- 1Research Lab of Translational Medicine, Hengyang Medical School, University of South China, Hengyang, China
- 2Key Laboratory for Arteriosclerology of Hunan Province, Human International Scientific and Technological Cooperation Base of Arteriosclerotic Disease, Institute of Cardiovascular Disease, Hengyang Medical College, University of South China, Hengyang, China
- 3Institution of Pathogenic Biology, Hengyang Medical School, University of South China, Hengyang, China
Stem cell transplantation (SCT) holds great promise for overcoming diseases by regenerating damaged cells, tissues and organs. The potential for self-renewal and differentiation is the key to SCT. RNA methylation, a dynamic and reversible epigenetic modification, is able to regulate the ability of stem cells to differentiate and regenerate. N6-methyladenosine (m6A) is the richest form of RNA methylation in eukaryotes and is regulated by three classes of proteins: methyltransferase complexes, demethylase complexes and m6A binding proteins. Through the coordination of these proteins, RNA methylation precisely modulates the expression of important target genes by affecting mRNA stability, translation, selective splicing, processing and microRNA maturation. In this review, we summarize the most recent findings on the regulation of m6A modification in embryonic stem cells, induced pluripotent stem cells and adult stem cells, hoping to provide new insights into improving SCT technology.
Introduction
Stem cells are unique cell populations possessing proliferation and differential ability, which have been identified in embryo, reprogrammed somatic cells and many adult tissues, including muscle, brain, bone marrow, blood and fat, and play a vital role in the field of regenerative medicine (Gurusamy et al., 2018; Belenguer et al., 2021; Ding et al., 2021). In vitro, stem cells can differentiate into specific cell lineages in response to special inducers, allowing for the replacement of lost cells, providing tropic support and modulating inflammation (Gnecchi et al., 2012; Mouhieddine et al., 2014; Ji et al., 2018; Cheng et al., 2019). With advances in stem cell transplantation (SCT) technology, the process of treating diseases associated with poor regenerative abilities and limited amounts of endogenous adult stem cells (ASCs), such as cardiovascular, nervous and sports system diseases, has progressed (Beitnes et al., 2011; Ma et al., 2013; Trounson and Mcdonald, 2015; Gallo et al., 2016; Gorecka et al., 2018). Increase of the ability to self-renewal and differentiate into specific cell lineages of stem cells will be of great significance for improving the SCT efficiency.
RNA methylation, which was first identified in Novikoff hepatoma cells in 1970, influences various cell processes (C. Desrosiers et al., 1974). In 2011, RNA methylation once again received the attention of scientists as a reversible and dynamic epigenetic modification (Jia et al., 2011). Although more than 100 types of chemical modifications, such as N1-methyladenosine (m1A), N6, 2′-O-dimethyladenosine (m6Am), as well as N6-methyladenosine (m6A), have been identified in cellular RNAs, m6A has been considered as the most abundant modification in eukaryotes (Roundtree et al., 2017), which is common in 3′ untranslated regions (UTRs), internal long exons and stop codons. It has been reported that the function of m6A is largely dependent on modifications around 3′ UTRs (Meyer et al., 2012; Ke et al., 2015; Lin et al., 2016). The proteins that regulate m6A modification levels have been named methyltransferase complexes (writer), demethylase complexes (eraser) and binding protein complexes (reader) (Figure 1). To date, three main methyltransferases, METTL3, METTL14 and WTAP, as well as a newly reported METTL5, have been found to catalyze RNA m6A (Ignatova et al., 2020). In addition, two kinds of important demethylases, FTO and ALKBH5, possess the ability to remove RNA m6A (Wu et al., 2019). Coordination of the writer and eraser allows for dynamic changes in m6A levels according to developmental stage and physiological conditions. However, the reader can only modulate cell processes when it binds m6A sites. To date, the YTH domain family, HNRNPC family and HNRNPA2B1 have been identified as binding proteins that play different roles in m6A functioning (Ji et al., 2018). Among YTH domain family members, YTHDF2 was the first found to bind with m6A-RNA to regulate mRNA degeneration (Mapperley et al., 2021). YTHDF1 and YTHDF3 can improve the translation efficiency of m6A-mRNA upon binding (Wang et al., 2015; Tiwary, 2020). In addition, YTHDC1 (initially referred as YT521-B), a binding protein in the cell nucleus, can regulate RNA splicing (Xu et al., 2014; Roundtree and He, 2016). HNRNPC family members include HNRNPC and HNRNPG, which can switch the secondary structure of mRNA (Liu et al., 2017). Unlike the YTH domain family and HNRNPC family, HNRNPA2B1 can recognize m6A sites and further regulate the splicing and processing of pri-miRNA (Alarcon et al., 2015).
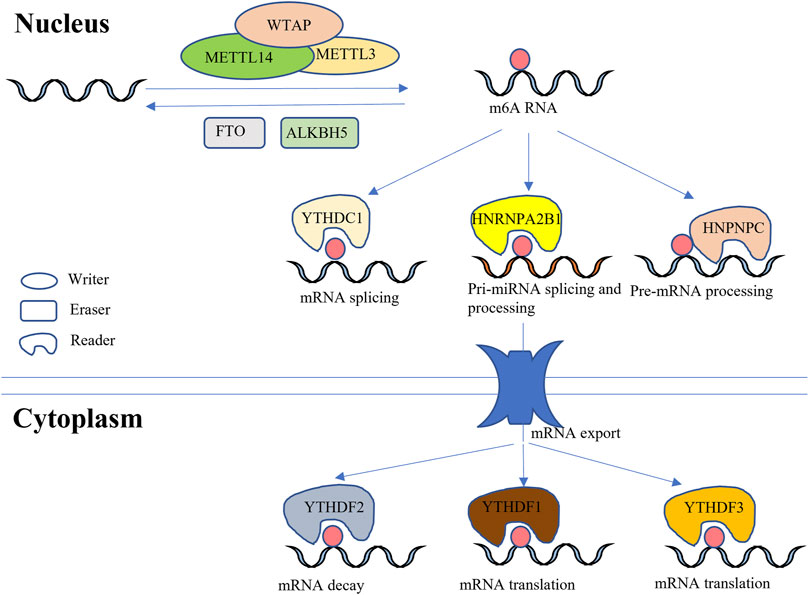
FIGURE 1. Coordination between writer, eraser and reader. The ellipse represents the writer, which possesses the ability to catalyze RNA into m6A-containing RNA. The rectangle indicates the eraser, which moves m6A, thereby downregulating m6A-RNA. Precise coordination between the writer and eraser allows for dynamic and reversible regulation of m6A levels in eukaryocytes. The irregular shape represents the reader, which binds m6A and modulates expression of important genes. Both the nucleus and cytoplasm contain reader, and different readers play unique roles in the physiological functioning of organisms. Some readers, notably YTHDF1 and YTHDF3, promote the expression of specific genes, whereas YTHDF2 has the opposite effect, decreasing mRNA levels.
M6A methylation have shown powerful bioregulatory functions in RNA metabolism including pre-mRNA splicing, mRNA translation, stability, and transport, 3′-end processing, and non-coding RNA processing (Sun et al., 2019; Oerum et al., 2021), which is involved in the regulation of a series of cellular processes such as cell fate decision, cell cycle regulation, cell proliferation and differentiation, indicating its pivotal roles in embryonic development, hematopoietic development, neurogenesis, stress responses, sex determination, tumorigenesis and so on (Liu et al., 2018; Jiang et al., 2021). Increasing evidences have displayed that m6A methylation possesses the ability to regulate proliferation and differentiation of stem cells by modulating the expression of pluripotency, proliferation or lineage-specific genes (Figure 2) (Lin and Gregory, 2014; Li J. et al., 2021), implying great application prospects for SCT. In this review, we provide a comprehensive summary of the biological functions of m6A in stem cell proliferation and differentiation to facilitate the improvement of SCT technology.
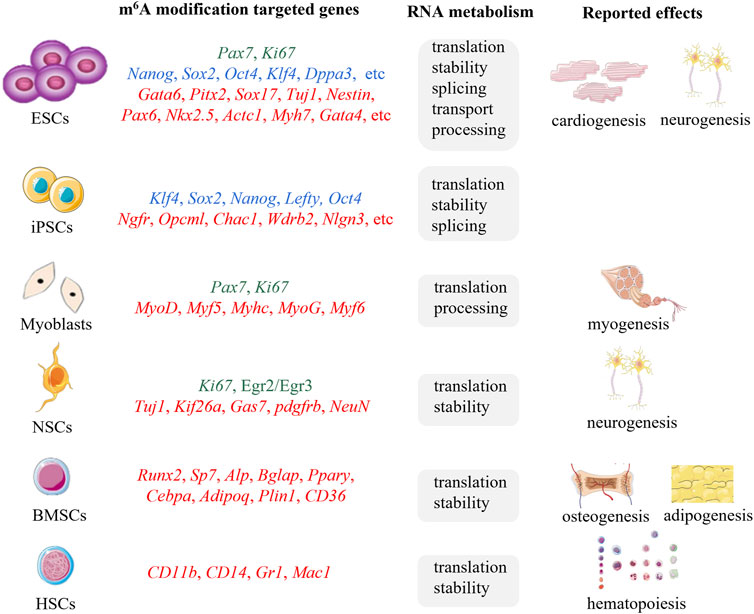
FIGURE 2. The m6A modifications targeted genes, associated RNA metabolic mechanisms, and reported effects of ESCs, iPSCs, myoblasts, NSCs, BMSCs, HSCs. Proliferation associated genes (green), pluripotency related genes (blue), differentiation associated genes (red).
THE Role of m6A in Embryonic Stem Cells
Embryonic stem cells (ESCs) are derived from the inner cell mass of the developing blastocyst possessing the ability to self-renewal and form any fully differentiated cell of the body (Biswas and Hutchins, 2007; Young, 2011). The cell fate of ESCs between proliferation and differentiation is regulated by multiple pathways that orchestrate gene expression (Table 1). The self-renewal capacity of ESCs is maintained by activating the pluripotency genes such as Oct4, Nanog, Sox2, Dppa3, and Klf4, as well as suppressing lineage-specific genes (Yeo and Ng, 2013). Ki67 is the most important marker associated with stem cells proliferation. Additionally, a few developmental regulators associated genes, including Gata6, Pitx2, Sox17, Cdx2 were involved in the ESCs differentiation except some lineage-specific ones (Wang et al., 2014). Many ESCs transcripts of developmental regulators and lineage priming regulators contain m6A, suggesting that it plays a vital role in the decision of ESC fate.
Writers
Early reports about the role of METTL3 and METTL14 in the self-renewal and differentiation of ESCs was somewhat controversial. Wang et al., reported that either Mettl3 or Mettl14 deficiency in mouse ESCs (mESCs) impaired self-renewal, and enhanced the expressional levels of developmental genes such as Fgf5 and Cdx2, suggesting the loss of m6A levels inhibited self-renewal, and promoted differentiation of mESCs (Wang et al., 2014). On the contrary, another study revealed that genetic inactivation or depletion of Mettl3 in mESCs and human ESCs (hESCs) prolonged Nanog expression upon differentiation to make the ESCs incapable of differentiating into specific lineages (Batista et al., 2014). However, the differences of m6A modification on proliferation and differentiation in ESCs were explained by Geula et al., and others that the m6A modification could decrease mRNA stability of naïve pluripotency-promoting genes or pro-differentiation genes depending on the pluripotent state of the ESCs, i.e., naive or primed (Geula et al., 2015; Zhao and He, 2015). In naïve pluripotent mESCs, Mettl3 knockout drive the transition from the naïve state into the primed epiblast stem cells (EpiSCs), which indicated a more differentiated state. Naïve mESCs presented increased pluripotency upon Mettl3 knockdown, whereas primed EpiSCs displayed enhanced propensity to lineage differentiation upon m6A depletion. Therefore, Mettl3 depletion blocked differentiation and promoted proliferation in mESCs, but played an opposite role in EpiSCs (Geula et al., 2015; Zhao and He, 2015). METTL3-mediated m6A modification of lincRNA 1281 is critically required for proper differentiation of mESCs by sequestering its interaction with pluripotency-related let-7 family microRNAs without influencing its proliferation (Yang et al., 2018). METTL5 also occupied an important role in m6A modification, which catalyzed m6A in 18S rRNA at position A1832 to enhance global translation rate, promoted the self-renewal and differentiation of mESCs (Ignatova et al., 2020).
Erasers
Chen et al. developed a targeted RNA m6A erasure system to demethylate RNAs site-specifically in hESCs, thus modulating individual m6A modification in ESCs with precise temporal control (Chen X. et al., 2021). The fusion of the catalytic domain of ALKBH5 with a stably transfected, doxycycline-inducible dCas13a precisely and reversibly demethylated the targeted m6A site of Sox2, which significantly increased the amount and stability of corresponding transcripts and facilitated the differentiation of hESCs without affecting the self-renewal capacity (Chen X. et al., 2021). In addition, overexpression of ALKBH5 remarkably blocked cardiomyocyte differentiation of hESCs by down-regulating the expression of KDM5B and RBBP5, two downstream targets for ALKBH5 in cardiac-committed hESCs (Han et al., 2021).
Readers
YTHDF1, YTHDF3, YTHDC1 and HNRNPA2/B1 are readers reported to be involved in the regulation of m6A in ESCs. Interestingly, YTHDF1 and YTHDF3 not only displayed differential roles in regulating the pluripotency of ESCs, but also showed converse contribution to the cardiac differentiation (Wang S. et al., 2021). Depletion of Ythdf3 in mESCs led to loss of pluripotency with accelerated cardiac differentiation through facilitating the cardiomyocyte-specific genes expression and displayed increased cell proliferation. While, Ythdf1 loss resulted in marked impairment of cardiomyocytes differentiation, as well as slightly decreased proliferation. Remarkably, YTHDF3 appears to mediate cellular differentiation partially by suppressing YTHDF1, demonstrating the contrasting but interrelated roles of YTHDF1 and YTHDF3 in determination of cell fate (Wang S. et al., 2021). Additionally, YTHDC1 could recognize the m6A-marked LINE1 RNAs in the nucleus and regulate the formation of the LINE1-NCL-KAP1 complex to modulate the RNA scaffold, thus ensuring the appropriate transcriptome and developmental potency of mESCs and early embryos (Chen C. et al., 2021). HNRNPA2/B1 was essential for early embryonic development by promoting the pluripotency-related gene expression depending on the METTL3-mediated m6A RNA methylation (Choi et al., 2013; Kwon et al., 2019).
Other Regulatory Proteins
The m6A modification level in ESCs were regulated by multiple proteins. Protein arginine methyltransferase 1 (PRMT1) can increase the global m6A RNA level (Liu X. et al., 2021; Wang Z. et al., 2021). PRMT1 was able to interact with and methylate METTL14 in its disordered C terminal region at R255 to enhance its RNA methylation activity and promoted the binding of the METTL3/METTL14 complex to substrate RNA, thereby triggering global m6A modification and mESCs endoderm differentiation (Liu X. et al., 2021; Wang Z. et al., 2021). Zinc-finger protein Zc3h13 played a negative role in the m6A methylation, which directly interacted with WTAP to form a Zc3h13-WTAP-Virilizer-Hakai complex to regulate RNA m6A methylation in the nucleus. Zc3h13 knockdown markedly impaired self-renewal and promoted mESCs differentiation (Wen et al., 2018). Interestingly, Zinc finger protein 217 (Zfp217) displayed significant discrepancy in affecting the level of m6A methylation owning to the cell state differences, which could modulate m6A deposition on their transcripts via sequestering the METTL3, thus hindering METTL3 binding to RNAs (Lee et al., 2016; Aguilo and Walsh, 2017). mESCs in the undifferentiated state showed high level of ZFP217, which significantly suppressed METTL3 methyltransferase activity, preventing core ESC transcripts from aberrant methylation. However, in differential mESCs, ZFP217 contents and its target genes promptly decreased, allowing the METTL3 to release and catalyze m6A methylation at the remaining pluripotency transcripts, thus triggering ESC differentiation (Aguilo et al., 2015; Lee et al., 2016).
The Role of m6A in Induced Pluripotent Stem Cells
Induced pluripotent stem cells (iPSCs) were originally generated in 2006 by successfully transferring four transcription factors Oct4, Sox2, Klf4, and c-Myc into adult somatic cells to reprogram them (Takahashi et al., 2007; Park et al., 2008). Compared to ESCs, iPSCs derived from adult somatic tissues, such as skin, blood, and urine, displayed no immune rejection when transplanted autologously, which made iPSCs an extraordinary candidate for personalized medicine. M6A modification remarkably influences the pluripotency and differentiation of iPSCs (Table 1).
Writers
Owning to the great similarity in genome and physiological characteristics, porcine iPSCs (piPSCs) have become an important study model. Mettl3 deficiency markedly inhibited self-renewal and promoted differentiation of piPSCs. Mechanistically, METTL3 silence caused lowered m6A levels of JAK2 and SOCS3, which induced the alteration of JAK2 and SOCS3 protein expression, thus inhibiting JAK2–STAT3 pathway to affect proliferation and differentiation of piPSCs (Wu et al., 2019).
Readers
YTHDF2 and YTHDF3 are two readers reported to be required for the reprogramming of somatic cells into iPSCs. Both the YTHDF2-CCR4-NOT and YTHDF3-PAN2-PAN3 deadenylase complex could promote the mRNA clearance of somatic genes, such as Tead2 and Tgfb1, thus conducting the reprograming (Liu et al., 2020). The cell proliferation of iPSCs was significantly decreased upon Ythdf2/3 deficiency (Liu et al., 2020). In addition, YTHDF2 could destabilize m6A-modified transcripts related with neural development, thus restraining differentiation in iPSCs (Heck et al., 2020).
Other Regulatory Proteins
SMAD2/3 and Matrin3 (MATR3) are two proteins associated with the m6A methylation in iPSCs. SMAD2/3 interacted with METTL3–METTL14–WTAP complex to mediate the conversion of adenosine to m6A on RNA, which facilitated the destabilization of SMAD2/3 targeted genes such as pluripotency factor gene Nanog, thus leading to timely exit from pluripotency and neuroectodermal differentiation (Bertero et al., 2018). MATR3 acted as a nuclear RNA/DNA-binding protein to regulate gene expression by stabilizing target RNAs. MATR3 maintained pluripotency in human iPSCs mainly in two ways. For one thing, MATR3 binded to the Oct4 and Ythdf1 promoters to facilitate their expression. For another, MATR3 was recruited on ribosomes to regulate the translation of various specific transcripts such as Nanog and Lin28a through direct binding to keep their stabilization. MATR3 deficiency hampered both the proliferation and differentiation of human iPSCs (Pollini et al., 2021).
The Role of m6A in ASCs
The Role of m6A in Myoblasts/Muscle Stem Cells
Skeletal muscle degeneration is associated with various conditions, including developmental disorders, muscular dystrophies, neuromuscular degenerative diseases, cardiac diseases, and aging (Emery, 2002; Negroni et al., 2015; Chal and Pourquie, 2017). Skeletal myoblasts derive from satellite cells (progenitor cells) which are responsible for skeletal muscle growth and repair. Following muscle damages, the satellite cells exit cell cycle to differentiate into myoblasts, which further proliferate and fuse to myotubes and fresh muscle fibers to repair tissues. The phase of myogenic differentiation mainly includes early stage and late stage. MyoD and Myf5 have been considered as early-stage markers, whereas MyoG, Mrf4 (Myf6), and MyHC have been regarded as later-stage markers (Chen et al., 2019). Myoblasts act as the first candidates for stem cell therapy in muscle repair owing to their easy to obtain from muscle biopsies and autologous transplantation characteristics (Negroni et al., 2015; Rikhtegar et al., 2019). Therefore, enhancing the myogenic capacity of myoblasts holds great promise for alleviating skeletal muscle degenerative diseases.
Writers
The discovery that m6A RNA methylation was directly involved in the regulation of myogenic differentiation was performed in murine C2C12 myoblasts. METTL3 possessed a vital role in controlling the transition of muscle stem cells/myoblasts to different cell states (Gheller et al., 2020). The m6A-specific RNA-sequencing revealed a distinct profile of m6A modification between proliferating and differentiating C2C12 myoblasts (Gheller et al., 2020). Overexpression of Mettl3 positively regulated m6A RNA methylation and further promoted myogenic differentiation in myoblasts (Chen et al., 2019), while knockdown of Mettl3 could reduce the mRNA levels of myogenic transcription factor MyoD (Kudou et al., 2017), indicating METTL3 is a positive regulator of skeletal muscle differentiation, which was also confirmed in mice higher or lower expression of METTL3 (Liang et al., 2021). However, a recent study demonstrated that METTL3 might possess negatively regulatory effect on skeletal muscle differentiation. The muscle specific miRNAs, important regulators of skeletal muscle development and muscle cell functions, were significantly repressed by Mettl3 in C2C12 myoblasts and in vivo model of mouse skeletal muscle regeneration after injury, exhibiting an anti-differentiation role of METTL3 in myogenic differentiation (Diao et al., 2021). The conflicting results observed above revealed that the effects of m6A modification on proliferation and differentiation in C2C12 myoblasts may also correlated with the cell state, which is in conformed to the observations in ESCs (Geula et al., 2015). Mechanical exploration revealed that the Notch signaling pathway is markedly involved in regulating the muscle stem cells and muscle regeneration of METTL3-mediated m6A modification (Liang et al., 2021). METTL14 is another key m6A methyltransferase verified to be involved in the prenatal myogenic differentiation (Zhang et al., 2020), which form a stable heterodimeric core complex with METTL3 to regulate m6A RNA methylation (Liu et al., 2014). Mettl14 knockdown could inhibit the differentiation and enhance the proliferation of C2C12 myoblast cells (Zhang et al., 2020).
Erasers
Increasing evidences demonstrated that the demethylase FTO significantly promoted myogenic differentiation. In Fto-silenced C2C12 myoblasts and Fto-deficient mice, the myoblasts differentiation and skeletal muscle development were impaired, respectively, which was associated with mTOR-PGC-1α pathway-mediated mitochondria biogenesis (Wang et al., 2017). In goat primary myoblasts, silencing Fto drastically reduced cyclin D1 expression via YTHDF2-mediated mRNA degradation, thus resulting in delayed G1 phase and impaired myoblast proliferation (Deng et al., 2021). Moreover, in myoblasts from female embryo, FTO promoted proliferation and myoblast differentiation through the focal adhesion pathway (Huang et al., 2020).
Readers
The m6A reader protein IGF2BP1 also participated in the regulation of prenatal myogenesis, which target the key marker genes MYH2 and MyoG to promote the differentiation and inhibit the proliferation of C2C12 myoblast cells (Zhang et al., 2020).
The Role of m6A in Neural Stem Cells
Neurodegenerative diseases, including Alzheimer’s disease, Parkinson’s disease, and Huntington’s disease, are highly disabling and greatly influence both patients and their caregivers. However, no curative treatment is available to stop or reverse the disease progression. Increasing evidences have confirmed that the neural stem cells (NSCs), a life-long source of neurons and glia, possess a remarkable promise for the treatment of neurodegenerative diseases because of its beneficial effects such as decrease of neuroinflammation, production of neurotrophic factors, augment of neuronal plasticity and cell replacement (Gage Fred and Temple, 2013; De Gioia et al., 2020). Differentiation of NSCs is labeled by immature differentiational marker Dcx, and mature differentiational markers NeuN and Tuj1 (Li et al., 2017; Wang Y. et al., 2018). M6A modification seems to be highest in the brain and significantly impacts the cell biological processes in nervous system (Meyer and Jaffrey, 2014; Li J. et al., 2019). Recent studies have implied that the disordered m6A modification may be greatly correlated with neurodegenerative diseases (Yen and Chen, 2021). Therefore, regulating the level of m6A in NSCs is of great potential for the treatment of neurodegenerative diseases (Table 1).
Writers
The m6A methyltransferase METTL3 and METTL14 formed a heterodimer to regulate neurogenesis and neuronal development. In 2018, Wang et al. found that the mouse NSCs (mNSCs) defected in Mettl14 displayed marked decrease in proliferation and premature differentiation, demonstrating that m6A modification augmented NSC self-renewal, which was correlated with the alteration of histone modifications including H3K27me3, H3K27ac and H3K4me3 (Wang Y. et al., 2018). In adult mNSCs, Mettl3 depletion suppressed neuronal development, and skewed the NSCs to differentiate more toward glial lineage by modulating histone methyltransferase Ezh2 and the consequent H3K27me3 levels (Chen J et al., 2019). In zebrafish spinal cord injury model, the m6A RNA methylation profiling and transcription level of METTL3 were both increased, which is consistent with the results observed in NSCs in mice with spinal cord injury, indicating METTL3 may contribute to the spinal cord regeneration (Xing et al., 2021).
Erasers
The demethylase FTO is abundant in the brain (Mctaggart et al., 2011), its knockout caused m6A over-accumulation and damaged neuronal activity (Hess et al., 2013). In Fto knockout mice, diminished adult NSCs pool and reduced proliferation and differentiation of adult NSCs was observed, leading to impaired learning and memory. Further exploration of the underlying mechanisms revealed that FTO-mediated m6A modification was associated with brain derived neurotrophic factor pathway (Li et al., 2017). However, a recent study revealed that the Fto ablation in adult NSCs transiently presented increased proliferation of NSCs and enhanced neuronal differentiation both in vitro and in vivo. But in a long time, the adult neurogenesis and neuronal development were inhibited by the Fto deficiency. Mechanical investigation showed that Fto ablation significantly promoted m6A modification in Pdgfra and Socs5, which synergistically modulated the phosphorylation of Stat3, therefore repairing the neurogenic deficits induced by Fto depletion (Cao et al., 2020).
Readers
FMRP is verified to affect the NSCs differentiation and neural development. both the Mettl14c and Fmr1 deficiency in mice caused the nuclear retention of m6A-modified FMRP target mRNAs, thus displaying delayed cycle progression of NSCs and extended maintenance of proliferating NSCs into postnatal stages, which indicates FMRP plays a critical role in mediating m6A-dependent mRNA nuclear export during neural differentiation (Edens et al., 2019).
The Role of m6A in Bone Marrow Mesenchymal Stem Cells
Bone marrow mesenchymal stem cells (BMSCs), the progenitors for osteoblasts and marrow adipocytes, are commonly obtained from bone marrow aspirates and have been widely considered as the most promising cellular source for tissue regeneration (Baker et al., 2015). Differentiation of BMSCs primarily include osteogenic differentiation and adipogenic differentiation, with Runx2, Sp7, Alp, and Bglap, and Ppary, Cebpa, Adipoq, Plin1, and CD36 as markers, respectively (Wu et al., 2018). With widespread ethical acceptability and relative accessibility, BMSCs are the most commonly used mesenchymal stem cells (MSCs) in clinical trials (Zhou et al., 2019).
Writers
The existed researches about the m6A RNA methylation in BMSCs mainly focused on their osteogenic and adipogenic differentiation capacity. METTL3-mediated m6A modification make significant contributions to the bone formation, osteogenic differentiation, and prevention of increased marrow adiposity via multiple mechanisms (Wu et al., 2018; Tian et al., 2019; Yan et al., 2020; Peng et al., 2022). Mettl3 knockout significantly decreased the translation efficiency of MSCs lineage allocator parathyroid hormone receptor-1 (Pth1r) and suppressed the parathyroid hormone (PTH)-caused osteogenic and adipogenic responses in mice, unveiling METTL3-mediated epigenetic m6A RNA methylation in BMSCs from an early phase regulated Pth1r translation and affected their responses to PTH during bone accrual (Wu et al., 2018). During osteogenic induction in BMSCs, it is METTL3 instead of METTL14, FTO and ALIBH5 that was upregulated. Further investigation revealed that Mettl3 knockdown reduced the level of bone formation-related genes, as well as the mineralized nodules formation, which might be correlated with the involvement of the phosphatidylinositol 3-kinase/AKT (PI3K-Akt) signaling pathway (Tian et al., 2019). In addition, Mettl3 knockdown also decreased the level of Vegfa and its splice variants in BMSCs, indicating METTL3 could promote angiogenesis to improve bone regeneration (Tian et al., 2019). The m6A methylation of precursor-miR-320/runt related transcription factor 2 (RUNX2) is another factor controlling the osteogenic differentiation of BMSCs. METTL3 enhanced, whereas silence or knockout of METTL3 decreased, the m6A RNA methylations of RUNX2, a key transcription factor for osteoblast differentiation and bone formation, and precursor-miR-320, which was rescued by the downregulation of mature miR-320 (Yan et al., 2020). Furthermore, METTL3 mediated m6A methylation of LINC00657, which served as a ceRNA to enhance the expression of BMPR1B via sponging miR-144-3p, could promote osteogenic differentiation of BMSCs (Peng et al., 2022). However, as a regulator of m6A modification, METTL3 also possessed a negative effect on osteogenic differentiation of BMSCs. METTL3 markedly enhanced m6A RNA methylation to MYD88 and subsequently activating NF-κB which is widely considered as a suppressor of osteogenesis (Yu et al., 2020). In the process of adipogenic differentiation, METTL3 expression negatively regulated adipogenic differentiation of porcine BMSCs via the JAK1/STAT5/C/EBPβ pathway in an m6A-YTHDF2-dependent manner (Yao et al., 2019). Moreover, METTL3 inhibited expression of AKT protein in BMSCs by mediating m6A RNA methylation of AKT1, thus reducing MSC adipogenesis and alleviating chemoresistance in AML cells (Pan et al., 2021). METTL14 was also involved in the proliferation and osteogenic differentiation of BMSCs, which triggered the m6A RNA methylation of PTPN6 to increase its mRNA stability and activated Wnt signaling pathway to enhance cell proliferation and osteogenic differentiation (Cheng et al., 2021).
Erasers
The m6A demethylase ALKBH5 presented a crucial role in inhibiting MSCs osteogenic capacity by improving the mRNA decay rate of protein arginine methyltransferase 6 (PRMT6) and discovered the PI3K/AKT pathway as a pivotal downstream target of the ALKBH5-PRMT6 axis (Li Z. et al., 2021). ALKBH5 could also dynamically reverse the METTL3-induced m6A methylation, and coordinated with METTL3 to regulate osteogenic differentiation via NF-κB signaling (Yu et al., 2020). Additionally, ALKBH5 promoted osteoblast differentiation through modulating Runx2 mRNA stability (Feng et al., 2021). The m6A demethylase FTO also possesses a crucial role in BMSCs differentiation, the expression of which was improved during adipocyte differentiation whereas its expression was reduced during osteoblast differentiation, indicating FTO facilitated BMSCs to differentiate into adipocytes compared with osteoblasts (Shen et al., 2018). Mechanical investigation demonstrated that FTO bound to the mRNA of Peroxisome proliferator-activated receptor gamma (PPARγ) and increased its expression, the knockdown of which further blocked the function of growth differentiation factor 11 (GDF11)-FTO to inhibit osteoblast differentiation in BMSCs. Therefore, downregulation of the GDF11-FTO-PPARγ axis might be beneficial for increasing bone formation instead of adipogenesis to treat osteoporosis (Shen et al., 2018). Furthermore, FTO could also trigger osteoporosis of BMSCs through demethylating Runx2 mRNA and suppressing osteogenic differentiation (Wang J. et al., 2021).
Readers
YTHDF1 and YTHDF2 are important readers involved in the proliferation and differentiation of BMSCs. YTHDF1 targeted the zinc finger protein ZNF839, which interacted with and augmented the transcription activity of Runx2, and further enhances osteogenic differentiation of BMSCs (Liu T. et al., 2021). YTHDF2 played a crucial role in METTL3-mediated adipogenic differentiation, which facilitated the degradation of JAK1 to inhibit JAK/STAT signaling pathway, thus suppressing the differentiation of BMSCs (Yao et al., 2019).
The Role of m6A in Hematopoietic Stem Cells
Hematopoietic stem cells (HSCs) are a population of cells possessing significant self-renewal ability and multipotent differentiation to all blood cell types (Li Z. et al., 2019). HSCs therefore act as an important cell type both in the clinic and in basic studies, where HSC transplantation is extensively used for a number of malignant and non-malignant diseases, including leukemia (Wilkinson and Nakauchi, 2020). Myeloid differentiation of HSCs is associated with various markers, including CD11b, CD14, Gr1, and Mac1 (Vu et al., 2017; Lee et al., 2019). Emerging evidences have emphasized the significance of m6A RNA modification in maintaining stem cell function in normal and abnormal hematopoiesis, indicating regulation of m6A RNA methylation in HSCs possessed pivotal role in determining its fate and further affecting its usage in treating diseases such as acute myeloid leukaemia (AML) (Weng et al., 2019; Wang P. et al., 2021).
Writers
In AML cells, expression of METTL3 mRNA and protein are more abundant than in healthy HSCs or other types of tumor cells, the depletion of which promotes cell differentiation and apoptosis and delays leukemia progression in mice (Vu et al., 2017). The direct target of m6A in HSCs was Myc, a marker of HSC symmetric and asymmetric commitment. The translation of c-Myc mRNAs could be improved in the human AML MOLM-13 cell line, while Mettl3-deficient HSCs failed to upregulate MYC expression following stimulation of differentiation (Vu et al., 2017; Lee et al., 2019). In addition, loss of Mettl3 also resulted in enhanced levels of phosphorylated AKT and further promotes the differential effects of Mettl3 depletion (Vu et al., 2017). Yao et, al. reported that METTL3 dominated the function of METTL3-METTL14 m6A methyltransferase complex in catalyzing m6A formation by promoting the expression of genes that maintain HSC quiescence. Nevertheless, METTL14 expression was also regarded to take pivotal role in normal myelopoiesis and AML pathogenesis, the knockdown of which significantly promoted HSCs differentiation into myeloid cells through SPI1-METTL14-MYB/MYC signaling axis (Weng et al., 2018).
Readers
YTHDF2 is a critical m6A reader associated with the self-renewal and regeneration of HSCs. Various studies demonstrated that suppression of YTHDF2 could bolster HSCs expansion without apparent signs of hematological malignancies, making it a promising way for HSC-based gene therapy (Wang H. et al., 2018; Huang and Broxmeyer, 2018; Li et al., 2018). Mechanical exploration revealed that YTHDF2 deficiency simultaneously inhibited the degradation of mRNAs of Wnt target genes as well as survival and proliferation-related genes, thus augmenting the regenerative capacity of HSCs (Wang H. et al., 2018; Huang and Broxmeyer, 2018; Li et al., 2018). Furthermore, in line with myeloid bias, YTHDF2 expression is induced by inflammatory stimulation in HSCs, and deletion of which leads to proinflammatory pathways activation (Mapperley et al., 2021).
Problems and Pespectives
The derivation of iPSCs has almost revolutionized stem-cell research, which not only possess the self-renewal and differentiate capacity like ESCs, but also display a set of advantages because of generating from somatic cells, including freedom from ethical debates, establishments of patient-derived models of disease for etiology research and disease treatment, and important resources for experimental transplantation therapies (Rowe and Daley, 2019). In comparison to ESCs and iPSCs, adult stem cells exhibit restricted potency, but display advantages such as less concern regarding the tumorigenicity and similar gene-expression pattern to adult cells (Blau and Daley, 2019).
Increasing evidences have proved that stem cells therapy is efficient in regenerating damaged tissues of diseases, including heart failure, diabetes, and cerebral palsy (Gurusamy et al., 2018; Blau and Daley, 2019). As reviewed above, m6A modification plays a critical role in facilitating SCT by regulating the proliferation and differentiation capacity of stem cells to affect embryonic development, osteogenesis, adipogenesis, myogenesis, hematopoiesis, neurogenesis and neuronal development (Cao et al., 2020; He and He, 2021). m6A marks a wide range of transcripts associated with self-renewal and differentiation of stem cells, the dynamic changes of which need not only maintain cells self-renewal capacity, but also require the flexibility to differentiate upon receiving corresponding stimulus (Batista et al., 2014; Geula et al., 2015). M6A modification always leads to different outcomes in the same or different types of stem cells owning to the differences in species or tissue sources, cell states, culture environment, and so on (Kudou et al., 2017; Chen et al., 2019; Gheller et al., 2020; Diao et al., 2021; Liang et al., 2021). Therefore, more researches are required to clarify the underlying mechanisms in different kinds of stem cells. M6A RNA modification regulates proliferation and differentiation of stem cells by various mechanisms, mainly affecting RNA metabolism such as RNA stability, translation, decay, splicing, transport, and processing. In overall, m6A methylation promotes the translation and decay, reduces the mRNA stability of targeted genes. Interestingly, some differences are observed in BMSCs that m6A modification increases the mRNA stability of targets in the process of adipogenic differentiation (Yao et al., 2019), while decreasing the mRNA stability of corresponding genes to trigger osteogenic differentiation (Yan et al., 2020; Cheng et al., 2021). However, more researches are required to verify the correctness and elucidate the behind mechanisms. Additionally, a series of fundamental questions should be in careful consideration in order to fully clarify the effect of m6A in various biological processes of stem cells: 1) New components associated with m6A modification, such as writers, erasers, readers, and regulatory proteins need to be identified to further clarify the networks of m6A. 2) The lineage-specific genes are closely associated with stem cells differentiation, which might be the targets of m6A modification to enable direct differentiation of stem cells into required lineages. For example, both the writer METTL3, and the erasers FTO and ALKBH5 can target the Runx2 mRNA of BMSCs to regulate osteogenesis, indicating the methylated level of Runx2 mRNA is pivotal for osteogenic differentiation (Yan et al., 2020; Wang J. et al., 2021; Feng et al., 2021). However, more efforts are needed to determine the key genes associated with the cells differentiation and clarify the potential mechanisms. 3) Although the m6A demethylase FTO and ALKBH5 are reported to facilitate leukemogenesis, their effects on proliferation and differentiation of HSCs have not been investigated. However, FTO and ALKBH5 demethylate m6A RNA to promote the mRNA stability and affect nuclei-to-cytoplasm transport of targets to regulate the proliferation and differentiation of stem cells such as ESCs and BMSCs (Chen X. et al., 2021; Li Z. et al., 2021; Han et al., 2021). More researches are in requirement to elucidate the role of FTO and ALKBH5 in the cell fate determination of HSCs, thus facilitating their application in treating diseases such as AML. 4) As the adipose-derived stem cells (ADSCs) and BMSCs are of increasing importance in the application of SCT to treat diseases of the cardiovascular, nervous and sports systems (Venkataramana et al., 2010; Ahmed et al., 2014; Xu et al., 2016) owning to their characteristics such as multi-directional differentiation, low immunogenicity and high portability (Miao et al., 2017), researches assessing the effects of m6A in ADSCs and BMSCs are more required; 5) The transition of islet cells, myocardial cells and chondrocytes from stem cells is of great significance for the treatment of diabetes, heart diseases and osteoarthritis, more studies are needed to determine the role of m6A in the above processes; 6) researches investigating the regulation of m6A in stem cells have been limited mainly to proliferation and differentiation, despite the fact that other functions of stem cells, such as migratory, homing, immunomodulatory, anti-inflammatory and anti-fibrotic functions, are of the utmost importance for SCT; 7) m6A modification is regulated by a variety of functional proteins. To date, limited research has been conducted on ALKBH5, YTHDF1, YTHDC1 and the HNRNPC family, which may modulate stem cell proliferation and differentiation.
Conclusion
In conclusion, dynamic changes in m6A sites or levels regulated by the coordination between methyltransferase complexes and demethylase complexes alter the m6A landscape in stem cells.
Elucidating the detailed mechanisms and functional roles of m6A RNA in the differentiation and self-renewal processes of stem cells will be essential for treating diseases. As reading proteins bind specific m6A-RNA, m6A indirectly or directly enhances or impedes the expression of key genes responsible for stem cell proliferation and differentiation (Ji et al., 2018). However, the study of m6A in SCT is still in its infancy, and much research remains to be conducted to facilitate its application.
Author Contributions
BW wrote the manuscript and designed the figures and tables. ND revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Undergraduate Innovation Program (grant No. S202110555105), the Hunan University students Innovation and Entrepreneurship training program project (grant No. S202010555133), the Scientific research project of Hunan Education Department (grant Nos 20B506 and 21B0404), the Natural Science Foundation for Young Scientists of Hunan Province (grant Nos. 2021JJ40478 and 2021JJ40471), and the National Natural Science Foundation of China (grant No. 82101869).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aguilo, F., and Walsh, M. J. (2017). The N6-Methyladenosine RNA Modification in Pluripotency and Reprogramming. Curr. Opin. Genet. Develop. 46, 77–82. doi:10.1016/j.gde.2017.06.006
Aguilo, F., Zhang, F., Sancho, A., Fidalgo, M., Di Cecilia, S., Vashisht, A., et al. (2015). Coordination of M 6 A mRNA Methylation and Gene Transcription by ZFP217 Regulates Pluripotency and Reprogramming. Cell Stem Cell 17, 689–704. doi:10.1016/j.stem.2015.09.005
Ahmed, H., Salem, A., Atta, H., Ghazy, M., and Aglan, H. (2014). RETRACTED: Do Adipose Tissue-Derived Mesenchymal Stem Cells Ameliorate Parkinson's Disease in Rat Model? Hum. Exp. Toxicol. 33, 1217–1231. doi:10.1177/0960327114524238
Alarcón, C. R., Goodarzi, H., Lee, H., Liu, X., Tavazoie, S., and Tavazoie, S. F. (2015). HNRNPA2B1 Is a Mediator of m6A-dependent Nuclear RNA Processing Events. Cell 162, 1299–1308. doi:10.1016/j.cell.2015.08.011
Baker, N., Boyette, L. B., and Tuan, R. S. (2015). Characterization of Bone Marrow-Derived Mesenchymal Stem Cells in Aging. Bone 70, 37–47. doi:10.1016/j.bone.2014.10.014
Batista, P. J., Molinie, B., Wang, J., Qu, K., Zhang, J., Li, L., et al. (2014). m6A RNA Modification Controls Cell Fate Transition in Mammalian Embryonic Stem Cells. Cell Stem Cell 15, 707–719. doi:10.1016/j.stem.2014.09.019
Beitnes, J. O., Lunde, K., Brinchmann, J. E., and Aakhus, S. (2011). Stem Cells for Cardiac Repair in Acute Myocardial Infarction. Expert Rev. Cardiovasc. Ther. 9, 1015–1025. doi:10.1586/erc.11.108
Belenguer, G., Duart-Abadia, P., Jordán-Pla, A., Domingo-Muelas, A., Blasco-Chamarro, L., Ferrón, S. R., et al. (2021). Adult Neural Stem Cells Are Alerted by Systemic Inflammation through TNF-α Receptor Signaling. Cell Stem Cell 28, 285–299. e289. doi:10.1016/j.stem.2020.10.016
Bertero, A., Brown, S., Madrigal, P., Osnato, A., Ortmann, D., Yiangou, L., et al. (2018). The SMAD2/3 Interactome Reveals that TGFβ Controls m6A mRNA Methylation in Pluripotency. Nature 555, 256–259. doi:10.1038/nature25784
Biswas, A., and Hutchins, R. (2007). Embryonic Stem Cells. Stem Cell Develop. 16, 213–222. doi:10.1089/scd.2006.0081
Blau, H. M., and Daley, G. Q. (2019). Stem Cells in the Treatment of Disease. N. Engl. J. Med. 380, 1748–1760. doi:10.1056/NEJMra1716145
Cao, Y., Zhuang, Y., Chen, J., Xu, W., Shou, Y., Huang, X., et al. (2020). Dynamic Effects of Fto in Regulating the Proliferation and Differentiation of Adult Neural Stem Cells of Mice. Hum. Mol. Genet. 29, 727–735. doi:10.1093/hmg/ddz274
Chal, J., and Pourquié, O. (2017). Making Muscle: Skeletal Myogenesis In Vivo and In Vitro. Development 144, 2104–2122. doi:10.1242/dev.151035
Chen, C, C., Liu, W., Guo, J., Liu, Y., Liu, X., Liu, J., et al. (2021). Nuclear m6A Reader YTHDC1 Regulates the Scaffold Function of LINE1 RNA in Mouse ESCs and Early Embryos. Protein Cell 12, 455–474. doi:10.1007/s13238-021-00837-8
Chen, J. N, J. N., Chen, Y., Wei, Y. Y., Raza, M. A., Zou, Q., Xi, X. Y., et al. (2019). Regulation of m6A RNA Methylation and its Effect on Myogenic Differentiation in Murine Myoblasts. Mol. Biol. 53, 384–392. doi:10.1134/s002689331903004x
Chen, J, J., Zhang, Y.-C., Huang, C., Shen, H., Sun, B., Cheng, X., et al. (2019). m6A Regulates Neurogenesis and Neuronal Development by Modulating Histone Methyltransferase Ezh2. Genomics, Proteomics & Bioinformatics 17, 154–168. doi:10.1016/j.gpb.2018.12.007
Chen, X, X., Zhao, Q., Zhao, Y. L., Chai, G. S., Cheng, W., Zhao, Z., et al. (2021). Targeted RNA N 6 ‐Methyladenosine Demethylation Controls Cell Fate Transition in Human Pluripotent Stem Cells. Adv. Sci. 8, 2003902. doi:10.1002/advs.202003902
Cheng, C., Zhang, H., Zheng, J., Jin, Y., Wang, D., and Dai, Z. (2021). METTL14 Benefits the Mesenchymal Stem Cells in Patients with Steroid-Associated Osteonecrosis of the Femoral Head by Regulating the m6A Level of PTPN6. Aging 13, 25903–25919. doi:10.18632/aging.203778
Cheng, Y., Luo, H., Izzo, F., Pickering, B. F., Nguyen, D., Myers, R., et al. (2019). m6A RNA Methylation Maintains Hematopoietic Stem Cell Identity and Symmetric Commitment. Cel Rep. 28, 1703–1716. doi:10.1016/j.celrep.2019.07.032
Choi, H. S., Lee, H. M., Jang, Y.-J., Kim, C.-H., and Ryu, C. J. (2013). Heterogeneous Nuclear Ribonucleoprotein A2/B1 Regulates the Self-Renewal and Pluripotency of Human Embryonic Stem Cells via the Control of the G1/S Transition. Stem Cells 31, 2647–2658. doi:10.1002/stem.1366
De Gioia, R., Biella, F., Citterio, G., Rizzo, F., Abati, E., Nizzardo, M., et al. (2020). Neural Stem Cell Transplantation for Neurodegenerative Diseases. Ijms 21, 3103. doi:10.3390/ijms21093103
Deng, K., Zhang, Z., Ren, C., Liang, Y., Gao, X., Fan, Y., et al. (2021). FTO Regulates Myoblast Proliferation by Controlling CCND1 Expression in an m6A-YTHDF2-dependent Manner. Exp. Cel Res. 401, 112524. doi:10.1016/j.yexcr.2021.112524
Desrosiers, R. R., Friderici, K. K., and Rottman, F. (1974). Identification of Methylated Nucleosides in Messenger RNA from Novikoff Hepatoma Cells. Proc. Natl. Acad. Sci. U.S.A. 71, 3971–3975. doi:10.1073/pnas.71.10.3971
Diao, L.-T., Xie, S.-J., Lei, H., Qiu, X.-S., Huang, M.-C., Tao, S., et al. (2021). METTL3 Regulates Skeletal Muscle Specific miRNAs at Both Transcriptional and post-transcriptional Levels. Biochem. Biophysical Res. Commun. 552, 52–58. doi:10.1016/j.bbrc.2021.03.035
Ding, N., Li, E., Ouyang, X., Guo, J., and Wei, B. (2021). The Therapeutic Potential of Bone Marrow Mesenchymal Stem Cells for Articular Cartilage Regeneration in Osteoarthritis. Cscr 16, 840–847. doi:10.2174/1574888X16666210127130044
Edens, B. M., Vissers, C., Su, J., Arumugam, S., Xu, Z., Shi, H., et al. (2019). FMRP Modulates Neural Differentiation through m6A-dependent mRNA Nuclear Export. Cel Rep. 28, 845–854. doi:10.1016/j.celrep.2019.06.072
Emery, A. E. (2002). The Muscular Dystrophies. The Lancet 359, 687–695. doi:10.1016/s0140-6736(02)07815-7
Feng, L., Fan, Y., Zhou, J., Li, S., and Zhang, X. (2021). The RNA Demethylase ALKBH5 Promotes Osteoblast Differentiation by Modulating Runx2 mRNA Stability. FEBS Lett. 595, 2007–2014. doi:10.1002/1873-3468.14145
Gage, F. H., and Temple, S. (2013). Neural Stem Cells: Generating and Regenerating the Brain. Neuron 80, 588–601. doi:10.1016/j.neuron.2013.10.037
Gallo, A., Solfrizzo, M., Epifani, F., Panzarini, G., and Perrone, G. (2016). Effect of Temperature and Water Activity on Gene Expression and Aflatoxin Biosynthesis in Aspergillus flavus on almond Medium. Int. J. Food Microbiol. 217, 162–169. doi:10.1016/j.ijfoodmicro.2015.10.026
Geula, S., Moshitch-Moshkovitz, S., Dominissini, D., Mansour, A. A., Kol, N., Salmon-Divon, M., et al. (2015). m 6 A mRNA Methylation Facilitates Resolution of Naïve Pluripotency toward Differentiation. Science 347, 1002–1006. doi:10.1126/science.1261417
Gheller, B. J., Blum, J. E., Fong, E. H. H., Malysheva, O. V., Cosgrove, B. D., and Thalacker-Mercer, A. E. (2020). A Defined N6-Methyladenosine (m6A) Profile Conferred by METTL3 Regulates Muscle Stem Cell/myoblast State Transitions. Cell Death Discov. 6, 95. doi:10.1038/s41420-020-00328-5
Gnecchi, M., Danieli, P., and Cervio, E. (2012). Mesenchymal Stem Cell Therapy for Heart Disease. Vasc. Pharmacol. 57, 48–55. doi:10.1016/j.vph.2012.04.002
Gorecka, A., Salemi, S., Haralampieva, D., Moalli, F., Stroka, D., Candinas, D., et al. (2018). Autologous Transplantation of Adipose-Derived Stem Cells Improves Functional Recovery of Skeletal Muscle without Direct Participation in New Myofiber Formation. Stem Cel Res Ther 9, 195. doi:10.1186/s13287-018-0922-1
Gurusamy, N., Alsayari, A., Rajasingh, S., and Rajasingh, J. (2018). Adult Stem Cells for Regenerative Therapy. Prog. Mol. Biol. Transl Sci. 160, 1–22. doi:10.1016/bs.pmbts.2018.07.009
Han, Z., Xu, Z., Yu, Y., Cao, Y., Bao, Z., Gao, X., et al. (2021). ALKBH5-mediated m6A mRNA Methylation Governs Human Embryonic Stem Cell Cardiac Commitment. Mol. Ther. - Nucleic Acids 26, 22–33. doi:10.1016/j.omtn.2021.05.019
He, P. C., and He, C. (2021). m 6 A RNA Methylation: from Mechanisms to Therapeutic Potential. EMBO J. 40, e105977. doi:10.15252/embj.2020105977
Heck, A. M., Russo, J., Wilusz, J., Nishimura, E. O., and Wilusz, C. J. (2020). YTHDF2 Destabilizes m6A-Modified Neural-specific RNAs to Restrain Differentiation in Induced Pluripotent Stem Cells. RNA 26, 739–755. doi:10.1261/rna.073502.119
Hess, M. E., Hess, S., Meyer, K. D., Verhagen, L. A. W., Koch, L., Brönneke, H. S., et al. (2013). The Fat Mass and Obesity Associated Gene (Fto) Regulates Activity of the Dopaminergic Midbrain Circuitry. Nat. Neurosci. 16, 1042–1048. doi:10.1038/nn.3449
Huang, H., Liu, L., Li, C., Liang, Z., Huang, Z., Wang, Q., et al. (2020). Fat Mass- and Obesity-Associated (FTO) Gene Promoted Myoblast Differentiation through the Focal Adhesion Pathway in Chicken. 3 Biotech. 10, 403. doi:10.1007/s13205-020-02386-z
Huang, X., and Broxmeyer, H. E. (2018). m6A Reader Suppression Bolsters HSC Expansion. Cell Res 28, 875–876. doi:10.1038/s41422-018-0081-z
Ignatova, V. V., Stolz, P., Kaiser, S., Gustafsson, T. H., Lastres, P. R., Sanz-Moreno, A., et al. (2020). The rRNA m6A Methyltransferase METTL5 Is Involved in Pluripotency and Developmental Programs. Genes Dev. 34, 715–729. doi:10.1101/gad.333369.119
Ji, P., Wang, X., Xie, N., and Li, Y. (2018). N6-methyladenosine in RNA and DNA: an Epitranscriptomic and Epigenetic Player Implicated in Determination of Stem Cell Fate. Stem Cell Int. 2018, 1–18. doi:10.1155/2018/3256524
Jia, G., Fu, Y., Zhao, X., Dai, Q., Zheng, G., Yang, Y., et al. (2011). N6-Methyladenosine in Nuclear RNA Is a Major Substrate of the Obesity-Associated FTO. Nat. Chem. Biol. 7, 885–887. doi:10.1038/nchembio.687
Jiang, X., Liu, B., Nie, Z., Duan, L., Xiong, Q., Jin, Z., et al. (2021). The Role of m6A Modification in the Biological Functions and Diseases. Sig Transduct Target. Ther. 6, 74. doi:10.1038/s41392-020-00450-x
Ke, S., Alemu, E. A., Mertens, C., Gantman, E. C., Fak, J. J., Mele, A., et al. (2015). A Majority of m6A Residues Are in the Last Exons, Allowing the Potential for 3′ UTR Regulation. Genes Dev. 29, 2037–2053. doi:10.1101/gad.269415.115
Kudou, K., Komatsu, T., Nogami, J., Maehara, K., Harada, A., Saeki, H., et al. (2017). The Requirement of Mettl3-Promoted MyoD mRNA Maintenance in Proliferative Myoblasts for Skeletal Muscle Differentiation. Open Biol. 7, 170119. doi:10.1098/rsob.170119
Kwon, J., Jo, Y.-J., Namgoong, S., and Kim, N.-H. (2019). Functional Roles of hnRNPA2/B1 Regulated by METTL3 in Mammalian Embryonic Development. Sci. Rep. 9, 8640. doi:10.1038/s41598-019-44714-1
Lee, D.-F., Walsh, M. J., and Aguiló, F. (2016). ZNF217/ZFP217 Meets Chromatin and RNA. Trends Biochem. Sci. 41, 986–988. doi:10.1016/j.tibs.2016.07.013
Lee, H., Bao, S., Qian, Y., Geula, S., Leslie, J., Zhang, C., et al. (2019). Stage-specific Requirement for Mettl3-dependent m6A mRNA Methylation during Haematopoietic Stem Cell Differentiation. Nat. Cel Biol 21, 700–709. doi:10.1038/s41556-019-0318-1
Li, J, J., Pei, Y., Zhou, R., Tang, Z., and Yang, Y. (2021). Regulation of RNA N6-Methyladenosine Modification and its Emerging Roles in Skeletal Muscle Development. Int. J. Biol. Sci. 17, 1682–1692. doi:10.7150/ijbs.56251
Li, J, J., Yang, X., Qi, Z., Sang, Y., Liu, Y., Xu, B., et al. (2019). The Role of mRNA m6A Methylation in the Nervous System. Cell Biosci 9, 66. doi:10.1186/s13578-019-0330-y
Li, L., Zang, L., Zhang, F., Chen, J., Shen, H., Shu, L., et al. (2017). Fat Mass and Obesity-Associated (FTO) Protein Regulates Adult Neurogenesis. Hum. Mol. Genet. 26, 2398–2411. doi:10.1093/hmg/ddx128
Li, Z, Z., He, X. C., and Li, L. (2019). Hematopoietic Stem Cells. Curr. Opin. Hematol. 26, 258–265. doi:10.1097/MOH.0000000000000506
Li, Z., Qian, P., Shao, W., Shi, H., He, X. C., Gogol, M., et al. (2018). Suppression of m6A Reader Ythdf2 Promotes Hematopoietic Stem Cell Expansion. Cel Res 28, 904–917. doi:10.1038/s41422-018-0072-0
Li, Z, Z., Wang, P., Li, J., Xie, Z., Cen, S., Li, M., et al. (2021). The N6-Methyladenosine Demethylase ALKBH5 Negatively Regulates the Osteogenic Differentiation of Mesenchymal Stem Cells through PRMT6. Cell Death Dis 12, 578. doi:10.1038/s41419-021-03869-4
Liang, Y., Han, H., Xiong, Q., Yang, C., Wang, L., Ma, J., et al. (2021). METTL3-Mediated m6A Methylation Regulates Muscle Stem Cells and Muscle Regeneration by Notch Signaling Pathway. Stem Cell Int. 2021, 1–13. doi:10.1155/2021/9955691
Lin, S., Choe, J., Du, P., Triboulet, R., and Gregory, R. I. (2016). The M 6 A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Mol. Cel. 62, 335–345. doi:10.1016/j.molcel.2016.03.021
Lin, S., and Gregory, R. I. (2014). Methyltransferases Modulate RNA Stability in Embryonic Stem Cells. Nat. Cel Biol 16, 129–131. doi:10.1038/ncb2914
Liu, J., Gao, M., Xu, S., Chen, Y., Wu, K., Liu, H., et al. (2020). YTHDF2/3 Are Required for Somatic Reprogramming through Different RNA Deadenylation Pathways. Cel Rep. 32, 108120. doi:10.1016/j.celrep.2020.108120
Liu, J., Yue, Y., Han, D., Wang, X., Fu, Y., Zhang, L., et al. (2014). A METTL3-METTL14 Complex Mediates Mammalian Nuclear RNA N6-Adenosine Methylation. Nat. Chem. Biol. 10, 93–95. doi:10.1038/nchembio.1432
Liu, N., Zhou, K. I., Parisien, M., Dai, Q., Diatchenko, L., and Pan, T. (2017). N 6-methyladenosine Alters RNA Structure to Regulate Binding of a Low-Complexity Protein. Nucleic Acids Res. 45, 6051–6063. doi:10.1093/nar/gkx141
Liu, T, T., Zheng, X., Wang, C., Wang, C., Jiang, S., Li, B., et al. (2021). The m6A "reader" YTHDF1 Promotes Osteogenesis of Bone Marrow Mesenchymal Stem Cells through Translational Control of ZNF839. Cel Death Dis 12. doi:10.1038/s41419-021-04312-4
Liu, X, X., Wang, H., Zhao, X., Luo, Q., Wang, Q., Tan, K., et al. (2021). Arginine Methylation of METTL14 Promotes RNA N6-Methyladenosine Modification and Endoderm Differentiation of Mouse Embryonic Stem Cells. Nat. Commun. 12, 3780. doi:10.1038/s41467-021-24035-6
Liu, Z.-X., Li, L.-M., Sun, H.-L., and Liu, S.-M. (2018). Link between m6A Modification and Cancers. Front. Bioeng. Biotechnol. 6, 89. doi:10.3389/fbioe.2018.00089
Ma, T., Gong, K., Ao, Q., Yan, Y., Song, B., Huang, H., et al. (2013). Intracerebral Transplantation of Adipose-Derived Mesenchymal Stem Cells Alternatively Activates Microglia and Ameliorates Neuropathological Deficits in Alzheimer's Disease Mice. Cel Transpl. 22 (Suppl. 1), 113–126. doi:10.3727/096368913X672181
Mapperley, C., Van De Lagemaat, L. N., Lawson, H., Tavosanis, A., Paris, J., Campos, J., et al. (2021). The mRNA m6A Reader YTHDF2 Suppresses Proinflammatory Pathways and Sustains Hematopoietic Stem Cell Function. J. Exp. Med. 218. doi:10.1084/jem.20200829
Mctaggart, J. S., Lee, S., Iberl, M., Church, C., Cox, R. D., and Ashcroft, F. M. F. (2011). FTO Is Expressed in Neurones throughout the Brain and its Expression Is Unaltered by Fasting. PLoS One 6, e27968. doi:10.1371/journal.pone.0027968
Meyer, K. D., and Jaffrey, S. R. (2014). The Dynamic Epitranscriptome: N6-Methyladenosine and Gene Expression Control. Nat. Rev. Mol. Cel Biol 15, 313–326. doi:10.1038/nrm3785
Meyer, K. D., Saletore, Y., Zumbo, P., Elemento, O., Mason, C. E., and Jaffrey, S. R. (2012). Comprehensive Analysis of mRNA Methylation Reveals Enrichment in 3′ UTRs and Near Stop Codons. Cell 149, 1635–1646. doi:10.1016/j.cell.2012.05.003
Miao, C., Lei, M., Hu, W., Han, S., and Wang, Q. (2017). A Brief Review: The Therapeutic Potential of Bone Marrow Mesenchymal Stem Cells in Myocardial Infarction. Stem Cel Res Ther 8. doi:10.1186/s13287-017-0697-9
Negroni, E., Gidaro, T., Bigot, A., Butler-Browne, G. S., Mouly, V., and Trollet, C. (2015). Invited Review: Stem Cells and Muscle Diseases: Advances in Cell Therapy Strategies. Neuropathol. Appl. Neurobiol. 41, 270–287. doi:10.1111/nan.12198
Oerum, S., Meynier, V., Catala, M., and Tisné, C. (2021). A Comprehensive Review of m6A/m6Am RNA Methyltransferase Structures. Nucleic Acids Res. 49, 7239–7255. doi:10.1093/nar/gkab378
Pan, Z. p., Wang, B., Hou, D. y., You, R. l., Wang, X. t., Xie, W. h., et al. (2021). METTL3 Mediates Bone Marrow Mesenchymal Stem Cell Adipogenesis to Promote Chemoresistance in Acute Myeloid Leukaemia. FEBS Open Bio 11, 1659–1672. doi:10.1002/2211-5463.13165
Park, I.-H., Zhao, R., West, J. A., Yabuuchi, A., Huo, H., Ince, T. A., et al. (2008). Reprogramming of Human Somatic Cells to Pluripotency with Defined Factors. Nature 451, 141–146. doi:10.1038/nature06534
Peng, J., Zhan, Y., and Zong, Y. (2022). METTL3-mediated LINC00657 Promotes Osteogenic Differentiation of Mesenchymal Stem Cells via miR-144-3p/BMPR1B axis. Cell Tissue Res. doi:10.1007/s00441-022-03588-y
Pollini, D., Loffredo, R., Maniscalco, F., Cardano, M., Micaelli, M., Bonomo, I., et al. (2021). Multilayer and MATR3-dependent Regulation of mRNAs Maintains Pluripotency in Human Induced Pluripotent Stem Cells. iScience 24, 102197. doi:10.1016/j.isci.2021.102197
Rikhtegar, R., Pezeshkian, M., Dolati, S., Safaie, N., Afrasiabi Rad, A., Mahdipour, M., et al. (2019). Stem Cells as Therapy for Heart Disease: iPSCs, ESCs, CSCs, and Skeletal Myoblasts. Biomed. Pharmacother. 109, 304–313. doi:10.1016/j.biopha.2018.10.065
Roundtree, I. A., Evans, M. E., Pan, T., and He, C. (2017). Dynamic RNA Modifications in Gene Expression Regulation. Cell 169, 1187–1200. doi:10.1016/j.cell.2017.05.045
Roundtree, I. A., and He, C. (2016). Nuclear M 6 A Reader YTHDC1 Regulates mRNA Splicing. Trends Genet. 32, 320–321. doi:10.1016/j.tig.2016.03.006
Rowe, R. G., and Daley, G. Q. (2019). Induced Pluripotent Stem Cells in Disease Modelling and Drug Discovery. Nat. Rev. Genet. 20, 377–388. doi:10.1038/s41576-019-0100-z
Shen, G.-S., Zhou, H.-B., Zhang, H., Chen, B., Liu, Z.-P., Yuan, Y., et al. (2018). The GDF11-FTO-Pparγ axis Controls the Shift of Osteoporotic MSC Fate to Adipocyte and Inhibits Bone Formation during Osteoporosis. Biochim. Biophys. Acta (Bba) - Mol. Basis Dis. 1864, 3644–3654. doi:10.1016/j.bbadis.2018.09.015
Sun, T., Wu, R., and Ming, L. (2019). The Role of m6A RNA Methylation in Cancer. Biomed. Pharmacother. 112, 108613. doi:10.1016/j.biopha.2019.108613
Takahashi, K., Tanabe, K., Ohnuki, M., Narita, M., Ichisaka, T., Tomoda, K., et al. (2007). Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell 131, 861–872. doi:10.1016/j.cell.2007.11.019
Tian, C., Huang, Y., Li, Q., Feng, Z., and Xu, Q. (2019). Mettl3 Regulates Osteogenic Differentiation and Alternative Splicing of Vegfa in Bone Marrow Mesenchymal Stem Cells. Ijms 20, 551. doi:10.3390/ijms20030551
Tiwary, S. (2020). YTHDF3 Induces the Translation of m6A-Enriched Gene Transcripts to Promote Breast Cancer Brain Metastasis. Cancer Cell 38.
Trounson, A., and Mcdonald, C. (2015). Stem Cell Therapies in Clinical Trials: Progress and Challenges. Cell Stem Cell 17, 11–22. doi:10.1016/j.stem.2015.06.007
Venkataramana, N. K., Kumar, S. K. V., Balaraju, S., Radhakrishnan, R. C., Bansal, A., Dixit, A., et al. (2010). Open-labeled Study of Unilateral Autologous Bone-Marrow-Derived Mesenchymal Stem Cell Transplantation in Parkinson's Disease. Translational Res. 155, 62–70. doi:10.1016/j.trsl.2009.07.006
Vu, L. P., Pickering, B. F., Cheng, Y., Zaccara, S., Nguyen, D., Minuesa, G., et al. (2017). The N6-Methyladenosine (m6A)-Forming Enzyme METTL3 Controls Myeloid Differentiation of normal Hematopoietic and Leukemia Cells. Nat. Med. 23, 1369–1376. doi:10.1038/nm.4416
Wang, H, H., Zuo, H., Liu, J., Wen, F., Gao, Y., Zhu, X., et al. (2018). Loss of YTHDF2-Mediated m6A-dependent mRNA Clearance Facilitates Hematopoietic Stem Cell Regeneration. Cel Res 28, 1035–1038. doi:10.1038/s41422-018-0082-y
Wang, J, J., Fu, Q., Yang, J., Liu, J.-L., Hou, S.-M., Huang, X., et al. (2021). RNA N6-Methyladenosine Demethylase FTO Promotes Osteoporosis through Demethylating Runx2 mRNA and Inhibiting Osteogenic Differentiation. Aging 13, 21134–21141. doi:10.18632/aging.203377
Wang, K. W., Mouhieddine, T., Nokkari, A., Itani, M., and Kobeissy, F. (2014). Stem Cells in Neuroinjury and Neurodegenerative Disorders: Challenges and Future Neurotherapeutic Prospects. Neural Regen. Res. 9, 901–906. doi:10.4103/1673-5374.133129
Wang, P, P., Feng, M., Han, G., Yin, R., Li, Y., Yao, S., et al. (2021). RNA m6A Modification Plays a Key Role in Maintaining Stem Cell Function in Normal and Malignant Hematopoiesis. Front. Cel Dev. Biol. 9, 710964. doi:10.3389/fcell.2021.710964
Wang, S, S., Zhang, J., Wu, X., Lin, X., Liu, X.-M., and Zhou, J. (2021). Differential Roles of YTHDF1 and YTHDF3 in Embryonic Stem Cell-Derived Cardiomyocyte Differentiation. RNA Biol. 18, 1–10. doi:10.1080/15476286.2020.1850628
Wang, X., Huang, N., Yang, M., Wei, D., Tai, H., Han, X., et al. (2017). FTO Is Required for Myogenesis by Positively Regulating mTOR-PGC-1α Pathway-Mediated Mitochondria Biogenesis. Cel Death Dis 8, e2702. doi:10.1038/cddis.2017.122
Wang, X., Zhao, B. S., Roundtree, I. A., Lu, Z., Han, D., Ma, H., et al. (2015). N6-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 161, 1388–1399. doi:10.1016/j.cell.2015.05.014
Wang, Y., Li, Y., Toth, J. I., Petroski, M. D., Zhang, Z., and Zhao, J. C. (2014). N6-methyladenosine Modification Destabilizes Developmental Regulators in Embryonic Stem Cells. Nat. Cel Biol 16, 191–198. doi:10.1038/ncb2902
Wang, Y, Y., Li, Y., Yue, M., Wang, J., Kumar, S., Wechsler-Reya, R. J., et al. (2018). N6-methyladenosine RNA Modification Regulates Embryonic Neural Stem Cell Self-Renewal through Histone Modifications. Nat. Neurosci. 21, 195–206. doi:10.1038/s41593-017-0057-1
Wang, Z, Z., Pan, Z., Adhikari, S., Harada, B. T., Shen, L., Yuan, W., et al. (2021). m 6 A Deposition Is Regulated by PRMT1‐mediated Arginine Methylation of METTL14 in its Disordered C‐terminal Region. EMBO J. 40, e106309. doi:10.15252/embj.2020106309
Wen, J., Lv, R., Ma, H., Shen, H., He, C., Wang, J., et al. (2018). Zc3h13 Regulates Nuclear RNA m6A Methylation and Mouse Embryonic Stem Cell Self-Renewal. Mol. Cel 69, 1028–1038. e1026. doi:10.1016/j.molcel.2018.02.015
Weng, H., Huang, H., and Chen, J. (2019). RNA N 6-Methyladenosine Modification in Normal and Malignant Hematopoiesis. Adv. Exp. Med. Biol. 1143, 75–93. doi:10.1007/978-981-13-7342-8_4
Weng, H., Huang, H., Wu, H., Qin, X., Zhao, B. S., Dong, L., et al. (2018). METTL14 Inhibits Hematopoietic Stem/Progenitor Differentiation and Promotes Leukemogenesis via mRNA m6A Modification. Cell Stem Cell 22, 191–205. doi:10.1016/j.stem.2017.11.016
Wilkinson, A. C., and Nakauchi, H. (2020). Stabilizing Hematopoietic Stem Cells In Vitro. Curr. Opin. Genet. Develop. 64, 1–5. doi:10.1016/j.gde.2020.05.035
Wu, R., Liu, Y., Zhao, Y., Bi, Z., Yao, Y., Liu, Q., et al. (2019). m6A Methylation Controls Pluripotency of Porcine Induced Pluripotent Stem Cells by Targeting SOCS3/JAK2/STAT3 Pathway in a YTHDF1/YTHDF2-Orchestrated Manner. Cel Death Dis 10, 171. doi:10.1038/s41419-019-1417-4
Wu, Y., Xie, L., Wang, M., Xiong, Q., Guo, Y., Liang, Y., et al. (2018). Mettl3-mediated m6A RNA Methylation Regulates the Fate of Bone Marrow Mesenchymal Stem Cells and Osteoporosis. Nat. Commun. 9. doi:10.1038/s41467-018-06898-4
Xing, L., Cai, Y., Yang, T., Yu, W., Gao, M., Chai, R., et al. (2021). Epitranscriptomic m6A Regulation Following Spinal Cord Injury. J. Neurosci. Res. 99, 843–857. doi:10.1002/jnr.24763
Xu, C., Wang, X., Liu, K., Roundtree, I. A., Tempel, W., Li, Y., et al. (2014). Structural Basis for Selective Binding of m6A RNA by the YTHDC1 YTH Domain. Nat. Chem. Biol. 10, 927–929. doi:10.1038/nchembio.1654
Xu, H. M., Liu, Y. R., Wan, Q. L., Li, Y. M., and Wu, C. W. (2016). PTEN-silenced Bone Marrow Mesenchymal Stem Cell Transplantation for Acute Myocardial Infarction. Chin. J. Tissue Eng. Res. 20, 3400–3406. doi:10.3969/j.issn.2095-4344.2016.23.008
Yan, G., Yuan, Y., He, M., Gong, R., Lei, H., Zhou, H., et al. (2020). m6A Methylation of Precursor-miR-320/runx2 Controls Osteogenic Potential of Bone Marrow-Derived Mesenchymal Stem Cells. Mol. Ther. - Nucleic Acids 19, 421–436. doi:10.1016/j.omtn.2019.12.001
Yang, D., Qiao, J., Wang, G., Lan, Y., Li, G., Guo, X., et al. (2018). N 6-Methyladenosine Modification of lincRNA 1281 Is Critically Required for mESC Differentiation Potential. Nucleic Acids Res. 46, 3906–3920. doi:10.1093/nar/gky130
Yao, Y., Bi, Z., Wu, R., Zhao, Y., Liu, Y., Liu, Q., et al. (2019). METTL3 Inhibits BMSC Adipogenic Differentiation by Targeting the JAK1/STAT5/C/EBPβ Pathway via an M 6 A‐YTHDF2-dependent Manner. FASEB j. 33, 7529–7544. doi:10.1096/fj.201802644R
Yen, Y.-P., and Chen, J.-A. (2021). The m6A Epitranscriptome on Neural Development and Degeneration. J. Biomed. Sci. 28, 40. doi:10.1186/s12929-021-00734-6
Yeo, J.-C., and Ng, H.-H. (2013). The Transcriptional Regulation of Pluripotency. Cel Res 23, 20–32. doi:10.1038/cr.2012.172
Young, R. A. (2011). Control of the Embryonic Stem Cell State. Cell 144, 940–954. doi:10.1016/j.cell.2011.01.032
Yu, J., Shen, L., Liu, Y., Ming, H., Zhu, X., Chu, M., et al. (2020). The m6A Methyltransferase METTL3 Cooperates with Demethylase ALKBH5 to Regulate Osteogenic Differentiation through NF-Κb Signaling. Mol. Cel Biochem 463, 203–210. doi:10.1007/s11010-019-03641-5
Zhang, X., Yao, Y., Han, J., Yang, Y., Chen, Y., Tang, Z., et al. (2020). Longitudinal Epitranscriptome Profiling Reveals the Crucial Role of N6-Methyladenosine Methylation in Porcine Prenatal Skeletal Muscle Development. J. Genet. Genomics 47, 466–476. doi:10.1016/j.jgg.2020.07.003
Zhao, B. S., and He, C. (2015). Fate by RNA Methylation: m6A Steers Stem Cell Pluripotency. Genome Biol. 16, 43. doi:10.1186/s13059-015-0609-1
Keywords: N6-methyladenosine, RNA modification, stem cell, proliferation, differentiation
Citation: Wei B, Zeng M, Yang J, Li S, Zhang J, Ding N and Jiang Z (2022) N6-Methyladenosine RNA Modification: A Potential Regulator of Stem Cell Proliferation and Differentiation. Front. Cell Dev. Biol. 10:835205. doi: 10.3389/fcell.2022.835205
Received: 14 December 2021; Accepted: 09 March 2022;
Published: 04 April 2022.
Edited by:
Prasad S. Koka, Biomedical Research Institute of Southern California, United StatesReviewed by:
Xiaoyong Li, Shanghai Jiao Tong University, ChinaKar-Tong Tan, Harvard University, United States
Copyright © 2022 Wei, Zeng, Yang, Li, Zhang, Ding and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nan Ding, bmFuZGluZzE5ODlAMTI2LmNvbQ==; Zhisheng Jiang, enNqaWFuZzIwMTZAMTYzLmNvbQ==
 Bo Wei
Bo Wei Meiyu Zeng1
Meiyu Zeng1 Nan Ding
Nan Ding