- 1Division of Neuroscience, San Raffaele Scientific Institute and Vita-Salute San Raffaele University, Milano, Italy
- 2Department of Cell Physiology and Metabolism, University of Geneva, Centre Médical Universitaire, Geneva, Switzerland
Focal adhesions are specialized integrin-dependent adhesion complexes, which ensure cell anchoring to the extracellular matrix. Focal adhesions also function as mechano-signaling platforms by perceiving and integrating diverse physical and (bio)chemical cues of their microenvironment, and by transducing them into intracellular signaling for the control of cell behavior. The fundamental biological mechanism of creating intracellular signaling in response to changes in tensional forces appears to be tightly linked to paxillin recruitment and binding to focal adhesions. Interestingly, the tension-dependent nature of the paxillin binding to adhesions, combined with its scaffolding function, suggests a major role of this protein in integrating multiple signals from the microenvironment, and accordingly activating diverse molecular responses. This minireview offers an overview of the molecular bases of the mechano-sensitivity and mechano-signaling capacity of core focal adhesion proteins, and highlights the role of paxillin as a key component of the mechano-transducing machinery based on the interaction of cells to substrates activating the β3 integrin-talin1-kindlin.
Introduction
Focal adhesions (FAs) are specialized integrin-dependent adhesion complexes, which mediate cell anchoring to the extracellular matrix (ECM) (Winograd-Katz et al., 2014). FAs also function as mechano-transducing machineries perceiving and integrating diverse physical and (bio)chemical environmental cues, and transducing them into intracellular signaling pathways (Zaidel-Bar et al., 2007a; Wehrle-Haller, 2012; Yu et al., 2012). Indeed, FAs control cellular programs as diverse as cell adhesion, migration, survival, growth, proliferation, and differentiation (Wehrle-Haller, 2012; Winograd-Katz et al., 2014). To accomplish these diverse regulatory functions, αβ heterodimeric integrin receptors (Hynes, 2002) cluster in the plasma membrane, recruit numerous proteins to their cytoplasmic tails, and give rise to a highly dynamic intracellular protein network which has been termed the “integrin adhesome” (Zaidel-Bar et al., 2007a; Winograd-Katz et al., 2014). The tight regulation of its protein composition ensures FAs functioning as mechanical anchoring points, as well as signaling platforms (Wozniak et al., 2004; Zaidel-Bar et al., 2007a; Geiger and Yamada, 2011; Winograd-Katz et al., 2014).
The extensive implication of integrins and FAs-dependent signaling in pathological conditions (Bachmann et al., 2019) pushes current research towards a better understanding of their functioning and spatiotemporal regulation (Wu et al., 2019; Su et al., 2020). A few years ago, interferometric photoactivated localization microscopy (iPALM) has revealed a layered organization of integrin-containing FAs (Kanchanawong et al., 2010; Case et al., 2015). This model, proposing the spatial segregation of specific adhesome components between a integrin signaling layer (closest to the membrane), a force transduction layer, and an actin regulatory layer (innermost), has been endorsed by studies making advantage of diverse techniques, such as single protein tracking microscopy, superresolution microscopy, proximity biotinylation, and bimolecular fluorescence complementation (Dong et al., 2016; Chastney et al., 2020; Legerstee and Houtsmuller, 2021; Orre et al., 2021; Ripamonti et al., 2021). Despite these great advances, the characterization of several structural and mechanical aspects of the sophisticated integrin-dependent protein network, a comprehensive understanding of the FA machinery is still far from being accomplished (Chastney et al., 2020; Legerstee and Houtsmuller, 2021). The decoding of how specific cellular responses can be provoked by a given physiological, pathological, or pharmacological stimulus is challenged by the interdependency of FA players, regulatory systems, including the plasma membrane and its composition, and the tension across integrin receptors (Vogel, 2006; Gauthier and Roca-Cusachs, 2018). In addition, a wide range of post-translational modifications and the expression of many FA protein splice variants and isoforms generate additional layers of complexity that need to be understood to identify specific versus more general functions of FAs (Anthis et al., 2010; Choi et al., 2011; Soto-Ribeiro et al., 2019).
This review offers an overview of the molecular basis of the mechano-sensitivity and mechano-signaling capacity of the core FA proteins β3 integrin, talin1, and kindlin (Figure 1) that enable mechano-transduction. The focus is on β3 integrins as a paradigm for paxillin- and mechano-dependent mechanisms that may be extended to other classes of integrins. We will first address β3 integrin receptors, their link to talin1 and kindlin, and how paxillin is recruited to this complex for further mechanical stabilization, as well as to elicit diverse signaling pathways. We will highlight the role of paxillin and its central position to integrate the structural changes of the β3 integrin-talin1-kindlin complex, and offer evidence that paxillin is not only a scaffold or signaling protein as previously described, but also a key component of the mechano-transducing machinery (Figure 2).
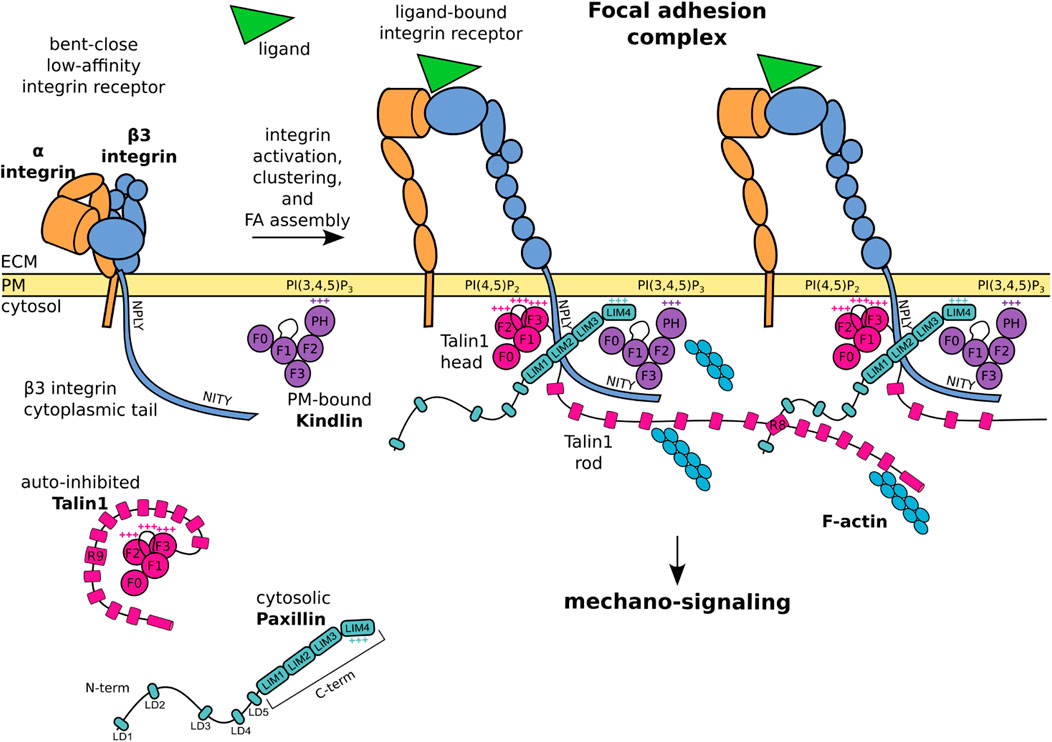
FIGURE 1. Schematic representation of core FA proteins and their interactions. The β3 integrin subunit is composed of an extracellular domain, a transmembrane domain, and a C-terminal cytoplasmic tail presenting the membrane proximal NPLY talin-binding and membrane distal NITY kindlin-binding sites. The heterodimeric integrin receptor in a bent-close low-affinity conformation (left) switches to an extended-open high-affinity state and binds ligands and intracellular proteins (right). Talin1 consists of a globular head, an unstructured linker, and a C-terminal rod domain which intramolecularly interacts with the head domain to keep cytosolic talin1 auto-inhibited. Upon integrin activation, the talin1 F2 domain binds to membrane phospholipid PI(4,5)P2, the talin F3 subdomain to the membrane proximal NPLY motif in the β3 integrin cytoplasmic tail, and the talin rod engages the F-actin network. Kindlin is similarly organized to talin-head but with the addition of a PH domain inserted within the F2 domain which recognizes membrane phosphoinositides, while the F3 domain binds to the membrane-distal NITY motif in the β3 integrin cytoplasmic tail. The paxillin amino-terminal half presents five short LD motifs and is followed by the carboxyl-terminal half composed of four LIM domains. The paxillin N-terminal LD1 and LD2 interact with the talin1 R8 domain, the LIM domains point towards the membrane proximal region, the positively charged LIM4 domain interacts with kindlin and the plasma membrane. One of the paxillin LIM domain could recognize the Y presented by the NPLY motif. +++ indicates positively charged regions. For representative purposes, the β3 integrin tail is outsized and some protein domains are simplified or omitted for clarity.
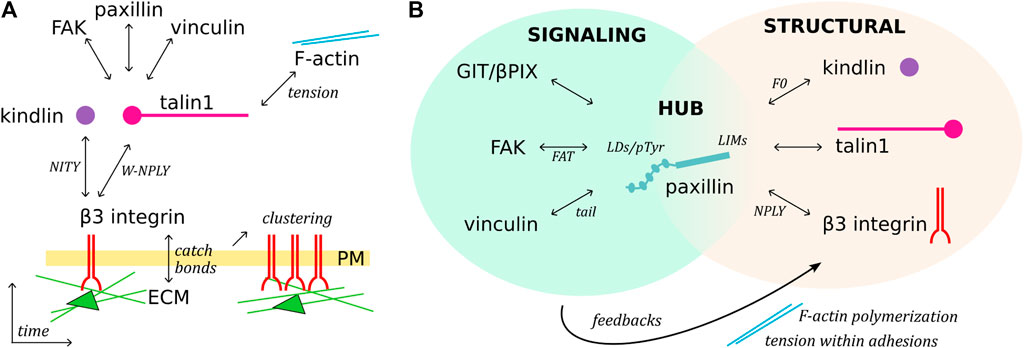
FIGURE 2. Temporal sequence of β3 integrin activation and paxillin-mediated organization of adhesions. (A), Schematic representation of FA assembly over time. Ligand, talin and kindlin binding to integrin receptors triggers their clustering, mediates the mechanical connection with the F-actin network, and recruits cytoplasmic proteins. (B), Paxillin is a structural, signaling and linker component of FAs. C-terminal LIM domains target paxillin to FAs, possibly directly interacting with talin, kindlin, and β3 integrin (putative domains indicated). The paxillin N-terminus functions as a signaling molecule, binding/recruiting different subsets of FA proteins, modulating F-actin polymerization and tension within adhesions, and therefore generating feedback signaling which can lead to FA turnover.
The αvβ3 Integrin Receptor
Integrins come in different flavors, ranging from diversity in ligand binding and exhibiting specific structural features (Hynes, 2002; Bachmann et al., 2019), which makes it impossible to cover the entire family in this review. For historical reasons the αvβ3 integrin is one of the best studied receptors, with implication in many pathophysiological settings (Zhu C. et al., 2019), representing a typical example of many integrin-dependent functions.
The β3 integrin receptors comprise two heterodimers originating from the pairing of the β3 subunit with either αIIb or αv chains, creating the αIIbβ3 and the αvβ3 heterodimers, respectively (Hynes, 2002; Bachmann et al., 2019). The αIIbβ3 integrin complex is a platelet-specific receptor which is activated by multiple signaling cascades to trigger platelet activation and aggregation (Ye et al., 2012; Huang et al., 2019). In contrast, the αvβ3 integrin has a wider expression and physiological functions related to tissue repair and inflammation in osteoclasts, platelets, megakaryocytes, kidney, vascular smooth muscles, endothelium, and placenta (Horton, 1997). In addition, αvβ3 is upregulated in endothelial cells undergoing tumor-induced angiogenesis (Mahabeleshwar et al., 2007), as well as in many tumor cells (Horton, 1997; Vonlaufen et al., 2001). The αvβ3 integrin recognizes the Arg-Gly-Asp (RGD)-tripeptide-containing sequence present in different ECM ligands (Pytela et al., 1985; Horton, 1997; Humphries et al., 2006), and preferentially binds to vitronectin and osteopontin, especially under low force conditions (Bachmann et al., 2020). Changes in mechanical cues of the microenvironment enlarge the ligand preference of αvβ3 integrin, and can induce mis-regulation of integrin-dependent signaling pathways (Bachmann et al., 2020), as in the case of pathological ECM stiffening in the tumor niche (Attieh et al., 2017). The αvβ3 receptor also plays a role in tumor progression and metastasis formation: by controlling the actin cytoskeleton (Havaki et al., 2007); by supporting tumor cell binding to, and transmigration across activated endothelia (Saalbach et al., 2005); by synergizing with VEGF-dependent pathways to promote angiogenesis (Mahabeleshwar et al., 2007); and by sustaining the activation of the Src kinase (Huveneers et al., 2007). Importantly, αvβ3 FAs localize proteolytically active matrix metalloproteinases at the cell surface (Brooks et al., 1996) and support efficient directed cell migration to promote metastasis formation (Ballestrem et al., 2001).
The β3 Integrin-talin1-Kindlin-Paxillin Complex
The assembly of FAs requires the conformational switch of the β3 integrin receptor from a bent-close low-affinity, to an extended-open high-affinity state (Figure 1). This activation can be triggered by the binding of the intracellular adapter proteins talin1 and kindlin to the cytoplasmic tail of the β3 integrin (Hytonen and Wehrle-Haller, 2014; Bachmann et al., 2019; Huang et al., 2019). These integrin activators are essential for integrin-dependent attachment and spreading: talin-null cells and kindlin-null cells display a non-adherent phenotype (Bottcher et al., 2017), suggesting a lack of transmission of mechanical signals and of cellular responses.
The progression from first integrin-adapter interactions and ECM-ligand binding toward FA maturation involves integrins clustering and their mechanical connection to the intracellular actin network (Thievessen et al., 2013; Hytonen and Wehrle-Haller, 2014) (Figure 1; Figure 2A). During this process, force is a key player acting at several steps. For example, catch bonds (i.e. force-dependent bonds strengthened by the force applied along the receptor) are formed at the level of the integrin-ligand interaction (Gauthier and Roca-Cusachs, 2018), while the mechanical tension along the integrin-adapters-actin axis leads to the exposure of cryptic binding sites in the talin C-terminal rod domain (Del Rio et al., 2009; Rahikainen et al., 2017), thus favoring the interaction with adapter proteins (e.g., paxillin, vinculin, and FAK) and the assembly of multiprotein signaling complexes (Hytonen and Vogel, 2008; Hytonen and Wehrle-Haller, 2014; Goult et al., 2018).
Talin1
By virtue of its structure, talin fulfils the role of a mechano-sensor of the extracellular rigidity, as well as of a mechano-transducer (Austen et al., 2015; Gough and Goult, 2018). The talin N-terminal head domain binds to integrins, induces conformational changes of the juxtamembrane- and ecto-domains of the integrin receptor, and stimulates integrin activation and clustering (Wegener et al., 2008; Saltel et al., 2009). Alongside, the binding of the talin C-terminal rod domain to the F-actin network directly transmits mechanical forces to the cellular cytoskeleton (Zhang X. et al., 2008; Rahikainen et al., 2017). As a feedback mechanism, the stretching of the talin rod domain reveals additional binding sites and ensures a tension-dependent recruitment of cytoplasmic proteins to the adhesion complexes (Figure 1; Figure 2A).
The talin N-terminal head domain consists of a globular FERM domain (F0 to F3 subdomains) (Zhang et al., 2020), connected by an unstructured linker to a C-terminal rod domain, which contains 13 α-helical bundles (R1 to R13) (Rahikainen et al., 2017) (Figure 1). The interaction of the talin head with the talin rod domain (via F3-R9) keeps cytosolic talin in a globular, autoinhibited conformation (Goksoy et al., 2008; Calderwood et al., 2013) (Figure 1). The recruitment of the talin head to the plasma membrane is controlled by two mechanisms: 1) the binding of the membrane-bound Rap1 GTPase to the F0 and F1-subdomains (Lagarrigue et al., 2020), and 2) the simultaneous binding of the F1-loop and the F2 subdomains to the membrane phospholipid PI(4,5)P2 (Anthis et al., 2009; Saltel et al., 2009; Goult et al., 2010). The concomitant membrane association of a basic loop in the talin F3 subdomain leads to the release of the autoinhibition, the detachment of the C-terminal domain, and the exposure of an additional β3 integrin tail binding site in the talin F3 subdomain, which assures binding to a juxtamembrane acidic motif, as well as the membrane proximal W-NPLY peptide, in the β3 integrin cytoplasmic tail (Cluzel et al., 2005; Wegener et al., 2008; Saltel et al., 2009; Zhang et al., 2020) (Figure 1). Full integrin activation and clustering however requires an F1-loop mediated interaction with the inner-membrane clasp to open the inhibitory salt-bridge formed between the αv and β3 integrin tails (Kukkurainen et al., 2020; Lagarrigue et al., 2020).
Once the auto-inhibited conformation of talin is released, talin engages the F-actin network: either directly through the two main actin-binding sites in the rod domain (Figure 1; Figure 2A); or indirectly, through the interaction with F-actin-bound vinculin (Humphries et al., 2007; Austen et al., 2015; Rahikainen et al., 2017; Gough and Goult, 2018; Atherton et al., 2020). When this mechanical connection is established, the application of tension results in the reversible unfolding of the talin rod, which reveals cryptic binding sites and allows the conversion of the tensional force on the talin rod into the recruitment of additional adapters (Yao et al., 2016; Rahikainen et al., 2017; Goult et al., 2018). Importantly, the role of talin in integrin activation is distinct from its mechano-transducing function (Austen et al., 2015; Rahikainen et al., 2017). In fact, the binding of a talin head only construct lacking actin binding capacity and the ability to transmit mechanical force is sufficient to induce integrin “inside-out” activation and clustering, in the absence of mechano-transmission and FA-dependent signaling (Cluzel et al., 2005; Zhang X. et al., 2008; Saltel et al., 2009; Rahikainen et al., 2017; Keeble et al., 2019; Kukkurainen et al., 2020).
Kindlin
Proteins of the kindlin family, also known as FERMT proteins, have a structure similar to the talin head, with F0, F1, F2 and F3 domains, and a largely unstructured F1-loop (Li et al., 2017; Zhang et al., 2020) (Figure 1). In addition, within the F2 domain of kindlin is inserted a PH (pleckstrin homology) domain that recognizes membrane phosphoinositides PIP2 and PIP3 (Liu et al., 2011; Liu et al., 2012). The F3 domain of kindlin binds to the membrane-distal NITY motif and the preceding ß-sheet of the β3 integrin cytoplasmic tail (Moser et al., 2008; Harburger et al., 2009; Li et al., 2017; Bachmann et al., 2019) (Figure 1). An indirect binding of kindlin to F-actin is mediated by the ILK/Pinch/Parvin (IPP) complex (Nikolopoulos and Turner, 2000; Honda et al., 2013; Kadry et al., 2018). However, a direct interaction of kindlin with F-actin was also suggested by pull down assays (Bledzka et al., 2016). Like talin, kindlin is essential for integrin activation (Montanez et al., 2008; Moser et al., 2008; Harburger et al., 2009; Theodosiou et al., 2016; Hirbawi et al., 2017; Li et al., 2017) and cell spreading, in a mechanism proposed to be mediated by its binding to paxillin (Theodosiou et al., 2016). Mechano-transduction of kindlin within FAs appears to be linked to its ability of inducing the talin head-mediated activation and clustering of integrins (Kukkurainen et al., 2020), an essential step in the assembly of FAs (Gao et al., 2017; Li et al., 2017). In the sequence of events leading to integrin activation and clustering, it is proposed that membrane-associated kindlin assures the initial integrin-recognition event (Figure 2A), which is followed by the talin recruitment and immobilisation of the integrin-talin1-kindlin complex within FAs (Orre et al., 2019).
Paxillin
Paxillin is a fundamental FA-associated adapter that connects structural and signaling components (Figure 2B), including tyrosine and serine/threonine kinases and GAPs/GEFs (Schaller, 2001). This hub protein coordinates integrin-downstream signaling pathways (Zaidel-Bar et al., 2007a; Green and Brown, 2019), contributing to cell spreading (Wade et al., 2002; Brimer et al., 2014; Pinon et al., 2014), migration, and proliferation (Deakin and Turner, 2008). In addition to its physiological role, paxillin sustains pathological processes in cancer progression (Lopez-Colome et al., 2017), including cell invasion (Iwasaki et al., 2002), metastasis (Ito et al., 2000) and angiogenesis (German et al., 2014).
Paxillin is composed of two modules (Figure 1): an unstructured amino-terminal half, comprising five leucine- and aspartic acid-rich motifs (with the consensus LDXLLXXL and thus named LD) forming short amphipathic α-helices (Bertolucci et al., 2005); and a carboxyl-terminal half composed of four LIM domains, each folded in two consecutive zinc fingers (Freyd et al., 1990; Velyvis et al., 2001). Recruitment of paxillin to FAs is mediated by the array of LIM domains, while its signaling capacity mostly relies on the N-terminal LD motif containing sequences (Brown et al., 1996; Ripamonti et al., 2021) (Figure 2B).
Although several interactions of paxillin with the elements of the β3 integrin-talin1-kindlin complex have been reported (Zacharchenko et al., 2016; Bottcher et al., 2017; Gao et al., 2017; Gough and Goult, 2018; Zhu C. et al., 2019; Atherton et al., 2020), a comprehensive understanding of paxillin engagement with this protein complex is missing. While the region of paxillin interacting with the talin F2/F3 subdomain remains unclear (Gao et al., 2017), the short helices of LD1 and LD2 can both pack against the side of the talin R8 four-helix bundle (Zacharchenko et al., 2016; Gough and Goult, 2018) (Figure 1). This mechanism of talin-binding is exploited also by other FA proteins (e.g., Rho GAP, DLC1) to interact with the talin rod, suggesting that competitive interactions among different LD-motif binding proteins, such as FAK, vinculin and talin can take place within FAs (Zacharchenko et al., 2016).
Several reports have suggested that paxillin also interacts with kindlin to promote integrin activation and cell spreading (Bottcher et al., 2017; Zhu L. et al., 2019). A direct binding of the paxillin LIM3 domain to the PH domain of kindlin was proposed, based on co-immunoprecipitation experiments, deletion mutagenesis and binding assays (Theodosiou et al., 2016). In addition, interactions between the N-terminal LD motifs of paxillin and the PH and F0 domains of kindlin2, as well as between the F0 domain and the paxillin LIM3-LIM4 domains, have been identified by lysine cross-linking proteomic experiments of recombinant kindlin2-paxillin complexes (Bottcher et al., 2017). These apparently conflicting data may represent different maturation stages of FAs. It is also possible that the exceptional abundance of lysine residues within the paxillin LD motifs and the LIM4 domain revealed interactions that are only short lived or not occurring in a physiological context. The NMR structure of the kindlin F0 domain complexed with paxillin LIM4 domain (Zhu L. et al., 2019) is consistent with the recently proposed orientation of paxillin within the FA complex, and with the interaction of its positively charged LIM4 domain with the plasma membrane (Kanchanawong et al., 2010; Ripamonti et al., 2021) (Figure 1). Interestingly, the disposition of proteins within adhesions has been also addressed by means of a proximity biotinylation assay (BioID), which revealed that the paxillin N-terminus could extend for ∼25 nm into the cytoplasm, and accommodate interactions within the intermediate zone of FAs, where are situated proteins that cannot be detected by using kindlin2 as BioID probe (Dong et al., 2016). All this is consistent with iPALM studies showing that N-terminally tagged paxillin is farther away from the PM compared to the C-terminally-tagged protein (Kanchanawong et al., 2010).
The interaction of paxillin with β3 integrin is still controversial: although reported two decades ago (Pfaff and Jurdic, 2001), several biochemical experiments failed to detect the direct binding of paxillin to the cytoplasmic tail of β3 (Brown et al., 1996; de Curtis and Malanchini, 1997; Tanaka et al., 2010). According to the tension-dependent recruitment of paxillin to FAs and stressed actin filaments (Sawada and Sheetz, 2002; Schiller et al., 2011; Sun et al., 2020; Winkelman et al., 2020), these results may be explained by the lack of tension and proper presentation of crucial integrin residues, required for paxillin binding (Pinon et al., 2014; Ripamonti et al., 2021). Different LIM domain-mediated protein-protein interactions involve the recognition of Tyr-containing motifs by the aromatic pocket of LIM domains (Wu and Gill, 1994; Wixler et al., 2000). By analogy, it was proposed that paxillin is binding to the membrane-proximal NPLY motif of β3 integrin (Pinon et al., 2014; Soto-Ribeiro et al., 2019; Ripamonti et al., 2021). Interestingly, modification of the talin1-binding NPLY sequence led to the loss of paxillin recruitment at FAs, and to a delay in cell spreading (Wegener et al., 2007; Pinon et al., 2014; Soto-Ribeiro et al., 2019).
The intricate interplay between paxillin and FA components is exemplified by the observation that none of the in vitro protein-protein interactions identified so far is strictly required or sufficient for paxillin recruitment to FAs in living cells (Ripamonti et al., 2021). Conversely, a multitude of low-affinity interactions could contribute to paxillin localization at FAs and/or nascent adhesions prior to tensional force generation. For instance: the kindlin F0 binding to paxillin LIM4 domain was proposed to mediate paxillin recruitment to the plasma membrane at sites of FA assembly (Zhu L. et al., 2019); similarly the dynamic and transient binding of the paxillin LIM4 domain to the plasma membrane was shown to stabilize paxillin docking to FAs (Ripamonti et al., 2021) (Figure 1). Furthermore, a tension-independent paxillin binding to talin was disclosed by the employment of a mitochondrial targeting assay (Atherton et al., 2020), and a solid-phase binding assay (Ripamonti et al., 2021). This interaction could be functionally similar to the binding of paxillin LIM3 to the PH domain of kindlin, which was suggested to drive paxillin recruitment into nascent adhesions but not into mature FAs (Theodosiou et al., 2016). To which extent each of these interactions contributes to the stable docking of paxillin within FAs was recently addressed by measuring the off-rate of engineered paxillin molecules photoactivated within FAs in living cells (Ripamonti et al., 2021). This study confirmed the presence of a multitude of low-affinity interactions leading to paxillin FA-localization, and a complex interplay of LIM1, LIM2 and LIM4 domains to get paxillin stabilized within mature FAs.
Paxillin as a Central Hub Mediating Mechano-Transduction at FAs
Integrin-mediated adhesions are described as mechano-sensitive because of their changes in response to mechanical stimuli (Hytonen and Wehrle-Haller, 2016; Gauthier and Roca-Cusachs, 2018). However, adhesions also fulfil the role of mechano-transducer, transmitting physical and mechanical signals from the ECM to the cytoskeleton, and converting them into cellular responses (Wehrle-Haller, 2012; Stutchbury et al., 2017). This function of FAs relies on the presence of intracellular proteins capable of sensing force-induced conformational changes, as observed for the talin and kindlin adapters (Stutchbury et al., 2017; Bachmann et al., 2019).
Several reports described the mechano-sensitivity of paxillin, although the molecular basis of its force sensing capacity is at the present not fully understood. The presence of talin and kindlin for the arrival of paxillin at nascent adhesions is necessary but not sufficient, since the development of force across the adhesion complex is also required (Cluzel et al., 2005; Hytonen and Wehrle-Haller, 2014) (Figure 2A). Along this line, it was shown that paxillin exhibits a stretch-dependent binding to the cytoskeleton (Sawada and Sheetz, 2002), as well as a remarkable ability of its LIM domains to detect mechanically strained stress fibers (Smith et al., 2013). Furthermore, several LIM domain-containing proteins that cluster at FAs (Kadrmas and Beckerle, 2004) are recruited in a myosin II-dependent fashion, suggesting that LIM domains could function as tension sensors of a strained F-actin network (Schiller et al., 2011; Sun et al., 2020; Winkelman et al., 2020).
The tension-dependent binding of paxillin to adhesions, combined to its hub function, suggests a major role of this protein in integrating signals from the integrin complex and in activating molecular pathways shaping cell behaviour (Green and Brown, 2019) (Figure 2B). The versatility of paxillin in the selection of binding partners is supported by the nature of its LD domains (Alam et al., 2020) that generally establish poorly selective, transient interactions, which require multiple layers of regulation (Alam et al., 2014). Due to the low binding affinity of single LD motifs, multiple simultaneous interactions are required to achieve stable complexes and elicit cellular responses (Alam et al., 2014). For example, opposite faces of the four-helix bundle in the FAT (Focal Adhesion Targeting) domain of FAK and in vinculin tail associate to paxillin LD2 and LD4 (Hoellerer et al., 2003).
Owing to its extraordinary connection with a plethora of adhesome components, paxillin is regarded as a unique protein capable of integrating the diverse functions of FAs (Green and Brown, 2019; Chastney et al., 2020). In other words, paxillin fulfils the crucial linker function connecting the core actin, the cell cortex, the signaling, and the regulatory modules constituted by subsets of FA proteins (Figure 2B) (Green and Brown, 2019; Chastney et al., 2020). For a complete understanding of paxillin interactions and functions, precise analyses considering FA protein isoforms and their post-translational modifications should be considered as well. The analysis of these aspects goes beyond the goal of this review, yet we can provide as an example the cell adhesion-triggered paxillin phosphorylation at Tyr31 and Tyr118 (Burridge et al., 1992) which modulates its binding to β3 integrin adhesions (Ripamonti et al., 2021), possibly by increasing paxillin affinity for FAK and vinculin (Zaidel-Bar et al., 2007b; Choi et al., 2011; Case et al., 2015). Noteworthy, the described paxillin-dependent nanoscale (re-)localization of vinculin within the FA architecture (Case et al., 2015) suggests that paxillin functions as a FA organizer beside its linker function (Figure 2B) (Green and Brown, 2019).
How tension and the assembly of the described β3 integrin-talin1-kindlin-paxillin complex at FAs is dynamically regulated during cell motility remains an open question. Adhesion remodelling directly and positively correlates with the ability of cells to migrate (Deakin and Turner, 2011), which physically relies on adhesion formation at the leading edge and adhesion disassembly at the cell rear (Webb et al., 2004; Cluzel et al., 2005). The latter was proposed to be under the control of Src-mediated phosphorylation of paxillin Tyr31/118 (Cortesio et al., 2011). Accordingly, Tyr-to-Phe mutations of these residues hampered adhesion turnover (Webb et al., 2004; Zaidel-Bar et al., 2007b) and inhibited tumor cell invasion (Mekhdjian et al., 2017). Consistent with these findings, sustained paxillin binding to FAs, phosphorylation of Tyr31/118, and FAK signaling can result in FA disassembly and turnover of its components (Webb et al., 2004). On the other hand, loss of paxillin phosphorylation was proposed to be responsible of hindering FA disassembly and support FA maturation toward fibrillar adhesions and their translocation to the cell center (Zaidel-Bar et al., 2007b; Bachmann et al., 2019).
Paxillin may be involved in the regulation of tension at the cell edge during migration on ECM ligands. In this direction, the complex between the ArfGAP and scaffold protein GIT1 (G-protein-coupled receptor-kinase interacting protein-1) and the guanine nucleotide exchange factor for Rac1 βPix has been implicated in the regulation of FAs and cell migration (Turner et al., 1999; Premont et al., 2000). GIT1 is recruited to FAs by direct binding of its FA-targeting domain to paxillin LD2 and LD4 motifs (Schmalzigaug et al., 2007; Zhang Z. M. et al., 2008; Wehrle-Haller and Bastmeyer, 2014). Recently, evidence has been provided for the formation of protein condensates of the GIT1/βPix complex driven by liquid-liquid phase separation (Zhu et al., 2020), a process involved in the organization and compartmentalization of several events occurring in the cytoplasm and the nucleus of eukaryotic cells (Banani et al., 2017; de Curtis, 2021). The results from the study of the Zhang’s group indicate that the formation of GIT1/βPix condensates and their targeting at FAs by paxillin are required to regulate cell migration (Zhu et al., 2020). Paxillin is shown to promote the formation of GIT1/βPix condensates, and one intriguing hypothesis is that paxillin-mediated formation and recruitment of GIT1/βPix condensates at FAs may modulate F-actin polymerization and tension within adhesions to modulate FAs turnover (Figure 2B). Also, dominant-active Rac1-transfected cells presented slower integrin turnover than control cells, indicating that βPix activity may locally stabilize the turnover of integrins, and arrest retrograde sliding adhesions (Ballestrem et al., 2001). These mechanisms could explain the paxillin-mediated rescue of unstable and rapidly sliding adhesions, indicating a role of paxillin in the control of the F-actin feedback loop (Ripamonti et al., 2021).
Conclusion and Perspectives
The gathering of structural and positional data led to the proposal of the slanted fence model of FAs, in which connections among neighbouring integrin-talin1-kindlin-paxillin units stabilize the complex (Sun et al., 2016; Bachmann et al., 2019; Ripamonti et al., 2021) (Figure 1). The proposed layered organization of FAs potentially bears the secret how integrin receptors support mechanical load and create intracellular signaling in response to changes in tensional forces (Kanchanawong et al., 2010; Bachmann et al., 2019). The detailed characterization of this key biological mechanism, tightly related to paxillin recruitment and binding to FAs (Figure 2) (Cluzel et al., 2005; Hytonen and Wehrle-Haller, 2014, 2016; Ripamonti et al., 2021), will help in the development of efficient integrin-targeting anti-cancer therapies, so far challenged by the complexity of the integrin system which has caused unexpected side effects (Su et al., 2020; Li et al., 2021).
Author Contributions
MR wrote the manuscript in collaboration with BW-H and IdC.
Funding
This work was supported by grant IG 20203 from AIRC (Associazione Italiana per la Ricerca sul Cancro) to IdC. MR is supported by a post-doctoral fellowship from Fondazione Umberto Veronesi.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Alam, T., Alazmi, M., Naser, R., Huser, F., Momin, A. A., Astro, V., et al. (2020). Proteome-level Assessment of Origin, Prevalence and Function of Leucine-Aspartic Acid (LD) Motifs. Bioinformatics 36, 1121–1128. doi:10.1093/bioinformatics/btz703
Alam, T., Alazmi, M., Gao, X., and Arold, S. T. (2014). How to Find a Leucine in a Haystack? Structure, Ligand Recognition and Regulation of Leucine-Aspartic Acid (LD) Motifs. Biochem. J. 460, 317–329. doi:10.1042/bj20140298
Anthis, N. J., Wegener, K. L., Critchley, D. R., and Campbell, I. D. (2010). Structural Diversity in Integrin/talin Interactions. Structure 18, 1654–1666. doi:10.1016/j.str.2010.09.018
Anthis, N. J., Wegener, K. L., Ye, F., Kim, C., Goult, B. T., Lowe, E. D., et al. (2009). The Structure of an Integrin/talin Complex Reveals the Basis of Inside-Out Signal Transduction. EMBO J. 28, 3623–3632. doi:10.1038/emboj.2009.287
Atherton, P., Lausecker, F., Carisey, A., Gilmore, A., Critchley, D., Barsukov, I., et al. (2020). Relief of Talin Autoinhibition Triggers a Force-independent Association with Vinculin. J. Cel Biol 219, e201903134. doi:10.1083/jcb.201903134
Attieh, Y., Clark, A. G., Grass, C., Richon, S., Pocard, M., Mariani, P., et al. (2017). Cancer-associated Fibroblasts lead Tumor Invasion through Integrin-β3-dependent Fibronectin Assembly. J. Cel Biol 216, 3509–3520. doi:10.1083/jcb.201702033
Austen, K., Ringer, P., Mehlich, A., Chrostek-Grashoff, A., Kluger, C., Klingner, C., et al. (2015). Extracellular Rigidity Sensing by Talin Isoform-specific Mechanical Linkages. Nat. Cel Biol 17, 1597–1606. doi:10.1038/ncb3268
Bachmann, M., Schäfer, M., Mykuliak, V. V., Ripamonti, M., Heiser, L., Weissenbruch, K., et al. (2020). Induction of Ligand Promiscuity of αVβ3 Integrin by Mechanical Force. J. Cel Sci 133, jcs242404. doi:10.1242/jcs.242404
Bachmann, M., Kukkurainen, S., Hytönen, V. P., and Wehrle-Haller, B. (2019). Cell Adhesion by Integrins. Physiol. Rev. 99, 1655–1699. doi:10.1152/physrev.00036.2018
Ballestrem, C., Hinz, B., Imhof, B. A., and Wehrle-Haller, B. (2001). Marching at the Front and Dragging behind. J. Cel Biol 155, 1319–1332. doi:10.1083/jcb.200107107
Banani, S. F., Lee, H. O., Hyman, A. A., and Rosen, M. K. (2017). Biomolecular Condensates: Organizers of Cellular Biochemistry. Nat. Rev. Mol. Cel Biol 18, 285–298. doi:10.1038/nrm.2017.7
Bertolucci, C. M., Guibao, C. D., and Zheng, J. (2005). Structural Features of the Focal Adhesion Kinase-Paxillin Complex Give Insight into the Dynamics of Focal Adhesion Assembly. Protein Sci. 14, 644–652. doi:10.1110/ps.041107205
Bledzka, K., Bialkowska, K., Sossey-Alaoui, K., Vaynberg, J., Pluskota, E., Qin, J., et al. (2016). Kindlin-2 Directly Binds Actin and Regulates Integrin Outside-In Signaling. J. Cel Biol 213, 97–108. doi:10.1083/jcb.201501006
Böttcher, R. T., Veelders, M., Rombaut, P., Faix, J., Theodosiou, M., Stradal, T. E., et al. (2017). Kindlin-2 Recruits Paxillin and Arp2/3 to Promote Membrane Protrusions during Initial Cell Spreading. J. Cel Biol 216, 3785–3798. doi:10.1083/jcb.201701176
Brimer, N., Wade, R., and Vande Pol, S. (2014). Interactions between E6, FAK, and GIT1 at Paxillin LD4 Are Necessary for Transformation by Bovine Papillomavirus 1 E6. J. Virol. 88, 9927–9933. doi:10.1128/jvi.00552-14
Brooks, P. C., Strömblad, S., Sanders, L. C., Von Schalscha, T. L., Aimes, R. T., Stetler-Stevenson, W. G., et al. (1996). Localization of Matrix Metalloproteinase MMP-2 to the Surface of Invasive Cells by Interaction with Integrin αvβ3. Cell 85, 683–693. doi:10.1016/s0092-8674(00)81235-0
Brown, M. C., Perrotta, J. A., and Turner, C. E. (1996). Identification of LIM3 as the Principal Determinant of Paxillin Focal Adhesion Localization and Characterization of a Novel Motif on Paxillin Directing Vinculin and Focal Adhesion Kinase Binding. J. Cel Biol 135, 1109–1123. doi:10.1083/jcb.135.4.1109
Burridge, K., Turner, C. E., and Romer, L. H. (1992). Tyrosine Phosphorylation of Paxillin and pp125FAK Accompanies Cell Adhesion to Extracellular Matrix: a Role in Cytoskeletal Assembly. J. Cel Biol 119, 893–903. doi:10.1083/jcb.119.4.893
Calderwood, D. A., Campbell, I. D., and Critchley, D. R. (2013). Talins and Kindlins: Partners in Integrin-Mediated Adhesion. Nat. Rev. Mol. Cel Biol 14, 503–517. doi:10.1038/nrm3624
Case, L. B., Baird, M. A., Shtengel, G., Campbell, S. L., Hess, H. F., Davidson, M. W., et al. (2015). Molecular Mechanism of Vinculin Activation and Nanoscale Spatial Organization in Focal Adhesions. Nat. Cel Biol 17, 880–892. doi:10.1038/ncb3180
Chastney, M. R., Lawless, C., Humphries, J. D., Warwood, S., Jones, M. C., Knight, D., et al. (2020). Topological Features of Integrin Adhesion Complexes Revealed by Multiplexed Proximity Biotinylation. J. Cel Biol 219, e202003038. doi:10.1083/jcb.202003038
Choi, C. K., Zareno, J., Digman, M. A., Gratton, E., and Horwitz, A. R. (2011). Cross-correlated Fluctuation Analysis Reveals Phosphorylation-Regulated Paxillin-FAK Complexes in Nascent Adhesions. Biophysical J. 100, 583–592. doi:10.1016/j.bpj.2010.12.3719
Cluzel, C., Saltel, F., Lussi, J., Paulhe, F., Imhof, B. A., and Wehrle-Haller, B. (2005). The Mechanisms and Dynamics of αvβ3 Integrin Clustering in Living Cells. J. Cel Biol 171, 383–392. doi:10.1083/jcb.200503017
Cortesio, C. L., Boateng, L. R., Piazza, T. M., Bennin, D. A., and Huttenlocher, A. (2011). Calpain-mediated Proteolysis of Paxillin Negatively Regulates Focal Adhesion Dynamics and Cell Migration. J. Biol. Chem. 286, 9998–10006. doi:10.1074/jbc.m110.187294
de Curtis, I. (2021). Biomolecular Condensates at the Front: Cell Migration Meets Phase Separation. Trends Cel Biol. 31, 145–148. doi:10.1016/j.tcb.2020.12.002
de Curtis, I., and Malanchini, B. (1997). Integrin-mediated Tyrosine Phosphorylation and Redistribution of Paxillin during Neuronal Adhesion. Exp. Cel Res. 230, 233–243. doi:10.1006/excr.1996.3423
Deakin, N. O., and Turner, C. E. (2011). Distinct Roles for Paxillin and Hic-5 in Regulating Breast Cancer Cell Morphology, Invasion, and Metastasis. MBoC 22, 327–341. doi:10.1091/mbc.e10-09-0790
Deakin, N. O., and Turner, C. E. (2008). Paxillin Comes of Age. J. Cel Sci 121, 2435–2444. doi:10.1242/jcs.018044
Del Rio, A., Perez-Jimenez, R., Liu, R., Roca-Cusachs, P., Fernandez, J. M., and Sheetz, M. P. (2009). Stretching Single Talin Rod Molecules Activates Vinculin Binding. Science 323, 638–641. doi:10.1126/science.1162912
Dong, J. M., Tay, F. P., Swa, H. L., Gunaratne, J., Leung, T., Burke, B., et al. (2016). Proximity Biotinylation Provides Insight into the Molecular Composition of Focal Adhesions at the Nanometer Scale. Sci. Signal. 9, rs4. doi:10.1126/scisignal.aaf3572
Freyd, G., Kim, S. K., and Horvitz, H. R. (1990). Novel Cysteine-Rich Motif and Homeodomain in the Product of the Caenorhabditis elegans Cell Lineage Gene Lin-II. Nature 344, 876–879. doi:10.1038/344876a0
Gao, J., Huang, M., Lai, J., Mao, K., Sun, P., Cao, Z., et al. (2017). Kindlin Supports Platelet Integrin αIIbβ3 Activation by Interacting with Paxillin. J. Cel Sci 130, 3764–3775. doi:10.1242/jcs.205641
Gauthier, N. C., and Roca-Cusachs, P. (2018). Mechanosensing at Integrin-Mediated Cell-Matrix Adhesions: from Molecular to Integrated Mechanisms. Curr. Opin. Cel Biol. 50, 20–26. doi:10.1016/j.ceb.2017.12.014
Geiger, B, and Yamada, KM (2011). Molecular architecture and function of matrix adhesions. Cold Spring Harb Perspect Biol 3, a005033. doi:10.1101/cshperspect.a005033
German, A. E., Mammoto, T., Jiang, E., Ingber, D. E., and Mammoto, A. (2014). Paxillin controls endothelial cell migration and tumor angiogenesis by altering neuropilin 2 expression. J Cell Sci 127, 1672–1683. doi:10.1242/jcs.132316
Goksoy, E., Ma, Y.-Q., Wang, X., Kong, X., Perera, D., Plow, E. F., and Qin, J. (2008). Structural basis for the autoinhibition of talin in regulating integrin activation. Molecular Cell 31, 124–133. doi:10.1016/j.molcel.2008.06.011
Gough, R. E., and Goult, B. T. (2018). The Tale of Two Talins - Two Isoforms to fine-tune Integrin Signalling. FEBS Lett. 592, 2108–2125. doi:10.1002/1873-3468.13081
Goult, B. T., Bouaouina, M., Elliott, P. R., Bate, N., Patel, B., Gingras, A. R., et al. (2010). Structure of a Double Ubiquitin-like Domain in the Talin Head: a Role in Integrin Activation. EMBO J. 29, 1069–1080. doi:10.1038/emboj.2010.4
Goult, B. T., Yan, J., and Schwartz, M. A. (2018). Talin as a Mechanosensitive Signaling Hub. J. Cel Biol 217, 3776–3784. doi:10.1083/jcb.201808061
Green, H. J., and Brown, N. H. (2019). Integrin Intracellular Machinery in Action. Exp. Cel Res. 378, 226–231. doi:10.1016/j.yexcr.2019.03.011
Harburger, D. S., Bouaouina, M., and Calderwood, D. A. (2009). Kindlin-1 and -2 Directly Bind the C-Terminal Region of β Integrin Cytoplasmic Tails and Exert Integrin-specific Activation Effects. J. Biol. Chem. 284, 11485–11497. doi:10.1074/jbc.m809233200
Havaki, S., Kouloukoussa, M., Amawi, K., Drosos, Y., Arvanitis, L. D., Goutas, N., et al. (2007). Altered Expression Pattern of Integrin Alphavbeta3 Correlates with Actin Cytoskeleton in Primary Cultures of Human Breast Cancer. Cancer Cel Int 7, 16. doi:10.1186/1475-2867-7-16
Hirbawi, J., Bialkowska, K., Bledzka, K. M., Liu, J., Fukuda, K., Qin, J., et al. (2017). The Extreme C-Terminal Region of Kindlin-2 Is Critical to its Regulation of Integrin Activation. J. Biol. Chem. 292, 14258–14269. doi:10.1074/jbc.m117.776195
Hoellerer, M. K., Noble, M. E. M., Labesse, G., Campbell, I. D., Werner, J. M., and Arold, S. T. (2003). Molecular Recognition of Paxillin LD Motifs by the Focal Adhesion Targeting Domain. Structure 11, 1207–1217. doi:10.1016/j.str.2003.08.010
Honda, S., Shirotani-Ikejima, H., Tadokoro, S., Tomiyama, Y., and Miyata, T. (2013). The Integrin-Linked Kinase-PINCH-Parvin Complex Supports Integrin αIIbβ3 Activation. PLoS One 8, e85498. doi:10.1371/journal.pone.0085498
Horton, M. A. (1997). The αvβ3 Integrin "vitronectin Receptor". Int. J. Biochem. Cel Biol. 29, 721–725. doi:10.1016/s1357-2725(96)00155-0
Huang, J., Li, X., Shi, X., Zhu, M., Wang, J., Huang, S., et al. (2019). Platelet Integrin αIIbβ3: Signal Transduction, Regulation, and its Therapeutic Targeting. J. Hematol. Oncol. 12, 26. doi:10.1186/s13045-019-0709-6
Humphries, J. D., Byron, A., and Humphries, M. J. (2006). Integrin Ligands at a Glance. J. Cel Sci 119, 3901–3903. doi:10.1242/jcs.03098
Humphries, J. D., Wang, P., Streuli, C., Geiger, B., Humphries, M. J., and Ballestrem, C. (2007). Vinculin Controls Focal Adhesion Formation by Direct Interactions with Talin and Actin. J. Cel Biol 179, 1043–1057. doi:10.1083/jcb.200703036
Huveneers, S., Van Den Bout, I., Sonneveld, P., Sancho, A., Sonnenberg, A., and Danen, E. H. J. (2007). Integrin αvβ3 Controls Activity and Oncogenic Potential of Primed C-Src. Cancer Res. 67, 2693–2700. doi:10.1158/0008-5472.can-06-3654
Hytönen, V. P., and Vogel, V. (2008). How Force Might Activate Talin's Vinculin Binding Sites: SMD Reveals a Structural Mechanism. Plos Comput. Biol. 4, e24. doi:10.1371/journal.pcbi.0040024
Hytönen, V. P., and Wehrle-Haller, B. (2016). Mechanosensing in Cell-Matrix Adhesions - Converting Tension into Chemical Signals. Exp. Cel Res. 343, 35–41. doi:10.1016/j.yexcr.2015.10.027
Hytönen, V. P., and Wehrle-Haller, B. (2014). Protein Conformation as a Regulator of Cell-Matrix Adhesion. Phys. Chem. Chem. Phys. 16, 6342–6357. doi:10.1039/c3cp54884h
Ito, A., Kataoka, T. R., Watanabe, M., Nishiyama, K., Mazaki, Y., Sabe, H., et al. (2000). A Truncated Isoform of the PP2A B56 Subunit Promotes Cell Motility through Paxillin Phosphorylation. EMBO J. 19, 562–571. doi:10.1093/emboj/19.4.562
Iwasaki, T., Nakata, A., Mukai, M., Shinkai, K., Yano, H., Sabe, H., et al. (2002). Involvement of Phosphorylation of Tyr-31 and Tyr-118 of Paxillin in MM1 Cancer Cell Migration. Int. J. Cancer 97, 330–335. doi:10.1002/ijc.1609
Kadrmas, J. L., and Beckerle, M. C. (2004). The LIM Domain: from the Cytoskeleton to the Nucleus. Nat. Rev. Mol. Cel Biol 5, 920–931. doi:10.1038/nrm1499
Kadry, Y. A., Huet-Calderwood, C., Simon, B., and Calderwood, D. A. (2018). Kindlin-2 Interacts with a Highly Conserved Surface of ILK to Regulate Focal Adhesion Localization and Cell Spreading. J. Cel Sci 131, jcs221184. doi:10.1242/jcs.221184
Kanchanawong, P., Shtengel, G., Pasapera, A. M., Ramko, E. B., Davidson, M. W., Hess, H. F., et al. (2010). Nanoscale Architecture of Integrin-Based Cell Adhesions. Nature 468, 580–584. doi:10.1038/nature09621
Keeble, A. H., Turkki, P., Stokes, S., Khairil Anuar, I. N. A., Rahikainen, R., Hytonen, V. P., et al. (2019). Approaching Infinite Affinity through Engineering of Peptide-Protein Interaction. Proc Natl Acad Sci U S A.
Kukkurainen, S., Azizi, L., Zhang, P., Jacquier, M. C., Baikoghli, M., Von Essen, M., et al. (2020). The F1 Loop of the Talin Head Domain Acts as a Gatekeeper in Integrin Activation and Clustering. J. Cel Sci 133, jcs239202. doi:10.1242/jcs.239202
Lagarrigue, F., Paul, D. S., Gingras, A. R., Valadez, A. J., Sun, H., Lin, J., et al. (2020). Talin-1 Is the Principal Platelet Rap1 Effector of Integrin Activation. Blood 136, 1180–1190. doi:10.1182/blood.2020005348
Legerstee, K., and Houtsmuller, A. B. (2021). A Layered View on Focal Adhesions. Biology (Basel) 10, 1189. doi:10.3390/biology10111189
Li, H., Deng, Y., Sun, K., Yang, H., Liu, J., Wang, M., et al. (2017). Structural Basis of Kindlin-Mediated Integrin Recognition and Activation. Proc. Natl. Acad. Sci. U.S.A. 114, 9349–9354. doi:10.1073/pnas.1703064114
Li, M., Wang, Y., Li, M., Wu, X., Setrerrahmane, S., and Xu, H. (2021). Integrins as Attractive Targets for Cancer Therapeutics. Acta Pharmaceutica Sinica B 11, 2726–2737. doi:10.1016/j.apsb.2021.01.004
Liu, J., Fukuda, K., Xu, Z., Ma, Y.-Q., Hirbawi, J., Mao, X., et al. (2011). Structural Basis of Phosphoinositide Binding to Kindlin-2 Protein Pleckstrin Homology Domain in Regulating Integrin Activation. J. Biol. Chem. 286, 43334–43342. doi:10.1074/jbc.m111.295352
Liu, Y., Zhu, Y., Ye, S., and Zhang, R. (2012). Crystal Structure of Kindlin-2 PH Domain Reveals a Conformational Transition for its Membrane Anchoring and Regulation of Integrin Activation. Protein Cell 3, 434–440. doi:10.1007/s13238-012-2046-1
López-Colomé, A. M., Lee-Rivera, I., Benavides-Hidalgo, R., and López, E. (2017). Paxillin: a Crossroad in Pathological Cell Migration. J. Hematol. Oncol. 10, 50. doi:10.1186/s13045-017-0418-y
Mahabeleshwar, G. H., Feng, W., Reddy, K., Plow, E. F., and Byzova, T. V. (2007). Mechanisms of Integrin-Vascular Endothelial Growth Factor Receptor Cross-Activation in Angiogenesis. Circ. Res. 101, 570–580. doi:10.1161/circresaha.107.155655
Mekhdjian, A. H., Kai, F., Rubashkin, M. G., Prahl, L. S., Przybyla, L. M., Mcgregor, A. L., et al. (2017). Integrin-mediated Traction Force Enhances Paxillin Molecular Associations and Adhesion Dynamics that Increase the Invasiveness of Tumor Cells into a Three-Dimensional Extracellular Matrix. MBoC 28, 1467–1488. doi:10.1091/mbc.e16-09-0654
Montanez, E., Ussar, S., Schifferer, M., Bösl, M., Zent, R., Moser, M., et al. (2008). Kindlin-2 Controls Bidirectional Signaling of Integrins. Genes Dev. 22, 1325–1330. doi:10.1101/gad.469408
Moser, M., Nieswandt, B., Ussar, S., Pozgajova, M., and Fässler, R. (2008). Kindlin-3 Is Essential for Integrin Activation and Platelet Aggregation. Nat. Med. 14, 325–330. doi:10.1038/nm1722
Nikolopoulos, S. N., and Turner, C. E. (2000). Actopaxin, a New Focal Adhesion Protein that Binds Paxillin LD Motifs and Actin and Regulates Cell Adhesion. J. Cel Biol 151, 1435–1448. doi:10.1083/jcb.151.7.1435
Orré, T., Joly, A., Karatas, Z., Kastberger, B., Cabriel, C., Böttcher, R. T., et al. (2021). Molecular Motion and Tridimensional Nanoscale Localization of Kindlin Control Integrin Activation in Focal Adhesions. Nat. Commun. 12, 3104. doi:10.1038/s41467-021-23372-w
Orré, T., Rossier, O., and Giannone, G. (2019). The Inner Life of Integrin Adhesion Sites: From Single Molecules to Functional Macromolecular Complexes. Exp. Cel Res. 379, 235–244. doi:10.1016/j.yexcr.2019.03.036
Pfaff, M., and Jurdic, P. (2001). Podosomes in Osteoclast-like Cells. J. Cel Sci 114, 2775–2786. doi:10.1242/jcs.114.15.2775
Pinon, P., Pärssinen, J., Vazquez, P., Bachmann, M., Rahikainen, R., Jacquier, M.-C., et al. (2014). Talin-bound NPLY Motif Recruits Integrin-Signaling Adapters to Regulate Cell Spreading and Mechanosensing. J. Cel Biol 205, 265–281. doi:10.1083/jcb.201308136
Premont, R. T., Claing, A., Vitale, N., Perry, S. J., and Lefkowitz, R. J. (2000). The GIT Family of ADP-Ribosylation Factor GTPase-Activating Proteins. J. Biol. Chem. 275, 22373–22380. doi:10.1074/jbc.275.29.22373
Pytela, R., Pierschbacher, M. D., and Ruoslahti, E. (1985). A 125/115-kDa Cell Surface Receptor Specific for Vitronectin Interacts with the Arginine-Glycine-Aspartic Acid Adhesion Sequence Derived from Fibronectin. Proc. Natl. Acad. Sci. U.S.A. 82, 5766–5770. doi:10.1073/pnas.82.17.5766
Rahikainen, R., Von Essen, M., Schaefer, M., Qi, L., Azizi, L., Kelly, C., et al. (2017). Mechanical Stability of Talin Rod Controls Cell Migration and Substrate Sensing. Sci. Rep. 7, 3571. doi:10.1038/s41598-017-03335-2
Ripamonti, M., Liaudet, N., Azizi, L., Bouvard, D., Hytönen, V. P., and Wehrle-Haller, B. (2021). Structural and Functional Analysis of LIM Domain-dependent Recruitment of Paxillin to αvβ3 Integrin-Positive Focal Adhesions. Commun. Biol. 4, 380. doi:10.1038/s42003-021-01886-9
Saalbach, A., Wetzel, A., Haustein, U.-F., Sticherling, M., Simon, J. C., and Anderegg, U. (2005). Interaction of Human Thy-1 (CD 90) with the Integrin αvβ3 (CD51/CD61): an Important Mechanism Mediating Melanoma Cell Adhesion to Activated Endothelium. Oncogene 24, 4710–4720. doi:10.1038/sj.onc.1208559
Saltel, F., Mortier, E., Hytönen, V. P., Jacquier, M.-C., Zimmermann, P., Vogel, V., et al. (2009). New PI(4,5)P2- and Membrane Proximal Integrin-Binding Motifs in the Talin Head Control β3-integrin Clustering. J. Cel Biol 187, 715–731. doi:10.1083/jcb.200908134
Sawada, Y., and Sheetz, M. P. (2002). Force Transduction by Triton Cytoskeletons. J. Cel Biol 156, 609–615. doi:10.1083/jcb.200110068
Schaller, M. D. (2001). Paxillin: a Focal Adhesion-Associated Adaptor Protein. Oncogene 20, 6459–6472. doi:10.1038/sj.onc.1204786
Schiller, H. B., Friedel, C. C., Boulegue, C., and Fässler, R. (2011). Quantitative Proteomics of the Integrin Adhesome Show a Myosin II‐dependent Recruitment of LIM Domain Proteins. EMBO Rep. 12, 259–266. doi:10.1038/embor.2011.5
Schmalzigaug, R., Garron, M., Roseman, J., Xing, Y., Davidson, C., Arold, S., et al. (2007). GIT1 Utilizes a Focal Adhesion Targeting-Homology Domain to Bind Paxillin. Cell Signal. 19, 1733–1744. doi:10.1016/j.cellsig.2007.03.010
Smith, M. A., Blankman, E., Deakin, N. O., Hoffman, L. M., Jensen, C. C., Turner, C. E., et al. (2013). LIM Domains Target Actin Regulators Paxillin and Zyxin to Sites of Stress Fiber Strain. PLoS One 8, e69378. doi:10.1371/journal.pone.0069378
Soto-Ribeiro, M., Kastberger, B., Bachmann, M., Azizi, L., Fouad, K., Jacquier, M. C., et al. (2019). β1D Integrin Splice Variant Stabilizes Integrin Dynamics and Reduces Integrin Signaling by Limiting Paxillin Recruitment. J. Cel Sci 132, jcs224493. doi:10.1242/jcs.224493
Stutchbury, B., Atherton, P., Tsang, R., Wang, D. Y., and Ballestrem, C. (2017). Distinct Focal Adhesion Protein Modules Control Different Aspects of Mechanotransduction. J. Cel Sci 130, 1612–1624. doi:10.1242/jcs.195362
Su, C.-y., Li, J.-q., Zhang, L.-l., Wang, H., Wang, F.-h., Tao, Y.-w., et al. (2020). The Biological Functions and Clinical Applications of Integrins in Cancers. Front. Pharmacol. 11, 579068. doi:10.3389/fphar.2020.579068
Sun, X., Phua, D. Y. Z., Axiotakis, L., Smith, M. A., Blankman, E., Gong, R., et al. (2020). Mechanosensing through Direct Binding of Tensed F-Actin by LIM Domains. Developmental Cel 55, 468–482. doi:10.1016/j.devcel.2020.09.022
Sun, Z., Guo, S. S., and Fässler, R. (2016). Integrin-mediated Mechanotransduction. J. Cel Biol 215, 445–456. doi:10.1083/jcb.201609037
Tanaka, T., Moriwaki, K., Murata, S., and Miyasaka, M. (2010). LIM Domain-Containing Adaptor, Leupaxin, Localizes in Focal Adhesion and Suppresses the Integrin-Induced Tyrosine Phosphorylation of Paxillin. Cancer Sci. 101, 363–368. doi:10.1111/j.1349-7006.2009.01398.x
Theodosiou, M., Widmaier, M., Böttcher, R. T., Rognoni, E., Veelders, M., Bharadwaj, M., et al. (2016). Kindlin-2 Cooperates with Talin to Activate Integrins and Induces Cell Spreading by Directly Binding Paxillin. Elife 5, e10130. doi:10.7554/eLife.10130
Thievessen, I., Thompson, P. M., Berlemont, S., Plevock, K. M., Plotnikov, S. V., Zemljic-Harpf, A., et al. (2013). Vinculin-actin Interaction Couples Actin Retrograde Flow to Focal Adhesions, but Is Dispensable for Focal Adhesion Growth. J. Cel Biol 202, 163–177. doi:10.1083/jcb.201303129
Turner, C. E., Brown, M. C., Perrotta, J. A., Riedy, M. C., Nikolopoulos, S. N., Mcdonald, A. R., et al. (1999). Paxillin LD4 Motif Binds PAK and PIX through a Novel 95-kD Ankyrin Repeat, ARF-GAP Protein: A Role in Cytoskeletal Remodeling. J. Cel Biol 145, 851–863. doi:10.1083/jcb.145.4.851
Velyvis, A., Yang, Y., Wu, C., and Qin, J. (2001). Solution Structure of the Focal Adhesion Adaptor PINCH LIM1 Domain and Characterization of its Interaction with the Integrin-Linked Kinase Ankyrin Repeat Domain. J. Biol. Chem. 276, 4932–4939. doi:10.1074/jbc.m007632200
Vogel, V. (2006). Mechanotransduction Involving Multimodular Proteins: Converting Force into Biochemical Signals. Annu. Rev. Biophys. Biomol. Struct. 35, 459–488. doi:10.1146/annurev.biophys.35.040405.102013
Vonlaufen, A., Wiedle, G., Borisch, B., Birrer, S., Luder, P., and Imhof, B. A. (2001). Integrin αvβ3 Expression in Colon Carcinoma Correlates with Survival. Mod. Pathol. 14, 1126–1132. doi:10.1038/modpathol.3880447
Wade, R., Bohl, J., and Vande Pol, S. (2002). Paxillin Null Embryonic Stem Cells Are Impaired in Cell Spreading and Tyrosine Phosphorylation of Focal Adhesion Kinase. Oncogene 21, 96–107. doi:10.1038/sj.onc.1205013
Webb, D. J., Donais, K., Whitmore, L. A., Thomas, S. M., Turner, C. E., Parsons, J. T., et al. (2004). FAK-src Signalling through Paxillin, ERK and MLCK Regulates Adhesion Disassembly. Nat. Cel Biol 6, 154–161. doi:10.1038/ncb1094
Wegener, K. L., Basran, J., Bagshaw, C. R., Campbell, I. D., Roberts, G. C. K., Critchley, D. R., et al. (2008). Structural Basis for the Interaction between the Cytoplasmic Domain of the Hyaluronate Receptor Layilin and the Talin F3 Subdomain. J. Mol. Biol. 382, 112–126. doi:10.1016/j.jmb.2008.06.087
Wegener, K. L., Partridge, A. W., Han, J., Pickford, A. R., Liddington, R. C., Ginsberg, M. H., et al. (2007). Structural Basis of Integrin Activation by Talin. Cell 128, 171–182. doi:10.1016/j.cell.2006.10.048
Wehrle-Haller, B., and Bastmeyer, M. (2014). Intracellular Signaling and Perception of Neuronal Scaffold through Integrins and Their Adapter Proteins. Prog. Brain Res. 214, 443–460. doi:10.1016/b978-0-444-63486-3.00018-9
Wehrle-Haller, B. (2012). Structure and Function of Focal Adhesions. Curr. Opin. Cel Biol. 24, 116–124. doi:10.1016/j.ceb.2011.11.001
Winkelman, J. D., Anderson, C. A., Suarez, C., Kovar, D. R., and Gardel, M. L. (2020). Evolutionarily Diverse LIM Domain-Containing Proteins Bind Stressed Actin Filaments through a Conserved Mechanism. Proc. Natl. Acad. Sci. U.S.A. 117, 25532–25542. doi:10.1073/pnas.2004656117
Winograd-Katz, S. E., Fässler, R., Geiger, B., and Legate, K. R. (2014). The Integrin Adhesome: from Genes and Proteins to Human Disease. Nat. Rev. Mol. Cel Biol 15, 273–288. doi:10.1038/nrm3769
Wixler, V., Geerts, D., Laplantine, E., Westhoff, D., Smyth, N., Aumailley, M., et al. (2000). The LIM-Only Protein DRAL/FHL2 Binds to the Cytoplasmic Domain of Several α and β Integrin Chains and Is Recruited to Adhesion Complexes. J. Biol. Chem. 275, 33669–33678. doi:10.1074/jbc.m002519200
Wozniak, M. A., Modzelewska, K., Kwong, L., and Keely, P. J. (2004). Focal Adhesion Regulation of Cell Behavior. Biochim. Biophys. Acta (Bba) - Mol. Cel Res. 1692, 103–119. doi:10.1016/j.bbamcr.2004.04.007
Wu, P. H., Opadele, A. E., Onodera, Y., and Nam, J. M. (2019). Targeting Integrins in Cancer Nanomedicine: Applications in Cancer Diagnosis and Therapy. Cancers (Basel) 11, 1783. doi:10.3390/cancers11111783
Wu, R. Y., and Gill, G. N. (1994). LIM Domain Recognition of a Tyrosine-Containing Tight Turn. J. Biol. Chem. 269, 25085–25090. doi:10.1016/s0021-9258(17)31502-8
Yao, M., Goult, B. T., Klapholz, B., Hu, X., Toseland, C. P., Guo, Y., et al. (2016). The Mechanical Response of Talin. Nat. Commun. 7, 11966. doi:10.1038/ncomms11966
Ye, F., Kim, C., and Ginsberg, M. H. (2012). Reconstruction of Integrin Activation. Blood 119, 26–33. doi:10.1182/blood-2011-04-292128
Yu, C.-h., Luo, W., and Sheetz, M. P. (2012). Spatial-temporal Reorganization of Activated Integrins. Cell Adhes. Migration 6, 280–284. doi:10.4161/cam.20753
Zacharchenko, T., Qian, X., Goult, B. T., Jethwa, D., Almeida, T. B., Ballestrem, C., et al. (2016). LD Motif Recognition by Talin: Structure of the Talin-DLC1 Complex. Structure 24, 1130–1141. doi:10.1016/j.str.2016.04.016
Zaidel-Bar, R., Itzkovitz, S., Ma'ayan, A., Iyengar, R., and Geiger, B. (2007a). Functional Atlas of the Integrin Adhesome. Nat. Cel Biol 9, 858–867. doi:10.1038/ncb0807-858
Zaidel-Bar, R., Milo, R., Kam, Z., and Geiger, B. (2007b). A Paxillin Tyrosine Phosphorylation Switch Regulates the Assembly and Form of Cell-Matrix Adhesions. J. Cel Sci 120, 137–148. doi:10.1242/jcs.03314
Zhang, P., Azizi, L., Kukkurainen, S., Gao, T., Baikoghli, M., Jacquier, M. C., et al. (2020). Crystal Structure of the FERM-Folded Talin Head Reveals the Determinants for Integrin Binding. Proc. Natl. Acad. Sci. U S A. 117 (51), 32402–32412. doi:10.1073/pnas.2014583117
Zhang, X., Jiang, G., Cai, Y., Monkley, S. J., Critchley, D. R., and Sheetz, M. P. (2008a). Talin Depletion Reveals independence of Initial Cell Spreading from Integrin Activation and Traction. Nat. Cel Biol 10, 1062–1068. doi:10.1038/ncb1765
Zhang, Z. M., Simmerman, J. A., Guibao, C. D., and Zheng, J. J. (2008b). GIT1 Paxillin-Binding Domain Is a Four-helix Bundle, and it Binds to Both Paxillin LD2 and LD4 Motifs. J. Biol. Chem. 283, 18685–18693. doi:10.1074/jbc.m801274200
Zhu, C., Kong, Z., Wang, B., Cheng, W., Wu, A., and Meng, X. (2019a). ITGB3/CD61: a Hub Modulator and Target in the Tumor Microenvironment. Am. J. Transl Res. 11, 7195–7208.
Zhu, J., Zhou, Q., Xia, Y., Lin, L., Li, J., Peng, M., et al. (2020). GIT/PIX Condensates Are Modular and Ideal for Distinct Compartmentalized Cell Signaling. Mol. Cel 79, 782–796. doi:10.1016/j.molcel.2020.07.004
Keywords: mechano-sensing, tensional force, lim domain, integrin activation, plasma membrane
Citation: Ripamonti M, Wehrle-Haller B and de Curtis I (2022) Paxillin: A Hub for Mechano-Transduction from the β3 Integrin-Talin-Kindlin Axis. Front. Cell Dev. Biol. 10:852016. doi: 10.3389/fcell.2022.852016
Received: 10 January 2022; Accepted: 14 March 2022;
Published: 05 April 2022.
Edited by:
Ben Goult, University of Kent, United KingdomReviewed by:
Jon Humphries, Manchester Metropolitan University, United KingdomIgor Barsukov, University of Liverpool, United Kingdom
Copyright © 2022 Ripamonti, Wehrle-Haller and de Curtis. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ivan de Curtis, ZGVjdXJ0aXMuaXZhbkBoc3IuaXQ=
 Marta Ripamonti
Marta Ripamonti Bernhard Wehrle-Haller
Bernhard Wehrle-Haller Ivan de Curtis
Ivan de Curtis