
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Cell Dev. Biol., 21 April 2023
Sec. Signaling
Volume 11 - 2023 | https://doi.org/10.3389/fcell.2023.1069256
This article is part of the Research TopicHomeostatic Regulation of Protein Synthesis, Folding and Secretion by Stress Response Pathways in EukaryotesView all 5 articles
The conventional early secretory pathway and autophagy are two essential interconnected cellular processes that are crucial for maintaining cellular homeostasis. The conventional secretory pathway is an anabolic cellular process synthesizing and delivering proteins to distinct locations, including different organelles, the plasma membrane, and the extracellular media. On the other hand, autophagy is a catabolic cellular process that engulfs damaged organelles and aberrant cytosolic constituents into the double autophagosome membrane. After fusion with the lysosome and autolysosome formation, this process triggers digestion and recycling. A growing list of evidence indicates that these anabolic and catabolic processes are mutually regulated. While knowledge about the molecular actors involved in the coordination and functional cooperation between these two processes has increased over time, the mechanisms are still poorly understood. This review article summarized and discussed the most relevant evidence about the key molecular players implicated in the interorganelle crosstalk between the early secretory pathway and autophagy under normal and stressful conditions.
The secretory pathway is responsible for synthesizing, folding, sorting, and delivering a variety of cellular proteins to distinct locations, such as different organelles, the plasma membrane, and the extracellular media. In the early secretory pathway, the endoplasmic reticulum (ER) ensures the correct entry of nascent proteins safeguarding their correct post-translational modifications and folding. Folded proteins are incorporated into ER-derived transport vesicles where they are delivered to the Golgi apparatus, the next station of the early anterograde transport pathway. On the other hand, ER-resident proteins that escape from the ER and cycle between the two early stations, need to constantly return from the Golgi to the ER, a process called the retrograde transport pathway (Barlowe and Miller, 2013). Both anterograde and retrograde early secretory pathways are critical to maintaining organelle structure, function, and cell homeostasis (Farhan et al., 2008; Cancino et al., 2014; Hanna et al., 2018).
Meanwhile, macroautophagy (hereafter referred to as autophagy) is a catabolic cellular process that engulfs damaged organelles and aberrant cytosolic components into a double membrane called the autophagosome. The autophagosome then fuses with the lysosomes to form a hybrid organelle called an autolysosome, which digests and recycles the molecular components (Mizushima et al., 2008; Pu et al., 2016; Mizushima, 2018). The process of forming an autophagosome begins with the formation of a pre-autophagosomal structure (PAS). The PAS initiates the nucleation of essential components that are necessary for the elongation, maturation, and sealing of the double membrane that forms the autophagosome (Kawamata et al., 2008; Suzuki and Ohsumi, 2010; Harada et al., 2019). The autophagosomes then fuse with lysosomes to form an autolysosome (Reggiori and Ungermann, 2017).
It is important to note that there is a mutual regulation between the early conventional secretory pathway and autophagy (summarized in Table 1). Recent research suggests that the early secretory pathway, including de ER and Golgi apparatus, plays an important role in the regulation of autophagy beyond providing membrane sources for autophagosome formation. Conversely, autophagy regulates important organelles and machinery of the early secretory pathways, such as the ER size through ER-phagy (Gubas and Dikic, 2022), turnover of the Golgi apparatus (Nthiga et al., 2021; Rahman et al., 2022), and the quantity and efficiency of the quality control machinery that defines efficient protein secretion, such as lectin binding proteins that recognize unfolded proteins for degradation (Park et al., 2014) and the proteasome through proteophagy (Albornoz et al., 2019; Quinet et al., 2020). Moreover, membrane composition alters both, the early secretory pathway, and autophagy in yeast (Li et al., 2020). The disruption of triacylglycerol synthesis led to diacylglycerol (DAG) accumulation disturbing the balance of ER-Golgi protein trafficking, resulting in ER swelling, loss of the Golgi apparatus, and autophagy inhibition.
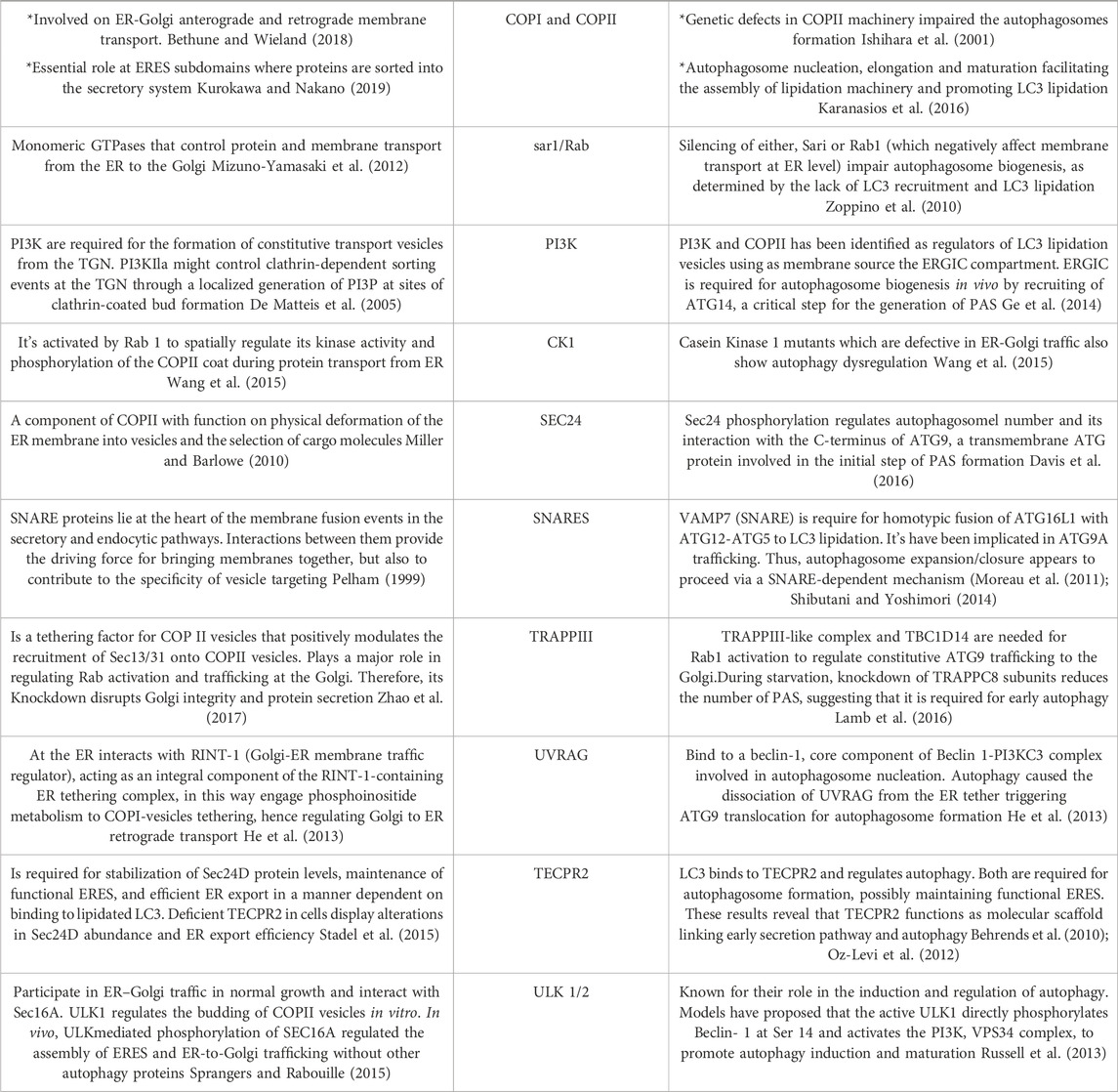
TABLE 1. Common regulators of the ER-Golgi early secretory pathway and autophagy. The main proteins described in this work were summarized in this table describing the role of those protein complexes and regulators on both, the secretory and autophagy pathways.
Here, we address several cellular and molecular aspects that demonstrate how the early conventional secretory pathway and autophagy are mutually regulated, including the growing literature about the key regulatory clues under normal and stressful scenarios and their impact on cellular homeostasis maintenance.
The two compartments of the early conventional secretory pathway, ER and Golgi apparatus, play a role in regulating autophagy at different levels, where the anterograde and retrograde pathways have complementary outcomes in this catabolic pathway. Concerning this, the coat protein complex II (COPII) and coat protein complex I (COPI) involved in ER-Golgi anterograde and retrograde Golgi to ER membrane transport, respectively (Bethune and Wieland, 2018) have specific contributions in how these two pathways control autophagy (Table 1).
COPII complex is recruited hierarchically to the specific ER membrane spots called ER exit sites (ERES). Functional, proteomic, and cytological analyses have shown that autophagosomes are spatially, physically, and functionally linked to ERES (Graef et al., 2013; Lamb et al., 2013; Suzuki et al., 2013; Sanchez-Wandelmer et al., 2015). The COPII recruitment is initiated by the small G-protein Sar1, which is regulated by the exchange of GDP for GTP catalyzed by the ER membrane protein SEC12, which activates Sar1 and recruits the SEC23 and SEC24 heterodimer (Nakano et al., 1988; Nakano and Muramatsu, 1989; Barlowe et al., 1994; Mancias and Goldberg, 2008). SEC24 is the cargo binding protein (Miller et al., 2002) interacting with several different ER export motifs through various domains (Miller et al., 2003). The SEC23/SEC24 heterodimer also provides the platform for the recruitment of the outer layer of the COPII coat, a heterodimer formed by SEC13 and SEC31 (Sato and Nakano, 2007; Bisnett et al., 2020).
The first evidence that suggested the COPII complex was regulating autophagy was found in yeast. They observed that the genetic ablation of the SEC24, a component of the COPII complex, abolished autophagosome formation (Ishihara et al., 2001). Recently, SEC23 and its mammalian counterpart SEC24C have been implicated in autophagy as receptors, selecting ER membranes and cargo for degradation (Cui et al., 2019). Additionally, studies in mammalian cells have shown that the COPII complex is also required for autophagosome formation (Shima et al., 2019). The precise mechanism by which COPII regulates autophagosome formation is not fully understood, but it is thought to involve the regulation of the phosphatidylinositol 3-kinase (PI3K) complex, which is involved in the initiation of autophagosome formation (Ge et al., 2014). Overall, the evidence suggests that the COPII complex plays a crucial role in the regulation of autophagy by providing the necessary membrane for autophagosome formation and regulating the PI3K complex. Recently, using a vesicle–labeling system in fluorescence and immunoelectron microscopy, it has been confirmed that COPII vesicles play a role as a membrane source for autophagosomes (Shima et al., 2019). In agreement with this finding, treatment with FLI06, a drug that prevents the loading of cargo to the COPII complex and causes a blockage in general secretion at the ER, triggers a decrease in the levels of ATG13, a factor required for autophagosome formation, and a significant reduction in the number of autophagosomes, similar to the phenotype observed in non-starved cells (Karanasios et al., 2016). In addition, another piece of evidence refers to the role of SEC24, a protein that has been shown to participate in the signaling required for the exit of cargo from the ER, called autoregulation of ER export (AREX). SEC24 acts as a sensor of folded cargo by working as a guanine nucleotide exchange factor (GEF) to promote cargo export at the ERES by activating the trimeric Gα12 GTPase (Subramanian et al., 2019) (Figure 1A). Interestingly, it has been reported that the absence of SEC24 is also required to initiate autophagosomal biogenesis (Ishihara et al., 2001). Additionally, because the SEC13/SEC31 heterodimer is not necessary for autophagy, critical components of the COPII complexes suggest that vesicle formation and autophagosomal biogenesis share some important machinery but not all the structural players. This also opens the possibility that SEC24 is a common player in these two pathways. However, the mechanism of how SEC24 is recruited to the ER in one direction, or another is not yet understood. One possible mechanism might be related to SEC24 post-translational modifications. In this regard, several phosphorylation sites have been reported on SEC24; among them, phosphorylation on T324, T325, and T328 residues is required for autophagy. The phosphorylation of SEC24 by the casein kinase 1 (Hrr25, CK1 in mammalian cells) promotes its interaction with Atg9 and induces autophagosome formation (Davis et al., 2016) (Figure 1B), post-translational modifications that are not required for ER-to-Golgi anterograde transport in yeast. Alternatively, another explanation could be related to the function of the Gαi3 GTPase (Petiot et al., 1999). Gαi3 is located at the ER and Golgi apparatus and controls ADP-ribosylation factor 1 (Arf1) signaling, serving as a crossroad between signals controlled by the trimeric G proteins and the Arf family of monomeric GTPases to regulate early conventional anterograde pathway (Lo et al., 2015) (Figure 1C). Importantly, it has been reported that Gαi3 GTPase acts as a regulator of autophagy, controlling autophagosome biogenesis (Gohla et al., 2007), by a mechanism dependent on insulin signaling and nutrients (Gohla et al., 2007).
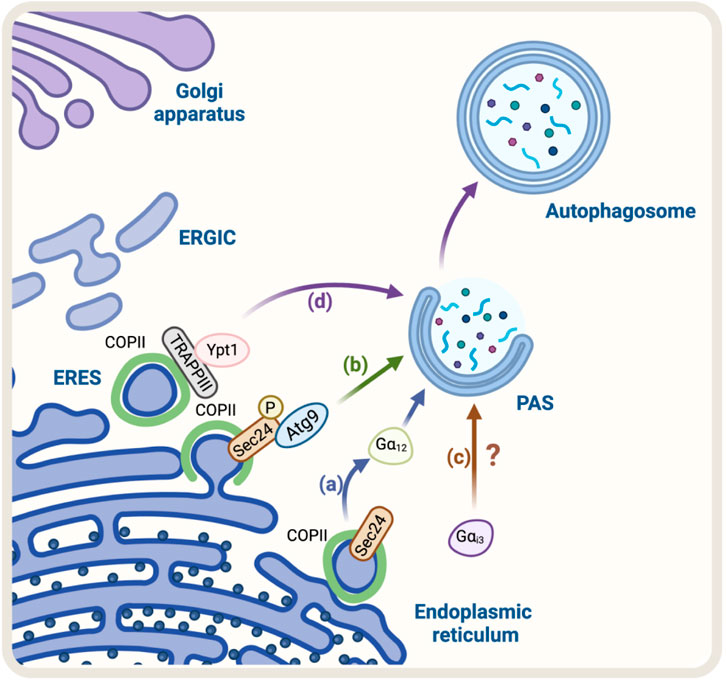
FIGURE 1. Schematic representation of the Regulators of ER-Golgi anterograde pathway and autophagy. (A) SEC24, a COPII protein, acts as a GEF activating the trimeric Gα12 GTPase to promote cargo export at the ERES. Gα12 activation is linked to autophagosome biogenesis. (B) Casein kinase 1 (Hrr25 in yeast) phosphorylates SEC24, promoting its interaction with Atg9 and autophagosome formation (C) Gαi3 regulates the initial steps of autophagosome formation. Still, the mechanism remains unknown (D) TRAPPIII, a transport complex that binds to the COPII complex, acts as a GEF for Rab GTPase Ypt1 (the yeast homolog of Rab1 in mammals). Ypt1 modulates PAS formation.
In addition, another protein mutually implicated is the transport protein particle complex (TRAPP), a tethering of COPII vesicles at the ER surface (Sacher et al., 2008). Mammalian cells have two forms of TRAPP complexes: TRAPPII and TRAPPIII (Yamasaki et al., 2009; Bassik et al., 2013; Wang et al., 2013). TRAPPIII plays a key role in the targeting and/or fusion of ER-to-Golgi transport vesicles with their acceptor compartment (Sacher et al., 1998; Scrivens et al., 2011). TRAPPIII, specifically its TRAPPC12 subunit, interacts with SEC13/SEC31, modulating positively the SEC13/SEC31 tetramer assembly in COPII-positive vesicles (Zhao et al., 2017). TRAPPC12 localizes to ERES and its deletion causes ER-to-Golgi transport delay (Zhao et al., 2017). Moreover, it has been proposed that TRAPPIII acts as a GEF of the Rab GTPase Ypt1, which is involved in the membrane tethering during the formation of the PAS. Ablation of Bet1, a component of TRAPPIII, can disrupt autophagy (Tan et al., 2013). These findings confirm that COPII and TRAPPIII complexes are also involved in autophagosome formation (Figure 1D).
ERES plays a role in autophagosome formation by mediating contact sites with ERGIC through TMED9 and SEC12 (Li et al., 2021). ERGIC-ERES contact promotes the formation of the COPII vesicle, a precursor of the autophagosome derived from ERGIC. This contact differs both physically and functionally from the TFG-mediated ERGIC-ERES interaction involved in the early secretory pathway (Witte et al., 2011; Johnson et al., 2015; Hanna et al., 2017). TMED9 RNAi, which inhibits SEC12 recruitment and autophagosomes formation, does not affect protein secretion (Li et al., 2022). This provides a mechanistic insight by which ERES can switch between autophagosomes formation and ER-to-Golgi cargo transport.
TMEM39A is an ER-localized transmembrane protein that regulates autophagy by affecting the distribution and levels of PI4P. It interacts with the ER-localized PI4P phosphatase SAC1, an integral membrane protein that cycles between the ER and Golgi (Del Bel and Brill, 2018), and promotes its ER-to-Golgi transport via COPII SEC23/SEC24 subunits. The knock-down of TMEM39A leads to SAC1 retention in the ER, resulting in increased autophagosome formation (Miao et al., 2020).
Altogether, these findings provide strong evidence for the existence of a close functional and interorganelle coordination between the conventional early secretory pathway and autophagy, which requires further investigation in mammals.
The Golgi-to-ER retrograde early secretory pathway has been shown to play a role in regulating autophagy (Tapia et al., 2019; Bustamante et al., 2020). In this context, a crucial protein complex implicated in this retrograde pathway is the coat protein complex COPI, responsible for forming a coat around transport vesicles, which helps to protect the contents of the vesicles during transport and directs them to specific destinations. This complex consists of seven core subunits α-COP, β′-COP, ε-COP, β-COP, δ-COP, γ-COP, and ζ-COP, the coatomer. The heptameric cytosolic coatomer is recruited to the Golgi membrane by the small GTPase Arf1 together with other Arf family members to form the COPI coat (Hara-Kuge et al., 1994; Popoff et al., 2011). Additionally, COPI has been linked with the autophagy pathway (Karanasios et al., 2016). In fact, under starvation conditions, a pool of COPI is relocated to the ER-Golgi intermediate compartment (ERGIC), a subcompartment of the ER located between the cis-Golgi and the medial-Golgi, which is thought to play a role in regulating autophagy (Razi et al., 2009; Karanasios et al., 2016). In this context, during starvation, a pool of COPI colocalizes with ERGIC53, a marker of the ERGIC compartment (Razi et al., 2009). Additionally, BFA, a fungal metabolite that blocks COPI-dependent trafficking (Lippincott-Schwartz et al., 1989), decreases autophagy induced by amino-acid starvation (Karanasios et al., 2016). When cells are treated with BFA, it interferes with the recruitment of Atg13 to autophagosomes, a component of the ULK1 complex required for the initiation of autophagy, where its recruitment to autophagosomes is necessary for the proper initiation of autophagy (Karanasios et al., 2016) (Figure 2A).
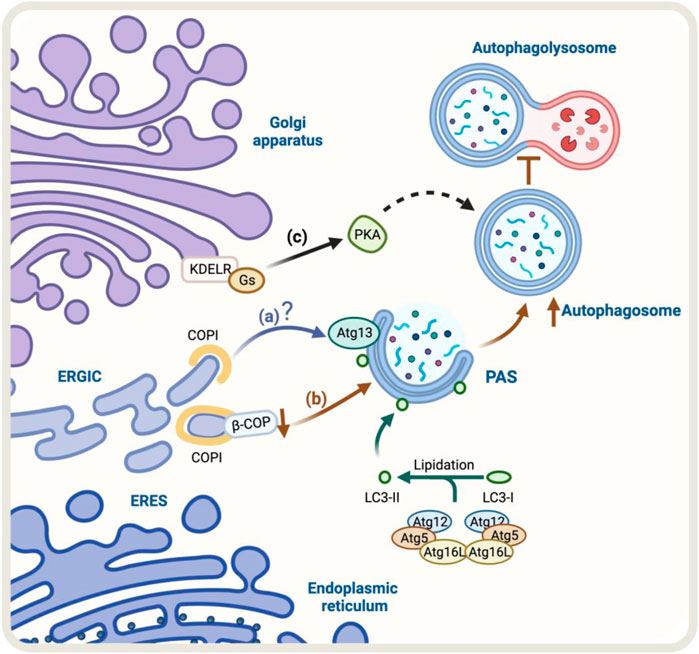
FIGURE 2. Schematic representation of the regulators of Golgi-to-ER retrograde pathway and autophagy. (A) Under starvation conditions, COPI localizes at the ERGIC. COPI-mediated trafficking may recruit Atg13, however, the molecular mechanism is still unclear (blue arrow). (B) The knock-down of β-COP, a protein of the COPI complex, results in an accumulation of autophagosomal structures. Still, it also partially hinders the fusion of the autophagosome with the lysosome and the degradation and recycling process (brown arrows) (C) KDEL/Gs signaling pathway, which includes PKA activation, may be involved in the upregulation of autophagy (black arrows). The green arrow represents the process of lipidation of LC3 protein by Atgs proteins that is necessary for substrate recognition and autophagosome formation.
The lipidation of LC3 to form LC3-II is an obligatory step for the expansion of the autophagosomal membrane (Tanida et al., 2008), a stage in which COPI participates (Karanasios et al., 2016). In contrast, COPI is not necessary for the steps upstream of LC3 lipidation, such as nucleation (Ge et al., 2013). When COPI function is lost, it leads to fragmentation of the Golgi apparatus, accumulation of immature autophagosomes, and a decrease in the overall process of autophagy (Shtutman and Roninson, 2011). Moreover, the reduction or elimination of specific components of COPI, such as β-COP, leads to an increase in the formation of autophagosomes. However, this also hinders the fusion of these autophagosomes with lysosomes and the subsequent degradation and recycling process (Claerhout et al., 2012) (Figure 2B). Although the COPI complex is required for autophagy, the precise role of COPI in this pathway is not yet fully understood, and further research is needed to determine its specific mechanisms of involvement in this process.
The Golgi apparatus can maintain and recover its normal composition and morphology upon cellular stress, such as changes in transport rates to and from Golgi (Mironov et al., 2001; Trucco et al., 2004). In this regard, the Golgi reassembly stacking protein of 55 kDa (GRASP55) is required for Golgi cisternae stacking, ribbon linking, and mitotic fragmentation (Shorter et al., 1999; Duran et al., 2008; Feinstein and Linstedt, 2008; Xiang and Wang, 2010). Although GRASP55 is viewed as a Golgi protein, it is also found at the ER-Golgi interface (Piao et al., 2017). GRASP55 is not strictly required for conventional protein secretion (Duran et al., 2008; Feinstein and Linstedt, 2008) but GRASP55 knock-down increases protein secretion (Xiang et al., 2013). On the other hand, GRASP55 knockdown and CRISPR-Cas9 deletion in HEK293T cells enhance LC3 puncta formation, indicating that GRASP55, required for ER-Golgi organization, restricts autophagosome formation (Liu et al., 2021).
One homeostatic mechanism that has been reported is based on the signaling properties of the KDEL receptor (KDELR) (Pulvirenti et al., 2008; Giannotta et al., 2012; Cancino et al., 2014). The KDELR shares functional properties with G protein-coupled receptors (Giannotta et al., 2012; Solis et al., 2017) and regulates transport out from the Golgi (Pulvirenti et al., 2008; Giannotta et al., 2012; Cancino et al., 2014). KDELR binds to and recycles chaperones that escape the ER to the Golgi using the Golgi-to-ER retrograde pathway in yeast (Semenza et al., 1990) and mammals (Lewis and Pelham, 1990). In this context, KDELR signaling in the early conventional secretory pathway controls trafficking through downstream signaling pathways that involve two heterotrimeric Gα proteins, Gs and Gq (Giannotta et al., 2012; Cancino et al., 2014). The activation of the KDELR/Gs signaling pathway plays an important role in maintaining the homeostasis of the Golgi apparatus. This signaling cascade involves the activation of protein kinase A (PKA) which phosphorylates the COPI subunits, increasing the dynamics of the COPI coat complex (Cancino et al., 2014). The activation of the KDELR/Gs signaling pathway has been shown to increase autophagy, by increasing the formation of autophagosomes and their fusion with lysosomes. This is a new function for KDELR (Tapia et al., 2019) (Figure 2C). However, it is not yet clear whether the PKA-dependent COPI phosphorylation is required for this response to take place, more research is needed to understand the underlying mechanism. Despite this, studies have shown that the KDELR pathway plays a role in promoting autophagy under stress conditions, making it a new and exciting area of research that might lead to a better understanding and regulation of the autophagic process. The accumulation of misfolded proteins related to neurodegenerative diseases increases KDELR mRNA. Additionally, overexpression of KDELR, but not the signaling-defective KDELR-D193N mutant, has been shown to induce autophagy to promote the clearance of protein associated with diseases, such as superoxide dismutase (SOD1), Parkinson’s disease-associated A53T alpha-synuclein, and Huntington’s disease-related expanded huntingtin (Wang et al., 2011). This response is not controlled by, but rather by the mitogen extracellular kinase 1 (MEK1). Therefore, this KDELR pathway seems to involve more than one signaling pathway.
In addition to COPI and KDELR-dependent signaling, another protein that regulates Golgi-to-ER retrograde pathway is PSMD14, a subunit of the 19S regulatory particle of the proteasome with deubiquitinating (DUB) enzyme activity (Bustamante et al., 2020). Importantly, pharmacological inhibition of PSMD14 with Capzimin (CZM) has been shown to act as a potent blocker of autophagy by a mechanism related to the decay in the Golgi-to-ER retrograde transport. Additionally, research suggests that PSMD14’s DUB activity plays non-canonical roles not coupled to the translocation of substrates into the core of the 20S proteasome (Bustamante et al., 2023). This opens the possibility that during stress conditions, the upregulation of autophagy may require an active Golgi-to-ER retrograde transport. Further studies such as kinetic studies to explore the speed of vesicle transport labeled with specific cargoes of the Golgi-to-ER retrograde transport could provide more insight into the role of PSMD14 in this process.
Autophagy adds another level of communication that includes its interplay with the conventional secretory pathway, and in recent years, several new findings confirm these functional connections. Upon starvation, the de novo formation of autophagosomes is upregulated and is driven by conserved machinery composed mostly of cytosolic complexes that are sequentially and hierarchically recruited to the ER (Ohsumi, 2001; Suzuki et al., 2007). Autophagy is a complex multi-step process that involves various proteins and molecular machinery. It is regulated by cytosolic proteins that control the initiation of autophagosome formation, such as the Ser/Thr kinase Unc-51-like kinase-1 complex (ULK1; Atg1-complex in yeasts) (Scott et al., 1996; Neufeld, 2007), the transmembrane protein Atg9 which is required for phagophore elongation (Zhou et al., 2017), and the class III PI 3-kinase complexes C1 and C2 (Ma et al., 2017). These complexes are differentiated based on the composition of their subunits and their specific functions. The PI3K3C-C1 complex, which includes the catalytic subunit Vps34 and the regulatory subunit Beclin-1, plays a crucial role in the initiation of autophagosome formation by inducing the production of phosphatidylinositol 3-phosphate (PtdIns3P) (Axe et al., 2008) that recruits other necessary proteins to the site of PAS formation such as WIPI 1-4, Atg8/LC3 family, and the Atg12 system (Simonsen and Tooze, 2009; Itakura and Mizushima, 2010). The PI3K3C-C2 complex, consisting of the catalytic subunit Vps34, the regulatory subunit Beclin-1, and the UV radiation resistance-associated gene protein (UVRAG), is involved in the maturation of autophagosomes. The inclusion of UVRAG instead of Atg14 in the PI3K3C-C1 complex leads to the formation of PI3K3C-C2, which contributes to autophagosome maturation (Liang et al., 2008). Moreover, growing evidence indicates that a functional integration between the early conventional secretory pathway and autophagy plays an important role in maintaining cellular homeostasis, sharing important molecular players (Lamb et al., 2013). Interestingly, the connection between these pathways is not limited to shared molecular players; structural and regulatory autophagy proteins also depend on membrane transport to function effectively (Mari et al., 2010; Kakuta et al., 2012; Orsi et al., 2012; Imai et al., 2016; Lamb et al., 2016; Lamb et al., 2017; Zhou et al., 2017)
The formation of autophagosomes begins at multiple PtdIns3P-enriched cup-like subdomains of the ER, a structure called PAS (Axe et al., 2008). Studies have demonstrated that COPII components are recruited to the ERGIC in a phosphatidylinositol kinase class III (PI3K)-dependent manner during autophagy (Ge et al., 2014). Concerning this, UVRAG is a PtdIns3P-binding protein. During autophagy induction, UVRAG coordinates Golgi-to-ER retrograde transport including the cargo ATG9A, by differential interactions with the ER tether and the BECLIN-1 complex (He et al., 2013) (Figure 3A). At the ER, UVRAG interacts with RINT-1 (Golgi-ER membrane traffic regulator); this way engages phosphoinositide metabolism to COPI-vesicle tethering, hence regulating Golgi-to-ER retrograde transport. Moreover, during autophagy induction, UVRAG dissociates from the ER tether, triggering ATG9A translocation for autophagosome formation (He et al., 2013) (Figure 3B).

FIGURE 3. Illustration of the regulators of autophagy pathway controlling the early conventional secretory pathway. (A) UVRAG binds to the PtdIns3P-enriched membranes at the ER where it interacts with RINT-1. During autophagy induction, UVRAG dissociates from the ER to become part of the complex PI3K3C-C2 (for simplicity, only the Beclin-1, Atg14, and UVRAG proteins are shown). The dissociation of UVRAG from the ER induces the translocation of the Atg9 vesicles, necessary to promote autophagosome formation (brown arrows) (B) The ULK1 complex can phosphorylate Beclin-1 inducing autophagy (purple arrow). (C) Additionally, ULK1 phosphorylates SEC16A promoting anterograde transport (blue arrow).
ULK1/2 is a serine/threonine protein kinase that regulates autophagy (Jung et al., 2009; Joo et al., 2016), a kinase that also has been shown to play a role in ER-to-Golgi anterograde pathway (Joo et al., 2016). By proteomics approach, SEC16A (molecular scaffold which participates in the organization of ERES), was identified as a ULK1/2 interaction partner. ULK1 activity regulates the formation of COPII vesicles in vitro. In vivo, ULK1-phosphorylates SEC16A regulates the assembly of ERES and, consequently, ER-to-Golgi anterograde transport (Sprangers and Rabouille, 2015) and does not require other autophagy proteins (Joo et al., 2016). ULK1 can also phosphorylate SEC23, an essential protein of the COPII complex, phosphorylation that alters the morphology of ERES reducing global cellular secretion (Gan et al., 2017) (Figure 3C).
While inhibition of COPII transport impairs starvation-induced autophagy (Zoppino et al., 2010; Graef et al., 2013), the role of ULK1/2-mediated phosphorylation of SEC16A in autophagy is unknown. Because SEC16A is a target of several kinases (Farhan et al., 2010; Zacharogianni et al., 2011; Tillmann et al., 2015), its posttranslational modification may provide a means of adjusting the regulation of COPII transport with the availability of nutrients (Farhan et al., 2017).
Collectively, these findings demonstrated that autophagy and membrane transport systems are closely related, mutually dependent, and functionally coordinated. However, the extent of this functional integration is poorly understood and will require deep investigation to establish its principles and molecular architecture.
The early conventional secretory pathway changes for cell survival in response to stress to maintain homeostasis and prevent damage to the cell. When the cell experiences stress, it triggers signaling pathways that alter the function of the secretory pathway, including the ER and the Golgi apparatus. These changes can include changes in protein folding and trafficking, as well as the activation of autophagy. Ultimately, these changes in the early secretory pathway aim to restore cellular balance and promote adaptation mechanisms for cell survival (Ding et al., 2007; Kroemer et al., 2010; Murrow and Debnath, 2013).
A good example of these cellular adaptations is the ER-phagy process that occurs under ER stress (Song et al., 2018). The ER-phagy process is a cellular adaptation that occurs under ER stress and involves the degradation of the endoplasmic reticulum (ER) by autophagy. In yeast, some components like Lst1/SEC24C-SEC23 that belong to the COPII complex play a role in ER degradation through ER-phagy. This is because they can interact with Atg40, a specific ER-phagy receptor, which binds to Atg8/LC3 (Kercsmar et al., 1988; Cui et al., 2019) (Figure 4A). Under stress, the COPII machinery may decrease to conserve energy, while autophagy may increase to recycle cellular components and provide energy. Altogether, the increase in Golgi-to-ER retrograde transport and formation of autophagosomes indicate strong communication between the conventional secretory pathway and autophagy during ER stress.
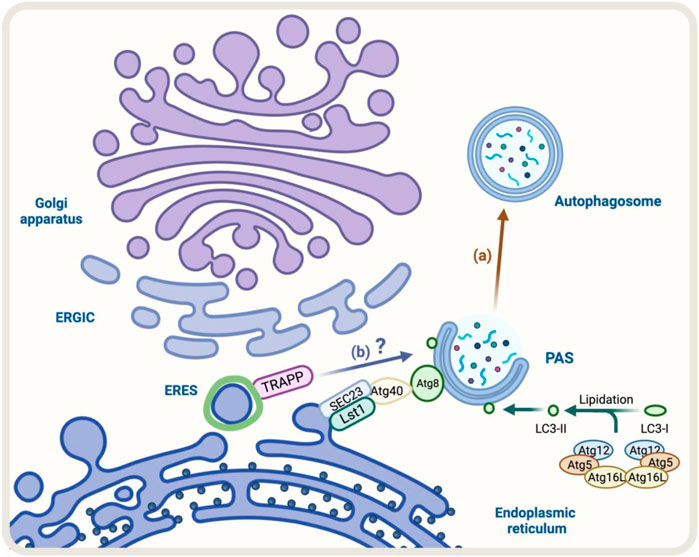
FIGURE 4. Diagram representation of the regulators of autophagy and early conventional secretory pathway under stress. (A) During ER-phagy, components of COPII complex such as Lst1/SEC24C-SEC23 can interact with Atg40, a specific ER-phagy receptor, which binds to Atg8/LC3 allowing the degradation of the ER by autophagy (brown arrow) (B) TRAPP components respond to Golgi stress with impact on autophagy, however, the molecular mechanism is not fully resolved (blue arrow).
The impact of osmotic stress on the early conventional secretory pathway has been studied. The data show that in a hypertonic treatment, the ER to Golgi transport is inhibited (Docherty and Snider, 1991). Conversely, a hypotonic treatment promotes the tubulation of the Golgi membranes (Lee and Linstedt, 1999). Similarly, hyperosmotic conditions induce the fragmentation and redistribution of the Golgi back to the ER, as seen with Brefeldin A (BFA) treatment (Lippincott-Schwartz et al., 1990; Sciaky et al., 1997; Lee and Linstedt, 1999). In contrast, hypo-osmotic conditions result in a reduction in the number of export sites on the ER, leading to less trafficking from the ER to the Golgi apparatus (Lee and Linstedt, 1999).
Regulation of intracellular trafficking involves Rab proteins, which are small GTPases that control the formation and movement of vesicles between intracellular compartments (Zerial and McBride, 2001; Grosshans et al., 2006). Rab1 and Rab6 have been found to play a role in the anterograde and retrograde traffic between the ER and Golgi apparatus, respectively (Ortiz Sandoval and Simmen, 2012). In yeast, Rab1 has been linked to the regulation of the UPR (Tsvetanova et al., 2012). However, in hippocampal cells during UPR, the levels of Rab1 decrease along with the cell viability. In contrast, Rab6 and UPR-associated protein increase in levels, suggesting a connection between vesicle trafficking and ER homeostasis. In the context of Alzheimer’s disease, ER stress causes neuronal death. The stress signals propagate from the ER to Golgi, affecting the transport and processing of AD-related proteins such as β-amyloid precursor protein (Suga et al., 2009). Regarding this, inhibition of the Golgi-to-ER retrograde pathway by silencing PSMD14 leads to a dramatic increase in the levels of the β-amyloid precursor protein (Bustamante et al., 2020). Additionally, ER stress increases the expression of the ER-Golgi SNARE protein Syntaxin5 (Syx5), which is crucial for the fusion of vesicles with early stations in the conventional secretory pathway, leading to a reduction in the secretion of amyloid β peptide (Suga et al., 2015). In this context, in the primary culture of rat hippocampal neurons, ER stress leads to an increase in the expression of Syx5 and Bet1 (another SNARE protein complex involved in ER-to-Golgi transport) through the novo synthesis (Donkervoort et al., 2021). The impact of these changes in SNARE proteins on autophagy during stress conditions is still unknown, but it has been suggested as a possibility. In yeast, Ufe1, a Qa/t-SNARE localized in the ER, participates in autophagy. During starvation, it is exported from the ER to intracellular locations containing autophagy markers Atg8 and Atg9, where it interacts with non-ER SNARE involved in autophagosome formation (Lemus et al., 2016). Data suggests that Ufe1 reaches autophagosome formation sites via specific COPII vesicles, connecting ER stress changes to autophagy.
Another factor that responds to cellular stress is TRAPPC13, a subunit of the TRAPP complex involved in ER-to-Golgi and intra-Golgi transport (Barrowman et al., 2010). During Golgi stress induced by BFA, the absence of TRAPPC13 protects cells from death caused by BFA treatment and is correlated with reduced autophagy, suggesting TRAPPC13’s role in autophagy and autophagy’s role in cell death due to prolonged Golgi stress (Ramirez-Peinado et al., 2017) (Figure 4B). Other TRAPP complex subunits (TRAPPC12, TRAPPC11, TRAPPC9, and TRAPPC3) also behave similarly to TRAPPC13 against BFA-induced toxicity, indicating multiple TRAPP complex components play a role in the Golgi stress response and autophagy (Barrowman et al., 2010).
STING1, a transmembrane protein that cycles between Golgi and ER via a COPI-dependent mechanism, is another example (Burdette et al., 2011; Mukai et al., 2021). During stress from pathogen infection (bacteria and viruses), STING1 positively regulates autophagy (Watson et al., 2012; Collins et al., 2015) and is reported to interact and co-localize with LC3 and ATG9 (Saitoh et al., 2009). These findings suggest different stress conditions can alter the normal functioning of early secretory pathway proteins, which in turn could directly influence the normal development of autophagy. An excellent example is the UVRAG-RINT-1 interaction. Under non-stressful scenarios, these proteins play an important role in Golgi-to-ER retrograde transport (He et al., 2013). However, under stress by nutritional privation, UVRAG dissociates from RINT-1, instead interacting with Beclin-1 to promote autophagosome biogenesis (He et al., 2013). Intriguingly, lack of beclin-1 can result in an enlarged Golgi apparatus with higher levels of PI4P and proteins that normally traffics in the Golgi-to-ER retrograde transport, such as β′-COP (Bork et al., 2022), suggesting beclin-1 is also a key protein in the early conventional secretory pathway.
Despite our understanding of these cellular adaptations, there is still a need for further research to uncover the full extent of changes that occur under various stress conditions. This will help us gain a deeper understanding of the mechanisms involved and develop more effective treatments in the future to treat diseases.
To understand cell survival strategies, particularly under stress, it's crucial to unlocking the connection between early conventional secretion and autophagy. The relationship between these pathways has important implications for cellular metabolism and helps us understand how cells adapt and survive in changing conditions. A comprehensive view of organelles such as lipid droplets, mitochondria, and lysosomes will enhance our understanding of the interactions between these pathways and their response to stressors. It’s crucial to expand our understanding of how cells respond to lipid imbalance and fatty acid (FAs) overload, given the recent global dietary changes. Cells respond to high energy demands by moving fatty acids to mitochondria for ATP production. As a result, intracellular membranes create MCSs to coordinate inter-organelle communication, enabling the cellular plasticity events needed for adaptation and survival. However, regulation of MCSs during protein secretion and the roles of autophagy and membrane transport machinery is still unknown. Gaining an understanding of the crosstalk between pathways involved in intracellular membrane networks will shed light on how cells coordinate stress responses, especially during periods of high protein secretion demand. The discovery of TIDeRS (traffic-induced degradation response for secretion) highlights the significance of inter-organelle coordination under different nutrient conditions, with a focus on the connection between lipid droplets and protein secretion. Finally, the mutual regulation between ER-Golgi transport, Golgi-to-ER retrograde pathway, autophagy, and its impact on each other raises many questions and offers future opportunities for developing therapeutic interventions in biomedical fields.
DT wrote the manuscript and prepared tables, VC wrote the manuscript and prepared figures, and PB and JC wrote and revised the manuscript. All authors read and approved the final manuscript.
This work was supported by grants from the Agencia Nacional de Investigación y Desarrollo de Chile (ANID), FONDECYT 1221374 (JC) and 1211261 (PB), Centro Ciencia & Vida FB210008 (PB), FONDECYT 3220485 (VC) and VRID fellowship from Universidad San Sebastián (DT).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
ER, endoplasmic reticulum; ERES, ER exit sites; ER Golgi intermediate 19 compartment, ERGIC; BFA, Brefeldin A; KDELR, KDEL receptor; PAS, pre-autophagosomal 20 structure; LD, lipid droplets; PKA, protein kinase A; PM, plasma membrane; TIDeRS, traffic-21 induced degradation response for secretion.
Albornoz, N., Bustamante, H., Soza, A., and Burgos, P. (2019). Cellular responses to proteasome inhibition: Molecular mechanisms and beyond. Int. J. Mol. Sci. 20, 3379. doi:10.3390/ijms20143379
Axe, E. L., Walker, S. A., Manifava, M., Chandra, P., Roderick, H. L., Habermann, A., et al. (2008). Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 182, 685–701. doi:10.1083/jcb.200803137
Barlowe, C. K., and Miller, E. A. (2013). Secretory protein biogenesis and traffic in the early secretory pathway. Genetics 193, 383–410. doi:10.1534/genetics.112.142810
Barlowe, C., Orci, L., Yeung, T., Hosobuchi, M., Hamamoto, S., Salama, N., et al. (1994). COPII: A membrane coat formed by sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell 77, 895–907. doi:10.1016/0092-8674(94)90138-4
Barrowman, J., Bhandari, D., Reinisch, K., and Ferro-Novick, S. (2010). TRAPP complexes in membrane traffic: Convergence through a common Rab. Nat. Rev. Mol. Cell Biol. 11, 759–763. doi:10.1038/nrm2999
Bassik, M. C., Kampmann, M., Lebbink, R. J., Wang, S., Hein, M. Y., Poser, I., et al. (2013). A systematic mammalian genetic interaction map reveals pathways underlying ricin susceptibility. Cell 152, 909–922. doi:10.1016/j.cell.2013.01.030
Behrends, C., Sowa, M. E., Gygi, S. P., and Harper, J. W. (2010). Network organization of the human autophagy system. Nature 466, 68–76. doi:10.1038/nature09204
Bethune, J., and Wieland, F. T. (2018). Assembly of COPI and COPII vesicular coat proteins on membranes. Annu. Rev. Biophys. 47, 63–83. doi:10.1146/annurev-biophys-070317-033259
Bisnett, B. J., Condon, B. M., Lamb, C. H., Georgiou, G. R., and Boyce, M. (2020). Export control: Post-transcriptional regulation of the COPII trafficking pathway. Front. Cell Dev. Biol. 8, 618652. doi:10.3389/fcell.2020.618652
Bork, T., Liang, W., Kretz, O., Lagies, S., Yamahara, K., Hernando-Erhard, C., et al. (2022). BECLIN1 is essential for podocyte secretory pathways mediating VEGF secretion and podocyte-endothelial crosstalk. Int. J. Mol. Sci. 23, 3825. doi:10.3390/ijms23073825
Burdette, D. L., Monroe, K. M., Sotelo-Troha, K., Iwig, J. S., Eckert, B., Hyodo, M., et al. (2011). STING is a direct innate immune sensor of cyclic di-GMP. Nature 478, 515–518. doi:10.1038/nature10429
Bustamante, H. A., Albornoz, N., Morselli, E., Soza, A., and Burgos, P. V. (2023). Novel insights into the non-canonical roles of PSMD14/POH1/Rpn11 in proteostasis and in the modulation of cancer progression. Cell Signal 101, 110490. doi:10.1016/j.cellsig.2022.110490
Bustamante, H. A., Cereceda, K., Gonzalez, A. E., Valenzuela, G. E., Cheuquemilla, Y., Hernandez, S., et al. (2020). The proteasomal deubiquitinating enzyme PSMD14 regulates macroautophagy by controlling golgi-to-ER retrograde transport. Cells 9, 777. doi:10.3390/cells9030777
Cancino, J., Capalbo, A., Di Campli, A., Giannotta, M., Rizzo, R., Jung, J. E., et al. (2014). Control systems of membrane transport at the interface between the endoplasmic reticulum and the Golgi. Dev. Cell 30, 280–294. doi:10.1016/j.devcel.2014.06.018
Claerhout, S., Dutta, B., Bossuyt, W., Zhang, F., Nguyen-Charles, C., Dennison, J. B., et al. (2012). Abortive autophagy induces endoplasmic reticulum stress and cell death in cancer cells. PLoS One 7, e39400. doi:10.1371/journal.pone.0039400
Collins, A. C., Cai, H., Li, T., Franco, L. H., Li, X. D., Nair, V. R., et al. (2015). Cyclic GMP-AMP synthase is an innate immune DNA sensor for Mycobacterium tuberculosis. Cell Host Microbe 17, 820–828. doi:10.1016/j.chom.2015.05.005
Cui, Y., Parashar, S., Zahoor, M., Needham, P. G., Mari, M., Zhu, M., et al. (2019). A COPII subunit acts with an autophagy receptor to target endoplasmic reticulum for degradation. Science 365, 53–60. doi:10.1126/science.aau9263
Davis, S., Wang, J., Zhu, M., Stahmer, K., Lakshminarayan, R., Ghassemian, M., et al. (2016). Sec24 phosphorylation regulates autophagosome abundance during nutrient deprivation. Elife 5, e21167. doi:10.7554/eLife.21167
De Matteis, M. A., Di Campli, A., and Godi, A. (2005). The role of the phosphoinositides at the Golgi complex. Biochim. Biophys. Acta 1744, 396–405. doi:10.1016/j.bbamcr.2005.04.013
Del Bel, L. M., and Brill, J. A. (2018). Sac1, a lipid phosphatase at the interface of vesicular and nonvesicular transport. Traffic 19, 301–318. doi:10.1111/tra.12554
Ding, W. X., Ni, H. M., Gao, W., Hou, Y. F., Melan, M. A., Chen, X., et al. (2007). Differential effects of endoplasmic reticulum stress-induced autophagy on cell survival. J. Biol. Chem. 282, 4702–4710. doi:10.1074/jbc.M609267200
Docherty, P. A., and Snider, M. D. (1991). Effects of hypertonic and sodium-free medium on transport of a membrane glycoprotein along the secretory pathway in cultured mammalian cells. J. Cell Physiol. 146, 34–42. doi:10.1002/jcp.1041460106
Donkervoort, S., Krause, N., Dergai, M., Yun, P., Koliwer, J., Gorokhova, S., et al. (2021). BET1 variants establish impaired vesicular transport as a cause for muscular dystrophy with epilepsy. EMBO Mol. Med. 13, e13787. doi:10.15252/emmm.202013787
Duran, J. M., Kinseth, M., Bossard, C., Rose, D. W., Polishchuk, R., Wu, C. C., et al. (2008). The role of GRASP55 in Golgi fragmentation and entry of cells into mitosis. Mol. Biol. Cell 19, 2579–2587. doi:10.1091/mbc.e07-10-0998
Farhan, H., Kundu, M., and Ferro-Novick, S. (2017). The link between autophagy and secretion: A story of multitasking proteins. Mol. Biol. Cell 28, 1161–1164. doi:10.1091/mbc.E16-11-0762
Farhan, H., Weiss, M., Tani, K., Kaufman, R. J., and Hauri, H. P. (2008). Adaptation of endoplasmic reticulum exit sites to acute and chronic increases in cargo load. EMBO J. 27, 2043–2054. doi:10.1038/emboj.2008.136
Farhan, H., Wendeler, M. W., Mitrovic, S., Fava, E., Silberberg, Y., Sharan, R., et al. (2010). MAPK signaling to the early secretory pathway revealed by kinase/phosphatase functional screening. J. Cell Biol. 189, 997–1011. doi:10.1083/jcb.200912082
Feinstein, T. N., and Linstedt, A. D. (2008). GRASP55 regulates Golgi ribbon formation. Mol. Biol. Cell 19, 2696–2707. doi:10.1091/mbc.e07-11-1200
Gan, W., Zhang, C., Siu, K. Y., Satoh, A., Tanner, J. A., and Yu, S. (2017). ULK1 phosphorylates Sec23A and mediates autophagy-induced inhibition of ER-to-Golgi traffic. BMC Cell Biol. 18, 22. doi:10.1186/s12860-017-0138-8
Ge, L., Melville, D., Zhang, M., and Schekman, R. (2013). The ER-Golgi intermediate compartment is a key membrane source for the LC3 lipidation step of autophagosome biogenesis. Elife 2, e00947. doi:10.7554/eLife.00947
Ge, L., Zhang, M., and Schekman, R. (2014). Phosphatidylinositol 3-kinase and COPII generate LC3 lipidation vesicles from the ER-Golgi intermediate compartment. Elife 3, e04135. doi:10.7554/eLife.04135
Giannotta, M., Ruggiero, C., Grossi, M., Cancino, J., Capitani, M., Pulvirenti, T., et al. (2012). The KDEL receptor couples to Gαq/11 to activate Src kinases and regulate transport through the Golgi. EMBO J. 31, 2869–2881. doi:10.1038/emboj.2012.134
Gohla, A., Klement, K., Piekorz, R. P., Pexa, K., Vom Dahl, S., Spicher, K., et al. (2007). An obligatory requirement for the heterotrimeric G protein Gi3 in the antiautophagic action of insulin in the liver. Proc. Natl. Acad. Sci. U. S. A. 104, 3003–3008. doi:10.1073/pnas.0611434104
Graef, M., Friedman, J. R., Graham, C., Babu, M., and Nunnari, J. (2013). ER exit sites are physical and functional core autophagosome biogenesis components. Mol. Biol. Cell 24, 2918–2931. doi:10.1091/mbc.E13-07-0381
Grosshans, B. L., Ortiz, D., and Novick, P. (2006). Rabs and their effectors: Achieving specificity in membrane traffic. Proc. Natl. Acad. Sci. U. S. A. 103, 11821–11827. doi:10.1073/pnas.0601617103
Gubas, A., and Dikic, I. (2022). ER remodeling via ER-phagy. Mol. Cell 82, 1492–1500. doi:10.1016/j.molcel.2022.02.018
Hanna, M. G., Peotter, J. L., Frankel, E. B., and Audhya, A. (2018). Membrane transport at an organelle interface in the early secretory pathway: Take your coat off and stay a while: Evolution of the metazoan early secretory pathway. Bioessays 40, e1800004. doi:10.1002/bies.201800004
Hanna, M. G. T., Block, S., Frankel, E. B., Hou, F., Johnson, A., Yuan, L., et al. (2017). TFG facilitates outer coat disassembly on COPII transport carriers to promote tethering and fusion with ER-Golgi intermediate compartments. Proc. Natl. Acad. Sci. U. S. A. 114, E7707–E7716. doi:10.1073/pnas.1709120114
Hara-Kuge, S., Kuge, O., Orci, L., Amherdt, M., Ravazzola, M., Wieland, F. T., et al. (1994). En bloc incorporation of coatomer subunits during the assembly of COP-coated vesicles. J. Cell Biol. 124, 883–892. doi:10.1083/jcb.124.6.883
Harada, K., Kotani, T., Kirisako, H., Sakoh-Nakatogawa, M., Oikawa, Y., Kimura, Y., et al. (2019). Two distinct mechanisms target the autophagy-related E3 complex to the pre-autophagosomal structure. Elife 8, e43088. doi:10.7554/eLife.43088
He, S., Ni, D., Ma, B., Lee, J. H., Zhang, T., Ghozalli, I., et al. (2013). PtdIns(3)P-bound UVRAG coordinates Golgi-ER retrograde and Atg9 transport by differential interactions with the ER tether and the beclin 1 complex. Nat. Cell Biol. 15, 1206–1219. doi:10.1038/ncb2848
Imai, K., Hao, F., Fujita, N., Tsuji, Y., Oe, Y., Araki, Y., et al. (2016). Atg9A trafficking through the recycling endosomes is required for autophagosome formation. J. Cell Sci. 129, 3781–3791. doi:10.1242/jcs.196196
Ishihara, N., Hamasaki, M., Yokota, S., Suzuki, K., Kamada, Y., Kihara, A., et al. (2001). Autophagosome requires specific early Sec proteins for its formation and NSF/SNARE for vacuolar fusion. Mol. Biol. Cell 12, 3690–3702. doi:10.1091/mbc.12.11.3690
Itakura, E., and Mizushima, N. (2010). Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy 6, 764–776. doi:10.4161/auto.6.6.12709
Johnson, A., Bhattacharya, N., Hanna, M., Pennington, J. G., Schuh, A. L., Wang, L., et al. (2015). TFG clusters COPII-coated transport carriers and promotes early secretory pathway organization. EMBO J. 34, 811–827. doi:10.15252/embj.201489032
Joo, J. H., Wang, B., Frankel, E., Ge, L., Xu, L., Iyengar, R., et al. (2016). The noncanonical role of ULK/ATG1 in ER-to-golgi trafficking is essential for cellular homeostasis. Mol. Cell 62, 982–506. doi:10.1016/j.molcel.2016.05.030
Jung, C. H., Jun, C. B., Ro, S. H., Kim, Y. M., Otto, N. M., Cao, J., et al. (2009). ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol. Biol. Cell 20, 1992–2003. doi:10.1091/mbc.e08-12-1249
Kakuta, S., Yamamoto, H., Negishi, L., Kondo-Kakuta, C., Hayashi, N., and Ohsumi, Y. (2012). Atg9 vesicles recruit vesicle-tethering proteins Trs85 and Ypt1 to the autophagosome formation site. J. Biol. Chem. 287, 44261–44269. doi:10.1074/jbc.M112.411454
Karanasios, E., Walker, S. A., Okkenhaug, H., Manifava, M., Hummel, E., Zimmermann, H., et al. (2016). Autophagy initiation by ULK complex assembly on ER tubulovesicular regions marked by ATG9 vesicles. Nat. Commun. 7, 12420. doi:10.1038/ncomms12420
Kawamata, T., Kamada, Y., Kabeya, Y., Sekito, T., and Ohsumi, Y. (2008). Organization of the pre-autophagosomal structure responsible for autophagosome formation. Mol. Biol. Cell 19, 2039–2050. doi:10.1091/mbc.e07-10-1048
Kercsmar, C. M., Martin, R. J., Chatburn, R. L., and Carlo, W. A. (1988). Bronchoscopic findings in infants treated with high-frequency jet ventilation versus conventional ventilation. Pediatrics 82, 884–887. doi:10.1542/peds.82.6.884
Kroemer, G., Marino, G., and Levine, B. (2010). Autophagy and the integrated stress response. Mol. Cell 40, 280–293. doi:10.1016/j.molcel.2010.09.023
Kurokawa, K., and Nakano, A. (2019). The ER exit sites are specialized ER zones for the transport of cargo proteins from the ER to the Golgi apparatus. J. Biochem. 165, 109–114. doi:10.1093/jb/mvy080
Lamb, C. A., Joachim, J., and Tooze, S. A. (2017). Quantifying autophagic structures in mammalian cells using confocal microscopy. Methods Enzymol. 587, 21–42. doi:10.1016/bs.mie.2016.09.051
Lamb, C. A., Nuhlen, S., Judith, D., Frith, D., Snijders, A. P., Behrends, C., et al. (2016). TBC1D14 regulates autophagy via the TRAPP complex and ATG9 traffic. EMBO J. 35, 281–301. doi:10.15252/embj.201592695
Lamb, C. A., Yoshimori, T., and Tooze, S. A. (2013). The autophagosome: Origins unknown, biogenesis complex. Nat. Rev. Mol. Cell Biol. 14, 759–774. doi:10.1038/nrm3696
Lee, T. H., and Linstedt, A. D. (1999). Osmotically induced cell volume changes alter anterograde and retrograde transport, Golgi structure, and COPI dissociation. Mol. Biol. Cell 10, 1445–1462. doi:10.1091/mbc.10.5.1445
Lemus, L., Ribas, J. L., Sikorska, N., and Goder, V. (2016). An ER-localized SNARE protein is exported in specific COPII vesicles for autophagosome biogenesis. Cell Rep. 14, 1710–1722. doi:10.1016/j.celrep.2016.01.047
Lewis, M. J., and Pelham, H. R. (1990). A human homologue of the yeast HDEL receptor. Nature 348, 162–163. doi:10.1038/348162a0
Li, D., Yang, S. G., He, C. W., Zhang, Z. T., Liang, Y., Li, H., et al. (2020). Excess diacylglycerol at the endoplasmic reticulum disrupts endomembrane homeostasis and autophagy. BMC Biol. 18, 107. doi:10.1186/s12915-020-00837-w
Li, S., Yan, R., Xu, J., Zhao, S., Ma, X., Sun, Q., et al. (2022). A new type of ERGIC-ERES membrane contact mediated by TMED9 and SEC12 is required for autophagosome biogenesis. Cell Res. 32, 119–138. doi:10.1038/s41422-021-00563-0
Li, S., Zhang, M., and Ge, L. (2021). A new type of membrane contact in the ER-Golgi system regulates autophagosome biogenesis. Autophagy 17, 4499–4501. doi:10.1080/15548627.2021.1972406
Liang, C., Lee, J. S., Inn, K. S., Gack, M. U., Li, Q., Roberts, E. A., et al. (2008). Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat. Cell Biol. 10, 776–787. doi:10.1038/ncb1740
Lippincott-Schwartz, J., Donaldson, J. G., Schweizer, A., Berger, E. G., Hauri, H. P., Yuan, L. C., et al. (1990). Microtubule-dependent retrograde transport of proteins into the ER in the presence of brefeldin A suggests an ER recycling pathway. Cell 60, 821–836. doi:10.1016/0092-8674(90)90096-w
Lippincott-Schwartz, J., Yuan, L. C., Bonifacino, J. S., and Klausner, R. D. (1989). Rapid redistribution of Golgi proteins into the ER in cells treated with brefeldin A: Evidence for membrane cycling from Golgi to ER. Cell 56, 801–813. doi:10.1016/0092-8674(89)90685-5
Liu, J. Y., Lin, Y. T., Leidal, A. M., Huang, H. H., Ye, J., Wiita, A. P., et al. (2021). GRASP55 restricts early-stage autophagy and regulates spatial organization of the early secretory network. Biol. Open 10, bio058736. doi:10.1242/bio.058736
Lo, I. C., Gupta, V., Midde, K. K., Taupin, V., Lopez-Sanchez, I., Kufareva, I., et al. (2015). Activation of Gαi at the Golgi by GIV/Girdin imposes finiteness in Arf1 signaling. Dev. Cell 33, 189–203. doi:10.1016/j.devcel.2015.02.009
Ma, M., Liu, J. J., Li, Y., Huang, Y., Ta, N., Chen, Y., et al. (2017). Cryo-EM structure and biochemical analysis reveal the basis of the functional difference between human PI3KC3-C1 and -C2. Cell Res. 27, 989–1001. doi:10.1038/cr.2017.94
Mancias, J. D., and Goldberg, J. (2008). Structural basis of cargo membrane protein discrimination by the human COPII coat machinery. EMBO J. 27, 2918–2928. doi:10.1038/emboj.2008.208
Mari, M., Griffith, J., Rieter, E., Krishnappa, L., Klionsky, D. J., and Reggiori, F. (2010). An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J. Cell Biol. 190, 1005–1022. doi:10.1083/jcb.200912089
Miao, G., Zhang, Y., Chen, D., and Zhang, H. (2020). The ER-localized transmembrane protein tmem39a/SUSR2 regulates autophagy by controlling the trafficking of the PtdIns(4)P phosphatase SAC1. Mol. Cell 77, 618–632.e5. doi:10.1016/j.molcel.2019.10.035
Miller, E. A., and Barlowe, C. (2010). Regulation of coat assembly--sorting things out at the ER. Curr. Opin. Cell Biol. 22, 447–453. doi:10.1016/j.ceb.2010.04.003
Miller, E. A., Beilharz, T. H., Malkus, P. N., Lee, M. C., Hamamoto, S., Orci, L., et al. (2003). Multiple cargo binding sites on the COPII subunit Sec24p ensure capture of diverse membrane proteins into transport vesicles. Cell 114, 497–509. doi:10.1016/s0092-8674(03)00609-3
Miller, E., Antonny, B., Hamamoto, S., and Schekman, R. (2002). Cargo selection into COPII vesicles is driven by the Sec24p subunit. EMBO J. 21, 6105–6113. doi:10.1093/emboj/cdf605
Mironov, A. A., Beznoussenko, G. V., Nicoziani, P., Martella, O., Trucco, A., Kweon, H. S., et al. (2001). Small cargo proteins and large aggregates can traverse the Golgi by a common mechanism without leaving the lumen of cisternae. J. Cell Biol. 155, 1225–1238. doi:10.1083/jcb.200108073
Mizuno-Yamasaki, E., Rivera-Molina, F., and Novick, P. (2012). GTPase networks in membrane traffic. Annu. Rev. Biochem. 81, 637–659. doi:10.1146/annurev-biochem-052810-093700
Mizushima, N. (2018). A brief history of autophagy from cell biology to physiology and disease. Nat. Cell Biol. 20, 521–527. doi:10.1038/s41556-018-0092-5
Mizushima, N., Levine, B., Cuervo, A. M., and Klionsky, D. J. (2008). Autophagy fights disease through cellular self-digestion. Nature 451, 1069–1075. doi:10.1038/nature06639
Moreau, K., Ravikumar, B., Renna, M., Puri, C., and Rubinsztein, D. C. (2011). Autophagosome precursor maturation requires homotypic fusion. Cell 146, 303–317. doi:10.1016/j.cell.2011.06.023
Mukai, K., Ogawa, E., Uematsu, R., Kuchitsu, Y., Kiku, F., Uemura, T., et al. (2021). Homeostatic regulation of STING by retrograde membrane traffic to the ER. Nat. Commun. 12, 61. doi:10.1038/s41467-020-20234-9
Murrow, L., and Debnath, J. (2013). Autophagy as a stress-response and quality-control mechanism: Implications for cell injury and human disease. Annu. Rev. Pathol. 8, 105–137. doi:10.1146/annurev-pathol-020712-163918
Nakano, A., Brada, D., and Schekman, R. (1988). A membrane glycoprotein, Sec12p, required for protein transport from the endoplasmic reticulum to the Golgi apparatus in yeast. J. Cell Biol. 107, 851–863. doi:10.1083/jcb.107.3.851
Nakano, A., and Muramatsu, M. (1989). A novel GTP-binding protein, Sar1p, is involved in transport from the endoplasmic reticulum to the Golgi apparatus. J. Cell Biol. 109, 2677–2691. doi:10.1083/jcb.109.6.2677
Neufeld, T. P. (2007). Contribution of Atg1-dependent autophagy to TOR-mediated cell growth and survival. Autophagy 3, 477–479. doi:10.4161/auto.4348
Nthiga, T. M., Shrestha, B. K., Bruun, J. A., Larsen, K. B., Lamark, T., and Johansen, T. (2021). Regulation of Golgi turnover by CALCOCO1-mediated selective autophagy. J. Cell Biol. 220, e202006128. doi:10.1083/jcb.202006128
Ohsumi, Y. (2001). Molecular dissection of autophagy: Two ubiquitin-like systems. Nat. Rev. Mol. Cell Biol. 2, 211–216. doi:10.1038/35056522
Orsi, A., Razi, M., Dooley, H. C., Robinson, D., Weston, A. E., Collinson, L. M., et al. (2012). Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, are required for autophagy. Mol. Biol. Cell 23, 1860–1873. doi:10.1091/mbc.E11-09-0746
Ortiz Sandoval, C., and Simmen, T. (2012). Rab proteins of the endoplasmic reticulum: Functions and interactors. Biochem. Soc. Trans. 40, 1426–1432. doi:10.1042/BST20120158
Oz-Levi, D., Ben-Zeev, B., Ruzzo, E. K., Hitomi, Y., Gelman, A., Pelak, K., et al. (2012). Mutation in TECPR2 reveals a role for autophagy in hereditary spastic paraparesis. Am. J. Hum. Genet. 91, 1065–1072. doi:10.1016/j.ajhg.2012.09.015
Park, S., Jang, I., Zuber, C., Lee, Y., Cho, J. W., Matsuo, I., et al. (2014). ERADication of EDEM1 occurs by selective autophagy and requires deglycosylation by cytoplasmic peptide N-glycanase. Histochem Cell Biol. 142, 153–169. doi:10.1007/s00418-014-1204-3
Pelham, H. R. (1999). SNAREs and the secretory pathway-lessons from yeast. Exp. Cell Res. 247, 1–8. doi:10.1006/excr.1998.4356
Petiot, A., Ogier-Denis, E., Bauvy, C., Cluzeaud, F., Vandewalle, A., and Codogno, P. (1999). Subcellular localization of the Gαi3 protein and G alpha interacting protein, two proteins involved in the control of macroautophagy in human colon cancer HT-29 cells. Biochem. J. 337 (2), 289–295. doi:10.1042/bj3370289
Piao, H., Kim, J., Noh, S. H., Kweon, H. S., Kim, J. Y., and Lee, M. G. (2017). Sec16A is critical for both conventional and unconventional secretion of CFTR. Sci. Rep. 7, 39887. doi:10.1038/srep39887
Popoff, V., Langer, J. D., Reckmann, I., Hellwig, A., Kahn, R. A., Brugger, B., et al. (2011). Several ADP-ribosylation factor (Arf) isoforms support COPI vesicle formation. J. Biol. Chem. 286, 35634–35642. doi:10.1074/jbc.M111.261800
Pu, J., Guardia, C. M., Keren-Kaplan, T., and Bonifacino, J. S. (2016). Mechanisms and functions of lysosome positioning. J. Cell Sci. 129, 4329–4339. doi:10.1242/jcs.196287
Pulvirenti, T., Giannotta, M., Capestrano, M., Capitani, M., Pisanu, A., Polishchuk, R. S., et al. (2008). A traffic-activated Golgi-based signalling circuit coordinates the secretory pathway. Nat. Cell Biol. 10, 912–922. doi:10.1038/ncb1751
Quinet, G., Gonzalez-Santamarta, M., Louche, C., and Rodriguez, M. S. (2020). Mechanisms regulating the UPS-als crosstalk: The role of proteaphagy. Molecules 25, 2352. doi:10.3390/molecules25102352
Rahman, A., Lorincz, P., Gohel, R., Nagy, A., Csordas, G., Zhang, Y., et al. (2022). GMAP is an Atg8a-interacting protein that regulates Golgi turnover in Drosophila. Cell Rep. 39, 110903. doi:10.1016/j.celrep.2022.110903
Ramirez-Peinado, S., Ignashkova, T. I., Van Raam, B. J., Baumann, J., Sennott, E. L., Gendarme, M., et al. (2017). TRAPPC13 modulates autophagy and the response to Golgi stress. J. Cell Sci. 130, 2251–2265. doi:10.1242/jcs.199521
Razi, M., Chan, E. Y., and Tooze, S. A. (2009). Early endosomes and endosomal coatomer are required for autophagy. J. Cell Biol. 185, 305–321. doi:10.1083/jcb.200810098
Reggiori, F., and Ungermann, C. (2017). Autophagosome maturation and fusion. J. Mol. Biol. 429, 486–496. doi:10.1016/j.jmb.2017.01.002
Russell, R. C., Tian, Y., Yuan, H., Park, H. W., Chang, Y. Y., Kim, J., et al. (2013). ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat. Cell Biol. 15, 741–750. doi:10.1038/ncb2757
Sacher, M., Jiang, Y., Barrowman, J., Scarpa, A., Burston, J., Zhang, L., et al. (1998). TRAPP, a highly conserved novel complex on the cis-Golgi that mediates vesicle docking and fusion. EMBO J. 17, 2494–2503. doi:10.1093/emboj/17.9.2494
Sacher, M., Kim, Y. G., Lavie, A., Oh, B. H., and Segev, N. (2008). The TRAPP complex: Insights into its architecture and function. Traffic 9, 2032–2042. doi:10.1111/j.1600-0854.2008.00833.x
Saitoh, T., Fujita, N., Hayashi, T., Takahara, K., Satoh, T., Lee, H., et al. (2009). Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc. Natl. Acad. Sci. U. S. A. 106, 20842–20846. doi:10.1073/pnas.0911267106
Sanchez-Wandelmer, J., Ktistakis, N. T., and Reggiori, F. (2015). Eres: Sites for autophagosome biogenesis and maturation? J. Cell Sci. 128, 185–192. doi:10.1242/jcs.158758
Sato, K., and Nakano, A. (2007). Mechanisms of COPII vesicle formation and protein sorting. FEBS Lett. 581, 2076–2082. doi:10.1016/j.febslet.2007.01.091
Sciaky, N., Presley, J., Smith, C., Zaal, K. J., Cole, N., Moreira, J. E., et al. (1997). Golgi tubule traffic and the effects of brefeldin A visualized in living cells. J. Cell Biol. 139, 1137–1155. doi:10.1083/jcb.139.5.1137
Scott, S. V., Hefner-Gravink, A., Morano, K. A., Noda, T., Ohsumi, Y., and Klionsky, D. J. (1996). Cytoplasm-to-vacuole targeting and autophagy employ the same machinery to deliver proteins to the yeast vacuole. Proc. Natl. Acad. Sci. U. S. A. 93, 12304–12308. doi:10.1073/pnas.93.22.12304
Scrivens, P. J., Noueihed, B., Shahrzad, N., Hul, S., Brunet, S., and Sacher, M. (2011). C4orf41 and TTC-15 are mammalian TRAPP components with a role at an early stage in ER-to-Golgi trafficking. Mol. Biol. Cell 22, 2083–2093. doi:10.1091/mbc.E10-11-0873
Semenza, J. C., Hardwick, K. G., Dean, N., and Pelham, H. R. (1990). ERD2, a yeast gene required for the receptor-mediated retrieval of luminal ER proteins from the secretory pathway. Cell 61, 1349–1357. doi:10.1016/0092-8674(90)90698-e
Shibutani, S. T., and Yoshimori, T. (2014). A current perspective of autophagosome biogenesis. Cell Res. 24, 58–68. doi:10.1038/cr.2013.159
Shima, T., Kirisako, H., and Nakatogawa, H. (2019). COPII vesicles contribute to autophagosomal membranes. J. Cell Biol. 218, 1503–1510. doi:10.1083/jcb.201809032
Shorter, J., Watson, R., Giannakou, M. E., Clarke, M., Warren, G., and Barr, F. A. (1999). GRASP55, a second mammalian GRASP protein involved in the stacking of Golgi cisternae in a cell-free system. EMBO J. 18, 4949–4960. doi:10.1093/emboj/18.18.4949
Shtutman, M., and Roninson, I. B. (2011). A subunit of coatomer protein complex offers a novel tumor-specific target through a surprising mechanism. Autophagy 7, 1551–1552. doi:10.4161/auto.7.12.17659
Simonsen, A., and Tooze, S. A. (2009). Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. J. Cell Biol. 186, 773–782. doi:10.1083/jcb.200907014
Solis, G. P., Bilousov, O., Koval, A., Luchtenborg, A. M., Lin, C., and Katanaev, V. L. (2017). Golgi-resident gαo promotes protrusive membrane dynamics. Cell 170, 939–955.e24. doi:10.1016/j.cell.2017.07.015
Song, S., Tan, J., Miao, Y., and Zhang, Q. (2018). Crosstalk of ER stress-mediated autophagy and ER-phagy: Involvement of UPR and the core autophagy machinery. J. Cell Physiol. 233, 3867–3874. doi:10.1002/jcp.26137
Sprangers, J., and Rabouille, C. (2015). SEC16 in COPII coat dynamics at ER exit sites. Biochem. Soc. Trans. 43, 97–103. doi:10.1042/BST20140283
Stadel, D., Millarte, V., Tillmann, K. D., Huber, J., Tamin-Yecheskel, B. C., Akutsu, M., et al. (2015). TECPR2 cooperates with LC3C to regulate COPII-dependent ER export. Mol. Cell 60, 89–104. doi:10.1016/j.molcel.2015.09.010
Subramanian, A., Capalbo, A., Iyengar, N. R., Rizzo, R., Di Campli, A., Di Martino, R., et al. (2019). Auto-regulation of secretory flux by sensing and responding to the folded cargo protein load in the endoplasmic reticulum. Cell 176, 1461–1476.e23. doi:10.1016/j.cell.2019.01.035
Suga, K., Saito, A., and Akagawa, K. (2015). ER stress response in NG108-15 cells involves upregulation of syntaxin 5 expression and reduced amyloid beta peptide secretion. Exp. Cell Res. 332, 11–23. doi:10.1016/j.yexcr.2015.01.001
Suga, K., Saito, A., Tomiyama, T., Mori, H., and Akagawa, K. (2009). The Syntaxin 5 isoforms Syx5 and Syx5L have distinct effects on the processing of {beta}-amyloid precursor protein. J. Biochem. 146, 905–915. doi:10.1093/jb/mvp138
Suzuki, K., Akioka, M., Kondo-Kakuta, C., Yamamoto, H., and Ohsumi, Y. (2013). Fine mapping of autophagy-related proteins during autophagosome formation in Saccharomyces cerevisiae. J. Cell Sci. 126, 2534–2544. doi:10.1242/jcs.122960
Suzuki, K., Kubota, Y., Sekito, T., and Ohsumi, Y. (2007). Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes 12, 209–218. doi:10.1111/j.1365-2443.2007.01050.x
Suzuki, K., and Ohsumi, Y. (2010). Current knowledge of the pre-autophagosomal structure (PAS). FEBS Lett. 584, 1280–1286. doi:10.1016/j.febslet.2010.02.001
Tan, D., Cai, Y., Wang, J., Zhang, J., Menon, S., Chou, H. T., et al. (2013). The EM structure of the TRAPPIII complex leads to the identification of a requirement for COPII vesicles on the macroautophagy pathway. Proc. Natl. Acad. Sci. U. S. A. 110, 19432–19437. doi:10.1073/pnas.1316356110
Tanida, I., Ueno, T., and Kominami, E. (2008). LC3 and autophagy. Methods Mol. Biol. 445, 77–88. doi:10.1007/978-1-59745-157-4_4
Tapia, D., Jimenez, T., Zamora, C., Espinoza, J., Rizzo, R., Gonzalez-Cardenas, A., et al. (2019). KDEL receptor regulates secretion by lysosome relocation- and autophagy-dependent modulation of lipid-droplet turnover. Nat. Commun. 10, 735. doi:10.1038/s41467-019-08501-w
Tillmann, K. D., Reiterer, V., Baschieri, F., Hoffmann, J., Millarte, V., Hauser, M. A., et al. (2015). Regulation of Sec16 levels and dynamics links proliferation and secretion. J. Cell Sci. 128, 670–682. doi:10.1242/jcs.157115
Trucco, A., Polishchuk, R. S., Martella, O., Di Pentima, A., Fusella, A., Di Giandomenico, D., et al. (2004). Secretory traffic triggers the formation of tubular continuities across Golgi sub-compartments. Nat. Cell Biol. 6, 1071–1081. doi:10.1038/ncb1180
Tsvetanova, N. G., Riordan, D. P., and Brown, P. O. (2012). The yeast Rab GTPase Ypt1 modulates unfolded protein response dynamics by regulating the stability of HAC1 RNA. PLoS Genet. 8, e1002862. doi:10.1371/journal.pgen.1002862
Wang, J., Davis, S., Menon, S., Zhang, J., Ding, J., Cervantes, S., et al. (2015). Ypt1/Rab1 regulates Hrr25/CK1δ kinase activity in ER-Golgi traffic and macroautophagy. J. Cell Biol. 210, 273–285. doi:10.1083/jcb.201408075
Wang, J., Menon, S., Yamasaki, A., Chou, H. T., Walz, T., Jiang, Y., et al. (2013). Ypt1 recruits the Atg1 kinase to the preautophagosomal structure. Proc. Natl. Acad. Sci. U. S. A. 110, 9800–9805. doi:10.1073/pnas.1302337110
Wang, P., Li, B., Zhou, L., Fei, E., and Wang, G. (2011). The KDEL receptor induces autophagy to promote the clearance of neurodegenerative disease-related proteins. Neuroscience 190, 43–55. doi:10.1016/j.neuroscience.2011.06.008
Watson, R. O., Manzanillo, P. S., and Cox, J. S. (2012). Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell 150, 803–815. doi:10.1016/j.cell.2012.06.040
Witte, K., Schuh, A. L., Hegermann, J., Sarkeshik, A., Mayers, J. R., Schwarze, K., et al. (2011). TFG-1 function in protein secretion and oncogenesis. Nat. Cell Biol. 13, 550–558. doi:10.1038/ncb2225
Xiang, Y., and Wang, Y. (2010). GRASP55 and GRASP65 play complementary and essential roles in Golgi cisternal stacking. J. Cell Biol. 188, 237–251. doi:10.1083/jcb.200907132
Xiang, Y., Zhang, X., Nix, D. B., Katoh, T., Aoki, K., Tiemeyer, M., et al. (2013). Regulation of protein glycosylation and sorting by the Golgi matrix proteins GRASP55/65. Nat. Commun. 4, 1659. doi:10.1038/ncomms2669
Yamasaki, A., Menon, S., Yu, S., Barrowman, J., Meerloo, T., Oorschot, V., et al. (2009). mTrs130 is a component of a mammalian TRAPPII complex, a Rab1 GEF that binds to COPI-coated vesicles. Mol. Biol. Cell 20, 4205–4215. doi:10.1091/mbc.e09-05-0387
Zacharogianni, M., Kondylis, V., Tang, Y., Farhan, H., Xanthakis, D., Fuchs, F., et al. (2011). ERK7 is a negative regulator of protein secretion in response to amino-acid starvation by modulating Sec16 membrane association. EMBO J. 30, 3684–3700. doi:10.1038/emboj.2011.253
Zerial, M., and Mcbride, H. (2001). Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2, 107–117. doi:10.1038/35052055
Zhao, S., Li, C. M., Luo, X. M., Siu, G. K., Gan, W. J., Zhang, L., et al. (2017). Mammalian TRAPPIII Complex positively modulates the recruitment of Sec13/31 onto COPII vesicles. Sci. Rep. 7, 43207. doi:10.1038/srep43207
Zhou, C., Ma, K., Gao, R., Mu, C., Chen, L., Liu, Q., et al. (2017). Regulation of mATG9 trafficking by Src- and ULK1-mediated phosphorylation in basal and starvation-induced autophagy. Cell Res. 27, 184–201. doi:10.1038/cr.2016.146
Keywords: endoplasmic reticulum (ER), KDEL receptor (KDELR), lipid droplets (LD), protein kinase A (PKA), plasma membrane (PM), traffic-induced degradation response for secretion (TIDeRS)
Citation: Tapia D, Cavieres VA, Burgos PV and Cancino J (2023) Impact of interorganelle coordination between the conventional early secretory pathway and autophagy in cellular homeostasis and stress response. Front. Cell Dev. Biol. 11:1069256. doi: 10.3389/fcell.2023.1069256
Received: 13 October 2022; Accepted: 07 April 2023;
Published: 21 April 2023.
Edited by:
Paolo Remondelli, University of Salerno, ItalyReviewed by:
Melat Gebru, California Life Company (Calico), United StatesCopyright © 2023 Tapia, Cavieres, Burgos and Cancino. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jorge Cancino, am9yZ2UuY2FuY2lub0B1c3MuY2w=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.