
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell Dev. Biol., 01 April 2022
Sec. Cellular Biochemistry
Volume 10 - 2022 | https://doi.org/10.3389/fcell.2022.839583
Pigs are important biomedical model animals for the study of human neurological diseases. Similar to human aggressive behavior in children and adolescents, weaned pigs also show more aggressive behavior after mixing, which has negative effects on animal welfare and growth performance. The identification of functional single-nucleotide polymorphisms (SNPs) related to the aggressive behavior of pigs would provide valuable molecular markers of the aggressive behavioral trait for genetic improvement program. The Rho GTPase–activating protein 24 (ARHGAP24) gene plays an important role in regulating the process of axon guidance, which may impact the aggressive behavior of pigs. By resequencing the entire coding region, partially adjacent introns and the 5′ and 3′ flanking regions, six and four SNPs were identified in the 5′ flanking region and 5′ untranslated region (UTR) of the porcine ARHGAP24 gene, respectively. Association analyses revealed that nine SNPs were significantly associated with aggressive behavioral traits (p = < 1.00 × 10–4–4.51 × 10–2), and their haplotypes were significantly associated with aggressive behavior (p = < 1.00 × 10–4–2.99 × 10–2). The core promoter region of the ARHGAP24 gene has been identified between −670 and −1,113 bp. Furthermore, the luciferase activity of allele A of rs335052970 was significantly less than that of allele G, suggesting that the transcriptional activity of the ARHGAP24 gene was inhibited by allele A of rs335052970. It was identified that the transcription factor p53 bound to the transcription factor binding sites (TFBSs) containing allele A of rs335052970. In porcine primary neural cells, p53 binds to the target promoter region of the ARHGAP24 gene, reduces its promoter transcriptional activity, and then reduces its messenger RNA (mRNA) and protein expression. The results demonstrated that the ARHGAP24 gene had significant genetic effects on aggressive behavioral traits of pigs. Therefore, rs335052970 in the ARHGAP24 gene can be used as a molecular marker to select the less aggressive pigs.
Aggressive behavior of pigs after mixing, an important animal welfare issue, causes negative impacts on the growth performance, feed conversion ratio, immunity, and meat quality, which affects the economic benefits of pig industries (Tuchscherer et al., 1998; D’Eath et al., 2010). Previous studies have shown that aggressive behavior was affected by environmental factors, such as stocking density (Arey, 1999), mixing (Greenwood et al., 2014), feeding space, and environmental enrichment (O'Connell et al., 2004). However, genetic factors also have an important impact on the aggressive behavior of pigs (Rohrer et al., 2013). Due to their complicated assessment process, aggressive behavioral traits are rarely included in traditional pig breeding selection programs. The phenotypic determination of individual animal aggressive behavior is challenging and limits the improvement of behavioral traits through genetic selection. Therefore, the identification of molecular genetic markers of aggressive behavior could contribute to the genetic selection of less aggressive pigs.
In the process of screening candidate genes for aggressive behavior in pigs, the porcine Rho GTPase–activating protein 24 (ARHGAP24) gene attracted our attention. The ARHGAP24 gene encodes Rho GTPase–activating protein (RhoGAP), which stimulates the GTPase activities of the Rho family of small GTPases and terminates the binding of Rho with GTP, thus inactivating the activity of Rho family proteins (Tcherkezian and Lamarche-Vane, 2007; Muller et al., 2020). Rho GTPase family members, including RhoA, Rac, and Cdc42 proteins (Bagci et al., 2020), play important roles during the development of the nervous system (Leslie et al., 2012; Antoine-Bertrand et al., 2016). RhoA and Rac1 are regulated by GTPase-activating proteins and serve as their downstream targets (Elvers et al., 2012). Previous studies also have shown that the ARHGAP24 gene is implicated in axon and dendrite outgrowth and branching (Nguyen et al., 2012). Meanwhile, it has been reported that aggressive behavior is associated with the axon guidance signaling pathway in humans (Viding et al., 2010; Zhang-James et al., 2019). Therefore, the ARHGAP24 gene may play an important role in regulating the process of axon guidance, which then impacts aggressive behavior. In addition, previous studies have found that the ARHGAP24 gene is associated with human depression (Watanabe et al., 2017) and childhood autism (Coutton et al., 2015) accompanied by aggressive behavior. The ARHGAP24 gene is also related to the growth performance of pigs (Meng et al., 2017). However, the role of the ARHGAP24 gene in the regulation of aggressive behavior in pigs has remained unclear.
In this study, we hypothesized that the aggressive behavior of pigs is associated with the expression and function of the ARHGAP24 gene. We aimed to identify the functional SNPs of the ARHGAP24 gene and investigate their molecular mechanisms for aggressive behavior regulation in weaned pigs after mixing. This research could provide valuable molecular markers of aggressive behavior for the genetic improvement of pigs.
This study was approved by the Animal Care and Use Committee of Nanjing Agricultural University (SYXK Su 2017-0007). A total of 500 piglets from 65 litters were selected in the Huaiyin pig breeding farm (Huaian, Jiangsu, China). The piglets were weaned at 35 days of age and moved into new empty pens with their original littermates in a nursery room 2 days before mixing. Then, nine or ten weaned pigs with the same sex and similar body weight from different litters were mixed in the pens of dimension 2.5 m × 2.2 m. The pens were equipped with slatted floors, stainless steel feeders, and nipple drinkers to allow ad libitum access to feed and water. The ear tissues of weaned piglets were collected, and genomic DNA was extracted by a standard phenol/chloroform method.
A digital video recording system (Hikvision DS-2CE56C2P-IT3 3.6 mm; Hikvision network hard disk video recorder DS-7808HW-E1/M; Hikvision Digital Technology Co. Ltd., Hangzhou, Zhejiang, China) was used to continuously record the behavior of piglets for 72 h after mixing. A video camera was installed over each pen. To individually identify pigs in the video recording, all pigs in each pen were marked with different numbers on their back using a spray paint (7CF, Shenzhen Zhaoxin Energy Co., Ltd., Shenzhen, Guangdong, China) before mixing. The definitions of aggressive behavioral traits used were described in our previous studies (Chen et al., 2019; Tong et al., 2019) with some additional new traits. Specifically, nine indicators were used to quantify aggression, and their description and definition are shown in Table 1. A fighting behavior was recorded when it lasted for more than 3 s. For the same pair of pigs, the intervening period between each fight event was at least 8 s (Stukenborg et al., 2011).
To identify the functional SNPs of the ARHGAP24 gene regulating the aggressive behavior of weaned pigs after mixing, specific primers (Supplementary Table S1) were used to amplify the ARHGAP24 gene, including the coding regions, partially adjacent introns, and the 5′- and 3′-flanking regions according to the reference genome sequence of pigs (GenBank accession number: NC_010450.4). The DNA sequences contained potential SNPs of the ARHGAP24 gene from 224 aggressive and docile pigs which were amplified by polymerase chain reactions (PCR). PCR was performed using 1.1 × T3 Super PCR Mix (TsingKe, Nanjing, Jiangsu, China), and the amplified PCR products were sequenced using the Sanger method. The DNA sequences of the porcine ARHGAP24 gene were aligned by DNAMAN (Lynnon Biosoft, Quebec, QC, Canada) and SnapGene Viewer software (BSL Biotech LLC, Chicago, IL, United States).
The extent of LD between the identified SNPs was estimated using Haploview 4.2 (the Broad Institute of MIT and Harvard, Cambridge, MA, United States). The association analyses for aggressive behavioral traits were performed using GLIMMIX procedure of SAS 9.4 with the following generalized linear mixed model:
The promoter region of the porcine ARHGAP24 gene was predicted by Promoter 2.0 (http://www.cbs.dtu.dk/services/Promoter/) (Knudsen, 1999) and Neural Network Promoter Prediction (https://www.fruitfly.org/seq_tools/promoter.html) (Reese, 2001). Putative transcriptional binding start sites caused by the SNP mutation in the 5’ flanking or the UTR region of the ARHGAP24 gene were predicted by using JASPAR 2020 (http://jaspar.genereg.net/) (Fornes et al., 2020) and AnimalTFDB 3.0 (http://bioinfo.life.hust.edu.cn/AnimalTFDB/#!/) (Hu et al., 2019).
The promoter region of the porcine ARHGAP24 gene was amplified by PCR using Vazyme LAmp Master Mix (Vazyme Biotech, Nanjing, Jiangsu, China). Subsequently, plasmids containing variable lengths of the truncated porcine ARHGAP24 promoter were individually amplified using different forward primers and a common reverse primer (ARHGAP24-P1: −33/+352; ARHGAP24-P2: −308/+352; ARHGAP24-P3: −670/+352; ARHGAP24-P4: −1,113/+352; ARHGAP24-P5: −1,572/+352; and ARHGAP24-P6: −1976/+352), and the primers contained MluI and XhoI recognition sequences, respectively (Supplementary Table S2). Subsequently, the amplified fragments were inserted into the multiple cloning sites of the pGL3-basic vector to generate luciferase reporter plasmids. Moreover, specific regions containing rs335052970, rs344700648, and rs339198696 were amplified using ARHGAP24-Haplotype primers contained the recognition sequences of MluI and XhoI (Supplementary Table S2). The DNA samples of piglets were amplified using primers (Supplementary Table S2) targeting the promoter region of the porcine ARHGAP24 gene containing the p53 transcription factor binding element (PBE) motif and then cloned into the pGL3-basic vector by MluI and XhoI. The plasmid with PBE was used as a DNA template and was amplified by point mutation primer-ARHGAP24-PBE-MUT (Supplementary Table S2). The cDNA fragments of p53 were amplified using the primer CDS-p53 (Supplementary Table S2) and connected to the eukaryotic expression vector pcDNA3.1 (+) (pcDNA3.1-p53). The plasmid structures were sequenced to confirm the integrity of the constructed fragments.
Human embryonic kidney 293T (HEK 293T) cells were used for promoter activity analysis. First, HEK 293T cells were cultured in an incubator at 37°C and 5% CO2. The cells were plated in 12-well plates with three wells for each group. On the following day, the plasmids contained the variable length of ARHGAP24 promoter fragments and the rs335052970 A or G allele and haplotypes were individually cotransfected into the HEK 293T cells with pRL-TK Renilla luciferase reporter vector (Promega, Madison, WI, United States) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, United States). The controls were the pGL3-basic and pGL3-control luciferase reporter gene vector. After 24 h, the cells were harvested with passive lysis buffer (Promega, Madison, WI, United States). The cell lysates were assayed for reporter gene activity using a dual-luciferase assay system (Promega, Madison, WI, United States) according to the manufacturer’s instructions.
Primary neural cells were prepared from cerebral cortices from a 1-day-old piglet, as previously described for rats (Yodoya et al., 2006). In brief, cerebral cortices were removed from the piglet’s brains. Then, the meninges and microvessels were carefully removed in ice-precooled D-Hanks’ balanced salt solution (HBSS, Gibco, Grand Island, NY, United States), and the brain tissues were minced into small pieces of about 1 mm3. After papain (Biofroxx, Einhausen, Germany) and DNase 1 (BioFroxx, Einhausen, Germany) were added, respectively, they were digested in an incubator at 37°C for 30 min. After the digestion was terminated with Dulbecco’s modified Eagle’s medium (DMEM, Gibco, Grand Island, NY, United States), they were sub-packed into 15-ml centrifuge tubes for centrifugation for 10 min. The porcine neural cells were cultured in DMEM supplemented with 20% fetal bovine serum (FBS, Gibco, Grand Island, NY, United States) at 37°C in a humidified atmosphere with 5% CO2 for 48 h. After 10 days of culture, the porcine neural cells were identified by immunofluorescence with anti–beta III tubulin (Tuj1) antibody (Abcam, Cambridge, United Kingdom). Endotoxin-free plasmids of pcDNA3.1-p53 and pcDNA3.1 were transfected into primary neural cells using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, United States). The siRNA of p53 and ARHGAP24 were designed and chemically synthesized by Shanghai Jima Pharmaceutical Technology Co., Ltd. The primer sequence is shown in Supplementary Table S2. Either scrambled siRNA or p53 siRNA plasmids was transfected into primary neural cells using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s instructions.
The cells were harvested at day 1 post transfection. The total RNA of porcine neural cells was extracted by using TRIzol reagent (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s protocol. The purity of RNA was detected by the NanoDrop 2000 (Thermo Fisher Scientific, Fremont, CA, United States). To quantify the mRNA expression level of ARHGAP24 and p53, total RNA was reverse-transcribed onto cDNA using the HiScript III RT SuperMix (Vazyme Biotech, Nanjing, Jiangsu, China). The RT-qPCR was performed on the Quantum Studio 5 quantitative PCR instrument (Applied Biosystems, Foster, CA, United States) using SYBR Green Master Mix (Vazyme Biotech, Nanjing, Jiangsu, China) and specific primers (Supplementary Table S2). Relative expression levels were calculated by using the 2−ΔΔCt method (Livak and Schmittgen, 2001). The coding gene expression levels were normalized to the expression of GAPDH. For the RT-qPCR reaction, each treatment had at least three biological replicates.
The cell protein lysates were harvested using 200 µL ice-cold radioimmunoprecipitation assay (RIPA) lysis buffer (Beyotime, Shanghai, China) with 1% phenylmethyl sulfonyl fluoride (PMSF, Beyotime, Shanghai, China). Total protein extracts were separated on 4–20% SDS–PAGE gels (GenScript Biotech, Nanjing, Jiangsu, China) and then blotted onto a polyvinylidene fluoride (PVDF) membrane (Bio-Rad, Hercules, CA, United States). After blocking with QuickBlock™ blocking buffer (Beyotime, Shanghai, China) for 30 min, the PVDF membranes were incubated overnight with the following primary antibodies: immunoreactive proteins were detected with a rabbit polyclonal antibody for anti-P53 (1:1,000; AF0879, Affinity, China), anti-ARHGAP24 (1:1,000; DF9858, Affinity, China), and anti-GAPDH (1:5,000; AF7021, Affinity, China). The appropriate antirabbit secondary antibody (1:8,000; S0001, Affinity, China) was used to incubate the membranes. The ECL peroxidase color development kit (Vazyme, Nanjing, Jiangsu, China) was used in chromogenic reaction according to the manufacturer’s instructions. The protein band visualization was performed by the ChemiDoc XRS + System (Bio-Rad, Hercules, CA, United States). The band density was analyzed using ImageJ software.
Data were analyzed using SAS 9.4 (SAS Institute Inc., Cary, NC, United States). Chi-square tests were applied to analyze the difference in allele frequency between the most aggressive and least aggressive pigs. The behavioral data were analyzed using the GLIMMIX procedure with a model option DIST = EXPO. The relative fluorescence activity value was normalized by the negative control pGL3-basic. The significance of luciferase activity statistics was analyzed by unpaired two-sided student’s t-test and one-way ANOVA. The results were presented as mean ± standard error of the mean (SEM), and p < 0.05 was considered statistically significant.
A total of 10 SNPs were identified by sequencing on the entire coding region, 5′- and 3′-flanking regions of the porcine ARHGAP24 gene in 178 pigs (Table 2). In total, six SNPs (rs339198696, rs344700648, rs335052970, rs344498203, rs323776551, and rs342083908) were located in the 5′-flanking region, four SNPs (rs333053350, rs342210686, rs328435752, and rs787973778) were located in the 5′-UTR of the porcine ARHGAP24 gene, and five SNPs (rs344700648, rs335052970, rs323776551, rs342083908, and rs787973778) showed significant difference (p < 0.05) in allele frequencies between the most aggressive and least aggressive pigs.
Association analyses between genotype and aggressive behaviors during the first 2, 24, 48, or 72 h after mixing are presented in Supplementary Table S3. A total of three SNPs (rs339198696, rs344700648, and rs335052970) in 5′-flanking regions were significantly associated with aggressive behaviors during the first 2, 24, 48, or 72 h after mixing (p < 0.05). Interestingly, all SNPs had a strong association with multiple aggressive behaviors during the first 2 h after mixing. Moreover, CAS, duration of active attacks, frequency of active attack, frequency of standoff, and win were greater in the pigs with the mutant AA genotype of rs335052970 than those in the pigs with the wild GG genotype during the first 2, 24, 48, or 72 h after mixing (p < 0.05, Figure 1). Similarly, four SNPs (rs333053350, rs342210686, rs328435752, and rs787973778) in the 5′-UTR were significantly associated with the duration of active attacks and duration of standoff (p < 0.05) (Supplementary Table S3 for details).
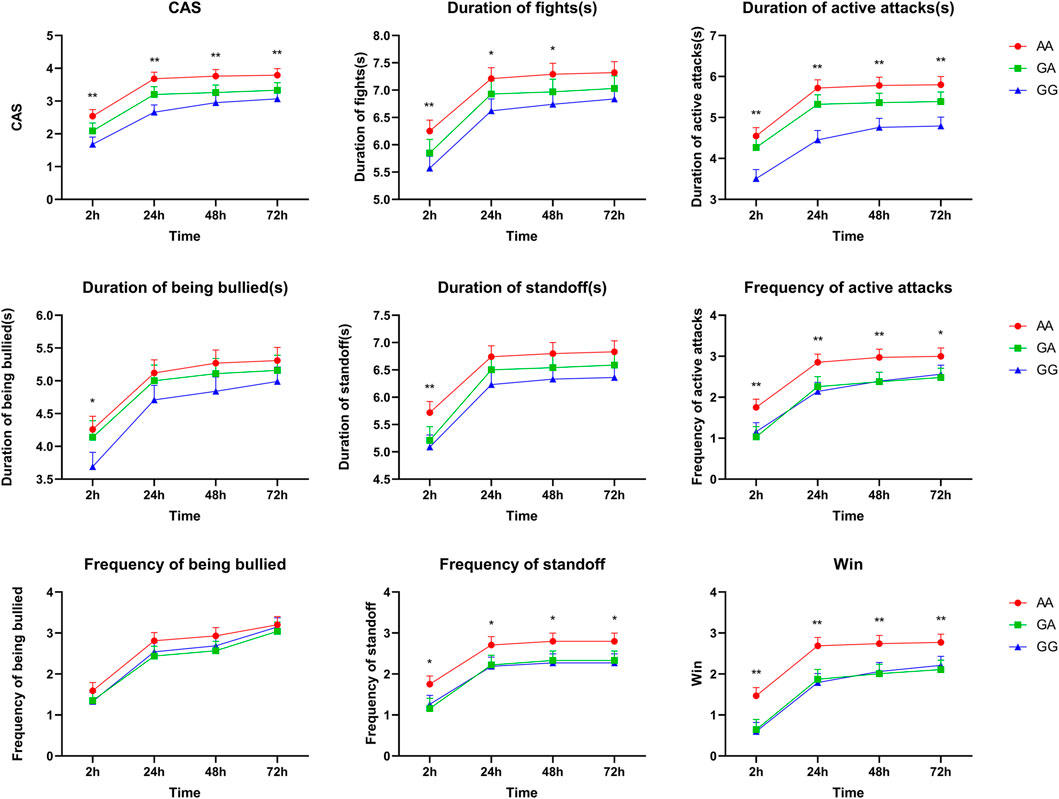
FIGURE 1. Associations of SNP rs335052970 in the ARHGAP24 gene with aggressive behavioral traits at the first 2, 24, 48, or 72 h after mixing in weaned pigs (LSM ± SE). *p < 0.05 and **p < 0.01 indicate that the difference is significant.
We estimated the LD among the 10 SNPs of the ARHGAP24 gene using Haploview 4.2. A total of seven SNPs (rs333053350, rs342210686, rs328435752, rs787973778, rs335052970, rs344700648, and rs339198696) were highly linked (D′ > 0.69; Figure 2) in two haplotype blocks. The first haplotype block (block 1) consisted of three haplotypes: H1 (GGAA), H2 (TAGC), and H3 (GAAA). The second haplotype block (block 2) consisted of three haplotypes: H1 (AAC), H2 (GAA), and H3 (GTA). The two haplotype blocks were significantly associated with aggressive behavior (p < 0.05) (Supplementary Table S4). In the haplotype block 1, pigs with haplotype H1 (GGAA) were more aggressive than pigs with haplotype H2 (TAGC) or H3 (GAAA). Similarly, pigs with haplotype H1 (AAC) in haplotype block 2 were more aggressive than those with haplotype H2 (GAA) or H3 (GTA) (p < 0.05).
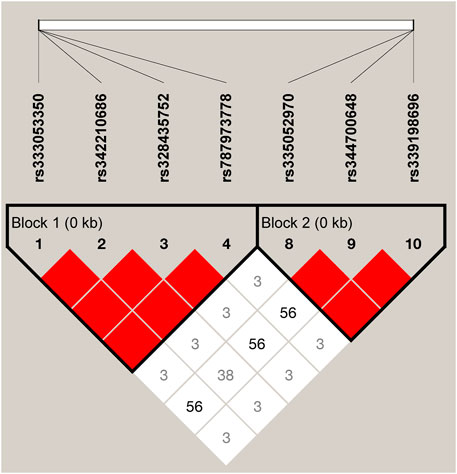
FIGURE 2. LD among the SNPs in the porcine ARHGAP24 gene (D’ = 0.03–1.00). It is to be noted that the blocks indicate haplotype blocks and the text before the horizontal numbers is the SNP name. The values in boxes are pairwise SNP correlations (D’), while bright red boxes without numbers represent complete LD (D’ = 1).
A total of two promoter regions (−1,364/−1,314 bp and +89/+139 bp) and three transcription initiation sites (−1,700, −800, and +100 bp) of the porcine ARHGAP24 gene were predicted by Promoter 2.0 and Neural Network Promoter Prediction. The transcription factor potential binding sites for RUNX2, RREB1, IRF2, IRF1, p53, CREBBP, POLR3A, and GLI1 were predicted in the 5′ flanking region of the porcine ARHGAP24 gene (Supplementary Table S5). In the promoter activity analyses, the luciferase activity of plasmids that contained the promoter fragments of the ARHGAP24 gene was greater than that of the pGL3-basic plasmid (p < 0.01) but less than that of the pGL3-control plasmid (p < 0.01). Moreover, the luciferase activity of pGL3-basic-P4, pGL3-basic-P5, and pGL3-basic-P6 was greater than that of pGL3-basic-P1, pGL3-basic-P2, and pGL3-basic-P3 (p < 0.01). The luciferase activity of pGL3-basic-P3 and pGL3-basic-P1 was greater than that of pGL3-basic-P2 (p < 0.05) (Figure 3A). These results revealed that the core promoter region of the ARHGAP24 gene is located between −670 and −1,113 bp, whereas a negative regulatory promoter region is located between −308 and −33 bp.
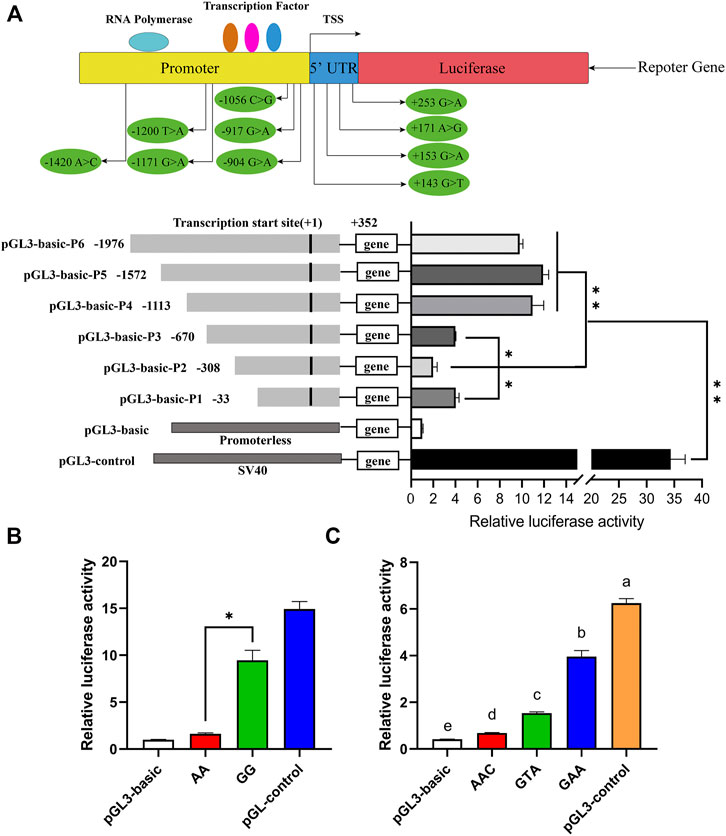
FIGURE 3. Luciferase assays for porcine ARHGAP24 promoter activity analyses. PGL3-basic as a negative control and PGL3-control as a positive control. (A) Gene promoter diagram and the location of SNPs in the promoter region and the 5′ UTR. A total of six luciferase reporter plasmids expressing successive truncations of the ARHGAP24 promoter sequence were constructed and transfected into HEK 293T cells. (B) Luciferase reporter gene assays of porcine ARHGAP24 alleles contained rs335052970 (−744G > A). (C) Luciferase activities of plasmids contained three haplotypes of the porcine ARHGAP24 gene. The relative luciferase activity values represent the mean ± SEM of three independent experiments. Statistical differences in luciferase activity were assessed using the one-way ANOVA; *p < 0.05 and **p < 0.01. Different letters (a, b, c, etc.) indicate that the difference is significant (p < 0.05).
The luciferase activity was greater in plasmids that contained the G allele of rs335052970 than that of plasmids containing the A allele (p < 0.01) (Figure 3B). Moreover, there are three linked SNPs (rs335052970, rs344700648, and rs339198696) in the core promoter region (−670/−1,113 bp) of the ARHGAP24 gene. They form only three haplotypes: H1 (AAC), H2 (GAA), and H3 (GTA). The luciferase activity of plasmids that contained the haplotypes of the core promoter region was greater than that of pGL3-basic but less than that of the pGL3-control (p < 0.0001). The relative luciferase activity of plasmids that contained haplotype GAA was the greatest, while that of plasmids that contained haplotype AAC was the least (p < 0.01) (Figure 3C). It indicates that the site affecting promoter activity is rs335052970 (−744G > A).
The effects of SNP rs335052970 (−744G > A) on the transcription factor binding sites (TFBSs) were predicted using the Animal TFDB online website. The allele A of rs335052970 was found to be located in the potential binding sequence of the transcription factor p53 (TP53) (Supplementary Table S5). To verify the binding sequence of the transcription factor p53 in the upstream region of the ARHGAP24 gene containing SNP rs335052970, a chromatin immunoprecipitation (ChIP) assay was used to demonstrate that p53 binds to the transcription factor binding element (PBE) motif directly in vivo (Figure 4A). To investigate whether p53 regulates the expression of ARHGAP24 through the PBE site, we cloned the PBE site into a pGL3 vector (Promega, United States) to construct PBE-allele A (pGL3-WT) and PBE-allele G (pGL3-MUT) reporter vectors (Figure 4B). Reporter vectors and p53 overexpression vector (pcDNA3.1-p53) were cotransfected into porcine neural cells. The results of immunofluorescence identification are shown in Figure 4C. Porcine primary neural cells treated with TuJ1 antibody showed red fluorescence, indicating that TuJ1 detection is positive, the cell neurites are connected with each other, and the dendritic contour is clearly visible. The mRNA expression level of p53 in the pcDNA3.1-p53 group was greater than that in the control group (p < 0.05) (Figure 5A). The mRNA expression level of p53 in the siRNA-p53 group was less than that in the scrambled group (p < 0.05) (Figure 5B). The luciferase activity of the pcDNA3.1-WT group was less than that of the pcDNA3.1-MUT group (p < 0.05) when p53 was overexpressed (Figure 5C). Moreover, the luciferase activity of the siRNA-p53 group was greater than that of the control group (p < 0.05) (Figure 5D). The overexpression of p53 reduced the mRNA and protein expression level of ARHGAP24 (p < 0.01) (Figures 5E,G), but interfering p53 increased the mRNA expression level of ARHGAP24 (p < 0.01) (Figures 5F,H).
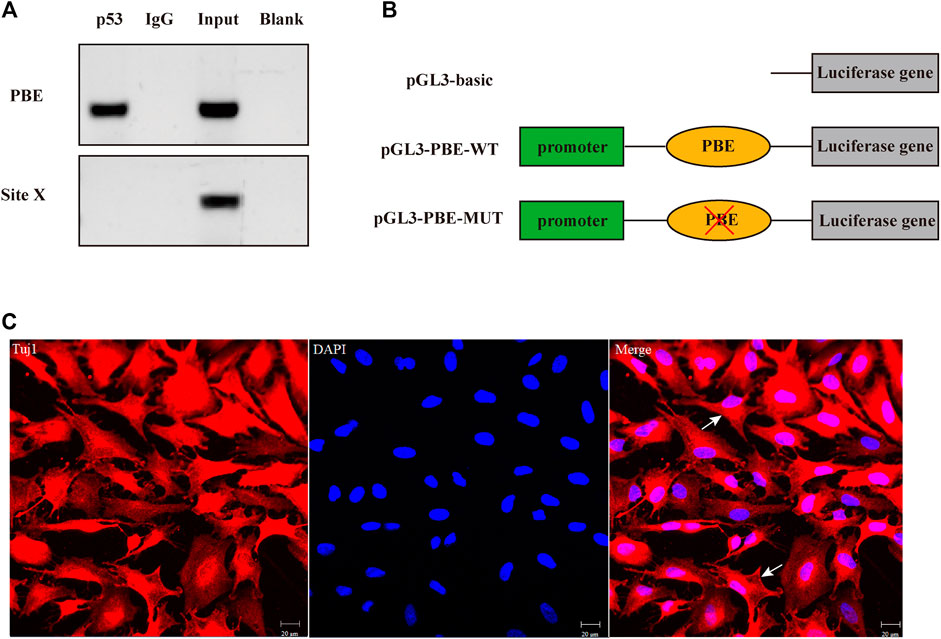
FIGURE 4. Transcription factor p53 directly targeted the binding element (PBE) motif of the ARHGAP24 gene in porcine neural cells. (A) Confirmation of the direct interaction between the p53 and ARHGAP24 promoter. ChIP-qPCR assay was performed with IgG as the negative control. Site X, a negative control locus, input, and total DNA from untreated cells. (B) Construction of luciferase activity reporter vectors containing wild-type (WT) and mutant-type (MUT) PBE on the promoter of the ARHGAP24 gene. Blue boxes represent the luciferase gene; green boxes represent pGL-3 promoter; orange ovals represent PBE; and red fork represents mutation. (C) Immunofluorescence identification of porcine primary neural cells. Immunofluorescence staining of porcine neural cells with Tuj1 (red) and DAPI (blue); the white arrow shows the primary porcine nerve cells; Scale bars represent 20 μm.
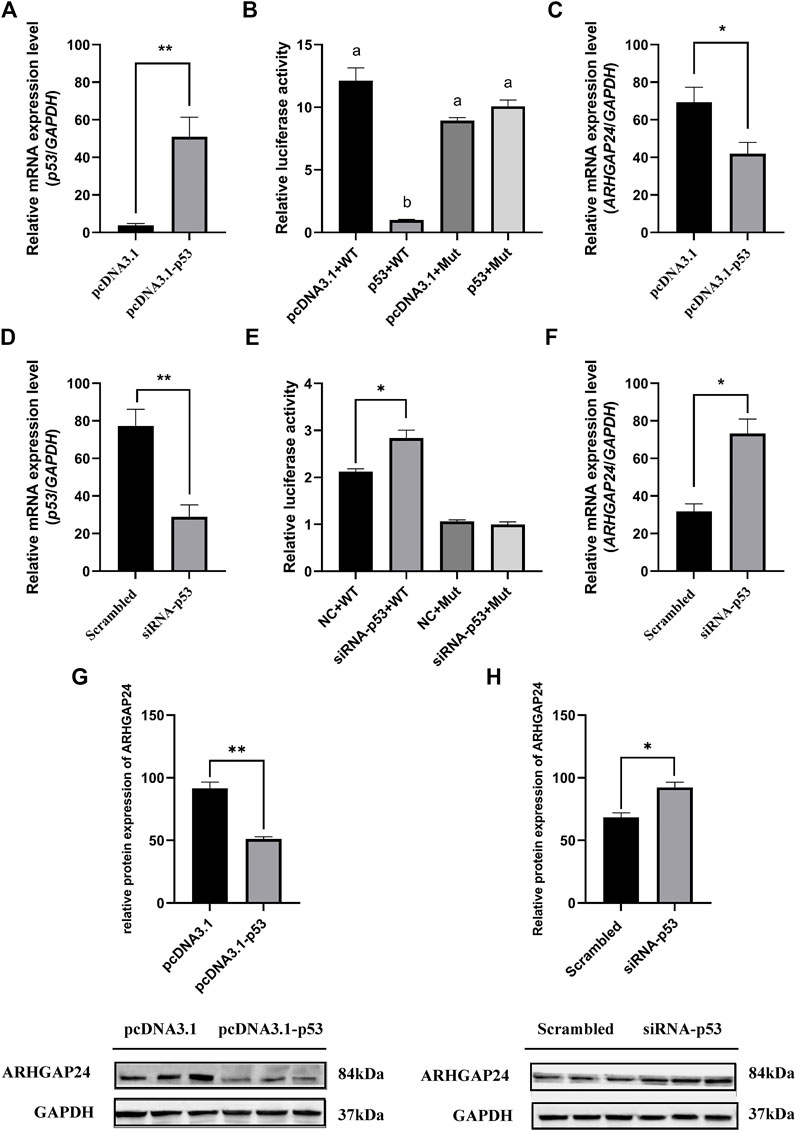
FIGURE 5. Transcription factor p53 regulates ARHGAP24 gene expression in porcine neural cells. (A) mRNA level of p53 after p53 overexpression. (B) mRNA level of p53 after p53 knockdown. (C) Luciferase activity of the ARHGAP24 promoter region after p53 overexpression (D) Luciferase activity of the ARHGAP24 promoter region after p53 knockdown (E) mRNA level of ARHGAP24 after p53 overexpression. (F) mRNA level of ARHGAP24 after p53 knockdown. (G) Western blot analyses of ARHGAP24 protein expression in porcine neural cells transfected with pcDNA3.1-p53 and pcDNA3.1 (+). (H) Western blot analyses of ARHGAP24 protein expression in porcine neural cells transfected with the scrambled and siRNA-p53 groups. The protein levels were normalized to GAPDH. *p < 0.05 and **p < 0.01. Different letters (a, b, c, etc.) before the columns indicate that the difference is significant (p < 0.05)
In order to explore how the signal pathway involved in the ARHGAP24 gene regulates aggressive behavior and the expression of related genes in the signal pathway when p53 is overexpressed or inhibited, we connected the eukaryotic expression vector pcDNA3.1 (+) (pcDNA3.1-p53) and chemically synthesized siRNA-p53 and siRNA-ARHGAP24 and then transfected into porcine neural cells to detect the expression level of related genes in the axon guidance pathway. The mRNA expression level of RHOA in the pcDNA3.1-p53 group was greater than that in the control group (p < 0.05) (Figure 6A), while the mRNA expression level of RHOA in the siRNA-p53 group was less than that in the scrambled group (p < 0.05) (Figure 6B). Similarly, the mRNA expression level of ROCK1 in the pcDNA3.1-p53 group had an increased tendency than that in the control group (p = 0.0567) (Figure 6C), while the mRNA expression level of ROCK1 in the siRNA-p53 group was less than that in the scrambled group (p < 0.05) (Figure 6D). By contrast, the mRNA expression level of RAC1 in the pcDNA3.1-p53 group was less than that in the control group (p < 0.05) (Figure 6E), while the mRNA expression level of RAC1 in the siRNA-p53 group was greater than that in the scrambled group (Figure 6F). Moreover, the mRNA expression level of RHOA and ROCK1 in the siRNA-ARHGAP24 group was greater than that in the scrambled group (p < 0.05) (Figures 6G,H). However, the mRNA expression level of RAC1 was not different between the siRNA-ARHGAP24 and scrambled groups (Figure 3I).
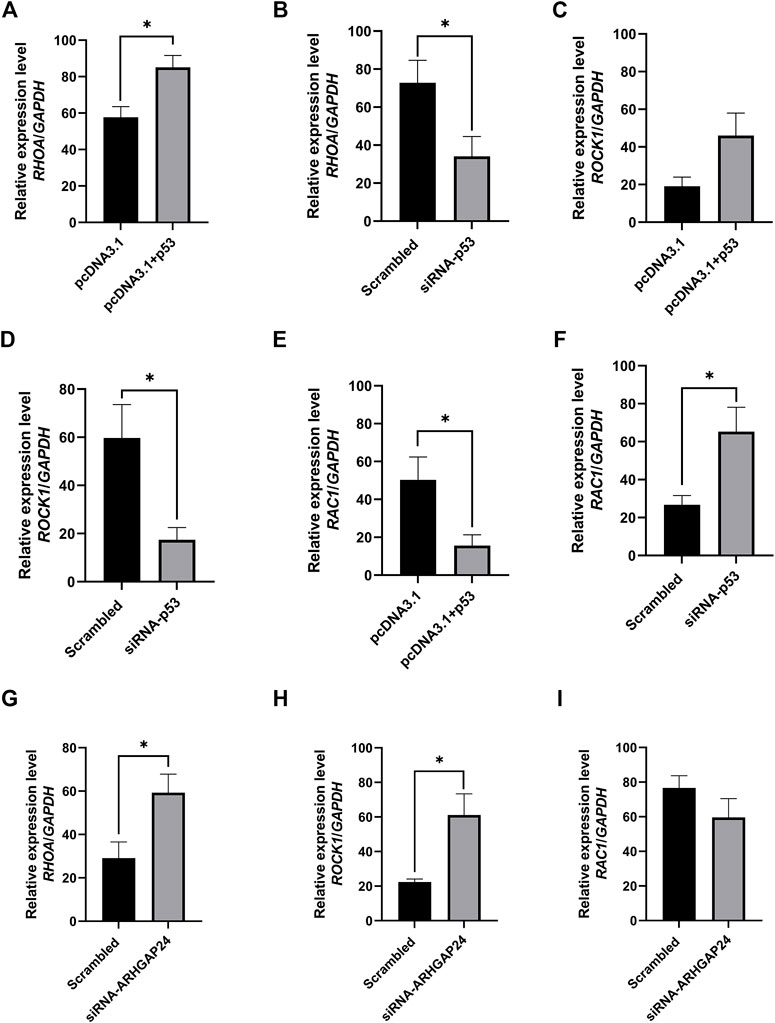
FIGURE 6. Transcription factor p53 regulates axon guidance pathway–related gene expression in porcine neural cells. (A) mRNA level of RHOA after p53 overexpression. (B) mRNA level of RHOA after p53 knockdown. (C) mRNA level of ROCK1 after p53 overexpression (D) mRNA level of ROCK1 after p53 knockdown (E) mRNA level of RAC1 after p53 overexpression. (F) mRNA level of RAC1 after p53 knockdown. (G) mRNA level of RHOA after ARHGAP24 knockdown. (H) mRNA level of ROCK1 after ARHGAP24 knockdown. (I) mRNA level of RAC1 after ARHGAP24 knockdown. *p < 0.05 and **p < 0.01. Scrambled: a negative control for siRNA as a scrambled sequence of the siRNA target sequence.
To our knowledge, the ARHGAP24 gene encodes a GTPase-activating protein. RhoGAPs are the important negative regulators of the Rho signaling pathway (Wang et al., 2019). Studies have also revealed that several members of the Rho family of GTPase activators have neuronal functions, including regulating dendritic morphology and synaptic plasticity (Ramakers, 2002). ARHGAP24 has been found to be a genetic marker to distinguish patients with major depression from healthy people (Watanabe et al., 2017). In this study, we first demonstrated that SNPs in the 5′-flanking region of the ARHGAP24 gene were associated with several aggressive behavioral traits. Among them, the difference of aggression among individuals with different genotypes at SNP rs335052970 is the most significant, which suggests that this SNP is worthy of further investigation. Four SNPs (rs333053350, rs342210686, rs328435752, and rs787973778) in the 5′-UTR were also significantly associated with aggressive behavioral traits. It is interesting that pigs with wild genotypes of this four linked SNPs in the ARHGAP24 gene were more aggressive than pigs with mutant genotypes. Next, we predicted the changes of TFBSs caused by them. The TFBSs for MECP2, REST, and NOTCH1 contain one of the four SNPs in the 5′-UTR. Previous studies showed that the genes activated by MECP2 caused many neuropsychiatric diseases (Chahrour et al., 2008). As a neural specific target, the transcription factor REST precisely regulated the transcription in the process of neuronal differentiation and development (Monestime et al., 2019). We herein hypothesized that MECP2, REST, and NOTCH1 could upregulate/downregulate the expression of ARHGAP24 to affect the aggressive behavioral traits. In addition, haplotypes were used as markers in association analysis to explain important genetic variation (Nothnagel and Rohde, 2005). LD was used to locate causal mutation sites that could not be precisely located by simple single-marker association (Hagenblad et al., 2004). Association analyses between haplotypes and aggressive behavior traits revealed that two haplotype blocks were all significantly associated with aggressive behavioral traits, which is consistent with the association between the SNPs and aggressive behavior trait in weaned pigs after mixing.
A promoter is necessary for the initiation of gene transcription and one of the upstream cis-acting elements for gene expression regulation (Civas et al., 2006; Yaniv, 2014). A core promoter initiates transcription, including transcription initiation sites (TSS) and upstream elements (Smale and Kadonaga, 2003). Based on luciferase activity analyses, the porcine ARHGAP24 gene had not only a positive regulatory promoter region (from −670 to −1,113 bp) but also a negative regulatory promoter region located (from −308 to −33 bp). The core promoter region has specific transcription factor binding sites and initiates the expression of downstream genes (Lubliner et al., 2015). In addition, the SNPs located in the core promoter region affected mRNA transcription by affecting the binding to transcription factors (Zubenko and Hughes, 2009). A previous study has shown that SNPs in the 5′-UTR of FGF13 interfered with the translation process of FGF13 and led to defects in the brain development and cognitive functions (Pan et al., 2021). In the present study, three SNPs (rs335052970, rs344700648, and rs339198696) in the core promoter region were in LD. Moreover, the relative luciferase activity of plasmids with haplotype AAC was the least than those with the other two haplotypes. A possible reason for lower transcriptional activity is that the allele A of rs335052970 present in the haplotype AAC promoter sequence promotes the potential binding site of the transcriptional repressors. Meanwhile, the luciferase activity was greater in plasmids with genotype GG than that of plasmids with genotype AA of rs335052970, implying that the promoter with allele G of rs335052970 might have higher transcriptional activity than the promoter with allele A. In conclusion, in view of the significant genetic effect on the aggressive behavior of SNP rs335052970 located in the core promoter region, it may regulate the expression of the ARHGAP24 gene by affecting promoter activity. Further research is needed to analyze how SNPs rs335052970 regulate aggressive behavior in pigs.
Previous studies revealed that SNPs located in the core promoter region changed the transcription factor binding sites (An et al., 2011; Vega et al., 2018). Transcription factors can activate or inhibit gene expression, which could result in a change of phenotype (Ameur et al., 2009; Kim et al., 2015). A previous study has shown that transcription factor YY1 binds to the promoter region of the Stx1a gene related to synaptic transmission and neurodevelopmental disorders and negatively regulates its transcription in a cell/tissue-specific manner (Nakayama et al., 2021). In the present study, the allele A of rs335052970 located in the core promoter region of the ARHGAP24 gene was predicted to invent the TFBSs for p53. Subsequently, a ChIP analysis demonstrated that p53 directly binds to the transcription factor binding element (PBE) motif containing allele A of rs335052970 in vitro. p53, a tumor suppressor gene (Boutelle and Attardi, 2021), is well-known for its functions as a transcription factor, which mediates transcriptional activation (Olivero et al., 2020) or repression (Wang et al., 2010). In the present study, the mRNA and protein expression level of ARHGAP24 was decreased after the overexpression of p53. Furthermore, the mRNA and protein expression level of ARHGAP24 was increased by interfering p53. A previous study presented that p53 acted as a repressor to downregulate PRR11-SKA2 to inhibit tumor formation (Wang et al., 2017). Furthermore, p53 acts as a transcription factor, represses the transcription of the PINK1 gene and then inhibits autophagy (Goiran et al., 2018), and represses antiapoptotic target genes (Castrogiovanni et al., 2018), which is similar to our present study. In addition, a recent study found that p53 may be a central regulator of neurodegeneration (Maor-Nof et al., 2021). Therefore, we speculated that p53 binds to TFBSs containing allele A of rs335052970 in the core promoter region of the ARHGAP24 gene, reduces the transcriptional activity of the promoter, and then inhibits the mRNA and protein expression level of the ARHGAP24 gene in porcine neural cells.
The pathways regulating aggressive behavior include the G-protein–coupled receptor (GPCR) signaling pathway, axon guidance, and ERK/MAPK signaling (Zhang-James et al., 2019). The small GTPase Rho, including RhoA, Rac, and Cdc42, as downstream regulators of RhoGAPs regulates the development of the nervous system by participating in the axon guidance pathway (Antoine-Bertrand et al., 2016). Rho kinase (ROCK), a downstream target of small GTPase Rho, is associated with a variety of neural functions, such as dendritic development and axon extension (Fujita and Yamashita, 2014). In the present study, the mRNA expression level of RHOA was increased after the overexpression of p53, while the mRNA expression level of RHOA and ROCK1 was decreased by interfering p53. While, the mRNA expression level of RAC1 was decreased after the overexpression of p53, it was increased by interfering p53. In general, Rac1 and Cdc42 are the positive regulators of axon growth and guidance, while RhoA is a negative regulator (Gonzalez-Billault et al., 2012). Moreover, the mRNA expression level of RHOA and ROCK1 was greater than that in the scrambled group when ARHGAP24 was inhibited in the present study. It has been reported that p53 was transcriptionally activated and participated in neural growth factor–mediated neurite growth (Brynczka et al., 2007; Brynczka and Merrick, 2008). Meanwhile, the deregulation of the ARHGAP24 gene inhibited the growth and branching of axons and dendrites (Nguyen et al., 2012). Repeated stress in rats resulted in atrophy of dendrites in hippocampal and medial prefrontal cortex neurons and increased aggression (Miller and McEwen, 2006). Therefore, p53 might reduce the axonal outgrowth and dendritic arborization by inhibiting the expression of the ARHGAP24 gene, which makes pigs more aggressive after weaning (Figure 7). Thus, SNP rs335052970 may be a potential causal mutation agent of porcine aggressive behavioral traits. It changes the transcriptional activity of the ARHGAP24 gene and regulates gene expression of axon guidance in combination with the transcription factor p53. However, further functional studies are needed to verify how the transcription factors affect the aggressive behavior of pigs.
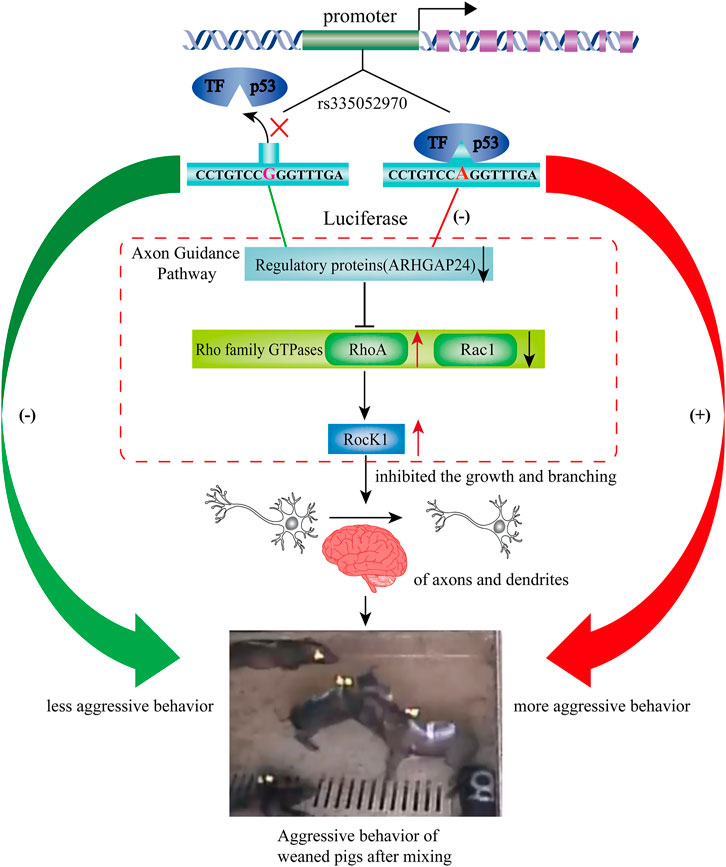
FIGURE 7. Based on the results, we hypothesized a regulating pathway by rs335052970 on the aggressive behavior of weaned pigs after mixing. Based on the results, we hypothesized a regulating pathway by rs335052970 in the ARHGAP24 gene on the aggressive behavior of pigs. In this diagram, rs335052970 binds to the transcription factor p53 to form a complex that regulates ARHGAP24 gene expression. Specifically, the promoter with allele A is more capable of binding to the transcriptional repressor p53 and downregulates ARHGAP24 gene expression. Since RhoA and RocK1 are the downstream targets of ARHGAP24, p53 also activates the expression of RhoA and Rock1. This might reduce the growth and branching of axons and dendrites, which make pigs more aggressive after weaning. Consequently, the promoter with allele A of rs335052970 upregulates the aggression of weaned pigs after mixing.
In conclusion, our results revealed the significant genetic effects of the ARHGAP24 gene on aggressive behavioral traits in weaned pigs after mixing. The functional SNP can be used for the genetic selection for less aggressive pigs. In addition, rs335052970 was highlighted as a functional mutation for aggressive behavioral traits that changed the transcriptional activity of the ARHGAP24 gene by affecting the binding of the transcription factor p53. Furthermore, functional verification is needed to find further scientific evidence on the regulation mechanism of the ARHGAP24 gene on aggressive behavioral traits in pigs.
The authors acknowledge that the data presented in this study must be deposited and made publicly available in an acceptable repository, prior to publication. Frontiers cannot accept a article that does not adhere to our open-data policies.
The animal study was reviewed and approved by the Animal Care and Use Committee of Nanjing Agricultural University (SYXK Su 2017-0007).
QX: conceptualization. BZ: conceptualization and data curation. QX: formal analysis and writing—original draft preparation. BZ: methodology; writing—review and editing; and funding acquisition. JZ and YG: data curation and investigation. ML: visualization. AS and BZ: writing—review and editing. All authors have read and agreed to the published version of the manuscript.
This study was supported by the National Natural Science Foundation of China (Grant Nos 32172786 and 31672465).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We are grateful to the Huaiyin Pig Breeding Farm for their participation and the National Natural Science Foundation of China for their support.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2022.839583/full#supplementary-material
ARHGAP24, Rho GTPase-activating protein 24; ChIP, chromatin immunoprecipitation; GPCR, G-protein-coupled receptor; HEK 293T, human embryonic kidney 293T; PBE, p53 transcription factor binding element; PCR, polymerase chain reactions; RhoGAP, GTPases activating protein; SNPs, single-nucleotide polymorphisms; TP53, transcription factor p53; TSS, transcription initiation sites; ROCK, Rho kinase; and UTR, untranslated region.
Ameur, A., Rada-Iglesias, A., Komorowski, J., and Wadelius, C. (2009). Identification of Candidate Regulatory SNPs by Combination of Transcription-Factor-Binding Site Prediction, SNP Genotyping and haploChIP. Nucleic Acids Res. 37, e85. doi:10.1093/nar/gkp381
An, Y., Ohnishi, H., Matsui, E., Funato, M., Kato, Z., Teramoto, T., et al. (2011). Genetic Variations in MyD88 Adaptor-Like Are Associated with Atopic Dermatitis. Int. J. Mol. Med. 27, 795–801. doi:10.3892/ijmm.2011.645
Antoine-Bertrand, J., Fu, M., and Lamarche-Vane, N. (2016). Direct Measurement of Oscillatory RhoA Activity in Embryonic Cortical Neurons Stimulated with the Axon Guidance Cue Netrin-1 Using Fluorescence Resonance Energy Transfer. Biol. Cel 108, 115–126. doi:10.1111/boc.201500077
Arey, D. S. (1999). Time Course for the Formation and Disruption of Social Organisation in Group-Housed Sows. Appl. Anim. Behav. Sci. 62, 199–207. doi:10.1016/S0168-1591(98)00224-X
Bagci, H., Sriskandarajah, N., Robert, A., Boulais, J., Elkholi, I. E., Tran, V., et al. (2020). Mapping the Proximity Interaction Network of the Rho-Family GTPases Reveals Signalling Pathways and Regulatory Mechanisms. Nat. Cel Biol 22, 120–134. doi:10.1038/s41556-019-0438-7
Boutelle, A. M., and Attardi, L. D. (2021). p53 and Tumor Suppression: It Takes a Network. Trends Cel Biol. 31, 298–310. doi:10.1016/j.tcb.2020.12.011
Brynczka, C., Labhart, P., and Merrick, B. A. (2007). NGF-Mediated Transcriptional Targets of P53 in PC12 Neuronal Differentiation. BMC Genomics 8, 139. doi:10.1186/1471-2164-8-139
Brynczka, C., and Merrick, B. A. (2008). The P53 Transcriptional Target Gene Wnt7b Contributes to NGF-Inducible Neurite Outgrowth in Neuronal PC12 Cells. Differentiation 76, 795–808. doi:10.1111/j.1432-0436.2007.00261.x
Camerlink, I., Arnott, G., Farish, M., and Turner, S. P. (2016). Complex Contests and the Influence of Aggressiveness in Pigs. Anim. Behav. 121, 71–78. doi:10.1016/j.anbehav.2016.08.021
Castrogiovanni, C., Waterschoot, B., De Backer, O., and Dumont, P. (2018). Serine 392 Phosphorylation Modulates P53 Mitochondrial Translocation and Transcription-Independent Apoptosis. Cell Death Differ 25, 190–203. doi:10.1038/cdd.2017.143
Chahrour, M., Jung, S. Y., Shaw, C., Zhou, X., Wong, S. T. C., Qin, J., et al. (2008). MeCP2, a Key Contributor to Neurological Disease, Activates and Represses Transcription. Science 320, 1224–1229. doi:10.1126/science.1153252
Chen, R., Chu, Q., Shen, C., Tong, X., Gao, S., Liu, X., et al. (2019). Identification of Single Nucleotide Polymorphisms in Porcine MAOA Gene Associated with Aggressive Behavior of Weaned Pigs after Group Mixing. Animals 9, 952. doi:10.3390/ani9110952
Civas, A., Génin, P., Morin, P., Lin, R., and Hiscott, J. (2006). Promoter Organization of the Interferon-A Genes Differentially Affects Virus-Induced Expression and Responsiveness to TBK1 and IKK∈. J. Biol. Chem. 281, 4856–4866. doi:10.1074/jbc.M506812200
Coutton, C., Dieterich, K., Satre, V., Vieville, G., Amblard, F., David, M., et al. (2015). Array-CGH in Children with Mild Intellectual Disability: A Population-Based Study. Eur. J. Pediatr. 174, 75–83. doi:10.1007/s00431-014-2367-6
D’Eath, R. B., Kurt, E., Evans, G., Thölking, L., Looft, H., Wimmers, K., et al. (2010). Pigs' Aggressive Temperament Affects Pre-Slaughter Mixing Aggression, Stress and Meat Quality. Animal 4, 604–616. doi:10.1017/S1751731109991406
Elvers, M., Beck, S., Fotinos, A., Ziegler, M., and Gawaz, M. (2012). The GRAF Family Member Oligophrenin1 Is a RhoGAP with BAR Domain and Regulates Rho GTPases in Platelets. Cardiovasc. Res. 94, 526–536. doi:10.1093/cvr/cvs079
Figler, M. H., Finkelstein, J. E., Twum, M., and Peeke, H. V. S. (2006). Intruding Male Red Swamp Crayfish, Procambarus C , Immediately Dominate Members of Established Communities of Smaller, Mixed‐Sex Conspecifics. Aggressive Behav. 21, 225–236. doi:10.1002/1098-2337(1995)21:3<225::aid-ab2480210305>3.0.co;2-#
Fornes, O., Castro-Mondragon, J. A., Khan, A., van der Lee, R., Zhang, X., Richmond, P. A., et al. (2020). JASPAR 2020: Update of the Open-Access Database of Transcription Factor Binding Profiles. Nucleic Acids Res. 48, D87–D92. doi:10.1093/nar/gkz1001
Fujita, Y., and Yamashita, T. (2014). Axon Growth Inhibition by RhoA/ROCK in the Central Nervous System. Front. Neurosci. 8, 338. doi:10.3389/fnins.2014.00338
Goiran, T., Duplan, E., Rouland, L., El Manaa, W., Lauritzen, I., Dunys, J., et al. (2018). Nuclear P53-Mediated Repression of Autophagy Involves PINK1 Transcriptional Down-Regulation. Cel Death Differ 25, 873–884. doi:10.1038/s41418-017-0016-0
Gonzalez-Billault, C., Muñoz-Llancao, P., Henriquez, D. R., Wojnacki, J., Conde, C., and Caceres, A. (2012). The Role of Small GTPases in Neuronal Morphogenesis and Polarity. Cytoskeleton 69, 464–485. doi:10.1002/cm.21034
Greenwood, E. C., Plush, K. J., van Wettere, W. H. E. J., and Hughes, P. E. (2014). Hierarchy Formation in Newly Mixed, Group Housed Sows and Management Strategies Aimed at Reducing its Impact. Appl. Anim. Behav. Sci. 160, 1–11. doi:10.1016/j.applanim.2014.09.011
Hagenblad, J., Tang, C., Molitor, J., Werner, J., Zhao, K., Zheng, H., et al. (2004). Haplotype Structure and Phenotypic Associations in the Chromosomal Regions Surrounding Two Arabidopsis T Flowering Time LociSequence Data from This Article Have Been Deposited with the EMBL/GenBank Data Libraries under Accession Nos. AY781906, AY785055. Genetics 168, 1627–1638. doi:10.1534/genetics.104.029470
Hu, H., Miao, Y.-R., Jia, L.-H., Yu, Q.-Y., Zhang, Q., and Guo, A.-Y. (2019). AnimalTFDB 3.0: A Comprehensive Resource for Annotation and Prediction of Animal Transcription Factors. Nucleic Acids Res. 47, D33–D38. doi:10.1093/nar/gky822
Kim, M., Kim, T., Johnson, R. L., and Lim, D.-S. (2015). Transcriptional Co-Repressor Function of the Hippo Pathway Transducers YAP and TAZ. Cel Rep. 11, 270–282. doi:10.1016/j.celrep.2015.03.015
Knudsen, S. (1999). Promoter2.0: for the Recognition of PolII Promoter Sequences. Bioinformatics 15, 356–361. doi:10.1093/bioinformatics/15.5.356
Langbein, J., and Puppe, B. (2004). Analysing Dominance Relationships by Sociometric Methods-A Plea for a More Standardised and Precise Approach in Farm Animals. Appl. Anim. Behav. Sci. 87, 293–315. doi:10.1016/j.applanim.2004.01.007
Leslie, J. R., Imai, F., Zhou, X., Lang, R. A., Zheng, Y., and Yoshida, Y. (2012). RhoA Is Dispensable for Axon Guidance of Sensory Neurons in the Mouse Dorsal Root Ganglia. Front. Mol. Neurosci. 5, 67. doi:10.3389/fnmol.2012.00067
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi:10.1006/meth.2001.1262
Lubliner, S., Regev, I., Lotan-Pompan, M., Edelheit, S., Weinberger, A., and Segal, E. (2015). Core Promoter Sequence in Yeast Is a Major Determinant of Expression Level. Genome Res. 25, 1008–1017. doi:10.1101/gr.188193.114
Maor-Nof, M., Shipony, Z., Lopez-Gonzalez, R., Nakayama, L., Zhang, Y.-J., Couthouis, J., et al. (2021). p53 Is a Central Regulator Driving Neurodegeneration Caused by C9orf72 Poly(PR). Cell 184, 689–708. doi:10.1016/j.cell.2020.12.025
Meng, Q., Wang, K., Liu, X., Zhou, H., Xu, L., Wang, Z., et al. (2017). Identification of Growth Trait Related Genes in a Yorkshire Purebred Pig Population by Genome Wide Association Studies. Asian-australas J. Anim. Sci. 30, 462–469. doi:10.5713/ajas.16.0548
Miller, M. M., and McEwen, B. S. (2006). Establishing an Agenda for Translational Research on PTSD. Ann. N. Y Acad. Sci. 1071, 294–312. doi:10.1196/annals.1364.023
Monestime, C. M., Taibi, A., Gates, K. P., Jiang, K., and Sirotkin, H. I. (2019). CoRest1 Regulates Neurogenesis in a Stage‐dependent Manner. Develop. Dyn. 248, 918–930. doi:10.1002/dvdy.86
Müller, P. M., Rademacher, J., Bagshaw, R. D., Wortmann, C., Barth, C., van Unen, J., et al. (2020). Systems Analysis of RhoGEF and RhoGAP Regulatory Proteins Reveals Spatially Organized RAC1 Signalling from Integrin Adhesions. Nat. Cel Biol 22, 498–511. doi:10.1038/s41556-020-0488-x
Nakayama, T., Fukutomi, T., Terao, Y., and Akagawa, K. (2021). Syntaxin 1A Gene Is Negatively Regulated in a Cell/Tissue Specific Manner by YY1 Transcription Factor, Which Binds to the −183 to −137 Promoter Region Together with Gene Silencing Factors Including Histone Deacetylase. Biomolecules 11, 146. doi:10.3390/biom11020146
Nguyen, L. S., Jolly, L., Shoubridge, C., Chan, W. K., Huang, L., Laumonnier, F., et al. (2012). Transcriptome Profiling of UPF3B/NMD-Deficient Lymphoblastoid Cells from Patients with Various Forms of Intellectual Disability. Mol. Psychiatry 17, 1103–1115. doi:10.1038/mp.2011.163
Nothnagel, M., and Rohde, K. (2005). The Effect of Single-Nucleotide Polymorphism Marker Selection on Patterns of Haplotype Blocks and Haplotype Frequency Estimates. Am. J. Hum. Genet. 77, 988–998. doi:10.1086/498175
O'Connell, N. E., Beattie, V. E., and Moss, B. W. (2004). Influence of Social Status on the Welfare of Growing Pigs Housed in Barren and Enriched Environments. Anim. Welfare 13, 425–431. doi:10.1017/s1752756200011984
Olivero, C. E., Martínez-Terroba, E., Zimmer, J., Liao, C., Tesfaye, E., Hooshdaran, N., et al. (2020). p53 Activates the Long Noncoding RNA Pvt1b to Inhibit Myc and Suppress Tumorigenesis. Mol. Cel 77, 761–774. doi:10.1016/j.molcel.2019.12.014
O’Malley, C. I., Wurtz, K., Steibel, J. P., Bates, R. O., Ernst, C. W., and Siegford, J. M. (2018). Relationships Among Aggressiveness, Fearfulness and Response to Humans in Finisher Pigs. Appl. Anim. Behav. Sci. 205, 194–201. doi:10.1016/j.applanim.2018.03.001
Pan, X., Zhao, J., Zhou, Z., Chen, J., Yang, Z., Wu, Y., et al. (2021). 5'-UTR SNP of FGF13 Causes Translational Defect and Intellectual Disability. Elife 10, e63021. doi:10.7554/eLife.63021
Ramakers, G. J. A. (2002). Rho Proteins, Mental Retardation and the Cellular Basis of Cognition. Trends Neurosciences 25, 191–199. doi:10.1016/s0166-2236(00)02118-4
Reese, M. G. (2001). Application of a Time-Delay Neural Network to Promoter Annotation in the Drosophila M Genome. Comput. Chem. 26, 51–56. doi:10.1016/s0097-8485(01)00099-7
Rohrer, G. A., Brown-Brandl, T., Rempel, L. A., Schneider, J. F., and Holl, J. (2013). Genetic Analysis of Behavior Traits in Swine Production. Livestock Sci. 157, 28–37. doi:10.1016/j.livsci.2013.07.002
Shen, C., Tong, X., Chen, R., Gao, S., Liu, X., Schinckel, A. P., et al. (2020). Identifying Blood-Based Biomarkers Associated with Aggression in Weaned Pigs after Mixing. Appl. Anim. Behav. Sci. 224, 104927. doi:10.1016/j.applanim.2019.104927
Smale, S. T., and Kadonaga, J. T. (2003). The RNA Polymerase II Core Promoter. Annu. Rev. Biochem. 72, 449–479. doi:10.1146/annurev.biochem.72.121801.161520
Stukenborg, A., Traulsen, I., Puppe, B., Presuhn, U., and Krieter, J. (2011). Agonistic Behaviour after Mixing in Pigs under Commercial Farm Conditions. Appl. Anim. Behav. Sci. 129, 28–35. doi:10.1016/j.applanim.2010.10.004
Tcherkezian, J., and Lamarche-Vane, N. (2007). Current Knowledge of the Large RhoGAP Family of Proteins. Biol. Cel 99, 67–86. doi:10.1042/BC20060086
Tong, X., Shen, C., Chen, R., Gao, S., Liu, X., Schinckel, A. P., et al. (2019). Reestablishment of Social Hierarchies in Weaned Pigs after Mixing. Animals 10, 36. doi:10.3390/ani10010036
Tuchscherer, M., Puppe, B., Tuchscherer, A., and Kanitz, E. (1998). Effects of Social Status after Mixing on Immune, Metabolic, and Endocrine Responses in Pigs. Physiol. Behav. 64, 353–360. doi:10.1016/s0031-9384(98)00084-5
Vega, W. H. O., Quirino, C. R., Bartholazzi-Junior, A., Rua, M. A. S., Serapião, R. V., and Oliveira, C. S. (2018). Variants in the CYP19A1 Gene Can Affect In Vitro Embryo Production Traits in Cattle. J. Assist. Reprod. Genet. 35, 2233–2241. doi:10.1007/s10815-018-1320-4
Viding, E., Hanscombe, K. B., Curtis, C. J. C., Davis, O. S. P., Meaburn, E. L., and Plomin, R. (2010). In Search of Genes Associated with Risk for Psychopathic Tendencies in Children: A Two-Stage Genome-Wide Association Study of Pooled DNA. J. Child. Psychol. Psychiatry 51, 780–788. doi:10.1111/j.1469-7610.2010.02236.x
Wang, B., Xiao, Z., Ko, H. L., and Ren, E.-C. (2010). The P53 Response Element and Transcriptional Repression. Cell Cycle 9, 870–879. doi:10.4161/cc.9.5.10825
Wang, L., Shen, S., Wang, M., Ding, F., Xiao, H., Li, G., et al. (2019). Rho GTPase Activating Protein 24 (ARHGAP24) Silencing Promotes Lung Cancer Cell Migration and Invasion by Activating β-Catenin Signaling. Med. Sci. Monit. 25, 21–31. doi:10.12659/MSM.911503
Wang, Y., Weng, H., Zhang, Y., Long, Y., Li, Y., Niu, Y., et al. (2017). The PRR11-SKA2 Bidirectional Transcription Unit Is Negatively Regulated by P53 through NF-Y in Lung Cancer Cells. Int. J. Mol. Sci. 18, 534. doi:10.3390/ijms18030534
Watanabe, S.-Y., Numata, S., Iga, J.-I., Kinoshita, M., Umehara, H., Ishii, K., et al. (2017). Gene Expression-Based Biological Test for Major Depressive Disorder: An Advanced Study. Neuropsychiatr. Dis. Treat. 13, 535–541. doi:10.2147/NDT.S120038
Yaniv, M. (2014). Chromatin Remodeling: From Transcription to Cancer. Cancer Genet. 207, 352–357. doi:10.1016/j.cancergen.2014.03.006
Yodoya, E., Wada, M., Shimada, A., Katsukawa, H., Okada, N., Yamamoto, A., et al. (2006). Functional and Molecular Identification of Sodium-Coupled Dicarboxylate Transporters in Rat Primary Cultured Cerebrocortical Astrocytes and Neurons. J. Neurochem. 97, 162–173. doi:10.1111/j.1471-4159.2006.03720.x
Zhang-James, Y., Fernàndez-Castillo, N., Hess, J. L., Malki, K., Glatt, S. J., Cormand, B., et al. (2019). An Integrated Analysis of Genes and Functional Pathways for Aggression in Human and Rodent Models. Mol. Psychiatry 24, 1655–1667. doi:10.1038/s41380-018-0068-7
Keywords: aggressive behavior, piglet, Rho GTPase–activating protein 24, transcription factor, axon guidance, animal welfare
Citation: Xu Q, Zhao J, Guo Y, Liu M, Schinckel AP and Zhou B (2022) A Single-Nucleotide Polymorphism in the Promoter of Porcine ARHGAP24 Gene Regulates Aggressive Behavior of Weaned Pigs After Mixing by Affecting the Binding of Transcription Factor p53. Front. Cell Dev. Biol. 10:839583. doi: 10.3389/fcell.2022.839583
Received: 20 December 2021; Accepted: 28 February 2022;
Published: 01 April 2022.
Edited by:
Venkaiah Betapudi, Department of Homeland Security, United StatesReviewed by:
Xianyong Lan, Northwest A&F University, ChinaCopyright © 2022 Xu, Zhao, Guo, Liu, Schinckel and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bo Zhou, emhvdWJvQG5qYXUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.