Abstract
Vesicle-associated membrane protein (VAMP)-associated proteins (VAPs) are ubiquitous ER-resident tail-anchored membrane proteins in eukaryotic cells. Their N-terminal major sperm protein (MSP) domain faces the cytosol and allows them to interact with a wide variety of cellular proteins. Therefore, VAP proteins are vital to many cellular processes, including organelle membrane tethering, lipid transfer, autophagy, ion homeostasis and viral defence. Here, we provide a timely overview of the increasing number of VAPA/B binding partners and discuss the role of VAPA/B in maintaining organelle-ER interactions and cooperation. Furthermore, we address how viruses and intracellular bacteria hijack VAPs and their binding partners to induce interactions between the host ER and pathogen-containing compartments and support pathogen replication. Finally, we focus on the role of VAP in human disease and discuss how mutated VAPB leads to the disruption of cellular homeostasis and causes amyotrophic lateral sclerosis.
Introduction
VAP was initially cloned from the marine mollusk Aplysia californica and named vesicle-associated membrane protein (VAMP)-associated protein of 33 kilodaltons (VAP-33) because of its ability to interact with vesicle fusion protein VAMP (also termed synaptobrevin) (Skehel et al., 1995). Since then, VAP proteins have been identified in all eukaryotic cells and reported to interact with a large number of intracellular proteins (Lev et al., 2008; Kamemura and Chihara, 2019). VAPs are C-tail-anchored (or type II) ER membrane proteins with a central coiled-coil domain and N-terminal major sperm (MSP) domain (∼125 residues), which faces the cytoplasmic side (Nishimura et al., 1999). This N-terminal domain consists of an immunoglobulin-like β-sheet and is named after the Ascaris suum protein MSP due to its 22% sequence identity (Bullock et al., 1996; Kaiser et al., 2005). The highly conserved VAP protein family consists in mammals of VAPA and VAPB, including the VAPB splice variant VAPC which lacks both the C-terminal transmembrane domain (TMD) and the coiled-coil domain (Weir et al., 1998; Nishimura et al., 1999). Five additional VAPB splice variants were detected at the mRNA level, although protein levels were undetectable in human tissue lysates by immunoblotting (Nachreiner et al., 2010). VAPA and VAPB share 63% sequence identity, mainly due to similarities in the MSP domain, and a clear functional difference between the paralogues has not been established. Recently, the VAP family was extended with the motile sperm domain-containing proteins 1, 2 and 3 (MOSPD1, MOSPD2 and MOSPD3), which also possess an MSP domain and share binding partners with VAPA/B, though with different affinities (Thaler et al., 2011; Mattia et al., 2018; Cabukusta et al., 2020).
VAP proteins are ubiquitously expressed in mammals (Weir et al., 1998; Nishimura et al., 1999; Skehel et al., 2000), with tissue-specific RNA expression patterns during development (Gabetta et al., 2003). They interact with a wide variety of proteins, imparting them with various functions, including organelle membrane tethering (Costello et al., 2017a), lipid transfer between organelles (Kawano et al., 2006; Ngo and Ridgway, 2009), regulation of calcium homeostasis (De Vos et al., 2012; Lindhout et al., 2019), autophagy (Zhao et al., 2018) and the unfolded protein response (UPR) (Kanekura et al., 2006; Gkogkas et al., 2008). VAP might also have extracellular functions through its cleaved and secreted MSP domain (Deidda et al., 2014), although this has been mainly studied in C. elegans and D. melanogaster (Tsuda et al., 2008; Han et al., 2012).
In this review, we provide a timely update of VAPA/B binding partners and discuss the role of VAPA/B in maintaining organelle-ER interactions and cooperation. This includes VAP hijacking by viruses and intracellular bacteria to induce interactions between the host ER and pathogen-containing compartments, and the recruitment of host proteins to these sites to support pathogen replication. Finally, we focus on how a mutation in VAPB leads to the disruption of cellular homeostasis, causing amyotrophic lateral sclerosis type 8 (ALS8).
VAP Binding Partners and the Functions of the Ensuing Complex
The VAP Interaction
The interaction of VAP with a multitude of diverse proteins means that the VAP proteins are important for many cellular processes, including organelle tethering, lipid transfer, autophagy, ion homeostasis and viral defence. Table 1 provides an overview of the current experimentally confirmed VAP interactors and the proposed functions of the ensuing complex. Many binding partners interact with the MSP domain of VAP via a “two phenylalanines in an acidic tract” (FFAT) motif, which consists of the core consensus sequence 1EFFDA-E7 flanked by acidic residues (Loewen et al., 2003). In the proposed interaction model, the acidic tract upstream of the core initially binds in a non-specific manner to the basic electropositive surface of the MSP domain followed by a second step, in which the core residues bind to the FFAT-binding site to form a stable complex (Kaiser et al., 2005; Furuita et al., 2010). The FFAT motif can vary in sequence quite considerably, whilst still allowing binding interaction, potentially giving proteins a different affinity for VAP (Murphy and Levine, 2016). Many known VAP interactors contain only one or no phenylalanine in the FFAT motif, and there is some redundancy in the sequence (seeTable 1). In addition, the acidic tract can vary in length and the number of acidic residues. Interestingly, the VAP family proteins MOSPD1 and MOSPD3 favour motifs with “two phenylalanines in a neutral tract” (FFNT) (Cabukusta et al., 2020). Furthermore, the interaction with VAP can be modulated on multiple levels; for instance, the FFAT-VAP binding can be strengthened and reduced by (de)phosphorylation of the FFAT motif (Kumagai et al., 2014; Johnson et al., 2018; Kirmiz et al., 2018; Di Mattia et al., 2020; Guillén-Samander et al., 2021; Kors et al., 2022). VAP dimerization via the TMD and coiled-coil domains might enhance the recruitment of pre-existing homodimers of FFAT proteins (e.g. OSBP) or bring together two unrelated FFAT-containing proteins, stabilizing the complex (Kaiser et al., 2005; Kim et al., 2010). Additionally, some proteins contain two FFAT motifs, suggesting that the interactor could bind to two VAPs at the same time (e.g. PTPIP51 and ORP3, seeTable 1). However, not all known VAP binding partners possess a FFAT motif–some proteins bind to the MSP in a different, FFAT-independent way, while other proteins mediate the interaction via their and VAP’s TMD (Table 1).
TABLE 1
| Complex | Interaction domain | Localisationa | MCS | Physiological role of the VAP complex | Reference | |
|---|---|---|---|---|---|---|
| Binding partner | VAP | |||||
| ACBD4-VAPB | FFAT motif (score 3.5) | MSP | Peroxisomes (TMD) | Peroxisome-ER | Organelle tethering function | Costello et al. (2017b), |
| 173RDLDSE VFCDS—LE QL | Kors et al. (2022) | |||||
| ACBD5-VAPA/B | FFAT motif (score 2.5) | MSP | Peroxisomes (TMD) | Peroxisome-ER | Organelle tethering function, implicated in: peroxisome motility; peroxisome membrane expansion; plasmalogen synthesis; maintenance of cholesterol levels | Costello et al. (2017a), |
| 259SDSDSE VYCDS—ME QF | Hua et al. (2017) | |||||
| α-Synuclein-VAPB | ? | MSP | Cytosol, nucleus, membranes | Disrupts the PTPIP51-VAPB interaction, hence mitochondria-ER MCS (affecting Ca2+ exchange) | Paillusson et al. (2017) | |
| ASNA1-VAPA/B | FFAT motif (score 2.0) | MSP | Cytosol, ER, nucleus | Mediating interaction with the transmembrane-domain recognition complex (TRC; insertion of tail anchored ER proteins) | Baron et al. (2014) | |
| 8WGVEAE EFEDAPD VE | ||||||
| ATF6-VAPA/B | ? | MSP | ER (TMD), nucleus (cleaved) | Modulates the activity of ATF6-regulated transcription of genes involved in the unfolded protein response (UPR) | Gkogkas et al. (2008) | |
| CALCOCO1-VAPA/B | FFAT-motif (score 3.0) | MSP | Nucleus, cytosol | Autophagosome-ER | Acts as ER-phagy receptor for degradation of the tubular ER, via ATG8 interaction | Nthiga et al. (2020) |
| 674DHMDGH FFFS——T—QD PF | ||||||
| CaSR-VAPA | FFAT motif (score 2.5)b | MSP | Plasma membrane (TMD) | Plasma membrane-ER | Ca2+ sensing; near surface CaSR expression | Gorkhali et al. (2021) |
| 755ELEDEI IFIT——CHE GS | ||||||
| CERT-VAPA/B | FFAT motif (score 1.0) | MSP | Cytosol, Golgi (PH domain) | Golgi-ER | Ceramide transfer from the ER to the Golgi apparatus, for sphingomyelin synthesis | Kawano et al. (2006), |
| 315SLINEE EFFDAVE AA | Saito et al. (2008), | |||||
| Kumagai et al. (2014) | ||||||
| CLN8-VAPA | ? | ? | ER (TMD), ER-Golgi intermediate compartment (ERGIC; TMD) | Possibly: Ceramide metabolism; endo-lysosomal dynamics | Passantino et al. (2013), | |
| Adhikari et al. (2019), | ||||||
| Pesaola et al. (2021) | ||||||
| CDIP1-VAPA/B | FFAT motif (score 6.0) | MSP | Endocytic compartments (MMD) | CDIP1-induced cell death | Inukai et al. (2021) | |
| 180IPCLIN DFKDVTH TC | ||||||
| FAF1-VAPA/B | FFAT motif (score 1.0) | MSP | Nucleus, cytosol | Binding of ubiquitinated proteins; recruiting p97 to the ER membrane (involved in ER-associated protein degradation (ERAD)) | Baron et al. (2014) | |
| 289SDSDGD DFEDATE FG | ||||||
| FAPP1-VAPA/B | C-terminus | ? | Golgi (PH domain) | Golgi-ER | Formation of the SAC1-FAPP1-VAP complex – binding of FAPP1 to the PI4P-phosphatase SAC1 promotes the phosphatase activity | Venditti et al. (2019) |
| FIP200-VAPA/B | FFAT motif 1 (score 3.0) | MSP | Cytosol, (pre-) autophagosomal structures, lysosomes, nucleus | Isolation membrane-ER | Formation/stabilization of the ULK1/FIP200-WIPI2 complex during isolation membrane expansion for autophagosome formation | Zhao et al. (2018) |
| 725AESPES DFMS——AVN EF | ||||||
| FFAT motif 2 (score 4.0) | ||||||
| 206ECLTRH SYRECLG RL | ||||||
| GLTP-VAPA | FFAT motif (score 3.5) | MSP | Cytosol | Possibly: Glycolipid transfer, glucosylceramide sensor | Tuuf et al. (2009), | |
| 26AVSHLP PFFDCLG SP | Backman et al. (2018) | |||||
| HCN2-VAPA/B | TMD | TMD | ER | Regulation of HCN channel Na+/K+ pacemaker currents; dendritic localization of HCN2 | Silbernagel et al. (2018) | |
| IFITM3-VAPA | TMD | TMD and CC | Endosomes, lysosomes, plasma membrane | Preventing the VAPA-OSBP association, which induces cholesterol accumulation, inhibiting viral entry | Amini-Bavil-Olyaee et al. (2013) | |
| JMY-VAPA | FFAT motif (score 1.5)b | MSP | Nucleus, cytoskeleton | Possibly: Vesicle based transport | Schlüter et al. (2014) | |
| 312ETDDPE EYYES—LS EL | ||||||
| Kv2.1-VAPA/B | FFAT motif (score 3.5) | MSP | Plasma membrane (TMD) | Plasma membrane-ER | Kv2 channel clustering; regulating proapoptotic K+ currents; phosphatidylinositol homeostasis (via NIR2 recruitment) | Johnson et al. (2018), |
| 584SMSSID SFIS——CAT DF | Kirmiz et al. (2018, 2019), | |||||
| Schulien et al. (2020) | ||||||
| Kv2.2-VAPA/B | FFAT motif (score 3.0) | MSP | Plasma membrane (TMD) | Plasma membrane-ER | Kv2 channel clustering | Johnson et al. (2018), |
| 599STSSID SFTS——CAT DF | Kirmiz et al. (2018) | |||||
| MIGA2-VAPA/B | FFAT motif (score 1.5) | MSP | Mitochondria (TMD), lipid droplets | Mitochondria-ER | Linking reactions of de novo lipogenesis in mitochondria to triglyceride production in the ER | Freyre et al. (2019) |
| 286SLTSED SFFS——ATE LF | ||||||
| NIR1-VAPB | FFAT motif (score 0.0) | MSP | Plasma membrane (LNS2 domain), cytosol | Plasma membrane-ER | Promoting NIR2 recruitment for phosphatidylinositol homeostasis | Amarilio et al. (2005), |
| 28VESSDD EFFDARE EM | Quintanilla et al. (2022) | |||||
| NIR2-VAPB | FFAT motif (score 0.0) | MSP | Golgi (LNS2 domain), plasma membrane (LNS2 domain), cytosol | Golgi-ER | Phosphatidylinositol transfer to the Golgi apparatus and phosphatidylcholine transfer to the ER (important for CERT and OSBP Golgi targeting/function) | Amarilio et al. (2005), |
| 343ENSSEE EFFDAHE GF | Plasma membrane-ER | Phosphatidylinositol transfer from the ER to the plasma membrane | Peretti et al. (2008),Chang et al. (2013),Chang and Liou (2015),Kirmiz et al. (2019) | |||
| NIR3-VAPB | FFAT motif (score 0.0) | MSP | Plasma membrane (LNS2 domain), cytosol | Plasma membrane-ER | Phosphatidylinositol transfer from the ER to the plasma membrane | Amarilio et al. (2005), |
| 338DESSDD EFFDAHE DL | Microtubule interaction | Chang and Liou, (2015) | ||||
| OSBP-VAPA | FFAT motif (score 0.0)352DEDDEN EFFDAPE II | MSP | Golgi (PH domain), endosomes (PH domain), lysosomes (PH domain), cytosol | Golgi-EREndosome-ERLysosome-ER | Cholesterol transfer from the ER to the Golgi apparatus in exchange for PI4PRegulation of PI4P levels on endosomesCholesterol transfer from the ER to lysosomes, regulating mTORC1 activation | Wyles et al. (2002),Loewen et al. (2003),Mesmin et al. (2013),Dong et al. (2016),Lim et al. (2019) |
| ORP1L-VAPA/B | FFAT motif (score 1.5) | MSP | Late endosomes/lysosomes (PH domain and ankyrin motif), autophagosome, phagolysosome | Late endosome/lysosome (LEL)-ER | Cholesterol transport from the LEL to the ER (high cholesterol) and vice versa (low cholesterol); endosome positioning | Rocha et al. (2009), |
| 469SILSED EFYDALS DS | Autophagosome-ER | Regulating autopagosome transport and maturation | Eden et al. (2016), | |||
| Phagolysosome-ER | PI4P transfer to the ER, for phagolysosome resolution | Wijdeven et al. (2016), | ||||
| Zhao and Ridgway (2017), | ||||||
| Levin-Konigsberg et al. (2019) | ||||||
| ORP2-VAPA | FFAT motif (score 1.5) | MSP | Lipid droplets, plasma membrane, cytosol | Lipid droplet-ER | Triglyceride metabolism | Weber-Boyvat et al. (2015b) |
| 1MNGEE EFFDAVT GF | ||||||
| ORP3-VAPA | FFAT motif 1 (score 1.0) | MSP | Plasma membrane (PH domain), cytosol | Plasma membrane-ER | Stimulating R-Ras signalling | Lehto et al. (2005), |
| 444ITDSLS EFFDAQE VL | Late endosome-nuclear envelope | The nuclear transfer of extracellular vesicle-derived materials | Weber-Boyvat et al. (2015a), | |||
| FFAT motif 2 (score 4.5) | Santos et al. (2018) | |||||
| 155FPHEVN HFFS——GST IT | ||||||
| ORP4L-VAPA | FFAT motif (score 0.0) | MSP | Golgi (PH domain), plasma membrane | Plasma membrane-ERc | Controlling the localization and activation of the phosphoinositide phospholipases C β3 (PLCβ3) at the plasma membrane | Pan et al. (2018), |
| 445EEDEDT EYFDAME DS | Golgi-ER | The maintenance of Golgi structure | Pietrangelo and Ridgway, (2018) | |||
| ORP6-VAP | FFAT motif (score 1.0)b | MSP | Plasma membrane (PH domain), cytosol | Plasma membrane-ER | PI4P turnover | Wyles and Ridgway (2004), |
| 488MSESVS EFFDAQE VL | Mochizuki et al. (2018) | |||||
| ORP7-VAP | FFAT motif (score 1.0)b | MSP | Plasma membrane (PH domain), cytosol | ? | Wyles and Ridgway (2004), | |
| 396LADSHT EFFDACE VL | Weber-Boyvat et al. (2015b) | |||||
| ORP9L-VAPA | FFAT motif (score 1.0) | MSP | Golgi (PH domain), cytosol | Golgi-ER | Golgi organization and protein transport; cholesterol transfer | Wyles and Ridgway (2004), |
| 294YSSSED EFYDADE FH | Ngo and Ridgway, (2009) | |||||
| PP2Cϵ-VAP | TMD | TMD | ER (TMD) | Dephosphorylating CERT, which promotes Golgi localisation of CERT and enhances the CERT-VAPA interaction | Saito et al. (2008) | |
| PRA1-VAP | FFAT motif (score 5.5)b | MSP | Golgi (TMD), ER (TMD) | Mitochondria-ER | ER retention of PRA1 | Abu Irqeba and Ogilvie, (2020) |
| 66RLVRNV EYYQS—NY VF | ||||||
| Prestin-VAPA | ? | ? | Plasma membrane (TMD) | Prestin translocation to the plasma membrane | Sengupta et al. (2010) | |
| Protrudin-VAPA/B | FFAT motif (score 1.0) | MSP, TMD | ER (TMD), endosomes (FYVE domain), plasma membrane (FYVE domain) | Endosome-ER | Endosome trafficking; stimulating process/neurite formation | Saita et al. (2009), |
| 280EAEPDE EFKDAIE ET | Matsuzaki et al. (2011), | |||||
| Raiborg et al. (2015), | ||||||
| Petrova et al. (2020) | ||||||
| PTPIP51-VAP | FFAT motif 1 (score 3.0) | MSP | Mitochondria (TMD) | Mitochondria-ER | Ca2+ delivery to mitochondria from ER stores, which regulates autophagy and synaptic function; phosphatidic acid transfer, important for mitochondrial cardiolipin synthesis | De Vos et al. (2012), |
| 151STGSSS VYFT——ASS GA | Stoica et al. (2014), | |||||
| FFAT motif 2 (score 3.5) | Gómez-Suaga et al. (2017), | |||||
| 160TASSGA TFTDAES EG | Gómez-Suaga et al. (2019), | |||||
| Yeo et al. (2021) | ||||||
| RAB3GAP1-VAPA/B | FFAT motif (score 0.5) | MSP | Cytosol | Regulating nuclear envelope formation through ERGIC | Baron et al. (2014), | |
| 578WSDSEE EFFECLS DT | Hantan et al. (2014) | |||||
| SCRN1-VAPA/B | FFAT motif (score 2.5) | MSP | Cytosol | Modulating Ca2+ homeostasis and synaptic vesicle cycling; ER dynamics | Lindhout et al. (2019) | |
| 394AEVGDL FYDCVD TE | ||||||
| SNX2- VAPB | FFAT motif 1 (score 3.5) | MSP | Endosomes (PX domain) | Endosome-ER | Retromer-/WASH-dependent actin nucleation (vesicle budding) of endosomes, with a role of PI4P (seeOSBP) | Dong et al. (2016) |
| 21LEDGED LFTS——T—VS TL | ||||||
| FFAT motif 2 (score 2.0) | ||||||
| 66DDDRED LFAEATE EV | ||||||
| STARD3-VAPA/B | FFAT motif (score 5.5) | MSP | Late endosomes (TMD) | Late endosome-ER | Cholesterol transport from the ER to endosome | Alpy et al. (2013), |
| 200GALSEG QFYS——PPE SF | Wilhelm et al. (2017), | |||||
| Di Mattia et al. (2020) | ||||||
| STARD3NL-VAPA/B | FFAT motif (score 5.5) | MSP | Late endosomes (TMD) | Late endosome-ER | Formation of endosomal tubules | Alpy et al. (2013) |
| 201GGLSDG QFYS——PPE SE | ||||||
| TRPC3-VAPB | FFAT motif (score 5.0) | MSP | Plasma membrane | Plasma membrane-ER | Controls TRPC3’s Ca2+ current and its receptor-mediated activation | Liu et al. (2022) |
| 140QELQDD DFYAYDE DG | ||||||
| TTC39B-VAPB | FFAT motif (score 2.0) | MSP | Cytosol | Stabilizing ER-membrane protein SCAP, involved in hepatic lipogenic gene expression | Hsieh et al. (2021) | |
| 76LEADED VFEDALE TI | ||||||
| ULK1-VAPA/B | FFAT motif 1 (score 5.5) | MSP | Cytosol, pre-autophagosomal structures | Isolation membrane-ER | Formation/stabilization of the ULK1/FIP200-WIPI2 complex during isolation membrane expansion for autophagy | Zhao et al. (2018) |
| 87SVYLVM EYCNGGD LA | ||||||
| FFAT motif 2 (score 5.5) | ||||||
| 74NIVALY DFQEMAN SV | ||||||
| Viperin-VAPA | C-terminus | C-terminus | ER, lipid droplets | Restricting Hepatitis C virus replication complex formation by promoting degradation of viral NS5A through VAPA | Wang et al. (2012), | |
| Ghosh et al. (2020) | ||||||
| VPS13A-VAPA/B | FFAT motif (score 1.0) | MSP | Mitochondria (ATG homology region, PH domain), lipid droplets (PH domain) | Mitochondria-ER | Mitochondria elongation; glycerolipid transfer between membranes | Kumar et al. (2018), |
| 836EDDSEE EFFDAPC SP | Lipid droplet-ER | Lipid droplet size and motility; glycerolipid transfer between membranes | Yeshaw et al. (2019) | |||
| VPS13C-VAPB | FFAT motif (score 0.0) | MSP | Late endosomes/lysosomes (WD40 module), lipid droplets (PH domain) | Endolysosome-ER | Glycerolipid transfer between membranes | Kumar et al. (2018) |
| 871ESESDD EYFDAED GE | Lipid droplet-ER | Glycerolipid transfer between membranes | ||||
| VPS13D-VAPB | FFAT motif (score 5.0) | MSP | Golgi, mitochondria | Mitochondria-ER | Bridging the organelle membranes via MIRO at the mitochondrial membrane (likely similar with peroxisomes); membrane lipid transfer | Guillén-Samander et al. (2021) |
| 761TQFSDD EYKT——PLA TP | ||||||
| WDR44-VAPA/B | FFAT motif (score 0.5) | MSP | Cytosol, endosomes, Golgi | Tubular endosome formation and/or stabilization | Baron et al. (2014), | |
| 3SESDTE EFYDAPE DV | Häsler et al. (2020) | |||||
| YIF1A-VAPB | TMD | MSP | ER-Golgi intermediate compartment (ERGIC; TMD); ER (TMD), Golgi (TMD) | Controls the shuttling of YIF1A between the ERGIC and the ER; promotes intracellular membrane delivery into dendrites | Kuijpers et al. (2013b) | |
An overview of experimentally confirmed VAP binding partners in mammalian cells.
FFAT motif scores were calculated using the FFAT scoring algorithm (best FFAT motif scores zero)c(Murphy and Levine, 2016). Phosphorylation of serine/threonine at position 4 (double underlined) of the core (bold) of Phospho-FFAT motifs is critical for VAP binding (Di Mattia et al., 2020). Phosphorylation of serine/threonine at position 5 (underlined) of FFAT motifs abolishes VAP binding (Kors et al., 2022). The cellular localisations of the binding partners are listed. The physiological role describes the function of the VAP complex (binding partners may also have been implicated in other non-VAP related processes, or functions might not have been directly linked to VAP yet).
ACBD4/5, acyl-CoA-binding domain-containing protein 4/5; ASNA1 (TRC40), arsenite-stimulated ATPase; ATF6, activating transcription factor 6; CALCOCO1, calcium-binding and coiled-coil domain-containing protein 1; CaSR, calcium-sensing receptor; CERT, ceramide transfer protein; CLN8, ceroid-lipofuscinosis neuronal protein 8; CDIP1, cell death-inducing p53-target protein 1; FAF1, FAS-associated factor 1 (ubiquitin-binding protein); FAPP1, phosphatidylinositol-four-phosphate adapter protein 1; FIP200, FAK family kinase-interacting protein of 200 kDa; GLTP, glycolipid transfer protein; HCN2, hyperpolarization-activated cyclic nucleotide-gated channel 2; IFITM3, interferon-inducible transmembrane protein 3; JMY, junction-mediating and -regulatory protein; Kv2, potassium voltage-gated channel subfamily B; MCS, membrane contact site; MIGA2, mitoguardin 2; MMD, monotopic integral membrane domain; MSP, major sperm protein; NIR, PYK2 N-terminal domain-interacting receptor; ORP, oxysterol-binding protein-related protein; OSBP, oxysterol-binding protein; PP2Cϵ, protein phosphatase 2Cϵ; PRA1, prenylated Rab acceptor 1; PTPIP51, protein tyrosine phosphatase-interacting protein 51; RAB3GAP1, RAB3 GTPase-activating protein catalytic subunit; SCRN1, secernin-1; SNX2, sorting nexin-2; STARD3, StAR-related lipid transfer protein 3; STARD3NL, STARD3 N-terminal like; TMD, transmembrane domain; TRPC3, transient receptor potential channel 3; TTC39B, tetratricopeptide repeat domain containing protein 39 B; ULK1, UNC-51-like autophagy-activating kinase 1; Viperin, Virus inhibitory protein, endoplasmic reticulum-associated, interferon-inducible; VPS13, Vacuolar protein sorting-associated protein 13; WDR44, WD repeat-containing protein 44; YIF1A, YIP1-interacting factor homologue A.
Due to their interaction with VAP, the proteins also localise at the ER (ER is only mentioned if the protein contains another ER targeting domain, e.g. TMD).
Predicted FFAT motif, but not confirmed.
The FFAT score does not indicate the definite binding strength.
Organelle Tethering
Organelles form membrane contact sites (MCS) for efficient cooperation (Silva et al., 2020). These MCS are mediated by tethering proteins that cross the two opposing membranes, bringing them in close proximity. Various proteins are attracted to these sites to fulfil and regulate specific functions, e.g. membrane lipid and calcium (Ca2+) transfer between the organelles. In this section, we will describe the organelle tethering function of VAP in more detail, focussing on the FFAT motif-containing binding partners PTPIP51 and ACBD5 as examples. However, other VAP interactors and functions–many of which relate to MCS–are known. There is an abundance of processes involving VAP at other organelle-ER contacts (Table 1), e.g. CERT transfers ceramide from the ER to the Golgi apparatus (Kawano et al., 2006); STARD3 transfers cholesterol from the ER to endosomes (Wilhelm et al., 2017; Di Mattia et al., 2020); NIR2 transfers phosphatidylinositol from the ER to both the Golgi and plasma membrane, and phosphatidylcholine from the Golgi to the ER (Peretti et al., 2008; Chang and Liou, 2015); while the interaction of VAP with potassium (K+) channel Kv2 at plasma membrane-ER contacts is important for Kv2 channel clustering and regulation of K+ currents (Johnson et al., 2018; Kirmiz et al., 2018; Schulien et al., 2020). Furthermore, other examples will be discussed in the sections on pathogens and amyotrophic lateral sclerosis type 8 (ALS8).
The membrane proteins PTPIP51 (also named RMDN3) and ACBD5 interact with VAPB, mediating mitochondria-ER and peroxisome-ER associations, respectively (Figure 1) (De Vos et al., 2012; Costello et al., 2017a; Hua et al., 2017). Knockdown of PTPIP51 or ACBD5 reduced the contacts between the respective organelle and the ER, while overexpression increased the associations (Stoica et al., 2014; Costello et al., 2017a). Whilst VAP itself does not appear to possess lipid binding capacity, many of its interacting partners have lipid binding properties, including PTPIP51 and ACBD5. PTPIP51 has a tetratricopeptide repeat (TPR) domain with which it can bind and transfer phosphatidic acid (PA) (Ito et al., 2021; Yeo et al., 2021). PA supply to mitochondria from the ER is required for the synthesis of cardiolipin, an important phospholipid of the inner mitochondrial membrane, which was decreased upon depletion of PTPIP51. This function was independent of the tethering function of PTPIP51 (Yeo et al., 2021). Mitochondria-ER contacts are also important for cellular Ca2+ homeostasis, with transport between the organelles mediated by the IP3R-GRP75-VDAC1 complex. Although not directly involved in Ca2+ transfer, the PTPIP51-VAPB interaction plays an important tethering role to allow the Ca2+ uptake by mitochondria from ER stores (De Vos et al., 2012). This PTPIP51-VAPB-regulated Ca2+ delivery modulates autophagosome formation and synaptic activity (Gómez-Suaga et al., 2017, 2019). Additionally, PTPIP51 was shown to be involved in the mitochondrial Ca2+ overload during cardiac ischemia/reperfusion, by increasing the mitochondria-sarcoplasmic reticulum contacts (Qiao et al., 2017).
FIGURE 1
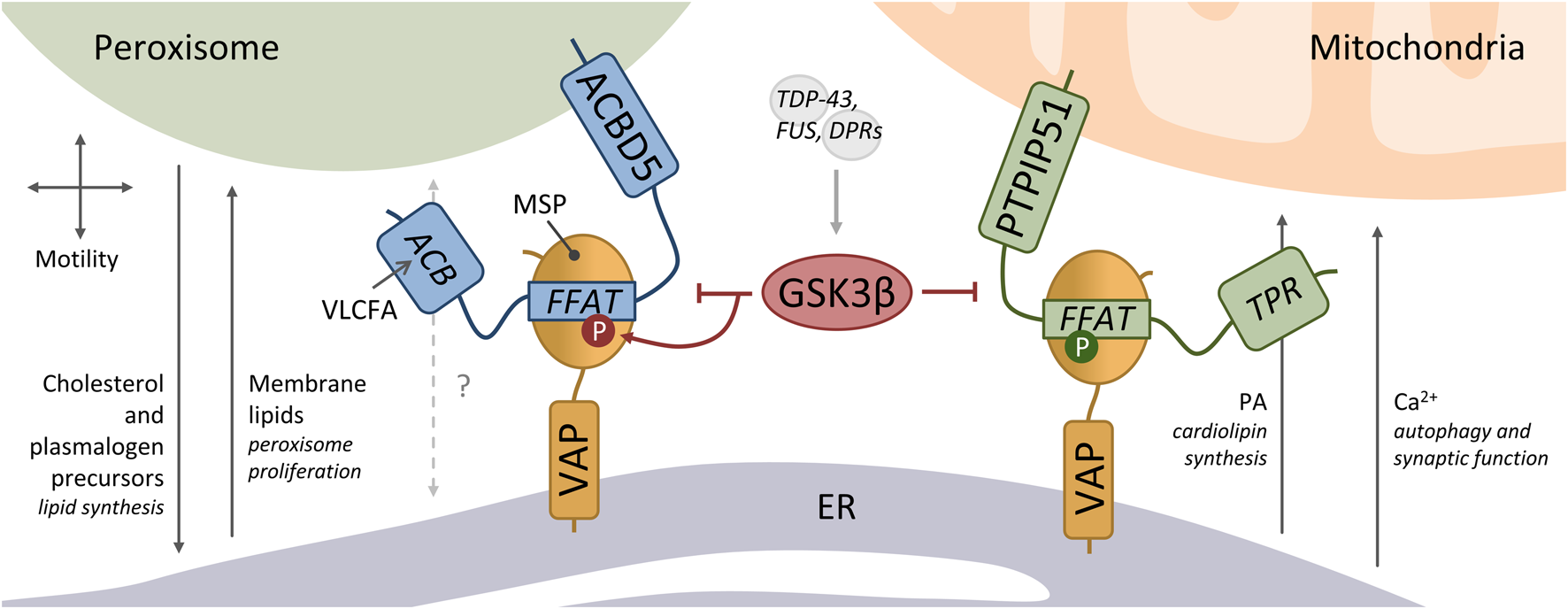
Peroxisome-ER and mitochondria-ER membrane contacts tethered by VAP. ACBD5 interacts via its FFAT motif to the major sperm (MSP domain) of VAP to mediate peroxisome-ER contacts. These peroxisome-ER contacts have been implicated in peroxisome motility, the transfer of cholesterol and plasmalogen precursors for further synthesis in the ER, and the transfer of membrane lipids for peroxisome proliferation. ACBD5 has an acyl-CoA binding (ACB) domain which likely binds very long chain fatty acids (VLCFA). PTPIP51 also binds to the VAP-MSP domain via a FFAT motif, which mediates mitochondria-ER contacts. PTPIP51 has a tetratricopeptide repeat (TPR) domain with which it can bind and transfer phosphatidic acid (PA) to the mitochondria - required for the synthesis of cardiolipin. Ca2+ uptake by mitochondria from ER stores at these contacts modulates autophagosome formation and synaptic activity. GSK3β negatively regulates both peroxisome-ER and mitochondria-ER associations. GSK3β acts on the ACBD5-VAP tether by directly phosphorylating the serine residue (S) at position 5 of the ACBD5 FFAT core (1VYCDSME7). GSK3β can be activated by the ALS-associated proteins TDP-43, FUS and C9orf72-derived dipeptide repeat polypeptides (DPR). Phosphorylation of PTPIP51 at position 4 of the FFAT core (1VYFTASS7) is critical for binding to VAP.
ACBD5 has an acyl-CoA binding (ACB) domain, which has been shown to have lipid/fatty acid binding capacity in vitro (Yagita et al., 2017), but it is not yet clear if it directly transfers lipids between peroxisomes and the ER. However, as ACBD5 deficient patients present with accumulation of very long chain fatty acids (VLCFA), it is suggested that ACBD5 facilitates VLCFA transport into peroxisomes for degradation via the peroxisomal ABC transporter for VLCFA (Ferdinandusse et al., 2017; Yagita et al., 2017; Herzog et al., 2018). The ACBD5-VAPB mediated peroxisome-ER contacts have also been implicated in the regulation of peroxisome motility and positioning, and the delivery of lipids for peroxisomal membrane expansion to maintain peroxisome biogenesis (Figure 1) (Costello et al., 2017a; Hua et al., 2017; Darwisch et al., 2020). ACBD5 and VAPB are also required to support the transfer of plasmalogen precursors, of which the synthesis is initiated in peroxisomes and completed in the ER, and for the maintenance of cholesterol levels (Hua et al., 2017; Herzog et al., 2018).
The examples above illustrate some of the various processes that occur at mitochondria-ER and peroxisome-ER contact sites. These processes appear to require contacts to be in a dynamic equilibrium, with reduced contacts reducing the required substrate transfer but increased contacts potentially also resulting in an excess of exchange. For example, whilst loss of PTPIP51-VAPB stimulates autophagy, increased PTPIP51-VAPB inhibits autophagy implying that dynamism in the mitochondria-ER interaction is required for this process (Gómez-Suaga et al., 2017)). In a similar way, whilst loss of ACBD5-VAPB tethering appears to limit peroxisomal membrane expansion, increased ACBD5 levels lead to peroxisomal elongation, potentially implying an excess membrane expansion (Costello et al., 2017a; Kors et al., 2022). Overall, this suggests that these organelle interactions involving VAP protein tethers are highly regulated. One way to regulate tethers would be to modulate the level of interaction between VAP and its interaction partners. In line with this, we revealed that the ACBD5-VAPB tether can be modulated by phosphorylation of serine/threonine residues within the acidic tract of the FFAT motif (Kors et al., 2022), a mechanism initially described for CERT (Kumagai et al., 2014). Phosphorylation of these residues mimics the canonical aspartic and glutamic acid residues, supporting the acidic environment and enhancing binding to VAPB. Notably, the acidic tract of PTPIP51 is also mainly composed of serine/threonine residues, suggesting that phosphorylation of these residues could as well modulate the binding of PTPIP51 to VAPB. Indeed, in vitro studies with PTPIP51 FFAT peptide and VAPB protein revealed a low affinity suggesting a minor contribution to mitochondria-ER tethering (Yeo et al., 2021). Although this may be different in vivo, phosphorylation of the acidic tract and the FFAT core of PTPIP51 (see below) could stengthen the interaction.
In addition to the acidic tract, phosphorylation of the core FFAT motif of both PTPIP51 and ACBD5 also regulates their interaction to VAPB. However, the different positions of the phosphorylated residues have opposing effects on the binding. Phosphorylation of PTPIP51 at position 4 of the FFAT core (1VYFTASS7) is critical for VAPB binding in vitro (Di Mattia et al., 2020), while phosphorylation of ACBD5 at position 5 of the FFAT core (1VYCDSME7) abolishes the interaction with VAPB (Kors et al., 2022). The canonical FFAT motif possesses aspartic acid (D) at position 4, which could be mimicked by phosphorylated threonine (T) at this position in PTPIP51 to enhance the VAPB binding. The residue at position 5 of the FFAT core–alanine (A) in the canonical motif–binds the VAP MSP domain in a hydrophobic pocket (Kaiser et al., 2005; Furuita et al., 2010). Adding a phosphate group to the serine (S) at this position in ACBD5 likely causes steric hindrance, blocking the interaction. We recently showed that GSK3β can directly phosphorylate this serine residue of the ACBD5 FFAT core (Figure 1). Accordingly, increased GSK3β activity inhibited the ACBD5-VAPB interaction and hence peroxisome-ER contacts, while reduced GSK3β activity increased the organelle associations (Kors et al., 2022). Interestingly, GSK3β also negatively regulates the PTPIP51-VAPB interaction and mitochondria-ER associations, although the precise mechanism is not known (Stoica et al., 2014, 2016; Gómez-Suaga et al., 2022). It was shown that the ALS-associated proteins TDP-43, FUS and C9orf72-derived dipeptide repeat polypeptides (DPR) activate GSK3β, causing disruption of the mitochondria-ER tether and membrane contacts. This suggests altered mitochondria-ER and peroxisome-ER MCS in TDP-43/FUS/C9orf72-induced pathologies.
Overall, there are three regulation mechanisms involving phosphorylation of FFAT motifs: 1) phosphorylation of residues in the acidic tract enhances the interaction with VAP, acting as a potential fine-tuning mechanism (Kumagai et al., 2014; Di Mattia et al., 2020; Kors et al., 2022); 2) phosphorylation of S/T in position 4 acts as a switch mechanism (OFF/ON), being critical for VAP binding and defines the so-called “Phospho-FFAT motif” (Kirmiz et al., 2018; Di Mattia et al., 2020; Guillén-Samander et al., 2021); and 3) phosphorylation of S/T in position 5 also acts as a switch mechanism (ON/OFF), but in this case the phosphorylated FFAT motif is not able to interact with VAP (Mikitova and Levine, 2012; Kors et al., 2022).
VAP Hijacking by Viruses and Bacteria
The VAP proteins are exploited by various viruses and intracellular bacteria for their replication. Some pathogens hijack VAP via FFAT motif mimicry, while others express pathogenic proteins that interact with VAP or VAP-interactors in other ways. Below we describe how different viruses and bacteria make use of the many functions of VAP for membrane remodelling, the formation of MCS between the host ER and pathogen-containing compartments, and targeting host MCS components to rewire the host lipid metabolism and other processes.
FFAT Motif-Containing Pathogenic Proteins
Chlamydia trachomatis
The bacterium Chlamydia trachomatis is an intracellular pathogen, causing non-congenital blindness, and is the most common sexually transmitted infection worldwide. The bacterium proliferates inside the cell in a membranous compartment, called an inclusion. The integral inclusion membrane protein IncV has been found to directly interact with VAPA/B via two FFAT motifs upon C. trachomatis infection (280DSSSSS EYMDALE TV; 256ESSSSS SFHTPPN SD; Figure 2A) (Stanhope et al., 2017). Overexpression of IncV in C. trachomatis-infected cells enhanced the recruitment of VAPA and the ER to the inclusion membrane, suggesting that IncV promotes the formation of inclusion-ER MCS. However, depletion of IncV had only a moderate impact on VAPA enrichment at the inclusion membrane, suggesting that other proteins contribute to the stability of inclusion-ER MCS (e.g. IncD-CERT-VAPA/B complex, see below). The two FFAT motifs of IncV both have an acidic tract consisting of multiple serine residues, suggesting that, like ACBD5, its interaction with VAPA/B could be regulated by phosphorylation of these residues, to mimic the negative charge of the conventional acidic residues (Stanhope et al., 2017). The exact contribution of the IncV-VAP interaction in Chlamydia pathogenesis remains to be determined.
FIGURE 2
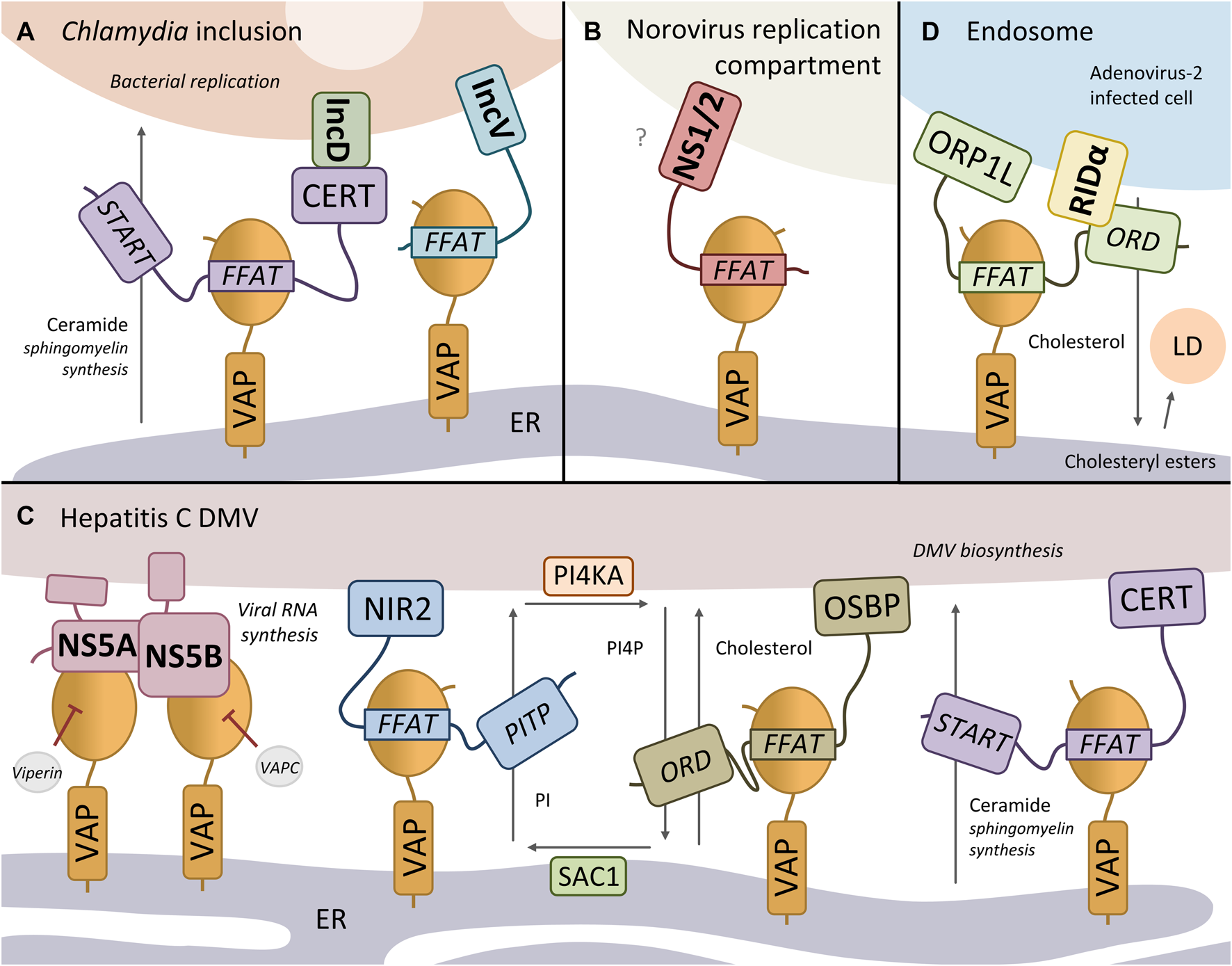
VAP hijacking by bacteria and viruses. (A) The Chlamydia integral inclusion membrane proteins IncV and IncD are both found in a complex with VAP. While IncV binds VAP directly via a FFAT motif, IncD interacts with VAP-interactor CERT, forming an IncD-CERT-VAP complex at inclusion-ER contact sites. CERT may facilitate ceramide transfer from the ER to the inclusion membrane for the synthesis of sphingomyelin, important for C. trachomatis replication. (B) The murine norovirus protein NS1/2 binds VAP via its FFAT motif, critical for viral replication. The NS1/2-VAP may tether the replication membrane to the ER. (C) VAP binds to the hepatitis C virus proteins NS5A and NS5B, important for viral RNA replication at double-membrane vesicles (DMVs). VAP supports the viral replication by recruiting host NIR2, OSBP, CERT to the DMV-ER contact site. NIR2 transfers PI from the ER membrane to the DMV membrane, where it is converted to PI4P by the PI4P-kinase PI4KA. The PI4P is then exchanged for cholesterol from the ER by OSBP. The PI4P-phosphatase SAC1 converts PI4P back to PI. CERT transports ceramide from the ER to the DMV, where it is converted to sphingomyelin, important for the biosynthesis of DMVs. (D) The Adenovirus-2 protein RIDα directly interacts and recruits ORP1L to maturing early endosomes to form endosome-ER MCS via ORP1L-VAP binding, which facilitates the transport of cholesterol from endosomes to the ER. Here, cholesterol is converted into cholesteryl esters, which are stored in lipid droplets (LD). Bacterial/viral proteins are indicated in bold.
In mammalian cells, ceramide is transported from the ER to the Golgi complex at MCS by transport protein CERT for the synthesis of the membrane lipid sphingomyelin (Table 1). Another C. trachomatis integral inclusion membrane protein, IncD, has been found to interact with host CERT, recruiting CERT and thus its binding partner VAPA/B to the inclusion membrane (Figure 2A) (Derré et al., 2011; Agaisse and Derré, 2014; Kumagai et al., 2018). In this way the IncD-CERT-VAPA/B complex may facilitate ceramide transfer from the ER to the inclusion membrane, where it is converted to sphingomyelin with the use of host and/or bacterial sphingomyelin synthases (Elwell et al., 2011; Tachida et al., 2020). This CERT-dependent sphingomyelin pathway is critical for C. trachomatis replication.
Another example of how C. Trachomatis hijacks components usually present at host ER-organelle MCS is STIM1. This ER-resident Ca2+ sensor protein colocalized with VAPB at inclusion-ER MCS (Agaisse and Derré, 2015). However, the plasma membrane Ca2+ channel ORAI1, the interaction partner of STIM1 at PM-ER MCS, did not associate with the inclusion membrane. Instead, STIM1 may work with another Ca2+ channel: IP3R, an ER protein present at mitochondria-ER MCS. IP3R has been found to bind both STIM1 (Santoso et al., 2011) and the inclusion membrane protein MrcA, presumably forming a Ca2+ signalling complex at the inclusion-ER MCS (Nguyen et al., 2018). Both STIM1 and ITPR3 are required for chlamydial release via extrusion of the inclusion. The regulation of local Ca2+ levels may influence the myosin motor complex, which promotes the extrusion.
Overall, the interaction of Chlamydia membrane protein IncV with VAPA/B promotes the formation of inclusion-ER MCS within cells. At these sites, C. trachomatis redirects several host proteins for sphingomyelin synthesis (e.g. CERT via IncD; important for bacterial replication) and Ca2+ signalling (e.g. STIM1 via MrcA; bacterial extrusion) to assist its pathogenicity.
Norovirus
Noroviruses are non-enveloped RNA viruses and the primary cause of gastroenteritis. The murine and human (GI) norovirus protein NS1/2 has been reported to interact with VAPA/B (Figure 2B) (Ettayebi and Hardy, 2003; McCune et al., 2017). Structural analysis revealed that the murine NS1/2-VAP interaction is mediated by a FFAT-motif mimic located in the N-terminal NS1 domain of NS1/2 (40ESEDEV NYMTPPE QE) (McCune et al., 2017). The FFAT-motif is conserved across murine norovirus strains, although the inherently disordered NS1 domain itself is not well conserved in contrast to the NS2 domain (Baker et al., 2012). Interestingly, NS1/2 has been found to form dimers, a property of many FFAT motif-containing proteins, which could stabilize the interaction with VAP-dimers. It would be interesting to determine whether the human NS1/2GI-VAP interaction is also mediated via a FFAT motif.
Strikingly, mutagenesis of the NS1/2 FFAT residues critical for VAP binding eliminated virus replication (McCune et al., 2017). Additionally, VAPA depletion in cells showed that VAPA was important in the early stage of norovirus replication. However, it is not clear how the NS1/2-VAP interaction contributes to the viral replication cycle. Localisation of NS1/2 to the ER might contribute to the formation of the membranous viral replication compartment, possibly by bridging the ER and replication membrane via its interaction with VAP and putative transmembrane domain (Baker et al., 2012).
VAP-Exploiting Pathogens
Hepatitis C Virus
Hepatitis C virus (HCV) is an enveloped RNA virus that predominantly infects liver cells, and can cause liver cirrhosis and cancer. Upon HCV infection, a so-called membranous web, consisting primarily of double-membrane vesicles (DMVs), is formed, that is thought to be the site of viral RNA replication. Three HCV proteins have been reported to associate with VAP. While a direct interaction for the viral NS3/4A protease was not examined (Ramage et al., 2015), structural studies have looked into the binding domains of HCV proteins NS5A and NS5B. The viral RNA-dependent RNA polymerase NS5B interacts via its C-terminal auto-regulatory motif with the MSP domain of VAPA/B (Figure 2C) (Gupta and Song, 2016). This C-terminal motif seems to define whether NS5B is in a folded, auto-inhibitory state, or in a disordered, active state that binds to VAP and initiates RNA synthesis. Additionally, several studies report an interaction between HCV protein NS5A and VAPB, although they attribute the interaction to different domains. One study reveals that NS5A forms a dynamic complex with VAP-MSP by interacting via its disordered C-terminal D3 domain (Gupta et al., 2012). However, other studies report that the coiled-coil domain and transmembrane domain of VAPA/B and other residues of NS5A are essential for NS5A-VAP binding (Tu et al., 1999; Hamamoto et al., 2005; Goonawardane et al., 2017; Wang and Tai, 2019). Phosphorylation of NS5A has been reported to regulate the interaction with VAP (Evans et al., 2004; Goonawardane et al., 2017).
Overexpression and knockdown studies show that the VAP proteins play an important, but yet undefined role in the formation of the HCV replication complex and in RNA replication (Gao et al., 2004; Hamamoto et al., 2005). Although the function of NS5A/B-VAP binding in HCV infection is not fully understood, recent studies are starting to decipher how VAPA/B supports the viral replication. It has been suggested that VAP, NIR2 and OSBP operate in a phosphoinositide cycle between the ER and HCV DMV membrane (Figure 2C). Both VAP and the VAP-interactor NIR2 are required to upregulate phosphatidylinositol-4-phosphate (PI4P) levels during HCV infection (Wang and Tai, 2019), indicating that the phosphatidylinositol (PI) transfer protein NIR2 transfers PI from the ER membrane to the DMV membrane, which is then used to generate PI4P by phosphatidylinositol 4-kinase III α (PI4KA) (Berger et al., 2011). Interestingly, NS5A was shown to associate with and stimulate PI4KA activity. The PI4P is then exchanged for cholesterol from the ER by the VAP-interactor OSBP (Wang et al., 2014). The PI4P enrichment of the DMV membrane can also recruit other PI4P-interacting proteins to the DMV-ER MCS such as the VAP-interactor CERT, which transports ceramide from the ER to the DMV, where it can be converted to sphingomyelin, important for the biosynthesis of DMVs (Gewaid et al., 2020). NIR2, OSBP and CERT normally function at the Golgi-ER MCS (Table 1).
To inhibit the replication of HCV, the cell has mechanisms to disrupt the NS5A/B-VAPA/B binding. The ER-associated virus inhibitory protein Viperin, which binds to both NS5A and the C-terminal region of VAPA (Table 1), promotes the degradation of NS5A, an effect that is enhanced by VAPA (Wang et al., 2012; Ghosh et al., 2020). VAPC, an unstructured VAPB splice variant, acts as an endogenous inhibitor by binding to NS5B, interrupting the interaction of NS5B with VAPA/B (Kukihara et al., 2009; Goyal et al., 2012). The ability of VAPC to negatively regulate HCV replication has been of interest in anti-HCV drug development (Wen et al., 2011). Another potential anti-HCV drug also acts via disrupting the viral-host protein interaction; bicyclol restricts HCV replication by upregulating FFAT-motif containing protein GLTP (Table 1), which interrupted the interaction between VAPA and NS5A (Huang et al., 2019).
Overall, it seems that the VAP proteins anchor the viral RNA replication machinery to the ER membrane via viral NS5A/B interaction, and recruit host VAP interactors (e.g. NIR2, OSBP, CERT) for the synthesis of cholesterol and sphingomyelin, important for HCV replication. Targeting VAP in this way allows pathogens to use a single degenerate and potentially regulatable FFAT motif to interact with a range of useful host proteins.
Other Pathogens and Strategies for Utilisation of VAP
In addition to HCV, several other viruses hijack cholesterol trafficking within the cell. The Aichi virus (AiV) proteins 2B, 2BC, 2C, 3A, and 3AB are found in a complex with VAPA/B, OSBP and other components of the cholesterol transport machinery at Golgi-ER MCS such as the PI4P-phosphatase SAC1 and ACBD3 (which recruits PI4KB) (Sasaki et al., 2012; Ishikawa-Sasaki et al., 2018). The proteins are recruited to AiV genome replication sites at the replication organelle (RO)-ER MCS, where cholesterol accumulates in the RO membrane. Knockdown of each component resulted in inhibition of AiV RNA replication. Other viruses that utilise the OSBP-cholesterol transport to facilitate RNA synthesis at RO membranes include poliovirus (Arita, 2014), rhinovirus (Roulin et al., 2014) and encephalomyocarditis virus (EMCV) (Dorobantu et al., 2015). Although no virus-VAP (interactor) complexes have been reported for these viruses, poliovirus and EMCV proteins bind to PI4KB and PI4KA respectively, which stimulates PI4P production and leads to recruitment of OSBP.
The Adenovirus-2 (Ad2) adopts a different mechanism to employ the host cholesterol transport pathway. The Ad2 membrane protein RIDα directly interacts and recruits sterol-binding protein ORP1L to maturing early endosomes to form endosome-ER MCS via ORP1L-VAP binding (Cianciola et al., 2017) (Figure 2D). RIDα stabilizes the interaction between ORP1L and VAP, which supports the transport of cholesterol from maturing endosomes to the ER under high cholesterol conditions. The RIDα-ORPL1-VAP interaction induces the conversion of cholesterol into cholesteryl esters, which are stored in lipid droplets. This change in cholesterol trafficking attenuates proinflammatory TLR4 signalling involved in the innate immune response. ORP1L is also hijacked by the intracellular bacterium Coxiella burnetii, which forms a lysosome-like parasitophorous vacuole (PV) in the host cell for its replication (Justis et al., 2017). ORP1L is recruited to the PV by an unknown PV membrane protein, while also associating with ER-VAP. Although the function of ORP1L at PV-ER MCS in C. burnetii pathogenicity is unclear, ORP1L is important for PV expansion.
Herpes simplex virus type-1 (HSV-1) replicates its DNA and assembles its capsids in the host cell nucleus. The virion then crosses the nuclear envelope for further maturation in the cytoplasm. VAPB contributes to this nuclear egress as knockdown led to nuclear virion accumulation, however its exact role in this is still unclear (Saiz-Ros et al., 2019). VAP also plays a role in the replication of another DNA virus, the human papillomavirus 16 (HPV-16). However, instead of a role in nuclear egress, VAP is important in the nuclear entry pathway of HPV-16 (Siddiqa et al., 2018). Virus particles enter the cell via endocytic uptake, disassemble into protein complexes that traffic to the trans-Golgi-network (TGN) and then access the nucleus during mitosis when the nuclear envelope breaks down. VAP is required for the endosome-to-Golgi viral protein delivery, as it is essential for the formation of endosomal tubules induced upon HPV-16 infection. Whether these viruses exploit VAP directly via viral protein interactions or via other mechanism needs to be further elucidated.
The genetic disease cystic fibrosis (CF) is caused by mutations in the CF transmembrane conductance regulator (CFTR) protein. Patients have an increased susceptibility to bacterial infections such as Pseudomonas aeruginosa infection, which aggravates CF. P. aeruginosa exploits VAPB’s mitochondrial tethering function for infection (Rimessi et al., 2020). The bacteria induced increased VAPB and PTPIP51 expression in CF bronchial cells, but not in non-CF cells. The consequent increase in mitochondria-ER contacts caused impairment of autophagy, inducing inflammation and disease progression.
Overall, a variety of different pathogens utilise VAP interaction and modulation to allow them increased access to host resources. This likely reflects the multifunctionality of VAP as a versatile access point (Murphy and Levine, 2016) to the ER membrane and also the diversity of its interaction partners, which have roles in many different cellular functions. Therapeutic strategies which attempt to prevent pathogen access to VAP could perhaps be feasible but would need to be carefully targeted as inhibition of VAP function itself has a dramatic effect on cellular function and is linked to numerous neuronal disorders, as addressed in the following section.
The Role of VAPB in Neuronal Disorders
VAPB has been linked to several neurological disorders, including amyotrophic lateral sclerosis (ALS), Alzheimer’s disease (AD) and the α-synucleinopathies, Parkinson’s disease (PD) and multiple system atrophy (MSA). This is via mutations in VAPB (ALS, PD) (Nishimura et al., 2004a; Kun-Rodrigues et al., 2015), disruption of VAPB’s interaction with PTPIP51 and hence ER-mitochondria contacts (ALS, AD, and PD via α-Synuclein binding, seeTable 1) (Paillusson et al., 2017; Lau et al., 2020; Gómez-Suaga et al., 2022) or reduced VAPB levels (ALS, AD, MSA) (Anagnostou et al., 2010; Lau et al., 2020; Mori et al., 2021). Recent findings on the role of mutated VAPB in the pathogenesis of ALS are discussed in more detail below.
Clinical Features of Amyotrophic Lateral Sclerosis Type 8 (ALS8)
An autosomal dominant missense mutation in VAPB, resulting in a substitution of proline to serine at codon 56 (P56S), was initially found in several Brazilian families (Nishimura et al., 2004a, 2004b; Marques et al., 2006). The patients presented with a heterogeneous phenotype of typical ALS, atypical ALS and late onset spinal muscular atrophy (SMA), and was termed ALS8 (OMIM 608627) (Nishimura et al., 2004a). Patients with ALS8 have predominant lower motor neuron involvement, with symptoms including progressive muscle weakness (mainly in the lower limbs), muscle atrophy, cramp, tremor, fasciculations, pain, abdominal protrusion, autonomic dysfunction (e.g. choking, constipation), and subtle cognitive and behavioural impairments (Nishimura et al., 2004a, 2004b; Marques et al., 2006; Funke et al., 2010; Kosac et al., 2013; Di et al., 2016; Chadi et al., 2017; Sun et al., 2017; Guber et al., 2018; de Alcântara et al., 2019; Trilico et al., 2020; Nunes Gonçalves et al., 2021; Temp et al., 2021; Leoni et al., 2022). ALS8’s clinical heterogeneity manifests not only in the symptoms but also in the age of onset (reported at 20–57 years) and the disease progression (rapid [<5 years] to slow [30+ years]). To understand the mechanisms behind this phenotypic variability, researchers compared gene expression profiles of iPSC (induced pluripotent stem cells)-derived motor neurons from mild and severe ALS8 patients (Oliveira et al., 2020). VAPB mRNA and protein levels were equally downregulated in mild and severe patients. The differentially expressed genes found in the study were associated with pathways involved in protein translation and protein targeting to the ER; pathways that may mitigate neurodegeneration in the mild ALS8 patients by maintaining proteostasis. Interestingly, a reduction in VAPB mRNA and protein levels was also observed in the spinal cord of sporadic and familiar (superoxide dismutase 1 (SOD1)-linked) ALS patients and mice, suggesting a role of VAPB in the pathogenesis of non-VAPB linked ALS as well (Teuling et al., 2007; Anagnostou et al., 2010). These reduced levels might be associated with SNPs (single-nucleotide polymorphisms) within the VAPB gene (Chen et al., 2010). It has even been suggested that VAPB aggregates can be used as a pathologic marker in the screening of sporadic non-VAPB linked ALS, as VAPB clusters were detected in peripheral blood mononuclear cells (PBMCs) and fibroblasts isolated from these patients (Cadoni et al., 2020).
Haplotype analysis showed a common Portuguese ancestor of the Brazilian families, with a founding event 23 generations ago, resulting in about 200 affected family members (Nishimura et al., 2005). Mutations in VAPB have not been associated with sporadic ALS (Conforti et al., 2006; Kirby et al., 2007) and the frequency of VAPB mutations is low in other populations (Tsai et al., 2011; Ingre et al., 2013; Kenna et al., 2013). However, the P56S mutation has also been identified in German, Japanese, Chinese and North American families displaying ALS8 symptoms, and have arisen independently from the Brazilian patients (Funke et al., 2010; Millecamps et al., 2010; Di et al., 2016; Guber et al., 2018). Another mutation in codon 56 of VAPB, in which proline is substituted for histidine (P56H), has also been found, in a Chinese family with similar clinical features as patients with P56S (Sun et al., 2017). Other mutations located in VAPB and associated with ALS are T46I, A145V and V234I (see Other Mutations in VAPB) (Chen et al., 2010; van Blitterswijk et al., 2012; Kabashi et al., 2013).
Although VAPB is ubiquitously expressed in the body and fulfils functions important for basal cell performance, it is mainly motor neuron dysfunction that is reported in the VAPB P56S/H patients. Electromyography and muscle/nerve biopsies revealed neurogenic damage with chronic denervation of muscles and reduced numbers of myelinated axons (Nishimura et al., 2004a; Marques et al., 2006; Kosac et al., 2013; Di et al., 2016; Sun et al., 2017; Guo et al., 2020). Additionally, neuroanatomical abnormalities were observed in ALS8 patients, including atrophy in the brainstem, globi pallida and upper cervical spinal cord (Leoni et al., 2022). The reason why VAPB mutations lead specifically to neurodegeneration is not well understood, although VAPB has been found to be highly abundant in motor neurons and different regions of the brain (Teuling et al., 2007; Larroquette et al., 2015; Leoni et al., 2022).
VAPB Aggregates
VAPB Aggregate Features and Formation
Several studies have reported that overexpression of VAPB P56S induces the formation of insoluble cytosolic aggregates in neuronal and non-neuronal cells (Nishimura et al., 2004b; Kanekura et al., 2006; Teuling et al., 2007), in culture as well as in transgenic mice and Drosophila ALS models (Chai et al., 2008; Ratnaparkhi et al., 2008; Qiu et al., 2013). The aggregation-prone VAPB P56S recruits wild-type VAPB and, to a lesser extent, VAPA to the aggregates, having a dominant-negative effect on normal VAP function (Kanekura et al., 2006; Teuling et al., 2007; Chai et al., 2008; Ratnaparkhi et al., 2008; Suzuki et al., 2009). The VAPB mutant has also been shown to sequesters ER-Golgi recycling protein YIF1A (via its TMD) to the aggregates, depleting the protein from these organelles (Kuijpers et al., 2013b). Nevertheless, VAPB P56S does not seem to induce “classical protein aggregates,” formed of insoluble fibrils, a hallmark of other neurodegenerative disorders like Huntington’s disease (huntingtin), PD (α-synuclein) and ALS (SOD1). For example, VAPB P56S forms aggregates rapidly after expression (<2 h), while the formation of SOD1 aggregates takes hours to days (Matsumoto et al., 2005; Fasana et al., 2010). Additionally, ultrastructural studies showed that overexpression of the mutant VAPB protein caused accumulation of large membranous aggregates, consisting of ribbons of stacked ER cisternae (Teuling et al., 2007; Fasana et al., 2010; Papiani et al., 2012). Live cell photobleaching experiments, using ER membrane-targeted GFP, revealed that VAPB P56S-ER subdomain inclusions are continuous with the rest of the ER (Fasana et al., 2010). However, there is some discrepancy between overexpression studies about the presence of proteins from the secretory pathway in the VAPB aggregates; for instance, ER luminal proteins calreticulin and PDI, and ER membrane protein calnexin associate with mutant VAPB aggregates is some studies, whilst others observed exclusion of these proteins (Kanekura et al., 2006; Teuling et al., 2007; Prosser et al., 2008; Fasana et al., 2010; Kuijpers et al., 2013a). This may be attributed to differences in cell lines, VAPB expression levels, and the exclusion of some (rough) ER membrane proteins from the aggregates (Fasana et al., 2010).
To understand how VAPB P56S induces aggregate formation, we will first discuss how the mutation affects the protein structure. VAPB proline 56 is conserved in VAPA, but mutating this residue does not seem to have such a significant effect, with some studies suggesting that no aggregation was observed whilst others observe minor levels of aberrant aggregation for VAPA P56S, notably in HeLa cells (Teuling et al., 2007; Prosser et al., 2008; Suzuki et al., 2009). VAPA P56S’s resistance to aggregation seems to rely on two other proline residues present in this region, whereas VAPB P56S has only one remaining proline residue (Nakamichi et al., 2011). Substituting one of the prolines in VAPA P56S to the equivalent in VAPB P56S (VAPA P56S/P63A), resulted in the formation of membranous aggregates indistinguishable from those observed with VAPB P56S. The three proline residues of VAPA are conserved in the yeast VAP protein Scs2p, which is also resistant to the ALS8-causing mutation, showing that the proline distribution is an important feature in the pathophysiology of ALS8 (Nakamichi et al., 2011).
P56 is located in the MSP domain of VAPB and is critical for the correct folding of the seven β-strands of the MSP domain (Figure 3A) (Shi et al., 2010). P56 stabilizes the cis-peptide bond within the S-shaped loop that connects strands D1 and D2 (Kaiser et al., 2005; Teuling et al., 2007; Shi et al., 2010). The P56S mutation induces a conformational change within the recombinant MSP domain, resulting in the exposure of hydrophobic patches, which may enhance oligomerization of the mutant VAPB protein under physiological conditions (Figure 3B) (Kim et al., 2010). However, studies with recombinant MSP P56S domains show differences in structural stability and solubility (Kim et al., 2010; Shi et al., 2010). P56S eliminates the native β-sheet structure in water, and the exposed hydrophobic patches seem to drive aggregation of recombinant MSP P56S, making the structure highly insoluble in various buffers (Shi et al., 2010; Qin et al., 2013a). This makes it difficult to understand exactly how the VAPB P56S structure behaves under physiological conditions. Nevertheless, it has been shown that MSP P56S retains its ability to bind to FFAT motif-containing proteins in HeLa cells, but the FFAT-binding of full-length VAPB P56S is perturbed (Kim et al., 2010). The aberrant oligomerization of full-length VAPB P56S may interfere with the binding of FFAT motifs to the MSP domain. In line with this, no FFAT-motif containing proteins were observed in pull-down assays using biotinylation-tagged VAPB P56S (Teuling et al., 2007). However, overexpression of a FFAT motif peptide rescued the aggregation phenotype of the mutant, suggesting protein stabilisation via FFAT motif-binding (Prosser et al., 2008). Additionally, VAPB P56S induces clustering of mitochondria and peroxisomes that colocalise with the VAPB aggregates (De Vos et al., 2012; Hua et al., 2017). The clustering of peroxisomes was dependent on the presence of ACBD5, suggesting that the mutant VAPB can sequester FFAT-motif containing proteins such as peroxisomal ACBD5 and possibly mitochondrial PTPIP51 (De Vos et al., 2012; Hua et al., 2017). However, FFAT-proteins ORP9 and NIR2 were not detectable in the VAPB aggregates (Kuijpers et al., 2013a). In summary, the P56S mutation causes conformational changes in the MSP domain and although this does not affect FFAT-binding to the domain on its own, in the presence of full-length VAPB, exposed hydrophobic patches cause enhanced oligomerization of the protein, which seems to reduce accessibility to the FFAT-binding site.
FIGURE 3

Structure of the VAP MSP domain. (A) Structure of the MSP domain of VAPB (PDB ID: 3IKK) and schematic representation of the domain architecture of VAPB, with the ALS-related mutations indicated. The two ALS-related residues that are located in the MSP domain (T46 and P56) are mapped onto the structure. (B) Structure of the MSP domain of VAPB P56S (PDB ID: 2MDK). The ALS-related mutation S56 is mapped onto the structure. (C) Structure of the MSP domain of VAPA in complex with the OSBP FFAT motif (358EFFDAPE I) (PDB ID: 2RR3). MSP residues T46 and P56 are indicated. The FFAT core residues of OSBP are written in italic. Images created with UCSF ChimeraX (Pettersen et al., 2021).
The disordered MSP P56S domain, but not the wild-type MSP domain, is able to interact with dodecylphosphocholine, a lipid commonly used to resemble membrane lipids, transforming the domain into a highly helical conformation (Qin et al., 2013b). This allows MSP P56S to be inserted into membrane environments (Qin et al., 2013a). Therefore, the interaction of VAPB P56S with lipids from the ER membranes could provide a mechanism for the formation of the membranous aggregates. The presence of membrane structures within the aggregates could also be attributed to VAPB being a tail-anchored protein. VAPB P56S has been shown to be efficiently post-translationally inserted into the ER membrane, after which it rapidly clusters (Fasana et al., 2010). This is further confirmed by a study showing that co-expression of a FFAT-containing peptide with VAPB P56S, partially restored the characteristic reticular ER pattern of VAPB (Prosser et al., 2008), suggesting that FFAT binding can maybe stabilise the mutant MSP structure and that the MSP domain/FFAT-interaction plays an important role in the formation of (membranous) aggregates.
Overall, VAPB P56S seems prone to aggregation due to instability of its MSP structure caused by the mutation. Because of its unaffected TMD, the mutant protein is still targeted to the ER membrane (Fasana et al., 2010), where it forms clusters, recruits wild-type VAPA/B and VAP interactors, and induces the formation of membranous clusters (Teuling et al., 2007; Fasana et al., 2010). Conceivably, newly synthesized mutant VAPB could also aggregate in the cytosol before its insertion into the ER membrane. This may depend on the rate of protein synthesis and levels/capacity of the chaperone machinery, which may differ between different cell types, but could then give rise to two different types of aggregates, cytosolic and membranous, which may explain some discrepancy between studies.
VAPB Aggregate Clearance
While overexpression studies show that VAPB P56S is aggregation prone, aggregate formation was also induced at physiological conditions at low levels of mutant VAPB, comparable to endogenous wild-type protein, in HeLa cells (Fasana et al., 2010; Papiani et al., 2012). However, more research is required to clarify in what extend these aggregates form in patients; it has been shown that iPSC-derived motor neurons from ALS8 patients have reduced levels of VAPB and no signs of aggregate accumulation (Mitne-Neto et al., 2011; Oliveira et al., 2020), while ALS8 patient-derived muscle biopsy and fibroblasts revealed VAPB aggregates (Tripathi et al., 2021). As HeLa cells also displayed aggregated VAPA P56S (Teuling et al., 2007), which was not observed in other cell types, it seems likely that different cell types show altered VAP aggregate accumulation.
Discrepancy in detection of aggregates in patients might be due to differences in clearance of mutant VAPB. VAPB P56S has been reported to be less stable than the wild-type protein in both cultured cells and transgenic mice (Papiani et al., 2012; Aliaga et al., 2013; Genevini et al., 2014). VAPB P56S was polyubiquitinated shortly after synthesis and degraded by the proteasome in inducibly-expressing HeLa and NSC34 (motoneuronal) cells, with no evident involvement of basal autophagy (although it can be targeted by stimulated autophagy) (Kanekura et al., 2006; Papiani et al., 2012; Genevini et al., 2014). Ubiquitination of VAPB P56S has also been observed in motor neurons and muscle of transgenic mice and flies (Ratnaparkhi et al., 2008; Tsuda et al., 2008; Tudor et al., 2010). The data further indicates that in the HeLa cells and transgenic mice, the mutant protein initially avoids degradation, clusters and is then cleared by the proteasome (Papiani et al., 2012; Kuijpers et al., 2013a). This comprises the involvement of ER membrane chaperone BAP31 and the ATPase chaperone p97/VCP, proteins involved in ER-associated protein degradation (ERAD), likely by extracting mutant VAPB from the ER membrane. However, a study reported that overexpression of both wild-type and mutant VAPB impaired proteasome activity, possibly by inducing ER stress (see below) (Moumen et al., 2011), although this might be attributed to the high levels of expressed VAPB in comparison to the inducible system. Interestingly, it has also been reported that VAPB P56S is resistant to proteolysis by an unidentified protease that releases the MSP domain from wild-type VAPB (Gkogkas et al., 2011).
Disruption of Cellular Homeostasis
Below we highlight some of the functions of VAPB and the effects that VAPB P56S has on ER stress responses and autophagy. But since VAPB has many functions and binding partners (seeTable 1), and the P56S mutation impacts the protein properties (see above), it is plausible that most processes involving VAP are in some extent impacted by mutant VAPB, including organelle tethering (Yamanaka et al., 2020) and regulation of PI4P levels (Wilson et al., 2021). We focus on how VAPB P56S affects motor neurons specifically. However, in addition to neurological problems, ALS8 patients also exhibit altered metabolic functions, such as dyslipidemia with increased cholesterol and triglyceride levels (Marques et al., 2006). VAPB P56S was found to suppresses adipocyte differentiation (Tokutake et al., 2015a) and VAPB is involved in different cholesterol and triglyceride pathways via its binding partners, as shown in Table 1.
ER Stress
The P56S mutant VAPB causes ER stress (Aliaga et al., 2013; Larroquette et al., 2015), altered ER domain properties (Fasana et al., 2010; Papiani et al., 2012; Yamanaka et al., 2020) and malfunction of the unfolded protein response (UPR), a physiological reaction to suppress accumulation of misfolded proteins in the ER (Kanekura et al., 2006; Suzuki et al., 2009). In mammalian cells, the three main signalling pathways of UPR are IRE1, ATF6, and PERK–with all three shown to be affected by mutant VAPB. VAPB P56S suppress the IRE1-XBP1 pathway that activates expression of UPR target genes, such as chaperones and ERAD components (Kanekura et al., 2006; Suzuki et al., 2009; Tokutake et al., 2015b). VAPB directly interacts with the ER-localized transcription factor ATF6 which, by acting as an ER stress sensor, regulates the transcription of genes encoding chaperones and other UPR transcription factors (Gkogkas et al., 2008). VAPB P56S was shown to attenuate the ATF6-mediated UPR transcription. On the other hand, VAPB P56S activates UPR via PERK-ATF4 which, by promoting the expression of the pro-apoptotic gene CHOP, initiates the cell apoptotic pathway under prolonged ER stress (Aliaga et al., 2013; Tokutake et al., 2015a). Increased basal ER stress and UPR activation has also been reported in the ALS8 patient-derived fibroblasts (Guber et al., 2018). Overall, if the UPR impacted by VAPB P56S cannot restore proteostasis, it might lead to apoptosis.
Autophagy
Mutant VAPB has been linked with dysfunctional autophagy (Zhao et al., 2018; Tripathi et al., 2021). The P56S mutation reduced VAPB’s interaction with early autophagy proteins ULK1 and FIP200, impairing autophagosome biogenesis (seeTable 1) (Zhao et al., 2018). Additionally, VAPB P56S accumulates in autophagosomes and impairs their clearance, showing that VAPB acts at different stages of autophagy (Larroquette et al., 2015; Tripathi et al., 2021). An accumulation and sequestering of autophagic markers p62 and LC3 at VAPB P56S aggregates was also observed in ALS8 patient fibroblasts and muscle biopsies (Tripathi et al., 2021). Impairment of autophagy by mutant VAPB can result in the aggregation of FUS, TDP-43 and Matrin 3 – mutations in which are associated with familial ALS–leading to the formation of stress granules (Tudor et al., 2010; Tripathi et al., 2021). Overexpression of FUS and TDP-43 have both been linked with disruption of the PTPIP51-VAPB association and hence, mitochondria-ER contacts (Stoica et al., 2014, 2016) (Figure 1). Loosening mitochondria-ER contacts via PTPIP51 or VAPB knockdown has been shown to stimulate autophagosome formation by disrupting the Ca2+ delivery to mitochondria from ER stores (Gómez-Suaga et al., 2017). VAPB P56S and TDP-43 may also co-operate in the pathogenesis of ALS by activating the mitochondrial apoptotic pathway (Suzuki and Matsuoka, 2011). VAPB is also involved in ER-phagy, a selective form of autophagy for degradation of the ER, via interaction with the soluble ER-phagy receptor CALCOCO1 which, via ATG8 binding, connects the ER and autophagosome membranes (seeTable 1) (Nthiga et al., 2020).
VAP in Neurones
Although VAPB is ubiquitously expressed and hence disruption caused by the P56S mutation would affect all cells in the body, ALS8 patients mainly present with (lower) motor neuron dysfunction and neurodegeneration. The large size and complex morphology of motor neurons make the maintenance of protein homeostasis and the distribution of organelles a greater challenge. Hence, motor neurons may be more vulnerable to the overall homeostatic disruption caused by aberrant VAPB. Several studies illustrate how VAPB P56S can affect neuron-specific processes and morphology. For instance, the mutant VAPB disrupts anterograde mitochondrial axonal transport by disrupting Ca2+ homeostasis in neurons (Mórotz et al., 2012). Peroxisomal movement in hippocampal neurones has also been shown to resemble that of mitochondria and be altered by levels of the peroxisome-ER tethering protein ACBD5 (Wang et al., 2018). However, unlike for mitochondria, this did not appear to be dependent upon VAPB interaction. A loss of the VAPB orthologue in Drosophila also resulted in abnormal organelle distribution in neuronal axons and dendrites, including mitochondria and the Golgi apparatus, which may have contributed to the altered dendrite morphology (Kamemura et al., 2021), indicating the importance of ER-tethering in organelle distribution. Furthermore, mitochondria-ER contacts, mediated by the PTPIP51-VAPB interaction, are present at synapses and regulate synaptic function (Gómez-Suaga et al., 2019). Loss of PTPIP51 or VAPB reduced synaptic function and altered dendritic morphology. VAPB P56S also sequesters VAP-interactor YIF1A (Table 1), which regulates membrane trafficking into dendrites and dendritic morphology (Kuijpers et al., 2013b). VAPB is also important for neurite extension of motor neurons (Genevini et al., 2014), possibly via its interaction with protrudin (Table 1) (Saita et al., 2009). Additionally, VAPB P56S led to a loss of HCN channel activity, important for neuronal and cardiac pacemaker currents (Silbernagel et al., 2018). These alterations in motor neurons may partly explain the neurodegeneration and muscle-related symptoms observed in ALS8 patients. VAPB P56S may also affect muscle cells more directly; the VAPB mutation disrupted the formation of multinuclear myotubes (muscle fibres) by mouse skeletal muscle cells (Tokutake et al., 2015b) and caused accumulation of ER Ca2+ sensor STIM1 at neuromuscular junctions (NMJ) in muscle fibres of ALS8 patients, suggesting altered intracellular Ca2+ homeostasis (Goswami et al., 2015). Interestingly, in Drosophila, VAPB regulates the number and size of synaptic boutons at NMJ (Pennetta et al., 2002; Chai et al., 2008). Additionally, VAPB deficient mice showed abnormal skeletal muscle energy metabolism upon fasting (Han et al., 2013). Impaired degradation pathways, accumulation/aggregation of misfolded proteins and disrupted Ca2+ homeostasis in motor neurons and muscle fibres may all contribute to ALS8 pathogenesis.
Acknowledging the various roles VAPB plays in many important physiological pathways, it is not surprising that disruption of the protein has a major effect on cellular homeostasis. Nevertheless, it is still under debate whether the P56S mutation in VAPB induces the symptoms of ALS8 patients by a loss of function (lost/reduced protein interactions), a toxic gain of function (aggregate formation, protein sequestering), or a dominant negative effect (wild-type VAP recruitment). VAPB P56S aggregates in the nervous system of transgenic mice did not cause motor neuron dysfunction, suggesting that aggregates are not sufficient to initiate pathogenesis (Tudor et al., 2010; Qiu et al., 2013), although, with a higher fold increase of VAPB P56S protein expression, mice developed abnormal motor behaviour and progressive degeneration of corticospinal motor neurons (Aliaga et al., 2013). A study using both homozygous and heterozygous VAPB P56S knock-in mice showed defects in motor behaviours, with accumulation of cytoplasmic inclusions selectively in motor neurons before onset of the defects, though the homozygous knock-in mice presented with a more severe phenotype, reflecting a dose-dependent effect of the mutant protein (Larroquette et al., 2015). On the other hand, VAPB knockdown was sufficient to lead to motor deficits in zebrafish and mild, late-onset motor deficits were observed in VAPB knockout mice, however, VAPB depletion was unable to induce a complete ALS phenotype (Kabashi et al., 2013). Thus, VAPB P56S abnormalities might be a combination of gained and lost functions, in a dominant and dose-dependent manner.
Other Mutations in VAPB
A second mutation located in the MSP domain of VAPB has also been associated with familial ALS. An amino acid change from threonine to isoleucine at codon 46 (T46I) was identified in a patient from the United Kingdom, with non-Brazilian kindred–affected family members were not available to screen (Chen et al., 2010). The patient presented with typical ALS, with onset of symptoms at the age of 73 years. Unlike the P65S mutation that completely eliminates the native MSP structure in various buffers (Shi et al., 2010), MSP T46I retains a structure highly similar to the native MSP domain, although with reduced stability (Lua et al., 2011). This makes the MSP domain more easily accessible to unfolded intermediates that are prone to aggregation as shown in vitro, in cultured cells as well as in vivo (Chen et al., 2010; Lua et al., 2011). T46 is part of the hydrophobic pocket that binds the side chain of FFAT motif residue 5 (A) and forms hydrogen bonds with the side chains of FFAT motif residues 2 (F) and 3 (F) (Figure 3C) (Kaiser et al., 2005; Furuita et al., 2010). The threonine to isoleucine substitution induced some dynamic changes of local regions within the MSP domain (Lua et al., 2011). These alterations seem to affect the ability of VAPB to bind FFAT-motif containing proteins, as illustrated with the NIR2 FFAT motif that showed a 3-fold decrease in binding affinity (Chen et al., 2010). Analysis of VAPB T46I in neuronal cells and D. melanogaster indicates similar cellular abnormalities as with the P56S mutation, such as wild-type VAPB sequestering, ER fragmentation and neurodegeneration.
Two VAPB mutations outside of the MSP domain have also been identified in ALS patients. An alanine to valine substitution at codon 145 (A145V) was identified (Kabashi et al., 2013), which is located in the region between the MSP and TMD of VAPB but little else is known about the pathogenicity of A145V. Furthermore, V234I was identified in a patient of Dutch origin, who also harboured a repeat expansion in C9orf72, an ALS causative gene (van Blitterswijk et al., 2012). Transgenic expression of the VAPB V234I orthologue in D. melanogaster was able to induce ALS hallmarks (Sanhueza et al., 2014). The valine to isoleucine substitution is located in the transmembrane domain of VAPB, and although it is close to the dimerization motif, it did not affect VAPB dimerization (Chattopadhyay and Sengupta, 2014). However, the V234I mutation seems to affect the ER-targeting of VAPB as it did not localize with ER-marker PDI. The V234I mutated VAPB did not form typical P56S aggregates, but formed small aggregates/granules in HeLa cells, which may lead to cell death (Chattopadhyay and Sengupta, 2014).
Conclusion
Here, we provided a timely summary of the constantly growing number of VAP interacting proteins, their FFAT motifs (if present) and interaction domains, which will present a helpful overview for future studies on VAP binding partners. We discussed new findings on the regulation of VAP binding by phosphorylation of the FFAT motif core, and the role of GSK3β in the regulation of both mitochondria-ER and peroxisome-ER membrane contact sites. How the interaction of VAP with tether proteins and other interaction partners is regulated, is still not well explored. Future studies may shed light on the regulation of those interactions and their impact on the multiple cellular functions of VAP proteins. An intriguing aspect is also the hijacking of VAP by bacteria and viruses and its role in pathogen infection. It will be interesting to investigate if and how the organelle-specific binding partners are influenced, and if those proteins are suitable new therapeutic targets to combat pathogen infection. Furthermore, the impact of VAP mutations on neurological disorders deserves further investigation. Although our knowledge about VAP and its binding partners at membrane contacts has increased, we do not yet fully understand the (patho)physiological consequences of altered ER-organelle contacts and how this would impact on neurological functions. Thus, VAP proteins and their interacting proteins will remain in the focus of fundamental, discovery-based research as well as biomedical studies.
Statements
Data availability statement
All datasets generated for this study are included in the article.
Author contributions
SK wrote the manuscript and created the figures and table. JC and MS conceived the project and wrote the manuscript.
Funding
This work was supported by the Biotechnology and Biological Sciences Research Council (BBSRC) (BB/T002255/1, BB/V018167/1 to MS and JC), an UK Research and Innovation Future Leader Fellowship Award (MR/T019409/1 to JC) and in part by grant MR/N0137941/1 for the GW4 BIOMED MRC DTP, awarded to the Universities of Bath, Bristol, Cardiff and Exeter from the Medical Research Council (MRC)/UKRI (PhD studentship SK). For the purpose of open access, the author has applied a Creative Commons Attribution (CC BY) licence to any Author Accepted Manuscript version arising.
Acknowledgments
The authors would like to thank R. E. Carmichael (University of Exeter) for critical reading of the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
ALS, amyotrophic lateral sclerosis; FFAT, two phenylalanines (FF) in an acidic tract; MCS, membrane contact sites; MSP, major sperm protein; PI4P, phosphatidylinositol-4-phosphate; TMD, transmembrane domain; VAP, vesicle-associated membrane protein (VAMP)-associated protein.
References
1
Abu IrqebaA.OgilvieJ. M. (2020). Di-arginine and FFAT-like Motifs Retain a Subpopulation of PRA1 at ER-Mitochondria Membrane Contact Sites. PLoS One15, e0243075. 10.1371/JOURNAL.PONE.0243075
2
AdhikariB.De SilvaB.MolinaJ. A.AllenA.PeckS. H.LeeS. Y. (2019). Neuronal Ceroid Lipofuscinosis Related ER Membrane Protein CLN8 Regulates PP2A Activity and Ceramide Levels. Biochimica Biophysica Acta (BBA) - Mol. Basis Dis.1865, 322–328. 10.1016/j.bbadis.2018.11.011
3
AgaisseH.DerréI. (2014). Expression of the Effector Protein IncD in Chlamydia trachomatis Mediates Recruitment of the Lipid Transfer Protein CERT and the Endoplasmic Reticulum-Resident Protein VAPB to the Inclusion Membrane. Infect. Immun.82, 2037–2047. 10.1128/IAI.01530-14
4
AgaisseH.DerréI. (2015). STIM1 Is a Novel Component of ER-Chlamydia trachomatis Inclusion Membrane Contact Sites. PLoS One10, e0125671. 10.1371/journal.pone.0125671
5
AliagaL.LaiC.YuJ.ChubN.ShimH.SunL. (2013). Amyotrophic Lateral Sclerosis-Related VAPB P56S Mutation Differentially Affects the Function and Survival of Corticospinal and Spinal Motor Neurons. Hum. Mol. Genet.22, 4293–4305. 10.1093/hmg/ddt279
6
AlpyF.RousseauA.SchwabY.LegueuxF.StollI.WendlingC.et al (2013). STARD3 or STARD3NL and VAP Form a Novel Molecular Tether between Late Endosomes and the ER. J. Cell Sci.126, 5500–5512. 10.1242/jcs.139295
7
AmarilioR.RamachandranS.SabanayH.LevS. (2005). Differential Regulation of Endoplasmic Reticulum Structure through VAP-Nir Protein Interaction. J. Biol. Chem.280, 5934–5944. 10.1074/jbc.M409566200
8
Amini-Bavil-OlyaeeS.ChoiY. J.LeeJ. H.ShiM.HuangI. C.FarzanM.et al (2013). The Antiviral Effector IFITM3 Disrupts Intracellular Cholesterol Homeostasis to Block Viral Entry. Cell Host Microbe13, 452–464. 10.1016/j.chom.2013.03.006
9
AnagnostouG.AkbarM. T.PaulP.AngelinettaC.SteinerT. J.de BellerocheJ. (2010). Vesicle Associated Membrane Protein B (VAPB) Is Decreased in ALS Spinal Cord. Neurobiol. Aging31, 969–985. 10.1016/j.neurobiolaging.2008.07.005
10
AritaM. (2014). Phosphatidylinositol-4 Kinase III Beta and Oxysterol-Binding Protein Accumulate Unesterified Cholesterol on Poliovirus-Induced Membrane Structure. Microbiol. Immunol.58, 239–256. 10.1111/1348-0421.12144
11
BackmanA. P. E.HalinJ.NurmiH.MöutsA.KjellbergM. A.MattjusP. (2018). Glucosylceramide Acyl Chain Length Is Sensed by the Glycolipid Transfer Protein. PLoS One13, e0209230. 10.1371/journal.pone.0209230
12
BakerE. S.LucknerS. R.KrauseK. L.LambdenP. R.ClarkeI. N.WardV. K. (2012). Inherent Structural Disorder and Dimerisation of Murine Norovirus NS1-2 Protein. PLoS One7, e30534. 10.1371/journal.pone.0030534
13
BaronY.PedrioliP. G.TyagiK.JohnsonC.WoodN. T.FountaineD.et al (2014). VAPB/ALS8 Interacts with FFAT-like Proteins Including the P97 Cofactor FAF1 and the ASNA1 ATPase. BMC Biol.12, 39. 10.1186/1741-7007-12-39
14
BergerK. L.KellyS. M.JordanT. X.TartellM. A.RandallG. (2011). Hepatitis C Virus Stimulates the Phosphatidylinositol 4-Kinase III Alpha-dependent Phosphatidylinositol 4-Phosphate Production that Is Essential for its Replication. J. Virol.85, 8870–8883. 10.1128/JVI.00059-11
15
BullockT. L.RobertsT. M.StewartM. (1996). 2.5 Å Resolution Crystal Structure of the Motile Major Sperm Protein (MSP) ofAscaris Suum. J. Mol. Biol.263, 284–296. 10.1006/JMBI.1996.0575
16
CabukustaB.BerlinI.van ElslandD. M.ForkinkI.SpitsM.de JongA. W. M.et al (2020). Human VAPome Analysis Reveals MOSPD1 and MOSPD3 as Membrane Contact Site Proteins Interacting with FFAT-Related FFNT Motifs. Cell Rep.33, 108475. 10.1016/j.celrep.2020.108475
17
CadoniM. P. L.BiggioM. L.ArruG.SecchiG.OrrùN.ClementeM. G.et al (2020). VAPB ER-Aggregates, a Possible New Biomarker in ALS Pathology. Cells9, 164. 10.3390/cells9010164
18
ChadiG.MaximinoJ. R.JorgeF. M. de. H.BorbaF. C. de.GilioJ. M.CallegaroD.et al (2017). Genetic Analysis of Patients with Familial and Sporadic Amyotrophic Lateral Sclerosis in a Brazilian Research Center. Amyotroph. Lateral Scler. Front. Degener.18, 249–255. 10.1080/21678421.2016.1254245
19
ChaiA.WithersJ.KohY. H.ParryK.BaoH.ZhangB.et al (2008). hVAPB, the Causative Gene of a Heterogeneous Group of Motor Neuron Diseases in Humans, Is Functionally Interchangeable with its Drosophila Homologue DVAP-33A at the Neuromuscular Junction. Hum. Mol. Genet.17, 266–280. 10.1093/hmg/ddm303
20
ChangC. L.HsiehT. S.YangT. T.RothbergK. G.AzizogluD. B.VolkE.et al (2013). Feedback Regulation of Receptor-Induced Ca2+ Signaling Mediated by E-Syt1 and Nir2 at Endoplasmic Reticulum-Plasma Membrane Junctions. Cell Rep.5, 813–825. 10.1016/j.celrep.2013.09.038
21
ChangC. L.LiouJ. (2015). Phosphatidylinositol 4, 5-bisphosphate Homeostasis Regulated by Nir2 and Nir3 Proteins at Endoplasmic Reticulum-Plasma Membrane Junctions. J. Biol. Chem.290, 14289–14301. 10.1074/jbc.M114.621375
22
ChattopadhyayD.SenguptaS. (2014). First Evidence of Pathogenicity of V234I Mutation of hVAPB Found in Amyotrophic Lateral Sclerosis. Biochem. Biophys. Res. Commun.448, 108–113. 10.1016/j.bbrc.2014.04.102
23
ChenH. J.AnagnostouG.ChaiA.WithersJ.MorrisA.AdhikareeJ.et al (2010). Characterization of the Properties of a Novel Mutation in VAPB in Familial Amyotrophic Lateral Sclerosis. J. Biol. Chem.285, 40266–40281. 10.1074/jbc.M110.161398
24
CianciolaN. L.ChungS.ManorD.CarlinC. R. (2017). Adenovirus Modulates Toll-like Receptor 4 Signaling by Reprogramming ORP1L-VAP Protein Contacts for Cholesterol Transport from Endosomes to the Endoplasmic Reticulum. J. Virol.91, 1904–1920. 10.1128/JVI.01904-16
25
ConfortiF. L.SprovieriT.MazzeiR.UngaroC.TessitoreA.TedeschiG.et al (2006). Sporadic ALS Is Not Associated with VAPB Gene Mutations in Southern Italy. J. Negat. Results Biomed.5, 7. 10.1186/1477-5751-5-7
26
CostelloJ. L.CastroI. G.HackerC.SchraderT. A.MetzJ.ZeuschnerD.et al (2017a). ACBD5 and VAPB Mediate Membrane Associations between Peroxisomes and the ER. J. Cell Biol.216, 331–342. 10.1083/jcb.201607055
27
CostelloJ. L.CastroI. G.SchraderT. A.IslingerM.SchraderM. (2017b). Peroxisomal ACBD4 Interacts with VAPB and Promotes ER-Peroxisome Associations. Cell Cycle16, 1039–1045. 10.1080/15384101.2017.1314422
28
DarwischW.von SpangenbergM.LehmannJ.SinginÖ.DeubertG.KühlH.et al (2020). Cerebellar and Hepatic Alterations in ACBD5-Deficient Mice Are Associated with Unexpected, Distinct Alterations in Cellular Lipid Homeostasis. Commun. Biol.3, 1–19. 10.1038/s42003-020-01442-x
29
de AlcântaraC.CruzeiroM. M.FrançaM. C.CamargosS. T.de SouzaL. C. (2019). Amyotrophic Lateral Sclerosis Type 8 Is Not a Pure Motor Disease: Evidence from a Neuropsychological and Behavioural Study. J. Neurol.266, 1980–1987. 10.1007/s00415-019-09369-y
30
De VosK. J.MórotzG. M.StoicaR.TudorE. L.LauK. F.AckerleyS.et al (2012). VAPB Interacts with the Mitochondrial Protein PTPIP51 to Regulate Calcium Homeostasis. Hum. Mol. Genet.21, 1299–1311. 10.1093/hmg/ddr559
31
DeiddaI.GalizziG.PassantinoR.CascioC.RussoD.CollettiT.et al (2014). Expression of Vesicle-Associated Membrane-Protein-Associated Protein B Cleavage Products in Peripheral Blood Leukocytes and Cerebrospinal Fluid of Patients with Sporadic Amyotrophic Lateral Sclerosis. Eur. J. Neurol.21, 478–485. 10.1111/ENE.12334
32
DerréI.SwissR.AgaisseH. (2011). The Lipid Transfer Protein CERT Interacts with the Chlamydia Inclusion Protein IncD and Participates to ER-Chlamydia Inclusion Membrane Contact Sites. PLoS Pathog.7, e1002092. 10.1371/journal.ppat.1002092
33
DiL.ChenH.DaY.WangS.ShenX. M. (2016). Atypical Familial Amyotrophic Lateral Sclerosis with Initial Symptoms of Pain or Tremor in a Chinese Family Harboring VAPB-P56s Mutation. J. Neurol.263, 263–268. 10.1007/s00415-015-7965-3
34
Di MattiaT.MartinetA.IkhlefS.McEwenA. G.NominéY.WendlingC.et al (2020). FFAT Motif Phosphorylation Controls Formation and Lipid Transfer Function of Inter‐organelle Contacts. EMBO J.39, e104369. 10.15252/embj.2019104369
35
DongR.SahekiY.SwarupS.LucastL.HarperJ. W.De CamilliP. (2016). Endosome-ER Contacts Control Actin Nucleation and Retromer Function through VAP-dependent Regulation of PI4P. Cell166, 408–423. 10.1016/j.cell.2016.06.037
36
DorobantuC. M.AlbulescuL.HarakC.FengQ.van KampenM.StratingJ. R. P. M.et al (2015). Modulation of the Host Lipid Landscape to Promote RNA Virus Replication: The Picornavirus Encephalomyocarditis Virus Converges on the Pathway Used by Hepatitis C Virus. PLoS Pathog.11, e1005185. 10.1371/JOURNAL.PPAT.1005185
37
EdenE. R.Sanchez-HerasE.TsaparaA.SobotaA.LevineT. P.FutterC. E. (2016). Annexin A1 Tethers Membrane Contact Sites that Mediate ER to Endosome Cholesterol Transport. Dev. Cell37, 473–483. 10.1016/J.DEVCEL.2016.05.005
38
ElwellC. A.JiangS.KimJ. H.LeeA.WittmannT.HanadaK.et al (2011). Chlamydia trachomatis Co-opts GBF1 and CERT to Acquire Host Sphingomyelin for Distinct Roles during Intracellular Development. PLoS Pathog.7, e1002198. 10.1371/journal.ppat.1002198
39
EttayebiK.HardyM. E. (2003). Norwalk Virus Nonstructural Protein P48 Forms a Complex with the SNARE Regulator VAP-A and Prevents Cell Surface Expression of Vesicular Stomatitis Virus G Protein. J. Virol.77, 11790–11797. 10.1128/jvi.77.21.11790-11797.2003
40
EvansM. J.RiceC. M.GoffS. P. (2004). Phosphorylation of Hepatitis C Virus Nonstructural Protein 5A Modulates its Protein Interactions and Viral RNA Replication. Proc. Natl. Acad. Sci.101, 13038–13043. 10.1073/PNAS.0405152101
41
FasanaE.FossatiM.RuggianoA.BrambillascaS.HoogenraadC. C.NavoneF.et al (2010). A VAPB Mutant Linked to Amyotrophic Lateral Sclerosis Generates a Novel Form of Organized Smooth Endoplasmic Reticulum. FASEB J.24, 1419–1430. 10.1096/fj.09-147850
42
FerdinandusseS.FalkenbergK. D.KosterJ.MooyerP. A.JonesR.van RoermundC. W. T.et al (2017). ACBD5 Deficiency Causes a Defect in Peroxisomal Very Long-Chain Fatty Acid Metabolism. J. Med. Genet.54, 330–337. 10.1136/jmedgenet-2016-104132
43
FreyreC. A. C.RauherP. C.EjsingC. S.KlemmR. W. (2019). MIGA2 Links Mitochondria, the ER, and Lipid Droplets and Promotes De Novo Lipogenesis in Adipocytes. Mol. Cell76, 811–825. e14. 10.1016/j.molcel.2019.09.011
44
FunkeA. D.EsserM.KrüttgenA.WeisJ.Mitne-NetoM.LazarM.et al (2010). The p.P56S Mutation in the VAPB Gene Is Not Due to a Single Founder: The First European Case. Clin. Genet.77, 302–303. 10.1111/j.1399-0004.2009.01319.x
45
FuruitaK.JeeJ. G.FukadaH.MishimaM.KojimaC. (2010). Electrostatic Interaction between Oxysterol-Binding Protein and VAMP-Associated Protein a Revealed by NMR and Mutagenesis Studies. J. Biol. Chem.285, 12961–12970. 10.1074/jbc.M109.082602
46
GabettaV.TrzynaW.PhielC.McHughK. M. (2003). Vesicle-associated Protein-A Is Differentially Expressed during Intestinal Smooth Muscle Cell Differentiation. Dev. Dyn.228, 11–20. 10.1002/dvdy.10349
47
GaoL.AizakiH.HeJ.-W.LaiM. M. C. (2004). Interactions between Viral Nonstructural Proteins and Host Protein hVAP-33 Mediate the Formation of Hepatitis C Virus RNA Replication Complex on Lipid Raft. J. Virol.78, 3480–3488. 10.1128/JVI.78.7.3480-3488.2004
48
GeneviniP.PapianiG.RuggianoA.CantoniL.NavoneF.BorgeseN. (2014). Amyotrophic Lateral Sclerosis-Linked Mutant VAPB Inclusions Do Not Interfere with Protein Degradation Pathways or Intracellular Transport in a Cultured Cell Model. PLoS One9, e113416. 10.1371/journal.pone.0113416
49
GewaidH.AoyagiH.AritaM.WatashiK.SuzukiR.SakaiS.et al (2020). Sphingomyelin Is Essential for the Structure and Function of the Double-Membrane Vesicles in Hepatitis C Virus RNA Replication Factories. J. Virol.94, e01080–20. 10.1128/JVI.01080-20
50
GhoshS.PatelA. M.GrunkemeyerT. J.DumbrepatilA. B.ZegaliaK.KennedyR. T.et al (2020). Interactions between Viperin, Vesicle-Associated Membrane Protein A, and Hepatitis C Virus Protein NS5A Modulate Viperin Activity and NS5A Degradation. Biochemistry59, 780–789. 10.1021/acs.biochem.9B01090
51
GkogkasC.MiddletonS.KremerA. M.WardropeC.HannahM.GillingwaterT. H.et al (2008). VAPB Interacts with and Modulates the Activity of ATF6. Hum. Mol. Genet.17, 1517–1526. 10.1093/hmg/ddn040
52
GkogkasC.WardropeC.HannahM.SkehelP. (2011). The ALS8-Associated Mutant VAPBP56S Is Resistant to Proteolysis in Neurons. J. Neurochem.117, 286–294. 10.1111/j.1471-4159.2011.07201.x
53
Gómez-SuagaP.Gábor|MórotzM.MarkovinovicA.Martín-GuerreroS. M.PrezaE.et al (2022). Disruption of ER-Mitochondria Tethering and Signalling in C9orf72-Associated Amyotrophic Lateral Sclerosis and Frontotemporal Dementia. Aging Cell00, e13549. 10.1111/ACEL.13549
54
Gómez-SuagaP.PaillussonS.StoicaR.NobleW.HangerD. P.MillerC. C. J. (2017). The ER-Mitochondria Tethering Complex VAPB-PTPIP51 Regulates Autophagy. Curr. Biol.27, 371–385. 10.1016/J.CUB.2016.12.038
55
Gómez-SuagaP.Pérez-NievasB. G.GlennonE. B.LauD. H. W.PaillussonS.MórotzG. M.et al (2019). The VAPB-PTPIP51 Endoplasmic Reticulum-Mitochondria Tethering Proteins Are Present in Neuronal Synapses and Regulate Synaptic Activity. Acta Neuropathol. Commun.7, 35. 10.1186/S40478-019-0688-4
56
GoonawardaneN.GebhardtA.BartlettC.PichlmairA.HarrisM. (2017). Phosphorylation of Serine 225 in Hepatitis C Virus NS5A Regulates Protein-Protein Interactions. J. Virol.91, e00805–17. 10.1128/JVI.00805-17
57
GorkhaliR.TianL.DongB.BagchiP.DengX.PawarS.et al (2021). Extracellular Calcium Alters Calcium-Sensing Receptor Network Integrating Intracellular Calcium-Signaling and Related Key Pathway. Sci. Rep.11, 20576. 10.1038/S41598-021-00067-2
58
GoswamiA.JesseC. M.ChandrasekarA.BushuvenE.VollrathJ. T.DreserA.et al (2015). Accumulation of STIM1 Is Associated with the Degenerative Muscle Fibre Phenotype in ALS and Other Neurogenic Atrophies. Neuropathol. Appl. Neurobiol.41, 304–318. 10.1111/NAN.12164
59
GoyalS.GuptaG.QinH.UpadyaM. H.TanY. J.ChowV. T. K.et al (2012). VAPC, an Human Endogenous Inhibitor for Hepatitis C Virus (HCV) Infection, Is Intrinsically Unstructured but Forms a “Fuzzy Complex” with HCV NS5B. PLoS One7, e40341. 10.1371/JOURNAL.PONE.0040341
60
GuberR. D.SchindlerA. B.BudronM. S.ChenK. L.LiY.FischbeckK. H.et al (2018). Nucleocytoplasmic Transport Defect in a North American Patient with ALS8. Ann. Clin. Transl. Neurol.5, 369–375. 10.1002/ACN3.515
61
Guillén-SamanderA.LeonzinoM.HannaM. G.TangN.ShenH.De CamilliP. (2021). VPS13D Bridges the ER to Mitochondria and Peroxisomes via Miro. J. Cell Biol.220, e202010004. 10.1083/jcb.202010004
62
GuoX.GangQ.MengL.LiJ.LiF.LiuJ.et al (2020). Peripheral Nerve Pathology in VAPB-Associated Amyotrophic Lateral Sclerosis with Dysautonomia in a Chinese Family. Clin. Neuropathol.39, 282–287. 10.5414/NP301281
63
GuptaG.QinH.SongJ. (2012). Intrinsically Unstructured Domain 3 of Hepatitis C Virus NS5A Forms a “Fuzzy Complex” with VAPB-MSP Domain Which Carries ALS-Causing Mutations. PLoS One7, e39261. 10.1371/JOURNAL.PONE.0039261
64
GuptaG.SongJ. (2016). C-terminal Auto-Regulatory Motif of Hepatitis C Virus NS5B Interacts with Human VAPB-MSP to Form a Dynamic Replication Complex. PLoS One11, e0147278. 10.1371/JOURNAL.PONE.0147278
65
HamamotoI.NishimuraY.OkamotoT.AizakiH.LiuM.MoriY.et al (2005). Human VAP-B Is Involved in Hepatitis C Virus Replication through Interaction with NS5A and NS5B. J. Virol.79, 13473–13482. 10.1128/JVI.79.21.13473-13482.2005
66
HanS. M.El OussiniH.Scekic-ZahirovicJ.VibbertJ.CotteeP.PrasainJ. K.et al (2013). VAPB/ALS8 MSP Ligands Regulate Striated Muscle Energy Metabolism Critical for Adult Survival in Caenorhabditis elegans. PLoS Genet.9, e1003738. 10.1371/JOURNAL.PGEN.1003738
67
HanS. M.TsudaH.YangY.VibbertJ.CotteeP.LeeS. J.et al (2012). Secreted VAPB/ALS8 Major Sperm Protein Domains Modulate Mitochondrial Localization and Morphology via Growth Cone Guidance Receptors. Dev. Cell22, 348–362. 10.1016/J.DEVCEL.2011.12.009
68
HantanD.YamamotoY.SakisakaT. (2014). VAP-B Binds to Rab3GAP1 at the ER: Its Implication in Nuclear Envelope Formation through the ER-Golgi Intermediate Compartment. Kobe J. Med. Sci.60, 48–56. 10.24546/81008726
69
HäslerS. L. A.VallisY.PascheM.McMahonH. T. (2020). GRAF2, WDR44, and MICAL1 Mediate Rab8/10/11–dependent Export of E-Cadherin, MMP14, and CFTR ΔF508. J. Cell Biol.219, e201811014. 10.1083/JCB.201811014/151714
70
HerzogK.Pras-RavesM. L.FerdinandusseS.VervaartM. A. T.LuyfA. C. M.van KampenA. H. C.et al (2018). Functional Characterisation of Peroxisomal β-oxidation Disorders in Fibroblasts Using Lipidomics. J. Inherit. Metab. Dis.41, 479–487. 10.1007/s10545-017-0076-9
71
HsiehJ.MoluskyM. M.McCabeK. M.FotakisP.XiaoT.TascauL.et al (2021). TTC39B Destabilizes Retinoblastoma Protein Promoting Hepatic Lipogenesis in a Sex-specific Fashion. J. Hepatol.76, 383–393. 10.1016/J.JHEP.2021.09.021
72
HuaR.ChengD.CoyaudÉ.FreemanS.Di PietroE.WangY.et al (2017). VAPs and ACBD5 Tether Peroxisomes to the ER for Peroxisome Maintenance and Lipid Homeostasis. J. Cell Biol.216, 367–377. 10.1083/jcb.201608128
73
HuangM. H.LiH.XueR.LiJ.WangL.ChengJ.et al (2019). Up-regulation of Glycolipid Transfer Protein by Bicyclol Causes Spontaneous Restriction of Hepatitis C Virus Replication. Acta Pharm. Sin. B9, 769–781. 10.1016/J.APSB.2019.01.013
74
IngreC.PintoS.BirveA.PressR.DanielssonO.De CarvalhoM.et al (2013). No Association between VAPB Mutations and Familial or Sporadic ALS in Sweden, Portugal and Iceland. Amyotroph. Lateral Scler. Front. Degener.14, 620–627. 10.3109/21678421.2013.822515
75
InukaiR.MoriK.KuwataK.SuzukiC.MakiM.TakaharaT.et al (2021). The Novel ALG-2 Target Protein CDIP1 Promotes Cell Death by Interacting with ESCRT-I and VAPA/B. Int. J. Mol. Sci.22, 1175. 10.3390/ijms22031175
76
Ishikawa-SasakiK.NagashimaS.TaniguchiK.SasakiJ. (2018). Model of OSBP-Mediated Cholesterol Supply to Aichi Virus RNA Replication Sites Involving Protein-Protein Interactions Among Viral Proteins, ACBD3, OSBP, VAP-A/B, and SAC1. J. Virol.92, 1952–1969. 10.1128/JVI.01952-17
77
ItoN.TakahashiT.ShiibaI.NagashimaS.InatomeR.YanagiS. (2021). MITOL Regulates Phosphatidic Acid-Binding Activity of RMDN3/PTPIP51. J. Biochem.1, mvab153. 10.1093/JB/MVAB153
78
JohnsonB.LeekA. N.SoléL.MaverickE. E.LevineT. P.TamkunM. M. (2018). Kv2 Potassium Channels Form Endoplasmic Reticulum/plasma Membrane Junctions via Interaction with VAPA and VAPB. Proc. Natl. Acad. Sci.115, E7331–E7340. 10.1073/pnas.1805757115
79
JustisA. V.HansenB.BeareP. A.KingK. B.HeinzenR. A.GilkS. D. (2017). Interactions between the Coxiella Burnetii Parasitophorous Vacuole and the Endoplasmic Reticulum Involve the Host Protein ORP1L. Cell. Microbiol.19, e12637. 10.1111/CMI.12637
80
KabashiE.El OussiniH.BercierV.Gros-LouisF.ValdmanisP. N.McdearmidJ.et al (2013). Investigating the Contribution of VAPB/ALS8 Loss of Function in Amyotrophic Lateral Sclerosis. Hum. Mol. Genet.22, 2350–2360. 10.1093/HMG/DDT080
81
KaiserS. E.BricknerJ. H.ReileinA. R.FennT. D.WalterP.BrungerA. T. (2005). Structural Basis of FFAT Motif-Mediated ER Targeting. Structure13, 1035–1045. 10.1016/j.str.2005.04.010
82
KamemuraK.ChenC. A.OkumuraM.MiuraM.ChiharaT. (2021). Amyotrophic Lateral Sclerosis-Associated Vap33 Is Required for Maintaining Neuronal Dendrite Morphology and Organelle Distribution in Drosophila. Genes Cells26, 230–239. 10.1111/GTC.12835
83
KamemuraK.ChiharaT. (2019). Multiple Functions of the ER-Resident VAP and its Extracellular Role in Neural Development and Disease. J. Biochem.165, 391–400. 10.1093/jb/mvz011
84
KanekuraK.NishimotoI.AisoS.MatsuokaM. (2006). Characterization of Amyotrophic Lateral Sclerosis-Linked P56S Mutation of Vesicle-Associated Membrane Protein-Associated Protein B (VAPB/ALS8). J. Biol. Chem.281, 30223–30233. 10.1074/jbc.M605049200
85
KawanoM.KumagaiK.NishijimaM.HanadaK. (2006). Efficient Trafficking of Ceramide from the Endoplasmic Reticulum to the Golgi Apparatus Requires a VAMP-Associated Protein-Interacting FFAT Motif of CERT. J. Biol. Chem.281, 30279–30288. 10.1074/jbc.M605032200
86
KennaK. P.McLaughlinR. L.ByrneS.ElaminM.HeverinM.KennyE. M.et al (2013). Delineating the Genetic Heterogeneity of ALS Using Targeted High-Throughput Sequencing. J. Med. Genet.50, 776–783. 10.1136/jmedgenet-2013-101795
87
KimS. H.LealS. S.HalevyD. Ben.GomesC. M.LevS. (2010). Structural Requirements for VAP-B Oligomerization and Their Implication in Amyotrophic Lateral Sclerosis-Associated VAP-B(P56S) Neurotoxicity. J. Biol. Chem.285, 13839–13849. 10.1074/jbc.M109.097345
88
KirbyJ.HewamaddumaC. A. A.HartleyJ. A.NixonH. C.EvansH.WadhwaR. R.et al (2007). Mutations in VAPB Are Not Associated with Sporadic ALS. Neurology68, 1951–1953. 10.1212/01.wnl.0000263195.50981.a6
89
KirmizM.GilliesT. E.DicksonE. J.TrimmerJ. S. (2019). Neuronal ER-Plasma Membrane Junctions Organized by Kv2-VAP Pairing Recruit Nir Proteins and Affect Phosphoinositide Homeostasis. J. Biol. Chem.294, 17735–17757. 10.1074/jbc.RA119.007635
90
KirmizM.VierraN. C.PalacioS.TrimmerJ. S. (2018). Identification of VAPA and VAPB as Kv2 Channel-Interacting Proteins Defining Endoplasmic Reticulum-Plasma Membrane Junctions in Mammalian Brain Neurons. J. Neurosci.38, 7562–7584. 10.1523/JNEUROSCI.0893-18.2018
91
KorsS.HackerC.BoltonC.MaierR.ReimannL.KitchenerE. J. A.et al (2022). Regulating Peroxisome–ER Contacts via the ACBD5-VAPB Tether by FFAT Motif Phosphorylation and GSK3β. J. Cell Biol.221, e202003143. 10.1083/JCB.202003143
92
KosacV.de FreitasM. R. G.PradoF. M.NascimentoO. J. M.BittarC. (2013). Familial Adult Spinal Muscular Atrophy Associated with the VAPB Gene: Report of 42 Cases in Brazil. Arq. Neuropsiquiatr.71, 788–790. 10.1590/0004-282X20130123
93
KuijpersM.van DisV.HaasdijkE. D.HarterinkM.VockingK.PostJ. A.et al (2013a). Amyotrophic Lateral Sclerosis (ALS)-associated VAPB-P56s Inclusions Represent an ER Quality Control Compartment. Acta Neuropathol. Commun.2, 1–19. 10.1186/2051-5960-1-24
94
KuijpersM.YuLouK.TeulingE.AkhmanovaA.JaarsmaD.HoogenraadC. C. (2013b). The ALS8 Protein VAPB Interacts with the ER–Golgi Recycling Protein YIF1A and Regulates Membrane Delivery into Dendrites. EMBO J.32, 2056–2072. 10.1038/emboj.2013.131
95
KukiharaH.MoriishiK.TaguwaS.TaniH.AbeT.MoriY.et al (2009). Human VAP-C Negatively Regulates Hepatitis C Virus Propagation. J. Virol.83, 7959–7969. 10.1128/JVI.00889-09
96
KumagaiK.ElwellC. A.AndoS.EngelJ. N.HanadaK. (2018). Both the N- and C- Terminal Regions of the Chlamydial Inclusion Protein D (IncD) Are Required for Interaction with the Pleckstrin Homology Domain of the Ceramide Transport Protein CERT. Biochem. Biophys. Res. Commun.505, 1070–1076. 10.1016/j.bbrc.2018.09.168
97
KumagaiK.Kawano-KawadaM.HanadaK. (2014). Phosphoregulation of the Ceramide Transport Protein CERT at Serine 315 in the Interaction with VAMP-Associated Protein (VAP) for Inter-organelle Trafficking of Ceramide in Mammalian Cells. J. Biol. Chem.289, 10748–10760. 10.1074/jbc.M113.528380
98
KumarN.LeonzinoM.Hancock-CeruttiW.HorenkampF. A.LiP. Q.LeesJ. A.et al (2018). VPS13A and VPS13C Are Lipid Transport Proteins Differentially Localized at ER Contact Sites. J. Cell Biol.217, 3625–3639. 10.1083/JCB.201807019
99
Kun-RodriguesC.GanosC.GuerreiroR.SchneiderS. A.SchulteC.LesageS.et al (2015). A Systematic Screening to Identify De Novo Mutations Causing Sporadic Early-Onset Parkinson’s Disease. Hum. Mol. Genet.24, 6711–6720. 10.1093/HMG/DDV376
100
LarroquetteF.SetoL.GaubP. L.KamalB.WallisD.LarivièreR.et al (2015). Vapb/Amyotrophic Lateral Sclerosis 8 Knock-In Mice Display Slowly Progressive Motor Behavior Defects Accompanying ER Stress and Autophagic Response. Hum. Mol. Genet.24, 6515–6529. 10.1093/HMG/DDV360
101
LauD. H. W.PaillussonS.HartoppN.RupawalaH.MórotzG. M.Gomez-SuagaP.et al (2020). Disruption of Endoplasmic Reticulum-Mitochondria Tethering Proteins in Post-mortem Alzheimer’s Disease Brain. Neurobiol. Dis.143, 105020. 10.1016/J.NBD.2020.105020
102
LehtoM.HynynenR.KarjalainenK.KuismanenE.HyvärinenK.OlkkonenV. M. (2005). Targeting of OSBP-Related Protein 3 (ORP3) to Endoplasmic Reticulum and Plasma Membrane Is Controlled by Multiple Determinants. Exp. Cell Res.310, 445–462. 10.1016/j.yexcr.2005.08.003
103
LeoniT. B.RezendeT. J. R.PeluzzoT. M.MartinsM. P.NetoA. R. C.Gonzalez-SalazarC.et al (2022). Structural Brain and Spinal Cord Damage in Symptomatic and Pre-symptomatic VAPB-Related ALS. J. Neurol. Sci.434, 120126. 10.1016/J.JNS.2021.120126
104
LevS.Ben HalevyD.PerettiD.DahanN. (2008). The VAP Protein Family: From Cellular Functions to Motor Neuron Disease. Trends Cell Biol.18, 282–290. 10.1016/j.tcb.2008.03.006
105
Levin-KonigsbergR.Montaño-RendónF.Keren-KaplanT.LiR.EgoB.MylvaganamS.et al (2019). Phagolysosome Resolution Requires Contacts with the Endoplasmic Reticulum and Phosphatidylinositol-4-Phosphate Signalling. Nat. Cell Biol.21, 1234–1247. 10.1038/s41556-019-0394-2
106
LimC. Y.DavisO. B.ShinH. R.ZhangJ.BerdanC. A.JiangX.et al (2019). ER–lysosome Contacts Enable Cholesterol Sensing by mTORC1 and Drive Aberrant Growth Signalling in Niemann–Pick Type C. Nat. Cell Biol.21, 1206–1218. 10.1038/s41556-019-0391-5
107
LindhoutF. W.CaoY.KevenaarJ. T.BodzętaA.StucchiR.BoumpoutsariM. M.et al (2019). VAP‐SCRN1 Interaction Regulates Dynamic Endoplasmic Reticulum Remodeling and Presynaptic Function. EMBO J.38, e101345. 10.15252/embj.2018101345
108
LiuH.LinW.-Y.LeibowS. R.MorateckA. J.AhujaM.MuallemS. (2022). TRPC3 Channel Gating by Lipids Requires Localization at the ER/PM Junctions Defined by STIM1. J. Cell Biol.221, e202107120. 10.1083/JCB.202107120
109
LoewenC. J. R.RoyA.LevineT. P. (2003). A Conserved ER Targeting Motif in Three Families of Lipid Binding Proteins and in Opi1p Binds VAP. EMBO J.22, 2025–2035. 10.1093/emboj/cdg201
110
LuaS.QinH.LimL.ShiJ.GuptaG.SongJ. (2011). Structural, Stability, Dynamic and Binding Properties of the ALS-Causing T46I Mutant of the hVAPB MSP Domain as Revealed by NMR and MD Simulations. PLoS One6, e27072. 10.1371/journal.pone.0027072
111
MarquesV. D.BarreiraA. A.DavisM. B.Abou-SleimanP. M.SilvaW. A.ZagoM. A.et al (2006). Expanding the Phenotypes of the Pro56SerVAPB Mutation: Proximal SMA with Dysautonomia. Muscle Nerve34, 731–739. 10.1002/mus.20657
112
MatsumotoG.StojanovicA.HolmbergC. I.KimS.MorimotoR. I. (2005). Structural Properties and Neuronal Toxicity of Amyotrophic Lateral Sclerosis-Associated Cu/Zn Superoxide Dismutase 1 Aggregates. J. Cell Biol.171, 75–85. 10.1083/jcb.200504050
113
MatsuzakiF.ShiraneM.MatsumotoM.NakayamaK. I. (2011). Protrudin Serves as an Adaptor Molecule that Connects KIF5 and its Cargoes in Vesicular Transport during Process Formation. Mol. Biol. Cell22, 4602–4620. 10.1091/mbc.e11-01-0068
114
MattiaT.WilhelmL. P.IkhlefS.WendlingC.SpehnerD.NominéY.et al (2018). Identification of MOSPD2, a Novel Scaffold for Endoplasmic Reticulum Membrane Contact Sites. EMBO Rep.19, e45453. 10.15252/EMBR.201745453
115
McCuneB. T.TangW.LuJ.EagleshamJ. B.ThorneL.MayerA. E.et al (2017). Noroviruses Co-opt the Function of Host Proteins VAPA and VAPB for Replication via a Phenylalanine–Phenylalanine- Acidic-Tract-Motif Mimic in Nonstructural Viral Protein NS1/2. MBio8, 668–685. 10.1128/mBio.00668-17
116
MesminB.BigayJ.Moser Von FilseckJ.Lacas-GervaisS.DrinG.AntonnyB. (2013). A Four-step Cycle Driven by PI(4)P Hydrolysis Directs sterol/PI(4)P Exchange by the ER-Golgi Tether OSBP. Cell155, 830–843. 10.1016/j.cell.2013.09.056
117
MikitovaV.LevineT. P. (2012). Analysis of the Key Elements of FFAT-like Motifs Identifies New Proteins that Potentially Bind VAP on the ER, Including Two AKAPs and FAPP2. PLoS One7, e30455. 10.1371/journal.pone.0030455
118
MillecampsS.SalachasF.CazeneuveC.GordonP.BrickaB.CamuzatA.et al (2010). SOD1, ANG, VAPB, TARDBP, and FUS Mutations in Familial Amyotrophic Lateral Sclerosis: Genotype-Phenotype Correlations. J. Med. Genet.47, 554–560. 10.1136/jmg.2010.077180
119
Mitne-NetoM.Machado-CostaM.MarchettoM. C. N.BengtsonM. H.JoazeiroC. A.TsudaH.et al (2011). Downregulation of VAPB Expression in Motor Neurons Derived from Induced Pluripotent Stem Cells of ALS8 Patients. Hum. Mol. Genet.20, 3642–3652. 10.1093/hmg/ddr284
120
MochizukiS.MikiH.ZhouR.KidoY.NishimuraW.KikuchiM.et al (2018). Oxysterol-binding Protein-Related Protein (ORP) 6 Localizes to the ER and ER-Plasma Membrane Contact Sites and Is Involved in the Turnover of PI4P in Cerebellar Granule Neurons. Exp. Cell Res.370, 601–612. 10.1016/J.YEXCR.2018.07.025
121
MoriF.MikiY.TanjiK.KonT.TomiyamaM.KakitaA.et al (2021). Role of VAPB and Vesicular Profiles in α-synuclein Aggregates in Multiple System Atrophy. Brain Pathol.31, e13001. 10.1111/BPA.13001
122
MórotzG. M.De VosK. J.VagnoniA.AckerleyS.ShawC. E.MillerC. C. J. (2012). Amyotrophic Lateral Sclerosis-Associated Mutant VAPBP56S Perturbs Calcium Homeostasis to Disrupt Axonal Transport of Mitochondria. Hum. Mol. Genet.21, 1979–1988. 10.1093/HMG/DDS011
123
MoumenA.VirardI.RaoulC. (2011). Accumulation of Wildtype and ALS-Linked Mutated VAPB Impairs Activity of the Proteasome. PLoS One6, e26066. 10.1371/journal.pone.0026066
124
MurphyS. E.LevineT. P. (2016). VAP, a Versatile Access Point for the Endoplasmic Reticulum: Review and Analysis of FFAT-like Motifs in the VAPome. Biochim. Biophys. Acta - Mol. Cell Biol. Lipids1861, 952–961. 10.1016/j.bbalip.2016.02.009
125
NachreinerT.EsserM.TentenV.TroostD.WeisJ.KrüttgenA. (2010). Novel Splice Variants of the Amyotrophic Lateral Sclerosis-Associated Gene VAPB Expressed in Human Tissues. Biochem. Biophys. Res. Commun.394, 703–708. 10.1016/j.bbrc.2010.03.055
126
NakamichiS.YamanakaK.SuzukiM.WatanabeT.KagiwadaS. (2011). Human VAPA and the Yeast VAP Scs2p with an Altered Proline Distribution Can Phenocopy Amyotrophic Lateral Sclerosis-Associated VAPB(P56S). Biochem. Biophys. Res. Commun.404, 605–609. 10.1016/j.bbrc.2010.12.011
127
NgoM.RidgwayN. D. (2009). Oxysterol Binding Protein-Related Protein 9 (ORP9) Is a Cholesterol Transfer Protein that Regulates Golgi Structure and Function. Mol. Biol. Cell20, 1388–1399. 10.1091/mbc.E08-09-0905
128
NguyenP. H.LutterE. I.HackstadtT. (2018). Chlamydia trachomatis Inclusion Membrane Protein MrcA Interacts with the Inositol 1,4,5-trisphosphate Receptor Type 3 (ITPR3) to Regulate Extrusion Formation. PLoS Pathog.14, e1006911. 10.1371/journal.ppat.1006911
129
NishimuraA. L.Al-ChalabiA.ZatzM. (2005). A Common Founder for Amyotrophic Lateral Sclerosis Type 8 (ALS8) in the Brazilian Population. Hum. Genet.118, 499–500. 10.1007/s00439-005-0031-y
130
NishimuraA. L.Mitne-NetoM.SilvaH. C. A.OliveiraJ. R. M.VainzofM.ZatzM. (2004a). A Novel Locus for Late Onset Amyotrophic Lateral Sclerosis/motor Neurone Disease Variant at 20q13. J. Med. Genet.41, 315–320. 10.1136/jmg.2003.013029
131
NishimuraA. L.Mitne-NetoM.SilvaH. C. A.Richieri-CostaA.MiddletonS.CascioD.et al (2004b). A Mutation in the Vesicle-Trafficking Protein VAPB Causes Late-Onset Spinal Muscular Atrophy and Amyotrophic Lateral Sclerosis. Am. J. Hum. Genet.75, 822–831. 10.1086/425287
132
NishimuraY.HayashiM.InadaH.TanakaT. (1999). Molecular Cloning and Characterization of Mammalian Homologues of Vesicle-Associated Membrane Protein-Associated (VAMP-Associated) Proteins. Biochem. Biophys. Res. Commun.254, 21–26. 10.1006/bbrc.1998.9876
133
NthigaT. M.Kumar ShresthaB.SjøttemE.BruunJ.Bowitz LarsenK.BhujabalZ.et al (2020). CALCOCO 1 Acts with VAMP ‐associated Proteins to Mediate ER ‐phagy. EMBO J.39, e103649. 10.15252/embj.2019103649
134
Nunes GonçalvesJ. P.LeoniT. B.MartinsM. P.PeluzzoT. M.DouradoM. E. T.SauteJ. A. M.et al (2021). Genetic Epidemiology of Familial ALS in Brazil. Neurobiol. Aging102, 227. 10.1016/j.neurobiolaging.2021.01.007
135
OliveiraD.Morales-VicenteD. A.AmaralM. S.LuzL.SertiéA. L.LeiteF. S.et al (2020). Different Gene Expression Profiles in iPSC-Derived Motor Neurons from ALS8 Patients with Variable Clinical Courses Suggest Mitigating Pathways for Neurodegeneration. Hum. Mol. Genet.29, 1465–1475. 10.1093/hmg/ddaa069
136
PaillussonS.Gomez-SuagaP.StoicaR.LittleD.GissenP.DevineM. J.et al (2017). α-Synuclein Binds to the ER–Mitochondria Tethering Protein VAPB to Disrupt Ca2+ Homeostasis and Mitochondrial ATP Production. Acta Neuropathol.134, 129–149. 10.1007/s00401-017-1704-z
137
PanG.CaoX.LiuB.LiC.LiD.ZhengJ.et al (2018). OSBP-related Protein 4L Promotes Phospholipase Cβ3 Translocation from the Nucleus to the Plasma Membrane in Jurkat T-Cells. J. Biol. Chem.293, 17430–17441. 10.1074/jbc.RA118.005437
138
PapianiG.RuggianoA.FossatiM.RaimondiA.BertoniG.FrancoliniM.et al (2012). Restructured Endoplasmic Reticulum Generated by Mutant Amyotrophic Lateral Sclerosis-Linked VAPB Is Cleared by the Proteasome. J. Cell Sci.125, 3601–3611. 10.1242/jcs.102137
139
PassantinoR.CascioC.DeiddaI.GalizziG.RussoD.SpedaleG.et al (2013). Identifying Protein Partners of CLN8, an ER-Resident Protein Involved in Neuronal Ceroid Lipofuscinosis. Biochim. Biophys. Acta - Mol. Cell Res.1833, 529–540. 10.1016/j.bbamcr.2012.10.030
140
PennettaG.HiesingerP. R.Fabian-FineR.MeinertzhagenI. A.BellenH. J. (2002). Drosophila VAP-33A Directs Bouton Formation at Neuromuscular Junctions in a Dosage-dependent Manner. Neuron35, 291–306. 10.1016/S0896-6273(02)00769-9
141
PerettiD.DahanN.ShimoniE.HirschbergK.LevS. (2008). Coordinated Lipid Transfer between the Endoplasmic Reticulum and the Golgi Complex Requires the VAP Proteins and Is Essential for Golgi-Mediated Transport. Mol. Biol. Cell19, 3871–3844. 10.1091/mbc.E0810.1091/mbc.e08-05-0498
142
PesaolaF.QuassolloG.VenierA. C.De PaulA. L.NoherI.BisbalM. (2021). The Neuronal Ceroid Lipofuscinosis-Related Protein CLN8 Regulates Endo-Lysosomal Dynamics and Dendritic Morphology. Biol. Cell113, 419–437. 10.1111/BOC.202000016
143
PetrovaV.PearsonC. S.ChingJ.TribbleJ. R.SolanoA. G.YangY.et al (2020). Protrudin Functions from the Endoplasmic Reticulum to Support Axon Regeneration in the Adult CNS. Nat. Commun.11, 1–15. 10.1038/s41467-020-19436-y
144
PettersenE. F.GoddardT. D.HuangC. C.MengE. C.CouchG. S.CrollT. I.et al (2021). UCSF ChimeraX: Structure Visualization for Researchers, Educators, and Developers. Protein Sci.30, 70–82. 10.1002/PRO.3943
145
PietrangeloA.RidgwayN. D. (2018). Golgi Localization of Oxysterol Binding Protein-Related Protein 4L (ORP4L) Is Regulated by Ligand Binding. J. Cell Sci.131, jcs215335. 10.1242/jcs.215335
146
ProsserD. C.TranD.GougeonP. Y.VerlyC.NgseeJ. K. (2008). FFAT Rescues VAPA-Mediated Inhibition of ER-To-Golgi Transport and VAPB-Mediated ER Aggregation. J. Cell Sci.121, 3052–3061. 10.1242/jcs.028696
147
QiaoX.JiaS.YeJ.FangX.ZhangC.CaoY.et al (2017). PTPIP51 Regulates Mouse Cardiac Ischemia/reperfusion through Mediating the Mitochondria-SR Junction. Sci. Rep.7, 1–14. 10.1038/srep45379
148
QinH.LimL.WeiY.GuptaG.SongJ. (2013a). Resolving the Paradox for Protein Aggregation Diseases: NMR Structure and Dynamics of the Membrane-Embedded P56S-MSP Causing ALS Imply a Common Mechanism for Aggregation-Prone Proteins to Attack Membranes. F1000Research2, 221. 10.12688/f1000research.2-221.v2
149
QinH.WangW.SongJ. (2013b). ALS-causing P56S Mutation and Splicing Variation on the hVAPB MSP Domain Transform its β-sandwich Fold into Lipid-Interacting Helical Conformations. Biochem. Biophys. Res. Commun.431, 398–403. 10.1016/j.bbrc.2013.01.039
150
QiuL.QiaoT.BeersM.TanW.WangH.YangB.et al (2013). Widespread Aggregation of Mutant VAPB Associated with ALS Does Not Cause Motor Neuron Degeneration or Modulate Mutant SOD1 Aggregation and Toxicity in Mice. Mol. Neurodegener.8, 1–16. 10.1186/1750-1326-8-1
151
QuintanillaC. G.LeeW.-R.LiouJ. (2022). Nir1 Constitutively Localizes at ER-PM Junctions and Promotes Nir2 Recruitment for PIP 2 Homeostasis. Mol. Biol. Cell33 (3), br2. 10.1091/MBC.E21-07-0356
152
RaiborgC.WenzelE. M.PedersenN. M.OlsvikH.SchinkK. O.SchultzS. W.et al (2015). Repeated ER-Endosome Contacts Promote Endosome Translocation and Neurite Outgrowth. Nature520, 234–238. 10.1038/nature14359
153
RamageH. R.KumarG. R.VerschuerenE.JohnsonJ. R.VonDollenJ.JohnsonT.et al (2015). A Combined Proteomics/Genomics Approach Links Hepatitis C Virus Infection with Nonsense-Mediated mRNA Decay. Mol. Cell57, 329–340. 10.1016/J.MOLCEL.2014.12.028
154
RatnaparkhiA.LawlessG. M.SchweizerF. E.GolshaniP.JacksonG. R. (2008). A Drosophila Model of ALS: Human ALS-Associated Mutation in VAP33A Suggests a Dominant Negative Mechanism. PLoS One3, e2334. 10.1371/journal.pone.0002334
155
RimessiA.PozzatoC.CarparelliL.RossiA.RanucciS.de FinoI.et al (2020). Pharmacological Modulation of Mitochondrial Calcium Uniporter Controls Lung Inflammation in Cystic Fibrosis. Sci. Adv.6, eaax9093. 10.1126/sciadv.aax9093
156
RochaN.KuijlC.Van Der KantR.JanssenL.HoubenD.JanssenH.et al (2009). Cholesterol Sensor ORP1L Contacts the ER Protein VAP to Control Rab7-RILP-p150Glued and Late Endosome Positioning. J. Cell Biol.185, 1209–1225. 10.1083/JCB.200811005/VIDEO-8
157
RoulinP. S.LötzerichM.TortaF.TannerL. B.Van KuppeveldF. J. M.WenkM. R.et al (2014). Rhinovirus Uses a Phosphatidylinositol 4-Phosphate/Cholesterol Counter-current for the Formation of Replication Compartments at the ER-Golgi Interface. Cell Host Microbe16, 677–690. 10.1016/J.CHOM.2014.10.003
158
SaitaS.ShiraneM.NatumeT.IemuraS. I.NakayamaK. I. (2009). Promotion of Neurite Extension by Protrudin Requires its Interaction with Vesicle-Associated Membrane Protein-Associated Protein. J. Biol. Chem.284, 13766–13777. 10.1074/jbc.M807938200
159
SaitoS.MatsuiH.KawanoM.KumagaiK.TomishigeN.HanadaK.et al (2008). Protein Phosphatase 2Cε Is an Endoplasmic Reticulum Integral Membrane Protein that Dephosphorylates the Ceramide Transport Protein CERT to Enhance its Association with Organelle Membranes. J. Biol. Chem.283, 6584–6593. 10.1074/jbc.M707691200
160
Saiz-RosN.CzapiewskiR.EpifanoI.StevensonA.SwansonS. K.DixonC. R.et al (2019). Host Vesicle Fusion Protein VAPB Contributes to the Nuclear Egress Stage of Herpes Simplex Virus Type-1 (HSV-1) Replication. Cells8, 120. 10.3390/CELLS8020120
161
SanhuezaM.ZechiniL.GillespieT.PennettaG. (2014). Gain-of-function Mutations in the ALS8 Causative Gene VAPB Have Detrimental Effects on Neurons and Muscles. Biol. Open3, 59–71. 10.1242/BIO.20137070
162
SantosM. F.RappaG.KarbanováJ.KurthT.CorbeilD.LoricoA. (2018). VAMP-associated Protein-A and Oxysterol-Binding Protein–Related Protein 3 Promote the Entry of Late Endosomes into the Nucleoplasmic Reticulum. J. Biol. Chem.293, 13834–13848. 10.1074/jbc.RA118.003725
163
SantosoN. G.CebotaruL.GugginoW. B. (2011). Polycystin-1, 2, and STIM1 Interact with IP3R to Modulate ER Ca2+ Release through the PI3K/Akt Pathway. Cell. Physiol. biochem.27, 715–726. 10.1159/000330080
164
SasakiJ.IshikawaK.AritaM.TaniguchiK. (2012). ACBD3-mediated Recruitment of PI4KB to Picornavirus RNA Replication Sites. EMBO J.31, 754–766. 10.1038/emboj.2011.429
165
SchlüterK.WaschbüschD.AnftM.HüggingD.KindS.HänischJ.et al (2014). JMY Is Involved in Anterograde Vesicle Trafficking from the Trans-golgi Network. Eur. J. Cell Biol.93, 194–204. 10.1016/j.ejcb.2014.06.001
166
SchulienA. J.YehC. Y.OrangB. N.PaO. J.HopkinM. P.MoutalA.et al (2020). Targeted Disruption of Kv2.1-VAPA Association Provides Neuroprotection against Ischemic Stroke in Mice by Declustering Kv2.1 Channels. Sci. Adv.6, eaaz8110. 10.1126/sciadv.aaz8110
167
SenguptaS.MillerK. K.HommaK.EdgeR.CheathamM. A.DallosP.et al (2010). Interaction between the Motor Protein Prestin and the Transporter Protein VAPA. Biochim. Biophys. Acta - Mol. Cell Res.1803, 796–804. 10.1016/j.bbamcr.2010.03.017
168
ShiJ.LuaS.TongJ. S.SongJ. (2010). Elimination of the Native Structure and Solubility of the hVAPB MSP Domain by the Pro56Ser Mutation that Causes Amyotrophic Lateral Sclerosis. Biochemistry49, 3887–3897. 10.1021/bi902057a
169
SiddiqaA.MassimiP.PimD.BroniarczykJ.BanksL. (2018). Human Papillomavirus 16 Infection Induces VAP-dependent Endosomal Tubulation. J. Virol.92, e01514–17. 10.1128/JVI.01514-17
170
SilbernagelN.WaleckiM.SchäferM. K. H.KesslerM.ZobeiriM.RinnéS.et al (2018). The VAMP-Associated Protein VAPB Is Required for Cardiac and Neuronal Pacemaker Channel Function. FASEB J.32, 6159–6173. 10.1096/FJ.201800246R
171
SilvaB. S. C.DiGiovanniL.KumarR.CarmichaelR. E.KimP. K.SchraderM. (2020). Maintaining Social Contacts: The Physiological Relevance of Organelle Interactions. Biochim. Biophys. Acta - Mol. Cell Res.1867, 118800. 10.1016/j.bbamcr.2020.118800
172
SkehelP. A.Fabian-FineR.KandelE. R. (2000). Mouse VAP33 Is Associated with the Endoplasmic Reticulum and Microtubules. Proc. Natl. Acad. Sci. U. S. A.97, 1101–1106. 10.1073/pnas.97.3.1101
173
SkehelP. A.MartinK. C.KandelE. R.BartschD. (1995). A VAMP-Binding Protein from Aplysia Required for Neurotransmitter Release. Sci. (80-269, 1580–1583. 10.1126/science.7667638
174
StanhopeR.FloraE.BayneC.DerréI. (2017). IncV, a FFAT Motif-Containing Chlamydia Protein, Tethers the Endoplasmic Reticulum to the Pathogen-Containing Vacuole. Proc. Natl. Acad. Sci.114, 12039–12044. 10.1073/pnas.1709060114
175
StoicaR.De VosK. J.PaillussonS.MuellerS.SanchoR. M.LauK.-F.et al (2014). ER-mitochondria Associations Are Regulated by the VAPB-PTPIP51 Interaction and Are Disrupted by ALS/FTD-associated TDP-43. Nat. Commun.5, 3996. 10.1038/ncomms4996
176
StoicaR.PaillussonS.Gomez-SuagaP.MitchellJ. C.LauD. H.GrayE. H.et al (2016). ALS/FTD-associated FUS Activates GSK-3β to Disrupt the VAPB-PTPIP51 Interaction and ER-Mitochondria Associations. EMBO Rep.17, 1326–1342. 10.15252/embr.201541726
177
SunY. M.DongY.WangJ.LuJ. H.ChenY.WujunJ. (2017). A Novel Mutation of VAPB in One Chinese Familial Amyotrophic Lateral Sclerosis Pedigree and its Clinical Characteristics. J. Neurol.264, 2387–2393. 10.1007/s00415-017-8628-3
178
SuzukiH.KanekuraK.LevineT. P.KohnoK.OlkkonenV. M.AisoS.et al (2009). ALS-linked P56S-VAPB, an Aggregated Loss-Of-Function Mutant of VAPB, Predisposes Motor Neurons to ER Stress-Related Death by Inducing Aggregation of Co-expressed Wild-type VAPB. J. Neurochem.108, 973–985. 10.1111/j.1471-4159.2008.05857.x
179
SuzukiH.MatsuokaM. (2011). Amyotrophic Lateral Sclerosis-Linked Mutant VAPB Enhances TDP-43-Induced Motor Neuronal Toxicity. J. Neurochem.119, 1099–1107. 10.1111/J.1471-4159.2011.07491.X
180
TachidaY.KumagaiK.SakaiS.AndoS.YamajiT.HanadaK. (2020). Chlamydia trachomatis ‐infected Human Cells Convert Ceramide to Sphingomyelin without Sphingomyelin Synthases 1 and 2. FEBS Lett.594, 519–529. 10.1002/1873-3468.13632
181
TempA. G. M.DyrbaM.KasperE.TeipelS.PrudloJ. (2021). Case Report: Cognitive Conversion in a Non-brazilian VAPB Mutation Carrier (ALS8). Front. Neurol.12, 668772. 10.3389/fneur.2021.668772
182
TeulingE.AhmedS.HaasdijkE.DemmersJ.SteinmetzM. O.AkhmanovaA.et al (2007). Motor Neuron Disease-Associated Mutant Vesicle-Associated Membrane Protein-Associated Protein (VAP) B Recruits Wild-type VAPs into Endoplasmic Reticulum-Derived Tubular Aggregates. J. Neurosci.27, 9801–9815. 10.1523/JNEUROSCI.2661-07.2007
183
ThalerR.RumplerM.SpitzerS.KlaushoferK.VargaF. (2011). Mospd1, a New Player in Mesenchymal versus Epidermal Cell Differentiation. J. Cell. Physiol.226, 2505–2515. 10.1002/JCP.22595
184
TokutakeY.GushimaK.MiyazakiH.ShimosatoT.YonekuraS. (2015a). ALS-associated P56S-VAPB Mutation Restrains 3T3-L1 Preadipocyte Differentiation. Biochem. Biophys. Res. Commun.460, 831–837. 10.1016/J.BBRC.2015.03.118
185
TokutakeY.YamadaK.OhataM.ObayashiY.TsuchiyaM.YonekuraS. (2015b). ALS-linked P56S-VAPB Mutation Impairs the Formation of Multinuclear Myotube in C2C12 Cells. Int. J. Mol. Sci.16, 18628–18641. 10.3390/IJMS160818628
186
TrilicoM. L. C.LorenzoniP. J.KayC. S. K.DucciR. D. P.FustesO. J. H.WerneckL. C.et al (2020). Characterization of the Amyotrophic Lateral Sclerosis-Linked P56S Mutation of the VAPB Gene in Southern Brazil. Amyotroph. Lateral Scler. Front. Degener.21, 286–290. 10.1080/21678421.2020.1738495
187
TripathiP.GuoH.DreserA.YamoahA.SechiA.JesseC. M.et al (2021). Pathomechanisms of ALS8: Altered Autophagy and Defective RNA Binding Protein (RBP) Homeostasis Due to the VAPB P56S Mutation. Cell Death Dis.125 (12), 1–18. 10.1038/s41419-021-03710-y
188
TsaiC. P.SoongB. W.LinK. P.TuP. H.LinJ. L.LeeY. C. (2011). FUS, TARDBP, and SOD1 Mutations in a Taiwanese Cohort with Familial ALS. Neurobiol. Aging32, 553. 10.1016/j.neurobiolaging.2010.04.009
189
TsudaH.HanS. M.YangY.TongC.LinY. Q.MohanK.et al (2008). The Amyotrophic Lateral Sclerosis 8 Protein VAPB Is Cleaved, Secreted, and Acts as a Ligand for Eph Receptors. Cell133, 963–977. 10.1016/j.cell.2008.04.039
190
TuH.GaoL.ShiS. T.TaylorD. R.YangT.MircheffA. K.et al (1999). Hepatitis C Virus RNA Polymerase and NS5A Complex with a SNARE-like Protein. Virology263, 30–41. 10.1006/VIRO.1999.9893
191
TudorE. L.GaltreyC. M.PerkintonM. S.LauK. F.De VosK. J.MitchellJ. C.et al (2010). Amyotrophic Lateral Sclerosis Mutant Vesicle-Associated Membrane Protein-Associated Protein-B Transgenic Mice Develop TAR-DNA-Binding Protein-43 Pathology. Neuroscience167, 774–785. 10.1016/j.neuroscience.2010.02.035
192
TuufJ.WistbackaL.MattjusP. (2009). The Glycolipid Transfer Protein Interacts with the Vesicle-Associated Membrane Protein-Associated Protein VAP-A. Biochem. Biophys. Res. Commun.388, 395–399. 10.1016/j.bbrc.2009.08.023
193
van BlitterswijkM.van EsM. A.KoppersM.van RheenenW.MedicJ.SchelhaasH. J.et al (2012). VAPB and C9orf72 Mutations in 1 Familial Amyotrophic Lateral Sclerosis Patient. Neurobiol. Aging33, 2950e1–2950. e4. 10.1016/j.neurobiolaging.2012.07.004
194
VendittiR.MasoneM. C.RegaL. R.TullioG.SantoroM.PolishchukE.et al (2019). The Activity of Sac1 across ER–TGN Contact Sites Requires the Four-Phosphate-Adaptor-Protein-1. J. Cell Biol.218, 783–797. 10.1083/JCB.201812021/VIDEO-1
195
WangH.PerryJ. W.LauringA. S.NeddermannP.De FrancescoR.TaiA. W. (2014). Oxysterol-Binding Protein Is a Phosphatidylinositol 4-Kinase Effector Required for HCV Replication Membrane Integrity and Cholesterol Trafficking. Gastroenterology146, 1373–1385. e11. 10.1053/J.GASTRO.2014.02.002
196
WangH.TaiA. W. (2019). Nir2 Is an Effector of VAPs Necessary for Efficient Hepatitis C Virus Replication and Phosphatidylinositol 4-Phosphate Enrichment at the Viral Replication Organelle. J. Virol.93, e00742–19. 10.1128/JVI.00742-19
197
WangS.WuX.PanT.SongW.WangY.ZhangF.et al (2012). Viperin Inhibits Hepatitis C Virus Replication by Interfering with Binding of NS5A to Host Protein hVAP-33. J. Gen. Virol.93, 83–92. 10.1099/VIR.0.033860-0/CITE/REFWORKS
198
WangY.MetzJ.CostelloJ. L.PassmoreJ.SchraderM.SchultzC.et al (2018). Intracellular Redistribution of Neuronal Peroxisomes in Response to ACBD5 Expression. PLoS One13, e0209507. 10.1371/journal.pone.0209507
199
Weber-BoyvatM.KentalaH.LiljaJ.VihervaaraT.HanninenR.ZhouY.et al (2015a). OSBP-related Protein 3 (ORP3) Coupling with VAMP-Associated Protein A Regulates R-Ras Activity. Exp. Cell Res.331, 278–291. 10.1016/j.yexcr.2014.10.019
200
Weber-BoyvatM.KentalaH.PeränenJ.OlkkonenV. M. (2015b1987). Ligand-dependent Localization and Function of ORP-VAP Complexes at Membrane Contact Sites. Cell. Mol. Life Sci.72, 1967. 10.1007/s00018-014-1786-x
201
WeirM. L.KlipA.TrimbleW. S. (1998). Identification of a Human Homologue of the Vesicle-Associated Membrane Protein (VAMP)-associated Protein of 33 kDa (VAP-33): A Broadly Expressed Protein that Binds to VAMP. Biochem. J.333, 247–251. 10.1042/bj3330247
202
WenX.AbeT.KukiharaH.TaguwaS.MoriY.TaniH.et al (2011). Elimination of Hepatitis C Virus from Hepatocytes by a Selective Activation of Therapeutic Molecules. PLoS One6, e15967. 10.1371/JOURNAL.PONE.0015967
203
WijdevenR. H.JanssenH.NahidiazarL.JanssenL.JalinkK.BerlinI.et al (2016). Cholesterol and ORP1L-Mediated ER Contact Sites Control Autophagosome Transport and Fusion with the Endocytic Pathway. Nat. Commun.71 (7), 1–14. 10.1038/ncomms11808
204
WilhelmL. P.WendlingC.VédieB.KobayashiT.ChenardM.-P.TomasettoC.et al (2017). STARD3 Mediates Endoplasmic Reticulum-To-Endosome Cholesterol Transport at Membrane Contact Sites. EMBO J.36, 1412–1433. 10.15252/EMBJ.201695917
205
WilsonC.VendittiR.De MatteisM. A. (2021). Deregulation of Phosphatidylinositol-4-Phosphate in the Development of Amyotrophic Lateral Sclerosis 8. Adv. Biol. Regul.79, 100779. 10.1016/J.JBIOR.2020.100779
206
WylesJ. P.McMasterC. R.RidgwayN. D. (2002). Vesicle-associated Membrane Protein-Associated Protein-A (VAP-A) Interacts with the Oxysterol-Binding Protein to Modify Export from the Endoplasmic Reticulum. J. Biol. Chem.277, 29908–29918. 10.1074/jbc.M201191200
207
WylesJ. P.RidgwayN. D. (2004). VAMP-associated Protein-A Regulates Partitioning of Oxysterol-Binding Protein-Related Protein-9 between the Endoplasmic Reticulum and Golgi Apparatus. Exp. Cell Res.297, 533–547. 10.1016/j.yexcr.2004.03.052
208
YagitaY.ShinoharaK.AbeY.NakagawaK.Al-OwainM.AlkurayaF. S.et al (2017). Deficiency of a Retinal Dystrophy Protein, Acyl-Coa Binding Domain-Containing 5 (ACBD5), Impairs Peroxisomal β-oxidation of Very-Long-Chain Fatty Acids. J. Biol. Chem.292, 691–705. 10.1074/jbc.M116.760090
209
YamanakaT.NishiyamaR.ShimogoriT.NukinaN. (2020). Proteomics-Based Approach Identifies Altered ER Domain Properties by ALS-Linked VAPB Mutation. Sci. Rep.10, 1–16. 10.1038/s41598-020-64517-z
210
YeoH. K.ParkT. H.KimH. Y.JangH.LeeJ.HwangG.-S.et al (2021). Phospholipid Transfer Function of PTPIP51 at Mitochondria-Associated ER Membranes. EMBO Rep.22, e51323. 10.15252/EMBR.202051323
211
YeshawW. M.van der ZwaagM.PintoF.LahayeL. L.FaberA. I. E.Gómez-SánchezR.et al (2019). Human VPS13A Is Associated with Multiple Organelles and Influences Mitochondrial Morphology and Lipid Droplet Motility. Elife8, e43561. 10.7554/eLife.43561
212
ZhaoK.RidgwayN. D. (2017). Oxysterol-Binding Protein-Related Protein 1L Regulates Cholesterol Egress from the Endo-Lysosomal System. Cell Rep.19, 1807–1818. 10.1016/J.CELREP.2017.05.028
213
ZhaoY. G.LiuN.MiaoG.ChenY.ZhaoH.ZhangH. (2018). The ER Contact Proteins VAPA/B Interact with Multiple Autophagy Proteins to Modulate Autophagosome Biogenesis. Curr. Biol.28, 1234–1245. e4. 10.1016/j.cub.2018.03.002
Summary
Keywords
VAPB, FFAT motif, membrane contact sites, endoplasmic reticulum, pathogen-host interactions, amyotrophic lateral scelerosis, VAPA, peroxisomes
Citation
Kors S, Costello JL and Schrader M (2022) VAP Proteins – From Organelle Tethers to Pathogenic Host Interactors and Their Role in Neuronal Disease. Front. Cell Dev. Biol. 10:895856. doi: 10.3389/fcell.2022.895856
Received
14 March 2022
Accepted
25 April 2022
Published
08 June 2022
Volume
10 - 2022
Edited by
Francisco Javier Martin-Romero, University of Extremadura, Spain
Reviewed by
Fabien Alpy, INSERM U964 Institut de Génétique et de Biologie Moléculaire et Cellulaire (IGBMC), France
Gabor Morotz, King’s College London, United Kingdom
Updates
Copyright
© 2022 Kors, Costello and Schrader.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Suzan Kors, s.kors@exeter.ac.uk; Michael Schrader, m.schrader@exeter.ac.uk
This article was submitted to Signaling, a section of the journal Frontiers in Cell and Developmental Biology
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.