- 1Division of Biosciences, College of Dentistry, The Ohio State University, Columbus, OH, United States
- 2Department of Cell Developmental and Integrative Biology, University of Alabama at Birmingham, Birmingham, AL, United States
- 3Department of Biomedical Informatics, The Ohio State University, Columbus, OH, United States
Transforming growth factor β (TGFβ) plays an important role in tooth morphogenesis and mineralization. During postnatal development, the dental pulp (DP) mesenchyme secretes neurotrophic factors that guide trigeminal nerve fibers into and throughout the DP. This process is tightly linked with dentin formation and mineralization. Our laboratory established a mouse model in which Tgfbr2 was conditionally deleted in DP mesenchyme using an Osterix promoter-driven Cre recombinase (Tgfbr2cko). These mice survived postnatally with significant defects in bones and teeth, including reduced mineralization and short roots. Hematoxylin and eosin staining revealed reduced axon-like structures in the mutant mice. Reporter imaging demonstrated that Osterix-Cre activity within the tooth was active in the DP and derivatives, but not in neuronal afferents. Immunofluorescence staining for β3 tubulin (neuronal marker) was performed on serial cryosections from control and mutant molars on postnatal days 7 and 24 (P7, P24). Confocal imaging and pixel quantification demonstrated reduced innervation in Tgfbr2cko first molars at both stages compared to controls, indicating that signals necessary to promote neurite outgrowth were disrupted by Tgfbr2 deletion. We performed mRNA-Sequence (RNA-Seq) and gene onotology analyses using RNA from the DP of P7 control and mutant mice to investigate the pathways involved in Tgfbr2-mediated tooth development. These analyses identified downregulation of several mineralization-related and neuronal genes in the Tgfbr2cko DP compared to controls. Select gene expression patterns were confirmed by quantitative real-time PCR and immunofluorescence imaging. Lastly, trigeminal neurons were co-cultured atop Transwell filters overlying primary Tgfbr2f/f DP cells. Tgfbr2 in the DP was deleted via Adenovirus-expressed Cre recombinase. Confocal imaging of axons through the filter pores showed increased axonal sprouting from neurons cultured with Tgfbr2-positive DP cells compared to neurons cultured alone. Axon sprouting was reduced when Tgfbr2 was knocked down in the DP cells. Immunofluorescence of dentin sialophosphoprotein in co-cultured DP cells confirmed reduced mineralization potential in cells with Tgfbr2 deletion. Both our proteomics and RNA-Seq analyses indicate that axonal guidance cues, particularly semaphorin signaling, were disrupted by Tgfbr2 deletion. Thus, Tgfbr2 in the DP mesenchyme appears to regulate differentiation and the cells’ ability to guide neurite outgrowth during tooth mineralization and innervation.
Introduction
The transforming growth factor β (TGFβ) superfamily regulates many developmental processes, including those that occur in the tooth and skeleton (Oka et al., 2007; Seo and Serra, 2009; Sohn et al., 2010; Wang et al., 2013; Zhen et al., 2013; Iwata et al., 2014; Choi et al., 2016; Peters et al., 2017; Niwa et al., 2018; Kawatsu et al., 2021; Zhang et al., 2021). Tooth development requires the coordination of epithelial, mesenchymal, hematopoietic, and neuronal cell populations to construct a functional sensory organ. Tooth development begins with an invagination of the ectodermal epithelium into the surrounding mesenchyme, around embryonic day 11 in the mouse. The epithelial bud then bifurcates and elongates. The underlying dental mesenchyme later gives rise to the dental pulp (DP) and dentin-producing odontoblasts. During postnatal development, afferent axons from the trigeminal ganglia (TG) penetrate into and throughout the tooth around the time dentin deposition begins [reviewed in (Pagella et al., 2014); (Shadad et al., 2019)]. Innervation rapidly progresses to arborize into sub-odontoblastic, odontoblastic, and predentinal axonal networks, with axons present in coronal dentin canals (Byers, 1980). This sensory innervation provides signals crucial to daily oral activities, such as eating and talking, and provides pain signals to protect the tooth organ (Hildebrand et al., 1995). The roles of TGFβ signaling in early aspects of tooth development and pathology were previously documented in mice with mutations or targeted deletions of genes encoding components of TGFβ signaling pathways (Ko et al., 2007; Zhao et al., 2008; Cho et al., 2013; Wang et al., 2013; Peters et al., 2017). However, Tgfbr2 deletion in the dental and skeletal mesenchyme or epithelium is embryonic or perinatal lethal (Dünker and Krieglstein, 2002; Ito et al., 2003; Baffi et al., 2004; Seo and Serra, 2007, 2009; Zhao et al., 2011; Iwata et al., 2012). Since axons from the trigeminal ganglion do not penetrate the DP until around P3 (Moe et al., 2012; Shadad et al., 2019), this lethality prevented comprehensive research into the roles of TGFβ signaling in tooth innervation.
To address this obstacle, our laboratory established an Osterix promoter-driven Cre recombinase mouse model in which Tgfbr2 was conditionally deleted in the osteoblast- and odontoblast-producing mesenchyme (Tgfbr2cko) (Wang et al., 2013; Peters et al., 2017; Corps et al., 2021). The Tgfbr2cko mice did not demonstrate skeletal or dental abnormalities at birth, but within the first week of life began to demonstrate reduced growth and mineralization in bones and teeth that worsened with development. The mutant phenotype included stunted root elongation and hypomineralization in molars (Wang et al., 2013; Peters et al., 2017; Corps et al., 2021). We stained P5, P7, P10, and P14 mandibular first molars (M1s) with hematoxylin and eosin (H&E) to investigate the short tooth root phenotype and noticed a reduction in axon-like structures in the mutant mice. Since Osterix-Cre should be restricted to the mesenchymal lineage (Rodda and McMahon, 2006; Rakian et al., 2013; Wang et al., 2013; Ono et al., 2016; Peters et al., 2017), neuronal alterations were unexpected. However, previous studies indicated that DP cells (DPCs) secrete neurotrophic factors to regulate tooth innervation (Kettunen, 2005; Kettunen et al., 2007; Moe et al., 2012; Sijaona et al., 2012; de Almeida et al., 2014) although little information was available regarding the signaling pathways that guide this phenomenon. Furthermore, many of the previous studies focused on embryonic stages when the DP secretes repellants to prevent neurite extensions from entering the tooth rather than postnatal stages when the DP secretes axonal attractants (Lillesaar and Fried, 2004; Fried et al., 2007). We hypothesized that Tgfbr2 in the DP mesenchyme regulates paracrine signals to guide postnatal tooth innervation.
We herein demonstrate by quantitation of a neuronal marker, β3 tubulin (β3T), that innervation was reduced in molars from Tgfbr2cko mice compared to controls starting at P7 and continuing to P24. We confirmed that the Cre expression was isolated to the dental pulp mesenchyme by crossing Osterix-Cre with the ROSA26mTmG reporter line (Rakian et al., 2013; Wang et al., 2013; Ono et al., 2016). To investigate the Tgfbr2-related pathways involved in tooth development, we performed an RNA-Seq analysis on DP tissue, including odontoblasts, from control and Tgfbr2cko P7 mice. Gene ontology analyses identified several overrepresented neuronal genes in the mutant mice compared to the controls, and these findings correlated with the phenotype seen in the H&E and β3T images. Subsequent qPCR analyses of RNA extracted from P7 first molars confirmed the findings for selected genes from the RNA-Seq. We then performed a co-culture using dispersed cells from the TG and primary DPCs and showed that neurite outgrowth was reduced when Tgfbr2 was deleted. Lastly, a global evaluation of our RNA-Seq, proteomics on cell culture media, and immunofluorescence images demonstrated the existence of a developmental program regulating tooth mineralization and innervation that involves semaphorin signaling (Lillesaar and Fried, 2004; Sijaona et al., 2012). These data reveal a novel function for Tgfbr2 in the development of mineralized, innervated teeth.
Materials and Methods
Mice
All experiments with mice were approved by the UAB and OSU Institutional Animal Care and Use Committees. The Osterix-Cre and Tgfbr2cko mice were described previously (Wang et al., 2013; Peters et al., 2017). Trigeminal neuron bundles for co-cultures were collected from 6-week-old B6.Cg-Tg (Thy1-YFP)16Jrs/J male and female mice. The Cre reporter strain Gt (ROSA)26SORtm4(ACTB-tdTomato,-EGFP) Luo/J (ROSA26mTmG) was crossed with Osterix-Cre mice to generate Osterix-Cre; ROSA26-mTmG mice with traceable Cre activity (Muzumdar et al., 2007). Equivalent numbers of males and females were used in all experiments.
Imaging of DP Structures
Postnatal mandibles were fixed in 4% paraformaldehyde for 1 h, followed by EDTA decalcification. Tissue was embedded and frozen in optimal cutting temperature compound and sectioned on the sagittal plane with a cryostat (Leica Biosystems, Buffalo Grove, IL, United States) at 10 μm thickness. For immunofluorescence studies of β3T and CD31, sections were permeabilized with cold acetone at −20°C for 10 min and rinsed twice with phosphate-buffered saline (PBS). The sections were immersed in hot citrate buffer for antigen retrieval and rinsed in PBS. Histology slides were similarly immersed in hot citrate buffer and then stained with hematoxylin and eosin. Permeabilization for myopalladin and Sema3a was performed with 0.5% Tween added to the blocking solution. For all immunofluorescence studies, slides were blocked with 10% bovine serum albumin (BSA), 5% goat serum and Mouse on Mouse blocking serum (Vector Laboratories, MKB-2213-1) in PBS-Tween for 1 h at room temperature, followed by incubation overnight at 4°C in antibody dilutions: mouse anti-β3T (1:100, R&D Systems MAB1195), rabbit anti-Sema3a (1:2000, Abcam ab199475), rabbit anti-Myopalladin (1:100, Bioss Bs-10366), or rat anti-CD31 Alexafluor 647 direct conjugate (1:250, Biolegend 102516). Sections were rinsed and incubated with goat anti-mouse, goat anti-rabbit, IgG (1:500, Invitrogen) or DAPI (1:1000, Thermo Scientific) overnight at 4°C. Ten-micron thick z-stacks were obtained by confocal microscopy and presented in maximum projection mode, quantified for total pixels and normalized to the DAPI pixels using FIJI software (Schindelin et al., 2012). Unpaired t-tests with significance levels set to p < 0.05 were used to evaluate the significance of differences in β3T levels among four biological replicates of each genotype for P7 and P24 first molars. These tests were also used to assess the differences in CD31 among three biological replicates at multiple regions in P7 first molars.
The Comparative Pathology and Digital Imaging Shared Resource of The Ohio State University Comprehensive Cancer Center performed immunohistochemistry (IHC) on heads from P24-P27 WT and Tgfbr2cko mice. Whole heads were fixed for a minimum of 48 h in 10% neutral buffered formalin. Following decalcification using Formical-2000 (StatLab, McKinney, TX, United States), the heads were embedded in paraffin and sectioned horizontally at 4 μm thickness. Slides were dried overnight, then deparaffinized and immersed in hot citrate buffer (Dako, S1699) for antigen retrieval. A Lab Vision 360 Autostainer (Thermo Scientific, Waltham, MA, United States) was used for IHC to apply rabbit-anti-CD31 (1:900, Abcam ab281583) diluted in Dako antibody diluent (S3022) followed by donkey-anti-rabbit IgG (1:500, Jackson Immunoresearch 711-065-152), VECTASTAIN Elite ABC-HRP Reagent Peroxidase (Vector Laboratories, PK-7100), and DAB (Dako, K3468) using a standard protocol. Nuclei were stained with hematoxylin and slides were mounted for imaging. A Lionheart LX microscope (BioTek, Winooski, VT, United States) was used for imaging at ×10 magnification. IHC Images were deconvoluted and thresholded according to a previous report (Crowe and Yue, 2019). Thresholded images were quantified for total pixels, normalized to DAPI using FIJI software and analyzed with an unpaired t-test. Significance was set to p < 0.05 for 5 WT and 6 Tgfbr2cko biological replicates.
RNA Isolation
Dental pulp was extracted from the mineralized tissue and enamel organ of M1s from P7 Osterix-Cre and Tgfbr2cko mice. The left and right M1s were compiled in lysis buffer (Qiagen, 79216) and dispersed with a motorized, sterile pestle, then RNA was extracted with a column isolation kit (Zymo Research, R2070) and analyzed in subsequent qPCR and RNA-Seq experiments.
mRNA-Sequence Analysis
The RNA Integrity Number (RIN) and quantification for total RNA were determined using a BioAnalyzer RNA 6000 Pico Kit (Agilent, 5067-1513) and Qubit RNA HS Assay kit (Invitrogen, Q32852), respectively. Samples with a RIN > 7 were used for global preamplification for the 1-ng low-input RNA-seq using the SMAR-seq HT kit (Clonetech, 634437). The resultant cDNA samples were used for library generation using the Nextera XT DNA Library Prep Kit (Illumina, 15031942) and sequenced on an Illumina NovaSeq SP flowcell for 12–15 million PF clusters per sample. This protocol was described in (Lucas et al., 2019; Walker et al., 2020). Data processing and analysis were performed using a previously published pipeline (Gadepalli et al., 2019). Briefly, raw reads were aligned to a mouse reference genome (GRCh38) using HISAT2 (Liao et al., 2014; Kim et al., 2015). Gene expression values were quantified using featureCounts with GENCODE M14 (mouse) as the transcript reference (GENCODE annotation). The data were processed using the CLEAR workflow, which identifies reliably quantifiable transcripts in low-input RNA-seq experiments (Walker et al., 2020). Processed counts were transformed and normalized by voom, and a differential gene expression analysis was performed using R package “limma” (Ritchie et al., 2015). The original contributions presented in the study are publicly available. This data can be found here: NCBI, GSE190582. A gene ontology analysis was subsequently performed using Panther GO biological process complete statistical overrepresentation testing (www.pantherdb.org), employing Fisher’s exact test with false discovery rates for genes with ≥ ± 2-fold difference and p < 0.05.
Quantitative Real-Time PCR
The qPCR analysis was performed according to the TaqMan® PreAmp Master Mix Kit Protocol (4384557B). In brief, 20 ng of total RNA was retrotranscribed using a High Capacity cDNA Reverse Transcription kit (Life Technologies, 4374967). The samples were enriched for a pool of selected targets of interest (including the housekeeping genes), reported in Supplementary Table S1, by mixing 2.5 μl of cDNA with a pool of the Gene Expression TaqMan assays, which were mixed and diluted to ×0.2 using ×1 TE Buffer and the TaqMan PreAmp Master Mix (×2, 4391128). A total of 14 preamplification cycles were applied, then the product was diluted 1:20 with ×1 TE Buffer. A 2 μl aliquot of amplified cDNA was used to perform the qPCR experiment, with samples run in triplicate, including no-template controls. All samples were run and analyzed using the QuantStudio 12K Flex Real-Time PCR System (ThermoFisher). The Ct Average of each triplicate sample was used to perform the relative quantification analysis. The samples were normalized using the housekeeping gene, beta 2 microglobulin (B2MG) and the relative expression was calculated using the comparative Ct method. qPCR was performed on 10 Osterix-Cre M1 pairs and 10 Tgfbr2cko M1 pairs. Primer information is available in Supplementary Table S1.
Co-Culture of TGNs and Primary DPCs
Co-culture assays were performed as previously described in (Barkley et al., 2020). Briefly, M1s from P5-8 Tgfbr2fl/fl mice were dispersed, plated and grown to confluence in alpha-minimum essential medium (Gibco, 12571063) supplemented with 10% heat-inactivated fetal bovine serum, L-glutamine (Gibco, 25030-081), and antibiotics. At confluence, the media were replaced with media containing Adenovirus-Cre-GFP (Ad-Cre) or Adenovirus-eGFP (Ad-GFP) in the presence of 10 mg/ml Polybrene (Millipore, TR-1003-G). After 48 h of infection, the media containing virus were removed and replaced with fresh media. NuncBlue (Invitrogen, 2272577) was added to label nuclei per the manufacturer’s instructions. Infection was confirmed by GFP fluorescence. The infection percentages were determined using NIS-Elements Advanced Research software in which an overlay of DAPI and GFP images was used to calculate the numbers of total and infected cells.
Cells from trigeminal nerve bundles were collected from 6-week-old B6. Cg-Tg (Thy1-YFP) 16Jrs/J male and female mice and dispersed with type II collagenase (Fisher, 17101015) followed by type II trypsin (Sigma, T-7409). Cells were counted using a standard hemocytometer. Transwell inserts (Greiner Thincerts, 3 μm porosity, 19380216) were coated with 10 μg/ml laminin (Sigma, L2020) and seeded with 50,000 cells/250 μl. Media were replaced with media containing 1 mM uridine (Sigma, BCCC 2099) and 15 mM 5′-fluor-2′deoxyuridine (Sigma, SLCB5571) for mitotic inhibition of mesenchymal cells within the mixture after 24 h. Co-culture continued for an additional 4 days.
Tgfbr2 knockdown in the co-culture model was confirmed using our previously-developed semi-qPCR assay (Seo and Serra, 2007; Peters et al., 2017) (forward 5′-TTA ACA GTG ATG TCA TGG CCA GCG-3′ and reverse 5′- AGA CTT CAT GCG GCT TCT CAC AGA-3′) and B2MG (forward 5′-GCT ATC CAG AAA ACC CCT CAA-3′ and reverse 5′- CAT GTC TCG ATC CCA GTA GAC-3′), which was performed for four separate experiments.
Primary DP Cell Staining, Imaging and Quantification
To determine the morphological responses to Tgfbr2 knockdown in the primary DPCs with Ad-Cre infection, cells were grown atop Poly-D-lysine-coated coverslips during the co-culture assay. For immunofluorescence, cells were fixed with 4% PFA followed by permeabilization with 0.5% Tween added to a blocking solution of 10% BSA, 5% goat serum and Mouse-on-Mouse blocking serum in PBS. Primary incubation with mouse anti-DSPP (1:100, Santa Cruz, sc-73632) proceeded overnight at 4°C. The following day, the coverslips were rinsed and incubated with goat anti-mouse secondary antibody (1:500, Invitrogen) and DAPI (1:1000, Thermo Scientific) overnight at 4°C. Fluorescent images were captured and quantified with FIJI software. DSPP was normalized to the cell number. A two-tailed, unpaired t-test was applied to four separate experiments with values p < 0.05 considered to be significant.
Assessment of Neurite Outgrowth: Staining, Imaging and Quantification
Transwell filters containing TG outgrowths were rinsed with ×1 PBS, fixed with 4% PFA, and blocked for 1 h at room temperature. Filters were then incubated at 4°C overnight with a 1:200 dilution of chicken anti-GFP primary antibody (1:200, Aves Lab, GFP-1010) to selectively stain the YFP+ neurons (due to the high conservation between GFP and YFP). Filters were then rinsed and incubated in anti-chicken IgY-H-L secondary antibody conjugated to Alexa-546 (1:500, Invitrogen, A32931). The high YFP levels in the late postnatal TG bundles resulted in high specificity for the antibody above the background fluorescence from mesenchymal cells and filter pores.
Subsequently, 100 μm-thick z-stacks were collected and stitched together in NIS-Elements. The resultant image was presented in maximum projection mode to demonstrate total neurite outgrowth throughout the entire Transwell filter. Images were auto-thresholded with the Intermodes auto-threshold function in the FIJI software, and the total numbers of pixels were quantified. A 2-way ANOVA was applied to four separate experiments with significance set to p < 0.05, and then a Tukey’s multiple comparison post-hoc test was performed.
Proteomics Analysis on Co-Culture Media
The global proteomic analysis was carried out as previously reported in (Ludwig et al., 2016). In brief, cell culture media was collected at the end of the 5 days assay. A 100 μl aliquot was passed through a bone serum albumin depletion column (Genway, GWB-BSAIGY) and 40 μg of post-depleted protein was subsequently used for each GelC run. Protein samples were reduced with DTT and denatured at 70°C for 10 min prior to loading onto 10% Bis-Tris Protein gels and being fully separated. The gels were stained overnight with colloidal Coomassie for visualization purposes, then each lane was cut into 4-MW fractions and equilibrated in 100 mM ammonium bicarbonate. Each gel plug was digested overnight with mass spectrometry-grade Trypsin Gold (Promega, V5280) following the manufacturer’s instructions. Peptide extracts were reconstituted in 0.1% Formic Acid/ddH2O at 0.1 μg/μl. Mass spectrometry was carried out, and the data were processed, searched, filtered, grouped, and quantified as previously reported (Ludwig et al., 2016). Unpaired t-tests were performed comparing the co-cultures, with significance set to p < 0.05. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [1] partner repository with the dataset identifier PXD031046 and 10.6019/PXD031046.
Results
Tgfbr2 Deletion is Isolated to the DP Mesenchyme
Tgfbr2cko mice demonstrated alterations in development of the M1s by P5 (Wang et al., 2013). To investigate the mechanisms responsible for the phenotypes observed in Tgfbr2cko teeth, we performed H&E staining on sections from P5, P7, P10 and P14 control and mutant M1s. These time points were chosen because tooth root elongation and dentin mineralization were both actively occurring on these days. Axon-like structures were reduced in mutant teeth relative to controls at all stages (Supplementary Figure S1).
We subsequently assessed whether the hypothesized reduction of innervation in the Tgfbr2cko mice was due to unexpected Cre activity in neurons by crossing Osterix-Cre and ROSA26-mTmG reporter mice. The ROSA26-mT/mG reporter line expresses a membrane-bound GFP (mG) tag in cells that express Cre at any point, or a Tomato red fluorescent (mT) tag in cells that never express Cre (Muzumdar et al., 2007). Osterix:GFP:Cre is also expressed in the nucleus (Rodda and McMahon, 2006). We collected P7 Osterix-Cre;ROSA26mTmG DP and performed immunofluorescence staining for β3T on 10 μm cryosections with a Cy5 secondary antibody, followed by confocal microscopy for mG, mT, and Cy5 to detect neuronal afferents. We found scattered nuclear GFP in the DP and membranous GFP (mG) in the odontoblast (OD) layers (Figures 1B,D), with mT isolated to DP cells without nuclear GFP (Figures 1A,D). These results agree with previous reports of Osterix-Cre being isolated to the odontoblast and DPC populations within the developing tooth (Rakian et al., 2013; Wang et al., 2013; Ono et al., 2016). There was no co-labelling of mT and mG, as expected (Muzumdar et al., 2007). The β3T labelling of microtubules in the neuronal afferents did not correlate to any GFP+ structures and localized in some regions with the membranous mT. This confirmed that Osterix-Cre was isolated to the DP mesenchymal lineage and that there was no off-target deletion of Tgfbr2 in the axonal afferents of Tgfbr2cko M1s.
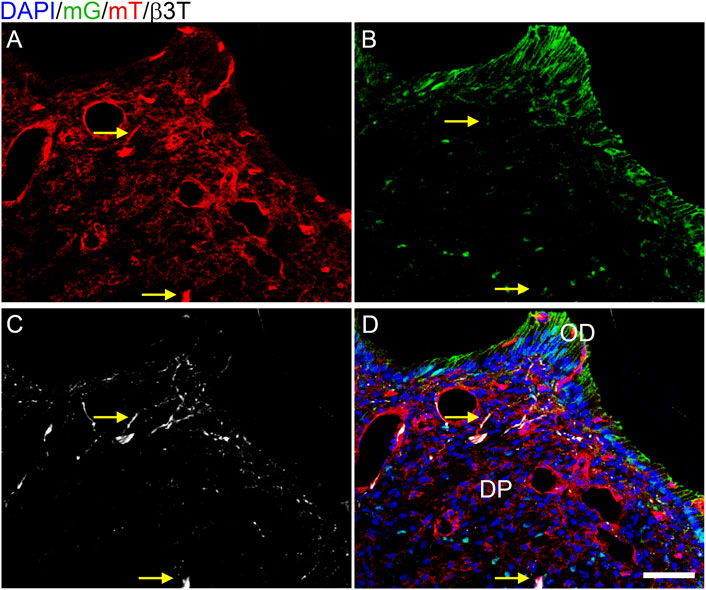
FIGURE 1. Cre activity is isolated to the mesenchyme in P7 Osterix-Cre;ROSA26mTmG mouse dental pulp. Z-stacks (10 μm thick) were collected and presented in maximum projection mode to examine the Cre expression in DP structures. (A) membranous tomato red (mT) indicating an absence of Cre activity; (B) membranous green (mG) and nuclear Osterix-GFP indicating Cre activity; (C) β3T immunofluorescence labelling axonal afferents in white. The β3T correlated with only mT (A–D) and never mG (B–D), demonstrating that Cre activity is isolated to the DP mesenchyme and odontoblasts (OD). Arrows highlight two particular regions of β3T and mT without mG, with a dense β3T+ structure indicated in the bottom center and a single-fiber β3T+ structure in the top center (C). N = 8 Osterix-Cre;ROSA26mTmG mice. The scale bar is 50 μm.
Afferent Innervation is Reduced in Tgfbr2cko Molars
To directly assess innervation, sections from P7 and P24 control and mutant molars were stained with pan-neuronal β3T, and the levels of fluorescence were compared. Nerve fascicles branched throughout both the control and mutant DP, with individual axons entering the predentin and dentin regions of the tooth organ (Figure 2). Maximum projections from confocal microscopy were used to evaluate and quantify fluorescence as a measure of tooth innervation. The total axon coverage was quantified as the pixel density of equivalent regions of the image normalized to DAPI. We found initial nerve penetration in both control and Tgfbr2cko second molars (M2s), with no significant difference in total innervation (Figure 2O). To investigate the difference between nerve penetration and neurite outgrowth, we compared the levels of β3T fluorescence in the densely innervated coronal regions of P7 and P24 control and mutant first molars. Nerve fascicles branched throughout both the control and mutant DP, with individual axons entering the predentin and dentin regions of the tooth organ (Figures 2D–G,K–N). We found a significant reduction in nerve fiber coverage in molars from P7 and P24 Tgfbr2cko mice compared to controls (Figure 2O), supporting the hypothesis that Tgfbr2 in the DP mesenchyme mediates tooth innervation. Similar nerve penetration was seen in the M2s, and that observation in tandem with the persistent low amount of innervation in Tgfbr2cko mice at P7 and P24, suggest that this phenomenon was not due to a developmental delay, but that the cellular mechanisms guiding neurite outgrowth in developing teeth were disrupted by the deletion of Tgfbr2. Since Tgfbr2cko mice die around 4 weeks of age (Wang et al., 2013; Peters et al., 2017), we were unable to investigate beyond this time point.
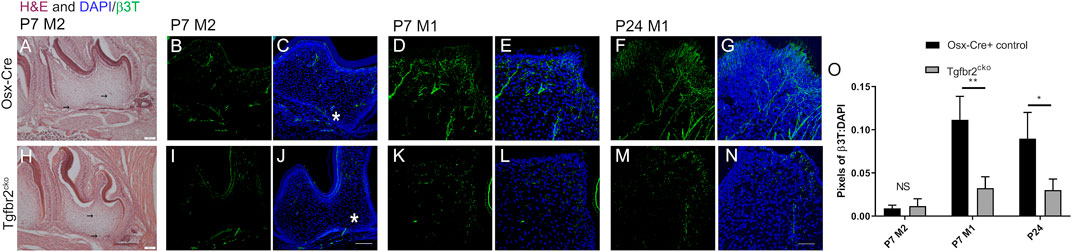
FIGURE 2. DP innervation is reduced in developing Tgfbr2cko molars. Innervation did not appear significantly disrupted during early postnatal tooth development. Representative images of H&E-stained mandibular M2s at P7 (A–H) with two black arrows in each image indicating hypothesized neuronal structures. Scale bar = 100 μm. N = 3 for control Osterix-Cre and 4 Tgfbr2cko samples. (B–G, I–N) Z-stacks (10 μm thick) of β3T-positive structures were collected and presented in maximum projection mode to demonstrate total afferent innervation. (B–C, I–J) Confocal images of control (B–C) and Tgfbr2cko (I–J) M2 DP demonstrated afferent structures throughout the entire tooth. The asterisk shows similar areas of nerve penetration in the apical region of the tooth. The scale bar is 100 μm. Coronal neurite outgrowth appeared to be more extensive in control mice, but the overall afferent structures were not significantly different between control and Tgfbr2cko mice as indicated by unpaired t-tests (O). (D–G, K–N) Confocal images of control (D–G) and Tgfbr2cko (K–N) M1 DP demonstrated that there were more β3T+ neuronal structures in P7 and P24 control mice than mutant mice. The scale bar is 50 μm. (O) The ratios of total pixels for neuronal structures normalized to the pixels for DAPI indicate that there was significantly more innervation per region in control mice than mutant mice at P7 and P24. *, p < 0.05, **p < 0.01 by unpaired t-tests, n = 4 for Osterix-Cre and 4 for Tgfbr2cko samples at each time point.
Vascularization is Not Altered in Tgfbr2cko Molars
Blood vessels and nerve fibers often run along parallel tracks with similar branching patterns in developing tissues [reviewed in (Carmeliet, 2003; Serini and Bussolino, 2004; Adams and Eichmann, 2010)]. We found that the β3T+ afferents in some regions resembled vasculature in the Osterix-Cre;ROSA26-mTmG mouse molars, making it difficult to determine whether Tgfbr2 deletion might also be reducing tooth vascularization. To address this possibility, we assessed vascularization in P7 and P24-27 control and mutant molars using CD31/PECAM immunofluorescence and IHC labelling, respectively. Because the tooth organ is much larger at P24, we sectioned the tissue on the horizontal plane and captured images at a lower objective to obtain a more comprehensive view of the vascularization. Figure 3 shows that there were no significant alterations in vascularization at either time point. These results confirm that tooth vascularization and innervation occur via different mechanisms during development (Shadad et al., 2019) and that this distinction remains intact within the Tgfbr2cko mice.
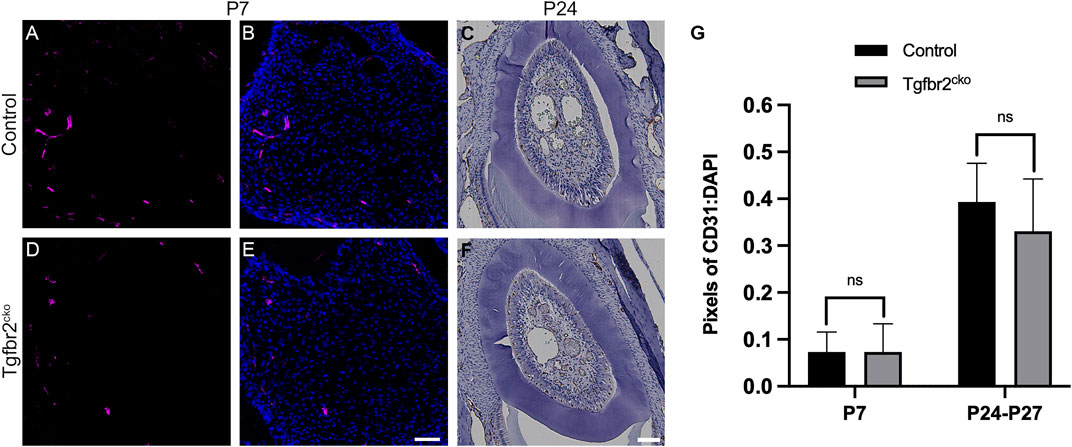
FIGURE 3. Vascularization is not significantly different between Tgfbr2cko and control Osterix-Cre P7 and P24-27 first molars. Representative images of vascular structures labelled by CD31 in P7 and P24 control and Tgfbr2cko M1s. (A,B,D,E) Representative confocal images of vasculature structures amidst DP cells (nuclei labelled by DAPI) in sagittal sections of P7 Osterix-Cre control (A,B) and Tgfbr2cko (D,E) first molars. (C–F) Representative brightfield images of CD31 IHC in horizontal sections of P24 control (C) and Tgfbr2cko (F) first molars. For quantification, confocal images (A,B,D,E) were thresholded using FIJI software to remove background fluorescence and the remaining pixels were quantified and normalized to DAPI. For P24-P27 IHC images (C–F), color deconvolution using FIJI preceded thresholding and quantification. For both time points, an unpaired t-test showed no significant difference between genotypes (G). For P7, n = 3 for each genotype; for P24-P27, n = 5 for control and 6 for Tgfbr2cko. The scale bars are 50 µm.
Downregulation of Osteogenic Genes in Tgfbr2cko Molars Confirms Hypomineralization Phenotype
To investigate the mechanisms responsible for the phenotypes observed in Tgfbr2cko teeth, a global analysis of gene expression was performed using RNA-Seq. We chose to perform this analysis at P7 because this was the earliest time point at which mineralization and innervation differences were apparent. A principal components analysis (PCA) was performed to cluster the samples associated with mouse genotypes to confirm the differences between the Osterix-Cre and Tgfbr2cko samples (Figure 4). Select genes from the RNA-Seq analysis were confirmed by qPCR (Supplementary Table S2). The expression of a specific transcript was considered to be altered in Tgfbr2cko tissue if the expression differed by at least ±1.5-fold relative to control with p < 0.05. These studies showed that several genes associated with osteo/odontogenesis and mineralization were dysregulated by Tgfrbr2 knockout. Our data confirmed the downregulation of select genes known to be associated with odontogenesis and mineralization: Bglap, Bmp1, Col1a1, Nes, and Wnt10a (Gorter de Vries et al., 1987; About et al., 2000; Yamashiro et al., 2007; Quispe-Salcedo et al., 2012; Bae et al., 2015; Andersson et al., 2017; Zhang et al., 2017; Foster et al., 2018; Niwa et al., 2018; Rosowski et al., 2019). The expression of DSPP trended toward downregulation as well, with p = 0.05. We also confirmed a lack of dysregulation of Dmp1 and NFIC (Supplementary Figure S2), as previously reported (Wang et al., 2013). Clusterin (Clu), a gene reportedly regulated by TGFβ signaling during postnatal tooth development (Khan et al., 2013), was upregulated in the mutant mice. There was also upregulation of Fibulin-7 (Fbln7), a gene involved in cellular adhesion during predentin and dentin formation (de Vega et al., 2007; Arany et al., 2009), in the Tgfbr2cko dental pulp compared to control teeth. Lastly, Tenascin N (Tnn) was upregulated, indicating a lack of differentiation of the dental pulp into mineralizing odontoblasts (Thesleff et al., 1987; Sahlberg et al., 2001; Imhof et al., 2021). Together, these data indicate that Tgfbr2 deletion led to developmental disruption resulting in tooth hypomineralization, confirming the previous reports (Wang et al., 2013; Peters et al., 2017; Corps et al., 2021).
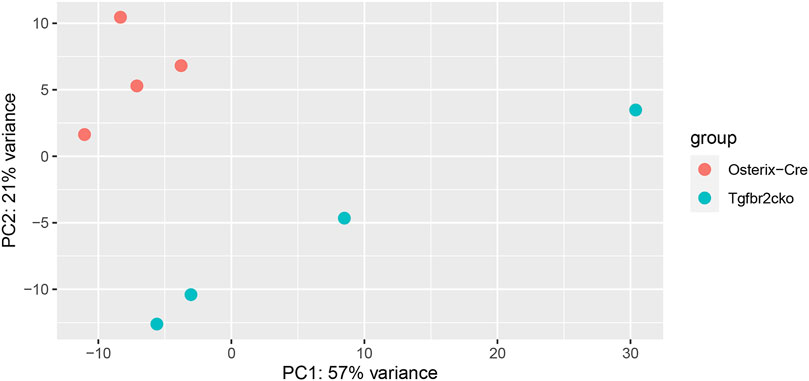
FIGURE 4. Results of principal components analysis (PCA) of RNA-Seq data from Osterix-Cre and Tgfbr2cko DP at P7 for all samples used in this study.
Neuronal Genetics are Disrupted in Tgfbr2cko Molars
A gene ontology analysis was performed using the lists of regulated transcripts (≥1.5-fold difference, p < 0.05) generated above. The analysis was performed using the Protein Analysis Through Evolutionary Relationship (Panther) biological processes tool (www.pantherdb.org) (Mi et al., 2010). This tool determines which genes are over-represented in a specific dataset relative to the whole transcriptome. Several neuronal transcriptomes were over-represented. The neuronal bodies that innervate teeth are located in the trigeminal nerve bundle outside of the teeth, so only axonal projections could contribute to the neuronal RNA in the DP. We therefore concentrated on the biological categories related to axon projection and guidance, and the findings are presented in Figure 5A. There were several over-represented neuronal transcripts identified by Panther, with select transcripts shown in the volcano plot of genes differentially expressed between Osterix-Cre and Tgfbr2cko samples (Figure 5B). The entire list of over-represented transcripts from these categories is presented in Supplementary Table S3.
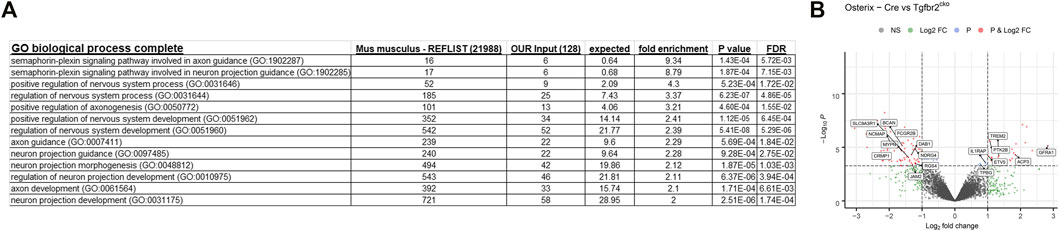
FIGURE 5. Panther gene ontology analyses identify several transcriptomic categories related to axon guidance that differ between the Osterix-Cre and Tgfbr2cko mice. (A) Several GO biological complete categories were identified as being over-represented based on the transcriptomes that were dysregulated by ≥1.5-fold with p < 0.05 in Tgfbr2cko mice, as identified by RNA-Seq. The Panther gene ontology analysis was performed using the Panther GO biological process complete for overrepresented genes with a Fisher’s exact test. (B) A Volcano plot of the RNA-Seq data demonstrating significant differences between Osterix-Cre and Tgfbr2cko samples and select overrepresented neuronal genes reported by Panther in the categories shown in (A).
We did not observe dysregulation of well-known neurotrophins, brain-derived neurotrophic factor (Bdnf), glial-cell derived neurotrophic factor (Gdnf), nerve growth factor (Ngf) or neurotrophin 3 (NT-3) in our RNA-Seq analysis (Supplementary Figure S2), despite several reports that these neurotrophins are released by DP cells and dentin to promote neurite outgrowth (de Almeida et al., 2014; Martens et al., 2014; Austah et al., 2019; Widbiller et al., 2019; Pagella et al., 2020; Sultan et al., 2020). Because this was surprising, we performed subsequent qPCR experiments to verify these findings. These experiments confirmed that there were no significant alterations of Bdnf, Gdnf, or Ngf in the Tgfbr2cko samples but confirmed downregulation of Mypn with qPCR (Supplementary Table S2). Myopalladin is structurally similar to palladin (Otey et al., 2005), a gene highly associated with axonal growth and maintenance (Boukhelifa et al., 2001; Hwang et al., 2001). Our RNA-Seq analysis also indicated that there was downregulation of several other neuronal transcripts. Synaptotagmin 5 (Syn5) and 15 (Syn15), which belong to the family of neurotransmitter release genes in axons (Norlin et al., 1999; Dean et al., 2012; Südhof, 2013), were both downregulated in the Tgfbr2cko mice. There was also downregulation of Fxyd2, Fxd6, TMEM178A, TMEM47, TMEM88, TMEM94, TMEM97, and TrpM3, all genes that were previously reported to be specifically expressed in the axonal afferents of mouse molars (Emrick et al., 2020). We confirmed our RNA-Seq could detect alterations in axonal-specific RNA by performing immunofluorescence and confocal imaging for MYPN. We found very specific labelling of MYPN in the axonal structures (Supplementary Figure S3). Thus, there was a reduction in the expression of numerous genes associated with axonal transcriptomes in mutant mouse molars, confirming the phenotypic reduced afferent innervation in the Tgfbr2cko mouse molars.
Neurite Outgrowth in DP-TGN Co-Cultures Requires Tgfbr2
We utilized co-cultures to directly assess the role of Tgfbr2 in neurite outgrowth in response to DP secretions (Figure 6). Transwell filters were seeded with TG cells, and DPCs were grown in the bottom wells. TGN from Thy1-YFP mice were used so that neurite outgrowth through the filter could be easily imaged. It was previously reported that Thy1-YFP levels become detectable around P6-P10 in neurons and increases exponentially throughout the nervous system during postnatal and adult life (Caroni, 1997; Alić et al., 2016). We observed the brightest staining of Thy1-YFP axons (using the cross-reactive GFP antibody) at 6 weeks of age, so TGN from 6-weeks old mice were used in our co-culture experiments. We performed confocal microscopy and image stitching of the axonal extensions throughout the entire filter and quantified auto-thresholded images. When the TGN were grown on filters in the absence of DPCs, very little neurite outgrowth occurred in comparison to when the TGN were grown above control DPCs (Figures 6A,C,G), confirming that PCs secrete paracrine factors that promote neurite growth (de Almeida et al., 2014; Pagella et al., 2020; Sultan et al., 2020). To examine whether Tgfbr2 from the DPCs regulates neurite outgrowth, Tgfbr2 was deleted using Ad-Cre-GFP. Cells infected with Ad-GFP were used as controls. Equivalent numbers of infected cells were used (as identified by GFP fluorescence; Figures 6D,E), and deletion of Tgfbr2 was confirmed by semi-qPCR (Figure 6F). Deletion of Tgfbr2 in the DPCs significantly decreased the neurite extension to a level similar to cultures without any DPCs (Figures 6A–C,G). We used DSPP as a marker to confirm that there was reduced odontoblast differentiation and mineralization (Chen et al., 2009; Dissanayaka et al., 2012; Hou et al., 2012) in cultures with Tgbr2 deletion in the DPCs due to Ad-Cre infection (Figures 6H,I). We collected media from the co-culture wells, TG cells cultured alone, and DP cells cultured alone (DPA) with and without Tgfbr2 deletion for proteomics analysis. Similar to our RNA-Seq data, we did not find significant alterations in NGF, BDNF, GDNF, or NT-3 (data not shown), indicating Tgfbr2 did not regulate secretion of these neurotrophins. The results of the co-culture experiments suggest that Tgfbr2 in the DP mesenchyme is required to promote both odontoblastic differentiation and subsequent neurite outgrowth.
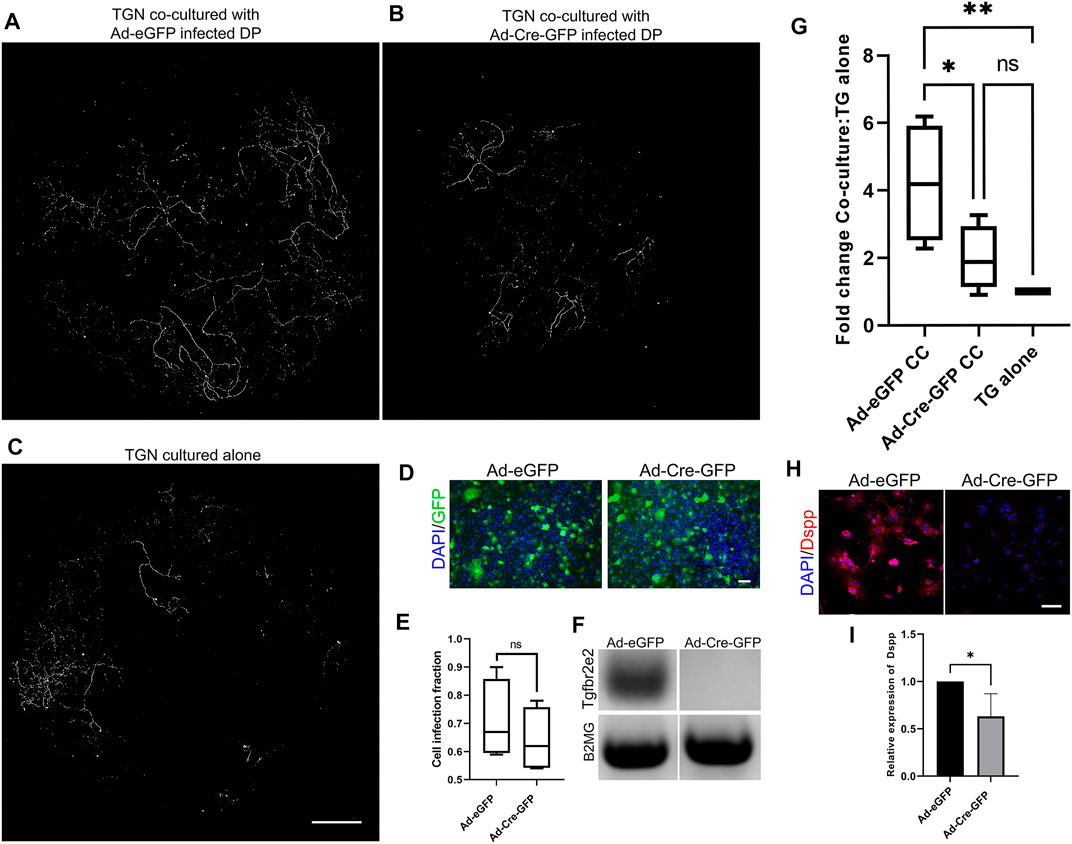
FIGURE 6. Tgfbr2 signaling in primary DP mediates trigeminal neurite outgrowth in co-culture. (A–C) Thy1-YFP trigeminal neurons were cultured in Transwell filters with 3 μm pores atop primary Tgfbr2f/f DPCs. Immunofluorescence was performed for the YFP protein using an anti-GFP antibody for highly specific staining of neuronal structures over the entire filter. Maximum projections of 100 μm z-stack confocal microscopy images at 10× were collected and stitched with stitching software. TG neurons demonstrated significantly more outgrowth when co-cultured with DPCs (A) than when cultured alone (C). Neurite outgrowth was not induced when neurons were co-cultured with DP infected with Ad-Cre-GFP to knock down Tgfbr2 (B). The scale bar is 1,000 μm for (A–C). (D) Adenovirus infection was confirmed with GFP expression and normalized to DAPI to confirm equivalent numbers of infected cells between control Ad-GFP and Ad-Cre-GFP DPCs (D,E) The scale bar is 100 μm for (D). Semi-quantitative PCR confirmed Tgfbr2 KD (F). Images of axonal outgrowth on the filters were auto-thresholded with FIJI software to remove background fluorescence and the remaining pixels were quantified and normalized to the TGN cultured alone from each experimental set (G). *p <0.05, **p < 0.01 by 2-way ANOVA tests with a Tukey’s multiple comparison post-hoc set. (H) Representative images of DPCs from co-culture demonstrating reduced expression of an odontoblast marker, DSPP. (I) Quantification of DSPP normalized to DAPI demonstrates a significant reduction in cells with Tgfbr2 deletion. *p < 0.05 by two-tailed unpaired t-test. All analyses are based on four separate experiments.
In Vivo and In Vitro Models Suggest Odontoblast Differentiation Coordinates Axonal Guidance Cues
We next searched the RNA-Seq data from the P7 M1s and proteomics data obtained for co-culture media to identify neurotrophic factors that were downregulated by Tgfbr2 deletion. We found that both the RNA-Seq and proteomics analyses identified several semaphorins, which are well-known axonal guidance cues (Kitsukawa et al., 1997; Lillesaar and Fried, 2004; Ding et al., 2007; Fried et al., 2007; Kolk et al., 2009; Sijaona et al., 2012; Moe et al., 2012; Sabag et al., 2014; Shrestha et al., 2014; Yoshida et al., 2016; Song et al., 2017; Yu et al., 2018; Bott et al., 2019; Coutiño and Mayor, 2021), were dysregulated with Tgfbr2 deletion (Figure 7). Our RNA-Seq identified Sema3f, 4a, 4b, and 7a were altered in the Tgfbr2cko molars. The RNA-Seq also identified dysregulation of semaphorin receptors, Plexin A2 (PlxnA2), Plexin A4 (PlxnA4), Plexin D1 (PlxnD1) and Neuropilin 1 (Nrp1) (Sijaona et al., 2012; Sabag et al., 2014; Yoshida et al., 2016; Song et al., 2017; Limoni, 2021) when Tgfbr2 was deleted (Figure 7A). Sema3a was expressed at levels too low for RNA-Seq statistics. We observed reduced Semaphorin 5B (SEMA5B) secretion by the DPCs into the media with Tgfbr2 deletion when they were cultured alone, suggesting that it is directly regulated by Tgfbr2 signals in the DPCs (Figure 7B). Interestingly, we did not find any Semaphorin 3a (SEMA3A) in the media of DPCs cultured alone whether or not Tgfbr2 was deleted (Figure 7C). These data suggest that the complex interplay between neuronal and mesenchymal cells could be regulated through coordinated semaphorin signaling.
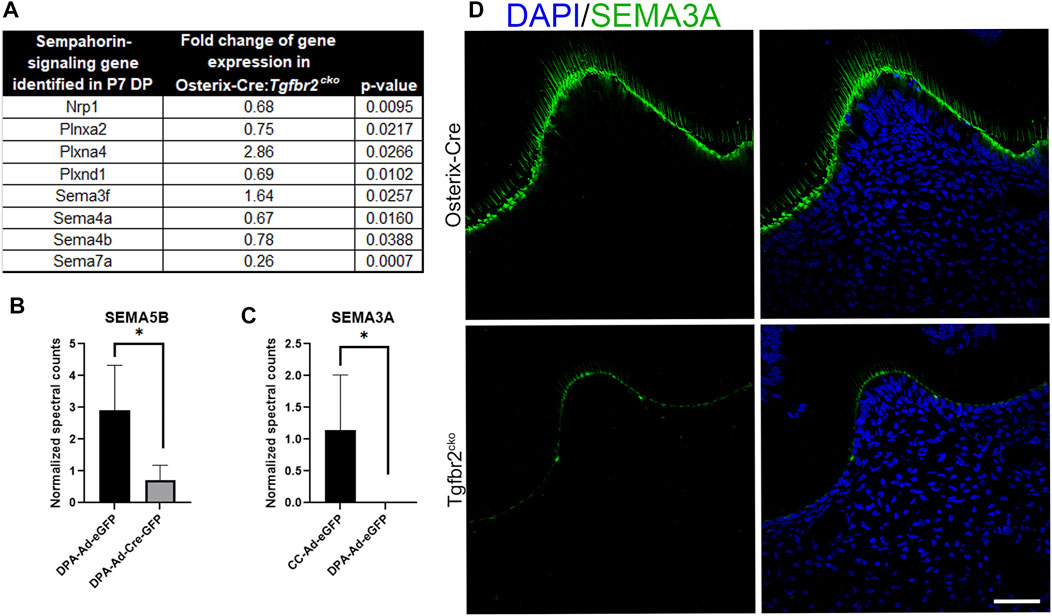
FIGURE 7. Tgfbr2 deletion in DP cells alters semaphorin signaling. (A) RNA Seq analysis of P7 M1 tissue indicates altered semaphorin signaling components. N = 5 for Osterix-Cre and Tgfbr2cko samples. (B,C) Proteomic analysis of co-culture (CC) and dental pulp alone (DPA) media. (B) Proteomics indicates decreased Semaphorin 5B present in the media with Tgfbr2 deletion in the DPCs. (C) Proteomics indicates Semaphorin 3A is only present in CC media and not when dental pulp cells were cultured alone. N = 4 media collected and analyzed from experiments including: co-culture samples with and without Tgfbr2 deletion, DPA samples with and without Tgfbr2 deletion, and TG cells cultured alone. *p < 0.05 by unpaired t-test. (D) Immunofluorescence of SEMA3A (green) demonstrates reduced chemoattraction signals in the Tgfbr2cko odontoblasts compared to controls. N = 4 for Osterix-Cre and 4 Tgfbr2cko samples. The scale bar is 50 μm.
We next questioned whether the reduced differentiation of the developing teeth and DPCs with Tgfbr2 knockout could be disrupting the necessary signals to support neurite outgrowth. Semaphorin signaling is known to regulate non-neuronal functions, including odonto-/osteogenic differentiation (Sijaona et al., 2012; Song et al., 2017; Yu et al., 2018), so we performed immunoflouresence for SEMA3A on P7 Osterix-Cre and Tgfbr2cko M1s to determine which cell population(s) demonstrated altered semaphorin signaling. We found reduced SEMA3A protein levels in the odontoblast layer of the Tgfbr2cko mice compared to the controls. Neither mouse genotype demonstrated detectable levels SEMA3A in the pulpal cells (Figure 7D), indicating that axonal guidance in the coronal regions of the teeth may be orchestrated by the odontoblast population.
In summary, our data demonstrate that deletion of Tgfbr2 in the DP of mice during TGN sprouting in the tooth changes inherent axonal guidance cues, resulting in reduced innervation in postnatal teeth. Co-culture experiments supported that Tgfbr2 in the DP is required for odontoblast differentiation and DP-mediated neurite outgrowth. We propose that Tgfbr2 regulates the signals necessary to promote the mineralization and innervation of the tooth organ into a hard tissue capable of the daily activities of the oral cavity.
Discussion
Tooth innervation serves unique roles that allow teeth to sense pressure, temperature, and infectious agents. Without these sensations, decay and normal oral activities would cause irreparable damage to the tooth and surrounding tissues. Teeth are primarily innervated by nociceptors extending from the trigeminal ganglia (Fried et al., 2007) that respond to signals from the dental mesenchyme during development (Lillesaar et al., 1999; Fried et al., 2005, 2007; Kettunen, 2005; Kettunen et al., 2007; Moe et al., 2012; Yu et al., 2012; de Almeida et al., 2014; Pagella et al., 2020; Sultan et al., 2020). Interestingly, the DP mesenchyme initially secretes repellant signals during embryogenesis to prevent axon entry into the developing tooth organ, which later shifts to the secretion of attraction factors as the tooth nears eruption (Lillesaar and Fried, 2004; Fried et al., 2007). While most organs are fully functional and innervated by the time of birth, tooth development extends into adult life, with tooth innervation and mineralization occurring in concert during different postnatal stages (Kollar and Lumsden, 1979; Moe et al., 2012). These previous reports clearly demonstrate that neuronal-mesenchymal crosstalk is required for tooth organogenesis and maintenance, but the details of these mechanisms remained largely unclear. We herein present the first in vivo and in vitro data showing neurite responses to the deletion of Tgfbr2 in the DP mesenchyme during postnatal development. Our results delineate a novel role for Tgfbr2 in the DP mesenchyme, where it regulates the development of sensory activity in teeth.
Of interest, neither our RNA-Seq nor proteomics data uncovered any significant alterations in NGF, BDNF, GDNF, or NT-3, which were previously reported to correlate with neurite outgrowth (Lillesaar et al., 2001; Lillesaar and Fried, 2004; Price et al., 2005; Kitisin et al., 2007; Austah et al., 2018, 2019; Widbiller et al., 2019; Pagella et al., 2020). The differences in the model systems used may underlie many of the observed differences. For example, some groups have used cultures of specific cell populations [e.g., stem cell apical papilla (SCAP) cells (de Almeida et al., 2014) or DPCs from rat incisors (Sultan et al., 2020)], while we included in vivo and in vitro studies of the entire pulp tissue from the molar tooth organ, including the odontoblasts. The different findings may reflect the differences in cell-cell interactions, the specific media and additives used for culture, or the differences between the in vitro and in vivo cellular milieu. It is also possible that the impact on the semaphorin pathway may have been overlooked in the previous studies due to their focus on the changes in the above markers. Our assays investigated global expression at both the RNA and protein levels, permitting the identification of novel mediators of neurite outgrowth.
We reported that several genes associated with odontoblast differentiation and mineralization were downregulated in the Tgfbr2cko DP, verifying previous reports of hypomineralized dentition when Tgfbr2 was knocked out (Wang et al., 2013; Corps et al., 2021). We also found upregulation of some genes related to mineralization, such as Fbln7, Clu, and Tnn. Fbln7 is expressed by preodontoblasts and odontoblasts in fibronectin-rich regions and is involved in dentin formation (de Vega et al., 2007; Arany et al., 2009). The increased expression of Fbln7 in the Tgfbr2cko dental pulp may be present to compensate for the decreased fibronectin-related genes, fibronectin domain containing protein 3a (Fndc3a) and fibronectin domain containing protein 3b (Fndc3b) (data not shown), in the Tgfbr2cko mice. Fndc3a and Fndc3b are both also called Factor for adipocyte differentiation 104 (Fad104), which is reportedly regulated by TGFβ signaling (Goto et al., 2017) and contributes to bone formation (Kishimoto et al., 2013; Cao et al., 2016). Additional studies are needed to elucidate how Fbln7 and Fad104 contribute to the cell-extracellular matrix interactions during tooth development. We also found upregulation of Clu, a gene highly upregulated during secretory odontogenesis in early postnatal tooth development (Khan et al., 2013). However, reports that Clu is also known to promote axonal outgrowth led us to postulate that the DP could be secreting excess Clusterin in the Tgfbr2cko mice in an attempt to overcome the lack of innervation (Kang et al., 2005; Wright et al., 2014). We hypothesized that some of the mineralization-related genes with altered expression levels might instead be regulating axonal outgrowth in the tooth. For instance, Wnt proteins are also known to play important roles in the nervous system. In support of this theory, we found downregulation of Wnt10a, which is thought to promote axon growth by activating small G-proteins (Matsukawa et al., 2018). We noted that there was upregulation of Tnn, which is often localized with Tenascin-C (Tnc) (Meloty-Kapella et al., 2006). This is particularly interesting because tenascins are developmentally regulated by TGFβ signaling in the tooth (Thesleff et al., 1987; Sahlberg et al., 2001). A recent report indicating that TNN deficiency in dental tissues resulted in increased axonal branching (Imhof et al., 2021) further suggested that tenascins promote neurite outgrowth (Wehrle and Chiquet, 1990; Chiquet and Wehrle-Haller, 1994; Meiners et al., 1999; Rigato et al., 2002; Michele and Faissner, 2009). In addition to our RNA-Seq data, we found that high levels of Tenascin and Tenascin-C were secreted by DPCs in our proteomics study, indicating that tenascins may play a role as a neurotrophic paracrine signals. Together, the phenotypic, genetic and proteomic data suggest that the differentiation of the DP mesenchyme is developmentally regulated, as are axonal guidance cues, and these developmental processes may utilize the same signals.
The semaphorin proteins are well known axonal guidance cues reported to be active in a variety of tissues (Kitsukawa et al., 1997; Pasterkamp et al., 2003; Ding et al., 2007; Kang et al., 2007; Kolk et al., 2009; Kopp et al., 2010; Coutiño and Mayor, 2021; Limoni, 2021) including dental pulp tissues (Lillesaar and Fried, 2004; Maurin et al., 2005; Moe et al., 2012; Sijaona et al., 2012; Shrestha et al., 2014; Kuchler-Bopp et al., 2018). One research group investigated the expression profiles of all the semaphorins in M1s from E13 through P9. They found that Semaphorin 3a, 3f, and 5b dramatically decreased through the postnatal stages, Semaphorin 4a and 4b remained at high levels in postnatal stages, and Semaphorin 7a dramatically increased from P5 to P9 (Lillesaar and Fried, 2004). This highlights the fact that semaphorins do not act in a linear fashion or in isolation, rather that the cooperation of signals regulates the axonal branching that occurs throughout coronal regions of the developing tooth. We found that several semaphorins and semaphorin receptors were altered in both our RNA-Seq and proteomics data. In particular, we saw reduced Sema4a, Sema4b, and Sema7a in the M1s and increased Sema3f. Sema3a was expressed at levels below the statistical cutoffs for RNA-Seq, but our immunofluorescence studies indicated stable protein levels in odontoblast cells that were clearly reduced in the Tgfbr2cko M1s. Our proteomic analyses indicated that DPCs do not secrete SEMA3A in the absence of TG cells, which highlights the importance of mesenchymal-neuronal populations during development. However, SEMA5B secretion was directly downregulated in isolated DPCs with Tgfbr2 deletion, suggesting that class 5 semaphorins may be more directly regulated by TGFβ signaling. Of particular note is that the Tgfbr2cko DP showed downregulation of Sema7a, which is reportedly regulated by TGFβ signals (Kang et al., 2007) to promote axon outgrowth (Pasterkamp et al., 2003). A previous study reported that the SEMA7A localization in the odontoblast layer correlated with axonal branching into the dentin-pulp complex. Co-cultures with transfected cells overexpressing Sema7a demonstrated increased neurite outgrowth toward the ganglia explant (Maurin et al., 2005). Our present study suggests the decreased Sema7a (and potentially other semaphorins) in our mutant mouse DP mesenchyme may have directly led to decreased axonal outgrowth in the dentin-pulp regions. We also saw an increase of Sema3f in the Tgfbr2cko M1s, which has been reported to act as a chemorepellent along with its receptor, Plexin A4 (Plxna4) (Sijaona et al., 2012). Since we also found increased expression of Plxna4 in our mutant M1s, we propose that this Sema3f-Plxn4a increase could be directly repelling axonal afferents in the Tgfbr2cko mice. Previous reports indicated that there is developmental regulation of class 3 semaphorin receptor, Neuropilin (Nrp1) (He and Tessier-Lavigne, 1997; Kitsukawa et al., 1997; Sijaona et al., 2012). We observed downregulation of Nrp1, which could reflect reduced SEMA3A signaling, particularly because the expression of the other Sema3a receptor, Plexin-A2 (Plxna2) (Sabag et al., 2014) and SEMA3A levels were also reduced in our mutant mice. Our current findings and previous publications suggest that semaphorin signaling plays an important role in neurite outgrowth throughout the coronal regions of the teeth.
Lastly, previous research indicates that semaphorin signaling is tied into odontoblastic differentiation, with SEMA7A and SEMA3A levels elevated in odontoblast regions (Maurin et al., 2005; Quispe-Salcedo et al., 2012). Exogenous SEMA3A has been shown to enhance osteogenesis (Yu et al., 2018) and odontogenesis (Yoshida et al., 2016) in vitro. Similarly, the Class III Semaphorin receptor, NRP1, stimulates odontogenesis in vitro (Song et al., 2017). We found that SEMA3A is reduced specifically in the odontoblast layer of our Tgfbr2cko mice (Figure 8), suggesting this may relate to the downregulated odontoblast differentiation with Tgfbr2 deletion in the dental mesenchyme. We hypothesize that semaphorin signaling may serve as a feedback mechanism for interactions between the DP mesenchyme and neuronal afferents. Future studies could include several co-culture semaphorin-rescue experiments and investigations into the tooth and bone mineralization in the SEMA7A null mouse. Lastly, in fully developed teeth, axons sprout after dentin injury and then signal to the DP to repair the damaged dentin (Taylor et al., 1988). This suggests that there is a neuronal-mesenchymal feedback loop guiding tooth maintenance that exists into adulthood. Future studies could be performed utilizing the inducible (tet-off) Osterix-GFP:Cre system (Wang et al., 2013) to study whether TGFβ-guided signals, including the semaphorins, are necessary and sufficient to maintain tooth vitality.
In summary, we found that Tgfbr2 deletion in the DP mesenchyme reduced the ability of DPCs to differentiate and provide the axonal guidance cues necessary to support neurite outgrowth. We propose that investigations of the neuronal structures in other mouse models with hypomineralized dentition would provide information key to understanding peripheral nerve branching and the developmental process of tooth innervation.
Data Availability Statement
The RNA-Seq data presented in the study are deposited in the NCBI repository, accession number GSE190582. The proteomics data presented in the study are deposited here: http://proteomecentral.proteomexchange.org/. The accession number is PXD031046.
Ethics Statement
The animal study was reviewed and approved by the University of Alabama at Birmingham and the Ohio State University Institutional Animal Care and Use Committees.
Author Contributions
SP and RS contributed to conception and design of the study. SP, MS, RS, and MP contributed to manuscript preparation. SP, MS, CB, AK, CA, and AS performed experiments and analysis. MP, AW, and YZ analyzed RNA-Sequence data analysis and AW performed GEO submission. SP, MS, MP, and AW created visual interpretations of the data. SS provided the mTmG reporter mouse line.
Funding
This work was supported by the 1) National Institutes of Health/NIAMS grant numbers R01 AR062507 and R01 AR053860 to RS, 2) University of Alabama at Birmingham Dental Academic Research Training (DART) grant T90DE022736 (PI MacDougall) to SP from the National Institute of Dental and Craniofacial Research/National Institutes of Health, 3) UAB Global Center for Craniofacial, Oral and Dental Disorders (GC-CODED) Pilot and Feasibility Grant to SP, 4) National Institute of Dental and Craniofacial Research/National Institutes of Health K99/R00 DE024406 grant to SP, and 5) award number UL1TR002733 from the National Center for Advancing Translational Sciences. These studies were supported in part by the OSU CCC and the National Institutes of Health grant P30 CA16058 and National Cancer Institute support provided for the GSR’s technical director, Pearlly Yan, under grant R50 CA211524. This work was supported in part by grant P30 CA16058 from the National Cancer Institute.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the Genomics Shared Resource at The Ohio State University Comprehensive Cancer Center (OSU CCC), Columbus, OH for conducting the qPCR and RNA Sequence analyses. We thank the Comparative Pathology and Digital Imaging Shared Resource of the OSU CCC for optimizing CD31 immunohistochemistry. We would also like to thank James Mobley from the O’Neil & CCC Mass Spectrometry & Proteomics Shared Resource at UAB for the proteomics analysis and uploading of our co-culture model, along with several technical discussions about best practice and data analysis. Lastly, we thank Hope Amm at UAB for her continued feedback and Elizabeth Rayburn from Magic City Medical Communications for editorial work on this manuscript. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center for Advancing Translational Sciences or the National Institutes of Health.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2022.834815/full#supplementary-material
Supplementary Figure S1 | Nerve structures in postnatal teeth appear to be reduced in Tgfbr2cko mice. Representative images of H&E-stained mandibular M1s from postnatal days 5, 7, 10, and 14. The black arrows in each image indicate hypothesized neuronal structures. Several other structures are present but are not indicated to prevent image crowding. N = 2 for P5 and N = 3 for P7, 10 and P14 for each genotype. The scale bar is 100 μm.
Supplementary Figure S2 | A volcano plot of the RNA-Seq data demonstrating that there were no significant differences in select genes between Osterix-Cre and Tgfbr2cko samples. The volcano plot visually demonstrates that the Ngf, Gdnf, Nt-3, Nfic, and Dmp1 transcripts were not altered, as reported in the manuscript. Bdnf was not reported in the RNA-Seq analysis. RNA-Seq was performed on 4 Osterix-Cre and 4 Tgfbr2cko samples.
Supplementary Figure S3 | Representative image of Mypn in Osterix-Cre and Tgfbr2cko P7 M1. Representative immunofluorescence images of myopalladin in P7 M1 Osterix-Cre and Tgfbr2cko mice confirming axonal-specific expression. N = 4 Osterix-Cre and Tgfbr2cko samples.
References
About, I., Laurent-Maquin, D., Lendahl, U., and Mitsiadis, T. A. (2000). Nestin Expression in Embryonic and Adult Human Teeth under Normal and Pathological Conditions. Am. J. Pathol. 157, 287–295. doi:10.1016/S0002-9440(10)64539-7
Adams, R. H., and Eichmann, A. (2010). Axon Guidance Molecules in Vascular Patterning. Cold Spring Harbor Perspect. Biol. 2, a001875. doi:10.1101/CSHPERSPECT.A001875
Alić, I., Kosi, N., Kapuralin, K., Gorup, D., Gajović, S., Pochet, R., et al. (2016). Neural Stem Cells from Mouse Strain Thy1 YFP-16 Are a Valuable Tool to Monitor and Evaluate Neuronal Differentiation and Morphology. Neurosci. Lett. 634, 32–41. doi:10.1016/j.neulet.2016.10.001
Andersson, K., Dahllöf, G., Lindahl, K., Kindmark, A., Grigelioniene, G., Åström, E., et al. (2017). Mutations in COL1A1 and COL1A2 and Dental Aberrations in Children and Adolescents with Osteogenesis Imperfecta - A Retrospective Cohort Study. PLoS One 12, e0176466. doi:10.1371/JOURNAL.PONE.0176466
Arany, S., Koyota, S., and Sugiyama, T. (2009). Nerve Growth Factor Promotes Differentiation of Odontoblast-like Cells. J. Cel. Biochem. 106, 539–545. doi:10.1002/jcb.22006
Austah, O., Widbiller, M., Tomson, P. L., and Diogenes, A. (2018). Expression of Neurotrophic Factors in Human Dentin and Their Regulation of Trigeminal Neurite Outgrowth. J. Endod. 45, 414–419. doi:10.1016/j.joen.2018.12.011
Austah, O., Widbiller, M., Tomson, P. L., and Diogenes, A. (2019). Expression of Neurotrophic Factors in Human Dentin and Their Regulation of Trigeminal Neurite Outgrowth. J. Endod. 45, 414–419. doi:10.1016/j.joen.2018.12.011
Bae, C. H., Kim, T. H., Ko, S. O., Lee, J. C., Yang, X., and Cho, E. S. (2015). Wntless Regulates Dentin Apposition and Root Elongation in the Mandibular Molar. J. Dent. Res. 94, 439–445. doi:10.1177/0022034514567198
Baffi, M. O., Slattery, E., Sohn, P., Moses, H. L., Chytil, A., and Serra, R. (2004). Conditional Deletion of the TGF-β Type II Receptor in Col2a Expressing Cells Results in Defects in the Axial Skeleton without Alterations in Chondrocyte Differentiation or Embryonic Development of Long Bones. Dev. Biol. 276, 124–142. doi:10.1016/j.ydbio.2004.08.027
Barkley, C., Serra, R., and Peters, S. B. (2020). A Co-Culture Method to Study Neurite Outgrowth in Response to Dental Pulp Paracrine Signals. JoVE. doi:10.3791/60809
Bott, C. J., Johnson, C. G., Yap, C. C., Dwyer, N. D., Litwa, K. A., and Winckler, B. (2019). Nestin in Immature Embryonic Neurons Affects Axon Growth Cone Morphology and Semaphorin3a Sensitivity. MBoC 30, 1214–1229. doi:10.1091/MBC.E18-06-0361
Boukhelifa, M., Parast, M. M., Valtschanoff, J. G., LaMantia, A. S., Meeker, R. B., and Otey, C. A. (2001). A Role for the Cytoskeleton-Associated Protein Palladin in Neurite Outgrowth. MBoC 12, 2721–2729. doi:10.1091/mbc.12.9.2721
Byers, M. R. (1980). Development of Sensory Innervation in Dentin. J. Comp. Neurol. 191, 413–427. doi:10.1002/cne.901910307
Cao, Y., Mitchell, E. B., Gorski, J. L., Hollinger, C., and Hoppman, N. L. (2016). Two Cases with de novo 3q26.31 Microdeletion Suggest a Role forFNDC3Bin Human Craniofacial Development. Am. J. Med. Genet. 170, 3276–3281. doi:10.1002/AJMG.A.37892
Carmeliet, P. (2003). Blood Vessels and Nerves: Common Signals, Pathways and Diseases. Nat. Rev. Genet. 4 (4), 710–720. doi:10.1038/nrg1158
Caroni, P. (1997). Overexpression of Growth-Associated Proteins in the Neurons of Adult Transgenic Mice. J. Neurosci. Methods 71, 3–9. doi:10.1016/s0165-0270(96)00121-5
Chen, S., Gluhak-Heinrich, J., Wang, Y. H., Wu, Y. M., Chuang, H. H., Chen, L., et al. (2009). Runx2, Osx, andDsppin Tooth Development. J. Dent. Res. 88, 904–909. doi:10.1177/0022034509342873
Chiquet, M., and Wehrle-Haller, B. (1994). Tenascin-C in Peripheral Nerve Morphogenesis. Perspect. Dev. Neurobiol. 2, 67–74. Available at: https://pubmed.ncbi.nlm.nih.gov/7530145/.
Cho, A., Haruyama, N., Hall, B., Danton, M. J. S., Zhang, L., Arany, P., et al. (2013). TGF-ß Regulates Enamel Mineralization and Maturation through KLK4 Expression. PLoS One 8, e82267. doi:10.1371/journal.pone.0082267
Choi, H., Ahn, Y.-H., Kim, T.-H., Bae, C.-H., Lee, J.-C., You, H.-K., et al. (2016). TGF-β Signaling Regulates Cementum Formation through Osterix Expression. Sci. Rep. 6, 1–11. doi:10.1038/srep26046
Corps, K., Stanwick, M., Rectenwald, J., Kruggel, A., and Peters, S. B. (2021). Skeletal Deformities in Osterix-Cre;tgfbr2f/f Mice May Cause Postnatal Death. Genes (Basel) 12, 975. doi:10.3390/GENES12070975/S1
Coutiño, B. C., and Mayor, R. (2021). The Mechanosensitive Channel Piezo1 Cooperates with Semaphorin to Control Neural Crest Migration. Development 148, dev200001, doi:10.1242/dev.200001
Crowe, A., and Yue, W. (2019). Semi-quantitative Determination of Protein Expression Using Immunohistochemistry Staining and Analysis: An Integrated Protocol. Bio-Protocol 9, e3465. doi:10.21769/BIOPROTOC.3465
de Almeida, J. F. A., Chen, P., Henry, M. A., and Diogenes, A. (2014). Stem Cells of the Apical Papilla Regulate Trigeminal Neurite Outgrowth and Targeting through a BDNF-dependent Mechanism. Tissue Eng. A 20, 3089–3100. doi:10.1089/ten.TEA.2013.0347
de Vega, S., Iwamoto, T., Nakamura, T., Hozumi, K., McKnight, D. A., Fisher, L. W., et al. (2007). TM14 Is a New Member of the Fibulin Family (Fibulin-7) that Interacts with Extracellular Matrix Molecules and Is Active for Cell Binding. J. Biol. Chem. 282, 30878–30888. doi:10.1074/jbc.M705847200
Dean, C., Dunning, F. M., Liu, H., Bomba-Warczak, E., Martens, H., Bharat, V., et al. (2012). Axonal and Dendritic Synaptotagmin Isoforms Revealed by a pHluorin-Syt Functional Screen. MBoC 23, 1715–1727. doi:10.1091/mbc.E11-08-0707
Ding, S., Luo, J.-h., and Yuan, X.-b. (2007). Semaphorin-3F Attracts the Growth Cone of Cerebellar Granule Cells through cGMP Signaling Pathway. Biochem. Biophys. Res. Commun. 356, 857–863. doi:10.1016/J.BBRC.2007.03.073
Dissanayaka, W. L., Zhan, X., Zhang, C., Hargreaves, K. M., Jin, L., and Tong, E. H. Y. (2012). Coculture of Dental Pulp Stem Cells with Endothelial Cells Enhances Osteo-/Odontogenic and Angiogenic Potential In Vitro. J. Endod. 38, 454–463. doi:10.1016/j.joen.2011.12.024
Dünker, N., and Krieglstein, K. (2002). Tgfß2 -/- Tgfß3 -/- Double Knockout Mice Display Severe Midline Fusion Defects and Early Embryonic Lethality. Anat. Embryol. 206, 73–83. doi:10.1007/s00429-002-0273-6
Emrick, J. J., von Buchholtz, L. J., and Ryba, N. J. P. (2020). Transcriptomic Classification of Neurons Innervating Teeth. J. Dent. Res. 99, 1478–1485. doi:10.1177/0022034520941837
Foster, B. L., Ao, M., Salmon, C. R., Chavez, M. B., Kolli, T. N., Tran, A. B., et al. (2018). Osteopontin Regulates Dentin and Alveolar Bone Development and Mineralization. Bone 107, 196–207. doi:10.1016/j.bone.2017.12.004
Fried, K., Sime, W., Lillesaar, C., Virtanen, I., Tryggvasson, K., and Patarroyo, M. (2005). Laminins 2 (α2β1γ1, Lm-211) and 8 (α4β1γ1, Lm-411) Are Synthesized and Secreted by Tooth Pulp Fibroblasts and Differentially Promote Neurite Outgrowth from Trigeminal Ganglion Sensory Neurons. Exp. Cel Res. 307, 329–341. doi:10.1016/j.yexcr.2005.04.009
Fried, K., Lillesaar, C., Sime, W., Kaukua, N., and Patarroyo, M. (2007). Target Finding of Pain Nerve Fibers: Neural Growth Mechanisms in the Tooth Pulp. Physiol. Behav. 92, 40–45. doi:10.1016/J.PHYSBEH.2007.05.032
Gadepalli, V. S., Ozer, H. G., Yilmaz, A. S., Pietrzak, M., and Webb, A. (2019). BISR-RNAseq: an Efficient and Scalable RNAseq Analysis Workflow with Interactive Report Generation. BMC Bioinformatics 20, 670. doi:10.1186/S12859-019-3251-1
Gorter de Vries, I., Quartier, E., Boute, P., Wisse, E., and Coomans, D. (1987). Immunocytochemical Localization of Osteocalcin in Developing Rat Teeth. J. Dent. Res. 66, 784–790. doi:10.1177/00220345870660031601
Goto, M., Osada, S., Imagawa, M., and Nishizuka, M. (2017). FAD104, a Regulator of Adipogenesis, Is a Novel Suppressor of TGF-β-Mediated EMT in Cervical Cancer Cells. Sci. Rep. 7, 1–12. doi:10.1038/s41598-017-16555-3
He, Z., and Tessier-Lavigne, M. (1997). Neuropilin Is a Receptor for the Axonal Chemorepellent Semaphorin III. Cell 90, 739–751. doi:10.1016/S0092-8674(00)80534-6
Hildebrand, C., Fried, K., Tuisku, F., and Johansson, C. S. (1995). Teeth and Tooth Nerves. Prog. Neurobiol. 45, 165–222. doi:10.1016/0301-0082(94)00045-J
Hou, C., Liu, Z. X., Tang, K. L., Wang, M. G., Sun, J., Wang, J., et al. (2012). Developmental Changes and Regional Localization of Dspp, Mepe, Mimecan and Versican in Postnatal Developing Mouse Teeth. J. Mol. Hist. 43, 9–16. doi:10.1007/s10735-011-9368-9
Hwang, S. J., Pagliardini, S., Boukhelifa, M., Parast, M. M., Otey, C. A., Rustioni, A., et al. (2001). Palladin Is Expressed Preferentially in Excitatory Terminals in the Rat central Nervous System. J. Comp. Neurol. 436, 211–224. doi:10.1002/cne.1062
Imhof, T., Balic, A., Heilig, J., Chiquet-Ehrismann, R., Chiquet, M., Niehoff, A., et al. (2021). Pivotal Role of Tenascin-W (-N) in Postnatal Incisor Growth and Periodontal Ligament Remodeling. Front. Immunol. 11, 608223. doi:10.3389/FIMMU.2020.608223/FULL
Ito, Y., Yeo, J. Y., Chytil, A., Han, J., Bringas, P., Nakajima, A., et al. (2003). Conditional Inactivation of Tgfbr2 in Cranial Neural Crest Causes Cleft Palate and Calvaria Defects. Development 130, 5269–5280. doi:10.1242/dev.00708
Iwata, J.-i., Hacia, J. G., Suzuki, A., Sanchez-Lara, P. A., Urata, M., and Chai, Y. (2012). Modulation of Noncanonical TGF-β Signaling Prevents Cleft Palate in Tgfbr2 Mutant Mice. J. Clin. Invest. 122, 873–885. doi:10.1172/JCI61498
Iwata, J.-i., Suzuki, A., Yokota, T., Ho, T.-V., Pelikan, R., Urata, M., et al. (2014). TGFβ Regulates Epithelial-Mesenchymal Interactions through WNT Signaling Activity to Control Muscle Development in the Soft Palate. Development 141, 909–917. doi:10.1242/dev.103093
Kang, S., Shin, Y., Shim, Y., Jeong, S., Park, I., and Min, B. (2005). Clusterin Interacts with SCLIP (SCG10-like Protein) and Promotes Neurite Outgrowth of PC12 Cells. Exp. Cel Res. 309, 305–315. doi:10.1016/j.yexcr.2005.06.012
Kang, H.-R., Lee, C. G., Homer, R. J., and Elias, J. A. (2007). Semaphorin 7A Plays a Critical Role in TGF-β1-Induced Pulmonary Fibrosis. J. Exp. Med. 204, 1083–1093. doi:10.1084/jem.20061273
Kawatsu, M., Takeshita, N., Takimoto, A., Yoshimoto, Y., Seiryu, M., Ito, A., et al. (2021). Scleraxis Upregulated by Transforming Growth Factor-β1 Signaling Inhibits Tension-Induced Osteoblast Differentiation of Priodontal Ligament Cells via Ephrin A2. Bone 149, 115969. doi:10.1016/J.BONE.2021.115969
Kettunen, P., Spencer-Dene, B., Furmanek, T., Kvinnsland, I. H., Dickson, C., Thesleff, I., et al. (2007). Fgfr2b Mediated Epithelial-Mesenchymal Interactions Coordinate Tooth Morphogenesis and Dental Trigeminal Axon Patterning. Mech. Dev. 124, 868–883. doi:10.1016/j.mod.2007.09.003
Kettunen, P., Løes, S., Furmanek, T., Fjeld, K., Kvinnsland, I. H., Behar, O., et al. (2005). Coordination of Trigeminal Axon Navigation and Patterning with Tooth Organ Formation: Epithelial-Mesenchymal Interactions, and Epithelial Wnt4 and Tgfβ1 Regulate Semaphorin 3a Expression in the Dental Mesenchyme. Development 132, 323–334. doi:10.1242/dev.01541
Khan, Q.-E. -S., Sehic, A., Khuu, C., Risnes, S., and Osmundsen, H. (2013). Expression ofCluandTgfb1during Murine Tooth Development: Effects of Iin-Vvivo Transfection with Anti-miR-214. Eur. J. Oral Sci. 121, 303–312. doi:10.1111/eos.12056
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: a Fast Spliced Aligner with Low Memory Requirements. Nat. Methods 12, 357–360. doi:10.1038/nmeth.3317
Kishimoto, K., Nishizuka, M., Katoh, D., Kato, A., Osada, S., and Imagawa, M. (2013). FAD104, a Regulatory Factor of Adipogenesis, Acts as a Novel Regulator of Calvarial Bone Formation. J. Biol. Chem. 288, 31772–31783. doi:10.1074/JBC.M113.452961
Kitisin, K., Saha, T., Blake, T., Golestaneh, N., Deng, M., Kim, C., et al. (2007). TGF-β Signaling in Development. Sci. STKE 2007, cm1. doi:10.1126/stke.3992007cm1
Kitsukawa, T., Shimizu, M., Sanbo, M., Hirata, T., Taniguchi, M., Bekku, Y., et al. (1997). Neuropilin-semaphorin III/D-mediated Chemorepulsive Signals Play a Crucial Role in Peripheral Nerve Projection in Mice. Neuron 19, 995–1005. doi:10.1016/s0896-6273(00)80392-x
Ko, S. O., Chung, I. H., Xu, X., Oka, S., Zhao, H., Cho, E. S., et al. (2007). Smad4 Is Required to Regulate the Fate of Cranial Neural Crest Cells. Dev. Biol. 312, 435–447. doi:10.1016/j.ydbio.2007.09.050
Kolk, S. M., Gunput, R.-A. F., Tran, T. S., Van Den Heuvel, D. M. A., Prasad, A. A., Hellemons, A. J. C. G. M., et al. (2009). Semaphorin 3F Is a Bifunctional Guidance Cue for Dopaminergic Axons and Controls Their Fasciculation, Channeling, Rostral Growth, and Intracortical Targeting. J. Neurosci. 29, 12542–12557. doi:10.1523/JNEUROSCI.2521-09.2009
Kollar, E. J., and Lumsden, A. G. (1979). Tooth Morphogenesis: the Role of the Innervation during Induction and Pattern Formation. J. Biol. Buccale 7, 49–60. Available at: http://www.ncbi.nlm.nih.gov/pubmed/285074 (Accessed May 1, 2017).
Kopp, M. A., Brommer, B., Gatzemeier, N., Schwab, J. M., and Prüss, H. (2010). Spinal Cord Injury Induces Differential Expression of the Profibrotic Semaphorin 7A in the Developing and Mature Glial Scar. Glia 58, 1748–1756. doi:10.1002/GLIA.21045
Kuchler‐Bopp, S., Bagnard, D., Van‐Der‐Heyden, M., Idoux‐Gillet, Y., Strub, M., Gegout, H., et al. (2018). Semaphorin 3A Receptor Inhibitor as a Novel Therapeutic to Promote Innervation of Bioengineered Teeth. J. Tissue Eng. Regen. Med. 12, e2151–e2161. doi:10.1002/term.2648
Liao, Y., Smyth, G. K., and Shi, W. (2014). featureCounts: an Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinformatics 30, 923–930. doi:10.1093/BIOINFORMATICS/BTT656
Lillesaar, C., and Fried, K. (2004). Neurites from Trigeminal Ganglion Explants Grown In Vitro Are Repelled or Attracted by Tooth-Related Tissues Depending on Developmental Stage. Neuroscience 125, 149–161. doi:10.1016/J.NEUROSCIENCE.2004.01.008
Lillesaar, C., Eriksson, C., Johansson, C. S., Fried, K., and Hildebrand, C. (1999). Tooth Pulp Tissue Promotes Neurite Outgrowth from Rat Trigeminal Ganglia In Vitro. J. Neurocytol. 28, 663–670. doi:10.1023/a:1007008815621
Lillesaar, C., Eriksson, C., and Fried, K. (2001). Rat Tooth Pulp Cells Elicit Neurite Growth from Trigeminal Neurones and Express mRNAs for Neurotrophic Factors In Vitro. doi:10.1016/S0304-3940(01)01997-8
Limoni, G. (2021). Modelling and Refining Neuronal Circuits with Guidance Cues: Involvement of Semaphorins. Ijms 22, 6111. doi:10.3390/IJMS22116111
Lucas, F., Rogers, K. A., Harrington, B. K., Pan, A., Yu, L., Breitbach, J., et al. (2019). Eμ-TCL1xMyc: A Novel Mouse Model for Concurrent CLL and B-Cell Lymphoma. Clin. Cancer Res. 25, 6260–6273. doi:10.1158/1078-0432.CCR-19-0273
Ludwig, M. R., Kojima, K., Bowersock, G. J., Chen, D., Jhala, N. C., Buchsbaum, D. J., et al. (2016). Surveying the Serologic Proteome in a Tissue-specific kras(G12D) Knockin Mouse Model of Pancreatic Cancer. Proteomics 16, 516–531. doi:10.1002/pmic.201500133
Martens, W., Sanen, K., Georgiou, M., Struys, T., Bronckaers, A., Ameloot, M., et al. (2014). Human Dental Pulp Stem Cells Can Differentiate into Schwann Cells and Promote and Guide Neurite Outgrowth in an Aligned Tissue‐engineered Collagen Construct In Vitro. FASEB j. 28, 1634–1643. doi:10.1096/FJ.13-243980
Matsukawa, T., Morita, K., Omizu, S., Kato, S., and Koriyama, Y. (2018). Mechanisms of RhoA Inactivation and CDC42 and Rac1 Activation during Zebrafish Optic Nerve Regeneration. Neurochem. Int. 112, 71–80. doi:10.1016/j.neuint.2017.11.004
Maurin, J., Delorme, G., Machucagayet, I., Couble, M., Magloire, H., Jurdic, P., et al. (2005). Odontoblast Expression of Semaphorin 7A during Innervation of Human Dentin. Matrix Biol. 24, 232–238. doi:10.1016/J.MATBIO.2005.03.005
Meiners, S., Mercado, M. L. T., Nur-e-Kamal, M. S. A., and Geller, H. M. (1999). Tenascin-C Contains Domains that Independently Regulate Neurite Outgrowth and Neurite Guidance. J. Neurosci. 19, 8443–8453. doi:10.1523/JNEUROSCI.19-19-08443.1999
Meloty-Kapella, C. V., Degen, M., Chiquet-Ehrismann, R., and Tucker, R. P. (2006). Avian Tenascin-W: Expression in Smooth Muscle and Bone, and Effects on Calvarial Cell Spreading and Adhesion In Vitro. Dev. Dyn. 235, 1532–1542. doi:10.1002/DVDY.20731
Mi, H., Dong, Q., Muruganujan, A., Gaudet, P., Lewis, S., and Thomas, P. D. (2010). PANTHER Version 7: Improved Phylogenetic Trees, Orthologs and Collaboration with the Gene Ontology Consortium. Nucleic Acids Res. 38, D204–D210. doi:10.1093/nar/gkp1019
Michele, M., and Faissner, A. (2009). Tenascin-C Stimulates Contactin-dependent Neurite Outgrowth via Activation of Phospholipase C. Mol. Cell Neurosci. 41, 397–408. doi:10.1016/J.MCN.2009.04.004
Moe, K., Sijaona, A., Shrestha, A., Kettunen, P., Taniguchi, M., and Luukko, K. (2012). Semaphorin 3A Controls Timing and Patterning of the Dental Pulp Innervation. Differentiation 84, 371–379. doi:10.1016/j.diff.2012.09.003
Muzumdar, M. D., Tasic, B., Miyamichi, K., Li, L., and Luo, L. (2007). A Global Double-Fluorescent Cre Reporter Mouse. genesis 45, 593–605. doi:10.1002/dvg.20335
Niwa, T., Yamakoshi, Y., Yamazaki, H., Karakida, T., Chiba, R., Hu, J. C.-C., et al. (2018). The Dynamics of TGF-β in Dental Pulp, Odontoblasts and Dentin. Sci. Rep. 8, 4450. doi:10.1038/s41598-018-22823-7
Norlin, T., Hilliges, M., and Brodin, L. (1999). Immunohistochemical Demonstration of Exocytosis-Regulating Proteins within Rat Molar Dentinal Tubules. Arch. Oral Biol. 44, 223–231. doi:10.1016/S0003-9969(98)00115-0
Oka, K., Oka, S., Sasaki, T., Ito, Y., Bringas, P., Nonaka, K., et al. (2007). The Role of TGF-β Signaling in Regulating Chondrogenesis and Osteogenesis during Mandibular Development. Dev. Biol. 303, 391–404. doi:10.1016/j.ydbio.2006.11.025
Ono, W., Sakagami, N., Nishimori, S., Ono, N., and Kronenberg, H. M. (2016). Parathyroid Hormone Receptor Signalling in Osterix-Expressing Mesenchymal Progenitors Is Essential for Tooth Root Formation. Nat. Commun. 7, 1–16. doi:10.1038/ncomms11277
Otey, C. A., Rachlin, A., Moza, M., Arneman, D., and Carpen, O. (2005). The Palladin/Myotilin/Myopalladin Family of Actin‐Associated Scaffolds. Int. Rev. Cytol. 246, 31–58. doi:10.1016/S0074-7696(05)46002-7
Pagella, P., Jiménez-Rojo, L., and Mitsiadis, T. A. (2014). Roles of Innervation in Developing and Regenerating Orofacial Tissues. Cell. Mol. Life Sci. 71, 2241–2251. doi:10.1007/s00018-013-1549-0
Pagella, P., Miran, S., Neto, E., Martin, I., Lamghari, M., and Mitsiadis, T. A. (2020). Human Dental Pulp Stem Cells Exhibit Enhanced Properties in Comparison to Human Bone Marrow Stem Cells on Neurites Outgrowth. FASEB j. 34, 5499–5511. doi:10.1096/fj.201902482R
Pasterkamp, R. J., Peschon, J. J., Spriggs, M. K., and Kolodkin, A. L. (2003). Semaphorin 7A Promotes Axon Outgrowth through Integrins and MAPKs. Nature 424, 398–405. doi:10.1038/nature01790
Peters, S. B., Wang, Y., and Serra, R. (2017). Tgfbr2 Is Required in Osterix Expressing Cells for Postnatal Skeletal Development. Bone 97, 54–64. doi:10.1016/j.bone.2016.12.017
Price, T. J., Louria, M. D., Candelario-Soto, D., Dussor, G. O., Jeske, N. A., Patwardhan, A. M., et al. (2005). Treatment of Trigeminal Ganglion Neurons In Vitro with NGF, GDNF or BDNF: Effects on Neuronal Survival, Neurochemical Properties and TRPV1-Mediated Neuropeptide Secretion. BMC Neurosci. 6, 4. doi:10.1186/1471-2202-6-4
Quispe-Salcedo, A., Ida-Yonemochi, H., Nakatomi, M., and Ohshima, H. (2012). Expression Patterns of Nestin and Dentin Sialoprotein during Dentinogenesis in Mice. Biomed. Res. 33, 119–132. doi:10.2220/BIOMEDRES.33.119
Rakian, A., Yang, W.-C., Gluhak-Heinrich, J., Cui, Y., Harris, M. A., Villarreal, D., et al. (2013). Bone Morphogenetic Protein-2 Gene Controls Tooth Root Development in Coordination with Formation of the Periodontium. Int. J. Oral Sci. 5, 75–84. doi:10.1038/ijos.2013.41
Rigato, F., Garwood, J., Calco, V., Heck, N., Faivre-Sarrailh, C., and Faissner, A. (2002). Tenascin-C Promotes Neurite Outgrowth of Embryonic Hippocampal Neurons through the Alternatively Spliced Fibronectin Type III BD Domains via Activation of the Cell Adhesion Molecule F3/contactin. J. Neurosci. 22, 6596–6609. doi:10.1523/jneurosci.22-15-06596.2002
Ritchie, M. E., Phipson, B., Wu, D., Hu, Y., Law, C. W., Shi, W., et al. (2015). Limma powers Differential Expression Analyses for RNA-Sequencing and Microarray Studies. Nucleic Acids Res. 43, e47. doi:10.1093/NAR/GKV007
Rodda, S. J., and McMahon, A. P. (2006). Distinct Roles for Hedgehog and Canonical Wnt Signaling in Specification,differentiation and Maintenance of Osteoblast Progenitors. Development 133, 3231–3244. doi:10.1242/dev.02480
Rosowski, J., Bräunig, J., Amler, A.-K., Strietzel, F. P., Lauster, R., and Rosowski, M. (2019). Emulating the Early Phases of Human Tooth Development In Vitro. Sci. Rep. 9, 1–14. doi:10.1038/s41598-019-43468-0
Sabag, A. D., Smolkin, T., Mumblat, Y., Ueffing, M., Kessler, O., Gloeckner, C. J., et al. (2014). The Role of the Plexin-A2 Receptor in semaphorin-3A and semaphorin-3B Signal Transduction. J. Cel. Sci. 127, 5240–5252. doi:10.1242/JCS.155960
Sahlberg, C., Aukhil, I., and Thesleff, I. (2001). Tenascin-C in Developing Mouse Teeth: Expression of Splice Variants and Stimulation by TGFβ and FGF. Eur. J. Oral Sci. 109, 114–124. doi:10.1034/J.1600-0722.2001.00990.X
Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., et al. (2012). Fiji: an Open-Source Platform for Biological-Image Analysis. Nat. Methods 9, 676–682. doi:10.1038/nmeth.2019
Seo, H.-S., and Serra, R. (2007). Deletion of Tgfbr2 in Prx1-Cre Expressing Mesenchyme Results in Defects in Development of the Long Bones and Joints. Dev. Biol. 310, 304–316. doi:10.1016/j.ydbio.2007.07.040
Seo, H.-S., and Serra, R. (2009). Tgfbr2 Is Required for Development of the Skull Vault. Dev. Biol. 334, 481–490. doi:10.1016/j.ydbio.2009.08.015
Serini, G., and Bussolino, F. (2004). Common Cues in Vascular and Axon Guidance. Physiology 19, 348–354. doi:10.1152/physiol.00021.2004
Shadad, O., Chaulagain, R., Luukko, K., and Kettunen, P. (2019). Establishment of Tooth Blood Supply and Innervation Is Developmentally Regulated and Takes Place through Differential Patterning Processes. J. Anat. 234, 465–479. doi:10.1111/joa.12950
Shrestha, A., Moe, K., Luukko, K., Taniguchi, M., and Kettunen, P. (2014). Sema3A Chemorepellant Regulates the Timing and Patterning of Dental Nerves during Development of Incisor Tooth Germ. Cell Tissue Res. 357, 15–29. doi:10.1007/s00441-014-1839-3
Sijaona, A., Luukko, K., Kvinnsland, I. H., and Kettunen, P. (2012). Expression Patterns of Sema3F, PlexinA4, -A3, Neuropilin1 and -2 in the Postnatal Mouse Molar Suggest Roles in Tooth Innervation and Organogenesis. Acta Odontol. Scand. 70, 140–148. doi:10.3109/00016357.2011.600708
Sohn, P., Cox, M., Chen, D., and Serra, R. (2010). Molecular Profiling of the Developing Mouse Axial Skeleton: a Role for Tgfbr2 in the Development of the Intervertebral Disc. BMC Dev. Biol. 10, 29. doi:10.1186/1471-213X-10-29
Song, Y., Liu, X., Feng, X., Gu, Z., Gu, Y., Lian, M., et al. (2017). NRP1 Accelerates Odontoblast Differentiation of Dental Pulp Stem Cells through Classical Wnt/β-Catenin Signaling. Cell. Reprogram. 19, 324–330. doi:10.1089/cell.2017.0020
Südhof, T. C. (2013). A Molecular Machine for Neurotransmitter Release: Synaptotagmin and beyond. Nat. Med. 19, 1227–1231. doi:10.1038/nm.3338
Sultan, N., Amin, L. E., Zaher, A. R., Grawish, M. E., and Scheven, B. A. (2020). Neurotrophic Effects of Dental Pulp Stem Cells on Trigeminal Neuronal Cells. Sci. Rep. 10, 1–13. doi:10.1038/s41598-020-76684-0
Taylor, P. E., Byers, M. R., and Redd, P. E. (1988). Sprouting of CGRP Nerve Fibers in Response to Dentin Injury in Rat Molars. Brain Res. 461, 371–376. doi:10.1016/0006-8993(88)90270-3
Thesleff, I., Mackie, E., Vainio, S., and Chiquet-Ehrismann, R. (1987). Changes in the Distribution of Tenascin during Tooth Development. Development 101, 289–296. doi:10.1242/dev.101.2.289
Walker, L. A., Sovic, M. G., Chiang, C. L., Hu, E., Denninger, J. K., Chen, X., et al. (2020). CLEAR: Coverage-Based Limiting-Cell experiment Analysis for RNA-Seq. J. Transl. Med. 18, 63–15. doi:10.1186/S12967-020-02247-6/TABLES/1
Wang, Y., Cox, M. K., Coricor, G., MacDougall, M., and Serra, R. (2013). Inactivation of Tgfbr2 in Osterix-Cre Expressing Dental Mesenchyme Disrupts Molar Root Formation. Dev. Biol. 382, 27–37. doi:10.1016/j.ydbio.2013.08.003
Wehrle, B., and Chiquet, M. (1990). Tenascin Is Accumulated along Developing Peripheral Nerves and Allows Neurite Outgrowth In Vitro. Development 110, 401–415. Available at: https://pubmed.ncbi.nlm.nih.gov/1723942/. (Accessed December 7, 2021). doi:10.1242/dev.110.2.401
Widbiller, M., Austah, O., Lindner, S. R., Sun, J., and Diogenes, A. (2019). Neurotrophic Proteins in Dentin and Their Effect on Trigeminal Sensory Neurons. J. Endod. 45, 729–735. doi:10.1016/j.joen.2019.02.021
Wright, M. C., Mi, R., Connor, E., Reed, N., Vyas, A., Alspalter, M., et al. (2014). Novel Roles for Osteopontin and Clusterin in Peripheral Motor and Sensory Axon Regeneration. J. Neurosci. 34, 1689–1700. doi:10.1523/JNEUROSCI.3822-13.2014
Yamashiro, T., Zheng, L., Shitaku, Y., Saito, M., Tsubakimoto, T., Takada, K., et al. (2007). Wnt10a Regulates Dentin Sialophosphoprotein mRNA Expression and Possibly Links Odontoblast Differentiation and Tooth Morphogenesis. Differentiation 75, 452–462. doi:10.1111/j.1432-0436.2006.00150.x
Yoshida, S., Wada, N., Hasegawa, D., Miyaji, H., Mitarai, H., Tomokiyo, A., et al. (2016). Semaphorin 3A Induces Odontoblastic Phenotype in Dental Pulp Stem Cells. J. Dent. Res. 95, 1282–1290. doi:10.1177/0022034516653085
Yu, R., Ma, L., Li, J., Gu, J., Shi, Q., and Yu, R. (2012). Effects of SEMA3G on Migration and Invasion of Glioma Cells. Oncol. Rep. 28, 269–275. doi:10.3892/or.2012.1796
Yu, H., Pei, T., Ren, J., Ding, Y., Wu, A., and Zhou, Y. (2018). Semaphorin 3A Enhances Osteogenesis of MG63 Cells through Interaction with Schwann Cells In Vitro. Mol. Med. Rep. 17, 6084–6092. doi:10.3892/MMR.2018.8628/HTML
Zhang, H., Jani, P., Liang, T., Lu, Y., and Qin, C. (2017). Inactivation of Bone Morphogenetic Protein 1 (Bmp1) and Tolloid-like 1 (Tll1) in Cells Expressing Type I Collagen Leads to Dental and Periodontal Defects in Mice. J. Mol. Hist. 48, 83–98. doi:10.1007/S10735-016-9708-X/FIGURES/12
Zhang, R., Lin, J., Liu, Y., Yang, S., He, Q., Zhu, L., et al. (2021). Transforming Growth Factor-β Signaling Regulates Tooth Root Dentinogenesis by Cooperation with Wnt Signaling. Front. Cell Dev. Biol. 9, 687099. doi:10.3389/FCELL.2021.687099
Zhao, H., Oka, K., Bringas, P., Kaartinen, V., and Chai, Y. (2008). TGF-β Type I Receptor Alk5 Regulates Tooth Initiation and Mandible Patterning in a Type II Receptor-independent Manner. Dev. Biol. 320, 19–29. doi:10.1016/j.ydbio.2008.03.045
Zhao, H., Li, S., Han, D., Kaartinen, V., and Chai, Y. (2011). Alk5-Mediated Transforming Growth Factor Signaling Acts Upstream of Fibroblast Growth Factor 10 to Regulate the Proliferation and Maintenance of Dental Epithelial Stem Cells. Mol. Cell Biol. 31, 2079–2089. doi:10.1128/MCB.01439-10
Keywords: TGFβ, semaphorins, dental pulp, tooth innervation, neurite outgrowth
Citation: Stanwick M, Barkley C, Serra R, Kruggel A, Webb A, Zhao Y, Pietrzak M, Ashman C, Staats A, Shahid S and Peters SB (2022) Tgfbr2 in Dental Pulp Cells Guides Neurite Outgrowth in Developing Teeth. Front. Cell Dev. Biol. 10:834815. doi: 10.3389/fcell.2022.834815
Received: 13 December 2021; Accepted: 24 January 2022;
Published: 21 February 2022.
Edited by:
Chisa Shukunami, Hiroshima University, JapanReviewed by:
Eumorphia Remboutsika, National and Kapodistrian University of Athens, GreeceYuji Mishina, University of Michigan, United States
Copyright © 2022 Stanwick, Barkley, Serra, Kruggel, Webb, Zhao, Pietrzak, Ashman, Staats, Shahid and Peters. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sarah B. Peters, cGV0ZXJzLjEwMjZAb3N1LmVkdQ==
 Monica Stanwick
Monica Stanwick Courtney Barkley
Courtney Barkley Rosa Serra
Rosa Serra Andrew Kruggel
Andrew Kruggel Amy Webb3
Amy Webb3 Maciej Pietrzak
Maciej Pietrzak Chandler Ashman
Chandler Ashman Allie Staats
Allie Staats Sarah B. Peters
Sarah B. Peters