
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell Dev. Biol. , 23 August 2022
Sec. Stem Cell Research
Volume 10 - 2022 | https://doi.org/10.3389/fcell.2022.945572
This article is part of the Research Topic Germline Development: From Germline Stem Cells to Gametes, Volume II View all 7 articles
 Houda Ouns Maaroufi1,2
Houda Ouns Maaroufi1,2 Lucie Pauchova1,2
Lucie Pauchova1,2 Yu-Hsien Lin1,2
Yu-Hsien Lin1,2 Bulah Chia-Hsiang Wu1,2
Bulah Chia-Hsiang Wu1,2 Lenka Rouhova1,2
Lenka Rouhova1,2 Lucie Kucerova1
Lucie Kucerova1 Ligia Cota Vieira1
Ligia Cota Vieira1 Marek Renner2
Marek Renner2 Hana Sehadova1,2
Hana Sehadova1,2 Miluse Hradilova3
Miluse Hradilova3 Michal Zurovec1,2*
Michal Zurovec1,2*Concentrative nucleoside transporters (Cnts) are unidirectional carriers that mediate the energy-costly influx of nucleosides driven by the transmembrane sodium gradient. Cnts are transmembrane proteins that share a common structural organization and are found in all phyla. Although there have been studies on Cnts from a biochemical perspective, no deep research has examined their role at the organismal level. Here, we investigated the role of the Drosophila melanogaster cnt1 gene, which is specifically expressed in the testes. We used the CRISPR/Cas9 system to generate a mutation in the cnt1 gene. The cnt1 mutants exhibited defects in the duration of copulation and spermatid maturation, which significantly impaired male fertility. The most striking effect of the cnt1 mutation in spermatid maturation was an abnormal structure of the sperm tail, in which the formation of major and minor mitochondrial derivatives was disrupted. Our results demonstrate the importance of cnt1 in male fertility and suggest that the observed defects in mating behavior and spermatogenesis are due to alterations in nucleoside transport and associated metabolic pathways.
Male fertility in Drosophila depends on a number of traits, including appropriate mating behavior and proper progression of spermatogenesis (Baptissart et al., 2013; Lotti and Maggi, 2018). Mating is a complex process that is essential for reproductive success. The male pursues the female while vibrating his wings to produce a courtship song, tapping the female’s abdomen with his forelegs, touching the female’s genitals with his mouthparts, and curving his abdomen for copulation (Pavlou and Goodwin, 2013; Ziegler et al., 2013). The transfer of mature and healthy sperm to the female genitalia also depends on proper spermatogenesis, which involves the proliferation and differentiation of spermatogonial stem cells (Wakimoto et al., 2004; Yuan et al., 2019).
Spermatogenesis in Drosophila begins with stem cell proliferation, wherein the stem cell undergoes four mitotic divisions with incomplete cytokinesis, forming a cyst with 16 interconnected spermatogonia. This is followed by cell growth and meiosis to produce a syncytial group of 64 spherical haploid spermatids (Steinhauer, 2015). Subsequently, these spermatids undergo elongation and individualization (Tokuyasu, 1975; Noguchi and Miller, 2003), a process known as Drosophila spermiogenesis, in which the spermatids are remodeled into spermatozoa (Alzyoud et al., 2021). Spermatogonia develop in an enclosure formed by two cyst cells of somatic origin that undergo extensive morphogenesis and eventually differentiate into the head and tail cysts during spermatid elongation (Lindsley and Tokuyasu, 1980; White-Cooper, 2004). During this process, the round-shaped spermatid nuclei become long and thin (needle-shaped nuclei), accompanied by chromatin condensation. The fully elongated spermatids begin the individualization process at the basal end of the testis in the region of the terminal epithelium (TE), where actin-based individualization complexes (ICs) form around the nuclei then migrate from the heads to the tip of the tails. As ICs move along the elongated spermatid, unnecessary organelles and additional cytoplasm are stripped off and form a cystic bulge (CB). Once the cyst detaches from the tail, it forms a rounder waste bag (WB), which is eventually degraded, leaving each mature spermatozoon enclosed in its own tight membrane (Bader et al., 2010; Fabian and Brill, 2012). At the end of spermatogenesis, each mature sperm group coils in the TE region and is released in the testis lumen before migrating from the TE region to the seminal vesicle (SV), where it is prepared for ejaculation (Fabian and Brill, 2012; Demarco et al., 2014).
More than 2000 mutations affecting male fertility have been discovered in Drosophila (Wakimoto et al., 2004), but most of these mutations are not well characterized. Moreover, a number of genes with specific expression in the testis have not been well studied (Wakimoto et al., 2004; Zhao et al., 2010; Vedelek et al., 2018; Witt et al., 2019). Previous studies have shown that some of the mutations that affect mating behavior are connected to Drosophila morphology (white and yellow (Zhang and Odenwald, 1995)), learning and memory (dunce and amnesiac (Siegel and Hall, 1979)), sex determination (fruitless (fru) (Baker et al., 2001; Rideout et al., 2007)), or copulation (Phosphodiesterase 1c (pde1) (Morton et al., 2010)). Most of the spermatogenesis-related mutations affect germline cells (e.g., zn finger homeodomain 1 and delta (Leatherman and Dinardo, 2008; Ng et al., 2019)), spermatocytes (e.g., cannonball (Chen et al., 2005)) and spermatids (e.g., driceless and dynein intermediate chain at 61B (dic61B)) (Huh et al., 2004; Fatima, 2011)). Other mutations may regulate the number (e.g., headcase (Resende et al., 2013)), morphology (e.g., salto (Augière et al., 2019)), or motility (e.g., hemingway (Soulavie et al., 2014)) at different cell stages.
We studied a gene encoding the Drosophila concentrative nucleoside transporter 1 (cnt1), which is expressed in a testis-specific manner (Larkin et al., 2021). Cnt1 is a unidirectional carrier that mediates the energy-costly influx of nucleosides driven by the transmembrane sodium gradient (Gray et al., 2004; Chintapalli et al., 2007; Larkin et al., 2021). Nucleoside transporters and the enzymatic activities of adenosine metabolism affect many physiological processes by influencing the adenosine levels as a signaling molecule, especially the role of the adenosine signal terminator at the respective receptor (Knight et al., 2010). Cnts are found in most organisms, including mammals and insects, and are fairly conserved among phyla. For example, Cnts in humans and Drosophila have 24–33% sequence similarity (Machado et al., 2007). The involvement of Drosophila Cnt1 and Cnt2 in adenosine transport was examined in a previous study that showed that knocking down their expression rescues Cl.8 + tissue culture cells from the cytotoxic effect of a high concentration of extracellular adenosine (Fleischmannova et al., 2012). The Cnt protein structure is characterized by 13 transmembrane domains (Young et al., 2013; Dos Santos-Rodrigues et al., 2015), and they seem to have limited tissue distribution within organisms. For example, in mammals, Cnts have been detected in specialized cells, particularly intestinal and hepatic epithelial cells and testis (Kato et al., 2005; Masino and Boison, 2013). Cnts have been identified in rat epididymis and shown to play a role in sperm maturation (Leung et al., 2001; Klein et al., 2017). In addition, in a recent study, the deficiency of human Cnt1 has been associated with a newly discovered inborn error of pyrimidine metabolism and organ failure in an infant (Pérez-Torras et al., 2019).
In the present study, we identified the cnt1 gene as an important component in mediating the crosstalk between fertility and systemic metabolism in Drosophila. We showed that Cnt1 is highly expressed in testes. To determine the role of cnt1 in Drosophila, we created a mutation in this gene using the CRISPR/Cas9 system and named the obtained allele cnt1{fertility defect} (cnt1FD). The mutation in cnt1 caused a defect in mating behavior (copulation duration) and spermatid maturation that significantly impaired male fertility. The prominent effect of the cnt1FD mutation in spermatid maturation involved an abnormal structure of the spermatid tail and low sperm count.
Fly strains were reared on a standard cornmeal medium at 25°C with 12/12 h light/dark cycle and 60% relative humidity. Cnt1FD mutant and cnt1::GFP were used and generated as described below. W1118 was used as a wild-type control. Flies containing protA::GFP were provided by Dr. R. Renkawitz-Pohl and Dr. Ch. Rathke (Jayaramaiah Raja and Renkawitz-Pohl, 2005), while the dj::GFP (BL 5417, USA) and Df(2R)BSC271 (BL 23167, USA) were obtained from Bloomington (BL5417, USA).
The mutation in the Drosophila cnt1 gene was generated through the CRISPR/cas9 gene editing approach (Kondo and Ueda, 2013). Two guide RNA (gRNA) [GGCAAAGCGAATCACCTATCTGG and GTGCCGCAATGACGCATACAAGG] were used to create a deletion. The first was located in the first coding exon and the second in the third coding exon. The double-strand break was corrected by non-homologous end-joining, which led to an open reading frame deletion of 477 bp and substitution of one amino acid at the break site (the entire cnt1 deletion comprises 752 bp, including coding exon 1, complete exon 2, part of coding exon 3, and two complete introns.). The cnt1FD mutants were generated by injecting the described construct into the embryos of nanos-cas9 expressing flies and examined through polymerase chain reaction (PCR). The flies were isogenized to the background of w1118 flies. Both male and female flies were homozygous viable for this deletion. We confirmed the mutation in cnt1 (cnt1FD) with the primers at the deletion site (Supplementary Table S1).
We generated flies containing a genomic sequence of cnt1 tagged with GFP (cnt1::GFP) (Ejsmont et al., 2011). We cloned the GFP sequence into a fosmid vector containing the cnt1 gene as a pre-tagging vector (Ejsmont et al., 2011; Sarov et al., 2016). The tagged clone was later injected into Drosophila embryos using GenetiVision services (Groth et al., 2004). The mutation in the flies was verified via PCR according to manufacturer protocol (Thermo Scientific Phusion High-Fidelity PCR Master Mix). The transgenic flies, cnt1::GFP, were used to trace the expression pattern of cnt1 through the integrated GFP and used as a rescuers by combining it with the cnt1FD mutant fly.
The RT-qPCR was performed as previously described (Lin et al., 2019). Total RNA was extracted from adult flies using TRIzol reagent (#1559026, Invitrogen, USA). Thereafter, the isolated RNAs were treated with DNase using the Promega RQ1 RNase-free DNase kit to prevent genomic DNA contamination. The DNase-treated RNAs were used for cDNA synthesis from 2 μg total RNA using a RevertAid H Minus First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Vilnius, Lithuania). The cDNA was amplified by RT-qPCR using the 5 × Hot FirePol EvaGreen qPCR Mix Plus with ROX (Solis BioDyne, Tartu, Estonia) and Rotor Gene Q Instrument (QIAGEN, Hilden, Germany). The primers used for cnt1 quantification are presented in Supplementary Table S1. αTub84B (FBgn0003884) and act5C (FBgn0000042) were used as reference housekeeping genes. The expression level was calculated using the 2–ΔΔCt method after normalizing the ct values of the target genes to the reference gene.
Virgin male and female flies were collected and separated for 3 days. Then, each of the adult male flies was placed in a vial along with 10 w1118 virgin female flies (3 days old) at 25°C. After 24 and 48 h, the females were placed in individual vials and examined for the appearance of offspring. The number of fertilized females per male was recorded.
For the copulation length assay, virgin male and female flies were collected and separated for 3 days. Thereafter, the flies were sedated, placed in individual vials, and left to recover for 1 h. Then, a single male and a female were placed in a common vial, and video recording was undertaken.
For the copulation frequency assay, virgin male and female flies were collected and separated for 3 days. Then, a single male and 10 female flies were placed in a vial. Afterwards, the number of copulations was recorded.
Immunohistochemical staining was performed as described previously with a few modifications (Lin et al., 2021). Drosophila adult testes were dissected in 1 × phosphate buffer saline (PBS) and fixed in 4% paraformaldehyde for 20 min. After washing with PBST (0.3% Triton in 1× PBS) three times, the testes were incubated in a blocking buffer (5% goat serum in 0.1% PBST) for 1 h. Next, the blocking buffer was replaced by primary antibodies mixed in PAT (1% BSA, 0.1% Triton and 0.01% sodium azide in 1× PBS) and the incubation lasted for overnight. Then, the testes were washed three times with 0.1% PBST and incubated in secondary antibodies mixed in 0.1% PBST for 1 h. After washing with 0.1% PBST three times, the tissues were stained with DAPI for 10 min and washed three times before mounting with Fluoromount-G. DAPI (#MBD0015, Sigma-Aldrich, Germany), DCP1 (#9578S, Cell signaling, USA).
Images of the stained testes were taken using a laser scanning confocal microscope FluoView™ FV1000 (Olympus, Tokyo, Japan). To quantify the canoe-stage spermatids, sperms, and waste bags, the area of the TE region was obtained using ImageJ software (Schneider et al., 2012).
The testes dissected in PBS were fixed in 2.5% glutaraldehyde for at least 4 h at room temperature (RT) or overnight at 4°C. The specimens were placed in a wash solution containing PBS with 4% glucose (three times for 15 min at RT) and then treated with a 1:1 mixture of PBS and 4% OsO4 solution (for 2 h at RT). After application of the wash solution (three times for 15 min at RT), the tissues were dehydrated in an acetone series (30, 50, 70, 80, 90, 95, and 100% for 15 min each). The dehydrated samples were embedded in Epon resin by gradually increasing the volume ratio of resin to acetone (1:2, 1:1, and 2:1 for 1 h each at RT). The specimens were left in the resin for 24 h (RT), and then the resin was polymerized at 62°C for 48 h. The ultrathin sections were cut with a diamond knife and stretched with chloroform. The sections were placed on copper meshes and contrasted with uranyl acetate and lead citrate, after which they were coated with carbon. The samples were imaged under the JEOL JSM-7401F transmission electron microscope (JEOL, Akishima, Japan).
Graphs were produced in GraphPad Prism 5 software. Statistical analysis of the data was executed using Statistica software (StarSoft CR) and Microsoft Excel. Significance was established using parametric tests: Student’s t-test (N.S., not significant, *p < 0.05, **p < 0.01, ***p < 0.001) and one-way ANOVA (combined with Tukey post-hoc test), or non-parametric test: Kruskal–Wallis (combined with comparisons of mean ranks of all pairs of groups post-hoc test). Statistical analysis of the RT-qPCR was done using log2 value (Supplementary Table S2).
The Drosophila testes were dissected and frozen in liquid nitrogen. Biological triplicates were prepared, and 40 individuals were dissected for each replicate. TRIzol (#1559026 Invitrogen, USA) was applied to each sample, and RNA was isolated according to manufacturer protocol. The isolated RNA was further purified and treated by cDNAse using a NucleoSpin RNA kit (#740955.250 Macherey-Nagel, Germany). The quality of total RNA was controlled using an RNA 6000 Nano Kit on an Agilent Bioanalyzer 2,100 and the quantity using an RNA BR Assay Kit on a Qubit 2.0 Fluorometer (Life Technologies, USA). The sequencing library was prepared from a 1,000 ng input of high-quality (RIN >7) total RNA using a KAPA mRNA HyperPrep Kit for Illumina Platforms (Roche, KK8580, Switzerland) according to manufacturer protocol, with fragmentation conditions set at 94°C and 7 min. The final library PCR amplification was set at nine cycles. Library length profiles and concentrations were monitored using an Agilent Bioanalyzer (High-Sensitivity DNA chip) and a Qubit 2.0 Fluorometer (dsDNA HS Assay Kit), respectively. A pool of libraries in equimolar ratio was generated and sequenced in single-end mode on the NextSeq 500 Illumina platform using the NextSeq 500/550 High Output Kit v2.5 (75 Cycles) with a loading molarity of 1,8 p.m. including 1% PhiX control. Data analysis was performed using Galaxy online software and its associated tutorial (Batut et al., 2018). Our SRA database has been released on NCBI under the accession: PRJNA838856.
Representative protein sequences of Cnt homologs, namely Sodium/Nucleoside Cotransporters or Solute Carrier Family 28 Members-3, covering the phylogeny of the genus Drosophila and the phylogeny of insects were identified using the Basic Local Alignment Search Tool (BLAST) in the GenBank database (Clark et al., 2016). Amino acid sequences were then aligned using the software MUSCLE (Edgar, 2004). Smart Model Selection, an online software (Lefort et al., 2017), was used to pick the best model according to the lowest Bayesian Information Criterion (BIC) score. Phylogenetic analyses were conducted through the Maximum Likelihood approach in PhyML 3.0 (Guindon et al., 2010). For each branch, statistical support was calculated as aBayes values. Trees were finalized with the aid of MEGA version X (Kumar et al., 2018).
The cnt genes are present in most living organisms, including eubacteria, suggesting that they are evolutionarily ancient (Young et al., 2013). Different taxonomic groups may differ in the number of cnt isoforms. For example, insects have one to three cnt isoforms, whereas mammals have three different cnt genes (Molina-Arcas and Pastor-Anglada, 2013; Young et al., 2013; Young, 2016). Moreover, among insects, Isoptera and Diptera seem to be the only insect orders where duplication events occur (Figure 1A). The duplication within the Isoptera appears to be a single origin event, whereas in Diptera, there seem to be multiple duplications and gene loss events. A closer look at the distribution of cnts within the genus Drosophila reveals a recent duplication of cnt in the common ancestor of Drosophila and Scaptodrosophila, with a secondary loss of cnt1 in the subgenus Drosophila (Figure 1B). Based on the alignments among insects, the Cnts differ mainly by the first 20 amino acids, while the rest of the sequences are well conserved (Supplementary Figure S1).
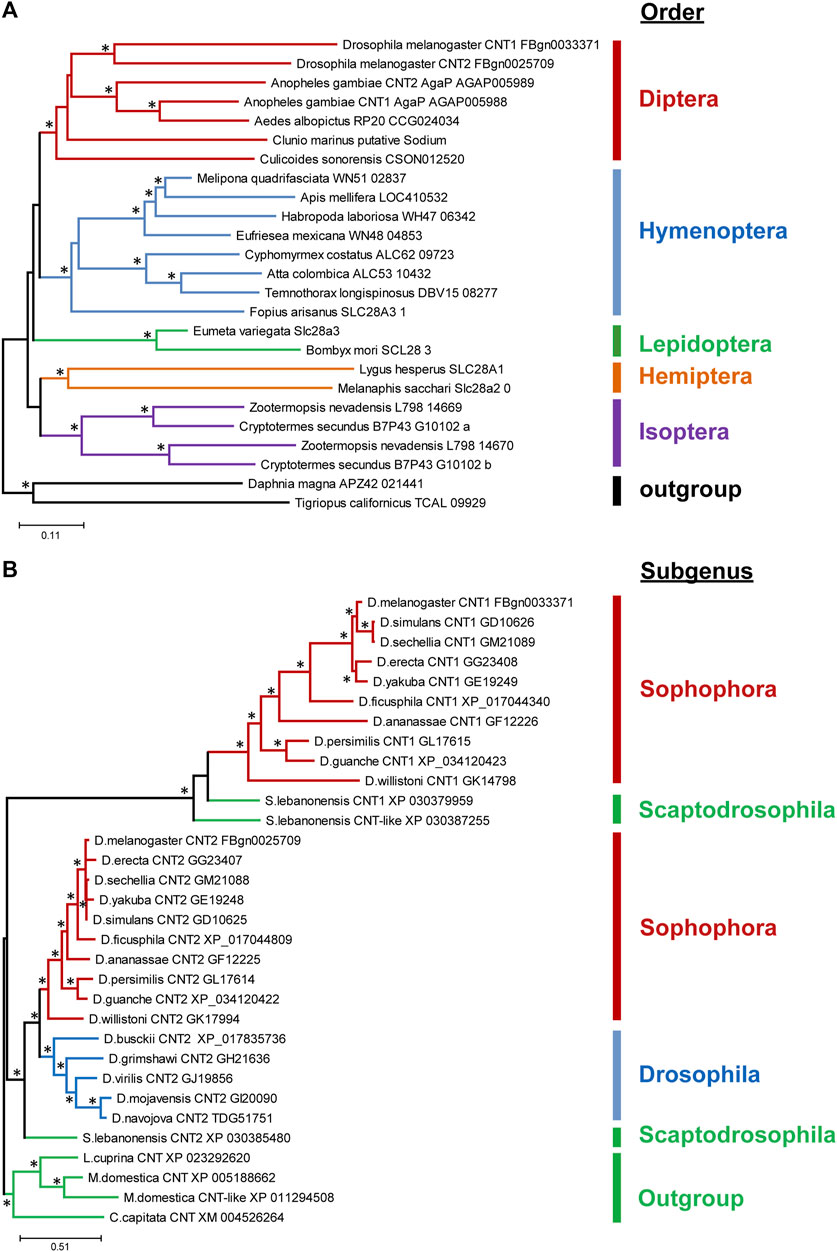
FIGURE 1. Distribution of Cnts in insects and family drosophilidae. (A) Simplified phylogenetic tree of Cnts among insects. Each color represents a particular order of insects. (B) Simplified phylogenetic tree of Cnt in Drosophila. Each color represents a subgenus of Drosophila. Branches with statistical support (aBayes values) greater than 70 are marked with asterisks.
The presence of Cnts in all species indicates that they perform important functions in organisms. The presence of multiple isoforms in some Drosophila species suggests that they may also be required in a tissue-specific manner.
According to FlyBase, cnt1 is highly expressed in the male testes (Larkin et al., 2021). To confirm this expression profile, we performed RT-qPCR using cnt1 gene-specific primers on the extracted reproductive organs and the rest of the body of w1118 male flies and females (Supplementary Table S1). Data analysis showed a high expression of cnt1 in males and especially in the testes rather than the rest of the body organs in males (Figure 2A). We also used the fosmid FlyFos transgene (Ejsmont et al., 2009) and analyzed the expression of GFP-tag (Sarov et al., 2016) (cnt1::GFP) in w1118 background. A well-defined GFP signal was found in the TE region of the testis in flies carrying the cnt1::GFP transgene but not in the control w1118 (Figure 2B and Supplementary Figure S2). This signal was located in the head of the canoe- and needle stage-spermatids and overlapped with the DNA staining. In addition, a weak GFP signal was detected in the spermatid tail (Figures 2B,C). Detailed analysis showed that Cnt1 expression begins during spermiogenesis when late canoe-stage spermatids start to form, and that the signal persists until the formation of needle-stage spermatids (Figure 2C). These results confirm the tissue specificity of Cnt1 expression.
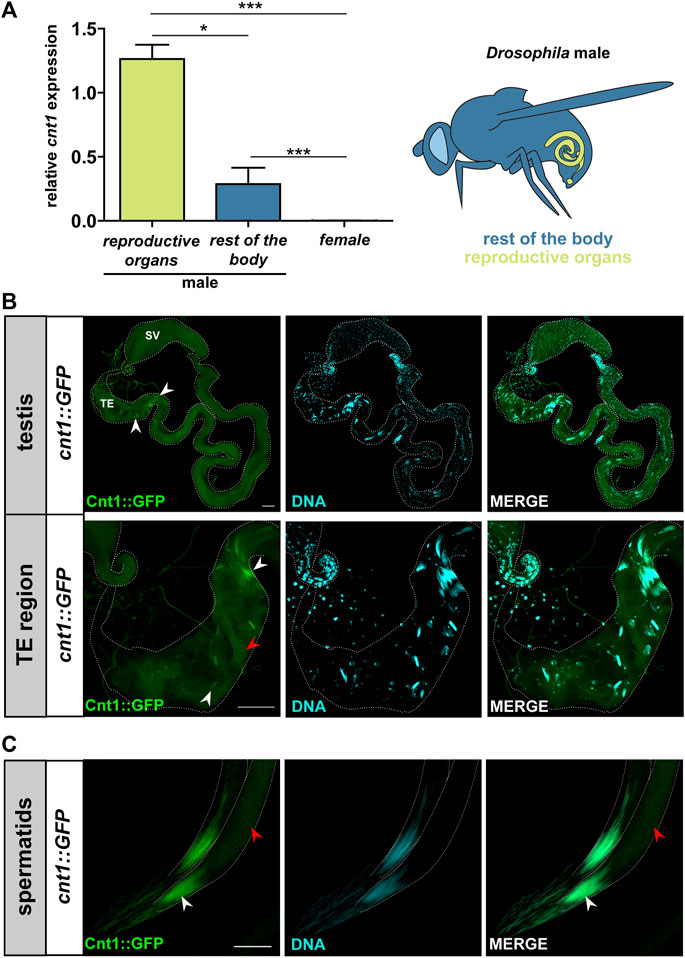
FIGURE 2. Cnt1 expression in the head and tail of spermatids. (A) The results of RT-qPCR analysis show a relative expression of cnt1 in the reproductive organs and the rest of the body of the w1118 male and female flies (see the illustration). Expressions were normalized to αTub84B and act5C transcripts (ΔΔCT). Significance was analyzed by one-way analysis of variance (ANOVA) and labeled as follows: *p < 0.05, ***p < 0.001; n = 5. Error bars are given as mean ± SEM. (B) Cnt1::GFP localization in Drosophila testes and its TE region. Scale bar: 40 µm. (C) A closer look at the localization of Cnt1::GFP in spermatid heads (needle-stage spermatids). Scale bar: 10 μm. Cnt1::GFP staining in green and DNA staining (DAPI) in cyan. Head of spermatids (white arrowhead), tail of spermatids (red arrowhead), terminal epithelium (TE), seminal vesicle (SV).
Using CRISPR-Cas9 technology (Kondo and Ueda, 2013), we deleted the internal part of the cnt1 gene using two guide RNAs (gRNA) targeting the first and third coding exons (Figure 3A). The double-strand breaks resulted in a deletion of 477 bp and the substitution of an amino acid at the break site (Figures 3B,C). The deletion did not affect the reading frame of the gene but removed three transmembrane domains out of 13. We confirmed the cnt1 mutation by sequencing and RT-qPCR (Figure 3D).
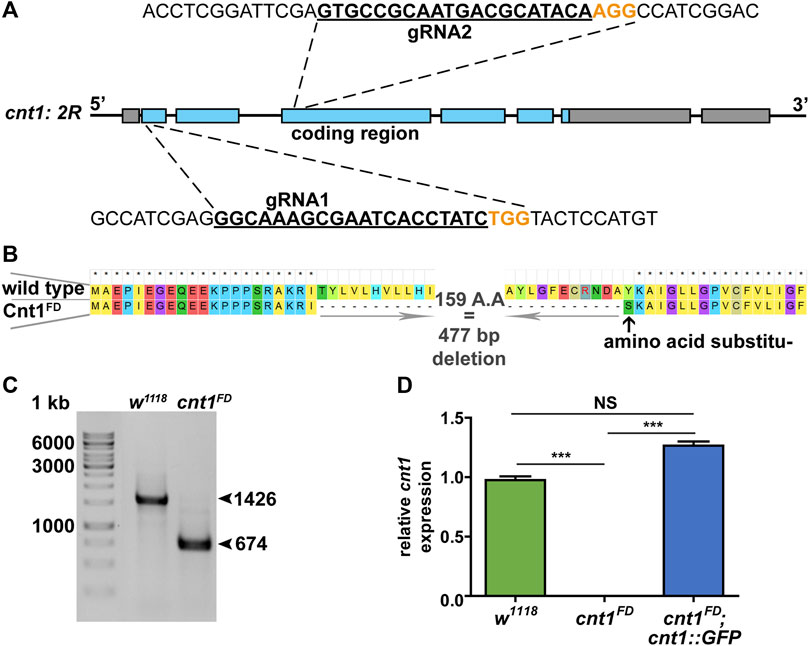
FIGURE 3. Generation of mutation in Drosophila cnt1. (A) Schematic representation of the cnt1 mutation using two guide RNAs (gRNA); the target of the first gRNA is located in the first coding exon and the second in the third coding exon. (B) Protein sequence alignment of Cnt1 in control w1118 and cnt1FD. The mutation caused a deletion of 477 bp (159 A.A) in the coding region of cnt1 gene (a total of 752 bp including introns) and a substitution of the tyrosine residue for serine. (C) The PCR products of the cnt1 mutant compared with its control. (D) Results of RT-qPCR analysis showing a relative expression of cnt1 in control flies, cnt1FD mutant flies, and rescue flies cnt1FD;cnt1::GFP. Expressions were normalized to αTub84B and act5C transcripts (ΔΔCT). Significance was analyzed by one-way analysis of variance (ANOVA) and labeled as follows: ***p < 0.001, NS > 0.05; n = 4. Error bars are shown as mean ± SEM.
The male and female mutants showed normal viability. Given that cnt1 expression is high in Drosophila testes, we investigated whether cnt1 mutation affects male fertility. Individual naive males of w1118 and cnt1FD were confined with 10 virgin w1118 females for 24 or 48 h (Figure 4A). The cnt1FD males were able to fertilize three times fewer females within 24 h than the control male flies (Figure 4B). Notably, extending the mating to 48 h showed no significant difference in fertility between the cnt1FD and w1118 males (Figure 4C), suggesting that cnt1FD males are less fertile and may require more time to inseminate all females.
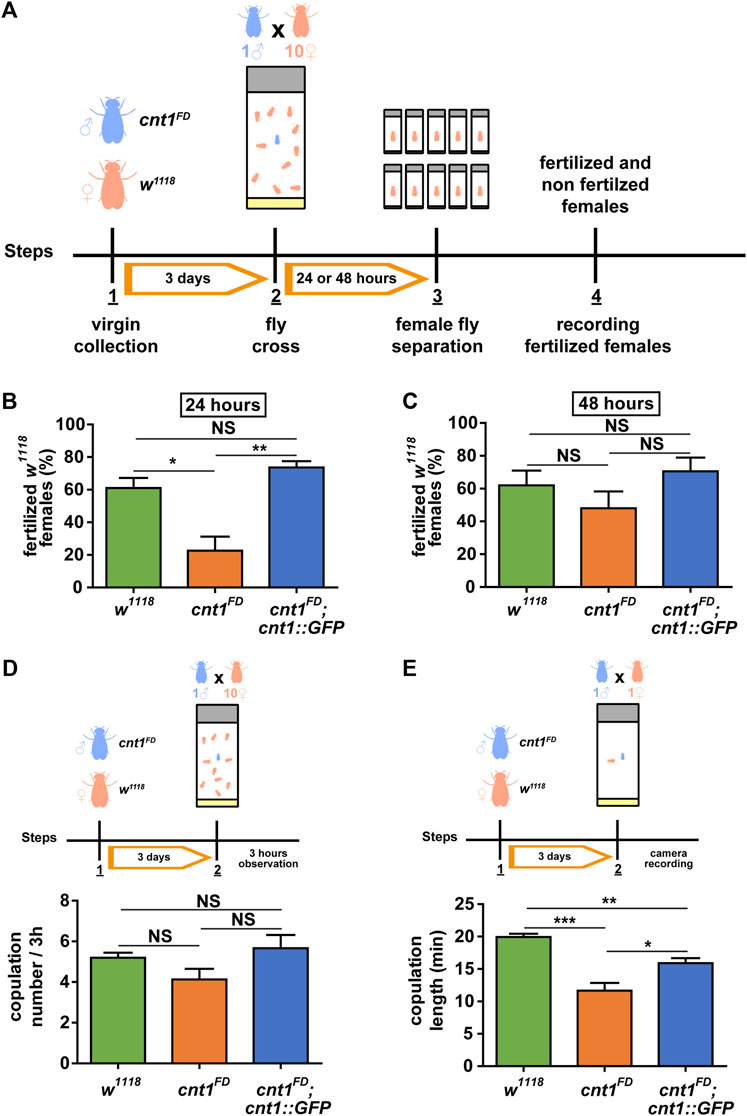
FIGURE 4. The cnt1 mutation causes partial sterility in Drosophila males. (A) Experimental procedure of the fertility assay. Virgin mutant males (cnt1FD) and control females (w1118) were collected. After 3 days, one male was placed in a vial with 10 females for 24 or 48 h. Subsequently, each female was placed separately in a single vial, and the presence or absence of larvae was recorded. (B) Percentage of fertilized females after 24 h of mating with cnt1FD males; n ≥ 10. (C) Percentage of fertilized females after 48 h of mating with cnt1FD males; n ≥ 10. (D) Number of copulations within 3 h of observation. Virgin males (cnt1FD) and females (w1118) were collected. After 3 days, one male was placed in a vial with 10 females, and their mating was observed; n = 15. Error bars are presented as mean ± SEM. (E) Duration of copulation and experimental procedure. Virgin males (cnt1FD) and females (w1118) were collected. After 3 days, a male was placed in a vial with a female, and mating was recorded with a camera. The calculated times of copulations are in minutes; n = 15. Significance was analyzed by one-way ANOVA (C) and Kruskal–Wallis (B,D,E); and labeled as follows: *p < 0.05, **p < 0.01, ***p < 0.001, NS > 0.05. Error bars are presented as mean ± SEM.
Previous experiments have shown that 75% of the proteins tagged with GFP are functional in vivo (Nagarkar-Jaiswal et al., 2015). To determine whether the tagged cnt1 was functional and test whether the observed cnt1FD phenotype was specific to cnt1FD mutation, we crossed the cnt1FD flies with flies carrying the cnt1::GFP transgene. As shown in Figures 4B,C, males of the rescue line cnt1FD;cnt1::GFP were as fertile as the control line. We also used hetero-allelic flies carrying a genomic deletion including cnt1 and the results show that after 24 h of mating the hetero-allelic male cnt1FD/Df(2R)BSC271 flies were able to fertilize a similar percentage of females as the homozygous male cnt1FD mutants but not as many as the control male w1118 flies (Supplementary Figure S3). These data confirm that the observed phenotypes are caused by the loss of cnt1 function.
Based on the above results, we tested whether the fertility disorder was related to abnormal mating behavior. To test whether mating frequency is affected by the cnt1 mutation, we performed a test in which a male was placed in a vial with 10 virgin females and mating was recorded for 3 h. The observations showed no significant difference in mating frequency among the cnt1FD flies, w1118 control flies, and cnt1FD;cnt1::GFP rescue flies (Figure 4D).
In the next experiment, single 3-day-old naïve males were paired with single virgin females, and their copulation time was recorded. Again, we observed that all cnt1FD males mated with the females at the same frequency, but the copulation time was shorter compared with the controls (Figure 4E). Thus, we conclude that the changes in copulation duration in cnt1 mutants likely contribute to the observed phenotype of lower fecundity.
The maximum cnt1 expression is detected in canoe- and needle-stage spermatids. We tested whether their numbers are affected by the cnt1 mutation. To test whether the reduced fertility of mutant males was caused by a defect in spermatid production, we used the protamine A (protA) fluorescent marker (protA::GFP) (Jayaramaiah Raja and Renkawitz-Pohl, 2005) to determine spermatid and sperm counts. ProtA is known to be expressed in the sperm heads from the end of spermiogenesis. We also used another marker, don Juan (dj) tagged with GFP (dj::GFP) (Santel et al., 1997; Santel et al., 1998), which is present in spermatids and sperm tails, to monitor possible defects in sperm flagella. Both protA::GFP and dj::GFP were used in the background of cnt1FD mutants, as well as in its control w1118. Analysis showed that, in contrast to the control, cnt1FD had a higher number of spermatid groups, including the needle-stage spermatid groups, which were scattered throughout the TE region (Figures 5A–C). The volume in this region was also increased (Figure 5D). Moreover, we counted the number of mature sperms released in the TE region and found it to be higher in cnt1FD mutants compared with the control (Figure 5E). The higher number of spermatid groups and mature sperms in the TE region of cnt1FD mutants could be due to their higher production or slow mature sperm displacement toward the SV.
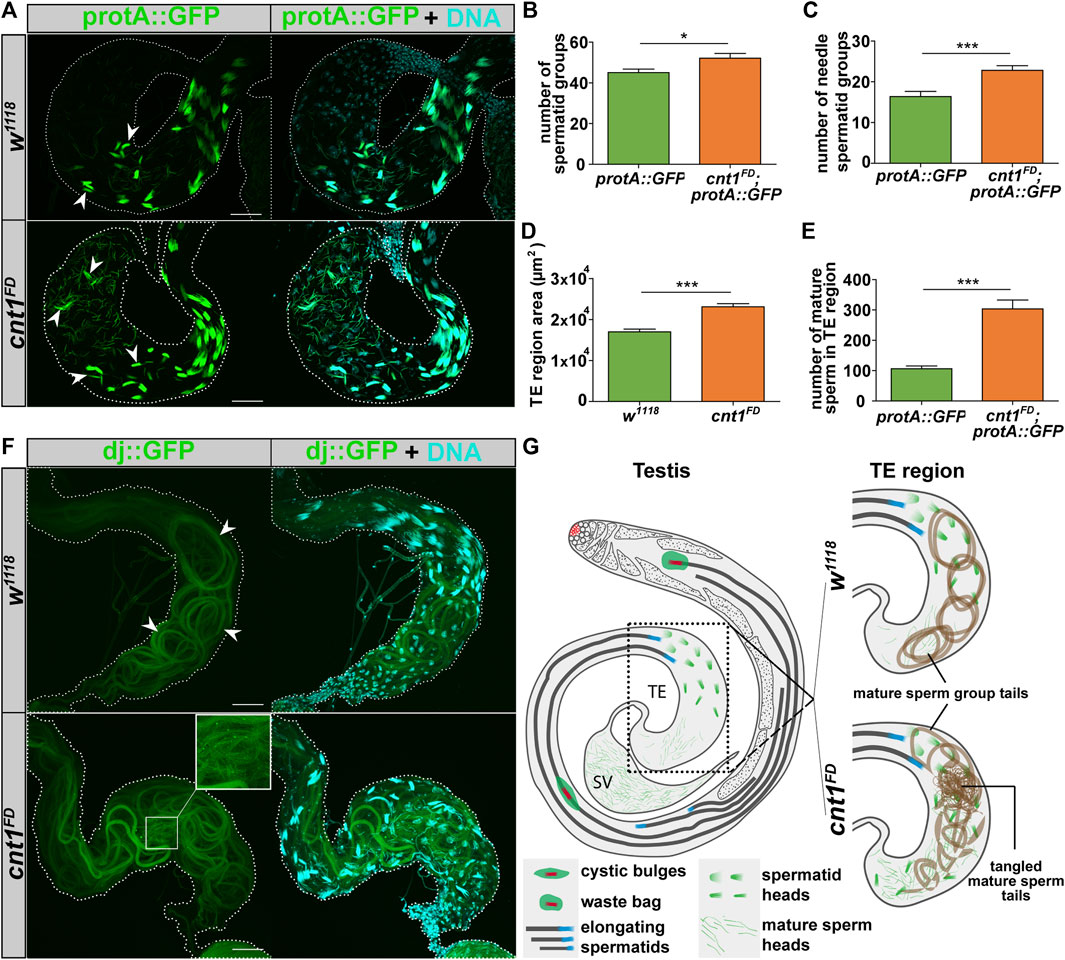
FIGURE 5. The cnt1 mutation increases the number of spermatid groups and spermatozoa in the TE region and causes tail miscoiling. (A) Microscopic observation of spermatids expressing protamine A signals in the region of the terminal epithelium of the testis. The arrowheads show examples of the needle spermatid groups. W1118 is used as a control. In w1118, spermatid groups expressing protamine A are observed in the first half of the TE region, whereas the spermatid groups are scattered throughout the TE region in the cnt1FD mutants. Protamine A (green), DNA (cyan). Scale bar: 40 µm. (B) Number of spermatid groups; n ≥ 10. (C) Number of needle-stage spermatid groups; n ≥ 10. (D) Size of the TE region; n ≥ 28. (E) Number of mature sperm in the TE region; n ≥ 10. Significance was analyzed with a one-tailed Student’s t-test and labeled as follows: *p < 0.05, ***p < 0.001. Error bars are shown as mean ± SEM. (F) Microscopic observation of coiled tails of mature sperm groups expressing a “dj” signal in the region of the terminal epithelium of the testis. W1118 is used as a control. In w1118, the tails of the mature sperm group were well organized and coiled (white arrowhead). In cnt1FD, the tails of mature sperm were poorly organized. Upon closer inspection, the tails were tangled in the region of the TE. Dj (green), DNA (cyan). Scale bar: 40 µm. (G) Summarized representation of protamine A and dj organization in mature sperm in w1118 and cnt1FD.
To distinguish whether the increased number of mature sperms is the result of their higher production or accumulation, we investigated whether the defect of the spermatid group number occurs in newly emerged males. The microscopic analysis of the TE regions from 24-hour-old males shows that cnt1FD already produces a high number of spermatid groups (Supplementary Figure S4). Thus, the effect of cnt1 on spermatid groups starts at an early age in adult males, possibly due to their higher production.
The alteration in spermatid group production was followed by the disorganization of the tails of the mature sperm groups in the TE region, which sometimes showed tangled tails. This phenomenon, most likely occurred during coiling (Figure 5F). In wild-type flies, the coiling process of mature sperm groups occurs with a well-defined circular pattern of flagella, whereas mature sperm groups from cnt1FD flies do not show a similar circular shape (Figures 5F,G). We also show that cnt1FD flies have a high proportion of miscoiled tails (87.8%) compared with w1118 (11.76%) and cnt1FD;cnt1::GFP (19.44%) (Supplementary Figure S5). This could be due to insufficient space in the TE region for new ICs to proceed with the remaining steps of spermatogenesis or because the tail of the Drosophila mature sperm has a defect in coiling due to the cnt1 mutation.
We also counted the number of spermatozoa in the SV to verify whether the observed tail defect in the TE region affected sperm migration. The results showed that sperm count was lower in the cnt1FD SV compared with the control. This may indicate that the fertility defect in cnt1FD males is partly due to insufficient sperm count in the SV (Supplementary Figure S6).
To further examine the reason for the accumulation of spermatids and the disorganization of mature sperms during coiling in the TE region of the cnt1 mutant, we tested whether the individualization process was affected. We used the death caspase-1 (dcp1) antibody, which stains CB and WB. In Drosophila, CB and WB are formed during the individualization of elongated spermatids. They are important for the removal of redundant cytoplasm and organelles that are degraded in the bag once they reach the end of the spermatid tail (Fabian and Brill, 2012). As shown in Figure 6A, cnt1FD has more WB than the control group. In addition, most of the CB did not have an oval shape but were irregular and left a trail as they moved toward the tail end of the spermatid (Figures 6B,C). This indicates that caspase signaling remained active in the individualized portion of the spermatids (Huh et al., 2004), and that the apoptotic-like process required for the elimination of excess organelles is impaired in cnt1FD mutants. This may be related to the disorganization of the tail of spermatids observed in cnt1FD in the TE region.
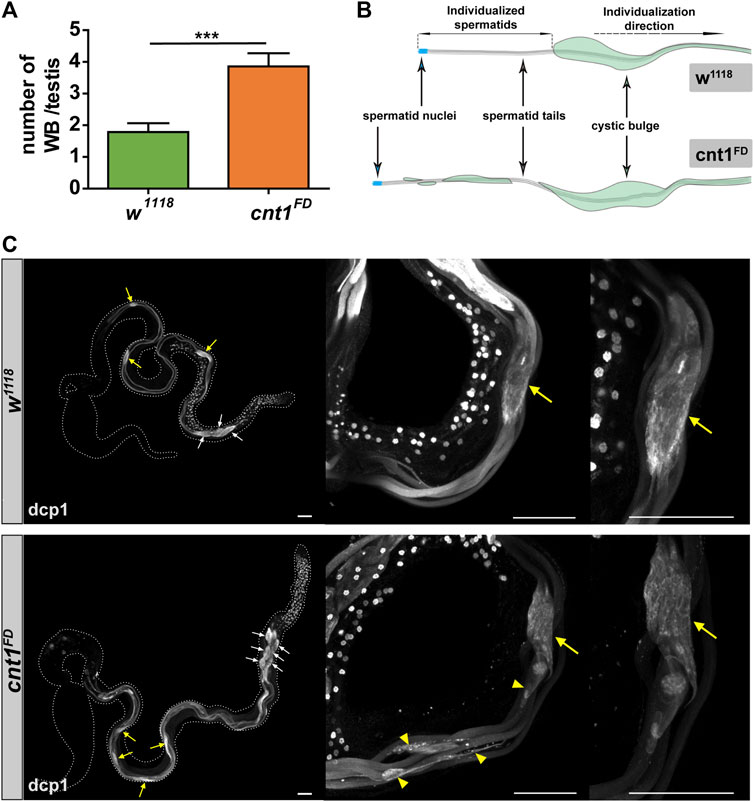
FIGURE 6. Cnt1 mutation impairs spermatid maturation during the spermatid individualization process. (A) Number of waste bags in w1118 and cnt1FD strains. Significance was analyzed using one-tailed Student’s t-test and labeled as follows: ***p < 0.001; n ≥ 10. Error bars are shown as mean ± SEM. (B) Illustration of the distorted shape of the cystic bulge in cnt1FD compared with w1118. (C) Testis staining with death caspase-1 antibody (white). Dcp1 signal is absent in the individualized part of the cyst in w1118, whereas it is present in cnt1 mutants (yellow arrowhead). The yellow arrows point to the cystic bulges, while the white arrows point to the waste bags. Scale bar: 50 µm.
To further explore the cause of the increased number of spermatids and the disorganization of their tails in the TE region, we checked the number of spermatids within their cyst group and the elongated spermatid shape under a transmission electron microscope in both control and mutant testes. This allowed us to examine the major spermatid structures, including the major and minor mitochondria and the axoneme. These components serve as structural support for the sustained elongation of the sperm tail and sperm motility in Drosophila (Porter, 1996; Noguchi et al., 2011). In the cross-section of the w1118 testis, the spermatid cysts revealed a group of 64 spermatids. Moreover, the structure of the elongated spermatids in the w1118 had all the components required to generate a healthy sperm (Figures 7A,B). By contrast, cnt1FD had a lower number of spermatids within a cyst, ranging from 47 to 64, compared to the controls (Figure 7A and Supplementary Figures S7A,B). Some of these cysts contained unusual shapes of elongated spermatids (Figures 7A,B and Supplementary Figure S7).
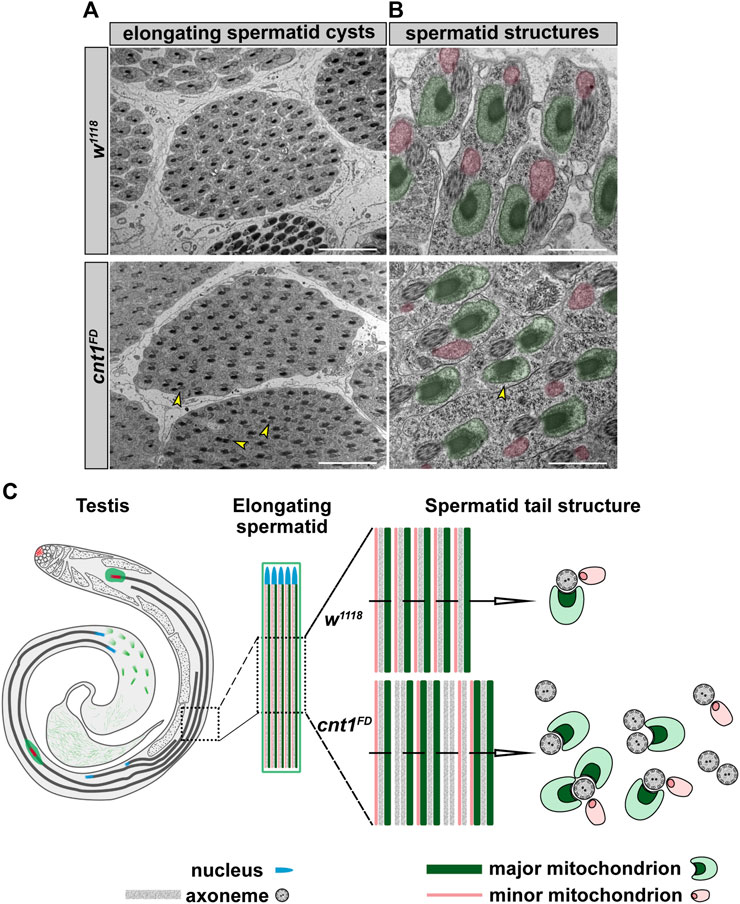
FIGURE 7. The cnt1 mutants exhibit mitochondrial defects. (A) Transversal section of w1118 and cnt1FD mutant testes showing a cyst of elongating spermatids. Scale bar: 5 µm. (B) W1118 shows a normal structure of an elongated spermatid tail with one axoneme, two major mitochondria, and one minor mitochondrion. cnt1FD shows an abnormal structure of an elongated spermatid tail with one or two axonemes and an abnormal number of mitochondrial subunits. Major mitochondria (highlighted in green), minor mitochondria (highlighted in pink). Scale bar: 1 µm. (C) Summary of observed mitochondrial phenotypes in the cnt1FD mutant compared with w1118.
The male cnt1FD mutant, in contrast to w1118 and cnt1FD;cnt1::GFP, exhibited spermatids with abnormal numbers of major and minor mitochondria (Figures 7B,C and Supplementary Figures S7A,C). In several cysts, some of the spermatid structures were in a common cytoplasm, indicating that they could not be separated from each other (Supplementary Figure S7A) (Huh et al., 2004). Fused spermatids and spermatids with altered number of mitochondria in cnt1FD were calculated and compared with those of control flies w1118 and cnt1FD;cnt1::GFP. We show that cnt1FD mutants have an average of two defective spermatids per cyst compared with the controls (Supplementary Figure S7D). These features may account for the failed coiling process and the presence of tangled sperm tails in the TE region, where they could not move toward SV. This could be due to abnormal mitochondrial activity and other physiological changes that could affect the energy balance of the spermatids.
To investigate the gene expression profile associated with the phenotype of the cnt1 mutant, we performed RNAseq of reproductive oragans cDNA libraries of cnt1FD mutants and the control w1118 group (three biological replicates for each cnt1FD and w1118 strain). RNAseq analysis identified 4,755 differentially expressed genes in the testes of the cnt1FD mutants, among which 2,472 were upregulated and 2,283 were downregulated (Supplementary Table S3). RNAseq data were validated through RT-qPCR (Supplementary Figure S8 and Supplementary Table S1). Ten representative genes were selected, and RT-qPCR showed concordant results with the RNA-sequencing data.
Pathview analysis of the differentially expressed genes (adj. p-value<0.05) revealed only one significant KEGG pathway, namely, the ribosomal pathway (dme03010). Two protein-coding ribosomal genes were upregulated, while 78 protein-coding ribosomal genes were downregulated (Supplementary Figure S9, Supplementary Table S3). By contrast, the expressions of genes encoding ribosomal RNAs of both subunits were stable (Supplementary Figure S9). The dysregulated genes were associated with both large and small ribosomal subunits involved in cytoplasmic translation (Supplementary Figures S9, S10). Subsequently, RNA sequencing data were further filtered for abs (log2FC) > 1 and annotated with GO terms from Drosophila melanogaster. The genes were sorted in a Venn diagram according to three main categories: reproduction (714 genes), immunity and Inflammation (346 genes), and metabolism (1733 genes) (Figure 8 and Supplementary Table S3). Among the genes in the common category, death regulator Nedd2-like caspase (dronc), and death-associated inhibitor of apoptosis 1 (diap1) were dysregulated. These genes have previously been linked to impaired apoptotic machinery during spermatid individualization (Huh et al., 2004). Data analysis also revealed altered expressions of a number of genes known from previous studies to be involved in male fertility, including sperm-specific dynein intermediate chain 4 (sdic4), a dynein gene essential for sperm tail motorization (Yeh et al., 2012).
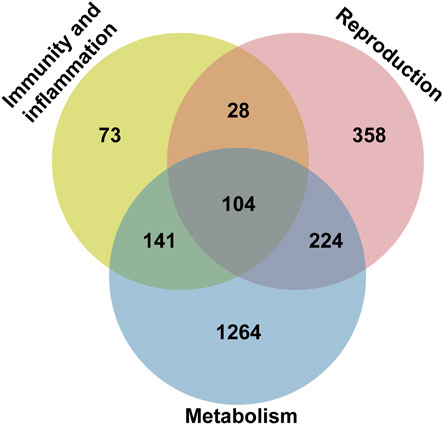
FIGURE 8. Functional classification of differentially regulated genes from the RNAseq data of cnt1FD and w1118 in a Venn diagram. The pie chart shows the distribution of genes in different categories with special emphasis on the reproduction domain. The Venn diagram shows genes with adj. p-value ≥ 0.05.
We have demonstrated that the cnt1 gene, which is almost entirely expressed in the testis, is required for male fertility in Drosophila. We created a mutation in cnt1 through targeted mutagenesis. The mutant showed alterations in reproductive behavior and defects in sperm maturation. Consequently, our characterization of the transcriptional responses to the cnt1 mutation revealed changes in the expression of key genes that were previously reported to play a role in the above-mentioned aspects of male fertility.
Our data indicate that male fertility caused by the cnt1 mutation is partly due to a behavioral defect involving a short copulation duration. Previous reports have shown that the duration of copulation in Drosophila is primarily controlled by males (MacBean and Parsons, 1967; Lee and Hall, 2001; Acebes et al., 2004), with a considerable period to ensure successful sperm transfer beginning 5 minutes after copulation (Gilchrist and Partridge, 2000; Crickmore and Vosshall, 2013). Abnormal copulation duration has been found in a number of mutants, including null mutants of the clock genes period and timeless (Beaver and Giebultowicz, 2004), and fru mutants (Lee et al., 2001). For example, fru mutation affect copulation by altering fru neurons. These neurons localized in the abdominal ganglion exhibit innervation at the SV, accessory gland, and end base of the testis. Interestingly, knocking down a subset of these neurons, called sAbg-1 using the UAS-tetanus toxin (TNT) resulted in shortened copulation duration (Jois et al., 2018).
We have previously shown that mutations in the equilibrative nucleoside transporter 2 (ent2) or the adenosine receptor (AdoR) of Drosophila impair synaptic transmission and memory (Knight et al., 2010). Therefore, we hypothesize that the cnt1FD induced loss of adenosine nucleoside transport in the testis may also impair adenosine signaling, thus affecting behavior (Leung et al., 2001). Accordingly, a recent study has shown that phosphodiesterase pde1, an enzyme that degrades cyclic AMP (a downstream signal of the adenosine receptor), causes male sterility and behavioral abnormalities that affect copulation in Drosophila (Morton et al., 2010). The adenosine pathway is known to cause neuromodulation in mammals and has been suggested to influence sex-dependent neuropsychiatric behavior and erection in humans (Phatarpekar et al., 2010; Osborne et al., 2018).
Our results further showed that the effects of the cnt1FD mutation are pleiotropic and include morphological changes during spermiogenesis, where spermatid maturation is defective. The most striking phenotype of the cnt1 mutation is a higher number of spermatid groups and mature sperms (Figure 5). We also showed that the tail coiling of mature sperm groups was disorganized and tangled, which probably prevented them from moving to the SV, where the sperm count was low. The higher number of spermatid groups contrasts with the low number of mature sperms in the SV, suggesting that spermatids accumulate in the TE region.
Thus, cnt1 appears to play an important role in regulating mechanical movements in maturing sperms, which reportedly depend on intact mitochondria and dynein proteins (Ranz et al., 2003; Yeh et al., 2012), the latter of which require high ATP concentrations for proper function (Xie et al., 2006). A previous study has shown that knockdown of ATP synthase results in male infertility and abnormal spermatogenesis in Drosophila testes (Yu et al., 2019). In addition, ATP synthase has been reported to affect the elongated shape of mitochondria when knocked out (Sawyer et al., 2017). This finding is consistent with our RNAseq data that shows the dysregulation of genes belonging to the mitochondrial respiratory chain (ATP synthase-coupling factor 6 (ATPsyn-Cf6), NADH dehydrogenase 75 (ND-75), cytochrome-c1 (cyt-c1), and cytochrome c distal (cyt-c-d)), of which cyt-c1 was confirmed by RT-qPCR to be downregulated in cnt1FD mutants (Supplementary Figure S8). In addition, previous studies have shown that mutations in dynein, such as dic61B, play a crucial role in sperm motility (Fatima, 2011). The dynein mutation in dic61B has been associated with impaired spermatid individualization and motility, which eventually led to male sterility. Furthermore, the dic61B mutation has also been reported to contribute to a defect in the formation of major and minor mitochondria derivatives (Fatima, 2011), supporting the idea that alterations in both dynein and mitochondria may also affect sperm axoneme movement in cnt1FD mutants. Among the dysregulated dynein genes in our RNAseq data, we found and confirmed that the gene-encoding sperm-specific dynein (sdic4) is upregulated in cnt1FD mutants. Taken together, mitochondria and dynein functions may be altered in cnt1FD mutants.
In addition, our data showed an increased number of waste bags, consistent with the observed high number of spermatid groups and low count of mature sperms in the TE region. Furthermore, these bags left a trail as they moved to the end of the spermatid tail, suggesting that the bags could not process their degradation properly. A similar phenotype to the cnt1FD mutant was previously observed in the driceless mutant and associated with unusual active caspase signaling (Huh et al., 2004). Further phenotypic analysis using electron microscopy confirmed that, similar to driceless mutants (Huh et al., 2004), the spermatid cysts in cnt1FD underwent partial individualization, with the spermatid cysts containing sheathed spermatids in a common cytoplasmic membrane, while the other spermatids have their own cytoplasmic membrane. These mutants also showed a defect in the cystic bulges with trailing edge (Huh et al., 2004). Consistently, our RNAseq data show a dysregulation of a key gene involved in the apoptotic machinery during spermatid individualization: dcp1 (Huh et al., 2004). It was reported in connection with the failure of the individualization process, like the cnt1FD mutant, in which multiple cysts contained many single spermatid units with excess cytoplasm.
Drosophila cnt1 and cnt2 are similar genes which most probably have the same biochemical function (nucleoside transport) but they differ by tissue specificity of expression. Their phenotypes seem to be quite independent. In our experiments, the removal of one copy of cnt2 did not enhance the cnt1 fertility phenotype the heteroallelic flies cnt1FD/Df(2R)BSC271 (this deletion lacks both cnt1 and cnt2) (Supplementary Figure S3).
Similar to Drosophila, the expressions of Cnt isoforms were detected in both human and rat testes, mainly in the Sertoli cells, which are essential for sperm nutrition (Klein et al., 2013; Hau et al., 2020). The pharmacological blockade of nucleoside transporters in Sertoli cells seems to interfere with the spermatid maturation process, suggesting that nucleoside transport may be important for providing the nucleosides essential for sperm maturation (Leung et al., 2001; Kato et al., 2005). Previous research in mice has shown the relationship between sperm motility and mitochondrial dysfunction (Cardullo and Baltz, 1991; Ruiz-Pesini et al., 1998). These findings argue for similarities in the regulation of spermatogenesis in Drosophila and mammals.
In summary, we show that cnt1 has a pleiotropic effect on mating behavior, spermatid maturation, and spermatid mitochondrial morphology in Drosophila. It affects the transcription of several key genes known from previous reports to be involved in these phenotypes, including dynein, ATP synthases, and apoptotic genes. Further work is needed to understand the relationship of these cnt1 phenotypes to adenosine signaling and related metabolic pathways.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: SRA data: PRJNA838856, https://www.ncbi.nlm.nih.gov/sra/PRJNA838856.
HOM conceived the project, performed the experiments, made the illustrations, and prepared the manuscript. LP performed the fertility assay and immunohistochemical staining. Y-HL generated the cnt1::GFP tagged flies. BC-HW and LR analyzed the RNAseq data. LK performed the phylogeny tree. LV and MR generated the mutants. HS performed the electron microscopy. MH performed the RNAseq. MZ supervised the project and manuscript preparation.
This work was supported by the European fund for regional development “Interreg Austria/Czech Republic” (REGGEN-ATCZ207) and the junior grant project GACR (19-13784Y to LK).
We thank Dr. Mihail Sarov (MPI-CBG) for providing us with the plasmids (tagging cassettes) used to generate the GFP tag (Sarov et al., 2016). We also acknowledge the core facility of the Laboratory of Electron Microscopy, Biology Centre CAS supported by MEYS CR (LM2015129 Czech‐BioImaging) and ERDF (No. CZ.02.1.01/0.0/0.0/13_013/0001445). We thank Jitka Pflegerova for her technical assistance and for preparing the TEM samples.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2022.945572/full#supplementary-material
Acebes, A., Grosjean, Y., Everaerts, C., and Ferveur, J. F. (2004). Cholinergic control of synchronized seminal emissions in Drosophila. Curr. Biol. 14, 704–710. doi:10.1016/j.cub.2004.04.003
Alzyoud, E., Vedelek, V., Réthi-Nagy, Z., Lipinszki, Z., and Sinka, R. (2021). Microtubule organizing centers contain testis-specific γ-turc proteins in spermatids of Drosophila. Front. Cell Dev. Biol. 9, 727264. doi:10.3389/fcell.2021.727264
Augière, C., Lapart, J.-A., Duteyrat, J.-L., Cortier, E., Maire, C., Thomas, J., et al. (2019). salto/CG13164 is required for sperm head morphogenesis in Drosophila. Mol. Biol. Cell 30, 636–645. doi:10.1091/mbc.E18-07-0429
Bader, M., Arama, E., and Steller, H. (2010). A novel F-box protein is required for caspase activation during cellular remodeling in Drosophila. Development 137, 1679–1688. doi:10.1242/dev.050088
Baker, B. S., Taylor, B. J., and Hall, J. C. (2001). Are complex behaviors specified by dedicated regulatory genes? Reasoning from Drosophila. Cell 105, 13–24. doi:10.1016/S0092-8674(01)00293-8
Baptissart, M., Vega, A., Martinot, E., and Volle, D. H. (2013). Male fertility: Is spermiogenesis the critical step for answering biomedical issues? Spermatogenesis 3, e24114. doi:10.4161/spmg.24114
Batut, B., Hiltemann, S., Bagnacani, A., Baker, D., Bhardwaj, V., Blank, C., et al. (2018). Community-driven data analysis training for biology. Cell Syst. 6, 752–758.e1. doi:10.1016/j.cels.2018.05.012
Beaver, L. M., and Giebultowicz, J. M. (2004). Regulation of copulation duration by period and timeless in Drosophila melanogaster. Curr. Biol. 14, 1492–1497. doi:10.1016/j.cub.2004.08.022
Cardullo, R. A., and Baltz, J. M. (1991). Metabolic regulation in mammalian sperm: Mitochondrial volume determines sperm length and flagellar beat frequency. Cell Motil. Cytoskelet. 19, 180–188. doi:10.1002/cm.970190306
Chen, X., Hiller, M., Sancak, Y., and Fuller, M. T. (2005). Tissue-specific TAFs counteract polycomb to turn on terminal differentiation. Science 310, 869–872. doi:10.1126/science.1118101
Chintapalli, V. R., Wang, J., and Dow, J. A. T. (2007). Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat. Genet. 39, 715–720. doi:10.1038/ng2049
Clark, K., Karsch-Mizrachi, I., Lipman, D. J., Ostell, J., and Sayers, E. W. (2016). GenBank. Nucleic Acids Res. 44, D67–D72. doi:10.1093/nar/gkv1276
Crickmore, M. A., and Vosshall, L. B. (2013). Opposing dopaminergic and GABAergic neurons control the duration and persistence of copulation in Drosophila. Cell 155, 881–893. doi:10.1016/j.cell.2013.09.055
Demarco, R. S., Eikenes, Å. H., Haglund, K., and Jones, D. L. (2014). Investigating spermatogenesis in Drosophila melanogaster. Methods 68, 218–227. doi:10.1016/j.ymeth.2014.04.020
Dos Santos-Rodrigues, A., Pereira, M. R., Brito, R., de Oliveira, N. A., and Paes-de-Carvalho, R. (2015). Adenosine transporters and receptors: Key elements for retinal function and neuroprotection. Vitam. Horm. 98, 487–523. doi:10.1016/bs.vh.2014.12.014
Edgar, R. C. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi:10.1093/nar/gkh340
Ejsmont, R. K., Ahlfeld, P., Pozniakovsky, A., Stewart, A. F., Tomancak, P., and Sarov, M. (2011). Recombination-mediated genetic engineering of large genomic DNA transgenes. Methods Mol. Biol. 772, 445–458. doi:10.1007/978-1-61779-228-1_26
Ejsmont, R. K., Sarov, M., Winkler, S., Lipinski, K. A., and Tomancak, P. (2009). A toolkit for high-throughput, cross-species gene engineering in Drosophila. Nat. Methods 6, 435–437. doi:10.1038/nmeth.1334
Fabian, L., and Brill, J. A. (2012). Drosophila spermiogenesis: Big things come from little packages. Spermatogenesis 2, 197–212. doi:10.4161/spmg.21798
Fatima, R. (2011). Drosophila Dynein intermediate chain gene, Dic61B, is required for spermatogenesis. PLoS One 6, e27822. doi:10.1371/journal.pone.0027822
Fleischmannova, J., Kucerova, L., Sandova, K., Steinbauerova, V., Broz, V., Simek, P., et al. (2012). Differential response of Drosophila cell lines to extracellular adenosine. Insect biochem. Mol. Biol. 42, 321–331. doi:10.1016/j.ibmb.2012.01.002
Gilchrist, A. S., and Partridge, L. (2000). Why it is difficult to model sperm displacement in Drosophila melanogaster: The relation between sperm transfer and copulation duration. Evolution 54, 534–542. doi:10.1111/j.0014-3820.2000.tb00056.x
Gray, J. H., Owen, R. P., and Giacomini, K. M. (2004). The concentrative nucleoside transporter family, SLC28. Pflugers Arch. 447, 728–734. doi:10.1007/s00424-003-1107-y
Groth, A. C., Fish, M., Nusse, R., and Calos, M. P. (2004). Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics 166, 1775–1782. doi:10.1534/genetics.166.4.1775
Guindon, S., Dufayard, J.-F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi:10.1093/sysbio/syq010
Hau, R. K., Miller, S. R., Wright, S. H., and Cherrington, N. J. (2020). Generation of a hTERT-immortalized human Sertoli cell model to study transporter dynamics at the blood-testis barrier. Pharmaceutics 12, E1005. doi:10.3390/pharmaceutics12111005
Huh, J. R., Vernooy, S. Y., Yu, H., Yan, N., Shi, Y., Guo, M., et al. (2004). Multiple apoptotic caspase cascades are required in nonapoptotic roles for Drosophila spermatid individualization. PLoS Biol. 2, E15. doi:10.1371/journal.pbio.0020015
Jayaramaiah Raja, S., and Renkawitz-Pohl, R. (2005). Replacement by Drosophila melanogaster protamines and Mst77F of histones during chromatin condensation in late spermatids and role of sesame in the removal of these proteins from the male pronucleus. Mol. Cell. Biol. 25, 6165–6177. doi:10.1128/MCB.25.14.6165-6177.2005
Jois, S., Chan, Y. B., Fernandez, M. P., and Leung, A. K.-W. (2018). Characterization of the sexually dimorphic fruitless neurons that regulate copulation duration. Front. Physiol. 9, 780. doi:10.3389/fphys.2018.00780
Kato, R., Maeda, T., Akaike, T., and Tamai, I. (2005). Nucleoside transport at the blood-testis barrier studied with primary-cultured Sertoli cells. J. Pharmacol. Exp. Ther. 312, 601–608. doi:10.1124/jpet.104.073387
Klein, D. M., Evans, K. K., Hardwick, R. N., Dantzler, W. H., Wright, S. H., and Cherrington, N. J. (2013). Basolateral uptake of nucleosides by Sertoli cells is mediated primarily by equilibrative nucleoside transporter 1. J. Pharmacol. Exp. Ther. 346, 121–129. doi:10.1124/jpet.113.203265
Klein, D. M., Harding, M. C., Crowther, M. K., and Cherrington, N. J. (2017). Localization of nucleoside transporters in rat epididymis. J. Biochem. Mol. Toxicol. 31, e21911. doi:10.1002/jbt.21911
Knight, D., Harvey, P. J., Iliadi, K. G., Klose, M. K., Iliadi, N., Dolezelova, E., et al. (2010). Equilibrative nucleoside transporter 2 regulates associative learning and synaptic function in Drosophila. J. Neurosci. 30, 5047–5057. doi:10.1523/JNEUROSCI.6241-09.2010
Kondo, S., and Ueda, R. (2013). Highly improved gene targeting by germline-specific Cas9 expression in Drosophila. Genetics 195, 715–721. doi:10.1534/genetics.113.156737
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi:10.1093/molbev/msy096
Larkin, A., Marygold, S. J., Antonazzo, G., Attrill, H., dos Santos, G., Garapati, P. V., et al. (2021). FlyBase: Updates to the Drosophila melanogaster knowledge base. Nucleic Acids Res. 49, D899–D907. doi:10.1093/nar/gkaa1026
Leatherman, J. L., and Dinardo, S. (2008). Zfh-1 controls somatic stem cell self-renewal in the Drosophila testis and nonautonomously influences germline stem cell self-renewal. Cell Stem Cell 3, 44–54. doi:10.1016/j.stem.2008.05.001
Lee, G., and Hall, J. C. (2001). Abnormalities of male-specific FRU protein and serotonin expression in the CNS of fruitless mutants in Drosophila. J. Neurosci. 21, 513–526. doi:10.1523/JNEUROSCI.21-02-00513.2001
Lee, G., Villella, A., Taylor, B. J., and Hall, J. C. (2001). New reproductive anomalies in fruitless-mutant Drosophila males: Extreme lengthening of mating durations and infertility correlated with defective serotonergic innervation of reproductive organs. J. Neurobiol. 47, 121–149. doi:10.1002/neu.1021
Lefort, V., Longueville, J.-E., and Gascuel, O. (2017). SMS: Smart model selection in PhyML. Mol. Biol. Evol. 34, 2422–2424. doi:10.1093/molbev/msx149
Leung, G. P., Ward, J. L., Wong, P. Y., and Tse, C. M. (2001). Characterization of nucleoside transport systems in cultured rat epididymal epithelium. Am. J. Physiol. Cell Physiol. 280, C1076–C1082. doi:10.1152/ajpcell.2001.280.5.C1076
Lin, Y.-H., Maaroufi, H. O., Ibrahim, E., Kucerova, L., and Zurovec, M. (2019). Expression of human mutant huntingtin protein in Drosophila hemocytes impairs immune responses. Front. Immunol. 10, 2405. doi:10.3389/fimmu.2019.02405
Lin, Y.-H., Maaroufi, H. O., Kucerova, L., Rouhova, L., Filip, T., and Zurovec, M. (2021). Adenosine receptor and its downstream targets, mod(mdg4) and Hsp70, work as a signaling pathway modulating cytotoxic damage in Drosophila. Front. Cell Dev. Biol. 9, 651367. doi:10.3389/fcell.2021.651367
Lindsley, D. I., and Tokuyasu, K. T. (1980). “Spermatogenesis,” in Genetics and Biology of Drosophila (New York: Academic Press), 225–294.
Lotti, F., and Maggi, M. (2018). Sexual dysfunction and male infertility. Nat. Rev. Urol. 15, 287–307. doi:10.1038/nrurol.2018.20
MacBean, I. T., and Parsons, P. A. (1967). Directional selection for duration of copulation in Drosophila melanogaster. Genetics 56, 233–239. doi:10.1093/genetics/56.2.233
Machado, J., Abdulla, P., Hanna, W. J. B., Hilliker, A. J., and Coe, I. R. (2007). Genomic analysis of nucleoside transporters in Diptera and functional characterization of DmENT2, a Drosophila equilibrative nucleoside transporter. Physiol. Genomics 28, 337–347. doi:10.1152/physiolgenomics.00087.2006
Masino, S., and Boison, D. (2013). Adenosine: A key link between metabolism and brain activity. New York, NY: Springer. doi:10.1007/978-1-4614-3903-5
Molina-Arcas, M., and Pastor-Anglada, M. (2013). Nucleoside transporters (SLC28 and SLC29) family. Pharmacogenomics of Human Drug Transporters. 11, 243–270. doi:10.1002/9781118353240.ch11
Morton, D. B., Clemens-Grisham, R., Hazelett, D. J., and Vermehren-Schmaedick, A. (2010). Infertility and male mating behavior deficits associated with Pde1c in Drosophila melanogaster. Genetics 186, 159–165. doi:10.1534/genetics.110.118018
Nagarkar-Jaiswal, S., Lee, P.-T., Campbell, M. E., Chen, K., Anguiano-Zarate, S., Cantu Gutierrez, M., et al. (2015). A library of MiMICs allows tagging of genes and reversible, spatial and temporal knockdown of proteins in Drosophila. Elife 4, e05338. doi:10.7554/eLife.05338
Ng, C. L., Qian, Y., and Schulz, C. (2019). Notch and Delta are required for survival of the germline stem cell lineage in testes of Drosophila melanogaster. PLoS One 14, e0222471. doi:10.1371/journal.pone.0222471
Noguchi, T., Koizumi, M., and Hayashi, S. (2011). Sustained elongation of sperm tail promoted by local remodeling of giant mitochondria in Drosophila. Curr. Biol. 21, 805–814. doi:10.1016/j.cub.2011.04.016
Noguchi, T., and Miller, K. G. (2003). A role of actin dynamics in individualization during spermatogenesis in Drosophila melanogaster. Development 130, 1805–1816. doi:10.1242/dev.00406
Osborne, D. M., Sandau, U. S., Jones, A. T., Vander Velden, J. W., Weingarten, A. M., Etesami, N., et al. (2018). Developmental role of adenosine kinase for the expression of sex-dependent neuropsychiatric behavior. Neuropharmacology 141, 89–97. doi:10.1016/j.neuropharm.2018.08.025
Pavlou, H. J., and Goodwin, S. F. (2013). Courtship behavior in Drosophila melanogaster: Towards a ‘courtship connectome. Curr. Opin. Neurobiol. 23, 76–83. doi:10.1016/j.conb.2012.09.002
Pérez-Torras, S., Mata-Ventosa, A., Drögemöller, B., Tarailo-Graovac, M., Meijer, J., Meinsma, R., et al. (2019). Deficiency of perforin and hCNT1, a novel inborn error of pyrimidine metabolism, associated with a rapidly developing lethal phenotype due to multi-organ failure. Biochim. Biophys. Acta. Mol. Basis Dis. 1865, 1182–1191. doi:10.1016/j.bbadis.2019.01.013
Phatarpekar, P. V., Wen, J., and Xia, Y. (2010). Role of adenosine signaling in penile erection and erectile disorders. J. Sex. Med. 7, 3553–3564. doi:10.1111/j.1743-6109.2009.01555.x
Porter, M. E. (1996). Axonemal dyneins: Assembly, organization, and regulation. Curr. Opin. Cell Biol. 8, 10–17. doi:10.1016/S0955-0674(96)80042-1
Ranz, J. M., Ponce, A. R., Hartl, D. L., and Nurminsky, D. (2003). Origin and evolution of a new gene expressed in the Drosophila sperm axoneme. Genetica 118, 233–244. doi:10.1023/A:1024186516554
Resende, L. P. F., Boyle, M., Tran, D., Fellner, T., and Jones, D. L. (2013). Headcase promotes cell survival and niche maintenance in the Drosophila testis. PLoS One 8, e68026. doi:10.1371/journal.pone.0068026
Rideout, E. J., Billeter, J.-C., and Goodwin, S. F. (2007). The sex-determination genes fruitless and doublesex specify a neural substrate required for courtship song. Curr. Biol. 17, 1473–1478. doi:10.1016/j.cub.2007.07.047
Ruiz-Pesini, E., Diez, C., Lapeña, A. C., Pérez-Martos, A., Montoya, J., Alvarez, E., et al. (1998). Correlation of sperm motility with mitochondrial enzymatic activities. Clin. Chem. 44, 1616–1620. doi:10.1093/clinchem/44.8.1616
Santel, A., Blümer, N., Kämpfer, M., and Renkawitz-Pohl, R. (1998). Flagellar mitochondrial association of the male-specific Don Juan protein in Drosophila spermatozoa. J. Cell Sci. 111 (2), 3299–3309. doi:10.1242/jcs.111.22.3299
Santel, A., Winhauer, T., Blümer, N., and Renkawitz-Pohl, R. (1997). The Drosophila don juan (dj) gene encodes a novel sperm specific protein component characterized by an unusual domain of a repetitive amino acid motif. Mech. Dev. 64, 19–30. doi:10.1016/s0925-4773(97)00031-2
Sarov, M., Barz, C., Jambor, H., Hein, M. Y., Schmied, C., Suchold, D., et al. (2016). A genome-wide resource for the analysis of protein localisation in Drosophila. Elife 5, e12068. doi:10.7554/eLife.12068
Sawyer, E. M., Brunner, E. C., Hwang, Y., Ivey, L. E., Brown, O., Bannon, M., et al. (2017). Testis-specific ATP synthase peripheral stalk subunits required for tissue-specific mitochondrial morphogenesis in Drosophila. BMC Cell Biol. 18, 16. doi:10.1186/s12860-017-0132-1
Schneider, C. A., Rasband, W. S., and Eliceiri, K. W. (2012). NIH image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. doi:10.1038/nmeth.2089
Siegel, R. W., and Hall, J. C. (1979). Conditioned responses in courtship behavior of normal and mutant Drosophila. Proc. Natl. Acad. Sci. U. S. A. 76, 3430–3434. doi:10.1073/pnas.76.7.3430
Soulavie, F., Piepenbrock, D., Thomas, J., Vieillard, J., Duteyrat, J.-L., Cortier, E., et al. (2014). Hemingway is required for sperm flagella assembly and ciliary motility in Drosophila. Mol. Biol. Cell 25, 1276–1286. doi:10.1091/mbc.e13-10-0616
Steinhauer, J. (2015). Separating from the pack: Molecular mechanisms of Drosophila spermatid individualization. Spermatogenesis 5, e1041345. doi:10.1080/21565562.2015.1041345
Tokuyasu, K. T. (1975). Dynamics of spermiogenesis in Drosophila melanogaster. VI. Significance of “onion” nebenkern formation. J. Ultrastruct. Res. 53, 93–112. doi:10.1016/S0022-5320(75)80089-X
Vedelek, V., Bodai, L., Grézal, G., Kovács, B., Boros, I. M., Laurinyecz, B., et al. (2018). Analysis of Drosophila melanogaster testis transcriptome. BMC Genomics 19, 697. doi:10.1186/s12864-018-5085-z
Wakimoto, B. T., Lindsley, D. L., and Herrera, C. (2004). Toward a comprehensive genetic analysis of male fertility in Drosophila melanogaster. Genetics 167, 207–216. doi:10.1534/genetics.167.1.207
White-Cooper, H. (2004). Spermatogenesis: Analysis of meiosis and morphogenesis. Methods Mol. Biol. 247, 45–75. doi:10.1385/1-59259-665-7:45
Witt, E., Benjamin, S., Svetec, N., and Zhao, L. (2019). Testis single-cell RNA-seq reveals the dynamics of de novo gene transcription and germline mutational bias in Drosophila. Elife 8, e47138. doi:10.7554/eLife.47138
Xie, P., Dou, S.-X., and Wang, P.-Y. (2006). Model for unidirectional movement of axonemal and cytoplasmic dynein molecules. Acta Biochim. Biophys. Sin. 38, 711–724. doi:10.1111/j.1745-7270.2006.00223.x
Yeh, S.-D., Do, T., Abbassi, M., and Ranz, J. M. (2012). Functional relevance of the newly evolved sperm dynein intermediate chain multigene family in Drosophila melanogaster males. Commun. Integr. Biol. 5, 462–465. doi:10.4161/cib.21136
Young, J. D. (2016). The SLC28 (CNT) and SLC29 (ENT) nucleoside transporter families: A 30-year collaborative odyssey. Biochem. Soc. Trans. 44, 869–876. doi:10.1042/BST20160038
Young, J. D., Yao, S. Y. M., Baldwin, J. M., Cass, C. E., and Baldwin, S. A. (2013). The human concentrative and equilibrative nucleoside transporter families, SLC28 and SLC29. Mol. Asp. Med. 34, 529–547. doi:10.1016/j.mam.2012.05.007
Yu, J., Chen, B., Zheng, B., Qiao, C., Chen, X., Yan, Y., et al. (2019). ATP synthase is required for male fertility and germ cell maturation in Drosophila testes. Mol. Med. Rep. 19, 1561–1570. doi:10.3892/mmr.2019.9834
Yuan, X., Zheng, H., Su, Y., Guo, P., Zhang, X., Zhao, Q., et al. (2019). Drosophila Pif1A is essential for spermatogenesis and is the homolog of human CCDC157, a gene associated with idiopathic NOA. Cell Death Dis. 10, 125. doi:10.1038/s41419-019-1398-3
Zhang, S. D., and Odenwald, W. F. (1995). Misexpression of the white (w) gene triggers male-male courtship in Drosophila. Proc. Natl. Acad. Sci. U. S. A. 92, 5525–5529. doi:10.1073/pnas.92.12.5525
Zhao, J., Klyne, G., Benson, E., Gudmannsdottir, E., White-Cooper, H., and Shotton, D. (2010). FlyTED: The Drosophila testis gene expression database. Nucleic Acids Res. 38, D710–D715. doi:10.1093/nar/gkp1006
Keywords: cnt1, gamete, spermatogenesis, testis, adenosine, copulation, mitochondria, male fertility
Citation: Maaroufi HO, Pauchova L, Lin Y-H, Wu BC-H, Rouhova L, Kucerova L, Vieira LC, Renner M, Sehadova H, Hradilova M and Zurovec M (2022) Mutation in Drosophila concentrative nucleoside transporter 1 alters spermatid maturation and mating behavior. Front. Cell Dev. Biol. 10:945572. doi: 10.3389/fcell.2022.945572
Received: 16 May 2022; Accepted: 27 July 2022;
Published: 23 August 2022.
Edited by:
Sung Min Han, University of Florida, United StatesReviewed by:
Paul Lasko, McGill University, CanadaCopyright © 2022 Maaroufi, Pauchova, Lin, Wu, Rouhova, Kucerova, Vieira, Renner, Sehadova, Hradilova and Zurovec. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michal Zurovec, enVyb3ZlY0BlbnR1LmNhcy5jeg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.