- 1Department of Neurobiology and Department of Neurosurgery of the Fourth Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, China
- 2Liangzhu Laboratory, MOE Frontier Science Center for Brain Science and Brain-machine Integration, State Key Laboratory of Brain-machine Intelligence, Zhejiang University, Hangzhou, China
- 3NHC and CAMS Key Laboratory of Medical Neurobiology, School of Medicine, Zhejiang University, Hangzhou, Zhejiang, China
Numerous taste receptors and related molecules have been identified in vertebrates and invertebrates. Otopetrin1 has recently been identified as mammalian sour taste receptor which is essential for acid sensation. However, whether other Otopetrin proteins are involved in PH-sensing remains unknown. In C. elegans, there are eight otopetrin homologous genes but their expression patterns and functions have not been reported so far. Through heterologous expression in HEK293T cells, we found that ceOTOP1a can be activated by acid in NMDG+ solution without conventional cations, which generated inward currents and can be blocked by zinc ions. Moreover, we found that Otopetrin channels are widely expressed in numerous tissues, especially in sensory neurons in the nematode. These results suggest that the biophysical characteristics of the Otopetrin channels in nematodes are generally conserved. However, a series of single gene mutations of otopetrins, which were constructed by CRISPR-Cas9 method, did not affect either calcium responses in ASH polymodal sensory neurons to acid stimulation or acid avoidance behaviors, suggesting that Otopetrin channels might have diverse functions among species. This study reveals that nematode Otopetrins are evolutionarily conserved acid-sensitive proton channels, and provides a framework for further revealing the function and mechanisms of Otopetrin channels in both invertebrates and vertebrates.
Introduction
Otopetrins are recently identified as proton-selective ion channels (Tu et al., 2018). There are numerous types of ion channels which selectively permeate the flow of sodium, potassium, calcium, or chloride ions (Gouaux and Mackinnon, 2005; Catterall et al., 2017), but the number of proton channels are relatively small in nature (Decoursey, 2003). The Matrix-2 (M2) proteins in the influenza A virus permeate protons into the interior of virions (Pinto et al., 1992). Prior to the discovery of Otopetrin channels, voltage-gated Hv1 was the only known proton channel in eukaryotes (DeCoursey, 1991; Ramsey et al., 2006; Sasaki et al., 2006), which functions in many physiological and pathophysiological processes such as regulating cellular pH in response to depolarization and maintaining pH in human neutrophils during phagocytosis (Morgan et al., 2009). Another recently discovered proton-selective channel in mammals, TMEM175, is proton-activated and expressed on lysosomal membranes mediating protonic fluid to maintain pH homeostasis (Hu et al., 2022).
Otopetrin1 (OTOP1) was initially uncovered by its role in the vestibular system of mice and zebrafish (Hurle et al., 2003; de Caprona et al., 2004; Hughes et al., 2004; Söllner et al., 2004). In the mammalian inner ear, otoliths are hard, calcium carbonate structures that sense gravity and linear acceleration (Hu et al., 2022). The zebrafish Otopetrin, zeOTOP1 is required for otolith formation (Hughes et al., 2004). As such, the protein was named as Otopetrin for its influence on the development of murine otolith. Mutant alleles, tlt and mlh, of otop1 cause abnormalities in the murine vestibular system (Hurle et al., 2003; Hughes et al., 2007), disrupting the interaction of otop1 with other proteins and membrane trafficking in the vestibular support cells (Kim et al., 2011). In addition, mutant mice exhibit abnormalities in forced swimming tests due to the degeneration of otolith (Ornitz et al., 1998). In sea urchins, Otopetrin functions as a proton channel to promote clearance of protons during calcium carbonate production in primary mesenchymal cells (Chang et al., 2021).
The otopetrin gene family is evolutionarily conserved from Caenorhabditis elegans to humans (Hurle et al., 2003; Hughes et al., 2008). There are three murine otop1 homologous genes in the Drosophila melanogaster genome (Hurle et al., 2003; Hughes et al., 2008) and eight otopetrin genes in C. elegans (Hurle et al., 2011). The murine Otopetrin family is widely expressed in various tissues (Tu et al., 2018), suggesting their involvement in physiological processes.
Heterologous expression of otopetrin cDNA from mouse, Drosophila, Zebrafish, and Xenopus in HEK293 cells induced a large inward proton current responding to extracellular acidification which can be blocked by Zn2+ (Tu et al., 2018; Chen et al., 2019; Saotome et al., 2019; Mi et al., 2021). In addition, Otopetrin channels mediate the flow of protons at resting potential to permeate the cytoplasm. The Otopetrin family and other types of ion channels are not structurally similar based on Cryo-EM structural analysis (Hughes et al., 2008; Chen et al., 2019; Saotome et al., 2019). Amino acid mutations in the predicted proton permeability sequences can significantly disturb the recorded proton channel currents (Chen et al., 2019; Saotome et al., 2019).
Over the past decades, numerous taste receptors and related molecules have been identified in vertebrates and invertebrates (Liman et al., 2014). Taste receptor cells respond to different taste stimuli which include sweet, salty, bitter, sour and umami tastes (Chandrashekar et al., 2006). Subsequent studies found that the receptor for salty taste is the sodium ion-permeable ENaC channel (Chandrashekar et al., 2010; Nomura et al., 2020). The discovery of sour taste receptors was relatively delayed until the Otopetrin family was identified as novel proton channels (Tu et al., 2018), then OTOP1 was confirmed to be the sour taste receptor after a series of experimentations (Liman and Kinnamon, 2021; Turner and Liman, 2022). Notably, in the study of sour taste there is a proton-sensitive conductance in taste receptor cells which is different from other taste stimuli (Chang et al., 2010). OTOP1 is specifically expressed in the sour taste receptor cells of mice, and its mutation inhibit the proton current generated by acid stimulation in sour taste receptor cells (Tu et al., 2018). Furthermore, Otop1 knockout mice have significantly reduced taste receptor neural responses to both hydrochloric acid and citric acid stimulation (Teng et al., 2019) indicating that OTOP1 is definitely the sour receptor in mice (Zhang et al., 2019). In addition, the Otopetrin-like protein OtopLA is also the sour taste receptor in Drosophila (Ganguly et al., 2021; Mi et al., 2021), suggesting that the Otopetrin1 may function as the universal sour taste receptors across species (Frank et al., 2022). However, it remains unknown whether other Otopetrin channels are required for acid sensation, or they are involved in avoidance behaviors in response to aversive acidic pH.
C. elegans can detect a variety of stimuli including chemical stimuli like acidic pH (Bargmann, 2006; Iliff and Xu, 2020). The ASH sensory neurons sense acidic stimulation and mediate acid avoidance behavior (Hilliard et al., 2004; Wang et al., 2016). The nematode consists of eight otopetrin genes. In this study, we explored the biophysical characteristics, the expression patterns and the functions of C. elegans Otopetrins.
Materials and methods
Worm cultivation
Worms were cultivated at 20°C on NGM plates seeded with OP50 E. Coli. The well-fed adult hermaphrodites were used in all experiments. Bristol N2 and osm-9(ky10) were obtained from the Caenorhabditis Genetics Center. KanIS8 [Psra-6::GCaMP5.0 + Psra-6::mCherry + Plin-44::GFP] stain was served as the control strain in recording of acid-evoked calcium responses and currents.
Molecular biology
Promoter fragments were designed and amplified from C. elegans N2 strain and then cloned into pBS77::mCherry vector using the In-Fusion PCR Cloning Kit (TaKaRa Inc.). The promoters used are as follow: otpl-1: 1,164 bp upstream of otpl-1 start codon; otpl-2: 2,506 bp upstream of otpl-2 start codon; otpl-3: −814 bp∼ + 132 bp; otpl-4: −614 bp−1 bp; otpl-5: −1,036 bp801 bp; otpl-6: −1,518 bp−1 bp; otpl-7: −2,502 bp−1 bp; and otpl-8: 2,698 bp−1 bp. Full-length Otop1a cDNA was amplified by PCR from template obtained from all-worm RNA reverse transcription and cloned into pEFGP-C3 vector.
Confocal imaging
Worms were immobilized using 200 mM sodium azide (NaN3). Images were captured by an inverted Olympus FV1000 laser scanning confocal microscope with filter setting for both GFP and mCherry.
Generation of otopetrin mutants by CRISPR-Cas9
Knockout otopetrin strains were obtained by CRISPR-Cas9 referring to Friedland’s method (Friedland et al., 2013). Briefly, exons located in front of the otopetrin genes were chosen as targets. Based on the selected sequences, specific guide RNAs (sgRNAs) were predicted and designed. Nematodes were then genotyped after microinjection of the plasmid to obtain the desired mutant strains of the target genes. CRISPR-Cas9 plasmids with eft-3 promoter were used to finally obtain whole worm knockout mutants. CRISPR-Cas9 knockout plasmids of dpy-10 were co-injected to produce chubby phenotypes as a pick-out marker and a system control. The otopetrin mutants selected in this study were all frameshift mutations. The sgRNAs for each otopetrin mutant and PCR primers used to genotype each otopetrin mutant are available in the Supplementary Material as TablesS1, S2, respectively.
Calcium imaging
Day 1 adult hermaphrodites were immersed in bath solution and glued on a glass coverslip. The bath solution contained 145 mM NaCl, 5 mM KCl, 1 mM MgCl2, 2 mM CaCl2, 20 mM glucose, 10 mM HEPES (pH 7.3 with NaOH). The acid stimulation solution contained 145 mM NaCl, 5 mM KCl, 1 mM MgCl2, 2 mM CaCl2, 20 mM glucose, 10 mM MES, which was adjusted to pH 2.5, 3 or 4 with HCl. GCaMP5.0 was used as the calcium indicator and images were acquired using an Olympus microscope (IX71) under a × 40 objective lens coupled with an Andor DL-604M EMCCD camera. Data were collected using the Micro-Manager software. GCaMP5.0 was excited by a ThorLabs blue light (460–480 nm) LED lamp. The average GCaMP5.0 signal in the first 10 s before stimulation was set as F0 and △F/F0 was calculated for each data point. All image stacks were analyzed using the Image-J software (Cheng et al., 2022).
Behavioral assays
A drop test was used to detect the avoidance behavior of nematodes. Day 1 adult hermaphrodites were transferred onto unseeded NGM plates. Using a micropipette, an acid stimulation solution (pH 3) droplet was delivered near the tail, flowed along the body and finally to the nose tip of nematode. Reversal behavior was defined as going backward for more than two body bends and less than 3 s responses. Worms presenting avoidance response was counted as 1, and no response was counted as 0. Each otopetrin mutant was tested on five different days, with approximately 30 worms tested each day. Wild-type and osm-9 mutants were employed as positive and negative controls. The avoidance response ratio of all the worms tested from each strain was calculated and statistically analyzed by One-way analysis of variance (ANOVA) and Dunnett’s test.
Transfection in HEK293T cells
Full-length ceOTOP1a cDNA was cloned into pEFGP-C3 vector. EGFP-ceOTOP1a was co-transfected with EGFP-empty (5:1) into HEK293T cells using Lipofectamine 2000 Transfection Reagent. Cells were detached by trypsin-EDTA 1 day before patch clamp recording and transferred onto coverslips coated with poly-lysine.
Patch-clamp electrophysiology
For HEK293T cells recording, the extracellular solution contained 160 mM NMDG, 2 mM CaCl2, 10 mM HEPES (pH 7.4 with HCl). Acidic solution contained 160 mM NMDG, 2 mM CaCl2, 10 mM MES (pH 4.5 with HCl), 1 mM ZnCl2 was added additionally for Zn2+-sensitivity test. The pipette solution contained 120 mM Cs-aspartate, 15 mM CsCl, 2 mM Mg-ATP, 5 mM EGTA, 2.4 mM CaCl2, 10 mM HEPES (pH 7.3, 290 mOsm).
For ASH in vivo recordings, the bath solution (NaCl, pH 7) contained 145 mM NaCl, 2.5 mM KCl, 5 mM CaCl2, 1 mM MgCl2, 20 mM glucose, 10 mM HEPES (pH 7.3, 325–335 mOsm). Extracellular solution (NMDG, pH 7) contained 145 mM NMDG, 2.5 mM KCl, 5 mM CaCl2, 1 mM MgCl2, 20 mM glucose, 10 mM HEPES (pH 7.3, 325–335 mOsm). Extracellular solution (NMDG, pH 3) contained 145 mM NMDG, 2.5 mM KCl, 5 mM CaCl2, 1 mM MgCl2, 20 mM glucose, 10 mM MES (pH 3, 325–335 mOsm), 1 mM ZnCl2 was added additionally for Zn2+-sensitivity test. The pipette solution contained 145 mM KCl, 5 mM MgCl2, 5 mM EGTA, 0.25 mM CaCl2, 10 mM HEPES, 10 mM glucose, 5 mM Na2ATP, 0.5 mM Na2GTP (pH 7.2, 315–325 mOsm).
Electrophysiological recordings were performed on an Olympus microscope (BX51WI) with an EPC-10 amplifier and Patchmaster software (HEKA). Day 2 adult worms were glued on the surface of sylgard-coated coverslips using cyanoacrylate-based glue. The soma of the ASH neurons were exposed with a dorsolateral incision made by a sharp glass pipettes. The membrane potential was clamped at −60 mV.
Statistical analysis
Data was analyzed using GraphPad Prism 5 and Igor Pro5.0 software (Wavemetrics). Error bars are presented as mean ± SEM.
Results
Expression patterns of nematode otopetrin gene family
We firstly sought to explore the expression patterns of the otopetrin genes, and constructed exogenous expression lines by microinjection. Using the promoters of otopetrins fused with mCherry, the expression patterns of all eight otopetrin genes in C. elegans were observed. The otopetrin genes otpl-1, otpl-2, otpl-4, and otpl-7 were expressed in sensory neurons in the head of the worms clearly co-localized with Psra-6::GFP, which marks chemosensory neurons ASH and ASI (Figure 1A). Four genes (otpl-1, otpl-2, otpl-4, and otpl-7) showed strong expression in ASI neurons, and otpl-2, otpl-4, and otpl-7 were also expressed in ASH neurons. The explicit expression of the otopetrin gene family in head sensory neurons suggests its underlying function might be related to acid sensation.
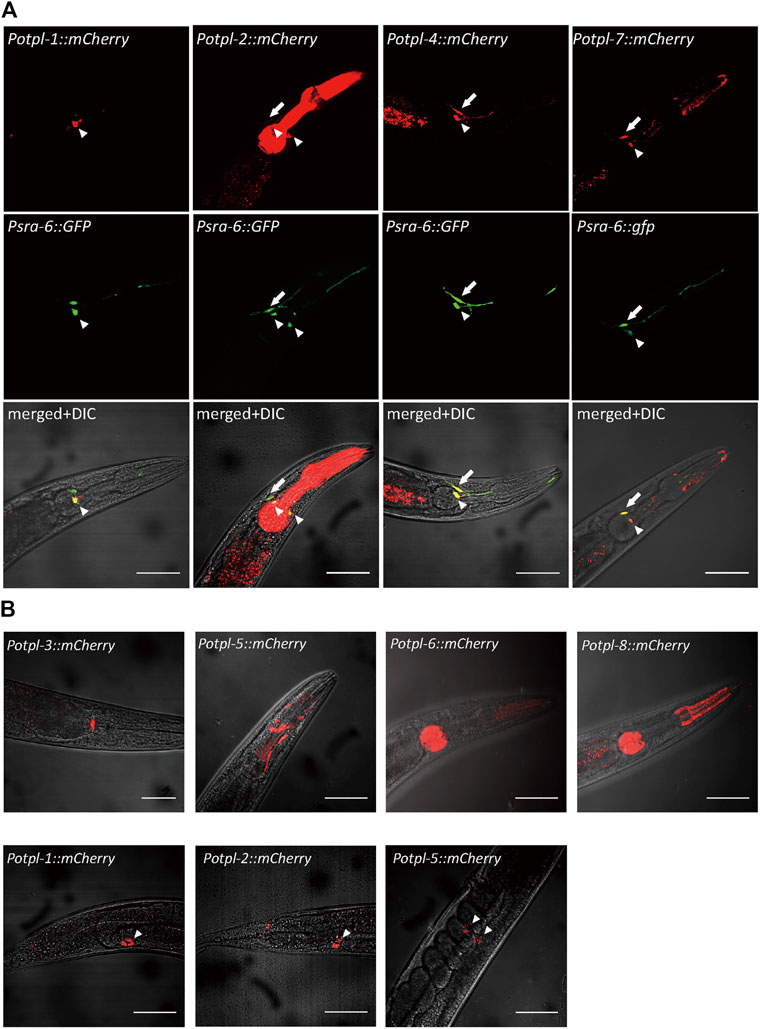
FIGURE 1. Expression patterns of C. elegans otopetrins. (A) Confocal images show four otopetrin gene co-located with head sensory neurons ASH and ASI neurons. Psra-6::GFP labeled ASH and ASI neurons with green fluorescent proteins. Arrowheads indicate ASI neurons, and arrows indicate ASH neurons. Scale bar, 50 μm. (B) The expression of otopetrin gene family in the nematode pharynx and head (upper) and in coelomocytes (lower). Scale bar, 50 μm.
In the pharynx, otpl-2, otpl-5, otpl-6, and otpl-8 were strongly expressed in different parts, and otpl-3 was observed at the junction of the pharynx and gut (Figure 1B). Further, otpl-5 was observed in the head and otpl-1, otpl-2, and otpl-5 were expressed in coelomocytes (Figure 1B). Several otopetrin genes were co-expressed in same cell, suggesting they may function cooperatively or redundantly.
ceOTOP1a mediates a zinc-sensitive acid-responsive current
Proton currents sensitive to zinc ions were recorded when the otopetrin gene family of mice, zebrafish, and chicken were heterologously expressed in HEK293 cells (Tu et al., 2018; Chen et al., 2019; Saotome et al., 2019; Mi et al., 2021). To explore the characteristics of C. elegans Otopetrin proteins, we heterologously expressed ceOTOP1a in HEK293T cell line. Full-length cDNA of worm OTOP1a was amplified and cloned into pEGFP-C3 vector. Cells marked by EGFP were chosen for patch clamp recording. Extracellular solutions were NMDG+-based, pH of the bath solution was 7.4 and pH of the acid stimulation solution was adjusted to 4.5. Large inward currents were recorded when the acid stimulation solution was given by a gravity perfusion system toward the cells (Figures 2A–C).
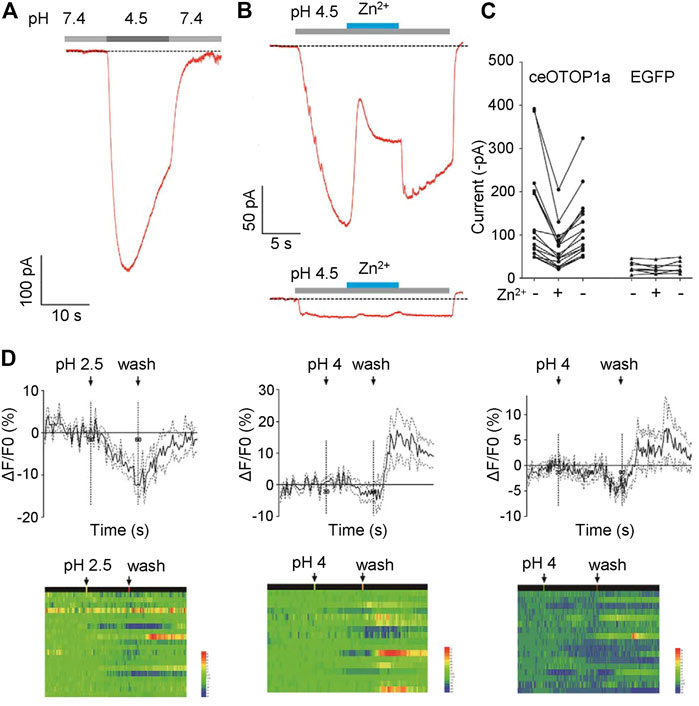
FIGURE 2. CeOTOP1a heterologous expression in HEK273T cells shows zinc ion-sensitive acid-responsive currents. (A) Acid-evoked currents were recorded by whole-cell patch clamp recording in HEK273T cells heterologously expressing nematode ceOTOP1a. The membrane potential was clamped at −80 mV. (B) Acid-evoked currents can be reversible by 1 mM Zn2+. The gravity perfusion system was used, switching channels to replace the solution. Membrane potential was clamped at −80 mV. Acid stimulation (grey), Zn2+ application (blue). (C) Statistics of the current amplitude. Membrane potential was clamped at −80 mV. (D) Acid stimulation does not induce significant calcium responses in ASI neurons.
In order to determine whether the acid-responsive current is sensitive to zinc ions, ZnCl2 was added to the acid-stimulated external solution, and the final concentration of zinc ion was 1 mM. We found that the acid activated current was reversibly inhibited by zinc ions (Figures 2B, C). These results show that ceOTOP1a is a zinc-sensitive proton channel, suggesting that the nematode otopetrin gene is evolutionarily conserved with other species.
Acid stimulation does not induce significant calcium response in ASI neurons
Given that otopetrin genes are expressed in ASI sensory neurons, we explore whether ASI neurons evoke calcium responses to acid stimulation. Stimuli were delivered in three different patterns; pH 2.5 for 30 s, pH 4 for 30 s, and pH 4 for 60 s. Under each stimulus, no significant calcium response was recorded during the time course (Figure 2D). Only a wash out response of about 10% was recorded, which was very weak compared to the response amplitude of ASI neurons to external stimuli such as Cu2+, suggesting that ASI may play a minor role in acid sensation.
Acid stimulation to otopetrin single-gene mutants does not affect the calcium response of ASH neurons
ASH neurons displayed robust calcium responses to acid stimulation when a pH 2.5 stimulus of 30 s duration was delivered (Figure 3A). The average amplitude of the response is about 50%. The pattern of the superimposed curve was generally that the amplitude increased after dosing, then reached a plateau, and the response amplitude decreased rapidly after elution (Figure 3A). The osm-9 mutant strain showed almost no obvious calcium response to acid stimulation, which was used as a negative control (Figure 3A).
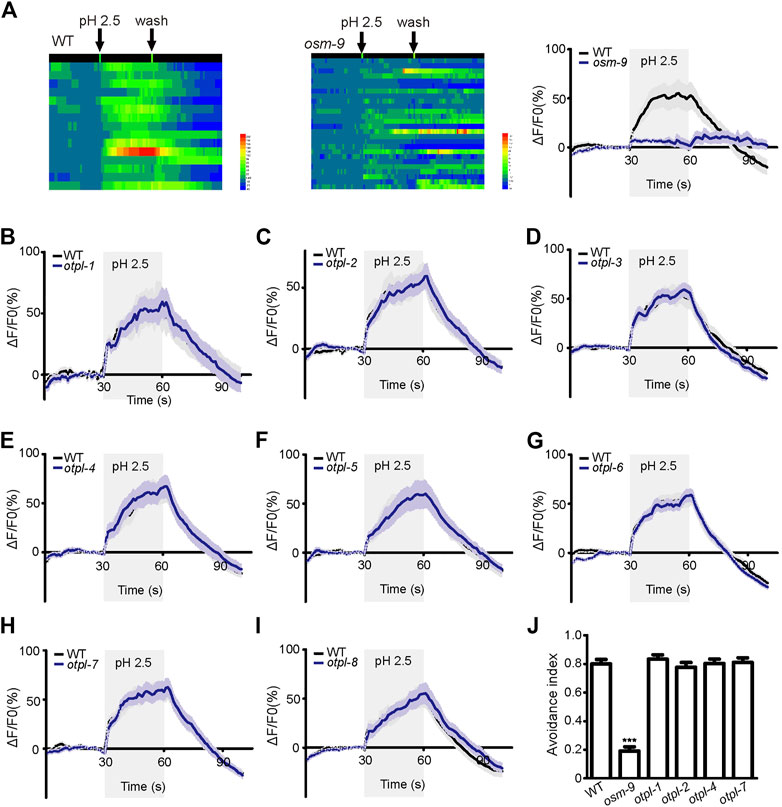
FIGURE 3. Otopetrin single gene mutations do not affect calcium responses of ASH neurons to acid stimulation and acid avoidance behavior in nematodes (A) ASH neurons were able to induce calcium responses to acid stimulation, osm-9 mutant was used as a negative control. (B–I) Calcium responses of ASH neurons to acid solution stimulation in all eight otopetrin single mutants, which were not significantly different from WT. (J) The mutant of four ASH neuron-expressing otopetrin genes did not affect acid avoidance behavior of nematodes. The number tested of each strain were 160 (WT), 173 (osm-9), 150 (otpl-1), 152 (otpl-2), 162 (otpl-4), and 142 (otpl-7). One-way ANOVA, Dunnett’s test, ***p <0.001.
We then used CRISPR-Cas9 system to edit otopetrin genes and obtained single-gene knockout nematode mutants. Otopetrin mutants finally selected are all frameshift mutations. All otopetrin knockout mutants did not show significant developmental differences from WT.
Each mutant of the eight genes of the otopetrin family did not significantly affect acid stimulation-evoked calcium increases in ASH (Figures 3B–I). Given that only strains of single-gene mutants were tested, it is unclear whether multiple otopetrins mediate the calcium response of ASH to acid stimulation simultaneously.
Otopetrin knockout mutants do not affect acid avoidance behavior in nematodes
Previous studies have showed that OTOP1 is the sour taste receptor in mice (Zhang et al., 2019) and OtopLA is the sour taste receptor in Drosophila (Ganguly et al., 2021; Mi et al., 2021). The functions of Otopetrins are highly consistent in various species, suggesting that Otopetrin1 may function as the universal sour taste receptor (Frank et al., 2022). In C. elegans, ASH neurons can sense acid stimulation and mediate avoidance response behaviors (Hilliard et al., 2004), which are impaired in osm-9 mutants (Wang et al., 2016). According to the expression patterns, four otopetrin genes (otpl-1, otpl-2, otpl-4, and otpl-7) were co-localized within ASH sensory neurons, then we tested whether mutants of these otopetrin genes affect acid avoidance behavior.
A pH 3 bath solution droplet was delivered using a micropipette near the tail, flowed along the body and finally to the nose tip of the worm. When the acidic solution is in contact with the nose tip, the worm reversed—avoidance behavior. Worm presenting avoidance response with 3 s was counted as 1, and no response was counted as 0. WT and osm-9 mutant strains were employed as positive and negative controls, respectively. The response of osm-9 to pH 3 stimulation was significantly attenuated, whereas each mutant of the four otopetrin genes exhibited normal avoidance behaviors to acid stimulation similar to that of WT (Figure 3J).
Single gene mutations of otopetrins do not influence the acid-evoked current of ASH neurons in nematodes
Since ASH neurons are sensitive to acidic stimulation, we then checked whether otopetrin mutants affect the acid-evoked currents of ASH recorded by in vivo patch clamp recordings. The pH 3-activated currents of ASH neurons were recorded, and 1 mM Zn2+ did not significantly inhibit the acid-sensing inward current (Figure 4A). Moreover, the current amplitudes from the otopetrin mutants (otpl-1, otpl-2, otpl-4, and otpl-7) were not significantly different from WT (Figure 4B). Single gene mutations of otopetrins did not affect the currents of ASH neurons in response to acidic stimulation, suggesting that multiple Otopetrins may mediate the acid-evoked currents of ASH simultaneously, or the acid-evoked currents of ASH are conducted by other molecules.
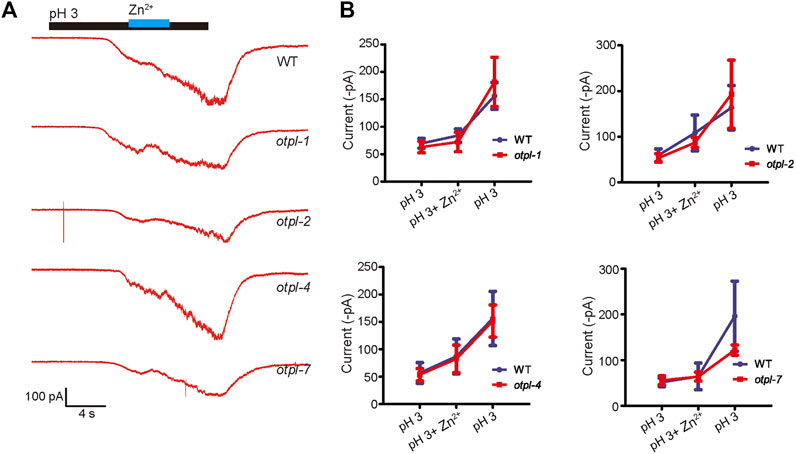
FIGURE 4. Single-gene otopetrin mutants do not affect in vivo acid-responsive currents of ASH neurons. (A) In vivo recording of ASH neurons in C. elegans, acid-sensing currents were not blocked by 1 mM Zn2+. The cation in solution is NMDG+. Gravity perfusion system was used to deliver acid stimulation, switching channels to replace the solution. Membrane potential was clamped at −60 mV. (B) Statistical values of the current. The membrane potential was clamped at −60 mV.
Discussion
Similar to other species, when ceOTOP1a is heterologously expressed in HEK293T cells, a sodium-independent acid-responsive current blocked by zinc ions can be recorded responding to extracellular acidification. The nematode ceOTOP1a exhibits common electrophysiological properties of the Otopetrin channel family, indicating that the C. elegans Otopetrins are functionally conserved as proton channels.
Expression patterns indicate that C. elegans otopetrins are located in sensory neurons and many tissues. Among them, there is exact expression in ASI and ASH neurons, suggesting the potential function of Otopetrin as the sensory receptor. ASI did not show significant calcium responses to strong acid stimulation. We mainly tested the role of otopetrin mutants in ASH sensory neurons. Otopetrin gene knockout does not affect acid avoidance behavior in nematodes, all mutants of the otopetrin family still had robust calcium response to acid stimulation, and the otopetrin mutation did not affects acid activated currents of ASH. These data showed that single-gene mutants of the otopetrin family did not affect the response of nematodes to acid stimulation. Given there are eight otopetrin genes in nematodes, it is still unclear whether the polygenic mutations of otopetrins affect the acid response of nematodes.
The biological function of otopetrin gene in C. elegans and other species still to be discovered. Because of the widespread tissue expression, and the characteristics of Otopetrin channel are conserved. The follow-up topics should focus on screening and discovering the physiological functions of various otopetrin genes on various tissue cells based on the nematode system, and exploring their roles and regulatory mechanisms. It will lay a valuable scientific foundation for the discovery of the physiological function and pharmacological mechanism of mammalian Otopetrin proteins.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author contributions
SL conducted experiments. SL, YC, and LK analyzed and interpreted results. SL, YC, and LK designed experiments. SL, UA-S, YC, and LK wrote the manuscript.
Funding
This work was supported by the Ministry of Science and Technology of China (STI2030-Major Projects, grant number 2021ZD020330); the National Foundation of Natural Science of China (grant numbers 32271017, 31771113, 31471023); and Zhejiang Provincial Natural Science Foundation (grant number LZ22C090001). We thank the Caenorhabditis Genetic Center (CGC), which is supported by the National Institutes of Health - Office of Research Infrastructure Programs (P40 OD010440), for strains.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2023.1133890/full#supplementary-material
References
Bargmann, C. I. (2006). Chemosensation in C. elegans. WormBook 123, 1–29. doi:10.1895/wormbook.1.123.1
Catterall, W. A., Wisedchaisri, G., and Zheng, N. (2017). The chemical basis for electrical signaling. Nat. Chem. Biol. 13, 455–463. doi:10.1038/nchembio.2353
Chandrashekar, J., Hoon, M. A., Ryba, N. J., and Zuker, C. S. (2006). The receptors and cells for mammalian taste. Nature 444, 288–294. doi:10.1038/nature05401
Chandrashekar, J., Kuhn, C., Oka, Y., Yarmolinsky, D. A., Hummler, E., Ryba, N. J. P., et al. (2010). The cells and peripheral representation of sodium taste in mice. Nature 464, 297–301. doi:10.1038/nature08783
Chang, R. B., Waters, H., and Liman, E. R. (2010). A proton current drives action potentials in genetically identified sour taste cells. Proc. Natl. Acad. Sci. U. S. A. 107, 22320–22325. doi:10.1073/pnas.1013664107
Chang, W. W., Matt, A. S., Schewe, M., Musinszki, M., Grussel, S., Brandenburg, J., et al. (2021). An otopetrin family proton channel promotes cellular acid efflux critical for biomineralization in a marine calcifier. Proc. Natl. Acad. Sci. U. S. A. 118, e2101378118. doi:10.1073/pnas.2101378118
Chen, Q., Zeng, W., and She, J. (2019). Structural and functional characterization of an otopetrin family proton channel. Elife 8, 46710. doi:10.7554/eLife.46710
Cheng, H., Al-Sheikh, U., Chen, D., Duan, D., and Kang, L. (2022). Protocol for glial Ca(2+) imaging in C. elegans following chemical, mechanical, or optogenetic stimulation. Star. Protoc. 3, 101169. doi:10.1016/j.xpro.2022.101169
de Caprona, M. D., Beisel, K. W., Nichols, D. H., and Fritzsch, B. (2004). Partial behavioral compensation is revealed in balance tasked mutant mice lacking otoconia. Brain Res. Bull. 64, 289–301. doi:10.1016/j.brainresbull.2004.08.004
DeCoursey, T. E. (1991). Hydrogen ion currents in rat alveolar epithelial cells. Biophys. J. 60, 1243–1253. doi:10.1016/s0006-3495(91)82158-0
Decoursey, T. E. (2003). Voltage-gated proton channels and other proton transfer pathways. Physiol. Rev. 83, 475–579. doi:10.1152/physrev.00028.2002
Frank, H. E. R., Amato, K., Trautwein, M., Maia, P., Liman, E. R., Nichols, L. M., et al. (2022). The evolution of sour taste. Proc. Biol. Sci. 289, 20211918. doi:10.1098/rspb.2021.1918
Friedland, A. E., Tzur, Y. B., Esvelt, K. M., Colaiacovo, M. P., Church, G. M., and Calarco, J. A. (2013). Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat. Methods 10, 741–743. doi:10.1038/nmeth.2532
Ganguly, A., Chandel, A., Turner, H., Wang, S., Liman, E. R., and Montell, C. (2021). Requirement for an Otopetrin-like protein for acid taste in Drosophila. Proc. Natl. Acad. Sci. U. S. A. 118, e2110641118. doi:10.1073/pnas.2110641118
Gouaux, E., and Mackinnon, R. (2005). Principles of selective ion transport in channels and pumps. Science 310, 1461–1465. doi:10.1126/science.1113666
Hilliard, M. A., Bergamasco, C., Arbucci, S., Plasterk, R. H. A., and Bazzicalupo, P. (2004). Worms taste bitter: ASH neurons, QUI-1, GPA-3 and ODR-3 mediate quinine avoidance in Caenorhabditis elegans. Embo J. 23, 1101–1111. doi:10.1038/sj.emboj.7600107
Hu, M., Li, P., Wang, C., Feng, X., Geng, Q., Chen, W., et al. (2022). Parkinson's disease-risk protein TMEM175 is a proton-activated proton channel in lysosomes. Cell 185, 2292–2308 e20. doi:10.1016/j.cell.2022.05.021
Hughes, I., Binkley, J., Hurle, B., Green, E. D., and Sidow, A. (2008). Identification of the Otopetrin Domain, a conserved domain in vertebrate otopetrins and invertebrate otopetrin-like family members. BMC Evol. Biol. 8, 41. doi:10.1186/1471-2148-8-41
Hughes, I., Blasiole, B., Huss, D., Warchol, M. E., Rath, N. P., Hurle, B., et al. (2004). Otopetrin 1 is required for otolith formation in the zebrafish Danio rerio. Dev. Biol. 276, 391–402. doi:10.1016/j.ydbio.2004.09.001
Hughes, I., Saito, M., Schlesinger, P. H., and Ornitz, D. M. (2007). Otopetrin 1 activation by purinergic nucleotides regulates intracellular calcium. Proc. Natl. Acad. Sci. U. S. A. 104, 12023–12028. doi:10.1073/pnas.0705182104
Hurle, B., Ignatova, E., Massironi, S. M., Mashimo, T., Rios, X., Thalmann, I., et al. (2003). Non-syndromic vestibular disorder with otoconial agenesis in tilted/mergulhador mice caused by mutations in otopetrin 1. Hum. Mol. Genet. 12, 777–789. doi:10.1093/hmg/ddg087
Hurle, B., Marques-Bonet, T., Antonacci, F., Hughes, I., and Ryan, J. F. (2011). Lineage-specific evolution of the vertebrate Otopetrin gene family revealed by comparative genomic analyses. BMC Evol. Biol. 11, 23. doi:10.1186/1471-2148-11-23
Iliff, A. J., and Xu, X. Z. S. (2020). C. elegans: A sensible model for sensory biology. J. Neurogenet. 34, 347–350. doi:10.1080/01677063.2020.1823386
Kim, E., Hyrc, K. L., Speck, J., Salles, F. T., Lundberg, Y. W., Goldberg, M. P., et al. (2011). Missense mutations in Otopetrin 1 affect subcellular localization and inhibition of purinergic signaling in vestibular supporting cells. Mol. Cell Neurosci. 46, 655–661. doi:10.1016/j.mcn.2011.01.005
Liman, E. R., and Kinnamon, S. C. (2021). Sour taste: Receptors, cells and circuits. Curr. Opin. Physiol. 20, 8–15. doi:10.1016/j.cophys.2020.12.006
Liman, E. R., Zhang, Y. V., and Montell, C. (2014). Peripheral coding of taste. Neuron 81, 984–1000. doi:10.1016/j.neuron.2014.02.022
Mi, T., Mack, J. O., Lee, C. M., and Zhang, Y. V. (2021). Molecular and cellular basis of acid taste sensation in Drosophila. Nat. Commun. 12, 3730. doi:10.1038/s41467-021-23490-5
Morgan, D., Capasso, M., Musset, B., Cherny, V. V., Rios, E., Dyer, M. J. S., et al. (2009). Voltage-gated proton channels maintain pH in human neutrophils during phagocytosis. Proc. Natl. Acad. Sci. U. S. A. 106, 18022–18027. doi:10.1073/pnas.0905565106
Nomura, K., Nakanishi, M., Ishidate, F., Iwata, K., and Taruno, A. (2020). All-Electrical Ca(2+)-independent signal transduction mediates attractive sodium taste in taste buds. Neuron 106, 816–829. doi:10.1016/j.neuron.2020.03.006
Ornitz, D. M., Bohne, B. A., Thalmann, I., Harding, G. W., and Thalmann, R. (1998). Otoconial agenesis in tilted mutant mice. Hear Res. 122, 60–70. doi:10.1016/s0378-5955(98)00080-x
Pinto, L. H., Holsinger, L. J., and Lamb, R. A. (1992). Influenza virus M2 protein has ion channel activity. Cell 69, 517–528. doi:10.1016/0092-8674(92)90452-i
Ramsey, I. S., Moran, M. M., Chong, J. A., and Clapham, D. E. (2006). A voltage-gated proton-selective channel lacking the pore domain. Nature 440, 1213–1216. doi:10.1038/nature04700
Saotome, K., Teng, B., Tsui, C. C. A., Lee, W. H., Tu, Y. H., Kaplan, J. P., et al. (2019). Structures of the otopetrin proton channels Otop1 and Otop3. Nat. Struct. Mol. Biol. 26, 518–525. doi:10.1038/s41594-019-0235-9
Sasaki, M., Takagi, M., and Okamura, Y. (2006). A voltage sensor-domain protein is a voltage-gated proton channel. Science 312, 589–592. doi:10.1126/science.1122352
Söllner, C., Schwarz, H., Geisler, R., and Nicolson, T. (2004). Mutated otopetrin 1 affects the Genesis of otoliths and the localization of Starmaker in zebrafish. Dev. Genes Evol. 214, 582–590. doi:10.1007/s00427-004-0440-2
Teng, B., Wilson, C. E., Tu, Y. H., Joshi, N. R., Kinnamon, S. C., and Liman, E. R. (2019). Cellular and neural responses to sour stimuli require the proton channel Otop1. Curr. Biol. 29, 3647–3656. doi:10.1016/j.cub.2019.08.077
Tu, Y. H., Cooper, A. J., Teng, B., Chang, R. B., Artiga, D. J., Turner, H. N., et al. (2018). An evolutionarily conserved gene family encodes proton-selective ion channels. Science 359, 1047–1050. doi:10.1126/science.aao3264
Turner, H. N., and Liman, E. R. (2022). The cellular and molecular basis of sour taste. Annu. Rev. Physiol. 84, 41–58. doi:10.1146/annurev-physiol-060121-041637
Wang, X., Li, G., Liu, J., and Xu, X. Z. S. (2016). TMC-1 mediates alkaline sensation in C. elegans through nociceptive neurons. Neuron 91, 146–154. doi:10.1016/j.neuron.2016.05.023
Keywords: acid sensation, Caenorhabditis elegans, sensory receptors, otopetrin channels, calcium imaging
Citation: Li S, Al-Sheikh U, Chen Y and Kang L (2023) Nematode homologs of the sour taste receptor Otopetrin1 are evolutionarily conserved acid-sensitive proton channels. Front. Cell Dev. Biol. 11:1133890. doi: 10.3389/fcell.2023.1133890
Received: 29 December 2022; Accepted: 16 January 2023;
Published: 26 January 2023.
Edited by:
Zhitao Hu, The University of Queensland, AustraliaCopyright © 2023 Li, Al-Sheikh, Chen and Kang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yili Chen, MzE5NDA4NkB6anUuZWR1LmNu; Lijun Kang, a2FuZ2xpanVuQHpqdS5lZHUuY24=
 Shitian Li1,2,3
Shitian Li1,2,3 Umar Al-Sheikh
Umar Al-Sheikh Lijun Kang
Lijun Kang