- Waksman Institute of Microbiology, Rutgers University, Piscataway, NJ, United States
Complementary forward and reverse genetic approaches in several model systems have resulted in a recent burst of fertilization gene discovery. The number of genetically validated gamete surface molecules have more than doubled in the last few years. All the genetically validated sperm fertilization genes encode transmembrane or secreted molecules. Curiously, the discovery of genes that encode oocyte molecules have fallen behind that of sperm genes. This review discusses potential experimental biases and inherent biological reasons that could slow egg fertilization gene discovery. Finally, we shed light on current strategies to identify genes that may result in further identification of egg fertilization genes.
1 Introduction
1.1 The big questions and fertilization gene discovery
Understanding the genetic underpinnings of fertilization is essential for developing infertility treatments, contraceptive targets, understanding speciation, and mechanisms of cell-cell interactions (Schultz and Williams, 2002; Krauchunas et al., 2016; Bhakta et al., 2019; Findlay et al., 2019). In the last several years there has been a rapid increase in the discovery of genetically validated fertilization molecules in several key model systems (Deneke and Pauli, 2021; Mei and Singson, 2021). While fertilization is studied in many model systems, forward and reverse genetic approaches in worms, zebrafish, and mice have recently led the charge. Sterile mutants obtained through mutant screens or genetic knockouts are the modern gold standard to demonstrate the requirement of a gene to encode a factor in fertilization. The progress from genetic screens and knockouts has also established that there are fertilization genes that are deeply conserved from nematode worms to mammals (Mei and Singson, 2021). For example, there are several immunoglobulin superfamily molecules that were independently identified with roles in fertilization in several different species (Ellerman et al., 2003; Krauchunas et al., 2016; Binner et al., 2022). The DCSTAMP sperm molecules spe-42 and spe-49 (Kroft et al., 2005; Wilson et al., 2018) have essentially the same sperm sterile mutant phenotype to DCST1/2 mutants in flies, fish, and mice (Inoue et al., 2021; Mei and Singson, 2021; Noda et al., 2022). Progress in fertility gene discovery has also uncovered a high degree of molecular complexity required for sperm egg interactions. This was the inspiration for the concept of the fertilization synapse as the intellectual framework to understand how many newly discovered fertility gene products work in conjunction at the interface of sperm and egg plasma membranes (Krauchunas et al., 2016). Sperm and egg interactions have parallels with other cellular synapses (neural and immune) that include specialized cell structures, integrated with adhesion, secretion, and cell signaling. Understanding the fertilization synapse will require knowing what molecules interact either in cis with other molecules on the same gamete or in trans with molecules of the opposite gamete. The realization of the fertilization synapse also opens new questions. How are all the sperm and egg components assembled into complexes and at the right time and place? Genes are being discovered that may impact the processing and assembly of the fertilization synapse (Gleason et al., 2006; Contreras et al., 2022; Schreier et al., 2022). For example, Frey can regulate the assembly of Izumo1, a key protein involved in fertilization on the surface of the sperm (Contreras et al., 2022). It does this by helping to assemble Izumo into the correct protein complex. The road to the identification of genes involved in fertilization faces many challenges which researchers must overcome.
1.2 Difficulties in determining genes involved in fertilization
Unlike some other biological processes, fertilization comprises multiple cellular processes including gamete activation, recognition, binding, and fusion. These processes must be executed very precisely to combine one sperm and one egg. This requires that the space, time, and the concentration of proteins during fertilization be exact. The transient interactions and combination of proteins at the correct level can be difficult to recapitulate in vitro. Genes must be expressed specifically on the surface of the cell and often shift after fertilization. As there are multiple processes that require very specific interactions, it is of no surprise that the sperm and egg also require multiple protein interactions. Beyond the transient interactions, redundancy, pleiotropy, and maintaining sterile mutants have been roadblocks for gene discovery on both gametes.
1.3 Current fertilization molecules and open questions
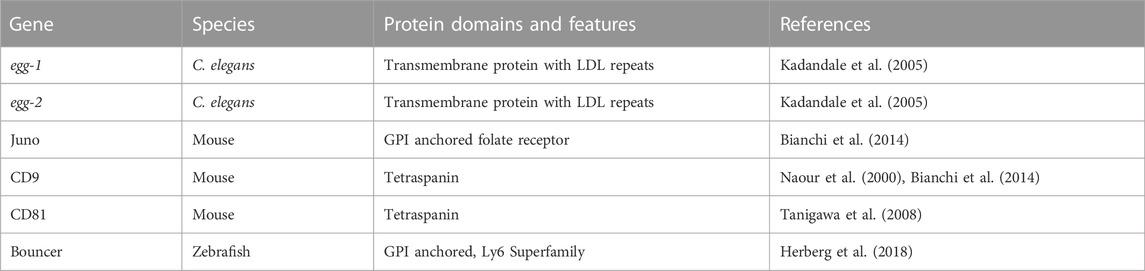
As of writing this review, going by the sterile mutant gold standard, there are ten mouse genes (Bianchi and Wright, 2020; Deneke and Pauli, 2021) and 12 C. elegans genes that encode transmembrane or secreted proteins that are required for fertilization and are components of the fertilization synapse (Mei and Singson, 2021) (Figure 1). For complete recent information about the fertilization proteins, please see recent reviews by Deneke and Pauli (2021); Mei and Singson (2021). Anyone would agree that when it comes to fertilization, it takes two to tango as sperm and egg are the ultimate in complementary cells. However, gazing at the molecules in the emerging fertilization synapse of worms, fish, and mouse shown in Figure 1 there is a striking asymmetry. Most currently known fertilization genes encode sperm factors. Ten of twelve genes in worms and seven of ten genes in mice are on the sperm surface (Figure 1). Here we review why we currently observe this gene discovery asymmetry between known sperm and egg genes. As of writing this review, only five egg surface molecules have been discovered: egg-1/2 in C. elegans, Bouncer in Zebrafish, and Juno and CD9 in mouse (Naour et al., 2000; Kadandale et al., 2005; Bianchi et al., 2014; Herberg et al., 2018) (Table 1). These genes encode a multitude of different types of proteins. Egg-1 and egg-2 have LDL receptor repeats, Bouncer a Ly6/uPAR protein, Juno, is related to folate receptors, and CD9 is a tetraspanin (Naour et al., 2000; Kadandale et al., 2005; Bianchi et al., 2014; Herberg et al., 2018). However, these genes are not sufficient for all the different functions of fertilization. Therefore, the search for egg genes continues. There are other proteins such as Phospholipid C zeta which is a sperm specific soluble enzyme that can trigger oocyte activation (Yoon et al., 2008; Yoon et al., 2012; Hachem et al., 2017; Sanders et al., 2018). This review in particular focuses only on proteins that are expressed on the surface of the gamete. This asymmetry underscores the importance of studies in females. Female processes have been historically understudied. This has been a focus of the NIH since 2016 when they published their policy on sex as a biological variable (Arnegard et al., 2020). The gap in egg gene discovery is an important subject that we, as researchers, should examine and work to close. In this review, we discuss different biological mechanisms such as redundancy, pleiotropy, and how the evolution of sperm expressed genes have impacted the identification of fertilization genes in the oocyte. A likely source of lagging egg gene discovery is also experimental bias either in methods or the model organisms. Potential experimental biases could include maintenance and propagation of sterile mutants, screening strategies, and difficulty in identifying fertilization phenotypes. While many of these experimental biases are not an exclusive challenge to discovering egg genes, we think that these are important ideas to bring to the forefront of the research community. Finally, we conclude by discussing new experimental approaches and options that could address the question of where are all the egg genes?
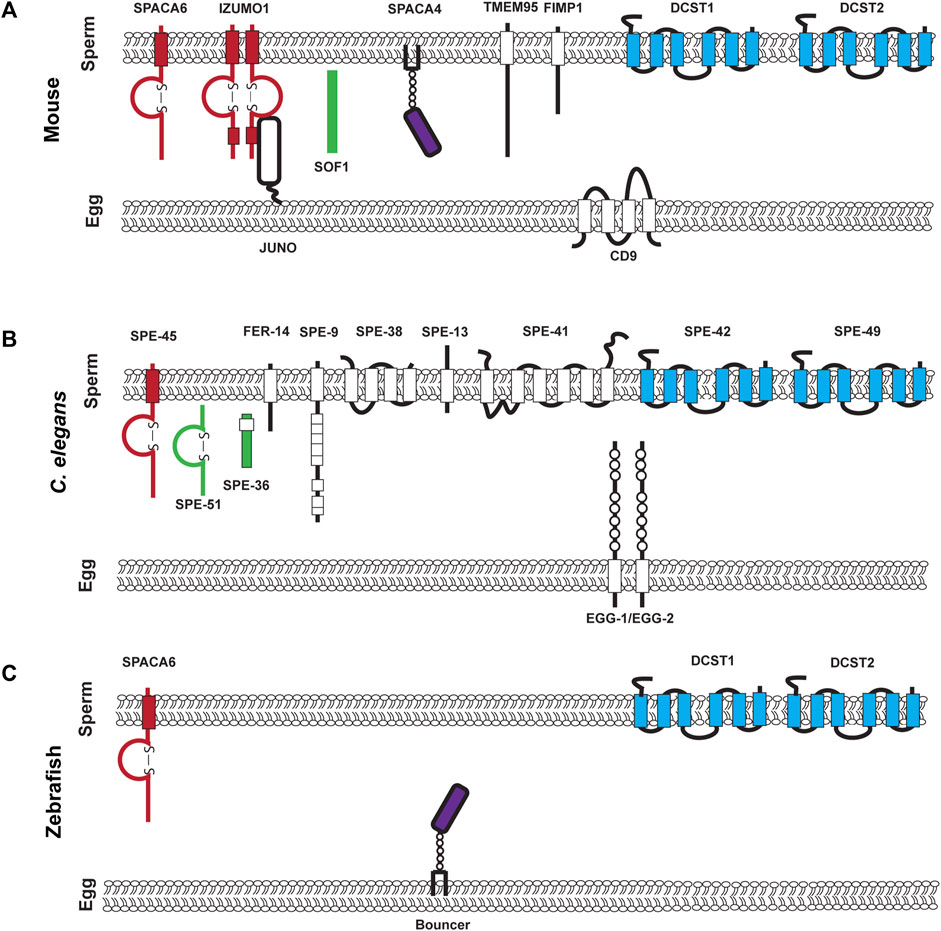
FIGURE 1. Known components of the mouse, C. elegans, and Zebrafish fertilization synapse. (A). Mouse fertilization synapse, (B). C. elegans fertilization synapse, and (C). Zebrafish fertilization synapse. For (A–C) each of the molecules that are denoted have been experimentally validated though loss of function mutations. Proteins on the left of each synapse in red contain an immunoglobulin domain, proteins in green are secreted, proteins in purple are in the uPAR/Ly6 family and on the right in blue are conserved DCSTAMP proteins.
2 Challenges in the identification of egg genes
2.1 Fertilization and redundant genes
Fertilization is an essential process and redundant genes can help protect against deleterious mutations. The same protection that redundancy provides may make it more difficult to identify egg genes with genetic approaches. For sexually reproducing organisms, loss of fertility is evolutionarily devastating, rendering animals unable to pass on their genes to future generations. One random mutation could render an animal sterile and subsequently bring their fitness level to zero. Therefore, like other genes that have essential functions, gene duplication and redundant copies can decrease the likelihood of extinction occurring over many generations. This has been observed across many different organisms in fertility and particularly in the previously identified egg genes. In C. elegans, egg-1 and egg-2 function redundantly with one another. The loss of egg-1 which encodes an LDL receptor repeat-containing protein that functions redundantly with egg-2 another LDL receptor. These genes encode a protein with 67% similarity (Kadandale et al., 2005). The initial observation by RNAi of egg-1 also knocks down egg-2 generating a sterile phenotype, however genetic loss of only egg-1 or egg-2 decreases fertility but does not cause sterility.
On the oocyte surface, one abundant group of proteins that were at the forefront of fertilization research for many years were integrins on the egg surface. Interest in this group of proteins arose due to a disintegrin domain in ADAM2/Fertilin that was found on the sperm surface (Blobel et al., 1992). Investigation of this class of integrin proteins which were expressed in eggs revealed 24 different integrin combinations. The complexity made this a difficult area to study (Evans, 2002). Individual knockouts of integrins generated modest decreases in fertility and multiple knockouts generated other fertility defects such as embryonic lethality making this challenging to elucidate their exact role during fertilization (Miller et al., 2000; Hynes, 2002; He et al., 2003; Vjugina and Evans, 2008). While redundancy is not the singular reason that it has been difficult to identify egg genes, we hypothesize that it is one of the most prevalent reasons.
On the sperm side of the equation, there have also been genes that act redundantly with one another. One example is the CRISP (Cysteine-RIch Secretory Protein) gene family which regulates calcium channels in fertilization. In mouse, there are four CRISP family members, knockout of one member of the family reduces fertility but does not completely abolish it, however knockout of three or four of the family members completely disrupts male fertility demonstrating their redundant or compensatory functions (Ros et al., 2008; Curci et al., 2020). Interestingly in humans there are three CRISP genes which are all located on the same chromosome, therefore it would take a large deletion or rare collection of point mutations in all three genes to remove all function rendering them infertile (Gibbs et al., 2008; Curci et al., 2020). Redundant or compensatory genes may be a big challenge in our field in discovering egg genes as well as additional sperm genes. With the rise of CRISPR/Cas9 gene editing, it may be valuable to look for homologues and potential gene families and then knockout multiple genes in these family groups to examine for fertility phenotypes.
2.2 Less gene expression and pleiotropy
Oocytes enter a period of transcriptional quiescence usually during which fertilization, egg activation, and early embryonic development must occur (Kim and You, 2022). This contrasts with sperm cells which have a short period of quiescence, however their function largely ends after fertilization. The oocyte must provide all the proteins necessary for these very diverse cellular processes. One way that this could be accomplished is through pleiotropy, where one gene regulates multiple functions. As the first genomes were sequenced, researchers were surprised that there are far fewer genes than previously predicted, pleiotropy is one explanation for this phenomenon (Lander et al., 2001).
A related reason that it might be difficult to identify egg genes is that there may be tissue or stage specific isoforms of genes. This is where one isoform of a gene functions in development and another during adulthood, or one isoform in a somatic cell and other functions in the germline. For example, Juno in mouse is widely expressed in tissues other than just oocytes including the thymus, spleen, and lung however loss of Juno in these tissues does not appear to solicit additional loss of function phenotypes (Spiegelstein et al., 2000; Bianchi et al., 2014). If multiple phenotypes are exhibited this can complicate our ability to interpret gene functions. For example, in C. elegans, nhr-23 is expressed during the process of molting which takes place during larval development and precedes much of germ cell differentiation. Later in development nhr-23 also functions as a transcription factor during spermatogenesis (Ragle et al., 2020). The earlier molting defects exhibited by nhr-23 had been established for many years prior to its function during spermatogenesis. The function during spermatogenesis was not as evident as animals arrested prior to adulthood (Kostrouchova et al., 1998; Kostrouchova et al., 2001). Finally, as mentioned before in Section 2.1, loss of integrins on the egg surface identified pleiotropic functions in oogenesis and embryonic development. Mutant phenotypes that are exhibited in other tissues or earlier in development can make the isolation of specific genes involved in fertilization more difficult and these pleiotropic defects may obscure specific genes roles in fertilization. The limited number of genes that are expressed in the oocyte make this a compelling reason why egg expressed gene roles have been difficult to identify.
2.3 Differential cell expression, opposing sex specific expression patterns, and homotypic interactions can make elucidating gene function difficult
Another reason that identifying egg genes could be difficult is that different proteins or protein domains maybe used in various species. Further it may be where two identical structural elements of individual monomers are interchanged to stabilize protein complexes (Bennett et al., 1994). The puzzle of molecular swapping between two gametes underscores the importance of studying fertilization in different organisms to understand the mechanism of cell-cell interactions between sperm and oocytes.
For example, Bouncer, a Ly6/uPAR protein in Zebrafish is essential for sperm-egg interactions which facilitate sperm binding to the oolemma when expressed on the egg surface (Herberg et al., 2018). SPACA4, the mammalian homologue of Bouncer is actually expressed on the sperm surface in mice and functions to penetrate the Zona Pellucida (ZP) for binding to the egg surface in mouse (Fujihara et al., 2021). At this point, the mechanism underlying opposing sex specific expression patterns for Bouncer and SPACA4 is not clear. Hypotheses surrounding this include it being due to different environments for fertilization as Zebrafish have a micropyle and therefore the sperm do not need to pass through the egg surface whereas mammals need penetration of the ZP and the acrosome reaction to occur. Another hypothesis is that this is a sperm specific loss of function that was retained in the egg during evolution.
In the same vein, homotypic interactions where either two proteins or protein domains interact with one on each of the opposing sperm and egg cells acting to signal for binding or fusion to occur. No homotypic interactions have been observed in fertilization thus far. However, this has been observed in cadherin signaling for polarity in Drosophila (Chen et al., 2008). Polarity is also required for fertilization and establishing egg activation. Another example related to cell-cell fusion is EFF-1 in C. elegans which mediates cell-cell fusion in the Soma (Segev et al., 2018). The EFF-1 interactions are structurally homologous to HAP2/GCS1 in flowering plants which is a sperm-egg fusogen in plants (Brukman et al., 2022). Homotypic interactions and sex specific expression patterns can make it difficult for identification of egg molecules as homotypic mutant animals would be sterile in both sexes and be very difficult to generate and maintain.
2.4 De novo gene formation in the testis and the co-evolution of fertilization receptors create complex genetic dynamics
Gene duplication and deletion events often drive evolution and establish new biological functions and phenotypes. Interestingly, it has been shown that de novo gene synthesis occurs much more often in the male lineage particularly through the testes (Kondo et al., 2017). This may also be referred to as the out of the testis hypothesis. It can be hypothesized that the abundant number of new genes in the male lineage may provide opportunities for these newly synthesized genes to be adopted into fertilization synapse complex working to stabilize this process and further define speciation. The male germline has several attributes that can facilitate new gene synthesis. These include histone modifications, demethylation of CpG islands, and increased expression of transcription associated proteins, as well as increased selective pressure from sperm competition (Kleene, 2001; Haerty et al., 2007; Kondo et al., 2017; Nyberg and Carthew, 2017). In contrast to gene synthesis, in gene loss, genes which no longer have biological functions can occur. Gene loss is less likely to occur in oocytes and primarily impacts multi-copy gene families (Assis, 2019). These results may demonstrate that while over time more sperm surface genes may be involved in fertilization, the genes on the oocyte side of the equation are less likely to change and may have strong levels of conservation and be involved in multiple processes such as both oogenesis and fertilization. It has also been hypothesized from observations in marine invertebrates that there is a co-evolution “arms race” of the male and female fertilization proteins in different species which may drive species specificity and could make identification of conserved fertilization molecules more difficult to identify (Wilburn and Swanson, 2016). The long-term evolutionary consequences of these changes in genes can both drive speciation and impact the genes that are involved in fertilization. These, however, are unlikely to be phenotypes that can be picked up in genetic screens but would require analysis of different species and genomes.
2.5 Sterile animals require active maintenance in the lab
As anyone who works in fertilization and gamete activation knows, maintaining mutant animals that are sterile or have lethal mutations requires active thought and coordination by researchers for each generation. To overcome these difficulties, there are a few very clever ways to maintain sterile animals for experimental analysis. One example is the identification conditional mutations. These conditional mutants can be temperature sensitive (ts) mutations that can be more easily screened for in microbes, worms, and flies. These ts animals are sterile when cultured at high temperatures but fertile when cultured at lower temperatures (also see Section 2.7 below) (Suzuki and Procunier, 1969; O’Connell et al., 1998; Mei and Singson, 2021). A second approach is through inducible systems such as Cre-Lox, GAL4/UAS, and Auxin Inducible Degrons which use either site specific recombinases, DNA binding activation sites, or chemical induction to induce sterile phenotypes (Brand and Perrimon, 1993; Zhang et al., 2015; Kim et al., 2018). Finally, we can use balancer chromosomes which are rearranged chromosomes often with morphological and fluorescent tags which can be used to maintain recessive lethal or sterile mutations as heterozygotes without recombination (Dejima et al., 2018; Miller et al., 2019). Unfortunately, all of these techniques have their limitations. Temperature sensitive mutations rely on protein misfolding at differential temperatures and there are relatively few model organisms where this is a practical approach (body temperature conforms to ambient temperature). Furthermore, not all genes are able to be mutated to become temperature sensitive, this favors proteins with hydrophobic residues and higher free energy levels (Varadarajan et al., 1996). Inducible systems require knowledge of the specific gene and favor reverse genetic approaches which can bias gene identification. Some balancer chromosomes rely on active maintenance of mutant animals as heterozygotes. Finally, collecting enough mutant animals for analysis may require specific breeding schemes as in the case of mice and the amount of space and resources required can limit the amount of exploration that can occur. For egg surface genes, extra care must be taken in order to maintain a set of fertile-heterozygotes in order to generate mutants for the next generation. This sibling selection may require constant costly and laborious molecular genotyping.
2.6 Gametogenesis and fertilization are often inherently temperature sensitive, egg genes might be more specifically unable to become temperature sensitive
One way that researchers working on worms, flies, and fish have managed to keep sterile animal lines going is through temperature sensitive mutations as described in Section 2.5 (Mei and Singson, 2021). While this has been an extremely useful tool, not all proteins can be mutated to become temperature sensitive. In fact, spermatogenesis and sperm activation are inherently more temperature sensitive than egg processes (Nakamura et al., 1987; Kurhanewicz et al., 2020; Hirano et al., 2022). The ability of oocytes to buffer temperature may make it more difficult to identify mutations but may also reveal information about the biology of oocytes. In many male-female organisms, oocytes are available in a limited number, therefore protecting the viability of oocytes becomes essential for an animal’s reproductive success, consequently our techniques of temperature stress may not be as efficient for egg specific genes.
2.7 Egg fertilization genes may be mischaracterized as embryonic lethal and meiosis mutants
Despite the difficulties in maintaining sterile animals, the field still engages in both forward and reverse genetic screens identify additional genes involved in fertilization. This has often required creative techniques such as temperature sensitive mutants, sibling selection, balancing all mutants prior to analysis, and using CRISPR/Cas9 to knock out all candidate genes to capture sterile alleles for future analysis (Mei and Singson, 2021). After mutagenesis one limitation is that it can be difficult to differentiate between genes that are embryonic lethal or have meiosis defects from fertilization specific genes at a high level since they often display similar terminal phenotypes. This can lead to a bottleneck in assessing mutations in a screen. Furthermore, on the flipside, when researchers are conducting screens for other phenotypes, they may encounter sterile mutants but not have the experimental tools to keep these mutations from getting dropped out of the population. It is possible that the egg surface genes that we are interested in may have been discarded in screens looking for embryonic lethal or meiosis genes.
2.8 Laboratory environments may not mimic environmental conditions that the animal might experience in the wild
Labs are often sterile environments that researchers work hard to keep free from contamination. However, this does not mimic the natural environment that our experimental organisms experience in the wild. Animals in laboratory environments do not face competition for food, contamination by parasites and bacteria, the temperature is stable, and light-dark cycles are controlled. These environmental stresses can often decrease fertility in the wild through changes in immune response, diet, and seasonality (Amaral et al., 2014). Therefore, in laboratory environments, we may be unable to recapitulate the environmental stressors that may impact fertility by modulating gene responses.
2.9 Egg fertility proteins may be sensitive to one mutant copy and quickly become sterile
In contrast to the idea that we’re unable to capture egg genes due to redundancy where there is no observable phenotype because there’s another copy of the gene which is compensating for the loss of one copy is a dominant sterile mutation. In a dominant sterile mutation, the loss of one copy of a gene through mutation renders an animal sterile or lethal (Erdélyi and Szabad, 1989). Capturing dominant sterile mutations may occur in genetic screens but lead to a dead end. It is incredibly difficult to keep dominant sterile strains alive. Maintaining a dominant sterile mutation requires the gene to have sex restricted expression where one sex is not sterile. For example, a dominant negative sperm mutation would not impact females, therefore the mutation could be carefully maintained in the maternal lineage and then crossed to a wild-type male and analyze their male progeny which would have one copy of the gene. One example of this is wee-1.3 in C. elegans. The wee-1.3 gene encodes a kinase that functions during spermatogenesis, a dominant mutation in this gene renders an animal infertile (Lamitina and L’Hernault, 2002). The hermaphrodite-male androdiecious reproductive mode in C. elegans is the only way that these mutant animals were able to be maintain. These limitations therefore preclude anything, that is, homotypic and expressed in both gametes. If prepared, it is possible to generate conditional mutants, however this excludes forward genetic screens and simple knockouts for analyses.
2.10 Small proteins are less likely to be mutated in genetic screens and less likely to be identified with biochemical methods
In fertilization small genes (less than 200 amino acids) have been shown to have important roles for fertilization. In Zebrafish, Bouncer, the Ly6/uPAR protein on the egg surface is 80 amino acids (Herberg et al., 2018). On the sperm side of the equation, spe-13 in C. elegans is 130 amino acids (Singson Lab unpublished observation) and fimp-1 in mouse with 111 amino acids are also small proteins involved in fertilization (Fujihara et al., 2020). Despite their important roles, small proteins are more likely to be missed in both genetic screens as well as mass spectrometry or ribosomal profiling (Jorgensen and Mango, 2002; Harney et al., 2019). In genetic screens the smaller number of nucleotides in each gene decreases the probability that individual base pairs will be mutated in the right region for sterility to occur. For mass spectrometry and ribosome profiling, this is also an issue as RNA based methods are more likely occurring with amplified RNA which can filter out smaller RNAs, and small transcripts may be beyond the sensitivity of the equipment (Ahrens et al., 2021). The emergence of the importance of small proteins may be a good avenue for future analysis.
2.11 Mutations in egg fertilization genes may generate extremely subfertile animals but not be labeled fertilization defects
The question of what constitutes a sterile phenotype may also mire the picture of what genes are involved with fertilization on the egg’s surface. Infertility is clinically defined as failure to conceive after 12 months of unprotected intercourse (Mélodie and Christine, 2018). This also discounts couples that may be able to have one child but then experience secondary infertility. A significant drop in the number of progeny that an animal produces due to a genetic mutation while not completely sterilizing an animal could make it difficult to determine if involvement is technically in fertilization. There is some debate in what should be considered subfertile or sterile, particularly in animals with larger broods and faster rates of ovulation. For example, mice ovulate at 8 times the rate of humans with more than one egg at a time, a decrease in litter size here could look like clinical infertility in humans (Vjugina and Evans, 2008). Similarly in C. elegans which have a brood size of ∼300 progeny in their reproductive lifetime, a decrease to two progeny in their lifetime would be very likely to be considered clinically infertile if modeled in humans. Thus, we encourage careful analysis of animals with fertility defects and a careful analysis of the phenotypes in determining their role at the molecular level.
2.12 Previous biochemical analysis has often been unable to be genetically validated
Previous biochemical analysis primarily in sea urchin and abalone identified a multitude of proteins such as Bindin, speract and resact, VERL and lysin through analysis of cell extracts using monoclonal antibodies to inhibit fertilization (Vacquier and Moy, 1977; Hansbrough and Garbers, 1981). These approaches were valiant and groundbreaking to our understanding of protein candidates for fertilization. However, when genetic knockouts were examined for many of these genes, the animals were still fertile (Deneke and Pauli, 2021). Monoclonal antibodies to gamete surface antigens were helpful in identifying and validating IZUMO (Okabe et al., 1988). CD81 was also identified through monoclonal antibodies but is not essential but still can contribute to fertilization (Takahashi et al., 2001; Rubinstein et al., 2006). As we continue our search for all the genes that are involved in fertilization these contributions help us to understand the redundancy of the process and the contributing factors.
2.13 Historical narratives surrounding the roles of females may impact current egg gene discovery
Both historically and currently, females are understudied (Ah-King et al., 2014). Despite awareness and advocacy of this issue from individual researchers to as well as organizations such as the NIH, this gap has persisted (Arnegard et al., 2020). Commonly described reasons for why females continue to be understudied include that there is a stronger interest in male processes of reproduction, and that eggs are presumed to be less complex and passive receivers of sperm whose biology has already been fully characterized (Méndez and Córdoba-Aguilar, 2004). These reasons are not evidence based and fail to take into account recent gene discovery in fertilization, egg activation, and blocks to polyspermy which still have largely unknown mechanisms and complex cell signaling and organization. Perpetuating these ideas minimizes the role of females and can be potentially harmful in the treatment of female reproductive health.
3 Addressing the sperm and egg gene discovery question
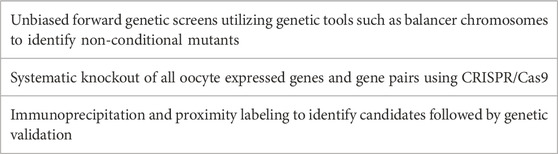
To address the difficulty in identifying egg genes (and in fact sperm genes), there are several approaches that can be and are currently being taken (Table 2). For example, in C. elegans we are employing an approach which will involve doing random mutagenesis and then using balancer chromosomes to generate stable balanced lines of mutants, in contrast to previous screens which have identified temperature sensitive mutants that are typically not genetic nulls. This approach in invertebrate systems can allow for higher throughput of mutations to be screened as well as help differentiate embryonic lethal and meiosis mutants. In parallel with characterization of genetic mutants, Whole Genome Sequencing (WGS) can be employed to identify the causative mutation. Another approach being undertaken is to use CRISPR-Cas9 to generate knockouts of all testes expressed genes as is being undertaken by several labs to ascertain which knockouts impact fertilization (Miyata et al., 2016; Abbasi et al., 2020). A similar approach could be taken to knock out all oocyte expressed genes. This is however an expensive approach and can miss redundant or pleiotropic genes. Redundancy and pleiotropy remain problems which are difficult to address. A bioinformatic approach identifying potential gene pairs could aid in prioritizing generating double mutants could partially address this issue. Finally, for pleiotropic genes, conditional systems such as those mentioned in Section 2.6 can be employed. One caveat to this approach is that it is limited in the number of candidates that this would be feasible for as these can be labor intensive strains to build. Complementary to these approaches, utilizing biochemical approaches such as immunoprecipitation and proximity labeling will help us to better understand other proteins that are involved in fertilization. Taking advantage of biochemical approaches now supported by complementary genetic approaches will bring all available tools to bear on our understanding of how nature does conception.
4 Conclusion
Is this question of where are all the egg genes an experimental problem or a true biological asymmetry? We imagine that it might be a combination of both possibilities. Will this asymmetry of sperm and egg molecules also be seen in other species such as fish, frogs, and humans? We have great faith in the determination and productivity of the reproductive biology community to continue with its rapid and transforming fertility gene discovery. Exciting new experimental approaches to aid in egg gene discovery are rapidly coming into play. The future discoveries of egg genes will greatly enhance our understanding of fertilization to aid in the development of infertility treatments and novel contraceptive techniques as well as provide further understanding of how two morphologically distinct cells interact with each other.
Author contributions
KM and AS prepared and revised the manuscript. KM generated the figures.
Funding
This work was supported by funding from an NIH Grant to AS (R01HD054681) and KM was also supported by an NIH Institutional Research and Academic Career Development Award (K12GM093854) Fellowship.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abbasi, F., Kodani, M., Emori, C., Kiyozumi, D., Mori, M., Fujihara, Y., et al. (2020). CRISPR/Cas9-Mediated genome editing reveals oosp family genes are dispensable for female fertility in mice. Cells 9, 821. doi:10.3390/cells9040821
Ah-King, M., Barron, A. B., and Herberstein, M. E. (2014). Genital evolution: Why are females still understudied? Plos Biol. 12, e1001851. doi:10.1371/journal.pbio.1001851
Ahrens, C. H., Wade, J. T., Champion, M. M., and Langer, J. D. (2021). A practical guide to small protein discovery and characterization using mass spectrometry. J. Bacteriol. 204, e0035321. doi:10.1128/jb.00353-21
Amaral, F. G., Castrucci, A. M., Cipolla-Neto, J., Poletini, M. O., Mendez, N., Richter, H. G., et al. (2014). Environmental control of biological rhythms: Effects on development, fertility and metabolism. J. Neuroendocrinol. 26, 603–612. doi:10.1111/jne.12144
Arnegard, M. E., Whitten, L. A., Hunter, C., and Clayton, J. A. (2020). Sex as a biological variable: A 5-year progress report and call to action. J. Women’s Heal 29, 858–864. doi:10.1089/jwh.2019.8247
Assis, R. (2019). Out of the testis, into the ovary: Biased outcomes of gene duplication and deletion in Drosophila. Evolution 73, 1850–1862. doi:10.1111/evo.13820
Bennett, M. J., Choe, S., and Eisenberg, D. (1994). Domain swapping: Entangling alliances between proteins. Proc. Natl. Acad. Sci. 91, 3127–3131. doi:10.1073/pnas.91.8.3127
Bhakta, H. H., Refai, F. H., and Avella, M. A. (2019). The molecular mechanisms mediating mammalian fertilization. Development 146, dev176966. doi:10.1242/dev.176966
Bianchi, E., Doe, B., Goulding, D., and Wright, G. J. (2014). Juno is the egg Izumo receptor and is essential for mammalian fertilization. Nature 508, 483–487. doi:10.1038/nature13203
Bianchi, E., and Wright, G. J. (2020). Find and fuse: Unsolved mysteries in sperm–egg recognition. Plos Biol. 18, e3000953. doi:10.1371/journal.pbio.3000953
Binner, M. I., Kogan, A., Panser, K., Schleiffer, A., Deneke, V. E., and Pauli, A. (2022). The sperm protein Spaca6 is essential for fertilization in zebrafish. Front. Cell Dev. Biol. 9, 806982. doi:10.3389/fcell.2021.806982
Blobel, C. P., Wolfsberg, T. G., Turck, C. W., Myles, D. G., Primakoff, P., and White, J. M. (1992). A potential fusion peptide and an integrin ligand domain in a protein active in sperm–egg fusion. Nature 356, 248–252. doi:10.1038/356248a0
Brand, A. H., and Perrimon, N. (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415. doi:10.1242/dev.118.2.401
Brukman, N. G., Li, X., and Podbilewicz, B. (2022). Fusexins, HAP2/GCS1 and evolution of gamete fusion. Front. Cell Dev. Biol. 9, 824024. doi:10.3389/fcell.2021.824024
Chen, W.-S., Antic, D., Matis, M., Logan, C. Y., Povelones, M., Anderson, G. A., et al. (2008). Asymmetric homotypic interactions of the atypical cadherin flamingo mediate intercellular polarity signaling. Cell 133, 1093–1105. doi:10.1016/j.cell.2008.04.048
Lander, E. S., Linton, L. M., Birren, B., Zody, M. C., Devon, K., Dewar, K., et al. (2001). Initial sequencing and analysis of the human genome. Nature 409, 860–921. doi:10.1038/35057062
Contreras, W., Wiesehöfer, C., Schreier, D., Leinung, N., Peche, P., Wennemuth, G., et al. (2022). C11orf94/Frey is a key regulator for male fertility by controlling Izumo1 complex assembly. Sci. Adv. 8, eabo6049. doi:10.1126/sciadv.abo6049
Curci, L., Brukman, N. G., Muñoz, M. W., Rojo, D., Carvajal, G., Sulzyk, V., et al. (2020). Functional redundancy and compensation: Deletion of multiple murine Crisp genes reveals their essential role for male fertility. Faseb J. 34, 15718–15733. doi:10.1096/fj.202001406r
Dejima, K., Hori, S., Iwata, S., Suehiro, Y., Yoshina, S., Motohashi, T., et al. (2018). An aneuploidy-free and structurally defined balancer chromosome toolkit for Caenorhabditis elegans. Cell Rep. 22, 232–241. doi:10.1016/j.celrep.2017.12.024
Deneke, V. E., and Pauli, A. (2021). The fertilization enigma: How sperm and egg fuse. Annu. Rev. Cell Dev. Bi 37, 391–414. doi:10.1146/annurev-cellbio-120219-021751
Ellerman, D. A., Ha, C., Primakoff, P., Myles, D. G., and Dveksler, G. S. (2003). Direct binding of the ligand PSG17 to CD9 requires a CD9 site essential for sperm-egg fusion. Mol. Biol. Cell 14, 5098–5103. doi:10.1091/mbc.e03-04-0244
Erdélyi, M., and Szabad, J. (1989). Isolation and characterization of dominant female sterile mutations of Drosophila melanogaster. I. Mutations on the third chromosome. Genetics 122, 111–127. doi:10.1093/genetics/122.1.111
Evans, J. P. (2002). The molecular basis of sperm–oocyte membrane interactions during mammalian fertilization. Hum. Reprod. Update 8, 297–311. doi:10.1093/humupd/8.4.297
Findlay, J. K., Holland, M. K., and Wong, B. B. M. (2019). Reproductive science and the future of the planet. Reproduction 1, R91–R96. doi:10.1530/rep-18-0640
Fujihara, Y., Herberg, S., Blaha, A., Panser, K., Kobayashi, K., Larasati, T., et al. (2021). The conserved fertility factor SPACA4/Bouncer has divergent modes of action in vertebrate fertilization. Proc. Natl. Acad. Sci. 118, e2108777118. doi:10.1073/pnas.2108777118
Fujihara, Y., Lu, Y., Noda, T., Oji, A., Larasati, T., Kojima-Kita, K., et al. (2020). Spermatozoa lacking Fertilization Influencing Membrane Protein (FIMP) fail to fuse with oocytes in mice. Proc. Natl. Acad. Sci. 117, 9393–9400. doi:10.1073/pnas.1917060117
Gibbs, G. M., Roelants, K., and O’Bryan, M. K. (2008). The CAP superfamily: Cysteine-rich secretory proteins, antigen 5, and pathogenesis-related 1 proteins—roles in reproduction, cancer, and immune defense. Endocr. Rev. 29, 865–897. doi:10.1210/er.2008-0032
Gleason, E. J., Lindsey, W. C., Kroft, T. L., Singson, A. W., and L’Hernault, S. W. (2006). spe-10 encodes a DHHC–CRD zinc-finger membrane protein required for endoplasmic reticulum/golgi membrane morphogenesis during Caenorhabditis elegans spermatogenesis. Genetics 172, 145–158. doi:10.1534/genetics.105.047340
Hachem, A., Godwin, J., Ruas, M., Lee, H. C., Buitrago, M. F., Ardestani, G., et al. (2017). PLCζ is the physiological trigger of the Ca2+ oscillations that induce embryogenesis in mammals but conception can occur in its absence. Development 144, 2914–2924. doi:10.1242/dev.150227
Haerty, W., Jagadeeshan, S., Kulathinal, R. J., Wong, A., Ram, K. R., Sirot, L. K., et al. (2007). Evolution in the fast lane: Rapidly evolving sex-related genes in Drosophila. Genetics 177, 1321–1335. doi:10.1534/genetics.107.078865
Hansbrough, J. R., and Garbers, D. L. (1981). Speract. Purification and characterization of a peptide associated with eggs that activates spermatozoa. J. Biol. Chem. 256, 1447–1452. doi:10.1016/s0021-9258(19)69983-7
Harney, D. J., Hutchison, A. T., Su, Z., Hatchwell, L., Heilbronn, L. K., Hocking, S., et al. (2019). Small-protein enrichment assay enables the rapid, unbiased analysis of over 100 low abundance factors from human plasma. Mol. Cell Proteomics 18, 1899–1915. doi:10.1074/mcp.tir119.001562
He, Z.-Y., Brakebusch, C., Fässler, R., Kreidberg, J. A., Primakoff, P., and Myles, D. G. (2003). None of the integrins known to be present on the mouse egg or to be ADAM receptors are essential for sperm–egg binding and fusion. Dev. Biol. 254, 226–237. doi:10.1016/s0012-1606(02)00043-x
Herberg, S., Gert, K. R., Schleiffer, A., and Pauli, A. (2018). The Ly6/uPAR protein Bouncer is necessary and sufficient for species-specific fertilization. Science 361, 1029–1033. doi:10.1126/science.aat7113
Hirano, K., Nonami, Y., Nakamura, Y., Sato, T., Sato, T., Ishiguro, K., et al. (2022). Temperature sensitivity of DNA double-strand break repair underpins heat-induced meiotic failure in mouse spermatogenesis. Commun. Biol. 5, 504. doi:10.1038/s42003-022-03449-y
Hynes, R. O. (2002). Integrins bidirectional, allosteric signaling machines. Cell 110, 673–687. doi:10.1016/s0092-8674(02)00971-6
Inoue, N., Hagihara, Y., and Wada, I. (2021). Evolutionarily conserved sperm factors, DCST1 and DCST2, are required for gamete fusion. Elife 10, e66313. doi:10.7554/elife.66313
Jorgensen, E. M., and Mango, S. E. (2002). The art and design of genetic screens: Caenorhabditis elegans. Nat. Rev. Genet. 3, 356–369. doi:10.1038/nrg794
Kadandale, P., Stewart-Michaelis, A., Gordon, S., Rubin, J., Klancer, R., Schweinsberg, P., et al. (2005). The egg surface LDL receptor repeat-containing proteins EGG-1 and EGG-2 are required for fertilization in Caenorhabditis elegans. Curr. Biol. 15, 2222–2229. doi:10.1016/j.cub.2005.10.043
Kim, H., Kim, M., Im, S.-K., and Fang, S. (2018). Mouse cre-LoxP system: General principles to determine tissue-specific roles of target genes. Lab. Anim. Res. 34, 147–159. doi:10.5625/lar.2018.34.4.147
Kim, J., and You, Y.-J. (2022). Oocyte quiescence: From formation to awakening. Endocrinology 163, bqac049. doi:10.1210/endocr/bqac049
Kleene, K. C. (2001). A possible meiotic function of the peculiar patterns of gene expression in mammalian spermatogenic cells. Mech. Dev. 106, 3–23. doi:10.1016/s0925-4773(01)00413-0
Kondo, S., Vedanayagam, J., Mohammed, J., Eizadshenass, S., Kan, L., Pang, N., et al. (2017). New genes often acquire male-specific functions but rarely become essential in Drosophila. Gene Dev. 31, 1841–1846. doi:10.1101/gad.303131.117
Kostrouchova, M., Krause, M., Kostrouch, Z., and Rall, J. E. (1998). CHR3: A Caenorhabditis elegans orphan nuclear hormone receptor required for proper epidermal development and molting. Development 125, 1617–1626. doi:10.1242/dev.125.9.1617
Kostrouchova, M., Krause, M., Kostrouch, Z., and Rall, J. E. (2001). Nuclear hormone receptor CHR3 is a critical regulator of all four larval molts of the nematode Caenorhabditis elegans. Proc. Natl. Acad. Sci. 98, 7360–7365. doi:10.1073/pnas.131171898
Krauchunas, A. R., Marcello, M. R., and Singson, A. (2016). The molecular complexity of fertilization: Introducing the concept of a fertilization synapse. Mol. Reprod. Dev. 83, 376–386. doi:10.1002/mrd.22634
Kroft, T. L., Gleason, E. J., and L’Hernault, S. W. (2005). The spe-42 gene is required for sperm–egg interactions during C. elegans fertilization and encodes a sperm-specific transmembrane protein. Dev. Biol. 286, 169–181. doi:10.1016/j.ydbio.2005.07.020
Kurhanewicz, N. A., Dinwiddie, D., Bush, Z. D., and Libuda, D. E. (2020). Elevated temperatures cause transposon-associated DNA damage in C. elegans spermatocytes. Curr. Biol. 30, 5007–5017. doi:10.1016/j.cub.2020.09.050
Lamitina, S. T., and L’Hernault, S. W. (2002). Dominant mutations in the Caenorhabditis elegans Myt1 ortholog wee-1.3 reveal a novel domain that controls M-phase entry during spermatogenesis. Development 129, 5009–5018. doi:10.1242/dev.129.21.5009
Mei, X., and Singson, A. W. (2021). The molecular underpinnings of fertility: Genetic approaches in Caenorhabditis elegans. Adv. Genet. 2, e10034. doi:10.1002/ggn2.10034
Mélodie, V. B., and Christine, W. (2018). Fertility and infertility: Definition and epidemiology. Clin. Biochem. 62, 2–10. doi:10.1016/j.clinbiochem.2018.03.012
Méndez, V., and Córdoba-Aguilar, A. (2004). Sexual selection and animal genitalia. Trends Ecol. Evol. 19, 224–225. doi:10.1016/j.tree.2004.03.012
Miller, B. J., Georges-Labouesse, E., Primakoff, P., and Myles, D. G. (2000). Normal fertilization occurs with eggs lacking the integrin alpha6beta1 and is CD9-dependent. J. Cell Biol. 149, 1289–1296. doi:10.1083/jcb.149.6.1289
Miller, D. E., Cook, K. R., and Hawley, R. S. (2019). The joy of balancers. Plos Genet. 15, e1008421. doi:10.1371/journal.pgen.1008421
Miyata, H., Castaneda, J. M., Fujihara, Y., Yu, Z., Archambeault, D. R., Isotani, A., et al. (2016). Genome engineering uncovers 54 evolutionarily conserved and testis-enriched genes that are not required for male fertility in mice. Proc. Natl. Acad. Sci. 113, 7704–7710. doi:10.1073/pnas.1608458113
Nakamura, M., Namiki, M., Okuyama, A., Matsui, T., Doi, Y., Takeyama, M., et al. (1987). Temperature sensitivity of human spermatogonia and spermatocytes in vitro. Arch. Androl. 19, 127–132. doi:10.3109/01485018708986808
Naour, F. L., Rubinstein, E., Jasmin, C., Prenant, M., and Boucheix, C. (2000). Severely reduced female fertility in CD9-deficient mice. Science 287, 319–321. doi:10.1126/science.287.5451.319
Noda, T., Blaha, A., Fujihara, Y., Gert, K. R., Emori, C., Deneke, V. E., et al. (2022). Sperm membrane proteins DCST1 and DCST2 are required for sperm-egg interaction in mice and fish. Commun. Biol. 5, 332. doi:10.1038/s42003-022-03289-w
Nyberg, K. G., and Carthew, R. W. (2017). Out of the testis: Biological impacts of new genes. Gene Dev. 31, 1825–1826. doi:10.1101/gad.307496.117
O’Connell, K. F., Leys, C. M., and White, J. G. (1998). A genetic screen for temperature-sensitive cell-division mutants of Caenorhabditis elegans. Genetics 149, 1303–1321. doi:10.1093/genetics/149.3.1303
Okabe, M., Yagasaki, M., Oda, H., Matzno, S., Kohama, Y., and Mimura, T. (1988). Effect of a monoclonal anti-mouse sperm antibody (OBF13) on the interaction of mouse sperm with zona-free mouse and hamster eggs. J. Reprod. Immunol. 13, 211–219. doi:10.1016/0165-0378(88)90002-2
Ragle, J. M., Aita, A. L., Morrison, K. N., Martinez-Mendez, R., Saeger, H. N., Ashley, G. A., et al. (2020). The conserved molting/circadian rhythm regulator NHR-23/NR1F1 serves as an essential co-regulator of C. elegans spermatogenesis. Development 147, dev193862. doi:10.1242/dev.193862
Ros, V. G. D., Maldera, J. A., Willis, W. D., Cohen, D. J., Goulding, E. H., Gelman, D. M., et al. (2008). Impaired sperm fertilizing ability in mice lacking Cysteine-RIch Secretory Protein 1 (CRISP1). Dev. Biol. 320, 12–18. doi:10.1016/j.ydbio.2008.03.015
Rubinstein, E., Ziyyat, A., Prenant, M., Wrobel, E., Wolf, J.-P., Levy, S., et al. (2006). Reduced fertility of female mice lacking CD81. Dev. Biol. 290, 351–358. doi:10.1016/j.ydbio.2005.11.031
Sanders, J. R., Ashley, B., Moon, A., Woolley, T. E., and Swann, K. (2018). PLCζ induced Ca2+ oscillations in mouse eggs involve a positive feedback cycle of Ca2+ induced InsP3 formation from cytoplasmic PIP2. Front. Cell Dev. Biol. 6, 36. doi:10.3389/fcell.2018.00036
Schreier, J., Dietz, S., Boermel, M., Oorschot, V., Seistrup, A.-S., Domingues, A. M. de J., et al. (2022). Membrane-associated cytoplasmic granules carrying the Argonaute protein WAGO-3 enable paternal epigenetic inheritance in Caenorhabditis elegans. Nat. Cell Biol. 24, 217–229. doi:10.1038/s41556-021-00827-2
Schultz, R. M., and Williams, C. J. (2002). The science of ART. Science 296, 2188–2190. doi:10.1126/science.1071741
Segev, N., Avinoam, O., and Podbilewicz, B. (2018). Fusogens. Curr. Biol. 28, R37–R380. doi:10.1016/j.cub.2018.01.024
Spiegelstein, O., Eudy, J. D., and Finnell, R. H. (2000). Identification of two putative novel folate receptor genes in humans and mouse. Gene 258, 117–125. doi:10.1016/s0378-1119(00)00418-2
Suzuki, D. T., and Procunier, D. (1969). Temperature-sensitive mutations in Drosophila melanogaster. 3. Dominant lethals and semilethals on chromosome 2*†. Proc. Natl. Acad. Sci. 62, 369–376. doi:10.1073/pnas.62.2.369
Takahashi, Y., Bigler, D., Ito, Y., and White, J. M. (2001). Sequence-specific interaction between the disintegrin domain of mouse ADAM 3 and murine eggs: Role of beta1 integrin-associated proteins CD9, CD81, and CD98. Mol. Biol. Cell 12, 809–820. doi:10.1091/mbc.12.4.809
Tanigawa, M., Miyamoto, K., Kobayashi, S., Sato, M., Akutsu, H., Okabe, M., et al. (2008). Possible involvement of CD81 in acrosome reaction of sperm in mice. Mol. Reprod. Dev. 75, 150–155. doi:10.1002/mrd.20709
Vacquier, V. D., and Moy, G. W. (1977). Isolation of bindin: The protein responsible for adhesion of sperm to sea urchin eggs. Proc. Natl. Acad. Sci. 74, 2456–2460. doi:10.1073/pnas.74.6.2456
Varadarajan, R., Nagarajaram, H. A., and Ramakrishnan, C. (1996). A procedure for the prediction of temperature-sensitive mutants of a globular protein based solely on the amino acid sequence. Proc. Natl. Acad. Sci. 93, 13908–13913. doi:10.1073/pnas.93.24.13908
Vjugina, U., and Evans, J. P. (2008). New insights into the molecular basis of mammalian sperm-egg membrane interactions. Front. Biosci. 13, 462–476. doi:10.2741/2693
Wilburn, D. B., and Swanson, W. J. (2016). From molecules to mating: Rapid evolution and biochemical studies of reproductive proteins. J. Proteomics 135, 12–25. doi:10.1016/j.jprot.2015.06.007
Wilson, L. D., Obakpolor, O. A., Jones, A. M., Richie, A. L., Mieczkowski, B. D., Fall, G. T., et al. (2018). The Caenorhabditis elegans spe-49 gene is required for fertilization and encodes a sperm-specific transmembrane protein homologous to SPE-42. Mol. Reprod. Dev. 85, 563–578. doi:10.1002/mrd.22992
Yoon, S.-Y., Eum, J. H., Lee, J. E., Lee, H. C., Kim, Y. S., Han, J. E., et al. (2012). Recombinant human phospholipase C zeta 1 induces intracellular calcium oscillations and oocyte activation in mouse and human oocytes. Hum. Reprod. 27, 1768–1780. doi:10.1093/humrep/des092
Yoon, S.-Y., Jellerette, T., Salicioni, A. M., Lee, H. C., Yoo, M., Coward, K., et al. (2008). Human sperm devoid of PLC, zeta 1 fail to induce Ca2+ release and are unable to initiate the first step of embryo development. J. Clin. Invest. 118, 3671–3681. doi:10.1172/jci36942
Keywords: egg, oocyte, fertilization, C. elegans, mouse, zebrafish, sperm
Citation: Maniates KA and Singson A (2023) Where are all the egg genes?. Front. Cell Dev. Biol. 11:1107312. doi: 10.3389/fcell.2023.1107312
Received: 24 November 2022; Accepted: 24 January 2023;
Published: 03 February 2023.
Edited by:
Shao-Chen Sun, Nanjing Agricultural University, ChinaReviewed by:
Jianqiang Bao, University of Science and Technology of China, ChinaLena Arévalo, University Hospital Bonn, Germany
Copyright © 2023 Maniates and Singson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Katherine A. Maniates, bWFuaWF0ZXNAd2Frc21hbi5ydXRnZXJzLmVkdQ==
 Katherine A. Maniates
Katherine A. Maniates Andrew Singson
Andrew Singson