- 1National Engineering Laboratory for Animal Breeding and Key Laboratory of Animal Genetics, Breeding and Reproduction, Ministry of Agriculture and Rural Affairs, College of Animal Science and Technology, China Agricultural University, Beijing, China
- 2Shenzhen Branch, Guangdong Laboratory of Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen, China
In birds, male gonads form on both sides whereas most females develop asymmetric gonads. Multiple early lines of evidence suggested that the right gonad fails to develop into a functional ovary, mainly due to differential expression of PITX2 in the gonadal epithelium. Despite some advances in recent years, the molecular mechanisms underlying asymmetric gonadal development remain unclear. Here, using bulk analysis of whole gonads, we established a relatively detailed profile of four representative stages of chicken gonadal development at the transcriptional and chromatin levels. We revealed that many candidate genes were significantly enriched in morphogenesis, meiosis and subcellular structure formation, which may be responsible for asymmetric gonadal development. Further chromatin accessibility analysis suggested that the transcriptional activities of the candidate genes might be regulated by nearby open chromatin regions, which may act as transcription factor (TF) binding sites and potential cis-regulatory elements. We found that LHX9 was a promising TF that bound to the left-biased peaks of many cell cycle-related genes. In summary, this study provides distinctive insights into the potential molecular basis underlying the asymmetric development of chicken gonads.
Introduction
The ovary, a significant organ in humans and animals, affects sexual maturity and reproductive performance. In the poultry industry, well-developed ovaries are of great significance for improving production performance and economic benefits. In addition, gonadogenesis is also a great biomedical model for studying infertility and sex differentiation disorders in humans (Arboleda et al., 2014; Guercio and Rey, 2014). Unlike mammals, most female birds develop asymmetric gonads, that is, only the left gonad develops into a functional ovary, whereas the right gonad remains rudimentary (Smith and Sinclair, 2004; González-Morán, 2011). Although several investigations have focused on this particularly intriguing phenomenon, the distinct developmental patterns that occur on the two sides are still elusive (Scheider et al., 2014; Wan et al., 2017; Yu et al., 2017; Zou et al., 2020). Thus, there is an urgent need to elucidate the molecular mechanisms underlying asymmetric gonadal development in female embryos.
In chicken embryos, gonadogenesis begins at approximately 72 h of development (Hamburger Hamilton Stage 23, HH23) (Hamburger and Hamilton, 1951). As development progresses, the morphology of the bilateral gonads in females is initially similar at embryonic days 5.5 (E5.5, HH28), but by E6.5 (HH30), the gonads are distinctly different (Guioli et al., 2014). It is widely accepted that the gonads contain two layers, namely, the cortex and medulla (Smith and Sinclair, 2004). The left gonad develops a side-dependent thicker cortex and ultimately differentiates into a functional ovary, while the right ovary is surrounded by a thin, flat epithelial layer and later undergoes developmental arrest (Smith et al., 2007; González-Morán, 2011; Intarapat and Stern, 2013). Almost all primordial germ cells (PGCs) are located in the cortex, and a few are scattered in the medulla. Indeed, due to the preferential migration, the number of PGCs present in the gonads exhibits an obvious sex-independent asymmetric distribution, and more PGCs are found on the left side at approximately 50 h (HH15) to E16 (HH42) and before hatching (Van Limborgh, 1968; Nakamura et al., 2007; Intarapat and Stern, 2013; de Melo Bernardo et al., 2015). Additionally, the PGCs in the cortex exhibit faster proliferation rates at E5-E6 (HH27-HH29) (Ishimaru et al., 2008). However, the PGCs in the right gonad fail to enter meiosis and disappear posthatching. This fate is similar to that of germ cells in the medulla of the left ovary (Guioli et al., 2014). Therefore, increased proliferation in the left cortex is primarily responsible for asymmetric ovarian development. Although this anatomical developmental process is relatively clear, the underlying functional drivers and regulatory network remain to be further studied.
The side-dependent morphological development of the embryonic gonad is regulated by multiple side-biased genes, which were described in a previous study (Monsoro-Burq and Levin, 2018). For instance, PITX2, which is strongly expressed in the gonadal epithelium, controls gonadal morphogenesis and rescues right gonad fate by affecting the RA pathway to regulate target gene expression and stimulate cell proliferation (Guioli and Lovell-Badge, 2007; Ishimaru et al., 2008; Rodríguez-León et al., 2008). In addition, several genes, such as FET-1, BMP7, R-Spondin1 and OVEX1, have been reported to be expressed in the developing gonads of chickens in a side-dependent manner (Reed and Sinclair, 2002; Hoshino et al., 2005; Smith et al., 2008b; Carré-Eusèbe et al., 2009). Despite the identification of putative candidate genes, the mechanisms by which these genes regulate asymmetric gonadal development are not clear. Accessible regions of the genome are considered primary regulatory elements, such as promoters and enhancers, and TFs binding sites to modulate gene expression and further determine cell fate (Buenrostro et al., 2013; Rivera and Ren, 2013; Burton and Torres-Padilla, 2014; Wu et al., 2016). Accordingly, it is essential to elucidate the dynamic patterns and regulatory functions of open chromatin regions across asymmetric gonadal development.
To obtain a better understanding of this progress, we explored the transcriptional and epigenetic changes that occur during asymmetric gonadal development by high-throughput paired-end RNA sequencing (RNA-seq) and Assay for Transposase Accessible Chromatin with high-throughput sequencing (ATAC-seq). The present work generated a high-resolution developmental profile of chicken gonads at the gene expression and chromatin accessibility levels, identified numerous candidate genes and cis-regulatory elements, and showed functional relationships between these elements. It should be noted that we proposed that a promising TF, LHX9, was enriched in left side-specific open chromatin regions and might be recruited by certain pioneer TFs to activate left ovary development; this hypothesis was initially supported by in vitro experiments. In summary, our findings provide important insight into the earlier steps of asymmetric gonadal development in female chicken embryos and will be beneficial for future studies on human infertility.
Materials and Methods
Ethics Statement
The experiments were approved by the Animal Welfare Committee of China Agricultural University and performed in accordance with the protocol outlined in the “Guide for Care and Use of Laboratory Animals” (China Agricultural University).
Embryo Tissue Collection
Fertilized eggs were obtained from a pure line of White Leghorns raised in the Experimental Base of Poultry Genetic Resources and Breeding, College of Animal Science and Technology, China Agricultural University. The eggs were incubated in an automated egg incubator at 37.8°C and 60% relative humidity, and the eggs were rotated every 2 h. Once the eggs reached E4.5 (HH25), E5.5 (HH28), E7 (HH31) and E10 (HH36), respectively, the left and right gonads of the chicken embryo were separately harvested. The remaining embryo tissues were collected to determine sex by a direct PCR kit (TransGen Biotech, Beijing, China) using CHD1 primers (Supplementary Table S1). Additionally, three biological replicate samples were collected from each side and each stage.
RNA Isolation, Library Preparation and Sequencing
The left and right gonads were dissected. One embryonic gonad was used for each biological replicate. The gonads were immediately placed in RNAlater™ Stabilization Solution (Thermo Fisher Scientific, Waltham, MA, United States) and stored at −20°C. Total RNA was extracted with TRIzol reagent (Thermo Fisher Scientific, Waltham, MA, United States) following the manufacturer’s instructions. The RNA concentration and purity were measured using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, United States). The integrity of the RNA was determined using an Agilent 2100 Bioanalyzer (Agilent Technologies, CA, United States). To construct the libraries, reverse transcription and amplification of the transcripts were performed following the Smart-seq2 protocol. The libraries were sequenced on the MGI-500 platform (150 bp paired-end, PE150).
ATAC Library Preparation and Sequencing
Each biological replicate for four developmental stages comprised one embryonic gonad. The samples were dissected as described above and then preserved in a cryopreservation tube with DMEM/F12 (GIBCO, Grand Island, NY, United States) supplemented with 10% fetal bovine serum (GIBCO, Grand Island, NY, United States) and 10% DMSO (Sigma-Aldrich). The cryopreservation tubes containing the samples were immediately placed in a cryopreservation box at 4°C for 1 h and then -80°C overnight. The nuclei were isolated from the frozen tissues according to previously described methods (Corces et al., 2017). The nuclei were pelleted at 500 RCF for 10 min at 4°C in a fixed angle centrifuge. The cell pellets were resuspended in 50 μL of transposition mixture containing 25 μL 2x TD buffer, 2.5 μL transposase (100 nM final), 16.5 μL phosphate-buffered saline (PBS; GIBCO, Grand Island, NY, United States), 0.5 μL 1% digitonin, 0.5 μL 10% Tween-20 and 5 μL H2O and incubated at 37°C for 30 min. A cleanup reaction was performed to obtain the DNA for the PCR amplification of transposed fragments with 29 cycles. The PCR products were quantified with the KAPA Library Quantification Kit (#KK4844; Kapa Biosystems) and then sequenced using PE150 reads on the Illumina Novaseq 6,000 platform.
RNA-Seq Data Analysis
Paired-end RNA-seq reads were aligned to the chicken reference genome (GRCg6a) using HISAT2 (Kim et al., 2015) with default parameters before using HTSeq-count (Anders et al., 2015) to count the reads mapped to individual genes. The generated count matrix was used as input for the DESeq2 (Love et al., 2014) package to identify DEGs between left and right gonads. Candidates with greater than 1.5-fold changes at adjusted p-values < 0.1 were considered significant DEGs.
ATAC-Seq Data Processing
The PE150 ATAC-seq reads were trimmed into PE50 using Perl scripts, and then, the PE50 ATAC-seq reads were mapped to the chicken reference genome (GRCg6a) assembly using Burrows–Wheeler Aligner (BWA) (Li and Durbin, 2009) with default parameters. SAM files were converted to the BAM format using Samtools (Li et al., 2009), and PCR duplicates were removed using the Picard MarkDuplicates option to generate filtered BAM files. The peaks were called using MACS2 (Zhang et al., 2008) in each sample and using the filtered BAM files with the parameters (-g 9.6e8 -q 0.01 -B --SPMR --nomodel -shift -100 -extsize 200). Referring to the proportion of the effective genome sizes of humans and mice, we adjusted the parameters of –g in chickens. All the alignment files were extended to 200 bp and scaled to RPKM-normalized read coverage files using deepTools (Ramírez et al., 2014) for visualization. The library size factors estimated by the DESeq2 package were applied to the RPKM values to compare binding profiles between different samples in an unbiased manner. DARs were detected using DESeq2 with a fold change less than 1.5 and q-value below 0.1. The HOMER tool (http://homer.salk.edu/homer/motif/) was used to detect the motifs. The BEDtools suite (https://bedtools.readthedocs.io/en/latest/content/bedtools-suite.html) was used to test overlap and enrichment between different intervals.
Functional Annotation
Using biomaRt, we identified homologs of chicken DEGs in humans. Functional analysis of these homologs was performed using the Metascape online tool (http://metascape.org). The Gene Ontology (GO) terms for biological process, cellular component, and molecular function categories, as well as Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, were enriched based on the Metascape online tool with default parameters.
Cell Separation and Culture
The left gonads were removed and placed in cold PBS when the fertile chicken eggs reached E7 (HH31). Samples were dispersed by incubation with trypsin 0.25% (GIBCO, Grand Island, NY, United States) at 37°C for 10 min with constant shaking. The digestions were stopped by the addition of culture medium containing DMEM/F12 with 10% (v/v) fetal bovine serum and 1% penicillin/streptomycin (GIBCO, Grand Island, NY, United States), and the samples were passed through a cell strainer (40 μm) and collected into 50-ml tubes. Subsequently, the cells were washed with PBS and seeded in cell culture plates. Sex identification was carried out by PCR as described above. Cells, derived from 15 gonads, were pooled for each biological replicate.
Plasmid Construction, Transfection and qRT-PCR
Gonadal cDNA was synthesized using the First-Strand Synthesis kit (Takara, Japan) and PrimeScript™ RT reagent kit with gDNA Eraser (Takara, Japan). The coding sequence of LHX9 was amplified with PCR primers (Supplementary Table S2) using Polymerase PrimeSTAR (Takara, Japan). The amplified fragments were digested with the restriction enzymes MluI and HindIII (NEB) and cloned into the pcDNA3.1 vector using T4 DNA ligase (NEB). When the cells reached 40% confluence, they were transfected with the plasmids by using Fugene HD (Promega, Madison, WI) according to the manufacturer’s instructions. After transfection for 48 h, the cells were collected for RNA extraction, and cDNA was synthesized as described above. qRT-PCR was performed with an ABI 7500 system (Applied Biosystems) using the TB Green® Premix Ex Taq™ Kit (Takara, Japan) according the manufacturer’s instructions. β-actin was used as the internal control, and the sequences of the gene-specific primers are listed in Supplementary Table S3.
Statistics Analysis
Statistical analyses were performed using the SPSS software (version 25.0; IBM, Chicago, IL, United States). The data are expressed as the mean ± SD (standard deviation) and were analyzed using a two-tailed Student’s t-test, and at least three replicates were conducted in multiple independent experiments. The differences were considered to be statistically significant at a p-value < 0.05.
Results
Asymmetric Development of Gonads
Gonadal morphological characteristics were assessed at four representative stages during embryonic development, including E4.5 (HH25), E5.5 (HH28), E7 (HH31) and E10 (HH36). To ensure a comprehensive understanding of asymmetric gonadal development, bilateral gonads were collected at each stage in triplicate for RNA-seq and ATAC-seq (Figure 1) The bilateral gonads emerge on the ventral surface of each mesonephros and continue to increase in size at different rates during embryonic development. The gonadal morphological appearance of the female chicken embryos was initially similar on the left and right sides at E4.5. Generally, bilateral gonads were not macroscopically evident until E5.5. As development progressed, the left gonads of the female chicken embryos were markedly larger than the right gonads at E7. Subsequently, the dimensions of the left gonads were almost 2.5 times larger than those of the right gonad at E10 (Supplementary Figures S1A–D).
FIGURE 1. Schematic representation of chicken embryos with left and right gonads dissected for RNA-seq and ATAC-seq at each time point in triplicate.
Transcriptional Profiling
After harvest, gonads were first processed for RNA-seq. The mapping statistics are displayed in Supplementary Table S4. Three replicates of RNA-seq analysis showed high correlation using the Pearson correlation coefficient (Supplementary Figure S2A). The principal component analysis (PCA) plot based on the gene expression profiles exhibited a clear separation between the left and right gonads in each stage (Figure 2A). To identify the differentially expressed genes (DEGs), we conducted a differential expression analysis between left and right gonads in each stage. A total of 855, 123, 378 and 301 left-biased DEGs (LBGs) and 515, 30, 423 and 230 right-biased DEGs (RBGs) were identified at E4.5, E5.5, E7 and E10, respectively (Supplementary Figure S2B; Supplementary Tables S5–S8).
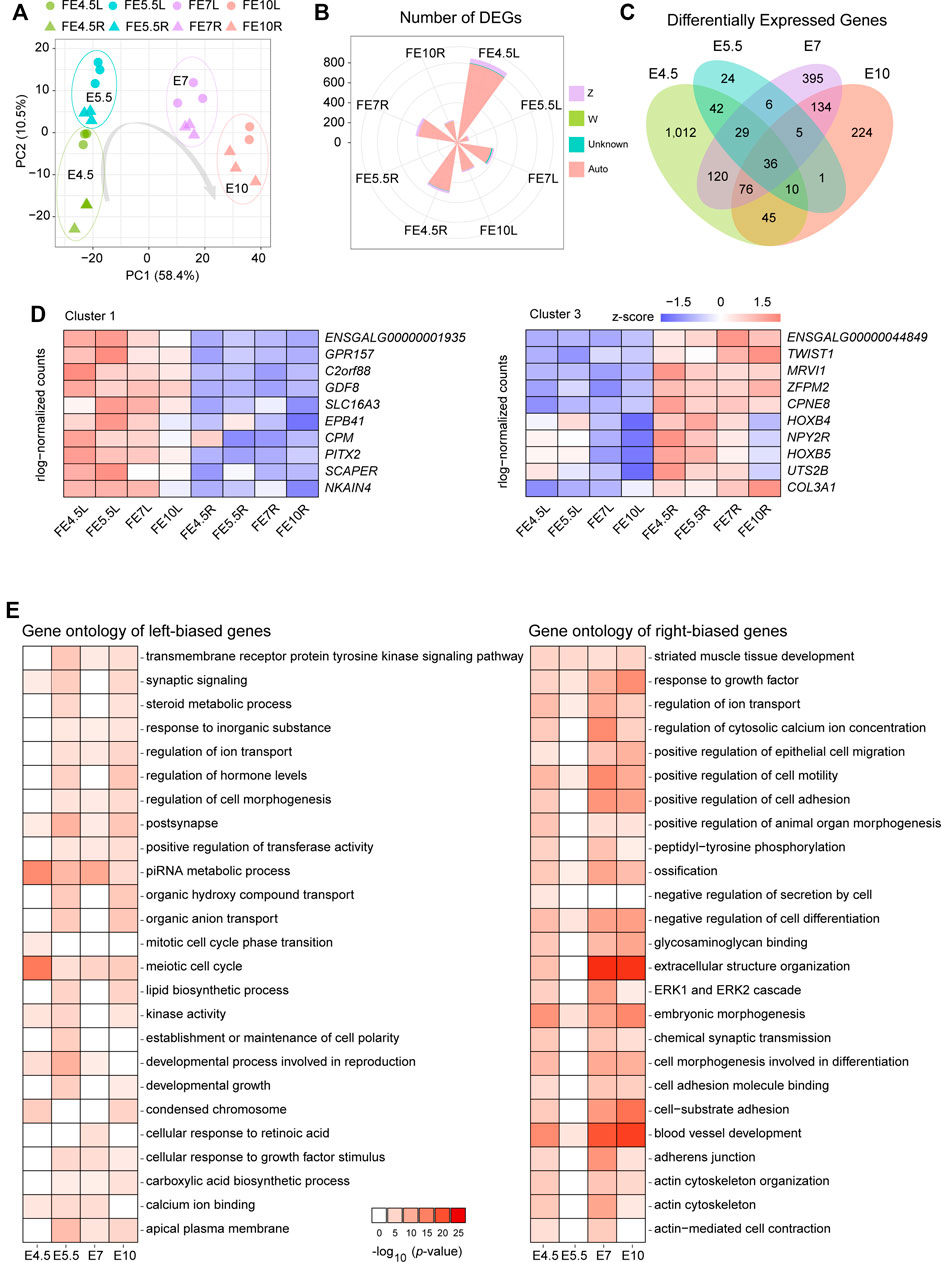
FIGURE 2. Analysis of RNA-seq data from the left and right gonads. (A) PCA plot of RNA-seq data of left and right gonads at different developmental stages; (B) The number of DEGs and chromosomal allocation in the left and right gonads; (C) Venn diagrams show the shared and unique DEGs obtained from each pairwise comparison between the left and right gonads, including four groups: left gonads and right gonads of female embryonic day 4.5(FE4.5L vs FE4.5R), FE5.5L vs. FE5.5R, FE7L vs. FE7R, and FE10L vs. FE10R); (D) The gene expression patterns of cluster 1 (left) and cluster 3 (right); (E) Top 25 significantly enriched terms of genes among the LBGs (left) and RBGs (right). The depth of the colors represents the size of the p-value.
The majority of the DEGs clearly resided on the autosome, and approximately 5% of DEGs were located on the Z chromosome (Figure 2B, Supplementary Figure S2C). The distribution of the DEGs between the bilateral gonads was revealed using a Venn diagram (Figure 2C, Supplementary Figures S1E,F), and 36 common DEGs were identified in the four stages and shown to be located on the autosome (Supplementary Table S9); these common DEGs included 8 TFs (CALR3, CPEB1, DMRTB1, HOXB4, HOXB5, PITX2, TWIST1 and ZFPM2). We identified three clusters of these 36 common DEGs based on a correlation analysis of gene expression (Figure 2D, Supplementary Figure S2D). The heatmap shows the gene expression pattern in the bilateral gonads across the four developmental stages. Clusters 1 and 2 comprise LBGs whose expression levels decreased or increased during the development of the left gonads, and cluster 3 mainly comprises RBGs. The changes in the gene expression CPM and SCAPER represent characteristic changes in cluster 1, while the dynamic changes in cluster 2 and cluster 3 are represented by changes in PIWIL1 and CPNE8 expression, respectively (Supplementary Figure S2E).
To better understand the functional consequences of all the DEGs, enrichment analysis of the LBGs and RBGs identified in all the stages was performed using Metascape software. A total of 25 significantly enriched gene ontology (GO) terms of these DEGs in the bilateral gonads were identified. The significant terms of the LBGs were related to meiotic cell cycle, piRNA metabolic process, regulation of cell morphogenesis, establishment or maintenance of cell polarity, and cellular response to retinoic acid, and these results indicated the role of these genes in sustaining growth and development in left gonads (Figure 2E). Moreover, the terms related to the RBGs were mainly enriched in extracellular structure organization, actin cytoskeleton, cell morphogenesis involved in differentiation and negative regulation of cell differentiation (Figure 2E).
Chromatin Accessibility Analysis
To understand how genomic regulatory elements control key genes that drive asymmetric gonadal development, we profiled the global chromatin accessibility patterns in both the left and right gonads during each stage using ATAC-seq. Our ATAC-seq data showed high quality in terms of both mapping rate and peak calling (Supplementary Table S10), and the three replicates examined from each stage exhibited great correlation (Figure 3A, Supplementary Figure S3A), indicating strong reproducibility. The PCA plot based on chromatin accessibility showed a clear separation between the left and right gonads in E5.5 and E7 (Figure 3B).
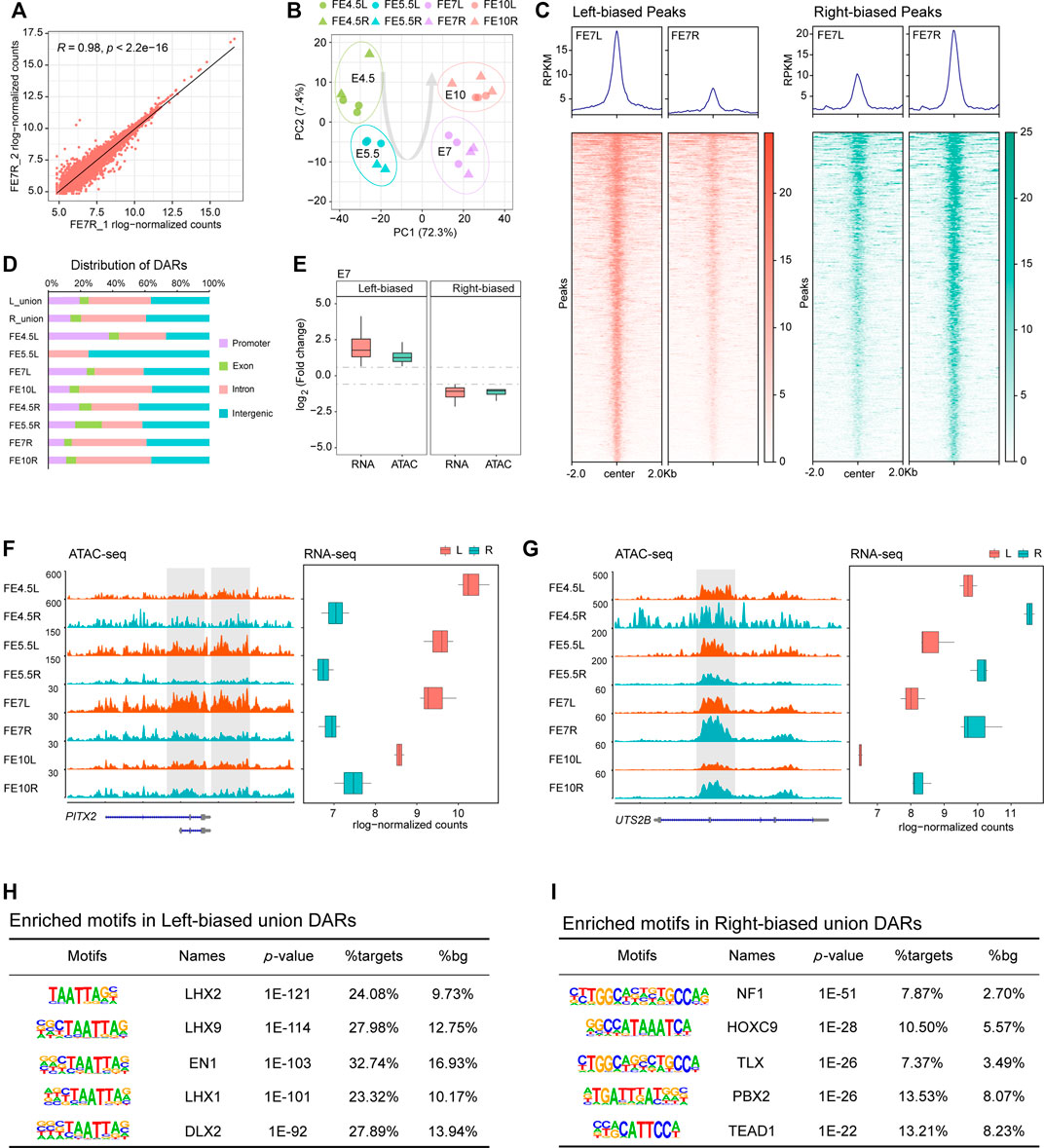
FIGURE 3. Differential chromatin accessibility between the left and right gonads. (A) Scatter plots of sample correlation show high similarities among right gonad duplicates at E7; (B) PCA plot of ATAC-seq data of left and right gonads at different developmental stages; (C) Bandplots (top) and heatmaps (bottom) showing the quantification of ATAC-seq data of left-biased and right-biased signals in gonads at E7; (D) The distribution of DARs in the genome; (E) Transcriptional changes in DEGs associated with DARs between the left and right gonads; (F,G) Example of left-biased DARs and their associated DEGs (PITX2), as well as right-biased DARs and their associated DEGs (UTS2B). ATAC-seq tracks are shown in the RPKM scale. The y axis of the RNA-seq boxplot shows the embryonic day, and the x axis shows the mean rlog-normalized counts; (H) Top enriched motifs in left union peaks; (I) Top enriched motifs in right union peaks.
Subsequently, we assessed the differentially accessible regions (DARs), indicated by ATAC-seq signals, between the bilateral gonads in each stage. We identified a total of 3,228 and 3,476 significant DARs in left and right gonadal development, respectively, which are shown by volcano plots (Supplementary Figure S3B). The heatmaps revealed that these significant DARs showed very clear side-specific patterns in each stage (Figure 3C, Supplementary Figures S3C,D). Because few DARs were identified at E5.5 (n = 18), only accessibility signals were displayed for the other three stages. Then, the genomic distribution of the DARs in the four developmental stages, the left-biased union DARs and the right-biased union DARs were annotated in their promoter, exon, intron and intergenic regions, and we found that the proportions of the left-biased and right-biased union DARs located in promoters were 19.5 and 13.8%, respectively (Figure 3D). More DARs were located in promoters in the left gonads, suggesting that more genes were activated.
We integrated ATAC-seq and RNA-seq to investigate the relationship between chromatin accessibility and gene expression during asymmetric gonadal development. As expected, we found a strong positive correlation between accessibility signatures and gene expression patterns based on calculated fold changes by assigning open chromatin regions to the nearest DEGs (Figure 3E, Supplementary Figures S3E,F). We found that PITX2 was highly expressed in the left gonads in the four developmental stages, which was accompanied by striking increases in chromatin accessibility (Figure 3F). Additionally, the gene expression and chromatin accessibility of UTS2B, a cell activity-related gene (Prosser et al., 2008), in the right gonads exhibited similar dynamic changes (Figure 3G).
Given that TFs are major factors that drive gene expression by binding to associated regulatory elements, we performed a comparative motif analysis between left-biased and right-biased union DARs using HOMER. We found that the HOXC9 and TEAD gene families were enriched the in right-biased DARs (Figure 3I). Among the left-biased union DARs, the LIM homeodomain (Lhx) family of transcription factors was significantly enriched. Notably, the motif of LHX9, which is related to ovary development in mammals (Birk et al., 2000), was identified as the second most highly enriched motif (Figure 3H).
The Analysis of LHX9 in Asymmetric Gonadal Development
Considering the important biological functions of LHX9, we first examined the changes in its gene expression in the RNA-seq data to validate our motif screening results. Transcription of the LHX9 gene remained high but was continuously downregulated in the bilateral differentiated gonads throughout embryonic development (Figure 4A). Notably, compared with the right gonads, the left gonads exhibited significantly lower LHX9 transcription at E7, suggesting crucial regulatory effects of LHX9 on the developmental trajectory from E5.5 to E7 during gonadogenesis. Moreover, we observed clear differences in chromatin accessibility in the gene body and promoter of LHX9 (Figure 4B). In light of the differential expression patterns and side-biased motif enrichment, we hypothesized that LHX9 might change its target genes over time by binding to open chromatin regions that change during development.
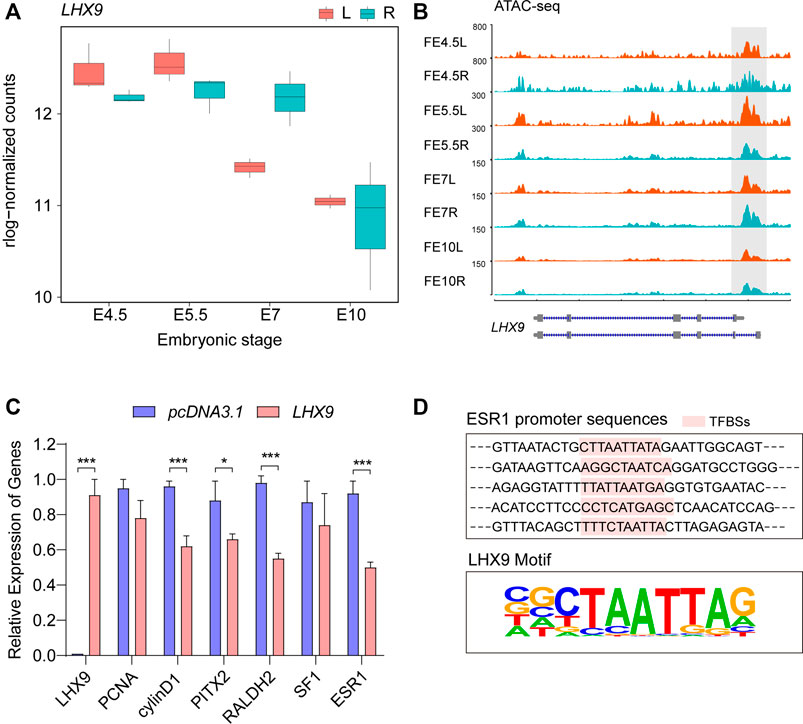
FIGURE 4. Comprehensive analysis of LHX9 expression during gonadal development. (A) The expression of LHX9 at each stage. The x axis shows the embryonic day, and the y axis shows the rlog-normalized counts; (B) Chromatin accessibility of the LHX9 gene at each stage; (C) Changes in the relative expression of genes related to asymmetric gonadal development in cells overexpressing LHX9; (D) The specific TF (LHX9) binding sites (TFBSs) in the promoter regions of ESR1.
To validate our motif enrichment results and further explore the role of LHX9 in gonadal development, we used an overexpression vector to regulate the expression of LHX9 in female left gonadal cells. The qPCR assay was performed to evaluate the effect of LHX9 on gonadal cells at 48 h. We also assessed the expression of PITX2, RALDH2, cyclin D1 and ESR1, which are involved in asymmetric gonadal development, in cells overexpressing LHX9 and found that their expression levels were significantly downregulated (Figure 4C). Given that ESR1 is a gene responsible for cortex formation of the left gonad (Guioli et al., 2020), we predicted the potential binding sites of LHX9 in the ESR1 promoter. Multiple potential LHX9 binding sites were found in the promoter region of ESR1 (Figure 4D). ESR1 is a left-biased gene throughout asymmetric gonadal development in female embryos, and chromatin accessibility of ESR1 promoter increases from E5 to E7 (Supplementary Figure S4). On the basis of these observations, we concluded that LHX9 could affect asymmetric gonadal development by regulating side-biased transcriptional and epigenetic patterns at E7.
Discussion
Chicken embryos are an excellent model for studying cell biology and embryonic organ development (Davey and Tickle, 2007; Vergara and Canto-Soler, 2012; Seal et al., 2019; Vilches-Moure, 2019). Gonadal development in female chicken embryos is asymmetric, and only the left gonad develops into a functional ovary (Smith and Sinclair, 2004; González-Morán, 2011), and the right gonad remains a steroidogenic organ (Narbaitz and Kolodny, 1964; Samar et al., 1983). This biological phenomenon suggests that regulatory factors related to ovarian development are silent in the right gonads. Therefore, studying the particular mechanisms underlying gonadal development in female chickens is of great significance for studies about sexual development disorders and infertility (Wilhelm and Koopman, 2006; Guercio and Rey, 2014). However, the molecular mechanisms underlying asymmetric gonadal development are still poorly understood. Here, we performed time-course RNA-seq and ATAC-seq experiments in bilateral gonads of female chicken embryos and provided a dynamic profile of transcriptional and chromatin accessibility patterns throughout development. The study elucidated the regulatory network of asymmetric gonadal development and identified several candidate genes and regulatory elements. Significantly, we showed that the LHX9 TF could drive left gonadal development by binding to left-biased DARs and preliminary verified its basic function. The present work provided unique insights into the regulatory mechanisms underlying asymmetric gonadal development.
Due to the limitations of sample availability and sequencing technologies (Wan et al., 2017; Yu et al., 2017; Zou et al., 2020), many previous studies failed to identify the functional genes and elucidate the regulatory mechanisms underlying asymmetric gonadal development. Here, bilateral gonads were harvested from female chicken embryos for sequencing at four developmental stages (E4.5, E5.5, E7 and E10). Each sample library was derived from a single gonad from an independent individual, and this approach is more refined than previous approaches involving the sequencing of pooled samples from multiple individuals. Notably, we obtained sequencing data of bilateral gonads at E4.5; thus, our work is a pioneering study that explored the earliest available stages of gonadal development. Despite the lack of clear differences in morphology between the left and right gonads at E4.5, we observed obvious transcriptional changes, suggesting that the underlying molecular changes that cause developmental discrepancies already occurred. By integrating RNA-seq and ATAC-seq data of female bilateral gonads from different developmental stages, active genes and open chromatin regions were characterized; these findings can contribute to clearly exploring functional genes and detailed regulatory patterns and can provide more accurate guidance for studies of asymmetric gonadal development.
Gonadal morphological differences mainly occur due to increased cell number, proliferation rate and meiosis in the left ovary (Ishimaru et al., 2008; Intarapat and Stern, 2013; de Melo Bernardo et al., 2015). Previous studies proposed that the germ cells were lost by cell death in the right embryonic gonads (Ukeshima, 1996). However, the number of PGCs in the right ovary increases until hatching (González-Morán, 2011), suggesting that any loss in PGC number is compensated by germ cell proliferation. GO analysis showed that the LBGs are mainly enriched in cell proliferation- and cycle-related items, which suggests that LBGs are related to organ development and actively drive gonadal development (Smith et al., 2008a; Rodríguez-León et al., 2008; Thomson and Lin, 2009; Yu et al., 2013; Rengaraj et al., 2014). Previous studies revealed hundreds of side-biased genes in asymmetric gonadal development at the embryonic (E6, E9, and E12) and posthatching stages (D1); these genes include DAZL, GDF8, PITX2, PIWIL1, and TDRD5 (Intarapat and Stern, 2013; Wan et al., 2017; Zou et al., 2020), and these findings are consistent with our work. As the factor that determines left-right bias in vertebrates (Amand et al., 1998; Piedra et al., 1998; Yoshioka et al., 1998; Schweickert et al., 2000; Dagle et al., 2003; Gormley and Nascone-Yoder, 2003; Grimes and Burdine, 2017; Hamada, 2020; Little and Norris, 2021), PITX2 has been proven to play a key role in gonadal lateralization and morphogenesis by regulating RALDH2, SF1 and cyclin D1 expression, promoting cell proliferation and reorganizing the cytoskeleton. Besides, overexpression of PITX2 can rescue the degeneration of the right ovary during gonadogenesis (Guioli and Lovell-Badge, 2007; Ishimaru et al., 2008; Rodríguez-León et al., 2008). Based on gene expression in the four developmental stages, we identified a cluster of genes among 36 common DEGs whose expression was most highly correlated with PITX2 expression. The heatmap of cluster 1 pattern revealed that the gene expression of left gonad increased during development, which may be indicative of its facilitation actions in left gonadal development. Among these genes, we found that the SCAPER, CPM and GDF8 genes were related to cell cycle progression, differentiation, and proliferation (Rehli et al., 1995; McPherron et al., 1997; Tsang et al., 2007), which further confirmed and narrowed the scope of candidate genes. Nevertheless, the exact roles of these candidate DEGs during asymmetric gonadal development require further investigations. In short, we provided a distinct regulatory network of asymmetric gonadal development.
Although some advances have been made in assessing core genes related to asymmetric gonadal development, the upstream regulatory elements remain largely unknown. To fill this knowledge gap, ATAC-seq experiments were used to identify key regulatory drivers responsible for gonadal development. Studying dynamic changes in chromatin accessibility in gene regulatory regions during transcriptional activation provides insight into changes in cell fate. Interestingly, the PC1 in both RNA-seq and ATAC-seq PCA plots separated the samples into undifferentiated and differentiated. Additionally, E5.5 showed fewer DEGs and DARs than the rest of them, which means that the ovarian asymmetry is strongly induced after the onset of sex differentiation (E6). A previous study showed that the formation of the embryonic gonadal cortex was induced by estrogen (Guioli et al., 2020). Indeed, the estrogen synthesis was induced by the aromatase in the female chick embryo (Nakabayashi et al., 1998; Lambeth et al., 2016). Aromatase shows sexual dimorphism and gonads begin morphological differentiation into testes or ovaries at E6 in the chicken embryos (Ayers et al., 2015), which means that the ovary of chicken embryos produced estrogen from the start of female-specific differentiation (Guioli et al., 2020). Thus, the unique side-dependent developmental pattern of female bird gonad was affected by sex determination or differentiation pathways. Integration of RNA-seq and ATAC-seq data can facilitate the characterization of the chromatin accessibility and gene expression patterns that are pivotal for asymmetric gonadal development. In this study, the RNA-seq data showed that more DEGs were identified in the left gonad, which is consistent with the high proportion of left-biased DARs in promoter regions as shown by ATAC-seq. This result indicated that more genes remained active in the left gonad, thereby promoting left gonadal development. In general, gene expression is regulated by TFs that bind to DNA in open chromatin regions (Thurman et al., 2012; MacNeil et al., 2015). Thus, we predicted putative TF binding motifs in DARs. HOXC9, which acts as a regulator of cell proliferation and cycle related gene expression and as a mediator of RA action in neuroblastoma cells (Mao et al., 2011; Stoll et al., 2011; Wang et al., 2013), was the second most highly enriched motif in the right-biased DARs. In addition, the TEAD gene family, which includes core members of the Hippo pathway, functions mainly to inhibit proliferation and to promote apoptosis, thereby limiting the overgrowth of organs (Harvey and Tapon, 2007; Harvey et al., 2013); members of this gene family were also enriched in the right-biased DARs. Notably, the TFs of the LIM homeobox gene family were enriched in the left-biased DARs, which suggested that the LHX family probably binds to open chromatin regions during gonadal development. Several previous works suggested that the LIM homeobox gene family plays key roles in regulating gene expression patterns throughout the body during development in invertebrates and vertebrates (Curtiss and Heilig, 1998).
Differential motif analysis confirmed that LHX9 preferentially bound to open chromatin regions during left gonadal development. Interestingly, LHX9, which is required for gonadogenesis in mice, promotes the proliferation of the early somatic cell population (Birk et al., 2000). However, previous studies did not sufficiently elucidate the function of LHX9 in asymmetric gonadal development (Guioli and Lovell-Badge, 2007; Ishimaru et al., 2008). A recent study showed that LHX9, which is strongly expressed in the cortex overlying chicken gonads, is a downstream target of hedgehog signaling (the upstream pathway most closely involved in triggering and orchestrating gonadogenesis in chickens) (Yoshino et al., 2016); these results indicated that LHX9 is required in the normal development of gonads. The expression of the TF that regulates LHX9 expression remains constant throughout bilateral gonadal development; however, the TF has a distinct preference for binding the genome in the left gonads, suggesting that chromatin accessibility determines local affinity for the LHX9 gene. Research has suggested that prior to the binding of other TFs, pioneer TFs recognize specific sequences in DNA binding sites and directly recruit chromatin-remodeling complexes, actively increase or decrease local chromatin accessibility, and then precisely control gene expression (Zaret and Carroll, 2011). The switch in LHX9 binding sites may explain how LHX9 performs different functions in bilateral gonads.
Our in vitro experiments revealed that genes related to asymmetric gonadal development were altered with LHX9 overexpression, which means that LHX9 may target these genes to regulate gonadal development at E7. Previous work showed that the ovarian L-R asymmetry is strongly induced by estrogen, which acts as the major promoting signal for cortex development. ESR1 (ERα), depending on its activity within the left epithelial cells, is the main transducer of estrogen signal in cortex formation of the left gonad (Guioli et al., 2020). Interestingly, ESR1 is a left-biased gene throughout asymmetric gonadal development in female embryos, and chromatin accessibility of ESR1 promoter increases from E5 to E7. As development progressed, the left gonads of the female chicken embryos were markedly larger than the right gonads until E7, which accompanied by the left gonads exhibited significantly lower LHX9 transcription. In addition, ERα and LHX9 co-localize only in cortical cells of the left gonad at E7 (Guioli and Lovell-Badge, 2007). Further, we predicted the potential binding sites of LHX9 in the ESR1 promoter using Homer. As expected, we found multiple LHX9 potential binding sites in the promoter regions of ESR1. ESR1 was downregulated by LHX9 in female gonad cells at E7, which suggested that LHX9 could act upstream of ESR1. Thus, decreased transcription of LHX9 on the left side, especially at E7, should be an important factor that drives asymmetric gonadal development via regulating ESR1 activity. It is necessary for future research to confirm the regulation and function of LHX9 in vivo in gonadal epithelium formation through overexpressing and knocking down/out experiments. Bulk RNA-seq mainly reflects the averaged gene expression across thousands of cells. Gonad RNA-seq could not reveal the subtle spatio-temporal transcriptional pattern of LHX9 and ESR1 in gonadal epithelial cells, since the two candidates might express in only a certain cell type. Single-cell RNA-sequencing technologies can study cell heterogeneity and analyze gene expression at individual cells, which may contribute to a better understanding of the role of LHX9 in chicken embryonic gonads. Further study at the single-cell level would be more informative to explore the distribution of LHX9 in different gonadal lineages. Integrating matched RNA-seq, ATAC-seq and single cell RNA-seq will be helpful to elucidate molecular basis underlying asymmetric gonadal development. Although the function of LHX9 in asymmetric gonadal development is preliminarily understood, the affinity of LHX9 for local chromatin is still not very clear. Therefore, follow-up work should focus on exploring the regulation of pioneer TFs that in turn regulate chromatin.
Previous studies have demonstrated the effect of transcription factors on asymmetric development. Here, we first revealed the contribution of dynamic changes in chromatin accessibility to these processes. In summary, we established a high-resolution profile of transcriptional and chromatin accessibility patterns during chicken gonadal development, revealed a functional relationship between chromatin accessibility and candidate gene expression, and identified a promising TF, LHX9, that may regulate the left gonad to develop into a functional ovary. Together, our findings will enable a better understanding of ovarian development after sex determination and the mechanisms underlying asymmetric patterns of development in vertebrates. Additionally, our findings can also serve as a potential guide for establishing biological models to probe the causes of ovary-related diseases in humans.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: National Center for Biotechnology Information (NCBI) BioProject database under accession number PRJNA756705.
Ethics Statement
The animal study was reviewed and approved by The Animal Welfare Committee of China Agricultural University. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author Contributions
NY conceived and designed the project. JbL, JZ, and JyL performed the experiments and collected samples. JbL, CS, and GY analyzed the data. JbL and GY wrote the manuscript. NY revised the manuscript. All authors read and approved the final draft.
Funding
This work was funded in part by grants from China Agriculture Research Systems (CARS-40).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, orclaim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2022.832132/full#supplementary-material
References
Amand, T. R. S., Ra, J., Zhang, Y., Hu, Y., Baber, S. I., Qiu, M., et al. (1998). Cloning and Expression Pattern of ChickenPitx2:A New Component in the SHH Signaling Pathway Controlling Embryonic Heart Looping. Biochem. Biophysical Res. Commun. 247 (1), 100–105. doi:10.1006/bbrc.1998.8740
Anders, S., Pyl, P. T., and Huber, W. (2015). HTSeq--a Python Framework to Work with High-Throughput Sequencing Data. Bioinformatics 31 (2), 166–169. doi:10.1093/bioinformatics/btu638
Arboleda, V. A., Sandberg, D. E., and Vilain, E. (2014). DSDs: Genetics, Underlying Pathologies and Psychosexual Differentiation. Nat. Rev. Endocrinol. 10 (10), 603–615. doi:10.1038/nrendo.2014.130
Ayers, K. L., Lambeth, L. S., Davidson, N. M., Sinclair, A. H., Oshlack, A., and Smith, C. A. (2015). Identification of Candidate Gonadal Sex Differentiation Genes in the Chicken Embryo Using RNA-Seq. BMC Genomics 16 (1), 704. doi:10.1186/s12864-015-1886-5
Birk, O. S., Casiano, D. E., Wassif, C. A., Cogliati, T., Zhao, L., Zhao, Y., et al. (2000). The LIM Homeobox Gene Lhx9 Is Essential for Mouse Gonad Formation. Nature 403 (6772), 909–913. doi:10.1038/35002622
Buenrostro, J. D., Giresi, P. G., Zaba, L. C., Chang, H. Y., and Greenleaf, W. J. (2013). Transposition of Native Chromatin for Fast and Sensitive Epigenomic Profiling of Open Chromatin, DNA-Binding Proteins and Nucleosome Position. Nat. Methods 10 (12), 1213–1218. doi:10.1038/nmeth.2688
Burton, A., and Torres-Padilla, M.-E. (2014). Chromatin Dynamics in the Regulation of Cell Fate Allocation during Early Embryogenesis. Nat. Rev. Mol. Cel Biol. 15 (11), 723–735. doi:10.1038/nrm3885
Carré-Eusèbe, D., Coudouel, N., and Magre, S. (2009). OVEX1, a Novel Chicken Endogenous Retrovirus with Sex-specific and Left-Right Asymmetrical Expression in Gonads. Retrovirology 6, 59. doi:10.1186/1742-4690-6-59
Corces, M. R., Trevino, A. E., Hamilton, E. G., Greenside, P. G., Sinnott-Armstrong, N. A., Vesuna, S., et al. (2017). An Improved ATAC-Seq Protocol Reduces Background and Enables Interrogation of Frozen Tissues. Nat. Methods 14 (10), 959–962. doi:10.1038/nmeth.4396
Curtiss, J., and Heilig, J. S. (1998). DeLIMiting Development. Bioessays 20 (1), 58–69. doi:10.1002/(sici)1521-1878(199801)20:1<58::aid-bies9>3.0.co;2-o
Dagle, J. M., Sabel, J. L., Littig, J. L., Sutherland, L. B., Kolker, S. J., and Weeks, D. L. (2003). Pitx2c Attenuation Results in Cardiac Defects and Abnormalities of Intestinal Orientation in Developing Xenopus laevis. Develop. Biol. 262 (2), 268–281. doi:10.1016/s0012-1606(03)00389-0
Davey, M. G., and Tickle, C. (2007). The Chicken as a Model for Embryonic Development. Cytogenet. Genome Res. 117 (1-4), 231–239. doi:10.1159/000103184
de Melo Bernardo, A., Heeren, A. M., van Iperen, L., Fernandes, M. G., He, N., Anjie, S., et al. (2015). Meiotic Wave Adds Extra Asymmetry to the Development of Female Chicken Gonads. Mol. Reprod. Dev. 82 (10), 774–786. doi:10.1002/mrd.22516
González-Morán, M. G. (2011). Histological and Stereological Changes in Growing and Regressing Chicken Ovaries during Development. Anat. Rec. 294 (5), 893–904. doi:10.1002/ar.21364
Gormley, J. P., and Nascone-Yoder, N. M. (2003). Left and Right Contributions to the Xenopus Heart: Implications for Asymmetric Morphogenesis. Develop. Genes Evol. 213 (8), 390–398. doi:10.1007/s00427-003-0337-5
Grimes, D. T., and Burdine, R. D. (2017). Left-Right Patterning: Breaking Symmetry to Asymmetric Morphogenesis. Trends Genet. 33 (9), 616–628. doi:10.1016/j.tig.2017.06.004
Guercio, G., and Rey, R. A. (2014). Fertility Issues in the Management of Patients with Disorders of Sex Development. Endocr. Dev. 27, 87–98. doi:10.1159/000363633
Guioli, S., and Lovell-Badge, R. (2007). PITX2 Controls Asymmetric Gonadal Development in Both Sexes of the Chick and Can rescue the Degeneration of the Right Ovary. Development 134 (23), 4199–4208. doi:10.1242/dev.010249
Guioli, S., Nandi, S., Zhao, D., Burgess-Shannon, J., Lovell-Badge, R., and Clinton, M. (2014). Gonadal Asymmetry and Sex Determination in Birds. Sex. Dev. 8 (5), 227–242. doi:10.1159/000358406
Guioli, S., Zhao, D., Nandi, S., Clinton, M., and Lovell-Badge, R. (2020). In the Chick Embryo, Estrogen Can Induce Chromosomally Male ZZ Left Gonad Epithelial Cells to Form an Ovarian Cortex, Which Supports Oogenesis. Development 147 (4), dev181693. doi:10.1242/dev.181693
Hamada, H. (2020). Molecular and Cellular Basis of Left-Right Asymmetry in Vertebrates. Proc. Jpn. Acad. Ser. B: Phys. Biol. Sci. 96 (7), 273–296. doi:10.2183/pjab.96.021
Hamburger, V., and Hamilton, H. L. (1951). A Series of normal Stages in the Development of the Chick Embryo. J. Morphol. 88 (1), 49–92. doi:10.1002/jmor.1050880104
Harvey, K. F., Zhang, X., and Thomas, D. M. (2013). The Hippo Pathway and Human Cancer. Nat. Rev. Cancer 13 (4), 246–257. doi:10.1038/nrc3458
Harvey, K., and Tapon, N. (2007). The Salvador-Warts-Hippo Pathway - an Emerging Tumour-Suppressor Network. Nat. Rev. Cancer 7 (3), 182–191. doi:10.1038/nrc2070
Hoshino, A., Koide, M., Ono, T., and Yasugi, S. (2005). Sex-specific and Left-Right Asymmetric Expression Pattern of Bmp7 in the Gonad of normal and Sex-Reversed Chicken Embryos. Dev. Growth Differ. 47 (2), 65–74. doi:10.1111/j.1440-169x.2004.00783.x
Intarapat, S., and Stern, C. D. (2013). Sexually Dimorphic and Sex-independent Left-Right Asymmetries in Chicken Embryonic Gonads. PloS one 8 (7), e69893. doi:10.1371/journal.pone.0069893
Ishimaru, Y., Komatsu, T., Kasahara, M., Katoh-Fukui, Y., Ogawa, H., Toyama, Y., et al. (2008). Mechanism of Asymmetric Ovarian Development in Chick Embryos. Development 135 (4), 677–685. doi:10.1242/dev.012856
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: a Fast Spliced Aligner with Low Memory Requirements. Nat. Methods 12 (4), 357–360. doi:10.1038/nmeth.3317
Lambeth, L. S., Morris, K. R., Wise, T. G., Cummins, D. M., O'Neil, T. E., Cao, Y., et al. (2016). Transgenic Chickens Overexpressing Aromatase Have High Estrogen Levels but Maintain a Predominantly Male Phenotype. Endocrinology 157 (1), 83–90. doi:10.1210/en.2015-1697
Li, H., and Durbin, R. (2009). Fast and Accurate Short Read Alignment with Burrows-Wheeler Transform. Bioinformatics 25 (14), 1754–1760. doi:10.1093/bioinformatics/btp324
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The Sequence Alignment/Map Format and SAMtools. Bioinformatics 25 (16), 2078–2079. doi:10.1093/bioinformatics/btp352
Little, R. B., and Norris, D. P. (2021). Right, Left and Cilia: How Asymmetry Is Established. Semin. Cel Develop. Biol. 110, 11–18. doi:10.1016/j.semcdb.2020.06.003
Love, M. I., Huber, W., and Anders, S. (2014). Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 15 (12), 550. doi:10.1186/s13059-014-0550-8
MacNeil, L. T., Pons, C., Arda, H. E., Giese, G. E., Myers, C. L., and Walhout, A. J. M. (2015). Transcription Factor Activity Mapping of a Tissue-specific In Vivo Gene Regulatory Network. Cel Syst. 1 (2), 152–162. doi:10.1016/j.cels.2015.08.003
Mao, L., Ding, J., Zha, Y., Yang, L., McCarthy, B. A., King, W., et al. (2011). HOXC9 Links Cell-Cycle Exit and Neuronal Differentiation and Is a Prognostic Marker in Neuroblastoma. Cancer Res. 71 (12), 4314–4324. doi:10.1158/0008-5472.Can-11-0051
McPherron, A. C., Lawler, A. M., and Lee, S.-J. (1997). Regulation of Skeletal Muscle Mass in Mice by a New TGF-P Superfamily Member. Nature 387 (6628), 83–90. doi:10.1038/387083a0
Monsoro-Burq, A. H., and Levin, M. (2018). Avian Models and the Study of Invariant Asymmetry: How the Chicken and the Egg Taught Us to Tell Right from Left. Int. J. Dev. Biol. 62 (1-2-3), 63–77. doi:10.1387/ijdb.180047ml
Nakabayashi, O., Kikuchi, H., Kikuchi, T., and Mizuno, S. (1998). Differential Expression of Genes for Aromatase and Estrogen Receptor during the Gonadal Development in Chicken Embryos. J. Mol. Endocrinol. 20 (2), 193–202. doi:10.1677/jme.0.0200193
Nakamura, Y., Yamamoto, Y., Usui, F., Mushika, T., Ono, T., Setioko, A. R., et al. (2007). Migration and Proliferation of Primordial Germ Cells in the Early Chicken Embryo. Poult. Sci. 86 (10), 2182–2193. doi:10.1093/ps/86.10.2182
Narbaitz, R., and Kolodny, L. (1964). ?5-3?-Hydroxysteroid Dehydrogenase in Differentiating Chick Gonads. Z. Zellforschung 63, 612–617. doi:10.1007/BF00339909
Piedra, M. E., Icardo, J. M., Albajar, M., Rodriguez-Rey, J. C., and Ros, M. A. (1998). Pitx2 Participates in the Late Phase of the Pathway Controlling Left-Right Asymmetry. Cell 94 (3), 319–324. doi:10.1016/s0092-8674(00)81475-0
Prosser, H. C. G., Forster, M. E., Richards, A. M., and Pemberton, C. J. (2008). Urotensin II and Urotensin II-Related Peptide (URP) in Cardiac Ischemia-Reperfusion Injury. Peptides 29 (5), 770–777. doi:10.1016/j.peptides.2007.08.013
Ramírez, F., Dündar, F., Diehl, S., Grüning, B. A., and Manke, T. (2014). deepTools: a Flexible Platform for Exploring Deep-Sequencing Data. Nucleic Acids Res. 42 (Web Server issue), W187–W191. doi:10.1093/nar/gku365
Reed, K. J., and Sinclair, A. H. (2002). FET-1: a Novel W-Linked, Female Specific Gene Up-Regulated in the Embryonic Chicken Ovary. Mech. Develop. 119 (Suppl. 1), S87–S90. doi:10.1016/s0925-4773(03)00097-2
Rehli, M., Krause, S. W., Kreutz, M., and Andreesen, R. (1995). Carboxypeptidase M Is Identical to the MAX.1 Antigen and its Expression Is Associated with Monocyte to Macrophage Differentiation. J. Biol. Chem. 270 (26), 15644–15649. doi:10.1074/jbc.270.26.15644
Rengaraj, D., Lee, S. I., Park, T. S., Lee, H. J., Kim, Y. M., Sohn, Y. A., et al. (2014). Small Non-coding RNA Profiling and the Role of piRNA Pathway Genes in the protection of Chicken Primordial Germ Cells. BMC Genomics 15 (1), 757. doi:10.1186/1471-2164-15-757
Rivera, C. M., and Ren, B. (2013). Mapping Human Epigenomes. Cell 155 (1), 39–55. doi:10.1016/j.cell.2013.09.011
Rodriguez-Leon, J., Esteban, C. R., Marti, M., Santiago-Josefat, B., Dubova, I., Rubiralta, X., et al. (2008). Pitx2 Regulates Gonad Morphogenesis. Proc. Natl. Acad. Sci. 105 (32), 11242–11247. doi:10.1073/pnas.0804904105
Samar, M. E., Avila, R. E., and de Fabro, S. P. (1983). Cytochemical Study of the Ovary Cells in the Chick Embryo. Folia Histochem. Cytochem. (Krakow) 21 (3-4), 173–180.
Scheider, J., Afonso-Grunz, F., Hoffmeier, K., Horres, R., Groher, F., Rycak, L., et al. (2014). Gene Expression of Chicken Gonads Is Sex- and Side-specific. Sex. Dev. 8 (4), 178–191. doi:10.1159/000362259
Schweickert, A., Campione, M., Steinbeisser, H., and Blum, M. (2000). Pitx2 Isoforms: Involvement of Pitx2c but Not Pitx2a or Pitx2b in Vertebrate Left-Right Asymmetry. Mech. Develop. 90 (1), 41–51. doi:10.1016/s0925-4773(99)00227-0
Seal, H. E., Lilian, S. J., Popratiloff, A., Hirsch, J. C., and Peusner, K. D. (2019). Implementing the Chick Embryo Model to Study Vestibular Developmental Disorders. J. Neurophysiol. 122 (6), 2272–2283. doi:10.1152/jn.00434.2019
Smith, C. A., Roeszler, K. N., Bowles, J., Koopman, P., and Sinclair, A. H. (2008a). Onset of Meiosis in the Chicken Embryo; Evidence of a Role for Retinoic Acid. BMC Develop. Biol. 8, 85. doi:10.1186/1471-213x-8-85
Smith, C. A., Roeszler, K. N., Hudson, Q. J., and Sinclair, A. H. (2007). Avian Sex Determination: what, when and where? Cytogenet. Genome Res. 117 (1-4), 165–173. doi:10.1159/000103177
Smith, C. A., Shoemaker, C. M., Roeszler, K. N., Queen, J., Crews, D., and Sinclair, A. H. (2008b). Cloning and Expression of R-Spondin1in Different Vertebrates Suggests a Conserved Role in Ovarian Development. BMC Dev. Biol. 8, 72. doi:10.1186/1471-213x-8-72
Smith, C. A., and Sinclair, A. H. (2004). Sex Determination: Insights from the Chicken. Bioessays 26 (2), 120–132. doi:10.1002/bies.10400
Stoll, S. J., Bartsch, S., Augustin, H. G., and Kroll, J. (2011). The Transcription Factor HOXC9 Regulates Endothelial Cell Quiescence and Vascular Morphogenesis in Zebrafish via Inhibition of Interleukin 8. Circ. Res. 108 (11), 1367–1377. doi:10.1161/circresaha.111.244095
Thomson, T., and Lin, H. (2009). The Biogenesis and Function of PIWI Proteins and piRNAs: Progress and prospect. Annu. Rev. Cel Dev. Biol. 25, 355–376. doi:10.1146/annurev.cellbio.24.110707.175327
Thurman, R. E., Rynes, E., Humbert, R., Vierstra, J., Maurano, M. T., Haugen, E., et al. (2012). The Accessible Chromatin Landscape of the Human Genome. Nature 489 (7414), 75–82. doi:10.1038/nature11232
Tsang, W. Y., Wang, L., Chen, Z., Sánchez, I., and Dynlacht, B. D. (2007). SCAPER, a Novel Cyclin A-Interacting Protein that Regulates Cell Cycle Progression. J. Cel Biol. 178 (4), 621–633. doi:10.1083/jcb.200701166
Ukeshima, A. (1996). Germ Cell Death in the Degenerating Right Ovary of the Chick Embryo. Zoolog. Sci. 13 (4), 559–563. doi:10.2108/zsj.13.559
Van Limborgh, J. (1968). [The First Sign of Sexual Differentiation of the Gonads in Chick Embryos]. Arch. Anat. Microsc. Morphol. Exp. 57 (1), 79–90.
Vergara, M. N., and Canto-Soler, M. V. (2012). Rediscovering the Chick Embryo as a Model to Study Retinal Development. Neural Dev. 7, 22. doi:10.1186/1749-8104-7-22
Vilches-Moure, J. G. (2019). Embryonic Chicken (Gallus gallus Domesticus) as a Model of Cardiac Biology and Development. Comp. Med. 69 (3), 184–203. doi:10.30802/aalas-cm-18-000061
Wan, Z., Lu, Y., Rui, L., Yu, X., Yang, F., Tu, C., et al. (2017). Gene Expression Profiling Reveals Potential Players of Left-Right Asymmetry in Female Chicken Gonads. Ijms 18 (6), 1299. doi:10.3390/ijms18061299
Wang, X., Choi, J.-H., Ding, J., Yang, L., Ngoka, L. C., Lee, E. J., et al. (2013). HOXC9 Directly Regulates Distinct Sets of Genes to Coordinate Diverse Cellular Processes during Neuronal Differentiation. BMC Genomics 14, 830. doi:10.1186/1471-2164-14-830
Wilhelm, D., and Koopman, P. (2006). The Makings of Maleness: towards an Integrated View of Male Sexual Development. Nat. Rev. Genet. 7 (8), 620–631. doi:10.1038/nrg1903
Wu, J., Huang, B., Chen, H., Yin, Q., Liu, Y., Xiang, Y., et al. (2016). The Landscape of Accessible Chromatin in Mammalian Preimplantation Embryos. Nature 534 (7609), 652–657. doi:10.1038/nature18606
Yoshino, T., Murai, H., and Saito, D. (2016). Hedgehog-BMP Signalling Establishes Dorsoventral Patterning in Lateral Plate Mesoderm to Trigger Gonadogenesis in Chicken Embryos. Nat. Commun. 7, 12561. doi:10.1038/ncomms12561
Yoshioka, H., Meno, C., Koshiba, K., Sugihara, M., Itoh, H., Ishimaru, Y., et al. (1998). Pitx2, a Bicoid-type Homeobox Gene, Is Involved in a Lefty-Signaling Pathway in Determination of Left-Right Asymmetry. Cell 94 (3), 299–305. doi:10.1016/s0092-8674(00)81473-7
Yu, J., Yan, L., Chen, Z., Li, H., Ying, S., Zhu, H., et al. (2017). Investigating Right Ovary Degeneration in Chick Embryos by Transcriptome Sequencing. J. Reprod. Develop. 63 (3), 295–303. doi:10.1262/jrd.2016-134
Yu, M., Yu, P., Leghari, I. H., Ge, C., Mi, Y., and Zhang, C. (2013). RALDH2, the Enzyme for Retinoic Acid Synthesis, Mediates Meiosis Initiation in Germ Cells of the Female Embryonic Chickens. Amino Acids 44 (2), 405–412. doi:10.1007/s00726-012-1343-6
Zaret, K. S., and Carroll, J. S. (2011). Pioneer Transcription Factors: Establishing Competence for Gene Expression. Genes Dev. 25 (21), 2227–2241. doi:10.1101/gad.176826.111
Zhang, Y., Liu, T., Meyer, C. A., Eeckhoute, J., Johnson, D. S., Bernstein, B. E., et al. (2008). Model-based Analysis of ChIP-Seq (MACS). Genome Biol. 9 (9), R137. doi:10.1186/gb-2008-9-9-r137
Zou, X., Wang, J., Qu, H., Lv, X. H., Shu, D. M., Wang, Y., et al. (2020). Comprehensive Analysis of miRNAs, lncRNAs, and mRNAs Reveals Potential Players of Sexually Dimorphic and Left-Right Asymmetry in Chicken Gonad during Gonadal Differentiation. Poult. Sci. 99 (5), 2696–2707. doi:10.1016/j.psj.2019.10.019
Keywords: chicken, gonad, asymmetry, RNA-seq, ATAC-seq, LHX9
Citation: Li J, Sun C, Zheng J, Li J, Yi G and Yang N (2022) Time-Course Transcriptional and Chromatin Accessibility Profiling Reveals Genes Associated With Asymmetrical Gonadal Development in Chicken Embryos. Front. Cell Dev. Biol. 10:832132. doi: 10.3389/fcell.2022.832132
Received: 09 December 2021; Accepted: 09 February 2022;
Published: 08 March 2022.
Edited by:
Harry Leitch, London Institute of Medical Sciences, United KingdomReviewed by:
Mike McGrew, University of Edinburgh, United KingdomMartin Andres Estermann, Monash University, Australia
Copyright © 2022 Li, Sun, Zheng, Li, Yi and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guoqiang Yi, eWlndW9xaWFuZ0BjYWFzLmNu; Ning Yang, bnlhbmdAY2F1LmVkdS5jbg==
 Jianbo Li
Jianbo Li Congjiao Sun
Congjiao Sun Jiangxia Zheng1
Jiangxia Zheng1 Junying Li
Junying Li Guoqiang Yi
Guoqiang Yi Ning Yang
Ning Yang