- 1Department of Regenerative Medicine, Cell Science Research Center, Royan Institute for Stem Cell Biology and Technology, ACECR, Tehran, Iran
- 2Gastroenterology and Liver Diseases Research Center, Research Institute for Gastroenterology and Liver Diseases, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 3Laboratory for Stem Cell and Regenerative Medicine, Natural and Medical Sciences Research Center, University of Nizwa, Nizwa, Oman
- 4Experimental Cancer Medicine, Institution for Laboratory Medicine, Karolinska Institute, Stockholm, Sweden
Familial hypercholesterolemia (FH) is a common monogenic disease which is associated with high serum levels of low-density lipoprotein cholesterol (LDL-C) and leads to atherosclerosis and cardiovascular disease (CVD). Early diagnosis and effective treatment strategy can significantly improve prognosis. Recently, non-coding RNAs (ncRNAs) have emerged as novel biomarkers for the diagnosis and innovative targets for therapeutics. Non-coding RNAs have essential roles in the regulation of LDL-C homeostasis, suggesting that manipulation and regulating ncRNAs could be a promising theranostic approach to ameliorate clinical complications of FH, particularly cardiovascular disease. In this review, we briefly discussed the mechanisms and pathophysiology of FH and novel therapeutic strategies for the treatment of FH. Moreover, the theranostic effects of different non-coding RNAs for the treatment and diagnosis of FH were highlighted. Finally, the advantages and disadvantages of ncRNA-based therapies vs. conventional therapies were discussed.
1 Background
Familial hypercholesterolemia (FH) is an autosomal co-dominant disorder that affects people of all ethnic backgrounds. The FH disease is characterized by abnormally high serum levels of low-density lipoprotein cholesterol (LDL-C), and if remains untreated, it can lead to atherosclerosis, cardiovascular diseases (CVDs), and even death (Chen, 2001; Hovingh et al., 2013). FH is a hereditary disorder in humans and is classified generally as heterozygous FH (HeFH) and Homozygous FH (HoFH) (Nordestgaard et al., 2013; Cuchel et al., 2014). The HeFH is associated with 2- to 3-folds higher cholesterol levels, but HoFH, the more aggressive and rare form of FH, is associated with 3- to 6-folds cholesterol levels higher than normal individuals. According to the previous research, the prevalence of HeFH is 1 case per 200–300 people (Goldstein et al., 1973), while the HoFH may affect up to 6 individuals per million (or 1 in 160,000–300,000) (Nordestgaard et al., 2013). Given the high prevalence of FH, feasibility in the measurement of LDL-C level, and considering the substantial socioeconomic burden of the disease, FH is severely underdiagnosed and undertreated (Nordestgaard et al., 2013; Watts et al., 2014; Gidding et al., 2015).
FH increases the risk of atherosclerosis and CVD, and approximately 2% of the patients with myocardial infarction at <50 years of age have a molecular diagnosis of FH (Do et al., 2015). Advanced age, elevated body mass index (BMI), elevated triglyceride levels, higher plasma LDL levels, reduced HDL cholesterol levels, type 2 diabetes, hypertension, and smoking are risk factors associated with CVD in HeFH, but these risk factors are not accompanied by HoFH (Wiegman et al., 2015; Pérez de Isla et al., 2016). Early diagnosis of the FH reduces the risk of CVD and is necessary for the use of cholesterol-reducing therapy and family screening to identify asymptomatic affected relatives (Umans-Eckenhausen et al., 2001). The first diagnostic approach is measuring LDL-Cholesterol level, which is clinically informative when it is > 4.9 mmol/L (190 mg/dl) in adults or 4.1 mmol/L (160 mg/dl) in children (Umans-Eckenhausen et al., 2001).
The gold standard for disease diagnosis is identifying a pathogenetic variant in the group of genes associated with FH. Because of high risk of CVD among individuals with pathogenetic variants, it could be argued that genetic testing should be performed in high risk individuals (Khera et al., 2016). Three important risk genes (LDLR, APOB, and PCSK9) are usually targeted for next-generation DNA sequencing. This method is the most commonly used one for molecular diagnosis, although bioinformatics analysis of whole-exome sequence data and the use of small non-coding RNAs (ncRNAs) focused on genes related to FH might be the methods of choice in the future (Iacocca and Hegele, 2017; Jiang et al., 2018).
2 Mechanisms and Pathophysiology of Familial Hypercholesterolemia
FH is associated with mutations in the genes that regulate the uptake of LDL particles from circulation. The HeFH and HoFH disorders are caused by autosomal dominant mutations in LDLR (LDL receptor), APOB (apolipoprotein B100), and PCSK9 (proprotein convertase subtilisin/kexin type 9) genes (Nordestgaard et al., 2013; Gidding et al., 2015). Mutations in the LDLR can reduce the number of LDLR molecules or the LDLR activity in hepatocytes, lowering the LDL-C clearance from the plasma (Goldstein and Brown, 2009). More than 2900 different variants have been identified in the LDLR (Leigh et al., 2017) and more than 90% of these variants are likely to be pathogenic (Usifo et al., 2012). The apoB protein is the major lipoprotein in LDL-C and serves as a ligand for the LDL-receptor. Therefore, LDL particles with mutated and consequently defective apoB100 fail to bind to LDLR, and then the LDL-C particles accumulate in the plasma (Marais, 2004). PCSK9 activation leads to internalization and lysosomal degradation of LDL receptors, resulting in reduced LDL-C clearance from the circulation (Horton et al., 2009; Lakoski et al., 2009). Gain of function mutations or overexpression of the PCSK9 gene is associated with rapid LDLR internalization and degradation and thus reduced the number of LDLR molecules in the cell membrane of hepatocytes (Goldstein et al., 1973). Some studies indicated that LDLR mutations are responsible for the highest mean levels of the LDL-C in most FH patients, whereas individuals with APOB or PCSK9 mutations usually have somewhat lower levels (Nordestgaard et al., 2013; Cuchel et al., 2014). Interestingly, it should be noticed that the mutations of LDLR, APOB, and PCSK9 are co-dominant, and on the other hand both mutated alleles are contributed to the disease phenotype; thus, gene-dosage and the association between the numbers of mutations can affect the intensity of the disease phenotype (Nordestgaard et al., 2013; Cuchel et al., 2014).
3 Management of Familial Hypercholesterolemia
The main goal of managing FH disease is to control the lipid levels in the patient’s sera. FH is being treated with different strategies, including lifestyle modifications and pharmacological therapy. The recommended goal for patients with FH is a 50% reduction of LDL-C in the sera (Defesche et al., 2017). According to the European Atherosclerosis Consensus panel, LDL-C values in HeFH should be < 3.5 mmol/L (135 mg/dl) in children and <2.5 mmol/L (100 mg/dl) in adults. The same LDL-C levels are also considered for HoFH patients, however, obtaining these LDL-C levels with currently available medications is challenging (Vallejo-Vaz et al., 2016). Furthermore, high-dose statins can reduce LDL-C levels by 10–25% in HeFH patients, while high-dose statins combined with ezetimibe can only reduce LDL-C levels by 20–30% in HeFH patients (Nordestgaard et al., 2013).
3.1 Novel Therapeutic Approaches
The US Food and Drug Administration (FDA) has approved four novel LDL-C-lowering drugs for HoFH and HeFH (Ajufo and Rader, 2018). Some monoclonal antibodies have been developed to inhibit enzymes or proteins involved in regulating LDL-C levels. Evolocumab, an anti-PCSK9 monoclonal antibody, was developed for HoFH patients in combination with other lipid-lowering therapies (Sabatine et al., 2017). Alirocumab is another anti-PCSK9 monoclonal antibody that has been developed for HeFH or clinical ASCVD patients (Robinson et al., 2015). Evinacumab, an inhibitor of ANGPTL3, is another monoclonal antibody that is in phase III clinical trial (Raal et al., 2020). Monoclonal antibodies also have some side effects; for example, infections of the upper respiratory tract, gastroenteritis, and nasopharyngitis, were the most common adverse effects of evolocumab (Ito and Watts, 2015).
Small molecules are new therapeutic agents that can inhibit specific targets in LDL-C metabolism. Due to ease of administration, low cost, long shelf life, and non-immunogenic response, small molecules are preferred drugs for the treatment of FH patients (Zhang et al., 2014a). Lomitapide (AEGR-733/BMS201038) is an orally administered small-molecule inhibitor of microsomal triglyceride transfer protein (MTP). It mediates the transfer of TG molecules to nascent apoB during the assembly of VLDL, and its inhibition decreases VLDL and eventually LDL assembly (Hooper et al., 2015). DS-9001a is a small biologic molecule (∼22 kDa) that inhibits PCSK9 binding to LDLR and attenuates LDLR degradation. Single intravenous injection of DS-9001a reduced LDL-C levels by ∼62%, and this effect was sustained for up to 21 days in the cynomolgus monkey (Masuda et al., 2018). 7030B-C5 is another small molecule that down-regulated PCSK9 expression and increased the total cellular LDLR protein and mediated LDL-C uptake by HepG2 cells (Wang et al., 2020). Despite some advantages discussed, small molecules have several drawbacks, including low selectivity, non-tissue specific effects, and severe side effects, which limit their use as a drug (Miyosawa et al., 2015).
Mimetic peptides are also new therapeutic molecules designed to mimic peptides of a target protein. Many mimetic peptides that inhibit the PCSK9 have been developed. They mimic the EGF-A or EGF-AB binding domains of LDLR and bind to the catalytic domain of PCSK9, thereby reducing the chance of interaction between PCSK9 and LDLR (Craik et al., 2013). LDLR EGFA domain mimetic peptide was designed to inhibit PCSK9-mediated degradation of LDLR and increase the LDL uptake in a dose-dependent manner (Zhang et al., 2007). Moreover, the smaller mimetic peptide has been synthesized to inhibit the binding of PCSK9 to LDLR (Schroeder et al., 2014).
Vaccination therapy is an alternative strategy for the long-term reduction of LDL-C levels in FH patients. An anti-PCSK9 vaccine based on a designed peptide has been developed. This vaccine stimulates the immune system to produce antibodies against PCSK9 protein. Active vaccination offers some advantages over passive administration of PCSK9 monoclonal antibodies because active vaccination needs fewer injections, lower doses of drug administration, and reasonable cost. Importantly monoclonal antibodies may be neutralized by the host immune system (Nishikido and Ray, 2019). Landlinger et al. developed AT04A, a vaccine that mimics the fragments of a mature human PCSK9 protein conjugated to a foreign carrier protein (Keyhole limpet hemocyanin) for the management of FH disease (Landlinger et al., 2017). Mice vaccinated with AT04A have lower total cholesterol and LDL-C levels up to 30 and 50%, respectively (Galabova et al., 2014). A new method for developing innovative vaccines against self-antigens such as the PCSK9 is to use virus-like particles (VLPs) containing self-antigen on their surfaces. Crossey et al. designed a VLP-based vaccine targeting PCSK9, and they showed that the level of LDL-C reduced 30–40% in Macaques when combined with statins (Crossey et al., 2015). Furthermore, long-term efficacy is a crucial advantage of FH vaccination therapy, but potential adverse effects include antibody-dependent cell-mediated cytotoxicity or complement-dependent cytotoxicity, and non-specific cell destruction (Civeira and Jarauta, 2017).
The clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 system is a novel therapeutic approach for the treatment of monogenic inherited diseases (Zhao et al., 2020). CRISPR technology has some advantages including simple production, lower cost, and high efficiency (Jiang et al., 2018). In the FH, the CRISPR/Cas9 system can be employed to repair mutations in, for example, the LDLR gene or to knock out some genes related to diseases, such as PCSK9, APOB, and ANGPTL3. Omer et al. (2017) used the CRISPR/Cas9 technology to edit the 3 base-pair homozygous deletion in the exon 4 of LDLR gene in the HoFH patient-derived induced pluripotent stem cells (Omer et al., 2017). In the future, the CRISPR/Cas9 technology could be a potential therapeutic tool for the treatment of FH disease.
4 Non-Coding RNAs for the Treatment and Diagnosis of Familial Hypercholesterolemia and Other Dyslipidemia
4.1 Long Non-Coding RNAs (lncRNAs)
LncRNAs are defined as RNAs longer than 200 nucleotides (Huarte, 2015). The biogenesis of these RNAs is similar to mRNAs, and they are also processed by splicing, capping, polyadenylation, and editing (Klingenberg et al., 2017). lncRNAs lack significant open reading frames and thus have limited protein-coding potential. LncRNAs are transcribed by RNA polymerase (RNAP) II, and their expression is tightly regulated in various tissues (Vučićević et al., 2015; Shafabakhsh et al., 2021). Human GENCODE suggests more than 16,000 lncRNA genes in the human genome, but other databases estimated even more (Uszczynska-Ratajczak et al., 2018). LncRNAs are classified as long intergenic non-coding (linc)-RNAs, long non-coding antisense transcripts, long intronic ncRNAs, and non-overlapping antisense lncRNAs (Derrien et al., 2012). These RNAs have various regulatory functions at the chromatin level, including alternative splicing, cell differentiation, and cell cycle regulation (Meng et al., 2020a). Also, lncRNAs have regulatory roles in many diseases (Meng et al., 2020a). Recently, it has been demonstrated that some lncRNAs such as MeXis, LeXis, and ApoA4-AS can control lipid metabolism in different tissues (Zhang et al., 2019).
4.1.1 Mechanism of Biogenesis
The biogenesis of lncRNAs is a multi-step process, and different mechanisms are involved, including RNaseP digestion (for the generation of mature ends), snoRNP complex formation (for insertion of a cap to the end of lncRNAs), and formation of circular structures (Chen, 2016; Dashti et al., 2021). Previously, scientists thought that the transcription and processing of lncRNAs were similar to mRNAs, but recently some studies revealed distinct features in transcription, processing, export, and turnover of lncRNAs (Statello et al., 2021). The co-transcriptionally splicing of lncRNAs is weakly performed in the nucleus and the transcription termination is independent of polyadenylation signals (Schlackow et al., 2017). Therefore, the lncRNAs accumulate in the nucleus and then are degraded by RNA exosomes (Schlackow et al., 2017). In comparison to mRNAs, lncRNA genes are less evolutionarily conserved, contain fewer exons, and are less abundantly expressed (Guo et al., 2020). The low expression of lncRNAs is probably dependent on repressive histone modifications at their promoters (Melé et al., 2017). Most lncRNAs are found in the nucleus due to the high levels of U1 small nuclear RNA binding sites on lncRNA (Yin et al., 2020), the loss of function of the Pol II-associated elongation factor SPT6 (Nojima et al., 2018), and differential expression of specific splicing regulators (Melé et al., 2017). Nevertheless, certain lncRNAs are exported to the cytosol with the same processing and export pathways as mRNAs (Statello et al., 2021). The nuclear RNA export factor 1 (NXF1) pathway is responsible for exporting long and A/U-rich transcripts of lncRNAs with one or two exons (Zuckerman et al., 2020). They undergo specific sorting processes and each lncRNAs distributed in a specific organelle or cytoplasm (Statello et al., 2021).
Circular RNAs (circRNAs) are single stranded RNAs that are covalently looped. There are three types of circRNAs based on different circularizing mechanisms, including exonic circRNAs, intronic circRNAs, and intron-retained circRNAs (Chen, 2016). Usually, circRNAs are originated from exons, but others are also generated from 3´UTR, 5´UTR, introns, intergenic regions, and antisense RNAs (Li et al., 2015a). These RNAs are less efficiently transcribed than linear RNAs, but they are stable, and thus their number in the cells are still substantial (Qu et al., 2015). The mechanism of biogenesis of circRNAs is not completely clear, but the back-splicing mechanism depends on complementary intron matches is involved (Zhang et al., 2014b). To generate circRNAs, the back-splicing occurs in reversed orientation that connects a downstream 3´ splice site to an upstream 5´ splice site (Zhang et al., 2016). Generation of intronic circRNA depends on the splicing mechanism, and the GU-rich sequences near the 5´ splice site and the C-rich sequences near the branch point are needed. During the back-splicing process, the two segments bind to form the circle and then the exonic and intronic sequences are cut out by the spliceosome. After all, the introns are pieced together to form mature circRNAs (Talhouarne and Gall, 2014; Chen, 2016).
4.1.2 Mechanism of Action
LncRNAs have various regulatory functions. At the chromatin level, they are involved in dosage compensation, genomic imprinting, chromatin modification, and remodeling (Kanduri, 2016). At the transcription level, lncRNAs regulate alternative splicing, change the activity of transcriptional factors, and modify the activity of RNAP II (Zhao et al., 2008). LncRNAs also have a role in regulating cell differentiation and cell cycle, as well as the incidence of many diseases (Kitagawa et al., 2013; Fatica and Bozzoni, 2014; Yang et al., 2014). The cis and trans-acting modulator function of lncRNAs is crucial for the expression of many protein-coding genes (Mao et al., 2011). Also, these RNAs recruit the chromatin-remodeling complex to a specific locus of the chromatin and help regulate epigenetic changes in the genome (Bhat et al., 2016). Some of the well-known lncRNAs involved in epigenetic changes are ANRIL, XIST, HOTAIR, and KCNQ1OT1 (Bhat et al., 2016). LncRNAs interact with RNA-binding factors such as hnRNPs to either promote or suppress gene transcription. These RNAs also recruit transcription factors to the adjacent target genes to enhance their transcription (Bhat et al., 2016). Some lncRNAs regulate transcription by binding to regulatory proteins, RNA, or DNA molecules and sequestering them from their target site (Chowdhary et al., 2021). Some lncRNAs can identify complementary sequences in target RNAs, enabling them to participate in post-transcriptional regulation. Particularly, some lncRNAs can act as a sponge and bind to miRNAs and prevent their interaction with their target mRNAs (Salmena et al., 2011). Interestingly, the miRNA Response Element (MRE) located in the 3′ end of most lncRNAs is in charge of capturing miRNAs, because this reign is complementary with the Ago binding sites which were presented in the most of miRNAs (Jalali et al., 2013). LncRNAs are also involved in the maturation, transportation, degradation, and stability of mRNAs. For example, the NAT lncRNA has an essential role in the regulation of mRNA dynamics (Kim et al., 2020).
circRNAs are important regulatory elements that regulate at both transcription and post-transcription levels. They act as miRNA sponges and compete with endogenous RNAs (ceRNAs). As ceRNAs, they can compete for miRNA-binding sites and regulate mRNA expression (Memczak et al., 2013). Also, circRNAs can bind to RBPs and regulate the transcription, and in some cases translation (Altesha et al., 2019). It was shown that a group of regulatory circRNAs, i.e., exon-intron circRNAs (EIciRNAs), play an important role in gene expression and transcription (Li et al., 2015b). The circRNAs encapsulated into exosomes have a role in cellular communications and physiological and pathological conditions (Rayner and Hennessy, 2013).
4.1.3 Theranostic Application of lncRNAs in Dyslipidemia and Familial Hypercholesterolemia
lncRNAs are involved in cholesterol homeostasis and lipid-related diseases such as cardiovascular diseases (Ou et al., 2020). Next-generation sequencing is one of the best techniques to recognize novel lncRNAs related to FH (Chowdhary et al., 2021). Other methods such as microarray and RNA-Seq technologies are used to investigate the effects of lncRNAs on lipid metabolisms (Chowdhary et al., 2021). Computational tools combined with experimental techniques are promising approaches for detecting novel lncRNAs and also for the recognition of their role in lipid metabolisms and related diseases (Jalali et al., 2015).
Many circRNAs have been identified that have role in FH, CHD, atherosclerosis, myocardial fibrosis (MF), and other cardiovascular diseases (Gabriel et al., 2020; Lim et al., 2020). A study conducted by Sun, Y. et al. reported that 447 circRNAs were upregulated and 219 circRNAs were downregulated in FH (Sun et al., 2020). circHRCR is another circRNA which was reported as an endogenous miR-223 sponge and has a role in the inhibition of cardiac hypertrophy and HF (Wang et al., 2016).
As a therapeutic approach, lncRNAs may serve as key regulators of lipid homeostasis. Here we briefly highlighted various lncRNAs involved in cholesterol metabolism and their potential therapeutic applications (Figure 1; Table 1).
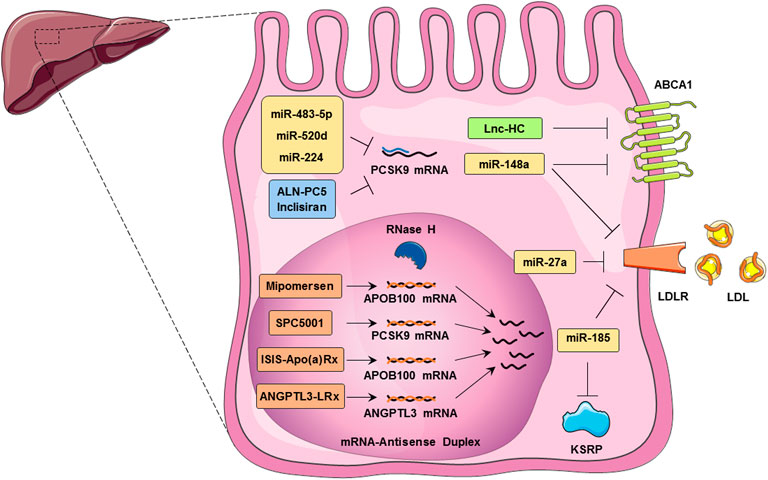
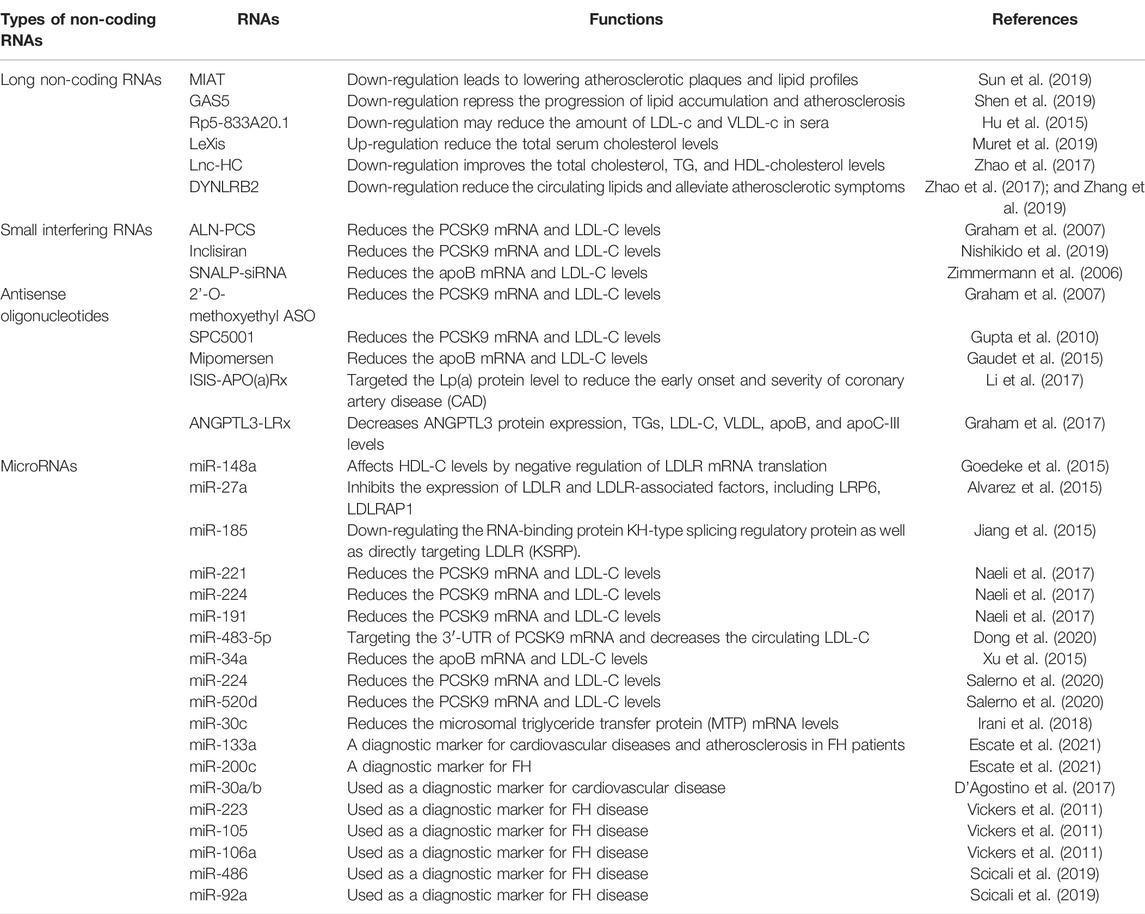
FIGURE 1. Noncoding RNAs regulate cholesterol homeostasis in hepatocytes. These RNAs, including, LncRNAs, miRNAs, ASOs, and siRNAs, control cholesterol accumulation, cholesterol efflux, cholesterol biosynthesis, and cholesterol metabolism. ABCA1 expression is regulated by both Lnc-HC and miR-148a in hepatocytes which leads to cholesterol efflux inhibition. MicroRNAs such as miR-483-5p, miR-520d, and miR-224 inhibit PCSK9 translation. miR-27a and miR-185 are involved in the inhibition of LDLR translation and thus reduce the LDL uptake by hepatocytes. Mipomersen, ISIS-Apo(a)Rx, and ANGPTL3-LRx are antisense oligonucleotides that regulate the APOB100 mRNA. SPC5001 induces the degradation of PCSK9 mRNA in the nucleus of the hepatocytes. Inclisiran, an approved siRNA, and ALN-PC5 inhibit the translation of PCSK9 in the cytoplasm of hepatocyte and hence LDLR internalization.
4.1.3.1 Myocardial Infarction-Associated Transcript
MIAT is a long non-coding RNA located on chromosome 22q12.1 and regulates the expression of genes at both transcriptional and post-transcriptional levels (Ghafouri-Fard et al., 2021). Many processes in target cells are influenced by the MIAT lncRNA, such as cell proliferation and apoptosis, cell cycle progression and migration, as well as lipid metabolism (Da et al., 2020). It has been reported that MIAT can promote the production of inflammatory factors such as IL-1β, IL-6, and TNF-α in atherosclerotic mice model via the PI3K/Akt signaling pathway (Sun et al., 2019). The PI3K/Akt signaling pathway activation can reduce reactive oxygen species and lipid deposits, which could suppress the formation of the atherosclerotic plaques (Luo et al., 2015). MIAT up-regulation activates the PI3K/Akt signaling pathway and thus aggravates the development of atherosclerosis in the mice model (Sun et al., 2019). Also, it is worth mentioning that MIAT is increased in patients with high Lp(a) (Fasolo et al., 2021). Therefore, down-regulation of this lncRNA can be a potential therapeutic target for lowering atherosclerotic plaques and lipid profiles (Figure 2).
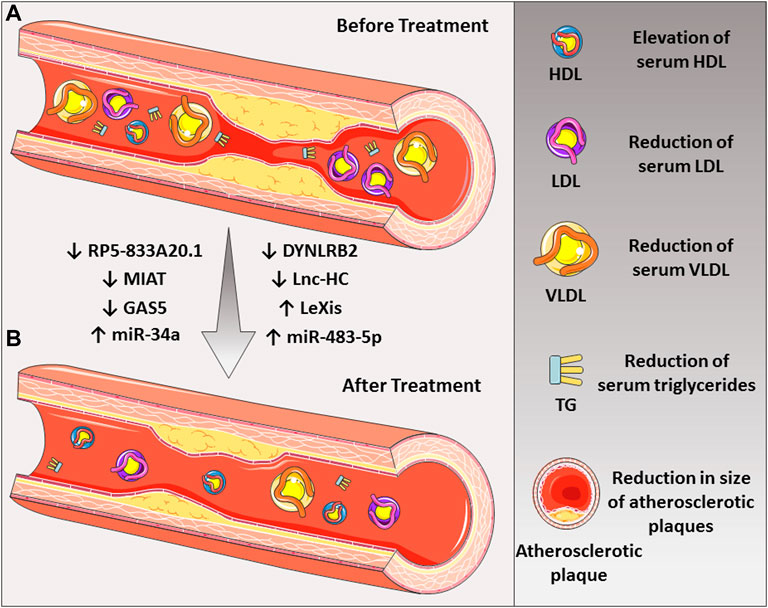
FIGURE 2. The roles of various noncoding RNAs in the treatment of atherosclerotic disease. (A) Down-regulation of some lncRNAs such as MIAT, GAS5, RP5-833A20.1, DYNLRB2, and Lnc-HC and (B) up-regulation of LeXis lncRNA could lead to the reduction of LDL-C and atherosclerotic plaque. Also, up-regulation of miR-34a and miR-483-5p could be beneficial in the treatment of the disease.
4.1.3.2 Growth Arrest-specific 5
GAS5 gene comprises 12 exons, located at 1q25.1. It encodes long noncoding RNA (Pickard and Williams, 2015). GAS5 is up-regulated in atherosclerosis plaque as compared with normal people (Chen et al., 2016a). LncRNA GAS5 binds to the zeste homolog 2 (EZH2) enhancer element, inhibiting the expression of the ABCA1 gene in THP-1 macrophages. In APOE-deficient mice, inhibiting ABCA1 reduces cholesterol efflux as well as the progression of atherosclerosis (Meng et al., 2020b). In the atherosclerotic and apoE−/− mice model, up-regulation of GAS5 increases the levels of LDL-C, total cholesterol, and aortic plaques (Meng et al., 2020b). In contrast, GAS5 silencing can repress the progression of lipid accumulation and atherosclerosis (Shen et al., 2019), suggesting that GAS5 targeting might be a promising approach for atherosclerosis (Figure 2).
4.1.3.3 RP5-833A20.1
The RP5-833A20.1 lncRNA is located in an opposite transcriptional direction of the Nuclear Factor IA (NFIA) gene (Kang et al., 2016). In macrophage-derived foam cells, up-regulation of the RP5-833A20.1 lncRNA results in down-regulation of the NFIA gene. It was shown that overexpression of the NFIA gene is associated with increased HDL-C circulation, decreased LDL-C cholesterol, and VLDL-C circulation in apoE−/− mice (Hu et al., 2015). Therefore, inhibition of RP5-833A20.1 expression may represent a therapeutic target to reduce LDL-c and VLDL-c in sera (Ye et al., 2021).
4.1.3.4 Liver-Expressed LXR-Induced Sequence (LeXis)
As a regulator of hepatic sterol content and serum cholesterol levels, LeXis lncRNA orchestrates the crosstalk between liver X receptor and SREBP transcription factors (Sallam et al., 2016). It was demonstrated that LeXis decrease cholesterol biosynthesis via binding to RALY (a heterogeneous ribonucleoprotein) and enhance cholesterol efflux in the liver (Muret et al., 2019). While LeXis lncRNA regulates RALY, it interacts with DNA as a transcription factor and inhibits the genes involved in the synthesis of cholesterol. It was shown that adenoviral administration of LeXis in mice results in reducing the total serum cholesterol levels (Sallam et al., 2015; Zhao et al., 2017).
4.1.3.5 Lnc-HC
Lnc-HC regulates hepatocyte cholesterol catabolism and is found in the liver and adipocytes. The lnc-HC forms the lncRNA-protein complex with hnRNPA2B1 and as a co-regulator binds to the mRNAs encoding Cyp7a1 and Abca1 and decreases their stability. Cyp7a1 and Abca1 are critical enzymes in cholesterol catabolism (Lan et al., 2016; Lan et al., 2019). Inhibition of Lnc-HC significantly increased the expression of CYP7a1 and ABCA1 in hepatocytes. It is now clear that lnc-HC knockdown improves the total cholesterol, TG, and HDL-cholesterol levels in rats (Zhao et al., 2017).
4.1.3.6 DYNLRB2
DYNLRB2 is a long intergenic non-coding RNA expressed in human macrophages in response to oxidized-LDL (ox-LDL) (Hu et al., 2014). In macrophage foam cells, overexpression of the DYNLRB2 lncRNA promotes the expression of the ABCA1 and G protein-coupled receptor 119 (GPR119) genes, which increase cholesterol efflux and decrease neutral lipid accumulation. Down-regulation of DYNLRB2 in mice decreased circulating lipids and alleviate atherosclerotic symptoms (Zhao et al., 2017; Zhang et al., 2019).
4.2 MicroRNAs
4.2.1 Mechanism of Biogenesis
As small but vitally regulatory modules, micro-RNAs that vary in length from 18 to 24 nucleotides belong to the ncRNAs class (Ergin and Çetinkaya, 2022). These molecules, which are dysregulated in many diseases and cancers, influence gene expression in the post-translational phases of numerous processes, including cell growth and development (Annese et al., 2020). In the first step, miRNA synthesis starts in the nucleus, and other consecutive steps occur in the cytoplasm. This pathway is multistep: in step 1, RNAP II and III execute transcription of miRNA genes that are generally assigned to miRNA-specific genes (Pourhanifeh et al., 2020; Dashti et al., 2021).
The hairpin primary miRNA sequence (pri-miRNA) produced at this stage is capable of producing one miRNA or, in some cases, more than one miRNA (Lin and Gregory, 2015). Then, in step 2, pri-miRNA cleaved to pre-miRNA by Drosha and DiGeorge critical region 8 (DGCR8). In step 3, Exportin 5 (Exp5) exports the produced pre-miRNA into the cytoplasm (Lee and Shin, 2018; Treiber et al., 2019). In the fourth step, the Dicer enzyme cleaves the pre-miRNA to miRNA duplex in the cytoplasm. Finally, one strand (guide strand) of duplex miRNA is linked to the Argonaute (Ago) protein to create the RNA-induced silencing complex (RISC), and miRNA was activated (Detassis et al., 2017). The nomenclature of the miRNA strand is due to the origination of 3′-end or 5′-end of pre-miRNA source (Zaporozhchenko et al., 2020).
In addition to canonical rout, there are some atypical miRNA biogenesis pathways that can be relevant to CVD. One class of miRNAs biosynthesized in these non-canonical pathways are miRtrons that are derived from introns. Due to the short length of a hairpin structure, after splicing in the nucleus and export to the cytoplasm, processes of pre-miRNA are done by Dicer instead of Drosha and DGCR8 (Lopez-Pedrera et al., 2020). Despite Dicer dependent-pathway, some miRNA are synthesized in a Dicer-independent manner. For instance, because of miR-451 precursor’s cleavage by Drosha/Dgcr8, the product is not long enough to cleave by Dicer. Hence, Pre-miR-451 was cleaved by unknown enzymes and then loaded into the Ago-RISC complex (Natarelli and Weber, 2022). Furthermore, there are some miRNAs localized in the nucleus through specific nucleotide motifs. These Atypical miRNAs such as miR-320, Let- 7d, and Let-7i can regulate the expression of genes at epigenetic levels. Besides, miR-126-5p and miR-133a as nuclear miRNAs implicates in atherosclerosis and CVD (Di Mauro et al., 2019; Santovito et al., 2020a). The microRNA duplexes with both active strands are another group of non-canonical miRNA. Conventionally one strand of cleaved miRNA loaded to RISC was named the guide strand, and another passenger strand was degraded. Interestingly, in microRNA duplexes such as miR-126, which are related to cardiovascular diseases, both guide and passenger strands are functional (Santovito and Weber, 2022).
4.2.2 Mechanism of Action
microRNAs as critical gene regulators can affect the function and developmental processes of human cells. miRNAs negatively regulate target genes expression. By cleaving target mRNA in the RISC complex (miRISC), it completely stops mRNA translation (Zhang et al., 2020). The GW182 factor, as well as its human homologs TNRC6A, B, and C, play an important role in the RISC function (Zaporozhchenko et al., 2020). Usually, target mRNA through complement nucleotides in its 3′ UTR region can interact with the seed sequence of miRNA (nucleotides 2-8 from the 5´-end of the miRNA). Translation of target mRNA is inhibited and then degraded by entirely complementary binding of miRNA and a specific region of 3′ UTR mRNA (perfect complementarity) (Annese et al., 2020; O'Brien et al., 2018). Additionally, especially in mammals, translational suppression is linked to partial complementary base pairing including mismatches and bulges (Lee et al., 2009; Ekimler and Sahin, 2014).
4.2.3 Role of miRNAs in Treatment of Familial Hypercholesterolemia
Currently, FH is treated with conventional medicines that lower LDL-C levels that have risen due to mutations in the LDLR gene or associated proteins (Pedro-Botet et al., 2021). However, the results suggest that using these medicines does not result in an acceptable level of LDL-C (Momtazi et al., 2018). Hence, it seems that regulating FH associated genes that participate in cholesterol hemostasis through miRNA could be a promising therapy (Ahangari et al., 2018). On the one hand, some silencing miRNA can attenuate the FH and related cardiovascular signs. For instance, in one way, miR-148a affects LDL-C levels by down-regulation of LDLR mRNA translation. In addition, the down-regulation of ABCA1 mRNA by miR-148a can lower the cholesterol (that affects circulating HDL) efflux to apoA1 in Huh7 cells. On the other hand, in vivo inhibition of mir-148a reverses the negative regulatory effect on LDLR and ABCA1 mRNA (Goedeke et al., 2015). Besides, ABCA1 and ABCG1 genes are targeted by miR-33, a well-known miRNA involved in cholesterol efflux. Inhibition of this miRNA influences cholesterol efflux and promotes HDL circulating levels (Rayner et al., 2010; Das et al., 2018). Despite that statins are being considered as an LDL-C reducing drug, it is worth mentioning that they can impact miR-33 expression. The result showed that treatment with atorvastatin and pitavastatin could lead to miR-33 overexpression which can cause ABCA1 down-regulation and lowering cholesterol efflux in Macrophages in a dose-dependent manner (Chen et al., 2016b; Tsilimigras et al., 2021). Ubilla and colleagues compared the levels of circulating miRNAs in hypercholesterolemia (HC) patients treated with atorvastatin and in the control group. The findings show that atorvastatin treatment (20 mg/day) causes a significant increase in miRNA-33b-5p levels in HC patients (Ubilla et al., 2021). In contrast, Santovito et al. confirmed that high doses of rosuvastatin (40 mg) up-regulate the ABCA1 protein in macrophages through down-regulation of miR-33b-5p in HC patients (Santovito et al., 2020b). miR-27a regulates cholesterol homeostasis both directly and indirectly. miR-27a inhibits the expression of LDLR and LDLR-associated factors, including LRP6, LDLRAP1, which are involved in LDL endocytosis, while promoting the expression of PCSK9 (Alvarez et al., 2015). miR-185 regulates LDL absorption via down-regulating the RNA-binding protein KH-type splicing regulatory protein (KSRP) as well as directly targeting LDLR. As a result, inhibiting miR-185 may be a potential method for treating atherosclerosis (Jiang et al., 2015). Overexpression of other miRNAs, on the other hand, is possibly effective in FH treatment. According to the findings, there is a negative association between the expression levels of miR-221, miR-224, and miR-191 with PCSK9 in hepatoma cells. The aforementioned miRNAs have been connected to hypercholesterolemia because they can interact directly with PCSK9's mRNA (Naeli et al., 2017). Intracellular PCSK9 can cause LDL-R breakdown in the lysosome and reduce LDL-C uptake. In humans, there is an inverse association between miR-483-5p levels and circulating LDL-C. Up-regulation of miR-483-5p resulted in a decrease in circulating LDL-C and increased LDL-R presence on the cell surface via robust targeting of the 3′-UTR of PCSK9 mRNA (Dong et al., 2020). ApoB protein was considerably down-regulated in HepG2 cells treated with miR-34a. miR-34a, a lipid metabolism regulator, modulates hepatic HNF4a expression both in vivo and in vitro. The results showed that miR-34a could reduce VLDL secretion while increasing hypolipidemia (Xu et al., 2015). Similar research has found that miR-224 and miR-520d, and miR-30c can inhibit PCSK9 and microsomal triglyceride transfer protein (MTP), respectively. These miRNAs can help with LDL clearance, lowering plasma LDL-C, and lessening the effects of hypercholesterolemia (Irani et al., 2018; Salerno et al., 2020) (Figure 1; Table 1).
4.2.4 Role of miRNAs in Diagnosis of Familial Hypercholesterolemia
miRNAs are intriguing new biomarkers that are crucial not only in FH therapeutic research but also in the diagnosis and prognosis of the disease. A cohort study was used to examine miRNAs in FH patients who had a cardiovascular event (FH-CVE) and FH patients who did not have a cardiovascular incident (FH-nCVE). miR-133a was a marker that was elevated in the FH-CVE population. Because miR-133a is linked to inflammatory cytokines, this could be used as a diagnostic marker for cardiovascular and atherosclerosis in FH patients (Escate et al., 2021). Notably, the increased level of the miR-133a was found in vulnerable human atherosclerotic plaques in dyslipidemic patients (Cipollone et al., 2011). Furthermore, researchers discovered that the level of miR-200C in plasma from children with FH was much higher than in healthy subjects. This miRNA's expression corresponds to the level of miR-30a/b, and it appears to be used as a diagnostic marker for cardiovascular disease susceptibility (D’Agostino et al., 2017). Many studies have tried to show HDL-miRNAs as disease biomarkers due to their accessibility in plasma (Vickers and Michell, 2021). Previously, FH patients and healthy people were tested for the whole HDL micro-transcriptome. The data showed a significant difference in miR-223, miR-105, and miR-106a expression between the two groups. Furthermore, miR-223 has a 3780.6-fold change in the FH population, suggesting that it has an impact on the gene expression profile of HDL-recipient cells (Vickers et al., 2011). Comparing HDL miRNA panel expression in the LDLR-null group to the LDLR-defective group, the result showed high expression of miR-486 and miR-92a in the former (Scicali et al., 2019) and these up-regulating miRNAs in HDL are related to CAD (Niculescu et al., 2015).
4.3 Small Interfering RNAs
siRNAs (20–30 nucleotide-long) are synthesized artificially and employed as novel therapeutic agents. These RNAs can bind to target mRNAs and help to silence disease-causing genes (Zimmermann et al., 2006). As compared with small molecules and monoclonal antibodies, siRNAs have some advantages. Small molecules and monoclonal antibodies need to bind to the spatial conformation of target molecules (for example, proteins), but siRNAs can bind to the target mRNA by just Watson–Crick base pairing pattern. As a result of base-pairing, siRNAs can target many mRNAs, enabling the treatment of a wide range of disorders (Watanabe et al., 2009). siRNAs have some disadvantages; for example, their delivery to the targeted tissues and their stability are still facing challenges (Rinaldi and Wood, 2018). Recently some siRNAs have been developed to treat FH by targeting various genes in this pathway.
4.3.1 Synthesis and Mechanism of Action of Small Interfering RNAs
Many siRNAs are developed for targeting various genes in different diseases. siRNAs mimic endogenous miRNA duplexes to silence gene expression. Commercially, siRNAs are synthesized via the solid-phase synthesis method (Crooke, 2017). siRNAs can be ordered as pre-formed duplexes or single-stranded RNA. In-vitro transcription of siRNAs and shRNAs is another way for synthesizing siRNAs, but due to the immuno-stimulatory effects, the 5′ triphosphate at the end of siRNA must be removed using phosphatases (Salomon et al., 2015). Some new siRNAs are chemically modified by substituting the 2′-OH with a 2′-O-methyl or 2′-methoxyethyl group (Crooke et al., 2007). Moreover, some siRNAs can be modified by substitution of specific nucleotides; for example, locked, unlocked, and glycol nucleotides can be inserted into siRNAs. These modifications can be useful for suppressing the immuno-stimulatory effects of siRNAs and enhancing their activity, specificity, and, reducing their off-target-induced toxicity (Rozema, 2017).
Dicer cleaves the long dsRNA in the target cell to form the siRNA duplex (Dias and Stein, 2002). Dicer is a member of the RNase III family that processes small RNA molecules to produce functional RNAs. The duplex siRNA has a passenger or sense strand and a guide or antisense strand. In the first step, RNA forms a pre-RISC complex with Dicer, which is then cleaved by this enzyme. Two proteins, the TRBP (HIV trans-activating response RNA-binding protein) and PACT (protein activator of PKR) are involved in capturing the RNA molecule for further processing (Watanabe et al., 2006). One of the captured RNA strands is then selected as the guide strand and binds to the Argonaute (AGO) protein to form the RNA-induced silencing complex (RISC) complex. In the RISC complex, the duplex RNA molecule is unwound and the passenger strand is degraded. The thermodynamic asymmetry properties of the end of RNA strands are the main factor in determining the guide or passenger strands. The guide strand usually has no stable 5’ end compared to the passenger strand (Geary et al., 2001). Then, the RISC complex was activated and recognized a specific target site in the single-stranded guide RNA molecule. Therefore, the RISC-siRNA complex is ready to bind to the particular sequence of the target RNA molecule and either inhibit the translation of the target messenger RNA or degrade the complementary RNA (Hamm et al., 2010).
4.3.2 Small Interfering RNAs in Familial Hypercholesterolemia
The ALN-PCS, which targets the PCSK9 mRNA, is one of the best examples of modified siRNAs used to treat FH disease. Treatment with this molecule reduces PCSK9 mRNA and LDL-C levels (Graham et al., 2007). Intravenous infusion of ALN-PCS in healthy volunteers reduced the PCSK9 mRNA level by 70% and the LDL-C levels by 40% after 3 days of infusion (Hudziak et al., 1996). Inclisiran (ALNPCSsc: ALN-60,212) is an improved version of ALN-PCS composed of nucleotides with 2′-deoxy, 2′-fluoro, and 2′-O-methyl groups. N-acetylgalactosamine (GalNAc) is conjugated to the 3′-end of Inclisiran to target the drug to the asialoglycoprotein receptors in the liver (Nishikido and Ray, 2019). This receptor is highly expressed in the hepatocytes and is a promising option for the effective uptake of inclisiran as well as a protective mechanism against off-target effects (Nishikido and Ray, 2019). Therefore, 24 h after subcutaneous injection, the drug's plasma level had drastically reduced to undetectable levels (Gupta et al., 2010). The results of the phase III study of inclisiran showed that twice administration (yearly) accompanied by statin therapy is a safe, effective, and well-tolerated regiment to lower LDL-C in adults with HeFH (van Poelgeest et al., 2015) (Figure 1).
In a study, 84 siRNA molecules specific to human and mouse ApoB mRNA were designed and synthesized using bioinformatic methods. The qRT–PCR and ELISA results showed that five siRNAs could reduce both mRNA and protein levels by 70% (van Poelgeest et al., 2013). In another study, ApoB-specific siRNA was encapsulated in Stable Nucleic Acid Lipid Particle (SNALP) and administered by intravenous injection. Results showed that single-dose injection reduced the ApoB mRNA expression in the liver as measured by stem-loop RT-qPCR. Further experiments showed that the siRNA encapsulated in SNALP significantly reduced the serum levels of cholesterol and LDL in LDL receptor knockout mice (Zimmermann et al., 2006). The combination of ApoB-specific siRNA and dendritic poly (L-lysine) administered intravenously suppressed APOB expression in C57BL/6 mice without causing hepatotoxicity. Results demonstrated that the level of LDL-C reduced significantly (Watanabe et al., 2009; Table 1).
4.4 Antisense Oligonucleotides
ASOs are synthetic small non-coding RNA molecules that influence RNA processing and protein expression and are used in the clinic as promising alternative to target specific genes (Rinaldi and Wood, 2018). Four mechanisms for the ASO-mediated down-regulation of target genes include, RNase H endonuclease mediated cleavage of the RNA-DNA hetero-duplex, inhibition of 5′ cap formation, alteration of splicing process (splice-switching), and steric hindrance of ribosomal activity (Bennett et al., 2017; Crooke, 2017). The RNase H endonuclease mechanism was described well and had three phases: pre-hybridization, hybridization, and post-hybridization. In the pre-hybridization step, the ASO enters the target cell and achieves effective concentration at the site of interest. Then, the ASO aligns with the cell’s nucleic acid and hybridizes with the cognitive sites in the hybridization step (Salomon et al., 2015). After hybridization, depending on the chemical modifications in the designed ASOs, different events may occur that change a specific gene’s expression (Crooke et al., 2007).
It seems that some limitations affect the use of ASOs in clinic as therapeutic agents. For instance, ASOs are very sensitive to endonucleases and are susceptible to rapid degradation. Thus, chemical modifications have now been applied to improve their half-life in the cells (Dias and Stein, 2002). Replacement of one non-bridging oxygen atom in the ASO backbone with sulfur is a suitable strategy for enhancing nuclease resistance and binding efficiency to serum proteins (Watanabe et al., 2006). To boost the stability of ASOs and improve their safety and efficacy profiles, chemical modifications at the 2 positions of ribose sugar are utilized, such as 2ʹ-O-methyl (2ʹ-OMe) and 2ʹ-O-methoxyethyl (2ʹ-MOE) (Rinaldi and Wood, 2018). Second-generation ASOs were developed using phosphorothioate backbone and 2ʹ-O modified nucleotides. It was observed that the biological activity and stability of these molecules increased, and the ASOs immuno-stimulatory reduced considerably (Geary et al., 2001; Hamm et al., 2010). Substitution of natural phosphoribose backbone with phosphorodiamidate morpholino oligomers (PMO) is another strategy to increase the resistance of ASOs against nuclease and protease activity (Hudziak et al., 1996). In the PMO, the deoxyribose moiety is replaced by a morpholine ring which is resistant to nucleases (Hudziak et al., 1996).
4.4.1 Antisense Oligonucleotides in Familial Hypercholesterolemia
ASOs are synthesized to regulate the level of some proteins involved in the LDL-c metabolism. To increase the LDL-C uptake in the hepatocytes, second-generation 2′-O-methoxyethyl ASO targeting PCSK9 mRNA was designed and explored. Administration of high doses of this ASO significantly decreased PCSK9 mRNA (92%), increased LDLR protein (2 fold), and reduced LDL-C in vivo (Graham et al., 2007). Because of insufficient binding affinity, the development of second-generation ASOs was terminated, and shorter ASOs based on locked nucleic acid technology such as SPC5001 were explored. These ASOs have higher stability, binding affinity, and specificity for PCSK9 mRNA than second-generation and longer oligonucleotides. Administration of ASO reduces the level of PCSK9 mRNA by 60% within 24 h post-injection, and the inhibitory effects remained for more than 16 days (Gupta et al., 2010). In healthy individuals, subcutaneous administration of SPC5001 decreased the levels of PCSK9 mRNA and LDL-C by ∼50 and 25%, respectively (Nishikido and Ray, 2019). Likewise, the SPC5001 ASOs reduced apoB levels and increased apoA1 in the target cells (van Poelgeest et al., 2015). Some side effects are observed following administration of SPC5001, such as transient renal tubular toxicity and injection site reactions, which terminated further clinical development (van Poelgeest et al., 2013). Another anti-PCSK9 ASO was developed with high potency and low toxicity. Administration of aforementioned anti-PCSK9 ASO in mice, twice a week for 6 weeks, showed that the level of PCSK9 mRNA and LDL-C decreased in a dose-dependent manner (Yamamoto et al., 2012) (Figure 1).
Mipomersen is an ASO that binds to ApoB mRNA in the liver and inhibits its translation (Gaudet and Brisson, 2015). In 2010, the randomized, double-blind, placebo-controlled, phase III study using 52 patients with HoFH was conducted. In these patients, 200 mg Mipomersen was administered once a week for 26 weeks, and the results showed that the LDL-C level was reduced by 24.7% (Raal et al., 2010). Likewise, in another study, 104 FH patients were treated with Mipomersen. After 1-year follow-up, the results showed that the mean LDL-c and Lp(a) levels were decreased by 28 and 16.6%, respectively (Duell et al., 2016). According to the results of a 2-year follow-up, the most frequently reported adverse effects were injection-site reactions, elevations of alanine aminotransaminase, and potential hepatic toxicity (Duell et al., 2016).
ISIS-APO(a)Rx is another ASO that targeted the Lp(a) protein level in FH patients to reduce the early onset and severity of coronary artery disease (CAD) (Li et al., 2017). In a phase I clinical trial, subcutaneous administration of 300 mg of ISIS-APO(a)Rx reduced the level of Lp(a) protein and associated oxidized phospholipid (OxPL) by 89 and 93%, respectively. The most common adverse effect associated with this ASO is mild injection-site reactions (Tsimikas et al., 2015).
Another ASO,ANGPTL3-LRx, reduces the level of ANGPTL3 protein, which lowers the risk of cardiovascular disease (Dewey et al., 2017). ANGPTL3 is a secretory protein that modulates plasma lipid levels by inhibiting postprandial lipoprotein lipase activity (Tikka and Jauhiainen, 2016). After 6 weeks of treatment, ANGPTL3-LRx decreases ANGPTL3 protein, TGs, LDL-C, VLDL, apoB, and apoC-III levels by up to 84.5, 63.1, 32.9, 60, 25.7, and 58.8%, respectively (Graham et al., 2017; Table 1).
5 The Challenges and Opportunities of RNA-Based Therapeutics for Dyslipidemia
Multiple conventional therapies are available to manage LDL-C levels in FH patients. Statins can up-regulate the LDLR level and reduce the LDL-C in HoFH patients lacking functional LDLR. Therefore, they are not generally effective (Marais et al., 2002). Combination of statins with bile acid sequesters or niacin reduces LDL-C, but potential adverse effects limited their administration (Cuchel et al., 2014). Lomitapide is a small molecule that binds to and inhibits the microsomal triglyceride transfer protein (MTP) in hepatocytes’ endoplasmic reticulum. MTP is an enzyme responsible for the transfer of triglycerides to nascent apoB and the assembly of VLDL and chylomicron (Alonso et al., 2019). MTP inhibition results in post-transcriptional degradation of apoB which leads to lowering the secretion of lipoprotein and then serum cholesterol as well as triglyceride levels (Burnett and Watts, 2007; Jiang et al., 2008). Lomitapide as a small molecule has some adverse effects such as gastrointestinal side effects, accumulation of liver fat, and elevations in alanine aminotransaminase levels (Ito and Watts, 2015). Some monoclonal antibodies were used to treat FH such as Evolocumab and Alirocumab. These anti-PCSK9 monoclonal antibodies were approved by FDA for the treatment of adults. These monoclonal antibodies have some adverse effects including upper respiratory tract infections, gastroenteritis, nasopharyngitis, and influenza (Ito and Watts, 2015). Also, it was shown that the response to evolocumab was dependent on LDLR status because no response was observed in patients with LDLR-negative mutations on both alleles (Raal et al., 2015). Small molecules are other therapeutic agents that can target specific points in LDL-C metabolism. They have some advantages such as ease of administration, low cost, long shelf life, and non-immunogenic response. However, because of some disadvantages including low selectivity, non-tissue specific effects, and severe side effects, their use as medications have faced some limitations (Miyosawa et al., 2015). Mimetic peptides are also new therapeutic molecules designed to mimic peptides of a target protein. They mimic, for example, the EGF-A or EGF-AB binding domains of LDLR or bind to and inhibit the catalytic domain of PCSK9, thereby they can reduce interaction between PCSK9 and LDLR (Craik et al., 2013). Like mAbs and because of limited oral bioavailability, mimetic peptides often require systemic delivery (Bergeron et al., 2015). Vaccination therapy is an alternative strategy for the long-term reduction of LDL-C levels in FH patients. For example, AT04A, a vaccine that mimics the fragments of PCSK9 protein conjugated to a foreign carrier protein (Keyhole limpet hemocyanin), was developed for the management of FH (Landlinger et al., 2017). Besides long-term efficacy of vaccination therapy in FH, potential adverse effects include antibody-dependent cell-mediated cytotoxicity or complement-dependent cytotoxicity, and non-specific cell destruction which have limited their application (Civeira and Jarauta, 2017).
Non-coding RNAs are used in novel theranostic applications to regulate the expression of some genes related to FH diseases. These RNAs can target mRNAs at various levels, both in the same or different pathways. Now, high-throughput technologies are developed to provide valuable information about different ncRNAs, their expression, and the mRNA transcripts which they might regulate (Akhtar et al., 2016). Several ncRNA-based therapies have been developed, and many have been described (Rupaimoole and Slack, 2017). Currently, at least 11 ncRNA-based drugs are approved by the Food and Drug Administration (FDA) or the European Medicines Agency (EMA) to treat different diseases. Moreover, many ncRNAs therapeutics are in clinical phases, but, until now, no ncRNA-based drugs have been considered in clinical settings (Winkle et al., 2021a). MicroRNAs are a large class of non-coding RNAs that play essential roles in regulating gene expression. miRNA-based therapeutics have at least two advantages; first, they are naturally processed and produced in human cells. Second, they act through targeting multiple genes within one pathway and thus have a broader and more specific response (Winkle et al., 2021a). Therefore, clinical application of natural miRNAs could be a promising approach for the treatment of some diseases and may potentially enhance the therapeutic effects as compared with siRNAs or other synthetic RNAs (Rupaimoole and Slack, 2017). Interestingly, using miRNAs analogs could increase the half-life and delivery of miRNAs to the target tissues; for example, in miR-30c analog, the passenger strand of miR-30c was modified which leads to stability enhancement and augment its delivery to liver cells (Yadav et al., 2022). Despite the importance and necessity of ncRNAs in preclinical investigations, their efficient and successful utilization in clinical trials has faced hurdles so far. For instance, there are difficulties in administering miRNAs in vivo since they are a subset of non-coding RNA. On one side, negative charges on miRNA and hydrophilic feature restricted its absorption by cells (Kapadia et al., 2020). On the other side, systemic treatment of miRNA and siRNA causes off-target effects (Bartoszewski and Sikorski, 2019). Aside from that, short half-lives and immune responses are critical issues that may limit miRNA clinical trials (Lee et al., 2019). Furthermore, non-coding RNA inhibitors may not have the desired impact on the complete body due to initial accumulation in specific organs (Atri et al., 2019). All in all, the goal of making these ncRNAs more efficient for therapeutic purposes is to reduce toxicity, optimize delivery to the target organ and cell in the first phase, and then recruit for specific cellular functions with high potency and stability (Winkle et al., 2021b).
However, there are some barriers to the use of ncRNA-based therapies in the clinic, including a) the specificity of targets involved in the development of the disease, b) targeted delivery of the drug at the subcellular level, c) stability of ncRNA-based therapeutics, and d) off-target effects (Hueso et al., 2021). For example, several fundamental disadvantages; including scrambling in the biogenesis of the endogenous miR, an evocation of the interferon response, and the off-target effects limit the therapeutic application of siRNA (Navarro et al., 2020). Recently, using improved algorithms for the development of more effective and specific ncRNAs, new chemistries for their stabilizing, and more efficient vehicles for enhancement of their delivery, lead to the development of new and effective ncRNA-based therapeutics.
6 Conclusion
Early detection and suitable intervention for FH in order to lower LDL is the standard goal to reduce the associated cardiovascular disease burden. Various non-coding RNAs, including lncRNAs, miRNAs, siRNAs, and ASOs interact with and regulate genes and their transcripts in lipid and cholesterol metabolism. As a therapeutic approach, some lncRNAs such as MIAT, GAS5, RP5-833A20.1, LeXis, Lnc-HC, and DYNLRB2 may serve as crucial regulators of cholesterol homeostasis. Small RNA interference drugs are used to silence disease-causing genes. Inclisiran (ALNPCSsc: ALN-60,212) is an excellent example of siRNAs used to reduce the level of LDL-C in FH. ASOs are another group of ncRNAs synthesized to regulate the level of some proteins involved in the LDL-c metabolism. Mipomersen is a well-known ASO used to inhibit the translation of ApoB mRNA. Also, miRNAs are another class of non-coding RNAs that could be promising tools for the early diagnosis and treatment of FH. Despite the importance and necessity of ncRNAs in preclinical investigations, their efficient and successful utilization in clinical trials has faced some challenges. These data suggest that non-coding RNAs could be promising tools for the early detection and treatment of FH disease, but more research is needed to overcome current challenges.
Author Contributions
HK drafted and edited manuscript, BS drafted and prepared figures, MP, HS, and NH involved in drafting, FJ, SA, and MH involved in conceptualization editing and proofreading, and MV involved in conceptualization, review, and editing the draft and final approval.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
Authors appreciate supportive colleagues in the regenerative medicine department, Royan institute.
References
Ahangari, N., Ghayour Mobarhan, M., Sahebkar, A., and Pasdar, A. (2018). Molecular Aspects of Hypercholesterolemia Treatment: Current Perspectives and Hopes. Ann. Med. 50 (4), 303–311. doi:10.1080/07853890.2018.1457795
Ajufo, E., and Rader, D. J. (2018). New Therapeutic Approaches for Familial Hypercholesterolemia. Annu. Rev. Med. 69, 113–131. doi:10.1146/annurev-med-051215-030943
Akhtar, M. M., Micolucci, L., Islam, M. S., Olivieri, F., and Procopio, A. D. (2016). Bioinformatic Tools for microRNA Dissection. Nucleic Acids Res. 44 (1), 24–44. doi:10.1093/nar/gkv1221
Alonso, R., Cuevas, A., and Mata, P. (2019). Lomitapide: a Review of its Clinical Use, Efficacy, and Tolerability. Ce 14, 19–30. doi:10.2147/ce.s174169
Altesha, M. A., Ni, T., Khan, A., Liu, K., and Zheng, X. (2019). Circular RNA in Cardiovascular Disease. J. Cell. Physiology 234 (5), 5588–5600. doi:10.1002/jcp.27384
Alvarez, M. L., Khosroheidari, M., Eddy, E., and Done, S. C. (2015). MicroRNA-27a Decreases the Level and Efficiency of the LDL Receptor and Contributes to the Dysregulation of Cholesterol Homeostasis. Atherosclerosis 242 (2), 595–604. doi:10.1016/j.atherosclerosis.2015.08.023
Annese, T., Tamma, R., De Giorgis, M., and Ribatti, D. (2020). microRNAs Biogenesis, Functions and Role in Tumor Angiogenesis. Front. Oncol. 10, 581007. doi:10.3389/fonc.2020.581007
Atri, C., Guerfali, F. Z., and Laouini, D. (2019). MicroRNAs in Diagnosis and Therapeutics. AGO-Driven Non-Coding RNAs 2019, 137–177. doi:10.1016/B978-0-12-815669-8.00006-3
Bartoszewski, R., and Sikorski, A. F. (2019). Editorial Focus: Understanding Off-Target Effects as the Key to Successful RNAi Therapy. Cell Mol. Biol. Lett. 24 (1), 69–23. doi:10.1186/s11658-019-0196-3
Bennett, C. F., Baker, B. F., Pham, N., Swayze, E., and Geary, R. S. (2017). Pharmacology of Antisense Drugs. Annu. Rev. Pharmacol. Toxicol. 57, 81–105. doi:10.1146/annurev-pharmtox-010716-104846
Bergeron, N., Phan, B. A. P., Ding, Y., Fong, A., and Krauss, R. M. (2015). Proprotein Convertase Subtilisin/Kexin Type 9 Inhibition. Circulation 132 (17), 1648–1666. doi:10.1161/circulationaha.115.016080
Bhat, S. A., Ahmad, S. M., Mumtaz, P. T., Malik, A. A., Dar, M. A., Urwat, U., et al. (2016). Long Non-coding RNAs: Mechanism of Action and Functional Utility. Non-coding RNA Res. 1 (1), 43–50. doi:10.1016/j.ncrna.2016.11.002
Burnett, J. R., and Watts, G. F. (2007). MTP Inhibition as a Treatment for Dyslipidaemias: Time to Deliver or Empty Promises? Expert Opin. Ther. targets 11 (2), 181–189. doi:10.1517/14728222.11.2.181
Chen, L.-L. (2016). The Biogenesis and Emerging Roles of Circular RNAs. Nat. Rev. Mol. Cell Biol. 17 (4), 205–211. doi:10.1038/nrm.2015.32
Chen, L., Yao, H., Hui, J.-y., Ding, S.-h., Fan, Y.-l., Pan, Y.-h., et al. (2016). Global Transcriptomic Study of Atherosclerosis Development in Rats. Gene 592 (1), 43–48. doi:10.1016/j.gene.2016.07.023
Chen, W.-M., Sheu, W. H.-H., Tseng, P.-C., Lee, T.-S., Lee, W.-J., Chang, P.-J., et al. (2016). Modulation of microRNA Expression in Subjects with Metabolic Syndrome and Decrease of Cholesterol Efflux from Macrophages via microRNA-33-Mediated Attenuation of ATP-Binding Cassette Transporter A1 Expression by Statins. PloS one 11 (5), e0154672. doi:10.1371/journal.pone.0154672
Chen, Y. (2001). The Metabolic and Molecular Bases of Inherited Disease. Glycogen storage Dis. 2001, 1521–1551.
Chowdhary, A., Satagopam, V., and Schneider, R. (2021). Long Non-coding RNAs: Mechanisms, Experimental, and Computational Approaches in Identification, Characterization, and Their Biomarker Potential in Cancer. Front. Genet. 12, 770. doi:10.3389/fgene.2021.649619
Cipollone, F., Felicioni, L., Sarzani, R., Ucchino, S., Spigonardo, F., Mandolini, C., et al. (2011). A Unique microRNA Signature Associated with Plaque Instability in Humans. Stroke 42 (9), 2556–2563. doi:10.1161/strokeaha.110.597575
Civeira, F., and Jarauta, E. (2017). Vaccine against PCSK9: the Natural Strategy from Passive to Active Immunization for the Prevention of Atherosclerosis. J. Thorac. Dis. 9 (11), 4291–4294. doi:10.21037/jtd.2017.10.18
Craik, D. J., Fairlie, D. P., Liras, S., and Price, D. (2013). The Future of Peptide-Based Drugs. Chem. Biol. drug Des. 81 (1), 136–147. doi:10.1111/cbdd.12055
Crooke, S. T., Vickers, T., Lima, W., and Wu, H. (2007). “Mechanisms of Antisense Drug Action, an Introduction,” in Antisense Drug Technology (Florida, US: CRC Press), 21–64.
Crooke, S. T. (2017). Molecular Mechanisms of Antisense Oligonucleotides. Nucleic acid. Ther. 27 (2), 70–77. doi:10.1089/nat.2016.0656
Crossey, E., Amar, M. J. A., Sampson, M., Peabody, J., Schiller, J. T., Chackerian, B., et al. (2015). A Cholesterol-Lowering VLP Vaccine that Targets PCSK9. Vaccine 33 (43), 5747–5755. doi:10.1016/j.vaccine.2015.09.044
Cuchel, M., Bruckert, E., Ginsberg, H. N., Raal, F. J., Santos, R. D., Hegele, R. A., et al. (2014). Homozygous Familial Hypercholesterolaemia: New Insights and Guidance for Clinicians to Improve Detection and Clinical Management. A Position Paper from the Consensus Panel on Familial Hypercholesterolaemia of the European Atherosclerosis Society. Eur. heart J. 35 (32), 2146–2157. doi:10.1093/eurheartj/ehu274
Da, C.-m., Gong, C.-Y., Nan, W., Zhou, K.-S., Wu, Z.-L., and Zhang, H.-H. (2020). The Role of Long Non-coding RNA MIAT in Cancers. Biomed. Pharmacother. 129, 110359. doi:10.1016/j.biopha.2020.110359
D’Agostino, M., Martino, F., Sileno, S., Barillà, F., Beji, S., Marchetti, L., et al. (2017). Circulating miR-200c Is Up-Regulated in Paediatric Patients with Familial Hypercholesterolaemia and Correlates with miR-33a/b Levels: Implication of a ZEB1-dependent Mechanism. Clin. Sci. (Lond) 131 (18), 2397–2408. doi:10.1042/CS20171121
Das, A., Samidurai, A., and Salloum, F. N. (2018). Deciphering Non-coding RNAs in Cardiovascular Health and Disease. Front. Cardiovasc. Med. 5, 73. doi:10.3389/fcvm.2018.00073
Dashti, F., Mirazimi, S. M. A., Rabiei, N., Fathazam, R., Rabiei, N., Piroozmand, H., et al. (2021). The Role of Non-coding RNAs in Chemotherapy for Gastrointestinal Cancers. Mol. Ther. - Nucleic Acids 26, 892–926. doi:10.1016/j.omtn.2021.10.004
Defesche, J. C., Gidding, S. S., Harada-Shiba, M., Hegele, R. A., Santos, R. D., and Wierzbicki, A. S. (2017). Familial Hypercholesterolaemia. Nat. Rev. Dis. Prim. 3 (1), 17093–17120. doi:10.1038/nrdp.2017.93
Derrien, T., Johnson, R., Bussotti, G., Tanzer, A., Djebali, S., Tilgner, H., et al. (2012). The GENCODE V7 Catalog of Human Long Noncoding RNAs: Analysis of Their Gene Structure, Evolution, and Expression. Genome Res. 22 (9), 1775–1789. doi:10.1101/gr.132159.111
Detassis, S., Grasso, M., Del Vescovo, V., Denti, M. A., and biology, d. (2017). microRNAs Make the Call in Cancer Personalized Medicine. Front. Cell Dev. Biol. 5, 86. doi:10.3389/fcell.2017.00086
Dewey, F. E., Gusarova, V., Dunbar, R. L., O’Dushlaine, C., Schurmann, C., Gottesman, O., et al. (2017). Genetic and Pharmacologic Inactivation of ANGPTL3 and Cardiovascular Disease. N. Engl. J. Med. 377 (3), 211–221. doi:10.1056/nejmoa1612790
Di Mauro, V., Crasto, S., Colombo, F. S., Di Pasquale, E., and Catalucci, D. (2019). Wnt Signalling Mediates miR-133a Nuclear Re-localization for the Transcriptional Control of Dnmt3b in Cardiac Cells. Sci. Rep. 9 (1), 9320–9415. doi:10.1038/s41598-019-45818-4
Dias, N., and Stein, C. A. (2002). Antisense Oligonucleotides: Basic Concepts and Mechanisms. Mol. Cancer Ther. 1 (5), 347–355.
Do, R., Stitziel, N. O., Stitziel, N. O., Won, H.-H., Jørgensen, A. B., Duga, S., et al. (2015). Exome Sequencing Identifies Rare LDLR and APOA5 Alleles Conferring Risk for Myocardial Infarction. Nature 518 (7537), 102–106. doi:10.1038/nature13917
Dong, J., He, M., Li, J., Pessentheiner, A., Wang, C., Zhang, J., et al. (2020). microRNA-483 Ameliorates Hypercholesterolemia by Inhibiting PCSK9 Production. JCI Insight 5 (23). doi:10.1172/jci.insight.143812
Duell, P. B., Santos, R. D., Kirwan, B.-A., Witztum, J. L., Tsimikas, S., and Kastelein, J. J. P. (2016). Long-term Mipomersen Treatment Is Associated with a Reduction in Cardiovascular Events in Patients with Familial Hypercholesterolemia. J. Clin. Lipidol. 10 (4), 1011–1021. doi:10.1016/j.jacl.2016.04.013
Ekimler, S., and Sahin, K. (2014). Computational Methods for microRNA Target Prediction. Genes 5 (3), 671–683. doi:10.3390/genes5030671
Ergin, K., and Çetinkaya, R. (2022). Regulation of MicroRNAs. Methods Mol. Biol. 2257, 1–32. doi:10.1007/978-1-0716-1170-8_1
Escate, R., Padró, T., Suades, R., Camino, S., Muñiz, O., Diaz-Diaz, J. L., et al. (2021). High miR-133a Levels in the Circulation Anticipates Presentation of Clinical Events in Familial Hypercholesterolaemia Patients. Cardiovasc Res. 117 (1), 109–122. doi:10.1093/cvr/cvaa039
Fasolo, F., Jin, H., Winski, G., Chernogubova, E., Pauli, J., Winter, H., et al. (2021). Long Noncoding RNA MIAT Controls Advanced Atherosclerotic Lesion Formation and Plaque Destabilization. Circulation 144 (19), 1567–1583. doi:10.1161/circulationaha.120.052023
Fatica, A., and Bozzoni, I. (2014). Long Non-coding RNAs: New Players in Cell Differentiation and Development. Nat. Rev. Genet. 15 (1), 7–21. doi:10.1038/nrg3606
Gabriel, A. F., Costa, M. C., and Enguita, F. J. (2020). Circular RNA-Centered Regulatory Networks in the Physiopathology of Cardiovascular Diseases. Int. J. Mol. Sci. 21 (2), 456. doi:10.3390/ijms21020456
Galabova, G., Brunner, S., Winsauer, G., Juno, C., Wanko, B., Mairhofer, A., et al. (2014). Peptide-Based Anti-PCSK9 Vaccines - an Approach for Long-Term LDLc Management. PLoS One 9 (12), e114469. doi:10.1371/journal.pone.0114469
Gaudet, D., and Brisson, D. (2015). Gene-based Therapies in Lipidology. Curr. Opin. Lipidol. 26 (6), 553–565. doi:10.1097/mol.0000000000000240
Geary, R. S., Yu, R. Z., and Levin, A. A. (2001). Pharmacokinetics of Phosphorothioate Antisense Oligodeoxynucleotides. Curr. Opin. Investig. Drugs 2 (4), 562–573.
Ghafouri-Fard, S., Azimi, T., and Taheri, M. (2021). Myocardial Infarction Associated Transcript (MIAT): Review of its Impact in the Tumorigenesis. Biomed. Pharmacother. 133, 111040. doi:10.1016/j.biopha.2020.111040
Gidding, S. S., Ann Champagne, M., de Ferranti, S. D., Defesche, J., Ito, M. K., Knowles, J. W., et al. (2015). The Agenda for Familial Hypercholesterolemia. Circulation 132 (22), 2167–2192. doi:10.1161/cir.0000000000000297
Goedeke, L., Rotllan, N., Canfrán-Duque, A., Aranda, J. F., Ramírez, C. M., Araldi, E., et al. (2015). MicroRNA-148a Regulates LDL Receptor and ABCA1 Expression to Control Circulating Lipoprotein Levels. Nat. Med. 21 (11), 1280–1289. doi:10.1038/nm.3949
Goldstein, J. L., and Brown, M. S. (2009). The LDL Receptor. Arterioscler. Thromb. Vasc. Biol. 29, 431–438. doi:10.1161/ATVBAHA.108.179564
Goldstein, J. L., Schrott, H. G., Hazzard, W. R., Bierman, E. L., and Motulsky, A. G. (1973). Hyperlipidemia in Coronary Heart Disease II. Genetic Analysis of Lipid Levels in 176 Families and Delineation of a New Inherited Disorder, Combined Hyperlipidemia. J. Clin. Invest. 52 (7), 1544–1568. doi:10.1172/jci107332
Graham, M. J., Lee, R. G., Brandt, T. A., Tai, L.-J., Fu, W., Peralta, R., et al. (2017). Cardiovascular and Metabolic Effects of ANGPTL3 Antisense Oligonucleotides. N. Engl. J. Med. 377, 222–232. doi:10.1056/nejmoa1701329
Graham, M. J., Lemonidis, K. M., Whipple, C. P., Subramaniam, A., Monia, B. P., Crooke, S. T., et al. (2007). Antisense Inhibition of Proprotein Convertase Subtilisin/kexin Type 9 Reduces Serum LDL in Hyperlipidemic Mice. J. lipid Res. 48 (4), 763–767. doi:10.1194/jlr.c600025-jlr200
Guo, C.-J., Ma, X.-K., Xing, Y.-H., Zheng, C.-C., Xu, Y.-F., Shan, L., et al. (2020). Distinct Processing of lncRNAs Contributes to Non-conserved Functions in Stem Cells. Cell 181 (3), 621–636. e22. doi:10.1016/j.cell.2020.03.006
Gupta, N., Fisker, N., Asselin, M.-C., Lindholm, M., Rosenbohm, C., Ørum, H., et al. (2010). A Locked Nucleic Acid Antisense Oligonucleotide (LNA) Silences PCSK9 and Enhances LDLR Expression In Vitro and In Vivo. PloS one 5 (5), e10682. doi:10.1371/journal.pone.0010682
Hamm, S., Latz, E., Hangel, D., Müller, T., Yu, P., Golenbock, D., et al. (2010). Alternating 2′-O-Ribose Methylation Is a Universal Approach for Generating Non-stimulatory siRNA by Acting as TLR7 Antagonist. Immunobiology 215 (7), 559–569. doi:10.1016/j.imbio.2009.09.003
Hooper, A. J., Burnett, J. R., and Watts, G. F. (2015). Contemporary Aspects of the Biology and Therapeutic Regulation of the Microsomal Triglyceride Transfer Protein. Circ. Res. 116 (1), 193–205. doi:10.1161/circresaha.116.304637
Horton, J. D., Cohen, J. C., and Hobbs, H. H. (2009). PCSK9: a Convertase that Coordinates LDL Catabolism. J. lipid Res. 50, S172–S177. doi:10.1194/jlr.r800091-jlr200
Hovingh, G. K., Davidson, M. H., Kastelein, J. J. P., and O'Connor, A. M. (2013). Diagnosis and Treatment of Familial Hypercholesterolaemia. Eur. heart J. 34 (13), 962–971. doi:10.1093/eurheartj/eht015
Hu, Y.-W., Yang, J.-Y., Ma, X., Chen, Z.-P., Hu, Y.-R., Zhao, J.-Y., et al. (2014). A lincRNA-DYNLRB2-2/gpr119/glp-1r/abca1-dependent Signal Transduction Pathway Is Essential for the Regulation of Cholesterol Homeostasis. J. lipid Res. 55 (4), 681–697. doi:10.1194/jlr.m044669
Hu, Y.-W., Zhao, J.-Y., Li, S.-F., Huang, J.-L., Qiu, Y.-R., Ma, X., et al. (2015). RP5-833A20.1/miR-382-5p/NFIA-Dependent Signal Transduction Pathway Contributes to the Regulation of Cholesterol Homeostasis and Inflammatory Reaction. Arterioscler. Thromb. Vasc. Biol. 35 (1), 87–101. doi:10.1161/atvbaha.114.304296
Huarte, M. (2015). The Emerging Role of lncRNAs in Cancer. Nat. Med. 21 (11), 1253–1261. doi:10.1038/nm.3981
Hudziak, R. M., Barofsky, E., Barofsky, D. F., Weller, D. L., Huang, S.-B., and Weller, D. D. (1996). Resistance of Morpholino Phosphorodiamidate Oligomers to Enzymatic Degradation. Antisense Nucleic Acid Drug Dev. 6 (4), 267–272. doi:10.1089/oli.1.1996.6.267
Hueso, M., Mallén, A., Suñé-Pou, M., Aran, J. M., Suñé-Negre, J. M., and Navarro, E. (2021). ncRNAs in Therapeutics: Challenges and Limitations in Nucleic Acid-Based Drug Delivery. Ijms 22 (21), 11596. doi:10.3390/ijms222111596
Iacocca, M. A., and Hegele, R. A. (2017). Recent Advances in Genetic Testing for Familial Hypercholesterolemia. Expert Rev. Mol. diagnostics 17 (7), 641–651. doi:10.1080/14737159.2017.1332997
Irani, S., Iqbal, J., Antoni, W. J., Ijaz, L., and Hussain, M. M. (2018). microRNA-30c Reduces Plasma Cholesterol in Homozygous Familial Hypercholesterolemic and Type 2 Diabetic Mouse Models. J. Lipid Res. 59 (1), 144–154. doi:10.1194/jlr.m081299
Ito, M. K., and Watts, G. F. (2015). Challenges in the Diagnosis and Treatment of Homozygous Familial Hypercholesterolemia. Drugs 75 (15), 1715–1724. doi:10.1007/s40265-015-0466-y
Jalali, S., Bhartiya, D., Lalwani, M. K., Sivasubbu, S., and Scaria, V. (2013). Systematic Transcriptome Wide Analysis of lncRNA-miRNA Interactions. PloS one 8 (2), e53823. doi:10.1371/journal.pone.0053823
Jalali, S., Kapoor, S., Sivadas, A., Bhartiya, D., and Scaria, V. (2015). Computational Approaches towards Understanding Human Long Non-coding RNA Biology. Bioinformatics 31 (14), 2241–2251. doi:10.1093/bioinformatics/btv148
Jiang, H., Zhang, J., Du, Y., Jia, X., Yang, F., Si, S., et al. (2015). microRNA-185 Modulates Low Density Lipoprotein Receptor Expression as a Key Posttranscriptional Regulator. Atherosclerosis 243 (2), 523–532. doi:10.1016/j.atherosclerosis.2015.10.026
Jiang, L., Wang, L.-Y., and Cheng, X.-s. (2018). Novel Approaches for the Treatment of Familial Hypercholesterolemia: Current Status and Future Challenges. Jat 25 (8), 665–673. doi:10.5551/jat.43372
Jiang, Z. G., Liu, Y., Hussain, M. M., Atkinson, D., and McKnight, C. J. (2008). Reconstituting Initial Events during the Assembly of Apolipoprotein B-Containing Lipoproteins in a Cell-free System. J. Mol. Biol. 383 (5), 1181–1194. doi:10.1016/j.jmb.2008.09.006
Kanduri, C. (2016). Long Noncoding RNAs: Lessons from Genomic Imprinting. Biochimica Biophysica Acta (BBA) - Gene Regul. Mech. 1859 (1), 102–111. doi:10.1016/j.bbagrm.2015.05.006
Kang, C.-M., Hu, Y.-W., Nie, Y., Zhao, J.-Y., Li, S.-F., Chu, S., et al. (2016). Long Non-coding RNA RP5-833A20.1 Inhibits Proliferation, Metastasis and Cell Cycle Progression by Suppressing the Expression of NFIA in U251 Cells. Mol. Med. Rep. 14 (6), 5288–5296. doi:10.3892/mmr.2016.5854
Kapadia, C. H., Luo, B., Dang, M. N., Irvin‐Choy, N. D., Valcourt, D. M., and Day, E. S. (2020). Polymer Nanocarriers for MicroRNA Delivery. J. Appl. Polym. Sci. 137 (25), 48651. doi:10.1002/app.48651
Khera, A. V., Won, H.-H., Peloso, G. M., Lawson, K. S., Bartz, T. M., Deng, X., et al. (2016). Diagnostic Yield and Clinical Utility of Sequencing Familial Hypercholesterolemia Genes in Patients with Severe Hypercholesterolemia. J. Am. Coll. Cardiol. 67 (22), 2578–2589. doi:10.1016/j.jacc.2016.03.520
Kim, J. H., Yoon, C. Y., Jun, Y., Lee, B. B., Lee, J. E., Ha, S. D., et al. (2020). NuA3 HAT Antagonizes the Rpd3S and Rpd3L HDACs to Optimize mRNA and lncRNA Expression Dynamics. Nucleic acids Res. 48 (19), 10753–10767. doi:10.1093/nar/gkaa781
Kitagawa, M., Kitagawa, K., Kotake, Y., Niida, H., and Ohhata, T. (2013). Cell Cycle Regulation by Long Non-coding RNAs. Cell. Mol. Life Sci. 70 (24), 4785–4794. doi:10.1007/s00018-013-1423-0
Klingenberg, M., Matsuda, A., Diederichs, S., and Patel, T. (2017). Non-coding RNA in Hepatocellular Carcinoma: Mechanisms, Biomarkers and Therapeutic Targets. J. hepatology 67 (3), 603–618. doi:10.1016/j.jhep.2017.04.009
Lakoski, S. G., Lagace, T. A., Cohen, J. C., Horton, J. D., and Hobbs, H. H. (2009). Genetic and Metabolic Determinants of Plasma PCSK9 Levels. J. Clin. Endocrinol. Metabolism 94 (7), 2537–2543. doi:10.1210/jc.2009-0141
Lan, X., Wu, L., Wu, N., Chen, Q., Li, Y., Du, X., et al. (2019). Long Noncoding RNA Lnc-HC Regulates PPARγ-Mediated Hepatic Lipid Metabolism through miR-130b-3p. Mol. Ther. - Nucleic Acids 18, 954–965. doi:10.1016/j.omtn.2019.10.018
Lan, X., Yan, J., Ren, J., Zhong, B., Li, J., Li, Y., et al. (2016). A Novel Long Noncoding RNA Lnc-HC Binds hnRNPA2B1 to Regulate Expressions of Cyp7a1 and Abca1 in Hepatocytic Cholesterol Metabolism. Hepatology 64 (1), 58–72. doi:10.1002/hep.28391
Landlinger, C., Pouwer, M. G., Juno, C., van der Hoorn, J. W. A., Pieterman, E. J., Jukema, J. W., et al. (2017). The AT04A Vaccine against Proprotein Convertase Subtilisin/kexin Type 9 Reduces Total Cholesterol, Vascular Inflammation, and Atherosclerosis in APOE*3Leiden.CETP Mice. Eur. heart J. 38 (32), 2499–2507. doi:10.1093/eurheartj/ehx260
Lee, D., and Shin, C. (2018). Emerging Roles of DROSHA beyond Primary microRNA Processing. RNA Biol. 15 (2), 186–193. doi:10.1080/15476286.2017.1405210
Lee, I., Ajay, S. S., Yook, J. I., Kim, H. S., Hong, S. H., Kim, N. H., et al. (2009). New Class of microRNA Targets Containing Simultaneous 5′-UTR and 3′-UTR Interaction Sites. Genome Res. 19 (7), 1175–1183. doi:10.1101/gr.089367.108
Lee, S. W. L., Paoletti, C., Campisi, M., Osaki, T., Adriani, G., Kamm, R. D., et al. (2019). MicroRNA Delivery through Nanoparticles. J. Control. Release 313, 80–95. doi:10.1016/j.jconrel.2019.10.007
Leigh, S., Futema, M., Whittall, R., Taylor-Beadling, A., Williams, M., den Dunnen, J. T., et al. (2017). The UCL Low-Density Lipoprotein Receptor Gene Variant Database: Pathogenicity Update. J. Med. Genet. 54 (4), 217–223. doi:10.1136/jmedgenet-2016-104054
Li, S., Wu, N.-Q., Zhu, C.-G., Zhang, Y., Guo, Y.-L., Gao, Y., et al. (2017). Significance of Lipoprotein(a) Levels in Familial Hypercholesterolemia and Coronary Artery Disease. Atherosclerosis 260, 67–74. doi:10.1016/j.atherosclerosis.2017.03.021
Li, Y., Zheng, Q., Bao, C., Li, S., Guo, W., Zhao, J., et al. (2015). Circular RNA Is Enriched and Stable in Exosomes: a Promising Biomarker for Cancer Diagnosis. Cell Res. 25 (8), 981–984. doi:10.1038/cr.2015.82
Li, Z., Huang, C., Bao, C., Chen, L., Lin, M., Wang, X., et al. (2015). Exon-intron Circular RNAs Regulate Transcription in the Nucleus. Nat. Struct. Mol. Biol. 22 (3), 256–264. doi:10.1038/nsmb.2959
Lim, T. B., Lavenniah, A., and Foo, R. S. (2020). Circles in the Heart and Cardiovascular System. Cardiovasc Res. 116 (2), 269–278. doi:10.1093/cvr/cvz227
Lin, S., and Gregory, R. I. (2015). MicroRNA Biogenesis Pathways in Cancer. Nat. Rev. Cancer 15 (6), 321–333. doi:10.1038/nrc3932
Lopez-Pedrera, C., Barbarroja, N., Patiño-Trives, A. M., Luque-Tévar, M., Torres-Granados, C., Aguirre-Zamorano, M. A., et al. (2020). Role of microRNAs in the Development of Cardiovascular Disease in Systemic Autoimmune Disorders. Int. J. Mol. Sci. 21 (6), 2012. doi:10.3390/ijms21062012
Luo, Y., Sun, G., Dong, X., Wang, M., Qin, M., Yu, Y., et al. (2015). Isorhamnetin Attenuates Atherosclerosis by Inhibiting Macrophage Apoptosis via PI3K/AKT Activation and HO-1 Induction. PLoS One 10 (3), e0120259. doi:10.1371/journal.pone.0120259
Mao, Y. S., Sunwoo, H., Zhang, B., and Spector, D. L. (2011). Direct Visualization of the Co-transcriptional Assembly of a Nuclear Body by Noncoding RNAs. Nat. Cell Biol. 13 (1), 95–101. doi:10.1038/ncb2140
Marais, A. D., Blom, D. J., and Firth, J. C. (2002). Statins in Homozygous Familial Hypercholesterolemia. Curr. Atheroscler. Rep. 4 (1), 19–25. doi:10.1007/s11883-002-0058-7
Masuda, Y., Yamaguchi, S., Suzuki, C., Aburatani, T., Nagano, Y., Miyauchi, R., et al. (2018). Generation and Characterization of a Novel Small Biologic Alternative to Proprotein Convertase Subtilisin/Kexin Type 9 (PCSK9) Antibodies, DS-9001a, Albumin Binding Domain-Fused Anticalin Protein. J. Pharmacol. Exp. Ther. 365 (2), 368–378. doi:10.1124/jpet.117.246652
Melé, M., Mattioli, K., Mallard, W., Shechner, D. M., Gerhardinger, C., and Rinn, J. L. (2017). Chromatin Environment, Transcriptional Regulation, and Splicing Distinguish lincRNAs and mRNAs. Genome Res. 27 (1), 27–37. doi:10.1101/gr.214205.116
Memczak, S., Jens, M., Elefsinioti, A., Torti, F., Krueger, J., Rybak, A., et al. (2013). Circular RNAs Are a Large Class of Animal RNAs with Regulatory Potency. Nature 495 (7441), 333–338. doi:10.1038/nature11928
Meng, Q., Pu, L., Luo, X., Wang, B., Li, F., and Liu, B. (2020). Regulatory Roles of Related Long Non-coding RNAs in the Process of Atherosclerosis. Front. Physiol. 11, 564604. doi:10.3389/fphys.2020.564604
Meng, X.-D., Yao, H.-H., Wang, L.-M., Yu, M., Shi, S., Yuan, Z.-X., et al. (2020). Knockdown of GAS5 Inhibits Atherosclerosis Progression via Reducing EZH2-Mediated ABCA1 Transcription in ApoE−/− Mice. Mol. Ther. - Nucleic Acids 19, 84–96. doi:10.1016/j.omtn.2019.10.034
Miyosawa, K., Watanabe, Y., Murakami, K., Murakami, T., Shibata, H., Iwashita, M., et al. (2015). New CETP Inhibitor K-312 Reduces PCSK9 Expression: a Potential Effect on LDL Cholesterol Metabolism. Am. J. Physiology-Endocrinology Metabolism 309 (2), E177–E190. doi:10.1152/ajpendo.00528.2014
Momtazi, A. A., Banach, M., Pirro, M., Stein, E. A., and Sahebkar, A. (2018). MicroRNAs: New Therapeutic Targets for Familial Hypercholesterolemia? Clin. Rev. Allerg. Immunol. 54 (2), 224–233. doi:10.1007/s12016-017-8611-x
Muret, K., Désert, C., Lagoutte, L., Boutin, M., Gondret, F., Zerjal, T., et al. (2019). Long Noncoding RNAs in Lipid Metabolism: Literature Review and Conservation Analysis across Species. BMC genomics 20 (1), 882–918. doi:10.1186/s12864-019-6093-3
Naeli, P., Mirzadeh Azad, F., Malakootian, M., Seidah, N. G., and Mowla, S. J. (2017). Post-transcriptional Regulation of PCSK9 by miR-191, miR-222, and miR-224. Front. Genet. 8, 189. doi:10.3389/fgene.2017.00189
Natarelli, L., and Weber, C. (2022). A Non-canonical Link between Non-coding RNAs and Cardiovascular Diseases. Biomedicines 10 (2), 445. doi:10.3390/biomedicines10020445
Navarro, E., Mallén, A., Cruzado, J. M., Torras, J., and Hueso, M. (2020). Unveiling ncRNA Regulatory Axes in Atherosclerosis Progression. Clin. Transl. Med. 9 (1), 5–18. doi:10.1186/s40169-020-0256-3
Niculescu, L. S., Simionescu, N., Sanda, G. M., Carnuta, M. G., Stancu, C. S., Popescu, A. C., et al. (2015). MiR-486 and miR-92a Identified in Circulating HDL Discriminate between Stable and Vulnerable Coronary Artery Disease Patients. PLoS ONE 10 (10), e0140958. doi:10.1371/journal.pone.0140958
Nishikido, T., and Ray, K. K. (2019). Non-antibody Approaches to Proprotein Convertase Subtilisin Kexin 9 Inhibition: siRNA, Antisense Oligonucleotides, Adnectins, Vaccination, and New Attempts at Small-Molecule Inhibitors Based on New Discoveries. Front. Cardiovasc. Med. 5, 199. doi:10.3389/fcvm.2018.00199
Nojima, T., Tellier, M., Foxwell, J., Ribeiro de Almeida, C., Tan-Wong, S. M., Dhir, S., et al. (2018). Deregulated Expression of Mammalian lncRNA through Loss of SPT6 Induces R-Loop Formation, Replication Stress, and Cellular Senescence. Mol. cell 72 (6), 970–984. e7. doi:10.1016/j.molcel.2018.10.011
Nordestgaard, B. G., Chapman, M. J., Humphries, S. E., Ginsberg, H. N., Masana, L., Descamps, O. S., et al. (2013). Familial Hypercholesterolaemia Is Underdiagnosed and Undertreated in the General Population: Guidance for Clinicians to Prevent Coronary Heart Disease: Consensus Statement of the European Atherosclerosis Society. Eur. heart J. 34 (45), 3478–3490. doi:10.1093/eurheartj/eht273
O'Brien, J., Hayder, H., Zayed, Y., and Peng, C. (2018). Overview of microRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 9, 402. doi:10.3389/fendo.2018.00402
Omer, L., Hudson, E. A., Zheng, S., Hoying, J. B., Shan, Y., and Boyd, N. L. (2017). CRISPR Correction of a Homozygous Low-Density Lipoprotein Receptor Mutation in Familial Hypercholesterolemia Induced Pluripotent Stem Cells. Hepatol. Commun. 1 (9), 886–898. doi:10.1002/hep4.1110
Ou, M., Li, X., Zhao, S., Cui, S., and Tu, J. (2020). Long Non-coding RNA CDKN2B-AS1 Contributes to Atherosclerotic Plaque Formation by Forming RNA-DNA Triplex in the CDKN2B Promoter. EBioMedicine 55, 102694. doi:10.1016/j.ebiom.2020.102694
Pedro-Botet, J., Climent, E., and Benaiges, D. (2021). Familial Hypercholesterolemia: Do HDL Play a Role? Biomedicines 9 (7), 810. doi:10.3390/biomedicines9070810
Pérez de Isla, L., Alonso, R., Mata, N., Saltijeral, A., Muñiz, O., Rubio-Marin, P., et al. (2016). Coronary Heart Disease, Peripheral Arterial Disease, and Stroke in Familial Hypercholesterolaemia: Insights from the SAFEHEART Registry (Spanish Familial Hypercholesterolaemia Cohort Study). Arterioscler. Thromb. Vasc. Biol. 36 (9), 2004–2010. doi:10.1161/ATVBAHA.116.307514
Pickard, M., and Williams, G. (2015). Molecular and Cellular Mechanisms of Action of Tumour Suppressor GAS5 LncRNA. Genes 6 (3), 484–499. doi:10.3390/genes6030484
Pourhanifeh, M. H., Vosough, M., Mahjoubin-Tehran, M., Hashemipour, M., Nejati, M., Abbasi-Kolli, M., et al. (2020). Autophagy-related microRNAs: Possible Regulatory Roles and Therapeutic Potential in and Gastrointestinal Cancers. Pharmacol. Res. 161, 105133. doi:10.1016/j.phrs.2020.105133
Qu, S., Yang, X., Li, X., Wang, J., Gao, Y., Shang, R., et al. (2015). Circular RNA: a New Star of Noncoding RNAs. Cancer Lett. 365 (2), 141–148. doi:10.1016/j.canlet.2015.06.003
Raal, F. J., Honarpour, N., Blom, D. J., Hovingh, G. K., Xu, F., Scott, R., et al. (2015). Inhibition of PCSK9 with Evolocumab in Homozygous Familial Hypercholesterolaemia (TESLA Part B): a Randomised, Double-Blind, Placebo-Controlled Trial. Lancet 385 (9965), 341–350. doi:10.1016/s0140-6736(14)61374-x
Raal, F. J., Rosenson, R. S., Reeskamp, L. F., Hovingh, G. K., Kastelein, J. J. P., Rubba, P., et al. (2020). Evinacumab for Homozygous Familial Hypercholesterolemia. N. Engl. J. Med. 383 (8), 711–720. doi:10.1056/nejmoa2004215
Raal, F. J., Santos, R. D., Blom, D. J., Marais, A. D., Charng, M.-J., Cromwell, W. C., et al. (2010). Mipomersen, an Apolipoprotein B Synthesis Inhibitor, for Lowering of LDL Cholesterol Concentrations in Patients with Homozygous Familial Hypercholesterolaemia: a Randomised, Double-Blind, Placebo-Controlled Trial. Lancet 375 (9719), 998–1006. doi:10.1016/s0140-6736(10)60284-x
Rayner, K. J., and Hennessy, E. J. (2013). Extracellular Communication via microRNA: Lipid Particles Have a New Message. J. lipid Res. 54 (5), 1174–1181. doi:10.1194/jlr.r034991
Rayner, K. J., Suárez, Y., Dávalos, A., Parathath, S., Fitzgerald, M. L., Tamehiro, N., et al. (2010). MiR-33 Contributes to the Regulation of Cholesterol Homeostasis. science 328 (5985), 1570–1573. doi:10.1126/science.1189862
Rinaldi, C., and Wood, M. J. A. (2018). Antisense Oligonucleotides: the Next Frontier for Treatment of Neurological Disorders. Nat. Rev. Neurol. 14 (1), 9–21. doi:10.1038/nrneurol.2017.148
Robinson, J. G., Farnier, M., Krempf, M., Bergeron, J., Luc, G., Averna, M., et al. (2015). Efficacy and Safety of Alirocumab in Reducing Lipids and Cardiovascular Events. N. Engl. J. Med. 372 (16), 1489–1499. doi:10.1056/nejmoa1501031
Rozema, D. B. (2017). The Chemistry of Oligonucleotide Delivery. Annu. Rep. Med. Chem. 50, 17–59. doi:10.1016/bs.armc.2017.07.003
Rupaimoole, R., and Slack, F. J. (2017). MicroRNA Therapeutics: towards a New Era for the Management of Cancer and Other Diseases. Nat. Rev. Drug Discov. 16 (3), 203–222. doi:10.1038/nrd.2016.246
Sabatine, M. S., Giugliano, R. P., Keech, A. C., Honarpour, N., Wiviott, S. D., Murphy, S. A., et al. (2017). Evolocumab and Clinical Outcomes in Patients with Cardiovascular Disease. N. Engl. J. Med. 376 (18), 1713–1722. doi:10.1056/nejmoa1615664
Salerno, A. G., Van Solingen, C., Scotti, E., Wanschel, A. C. B. A., Afonso, M. S., Oldebeken, S. R., et al. (2020). LDL Receptor Pathway Regulation by miR-224 and miR-520d. Front. Cardiovasc. Med. 7, 81. doi:10.3389/fcvm.2020.00081
Sallam, T., Jones, M., Gilliland, T., Li, Z., Wu, X., Eskin, A., et al. (2015). Feedback Regulation of Cholesterol Metabolism by Lexis, a Lipid-Responsive Non-coding RNA. Circulation 132 (Suppl. l_3), A13674–A. doi:10.1038/nature17674
Sallam, T., Jones, M. C., Gilliland, T., Zhang, L., Wu, X., Eskin, A., et al. (2016). Feedback Modulation of Cholesterol Metabolism by the Lipid-Responsive Non-coding RNA LeXis. Nature 534 (7605), 124–128. doi:10.1038/nature17674
Salmena, L., Poliseno, L., Tay, Y., Kats, L., and Pandolfi, P. P. (2011). A ceRNA Hypothesis: the Rosetta Stone of a Hidden RNA Language? Cell 146 (3), 353–358. doi:10.1016/j.cell.2011.07.014
Salomon, W. E., Jolly, S. M., Moore, M. J., Zamore, P. D., and Serebrov, V. (2015). Single-molecule Imaging Reveals that Argonaute Reshapes the Binding Properties of its Nucleic Acid Guides. Cell 162 (1), 84–95. doi:10.1016/j.cell.2015.06.029
Santovito, D., Egea, V., Bidzhekov, K., Natarelli, L., Mourão, A., Blanchet, X., et al. (2020). Noncanonical Inhibition of Caspase-3 by a Nuclear microRNA Confers Endothelial Protection by Autophagy in Atherosclerosis. Sci. Transl. Med. 12 (546), eaaz2294. doi:10.1126/scitranslmed.aaz2294
Santovito, D., and Weber, C. (2022). Non-canonical Features of microRNAs: Paradigms Emerging from Cardiovascular Disease. Nat. Rev. Cardiol. 2022, 1–19. doi:10.1038/s41569-022-00680-2
Santovito, D., Marcantonio, P., Mastroiacovo, D., Natarelli, L., Mandolini, C., De Nardis, V., et al. (2020). High Dose Rosuvastatin Increases ABCA1 Transporter in Human Atherosclerotic Plaques in a Cholesterol-independent Fashion. Int. J. Cardiol. 299, 249–253. doi:10.1016/j.ijcard.2019.07.094
Schlackow, M., Nojima, T., Gomes, T., Dhir, A., Carmo-Fonseca, M., and Proudfoot, N. J. (2017). Distinctive Patterns of Transcription and RNA Processing for Human lincRNAs. Mol. cell 65 (1), 25–38. doi:10.1016/j.molcel.2016.11.029
Schroeder, C. I., Swedberg, J. E., Withka, J. M., Rosengren, K. J., Akcan, M., Clayton, D. J., et al. (2014). Design and Synthesis of Truncated EGF-A Peptides that Restore LDL-R Recycling in the Presence of PCSK9 In Vitro. Chem. Biol. 21 (2), 284–294. doi:10.1016/j.chembiol.2013.11.014
Scicali, R., Di Pino, A., Pavanello, C., Ossoli, A., Strazzella, A., Alberti, A., et al. (2019). Analysis of HDL-microRNA Panel in Heterozygous Familial Hypercholesterolemia Subjects with LDL Receptor Null or Defective Mutation. Sci. Rep. 9 (1), 20354–20359. doi:10.1038/s41598-019-56857-2
Shafabakhsh, R., Arianfar, F., Vosough, M., Mirzaei, H. R., Mahjoubin-Tehran, M., khanbabaei, H., et al. (2021). Autophagy and Gastrointestinal Cancers: the behind the Scenes Role of Long Non-coding RNAs in Initiation, Progression, and Treatment Resistance. Cancer Gene Ther. 28 (12), 1229–1255. doi:10.1038/s41417-020-00272-7
Shen, S., Zheng, X., Zhu, Z., Zhao, S., Zhou, Q., Song, Z., et al. (2019). Silencing of GAS5 Represses the Malignant Progression of Atherosclerosis through Upregulation of miR-135a. Biomed. Pharmacother. 118, 109302. doi:10.1016/j.biopha.2019.109302
Statello, L., Guo, C.-J., Chen, L.-L., and Huarte, M. (2021). Gene Regulation by Long Non-coding RNAs and its Biological Functions. Nat. Rev. Mol. Cell Biol. 22 (2), 96–118. doi:10.1038/s41580-020-00315-9
Sun, G., Li, Y., and Ji, Z. (2019). Up-regulation of MIAT Aggravates the Atherosclerotic Damage in Atherosclerosis Mice through the Activation of PI3K/Akt Signaling Pathway. Drug Deliv. 26 (1), 641–649. doi:10.1080/10717544.2019.1628116
Sun, Y., Jiang, X., Lv, Y., Liang, X., Zhao, B., Bian, W., et al. (2020). Circular RNA Expression Profiles in Plasma from Patients with Heart Failure Related to Platelet Activity. Biomolecules 10 (2), 187. doi:10.3390/biom10020187
Talhouarne, G. J. S., and Gall, J. G. (2014). Lariat Intronic RNAs in the Cytoplasm of Xenopus Tropicalis Oocytes. Rna 20 (9), 1476–1487. doi:10.1261/rna.045781.114
Tikka, A., and Jauhiainen, M. (2016). The Role of ANGPTL3 in Controlling Lipoprotein Metabolism. Endocrine 52 (2), 187–193. doi:10.1007/s12020-015-0838-9
Treiber, T., Treiber, N., and Meister, G. (2019). Regulation of microRNA Biogenesis and its Crosstalk with Other Cellular Pathways. Nat. Rev. Mol. Cell Biol. 20 (1), 5–20. doi:10.1038/s41580-018-0059-1
Tsilimigras, D. I., Bibli, S.-I., Siasos, G., Oikonomou, E., Perrea, D. N., Filis, K., et al. (2021). Regulation of Long Non-coding RNAs by Statins in Atherosclerosis. Biomolecules 11 (5), 623. doi:10.3390/biom11050623
Tsimikas, S., Viney, N. J., Hughes, S. G., Singleton, W., Graham, M. J., Baker, B. F., et al. (2015). Antisense Therapy Targeting Apolipoprotein(a): a Randomised, Double-Blind, Placebo-Controlled Phase 1 Study. Lancet 386 (10002), 1472–1483. doi:10.1016/s0140-6736(15)61252-1
Ubilla, C. G., Prado, Y., Angulo, J., Obreque, I., Paez, I., Saavedra, N., et al. (2021). MicroRNA-33b Is a Potential Non-invasive Biomarker for Response to Atorvastatin Treatment in Chilean Subjects with Hypercholesterolemia: A Pilot Study. Front. Pharmacol. 12, 674252. doi:10.3389/fphar.2021.674252
Umans-Eckenhausen, M. A., Defesche, J. C., Sijbrands, E. J., Scheerder, R. L., and Kastelein, J. J. (2001). Review of First 5 Years of Screening for Familial Hypercholesterolaemia in the Netherlands. Lancet 357 (9251), 165–168. doi:10.1016/s0140-6736(00)03587-x
Usifo, E., Leigh, S. E. A., Whittall, R. A., Lench, N., Taylor, A., Yeats, C., et al. (2012). Low-Density Lipoprotein Receptor Gene Familial Hypercholesterolemia Variant Database: Update and Pathological Assessment. Ann. Hum. Genet. 76 (5), 387–401. doi:10.1111/j.1469-1809.2012.00724.x
Uszczynska-Ratajczak, B., Lagarde, J., Frankish, A., Guigó, R., and Johnson, R. (2018). Towards a Complete Map of the Human Long Non-coding RNA Transcriptome. Nat. Rev. Genet. 19 (9), 535–548. doi:10.1038/s41576-018-0017-y
Vallejo-Vaz, A. J., Akram, A., Kondapally Seshasai, S. R., Cole, D., Watts, G. F., Hovingh, G. K., et al. (2016). Pooling and Expanding Registries of Familial Hypercholesterolaemia to Assess Gaps in Care and Improve Disease Management and Outcomes: Rationale and Design of the Global EAS Familial Hypercholesterolaemia Studies Collaboration. Atheroscler. Suppl. 22, 1–32. doi:10.1016/j.atherosclerosissup.2016.10.001
van Poelgeest, E. P., Hodges, M. R., Moerland, M., Tessier, Y., Levin, A. A., Persson, R., et al. (2015). Antisense-mediated Reduction of Proprotein Convertase Subtilisin/kexin Type 9 (PCSK9): a First-In-Human Randomized, Placebo-Controlled Trial. Br. J. Clin. Pharmacol. 80 (6), 1350–1361. doi:10.1111/bcp.12738
van Poelgeest, E. P., Swart, R. M., Betjes, M. G. H., Moerland, M., Weening, J. J., Tessier, Y., et al. (2013). Acute Kidney Injury during Therapy with an Antisense Oligonucleotide Directed against PCSK9. Am. J. kidney Dis. 62 (4), 796–800. doi:10.1053/j.ajkd.2013.02.359
Vickers, K. C., and Michell, D. L. (2021). HDL-small RNA Export, Transport, and Functional Delivery in Atherosclerosis. Curr. Atheroscler. Rep. 23 (7), 1–10. doi:10.1007/s11883-021-00930-7
Vickers, K. C., Palmisano, B. T., Shoucri, B. M., Shamburek, R. D., and Remaley, A. T. (2011). MicroRNAs Are Transported in Plasma and Delivered to Recipient Cells by High-Density Lipoproteins. Nat. Cell Biol. 13 (4), 423–433. doi:10.1038/ncb2210
Vučićević, D., Corradin, O., Ntini, E., Scacheri, P. C., and Ørom, U. A. (2015). Long ncRNA Expression Associates with Tissue-specific Enhancers. Cell cycle 14 (2), 253–260. doi:10.4161/15384101.2014.977641
Wang, K., Long, B., Liu, F., Wang, J.-X., Liu, C.-Y., Zhao, B., et al. (2016). A Circular RNA Protects the Heart from Pathological Hypertrophy and Heart Failure by Targeting miR-223. Eur. Heart J. 37 (33), 2602–2611. doi:10.1093/eurheartj/ehv713
Wang, X., Chen, X., Zhang, X., Su, C., Yang, M., He, W., et al. (2020). A Small-Molecule Inhibitor of PCSK9 Transcription Ameliorates Atherosclerosis through the Modulation of FoxO1/3 and HNF1α. EBioMedicine 52, 102650. doi:10.1016/j.ebiom.2020.102650
Watanabe, K., Harada-Shiba, M., Suzuki, A., Gokuden, R., Kurihara, R., Sugao, Y., et al. (2009). In Vivo siRNA Delivery with Dendritic Poly(l-Lysine) for the Treatment of Hypercholesterolemia. Mol. Biosyst. 5 (11), 1306–1310. doi:10.1039/b900880b
Watanabe, T. A., Geary, R. S., and Levin, A. A. (2006). Plasma Protein Binding of an Antisense Oligonucleotide Targeting Human ICAM-1 (ISIS 2302). Oligonucleotides 16 (2), 169–180. doi:10.1089/oli.2006.16.169
Watts, G. F., Gidding, S., Wierzbicki, A. S., Toth, P. P., Alonso, R., Brown, W. V., et al. (2014). Integrated Guidance on the Care of Familial Hypercholesterolaemia from the International FH Foundation. Int. J. Cardiol. 171 (3), 309–325. doi:10.1016/j.ijcard.2013.11.025
Wiegman, A., Gidding, S. S., Watts, G. F., Chapman, M. J., Ginsberg, H. N., Cuchel, M., et al. (2015). Familial Hypercholesterolaemia in Children and Adolescents: Gaining Decades of Life by Optimizing Detection and Treatment. Eur. Heart J. 36 (36), 2425–2437. doi:10.1093/eurheartj/ehv157
Winkle, M., El-Daly, S. M., Fabbri, M., and Calin, G. A. (2021). Noncoding RNA Therapeutics—Challenges and Potential Solutions. Nat. Rev. Drug Discov. 20, 629–651. doi:10.1038/s41573-021-00219-z
Winkle, M., El-Daly, S. M., Fabbri, M., and Calin, G. A. (2021). Noncoding RNA Therapeutics - Challenges and Potential Solutions. Nat. Rev. Drug Discov. 20, 629–651. doi:10.1038/s41573-021-00219-z
Xu, Y., Zalzala, M., Xu, J., Li, Y., Yin, L., and Zhang, Y. (2015). A Metabolic Stress-Inducible miR-34a-Hnf4α Pathway Regulates Lipid and Lipoprotein Metabolism. Nat. Commun. 6 (1), 7466–7511. doi:10.1038/ncomms8466
Yadav, P. K., Haruehanroengra, P., Irani, S., Wang, T., Ansari, A., Sheng, J., et al. (2022). Novel Efficacious microRNA-30c Analogs Reduce Apolipoprotein B Secretion in Human Hepatoma and Primary Hepatocyte Cells. J. Biol. Chem. 298 (4), 101813. doi:10.1016/j.jbc.2022.101813
Yamamoto, T., Harada-Shiba, M., Nakatani, M., Wada, S., Yasuhara, H., Narukawa, K., et al. (2012). Cholesterol-lowering Action of BNA-Based Antisense Oligonucleotides Targeting PCSK9 in Atherogenic Diet-Induced Hypercholesterolemic Mice. Mol. Ther. - Nucleic Acids 1, e22. doi:10.1038/mtna.2012.16
Yang, G., Lu, X., and Yuan, L. (2014). LncRNA: a Link between RNA and Cancer. Biochimica Biophysica Acta (BBA) - Gene Regul. Mech. 1839 (11), 1097–1109. doi:10.1016/j.bbagrm.2014.08.012
Ye, W-C., Huang, S-F., Hou, L-J., Long, H-J., Yin, K., Hu, C. Y., et al. (2021). Potential Therapeutic Targeting of lncRNAs in Cholesterol Homeostasis. Front. Cardiovasc. Med. 8 (585). doi:10.3389/fcvm.2021.688546
Yin, Y., Lu, J. Y., Zhang, X., Shao, W., Xu, Y., Li, P., et al. (2020). U1 snRNP Regulates Chromatin Retention of Noncoding RNAs. Nature 580 (7801), 147–150. doi:10.1038/s41586-020-2105-3
Zaporozhchenko, I. A., Rykova, E. Y., and Laktionov, P. P. (2020). The Fundamentals of miRNA Biology: Structure, Biogenesis, and Regulatory Functions. Russ. J. Bioorg Chem. 46 (1), 1–13. doi:10.1134/s106816202001015x
Zhang, B., You, C., Zhang, Y., Zeng, L., Hu, J., Zhao, M., et al. (2020). Linking Key Steps of microRNA Biogenesis by TREX-2 and the Nuclear Pore Complex in Arabidopsis. Nat. Plants 6 (8), 957–969. doi:10.1038/s41477-020-0726-z
Zhang, D.-W., Lagace, T. A., Garuti, R., Zhao, Z., McDonald, M., Horton, J. D., et al. (2007). Binding of Proprotein Convertase Subtilisin/kexin Type 9 to Epidermal Growth Factor-like Repeat A of Low Density Lipoprotein Receptor Decreases Receptor Recycling and Increases Degradation. J. Biol. Chem. 282 (25), 18602–18612. doi:10.1074/jbc.m702027200
Zhang, X.-O., Dong, R., Zhang, Y., Zhang, J.-L., Luo, Z., Zhang, J., et al. (2016). Diverse Alternative Back-Splicing and Alternative Splicing Landscape of Circular RNAs. Genome Res. 26 (9), 1277–1287. doi:10.1101/gr.202895.115
Zhang, X.-O., Wang, H.-B., Zhang, Y., Lu, X., Chen, L.-L., and Yang, L. (2014). Complementary Sequence-Mediated Exon Circularization. Cell 159 (1), 134–147. doi:10.1016/j.cell.2014.09.001
Zhang, X., Price, N. L., and Fernández-Hernando, C. (2019). Non-coding RNAs in Lipid Metabolism. Vasc. Pharmacol. 114, 93–102. doi:10.1016/j.vph.2018.06.011
Zhang, Y., Eigenbrot, C., Zhou, L., Shia, S., Li, W., Quan, C., et al. (2014). Identification of a Small Peptide that Inhibits PCSK9 Protein Binding to the Low Density Lipoprotein Receptor. J. Biol. Chem. 289 (2), 942–955. doi:10.1074/jbc.m113.514067
Zhao, H., Li, Y., He, L., Pu, W., Yu, W., Li, Y., et al. (2020). In Vivo AAV-CRISPR/Cas9-Mediated Gene Editing Ameliorates Atherosclerosis in Familial Hypercholesterolemia. Circulation 141 (1), 67–79. doi:10.1161/circulationaha.119.042476
Zhao, J., Sun, B. K., Erwin, J. A., Song, J.-J., and Lee, J. T. (2008). Polycomb Proteins Targeted by a Short Repeat RNA to the Mouse X Chromosome. Science 322 (5902), 750–756. doi:10.1126/science.1163045
Zhao, Y., Wu, J., Liangpunsakul, S., and Wang, L. (2017). Long Non-coding RNA in Liver Metabolism and Disease: Current Status. Liver Res. 1 (3), 163–167. doi:10.1016/j.livres.2017.09.001
Zimmermann, T. S., Lee, A. C. H., Akinc, A., Bramlage, B., Bumcrot, D., Fedoruk, M. N., et al. (2006). RNAi-mediated Gene Silencing in Non-human Primates. Nature 441 (7089), 111–114. doi:10.1038/nature04688
Keywords: familial hypercholesterolemia, low-density lipoprotein cholesterol, cardiovascular disease, advanced therapy, ncRNA-based therapy, ncRNA-based diagnosis
Citation: Keshavarz Alikhani H, Pourhamzeh M, Seydi H, Shokoohian B, Hossein-khannazer N, Jamshidi-adegani F, Al-Hashmi S, Hassan M and Vosough M (2022) Regulatory Non-Coding RNAs in Familial Hypercholesterolemia, Theranostic Applications. Front. Cell Dev. Biol. 10:894800. doi: 10.3389/fcell.2022.894800
Received: 12 March 2022; Accepted: 01 June 2022;
Published: 23 June 2022.
Edited by:
Hani Hosseini Far, Hudson Institute of Medical Research, AustraliaReviewed by:
Donato Santovito, Institute of Genetic and Biomedical Research, (CNR), ItalyAbulaish Ansari, NYU Langone Health, United States
Copyright © 2022 Keshavarz Alikhani, Pourhamzeh, Seydi, Shokoohian, Hossein-khannazer, Jamshidi-adegani, Al-Hashmi, Hassan and Vosough. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Massoud Vosough, bWFzdm9zQFJveWFuaW5zdGl0dXRlLm9yZw==
 Hani Keshavarz Alikhani1
Hani Keshavarz Alikhani1 Fatemeh Jamshidi-adegani
Fatemeh Jamshidi-adegani Moustapha Hassan
Moustapha Hassan Massoud Vosough
Massoud Vosough