
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol., 11 August 2022
Sec. Morphogenesis and Patterning
Volume 10 - 2022 | https://doi.org/10.3389/fcell.2022.944016
This article is part of the Research TopicPattern Formation in BiologyView all 13 articles
Vertebrate embryo somitogenesis is the earliest morphological manifestation of the characteristic patterned structure of the adult axial skeleton. Pairs of somites flanking the neural tube are formed periodically during early development, and the molecular mechanisms in temporal control of this early patterning event have been thoroughly studied. The discovery of a molecular Embryo Clock (EC) underlying the periodicity of somite formation shed light on the importance of gene expression dynamics for pattern formation. The EC is now known to be present in all vertebrate organisms studied and this mechanism was also described in limb development and stem cell differentiation. An outstanding question, however, remains unanswered: what sets the different EC paces observed in different organisms and tissues? This review aims to summarize the available knowledge regarding the pace of the EC, its regulation and experimental manipulation and to expose new questions that might help shed light on what is still to unveil.
• The vertebrate Embryo Clock oscillates with species-specific periodicity
• Embryo Clock periodicity is tissue-specific within the same organism
• A comprehensive concept of the Embryo Clock is presented
Vertebrate embryo development comprises several processes that are highly regulated in time. One such process is somitogenesis, which is characterized by the periodic formation of metameric structures, the somites, along the anterior-to-posterior (A-P) axis of the early embryonic body. Somites are formed in pairs from the anterior-most portion of the presomitic mesoderm (PSM), on each side of the neural tube, and they are the first morphological manifestation of the characteristic segmented structure of the adult vertebrate axial skeleton. In fact, somites not only give rise to the axial skeleton and skeletal musculature, but also impose the segmented organization of the peripheral nervous system (Keynes and Stern, 1988). Most importantly to the subject of this review, somite pairs are formed sequentially, over time, while the embryonic body is elongating in an A-P direction. This is characteristic of all vertebrates, although the pace at which somites are formed varies among species (Table 1).
In 1976, Cooke and Zeeman proposed a theoretical model that aimed to explain the formation of periodic structures during vertebrate development. In their Clock and Wavefront model (Cooke and Zeeman, 1976), the authors proposed the existence of two players: a molecular oscillator (clock), responsible for the rhythmic generation of a cell responsive state, and a maturation wavefront, moving slowly in an anterior-to-posterior direction. Exposure of a clock-induced cell population to the wavefront signal would promote a rapid change in cell properties, leading to the formation of a somite. Together, these two components would translate temporal information into a spatial pattern. According to this model, somite size and number are jointly determined by the period of the clock’s oscillations and the speed of the moving wavefront (Cooke and Zeeman, 1976; Oates et al., 2012). However, breakthroughs regarding the identity of the molecules comprising the Clock and the Wavefront were only made 20 years later.
The Embryo Clock (EC)—or the developmental clock, as it was first termed–arose from the discovery that the mRNA of chick hairy1 (now termed hes4), a member of the Hairy Enhancer of Split (HES) transcription factor family, oscillated in the chicken embryo PSM with a 90 min periodicity, concomitant with the formation of a new pair of somites (Palmeirim et al., 1997). In their study, the authors first observed that chicken embryos with the same number of somites (i.e., within the same developmental stage) displayed very different patterns of hairy1 expression, leading them to hypothesize that its expression could be cyclic. Indeed, by bisecting the embryo, and culturing one half for a given time while the other was immediately fixed, hairy1 expression recapitulated after 90 min. Moreover, hairy1 oscillations in the PSM were found to be an intrinsic property of the system, as they were maintained even when the PSM was sectioned in smaller pieces or isolated from the surrounding tissues (Palmeirim et al., 1997). Since then, many genes that display an oscillatory behaviour during somitogenesis have been identified in multiple organisms, evidencing that the EC underlying somitogenesis is a conserved mechanism among vertebrates (Krol et al., 2011).
The first evidence for a Wavefront in control of somite formation was provided soon after (Dubrulle et al., 2001 in chick; Sawada et al., 2001 in zebrafish). A gradient of fgf8 mRNA (chick) and signalling activity (zebrafish) was described, with high levels at the embryo tail bud decreasing towards the anterior PSM. Local inhibition of FGF8 signalling in the anterior PSM resulted in longer somites, suggesting an instructive role for FGF signalling in positioning the somitic boundary (Dubrulle et al., 2001; Sawada et al., 2001). This was consistent with what was previously proposed for the wavefront activity (Cooke and Zeeman, 1976). Further studies elucidated that the chick fgf8 mRNA gradient resulted from the production of stable mRNA transcripts in the tail bud region alone, that degraded over time as the embryo elongated posteriorly, leading to less mRNA levels in the anterior PSM relative to the posterior region (Dubrulle and Pourquié, 2004). Graded Wnt activity and an opposing, anterior-to-posterior gradient of retinoic acid signalling were further shown to have wavefront activity in defining somite boundary positioning (reviewed in Resende et al., 2014).
This review reunites and summarizes key findings on Embryo Clock operation over the last 25 years, since hes4 oscillations in the chicken embryo were first described (Palmeirim et al., 1997). Originally termed “developmental clock” (Palmeirim et al., 1997), then “segmentation clock” (McGrew et al., 1998) and “somitogenesis clock” (Leimeister et al., 2000), herein we employ a more comprehensive concept of “Embryo Clock,” since oscillations in clock gene expression have been described in cells and developmental stages that are not associated with embryo somite formation (discussed below). We propose the term Embryo Clock to refer to the system of molecular oscillators operating in embryonic cells undergoing temporally controlled morphogenetic processes and/or cell fate specification. These genetic (or, in some cases, biochemical) oscillators exhibit periodic alterations (in contrast to stochastic pulses) that are maintained by negative feedback regulation. Due to the extensive and growing number of studies performed on the subject, we have focused our attention on the temporal dynamics of the EC. We aim to provide an overview of the main factors contributing to the exquisite temporal properties of this biological oscillator and anticipate this will be a useful roadmap for researchers interested in this increasingly exciting scientific field.
After the description of the first segmentation clock gene, hairy1, in the chicken embryo (Palmeirim et al., 1997), similar oscillatory patterns of expression were identified for other genes, and in multiple organisms (Table 2). The use of genome-wide approaches identified a wide range of genes with oscillatory gene expression during somitogenesis and evidenced that the embryo clock is an intricate oscillatory genetic network, that comprises genes belonging to multiple signalling pathways, notably, Notch, Wnt and FGF (Dequéant et al., 2006; Krol et al., 2011) (Figure 1A). These include Wnt-dependent Axin2, FGF signalling pathway genes Dusp1/2/4/6, Snail1/2, Spry2/4, and Notch pathway genes of the Already defined earlier. HES family, Lfng and Nrarp, among others. Strikingly, only two genes–the Hes1 and Hes5 orthologs–were conserved in mouse, chicken and zebrafish. Otherwise, the identity of the pathway-specific oscillating genes varied considerably, evidencing evolutionary plasticity of the conserved oscillations in signalling pathway activity (Krol et al., 2011). Several studies have shown that these intercellular communication pathways cooperate during embryo body segmentation. Niwa et al. (2007), (2011) showed that the onset of Hes7 expression in the mouse tailbud is FGF-dependent, while its maintenance and propagation throughout the PSM requires Notch signalling. A gradient of nuclear Wnt-related β-catenin was shown to control key features of PSM maturation and somite formation (Aulehla et al., 2008). Notch- and Wnt-dependent gene expression oscillations are coupled in the PSM and undergo a phase shift towards the anterior PSM. Inhibition of this phase shift in an in vitro setting delayed the arrest of EC waves and impaired tissue segmentation (Sonnen et al., 2018). Identification of EC oscillatory dynamics at the protein level has lagged behind, mostly due to the lack of appropriate antibodies and because it is technically more challenging than the well-established in situ hybridization protocols for RNA detection. However, corresponding cycles of protein expression with the same periodicity have been reported and are summarized in Table 3.
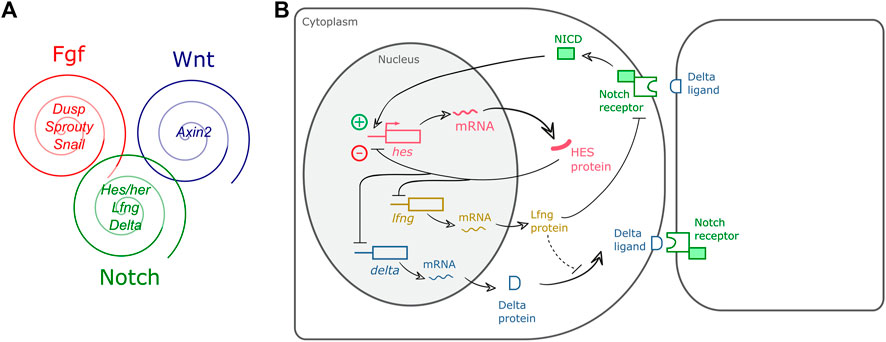
FIGURE 1. Embryo Clock (EC) gene expression oscillations. (A) The EC encompasses oscillatory genes belonging to the Fgf, Wnt and Notch signalling pathways (representative genes are indicated); (B) Negative feedback regulation of hairy-enhancer-of-split (HES) oscillations. In PSM cells, hes transcription is induced by pulses of intercellular Notch-Delta signalling, leading to HES protein production. HES protein enters the nucleus and represses its own promoter. HES protein and mRNA are rapidly degraded allowing for a new cycle of expression. HES also inhibits delta and lfng expression ensuring coupled oscillations in neighbour cells of the tissue. Dashed line represents a delay imposed on Delta integration in the cell membrane (Yoshioka-Kobayashi et al., 2020). NICD: Notch intracellular domain.
Several studies have identified negative feedback regulation as a fundamental feature of EC oscillations (Hirata et al., 2002; Bessho et al., 2003; Lewis, 2003; Chen et al., 2005). Figure 1B presents a simplified view of the negative feedback regulatory mechanisms underlying HES gene expression oscillations, whereby hes oscillations are maintained by an inhibitory action of the HES protein on its own promoter. HES also inhibits delta expression and/or expression of the Notch-modulator Lunatic fringe–Lfng, which contributes to synchronized oscillations of Notch-dependent gene expression in neighbour cells. Rapid degradation of the molecular products produced, mRNAs and proteins, ensures propagation of the oscillatory behaviour (reviewed in Kageyama et al., 2012).
Using mathematical modelling, Lewis (2003), Monk (2003) and Jensen et al. (2003) independently postulated that oscillations in gene expression are influenced by delays in the various steps of the regulatory negative feedback loop. Indeed, further experimental evidence showed that the EC mechanism depends on Delayed Negative Feedback loops and that the temporal delays are introduced in multiple steps of the process. Namely:
Lewis (2003) proposed that the time it takes to synthesize a transcript was one of the major accountants for transcriptional delay, so one would assume lengthier genes would have larger transcriptional delays. Elongation was, however, not found to have a major contribution to these delays–RNA polymerase II elongation rate measured in intact zebrafish embryos showed that the time needed to transcribe her1 and her7 is negligible and elongation kinetics of Hes7 and Lfng determined using mouse cells also occurred at a fast rate (Hanisch et al., 2013; Hoyle and Ish-Horowicz, 2013). Besides the elongation rate, however, there are other factors that significantly influence the transcriptional delay, namely mRNA nuclear export and mRNA splicing. Giudicelli et al. (2007) experimentally observed a delay from nuclear mRNA production to mature mRNA detection in the cytoplasm of her1, her7 and deltaC–zebrafish’s key clock components. Takashima et al. (2011) addressed the contribution of mRNA splicing to gene expression oscillations. They used transgenic mice carrying all or none of Hes7 introns, together with a luciferase reporter, and assessed the time of Hes7 transcription and protein production in both conditions. Mice carrying all Hes7 introns showed a delay of approximately 19 min in Hes7 expression, when compared to the intron-null mice. When abolishing this delay in a mathematical model, Hes7 oscillations were abolished, and this was confirmed in the mutant animals. Hoyle and Ish-Horowicz (2013) corroborated that mRNA splicing and nuclear export account for most of the EC transcriptional delay. Additionally, they compared the splicing and export delays in mouse, chicken and zebrafish, and concluded that organisms that have longer delays in these processes also present longer clock periods.
Another aspect to take into consideration are the half-life times of EC mRNAs and proteins. Due to the inhibitory action of EC products on their own transcription, the time required for their clearance from the cell will directly impact the rate at which a new cycle of gene expression is initiated. Multiple factors that contribute to differential mRNA stability were experimentally assessed for their involvement in EC regulation. These include the mRNA 3′ untranslated region (3′UTR), polyA tail length and microRNA-mediated degradation.
While studying the mechanisms that control segmental gene expression in Xenopus, Davis et al. (2001) found that hes4 (formerly known as hairy2a) expression dynamics was influenced by its 3′UTR sequence. When the 3′UTR of hes4 was substituted by the 3′UTR of other hes genes (either from Xenopus or other vertebrate species), hes4 expression retained its characteristic striped pattern in the PSM, unlike what happened when the 3′UTR of constitutively expressed genes was used. The authors further identified a phylogenetically conserved 25 bp sequence in the 3′UTR of EC genes which was necessary and sufficient to confer instability to these transcripts (Davis et al., 2001). Similar findings were reported by Hilgers and colleagues (2005) using an in vivo inducible system to halt transcription and measure mRNA degradation rate in the chicken embryo. They clearly showed that the 3′UTR of the EC gene Lfng promoted rapid mRNA decay, while the 3′UTR of fgf8 mRNA contributed to stabilization of the reporter mRNA (Hilgers et al., 2005), which is compatible with fgf8 graded expression pattern in the PSM (Dubrulle and Pourquié, 2004). Similar findings were reported for zebrafish EC genes (Fujino et al., 2018), evidencing that 3′UTR-mediated regulation of EC gene expression oscillations is a conserved feature in vertebrates. The Amacher lab went on to specify that mRNA decay of both zebrafish her1 and deltaC relies on the Pumilio response- and AU-rich-elements present in their distal 3′UTRs, in a Pnrc2-dependent manner (Gallagher et al., 2017; Tietz et al., 2020).
Different EC genes with the same periodicity in the PSM can nevertheless present very different expression patterns. Nitanda et al. (2014) explored this feature focussing on Hes7 and Lfng in the mouse PSM. After bisecting the PSM and culturing one half in actinomycin D to inhibit transcription, while the other was immediately fixed, quantitative PCR analysis showed that Lfng mRNA is less stable than Hes7 mRNA. This was attributed to the 3′UTR, as demonstrated using cells transfected with a reporter vector containing either Hes7 or Lfng 3′UTRs and monitoring mRNA degradation. The authors then generated transgenic mice lines, both containing a reporter gene driven by the Hes7 promoter, but with different 3′UTRs–one from Hes7, and another from Lfng. The transgenic line with the Lfng 3′UTR showed a severe reduction in reporter mRNA, further confirming the role of the 3′UTR in promoting rapid mRNA decay. Importantly, the reporter mRNA presented the same expression pattern as its 3′UTR-donor gene, i.e., the Lfng 3′UTR-reporter displayed the same pattern as endogenous Lfng, and this was also true for the Hes7 3′UTR-transgene (Nitanda et al., 2014). These results strongly suggest that 3′UTR-mediated mRNA stability defines both the temporal and spatial properties of EC oscillations in the PSM.
Fujimuro et al. (2014) showed that Hes7 3′UTR is also required for the production of proper amounts of Hes7 protein to maintain oscillations. In the absence of the 3′UTR, Hes7 mRNA no longer displayed cyclic expression patterns. The authors found that transcription levels of Hes7 mRNA were reduced, and that Hes7 protein was hardly detectable in the mouse PSM, compared to wild-type embryos. As expected, since the protein was not being correctly produced, Hes7 transcription inhibition was impaired, which compromised the maintenance of the oscillations (Fujimuro et al., 2014).
Work performed by Fujino and colleagues (2018) suggested that poly(A) tail length could also be important for EC mRNA rapid turnover. These authors measured the lengths of the poly(A) tails of zebrafish her1, her7 and hes6, and observed that the first two genes, that display cyclic expression in the PSM, have shorter poly(A) tails, while hes6 that is expressed in a gradient has a longer one. Through the inhibition of the deadenylase complex CCR4-NOT, the authors were able to lengthen the poly(A) tails of her1 and her7, and this resulted in a 2-3-fold increase in mRNA levels, indicating an increase in mRNA stability (Fujino et al., 2018).
Finally, EC mRNA degradation rate is also regulated by microRNAs (miRNAs). Xie et al. (2007) were the first to theoretically propose a role for miRNAs in EC delayed negative feedback regulation. Experimental evidence for oscillatory gene modulation by miRNAs was provided by Bonev et al. (2012), who reported that mouse Hes1 mRNA is a direct target of microRNA-9 (miR-9). Hes1 oscillations were dampened either when mir-9 was overexpressed or its binding to Hes1 was inhibited, suggesting that Hes1 oscillations are maintained within a certain range of miR-9 levels. This is ensured by negative feedback of Hes1 on the production of miR-9 primary transcripts, generating a double-negative feedback loop. Although the pri-miR-9 and pre-miR-9 are processed and cleared at a fast rate, the same is not true for the mature miR-9 which accumulates in the cell over time. Hence, a self-limiting oscillator model was proposed, whereby when miR-9 levels reach a certain threshold, Hes1 is permanently downregulated and NPC differentiation occurs (Bonev et al., 2012). Similar findings were further reported in zebrafish hindbrain development. Here, miR-9 acts on her6 to ensure robust oscillatory expression during neural progenitor cell differentiation (Soto et al., 2020).
During somitogenesis, miR-125a-5p is expressed in the chicken PSM where it targets the Lfng 3′UTR (Riley et al., 2013). Inhibition of chicken miR-125a-5p activity resulted in abnormal somite segmentation, resembling the phenotype obtained when Lfng was ubiquitously expressed in the chicken PSM (Dale et al., 2003). This is consistent with a role for miR-125a-5p in promoting Lfng mRNA decay. Moreover, Lfng and hairy1 lost their oscillatory expression pattern, further evidencing that miRNA-mediated regulation is necessary for EC gene expression oscillations (Riley et al., 2013). A regulatory action of miR-125a-5p on Lfng mRNA degradation and expression dynamics was also documented in the mouse embryo (Wahi et al., 2017). Mathematical modelling performed by Jing et al. (2015) provided important insights regarding miRNA role in the segmentation clock. Their work suggests that the interaction between Lfng and miR-125a-5p affects both the amplitude and period of the oscillations, thus acting as a fine-tuning mechanism to Notch activity during somitogenesis.
Despite the established importance of miRNAs for mRNA decay, the extent of their relevance for EC oscillations is still unclear. Recent work from our group showed that different miRNA species are expressed in the PSM and in the forelimb distal cyclic domain. These tissues have very different EC periodicities (discussed below), which suggests that miRNAs may play a role in establishing different paces of the EC (Duarte et al., 2022).
Even though translational delays are not accounted to influence oscillations (Hoyle and Ish-Horowicz, 2013), protein stability plays a crucial role. Hirata et al. (2004) addressed what would happen if Hes7 protein half-life time increased from 20 min (wild-type conditions) to 30 min and found that this provokes a dampening in both Hes7 mRNA and protein oscillations over time. Interestingly, lysine residues were found to play a key role in Hes7 protein stability. The authors generated Hes7 protein mutants, by introducing lysine-to-arginine mutations for each of the seven lysine residues in Hes7 sequence and found that different mutations gave rise to proteins with a half-life that differed from the wild-type. Ishii et al. (2008) reported that some of the lysine mutants lost transcriptional repressor activity, although they were more stable than the wild-type counterpart, thus evidencing the role of these lysine residues in Hes7 protein stability. Studies done by Lewis (2003) and Giudicelli et al. (2007) also stated that her protein half-life time should be short, compared to the zebrafish’s segmentation clock pace. Mathematical modelling performed by Ay et al. (2013) reiterated the finding that proteins with a short half-life time are an essential requirement for the maintenance of the period of oscillations in the wild-type zebrafish segmentation clock. They further confirmed this by determining that Her7 protein has a half-life time ∼10 times inferior to the zebrafish segmentation clock period (Ay et al., 2013).
Embryo Clock gene expression oscillations are a cell autonomous property. This was first hinted by dissecting the chicken PSM in multiple portions and observing that the overall expression pattern of hes4 (Palmeirim et al., 1997) and Lfng (Maroto et al., 2005) remained intact. The same was observed in dissected mouse PSMs (Masamizu et al., 2006). Cyclic Lfng gene expression even persisted in dissociated chicken PSM cells, but it occurred asynchronously among cells, evidencing the need for cell-cell contact to ensure synchrony and establish robust cyclic expression patterns at the tissue level (Maroto et al., 2005). In dissociated mouse PSM cells, Hes1 oscillations also occur cell-autonomously (Masamizu et al., 2006) and Webb et al. (2016) further reported that zebrafish her1 gene retains oscillatory expression in isolated tailbud cells. In this case, oscillations in individual cells presented a longer period and were less robust, compared with the intact tissue. Altogether, these results suggest that cell-cell communication is a key requirement for oscillations to be in phase within the vertebrate PSM tissue. This was corroborated by the work of Tsiairis and Aulehla (2016), that showed that a re-aggregation of mouse PSM cells is able to synchronize oscillations. Cell-autonomous EC gene expression oscillations have also been described in other cell types, such as mouse embryonic stem cells (ESC) (Kobayashi et al., 2009), individual fibroblasts (Masamizu et al., 2006) and neural progenitor cells (Shimojo et al., 2008; Bonev et al., 2012; Manning et al., 2019). These can be synchronized in vitro by the application of a serum shock or by Notch activation (Hirata et al., 2002). However, cell-specific distinct phases of EC oscillations may also play important roles in vivo. This will be discussed further below.
EC synchronization between PSM cells is required for proper somite formation. Local synchrony within the PSM tissue is achieved through Delta-Notch signalling (Jiang et al., 2000; Horikawa et al., 2006; Riedel-Kruse et al., 2007), which also functions to overcome the effect of “noise” introduced by other biological processes, such as cell division (Horikawa et al., 2006; Riedel-Kruse et al., 2007). Riedel-Kruse and colleagues found that EC synchrony in the zebrafish embryo is achieved by simultaneous (Notch-independent) activation immediately prior to gastrulation and is then maintained by Notch-dependent self-organized synchronization. The latter was elegantly shown by incubating embryos with the Notch-inhibitor DAPT until complete EC desynchronization. Then, DAPT washout alone was sufficient to completely restore both dlc oscillations and somite formation (Riedel-Kruse et al., 2007). Delaune et al. (2012) applied single cell live-imaging of her1 expression in zebrafish wild-type and mutant embryos for deltaC, deltaD and notch1a to study the role of Delta-Notch signalling in EC synchronization. In the mutants, her1 dynamics persisted in PSM neighbour cells, but in different oscillation phases (Delaune et al., 2012). Interestingly, deltaC and deltaD work together to ensure synchrony of the zebrafish segmentation clock in distinct portions of the PSM. While deltaD is responsible for the onset of the oscillations at the tailbud level, deltaC plays in role in maintaining and amplifying the oscillations in adjacent cells along the PSM tissue (Mara et al., 2007).
Soza-Ried et al. (2014) provided conclusive evidence for the role of oscillations of Notch-Delta signalling in maintaining the EC synchronized in neighbour cells for somite segmentation. Using a deltaC zebrafish mutant line, the authors were able to rescue both her1 oscillations and somite formation by applying short artificial pulses of deltaC expression, evidencing that Notch signalling is indeed maintaining cell synchrony during somitogenesis. Accordingly, longer intervals between deltaC pulses generated larger somites (Soza-Ried et al., 2014). This was confirmed by Isomura et al. (2017) who developed an optogenetics-based system to monitor Notch-Delta signalling dynamics in neighbour cells. Light-induced Dll1 pulses in sender cells were able to generate synchronized oscillations of Hes1 expression in receiver cells. Furthermore, they were able to determine the time from the induction of Dll1 to the cleavage of NICD, which was ∼50.9 min, followed by an additional ∼77 min until maximum Hes1 levels were reached (Isomura et al., 2017). More recently, Yoshioka-Kobayashi et al. (2020) used this system to show that LFNG in sender cells introduces a 15 min-delay in the transport of Dll1 protein to the cell membrane, without which HES7 oscillations are severely dampened in individual cells of the PSM. These studies corroborate the importance of delays in cell-cell communication for EC oscillations and illustrate the power of optogenetics-based tools for dissecting these intricate regulatory mechanisms.
Notch-Delta signalling was shown to require non-muscle myosin II (NM II)-dependent contractility in both signal-sending and -receiving cells (Hunter et al., 2019). Recently, our lab evaluated the importance of fibronectin (FN) extracellular matrix assembly and signalling through the integrin-ROCK-NM II axis for somite segmentation and EC oscillations in chick PSM. We found that experimental treatments targeting FN matrix assembly, cell-FN interactions and actomyosin contractility significantly perturbed somite formation and EC gene expression, highlighting the importance of the PSM tissue’s mechanical properties for EC oscillations (Gomes de Almeida et al., 2022).
Hes7 and her1 oscillations slowdown in the anterior PSM in mouse (about 1.5-fold) (Niwa et al., 2011) and in zebrafish (Giudicelli et al., 2007; Delaune et al., 2012). Shih et al. (2015) corroborated these findings using live imaging in a transgenic zebrafish line with a her1-venus reporter. They saw that the periodicity of the segmentation clock increases by 1.5-fold in the anterior PSM, comparatively to the posterior PSM. SSoroldoni et al. (2014) had previously described this as a Dynamic Wavelength effect that, together with a Doppler effect resulting from the relative motion of the anterior PSM towards the posterior end due to tissue shortening over time, explains the rhythm of embryo body segmentation. To better understand the dynamics of EC deacceleration in the anterior PSM, Shih et al. (2015) assessed her1-venus reporter expression in cells that would form either side of a somite boundary. Within the same presumptive somite, clock oscillations were arrested in a posterior-to-anterior direction, i.e., cells that were incorporated in a posterior somite boundary ceased oscillations prior to cells that were incorporated in the anterior boundary of the same somite. Moreover, the authors reported that cells at a one-somite distance are initially synchronized in the posterior PSM and, as the clock slows down in the anterior PSM, they assume opposite phases of EC expression (Shih et al., 2015).
Another important alteration that cells experience as they transition from the posterior to anterior PSM is the relative timing of Notch- and Wnt-dependent EC oscillators (Sonnen et al., 2018). In fact, Lfng (Notch) and Axin2 (Wnt) oscillated out-of-phase in the posterior PSM and were progressively coupled towards the anterior PSM, where their synchronization was critical for somite segmentation. This was shown using an ingenious microfluidics-based system, which allowed for precise manipulation of gene expression oscillations by applying temporally controlled pulses of Wnt/Notch-specific activator molecules (Sonnen et al., 2018). Recently, it was shown that the distinct levels of FGF signalling experienced in the anterior and posterior regions of the PSM could underlie the differences in EC dynamics observed in these cells (Diaz Cuadros et al., 2020; Yaman et al., 2022). Yaman et al. (2022) further reported that the posterior-to-anterior FGF gradient in the PSM, classically solely associated with the wavefront activity, is also controlling the anterior propagation of the EC oscillations (Yaman et al., 2022).
There is also evidence of significant spatiotemporal metabolic changes along the PSM axis. Microarray analysis performed in PSM and tailbud samples of zebrafish embryos revealed that cell cycle/DNA metabolic functions are enriched in the posterior PSM, while translation/oxidative metabolism is enriched in anterior PSM and somites. The authors also reported a 2-fold increase in ATP content, as well as 2.5-fold decrease of Cytochrome C oxidase activity in the posterior PSM compared to anterior tissues (Özbudak et al., 2010). A posterior-to-anterior gradient of glycolytic activity was also linked to presomitic mesoderm development in mouse (Bulusu et al., 2017) and chicken (Oginuma et al., 2017) embryos. To test the functional relevance of these metabolic differences, PSM explants were cultured in glucose- or pyruvate-supplemented medium. While explants cultured in glucose supplemented medium developed normally, pyruvate-cultured explants displayed several defects concomitantly with loss of Lfng gene expression in the posterior PSM. Using a genetically encoded sensor for pyruvate to monitor metabolic transitions during PSM differentiation in real-time, the authors reported that pyruvate levels, i.e., glycolytic activity, decreased as cells transited towards an anterior PSM-like, more differentiated, state. Consistent with these findings, chicken embryos treated with 2-deoxy-D-glucose (2DG), a competitive inhibitor of the glycolytic enzyme hexokinase, displayed severe elongation defects, even though somite formation occurred normally. On the other hand, embryos treated with sodium azide (NaN3)—a respiration inhibitor, had impaired somite segmentation (Oginuma et al., 2017).
The time each pair of somites takes to form is species-specific and displays great variability between organisms, ranging from 30 min in zebrafish to approximately 5 h in Human (Table 1). Similarly, the expression of segmentation clock genes oscillates with a periodicity characteristic of each species, which closely matches the time of somite formation (Figure 2; Table 2). The signalling pathways that comprise these genes are conserved; however, data suggests that cyclic genes display an evolutionary plasticity, since the specific genes involved in each pathway differ in the studied organisms (Krol et al., 2011).
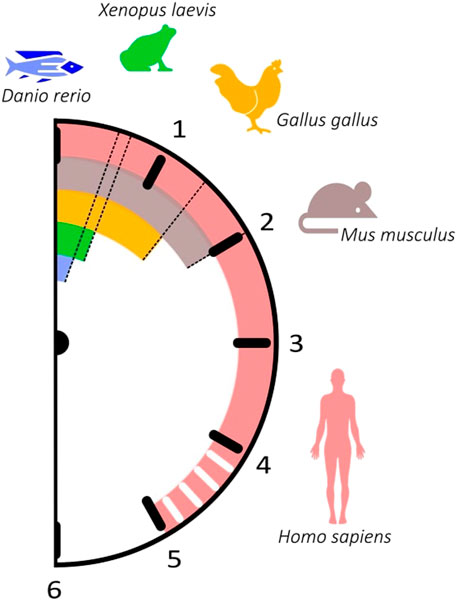
FIGURE 2. The somitogenesis clock ticks with different paces among vertebrates. The periodicity of somitogenesis clock gene expression in different organisms correlates with somite formation time. Danio rerio: 30 min (blue); Xenopus laevis: 40 min (green); Gallus gallus: 90 min (orange); Mus musculus: 120 min (brown); Homo sapiens: ∼5 h (pink).
A curious aspect of somite formation time is that the size of the organism does not significantly influence the time a pair of somites takes to form. For instance, somites have a relatively similar time of formation in the chicken and the emu, although dimension-wise these two birds are very distinct (Nagai et al., 2011). Likewise, the time of somite formation does not depend on phylogenetic relationships, since vertebrates belonging to different phyla can have the same somitogenesis period: 60 min for medaka and the house snake (Gomez et al., 2008).
Recent work started shedding light into the molecular mechanisms underlying divergent EC periodicity among organisms. Using in vitro models to compare EC gene expression dynamics in mouse vs. human cells, two independent studies found that the near 2-fold difference in oscillation periodicity could be explained by the different speeds in biochemical reactions within human and mouse cells, in particular mRNA and/or protein decay rates (Matsuda et al., 2020a; Rayon et al., 2020). This was documented in different cell types, which suggests that global temporal scaling mechanisms are a cell-autonomous property of the organism (Rayon and Briscoe, 2021).
EC oscillations underlying the formation of somites positioned at different A-P levels of the vertebrate body axis have different periodicities. In the chicken embryo, somitogenesis and EC oscillations occur with a 90 min-periodicity for somites 15–20 (HH12-13+) (Palmeirim et al., 1997). However, the final 5-8 somites (HH23) form with a periodicity of 150 min, matched by correspondingly slower cycles of Lfng gene expression (Gibb et al., 2009; Tenin et al., 2010) (Figure 3). In the opposite end, knowledge on the EC in the formation of the anterior-most somites is scarce. Rodrigues et al. (2006) characterized the expression of Notch-related EC genes in somites 1–10 and reported that, while they were dynamically expressed in the PSM, they did not present somite A-P polarity, as occurs in caudal somites. The EC was proposed to already be active even earlier in development, during gastrulation. In fact, Jouve et al. (2002) reported the existence of pulses of gene expression of hairy2 and Lfng in the prospective PSM of early chicken embryos. In zebrafish gastrulation stages, the EC already oscillates with 30-min periodicity (Riedel-Kruse et al., 2007) and in mouse, with a ∼2-h period (Falk et al., 2022). What triggers the onset of the Embryo Clock, and the existence of a clear periodicity in the early developmental stages of chick development, however, remains elusive.
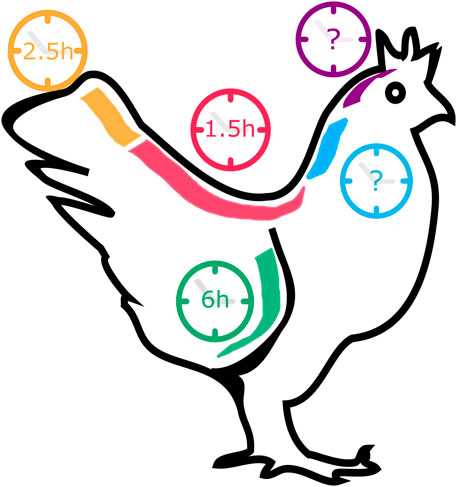
FIGURE 3. The pace of Gallus gallus Embryo Clock (EC) oscillations in different tissues. During somitogenesis (red and orange), the EC pace ranges from 1.5 to 2.5 h, while during forelimb development (green) a cycle lasts 6 h. EC dynamics in the early stages of somitogenesis (blue) and in neural development (purple) remain unknown.
Differences regarding segmentation clock operation in anterior and posterior regions of the zebrafish body axis have also been described (Choorapoikayil et al., 2012; Schröter et al., 2012; Hanisch et al., 2013). Although the double-mutant and -morphant for her1 and her7 exhibit defects throughout the entire body axis (Henry et al., 2002; Oates and Ho, 2002; Lleras-Forero et al., 2018), disrupting her1 or her7 gene expression has a different impact depending on the somites that are being formed. In fact, segmentation defects in her1 mutants are restricted to the anterior trunk, while in her7 mutants somites become defective only posterior to somite 8 (Choorapoikayil et al., 2012; Schröter et al., 2012; Hanisch et al., 2013).
Throughout development, the same gene regulatory networks can be employed in different tissues to produce multiple outcomes. Besides the segmentation of the axial vertebrate body plan, EC oscillations also participate in other developmental processes, where their dynamics differs from the one presented in the PSM during somitogenesis.
Embryonic stem cells (ESC) differentiate into cell types belonging to all three germ layers–mesoderm, endoderm and ectoderm. Evidence that oscillatory gene expression played an important role in ESC differentiation was first provided by Kobayashi et al. (2009). The authors identified unsynchronized cycles of Hes1 gene expression in mouse ESCs with a periodicity of 3–5 h–longer than the 2-h Hes1 period described for other cell types from this organism (see Table 2). Under the same culture conditions, Hes1-high cells differentiated more efficiently into mesodermal cells, while Hes1-low cells into neurons, suggesting that unsynchronized gene expression oscillations might potentiate heterogeneous cell fate specification within the same population of cells (Kobayashi et al., 2009).
Oscillatory EC expression was also described in mouse neural progenitor cells (NPC). Using real-time imaging, Hes5 and Dll1 mRNA were determined to oscillate with a 2 h-periodicity (Imayoshi et al., 2013; Shimojo et al., 2008, 2016), and for Hes1 this period was 2–3 h (Shimojo et al., 2008; Imayoshi et al., 2013) (Table 2). Likewise, Hes1 and Delta1 proteins display an identical period of oscillation (Table 3; Imayoshi et al., 2013). Studies performed in mouse NPCs reiterated the importance of gene expression oscillations for cell fate determination. During neurogenesis, the NPC population is maintained due to the repression of neural fate determination factors, such as Neurogenin2, Ascl1/Mash1 and Olig2 by Hes1 and Hes5 oscillatory expression levels. As a consequence, Neurogenin2 and Ascl1 also display oscillatory mRNA expression (Shimojo et al., 2008; Imayoshi et al., 2013). Unlike somitogenesis, oscillations during neural development are asynchronous. While undergoing differentiation, neural cells impede their neighbours to differentiate into the same cell type through lateral inhibition mediated by the Notch signalling pathway. Imayoshi et al. (2013) reported that the expression level of gene oscillations plays an important role in NPC differentiation. Upon segregating NPC according to their levels of expression of Hes1, Ascl1 and Olig2 and culturing them in differentiation medium, the authors found that high or low EC expression levels dictated different differentiation outcomes. For instance, Hes1-high NPCs differentiated into an astrocyte lineage, while Hes1-low NPCs into neurons (Imayoshi et al., 2013).
Similar to what is observed during neural development, Hes1 expression is required for the maintenance of multipotency in pancreatic progenitors, and undifferentiated neighbour cells undergo a mechanism of lateral inhibition to give rise to different cell types (de Lichtenberg et al., 2018). Even though pancreas development shares common players with somitogenesis and neural development, it was unknown if they displayed an oscillatory behaviour in this tissue. Recently, Seymour and colleagues (2020) reported that Hes1 and Dll1 proteins oscillate with a 90-min periodicity in cultured mouse pancreatic explants, and that this stimulates progenitor proliferation. The periodicity is different from the average 150 min period of Hes1 oscillations in NPC, which could be explained by lower levels of Notch activation in pancreatic progenitors. Importantly, extending the Hes1 oscillation period to ∼120 min by inhibiting NICD degradation altered cell fate specification (Seymour et al., 2020).
Oscillatory gene expression was also reported during chicken limb development (Pascoal et al., 2007) (Figure 3). Oscillations of hairy2 expression were first described in the chicken PSM, with the same periodicity as somite formation–90 min (Jouve et al., 2000). To study hairy2 expression dynamics in the developing chick forelimb, Pascoal et al. (2007) microsurgically removed one limb from HH22-26 embryos in ovo and reincubated the embryo for different periods of time. hairy2 expression was then assessed in each limb pair using in situ hybridization, revealing that hairy2 has very dynamic expression in the distal limb field, that is recapitulated every 6 h. The authors then determined that the time required to form a new autopod skeletal element is 12 h, suggesting that the limb chondrogenic precursor cells undergo two cycles of hairy2 expression for the formation of each autopod segmented element (Pascoal et al., 2007). This was the first evidence that a molecular clock is operating during limb development, a process where temporal control is also fundamental. hes4 is also expressed in the distal mesenchyme of the avian limb (Vasiliauskas et al., 2003) and recent work suggests that its expression is also cyclic during limb development Bhat et al. (2019). cultured cells from chicken pre-cartilage leg mesenchyme and observed oscillations of hes4 expression with a period of 6 h, suggesting that EC periodicity is a tissue-specific property.
The cases mentioned above clearly exemplify that the EC can play very distinct roles in different cells and tissues. For the formation of segmented structures such as somites and autopod limb elements, cells need to be synchronized to aggregate and give rise to a new segment. In multipotent cells, such as ESC, NPC and pancreatic progenitors, asynchronous EC oscillations function to allow heterogeneous cell fate responses of the population to a differentiation signal, ensuring the simultaneous specification of multiple cell types required for normal development.
Many attempts have been made to manipulate EC gene expression levels and/or temporal dynamics in order to obtain a clear understanding of the mechanisms underlying ultradian biological rhythms and their impact on embryo development. Although EC periodicity can be significantly altered throughout the developmental program (Figure 3)–chicken hairy2 oscillates with a periodicity of 90 min in embryos with 48 h (PSM) (Palmeirim et al., 1997) and 6 h in the forelimb of older embryos (4–5 days) (Pascoal et al., 2007)–it has been extremely challenging to produce such significant alterations in an experimental setting. Most of the attempts to date completely disrupted EC expression or oscillatory dynamics (Table 4). In the most cases, only slight alterations to its rhythmicity were obtained (Table 5). The knowledge gained by such approaches, however, has been invaluable, and is patent in the topics described in the previous sections of this review.
Genetic manipulation of EC genes and associated intercellular signalling pathways provided the main framework of what we know today. Figure 4 offers a graphical overview of the alterations to EC gene expression imposed by genetic manipulation in the mouse and zebrafish models (references listed in Tables 4, 5). An interesting observation is that manipulation of Notch-dependent EC genes has limited impact on the dynamics of oscillatory genes associate with the FGF or Wnt signalling pathways, while the other Notch-EC genes are significantly altered. The major effects on FGF clock genes were observed when Hes7 or Lfng were expressed at constant levels and only the latter altered Wnt-related Axin2 oscillations. On the contrary, modulation of key components of FGF and Wnt pathways significantly impacted the expression of EC genes pertaining to all signalling pathways (Figure 4). The available information in zebrafish regards only to Notch-pathway EC genes and provides complementary knowledge to what is described for mouse. As can be easily perceived from Figure 4, many more studies are required to make full sense of the information gathered to date and to allow a clear inter-species comparison of the EC mechanism. It is worth highlighting that conclusive evidence for the functional relevance of the dynamic nature of EC gene expression, in opposition to EC expression levels, was provided by Shimojo et al. (2016). These authors succeeded in abolishing Dll1 oscillations while ensuring physiological expression levels of the protein and this led to defective somitogenesis.
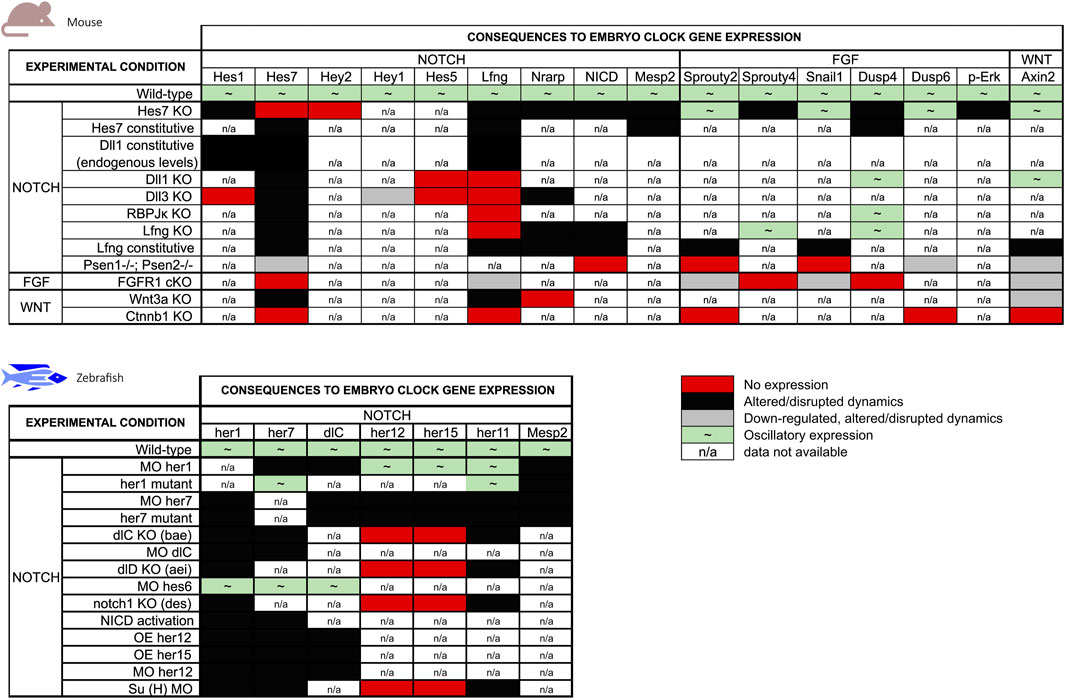
FIGURE 4. Summary of the effects of genetic manipulation on mouse and zebrafish EC gene expression. EC genes were grouped by the main signalling pathways they are associated with.
Besides genetic manipulation, other factors, such as environmental hypoxia or alterations to reactive oxygen species (ROS) levels, can impact EC operation in the PSM. Exposure of pregnant mice to mild hypoxia disrupted EC oscillations and induced vertebral malformations in heterozygous pups for Hes7 or Mesp2, which otherwise developed normally (Sparrow et al., 2012). Ventre et al. (2015) suggested that this effect could be mediated by ROS, since pharmacological modulation of ROS levels in Medaka (Oryzias latipes) impaired somite formation and downregulated her4 and hey1, two EC genes in this organism (Ventre et al., 2015). Recent studies using in vitro-derived PSM-like tissues showed that graded levels of Fgf ligands are required to ensure the EC dynamics (pace, amplitude and phase) (Diaz Cuadros et al., 2020; Yaman et al., 2022) and directionality of EC oscillation waves in the PSM (Yaman et al., 2022).
Sheeba et al. (2014) studied the regulation of hairy2 oscillations in the chick distal forelimb. When the Apical Ectodermal Ridge (AER) or the Zone of Polarizing Activity (ZPA), the key sources of FGF and Shh, respectively, were surgically removed from developing limbs, hairy2 expression was abolished from the distal cyclic domain. This suggested that the EC could represent a functional intersection of these key molecules for limb proximal-distal outgrowth and patterning.
As described above, most attempts to modify the EC led to a complete disruption of the oscillations. However, an increasing number of experimental approaches have succeeded in altering the pace of the EC, which is key to understanding how this biological oscillator is regulated and the functional significance of its temporal dynamics (Table 5).
Following the work of by Takashima et al. (2011), Harima and colleagues generated a mouse mutant lacking only the first two introns of the Hes7 gene. This resulted in Hes7 oscillations with an 11 min-faster periodicity than the WT mice. This shorter cycle did not abolish EC oscillations nor somite formation. As predicted by the Clock and Wavefront model, since the EC presented a faster pace, this culminated in more and smaller somites (Harima et al., 2013). Recently, a similar approach was used to modify the tempo of Hes1 oscillations in NPCs (Ochi et al., 2020). Deleting all the introns of the mouse Hes1 gene accelerated expression oscillations by 13.6 min. On the other hand, by substantially increasing the primary transcript length the authors obtained exactly the opposite result, delayed EC in 13.5 min (Ochi et al., 2020). These are powerful examples of how transcriptional delays of EC genes can be modulated to tinker gene expression dynamics.
In the EC negative feedback regulatory loop, HES proteins are imported to the nucleus and repress their own transcription. Repression is lifted upon protein degradation, allowing a new transcription cycle to begin. Hence, by changing protein stability the repressive time on the gene promoter also changes, culminating in an overall alteration in the tempo of the EC. Through replacement of a Lysine for an Arginine in position 14, Hirata and colleagues were able to increase the half-life of the HES7 protein by 10 min, which led to an increase of 10,2 min in the global pace of the Embryo Clock (Hirata et al., 2004). A similar approach was applied by Kim et al. (2011) by knocking-out Nrarp, a negative effector of Notch signalling. This approach, suggested to delay NICD turnover, extended the EC pace by 5 min and resulted in the formation of fewer and defective vertebrae (Kim et al., 2011). Wiedermann et al. (2015) also accomplished to delay EC oscillations by stabilising NICD in the chick embryo, further corroborating these findings.
A wealth of knowledge on EC pace manipulation has also been provided using the zebrafish model. The Oates lab produced mutants in genes belonging to the Notch signalling pathway that displayed slower EC oscillations (Herrgen et al., 2010; Schröter and Oates, 2010). Knock-out of notch1a, mib1, and deltaD slowed the EC pace by 7%, 19%, and 23%, respectively (Herrgen et al., 2010), and mutating hes6 delayed the EC by 6.5% (Schröter and Oates, 2010). Finally, chemical inhibition of the notch signalling pathway using DAPT delayed the EC by 18% (Herrgen et al., 2010). Corresponding delays in somitogenesis periodicity and a reduced final somite number corroborated the importance of Delta-Notch coupled oscillations for timely embryo body segmentation. Accordingly, elevation of Delta-Notch signalling accelerated EC oscillations and somite formation (Liao et al., 2016). Liao et al. (2016) created fish lines with 7 (Dover) or 100 (Damascus) extra copies of deltaD. Only the Damascus mutant displayed alterations to the EC, where oscillations were 1.6 min (6.4%) faster than in the wild-type. This increased the number of trunk segments by 7,6% and, despite the dramatic overexpression of deltaD, segmentation defects were rarely observed (Liao et al., 2016).
Other intercellular communication pathways contribute to the proper timing of the Embryo Clock. Sonic hedgehog (Shh) was shown to participate in EC tempo regulation. By comparing chicken PSM explants cultured with and without a source of Shh (notochord tissue and/or SHH-expressing cells), Resende et al. (2010) showed that the absence of Shh significantly delayed both EC oscillations and somite formation. Absence of Shh signalling led to an 85% increase of the EC period, from 90 min to approximately 2 h and 45 min. Similar experimental approaches showed that Wnt pathway inhibition by CKI-7 extended the EC pace from 90 to 120 min (Gibb et al., 2009). Comparable results were further obtained in the mouse model. Here, both CKI-7 treatment and activation of Wnt signalling using LiCl delayed Hes7 oscillations (González et al., 2013). Using PSM-like tissues induced from mouse ESC, Yoshioka-Kobayashi et al. (2020) performed a high throughput chemical library screening and identified multiple small compounds capable of altering the period of Hes7 oscillations by up to 40 min (Yoshioka-Kobayashi et al., 2020). These included modulators of a wide range of cellular processes and signalling pathways and further characterization of these alterations will surely improve our knowledge on EC operation.
Recently, we showed that the fibronectin-integrin-ROCK-NM II signalling axis regulates EC dynamics in the chicken PSM. Importantly, inhibition of actomyosin-mediated contractility delayed the period of hairy1 (hes4) oscillations from 90 to 120 min (Gomes de Almeida et al., 2022), unveiling a previously unappreciated biomechanical regulation of the EC periodicity.
Great attention has classically been dedicated to studying the molecular mechanisms involved in correct spatial positioning of cells/tissues/organs during embryo development, while the dynamics of gene expression over time was an under-represented concern. The discovery of a molecular Embryo Clock underlying somite formation gave way to a dramatic shift in this trend. Since it was first described in 1997, the EC has been characterized in multiple vertebrate species, evidencing a phylogenetically conserved mechanism. However, there are two aspects that differ depending on the organism: the pace of the EC and the specific oscillatory genes, although common signalling pathways are involved. The EC biological function has been tightly correlated with the segmentation of paraxial mesoderm, and mutations in Human EC genes give rise to severe congenital malformations of the axial skeleton, such as the phenotypes associated with spondylocostal dysostosis (Sparrow et al., 2012; Nobrega et al., 2021).
There is great interest in clarifying the EC clock dynamics and regulatory mechanisms in tissues other than the paraxial mesoderm and in different species, since this should help evidence what constitutes the central mechanism(s) of the clock, and which components are species/tissue-specific. Hairy-enhancer-of-split oscillatory expression is conserved in all species and tissues analysed, which has suggested their role as “core” members of the EC, but conclusive evidence for such fundamental clock components remains elusive. Studies on the mechanism(s) associated with the onset of gene expression oscillations during development might help elucidate whether EC operation is the output of a limited set of “core” clock genes or if it is an emergent property of the developing biological system, reverberating the oscillatory nature of the very first events during fertilization (e.g., Ca2+ oscillations induced upon sperm-oocyte fusion).
For many years, only hairy2 was described to have cyclic expression in the chicken limb bud. More recently, Bhat et al. (2019) reported oscillations of hes4 expression in chick limb micromass cultures. Here, hes4 oscillates with a 6 h periodicity (Bhat et al., 2019), which matches the rate of limb hairy2 oscillations in vivo (Pascoal et al., 2007). This suggests that the expression dynamics of both hairy2 and hes4 may be regulated by common mechanisms in the developing limb, further reinforcing the existence an EC-like mechanism operating during limb development (Sheeba et al., 2016). However, it is still unknown if this is conserved in other vertebrates and if altering hairy2 or hes4 expression may impact limb outgrowth and patterning. Clues arise from recent work evidencing that Hes1 is a critical downstream effector of the Shh/Gli3 pathway in mouse limb development, where it regulates mesenchymal cell proliferation (Sharma et al., 2021). Importantly, Hes1 overexpression promoted supernumerary digit formation and the authors concluded that Hes1 regulates anterior boundary formation for digit development. Together, these studies suggest that synchronized Hes oscillations in the distal limb field could be functioning to prepattern the tissue for segment (digit) formation, which is reminiscent of the EC function in the PSM. Hence, the developing limb bud represents an additional extraordinary model system to further study EC regulation and function.
Despite the effort put into characterizing the EC, many fundamental questions remain unanswered. What triggers the onset of EC oscillations? What sets the tempo of the clock? What is the functional relevance of EC oscillations in different cell types and embryonic tissues? Is there a core component common to all vertebrates? Answering these and other pressing questions would allow us to understand how TIME is set and perceived for pattern formation during embryo development. Currently, there is a growing number of researchers employing novel experimental in vitro model systems that bring great promise to dissecting the EC mechanism in ways that have been hindered in vivo (reviewed in Diaz-Cuadros and Pourquié, 2021). These include the recently described Human somitoids derived from induced pluripotent stem cells (iPSC), which display segment formation and Hes7 gene expression oscillations with the same periodicity as that previously describe for Human somitogenesis: ∼5 h (Sanaki-Matsumiya et al., 2022). After 25 years since the somitogenesis Embryo Clock was first described, the scientific community is more aware than ever of the existing knowledge gaps, but is also more equipped than ever to tackle the challenges ahead.
GC, AM-J, and RPA conceptualized, wrote, and revised the manuscript. GC and AM-J created figures and tables. RPA provided supervision and guidance on the manuscript organization.
This study was supported by the Portuguese Fundação para a Ciência e Tecnologia (FCT) (scholarships SFRH/BD/101609/2014 and PTDC/BEX-BID/5410/2014 to GC and AM-J, respectively) and by AD-ABC and Município de Loulé to RPA.
The authors are grateful for the careful analysis and critical discussion with the reviewers that greatly improved our final manuscript. We also acknowledge Letícia Leite and Betina Silva for their contributions to Figure 1 construction.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aulehla, A., and Johnson, R. L. (1999). Dynamic expression of lunatic fringe suggests a link between notch signaling and an autonomous cellular oscillator driving somite segmentation. Dev. Biol. 207, 49–61. doi:10.1006/dbio.1998.9164
Aulehla, A., Wehrle, C., Brand-Saberi, B., Kemler, R., Gossler, A., Kanzler, B., et al. (2003). Wnt3a plays a major role in the segmentation clock controlling somitogenesis. Dev. Cell 4, 395–406. doi:10.1016/s1534-5807(03)00055-8
Aulehla, A., Wiegraebe, W., Baubet, V., Wahl, M. B., Deng, C., Taketo, M., et al. (2008). A β-catenin gradient links the clock and wavefront systems in mouse embryo segmentation. Nat. Cell Biol. 10, 186–193. doi:10.1038/ncb1679
Ay, A., Knierer, S., Sperlea, A., Holland, J., and Özbudak, E. M. (2013). Short-lived her proteins drive robust synchronized oscillations in the Zebrafish segmentation clock. Development 140, 3244–3253. doi:10.1242/dev.093278
Barrantes, I. D. B., Elia, A. J., Wünsch, K., De Angelis, M. H., Mak, T. W., Rossant, J., et al. (1999). Interaction between Notch signalling and Lunatic fringe during somite boundary formation in the mouse. Curr. Biol. 9, 470–480. doi:10.1016/S0960-9822(99)80212-7
Bessho, Y., Hirata, H., Masamizu, Y., and Kageyama, R. (2003). Periodic repression by the bHLH factor Hes7 is an essential mechanism for the somite segmentation clock. Genes Dev. 17, 1451–1456. doi:10.1101/gad.1092303
Bessho, Y., Sakata, R., Komatsu, S., Shiota, K., Yamada, S., Kageyama, R., et al. (2001). Dynamic expression and essential functions of Hes7 in somite segmentation. Genes Dev. 15, 2642–2647. doi:10.1101/gad.930601
Bhat, R., Glimm, T., Linde-medina, M., Cui, C., and Newman, S. A. (2019). Synchronization of Hes1 oscillations coordinates and refines condensation formation and patterning of the avian limb skeleton. Mech. Dev. 156, 41–54. doi:10.1016/j.mod.2019.03.001
Bone, R. A., Bailey, C. S. L., Wiedermann, G., Ferjentsik, Z., Appleton, P. L., Murray, P. J., et al. (2014). Spatiotemporal oscillations of Notch1, Dll1 and NICD are coordinated across the mouse PSM. Development 141, 4806–4816. doi:10.1242/dev.115535
Bonev, B., Stanley, P., and Papalopulu, N. (2012). MicroRNA-9 modulates hes1 ultradian oscillations by forming a double-negative feedback loop. Cell Rep. 2, 10–18. doi:10.1016/j.celrep.2012.05.017
Bulusu, V., Prior, N., Snaebjornsson, M. T., Schultz, C., Sauer, U., Aulehla, A., et al. (2017). Spatiotemporal analysis of a glycolytic activity gradient linked to mouse embryo mesoderm development. Dev. Cell 40, 331–341. doi:10.1016/j.devcel.2017.01.015
Chen, J., Kang, L., and Zhang, N. (2005). Negative feedback loop formed by lunatic fringe and Hes7 controls their oscillatory expression during somitogenesis. Genesis 43, 196–204. doi:10.1002/gene.20171
Choorapoikayil, S., Willems, B., Ströhle, P., and Gajewski, M. (2012). Analysis of her1 and her7 mutants reveals a spatio temporal separation of the somite clock module. PLoS One 7, e39073. doi:10.1371/journal.pone.0039073
Chu, L.-F., Mamott, D., Ni, Z., Bacher, R., Liu, C., Swanson, S., et al. (2019). An in vitro human segmentation clock model derived from embryonic stem cells. Cell Rep. 28, 2247–2255. e5. doi:10.1016/j.celrep.2019.07.090
Cooke, J., and Zeeman, E. C. (1976). A clock and wavefront model for control of the number of repeated structures during animal morphogenesis. J. Theor. Biol. 58, 455–476. doi:10.1016/S0022-5193(76)80131-2
Dale, J. K., Malapert, P., Chal, J., Vilhais-Neto, G., Johnson, T., Maroto, M., et al. (2006). Oscillations of the Snail genes in the presomitic mesoderm coordinate segmental patterning and morphogenesis in vertebrate somitogenesis. Dev. Cell 10, 355–366. doi:10.1016/j.devcel.2006.02.011
Dale, J. K., Maroto, M., Dequeant, M. L., Malapert, P., McGrew, M., Pourquie, O., et al. (2003). Periodic Notch inhibition by lunatic fringe underlies the chick segmentation clock. Nature 421, 275–278. doi:10.1038/nature01244
Davis, R. L., Turner, D. L., Evans, L. M., and Kirschner, M. W. (2001). Molecular targets of vertebrate segmentation: Two mechanisms control segmental expression of Xenopus hairy2 during somite formation. Dev. Cell 1, 553–565. doi:10.1016/S1534-5807(01)00054-5
De Lichtenberg, K. H., Seymour, P. A., Jørgensen, M. C., Kim, Y.-H., Grapin-Botton, A., Magnuson, M. A., et al. (2018). Notch controls multiple pancreatic cell fate regulators through direct hes1-mediated repression. bioRxiv, 336305. doi:10.1101/336305
Delaune, E. A., François, P., Shih, N. P., and Amacher, S. L. (2012). Single-cell-resolution imaging of the impact of notch signaling and mitosis on segmentation clock dynamics. Dev. Cell 23, 995–1005. doi:10.1016/j.devcel.2012.09.009
Dequéant, M.-L., Glynn, E., Gaudenz, K., Wahl, M., Chen, J., Mushegian, A., et al. (2006). A complex oscillating network of signaling genes underlies the mouse segmentation clock. Sci. (80-. ) 314, 1595–1598. doi:10.1126/science.1133141
Diaz-cuadros, M., and Pourquie, O. (2021). In vitro systems: A new window to the segmentation clock. Dev. Growth Differ. 63, 140–153. doi:10.1111/dgd.12710
Diaz-cuadros, M., Wagner, D. E., Budjan, C., Hubaud, A., Tarazona, O. A., Donelly, S., et al. (2020). In vitro characterization of the human segmentation clock. Nature 580, 113–118. doi:10.1038/s41586-019-1885-9
Duarte, I., Carraco, G., de Azevedo, N. T. D., Benes, V., and Andrade, R. P. (2022). gga-miRNOME, a microRNA-sequencing dataset from chick embryonic tissues. Sci. Data 9, 29. doi:10.1038/s41597-022-01126-7
Dubrulle, J., Mcgrew, M. J., and Pourquié, O. (2001). FGF signaling controls somite boundary position and regulates segmentation clock control of spatiotemporal hox gene activation. Cell 106, 219–232. doi:10.1016/S0092-8674(01)00437-8
Dubrulle, J., and Pourquie, O. (2004). fgf8 mRNA decay establishes a gradient that couples axial elongation to patterning in the vertebrate embryo. Nature 427, 419–422. doi:10.1038/nature02216
Dunty, W. C., Biris, K. K., Chalamalasetty, R. B., Taketo, M. M., Lewandoski, M., Yamaguchi, T. P., et al. (2008). Wnt3a/beta-catenin signaling controls posterior body development by coordinating mesoderm formation and segmentation. Development 135, 85–94. doi:10.1242/dev.009266
Dunwoodie, S. L., Clements, M., Sparrow, D. B., Sa, X., Conlon, R. A., Beddington, R. S. P., et al. (2002). Axial skeletal defects caused by mutation in the spondylocostal dysplasia/pudgy gene Dll3 are associated with disruption of the segmentation clock within the presomitic mesoderm. Development 129, 1795–1806. doi:10.1242/dev.129.7.1795
Elmasri, H., Liedtke, D., Lücking, G., Volff, J. N., Gessler, M., Winkler, C., et al. (2004). Her7 and hey1, but not lunatic fringe show dynamic expression during somitogenesis in medaka (Oryzias latipes). Gene Expr. Patterns 4, 553–559. doi:10.1016/j.modgep.2004.02.003
Falk, H. J., Tomita, T., Mönke, G., McDole, K., and Aulehla, A. (2022). Imaging the onset of oscillatory signaling dynamics during mouse embryo gastrulation. Development 149 (13), 1027–1030. doi:10.1242/dev.200083
Ferjentsik, Z., Hayashi, S., Dale, J. K., Bessho, Y., Herreman, A., De Strooper, B., et al. (2009). Notch is a critical component of the mouse somitogenesis oscillator and is essential for the formation of the somites. PLoS Genet. 5, e1000662. doi:10.1371/journal.pgen.1000662
Forsberg, H., Crozet, F., and Brown, N. A. (1998). Waves of mouse Lunatic fringe expression, in four-hour cycles at two-hour intervals, precede somite boundary formation. Curr. Biol. 8, 1027–1030. doi:10.1016/S0960-9822(07)00424-1
Fujimuro, T., Matsui, T., Nitanda, Y., Matta, T., Sakumura, Y., Saito, M., et al. (2014). Hes7 3’UTR is required for somite segmentation function. Sci. Rep. 4, 6462. doi:10.1038/srep06462
Fujino, Y., Yamada, K., Sugaya, C., Ooka, Y., Ovara, H., Ban, H., et al. (2018). Deadenylation by the CCR4-NOT complex contributes to the turnover of hairy-related mRNAs in the zebrafish segmentation clock. FEBS Lett. 592, 3388–3398. doi:10.1002/1873-3468.13261
Gajewski, M., Elmasri, H., Girschick, M., Sieger, D., and Winkler, C. (2006). Comparative analysis of her genes during fish somitogenesis suggests a mouse/chick-like mode of oscillation in medaka. Dev. Genes Evol. 216, 315–332. doi:10.1007/s00427-006-0059-6
Gajewski, M., Sieger, D., Alt, B., Leve, C., Hans, S., Wolff, C., et al. (2003). Anterior and posterior waves of cyclic her1 gene expression are differentially regulated in the presomitic mesoderm of zebrafish. Development 130, 4269–4278. doi:10.1242/dev.00627
Gallagher, T. L., Tietz, K. T., Morrow, Z. T., Mccammon, J. M., Goldrich, M. L., Derr, N. L., et al. (2017). Pnrc2 regulates 3’UTR-mediated decay of segmentation clock-associated transcripts during zebrafish segmentation. Dev. Biol. 429, 225–239. doi:10.1016/j.ydbio.2017.06.024
Gibb, S., Zagorska, A., Melton, K., Tenin, G., Vacca, I., Trainor, P., et al. (2009). Interfering with Wnt signalling alters the periodicity of the segmentation clock. Dev. Biol. 330, 21–31. doi:10.1016/j.ydbio.2009.02.035
Giudicelli, F., Özbudak, E. M., Wright, G. J., and Lewis, J. (2007). Setting the tempo in development: An investigation of the zebrafish somite clock mechanism. PLoS Biol. 5, e150. doi:10.1371/journal.pbio.0050150
Gomes de Almeida, P., Rifes, P., Martins-Jesus, A. P., Pinheiro, G. G., Andrade, R. P., Thorsteinsdóttir, S., et al. (2022). Cell-fibronectin interactions and actomyosin contractility regulate the segmentation clock and spatio-temporal somite cleft formation during chick embryo somitogenesis. Cells 11, 2003. doi:10.3390/cells11132003
Gomez, C., Özbudak, E. M., Wunderlich, J., Baumann, D., Lewis, J., Pourquié, O., et al. (2008). Control of segment number in vertebrate embryos. Nature 454, 335–339. doi:10.1038/nature07020
González, A., Manosalva, I., Liu, T., and Kageyama, R. (2013). Control of Hes7 expression by Tbx6, the Wnt pathway and the chemical Gsk3 inhibitor LiCl in the mouse segmentation clock. PLOS ONE 8, e53323. doi:10.1371/journal.pone.0053323
Hanisch, A., Holder, M. V., Choorapoikayil, S., Gajewski, M., Özbudak, E. M., Lewis, J., et al. (2013). The elongation rate of RNA polymerase II in zebrafish and its significance in the somite segmentation clock. Development 453, 444–453. doi:10.1242/dev.077230
Harima, Y., Takashima, Y., Ueda, Y., Ohtsuka, T., and Kageyama, R. (2013). Accelerating the tempo of the segmentation clock by reducing the number of introns in the Hes7 gene. Cell Rep. 3, 1–7. doi:10.1016/j.celrep.2012.11.012
Hayashi, S., Shimoda, T., Nakajima, M., Tsukada, Y., Sakumura, Y., Dale, J. K., et al. (2009). Sprouty4, an FGF inhibitor, displays cyclic gene expression under the control of the notch segmentation clock in the mouse PSM. PloS One 4, e5603. doi:10.1371/journal.pone.0005603
Henry, C. A., Urban, M. K., Dill, K. K., Merlie, J. P., Page, M. F., Kimmel, C. B., et al. (2002). Two linked hairy/Enhancer of split-related zebrafish genes, her1 and her7, function together to refine alternating somite boundaries. Development 129, 3693–3704. doi:10.1242/dev.129.15.3693
Herrgen, L., Ares, S., Morelli, L. G., Schroter, C., Julicher, F., Oates, A. C., et al. (2010). Intercellular coupling regulates the period of the segmentation clock. Curr. Biol. 20, 1244–1253. doi:10.1016/j.cub.2010.06.034
Hilgers, V., Pourquié, O., and Dubrulle, J. (2005). In vivo analysis of mRNA stability using the Tet-Off system in the chicken embryo. Dev. Biol. 284, 292–300. doi:10.1016/j.ydbio.2005.05.021
Hirata, H., Bessho, Y., Kokubu, H., Masamizu, Y., Yamada, S., Lewis, J., et al. (2004). Instability of Hes7 protein is crucial for the somite segmentation clock. Nat. Genet. 36, 750–754. doi:10.1038/ng1372
Hirata, H., Yoshiura, S., Ohtsuka, T., Bessho, Y., Harada, T., Yoshikawa, K., et al. (2002). Oscillatory expression of the BHLH factor Hes1 regulated by a negative feedback loop. Science 298, 840–843. doi:10.1126/science.1074560
Holley, S. A., Geisler, R., and Nüsslein-Volhard, C. (2000). Control of her1 expression during zebrafish somitogenesis by a Delta-dependent oscillator and an independent wave-front activity. Genes Dev. 14, 1678–1690. doi:10.1101/gad.14.13.1678
Holley, S. A., Jülich, D. R., Rauch, G.-J. R., Geisler, R., and Nüsslein-Volhard, C. (2002). her1 and the notch pathway function within the oscillator mechanism that regulates zebrafish somitogenesis. Development 129, 1175–1183. doi:10.1242/dev.129.5.1175
Horikawa, K., Ishimatsu, K., Yoshimoto, E., Kondo, S., and Takeda, H. (2006). Noise-resistant and synchronized oscillation of the segmentation clock. Nature 441, 719–723. doi:10.1038/nature04861
Hoyle, N. P., and Ish-Horowicz, D. (2013). Transcript processing and export kinetics are rate-limiting steps in expressing vertebrate segmentation clock genes. Proc. Natl. Acad. Sci. U. S. A. 110, E4316–E4324. doi:10.1073/pnas.1308811110
Hunter, G. L., He, L., Perrimon, N., Charras, G., Giniger, E., Baum, B., et al. (2019). A role for actomyosin contractility in Notch signaling. BMC Biol. 17 (1), 12. doi:10.1186/s12915-019-0625-9
Huppert, S. S., Ilagan, M. X. G., De Strooper, B., and Kopan, R. (2005). Analysis of notch function in presomitic mesoderm suggests a γ-secretase-independent role for presenilins in somite differentiation. Dev. Cell 8, 677–688. doi:10.1016/j.devcel.2005.02.019
Imayoshi, I., Isomura, A., Harima, Y., Kawaguchi, K., Kori, H., Miyachi, H., et al. (2013). Oscillatory control of factors determining multipotency and fate in mouse neural progenitors. Science 342, 1203–1208. doi:10.1126/science.1242366
Ishii, A., Kobayashi, T., and Kageyama, R. (2008). Requirement of multiple lysine residues for the transcriptional activity and the instability of Hes7. Biochem. Biophys. Res. Commun. 372, 142–146. doi:10.1016/j.bbrc.2008.05.015
Ishikawa, A., Kitajima, S., Takahashi, Y., Kokubo, H., Kanno, J., Inoue, T., et al. (2004). Mouse Nkd1, a Wnt antagonist, exhibits oscillatory gene expression in the PSM under the control of Notch signaling. Mech. Dev. 121, 1443–1453. doi:10.1016/j.mod.2004.08.003
Isomura, A., Ogushi, F., Kori, H., and Kageyama, R. (2017). Optogenetic perturbation and bioluminescence imaging to analyze cell-to-cell transfer of oscillatory information. Genes Dev. 31, 524–535. doi:10.1101/gad.294546.116
Iwamatsu, T. (2004). Stages of normal development in the medaka Oryzias latipes. Mech. Dev. 121, 605–618. doi:10.1016/j.mod.2004.03.012
Jensen, M. H., Sneppen, K., and Tiana, G. (2003). Sustained oscillations and time delays in gene expression of protein Hes1. FEBS Lett. 541, 176–177. doi:10.1016/S0014-5793(03)00279-5
Jiang, Y.-J., Aerne, B. L., Smithers, L., Haddon, C., Ish-Horowicz, D., Lewis, J., et al. (2000). Notch signalling and the synchronization of the somite segmentation clock. Nature 408, 475–479. doi:10.1038/35044091
Jing, B., Yuan, J., Yin, Z., Lv, C., Lu, S., Xiong, H., et al. (2015). Dynamic properties of the segmentation clock mediated by microRNA. Int. J. Clin. Exp. Pathol. 8, 196–206.
Jouve, C., Iimura, T., and Pourquié, O. (2002). Onset of the segmentation clock in the chick embryo: Evidence for oscillations in the somite precursors in the primitive streak. Development 129, 1107–1117. doi:10.1242/dev.129.5.1107
Jouve, C., Palmeirim, I., Henrique, D., Beckers, J., Gossler, A., Ish-Horowicz, D., et al. (2000). Notch signalling is required for cyclic expression of the hairy-like gene HES1 in the presomitic mesoderm. Development 127, 1421–1429. doi:10.1242/dev.127.7.1421
Kageyama, R., Niwa, Y., Isomura, A., González, A., and Harima, Y. (2012). Oscillatory gene expression and somitogenesis, 1. Wiley Interdiscip Rev Dev Biol, 629–641. doi:10.1002/wdev.46
Keynes, R. J., and Stern, C. D. (1988). Mechanisms of vertebrate segmentation. Development 103, 413–429. doi:10.1242/dev.103.3.413
Kim, W., Matsui, T., Yamao, M., Ishibashi, M., Tamada, K., Takumi, T., et al. (2011). The period of the somite segmentation clock is sensitive to Notch activity. Mol. Biol. Cell 22, 3541–3549. doi:10.1091/mbc.E11-02-0139
Kimmel, C. B., Ballard, W. W., Kimmel, S. R., Ullmann, B., and Schilling, T. F. (1995). Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253–310. doi:10.1002/aja.1002030302
Kobayashi, T., Mizuno, H., Imayoshi, I., Furusawa, C., Shirahige, K., Kageyama, R., et al. (2009). The cyclic gene Hes1 contributes to diverse differentiation responses of embryonic stem cells. Genes Dev. 23, 1870–1875. doi:10.1101/gad.1823109
Krol, A. J., Roelling, D., Dequéant, M.-L., Tassy, O., Glynn, E., Hattem, G., et al. (2011). Evolutionary plasticity of segmentation clock networks. Development 138, 2783–2792. doi:10.1242/dev.063834
Leimeister, C., Dale, K., Fischer, A., Klamt, B., Hrabe de Angelis, M., Radtke, F., et al. (2000). Oscillating expression of c-Hey2 in the presomitic mesoderm suggests that the segmentation clock may use combinatorial signaling through multiple interacting bHLH factors. Dev. Biol. 227, 91–103. doi:10.1006/dbio.2000.9884
Lewis, J. (2003). Autoinhibition with transcriptional delay: A simple mechanism for the zebrafish somitogenesis oscillator. Curr. Biol. 13, 1398–1408. doi:10.1016/S0960-9822(03)00534-7
Li, Y., Fenger, U., Niehrs, C., and Pollet, N. (2003). Cyclic expression of esr9 gene in Xenopus presomitic mesoderm. Differentiation. 71, 83–89. doi:10.1046/j.1432-0436.2003.700608.x
Liao, B. K., Jorg, D. J., and Oates, A. C. (2016). Faster embryonic segmentation through elevated Delta-Notch signalling. Nat. Commun. 7, 11861. doi:10.1038/ncomms11861
Lleras Forero, L., Narayanan, R., Huitema, L. F., VanBergen, M., Apschner, A., Peterson-Maduro, J., et al. (2018). Segmentation of the zebrafish axial skeleton relies on notochord sheath cells and not on the segmentation clock, 7. doi:10.7554/eLife.33843
Manning, C. S., Biga, V., Boyd, J., Kursawe, J., Ymisson, B., Spiller, D. G., et al. (2019). Quantitative single-cell live imaging links HES5 dynamics with cell-state and fate in murine neurogenesis. Nat. Commun. 10, 2835. doi:10.1038/s41467-019-10734-8
Mara, A., Schroeder, J., Chalouni, C., and Holley, S. A. (2007). Priming, initiation and synchronization of the segmentation clock by deltaD and deltaC. Nat. Cell Biol. 9, 523–530. doi:10.1038/ncb1578
Maroto, M., Dale, J. K., Dequéant, M. L., Petit, A. C., and Pourquié, O. (2005). Synchronised cycling gene oscillations in presomitic mesoderm cells require cell-cell contact. Int. J. Dev. Biol. 49, 309–315. doi:10.1387/ijdb.041958mm
Maruhashi, M., Van De Putte, T., Huylebroeck, D., Kondoh, H., and Higashi, Y. (2005). Involvement of SIP1 in positioning of somite boundaries in the mouse embryo. Dev. Dyn. 234, 332–338. doi:10.1002/dvdy.20546
Masamizu, Y., Ohtsuka, T., Takashima, Y., Nagahara, H., Takenaka, Y., Yoshikawa, K., et al. (2006). Real-time imaging of the somite segmentation clock: Revelation of unstable oscillators in the individual presomitic mesoderm cells. Proc. Natl. Acad. Sci. U. S. A. 103, 1313–1318. doi:10.1073/pnas.0508658103
Matsuda, M., Hayashi, H., Garcia-ojalvo, J., Yoshioka-kobayashi, K., Kageyama, R., Yamanaka, Y., et al. (2020a). Species-specific segmentation clock periods are due to differential biochemical reaction speeds. Science 369, 1450–1455. doi:10.1126/science.aba7668
Matsuda, M., Yamanaka, Y., Uemura, M., Osawa, M., Saito, M. K., Nagahashi, A., et al. (2020b). Recapitulating the human segmentation clock with pluripotent stem cells. Nature 580, 124–129. doi:10.1038/s41586-020-2144-9
Matsumiya, M., Tomita, T., Yoshioka-Kobayashi, K., Isomura, A., and Kageyama, R. (2018). ES cell-derived presomitic mesoderm-like tissues for analysis of synchronized oscillations in the segmentation clock. Development 145, dev156836. doi:10.1242/dev.156836
McGrew, M. J., Dale, J. K., Fraboulet, S., and Pourquié, O. (1998). The lunatic Fringe gene is a target of the molecular clock linked to somite segmentation in avian embryos. Curr. Biol. 8, 979–982. doi:10.1016/S0960-9822(98)70401-4
Monk, N. A. M. (2003). Oscillatory expression of Hes1, p53, and NF-kappaB driven by transcriptional time delays. Curr. Biol. 13, 1409–1413. doi:10.1016/S0960-9822(03)00494-9
Morimoto, M., Takahashi, Y., Endo, M., and Saga, Y. (2005). The Mesp2 transcription factor establishes segmental borders by suppressing Notch activity. Nature 435, 354–359. doi:10.1038/nature03591
Müller, F., and O’Rahilly, R. (1986). Somitic‐vertebral correlation and vertebral levels in the human embryo. Am. J. Anat. 177, 3–19. doi:10.1002/aja.1001770103
Nagai, H., Mak, S. S., Weng, W., Nakaya, Y., Ladher, R., Sheng, G., et al. (2011). Embryonic development of the emu, Dromaius novaehollandiae. Dev. Dyn. 240, 162–175. doi:10.1002/dvdy.22520
Nakaya, M. A., Biris, K., Tsukiyama, T., Jaime, S., Rawls, J. A., Yamaguchi, T. P., et al. (2005). Wnt3a links left-right determination with segmentation and anteroposterior axis elongation. Development 132, 5425–5436. doi:10.1242/dev.02149
Nakayama, K., Satoh, T., Igari, A., Kageyama, R., and Nishida, E. (2008). FGF induces oscillations of Hes1 expression and Ras/ERK activation. Curr. Biol. 18, 332–334. doi:10.1016/j.cub.2008.03.013
Nitanda, Y., Matsui, T., Matta, T., Higami, A., Kohno, K., Nakahata, Y., et al. (2014). 3′-UTR-dependent regulation of mRNA turnover is critical for differential distribution patterns of cyclic gene mRNAs. FEBS J. 281, 146–156. doi:10.1111/febs.12582
Niwa, Y., Masamizu, Y., Liu, T., Nakayama, R., Deng, C. X., Kageyama, R., et al. (2007). The initiation and propagation of Hes7 oscillation are cooperatively regulated by fgf and notch signaling in the somite segmentation clock. Dev. Cell 13, 298–304. doi:10.1016/j.devcel.2007.07.013
Niwa, Y., Shimojo, H., Isomura, A., González, A., Miyachi, H., Kageyama, R., et al. (2011). Different types of oscillations in notch and Fgf signaling regulate the spatiotemporal periodicity of somitogenesis. Genes Dev. 25, 1115–1120. doi:10.1101/gad.2035311
Nobrega, A., Maia-Fernandes, A. C., and Andrade, R. P. (2021). Altered cogs of the clock: Insights into the embryonic etiology of spondylocostal dysostosis. J. Dev. Biol. 9, 5. doi:10.3390/jdb9010005
Oates, A. C., and Ho, R. K. (2002). Hairy/E(spl)-related (Her) genes are central components of the segmentation oscillator and display redundancy with the Delta/Notch signaling pathway in the formation of anterior segmental boundaries in the zebrafish. Development 129, 2929–2946. doi:10.1242/dev.129.12.2929
Oates, A. C., Morelli, L. G., and Ares, S. (2012). Patterning embryos with oscillations: Structure, function and dynamics of the vertebrate segmentation clock. Development 139 (4), 625–639. doi:10.1242/dev.063735
Ochi, S., Imaizumi, Y., Shimojo, H., Miyachi, H., and Kageyama, R. (2020). Oscillatory expression of Hes1 regulates cell proliferation and neuronal differentiation in the embryonic brain. Development 147, dev182204. doi:10.1242/dev.182204
Oginuma, M., Moncuquet, P., Xiong, F., Karoly, E., Guevorkian, K., and Pourquié, O. (2017). A gradient of glycolytic activity coordinates FGF and Wnt signaling during elongation of the body Axis in amniote embryos. Dev. Cell 40, 342–353. doi:10.1016/j.devcel.2017.02.001
Özbudak, E. M., and Lewis, J. (2008). Notch signalling synchronizes the zebrafish segmentation clock but is not needed to create somite boundaries. PLoS Genet. 4, e15. doi:10.1371/journal.pgen.0040015
Özbudak, E. M., Tassy, O., and Pourquié, O. (2010). Spatiotemporal compartmentalization of key physiological processes during muscle precursor differentiation. Proc. Natl. Acad. Sci. U. S. A. 107, 4224–4229. doi:10.1073/pnas.0909375107
Packard, D. S. J. (1980). Somitogenesis in cultured embryos of the Japanese quail, Coturnix coturnix japonica. Am. J. Anat. 158, 83–91. doi:10.1002/aja.1001580108
Palmeirim, I., Henrique, D., Ish-Horowicz, D., and Pourquié, O. (1997). Avian hairy gene expression identifies a molecular clock linked to vertebrate segmentation and somitogenesis. Cell 91, 639–648. doi:10.1016/S0092-8674(00)80451-1
Pascoal, S., Carvalho, C. R., Rodriguez-León, J., Delfini, M. C., Duprez, D., Thorsteinsdóttir, S., et al. (2007). A molecular clock operates during chick autopod proximal-distal outgrowth. J. Mol. Biol. 368, 303–309. doi:10.1016/j.jmb.2007.01.089
Prajapati, R. S., Mitter, R., Vezzaro, A., and Ish-Horowicz, D. (2020). Greb1 is required for axial elongation and segmentation in vertebrate embryos. Biol. Open 9, bio047290. doi:10.1242/bio.047290
Rayon, T., and Briscoe, J. (2021). Cross-species comparisons and in vitro models to study tempo in development and homeostasis. Interface Focus 11, 20200069. doi:10.1098/rsfs.2020.0069
Rayon, T., Stamataki, D., Perez-carrasco, R., Garcia-perez, L., Barrington, C., Melchionda, M., et al. (2020). Species-specific pace of development is associated with differences in protein stability. Science 369, eaba7667. doi:10.1126/science.aba7667
Resende, T. P., Andrade, R. P., and Palmeirim, I. (2014). Timing embryo segmentation: Dynamics and regulatory mechanisms of the vertebrate segmentation clock. Biomed. Res. Int. 2014, 718683. doi:10.1155/2014/718683
Resende, T. P., Ferreira, M., Teillet, M.-A., Tavares, A. T., Andrade, R. P., Palmeirim, I., et al. (2010). Sonic hedgehog in temporal control of somite formation. Proc. Natl. Acad. Sci. U. S. A. 107, 12907–12912. doi:10.1073/pnas.1000979107
Riedel-Kruse, I. H., Müller, C., and Oates, A. C. (2007). Synchrony dynamics during initiation, failure, and rescue of the segmentation clock. Science 317 (5846), 1911–1915. doi:10.1126/science.1142538
Riley, M. F., Bochter, M. S., Wahi, K., Nuovo, G. J., and Cole, S. E. (2013). Mir-125a-5p-Mediated regulation of lfng is essential for the avian segmentation clock. Dev. Cell 24, 554–561. doi:10.1016/j.devcel.2013.01.024
Rodrigues, S., Santos, J., and Palmeirim, I. (2006). Molecular characterization of the rostral-most somites in early somitic stages of the chick embryo. Gene Expr. Patterns 6, 673–677. doi:10.1016/j.modgep.2006.01.004
Sieger, D., Tautz, D., and Gajewski, M. (2004). Her11 is involved in the somitogenesis clock in zebrafish. Dev. Genes Evol. 214, 393–406. doi:10.1007/s00427-004-0427-z
Sanaki-matsumiya, M., Matsuda, M., Gritti, N., Nakaki, F., Sharpe, J., Trivedi, V., et al. (2022). Periodic formation of epithelial somites from human pluripotent stem cells. Nat. Commun. 13, 2325. doi:10.1038/s41467-022-29967-1
Sawada, A., Shinya, M., Jiang, Y.-J., Kawakami, A., Kuroiwa, A., Takeda, H., et al. (2001). Fgf/MAPK signalling is a crucial positional cue in somite boundary formation. Development 128 (23), 4873–4880. doi:10.1242/dev.128.23.4873
Schröter, C., Ares, S., Morelli, L. G., Isakova, A., Hens, K., Soroldoni, D., et al. (2012). Topology and dynamics of the zebrafish segmentation clock core circuit. PLoS Biol. 10, e1001364. doi:10.1371/journal.pbio.1001364
Schröter, C., and Oates, A. C. (2010). Segment number and axial identity in a segmentation clock period mutant. Curr. Biol. 20, 1254–1258. doi:10.1016/j.cub.2010.05.071
Serth, K., Schuster-gossler, K., Cordes, R., and Gossler, A. (2003). Transcriptional oscillation of lunatic fringe is essential for somitogenesis. Genes Dev. 17, 912–925. doi:10.1101/gad.250603
Sewell, W., Sparrow, D. B., Smith, A. J., Gonzalez, D. M., Rappaport, F., Dunwoodie, S. L., et al. (2009). Cyclical expression of the Notch/Wnt regulator Nrarp requires modulation by Dll3 in somitogenesis. Dev. Biol. 329, 400–409. doi:10.1016/j.ydbio.2009.02.023
Seymour, P. A., Collin, C. A., Egeskov-madsen, A. R., Jørgensen, M. C., Shimojo, H., Imayoshi, I., et al. (2020). Jag1 modulates an oscillatory dll1-notch-hes1 signaling module to coordinate growth and fate of pancreatic progenitors. Dev. Cell 52, 731–747. e8. doi:10.1016/j.devcel.2020.01.015
Shankaran, S. S., Sieger, D., Schröter, C., Czepe, C., Pauly, M. C., Laplante, M. A., et al. (2007). Completing the set of h/E(spl) cyclic genes in zebrafish: her12 and her15 reveal novel modes of expression and contribute to the segmentation clock. Dev. Biol. 304, 615–632. doi:10.1016/j.ydbio.2007.01.004
Sharma, D., Mirando, A. J., Leinroth, A., Long, J. T., Karner, C. M., Hilton, M. J., et al. (2021). HES1 is a novel downstream modifier of the SHH-GLI3 Axis in the development of preaxial polydactyly. PLoS Genet. 17, e1009982. doi:10.1371/journal.pgen.1009982
Sheeba, C. J., Andrade, R. P., and Palmeirim, I. (2012). Joint interpretation of AER/FGF and ZPA/SHH over time and space underlies hairy2 expression in the chick limb. Biol. Open 1, 1102–1110. doi:10.1242/bio.20122386
Sheeba, C. J., Andrade, R. P., and Palmeirim, I. (2014). Limb patterning: From signaling gradients to molecular oscillations. J. Mol. Biol. 426, 780–784. doi:10.1016/j.jmb.2013.11.022
Sheeba, C. J., Andrade, R. P., and Palmeirim, I. (2016). Mechanisms of vertebrate embryo segmentation: Common themes in trunk and limb development. Semin. Cell Dev. Biol. 49, 125–134. doi:10.1016/j.semcdb.2016.01.010
Shifley, E. T., Vanhorn, K. M., Perez-balaguer, A., Franklin, J. D., Weinstein, M., Cole, S. E., et al. (2008). Oscillatory lunatic fringe activity is crucial for segmentation of the anterior but not posterior skeleton. Development 135, 899–908. doi:10.1242/dev.006742
Shih, N. P., Francois, P., Delaune, E. A., and Amacher, S. L. (2015). Dynamics of the slowing segmentation clock reveal alternating two-segment periodicity. Development 142, 1785–1793. doi:10.1242/dev.119057
Shimojo, H., Isomura, A., Ohtsuka, T., Kori, H., Miyachi, H., Kageyama, R., et al. (2016). Oscillatory control of Delta-like1 in cell interactions regulates dynamic gene expression and tissue morphogenesis. Genes Dev. 30, 102–116. doi:10.1101/gad.270785.115
Shimojo, H., Ohtsuka, T., and Kageyama, R. (2008). Oscillations in notch signaling regulate maintenance of neural progenitors. Neuron 58, 52–64. doi:10.1016/j.neuron.2008.02.014
Sieger, D., Tautz, D., and Gajewski, M. (2003). The role of Suppressor of Hairless in Notch mediated signalling during zebrafish somitogenesis. Mech. Dev. 120, 1083–1094. doi:10.1016/S0925-4773(03)00154-0
Sonnen, K. F., Lauschke, V. M., Uraji, J., Falk, H. J., Petersen, Y., Funk, M. C., et al. (2018). Modulation of phase shift between Wnt and notch signaling oscillations controls mesoderm segmentation. Cell 172, 1079–1090. e12. doi:10.1016/j.cell.2018.01.026
Soroldoni, D., Jörg, D. J., Morelli, L. G., Richmond, D. L., Schindelin, J., Jülicher, F., et al. (2014). Genetic oscillations. A Doppler effect in embryonic pattern formation. Sci. (80-. ) 345, 222–225. doi:10.1126/science.1253089
Soto, X., Biga, V., Kursawe, J., Lea, R., Doostdar, P., Thomas, R., et al. (2020). Dynamic properties of noise and Her6 levels are optimized by miR-9, allowing the decoding of the Her6 oscillator. EMBO J. 39, e103558. doi:10.15252/embj.2019103558
Soza-Ried, C., Ozturk, E., Ish-Horowicz, D., and Lewis, J. (2014). Pulses of Notch activation synchronise oscillating somite cells and entrain the zebrafish segmentation clock. Development 141, 1780–1788. doi:10.1242/dev.102111
Sparrow, D. B., Chapman, G., Smith, A. J., Mattar, M. Z., Major, J. A., O’Reilly, V. C., et al. (2012). A mechanism for gene-environment interaction in the etiology of congenital scoliosis. Cell 149, 295–306. doi:10.1016/j.cell.2012.02.054
Suriben, R., Fisher, D. A., and Cheyette, B. N. R. (2006). Dact1 presomitic mesoderm expression oscillates in phase with Axin2 in the somitogenesis clock of mice. Dev. Dyn. 235, 3177–3183. doi:10.1002/dvdy.20968
Takashima, Y., Ohtsuka, T., González, A., Miyachi, H., and Kageyama, R. (2011). Intronic delay is essential for oscillatory expression in the segmentation clock. Proc. Natl. Acad. Sci. U. S. A. 108, 3300–3305. doi:10.1073/pnas.1014418108
Tam, P. P. L. (1981). The control of somitogenesis in mouse embryos. J. Embryol. Exp. Morphol. 65 103–128. doi:10.1242/dev.65.Supplement.103
Tenin, G., Right, D., Ferjentsik, Z., Bone, R., McGrew, M. J., Maroto, M., et al. (2010). The chick somitogenesis oscillator is arrested before all paraxial mesoderm is segmented into somites. BMC Dev. Biol. 10, 24. doi:10.1186/1471-213x-10-24
Tietz, K. T., Gallagher, T. L., Mannings, M. C., Morrow, Z. T., Derr, N. L., Amacher, S. L., et al. (2020). Pumilio response and AU-rich elements drive rapid decay of Pnrc2-regulated cyclic gene transcripts. Dev. Biol. 462, 129–140. doi:10.1016/j.ydbio.2020.03.017
Trofka, A., Schwendinger-schreck, J., Brend, T., Pontius, W., Emonet, T., Holley, S. A., et al. (2012). The Her7 node modulates the network topology of the zebrafish segmentation clock via sequestration of the Hes6 hub. Development 139, 940–947. doi:10.1242/dev.073544
Tsiairis, C. D., and Aulehla, A. (2016). Self-organization of embryonic genetic oscillators into spatiotemporal wave patterns. Cell 164, 656–667. doi:10.1016/j.cell.2016.01.028
Vasiliauskas, D., Laufer, E., and Stern, C. D. (2003). A role for hairy1 in regulating chick limb bud growth. Dev. Biol. 262, 94–106. doi:10.1016/S0012-1606(03)00360-9
Ventre, S., Indrieri, A., Fracassi, C., Franco, B., Conte, I., Cardone, L., et al. (2015). Metabolic regulation of the ultradian oscillator Hes1 by reactive oxygen species. J. Mol. Biol. 427, 1887–1902. doi:10.1016/j.jmb.2015.03.007
Wahi, K., Friesen, S., Coppola, V., and Cole, S. E. (2017). Putative binding sites for mir-125 family miRNAs in the mouse Lfng 3′UTR affect transcript expression in the segmentation clock, but mir-125a-5p is dispensable for normal somitogenesis. Dev. Dyn. 246, 740–748. doi:10.1002/dvdy.24552
Wahl, M. B., Deng, C., Lewandowski, M., and Pourquié, O. (2007). FGF signaling acts upstream of the NOTCH and WNT signaling pathways to control segmentation clock oscillations in mouse somitogenesis. Development 134, 4033–4041. doi:10.1242/dev.009167
Wanglar, C., Takahashi, J., Yabe, T., and &Takada, S. (2014). Tbx protein level critical for clock-mediated somite positioning is regulated through interaction between tbx and ripply. PloS One 9, e107928. doi:10.1371/journal.pone.0107928
Webb, A. B., Lengyel, I. M., Jörg, D. J., Valentin, G., Jülicher, F., Morelli, L. G., et al. (2016). Persistence, period and precision of autonomous cellular oscillators from the zebrafish segmentation clock. Elife 5, e08438. doi:10.7554/eLife.08438
Wiedermann, G., Bone, R. A., Silva, J. C., Bjorklund, M., Murray, P. J., Dale, J. K., et al. (2015). A balance of positive and negative regulators determines the pace of the segmentation clock. eLife 4, e05842. doi:10.7554/eLife.05842
William, D. A., Saitta, B., Gibson, J. D., Traas, J., Markov, V., Gonzalez, D. M., et al. (2007). Identification of oscillatory genes in somitogenesis from functional genomic analysis of a human mesenchymal stem cell model. Dev. Biol. 305, 172–186. doi:10.1016/j.ydbio.2007.02.007
Williams, D. R., Shifley, E. T., Braunreiter, K. M., and Cole, S. E. (2016). Disruption of somitogenesis by a novel dominant allele of Lfng suggests important roles for protein processing and secretion. Development 143, 822–830. doi:10.1242/dev.128538
Winkler, C., Elmasri, H., Klamt, B., Volff, J.-N., and Gessler, M. (2003). Characterization of hey bHLH genes in teleost fish. Dev. Genes Evol. 213, 541–553. doi:10.1007/s00427-003-0360-6
Wright, D., Ferjentsik, Z., Chong, S.-W., Qiu, X., Yun-Jin, J., Malapert, P., et al. (2009). Cyclic Nrarp mRNA expression is regulated by the somitic oscillator but Nrarp protein levels do not oscillate. Dev. Dyn. 238, 3043–3055. doi:10.1002/dvdy.22139
Xie, Z. R., Yang, H., and Hwang, M. J. (2007). The role of microRNA in the delayed negative feedback regulation of gene expression. Biochem. Biophys. Res. Commun. 358, 722–726. doi:10.1016/j.bbrc.2007.04.207
Yaman, Y. I., Huang, R., and Ramanathan, S. (2022). Coupled organoids reveal that signaling gradients drive traveling segmentation clock waves during human axial morphogenesis. bioRxiv 2005, 491359. doi:10.1101/2022.05.10.491359
Yoshioka-Kobayashi, K., Matsumiya, M., Niino, Y., Isomura, A., Kori, H., Miyawaki, A., et al. (2020). Coupling delay controls synchronized oscillation in the segmentation clock. Nature 580 (7801), 119–123. doi:10.1038/s41586-019-1882-z
Yoshiura, S., Ohtsuka, T., Takenaka, Y., Nagahara, H., Yoshikawa, K., Kageyama, R., et al. (2007). Ultradian oscillations of Stat, Smad, and Hes1 expression in response to serum. Proc. Natl. Acad. Sci. U. S. A. 104, 11292–11297. doi:10.1073/pnas.0701837104
Zhang, N., Norton, C. R., and Gridley, T. (2002). Segmentation defects of Notch pathway mutants and absence of a synergistic phenotype in lunatic fringe/radical fringe double mutant mice. Genesis 33, 21–28. doi:10.1002/gene.10081
Keywords: temporal control, embryo clock, somitogenesis, negative feedback regulation, notch signalling, HES
Citation: Carraco G, Martins-Jesus AP and Andrade RP (2022) The vertebrate Embryo Clock: Common players dancing to a different beat. Front. Cell Dev. Biol. 10:944016. doi: 10.3389/fcell.2022.944016
Received: 14 May 2022; Accepted: 11 July 2022;
Published: 11 August 2022.
Edited by:
David M. Holloway, British Columbia Institute of Technology, CanadaReviewed by:
Nick Monk, The University of Sheffield, United KingdomCopyright © 2022 Carraco, Martins-Jesus and Andrade. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Raquel P. Andrade, cmdhbmRyYWRlQHVhbGcucHQ=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.