.2020.09.022
Molinie, B., Wang, J., Lim, K. S., Hillebrand, R., Lu, Z. X., Van Wittenberghe, N., et al. (2016). m(6 A-LAIC-seq reveals the census and complexity of the m(6)A epitranscriptome. Nat. Methods 13 (8), 692–698. doi:10.1038/nmeth.3898
Mu, H., Li, H., Liu, Y., Wang, X., Mei, Q., and Xiang, W. (2022). N6-Methyladenosine modifications in the female reproductive system: Roles in gonad development and diseases. Int. J. Biol. Sci. 18 (2), 771–782. doi:10.7150/ijbs.66218
Nelissen, E. C., van Montfoort, A. P., Dumoulin, J. C., and Evers, J. L. (2011). Epigenetics and the placenta. Hum. Reprod. Update 17 (3), 397–417. doi:10.1093/humupd/dmq052
Ping, X. L., Sun, B. F., Wang, L., Xiao, W., Yang, X., Wang, W. J., et al. (2014). Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 24 (2), 177–189. doi:10.1038/cr.2014.3
Pinto, R., Vagbo, C. B., Jakobsson, M. E., Kim, Y., Baltissen, M. P., O'Donohue, M. F., et al. (2020). The human methyltransferase ZCCHC4 catalyses N6-methyladenosine modification of 28S ribosomal RNA. Nucleic Acids Res. 48 (2), 830–846. doi:10.1093/nar/gkz1147
Pringle, K. G., Kind, K. L., Sferruzzi-Perri, A. N., Thompson, J. G., and Roberts, C. T. (2010). Beyond oxygen: Complex regulation and activity of hypoxia inducible factors in pregnancy. Hum. Reprod. Update 16 (4), 415–431. doi:10.1093/humupd/dmp046
Prins, J. R., Schoots, M. H., Wessels, J. I., Campmans-Kuijpers, M. J. E., Navis, G. J., van Goor, H., et al. (2022). The influence of the dietary exposome on oxidative stress in pregnancy complications. Mol. Asp. Med. 87, 101098. doi:10.1016/j.mam.2022.101098
Qiu, W., Zhou, Y., Wu, H., Lv, X., Yang, L., Ren, Z., et al. (2021). RNA demethylase FTO mediated RNA m(6)A modification is involved in maintaining maternal-fetal interface in spontaneous abortion. Front. Cell Dev. Biol. 9, 617172. doi:10.3389/fcell.2021.617172
Ren, W., Lu, J., Huang, M., Gao, L., Li, D., Wang, G. G., et al. (2019). Structure and regulation of ZCCHC4 in m(6)A-methylation of 28S rRNA. Nat. Commun. 10 (1), 5042. doi:10.1038/s41467-019-12923-x
Roberts, J. M. (2014). Pathophysiology of ischemic placental disease. Semin. Perinatol. 38 (3), 139–145. doi:10.1053/j.semperi.2014.03.005
Roundtree, I. A., Luo, G. Z., Zhang, Z., Wang, X., Zhou, T., Cui, Y., et al. (2017). YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs. Elife 6, e31311. doi:10.7554/eLife.31311
Saletore, Y., Meyer, K., Korlach, J., Vilfan, I. D., Jaffrey, S., and Mason, C. E. (2012). The birth of the epitranscriptome: Deciphering the function of RNA modifications. Genome Biol. 13 (10), 175. doi:10.1186/gb-2012-13-10-175
Schwartz, S., Agarwala, S. D., Mumbach, M. R., Jovanovic, M., Mertins, P., Shishkin, A., et al. (2013). High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell 155 (6), 1409–1421. doi:10.1016/j.cell.2013.10.047
Sepich-Poore, C., Zheng, Z., Schmitt, E., Wen, K., Zhang, Z. S., Cui, X. L., et al. (2022). The METTL5-TRMT112 N(6)-methyladenosine methyltransferase complex regulates mRNA translation via 18S rRNA methylation. J. Biol. Chem. 298 (3), 101590. doi:10.1016/j.jbc.2022.101590
Shafik, A. M., Allen, E. G., and Jin, P. (2020). Dynamic N6-methyladenosine RNA methylation in brain and diseases. Epigenomics-Uk 12 (4), 371–380. doi:10.2217/epi-2019-0260
She, J., Tan, K., Liu, J., Cao, S., Li, Z., Peng, Y., et al. (2022). The alteration of m(6)A modification at the transcriptome-wide level in human villi during spontaneous abortion in the first trimester. Front. Genet. 13, 861853. doi:10.3389/fgene.2022.861853
Shen, W. B., Ni, J., Yao, R., Goetzinger, K. R., Harman, C., Reece, E. A., et al. (2022). Maternal obesity increases DNA methylation and decreases RNA methylation in the human placenta. Reprod. Toxicol. 107, 90–96. doi:10.1016/j.reprotox.2021.12.002
Shima, H., Matsumoto, M., Ishigami, Y., Ebina, M., Muto, A., Sato, Y., et al. (2017). S-adenosylmethionine synthesis is regulated by selective N(6)-adenosine methylation and mRNA degradation involving METTL16 and YTHDC1. Cell Rep. 21 (12), 3354–3363. doi:10.1016/j.celrep.2017.11.092
Shu, X., Cao, J., Cheng, M., Xiang, S., Gao, M., Li, T., et al. (2020). A metabolic labeling method detects m(6)A transcriptome-wide at single base resolution. Nat. Chem. Biol. 16 (8), 887–895. doi:10.1038/s41589-020-0526-9
Song, T., Lu, J., Deng, Z., Xu, T., Yang, Y., Wei, H., et al. (2018). Maternal obesity aggravates the abnormality of porcine placenta by increasing N(6)-methyladenosine. Int. J. Obes. (Lond). 42 (10), 1812–1820. doi:10.1038/s41366-018-0113-2
Sun, C. Y., Cao, D., Du, B. B., Chen, C. W., and Liu, D. (2022). The role of Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs) as m(6)A readers in cancer. Int. J. Biol. Sci. 18 (7), 2744–2758. doi:10.7150/ijbs.70458
Sun, R., Yuan, L., Jiang, Y., Wan, Y., Ma, X., Yang, J., et al. (2023). ALKBH5 activates FAK signaling through m6A demethylation in ITGB1 mRNA and enhances tumor-associated lymphangiogenesis and lymph node metastasis in ovarian cancer. Theranostics 13 (2), 833–848. doi:10.7150/thno.77441
Tang, C., Klukovich, R., Peng, H., Wang, Z., Yu, T., Zhang, Y., et al. (2018). ALKBH5-dependent m6A demethylation controls splicing and stability of long 3'-UTR mRNAs in male germ cells. Proc. Natl. Acad. Sci. U. S. A. 115 (2), E325–E333. doi:10.1073/pnas.1717794115
Tang, C., Xie, Y., Yu, T., Liu, N., Wang, Z., Woolsey, R. J., et al. (2020). m(6 A-dependent biogenesis of circular RNAs in male germ cells. Cell Res. 30 (3), 211–228. doi:10.1038/s41422-020-0279-8
Taniguchi, K., Kawai, T., Kitawaki, J., Tomikawa, J., Nakabayashi, K., Okamura, K., et al. (2020). Epitranscriptomic profiling in human placenta: N6-methyladenosine modification at the 5'-untranslated region is related to fetal growth and preeclampsia. Faseb J. 34 (1), 494–512. doi:10.1096/fj.201900619RR
Tegowski, M., Flamand, M. N., and Meyer, K. D. (2022). scDART-seq reveals distinct m(6)A signatures and mRNA methylation heterogeneity in single cells. Mol. Cell 82 (4), 868–878.e10. doi:10.1016/j.molcel.2021.12.038
Tsui, M. H., Pang, M. W., Melender, H. L., Xu, L., Lau, T. K., and Leung, T. N. (2006). Maternal fear associated with pregnancy and childbirth in Hong Kong Chinese women. Women Health 44 (4), 79–92. doi:10.1300/j013v44n04_05
Ueda, Y., Ooshio, I., Fusamae, Y., Kitae, K., Kawaguchi, M., Jingushi, K., et al. (2017). AlkB homolog 3-mediated tRNA demethylation promotes protein synthesis in cancer cells. Sci. Rep. 7, 42271. doi:10.1038/srep42271
Valero, P., Fuentes, G., Cornejo, M., Vega, S., Grismaldo, A., Pardo, F., et al. (2022). Exposome and foetoplacental vascular dysfunction in gestational diabetes mellitus. Mol. Asp. Med. 87, 101019. doi:10.1016/j.mam.2021.101019
Walters, B. J., Mercaldo, V., Gillon, C. J., Yip, M., Neve, R. L., Boyce, F. M., et al. (2017). The role of the RNA demethylase FTO (fat mass and obesity-associated) and mRNA methylation in hippocampal memory formation. Neuropsychopharmacol 42 (7), 1502–1510. doi:10.1038/npp.2017.31
Wang, C., Sun, H., Jiang, X., Guan, X., Gao, F., and Shi, B. (2022). Maternal oxidized soybean oil administration in rats during pregnancy and lactation alters the intestinal DNA methylation in offspring. J. Agric. Food Chem. 70 (20), 6224–6238. doi:10.1021/acs.jafc.2c01100
Wang, J., Wang, K., Liu, W., Cai, Y., and Jin, H. (2021). m6A mRNA methylation regulates the development of gestational diabetes mellitus in Han Chinese women. Genomics 113 (3), 1048–1056. doi:10.1016/j.ygeno.2021.02.016
Wang, P., Doxtader, K. A., and Nam, Y. (2016). Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol. Cell 63 (2), 306–317. doi:10.1016/j.molcel.2016.05.041
Wang, Q., Pan, M., Zhang, T., Jiang, Y., Zhao, P., Liu, X., et al. (2022). Fear stress during pregnancy affects placental m6A-modifying enzyme expression and epigenetic modification levels. Front. Genet. 13, 927615. doi:10.3389/fgene.2022.927615
Wang, S., Gao, S., Zeng, Y., Zhu, L., Mo, Y., Wong, C. C., et al. (2022). N6-Methyladenosine reader YTHDF1 promotes ARHGEF2 translation and RhoA signaling in colorectal cancer. Gastroenterology 162 (4), 1183–1196. doi:10.1053/j.gastro.2021.12.269
Wang, X., Lu, Z., Gomez, A., Hon, G. C., Yue, Y., Han, D., et al. (2014). N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505 (7481), 117–120. doi:10.1038/nature12730
Wang, X., Zhao, B. S., Roundtree, I. A., Lu, Z., Han, D., Ma, H., et al. (2015). N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell 161 (6), 1388–1399. doi:10.1016/j.cell.2015.05.014
Wang, Y., Li, Y., Toth, J. I., Petroski, M. D., Zhang, Z., and Zhao, J. C. (2014). N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 16 (2), 191–198. doi:10.1038/ncb2902
Wang, Y., Li, Y., Yue, M., Wang, J., Kumar, S., Wechsler-Reya, R. J., et al. (2018). N(6)-methyladenosine RNA modification regulates embryonic neural stem cell self-renewal through histone modifications. Nat. Neurosci. 21 (2), 195–206. doi:10.1038/s41593-017-0057-1
Wang, Y., Xiao, Y., Dong, S., Yu, Q., and Jia, G. (2020). Antibody-free enzyme-assisted chemical approach for detection of N(6)-methyladenosine. Nat. Chem. Biol. 16 (8), 896–903. doi:10.1038/s41589-020-0525-x
Warda, A. S., Kretschmer, J., Hackert, P., Lenz, C., Urlaub, H., Hobartner, C., et al. (2017). Human METTL16 is a N(6)-methyladenosine (m(6)A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. Embo Rep. 18 (11), 2004–2014. doi:10.15252/embr.201744940
Wen, J., Lv, R., Ma, H., Shen, H., He, C., Wang, J., et al. (2018). Zc3h13 regulates nuclear RNA m6A methylation and mouse embryonic stem cell self-renewal. Mol. Cell 69 (6), 1028–1038. doi:10.1016/j.molcel.2018.02.015
Widagdo, J., Zhao, Q. Y., Kempen, M. J., Tan, M. C., Ratnu, V. S., Wei, W., et al. (2016). Experience-dependent accumulation of N6-methyladenosine in the prefrontal cortex is associated with memory processes in mice. J. Neurosci. 36 (25), 6771–6777. doi:10.1523/JNEUROSCI.4053-15.2016
Xiao, S., Cao, S., Huang, Q., Xia, L., Deng, M., Yang, M., et al. (2019). The RNA N(6)-methyladenosine modification landscape of human fetal tissues. Nat. Cell Biol. 21 (5), 651–661. doi:10.1038/s41556-019-0315-4
Xiao, W., Adhikari, S., Dahal, U., Chen, Y. S., Hao, Y. J., Sun, B. F., et al. (2016). Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Mol. Cell 61 (4), 507–519. doi:10.1016/j.molcel.2016.01.012
Xu, K., Yang, Y., Feng, G. H., Sun, B. F., Chen, J. Q., Li, Y. F., et al. (2017). Mettl3-mediated m(6)A regulates spermatogonial differentiation and meiosis initiation. Cell Res. 27 (9), 1100–1114. doi:10.1038/cr.2017.100
Xu, Z., Tian, P., Guo, J., Mi, C., Liang, T., Xie, J., et al. (2021). Lnc-HZ01 with m6A RNA methylation inhibits human trophoblast cell proliferation and induces miscarriage by up-regulating BPDE-activated lnc-HZ01/MXD1 positive feedback loop. Sci. Total Environ. 776, 145950. doi:10.1016/j.scitotenv.2021.145950
Yu, J., Chen, M., Huang, H., Zhu, J., Song, H., Zhu, J., et al. (2018). Dynamic m6A modification regulates local translation of mRNA in axons. Nucleic Acids Res. 46 (3), 1412–1423. doi:10.1093/nar/gkx1182
Yuan, W., Chen, S., Li, B., Han, X., Meng, B., Zou, Y., et al. (2022). The N6-methyladenosine reader protein YTHDC2 promotes gastric cancer progression via enhancing YAP mRNA translation. Transl. Oncol. 16, 101308. doi:10.1016/j.tranon.2021.101308
Yue, Y., Liu, J., Cui, X., Cao, J., Luo, G., Zhang, Z., et al. (2018). VIRMA mediates preferential m(6)A mRNA methylation in 3'UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 4, 10. doi:10.1038/s41421-018-0019-0
Zarkovic, M., Hufsky, F., Markert, U. R., and Marz, M. (2022). The role of non-coding RNAs in the human placenta. Cells-Basel 11 (9), 1588. doi:10.3390/cells11091588
Zhang, C., Chen, Y., Sun, B., Wang, L., Yang, Y., Ma, D., et al. (2017). m(6 A modulates haematopoietic stem and progenitor cell specification. Nature 549 (7671), 273–276. doi:10.1038/nature23883
Zhang, H., Shi, X., Huang, T., Zhao, X., Chen, W., Gu, N., et al. (2020). Dynamic landscape and evolution of m6A methylation in human. Nucleic Acids Res. 48 (11), 6251–6264. doi:10.1093/nar/gkaa347
Zhang, S., Meng, P., Cheng, S., Jiang, X., Zhang, J., Qin, X., et al. (2020). Pregnancy exposure to carbon black nanoparticles induced neurobehavioral deficits that are associated with altered m(6)A modification in offspring. Neurotoxicology 81, 40–50. doi:10.1016/j.neuro.2020.07.004
Zhang, S., Zhao, B. S., Zhou, A., Lin, K., Zheng, S., Lu, Z., et al. (2017). m(6 A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell 31 (4), 591–606. doi:10.1016/j.ccell.2017.02.013
Zhang, X., Dai, X. Y., Qian, J. Y., Xu, F., Wang, Z. W., Xia, T., et al. (2022). SMC1A regulated by KIAA1429 in m6A-independent manner promotes EMT progress in breast cancer. Mol. Ther. Nucleic Acids 27, 133–146. doi:10.1016/j.omtn.2021.08.009
Zhang, Y., Yang, H., Long, Y., Zhang, Y., Chen, R., Shi, J., et al. (2021). circRNA N6-methyladenosine methylation in preeclampsia and the potential role of N6-methyladenosine-modified circPAPPA2 in trophoblast invasion. Sci. Rep. 11 (1), 24357. doi:10.1038/s41598-021-03662-5
Zhang, Z., Chen, L. Q., Zhao, Y. L., Yang, C. G., Roundtree, I. A., Zhang, Z., et al. (2019). Single-base mapping of m6A by an antibody-independent method. Sci. Adv. 5 (7), eaax0250. doi:10.1126/sciadv.aax0250
Zhao, B. S., Wang, X., Beadell, A. V., Lu, Z., Shi, H., Kuuspalu, A., et al. (2017). m(6 A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. Nature 542 (7642), 475–478. doi:10.1038/nature21355
Zhao, X., Yang, Y., Sun, B. F., Shi, Y., Yang, X., Xiao, W., et al. (2014). FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 24 (12), 1403–1419. doi:10.1038/cr.2014.151
Zhao, Z., Ju, Q., Ji, J., Li, Y., and Zhao, Y. (2022). N6-Methyladenosine methylation regulator RBM15 is a potential prognostic biomarker and promotes cell proliferation in pancreatic adenocarcinoma. Front. Mol. Biosci. 9, 842833. doi:10.3389/fmolb.2022.842833
Zheng, G., Dahl, J. A., Niu, Y., Fedorcsak, P., Huang, C. M., Li, C. J., et al. (2013). ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49 (1), 18–29. doi:10.1016/j.molcel.2012.10.015
Keywords: N6-methyladenosine, maternal-fetal crosstalk, gestational diseases, epigenetics, fetal growth
Citation: Wu S, Liu K, Zhou B and Wu S (2023) N6-methyladenosine modifications in maternal-fetal crosstalk and gestational diseases. Front. Cell Dev. Biol. 11:1164706. doi: 10.3389/fcell.2023.1164706
Received: 13 February 2023; Accepted: 08 March 2023;
Published: 16 March 2023.
Edited by:
Daniela Bebbere, University of Sassari, ItalyReviewed by:
Xiao-Min Liu, China Pharmaceutical University, ChinaCopyright © 2023 Wu, Liu, Zhou and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bingyan Zhou, emhvdWJpbmd5YW5fMjAxMUAxNjMuY29t; Suwen Wu, d3VzdXdlbmh1c3RAMTYzLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.