- RIKEN Center for Advanced Photonics, Wako, Japan
The Golgi apparatus represents a central compartment of membrane traffic. Its apparent architecture, however, differs considerably among species, from unstacked and scattered cisternae in the budding yeast Saccharomyces cerevisiae to beautiful ministacks in plants and further to gigantic ribbon structures typically seen in mammals. Considering the well-conserved functions of the Golgi, its fundamental structure must have been optimized despite seemingly different architectures. In addition to the core layers of cisternae, the Golgi is usually accompanied by next-door compartments on its cis and trans sides. The trans-Golgi network (TGN) can be now considered as a compartment independent from the Golgi stack. On the cis side, the intermediate compartment between the ER and the Golgi (ERGIC) has been known in mammalian cells, and its functional equivalent is now suggested for yeast and plant cells. High-resolution live imaging is extremely powerful for elucidating the dynamics of these compartments and has revealed amazing similarities in their behaviors, indicating common mechanisms conserved along the long course of evolution. From these new findings, I would like to propose reconsideration of compartments and suggest a new concept to describe their roles comprehensively around the Golgi and in the post-Golgi trafficking.
Introduction
The Golgi apparatus has been recognized as a central station of intracellular membrane traffic because it stands at the intersection of secretory, lysosomal/vacuolar, and endosomal transport routes (Jamieson and Palade, 1966; Farquhar and Palade, 1998). Its beautiful structure comprising a layer of cisternae has attracted many cell biologists. However, the more we know about its functions, the more questions confront us leading to very active debates.
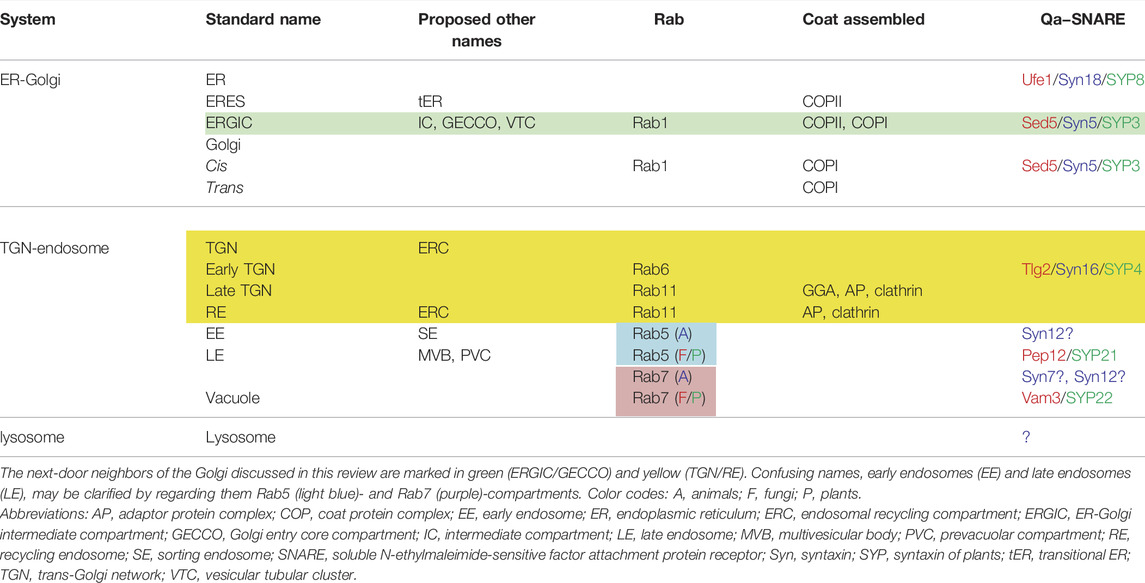
As will be discussed below, the organization of the Golgi differs considerably among different species, making it difficult to draw a comprehensive picture of this organelle (Figure 1). A simple model shown in many textbooks describes a stack of several flattened cisternae with a polarity from cis to trans, but such a typical structure is not always seen in living cells.
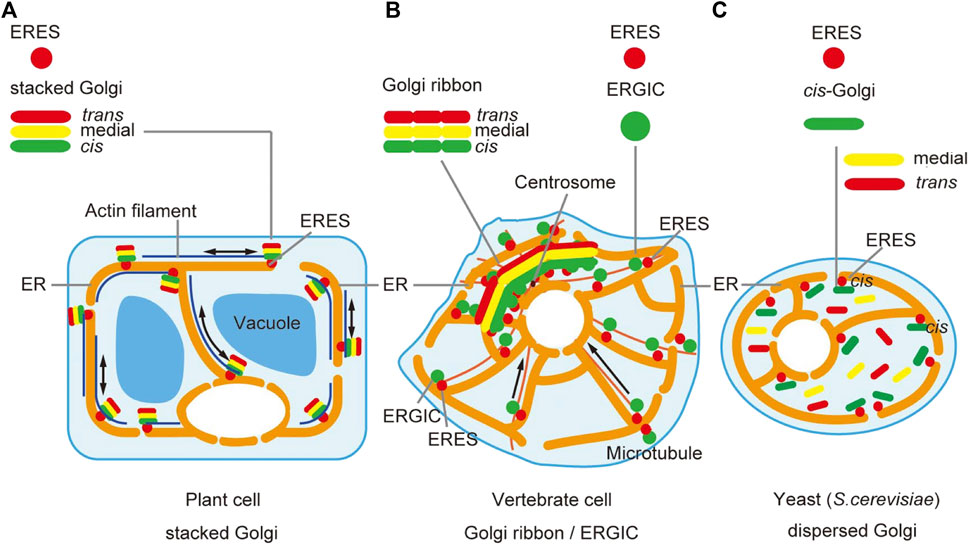
FIGURE 1. Spatial organization of the Golgi. The spatial arrangement of the Golgi apparatus looks different among species. (A) In plant cells, Golgi cisternae (cis, medial, and trans) are beautifully layered in single stack units, which are scattered in the cytoplasm. Model animals of invertebrates, Drosophila and C. elegans, show a similar pattern. (B) In vertebrate animals like mammals, Golgi stacks are concentrated at the perinuclear centrosomal region often in an interconnected fashion to form a gigantic Golgi ribbon structure. (C) In the budding yeast S. cerevisiae, Golgi cisternae do not stack and are individually scattered in the cytoplasm. Note that ERES (ER exit sites) are very frequently observed in the very vicinity of the Golgi stack (A) or ERGIC (ER-Golgi intermediate compartment) (B) or cis-Golgi (C) [modified from Kurokawa et al. (2019)].
The best-known Golgi function is its role as a glycosylation factory. Many secretory proteins have N- and/or O-linked oligosaccharide chains, which are subjected to complicated modification reactions (Berger and Rohrer, 2008; Stanley, 2011). In plants, the synthesis of cell wall polysaccharides is also an important task of the Golgi (Driouich et al., 1993; Hoffmann et al., 2021). To fulfill these biochemical reactions, the array of Golgi cisternae is differentiated into sub-compartments and zones to achieve the best performance (Dunphy and Rothman, 1985; Tie et al., 2018; Tojima et al., 2019).
In addition to the important function of glycosylation, the Golgi is known to act as a sorting platform. It receives a variety of proteins from the ER and recycles some of them back to the ER (Nilsson et al., 1989; Pelham and Munro, 1993; Letourneur et al., 1994; Sato et al., 1995, 1997; Sato et al., 1996). There are a variety of destinations for cargo transiting the Golgi, involving many different carriers to the plasma membrane and other organelles (Griffiths and Simons, 1986; Weiss and Nilsson, 2000; Rodriguez-Boulan and Müsch, 2005; Bonifacino, 2014; Dell’Angelica and Bonifacino, 2019). Recent studies have indicated that these sorting functions should be mostly ascribed to specialized regions proximal to and distal to the main body of the Golgi. Names given to these compartments are IC (intermediate compartment) or ERGIC (ER-Golgi intermediate compartment) on the cis-side (Schweizer et al., 1990; Saraste and Svensson, 1991; Plutner et al., 1992; Klumperman et al., 1998) and TGN (trans-Golgi network) on the trans-side (Roth et al., 1985; Griffiths and Simons, 1986; Taatjes and Roth, 1986; Geuze and Morré, 1991). The purpose of this review is to provide insights into the common features of these neighboring compartments, from a comparative view of yeast, plant, and animal cells.
One famous and memorable debate regarding trafficking in the Golgi dealt with the question of how cargo molecules are conveyed within the Golgi (Mellman and Simons, 1992; Glick and Malhotra, 1998; Pelham and Rothman, 2000). Live imaging of the budding yeast provided strong support for the cisternal maturation model (Losev et al., 2006; Matsuura-Tokita et al., 2006; Glick and Nakano, 2009; Nakano and Luini, 2009; Glick and Luini, 2011), but many unanswered questions remain. A group of researchers working on such critical Golgi issues gathered in Barcelona in 2009, mutually exchanging ideas and proposing future directions. The summary of the meeting review (Emr et al., 2009) concluded that “advances in high-resolution microscopy and biochemical techniques will help to clarify this issue.” As one of those who were present, and one who has seriously pursued the development of high-speed and super-resolution live imaging microscopy, I would like to summarize what we know now and propose some concepts emerging from the new knowledge. The Golgi and its next-door neighbors, as well as selected aspects of post-Golgi trafficking will be discussed in this review article. For the details of our imaging technology (SCLIM; super-resolution confocal live imaging microscopy) (see Footnote for abbreviations), I refer to our reviews published elsewhere (Nakano, 2002; Kurokawa et al., 2013; Kurokawa and Nakano, 2020; Tojima et al., 2022).
Spatial Organization of the Golgi
Many studies on membrane trafficking have been performed on mammalian cells, whose Golgi typically forms a giant structure called the Golgi ribbon in the perinuclear area near the centrosome (Ladinsky et al., 1999; Thyberg and Moskalewski, 1999; Rios and Bornens, 2003) (Figure 1B). Formation of Golgi ribbon depends on dynein-motor-driven gathering of membranes along microtubules and thus depolymerization of microtubules causes the scattering of hundreds of “Golgi ministacks” in the cytoplasm. This kind of organization has been thought specific to vertebrates, which have evolved radial patterning of microtubules. Indeed, invertebrate model animals, such as Drosophila and C. elegans, have scattered ministack-type of Golgi. Concentration of the Golgi near the centrosome is considered advantageous for massive protein secretion in a polarized fashion, for example, during cell migration, but why it forms the ribbon structure remains elusive (Terasaki, 2000; Marra et al., 2007; Yadav et al., 2009; Wei and Seemann, 2010; Yadav et al., 2011; Ferraro et al., 2014; Saraste and Prydz, 2019). The gigantic Golgi ribbon structure makes it sometimes difficult to apply detailed live imaging.
Plant cells, in contrast, provide ideal systems to study the Golgi at high resolution (Ito et al., 2014) (Figure 1A). Beautiful Golgi stacks are separately seen in the cytoplasm in their natural state and EM studies reveal details of their structure (Zhang and Staehelin, 1992; Staehelin and Kang, 2008; Donohoe et al., 2013; Robinson, 2020). Furthermore, we have relied on this advantage of plant cells to apply live imaging of Golgi and its neighboring compartments, as will be discussed in detail in this review.
The budding yeast S. cerevisiae is another extreme example (Figure 1C). The Golgi in this organism does not form stacks but exists as individual cisternae scattered in the cytoplasm (Preuss et al., 1992; Rambourg et al., 2001; Beznoussenko et al., 2016). This feature gave a great advantage for the demonstration of cisternal maturation by live imaging (Losev et al., 2006; Matsuura-Tokita et al., 2006). It appears that S. cerevisiae has somehow abandoned stacking of Golgi cisternae, because there exist species of budding yeast that have stacked Golgi (Rambourg et al., 1995; Glick, 1996; Rossanese et al., 1999). Nevertheless, S. cerevisiae has a very efficient secretory activity and why it copes without stacking remains a big mystery. Although the mechanisms and meanings of Golgi cisternal stacking are not amenable to study in this organism, it is still a fascinating choice to work with because of the invaluable molecular knowledge accumulated on trafficking machinery and the availability of various techniques including powerful genetics (Rothblatt and Schekman, 1989; Schekman, 1992; Nakano, 2008; Jackson, 2009; Papanikou and Glick 2009; Suda and Nakano, 2012).
The comparison of typical spatial arrangement of the Golgi in plant, animal, and yeast cells is summarized in Figure 1, giving an impression of how much they are different. However, despite the architectural differences, the important message of this review emphasizes how conserved the trafficking mechanisms are among them.
ER-Golgi Interface
Let’s think about the cargo delivery process from the ER to the Golgi. It is well known that the assembly of the coat protein complex II (COPII) plays an essential role in the formation of the vesicles (COPII vesicles) that export cargo proteins from the ER (Barlowe et al., 1994; Sato and Nakano, 2007). Activation of Sar1 GTPase by its guanine-nucleotide exchange factor (GEF) Sec12 is thought to be the most upstream reaction in the vesicular journey of the secretory pathway (Nakano et al., 1988; Nakano and Muramatsu, 1989; Barlowe and Schekman, 1993). The specialized places on the ER forming COPII vesicles are called transitional ER (tER) or ER exit sites (ERES) (Palade, 1975; Hughes and Stephens, 2008; Budnik and Stephens, 2009; Okamoto et al., 2012; Kurokawa and Nakano, 2019). In a simple scenario, the COPII vesicles released from the ERES into the cytoplasm somehow find and reach the Golgi to hand over cargo. However, this is not easy in a large mammalian cell, because a large number of ERES are present at the cell periphery and the Golgi is concentrated near the centrosome. There is a very long way to go.
The intermediate compartment between the ER and Golgi (IC/ERGIC) was first defined as a compartment where a unique marker protein (rat p58 or human ERGIC53) is present (Schweizer et al., 1990; Saraste and Svensson, 1991; Plutner et al., 1992; Klumperman et al., 1998; Hauri et al., 2000; Ying et al., 2000; Ben-Tekaya et al., 2005; Appenzeller-Herzog and Hauri, 2006). A complex structure containing both COPII and COPI (coat protein complex I) was discovered by immuno-EM studies and named vesicular tubular cluster (VTC) and proposed to be the place where cargo sorting takes place between the ER and the Golgi (Bannykh et al., 1996; Martínez-Menàrguez et al., 1999). While ERGIC appeared to be a stable compartment and VTC was first regarded as a transient structure, they are most likely referring to the same intermediate compartment connecting the traffic from the ERES to the cis face of the Golgi. Such an intermediate compartment was not defined in yeast and plant cells until recently. It should be noted here that ERGIC is frequently observed in the vicinity of ERES (Sannerud et al., 2006; Budnik and Stephens, 2009; Saraste and Marie, 2018).
Experiments using brefeldin A (BFA), an inhibitor of GEF toward Arf GTPases, give us hints on the commonness of trafficking mechanisms (Takatsuki and Tamura, 1985; Klausner et al., 1992). Arf GTPases are relatives of Sar1 and are required for formation of the vesicles coated with COPI, adapter protein complexes (AP-1∼5), and GGAs (Golgi-localized, γ-ear-containing, Arf-binding family of proteins) (Jackson and Casanova, 2000; Pucadyil and Schmid, 2009; Dacks and Robinson, 2017; Dell’Angelica and Bonifacino, 2019). COPI vesicles are involved in retrograde traffic in the Golgi and in the Golgi-to-ER recycling (Letourneur et al., 1994; Rabouille and Klumperman, 2005; Donohoe et al., 2007). Treatment with BFA causes blockade of COPI vesicle formation and induces enormous reorganization of endomembrane systems. Remarkable studies on mammalian cells by Lippincott-Schwartz et al. (1990), Lippincott-Schwartz et al. (1991) demonstrated that the Golgi was almost completely absorbed into the ER upon BFA treatment.
Similar effects of BFA on the Golgi have been seen in plant cells as well (Ritzenthaler et al., 2002; Langhans et al., 2007); that is, most of the Golgi components are relocated to the ER. However, Ito et al. (2012) discovered that in tobacco BY-2 cells some cis-components such as SYP31 (plant counterpart of yeast Sed5 and animal Syntaxin-5) and RER1 (Golgi-to-ER retrieval receptor) remain associated with small punctate structures on BFA treatment, which act as the scaffold of Golgi regeneration when BFA is washed out (its action is known to be reversible) (Figure 2). Since components of the main Golgi body traverse this specialized cis-most compartment during Golgi regeneration, the name GECCO, standing for Golgi entry core compartment, was proposed for this compartment (Ito et al., 2018b; Ito and Boutté, 2020). The role of GECCO can be regarded similar to that of ERGIC. In fact, in mammals, ERGIC is known to remain unabsorbed to the ER upon BFA treatment (Lippincott-Schwartz et al., 1990; Saraste and Svensson, 1991; Tang et al., 1993; Füllekrug et al., 1997). In the case of plant cells, the Golgi is almost always located in the vicinity of ERES (daSilva et al., 2004; Sparkes et al., 2009; Takagi et al., 2020) and thus a long-way connection is not necessary (Robinson et al., 2015). The existence of such similar compartments at the interface between the ER (ERES) and the Golgi (cis-Golgi) suggests the conservation of fundamental architecture.
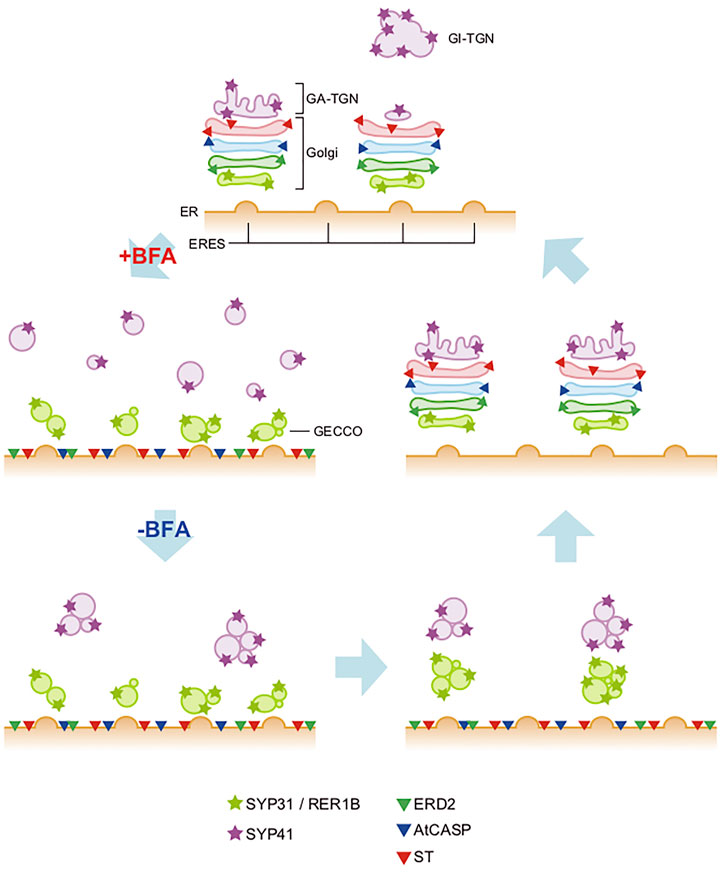
FIGURE 2. Behaviors of Golgi and TGN upon BFA treatment and washout in plant cells. In tobacco BY-2 cells, BFA-treatment gives distinct effects to the cis-most Golgi, the Golgi main body, and TGN (+BFA). The main-body Golgi (with the resident proteins shown by triangles) is completely absorbed into the ER, whereas cis-most Golgi, marked by SYP31 and RER1B (light green stars), remains as punctate structures near the ERES, which we designate GECCO (Golgi entry core compartment). TGN (marked by purple stars; SYP41), on the other hand, gets fragmented into small vesicles and dispersed in the cytoplasm. When BFA is washed out (-BFA), these components regenerate the original structures but their processes are again quite different. The Golgi main body reforms stacks in the cis-to-trans direction using the GECCO as the regeneration scaffold. TGN reassembles by itself independent of the GECCO and the Golgi main body, but finally meets Golgi and starts staying side by side (Ito et al., 2012; Ito et al., 2017; Ito et al., 2018b). GA-TGN, Golgi-associated TGN; GI-TGN, Golgi-independent TGN (see Figure 3).
Recently, Fujii et al. (2020b) showed in Drosophila cells that knockdown of Arf GEF Garz also induces dynamic relocalization of Golgi proteins to the ER, while leaving behind GM130, a component of cis-Golgi, in the cytoplasm, again suggesting the existence of a GECCO-like compartment in invertebrate cells (see Figure 4C). It should be mentioned here that mammalian GM130 was also shown to stay in BFA-resistant structures together with ERGIC (Nakamura et al., 1995).
What about the budding yeast then? As the yeast S. cerevisiae does not stack its Golgi cisternae, it is not easy to describe the behavior of the Golgi as a whole, but we noticed that Sed5, one of the cis-most Golgi components, showed repeated approach and contact to the ERES (Kurokawa et al., 2014). During this action, which we named hug and kiss, cargo is handed over from the ERES to the cis-Golgi. This finding also suggests that the cis-most compartment of the yeast Golgi has a special role for the delivery of cargo from the ERES, like GECCO in plant cells and ERGIC in animal cells.
The effects of BFA may be also informative to define such a compartment. Indeed, our preliminary experiments on yeast indicate that there is a group of cis-Golgi proteins that do not relocalize to the ER by BFA (Tojima and Nakano, unpublished). These proteins would be good candidates to constitute the yeast counterpart of GECCO.
On the ERES side, COPII proteins are considered to play roles in the selection of cargo (Aridor et al., 2001; Sato and Nakano, 2005; Miller and Barlowe, 2010). As activation of Sar1 GTPase is essential for the COPII assembly, Sar1 was long thought to be present all over the COPII lattice (Miller and Barlowe, 2010; Gillon et al., 2012). However, recent in vivo and in vitro studies (Kurokawa et al., 2016; Iwasaki et al., 2017) have shown that Sar1 is mostly excluded from the completed COPII cage and present only at the rim or edge regions. This suggests that the uncoating of COPII could take place independently of the vesicle release from the ER (Raote and Malhotra, 2019).
Recent studies also indicate that the coated cage of COPII is very flexible (Mironov et al., 2003; Palmer and Stephens, 2004; Gillon et al., 2012; Raote et al., 2021; Shomron et al., 2021) and can accommodate very large cargo such as collagen or chylomicron in mammalian cells (Raote and Malhotra, 2021). The COPII buds may even be capable of conveying cargo to the next compartments (ERGIC/GECCO and further to the Golgi) before they physically detach from the ER (Raote and Malhotra, 2019). The wonderful morphological work on HeLa cells by Weigel et al. (2021) demonstrated amazing structures extending from the ERES, which indicated VTC-like profiles on the top of ERES. Pearled tubular membrane intermediates are occasionally seen, which probably connect to the Golgi, suggesting a very dynamic nature of the ERGIC compartment.
While admitting the importance of the long-distance transport from the peripheral ERES to the perinuclear Golgi ribbon, we can observe even in mammalian cells that ERES and ERGIC are very abundant in the vicinity of the Golgi ribbon. The cargo trafficking in mammals may actually be performed largely in the pericentrosomal region where the ER and the Golgi are very close to each other. We should not underestimate the short-distance processes (Saraste and Marie, 2018).
An intriguing question is whether GECCO/ERGIC is a part of the Golgi apparatus or an independent organelle (see Marie et al., 2012; Ito and Boutté, 2020). Considering the common role of this compartment, I suppose that the position in very close vicinity to both the ER and the Golgi is the classic, default pattern, that is conserved during evolution. The fact that the ministacks of mammalian Golgi produced by depolymerization of microtubules become located in front of ERES (Cole et al., 1996) supports this idea.
The Main Body of the Golgi
Here I am not going to discuss the mechanism of cargo transport within the Golgi. It is widely accepted now that cisternal maturation is the major mechanism for anterograde movement of cargo (Glick and Nakano, 2009; Nakano and Luini, 2009). The roles of COPI and tubular connections (Yang et al., 2011; Ishii et al., 2016) have been shown, but disputes remain. Their clarification will need more critical experiments. Recent yeast studies by precise live imaging revealed that secretory cargo sometimes shows backward movement, suggesting cargo recycling in or around the Golgi (Casler et al., 2019; Kurokawa et al., 2019). To understand the underlying mechanisms, it is probably necessary to consider the roles of TGN.
Enzymes involved in glycosylation/modification reactions are considered as regular members of the main body of the Golgi. It is probably safe to postulate that sequential reactions go on when cargo proteins proceed across local regions from cisterna to cisterna. However, there are multiple glycosylation reactions that could occur in parallel, for example, N-linked and O-linked modifications. Whether such different series of reactions take different paths in the Golgi is another exciting question to be pursued (Yano et al., 2005; D’Angelo et al., 2013).
Besides glycosylation enzymes, some non-enzyme components have been mapped in the trans region of the Golgi. They include lipid-transferring proteins such as OSBP (oxysterol-binding protein), CERT (ceramide transfer protein) etc. (Levine and Munro, 2002; Hanada et al., 2007), and perhaps some Arl and Rab GTPases. As these protein functions could be executed in TGN as well, careful examination may be necessary when discussing their exact localizations.
Trans-Golgi Network
The name trans-Golgi network was given to the morphologically prominent reticular structure found at the trans side of the Golgi stack (Roth et al., 1985; Griffiths and Simons, 1986; Taatjes and Roth, 1986; Geuze and Morré, 1991), which was regarded as a part of the Golgi for some time. A variety of cargo carriers including clathrin-coated vesicles were found in this area, and the concept that this compartment is important for sorting of cargo for different destinations was established (Griffiths et al., 1989; Keller and Simons, 1997; Hinners and Tooze, 2003; Bard and Malhotra, 2006; De Matteis and Luini, 2008; Gonzalez and Rodriguez-Boulan, 2009; Chen et al., 2017).
Now, accumulating evidence tells us that TGN should be considered as an organelle independent of the Golgi. The most striking discovery was that TGN can exist away from the Golgi in plant cells (Uemura et al., 2004; Staehelin and Kang, 2008; Viotti et al., 2010; Uemura and Nakano, 2013). SYP4 and SYP6 subfamilies of plant SNARE proteins (corresponding to Tlg2 and Tlg1 proteins in yeast and Syntaxin-16 and Syntaxin-6 proteins in mammals, respectively) are good markers of TGN. Uemura et al. (2004) realized that when transiently expressed in Arabidopsis protoplasts they mark structures clearly distinct from the Golgi. Such distinction of Golgi/TGN was also seen in living Arabidopsis root tissues, although the extent of displacement depends on the developmental stages of cells (Uemura et al., 2012; Uemura et al., 2014). Furthermore, the live imaging of Arabidopsis TGN demonstrated that the Golgi-independent free TGN can dissociate from the Golgi-associated TGN and repeat reattachment to and dissociation from the Golgi (Uemura et al., 2014) (Figure 3).
FIGURE 3. Trans-Golgi network of plant cells. In plant cells, TGN (trans-Golgi network) takes two statuses, one being attached to the trans face of the Golgi (Golgi-associated TGN) and the other detached from the Golgi and free in the cytoplasm (Golgi-independent TGN) (Staehelin and Kang, 2008; Uemura et al., 2014). These two statuses are interchangeable; Golgi-independent TGN can dissociate from the Golgi and reassociate with it.
Another surprise about the plant TGN was that it acts as a very early compartment of endocytosis. A lipidic dye FM4-64, a popular marker for endocytosis, was shown to reach the TGN much earlier than other compartments (Dettmer et al., 2006; Lam et al., 2007; Chow et al., 2008). FM4-64 was also recognized to label punctate structures in yeast very early after entry (Vida and Emr, 1995; Best et al., 2020), which turned out to correspond to TGN (Day et al., 2018). Receptor-mediated endocytosis was also shown to take the direct route from the plasma membrane to the TGN in yeast (Day et al., 2018; Nagano et al., 2019) and plant cells (Viotti et al., 2010). These findings indicate very important roles of TGN during the early steps of endocytosis, at least for plant and yeast cells.
In mammalian cells, endocytic trafficking involves different routes of recycling, very quickly from the early or sorting endosomes and in a more delayed fashion from the recycling endosomes (RE) (Spang, 2009; Poteryaev et al., 2010; Taguchi, 2013; Scott et al., 2014). I will discuss the close relationship between RE and TGN later.
A group of bacterial toxins are known to take a retrograde trafficking route from the plasma membrane to the Golgi and further to the ER (Sandvig and van Deurs, 1996; Bujny et al., 2007). An early process of this retrograde pathway involves transport from the early endosomes to TGN, which is also utilized for recycling of a variety of cargo receptors. Retromer (and its functional homologs) and GARP (Golgi-associated retrograde protein complex) are important players in this traffic and very well conserved from yeast to plants and animals (Seaman et al., 1998; Sannerud et al., 2003; Bonifacino and Rojas, 2006; Bonifacino, 2014; Schindler et al., 2015; Cullen and Steinberg, 2018; Heucken and Ivanov, 2018; Ishida and Bonifacino, 2019; Ma and Burd, 2020).
All these findings indicate that the role of TGN as the trafficking platform is very important not only for exocytosis/secretion but also for endocytic recycling. To achieve such complicated roles of cargo sorting, TGN must have differentiated domains or zones to perform individual functions.
We have been trying to dissect compartments within TGN in the budding yeast by applying 3D and 4D live imaging approaches, which were powerful in demonstration of cisternal maturation (Kurokawa et al., 2013; Kurokawa and Nakano, 2020). By examining the transition of residents in individual cisternae, we can define the order of events in Golgi and TGN. By the extensive analysis of more than 20 constituents of yeast Golgi and TGN (Tojima et al., 2019; Tojima and Nakano, unpublished), we have proposed to classify yeast TGN proteins into two groups, the early TGN for cargo reception and the late TGN for carrier formation. Tlg2 (the homolog of mammalian Syntaxin-16 and plant SYP41/42/43) is the representative of the early TGN, where a variety of cargo flow in, not only from the Golgi but also from endosomes and the plasma membrane. The late TGN produces carriers with different destinations, marked by clathrin, AP-1, GGA, and exomer, and the Arf GEF Sec7 appears to be mostly overlapping with this area.
In distinguishing TGN from Golgi, BFA is again a good tool. Early studies on mammalian cells showed that BFA caused the absorption of Golgi proteins to the ER but did not affect TGN and endosomes in that way and gave rise to tubular clusters independent of the ER/Golgi hybrid compartment (Chege and Pfeffer, 1990; Lippincott-Schwartz et al., 1991; Wood et al., 1991). In later studies on plants, Ito et al. (2017) showed that BFA treatment of tobacco BY-2 cells caused relocation to the ER of most of proteins of the Golgi main body, leaving GECCO compartment behind. Under the same conditions, TGN proteins such as SYP41 (Tlg2 homolog) were not absorbed to the ER but became dispersed in the cytoplasm as numerous small vesicles, which is not exactly the same but similar to the situation seen in mammalian cells (Figure 2). As the effect of BFA is reversible, regeneration of Golgi and TGN was observed after BFA washout, but their behaviors turned out quite distinct. They gradually reformed Golgi and TGN, but in an independent fashion without significantly overlapping for a certain period of time. After a long time of recovery, they meet again and stay side by side, re-establishing the next-door relationship (Figure 2). Thus, we concluded that the Golgi stack and the TGN have quite different origins and properties but somehow collaborate with each other (Ito et al., 2017).
Regarding the positional properties of Golgi and TGN, plant TGN shows two statuses, one associated with Golgi and the other independent of Golgi (Uemura and Nakano, 2013) as mentioned above, and these two statuses appear to be interchangeable (Uemura et al., 2014) (Figure 3).
Fujii et al. (2020a) recently reported that, in Drosophila S2 cells and nocodazole-treated HeLa cells, Golgi and RE exhibit repeated attachment and detachment (Figure 4A). They called the two states of RE, Golgi-associated RE and free RE. This finding manifests amazing similarity to the case of plant Golgi and TGN. Indeed, markers of TGN and RE show close localization and even partial overlap in Drosophila cells (Fujii et al., 2020a; Fujii et al., 2020b).
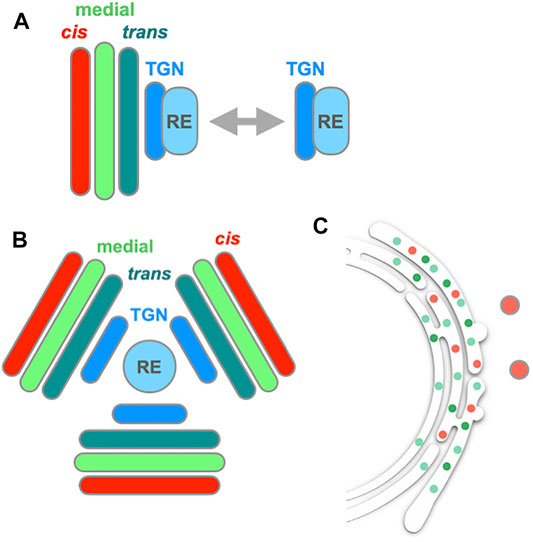
FIGURE 4. | Behavior of the Golgi, TGN and RE in Drosophila cells. (A) In Drosophila S2 cells, Golgi stack (cis, medial, and trans) and RE (recycling endosome) show attachment and detachment repeatedly (Fujii et al., 2020a). TGN appears to behave together with RE. Similar behavior of RE is also seen in nocodazole-treated HeLa cells. (B) When S2 cells are treated with BFA, which inhibits Arf GEF Sec71, Golgi, TGN and RE form a large aggregate (BFA body). In the BFA body, RE sits at the center, TGN follows next and Golgi cisternae align from trans to cis toward the periphery. (C) When the other Arf GEF of Drosophila Garz, which is insensitive to BFA, is knocked down, the Golgi main body is mostly absorbed into the ER, while leaving behind a cis component (GM130, red spots) (Fujii et al., 2020b).
BFA treatment of Drosophila cells also gave very interesting results (Fujii et al., 2020b) (Figures 4B,C). Drosophila has two Arf GEFs, called Sec71 (homolog of mammalian BIG and yeast Sec7, which functions in TGN) and Garz (homolog of mammalian GBF1 and yeast Gea1, which functions in cis-Golgi). Of the two only Sec71 is sensitive to BFA. When Drosophila S2 cells were treated with BFA, a few gigantic structures emerged, containing Golgi, TGN and RE components (Figure 4B). This kind of BFA-induced large structures are also seen in plant Arabidopsis cells and called BFA bodies (Geldner et al., 2003; Langhans et al., 2011; Nielsen et al., 2012). It should be noted here that the situation is somewhat complicated in plants. Tobacco and Arabidopsis have different sets of Arf GEFs, some of which are BFA-sensitive and others insensitive. GNOM and GNOM-LIKE1 (GNL1) are two major Arf GEFs functioning in and around the Golgi; GNOM is BFA-sensitive in both tobacco and Arabidopsis, while GNL1 is BFA-sensitive in tobacco but insensitive in Arabidopsis (Richter et al., 2007; Teh and Moore, 2007; Robinson et al., 2008; Naramoto et al., 2014). Their sensitivities can be artificially converted by amino acid point mutations (Geldner et al., 2003), but I skip details here to avoid confusion.
Interestingly, detailed examination of such BFA-induced structures in S2 cells reveals that RE is at their center, TGN follows next and Golgi cisternae align from trans to cis toward their periphery (Figure 4B). Very similar alignment is seen in the BFA bodies of Arabidopsis cells (Langhans et al., 2011). On the other hand, when the other Arf GEF, Garz, is knocked down in Drosophila, most of the Golgi markers are absorbed into the ER (Figure 4C), like the case with mammalian cells (Fujii et al., 2020b). As GNL1, the Arf GEF working in the ER-Golgi traffic in Arabidopsis, is insensitive to BFA, the BFA body formation in Arabidopsis and Drosophila is most likely based on the inhibition of Arf activation only in the post-Golgi processes, leading to the aggregation of the TGN-RE system. The absorption of Golgi into the ER takes place when the Arf activation is inhibited early in the ER-Golgi traffic. These findings imply that the endomembrane system could be classified into two groups, the ER-Golgi system and the TGN-endosome system (see also Lippincott-Schwartz et al., 1991; Manolea et al., 2008; Marie et al., 2009), and the failure to control their organization by compromising Arf activation leads to the jumbling of membranes within each system. If so, there must be a boundary between the main body of Golgi and TGN.
Maturation of compartments appears to go on rather smoothly in the budding yeast and it is not easy to draw a line between the main Golgi body and the TGN. As the order of events we have analyzed relates only to the temporal overlapping and separation of residents, we will probably need to consider spatial segregation of the zones involved in different sorting reactions even in the regions that show up at similar timing.
At the crossroad of multiple pathways, machineries in TGN are waiting for coming cargo and will then sort them out to different destinations. To gain insights into such differentiation of sorting machineries, the high-speed and super-resolution live imaging technology we have developed (SCLIM) is extremely useful (Kurokawa et al., 2013; Kurokawa and Nakano, 2020; Tojima et al., 2022). We have recently demonstrated that Arabidopsis TGN harbors at least two distinct zones, one containing AP-1 and clathrin for secretory trafficking and the other containing AP-4 for vacuolar trafficking (Shimizu et al., 2021) (Figure 5). This kind of state-of-the-art visualization analysis is expected to unveil dynamic sorting events going on real time in living cells.
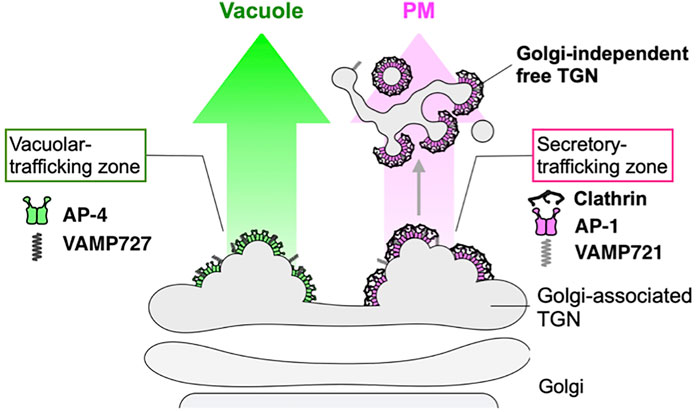
FIGURE 5. Sorting zones in plant TGN differentiated for secretory and vacuolar trafficking. Cargo sorting events in root epidermal cells of Arabidopsis have been analyzed in detail by state-of-the-art live imaging (SCLIM) (Shimizu et al., 2021). TGN-localized proteins exhibit spatially and temporally distinct distribution. VAMP721 (R-SNARE), AP−1, and clathrin which are involved in secretory trafficking compose an exclusive subregion, whereas VAMP727 (R-SNARE) and AP-4 involved in vacuolar trafficking compose another subregion on the same TGN. These findings indicate that the single TGN has at least two subregions, or “zones”, responsible for distinct cargo sorting: the secretory-trafficking zone destined to the plasma membrane (PM) and the vacuolar-trafficking zone.
An Attempt to Draw a Simple Common Model
As has been discussed above, despite the seemingly big differences in the trafficking architectural systems between animal, plant and yeast cells, there are also significant similarities in many aspects. In Figure 6A, I summarize the routes connecting the Golgi and its next-door neighbors as well as post-Golgi trafficking to vacuole via the prevacuolar compartment (PVC), which can explain most of the observations I have described above. What makes things confusing may be the names of compartments adapted in different systems. A good example is endosomes. Early, late and recycling endosomes do not necessarily refer to the compartments comparable among different species. Let me try to simplify from a different point of view.
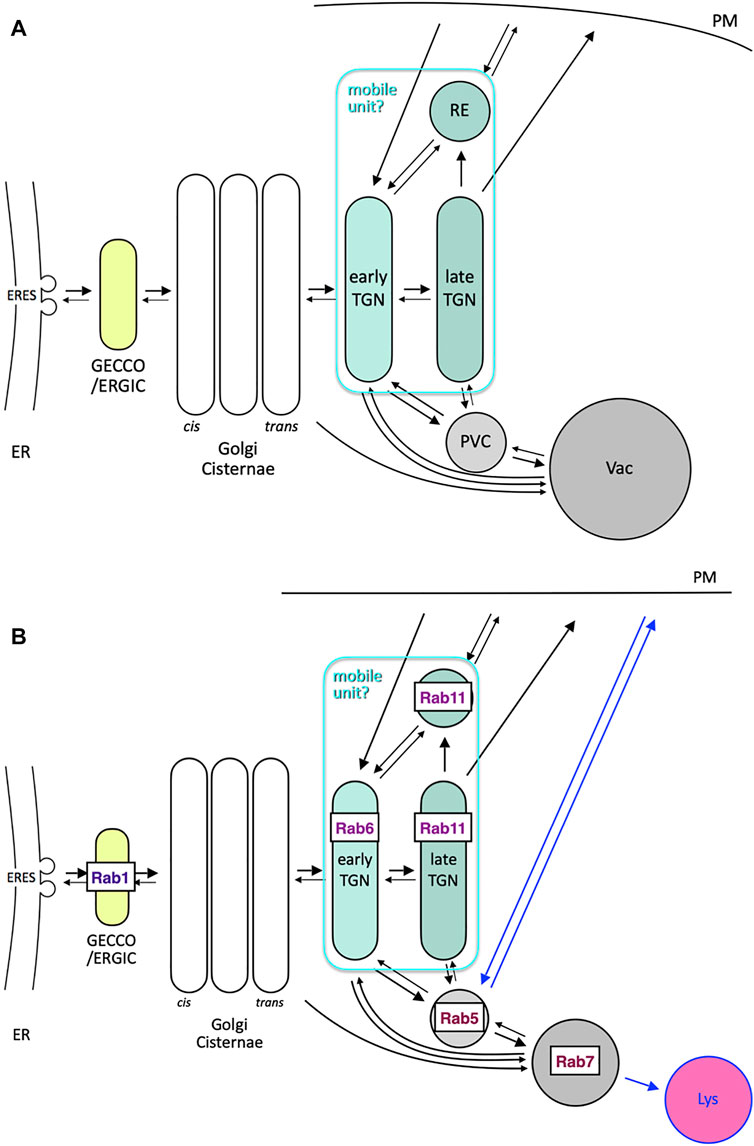
FIGURE 6. Comprehensive models of trafficking around Golgi and post Golgi. (A) Trafficking pathways around Golgi and its next-door neighbors (GECCO/ERGIC and TGN/RE) as well as post-Golgi trafficking to vacuole (Vac) via prevacuolar compartment (PVC). Arrows are drawn in an attempt to explain the observations described in this review. Note that early TGN, late TGN, and RE can behave together as a mobile unit, which takes Golgi-associated and Golgi-independent statuses. (B) Consideration of conserved Rab GTPases as signboards of compartments, which may be common to yeast, plant, and animal cells. Rab1 (Ypt1 in yeast and RABDs in plants) sits on GECCO/ERGIC, Rab6 (Ypt6 in yeast and RABHs in plants) is probably a good marker of early TGN, and Rab11 (Ypt31/32 in yeast and RABAs in plants) is present in late TGN and RE. The cases of Rab5 (Vps21 in yeast and RABFs in plants) and Rab7 (Ypt7 in yeast and RABGs in plants) are complicated. They mark PVC (MVB) and vacuole, respectively, in yeast and plant cells, but are considered to reside in early and late endosomes, respectively, in mammalian cells (see also Table 1). Blue arrows, i.e., back and forth between plasma membrane and Rab5 compartment and from Rab7 compartment to lysosome (Lys), could be considered inventions in mammals.
Here I propose to consider the localization of major Rab GTPases (Figure 6B). In contrast to Sar/Arf GTPases, which are necessary for vesicle budding, Rab GTPases are generally thought to play roles in targeting and fusion of vesicles (Segev, 2001). In addition, as they collaborate with tethering and fusion machinery, they are often good markers of target compartments (Segev, 2001; Zerial and McBride, 2001).
Let’s pay attention to two Rabs involved in the endocytic pathway, Rab5 and Rab7. These Rab GTPases are extremely well conserved among species. In mammalian cells, Rab5 marks early endosomes and Rab7 is on late endosomes. In yeast and plant cells, however, the situation is quite different. As described above, accumulating evidence indicates that TGN functions as an early endosome. Rab5 is located at PVC and Rab7 is on the vacuole (Saito and Ueda, 2009; Borchers et al., 2021). The transition from the Rab5-to the Rab7-compartment has been well investigated not only in mammals but also in yeast and plants (Rink et al., 2005; Sriram et al., 2006; Balderhaar and Ungermann, 2013; Ebine et al., 2014; Takemoto et al., 2018; Casler and Glick, 2020; Borchers et al., 2021). A big difference between mammals and plants/fungi is the presence of lysosomes (de Duve, 1983; de Duve, 2005). If lysosomes are regarded as an organelle specialized in very efficient degradation in an extremely acidic milieu, the common roles of Rab5 and Rab7 compartments could be conserved in all species.
Mechanisms of intraluminal vesicle (ILV) formation by ESCRT (endosomal sorting complex required for transport) machinery are well understood (Raiborg and Stenmark, 2009; Henne et al., 2011). It gives rise to multivesicular bodies (MVB) or multivesicular endosomes. In mammals, MVB sounds like a synonym of late endosomes (Rab7 compartment), but we should remember that ILV formation begins already in early endosomes (Rab5 compartment). In yeast and plant cells, Rab5 compartment is the MVB (Goh et al., 2007; Haas et al., 2007; Russell et al., 2012). Furthermore, endocytosed cargo is first delivered to TGN and then transferred to the Rab5 compartment, leading to the concept that Rab5 resides in late endosomes in yeast and plants (Cui et al., 2016; Day et al., 2018). Plant Rab5 is further diversified into two types, one directing the route to the vacuole and the other for recycling back to the plasma membrane (Ueda et al., 2001; Ebine et al., 2011; Sakurai et al., 2016; Ito et al., 2018a). Why has the Rab5 compartment changed its role so much during evolution?
In and around the Golgi, several other Rab GTPases are also very important. Rab1 (Ypt1 in yeast) is essential for ER-to-Golgi transport, although it may also be involved in a later process within the Golgi (Segev et al., 1988; Saraste and Goud, 2007; Suda et al., 2013; Gustafson and Fromme, 2017; Suda et al., 2018; Tojima and Nakano, unpublished). The role of Rab6 is somewhat controversial but it is important for receiving cargo in the late Golgi or TGN from endosomes (Liu and Storrie, 2012, 2015; Suda et al., 2013; Thomas et al., 2021). Rab11 is recognized as a good marker of RE (Welz et al., 2014), but its yeast homolog Ypt31/32 is essential for export of secretory cargo from TGN to the plasma membrane (Jedd et al., 1997). Rab11-positive RE is also shown to serve as an intermediate of anterograde Golgi-to-plasma membrane traffic in Drosophila and mammalian cells (Ang et al., 2004; Satoh et al., 2005; Taguchi, 2013). In plant cells, the Rab11 group has been diversified remarkably. Arabidopsis has 26 Rab11 members out of 57 Rab proteins (Rutherford and Moore, 2002; Nielsen et al., 2008; Saito and Ueda, 2009). This suggests that the Rab11 group can fulfill a variety of functions depending on situations (Feraru et al., 2012; Asaoka et al., 2013; Choi et al., 2013), but the conservation of this group also supports its very fundamental role.
Considering the role of Rab11 in sorting cargo into the secretory pathway, its location best fits with the late TGN we defined in yeast. Rab6, on the other hand, is probably working in the early TGN or at the boundary between Golgi and TGN. The relationships of compartments linked to specific Rab GTPases are summarized in Table 1.
From these considerations, I propose a basic principle of Rab-directed compartmentalization as shown in Figure 6B. Rab1, Rab6, Rab11, Rab5, and Rab7 GTPases are selected as the major Rab members very well conserved in evolution. I would like to place Rab6 and Rab11 in early and late TGN, respectively, but the Rab11 compartment could also be called RE. As described above, the behaviors of late TGN and RE are often very similar (Ang et al., 2004; Taguchi, 2013; Bonifacino, 2014). They might be regarded as different sub-compartments or zones of a kind of continuous large platform. The fact that retrograde trafficking from the Rab5 compartment involves tether complexes GARP and EARP (endosome-associated recycling protein complex) for TGN and RE, respectively, which are slightly different in their subunit composition, may support this idea (Schindler et al., 2015). Since both TGN and RE show very dynamic movement back and forth to the Golgi in animal and plant cells, they can be regarded as a large-sized mobile unit as a whole.
Now, a big difference remains regarding the role of Rab5 compartment. Plenty of evidence exists in mammalian cells showing that this is the earliest compartment where newly endocytosed cargo comes in (Chavrier, et al., 1990; Spang, 2009; Scott et al., 2014). In plant and yeast cells, it operates much later in the endocytic pathway (Cui et al., 2016; Day et al., 2018). On the other hand, there is a significant similarity that retrograde trafficking involving retromer and GARP/EARP takes place from the Rab5 compartment to TGN/RE (Bonifacino and Rojas, 2006; Cullen and Steinberg, 2018; Heucken and Ivanov, 2018; Ishida and Bonifacino, 2019; Ma and Burd, 2020). Should this traffic to TGN/RE be considered as that from early endosomes or from late endosomes?
One explanation may be that mammalian cells have somehow evolved to bypass the TGN/RE route to reach the Rab5 compartment. If so, the classic TGN/RE route for endocytosis might also remain. The presence of the mammalian Rab5 compartment as the early endosome has been so prominent; therefore, other possibilities could have been overlooked. Careful revisiting of old data may uncover new findings. To achieve rapid recycling from the early endosome (Rab5 compartment), mammalian cells utilize Rab4, which is not conserved in plant and yeast cells (Sönnichsen et al., 2000; de Renzis et al., 2002; Franke et al., 2019). The need for rapid endocytosis/recycling could be a driving force to take a shortcut and evade the conventional TGN/RE route. Detailed examination of endocytic processes in other model animals would also be interesting.
Author Contributions
AN wrote the whole manuscript.
Funding
This research was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (Grant numbers: 25221103, 17H06420, and 18H05275) to AN.
Conflict of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The concepts I describe in this review are derived from very passionate and fruitful discussions with my friends and colleagues. I am particularly grateful to Kazuo Kurokawa, Takuro Tojima, Yasuyuki Suda, Tomohiro Uemura, Emi Ito, Yoko Ito, Yutaro Shimizu, Akiko K. Satoh, Jiro Toshima, and Jaakko Saraste for their invaluable comments.
References
Ang, A. L., Taguchi, T., Francis, S., Fölsch, H., Murrells, L. J., Pypaert, M., et al. (2004). Recycling Endosomes Can Serve as Intermediates during Transport from the Golgi to the Plasma Membrane of MDCK Cells. J. Cel Biol. 167, 531–543. doi:10.1083/jcb.200408165
Appenzeller-Herzog, C., and Hauri, H.-P. (2006). The ER-Golgi Intermediate Compartment (ERGIC): in Search of its Identity and Function. J. Cel Sci. 119, 2173–2183. doi:10.1242/jcs.03019
Aridor, M., Fish, K. N., Bannykh, S., Weissman, J., Roberts, T. H., Lippincott-Schwartz, J., et al. (2001). The Sar1 GTPase Coordinates Biosynthetic Cargo Selection with Endoplasmic Reticulum export Site Assembly. J. Cel Biol. 152, 213–230. doi:10.1083/jcb.152.1.213
Asaoka, R., Uemura, T., Ito, J., Fujimoto, M., Ito, E., Ueda, T., et al. (2013). Arabidopsis RABA1 GTPases Are Involved in Transport between Thetrans-Golgi Network and the Plasma Membrane, and Are Required for Salinity Stress Tolerance. Plant J. 73, 240–249. doi:10.1111/tpj.12023
Balderhaar, H. J. k., and Ungermann, C. (2013). CORVET and HOPS Tethering Complexes-Coordinators of Endosome and Lysosome Fusion. J. Cel Sci. 126, 1307–1316. doi:10.1242/jcs.107805
Bannykh, S. I., Rowe, T., and Balch, W. E. (1996). The Organization of Endoplasmic Reticulum export Complexes. J. Cel Biol. 135, 19–35. doi:10.1083/jcb.135.1.19
Bard, F., and Malhotra, V. (2006). The Formation of TGN-To-Plasma-Membrane Transport Carriers. Annu. Rev. Cel Dev. Biol. 22, 439–455. doi:10.1146/annurev.cellbio.21.012704.133126
Barlowe, C., Orci, L., Yeung, T., Hosobuchi, M., Hamamoto, S., Salama, N., et al. (1994). COPII: a Membrane Coat Formed by Sec Proteins that Drive Vesicle Budding from the Endoplasmic Reticulum. Cell 77, 895–907. doi:10.1016/0092-8674(94)90138-4
Barlowe, C., and Schekman, R. (1993). SEC12 Encodes a Guanine-Nucleotide-Exchange Factor Essential for Transport Vesicle Budding from the ER. Nature 365, 347–349. doi:10.1038/365347a0
Ben-Tekaya, H., Miura, K., Pepperkok, R., and Hauri, H.-P. (2005). Live Imaging of Bidirectional Traffic from the ERGIC. J. Cel Sci. 118, 357–367. doi:10.1242/jcs.01615
Berger, E., and Rohrer, J. (2008). “Golgi Glycosylation Enzymes,” in The Golgi Apparatus, State of the Art 110 Years after Camillo Golgi’s Discovery. Editors A. Mironov, and M. Pavelka (Wien: Springer-Verlag), 161–189.
Best, J. T., Xu, P., McGuire, J. G., Leahy, S. N., and Graham, T. R. (2020). Yeast Synaptobrevin, Snc1, Engages Distinct Routes of Postendocytic Recycling Mediated by a Sorting Nexin, Rcy1-COPI, and Retromer. MBoC 31, 944–962. doi:10.1091/mbc.e19-05-0290
Beznoussenko, G. V., Ragnini-Wilson, A., Wilson, C., and Mironov, A. A. (2016). Three-dimensional and Immune Electron Microscopic Analysis of the Secretory Pathway in Saccharomyces cerevisiae. Histochem. Cel Biol. 146, 515–527. doi:10.1007/s00418-016-1483-y
Bonifacino, J. S. (2014). Adaptor Proteins Involved in Polarized Sorting. J. Cel Biol. 204, 7–17. doi:10.1083/jcb.201310021
Bonifacino, J. S., and Rojas, R. (2006). Retrograde Transport from Endosomes to the Trans-golgi Network. Nat. Rev. Mol. Cel Biol. 7, 568–579. doi:10.1038/nrm1985
Borchers, A. C., Langemeyer, L., and Ungermann, C. (2021). Who's in Control? Principles of Rab GTPase Activation in Endolysosomal Membrane Trafficking and beyond. J. Cel Biol. 220, e202105120. doi:10.1083/jcb.202105120
Budnik, A., and Stephens, D. J. (2009). ER Exit Sites-Llocalization and Control of COPII Vesicle Formation. FEBS Lett. 583, 3796–3803. doi:10.1016/j.febslet.2009.10.038
Bujny, M. V., Popoff, V., Johannes, L., and Cullen, P. J. (2007). The Retromer Component Sorting Nexin-1 Is Required for Efficient Retrograde Transport of Shiga Toxin from Early Endosome to the Trans Golgi Network. J. Cel Sci. 120, 2010–2021. doi:10.1242/jcs.003111
Casler, J. C., and Glick, B. S. (2020). A Microscopy-Based Kinetic Analysis of Yeast Vacuolar Protein Sorting. eLife 9, e56844. doi:10.7554/eLife.56844
Casler, J. C., Papanikou, E., Barrero, J. J., and Glick, B. S. (2019). Maturation-driven Transport and AP-1-dependent Recycling of a Secretory Cargo in the Golgi. J. Cel Biol. 218, 1582–1601. doi:10.1083/jcb.201807195
Chavrier, P., Parton, R. G., Hauri, H. P., Simons, K., and Zerial, M. (1990). Localization of Low Molecular Weight GTP Binding Proteins to Exocytic and Endocytic Compartments. Cell 62, 317–329. doi:10.1016/0092-8674(90)90369-p
Chege, N. W., and Pfeffer, S. R. (1990). Compartmentation of the Golgi Complex: Brefeldin-A Distinguishes Trans-golgi Cisternae from the Trans-golgi Network. J. Cel Biol. 111, 893–899. doi:10.1083/jcb.111.3.893
Chen, Y., Gershlick, D. C., Park, S. Y., and Bonifacino, J. S. (2017). Segregation in the Golgi Complex Precedes export of Endolysosomal Proteins in Distinct Transport Carriers. J. Cel Biol. 216, 4141–4151. doi:10.1083/jcb.201707172
Choi, S.-W., Tamaki, T., Ebine, K., Uemura, T., Ueda, T., and Nakano, A. (2013). RABA Members Act in Distinct Steps of Subcellular Trafficking of the FLAGELLIN SENSING2 Receptor. Plant Cell 25, 1174–1187. doi:10.1105/tpc.112.108803
Chow, C.-M., Neto, H., Foucart, C., and Moore, I. (2008). Rab-A2 and Rab-A3 GTPases Define Atrans-Golgi Endosomal Membrane Domain inArabidopsisThat Contributes Substantially to the Cell Plate. Plant Cell 20, 101–123. doi:10.1105/tpc.107.052001
Cole, N. B., Sciaky, N., Marotta, A., Song, J., and Lippincott-Schwartz, J. (1996). Golgi Dispersal during Microtubule Disruption: Regeneration of Golgi Stacks at Peripheral Endoplasmic Reticulum Exit Sites. MBoC 7, 631–650. doi:10.1091/mbc.7.4.631
Cui, Y., Shen, J., Gao, C., Zhuang, X., Wang, J., and Jiang, L. (2016). Biogenesis of Plant Prevacuolar Multivesicular Bodies. Mol. Plant 9, 774–786. doi:10.1016/j.molp.2016.01.011
Cullen, P. J., and Steinberg, F. (2018). To Degrade or Not to Degrade: Mechanisms and Significance of Endocytic Recycling. Nat. Rev. Mol. Cel Biol 19, 679–696. doi:10.1038/s41580-018-0053-7
D'Angelo, G., Uemura, T., Chuang, C. C., Polishchuk, E., Santoro, M., Ohvo-Rekilä, H., et al. (2013). Vesicular and Non-vesicular Transport Feed Distinct Glycosylation Pathways in the Golgi. Nature 501, 116–120. doi:10.1038/nature12423
Dacks, J. B., and Robinson, M. S. (2017). Outerwear through the Ages: Evolutionary Cell Biology of Vesicle coats. Curr. Opin. Cel Biol. 47, 108–116. doi:10.1016/j.ceb.2017.04.001
daSilva, L. L. P., Snapp, E. L., Denecke, J., Lippincott-Schwartz, J., Hawes, C., and Brandizzi, F. (2004). Endoplasmic Reticulum Export Sites and Golgi Bodies Behave as Single Mobile Secretory Units in Plant Cells[W]. Plant Cell 16, 1753–1771. doi:10.1105/tpc.022673
Day, K. J., Casler, J. C., and Glick, B. S. (2018). Budding Yeast Has a Minimal Endomembrane System. Developmental Cel 44, 56–72. doi:10.1016/j.devcel.2017.12.014
de Duve, C. (1983). Lysosomes Revisited. Eur. J. Biochem. 137, 391–397. doi:10.1111/j.1432-1033.1983.tb07841.x
De Matteis, M. A., and Luini, A. (2008). Exiting the Golgi Complex. Nat. Rev. Mol. Cel Biol. 9, 273–284. doi:10.1038/nrm2378
de Renzis, S., Sönnichsen, B., and Zerial, M. (2002). Divalent Rab Effectors Regulate the Sub-compartmental Organization and Sorting of Early Endosomes. Nat. Cel Biol 4, 124–133. doi:10.1038/ncb744
Dell’Angelica, E. C., and Bonifacino, J. S. (2019). Coatopathies: Genetic Disorders of Protein coats. Annu. Rev. Cel Dev. Biol. 35, 131–168.
Dettmer, J., Hong-Hermesdorf, A., Stierhof, Y.-D., and Schumacher, K. (2006). Vacuolar H+-ATPase Activity Is Required for Endocytic and Secretory Trafficking inArabidopsis. Plant Cell 18, 715–730. doi:10.1105/tpc.105.037978
Donohoe, B. S., Kang, B.-H., Gerl, M. J., Gergely, Z. R., McMichael, C. M., Bednarek, S. Y., et al. (2013). cis-Golgi Cisternal Assembly and Biosynthetic Activation Occur Sequentially in Plants and Algae. Traffic 14, 551–567. doi:10.1111/tra.12052
Donohoe, B. S., Kang, B.-H., and Staehelin, L. A. (2007). Identification and Characterization of COPIa- and COPIb-type Vesicle Classes Associated with Plant and Algal Golgi. Proc. Natl. Acad. Sci. U.S.A. 104, 163–168. doi:10.1073/pnas.0609818104
Driouich, A., Faye, L., and Staehelin, A. (1993). The Plant Golgi Apparatus: a Factory for Complex Polysaccharides and Glycoproteins. Trends Biochem. Sci. 18, 210–214. doi:10.1016/0968-0004(93)90191-o
Dunphy, W., and Rothman, J. (1985). Compartmental Organization of the Golgi Stack. Cell 42, 13–21. doi:10.1016/s0092-8674(85)80097-0
Ebine, K., Fujimoto, M., Okatani, Y., Nishiyama, T., Goh, T., Ito, E., et al. (2011). A Membrane Trafficking Pathway Regulated by the Plant-specific RAB GTPase ARA6. Nat. Cel Biol. 13, 853–859. doi:10.1038/ncb2270
Ebine, K., Inoue, T., Ito, J., Ito, E., Uemura, T., Goh, T., et al. (2014). Plant Vacuolar Trafficking Occurs through Distinctly Regulated Pathways. Curr. Biol. 24, 1375–1382. doi:10.1016/j.cub.2014.05.004
Emr, S., Glick, B. S., Linstedt, A. D., Lippincott-Schwartz, J., Luini, A., Malhotra, V., et al. (2009). Journeys through the Golgi-Taking Stock in a new era. J. Cel Biol. 187, 449–453. doi:10.1083/jcb.200909011
Farquhar, M. G., and Palade, G. E. (1998). The Golgi Apparatus: 100 Years of Progress and Controversy. Trends Cel Biol. 8, 2–10. doi:10.1016/s0962-8924(97)01187-2
Feraru, E., Feraru, M. I., Asaoka, R., Paciorek, T., De Rycke, R., Tanaka, H., et al. (2012). BEX5/RabA1b Regulates Trans-golgi Network-To-Plasma Membrane Protein Trafficking in Arabidopsis. Plant Cell 24, 3074–3086. doi:10.1105/tpc.112.098152
Ferraro, F., Kriston-Vizi, J., Metcalf, D. J., Martin-Martin, B., Freeman, J., Burden, J. J., et al. (2014). A Two-Tier Golgi-Based Control of Organelle Size Underpins the Functional Plasticity of Endothelial Cells. Developmental Cel 29, 292–304. doi:10.1016/j.devcel.2014.03.021
Franke, C., Repnik, U., Segeletz, S., Brouilly, N., Kalaidzidis, Y., Verbavatz, J. M., et al. (2019). Correlative Single‐molecule Localization Microscopy and Electron Tomography Reveals Endosome Nanoscale Domains. Traffic 20, 601–617. doi:10.1111/tra.12671
Fujii, S., Kurokawa, K., Inaba, R., Hiramatsu, N., Tago, T., Nakamura, Y., et al. (2020a). Recycling Endosomes Attach to the Trans-side of Golgi Stacks in Drosophila and Mammalian Cells. J. Cel Sci. 133, jcs236935. doi:10.1242/jcs.236935
Fujii, S., Kurokawa, K., Tago, T., Inaba, R., Takiguchi, A., Nakano, A., et al. (2020b). Sec71 Separates Golgi Stacks in Drosophila S2 Cells. J. Cel Sci. 133, jcs245571. doi:10.1242/jcs.245571
Füllekrug, J., Sönnichsen, B., Schäfer, U., Van, P. N., Söling, H.-D., and Mieskes, G. (1997). Characterization of Brefeldin A Induced Vesicular Structures Containing Cycling Proteins of the Intermediate Compartment/cis-Golgi Network. FEBS Lett. 404, 75–81.
Geldner, N., Anders, N., Wolters, H., Keicher, J., Kornberger, W., Muller, P., et al. (2003). The Arabidopsis GNOM ARF-GEF Mediates Endosomal Recycling, Auxin Transport, and Auxin-dependent Plant Growth. Cell 112, 219–230. doi:10.1016/s0092-8674(03)00003-5
Geuze, H. J., and Morré, D. J. (1991). Trans-Golgi Reticulum. J. Elec. Microsc. Tech. 17, 24–34. doi:10.1002/jemt.1060170105
Gillon, A. D., Latham, C. F., and Miller, E. A. (2012). Vesicle-mediated ER export of Proteins and Lipids. Biochim. Biophys. Acta (Bba) - Mol. Cel Biol. Lipids 1821, 1040–1049. doi:10.1016/j.bbalip.2012.01.005
Glick, B. S. (1996). Cell Biology: Alternatives to baker's Yeast. Curr. Biol. 6, 1570–1572. doi:10.1016/s0960-9822(02)70774-4
Glick, B. S., and Luini, A. (2011). Models for Golgi Traffic: a Critical Assessment. Cold Spring Harbor Perspect. Biol. 3, a005215. doi:10.1101/cshperspect.a005215
Glick, B. S., and Malhotra, V. (1998). The Curious Status of the Golgi Apparatus. Cell 95, 883–889. doi:10.1016/s0092-8674(00)81713-4
Glick, B. S., and Nakano, A. (2009). Membrane Traffic within the Golgi Apparatus. Annu. Rev. Cel Dev. Biol. 25, 113–132. doi:10.1146/annurev.cellbio.24.110707.175421
Goh, T., Uchida, W., Arakawa, S., Ito, E., Dainobu, T., Ebine, K., et al. (2007). VPS9a, the Common Activator for Two Distinct Types of Rab5 GTPases, Is Essential for the Development of Arabidopsis thaliana. Plant Cell 19, 3504–3515. doi:10.1105/tpc.107.053876
Gonzalez, A., and Rodriguez-Boulan, E. (2009). Clathrin and AP1B: Key Roles in Basolateral Trafficking through Trans-endosomal Routes. FEBS Lett. 583, 3784–3795. doi:10.1016/j.febslet.2009.10.050
Griffiths, G., Fuller, S. D., Back, R., Hollinshead, M., Pfeiffer, S., and Simons, K. (1989). The Dynamic Nature of the Golgi Complex. J. Cel Biol. 108, 277–297. doi:10.1083/jcb.108.2.277
Griffiths, G., and Simons, K. (1986). The Trans Golgi Network: Sorting at the Exit Site of the Golgi Complex. Science 234, 438–443. doi:10.1126/science.2945253
Gustafson, M. A., and Fromme, J. C. (2017). Regulation of Arf Activation Occurs via Distinct Mechanisms at Early and Late Golgi Compartments. MBoC 28, 3660–3671. doi:10.1091/mbc.e17-06-0370
Haas, T. J., SliwinskiSliwinski, M. K. M. K., Martínez, D. E., Preuss, M., Ebine, K., Ueda, T., et al. (2007). TheArabidopsisAAA ATPase SKD1 Is Involved in Multivesicular Endosome Function and Interacts with its Positive Regulator LYST-INTERACTING PROTEIN5. Plant Cell 19, 1295–1312. doi:10.1105/tpc.106.049346
Hanada, K., Kumagai, K., Tomishige, N., and Kawano, M. (2007). CERT and Intracellular Trafficking of Ceramide. Biochim. Biophys. Acta (Bba) - Mol. Cel Biol. Lipids 1771, 644–653. doi:10.1016/j.bbalip.2007.01.009
Hauri, H. P., Kappeler, F., Andersson, H., and Appenzeller, C. (2000). ERGIC-53 and Traffic in the Secretory Pathway. J. Cel Sci. 113, 587–596. doi:10.1242/jcs.113.4.587
Henne, W. M., Buchkovich, N. J., and Emr, S. D. (2011). The ESCRT Pathway. Developmental Cel 21, 77–91. doi:10.1016/j.devcel.2011.05.015
Heucken, N., and Ivanov, R. (2018). The Retromer, Sorting Nexins and the Plant Endomembrane Protein Trafficking. J. Cel Sci. 131, jcs203695. doi:10.1242/jcs.203695
Hinners, I., and Tooze, S. A. (2003). Changing Directions: Clathrin-Mediated Transport between the Golgi and Endosomes. J. Cel Sci. 116, 763–771. doi:10.1242/jcs.00270
Hoffmann, N., King, S., Samuels, A. L., and McFarlane, H. E. (2021). Subcellular Coordination of Plant Cell wall Synthesis. Developmental Cel 56, 933–948. doi:10.1016/j.devcel.2021.03.004
Hughes, H., and Stephens, D. J. (2008). Assembly, Organization, and Function of the COPII Coat. Histochem. Cel Biol. 129, 129–151. doi:10.1007/s00418-007-0363-x
Ishida, M., and Bonifacino, J. S. (2019). ARFRP1 Functions Upstream of ARL1 and ARL5 to Coordinate Recruitment of Distinct Tethering Factors to the Trans-golgi Network. J. Cel Biol. 218, 3681–3696. doi:10.1083/jcb.201905097
Ishii, M., Suda, Y., Kurokawa, K., and Nakano, A. (2016). COPI Is Essential for Golgi Cisternal Maturation and Dynamics. J. Cel Sci. 129, 3251–3261. doi:10.1242/jcs.193367
Ito, E., Ebine, K., Choi, S. W., Ichinose, S., Uemura, T., Nakano, A., et al. (2018a). Integration of Two RAB5 Groups during Endosomal Transport in Plants. eLife 7, e34064. doi:10.7554/eLife.34064
Ito, Y., Uemura, T., and Nakano, A. (2018b). The Golgi Entry Core Compartment Functions as a COPII-independent Scaffold for ER-To-Golgi Transport in Plant Cells. J. Cel Sci 131, jcs203893. doi:10.1242/jcs.203893
Ito, Y., and Boutté, Y. (2020). Differentiation of Trafficking Pathways at Golgi Entry Core Compartments and post-Golgi Subdomains. Front. Plant Sci. 11, 609516. doi:10.3389/fpls.2020.609516
Ito, Y., Toyooka, K., Fujimoto, M., Ueda, T., Uemura, T., and Nakano, A. (2017). The Trans-golgi Network and the Golgi Stacks Behave Independently during Regeneration after Brefeldin A Treatment in Tobacco BY-2 Cells. Plant Cel Physiol 58, 811–821. doi:10.1093/pcp/pcx028
Ito, Y., Uemura, T., and Nakano, A. (2014). Formation and Maintenance of the Golgi Apparatus in Plant Cells. Intl. Rev. Cel Mol. Biol. 310, 221–287. doi:10.1016/b978-0-12-800180-6.00006-2
Ito, Y., Uemura, T., Shoda, K., Fujimoto, M., Ueda, T., and Nakano, A. (2012). cis-Golgi Proteins Accumulate Near the ER Exit Sites and Act as the Scaffold for Golgi Regeneration after Brefeldin A Treatment in Tobacco BY-2 Cells. MBoC 23, 3203–3214. doi:10.1091/mbc.e12-01-0034
Iwasaki, H., Yorimitsu, T., and Sato, K. (2017). Microscopy Analysis of Reconstituted COPII Coat Polymerization and Sec16 Dynamics. J. Cel Sci. 130, 2893–2902. doi:10.1242/jcs.203844
Jackson, C. L., and Casanova, J. E. (2000). Turning on ARF: the Sec7 Family of Guanine-Nucleotide-Exchange Factors. Trends Cel Biol. 10, 60–67. doi:10.1016/s0962-8924(99)01699-2
Jackson, C. L. (2009). Mechanisms of Transport through the Golgi Complex. J. Cel Sci. 122, 443–452. doi:10.1242/jcs.032581
Jamieson, J. D., and Palade, G. E. (1966). Role of the Golgi Complex in the Intracellular Transport of Secretory Proteins. Proc. Natl. Acad. Sci. U.S.A. 55, 424–431. doi:10.1073/pnas.55.2.424
Jedd, G., Mulholland, J., and Segev, N. (1997). Two New Ypt GTPases Are Required for Exit from the Yeast Trans-golgi Compartment. J. Cel Biol. 137, 563–580. doi:10.1083/jcb.137.3.563
Keller, P., and Simons, K. (1997). Post-golgi Biosynthetic Trafficking. J. Cel Sci. 110, 3001–3009. doi:10.1242/jcs.110.24.3001
Klausner, R. D., Donaldson, J. G., and Lippincott-Schwartz, J. (1992). Brefeldin A: Insights into the Control of Membrane Traffic and Organelle Structure. J. Cel Biol. 116, 1071–1080. doi:10.1083/jcb.116.5.1071
Klumperman, J., Schweizer, A., Clausen, H., Tang, B. L., Hong, W., Oorschot, V., et al. (1998). The Recycling Pathway of Protein ERGIC-53 and Dynamics of the ER-Golgi Intermediate Compartment. J. Cel Sci. 111, 3411–3425. doi:10.1242/jcs.111.22.3411
Kurokawa, K., and Nakano, A. (2020). Live-cell Imaging by Super-resolution Confocal Live Imaging Microscopy (SCLIM): Simultaneous Three-Color and Four-Dimensional Live Cell Imaging with High Space and Time Resolution. Bio Protoc. 10, e3732. doi:10.21769/BioProtoc.3732
Kurokawa, K., Suda, Y., and Nakano, A. (2016). Sar1 Localizes at the Rims of COPII-Coated Membranes In Vivo. J. Cel SciJ. Cel Sci. 129, 3231–3237. doi:10.1242/jcs.189423
Kurokawa, K., Ishii, M., Suda, Y., Ichihara, A., and Nakano, A. (2013). Live Cell Visualization of Golgi Membrane Dynamics by Super-resolution Confocal Live Imaging Microscopy. Methods Cel Biol 118, 235–242. doi:10.1016/b978-0-12-417164-0.00014-8
Kurokawa, K., and Nakano, A. (2019). The ER Exit Sites Are Specialized ER Zones for the Transport of Cargo Proteins from the ER to the Golgi Apparatus. J. Biochem. 165, 109–114. doi:10.1093/jb/mvy080
Kurokawa, K., Okamoto, M., and Nakano, A. (2014). Contact of Cis-Golgi with ER Exit Sites Executes Cargo Capture and Delivery from the ER. Nat. Commun. 5, 3653. doi:10.1038/ncomms4653
Kurokawa, K., Osakada, H., Kojidani, T., Waga, M., Suda, Y., Asakawa, H., et al. (2019). Visualization of Secretory Cargo Transport within the Golgi Apparatus. J. Cel Biol. 218, 1602–1618. doi:10.1083/jcb.201807194
Ladinsky, M. S., Mastronarde, D. N., McIntosh, J. R., Howell, K. E., and Staehelin, L. A. (1999). Golgi Structure in Three Dimensions: Functional Insights from the normal Rat Kidney Cell. J. Cel Biol. 144, 1135–1149. doi:10.1083/jcb.144.6.1135
Lam, S. K., Siu, C. L., Hillmer, S., Jang, S., An, G., Robinson, D. G., et al. (2007). Rice SCAMP1 Defines Clathrin-Coated,trans-Golgi-Located Tubular-Vesicular Structures as an Early Endosome in Tobacco BY-2 Cells. Plant Cell 19, 296–319. doi:10.1105/tpc.106.045708
Langhans, M., Förster, S., Helmchen, G., and Robinson, D. G. (2011). Differential Effects of the Brefeldin A Analogue (6R)-Hydroxy-BFA in Tobacco and Arabidopsis. J. Exp. Bot. 62, 2949–2957. doi:10.1093/jxb/err007
Langhans, M., Hawes, C., Hillmer, S., Hummel, E., and Robinson, D. G. (2007). Golgi Regeneration after Brefeldin A Treatment in BY-2 Cells Entails Stack Enlargement and Cisternal Growth Followed by Division. Plant Physiol. 145, 527–538. doi:10.1104/pp.107.104919
Letourneur, F., Gaynor, E. C., Hennecke, S., Démollière, C., Duden, R., Emr, S. D., et al. (1994). Coatomer Is Essential for Retrieval of Dilysine-Tagged Proteins to the Endoplasmic Reticulum. Cell 79, 1199–1207. doi:10.1016/0092-8674(94)90011-6
Levine, T. P., and Munro, S. (2002). Targeting of Golgi-specific Pleckstrin Homology Domains Involves Both PtdIns 4-kinase-dependent and -independent Components. Curr. Biol. 12, 695–704. doi:10.1016/s0960-9822(02)00779-0
Lippincott-Schwartz, J., Yuan, L. C., Bonifacino, J. S., and Klausner, R. D. (1989). Rapid Redistribution of Golgi Proteins into the ER in Cells Treated with Brefeldin A: Evidence for Membrane Cycling from Golgi to ER. Cell 56, 801–813. doi:10.1016/0092-8674(89)90685-5
Lippincott-Schwartz, J., Donaldson, J. G., Schweizer, A., Berger, E. G., Hauri, H.-P., Yuan, L. C., et al. (1990). Microtubule-dependent Retrograde Transport of Proteins into the ER in the Presence of Brefeldin A Suggests an ER Recycling Pathway. Cell 60, 821–836. doi:10.1016/0092-8674(90)90096-w
Lippincott-Schwartz, J., Yuan, L., Tipper, C., Amherdt, M., Orci, L., and Klausner, R. D. (1991). Brefeldin A's Effects on Endosomes, Lysosomes, and the TGN Suggest a General Mechanism for Regulating Organelle Structure and Membrane Traffic. Cell 67, 601–616. doi:10.1016/0092-8674(91)90534-6
Liu, S., and Storrie, B. (2012). Are Rab Proteins the Link between Golgi Organization and Membrane Trafficking? Cell. Mol. Life Sci. 69, 4093–4106. doi:10.1007/s00018-012-1021-6
Liu, S., and Storrie, B. (2015). How Rab Proteins Determine Golgi Structure. Int. Rev. Cel Mol. Biol. 315, 1–22. doi:10.1016/bs.ircmb.2014.12.002
Losev, E., Reinke, C. A., Jellen, J., Strongin, D. E., Bevis, B. J., and Glick, B. S. (2006). Golgi Maturation Visualized in Living Yeast. Nature 441, 1002–1006. doi:10.1038/nature04717
Ma, M., and Burd, C. G. (2020). Retrograde Trafficking and Plasma Membrane Recycling Pathways of the Budding yeastSaccharomyces Cerevisiae. Traffic 21, 45–59. doi:10.1111/tra.12693
Manolea, F., Claude, A., Chun, J., Rosas, J., and Melançon, P. (2008). Distinct Functions for Arf Guanine Nucleotide Exchange Factors at the Golgi Complex: GBF1 and BIGs Are Required for Assembly and Maintenance of the Golgi Stack Andtrans-Golgi Network, Respectively. MBoC 19, 523–535. doi:10.1091/mbc.e07-04-0394
Marie, M., Dale, H. A., Kouprina, N., and Saraste, J. (2012). Division of the Intermediate Compartment at the Onset of Mitosis Provides a Mechanism for Golgi Inheritance. J. Cel Sci. 125, 5403–5416. doi:10.1242/jcs.108100
Marie, M., Dale, H. A., Sannerud, R., and Saraste, J. (2009). The Function of the Intermediate Compartment in Pre-golgi Trafficking Involves its Stable Connection with the Centrosome. MBoC 20, 4458–4470. doi:10.1091/mbc.e08-12-1229
Marra, P., Salvatore, L., Mironov, A., Di Campli, A., Di Tullio, G., Trucco, A., et al. (2007). The Biogenesis of the Golgi Ribbon: the Roles of Membrane Input from the ER and of GM130. Mol. Biol. Cel 18, 1595–1608. doi:10.1091/mbc.e06-10-0886
Martínez-Menárguez, J. A., Geuze, H. J., Slot, J. W., and Klumperman, J. (1999). Vesicular Tubular Clusters between the ER and Golgi Mediate Concentration of Soluble Secretory Proteins by Exclusion from COPI-Coated Vesicles. Cell 98, 81–90. doi:10.1016/S0092-8674(00)80608-X
Matsuura-Tokita, K., Takeuchi, M., Ichihara, A., Mikuriya, K., and Nakano, A. (2006). Live Imaging of Yeast Golgi Cisternal Maturation. Nature 441, 1007–1010. doi:10.1038/nature04737
Mellman, I., and Simons, K. (1992). The Golgi Complex: In Vitro Veritas? Cell 68, 829–840. doi:10.1016/0092-8674(92)90027-a
Miller, E. A., and Barlowe, C. (2010). Regulation of Coat Assembly-Sorting Things Out at the ER. Curr. Opin. Cel Biol. 22, 447–453. doi:10.1016/j.ceb.2010.04.003
Mironov, A. A., Mironov, A. A., Beznoussenko, G. V., Trucco, A., Lupetti, P., Smith, J. D., et al. (2003). ER-to-Golgi carriers arise through direct en bloc protrusion and multistage maturation of specialized ER exit domains. Developmental Cel 5, 583–594. doi:10.1016/s1534-5807(03)00294-6
Nagano, M., Toshima, J. Y., Siekhaus, D. E., and Toshima, J. (2019). Rab5-mediated Endosome Formation Is Regulated at the Trans-golgi Network. Commun. Biol. 2, 419. doi:10.1038/s42003-019-0670-5
Nakamura, N., Rabouille, C., Watson, R., Nilsson, T., Hui, N., Slusarewicz, P., et al. (1995). Characterization of a Cis-Golgi Matrix Protein, GM130. J. Cel Biol. 131, 1715–1726. doi:10.1083/jcb.131.6.1715
Nakano, A., and Luini, A. (2009). Passage through the Golgi. Curr. Opin. Cel Biol. 22, 471–478. doi:10.1016/j.ceb.2010.05.003
Nakano, A., Brada, D., and Schekman, R. (1988). A Membrane Glycoprotein, Sec12p, Required for Protein Transport from the Endoplasmic Reticulum to the Golgi Apparatus in Yeast. J. Cel Biol. 107, 851–863. doi:10.1083/jcb.107.3.851
Nakano, A., and Muramatsu, M. (1989). A Novel GTP-Binding Protein, Sar1p, Is Involved in Transport from the Endoplasmic Reticulum to the Golgi Apparatus. J. Cel Biol. 109, 2677–2691. doi:10.1083/jcb.109.6.2677
Nakano, A. (2002). Spinning-disk Confocal Microscopy. A Cutting-Edge Tool for Imaging of Membrane Traffic. Cell Struct. Funct. 27, 349–355. doi:10.1247/csf.27.349
Nakano, A. (2008). “Yeast Golgi Apparatus,” in The Golgi Apparatus, State of the Art 110 Years after Camillo Golgi’s Discovery. Editors A. Mironov, and M. Pavelka (Wien: Springer-Verlag), 623–629.
Naramoto, S., Otegui, M. S., Kutsuna, N., de Rycke, R., Dainobu, T., Karampelias, M., et al. (2014). Insights into the Localization and Function of the Membrane Trafficking Regulator GNOM ARF-GEF at the Golgi Apparatus in Arabidopsis. Plant Cell 26, 3062–3076. doi:10.1105/tpc.114.125880
Nielsen, E., Cheung, A. Y., and Ueda, T. (2008). The Regulatory RAB and ARF GTPases for Vesicular Trafficking. Plant Physiol. 147, 1516–1526. doi:10.1104/pp.108.121798
Nielsen, M. E., Feechan, A., Böhlenius, H., Ueda, T., and Thordal-Christensen, H. (2012). Arabidopsis ARF-GTP Exchange Factor, GNOM, Mediates Transport Required for Innate Immunity and Focal Accumulation of Syntaxin PEN1. Proc. Natl. Acad. Sci. U.S.A. 109, 11443–11448. doi:10.1073/pnas.1117596109
Nilsson, T., Jackson, M., and Peterson, P. A. (1989). Short Cytoplasmic Sequences Serve as Retention Signals for Transmembrane Proteins in the Endoplasmic Reticulum. Cell 58, 707–718. doi:10.1016/0092-8674(89)90105-0
Okamoto, M., Kurokawa, K., Matsuura-Tokita, K., Saito, C., Hirata, R., and Nakano, A. (2012). High-curvature Domains of the ER Are Important for the Organization of ER Exit Sites in Saccharomyces cerevisiae. J. Cel Sci. 125, 3412–3420. doi:10.1242/jcs.100065
Palade, G. (1975). Intracellular Aspects of the Process of Protein Synthesis. Science 189, 347–358. doi:10.1126/science.1096303
Palmer, K. J., and Stephens, D. J. (2004). Biogenesis of ER-To-Golgi Transport Carriers: Complex Roles of COPII in ER export. Trends Cel Biol. 14, 57–61. doi:10.1016/j.tcb.2003.12.001
Papanikou, E., and Glick, B. S. (2009). The Yeast Golgi Apparatus: Insights and Mysteries. FEBS Lett. 583, 3746–3751. doi:10.1016/j.febslet.2009.10.072
Pelham, H., and Munro, S. (1993). Sorting of Membrane Proteins in the Secretory Pathway. Cell 75, 603–605. doi:10.1016/0092-8674(93)90479-a
Pelham, H. R. B., and Rothman, J. E. (2000). The Debate about Transport in the Golgi-Two Sides of the Same Coin? Cell 102, 713–719. doi:10.1016/s0092-8674(00)00060-x
Plutner, H., Davidson, H. W., Saraste, J., and Balch, W. E. (1992). Morphological Analysis of Protein Transport from the ER to Golgi Membranes in Digitonin-Permeabilized Cells: Role of the P58 Containing Compartment. J. Cel Biol. 119, 1097–1116. doi:10.1083/jcb.119.5.1097
Poteryaev, D., Datta, S., Ackema, K., Zerial, M., and Spang, A. (2010). Identification of the Switch in Early-To-Late Endosome Transition. Cell 141, 497–508. doi:10.1016/j.cell.2010.03.011
Preuss, D., Mulholland, J., Franzusoff, A., Segev, N., and Botstein, D. (1992). Characterization of the Saccharomyces Golgi Complex through the Cell Cycle by Immunoelectron Microscopy. MBoC 3, 789–803. doi:10.1091/mbc.3.7.789
Pucadyil, T. J., and Schmid, S. L. (2009). Conserved Functions of Membrane Active GTPases in Coated Vesicle Formation. Science 325, 1217–1220. doi:10.1126/science.1171004
Rabouille, C., and Klumperman, J. (2005). The Maturing Role of COPI Vesicles in Intra-golgi Transport. Nat. Rev. Mol. Cel Biol. 6, 812–817. doi:10.1038/nrm1735
Raiborg, C., and Stenmark, H. (2009). The ESCRT Machinery in Endosomal Sorting of Ubiquitylated Membrane Proteins. Nature 458, 445–452. doi:10.1038/nature07961
Rambourg, A., Clermont, Y., Ovtracht, L., and Képès, F. (1995). Three-dimensional Structure of Tubular Networks, Presumably Golgi in Nature, in Various Yeast Strains: a Comparative Study. Anat. Rec. 243, 283–293. doi:10.1002/ar.1092430302
Rambourg, A., Jackson, C. L., and Clermont, Y. (2001). Three Dimensional Configuration of the Secretory Pathway and Segregation of Secretion Granules in the Yeast Saccharomyces cerevisiae. J. Cel Sci. 114, 2231–2239. doi:10.1242/jcs.114.12.2231
Raote, I., and Malhotra, V. (2019). Protein Transport by Vesicles and Tunnels. J. Cel Biol. 218, 737–739. doi:10.1083/jcb.201811073
Raote, I., and Malhotra, V. (2021). Tunnels for Protein export from the Endoplasmic Reticulum. Annu. Rev. Biochem. 90, 605–630. doi:10.1146/annurev-biochem-080120-022017
Raote, I., Saxena, S., Campelo, F., and Malhotra, V. (2021). TANGO1 Marshals the Early Secretory Pathway for Cargo export. Biochim. Biophys. Acta (Bba) - Biomembranes 1863, 183700. doi:10.1016/j.bbamem.2021.183700
Richter, S., Geldner, N., Schrader, J., Wolters, H., Stierhof, Y.-D., Rios, G., et al. (2007). Functional Diversification of Closely Related ARF-GEFs in Protein Secretion and Recycling. Nature 448, 488–492. doi:10.1038/nature05967
Rink, J., Ghigo, E., Kalaidzidis, Y., and Zerial, M. (2005). Rab Conversion as a Mechanism of Progression from Early to Late Endosomes. Cell 122, 735–749. doi:10.1016/j.cell.2005.06.043
Rios, R. M., and Bornens, M. (2003). The Golgi Apparatus at the Cell centre. Curr. Opin. Cel Biol. 15, 60–66. doi:10.1016/s0955-0674(02)00013-3
Ritzenthaler, C., Nebenführ, A., Movafeghi, A., Stussi-Garaud, C., Behnia, L., Pimpl, P., et al. (2002). Reevaluation of the Effects of Brefeldin A on Plant Cells Using Tobacco Bright Yellow 2 Cells Expressing Golgi-Targeted green Fluorescent Protein and COPI Antisera. Plant Cell 14, 237–261. doi:10.1105/tpc.010237
Robinson, D. G., Brandizzi, F., Hawes, C., and Nakano, A. (2015). Vesicles versus Tubes: Is Endoplasmic Reticulum-Golgi Transport in Plants Fundamentally Different from Other Eukaryotes? Plant Physiol. 168, 393–406. doi:10.1104/pp.15.00124
Robinson, D. G., Langhans, M., Saint-Jore-Dupas, C., and Hawes, C. (2008). BFA Effects Are Tissue and Not Just Plant Specific. Trends Plant Sci. 13, 405–408. doi:10.1016/j.tplants.2008.05.010
Rodriguez-Boulan, E., and Müsch, A. (2005). Protein Sorting in the Golgi Complex: Shifting Paradigms. Biochim. Biophys. Acta (Bba) - Mol. Cel Res. 1744, 455–464. doi:10.1016/j.bbamcr.2005.04.007
Rossanese, O. W., Soderholm, J., Bevis, B. J., Sears, I. B., O'Connor, J., Williamson, E. K., et al. (1999). Golgi Structure Correlates with Transitional Endoplasmic Reticulum Organization in Pichia pastoris and Saccharomyces cerevisiae. J. Cel Biol. 145, 69–81. doi:10.1083/jcb.145.1.69
Roth, J., Taatjes, D. J., Lucocq, J. M., Weinstein, J., and Paulson, J. C. (1985). Demonstration of an Extensive Trans-tubular Network Continuous with the Golgi Apparatus Stack that May Function in Glycosylation. Cell 43, 287–295. doi:10.1016/0092-8674(85)90034-0
Rothblatt, J., and Schekman, R. (1989). Chapter 1 A Hitchhiker's Guide to Analysis of the Secretory Pathway in Yeast. Methods Cel Biol 32, 3–36. doi:10.1016/s0091-679x(08)61165-6
Russell, M. R., Shideler, T., Nickerson, D. P., West, M., and Odorizzi, G. (2012). Class E Compartments Form in Response to ESCRT Dysfunction in Yeast Due to Hyperactivity of the Vps21 Rab GTPase. J. Cel Sci. 125, 5208–5220. doi:10.1242/jcs.111310
Rutherford, S., and Moore, I. (2002). The Arabidopsis Rab GTPase Family: Another enigma Variation. Curr. Opin. Plant Biol. 5, 518–528. doi:10.1016/s1369-5266(02)00307-2
Saito, C., and Ueda, T. (2009). Chapter 4 Functions of RAB and SNARE Proteins in Plant Life. Int. Rev. Cel Mol. Biol. 274, 183–233. doi:10.1016/s1937-6448(08)02004-2
Sakurai, H. T., Inoue, T., Nakano, A., and Ueda, T. (2016). ENDOSOMAL RAB EFFECTOR WITH PX-DOMAIN, an Interacting Partner of RAB5 GTPases, Regulates Membrane Trafficking to Protein Storage Vacuoles in Arabidopsis. Plant Cell 28, 1490–1503. doi:10.1105/tpc.16.00326
Sandvig, K., and van Deurs, B. (1996). Endocytosis, Intracellular Transport, and Cytotoxic Action of Shiga Toxin and Ricin. Physiol. Rev. 76, 949–966. doi:10.1152/physrev.1996.76.4.949
Sannerud, R., Marie, M., Nizak, C., Dale, H. A., Pernet-Gallay, K., Perez, F., et al. (2006). Rab1 Defines a Novel Pathway Connecting the Pre-golgi Intermediate Compartment with the Cell Periphery. MBoC 17, 1514–1526. doi:10.1091/mbc.e05-08-0792
Sannerud, R., Saraste, J., and Goud, B. (2003). Retrograde Traffic in the Biosynthetic-Secretory Route: Pathways and Machinery. Curr. Opin. Cel Biol. 15, 438–445. doi:10.1016/s0955-0674(03)00077-2
Saraste, J., and Goud, B. (2007). Functional Symmetry of Endomembranes. MBoC 18, 1430–1436. doi:10.1091/mbc.e06-10-0933
Saraste, J., and Marie, M. (2018). Intermediate Compartment (IC): from Pre-golgi Vacuoles to a Semi-autonomous Membrane System. Histochem. Cel Biol 150, 407–430. doi:10.1007/s00418-018-1717-2
Saraste, J., and Prydz, K. (2019). A New Look at the Functional Organization of the Golgi Ribbon. Front. Cel Dev. Biol. 7, 171. doi:10.3389/fcell.2019.00171
Saraste, J., and Svensson, K. (1991). Distribution of the Intermediate Elements Operating in ER to Golgi Transport. J. Cel Sci. 100, 415–430. doi:10.1242/jcs.100.3.415
Sato, K., and Nakano, A. (2005). Dissection of COPII Subunit-Cargo Assembly and Disassembly Kinetics during Sar1p-GTP Hydrolysis. Nat. Struct. Mol. Biol. 12, 167–174. doi:10.1038/nsmb893
Sato, K., and Nakano, A. (2007). Mechanisms of COPII Vesicle Formation and Protein Sorting. FEBS Lett. 581, 2076–2082. doi:10.1016/j.febslet.2007.01.091
Sato, K., Nishikawa, S., and Nakano, A. (1995). Membrane Protein Retrieval from the Golgi Apparatus to the Endoplasmic Reticulum (ER): Characterization of the RER1 Gene Product as a Component Involved in ER Localization of Sec12p. MBoC 6, 1459–1477. doi:10.1091/mbc.6.11.1459
Sato, K., Sato, M., and Nakano, A. (1997). Rer1p as Common Machinery for the Endoplasmic Reticulum Localization of Membrane Proteins. Proc. Natl. Acad. Sci. U.S.A. 94, 9693–9698. doi:10.1073/pnas.94.18.9693
Sato, M., Sato, K., and Nakano, A. (1996). Endoplasmic Reticulum Localization of Sec12p Is Achieved by Two Mechanisms: Rer1p-dependent Retrieval that Requires the Transmembrane Domain and Rer1p-independent Retention that Involves the Cytoplasmic Domain. J. Cel Biol. 134, 279–293. doi:10.1083/jcb.134.2.279
Satoh, A. K., O'Tousa, J. E., Ozaki, K., and Ready, D. F. (2005). Rab11 Mediates post-Golgi Trafficking of Rhodopsin to the Photosensitive Apical Membrane ofDrosophilaphotoreceptors. Development 132, 1487–1497. doi:10.1242/dev.01704
Schekman, R. (1992). Genetic and Biochemical Analysis of Vesicular Traffic in Yeast. Curr. Opin. Cel Biol. 4, 587–592. doi:10.1016/0955-0674(92)90076-o
Schindler, C., Chen, Y., Pu, J., Guo, X., and Bonifacino, J. S. (2015). EARP Is a Multisubunit Tethering Complex Involved in Endocytic Recycling. Nat. Cel Biol. 17, 639–650. doi:10.1038/ncb3129
Schweizer, A., Fransen, J. A., Matter, K., Kreis, T. E., Ginsel, L., and Hauri, H. P. (1990). Identification of an Intermediate Compartment Involved in Protein Transport from Endoplasmic Reticulum to Golgi Apparatus. Eur. J. Cel Biol. 53, 185–196.
Scott, C. C., Vacca, F., and Gruenberg, J. (2014). Endosome Maturation, Transport and Functions. Semin. Cel Developmental Biol. 31, 2–10. doi:10.1016/j.semcdb.2014.03.034
Seaman, M. N., McCaffery, J. M., and Emr, S. D. (1998). A Membrane Coat Complex Essential for Endosome-To-Golgi Retrograde Transport in Yeast. J. Cel Biol. 142, 665–681. doi:10.1083/jcb.142.3.665
Segev, N., Mulholland, J., and Botstein, D. (1988). The Yeast GTP-Binding YPT1 Protein and a Mammalian Counterpart Are Associated with the Secretion Machinery. Cell 52, 915–924. doi:10.1016/0092-8674(88)90433-3
Segev, N. (2001). Ypt and Rab GTPases: Insight into Functions through Novel Interactions. Curr. Opin. Cel Biol. 13, 500–511. doi:10.1016/s0955-0674(00)00242-8
Shimizu, Y., Takagi, J., Ito, E., Ito, Y., Ebine, K., Komatsu, Y., et al. (2021). Cargo Sorting Zones in the Trans-golgi Network Visualized by Super-resolution Confocal Live Imaging Microscopy in Plants. Nat. Commun. 12, 1901. doi:10.1038/s41467-021-22267-0
Shomron, O., Nevo-Yassaf, I., Aviad, T., Yaffe, Y., Zahavi, E. E., Dukhovny, A., et al. (2021). COPII Collar Defines the Boundary between ER and ER Exit Site and Does Not Coat Cargo Containers. J. Cel Biol. 220, e201907224. doi:10.1083/jcb.201907224
Sönnichsen, B., De Renzis, S., Nielsen, E., Rietdorf, J., and Zerial, M. (2000). Distinct Membrane Domains on Endosomes in the Recycling Pathway Visualized by Multicolor Imaging of Rab4, Rab5, and Rab11. J. Cel Biol. 149, 901–914. doi:10.1083/jcb.149.4.901
Sparkes, I. A., Ketelaar, T., De Ruijter, N. C. A., and Hawes, C. (2009). Grab a Golgi: Laser Trapping of Golgi Bodies Revealsin vivoInteractions with the Endoplasmic Reticulum. Traffic 10, 567–571. doi:10.1111/j.1600-0854.2009.00891.x
Sriram, V., Krishnan, K. S., and Mayor, S. (2006). Biogenesis and Function of Late Endosomes and Lysosomes. Proc. Indian Natl. Sci. Acad. 1, 19–42.
Staehelin, L. A., and Kang, B.-H. (2008). Nanoscale Architecture of Endoplasmic Reticulum Export Sites and of Golgi Membranes as Determined by Electron Tomography. Plant Physiol. 147, 1454–1468. doi:10.1104/pp.108.120618
Stanley, P. (2011). Golgi Glycosylation. Cold Spring Harbor Perspect. Biol. 3, a005199. doi:10.1101/cshperspect.a005199
Suda, Y., Kurokawa, K., Hirata, R., and Nakano, A. (2013). Rab GAP cascade Regulates Dynamics of Ypt6 in the Golgi Traffic. Proc. Natl. Acad. Sci. U.S.A. 110, 18976–18981. doi:10.1073/pnas.1308627110
Suda, Y., Kurokawa, K., and Nakano, A. (2018). Regulation of ER-Golgi Transport Dynamics by GTPases in Budding Yeast. Front. Cel Dev. Biol. 5, 122. doi:10.3389/fcell.2017.00122
Suda, Y., and Nakano, A. (2012). The Yeast Golgi Apparatus. Traffic 13, 505–510. doi:10.1111/j.1600-0854.2011.01316.x
Taatjes, D. J., and Roth, J. (1986). The Trans-tubular Network of the Hepatocyte Golgi Apparatus Is Part of the Secretory Pathway. Eur. J. Cel Biol. 42, 344–350.
Taguchi, T. (2013). Emerging Roles of Recycling Endosomes. J. Biochem. 153, 505–510. doi:10.1093/jb/mvt034
Takagi, J., Kimori, Y., Shimada, T., and Hara-Nishimura, I. (2020). Dynamic Capture and Release of Endoplasmic Reticulum Exit Sites by Golgi Stacks in Arabidopsis. iScience 23, 101265. doi:10.1016/j.isci.2020.101265
Takatsuki, A., and Tamura, G. (1985). Brefeldin A, a Specific Inhibitor of Intracellular Translocation of Vesicular Stomatitis Virus G Protein: Intracellular Accumulation of High-Mannose Type G Protein and Inhibition of its Cell Surface Expression. Agric. Biol. Chem. 49, 899–902. doi:10.1080/00021369.1985.10866826
Takemoto, K., Ebine, K., Askani, J. C., Krüger, F., Gonzalez, Z. A., Ito, E., et al. (2018). Distinct Sets of Tethering Complexes, SNARE Complexes, and Rab GTPases Mediate Membrane Fusion at the Vacuole in Arabidopsis. Proc. Natl. Acad. Sci. U S A. 115, E2457–E2466. doi:10.1073/pnas.1717839115
Tang, B. L., Wong, S. H., Qi, X. L., Low, S. H., and Hong, W. (1993). Molecular Cloning, Characterization, Subcellular Localization and Dynamics of P23, the Mammalian KDEL Receptor. J. Cel Biol. 120, 325–338. doi:10.1083/jcb.120.2.325
Teh, O.-K., and Moore, I. (2007). An ARF-GEF Acting at the Golgi and in Selective Endocytosis in Polarized Plant Cells. Nature 448, 493–496. doi:10.1038/nature06023
Terasaki, M. (2000). Dynamics of the Endoplasmic Reticulum and Golgi Apparatus during Early Sea Urchin Development. MBoC 11, 897–914. doi:10.1091/mbc.11.3.897
Thomas, L. L., Highland, C. M., and Fromme, J. C. (2021). Arf1 Orchestrates Rab GTPase Conversion at the Trans-golgi Network. Mol. Biol. Cel 32, 1104–1120. doi:10.1091/mbc.E20-10-0664
Thyberg, J., and Moskalewski, S. (1999). Role of Microtubules in the Organization of the Golgi Complex. Exp. Cel Res. 246, 263–279. doi:10.1006/excr.1998.4326
Tie, H. C., Ludwig, A., Sandin, S., and Lu, L. (2018). The Spatial Separation of Processing and Transport Functions to the interior and Periphery of the Golgi Stack. eLife 7, e41301. doi:10.7554/eLife.41301
Tojima, T., Suda, Y., Ishii, M., Kurokawa, K., and Nakano, A. (2019). Spatiotemporal Dissection of the Trans-golgi Network in Budding Yeast. J. Cel Sci 132, jcs231159. doi:10.1242/jcs.231159
Tojima, T., Miyashiro, D., Kosugi, Y., and Nakano, A. (2022). Super-resolution Live Imaging of Cargo Traffic through the Golgi Apparatus in Mammalian Cells. Methods Mol. Biol. in press.
Ueda, T., Yamaguchi, M., Uchimiya, H., and Nakano, A. (2001). Ara6, a Plant-Unique Novel Type Rab GTPase, Functions in the Endocytic Pathway of Arabidopsis thaliana. EMBO J. 20, 4730–4741. doi:10.1093/emboj/20.17.4730
Uemura, T., Kim, H., Saito, C., Ebine, K., Ueda, T., Schulze-Lefert, P., et al. (2012). Qa-SNAREs Localized to the Trans -Golgi Network Regulate Multiple Transport Pathways and Extracellular Disease Resistance in Plants. Proc. Natl. Acad. Sci. U.S.A. 109, 1784–1789. doi:10.1073/pnas.1115146109
Uemura, T., and Nakano, A. (2013). Plant TGNs: Dynamics and Physiological Functions. Histochem. Cel Biol. 140, 341–345. doi:10.1007/s00418-013-1116-7
Uemura, T., Suda, Y., Ueda, T., and Nakano, A. (2014). Dynamic Behavior of the Trans-golgi Network in Root Tissues of Arabidopsis Revealed by Super-resolution Live Imaging. Plant Cel Physiol 55, 694–703. doi:10.1093/pcp/pcu010
Uemura, T., Ueda, T., Ohniwa, R. L., Nakano, A., Takeyasu, K., and Sato, M. H. (2004). Systematic Analysis of SNARE Molecules in Arabidopsis: Dissection of the post-Golgi Network in Plant Cells. Cel Struct. Funct. 29, 49–65. doi:10.1247/csf.29.49
Vida, T. A., and Emr, S. D. (1995). A New Vital Stain for Visualizing Vacuolar Membrane Dynamics and Endocytosis in Yeast. J. Cel Biol. 128, 779–792. doi:10.1083/jcb.128.5.779
Viotti, C., Bubeck, J., Stierhof, Y.-D., Krebs, M., Langhans, M., van den Berg, W., et al. (2010). Endocytic and Secretory Traffic in Arabidopsis Merge in the Trans-golgi Network/early Endosome, an Independent and Highly Dynamic Organelle. Plant Cell 22, 1344–1357. doi:10.1105/tpc.109.072637
Wei, J.-H., and Seemann, J. (2010). Unraveling the Golgi Ribbon. Traffic 11, 1391–1400. doi:10.1111/j.1600-0854.2010.01114.x
Weigel, A. V., Chang, C.-L., Shtengel, G., Xu, C. S., Hoffman, D. P., Freeman, M., et al. (2021). ER-to-Golgi Protein Delivery through an Interwoven, Tubular Network Extending from ER. Cell 184, 1–18. doi:10.1016/j.cell.2021.03.035
Weiss, M., and Nilsson, T. (2000). Protein Sorting in the Golgi Apparatus: a Consequence of Maturation and Triggered Sorting. FEBS Lett. 486, 2–9. doi:10.1016/s0014-5793(00)02155-4
Welz, T., Wellbourne-Wood, J., and Kerkhoff, E. (2014). Orchestration of Cell Surface Proteins by Rab11. Trends Cel Biol. 24, 407–415. doi:10.1016/j.tcb.2014.02.004
Wood, S. A., Park, J. E., and Brown, W. J. (1991). Brefeldin A Causes a Microtubule-Mediated Fusion of the Trans-golgi Network and Early Endosomes. Cell 67, 591–600. doi:10.1016/0092-8674(91)90533-5
Yadav, S., and Linstedt, A. D. (2011). Golgi Positioning, Cold Spring Harb. Perspect. 3, a005322. doi:10.1101/cshperspect.a005322Biol
Yadav, S., Puri, S., and Linstedt, A. D. (2009). A Primary Role for Golgi Positioning in Directed Secretion, Cell Polarity, and Wound Healing. MBoC 20, 1728–1736. doi:10.1091/mbc.e08-10-1077
Yang, J.-S., Valente, C., Polishchuk, R. S., Turacchio, G., Layre, E., Moody, D. B., et al. (2011). COPI Acts in Both Vesicular and Tubular Transport. Nat. Cel Biol 13, 996–1003. doi:10.1038/ncb2273
Yano, H., Yamamoto-Hino, M., Abe, M., Kuwahara, R., Haraguchi, S., Kusaka, I., et al. (2005). Distinct Functional Units of the Golgi Complex in Drosophila Cells. Proc. Natl. Acad. Sci. U.S.A. 102, 13467–13472. doi:10.1073/pnas.0506681102
Ying, M., Flatmark, T., and Saraste, J. (2000). The P58-Positive Pre-golgi Intermediates Consist of Distinct Subpopulations of Particles that Show Differential Binding of COPI and COPII coats and Contain Vacuolar H(+)-ATPase. J. Cel Sci. 113, 3623–3638. doi:10.1242/jcs.113.20.3623
Zerial, M., and McBride, H. (2001). Rab Proteins as Membrane Organizers. Nat. Rev. Mol. Cel Biol. 2, 107–117. doi:10.1038/35052055
Zhang, G. F., and Staehelin, L. A. (1992). Functional Compartmentation of the Golgi Apparatus of Plant Cells. Plant Physiol. 99, 1070–1083. doi:10.1104/pp.99.3.1070
Glossary
AP adaptor protein complex
BFA brefeldin A
CERT ceramide transfer protein
COP coat protein complex
EARP endosome-associated recycling protein complex
EE early endosome
EM electron microscopy
ER endoplasmic reticulum
ERC endocytic recycling compartment
ERES ER exit site
ERGIC ER-Golgi intermediate compartment
ESCRT endosomal sorting complex required for transport
GARP Golgi-associated retrograde protein complex
GECCO Golgi entry core compartment
GEF guanine-nucleotide exchange factor
GGA Golgi-localized, γ-ear-containing, Arf-binding family of protein
IC intermediate compartment
ILV intraluminal vesicle
LE late endosome
MVB multivesicular body
OSBP oxysterol-binding protein
PM plasma membrane
PVC prevacuolar compartment
RE recycling endosome
SCLIM super-resolution confocal live imaging microscopy
SE sorting endosome
SNARE soluble N-ethylmaleimide-sensitive factor attachment protein receptor
Syn syntaxin
SYP syntaxin of plants
tER transitional ER
TGN trans-Golgi network
VTC vesicular tubular cluster.
Keywords: Golgi, membrane traffic, ERGIC, GECCO, trans-Golgi network, recycling endosome, live imaging, SCLIM
Citation: Nakano A (2022) The Golgi Apparatus and its Next-Door Neighbors. Front. Cell Dev. Biol. 10:884360. doi: 10.3389/fcell.2022.884360
Received: 26 February 2022; Accepted: 28 March 2022;
Published: 28 April 2022.
Edited by:
Daniel Ungar, University of York, United KingdomReviewed by:
J. Christopher Fromme, Cornell University, United StatesBrian Storrie, University of Arkansas for Medical Sciences, United States
Benjamin S. Glick, The University of Chicago, United States
Copyright © 2022 Nakano. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Akihiko Nakano, bmFrYW5vQHJpa2VuLmpw
 Akihiko Nakano
Akihiko Nakano