- 1State Key Laboratory of Stem Cell and Reproductive Biology, Institute of Zoology, Chinese Academy of Sciences, Beijing, China
- 2Institute for Stem Cell and Regeneration, Chinese Academy of Sciences, Beijing, China
- 3Beijing Institute for Stem Cell and Regenerative Medicine, Beijing, China
- 4Savaid Medical School, University of Chinese Academy of Sciences, Beijing, China
MicroRNAs (miRNAs) are believed to play important roles in mammalian spermatogenesis mainly because spermatogenesis is more or less disrupted when genes encoding key enzymes for miRNA biogenesis are mutated. However, it is challenging to study the functions of individual miRNAs due to their family-wise high sequence similarities and the clustered genomic distributions of their genes, both of which expose difficulties in using genetic methods. Accumulating evidence shows that a number of miRNAs indeed play important roles in mammalian spermatogenesis and the underlying mechanisms start to be understood. In this mini review, we focus on highlighting the roles of miRNAs in mammalian spermatogenesis elucidated mainly by using in vivo genetic methods and on discussing the underlying mechanisms. We propose that studies on the roles of miRNAs in spermatogenesis should and can be conducted in a more fruitful way given the progress in traditional methods and the birth of new technologies.
Introduction
Spermatogenesis is a cellular developmental process starting from the differentiation of spermatogonial stem cells (SSCs) and ending up with the production of spermatozoa via the sequential generation of many intermediate cell types including differentiating spermatogonia, meiotic spermatocytes, and haploid spermatids (Figure 1) (Handel and Schimenti, 2010; Griswold, 2016). Like other developmental processes, spermatogenesis is driven by the dynamic gene expression that is regulated at different levels along the path depicted by the central dogma. One of the unique features of spermatogenesis is that the complexity of transcriptome in spermatogenic cells is the highest compared with those in other cell types (Handel and Schimenti, 2010; Xia et al., 2020). Moreover, a large proportion of transcripts including mRNAs, miRNAs, piRNAs, lncRNAs, and circular RNAs are specifically expressed in spermatogenic cells (Soumillon et al., 2013; Lin et al., 2016). However, to what extent all these transcripts are functional remains an open question.
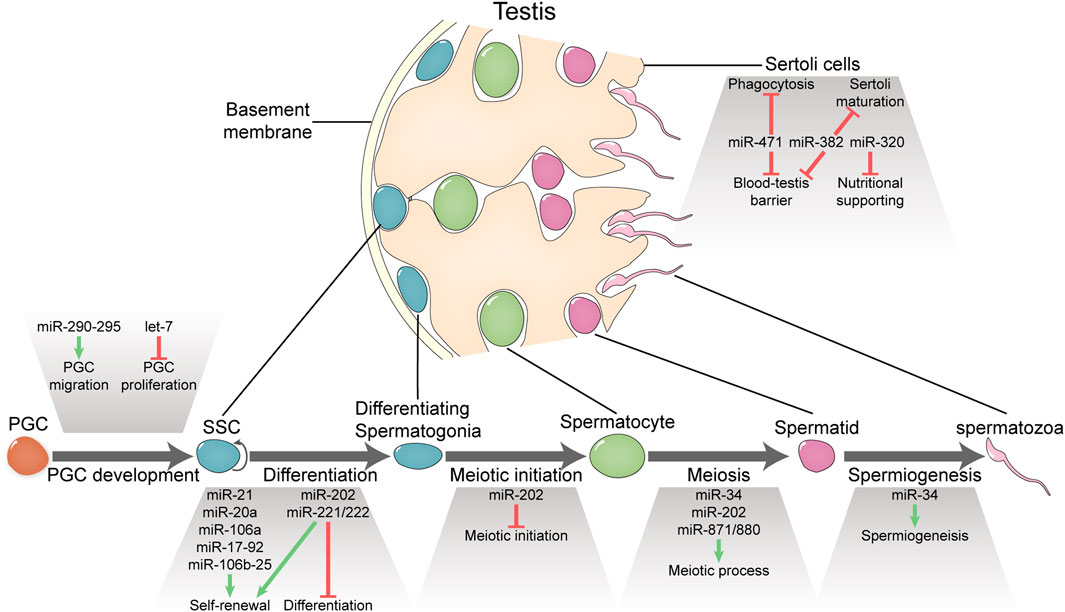
FIGURE 1. Functional miRNAs involved in spermatogenesis and male fertility. MicroRNAs portray essential or important roles in PGC development, spermatogenesis, and Sertoli functions.
MicroRNAs are small RNAs that are evolutionarily conserved and play important roles in many developmental processes of metazoans (Bartel, 2018). Conventionally, primary miRNA transcripts are transcribed by RNA polymerase II and then processed by two RNase III enzymes, DROSHA and DICER, into mature miRNAs to evoke mRNA degradation and translational repression (Czech and Hannon, 2011). Global miRNA loss caused by mutations in key miRNA-biogenesis regulators generates severe defects in almost all tissues examined, whereas individual miRNA deletion often lacks dramatic phenotypic consequences, likely due to the functional redundancy of miRNAs (Park et al., 2012). miRNAs are believed to be also important for mammalian spermatogenesis as indicated by the disrupted spermatogenesis in mice that are mutants for genes encoding key enzymes involved in miRNA biogenesis (Hayashi et al., 2008; Maatouk et al., 2008; Korhonen et al., 2011; Romero et al., 2011; Greenlee et al., 2012; Wu et al., 2012; Zimmermann et al., 2014; Modzelewski et al., 2015; Hilz et al., 2016). Out of the 1,200 miRNAs that have been identified in mice, at least 300 are expressed in at least one of three spermatogenic cell types—type A spermatogonia, pachytene spermatocytes, and round spermatids (Chen et al., 2017).
It has been challenging to define the functions of these miRNAs in spermatogenesis for two reasons. First, most miRNAs are functionally redundant as predicted by high sequence similarities of miRNAs belonging to the same family. Second, miRNA genes usually distribute in the genome as clusters. Both features of miRNAs prevent effective investigation of their functions using traditional genetic perturbation method. Many studies about miRNA functions in spermatogenesis take indirect methods. Some infer miRNA functions from their expression patterns and/or their predicted targets while others are based on results in cultured cells. It is very suspicious how reliable conclusions based on such indirect methods can be extrapolated to in vivo conditions.
In this mini review, we summarize observations on miRNA functions at different stages of mammalian spermatogenesis and the underlying mechanisms if available derived from studies using gene knockout/overexpression mice and in vitro cultured SSCs upon being transplanted (Figure 1; Table 1). As spermatogenic cells are progeny of primordial germ cells (PGCs) (Saitou et al., 2012), we also include studies about miRNA function in PGCs. At the end, we discuss how miRNA functional studies can be conducted more effectively with traditional method and new techniques such as CRISPR-based procedures.
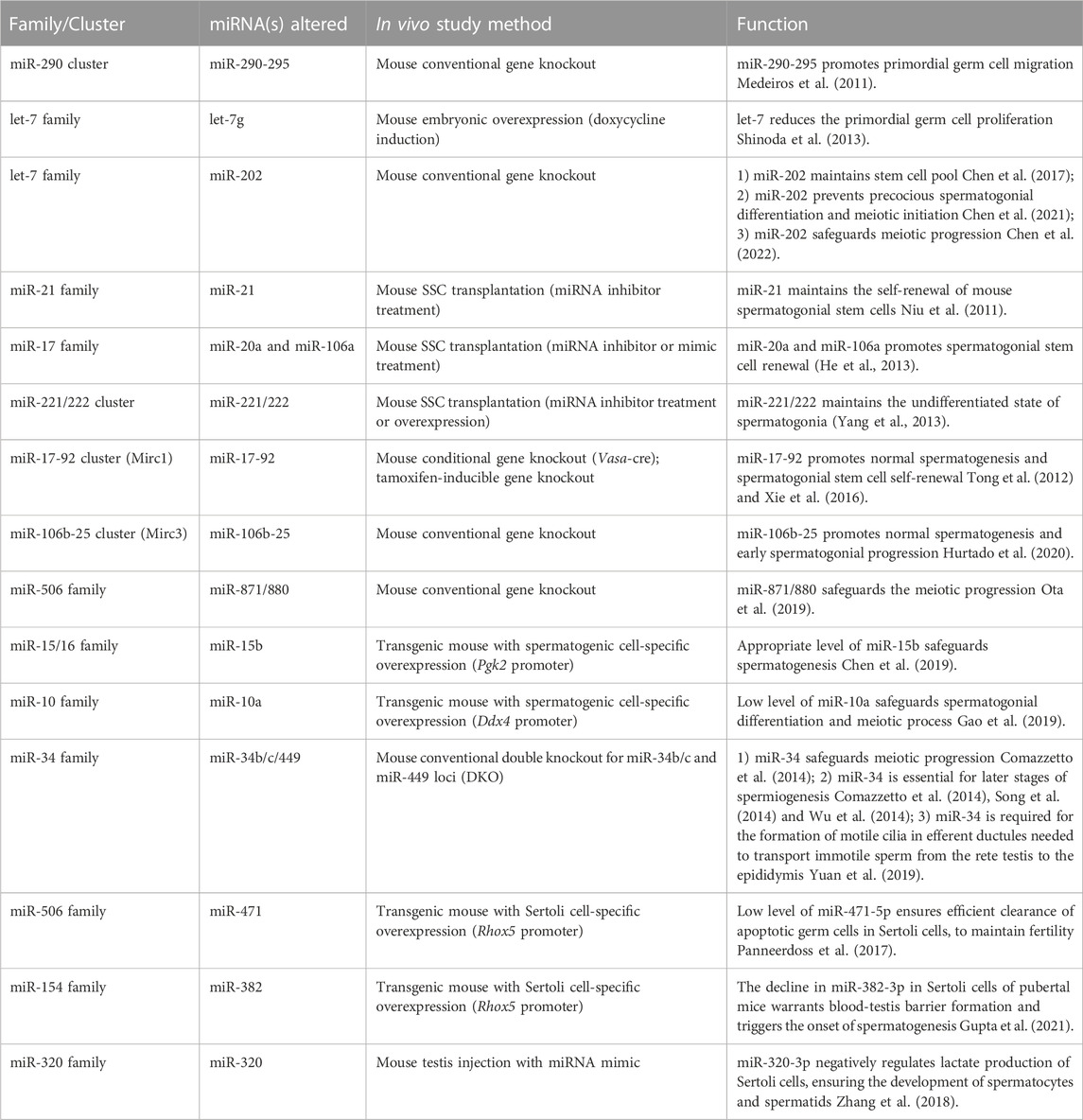
TABLE 1. Abnormal phenotypes observed in spermatogenesis in mice after knocking out or overexpressing one or more members of miRNA family/cluster.
miRNAs involved in PGC development and SSC fate decision
The first piece of evidences implying miRNA functions in germ cells came from a study by Hayashi et al., in which the Dicer1 was conditionally deleted using Tnap-Cre as the deletion initiator (Hayashi et al., 2008). In that study, poor proliferation of PGCs and spermatogonia was observed in the knockout mice. As the first direct evidence for the role of miRNAs in germ cell development, Medeiros et al. used conventional gene targeting method to delete the mammalian-specific miR-290-295 gene cluster, from which miRNAs are specifically expressed in early embryos and embryonic germ cells, and found that the number of post-migratory PGCs were reduced in both male and female mice mostly likely due to mislocalization of a subpopulation of migrating PGCs although the effect of a slower proliferation kinetics could not be excluded. Interestingly, female but not male fertility is compromised because male germ cells are able to recover from this initial germ cell loss due to their extended proliferative lifespan while female ones are unable to recover due to the lack of the mitotic amplification phase in fetal gonads (Medeiros et al., 2011).
let-7 family miRNAs are highly conserved in bilaterian animals and play important roles in many developmental and disease processes (Roush and Slack, 2008). The involvement of let-7 in germ cells development is first inferred by the roles of its negative regulator LIN28 in PGC proliferation (West et al., 2009), and then validated by the effect of induced overexpression of let-7 in transgenic mice (Shinoda et al., 2013). More specifically, LIN28A protein is detected in both gonocytes and undifferentiated spermatogonia and its conditional gene knockout in germ cells using Ddx4-Cre compromises PGC proliferation, the size of germ cell pool, and fertility in both male and female mice. Consistently, overexpression of let-7 in the doxycycline inducible iLet-7 mice also reduces the germ cell pool (Shinoda et al., 2013). These results suggest that the suppression of let-7 is important for PGC proliferation. These results are also consistent with earlier observations that let-7 directly regulates BLIMP1 (Nie et al., 2008), a key player in PGC specification and early development, and that BLIMP1 can rescue the effect of LIN28 deficiency during PGC development (West et al., 2009).
The intergenic miR-202 is distantly related to the let-7 family (Roush and Slack, 2008) and generates two complementary miRNAs, miR-202-5p and miR-202-3p, both of which are highly and specifically expressed in testes (Chen et al., 2017). By using an inducible CRISPR-Cas9 knockout platform in cultured SSCs and transplantation assays, we first showed that miR-202 is required to maintain stem cell pools. Using iTRAQ-based proteomic analysis and RNA sequencing, the target genes are identified and enriched with cell cycle regulators and RNA-binding proteins. Moreover, the RNA-binding protein RBFOX2, which is directly regulated by miR-202-3p, exhibits a regulatory role in meiotic initiation (Chen et al., 2017). Further, using knockout mice generated by the CRISPR-Cas9 technology, we showed that miR-202 prevents precocious spermatogonial differentiation and meiotic initiation by inhibiting the expression of key meiosis-initiating regulators—STRA8 and DMRT6. Moreover, Dmrt6 mRNA is a direct target of miR-202-5p and Dmrt6 knockdown could partially rescue the defects caused by miR-202 knockout in an in vitro assay. These findings add the miR-202/DMRT6 axis to the regulatory network of meiotic initiation (Chen et al., 2021). Notably, miR-202 knockout exerts no effects on Sertoli cell function (Chen et al., 2021).
Niu et al. first used high-throughput sequencing to compare miRNA profiles in Thy1+ SSC-enriched cells and Thy1- somatic cell-enriched cells from pre-pubertal mice, and identified a panel of miRNAs that are preferentially expressed in the Thy1+ population. Out of this panel, miR-21 was selected for further study and its gene was found to be directly targeted and activated by transcription factor ETV5, which is critical for SSC self-renewal. More importantly, transient inhibition of miR-21 in cultured SSCs followed by transplantation assay indicated that miR-21 prevents germ cell apoptosis and maintains stem cell activity of the cultured SSCs (Niu et al., 2011). Using the same SSC perturbation/transplantation strategy, more miRNAs that maintain SSC/undifferentiated spermatogonium identity are reported (He et al., 2013; Yang et al., 2013). These include miR-20 and miR-106a belonging to miR-17 family originated from the miR-17-92 cluster that were initially found to be highly expressed in PGCs (Hayashi et al., 2008), and miR-221/222 from an X chromosome cluster. These studies also made effort to identify direct targets of miRNAs based on bioinformatics prediction and experimental validation. In this sense, Stat3 and Ccnd1 transcripts are likely direct targets of miR-20 and miR-106a (He et al., 2013) while Kit translation is directly suppressed by miR-221/222, indicating a role for miR-221/222 in preventing transition to a KIT+ differentiation state in spermatogonia (Yang et al., 2013).
The miR-17-92 cluster (Mirc1) and its paralogous clusters miR-106a-363 (Mirc2) and miR-106b-25 (Mirc3) generate miRNAs belonging to four families: the miR-17, miR-18, miR-19, and the miR-25 families (Mendell, 2008). These miRNAs, highly conserved among vertebrates, not only act as oncogenes to promote cell proliferation, suppress apoptosis of cancer cells, and induce tumor angiogenesis but also play essential roles during normal development of the heart, lungs, and immune system (Mendell, 2008). miRNAs from Mirc1 are highly expressed in PGCs and Thy1+ undifferentiated spermatogonia (Hayashi et al., 2008; Tong et al., 2012). Tong et al. deleted the Mirc1 cluster in mouse germ cells using Vasa-Cre and found that spermatogenesis is disrupted with reduced testis weight, lower sperm count, and appearance of Sertoli cell-only tubules (Tong et al., 2012). Xie et al. (2016) also showed that miR-17-92 knockout progressively results in smaller testes and reduced numbers of spermatogonia and SSCs, all of which are mediated by the activation of the mTOR-signaling pathway, indicating that Mirc1 miRNAs contribute to maintain the undifferentiated state of spermatogonia. A recent study demonstrates that the conventional knockout of the Mirc3 cluster results in similar phenotypes as the Mirc1 mutants in spermatogenesis, i.e., reduced testis size, altered spermatogenesis, and low sperm count (Hurtado et al., 2020). Interestingly, fertility of either Mirc1 or Mirc3 knockout mice is not significantly reduced compared with wild-type controls but is severely reduced in Mirc1+/−/Mirc3−/− double knockout mice suggesting the dosage effect of miRNAs from these clusters (Hurtado et al., 2020).
miRNAs involved in meiotic progression and postmeiotic development
Conditional knockout of Dicer, Drosha or Dgcr8 in mouse germ lines after embryonic day 18 (driven by Ddx4, Stra8 or Ngn3 promoter) demonstrates that miRNAs are essential for meiotic progression, and spermiogenesis (Korhonen et al., 2011; Romero et al., 2011; Greenlee et al., 2012; Wu et al., 2012; Zimmermann et al., 2014; Modzelewski et al., 2015). Ota et al. identified three germ-cell specific miRNAs, miR-741-3p, miR-871-3p, and miR-880-3p, whose genes are contiguously located on the X chromosome, and generated single, double, and triple mutant mice (Ota et al., 2019). Spermatogenesis is normal in any of the three single mutant mouse lines but disrupted in the miR-871/880 double mutants and triple mutants. Numbers of undifferentiated spermatogonia and early-stage spermatocytes were not significantly changed, but incomplete meiosis arrest at the pachynema/diplonema transition was observed in the triple mutants. Surprisingly, a recent study reported that no phenotypes were observed when 18 out of the 21 X-linked miR-506 family miRNAs including the above three miRNAs were knocked out (Wang et al., 2020). This may be explained by the different mouse genetic backgrounds or counteracting effects of these miRNAs.
miR-202 is also involved in the regulation of meiotic progression. Loss of miR-202 in mice causes spermatocyte apoptosis and perturbation of the zygonema-to-pachynema transition. Multiple defects occur during meiosis prophase I in knockout mice, including disrupted synapsis and crossover formation, and inter-sister chromatid synapses. REC8 plays an essential role in homologous synapsis during meiotic prophase I and is cleaved by SEPARASE afterward. We provide evidence that miR-202-3p directly targets Separase mRNA, and miR-202 upregulates REC8 by repressing Separase expression (Chen et al., 2022). Therefore, miR-202 regulates meiotic progress by acting on the established SEPARASE-REC8 axis.
Several studies also used miRNA overexpression in transgenic mice to investigate their functions. For example, the overexpression of miR-15b, which is primarily expressed in spermatocytes, was reported to impair spermatogenesis (Chen et al., 2019). However, the affected cell types and stages were not clear. A similar study reported that overexpression of miR-10a in transgenic mice results in male sterility, meiotic arrest, and aberrant spermatogonial differentiation. The Rad51 mRNA, which encodes a protein playing a critical role in meiotic recombination, was validated to be the direct target of miR-10a (Gao et al., 2019).
The members in the miR-34 family (miR-34a, 34b, 34c, 449a, 449b, and 449c) are conserved in vertebrates and are generated from three genomic loci—miR-34a, miR-34b/c, and miR-449, which are actively transcribed in the testes (Bouhallier et al., 2010; Bao et al., 2012; Concepcion et al., 2012). miR-34b/c and miR-449 are mainly expressed in pachytene spermatocytes and spermatids, whereas miR-34a is restricted to spermatogonia (Bouhallier et al., 2010; Bao et al., 2012; Concepcion et al., 2012). Surprisingly, mice with each of the three loci deleted and mice with both the miR-34a/miR-34b/c loci deleted revealed no discernible defects in spermatogenesis and fertility (Choi et al., 2011; Bao et al., 2012; Concepcion et al., 2012; Song et al., 2014). In contrast, oligoasthenoteratozoospermia and infertility were observed in mice with the miR-449/miR-34b/c double-knockout (DKO) or triple-knockout (Comazzetto et al., 2014; Song et al., 2014; Wu et al., 2014), indicating a redundant function of miR-449 and miR-34b/c in spermatogenesis. Interestingly, germ cell loss in DKO occurs at two distinct phases: during pachytene stage (Comazzetto et al., 2014) and later during spermatid elongation (Comazzetto et al., 2014; Song et al., 2014; Wu et al., 2014). Moreover, DKO leads to loss of the motile cilia in efferent ductules needed to transport immotile sperm from the rete testis to the epididymis (Yuan et al., 2019), reminiscent of previous observations that the formation of motile cilia in multiciliated epithelia is disrupted in DKO mice (Song et al., 2014; Wu et al., 2014; Yuan et al., 2015).
miRNAs involved in Sertoli functions
Sertoli cells constitute a critical component in spermatogenesis and miRNA biogenesis is essential for Sertoli cells to mature, survive, and ultimately support spermatogenesis (Papaioannou et al., 2009; Kim et al., 2010). Several studies used transgenic mice to investigate the in vivo miRNA functions in Sertoli cells. miR-471 belongs to the miR-506 family (Wang et al., 2020), and miR-471-5p is highly expressed in Sertoli cells in an androgen-dependent manner. Overexpression of miR-471-5p in Sertoli cells increases germ cell apoptosis and impairs male fertility due to disrupted integrity of blood-testis barrier (BTB), and defective engulfment and clearance of apoptotic germ cells. miR-471-5p directly regulates Dock180 which interacts with autophagy member proteins to constitute a functional phagocytic complex, revealing an essential role for miRNAs in the efficient clearance of apoptotic germ cells by Sertoli cells (Panneerdoss et al., 2017). In rat, miR-382-3p level exhibits dramatic decline in pubertal Sertoli cells compared to infant ones. Prevention of the decline in miR-382-3p in Sertoli cells of pubertal mice leads to severe testicular defects, low epididymal sperm counts, and infertility due to defective BTB formation, impaired Sertoli maturation, and compromised androgen receptor signaling. This implies that the decline in miR-382-3p levels at puberty is essential for triggering the onset of robust spermatogenesis. Interestingly, miR-382-3p directly represses AR and WT1, which are crucial for functional competence of Sertoli cells (Gupta et al., 2021). miR-320-3p is exclusively expressed in mouse Sertoli cells, and miR-320-3p overexpression in Sertoli cells compromises male fertility and leads to oligozoospermia and reduced sperm mobility. These defects may be caused by dysregulation of the nutritional supporting function of Sertoli cells by reducing lactate production through directly inhibiting glucose transporter 3 (GLUT3) expression (Zhang et al., 2018).
Discussion
Since the report of the first miRNA lin-4 in C. elegans in 1993, three decades have passed with big progresses in understanding the mechanism of biogenesis, the function, the underlying mechanism, and the regulated expression of miRNAs as a whole class of small RNAs (Bartel, 2018). These progresses are made by investigations using various cell types. Spermatogenic cells, possessing the most complex transcriptome and undergoing many unique processes, serve as ideal models to study gene regulation at different levels involving diverse regulators including miRNAs (Soumillon et al., 2013; Lin et al., 2016). Compared with other types of RNAs expressed in spermatogenic cells, miRNAs form a relatively smaller family and are supposed to be more tractable in their functional studies. However, as can be seen from the above review, we still know very little in every aspect.
We have only studied the function of a small number of miRNAs out of the more than 1,000 mammalian miRNAs using the mouse gene knockout/overexpression technique or the cultured SSC/transplantation method. The number of validated miRNA target genes is limited. And we know little about how miRNAs regulate biological processes through certain pathways—what pathways they regulate and through which pathways they are regulated. Despite the lack of in-depth knowledge about miRNAs, several vague patterns about their actions in spermatogenesis appear. The first one is that, like what has been observed in other biological processes, miRNAs in spermatogenesis also seem to be functionally redundant and their effects are minor and accumulative. Abolishment of the expression of a single miRNA seldom disrupts spermatogenesis, and if spermatogenesis is disrupted by perturbing the expression of several miRNA genes, the effect is still not dramatic and fertility is usually not affected. Second, miRNAs seem to execute functions more often during spermatogonial proliferation than during meiotic and post-meiotic development. This is consistent with what was observed that miRNAs are much more abundant in type A spermatogonia than in spermatocytes and spermatids in which piRNAs dominate (Gan et al., 2011). Third, miRNA targeted genes are functionally diversified compared with those of piRNAs, which are mainly transcripts of retrotransposons. Although piRNAs also target mRNAs for their regulated degradation, how general this theme remains to be further investigated (Gou et al., 2014; Zhang et al., 2015).
The challenges in studying miRNA function in spermatogenesis is not unique to spermatogenesis per se. In contrast, as can be seen from above review, spermatogenic cells possess several unique advantages for elucidating miRNA functions and the underlying mechanisms. First, SSCs can be amplified to a large population by long term culture, which provides sufficient materials for genetic manipulation and mechanistic studies. Second, cultured SSCs initiate meiosis by RA treatment or co-culture with Sertoli cells, providing a preliminary in vitro model for studying genes involved in meiotic initiation (Wang et al., 2016). Third, genetically modified SSCs can be transplanted into testes for in vivo phenotypic evaluation. This semi-in vivo method in miRNA studies not only is cost-effective but also allows researchers to focus on studying functions of genes in spermatogenesis without concerns brought about by traditional gene knockout techniques.
Notably, there are examples showing some contrasting findings regarding the phenotypic differences for miRNAs between knockdown and knockout models. For example, using knockout mice, Yuan et al. showed that miR-34b/c and miR-449a/b/c were not required for fertilization, first cleavage division, or subsequent development (Yuan et al., 2015), whereas Liu et al. revealed that injection of miR-34c inhibitor into zygotes significantly attenuated the first cleavage division after fertilization (Liu et al., 2012). The differences may come from the redundant manners of miRNA effects. The long-lasting fine-tuning functions of miRNAs could be compensated by other miRNA members in the knockout models, but not in the knockdown models, where miRNA inhibitors are usually designed to block the seed sequences of miRNAs and possibly inhibit all members of the families.
The early gene knockout method was mainly based on gene targeting technique. Perturbation of the expression of miRNAs, which are either closely clustered or transcribed as polycistronic units, is a truly challenging task. With the development of new technologies such as the CRISPR-based ones, this demanding work may be alleviated. For example, seed sequences of miRNA genes in a cluster can be separately targeted by carefully designed sgRNAs (Ota et al., 2019). As target identification and characterization of miRNA-mRNA networks are keys to elucidate the functions and mechanisms of miRNAs, the recent development of the high-resolution Halo-Enhanced AGO2 pull-down technique is promising in the new era of in vivo miRNA explorations (Li et al., 2020).
Author contributions
JC and CH wrote the manuscript.
Funding
This work was supported by grants from the National Key R&D Program of China (2018YFE0201100), and the National Natural Science Foundation of China (31970795, 31771631).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Bao, J., Li, D., Wang, L., Wu, J., Hu, Y., Wang, Z., et al. (2012). MicroRNA-449 and microRNA-34b/c function redundantly in murine testes by targeting E2F transcription factor-retinoblastoma protein (E2F-pRb) pathway. J. Biol. Chem. 287 (26), 21686–21698. doi:10.1074/jbc.M111.328054
Bouhallier, F., Allioli, N., Lavial, F., Chalmel, F., Perrard, M. H., Durand, P., et al. (2010). Role of miR-34c microRNA in the late steps of spermatogenesis. RNA 16 (4), 720–731. doi:10.1261/rna.1963810
Chen, J., Cai, T., Zheng, C., Lin, X., Wang, G., Liao, S., et al. (2017). MicroRNA-202 maintains spermatogonial stem cells by inhibiting cell cycle regulators and RNA binding proteins. Nucleic Acids Res. 45 (7), 4142–4157. doi:10.1093/nar/gkw1287
Chen, J., Gao, C., Lin, X., Ning, Y., He, W., Zheng, C., et al. (2021). The microRNA miR-202 prevents precocious spermatogonial differentiation and meiotic initiation during mouse spermatogenesis. Development 148 (24), dev199799. doi:10.1242/dev.199799
Chen, J., Gao, C., Luo, M., Zheng, C., Lin, X., Ning, Y., et al. (2022). MicroRNA-202 safeguards meiotic progression by preventing premature SEPARASE-mediated REC8 cleavage. EMBO Rep. 23 (8), e54298. doi:10.15252/embr.202154298
Chen, Y., Li, X., Liao, H., Leung, X., He, J., Wang, X., et al. (2019). CFTR mutation compromises spermatogenesis by enhancing miR-15b maturation and suppressing its regulatory target CDC25A. Biol. reproduction 101, 50–62. doi:10.1093/biolre/ioz062
Choi, Y. J., Lin, C. P., Ho, J. J., He, X., Okada, N., Bu, P., et al. (2011). miR-34 miRNAs provide a barrier for somatic cell reprogramming. Nat. Cell Biol. 13 (11), 1353–1360. doi:10.1038/ncb2366
Comazzetto, S., Di Giacomo, M., Rasmussen, K. D., Much, C., Azzi, C., Perlas, E., et al. (2014). Oligoasthenoteratozoospermia and infertility in mice deficient for miR-34b/c and miR-449 loci. PLoS Genet. 10 (10), e1004597. doi:10.1371/journal.pgen.1004597
Concepcion, C. P., Han, Y. C., Mu, P., Bonetti, C., Yao, E., D'Andrea, A., et al. (2012). Intact p53-dependent responses in miR-34-deficient mice. PLoS Genet. 8 (7), e1002797. doi:10.1371/journal.pgen.1002797
Czech, B., and Hannon, G. J. (2011). Small RNA sorting: Matchmaking for argonautes. Nat. Rev. Genet. 12 (1), 19–31. doi:10.1038/nrg2916
Gan, H., Lin, X., Zhang, Z., Zhang, W., Liao, S., Wang, L., et al. (2011). piRNA profiling during specific stages of mouse spermatogenesis. RNA 17 (7), 1191–1203. doi:10.1261/rna.2648411
Gao, H., Wen, H., Cao, C., Dong, D., Yang, C., Xie, S., et al. (2019). Overexpression of MicroRNA-10a in germ cells causes male infertility by targeting Rad51 in mouse and human. Front. Physiol. 10, 765. doi:10.3389/fphys.2019.00765
Gou, L. T., Dai, P., Yang, J. H., Xue, Y. C., Hu, Y. P., Zhou, Y., et al. (2014). Pachytene piRNAs instruct massive mRNA elimination during late spermiogenesis. Cell Res. 24 (6), 680–700. doi:10.1038/cr.2014.41
Greenlee, A. R., Shiao, M. S., Snyder, E., Buaas, F. W., Gu, T., Stearns, T. M., et al. (2012). Deregulated sex chromosome gene expression with male germ cell-specific loss of Dicer1. PloS one 7 (10), e46359. doi:10.1371/journal.pone.0046359
Griswold, M. D. (2016). Spermatogenesis: The commitment to meiosis. Physiol. Rev. 96 (1), 1–17. doi:10.1152/physrev.00013.2015
Gupta, A., Mandal, K., Singh, P., Sarkar, R., and Majumdar, S. S. (2021). Declining levels of miR-382-3p at puberty trigger the onset of spermatogenesis. Mol. Ther. Nucleic Acids 26, 192–207. doi:10.1016/j.omtn.2021.07.001
Handel, M. A., and Schimenti, J. C. (2010). Genetics of mammalian meiosis: Regulation, dynamics and impact on fertility. Nat. Rev. Genet. 11 (2), 124–136. doi:10.1038/nrg2723
Hayashi, K., Chuva de Sousa Lopes, S. M., Kaneda, M., Tang, F., Hajkova, P., Lao, K., et al. (2008). MicroRNA biogenesis is required for mouse primordial germ cell development and spermatogenesis. PloS one 3 (3), e1738. doi:10.1371/journal.pone.0001738
He, Z., Jiang, J., Kokkinaki, M., Tang, L., Zeng, W., Gallicano, I., et al. (2013). MiRNA-20 and mirna-106a regulate spermatogonial stem cell renewal at the post-transcriptional level via targeting STAT3 and Ccnd1. Stem Cells 31 (10), 2205–2217. doi:10.1002/stem.1474
Hilz, S., Modzelewski, A. J., Cohen, P. E., and Grimson, A. (2016). The roles of microRNAs and siRNAs in mammalian spermatogenesis. Development 143 (17), 3061–3073. doi:10.1242/dev.136721
Hurtado, A., Palomino, R., Georg, I., Lao, M., Real, F. M., Carmona, F. D., et al. (2020). Deficiency of the onco-miRNA cluster, miR-106b∼25, causes oligozoospermia and the cooperative action of miR-106b∼25 and miR-17∼92 is required to maintain male fertility. Mol. Hum. Reprod. 26 (6), 389–401. doi:10.1093/molehr/gaaa027
Kim, G. J., Georg, I., Scherthan, H., Merkenschlager, M., Guillou, F., Scherer, G., et al. (2010). Dicer is required for Sertoli cell function and survival. Int. J. Dev. Biol. 54 (5), 867–875. doi:10.1387/ijdb.092874gk
Korhonen, H. M., Meikar, O., Yadav, R. P., Papaioannou, M. D., Romero, Y., Da Ros, M., et al. (2011). Dicer is required for haploid male germ cell differentiation in mice. PloS one 6 (9), e24821. doi:10.1371/journal.pone.0024821
Li, X., Pritykin, Y., Concepcion, C. P., Lu, Y., La Rocca, G., Zhang, M., et al. (2020). High-resolution in vivo identification of miRNA targets by halo-enhanced Ago2 pull-down. Mol. Cell 79 (1), 167–179. doi:10.1016/j.molcel.2020.05.009
Lin, X., Han, M., Cheng, L., Chen, J., Zhang, Z., Shen, T., et al. (2016). Expression dynamics, relationships, and transcriptional regulations of diverse transcripts in mouse spermatogenic cells. RNA Biol. 13 (10), 1011–1024. doi:10.1080/15476286.2016.1218588
Liu, W. M., Pang, R. T., Chiu, P. C., Wong, B. P., Lao, K., Lee, K. F., et al. (2012). Sperm-borne microRNA-34c is required for the first cleavage division in mouse. Proc. Natl. Acad. Sci. U. S. A. 109 (2), 490–494. doi:10.1073/pnas.1110368109
Maatouk, D. M., Loveland, K. L., McManus, M. T., Moore, K., and Harfe, B. D. (2008). Dicer1 is required for differentiation of the mouse male germline. Biol. reproduction 79 (4), 696–703. doi:10.1095/biolreprod.108.067827
Medeiros, L. A., Dennis, L. M., Gill, M. E., Houbaviy, H., Markoulaki, S., Fu, D., et al. (2011). Mir-290-295 deficiency in mice results in partially penetrant embryonic lethality and germ cell defects. Proc. Natl. Acad. Sci. U. S. A. 108 (34), 14163–14168. doi:10.1073/pnas.1111241108
Mendell, J. T. (2008). miRiad roles for the miR-17-92 cluster in development and disease. Cell 133 (2), 217–222. doi:10.1016/j.cell.2008.04.001
Modzelewski, A. J., Hilz, S., Crate, E. A., Schweidenback, C. T., Fogarty, E. A., Grenier, J. K., et al. (2015). Dgcr8 and Dicer are essential for sex chromosome integrity during meiosis in males. J. Cell Sci. 128 (12), 2314–2327. doi:10.1242/jcs.167148
Nie, K., Gomez, M., Landgraf, P., Garcia, J. F., Liu, Y., Tan, L. H., et al. (2008). MicroRNA-mediated down-regulation of PRDM1/blimp-1 in hodgkin/reed-sternberg cells: A potential pathogenetic lesion in hodgkin lymphomas. Am. J. Pathol. 173 (1), 242–252. doi:10.2353/ajpath.2008.080009
Niu, Z., Goodyear, S. M., Rao, S., Wu, X., Tobias, J. W., Avarbock, M. R., et al. (2011). MicroRNA-21 regulates the self-renewal of mouse spermatogonial stem cells. Proc. Natl. Acad. Sci. U. S. A. 108 (31), 12740–12745. doi:10.1073/pnas.1109987108
Ota, H., Ito-Matsuoka, Y., and Matsui, Y. (2019). Identification of the X-linked germ cell specific miRNAs (XmiRs) and their functions. PloS one 14 (2), e0211739. doi:10.1371/journal.pone.0211739
Panneerdoss, S., Viswanadhapalli, S., Abdelfattah, N., Onyeagucha, B. C., Timilsina, S., Mohammad, T. A., et al. (2017). Cross-talk between miR-471-5p and autophagy component proteins regulates LC3-associated phagocytosis (LAP) of apoptotic germ cells. Nat. Commun. 8 (1), 598. doi:10.1038/s41467-017-00590-9
Papaioannou, M. D., Pitetti, J-L., Ro, S., Park, C., Aubry, F., Schaad, O., et al. (2009). Sertoli cell Dicer is essential for spermatogenesis in mice. Dev. Biol. 326 (1), 250–259. doi:10.1016/j.ydbio.2008.11.011
Park, C. Y., Jeker, L. T., Carver-Moore, K., Oh, A., Liu, H. J., Cameron, R., et al. (2012). A resource for the conditional ablation of microRNAs in the mouse. Cell Rep. 1 (4), 385–391. doi:10.1016/j.celrep.2012.02.008
Romero, Y., Meikar, O., Papaioannou, M. D., Conne, B., Grey, C., Weier, M., et al. (2011). Dicer1 depletion in male germ cells leads to infertility due to cumulative meiotic and spermiogenic defects. PloS one 6 (10), e25241. doi:10.1371/journal.pone.0025241
Roush, S., and Slack, F. J. (2008). The let-7 family of microRNAs. Trends Cell Biol. 18 (10), 505–516. doi:10.1016/j.tcb.2008.07.007
Saitou, M., Kagiwada, S., and Kurimoto, K. (2012). Epigenetic reprogramming in mouse pre-implantation development and primordial germ cells. Development 139 (1), 15–31. doi:10.1242/dev.050849
Shinoda, G., De Soysa, T. Y., Seligson, M. T., Yabuuchi, A., Fujiwara, Y., Huang, P. Y., et al. (2013). Lin28a regulates germ cell pool size and fertility. Stem Cells 31 (5), 1001–1009. doi:10.1002/stem.1343
Song, R., Walentek, P., Sponer, N., Klimke, A., Lee, J. S., Dixon, G., et al. (2014). miR-34/449 miRNAs are required for motile ciliogenesis by repressing cp110. Nature 510 (7503), 115–120. doi:10.1038/nature13413
Soumillon, M., Necsulea, A., Weier, M., Brawand, D., Zhang, X., Gu, H., et al. (2013). Cellular source and mechanisms of high transcriptome complexity in the mammalian testis. Cell Rep. 3 (6), 2179–2190. doi:10.1016/j.celrep.2013.05.031
Tong, M. H., Mitchell, D. A., McGowan, S. D., Evanoff, R., and Griswold, M. D. (2012). Two miRNA clusters, Mir-17-92 (Mirc1) and Mir-106b-25 (Mirc3), are involved in the regulation of spermatogonial differentiation in mice. Biol. reproduction 86 (3), 72. doi:10.1095/biolreprod.111.096313
Wang, S., Wang, X., Ma, L., Lin, X., Zhang, D., Li, Z., et al. (2016). Retinoic acid is sufficient for the in vitro induction of mouse spermatocytes. Stem Cell Rep. 7 (1), 80–94. doi:10.1016/j.stemcr.2016.05.013
Wang, Z., Xie, Y., Wang, Y., Morris, D., Wang, S., Oliver, D., et al. (2020). X-linked miR-506 family miRNAs promote FMRP expression in mouse spermatogonia. EMBO Rep. 21 (1), e49024. doi:10.15252/embr.201949024
West, J. A., Viswanathan, S. R., Yabuuchi, A., Cunniff, K., Takeuchi, A., Park, I. H., et al. (2009). A role for Lin28 in primordial germ-cell development and germ-cell malignancy. Nature 460 (7257), 909–913. doi:10.1038/nature08210
Wu, J., Bao, J., Kim, M., Yuan, S., Tang, C., Zheng, H., et al. (2014). Two miRNA clusters, miR-34b/c and miR-449, are essential for normal brain development, motile ciliogenesis, and spermatogenesis. Proc. Natl. Acad. Sci. U. S. A. 111 (28), E2851–E2857. doi:10.1073/pnas.1407777111
Wu, Q., Song, R., Ortogero, N., Zheng, H., Evanoff, R., Small, C. L., et al. (2012). The RNase III enzyme DROSHA is essential for microRNA production and spermatogenesis. J. Biol. Chem. 287 (30), 25173–25190. doi:10.1074/jbc.M112.362053
Xia, B., Yan, Y., Baron, M., Wagner, F., Barkley, D., Chiodin, M., et al. (2020). Widespread transcriptional scanning in the testis modulates gene evolution rates. Cell 180 (2), 248–262. doi:10.1016/j.cell.2019.12.015
Xie, R., Lin, X., Du, T., Xu, K., Shen, H., Wei, F., et al. (2016). Targeted disruption of miR-17-92 impairs mouse spermatogenesis by activating mTOR signaling pathway. Med. Baltim. 95 (7), e2713. doi:10.1097/MD.0000000000002713
Yang, Q. E., Racicot, K. E., Kaucher, A. V., Oatley, M. J., and Oatley, J. M. (2013). MicroRNAs 221 and 222 regulate the undifferentiated state in mammalian male germ cells. Development 140 (2), 280–290. doi:10.1242/dev.087403
Yuan, S., Liu, Y., Peng, H., Tang, C., Hennig, G. W., Wang, Z., et al. (2019). Motile cilia of the male reproductive system require miR-34/miR-449 for development and function to generate luminal turbulence. Proc. Natl. Acad. Sci. U. S. A. 116 (9), 3584–3593. doi:10.1073/pnas.1817018116
Yuan, S., Tang, C., Zhang, Y., Wu, J., Bao, J., Zheng, H., et al. (2015). mir-34b/c and mir-449a/b/c are required for spermatogenesis, but not for the first cleavage division in mice. Biol. Open 4 (2), 212–223. doi:10.1242/bio.201410959
Zhang, L. L., Ma, J., Yang, B., Zhao, J., Yan, B. Y., Zhang, Y. Q., et al. (2018). Interference with lactate metabolism by mmu-miR-320-3p via negatively regulating GLUT3 signaling in mouse Sertoli cells. Cell death Dis. 9 (10), 964. doi:10.1038/s41419-018-0958-2
Zhang, P., Kang, J. Y., Gou, L. T., Wang, J. J., Xue, Y. C., Skogerboe, G., et al. (2015). MIWI and piRNA-mediated cleavage of messenger RNAs in mouse testes. Cell Res. 25 (2), 193–207. doi:10.1038/cr.2015.4
Zimmermann, C., Romero, Y., Warnefors, M., Bilican, A., Borel, C., Smith, L. B., et al. (2014). Germ cell-specific targeting of DICER or DGCR8 reveals a novel role for endo-siRNAs in the progression of mammalian spermatogenesis and male fertility. PloS one 9 (9), e107023. doi:10.1371/journal.pone.0107023
Keywords: miRNA, spermatogenesis, mammal, in vivo, male fertility
Citation: Chen J and Han C (2023) In vivo functions of miRNAs in mammalian spermatogenesis. Front. Cell Dev. Biol. 11:1154938. doi: 10.3389/fcell.2023.1154938
Received: 31 January 2023; Accepted: 21 April 2023;
Published: 05 May 2023.
Edited by:
Hengbin Wang, Virginia Commonwealth University, United StatesReviewed by:
Jianqiang Bao, University of Science and Technology of China, ChinaCopyright © 2023 Chen and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chunsheng Han, aGFuY3NAaW96LmFjLmNu
 Jian Chen
Jian Chen Chunsheng Han1,2,3,4*
Chunsheng Han1,2,3,4*