
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Cell Dev. Biol., 14 June 2022
Sec. Stem Cell Research
Volume 10 - 2022 | https://doi.org/10.3389/fcell.2022.917166
This article is part of the Research TopicCell Biology of Brain Development and EvolutionView all 10 articles
The acquisition of higher intellectual abilities that distinguish humans from their closest relatives correlates greatly with the expansion of the cerebral cortex. This expansion is a consequence of an increase in neuronal cell production driven by the higher proliferative capacity of neural progenitor cells, in particular basal radial glia (bRG). Furthermore, when the proliferation of neural progenitor cells is impaired and the final neuronal output is altered, severe neurodevelopmental disorders can arise. To effectively study the cell biology of human bRG, genetically accessible human experimental models are needed. With the pioneering success to isolate and culture pluripotent stem cells in vitro, we can now routinely investigate the developing human cerebral cortex in a dish using three-dimensional multicellular structures called organoids. Here, we will review the molecular and cell biological features of bRG that have recently been elucidated using brain organoids. We will further focus on the application of this simple model system to study in a mechanistically actionable way the molecular and cellular events in bRG that can lead to the onset of various neurodevelopmental diseases.
The temporal series of events that leads to the acquisition of specific structural and functional features of different organs in the human body is a fascinating, yet not fully understood phenomenon known as organogenesis. Heart, brain, skin and liver are all very distinct and specific organs with their own distinct functions, yet they developmentally originate from a single cell. Unveiling the cascade of steps leading from such a simple disordered system to an ordered complexity is not only essential from a developmental biology perspective but also for establishing therapeutic approaches in the context of regenerative medicine.
Whereas the classical in vivo model systems, such as Drosophila, zebrafish and mouse provided fundamental insight into the basic animal, vertebrate and mammalian development respectively, certain aspects of the complexity observed in humans can only be studied in the human model system. Hence, 2D in vitro and ex vivo systems, such as organotypic cultures, have been valuable to reveal human-specific features of organ development and pathology (Shamir and Ewald, 2014). However, they lack either the spatial complexity of the tissue or the ability to study the development for prolonged time periods. This raised the need to establish a human model system that would mimic human organogenesis with a sufficient level of spatio-temporal complexity.
The first step towards this goal was provided by the pioneering work of somatic cells reprogramming into pluripotent stem cells (PSCs) (Takahashi and Yamanaka, 2006). The subsequent ability to grow PSCs enabled the exploitation of this technology to generate human stem cell derived cultures (Takahashi et al., 2007). Additionally, cultivation of PSCs in a 3D configuration enabled the cell-cell and cell-extracellular matrix (ECM) communication which would otherwise be absent in a 2D culture (Blau and Miki, 2019). In 2008 a remarkable work conducted by Eiraku and others established for the first time a 3D polarised cortical tissue from embryonic stem cells (ESCs) (Eiraku et al., 2008). This paved the road towards the use of organoids, as multicellular structures that exhibit the capacity to self-organise into a complex system, to study organogenesis in a dish. The term “organoid” was consolidated by Sato and others who established for the first time intestinal organoids from single adult stem cells (Sato et al., 2009). Together, these fundamental studies led to the widespread application of organoid technology in developmental biology (Kaluthantrige Don and Huch, 2021).
Organoids contain the genetic background along with the cell-cell and cell-ECM interactions similar to those in vivo, however, they are grown in a simpler and fairly controllable environment (Shamir and Ewald, 2014). On the one hand this is a limiting factor in recapitulating the physiological features of organogenesis. On the other hand, this is an opportunity to dissect in depth the biology of cells of interest within an environment that can be readily controlled. For instance, hepatocyte organoids, that recapitulate the spectacular ability of the liver to regenerate upon a partial resection, can be used as a magnifying glass to study the cell types underlying this regeneration (Hu et al., 2018; Peng et al., 2018). In the context of human pathologies, organoids hold potential to treat various diseases, such as acute kidney injury or diabetes (Lancaster and Huch, 2019). Recently, kidney organoids transplanted under the renal capsule of mice acquired de novo vascularisation and tubular maturation (van den Berg et al., 2018), allowing future applications for the treatment of renal failure. Furthermore, the use of pancreatic islet organoids as a source of β-cells in vitro may potentially be an alternative cell therapy for diabetes (Wang et al., 2020). Comparison between human fetal retina and retinal organoids showed considerable similarities, thus anticipating a potential role of retinal organoids as cell source for transplantation in cell therapy (Sridhar et al., 2020).
A striking example of a model system that successfully simplifies a highly complex organ, but however mimics the key aspects of the development, is a brain organoid (see section 1.3 for discussion on brain organoids) (Heide et al., 2018; Benito-Kwiecinski and Lancaster, 2020; Lopez-Tobon et al., 2020; Marton and Pașca, 2020; Velasco et al., 2020; Sidhaye and Knoblich, 2021). Untangling the functional dynamics of distinct brain cells using animal models is a very tedious process due to the remarkable complexity stemming from the interaction of the brain with the other organs and the environment. One fundamental question is to understand how this complexity arises during development.
The cerebral cortex, and its evolutionary most recent part, the neocortex, arise from the forebrain region of the neural tube. It is arguably considered that the higher cognitive abilities of humans compared to other mammals are reflected by the size and the cytoarchitectural organisation of the human neocortex (Molnár et al., 2019; Rakic, 1988, 1995). Development of the neocortex initiates with the proliferation of neuroepithelial cells lining the neural tube. Transition from a proliferative state into a neurogenic state gives rise to apical radial glia (aRG), the chief parental progenitor cells that will initiate the neurogenesis, that is the series of events that lead to the production of neurons (Götz and Huttner, 2005; Taverna et al., 2014). The identity of the progenitors is defined based on the location of their mitosis (Taverna et al., 2014), which highlights the importance of the microenvironment for the cell fate specification. Indeed, proliferation of aRGs occurs on the ventricular (apical) surface and these cells form the apical-most neocortical histological layer, known as the ventricular zone (VZ) (Götz and Huttner, 2005). Moving along the apicobasal axis, asymmetrical divisions of aRGs give rise to basal progenitors that populate the second germinal layer, the subventricular zone (SVZ). In species with an expanded neocortex, the SVZ is divided into two distinct zones: the inner and the outer SVZ (ISVZ and OSVZ, respectively) (Smart, 2002; Dehay et al., 2015).
The neocortical expansion in mammals has been widely associated with a subpopulation of basal progenitors called basal or outer radial glia (bRG or oRG) (Fietz et al., 2010; Hansen et al., 2010; Reillo et al., 2011). The abundance of bRG and their proliferative capacity are strikingly increased in species with an expanded neocortex, such as human, macaque or ferret (Fietz et al., 2010; Hansen et al., 2010; Reillo et al., 2011; Betizeau et al., 2013; Kalebic et al., 2019), compared to species with a small brain, such as mouse (Wang et al., 2011; Wong et al., 2015). This results in an increased production of neurons, which in turn is associated with the expansion and folding of the neocortex. Hence, bRG are considered to be a key cell type underlying human neocortex development (Penisson et al., 2019; Kawaguchi, 2020; Pinson and Huttner, 2021; Del-Valle-Anton and Borrell, 2022). An additional layer of cellular complexity within the bRGs lies in their morphological heterogeneity (Kalebic and Huttner, 2020). We have identified that an increasing number of basal processes within the human bRGs coincides with an increase in their proliferative capacity (Kalebic et al., 2019). Interestingly, such bRGs complexity and proliferative capacity are absent in the mouse cortex, further corroborating the role of bRGs as chief cells underlying mammalian neocortical expansion.
As many studies have started to focus on this fascinating population of cells, multiple outstanding questions remain to be addressed. For example, what is the molecular and cell biological heterogeneity of bRG across different species and what is their contribution to the onset of human intellectual disabilities. The use of a reductionist system containing abundant bRGs such as the organoids may elucidate the mechanisms that lead towards the complexity of the neocortex organogenesis and pathogenesis. This review will focus on the diverse modalities to generate brain organoids and how we can exploit this technology in the context of neocortex development and pathologies, specifically focusing on bRGs.
During embryogenesis, the interplay of diverse signalling pathways leads to the differentiation into neuronal fate. Initial inhibition of the the bone morphogenic proteins (BMP) signalling is needed for the differentiation into the neuroectoderm which then invaginates to give rise to the neural tube (Muñoz-Sanjuán and Brivanlou, 2002; Sadler, 2005). Patterning of the neural tube into different regional identities is achieved through the regulated activity of WNT, fibroblast growth factor and retinoic acid pathways (Muñoz-Sanjuán and Brivanlou, 2002; Molotkova et al., 2005). To model such embryonic development in vitro and building on the earlier pioneering work, two studies reported the generation of brain organoids in 2013 (Kadoshima et al., 2013; Lancaster et al., 2013). Two distinct approaches have been applied for generating brain organoids: 1) the unguided method, which directs the generation of organoids with multiple regional identities; and 2) the guided method, which promotes the acquisition of specific regional identity through a step-wise time-dependent exogenous signalling (Table 1) (Lancaster and Knoblich, 2014; Di Lullo and Kriegstein, 2017). Each approach starts with the generation of 3D aggregates named embryoid bodies (EBs), which have the potential to differentiate into all three germ layers (Zhang et al., 2001). The first cerebral organoids were generated from EBs following the intrinsic program of neuroepithelial cells to differentiate into neural progenitor cells (Lancaster et al., 2013). Such unguided protocol results in a stochastic development of various and multiple regional identities (Ormel et al., 2018). Alternatively, the differentiation can be directed towards the acquisition of a specific regional identity, such as the dorsal forebrain. The latter can be achieved through the manipulation of the transforming growth factor-beta (TGF-β) signalling pathway and BMPs (Chambers et al., 2009; Kadoshima et al., 2013; Paşca et al., 2015; Qian et al., 2018, 2020; Sloan et al., 2018). Of note, the time dependent addition of small molecules in presence (Kadoshima et al., 2013; Lancaster et al., 2013; Qian et al., 2018, 2020; Sloan et al., 2018) or absence (Mariani et al., 2015) of Matrigel, a commercially available basement membrane matrix from mouse sarcoma (Li et al., 1987), shows a remarkable difference in the developmental timelines, prompting a question on how it might affect the progenitor biology (Table 1).
Although the timeline in the induction of neurogenesis is different, all protocols partially recapitulate the series of events known to occur in the developing human neocortex (Figure 1). Indeed, organoids readily contain aRGs marked by the expression of the transcription factors SOX2 and PAX6 (Kadoshima et al., 2013; Lancaster et al., 2013; Qian et al., 2018, 2020). Both subtypes of basal progenitors, bRG and the intermediate progenitors can be observed in organoids. bRG are marked by the presence of HOPX, SOX2 and PAX6, along with the absence of TBR2 (Pollen et al., 2015; Qian et al., 2016, 2020). Moreover, organoids also display the presence of early born (CTIP2+ or TBR1+) and late born neurons (SATB2+ and/or BRN2+) (Figure 1) (Paşca et al., 2015; Lancaster et al., 2017; Sloan et al., 2018; Qian et al., 2020; Giandomenico et al., 2021). Transcriptomic analysis identified that mature brain organoids between days 250 and 300 correspond to postnatal stages of human brain development (Gordon et al., 2021). However, the timing of expression of different cell types and formation of specific cortical layers differs from protocol to protocol. For example, the organoids produced by the Lancaster protocol show the presence of neurons already at day 30 (Lancaster et al., 2013). This contrasts the organoids generated by the Kadoshima protocol, which start neurogenesis after 70 days (Kadoshima et al., 2013). Thus, the caveat of timing across protocols needs to be considered when comparing the fetal human development.
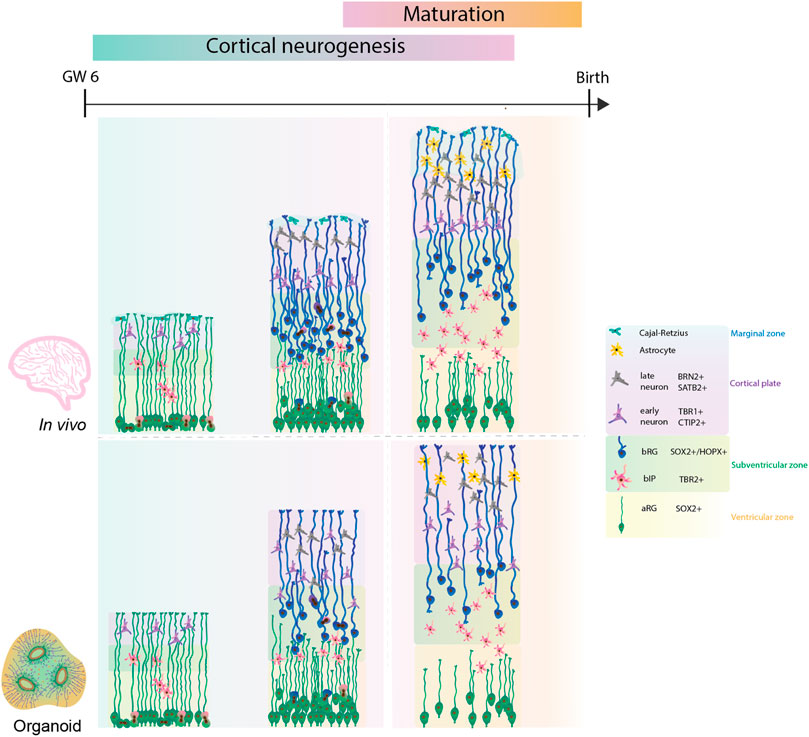
FIGURE 1. Comparison of human cortical development between human fetal neocortex and human forebrain organoids. Cortical neurogenesis in human fetal development (upper panel) starts with proliferation of neural progenitor cells. In this process, bRG are highly abundant and give a key contribution to the final neuronal output. Consequently, the neocortex expands into specific cytoarchitectural layers with formation of cortical folding on the basal side. Following neurogenesis, functional maturation of neurons and glia takes place. Human forebrain organoids (lower panel) recapitulate the cell diversity and developmental lineages, however further improvements are required to achieve bRG abundance, improved cytoarchitectural organisation, neuronal and glial maturation, and cortical folding similar to the ones observed during the human fetal neocortex.
Most of the initial organoid protocols showed similar limitations as they poorly recapitulated the tissue architecture, notably cortical layering, the bRG abundance, the presence of all developmental lineages and neuronal maturation. To address these limitations and to improve the nutrient and oxygen exchange within the organoid, several improvements of the initial protocols were reported. For example, adding microfilaments and culturing organoids at air-liquid interfaces advanced the original Lancaster protocol (Lancaster et al., 2017; Giandomenico et al., 2021). Recently, the use of external magnetic nanoparticles or inclusion of signalling gradients showed enhanced local patterning of brain organoids (Cederquist et al., 2019; Fattah et al., 2022). It is tempting to speculate that such methods could direct an improved cytoarchitectural organisation of forebrain organoids (Cederquist et al., 2019; Fattah et al., 2022). Further, repeated slicing of Qian organoids facilitated an expansion of cortical layers and an increased expression of HOPX + bRG cells at day 80, reminiscent of the OSVZ (Qian et al., 2020).
Another strategy to improve brain organoid maturation resulted in the fusion of phenotypically independent dorsal and ventral organoids, termed assembloid (Bagley et al., 2017; Sloan et al., 2018). The latter is particularly interesting because the ventral part of the forebrain is the principal origin of human interneurons, that subsequently migrate into the dorsal regions to integrate into the cortical circuits (Bandler et al., 2017; Hu et al., 2017; Lim et al., 2018). Interneurons are generated by the radial glia of the ventral forebrain, which appear to be more similar to the dorsal aRG than bRG (Velmeshev et al., 2021). Taken together, dorsal-ventral assembloids provide a model system to study generation, migration and integration of interneurons, which enables a more complete modelling of the human cortical development (Marton and Pașca, 2020).
Finally, several strategies have been adopted to improve vascularization. One approach consists in the co-culture of brain organoids with vascular cells such as human umbilical vascular endothelial cells (HUVECs). This resulted in a reduced hypoxic core and improved neuronal maturation (Shi et al., 2020). Additional strategy transplanted organoids into vascularised tissue of immunodeficient mice and showed functional blood circulation and improved organoid viability (Mansour et al., 2018). Implementation of vascularisation in forebrain organoids could enhance the viability and potentially promote neuronal maturation.
As mentioned above, bRG are considered to be the key cell type underlying human neocortical development. Human bRG are highly proliferative, likely generate most of the human neurons and serve as the scaffold for the neuronal migration to the cortical plate (Fietz et al., 2010; Hansen et al., 2010; Reillo et al., 2011; LaMonica et al., 2013). bRGs biology has been poorly assessed since the abundance and behaviour of this cell type is strikingly low in the key animal model, the mouse (Wang et al., 2011; Wong et al., 2015). Although the abundance of bRGs in cerebral organoids is still not comparable to the numbers present in fetal human tissue, organoids hold great promise to be a suitable in vitro system to study human bRGs.
To understand the extent at which the organoid system recapitulates the bRG identity observed in human fetal tissue, initial studies examined the transcriptomic profiles of both model systems. This revealed similar lineage relationships between aRG and bRG in both systems (Camp et al., 2015; Pollen et al., 2015). Subsequent studies readily confirmed the existence of a cell population with a transcriptomic signature of bRG (Bershteyn et al., 2017; Giandomenico et al., 2019; Pollen et al., 2019; Velasco et al., 2019; Cheroni et al., 2022). A recent work aimed to understand the reproducibility of organoids, identified consistent generation of diverse cell types, including bRG, in multiple forebrain organoids (Velasco et al., 2019). Subsequent work combined the latter dataset with spatial transcriptomics and identified a spatial patterning of different cell types within the organoid, with bRG being superficially positioned with respect to aRG (Uzquiano et al., 2022).
One striking characteristic of brain organoids compared to other organoids, such as the liver, is their outstanding increase in size during maturation. However, this can result in poor oxygenation and nutrient exchange within the organoid core causing a systematic cellular stress (Bhaduri et al., 2020). A recent analysis, however, suggested that the cellular stress is a feature of a subpopulation of cells, which can be removed in subsequent computational analyses (Vértesy et al., 2022).
Additional transcriptomic studies extended the role of brain organoids not only as a promising tool to tackle human cortical development but also showed the valuable use of organoids in modelling human brain evolution (Heide et al., 2018; Muchnik et al., 2019). For example, CTCL, a fusion transcript and a Wnt signalling modulator, which is expressed in human but not mouse developing brain, has recently been shown to be implicated in the proliferative capacity of bRG in human organoids (Ou et al., 2021). Comparison between human and non-human primate brain organoids pointed at the increased activation of another key signalling pathway, the PI3K-AKT-mTOR, in human bRG (Pollen et al., 2019; Andrews et al., 2020). Moreover, mTOR signalling in human organoids was shown to regulate bRG morphology and behaviour (Pollen et al., 2019; Andrews et al., 2020). Building on earlier findings that identified the role of Notch signalling in promoting human bRGs proliferation (Hansen et al., 2010), recent work conducted in brain organoids, identified the role of a human-specific gene NOTCH2NL to enhance the activity of Notch signalling and to delay the neural differentiation of bRG (Fiddes et al., 2018). The second human-specific gene implicated in neocortical expansion and known to operate in bRG, ARHGAP11B (Florio et al., 2015; Kalebic et al., 2018; Heide et al., 2020), has been introduced to chimpanzee organoids where it promoted bRG proliferation (Fischer et al., 2020). Kanton and others performed a comprehensive cell-type specific analysis of gene expression in human, chimpanzee and macaque organoids and further revealed the molecular mechanisms underlying the differences in gene expression across these species (Kanton et al., 2019; Muchnik et al., 2019). Their ATAC-seq analysis showed divergence in chromatin accessibility between human and chimpanzee organoids, which could be associated with the human-specific gene expression (Kanton et al., 2019; Muchnik et al., 2019). Additionally, organoids also offer the possibility to compare the differences in brain development between modern humans and ancestral species such as Neandertals. For instance, Muotri and others modelled Neandertal brains in organoids by introducing an archaic variant gene called Neuro-oncological ventral antigen 1 (NOVA1) (Trujillo et al., 2021). These organoids exhibited changes in organoid morphology and neuronal activity (Trujillo et al., 2021). Recently, Mora-Bermudez and others introduced specific ancestral variants involved in mitotic spindle and kinetochore function in organoids and showed shorter metaphase of apical progenitors compared to the longer metaphase of non-mutated modern human organoids (Mora-Bermúdez et al., 2022). It is interesting to speculate that such evolutionary differences between modern humans and ancestral human species might also be linked to bRG development and function.
The transcriptomic studies described above have shown that organoids successfully recapitulate the diversity of cell types and their lineage relationships. Combined with the existence of the organoid polarity, it allows us to use this model system to also study the cell biological features of bRG (Figure 2). Previous identification on the role of mTOR signalling pathway (Nowakowski et al., 2017) in human bRG led to its deeper analysis using organoids (Pollen et al., 2019). Upon the pharmacological inhibition of mTOR signalling in organoids, bRG exhibited a shorter basal process (Andrews et al., 2020). Interestingly, bRG morphology could be rescued by the activation of the Rho-GTPases CDC42/RAC1 in cortical tissue (Figure 2) (Andrews et al., 2020). CDC42 in particular has a very important role in radial glia morphology, as it has been found to affect polarisation, proliferation and migration of aRG (Cappello et al., 2006; Yokota et al., 2010). Another determinant of cell polarity involved in maintenance of the radial scaffold is GSK3 (Yokota et al., 2010). Recently, a pharmacological inhibition of GSK3 in organoids led to a reduction in the abundance of bRG and production of neurons, further emphasising the role of bRG polarity for normal neurogenesis (López-Tobón et al., 2019). Although bRG lack classical apicobasal polarity and the contact with the ventricular surface, they do possess a series of features that we have previously termed pseudo-apicobasal polarity (Kalebic and Namba, 2021). A key manifestation of such polarity is their morphology. Previous findings identified different bRG morphotypes in mouse, ferret, macaque and human developing cortex (Betizeau et al., 2013; Reillo et al., 2017; Kalebic et al., 2019), suggesting that an increased number of basal processes coincides with the proliferative capacity of bRG (Kalebic and Huttner, 2020). It would be interesting to identify these diverse bRG morphotypes in organoids and understand how morphology might be an important component for neurogenesis progression.
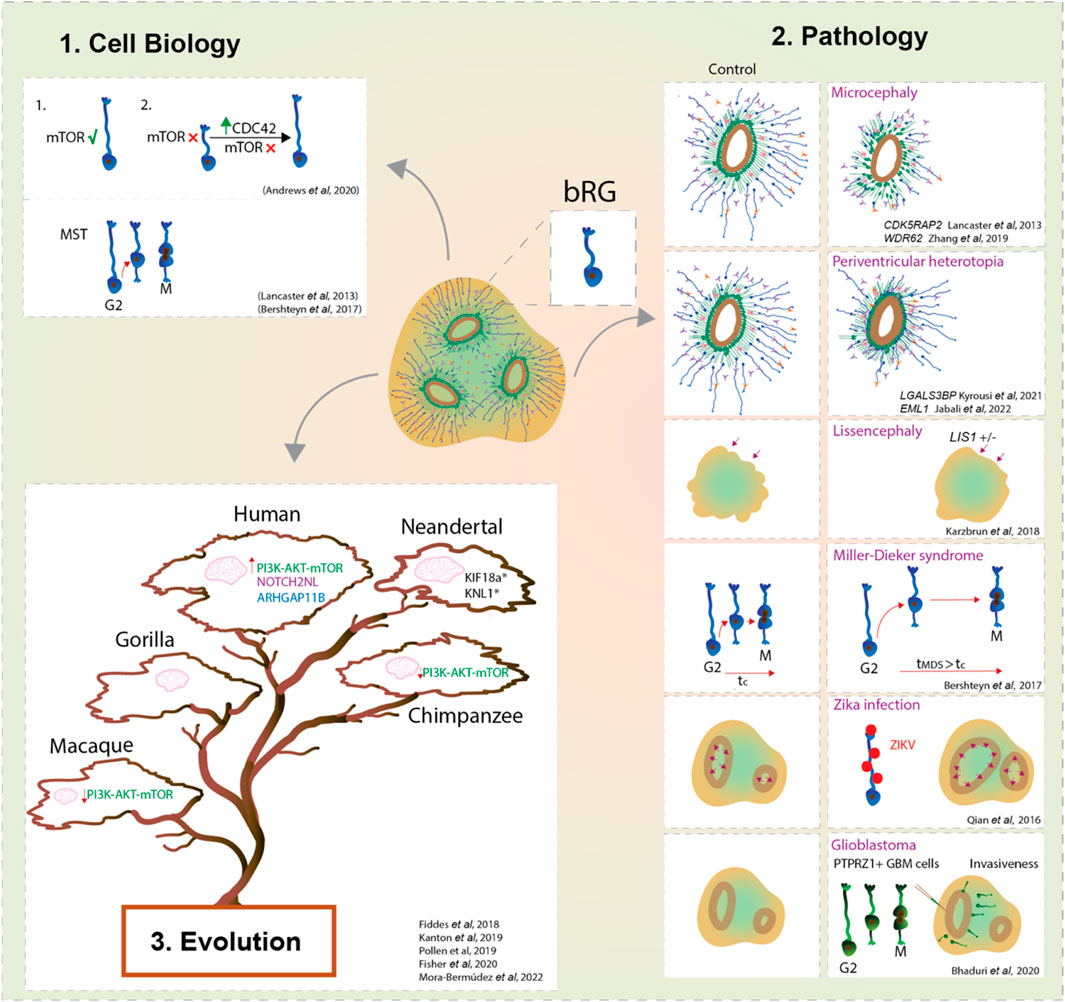
FIGURE 2. bRG in development, evolution and pathology using forebrain organoids. Forebrain organoids enable us to study the cell biological features and cell behaviours that characterise bRG (1); their role in the onset of malformations of cortical development, brain cancers and viral diseases (2); and their contribution to the neocortex expansion of modern humans compared to ancestral human species and non-human primates (3). Overall, organoids provide a new window into bRG and their link with the expansion of the neocortex.
Another key advantage of the organoid system is that it enables the studies of bRG structural and temporal dynamics without the complexity inherent to the in vivo and ex vivo systems. Prior to mitosis, bRG exhibit a distinctive saltatory migrational behaviour named mitotic somal translocation (MST) (Hansen et al., 2010; LaMonica et al., 2013; Ostrem et al., 2014; Ostrem et al., 2017). Remarkably, organoid studies based on GFP-electroporated radial glia identified this unique feature in cells localised away from the VZ (Lancaster et al., 2013; Otani et al., 2016) The importance of MST for human cortical development is obvious when observing a form of human lissencephaly called Miller-Dieker syndrome (MSD). MSD brain organoids showed prolonged mitosis and longer MST distances (Figure 2) (Bershteyn et al., 2017), suggesting that defects in bRG mode of division could lead to premature neurogenesis in human lissencephaly.
Organoids, especially when derived from patients’ cells, are a par excellence platform to dissect the pathogenesis of neurodevelopmental diseases (Figure 2). One of the first examples comes from the work conducted by Lancaster and others who generated patient-derived microcephalic cerebral organoids carrying a mutation in the centrosomal protein CDK5RAP2 (Lancaster et al., 2013). They showed an increase of asymmetric cell divisions in neural progenitors which led to their premature differentiation (Lancaster et al., 2013). The second principal way to model neurodevelopmental disorders in organoids is to introduce disease-causing mutations via genome editing in PSCs. For example, deletion of WDR62, another key gene causing microcephaly, resulted in a reduction of bRG proliferation, which in turn led to reduced organoid size (Zhang et al., 2019).
In addition to microcephaly, organoids have been useful to model periventricular heterotopia (Figure 2). A recent study used both patient-derived and genome-edited PSCs to study EML1 deficiency in cortical organoids (Jabali et al., 2022). The analyses revealed defects in the primary cilium structure and mitotic spindle orientation of aRG, which led to an increase in aRG delamination and subsequent formation of ectopic neural progenitors and heterotopic neurons (Jabali et al., 2022). Interestingly, deeper characterization identified that the majority of these ectopic progenitor cells are bRG with an unusual morphology (Jabali et al., 2022), linking bRG morphology with neurogenesis. Phenotypes of periventricular heterotopia were successfully recapitulated in human brain organoids also by manipulation of the expression levels of ECE2 and PLEKHG6 (O’Neill et al., 2018; Buchsbaum et al., 2020). Another key gene that has been recently described to be enriched in human bRG (Pollen et al., 2015), while being linked to periventricular nodular heterotopia, is LGALS3BP. Studies using organoids showed that LGALS3BP expression is essential for proper positioning of bRG, whereas altered LGALS3BP expression resulted in neuronal heterotopia and defects in local gyrification, emphasising once again a potential role of bRG in disease (Kyrousi et al., 2021).
Further studies identified a role of bRG in the pathogenesis of Pretzel syndrome (polyhydramnios, megalencephaly, symptomatic epilepsy; PMSE) derived from mutations in the STRADA gene, part of the mTOR pathway. PMSE organoids showed an increase of HOPX + bRG which could be linked with the megalencephaly observed in PMSE individuals (Dang et al., 2021). This also further strengthens the role of the mTOR pathway in the regulation of bRG (Nowakowski et al., 2017; Andrews et al., 2020). Mutation of CHD8 (chromodomain helicase DNA-binding 8) in cerebral organoids resulted in an increased proliferation of a population of radial glial cells which translated into altered neurodevelopmental trajectories (Villa et al., 2022).
Several studies modelled cortical folding using organoids (Figure 2). Activation of the PI3K-AKT signalling is known to be involved in increased proliferation of BPs (Kalebic et al., 2019) and its dysfunction is associated with brain overgrowth disorders (Hevner, 2015). Genetic ablation of PTEN, a regulator of PI3K, in human organoids showed an increase of HOPX + bRG with subsequent formation of cortical folding (Li et al., 2017). Interestingly, both PTEN mutant mice and human organoids showed an increase of brain or organoid volume, but only human organoids showed folding (Li et al., 2017). This suggests the importance of specific molecular and/or cellular features in humans, but not in mice, to direct cortical folding. Nevertheless, control human brain organoids lack the ability to achieve cortical folding, suggesting that they exhibit insufficient neuronal maturation and/or lack the mechanical signals from the microenvironment (Borrell, 2018; Kroenke and Bayly, 2018). Gyrification is important for the development of the neocortex as it maximises the surface to pack neurons relative to the brain size. Karzbrun et al. reported an induction of folding in organoids by physically constraining brain organoids using a chip (Karzbrun et al., 2018). Together with the mechanical forces from the cytoskeleton contraction and cell migration this induced wrinkles in organoids that are reminiscent of cortical folding (Karzbrun et al., 2018). In contrast, lissencephalic organoids (LIS-1 mutant) showed changes in the cytoskeleton and ECM that resulted in a reduced organoid wrinkling (Karzbrun et al., 2018). It would be interesting to apply the chip device to organoids whose age corresponds to the onset of bRG neurogenesis and examine a link between bRG and the mechanisms of cortical folding. Interestingly, in lissencephalic organoids modelling Miller-Dieker syndrome, bRG showed mitotic defects, suggesting a role of bRG in pathogenesis of lissencephaly (Bershteyn et al., 2017).
Apart from neurodevelopmental disorders, the use of brain organoids was beneficial to elucidate a role of bRG-like cells in malignant brain tumours such as glioblastoma (Figure 2). Live imaging on primary resected tumours displayed a population of bRG-like cells undergoing MST (Bhaduri et al., 2020). Upon transplantation into cortical organoids, these cells exhibited typical invasiveness and expansion of tumour-like cells (Bhaduri et al., 2020). This highlights an important role of bRG biology not only during brain development but also in the context of cancer progression.
Finally, brain organoids have a potential to mimic viral infectious diseases (Figure 2) (Harschnitz and Studer, 2021). An outstanding example was given in response to the outbreak of Zika virus (ZIKAV), in which ZIKAV induced microcephalic organoids were generated (Qian et al., 2016; Krenn et al., 2021). These organoids exhibited increased apoptosis, reduced proliferation with subsequent reduction of organoid size. Interestingly, the authors showed that bRG were readily infected by the Zika virus (Qian et al., 2016), hence indicating the advantage of using organoids to understand the contribution of different cell types, such as bRG, in the disease aetiology (Figure 2).
Brain organoids have hence provided invaluable insight into the role of bRG for human neurodevelopmental pathologies. Since rodent models poorly recapitulate features of human bRG, ferrets and primates are typical species of choice for in vivo exploration of the bRG role in neurodevelopmental disorders (Feng et al., 2020; Gilardi and Kalebic, 2021). Although they recapitulate well the key aspects of bRG biology, these models require substantial time and resources in addition to important ethical considerations for disease modelling. Hence the application of organoids, and particularly patient-derived organoids, has been instrumental for the advancement of knowledge regarding neurodevelopmental diseases and role of bRG in this context.
The ability to recapitulate organogenesis outside the embryo makes the organoid system a fascinating and useful technology. Although brain organoids differ from the brain in vivo, their ability to reproduce the diverse cell types and lineage trajectories comparable to human fetal cortex, makes the organoids a promising tool to address fundamental questions in neocortical development and pathologies. This is particularly relevant for bRG, a key progenitor cell type underlying human brain development. Future research will likely focus on further cell biological characterization of bRG in organoids and will better dissect the steps along the developmental trajectories examining the contribution of bRG for neocortical development. Given that organoids are becoming a key model system to study differences in brain development between modern humans and ancestral species, it is likely that further efforts in this direction will elucidate the contribution of specific genetic changes between these species for the biology of basal progenitors. Finally, disease modelling has been one of the principal directions of organoid-based research. Future efforts in this domain are expected to further develop in the direction of an ever-more personalised medicine combining patient-derived organoids with genetic and pharmacological screens. An elegant example of a genetic screen has been performed by Esk and others who tested 173 microcephaly-related genes in human brain organoids using CRISPR/Cas9-mediated genome editing (Esk et al., 2020). Future approaches can be used to study candidate genes of other neurodevelopmental pathologies, genes that have more subtle differences in expression level between control and pathological development and, finally, genes whose phenotype is likely to be pertinent to the later stages of organoid development, when bRG become more dominant. Hence, although brain organoids still do not recapitulate all the features of human cortical development, further advancement of the technology and/or combination with xenografting into animal models, are likely to pave the way for an ever-increasing use of this model system to study neurodevelopmental pathologies and human brain evolution.
Both authors wrote the manuscript, approved it for publication and agreed to be accountable for the content of the work.
Authors FD and NK were employed by Human Technopole.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Oliver Harschnitz (HT) for critical reading of the manuscript and all the members of the Kalebic group for helpful discussions.
aRG, apical radial glia; bIP, basal intermediate progenitor; bRG, basal radial glia; GW, gestational week.
Andrews, M. G., Subramanian, L., and Kriegstein, A. R. (2020). mTOR Signaling Regulates the Morphology and Migration of Outer Radial Glia in Developing Human Cortex. Elife 9. doi:10.7554/eLife.58737
Bagley, J. A., Reumann, D., Bian, S., Lévi-Strauss, J., and Knoblich, J. A. (2017). Fused Cerebral Organoids Model Interactions between Brain Regions. Nat. Methods 14, 743–751. doi:10.1038/nmeth.4304
Bandler, R. C., Mayer, C., and Fishell, G. (2017). Cortical Interneuron Specification: the Juncture of Genes, Time and Geometry. Curr. Opin. Neurobiol. 42, 17–24. doi:10.1016/j.conb.2016.10.003
Benito-Kwiecinski, S., and Lancaster, M. A. (2020). Brain Organoids: Human Neurodevelopment in a Dish. Cold Spring Harb. Perspect. Biol. 12. doi:10.1101/cshperspect.a035709
Bershteyn, M., Nowakowski, T. J., Pollen, A. A., Di Lullo, E., Nene, A., Wynshaw-Boris, A., et al. (2017). Human iPSC-Derived Cerebral Organoids Model Cellular Features of Lissencephaly and Reveal Prolonged Mitosis of Outer Radial Glia. Cell Stem Cell 20, 435–449. e4. doi:10.1016/j.stem.2016.12.007
Betizeau, M., Cortay, V., Patti, D., Pfister, S., Gautier, E., Bellemin-Ménard, A., et al. (2013). Precursor Diversity and Complexity of Lineage Relationships in the Outer Subventricular Zone of the Primate. Neuron 80, 442–457. doi:10.1016/j.neuron.2013.09.032
Bhaduri, A., Andrews, M. G., Mancia Leon, W., Jung, D., Shin, D., Allen, D., et al. (2020). Cell Stress in Cortical Organoids Impairs Molecular Subtype Specification. Nature 578, 142–148. doi:10.1038/s41586-020-1962-0
Blau, B. J., and Miki, T. (2019). The Role of Cellular Interactions in the Induction of Hepatocyte Polarity and Functional Maturation in Stem Cell-Derived Hepatic Cells. Differentiation 106, 42–48. doi:10.1016/j.diff.2019.02.006
Borrell, V. (2018). How Cells Fold the Cerebral Cortex. J. Neurosci. 38, 776–783. doi:10.1523/jneurosci.1106-17.2017
Buchsbaum, I. Y., Kielkowski, P., Giorgio, G., O'Neill, A. C., Di Giaimo, R., Kyrousi, C., et al. (2020). ECE2 Regulates Neurogenesis and Neuronal Migration during Human Cortical Development. EMBO Rep. 21, e48204. doi:10.15252/embr.201948204
Camp, J. G., Badsha, F., Florio, M., Kanton, S., Gerber, T., Wilsch-Bräuninger, M., et al. (2015). Human Cerebral Organoids Recapitulate Gene Expression Programs of Fetal Neocortex Development. Proc. Natl. Acad. Sci. U.S.A. 112, 15672–15677. doi:10.1073/pnas.1520760112
Cappello, S., Attardo, A., Wu, X., Iwasato, T., Itohara, S., Wilsch-Bräuninger, M., et al. (2006). The Rho-GTPase Cdc42 Regulates Neural Progenitor Fate at the Apical Surface. Nat. Neurosci. 9, 1099–1107. doi:10.1038/nn1744
Cederquist, G. Y., Asciolla, J. J., Tchieu, J., Walsh, R. M., Cornacchia, D., Resh, M. D., et al. (2019). Specification of Positional Identity in Forebrain Organoids. Nat. Biotechnol. 37, 436–444. doi:10.1038/s41587-019-0085-3
Chambers, S. M., Fasano, C. A., Papapetrou, E. P., Tomishima, M., Sadelain, M., and Studer, L. (2009). Highly Efficient Neural Conversion of Human ES and iPS Cells by Dual Inhibition of SMAD Signaling. Nat. Biotechnol. 27, 275–280. doi:10.1038/nbt.1529
Cheroni, C., Trattaro, S., Caporale, N., López-Tobón, A., Tenderini, E., Troglio, F., et al. (2022). Benchmarking Brain Organoid Recapitulation of Fetal Corticogenesis. biorxiv. doi:10.1101/2022.04.22.488753
Dang, L. T., Vaid, S., Lin, G., Swaminathan, P., Safran, J., Loughman, A., et al. (2021). STRADA ‐mutant Human Cortical Organoids Model Megalencephaly and Exhibit Delayed Neuronal Differentiation. Dev. Neurobiol. 81, 696–709. doi:10.1002/dneu.22816
Dehay, C., Kennedy, H., and Kosik, K. S. (2015). The Outer Subventricular Zone and Primate-specific Cortical Complexification. Neuron 85, 683–694. doi:10.1016/j.neuron.2014.12.060
Del-Valle-Anton, L., and Borrell, V. (2022). Folding Brains: from Development to Disease Modeling. Physiol. Rev. 102, 511–550. doi:10.1152/physrev.00016.2021
Di Lullo, E., and Kriegstein, A. R. (2017). The Use of Brain Organoids to Investigate Neural Development and Disease. Nat. Rev. Neurosci. 18, 573–584. doi:10.1038/nrn.2017.107
Eiraku, M., Watanabe, K., Matsuo-Takasaki, M., Kawada, M., Yonemura, S., Matsumura, M., et al. (2008). Self-organized Formation of Polarized Cortical Tissues from ESCs and its Active Manipulation by Extrinsic Signals. Cell Stem Cell 3, 519–532. doi:10.1016/j.stem.2008.09.002
Esk, C., Lindenhofer, D., Haendeler, S., Wester, R. A., Pflug, F., Schroeder, B., et al. (2020). A Human Tissue Screen Identifies a Regulator of ER Secretion as a Brain-Size Determinant. Science 370, 935–941. doi:10.1126/science.abb5390
Fattah, A. R. A., Kolaitis, N., Van Daele, K., Rustandi, A. G., and Ranga, A. (2022). Local Actuation of Organoids by Magnetic Nanoparticles. BioRxiv 2022. doi:10.1101/2022.03.17.484696
Feng, G., Jensen, F. E., Greely, H. T., Okano, H., Treue, S., Roberts, A. C., et al. (2020). Opportunities and Limitations of Genetically Modified Nonhuman Primate Models for Neuroscience Research. Proc. Natl. Acad. Sci. U.S.A. 117, 24022–24031. doi:10.1073/pnas.2006515117
Fiddes, I. T., Lodewijk, G. A., Mooring, M., Bosworth, C. M., Ewing, A. D., Mantalas, G. L., et al. (2018). Human-Specific NOTCH2NL Genes Affect Notch Signaling and Cortical Neurogenesis. Cell 173, 1356–1369. e22. doi:10.1016/j.cell.2018.03.051
Fietz, S. A., Kelava, I., Vogt, J., Wilsch-Bräuninger, M., Stenzel, D., Fish, J. L., et al. (2010). OSVZ Progenitors of Human and Ferret Neocortex Are Epithelial-like and Expand by Integrin Signaling. Nat. Neurosci. 13, 690–699. doi:10.1038/nn.2553
Fischer, J., Peters, J., Namba, T., Huttner, W. B., and Heide, M. (2020). Human-specific ARHGAP11B Is Necessary and Sufficient for Human-type Basal Progenitor Levels in Primate Brain Organoids. BioRxiv 2020. doi:10.1126/science.aaa1975
Florio, M., Albert, M., Taverna, E., Namba, T., Brandl, H., Lewitus, E., et al. (2015). Human-specific Gene ARHGAP11B Promotes Basal Progenitor Amplification and Neocortex Expansion. Science 347, 1465–1470. doi:10.1126/science.aaa1975
Giandomenico, S. L., Mierau, S. B., Gibbons, G. M., Wenger, L. M. D., Masullo, L., Sit, T., et al. (2019). Cerebral Organoids at the Air-Liquid Interface Generate Diverse Nerve Tracts with Functional Output. Nat. Neurosci. 22, 669–679. doi:10.1038/s41593-019-0350-2
Giandomenico, S. L., Sutcliffe, M., and Lancaster, M. A. (2021). Generation and Long-Term Culture of Advanced Cerebral Organoids for Studying Later Stages of Neural Development. Nat. Protoc. 16, 579–602. doi:10.1038/s41596-020-00433-w
Gilardi, C., and Kalebic, N. (2021). The Ferret as a Model System for Neocortex Development and Evolution. Front. Cell Dev. Biol. 9, 661759. doi:10.3389/fcell.2021.661759
Gordon, A., Yoon, S.-J., Tran, S. S., Makinson, C. D., Park, J. Y., Andersen, J., et al. (2021). Long-term Maturation of Human Cortical Organoids Matches Key Early Postnatal Transitions. Nat. Neurosci. 24, 331–342. doi:10.1038/s41593-021-00802-y
Götz, M., and Huttner, W. B. (2005). The Cell Biology of Neurogenesis. Nat. Rev. Mol. Cell Biol. 6, 777–788.
Hansen, D. V., Lui, J. H., Parker, P. R. L., and Kriegstein, A. R. (2010). Neurogenic Radial Glia in the Outer Subventricular Zone of Human Neocortex. Nature 464, 554–561. doi:10.1038/nature08845
Harschnitz, O., and Studer, L. (2021). Human Stem Cell Models to Study Host-Virus Interactions in the Central Nervous System. Nat. Rev. Immunol. 21, 441–453. doi:10.1038/s41577-020-00474-y
Heide, M., Haffner, C., Murayama, A., Kurotaki, Y., Shinohara, H., Okano, H., et al. (2020). Human-specific ARHGAP11B Increases Size and Folding of Primate Neocortex in the Fetal Marmoset. Science 369, 546–550. doi:10.1126/science.abb2401
Heide, M., Huttner, W. B., and Mora-Bermúdez, F. (2018). Brain Organoids as Models to Study Human Neocortex Development and Evolution. Curr. Opin. Cell Biol. 55, 8–16. doi:10.1016/j.ceb.2018.06.006
Hevner, R. F. (2015). Brain Overgrowth in Disorders of RTK-Pi3k-AKT Signaling: A Mosaic of Malformations. Seminars Perinatology 39, 36–43. doi:10.1053/j.semperi.2014.10.006
Hu, H., Gehart, H., Artegiani, B., LÖpez-Iglesias, C., Dekkers, F., Basak, O., et al. (2018). Long-Term Expansion of Functional Mouse and Human Hepatocytes as 3D Organoids. Cell 175, 1591–1606. e19. doi:10.1016/j.cell.2018.11.013
Hu, J. S., Vogt, D., Sandberg, M., and Rubenstein, J. L. (2017). Cortical Interneuron Development: a Tale of Time and Space. Development 144, 3867–3878. doi:10.1242/dev.132852
Jabali, A., Hoffrichter, A., Uzquiano, A., Marsoner, F., Wilkens, R., Siekmann, M., et al. (2022). Human Cerebral Organoids Reveal Progenitor Pathology in EML1-Linked Cortical Malformation. EMBO Rep., e54027.
Kadoshima, T., Sakaguchi, H., Nakano, T., Soen, M., Ando, S., Eiraku, M., et al. (2013). Self-organization of Axial Polarity, Inside-Out Layer Pattern, and Species-specific Progenitor Dynamics in Human ES Cell-Derived Neocortex. Proc. Natl. Acad. Sci. U.S.A. 110, 20284–20289. doi:10.1073/pnas.1315710110
Kalebic, N., Gilardi, C., Albert, M., Namba, T., Long, K. R., Kostic, M., et al. (2018). Human-specific ARHGAP11B Induces Hallmarks of Neocortical Expansion in Developing Ferret Neocortex. Elife 7. doi:10.7554/eLife.41241
Kalebic, N., Gilardi, C., Stepien, B., Wilsch-Bräuninger, M., Long, K. R., Namba, T., et al. (2019). Neocortical Expansion Due to Increased Proliferation of Basal Progenitors Is Linked to Changes in Their Morphology. Cell Stem Cell 24, 535–550. e9. doi:10.1016/j.stem.2019.02.017
Kalebic, N., and Huttner, W. B. (2020). Basal Progenitor Morphology and Neocortex Evolution. Trends Neurosci. 43, 843–853. doi:10.1016/j.tins.2020.07.009
Kalebic, N., and Namba, T. (2021). Inheritance and Flexibility of Cell Polarity: a Clue for Understanding Human Brain Development and Evolution. Development 148. doi:10.1242/dev.199417
Kaluthantrige Don, F., and Huch, M. (2021). Organoids, where We Stand and where We Go. Trends Mol. Med. 27, 416–418. doi:10.1016/j.molmed.2021.03.001
Kanton, S., Boyle, M. J., He, Z., Santel, M., Weigert, A., Sanchís-Calleja, F., et al. (2019). Organoid Single-Cell Genomic Atlas Uncovers Human-specific Features of Brain Development. Nature 574, 418–422. doi:10.1038/s41586-019-1654-9
Karzbrun, E., Kshirsagar, A., Cohen, S. R., Hanna, J. H., and Reiner, O. (2018). Human Brain Organoids on a Chip Reveal the Physics of Folding. Nat. Phys. 14, 515–522. doi:10.1038/s41567-018-0046-7
Kawaguchi, A. (2020). Neuronal Delamination and Outer Radial Glia Generation in Neocortical Development. Front. Cell Dev. Biol. 8, 623573. doi:10.3389/fcell.2020.623573
Krenn, V., Bosone, C., Burkard, T. R., Spanier, J., Kalinke, U., Calistri, A., et al. (2021). Organoid Modeling of Zika and Herpes Simplex Virus 1 Infections Reveals Virus-specific Responses Leading to Microcephaly. Cell Stem Cell 28, 1362–1379. e7. doi:10.1016/j.stem.2021.03.004
Kroenke, C. D., and Bayly, P. V. (2018). How Forces Fold the Cerebral Cortex. J. Neurosci. 38, 767–775. doi:10.1523/jneurosci.1105-17.2017
Kyrousi, C., O'Neill, A. C., Brazovskaja, A., He, Z., Kielkowski, P., Coquand, L., et al. (2021). Extracellular LGALS3BP Regulates Neural Progenitor Position and Relates to Human Cortical Complexity. Nat. Commun. 12, 1–22. doi:10.1038/s41467-021-26447-w
LaMonica, B. E., Lui, J. H., Hansen, D. V., and Kriegstein, A. R. (2013). Mitotic Spindle Orientation Predicts Outer Radial Glial Cell Generation in Human Neocortex. Nat. Commun. 4, 1665. doi:10.1038/ncomms2647
Lancaster, M. A., and Huch, M. (2019). Disease Modelling in Human Organoids. Dis. Model. Mech. 12. doi:10.1242/dmm.039347
Lancaster, M. A., Corsini, N. S., Wolfinger, S., Gustafson, E. H., Phillips, A. W., Burkard, T. R., et al. (2017). Guided Self-Organization and Cortical Plate Formation in Human Brain Organoids. Nat. Biotechnol. 35, 659–666. doi:10.1038/nbt.3906
Lancaster, M. A., and Knoblich, J. A. (2014). Organogenesis in a Dish: Modeling Development and Disease Using Organoid Technologies. Science 345, 1247125. doi:10.1126/science.1247125
Lancaster, M. A., Renner, M., Martin, C.-A., Wenzel, D., Bicknell, L. S., Hurles, M. E., et al. (2013). Cerebral Organoids Model Human Brain Development and Microcephaly. Nature 501, 373–379. doi:10.1038/nature12517
Li, M. L., Aggeler, J., Farson, D. A., Hatier, C., Hassell, J., and Bissell, M. J. (1987). Influence of a Reconstituted Basement Membrane and its Components on Casein Gene Expression and Secretion in Mouse Mammary Epithelial Cells. Proc. Natl. Acad. Sci. U.S.A. 84, 136–140. doi:10.1073/pnas.84.1.136
Lim, L., Mi, D., Llorca, A., and Marín, O. (2018). Development and Functional Diversification of Cortical Interneurons. Neuron 100, 294–313. doi:10.1016/j.neuron.2018.10.009
López-Tobón, A., Villa, C. E., Cheroni, C., Trattaro, S., Caporale, N., Conforti, P., et al. (2019). Human Cortical Organoids Expose a Differential Function of GSK3 on Cortical Neurogenesis. Stem Cell Rep. 13, 847–861. doi:10.1016/j.stemcr.2019.09.005
Lopez-Tobon, A., Caporale, N., Trattaro, S., and Testa, G. (2020). Three-dimensional Models of Human Brain Development. Stem Cell Epigenetics, 257–278. doi:10.1016/b978-0-12-814085-7.00011-8
Mansour, A. A., Gonçalves, J. T., Bloyd, C. W., Li, H., Fernandes, S., Quang, D., et al. (2018). An In Vivo Model of Functional and Vascularized Human Brain Organoids. Nat. Biotechnol. 36, 432–441. doi:10.1038/nbt.4127
Mariani, J., Coppola, G., Zhang, P., Abyzov, A., Provini, L., Tomasini, L., et al. (2015). FOXG1-Dependent Dysregulation of GABA/Glutamate Neuron Differentiation in Autism Spectrum Disorders. Cell 162, 375–390. doi:10.1016/j.cell.2015.06.034
Marton, R. M., and Pașca, S. P. (2020). Organoid and Assembloid Technologies for Investigating Cellular Crosstalk in Human Brain Development and Disease. Trends Cell Biol. 30, 133–143. doi:10.1016/j.tcb.2019.11.004
Molnár, Z., Clowry, G. J., Šestan, N., Alzu’bi, A., Bakken, T., Hevner, R. F., et al. (2019). New Insights into the Development of the Human Cerebral Cortex. J. Anat. 235, 432–451.
Molotkova, N., Molotkov, A., Sirbu, I. O., and Duester, G. (2005). Requirement of Mesodermal Retinoic Acid Generated by Raldh2 for Posterior Neural Transformation. Mech. Dev. 122, 145–155. doi:10.1016/j.mod.2004.10.008
Mora-Bermúdez, F., Kanis, P., Macak, D., Peters, J., Naumann, R., Sarov, M., et al. (2022). Longer Metaphase and Fewer Chromosome Segregation Errors in Modern Human Than Neandertal Brain Development. BioRxiv. doi:10.1101/2022.03.30.486431
Muchnik, S. K., Lorente-Galdos, B., Santpere, G., and Sestan, N. (2019). Modeling the Evolution of Human Brain Development Using Organoids. Cell 179, 1250–1253. doi:10.1016/j.cell.2019.10.041
Muñoz-Sanjuán, I., and Brivanlou, A. H. (2002). Neural Induction, the Default Model and Embryonic Stem Cells. Nat. Rev. Neurosci. 3, 271–280.
Nowakowski, T. J., Bhaduri, A., Pollen, A. A., Alvarado, B., Mostajo-Radji, M. A., Di Lullo, E., et al. (2017). Spatiotemporal Gene Expression Trajectories Reveal Developmental Hierarchies of the Human Cortex. Science 358, 1318–1323. doi:10.1126/science.aap8809
O'Neill, A. C., Kyrousi, C., Klaus, J., Leventer, R. J., Kirk, E. P., Fry, A., et al. (2018). A Primate-specific Isoform of PLEKHG6 Regulates Neurogenesis and Neuronal Migration. Cell Rep. 25, 2729–e6. doi:10.1016/j.celrep.2018.11.029
Ormel, P. R., Vieira de Sá, R., van Bodegraven, E. J., Karst, H., Harschnitz, O., Sneeboer, M. A. M., et al. (2018). Microglia Innately Develop within Cerebral Organoids. Nat. Commun. 9, 4167. doi:10.1038/s41467-018-06684-2
Ostrem, B., Di Lullo, E., and Kriegstein, A. (2017). oRGs and Mitotic Somal Translocation - a Role in Development and Disease. Curr. Opin. Neurobiol. 42, 61–67. doi:10.1016/j.conb.2016.11.007
Ostrem, B. E. L., Lui, J. H., Gertz, C. C., and Kriegstein, A. R. (2014). Control of Outer Radial Glial Stem Cell Mitosis in the Human Brain. Cell Rep. 8, 656–664. doi:10.1016/j.celrep.2014.06.058
Otani, T., Marchetto, M. C., Gage, F. H., Simons, B. D., and Livesey, F. J. (2016). 2D and 3D Stem Cell Models of Primate Cortical Development Identify Species-specific Differences in Progenitor Behavior Contributing to Brain Size. Cell Stem Cell 18, 467–480. doi:10.1016/j.stem.2016.03.003
Ou, M.-Y., Xiao, Q., Ju, X.-C., Zeng, P.-M., Huang, J., Sheng, A.-L., et al. (2021). The CTNNBIP1-CLSTN1 Fusion Transcript Regulates Human Neocortical Development. Cell Rep. 35, 109290. doi:10.1016/j.celrep.2021.109290
Paşca, A. M., Sloan, S. A., Clarke, L. E., Tian, Y., Makinson, C. D., Huber, N., et al. (2015). Functional Cortical Neurons and Astrocytes from Human Pluripotent Stem Cells in 3D Culture. Nat. Methods 12, 671–678. doi:10.1038/nmeth.3415
Peng, W. C., Logan, C. Y., Fish, M., Anbarchian, T., Aguisanda, F., Álvarez-Varela, A., et al. (2018). Inflammatory Cytokine TNFα Promotes the Long-Term Expansion of Primary Hepatocytes in 3D Culture. Cell 175, 1607–1619. e15. doi:10.1016/j.cell.2018.11.012
Penisson, M., Ladewig, J., Belvindrah, R., and Francis, F. (2019). Genes and Mechanisms Involved in the Generation and Amplification of Basal Radial Glial Cells. Front. Cell. Neurosci. 13. doi:10.3389/fncel.2019.00381
Pinson, A., and Huttner, W. B. (2021). Neocortex Expansion in Development and Evolution-From Genes to Progenitor Cell Biology. Curr. Opin. Cell Biol. 73, 9–18. doi:10.1016/j.ceb.2021.04.008
Pollen, A. A., Bhaduri, A., Andrews, M. G., Nowakowski, T. J., Meyerson, O. S., Mostajo-Radji, M. A., et al. (2019). Establishing Cerebral Organoids as Models of Human-specific Brain Evolution. Cell 176, 743–756. e17. doi:10.1016/j.cell.2019.01.017
Pollen, A. A., Nowakowski, T. J., Chen, J., Sandoval-Espinosa, C., Nicholas, C. R., Shuga, C. R., et al. (2015). Molecular Identity of Human Outer Radial Glia during Cortical Development. Cell 163, 55–67. doi:10.1016/j.cell.2015.09.004
Qian, X., Jacob, F., Song, M. M., Nguyen, H. N., Song, H., and Ming, G.-L. (2018). Generation of Human Brain Region-specific Organoids Using a Miniaturized Spinning Bioreactor. Nat. Protoc. 13, 565–580. doi:10.1038/nprot.2017.152
Qian, X., Nguyen, H. N., Song, M. M., Hadiono, C., Ogden, S. C., Hammack, C., et al. (2016). Brain-Region-Specific Organoids Using Mini-Bioreactors for Modeling ZIKV Exposure. Cell 165, 1238–1254. doi:10.1016/j.cell.2016.04.032
Qian, X., Su, Y., Adam, C. D., Deutschmann, A. U., Pather, S. R., Goldberg, E. M., et al. (2020). Sliced Human Cortical Organoids for Modeling Distinct Cortical Layer Formation. Cell Stem Cell 26, 766–781. e9. doi:10.1016/j.stem.2020.02.002
Rakic, P. (1995). A Small Step for the Cell, a Giant Leap for Mankind: a Hypothesis of Neocortical Expansion during Evolution. Trends Neurosci. 18, 383–388. doi:10.1016/0166-2236(95)93934-p
Rakic, P. (1988). Specification of Cerebral Cortical Areas. Science 241, 170–176. doi:10.1126/science.3291116
Reillo, I., de Juan Romero, C., Cárdenas, A., Clascá, F., Martínez-Martinez, M. Á., and Borrell, V. (2017). A Complex Code of Extrinsic Influences on Cortical Progenitor Cells of Higher Mammals. Cereb. Cortex 27, 4586–4606. doi:10.1093/cercor/bhx171
Reillo, I., de Juan Romero, C., García-Cabezas, M. Á., and Borrell, V. (2011). A Role for Intermediate Radial Glia in the Tangential Expansion of the Mammalian Cerebral Cortex. Cereb. Cortex 21, 1674–1694. doi:10.1093/cercor/bhq238
Sadler, T. W. (2005). Embryology of Neural Tube Development. Am. J. Med. Genet. 135C, 2–8. doi:10.1002/ajmg.c.30049
Sato, T., Vries, R. G., Snippert, H. J., van de Wetering, M., Barker, N., Stange, D. E., et al. (2009). Single Lgr5 Stem Cells Build Crypt-Villus Structures In Vitro without a Mesenchymal Niche. Nature 459, 262–265. doi:10.1038/nature07935
Shamir, E. R., and Ewald, A. J. (2014). Three-dimensional Organotypic Culture: Experimental Models of Mammalian Biology and Disease. Nat. Rev. Mol. Cell Biol. 15, 647–664. doi:10.1038/nrm3873
Shi, Y., Sun, L., Wang, M., Liu, J., Zhong, S., Li, R., et al. (2020). Vascularized Human Cortical Organoids (vOrganoids) Model Cortical Development In Vivo. PLoS Biol. 18, e3000705. doi:10.1371/journal.pbio.3000705
Sidhaye, J., and Knoblich, J. A. (2021). Brain Organoids: an Ensemble of Bioassays to Investigate Human Neurodevelopment and Disease. Cell Death Differ. 28, 52–67. doi:10.1038/s41418-020-0566-4
Sloan, S. A., Andersen, J., Pașca, A. M., Birey, F., and Pașca, S. P. (2018). Generation and Assembly of Human Brain Region-specific Three-Dimensional Cultures. Nat. Protoc. 13, 2062–2085. doi:10.1038/s41596-018-0032-7
Smart, I. H. M. (2002). Unique Morphological Features of the Proliferative Zones and Postmitotic Compartments of the Neural Epithelium Giving Rise to Striate and Extrastriate Cortex in the Monkey. Cereb. Cortex 12, 37–53. doi:10.1093/cercor/12.1.37
Sridhar, A., Hoshino, A., Finkbeiner, C. R., Chitsazan, A., Dai, L., Haugan, A. K., et al. (2020). Single-Cell Transcriptomic Comparison of Human Fetal Retina, hPSC-Derived Retinal Organoids, and Long-Term Retinal Cultures. Cell Rep. 30, 1644–1659. e4. doi:10.1016/j.celrep.2020.01.007
Takahashi, K., Tanabe, K., Ohnuki, M., Narita, M., Ichisaka, T., Tomoda, K., et al. (2007). Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell 131, 861–872. doi:10.1016/j.cell.2007.11.019
Takahashi, K., and Yamanaka, S. (2006). Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell 126, 663–676. doi:10.1016/j.cell.2006.07.024
Taverna, E., Götz, M., and Huttner, W. B. (2014). The Cell Biology of Neurogenesis: toward an Understanding of the Development and Evolution of the Neocortex. Annu. Rev. Cell Dev. Biol. 30, 465–502. doi:10.1146/annurev-cellbio-101011-155801
Trujillo, C. A., Rice, E. S., Schaefer, N. K., Chaim, I. A., Wheeler, E. C., Madrigal, A. A., et al. (2021). Reintroduction of the Archaic Variant of NOVA1 in Cortical Organoids Alters Neurodevelopment. Science 371. doi:10.1126/science.aax2537
Uzquiano, A., Kedaigle, A. J., Pigoni, M., Paulsen, B., Adiconis, X., Kim, K., et al. (2022). Single-cell Multiomics Atlas of Organoid Development Uncovers Longitudinal Molecular Programs of Cellular Diversification of the Human Cerebral Cortex. BioRxiv. doi:10.1101/2022.03.17.484798
van den Berg, C. W., Ritsma, L., Avramut, M. C., Wiersma, L. E., van den Berg, B. M., Leuning, D. G., et al. (2018). Renal Subcapsular Transplantation of PSC-Derived Kidney Organoids Induces Neo-Vasculogenesis and Significant Glomerular and Tubular Maturation In Vivo. Stem Cell Rep. 10, 751–765. doi:10.1016/j.stemcr.2018.01.041
Velasco, S., Kedaigle, A. J., Simmons, S. K., Nash, A., Rocha, M., Quadrato, G., et al. (2019). Individual Brain Organoids Reproducibly Form Cell Diversity of the Human Cerebral Cortex. Nature 570, 523–527. doi:10.1038/s41586-019-1289-x
Velasco, S., Paulsen, B., and Arlotta, P. (2020). 3D Brain Organoids: Studying Brain Development and Disease outside the Embryo. Annu. Rev. Neurosci. 43, 375–389. doi:10.1146/annurev-neuro-070918-050154
Velmeshev, D., Chavali, M., Nowakowski, T. J., Bhade, M., Mayer, S., Goyal, N., et al. (2021). Molecular Diversity and Lineage Commitment of Human Interneuron Progenitors. BioRxiv. doi:10.1101/2021.05.13.444045
Vértesy, Á., Eichmueller, O. L., Naas, J., Novatchkova, M., Esk, C., Balmaña, M., et al. (2022). Cellular Stress in Brain Organoids Is Limited to a Distinct and Bioinformatically Removable Subpopulation. BioRxiv. doi:10.1101/2022.03.11.483643
Villa, C. E., Cheroni, C., Dotter, C. P., López-Tóbon, A., Oliveira, B., Sacco, R., et al. (2022). CHD8 Haploinsufficiency Links Autism to Transient Alterations in Excitatory and Inhibitory Trajectories. Cell Rep. 39, 110615. doi:10.1016/j.celrep.2022.110615
Wang, D., Wang, J., Bai, L., Pan, H., Feng, H., Clevers, H., et al. (2020). Long-Term Expansion of Pancreatic Islet Organoids from Resident Procr+ Progenitors. Cell 180, 1198–1211. e19. doi:10.1016/j.cell.2020.02.048
Wang, X., Tsai, J.-W., LaMonica, B., and Kriegstein, A. R. (2011). A New Subtype of Progenitor Cell in the Mouse Embryonic Neocortex. Nat. Neurosci. 14, 555–561. doi:10.1038/nn.2807
Wong, F. K., Fei, J.-F., Mora-Bermúdez, F., Taverna, E., Haffner, C., Fu, J., et al. (2015). Sustained Pax6 Expression Generates Primate-like Basal Radial Glia in Developing Mouse Neocortex. PLoS Biol. 13, e1002217. doi:10.1371/journal.pbio.1002217
Yokota, Y., Eom, T.-Y., Stanco, A., Kim, W.-Y., Rao, S., Snider, W. D., et al. (2010). Cdc42 and Gsk3 Modulate the Dynamics of Radial Glial Growth, Inter-radial Glial Interactions and Polarity in the Developing Cerebral Cortex. Development 137, 4101–4110. doi:10.1242/dev.048637
Zhang, S.-C., Wernig, M., Duncan, I. D., Brüstle, O., and Thomson, J. A. (2001). In Vitro differentiation of Transplantable Neural Precursors from Human Embryonic Stem Cells. Nat. Biotechnol. 19, 1129–1133. doi:10.1038/nbt1201-1129
Keywords: neural progenitor cells, neural stem cells, neurodevelopmental disease, brain evolution, cerebral organoid
Citation: Kaluthantrige Don F and Kalebic N (2022) Forebrain Organoids to Model the Cell Biology of Basal Radial Glia in Neurodevelopmental Disorders and Brain Evolution. Front. Cell Dev. Biol. 10:917166. doi: 10.3389/fcell.2022.917166
Received: 10 April 2022; Accepted: 23 May 2022;
Published: 14 June 2022.
Edited by:
Felipe Mora-Bermúdez, Max Planck Society, GermanyReviewed by:
Leonardo Beccari, INSERM U1217 Institut NeuroMyoGène, FranceCopyright © 2022 Kaluthantrige Don and Kalebic. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nereo Kalebic, bmVyZW8ua2FsZWJpY0BmaHQub3Jn
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.