- Department of Biology, University of Rochester, Rochester, NY, United States
Epithelial tissues are vital to the function of most organs, providing critical functions such as secretion, protection, and absorption. Cells within an epithelial layer must coordinate to create functionally distinct apical, lateral, and basal surfaces in order to maintain proper organ function and organism viability. This is accomplished through the careful targeting of polarity factors to their respective locations within the cell, as well as the strategic placement of post-mitotic cells within the epithelium during tissue morphogenesis. The process of establishing and maintaining epithelial tissue integrity is conserved across many species, as important polarity factors and spindle orientation mechanisms can be found in many phyla. However, most of the information gathered about these processes and players has been investigated in bilaterian organisms such as C. elegans, Drosophila, and vertebrate species. This review discusses the advances made in the field of epithelial polarity establishment from more basal organisms, and the advantages to utilizing these simpler models. An increasing number of cnidarian model organisms have been sequenced in recent years, such as Hydra vulgaris and Nematostella vectensis. It is now feasible to investigate how polarity is established and maintained in basal organisms to gain an understanding of the most basal requirements for epithelial tissue morphogenesis.
Introduction
Cell polarity defines specific spatial and functional domains within a cell through the asymmetric positioning of cellular components such as proteins, organelles, and cytoskeletal components. Epithelia are polarized tissues that perform specialized functions, typically at the boundary between an organ and the external environment. Cell polarity establishment and maintenance is vital for these functions; directional processes such as secretion, nutrient uptake, and signaling require a defined apical and basal surface to occur successfully (Humbert et al., 2008; St Johnston and Ahringer 2010). The importance of epithelial polarity is also highlighted by the observation that its loss is a common feature of malignancy (Bergstralh and St Johnston 2012; Williams et al., 2017; Jung et al., 2019; Tenvooren et al., 2019; Catterall, Lelarge, and McCaffrey 2020; Che et al., 2021; Tilston-Lunel et al., 2021).
For decades, studies of epithelialization and polarity establishment have focused largely on well-established bilaterian models, namely C. elegans, Drosophila, and mammalian systems. As with all biological model systems, however, each of these organisms comes with its own set of technical and genetic caveats. In this brief article we highlight the utility of cnidarian animals to study the process of epithelial polarity establishment and maintenance (Figure 1). We argue that these animals represent a promising yet under-utilized class of model organisms. Cnidaria and Bilateria are both phyla under the larger Eumetazoan subkingdom. Consistent with previous work, we show here that cnidarians share polarity establishment factors with bilaterians, within a simpler body plan. They also possess regenerative capabilities and a robust ability to reorganize upon dissociation, furthering their potential to push the field of polarity establishment and maintenance forward.
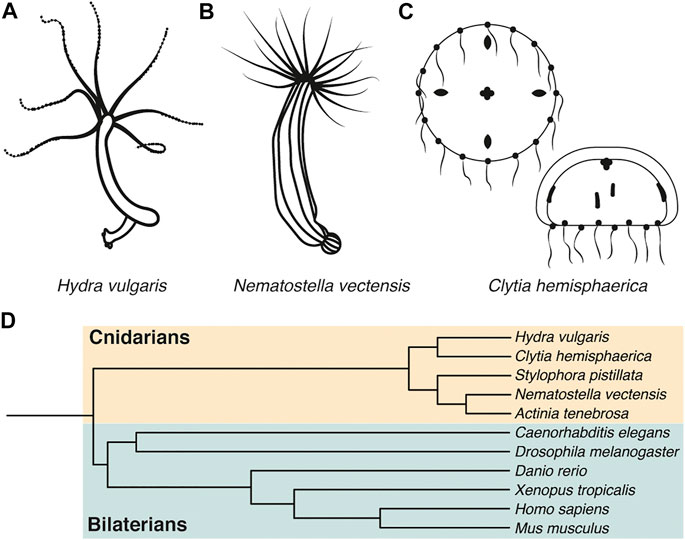
FIGURE 1. H. vulgaris, N. vectensis, and C. hemisphaerica are emerging model systems for polarity studies. (A-C) Representative drawings of Hydra vulgaris, Nematostella vectensis, and Clytia hemisphaerica. (D) Evolutionary tree depicting cnidarian and bilaterian model organisms.
Advantages to Phylogenetically Basal Model Organisms and Examples
There are some significant technical advantages to cnidarian models, an example of basal metazoans that diverged from bilaterians millions of years ago. Firstly, these simple organisms have robust regeneration capabilities that can be harnessed for use in studying polarity and cell sorting mechanisms (Seybold, Salvenmoser, and Hobmayer 2016; Cochet-Escartin et al., 2017; Skokan, Vale, and McKinley 2020). Hydra vulgaris and other cnidarians can completely regenerate from dissected tissue over the span of several days (Tucker and Adams 2014). Cnidarians are also able to reassemble from a completely dissociated cell suspension after mechanical or enzymatic dissociation (Tucker and Adams 2014; Cochet-Escartin et al., 2017). This provides an opportunity to follow the process of polarity establishment from a dissociated group of cells to a functional multicellular tissue in an in vivo animal context. This has previously been accomplished in polarized cell culture models such as MDCK cysts (Rodriguez-Boulan, Kreitzer, and Musch 2005; Martin-Belmonte et al., 2007; Mellman and Nelson 2008; Bryant et al., 2010) and three-dimensional organoid culture systems (Li et al., 2018; Serra et al., 2019; Lukonin et al., 2020; Rosenbluth et al., 2020; Schuster et al., 2020; Hendriks et al., 2021), however these models would not develop into a fully functional organism like an animal model such as Hydra vulgaris.
Cnidarian model systems can also be tailored to investigate how polarity establishment mechanisms differ between tissues. In addition to total dissociation protocols, there are established techniques to isolate particular tissues for site-specific studies. For example, Hydra mesoglea has been isolated through a detergent extraction and freezing protocol in order to investigate the extracellular matrix proteins within (Tucker and Adams 2014). Since extracellular matrix proteins can influence polarity establishment (reviewed in (Manninen 2015)), this protocol could be used to determine how extracellular matrix influences polarity establishment in Hydra, and possibly modified to investigate additional structures within the Hydra. Additionally, primary cell cultures can be created from cnidarian tissues for more in-depth studies, such as those generated from the cnidarian Anemonia viridis for use in tissue-specific and pluripotency marker expression studies (Ventura et al., 2018), providing another manner in which to study polarity establishment in specific cnidarian tissues.
Lastly, cnidarians such as Hydra vulgaris (Chapman et al., 2010; Siebert et al., 2019), the anemone Nematostella vectensis (Putnam et al., 2007; Sebe-Pedros et al., 2018), and jellyfish Clytia hemisphaerica (Leclere et al., 2019) have been genetically sequenced and/or transcriptionally characterized. This important factor increases the number of genetic tools available for use in these organisms such as CRISPR knockout technology (Ikmi et al., 2014; Lommel et al., 2017; Momose et al., 2018). Components of the Par, Crumbs, and Scribble polarity complexes have been identified and characterized in cnidarian organisms (Figures 1–4), and their high level of conservation with bilaterian systems suggests that studies in cnidarian model systems could contribute to the field of polarity establishment.
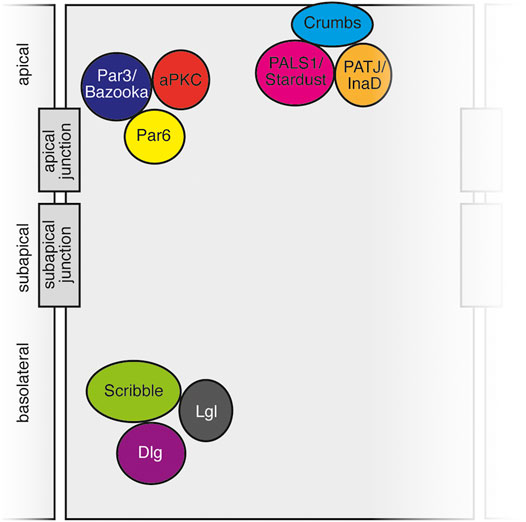
FIGURE 2. Schematic of polarity complexes in vertebrate and invertebrate cells. Apical Crumbs complex, subapical Par complex, and basolateral Scribble complex depicted with apical junctions (vertebrate tight junctions, invertebrate septate junctions) and subapical junctions (adherens junctions) denoted.
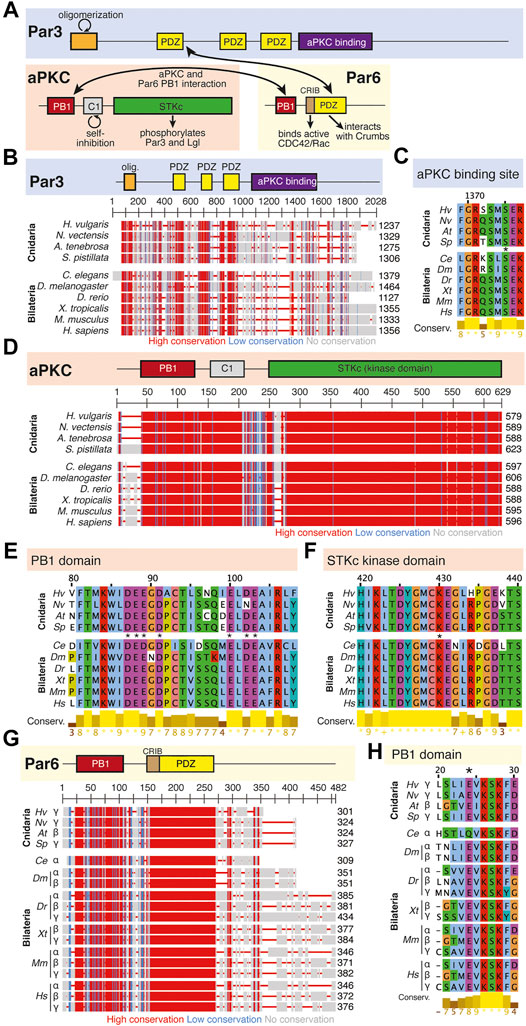
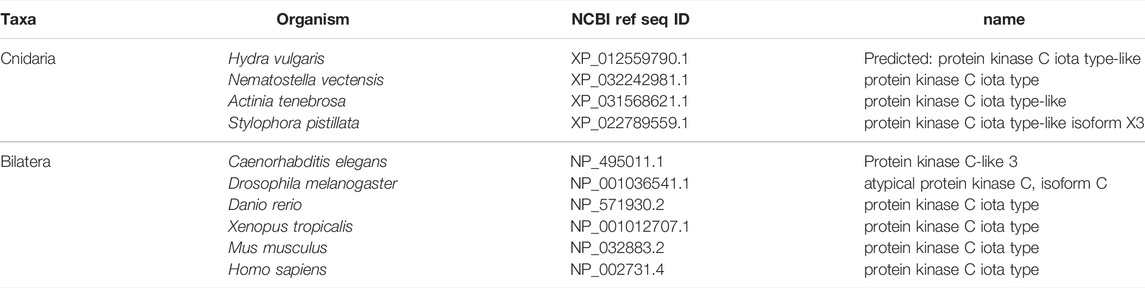
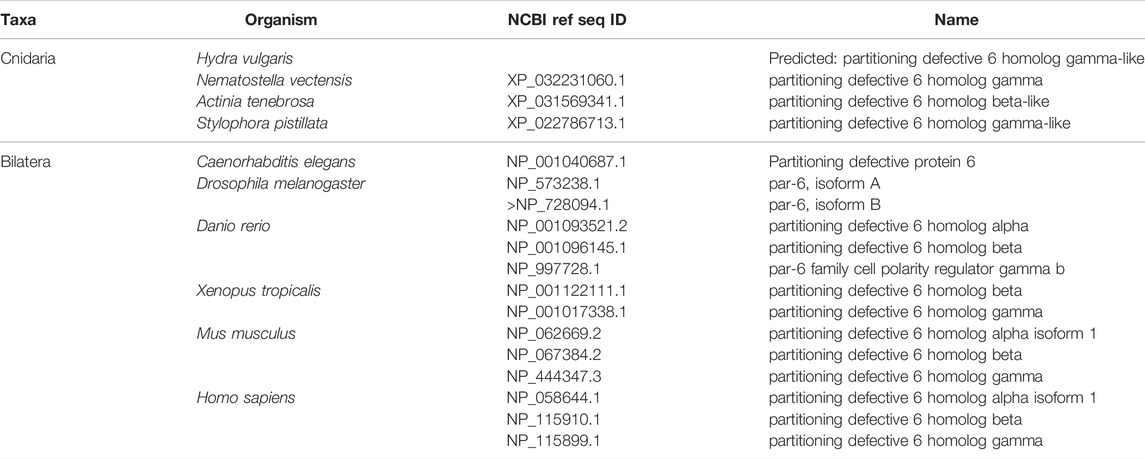
FIGURE 3. The Par complex defines the subapical domain and is conserved across bilaterians and cnidarians. (A) Representation of the human Par complex with Par3, Par6, and aPKC pictured. Functional domains and examples of protein-protein interactions denoted. (B) Full protein alignment of Par3 with functional domains depicted in their respective positions. (C) Amino acid alignment of the aPKC binding region of Par3. (D) Full protein alignment of aPKC with functional domains depicted in their respective positions. (E-F) Amino acid alignment of aPKC PB1 domain (E) and STKc kinase domain (F). Critical residues marked with asterisk. (G) Full protein alignment of Par6 with functional domains depicted in their respective positions. (H) Amino acid alignment of Par6 PB1 domain. Critical residues marked with asterisk. For all alignments: COBALT used for all protein alignments. High (red), low (blue), and no conservation (gray) regions denoted. FASTA sequences from COBALT visualized using JalView for amino acid alignments. Hydrophobic (blue), positively charged (red), negatively charged (magenta), aromatic (cyan), and polar (green) amino acids denoted by color, as well as cysteines (pink), glycines (orange), prolines (yellow). Conservation denoted on bottom of alignment. Cnidarians: Hydra vulgaris (Hv), Nematostella vectensis (Nv), Actinia tenebrosa (At), Stylophora pistillata (Sp). Bilaterians: Caenorhabditis elegans (Ce), Drosophila melanogaster (Dm), Danio rerio (Dr), Xenopus tropicalis (Xt), Mus musculus (Mm), Homo sapiens (Hs). Refer to Tables 1–3 for information regarding sequences used in this figure.
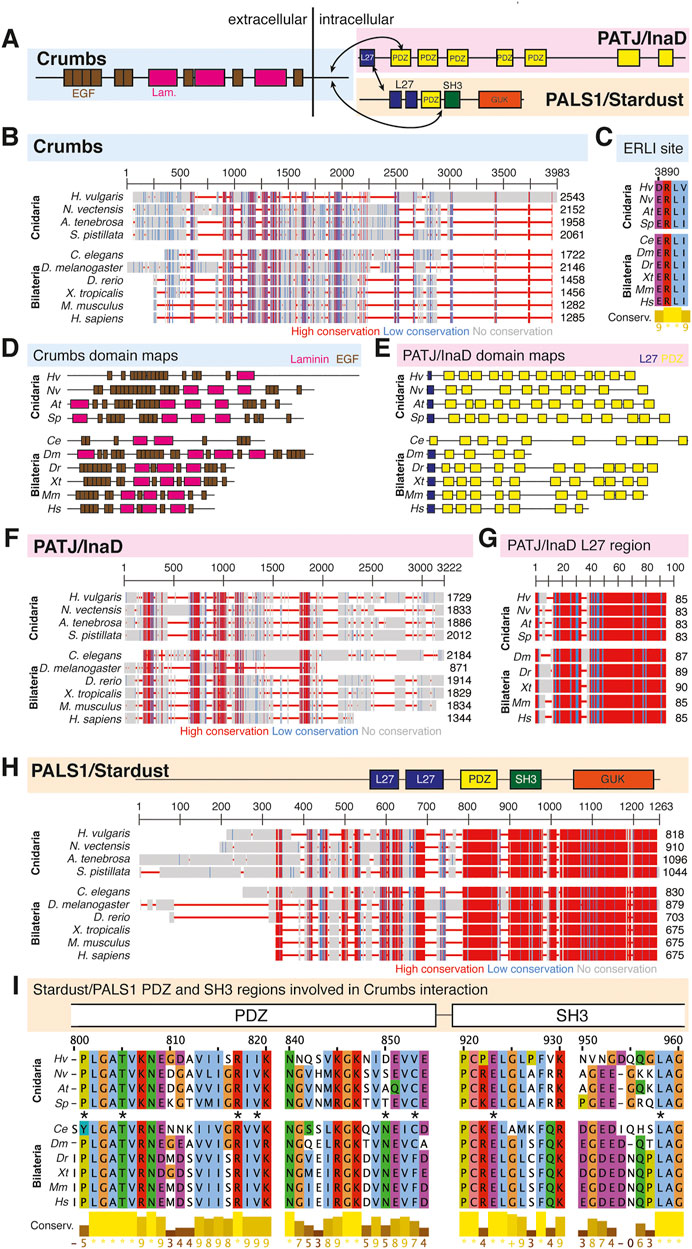
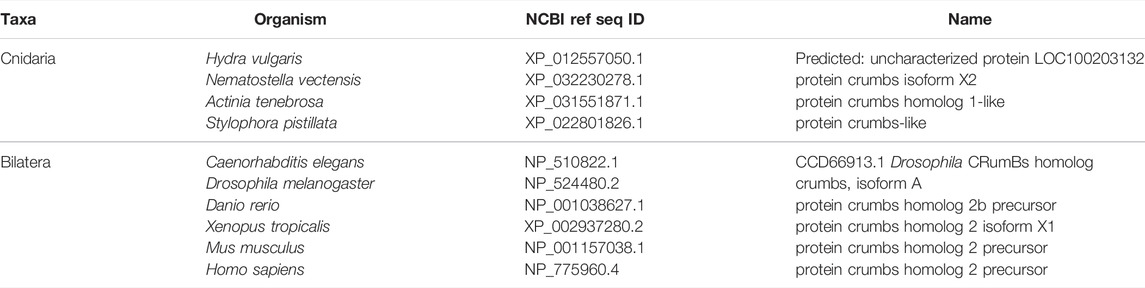
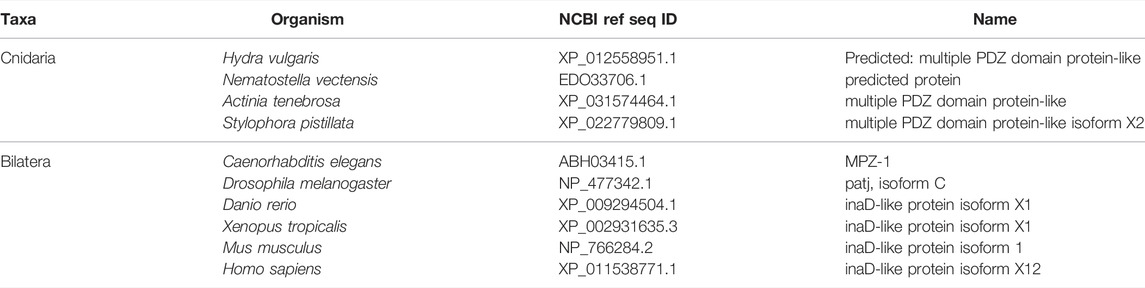
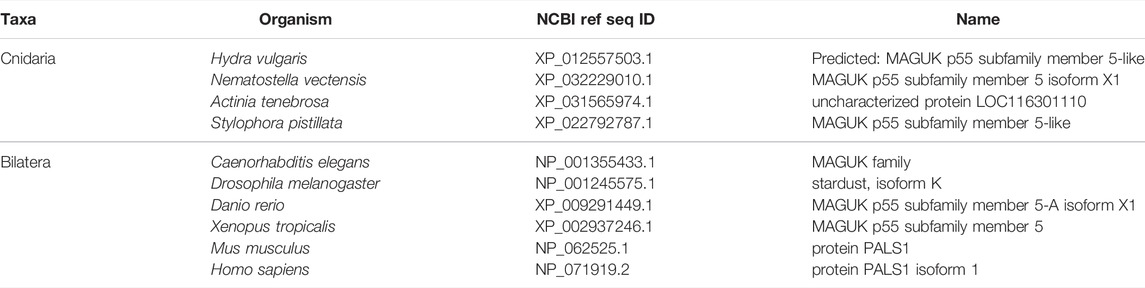
FIGURE 4. The Crumbs complex defines the apical domain and is conserved across bilaterians and cnidarians. (A) Representation of the human Crumbs complex with Crumbs, PALS1/Stardust, PATJ/InaD pictured. Functional domains and examples of protein-protein interactions denoted. (B) Full protein alignment of Crumbs. (C) Amino acid alignment of the ERLI motif at the Crumbs C-terminus. (D-E) Domains maps of Crumbs (D) and PATJ/InaD (E) depicting laminin (magenta, D), EGF (brown, D), L27 (navy, E), and PDZ (yellow, E) domain positions. Protein lengths drawn to scale. (F) Full protein alignment of PATJ/InaD. (G) Protein alignment of PATJ/InaD L27 domain magnified from (F). (H) Full protein alignment of PALS1/Stardust with functional domains depicted in their respective positions. (I) Amino acid alignment of regions within PALS1/Stardust PDZ and SH3 domains. Critical residues involved in binding Crumbs marked with asterisk. For all alignments: COBALT used for all protein alignments. High (red), low (blue), and no conservation (gray) regions denoted. FASTA sequences from COBALT visualized using JalView for amino acid alignments. Hydrophobic (blue), positively charged (red), negatively charged (magenta), aromatic (cyan), and polar (green) amino acids denoted by color, as well as cysteines (pink), glycines (orange), prolines (yellow). Conservation denoted on bottom of alignment. Cnidarians: Hydra vulgaris (Hv), Nematostella vectensis (Nv), Actinia tenebrosa (At), Stylophora pistillata (Sp). Bilaterians: Caenorhabditis elegans (Ce), Drosophila melanogaster (Dm), Danio rerio (Dr), Xenopus tropicalis (Xt), Mus musculus (Mm), Homo sapiens (Hs). Refer to Tables 4–6 for information regarding sequences used in this figure.
Beyond technical advantages, choosing to utilize a cnidarian or other basal model organism provides a simplified setting to investigate research questions within the context of a whole, functional animal. For example, Hydra vulgaris features a body plan with two main tissue layers, the ectoderm and endoderm, separated by the mesoglea and interstitial cells. This provides a setting to study processes such as tissue morphogenesis (Hicklin and Wolpert 1973; Maroudas-Sacks et al., 2021), cell differentiation and lineage tracing (Plickert and Kroiher 1988; Takashima, Gold, and Hartenstein 2013; Siebert et al., 2019), and asexual budding mechanisms (Clarkson and Wolpert 1967; Webster and Hamilton 1972) in a simplified, yet multi-tissue, context. Within the wide spectrum of biological research models to choose from, cnidarians and other basal animals represent an important niche between ex vivo cultured systems and more complicated in vivo bilaterian model organisms such as mouse, zebrafish, or Drosophila.
Cnidarians occupy an advantageous position within the evolutionary tree in a group historically referred to as Epitheliozoa. This subset of organisms is comprised of bilaterians, cnidarians, ctenophores, and placozoans, all of which are considered animals that have “true tissues” (Ax 1995). The exact definition of an epitheliozoan seems to be controversial in the literature. Some sources cite the presence of belt desmosomes as a requirement for this grouping (Ax 1995; Dohrmann and Worheide 2013), while others define “true tissues” as those that are connected by tight junctions (septate junctions in invertebrates) (Ganot et al., 2015). It is also debated whether they have the appropriate proteins or the ability to properly build these junctional complexes (Sperling, Peterson, and Pisani 2009). For example, sponges can be excluded since they do not have proper belt desmosomes (Leys and Riesgo 2012). Additionally, poriferans only have one tissue type within their body, raising the argument that they cannot have true epithelial tissues if they are not creating barriers between different tissue types. While the poriferan group is controversial when it comes to its relationship to epitheliozoans, cnidarians have been placed in this clade through a variety of genomic and phenotypic analyses (Ax 1995; Zrzavy et al., 1998; Sperling, Peterson, and Pisani 2009; Dohrmann and Worheide 2013; Ganot et al., 2015). Epithelia are therefore an ancient characteristic and important enough in biological evolution that a phylogenetic grouping has been created for organisms with epithelial tissues. Within this group, cnidarians are a more basal group of organisms, making them an option to study epithelial polarity establishment and maintenance in a simple epitheliozoan.
Polarity and Epithelialization Studies in Cnidarian Models
Mechanisms controlling planar cell polarity are conserved in cnidarians. Briefly, the core planar cell polarity pathway involves the asymmetric distribution of several critical proteins to distinguish one side of the cell from the other along the larger-scale axis of the tissue and embryo. Some of these proteins include Frizzled, Strabismus/Van Gogh, Flamingo, Dishevelled, Prickle, and Diego (Devenport 2014). These evolutionarily conserved proteins are required for proper morphogenesis and development in several cnidarian species. For example, Strabismus, Frizzled, and Dishevelled are required for Nematostella vectensis invagination (Kumburegama, Wijesena, and Wikramanayake 2008; Kumburegama et al., 2011; Wijesena and Martindale 2018; Technau 2020; Wijesena et al., 2022), ciliated epithelium development in Clytia hemispherica (Momose and Houliston 2007; Momose, Derelle, and Houliston 2008; Momose, Kraus, and Houliston 2012; Lapebie et al., 2014), and tissue evagination dependent on Strabismus, Dishevelled, and Frizzled in Hydra vulgaris (Philipp et al., 2009). Additionally, the Fat-Dachsous polarity pathway utilizes an asymmetry in the localization of the protocadherins Fat and Dachsous to create cellular and tissue polarity (Devenport 2014), and these components have been similarly identified and studied in Hydra and Nematostella (Magie and Martindale 2008; Hulpiau and van Roy 2011; Tucker and Adams 2014; Gul et al., 2017). While fewer studies concern the mechanisms driving apicobasal polarity cnidarians, the conservation of PCP factors suggests that apicobasal polarity complexes and mechanisms are also likely to be conserved in cnidarians.
Hydra vulgaris Hydra are the oldest cnidarian model system used in research (Figure 1A), first studied by Abraham Trembley in 1744 (Trembley, 1744). The Hydra body column is largely comprised of two epithelial cell layers, the ectoderm and endoderm, with various types of interstitial cells positioned between. Additional cell types are interspersed between the epithelial cells, including gland cells in endoderm, nematocytes and nematoblasts in the ectoderm, and various neural cell types found in both layers (Technau and Steele 2011). The mesoglea is positioned between the two layers, composed of a thick extracellular matrix secreted by the endoderm and ectoderm (Epp, Smid, and Tardent 1986; Sarras et al., 1993). Basal myofibrils present in both cell layers run in orthogonal directions, with those in the endoderm running circumferentially around the body column, and those of the ectoderm running longitudinally along the body axis (Otto 1977). The endoderm is sometimes referred to as the gastrodermis due to the presence of gland cells that aid in digestion. Hydra is even referred to as a “living gut” due to their simple digestive system that spans the body column, and therefore much of the Hydra body plan in general (Vogg, Galliot, and Tsiairis 2019). Hydra feature a robust asexual reproduction process called budding, which has been used to study developmental programming and head determination. Epithelialization is an important process in both budding and regeneration, which is one of the main reasons that Hydra is a promising model for the study of polarity establishment and cell sorting (Seybold, Salvenmoser, and Hobmayer 2016).
One of the main advantages of the Hydra model system is their ability to reassemble after dissociation and subsequent reaggregation. This reaggregation can occur with minimal cell numbers, as only approximately 5–15 cells are necessary to create a head organizer within the population, allowing for a full Hydra to grow out of the small cell cluster (Technau et al., 2000). The patterning mechanism that defines the head, foot, and tentacle regions of the Hydra have been modelled using simulations such as the Meinhardt reaction-diffusion model, where these regions positioned with respect to one another by gradients of inhibitory or activating signals that determine the Hydra body axis (MacWilliams 1982; Meinhardt 1993).
Hydra reaggregation experiments have been used to understand how cells sort within a mixed population. Such studies suggest that factors such as the capacity for epithelialization (Skokan, Vale, and McKinley 2020), cell surface tension (Cochet-Escartin et al., 2017), interfacial tension and cell-cell adhesiveness (Technau and Holstein 1992) dictate how cells will sort into layers within an aggregate. Ectodermal engulfment of the endoderm in Hydra is compared to the morphogenetic process of epiboly in other organisms, where epithelial tissue undergoes a spreading process in the early development of vertebrates such as fish and amphibians (Piccolo 2013). Similarly, the Hydra ectodermal cells engulfed the endodermal cell cluster when they came into contact, reforming the bilayer epithelium that is typically found in Hydra (Kishimoto, Murate, and Sugiyama 1996). As shown in these examples, the ability to completely dissociate Hydra tissues provides a setting to study the physical and biochemical characteristics of these cells to determine how they will behave when combined in a tissue with other cell populations, and how that multicellular tissue behaves as a whole during development.
Epithelialization as it relates to cell-cell adhesion establishment has also been extensively studied using Hydra reaggregation experiments. The process of cell-cell adhesion reestablishment was documented through electron microscopy to determine the order in which adhesion complexes are created. In this context, the apical-basal axis of developing epithelial cells elongates, followed by septate junction, gap junction, mesoglea, and hemidesmosome-like junction development. Interestingly, the process of planar cell polarity establishment begins before apical-basal polarity establishment has completed (Seybold, Salvenmoser, and Hobmayer 2016). Apicobasal polarity establishment both influences and is influenced by the setup of cell-cell adhesions, and these molecular players have been identified and studied in Hydra (Buzgariu et al., 2015).
Nematostella vectensis The sea anemone Nematostella vectensis is one of the more commonly utilized cnidarian model systems outside of hydroids (Figure 1B). Nematostella was the first cnidarian to be genetically sequenced and while it has a relatively small genome, there are remarkable similarities to the genomes of humans and other bilaterian vertebrates (Putnam et al., 2007). For example, almost half of 27,000 predicted protein coding transcripts have clear orthologs to protostomes, deuterostomes, or both, and the number of exons and splice sites are nearly identical to humans (Tucker and Adams 2014). Interestingly, a comprehensive single-cell analysis of whole Nematostella animals found that many genetic similarities are shared between cnidarians and vertebrate bilaterians that are not present in invertebrate bilaterians, which include common model systems such as C. elegans and Drosophila (Putnam et al., 2007; Sebe-Pedros et al., 2018). For example, DNA methylation is absent in both Drosophila and C. elegans but occurs in Nematostella and other invertebrate organisms (Feng et al., 2010; Zemach et al., 2010; Zemach and Zilberman 2010; Schwaiger et al., 2014).
Synchronous cell divisions increase the cell mass of the embryo during early Nematostella development. The localization of Par3/Bazooka and Par6 oscillates in time with these cell divisions, moving to cell surfaces and cell-cell interfaces between divisions and away from this location during divisions (Ragkousi et al., 2017; Doerr and Ragkousi 2019). Par6 localizes to the apical cortex during interphase, and Par3/Bazooka can be found at subapical cell-cell contacts during this time. However, consistent with previous work in Drosophila (Bergstralh, Lovegrove, and St Johnston 2013), neither protein is detected at these sites during mitosis (Ragkousi et al., 2017; Doerr and Ragkousi 2019). This points to a mechanism that can quickly dismantle and reestablish epithelial polarity between cell divisions to preserve epithelial integrity during tissue growth. Interestingly, components of the Par system are not present in the endomesodermal epithelial tissue during gastrulation in Nematostella, despite being present in blastula cells earlier in development (Salinas-Saavedra et al., 2015; Salinas-Saavedra, Rock, and Martindale 2018; Salinas-Saavedra and Martindale 2020). This coincides with the absence of adherens junctions in this same tissue, suggesting different mechanisms of cell adhesion in these adjacent tissues (Salinas-Saavedra, Rock, and Martindale 2018). This loss of both cell polarity and cell-cell adhesions points to EMT occurring in this specific population of cells during this stage of development (Whiteman et al., 2008; Lim and Thiery 2012). Furthermore, apical polarity proteins such as Par1, Par3/Bazooka, Par6, aPKC, and Lgl only become asymmetrically distributed to their respective membrane domains later on in development, whereas they are localized along the cytoplasm and microtubule cytoskeleton during early developmental stages (Salinas-Saavedra et al., 2015).
Additional studies have determined the role of cadherins during Nematostella tissue morphogenesis. The cadherin-catenin complex is conserved in Nematostella and is required at the adherens junctions for proper embryo development and later tissue morphogenesis (Clarke et al., 2016; Nathaniel Clarke, Lowe, and James Nelson 2019). For example, Cadherin1 and Cadherin3 are expressed at different times during development, marking the transition from blastoderm to distinct germ layer formation. Disruption of this cadherin expression pattern results in a loss of tissue integrity and improper embryogenesis (Pukhlyakova et al., 2019).
Clytia hemisphaerica The jellyfish Clytia hemisphaerica (Figure 1C) is an increasingly popular cnidarian model for the study of tissue development, wound healing, and regeneration (Kamran et al., 2017; Malamy and Shribak 2018; Kraus, Chevalier, and Houliston 2020; Sinigaglia et al., 2020). Its genome has been recently sequenced, and previous studies have also characterized the Clytia transcriptome through single cell RNA-sequencing (Leclere et al., 2019; Chari et al., 2021). Clytia are frequently used to study wound healing, a component of which is epithelialization. The epithelial layer covering the surface of Clytia medusa is composed of a flat, squamous monolayer, allowing for imaging with DIC (differential interference contrast) microscopy (Malamy and Shribak 2018). As a result, the migration of epithelial cells during development, wound healing, and regeneration has been carefully analyzed to determine the physical steps and chemical mechanisms involved (Malamy and Shribak 2018; Kraus, Chevalier, and Houliston 2020; Sinigaglia et al., 2020). These instances of epithelial-to-mesenchymal transitions, as well as the dedifferentiation events that occur in other jellyfish species (Lin, Grigoriev, and Spencer 2000; Estephane and Anctil 2010), make Clytia a promising model system for use in the study of both the establishment and breakdown of apical cell polarity. An additional advantage to this system is that Clytia components can be cultured outside the body for additional technical assays. For example, the female gonads of Clytia have been cultured to determine the mRNA gradients required to set up the body axes of the developing animal (Amiel and Houliston 2009).
Studies using Clytia have investigated the intersection between apicobasal polarity establishment and body axis establishment. Rapid synchronous cell divisions during early development increase embryonic cell mass prior to tissue layer specification. After the midblastula stage, cell divisions become asynchronous as cells begin to polarize and adopt a more columnar shape with nuclei that begin to localize towards the apical surface. At this point, ingression and gastrulation begin and the ectoderm and endoderm start to take form. This occurs through a population of cells called bottle cells that undergo EMT (epithelial to mesenchymal transition), change their shape to detach from the epithelial layer into the blastocoel, and adopt a mesenchymal morphology. These bottle cells do not undergo EMT and inward migration at the same time, allowing for many cells at varying stages of this process to be observed and characterized simultaneously. After 24 h post-ingression, these cells have reorganized to create the endodermal epithelial layer inside the developing embryo (Kraus, Chevalier, and Houliston 2020). This creates an opportunity to effectively study the process of polarity establishment in the early Clytia embryo, as well as its reverse process in EMT later during gastrulation.
Apicobasal Polarity Establishment and Maintenance
While the exact mechanism of polarity establishment on a molecular level is not completely understood, there are a few important steps that are consistent across species. Firstly, three cortical polarity complexes (Par, Crumbs, and Scribble) interact to eventually localize properly to their respective domains (Figure 2). This triggers downstream pathways to continue setting up the necessary molecular components at each cellular surface. Secondly, cell-cell adhesions are established to attach cells to one another and provide tissue structural integrity, as well as an avenue for communication and materials transport between cells. This includes tight junctions (known as septate junctions in invertebrates), adherens junctions, and desmosomal junctions. The organization of polarity complexes and creation of cell-cell junctions are interconnected processes, with each process influencing and being influenced by the other (Rodriguez-Boulan and Macara 2014). While this review focuses mainly on the role of the Crumbs, Par, and Scribble complexes, a comprehensive review of cell-cell adhesion molecules has been published by Tepass et al. (2001).
Cell polarity in epithelia is driven by mutual antagonism between cortical factors. Work across multiple systems has demonstrated the importance of at least three conserved protein complexes: the Par (Par6, aPKC, Par3/Bazooka) and Crumbs (Crumbs, Stardust, PATJ) complexes that are found at the apical and subapical surface, respectively, and the Scribble module (Discs Large, Lethal Giant Larvae, Scribble) that is sequestered to the basolateral membrane (Tepass, Theres, and Knust 1990; Knust, Tepass, and Wodarz 1993; Etemad-Moghadam, Guo, and Kemphues 1995; Tabuse et al., 1998; Bilder, Li, and Perrimon 2000; Bilder and Perrimon 2000). While the exact relationships between individual complex members continue to be elucidated, an emerging theme is that polarity is maintained by an exceedingly complex network of mutual antagonism. This also includes proteins outside the canonical Par, Crumbs, and Scribble complexes, such as Yurt (Laprise et al., 2006; Laprise et al., 2009) and Par1 (Benton and St Johnston 2003).
The Par complex includes Par6, Par3/Bazooka, and aPKC (atypical protein kinase C, Figure 3). The spatial relationship between Par proteins is conserved throughout animals, but interactions between the components are complex and historically have been difficult to parse out. In Drosophila, Par3/Bazooka, Par6, and aPKC localize apically (Kuchinke, Grawe, and Knust 1998; Wodarz et al., 2000; Petronczki and Knoblich 2001; Morais-de-Sa, Mirouse, and St Johnston 2010). The size of the apical domain is regulated through a negative feedback mechanism between Crumbs and Yurt, a basolateral protein that is recruited to apical membranes towards the end of epithelial development (Laprise et al., 2006; Laprise et al., 2009).
The Crumbs complex has four known components, Crumbs, Stardust/PALS1 (protein associated with Lin7 1), and PATJ/InaD (PALS1-associated tight junction protein/inactivation no afterpotential D, Figure 4) (Bachmann et al., 2001; Bachmann et al. 2004; Bachmann et al. 2008). Crumbs is partially responsible for the establishment of the apical domain in epithelial cells (Wodarz et al., 1995), and all components of the Crumbs complex are thought to be required for tight junction formation in mammals (Tan et al., 2020).
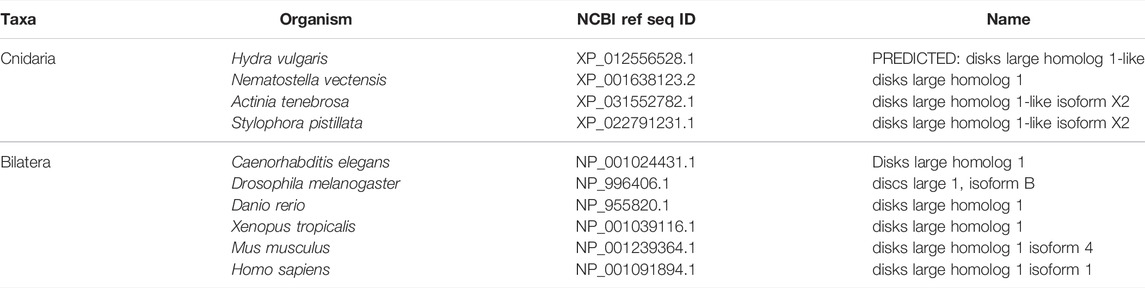
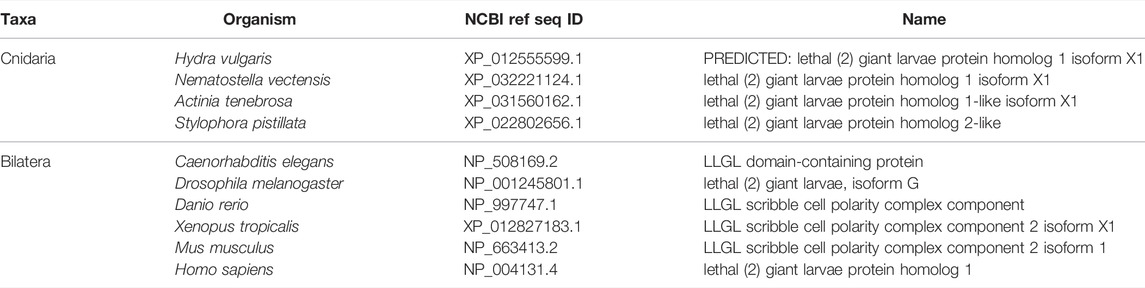
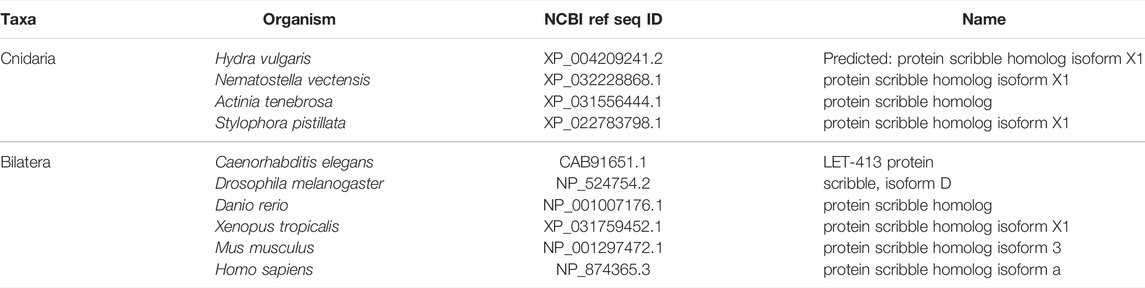
Scribble module factors localize to the basolateral membrane (Figure 5) (Bilder and Perrimon 2000; St Johnston and Ahringer 2010). In the Drosophila follicular epithelium, Discs Large (Dlg) localizes Scribble to the cortex via the Dlg SH3 domain. Lethal Giant Larvae (Lgl) is a known inhibitor of Par complex component aPKC, and this inhibition can occur here through the interaction of Lgl with Dlg and Scribble. This mechanism then confines the Scribble complex components to the basolateral domain, and aPKC along with other Par complex components to the apical domain above (Khoury and Bilder 2020; Ventura et al., 2020). Lgl is involved in mutual antagonism with aPKC, which phosphorylates Lgl to exclude it from the apical cortex. Conversely, Lgl inhibits aPKC from the lateral domain (Lee, Robinson, and Doe 2006; Atwood and Prehoda 2009; Ventura et al., 2020).
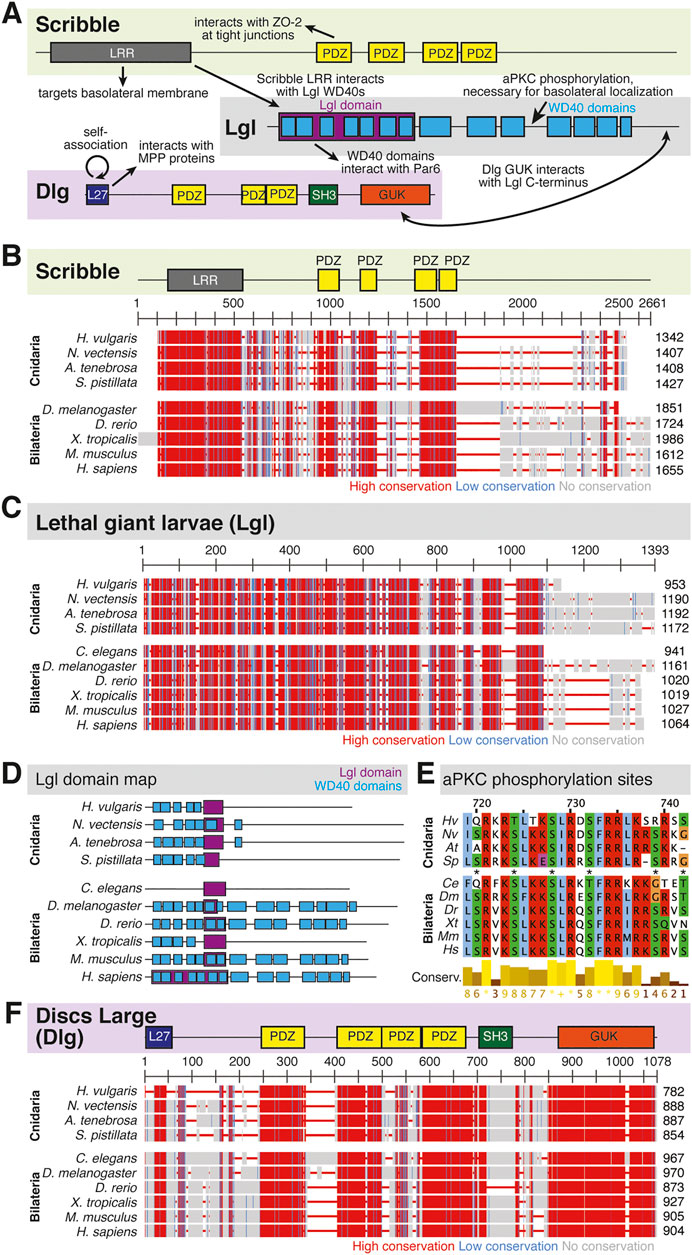
FIGURE 5. The Scribble complex defines the basolateral domain and is conserved across bilaterians and cnidarians. (A) Representation of the human Scribble complex with Scribble, Discs Large (Dlg), and Lethal Giant Larvae (Lgl) pictured. Functional domains and examples of protein-protein interactions denoted. (B,C) Full protein alignments of Scribble (B) and Lethal Giant Larvae (Lgl, C). Scribble functional domains depicted in their respective positions within alignment. (D) Lgl domains maps depicting WD40 (cyan) and Lgl (violet) positions. Protein lengths drawn to scale. (E) Amino acid alignment of Lgl region phosphorylated by aPKC. Critical residues depicted with asterisks. (F) Full protein alignment of Discs Large (Dlg). functional domains depicted in their respective positions within alignment. For all alignments: COBALT used for all protein alignments. High (red), low (blue), and no conservation (gray) regions denoted. FASTA sequences from COBALT visualized using JalView for amino acid alignments. Hydrophobic (blue), positively charged (red), negatively charged (magenta), aromatic (cyan), and polar (green) amino acids denoted by color, as well as cysteines (pink), glycines (orange), prolines (yellow). Conservation denoted on bottom of alignment. Cnidarians: Hydra vulgaris (Hv), Nematostella vectensis (Nv), Actinia tenebrosa (At), Stylophora pistillata (Sp). Bilaterians: Caenorhabditis elegans (Ce), Drosophila melanogaster (Dm), Danio rerio (Dr), Xenopus tropicalis (Xt), Mus musculus (Mm), Homo sapiens (Hs). Refer to Tables 7–9 for information regarding sequences used in this figure.
Cell-cell adhesion molecules also play an important role in polarity establishment and maintenance in cooperation with the three polarity complexes mentioned above. For example, adherens junctions can drive apical polarity establishment (Nejsum and Nelson 2007; Desai, Harmon, and Green 2009) in cells, but cell polarity can also govern adherens junction formation in other settings (Qin et al., 2005). While this complicated relationship is still being investigated, it is clear that both cell-cell adhesion and cell polarity establishment are interconnected processes during epithelial tissue morphogenesis and development (Coopman and Djiane 2016).
Most information in the field of polarity establishment and maintenance have come from studies in complex animal systems like Drosophila and C. elegans, or in cultured systems such as MDCK cells (Etemad-Moghadam, Guo, and Kemphues 1995; Wodarz et al., 1995; Kuchinke, Grawe, and Knust 1998; Tabuse et al., 1998; Wodarz et al., 2000; Petronczki and Knoblich 2001; Martin-Belmonte et al., 2007; Bryant et al., 2010; Morais-de-Sa, Mirouse, and St Johnston 2010; Bergstralh, Lovegrove, and St Johnston 2013; Khoury and Bilder 2020; Ventura et al., 2020). While these studies have contributed a great amount of information to our understanding of epithelial polarization, the addition of more diverse model systems to this body of work would continue to push this field forward. Therefore, it is advantageous to consider cnidarian models and others outside the bilaterian group for future studies.
Identification and Function of Polarity Regulators in Cnidarians
To demonstrate the utility of cnidarians as a model system for the study of polarity, we and others have undertaken phylogenetic analysis to test whether polarity regulators are conserved. Components of the Par, Crumbs, and Scribble complex have been identified in many organisms outside the bilaterian clade, including several cnidarian species such as Hydra vulgaris and Nematostella vectensis (Ragkousi et al., 2017; Doerr and Ragkousi 2019; Schiller and Bergstralh 2021). Several important functional domains within these three polarity complexes are conserved between bilaterian and cnidarian organisms, making cnidarian organisms promising models for the study of polarity establishment and maintenance.
Within the Par Complex, aPKC, Par6, and Par3/Bazooka are highly conserved across several bilaterian and cnidarian species (Figure 3). Par3/Bazooka features a specific serine residue within the aPKC binding region that is the site of aPKC phosphorylation, which is vital for the function of the whole Par complex (Soriano et al., 2016; Nagai-Tamai et al., 2002). Although the rest of the protein alignment denotes increased variability, this specific serine residue is present in all organisms tested (Figure 3B, C, S1375). High levels of conservation were found across the alignment of aPKC (Figure 3D). Within aPKC, several specific residues are conserved that are required for the function of the PB1 domain, which interacts with Par6 (Figure 3E). Additionally, a specific lysine residue is required for the kinase function of the STKc (serine/threonine protein kinase catalytic) domain (Li et al., 1995). In addition to high levels of conservation across the whole domain (Figure 3D), this specific invariable lysine residue is conserved across the ten organisms investigated (Figure 3F). Par6 is also highly conserved across both the ten species investigated as well as multiple Par6 isoforms (Figure 3G), including the PB1 region required for interaction with aPKC (Figure 3H). This suggests that the molecular mechanisms driving Par complex function during polarity establishment are conserved across bilaterian and cnidarian organisms.
Overall, components of the Crumbs complex appear to be conserved between the bilaterian and cnidarian species investigated (Figure 4A). Although a protein alignment of Crumbs seems to suggest a low level of sequence conservation (Figure 4B), further examination revealed that the vital EGF and laminin G-like domains are present in all ten species, but in different locations and numbers (Figure 4D). These extracellular domains facilitate protein-protein interactions within the Crumbs complex (Tepass, Theres, and Knust 1990; Bulgakova and Knust 2009; Thompson, Pichaud, and Roper 2013; Rothberg et al., 1988; den Hollander et al., 1999; Sasaki et al., 1988; Omori and Malicki 2006). Additionally, the ERLI motif located at the C-terminus of the Crumbs protein is highly conserved across both cnidarians and bilaterians. This sequence allows for the interaction of Crumbs with the other components of the Crumbs complex, Stardust/PALS1 and PATJ/InaD. Similarly, PATJ/InaD features a highly conserved L27 domain, but the number of subsequent PDZ domains differs from species to species (Figures 4E–G). Lastly, Stardust/PALS1 demonstrates high levels of protein conservation, especially in the region of the PDZ and SH3 domains that facilitate its interaction with Crumbs (Figures 4H, I). These results show that although there is variability between Crumbs isoforms in various species, the functional domains are largely conserved. Therefore, it is likely that Crumbs function in apical polarity establishment and maintenance is conserved as well.
Components of the Scribble complex are highly conserved across bilaterian and cnidarian organisms (Schiller and Bergstralh 2021) (Figure 5A). Both Scribble and Dlg contain multiple PDZ domains with high levels of conservation. These PDZ domains are vital to protein-protein interactions that include Scribble and Dlg within the polarity establishment pathway and others (Kallay et al., 2006; Takizawa et al., 2006; How et al., 2019; Bilder 2003; Bilder, Li, and Perrimon 2000; Bilder, Schober, and Perrimon 2003; Sotelo et al., 2012; Matsumine et al., 1996; Makino et al., 1997; Subbaiah et al., 2012). Lgl displays varying positions of its LGL domain, as well as different numbers of WD40 domains between species (Figures 5C,D). Despite these differences, the aPKC phosphorylation site towards the middle of the protein is highly conserved (Figure 5E). Additionally, the domains of Dlg are highly conserved including the vital GUK domain, which facilitates interaction between Dlg and phosphorylated Pins/LGN/GPSM2 among other possible functions (Johnston et al., 2009; Johnston et al., 2011; Anderson et al., 2016; Schiller and Bergstralh 2021) (Figure 5F). This suggests that both the polarity and spindle orientation mechanisms of Dlg are evolutionarily conserved, as well as the overall function of the Scribble complex.
Conclusion
Despite the extensive number of epithelialization studies in the past several decades, the information obtained has come from a limited pool of model systems. While bilaterian systems have been incredibly useful in identifying the first polarity proteins and their respective pathways, the complexity of these organisms has mostly limited these studies to early embryogenesis. Alternatively, cultured settings have given a simplified model in which to test the role of these polarity factors during epithelialization, however these settings are not physiologically representative of what may occur in a whole, living organism. Therefore, it would be advantageous to look outside the bilaterian clade for candidate model systems in which to continue and supplement these existing studies. This review has highlighted the technical, genetic, and evolutionary evidence that supports the use of cnidarian model organisms in future studies of cell polarity establishment and maintenance.
Author Contributions
LR, CE: writing, editing, data collection and analysis, DB: editing.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Amiel, A., and Houliston, E. (2009). Three Distinct RNA Localization Mechanisms Contribute to Oocyte Polarity Establishment in the Cnidarian Clytia Hemisphærica. Developmental Biol. 327 (1), 191–203. doi:10.1016/j.ydbio.2008.12.007
Anderson, D. P., Whitney, D. S., Hanson-Smith, V., Woznica, A., Campodonico-Burnett, W., Volkman, B. F., et al. (2016). Evolution of an Ancient Protein Function Involved in Organized Multicellularity in Animals. Elife 5, e10147. doi:10.7554/eLife.10147
Atwood, S. X., and Prehoda, K. E. (2009). aPKC Phosphorylates Miranda to Polarize Fate Determinants during Neuroblast Asymmetric Cell Division. Curr. Biol. 19 (9), 723–729. doi:10.1016/j.cub.2009.03.056
Bachmann, A., Grawe, F., Johnson, K., and Knust, E. (2008). Drosophila Lin-7 Is a Component of the Crumbs Complex in Epithelia and Photoreceptor Cells and Prevents Light-Induced Retinal Degeneration. Eur. J. Cel Biol. 87 (3), 123–136. doi:10.1016/j.ejcb.2007.11.002
Bachmann, A., Schneider, M., Theilenberg, E., Grawe, F., and Knust, E. (2001). Drosophila Stardust Is a Partner of Crumbs in the Control of Epithelial Cell Polarity. Nature 414 (6864), 638–643. doi:10.1038/414638a
Bachmann, A., Timmer, M., Sierralta, J., Pietrini, G., Gundelfinger, E. D., Knust, E., et al. (2004). Cell Type-specific Recruitment of DrosophilaLin-7 to Distinct MAGUK-Based Protein Complexes Defines Novel Roles for Sdt and Dlg-S97. J. Cel Sci 117 (Pt 10), 1899–1909. doi:10.1242/jcs.01029
Benton, R., and Johnston, D. S. (2003). Drosophila PAR-1 and 14-3-3 Inhibit Bazooka/PAR-3 to Establish Complementary Cortical Domains in Polarized Cells. Cell 115 (6), 691–704. doi:10.1016/s0092-8674(03)00938-3
Bergstralh, D. T., Lovegrove, H. E., and St Johnston, D. (2013). Discs Large Links Spindle Orientation to Apical-Basal Polarity in Drosophila Epithelia. Curr. Biol. 23 (17), 1707–1712. doi:10.1016/j.cub.2013.07.017
Bergstralh, D. T., and St Johnston, D. (2012). Epithelial Cell Polarity: what Flies Can Teach Us about Cancer. Essays Biochem. 53, 129–140. doi:10.1042/bse0530129
Bilder, D., Li, M., and Perrimon, N. (2000). Cooperative Regulation of Cell Polarity and Growth by Drosophila Tumor Suppressors. Science 289 (5476), 113–116. doi:10.1126/science.289.5476.113
Bilder, D. (2003). PDZ Domain Polarity Complexes. Curr. Biol. 13 (17), R661–R662. doi:10.1016/s0960-9822(03)00599-2
Bilder, D., and Perrimon, N. (2000). Localization of Apical Epithelial Determinants by the Basolateral PDZ Protein Scribble. Nature 403 (6770), 676–680. doi:10.1038/35001108
Bilder, D., Schober, M., and Perrimon, N. (2003). Integrated Activity of PDZ Protein Complexes Regulates Epithelial Polarity. Nat. Cel Biol 5 (1), 53–58. doi:10.1038/ncb897
Bryant, D. M., Datta, A., Rodríguez-Fraticelli, A. E., Peränen, J., Martín-Belmonte, F., and Mostov, K. E. (2010). A Molecular Network for De Novo Generation of the Apical Surface and Lumen. Nat. Cel Biol 12 (11), 1035–1045. doi:10.1038/ncb2106
Bulgakova, N. A., and Knust, E. (2009). The Crumbs Complex: from Epithelial-Cell Polarity to Retinal Degeneration. J. Cel Sci 122 (Pt 15), 2587–2596. doi:10.1242/jcs.023648
Buzgariu, W., Al Haddad, S., Tomczyk, S., Wenger, Y., and Galliot, B. (2015). Multi-functionality and Plasticity Characterize Epithelial Cells inHydra. Tissue Barriers 3 (4), e1068908. doi:10.1080/21688370.2015.1068908
Catterall, R., Lelarge, V., and McCaffrey, L. (2020). Genetic Alterations of Epithelial Polarity Genes Are Associated with Loss of Polarity in Invasive Breast Cancer. Int. J. Cancer 146 (6), 1578–1591. doi:10.1002/ijc.32691
Chapman, J. A., Kirkness, E. F., Simakov, O., Hampson, S. E., Mitros, T., Weinmaier, T., et al. (2010). The Dynamic Genome of Hydra. Nature 464 (7288), 592–596. doi:10.1038/nature08830
Chari, T., Weissbourd, B., Gehring, J., Ferraioli, A., Leclère, L., Herl, M., et al. (2021). Whole-animal Multiplexed Single-Cell RNA-Seq Reveals Transcriptional Shifts across Clytia Medusa Cell Types. Sci. Adv. 7 (48), eabh1683. doi:10.1126/sciadv.abh1683
Che, J., Wang, J., Li, H., Zhen, H., Shang, K., Yang, Y., et al. (2021). Decreased Expression of Dlg5 Is Associated with a Poor Prognosis and Epithelial-Mesenchymal Transition in Squamous Cell Lung Cancer. J. Thorac. Dis. 13 (5), 3115–3125. doi:10.21037/jtd-21-752
Clarke, D. N., Miller, P. W., Lowe, C. J., Weis, W. I., and Nelson, W. J. (2016). Characterization of the Cadherin-Catenin Complex of the Sea AnemoneNematostella Vectensisand Implications for the Evolution of Metazoan Cell-Cell Adhesion. Mol. Biol. Evol. 33 (8), 2016–2029. doi:10.1093/molbev/msw084
Clarkson, S. G., and Wolpert, L. (1967). Bud Morphogenesis in hydra. Nature 214 (5090), 780–783. doi:10.1038/214780a0
Cochet-Escartin, O., Locke, T. T., Shi, W. H., Steele, R. E., and Collins, E.-M. S. (2017). Physical Mechanisms Driving Cell Sorting in Hydra. Biophysical J. 113 (12), 2827–2841. doi:10.1016/j.bpj.2017.10.045
Coopman, P., and Djiane, A. (2016). Adherens Junction and E-Cadherin Complex Regulation by Epithelial Polarity. Cell. Mol. Life Sci. 73 (18), 3535–3553. doi:10.1007/s00018-016-2260-8
den Hollander, A. I., ten Brink, J. B., de Kok, Y. J. M., van Soest, S., van den Born, L. I., van Driel, M. A., et al. (1999). Mutations in a Human Homologue of Drosophila Crumbs Cause Retinitis Pigmentosa (RP12). Nat. Genet. 23 (2), 217–221. doi:10.1038/13848
Desai, B. V., Harmon, R. M., and Green, K. J. (2009). Desmosomes at a Glance. J. Cel Sci 122 (Pt 24), 4401–4407. doi:10.1242/jcs.037457
Devenport, D. (2014). The Cell Biology of Planar Cell Polarity. J. Cel Biol 207 (2), 171–179. doi:10.1083/jcb.201408039
Doerr, S., and Ragkousi, K. (2019). Cell Polarity Oscillations in Mitotic Epithelia. Curr. Opin. Genet. Dev. 57, 47–53. doi:10.1016/j.gde.2019.07.007
Dohrmann, M., and Worheide, G. (2013). Novel Scenarios of Early Animal Evolution-Iis it Time to Rewrite Textbooks? Integr. Comp. Biol. 53 (3), 503–511. doi:10.1093/icb/ict008
Epp, L., Smid, I., and Tardent, P. (1986). Synthesis of the Mesoglea by Ectoderm and Endoderm in Reassembled hydra. J. Morphol. 189 (3), 271–279. doi:10.1002/jmor.1051890306
Estephane, D., and Anctil, M. (2010). Retinoic Acid and Nitric Oxide Promote Cell Proliferation and Differentially Induce Neuronal Differentiation In Vitro in the Cnidarian Renilla Koellikeri. Dev. Neurobiol. 70 (12), 842–852. doi:10.1002/dneu.20824
Etemad-Moghadam, B., Guo, S., and Kemphues, K. J. (1995). Asymmetrically Distributed PAR-3 Protein Contributes to Cell Polarity and Spindle Alignment in Early C. elegans Embryos. Cell 83 (5), 743–752. doi:10.1016/0092-8674(95)90187-6
Feng, S., Cokus, S. J., Zhang, X., Chen, P. Y., Bostick, M., Goll, M. G., et al. (2010). Conservation and Divergence of Methylation Patterning in Plants and Animals. Proc. Natl. Acad. Sci. U S A. 107 (19), 8689–8694. doi:10.1073/pnas.1002720107
Ganot, P., Zoccola, D., Tambutte, E., Voolstra, C. R., Aranda, M., Allemand, D., et al. (2015). Structural Molecular Components of Septate Junctions in Cnidarians point to the Origin of Epithelial Junctions in Eukaryotes. Mol. Biol. Evol. 32 (1), 44–62. doi:10.1093/molbev/msu265
Gul, I. S., Hulpiau, P., Saeys, Y., and van Roy, F. (2017). Evolution and Diversity of Cadherins and Catenins. Exp. Cel Res 358 (1), 3–9. doi:10.1016/j.yexcr.2017.03.001
Hendriks, D., Artegiani, B., Hu, H., Chuva de Sousa Lopes, S., and Clevers, H. (2021). Establishment of Human Fetal Hepatocyte Organoids and CRISPR-Cas9-Based Gene Knockin and Knockout in Organoid Cultures from Human Liver. Nat. Protoc. 16 (1), 182–217. doi:10.1038/s41596-020-00411-2
Hicklin, J., and Wolpert, L. (1973). Positional Information and Pattern Regulation in hydra: the Effect of Gamma-Radiation. J. Embryol. Exp. Morphol. 30 (3), 741–752. doi:10.1242/dev.30.3.741
How, J. Y., Caria, S., Humbert, P. O., and Kvansakul, M. (2019). Crystal Structure of the Human Scribble PDZ1 Domain Bound to the PDZ-Binding Motif of APC. FEBS Lett. 593 (5), 533–542. doi:10.1002/1873-3468.13329
Hulpiau, P., and van Roy, F. (2011). New Insights into the Evolution of Metazoan Cadherins. Mol. Biol. Evol. 28 (1), 647–657. doi:10.1093/molbev/msq233
Humbert, P. O., Grzeschik, N. A., Brumby, A. M., Galea, R., Elsum, I., and Richardson, H. E. (2008). Control of Tumourigenesis by the Scribble/Dlg/Lgl Polarity Module. Oncogene 27 (55), 6888–6907. doi:10.1038/onc.2008.341
Ikmi, A., McKinney, S. A., Delventhal, K. M., and Gibson, M. C. (2014). TALEN and CRISPR/Cas9-mediated Genome Editing in the Early-Branching Metazoan Nematostella vectensis. Nat. Commun. 5, 5486. doi:10.1038/ncomms6486
Johnston, C. A., Hirono, K., Prehoda, K. E., and Doe, C. Q. (2009). Identification of an Aurora-A/PinsLINKER/Dlg Spindle Orientation Pathway Using Induced Cell Polarity in S2 Cells. Cell 138 (6), 1150–1163. doi:10.1016/j.cell.2009.07.041
Johnston, C. A., Whitney, D. S., Volkman, B. F., Doe, C. Q., and Prehoda, K. E. (2011). Conversion of the Enzyme Guanylate Kinase into a Mitotic-Spindle Orienting Protein by a Single Mutation that Inhibits GMP-Induced Closing. Proc. Natl. Acad. Sci. U S A. 108 (44), E973–E978. doi:10.1073/pnas.1104365108
Jung, H. Y., Fattet, L., Tsai, J. H., Kajimoto, T., Chang, Q., Newton, A. C., et al. (2019). Apical-basal Polarity Inhibits Epithelial-Mesenchymal Transition and Tumour Metastasis by PAR-Complex-Mediated SNAI1 Degradation. Nat. Cel Biol 21 (3), 359–371. doi:10.1038/s41556-019-0291-8
Kallay, L. M., McNickle, A., Brennwald, P. J., Hubbard, A. L., and Braiterman, L. T. (2006). Scribble Associates with Two Polarity Proteins, Lgl2 and Vangl2, via Distinct Molecular Domains. J. Cel Biochem 99 (2), 647–664. doi:10.1002/jcb.20992
Kamran, Z., Zellner, K., Kyriazes, H., Kraus, C. M., Reynier, J. B., and Malamy, J. E. (2017). In Vivo imaging of Epithelial Wound Healing in the Cnidarian Clytia Hemisphaerica Demonstrates Early Evolution of Purse String and Cell Crawling Closure Mechanisms. BMC Dev. Biol. 17 (1), 17. doi:10.1186/s12861-017-0160-2
Khoury, M. J., and Bilder, D. (2020). Distinct Activities of Scrib Module Proteins Organize Epithelial Polarity. Proc. Natl. Acad. Sci. U S A. 117 (21), 11531–11540. doi:10.1073/pnas.1918462117
Kishimoto, Y., Murate, M., and Sugiyama, T. (1996). Hydra Regeneration from Recombined Ectodermal and Endodermal Tissue. I. Epibolic Ectodermal Spreading Is Driven by Cell Intercalation. J. Cel Sci 109 (Pt 4), 763–772. doi:10.1242/jcs.109.4.763
Knust, E., Tepass, U., and Wodarz, A. (1993). Crumbs and Stardust, Two Genes of Drosophila Required for the Development of Epithelial Cell Polarity. Dev. Suppl., 261–268. doi:10.1242/dev.119.supplement.261
Kraus, Y., Chevalier, S., and Houliston, E. (2020). Cell Shape Changes during Larval Body Plan Development in Clytia Hemisphaerica. Dev. Biol. 468 (1-2), 59–79. doi:10.1016/j.ydbio.2020.09.013
Kuchinke, U., Grawe, F., and Knust, E. (1998). Control of Spindle Orientation in Drosophila by the Par-3-Related PDZ-Domain Protein Bazooka. Curr. Biol. 8 (25), 1357–1365. doi:10.1016/s0960-9822(98)00016-5
Kumburegama, S., Wijesena, N., and Wikramanayake, A. H. (2008). Detecting Expression Patterns of Wnt Pathway Components in Nematostella vectensis Embryos. Methods Mol. Biol. 469, 55–67. doi:10.1007/978-1-60327-469-2_6
Kumburegama, S., Wijesena, N., Xu, R., and Wikramanayake, A. H. (2011). Strabismus-mediated Primary Archenteron Invagination Is Uncoupled from Wnt/beta-catenin-dependent Endoderm Cell Fate Specification in Nematostella vectensis (Anthozoa, Cnidaria): Implications for the Evolution of Gastrulation. Evodevo 2 (1), 2. doi:10.1186/2041-9139-2-2
Lapebie, P., Ruggiero, A., Barreau, C., Chevalier, S., Chang, P., Dru, P., et al. (2014). Differential Responses to Wnt and PCP Disruption Predict Expression and Developmental Function of Conserved and Novel Genes in a Cnidarian. Plos Genet. 10 (9), e1004590. doi:10.1371/journal.pgen.1004590
Laprise, P., Beronja, S., Silva-Gagliardi, N. F., Pellikka, M., Jensen, A. M., McGlade, C. J., et al. (2006). The FERM Protein Yurt Is a Negative Regulatory Component of the Crumbs Complex that Controls Epithelial Polarity and Apical Membrane Size. Dev. Cel 11 (3), 363–374. doi:10.1016/j.devcel.2006.06.001
Laprise, P., Lau, K. M., Harris, K. P., Silva-Gagliardi, N. F., Paul, S. M., Beronja, S., et al. (2009). Yurt, Coracle, Neurexin IV and the Na(+),K(+)-ATPase Form a Novel Group of Epithelial Polarity Proteins. Nature 459 (7250), 1141–1145. doi:10.1038/nature08067
Leclere, L., Horin, C., Chevalier, S., Lapebie, P., Dru, P., Peron, S., et al. (2019). The Genome of the Jellyfish Clytia Hemisphaerica and the Evolution of the Cnidarian Life-Cycle. Nat. Ecol. Evol. 3 (5), 801–810. doi:10.1038/s41559-019-0833-2
Lee, C. Y., Robinson, K. J., and Doe, C. Q. (2006). Lgl, Pins and aPKC Regulate Neuroblast Self-Renewal versus Differentiation. Nature 439 (7076), 594–598. doi:10.1038/nature04299
Leys, S. P., and Riesgo, A. (2012). Epithelia, an Evolutionary novelty of Metazoans. J. Exp. Zool B Mol. Dev. Evol. 318 (6), 438–447. doi:10.1002/jez.b.21442
Li, W., Yu, J. C., Shin, D. Y., and Pierce, J. H. (1995). Characterization of a Protein Kinase C-delta (PKC-delta) ATP Binding Mutant. An Inactive Enzyme that Competitively Inhibits Wild Type PKC-delta Enzymatic Activity. J. Biol. Chem. 270 (14), 8311–8318. doi:10.1074/jbc.270.14.8311
Li, X., Francies, H. E., Secrier, M., Perner, J., Miremadi, A., Galeano-Dalmau, N., et al. (2018). Organoid Cultures Recapitulate Esophageal Adenocarcinoma Heterogeneity Providing a Model for Clonality Studies and Precision Therapeutics. Nat. Commun. 9 (1), 2983. doi:10.1038/s41467-018-05190-9
Lim, J., and Thiery, J. P. (2012). Epithelial-mesenchymal Transitions: Insights from Development. Development 139 (19), 3471–3486. doi:10.1242/dev.071209
Lin, Y. C., Grigoriev, N. G., and Spencer, A. N. (2000). Wound Healing in Jellyfish Striated Muscle Involves Rapid Switching between Two Modes of Cell Motility and a Change in the Source of Regulatory Calcium. Dev. Biol. 225 (1), 87–100. doi:10.1006/dbio.2000.9807
Lommel, M., Tursch, A., Rustarazo-Calvo, L., Trageser, B., and Holstein, T. W. (2017). Genetic Knockdown and Knockout Approaches in Hydra. bioRxiv. doi:10.1101/230300
Lukonin, I., Serra, D., Challet Meylan, L., Volkmann, K., Baaten, J., Zhao, R., et al. (2020). Phenotypic Landscape of Intestinal Organoid Regeneration. Nature 586 (7828), 275–280. doi:10.1038/s41586-020-2776-9
MacWilliams, H. K. (1982). Numerical Simulations of hydra Head Regeneration Using a Proportion-Regulating Version of the Gierer-Meinhardt Model. J. Theor. Biol. 99 (4), 681–703. doi:10.1016/0022-5193(82)90194-1
Magie, C. R., and Martindale, M. Q. (2008). Cell-cell Adhesion in the Cnidaria: Insights into the Evolution of Tissue Morphogenesis. Biol. Bull. 214 (3), 218–232. doi:10.2307/25470665
Makino, K., Kuwahara, H., Masuko, N., Nishiyama, Y., Morisaki, T., Sasaki, J., et al. (1997). Cloning and Characterization of NE-dlg: a Novel Human Homolog of the Drosophila Discs Large (Dlg) Tumor Suppressor Protein Interacts with the APC Protein. Oncogene 14 (20), 2425–2433. doi:10.1038/sj.onc.1201087
Malamy, J. E., and Shribak, M. (2018). An Orientation-independent DIC Microscope Allows High Resolution Imaging of Epithelial Cell Migration and Wound Healing in a Cnidarian Model. J. Microsc. 270 (3), 290–301. doi:10.1111/jmi.12682
Manninen, A. (2015). Epithelial Polarity-Ggenerating and Integrating Signals from the ECM with Integrins. Exp. Cel Res 334 (2), 337–349. doi:10.1016/j.yexcr.2015.01.003
Maroudas-Sacks, Y., Garion, L., Shani-Zerbib, L., Livshits, A., Braun, E., and Keren, K. 2021. "Topological Defects in the Nematic Order of Actin Fibres as Organization Centres of Hydra Morphogenesis." Nat. Phys. 17 (2):251. doi:10.1038/s41567-020-01083-1
Martin-Belmonte, F., Gassama, A., Datta, A., Yu, W., Rescher, U., Gerke, V., et al. (2007). PTEN-mediated Apical Segregation of Phosphoinositides Controls Epithelial Morphogenesis through Cdc42. Cell 128 (2), 383–397. doi:10.1016/j.cell.2006.11.051
Matsumine, A., Ogai, A., Senda, T., Okumura, N., Satoh, K., Baeg, G. H., et al. (1996). Binding of APC to the Human Homolog of the Drosophila Discs Large Tumor Suppressor Protein. Science 272 (5264), 1020–1023. doi:10.1126/science.272.5264.1020
Meinhardt, H. (1993). A Model for Pattern Formation of Hypostome, Tentacles, and Foot in hydra: How to Form Structures Close to Each Other, How to Form Them at a Distance. Dev. Biol. 157 (2), 321–333. doi:10.1006/dbio.1993.1138
Mellman, I., and Nelson, W. J. (2008). Coordinated Protein Sorting, Targeting and Distribution in Polarized Cells. Nat. Rev. Mol. Cel Biol 9 (11), 833–845. doi:10.1038/nrm2525
Momose, T., De Cian, A., Shiba, K., Inaba, K., Giovannangeli, C., and Concordet, J. P. (2018). High Doses of CRISPR/Cas9 Ribonucleoprotein Efficiently Induce Gene Knockout with Low Mosaicism in the Hydrozoan Clytia Hemisphaerica through Microhomology-Mediated Deletion. Sci. Rep. 8 (1), 11734. doi:10.1038/s41598-018-30188-0
Momose, T., Derelle, R., and Houliston, E. (2008). A Maternally Localised Wnt Ligand Required for Axial Patterning in the Cnidarian Clytia Hemisphaerica. Development 135 (12), 2105–2113. doi:10.1242/dev.021543
Momose, T., and Houliston, E. (2007). Two Oppositely Localised Frizzled RNAs as axis Determinants in a Cnidarian Embryo. Plos Biol. 5 (4), e70. doi:10.1371/journal.pbio.0050070
Momose, T., Kraus, Y., and Houliston, E. (2012). A Conserved Function for Strabismus in Establishing Planar Cell Polarity in the Ciliated Ectoderm during Cnidarian Larval Development. Development 139 (23), 4374–4382. doi:10.1242/dev.084251
Morais-de-Sa, E., Mirouse, V., and St Johnston, D. (2010). aPKC Phosphorylation of Bazooka Defines the Apical/lateral Border in Drosophila Epithelial Cells. Cell 141 (3), 509–523. doi:10.1016/j.cell.2010.02.040
Nagai-Tamai, Y., Mizuno, K., Hirose, T., Suzuki, A., and Ohno, S. (2002). Regulated Protein-Protein Interaction between aPKC and PAR-3 Plays an Essential Role in the Polarization of Epithelial Cells. Genes Cells 7 (11), 1161–1171. doi:10.1046/j.1365-2443.2002.00590.x
Nathaniel Clarke, D., Lowe, C. J., and James Nelson, W. (2019). The Cadherin-Catenin Complex Is Necessary for Cell Adhesion and Embryogenesis in Nematostella vectensis. Dev. Biol. 447 (2), 170–181. doi:10.1016/j.ydbio.2019.01.007
Nejsum, L. N., and Nelson, W. J. (2007). A Molecular Mechanism Directly Linking E-Cadherin Adhesion to Initiation of Epithelial Cell Surface Polarity. J. Cel Biol 178 (2), 323–335. doi:10.1083/jcb.200705094
Omori, Y., and Malicki, J. (2006). Oko Meduzy and Related Crumbs Genes Are Determinants of Apical Cell Features in the Vertebrate Embryo. Curr. Biol. 16 (10), 945–957. doi:10.1016/j.cub.2006.03.058
Otto, J. J. (1977). Orientation and Behavior of Epithelial Cell Muscle Processes during Hydra Budding. J. Exp. Zool 202 (3), 307–322. doi:10.1002/jez.1402020303
Petronczki, M., and Knoblich, J. A. (2001). DmPAR-6 Directs Epithelial Polarity and Asymmetric Cell Division of Neuroblasts in Drosophila. Nat. Cel Biol 3 (1), 43–49. doi:10.1038/35050550
Philipp, I., Aufschnaiter, R., Ozbek, S., Pontasch, S., Jenewein, M., Watanabe, H., et al. (2009). Wnt/beta-catenin and Noncanonical Wnt Signaling Interact in Tissue Evagination in the Simple Eumetazoan Hydra. Proc. Natl. Acad. Sci. U S A. 106 (11), 4290–4295. doi:10.1073/pnas.0812847106
Piccolo, S. (2013). Developmental Biology: Mechanics in the Embryo. Nature 504 (7479), 223–225. doi:10.1038/504223a
Plickert, G., and Kroiher, M. (1988). Proliferation Kinetics and Cell Lineages Can Be Studied in Whole Mounts and Macerates by Means of BrdU/anti-BrdU Technique. Development 103 (4), 791–794. doi:10.1242/dev.103.4.791
Pukhlyakova, E. A., Kirillova, A. O., Kraus, Y. A., Zimmermann, B., and Technau, U. (2019). A Cadherin Switch marks Germ Layer Formation in the Diploblastic Sea Anemone Nematostella vectensis. Development 146 (20). doi:10.1242/dev.174623
Putnam, N. H., Srivastava, M., Hellsten, U., Dirks, B., Chapman, J., Salamov, A., et al. (2007). Sea Anemone Genome Reveals Ancestral Eumetazoan Gene Repertoire and Genomic Organization. Science 317 (5834), 86–94. doi:10.1126/science.1139158
Qin, Y., Capaldo, C., Gumbiner, B. M., and Macara, I. G. (2005). The Mammalian Scribble Polarity Protein Regulates Epithelial Cell Adhesion and Migration through E-Cadherin. J. Cel Biol 171 (6), 1061–1071. doi:10.1083/jcb.200506094
Ragkousi, K., Marr, K., McKinney, S., Ellington, L., and Gibson, M. C. (2017). Cell-Cycle-Coupled Oscillations in Apical Polarity and Intercellular Contact Maintain Order in Embryonic Epithelia. Curr. Biol. 27 (9), 1381–1386. doi:10.1016/j.cub.2017.03.064
Rodriguez-Boulan, E., Kreitzer, G., and Musch, A. (2005). Organization of Vesicular Trafficking in Epithelia. Nat. Rev. Mol. Cel Biol 6 (3), 233–247. doi:10.1038/nrm1593
Rodriguez-Boulan, E., and Macara, I. G. (2014). Organization and Execution of the Epithelial Polarity Programme. Nat. Rev. Mol. Cel Biol 15 (4), 225–242. doi:10.1038/nrm3775
Rosenbluth, J. M., Schackmann, R. C. J., Gray, G. K., Selfors, L. M., Li, C. M., Boedicker, M., et al. (2020). Organoid Cultures from normal and Cancer-Prone Human Breast Tissues Preserve Complex Epithelial Lineages. Nat. Commun. 11 (1), 1711. doi:10.1038/s41467-020-15548-7
Rothberg, J. M., Hartley, D. A., Walther, Z., and Artavanis-Tsakonas, S. (1988). Slit: an EGF-Homologous Locus of D. melanogaster Involved in the Development of the Embryonic central Nervous System. Cell 55 (6), 1047–1059. doi:10.1016/0092-8674(88)90249-8
Salinas-Saavedra, M., and Martindale, M. Q. (2020). Par Protein Localization during the Early Development of Mnemiopsis leidyi Suggests Different Modes of Epithelial Organization in the Metazoa. Elife 9. doi:10.7554/eLife.54927
Salinas-Saavedra, M., Rock, A. Q., and Martindale, M. Q. (2018). Germ Layer-specific Regulation of Cell Polarity and Adhesion Gives Insight into the Evolution of Mesoderm. Elife 7. doi:10.7554/eLife.36740
Salinas-Saavedra, M., Stephenson, T. Q., Dunn, C. W., and Martindale, M. Q. (2015). Par System Components Are Asymmetrically Localized in Ectodermal Epithelia, but Not during Early Development in the Sea Anemone Nematostella vectensis. Evodevo 6, 20. doi:10.1186/s13227-015-0014-6
Sarras, M. P., Zhang, X., Huff, J. K., Accavitti, M. A., St John, P. L., and Abrahamson, D. R. (1993). Extracellular Matrix (Mesoglea) of Hydra vulgaris III. Formation and Function during Morphogenesis of hydra Cell Aggregates. Dev. Biol. 157 (2), 383–398. doi:10.1006/dbio.1993.1143
Sasaki, M., Kleinman, H. K., Huber, H., Deutzmann, R., and Yamada, Y. (1988). Laminin, a Multidomain Protein. The A Chain Has a Unique Globular Domain and Homology with the Basement Membrane Proteoglycan and the Laminin B Chains. J. Biol. Chem. 263 (32), 16536–16544. doi:10.1016/s0021-9258(18)37424-6
Schiller, E. A., and Bergstralh, D. T. (2021). Interaction between Discs Large and Pins/LGN/GPSM2: a Comparison across Species. Biol. Open 10 (11). doi:10.1242/bio.058982
Schuster, B., Junkin, M., Kashaf, S. S., Romero-Calvo, I., Kirby, K., Matthews, J., et al. (2020). Automated Microfluidic Platform for Dynamic and Combinatorial Drug Screening of Tumor Organoids. Nat. Commun. 11 (1), 5271. doi:10.1038/s41467-020-19058-4
Schwaiger, M., Schonauer, A., Rendeiro, A. F., Pribitzer, C., Schauer, A., Gilles, A. F., et al. (2014). Evolutionary Conservation of the Eumetazoan Gene Regulatory Landscape. Genome Res. 24 (4), 639–650. doi:10.1101/gr.162529.113
Sebe-Pedros, A., Saudemont, B., Chomsky, E., Plessier, F., Mailhe, M. P., Renno, J., et al. (2018). Cnidarian Cell Type Diversity and Regulation Revealed by Whole-Organism Single-Cell RNA-Seq. Cell 173 (6), 1520–1534. doi:10.1016/j.cell.2018.05.019
Serra, D., Mayr, U., Boni, A., Lukonin, I., Rempfler, M., Challet Meylan, L., et al. (2019). Self-organization and Symmetry Breaking in Intestinal Organoid Development. Nature 569 (7754), 66–72. doi:10.1038/s41586-019-1146-y
Seybold, A., Salvenmoser, W., and Hobmayer, B. (2016). Sequential Development of Apical-Basal and Planar Polarities in Aggregating Epitheliomuscular Cells of Hydra. Dev. Biol. 412 (1), 148–159. doi:10.1016/j.ydbio.2016.02.022
Siebert, S., Farrell, J. A., Cazet, J. F., Abeykoon, Y., Primack, A. S., Schnitzler, C. E., et al. (2019). Stem Cell Differentiation Trajectories in Hydra Resolved at Single-Cell Resolution. Science 365 (6451). doi:10.1126/science.aav9314
Sinigaglia, C., Peron, S., Eichelbrenner, J., Chevalier, S., Steger, J., Barreau, C., et al. (2020). Pattern Regulation in a Regenerating Jellyfish. Elife 9. doi:10.7554/eLife.54868
Skokan, T. D., Vale, R. D., and McKinley, K. L. (2020). Cell Sorting in Hydra vulgaris Arises from Differing Capacities for Epithelialization between Cell Types. Curr. Biol. 30 (19), 3713–3723. doi:10.1016/j.cub.2020.07.035
Soriano, E. V., Ivanova, M. E., Fletcher, G., Riou, P., Knowles, P. P., Barnouin, K., et al. (2016). aPKC Inhibition by Par3 CR3 Flanking Regions Controls Substrate Access and Underpins Apical-Junctional Polarization. Dev. Cel 38 (4), 384–398. doi:10.1016/j.devcel.2016.07.018
Sotelo, N. S., Valiente, M., Gil, A., and Pulido, R. (2012). A Functional Network of the Tumor Suppressors APC, hDlg, and PTEN, that Relies on Recognition of Specific PDZ-Domains. J. Cel Biochem 113 (8), 2661–2670. doi:10.1002/jcb.24141
Sperling, E. A., Peterson, K. J., and Pisani, D. (2009). Phylogenetic-signal Dissection of Nuclear Housekeeping Genes Supports the Paraphyly of Sponges and the Monophyly of Eumetazoa. Mol. Biol. Evol. 26 (10), 2261–2274. doi:10.1093/molbev/msp148
St Johnston, D., and Ahringer, J. (2010). Cell Polarity in Eggs and Epithelia: Parallels and Diversity. Cell 141 (5), 757–774. doi:10.1016/j.cell.2010.05.011
Subbaiah, V. K., Narayan, N., Massimi, P., and Banks, L. (2012). Regulation of the DLG Tumor Suppressor by Beta-Catenin. Int. J. Cancer 131 (10), 2223–2233. doi:10.1002/ijc.27519
Tabuse, Y., Izumi, Y., Piano, F., Kemphues, K. J., Miwa, J., and Ohno, S. (1998). Atypical Protein Kinase C Cooperates with PAR-3 to Establish Embryonic Polarity in Caenorhabditis elegans. Development 125 (18), 3607–3614. doi:10.1242/dev.125.18.3607
Takashima, S., Gold, D., and Hartenstein, V. (2013). Stem Cells and Lineages of the Intestine: a Developmental and Evolutionary Perspective. Dev. Genes Evol. 223 (1-2), 85–102. doi:10.1007/s00427-012-0422-8
Takizawa, S., Nagasaka, K., Nakagawa, S., Yano, T., Nakagawa, K., Yasugi, T., et al. (2006). Human Scribble, a Novel Tumor Suppressor Identified as a Target of High-Risk HPV E6 for Ubiquitin-Mediated Degradation, Interacts with Adenomatous Polyposis Coli. Genes Cells 11 (4), 453–464. doi:10.1111/j.1365-2443.2006.00954.x
Tan, B., Yatim, Smjm., Peng, S. J. Gunaratne, W. Hunziker., and Ludwig, A. (2020). The Mammalian Crumbs Complex Defines a Distinct Polarity Domain Apical of Epithelial Tight Junctions. Curr. Biol. 30 (14), 2791–2804. e6. doi:10.1016/j.cub.2020.05.032
Technau, U., Cramer von Laue, C., Rentzsch, F., Luft, S., Hobmayer, B., Bode, H. R., et al. (2000). Parameters of Self-Organization in Hydra Aggregates. Proc. Natl. Acad. Sci. U S A. 97 (22), 12127–12131. doi:10.1073/pnas.97.22.12127
Technau, U. (2020). Gastrulation and Germ Layer Formation in the Sea Anemone Nematostella vectensis and Other Cnidarians. Mech. Dev. 163, 103628. doi:10.1016/j.mod.2020.103628
Technau, U., and Holstein, T. W. (1992). Cell Sorting during the Regeneration of Hydra from Reaggregated Cells. Dev. Biol. 151 (1), 117–127. doi:10.1016/0012-1606(92)90219-7
Technau, U., and Steele, R. E. (2011). Evolutionary Crossroads in Developmental Biology: Cnidaria. Development 138 (8), 1447–1458. doi:10.1242/dev.048959
Tenvooren, I., Jenks, M. Z., Rashid, H., Cook, K. L., Muhlemann, J. K., Sistrunk, C., et al. (2019). Elevated Leptin Disrupts Epithelial Polarity and Promotes Premalignant Alterations in the Mammary Gland. Oncogene 38 (20), 3855–3870. doi:10.1038/s41388-019-0687-8
Tepass, U., Tanentzapf, G., Ward, R., and Fehon, R. (2001). Epithelial Cell Polarity and Cell Junctions in Drosophila. Annu. Rev. Genet. 35, 747–784. doi:10.1146/annurev.genet.35.102401.091415
Tepass, U., Theres, C., and Knust, E. (1990). Crumbs Encodes an EGF-like Protein Expressed on Apical Membranes of Drosophila Epithelial Cells and Required for Organization of Epithelia. Cell 61 (5), 787–799. doi:10.1016/0092-8674(90)90189-l
Thompson, B. J., Pichaud, F., and Roper, K. (2013). Sticking Together the Crumbs - an Unexpected Function for an Old Friend. Nat. Rev. Mol. Cel Biol 14 (5), 307–314. doi:10.1038/nrm3568
Tilston-Lunel, A., Mazzilli, S., Kingston, N. M., Szymaniak, A. D., Hicks-Berthet, J., Kern, J. G., et al. (2021). Aberrant Epithelial Polarity Cues Drive the Development of Precancerous Airway Lesions. Proc. Natl. Acad. Sci. U S A. 118 (18). doi:10.1073/pnas.2019282118
Trembley, A. (1744). Mémoires pour servir à l'histoire d'un genre de ploypes d'eau douce, à bras en forme de cornes, 2. Paris: Chez Durand. doi:10.5962/bhl.title.64073
Tucker, R. P., and Adams, J. C. (2014). Adhesion Networks of Cnidarians: a Postgenomic View. Int. Rev. Cel Mol Biol 308, 323–377. doi:10.1016/B978-0-12-800097-7.00008-7
Ventura, G., Moreira, S., Barros-Carvalho, A., Osswald, M., and Morais-de-Sa, E. (2020). Lgl Cortical Dynamics Are Independent of Binding to the Scrib-Dlg Complex but Require Dlg-dependent Restriction of aPKC. Development 147 (15). doi:10.1242/dev.186593
Ventura, P., Toullec, G., Fricano, C., Chapron, L., Meunier, V., Rottinger, E., et al. (2018). Cnidarian Primary Cell Culture as a Tool to Investigate the Effect of Thermal Stress at Cellular Level. Mar. Biotechnol. (Ny) 20 (2), 144–154. doi:10.1007/s10126-017-9791-3
Vogg, M. C., Galliot, B., and Tsiairis, C. D. (2019). Model Systems for Regeneration: Hydra. Development 146 (21). doi:10.1242/dev.177212
Webster, G., and Hamilton, S. (1972). Budding in hydra: the Role of Cell Multiplication and Cell Movement in Bud Initiation. J. Embryol. Exp. Morphol. 27 (2), 301–316. doi:10.1242/dev.27.2.301
Whiteman, E. L., Liu, C. J., Fearon, E. R., and Margolis, B. (2008). The Transcription Factor Snail Represses Crumbs3 Expression and Disrupts Apico-Basal Polarity Complexes. Oncogene 27 (27), 3875–3879. doi:10.1038/onc.2008.9
Wijesena, N., and Martindale, M. Q. (2018). Reengineering the Primary Body axis by Ectopic cWnt Signaling. Curr. Biol. 28 (5), R206–R207. doi:10.1016/j.cub.2018.01.042
Wijesena, N., Sun, H., Kumburegama, S., and Wikramanayake, A. H. (2022). Distinct Frizzled Receptors Independently Mediate Endomesoderm Specification and Primary Archenteron Invagination during Gastrulation in Nematostella. Dev. Biol. 481, 215–225. doi:10.1016/j.ydbio.2021.11.002
Williams, E., Villar-Prados, A., Bowser, J., Broaddus, R., and Gladden, A. B. (2017). Loss of Polarity Alters Proliferation and Differentiation in Low-Grade Endometrial Cancers by Disrupting Notch Signaling. PLoS One 12 (12), e0189081. doi:10.1371/journal.pone.0189081
Wodarz, A., Hinz, U., Engelbert, M., and Knust, E. (1995). Expression of Crumbs Confers Apical Character on Plasma Membrane Domains of Ectodermal Epithelia of Drosophila. Cell 82 (1), 67–76. doi:10.1016/0092-8674(95)90053-5
Wodarz, A., Ramrath, A., Grimm, A., and Knust, E. (2000). Drosophila Atypical Protein Kinase C Associates with Bazooka and Controls Polarity of Epithelia and Neuroblasts. J. Cel Biol 150 (6), 1361–1374. doi:10.1083/jcb.150.6.1361
Zemach, A., McDaniel, I. E., Silva, P., and Zilberman, D. (2010). Genome-wide Evolutionary Analysis of Eukaryotic DNA Methylation. Science 328 (5980), 916–919. doi:10.1126/science.1186366
Zemach, A., and Zilberman, D. (2010). Evolution of Eukaryotic DNA Methylation and the Pursuit of Safer Sex. Curr. Biol. 20 (17), R780–R785. doi:10.1016/j.cub.2010.07.007
Keywords: cnidaria, epithelia, polarity, model organisms, apical-basal cell polarity
Citation: Rathbun LI, Everett CA and Bergstralh DT (2022) Emerging Cnidarian Models for the Study of Epithelial Polarity. Front. Cell Dev. Biol. 10:854373. doi: 10.3389/fcell.2022.854373
Received: 13 January 2022; Accepted: 01 March 2022;
Published: 01 April 2022.
Edited by:
Alexander Ludwig, Nanyang Technological University, SingaporeReviewed by:
Lucas Leclere, UMR7009 Laboratoire de Biologie du Développement de Villefranche sur Mer, FranceToshio Takahashi, Suntory Foundation for Life Sciences, Japan
Copyright © 2022 Rathbun, Everett and Bergstralh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dan T. Bergstralh, ZGFuLmJlcmdzdHJhbGhAcm9jaGVzdGVyLmVkdQ==
 Lindsay I. Rathbun
Lindsay I. Rathbun Coralee A. Everett
Coralee A. Everett Dan T. Bergstralh
Dan T. Bergstralh