- Key Laboratory of the Ministry of Education for Medicinal Plant Resources and Natural Pharmaceutical Chemistry, National Engineering Laboratory for Resource Development of Endangered Crude Drugs in the Northwest of China, College of Life Sciences, Shaanxi Normal University, Xi’an, China
The human body generates 10–100 billion cells every day, and the same number of cells die to maintain homeostasis. The genetically controlled, autonomously ordered cell death mainly proceeds by apoptosis. Apoptosis is an important way of programmed cell death in multicellular organisms, timely and effective elimination of apoptotic cells plays a key role in the growth and development of organisms and the maintenance of homeostasis. During the clearance of apoptotic cells, transcription factors bind to specific target promoters and act as activators or repressors to regulate multiple genes expression, how transcription factors regulate apoptosis is an important and poorly understood aspect of normal development. This paper summarizes the regulatory mechanisms of transcription factors in the clearance of apoptotic cells to date.
1 Introduction
In multicellular organisms, billions of cells are fated to be dead and cleared away for the normal development, and the excessive or harmful cells undergo a programmed progress which is activated and regulated by a series of genes (Fuchs and Steller, 2011). Programmed cell death (PCD) is a highly evolutionarily conserved process, which involves in embryonic development, maintenance of homeostasis and immune system development (Jacobson et al., 1997). At present, apoptosis is one of the typical types of PCD, which is regulated by mutiple molecular mechanisms and results in tissue remodeling or killing pathogens (Toda et al., 2015). This review summarizes the underlying mechanisms of the transcriptional control in apoptosis or apoptotic cell clearance.
2 Apoptosis
Apoptosis is an important cell biological process in animal development, which is considered be essensial for organogenesis and maintaining daily tissue homeostasis. Once the apoptosis program is initiated, the signals released from ACs rapidly recruit and be captured by the professional phagocytes or neighboring cells, ultimately promoting the engulfment and elimination of cell corpses (Elliott and Ravichandran, 2016). Generally, the occurrence of apoptosis involves three main steps: the activation and transduction of apoptotic signals, the initiation of apoptotic program, finally the clearance of ACs. Apoptosis occurs with a range of biochemical and morphological characteristics changes, like membrane shrinkage, DNA disassembly, production of apoptotic bodies and engulfment by phagocytes. The pronounced expression of cell surface apoptotic markers initiates early migration and recognition, triggering timely and efficient phagocytosis to avoid damaging neighboring tissues.
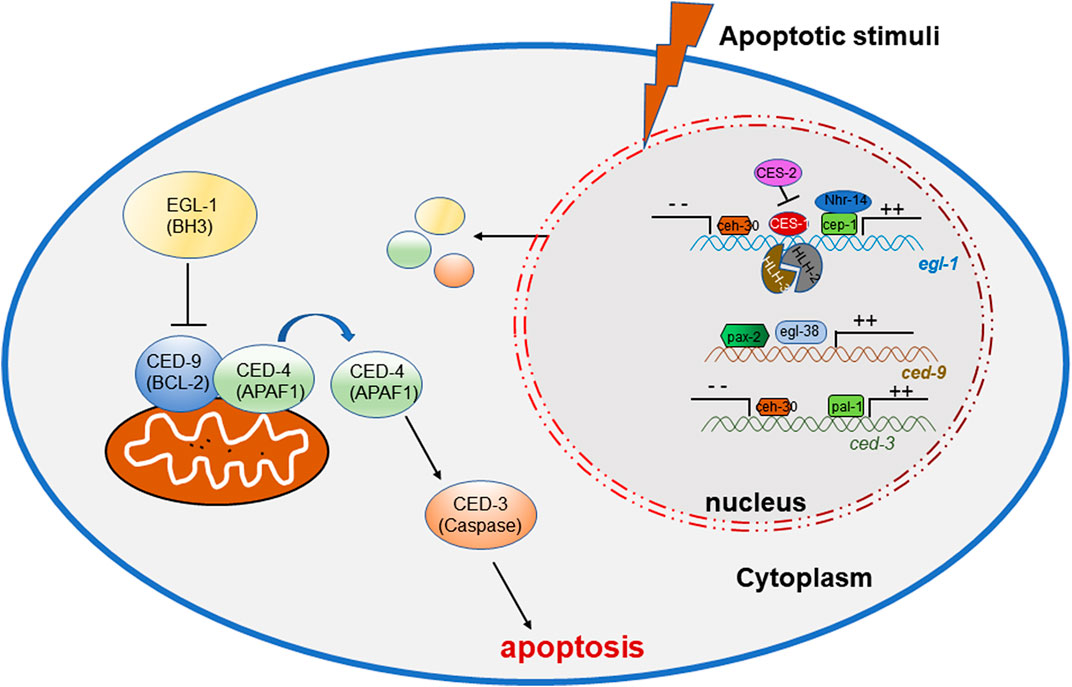
The original study of apoptosis molecular mechanism is started by the accurate description of the cell lineage during ontogenesis of Caenorhabditis elegans, in which there are 1090 somatic cells produced and 131 cells dead through apoptosis procedure. Observation on mutants which affected the 131 somatic cell deaths during worm development discovered a series of specific genes controlled the apoptosis process, which were proved to exist and play important roles as a similar manner in mammals (Sulston and Horvitz, 1977). In cells fated to die, EGL-1 binds to CED-9 and directly inhibits the interaction between CED-9 and CED-4, resulting in translocation of CED-4 from the mitochondrial surface to the perinucleus, thus promotes the activation of the CED-3 caspase and cell death (15) (Conradt and Horvitz, 1998).
Since the researchers have proved the apoptosis process is complicated yet evolutionarily conserved from invertebrates to mammals, which is mainly mediated by two independent and classical pathways: the death receptor (DR) extrinsic apoptotic pathway and the mitochondria-triggered endogenous pathway (Wang and Su, 2018; Xie et al., 2018). The extrinsic pathway is initiated by DRs members of the tumor necrosis factor-α (TNF-α) receptor superfamily, which bind to the exogenous ligands through their extracellular cyteine-rich domains, and transduct the apoptosis signal with the cytoplasmic domain called “death domain”. Generally, the death domains are essential for the downstream activation of apoptosis signal, which have high homology among different DRs. Current well-researched DR, Fas forms a tripo lymer in the cell membrane to recruit the adaptor protein FADD and proenzyme caspase 8, producing a protein complex called death inducing signaling complex (DISC) to initiate an apoptotic cascade (Guicciardi and Gores, 2009). In the intrinsic signaling pathway of apoptosis, mitochondria plays an critical role, and the release of cytochrome c is the key event mediated by the Bcl-2 family, in which process alters the permeability of cell membrane (Martinou and Youle, 2011). The two apoptotic pathways above seem to have totally different regulatory mechanisms, but they are both caspase dependent, where the caspases are cleavaged and activated to initiate the apoptosis through the amplified caspase cascade (Elmore, 2007; Abdolmaleki et al., 2018).
3 Apoptotic cell clearance
ACs that undergo programmed cell death are usually eugulfed by “professional” phagocytes or other cells timely and effectively, to avoid inflammatory responses or maintain homeostasis. Failure in this process would cause redundant ACs accumulation in the organ, ultimately lead to the development of inflammatory autoimmune or neurodegenerative diseases. Therefore, timely and effective clearance of apoptotic cells in multicellular organisms is of great importance for the homeostasis of the organism (Doran et al., 2020).
Before being recognized by phagocytes, ACs release or expose some different signal molecules, which may in turn promote the recognition and elimination of ACs. The process of clearing ACs by macrophages can be generally divided into four processes: recruitment, recognition, phagocytosis, and degradation (Wang and Yang, 2016). Through the study of model organisms such as C.elegans, Drosophila melanogaster, researchers have uncovered the molecular mechanism of ACs clearance by phagocytes. Differently, mammalian macrophages are divided into two sub groups according to their functions and levels of inflammatory cytokines (Roszer, 2015). The M1 macrophages (classically activated macrophages) are mainly activated by lipopolysaccharide produced by bacteria and cytokines, promoting to kill bacteria or inflammation (Rőszer, 2015). During clearance of ACs, macrophages are influenced by several factors (such as IL4) and polarize towards the M2 state. While M2 activation is involved in apoptotic cell clearance, wound healing is promoted and inflammation is inhibited (Lawrence and Natoli, 2011).
Apoptosis is also a ubiquitous process during epithelial morphogenesis and homeostasis, and typical mechanisms to maintain homeostasis include apical extrusion of apoptotic cells and phagocytes removal of residual cellular debris (Rosenblatt et al., 2001; Grieve and Rabouille, 2014). Epithelial cells and fibroblasts, as two typical non-professional macrophages, can absorb a limited range of apoptotic particles and play a certain degree of clearance (Rabinovitch, 1995). In recent years, such clearance mechanisms have been observed in the epithelial tissue areas of the lungs, thymus, and breasts (Cao et al., 2004; Monks et al., 2005; Juncadella et al., 2013). Unlike typical professional macrophages, epithelial cells perceive and process their environment more often from changes in actin and cadherin signals, rather than from typical transcription factors: the actin regulatory factor Coronin 1B promotes the formation of integrated actin networks through adhesion and recruitment of E-cadherin during biogenesis at the cell-cell junction to help it achieve optimal contraction and cyclic ability (Lubkov and Bar-Sagi, 2014; Michael et al., 2016).
3.1 Recruitment
When the process of apoptosis occurs, the ACs will generate and release some chemokines or other signals to recruit phagocytic cells. Meanwhile, in mammals, in addition to releasing find me to attract certain phagocytic cells, ACs also release “keep out” signaling factors to block the approach of inflammatory cells such as neutrophils. Lactoferrin, a multifunctional glycoprotein, is the only protein identified to date that acts as a “keep out” signal. “Find me” signalincludes lysophosphatidylcholine (LPC) (Lauber et al., 2003) and sphingosine 1-phosphate (S1P), nucleotides (including ATP and UTP 16), and chemokine CX3CL1 (fractalkine), etc. It has been shown that H2O2 may act as a “find me” signal in Drosophila embryos, which is required for the recruitment of blood cells to the wound area (Moreira et al., 2010). During the H2O2-induced wounding response, Src42A-Draper-Shark signaling was found to be essential for the hemocyte recruitment (Evans et al., 2015).
3.2 Identification
The cellular environment is very complex, including healthy cells, ACs, lymphocytes, and phagocytic cells. Therefore, the recognition of ACs by phagocytes is very crucial, and this process mainly depends on the molecules exposed to the apoptotic cell surface such as phosphatidylserine, Intercellular adhesionmolecule 3 (ICAM3) (Kristóf et al., 2013), Calreticulin (Gardai et al., 2005), oxidized low-density lipoprotein (Di and Maiseyeu, 2021), glycosylated surface proteins, etc. These signal molecules can be usually perceived as “eat me” signals, which distinguish ACs from healthy cells are easily recognized by phagocytes.
The most well-known and highly conserved “eat me” signal is phosphatidylserine (PS), phagocytes directly or indirectly recognize and bind these signals through their own PS recognizing membrane receptor (PSR), thereby promoting the recognition and engulfment during apoptosis and preventing inflammation. The trigger receptor and advanced glycation end product receptor of Brain Angiogenesis Inhibitor 1/3(BAI1/3), T cell immunoglobulin mucin receptor 4 (TIM4), Stabilin-1/2 (Stab2), (TREM)-like protein 2 (TLT2) are identified as directly binding to PS (Miyanishi et al., 2007; Park et al., 2007; Park et al., 2008; He et al., 2011). The recognition between PS and phagocytic receptors can also be mediated by bridging molecules, such as phagocytic receptor α v β3 integrin binds to ACs through PS-dependent bridging molecule MFG-E8; TAM receptor (receptor tyrosine kinase Mer Tyro3 and Axl, known as TAM receptors) recognizes PS through interacting with growth arrest-specific gene 6 (Gas6) or protein S (Savill et al., 1990; Nakano et al., 1997; Scott et al., 2001; Hanayama et al., 2002). Other bridging molecules including C1q, MBL, TSP-1 and TTR52 bind to phagocytic receptors LRP1, CRT, CD36 and MEGF10, respectively, in order to recognize PS (Savill et al., 1992; Ogden et al., 2001; Fond and Ravichandran, 2016). These receptors facilitate individually or coordinate with bridging molecules to promote the recognition of ACs.
We found that receptors on the surface of phagocytes are also highly conserved, such as in Drosophila Draper (Drpr, MEGF10 in mammals, CED-1 in worms), integrin and Croquemort (Crq, CD36 in mammals) and other phagocytosis receptors, which were found to function in different ways during phagocytosis: BAI1 is essential for phagosome formation, while TIM-4 stabilizes phagosomes (Mazaheri et al., 2014). In addition, healthy cells have a “Don’t eat me” signal that acts as an inhibitor to prevent phagocytosis by phagocytes, such as CD31, CD46 and CD47 (Poon et al., 2014).
Then macrophages integrate various signals from ACs and transmit them to the downstream, promoting a series of processes to recognize and engulf ACs.
3.3 Engulfment
Macrophages integrate signals from ACs to promote cytoskeletal rearrangement, a process that has been studied in the model organisms C. elegans and Drosophila, as well as in mammals. In C. elegans, upstream signals were found to converge in two parallel and independent signaling pathways:CED-2, CED-5, CED-12 pathway and CED-1, CED-6, CED-7 pathway which both subsequently activate CED-10, an evolutionarily highly conserved GTPase (Park and Kim, 2017) and thus stimulates skeletal rearrangement to form phagocytic vesicles (Wu and Horvitz, 1998a; Wu and Horvitz, 1998b; Liu and Hengartner, 1998; Reddien and Horvitz, 2000; Gumienny et al., 2001). CED-2/CED-5/CED-12 homologous signaling pathways in Drosophila and mouse were CG1587/myoblast city/Dmel, RKII/Dock180/ELMO1, and CED-1/CED-6 homologous signaling pathways were Drpr/dCed-6, MEGF10/GULP1 (Zheng et al., 2017), which are are highly conserved to regulate ACs clearance. Interaction between ABL-1 and ABI-1 inhibits ABI-1 and thus negatively regulates phagocytosis, but the study of this pathway in Drosophila and mammals remains to be determined (Hurwitz et al., 2009).
3.4 Degradation
The dynamics of PtdIns (4,5) P2 and PtdIns3P phosphatidylinositol is a very important event in the sealing of phagocytic vesicles (Flannagan et al., 2012; Cheng et al., 2015; Wang and Yang, 2016). PtdIns (4,5) P2, PtdIns3P are abundantly present in unconfined and confined phagosomes respectively. The model organism C. elegans has been relatively well studied in this regard. With the accumulation of PtdIns3P, the phagosome recruits SNX9 family protein LST-4 to the phagosome, and SNX9 further recruits DYN-1 to complete phagocytosis. Researches in C. elegans, Drosophila and mammalian cells show that abnormal Dyn-1 function results in stalled phagosome maturation and aggregation of apoptotic bodies within phagosomes, indicating that it plays a key role in phagosome maturation (Kinchen et al., 2008; Yu et al., 2008; Shklover et al., 2015).
The maturation of phagosomes undergo several processes, including early endosome, late phagosome and phagolysosome formation (Almendinger et al., 2011; Cheng et al., 2015). The different forms of membranous vesicles require a series of Rab GTPases proteins, which also participate in the acidification of phagocytic lysosomes. For example, the GTPases RAB-5 and RAB-7 bind to early and late endosomes respectively to mediate the processes (Li et al., 2009). It was shown that early phagosome recruits RAB-5 protein to assemble downstream factors while RAB-7 participates in the later stages of phagosome maturation and mediates phagocytosis and lysosomal fusion (Vieira et al., 2002; Kinchen and Ravichandran, 2008). In mammalian the Mon1 interacts with Rab5, and the Mon1-Ccz1 complex binds Rab7 and may affect Rab7 activation, so Mon1-Ccz1 may facilitate the transition from Rab5 to Rab7 through a Rab exchange mechanism, but whether SAND-1-CCZ-1 (Mon1-Ccz1) uses a similar mechanism to regulate phagosome formation in S. hidradiata remains to be determined (Kinchen and Ravichandran, 2010; Zheng et al., 2021).
Not only RAB-5, RAB-7 and LAMP-1 are recruited during the process of phagosome maturation, but also V-ATPase and other factors such as histone proteases. In nematode and zebrafish studies, V-ATPase was revealed to play a role in different stages of phagosome formation. In zebrafish V-ATPase is not only involved in the acidification of lysosomes, but may also play a role in phagosome maturation. In nematodes, it is required in the early stages of phagosome maturation (Zheng et al., 2021). After Rab7 recruitment to the phagosome, the HOPS complex begins to recruit, thereby activating Rab7 and facilitating its eventual fusion with the lysosomal structure (Kinchen and Ravichandran, 2008).
After phagocytic lysosome formation, the phagocytic lysosome recruits proteins to the surface and releases acid hydrolases, among others, to degrade ACs. In C. elegans, LAAT-1, the lysosomal lysine/arginine transporter maintains lysosomal persistence and releases lysosomal histone protease L (CPL-1) and DNase II(NUC-1) to control the digestion and degradation of ACs (Liu et al., 2012; Xu et al., 2014).
Finally, when apoptotic cells are degraded within the phagolysosomes, a large amount of metabolic cargo is produced, such as amino acids, lipids, nucleic acids and some other potentially cytotoxic macromolecules, so macrophages induce ABCA1 expression, allowing cholesterol efflux and reducing damage to membranes by harmful substances (Kiss et al., 2006; Han and Ravichandran, 2011). During the clearance of apoptotic cells, the vesicular cycle in the cell is also not negligible, and the process is regulated by the RAB family of GTPases. RAB17 is recruited into the phagosome containing apoptotic cells, thus mediating a vesicular cycle from the phagosome to the recycling endosome for the purpose of returning the cell surface area (Yin et al., 2019).
4 Transcription factor regulating apoptosis
In the past decades, we have made great progress in the study of apoptosis and ACs clearance, both in terms of mechanism and human diseases. However, in the large environment of the organism, there are multiple genes regulated by each other, protein-molecule interactions and signaling pathways to jointly maintain the homeostasis of the organism, so the research in this area still needs to be explored and exploited. As a hot topic of research in recent years, transcription factors play a crucial role in regulating the functions of organisms through the regulation of gene expression. Therefore, further study of apoptosis and apoptotic cell clearance by transcription factors has become an important direction to broaden the regulatory function of organisms.
Transcription factors are a class of proteins, interacting with cis-acting factors, which act as enhancers or silencers upstream of the transcription initiation region to enhance or inhibit gene expression respectively. The typical transcription factor contains three functional structural domains, namely the DNA binding domain, the transcriptional regulatory domain and the regulatory domain of other regulatory proteins, and individual transcription factors also have post-transcriptional regulatory domains (Ciarapica et al., 2003).
Generally, there are two categories of transcription factors classified by their action characteristics. The first category is general transcription factors, which can bind to RNA polymerase II to form transcription initiation complexes to enable downstream gene transcription and expression (Müller, 2001; Burley and Kamada, 2002). The second category of transcription factors is specific transcription factors, i.e., specific transcription factors required for individual gene expression. Transcription factors can be classified as zinc finger motif, helix-turn-helix (HTH), leucine zipper region (bZIP), homologous structural domains, nuclear receptors, etc. Based on the DNA binding domain, these categories account for more than 80% of human transcription factors (Vaquerizas et al., 2009; Lambert et al., 2018).
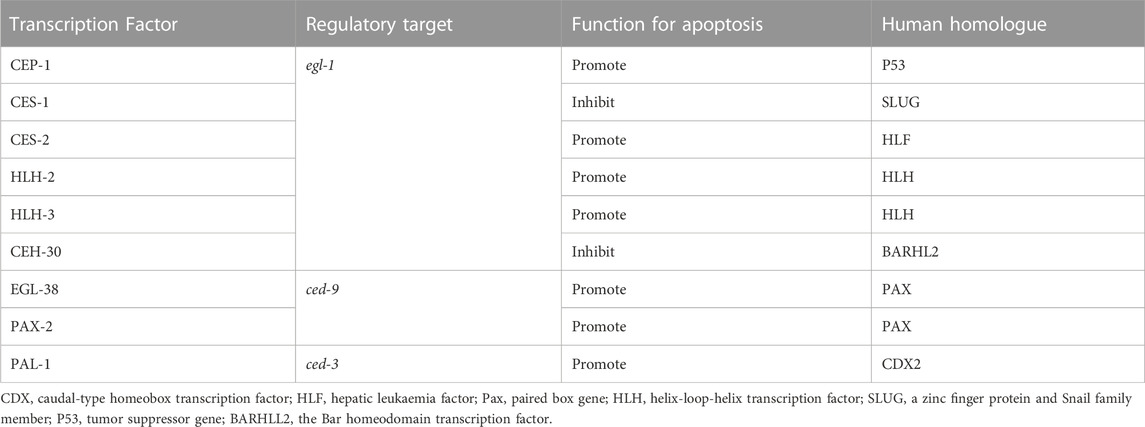
During the studies of apoptosis, researchers are seeking the roles of transcription factors and several key factors have been found to regulate the process. During the study of C. elegans, there are numerous correlations reporting the involvement of transcription factors in the apoptogenesis of ACs. Important factors in apoptosis, such as ced-3, ced-4, ced-9 and egl-1, are activated or suppressed by multiple transcription factors (Figure 1) (Wang and Yang, 2016), to initiate the normal occurrence of apoptosis, promoting the normal development and maintaining the homeostasis of the organism (Table 1).
In C. elegans, the p53 homolog CEP-1 is required for DNA damage-induced germ cell death by directly regulating the egl-1 transcripts (Hofmann et al., 2002), and NHR-14, which belongs to nuclear hormone receptors (NHRs), regulates egl-1 cooperating with CEP-1 to mediate germ cell apoptosis (Sang et al., 2022). During development, CES-1 inhibits the transcriptional expression of egl-1 through binding the Snail-binding sites in the elg-1 promoter, thus blocks programmed cell death in specific neurons. In C. elegans, ces-1 encodes a C2H2-type zinc finger transcription factor which belongs to the Snail family proteins, which transcription is negatively regulated by the bZIP transcription factor CES-2 by binding CES-1 upstream sequences in vitro and thus may directly repress ces-1 transcription in vivo (Metzstein and Horvitz, 1999). In regulating EGL-1 expression, the bHLH transcription factors HLH-2 and HLH-3 forms a heterodimer to bind to Snail-binding sites of the egl-1 locus in vitro and regulates the death of the NSM sister cells (Thellmann et al., 2003). ceh-30 is a Bar homeodomain transcription factor, encoding a homeodomain protein most similar to Drosophila and mammalian BarH1. CEH-30 blocks the death of the four male-specific cephalic companion neurons (CEMs) by repressing the transcription of both the egl-1 and ced-3 genes (Nehme et al., 2010).
The C. elegans gene egl-38 and pax-2 encodes a Pax transcription factor that is most similar to the mammalian Pax2/5/8 subclass of factors (Chamberlin et al., 1997), which have been proved to influence cell death and promote cell survival. Work in C. elegans shows that egl-38 and pax-2 act as a positive transcriptional regulators of ced-9 by directly binding to regulatory sequences upstream, thus influence both somatic and germline cell death (Park et al., 2006).
PAL-1, the C. elegans homolog of the mammalian tumor suppressor gene Cdx2, can bind to the ced-3 promoter sites and directly activate ced-3 transcription (Maurer et al., 2007), in order to control ced-3 expression and cell death in worm tail-spike cells.
Meanwhile, previous reports in mammals demonstrate that transcription factors promote normal cell reproduction and physiological activities by regulating various genes expression. For example, in response to different apoptotic stimuli, different genes are activated through transcription factors binding to specific DNA sequences, in order to promote or repress their expression. DNA damage gives rise to the production of the tumor supressor gene p53 (Gottlieb and Oren, 1998; Yu and Zhang, 2005), which activates the transcription of many pro-apoptotic genes and restain expression of oncogenes, such as c-Myc and E2F1, ultimitely leads to apoptosis (Thompson, 1998; Stanelle and Pützer, 2006). Pro-inflammatory cytokines or growth factors through NF-κB, IRF, STAT (signal transducer and activator of transcription) or FOXO family transcription factors are also proved to involve in apoptosis (de Martin et al., 1999; Fu, 1999; Zhang et al., 2011); transcription factors involved in apoptosis such as STAT92E, p53 and NF-κB have also been found in Drosophila (Betz et al., 2008; Tavignot et al., 2017; Zhou, 2019). Therefore, the study of transcription factors in apopptosis is of great importance in immune regulation, body homeostasis, and human disease treatment.
5 Transcriptional regulation of apoptotic cells clearance
Transcription factors are also involved in the clearance process of ACs. The current research on transcription factors is mainly focused on the process of macrophage maturity and activation, but the other processes are less involved at present.
5.1 Transcription factors regulate “find me” signaling
During efferocytosis, ACs can attract phagocytes by releasing “find me” signals, such as nucleotides, chemokines and their modified membranes. These molecules can stimulate the migration of macrophages to ACs, but the recruitment of find me signals to macrophages depends on many other factors, such as the type of phagocytes and ACs and the stimulation of apoptosis (Ravichandran, 2010).
S1P, a type of lysophospholipid, is a sphingosine metabolite produced by sphingosine kinase (SphK, SphK1/SphK2) acting on sphingosine as an important “find me” signal to recruit phagocytes (Gude et al., 2008; Luo et al., 2016). S1PR, the receptor for S1P, which belongs to the G protein-coupled receptor family, is required for cell survival, cell migration, apoptosis, and inflammation through binding to S1P. During apoptosis induced by DNA damage, p53 accumulates which in turn activates the lysosomal pathway as well as the mitochondrial pathway. The mitochondrial pathway causes caspase enzyme activation and the lysosomal pathway releases cathepsins into the cytoplasm. The proteases released by both pathways cause downregulation of SphK1, which in turn reduces intracellular S1P levels (Taha et al., 2004). However, it is not clear whether the regulation of SphK1 by proteases occurs through direct cleavage or some other indirect mechanism. Binding of extracellular S1P to S1PR reduces the production of pro-inflammatory factors and anti-inflammatory factors (e.g., IL-10), vascular endothelial growth factor (VEGF), nuclear transcription factor peroxisome proliferator-activated receptor λ (PPARλ), and erythropoietin EPO is also upregulated, thereby stimulating the anti-inflammatory macrophage phenotype, promoting the polarization of macrophage M2, and enhancing efferocytosis (Cheng et al., 2015). S1P also inhibits macrophage death, induces COX-2 expression, promotes cAMP production, and inhibites NF-κB signaling. In addition, binding of extracellular S1P to S1PR1 promoted phagocytic vesicle maturation after pathogen uptake (Weigert et al., 2009).
Nucleotides can also be used as find me signals to recruit phagocytes (Elliott et al., 2009). Nucleotides are released extracellularly in a time- and caspase-dependent manner at low levels of ATP as well as UTP. In ACs, the plasma membrane channel Pannexin-1 (PANX1) mediates the release of ATP and UTP by forming hexameric channels in a caspase-dependent manner, promoting the releasing of nucleotides, thus mediates the recruitment of macrophages by binding to the nucleotide receptor P2Y2 (Chekeni et al., 2010). Previous study showed the expression of Panx1 was initiated and activated by transcription factors CREB and ETV4 in the rat epididymis (Dufresne and Cyr, 2014), which may give us an inspiration on the regulatory pattern of find me signals. The conversion of extracellular ATP into adenosine, which is the result of the interaction between CD39 and CD73 (Idzko et al., 2014). Adenosine binds to the adenosine A2A receptor on the surface of macrophages and subsequently inhibits NF-κB signaling and upregulates the expression of Thbs1 and the nuclear receptor gene Nr4a. Thbs1 is a major activator of TGFβ, while members of the Nr4a family restrain the level of pro-inflammatory cytokines such as TNFα and IL-8 in macrophages and promote the efferocytosis process (Elliott et al., 1950; Yamaguchi et al., 2014) (Figure 2).
Since the research on find me signaling is relatively limited, the specific release mechanism of “find me” signaling during recruitment and how transcription factors regulate the process of recruiting macrophages remain to be discovered, which means that this area is well worth investigating.
5.2 Transcription factors regulate “eat-me” signaling
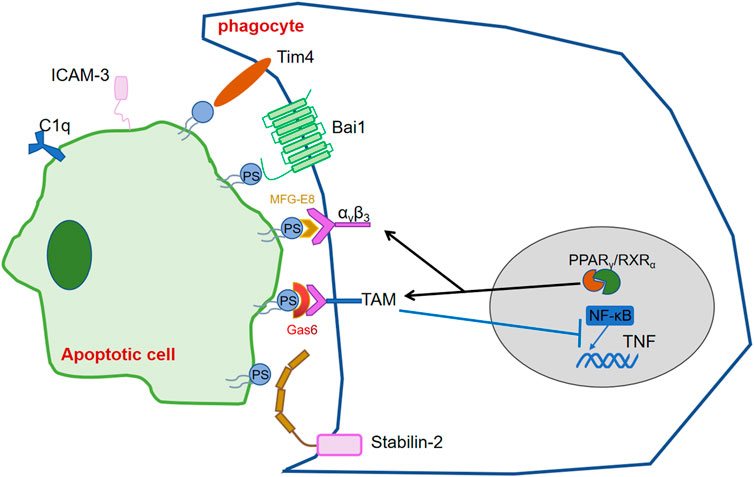
The migration and proximity of phagocytes to ACs depends on find me signals, while the specific recognition and binding to phagocytes depends on the exposure of eat me signals. The most studied and well-known eat me signal- PS, exposed on the cell surface of ACs by the co-regulation of phospholipid scramblase and flippase depending on the caspases activity (Segawa and Nagata, 2015).
Phagocytes recognize and bind these signals via their own PS recognition membrane receptors directly or indirectly via bridging molecules, thereby promote the clearance of ACs and prevent inflammation. Over the past few years, scientists realized that various receptors on phagocytes can recognize PS exposed on ACs. Tim4 expressed in lymphocytes of various mouse tissues, was found to bound and engulf ACs through recognizing PS by its immunoglobulin domain (Miyanishi et al., 2007). BAI1, which belongs to the adhesion-type G-protein-coupled receptor family, was reported as a PS recognition receptor, which formed a trimeric complex with ELMO and Dock180 to facilitate engulfment of ACs (Park et al., 2007). Stab2 which expresses in human monocyte-derived macrophages, mediates the clerance of aged red blood cells and ACs by recognizing PS (Park et al., 2008). MFG-E8 can simultaneously recruit integrin αvβ3 on phagocytes and recognize PS to uptake thre ACs. Gas6 and protein S, are involved in concatenate PS exposed on ACs to TAM receptors on phagocytes as bridging molecules (Anderson et al., 2003; Hanayama et al., 2004; Toda et al., 2012).
The expression of these receptors, and bridging molecules, enhances phagocyte recognition of ACs. Transcription factors such as the nuclear receptor super-family, STAT family and NF-κB have been found to be involved in the regulation of PSR, complement molecules and other eat me signals, which are important for the regulation of inflammation, the efficient and timely elimination of ACs and the stability of body’s immune system.
The peroxisome proliferator-activated receptor (PPAR) and the liver x receptor (LXR), which belong to the nuclear receptor superfamily, are involved in cellular lipid homeostasis. PPAR (heterodimers α, β/δ and γ) and LXR (heterodimers α, β) are ligand-activated transcriptional activators with different tissue expression. After binding to ligands such as fatty acids and oxysterols, PPAR and LXR then form heterodimers with the retinoid X receptor (RXR) and recruit co-activators to induce transcription of various genes involved in lipid and cholesterol metabolism (Han and Ravichandran, 2011). During ACs clearance, LXR and PPAR activate form heterodimers with RXR and upregulate phagocytic receptors and regulators. Existing studies have shown that activation of LXR and PPAR during apoptotic cell clearance leads to upregulation of phagocytic receptors (e.g.,: Mer) as well as of modulators (C1qb,Gas6,MFG-E8d) (A-Gonzalez et al., 2009; Mukundan et al., 2009). Here the clearance of apoptotic cells is influenced by two main pathways. In the first pathway, phagocytosis of apoptotic cells leads to the activation of LXR as well as PPAR, which upon activation regulates the expression of relevant phagocytic receptors as well as regulators. In the second pathway, recognition and binding of phagocytic receptors to phosphatidylserine activates LXR and PPAR, which in turn promotes the expression of Abca1, which not only induces cholesterol efflux but also promotes efferocytosis (Hamon et al., 2002). However, it is still unclear what ligands LXR and PPAR bind, how LXR and PPAR receive signals delivered by phosphatidylserine, and how they are transformed into macrophage lipid metabolism. Deficiency of RXR affects the transcription of cell surface receptors (e.g., CD36, Fcgr1, MERTK, Axl, etc.), regulators (e.g., C1qa, C1qb, C1qc, etc.) and transglutaminase-2 (Tgm2), which play important roles in cytokinesis and other macrophage functions.
PPAR-δ, PPAR-γ and RXRα have been demonstrated to upregulate MERTK and AXL transcriptional levels in macrophages (Han and Ravichandran, 2011). Macrophages lacking PPARγ, PPARβ/δ or RXRα (Vaquerizas et al., 2009; Gutiérrez-González et al., 2019), or PPARγ (Naeini et al., 2020) affect Axl transcription and inhibit ACs uptake (Röszer, 2017), causing macrophage adhesion and migration to be blocked (Garabuczi et al., 2015; Röszer, 2017). During efferocytosis, Axl and Mer signaling pathways directly inhibit Toll-like receptor (TLR) and type I IFN-driven inflammatory signaling pathways by different mechanisms. In dendritic cells, activation of Mer by ACs inhibits the IκB kinase IKK activity downstream of TLR4, suppressing NF-κB and reducing its binding to the TNF promoter. In addition, the reduction of TNFα is mediated by activation of Axl receptor tyrosine kinase and inducing of Twist transcriptional repressor, which binds to the E box region of the TNF promoter and inhibits NF-κB-dependent transcription (Sen et al., 2007).
MERTK is indirectly controlled by the glucocorticoid GC, which has been shown to upregulate LXR/RXR expression and eventually increase the uptake of ACs. Glucocorticoids increase the phagocytic ability to ACs both in short-term and subsequent phagocytosis. Short-term phagocytosis is mainly elevated by increasing of MERTK and C1q expression levels, whereas sustained phagocytosis acts by promoting the expression of LXR, PPARδ and UCP2 (Garabuczi et al., 2015) (Figure 3).
5.3 Transcription factors regulate apoptotic cell clearance and degradation
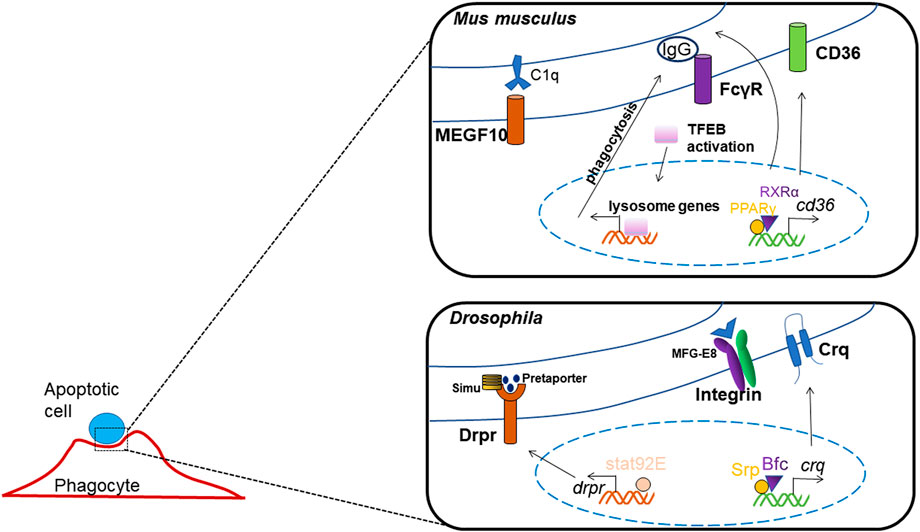
Macrophages internalize pathogens or ACs through enveloping them into vesicles called phagosomes, which then fuse with lysosomes to mature into phagolysosomes and ultimately degrade the pathogens or ACs. The cellular mechanism has been revealed relatively clear and detailed, yet the roles of transcription factors in controlling apoptotic cell clearance are less reported (Figure 4). The most studied transcription factors, which regulate apoptotic cell clearance are PPARs, LXR, RAR, RXR and GR.
Researchers have found that the activation of Fcγ-receptor which mediated phagocytosis and endocytosis, causes nuclear transposition of TFEB, which enhances the expression of lysosome genes, TFEB silencing reduces the enhancements in degradation and bacterial killing mediated by Fcγ-receptor (Gray et al., 2016).
The expression of the class B scavenger receptor CD36 was regulated by transcription factors PPARγ and RXRα, as the Cd36 promoter includes a interaction element for PPARγ/RXRα heterodimers, thus activates the process of pathogen elimination and apoptotic cell recognition (Silverstein and Febbraio, 2009; Röszer, 2017). And, the homolog of CD36 in Drosophila, crq was found to be transcriptional regulated by GATA factor and its co-factor, Bfc, which interacts with Srp zinc finger domain to strengthen this binding; thus, they function together in boosting crq expression and efferocytosis (Zheng et al., 2021). In Drosophila, it has also been shown that Srp may regulate the expression of factors mediating phagosome maturation and apoptotic cell degradation, thus the process of ACs clearance may be influenced by its deletion (Shlyakhover et al., 2018).
Another phagocytotic receptor in Drosophia, Drpr, has been reported to be regulated by transcription factor Stat92E, which can directly binds to the promoter of drpr, and mediates glial phagocytosis to axonal debris (Doherty et al., 2014).
In studies of TIM-1-mediated acute kidney injury, binding of TIM-1 to ACs triggers TIM-1 phosphorylation and the recruitment of p85, which interact with each other to block TLR4 expression, or the phosphorylation and activation of NF-κB, resulting in an anti-inflammatory phenotype and phagocytosis (Yang et al., 2015).
Immunoglobulins G and M (IgG, IgM) and complement binding to ACs provide eat me signals for macrophages. Increased PPARγ and RXR ligands promote IgG or IgM recognition by ACs, while the absence of PPARγ or RXRα reduces the expression of complement factors (e.g., C1q), thus inhibits the binding and uptake of ACs by macrophages (Roszer et al., 1950; Mukundan et al., 2009).
6 Conclusion
In the last few decades, we have made great progress in the study of apoptotic cell clearance. In living organisms, molecules, proteins and signalling pathways interact with each other, and the signalling pathways and functions involved are complex and diverse. However, there are still many regulators involved in apoptotic cell clearance waiting to be discovered and clarified. At the same time, the presence of transcription factors, a hot topic of research in recent years, regulates gene expression and affects the function of the organism.
The regulatory role of transcription factors is not entirely point-to-point; it may be a one-to-many or many-to-one process, hence the complexity of its study. In previous studies, numerous transcription factors such as the nuclear receptor superfamily, IRF, AP-1, NF-κB and STAT family have been identified as regulating the clearance of ACs. Through the regulation of genes participating in the clearance of ACs, transcription factors directly or indirectly influence the recognition of ACs, the maturation of macrophages and the degradation of ACs, leading to the development of organs and maintenance of the immune system. As impaired clearance of apoptotic cells leads to human diseases, the NR superfamily and transcription factors such as AhR have been documented as important targets for the prevention and treatment of hyperlipidaemia, diabetes and chronic inflammatory diseases including atherosclerosis, as well as autoimmune diseases.
In conclusion, transcription factors play important roles in the regulation of ACs clearance. There are still some unexplored signaling pathways between ACs and macrophages, and the current articles on the involvement of transcription factors in the process of apoptotic cell clearance are relatively superficial, and there are still many questions waiting to be explored, such as how transcription factors regulate how transcription factors regulate the shutdown of efferocytosis when apoptotic cell clearance is completed and how the high load/continuous b efferocytosis is transcriptionally regulated, etc. Therefore, finding new signaling pathways through transcription factors would be a valuable approach to broaden the field of apoptotic cell clearance. At the same time, with the advances in big data analysis and experimental techniques, it is hopeful that researchers will be able to broaden the field of research while working together to provide new insights and avenues to treat human diseases. There is an intriguing, but still not fully understood transcriptional mechanism between ACs and macrophages, which requires for further studies in the revelation of transcriptional regulation in ACs clearance and its therapeutic use.
Author contributions
HX and QZ conceived the study. YG and YJ wrote the manuscript and drew the figures in the text. XG and JL did the references searching.
Funding
This work was partially supported by the National Natural Science Foundation of China (Grant No. 31871387 to HX), Natural Science Foundation of Shaanxi Province Youth Program, China (Grant No. 2022JQ-208 to QZ).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
A-Gonzalez, N., Bensinger, S. J., Hong, C., Beceiro, S., Bradley, M. N., Zelcer, N., et al. (2009). Apoptotic cells promote their own clearance and immune tolerance through activation of the nuclear receptor LXR. Immunity 31 (2), 245–258. doi:10.1016/j.immuni.2009.06.018
Abdolmaleki, F., Farahani, N., Gheibi Hayat, S. M., Pirro, M., Bianconi, V., Barreto, G. E., et al. (2018). The role of efferocytosis in autoimmune diseases. Front. Immunol. 9, 1645. doi:10.3389/fimmu.2018.01645
Almendinger, J., Doukoumetzidis, K., Kinchen, J. M., Kaech, A., Ravichandran, K. S., and Hengartner, M. O. (2011). A conserved role for SNX9-family members in the regulation of phagosome maturation during engulfment of apoptotic cells. PloS One 6 (4), e18325. doi:10.1371/journal.pone.0018325
Anderson, H. A., Maylock, C. A., Williams, J. A., Paweletz, C. P., Shu, H., and Shacter, E. (2003). Serum-derived protein S binds to phosphatidylserine and stimulates the phagocytosis of apoptotic cells. Nat. Immunol. 4 (1), 87–91. doi:10.1038/ni871
Betz, A., Ryoo, H. D., Steller, H., and Darnell, J. E. (2008). STAT92E is a positive regulator of Drosophila inhibitor of apoptosis 1 (DIAP/1) and protects against radiation-induced apoptosis. Proc. Natl. Acad. Sci. U. S. A. 105 (37), 13805–13810. doi:10.1073/pnas.0806291105
Burley, S. K., and Kamada, K. (2002). Transcription factor complexes. Curr. Opin. Struct. Biol. 12 (2), 225–230. doi:10.1016/s0959-440x(02)00314-7
Cao, W. M., Murao, K., ImacHi, H., Hiramine, C., Abe, H., Yu, X., et al. (2004). Phosphatidylserine receptor cooperates with high-density lipoprotein receptor in recognition of apoptotic cells by thymic nurse cells. J. Mol. Endocrinol. 32 (2), 497–505. doi:10.1677/jme.0.0320497
Chamberlin, H. M., Palmer, R. E., Newman, A. P., Sternberg, P. W., Baillie, D. L., and Thomas, J. H. (1997). The PAX gene egl-38 mediates developmental patterning in Caenorhabditis elegans. Development 124 (20), 3919–3928. doi:10.1242/dev.124.20.3919
Chekeni, F. B., Elliott, M. R., Sandilos, J. K., Walk, S. F., Kinchen, J. M., Lazarowski, E. R., et al. (2010). Pannexin 1 channels mediate 'find-me' signal release and membrane permeability during apoptosis. Nature 467 (7317), 863–867. doi:10.1038/nature09413
Cheng, S., Wang, K., Zou, W., Miao, R., Huang, Y., Wang, H., et al. (2015). PtdIns(4, 5)P₂ and PtdIns3P coordinate to regulate phagosomal sealing for apoptotic cell clearance. J. Cell Biol. 210 (3), 485–502. doi:10.1083/jcb.201501038
Ciarapica, R., Rosati, J., Cesareni, G., and Nasi, S. (2003). Molecular recognition in helix-loop-helix and helix-loop-helix-leucine zipper domains. Design of repertoires and selection of high affinity ligands for natural proteins. J. Biol. Chem. 278 (14), 12182–12190. doi:10.1074/jbc.M211991200
Conradt, B., and Horvitz, H. R. (1998). The C. elegans protein EGL-1 is required for programmed cell death and interacts with the Bcl-2-like protein CED-9. Cell 93 (4), 519–529. doi:10.1016/s0092-8674(00)81182-4
de Martin, R., Schmid, J. A., and Hofer-Warbinek, R. (1999). The NF-kappaB/Rel family of transcription factors in oncogenic transformation and apoptosis. Mutat. Res. 437 (3), 231–243. doi:10.1016/s1383-5742(99)00089-7
Di, L., and Maiseyeu, A. (2021). Low-density lipoprotein nanomedicines: Mechanisms of targeting, biology, and theranostic potential. Drug Deliv. 28 (1), 408–421. doi:10.1080/10717544.2021.1886199
Doherty, J., Sheehan, A. E., Bradshaw, R., Fox, A. N., Lu, T. Y., and Freeman, M. R. (2014). PI3K signaling and Stat92E converge to modulate glial responsiveness to axonal injury. PLoS Biol. 12 (11), e1001985. doi:10.1371/journal.pbio.1001985
Doran, A. C., Yurdagul, A., and Tabas, I. (2020). Efferocytosis in health and disease. Nat. Rev. Immunol. 20 (4), 254–267. doi:10.1038/s41577-019-0240-6
Dufresne, J., and Cyr, D. G. (2014). Regulation of the pannexin-1 promoter in the rat epididymis. Biol. Reprod. 91 (6), 143. doi:10.1095/biolreprod.114.122168
Elliott, M. R., Chekeni, F. B., Trampont, P. C., Lazarowski, E. R., Kadl, A., Walk, S. F., et al. (2009). Nucleotides released by apoptotic cells act as a find-me signal to promote phagocytic clearance. Nature 461 (7261), 282–286. doi:10.1038/nature08296
Elliott, M. R., Koster, K. M., and Murphy, P. S. (1950). Efferocytosis signaling in the regulation of macrophage inflammatory responses, J. Immunol. 198, 1387–1394. doi:10.4049/jimmunol.1601520
Elliott, M. R., and Ravichandran, K. S. (2016). The dynamics of apoptotic cell clearance. Dev. Cell 38 (2), 147–160. doi:10.1016/j.devcel.2016.06.029
Elmore, S. (2007). Apoptosis: A review of programmed cell death. Toxicol. Pathol. 35 (4), 495–516. doi:10.1080/01926230701320337
Evans, I. R., Rodrigues, F. S. L. M., Armitage, E. L., and Wood, W. (2015). Draper/CED-1 mediates an ancient damage response to control inflammatory blood cell migration in vivo. Curr. Biol. CB 25 (12), 1606–1612. doi:10.1016/j.cub.2015.04.037
Flannagan, R. S., Jaumouillé, V., and Grinstein, S. (2012). The cell biology of phagocytosis. Annu. Rev. Pathology 7, 61–98. doi:10.1146/annurev-pathol-011811-132445
Fond, A. M., and Ravichandran, K. S. (2016). Clearance of dying cells by phagocytes: Mechanisms and implications for disease pathogenesis. Adv. Exp. Med. Biol. 930, 25–49. doi:10.1007/978-3-319-39406-0_2
Fu, X. Y. (1999). From PTK-STAT signaling to caspase expression and apoptosis induction. Cell Death Differ. 6 (12), 1201–1208. doi:10.1038/sj.cdd.4400613
Fuchs, Y., and Steller, H. (2011). Programmed cell death in animal development and disease. Cell 147 (4), 742–758. doi:10.1016/j.cell.2011.10.033
Garabuczi, É., Sarang, Z., and Szondy, Z. (2015). Glucocorticoids enhance prolonged clearance of apoptotic cells by upregulating liver X receptor, peroxisome proliferator-activated receptor-δ and UCP2. Biochimica Biophysica Acta 1853 (3), 573–582. doi:10.1016/j.bbamcr.2014.12.014
Gardai, S. J., McPhillips, K. A., Frasch, S. C., Janssen, W. J., Starefeldt, A., Murphy-Ullrich, J. E., et al. (2005). Cell-surface calreticulin initiates clearance of viable or apoptotic cells through trans-activation of LRP on the phagocyte. Cell 123 (2), 321–334. doi:10.1016/j.cell.2005.08.032
Gottlieb, T. M., and Oren, M. (1998). p53 and apoptosis. Seminars Cancer Biol. 8 (5), 359–368. doi:10.1006/scbi.1998.0098
Gray, M. A., Choy, C. H., Dayam, R. M., Ospina-Escobar, E., Somerville, A., Xiao, X., et al. (2016). Phagocytosis enhances lysosomal and bactericidal properties by activating the transcription factor TFEB. Curr. Biol. 26 (15), 1955–1964. doi:10.1016/j.cub.2016.05.070
Grieve, A. G., and Rabouille, C. (2014). Extracellular cleavage of E-cadherin promotes epithelial cell extrusion. J. Cell Sci. 127 (15), 3331–3346. doi:10.1242/jcs.147926
Gude, D. R., Alvarez, S. E., Paugh, S. W., Mitra, P., Yu, J., Griffiths, R., et al. (2008). Apoptosis induces expression of sphingosine kinase 1 to release sphingosine-1-phosphate as a "come-and-get-me" signal. FASEB J. Official Publ. Fed. Am. Soc. Exp. Biol. 22 (8), 2629–2638. doi:10.1096/fj.08-107169
Guicciardi, M. E., and Gores, G. J. (2009). Life and death by death receptors. FASEB J. 23 (6), 1625–1637. doi:10.1096/fj.08-111005
Gumienny, T. L., BrugnEra, E., Tosello-Trampont, A. C., Kinchen, J. M., Haney, L. B., NishiwaKi, K., et al. (2001). CED-12/ELMO, a novel member of the CrkII/Dock180/Rac pathway, is required for phagocytosis and cell migration. Cell 107 (1), 27–41. doi:10.1016/s0092-8674(01)00520-7
Gutiérrez-González, M., Latorre, Y., Zuniga, R., Aguillon, J. C., Molina, M. C., and Altamirano, C. (2019). Transcription factor engineering in CHO cells for recombinant protein production. Crit. Rev. Biotechnol. 39 (5), 665–679. doi:10.1080/07388551.2019.1605496
Hamon, Y., Chambenoit, O., and Chimini, G. (2002). ABCA1 and the engulfment of apoptotic cells. Biochimica Biophysica Acta 1585 (2-3), 64–71. doi:10.1016/s1388-1981(02)00325-6
Han, C. Z., and Ravichandran, K. S. (2011). Metabolic connections during apoptotic cell engulfment. Cell 147 (7), 1442–1445. doi:10.1016/j.cell.2011.12.006
Hanayama, R., Tanaka, M., Miwa, K., Shinohara, A., Iwamatsu, A., and Nagata, S. (2002). Identification of a factor that links apoptotic cells to phagocytes. Nature 417 (6885), 182–187. doi:10.1038/417182a
Hanayama, R., Tanaka, M., Miyasaka, K., Aozasa, K., Koike, M., Uchiyama, Y., et al. (2004). Autoimmune disease and impaired uptake of apoptotic cells in MFG-E8-deficient mice. Science 304 (5674), 1147–1150. doi:10.1126/science.1094359
He, M., Kubo, H., Morimoto, K., Fujino, N., Suzuki, T., Takahasi, T., et al. (2011). Receptor for advanced glycation end products binds to phosphatidylserine and assists in the clearance of apoptotic cells. EMBO Rep. 12 (4), 358–364. doi:10.1038/embor.2011.28
Hofmann, E. R., Milstein, S., Boulton, S. J., Ye, M., Hofmann, J. J., Stergiou, L., et al. (2002). Caenorhabditis elegans HUS-1 is a DNA damage checkpoint protein required for genome stability and EGL-1-mediated apoptosis. Curr. Biol. 12 (22), 1908–1918. doi:10.1016/s0960-9822(02)01262-9
Hurwitz, M. E., Vanderzalm, P. J., Bloom, L., Goldman, J., Garriga, G., and Horvitz, H. R. (2009). Abl kinase inhibits the engulfment of apoptotic [corrected] cells in Caenorhabditis elegans. PLoS Biol. 7 (4), e99. doi:10.1371/journal.pbio.1000099
Idzko, M., Ferrari, D., and Eltzschig, H. K. (2014). Nucleotide signalling during inflammation. Nature 509 (7500), 310–317. doi:10.1038/nature13085
Jacobson, M. D., Weil, M., and Raff, M. C. (1997). Programmed cell death in animal development. Cell 88 (3), 347–354. doi:10.1016/s0092-8674(00)81873-5
Juncadella, I. J., Kadl, A., Sharma, A. K., Shim, Y. M., Hochreiter-Hufford, A., Borish, L., et al. (2013). Apoptotic cell clearance by bronchial epithelial cells critically influences airway inflammation. Nature 493 (7433), 547–551. doi:10.1038/nature11714
Kinchen, J. M., Doukoumetzidis, K., Almendinger, J., Stergiou, L., Tosello-Trampont, A., Sifri, C. D., et al. (2008). A pathway for phagosome maturation during engulfment of apoptotic cells. Nat. Cell Biol. 10 (5), 556–566. doi:10.1038/ncb1718
Kinchen, J. M., and Ravichandran, K. S. (2010). Identification of two evolutionarily conserved genes regulating processing of engulfed apoptotic cells. Nature 464 (7289), 778–782. doi:10.1038/nature08853
Kinchen, J. M., and Ravichandran, K. S. (2008). Phagosome maturation: Going through the acid test. Nat. Rev. Mol. Cell Biol. 9 (10), 781–795. doi:10.1038/nrm2515
Kiss, R. S., Elliott, M. R., Ma, Z., Marcel, Y. L., and Ravichandran, K. S. (2006). Apoptotic cells induce a phosphatidylserine-dependent homeostatic response from phagocytes. Curr. Biol. 16 (22), 2252–2258. doi:10.1016/j.cub.2006.09.043
Kristóf, E., Zahuczky, G., Katona, K., Doro, Z., Nagy, E., and Fesus, L. (2013). Novel role of ICAM3 and LFA-1 in the clearance of apoptotic neutrophils by human macrophages. Apoptosis Int. J. Program. Cell Death 18 (10), 1235–1251. doi:10.1007/s10495-013-0873-z
Lambert, S. A., Jolma, A., Campitelli, L. F., Das, P. K., Yin, Y., Albu, M., et al. (2018). The human transcription factors. Cell 172 (4), 650–665. doi:10.1016/j.cell.2018.01.029
Lauber, K., Bohn, E., Krober, S. M., Xiao, Y. j., Blumenthal, S. G., Lindemann, R. K., et al. (2003). Apoptotic cells induce migration of phagocytes via caspase-3-mediated release of a lipid attraction signal. Cell 113 (6), 717–730. doi:10.1016/s0092-8674(03)00422-7
Lawrence, T., and Natoli, G. (2011). Transcriptional regulation of macrophage polarization: Enabling diversity with identity. Nat. Rev. Immunol. 11 (11), 750–761. doi:10.1038/nri3088
Li, W., Zou, W., Zhao, D., Yan, J., Zhu, Z., Lu, J., et al. (2009). C. elegans Rab GTPase activating protein TBC-2 promotes cell corpse degradation by regulating the small GTPase RAB-5. Development 136 (14), 2445–2455. doi:10.1242/dev.035949
Liu, B., Du, H., Rutkowski, R., Gartner, A., and Wang, X. (2012). LAAT-1 is the lysosomal lysine/arginine transporter that maintains amino acid homeostasis. Science 337 (6092), 351–354. doi:10.1126/science.1220281
Liu, Q. A., and Hengartner, M. O. (1998). Candidate adaptor protein CED-6 promotes the engulfment of apoptotic cells in C. elegans. Cell 93 (6), 961–972. doi:10.1016/s0092-8674(00)81202-7
Lubkov, V., and Bar-Sagi, D. (2014). E-cadherin-mediated cell coupling is required for apoptotic cell extrusion. Curr. Biol. 24 (8), 868–874. doi:10.1016/j.cub.2014.02.057
Luo, B., Gan, W., Liu, Z., Shen, Z., Wang, J., Shi, R., et al. (2016). Erythropoeitin signaling in macrophages promotes dying cell clearance and immune tolerance. Immunity 44 (2), 287–302. doi:10.1016/j.immuni.2016.01.002
Martinou, J. C., and Youle, R. J. (2011). Mitochondria in apoptosis: Bcl-2 family members and mitochondrial dynamics. Dev. Cell 21 (1), 92–101. doi:10.1016/j.devcel.2011.06.017
Maurer, C. W., Chiorazzi, M., and Shaham, S. (2007). Timing of the onset of a developmental cell death is controlled by transcriptional induction of the C. elegans ced-3 caspase-encoding gene. Development 134 (7), 1357–1368. doi:10.1242/dev.02818
Mazaheri, F., Breus, O., Durdu, S., Haas, P., Wittbrodt, J., Gilmour, D., et al. (2014). Distinct roles for Bai1 and TIM-4 in the engulfment of dying neurons by microglia. Nat. Commun. 5, 4046. doi:10.1038/ncomms5046
Metzstein, M. M., and Horvitz, H. R. (1999). The C. elegans cell death specification gene ces-1 encodes a snail family zinc finger protein. Mol. Cell 4 (3), 309–319. doi:10.1016/s1097-2765(00)80333-0
Michael, M., Meiring, J. C. M., Acharya, B. R., Matthews, D. R., Verma, S., Han, S. P., et al. (2016). Coronin 1B reorganizes the architecture of F-actin networks for contractility at steady-state and apoptotic adherens junctions. Dev. Cell 37 (1), 58–71. doi:10.1016/j.devcel.2016.03.008
Miyanishi, M., Tada, K., Koike, M., Uchiyama, Y., Kitamura, T., and Nagata, S. (2007). Identification of Tim4 as a phosphatidylserine receptor. Nature 450 (7168), 435–439. doi:10.1038/nature06307
Monks, J., Rosner, D., Geske, F. J., Lehman, L., Hanson, L., Neville, M. C., et al. (2005). Epithelial cells as phagocytes: Apoptotic epithelial cells are engulfed by mammary alveolar epithelial cells and repress inflammatory mediator release. Cell Death Differ. 12 (2), 107–114. doi:10.1038/sj.cdd.4401517
Moreira, S., Stramer, B., Evans, I., Wood, W., and Martin, P. (2010). Prioritization of competing damage and developmental signals by migrating macrophages in the Drosophila embryo. Curr. Biol. CB 20 (5), 464–470. doi:10.1016/j.cub.2010.01.047
Mukundan, L., Odegaard, J. I., Morel, C. R., Heredia, J. E., Mwangi, J. W., Ricardo-Gonzalez, R. R., et al. (2009). PPAR-delta senses and orchestrates clearance of apoptotic cells to promote tolerance. Nat. Med. 15 (11), 1266–1272. doi:10.1038/nm.2048
Müller, C. W. (2001). Transcription factors: Global and detailed views. Curr. Opin. Struct. Biol. 11 (1), 26–32. doi:10.1016/s0959-440x(00)00163-9
Naeini, M. B., Bianconi, V., Pirro, M., and Sahebkar, A. (2020). The role of phosphatidylserine recognition receptors in multiple biological functions. Cell. Mol. Biol. Lett. 25, 23. doi:10.1186/s11658-020-00214-z
Nakano, T., Ishimoto, Y., Kishino, J., UmedaM., , Inoue, K., Nagata, K., et al. (1997). Cell adhesion to phosphatidylserine mediated by a product of growth arrest-specific gene 6. J. Biol. Chem. 272 (47), 29411–29414. doi:10.1074/jbc.272.47.29411
Nehme, R., Grote, P., Tomasi, T., LoSer, S., Holzkamp, H., Schnabel, R., et al. (2010). Transcriptional upregulation of both egl-1 BH3-only and ced-3 caspase is required for the death of the male-specific CEM neurons. Cell Death Differ. 17 (8), 1266–1276. doi:10.1038/cdd.2010.3
Ogden, C. A., deCAthelineAu, A., Hoffmann, P. R., Bratton, D., GheBrehiwet, B., Fadok, V. A., et al. (2001). C1q and mannose binding lectin engagement of cell surface calreticulin and CD91 initiates macropinocytosis and uptake of apoptotic cells. J. Exp. Med. 194 (6), 781–795. doi:10.1084/jem.194.6.781
Park, D., Jia, H., Rajakumar, V., and Chamberlin, H. M. (2006). Pax2/5/8 proteins promote cell survival in C. elegans. Development 133 (21), 4193–4202. doi:10.1242/dev.02614
Park, D., Tosello-Trampont, A. C., Elliott, M. R., Lu, M., Haney, L. B., Ma, Z., et al. (2007). Bai1 is an engulfment receptor for apoptotic cells upstream of the ELMO/Dock180/Rac module. Nature 450 (7168), 430–434. doi:10.1038/nature06329
Park, S.-Y., and Kim, I.-S. (2017). Engulfment signals and the phagocytic machinery for apoptotic cell clearance. Exp. Mol. Med. 49 (5), e331. doi:10.1038/emm.2017.52
Park, S. Y., Jung, M. Y., Kim, H. J., Lee, S. J., Kim, S. Y., Lee, B. H., et al. (2008). Rapid cell corpse clearance by stabilin-2, a membrane phosphatidylserine receptor. Cell Death Differ. 15 (1), 192–201. doi:10.1038/sj.cdd.4402242
Poon, I. K. H., Lucas, C. D., Rossi, A. G., and Ravichandran, K. S. (2014). Apoptotic cell clearance: Basic biology and therapeutic potential. Nat. Rev. Immunol. 14 (3), 166–180. doi:10.1038/nri3607
Rabinovitch, M. (1995). Professional and non-professional phagocytes: An introduction. Trends Cell Biol. 5 (3), 85–87. doi:10.1016/s0962-8924(00)88955-2
Ravichandran, K. S. (2010). Find-me and eat-me signals in apoptotic cell clearance: Progress and conundrums. J. Exp. Med. 207 (9), 1807–1817. doi:10.1084/jem.20101157
Reddien, P. W., and Horvitz, H. R. (2000). CED-2/CrkII and CED-10/Rac control phagocytosis and cell migration in Caenorhabditis elegans. Nat. Cell Biol. 2 (3), 131–136. doi:10.1038/35004000
Rosenblatt, J., Raff, M. C., and Cramer, L. P. (2001). An epithelial cell destined for apoptosis signals its neighbors to extrude it by an actin- and myosin-dependent mechanism. Curr. Biol. 11 (23), 1847–1857. doi:10.1016/s0960-9822(01)00587-5
Roszer, T., Menendez-Gutierrez, M. P., Lefterova, M. I., Alameda, D., Nunez, V., Lazar, M. A., et al. (1950). Autoimmune kidney disease and impaired engulfment of apoptotic cells in mice with macrophage peroxisome proliferator-activated receptor gamma or retinoid X receptor alpha deficiency. J. Immunol. Baltim. Md 186 (1), 621–631. doi:10.4049/jimmunol.1002230
Röszer, T. (2017). Transcriptional control of apoptotic cell clearance by macrophage nuclear receptors. Apoptosis Int. J. Program. Cell Death 22 (2), 284–294. doi:10.1007/s10495-016-1310-x
Roszer, T. (2015). Understanding the mysterious M2 macrophage through activation markers and effector mechanisms. Mediat. Inflamm. 2015, 816460. doi:10.1155/2015/816460
Sang, L., Dong, R., Liu, R., Hao, Q., Bai, W., and Sun, J. (2022). Caenorhabditis elegans NHR-14/HNF4α regulates DNA damage-induced apoptosis through cooperating with cep-1/p53. Cell Commun. Signal 20 (1), 135. doi:10.1186/s12964-022-00920-5
Savill, J., DransfIeld, I., HoggN., , and Haslett, C. (1990). Vitronectin receptor-mediated phagocytosis of cells undergoing apoptosis. Nature 343 (6254), 170–173. doi:10.1038/343170a0
Savill, J., HoggN., , Ren, Y., and Haslett, C. (1992). Thrombospondin cooperates with CD36 and the vitronectin receptor in macrophage recognition of neutrophils undergoing apoptosis. J. Clin. Investigation 90 (4), 1513–1522. doi:10.1172/JCI116019
Scott, R. S., McMahon, E. J., Pop, S. M., Reap, E. A., CaRicchio, R., Cohen, P. L., et al. (2001). Phagocytosis and clearance of apoptotic cells is mediated by MER. Nature 411 (6834), 207–211. doi:10.1038/35075603
Segawa, K., and Nagata, S. (2015). An apoptotic 'eat me' signal: Phosphatidylserine exposure. Trends Cell Biol. 25 (11), 639–650. doi:10.1016/j.tcb.2015.08.003
Sen, P., Wallet, M. A., Yi, Z., Huang, Y., Henderson, M., Mathews, C. E., et al. (2007). Apoptotic cells induce Mer tyrosine kinase-dependent blockade of NF-kappaB activation in dendritic cells. Blood 109 (2), 653–660. doi:10.1182/blood-2006-04-017368
Shklover, J., MishnaevsKi, K., Levy-AdamF., , and Kurant, E. (2015). JNK pathway activation is able to synchronize neuronal death and glial phagocytosis in Drosophila. Cell Death Dis. 6, e1649. doi:10.1038/cddis.2015.27
Shlyakhover, E., Shklyar, B., Hakim-Mishnaevski, K., Levy-Adam, F., and Kurant, E. (2018). Drosophila GATA factor serpent establishes phagocytic ability of embryonic macrophages. Front. Immunol. 9, 266. doi:10.3389/fimmu.2018.00266
Silverstein, R. L., and Febbraio, M. (2009). CD36, a scavenger receptor involved in immunity, metabolism, angiogenesis, and behavior. Sci. Signal. 2 (72), re3. doi:10.1126/scisignal.272re3
Stanelle, J., and Pützer, B. M. (2006). E2F1-induced apoptosis: Turning killers into therapeutics. Trends Mol. Med. 12 (4), 177–185. doi:10.1016/j.molmed.2006.02.002
Sulston, J. E., and Horvitz, H. R. (1977). Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 56 (1), 110–156. doi:10.1016/0012-1606(77)90158-0
Taha, T. A., Osta, W., Kozhaya, L., Bielawski, J., Johnson, K. R., Gillanders, W. E., et al. (2004). Down-regulation of sphingosine kinase-1 by DNA damage: Dependence on proteases and p53. J. Biol. Chem. 279 (19), 20546–20554. doi:10.1074/jbc.M401259200
Tavignot, R., Chaduli, D., Djitte, F., Charroux, B., and Royet, J. (2017). Inhibition of a NF-κB/Diap1 pathway by PGRP-LF is required for proper apoptosis during Drosophila development. PLoS Genet. 13 (1), e1006569. doi:10.1371/journal.pgen.1006569
Thellmann, M., Hatzold, J., and Conradt, B. (2003). The Snail-like CES-1 protein of C. elegans can block the expression of the BH3-only cell-death activator gene egl-1 by antagonizing the function of bHLH proteins. Development 130 (17), 4057–4071. doi:10.1242/dev.00597
Thompson, E. B. (1998). The many roles of c-Myc in apoptosis. Annu. Rev. Physiology 60, 575–600. doi:10.1146/annurev.physiol.60.1.575
Toda, S., Hanayama, R., and Nagata, S. (2012). Two-step engulfment of apoptotic cells. Mol. Cell Biol. 32 (1), 118–125. doi:10.1128/MCB.05993-11
Toda, S., Nishi, C., Yanagihashi, Y., Segawa, K., and Nagata, S. (2015). Clearance of apoptotic cells and pyrenocytes. Curr. Top. Dev. Biol. 114, 267–295. doi:10.1016/bs.ctdb.2015.07.017
Vaquerizas, J. M., Kummerfeld, S. K., Teichmann, S. A., and Luscombe, N. M. (2009). A census of human transcription factors: Function, expression and evolution. Nat. Rev. Genet. 10 (4), 252–263. doi:10.1038/nrg2538
Vieira, O. V., Botelho, R. J., and Grinstein, S. (2002). Phagosome maturation: Aging gracefully. Biochem. J. 366 (3), 689–704. doi:10.1042/BJ20020691
Wang, M., and Su, P. (2018). The role of the Fas/FasL signaling pathway in environmental toxicant-induced testicular cell apoptosis: An update. Syst. Biol. Reproductive Med. 64 (2), 93–102. doi:10.1080/19396368.2017.1422046
Wang, X., and Yang, C. (2016). Programmed cell death and clearance of cell corpses in Caenorhabditis elegans. Cell. Mol. Life Sci. CMLS 73 (11-12), 2221–2236. doi:10.1007/s00018-016-2196-z
Weigert, A., Weis, N., and Brüne, B. (2009). Regulation of macrophage function by sphingosine-1-phosphate. Immunobiology 214 (9-10), 748–760. doi:10.1016/j.imbio.2009.06.003
Wu, Y. C., and Horvitz, H. R. (1998). C. elegans phagocytosis and cell-migration protein CED-5 is similar to human DOCK180. Nature 392 (6675), 501–504. doi:10.1038/33163
Wu, Y. C., and Horvitz, H. R. (1998). The C. elegans cell corpse engulfment gene ced-7 encodes a protein similar to ABC transporters. Cell 93 (6), 951–960. doi:10.1016/s0092-8674(00)81201-5
Xie, L.-L., Shi, F., Tan, Z., Li, Y., Bode, A. M., and Cao, Y. (2018). Mitochondrial network structure homeostasis and cell death. Cancer Sci. 109 (12), 3686–3694. doi:10.1111/cas.13830
Xu, M., Liu, Y., Zhao, L., Gan, Q., Wang, X., and Yang, C. (2014). The lysosomal cathepsin protease CPL-1 plays a leading role in phagosomal degradation of apoptotic cells in Caenorhabditis elegans. Mol. Biol. Cell 25 (13), 2071–2083. doi:10.1091/mbc.E14-01-0015
Yamaguchi, H., Maruyama, T., Urade, Y., and Nagata, S. (2014). Immunosuppression via adenosine receptor activation by adenosine monophosphate released from apoptotic cells. ELife 3, e02172. doi:10.7554/eLife.02172
Yang, L., Brooks, C. R., Xiao, S., Sabbisetti, V., Yeung, M. Y., Hsiao, L. L., et al. (2015). KIM-1-mediated phagocytosis reduces acute injury to the kidney. J. Clin. Investigation 125 (4), 1620–1636. doi:10.1172/JCI75417
Yin, C., Argintaru, D., and Heit, B. (2019). Rab17 mediates intermixing of phagocytosed apoptotic cells with recycling endosomes. Small GTPases 10 (3), 218–226. doi:10.1080/21541248.2017.1308852
Yu, J., and Zhang, L. (2005). The transcriptional targets of p53 in apoptosis control. Biochem. Biophysical Res. Commun. 331 (3), 851–858. doi:10.1016/j.bbrc.2005.03.189
Yu, X., Lu, N., and Zhou, Z. (2008). Phagocytic receptor CED-1 initiates a signaling pathway for degrading engulfed apoptotic cells. PLoS Biol. 6 (3), e61. doi:10.1371/journal.pbio.0060061
Zhang, X., Tang, N., Hadden, T. J., and Rishi, A. K. (2011). Akt, FoxO and regulation of apoptosis. Biochimica Biophysica Acta 1813 (11), 1978–1986. doi:10.1016/j.bbamcr.2011.03.010
Zheng, Q., Gao, N., Sun, Q., Li, X., Wang, Y., and Xiao, H. (2021). bfc, a novel serpent co-factor for the expression of croquemort, regulates efferocytosis in Drosophila melanogaster. PLoS Genet. 17 (12), e1009947. doi:10.1371/journal.pgen.1009947
Zheng, Q., Ma, A., Yuan, L., Gao, N., Feng, Q., Franc, N. C., et al. (2017). Apoptotic cell clearance in Drosophila melanogaster. Front. Immunol. 8, 1881. doi:10.3389/fimmu.2017.01881
Zhou, L. (2019). P53 and apoptosis in the Drosophila model. Adv. Exp. Med. Biol. 1167, 105–112. doi:10.1007/978-3-030-23629-8_6
Glossary
PCD Programmed cell death
TNF tumor necrosis factor
M1 classically activated macrophages
M2 alternative activated macrophages
LPC lysophosphatidylcholine
S1P sphingosine 1-phosphate
ICAM3 Intercellular adhesionmolecule 3
PS phosphatidylserine
PSR PS recognizing membrane receptor
BAI1/3 Brain Angiogenesis Inhibitor 1/3
TIM4 T cell immunoglobulin mucin receptor 4
Stab2 Stabilin-1/2
TLT2 (TREM)-like protein 2
TAM receptor tyrosine kinase Mer Tyro3 and Axl
Gas6 growth arrest-specific gene 6
Drpr Draper
Crq Croquemort
HTH helix-turn-helix
bZIP leucine zipper region
STAT signal transducer and activator of transcription
SphK sphingosine kinase
VEGF vascular endothelial growth factor
PPARλ peroxisome proliferator-activated receptor λ
PANX1 Pannexin-1
LXR liver x receptor
RXR retinoid X receptor
Tgm2 transglutaminase-2
TLR Toll-like receptor
AP-1 activator protein 1
IRF interferon regulatory factor
SOCS suppressors of cytokine signaling
IFNAR type Ⅰ interferon receptor
IFNBR type II interferon receptor
CSF colony-stimulating factors
JAK Janus kinase
RAR retinoid receptor
GR glucocorticoid receptor
RR retinoid-like receptors
ATRA all-trans retinoic acid
CRA cis-retinoic acid
iNOS inducible nitric oxide synthase
AhR Aryl hydrocarbon receptor
IgG, IgM Immunoglobulins G and M
Keywords: transcription factor, apoptosis, clearance of apoptotic cells, regulatory mechanisms, signal pathway
Citation: Gao Y, Jiao Y, Gong X, Liu J, Xiao H and Zheng Q (2023) Role of transcription factors in apoptotic cells clearance. Front. Cell Dev. Biol. 11:1110225. doi: 10.3389/fcell.2023.1110225
Received: 28 November 2022; Accepted: 09 January 2023;
Published: 19 January 2023.
Edited by:
Baojun Zhang, Xi’an Jiaotong University, ChinaReviewed by:
Bishuang Cai, Icahn School of Medicine at Mount Sinai, United StatesKohki Kawane, Kyoto Sangyo University, Japan
Copyright © 2023 Gao, Jiao, Gong, Liu, Xiao and Zheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hui Xiao, aHVpeGlhb0Bzbm51LmVkdS5jbg==; Qian Zheng, enFpYW5Ac25udS5lZHUuY24=
†These authors have contributed equally to this work
 Yuqiong Gao†
Yuqiong Gao† Hui Xiao
Hui Xiao Qian Zheng
Qian Zheng