
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell Dev. Biol. , 02 September 2022
Sec. Stem Cell Research
Volume 10 - 2022 | https://doi.org/10.3389/fcell.2022.940248
Osteoblasts are indispensable for skeletal growth and maintenance. Bone marrow-derived mesenchymal stem cells (BMSCs) are useful in studying osteogenesis. In this study, BMSCs isolated from White Leghorns were differentiated into osteoblasts in vitro. Cells induced for -1, 0, 1, 11, and 22 d were used for transcriptomic analyses using the HISAT2-Stringtie-DESeq2 pipeline. Weighted correlation network analysis was processed to investigate significant modules, including differentially expressed genes (DEGs), correlated with osteogenic differentiation. Gene ontology and pathway enrichment analyses of DEGs were performed to elucidate the mechanisms of osteoblast differentiation. A total of 534, 1,144, 1,077, and 337 DEGs were identified between cells induced for -1 and 0, 0 and 1, 1 and 11, and 11 and 22 d, respectively (|log2FC| > 1.0, FDR <0.05). DEGs were mainly enriched in pathways related to cell proliferation in the early stage of osteogenic differentiation and pathways, such as the TGF-β signaling pathway, in the middle and late stages of osteogenic differentiation. A protein–protein interaction network of the 87 DEGs in the MEturquoise module within top 5-%-degree value was built utilizing the STRING database. This study is the first to elucidate the transcriptomic changes in the osteogenic differentiation of BMSCs isolated from White Leghorns at different times. Our results provide insight into the dynamic transcriptome changes during BMSC differentiation into osteoblasts in chicken.
Commercially bred chickens are reared in high-density feeding environments to meet the demand for meat and egg production (Stratmann et al., 2015). Rapid weight gain (Bradshaw et al., 2002) and prolonged lack of exercise (Webster, 2004; Lay et al., 2011) increase the burden on the chicken skeletal system, leading to abnormal bone development, fractures, and even death. Skeletal problems in poultry are particularly important economic and animal welfare concerns. Therefore, bone properties are important indicators used for evaluating poultry quality and cultivating new poultry varieties in many countries (Rubin et al., 2007; Gonzalez-Ceron et al., 2015).
Bone is composed of a mineral matrix, mainly hydroxyapatite, and an organic matrix containing collagen, lipids, proteoglycans, and other bone structural proteins (Johnsson et al., 2015). A reduction of the mineralized bone mass increases the incidence of bone fragility and fractures in chickens, similar to humans and mammals (Rubin et al., 2007). Previous studies have noted that the skeletal system of chickens has a complex phenotype influenced by genetics, nutrition, hormones, and housing type (Reid, 1998; Rath et al., 2000; Whitehead & Fleming, 2000). Among these factors, genetic factors are the most fundamental (Rubin et al., 2007; Gonzalez-Ceron et al., 2015).
Osteoblasts account for 4–6% of the total residential bone cells and maintain bone mass by interacting with osteoclasts and osteocytes (Capulli et al., 2014). Osteoblasts derived from pluripotent mesenchymal stem cells (MSCs) have self-renewal capacity (Shanti et al., 2007) and can be obtained from different origins, such as bone marrow and adipose tissue (Beane & Darling, 2012). Current studies on bone marrow-derived MSCs (BMSCs) mostly use cells isolated from mammals, such as humans (Baghaei et al., 2017), rats (Li et al., 2013), mice (Huang et al., 2015), and pigs (Feyen et al., 2016). Chicken BMSCs have been successfully isolated from chicken embryos (Young et al., 1993) and postnatal chicken bone marrow (Bai et al., 2013). Alkaline phosphatase (ALP) staining was positive and alizarin red staining demonstrated the formation of mineralized calcium nodules when the osteogenic differentiation of BMSCs into osteoblasts was induced in vitro.
To date, studies on chicken skeletal system development and gene regulation mechanisms have been incomplete. In this study, dynamic changes in the transcriptome during osteogenic differentiation of BMSCs isolated from White Leghorn chicken embryos incubated for 15 d were analyzed. To our knowledge, this is the first study to analyze osteogenic differentiation of chicken BMSCs using transcriptome sequencing. Therefore, our study contributes a novel approach for further research on bone development in chicken by improving the gene regulatory network.
Unless otherwise stated, all chemicals used in the experiments were purchased from Sigma-Aldrich (St. Louis, MO, USA). All animal experiments followed the principles formulated by the Ministry of Agriculture of China and were performed in accordance with the guidelines of the Animal Care Committee of China Agricultural University (permit number: AW01111202-1-2).
White Leghorn eggs, hatched after incubation in an environment with a temperature of 37°C, 5% CO2, and 60% humidity for 15 d, were provided by Poultry Genetic Resources and Breeding Experimental Base of China Agricultural University. After euthanasia, the thighs were separated from the chicken embryos and soaked in 75% alcohol for 5 min under sterile conditions. The tibia was obtained after removing the attached muscle tissue and rinsing with phosphate-buffered saline (PBS) containing 5% penicillin-streptomycin (Gibco) three times. The metaphyseal was resected, and bone marrow was collected from the tibia by flushing with complete medium containing DMEM/F-12 (Gibco), 10% fetal bovine serum (FBS) (Gibco), and 1% penicillin-streptomycin (Gibco). The cell suspension was then centrifuged and cells were washed three times at 100 g for 10 min in complete medium. Then, the cells were resuspended in complete medium and plated into a 25 cm2 culture flask (Corning, Glendale, AZ, USA). The medium was refreshed for the first time after 14 h and every 2 days thereafter.
Cells were subcultured at a ratio of 1:2 when the cell confluence reached approximately 80% and purified using the differential adhesion method. After dissociation with 0.25% trypsin-EDTA (Gibco) and centrifugation at 100 g for 5 min, cells were suspended in complete medium and plated into culture flasks. Nonadherent cells were removed after 30 min and replaced with fresh medium. Adherent cells were labeled as the P1 generation. The procedure was repeated and P2, P3,..., Pn generations were obtained for further experiments.
Generation P3 was used for immunofluorescence staining and RT-PCR analysis of cell surface markers. Immunofluorescence assays was adopted to analyzed the expressions of CD29, CD44 and CD45. Cells were plated and fixed with 4% paraformaldehyde (Biosharp, Hefei, Anhui, China) and penetrated with 0.5% Triton X-100 (Solarbio, Beijing, China). The detections of P3 BMSCs CD29 (Proteintech, Wuhan, Hubei, China), CD44 (Bioss, Beijing, China), and CD45 (Proteintech, Wuhan, Hubei, China) were performed according to the instructions of antibodies. The nucleus was stained with DAPI (Solarbio, Beijing, China). Total RNA was extracted from P3 BMSCs using the Total RNA Extraction Kit (Omega BioTek, Norcross, GA, USA), and cDNA was obtained using the PrimeScript RT reagent Kit (TaKaRa Bio, Shiga, Japan) following the manufacturer’s manual. Primers for CD29, CD44, CD45, CD71, and GAPDH were specifically designed for PCR amplification (Table 1).
Generation P3 BMSCs were plated in 12-well plates and 75 cm2 culture flasks for osteogenic differentiation. Osteogenic induction medium, containing DMEM/F-12, 10% FBS, 1% penicillin-streptomycin, 10 nmol/L dexamethasone, 10 mmol/L β-glycerophosphate, and 50 μg/mL L-ascorbic acid, was added to the osteogenic induction groups to induce osteogenic differentiation of BMSCs. BMSCs at a cell confluence of 80% were labeled as the -1 day group, for studying changes in gene expression profile between varying stages of proliferation and levels of contact inhibition. After 24 h, BMSCs at a cell confluence of approximately 90% were labeled as the day zero group. A total of five osteogenic induction groups were set in this experiment depending on the number of days of osteogenic induction (-1, 0, 1, 11, and 22 d). The induction experiments were replicated three times for each group. During osteogenic differentiation, the induction medium was replaced every 3 days. Cells in culture flasks were collected for mRNA-seq on the corresponding days of induction. Concurrently, cells from these osteogenic induction groups in the 12-well plates were used for ALP staining (Beyotime Biotechnology, Shanghai, China) and Alizarin Red staining (Solarbio, Beijing, China), according to the manufacturer’s instructions.
Cells from the different treatment groups were collected in 2-ml tubes. The total RNA of each sample was extracted using a Total RNA Kit (Omega) according to the manufacturer’s instructions. The quality and concentration of the RNA samples were confirmed using NanoDrop-2000 (Thermo Fisher Scientific, Waltham, MA, USA). All RNA samples with ultraviolet absorbance ratios (A260/A280) between 1.8 and 2.0 were selected for RNA sequencing library preparation. cDNA library construction was performed using the Illumina NovaSeq PE150 platform (Beijing Youji Technology, Beijing, China). Clean reads used for further analysis were filtered and generated according to the following criteria: 1) reads with adapter pollution or N ratio of >5%; 2) reads with low quality of Q-value <20. The RNA-seq raw data were deposited in the NCBI SRA database under the accession number PRJNA824833.
Read mapping and transcript expression level quantification were performed using the HISAT2, StringTie, and DESeq2 workflows (Pertea et al., 2016). The high-quality reads obtained were aligned to the chicken reference genome (GRC6a) using HISAT2 (v2.1.0) (Kim et al., 2015). Transcript assembly, GTF document merging, and transcript quantification were performed using StringTie (v1.3.5) (Pertea et al., 2015). Statistical analysis of DEGs was performed using DESeq2 (v) with the criteria of |log2FC| >1.0 and FDR <0.05. Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses of DEGs were performed using DAVID (https://david.ncifcrf.gov/home.jsp) with default parameters. Weighted correlation network analysis (WGCNA) (Langfelder & Horvath, 2008) was performed to identify the modules of highly correlated genes during osteogenic differentiation. STRING (https://string-db.org/) and CytoNCA (Tang et al., 2015) were used to analyze and visualize the protein–protein interaction network of genes in the identified modules with high correlation.
After using the differential adhesion method for BMSC purification three times, immunofluorescence staining was applied to detected cell surface markers. Immunofluorescence showed that BMSCs were positive staining for CD29 and CD44, but negative for CD45 (Figure 1A). RNA was extracted from P3 BMSCs, reverse transcribed to cDNA, and RT-PCR was used to identify BMSCs. The results showed that P3 BMSCs isolated from White Leghorns were positive for CD29, CD44, and CD71, but negative for CD45 (Figure 1B).
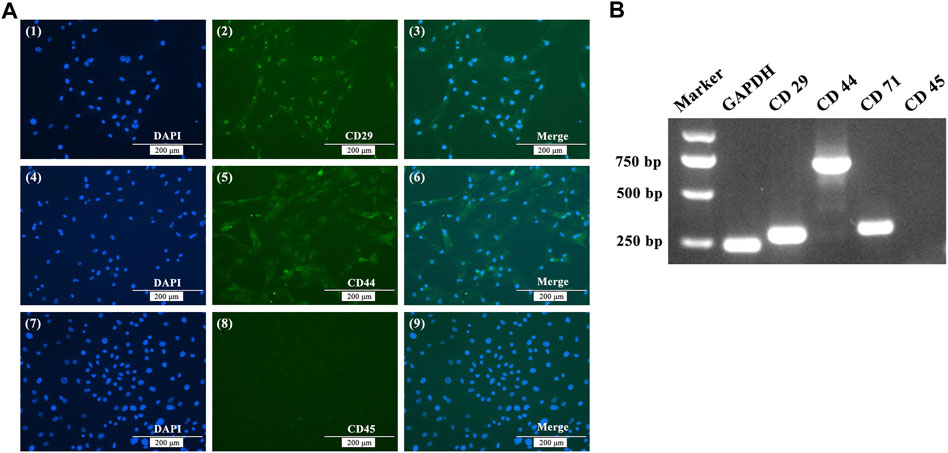
FIGURE 1. Immunofluorescence staining (A) and RT-PCR (B) analysis of BMSC cell surface markers. (A) Immunofluorescence showing positive staining for CD29 and CD44, but negative for CD45. (B) RT-PCR assays showed that the expression of GADPH, CD29, CD44 and CD71 were positive, while the expression of CD45 was negative.
Under the same experimental conditions, Alizarin Red staining and ALP staining were performed on BMSCs after osteogenic differentiation in a 12-well plate at different time points. As shown in Figure 2A, ALP staining and Alizarin Red staining of BMSCs were detected 22 d after induction. These results suggest that chicken BMSCs successfully differentiated into osteoblasts.
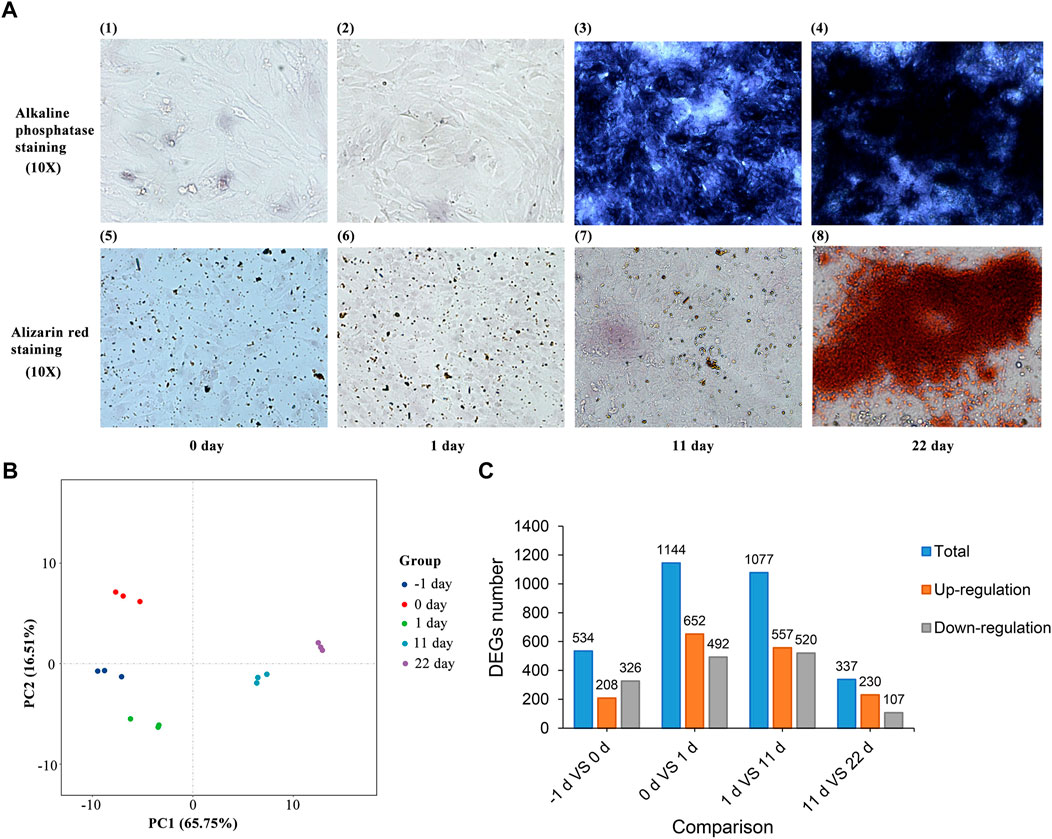
FIGURE 2. (A) ALP staining and Alizarin Red staining of BMSC osteogenic induction. (B) Principal component analysis of BMSCs induced for -1, 0, 1, 11 and 22 d. (C) Statistics of the differentially expressed genes between adjacent induction groups.
After processing with StringTie, principal component analysis (PCA) of all the samples was performed (Figure 2B). The results showed that differences between and within groups were appropriate for subsequent analysis. DEGs between groups with different induction times were identified after analysis using DESeq2. A total of 534, 1,144, 1,077, and 337 DEGs (Figure 2C and Supplementary Table S1) were identified between the groups induced for -1 and 0, 0 and 1, 1 and 11, and 11 and 22 d, respectively (|log2FC| > 1.0, FDR <0.05).
All the DEGs identified in the present study may influence the osteogenic differentiation of BMSCs. Functional annotation was performed on all DEGs to ascertain the functions of these genes. The results of the GO enrichment analysis and KEGG pathway analysis of up-regulated and down-regulated DEGs between two adjacent groups are displayed in Supplementary Table S2 and Supplementary Table S3, respectively. To explicitly show all the DEGs at the different times of the osteogenesis induction, the results of the KEGG pathway (p < 0.05) are presented in Figure 3. In the early stage of osteogenic differentiation (Figures 3A,B), the DEGs were enriched in pathways including the cell cycle, DNA replication, homologous recombination, and mismatch repair. The DEGs between 11 and 1 d of osteogenic differentiation (Figure 3C) were enriched in steroid biosynthesis, cell cycle, calcium signaling pathway, ECM–receptor interaction, and the MAPK signaling pathway. The DEGs between 22 and 11 d of osteogenic differentiation (Figure 3D) were mostly enriched in the TGF-β signaling pathway, MAPK signaling pathway, PPAR signaling pathway, Wnt signaling pathway, and cytokine–cytokine receptor interaction.
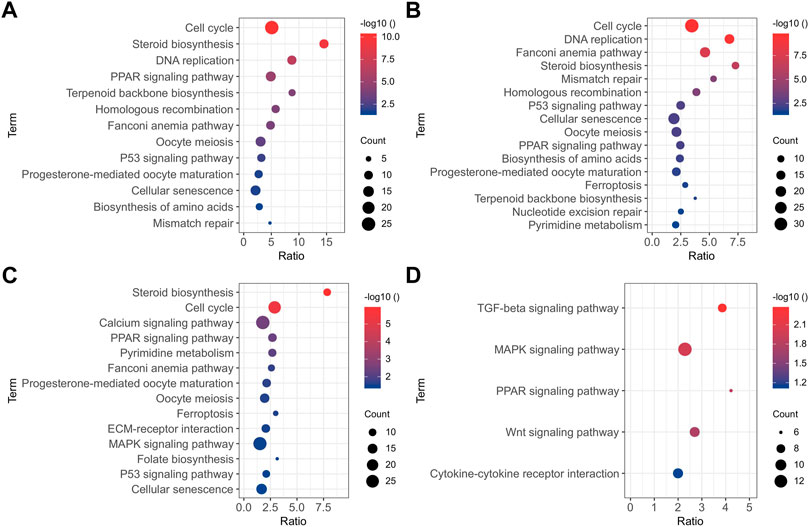
FIGURE 3. KEGG pathway analysis (p < 0.05) of DEGs between 0 and -1 (A), 1 and 0 (B), 11 and 1 (C), and 22 and 11 (D) d of osteogenic induction.
To gain a deeper understanding of the gene interactions occurring during osteogenic differentiation of BMSCs, WGCNA was performed to identify significant modules including DEGs that are highly correlated to osteogenic differentiation. A total of 4,577 nonredundant DEGs (Supplementary Table S4) was used to evaluate availability and construct a gene co-expression network (Figure 4A). The results of module-trait relationships (Figure 4B) showed that the MEblue module on day zero of induction, genes in the MEbrown module on day one of induction, and genes in the MEturquoise module at 11 and 22 days of induction were correlated with osteogenic differentiation, indicating that the MEblue and MEbrown modules are related with the early stage of osteogenic differentiation and that the MEturquoise module is related with the middle and late stages of osteogenic differentiation. The gene sets in the above induction times and corresponding modules were extracted for GO enrichment (Supplementary Table S5) and KEGG pathway (Table 2) analyses. The results of KEGG pathway analysis showed that the genes in the early stage of osteogenic differentiation were mainly enriched in pathways related to cell proliferation, such as DNA replication (gga03030), homologous recombination (gga03440), and cell cycle (gga04110), and the corresponding genes in the middle and late stages of osteogenic differentiation were mainly enriched in the PPAR signaling pathway (gga03320), MAPK signaling pathway (gga04010), TGF-β signaling pathway (gga04350), Wnt signaling pathway (Wnt signaling pathway), and others. The expression of the genes RUNX2, SPP1, and BGLAP (Figure 4C) and the correlation between genes and modules (Figure 4D) were analyzed, and the results showed that the turquoise module was significantly correlated (0.87–0.9, p-value < 0.01) with these three osteogenic marker genes.
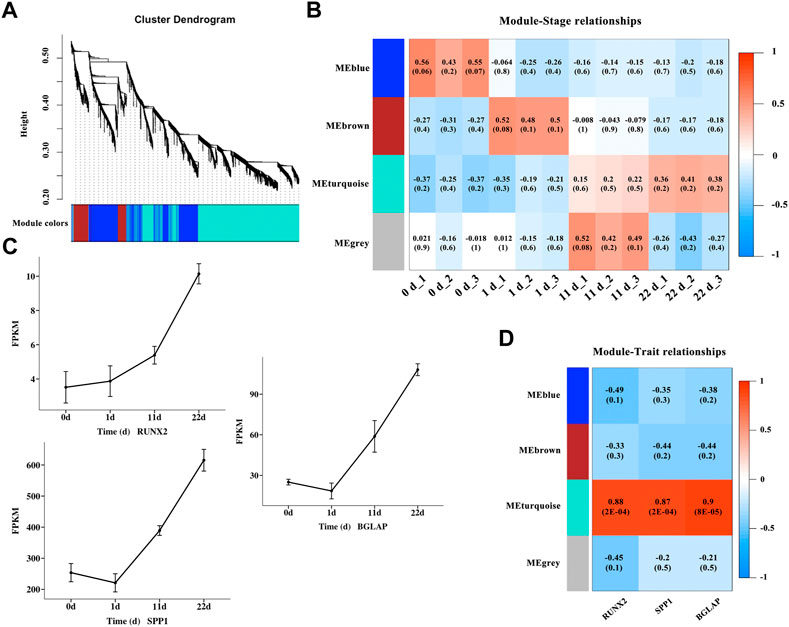
FIGURE 4. Identification of modules associated with osteogenic differentiation in different induction stages. (A) Clustering dendrogram of DEGs in different osteogenic induction groups. As a result, 4 modules with different colors were constructed. (B) Module-trait association and the corresponding p-value with all the DEGs (listed in Supplementary Table S4) at different osteogenic induction time. (C) The mRNA variation tendency of genes RUNX2, SPP1 and BGLAP. Horizontal coordinate represents the induction time. The ordinate represents the changes of fragments per kilobase per million (FPKM). (D) Module-gene correlations and the corresponding p-value of genes RUNX2, SPP1 and BGLAP.
A protein–protein interaction (PPI) network was constructed for DEGs in the MEturquoise module with STRING database and Cytoscape. A total of 87 nodes with the top 5%-degree value calculated by CytoNCA (Supplementary Table S6) were mapped to the PPI network (Figure 5), including JUN, FN1, IL6, MET, PTEN, ERBB4, ITGB2, ITGB3, BMP4, RUNX2, and SPP1.
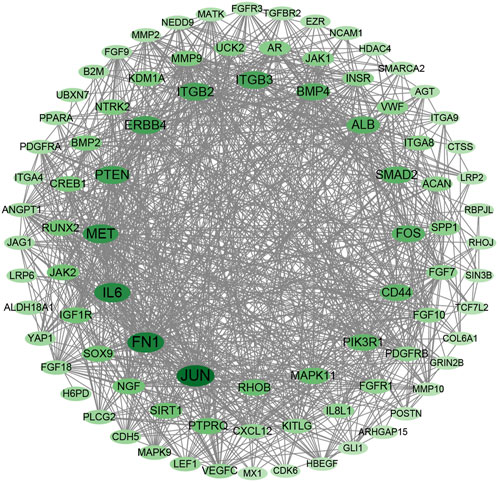
FIGURE 5. Protein–protein interaction network with DEGs in MEturquoise module within the top 5-%-degree value (Supplementary Table S6).
Over the long process of domestication, artificial selection has led to phenotypic differences between domestic chickens and Gallus (Talaty et al., 2010). The bone health of commercial breeds of laying hens (Hocking et al., 2004) and broilers (Silversides et al., 2006) varies considerably and the bone of these breeds is generally weaker than that of traditional breeds. Osteoporosis and fractures worsen the welfare and economic concerns in the chicken industry. Bone maturation involves various complicated molecular and biochemical changes in osteoblasts and osteoclasts. After basic structural development and mineralization are completed, the mature bone has physical and biological mechanical properties (Rath et al., 2000). Factors including bone mineral density, bone microarchitecture, bone geometry, and bone material properties are relevant to bone strength (Friedman, 2006). Studies have shown that the development of the skeletal system, like that of other systems and traits, is controlled by complex genetic networks (Bishop et al., 2000; Schreiweis et al., 2005; Raymond et al., 2018). Genetic selection has significant potential for improving bone strength and reducing osteoporosis (Mandour et al., 1989). Understanding the mechanisms of bone metabolism, such as mineralization during bone development, will provide novel insights into improving poultry bone performance and industrial efficiency in the future.
BMSCs have the potential for multi-lineage differentiation, such as osteogenic and adipogenic differentiation, making them useful tools for studying mineralization during osteogenesis. Many studies have demonstrated the potential of BMSCs for treating osteoporosis and osteoarthritis in humans (Kiernan et al., 2016; Liu et al., 2021). However, collection of human BMSCs requires general anesthesia, limiting its research and clinical applications (Mushahary et al., 2018). Based on previous research, MSCs from other tissues, such as adipose tissues and dental tissues, have become viable substitutes. At present, research on BMSCs in humans and mice mostly focus on the function of one or several genes (Frank et al., 2002; Wang et al., 2021) or microRNA (Lin et al., 2019) in the process of osteogenic differentiation. Zhang et al. analyzed the functional network of differential long noncoding RNA/mRNA after 7 days of osteogenic differentiation of human BMSCs (Zhang et al., 2017). Their results showed that DEGs were enriched in GO terms including cell adhesion and skeletal system development and pathways including cytokine-cytokine receptor interaction, ECM-receptor interaction and TGF-beta signaling pathway, which is consistent with the results of our study. There are few studies systematically analyzed RNA expression changes during different stages of osteogenic differentiation. This study analyzed the transcriptome changes of chicken BMSCs differentiation into osteoblasts at different time point in detail, providing research basis and experimental data for the future study on the osteogenic differentiation process.
The surface markers of BMSCs from different sources differ. CD29 is a member of the integrin gene superfamily that participates in cell-matrix and cell-cell adhesion as an integrin receptor (Jiménez-Marín et al., 2000). CD44 is a multi-structural and multifunctional cell surface glycoprotein that is involved in cell proliferation and differentiation, the docking of proteases on cell membranes, and signal transmission for cell survival (Naor et al., 2002). CD44 provides growth anchor sites for MSCs owing to the binding sites for type I collagen and fibronectin (Bai et al., 2013). CD71 is also known as a marker of BMSC cytoplasts and mediates cellular iron uptake. CD45 is a hematopoietic cell-specific tyrosine phosphatase that is expressed in all leukocytes (Tchilian & Beverley, 2006). RT-PCR assays showed that chicken BMSCs in this experiment were CD29−, CD44−, and CD71-positive, but CD45-negative, which is consistent with the findings of previous studies (Bai et al., 2013; Bhuvanalakshmi et al., 2014). BMSCs have a vast differentiation potential and are easily accessible in chicken. With the continuous in-depth study of isolation methods and differentiation potential, BMSCs can provide prospects for future research on avian developmental biology and molecular breeding.
Bone characteristics have become an important index for measuring poultry quality during breeding at home and abroad. Osteogenesis is a complex physiological process that has not yet been fully elucidated, especially in poultry. Understanding the process of BMSC differentiation into osteoblasts might help reveal the mechanisms, key genes, and pathways involved in osteoblast differentiation. In this study, BMSCs from White Leghorns were isolated and cultured to differentiate into osteoblasts. Many DEGs associated with osteogenic differentiation were analyzed using transcriptome sequencing of cells collected at different osteogenic differentiation time points. The key gene sets of the different differentiation times were further explored using WGCNA analysis and found to be enriched in several pathways, including DNA replication, cell cycle, Wnt signaling pathway, TGF-β signaling pathway and ECM-receptor interaction. Bones are dynamic organs and are replaced with newly deposited bone in humans and other vertebrates as they age or get damaged (Boskey, 2006). In this study, the DEGs of BMSCs during osteogenic differentiation were partially enriched in GO terms involving the cell surface, such as extracellular matrix, cell surface and cell adhesion, and vascular-related GO terms, including angiogenesis and blood vessel development. Bone marrow contains complex vascular networks and a variety of cell types including hematopoietic stem cells and MSCs. Stromal cells regulate hematopoietic cells by establishing highly specific interactions through niches (Gomariz & Helbling, 2018). BMSCs are found in perivascular locations, and blood vessels play an essential role in maintaining tissue functions, especially for the survival of stem cells within tissues (Sivan et al., 2019).
Genes enriched in DNA replication and the cell cycle may also influence the process of osteoblastic differentiation. Minichromosome maintenance proteins (MCMs) are DNA-dependent ATPases that are loaded onto replication regions and allow them to support the initiation of DNA replication (Sedlackova et al., 2020). The MCM complex consists of six subunits, MCM2–7. MCM3, which serves as a licensing factor, is essential for the initiation of DNA replication in eukaryotic cells. Yamamoto et al. reported that low-level laser irradiation may influence cell proliferation of osteoblasts through the enhancement of MCM family gene expression, such as MCM3 gene expression (Yamamoto et al., 2001). CDK2 is a catalytic subunit of the cyclin-dependent protein kinase complex that is essential for the G1/S phase of the cell cycle (Fei et al., 2010). Osteogenic growth peptide, one of the stimulators of bone formation, stimulates MSC proliferation and increases the expression of cyclin A in MSCs at both the mRNA and protein levels (Fei et al., 2010).
Energy metabolism plays important role in BMSC survival and differentiation. The intrinsic mechanism of BMSC differentiation remains incompletely understood. Glucose and fatty acid metabolism are considerable regulators in BMSC differentiation (Gastel & Carmeliet, 2021). DEGs during BMSC osteogenic differentiation are enriched in GO terms and KEGG pathways including ATP binding, metabolic pathways, and fatty acid metabolism. Huang et al. identified that the inhibition of HMGCR increased glycolysis, and observed the alteration in HMGCR gene expression during osteoclast differentiation (Huang et al., 2021). HMGCR activity is a key factor in cholesterol biosynthesis and bone formation in rodents (Mundy et al., 1999). Fatty acid levels in the bone microenvironment are correlated with diseases such as periodontitis, osteoporosis, bone fracture, and tumor-associated bone destruction (Bao et al., 2020). Short-chain fatty acids have been proved to play a crucial role in bone metabolism and immune responses in reducing the severity of arthritis in mice (Lucas et al., 2018). Palmitate, a fatty acid, promotes energy production during osteoblast differentiation and accelerates bone formation (Lee et al., 2017). The generation of ATP is mainly completed by β-oxidation of fatty acids in the mitochondria. Therefore, fatty acid synthesis is an effective way to store carbohydrates. Fatty acids are shuttled and stored in lipid droplets in the form of triglycerides or cholesteryl esters in almost all types of cells for later use (Kushwaha et al., 2018). Fatty acids and lipids can be available for producing ATP for osteoblastic requirements through the citric acid cycle (Lee et al., 2017). Studies have shown that osteoblasts absorb fatty acids; however, their mechanism of action has not been clearly described. The expression of FADS2, ACAA2, ACSL5, and ACAT2, which are enriched in fatty acid metabolism, was significantly increased at the early stage of osteogenic differentiation. Genetic variation of a few fatty acids at the plasma level and variants in the FADS1-FADS2 gene regions were correlated with the estimated bone mineral density and fracture risk in summary-level data from up to 426,824 individuals in the UK Biobank (Yuan et al., 2020). Evidence on the association of individual fatty acids with bone mineral density and fracture risk in chickens is scarce and is a prospective research direction. Energy metabolism enables BMSCs to match the different demands of osteo-adipogenic differentiation. Additional studies are needed to fully understand the metabolism in osteogenesis.
Wnt proteins are secreted morphogens that are indispensable for basic developmental processes and are conserved in species from nematodes to mammals (Maeda & Kobayashi, 2019). A total of 19 types of Wnt proteins have been identified in humans. Secreted Wnt proteins activate the Wnt signaling pathway in target cells. The β-catenin-mediated canonical Wnt signaling pathway and β-catenin-independent non-canonical Wnt signaling pathways are two types of Wnt signaling pathways. The β-catenin protein is involved in various physiological processes, such as cell adhesion and transcriptional regulation, and interacts with Wnt ligands. Wnt/β-catenin signaling is an important regulator of MSC fate, and Wnt family members Wnt6 and Wnt10a are the most promising candidates for bone formation (Cawthorn et al., 2012; Feng et al., 2020). In our study, Wnt6 was also one of the DEGs in the late stage of osteogenic differentiation, which is consistent with the results of Cawthorn. et al. The bone mass of transgenic mice overexpressing Wnt10b in adipose tissue was increased (Bennett et al., 2005). The role of Wnt signaling in osteogenesis has been extensively studied in mammals and humans.
The TGF-β signaling pathway and the bone morphogenetic protein (BMP) signaling pathway, play multiple roles in cell proliferation, differentiation, adhesion, and senescence (Zhao et al., 2020). To date, more than 20 BMPs have been documented, which are members of the TGF-β superfamily, except for BMP1 (Wozney et al., 1989). The growth factor BMPs mediate a variety of biological processes, such as embryo development and skeletal morphogenesis (Wan & Cao, 2005). BMP2, one of the DEGs involved in late osteogenic differentiation in our study, is related to skeletal development, postnatal bone growth, and fracture repair. TGF-β receptor II (TGFBR2) is also necessary for postnatal bone development in osterix-expressing cells (Peters et al., 2017). BMP3 can act as an endogenous antagonist of the BMP signaling pathway in bone formation owing to its similarity and abundance of amino acids in the bone extracellular matrix (ECM) (Lowery & Rosen, 2018). Collagen type IV alpha 2 (COL4A2), a member of the ECM-receptor interaction that is enriched in the late stage of osteogenic differentiation, is the main structural component of the basement membrane. COL4A2 is likely a potential biomarker in esophageal cancer (Warnecke-Eberz et al., 2016), lung cancer (Ilhan-Mutlu et al., 2016), and breast cancer (Huan et al., 2014). Few studies have clearly described the direct relationship between COL4A2 and bone development. A previous study showed that an atherogenic diet decreases bone density and increases serum cholesterol levels in male mice. Oxidized low-density lipoprotein increased the size and number of osteoclasts, the expression of CD36 and liver X receptors (LXRα) in osteoclasts, and low-density lipoprotein receptor and LXRα in osteoblasts, demonstrating that cholesterol could induce bone loss by increasing the activity of osteoclasts in mice (Sul et al., 2020). Interestingly, we also found that UCHL1, ATP8A2, CCK, and TRH were enriched in eating behavior (GO:0042755), a biological process, in the GO term. Dietary habits may cause changes in cholesterol and other substances in vivo, thereby affecting bone properties, such as bone mass. In addition, the expansion and survival of BMSCs and osteoblasts isolated from CD36-knockout mice decreased in vitro, as did the expression of RUNX2, osteogenic gene (OSX), bone sialoprotein (BSP), and osteocalcin (OCN), indicating that CD36 is essential for sufficient bone metabolism and that it mediates bone formation through osteoblasts (Kevorkova et al., 2013).
The coordinated and precise expression of the genome maintains the normal development of multicellular organisms (Foletta, 1996). Transcription factors regulate the expression of target genes involved in proliferation and differentiation (Sevilla et al., 2001). RUNX2 is the first essential transcription factor guiding MSCs to an osteoblastic lineage and inhibiting their differentiation into adipocytes and chondrocytes (Komori, 2010). The gene expression of RUNX2 increased gradually in our study, similar to the results of Komori (2008), verifying that RUNX2 keeps osteoblasts in an immature stage with low mineralization. However, further research with the extension of osteogenic induction time of chicken BMSCs is needed to verify whether the expression of RUNX2 is inhibited. The activator protein-1 (AP-1) is a dimeric transcription factor composed of JUN, FOS, and activating transcription factor subunits (Karin et al., 1997). JUN and FOS were DEGs within the MEturquoise module, and JUN protein exhibited the highest degree value in the PPI network. Prolonged expression of c-FOS results in potentiation of osteoclast differentiation (Miyauchi et al., 1994). FOS and JUN proteins are initially highly expressed and then inhibited during mineralization, which is essential to osteoblast differentiation in mice (Sandberg et al., 1988). Adequate dietary supplementation of calcium, vitamin D, and phosphorus affects bone strength in poultry. The gastrointestinal tract is essential for the absorption of nutrients and health in humans and animals. Distinct FOS/JUN members of gastrointestinal epithelium in the chicken embryonic digestive tract were analyzed by in situ hybridization and were important in preserving epithelial–mesenchymal interactions (Matsumoto et al., 1998). Therefore, FOS/JUN proteins, which are well-studied in osteogenic differentiation, can serve as candidate transcription factors affecting bone development in chickens by influencing the absorption of nutrients in intestinal epithelial cells.
In this study, dynamic changes in the transcriptome during osteogenic differentiation of BMSCs in White Leghorns were outlined. The DEGs from the gene set related to cell proliferation, such as cell cycle and DNA replication in the early stage of osteogenic differentiation, to the gene set enriched in the Wnt and TGF-β signaling pathways in the late stage of osteogenic differentiation, were analyzed and discussed. A DEG-related PPI network was constructed using the STRING database. The results of this study detail the transcriptome changes during the differentiation of BMSCs into osteoblasts, as well as the genes and pathways associated with bone development processes, such as osteoblast differentiation and mineralization.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI BioProject accession number: PRJNA824833.
The animal study was reviewed and approved by the Animal Care Committee of China Agricultural University.
SC and HL constructed this study. HL and TW performed the experiment and RNA-seq analysis. HL, TW, and SZ visualized the results. HL wrote the manuscript draft. SC and ZH revised the manuscript. All authors contributed to the article and approved the submitted version.
This research was funded by Fund of China Agricultural University Education Foundation, grant number SJ2021002003-2421006, Primary Research & Development Plan of Jiangsu Province, grant umber BE2017309, National Natural Science Foundation of China, grant number U1702232-1 and Programs for Changjiang Scholars and Innovative Research in University, grant number IRT15R62.
The authors wish to thank Fan Zhang and Qiangsen Zhao for advice on data analysis.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2022.940248/full#supplementary-material
Baghaei, K., Hashemi, S. M., Tokhanbigli, S., Asadi Rad, A., Assadzadeh-Aghdaei, H., Sharifian, A., et al. (2017). Isolation, differentiation, and characterization of mesenchymal stem cells from human bone marrow. Gastroenterol. Hepatol. Bed Bench 10, 208–213.
Bai, C., Hou, L., Ma, Y., Chen, L., Zhang, M., and Guan, W. (2013). Isolation and characterization of mesenchymal stem cells from chicken bone marrow. Cell Tissue Bank. 14, 437–451. doi:10.1007/s10561-012-9347-8
Bao, M., Zhang, K., Wei, Y., Hua, W., Gao, Y., Li, X., et al. (2020). Therapeutic potentials and modulatory mechanisms of fatty acids in bone. Cell Prolif. 53, e12735. doi:10.1111/cpr.12735
Beane, O. S., and Darling, E. M. (2012). Isolation, characterization, and differentiation of stem cells for cartilage regeneration. Ann. Biomed. Eng. 40, 2079–2097. doi:10.1007/s10439-012-0639-8
Bennett, C. N., Longo, K. A., Wright, W. S., Suva, L. J., Lane, T. F., Hankenson, K. D., et al. (2005). Regulation of osteoblastogenesis and bone mass by Wnt10b. Proc. Natl. Acad. Sci. U. S. A. 102, 3324–3329. doi:10.1073/pnas.0408742102
Bhuvanalakshmi, G., Arfuso, F., Dharmarajan, A., and Warrier, S. (2014). Multifunctional properties of chicken embryonic prenatal mesenchymal stem cells- pluripotency, plasticity, and tumor suppression. Stem Cell Rev. Rep. 10, 856–870. doi:10.1007/s12015-014-9530-3
Bishop, S. C., Fleming, R. H., McCormack, H. A., Flock, D. K., and Whitehead, C. C. (2000). Inheritance of bone characteristics affecting osteoporosis in laying hens. Br. Poult. Sci. 41, 33–40. doi:10.1080/00071660086376
Boskey, A. L. (2006). Mineralization, structure and function of bone. Dynamics of Bone and cartilage metabolism. Second Edition. Burlington: Academic Press, 201–212. doi:10.1016/B978-012088562-6/50013-3
Bradshaw, R. H., Kirkden, R. D., and Broom, D. M. (2002). A review of the aetiology and pathology of leg weakness in broilers in relation to welfare. Avian Poul. Biolog. Rev. 13, 45–103. doi:10.3184/147020602783698421
Capulli, M., Paone, R., and Rucci, N. (2014). Osteoblast and osteocyte: Games without frontiers. Arch. Biochem. Biophys. 561, 3–12. doi:10.1016/j.abb.2014.05.003
Cawthorn, W. P., Bree, A. J., Yao, Y., Du, B., Hemati, N., Martinez-Santibañez, G., et al. (2012). Wnt6, Wnt10a and Wnt10b inhibit adipogenesis and stimulate osteoblastogenesis through a β-catenin-dependent mechanism. Bone 50, 477–489. doi:10.1016/j.bone.2011.08.010
Fei, Q., Guo, C., Xu, X., Gao, J., Zhang, J., Chen, T., et al. (2010). Osteogenic growth peptide enhances the proliferation of bone marrow mesenchymal stem cells from osteoprotegerin-deficient mice by CDK2/cyclin A. Acta Biochim. Biophys. Sin. 42, 801–806. doi:10.1093/abbs/gmq086
Feng, L., Zhang, J. F., Shi, L., Yang, Z. M., Wu, T. Y., Wang, H. X., et al. (2020). MicroRNA-378 suppressed osteogenesis of MSCs and impaired bone formation via inactivating wnt/β-catenin signaling. Mol. Ther. Nucleic Acids 21, 1017–1028. doi:10.1016/j.omtn.2020.07.018
Feyen, D. A., van den Akker, F., Noort, W., Chamuleau, S. A., Doevendans, P. A., and Sluijter, J. P. (2016). Isolation of pig bone marrow-derived mesenchymal stem cells. Methods Mol. Biol. 1416, 225–232. doi:10.1007/978-1-4939-3584-0_12
Foletta, V. C. (1996). Transcription factor AP-1, and the role of Fra-2. Immunol. Cell Biol. 74, 121–133. doi:10.1038/icb.1996.17
Frank, O., Heim, M., Jakob, M., Barbero, A., Schäfer, D., Bendik, I., et al. (2002). Real-time quantitative RT-PCR analysis of human bone marrow stromal cells during osteogenic differentiation in vitro. J. Cell. Biochem. 85, 737–746. doi:10.1002/jcb.10174
Friedman, A. W. (2006). Important determinants of bone strength: Beyond bone mineral density. J. Clin. Rheumatol. 12, 70–77. doi:10.1097/01.rhu.0000208612.33819.8c
Gastel, N. V., and Carmeliet, G. (2021). Metabolic regulation of skeletal cell fate and function in physiology and disease. Nat. Metab. 3, 11–20. doi:10.1038/s42255-020-00321-3
Gomariz, A., Helbling, P. M., Isringhausen, S., Suessbier, U., Becker, A., Boss, A., et al. (2018). Quantitative spatial analysis of haematopoiesis-regulating stromal cells in the bone marrow microenvironment by 3D microscopy. Nat. Commun. 9, 2532. doi:10.1038/s41467-018-04770-z
Gonzalez-Ceron, F., Rekaya, R., and Aggrey, S. E. (2015). Genetic analysis of bone quality traits and growth in a random mating broiler population. Poult. Sci. 94, 883–889. doi:10.3382/ps/pev056
Hocking, P. M., Channing, C. E., Robertson, G. W., Edmond, A., and Jones, R. B. (2004). Between breed genetic variation for welfare-related behavioural traits in domestic fowl. Appl. Animal Behav. Sci. 89, 85–105. doi:10.1016/j.applanim.2004.03.014
Huan, J. L., Gao, X., Xing, L., Qin, X. J., Qian, H. X., Zhou, Q., et al. (2014). Screening for key genes associated with invasive ductal carcinoma of the breast via microarray data analysis. Genet. Mol. Res. 13, 7919–7925. doi:10.4238/2014.September.29.5
Huang, J., Zhao, X., Li, X., Peng, J., Yang, W., and Mi, S. (2021). HMGCR inhibition stabilizes the glycolytic enzyme PKM2 to support the growth of renal cell carcinoma. PLoS Biol. 19, e3001197. doi:10.1371/journal.pbio.3001197
Huang, S., Xu, L., Sun, Y., Wu, T., Wang, K., and Li, G. (2015). An improved protocol for isolation and culture of mesenchymal stem cells from mouse bone marrow. J. Orthop. Transl. 3, 26–33. doi:10.1016/j.jot.2014.07.005
Ilhan-Mutlu, A., Siehs, C., Berghoff, A. S., Ricken, G., Widhalm, G., Wagner, L., et al. (2016). Expression profiling of angiogenesis-related genes in brain metastases of lung cancer and melanoma. Tumour Biol. 37, 1173–1182. doi:10.1007/s13277-015-3790-7
Jiménez-Marín, A., Garrido, J. J., de Andrés-Cara, D. F., Morera, L., Barbancho, M. J., Llanes, D., et al. (2000). Molecular cloning and characterization of the pig homologue to human CD29, the integrin beta1 subunit. Transplantation 70, 649–655. doi:10.1097/00007890-200008270-00019
Johnsson, M., Jonsson, K. B., Andersson, L., Jensen, P., and Wright, D. (2015). Genetic regulation of bone metabolism in the chicken: Similarities and differences to mammalian systems. PLoS Genet. 11, e1005250. doi:10.1371/journal.pgen.1005250
Karin, M., Liu, Z., and Zandi, E. (1997). AP-1 function and regulation. Curr. Opin. Cell Biol. 9, 240–246. doi:10.1016/s0955-0674(97)80068-3
Kevorkova, O., Martineau, C., Martin-Falstrault, L., Sanchez-Dardon, J., Brissette, L., and Moreau, R. (2013). Low-bone-mass phenotype of deficient mice for the cluster of differentiation 36 (CD36). PLoS One 8, e77701. doi:10.1371/journal.pone.0077701
Kiernan, J., Hu, S., Grynpas, M., Davies, J. E., and Stanford, W. L. (2016). Systemic mesenchymal stromal cell transplantation prevents functional bone loss in a mouse model of age-related osteoporosis. Stem Cells Transl. Med. 5, 683–693. doi:10.5966/sctm.2015-0231
Kim, D., Langmead, B., and Salzberg, S. L. (2015). Hisat: A fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi:10.1038/nmeth.3317
Komori, T. (2008). Regulation of bone development and maintenance by Runx2. Front. Biosci. 13, 898–903. doi:10.2741/2730
Komori, T. (2010). Regulation of osteoblast differentiation by Runx2. Adv. Exp. Med. Biol. 658, 43–49. doi:10.1007/978-1-4419-1050-9_5
Kushwaha, P., Wolfgang, M. J., and Riddle, R. C. (2018). Fatty acid metabolism by the osteoblast. Bone 115, 8–14. doi:10.1016/j.bone.2017.08.024
Langfelder, P., and Horvath, S. (2008). Wgcna: an R package for weighted correlation network analysis. BMC Bioinforma. 9, 559. doi:10.1186/1471-2105-9-559
Lay, D. C., Fulton, R. M., Hester, P. Y., Karcher, D. M., Kjaer, J. B., Mench, J. A., et al. (2011). Hen welfare in different housing systems. Poult. Sci. 90, 278–294. doi:10.3382/ps.2010-00962
Lee, W. C., Guntur, A. R., Long, F., and Rosen, C. J. (2017). Energy metabolism of the osteoblast: Implications for osteoporosis. Endocr. Rev. 38, 255–266. doi:10.1210/er.2017-00064
Li, X., Zhang, Y., and Qi, G. (2013). Evaluation of isolation methods and culture conditions for rat bone marrow mesenchymal stem cells. Cytotechnology 65, 323–334. doi:10.1007/s10616-012-9497-3
Lin, Z., He, H., Wang, M., and Liang, J. (2019). MicroRNA-130a controls bone marrow mesenchymal stem cell differentiation towards the osteoblastic and adipogenic fate. Cell Prolif. 52, e12688. doi:10.1111/cpr.12688
Liu, F., Yuan, Y., Bai, L., Yuan, L., Zhang, J., Liu, J., et al. (2021). LRRc17 controls BMSC senescence via mitophagy and inhibits the therapeutic effect of BMSCs on ovariectomy-induced bone loss. Redox Biol. 43, 101963. doi:10.1016/j.redox.2021.101963
Lowery, J. W., and Rosen, V. (2018). The BMP pathway and its inhibitors in the skeleton. Physiol. Rev. 98, 2431–2452. doi:10.1152/physrev.00028.2017
Lucas, S., Omata, Y., Hofmann, J., Böttcher, M., Iljazovic, A., Sarter, K., et al. (2018). Short-chain fatty acids regulate systemic bone mass and protect from pathological bone loss. Nat. Commun. 9, 55. doi:10.1038/s41467-017-02490-4
Maeda, K., Kobayashi, Y., Koide, M., Uehara, S., Okamoto, M., Ishihara, A., et al. (2019). The regulation of bone metabolism and disorders by Wnt signaling. Int. J. Mol. Sci. 20, E5525. doi:10.3390/ijms20225525
Mandour, M. A., Nestor, K. E., Sacco, R. E., Polley, C. R., and Havenstein, G. B. (1989). Selection for increased humerus strength of cage-reared broilers. Poult. Sci. 68, 1168–1173. doi:10.3382/ps.0681168
Matsumoto, K., Saitoh, K., Koike, C., Narita, T., Yasugi, S., and Iba, H. (1998). Differential expression of fos and jun family members in the developing chicken gastrointestinal tract. Oncogene 16, 1611–1616. doi:10.1038/sj.onc.1201675
Miyauchi, A., Kuroki, Y., Fukase, M., Fujita, T., Chihara, K., and Shiozawa, S. (1994). Persistent expression of proto-oncogene c-fos stimulates osteoclast differentiation. Biochem. Biophys. Res. Commun. 205, 1547–1555. doi:10.1006/bbrc.1994.2843
Mundy, G., Garrett, R., Harris, S., Chan, J., Chen, D., Rossini, G., et al. (1999). Stimulation of bone formation in vitro and in rodents by statins. Science 286, 1946–1949. doi:10.1126/science.286.5446.1946
Mushahary, D., Spittler, A., Kasper, C., Weber, V., and Charwat, V. (2018). Isolation, cultivation, and characterization of human mesenchymal stem cells. Cytom. A 93, 19–31. doi:10.1002/cyto.a.23242
Naor, D., Nedvetzki, S., Golan, I., Melnik, L., and Faitelson, Y. (2002). CD44 in cancer. Crit. Rev. Clin. Lab. Sci. 39, 527–579. doi:10.1080/10408360290795574
Pertea, M., Kim, D., Pertea, G. M., Leek, J. T., and Salzberg, S. L. (2016). Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11, 1650–1667. doi:10.1038/nprot.2016.095
Pertea, M., Pertea, G. M., Antonescu, C. M., Chang, T. C., Mendell, J. T., and Salzberg, S. L. (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295. doi:10.1038/nbt.3122
Peters, S. B., Wang, Y., and Serra, R. (2017). Tgfbr2 is required in osterix expressing cells for postnatal skeletal development. Bone 97, 54–64. doi:10.1016/j.bone.2016.12.017
Rath, N. C., Huff, G. R., Huff, W. E., and Balog, J. M. (2000). Factors regulating bone maturity and strength in poultry. Poult. Sci. 79, 1024–1032. doi:10.1093/ps/79.7.1024
Raymond, B., Johansson, A. M., McCormack, H. A., Fleming, R. H., Schmutz, M., Dunn, I. C., et al. (2018). Genome-wide association study for bone strength in laying hens. J. Anim. Sci. 96, 2525–2535. doi:10.1093/jas/sky157
Reid, I. R. (1998). Glucocorticoid-induced osteoporosis: Assessment and treatment. J. Clin. Densitom. 1, 65–73. doi:10.1385/jcd:1:1:65
Rubin, C. J., Brandstrom, H., Wright, D., Kerje, S., Gunnarsson, U., Schutz, K., et al. (2007). Quantitative trait loci for BMD and bone strength in an intercross between domestic and wildtype chickens. J. Bone Min. Res. 22, 375–384. doi:10.1359/jbmr.061203
Sandberg, M., Vuorio, T., Hirvonen, H., Alitalo, K., and Vuorio, E. (1988). Enhanced expression of TGF-β and c-fos mRNAs in the growth plates of developing human long bones. Development 102, 461–470. doi:10.1242/dev.102.3.461
Schreiweis, M. A., Hester, P. Y., and Moody, D. E. (2005). Identification of quantitative trait loci associated with bone traits and body weight in an F2 resource population of chickens. Genet. Sel. Evol. 37, 677–698. doi:10.1186/1297-9686-37-7-677
Sedlackova, H., Rask, M. B., Gupta, R., Choudhary, C., Somyajit, K., and Lukas, J. (2020). Equilibrium between nascent and parental MCM proteins protects replicating genomes. Nature 587, 297–302. doi:10.1038/s41586-020-2842-3
Sevilla, L., Zaldumbide, A., Pognonec, P., and Boulukos, K. E. (2001). Transcriptional regulation of the bcl-x gene encoding the anti-apoptotic Bcl-xL protein by Ets, Rel/NFkappaB, STAT and AP1 transcription factor families. Histol. Histopathol. 16, 595–601. doi:10.14670/hh-16.595
Shanti, R. M., Li, W. J., Nesti, L. J., Wang, X., and Tuan, R. S. (2007). Adult mesenchymal stem cells: Biological properties, characteristics, and applications in maxillofacial surgery. J. Oral Maxillofac. Surg. 65, 1640–1647. doi:10.1016/j.joms.2007.04.008
Silversides, F. G., Korver, D. R., and Budgell, K. L. (2006). Effect of strain of layer and age at photostimulation on egg production, egg quality, and bone strength. Poult. Sci. 85, 1136–1144. doi:10.1093/ps/85.7.1136
Sivan, U., De Angelis, J., and Kusumbe, A. P. (2019). Role of angiocrine signals in bone development, homeostasis and disease. Open Biol. 9, 190144. doi:10.1098/rsob.190144
Stratmann, A., Frohlich, E. K., Harlander-Matauschek, A., Schrader, L., Toscano, M. J., Wurbel, H., et al. (2015). Soft perches in an aviary system reduce incidence of keel bone damage in laying hens. PLoS One 10, e0122568. doi:10.1371/journal.pone.0122568
Sul, O. J., Kim, J. E., Ke, K., Suh, J. H., and Choi, H. S. (2020). Atherogenic diet-induced bone loss is primarily due to increased osteoclastogenesis in mice. J. Nutr. Biochem. 79, 108337. doi:10.1016/j.jnutbio.2019.108337
Talaty, P. N., Katanbaf, M. N., and Hester, P. Y. (2010). Bone mineralization in male commercial broilers and its relationship to gait score. Poult. Sci. 89, 342–348. doi:10.3382/ps.2009-00382
Tang, Y., Li, M., Wang, J., Pan, Y., and Wu, F. X. (2015). CytoNCA: A cytoscape plugin for centrality analysis and evaluation of protein interaction networks. Biosystems. 127, 67–72. doi:10.1016/j.biosystems.2014.11.005
Tchilian, E. Z., and Beverley, P. C. (2006). Altered CD45 expression and disease. Trends Immunol. 27, 146–153. doi:10.1016/j.it.2006.01.001
Wan, M., and Cao, X. (2005). BMP signaling in skeletal development. Biochem. Biophys. Res. Commun. 328, 651–657. doi:10.1016/j.bbrc.2004.11.067
Wang, K., Zhao, Z., Wang, X., and Zhang, Y. (2021). BRD4 induces osteogenic differentiation of BMSCs via the Wnt/β-catenin signaling pathway. Tissue Cell 72, 101555. doi:10.1016/j.tice.2021.101555
Warnecke-Eberz, U., Metzger, R., Hölscher, A. H., Drebber, U., and Bollschweiler, E. (2016). Diagnostic marker signature for esophageal cancer from transcriptome analysis. Tumour Biol. 37, 6349–6358. doi:10.1007/s13277-015-4400-4
Webster, A. B. (2004). Welfare implications of avian osteoporosis. Poult. Sci. 83, 184–192. doi:10.1093/ps/83.2.184
Whitehead, C. C., and Fleming, R. H. (2000). Osteoporosis in cage layers. Poult. Sci. 79, 1033–1041. doi:10.1093/ps/79.7.1033
Wozney, J. M., Rosen, V., Celeste, A. J., Mitsock, L. M., Wang, E. A., Kriz, R. W., et al. (1989). Novel regulators of bone formation: Molecular clones and activities. Science 242, 1528–1534. doi:10.1126/science.3201241
Yamamoto, M., Tamura, K., Hiratsuka, K., and Abiko, Y. (2001). Stimulation of MCM3 gene expression in osteoblast by low level laser irradiation. Lasers Med. Sci. 16, 213–217. doi:10.1007/pl00011357
Young, H. E., Ceballos, E. M., Smith, J. C., Mancini, M. L., Wright, R. P., Ragan, B. L., et al. (1993). Pluripotent mesenchymal stem cells reside within avian connective tissue matrices. Vitro Cell. Dev. Biol. Anim. 29a, 723–736. doi:10.1007/bf02631429
Yuan, S., Lemming, E. W., Michaëlsson, K., and Larsson, S. C. (2020). Plasma phospholipid fatty acids, bone mineral density and fracture risk: Evidence from a Mendelian randomization study. Clin. Nutr. 39, 2180–2186. doi:10.1016/j.clnu.2019.09.005
Zhang, W., Dong, R., Diao, S., Du, J., Fan, Z., and Wang, F. (2017). Differential long noncoding RNA/mRNA expression profiling and functional network analysis during osteogenic differentiation of human bone marrow mesenchymal stem cells. Stem Cell Res. Ther. 8, 30. doi:10.1186/s13287-017-0485-6
Keywords: bone marrow mesenchymal stem cells, osteoblastic differentiation, RNA-seq transcriptome analysis, differentially expressed gene, chicken
Citation: Lv H, Wang T, Zhai S, Hou Z and Chen S (2022) Dynamic transcriptome changes during osteogenic differentiation of bone marrow-derived mesenchymal stem cells isolated from chicken. Front. Cell Dev. Biol. 10:940248. doi: 10.3389/fcell.2022.940248
Received: 10 May 2022; Accepted: 12 August 2022;
Published: 02 September 2022.
Edited by:
Ming Li, Osaka University, JapanReviewed by:
Anjali P. Kusumbe, University of Oxford, United KingdomCopyright © 2022 Lv, Wang, Zhai, Hou and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sirui Chen, Y3NyQGNhdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.