
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell Dev. Biol., 16 February 2023
Sec. Molecular and Cellular Pathology
Volume 11 - 2023 | https://doi.org/10.3389/fcell.2023.1113675
 Manxiu Ma1
Manxiu Ma1 Alyssa A. Brunal1,2
Alyssa A. Brunal1,2 Kareem C. Clark1
Kareem C. Clark1 Carleigh Studtmann1,2
Carleigh Studtmann1,2 Katelyn Stebbins1,2,3
Katelyn Stebbins1,2,3 Shin-ichi Higashijima4
Shin-ichi Higashijima4 Y. Albert Pan1,5,6*
Y. Albert Pan1,5,6*The corticotropin-releasing hormone (CRH)-expressing neurons in the hypothalamus are critical regulators of the neuroendocrine stress response pathway, known as the hypothalamic-pituitary-adrenal (HPA) axis. As developmental vulnerabilities of CRH neurons contribute to stress-associated neurological and behavioral dysfunctions, it is critical to identify the mechanisms underlying normal and abnormal CRH neuron development. Using zebrafish, we identified Down syndrome cell adhesion molecule like-1 (dscaml1) as an integral mediator of CRH neuron development and necessary for establishing normal stress axis function. In dscaml1 mutant animals, hypothalamic CRH neurons had higher crhb (the CRH homolog in zebrafish) expression, increased cell number, and reduced cell death compared to wild-type controls. Physiologically, dscaml1 mutant animals had higher baseline stress hormone (cortisol) levels and attenuated responses to acute stressors. Together, these findings identify dscaml1 as an essential factor for stress axis development and suggest that HPA axis dysregulation may contribute to the etiology of human DSCAML1-linked neuropsychiatric disorders.
The hypothalamic corticotropin-releasing hormone (CRH)-expressing neurons are the central regulators of the neuroendocrine stress response pathway, known as the hypothalamic-pituitary-adrenal (HPA) axis in mammals or the hypothalamic-pituitary-interrenal (HPI) axis in fish (Denver, 2009). Upon exposure to environmental disturbances (i.e., stressors), stress-related neural inputs converge on hypothalamic CRH neurons to activate a hormonal cascade that ultimately leads to the release of glucocorticoids, which broadly affects cognitive, affective, metabolic, and immune functions (Spencer and Deak, 2017; McEwen and Akil, 2020).
The development of CRH neurons has profound effects on the function of the HPA and HPI axis (collectively referred to as the stress axis). Developmental perturbations of CRH neurons, particularly in early-life periods, lead to long-term changes in CRH neuron function (Regev and Baram, 2014). Additionally, rodent models demonstrate that dysregulation of CRH neurons increases anxiety- and depressive-like phenotypes (Keen-Rhinehart et al., 2009; Kolber et al., 2010). These studies underscore the need to identify the genes and molecules mediating CRH-neuron development and determine how developmental perturbations affect stress axis function.
During early development, hypothalamic CRH neurons are generated from progenitors in the ventral diencephalon (Alvarez-Bolado, 2019; Nagpal et al., 2019; Placzek et al., 2020). Secreted factors including FGF10, SHH, BMPs, and Nodal first define the anterior-dorsal hypothalamic domain; within this domain, CRH neurons are progressively specified by a combination of key transcription factors, including Fezf2, Otp, Sim1, Arnt2, and Brn2. Once specified, CRH neurons undergo further differentiation, such as neuronal morphogenesis, synaptogenesis, epigenetic programming, and cell death, to establish a functional stress-responsive neural circuit. These later stages of neuronal differentiation are shaped by intercellular interactions mediated by membrane-localized cell-adhesion molecules (Moreland and Poulain, 2022). The specific cell-adhesion molecules that mediate developmental signaling in CRH neurons, however, are still unknown.
To address this knowledge gap, we utilized zebrafish (Danio rerio) as the model. The structure and function of the mammalian HPA axis and the teleostean HPI axis are highly conserved (Wendelaar Bonga, 1997; Lohr and Hammerschmidt, 2011). In mammals, the CRH neurons that are principally involved in HPA-axis activation are in the hypothalamic paraventricular nucleus (PVN). In teleosts (ray-finned fish), the neuroendocrine preoptic area (NPO) is ontogenically equivalent to the PVN, and CRH neurons within the NPO perform similar roles as their mammalian counterparts (Herget and Ryu, 2015). CRH neurons in other hypothalamic regions in the zebrafish (e.g., intermediate hypothalamus) may also regulate the HPI axis (Wagle et al., 2022). A unique advantage of the zebrafish system is its rapid and external development. The development of the HPI axis begins between 1 and 2 days post fertilization (dpf), and stress-induced cortisol signaling and behaviors can be observed at 4–5 dpf (Alsop and Vijayan, 2008; Alderman and Bernier, 2009; Bai et al., 2016). The rapid development and translucency of zebrafish allow direct microscopic observation of CRH neuron development in intact, developing animals.
In this study, we investigated the roles of a conserved neuronal signaling molecule—Down syndrome cell adhesion molecule-like 1 (zebrafish gene: dscaml1; mouse gene: Dscaml1; Human gene: DSCAML1; Protein: DSCAML1). DSCAML1 is one of two DSCAM family members in vertebrates, the other being DSCAM (Garrett et al., 2012). Unlike invertebrate DSCAMs, vertebrate DSCAMs are not significantly alternatively spliced (Sanes and Zipursky, 2020). In the mammalian retina, DSCAML1 prevents excessive aggregation between cells and promotes developmental cell death (Fuerst et al., 2009; Garrett et al., 2016). DSCAML1 also acts to refine synaptic specificity and synapse number (Yamagata and Sanes, 2008; Sachse et al., 2019). In humans, rare variants in DSCAML1 are associated with several neurodevelopmental disorders, including autism spectrum disorder, cortical abnormality, and epilepsy (Iossifov et al., 2014; Karaca et al., 2015; Hayase et al., 2020; Ogata et al., 2021). Genetic and epigenetic studies also implicate DSCAML1 in the stress response to violent experiences (Caramillo et al., 2015; Saadatmand et al., 2021). However, the relationship between DSCAML1 and the stress axis remains unknown.
Using zebrafish, we previously explored how DSCAML1 affects neural pathways and systemic functions (Ma et al., 2020a; Ma et al., 2020b). We found that dscaml1 deficiency resulted in various physiological and behavioral deficits, including darker pigmentation, slower light adaptation, and slower eye movements (saccades) (Ma et al., 2020b). Interestingly, darker pigmentation and slower light adaptation can also be caused by abnormal glucocorticoid receptor signaling, as seen in the zebrafish glucocorticoid receptor (gr) mutants, suggesting that the stress axis may be dysfunctional in dscaml1 mutants (Griffiths et al., 2012; Muto et al., 2013).
Here, we report that dscaml1 deficiency in zebrafish perturbs hypothalamic CRH neuron development and impairs the normal function of the HPI axis. These findings show that DSCAML1 is necessary for stress axis development and raise the possibility that stress dysfunction contributes to human DSCAML1-linked disorders.
To gain an unbiased view of the molecular changes resulting from dscaml1 deficiency, we compared the transcriptomic profiles between dscaml1 homozygous mutant (dscaml1−/−) and control (wild type) siblings using RNA sequencing (RNA-seq). cDNA from whole 3.5–4 days post-fertilization (dpf) dscaml1−/− and control larvae were sequenced using an Illumina next-generation sequencer. Using a threshold of at least two-fold change and an adjusted p-value of less than 0.01, we identified 25 upregulated and 79 downregulated genes (Figure 1A, Supplementary Datasheet S1).
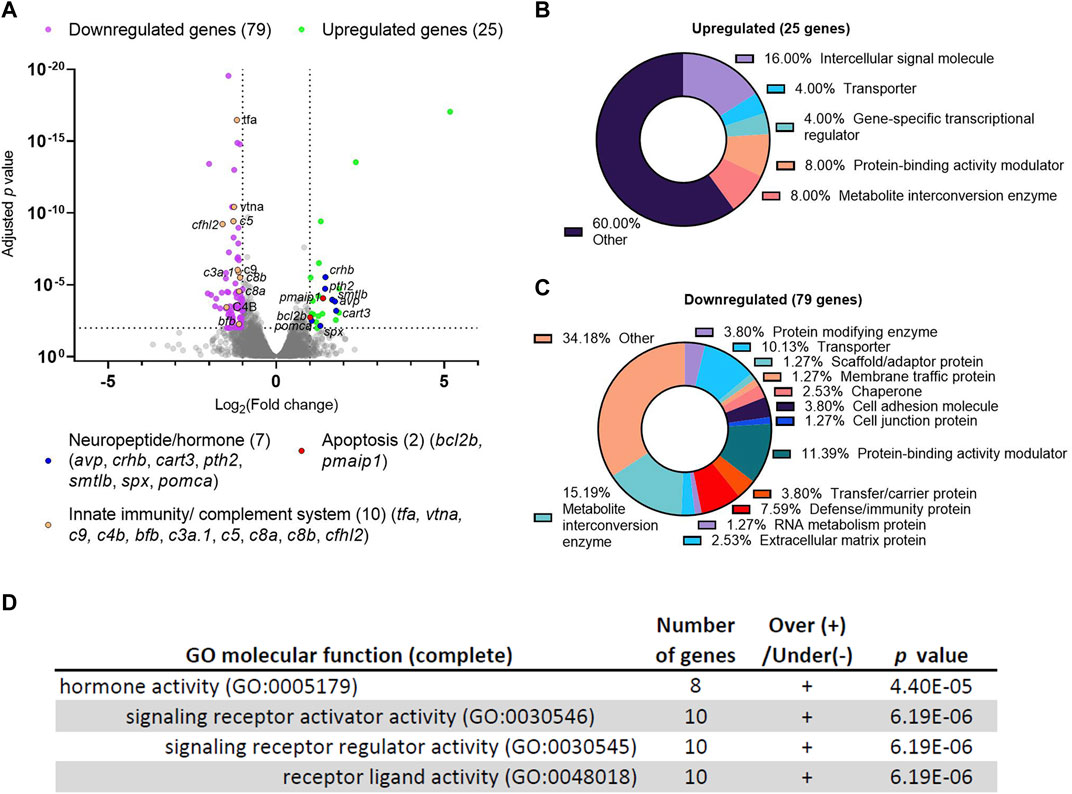
FIGURE 1. Differential gene expression analysis of dscaml1 deficient zebrafish. (A) Volcano plot of relative gene expression in dscaml1−/− versus control animals. Each dot represents an individual gene, with colored dots representing gene groups as indicated on the graph. The dotted lines show the significance level (adjusted p < 0.01) and fold change (increase or decrease by two-fold or more) thresholds (B, C) Protein class categorization analysis for upregulated (B) and downregulated (C) genes. (D) Table of significantly enriched (p < 0.05) GO terms for molecular function.
Among the 25 upregulated genes in the dscaml1−/− animals, seven were secreted neuropeptides/hormones expressed in the hypothalamus or pituitary: corticotropin-releasing hormone b (crhb), parathyroid hormone 2 (pth2), somatolactin beta (smtlb), cocaine- and amphetamine-regulated transcript 3 (cart3), proopiomelanocortin a (pomca), arginine vasopressin (avp), and spexin hormone (spx). Among them, three (crhb, avp, pomca) are core regulators of the HPI axis (Alsop and Vijayan, 2009; Lohr and Hammerschmidt, 2011). crhb encodes the zebrafish CRH; avp encodes the neuropeptide AVP that controls osmolarity, blood pressure, and synergizes with CRH to promote cortisol release; pomca encodes the adrenocorticotropic hormone (ACTH), the primary pituitary hormone that triggers glucocorticoid release. Additionally, two genes involved in apoptosis are upregulated: BCL2 apoptosis regulator b (bcl2b) and phorbol-12-myristate-13-acetate-induced protein 1 (pmaip1/noxa). Protein class categorization analysis with PANTHER revealed “intercellular signal molecule” as the largest class (4 genes, 16%) (Figure 1B) (Mi et al., 2021).
The 79 genes downregulated in the dscaml1−/− animals are more diverse in function compared to the upregulated genes. Protein class categorization analysis with PANTHER identified the largest protein classes as metabolite interconversion enzymes (12 genes, 15.19%), protein-binding activity modulators (9 genes, 11.39%), transporters (8 genes, 10.13%), and defense/immunity protein (6 genes, 7.59%) (Figure 1C). We noted that 30 of the 79 (38%) downregulated genes are highly expressed in the liver, and 10 of these genes are involved in innate immunity and the complement cascade (Figure 1A, Supplementary Datasheet S2). These results suggest that liver function and innate immunity may be suppressed in the dscaml1 mutants.
To identify the signaling pathways affected by dscaml1 deficiency, we analyzed all differentially expressed genes (p < 0.01, 210 mapped genes) using the statistical enrichment test (PANTHER Classification System, version 17.0) (Mi et al., 2021). All significantly enriched (FDR < 0.05) Gene Ontogeny (GO) terms for molecular function relate to the parent GO term hormone activity (Figure 1D). The PANTHER pathway analysis also identified another significant stress-modulating neuropeptide, adenylate cyclase activating polypeptide 1b (adcyap1b, also known as PACAP) that is significantly upregulated (0.66 fold change, adjusted p < 0.0001) (Stroth et al., 2011).
Together, our transcriptomic analyses indicate that dscaml1 deficiency results in the upregulation of neuropeptide/hormonal signaling and the downregulation of liver and innate immune function. Suppression of liver and immune function is a hallmark of stress axis activation, which is consistent with the overexpression of the principal neuropeptides involved in the stress axis (crhb, avp, pomca, and adcyap1b).
To further investigate whether the stress axis is perturbed in dscaml1 mutants, we first examined the development of CRH neurons in the NPO (CRHNPO neurons) (Figure 2A), which are localized in the dorsal preoptic area and characterized by their expression of crhb (Herget et al., 2014; Vom Berg-Maurer et al., 2016). We focused on three developmental stages (2, 3, and 5 dpf) that span the period between the first appearance of crhb + neurons in the NPO (2 dpf) and the onset of stress axis responsivity (4–5 dpf) (Chandrasekar et al., 2007; Alderman and Bernier, 2009; Clark et al., 2011). Sibling controls were used for all comparisons.
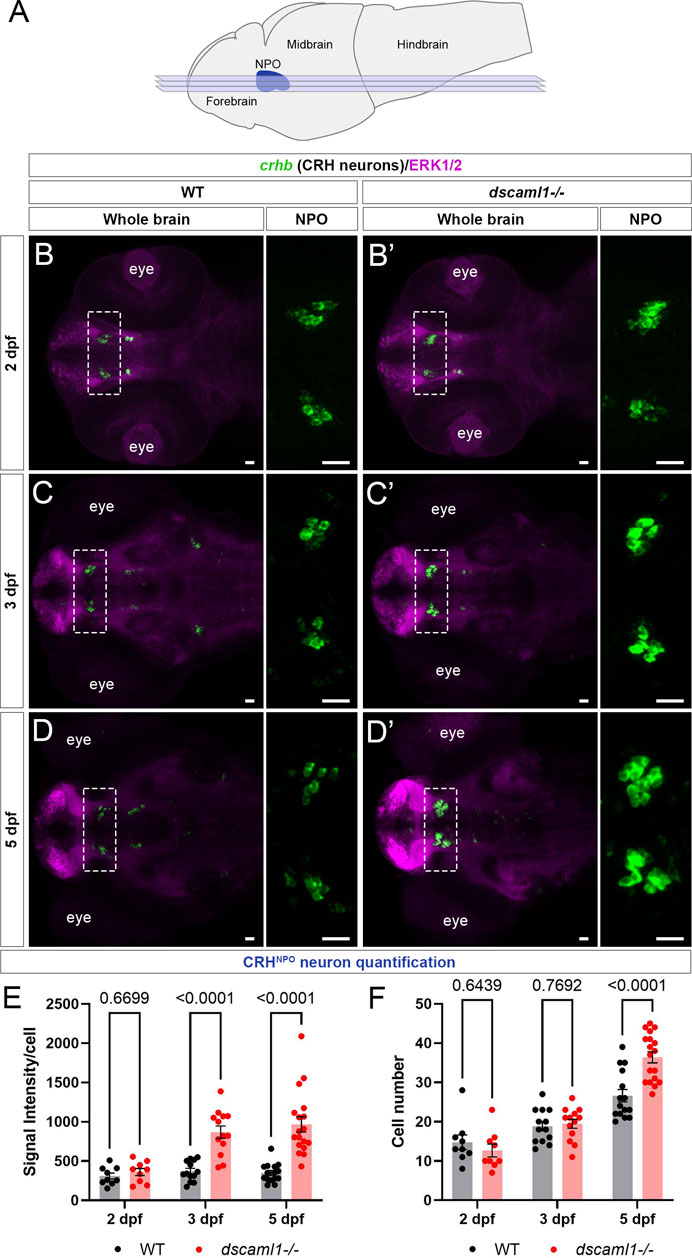
FIGURE 2. dscaml1 deficiency alters the development of CRHNPO neurons. (A) Illustration of the larval zebrafish brain (lateral view) with rostral to the left. The NPO is highlighted in blue. Imaging planes used for CRHNPO neuron analysis are shown (light blue planes) (B-D') Developmental trajectory of CRHNPO neurons, labeled by crhb FISH (green) and co-stained with anti-ERK1/2 (magenta). At each developmental stage, a representative confocal sub-stack containing the NPO is shown, with the NPO (boxed region) enlarged and shown in the right panel without the ERK1/2 co-stain. Wild-type (WT) animals are shown in panels B, C, and (D) dscaml1−/− animals are shown in panels B', C', and D' (E–F) Quantification of the signal intensity per cell (E) and cell number (F). Multiple-comparison corrected p values are as shown. WT: n = 9 (2 dpf), 13 (3 dpf), 15 (5 dpf). dscaml1−/−: n = 9 (2 dpf), 13 (3 dpf), 18 (5 dpf). Scale bars are 20 μm. Mean, standard error, and corrected p values are shown.
Using fluorescent in situ hybridization (FISH), we found that crhb expression pattern was initially similar between dscaml1−/− and wild-type (WT) animals at 2 dpf (Figure 2B, B'). At 3 dpf, we began to see higher crhb expression in the NPO of dscaml1−/− animals (Figure 2C, C'), and crhb expression in the NPO remains higher in dscaml1−/− animals at 5 dpf (Figure 2D, D”). Quantification of crhb FISH signal intensity among CRHNPO neurons showed that crhb expression is higher in dscaml1−/− animals, compared to WT animals (Figure 2E). There was a significant difference in signal intensity per cell by developmental stage and genotype, and a significant interaction between stage and genotype (two-way ANOVA, Supplementary Table SI). Pair-wise comparisons with Holm-Sidak correction found a significant increase at 3 and 5 dpf but not at 2 dpf (Figure 2E).
In addition to changes in crhb expression levels, dscaml1 deficiency also increased the number of crhb-expressing CRHNPO neurons. In WT animals, the number of CRHNPO neurons increased over time, from 14.78 cells (2 dpf) to 18.85 cells (3 dpf) to 26.60 cells (5 dpf) per animal (Figures 2B–D, F). In dscaml1−/− animals, the number of CRHNPO neurons increased at a higher rate, from 12.67 cells (2 dpf) to 19.46 cells (3 dpf) to 36.39 cells (5 dpf) per animal (Figure 2B', C', D', F). There was a significant difference in cell number by developmental stage and genotype, and there was a significant interaction between stage and genotype (Two-way ANOVA, Supplementary Table SI). Pair-wise comparisons with Holm-Sidak correction found a significant increase in cell number between dscaml1−/− and WT animals at 5 dpf but not at 2 and 3 dpf (Figure 2F, adjusted p values as shown).
Overall, we found significant increases in crhb expression and cell number in CRHNPO neurons in dscaml1−/− mutants as compared to WT. These phenotypes were not due to the visual deficits in dscaml1−/− animals (Ma et al., 2020b), as similar phenotypes were observed in animals raised in the dark (Supplementary Figure S1). Together, these findings show that dscaml1 is essential for the normal developmental trajectory of CRHNPO neurons.
To test whether other hypothalamic CRH neurons are affected similarly to CRHNPO neurons, we examined two additional crhb-expressing hypothalamic regions. In zebrafish, the intermediate hypothalamus (IH) CRH neurons (CRHIH neurons) regulate stress response and the perception of the emotional valence of light (Figure 3A) (Wagle et al., 2022). In the IH at 5 dpf, we did not observe any significant difference in crhb expression levels (FISH intensity) between dscaml1−/− and WT animals (Figures 3B–D). However, the number of CRHIH neurons was significantly higher in dscaml1−/− animals (unpaired t-test, Figure 3E).
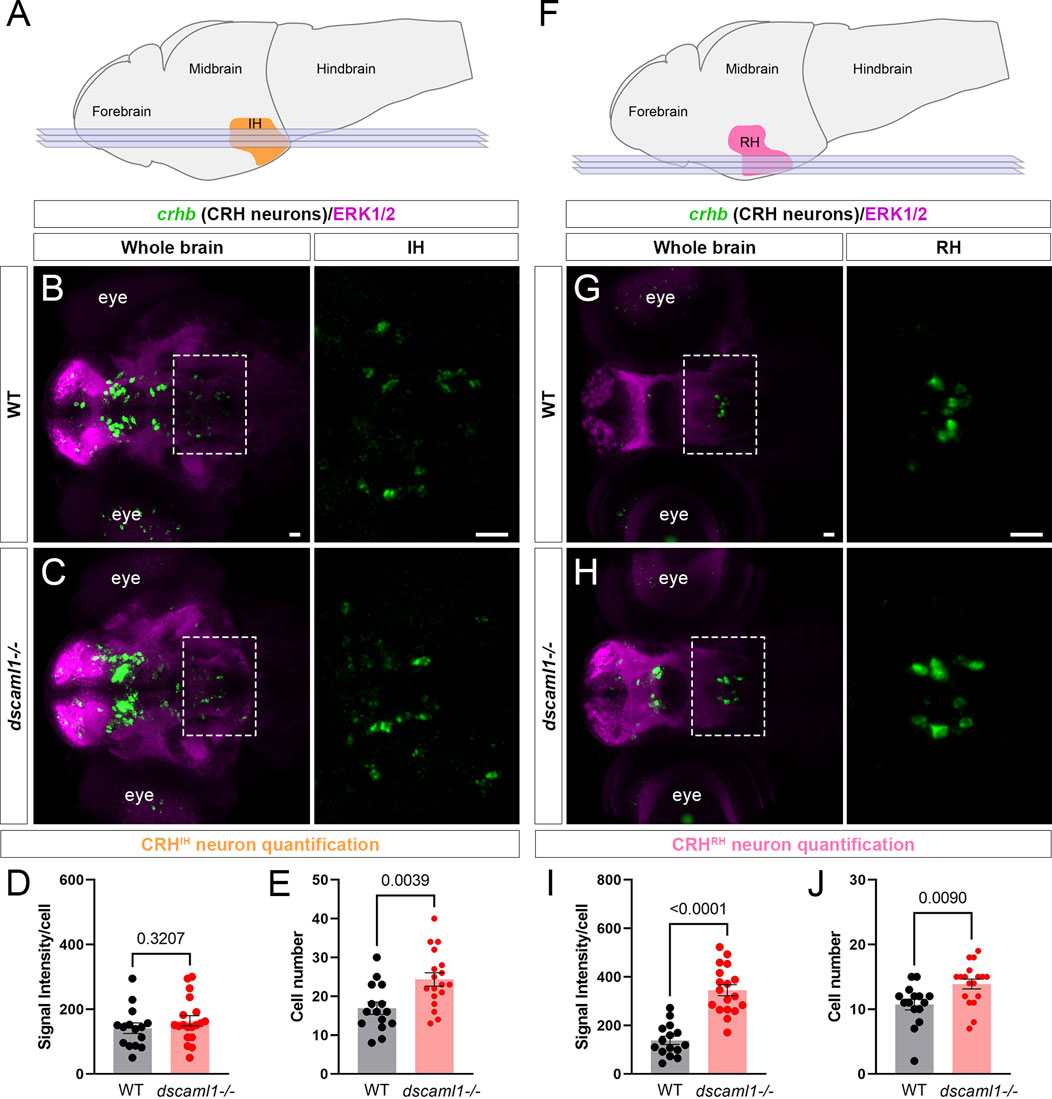
FIGURE 3. dscaml1 deficiency alters the development of CRH neurons in the intermediate and rostral hypothalamus. (A) Illustration of the larval brain, with the IH highlighted in orange. Imaging planes used for CRHIH neuron analysis are shown (light blue planes). (B, C) Labeling of CRHIH neurons by crhb FISH (green) and co-stained with anti-ERK1/2 (magenta). Representative confocal sub-stacks containing the IH are shown, with the IH (boxed region) enlarged and shown in the right panel without the ERK1/2 co-stain. WT brain is shown in panel B and dscaml1−/− brain is shown in panel C. (D–E) Quantification of the signal intensity per cell (D) and cell number (E). (F) Illustration of the larval brain, with the RH highlighted in pink. Imaging planes used for CRHRH neuron analysis are shown (light blue planes). (G–H) Labeling of CRHRH neurons by crhb FISH (green) and co-stained with anti-ERK1/2 (magenta). Representative confocal sub-stacks containing the RH are shown, with the RH (boxed region) enlarged and shown in the right panel without the ERK1/2 co-stain. WT brain is shown in panel G and dscaml1−/− brain is shown in panel H. (I–J) Quantification of the signal intensity per cell (I) and cell number (J). WT: n = 15. dscaml1−/−: n = 18. Scale bars are 20 μm. Mean, standard error, and p values are shown.
In addition to the IH, we have found consistent crhb expression in the rostral hypothalamus (RH) (Figure 3F). The roles of CRH neurons in the RH (CRHRH neurons) are still unknown, but they may contribute to the rostral hypothalamus-optic tectum pathway (Heap et al., 2017). In the RH at 5 dpf, both the crhb expression levels and the CRHRH cell number were higher in dscaml1−/− animals compared to WT (unpaired t-test, Figures 3G–J). Thus, dscaml1 is required broadly to regulate CRH neuron cell number in the developing hypothalamus, but dscaml1 deficiency only affects crhb expression in the RH and NPO.
DSCAM family proteins are known to reduce neuronal cell number by promoting PCD in the mammalian retina (Fuerst et al., 2012; Garrett et al., 2016). Additionally, our transcriptomic analysis showed that dscaml1 mutants express higher levels of apoptosis-regulating genes, bcl2b and pmaip1 (Figure 1A). Therefore, we hypothesized that dscaml1 deficiency might impair PCD in the developing hypothalamus.
To test this, we used an anti-activated caspase 3 (AC3) antibody to label apoptotic cells (Pyati et al., 2011). In wild-type animals, we saw widespread apoptosis in the nervous system, peaking at 3 dpf (Figure 4A–A”), consistent with previous findings using TUNEL (Cole and Ross, 2001). We found a decrease in AC3-positive cells in dscaml1−/− animals at 3 and 5 dpf, compared to wild-type sibling controls (Figure 4B–B”). To quantify the amount of cell death in specific hypothalamic regions, we used the Map-map calculation and the Z-Brain anatomical atlas (Randlett et al., 2015). In this method, individual confocal image stacks are registered to the Z-Brain reference brain; registered stacks are then grouped by genotype for voxel-wise comparisons using the Mann-Whitney U-statistic (WT n = 14; dscaml1−/− n = 21). The voxels with significant Z-scores, either higher in WT (WT over dscaml1) or higher in dscaml1−/− (dscaml1 over WT), were mapped to regions of interest (ROIs) and the mean voxel values in each ROI were calculated and displayed using a green-magenta color scale (green: higher in WT; magenta: higher in dscaml1−/−) (Figure 4C).
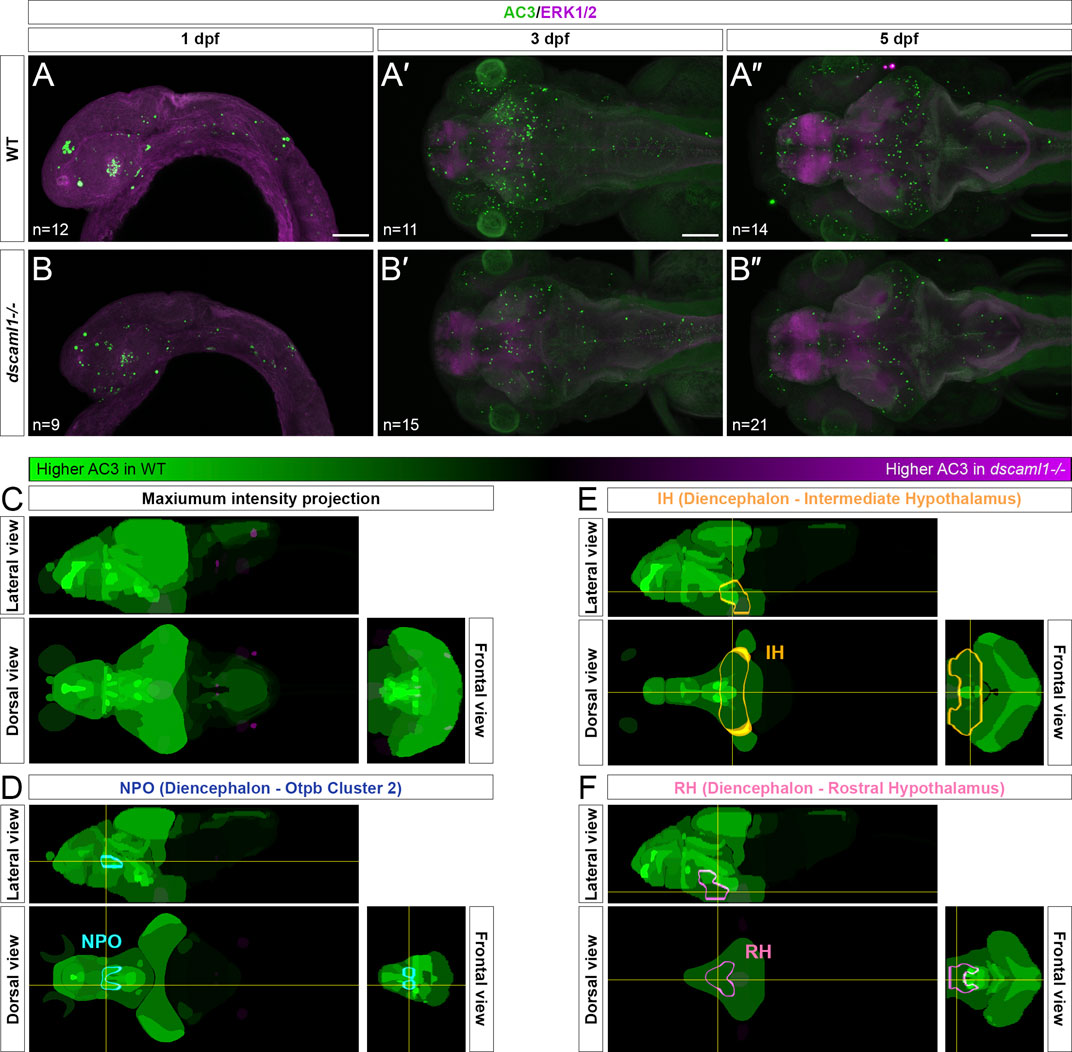
FIGURE 4. Programmed cell death in the brain is reduced in dscaml1 mutants. (A–B”) Immunolabeling with anti-AC3 (green) and counterstained with anti-ERK1/2 (magenta). 1 dpf embryos are shown laterally, and 3 and 5 dpf larvae dorsally. Wild-type (WT) animals are shown in panels A–A” and dscaml1−/− animals are shown in panels B–B”. (C) 5 dpf Z-Brain mapping of brain regions, showing regions with higher AC3 labeling in WT (green) or dscaml1−/− (magenta). Orthogonal maximum intensity projections are shown. (D) Z-Brain mapping in the NPO. Z-Brain ROI name is indicated in the parenthesis. Orthogonal sections are shown. (E) Z-Brain mapping in the IH. Z-Brain ROI name is indicated in the parenthesis. Orthogonal sections are shown. (F) Z-Brain mapping in the RH. Z-Brain ROI name is indicated in the parenthesis. Orthogonal sections are shown. Anterior is to the left for all images. Scale bars are 100 μm.
Overall, AC3 signal is higher in WT throughout the brain, compared to dscaml1−/− (i.e., many green regions and very few magenta ROIs). Out of 293 ROIs annotated in Z-Brain, 178 had voxels with significant WT over dscaml1 signal (mean signal = 1,384.90) (WT over dscaml1 sheet, Supplementary Datasheet S3). In contrast, only 65 ROIs had voxels with significant dscaml1 over WT signal (mean signal = 84.23) (dscaml1 over WT sheet, Supplementary Datasheet S3).
In the ROIs corresponding to the hypothalamic crhb expressing areas, the NPO (Z-Brain ROI: Diencephalon—Otpb Cluster 2) (WT over dscaml1 signal = 5,688.06; dscaml1 over WT signal = 0), IH (Diencephalon - Intermediate Hypothalamus) (WT over dscaml1 signal = 673.56; dscaml1 over WT signal = 36.40), and RH (Diencephalon - Rostral Hypothalamus) (WT over dscaml1 signal = 916.95; dscaml1 over WT signal = 0.70) all had higher AC3 signal in WT, compared to dscaml1−/− (Figures 4D–F). Together, these results suggest that reduced PCD in dscaml1−/− animals may contribute to the higher number of CRH neurons in the hypothalamus.
To further determine whether dscaml1 affects the survival of hypothalamic CRH neurons, we tracked the fate of individual CRHNPO neurons using in vivo time-lapse imaging at 3 to 5 dpf, when cell number begins to diverge between dscaml1−/− and WT animals. To visualize CRHNPO neurons in live zebrafish, we generated a crhb knock-in fluorescent reporter line using CRISPR-mediated genomic insertion (Figure 5A) (Kimura et al., 2014). A Cre-switchable hsp-LoxP-RFP-LoxP-GFP cassette was inserted 35 base pairs upstream of the first exon of crhb so that the expression of RFP (default) or GFP (with Cre-mediate recombination) would mark the endogenous crhb-expressing cells (Figure 5B). The resulting transgenic line, crhb:LoxP-RFP-LoxP-GFP (crhb:LRLG), has RFP and GFP expression pattern that matches the endogenous crhb transcript expression patterns reported by others (Figures 5C, D) (Shainer et al., 2022). To validate the fidelity of fluorescent reporter expression, we examined whether crhb:LRLG-labeled cells in the NPO express CRH protein. In animals not exposed to Cre (default RFP expression), we found that most RFP+ neurons are CRH immunopositive (84.71% ± 2.19%, Supplementary Figure S2A–B). Together, these results indicate that the crhb:LRLG line reliably labels CRHNPO neurons.
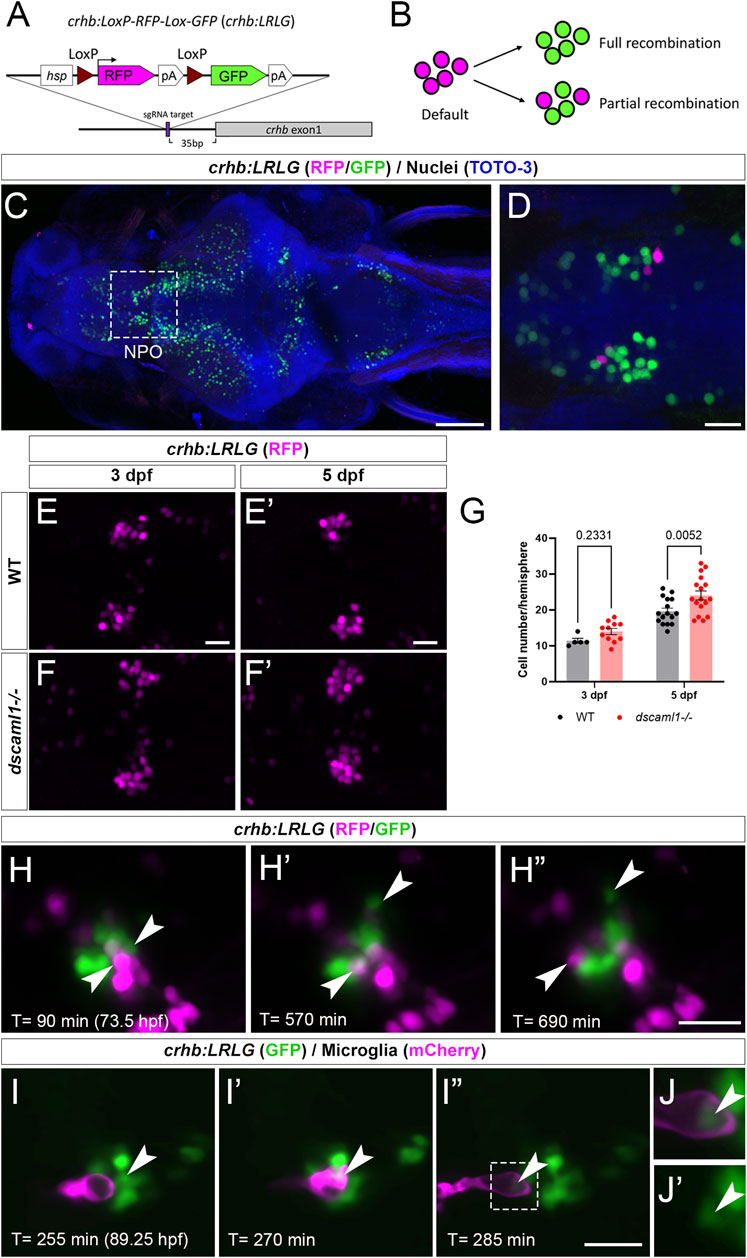
FIGURE 5. Fluorescent labeling and live imaging of CRHNPO neurons. (A) Schematic of CRISPR -mediated knock-in of the hsp-Lox-RFP-Lox-GFP cassette at the sgRNA target site, located 35 bp upstream of exon 1 of crhb. The orientation and junctional structure of insertion have not been determined. (B) Schematic of crhb:LRLG expression. Each circle represents a fluorescent cell. Without Cre (default), RFP is expressed in all cells. With full recombination, all cells express GFP. Partial recombination results in mosaic RFP and GFP labeling. (C) Dorsal view of a fixed 5 dpf crhb:LRLG larvae with partial recombination stained with anti-RFP (magenta) and anti-GFP (green). The boxed area marks the NPO. (D) Higher magnification image of the NPO. Both RFP and GFP-positive neurons can be seen. (E-F') Images of anti-RFP stained crh:LRLG animals without recombination. Representative WT (E-E') and dscaml1−/− (F-F') NPO neurons are shown. (G) Quantification of RFP-positive cells at 3 and 5 dpf. WT: n = 5 (3 dpf), 16 (5 dpf). dscaml1−/−: n = 11 (3 dpf), 17 (5 dpf). Mean, standard error, and corrected p values are shown. (H–H”) Live crhb:LRLG larvae with partial recombination were imaged from 72 to 84 hpf. Three time points are shown here. Two cells (arrowheads, one green and one magenta) move away over time. (I–I”) Live crhb:LRLG;mpeg1:Gal4;UAS:NTR-mCherry larvae with CRH neurons labeled with GFP (green) and microglia labeled with mCherry (magenta). In this image series, one CRH neuron (arrowhead) is engulfed (I') and then removed (I”) by a microglial cell. Images are confocal optical sections. (J, J') Panels showing enlarged views of the boxed area in (I”), with (J) or without (J') the mCherry channel. The remnant of the CRH neuron can still be seen inside the microglia (arrowheads). Scale bars are 100 μm (panel C) or 20 μm (all other images).
In crhb:LRLG animals, dscaml1 deficiency increased the number of RFP-labeled CRHNPO neurons (Figure 5E–F'). There were significant differences by developmental stage and genotype, with no significant interaction (two-way ANOVA, Supplementary Table SI). There was a significantly higher number of RFP-positive neurons in the dscaml1 mutants at 5 dpf but not 3 dpf (Figure 5G, multiple comparison test with Holm-Sidak correction). This result corroborates our crhb FISH results that show increased CRHNPO neuron number in dscaml1 mutants (Figure 2F).
Using the crhb:LRLG line, we first performed time-lapse imaging in anesthetized animals. We induced partial Cre-mediated recombination by injecting CreER mRNA into crhb:LRLG animals at the 1-cell stage and adding 4-hydroxytamoxifen (4-OHT) to activate CreER at 6–24 hpf (Figure 5B). At 3–4 dpf, intermingled GFP+ and RFP+ cells can be seen in the NPO by confocal imaging (Figure 5H–H” and Supplementary Video S1). Over time, some labeled cells moved away from the CRHNPO neuron cluster. These cells were likely dying cells being carried away by microglia, as seen in previous zebrafish studies (Mazaheri et al., 2014). To confirm this, we induced GFP expression (by injecting codon-optimized Cre mRNA) (Horstick et al., 2015) in all crhb:LRLG-labeled cells and labeled microglia with the mpeg1:mCherry transgene (Figure 5I–I” and Supplementary Video S2) (Espenschied et al., 2019). Indeed, mCherry+ microglia migrated toward the GFP+ cell cluster, engulfed GFP+ CRHNPO neurons, and carried the engulfed cells away. The remnant of the engulfed cell can be seen inside a large vacuole within the microglia (Figure 5J, J'). This type of engulfment events was seen in three out of five animals between 3.5-4 dpf. These results suggest that CRHNPO neurons undergo PCD and that dying cells are rapidly removed by microglia.
Next, to track the fate of individual CRHNPO neurons, we performed two-photon imaging from 3 to 5 dpf on crhb:LRLG animals with partial Cre-mediated recombination (Figure 6A). To minimize the potential effects of stress on PCD (Irles et al., 2014), we anesthetized and immobilized the animals during imaging. In between imaging sessions, each animal is allowed to recover in individual wells of 12-well plates, under normal light-dark cycles. Cells that are present at the first time point (78 hpf) were tracked at four subsequent time points (84, 96, 108, and 120 hpf) and categorized as either persisting (present at the last time point) or lost (lost at any of the following time points) (Figure 6B–B”). Overall, we observe a trend of reduced cell loss in the dscaml1+/− and dscaml1−/− animals (Chi-square test for trend, p = 0.0398) (Figure 6C, total number of tracked cells as indicated). In WT animals, 16.41% of cells are lost, versus 10.91% and 7.53% for dscaml1+/− and dscaml1−/−, respectively.
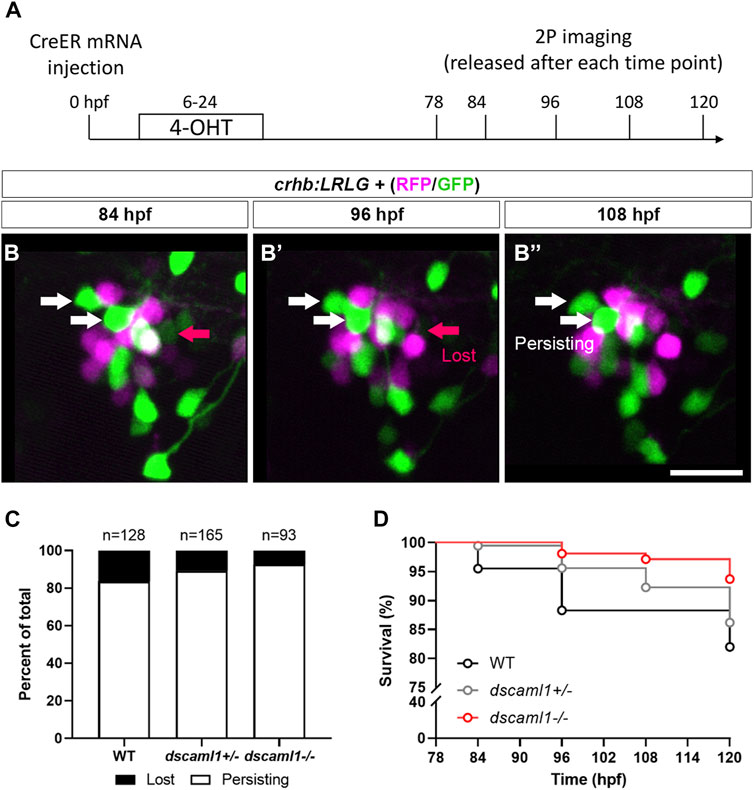
FIGURE 6. Live tracking of CRHNPO neuron cell fate. (A) Timeline of time-lapse two-photon (2P) imaging experiment. Partial recombination of crhb:LRLG was induced by 4-OHT at 6–24 hpf, and imaging was performed at 78, 84, 96, 108, and 120 hpf. Animals were briefly anesthetized during imaging and allowed to recover in between imaging sessions. (B–B”) Tracking of individual CRHNPO neurons. Three example time frames are shown. Individual fluorescent cells can be tracked over time and are divided into two categories: persisting (white arrows) or lost (pink arrow). The scale bar is 20 μm. (C) Quantification of the percentage of persisting versus lost CRHNPO neurons. Sample size (cell number) as indicated for each genotype. (D) Survival curve of individual CRHNPO neurons in each genotypic group.
Finally, considering the timing of cell loss, we plotted the survival curve of CRHNPO neurons for each genotype. Again, there was a significant difference in the trends of cell loss (Logrank test for trend, p = 0.0098), with the dscaml1−/− animals consistently showing a higher survival rate (Figure 6D). Together, these results show that dscaml1 deficiency reduces PCD of CRHNPO neurons.
Given the developmental perturbation of hypothalamic CRH neurons as well as the global changes in gene expression related to stress axis activation, we next determined whether the hormonal output of the stress axis—cortisol—is altered. Cortisol levels were measured using an enzyme-linked immunosorbent assay (ELISA) on homogenates made from pools of 5 dpf animals (30 animals per sample, 6 samples per condition). All animals were raised under standardized conditions, at the same density, and with a normal circadian cycle (14 h day/10 h night) (Yeh et al., 2013). Under this circadian cycle, dscaml1 mutant animals exhibit similar diurnal locomotor rhythms as wild-type animals (Ma et al., 2020b).
Baseline and stressed conditions were tested to evaluate potential alterations of cortisol in control (WT and dscaml1+/−) and dscaml1−/− siblings at 5 dpf. To measure baseline cortisol, we collected unperturbed animals within 30 min after light onset (zeitgeber time 0, ZT0) and in the afternoon (ZT6-8). To measure stress-induced cortisol, animals were exposed to either stirring stress (swirling water, 5 min) (Castillo-Ramirez et al., 2019) or hyperosmotic stress (250 mM NaCl, 20 min) at ZT6-8 (Yeh et al., 2013). These acute stressors are well-characterized and are comparable to the water current and salinity changes experienced by zebrafish larvae in their natural habitat (Clark et al., 2011).
At baseline, dscaml1−/− animals had significantly higher cortisol levels than control animals (Multiple Mann-Whitney test with Holm-Sidak correction. Adjusted p values shown in Figure 7A). At ZT0, the dscaml1 mutant baseline cortisol levels were 2.7-fold higher than that of controls (median 350.06 pg/mL versus 129.80 pg/mL). The difference in cortisol was less pronounced at ZT6-8, but dscaml1 mutants still had 2-fold higher cortisol levels than controls at baseline (median 238.51 pg/mL versus 118.52 pg/mL).
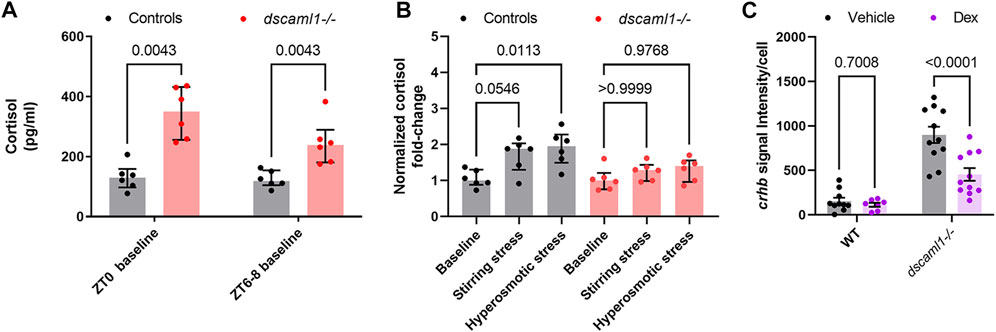
FIGURE 7. Cortisol levels and response to exogenous glucocorticoids. (A–B) Cortisol profile for 5 dpf larvae. For each sample (dot), cortisol was extracted from a pool of 30 animals. n = 6 for all groups. Median, interquartile range, and corrected p values are shown. (A) Baseline cortisol in control (black) and dscaml1−/− (red) animals. (B) Baseline-normalized cortisol fold change in control (black) and dscaml1−/− (red) animals. (C) Quantification of crhb signal intensity per cell in CRHNPO neurons. Vehicle: n = 10 (WT), 11 (dscaml1−/−). Dex: n = 7 (WT), 11 (dscaml1−/−). Mean, standard error, and corrected p values are shown.
After acute exposure to stressors, we found that dscaml1 mutant animals exhibited attenuated cortisol induction. We assessed the extent of stress-induced cortisol production by normalizing cortisol levels to the baseline cortisol of the same genotype at the same circadian time (ZT6-8). In the control group, stirring stress and hyperosmotic stress produced 1.88 and 1.95-fold increases in cortisol over the control baseline, respectively (grey bars, Figure 7B) (non-normalized cortisol data are shown in Supplementary Figure S3). The response to hyperosmotic stress was more robust (p = 0.0113, Multiple Mann-Whitney tests with Holm-Sidak correction) than that generated by stirring stress (p = 0.0546). In the dscaml1−/− group, stirring stress and hyperosmotic stress only produced 1.29 and 1.4-fold increases in cortisol over the dscaml1−/− baseline, respectively (red bars, Figure 7B). These increases were not statistically significant (p > 0.9999 for stirring stress, p = 0.9768 for hyperosmotic stress).
Together, these results show that dscaml1 deficiency elevates baseline cortisol levels and impair responses to acute stressors. These findings indicate that dscaml1 is critical for establishing the normal function of the stress axis.
Some aspects of the stress axis-related phenotypes in dscaml1 mutants resemble the zebrafish glucocorticoid receptor (gr) mutants. In particular, both gr and dscaml1 mutants exhibit elevated baseline cortisol and crhb (Ziv et al., 2013). This resemblance raises the possibility that the dscaml1 mutant phenotypes may result from insufficient glucocorticoid receptor-mediated signaling. To test this, we examined whether dscaml1 mutants can respond transcriptionally to exogenously applied glucocorticoids. We examined the expression of crhb in the NPO, as it is under feedback control from glucocorticoid-glucocorticoid receptor signaling (Watts, 2005). Wild-type siblings were used for controls.
A synthetic glucocorticoid receptor agonist, dexamethasone (Dex), was added to the embryo media at a final concentration of 2 μM from 4 to 5 dpf (24 h). Vehicle (0.02% ethanol) treated siblings were used for comparison. For crhb transcript level in CRHNPO neurons, we found significant differences caused by Dex treatment and genotype, with significant interaction between the two (two-way ANOVA, Supplementary Table SI). Multiple comparison tests found that Dex significantly reduced crhb transcript level in dscaml1−/− animals but not in wild-type animals (Holm-Sidak correction, Figure 7C with p values as indicated). These results show that dscaml1 deficiency does not result in a loss of glucocorticoid responsivity in CRHNPO neurons. Instead, dscaml1 deficiency may render animals more sensitive to the inhibitory effects of glucocorticoids.
The present study provides evidence that dscaml1 regulates the development of hypothalamic CRH neurons and is necessary for normal stress axis function. At the transcriptome level, dscaml1 deficiency in zebrafish results in gene expression changes that suggest stress axis hyperactivation. At the cellular level, we find that dscaml1−/− hypothalamic CRH neurons have increased stress axis-associated neuropeptide (crhb) expression, increased cell number, and reduced cell death. Physiologically, dscaml1 deficiency impairs normal neuroendocrine stress axis function, which is potentially caused by developmental deficits in hypothalamic CRH neurons as well as systemic changes in hormone/neuropeptide signaling. Together, these findings link DSCAML1 to the development of the stress axis and shed new light on the potential etiology of human DSCAML1-linked mental health conditions.
A major finding of this study is that DSCAML1 is necessary for the development of hypothalamic CRH neurons. To our knowledge, DSCAML1 is the first intercellular signaling molecule to be implicated in CRH neuron development. Our finding also provides the first link between DSCAML1 and hypothalamus development. We show that, similar to retinal neurons, the regulation of cell number by PCD is a DSCAML1-mediated process in hypothalamic CRH neurons (Garrett et al., 2016). Further investigations are required to determine whether other aspects of DSCAML1 function, such as the regulation of cellular spacing and synaptogenesis, are involved in CRH neuron development (Fuerst et al., 2009; Yamagata and Sanes, 2010; Sachse et al., 2019). Additionally, given that dscaml1 is expressed broadly in the nervous system (Ma et al., 2020b), including CRH neurons and non-CRH neurons (Supplementary Figure S4), it will be important to address whether DSCAML1 acts cell-autonomously in CRHNPO neurons.
During development, PCD is critical for eliminating transient cell types, matching input and output cell populations, and maintaining cellular spacing (Yamaguchi and Miura, 2015; Wong and Marin, 2019). It has been hypothesized that PCD in the hypothalamus may specify neural circuit assembly and shape output activity (Simerly, 2002; Forger, 2009). Congruent with this idea, reduced hypothalamic PCD caused by early-life stress is associated with increased sensitivity to acute stressors in adulthood (Zhang et al., 2012; Irles et al., 2014).
As an intercellular signaling molecule, DSCAML1 may act to transduce extracellular cues for PCD. One potential cue is synaptic activity, which is critical for neuronal survival during development (Wong and Marin, 2019). In culture, DSCAML1 is localized to excitatory synapses (Yamagata and Sanes, 2010) and inhibits excitatory synaptogenesis when overexpressed (Sachse et al., 2019). It remains to be determined whether DSCAML1 deficiency increases excitatory synaptic transmission and activates activity-dependent cell survival pathways in CRH neurons (Wong and Marin, 2019).
It is worth noting that CRH neuron cell death may not be the sole mechanism by which dscaml1 affects CRH neuron number. Changes in the number or proliferative capacity of neuroprogenitor cells may contribute to the increase CRH neuron number we observed in dscaml1 mutants. Additionally, the higher crhb expression induced by dscaml1 deficiency may lead to more cells being identified as CRH neurons. Further work is needed to determine how these factors control CRH neuron number during development.
dscaml1−/− animals exhibit multiple signs of stress axis activation at baseline, including cortisol elevation and the suppression of immunity-associated genes. The elevated baseline cortisol levels likely resulted in attenuated responses to acute stressors, similar to animals under chronic cortisol administration (Barton et al., 1987; Johnson et al., 2006). A likely cause of these phenotypes is the overexpression of crhb. In mice, broad overexpression of CRH leads to elevated corticosterone (the stress glucocorticoid in rodents) and produces phenotypes similar to Cushing’s syndrome, a human disorder caused by the overproduction of cortisol (Stenzel-Poore et al., 1992; Arnett et al., 2016).
Beyond dysfunction of hypothalamic neurons and CRH signaling, a broader neurological imbalance can also activate the stress axis. For example, seizures have been shown to activate the HPA axis, which increases the likelihood of future seizures (O'Toole et al., 2014; Hooper et al., 2018). It has been reported that Dscaml1 mutant rats and human patients with DSCAML1 loss-of-function variants exhibit neuronal hyperactivation and seizures (Hayase et al., 2020). It is, therefore, possible that excitation-inhibition imbalance in extra-hypothalamic regions may result in increased hypothalamic CRH neuron firing in zebrafish dscaml1 mutants. Further studies on the cell-type-specific functions of dscaml1 are needed to understand the precise cause of stress axis hyperactivation in dscaml1−/− animals.
Facilitation and feedback are signature features of the stress axis (Dallman et al., 1992; Spencer and Deak, 2017). While CRH neurons control cortisol levels, cortisol signaling also affects the development of CRH neurons. Zebrafish gr mutants have phenotypes similar to that of dscaml1−/− animals, including slow visual-background adaptation, sluggish light onset response, elevated cortisol, and increased expression of crhb (Griffiths et al., 2012; Muto et al., 2013; Ziv et al., 2013). Surprisingly, rather than decreased glucocorticoid receptor signaling (as in gr mutants), dscaml1−/− animals have intact glucocorticoid receptor signaling, with dexamethasone exerting strong suppression of crhb expression in the NPO. Thus, despite the superficial phenotypic similarity, the underlying signaling mechanisms are distinct between dscaml1 and gr mutants.
Given the known roles of DSCAML1 in regulating synapse formation (Sachse et al., 2019), the increase in crhb expression may be caused by the hyper-activation of upstream stress-related neural inputs. In rodents, various paradigms of chronic stress usually result in an upregulation of CRH in the PVN (Herman and Tasker, 2016). The transcriptional activation caused by stress-related synaptic activity may overcome the negative feedback mediated by glucocorticoid receptor signaling. Additional work is needed to disambiguate the relationship between cortisol disturbances and developmental deficits. A possible approach would be to normalize cortisol levels by genetically ablating interrenal cells (i.e., genetic adrenalectomy) and supplementing with constant levels of exogenous cortisol (Gutierrez-Triana et al., 2015).
In conclusion, this work shows that DSCAML1 is integral for developing the hypothalamic neurons that regulate the neuroendocrine stress axis. Using zebrafish as a vertebrate model for the ontogenesis of the stress axis, we found that dscaml1 deficiency results in CRH neuron deficits and dysfunction of the stress axis. Genetic perturbations of DSCAML1 are seen in patients suffering from a wide range of mental health disorders, including intellectual disability, autism spectrum disorder, schizophrenia, epilepsy, and stress disorder (Iossifov et al., 2014; Caramillo et al., 2015; Karaca et al., 2015; Hayase et al., 2020; Ogata et al., 2021; Saadatmand et al., 2021). Developmental deficits in the stress axis may contribute to the etiology of these disorders.
Zebrafish (all ages) were raised under a 14/10 light/dark cycle at 28.5°C. Embryos and larvae were raised in E3 buffer (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4) (Nüsslein-Volhard et al., 2002). All zebrafish used in this study were in a mixed background of AB and TL wild-type strains (Zebrafish International Resource Center). Sex was not a relevant variable for the stages used in this study (0–6 dpf), as laboratory zebrafish remain sexually undifferentiated until 2 weeks of age (Maack and Segner, 2003; Wilson et al., 2014).
The dscaml1vt1 loss-of-function allele contains a seven base pair deletion that results in frame shift and premature stop codon (Ma et al., 2020b). Animals used for live imaging were in homozygous nacre (mitfa) mutant background to prevent pigment formation (Lister et al., 1999). The microglia RFP line [Tg(mpeg1:Gal4;UAS:NTR-mCherry)] was obtained from Dr. John Rawls at Duke University (Espenschied et al., 2019). The crhb:LoxP-RFP-LoxP-GFP line was generated using CRISPR-mediated knock-in, as described by Kimura et al. (Kimura et al., 2014). The sgRNA sequence for the crhb knock-in locus is AGCTCGCGTCTGCGCAGAG. All group-wise comparisons (mutants versus controls) were done between siblings.
Progenies from heterozygous dscaml1 mutant parents were anesthetized and harvested at 3.5-4 dpf. The anterior half of the animal was used for RNA preparation using the RNA Miniprep Kit (Zymo). The posterior half was used for genotyping. Three biological replicates for each group were analyzed, each containing RNA from 6–11 animals. All samples had RIN ≥8.0 and were converted into a strand-specific library using Illumina’s TruSeq Stranded mRNA HT Sample Prep Kit (RS-122–2,103; Illumina) for subsequent cluster generation and sequencing on Illumina’s NextSeq 75 sequencer. Sequence data processing, alignment, read count, mapping, and quality control were performed as previously described (Ates et al., 2020). Differential expression was tested for significance using the false discovery rate (FDR) (Benjamini–Hochberg) corrected Likelihood Ratio Test (LRT) in the R-package DESeq2 (Love et al., 2014). 238 and 116 genes showed a significant difference in read counts at FDR < 0.01 and 0.001, respectively. Original sequence data have been deposited in NCBI’s Gene Expression Omnibus (Edgar et al., 2002) and accessible through GEO Series accession number GSE213858.
Single and double whole-mount fluorescent in situ hybridization (FISH) was performed using protocols described previously (Pan et al., 2012). Probes were synthesized by in vitro transcription using the DIG and Fluorescein RNA Labeling Mix (Roche). DIG and fluorescein-labeled probes were detected with anti-DIG or anti-Fluorescein POD-conjugated Fab fragments (Roche) and Cy3 or Fluorescein TSA-plus Reagent (Akoya Biosciences). Plasmid template for crhb (Lohr et al., 2009) was provided by Dr. David Prober at Caltech. The dscaml1 probe was generated as described previously (Ma et al., 2020b).
Immunohistochemistry was performed as described previously (Ma et al., 2020a). ERK1/2 was stained with mouse anti-ERK1/2 (4696S; Cell Signaling Technology). Activated caspase three was stained with rabbit anti-cleaved caspase 3 (559565; BD Biosciences). Nuclei were stained with TOTO-3 Iodide (ThermoFisher). RFP was stained with chicken anti-RFP (600-901-379S, Rockland) or rabbit anti-RFP (PM005, MBL Life Science). GFP was stained with rabbit anti-GFP (598, MBL Life Science). CRH was stained with rabbit anti-CRH (PBL rC68) provided by P. Sawchenko and J. Vaughan from the Salk Institute. All FISH and immunohistochemistry samples were mounted in 1.5% low-melt agarose in glass-bottomed Petri dishes (P50G-1.5–14-F; MatTek) and imaged using a Nikon A1 upright confocal microscope.
A detailed protocol for cortisol extraction and ELISA is provided online (Supplementary Datasheet S4). At 4.5 dpf, dscaml1−/− and control animals were separated based on the darker pigmentation of the dscaml1−/− animal (Ma et al., 2020b). 30–35 animals were placed in each petri dish for cortisol extraction. The morning baseline (unstressed) sample collections were done at 15–30 min after light onset (08:15–08:45), and the afternoon sample collections were done between 14:30–16:00. Hyperosmotic stress and stirring stress experiments were done between 14:30–15:30. 6 biological duplicates—each containing a pool of 30 animals—were collected for each genotype (dscaml1−/− and control) and stress condition (morning baseline, afternoon baseline, stirring stress, osmotic stress). Mutants and control animals are tested side by side for each experiment.
Stirring stress was induced by creating a vortex water flow with a spinning magnetic stir bar (Castillo-Ramirez et al., 2019). A small magnetic stir bar was placed into a 100 mm petri dish containing 35 animals and 20 mL of E3 media. The stir bar was rotated at 300 rpm with a stirring microplate for 5 min. Hyperosmotic stress was induced by increasing salt concentration in the media (Yeh et al., 2013). 30 animals were placed in 8 mL of E3 media. Then, 2 mL of prewarmed 1.25 M NaCl was added to the media for a final concentration of 250 mM for 20 min.
Sample homogenization and cortisol extraction were performed as described by Yeh et al. (Yeh et al., 2013). Briefly, 5 dpf larvae were rapidly immobilized with ice-cold E3 media and then flash-frozen at −80°C. Once all samples were collected, cortisol from the frozen samples was extracted with ethyl acetate (33211-1L-R; Sigma-Aldrich). Cortisol concentration was measured using a commercial ELISA kit, following the manufacturer’s instructions (500360; Cayman Chemical). Sample plates were read with a microplate reader (FilterMax F3; MicroDevices) 90–120 min after initial development.
Cre-mediated recombination of the crhb:LRLG transgene was induced by injecting Cre mRNA into the embryo at the 1-cell stage. To achieve partial Cre-mediated recombination, ∼30 pg of CreER mRNA was injected at the 1-cell stage and 4-Hydroxytamoxifen (4-OHT) was added to the embryo media at 6 hpf (10 μM), followed by washout with E3 media at 24 hpf. To achieve complete Cre-mediated recombination, ∼50 pg of in vitro transcribed Cre. zf1 mRNA (#61391, Addgene) was injected into the embryos (Horstick et al., 2015).
To perform confocal live imaging, 3 dpf animals were anesthetized with 0.01% tricaine methanesulfonate (MS-222, Sigma) and embedded in 1% low-melting point agarose, with the dorsal side resting on the glass surface inside the glass-bottomed petri dish (P50G-1.5–14-F; MatTek) (Beier et al., 2016). The petri dish was then filled with E3 media containing 0.01% tricaine methanesulfonate (MS-222, Sigma). Confocal z-stacks were acquired every 15 or 30 min for 12 h on a Nikon A1 confocal microscope.
Two-photon live imaging was done on a custom Bruker two-channel two-photon microscope. A tuneable Ti:Saphire laser (Chameleon Vision II; Coherent) was tuned to 980 nm to excite RFP and GFP simultaneously. 78 hpf animals were anesthetized and embedded the same way as confocal live imaging, but with the dorsal side away from the cover glass. Each animal was imaged at 78, 84, 96, 108, and 120 hpf (Figure 6A). After each time point, imaged larvae were gently removed from the agarose and recovered in E3 media at 28.5°C under normal day/night cycles. After the last time point, genomic DNA was prepared for all imaged animals and genotyped.
Images were processed using Fiji—an open-source image processing software (Schindelin et al., 2012). For FISH images, images were convolved (kernel = 12) to enhance cell boundaries, and the center of each cell was manually tagged using the ROI manager tool. The cell number equals the number of ROIs in each animal. The signal intensity per cell was defined as the median signal intensity of all ROIs in a given animal. The number of RFP+ cells in crhb:LRLG animals were counted using the ROI manager tool without convolution. For confocal live imaging, images were pre-processed using the Denoise. AI function in the Nikon Elements software. For two-photon imaging, z-stacks from different time points were aligned using the “Correct 3D drift” function. Spectral overlap between the RFP and GFP channels was linearly unmixed. Individual cells were tracked with the MTrackJ plugin in Fiji (Meijering et al., 2012).
All statistical analyses were performed in GraphPad Prism (Version 9). For normally-distributed data, parametric tests (t-test or ANOVA) were used. For non-normally distributed data, non-parametric tests (Mann-Whitney) were used. The Holm-Sidak post-test was used to correct for multiple comparisons, and the adjusted p values are shown. All values are expressed as mean ± standard error, unless otherwise noted. Statistical tests were considered significant when p < 0.05.
Quantification of apoptotic (AC3+) cells was done by the MAP-map quantitation and Z-Brain annotation method, but Anti-AC3 was used in place of anti-phosphorylated ERK1/2 (Randlett et al., 2015; Brunal et al., 2020). All MAP-map processing procedures (e.g., CMTK, MATLAB scripts) were performed as described by Randlett et al. The NPO is not annotated as such in Z-Brain, but the “Diencephalon - Otpb Cluster 2” ROI fits the definition of the NPO, as it delineate the dorsal region of the preoptic area that expresses otpb (Herget et al., 2014).
Next-generation sequencing data utilized in this publication are available from the Gene Expression Omnibus (accession code GSE213858). All other data related to this study will be provided by the authors upon reasonable request.
All procedures were performed according to protocols approved by the Institutional Animal Care and Use Committee at Virginia Tech and the National Institute for Basic Biology.
All authors contributed to the article and approved the submitted version. MM and YAP conceived and designed the experiments. MM and AAB prepared samples for RNA sequencing. YAP analyzed the RNA sequencing data. MM performed ELISA and histochemistry experiments and analyzed the data. MM and KCC performed in situ hybridization experiments and analyzed the data with contributions from CS and KS. SH generated the crhbl:LRLG transgenic line. MM and YAP wrote the manuscript. YAP provided project administration and acquired funding.
This work was supported by the Commonwealth Research Commercialization Fund (ER14S-001LS to YAP), the Virginia-Maryland College of Veterinary Medicine Intramural Research Fund, the Commonwealth Health Research Board Grant (#208–06-21 to YAP), the National Institutes of Health (R01MH131820), and funding from Virginia Tech.
We thank the animal care staff and veterinarians for animal husbandry; members of the Pan laboratory for helpful discussions; R. Settlage for computational analysis of RNA-seq data; S. Ryu and C. Yeh for sharing unpublished data; S. Imani for help with the quantification of IHC data; M. Wagle and S. Guo for advice on cortisol extraction procedures; M. Fox and A. Morozov for constructive feedback on the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2023.1113675/full#supplementary-material
Supplementary Video S1 | Time-lapse movie of crhb:LRLG larvae. CRH neurons were labeled with RFP (magenta) and GFP (green). Two cells (arrowheads, one green and one magenta), the same ones as shown in Figures 3H–H”, move away over time.
Supplementary Video S2 | Time-lapse movie of larvae with CRH neurons labeled with GFP (green) and microglia labeled with mCherry (magenta). The red “X” marks the cell indicated in Figures 3I–J'. Maximum projection of z-stack for each time point is shown.
Alderman, S. L., and Bernier, N. J. (2009). Ontogeny of the corticotropin-releasing factor system in zebrafish. Gen. Comp. Endocrinol. 164, 61–69. doi:10.1016/j.ygcen.2009.04.007
Alsop, D., and Vijayan, M. (2009). The zebrafish stress axis: molecular fallout from the teleost-specific genome duplication event. Gen. Comp. Endocrinol. 161, 62–66. doi:10.1016/j.ygcen.2008.09.011
Alsop, D., and Vijayan, M. M. (2008). Development of the corticosteroid stress axis and receptor expression in zebrafish. Am. J. Physiol. Regul. Integr. Comp. Physiol. 294, R711–R719. doi:10.1152/ajpregu.00671.2007
Alvarez-Bolado, G. (2019). Development of neuroendocrine neurons in the mammalian hypothalamus. Cell Tissue Res. 375, 23–39. doi:10.1007/s00441-018-2859-1
Arnett, M. G., Muglia, L. M., Laryea, G., and Muglia, L. J. (2016). Genetic approaches to hypothalamic-pituitary-adrenal Axis regulation. Neuropsychopharmacology 41, 245–260. doi:10.1038/npp.2015.215
Ates, K. M., Wang, T., Moreland, T., Veeranan-Karmegam, R., Ma, M., Jeter, C., et al. (2020). Deficiency in the endocytic adaptor proteins PHETA1/2 impairs renal and craniofacial development. Dis. models Mech. 13, dmm041913. doi:10.1242/dmm.041913
Bai, Y., Liu, H., Huang, B., Wagle, M., and Guo, S. (2016). Identification of environmental stressors and validation of light preference as a measure of anxiety in larval zebrafish. BMC Neurosci. 17, 63. doi:10.1186/s12868-016-0298-z
Barton, B. A., Schreck, C. B., and Barton, L. D. (1987). Effects of chronic cortisol administration and daily acute stress on growth, physiological conditions, and stress responses in juvenile rainbow trout. Dis. aquatic Org. 2, 173–185. doi:10.3354/dao002173
Beier, K. T., Mundell, N. A., Pan, Y. A., and Cepko, C. L. (2016). Anterograde or retrograde transsynaptic circuit tracing in vertebrates with vesicular stomatitis virus vectors. Curr. Protoc. Neurosci. 74, 1.26.1–1.26.27. doi:10.1002/0471142301.ns0126s74
Brunal, A. A., Clark, K. C., Ma, M., Woods, I. G., and Pan, Y. A. (2020). Effects of constitutive and acute connexin 36 deficiency on brain-wide susceptibility to PTZ-induced neuronal hyperactivity. Front. Mol. Neurosci. 13, 587978. doi:10.3389/fnmol.2020.587978
Caramillo, E. M., Khan, K. M., Collier, A. D., and Echevarria, D. J. (2015). Modeling PTSD in the zebrafish: are we there yet? Behav. Brain Res. 276, 151–160. doi:10.1016/j.bbr.2014.05.005
Castillo-Ramirez, L. A., Ryu, S., and De Marco, R. J. (2019). Active behaviour during early development shapes glucocorticoid reactivity. Sci. Rep. 9, 12796. doi:10.1038/s41598-019-49388-3
Chandrasekar, G., Lauter, G., and Hauptmann, G. (2007). Distribution of corticotropin-releasing hormone in the developing zebrafish brain. J. Comp. Neurol. 505, 337–351. doi:10.1002/cne.21496
Clark, K. J., Boczek, N. J., and Ekker, S. C. (2011). Stressing zebrafish for behavioral genetics. Rev. Neurosci. 22, 49–62. doi:10.1515/RNS.2011.007
Cole, L. K., and Ross, L. S. (2001). Apoptosis in the developing zebrafish embryo. Dev. Biol. 240, 123–142. doi:10.1006/dbio.2001.0432
Dallman, M. F., Akana, S. F., Scribner, K. A., Bradbury, M. J., Walker, C. D., Strack, A. M., et al. (1992). Stress, feedback and facilitation in the hypothalamo-pituitary-adrenal Axis. J. Neuroendocrinol. 4, 517–526. doi:10.1111/j.1365-2826.1992.tb00200.x
Denver, R. J. (2009). Structural and functional evolution of vertebrate neuroendocrine stress systems. Ann. N. Y. Acad. Sci. 1163, 1–16. doi:10.1111/j.1749-6632.2009.04433.x
Edgar, R., Domrachev, M., and Lash, A. E. (2002). Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30 (1), 207–210. doi:10.1093/nar/30.1.207
Espenschied, S. T., Cronan Mark, R., Matty Molly, A., Mueller, O., Redinbo Matthew, R., Tobin David, M., et al. (2019). Epithelial delamination is protective during pharmaceutical-induced enteropathy. Proc. Natl. Acad. Sci. 116, 16961–16970. doi:10.1073/pnas.1902596116
Forger, N. G. (2009). Control of cell number in the sexually dimorphic brain and spinal cord. J. Neuroendocrinol. 21, 393–399. doi:10.1111/j.1365-2826.2009.01825.x
Fuerst, P. G., Bruce, F., Rounds, R. P., Erskine, L., and Burgess, R. W. (2012). Cell autonomy of DSCAM function in retinal development. Dev. Biol. 361, 326–337. doi:10.1016/j.ydbio.2011.10.028
Fuerst, P. G., Bruce, F., Tian, M., Wei, W., Elstrott, J., Feller, M. B., et al. (2009). DSCAM and DSCAML1 function in self-avoidance in multiple cell types in the developing mouse retina. Neuron 64, 484–497. doi:10.1016/j.neuron.2009.09.027
Garrett, A. M., Tadenev, A. L., and Burgess, R. W. (2012). DSCAMs: restoring balance to developmental forces. Front. Mol. Neurosci. 5, 86. doi:10.3389/fnmol.2012.00086
Garrett, A. M., Tadenev, A. L., Hammond, Y. T., Fuerst, P. G., and Burgess, R. W. (2016). Replacing the PDZ-interacting C-termini of DSCAM and DSCAML1 with epitope tags causes different phenotypic severity in different cell populations. eLife 5, e16144. doi:10.7554/eLife.16144
Griffiths, B. B., Schoonheim, P. J., Ziv, L., Voelker, L., Baier, H., and Gahtan, E. (2012). A zebrafish model of glucocorticoid resistance shows serotonergic modulation of the stress response. Front. Behav. Neurosci. 6, 68. doi:10.3389/fnbeh.2012.00068
Gutierrez-Triana, J. A., Herget, U., Castillo-Ramirez, L. A., Lutz, M., Yeh, C. M., De Marco, R. J., et al. (2015). Manipulation of interrenal cell function in developing zebrafish using genetically targeted ablation and an optogenetic tool. Endocrinology 156, 3394–3401. doi:10.1210/EN.2015-1021
Hayase, Y., Amano, S., Hashizume, K., Tominaga, T., Miyamoto, H., Kanno, Y., et al. (2020). Down syndrome cell adhesion molecule like-1 (DSCAML1) links the GABA system and seizure susceptibility. Acta Neuropathol. Commun. 8, 206. doi:10.1186/s40478-020-01082-6
Heap, L. A., Vanwalleghem, G. C., Thompson, A. W., Favre-Bulle, I., Rubinsztein-Dunlop, H., and Scott, E. K. (2017). Hypothalamic projections to the optic tectum in larval zebrafish. Front. Neuroanat. 11, 135. doi:10.3389/fnana.2017.00135
Herget, U., and Ryu, S. (2015). Coexpression analysis of nine neuropeptides in the neurosecretory preoptic area of larval zebrafish. Front. Neuroanat. 9, 2. doi:10.3389/fnana.2015.00002
Herget, U., Wolf, A., Wullimann, M. F., and Ryu, S. (2014). Molecular neuroanatomy and chemoarchitecture of the neurosecretory preoptic-hypothalamic area in zebrafish larvae. J. Comp. Neurol. 522, 1542–1564. doi:10.1002/cne.23480
Herman, J. P., and Tasker, J. G. (2016). Paraventricular hypothalamic mechanisms of chronic stress adaptation. Front. Endocrinol. 7, 137. doi:10.3389/fendo.2016.00137
Hooper, A., Paracha, R., and Maguire, J. (2018). Seizure-induced activation of the HPA axis increases seizure frequency and comorbid depression-like behaviors. Epilepsy Behav. E&B 78, 124–133. doi:10.1016/j.yebeh.2017.10.025
Horstick, E. J., Jordan, D. C., Bergeron, S. A., Tabor, K. M., Serpe, M., Feldman, B., et al. (2015). Increased functional protein expression using nucleotide sequence features enriched in highly expressed genes in zebrafish. Nucleic acids Res. 43, e48. doi:10.1093/nar/gkv035
Iossifov, I., O'Roak, B. J., Sanders, S. J., Ronemus, M., Krumm, N., Levy, D., et al. (2014). The contribution of de novo coding mutations to autism spectrum disorder. Nature 515, 216–221. doi:10.1038/nature13908
Irles, C., Nava-Kopp, A. T., Morán, J., and Zhang, L. (2014). Neonatal maternal separation up-regulates protein signalling for cell survival in rat hypothalamus. Stress 17, 275–284. doi:10.3109/10253890.2014.913017
Johnson, S. A., Fournier, N. M., and Kalynchuk, L. E. (2006). Effect of different doses of corticosterone on depression-like behavior and HPA axis responses to a novel stressor. Behav. Brain Res. 168, 280–288. doi:10.1016/j.bbr.2005.11.019
Karaca, E., Harel, T., Pehlivan, D., Jhangiani, S. N., Gambin, T., Coban Akdemir, Z., et al. (2015). Genes that affect brain structure and function identified by rare variant analyses of mendelian neurologic disease. Neuron 88, 499–513. doi:10.1016/j.neuron.2015.09.048
Keen-Rhinehart, E., Michopoulos, V., Toufexis, D. J., Martin, E. I., Nair, H., Ressler, K. J., et al. (2009). Continuous expression of corticotropin-releasing factor in the central nucleus of the amygdala emulates the dysregulation of the stress and reproductive axes. Mol. Psychiatry 14, 37–50. doi:10.1038/mp.2008.91
Kimura, Y., Hisano, Y., Kawahara, A., and Higashijima, S. (2014). Efficient generation of knock-in transgenic zebrafish carrying reporter/driver genes by CRISPR/Cas9-mediated genome engineering. Sci. Rep. 4, 6545. doi:10.1038/srep06545
Kolber, B. J., Boyle, M. P., Wieczorek, L., Kelley, C. L., Onwuzurike, C. C., Nettles, S. A., et al. (2010). Transient early-life forebrain corticotropin-releasing hormone elevation causes long-lasting anxiogenic and despair-like changes in mice. J. Neurosci. 30, 2571–2581. doi:10.1523/JNEUROSCI.4470-09.2010
Lister, J. A., Robertson, C. P., Lepage, T., Johnson, S. L., and Raible, D. W. (1999). Nacre encodes a zebrafish microphthalmia-related protein that regulates neural-crest-derived pigment cell fate. Development 126, 3757–3767. doi:10.1242/dev.126.17.3757
Lohr, H., and Hammerschmidt, M. (2011). Zebrafish in endocrine systems: recent advances and implications for human disease. Annu. Rev. Physiol. 73, 183–211. doi:10.1146/annurev-physiol-012110-142320
Lohr, H., Ryu, S., and Driever, W. (2009). Zebrafish diencephalic A11-related dopaminergic neurons share a conserved transcriptional network with neuroendocrine cell lineages. Development 136, 1007–1017. doi:10.1242/dev.033878
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi:10.1186/s13059-014-0550-8
Ma, M., Kler, S., and Pan, Y. A. (2020a). Structural neural connectivity analysis in zebrafish with restricted anterograde transneuronal viral labeling and quantitative brain mapping. Front. Neural circuits 13, 85. doi:10.3389/fncir.2019.00085
Ma, M., Ramirez, A. D., Wang, T., Roberts, R. L., Harmon, K. E., Schoppik, D., et al. (2020b). Zebrafish dscaml1 deficiency impairs retinal patterning and oculomotor function. J. Neurosci. 40, 143–158. doi:10.1523/JNEUROSCI.1783-19.2019
Maack, G., and Segner, H. (2003). Morphological development of the gonads in zebrafish. J. Fish Biol. 62, 895–906. doi:10.1046/j.1095-8649.2003.00074.x
Mazaheri, F., Breus, O., Durdu, S., Haas, P., Wittbrodt, J., Gilmour, D., et al. (2014). Distinct roles for BAI1 and TIM-4 in the engulfment of dying neurons by microglia. Nat. Commun. 5, 4046. doi:10.1038/ncomms5046
McEwen, B. S., and Akil, H. (2020). Revisiting the stress concept: Implications for affective disorders. J. Neurosci. 40, 12–21. doi:10.1523/JNEUROSCI.0733-19.2019
Meijering, E., Dzyubachyk, O., and Smal, I. (2012). Methods for cell and particle tracking. Methods Enzymol. 504, 183–200. doi:10.1016/B978-0-12-391857-4.00009-4
Mi, H., Ebert, D., Muruganujan, A., Mills, C., Albou, L-P., Mushayamaha, T., et al. (2021). PANTHER version 16: a revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucleic Acids Res. 49, D394–D403. doi:10.1093/nar/gkaa1106
Moreland, T., and Poulain, F. E. (2022). To stick or not to stick: The multiple roles of cell adhesion molecules in neural circuit assembly. Front. Neurosci. 16, 889155. doi:10.3389/fnins.2022.889155
Muto, A., Taylor, M. R., Suzawa, M., Korenbrot, J. I., and Baier, H. (2013). Glucocorticoid receptor activity regulates light adaptation in the zebrafish retina. Front. Neural Circuits 7, 145. doi:10.3389/fncir.2013.00145
Nagpal, J., Herget, U., Choi, M. K., and Ryu, S. (2019). Anatomy, development, and plasticity of the neurosecretory hypothalamus in zebrafish. Cell Tissue Res. 375, 5–22. doi:10.1007/s00441-018-2900-4
Nüsslein-Volhard, C., Gilmour, D., and Dahm, R. (2002). “Introduction: zebrafish as a system to study development and organogenesis,” in Zebrafish: A practical approach, 1–5.
O'Toole, K. K., Hooper, A., Wakefield, S., and Maguire, J. (2014). Seizure-induced disinhibition of the HPA axis increases seizure susceptibility. Epilepsy Res. 108, 29–43. doi:10.1016/j.eplepsyres.2013.10.013
Ogata, S., Hashizume, K., Hayase, Y., Kanno, Y., Hori, K., Balan, S., et al. (2021). Potential involvement of DSCAML1 mutations in neurodevelopmental disorders. Genes Cells 26, 136–151. doi:10.1111/gtc.12831
Pan, Y. A., Choy, M., Prober, D. A., and Schier, A. F. (2012). Robo2 determines subtype-specific axonal projections of trigeminal sensory neurons. Development 139, 591–600. doi:10.1242/dev.076588
Placzek, M., Fu, T., and Towers, M. (2020). “Development of the neuroendocrine hypothalamus,” in Developmental neuroendocrinology. Editors S. Wray, and S. Blackshaw (Cham: Springer International Publishing), 3–30.
Pyati, U. J., Gjini, E., Carbonneau, S., Lee, J. S., Guo, F., Jette, C. A., et al. (2011). p63 mediates an apoptotic response to pharmacological and disease-related ER stress in the developing epidermis. Dev. Cell 21, 492–505. doi:10.1016/j.devcel.2011.07.012
Randlett, O., Wee, C. L., Naumann, E. A., Nnaemeka, O., Schoppik, D., Fitzgerald, J. E., et al. (2015). Whole-brain activity mapping onto a zebrafish brain atlas. Nat. Methods 12, 1039–1046. doi:10.1038/nmeth.3581
Regev, L., and Baram, T. Z. (2014). Corticotropin releasing factor in neuroplasticity. Front. Neuroendocrinol. 35, 171–179. doi:10.1016/j.yfrne.2013.10.001
Saadatmand, F., Gurdziel, K., Jackson, L., Kwabi-Addo, B., and Ruden, D. M. (2021). DNA methylation and exposure to violence among African American young adult males. Brain, Behav. Immun. - Health 14, 100247. doi:10.1016/j.bbih.2021.100247
Sachse, S. M., Lievens, S., Ribeiro, L. F., Dascenco, D., Masschaele, D., Horre, K., et al. (2019). Nuclear import of the DSCAM-cytoplasmic domain drives signaling capable of inhibiting synapse formation. EMBO J. 38, e99669. doi:10.15252/embj.201899669
Sanes, J. R., and Zipursky, S. L. (2020). Synaptic specificity, recognition molecules, and assembly of neural circuits. Cell 181, 536–556. doi:10.1016/j.cell.2020.04.008
Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. doi:10.1038/nmeth.2019
Shainer, I., Kuehn, E., Laurell, E., Al Kassar, M., Mokayes, N., Sherman, S., et al. (2022). A single-cell resolution gene expression atlas of the larval zebrafish brain. bioRxiv [Preprint]. doi:10.1101/2022.02.11.479024
Simerly, R. B. (2002). Wired for reproduction: organization and development of sexually dimorphic circuits in the mammalian forebrain. Annu. Rev. Neurosci. 25, 507–536. doi:10.1146/annurev.neuro.25.112701.142745
Spencer, R. L., and Deak, T. (2017). A users guide to HPA axis research. Physiol. Behav. 178, 43–65. doi:10.1016/j.physbeh.2016.11.014
Stenzel-Poore, M. P., Cameron, V. A., Vaughan, J., Sawchenko, P. E., and Vale, W. (1992). Development of Cushing's syndrome in corticotropin-releasing factor transgenic mice. Endocrinology 130, 3378–3386. doi:10.1210/endo.130.6.1597149
Stroth, N., Holighaus, Y., Ait-Ali, D., and Eiden, L. E. (2011). PACAP: a master regulator of neuroendocrine stress circuits and the cellular stress response. Ann. N. Y. Acad. Sci. 1220, 49–59. doi:10.1111/j.1749-6632.2011.05904.x
Vom Berg-Maurer, C. M., Trivedi, C. A., Bollmann, J. H., De Marco, R. J., and Ryu, S. (2016). The severity of acute stress is represented by increased synchronous activity and recruitment of hypothalamic CRH neurons. J. Neurosci. 36, 3350–3362. doi:10.1523/JNEUROSCI.3390-15.2016
Wagle, M., Zarei, M., Lovett-Barron, M., Poston, K. T., Xu, J., Ramey, V., et al. (2022). Brain-wide perception of the emotional valence of light is regulated by distinct hypothalamic neurons. Mol. Psychiatry 27, 3777–3793. doi:10.1038/s41380-022-01567-x
Watts, A. G. (2005). Glucocorticoid regulation of peptide genes in neuroendocrine CRH neurons: A complexity beyond negative feedback. Front. Neuroendocrinol. 26, 109–130. doi:10.1016/j.yfrne.2005.09.001
Wendelaar Bonga, S. E. (1997). The stress response in fish. Physiol. Rev. 77, 591–625. doi:10.1152/physrev.1997.77.3.591
Wilson, C. A., High, S. K., McCluskey, B. M., Amores, A., Yan, Y. L., Titus, T. A., et al. (2014). Wild sex in zebrafish: loss of the natural sex determinant in domesticated strains. Genetics 198, 1291–1308. doi:10.1534/genetics.114.169284
Wong, F. K., and Marin, O. (2019). Developmental cell death in the cerebral cortex. Annu. Rev. Cell Dev. Biol. 35, 523–542. doi:10.1146/annurev-cellbio-100818-125204
Yamagata, M., and Sanes, J. R. (2008). Dscam and Sidekick proteins direct lamina-specific synaptic connections in vertebrate retina. Nature 451, 465–469. doi:10.1038/nature06469
Yamagata, M., and Sanes, J. R. (2010). Synaptic localization and function of Sidekick recognition molecules require MAGI scaffolding proteins. J. Neurosci. 30, 3579–3588. doi:10.1523/JNEUROSCI.6319-09.2010
Yamaguchi, Y., and Miura, M. (2015). Programmed cell death in neurodevelopment. Dev. Cell 32, 478–490. doi:10.1016/j.devcel.2015.01.019
Yeh, C. M., Glock, M., and Ryu, S. (2013). An optimized whole-body cortisol quantification method for assessing stress levels in larval zebrafish. PLoS One 8, e79406. doi:10.1371/journal.pone.0079406
Zhang, L., Hernández, V. S., Liu, B., Medina, M. P., Nava-Kopp, A. T., Irles, C., et al. (2012). Hypothalamic vasopressin system regulation by maternal separation: its impact on anxiety in rats. Neuroscience 215, 135–148. doi:10.1016/j.neuroscience.2012.03.046
Keywords: HPA (hypothalamic-pituitary-adrenal) axis, zebrafish, CRH neuron, hypothalamus, development
Citation: Ma M, Brunal AA, Clark KC, Studtmann C, Stebbins K, Higashijima S-i and Pan YA (2023) Deficiency in the cell-adhesion molecule dscaml1 impairs hypothalamic CRH neuron development and perturbs normal neuroendocrine stress axis function. Front. Cell Dev. Biol. 11:1113675. doi: 10.3389/fcell.2023.1113675
Received: 01 December 2022; Accepted: 30 January 2023;
Published: 16 February 2023.
Edited by:
Arjun Singh, Memorial Sloan Kettering Cancer Center, United StatesReviewed by:
Caroline Wee, Institute of Molecular and Cell Biology (A∗STAR), SingaporeCopyright © 2023 Ma, Brunal, Clark, Studtmann, Stebbins, Higashijima and Pan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Y. Albert Pan, eWFwYW5AdnRjLnZ0LmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.