- 1Paul M. Rady Department of Mechanical Engineering, University of Colorado Boulder, Boulder, CO, United States
- 2Biomedical Engineering Program, University of Colorado Boulder, Boulder, CO, United States
- 3BioFrontiers Institute, University of Colorado Boulder, Boulder, CO, United States
Phenotypic plasticity, or adaptability, of a cell determines its ability to survive and function within changing cellular environments. Changes in the mechanical environment, ranging from stiffness of the extracellular matrix (ECM) to physical stress such as tension, compression, and shear, are critical environmental cues that influence phenotypic plasticity and stability. Furthermore, an exposure to a prior mechanical signal has been demonstrated to play a fundamental role in modulating phenotypic changes that persist even after the mechanical stimulus is removed, creating stable mechanical memories. In this mini review, our objective is to highlight how the mechanical environment alters both phenotypic plasticity and stable memories through changes in chromatin architecture, mainly focusing on examples in cardiac tissue. We first explore how cell phenotypic plasticity is modulated in response to changes in the mechanical environment, and then connect the changes in phenotypic plasticity to changes in chromatin architecture that reflect short-term and long-term memories. Finally, we discuss how elucidating the mechanisms behind mechanically induced chromatin architecture that lead to cell adaptations and retention of stable mechanical memories could uncover treatment methods to prevent mal-adaptive permanent disease states.
Introduction
Phenotypic plasticity in response to external signals is crucial for a cell to survive and maintain homeostasis. Phenotypic plasticity is defined as the ability of cells to adapt (e.g., changes in morphology, physiology, or behavior) in response to intrinsic or external cues (Nemec and Kilian, 2020). A cell’s adaptation relies upon a myriad of mechanical and biochemical stimuli, resulting in the activation or repression of numerous gene expression profiles that lead to cellular remodeling. At the same time, cells must maintain developmental stability, or a phenotype consistent to the developmental trajectory specific to the cell type, to carry out specific roles in the human body. Hence, the balance between stability and phenotypic plasticity is vital to maintaining a functioning organ system.
The mechanical plasticity of the cell is largely a function of how the cell senses and responds to mechanical signals through mechanotransduction pathways (Lammerding et al., 2004a). Mechanotransduction pathways are stimulated by many external physical cues such as matrix stiffness and tensile, compressive, or shear stress. Membrane level proteins, such as stretch activated ion channels (e.g., Piezo, TRP, TREK/TRACK channels) (Jin et al., 2020), focal adhesion complexes (e.g., focal adhesion kinase, FAK), and cell-cell junctions (e.g., cadherins) are sensors that initiate cascading pathways resulting in changes in transcription in the nucleus. For example, cells sense increased stiffness of the environment through focal adhesions which initiates signaling cascades, such as Rho/ROCK, PI3K/AKT, and MAPK, resulting in changes in gene expression through activation of transcription factors (Martino et al., 2018). Examples of well-studied mechanosensitive molecules regulating transcription include the serum response factor (SRF) and the yes associated protein (YAP) (Dupont et al., 2011). These and other mechanotransduction pathways have been extensively explored and reviewed (Jaalouk and Lammerding, 2009; Martino et al., 2018; Wagh et al., 2021; Zuela-Sopilniak and Lammerding, 2022). Forces applied to the membrane can also directly influence the genetic programs that dictate the phenotypic response of the cell since the cytoskeletal network is linked to the nuclear structure containing chromatin and DNA through the linker of nucleoskeleton and cytoskeleton (LINC) complex (Lombardi et al., 2011), which also has been reviewed extensively (Lombardi and Lammerding, 2011; Bouzid et al., 2019; Lityagina and Dobreva, 2021; Wong et al., 2021). Although the mechanisms of mechanotransduction that mediate phenotypic plasticity have been well characterized in many systems, what is less known is how the mechanical environment alters cell fate and establishes a stable response, or memory.
Recent literature shows that alterations in the mechanical environment can lead to stable responses that persist even after the mechanical stimulus is removed, a phenomenon called mechanical memory (Kanoldt et al., 2018; Dai et al., 2020). Although the mechanisms of mechanical memory remain largely unexplored, recent studies focus on how changes in chromatin architecture retain responses to the mechanical environment altering the long-term cell fate (Heo et al., 2015; Killaars et al., 2019; Walker et al., 2021). By using cardiac physiology and pathology as an example, this mini review explores how the mechanical environment influences phenotypic plasticity and stable mechanical memories, specifically by altering the chromatin architecture of cells.
Mechanical plasticity for maintaining organ-level and cell-level homeostasis
Often the plastic response of a cell allows for beneficial adaptations that lead to maintaining physiological homeostasis. When there is a mechanical perturbation that disrupts this stability, the cell will adapt to this change in an attempt to return the system to a mechanically stable state, or mechanostasis (Chan et al., 2011). The heart is a highly plastic organ that demonstrates the transition between alternate stable states. In cases of increased hemodynamic load (such as pregnancy, repeated exercise, or postnatal growth) the heart muscle will undergo hypertrophic growth to accommodate the new output requirements (Tingare et al., 2013). The left ventricle of well-trained athletes can exceed the mass of non-athletes by as much as 60% (Hill and Olson, 2008). Despite this tremendous alteration, the cells maintain their plasticity; the physiological hypertrophy is reversible when the increased demand is removed. The end of an intensive training schedule leads to cardiac atrophy of 10%–22% in humans (Perhonen et al., 2001). This is also observed after pregnancy when a woman’s heart will decrease in size and return to the pre-pregnancy mass (Tingare et al., 2013).
These highly plastic responses are enabled by cellular proteins that sense the changes in mechanical cues and adapt accordingly. In the case of cardiac muscle size and function, the plastic response arises from remodeling of cardiac cells and the surrounding ECM. For example, increased mechanical loading due to pressure overload has been shown to cause sarcomere deposition in parallel, leading to the increase in myocyte cross-sectional area ultimately causing concentric hypertrophy. In contrast, volume overload often results in the elongation of myocytes due to the deposition of sarcomeres in series, which leads to dilation of the ventricle (Lammerding et al., 2004a; Lyon et al., 2015). Specifically, a change in the mechanical stretch is sensed by many mechanosensors in cardiac tissue: stretch activated ion channels (Yamazaki et al., 1998; Reed et al., 2014), integrins and integrin associated proteins (melusin, integrin-linked kinase) (White et al., 2006), cell surface receptors (G-protein-coupled receptors and angiotensin II type receptors), cytoskeletal and sarcomeric proteins (titin, myosin, or the small LIM domain protein MLP) (Lyon et al., 2015), and the nucleus (Lammerding et al., 2004a). Ultimately, the mechanical signal alters the cardiomyocyte phenotype via changes in gene expression resulting from changes in calcium concentration and activation of transcription pathways such as MAPK, JAK/STAT, PKC, PI3K, and Hippo-YAP/TAZ (Ruwhof and van der Laarse, 2000; Lammerding et al., 2004a; Saucerman et al., 2019). The focal adhesion-integrin complex is a main mechanosensor in cardiac fibroblasts and is known to activate many pathways leading to the production of ECM and fibrosis, including MAPK and p38 pathways (Davis and Molkentin, 2014; Saucerman et al., 2019). In addition to the activation of transcription factors, epigenetic mechanisms that regulate gene transcription could also provide another layer of gene transcription regulation by external mechanical cues (Saucerman et al., 2019).
Epigenetic landscape: Chromatin architecture changes in plastic verses stable responses
Epigenetic regulation, or the processes involving chemical modifications of chromatin that influence gene transcription, allows for both phenotypic plasticity in response to environmental cues and the formation of stable memories. Epigenetic mechanisms, such as histone modifications or DNA methylation, are reversible (although DNA methylation is typically more stable than histone modifications) (Tzelepis et al., 2017). At the same time, epigenetic mechanisms can be inherited through cell division, thus maintaining a stable epigenetic memory across generations (Henikoff and Greally, 2016; Kim and Costello, 2017). On a single gene level, the epigenetic modifications along with the spatial organization of chromatin by architectural proteins, give rise to the overall 3-dimensional (3D) chromatin architecture. The spatial organization of chromosomes is dynamic and adaptable to environmental changes (Solovei et al., 2009; Uhler and Shivashankar, 2017) and has been shown to be altered in disease states (Seelbinder et al., 2021; Walker et al., 2021; Heo et al., 2022). However, 3D architectures are also known to be cell type specific and thus demonstrate developmental stability (Seelbinder et al., 2021; Winick-Ng et al., 2021). Therefore, the cellular epigenetic regulation and alterations in chromatin architecture that determine the epigenetic landscape can control both the stability and phenotypic plasticity of cells.
Conrad Waddington proposed the metaphor of the “epigenetic landscape” to describe the changes in cell phenotypic plasticity and stability with changes in epigenetic regulation (Figure 1A). The action of balls rolling down a hill represents the accumulation of epigenetic changes during development that lead to stable differentiated cell states (Goldberg et al., 2007) (Figure 1A). At the same time, the metaphorical balls can roll into valleys adjacent to the current position. This transition represents cell phenotypic changes in response to external cues (e.g., mechanical cues), which can alter the chromatin architecture of the cell (Figures 1A, B). For example, cardiac fibroblasts will become activated into a myofibroblast state in response to a stiffening environment (Wang et al., 2003), which has been shown to change the chromatin architecture (Walker et al., 2021). The phenotypic plasticity or stable memory of the altered state of chromatin architecture is denoted by the depth of the valley in Waddington’s epigenetic landscape.
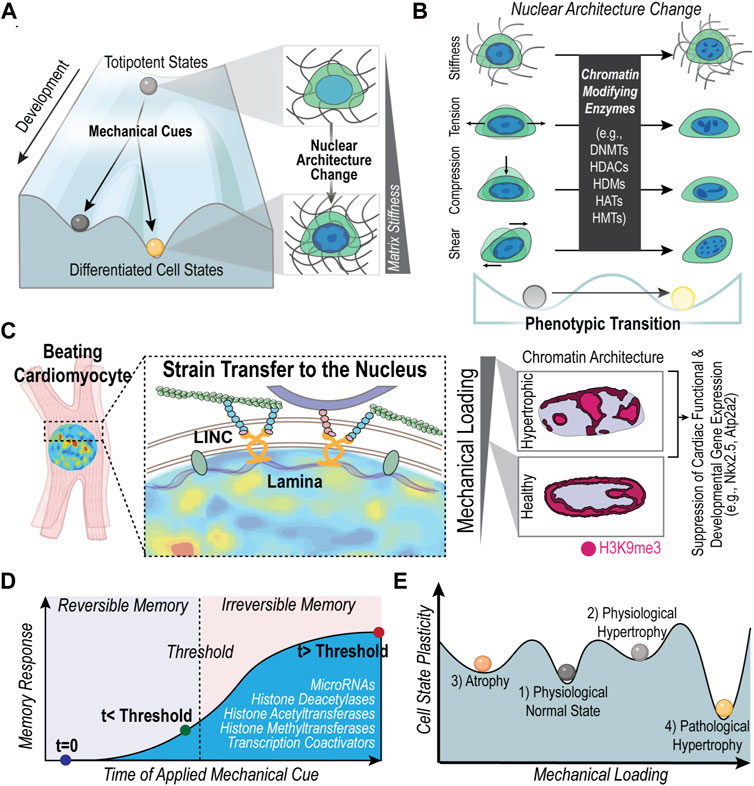
FIGURE 1. Mechanically induced nuclear architecture changes along Waddington’s landscape of epigenetic regulation. (A) The action of balls rolling downhill represents the increasing epigenetic changes that restrict the genetic landscape and ultimately lead to stable phenotypes. Environmental cues, such as the mechanical environment, can dictate cell fate changes and the overall differentiation process. As cells differentiate, the chromatin architecture changes (Peric-Hupkes et al., 2010; Zheng and Xie, 2019) and reflects a different state on the epigenetic landscape. (B) Changes in epigenetics due to mechanical cues (e.g., matrix mechanical properties, compression, tension, and shear forces) also alter the phenotypic state of cells, demonstrating that epigenetic regulation contributes to both the developmental stability and phenotypic plasticity of cells (Tzelepis et al., 2017). Chromatin architecture changes are also known to indicate a phenotypic change, especially in disease states (Seelbinder et al., 2021; Walker et al., 2021; Heo et al., 2022). (C) Previous work has demonstrated how a pathologically stiff mechanical environment in cardiac tissue is associated with a disruption of the chromatin architecture of repressive histone modification, H3K9me3, which has been linked to the dysregulation of cardiomyocyte differentiation and function (Seelbinder et al., 2021). Alterations in chromatin architecture of cardiomyocytes were associated with changes in nuclear deformation. Key molecular players that influence nuclear deformation of cardiomyocytes include nesprin proteins in the LINC complex (Ghosh et al., 2019). Literature also supports that lamin proteins are key regulators of nuclear mechanics and are associated with a loss of cardiac function (Lammerding et al., 2004b; Ghosh et al., 2019). (D) Cells exhibit mechanical memory; the history of mechanical priming can influence cell fate in a reversible and non-reversible manner depending on the exposure time to the mechanical cue. Molecules involved in storing mechanical memory include microRNAs, histone modifications, histone modifying enzymes and transcriptional coactivators (e.g., YAP). (E) As the mechanical load on heart cells increases or decreases the cell shifts to a new state of mechanostasis, shown as an equilibrium ‘valley’ on the graph. The cells in each state have different degrees of plasticity. The depth of the ‘valleys’ is proportional to the inability to adapt, or the lower degree of plasticity. For example, because the physiological hypertrophic state (2) is known to be highly reversible, its state of mechanostasis is depicted as a shallow ‘valley.’ Created with adapted graphics from BioRender.com.
How mechanical cues alter the epigenetic landscape and chromatin architecture
An increasing body of literature demonstrates the connections between the mechanical environment and the epigenetic landscape of cells (Spagnol et al., 2016; Miroshnikova et al., 2017; Wagh et al., 2021; Dupont and Wickström, 2022). Seminal work in the field demonstrated that substrate stiffness guides differentiation of mesenchymal stem cells (MSCs), highlighting the influence of the mechanical environment on how stem cells “roll” down the epigenetic landscape (Engler et al., 2006) (Figure 1A). In fact, chromatin condensation upon dynamic loading of MSCs was shown to be mediated by the cytoskeleton (Heo et al., 2015; Heo et al., 2016). Furthermore, the chromatin remodeling of MSCs in response to stiff culture environments occurs through the increase in histone acetyltransferase (HATs) and a decrease in histone deacetylases (HDACs) (Killaars et al., 2019; Killaars et al., 2020) (Figure 1B). Such chromatin remodeling through upregulation or downregulation of chromatin modifiers has also been observed in response to compressive forces (Damodaran et al., 2018) and hydrostatic pressure (Maki et al., 2021). In addition, chromosome positioning within the nucleus is also influenced by mechanical cues of cell geometry, which in turn affects the spatial organization of genes thereby regulating gene expression (Maharana et al., 2016; Wang et al., 2017; Alisafaei et al., 2019). Interestingly, recent studies have shown that mechanical cues can even induce cellular reprogramming via chromatin modifications. For instance, substrate topological cues, such as the presence of microgrooves, can influence the epigenetic state of fibroblasts and enhance their reprogramming to iPSCs through inducing changes in H3 acetylation and methylation (Downing et al., 2013). Also, nuclear deformation in fibroblasts through their confinement in microfluidic channels has shown to increase the efficiency of their reprogramming to neurons via decreased levels of H3K9me3 and DNA methylation (Song et al., 2022). Together, these studies demonstrate that the physical environment can alter the slope of the hill in the epigenetic landscape, either accelerating the “rolling down” of differentiating cells or allowing cells to “roll back up” the landscape in reprogramming processes (Figure 1A).
Although the mechanisms of how the epigenetic landscape is influenced by mechanical cues are not completely understood, recent studies focus on 1) indirect biochemical cues arising from the activation of mechanosensitive proteins and 2) the direct transmission of force to chromatin influencing a change in accessibility of transcription (Kirby and Lammerding, 2018; Dupont and Wickström, 2022; Kalukula et al., 2022). Deformation of stretch activated ion channels, such as Piezo1 found in the perinuclear endoplasmic reticulum, have been shown to decrease levels of H3K9me3 marked heterochromatin in response to cell stretching, while levels of H3K9me3 remained unchanged in response to stretch with Piezo1 depleted cells (Nava et al., 2020). Mechanosensitive nuclear membrane protein, Emerin, enrichment around the nuclear membrane can lead to the alteration of heterochromatin anchoring to the lamina associated domain through influencing nuclear G-actin levels (Le et al., 2016). Additionally, deformation of nuclear pores can allow nuclear shuttling of chromatin modifying enzymes, such as histone deacetylase 3 (HDAC3) from the cytoplasm (Zuela-Sopilniak and Lammerding, 2022). Alternatively, the physical links through the cytoskeleton and the LINC complex can allow for physical deformation of chromatin through nuclear strain transfer, influencing gene transcription (Lombardi et al., 2011; Lombardi and Lammerding, 2011; Neelam et al., 2015; Ghosh et al., 2017; Ghosh et al., 2019; Ghosh et al., 2021; Jones et al., 2022). For example, magnetic twisting of cell surface proteins induced transcription of a fluorescently tagged transgene depending on the direction and magnitude of the applied load (Tajik et al., 2016).
In the case of cardiac cells also, there is recent literature that highlights such mechanical regulation of the epigenetic landscape and its importance in cardiomyocyte development and function (Tingare et al., 2013; Stratton and McKinsey, 2016; Jarrell et al., 2019; Lityagina and Dobreva, 2021; Ross and Stroud, 2021; Powers and McCulloch, 2022). Connections from the cardiomyocyte nucleus to the external environment through microtubules and desmin intermediate filaments is required for the maintenance of the nuclear morphology and the loss of these connections results in aberrant gene expression and DNA damage (Heffler et al., 2020). Similarly, previous work has associated disrupted nuclear morphology with cardiac pathology (dilated cardiomyopathy) (Zhou et al., 2020) and the effects of transverse aortic constriction surgery (Karbassi et al., 2019). Cardiomyocyte nuclear morphology can be altered through both external cues and changes of internal cellular proteins, such as lamin proteins and proteins in the LINC complex, such as nesprins (Lammerding et al., 2004b; Ghosh et al., 2019) (Figure 1C). Work from our lab shows that when cardiomyocyte nuclear morphology is disrupted through culture on stiff substrates or through disruption of the LINC complex, the architecture of trimethylated H3K9 marked chromatin remodels, which we linked to dysregulation of the cardiomyocyte development and function (Seelbinder et al., 2021) (Figure 1C). Our work also supports a connection between nuclear strain and heterochromatin organization since regions of higher tensile strain in cardiomyocytes were more likely to overlap with regions of trimethylated H3K9 marked chromatin (Ghosh et al., 2019; Seelbinder et al., 2021). Additionally, stiff environments have been shown to induce changes in the chromatin architecture of cardiac interstitial cells that is linked to a persisting fibrotic phenotype (Walker et al., 2021). Key regulators of the chromatin changes of these cardiac interstitial cells included the CREB Binding Protein (Walker et al., 2022) and HDACs (Walker et al., 2021), while detachment of the nucleus from the cytoskeleton prevented the chromatin architecture changes. Taken together, these results support how the mechanical environment regulates the chromatin architecture, which in turn influences phenotypic changes of cardiac cells (Figures 1B, C).
Mechanical memory
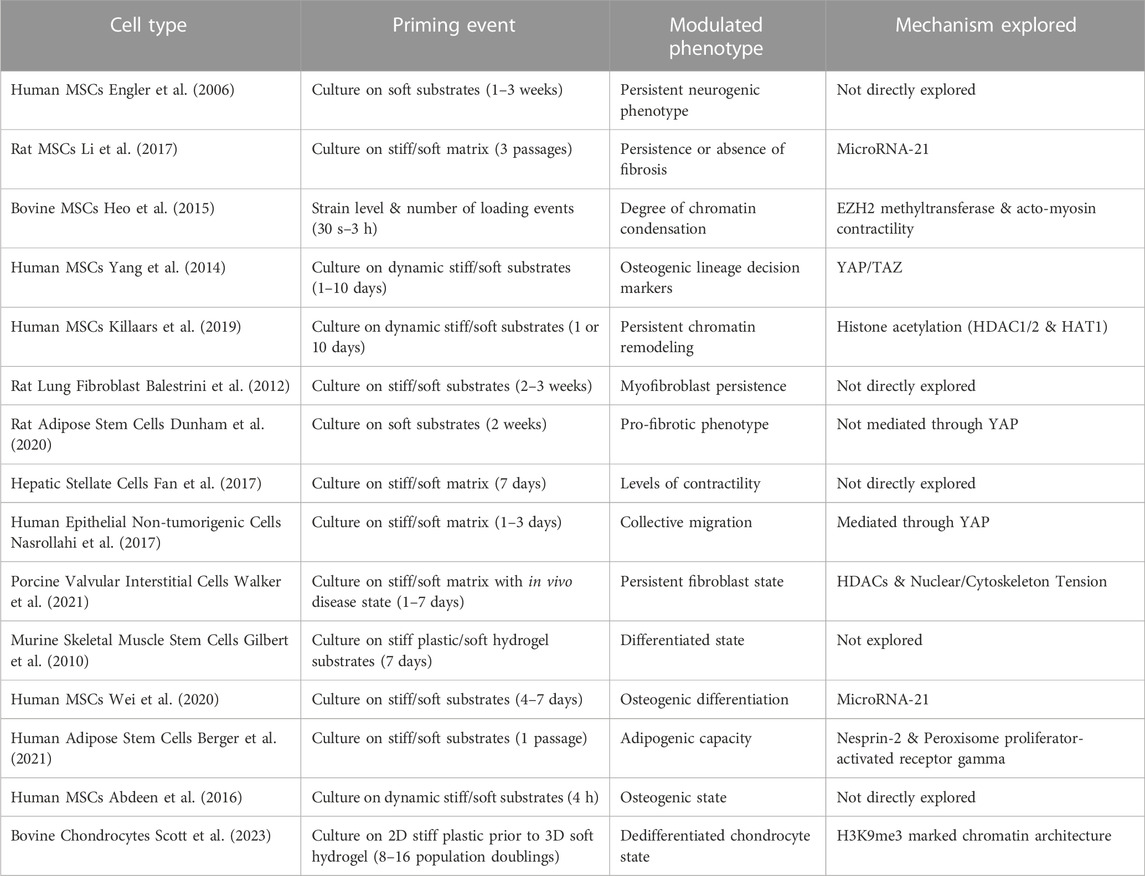
Recent studies have found that mechanical environments can cause phenotypic transitions that are maintained, even after the mechanical stimulus is removed demonstrating a stable mechanical memory (Figure 1D; Table 1) (Tingare et al., 2013). A critical factor observed to be an important determinant of mechanical memory is the prolonged exposure to the mechanical stimulus. For example, Balestrini et al. have shown that when lung fibroblasts are primed in a pathologically stiff culture environment for 3 weeks, they display a myofibroblast phenotype for 2 weeks after transfer to healthy soft substrates (Balestrini et al., 2012). In the reverse experiment, lung fibroblasts cultured on healthy soft substrates for 3 weeks were partially protected from myofibroblast activation after shifting to the pathologically stiff substrates (Balestrini et al., 2012). Similarly, Yang et al. showed that hMSC exposure to a relatively high stiffness of 10 kPa for 10 days before lowering the stiffness to 2 kPa, led to a permanent (>10 days) nuclear localization of RUNX2, a pre-osteogenic transcription factor, and YAP/TAZ, the transcriptional regulator of differentiation induced by ECM stiffness. Interestingly, experiments probing the mechanism of mechanical memory showed that YAP/TAZ acts as a “rheostat” that regulates the reversibility or irreversibility of the osteogenic response depending on whether the mechanical dosing time exceeded a critical memory threshold (Yang et al., 2014) (Figure 1D). Another molecule identified to play a role in preserving the fibrotic mechanical memory in rat MSCs is microRNA-21. In fact, knocking down microRNA-21 was sufficient to erase accumulated mechanical memory from stiff priming (Li et al., 2017).
In addition to the regulation of microRNAs and transcription factors, recent studies focus on chromatin remodeling induced by histone modifications as a mechanism for retaining mechanical memory (Turner, 2002; Hathaway et al., 2012; Heo et al., 2015; Fan et al., 2017) (Figure 1D). In fact, a theoretical framework to model mechanical memory required the modulation of epigenetic plasticity due to long term mechanical priming that locks in mechanical memory (Peng et al., 2017; Mathur et al., 2020). Dynamic loading applied for 150 s to 3 h resulted in increased levels of chromatin condensation in MSCs. The increased levels of chromatin condensation only persisted in cells dynamically loaded for 3 h, while chromatin condensation of cells that were loaded for less time (less than 1 h) was reversible (Heo et al., 2015). The persistence of condensed chromatin could be prevented by using pharmacological treatments to inhibit EZH2. Similarly, work from our lab shows that remodeling of H3K9me3 marked chromatin occurs when chondrocytes are exposed to stiff substrates and is later remembered when chondrocytes are transferred to a soft 3D environment. However, the remodeling of H3K9me3 can be partially disrupted with inhibitors of epigenetic modifiers (KDM4, EZH2) (Scott et al., 2023; Scott et al., 2022). Many of these examples of mechanical memory and others (listed in Table 1) highlight how changes in chromatin architecture can persist, maintaining stable memories. Therefore, not only are the mechanically induced changes in chromatin architecture involved in the plastic response of the cell, but such changes can also persist after removal of the stimulus, contributing to the stable mechanical memory of the cell.
Phenotypic stability of mechanically induced disease states
The effort to maintain mechanostasis in the presence of certain mechanical stimuli may lead to maladaptive responses that decrease the cell’s ability to function in the new environment. For example, a continuous excessive stress on the heart muscle, as caused by high blood pressure, often leads to thickening of the left ventricle walls of the heart, known as maladaptive cardiac hypertrophy (Dorn and Force, 2005; Shyu, 2009). Unlike the highly-reversible physiological hypertrophy, the hypertrophy induced by prolonged stress is usually followed by apoptosis or necrosis and has not been shown to be easily reversed (Jaalouk and Lammerding, 2009; Tingare et al., 2013). Regression of left ventricular remodeling due to pressure overload has been demonstrated with the removal of aortic band constrictions (Gao et al., 2005; Stansfield et al., 2007; Zhao et al., 2013; Yoshida et al., 2020) in animals as well as clinically with treatment of aortic stenosis after aortic valve replacements (Krayenbuehl et al., 1989; Matsumura et al., 2008; Hahn et al., 2013), demonstrating some degree of left ventricular remodeling plasticity. Additionally, regression of left ventricular remodeling has also been seen after weight loss (Kardassis et al., 2012). However, left ventricle dysfunction sometimes persists even after 3 years following weight loss (Alexander and Peterson, 1972). Aortic banding and de-banding studies suggest that exposure time to pressure overload may influence the regression and recovery of left ventricle remodeling (Zhao et al., 2013). Further exploration is needed to identify the conditions or mechanical threshold in which cardiac remodeling leads to heart failure (irreversible memory) and when there is potential for reversal of cardiac remodeling (reversible memory) (Figure 1D).
We propose a depiction of the cardiomyocyte response to loading in Figure 1E to demonstrate how the plastic response of cardiomyocytes changes with increased loading. Each valley in Figure 1E represents an alternate mechanically stable state, such as a physiological normal cardiomyocyte or a pathological hypertrophic cardiomyocyte. The depth of the valley is proportional to the inability to change phenotype. By exploring short and long-term changes in chromatin architecture of cardiac cells in response to various exposure times and degrees of mechanical loading, we may gain a more in-depth understanding of how plasticity or long-term disease states arise in the heart.
Discussion
Here, we have reviewed examples of how mechanical environments influence both the phenotypic plasticity and the stability of phenotypic states through changes in chromatin architecture, but many questions remain. What determines the threshold between a temporary adaptation to the physical environment and a stable memory? Perhaps 3D gene positioning relative to the areas of repressed heterochromatin in the nuclear periphery and areas of active transcriptional machinery dictate the degree of plasticity for the expression of gene programs. Alternatively, the overall 3D architecture of persistent states could be more energetically favorable from the perspective of phase separation models of chromatin (Erdel and Rippe, 2018). To what extent does cell division influence the phenotypic plasticity of cells in response to the mechanical environment? Does cell division present a window of opportunity (Probst et al., 2009) to alter cell fate in response to the surrounding mechanical environment? Furthermore, the phenomena of mechanical memory has only been demonstrated due to the exposure time to in vitro cultures. Can mechanical memories develop from changing mechanical environments in vivo? How does mechanical memory of cell states translate to organ level memory? Multi-scale computational models (Weinberg and Mofrad, 2007; Yoshida et al., 2022) incorporating cell models of mechanical memory (Nasrollahi et al., 2017) may begin to answer this question to understand how pathologies reach a point of no return. Overall, we propose that by further exploring these questions we could provide insight into methods to prevent long-term maladaptive disease states.
Author contributions
Conceptualization, AS, MR, and CN; Writing—original draft, AS, MR; Writing—review and editing, All authors; Funding acquisition, CN.
Funding
We are grateful for the support for this work in part by grant Nos. NIH R01 AR063712, NSF CAREER 1349735, and NSF 2212121 (CN).
Acknowledgments
We would like to acknowledge the Mechanobiology class taught by CN in the Paul M. Rady Department of Mechanical Engineering at the University of Colorado Boulder, as this class inspired the foundation of this review paper. Specifically, we would like to thank Luke Woolley and Kory Pearson for their help in planning the initial stages of this work, their feedback, and their edits.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdeen, A. A., Lee, J., Bharadwaj, N. A., Ewoldt, R. H., and Kilian, K. A. (2016). Temporal modulation of stem cell activity using magnetoactive hydrogels. Adv. Healthc. Mater 5, 2536–2544. doi:10.1002/adhm.201600349
Alexander, J. K., and Peterson, K. L. (1972). Cardiovascular effects of weight reduction. Circulation 45, 310–318. doi:10.1161/01.cir.45.2.310
Alisafaei, F., Jokhun, D. S., Shivashankar, G. v., and Shenoy, V. B. (2019). Regulation of nuclear architecture, mechanics, and nucleocytoplasmic shuttling of epigenetic factors by cell geometric constraints. Proc. Natl. Acad. Sci. U. S. A. 116, 13200–13209. doi:10.1073/pnas.1902035116
Balestrini, J. L., Chaudhry, S., Sarrazy, V., Koehler, A., and Hinz, B. (2012). The mechanical memory of lung myofibroblasts. Integr. Biol. 4, 410–421. doi:10.1039/c2ib00149g
Berger, A. J., Anvari, G., and Bellas, E. (2021). Mechanical memory impairs adipose-derived stem cell (ASC) adipogenic capacity after long-term in vitro expansion. Cell. Mol. Bioeng. 14, 397–408. doi:10.1007/s12195-021-00705-9
Bouzid, T., Kim, E., Riehl, B. D., Esfahani, A. M., Rosenbohm, J., Yang, R., et al. (2019). The LINC complex, mechanotransduction, and mesenchymal stem cell function and fate. J. Biol. Eng. 13, 68–12. doi:10.1186/s13036-019-0197-9
Chan, D. D., Van Dyke, W. S., Bahls, M., Connell, S. D., Critser, P., Kelleher, J. E., et al. (2011). Mechanostasis in apoptosis and medicine. Prog. Biophysics Mol. Biol. 106, 517–524. doi:10.1016/j.pbiomolbio.2011.08.002
Dai, E. N., Heo, S., and Mauck, R. L. (2020). “Looping in” mechanics: Mechanobiologic regulation of the nucleus and the epigenome. Adv. Healthc. Mater 9, 2000030. doi:10.1002/adhm.202000030
Damodaran, K., Venkatachalapathy, S., Alisafaei, F., Radhakrishnan, A. V., Sharma Jokhun, D., Shenoy, V. B., et al. (2018). Compressive force induces reversible chromatin condensation and cell geometry–dependent transcriptional response. Mol. Biol. Cell. 29, 3039–3051. doi:10.1091/mbc.E18-04-0256
Davis, J., and Molkentin, J. D. (2014). Myofibroblasts: Trust your heart and let fate decide. J. Mol. Cell. Cardiol. 70, 9–18. doi:10.1016/j.yjmcc.2013.10.019
Dorn, G. W., and Force, T. (2005). Protein kinase cascades in the regulation of cardiac hypertrophy. J. Clin. Investigation 115, 527–537. doi:10.1172/JCI24178
Downing, T. L., Soto, J., Morez, C., Houssin, T., Fritz, A., Yuan, F., et al. (2013). Biophysical regulation of epigenetic state and cell reprogramming. Nat. Mater 12, 1154–1162. doi:10.1038/nmat3777
Dunham, C., Havlioglu, N., Chamberlain, A., Lake, S., and Meyer, G. (2020). Adipose stem cells exhibit mechanical memory and reduce fibrotic contracture in a rat elbow injury model. FASEB J. 34, 12976–12990. doi:10.1096/fj.202001274R
Dupont, S., Morsut, L., Aragona, M., Enzo, E., Giulitti, S., Cordenonsi, M., et al. (2011). Role of YAP/TAZ in mechanotransduction. Nature 474 (7350), 179–183. doi:10.1038/nature10137
Dupont, S., and Wickström, S. A. (2022). Mechanical regulation of chromatin and transcription. Nat. Rev. Genet. 23 (10), 624–643. doi:10.1038/s41576-022-00493-6
Engler, A. J., Sen, S., Sweeney, H. L., and Discher, D. E. (2006). Matrix elasticity directs stem cell lineage specification. Cell. 126, 677–689. doi:10.1016/j.cell.2006.06.044
Erdel, F., and Rippe, K. (2018). Formation of chromatin subcompartments by phase separation. Biophys. J. 114, 2262–2270. doi:10.1016/j.bpj.2018.03.011
Fan, X., Zhu, L., Wang, K., Wang, B., Wu, Y., Xie, W., et al. (2017). Stiffness-controlled thermoresponsive hydrogels for cell harvesting with sustained mechanical memory. Adv. Healthc. Mater 6, 1601152. doi:10.1002/adhm.201601152
Gao, X. M., Kiriazis, H., Moore, X. L., Feng, X. H., Sheppard, K., Dart, A., et al. (2005). Regression of pressure overload-induced left ventricular hypertrophy in mice. Am. J. Physiol. Heart Circ. Physiol. 288, H2702–H2707. doi:10.1152/ajpheart.00836.2004
Ghosh, S., Cimino, J. G., Scott, A. K., Damen, F. W., Phillips, E. H., Veress, A. I., et al. (2017). In vivo multiscale and spatially-dependent biomechanics reveals differential strain transfer hierarchy in skeletal muscle. ACS Biomater. Sci. Eng. 3, 2798–2805. doi:10.1021/acsbiomaterials.6b00772
Ghosh, S., Scott, A. K., Seelbinder, B., Barthold, J. E., Martin, B. M. S., Kaonis, S., et al. (2021). Dedifferentiation alters chondrocyte nuclear mechanics during in vitro culture and expansion. Biophys. J. 121, 131–141. doi:10.1016/J.BPJ.2021.11.018
Ghosh, S., Seelbinder, B., Henderson, J. T., Watts, R. D., Scott, A. K., Veress, A. I., et al. (2019). Deformation microscopy for dynamic intracellular and intranuclear mapping of mechanics with high spatiotemporal resolution. Cell. Rep. 27, 1607–1620.e4. doi:10.1016/j.celrep.2019.04.009
Gilbert, P. M., Havenstrite, K. L., Magnusson, K. E. G., Sacco, A., Leonardi, N. A., Kraft, P., et al. (2010). Substrate elasticity regulates skeletal muscle stem cell self-renewal in culture. Science 329, 1078–1081. doi:10.1126/science.1191035
Goldberg, A. D., Allis, C. D., and Bernstein, E. (2007). Epigenetics: A landscape takes shape. Cell. 128, 635–638. doi:10.1016/j.cell.2007.02.006
Hahn, R. T., Pibarot, P., Stewart, W. J., Weissman, N. J., Gopalakrishnan, D., Keane, M. G., et al. (2013). Comparison of transcatheter and surgical aortic valve replacement in severe aortic stenosis: A longitudinal study of echocardiography parameters in cohort A of the partner trial (placement of aortic transcatheter valves). J. Am. Coll. Cardiol. 61, 2514–2521. doi:10.1016/j.jacc.2013.02.087
Hathaway, N. A., Bell, O., Hodges, C., Miller, E. L., Neel, D. S., and Crabtree, G. R. (2012). Dynamics and memory of heterochromatin in living cells. Cell. 149, 1447–1460. doi:10.1016/j.cell.2012.03.052
Heffler, J., Shah, P. P., Robison, P., Phyo, S., Veliz, K., Uchida, K., et al. (2020). A balance between intermediate filaments and microtubules maintains nuclear architecture in the cardiomyocyte. Circ. Res. 126, e10–e26. doi:10.1161/CIRCRESAHA.119.315582
Henikoff, S., and Greally, J. M. (2016). Epigenetics, cellular memory and gene regulation. Curr. Biol. 26, R644–R648. doi:10.1016/j.cub.2016.06.011
Heo, S. J., Han, W. M., Szczesny, S. E., Cosgrove, B. D., Elliott, D. M., Lee, D. A., et al. (2016). Mechanically induced chromatin condensation requires cellular contractility in mesenchymal stem cells. Biophys. J. 111, 864–874. doi:10.1016/j.bpj.2016.07.006
Heo, S. J. J., Thorpe, S. D., Driscoll, T. P., Duncan, R. L., Lee, D. A., and Mauck, R. L. (2015). Biophysical regulation of chromatin architecture instills a mechanical memory in mesenchymal stem cells. Sci. Rep. 5, 16895. doi:10.1038/srep16895
Heo, S. J., Thakur, S., Chen, X., Loebel, C., Xia, B., McBeath, R., et al. (2022). Aberrant chromatin reorganization in cells from diseased fibrous connective tissue in response to altered chemomechanical cues. Nat. Biomed. Eng. 7, 177–191. doi:10.1038/s41551-022-00910-5
Hill, J. A., and Olson, E. N. (2008). Cardiac plasticity. N. Engl. J. Med. 358, 1370–1380. doi:10.1056/NEJMra072139
Jaalouk, D. E., and Lammerding, J. (2009). Mechanotransduction gone awry. Nat. Rev. Mol. Cell. Biol. 10 (1), 63–73. doi:10.1038/nrm2597
Jarrell, D. K., Lennon, M. L., and Jacot, J. G. (2019). Epigenetics and Mechanobiology in heart development and congenital heart disease. Diseases 7, 52. doi:10.3390/diseases7030052
Jin, P., Jan, L. Y., and Jan, Y. N. (2020). Mechanosensitive ion channels: Structural features relevant to mechanotransduction mechanisms. Annu. Rev. Neurosci. 43, 207–229. doi:10.1146/annurev-neuro-070918-050509
Jones, M. L., Dahl, K. N., Lele, T. P., Conway, D. E., Shenoy, V., Ghosh, S., et al. (2022). The elephant in the cell: Nuclear mechanics and Mechanobiology. J. Biomech. Eng. 144, 080802–080803. doi:10.1115/1.4053797
Kalukula, Y., Stephens, A. D., Lammerding, J., and Gabriele, S. (2022). Mechanics and functional consequences of nuclear deformations. Nat. Rev. Mol. Cell. Biol. 23 (9), 583–602. doi:10.1038/s41580-022-00480-z
Kanoldt, V., Fischer, L., and Grashoff, C. (2018). Unforgettable force – crosstalk and memory of mechanosensitive structures. Biol. Chem. 400, 687–698. doi:10.1515/hsz-2018-0328
Karbassi, E., Rosa-Garrido, M., Chapski, D. J., Wu, Y., Ren, S., Wang, Y., et al. (2019). Direct visualization of cardiac transcription factories reveals regulatory principles of nuclear architecture during pathological remodeling. J. Mol. Cell. Cardiol. 128, 198–211. doi:10.1016/j.yjmcc.2019.02.003
Kardassis, D., Bech-Hanssen, O., Schönander, M., Sjöström, L., and Karason, K. (2012). The influence of body composition, fat distribution, and sustained weight loss on left ventricular mass and geometry in obesity. Obes. (Silver Spring) 20, 605–611. doi:10.1038/oby.2011.101
Killaars, A. R., Grim, J. C., Walker, C. J., Hushka, E. A., Brown, T. E., and Anseth, K. S. (2019). Extended exposure to stiff microenvironments leads to persistent chromatin remodeling in human mesenchymal stem cells. Adv. Sci. 6, 1801483. doi:10.1002/advs.201801483
Killaars, A. R., Walker, C. J., and Anseth, K. S. (2020). Nuclear mechanosensing controls MSC osteogenic potential through HDAC epigenetic remodeling. Proc. Natl. Acad. Sci. 117, 21258–21266. doi:10.1073/pnas.2006765117
Kim, M., and Costello, J. (2017). DNA methylation: An epigenetic mark of cellular memory. Exp. Mol. Med. 49, e322. doi:10.1038/emm.2017.10
Kirby, T. J., and Lammerding, J. (2018). Emerging views of the nucleus as a cellular mechanosensor. Nat. Cell. Biol. 20 (4), 373–381. doi:10.1038/s41556-018-0038-y
Krayenbuehl, H. P., Hess, O. M., Monrad, E. S., Schneider, J., Mall, G., and Turina, M. (1989). Left ventricular myocardial structure in aortic valve disease before, intermediate, and late after aortic valve replacement. Circulation 79, 744–755. doi:10.1161/01.cir.79.4.744
Lammerding, J., Kamm, R. D., and Lee, R. T. (2004). Mechanotransduction in cardiac myocytes. Ann. N. Y. Acad. Sci. 1015, 53–70. doi:10.1196/annals.1302.005
Lammerding, J., Schulze, P. C., Takahashi, T., Kozlov, S., Sullivan, T., Kamm, R. D., et al. (2004). Lamin A/C deficiency causes defective nuclear mechanics and mechanotransduction. J. Clin. Invest. 113, 370–378. doi:10.1172/JCI19670
Le, H. Q., Ghatak, S., Yeung, C. Y. C., Tellkamp, F., Günschmann, C., Dieterich, C., et al. (2016). Mechanical regulation of transcription controls Polycomb-mediated gene silencing during lineage commitment. Nat. Cell. Biol. 18, 864–875. doi:10.1038/ncb3387
Li, C. X., Talele, N. P., Boo, S., Koehler, A., Knee-Walden, E., Balestrini, J. L., et al. (2017). MicroRNA-21 preserves the fibrotic mechanical memory of mesenchymal stem cells. Nat. Mater 16, 379–389. doi:10.1038/nmat4780
Lityagina, O., and Dobreva, G. (2021). The LINC between mechanical forces and chromatin. Front. Physiol. 12, 710809. doi:10.3389/fphys.2021.710809
Lombardi, M. L., Jaalouk, D. E., Shanahan, C. M., Burke, B., Roux, K. J., and Lammerding, J. (2011). The interaction between nesprins and sun proteins at the nuclear envelope is critical for force transmission between the nucleus and cytoskeleton. J. Biol. Chem. 286, 26743–26753. doi:10.1074/jbc.M111.233700
Lombardi, M. L., and Lammerding, J. (2011). Keeping the LINC: The importance of nucleo-cytoskeletal coupling in intracellular force transmission and cellular function. Biochem. Soc. Trans. 39, 1729–1734. doi:10.1042/BST20110686
Lyon, R. C., Zanella, F., Omens, J. H., and Sheikh, F. (2015). Mechanotransduction in cardiac hypertrophy and failure. Circ. Res. 116, 1462–1476. doi:10.1161/CIRCRESAHA.116.304937
Maharana, S., Iyer, K. V., Jain, N., Nagarajan, M., Wang, Y., and Shivashankar, G. V. (2016). Chromosome intermingling—The physical basis of chromosome organization in differentiated cells. Nucleic Acids Res. 44, 5148–5160. doi:10.1093/nar/gkw131
Maki, K., Nava, M. M., Villeneuve, C., Chang, M., Furukawa, K. S., Ushida, T., et al. (2021). Hydrostatic pressure prevents chondrocyte differentiation through heterochromatin remodeling. J. Cell. Sci. 134, jcs247643. doi:10.1242/jcs.247643
Martino, F., Perestrelo, A. R., Vinarský, V., Pagliari, S., and Forte, G. (2018). Cellular mechanotransduction: From tension to function. Front. Physiol. 9, 824. doi:10.3389/fphys.2018.00824
Mathur, J., Shenoy, V., and Pathak, A. (2020). Mechanical memory in cells emerges from mechanotransduction with transcriptional feedback and epigenetic plasticity. 2020.03.20.000802. bioRxiv. doi:10.1101/2020.03.20.000802
Matsumura, Y., Gillinov, A. M., Toyono, M., Wada, N., Yamano, T., Thomas, J. D., et al. (2008). Usefulness of left ventricular shape to predict the early recovery of left ventricular function after isolated aortic valve replacement for aortic valve stenosis. Am. J. Cardiol. 102, 1530–1534. doi:10.1016/j.amjcard.2008.07.044
Miroshnikova, Y. A., Nava, M. M., and Wickström, S. A. (2017). Emerging roles of mechanical forces in chromatin regulation. J. Cell. Sci. 130, 2243–2250. doi:10.1242/jcs.202192
Nasrollahi, S., Walter, C., Loza, A. J., Schimizzi, G. V., Longmore, G. D., and Pathak, A. (2017). Past matrix stiffness primes epithelial cells and regulates their future collective migration through a mechanical memory. Biomaterials 146, 146–155. doi:10.1016/j.biomaterials.2017.09.012
Nava, M. M., Miroshnikova, Y. A., Biggs, L. C., Whitefield, D. B., Metge, F., Boucas, J., et al. (2020). Heterochromatin-driven nuclear softening protects the genome against mechanical stress-induced damage. Cell. 181, 800–817.e22. doi:10.1016/j.cell.2020.03.052
Neelam, S., Chancellor, T. J., Li, Y., Nickerson, J. A., Roux, K. J., Dickinson, R. B., et al. (2015). Direct force probe reveals the mechanics of nuclear homeostasis in the mammalian cell. Proc. Natl. Acad. Sci. U. S. A. 112, 5720–5725. doi:10.1073/pnas.1502111112
Nemec, S., and Kilian, K. A. (2020). Materials control of the epigenetics underlying cell plasticity. Nat. Rev. Mater 6, 69–83. doi:10.1038/s41578-020-00238-z
Peng, T., Liu, L., MacLean, A. L., Wong, C. W., Zhao, W., and Nie, Q. (2017). A mathematical model of mechanotransduction reveals how mechanical memory regulates mesenchymal stem cell fate decisions. BMC Syst. Biol. 11, 55. doi:10.1186/s12918-017-0429-x
Perhonen, M. A., Franco, F., Lane, L. D., Buckey, J. C., Blomqvist, C. G., Zerwekh, J. E., et al. (2001). Cardiac atrophy after bed rest and spaceflight. J. Appl. Physiol. 91, 645–653. doi:10.1152/jappl.2001.91.2.645
Peric-Hupkes, D., Meuleman, W., Pagie, L., Bruggeman, S. W. M., Solovei, I., Brugman, W., et al. (2010). Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol. Cell. 38, 603–613. doi:10.1016/j.molcel.2010.03.016
Powers, J. D., and McCulloch, A. D. (2022). Biomechanical signals regulating the structure of the heart. Curr. Opin. Physiol. 25, 100482. doi:10.1016/j.cophys.2021.100482
Probst, A. v., Dunleavy, E., and Almouzni, G. (2009). Epigenetic inheritance during the cell cycle. Nat. Rev. Mol. Cell. Biol. 10, 192–206. doi:10.1038/nrm2640
Reed, A., Kohl, P., and Peyronnet, R. (2014). Molecular candidates for cardiac stretch-activated ion channels. Glob. Cardiol. Sci. Pract. 9, 9–25. doi:10.5339/gcsp.2014.19
Ross, J. A., and Stroud, M. J. (2021). The nucleus: Mechanosensing in cardiac disease. Int. J. Biochem. Cell. Biol. 137, 106035. doi:10.1016/j.biocel.2021.106035
Ruwhof, C., and van der Laarse, A. (2000). Mechanical stress-induced cardiac hypertrophy: Mechanisms and signal transduction pathways. Cardiovasc Res. 47, 23–37. doi:10.1016/s0008-6363(00)00076-6
Saucerman, J. J., Tan, P. M., Buchholz, K. S., McCulloch, A. D., and Omens, J. H. (2019). Mechanical regulation of gene expression in cardiac myocytes and fibroblasts. Nat. Rev. Cardiol. 16, 361–378. doi:10.1038/s41569-019-0155-8
Scott, A. K., Gallagher, K. M., Schneider, S. E., Kurse, A., and Neu, C. P. (2022). Epigenetic priming enhances chondrogenic potential of expanded chondrocytes for cartilage repair. 2022.10.23.513439. bioRxiv. doi:10.1101/2022.10.23.513439
Scott, A. K., Casas, E., Schneider, S. E., Swearingen, A. R., Van Den Elzen, C. L., Seelbinder, B., et al. (2023). Mechanical memory stored through epigenetic remodeling reduces cell therapeutic potential. Biophys. J. doi:10.1016/j.bpj.2023.03.004
Seelbinder, B., Ghosh, S., Schneider, S. E., Scott, A. K., Berman, A. G., Goergen, C. J., et al. (2021). Nuclear deformation guides chromatin reorganization in cardiac development and disease. Nat. Biomed. Eng. 5, 1500–1516. doi:10.1038/S41551-021-00823-9
Shyu, K. G. (2009). Cellular and molecular effects of mechanical stretch on vascular cells and cardiac myocytes. Clin. Sci. 116, 377–389. doi:10.1042/CS20080163
Solovei, I., Kreysing, M., Lanctôt, C., Kösem, S., Peichl, L., Cremer, T., et al. (2009). Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell. 137, 356–368. doi:10.1016/j.cell.2009.01.052
Song, Y., Soto, J., Chen, B., Hoffman, T., Zhao, W., Zhu, N., et al. (2022). Transient nuclear deformation primes epigenetic state and promotes cell reprogramming. Nat. Mater. 21 (10), 1191–1199. doi:10.1038/s41563-022-01312-3
Spagnol, S. T., Armiger, T. J., and Dahl, K. N. (2016). Mechanobiology of chromatin and the nuclear interior. Cell. Mol. Bioeng. 9, 268–276. doi:10.1007/s12195-016-0444-9
Stansfield, W. E., Rojas, M., Corn, D., Willis, M., Patterson, C., Smyth, S. S., et al. (2007). Characterization of a model to independently study regression of ventricular hypertrophy. J. Surg. Res. 142, 387–393. doi:10.1016/j.jss.2007.01.037
Stratton, M. S., and McKinsey, T. A. (2016). Epigenetic regulation of cardiac fibrosis. J. Mol. Cell. Cardiol. 92, 206–213. doi:10.1016/j.yjmcc.2016.02.011
Tajik, A., Zhang, Y., Wei, F., Sun, J., Jia, Q., Zhou, W., et al. (2016). Transcription upregulation via force-induced direct stretching of chromatin. Nat. Mater 15, 1287–1296. doi:10.1038/nmat4729
Tingare, A., Thienpont, B., and Roderick, H. L. (2013). Epigenetics in the heart: The role of histone modifications in cardiac remodelling. Biochem. Soc. Trans. 41, 789–796. doi:10.1042/BST20130012
Turner, B. M. (2002). Cellular memory and the histone code. Cell. 111, 285–291. doi:10.1016/s0092-8674(02)01080-2
Tzelepis, I., Martino, M., and Göndör, A. (2017). “A brief introduction to chromatin regulation and dynamics,” in Chromatin regulation and dynamics (Amsterdam: Elsevier Inc.), 1–34. doi:10.1016/B978-0-12-803395-1.00001-0
Uhler, C., and Shivashankar, G. V. (2017). Regulation of genome organization and gene expression by nuclear mechanotransduction. Nat. Rev. Mol. Cell. Biol. 18, 717–727. doi:10.1038/nrm.2017.101
Wagh, K., Ishikawa, M., Garcia, D. A., Stavreva, D. A., Upadhyaya, A., and Hager, G. L. (2021). Mechanical regulation of transcription: Recent advances. Trends Cell. Biol. 31, 457–472. doi:10.1016/j.tcb.2021.02.008
Walker, C. J., Batan, D., Bishop, C. T., Ramirez, D., Aguado, B. A., Schroeder, M. E., et al. (2022). Extracellular matrix stiffness controls cardiac valve myofibroblast activation through epigenetic remodeling. Bioeng. Transl. Med. 7, e10394. doi:10.1002/btm2.10394
Walker, C. J., Crocini, C., Ramirez, D., Killaars, A. R., Grim, J. C., Aguado, B. A., et al. (2021). Nuclear mechanosensing drives chromatin remodelling in persistently activated fibroblasts. Nat. Biomed. Eng. 5, 1485–1499. doi:10.1038/s41551-021-00709-w
Wang, J., Chen, H., Seth, A., and McCulloch, C. A. (2003). Mechanical force regulation of myofibroblast differentiation in cardiac fibroblasts. Am. J. Physiol. Heart Circ. Physiol. 285, 1871–1881. doi:10.1152/ajpheart.00387.2003
Wang, Y., Nagarajan, M., Uhler, C., and Shivashankar, G. V. (2017). Orientation and repositioning of chromosomes correlate with cell geometry-dependent gene expression. Mol. Biol. Cell. 28, 1997–2009. doi:10.1091/mbc.E16-12-0825
Wei, D., Liu, A., Sun, J., Chen, S., Wu, C., Zhu, H., et al. (2020). Mechanics-controlled dynamic cell niches guided osteogenic differentiation of stem cells via preserved cellular mechanical memory. ACS Appl. Mater Interfaces 12, 260–274. doi:10.1021/acsami.9b18425
Weinberg, E. J., and Mofrad, M. R. K. (2007). Transient, three-dimensional, multiscale simulations of the human aortic valve. Cardiovasc Eng. 7, 140–155. doi:10.1007/s10558-007-9038-4
White, D. E., Coutu, P., Shi, Y. -F., Tardif, J. -C., Nattel, S., and Arnaud, R. S. (2006). Targeted ablation of ILK from the murine heart results in dilated cardiomyopathy and spontaneous heart failure. Genes Dev. 20, 2355–2360. doi:10.1101/gad.1458906
Winick-Ng, W., Kukalev, A., Harabula, I., Zea-Redondo, L., Szabó, D., Meijer, M., et al. (2021). Cell-type specialization is encoded by specific chromatin topologies. Nature 599 (7886), 684–691. doi:10.1038/s41586-021-04081-2
Wong, X., Loo, T. H., and Stewart, C. L. (2021). LINC complex regulation of genome organization and function. Curr. Opin. Genet. Dev. 67, 130–141. doi:10.1016/j.gde.2020.12.007
Yamazaki, T., Komuro, I., Kudoh, S., Zou, Y., Nagai, R., Aikawa, R., et al. (1998). Role of ion channels and exchangers in mechanical stretch–induced cardiomyocyte hypertrophy. Circ. Res. 82, 430–437. doi:10.1161/01.res.82.4.430
Yang, C., Tibbitt, M. W., Basta, L., and Anseth, K. S. (2014). Mechanical memory and dosing influence stem cell fate. Nat. Mater 13, 645–652. doi:10.1038/nmat3889
Yoshida, K., McCulloch, A. D., Omens, J. H., and Holmes, J. W. (2020). Predictions of hypertrophy and its regression in response to pressure overload. Biomech. Model. Mechanobiol. 19, 1079–1089. doi:10.1007/s10237-019-01271-w
Yoshida, K., Saucerman, J. J., and Holmes, J. W. (2022). Multiscale model of heart growth during pregnancy: Integrating mechanical and hormonal signaling. Biomech. Model. Mechanobiol. 21, 1267–1283. doi:10.1007/s10237-022-01589-y
Zhao, Z., Chen, L., Xiao, Y. B., Hao, J., Tang, C. Z., and Zheng, D. Z. (2013). A rabbit model to study regression of ventricular hypertrophy. Heart Lung Circ. 22, 373–382. doi:10.1016/j.hlc.2012.11.021
Zheng, H., and Xie, W. (2019). The role of 3D genome organization in development and cell differentiation. Nat. Rev. Mol. Cell. Biol. 20 (9), 535–550. doi:10.1038/s41580-019-0132-4
Zhou, Y., Chen, Z., Zhang, L., Zhu, M., Tan, C., Zhou, X., et al. (2020). Loss of filamin c is catastrophic for heart function. Circulation 141, 869–871. doi:10.1161/CIRCULATIONAHA.119.044061
Keywords: cellular plasticity, mechanotransduction, mechanical memory, chromatin, epigenetics
Citation: Scott AK, Rafuse M and Neu CP (2023) Mechanically induced alterations in chromatin architecture guide the balance between cell plasticity and mechanical memory. Front. Cell Dev. Biol. 11:1084759. doi: 10.3389/fcell.2023.1084759
Received: 30 October 2022; Accepted: 07 April 2023;
Published: 18 April 2023.
Edited by:
Xiang Wang, National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIH), United StatesReviewed by:
Aprotim Mazumder, Tata Institute of Fundamental Research (Hyderabad), IndiaEkta Makhija, Singapore-MIT Alliance for Research and Technology (SMART), Singapore
Copyright © 2023 Scott, Rafuse and Neu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Adrienne K. Scott, YWRyaWVubmUuc2NvdHRAY29sb3JhZG8uZWR1
 Adrienne K. Scott
Adrienne K. Scott Michael Rafuse
Michael Rafuse Corey P. Neu
Corey P. Neu