- Department of Life Sciences, Sir Alexander Fleming Building, Imperial College London, London, United Kingdom
The African trypanosome Trypanosoma brucei is a parasite of the mammalian bloodstream and tissues, where an antigenically variable Variant Surface Glycoprotein (VSG) coat protects it from immune attack. This dense layer comprised of ∼107 VSG proteins, makes VSG by far the most abundant mRNA (7–10% total) and protein (∼10% total) in the bloodstream form trypanosome. How can such prodigious amounts of VSG be produced from a single VSG gene? Extremely high levels of RNA polymerase I (Pol I) transcription of the active VSG provide part of the explanation. However, recent discoveries highlight the role of pre-mRNA processing, both in maintaining high levels of VSG transcription, as well as its monoallelic expression. Trypanosome mRNAs are matured through trans-splicing a spliced leader (SL) RNA to the 5’ end of precursor transcripts, meaning abundant SL RNA is required throughout the nucleus. However, requirement for SL RNA in the vicinity of the active VSG gene is so intense, that the cell reconfigures its chromatin architecture to facilitate interaction between the SL RNA genes and the active VSG. This presumably ensures that sufficient localised SL RNA is available, and not limiting for VSG mRNA expression. Recently, novel nuclear splicing bodies which appear to provide essential trans-splicing components, have been identified associating with the active VSG. These observations highlight the underappreciated role of pre-mRNA processing in modulating gene expression in trypanosomes. Dissecting the function of these nuclear RNA processing bodies should help us elucidate the mechanisms of both VSG expression and monoallelic exclusion in T. brucei.
Introduction
Nuclear bodies are increasingly being shown to be essential for the regulation and compartmentalisation of gene expression (Shin et al., 2018). These membraneless nuclear condensates facilitate vital functions in different organisms and cell types (Banani et al., 2017). They self-assemble through phase separation, functioning as “hot-spots” for specific nuclear processes. Through sequestering and concentrating proteins and RNA, they can increase reaction kinetics, and can co-ordinate inter-chromosomal interactions (Shin and Brangwynne, 2017). Cajal bodies for example, are specialised in the modification of small nuclear RNAs (snRNAs), and the assembly of splicing small nuclear ribonucleoproteins (snRNPs). The nucleolus is specialised in RNA polymerase I (Pol I) transcription of the ribosomal DNA (rDNA), processing and modification of rRNA, and ribosome assembly.
The African sleeping sickness parasite Trypanosoma brucei contains a Pol I-enriched nucleolus. However, it is unique among eukaryotes in using Pol I to transcribe some protein coding genes, including VSG. T. brucei contains thousands of VSG genes, of which one is transcribed at a time from one of ∼15 expression site (ES) transcription units (Hertz-Fowler et al., 2008; Cross et al., 2014). The active ES is located within an extra-nucleolar Pol I body called the Expression Site Body (ESB) (Navarro and Gull, 2001) (Figure 1). There has been intense interest in factors facilitating high levels of ES transcription, and numerous chromatin proteins have been identified (Pena et al., 2017). However, we are increasingly realising the importance of pre-mRNA processing both in maintaining high levels of VSG expression, as well as its monoallelic control. The two processes of transcription and splicing appear to be interconnected at the active ES, whereby blocking splicing results in strongly reduced processive transcription (Budzak et al., 2022). In this review, we discuss the role of nuclear bodies in facilitating high VSG expression levels.
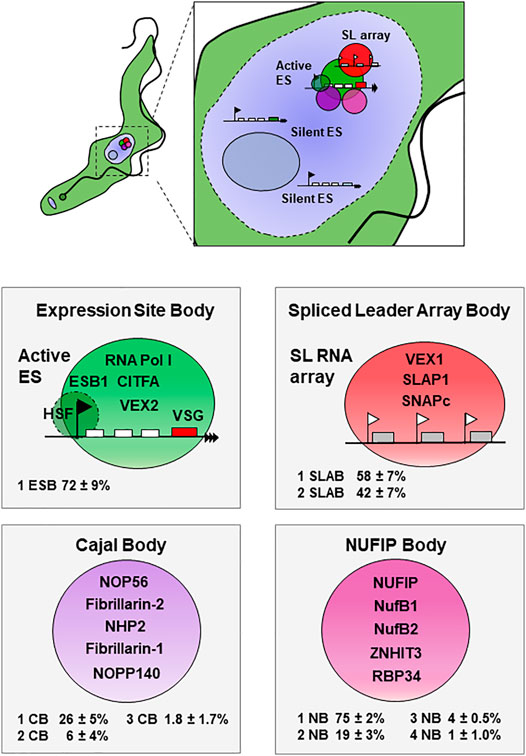
FIGURE 1. An assembly of nuclear bodies congregate at the active VSG expression site At the top a bloodstream form (BF) trypanosome is shown with the nucleus (dashed circle) magnified. This contains an assembly of nuclear bodies (coloured circles) at the active VSG expression site (ES), as well as a nucleolus (blue circle). The active ES has a Pol I promoter indicated with a black flag, various Expression site associated genes (ESAGs) with white boxes, and the VSG with a red box (not drawn to scale). The active ES associates with at least one of the spliced leader (SL) RNA gene arrays, shown with individual SL RNA genes (grey boxes) transcribed from Pol II promoters (white flags). Nuclear bodies are shown below as large coloured circles with selected associated protein components indicated. The Expression site body (ESB) (large green circle) at the active ES is associated with a highly SUMO-ylated focus (HSF) (dark green circle). The Spliced Leader Array Body (SLAB) (red circle) associates with the SL RNA gene arrays, of which at least one is associated with the active ES. Two additional nuclear splicing bodies associating with the active ES include the Cajal Body (violet circle) and a novel NUFIP body (purple circle). These nuclear bodies (with the exception of the ESB) are also found in procyclic form T. brucei. The percentage of BF T. brucei cells in G1 containing one or more of these nuclear bodies is shown in the respective panels [data from (Budzak et al., 2022)].
The ESB Facilitates Extremely High Rates of Monoallelic Transcription of VSG
The ESB is the first nuclear body shown to be key for monoallelic VSG expression (Navarro and Gull, 2001). A stringent restriction ensures that maximally one ESB is stably present within the bloodstream form (BF) trypanosome. Forced activation of a second ES, results in two ESs sharing the same ESB (Chaves et al., 1999; Budzak et al., 2019). VSG is essential for BF T. brucei, and extraordinarily high levels of continuous VSG mRNA production are required for proliferation (Sheader et al., 2005; Ridewood et al., 2017). VSG is by far the most abundant mRNA (∼7–10% total) in BF T. brucei. An estimated 666.7 VSG mRNA molecules are generated per hour from a single VSG gene, compared with 1.3 mRNA molecules per hour from a typical Pol II transcribed gene (Budzak et al., 2022) (Figure 2). This staggering 512-fold higher rate of mRNA production, enables generation of the vast amounts of Variant Surface Glycoprotein (VSG) (107 molecules, ∼10% total protein) necessary to form a fully protective surface coat (Bartossek et al., 2017; Maudlin et al., 2021). The ESB can therefore be considered a specialised transcription factory allowing enormous amounts of VSG pre-mRNA to be transcribed.
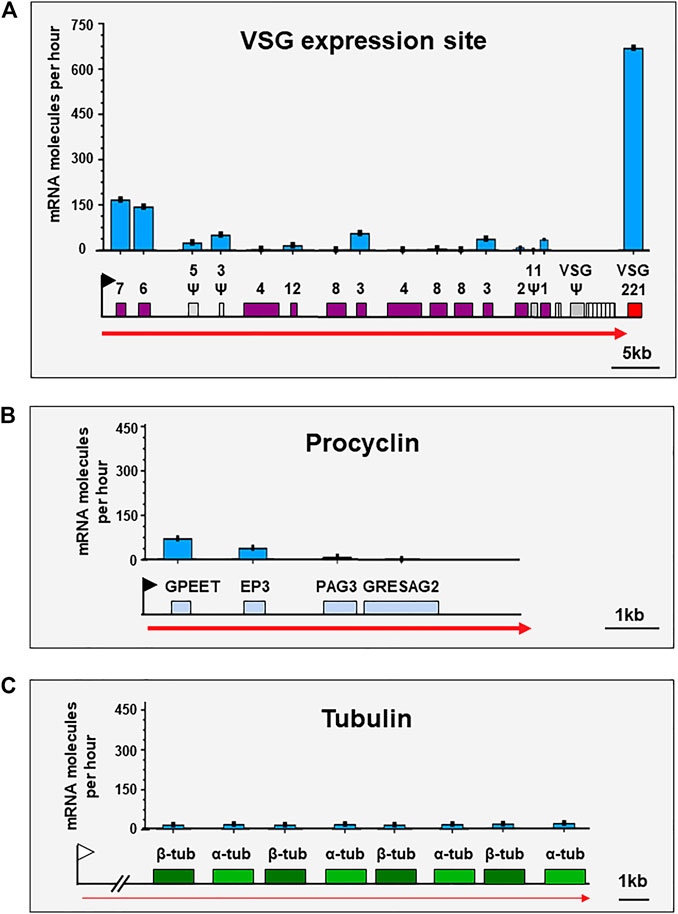
FIGURE 2. VSG is the mRNA generated at the highest rate in bloodstream form T. brucei. Schematics show the estimated number of mRNA molecules produced per hour from genes in different genomic loci in bloodstream form Trypanosoma brucei. (A) Schematic of an active VSG expression site containing the most abundant mRNA expressed in bloodstream form T. brucei; VSG. The Pol I ES promoter is indicated with a black flag, and high amounts of Pol I transcription with a thick red arrow. Expression Site Associated Genes (ESAGs) and VSG221 are indicated with coloured boxes, pseudogenes with Ψ (grey boxes), and 70 bp repeats with striped boxes. (B) Schematic showing one of the four procyclin loci, which contain the most abundant mRNAs expressed in procyclic form T. brucei. The Pol I promoter is indicated with a black flag, high levels of Pol I transcription with a thick red arrow, and relevant genes with coloured boxes. (C) Schematic of the tubulin locus, which contains the most abundant mRNAs transcribed by Pol II in T. brucei. The upstream Pol II promoter is indicated with a white flag, and low levels of Pol II transcription with a thin red arrow. Three pairs of alternating α-tubulin and ß-tubulin genes (out of a total of eight per locus) are indicated with coloured boxes. For all graphs, the y-axes are the same scale as in panel (A). Values for mRNA molecules generated per hour were derived from Supplementary Tables 1, 2 in Budzak et al. (2022).
Such an extreme requirement for mRNA production does not occur in many cell types. Something similar is seen in antibody secreting plasma cells. Antibody production in B cells can reach 108 immunoglobulin molecules per hour (Hibi and Dosch, 1986), with an extraordinary 70% of the mRNA transcriptome comprised solely of immunoglobulin (IgG) mRNAs (Shi et al., 2015). To achieve this, transcription of immunoglobulin heavy chain genes is boosted by super-enhancers (Le Noir et al., 2017). Genes present on different chromosomes colocalise in transcription factories located in activating regions at the nuclear periphery, thereby facilitating enhancer interactions (Park et al., 2014).
African trypanosomes have evolved their own unusual adaptations to cope with the extreme biology of producing such large amounts of mRNA from a single VSG gene. One of these is the incredible stability of the VSG mRNA. VSG mRNA has one of the longest half-lives in the cell, which is partly conferred by a specific RNA binding protein (CFB2), which recognises conserved elements in the VSG 3′UTR (Ridewood et al., 2017; Melo do Nascimento et al., 2021). N6-methyladenosine modification of the VSG poly(A) tails has also been proposed as a mechanism for increasing VSG mRNA stability (Viegas et al., 2020). An additional important adaptation is the use of Pol I for ES transcription (Gunzl et al., 2003). In mammalian cells, Pol I initiates transcription at significantly higher rates than Pol II, with a reinitiation interval of one polymerase per ∼1.4 s (Dundr et al., 2002), compared with an initiation rate for Pol II which is frequently less than ∼1-2 per minute (Darzacq et al., 2007; Fuchs et al., 2014). In eukaryotes Pol I normally exclusively transcribes rDNA, as it generates uncapped, and therefore untranslateable transcripts (Grummt and Skinner, 1985). In trypanosomes however, as trans-splicing adds capped Pol II derived spliced leader (SL) RNA to the 5′ end of mRNA precursors (Gunzl, 2010), Pol I derived mRNAs are rendered translateable. In T. brucei it is estimated that a Pol I transcribed gene is expressed at a more than ∼10-fold higher rate than one transcribed by Pol II (Biebinger et al., 1996). Trypanosomes therefore appear to have co-opted the highest initiating polymerase in the cell (Pol I), to maximise transcription of VSG.
Both Pol I bodies, the ESB and the nucleolus, are enriched for Pol I transcription factors including CITFAs and the architectural chromatin protein TDP1 (Narayanan and Rudenko, 2013; Nguyen et al., 2014). However, the ESB also contains components not found in the nucleolus including VEX2, which contains homology to the RNA helicase UPF1. Interestingly, depletion of VEX2 results in upregulation of ESAGs from the active ES, suggesting it may play a role in suppressing excessive ESAG mRNA production (Faria et al., 2019). In addition, the first ESB-specific protein (ESB1) has now been discovered, which could be involved in protein ubiquitinylation (Escobar et al., 2021). Both VEX2 and ESB1 are important for monoallelic VSG expression, as their perturbation results in upregulation of silent ESs. A highly SUMOylated focus is also observed at the active ES, but not the nucleolus, and could play a role in ESB stabilisation (Lopez-Farfan et al., 2014).
The ESB appears to be a specialised Pol I factory, facilitating extremely high levels of transcription, and monoallelic expression of VSG. However, we are increasingly realising the importance of highly efficient pre-mRNA processing in both of these processes. ES precursor transcripts must be rapidly trans-spliced and polyadenylated, and blocking trans-splicing using chemical inhibitors or anti-U2 Morpholinos results in a radical reduction in processive ES transcription (Budzak et al., 2022). In mammalian cells there is an extensive literature on the feedback between Pol II transcription and splicing, whereby splicing can enhance Pol II transcription elongation (Tellier et al., 2020). This can operate through interaction of the phosphorylated C-terminus of Pol II with different components of the splicing machinery (Kornblihtt et al., 2004; Maita and Nakagawa, 2020). However, in T. brucei this would require interaction of the splicing machinery with Pol I exclusively at the active ES and not the Pol I transcribed rDNA. However alternatively, blocking trans-splicing could result in unspliced ES precursor transcripts remaining associated with the extending Pol I molecules, thereby facilitating polymerase removal from the template by RNA-degradation factors. This would make processing of ES precursor transcripts through trans-splicing of the SL exon essential for maintaining high levels of transcription by allowing unimpeded elongation. It now appears that the sheer amount of localised SL RNA required in the vicinity of the active ES has necessitated the trypanosome to reconfigure its nucleus to accommodate this.
The Spliced Leader Array Body Interacts With the Active VSG
T. brucei mRNAs are generated through coupled trans-splicing of a capped SL RNA exon and polyadenylation (Matthews et al., 1994; Clayton, 2019). This allows the production of translateable mRNA from precursor transcripts from the extensive polycistronic transcription units comprising most of the T. brucei genome (Berriman et al., 2005). The large amount of SL RNA required is generated from two arrays of over 100 SL RNA genes, each with its own Pol II promoter (Gilinger and Bellofatto, 2001). SL RNA transcription is mediated by a specialised SNAP transcription factor complex (Schimanski et al., 2005). Two additional proteins localising at the SL RNA array in both BF and procyclic form (PF) T. brucei are VEX1 and Spliced Leader Array Protein 1 (SLAP1), (Faria et al., 2021) (Budzak et al., 2022). VEX1 is important for maintenance of monoallelic expression of VSG, but is nonessential (Glover et al., 2016). SLAP1 is essential, and its knockdown results in decreased amounts of the SL RNA intron, as well as defective splicing (Budzak et al., 2022).
The proteins colocalising at the SL RNA array allow identification of the Spliced Leader Array Body (SLAB), of which there are one or two, in both BF and PF T. brucei (Faria et al., 2021; Budzak et al., 2022) (Figure 1). The SLAB associates with the active ES in BF T. brucei, and when cells have two SLAB, at least one is typically within 350 nm of the ESB (Glover et al., 2016; Escobar et al., 2021; Faria et al., 2021; Budzak et al., 2022). This agrees with the surprising discovery made using Hi-C chromosome conformation capture experiments, that there is a robust inter-chromosomal interaction between the SL RNA array and the active ES (Faria et al., 2021). This contact could be mediated by the VEX1-VEX2 complex acting as a bridge between the SL-RNA array and the active VSG ES. However, only depletion of VEX2 (and not VEX1) disrupts this interaction, (Faria et al., 2021) (Budzak et al., 2022). This could indicate additional bridging molecules facilitate interaction of these two loci.
The Cajal and NUFIP Bodies
SL RNA trans-splicing in trypanosomes requires spliceosomal small nuclear ribonucleoproteins (snRNPs) (Gunzl, 2010), which are modified and assembled in Cajal bodies (Morris, 2008; Meier, 2017). Cajal bodies, while absent in many cell types, are typically found in rapidly dividing embryonic or cancer cells. Their presence and abundance is thought to be correlated to splicing rates (Young et al., 2000; Morris, 2008). They can be dispensable, as knockdown of the Cajal body scaffolding protein coilin, results in defective Cajal bodies but viable mice (Tucker et al., 2001). Cajal bodies are frequently associated with highly expressed loci (Wang et al., 2016). Their function therefore appears to be the concentration of essential splicing components, thereby catalysing processes which would otherwise be rate limiting (Sawyer et al., 2016).
The Cajal body has been elusive in T. brucei, as coilin, the canonical Cajal body marker, is not readily identifiable in the genome. However recently, a number of new nuclear bodies present in both BF and PF T. brucei were identified using the TrypTag database of T. brucei proteins tagged with mNeonGreen (Dean et al., 2017; Budzak et al., 2022). These include a Cajal body, containing conserved extra-nucleolar Cajal body proteins. Additionally, a novel nuclear body was identified, which was called the NUFIP body (Figure 1). This contains the highly conserved NUFIP and ZNHIT3 proteins, which mediate snRNP assembly in mammalian cells (Rothe et al., 2014; Bizarro et al., 2015). The NUFIP body appears to be important in BF T. brucei, as 75 ± 2% of G1 cells have one NUFIP body, with most of the rest containing two. In contrast, a Cajal body is present in only 26 ± 5% BF T. brucei in G1 (Budzak et al., 2022), making it unclear if it is essential. In PF T. brucei the relative abundance of these two nuclear bodies is shifted, with fewer G1 cells (45 ± 5.5%) containing minimally one NUFIP body, and more (36 ± 3.5%) containing at least one Cajal body.
Similar to the SLAB, when one or more NUFIP or Cajal bodies are visible in BF T. brucei, minimally one of these bodies is near the active ES (Figure 1) (Budzak et al., 2022). Another feature which is similar to the SLAB, is that the nuclear positioning of the NUFIP body is determined by ES activity, as when cells switch between ESs, these bodies interact with the newly activated ES (Faria et al., 2021; Budzak et al., 2022). Despite the importance of the SLAB in facilitating trans-splicing, the NUFIP body appears to be located even closer to the VSG at the telomere of the active ES, with an average distance of 240 nm compared with 350 nm for the SLAB (Budzak et al., 2022).
Although the function of conserved Cajal body proteins has been investigated in trypanosomes (Barth et al., 2008; Jae et al., 2011), the function of the NUFIP body is unclear. The conserved NUFIP and ZNHIT3 proteins in the T. brucei NUFIP body play a role in snRNP assembly in yeast and mammals (Rothe et al., 2014; Bizarro et al., 2015). However, the other three NUFIP body proteins identified do not have known functions, other than that they contain RNA recognition motifs (Budzak et al., 2022). Several NUFIP body components have been shown to co-immunoprecipitate with CRK9, which is involved in RNA modification of the SL RNA, and is essential for trans-splicing in T. brucei (Badjatia et al., 2016). Cajal bodies frequently associate with highly transcribed loci, including the U1 and U2 snRNA genes in mammalian cells (Smith et al., 1995). In both BF and PF T. brucei, the NUFIP body is frequently in close proximity to the SLAB (associating with the highly transcribed SL RNA genes), indicating possible transfer of splicing components (Budzak et al., 2022). A NUFIP body has not yet been identified in other organisms. Therefore, an attractive hypothesis is that the NUFIP body is a novel type of nuclear body similar to the Cajal body, but specifically dedicated to facilitating trans-splicing in Kinetoplastid protozoa. The presence of at least one NUFIP body in BF T. brucei cells argues that it is an essential structure.
Surprisingly, some NUFIP body proteins were previously identified associating with kinetochore proteins localising to the outer kinetochore during mitosis (Nerusheva et al., 2019; Brusini et al., 2021). Possibly the NUFIP body transiently interacts with the kinetochore during mitosis, indicating that T. brucei centromere function requires some aspect of RNA biology. Splicing factors could sometimes associate with kinetochores in mammalian cells, where they have been postulated to have additional secondary functions in the regulation of mitosis (Pellacani et al., 2018; Somma et al., 2020). Further studies are required to characterise the precise function(s) of the NUFIP body in T. brucei. However, if proximity of a NUFIP body to the active ES is required for efficient splicing, as most BF T. brucei have only one NUFIP body, this could provide an important restriction behind the monoallelic exclusion operating at the active VSG expression site.
Formation of a Nuclear Body Assembly at the Active VSG Expression Site
Why are all four nuclear bodies in the proximity of the active ES? Presumably the trypanosome can only keep up with the phenomenal demand for VSG mRNA by concentrating both transcription and splicing machineries at the active ES. The dynamic association of the splicing bodies with the active ES could explain how the trypanosome can maintain a VSG splicing rate which is 512-fold higher than at a typical Pol II gene (Budzak et al., 2022). If the ESB is considered a Pol I transcription factory, the SLAB, NUFIP and Cajal bodies could be considered mobile splicing component factories, each of which might be optimised to produce different required components. This nuclear body assembly associating at the active ES, could be analogous to a VSG mRNA super-factory with mobile subunits. The mobility of these splicing bodies presumably allows them to dynamically provide splicing components throughout the nucleus, collecting in regions of greatest need (Faria et al., 2021). It is unclear why trypanosomes require so many different splicing bodies. Possibly the NUFIP and Cajal bodies are where T. brucei performs the final stages of snRNP assembly to generate splicing competent particles. This could create an assembly line, where VSG pre-mRNA is generated and processed in a way optimised for maximal expression.
An important question which arises from this model, is how does VSG achieve such high expression levels, while ESAGs transcribed from the same ES are expressed at significantly lower levels? Possibly ESAG mRNAs are selectively degraded at a higher rate than VSG mRNA. Knockdown of VEX2 results in upregulation of ESAG mRNA from the active ES, suggesting that active RNA degradation suppresses maximal ESAG expression (Faria et al., 2019). However, how such selective degradation would occur is unclear. In contrast, VSG mRNA is stabilised through RNA binding proteins and RNA modification, which could prevent degradation immediately after transcription (Ridewood et al., 2017; Viegas et al., 2020; Melo do Nascimento et al., 2021). In addition, the SLAB, NUFIP and Cajal bodies appeared to be positioned closer to the ES telomere rather than the promoter, possibly facilitating particularly efficient VSG mRNA splicing (Budzak et al., 2022). Collectively, these different mechanisms could be used to achieve higher levels of production of VSG compared with ESAG mRNA.
This nuclear body assembly could also be important for monoallelic expression of VSG. Only one ESB nuclear body assembly is stably present in BF T. brucei, where the active ES is transcribed at a very high rate (Budzak et al., 2019; Budzak et al., 2022). Only low levels of transcription are observed immediately downstream of the 14 “silent” ES promoters (Kassem et al., 2014), and these transcripts are not spliced or polyadenylated efficiently (Vanhamme et al., 2000). Silent ESs are not in the same nuclear location as the active ES (Muller et al., 2018; Budzak et al., 2019). If proximity of the active ES to an assembly of transcription and splicing bodies is important for its activation, then exclusion of silent ESs from the ESB nuclear body assembly could maintain them in a silent state. As there is feedback between splicing and transcription elongation at the active ES, increased access of a “silent” ES to pre-mRNA processing machinery, possibly stimulates transcription elongation and therefore its activation. The limited number of nuclear splicing bodies within the BF trypanosome (only one or two of each type) could therefore be a restriction facilitating monoallelic expression of the active ES.
In mammalian cells, nuclear splicing bodies including Cajal bodies are preferentially located at highly transcribed loci, and Cajal body disruption leads to decreased gene expression (Wang et al., 2016). This is also the case for structures called “nuclear speckles”, which are enriched for pre-mRNA splicing factors as well as proteins involved in transcription and post-translational modifications (Galganski et al., 2017; Kim et al., 2020). Analysis of the 3D organisation of the mammalian nucleus showed that genes within transcription “hot zones” are in the proximity of nuclear speckles (Chen et al., 2018). In T. brucei, the reorganisation of the nuclear architecture appears to be more extreme, as a collection of up to four nuclear bodies assemble at a single locus. Further work will be required to determine the mechanisms which target these nuclear bodies to the active ES, and whether this targeting is a cause or a consequence of high levels of RNA processing. Although recent discoveries highlight the presence of splicing bodies at the active ES, other components of the pre-mRNA processing machinery involved in polyadenylation or RNA modification may also be concentrated at the active ES. However, this remains to be investigated. It is also unclear how minimally one of the SL RNA gene arrays interacts with the active VSG ES.
Conclusion
In summary, T. brucei continues to provide unique molecular solutions for its problems, including how to produce enough VSG (∼10% total protein) for a protective coat, from a single active VSG gene. The unusual use of Pol I allows extremely high levels of VSG transcription within an ESB. However, extraordinarily high levels of trans-splicing are also required at the active VSG. This appears to be facilitated by the recruitment of the SL RNA genes, as well as three splicing related nuclear bodies (SLAB, NUFIP and Cajal) to the vicinity of the active ES. These constitute an ES nuclear body assembly functioning as a VSG super-factory. This allows the trypanosome to push the limits of what is possible, to achieve phenomenal levels of expression from a single copy gene.
Author Contributions
JB and GR wrote the manuscript and designed the figures.
Funding
GR is a Wellcome Senior Fellow in the Basic Biomedical Sciences funded by the Wellcome Trust (212211/Z/18/Z).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank Calvin Tiengwe, Nikolay Kolev, Agustina Berazategui and Dominic Marshall Sabey for discussions and comments on the manuscript.
References
Badjatia, N., Park, S. H., Ambrósio, D. L., Kirkham, J. K., and Günzl, A. (2016). Cyclin-Dependent Kinase CRK9, Required for Spliced Leader Trans Splicing of Pre-mRNA in Trypanosomes, Functions in a Complex with a New L-type Cyclin and a Kinetoplastid-specific Protein. Plos Pathog. 12, e1005498. doi:10.1371/journal.ppat.1005498
Banani, S. F., Lee, H. O., Hyman, A. A., and Rosen, M. K. (2017). Biomolecular Condensates: Organizers of Cellular Biochemistry. Nat. Rev. Mol. Cel Biol 18, 285–298. doi:10.1038/nrm.2017.7
Barth, S., Shalem, B., Hury, A., Tkacz, I. D., Liang, X.-h., Uliel, S., et al. (2008). Elucidating the Role of C/D snoRNA in rRNA Processing and Modification in Trypanosoma Brucei. Eukaryot. Cel 7, 86–101. doi:10.1128/ec.00215-07
Bartossek, T., Jones, N. G., Schäfer, C., Cvitković, M., Glogger, M., Mott, H. R., et al. (2017). Structural Basis for the Shielding Function of the Dynamic Trypanosome Variant Surface Glycoprotein Coat. Nat. Microbiol. 2, 1523–1532. doi:10.1038/s41564-017-0013-6
Berriman, M., Ghedin, E., Hertz-Fowler, C., Blandin, G., Renauld, H., Bartholomeu, D. C., et al. (2005). The Genome of the African Trypanosome Trypanosoma Brucei. Science 309, 416–422. doi:10.1126/science.1112642
Biebinger, S., Rettenmaier, S., Flaspohler, J., Hartmann, C., Pena-Diaz, J., Wirtz, L. E., et al. (1996). The PARP Promoter of Trypanosoma Brucei Is Developmentally Regulated in a Chromosomal Context. Nucleic Acids Res. 24, 1202–1211. doi:10.1093/nar/24.7.1202
Bizarro, J., Dodré, M., Huttin, A., Charpentier, B., Schlotter, F., Branlant, C., et al. (2015). NUFIP and the HSP90/R2TP Chaperone Bind the SMN Complex and Facilitate Assembly of U4-specific Proteins. Nucleic Acids Res. 43, 8973–8989. doi:10.1093/nar/gkv809
Brusini, L., D’Archivio, S., Mcdonald, J., and Wickstead, B. (2021). Trypanosome KKIP1 Dynamically Links the Inner Kinetochore to a Kinetoplastid Outer Kinetochore Complex. Front. Cel. Infect. Microbiol. 11, 641174. doi:10.3389/fcimb.2021.641174
Budzak, J., Jones, R., Tschudi, C., Kolev, N. G., and Rudenko, G. (2022). An Assembly of Nuclear Bodies Associates with the Active VSG Expression Site in African Trypanosomes. Nat. Commun. 13, 101. doi:10.1038/s41467-021-27625-6
Budzak, J., Kerry, L. E., Aristodemou, A., Hall, B. S., Witmer, K., Kushwaha, M., et al. (2019). Dynamic Colocalization of 2 Simultaneously Active VSG Expression Sites within a Single Expression-Site Body in Trypanosoma Brucei. Proc. Natl. Acad. Sci. U.S.A. 116, 16561–16570. doi:10.1073/pnas.1905552116
Chaves, I., Rudenko, G., Dirks-Mulder, A., Cross, M., and Borst, P. (1999). Control of Variant Surface Glycoprotein Gene-Expression Sites in Trypanosoma Brucei. Embo J. 18, 4846–4855. doi:10.1093/emboj/18.17.4846
Chen, Y., Zhang, Y., Wang, Y., Zhang, L., Brinkman, E. K., Adam, S. A., et al. (2018). Mapping 3D Genome Organization Relative to Nuclear Compartments Using TSA-Seq as a Cytological Ruler. J. Cel Biol 217, 4025–4048. doi:10.1083/jcb.201807108
Clayton, C. (2019). Regulation of Gene Expression in Trypanosomatids: Living with Polycistronic Transcription. Open Biol. 9, 190072. doi:10.1098/rsob.190072
Cross, G. A. M., Kim, H.-S., and Wickstead, B. (2014). Capturing the Variant Surface Glycoprotein Repertoire (The VSGnome) of Trypanosoma Brucei Lister 427. Mol. Biochem. Parasitol. 195, 59–73. doi:10.1016/j.molbiopara.2014.06.004
Darzacq, X., Shav-Tal, Y., De Turris, V., Brody, Y., Shenoy, S. M., Phair, R. D., et al. (2007). In Vivo dynamics of RNA Polymerase II Transcription. Nat. Struct. Mol. Biol. 14, 796–806. doi:10.1038/nsmb1280
Dean, S., Sunter, J. D., and Wheeler, R. J. (2017). TrypTag.org: A Trypanosome Genome-wide Protein Localisation Resource. Trends Parasitol. 33, 80–82. doi:10.1016/j.pt.2016.10.009
Dundr, M., Hoffmann-Rohrer, U., Hu, Q., Grummt, I., Rothblum, L. I., Phair, R. D., et al. (2002). A Kinetic Framework for a Mammalian RNA Polymerase In Vivo. Science 298, 1623–1626. doi:10.1126/science.1076164
Escobar, L. L., Hanisch, B., Halliday, C., Dean, S., Sunter, J. D., Wheeler, R. J., et al. (2021). Monoallelic Antigen Expression in Trypanosomes Requires a Stage-specific Transcription Activator. bioRXiv. doi:10.1101/2021.05.06.442931
Faria, J., Glover, L., Hutchinson, S., Boehm, C., Field, M. C., and Horn, D. (2019). Monoallelic Expression and Epigenetic Inheritance Sustained by a Trypanosoma Brucei Variant Surface Glycoprotein Exclusion Complex. Nat. Commun. 10, 3023. doi:10.1038/s41467-019-10823-8
Faria, J., Luzak, V., Müller, L. S. M., Brink, B. G., Hutchinson, S., Glover, L., et al. (2021). Spatial Integration of Transcription and Splicing in a Dedicated Compartment Sustains Monogenic Antigen Expression in African Trypanosomes. Nat. Microbiol. 6, 289–300. doi:10.1038/s41564-020-00833-4
Fuchs, G., Voichek, Y., Benjamin, S., Gilad, S., Amit, I., and Oren, M. (2014). 4sUDRB-seq: Measuring Genomewide Transcriptional Elongation Rates and Initiation Frequencies within Cells. Genome Biol. 15, R69. doi:10.1186/gb-2014-15-5-r69
Galganski, L., Urbanek, M. O., and Krzyzosiak, W. J. (2017). Nuclear Speckles: Molecular Organization, Biological Function and Role in Disease. Nucleic Acids Res. 45, 10350–10368. doi:10.1093/nar/gkx759
Gilinger, G., and Bellofatto, V. (2001). Trypanosome Spliced Leader RNA Genes Contain the First Identified RNA Polymerase II Gene Promoter in These Organisms. Nucleic Acids Res. 29, 1556–1564. doi:10.1093/nar/29.7.1556
Glover, L., Hutchinson, S., Alsford, S., and Horn, D. (2016). VEX1 Controls the Allelic Exclusion Required for Antigenic Variation in Trypanosomes. Proc. Natl. Acad. Sci. U.S.A. 113, 7225–7230. doi:10.1073/pnas.1600344113
Grummt, I., and Skinner, J. A. (1985). Efficient Transcription of a Protein-Coding Gene from the RNA Polymerase I Promoter in Transfected Cells. Proc. Natl. Acad. Sci. U.S.A. 82, 722–726. doi:10.1073/pnas.82.3.722
Günzl, A., Bruderer, T., Laufer, G., Schimanski, B., Tu, L. C., Chung, H. M., et al. (2003). RNA Polymerase I Transcribes Procyclin Genes and Variant Surface Glycoprotein Gene Expression Sites in Trypanosoma Brucei. Eukaryot. Cel 2, 542–551. doi:10.1128/ec.2.3.542-551.2003
Günzl, A. (2010). The Pre-mRNA Splicing Machinery of Trypanosomes: Complex or Simplified? Eukaryot. Cel 9, 1159–1170. doi:10.1128/ec.00113-10
Hertz-Fowler, C., Figueiredo, L. M., Quail, M. A., Becker, M., Jackson, A., Bason, N., et al. (2008). Telomeric Expression Sites Are Highly Conserved in Trypanosoma Brucei. PLoS One 3, e3527. doi:10.1371/journal.pone.0003527
Hibi, T., and Dosch, H.-M. (1986). Limiting Dilution Analysis of the B Cell Compartment in Human Bone Marrow. Eur. J. Immunol. 16, 139–145. doi:10.1002/eji.1830160206
Jaé, N., Preusser, C., Krüger, T., Tkacz, I. D., Engstler, M., Michaeli, S., et al. (2011). snRNA-specific Role of SMN in Trypanosome snRNP Biogenesis In Vivo. RNA Biol. 8, 90–100. doi:10.4161/rna.8.1.13985
Kassem, A., Pays, E., and Vanhamme, L. (2014). Transcription Is Initiated on Silent Variant Surface Glycoprotein Expression Sites Despite Monoallelic Expression in Trypanosoma Brucei. Proc. Natl. Acad. Sci. U.S.A. 111, 8943–8948. doi:10.1073/pnas.1404873111
Kim, J., Venkata, N. C., Hernandez Gonzalez, G. A., Khanna, N., and Belmont, A. S. (2020). Gene Expression Amplification by Nuclear Speckle Association. J. Cel Biol 219, 201904046. doi:10.1083/jcb.201904046
Kornblihtt, A. R., De La Mata, M., Fededa, J. P., Muñoz, M. J., and Nogués, G. (2004). Multiple Links between Transcription and Splicing. RNA 10, 1489–1498. doi:10.1261/rna.7100104
Le Noir, S., Boyer, F., Lecardeur, S., Brousse, M., Oruc, Z., Cook-Moreau, J., et al. (2017). Functional Anatomy of the Immunoglobulin Heavy Chain 3΄ Super-enhancer Needs Not Only Core Enhancer Elements but Also Their Unique DNA Context. Nucleic Acids Res. 45, 5829–5837. doi:10.1093/nar/gkx203
López-Farfán, D., Bart, J.-M., Rojas-Barros, D. I., and Navarro, M. (2014). SUMOylation by the E3 Ligase TbSIZ1/PIAS1 Positively Regulates VSG Expression in Trypanosoma Brucei. Plos Pathog. 10, e1004545. doi:10.1371/journal.ppat.1004545
Maita, H., and Nakagawa, S. (2020). What Is the Switch for Coupling Transcription and Splicing? RNA Polymerase II C-Terminal Domain Phosphorylation, Phase Separation and beyond. Wiley Interdiscip. Rev. RNA 11, e1574. doi:10.1002/wrna.1574
Matthews, K. R., Tschudi, C., and Ullu, E. (1994). A Common Pyrimidine-Rich Motif Governs Trans-splicing and Polyadenylation of Tubulin Polycistronic Pre-mRNA in Trypanosomes. Genes Dev. 8, 491–501. doi:10.1101/gad.8.4.491
Maudlin, I. E., Kelly, S., Schwede, A., and Carrington, M. (2021). VSG mRNA Levels Are Regulated by the Production of Functional VSG Protein. Mol. Biochem. Parasitol. 241, 111348. doi:10.1016/j.molbiopara.2020.111348
Meier, U. T. (2017). RNA Modification in Cajal Bodies. RNA Biol. 14, 693–700. doi:10.1080/15476286.2016.1249091
Melo Do Nascimento, L., Egler, F., Arnold, K., Papavasiliou, N., Clayton, C., and Erben, E. (2021). Functional Insights from a Surface Antigen mRNA-Bound Proteome. Elife 10, e68136. doi:10.7554/eLife.68136
Morris, G. E. (2008). The Cajal Body. Biochim. Biophys. Acta (Bba) - Mol. Cel Res. 1783, 2108–2115. doi:10.1016/j.bbamcr.2008.07.016
Müller, L. S. M., Cosentino, R. O., Förstner, K. U., Guizetti, J., Wedel, C., Kaplan, N., et al. (2018). Genome Organization and DNA Accessibility Control Antigenic Variation in Trypanosomes. Nature 563, 121–125. doi:10.1038/s41586-018-0619-8
Narayanan, M. S., and Rudenko, G. (2013). TDP1 Is an HMG Chromatin Protein Facilitating RNA Polymerase I Transcription in African Trypanosomes. Nucleic Acids Res. 41, 2981–2992. doi:10.1093/nar/gks1469
Navarro, M., and Gull, K. (2001). A Pol I Transcriptional Body Associated with VSG Mono-Allelic Expression in Trypanosoma Brucei. Nature 414, 759–763. doi:10.1038/414759a
Nerusheva, O. O., Ludzia, P., and Akiyoshi, B. (2019). Identification of Four Unconventional Kinetoplastid Kinetochore Proteins KKT22-25 in Trypanosoma Brucei. Open Biol. 9, 190236. doi:10.1098/rsob.190236
Nguyen, T. N., Müller, L. S. M., Park, S. H., Siegel, T. N., and Günzl, A. (2014). Promoter Occupancy of the Basal Class I Transcription Factor A Differs Strongly between Active and Silent VSG Expression Sites in Trypanosoma Brucei. Nucleic Acids Res. 42, 3164–3176. doi:10.1093/nar/gkt1301
Park, S.-K., Xiang, Y., Feng, X., and Garrard, W. T. (2014). Pronounced Cohabitation of Active Immunoglobulin Genes from Three Different Chromosomes in Transcription Factories during Maximal Antibody Synthesis. Genes Dev. 28, 1159–1164. doi:10.1101/gad.237479.114
Pellacani, C., Bucciarelli, E., Renda, F., Hayward, D., Palena, A., Chen, J., et al. (2018). Splicing Factors Sf3A2 and Prp31 Have Direct Roles in Mitotic Chromosome Segregation. Elife 7. doi:10.7554/eLife.40325
Pena, A. C., Aresta-Branco, F., and Figueiredo, L. M. (2017). “Epigenetic Regulation in T. Brucei: Changing coats Is a Chance to Survive,” in Epigenetics of Infectious Diseases. Editors W. Doerfler, and J. Casadesus (Springer International Publishing), 221–241. doi:10.1007/978-3-319-55021-3_10
Ridewood, S., Ooi, C. P., Hall, B., Trenaman, A., Wand, N. V., Sioutas, G., et al. (2017). The Role of Genomic Location and Flanking 3'UTR in the Generation of Functional Levels of Variant Surface Glycoprotein in Trypanosoma Brucei. Mol. Microbiol. 106 (4), 614–634. doi:10.1111/mmi.13838
Rothé, B., Back, R., Quinternet, M., Bizarro, J., Robert, M.-C., Blaud, M., et al. (2014). Characterization of the Interaction between Protein Snu13p/15.5K and the Rsa1p/NUFIP Factor and Demonstration of its Functional Importance for snoRNP Assembly. Nucleic Acids Res. 42, 2015–2036. doi:10.1093/nar/gkt1091
Sawyer, I. A., Sturgill, D., Sung, M.-H., Hager, G. L., and Dundr, M. (2016). Cajal Body Function in Genome Organization and Transcriptome Diversity. Bioessays 38, 1197–1208. doi:10.1002/bies.201600144
Schimanski, B., Nguyen, T. N., and Gunzl, A. (2005). Characterization of a Multisubunit Transcription Factor Complex Essential for Spliced-Leader RNA Gene Transcription in Trypanosoma Brucei. Mol. Cel Biol 25, 7303–7313. doi:10.1128/mcb.25.16.7303-7313.2005
Sheader, K., Vaughan, S., Minchin, J., Hughes, K., Gull, K., and Rudenko, G. (2005). Variant Surface Glycoprotein RNA Interference Triggers a Precytokinesis Cell Cycle Arrest in African Trypanosomes. Proc. Natl. Acad. Sci. U.S.A. 102, 8716–8721. doi:10.1073/pnas.0501886102
Shi, W., Liao, Y., Willis, S. N., Taubenheim, N., Inouye, M., Tarlinton, D. M., et al. (2015). Transcriptional Profiling of Mouse B Cell Terminal Differentiation Defines a Signature for Antibody-Secreting Plasma Cells. Nat. Immunol. 16, 663–673. doi:10.1038/ni.3154
Shin, Y., and Brangwynne, C. P. (2017). Liquid Phase Condensation in Cell Physiology and Disease. Science 357, eaaf4382. doi:10.1126/science.aaf4382
Shin, Y., Chang, Y.-C., Lee, D. S. W., Berry, J., Sanders, D. W., Ronceray, P., et al. (2018). Liquid Nuclear Condensates Mechanically Sense and Restructure the Genome. Cell 175, 1481–1491. doi:10.1016/j.cell.2018.10.057
Smith, K. P., Carter, K. C., Johnson, C. V., and Lawrence, J. B. (1995). U2 and U1 snRNA Gene Loci Associate with Coiled Bodies. J. Cel. Biochem. 59, 473–485. doi:10.1002/jcb.240590408
Somma, M. P., Andreyeva, E. N., Pavlova, G. A., Pellacani, C., Bucciarelli, E., Popova, J. V., et al. (2020). Moonlighting in Mitosis: Analysis of the Mitotic Functions of Transcription and Splicing Factors. Cells 9, 1554. doi:10.3390/cells9061554
Tellier, M., Maudlin, I., and Murphy, S. (2020). Transcription and Splicing: A Two-Way Street. Wiley Interdiscip. Rev. RNA 11, e1593. doi:10.1002/wrna.1593
Tucker, K. E., Berciano, M. T., Jacobs, E. Y., Lepage, D. F., Shpargel, K. B., Rossire, J. J., et al. (2001). Residual Cajal Bodies in Coilin Knockout Mice Fail to Recruit Sm snRNPs and SMN, the Spinal Muscular Atrophy Gene Product. J. Cel Biol 154, 293–308. doi:10.1083/jcb.200104083
Vanhamme, L., Poelvoorde, P., Pays, A., Tebabi, P., Van Xong, H., and Pays, E. (2000). Differential RNA Elongation Controls the Variant Surface Glycoprotein Gene Expression Sites of Trypanosoma Brucei. Mol. Microbiol. 36, 328–340. doi:10.1046/j.1365-2958.2000.01844.x
Viegas, I. J., De Macedo, J. P., De Niz, M., Rodrigues, J. A., Aresta-Branco, F., Jaffrey, S. R., et al. (2020). N6-methyladenosine in Poly(A) Tails stabilizeVSGtranscripts. bioRxiv. doi:10.1101/2020.01.30.925776
Wang, Q., Sawyer, I. A., Sung, M.-H., Sturgill, D., Shevtsov, S. P., Pegoraro, G., et al. (2016). Cajal Bodies Are Linked to Genome Conformation. Nat. Commun. 7, 10966. doi:10.1038/ncomms10966
Keywords: variant surface glycoprotein, antigenic variation, nuclear bodies, trans-splicing, Trypanosoma brucei, nuclear architecture
Citation: Budzak J and Rudenko G (2022) Pedal to the Metal: Nuclear Splicing Bodies Turbo-Charge VSG mRNA Production in African Trypanosomes. Front. Cell Dev. Biol. 10:876701. doi: 10.3389/fcell.2022.876701
Received: 15 February 2022; Accepted: 18 March 2022;
Published: 20 April 2022.
Edited by:
Luisa M. Figueiredo, Universidade de Lisboa, PortugalReviewed by:
Susanne Kramer, Julius Maximilian University of Würzburg, GermanyJoana Raquel Correia Faria, University of York, United Kingdom
Copyright © 2022 Budzak and Rudenko. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gloria Rudenko, Z2xvcmlhLnJ1ZGVua29AaW1wZXJpYWwuYWMudWs=
 James Budzak
James Budzak Gloria Rudenko
Gloria Rudenko