
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell Dev. Biol. , 01 February 2023
Sec. Stem Cell Research
Volume 11 - 2023 | https://doi.org/10.3389/fcell.2023.1111705
This article is part of the Research Topic Cell-Based Neurodegenerative Disease Modeling View all 6 articles
The generation of midbrain dopaminergic neurons (mDAs) from pluripotent stem cells (hPSC) holds much promise for both disease modelling studies and as a cell therapy for Parkinson’s disease (PD). Generally, dopaminergic neuron differentiation paradigms rely on inhibition of smad signalling for neural induction followed by hedgehog signalling and an elevation of β-catenin to drive dopaminergic differentiation. Post-patterning, differentiating dopaminergic neuron cultures are permitted time for maturation after which the success of these differentiation paradigms is usually defined by expression of tyrosine hydroxylase (TH), the rate limiting enzyme in the synthesis of dopamine. However, during maturation, culture media is often supplemented with additives to promote neuron survival and or promote cell differentiation. These additives include dibutyryl cyclic adenosine monophosphate (dbcAMP), transforming growth factor β3 (TGFβ3) and or the γ-secretase inhibitor (DAPT). While these factors are routinely added to cultures, their impact upon pluripotent stem cell-derived mDA phenotype is largely unclear. In this study, we differentiate pluripotent stem cells toward a dopaminergic phenotype and investigate how the omission of dbcAMP, TGFβ3 or DAPT, late in maturation, affects the regulation of multiple dopaminergic neuron phenotype markers. We now show that the removal of dbcAMP or TGFβ3 significantly and distinctly impacts multiple markers of the mDA phenotype (FOXA2, EN1, EN2, FOXA2, SOX6), while commonly increasing both MSX2 and NEUROD1 and reducing expression of both tyrosine hydroxylase and WNT5A. Removing DAPT significantly impacted MSX2, OTX2, EN1, and KCNJ6. In the absence of any stressful stimuli, we suggest that these culture additives should be viewed as mDA phenotype-modifying, rather than neuroprotective. We also suggest that their addition to cultures is likely to confound the interpretation of both transplantation and disease modelling studies.
Parkinson’s disease (PD) is a progressive nervous system disorder with multiple impacts across the CNS. The characteristic motor impairments of PD, including bradykinesia, rigidity, and resting tremor, have been linked to the loss of the A9 dopaminergic neurons of the substantia nigra pars compacta. Midbrain dopaminergic (mDA) neurons offer promise for both Parkinson’s disease treatment and disease modelling. As a consequence, a number of protocols for differentiating mDAs have been proposed, all with a basic approximation of the embryonic developmental processes (Fasano et al., 2010; Kriks et al., 2011; Takazawa et al., 2012; Qi et al., 2017). Thus, neural induction is followed by midbrain patterning using sonic hedgehog (SHH) and Wnt signalling activators/mimetics. The resultant dopaminergic neurons have been shown to efficiently engraft in animal models of Parkinson’s disease (Fasano et al., 2010; Kriks et al., 2011; Qi et al., 2017) as well as provide a platform for studies of disease mechanisms (Haynes et al., 2021). Commonly, the authentication of mDA cultures is reliant upon the expression of a handful of genes that are essential for development and maturation. Thus, early in differentiation, developing cultures should express floor plate markers such as Foxa2 and Otx2, followed by En1/2 and Lmx1a (Nakatani et al., 2010; Aguila et al., 2014; Arenas et al., 2015). As neurons mature, markers such as Nr4a2, Pitx3 and Th (Nunes et al., 2003; Volpicelli et al., 2012) and finally, the dopamine transporters Slc18a2 and Slc6a3 (Miller et al., 1999; Ásgrímsdóttir and Arenas, 2020) are expressed. In addition, mDA neuron markers such as Kcnj6, Aldh1a1 and Sox6, and Calb1 and Otx2 are used to discriminate between substantia nigra and ventral tegmental area dopaminergic neurons, respectively. During maturation, multiple neurotrophic/survival factors are routinely added to the cultures. These additives include brain-derived neurotrophic factor (BDNF) (Binder and Scharfman, 2004), glial cell-derived neurotrophic factor (GDNF) (Rakowicz et al., 2002), dibutyryl cyclic AMP (dbcAMP) (Mena et al., 1995), N-[N-(3,5-difluorophen-acetyl)-l-alanyl]-S-phenylglycine t-butyl ester (DAPT) (Crawford and Roelink, 2007) and transforming growth factor (commonly TGFβ3) (Luo et al., 2016). BDNF and GDNF are neurotrophic factors known to enhance the survival and differentiation of neural progenitor cells toward the dopaminergic phenotype, protect injured nigrostriatal neurons, and stimulate dopamine turnover and release in rescued neurons (Rakowicz et al., 2002; Binder and Scharfman, 2004). DbcAMP, a cell-permeant analogue of cyclic AMP, has been shown to increase the number of Th-positive cells in fetal midbrain cultures without necessarily increasing neuron survival (Mena et al., 1995). Cultures may also contain DAPT, a Notch signalling inhibitor that blocks γ-secretase to promote differentiation of neurons from human embryonic stem cells (Crawford and Roelink, 2007). Previous work has shown that Notch signalling has no function in the specification of mesencephalic dopaminergic neural precursor cells but plays a vital role in regulating their expansion and differentiation into neurons (Trujillo-Paredes et al., 2016). Lastly, TGFβ3 is a member of the transforming growth factor-β superfamily of multifunctional cytokines that is commonly added to cultures, presumably to stimulate dopaminergic neuron survival (Flanders et al., 1998). The signalling is TGFβ receptor-mediated, involving the Smad and p38 mitogen-activated protein kinase pathways (Airaksinen and Saarma, 2002). Multiple lines of evidence suggest signalling of classic neurotrophic factors such as BDNF and GDNF may be affected by TGFβ (Schober et al., 2007; Luo et al., 2016) which also plays a critical role in astrocyte and microglial function under physiological and injury conditions (Bialas and Stevens, 2013; Butovsky et al., 2014). While these factors are routinely added for developing mDA cultures, their ability to affect cultures beyond commonly under-justified, short-term effects on survival is unclear.
In this study, we maintain mature mDA neurons in the presence of GDNF, BDNF, dbcAMP, DAPT, and TGFβ3 and assess the impact that the removal of dbcAMP, DAPT or TGFβ3 has upon mDA neuron culture development. We now show that a loss of dbcAMP downregulates transcripts associated with a neuronal midbrain phenotype (EN1, TH, PITX3, but not NR4A2; SLC6A3 or SLC18A2). In contrast, DAPT removal downregulates MSX2, OTX2, KCNJ6 and nestin while upregulating the astrocyte markers GFAP and S100B. TGFβ3 removal regulates the expression of early markers (OTX2, MSX2 and NEUROD1), as well as TH and the astrocyte marker S100B. In conclusion, we found that culture additives play a role in maintaining what might be construed as a mature mDA phenotype. We speculate that additive-induced promotion of a dopaminergic phenotype may i) mask inefficient patterning and therefore contribute to the equivocal outcomes of transplantation therapies and ii) impact the use of these neurons as disease models.
A modified H9 pluripotent stem cell line where eGFP was expressed under the control of LMX1A promoters (LMX1A-eGFP) (Niclis et al., 2017) was used to track the expression of LMX1A. All plates or flasks were pre-coated with 0.5 µg/cm2 Laminin-521 (Life Technologies, Australia). Undifferentiated cells were grown using a slightly modified method of (Watmuff et al., 2015). Briefly, pluripotent stem cells were seeded at a density of 3,000 cells per cm2 and cultivated in Essential 8 Medium with Essential 8 supplement (Thermo Fisher Scientific, Australia) and Penicillin/Streptomycin (Life Technologies, Australia) at 37°C in a humidified incubator containing 5% CO2. Cells were passaged at ∼80% confluence (ReLeSR™, Stemcell technologies, Australia). To enhance the survival of cells seeded at low-density, the ROCK inhibitor (10 μM Y-27632) was added to the medium for the first 24 h after passaging. Cells were maintained for a maximum 10 passages.
On day one of differentiation, the culture medium was replaced with differentiation medium 1 (D1): Knockout DMEM (Life Technologies, Australia), 15% knockout serum replacement (KSR, Life Technologies, Australia), 1% MEM Non-Essential Amino Acids Solution (Life Technologies, Australia), 2 µM GlutaMAX-I (Life Technologies, Australia), 100U/mL Penicillin/Streptomycin (Life Technologies, Australia) and 0.1 mM β-mercaptoethanol (Life Technologies, Australia) on Matrigel™ (BD Biosciences, Australia) until fully confluent (around 1.5 × 105 cells/cm2). From day five of differentiation D1 was gradually changed to differentiation medium 2 (D2): DMEM: F12 (Life Technologies, Australia), N2 supplement (Life Technologies, Australia), 100U/mL Penicillin/Streptomycin, 50 mg/mL human apo-transferrin (Sigma Aldrich) and 0.48 mg/mL human insulin (Sigma Aldrich). The change in media occurred over 6 days, with the D1:D2 ratios of 3:1 (days five and six), 1:1 (days seven and eight) and 1:3 (days nine and ten). From day 11 onwards, media was changed to neurobasal medium (NBM): Neurobasal® medium (Thermo Fisher Scientific, Australia); B27 supplement (Life Technologies, Australia); 100 U/mL Penicillin/Streptomycin; 2 mM GlutaMAX-I; 20 ng/mL recombinant GDNF (Lonza Peprotech, Australia), 20 ng/mL recombinant BDNF (Lonza Peprotech, Australia), 200 nM ascorbic acid (Sigma Aldrich, Australia). Specific cultures also contained combinations of 10 nM DAPT (Jomar Life Research, Australia), 0.5 mM dbcAMP (Sigma Aldrich, Australia) and 1 ng/mL recombinant TGF-β3 (Lonza Peprotech, Australia). Additive removal occurred from Day 40 onwards.
On day 65 of differentiation, cells were fixed with 4% paraformaldehyde and permeabilized with permeabilization solution 1 containing 0.1% (v/v) x-100, 0.1% (v/v) Tween-20, 20% (v/v) Hybri-Max dimethylsulphoxide (DMSO) in DPBS without calcium or magnesium (DPBS −/−). Then further incubated with permeabilization solution 2 containing 0.1% (v/v) Tween-20, 0.1% (v/v) Triton X-100, 0.1% deoxycholate, 0.1% tergitol solution, 20% (v/v) DMSO in DPBS (−/−). Cells were blocked with 3% (v/v) donkey serum in permeabilization solution 2 following incubation with primary antibody rabbit anti-PITX3 (1:100 Invitrogen), sheep anti-TH (1:500 Abcam), goat anti-GIRK2 (1:100 Abcam), rabbit anti-WNT5A (1:200 Abcam), mouse anti-TUBB3/chicken anti-MAP2 (1:1000 Abcam) in 3% donkey serum (1:100 dilution) overnight at 4°C. Then cells were stained with a secondary antibody (Supplementary Table S1) in 3% donkey serum (1:1000 dilution) for 4 h at room temperature. On the day of imaging, cells were stained with DAPI (1:5000 dilution) in DPBS (−/−). Fluorescence was detected using a Nikon A1R Confocal Microscope.
On day 65 of differentiation, total RNA was extracted from cultures using the Bioline RNA Micro Kit (Bioline, Australia) according to the manufacturer’s instructions. Samples were analyzed for RNA content using a Nanodrop ND-1000 (Thermo Fisher, Australia) spectrophotometer. RNA samples were converted to cDNA using SensiFAST™ cDNA Synthesis Kit (Bioline, Australia). cDNA samples were then diluted with an appropriate amount of DNase RNAse-free water and TaqMan® probes and mixed with SensiFAST Probe No-ROX Kit. RT-qPCR was performed in triplicate using the CFX96 Real-Time PCR detection system. HPRT1 and TBP were used as reference genes. Relative gene expression was expressed as target gene Ct values to reference genes. A list of Taqman probes is shown in Supplementary Table S2, while Supplementary Table S3 shows delta delta Ct values for each replicate.
Cells were lysed in RIPA lysis buffer (Thermo Fisher, Australia). Protein concentrations were determined by BCA assay (Thermo Fisher, Australia). Then, 20 ug protein was resolved using 4%–12% SDS polyacrylamide gel and transferred to a PVDF membrane (Thermo Fisher, Australia). The membranes were incubated overnight at 4 °C in a blocking solution containing 5% (w/v) non-fat dry milk in PBS with 0.1% Tween-20. Subsequently, membranes were incubated with primary antibodies, followed by an incubation either with fluorescence or horseradish peroxidases-secondary antibody using SuperSignal West Pico Chemiluminescent Substrate (Pierce, Thermo Fisher). GAPDH was used as an internal loading control. Detection of fluorescence was performed by Amersham™ Typhoon™ Biomolecular Imager (GE Healthcare) and HPR was performed by Gel Doc TM (BioRad). Quantitative data analysis was performed using ImageJ. All bands were normalized to GAPDH expression.
Cell fluorescence was thresholded using ImageJ (Supplementary Table S4) to enable estimations of cell protein fluorescence: DAPI fluorescence ratios.
Using MotifMap (Daily et al., 2011), we identified potential gene regulatory motifs, an arbitrary 10,000 base pairs up and 2000 base pairs downstream of the transcription start sites with FDR ≤0.5. We sorted the list of putative binding sites by the number of sites (the highest number came first in the list) and analyzed the common transcription putative binding site of several genes associated with mDA development whose expression is affected by additives added during mDA neuron maturation (Supplementary Table S5).
Unless otherwise stated, results from experiments are presented as the mean ± standard error of the mean (SEM) of at least three biological replicate experiments (each with at least two technical replicates). Statistical analysis was performed on RT-qPCR raw data with a one-sample t-test, or One-way ANOVA, followed by post hoc Dunnett’s test to show effects versus control for immunocytochemistry results. All analyses were performed using PRISM v8.00 (GraphPad Software, CA, United States). In all cases, a p < 0.05 was considered significant.
We studied the effect of the common tissue culture additives dbcAMP, TGFβ3 and DAPT upon the regulation of key dopaminergic neuron phenotype markers of early and late development, as well as A9 subtype mDA neuron specification. We maintained mature mDA neurons in “normal” media (i.e., in the presence of GDNF, BDNF, dbcAMP, DAPT, and TGFβ3) and assessed, at day 65 of differentiation, the impact that removal of dbcAMP, DAPT or TGFβ3 had upon genes expressed early in dopaminergic development (FOXA2, MSX2, LMX1A), in maturing (PITX3, NR4A2, TH) and mature cultures (SLC18A2, SLC6A3). We also looked for changes to the general character of the cultures (nestin, TUBB3), astrocyte markers (GFAP, S100B) and endogenous WNTs (WNT1, WNT5A).
The absence of dbcAMP, a membrane-permeable analogue of cyclic-AMP, promoted significant changes across dopaminergic neuron-containing cultures. Notably, decreased expression of a number of markers associated with dopaminergic neuron development and maturation, including EN1, EN2, PITX3, and most notably, TH, with no change in NR4A2, SLC6A3, and SLC18A2 expression. There was also a small decrease in a generic neural marker TUBB3 (Figure 1).
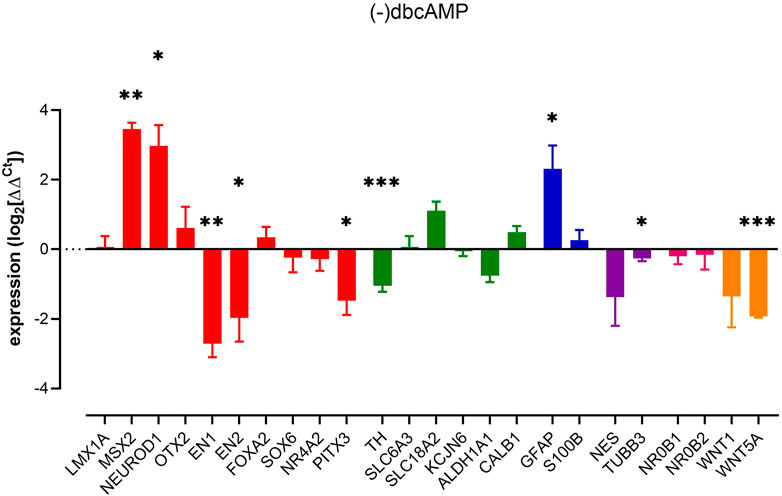
FIGURE 1. Impact of removal of dbcAMP upon pluripotent stem cell-derived dopaminergic neuron cultures. Columns show mean (+s.e. mean) data normalized to HPRT1 and TBP and expressed as fraction of (+) dbcAMP control (from matched differentiations). Statistical analysis was via one sample t-tests (n = 3–6 biological replicates). Graphs show mean +/- s. e. mean changes; * = p < 0.05, ** = p < 0.01 and *** = p < 0.001.
Given the roles that Wnt1 and Wnt5a play in regulating the genes involved in mDA development (Castelo-Branco et al., 2003; Prakash et al., 2006; Bryja et al., 2007; Joksimovic et al., 2009; Andersson et al., 2013), we assessed the expression of these WNT family ligands, WNT1 and WNT5A. RT-qPCR analysis showed significant downregulation of WNT5A (β-catenin-independent) but not β-catenin-dependent WNT1 (Figure 1). That WNT5A can be regulated by removal of dbcAMP likely indicates an endogenous WNT signalling present in mature cultures.
Other interesting observations were the elevations of MSX2, NEUROD1 and GFAP brought on by the removal of dbcAMP. The increase of GFAP, a type III intermediate filament protein present in mature astrocytes (Brenner and Messing, 2021) indicated an increase in reactive gliosis. This is consistent with previous studies showing that many astroglia genes are differentially regulated by cAMP signalling (Horvat and Vardjan, 2019), including the downregulation of reactive astrocyte markers (Paco et al., 2016).
Next, we investigated the impact of removal of TGFβ3 from our cultures. We observed changes in several key markers, often, but not exclusively associated, with neuronal-lineage commitment. Thus, we observed upregulations of nestin, MSX2, NEUROD1, OTX2, and SOX6. In contrast, we observed reduced expression of markers commonly associated with a more committed dopaminergic phenotype, FOXA2 and TH (Figure 2). Interestingly, the astrocyte-related gene S100B was increased significantly without an increase in the reactive astroglial marker GFAP, possibly indicating astroglial genesis rather than activation. The removal of TGFβ3, similar to the removal of dbcAMP, led to a decrease in both WNT5A and TH expression and a profound loss of the neural progenitor marker, nestin (Figure 2).
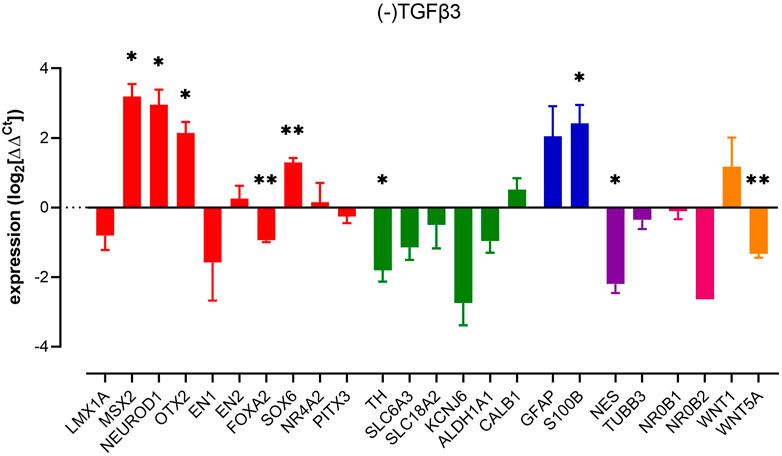
FIGURE 2. Impact of removal of TGFβ3 upon pluripotent stem cell-derived dopaminergic neuron cultures. Columns show mean (+s.e. mean) data normalized to HPRT1 and TBP and expressed as fraction of (+) TGFβ3 control (from matched differentiations). Statistical analysis was via one sample t-tests (n = 3 biological replicates). Graphs show mean +/- s. e. mean changes; * = p < 0.05, ** = p < 0.01 and *** = p < 0.001.
The removal of DAPT (activating Notch signalling) increased the expression of MSX2, OTX2 and astrocyte markers GFAP and S100B. In contrast, EN1, nestin and KCNJ6 showed reduced expression (Figure 3). These data indicate that, just as with the removal of dbcAMP and TGFβ3, the factors that may be construed to be part of the regulatory framework of dopaminergic neuron phenotype can be impacted selectively, and late in differentiation.
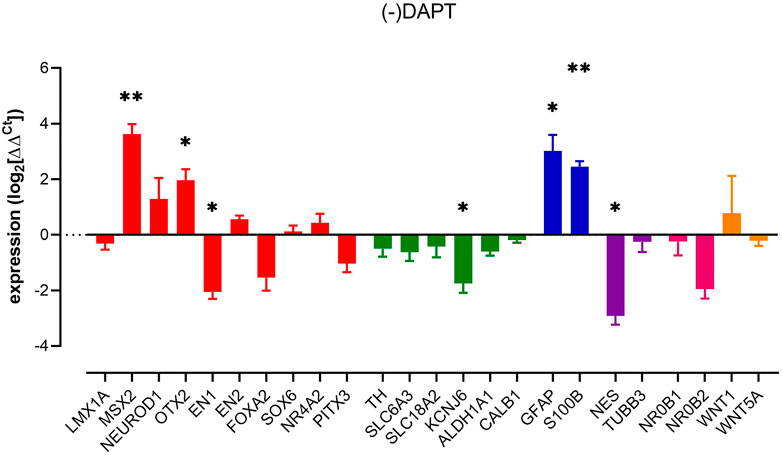
FIGURE 3. Impact of removal of DAPT upon pluripotent stem cell-derived dopaminergic neuron cultures. Columns show mean (+s.e. mean) data normalized to HPRT1 and TBP and expressed as fraction of (+) DAPT control (from matched differentiations). Statistical analysis was via one sample t-tests (n = 3 biological replicates). Graphs show mean +/- s. e. mean changes; * = p < 0.05, ** = p < 0.01, and *** = p < 0.001.
Increased expression of GFAP and S100B in our culture was anticipated as the Notch pathway has a prominent role in controlling neuronal morphology and determining astrocyte fate (Wang et al., 2019). In addition, DAPT suppresses the activation of astrocytes (Wang et al., 2019), (Qian et al., 2019) leading us to believe that notch signalling contributes to astrocyte function in this culture paradigm.
We undertook fluorescence imaging (eGFP) along with immunolabelling and western blotting to establish changes in protein expression.
Analysis of LMX1A-eGFP expression showed bright widespread eGFP across cultures, not necessarily localized to TUBB3-MAP2 immunolabelling, and not greatly impacted by removal of DAPT, dbcAMP or TGFβ3 (Figures 4A–D). Quantification of eGFP fluorescence (expressed as a percentage of DAPI fluorescence) showed that, consistent with the RT-qPCR studies, the removal of additives has little impact upon LMX1A-eGFP fluorescence (Figure 4E). In contrast PITX3 protein expression (Figure 4F shows typical blotting) was particularly impacted by removal of DAPT and dbcAMP (Figure 4G).
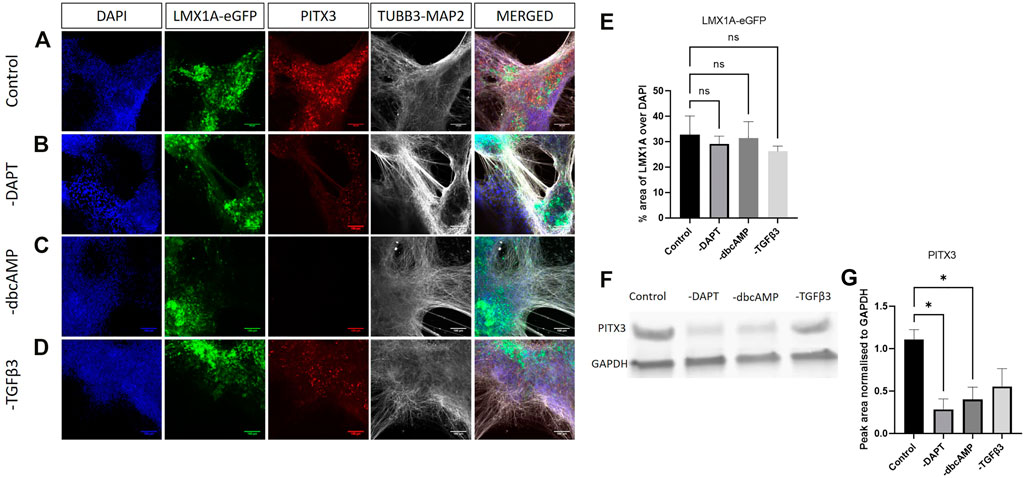
FIGURE 4. Immunolabelling of PITX3, TUBB3/MAP2 in LMX1A-eGFP cultures. Panel (A) shows, from left to right, DAPI nuclear labelling (blue), LMX1A-eGFP fluorescence (green), PITX3 (red) and TUBB3/MAP2 in the same channel (white) and a color combined image (merged). Panels (B–D) show the corresponding data, but following cultivation in the absence of DAPT, dbcAMP and TGFβ3, respectively. Scale bar indicates 100 μm. Panel (E) shows LMX1A-eGFP fluorescence as a fraction of DAPI fluorescence (n = 3). Panel (F) shows a typical Western blot while panel G shows changes in PITX3 expression following removal of additives (n = 3). Bar graphs show mean +/- s. e. mean; * = p < 0.05.
We labelled for TH to observe changes in expression and evidence of colocalization with LMX1A-eGFP positive cells following the removal of culture additives (Figures 5A–H, show lower magnification images of the same fields of view). While the higher magnification images showed, regardless of additive removal, the presence of TH immunofluorescence, low power magnification indicated a widespread reduction of TH across cultures in response to the removal of dbcAMP (Figure 5G). This finding was confirmed using immunoblotting (Figure 5I shows typical labelling) that showed a loss of TH protein across cultures following the removal of dbcAMP and TGFβ3 (Figure 5J).
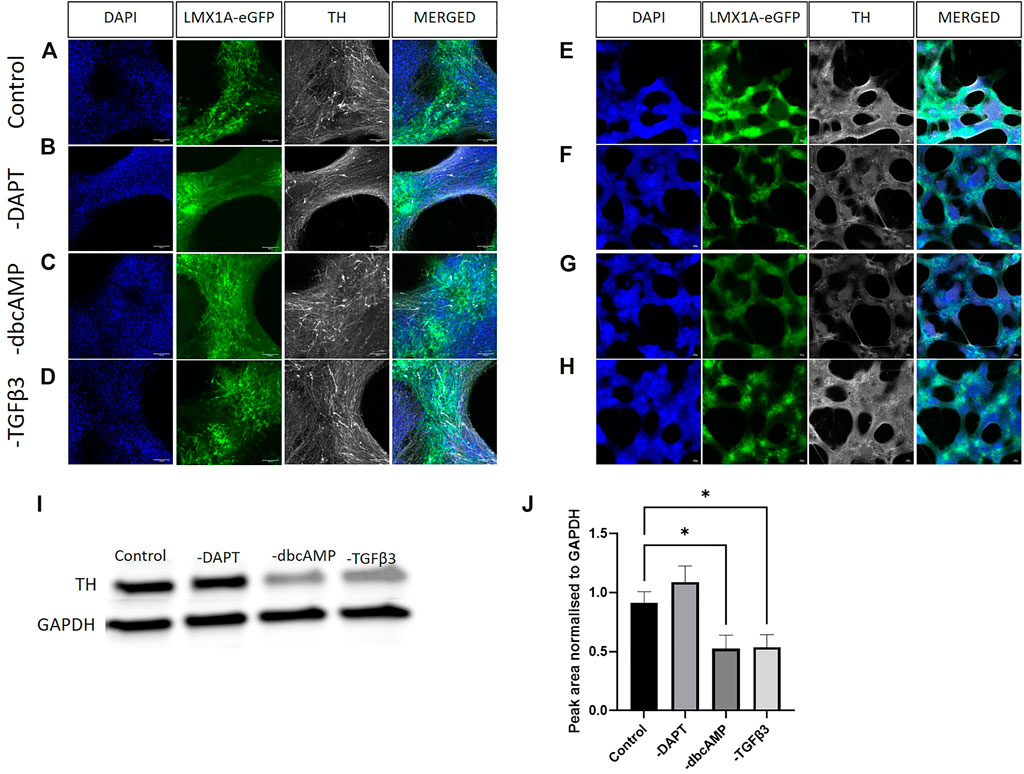
FIGURE 5. Immunolabelling of tyrosine hydroxylase in LMX1A-eGFP cultures. Panels [(A–D), ×20 objective) show, from left to right, DAPI nuclear labelling (blue), LMX1A-eGFP fluorescence (green), TH (white) in control (panel A), and after removal of DAPT [panel (B)], dbcAMP [panel (C)] and TGFβ3 [panel (D)], respectively. Panels (E–H) show corresponding low power images (×4 objective). Scale bar indicates 100 μm. Panel (I) shows typical Western blotting, while panel (J) shows quantification of TH (n = 3) following the removal of additives. Bar graphs show mean +/- s. e. mean; * = p < 0.05.
The widely used marker of A9 phenotype, KCNJ6, showed abundant expression in neuron-like cells across cultures, although the strongest expression, was not particularly colocalized with LMX1A-eGFP (Figure 6A). KCNJ6 expression was, in contrast with RT-qPCR data, impacted by the removal of all additives (Figures 6A–E, shows thresholding analysis of KCNJ6 fluorescence as a fraction of DAPI labelling).
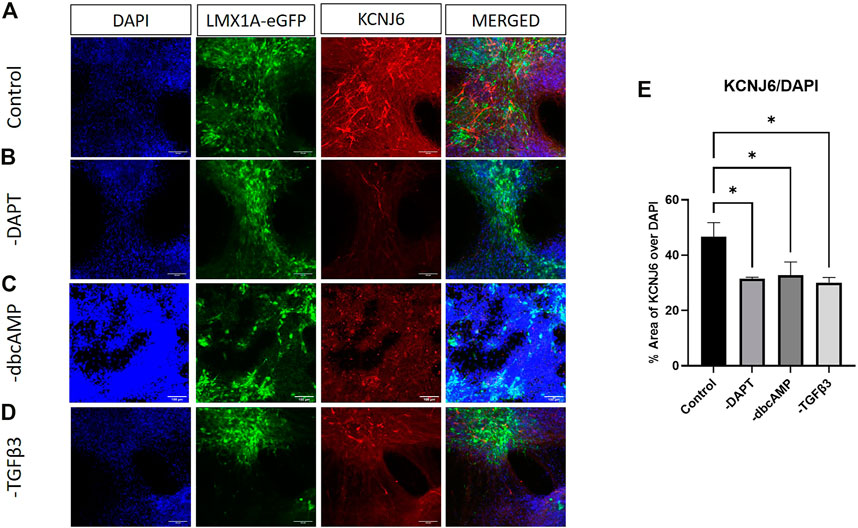
FIGURE 6. Immunolabelling of KCNJ6 in LMX1A-eGFP cultures. Panel (A) shows, from left to right, DAPI nuclear labelling (blue), LMX1A-eGFP fluorescence (green), KCNJ6 (red) and a color combined image (merged). Panels (B–D) show the corresponding data, but following cultivation in the absence of DAPT, dbcAMP and TGFβ3, respectively. Scale bar indicates 100 μm. Panel (E) shows KCNJ6 fluorescence as a fraction of DAPI fluorescence (n = 3). Bar graphs show mean +/- s. e. mean; * = p < 0.05.
GFAP and S100B showed considerable colocalization in cultures, although the most intense GFAP labelling was usually associated with lower levels of S100B expression (Figure 7A). Increases in GFAP in response to the removal of DAPT, dbcAMP or TGFβ3 were clearly evident in cultures (Figures 7B–D). There were widespread elevations of S100B apparent after the removal of DAPT and TGFβ3 (Figure 7E).
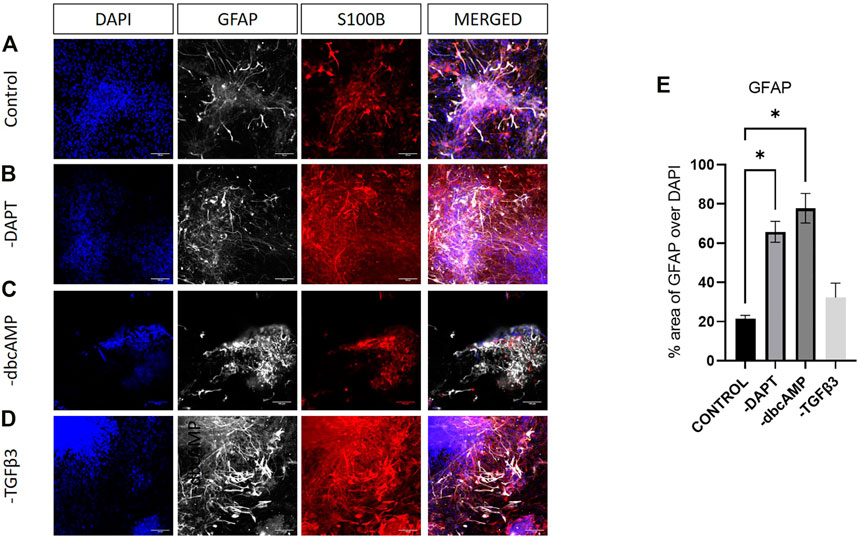
FIGURE 7. Immunolabelling of GFAP and S100B in LMX1A-eGFP cultures. Panel (A) shows, from left to right, DAPI nuclear labelling (blue), GFAP (white), S100B (red) and a color combined image (merged). Panels (B–D) show the corresponding data, but following cultivation in the absence of DAPT, dbcAMP and TGFβ3, respectively. Scale bar indicates 100 μm. Panel (E) shows GFAP fluorescence as a fraction of DAPI fluorescence (n = 3). Bar graphs show mean +/- s. e. mean; * = p < 0.05.
Given the modulation of WNT5A by removing dbcAMP and TGFβ3, we looked for WNT5A protein in cultures. Surprisingly WNT5A expression was both intense and widespread across cultures, evident in LMX1A-eGFP positive and other cell types, but largely restricted to nuclear regions (Figure 8A). WNT5A expression was clearly reduced following the removal of dbcAMP and TGFβ3 (Figures 8B–D). Quantifying culture-wide fluorescence showed reductions in WNT5A protein expression following the removal of all additives (Figure 8E).
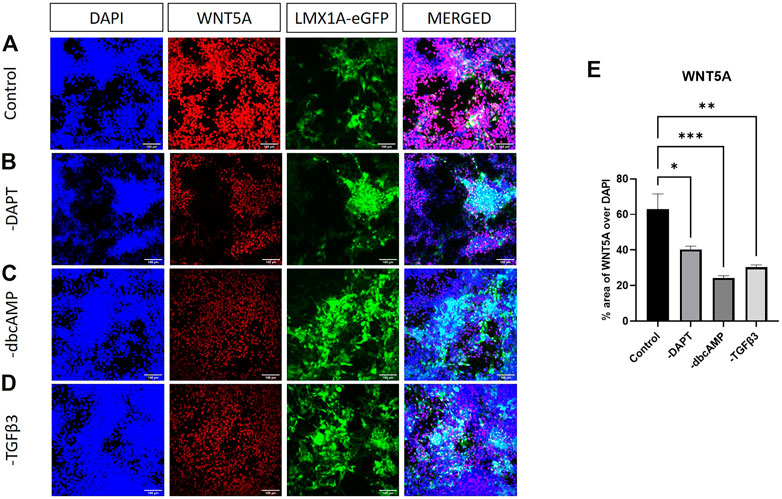
FIGURE 8. Immunolabelling of WNT5A in LMX1A-eGFP cultures. Panel (A) shows, from left to right, DAPI nuclear labelling (blue), WNT5A (red), LMX1A-eGFP fluorescence (green) and a color combined image (merged). Panels (B–D) show the corresponding data, but following cultivation in the absence of DAPT, dbcAMP and TGFβ3, respectively. Panel (E) shows WNT5A fluorescence as a fraction of DAPI fluorescence (n = 3). Scale bar indicates 100 μm. Bar graphs show mean +/- s. e. mean; * = p < 0.05, ** = p < 0.01, and *** = p < 0.001.
We turned to MotifMap analysis to investigate candidate regulatory motif sites involved in the changes of transcript observed in the culture. The top five motifs with FDR ≤0.5 (10,000 upstream and 2000 downstream, Supplementary Table S4) ranked by a number of sites revealed common putative transcription factor binding sites associated with the mDA development, namely NEUROD1, MZF 1_4, HNF4A, LEF-1 and YY1. We then searched for motifs mainly involved in the cAMP pathway, the SMAD pathway and the Notch signalling pathway in the hope of revealing candidate motifs affected by the additives. Intriguingly, SLC18A2 and NR4A2, which possess many putative CREB binding sites, seem unaffected by the absence of dbcAMP. Conversely, EN1 and PITX3, which possess few putative CREB binding sites, were most affected. Moreover, none of the genes was affected by the removal of TGFβ3 (e.g. FOXA2, MSX2, NESTIN, NEUROD1, TH, WNT5A) which have many putative SMAD binding sites, the main signal transducer for receptors of the transforming growth factor beta superfamily. Similarly, the genes affected by blocking notch signalling, MSX2, OTX2, KCNJ6, GFAP, S100B, and nestin, possess few putative RBPJ binding sites.
The acquisition of functional maturity by postmitotic neurons relies on the complex interactions of multiple intracellular signalling pathways (Lepski et al., 2011). In vitro, supplements and growth factors (i.e., dbcAMP, TGFβ3 and DAPT) are driving these pathways to ensure the survival of the neurons and/or accelerate maturation/survival (Kriks et al., 2011; Kirkeby et al., 2012; Arenas et al., 2015). Indeed, our earlier work has shown how these cultures develop over time and the profound impact that dysregulation of WNT signalling has upon phenotype (Haynes et al., 2021). However, in spite of an extensive amount of empirical data, the factors involved in maturation, as well as the general mechanisms regulating the timing of neuronal maturation, are unclear (Takazawa et al., 2012; Telias et al., 2014). In this study, we patterned and cultured mDA conventionally for 40 days prior to the removal particular additives. We then analyzed the expression of early and late markers of neuron and glial identity by RT-qPCR, Western blotting and immunostaining at day 65 of differentiation, when cultures are largely mature (Watmuff et al., 2015).
The most consistent observation following the removal of dbcAMP, TGFβ3 or DAPT was an increase in expression of the transcriptional repressor, MSX2. MSX2/Msx2 (muscle segment homeobox 2) is a homeobox transcription factor of the msh family critical for mesendoderm differentiation. It is a direct target gene of the BMP pathway and can be synergistically activated by Wnt signals via LEF1 during mesendoderm induction (Wu et al., 2015). Msx2 is expressed in mouse neural progenitors in the developing ventricular zone (La Manno et al., 2016) in the dopaminergic subset of Lmx1a-positive neural progenitors (Kee et al., 2017). MSX2 regulates various cellular processes including cellular proliferation and differentiation during embryonic development (Han et al., 2007; Bernal and Arranz, 2018). These observations are consistent with work showing that Notch1 activity inhibits BMP signalling at the level of Msx2 expression in embryonic cerebellar development (Machold et al., 2007) and that Notch signalling induces direct transcriptional activation of the MSX2 gene (Shimizu et al., 2009). In the developing nervous system, the Notch pathway has a prominent role in controlling neuronal morphology and determining astrocyte fate (Wang et al., 2019), which directs an irreversible switch from neurogenic to gliogenic activity (Morrison et al., 2000). Such activity may explain the elevations of S100B and GFAP following the removal of DAPT from cultures.
The removal of both dbcAMP and TGFβ3 elevated the expression of NEUROD1. NEUROD1 is a key regulator of neuron formation and function, it is expressed from early in mDA development into adulthood (Mesman et al., 2018). Its expression is enhanced directly by forskolin (Paldino et al., 2014) but, in our hands, the loss of dbcAMP promotes an elevation of NEUROD1. However, while this WNT-signalling-dependent transcription factor is essential for neural progenitor survival and neurogenesis (Kuwabara et al., 2009), it is also required for ERK-regulated pro-neural activity (Lee et al., 2020); indicating multiple levels of regulatory control. Consistent with our work, both NeuroD1 and Msx2 can be regulated by β-catenin-dependent WNT signalling (Kuwabara et al., 2009), and the marked decrease in expression of WNT5A in response to the removal of TGFβ3 and dbcAMP may be consistent with increased β-catenin-dependent signalling in these cultures, as proposed previously (Haynes et al., 2021).
While DAPT is routinely included in cultures to encourage differentiation, dbcAMP to increase TH expression (sometimes taken for promoting survival) and TGFβ3 to improve survival, the ability of these three factors to regulate markers of an immature phenotype; MSX2, NEUROD1 and nestin indicate that their removal, even at late stages of culture, has profound effects upon culture phenotype. Perhaps most significantly, with respect to TGFβ3 and DAPT, removal of these factors may lead to increased glial commitment while possibly also contributing to other markers of phenotype specification.
Generally, the floor plate markers LMX1A, FOXA2, EN1/2 and OTX2 (Nakatani et al., 2010; Aguila et al., 2014; Arenas et al., 2015) were robustly expressed throughout differentiation up until termination at ∼ day 65. Overall, LMX1A was the least impacted transcription factor as it was unaffected by any culture manipulation. We have, however, already shown LMX1A expression to be negatively influenced by the removal of β-catenin signalling (Haynes et al., 2021) which is broadly consistent with the idea that endogenous WNT, or at least β-catenin signalling networks are intact and functional in these cultures. FOXA2 was, however, positively under the influence of TGFβ3 since its removal promoted a loss of transcript, a finding consistent with SMAD-dependent changes in Foxa2 (Cai et al., 2013). This downregulation of FOXA2 was anticipated as previous studies have shown TGFβ signaling regulates the expression lncRNA DEANR1 (Definitive Endoderm Associated long Non-coding RNA1) by positively regulating endoderm factor FOXA2 expression (Mullen and Wrana, 2017).
In contrast to FOXA2, the engrailed genes (EN1 and EN2) were unaffected by the removal of TGFβ3 signalling but greatly impacted by the loss of dbcAMP. These genes are key players in mDA neuron patterning, axonal guidance, neuron specification and maintenance (Alves dos Santos and Smidt, 2011) (Danielian and McMahon, 1996). These genes are generally considered as WNT-dependent transcription factors (Danielian and McMahon, 1996) and are expressed in multiple dopaminergic neuronal subtypes (La Manno et al., 2016). There is, however, significant overlap between WNT and protein kinase A signalling since PKA phosphorylation of β-catenin at Ser522, a canonical WNT signalling independent phosphorylation site (Luckert et al., 2011) promotes β-catenin accumulation (Cognard et al., 2013). En1 (along with Wnt1) expression has been shown to be dependent upon Ras activation (Vennemann et al., 2008), a finding supported by the direct induction of En1 expression by FGF8 (Harada et al., 2016), EN1 expression can also be induced by TGFβ in a SMAD3-dependent manner (Gyorfi et al., 2021). Although TGFβ3 had no effect upon EN1 expression in our cultures, protein kinase A can facilitate ERK signalling through phosphorylation of the scaffold protein, KSR-1 (Smith et al., 2012), indicating a potential for TGFβ3 pathway-dependent expression to be modulated by PKA (i.e., dbcAMP). That EN2 could be regulated only by the removal of dbcAMP may indicate that similar mechanisms to those regulating EN1 are at play. However, one major point of difference is that EN1, but not EN2, is also downregulated by the removal of DAPT and presumably the activation of notch signalling. While this may be consistent with the differences in putative transcription factor binding site regulation of EN1 and EN2 expression, how this occurs is not immediately clear.
Lastly, OTX2 expression was downregulated by the removal of DAPT and TGFβ3. OTX2 can be driven by both notch signalling (Dvoriantchikova et al., 2015) and also by MAPK (Hongo and Okamoto, 2022). Moreover, a recent study reported reduced OTX2 expression in iPSC-derived spheroids incubated with WNT and DAPT (CHIR + DAPT) (Bejoy et al., 2020). While we observed no change in the expression of WNT1, previous literature reported that the Notch modulator Lfng and the Notch ligand Ser1 are also expressed in a compartment-restricted fashion at the midbrain-hindbrain boundary (MHB), and have a function in stabilization of the boundary and in the regulation of expression of the organizer genes Fgf8 and Wnt1 (Tossell et al., 2011). Whether these pathways act independently or interact to control expression is unknown.
These studies indicate that cultures maintain the expression of early markers of neural and floorplate phenotypes. While these markers are generally thought of as early factors in differentiation programs, they do persist late in cultures, and their expression patterns are modified by the removal of a variety of factors which broadly impacts the overall phenotype of cultures.
Even though SOX6 appears early in development it is often thought of for its association with the A9 subset of dopaminergic neurons (OTX2 is often considered in association with A10 neurons) (Panman et al., 2014). That SOX6 expression increases after the removal of TGFβ3 is somewhat at odds with the idea that SOX6 is an important downstream mediator of BMP-2 signalling in chondrogenesis (Fernández-Lloris et al., 2003), although we have previously shown SOX6 expression to be regulated by WNT signalling (albeit in the presence of TGFβ3, dbcAMP and DAPT) (Haynes et al., 2021).
PITX3 and NR4A2 are more typically expressed later in differentiation and in both A9 and A10 neurons (Mesman and Smidt, 2020). There is evidence of a great deal of interplay between PITX3, EN1 and NR4A2. Pitx3 has been proposed as a regulator of mDA (Saucedo-Cardenas et al., 1998; Lebel et al., 2001) and may cooperate with Nr4a2 to promote survival and terminal differentiation (Nunes et al., 2003; Jacobs et al., 2009; Chakrabarty et al., 2012), although other studies have also shown that Nr4a2 knockout has no influence on Pitx3 expression, suggesting independence of the two pathways (Zetterstrom et al., 1997; Castillo et al., 1998; Saucedo-Cardenas et al., 1998). Consistent with the idea of pathway independence we saw no reduction of NR4A2 expression when PITX3 was reduced following the removal of dbcAMP. Thus, while NR4A2 is thought to be necessary in establishing a Th-positive dopaminergic phenotype (Zetterstrom et al., 1997; Saucedo-Cardenas et al., 1998) our evidence supports the idea that PITX3 and NR4A2 expression occur independently; at least with respect to effects of dbcAMP. En1−/− mice embryos show (rostrolateral but not mediocaudal) reductions in Pitx3, Th and Slc6a3, but no changes in Nr4a2; Veenvliet and others speculated that En1 and Pitx3 cooperatively drive expression of Nr4a2 target genes Slc6a3 and Th in a rostrolateral subset of mdDA neurons but where Pitx3 represses some En1-driven genes (Veenvliet et al., 2013). In the absence of dbcAMP, we saw reductions in EN1, PITX3 and TH, but not SLC6A3 or NR4A2. Interestingly, the removal of DAPT led to a reduction in EN1 and KCNJ6 expression but not TH or PITX3, possibly indicating what may be construed as a loss of the A9, but not dopaminergic neuron phenotype. This might be consistent with an increase in the A10 marker, OTX2, but it is not consistent with the lack of change in expression of the other VTA marker, CALB1 indicating distinct regulatory mechanisms.
The effects of TGFβ3 and dbcAMP upon TH expression are not surprising as a substantial body of evidence shows the impact modulating cAMP and TGFβ has on cultures. For example, dbcAMP accelerates neuron maturation (Lepski et al., 2013), promotes dendritic arborization and increases density of high-affinity dopamine uptake sites and choline acetyltransferase activity (Mena et al., 1995). Cyclic AMP is a powerful inducer of TH gene transcription via transcription factors such as CREB and ATF1, which, in conjunction with the coactivator cyclic AMP response element-binding protein (CBP)/p300 control transcriptional regulation of the TH gene (Mena et al., 1995; Lim et al., 2000). A recent study reported that increasing cAMP levels by inhibiting phosphodiesterase activity is sufficient to support the rapid functional maturation of neuronal progenitors into fully functional neurons (Lepski et al., 2013). In our study, the removal of dbcAMP from cultures relatively late in maturation produced profound changes phenotype, without necessarily promoting much change in neuronal phenotype (although we saw a small reduction in TUBB3 expression). Similarly, our finding that mDA phenotype was compromised by the loss of TGFβ3 is consistent with data showing that blocking BMP signalling results in a loss of TH-positive mDA neurons (Jovanovic et al., 2018), as well as data showing that Tgfβ2−/− and Tgfβ3−/− mice have a severe reduction in mDA neuron numbers (Airaksinen and Saarma, 2002). Transforming growth factor-βs are widely recognized as prototypical multifunctional growth factors and master switches that regulate key events in development, disease, and repair (Krieglstein et al., 2002). BMP and TGFβ pathway mediators are critical upstream regulators of Wnt signalling during midbrain dopaminergic differentiation (Cai et al., 2013). TGFβ directly increases the number of Nr4a2-and Th- immunoreactive cells, which can be further increased when cultures are incubated with TGFβ in combination with Shh and fibroblast growth factor 8 (FGF8) (Airaksinen and Saarma, 2002). TGFβ3 may also ectopically induce TH-immunopositive cells in dorsal mesencephalon in vitro, in a Shh- and FGF8-independent manner (Roussa et al., 2006), a finding consistent with our data showing that the removal of TGFβ3 from our cultures impacted TH expression, albeit without effect upon NR4A2. There is evidence that the effect of TGFβ3 in maintaining TH expression in mature neurons may be via an indirect effect upon BDNF and GDNF; neurotrophic factors that belong to the TGFβ superfamily (Airaksinen and Saarma, 2002; Binder and Scharfman, 2004). Since GDNF and BDNF are still present in our cultures this remains a possibility.
WNT5A expression was greatly impacted by the removal of dbcAMP or TGFβ3. WNT5A is predominantly a non-β-catenin-dependent signalling member of the WNT family, which regulates a number of essential processes in early development (Topol et al., 2003). In mice, Wnt5a predominantly increases differentiation of Nr4a2 precursors into mDA neurons (Castelo-Branco et al., 2003), while Wnt5a deletion increases progenitor proliferation via activation of the GTPase, Rac1 (Andersson et al., 2008). Moreover, WNT5A regulates mDA axon growth and guidance both in vitro and in vivo (Blakely et al., 2011) and improves the differentiation and functional integration of stem cell-derived DA neurons in vivo (Parish et al., 2008). In our hands, a significant loss of WNT5A but not WNT1 was also observed in the absence of either dbcAMP or TGFβ3. There is clear evidence of crosstalk between β-catenin dependent WNT and cAMP/PKA signalling pathways (Hino et al., 2005; Taurin et al., 2006). Protein kinase A (PKA) phosphorylates β -catenin, promoting binding to its transcriptional coactivator, CREB-binding protein (Taurin et al., 2006). Furthermore, cAMP activation by parathyroid hormone (PTH) and ATP also inhibits the destruction complex, thereby reducing β-catenin degradation and promoting transcriptional activity (Taurin et al., 2006). Both pathways are known to stimulate cell proliferation (Takahashi et al., 2004; Clevers et al., 2014; Li et al., 2018). Similarly, TGFβ pathway mediators are critical upstream regulators of Wnt1-Lmx1a signalling in mDA progenitors (Cai et al., 2013) (Anderegg et al., 2013) (Claude et al., 2019). However, TGFβ directly regulates the expression of Wnt5a in the mammary gland (Serra et al., 2011) and Wnt5a potentiates TGFβ Signaling to promote colonic crypt regeneration after tissue injury in the mammalian intestinal epithelium (Miyoshi et al., 2012). Thus the interactions between cAMP, TGFβ, and WNT signalling systems; where cAMP-dependent suppression of TGFβ signalling (Jia et al., 2009) as well as a loss of Wnt5a or TGFβ resulting in a stabilization of β-catenin (Jia et al., 2009; Estarás et al., 2012) confound interpretation of this data. In the absence of dbcAMP, we found a profound loss of WNT5A, which supports the idea that WNT signalling is compromised and leads to changes in mDA phenotype even though our WNT5A immunolabelling revealed that WNT5A was abundantly expressed in most cell types, not just LMX1A-eGFP positive cells. These studies highlight the problems with ascribing activities to the removal of particular factors in as much as the resultant effects may be mediated via third-party signalling systems.
In conclusion, our findings demonstrate that common media additives are critical to maintaining the appearance of a widespread mDA neuron phenotype. These additives selectively affect many of the genes associated with mDA progenitor and mature neuron phenotype (NES, NEUROD1, FOXA2, SOX6, KCNJ6, PITX3, EN1/2, OTX2 and TH) as well as astroglial differentiation and activation (GFAP and S100B). Generally, the removal of any one of dbcAMP, TGFβ3 or DAPT is likely to give the impression of a loss of some aspect of an A9 or mDA phenotype from cultures. That dbcAMP and TGFβ3 regulated WNT5A expression indicates a dynamic endogenous WNT signalling network present in cultures. Consistent with this idea we observed a considerable lack of consistency between the putative transcription factors regulating expression of genes (Supplementary Table S5) and the impacts that removal of the activators of these signalling systems (ie. dbcAMP, TGFβ3 and DAPT) have upon gene expression. Understanding that these media supplements serve to elevate the expression of mDA (while suppressing astroglial) markers is of great significance to understand the outcomes of progenitor transplantation for Parkinson’s Disease. Thus, our cultures appear quite plastic with respect to the expression of markers of a dopaminergic neuronal phenotype, and we would argue that much of this culture-wide expression is dependent upon the addition of exogenous additives as well as the regulation of endogenous signalling systems (especially WNTS). We believe that this raises an important issue about the success of transplantation therapies where, for example the transplanted progenitors are under the influence of brain chemistry rather than exogenously applied factors designed to promote TH expression. Similarly, the influence of these factors is also likely to permeate into in vitro studies of survival and function.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
SS: method development, experimentation, analysis, writing. JH: investigation. CP: funding, administration, supervision. JMH: conceptualization, analysis, writing, supervision.
This work was supported by the Indonesian Endowment Fund for Education/Lembaga Pengelola Dana Pendidikan and National Agency of Drug and Food Control of Indonesia/Badan POM. The Australian Research Council (ARC SR1101002). The National Health and Medical Research Council (NHMRC APP1099372).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2023.1111705/full#supplementary-material
Aguila, J. C., Blak, A., van Arensbergen, J., Sousa, A., Vázquez, N., Aduriz, A., et al. (2014). Selection based on FOXA2 expression is not sufficient to enrich for dopamine neurons from human pluripotent stem cells. Stem Cells Transl. Med. 3, 1032–1042. doi:10.5966/sctm.2014-0011
Airaksinen, M. S., and Saarma, M. (2002). The GDNF family: Signalling, biological functions and therapeutic value. Nat. Rev. Neurosci. 3, 383–394. doi:10.1038/nrn812
Alves dos Santos, M. T., and Smidt, M. P. (2011). En1 and Wnt signaling in midbrain dopaminergic neuronal development. Neural Dev. 6, 23. doi:10.1186/1749-8104-6-23
Anderegg, A., Lin, H. P., Chen, J. A., Caronia-Brown, G., Cherepanova, N., Yun, B., et al. (2013). An Lmx1b-miR135a2 regulatory circuit modulates Wnt1/Wnt signaling and determines the size of the midbrain dopaminergic progenitor pool. PLoS Genet. 9, e1003973. doi:10.1371/journal.pgen.1003973
Andersson, E. R., Prakash, N., Cajanek, L., Minina, E., Bryja, V., Bryjova, L., et al. (2008). Wnt5a regulates ventral midbrain morphogenesis and the development of A9-A10 dopaminergic cells in vivo. PLoS One 3, e3517. doi:10.1371/journal.pone.0003517
Andersson, E. R., Salto, C., Villaescusa, J. C., Cajanek, L., Yang, S., Bryjova, L., et al. (2013). Wnt5a cooperates with canonical Wnts to generate midbrain dopaminergic neurons in vivo and in stem cells. Proc. Natl. Acad. Sci. U. S. A. 110, E602–E610. doi:10.1073/pnas.1208524110
Arenas, E., Denham, M., and Villaescusa, J. C. (2015). How to make a midbrain dopaminergic neuron. Development 142, 1918–1936. doi:10.1242/dev.097394
Ásgrímsdóttir, E. S., and Arenas, E. (2020). Midbrain dopaminergic neuron development at the single cell level: In vivo and in stem cells. Front. Cell Dev. Biol. 8, 463. doi:10.3389/fcell.2020.00463
Bejoy, J., Bijonowski, B., Marzano, M., Jeske, R., Ma, T., and Li, Y. (2020). Wnt-notch signaling interactions during neural and astroglial patterning of human stem cells. Tissue Eng. Part A 26, 419–431. doi:10.1089/ten.TEA.2019.0202
Bernal, A., and Arranz, L. (2018). Nestin-expressing progenitor cells: Function, identity and therapeutic implications. Cell Mol. Life Sci. 75, 2177–2195. doi:10.1007/s00018-018-2794-z
Bialas, A. R., and Stevens, B. (2013). TGF-β signaling regulates neuronal C1q expression and developmental synaptic refinement. Nat. Neurosci. 16, 1773–1782. doi:10.1038/nn.3560
Binder, D. K., and Scharfman, H. E. (2004). Brain-derived neurotrophic factor. Growth 22, 123–131. doi:10.1080/08977190410001723308
Blakely, B. D., Bye, C. R., Fernando, C. V., Horne, M. K., Macheda, M. L., Stacker, S. A., et al. (2011). Wnt5a regulates midbrain dopaminergic axon growth and guidance. PLoS One 6, e18373. doi:10.1371/journal.pone.0018373
Brenner, M., and Messing, A. (2021). Regulation of GFAP expression. ASN Neuro 13, 1759091420981206. doi:10.1177/1759091420981206
Bryja, V., Schulte, G., Rawal, N., Grahn, A., and Arenas, E. (2007). Wnt-5a induces Dishevelled phosphorylation and dopaminergic differentiation via a CK1-dependent mechanism. J. Cell Sci. 120, 586–595. doi:10.1242/jcs.03368
Butovsky, O., Jedrychowski, M. P., Moore, C. S., Cialic, R., Lanser, A. J., Gabriely, G., et al. (2014). Identification of a unique TGF-β-dependent molecular and functional signature in microglia. Nat. Neurosci. 17, 131–143. doi:10.1038/nn.3599
Cai, J., Schleidt, S., Pelta-Heller, J., Hutchings, D., Cannarsa, G., and Iacovitti, L. (2013). BMP and TGF-β pathway mediators are critical upstream regulators of Wnt signaling during midbrain dopamine differentiation in human pluripotent stem cells. Dev. Biol. 376, 62–73. doi:10.1016/j.ydbio.2013.01.012
Castelo-Branco, G., Wagner, J., Rodriguez, F. J., Kele, J., Sousa, K., Rawal, N., et al. (2003). Differential regulation of midbrain dopaminergic neuron development by Wnt-1, Wnt-3a, and Wnt-5a. Proc. Natl. Acad. Sci. U. S. A. 100, 12747–12752. doi:10.1073/pnas.1534900100
Castillo, S. O., Baffi, J. S., Palkovits, M., Goldstein, D. S., Kopin, I. J., Witta, J., et al. (1998). Dopamine biosynthesis is selectively abolished in substantia nigra/ventral tegmental area but not in hypothalamic neurons in mice with targeted disruption of the Nurr1 gene. Mol. Cell Neurosci. 11, 36–46. doi:10.1006/mcne.1998.0673
Chakrabarty, K., Von Oerthel, L., Hellemons, A., Clotman, F., Espana, A., Groot Koerkamp, M., et al. (2012). Genome wide expression profiling of the mesodiencephalic region identifies novel factors involved in early and late dopaminergic development. Biol. Open 1, 693–704. doi:10.1242/bio.20121230
Claude, B., Blaess, S., Partanen, J., and Prakash, N. (2019). Crosstalk of intercellular signaling pathways in the generation of midbrain dopaminergic neurons in vivo and from stem cells. J. Dev. Biol. 7, 3. doi:10.3390/jdb7010003
Clevers, H., Loh, K. M., and Nusse, R. (2014). Stem cell signaling. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science 346, 1248012. doi:10.1126/science.1248012
Cognard, E., Dargaville, C. G., Hay, D. L., and Shepherd, P. R. (2013). Identification of a pathway by which glucose regulates beta-catenin signalling via the cAMP/protein kinase A pathway in beta-cell models. Biochem. J. 449, 803–811. doi:10.1042/BJ20121454
Crawford, T. Q., and Roelink, H. (2007). The notch response inhibitor DAPT enhances neuronal differentiation in embryonic stem cell-derived embryoid bodies independently of sonic hedgehog signaling. Dev. Dyn. 236, 886–892. doi:10.1002/dvdy.21083
Daily, K., Patel, V. R., Rigor, P., Xie, X., and Baldi, P. (2011). MotifMap: Integrative genome-wide maps of regulatory motif sites for model species. BMC Bioinforma. 12, 495. doi:10.1186/1471-2105-12-495
Danielian, P. S., and McMahon, A. P. (1996). Engrailed-1 as a target of the Wnt-1 signalling pathway in vertebrate midbrain development. Nature 383, 332–334. doi:10.1038/383332a0
Dvoriantchikova, G., Perea-Martinez, I., Pappas, S., Barry, A. F., Danek, D., Dvoriantchikova, X., et al. (2015). Molecular characterization of Notch1 positive progenitor cells in the developing retina. PLoS One 10, e0131054. doi:10.1371/journal.pone.0131054
Estarás, C., Akizu, N., García, A., Beltrán, S., de la Cruz, X., and Martínez-Balbás, M. A. (2012). Genome-wide analysis reveals that Smad3 and JMJD3 HDM co-activate the neural developmental program. Development 139, 2681–2691. doi:10.1242/dev.078345
Fasano, C. A., Chambers, S. M., Lee, G., Tomishima, M. J., and Studer, L. (2010). Efficient derivation of functional floor plate tissue from human embryonic stem cells. Cell Stem Cell 6, 336–347. doi:10.1016/j.stem.2010.03.001
Fernández-Lloris, R., Viñals, F., López-Rovira, T., Harley, V., Bartrons, R., Rosa, J. L., et al. (2003). Induction of the Sry-related factor SOX6 contributes to bone morphogenetic protein-2-induced chondroblastic differentiation of C3H10T1/2 cells. Mol. Endocrinol. 17, 1332–1343. doi:10.1210/me.2002-0254
Flanders, K. C., Ren, R. F., and Lippa, C. F. (1998). Transforming growth factor-betas in neurodegenerative disease. Prog. Neurobiol. 54, 71–85. doi:10.1016/s0301-0082(97)00066-x
Gyorfi, A. H., Matei, A. E., Fuchs, M., Liang, C., Rigau, A. R., Hong, X., et al. (2021). Engrailed 1 coordinates cytoskeletal reorganization to induce myofibroblast differentiation. J. Exp. Med. 218, e20201916. doi:10.1084/jem.20201916
Han, J., Ishii, M., Bringas, P., Maas, R. L., Maxson, R. E., and Chai, Y. (2007). Concerted action of Msx1 and Msx2 in regulating cranial neural crest cell differentiation during frontal bone development. Mech. Dev. 124, 729–745. doi:10.1016/j.mod.2007.06.006
Harada, H., Sato, T., and Nakamura, H. (2016). Fgf8 signaling for development of the midbrain and hindbrain. Dev. Growth Differ. 58, 437–445. doi:10.1111/dgd.12293
Haynes, J. M., Sibuea, S. M., Aguiar, A. A., Li, F., Ho, J. K., and Pouton, C. W. (2021). Inhibition of β-catenin dependent WNT signalling upregulates the transcriptional repressor NR0B1 and downregulates markers of an A9 phenotype in human embryonic stem cell-derived dopaminergic neurons: Implications for Parkinson's disease. PLoS One 16, e0261730. doi:10.1371/journal.pone.0261730
Hino, S., Tanji, C., Nakayama, K. I., and Kikuchi, A. (2005). Phosphorylation of beta-catenin by cyclic AMP-dependent protein kinase stabilizes beta-catenin through inhibition of its ubiquitination. Mol. Cell Biol. 25, 9063–9072. doi:10.1128/MCB.25.20.9063-9072.2005
Hongo, I., and Okamoto, H. (2022). FGF/MAPK/Ets signaling in Xenopus ectoderm contributes to neural induction and patterning in an autonomous and paracrine manner, respectively. Cells Dev. 170, 203769. doi:10.1016/j.cdev.2022.203769
Horvat, A., and Vardjan, N. (2019). Astroglial cAMP signalling in space and time. Neurosci. Lett. 689, 5–10. doi:10.1016/j.neulet.2018.06.025
Jacobs, F. M., van Erp, S., van der Linden, A. J., von Oerthel, L., Burbach, J. P., and Smidt, M. P. (2009). Pitx3 potentiates Nurr1 in dopamine neuron terminal differentiation through release of SMRT-mediated repression. Development 136, 531–540. doi:10.1242/dev.029769
Jia, S., Wu, D., Xing, C., and Meng, A. (2009). Smad2/3 activities are required for induction and patterning of the neuroectoderm in zebrafish. Dev. Biol. 333, 273–284. doi:10.1016/j.ydbio.2009.06.037
Joksimovic, M., Yun, B. A., Kittappa, R., Anderegg, A. M., Chang, W. W., Taketo, M. M., et al. (2009). Wnt antagonism of Shh facilitates midbrain floor plate neurogenesis. Nat. Neurosci. 12, 125–131. doi:10.1038/nn.2243
Jovanovic, V. M., Salti, A., Tilleman, H., Zega, K., Jukic, M. M., Zou, H., et al. (2018). BMP/SMAD pathway promotes neurogenesis of midbrain dopaminergic neurons in vivo and in human induced pluripotent and neural stem cells. J. Neurosci. 38, 1662–1676. doi:10.1523/JNEUROSCI.1540-17.2018
Kee, N., Volakakis, N., Kirkeby, A., Dahl, L., Storvall, H., Nolbrant, S., et al. (2017). Single-cell analysis reveals a close relationship between differentiating dopamine and subthalamic nucleus neuronal lineages. Cell Stem Cell 20, 29–40. doi:10.1016/j.stem.2016.10.003
Kirkeby, A., Grealish, S., Wolf, D. A., Nelander, J., Wood, J., Lundblad, M., et al. (2012). Generation of regionally specified neural progenitors and functional neurons from human embryonic stem cells under defined conditions. Cell Rep. 1, 703–714. doi:10.1016/j.celrep.2012.04.009
Krieglstein, K., Strelau, J., Schober, A., Sullivan, A., and Unsicker, K. (2002). TGF-beta and the regulation of neuron survival and death. J. Physiol. Paris 96, 25–30. doi:10.1016/s0928-4257(01)00077-8
Kriks, S., Shim, J. W., Piao, J., Ganat, Y. M., Wakeman, D. R., Xie, Z., et al. (2011). Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson's disease. Nature 480, 547–551. doi:10.1038/nature10648
Kuwabara, T., Hsieh, J., Muotri, A., Yeo, G., Warashina, M., Lie, D. C., et al. (2009). Wnt-mediated activation of NeuroD1 and retro-elements during adult neurogenesis. Nat. Neurosci. 12, 1097–1105. doi:10.1038/nn.2360
La Manno, G., Gyllborg, D., Codeluppi, S., Nishimura, K., Salto, C., Zeisel, A., et al. (2016). Molecular diversity of midbrain development in mouse, human, and stem cells. Cell 167, 566–580. doi:10.1016/j.cell.2016.09.027
Lebel, M., Gauthier, Y., Moreau, A., and Drouin, J. (2001). Pitx3 activates mouse tyrosine hydroxylase promoter via a high-affinity binding site. J. Neurochem. 77, 558–567. doi:10.1046/j.1471-4159.2001.00257.x
Lee, T. Y., Cho, I. S., Bashyal, N., Naya, F. J., Tsai, M. J., Yoon, J. S., et al. (2020). ERK regulates NeuroD1-mediated neurite outgrowth via proteasomal degradation. Exp. Neurobiol. 29, 189–206. doi:10.5607/en20021
Lepski, G., Jannes, C. E., Nikkhah, G., and Bischofberger, J. (2013). cAMP promotes the differentiation of neural progenitor cells in vitro via modulation of voltage-gated calcium channels. Front. Cell Neurosci. 7, 155. doi:10.3389/fncel.2013.00155
Lepski, G., Maciaczyk, J., Jannes, C. E., Maciaczyk, D., Bischofberger, J., and Nikkhah, G. (2011). Delayed functional maturation of human neuronal progenitor cells in vitro. Mol. Cell Neurosci. 47, 36–44. doi:10.1016/j.mcn.2011.02.011
Li, C., Li, Z., Zhang, Y., Fathy, A. H., and Zhou, M. (2018). The role of the Wnt/β-catenin signaling pathway in the proliferation of gold nanoparticle-treated human periodontal ligament stem cells. Stem Cell Res. Ther. 9, 214. doi:10.1186/s13287-018-0954-6
Lim, J., Yang, C., Hong, S. J., and Kim, K. S. (2000). Regulation of tyrosine hydroxylase gene transcription by the cAMP-signaling pathway: Involvement of multiple transcription factors. Mol. Cell Biochem. 212, 51–60. doi:10.1023/a:1007148719497
Luckert, K., Gotschel, F., Sorger, P. K., Hecht, A., Joos, T. O., and Potz, O. (2011). Snapshots of protein dynamics and post-translational modifications in one experiment--beta-catenin and its functions. Mol. Cell Proteomics 10, 007377. doi:10.1074/mcp.M110.007377
Luo, S. X., Timbang, L., Kim, J. I., Shang, Y., Sandoval, K., Tang, A. A., et al. (2016). TGF-Β signaling in dopaminergic neurons regulates dendritic growth, excitatory-inhibitory synaptic balance, and reversal learning. Cell Rep. 17, 3233–3245. doi:10.1016/j.celrep.2016.11.068
Machold, R. P., Kittell, D. J., and Fishell, G. J. (2007). Antagonism between Notch and bone morphogenetic protein receptor signaling regulates neurogenesis in the cerebellar rhombic lip. Neural Dev. 2, 5. doi:10.1186/1749-8104-2-5
Mena, M. A., Casarejos, M. J., Bonin, A., Ramos, J. A., and Garcia Yebenes, J. (1995). Effects of dibutyryl cyclic AMP and retinoic acid on the differentiation of dopamine neurons: Prevention of cell death by dibutyryl cyclic AMP. J. Neurochem. 65, 2612–2620. doi:10.1046/j.1471-4159.1995.65062612.x
Mesman, S., Kruse, S. J., and Smidt, M. P. (2018). Expression analyzes of early factors in midbrain differentiation programs. Gene Expr. Patterns 27, 8–15. doi:10.1016/j.gep.2017.09.001
Mesman, S., and Smidt, M. P. (2020). Acquisition of the midbrain dopaminergic neuronal identity. Int. J. Mol. Sci. 21, 4638. doi:10.3390/ijms21134638
Miller, G. W., Gainetdinov, R. R., Levey, A. I., and Caron, M. G. (1999). Dopamine transporters and neuronal injury. Trends Pharmacol. Sci. 20, 424–429. doi:10.1016/s0165-6147(99)01379-6
Miyoshi, H., Ajima, R., Luo, C. T., Yamaguchi, T. P., and Stappenbeck, T. S. (2012). Wnt5a potentiates TGF-β signaling to promote colonic crypt regeneration after tissue injury. Science 338, 108–113. doi:10.1126/science.1223821
Morrison, S. J., Perez, S. E., Qiao, Z., Verdi, J. M., Hicks, C., Weinmaster, G., et al. (2000). Transient Notch activation initiates an irreversible switch from neurogenesis to gliogenesis by neural crest stem cells. Cell 101, 499–510. doi:10.1016/s0092-8674(00)80860-0
Mullen, A. C., and Wrana, J. L. (2017). TGF-Β family signaling in embryonic and somatic stem-cell renewal and differentiation. Cold Spring Harb. Perspect. Biol. 9, a022186. doi:10.1101/cshperspect.a022186
Nakatani, T., Kumai, M., Mizuhara, E., Minaki, Y., and Ono, Y. (2010). Lmx1a and Lmx1b cooperate with Foxa2 to coordinate the specification of dopaminergic neurons and control of floor plate cell differentiation in the developing mesencephalon. Dev. Biol. 339, 101–113. doi:10.1016/j.ydbio.2009.12.017
Niclis, J. C., Gantner, C. W., Alsanie, W. F., McDougall, S. J., Bye, C. R., Elefanty, A. G., et al. (2017). Efficiently specified ventral midbrain dopamine neurons from human pluripotent stem cells under xeno-free conditions restore motor deficits in parkinsonian rodents. Stem Cells Transl. Med. 6, 937–948. doi:10.5966/sctm.2016-0073
Nunes, I., Tovmasian, L. T., Silva, R. M., Burke, R. E., and Goff, S. P. (2003). Pitx3 is required for development of substantia nigra dopaminergic neurons. Proc. Natl. Acad. Sci. U. S. A. 100, 4245–4250. doi:10.1073/pnas.0230529100
Paco, S., Hummel, M., Plá, V., Sumoy, L., and Aguado, F. (2016). Cyclic AMP signaling restricts activation and promotes maturation and antioxidant defenses in astrocytes. BMC Genomics 17, 304. doi:10.1186/s12864-016-2623-4
Paldino, E., Cenciarelli, C., Giampaolo, A., Milazzo, L., Pescatori, M., Hassan, H. J., et al. (2014). Induction of dopaminergic neurons from human Wharton's jelly mesenchymal stem cell by forskolin. J. Cell Physiol. 229, 232–244. doi:10.1002/jcp.24442
Panman, L., Papathanou, M., Laguna, A., Oosterveen, T., Volakakis, N., Acampora, D., et al. (2014). Sox6 and Otx2 control the specification of substantia nigra and ventral tegmental area dopamine neurons. Cell Rep. 8, 1018–1025. doi:10.1016/j.celrep.2014.07.016
Parish, C. L., Castelo-Branco, G., Rawal, N., Tonnesen, J., Sorensen, A. T., Salto, C., et al. (2008). Wnt5a-treated midbrain neural stem cells improve dopamine cell replacement therapy in parkinsonian mice. J. Clin. Invest. 118, 149–160. doi:10.1172/JCI32273
Prakash, N., Brodski, C., Naserke, T., Puelles, E., Gogoi, R., Hall, A., et al. (2006). A Wnt1-regulated genetic network controls the identity and fate of midbrain-dopaminergic progenitors in vivo. Development 133, 89–98. doi:10.1242/dev.02181
Qi, Y., Zhang, X. J., Renier, N., Wu, Z., Atkin, T., Sun, Z., et al. (2017). Combined small-molecule inhibition accelerates the derivation of functional cortical neurons from human pluripotent stem cells. Nat. Biotechnol. 35, 154–163. doi:10.1038/nbt.3777
Qian, D., Li, L., Rong, Y., Liu, W., Wang, Q., Zhou, Z., et al. (2019). Blocking Notch signal pathway suppresses the activation of neurotoxic A1 astrocytes after spinal cord injury. Cell Cycle 18, 3010–3029. doi:10.1080/15384101.2019.1667189
Rakowicz, W. P., Staples, C. S., Milbrandt, J., Brunstrom, J. E., and Johnson, E. M. (2002). Glial cell line-derived neurotrophic factor promotes the survival of early postnatal spinal motor neurons in the lateral and medial motor columns in slice culture. J. Neurosci. 22, 3953–3962. doi:10.1523/JNEUROSCI.22-10-03953.2002
Roussa, E., Wiehle, M., Dünker, N., Becker-Katins, S., Oehlke, O., and Krieglstein, K. (2006). Transforming growth factor beta is required for differentiation of mouse mesencephalic progenitors into dopaminergic neurons in vitro and in vivo: Ectopic induction in dorsal mesencephalon. Stem Cells 24, 2120–2129. doi:10.1634/stemcells.2005-0514
Saucedo-Cardenas, O., Quintana-Hau, J. D., Le, W. D., Smidt, M. P., Cox, J. J., De Mayo, F., et al. (1998). Nurr1 is essential for the induction of the dopaminergic phenotype and the survival of ventral mesencephalic late dopaminergic precursor neurons. Proc. Natl. Acad. Sci. U. S. A. 95, 4013–4018. doi:10.1073/pnas.95.7.4013
Schober, A., Peterziel, H., von Bartheld, C. S., Simon, H., Krieglstein, K., and Unsicker, K. (2007). GDNF applied to the MPTP-lesioned nigrostriatal system requires TGF-beta for its neuroprotective action. Neurobiol. Dis. 25, 378–391. doi:10.1016/j.nbd.2006.10.005
Serra, R., Easter, S. L., Jiang, W., and Baxley, S. E. (2011). Wnt5a as an effector of TGFβ in mammary development and cancer. J. Mammary Gland. Biol. Neoplasia 16, 157–167. doi:10.1007/s10911-011-9205-5
Shimizu, T., Tanaka, T., Iso, T., Doi, H., Sato, H., Kawai-Kowase, K., et al. (2009). Notch signaling induces osteogenic differentiation and mineralization of vascular smooth muscle cells: Role of Msx2 gene induction via notch-RBP-jk signaling. Arterioscler. Thromb. Vasc. Biol. 29, 1104–1111. doi:10.1161/ATVBAHA.109.187856
Smith, Y., Wichmann, T., Factor, S. A., and DeLong, M. R. (2012). Parkinson's disease therapeutics: New developments and challenges since the introduction of levodopa. Neuropsychopharmacology 37, 213–246. doi:10.1038/npp.2011.212
Takahashi, H., Honma, M., Miyauchi, Y., Nakamura, S., Ishida-Yamamoto, A., and Iizuka, H. (2004). Cyclic AMP differentially regulates cell proliferation of normal human keratinocytes through ERK activation depending on the expression pattern of B-Raf. Arch. Dermatol Res. 296, 74–82. doi:10.1007/s00403-004-0478-z
Takazawa, T., Croft, G. F., Amoroso, M. W., Studer, L., Wichterle, H., and Macdermott, A. B. (2012). Maturation of spinal motor neurons derived from human embryonic stem cells. PLoS One 7, e40154. doi:10.1371/journal.pone.0040154
Taurin, S., Sandbo, N., Qin, Y., Browning, D., and Dulin, N. O. (2006). Phosphorylation of beta-catenin by cyclic AMP-dependent protein kinase. J. Biol. Chem. 281, 9971–9976. doi:10.1074/jbc.M508778200
Telias, M., Segal, M., and Ben-Yosef, D. (2014). Electrical maturation of neurons derived from human embryonic stem cells. F1000Res 3, 196. doi:10.12688/f1000research.4943.1
Topol, L., Jiang, X., Choi, H., Garrett-Beal, L., Carolan, P. J., and Yang, Y. (2003). Wnt-5a inhibits the canonical Wnt pathway by promoting GSK-3-independent beta-catenin degradation. J. Cell Biol. 162, 899–908. doi:10.1083/jcb.200303158
Tossell, K., Kiecker, C., Wizenmann, A., Lang, E., and Irving, C. (2011). Notch signalling stabilises boundary formation at the midbrain-hindbrain organiser. Development 138, 3745–3757. doi:10.1242/dev.070318
Trujillo-Paredes, N., Valencia, C., Guerrero-Flores, G., Arzate, D. M., Baizabal, J. M., Guerra-Crespo, M., et al. (2016). Regulation of differentiation flux by Notch signalling influences the number of dopaminergic neurons in the adult brain. Biol. Open 5, 336–347. doi:10.1242/bio.013383
Veenvliet, J. V., Dos Santos, M. T., Kouwenhoven, W. M., von Oerthel, L., Lim, J. L., van der Linden, A. J., et al. (2013). Specification of dopaminergic subsets involves interplay of En1 and Pitx3. Development 140, 3373–3384. doi:10.1242/dev.094565
Vennemann, A., Agoston, Z., and Schulte, D. (2008). Differential and dose-dependent regulation of gene expression at the mid-hindbrain boundary by Ras-MAP kinase signaling. Brain Res. 1206, 33–43. doi:10.1016/j.brainres.2008.01.100
Volpicelli, F., De Gregorio, R., Pulcrano, S., Perrone-Capano, C., di Porzio, U., and Bellenchi, G. C. (2012). Direct regulation of Pitx3 expression by Nurr1 in culture and in developing mouse midbrain. PLoS One 7, e30661. doi:10.1371/journal.pone.0030661
Wang, J. J., Zhu, J. D., Zhang, X. H., Long, T. T., Ge, G., and Yu, Y. (2019). Neuroprotective effect of Notch pathway inhibitor DAPT against focal cerebral ischemia/reperfusion 3 hours before model establishment. Neural Regen. Res. 14, 452–461. doi:10.4103/1673-5374.245469
Watmuff, B., Hartley, B. J., Hunt, C. P., Fabb, S. A., Pouton, C. W., and Haynes, J. M. (2015). Human pluripotent stem cell derived midbrain PITX3(eGFP/w) neurons: A versatile tool for pharmacological screening and neurodegenerative modeling. Front. Cell Neurosci. 9, 104. doi:10.3389/fncel.2015.00104
Wu, Q., Zhang, L., Su, P., Lei, X., Liu, X., Wang, H., et al. (2015). MSX2 mediates entry of human pluripotent stem cells into mesendoderm by simultaneously suppressing SOX2 and activating NODAL signaling. Cell Res. 25, 1314–1332. doi:10.1038/cr.2015.118
Keywords: Parkinson’s disease, human embryonic stem cells (hESCs), midbrain dopaminergic neurons, dibutyryl cAMP, transforming growth factor–beta, DAPT (PubChem: 5311272)
Citation: Sibuea S, Ho JK, Pouton CW and Haynes JM (2023) TGFβ3, dibutyryl cAMP and a notch inhibitor modulate phenotype late in stem cell-derived dopaminergic neuron maturation. Front. Cell Dev. Biol. 11:1111705. doi: 10.3389/fcell.2023.1111705
Received: 30 November 2022; Accepted: 19 January 2023;
Published: 01 February 2023.
Edited by:
Yohan Oh, Hanyang University, South KoreaReviewed by:
Claude Brodski, Ben-Gurion University of the Negev, IsraelCopyright © 2023 Sibuea, Ho, Pouton and Haynes. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: John M. Haynes, am9obi5oYXluZXNAbW9uYXNoLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.