- 1Department of Biological Sciences, University of North Carolina, Charlotte, NC, United States
- 2Department of Plant Biology and Carnegie Mass Spectrometry Facility, Carnegie Institution for Science, Stanford, CA, United States
- 3Center for Gene Regulation in Health and Disease, Department of Biological, Geological, and Environmental Sciences, College of Arts and Sciences, Cleveland State University, Cleveland, OH, United States
- 4Department of Molecular Genetics and Cell Biology, The University of Chicago, Chicago, IL, United States
Telomerase is a ribonucleoprotein enzyme responsible for maintaining the telomeric end of the chromosome. The telomerase enzyme requires two main components to function: the telomerase reverse transcriptase (TERT) and the telomerase RNA (TR), which provides the template for telomeric DNA synthesis. TR is a long non-coding RNA, which forms the basis of a large structural scaffold upon which many accessory proteins can bind and form the complete telomerase holoenzyme. These accessory protein interactions are required for telomerase activity and regulation inside cells. The interacting partners of TERT have been well studied in yeast, human, and Tetrahymena models, but not in parasitic protozoa, including clinically relevant human parasites. Here, using the protozoan parasite, Trypanosoma brucei (T. brucei) as a model, we have identified the interactome of T. brucei TERT (TbTERT) using a mass spectrometry-based approach. We identified previously known and unknown interacting factors of TbTERT, highlighting unique features of T. brucei telomerase biology. These unique interactions with TbTERT, suggest mechanistic differences in telomere maintenance between T. brucei and other eukaryotes.
1 Introduction
Telomeres are the nucleoprotein structures found at the ends of eukaryotic chromosomes. Conventional DNA polymerases are unable to fully replicate the ends of linear DNA molecules, which leads to progressive telomere shortening after every cell division (Shay and Wright, 2019). This problem is solved by the ribonucleoprotein enzyme, telomerase. Proper maintenance of the telomeric end is critical for maintaining genome integrity in eukaryotes. The telomerase enzyme has two essential components: the telomerase RNA (TR), which provides the template required for telomeric DNA synthesis (Greider and Blackburn, 1989); and the catalytic protein telomerase reverse transcriptase (TERT) that catalyzes the de novo synthesis of the telomere G-rich strand. The action of telomerase counteracts progressive telomere shortening after every cell division.
TERT and TR are the minimum components required for the telomerase activity in vitro (Chen et al., 2000; Collins, 2006). TR can form a large structural scaffold upon which many accessory proteins can bind to and form the complete telomerase holoenzyme in vivo. These accessory proteins are required for in vivo telomerase activity and regulation (Wang et al., 2019). Interacting partners of the TERT protein have been extensively characterized in yeast, human, and Tetrahymena systems, but they have not been extensively studied in parasitic protozoa including clinically relevant human parasites, such as Trypanosoma brucei (T. brucei). T. brucei is the parasite that causes African sleeping sickness in humans. During its life cycle, T. brucei will shuttle between an insect vector and a mammalian host. During this time, the parasite will differentiate into distinct developmental stages: Bloodstream form (BF) parasites proliferate in the mammalian host, and Procyclic form (PF) parasites proliferate in the midgut of the insect vector. Unlike human somatic cells, which tightly regulate telomerase activity, T. brucei telomerase is constantly active, enabling continued cell division in their hosts, leading to a chronic infection. BF T. brucei also has the advantage of evading its host immune response through antigenic variation. During this process, T. brucei regularly switches to express different Variant Surface Glycoproteins (VSGs), its major surface antigen. T. brucei has a very large VSG gene pool, but only one VSG gene is expressed at any given time. VSGs are expressed exclusively from VSG expression sites (ESs), which are large polycistronic transcription units located at subtelomeric loci of the parasite’s genome within 2 kb of telomeres (De Lange and Borst, 1982; Hertz-Fowler et al., 2008). Therefore, telomerase activity is critical to maintain the integrity of these VSG genes. In cells where the TERT protein has been deleted (TbTERT−/−), extremely short telomeres adjacent to the active ES leads to an increase in VSG switching frequency (Dreesen and Cross, 2006; Hovel-Miner et al., 2012). Telomeric binding proteins have also been shown to affect VSG silencing and switching (Yang et al., 2009; Benmerzouga et al., 2013; Jehi et al., 2014; Jehi et al., 2016; Nanavaty et al., 2017; Afrin et al., 2020; Rabbani et al., 2022). Telomerase mediated telomere maintenance in T. brucei is required for the maintenance of subtelomeric VSG genes. Because of this, studies of telomerase function and regulation in T. brucei could give novel insights into the pathogenicity of this parasite.
The RNA component of T. brucei telomerase (TbTR) has a unique structure and sequence composition compared to higher eukaryotes (Sandhu et al., 2013; Podlevsky et al., 2016; Dey et al., 2021). Our recent study on TbTR suggests mechanistic differences in telomere maintenance between T. brucei and higher eukaryotes (Dey et al., 2021; Rabbani et al., 2022, underscoring the importance of investigating the functional interactome of T. brucei telomerase. Previous proteomic studies on ciliated single-cell protozoan Tetrahymena identified several RNA binding proteins, including p65 that copurifies with TR and TERT and promotes proper folding of TR for telomerase holoenzyme assembly and activity (Witkin and Collins, 2004; Berman et al., 2010; Singh et al., 2012; Upton et al., 2017; He et al., 2021). Although homologs of p65 have not been found outside ciliate ancestry, the complex of dyskerin, NHP2, NOP10, and GAR1 that bind the H/ACA domain of human TR are thought to be the functional analog of the p65 chaperone in human telomerase (Berman et al., 2010; Roake and Artandi, 2020). Indeed, these H/ACA binding proteins are known to facilitate human TR folding by enabling the human CR4/5 domain to adopt a particular conformation that interacts with TERT (Egan and Collins, 2012; Chen et al., 2018). Interestingly, in TbTR, the human H/ACA type snoRNP binding domain is replaced by a unique C/D box snoRNA domain (Gupta et al., 2013). In addition, TCAB1, another telomerase RNA-binding protein, was previously discovered as a part of human telomerase complex by mass spectrometry, which is important for intracellular trafficking (Venteicher et al., 2008) and regulating the folding of CR4/5 domain of human TR and telomerase activation. However, despite the fact that TbTR possesses stage-specific structural changes in an active telomerase complex (Dey et al., 2021), major interactors in this complex remain unidentified.
In addition to the canonical function of the TERT protein to protect telomeric ends of chromosomes, emerging evidences also suggest that telomerase can contribute to oxidative stress response in a telomere-independent manner. TERT has been shown to shuttle to mitochondria under increased oxidative stress and influence processes related to DNA damage and cell death (Santos et al., 2004; Ahmed et al., 2008; Indran et al., 2011). There are 5 respiratory chain complexes in human mitochondria that can regulate redox processes and hTERT enhances complex I activity (Ale-Agha et al., 2021). Beyond this, very little is known about the involvement of mitochondrial proteins in TERT function. Interestingly, T. brucei protein TbUMSBP2 (Klebanov-Akopyan et al., 2018), which binds to single-stranded G-rich sequence at the replication origins of the mitochondrial DNA of trypanosomatids, colocalizes with telomeres at the nucleus, but whether this activity is coordinated by telomerase mediated DNA repair is not known.
In order to gain a global view and mechanistic insight into telomerase function and regulation in T. brucei, we identified the interacting factors of T. brucei telomerase reverse transcriptase (TbTERT) using an affinity-purification based mass spectrometry approach. We identified previously known and novel interactors of TbTERT and validated several key interactions. Studying the interactome of TbTERT lays the foundation for future studies of telomerase regulation in T. brucei.
2 Materials and methods
2.1 Culture of bloodstream form (BF) T. brucei cells
T. brucei Lister strain 427 was used throughout this study. All BF cells were grown in HMI-9 media supplemented with 10% heat-inactivated Fetal Bovine Serum (FBS) at 37°C and 5% CO2. T. brucei Lister 427 strain expressing the T7 polymerase and Tet repressor (single marker, AKA SM) (Wirtz et al., 1999) was grown in media containing 2 μg/mL of G418; TbTERT-FLAG-HA-HA (F2H) cells grown with 2 μg/mL of G418, 0.1 μg/mL Puromycin; TbTR ∆C/D box mutant cells were grown with 2 μg/mL of G418, 4 μg/mL of Hygromycin, 2.5 μg/mL of Phleomycin, 5 μg/mL of Blasticidin, 0.1 μg/mL of Puromycin and 0.1 μg/mL of Doxycycline to constitutively induce the TbTR mutations.
2.2 Plasmids
The TbTR WT gene without 3’ C/D box region (nt 841–943) together with 400 bp upstream and 380 bp of downstream TbTR flanking sequences were cloned into the pLew111 plasmid to generate pLew111-TbTR ΔC/D box plasmid.
2.3 Generation of the BF T. brucei ∆C/D box mutant strain
To generate the TbTR ΔC/D box mutant strain, pLew111-TbTR ΔC/D box plasmid (nt 841–943 deleted) was digested with NotI and targeted to an rDNA spacer in the SM/TbTR−/− cells under the phleomycin selection. Clones were confirmed by northern analysis. XhoI digested pSK-TbTERT-3C-FLAG-HA-HA-PUR plasmid (Dey et al., 2021) was subsequently transfected into the same cells under the puromycin selection to generate the SM/TbTR ΔC/D box/TbTERT+/F2H strain. Clones were confirmed by western and Southern blotting.
2.4 Immunopurification of T. brucei telomerase complexes
Immunoprecipitation of T. brucei telomerase was performed using a custom made anti-TbTERT antibody (Dey et al., 2021) to purify native telomerase complexes from BF Wild-type (WT) and BF TbTR ∆C/D box cells. Approximately, 5 × 108 cells/300 mL were collected by centrifugation at 1900 RPM for 6 min. Following centrifugation, cells were lysed by homogenization in 500 µL of 1X immunopurified (IP) lysis buffer (25 mM Tris-HCl pH 7.5, 150 mM KCl, 1 mM EDTA, 10 mM MgCl2, 0.5% IGEPAL CA630, 1X protease cocktail inhibitor, and 20 units of Ribolock RNase inhibitor). Lysate was then cleared of cell debris by centrifugation at 3000 RPM for 5 min at 4°C. The lysate was then pre-cleared by incubated with 50 µL of pre-washed Dynabeads protein G (10003D) for 1 h at 4°C on rotation. Pre-cleared lysates were then incubated overnight at 4°C on rotation with 5 µg of a custom anti-TbTERT antibody and an IgG antibody was added to the control (Dey et al., 2021). The next day, 50 µL of pre-washed Dynabeads protein G was added to the lysate antibody mixture and incubated at 4°C for 2 h on rotation. After incubation, the beads were collected in a magnetic stand and washed twice in ×1 IP lysis buffer. After washing, the bound protein was eluted off the beads by boiling in 100 µL of 1X SDS-PAGE dye for 5 min at 95°C. Eluted proteins were then stored in −80°C until further use. Each experiment was performed in biological triplicate (3 IPs and 3 IgG controls). Bound complexes were assayed for the presence of TbTERT by using an anti-FLAG antibody as the BF WT cells were TbTERT-FLAG-HA-HA tagged. Briefly, 4 µL of 100 µL of sample was loaded onto 4%–12% Novex Tris-glycine gel (Invitrogen, XP04120BOX). Western blotting was done using an anti-FLAG antibody, diluted 1:500, and the VeriBlot for IP Detection Reagent (HRP, ab131366) diluted to 1:10,000. Detection was then done using Pierce ECL Plus Chemiluminescence kit (Thermo Fisher Scientific, 32,106). Imaging was then done using Bio-Rad ChemiDoc MP system.
Immunopurification was also performed using Pierce Anti-DYKDDDDK magnetic beads (A36797). Approximately 6 × 108 cells/300 mL were harvested and lysed in 300 μL of immunopurified (IP) lysis buffer (25 mM Tris-HCl pH 7.5, 150 mM KCl, 25 mM NaCl, 1 mM EDTA, 10 mM MgCl2, 0.5% IGEPAL CA630, 1× protease cocktail inhibitor and 20 units of Ribolock RNase inhibitor). Lysate was cleared of debris by centrifugation at 3,000 rpm for 5 min at 4°C and incubated with pre-washed 50 μL of Pierce Anti-DYKDDDDK magnetic beads (A36797) at 4°C for 2 h with rotation. Following incubation, the beads were washed twice by ice cold IP buffer and once with ice cold DEPC water. The beads were then resuspended in 50 μL of RNAse free water.
2.5 LC-MS/MS analysis
Proteins were separated by SDS-PAGE and Gel segments were cut and subjected to in-gel digestion using trypsin. Peptides were desalted using C18 ZipTips (Millipore). Peptides were analyzed on a Q-Exactive HF hybrid quadrupole-Orbitrap mass spectrometer (Thermo Fisher) equipped with an Easy LC 1200 UPLC liquid chromatography system (Thermo Fisher). Peptides were first trapped using trapping column Acclaim PepMap 100 (75 uM x 2 cm, nanoViper 2Pk, C18, 3 μm, 100A), then separated using analytical column Acclaim PepMap RSLC (75 um × 25 cm, nanoViper, C18, 2 μm, 100A) (Thermo Fisher). The flow rate was 300 nL/min, and a 120-min gradient was used. Peptides were eluted by a gradient from 3% to 28% solvent B (80% (v/v) acetonitrile/0.1% (v/v) formic acid) over 100 min and from 28% to 44% solvent B over 20 min, followed by a short wash at 90% solvent B. For DDA acquisition, the precursor scan was from mass-to-charge ratio (m/z) 375 to 1,600 and the top 20 most intense multiply charged precursors were selected for fragmentation. Peptides were fragmented with higher-energy collision dissociation (HCD) with normalized collision energy (NCE) 27.
2.6 LC-MS/MS data and statistical analysis
The resulting raw data files were searched against a concatenated library (TbruceiTREU927 release 32 databases with 11,202 entries) using MaxQuant (Tyanova et al., 2016). Carbamidomethyl Cysteine was set as a fixed modification. Oxidation of methionine and N-terminal acetylation were set as variable modifications. Tolerance for precursor ions was set to 4.5 ppm and 20 ppm for fragment ions. A maximum of two missed cleavages was allowed. MaxQuant was set to match in between runs and report LFQ. All other parameters were at the default setting. The proteinGroups.txt file generated by MaxQuant was further processed using Perseus. Reverse and possible contaminants were removed from the protein groups. Samples were separated into an experimental group consisting of the pull downs of isotype matched IgG (control) and a group consisting of the TbTERT Immuno-Precipitation pull downs (3 IPs and 3 IgG controls). Protein groups were filtered to contain at least three quantifications in one experimental group. The remaining missing quantifications were imputed with random numbers from a normal distribution (width 0.3, shift = 1.8). A two-sided Student’s t-test was performed across replicates between each experimental group.
2.7 MS bioinformatics analysis
The mass spectrometry proteomic data was analyzed by a range of approaches. Volcano plot was generated using GraphPad Prism software version 9.3.1. The STRING database was used for classifying proteins based on functional categories and gene ontology (GO) terms. Protein-protein interaction network analysis was done using STRING version 11.5 (https://string-db.org/) and visualized by using the Cytoscape software version 3.9.1.
2.8 Structure-guided predictions
Proteins identified by mass spectrometry contained several hits that have very little primary sequence identity with proteins from other phyla, so their functional orthologs were not entirely evident from simple sequence homology. To identify whether structural homology exists between the local folds of these proteins and those reported in other organisms, we obtained predicted structure models of these T. brucei proteins from AlphaFold (Jumper et al., 2021) using their Uniprot IDs and then queried these ‘PDB’ entries using programs PDBeFold (Krissinel and Henrick, 2004) and DaliLite (Holm and Rosenström, 2010). The top scoring and only relevant hit with a DaliLite Z score cut-off of >2 is considered as biologically informative structural neighbor of the protein of interest (Supplementary Table S1).
2.9 Western blotting and SDS-PAGE analysis
All TbTERT western blots were done using either an anti-FLAG antibody (1:500) or a custom anti-TbTERT C terminus antibody (1:500) unless otherwise indicated. Nucleolar protein 58 (NOP58) was detected using an anti-NOP58 antibody (Thermo Fisher Scientific, PA5-54321) diluted 1:500 and an anti-Rabbit HRP conjugated secondary antibody diluted to 1:10,000. To qualitatively check protein levels, 4 µL of IP eluate was separated on a 4%–12% Novex Tris-glycine gel and stained with Coomassie Brilliant Blue R-250 Dye (Thermo Scientific, 20,278) for 30 min. The gel was then destained until bands were visible in destain solution (40% MeOH, 10% acetic acid).
2.10 TbTR detection and telomerase activity assay
TbTERT IP was performed as described in the methods Section 2.4. To detect the presence of TbTR in the IPed complex, total RNA was isolated from the protein G magnetic beads using the TRIzol reagent (Thermo Fisher Scientific, 15596026) following the manufacturers protocol. 100 ng of isolated RNA was then used for cDNA synthesis utilizing the SuperScript II reverse transcriptase (Thermo Fisher Scientific, 18064022) following the manufacturers protocol. The generated cDNA was then used for qRT-PCR analysis using TbTR specific primers (Fwd: CTGTGGAAATTTGTCGTAAGTG, Rev: AGTAGGGTTAGGGATCGTATAG).
To determine the activity of the Immunopurified T. brucei telomerase complex, a modified version of the exponential isothermal amplification of telomere repeat (EXPIATR) assay was performed (Tian and Weizmann, 2013; Dey et al., 2021)Briefly, A master mix was prepared on ice consisting of Nicking Telomerase Substrate (NTS, GTGCGTGAGAGCTCTTCCAATCCGTCGAGCAGAGTT), Nicking Probe (NP, AGCAGGAAGCGCTCTTCCTGCTCCCTAACCCTAACCC), 1X EXPIATR buffer (30 mM Tris-HCl, pH 8.3, 1.5 mM MgCl2, 100 mM KCl, 1 mM EGTA, 0.05% v/v Tween20), 200 µM dNTPs, Bst 2.0 Warm start DNA polymerase (0.96 units) and Nt. BspQ1 NEase (5 units). 17 μL of the master mix was aliquoted to PCR tubes containing, 3 µL of anti-FLAG bead-bound T. brucei telomerase, RNase A treated or heat-inactivated telomerase RNP bound beads, telomerase positive control (TPC8, GTGCGTGAGAGCTCTTCCAATCCGTCGAGCAGAGTTAGGGTTAGGGTTAGGGTTAGGGTTAGGGTTAGGGTTAGGGTTAGGG) (0.5 μM) and blank beads as a negative control. Telomerase activity was initiated by initial incubation of tubes at 28°C for 45 min for Nicking telomerase substrate (NTS) extension followed by amplification of resultant telomerase products at 55°C for 30 min. The amplified products were then analyzed on 12% Native PAGE gel by loading 10 μL of the reaction mixture.
3 Results
3.1 Affinity-purification mass spectrometry (AP-MS) of BF T. brucei telomerase reverse transcriptase
Characterizing the global interactors of a protein of interest can be done through affinity-purification mass spectrometry (AP-MS). Identifying the interactome of a protein is key in understanding its function in the cell and how it is regulated. To identify the global protein interactors of T. brucei telomerase, we utilized AP-MS to identify the global interactome of TbTERT at the BF stage. We first immunopurified (IP) TbTERT using a custom anti-TbTERT antibody along with its associated proteins and performed LC-MS/MS (Figure 1A). For immuno-affinity purification, 500 ul of lysate containing 1 mg of total protein was used per IP sample. TbTERT was pulled down from the lysate using a custom anti-C terminus TbTERT antibody (Dey et al., 2021). In addition to verifying the specificity of the custom antibody for binding to TbTERT, the presence of some non-specific cross-reactive bands were also observed. The presence of the immunopurified TbTERT was confirmed using western blotting and SDS-PAGE analysis (Figures 1B–D). Immunoblot analysis (anti-FLAG antibody) of the IP fractions from TbTERT-F2H cells showed that TbTERT was enriched in the pulldown products from anti-TbTERT C terminus antibody IPs but not from control groups. The anti-TbTERT C-terminus antibody IP samples were then subjected to SDS-PAGE and in-gel protease digestion, followed by mass spectrometry as described in ‘Materials and Methods’. To further validate the presence of telomerase components in the IP, RNA was extracted from IP beads and subjected to qRT-PCR analysis to confirm the presence of the telomerase RNA in the complex (Figure 1E). To determine if the immunopurified telomerase complex was catalytically active, we performed a telomerase activity assay (EXPIATR) using the IPed complex (Figure 1F). The result confirmed that the immunopurified T. brucei telomerase complex was catalytically active. To independently validate our proteomic screen, another set of affinity enrichment of TbTERT protein using an anti-FLAG antibody was performed in duplicates in TbTERT-F2H cells (two biological replicates) and screened for known TbTERT interactors. Both the TbTERT IP-MS and TbTERT FLAG pulldown MS data were compared. Since TbTERT and several other proteins that were previously linked to telomerase were identified in both of the FLAG and C-terminus IP mass spec data sets (Figure 2; Supplementary Table S1), it was apparent that the ribonucleoprotein complex identified by this approach is biologically relevant to telomerase function.
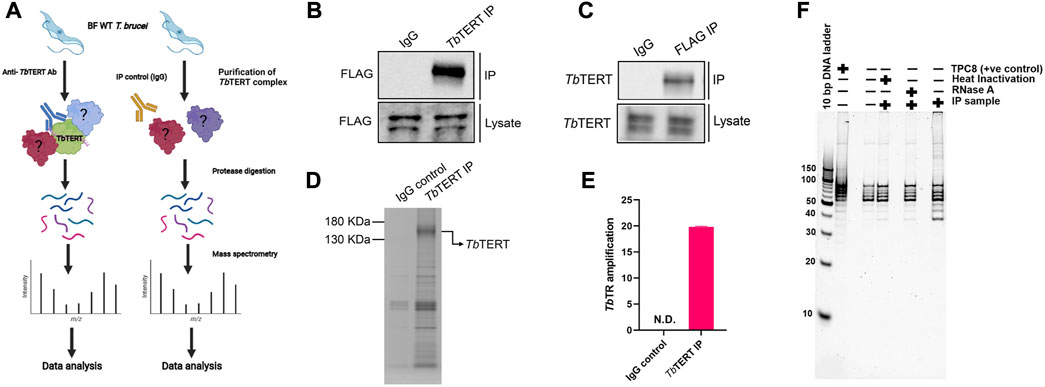
FIGURE 1. Affinity-purification mass spectrometry of bloodstream form T. brucei telomerase reverse transcriptase. (A) Experimental workflow for proteomic analysis. BF T. brucei cells expressing a FLAG tagged version of TbTERT were grown and TbTERT complexes were purified using a custom anti-TbTERT C terminus antibody. An IgG isotype antibody was used as a control. Purified TbTERT complexes were then digested with trypsin. These peptides were then analyzed by LC-MS/MS. (B) Western blot confirming the presence of immunopurified TbTERT. IP samples were obtained and run on SDS-PAGE gels and immunoblotted with anti-FLAG antibody to detect TbTERT. (C) Western blot confirming the presence of TbTERT. IP samples were obtained and run on SDS-PAGE gels and immunoblotted with an anti-TbTERT C terminus antibody to detect TbTERT. (D) SDS-PAGE analysis of immunopurified TbTERT. A small aliquot was also resolved on SDS-PAGE and stained with Coomassie stain to qualitatively check TbTERT protein levels. (E) RT-qPCR detection of TbTR from Immunopurified TbTERT complexes. (F) Telomerase activity of the bead-bound telomerase enzyme was analyzed by telomerase primer extension assay.
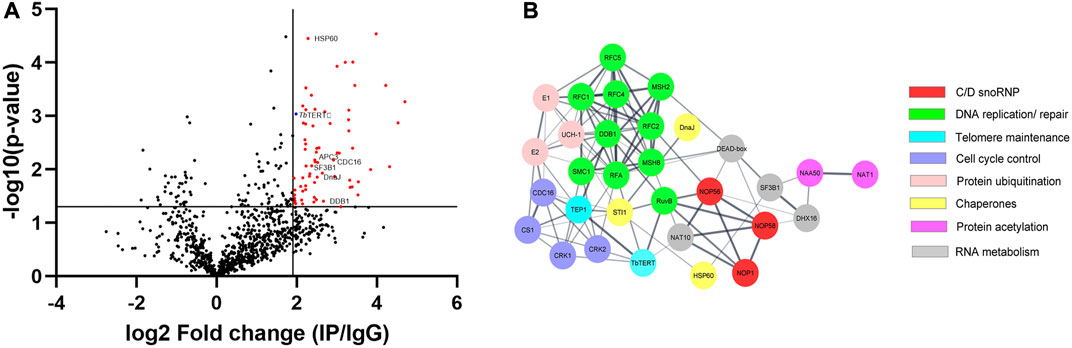
FIGURE 2. (A) Volcano plot was performed with an x-axis representing the difference in logarithmic protein intensities between the TbTERT immunoprecipitation elution and the isotype matched IgG control (Elution and Control experimental groups). The y-axis is the negative log of the two-sided Student’s t-test. The volcano plot serves as a visual representation of the protein groups that are significantly enriched between the elution and control groups. These enriched groups contain the bait protein TbTERT and several candidates interacting with a p-value ≤ 0.05. (B) Protein-protein interaction network of relevant TbTERT hits identified by MS. Network was generated using the STRING database and visualized using Cytoscape. Colors of nodes represent the protein’s biological function. The thickness of the lines denotes the strength of the interaction (confidence PPI, threshold: 0.4, medium confidence).
3.2 Proteomic analysis of BF T. brucei telomerase reverse transcriptase
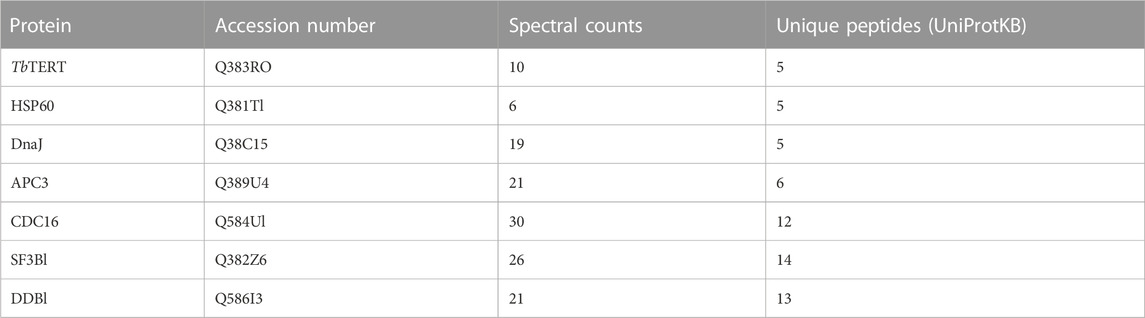
We identified 1,056 proteins with 2 or more peptides. After stringent filtering of this dataset for those proteins highly enriched (Log2(fold change) > 1.9) in the IP vs. control (Supplementary Table S1), 66 high-confidence proteins remained. To study the interactome of TbTERT, the relative abundance (log2(fold change)) and statistical significance -log10(P-value) of the proteins from the IP samples and controls were calculated (3 IP samples and 3 IgG controls samples) (Figure 2A). This resulted in the enrichment of 56 proteins for the anti-TbTERT IP samples (Supplementary Table S2). The protein samples that were significantly enriched in the IP samples versus the controls (Student’s t-test, -log10(P-value) > 1.3) included both nuclear and mitochondrial proteins that have previously been shown to interact with TERT or play a role in telomere maintenance, such as T. brucei telomerase reverse transcriptase (TbTERT, Tb927.11.10190), yeast telomerase cell cycle turnover-related (anaphase promoting complex) proteins, CDC16 (Tb927.6.2150) and CDC27 (Tb927.10.10330) homologs (Sealey et al., 2011; Ferguson et al., 2013), mitochondrial stress-response protein, human HSP60 homolog (Tb927.11.15040) which is known to accumulate with hTERT in the same fractions of human mitochondria (Sharma et al., 2012), Splicing factor 3B subunit 1 (SF3B1) homolog (Tb927.11.11850) involved in various cellular functions including DNA damage response (Te Raa et al., 2015) and telomere maintenance (Wang et al., 2016), damage specific DNA binding protein 1 (DDB1) homolog (Tb927.6.5110), involved in ubiquitin-mediated TERT protein degradation (Jung et al., 2013), and several other proteins involved in telomerase and telomere metabolism (selected proteins shown in Figure 2A; Table 1).
To highlight the connectivity of candidate TbTERT interactors, we used the STRING database and Cytoscape to generate a functional protein-protein interaction network of TbTERT (Figure 2B; Supplementary Figure S1). To further determine the functions of proteins in the network, STRING GO analysis was done and proteins were grouped by biological process, and cellular component (Figures 3A, B). Terms for biological process that were enriched included, telomere maintenance by telomerase, cell cycle control, DNA repair, and response to stress. Enriched cellular component terms included, box C/D snoRNP complex, DNA replication factor C, anaphase-promoting complex, and cullin-RING ubiquitin ligase complex. Notably, proteins that are important for telomerase RNA biogenesis, processing and trafficking were significantly enriched in these GO terms.
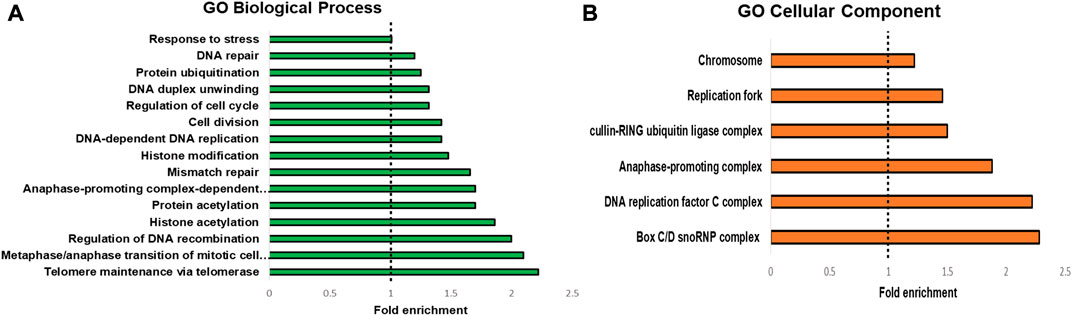
FIGURE 3. GO analysis of T. brucei TERT interactome. STRING GO analysis (A) Enrichment by Biological process. The top 15 enriched GO terms are shown. (B) enrichment by Cellular component. The top 9 enriched GO terms are shown.
An independent set of IP-MS using an anti-FLAG antibody to IP TbTERT also validated a majority of the proteins identified in the mass spec run described above from the anti-C terminus TbTERT IP because these proteins were identified with both IP approaches. A false discovery rate (FDR) of 1% was used as cut off for this data (Supplementary Table S3).
3.3 Known and novel interactors are part of the active telomerase RNP in T. brucei
Homologs of telomerase-associated proteins which are known regulators of telomerase functions were found to be part of the IP telomerase complex in T. brucei. Typically, three types of known telomerase-associated proteins were reported to co-purify with TERT. The foremost of the three are the telomerase RNA-binding proteins that are involved in TR biogenesis, trafficking and TR-TERT assembly. These include, for example, dyskerin in vertebrates (Mitchell and Collins, 2000; Mochizuki et al., 2004), Sm proteins in yeasts (Tang et al., 2012), and La-motif proteins, such as p65, in ciliates (Singh et al., 2012). Interestingly, dyskerin binding H/ACA domain of human TR is replaced by a novel C/D box domain in T. brucei (Gupta et al., 2013). Several unique C/D box snoRNA binding proteins (snoRNPs) were identified in our TbTERT immunopurified complex which are described in the next section. In addition, a T. brucei La protein, Tb927.10.2370, which shows 24% amino acid sequence identity and a Z-score of 8.6 with Tetrahymena thermophila, TR binding protein p65 was also identified in the IP-MS. Telomerase RNP assembly also requires molecular chaperones, such as AAA+ family of ATPases, known as Pontin and Reptin, which can directly interact with TERT and play critical roles in telomerase RNP accumulation (Venteicher et al., 2008). Both the Pontin (Tb927.4.1270) and Reptin (Tb927.4.2000) homologs, annotated as RuvB-like DNA helicases in the T. brucei genome database, were also identified by this TbTERT AP-MS analysis. Additionally, another AAA+ ATPase protein, a yeast CDC48 homolog, Tb927.10.5770, was also identified. CDC48, which was previously identified in a molecular complex that recognized and bound ubiquitinated proteins (Schuberth et al., 2004), was also found to be associated with yeast telomerase as a novel regulator of telomere length homeostasis. Notably, TERT turnover is dependent on ubiquitin-proteasome mediated degradation process (Jung et al., 2013) and therefore proteins that are important for ubiquitination were previously identified in the telomerase complexes, such as several isoforms of E3 ubiquitin ligases (Lin et al., 2015). Interestingly, several ubiquitin-family proteins were also identified in the IP-MS data including ubiquitin ligases, although the roles of these proteins in T. brucei telomerase biology remains uncertain until the ubiquitination status of TbTERT is determined.
Mammalian studies have identified chaperone proteins p23 and HSP90 as two important proteins that are physically and functionally associated with telomerase activity (Holt et al., 1999). The proteomic mapping also identified a mammalian HSP90 homolog, Tb927.3.3580, with enrichment of several unique peptides in TbTERT IP samples identified by MS. Poly(A)-specific ribonuclease (PARN) is a 3′-exoribonuclease that is known to play important role in the maturation of telomerase RNA (Moon et al., 2015). A T. brucei homolog of PARN, Tb927.9.13510 was identified in this proteomic mapping data that may relate with the fact that T. brucei telomerase RNA is a Pol II transcript (Sandhu et al., 2013) that may require PARN processing for maturation. All these known telomerase homologs of T. brucei are listed in Supplementary Table S1. In terms of proteomic identification, it should be noted that several of the above proteins were identified in the range of low scoring functions or higher FDR%, however, these proteins are identified in all four of the biological replicates analyzed by AP-MS and therefore could be biologically relevant. Importantly, all the above proteins identified were part of the IP sample that was able to extend T. brucei telomeric repeats using synthetic TTAGGG as substrates in activity assays (Figure 1F), indicating that the proteins in this IP are potentially part of an active telomerase complex.
3.4 The unique C/D box domain in T. brucei telomerase RNA is bound by snoRNPs
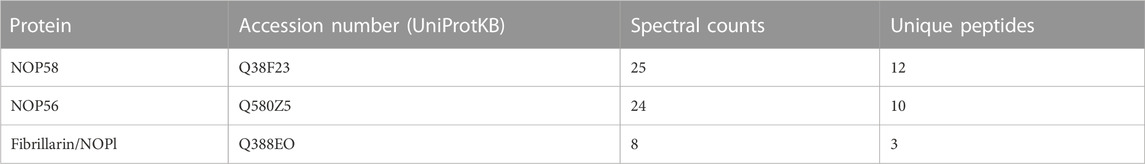
In contrast to hTR, the telomerase RNA in T. brucei contains a unique C/D snoRNA-like domain (Figure 4A bottom). Core C/D box RNPs, like NOP58, have previously been shown to interact with TbTR (Gupta et al., 2013). NOP58 is a 57 KDa protein that contains a coiled-coil (CC) domain and a NOP domain (Figure 4A top). NOP58 is highly conserved across eukaryotes and plays important roles in ribosomal RNA (rRNA) processing (Barth et al., 2008). In our AP-MS analysis of TbTERT, we identified three core C/D box binding proteins: NOP58, NOP56, and Fibrillarin (NOP1) (Table 2; Supplementary Table S3). To validate the interaction with NOP58, we performed a co-immunoprecipitation (Co-IP) assay of TbTERT and detected TbTERT and NOP58 through western blotting (Figure 4B top). For further confirmation of this interaction, we performed Co-IPs of TbTERT in both WT and ∆C/D box mutant cell lines. In the WT cells, NOP58 is present in the IP product, while in the ∆C/D mutant cells, the interaction with NOP58 is greatly diminished (Figure 4B bottom). This data supports the fact that the C/D box motif is important for the interaction between TbTR and NOP58.
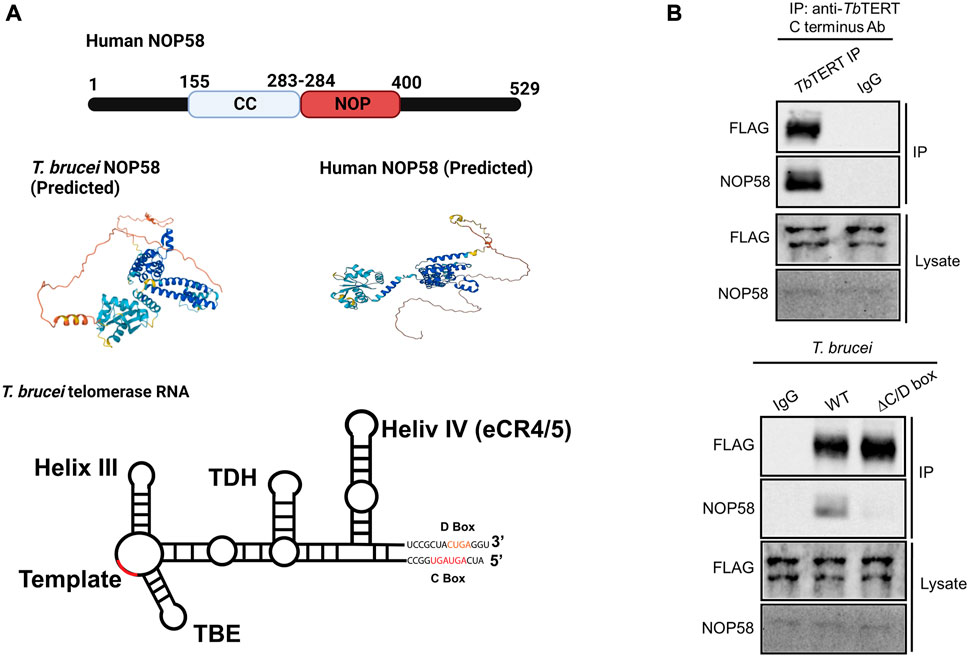
FIGURE 4. NOP58 interacts with the T. brucei telomerase complex. (A) Domain structure of human NOP58. Predicted secondary structure models for T. brucei and human NOP58 obtained from AlphaFold (Jumper et al., 2021) Predicted protein structures are shown in the same orientation. Dark blue represents a very high model confidence (pLDDT >90), light blue confident (90 > pLDDT >70), yellow low confidence (70 > pLDDT >50), orange very low confidence (pLDDT <50). Secondary structure model of T. brucei telomerase RNA. The C/D box binding motif is highlighted. [(B), top] Co-IP assay using WT T. brucei cell lysate. IP antibody: anti-TbTERT C terminus; Western blot antibodies: anti-FLAG and anti-NOP58. [(B), bottom] Co-IP using both WT and ∆C/D box mutant cells. IP antibody: anti-TbTERT C terminus; western blot antibodies: anti-FLAG and anti-NOP58.
4 Discussion
Eukaryotic microbes, such as T. brucei, rely on constitutive telomerase activity to sustain their proliferation in their hosts and to maintain the integrity of their subtelomeric virulence genes. Studying these interacting partners of telomerase in T. brucei is necessary to characterize the mechanism of telomerase mediated telomere maintenance in these parasites. Like any other RNA component of telomerase, T. brucei telomerase RNA in the holoenzyme acts as a structural scaffold that accessory proteins can bind to (Dey and Chakrabarti, 2018). Interacting proteins of telomerase in T. brucei have not been extensively studied. We have utilized a mass spectrometry approach to identify and characterize endogenous interactions of TbTERT in BF T. brucei cells. However, characterization of dynamic RNA-protein interactions like the one in telomerase complex comes with a challenge – that several of these interaction partners could be only transiently bound and therefore may not represent the complex interactions in its entirety. Non-etheless, this affinity purification based proteomic characterization of T. brucei telomerase RNP complex provides a global view of the cellular protein interactome landscape that can be used for in-depth functional characterization of telomerase complex proteins in parasites.
Some potential limitations of this study are the use of affinity purification methods. These methods are good at isolating strong interactions of our bait protein TbTERT, but as mentioned above, weaker or more transient interactions may be missed by this analysis. Also, proteins of low abundance, such as TERT, could be difficult to enrich in affinity purified complexes even by targeted approaches like RNA-targeted APEX based proteomic approach recently employed for human telomerase (Han et al., 2020). For that reason, it is possible that several TbTR and TbTERT associated proteins that are identified in this proteomic screen showed low level of enrichment in the IP complex, as evident from MS. Since IP experiments using MS provide a sensitive and accurate way of characterizing protein complexes, the quality of antibody may also play a role in isolating and analyzing specific interactions. The custom anti-TbTERT polyclonal antibody used in the IP experiments was cross-reactive to other proteins (data not shown), however, the binding specificity to the endogenous bait protein TbTERT was successfully confirmed using Co-IP and Western blot as TbTERT was detected as a single, discrete band. Additionally, detection of the telomerase-associated proteins in all biological replicates added confidence to the current approach.
Our proteomics experiments also identified proteins that have no known relationship with telomerase and therefore these hits could be false positives or newly discovered interactors of TbTERT. For example, an RNA cytidine acetyltransferase, NAT10 homolog (Tb927.5.2530) was found enriched in all proteomic datasets. NAT10 was previously shown to have predominantly nucleolar localization, association with human telomerase, and is primarily involved in telomerase RNA biogenesis (Fu and Collins, 2007). One more example is the putative NOT1 deadenylase (Tb927.10.1510), part of the CCR4-NOT deadenylase complex, which plays important regulatory roles both at the transcriptional and post-transcriptional levels, such as heterochromatic repression of sub-telomeric genes in fission yeast (Cotobal et al., 2015) and rapid deadenylation of m6A-containing RNAs by the CCR4–NOT deadenylase complex in mammalian cells (Du et al., 2016). Given that a majority of expressed virulence genes (VSGs) in T. brucei are subtelomeric (Saha et al., 2020) and human telomerase RNAs are known to contain m6A signatures (Han et al., 2020), the role of Tb927.10.1510 remains unexplored but relevant to T. brucei telomere biology.
In our proteomics screen, several mitochondrial proteins were highly enriched, including TbUMSBP2, which has been shown to associate with telomeres in T. brucei (Klebanov-Akopyan et al., 2018). This protein’s canonical function is the replication and segregation of T. brucei’s mitochondrial DNA, but it has been shown to also play a role in chromosome end protection in T. brucei (Klebanov-Akopyan et al., 2018). Significantly enriched GO terms from the analysis of the TbTERT interactome included, telomere maintenance via telomerase, cell cycle control, and chaperone binding. Similar terms and protein interactors have been previously observed for Saccharomyces cerevisiae telomerase (Lin et al., 2015). Telomere maintenance via telomerase is consistent with TbTERT’s known role in extending telomeres (Dreesen et al., 2005). Enrichment of proteins involved in cell cycle control highlight potential factors involved in the cell cycle specific regulation of TbTERT. Specifically, APC3 and CDC16 were significantly enriched in the TbTERT interactome. APC3 and CDC16 are core components of the anaphase-promoting complex (APC), which is a 1.5 MDa ubiquitin ligase complex that regulates sister-chromatid separation and the cells exit from mitosis (Peters, 2006). In S. cerevisiae, the APC has been shown to degrade the telomerase recruitment subunit, Est1p to regulate telomere maintenance (Ferguson et al., 2013). Whether an analogous mechanism exists in T. brucei remains to be explored.
Chaperone proteins such as DnaJ, which is a major co-chaperone for HSP70 and HSP60 were also found to be significantly enriched in the TbTERT interactome. Both DnaJ and HSP60 have previously been found to associate with human telomeres (Nittis et al., 2010). HSP60 is a predominately mitochondrial chaperone, where it works to maintain protein homeostasis (Caruso Bavisotto et al., 2020). In human cells, TERT has been previously reported to localize to the mitochondria and guard cells against oxidative stress (Ahmed et al., 2008). Human TERT has also been shown to associate with HSP60 and act independently of the TR in the mitochondria (Sharma et al., 2012). TbTERT’s association with HSP60 suggests a pool of T. brucei telomerase may also be localized in the mitochondrion.
In addition to proteins involved in cell cycle control and chaperones, core C/D snoRNP proteins, NOP58, NOP56, and Fibrillarin (NOP1) were also identified in the TbTERT interactome. Our co-IP western blot data validates the interaction of NOP58 with the T. brucei telomerase complex. NOP58 interacts with NOP56 and NOP1 to form a subcomplex, which participates in rRNA processing (Barth et al., 2008). Core C/D box binding proteins, like NOP58, have been previously shown to interact with the TbTR (Gupta et al., 2013). Our study supports these findings and shows that NOP58 interacts with the C/D box motif in the TbTR. The C/D box motif in TbTR is unique and lacking in higher eukaryotes. The TR in Leishmania also contains a C/D box motif (Vasconcelos et al., 2014). The conservation of the C/D box motif in the TR of these parasites could indicate a novel mechanism for telomerase biogenesis and processing, mediated by C/D box binding proteins, in these kinetoplastid parasites.
The work described here provides the first analysis of the TbTERT interactome. We have identified previously known and novel interactors of TbTERT. We were able to confirm NOP58’s interaction with the T. brucei telomerase complex, which supports earlier studies (Gupta el al., 2013). Taken together our study lays the foundation for future studies into the mechanism of telomerase mediated telomere maintenance in T. brucei. Future improvements are needed to develop a telomerase RNA -tagged proteomic mapping approach in T. brucei to validate endogenous interactions identified by this method and also detect new RNA-specific interactions. Future studies should also benefit from investigating interactomes from other T. brucei developmental stages since it appears from our recent study that T. brucei telomerase function is developmentally regulated (Dey et al., 2021). Therefore, characterizing stage-specific interactomes can provide novel insights into regulatory mechanisms that can affect rate of proliferation and telomerase activity in T. brucei.
Data availability statement
The data presented in the study are deposited in the ProteomeXchange (PRIDE) repository, accession number: PXD038235.
Author contributions
JD, BL, and KC designed experiments. JD, AR, Nitika, and AS conducted experiments. S-LX, AT, BL, and KC contributed critical reagents and suggestions. JD, AR, Nitika, and DW analyzed data. JD and KC wrote the manuscript. All authors revised and approved the final manuscript.
Funding
This work was funded by National Science Foundation (NSF) grant MCB-1764273 to KC, and MCB-1615896 to BL and by National Institute of Health (NIH) grant 1R15AI166764-01A1 to KC, and NIH grant 1R01GM139885 to AWT.
Acknowledgments
We thank KC lab members at UNC Charlotte and Li lab members at the Cleveland State University for their help and support with this work and Dr. Jun-tao Guo from the Department of Bioinformatics and Genomics at UNC Charlotte for help with structure-based prediction analysis using PDBeFold and DaliLite. Figures were created using BioRender.com.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2023.1110423/full#supplementary-material
References
Afrin, M., Gaurav, A. K., Yang, X., Pan, X., Zhao, Y., and Li, B ( (2020). Tb RAP1 has an unusual duplex DNA binding activity required for its telomere localization and VSG silencing. Sci. Adv. 6 (38), eabc4065. doi:10.1126/sciadv.abc4065
Ahmed, S., Passos, J. F., Birket, M. J., Beckmann, T., Brings, S., Peters, H., et al. (2008). Telomerase does not counteract telomere shortening but protects mitochondrial function under oxidative stress. J. Cell Sci. 121 (7), 1046–1053. doi:10.1242/jcs.019372
Ale-Agha, N., Jakobs, P., Goy, C., Zurek, M., Rosen, J., Dyballa-Rukes, N., et al. (2021). Mitochondrial telomerase reverse transcriptase protects from myocardial ischemia/reperfusion injury by improving complex I composition and function. Circulation 144 (23), 1876–1890. doi:10.1161/CIRCULATIONAHA.120.051923
Barth, S., Shalem, B., Hury, A., Tkacz, I. D., Liang, X. H., Uliel, S., et al. (2008). Elucidating the role of C/D snoRNA in rRNA processing and modification in Trypanosoma brucei. Eukaryot. Cell 7 (1), 86–101. doi:10.1128/EC.00215-07
Benmerzouga, I., Concepción-Acevedo, J., Kim, H. S., Vandoros, A. V., Cross, G. A., Klingbeil, M. M., et al. (2013). T rypanosoma brucei Orc 1 is essential for nuclear DNA replication and affects both VSG silencing and VSG switching. Mol. Microbiol. 87 (1), 196–210. doi:10.1111/mmi.12093
Berman, A. J., Gooding, A. R., and Cech, T. R. (2010). Tetrahymena telomerase protein p65 induces conformational changes throughout telomerase RNA (TER) and rescues telomerase reverse transcriptase and TER assembly mutants. J. Mol. Cell. Biol. 30 (20), 4965–4976. doi:10.1128/MCB.00827-10
Caruso Bavisotto, C., Alberti, G., Vitale, A. M., Paladino, L., Campanella, C., Rappa, F., et al. (2020). Hsp60 post-translational modifications: Functional and pathological consequences. Front. Mol. Biosci. 7, 95. doi:10.3389/fmolb.2020.00095
Chen, J. L., Blasco, M. A., and Greider, C. W. (2000). Secondary structure of vertebrate telomerase RNA. Cell 100 (5), 503–514. doi:10.1016/s0092-8674(00)80687-x
Chen, L., Roake, C. M., Freund, A., Batista, P. J., Tian, S., Yin, Y. A., et al. (2018). An activity switch in human telomerase based on RNA conformation and shaped by TCAB1. Cell 174 (1), 218–230. doi:10.1016/j.cell.2018.04.039
Collins, K. (2006). The biogenesis and regulation of telomerase holoenzymes. Nat. Rev. Mol. Cell Biol. 7 (7), 484–494. doi:10.1038/nrm1961
Cotobal, C., Rodríguez-López, M., Duncan, C., Hasan, A., Yamashita, A., Yamamoto, M., et al. (2015). Role of Ccr4-Not complex in heterochromatin formation at meiotic genes and subtelomeres in fission yeast. Epigenetics Chromatin 8 (1), 28–16. doi:10.1186/s13072-015-0018-4
De Lange, T., and Borst, P. (1982). Genomic environment of the expression-linked extra copies of genes for surface antigens of Trypanosoma brucei resembles the end of a chromosome. Nature 299 (5882), 451–453. doi:10.1038/299451a0
Dey, A., and Chakrabarti, K. (2018). Current perspectives of telomerase structure and function in eukaryotes with emerging views on telomerase in human parasites. Int. J. Mol. Sci. 19 (2), 333. doi:10.3390/ijms19020333
Dey, A., Monroy-Eklund, A., Klotz, K., Saha, A., Davis, J., Li, B., et al. (2021). In vivo architecture of the telomerase RNA catalytic core in Trypanosoma brucei. Nucleic Acids Res. 49 (21), 12445–12466. doi:10.1093/nar/gkab1042
Dreesen, O., and Cross, G. A. (2006). Telomerase-independent stabilization of short telomeres in Trypanosoma brucei. Mol. Cell Biol. 26 (13), 4911–4919. doi:10.1128/MCB.00212-06
Dreesen, O., Li, B., and Cross, G. A. (2005). Telomere structure and shortening in telomerase-deficient Trypanosoma brucei. Nucleic Acids Res. 33 (14), 4536–4543. doi:10.1093/nar/gki769
Du, H., Zhao, Y., He, J., Zhang, Y., Xi, H., Liu, M., et al. (2016). YTHDF2 destabilizes m6A-containing RNA through direct recruitment of the CCR4–NOT deadenylase complex. Nat. Commun. 7 (1), 12626–12637. doi:10.1038/ncomms12626
Egan, E. D., and Collins, K. (2012). An enhanced H/ACA RNP assembly mechanism for human telomerase RNA. Mol. Cell Biol. 32 (13), 2428–2439. doi:10.1128/MCB.00286-12
Ferguson, J. L., Chao, W. C. H., Lee, E., and Friedman, K. L. (2013). The anaphase promoting complex contributes to the degradation of the S. cerevisiae telomerase recruitment subunit Est1p. PLoS One 8 (1), e55055. doi:10.1371/journal.pone.0055055
Fu, D., and Collins, K. (2007). Purification of human telomerase complexes identifies factors involved in telomerase biogenesis and telomere length regulation. Mol. Cell 28 (5), 773–785. doi:10.1016/j.molcel.2007.09.023
Greider, C. W., and Blackburn, E. H. (1989). A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature 337 (6205), 331–337. doi:10.1038/337331a0
Gupta, S. K., Kolet, L., Doniger, T., Biswas, V. K., Unger, R., Tzfati, Y., et al. (2013). The Trypanosoma brucei telomerase RNA (TER) homologue binds core proteins of the C/D snoRNA family. FEBS Lett. 587 (9), 1399–1404. doi:10.1016/j.febslet.2013.03.017
Han, S., Zhao, B. S., Myers, S. A., Carr, S. A., He, C., and Ting, A. Y. (2020). RNA–protein interaction mapping via MS2-or Cas13-based APEX targeting. Proc. Natl. Acad. Sci. USA 117 (36), 22068–22079. doi:10.1073/pnas.2006617117
He, Y., Wang, Y., Liu, B., Helmling, C., Sušac, L., Cheng, R., et al. (2021). Structures of telomerase at several steps of telomere repeat synthesis. Nature 593 (7859), 454–459. doi:10.1038/s41586-021-03529-9
Hertz-Fowler, C., Figueiredo, L. M., Quail, M. A., Becker, M., Jackson, A., Bason, N., et al. (2008). Telomeric expression sites are highly conserved in Trypanosoma brucei. PLoS One 3 (10), e3527. doi:10.1371/journal.pone.0003527
Holm, L., and Rosenström, P. (2010). Dali server: Conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549. doi:10.1093/nar/gkq366
Holt, S. E., Aisner, D. L., Baur, J., Tesmer, V. M., Dy, M., Ouellette, M., et al. (1999). Functional requirement of p23 and Hsp90 in telomerase complexes. Genes Dev. 13 (7), 817–826. doi:10.1101/gad.13.7.817
Hovel-Miner, G. A., Boothroyd, C. E., Mugnier, M., Dreesen, O., Cross, G. A., and Papavasiliou, F. N. (2012). Telomere length affects the frequency and mechanism of antigenic variation in Trypanosoma brucei. PLoS Pathog. 8 (8), e1002900. doi:10.1371/journal.ppat.1002900
Indran, I. R., Hande, M. P., and Pervaiz, S. (2011). hTERT overexpression alleviates intracellular ROS production, improves mitochondrial function, and inhibits ROS-mediated apoptosis in cancer cells. Cancer Res. 71 (1), 266–276. doi:10.1158/0008-5472.CAN-10-1588
Jehi, S. E., Nanavaty, V., and Li, B. (2016). Trypanosoma brucei TIF2 and TRF suppress VSG switching using overlapping and independent mechanisms. PLoS One 11 (6), e0156746. doi:10.1371/journal.pone.0156746
Jehi, S. E., Wu, F., and Li, B. (2014). Trypanosoma brucei TIF2 suppresses VSG switching by maintaining subtelomere integrity. Cell Res. 24 (7), 870–885. doi:10.1038/cr.2014.60
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596 (7873), 583–589. doi:10.1038/s41586-021-03819-2
Jung, H.-Y., Wang, X., Jun, S., and Park, J.-I. (2013). Dyrk2-associated EDD-DDB1-VprBP E3 ligase inhibits telomerase by TERT degradation. J. Biol. Chem. 288 (10), 7252–7262. doi:10.1074/jbc.M112.416792
Klebanov-Akopyan, O., Mishra, A., Glousker, G., Tzfati, Y., and Shlomai, J. (2018). Trypanosoma brucei UMSBP2 is a single-stranded telomeric DNA binding protein essential for chromosome end protection. Nucleic Acids Res. 46 (15), 7757–7771. doi:10.1093/nar/gky597
Krissinel, E., and Henrick, K. (2004). Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D. Biol. Crystallogr. 60 (12), 2256–2268. doi:10.1107/S0907444904026460
Lin, K.-W., McDonald, K. R., Guise, A. J., Chan, A., Cristea, I. M., and Zakian, V. A. (2015). Proteomics of yeast telomerase identified Cdc48-Npl4-Ufd1 and Ufd4 as regulators of Est1 and telomere length. Nat. Commun. 6 (1), 8290–8304. doi:10.1038/ncomms9290
Mitchell, J. R., and Collins, K. (2000). Human telomerase activation requires two independent interactions between telomerase RNA and telomerase reverse transcriptase. Mol. Cell 6 (2), 361–371. doi:10.1016/s1097-2765(00)00036-8
Mochizuki, Y., He, J., Kulkarni, S., Bessler, M., and Mason, P. J. (2004). Mouse dyskerin mutations affect accumulation of telomerase RNA and small nucleolar RNA, telomerase activity, and ribosomal RNA processing. Proc. Natl. Acad. Sci. USA 101 (29), 10756–10761. doi:10.1073/pnas.0402560101
Moon, D. H., Segal, M., Boyraz, B., Guinan, E., Hofmann, I., Cahan, P., et al. (2015). Poly (A)-specific ribonuclease (PARN) mediates 3′-end maturation of the telomerase RNA component. Nat. Genet. 47 (12), 1482–1488. doi:10.1038/ng.3423
Nanavaty, V., Sandhu, R., Jehi, S. E., Pandya, U. M., and Li, B. (2017). Trypanosoma brucei RAP1 maintains telomere and subtelomere integrity by suppressing TERRA and telomeric RNA: DNA hybrids. Nucleic Acids Res. 45 (10), 5785–5796. doi:10.1093/nar/gkx184
Nittis, T., Guittat, L., LeDuc, R. D., Dao, B., Duxin, J. P., Rohrs, H., et al. (2010). Revealing novel telomere proteins using in vivo cross-linking, tandem affinity purification, and label-free quantitative LC-FTICR-MS. Mol. Cell Proteomics 9 (6), 1144–1156. doi:10.1074/mcp.M900490-MCP200
Peters, J.-M. (2006). The anaphase promoting complex/cyclosome: A machine designed to destroy. Nat. Rev. Mol. Cell Biol. 7 (9), 644–656. doi:10.1038/nrm1988
Podlevsky, J. D., Li, Y., and Chen, J. J. (2016). The functional requirement of two structural domains within telomerase RNA emerged early in eukaryotes. Nucleic Acids Res. 44 (20), 9891–9901. doi:10.1093/nar/gkw605
Rabbani, M., Tonini, M. L., Afrin, M., and Li, B. (2022). POLIE suppresses telomerase-mediated telomere G-strand extension and helps ensure proper telomere C-strand synthesis in trypanosomes. Nucleic Acids Res. 50 (4), 2036–2050. doi:10.1093/nar/gkac023
Roake, C. M., and Artandi, S. E. (2020). Regulation of human telomerase in homeostasis and disease. Nat. Rev. Mol. Cell Biol. 21 (7), 384–397. doi:10.1038/s41580-020-0234-z
Saha, A., Nanavaty, V. P., and Li, B. (2020). Telomere and subtelomere R-loops and antigenic variation in trypanosomes. J. Mol. Biol. 432 (15), 4167–4185. doi:10.1016/j.jmb.2019.10.025
Sandhu, R., Sanford, S., Basu, S., Park, M., Pandya, U. M., Li, B., et al. (2013). A trans-spliced telomerase RNA dictates telomere synthesis in Trypanosoma brucei. Cell Res. 23 (4), 537–551. doi:10.1038/cr.2013.35
Santos, J. H., Meyer, J. N., Skorvaga, M., Annab, L. A., and Van Houten, B. (2004). Mitochondrial hTERT exacerbates free-radical-mediated mtDNA damage. Aging Cell 3 (6), 399–411. doi:10.1111/j.1474-9728.2004.00124.x
Schuberth, C., Richly, H., Rumpf, S., and Buchberger, A. (2004). Shp1 and Ubx2 are adaptors of Cdc48 involved in ubiquitin-dependent protein degradation. EMBO Rep. 5 (8), 818–824. doi:10.1038/sj.embor.7400203
Sealey, D. C., Kostic, A. D., LeBel, C., Pryde, F., and Harrington, L. (2011). The TPR-containing domain within Est1 homologs exhibits species-specific roles in telomerase interaction and telomere length homeostasis. BMC Mol. Biol. 12 (1), 45–15. doi:10.1186/1471-2199-12-45
Sharma, N. K., Reyes, A., Green, P., Caron, M. J., Bonini, M. G., Gordon, D. M., et al. (2012). Human telomerase acts as a hTR-independent reverse transcriptase in mitochondria. Nucleic Acids Res. 40 (2), 712–725. doi:10.1093/nar/gkr758
Shay, J. W., and Wright, W. E. (2019). Telomeres and telomerase: Three decades of progress. Nat. Rev. Genet. 20 (5), 299–309. doi:10.1038/s41576-019-0099-1
Singh, M., Wang, Z., Koo, B.-K., Patel, A., Cascio, D., Collins, K., et al. (2012). Structural basis for telomerase RNA recognition and RNP assembly by the holoenzyme La family protein p65. Mol. Cell 47 (1), 16–26. doi:10.1016/j.molcel.2012.05.018
Tang, W., Kannan, R., Blanchette, M., and Baumann, P. (2012). Telomerase RNA biogenesis involves sequential binding by Sm and Lsm complexes. Nature 484 (7393), 260–264. doi:10.1038/nature10924
Te Raa, G., Derks, I., Navrkalová, V., Skowronska, A., Moerland, P., Van Laar, J., et al. (2015). The impact of SF3B1 mutations in CLL on the DNA-damage response. Leukemia 29 (5), 1133–1142. doi:10.1038/leu.2014.318
Tian, L., and Weizmann, Y. (2013). Real-time detection of telomerase activity using the exponential isothermal amplification of telomere repeat assay. J. Am. Chem. Soc. 135 (5), 1661–1664. doi:10.1021/ja309198j
Tyanova, S., Temu, Tikira., and Cox, Juergen. (2016). The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 11, 2301–2319. doi:10.1038/nprot.2016.136
Upton, H. E., Chan, H., Feigon, J., and Collins, K. J. J. o. B. C. (2017). Shared subunits of Tetrahymena telomerase holoenzyme and replication protein A have different functions in different cellular complexes. J. Biol. Chem. 292 (1), 217–228. doi:10.1074/jbc.M116.763664
Vasconcelos, E. J., Nunes, V. S., da Silva, M. S., Segatto, M., Myler, P. J., and Cano, M. I. N. (2014). The putative Leishmania telomerase RNA (Leish TER) undergoes trans-splicing and contains a conserved template sequence. PLoS One 9 (11), e112061. doi:10.1371/journal.pone.0112061
Venteicher, A. S., Meng, Z., Mason, P. J., Veenstra, T. D., and Artandi, S. E. (2008). Identification of ATPases pontin and reptin as telomerase components essential for holoenzyme assembly. Cell 132 (6), 945–957. doi:10.1016/j.cell.2008.01.019
Wang, L., Brooks, A. N., Fan, J., Wan, Y., Gambe, R., Li, S., et al. (2016). Transcriptomic characterization of SF3B1 mutation reveals its pleiotropic effects in chronic lymphocytic leukemia. Cancer Cell 30 (5), 750–763. doi:10.1016/j.ccell.2016.10.005
Wang, Y., Susac, L., and Feigon, J. (2019). Structural biology of telomerase. Cold Spring Harb. Perspect. Biol. 11 (12), a032383. doi:10.1101/cshperspect.a032383
Wirtz, E., Leal, S., Ochatt, C., and Cross, G. A. (1999). A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem Parasitol. 99 (1), 89–101. doi:10.1016/s0166-6851(99)00002-x
Witkin, K. L., and Collins, K. (2004). Holoenzyme proteins required for the physiological assembly and activity of telomerase. Genes Dev. 18 (10), 1107–1118. doi:10.1101/gad.1201704
Keywords: Trypanosoma brucei, cell proliferation, telomerase, telomere, cell growth, parasite, interactome, ribonucleoprotein (RNP)
Citation: Davis JA, Reyes AV, Nitika , Saha A, Wolfgeher DJ, Xu S-L, Truman AW, Li B and Chakrabarti K (2023) Proteomic analysis defines the interactome of telomerase in the protozoan parasite, Trypanosoma brucei. Front. Cell Dev. Biol. 11:1110423. doi: 10.3389/fcell.2023.1110423
Received: 28 November 2022; Accepted: 03 March 2023;
Published: 16 March 2023.
Edited by:
Ziyin Li, University of Texas Health Science Center at Houston, United StatesReviewed by:
Galadriel Hovel-Miner, George Washington University, United StatesMark Woodford, Upstate Medical University, United States
Copyright © 2023 Davis, Reyes, Nitika, Saha, Wolfgeher, Xu, Truman, Li and Chakrabarti. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kausik Chakrabarti, ay5jaGFrcmFiYXJ0aUB1bmNjLmVkdQ==
†Present address: Arpita Saha, Telomeres and Telomerase Group, Molecular Oncology Program, Spanish National Cancer Centre (CNIO), Madrid, Spain
 Justin A. Davis
Justin A. Davis Andres V. Reyes
Andres V. Reyes Nitika
Nitika Arpita Saha3†
Arpita Saha3† Shou-Ling Xu
Shou-Ling Xu Andrew W. Truman
Andrew W. Truman Bibo Li
Bibo Li Kausik Chakrabarti
Kausik Chakrabarti