- Department of Oncology, Shengjing Hospital of China Medical University, Shenyang, China
The modulation of the function and expression of epigenetic regulators of RNA modification has gradually become the hotspot of cancer research. Studies have shown that alteration of epigenetic modifications can promote the development and metastasis of breast cancer. This review highlights the progress in characterization of the link between RNA modification and the prognosis, carcinogenesis and treatment of breast cancer, which may provide a new theoretical basis for development of effective strategies for monitoring of breast cancer based on epigenetics.
Introduction
Breast cancer is the most common female cancer in the world (Harbeck et al., 2019). Current treatments for breast cancer include surgery, chemotherapy, radiotherapy, hormone therapy and targeted therapy (Bray et al., 2018). However, comprehensive treatment strategies for breast cancer are still limited. Therefore, eradication of breast cancer remains a significant challenge, and there is an urgent need for new treatment strategies (Pedrosa et al., 2018). All biological macromolecules require post-synthesis and covalent modifications (Maresca and Wismayer, 2016). Over 100 different kinds of post-synthetic modifications have been identified to exist in RNA, and the four kinds of RNA bases and ribose can be targets for modification (He et al., 2019). Studies have shown that RNA can exert functional effects on the expression of gene in addition to acting as an effector of protein synthesis. Therefore, the importance of RNA modification has received increased attention, and studies have shown that dysregulation of RNA modification may be associated with human diseases, including breast cancer (Huang et al., 2020; Boccaletto et al., 2022). Herein, we review the progress of research focused on RNA modification and regulators of RNA modification in breast cancer.
Ribonucleic Acid Modification Regulatory Proteins in Breast Cancer
Eight different internal RNA modifications associated with a variety of cancers have been characterized: methylation of adenosine on position 6 on RNA to generate N 6-methyladenosine (m6A); methylation of cytidine on position 5 to produce RNA with 5-methylcytosine (m5C); methylation at position 1 of adenosine on tRNA to formN-1-methyladenosine (m1A); 7-methylguanosine (m7G); pseudouridylation of RNA to produce pseudouridine; editing of RNA adenosine to inosine; U34 modification of tRNA; N4-acetylcytidine (ac4C) (Rong et al., 2021). Modification of RNA is a dynamic process that includes insertions, deletions, and recognition via specific cellular components called “writers,” “erasers” and “readers” respectively.
N6-Methyladenosine Methyltransferases
Dynamic and reversible RNA modification plays a key role in maintaining RNA balance, and can affect splicing, translation, degradation, and localization of RNA, resulting in the regulation of various biological functions in human disease (Li and Mason, 2014). Ribosomal RNA (rRNA) and transport RNA (tRNA) are the two most abundant RNAs. Post-transcriptional modifications are very common on rRNA and tRNA (Frye et al., 2018). Continuous development of technology to detect RNA modifications has allowed for identification of post-transcriptional modifications of messenger RNA (mRNA) and non-coding RNA (ncRNA) (Zhao et al., 2017). N6-methyladenosine (m6A) is the most common mRNA modification in mammals. In addition, m6A was shown to play an important role in stem cell self-renewal, metabolism and metastasis in multiple cancers (Dong et al., 2021; Wood et al., 2021). The methyltransferase complex (MTC), also known as the m6A “writer,” catalyzes m6A modification of adenylate on mRNA, and includes methyltransferase-like 3 (METTL3), methyltransferase-like 14 (METTL14), Wilms tumor 1 associated protein (WTAP), RNA binding motif protein 15/15B (RBM15/15B), Cbl proto-oncogene like 1 (CBLL1), zinc finger CCCH type containing 13 (ZC3H13), KIAA1429, CCHC-type zinc finger protein (ZCCHC4) and methyltransferase-like 16 (METTL16). METTL3 is a major catalytic enzyme in the N6-adenine methyltransferase system. The expression level of METTL3 is not consistent in each subtype of breast cancer (Yang et al., 2020). It was reported to play a tumor-suppressive role in triple-negative breast cancer (TNBC) while play an oncogenic role in other subtypes (Shi et al., 2020; He et al., 2021; Ruan et al., 2021). METTL14 stabilizes METTL3 and recognizes target RNA, which is found to be an oncogene or a tumor suppressor gene in breast cancer (Gong et al., 2020; Sun et al., 2020). WTAP is the main regulatory component of the m6A methylation complex, and has mutual effects with METTL3 and METTL14 to aid in nuclear localization. The expression of WTAP varied in different in breast cancer studies (Wu et al., 2019; Wang et al., 2022). RBM15/RBM15B interacts with spliceosome components to participate in the modulation of m6A modification in a WTAP-dependent manner. RBM15 was identified to be significantly high in TNBC (Yang et al., 2020). ZC3H13 is critical for anchoring regulatory complex in the nucleus. It was recognized as a tumorsupressor which positively related with tumor infiltrating lymphocytes (TILs) in the breast cancer (Gong et al., 2020). KIAA1429 is essential in the methylation process as a candidate new subunit in the methylase complex. High expression of KIAA1429 was associated with a poor prognosis in breast cancer (Zhang et al., 2022). CBLL1, as a co-regulator of m6A methylation, was proved to promote the apoptosis in breast cancer (Zheng F. et al., 2021). ZCCHC4, a novel methyltransferase in the mediating of ribosome methylation, has a high expression in the breast lesion compared with pancancerous tissue (Pinto et al., 2020). METTL16 targets ncRNAs, lncRNAs and pre-mRNAs which is critical in splicing regulation (Su et al., 2022).
N6-Methyladenosine Demethylases
The demethylases ALKB homolog 5 (ALKBH5) and fat mass and obesity-related protein (FTO), also known as “m6A erasers,” remove m6A using ferrous iron as a cofactor and α-ketoglutarate as a co-substrate (Jia et al., 2011; Zheng et al., 2013). ALKBH5 was higher in breast cancer tissue than in adjacent normal tissue of TNBC (Wang S. et al., 2020). FTO can oxidize m6A to N6-hydroxymethylsalicylic acid and N6-formyl adenosine, which can be hydrolyzed to adenine (Fu et al., 2013). The expression of FTO varied in different breast cancer studies. Most of studies show that down-regulation of FTO enhanced the phenotype of invasiveness, migration and EMT in breast cancer (Jeschke et al., 2021). But in other cases, FTO played an oncogenic role with a high expression in breast cancer (Niu et al., 2019).
N6-Methyladenosine Readers
The “readers” mainly include the YTH domain family (YTHDF) and heterogeneous nuclear ribonucleoproteins (hnRNPs) family, Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs) and YTH domain-containing protein (YTHDC) increase the translation levels of modified RNAs which recognize m6A, bind RNA and participate in regulatory functions (Huang et al., 2018; Xing et al., 2019; Dai X.-Y. et al., 2021). The YTHDF family includes three paralogs YTHDF1, YTHDF2 and YTHDF3, which can also be referred to as DF1, DF2, and DF3. DF1 promotes mRNA translation, DF2 promotes mRNA degradation, and DF3 promotes translation and degradation (Zaccara and Jaffrey, 2020). YTHDF1 and YTHDF3 were also found to overexpress in breast cancer (Chen et al., 2022; Lin et al., 2022). IGF2BP 2/3 and YTHDC2 were highly expressed in basal-like breast cancer (Yang et al., 2020). The overexpression of hnRNPc were related to poor prognosis in patients (Lv et al., 2021a), but hnRNPc A2/B1 was reported to negatively regulate the metastasis of breast cancer (Liu Y. et al., 2020). Although various readers, writers, and erasers may be independently associated with numerous changes in signaling pathways of cancer, there is evidence that writers, erasers and readers may have interplay with each other in cancer. Regulators in the same functional category show significant genetic changes and highly correlated expression patterns in cancer (Li et al., 2019). In addition, m6A methylation was involved in regulation of the malignant phenotypes of tumors by controlling the expression of tumor-related genes in breast cancer (Barbieri and Kouzarides, 2020; Zhang et al., 2020). Recent studies have shown that m5C, m1A, m7G, and recently discovered ac4C modifications, also play important roles in RNA processing and metabolism. For example, m5C could promote enucleation of mRNA through binding to its reader protein Aly/REF export factor (ALYREF) (Yang et al., 2017), m1A can affect the translation efficiency of its modified mRNA (Li et al., 2017; Safra et al., 2017), and ac4C stabilizes its modified mRNA and enhances translation efficiency (Arango et al., 2018).
5-Methylcytosine
The m5C modification is involved in the metastasis and proliferation of cancer cells, and the development of cancer stem cells. The currently identified writers of m5C genes include NOP2/Sun RNA methyltransferase 2 (NSUN2), NSUN6, tRNA aspartic acid methyltransferase 1 (TRDMT1), tRNA-specific methyltransferase 4B (TRM4B) and OsNSUN2 (Bujnicki et al., 2004; Moon and Redman, 2014; Liu et al., 2017; Muller et al., 2019; Tang et al., 2020; Li H. et al., 2021). The “readers” include ALYREF, DNA repair protein RAD52 homolog (RAD52) and Y-box binding protein 1 (YBX1) (Yang et al., 2017; Chen et al., 2020; Xue et al., 2021).
N1-Methyladenosine
The main modification of tRNA is m1A, which has also been found in 28SrRNA. The tRNA methyltransferase 10 homologue A (TRM)-TRM61 complex is the only known methyltransferase that catalyzes m1A modification (Saikia et al., 2010), and YTH protein family is a potential reader of m1A modifications (Dai et al., 2018). In addition, ALKBH3 is an eraser of m1A (Li et al., 2016).
7-Methylguanosine
The m7G modification was illustrated as part of the type O’ cap structure of mRNA and was also observed in rRNA and tRNA. The m7G maintained the integrity of structure mediated by the METTL1-WDR4 complex (Dai Z. et al., 2021). In addition, the m7G modification on rRNA is induced by Williams Beuren syndrome chromosome 22 region protein (WBSCR22) (Haag et al., 2015). Up-regulation of METTL1/WDR4 can promote the level of m7G modification on tRNAs, which in turn promotes the stability of tRNAs and the translation of mRNAs (Katsara and Schneider, 2021).
Pseudouridine
Pseudouridine can maintain the structure and stability of tRNA. The most-studied regulatory factor related to pseudouridine modification is Dyskerin Pseudouridine Synthase 1 (DKC1), which is a component of a small nucleolar ribonucleoprotein complex, needs RNA guidance to exert its catalytic activity, is overexpressed in various types of cancer.
Adenosine-to-Inosine Editing
Adenosine deaminases targeting RNA (ADARs) are effective in RNA editing, and are particularly important in the process of converting adenosine residues in double stranded RNA to creatinine (Ota et al., 2013). The ADAR1p110 subtype can regulate the stability of the chromosome terminal genome, and is required for continuous proliferation of cancer cells (Shiromoto et al., 2021).
U34 on Transport Ribonucleic Acid
Establishment of the U34 modification results from three steps: modification of U34 with an extender complex to produce 5-carboxymethyluridine (cm5U), transformation of cm5u to 5-methoxycarbonylmethyluridine (mcm5U) mediated by ALKBH8. Finally, thiolase, cytoplasmic trna2 thiolated protein 1 (CTU1), and CTU2 promote the formation of 5-methoxycarbonylmethyl-2-thiouridine (mcm5s2U) on specific tRNA (tRNAUUULys, tRNArUCGlu and tRNAAUGln) (Rapino et al., 2017).
N4-Acetylcytidine
N4-acetylcytodine (ac4C) is a conserved chemical modification in eukaryotes and prokaryotes. Early studies suggested that ac4C modifications mainly occurred on tRNA and 18SrRNA. Recent studies showed extensive ac4C modifications on mRNA, with similar abundance to the m7G cap modification on mRNA. To date, N-acetyltransferase 10 (NAT10) is the only protein known to have both an acetylase domain and an RNA-binding domain, and is therefore considered an RNA ac4C-modifying enzyme (Sas-Chen et al., 2020; Yang C. et al., 2021).
Association of Ribonucleic Acid Modification and Breast Cancer Prognosis
Data from public databases and clinical studies have indicated that levels of RNA modification regulators have prognostic value for breast cancer (Zheng F. et al., 2021). Low expression of METTL3, METTL14, WTAP and FTO was shown to correlate with relapse-free survival in breast cancer (Wu et al., 2019). METTL3 was also demonstrated to be related with a poor survival rate in breast cancer (Wang H. et al., 2020). METTL14 and ZC3H13 were associated with favorable prognosis, and correlated with adenomatous polyposis coli (APC). Furthermore, ZC3H13, METTL14 and APC expression levels were positively related with the number of infiltrating immune cells in breast cancer (Gong et al., 2020). The regulators YTHDF1, YTHDF3 and KIAA1429 were found to be upregulated in breast cancer, and were associated with the metastasis of lymph nodes, breast cancer progression, and also were predictors of poor prognosis (Liu et al., 2019; Anita et al., 2020; He et al., 2021). The demethylase ALKBH5 was found to be associated with poor prognosis in patients with TNBC (Wang S. et al., 2020). FTO was associated with short survival in Her-2 positive breast cancer (Xu et al., 2020). Non-coding RNAs such as miRNA, lncRNA, and circRNA, can undergo m6A modification, which regulates their expression and function. Ten m6A-modified lncRNAs-LINC00571, ANKRD10- IT1, LINC00593, miR-205HG, CIRBP- AS1, BLACAT1, SUCLG2- AS1, SAMD12- AS1, BVES-AS1, a18SrRNA nd HOXB-AS1 were used to construct a prognostic score model, and may be potential predictors of survival in patients with TNBC (Wu et al., 2021). A prognostic risk model comprised of six m6A-regulated lncRNAs-Z68871.1, AL122010.1, AL138724.1,OTUD6B-AS1, AC090948.3 and eosinophil granule ontogeny transcript (EGOT) for high-risk patients with tumor-infiltrating immune cells, indicated that m6A-regulated lncRNAs may modulate the immune microenvironment in breast cancer (Lv et al., 2021b). High expression of the m6A regulator hnRNPC, and low expression of hsa-miR-944, are associated with advanced stage breast cancer and poor prognosis (Lv et al., 2021a). Basal-like subtypes and other breast cancer subtypes are associated with the m6A regulators YTHDC2, IGF2BP2, IGF2BP3 and RBM15, and luminal A and B subtypes are classified into two clusters according to the methylation status of these four regulators. In addition, cluster1 has been found to be associated with cell adhesion signaling pathways and immune-associated genes of TILs. Furthermore, cluster1 was related to poor prognosis among patients with stage II and luminal B of breast cancer. The accuracy of diagnosis and efficacy of treatment may be improved by using m6A regulators as biomarkers of different subtypes (Yang et al., 2020). These studies indicated that METTL3, METTL14, WTAP, FTO, ALKBH5, and other N6-methyladenosine-related lncRNAs were associated with progression of breast cancer, and may be prognostic indicators. Changes in expression and activity of m6A modulators may promote breast cancer progression (Chen and Du, 2019; Lv et al., 2021b; Zhang et al., 2021) (Table 1). Few studies have mentioned m5C modifications and breast cancer, and most have focused on NSUN2. It was reported that NSUN2 expression was associated with tumor stage and pathological subtype of breast cancer. The m5C RNA methylation regulators NSUN2 and NSUN6 were predictors of survival and affected the progression and tumor immune microenvironment in TNBC (Huang Z. et al., 2021). Low expression of DKC1, rRNA pseudouridine modification, and decreased intrinsic ribosomal activity are associated with better breast cancer prognosis (Elsharawy et al., 2020; Guerrieri et al., 2020). In addition, the U34 modification enzymes ELP3, CTU1, and CTU2 were shown to be upregulated in breast cancer (Delaunay et al., 2016) (Table 1). Determination of the predictive value of mRNA m7G and m1A modifications, editing of RNA adenosine to inosine, U34 modification of tRNA, or ac4C-related effectors for tumor prognosis require further study.
Roles of the Ribonucleic Acid Modification in the Carcinogenesis of Breast Cancer
Previous studies have proven that m6A levels were strongly associated with cancer, which indicated that m6A may play a crucial role in the occurrence or inhibition of malignant tumors (Helm and Motorin, 2017; Mohammad et al., 2019; Gu et al., 2020).
Ribonucleic Acid Modification Regulators in the Proliferation, Invasion and Metastasis of Breast Cancer
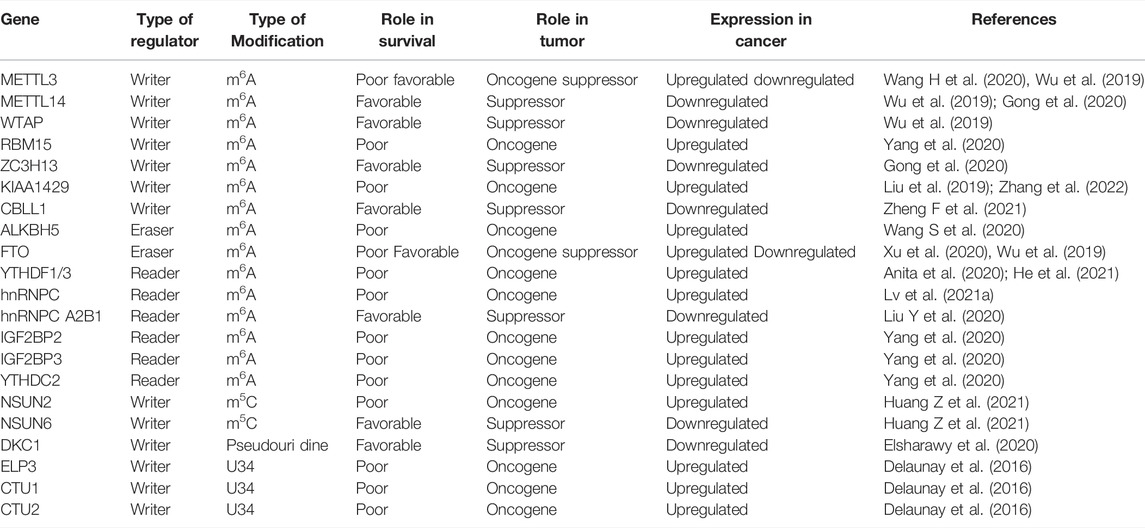
The writer KIAA1429 promotes proliferation and metastasis of breast cancer by modulating cyclin-dependent kinase 1 (CDK1) (Qian et al., 2019). Studies showed that the increasing of METTL3 promoted proliferation and inhibited apoptosis in breast cancer by targeting Bcl-2 (Wang H. et al., 2020). Hepatitis B X-interacting protein (HBXIP) upregulated the expression of METTL3 by inhibiting the miRNA let-7g in another study. In addition, METTL3 activated HBXIP via m6A modification, which promoted cell proliferation in breast cancer as part of a positive feedback loop (Cai et al., 2018). On the contrary, METTL3 played an anti-tumor role by COL3A1 and circMETTL3/miR-34c-3p in TNBC (Shi et al., 2020; Ruan et al., 2021). The expression of circMETTL3 was also found to be increased in breast cancer, and promoted migration, proliferation and invasion of breast cancer cells by targeting miR-31-5p/CDK1 (Li Z. et al., 2021). A further study showed that the m6A levels were significantly upregulated in lung metastatic breast cancer cells, which promoted the translation, elongation, and mRNA stability of keratin 7 (KRT7), a key epithelial-to-mesenchymal transition (EMT)-associated protein, by targeting FTO and METTL3, thereby promoting lung metastasis of breast cancer cells. LINC00675 m6A methylation was increased by METTL3, which resulted in the interaction with miR-513b-5p and inhibition of cancerous properties of breast cancer (Fan and Wang, 2021). LNC942 directly bound to METTL14 and promoted the expression of METTL14 protein through a specific binding domain (+176 to +265), resulting in the regulation of m6A methylation of C-X-C motif chemokine receptor 4 (CXCR4) and cytochrome P450 family 1 subfamily B member 1 (CYP1B) to stabilize their expression and translation and mediate the onset and development of breast cancer (Sun et al., 2020). It was showed that METTL14 increased the expression of has-miR-146a-5p and promoted the invasion and migration of breast cancer (Yi et al., 2020). High level of FTO enhanced the expression of ARL5B by down-regulating miR-181b-3p to promote the invasion and migration of Her-2 positive breast cancer (Xu et al., 2020). FTO mediated m6A demethylation in a YTHDF2-dependent manner and promoted the proliferation and metastasis of breast cancer via inhibiting BCL2 interacting protein 3 (BNIP3) (Niu et al., 2019). IGF2BP1 was shown to bind to LINC00483 and promote the proliferation of breast cancer cells (Qiao et al., 2021). Furthermore, the overexpression of NSUN2 induced by DNA hypomethylation promoted the proliferation, invasiveness and migration of breast cancer cells (Yi et al., 2017). Little is known about the functional mechanisms of m1A-modified RNA. Therefore, epigenetic transcriptome research should focus on the function of m1A-modified RNA. The up-regulation of m1A demethylase ALKBH3 was shown to be involved in decay of macrophage-colony stimulating factor-1 (CSF-1) mRNA, which resulted in promoting breast cancer cell invasiveness (Woo and Chambers, 2019) (Figure 1).
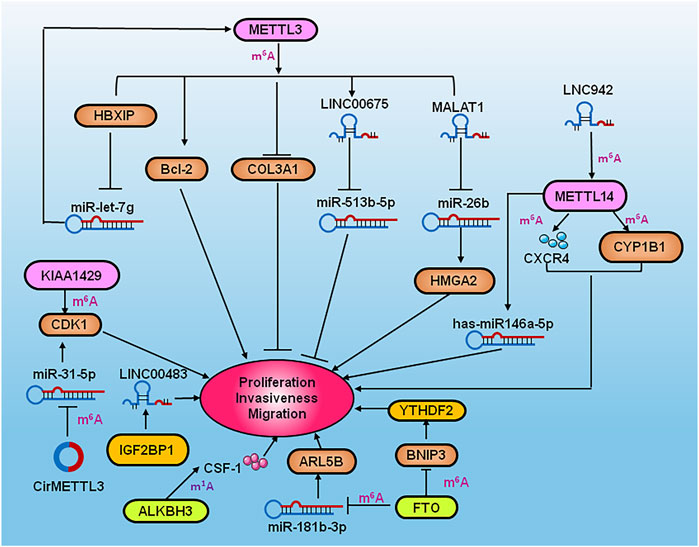
FIGURE 1. Diagram of RNA modification regulators playing a vital role in the proliferation, invasion and metastasis of breast cancer. METTL3 promoted the proliferation, invasion and metastasis by targeting Bcl-2, while it reduced the expression of COL3A1 to inhibit the metastasis of breast cancer. LINC00675 m6A methylation induced by METTL3 resulted in the inhibiting miR-513b-5p to suppress malignant phenotype breast cancer. LncMALAT1 increased/HMGA2 to facilitate the proliferation, invasion and metastasis of breast cancer cells by inhibiting miR-26b. LNC942 directly bound to METTL14 resulting in regulation of m6A methylation of CXCR4 and CYP1B1 and mediate the onset and development of breast cancer. METTL14 also increased the expression of has-miR-146a-5p to promote invasion and migration of breast cancer. The writer KIAA1429 promoted the proliferation and metastasis of breast cancer by regulating CDK1, whereas circMETTL3 promoted the progression of breast cancer cells by targeting miR-31-5p/CDK1. FTO enhanced ARL5B by down-regulating miR-181b-3p to promote the invasion and migration of breast cancer. It also mediated m6A demethylation by YTHDF2 to enhance the proliferation and metastasis of breast cancer via inhibiting BNIP3. IGF2BP1 promote proliferation of breast cancer by binding to LINC00483. ALKBH3 induced the decay of CSF-1 to promote breast cancer cell invasiveness.
Ribonucleic Acid Modification Regulators in the Breast Cancer Stem-Like Cells, Metastasis, Epithelial-to-Mesenchymal Transition, Glycosis and Immune Escape of Breast Caner
METTL3 was shown to methylate adenine 877 on the antisense nucleotide chain KRT7-AS of KRT7, which was recognized by IGF2BP1 and recruited the effector molecule HuR to increase the stability of the KRT7 and KRT7-AS complexes (Chen et al., 2021). METTL3 was demonstrated to upregulate PD-L1 expression via IGF2BP3 by m6A-dependent manner to modulate immune surveillance in breast cancer (Wan et al., 2022). The high level of METTL3 induced EMT, invasion and migration by targeting MALAT1/miR-26b/HMGA2 axis (Li et al., 2022). DROSHA RNase III was upregulated in a number of cancers and interacted with β-catenin to activate stanniocalcin 1 (STC1) in an RNA cleavage-independent manner, which in turn contributed to the properties of breast cancer stem-like cells (BCSCs). Aurora kinase A (AURKA)-induced m6A modification in BCSCs enhanced DROSHA mRNA stability. In addition, AURKA stabilized METTL14 by inhibiting its ubiquitination and degradation, thereby promoting methylation of DROSHA mRNA. Furthermore, binding of AURKA to DROSHA transcripts induced by IGF2BP2 to stabilize m6A-modified DROSHA, which enhanced BCSC stemness (Peng et al., 2021). Complement C5a receptor 1 (C5aR1)-positive neutrophils secreted IL (Interleukin) 1β and tumor necrosis factor α (TNFα) to synergistically activate ERK1/2, which resulted in phosphorylation of WTAP at serine 341, thereby stabilizing WTAP protein to promote RNA m6A methylation of enolase 1 (ENO1) and affected the glycolysis of breast cancer cells (Ou et al., 2021). The overexpression of writer KIAA1429 was shown to bind the 3′-UTR of structural maintenance of chromosomes 1A (SMC1A) to promote EMT in breast cancer (Zhang et al., 2022). Down-regulation of FTO was shown to increase adenine methylation at position 950 on KRT7 mRNA, and enhanced the elongation efficiency of translation by recruiting the effector molecule eEF-1 through the recognition protein YTHDF1. The overexpression of FTO and knockdown of METTL3 and KRT7 reduced lung metastasis (Chen et al., 2021). ALKBH5 or ZNF217 mediated demethylation of m6A in Nanog and KLF4 mRNA. The depleting of ALKBH5 reversed the pluripotency of breast cancer by inhibiting Nanog under hypoxic condition (Zhang et al., 2016). YTHDF3 enhanced the translation of m6A-enriched transcripts of ST6 beta-galactoside alpha-2, 6-sialyltransferase 5 (ST6GALNAC5), gap junction protein alpha 1 (GJA1), epidermal growth factor receptor (EGFR) and vascular endothelial growth factor (VEGF), which promoted breast cancer metastasis to the brain (Chang et al., 2020). Apoptosis was shown to be triggered by the inhibition of YTHDF2-dependent mRNA degradation in TNBC through MAPK pathway-dependent induction of the EMT, and increased the global translation of mRNA synthesis in MYC-driven breast cancers (Einstein et al., 2021). The Lnc RNA KB-1980E6.3 facilitated BCSC self-renewal and carcinogenesis under hypoxic condition. In addition, IGF2BP1 was shown to be recruited by LncRNA KB-1980E6.3 to strengthen the stability of c-Myc mRNA (Zhu et al., 2021). A study showed that CircBACH2 sponged hsa-miR-944, which resulted in MAPK signaling pathway-dependent up-regulation of hnRNPC expression and promotion of breast cancer cell proliferation (Lv et al., 2021a) (Figure 2).
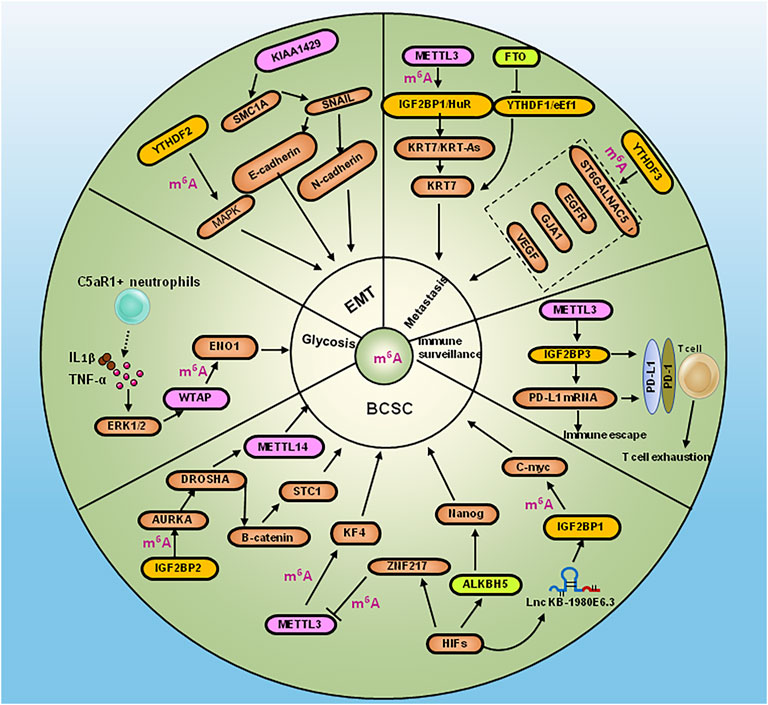
FIGURE 2. Diagram summarizing RNA modification regulators in the pathogensis of BCSC, metastasis, EMT, glycosis and immune escape of breast caner. BCSC: Binding of AURKA to DROSHA transcripts induced by IGF2BP2 to enhance BCSC stemness, meanwhile DROSHA interacted with β-catenin to contribute to the BCSC property by activating STC1. ALKBH5 or ZNF21 increased Nanog and KLF4 mRNA expression by m6A methylation, which led to pluripotency of breast cancer under hypoxic condition. The Lnc RNA KB-1980E6.3 facilitated BCSC self-renewal by IGF2BP1/c-Myc axis under hypoxic condition. Metastasis:YTHDF3 enhanced translation of ST6GALNAC5, GJA1, EGFR and VEFG to promote breast cancer metastasis to the brain. METTL3 and FTO promoted lung metastasis of breast cancer by KRT7 via suppressing YTHDF1/eEf1 complex and increasing IGF2BP1/HuR complex. EMT: YTHDF2 induced EMT by activating MAPK pathway. KIAA1429 promoted EMT by SMC1A/SNAIL in breast cancer. Glysosis: C5aR1-positive neutrophils secreted IL-1β and TNFα to synergistically activate ERK1/2, which resulted in the stabilizing WTAP to affect the glycolysis of breast cancer via ENO1. Immune surveillance: METTL3 upregulate PD-L1 via IGF2BP3 by m6A-dependent manner to modulate immune escape and T cell exhausition in breast cancer.
The effects of RNA modification of target genes on progression of breast cancer depends on three factors: 1) the gene is a suppressor or an oncogene; 2) abnormal levels of RNA methylation in cancer; 3) Regulation of target mRNA modification. Taken together, the current study of m6A RNA methylation in tumors is still at an early stage. RNA modification and its regulators seem to act as a “double-edged sword” in the tumor development, so it is challenging to rationally interpret the controversial findings. It is the functional versatility and tunability of this modification that underscores the important role of the environment in biological process. Therefore, the function of RNA modification may be more complex and extensive than the existing reports, and further exploration of its role in different cancers is expected to provide in-depth insights into tumorigenesis and development.
Ribonucleic Acid Modifications as Potential Drug Targets in Breast Cancer
Modification of RNA connects epigenetic transcriptomics with tumorigenesis and progression, and affects the processes of stem cell self-renewal and differentiation, proliferation and apoptosis, invasion and metastasis, drug resistance, and immunosuppression. Therefore, the key proteins involved in RNA modification are expected to become potential molecular targets for cancer diagnosis and treatment. To date, a number of small-molecule inhibitors that specifically target regulators of RNA methylation have shown great potential for suppression of carcinogenesis. For example, METTL3, METTL14 and WTAP were shown to be predictors of response to chemotherapy and hormone treatment (Song et al., 2021). S-adenosylhomocysteine (SAH) can be hydrolyzed to produce adenosine (adenine) and homocysteine, which can inhibit cellular methyltransferase activity through substrate inhibition, and regulates transmethylation through inhibition of METTL3-METTL14 activity (Eckert et al., 2019). The expression of MALAT1 was shown to be enhanced by METTL3 through recruitment of E2F transcription factor 1 (E2F1), resulting in transcription of anterior gradient 2 (AGR2), and subsequent adriamycin resistance in breast cancer (Li et al., 2022). In a further study, METTL3 also promoted maturation of miRNA-221-3p in an m6A-dependent manner, which negatively regulated HIPK2, upregulated the target gene Che-1, and induced chemoresistance of breast cancer cells to doxorubicin (Pan et al., 2021).
Adenylate kinase 4 (AK4) and the m6A writer METTL3 are highly expressed in tamoxifen-resistant breast cancer cell lines, and METTL3 was shown to promote tamoxifen resistance in breast cancer by promoting AK4 expression, reducing the production level of reactive oxygen species (ROS), and decreasing the activity of p38 (Liu X. et al., 2020). Metformin was found to inhibit the proliferation of breast cancer cells through upregulation of P21 in an m6A-dependent manner via METTL3 (Cheng et al., 2021). STM2457 is an orally bioavailable small molecule METTL3 inhibitor that are slated for human clinical trials by targeting a novel mechanism for the treatment of acute myeloid leukemia and other solid and hematological cancers (Yankova et al., 2021). In addition, WTAP binds to the m6A modified site of lncRNA DLGAP1 antisense RNA 1 (DLGAP1-AS1) to sponge miR-299-3p, resulting in adriamycin resistance in breast cancer (Huang T. et al., 2021). The inhibitor of 2-oxoglutarate oxygenase (OG) oxidase, IOX1, significantly inhibited ALKBH5 activity. Protein arginine methyltransferase 5 (PRMT5) inhibits doxorubicin-treated RNA m6A modification by promoting nuclear translocation of ALKBH5 (Wu et al., 2022). The applying of PRMT5 inhibitor tadalafil improves the chemosensitivity of Doxorubicin in breast cancer by modulating RNA methylation (Wu et al., 2022). The most widely studied RNA methylation regulator is FTO (Chen and Du, 2019). A few potent inhibitors of FTO have been reported in the literature, namely FG-2216/IOX3, FB23-2, rhein, meclofenamic acid (MA), entacapone, bisantren and brequinar (Mcmurray et al., 2015; Van Der Werf and Jamieson, 2019; Su et al., 2020; Xiao et al., 2020; Yang B. et al., 2021; Lv et al., 2022). FTO was shown to promote tumor glycolysis and limit the response of T cells. The FTO inhibitor Dac51 increased CD8+ T cell infiltration and acted in synergy with anti-PD-L1 blockade (Liu Y. et al., 2021). MA is a highly selective FTO inhibitor relative to ALKBH5 by using high-throughput fluorescence polarization analysis (Zheng Q.-K. et al., 2021). The overexpression of m6A reader hnRNPA2B1 (A2B1) resulted in tamoxifen and fulvestrant resistance, and decreased migration and invasion in TAM-resistant cells through activation of the protein kinase B (AKT) and mitogen-activated protein kinase (MAPK) signaling pathways (Petri et al., 2021). Transcription factor 3 (ATF3) was highly expressed in tamoxifen-resistant breast cancer, and was regulated by low expression of YTHDF2. Moreover, ATF3 enhanced the expression of ATP binding cassette subfamily B member 1 (ABCB1), which promotes tamoxifen resistance (Liu X. et al., 2021).
Through regulation by NSUN2, m5C modifications were shown to be involved in the onset of various cancers, and may be potential targets for cancer treatment (Huang Z. et al., 2021; Hu et al., 2021). The expression of NSUN2 could be reduced by the inhibition of sphingosine kinase (SPHK), which is involved in sphingolipid metabolism in cell growth. Therefore, the SPHK1 inhibitor SK1 may be a latent drug for treatment through modulation of NSUN2 expression (Guo et al., 2021). In addition, the m5C “reader” Y-box-binding-protein 1 (YBX1) is highly expressed in certain cisplatin-resistant cancers. A study showed that the YBX1 phosphorylation inhibitors including TAS0612 (multikinase inhibitor) and everolimus (rapamycin complex 1 inhibitor) mitigated antiestrogen resistance in breast cancer (Shibata et al., 2020). However, the effects of YBX1 inhibitors on drug resistance in breast cancer require further investigation (Jiang et al., 2022). Three inhibitors were developed based on the interaction between DKC1 and TERC to inhibit telomerase activity in breast cancer cell lines, which may aid in development of pseudouridine synthase inhibitors for treatment of cancer (Armando et al., 2018).
Modulating abnormal RNA modification levels can inhibit the occurrence and development of tumors (Figure 3). Although some RNA modification enzyme inhibitors have shown potential inhibitory effects in a variety of cancers (Table 2), more drugs and new therapeutic strategies related to RNA modification remain to be explored and requested in the clinical trials.
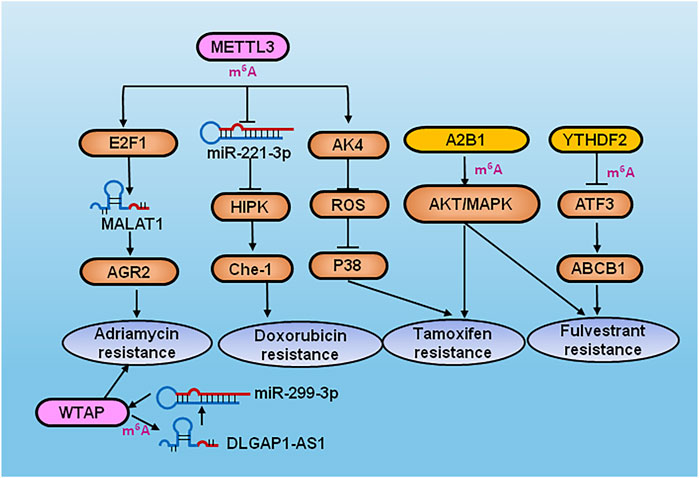
FIGURE 3. Diagram of RNA modification regulators inducing drugs resistance in breast cancer. METTL3 increased the expression of MALAT1 to activate AGR2 by recruiting E2F1 and subsequent adriamycin resistance in breast cancer. METTL3 also promoted maturation of miRNA-221-3p in a m6A-dependent manner and negatively regulated HIPK2 and upregulated Che-1 which induced chemoresistance of breast cancer cells to doxorubicin. METTL3 promoted tamoxifen resistance by promoting AK4 expression, reducing the production of ROS and decreasing the activity of p38. WTAP binds to DLGAP1-AS1 and sponged miR-299-3p to confer adriamycin resistance in breast cancer. A2B1 induced tamoxifen and fulvestrant resistance by AKT/MAPK signaling pathways. YTHDF2 downregulated ATF3 to activate ABCB1, resulting in tamoxifen resistance of breast cancer.
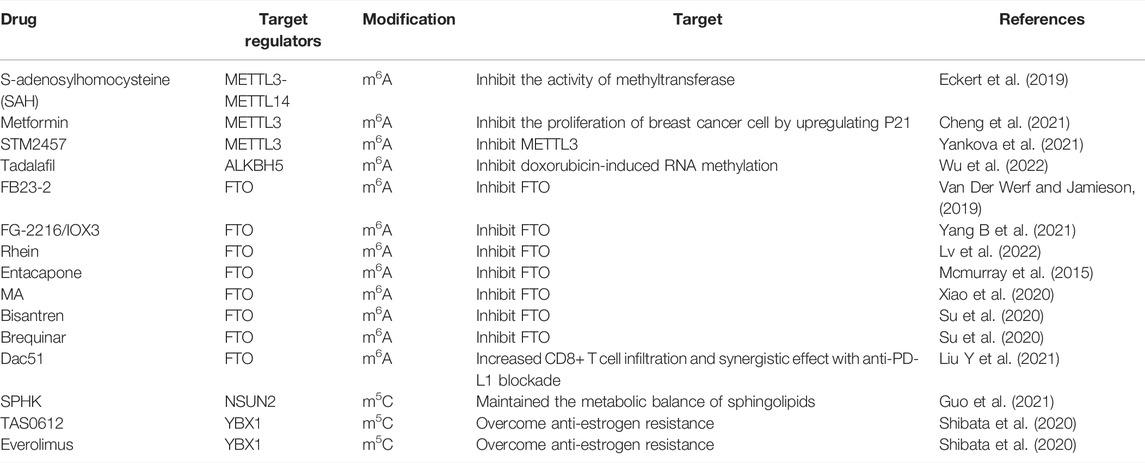
TABLE 2. The potential drugs applied in the treatment of breast cancer based on the RNA modification.
Future Directions
Research on tumor-related RNA modification is still in its infancy. Increasing number of novel RNA modifications are gradually discovered, such as RNA glycosylation modification, which is remarkably suggested that glycoRNA may play an important role in physiological and pathological processes including host immune defense, tumor immune escape, and autoimmune diseases (Flynn et al., 2021). It is also necessary to develop new technologies to discover new type of RNA modification. Further studies on the role of RNA methylation in the immune response will provide broader prospects for immunotherapy and prevention of tumor drug resistance. In terms of clinical application, it is of great significance to continue to explore whether RNA modification-related proteins could be potential diagnostic and therapeutic targets. Development of more specific and effective regulators of RNA modification is expected to result in new options for tumor treatment. In the context of disease treatment, small molecule inhibitors that can target RNA methylation-related effector proteins may have great promise. Demonstration of preclinical efficacy of these targeted drugs may result in future clinical use of RNA epigenetic drugs.
Conclusion
RNA methylation has been shown to exert tumor-promoting or tumor-suppressive activities, and is involved in the onset, development, and metastasis of breast cancer. The critical role of tumor-specific effects of RNA methylation provides insights into prognosis, pathogenesis, and treatment response in breast cancer. Design of novel therapeutics through targeted RNA modifications is an international research hotspot and may have profound implications in translational medicine application in breast cancer.
Author Contributions
Conception and design: XG and YL. Drafting of the manuscript: TZ, YL and JB. Drawing of figures: YJ, JB, and XZ. Conceiving and Critical revision of the manuscript for important intellectual content: XG and YL. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science foundation of China (Grant No. 82103468 to YL), Natural Science foundation of Liaoning Province of China (Grant No. 2020-MS-178 to XG) and 345 Talent Project of Shengjing Hospital of China Medical University (M0961 to YL).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Charlesworth Author Service (https://www.cwauthors.com.cn/) for editing this manuscript.
References
Anita, R., Paramasivam, A., Priyadharsini, J. V., and Chitra, S. (2020). The m6A Readers YTHDF1 and YTHDF3 Aberrations Associated with Metastasis and Predict Poor Prognosis in Breast Cancer Patients. Am. J. Cancer Res. 10, 2546–2554.
Arango, D., Sturgill, D., Alhusaini, N., Dillman, A. A., Sweet, T. J., Hanson, G., et al. (2018). Acetylation of Cytidine in mRNA Promotes Translation Efficiency. Cell 175, 1872–1886. e1824. doi:10.1016/j.cell.2018.10.030
Armando, R. G., Mengual Gómez, D. L., Juritz, E. I., Lorenzano Menna, P., and Gomez, D. E. (2018). Homology Model and Docking-Based Virtual Screening for Ligands of Human Dyskerin as New Inhibitors of Telomerase for Cancer Treatment. Int. J. Mol. Sci. 19, 3216. doi:10.3390/ijms19103216
Barbieri, I., and Kouzarides, T. (2020). Role of RNA Modifications in Cancer. Nat. Rev. Cancer 20, 303–322. doi:10.1038/s41568-020-0253-2
Boccaletto, P., Stefaniak, F., Ray, A., Cappannini, A., Mukherjee, S., Purta, E., et al. (2022). MODOMICS: a Database of RNA Modification Pathways. 2021 Update. Nucleic Acids Res. 50, D231–D235. doi:10.1093/nar/gkab1083
Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R. L., Torre, L. A., and Jemal, A. (2018). Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A Cancer J. Clin. 68, 394–424. doi:10.3322/caac.21492
Bujnicki, J. M., Feder, M., Ayres, C. L., and Redman, K. L. (2004). Sequence-structure-function Studies of tRNA:m5C Methyltransferase Trm4p and its Relationship to DNA:m5C and RNA:m5U Methyltransferases. Nucleic Acids Res. 32, 2453–2463. doi:10.1093/nar/gkh564
Cai, X., Wang, X., Cao, C., Gao, Y., Zhang, S., Yang, Z., et al. (2018). HBXIP-Elevated Methyltransferase METTL3 Promotes the Progression of Breast Cancer via Inhibiting Tumor Suppressor Let-7g. Cancer Lett. 415, 11–19. doi:10.1016/j.canlet.2017.11.018
Chang, G., Shi, L., Ye, Y., Shi, H., Zeng, L., Tiwary, S., et al. (2020). YTHDF3 Induces the Translation of m6A-Enriched Gene Transcripts to Promote Breast Cancer Brain Metastasis. Cancer Cell 38, 857–871. doi:10.1016/j.ccell.2020.10.004
Chen, F., Chen, Z., Guan, T., Zhou, Y., Ge, L., Zhang, H., et al. (2021). N6-Methyladenosine Regulates mRNA Stability and Translation Efficiency of KRT7 to Promote Breast Cancer Lung Metastasis. Cancer Res. 81, 2847–2860. doi:10.1158/0008-5472.can-20-3779
Chen, H., Yang, H., Zhu, X., Yadav, T., Ouyang, J., Truesdell, S. S., et al. (2020). m5C Modification of mRNA Serves a DNA Damage Code to Promote Homologous Recombination. Nat. Commun. 11, 2834. doi:10.1038/s41467-020-16722-7
Chen, H., Yu, Y., Yang, M., Huang, H., Ma, S., Hu, J., et al. (2022). YTHDF1 Promotes Breast Cancer Progression by Facilitating FOXM1 Translation in an m6A-Dependent Manner. Cell Biosci. 12, 19. doi:10.1186/s13578-022-00759-w
Chen, J., and Du, B. (2019). Novel Positioning from Obesity to Cancer: FTO, an m6A RNA Demethylase, Regulates Tumour Progression. J. Cancer Res. Clin. Oncol. 145, 19–29. doi:10.1007/s00432-018-2796-0
Cheng, L., Zhang, X., Huang, Y.-Z., Zhu, Y.-L., Xu, L.-Y., Li, Z., et al. (2021). Metformin Exhibits Antiproliferation Activity in Breast Cancer via miR-483-3p/METTL3/m6A/p21 Pathway. Oncogenesis 10, 7. doi:10.1038/s41389-020-00290-y
Dai, X.-Y., Shi, L., Li, Z., Yang, H.-Y., Wei, J.-F., and Ding, Q. (2021). Main N6-Methyladenosine Readers: YTH Family Proteins in Cancers. Front. Oncol. 11, 635329. doi:10.3389/fonc.2021.635329
Dai, Z., Liu, H., Liao, J., Huang, C., Ren, X., Zhu, W., et al. (2021). N7-Methylguanosine tRNA Modification Enhances Oncogenic mRNA Translation and Promotes Intrahepatic Cholangiocarcinoma Progression. Mol. Cell 81, 3339–3355. e3338. doi:10.1016/j.molcel.2021.07.003
Dai, X., Wang, T., Gonzalez, G., and Wang, Y. (2018). Identification of YTH Domain-Containing Proteins as the Readers for N1-Methyladenosine in RNA. Anal. Chem. 90, 6380–6384. doi:10.1021/acs.analchem.8b01703
Delaunay, S., Rapino, F., Tharun, L., Zhou, Z., Heukamp, L., Termathe, M., et al. (2016). Elp3 Links tRNA Modification to IRES-Dependent Translation of LEF1 to Sustain Metastasis in Breast Cancer. J. Exp. Med. 213, 2503–2523. doi:10.1084/jem.20160397
Dong, S., Wu, Y., Liu, Y., Weng, H., and Huang, H. (2021). N 6 ‐methyladenosine Steers RNA Metabolism and Regulation in Cancer. Cancer Commun. 41, 538–559. doi:10.1002/cac2.12161
Eckert, M. A., Coscia, F., Chryplewicz, A., Chang, J. W., Hernandez, K. M., Pan, S., et al. (2019). Proteomics Reveals NNMT as a Master Metabolic Regulator of Cancer-Associated Fibroblasts. Nature 569, 723–728. doi:10.1038/s41586-019-1173-8
Einstein, J. M., Perelis, M., Chaim, I. A., Meena, J. K., Nussbacher, J. K., Tankka, A. T., et al. (2021). Inhibition of YTHDF2 Triggers Proteotoxic Cell Death in MYC-Driven Breast Cancer. Mol. Cell 81, 3048–3064. e3049. doi:10.1016/j.molcel.2021.06.014
Elsharawy, K. A., Mohammed, O. J., Aleskandarany, M. A., Hyder, A., El-Gammal, H. L., Abou-Dobara, M. I., et al. (2020). The Nucleolar-Related Protein Dyskerin Pseudouridine Synthase 1 (DKC1) Predicts Poor Prognosis in Breast Cancer. Br. J. Cancer 123, 1543–1552. doi:10.1038/s41416-020-01045-7
Fan, S., and Wang, L. (2021). N6-Methyladenosine-Regulated LINC00675 Suppress the Proliferation, Migration and Invasion of Breast Cancer Cells via Inhibiting miR-513b-5p. Bioengineered 12, 10690–10702. doi:10.1080/21655979.2021.2001905
Flynn, R. A., Pedram, K., Malaker, S. A., Batista, P. J., Smith, B. A. H., Johnson, A. G., et al. (2021). Small RNAs Are Modified with N-Glycans and Displayed on the Surface of Living Cells. Cell 184, 3109–3124. doi:10.1016/j.cell.2021.04.023
Frye, M., Harada, B. T., Behm, M., and He, C. (2018). RNA Modifications Modulate Gene Expression during Development. Science 361, 1346–1349. doi:10.1126/science.aau1646
Fu, Y., Jia, G., Pang, X., Wang, R. N., Wang, X., Li, C. J., et al. (2013). FTO-mediated Formation of N6-Hydroxymethyladenosine and N6-Formyladenosine in Mammalian RNA. Nat. Commun. 4, 1798. doi:10.1038/ncomms2822
Gong, P.-J., Shao, Y.-C., Yang, Y., Song, W.-J., He, X., Zeng, Y.-F., et al. (2020). Analysis of N6-Methyladenosine Methyltransferase Reveals METTL14 and ZC3H13 as Tumor Suppressor Genes in Breast Cancer. Front. Oncol. 10, 578963. doi:10.3389/fonc.2020.578963
Gu, C., Shi, X., Dai, C., Shen, F., Rocco, G., Chen, J., et al. (2020). RNA m6A Modification in Cancers: Molecular Mechanisms and Potential Clinical Applications. InnovationInnovation (N Y) 1, 100066. doi:10.1016/j.xinn.2020.100066
Guerrieri, A. N., Zacchini, F., Onofrillo, C., Di Viggiano, S., Penzo, M., Ansuini, A., et al. (2020). DKC1 Overexpression Induces a More Aggressive Cellular Behavior and Increases Intrinsic Ribosomal Activity in Immortalized Mammary Gland Cells. Cancers (Basel) 12, 3512. doi:10.3390/cancers12123512
Guo, G., Pan, K., Fang, S., Ye, L., Tong, X., Wang, Z., et al. (2021). Advances in mRNA 5-methylcytosine Modifications: Detection, Effectors, Biological Functions, and Clinical Relevance. Mol. Ther. - Nucleic Acids 26, 575–593. doi:10.1016/j.omtn.2021.08.020
Haag, S., Kretschmer, J., and Bohnsack, M. T. (2015). WBSCR22/Merm1 Is Required for Late Nuclear Pre-ribosomal RNA Processing and Mediates N7-Methylation of G1639 in Human 18S rRNA. RNA 21, 180–187. doi:10.1261/rna.047910.114
Harbeck, N., Penault-Llorca, F., Cortes, J., Gnant, M., Houssami, N., Poortmans, P., et al. (2019). Breast Cancer. Nat. Rev. Dis. Prim. 5, 66. doi:10.1038/s41572-019-0111-2
He, L., Li, H., Wu, A., Peng, Y., Shu, G., and Yin, G. (2019). Functions of N6-Methyladenosine and its Role in Cancer. Mol. Cancer 18, 176. doi:10.1186/s12943-019-1109-9
He, X., Tan, L., Ni, J., and Shen, G. (2021). Expression Pattern of m6A Regulators Is Significantly Correlated with Malignancy and Antitumor Immune Response of Breast Cancer. Cancer Gene Ther. 28, 188–196. doi:10.1038/s41417-020-00208-1
Helm, M., and Motorin, Y. (2017). Detecting RNA Modifications in the Epitranscriptome: Predict and Validate. Nat. Rev. Genet. 18, 275–291. doi:10.1038/nrg.2016.169
Hu, Y., Chen, C., Tong, X., Chen, S., Hu, X., Pan, B., et al. (2021). NSUN2 Modified by SUMO-2/3 Promotes Gastric Cancer Progression and Regulates mRNA m5C Methylation. Cell Death Dis. 12, 842. doi:10.1038/s41419-021-04127-3
Huang, H., Weng, H., and Chen, J. (2020). m6A Modification in Coding and Non-coding RNAs: Roles and Therapeutic Implications in Cancer. Cancer Cell 37, 270–288. doi:10.1016/j.ccell.2020.02.004
Huang, H., Weng, H., Sun, W., Qin, X., Shi, H., Wu, H., et al. (2018). Recognition of RNA N6-Methyladenosine by IGF2BP Proteins Enhances mRNA Stability and Translation. Nat. Cell Biol. 20, 285–295. doi:10.1038/s41556-018-0045-z
Huang, T., Cao, L., Feng, N., Xu, B., Dong, Y., and Wang, M. (2021). N6-methyladenosine (m6A)-Mediated lncRNA DLGAP1-AS1enhances Breast Canceradriamycin Resistance through miR-299-3p/WTAP Feedback Loop. Bioengineered 12, 10935–10944. doi:10.1080/21655979.2021.2000198
Huang, Z., Pan, J., Wang, H., Du, X., Xu, Y., Wang, Z., et al. (2021). Prognostic Significance and Tumor Immune Microenvironment Heterogenicity of m5C RNA Methylation Regulators in Triple-Negative Breast Cancer. Front. Cell Dev. Biol. 9, 657547. doi:10.3389/fcell.2021.657547
Jeschke, J., Collignon, E., Al Wardi, C., Krayem, M., Bizet, M., Jia, Y., et al. (2021). Downregulation of the FTO m6A RNA Demethylase Promotes EMT-Mediated Progression of Epithelial Tumors and Sensitivity to Wnt Inhibitors. Nat. Cancer 2, 611–628. doi:10.1038/s43018-021-00223-7
Jia, G., Fu, Y., Zhao, X., Dai, Q., Zheng, G., Yang, Y., et al. (2011). N6-methyladenosine in Nuclear RNA Is a Major Substrate of the Obesity-Associated FTO. Nat. Chem. Biol. 7, 885–887. doi:10.1038/nchembio.687
Jiang, D., Qiu, T., Peng, J., Li, S., TalaRen, W., Yang, C., et al. (2022). YB-1 Is a Positive Regulator of KLF5 Transcription Factor in Basal-like Breast Cancer. Cell Death Differ. doi:10.1038/s41418-021-00920-x
Katsara, O., and Schneider, R. J. (2021). m7G tRNA Modification Reveals New Secrets in the Translational Regulation of Cancer Development. Mol. Cell 81, 3243–3245. doi:10.1016/j.molcel.2021.07.030
Li, H., Zhu, D., Wu, J., Ma, Y., Cai, C., Chen, Y., et al. (2021). New Substrates and Determinants for tRNA Recognition of RNA Methyltransferase DNMT2/TRDMT1. RNA Biol. 18, 2531–2545. doi:10.1080/15476286.2021.1930756
Li, Z., Yang, H.-Y., Dai, X.-Y., Zhang, X., Huang, Y.-Z., Shi, L., et al. (2021). CircMETTL3, Upregulated in a m6A-dependent Manner, Promotes Breast Cancer Progression. Int. J. Biol. Sci. 17, 1178–1190. doi:10.7150/ijbs.57783
Li, S., Jiang, F., Chen, F., Deng, Y., and Pan, X. (2022). Effect of m6A Methyltransferase METTL3 -mediated MALAT1/E2F1/AGR2 axis on Adriamycin Resistance in Breast Cancer. J. Biochem. Mol. Toxicol. 36, e22922. doi:10.1002/jbt.22922
Li, S., and Mason, C. E. (2014). The Pivotal Regulatory Landscape of RNA Modifications. Annu. Rev. Genom. Hum. Genet. 15, 127–150. doi:10.1146/annurev-genom-090413-025405
Li, X., Xiong, X., Wang, K., Wang, L., Shu, X., Ma, S., et al. (2016). Transcriptome-wide Mapping Reveals Reversible and Dynamic N(1)-Methyladenosine Methylome. Nat. Chem. Biol. 12, 311–316. doi:10.1038/nchembio.2040
Li, X., Xiong, X., Zhang, M., Wang, K., Chen, Y., Zhou, J., et al. (2017). Base-Resolution Mapping Reveals Distinct m1A Methylome in Nuclear- and Mitochondrial-Encoded Transcripts. Mol. Cell 68, 993–1005. doi:10.1016/j.molcel.2017.10.019
Li, Y., Xiao, J., Bai, J., Tian, Y., Qu, Y., Chen, X., et al. (2019). Molecular Characterization and Clinical Relevance of m6A Regulators across 33 Cancer Types. Mol. Cancer 18, 137. doi:10.1186/s12943-019-1066-3
Lin, Y., Jin, X., Nie, Q., Chen, M., Guo, W., Chen, L., et al. (2022). YTHDF3 Facilitates Triple-Negative Breast Cancer Progression and Metastasis by Stabilizing ZEB1 mRNA in an m6A-dependent Manner. Ann. Transl. Med. 10, 83. doi:10.21037/atm-21-6857
Liu, L., Liu, X., Dong, Z., Li, J., Yu, Y., Chen, X., et al. (2019). N6-methyladenosine-related Genomic Targets Are Altered in Breast Cancer Tissue and Associated with Poor Survival. J. Cancer 10, 5447–5459. doi:10.7150/jca.35053
Liu, R.-J., Long, T., Li, J., Li, H., and Wang, E.-D. (2017). Structural Basis for Substrate Binding and Catalytic Mechanism of a Human RNA:m5C Methyltransferase NSun6. Nucleic Acids Res. 45, 6684–6697. doi:10.1093/nar/gkx473
Liu, X., Gonzalez, G., Dai, X., Miao, W., Yuan, J., Huang, M., et al. (2020). Adenylate Kinase 4 Modulates the Resistance of Breast Cancer Cells to Tamoxifen through an m6A-Based Epitranscriptomic Mechanism. Mol. Ther. 28, 2593–2604. doi:10.1016/j.ymthe.2020.09.007
Liu, Y., Li, H., Liu, F., Gao, L.-B., Han, R., Chen, C., et al. (2020). Heterogeneous Nuclear Ribonucleoprotein A2/B1 Is a Negative Regulator of Human Breast Cancer Metastasis by Maintaining the Balance of Multiple Genes and Pathways. EBioMedicine 51, 102583. doi:10.1016/j.ebiom.2019.11.044
Liu, X., Yuan, J., Zhang, X., Li, L., Dai, X., Chen, Q., et al. (2021). ATF3 Modulates the Resistance of Breast Cancer Cells to Tamoxifen through an N6-Methyladenosine-Based Epitranscriptomic Mechanism. Chem. Res. Toxicol. 34, 1814–1821. doi:10.1021/acs.chemrestox.1c00206
Liu, Y., Liang, G., Xu, H., Dong, W., Dong, Z., Qiu, Z., et al. (2021). Tumors Exploit FTO-Mediated Regulation of Glycolytic Metabolism to Evade Immune Surveillance. Cell Metab. 33, 1221–1233. e1211. doi:10.1016/j.cmet.2021.04.001
Lv, D., Ding, S., Zhong, L., Tu, J., Li, H., Yao, H., et al. (2022). M(6)A Demethylase FTO-Mediated Downregulation of DACT1 mRNA Stability Promotes Wnt Signaling to Facilitate Osteosarcoma Progression. Oncogene 41 (12), 1727–1741. doi:10.1038/s41388-022-02214-z
Lv, W., Tan, Y., Xiong, M., Zhao, C., Wang, Y., Wu, M., et al. (2021a). Analysis and Validation of m6A Regulatory Network: a Novel circBACH2/has-miR-944/HNRNPC axis in Breast Cancer Progression. J. Transl. Med. 19, 527. doi:10.1186/s12967-021-03196-4
Lv, W., Wang, Y., Zhao, C., Tan, Y., Xiong, M., Yi, Y., et al. (2021b). Identification and Validation of m6A-Related lncRNA Signature as Potential Predictive Biomarkers in Breast Cancer. Front. Oncol. 11, 745719. doi:10.3389/fonc.2021.745719
Maresca, G., and Wismayer, P. S. (2016). DNA Methylation and Cancer: Identifying and Targeting Epigenetic Modifications May Be the Future of Cancer Therapy? Gulf J. Oncol. 1, 77–83.
Mcmurray, F., Demetriades, M., Aik, W., Merkestein, M., Kramer, H., Andrew, D. S., et al. (2015). Pharmacological Inhibition of FTO. Plos One 10, e0121829. doi:10.1371/journal.pone.0121829
Mohammad, H. P., Barbash, O., and Creasy, C. L. (2019). Targeting Epigenetic Modifications in Cancer Therapy: Erasing the Roadmap to Cancer. Nat. Med. 25, 403–418. doi:10.1038/s41591-019-0376-8
Moon, H. J., and Redman, K. L. (2014). Trm4 and Nsun2 RNA:m5C Methyltransferases Form Metabolite-dependent, Covalent Adducts with Previously Methylated RNA. Biochemistry 53, 7132–7144. doi:10.1021/bi500882b
Müller, M., Samel-Pommerencke, A., Legrand, C., Tuorto, F., Lyko, F., and Ehrenhofer-Murray, A. E. (2019). Division of Labour: tRNA Methylation by the NSun2 tRNA Methyltransferases Trm4a and Trm4b in Fission Yeast. RNA Biol. 16, 249–256. doi:10.1080/15476286.2019.1568819
Niu, Y., Lin, Z., Wan, A., Chen, H., Liang, H., Sun, L., et al. (2019). RNA N6-Methyladenosine Demethylase FTO Promotes Breast Tumor Progression through Inhibiting BNIP3. Mol. Cancer 18, 46. doi:10.1186/s12943-019-1004-4
Ota, H., Sakurai, M., Gupta, R., Valente, L., Wulff, B.-E., Ariyoshi, K., et al. (2013). ADAR1 Forms a Complex with Dicer to Promote microRNA Processing and RNA-Induced Gene Silencing. Cell 153, 575–589. doi:10.1016/j.cell.2013.03.024
Ou, B., Liu, Y., Yang, X., Xu, X., Yan, Y., and Zhang, J. (2021). C5aR1-positive Neutrophils Promote Breast Cancer Glycolysis through WTAP-dependent m6A Methylation of ENO1. Cell Death Dis. 12, 737. doi:10.1038/s41419-021-04028-5
Pan, X., Hong, X., Li, S., Meng, P., and Xiao, F. (2021). METTL3 Promotes Adriamycin Resistance in MCF-7 Breast Cancer Cells by Accelerating Pri-microRNA-221-3p Maturation in a m6A-dependent Manner. Exp. Mol. Med. 53, 91–102. doi:10.1038/s12276-020-00510-w
Pedrosa, R. M. S. M., Mustafa, D. A., Soffietti, R., and Kros, J. M. (2018). Breast Cancer Brain Metastasis: Molecular Mechanisms and Directions for Treatment. Neuro Oncol. 20, 1439–1449. doi:10.1093/neuonc/noy044
Peng, F., Xu, J., Cui, B., Liang, Q., Zeng, S., He, B., et al. (2021). Oncogenic AURKA-Enhanced N6-Methyladenosine Modification Increases DROSHA mRNA Stability to Transactivate STC1 in Breast Cancer Stem-like Cells. Cell Res. 31, 345–361. doi:10.1038/s41422-020-00397-2
Petri, B. J., Piell, K. M., South Whitt, G. C., Wilt, A. E., Poulton, C. C., Lehman, N. L., et al. (2021). HNRNPA2B1 Regulates Tamoxifen- and Fulvestrant-Sensitivity and Hallmarks of Endocrine Resistance in Breast Cancer Cells. Cancer Lett. 518, 152–168. doi:10.1016/j.canlet.2021.07.015
Pinto, R., Vågbø, C. B., Jakobsson, M. E., Kim, Y., Baltissen, M. P., O’Donohue, M.-F., et al. (2020). The Human Methyltransferase ZCCHC4 Catalyses N6-Methyladenosine Modification of 28S Ribosomal RNA. Nucleic Acids Res. 48, 830–846. doi:10.1093/nar/gkz1147
Qian, J.-Y., Gao, J., Sun, X., Cao, M.-D., Shi, L., Xia, T.-S., et al. (2019). KIAA1429 Acts as an Oncogenic Factor in Breast Cancer by Regulating CDK1 in an N6-methyladenosine-independent Manner. Oncogene 38, 6123–6141. doi:10.1038/s41388-019-0861-z
Qiao, Y. S., Zhou, J. H., Jin, B. H., Wu, Y. Q., and Zhao, B. (2021). LINC00483 Is Regulated by IGF2BP1 and Participates in the Progression of Breast Cancer. Eur. Rev. Med. Pharmacol. Sci. 25, 1379–1386. doi:10.26355/eurrev_202102_24846
Rapino, F., Delaunay, S., Zhou, Z., Chariot, A., and Close, P. (2017). tRNA Modification: Is Cancer Having a Wobble? Trends Cancer 3, 249–252. doi:10.1016/j.trecan.2017.02.004
Rong, D., Sun, G., Wu, F., Cheng, Y., Sun, G., Jiang, W., et al. (2021). Epigenetics: Roles and Therapeutic Implications of Non-coding RNA Modifications in Human Cancers. Mol. Ther. - Nucleic Acids 25, 67–82. doi:10.1016/j.omtn.2021.04.021
Ruan, H.-g., Gu, W.-c., Xia, W., Gong, Y., Zhou, X.-l., Chen, W.-y., et al. (2021). METTL3 Is Suppressed by Circular RNA circMETTL3/miR-34c-3p Signaling and Limits the Tumor Growth and Metastasis in Triple Negative Breast Cancer. Front. Oncol. 11, 778132. doi:10.3389/fonc.2021.778132
Safra, M., Sas-Chen, A., Nir, R., Winkler, R., Nachshon, A., Bar-Yaacov, D., et al. (2017). The m1A Landscape on Cytosolic and Mitochondrial mRNA at Single-Base Resolution. Nature 551, 251–255. doi:10.1038/nature24456
Saikia, M., Fu, Y., Pavon-Eternod, M., He, C., and Pan, T. (2010). Genome-wide Analysis of N1-Methyl-Adenosine Modification in Human tRNAs. RNA 16, 1317–1327. doi:10.1261/rna.2057810
Sas-Chen, A., Thomas, J. M., Matzov, D., Taoka, M., Nance, K. D., Nir, R., et al. (2020). Dynamic RNA Acetylation Revealed by Quantitative Cross-Evolutionary Mapping. Nature 583, 638–643. doi:10.1038/s41586-020-2418-2
Shi, Y., Zheng, C., Jin, Y., Bao, B., Wang, D., Hou, K., et al. (2020). Reduced Expression of METTL3 Promotes Metastasis of Triple-Negative Breast Cancer by m6A Methylation-Mediated COL3A1 Up-Regulation. Front. Oncol. 10, 1126. doi:10.3389/fonc.2020.01126
Shibata, T., Watari, K., Kawahara, A., Sudo, T., Hattori, S., Murakami, Y., et al. (2020). Targeting Phosphorylation of Y-Box-Binding Protein YBX1 by TAS0612 and Everolimus in Overcoming Antiestrogen Resistance. Mol. Cancer Ther. 19, 882–894. doi:10.1158/1535-7163.mct-19-0690
Shiromoto, Y., Sakurai, M., Minakuchi, M., Ariyoshi, K., and Nishikura, K. (2021). ADAR1 RNA Editing Enzyme Regulates R-Loop Formation and Genome Stability at Telomeres in Cancer Cells. Nat. Commun. 12, 1654. doi:10.1038/s41467-021-21921-x
Song, Y., Zheng, C., Tao, Y., Huang, R., and Zhang, Q. (2021). N6-Methyladenosine Regulators Are Involved in the Progression of and Have Clinical Impact on Breast Cancer. Med. Sci. Monit. 27, e929615. doi:10.12659/msm.929615
Su, R., Dong, L., Li, Y., Gao, M., Han, L., Wunderlich, M., et al. (2020). Targeting FTO Suppresses Cancer Stem Cell Maintenance and Immune Evasion. Cancer Cell 38, 79–96. doi:10.1016/j.ccell.2020.04.017
Su, R., Dong, L., Li, Y., Gao, M., He, P. C., Liu, W., et al. (2022). METTL16 Exerts an m6A-independent Function to Facilitate Translation and Tumorigenesis. Nat. Cell Biol. 24, 205–216. doi:10.1038/s41556-021-00835-2
Sun, T., Wu, Z., Wang, X., Wang, Y., Hu, X., Qin, W., et al. (2020). LNC942 Promoting METTL14-Mediated m6A Methylation in Breast Cancer Cell Proliferation and Progression. Oncogene 39, 5358–5372. doi:10.1038/s41388-020-1338-9
Tang, Y., Gao, C.-C., Gao, Y., Yang, Y., Shi, B., Yu, J.-L., et al. (2020). OsNSUN2-Mediated 5-Methylcytosine mRNA Modification Enhances Rice Adaptation to High Temperature. Dev. Cell 53, 272–286. doi:10.1016/j.devcel.2020.03.009
Van Der Werf, I., and Jamieson, C. (2019). The Yin and Yang of RNA Methylation: An Imbalance of Erasers Enhances Sensitivity to FTO Demethylase Small-Molecule Targeting in Leukemia Stem Cells. Cancer Cell 35, 540–541. doi:10.1016/j.ccell.2019.03.011
Wan, W., Ao, X., Chen, Q., Yu, Y., Ao, L., Xing, W., et al. (2022). METTL3/IGF2BP3 axis Inhibits Tumor Immune Surveillance by Upregulating N6-Methyladenosine Modification of PD-L1 mRNA in Breast Cancer. Mol. Cancer 21, 60. doi:10.1186/s12943-021-01447-y
Wang, C.-Q., Tang, C.-H., Wang, Y., Huang, B.-F., Hu, G.-N., Wang, Q., et al. (2022). Upregulated WTAP Expression Appears to Both Promote Breast Cancer Growth and Inhibit Lymph Node Metastasis. Sci. Rep. 12, 1023. doi:10.1038/s41598-022-05035-y
Wang, H., Xu, B., and Shi, J. (2020). N6-methyladenosine METTL3 Promotes the Breast Cancer Progression via Targeting Bcl-2. Gene 722, 144076. doi:10.1016/j.gene.2019.144076
Wang, S., Zou, X., Chen, Y., Cho, W. C., and Zhou, X. (2020). Effect of N6-Methyladenosine Regulators on Progression and Prognosis of Triple-Negative Breast Cancer. Front. Genet. 11, 580036. doi:10.3389/fgene.2020.580036
Woo, H.-H., and Chambers, S. K. (2019). Human ALKBH3-Induced m1A Demethylation Increases the CSF-1 mRNA Stability in Breast and Ovarian Cancer Cells. Biochimica Biophysica Acta (BBA) - Gene Regul. Mech. 1862, 35–46. doi:10.1016/j.bbagrm.2018.10.008
Wood, S., Willbanks, A., and Cheng, J. X. (2021). The Role of RNA Modifications and RNA-Modifying Proteins in Cancer Therapy and Drug Resistance. Ccdt 21, 326–352. doi:10.2174/1568009621666210127092828
Wu, J., Cai, Y., Zhao, G., and Li, M. (2021). A Ten N6-Methyladenosine-Related Long Non-coding RNAs Signature Predicts Prognosis of Triple-Negative Breast Cancer. J. Clin. Lab. Anal. 35, e23779.
Wu, L., Wu, D., Ning, J., Liu, W., and Zhang, D. (2019). Changes of N6-Methyladenosine Modulators Promote Breast Cancer Progression. BMC Cancer 19, 326. doi:10.1186/s12885-019-5538-z
Wu, Y., Wang, Z., Han, L., Guo, Z., Yan, B., Guo, L., et al. (2022). PRMT5 Regulates RNA m6A Demethylation for Doxorubicin Sensitivity in Breast Cancer. Mol. Ther. S1525–0016 (22), 00160–00165. doi:10.1016/j.ymthe.2022.03.003
Xiao, L., Li, X., Mu, Z., Zhou, J., Zhou, P., Xie, C., et al. (2020). FTO Inhibition Enhances the Antitumor Effect of Temozolomide by Targeting MYC-miR-155/23a Cluster-MXI1 Feedback Circuit in Glioma. Cancer Res. 80, 3945–3958. doi:10.1158/0008-5472.can-20-0132
Xing, S., Li, Z., Ma, W., He, X., Shen, S., Wei, H., et al. (2019). DIS3L2 Promotes Progression of Hepatocellular Carcinoma via hnRNP U-Mediated Alternative Splicing. Cancer Res. 79, 4923–4936. doi:10.1158/0008-5472.can-19-0376
Xu, Y., Ye, S., Zhang, N., Zheng, S., Liu, H., Zhou, K., et al. (2020). The FTO/miR‐181b‐3p/ARL5B Signaling Pathway Regulates Cell Migration and Invasion in Breast Cancer. Cancer Commun. 40, 484–500. doi:10.1002/cac2.12075
Xue, C., Zhao, Y., Li, G., and Li, L. (2021). Multi-Omic Analyses of the m5C Regulator ALYREF Reveal its Essential Roles in Hepatocellular Carcinoma. Front. Oncol. 11, 633415. doi:10.3389/fonc.2021.633415
Yang, B., Wang, J.-Q., Tan, Y., Yuan, R., Chen, Z.-S., and Zou, C. (2021). RNA Methylation and Cancer Treatment. Pharmacol. Res. 174, 105937. doi:10.1016/j.phrs.2021.105937
Yang, C., Wu, T., Zhang, J., Liu, J., Zhao, K., Sun, W., et al. (2021). Prognostic and Immunological Role of mRNA ac4C Regulator NAT10 in Pan-Cancer: New Territory for Cancer Research? Front. Oncol. 11, 630417. doi:10.3389/fonc.2021.630417
Yang, L., Wu, S., Ma, C., Song, S., Jin, F., Niu, Y., et al. (2020). RNA m6A Methylation Regulators Subclassify Luminal Subtype in Breast Cancer. Front. Oncol. 10, 611191. doi:10.3389/fonc.2020.611191
Yang, X., Yang, Y., Sun, B.-F., Chen, Y.-S., Xu, J.-W., Lai, W.-Y., et al. (2017). 5-methylcytosine Promotes mRNA Export - NSUN2 as the Methyltransferase and ALYREF as an m5C Reader. Cell Res. 27, 606–625. doi:10.1038/cr.2017.55
Yankova, E., Blackaby, W., Albertella, M., Rak, J., De Braekeleer, E., Tsagkogeorga, G., et al. (2021). Small-molecule Inhibition of METTL3 as a Strategy against Myeloid Leukaemia. Nature 593, 597–601. doi:10.1038/s41586-021-03536-w
Yi, D., Wang, R., Shi, X., Xu, L., Yilihamu, Y., and Sang, J. (2020). METTL14 Promotes the Migration and Invasion of Breast Cancer Cells by Modulating N6-methyladenosine and hsamiR146a5p Expression. Oncol. Rep. 43, 1375–1386. doi:10.3892/or.2020.7515
Yi, J., Gao, R., Chen, Y., Yang, Z., Han, P., Zhang, H., et al. (2017). Overexpression of NSUN2 by DNA Hypomethylation Is Associated with Metastatic Progression in Human Breast Cancer. Oncotarget 8, 20751–20765. doi:10.18632/oncotarget.10612
Zaccara, S., and Jaffrey, S. R. (2020). A Unified Model for the Function of YTHDF Proteins in Regulating m6A-Modified mRNA. Cell 181, 1582–1595. doi:10.1016/j.cell.2020.05.012
Zhang, B., Gu, Y., and Jiang, G. (2020). Expression and Prognostic Characteristics of M6 A RNA Methylation Regulators in Breast Cancer. Front. Genet. 11, 604597. doi:10.3389/fgene.2020.604597
Zhang, C., Zhi, W. I., Lu, H., Samanta, D., Chen, I., Gabrielson, E., et al. (2016). Hypoxia-inducible Factors Regulate Pluripotency Factor Expression by ZNF217- and ALKBH5-Mediated Modulation of RNA Methylation in Breast Cancer Cells. Oncotarget 7, 64527–64542. doi:10.18632/oncotarget.11743
Zhang, J., Shan, B., Lin, L., Dong, J., Sun, Q., Zhou, Q., et al. (2021). Dissecting the Role of N6-Methylandenosine-Related Long Non-coding RNAs Signature in Prognosis and Immune Microenvironment of Breast Cancer. Front. Cell Dev. Biol. 9, 711859. doi:10.3389/fcell.2021.711859
Zhang, X., Dai, X.-Y., Qian, J.-Y., Xu, F., Wang, Z.-W., Xia, T., et al. (2022). SMC1A Regulated by KIAA1429 in m6A-independent Manner Promotes EMT Progress in Breast Cancer. Mol. Ther. - Nucleic Acids 27, 133–146. doi:10.1016/j.omtn.2021.08.009
Zhao, B. S., Roundtree, I. A., and He, C. (2017). Post-transcriptional Gene Regulation by mRNA Modifications. Nat. Rev. Mol. Cell Biol. 18, 31–42. doi:10.1038/nrm.2016.132
Zheng F, F., Du, F., Qian, H., Zhao, J., Wang, X., Yue, J., et al. (2021). Expression and Clinical Prognostic Value of m6A RNA Methylation Modification in Breast Cancer. Biomark. Res. 9, 28. doi:10.1186/s40364-021-00285-w
Zheng, Q.-K., Ma, C., Ullah, I., Hu, K., Ma, R.-J., Zhang, N., et al. (2021). Roles of N6‐Methyladenosine Demethylase FTO in Malignant Tumors Progression. Onco Targets Ther. 14, 4837–4846. doi:10.2147/ott.s329232
Zheng, G., Dahl, J. A., Niu, Y., Fedorcsak, P., Huang, C.-M., Li, C. J., et al. (2013). ALKBH5 Is a Mammalian RNA Demethylase that Impacts RNA Metabolism and Mouse Fertility. Mol. Cell 49, 18–29. doi:10.1016/j.molcel.2012.10.015
Zhu, P., He, F., Hou, Y., Tu, G., Li, Q., Jin, T., et al. (2021). A Novel Hypoxic Long Noncoding RNA KB-1980E6.3 Maintains Breast Cancer Stem Cell Stemness via Interacting with IGF2BP1 to Facilitate C-Myc mRNA Stability. Oncogene 40, 1609–1627. doi:10.1038/s41388-020-01638-9
Glossary
APC adenomatous polyposis coli
ABCB1 ATP binding cassette subfamily B member 1
ARL5B ADP ribosylation factor like GTPase5B
AGR2 anterior gradient 2
AK4 adenylate kinase 4
ADARs adenosine deaminases acting on RNA
ALKBH5 Demethylases ALKB homolog 5
ac4C N4-acetylcytodine
AURKA aurora kinase a
ATF3 transcription factor 3
BNIP3 BCL2 Interacting Protein 3
C5aR1 C5a Receptor 1
COL3A1 collagen type III alpha 1 chain
CTU1 cytoplasmic trna2 thiolated protein 1
CDS coding sequence
CDK1 cyclin-dependent kinase 1
CYP1B1 cytochrome p450 family 1 subfamily B member 1
CSF-1 macrophage-colony stimulating factor-1
DLGAP1-AS1 lncRNA DLGAP1 antisense RNA 1
eIF3 eukaryotic initiation factor 3
EGFR epidermal growth factor receptor
EMT epithelial-to-mesenchymal transition
ENO1 enolase 1
EGOT eosinophil granule ontogeny transcript
E2F1 E2F transcription factor 1
FTO Fat mass and obesity-related protein
HBXIP hepatitis B X-interacting protein
hnRNP heterogeneous nuclear ribonucleoprotein
IL interlukin
IGF2BP1 insulin-like growth factor 2 mRNA-binding protein 1
KRT7 keratin 7
NSUN2 NOP2/sun RNA methyltransferase 2
GJA1 gap junction protein alpha 1
GRM3 glutamate metabotropic receptor 3
MALAT1 metastasis associated lung adenocarcinoma transcript 1
MTC methyltransferase complex
METTL3 methyltransferase-like 3
METT14 methyltransferase-like 14
MAPK mitogen-activated protein kinase
OG oxoglutarate oxygenase
rRNA ribosomal RNA
tRNA transport RNA
mRNA messenger RNA
ncRNA non coding RNA
m6A N6-methyladenosine
3’UTR 3’untranslated region
mcm5s2U 5-methoxycarbonylmethyl-2-thiouridine
RBM15 RNA binding motif protein 15
STC1 stanniocalcin 1
TNBC triple-negative breast cancer
TNF tumor-necrosis-factor
ROS reactive oxygen species
SMC1A chromosomes 1a
ST6GALNAC5 ST6 beta-galactoside alpha-2,6-sialyltransferase
RAD52 DNA repair protein RAD52 homolog
SAH S-adenosylhomocysteine
SPHK sphingosine kinase
TRM tRNA methyltransferase 10 homologue A
WATP wilms tumor 1 associated protein
ZC3H13 zinc finger CCCHType Containing 13
METTL16 methyltransferase like 16
YTHDF YTH domain family
YTHDC YTH domain-containing protein
TRDMT1 tRNA aspartic acid methyltransferase 1
TRM4B tRNA-specific methyltransferase 4B
YBX1 Y-box binding protein 1
WBSCR22 Williams Beuren syndrome chromosome 22 region protein
YBX1 Y-box binding protein 1
Keywords: RNA modification, breast cancer, m6A, carcinogenesis, treatment
Citation: Liu Y, Zhu T, Jiang Y, Bu J, Zhu X and Gu X (2022) The Key Role of RNA Modification in Breast Cancer. Front. Cell Dev. Biol. 10:885133. doi: 10.3389/fcell.2022.885133
Received: 27 February 2022; Accepted: 25 April 2022;
Published: 01 June 2022.
Edited by:
Hengyou Weng, Guangzhou Regenerative Medicine and Health Guangdong Laboratory, ChinaReviewed by:
Shuibin Lin, Sun Yat-sen University, ChinaDanny Misiak, Martin Luther University of Halle-Wittenberg, Germany
Copyright © 2022 Liu, Zhu, Jiang, Bu, Zhu and Gu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xi Gu, amFkZWd4QDE2My5jb20=
 Yang Liu
Yang Liu Tong Zhu
Tong Zhu Yi Jiang
Yi Jiang Jiawen Bu
Jiawen Bu Xudong Zhu
Xudong Zhu Xi Gu
Xi Gu