- Department of Orthopaedics, the Hospital of Shunyi District, Beijing, China
Intervertebral discdegeneration (IDD) is the most common cause of lower back pain, but the exact molecular mechanism of IDD is still unknown. Recently, studies have shown that circular RNAs (circRNAs) regulate diverse biological procedures such as cell metastasis, growth, metabolism, migration, apoptosis, and invasion. We demonstrated that IL-1β and TNF-α induced circ_0005918 expression in the NP cell, and circ_0005918 was overexpressed in the IDD group compared with the control group. Moreover, the upregulated expression of circ_0005918 was associated with disc degeneration degree. The elevated expression of circ_0005918 promoted cell growth and ECM degradation, and it induced secretion of inflammatory cytokines including IL-1β, IL-6, and TNF-α. Moreover, we found that circ_0005918 sponged miR-622 in the NP cell. In addition, the exposure to IL-1β and TNF-α suppressed the expression of miR-622, which was downregulated in the IDD group compared with the control group. Furthermore, the downregulated expression of miR-622 was associated with disc degeneration degree. The expression level of miR-622 was negatively associated with circ_0005918 expression in the IDD group. In conclusion, circ_0005918 regulated cell growth, ECM degradation, and secretion of inflammatory cytokines by regulating miR-622 expression. These data suggested that circ_0005918 played important roles in the development of IDD via sponging miR-622.
Introduction
Intervertebral disc degeneration (IDD) is a musculoskeletal disease, and is the most common cause of lower back pain, which affects a large portion of the population worldwide (Tan et al., 2018; Zhan et al., 2019; Zhao et al., 2019; Cao S. et al., 2021). IDD affects not only the quality of life but also causes economic and medical burdens to society (Chen W. et al., 2021; Li et al., 2021a; Tan et al., 2021). IDD is usually considered a natural disc aging procedure; however, a lot of patients demonstrate accelerated disc degeneration due to genetic and environmental factors (Chen et al., 2017; Wang et al., 2020; Sun et al., 2021; Zhan et al., 2021). The etiology and molecular mechanism of IDD are complicated and increasingly, evidence has shown that gender, age, inflammation, and behavioral influences are associated with the risk of IDD (Kraus et al., 2019; Jing and Liu, 2021; Tan et al., 2021). The exact molecular mechanism of IDD is still unknown. It is urgent to elucidate the underlying pathogenesis and find novel treatment strategies for IDD.
Circular RNAs (circRNAs), a new type of non-coding RNA, consist of exon transcripts and exert crucial roles in regulating the downstream gene expression (Liang et al., 2019; Ma et al., 2020; Ding et al., 2021; Yang et al., 2021). Increasing evidence has indicated that circRNAs regulate diverse cell biological procedures such as cell metastasis, growth, metabolism, migration, apoptosis, and invasion (Tian et al., 2019; Liu et al., 2020; Nie et al., 2020; Pu et al., 2020; Zhou et al., 2020). Recent studies have suggested that circRNAs participated in many diseases including congenital diseases, metabolic diseases, infection, and cancer (Li et al., 2019; Li et al., 2020a; Li et al., 2020b; Li et al., 2020c; Zhao et al., 2021). CircRNAs are involved in various orthopedic diseases such as osteoarthritis, femoral head necrosis, scoliosis, osteosarcoma, and also IDD (Guo et al., 2020a; Yan et al., 2020; Zhang et al., 2021a; Hao et al., 2021). For example, Hu et al. (2022) demonstrated that circ_0022382 suppressed IDD progression by modulating TGF-beta3/miR-4726-5p expression. Wang et al. (2021) showed that circARL15 played crucial roles in IDD development by sponging the miR-431-5p/DISC1 axis. Recently Guo et al. (2021) performed microarray hybridization and qRT-PCR assay to study differentially expressed circRNAs and identified that the expression of circ_0005918 was upregulated in the IDD group. However, its function in IDD remains unknown.
In our study, we studied the expression of circ_0005918 in IDD tissues and found that IL-1β and TNF-α induced circ_0005918 expression in the NP cell, and circ_0005918 was overexpressed in the IDD group compared with the control group. The elevated expression of circ_0005918 promoted cell growth and ECM degradation, and induced secretion of inflammatory cytokines including IL-1β, IL-6, and TNF-α.
Materials and Methods
Tissue Specimens
Our study was approved by the Clinical Ethics Committee of Shunyi Hospital, Beijing, China, and we followed the rules of the Helsinki Declaration. Participants gave written informed consent. Intervertebral disc specimens were collected from scoliosis patients who were undergoing osteotomy and spinal fusion as controls and intervertebral disc specimens from IDD patients who underwent surgery were collected in our hospital.
Cell Culture and Cell Transfection
Eight NP samples were obtained from IDD that were used for isolating and culturing NP cells. The human NP cells were cultured according to previous references (Gao et al., 2020; Chen Y. et al., 2021). The NP cells were then passaged two times to apply for other experiments. Cell transfection was performed using Lipofectamine 2000 (Invitrogen). pcDNA3.1-circ_0005918, miR-622 mimic, and an empty vector (used as a negative control) were purchased from Invitrogen.
Real-Time PCR
Total RNA was isolated from the NP samples or cells using a Trizol kit (Beyotime Biotechnology, China). The cDNA synthesis reagent (Sangon, Shanghai, China) was used for cDNA synthesis. The level of circ_0005918 and miR-622 was detected by real-time qRT-PCR using SYBR green (Applied Biosystems, Thermo) on the ABI 7500 system following the manufacturer’s protocols. The primer sequences were shown as follows: human circ_0005918 forward, 5′-AAAGCTACCCACGCAAGGAA-3′ and reverse, 5′-CTTTGTCAACTGGTCCACACAC-3'; miR-622 forward, 5′-ATCCC AGGGA GACAG AGATC GAGG-3′ and reverse, 5′-AAGCT TGGTG GTGGA CTTTT GGTTGT-3′; GAPHD: forward, 5′-GCACC GTCAA GGCTG AGAAC-3′ and reverse, 5′-TGGTG AAGAC GCCAG TGGA-3′; U6: forward, 5′-GGTGA AGCAG GCGTC GGAGG-3′ and reverse, 5′-GAGGG CAATG CCAGC CCCAG-3′. The relative level of these genes was determined by 2 −ΔΔCt, and GAPDH was performed as an internal reference for mRNA and circRNA and U6 as an internal reference for miRNA.
Cell Growth and ELISA
The NP cells were cultured in a 96-well culture dish at the density of 2*103 per dish. The cell proliferation rate was measured for 0, 24, 48, and 72h, and 10 μL CCK-8 reagent was added to each dish for incubation for 2 h. The absorbance at 450 nm was detected using a microplate reader. The concentrations of IL-6, TNF-α, and IL-1β in the cell supernatants were detected using an ELISA kit following the kit’s instructions (Boste, China).
Western Blot Analysis
Total protein from samples or cells was extracted with RIPA buffer. The concentration of protein was analyzed with a BCA protein assay. An equivalent quantity of protein was resolved on 12% SDS-PAGE (sodium dodecyl sulfate-polyacrylamide gel electrophoresis) and then transferred to the PVDF membrane (Bio-Rad). After blocking with bovine serum, the PVDF membrane was incubated with anti-GAPDH or anti-MMP9 overnight at 4°C. The antibody binding was measured with peroxidase-conjugated IgG and visualized using ECL chemiluminescent system (Pierce Biotechnology, IL) following the manufacturer’s description.
Luciferase Assay
The sequences of circ_0005918 containing miR-622 complementary sites were cloned into the pGL3 luciferase vector, which was called circ_0005918-WT or circ_0005918-mut. The cells were cultured in the 96-well plate and co-transfected with miR-622 mimic or control and circ_0005918-WT or circ_0005918-mut. After incubation for 48 h, luciferase activity was analyzed by a dual-luciferase reporter kit (Promega, WI, United States).
Statistical Analysis
Statistical assay was carried out by SPSS 20.0, and data between two groups were detected utilizing the Student’s test. p-value <0.05 was considered significantly different.
Results
Exposure to IL-1β and TNF-α Induced circ_0005918 Expression
NP cells were treated with different doses of IL-1β and TNF-α for 48 h, and the expression of circ_0005918 was detected via qRT-PCR. As shown in Figures 1A,C, circ_0005918 expression was upregulated in the IL-1β and TNF-α groups compared with the control groups. Moreover, we showed that IL-1β and TNF-α induced circ_0005918 expression in a time-dependent manner (Figures 1B,D).
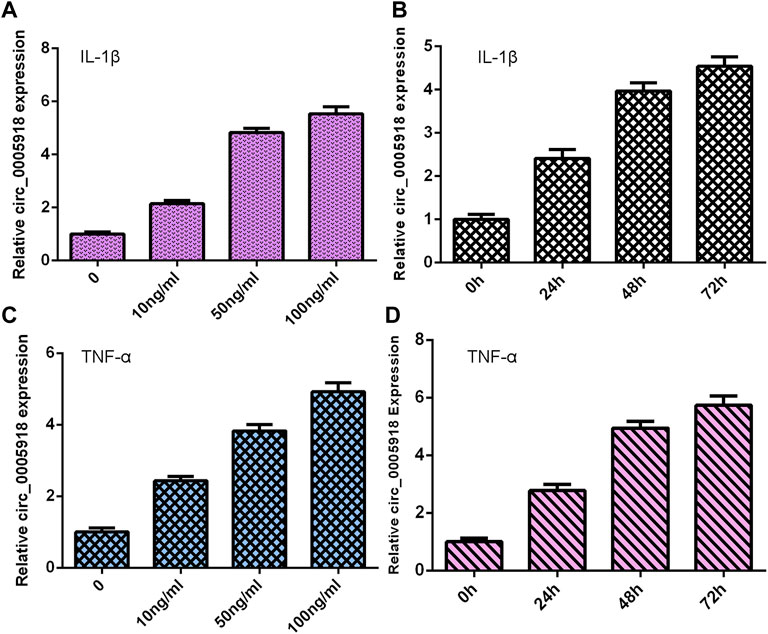
FIGURE 1. Exposure to IL-1β and TNF-α induced circ_0005918 expression. (A) Expression of circ_0005918 was measured by qRT-PCR assay. (B) IL-1β induced circ_0005918 expression in a time-dependent manner. (C) The level of circ_0005918 was detected via qRT-PCR assay. (D) TNF-α induced circ_0005918 expression in a time-dependent manner.
Upregulated circ_0005918 Levels Were Associated With IDD
Then, we measured circ_0005918 expression in the IVD samples. First, we showed that the circ_0005918 level was upregulated in NP cells from the IDD group compared with the control group (Figure 2A). Then, circ_0005918 was overexpressed in the IDD group compared with the control group (Figure 2B). Moreover, the upregulated expression of circ_0005918 was associated with disc degeneration degree (Figure 2C).
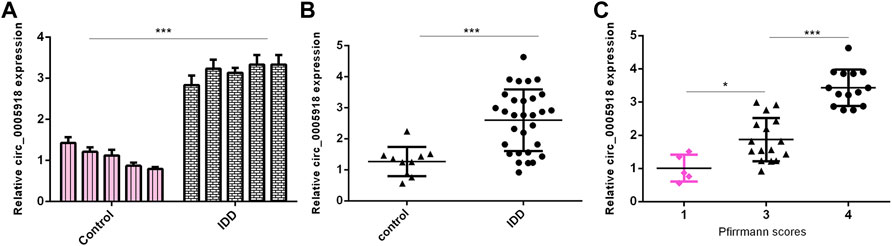
FIGURE 2. Upregulated circ_0005918 levels were associated with IDD. (A) The circ_0005918 level was upregulated 5 NP cells from IDD compared to control. (B) Circ_0005918 was overexpressed in the IDD group compared to the control group. (C) The upregulated expression of circ_0005918 was associated with disc degeneration levels. *p < 0.05 and ***p < 0.001.
Circ_0005918 Induced Cell Growth and ECM Degradation
Circ_0005918 was remarkably overexpressed in the NP cell after being transfected with pcDNA-circ_0005918 (Figure 3A). The elevated expression of circ_0005918 promoted cell growth in the NP cell (Figure 3B). The overexpression of circ_0005918 promoted MMP-9 expression in the NP cell (Figure 3C). Ectopic expression of circ_0005918 induced MMP-13 expression in the NP cell (Figure 3D). However, the overexpression of circ_0005918 suppressed collagen II expression in the NP cell (Figure 3E).
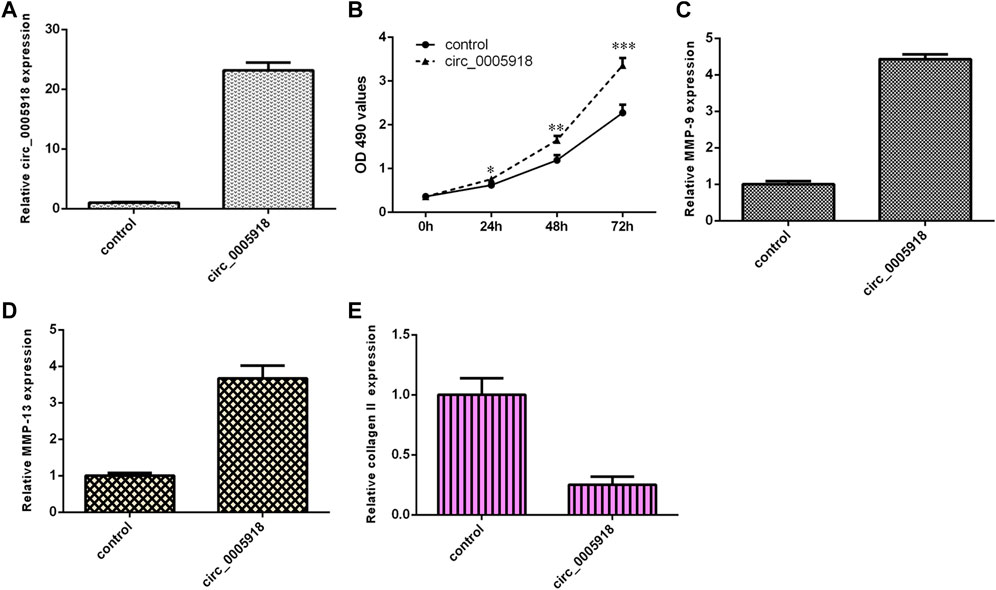
FIGURE 3. Circ_0005918 induced cell growth and ECM degradation. (A) The expression of circ_0005918 was measured by qRT-PCR assay. (B) The elevated expression of circ_0005918 promoted cell growth in the NP cell. (C) The overexpression of circ_0005918 promoted MMP-9 expressions in the NP cell. (D) The ectopic expression of circ_0005918 induced MMP-13 expression in the NP cell. (E) The overexpression of circ_0005918 suppressed collagen II expression in the NP cell. *p < 0.05, **p < 0.01, ***p < 0.001.
Circ_0005918 Induced Secretion of Inflammatory Cytokines
We showed that the ectopic expression of circ_0005918 promoted the secretion of three inflammatory cytokines including IL-1β (Figure 4A), IL-6 (Figure 4B), and TNF-α (Figure 4C).
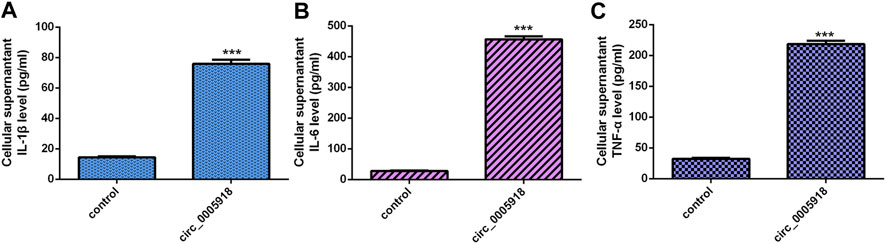
FIGURE 4. Circ_0005918 induced inflammatory cytokines secretion. (A) The ectopic expression of circ_0005918 promoted IL-1β expression. (B) The level of IL-6 in the cell supernatants was measured by ELISA. (C) The level of TNF-α in the cell supernatants was measured by ELISA. ***p < 0.001.
Circ_0005918 Regulated miR-622 Expression in NP Cell
By using Circular RNA Interactome software, we showed that circ_0005918 may serve as a miR-622 sponge (Figure 5A). MiR-622 was significantly upregulated in the miR-622 mimic group compared with the control group (Figure 5B). The luciferase reporter indicated ectopic expression of miR-622 suppressed the luciferase reporter value of circ_0005918 wild reporter vector but did not change the value of the mutant reporter (Figure 5C). The overexpression of circ_0005918 inhibited miR-622 expression (Figure 5D).
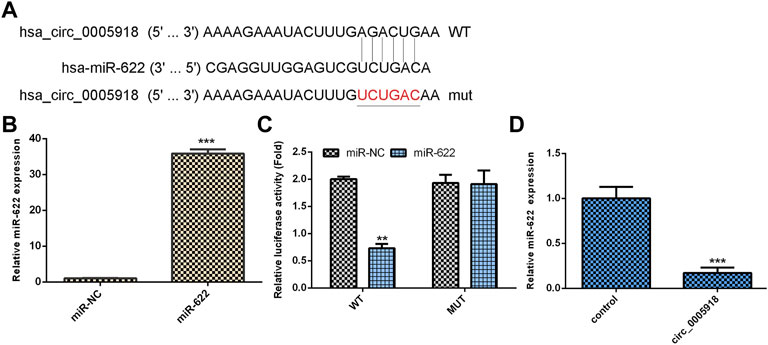
FIGURE 5. Circ_0005918 regulated miR-622 expression in NP cell. (A) By using Circular RNA Interactome software, it shows that circ_0005918 may sponge miR-622 expression. (B) The expression of miR-622 was measured by qRT-PCR assay. (C) The luciferase reporter indicated the ectopic expression of miR-622 suppressed the luciferase reporter value of circ_0005918 wild reporter vector but did not change the value of the mutant reporter. (D) The overexpression of circ_0005918 inhibited miR-622 expression. **p < 0.01, ***p < 0.001.
Exposure to IL-1β and TNF-α Suppressed miR-622 Expression
The NP cells were treated with different doses of IL-1β and TNF-α for 48 h, and the expression of miR-622 was detected via qRT-PCR. As shown in Figures 6A,C, the miR-622 expression was downregulated in the IL-1β and TNF-α groups compared with the control group. Moreover, we showed that IL-1β and TNF-α induced miR-622 expression in a time-dependent manner (Figures 6B,D).
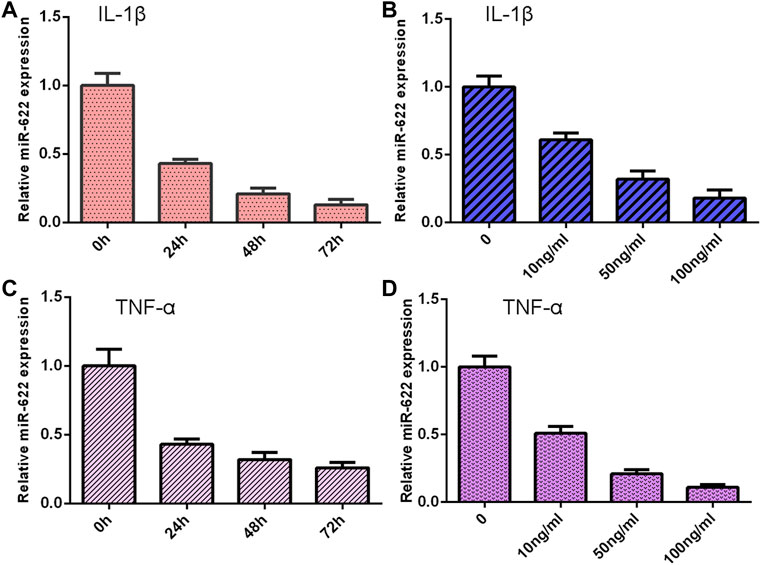
FIGURE 6. Exposure to IL-1β and TNF-α suppressed miR-622 expression. (A) The expression of miR-622 was measured by qRT-PCR assay. (B) IL-1β suppressed miR-622 expression in a time-dependent manner. (C) The level of miR-622 was detected via qRT-PCR assay. (D) TNF-α inhibited miR-622 expression in a time-dependent manner.
Downregulated miR-622 Level Was Associated With Degeneration Level in IDD
Then, we detected miR-622 expression in the IVD samples. First, we showed that the miR-622 level was downregulated by 5 NP cells from IDD compared with the control group (Figure 7A). Then, the miR-622 expression was lower in the IDD group compared with that the control group (Figure 7B). Moreover, the downregulated expression of miR-622 was associated with disc degeneration levels (Figure 7C). The level of miR-622 was negatively associated with circ_0005918 expression in the IDD group (Figure 7D).
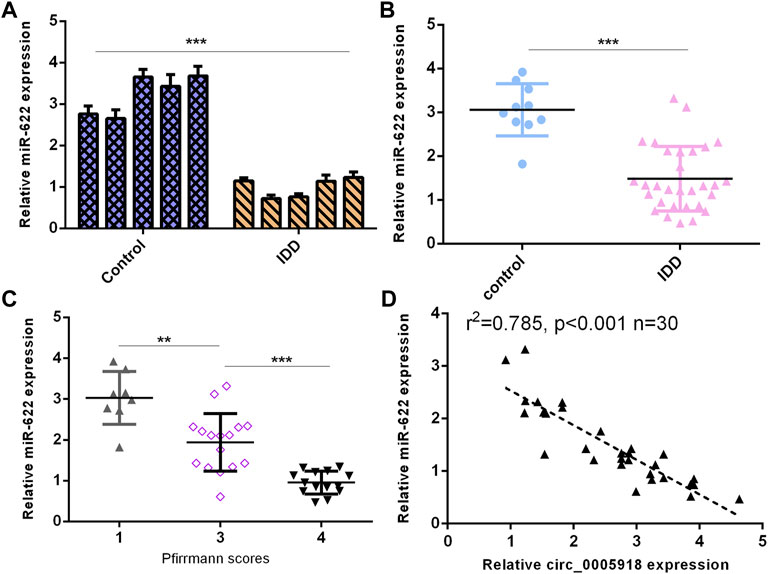
FIGURE 7. Downregulated miR-622 levels were associated with IDD. (A) miR-622 level was downregulated 5 NP cells from IDD compared to control. (B) miR-622 was lower in the IDD group than the control group. (C) The downregulated expression of miR-622 was associated with disc degeneration levels. (D) The level of miR-622 was negatively associated with circ_0005918 expression in the IDD group. **p < 0.01, ***p < 0.001.
Circ_0005918 Regulated Cell Growth, ECM Degradation Through Regulating miR-622
As circ_0005918 sponged miR-622, we next studied whether circ_0005918 regulated cell function by modulating miR-622. The overexpression of circ_0005918 induced cell proliferation in NP cells. However, miR-622 mimic decreased this function (Figure 8A). The ectopic expression of miR-622 suppressed the expression of MMP-9 (Figure 8B) and MMP-13 (Figure 8C), and enhanced the expression of collagen II (Figure 8D) in the circ_0005918-overexpressing NP cell.
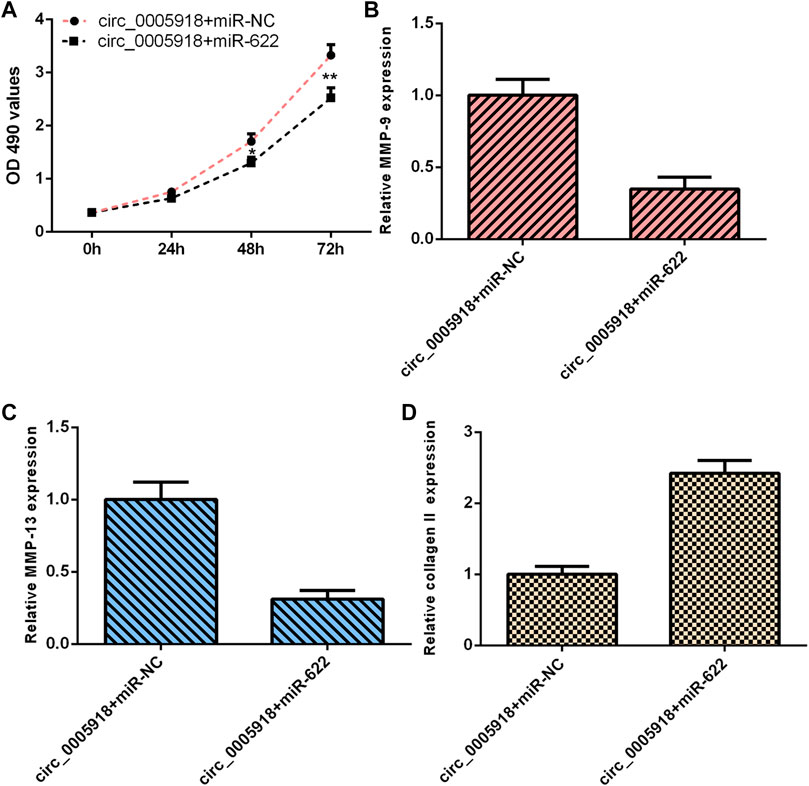
FIGURE 8. Circ_0005918 regulated the cell growth and ECM degradation by regulating miR-622. (A) Cell proliferation was detected by CCK-8 assay. (B) The ectopic expression of miR-622 suppressed MMP-9 expression in the circ_0005918-overexpressing NP cell. (C) The level of MMP-13 was measured by qRT-PCR analysis. (D) The level of collagen II was measured by qRT-PCR analysis. *p < 0.05, **p < 0.01.
Circ_0005918 Regulated Inflammatory Cytokines Secretion Through Regulating miR-622
The elevated expression of circ_0005918 induced the secretion of 3 inflammatory cytokines including IL-1β (Figure 9A), IL-6 (Figure 9B), and TNF-α (Figure 9C), while miR-622 mimic suppressed this function in the NP cell.
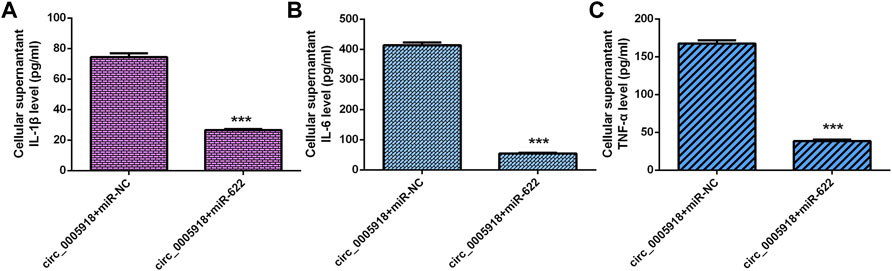
FIGURE 9. Circ_0005918 regulated the inflammatory cytokines secretion by regulating miR-622. (A) Elevated expression of circ_0005918 induced IL-1β expression, while miR-622 mimic suppressed this function in the NP cell. (B) The level of IL-6 in the cell supernatants was measured by ELISA. (C) The level of TNF-α in the cell supernatants was measured by ELISA. ***p < 0.001.
Discussion
In our study, we demonstrated that IL-1β and TNF-α induced circ_0005918 expression in the NP cell and circ_0005918 was overexpressed in the IDD group compared with the control group. Moreover, the upregulated expression of circ_0005918 was associated with disc degeneration levels. The elevated expression of circ_0005918 promoted cell growth, ECM degradation, and induced secretion of inflammatory cytokines, including IL-1β, IL-6, and TNF-α. Moreover, we found that circ_0005918 sponged miR-622 in the NP cell. In addition, the exposure to IL-1β and TNF-α suppressed miR-622 expression, and miR-622 was downregulated in the IDD group compared with the control group. Furthermore, the downregulated expression of miR-622 was associated with disc degeneration levels. The level of miR-622 was negatively associated with circ_0005918 expression in the IDD group. Circ_0005918 regulated cell growth, ECM degradation, and inflammatory cytokines secretion by regulating the miR-622 expression. These data suggested that circ_0005918 plays important roles in the development of IDD via sponging miR-622.
Growing evidence has shown that circRNAs play important roles in the development of IDD (Guo et al., 2020a; Guo et al., 2020b; Cui and Zhang, 2020). For instance, Huang et al. (2021a) demonstrated that circSPG21 decreased IDD development by regulating the miR-1197/ATP1B3 axis. Zhang et al. 2021b) found that circSNHG5 protected cartilage endplate degradation by modulating the miR-495-3p/CITED2 axis. Wang et al. (2021) showed that circARL15 was decreased in the IDD group and miR-431-5p, directly bound to DISC1 and circARL15. The elevated expression of circARL15 suppressed NP cell apoptosis but induced NP cell growth by regulating the miR-431-5p/DISC1 axis. Zhang et al. (2021c) demonstrated that circular RNA ITCH induced ECM degradation by promoting the Wnt/beta-catenin pathway in the development of IDD. Huang et al. (2021b) found that circPKNOX1 regulated IDD progression by modulating the miR-370-3p/KIAA0355 axis. Recently, Guo et al. (2021) performed microarray hybridization and qRT-PCR assay to study differentially expressed circRNA. They identified that the expression of circ_0005918 was upregulated in the IDD group. However, the function of circ_0005918 remains unknown. Therefore, we chose circ_0005918 to study and showed that circ_0005918 was overexpressed in the IDD group compared with the control group. Moreover, the upregulated expression of circ_0005918 was associated with disc degeneration degree. The elevated expression of circ_0005918 promoted cell growth, ECM degradation, and secretion of inflammatory cytokines including IL-1β, IL-6, and TNF-α.
Previous studies demonstrated that miRNAs are involved in the development of IDD. For example, Cao J. et al. (2021) showed that miR-200c-3p inhibited IDD development by regulating the RAP2C/ERK pathway. Lei et al. (2021) showed that miR-25 inhibited NP cell apoptosis by regulating SUMO2 in the IDD. Du et al. (2021) demonstrated that IL-1β induced miR-133a-5p expression to promote IDD development through regulating FBXO6. Rescuing FBXO6 and inhibiting the expression of miR-133a-5p may be potential strategies for IDD treatment. Recently, studies have shown that circRNAs might play their roles via sponging miRNAs. For instance, Li Y. et al. (2021) demonstrated that circ_0040039 induced IDD progression by modulating the MiR-874-3p/ESR1 pathway axis. Xiang et al. (2020) showed that downregulation of circCOG8 regulated the compression-induced IDD development by sponging the miR-182-5p/FOXO3 axis. In our study, by using Circular RNA Interactome software, we showed that circ_0005918 may sponge miR-622. The luciferase reporter indicated that ectopic expression of miR-622 suppressed the luciferase reporter value of circ_0005918 wild reporter vector but did not change the value of the mutant reporter. In addition, exposure to IL-1β and TNF-α suppressed miR-622 expression, and miR-622 was downregulated in the IDD group compared with the control group. Furthermore, the downregulated expression of miR-622 was associated with disc degeneration degree. The level of miR-622 was negatively associated with circ_0005918 expression in the IDD group. To conclude, circ_0005918 regulated cell growth, ECM degradation, and inflammatory cytokines secretion by regulating miR-622 expression.
In conclusion, our data demonstrated that IL-1β and TNF-α induced circ_0005918 expression in the NP cell. Circ_0005918 was overexpressed in IDD, and was associated with disc degeneration degree. Circ_0005918 regulated cell growth, ECM degradation, and inflammatory cytokines secretion by regulating miR-622 expression. We suggested that circ_0005918 plays important roles in the development of IDD via sponging miR-622.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding author.
Ethics Statement
The studies involving human participants were reviewed and approved by the Hospital of Shunyi District. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
YC, XZ, and YW collected the related studies, drafted, and wrote the manuscript. YC, XZ, and YW participated in the design of experiments. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Cao, J., Jiang, M., Ren, H., and Xu, K. (2021). MicroRNA-200c-3p -s-uppresses -i-ntervertebral -d-isc -d-egeneration by -t-argeting RAP2C/ERK -s-ignaling. Mol. Med. Rep. 24 (6). doi:10.3892/mmr.2021.12505
Cao, S., Li, J., Yang, K., and Li, H. (2021). Major ceRNA Regulation and Key Metabolic Signature Analysis of Intervertebral Disc Degeneration. BMC Musculoskelet. Disord. 22 (1), 249. doi:10.1186/s12891-021-04109-8
Chen, J., Jia, Y.-S., Liu, G.-Z., Sun, Q., Zhang, F., Ma, S., et al. (2017). Role of LncRNA TUG1 in Intervertebral Disc Degeneration and Nucleus Pulposus Cells via Regulating Wnt/β-Catenin Signaling Pathway. Biochem. Biophysical Res. Commun. 491 (3), 668–674. doi:10.1016/j.bbrc.2017.07.146
Chen, W., Li, S., and Zhang, F. (2021). Role of lncRNA XIST/microRNA-19/PTEN Network in Autophagy of Nucleus Pulposus Cells in Intervertebral Disc Degeneration via the PI3K/Akt Signaling Pathway. Cell Cycle 20, 1629–1641. doi:10.1080/15384101.2021.1924450
Chen, Y., Wu, Y., Chen, R., Xu, C., and Chen, Q. (2021). LncRNA LINC00324 Is Upregulated in Intervertebral Disk Degeneration and Upregulates FasL in Nucleus Pulposus Cells. Mol. Cell Biochem. 476 (5), 1995–2000. doi:10.1007/s11010-021-04058-9
Cui, S., and Zhang, L. (2020). circ_001653 Silencing Promotes the Proliferation and ECM Synthesis of NPCs in IDD by Downregulating miR-486-3p-Mediated CEMIP. Mol. Ther. - Nucleic Acids 20, 385–399. doi:10.1016/j.omtn.2020.01.026
Ding, W., Shi, Y., and Zhang, H. (2021). Circular RNA circNEURL4 Inhibits Cell Proliferation and Invasion of Papillary Thyroid Carcinoma by Sponging miR-1278 and Regulating LATS1 Expression. Am. J. Transl. Res. 13 (6), 5911–5927.
Du, X.-F., Cui, H.-T., Pan, H.-H., Long, J., Cui, H.-W., Chen, S.-L., et al. (2021). Role of the miR-133a-5p/FBXO6 axis in the Regulation of Intervertebral Disc Degeneration. J. Orthop. Transl. 29, 123–133. doi:10.1016/j.jot.2021.05.004
Gao, D., Hao, L., and Zhao, Z. (2020). Long Non-coding RNA PART1 Promotes Intervertebral Disc Degeneration through Regulating the miR93/MMP2 Pathway in Nucleus Pulposus Cells. Int. J. Mol. Med. 46 (1), 289–299. doi:10.3892/ijmm.2020.4580
Guo, W., Mu, K., Zhang, B., Sun, C., Zhao, L., Dong, Z.-Y., et al. (2020). The Circular RNA FAM169A Functions as a Competitive Endogenous RNA and Regulates Intervertebral Disc Degeneration by Targeting miR-583 and BTRC. Cell Death Dis. 11 (5), 315. doi:10.1038/s41419-020-2543-8
Guo, W., Mu, K., Zhang, B., Sun, C., Zhao, L., Li, H.-R., et al. (2020). The Circular RNA Circ-GRB10 Participates in the Molecular Circuitry Inhibiting Human Intervertebral Disc Degeneration. Cell Death Dis. 11 (8), 612. doi:10.1038/s41419-020-02882-3
Guo, Z. L., Liu, Y. Y., Gao, Y., Guan, X. M., Li, H., and Cheng, M. (2021). Circ-RNA Expression Pattern and Circ-RNA-miRNA-mRNA Network in the Pathogenesis of Human Intervertebral Disc Degeneration. Cell J. 23 (2), 218–224. doi:10.22074/cellj.2021.6832
Hao, Y., Lu, C., Zhang, B., Xu, Z., Guo, H., and Zhang, G. (2021). CircPVT1 Up‐regulation Attenuates Steroid‐induced Osteonecrosis of the Femoral Head through Regulating miR‐21‐5p‐mediated Smad7/TGFβ Signalling Pathway. J. Cell Mol. Med. 25 (10), 4608–4622. doi:10.1111/jcmm.16294
Hu, B., Xiao, L., Wang, C., Liu, C., Zhang, Y., Ding, B., et al. (2022). Circ_0022382 Ameliorated Intervertebral Disc Degeneration by Regulating TGF-β3 Expression through Sponge Adsorption of miR-4726-5p. Bone 154, 116185. doi:10.1016/j.bone.2021.116185
Huang, Y., Gao, J., Wang, J., Ye, H., Yao, T., Xu, Y., et al. (2021). Inhibition of Intervertebral Disc Disease Progression via the circPKNOX1-miR-370-3p-KIAA0355 axis. Cell Death Discov. 7 (1), 39. doi:10.1038/s41420-021-00420-4
Huang, Y., Zhang, Z., Wang, J., Shen, S., Yao, T., Xu, Y., et al. (2021). circSPG21 Protects against Intervertebral Disc Disease by Targeting miR-1197/ATP1B3. Exp. Mol. Med. 53 (10), 1547–1558. doi:10.1038/s12276-021-00674-z
Jing, W., and Liu, W. (2021). HOXC13-AS Induced Extracellular Matrix Loss via Targeting miR-497-5p/ADAMTS5 in Intervertebral Disc. Front. Mol. Biosci. 8, 643997. doi:10.3389/fmolb.2021.643997
Kraus, P., Sivakamasundari, V., Olsen, V., Villeneuve, V., Hinds, A., and Lufkin, T. (2019). Klhl14 Antisense RNA Is a Target of Key Skeletogenic Transcription Factors in the Developing Intervertebral Disc. Spine (Phila Pa 1976) 44 (5), E260–E268. doi:10.1097/BRS.0000000000002827
Lei, C., Li, J., Tang, G., and Wang, J. (2021). MicroRNA-25 -p-rotects -n-ucleus -p-ulposus -c-ells against -a-poptosis via -t-argeting SUMO2 in -i-ntervertebral -d-isc -d-egeneration. Mol. Med. Rep. 24 (4). doi:10.3892/mmr.2021.12363
Li, J., Qin, X., Wu, R., Wan, L., Zhang, L., and Liu, R. (2020). Circular RNA circFBXO11 Modulates Hepatocellular Carcinoma Progress and Oxaliplatin Resistance through miR‐605/FOXO3/ABCB1 axis. J. Cell. Mol. Medi 24 (9), 5152–5161. doi:10.1111/jcmm.15162
Li, S., Pei, Y., Wang, W., Liu, F., Zheng, K., and Zhang, X. (2020). Extracellular Nanovesicles‐transmitted Circular RNA Has_circ_0000190 Suppresses Osteosarcoma Progression. J. Cell Mol. Med. 24 (3), 2202–2214. doi:10.1111/jcmm.14877
Li, Y., Wang, X., Xu, H., Li, G., Huo, Z., Du, L., et al. (2021). Circ_0040039 May Aggravate Intervertebral Disk Degeneration by Regulating the MiR-874-3p-ESR1 Pathway. Front. Genet. 12, 656759. doi:10.3389/fgene.2021.656759
Li, Z., Li, X., Shen, J., Zhang, L., Chan, M. T. V., and Wu, W. K. K. (2020c). Emerging Roles of Non‐coding RNAs in Scoliosis. Cell Prolif. 53 (2), e12736. doi:10.1111/cpr.12736
Li, Z., Ma, J., Shen, J., Chan, M. T. V., Wu, W. K. K., and Wu, Z. (2019). Differentially Expressed Circular RNAs in Air Pollution-Exposed Rat Embryos. Environ. Sci. Pollut. Res. 26 (33), 34421–34429. doi:10.1007/s11356-019-06489-w
Li, Z., Sun, Y., He, M., and Liu, J. (2021). Differentially-expressed mRNAs, microRNAs and Long Noncoding RNAs in Intervertebral Disc Degeneration Identified by RNA-Sequencing. Bioengineered 12 (1), 1026–1039. doi:10.1080/21655979.2021.1899533
Liang, M., Huang, G., Liu, Z., Wang, Q., Yu, Z., Liu, Z., et al. (2019). Elevated Levels of Hsa_circ_006100 in Gastric Cancer Promote Cell Growth and Metastasis via miR‐195/GPRC5A Signalling. Cell Prolif. 52 (5), e12661. doi:10.1111/cpr.12661
Liu, T., Ye, P., Ye, Y., Lu, S., and Han, B. (2020). Circular RNA hsa_circRNA_002178 Silencing Retards Breast Cancer Progression via microRNA‐328‐3p‐mediated Inhibition of COL1A1. J. Cell Mol. Med. 24 (3), 2189–2201. doi:10.1111/jcmm.14875
Ma, D., Liu, H., Qin, Y., Li, D., Cui, Y., Li, L., et al. (2020). Circ_0007142/miR-186/FOXK1 axis Promoted Lung Adenocarcinoma Progression. Am. J. Transl. Res. 12 (8), 4728–4738.
Nie, H., Wang, Y., Liao, Z., Zhou, J., and Ou, C. (2020). The Function and Mechanism of Circular RNAs in Gastrointestinal Tumours. Cell Prolif. 53 (7), e12815. doi:10.1111/cpr.12815
Pu, J., Wang, J., Li, W., Lu, Y., Wu, X., Long, X., et al. (2020). hsa_circ_0000092 Promotes Hepatocellular Carcinoma Progression through Up‐regulating HN1 Expression by Binding to microRNA‐338‐3p. J. Cell Mol. Med. Online ahead of print. doi:10.1111/jcmm.15010
Sun, Z., Tang, X., Wang, H., Sun, H., Chu, P., Sun, L., et al. (2021). LncRNA H19 Aggravates Intervertebral Disc Degeneration by Promoting the Autophagy and Apoptosis of Nucleus Pulposus Cells through the miR-139/CXCR4/NF-κB Axis. Stem Cells Dev. 30 (14), 736–748. doi:10.1089/scd.2021.0009
Tan, H., Zhao, L., Song, R., Liu, Y., and Wang, L. (2018). The Long Noncoding RNA SNHG1 Promotes Nucleus Pulposus Cell Proliferation through Regulating miR-326 and CCND1. Am. J. Physiology-Cell Physiology 315 (1), C21–C27. doi:10.1152/ajpcell.00220.2017
Tan, L., Xie, Y., Yuan, Y., and Hu, K. (2021). LncRNA GAS5 as miR-26a-5p Sponge Regulates the PTEN/PI3K/Akt Axis and Affects Extracellular Matrix Synthesis in Degenerative Nucleus Pulposus Cells In Vitro. Front. Neurol. 12, 653341. doi:10.3389/fneur.2021.653341
Tian, F., Yu, C., Wu, M., Wu, X., Wan, L., and Zhu, X. (2019). MicroRNA‐191 Promotes Hepatocellular Carcinoma Cell Proliferation by has_circ_0000204/miR‐191/KLF6 axis. Cell Prolif. 52 (5), e12635. doi:10.1111/cpr.12635
Wang, H., Zhu, Y., Cao, L., Guo, Z., Sun, K., Qiu, W., et al. (2021). circARL15 Plays a Critical Role in Intervertebral Disc Degeneration by Modulating miR-431-5p/DISC1. Front. Genet. 12, 669598. doi:10.3389/fgene.2021.669598
Wang, X., Li, D., Wu, H., Liu, F., Liu, F., Zhang, Q., et al. (2020). LncRNA TRPC7-AS1 Regulates Nucleus Pulposus Cellular Senescence and ECM Synthesis via Competing with HPN for miR-4769-5p Binding. Mech. Ageing Dev. 190, 111293. doi:10.1016/j.mad.2020.111293
Xiang, Q., Kang, L., Zhao, K., Wang, J., Hua, W., Song, Y., et al. (2020). CircCOG8 Downregulation Contributes to the Compression-Induced Intervertebral Disk Degeneration by Targeting miR-182-5p and FOXO3. Front. Cell Dev. Biol. 8, 581941. doi:10.3389/fcell.2020.581941
Yan, M., Gao, H., Lv, Z., Liu, Y., Zhao, S., Gong, W., et al. (2020). Circular RNA PVT1 Promotes Metastasis via Regulating of miR‐526b/FOXC2 Signals in OS Cells. J. Cell Mol. Med. 24 (10), 5593–5604. doi:10.1111/jcmm.15215
Yang, L., Liu, Z., Ma, J., Wang, H., Gao, D., Zhang, C., et al. (2021). CircRPPH1 Serves as a Sponge for miR-296-5p to Enhance Progression of Breast Cancer by Regulating FOXP4 Expression. Am. J. Transl. Res. 13 (7), 7556–7573.
Zhan, J., Wang, S., Wei, X., Feng, M., Yin, X., Yu, J., et al. (2021). Systematic Analysis of Long Non-coding RNAs Reveals Diagnostic Biomarkers and Potential Therapeutic Drugs for Intervertebral Disc Degeneration. Bioengineered 12 (1), 5069–5084. doi:10.1080/21655979.2021.1950258
Zhan, S., Wang, K., Song, Y., Li, S., Yin, H., Luo, R., et al. (2019). Long Non-coding RNA HOTAIR Modulates Intervertebral Disc Degenerative Changes via Wnt/β-Catenin Pathway. Arthritis Res. Ther. 21 (1), 201. doi:10.1186/s13075-019-1986-8
Zhang, F., Lin, F., Xu, Z., and Huang, Z. (2021). Circular RNA ITCH Promotes Extracellular Matrix Degradation via Activating Wnt/β-Catenin Signaling in Intervertebral Disc Degeneration. Aging 13 (10), 14185–14197. doi:10.18632/aging.203036
Zhang, J., Cheng, F., Rong, G., Tang, Z., and Gui, B. (2021). Circular RNA Hsa_circ_0005567 Overexpression Promotes M2 Type Macrophage Polarization through miR-492/SOCS2 axis to Inhibit Osteoarthritis Progression. Bioengineered 12 (1), 8920–8930. doi:10.1080/21655979.2021.1989999
Zhang, J., Hu, S., Ding, R., Yuan, J., Jia, J., Wu, T., et al. (2021). CircSNHG5 Sponges Mir-495-3p and Modulates CITED2 to Protect Cartilage Endplate from Degradation. Front. Cell Dev. Biol. 9, 668715. doi:10.3389/fcell.2021.668715
Zhao, C.-x., Yan, Z.-x., Wen, J.-j., Fu, D., Xu, P.-p., Wang, L., et al. (2021). CircEAF2 Counteracts Epstein-Barr Virus-Positive Diffuse Large B-Cell Lymphoma Progression via miR-BART19-3p/APC/β-catenin axis. Mol. Cancer 20 (1), 153. doi:10.1186/s12943-021-01458-9
Zhao, K., Zhang, Y., Yuan, H., Zhao, M., and Zhao, D. (2019). Long Noncoding RNA LINC00958 Accelerates the Proliferation and Matrix Degradation of the Nucleus Pulposus by Regulating miR-203/SMAD3. Aging 11 (23), 10814–10825. doi:10.18632/aging.102436
Keywords: intervertebral disc degeneration, circular RNAs, circ_0005918, miR-622, cancers
Citation: Cui Y, Zhao X and Wu Y (2022) Circ_0005918 Sponges miR-622 to Aggravate Intervertebral Disc Degeneration. Front. Cell Dev. Biol. 10:905213. doi: 10.3389/fcell.2022.905213
Received: 26 March 2022; Accepted: 16 May 2022;
Published: 08 July 2022.
Edited by:
Xingye Li, Beijing Jishuitan Hospital, ChinaReviewed by:
Xuesong Wang, Qingdao Central Hospital, ChinaKunxiang Wu, Nanyang Second General Hospital, China
Copyright © 2022 Cui, Zhao and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yangang Wu, YmpzeXd0Z0AxNjMuY29t
 Yan Cui
Yan Cui Yangang Wu
Yangang Wu