- 1Key Laboratory of Arrhythmias, Ministry of Education, East Hospital, Department of Anatomy and Neurobiology, Tongji University School of Medicine, Shanghai, China
- 2Department of Laboratory Animal Science, Fudan University, Shanghai, China
- 3State Key Laboratory of Medical Neurobiology, MOE Frontiers Center for Brain Science, Institutes of Brain Science, Fudan University, Shanghai, China
- 4State Key Laboratory of Genetic Resources and Evolution, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, China
Rnf220 is reported to regulate the patterning of the ventral spinal neural tube in mice. The brainstem has divergent connections with peripheral and central targets and contains unique internal neuronal groups, but the role of Rnf220 in the early development of the hindbrain has not been explored. In this study, Nestin-Cre-mediated conditional knockout (Rnf220Nestin CKO) mice were used to examine if Rnf220 is involved in the early morphogenesis of the hindbrain. Rnf220 showed restricted expression in the ventral half of ventricular zone (VZ) of the hindbrain at embryonic day (E) 10.5, and as development progressed, Rnf220-expressing cells were also present in the mantle zone outside the VZ at E12.5. In Rnf220Nestin CKO embryos, alterations of progenitor domains in the ventral VZ were observed at E10.5. There were significant reductions of the p1 and p2 domains shown by expression of Dbx1, Olig2, and Nkx6.1, accompanied by a ventral expansion of the Dbx1+ p0 domain and a dorsal expansion of the Nkx2.2+ p3 domain. Different from the case in the spinal cord, the Olig2+ pMN (progenitors of somatic motor neuron) domain shifted and expanded dorsally. Notably, the total range of the ventral VZ and the extent of the dorsal tube were unchanged. In addition, the post-mitotic cells derived from their corresponding progenitor domain, including oligodendrocyte precursor cells (OPCs) and serotonergic neurons (5-HTNs), were also changed in the same trend as the progenitor domains do in the CKO embryos at E12.5. In summary, our data suggest similar functions of Rnf220 in the hindbrain dorsoventral (DV) patterning as in the spinal cord with different effects on the pMN domain. Our work also reveals novel roles of Rnf220 in the development of 5-HTNs and OPCs.
Introduction
Ubiquitination is an important post-translational modification in eukaryotic cells, involved in a variety of cellular processes, including signal transduction and transcriptional regulation (Pickart, 2001; Pickart and Eddins, 2004; Mukhopadhyay and Riezman, 2007). The entire course of ubiquitination is catalyzed by the ubiquitin-activating enzyme, the ubiquitin-conjugating enzyme and the ubiquitin ligase (E3), successively (Morreale and Walden, 2016). There are about 600 E3 ubiquitin ligases in human, and Rnf220 is a newly discovered one of them (Deshaies and Joazeiro, 2009).
As a member of the RING finger protein family, Rnf220 is highly conserved among different species. Rnf220 was first identified as an E3 ubiquitin ligase in 2010, which interacts with and promotes ubiquitination and proteasomal degradation of Sin3B (Kong et al., 2010). Our previous studies find that Rnf220 works as a novel modulator of sonic hedgehog (Shh)/Gli signaling gradient and is a key regulator of the patterning of the ventral spinal neural tube in mice (Ma et al., 2019; Ma et al., 2020). Furthermore, the development of noradrenergic neurons in locus coeruleus of the pons requires Rnf220/Zc4h2-mediated monoubiquitylation of transcription factors Phox2a and Phox2b (Song et al., 2020). However, the expression pattern of Rnf220 in the hindbrain and its potential function in the early development of the hindbrain remain unclear.
The procedure of embryonic cells forming different tissues and organs and comprising orderly spatial structures is termed pattern formation. The DV pattern formation of the hindbrain is similar to that of the spinal cord. Along the DV axis, the progenitors in the hindbrain are divided into dp1-dp6, p0, p1, p2, pMN, and p3 domains. The post-mitotic cells generated from the dorsal neural tube are dl1-dl6, and those from the ventral neural tube are distributed into V0, V1, V2, sMNs (somatic motor neurons), and V3 domains in the mantle zone from dorsal to ventral (Lebel et al., 2007; Le Dréau and Martí, 2012; Carcagno et al., 2014).
Unlike the spinal neural tube, the neurons generated from the p3 domain of the hindbrain are the visceral motor neurons (vMNs) and 5-HTNs instead of glutamatergic neurons (Carcagno et al., 2014), and this made us interested in the development of 5-HTNs in the absence of Rnf220. During embryonic development, 5-HTNs are generated from E10.5, and vMNs are generated before E10.5 (Ding et al., 2003; Pattyn et al., 2003). In addition, lineage-tracing experiments show that the progenitors from the Olig2+ pMN domain also give rise to OPCs and then generate oligodendrocytes at later embryonic stages (Masahira et al., 2006). Mature oligodendrocytes express myelin basic protein (Mbp) and proteolipid protein 1 (Plp1) and are critical for the myelination of axons and involved in neurodegenerative diseases in brain (Boggs, 2006; Simons and Nave, 2015; Berry et al., 2020).
The Shh signal is required for the ventral neural tube patterning along the entire neural tube, but different phenotypes are reported in the mouse hindbrain compared with the spinal cord when Shh signaling was interfered as shown by the fact that the Olig2+ domain is expanded ventrally in the spinal cord but disappeared in the hindbrain of Gli2−/− mice (Lebel et al., 2007). In this study, we examine the expression of Rnf220 in the ventral neural tube of the hindbrain and explore the territory of different progenitor domains by the examination of the domain-specific gene expression in Rnf220Nestin CKO mice. Overall, there is an expansion of the dorsal and ventral domains at the expense of the middle domains in the ventral neural tube with an unchanged range (territory) of the ventral tube. Our work also establishs clear roles of Rnf220 in the development of 5-HTNs and OPCs in the hindbrain.
Materials and Methods
Animals, Staging, and Genotyping
All mice were maintained and handled according to guidelines approved by the Animal Committee of Tongji University School of Medicine, Shanghai, China. All mice were maintained on a C57BL/6 background. Analysis was performed only after lines were crossed to C57BL/6 for at least three generations.
The stage of mouse embryos was determined by taking the morning when the copulation plug was seen as E0.5. All genotypes described were confirmed by PCR. Rnf220 alleles were genotyped using genome DNA prepared from tail tips. PCR primers were used as described in a previous report (Ma et al., 2019). PCR amplified DNA was analyzed on 1.5% TAE agarose gel.
In situ Hybridization Assays
Embryos were fixed in 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS, pH 7.4) for 24 h and cryoprotected with 30% sucrose in PBS. Then, 20-μm-thick transverse sections were cut on a cryostat (Leica), and in situ hybridization was performed as described previously (Song et al., 2011). Briefly, RNA probes for detecting Pax3, Axin2, Msx1, Phox2b, serotonin transporter (Sert), tryptophan 5-hydroxylase 2 (Tph2), vesicular monoamine transporter 2 (Vmat2), monoamine oxidase A (MaoA), Sex determining region Y-box 10 (Sox10), Mbp and Plp1 were generated according to the description on the website of Allen Brain Atlas (http://portal.brain-map.org/). Probes against Rnf220, Shh, Pax6, Dbx1, Dbx2, Hb9, Chx10, En1, and Evx1 were used as described previously (Ma et al., 2019). All probes were cloned into pGEM-T vector (Promega) and transcribed by T7 or SP6 in vitro transcription kit (Ambion). Sections were observed and images were captured using epifluorescence microscope (80i; Nikon).
Immunohistochemistry Analysis
For immunostaining, 20-μm-thick transverse sections were used, and the procedure is described in our previous report (Song et al., 2011). Antibodies used are listed in the Key Resources table. For double labeling using immunostaining and in situ hybridization, sections underwent the in situ hybridization procedure first. After visualization for mRNA, sections were incubated with primary antibody at 4°C overnight, followed by appropriate secondary antibody for 3 h. The sections were then processed using a Vectastain Elite ABC kit (Vector Laboratories) for 1 h, and immunoreactivity was visualized by incubation with diaminobenzidine (DAB) and H202.
Bromodeoxyuridine (BrdU) Labeling
For BrdU pulse labeling experiments to analyze cell proliferation, pregnant mice received a single injection of BrdU at 50 mg/kg body weight and were sacrificed 1 h later. Sections were immersed in 0.01 M citrate buffer at 95°C for 20 min, 0.5 M HCl at 55°C for 10 min, and then washed in PBS. Treated sections were immunostained with anti-BrdU antibody as described above.
Quantification and Statistical Analysis
To quantify the sizes of progenitor domains (Shh+ for floor plate, Nkx2.2+ for p3 domain, Olig2+ for pMN domain, Nkx6.1+ region located dorsal to Olig2+ for p2 domain, the gap between Dbx1+ and Nkx6.1+ for p1 domain, Dbx1+ for p0 domain and Pax3+ for dp1-dp6 domain) and the numbers of the ventral post-mitotic neurons (5-HT+, Sert+, Phox2b+, Hb9+, Chx10+, En1+, and Evx1+) and OPCs (Sox10+), serial sections of the E10.5 or E12.5 hindbrain neural tubes were immunostained with the indicated antibodies or processed with antisense RNA probes. At least three sections around the rhombomere 5 (r5) level determined by the presence of otic vesicles in transverse sections or at indicated hindbrain level were included from each embryo. The size of each domain in images was measured using ImageJ with the Segmented Line Tool, and the percentage of each domain to the whole DV extent of the VZ was calculated. The number of post-mitotic cells indicated with different markers and BrdU+ and Cleaved Caspase-3+ cells were counted using the Multi-point Tool of ImageJ. Data are expressed by mean ± SEM, and two-tailed Student’s t-test was used for each comparison. GraphPad software was used for statistical analysis. p-values less than 0.05 were considered statistically significant.
Results
Rnf220 is Expressed in the Ventral Half of the Hindbrain Neural Tube
Our previous study reports that Rnf220 is expressed in the ventral half of the spinal neural tube (Ma et al., 2019), and we assumed that it is also the case for the hindbrain. To test this, we carried out in situ hybridization in wild-type embryos at E10.5 and E12.5. As expected, intense expression of Rnf220 was present in the VZ of the ventral neural tube and weak expression was observed in the floor plate at E10.5 (Figures 1A–C). In the E12.5 hindbrain, Rnf220 expression was also localized in many post-mitotic cells in the ventral mantle zone (Figures 1D–F). However, in the VZ region at the level of r5, where Hb9+ sMNs are generated, much less Rnf220 expression was present at E10.5 (Figures 1G–I), and the corresponding VZ region containing Phox2b+ vMNs at E12.5 had weak, if any, Rnf220 transcripts (Figures 1J–L). To further localize its distribution pattern, we performed in situ hybridization for Rnf220 and Dbx1, a p0 domain marker (Pierani et al., 2001), and also for Pax3, a dp1-dp6 domain marker (Moore et al., 2013), on adjacent sections at E10.5. The result shows that the uppermost territory of Rnf220 expression domain corresponds well with that of Dbx1 (Figures 1M–O, S) and is adjacent to that of Pax3 (Figures 1P–R, S), confirming that Rnf220 is restrictively expressed in the ventral half of the hindbrain neural tube, covering ventral p0-p2, pMN, p3 domains, and the floor plate (Figure 1S).
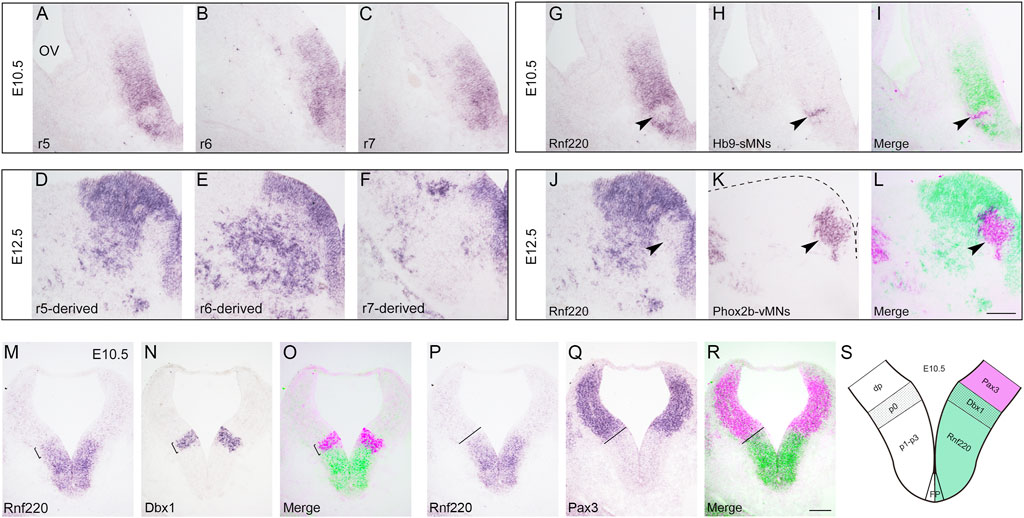
FIGURE 1. Expression of Rnf220 in the Developing Hindbrain. (A–C) In situ hybridization for Rnf220 of wild-type embryos at E10.5. Rnf220 is expressed in the VZ of the ventral neural tube and a few post-mitotic cells in the mantle zone. OV, otic vesicle; r5-r7, section at rhombomere 5-7 levels. (D–F) In situ hybridization for Rnf220 of wild-type embryos at E12.5. Rnf220 is expressed in the VZ of the ventral neural tube and many post-mitotic cells outside the VZ. (G–I) In situ hybridization for Rnf220 and Hb9 on adjacent sections at the r5 level of wild-type embryos at E10.5. Rnf220 (green) and Hb9 (magenta) are shown by pseudocolor (I). Few or very weak Rnf220 expression is present in Hb9+ sMNs (arrowheads). sMNs, somatic motor neurons. (J–L) In situ hybridization for Rnf220 and Phox2b on adjacent sections at the r5-derived level of wild-type embryos at E12.5. Rnf220 (green) and Phox2b (magenta) are shown by pseudocolor (L). Rnf220 is not expressed in Phox2b+ vMNs (arrowheads). Dashed line in (K) shows the boundary of the hindbrain. vMNs, visceral motor neurons. Scale bar, 100 μm (L); also applies to A–K). (M–O) In situ hybridization for Rnf220 and Dbx1 (a p0 domain marker) on adjacent slides of E10.5 wild-type embryos. Dbx1 (magenta) and Rnf220 (green) are shown by pseudocolor (O). The uppermost territory of Rnf220 expression domain corresponds well with that of Dbx1 (brackets). (P–R) In situ hybridization for Rnf220 and Pax3 (a dp1-dp6 domain marker) on adjacent slides of E10.5 wild-type embryos. The expression domain of Rnf220 is adjacent to that of Pax3 (dashed lines). Pax3 (magenta) and Rnf220 (green) are shown by pseudocolor (R). Scale bar, 100 μm (R); also applies to (M–Q). (S) Diagram showing the expression pattern of Rnf220 in the E10.5 hindbrain. Rnf220 is expressed from p0 to p3 domain and in the floor plate (FP), floor plate.
Progenitor Domains are Altered in the Ventral Hindbrain Neural Tube of Rnf220Nestin CKO Embryos
To investigate the function of Rnf220 in the early development of the hindbrain, we crossed Rnf220flox/flox (Ma et al., 2019) with a Nestin-Cre line (Tronche et al., 1999), which expresses Cre recombinase in the neuronal and glial cell precursors from E10.5. Nestin-Cre:Rnf220flox/+ mice were crossed with Rnf220flox/flox or Rnf220flox/+ mice, and the resulting embryos were genotyped using PCR and processed for phenotypic analysis. Rnf220Nestin CKO (Nestin-Cre:Rnf220flox/flox) embryos were obtained, and littermates with other genotypes showing no detectable phenotypes were used as controls. The Rnf220Nestin CKO embryos were found to be neonatal lethal, and the deletion of Rnf220 expression in these embryos was confirmed by in situ hybridization using an exon2-specific probe (Supplementary Figure S1). Residual weak Rnf220 expression in the ventral-most hindbrain is likely caused by incomplete or absent Cre activity in these cells.
A group of genes was used to determine whether the deletion of Rnf220 affects the progenitor domains in the ventral hindbrain neural tube at E10.5. In situ hybridization for Dbx1 and immunostaining for Nkx6.1 (a marker for p2-pMN and p3 domains) showed that the p0 domain was expanded ventrally at the expense of the p1 domain (a gap between the Nkx6.1+ and Dbx1+ domains) in Rnf220Nestin CKO embryos (Figures 2A,B, M,N). In addition, the Dbx2+ p0-p1 and Nkx6.1+ domains were located next to each other without overlapping in control neural tubes (Figure 2C). In contrast, there was an increase of Dbx2 expression in the p0 and p1 domains and weak Dbx2 expression was expanded ventrally into the Nkx6.1+ domain in Rnf220Nestin CKO embryos (Figure 2D). Double immunostaining for Olig2 (a pMN domain marker) and Nkx2.2 (a p3 domain marker) showed that both the p3 and pMN domains were dorsally expanded in the CKO embryos compared with controls (Figures 2E,F, M,N). The increase of the pMN and p3 domains were further confirmed by double immunostaining for Olig2 and Nkx6.1 (Figures 2G,H). The reduction of p2 domain was confirmed as shown by the Nkx6.1+ region located dorsal to the Olig2+/Nkx6.1+ pMN domain (Figures 2G,H, M,N), and the double staining also provided evidence showing the pMN domain was shifted dorsally (Figures 2G,H, N). Note that the extent of the ventral neural tube from the p0 domain to the floor plate was not changed (Figures 2M,N).
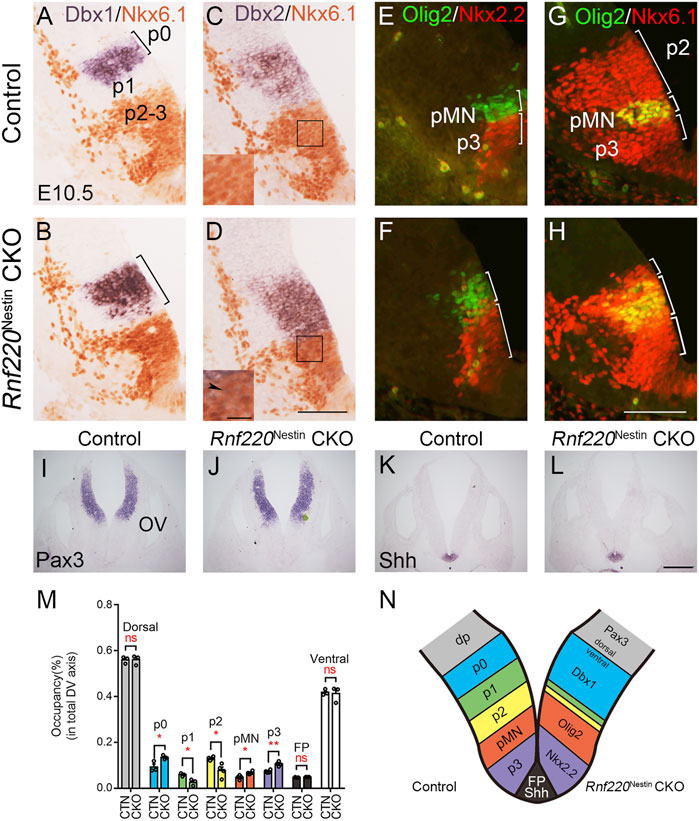
FIGURE 2. The Progenitor Domains are Altered in the Developing Ventral Hindbrain of Rnf220Nestin CKO Embryos at E10.5. (A,B) In situ hybridization for Dbx1 and immunostaining for Nkx6.1 show that Dbx1 expression is expanded ventrally to the dorsal boundary of the Nkx6.1+ domain in Rnf220Nestin CKO embryos (B), whereas there is a gap region (i.e., the p1 domain) between the Nkx6.1+ and Dbx1+ domains in control embryos (A). (C,D) In situ hybridization for Dbx2 and immunostaining for Nkx6.1 show that Dbx2 expression is expanded ventrally into the Nkx6.1+ domain in Rnf220Nestin CKO embryos (D) (arrowhead), whereas a clear boundary between the Nkx6.1+ and Dbx2+ domains is present in the control neural tubes (C). Scale bar, 100 μm (D); also applies to (A–C). Insets are high-magnification images of boxed areas in (C,D). Scale bar, 25 μm. (E,F) Double immunostaining shows that both the Nkx2.2+ (red) p3 domain and the Olig2+ (green) pMN domain are expanded along the DV axis of the VZ in Rnf220Nestin CKO embryos (F) relative to controls (E). (G,H) Double immunostaining for Nkx6.1 (red) and Olig2 (green) shows a dorsal shift of the Nkx6.1+/Olig2+ pMN domain and a reduction of the p2 domain in the Rnf220Nestin CKO neural tubes (H) compared with controls (G). The p2 domain corresponds to the Nkx6.1+ region located dorsal to the Nkx6.1+/Olig2+ pMN domain. Scale bar, 100 μm (H); also applies to (E-G). (I,J) In situ hybridization for Pax3 of control (I) and CKO embryos (J) at E10.5. OV, otic vesicle. (K,L) In situ hybridization for Shh of control (K) and CKO embryos (L) at E10.5. Scale bar, 100 μm (L); also applies to (I–K). (M) Quantification of the sizes of each progenitor domain in the neural tube of control and Rnf220Nestin CKO embryos. The size is shown by its proportion in the whole extent of the VZ along the DV axis. CTN, control; CKO, Rnf220Nestin CKO. Dorsal, Pax3+ domain; p0, Dbx1+ domain; p1, gap region between the Dbx1+ and Nkx6.1+ domains; p2, Olig2- domain in the dorsal part of the Nkx6.1+ region; pMN, Olig2+ domain; p3, Nkx2.2+ domain; FP, floor plate, Shh+ domain. Data are expressed by mean ± SEM. Student’s t-test (n≥3 for each). *, p<0.05; **, p<0.01; ns, no significant difference. (N) Diagram showing the changes of the progenitor domains of the ventral hindbrain in Rnf220Nestin CKO embryos at E10.5. The loss of Rnf220 leads to the expansion of the Dbx1+ p0, Olig2+ pMN, and Nkx2.2+ p3 domains in the ventral hindbrain neural tube at the expense of the middle domains (i.e., the p1 and p2 domains) without affecting the extent of the dorsal neural tube.
We next moved to examine if the floor plate and dorsal neural tube was affected in Rnf220Nestin CKO embryos. The floor plate shown by the expression of Shh and the dorsal neural tube shown by Pax3 were not obviously changed (Figures 2I–L). Taken together, we conclude that the loss of Rnf220 led to the expansions of the dorsal-most (p0) and ventral-most (pMN and p3) domains at the expense of the middle domains (i.e., the p1 and p2 domains) in the ventral hindbrain neural tube (Figure 2N).
No Obvious Changes in Proliferation and Cell Death in the Developing Ventral Hindbrain of Rnf220Nestin CKO Embryos at E10.5
The loss of Rnf220 resulted in alterations of progenitor domains of the ventral hindbrain neural tube. We wondered if the changes in progenitor domains were due to abnormal proliferation or cell death. To explore such a possibility, we performed pulse labeling of BrdU and immunostaining of Cleaved Caspase-3 to label cell proliferation and cell death, respectively. Overall, BrdU labeling was not obviously different between the two groups (Figures 3A,B). To clearly define the territory of the pMN and p3 domains for quantification of BrdU+ cells, double staining for BrdU and Olig2 was carried out. Because the DV extents of the pMN and p3 domains were increased, we compared the numbers of BrdU+ cells per 100 μm-length in the two domains and found no significant difference between Rnf220Nestin CKO and control embryos (Figure 3C). Next, we compared percentages of BrdU/Olig2 double-labeled cells in Olig2+ cells in the pMN domain and revealed no significant difference either (Figure 3D). In addition, no significant difference was detected in the numbers of Cleaved Caspase-3+ cells between the two groups (Figures 3E–G). However, proliferation and cell death were altered at E12.5. Rnf220Nestin CKO embryos showed lower density of BrdU+ cells in the ventral VZ than control embryos (Supplementary Figures S2A–C), and more Cleaved Caspase-3+ cells were detected in the ventral hindbrain of CKO embryos (Supplementary Figures S2D–F). These data suggest that the changes of progenitor domains in Rnf220Nestin CKO embryos at E10.5 are not due to abnormal proliferation or cell death, but it remains to be established how Rnf220 mutation affects neural stem cell proliferation and cell death at later stages.
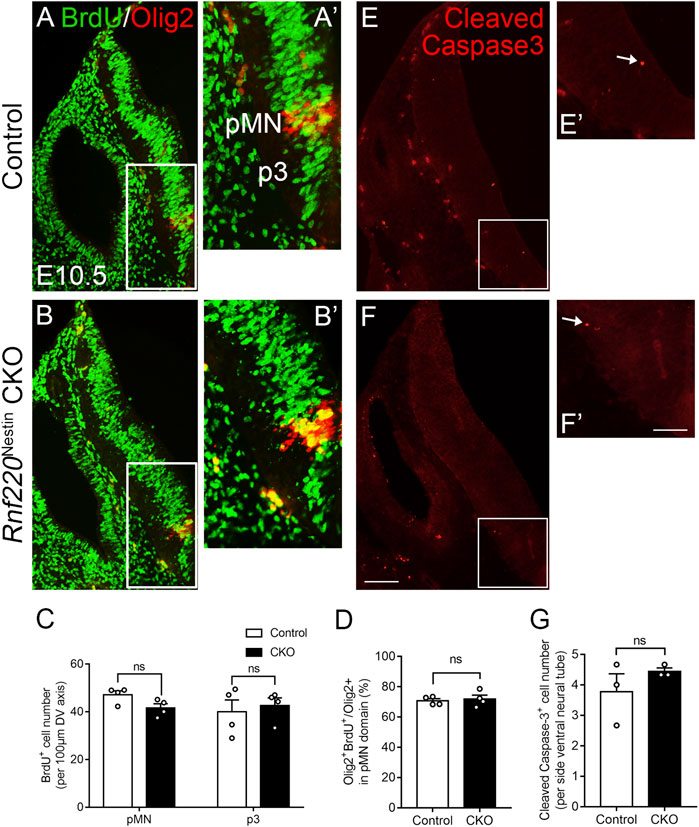
FIGURE 3. No Significant Changes in Proliferation and Cell Death in the Ventral Hindbrain of Rnf220Nestin CKO Embryos at E10.5. (A–B) Double immunostaining for BrdU (green) and Olig2 (red) shows no obvious changes of proliferation in the ventral VZ of the hindbrain between Rnf220Nestin CKO embryos and controls. (A’,B’) are high-magnification images of boxed areas in (A,B), respectively. (C) Quantification of the numbers of BrdU+ cells per 100 μm-length in the Olig2+ pMN domain and p3 domain of control and Rnf220Nesin CKO embryos at E10.5. The p3 domain corresponds to the BrdU+ region located ventral to the Olig2+ pMN domain. ns, no significant difference. (D) Quantification of the percentages of BrdU/Olig2 double-labeled cells in Olig2+ cells in the pMN domain of control and Rnf220Nesin CKO embryos at E10.5. (E,F) Immunostaining for Cleaved Caspase-3 shows similar cell death in Rnf220Nestin CKO embryos compared with controls. (E’,F’) are high-magnification images of boxed areas in (E,F), respectively. Scale bar, 100 μm (F); also applies to (A,B,E); 50 μm (F’); also applies to (A’,B’,E’). (G) Quantification of the numbers of Cleaved Caspase-3+ cells in the ventral neural tube in the two groups at E10.5. Data are expressed by mean ± SEM. Student’s t-test (n ≥ 3 for each).
Alterations of Post-mitotic Neurons in the Developing Ventral Hindbrain of Rnf220Nestin CKO Embryos
Having observed dramatic changes in the progenitor domains of the ventral hindbrain neural tube, it is likely that their progeny may also be altered in the absence of Rnf220. The progenitor domains that express Rnf220 within the ventral hindbrain neural tube of control embryos include p0, p1, p2, pMN, and p3 domains, which give rise to V0, V1, V2 interneurons; sMNs; OPCs; vMNs; and 5-HTNs, respectively. As expected, more post-mitotic neurons from the p0 domain were observed as shown by the increased expression of Evx1 in Rnf220Nestin CKO embryos relative to its expression in the control embryos at E12.5 (Figures 4A,B,K). More ventrally, the En1+ V1 and Chx10+ V2 interneurons were markedly reduced or abolished in CKO embryos (Figures 4C–F,K). The sMNs from the pMN domain express Hb9, which were significantly increased in Rnf220Nestin CKO embryos (Figures 4G,H,K). Unexpectedly, Phox2b+ vMNs from the p3 domain in Rnf220Nestin CKO embryos were slightly reduced compared with those in the control group (Figures 4I–K) although the p3 domain was increased in Rnf220Nestin CKO embryos (Figures 2E–H, M,N). In addition, there was ectopic distribution of Hb9+ cells at the r6-derived level of the hindbrain in Rnf220Nestin CKO embryos, whereas they were only present at the r5- and r7-derived hindbrain in control mice (Supplementary Figure S3). This is perhaps due to the migration defect of r5-derived neurons of the abducens nerve and r7-derived neurons associated with the hypoglossal nerve, but further experiment is needed. Thus, the deletion of Rnf220 not only changed the progenitor domains, but also altered their progeny in the ventral hindbrain correspondingly.
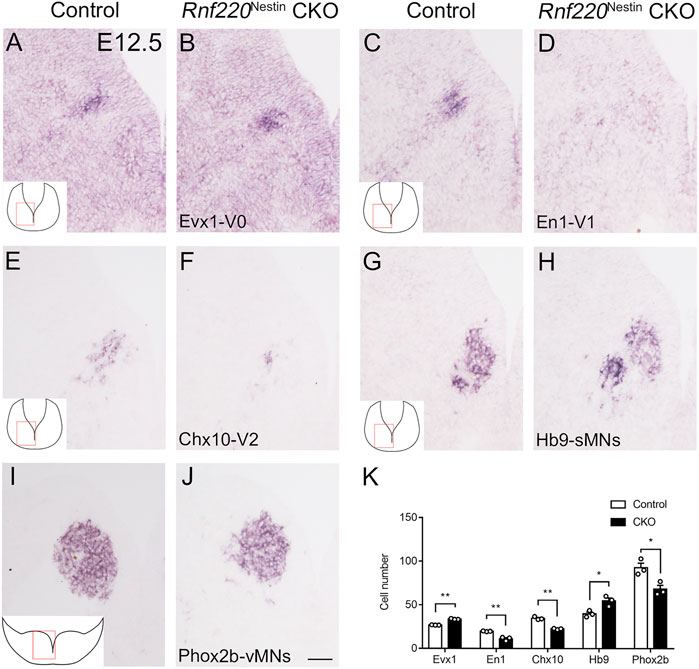
FIGURE 4. Changes in the Domains of Post-mitotic Neurons in the Ventral Hindbrain Neural Tube of Rnf220Nestin CKO Embryos at E12.5. (A,B) More Evx1+ V0 neurons are observed in Rnf220Nestin CKO embryos (B) than in controls (A). (C,D) The En1+ V1 neurons are hardly detected in Rnf220Nestin CKO embryos (D) compared with the V1 neuronal domain in controls (C). (E,F) The number of Chx10+ V2 neurons is reduced in Rnf220Nestin CKO embryos (F) compared with controls (E). (G,H) Hb9+ somatic motor neurons (sMNs) are increased in Rnf220Nestin CKO embryos (H) compared with controls (G). (I,J) Phox2b+ visceral motor neurons (vMNs) are reduced in Rnf220Nestin CKO embryos (J) compared with controls (I). Scale bar, 50 μm (J); also applies to (A–I). Insets (A,C,E,G,I) are schematic diagrams of the hindbrain level and position shown in (A–J). (K) Quantification of different groups of post-mitotic neurons in the ventral hindbrain neural tube of control and Rnf220Nestin CKO embryos. Data are expressed by mean ± SEM. Student’s t-test (n=3 for each). *, p<0.05; **, p<0.01.
5-HTNs are Increased in Rnf220Nestin CKO Mice
Neurons that are capable of synthesis and release the essential neurotransmitter 5-HT are called 5-HTNs. 5-HT is synthesized by Tph2 (Walther et al., 2003) and aromatic L-amino-acid decarboxylase (Aadc) (Albert et al., 1987) from tryptophan. Then, 5-HT is packaged into synaptic vesicles by Vmat2 (Weihe et al., 1994). Extracellular 5-HT can be transported back into the cells by Sert (Hoffman et al., 1991). In the cells, 5-HT is broken down by MaoA and MaoB (Luque et al., 1995). As 5-HTNs are generated from the p3 domain, which was increased in Rnf220Nestin CKO embryos, we examined whether the development of 5-HTNs was affected by knocking out Rnf220. In situ hybridization for Sert and immunostaining for 5-HT showed that the number of 5-HTNs was increased in Rnf220Nestin CKO embryos at E12.5 (Figures 5A–D,K). To further confirm this, in situ hybridization for Tph2, Vmat2, and MaoA were performed (Figures 5E–J), and expressions of all these genes were increased. We also detected the numbers of 5-HTNs at E14.5 (Figures 5L–O) and postnatal day 0 (Figures 5P,Q), which were still increased in Rnf220Nestin CKO mice.
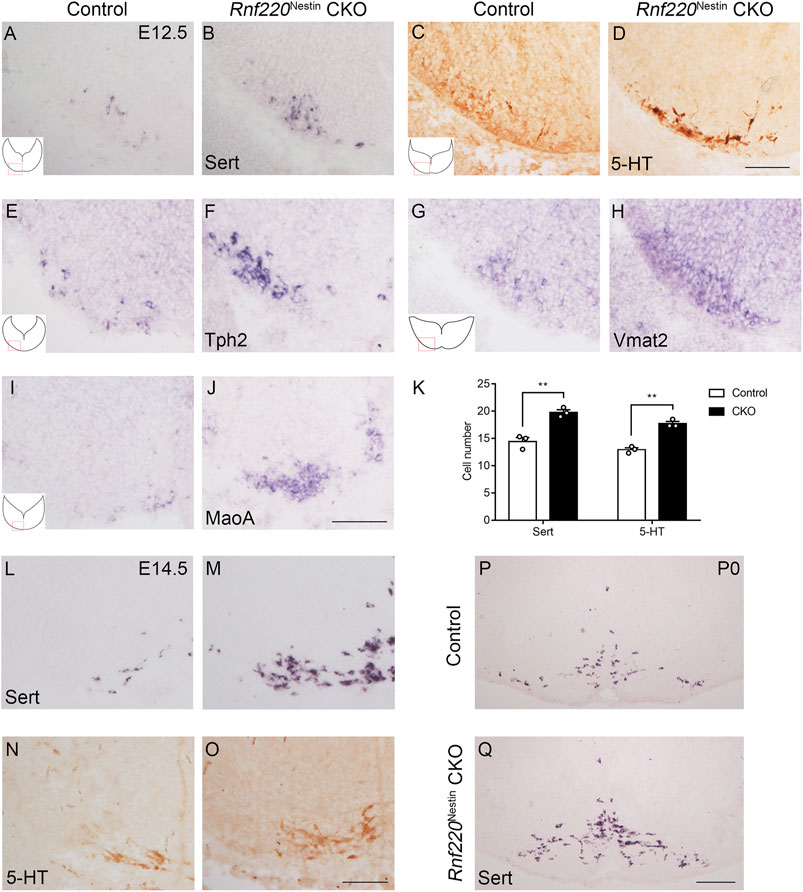
FIGURE 5. The Number of 5-HTNs is Increased in the Hindbrain of Rnf220Nestin CKO Mice. (A,B) In situ hybridization for Sert shows increased number of 5-HTNs in Rnf220Nestin CKO embryos (B) compared with controls (A) at E12.5. (C,D) DAB staining for 5-HT shows more 5-HTNs in Rnf220Nestin CKO embryos (D) than in controls (C) at E12.5. Scale bar, 100 μm (D); also applies to (A–C). (E–J) In situ hybridization for Tph2, Vmat2 and MaoA shows increased expressions of these genes in Rnf220Nestin CKO embryos (F,H,J) compared with controls (E,G,I). Scale bar, 50 μm (J); also applies to (E–I). Insets (A,C,E,G,I) are schematic diagrams of the hindbrain level and position shown in (A–J). (K) Quantification of Sert+ and 5-HT+ neurons. Data are expressed by mean ± SEM. Student’s t-test (n=3 for each). **, p<0.01. (L,M) In situ hybridization for Sert shows increased number of 5-HTNs in Rnf220Nestin CKO embryos (M) compared with controls (L) at E14.5. (N,O) DAB staining for 5-HT shows more 5-HTNs in Rnf220Nestin CKO embryos (O) than in controls (N) at E14.5. Scale bar, 100 μm (O); also applies to (L–N). (P,Q) In situ hybridization for Sert shows more 5-HTNs in Rnf220Nestin CKO mice (Q) than in controls (P) at postnatal day 0 (P0). Scale bar, 200 μm (Q); also applies to (P).
OPCs are also Increased in Rnf220Nestin CKO Embryos
OPCs are generated in the ventral hindbrain at E12.5 from the Olig2+ pMN domain (Richardson et al., 2006), which was shifted and expanded dorsally in Rnf220Nestin CKO embryos as mentioned above. This prompted us to examine if the generation of OPCs was altered in the CKO embryos. We observed that Sox10+ OPCs in the ventral hindbrain were increased and shifted dorsally at E12.5 (Figures 6A,B,E). As development progressed, mature oligodendrocytes, marked by Mbp and Plp1, were located in a narrow region in the mantle zone along the VZ in control embryos, whereas they displayed a dispersed distribution pattern in the Rnf220Nestin CKO hindbrain at E14.5 (Supplementary Figure S4). In addition, OPCs are also generated in the ventral spinal cord, but its development in Rnf220-deficient mice has not been examined. OPCs in the spinal cord, however, were diminished in CKO embryos at E12.5 (Figures 6C,D,F). Note that the reduction coincides with the decrease of the Olig2+ domain of the Rnf220−/− spinal neural tube (Ma et al., 2019). Thus, there is a different phenotype in the ventral neural tube patterning in the absence of Rnf220: An increased Olig2+ domain led to an increase of OPCs in the hindbrain, but a reduced Olig2+ domain resulted in a decrease of OPCs in the spinal cord.
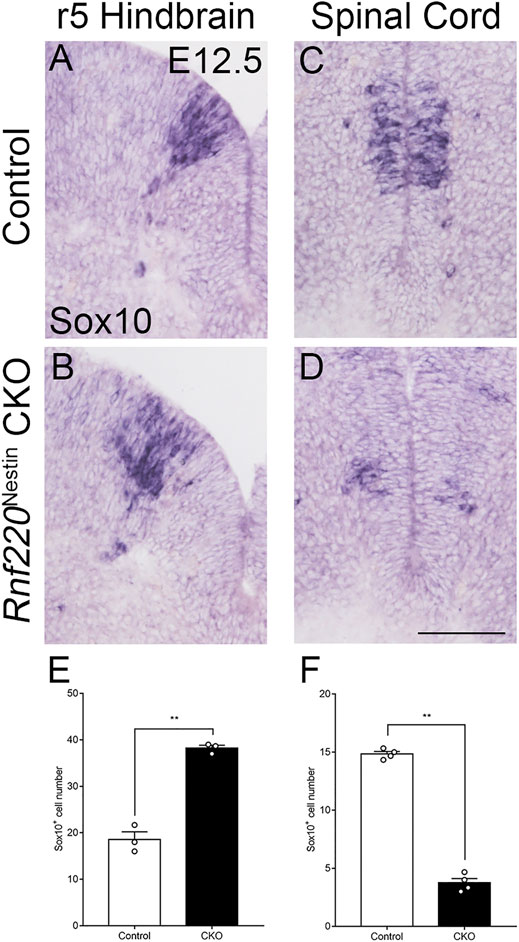
FIGURE 6. OPCs are Increased in the Hindbrain but not the Spinal Cord of Rnf220Nestin CKO Embryos. (A–D) In situ hybridization for Sox10 at E12.5. At the r5-derived level of the hindbrain, the number of the OPCs are increased in Rnf220Nestin CKO embryos (B) compared with controls (A), whereas the number is decreased in CKO (D) compared with that of the control (C) in the spinal cord. Scale bar, 100 μm (D); also applies to (A–C). (E) Quantification of Sox10+ cells of the r5 hindbrain at E12.5. (F) Quantification of Sox10+ cells of the spinal cord at E12.5. Data are expressed by mean ± SEM. Student’s t-test (n ≥ 3 for each). **, p<0.01.
Discussion
Our and other previous studies show that Rnf220 is expressed in the ventral spinal neural tube (Kim et al., 2018; Ma et al., 2019). The present study shows that this restricted expression pattern is also present in the hindbrain neural tube with the exception of Hb9+ sMNs and Phox2b+ vMNs, where few or weak Rnf220 transcripts are distributed (Figure 1). Consistent with the expression pattern, our data from Rnf220Nestin CKO embryos demonstrates that Rnf220 is involved in the DV patterning of the hindbrain neural tube in mice. The loss of Rnf220 alters neural progenitor domains: the p0, pMN, and p3 domains located at the two ends of the ventral tube are increased, and the p1 and p2 domains located in the middle are decreased; their progeny show similar changes (Figures 2, 7).
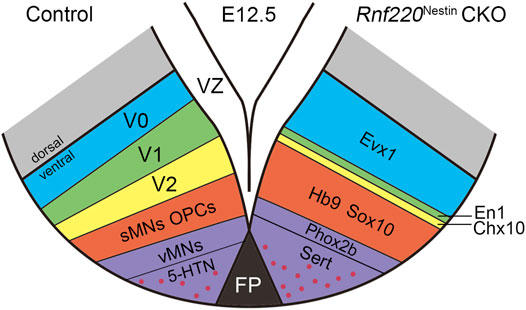
FIGURE 7. Diagram Showing the Changes of Post-mitotic Cells in the Ventral Hindbrain of Rnf220Nestin CKO Embryos at E12.5. The loss of Rnf220 leads to the expansion of the Evx1+ V0, Hb9+ sMNs and Sox10+ OPCs domains, whereas the En1+ V1 and Chx10+ V2 domains are reduced. Within the domain differentiated from p3, the Phox2b+ vMNs domain decreases slightly but the number of 5-HTNs is increased.
Shh signaling plays crucial roles in patterning the ventral neural tube of both the spinal cord and hindbrain by controlling the expressions of several patterning genes (e.g., Olig2, Nkx2.2, and Nkx6.1) through opposing gradients of repressor and activator forms of Gli transcription factors (Ruiz i Altaba, 1998; Cohen et al., 2015; Ma et al., 2019). Our previous study shows that the deletion of Rnf220 does not affect Shh expression in the notochord or the floor plate in the spinal cord but affects the functional balance between two forms of Glis via the K63-linked ubiquitination mechanism (Ma et al., 2019). The deletion of Rnf220 in the spinal neural tube leads to the expansions of the two end domains (p0 and p3) with reductions of those in the middle (p1, p2, and pMN) and intact expression of Shh (Ma et al., 2019). Generally, the defective neural tube patterning in the hindbrain of Rnf220Nestin CKO embryos is very similar to that in the spinal neural tube, but there is a difference in the pMN domain: reduced in the spinal cord but expanded in the hindbrain. The alterations led to similar changes in the numbers of sMNs: increased in the hindbrain (Figure 4) but reduced in the spinal cord (Ma et al., 2019). Overall, Rnf220 is homogeneously expressed in the VZ of the ventral hindbrain, but its expression is much reduced in the pMN domain at the level of generation of sMNs shown by Hb9 and devoid in the progenitors of the visceral motor neuron domain at the level of generation of vMNs indicated by Phox2b (Figure 1). Both Shh and Glis are proposed to work in concentration-associated gradient fashions, and the discontinued expression of Rnf220 in the ventral VZ may affect the shape of the gradients leading to the distinct alteration of the pMN domain in the hindbrain of Rnf220Nestin CKO embryos.
Although the neural tube patterning of the ventral hindbrain is altered in the absence of Rnf220, the proliferation of neural progenitors in the VZ seems not to be affected as shown by BrdU labeling at E10.5 (Figures 3A–D). However, BrdU+ cells are significantly reduced in the VZ of Rnf220Nestin CKO embryos at E12.5 (Supplementary Figures S2A–C), suggesting the proliferation capability is reduced during the embryonic development. Our previous studies show that the roles of Rnf220 in regulating neural development is achieved by controlling the stability and activity of some key genes (e.g., transcription factors) via ubiquitylation mechanisms (Ma et al., 2019; Song et al., 2020), and thus, exploring possible interactions between Rnf220 and genes involved in the proliferation of neural stem cells is required in future studies.
Different progenitor domains give rise to distinct types of neurons. Consistent with the changes in progenitor domains, more V0 interneurons and sMNs are observed, whereas V1 and V2 interneurons are significantly reduced or lost in Rnf220Nestin CKO embryos at later embryonic stages (Figures 4, 7). The loss of hindbrain V2 neurons might account for the neonatal death of the Rnf220Nestin CKO mice as the medullary V2a neurons are required for central respiratory rhythm generation in mice (Crone et al., 2012). Different from the spinal neural tube, the p3 domain in the hindbrain successively gives rise to vMNs and 5-HTNs (Pattyn et al., 2003). Interestingly, the number of vMNs is slightly reduced, and the number of 5-HTNs is significantly increased (Figures 4, 5, 7). Thus, although the p3 domain is expanded, the successively generated neurons from this domain are not equally affected in the absence of Rnf220. Future research is required to clarify whether the progenitor identity within the p3 domain was changed in Rnf220Nestin CKO embryos.
The pMN domain gives rise to sMNs first and OPCs at later embryonic stages in both the spinal and hindbrain neural tube (Zhou et al., 2000; Novitch et al., 2001; Zannino and Appel, 2009). Intriguingly, Hb9+ sMNs and Sox10+ OPCs are increased in the hindbrain, but they are decreased in the spinal cord (Figures 4, 6; Ma et al., 2019). Further studies are needed to explore the mechanism concerning the different effects of Rnf220 in the production of sMNs and OPCs between the hindbrain and spinal cord. It is also of interest to study if Rnf220 is implicated in the maintenance of 5-HTNs and oligodendrocytes in the adult brain as these two types of cells are involved in mental disorders and neurodegenerative diseases.
Dysfunctions of the central 5-HT system are implicated in psychiatric disorders in human and abnormal behaviors in mice (Stein and Stahl, 2000; Dai et al., 2008; Dalley and Roiser, 2012; Jia et al., 2014; Song et al., 2016; Daut and Fonken, 2019; Chen et al., 2021). Our data suggest that Rnf220 might play an important role in maintaining the homeostasis of the 5-HT system, which infers that Rnf220 might be a risk gene in mental diseases.
In summary, Rnf220 plays an indispensable role in the early development of the hindbrain, which is evidenced by drastic alterations of neural progenitor domains in the ventral hindbrain of Rnf220Nestin CKO mice. These results also reveal a conserved function of Rnf220 in regulating neural tube patterning in the early development of the spinal cord and hindbrain with difference in affecting the generation of post-mitotic cells such as sMNs, OPCs, and those located only in the hindbrain (i.e., 5-HTNs).
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Ethics Statement
The animal study was reviewed and approved by Animal Committee of Tongji University School of Medicine, Shanghai, China.
Author Contributions
BM and Y-QD conceived and designed the project. BM and PM generated the mice. Y-BW and N-NS carried out and analyzed data. LZ and LH helped with data analysis. YH, J-YC, and Y-BW helped with the mouse work. Y-BW, BM, and Y-QD wrote the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (31771134 to N-NS; 31671061 to LZ; 32170965 to PM, 81221002, 81571332, and 91232724 to Y-QD), National Key R&D Program of China (2017YFA0104002 to Y-QD), “Strategic Priority Research Program” of the Chinese Academy of Sciences (Grant XDB13000000 to BM), Shanghai Pujiang Program (20PJ1413300 to LZ), Collaborative Innovation Program of Shanghai Municipal Health Commission (2020CXJQ01 to N-NS and Y-QD), Shanghai Municipal Science and Technology Major Project (2018SHZDZX01), Zhangjiang (ZJ) Lab, and Shanghai Center for Brain Science and Brain-Inspired Technology (to N-NS and Y-QD).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2022.831365/full#supplementary-material
References
Albert, V. R., Allen, J. M., and Joh, T. H. (1987). A Single Gene Codes for Aromatic L-Amino Acid Decarboxylase in Both Neuronal and Non-neuronal Tissues. J. Biol. Chem. 262 (19), 9404–9411. doi:10.1016/s0021-9258(18)48094-5
Berry, K., Wang, J., and Lu, Q. R. (2020). Epigenetic Regulation of Oligodendrocyte Myelination in Developmental Disorders and Neurodegenerative Diseases. F1000Res 9, 105. doi:10.12688/f1000research.20904.1
Boggs, J. M. (2006). Myelin Basic Protein: a Multifunctional Protein. Cell. Mol. Life Sci. 63 (17), 1945–1961. doi:10.1007/s00018-006-6094-7
Carcagno, A. L., Di Bella, D. J., Goulding, M., Guillemot, F., and Lanuza, G. M. (2014). Neurogenin3 Restricts Serotonergic Neuron Differentiation to the Hindbrain. J. Neurosci. 34 (46), 15223–15233. doi:10.1523/jneurosci.3403-14.2014
Chen, L., Gong, W.-K., Yang, C.-p., Shao, C.-C., Song, N.-N., Chen, J.-Y., et al. (2021). Pten Is a Key Intrinsic Factor Regulating Raphe 5-HT Neuronal Plasticity and Depressive Behaviors in Mice. Transl Psychiatry 11 (1), 186. doi:10.1038/s41398-021-01303-z
Cohen, M., Kicheva, A., Ribeiro, A., Blassberg, R., Page, K. M., Barnes, C. P., et al. (2015). Ptch1 and Gli Regulate Shh Signalling Dynamics via Multiple Mechanisms. Nat. Commun. 6, 6709. doi:10.1038/ncomms7709
Crone, S. A., Viemari, J.-C., Droho, S., Mrejeru, A., Ramirez, J.-M., and Sharma, K. (2012). Irregular Breathing in Mice Following Genetic Ablation of V2a Neurons. J. Neurosci. 32 (23), 7895–7906. doi:10.1523/jneurosci.0445-12.2012
Dai, J.-X., Han, H.-L., Tian, M., Cao, J., Xiu, J.-B., Song, N.-N., et al. (2008). Enhanced Contextual Fear Memory in central Serotonin-Deficient Mice. Proc. Natl. Acad. Sci. 105 (33), 11981–11986. doi:10.1073/pnas.0801329105
Dalley, J. W., and Roiser, J. P. (2012). Dopamine, Serotonin and Impulsivity. Neuroscience 215, 42–58. doi:10.1016/j.neuroscience.2012.03.065
Daut, R. A., and Fonken, L. K. (2019). Circadian Regulation of Depression: A Role for Serotonin. Front. Neuroendocrinology 54, 100746. doi:10.1016/j.yfrne.2019.04.003
Deshaies, R. J., and Joazeiro, C. A. P. (2009). RING Domain E3 Ubiquitin Ligases. Annu. Rev. Biochem. 78, 399–434. doi:10.1146/annurev.biochem.78.101807.093809
Ding, Y.-Q., Marklund, U., Yuan, W., Yin, J., Wegman, L., Ericson, J., et al. (2003). Lmx1b Is Essential for the Development of Serotonergic Neurons. Nat. Neurosci. 6 (9), 933–938. doi:10.1038/nn1104
Hoffman, B. J., Mezey, E., and Brownstein, M. J. (1991). Cloning of a Serotonin Transporter Affected by Antidepressants. Science 254 (5031), 579–580. doi:10.1126/science.1948036
Jia, Y.-F., Song, N.-N., Mao, R.-R., Li, J.-N., Zhang, Q., Huang, Y., et al. (2014). Abnormal Anxiety- and Depression-like Behaviors in Mice Lacking Both central Serotonergic Neurons and Pancreatic Islet Cells. Front. Behav. Neurosci. 8, 325. doi:10.3389/fnbeh.2014.00325
Kim, J., Choi, T.-I., Park, S., Kim, M. H., Kim, C.-H., and Lee, S. (2018). Rnf220 Cooperates with Zc4h2 to Specify Spinal Progenitor Domains. Development 145 (17), dev165340. doi:10.1242/dev.165340
Kong, Q., Zeng, W., Wu, J., Hu, W., Li, C., and Mao, B. (2010). RNF220, an E3 Ubiquitin Ligase that Targets Sin3B for Ubiquitination. Biochem. Biophysical Res. Commun. 393 (4), 708–713. doi:10.1016/j.bbrc.2010.02.066
Le Dréau, G., and Martí, E. (2012). Dorsal-ventral Patterning of the Neural Tube: a Tale of Three Signals. Devel Neurobio 72 (12), 1471–1481. doi:10.1002/dneu.22015
Lebel, M., Mo, R., Shimamura, K., and Hui, C.-c. (2007). Gli2 and Gli3 Play Distinct Roles in the Dorsoventral Patterning of the Mouse Hindbrain. Dev. Biol. 302 (1), 345–355. doi:10.1016/j.ydbio.2006.08.005
Luque, J. M., Kwan, S.-W., Abell, C. W., Prada, M. D., and Richards, J. G. (1995). Cellular Expression of mRNAs Encoding Monoamine Oxidases A and B in the Rat central Nervous System. J. Comp. Neurol. 363 (4), 665–680. doi:10.1002/cne.903630410
Ma, P., Song, N.-N., Cheng, X., Zhu, L., Zhang, Q., Zhang, L. l., et al. (2020). ZC4H2 Stabilizes RNF220 to Pattern Ventral Spinal Cord through Modulating Shh/Gli Signaling. J. Mol. Cell Biol 12 (5), 337–344. doi:10.1093/jmcb/mjz087
Ma, P., Song, N.-N., Li, Y., Zhang, Q., Zhang, L., Zhang, L., et al. (2019). Fine-tuning of Shh/Gli Signaling Gradient by Non-proteolytic Ubiquitination during Neural Patterning. Cell Rep. 28 (2), 541–553. doi:10.1016/j.celrep.2019.06.017
Masahira, N., Takebayashi, H., Ono, K., Watanabe, K., Ding, L., Furusho, M., et al. (2006). Olig2-positive Progenitors in the Embryonic Spinal Cord Give Rise Not Only to Motoneurons and Oligodendrocytes, but Also to a Subset of Astrocytes and Ependymal Cells. Dev. Biol. 293 (2), 358–369. doi:10.1016/j.ydbio.2006.02.029
Moore, S., Ribes, V., Terriente, J., Wilkinson, D., Relaix, F., and Briscoe, J. (2013). Distinct Regulatory Mechanisms Act to Establish and Maintain Pax3 Expression in the Developing Neural Tube. Plos Genet. 9 (10), e1003811. doi:10.1371/journal.pgen.1003811
Morreale, F. E., and Walden, H. (2016). Types of Ubiquitin Ligases. Cell 165 (1), 248. doi:10.1016/j.cell.2016.03.003
Mukhopadhyay, D., and Riezman, H. (2007). Proteasome-independent Functions of Ubiquitin in Endocytosis and Signaling. Science 315 (5809), 201–205. doi:10.1126/science.1127085
Novitch, B. G., Chen, A. I., and Jessell, T. M. (2001). Coordinate Regulation of Motor Neuron Subtype Identity and Pan-Neuronal Properties by the bHLH Repressor Olig2. Neuron 31 (5), 773–789. doi:10.1016/s0896-6273(01)00407-x
Pattyn, A., Vallstedt, A., Dias, J. M., Samad, O. A., Krumlauf, R., Rijli, F. M., et al. (2003). Coordinated Temporal and Spatial Control of Motor Neuron and Serotonergic Neuron Generation from a Common Pool of CNS Progenitors. Genes Dev. 17 (6), 729–737. doi:10.1101/gad.255803
Pickart, C. M., and Eddins, M. J. (2004). Ubiquitin: Structures, Functions, Mechanisms. Biochim. Biophys. Acta (Bba) - Mol. Cell Res. 1695 (1-3), 55–72. doi:10.1016/j.bbamcr.2004.09.019
Pickart, C. M. (2001). Mechanisms Underlying Ubiquitination. Annu. Rev. Biochem. 70, 503–533. doi:10.1146/annurev.biochem.70.1.503
Pierani, A., Moran-Rivard, L., Sunshine, M. J., Littman, D. R., Goulding, M., and Jessell, T. M. (2001). Control of Interneuron Fate in the Developing Spinal Cord by the Progenitor Homeodomain Protein Dbx1. Neuron 29 (2), 367–384. doi:10.1016/S0896-6273(01)00212-4
Richardson, W. D., Kessaris, N., and Pringle, N. (2006). Oligodendrocyte Wars. Nat. Rev. Neurosci. 7 (1), 11–18. doi:10.1038/nrn1826
Ruiz i Altaba, A. (1998). Combinatorial Gli Gene Function in Floor Plate and Neuronal Inductions by Sonic Hedgehog. Development 125 (12), 2203–2212. doi:10.1242/dev.125.12.2203
Simons, M., and Nave, K.-A. (2015). Oligodendrocytes: Myelination and Axonal Support. Cold Spring Harb Perspect. Biol. 8 (1), a020479. doi:10.1101/cshperspect.a020479
Song, N.-N., Jia, Y.-F., Zhang, L., Zhang, Q., Huang, Y., Liu, X.-Z., et al. (2016). Reducing central Serotonin in Adulthood Promotes Hippocampal Neurogenesis. Sci. Rep. 6, 20338. doi:10.1038/srep20338
Song, N.-N., Ma, P., Zhang, Q., Zhang, L., Wang, H., Zhang, L., et al. (2020). RNF220/ZC4H2-mediated Monoubiquitination of Phox2 Is Required for Noradrenergic Neuron Development. Development 147 (6), dev.185199. doi:10.1242/dev.185199
Song, N.-N., Xiu, J.-B., Huang, Y., Chen, J.-Y., Zhang, L., Gutknecht, L., et al. (2011). Adult Raphe-specific Deletion of Lmx1b Leads to central Serotonin Deficiency. PLoS One 6 (1), e15998. doi:10.1371/journal.pone.0015998
Stein, D. J., and Stahl, S. (2000). Serotonin and Anxiety: Current Models. Int. Clin. Psychopharmacol. 15 (Suppl. 2), S1–S6. doi:10.1097/00004850-200008002-00002
Tronche, F., Kellendonk, C., Kretz, O., Gass, P., Anlag, K., Orban, P. C., et al. (1999). Disruption of the Glucocorticoid Receptor Gene in the Nervous System Results in Reduced Anxiety. Nat. Genet. 23 (1), 99–103. doi:10.1038/12703
Walther, D. J., Peter, J.-U., Bashammakh, S., Hörtnagl, H., Voits, M., Fink, H., et al. (2003). Synthesis of Serotonin by a Second Tryptophan Hydroxylase Isoform. Science 299 (5603), 76. doi:10.1126/science.1078197
Weihe, E., Schäfer, M. K.-H., Erickson, J. D., and Eiden, L. E. (1994). Localization of Vesicular Monoamine Transporter Isoforms (VMAT1 and VMAT2) to Endocrine Cells and Neurons in Rat. J. Mol. Neurosci. 5 (3), 149–164. doi:10.1007/bf02736730
Zannino, D. A., and Appel, B. (2009). Olig2+ Precursors Produce Abducens Motor Neurons and Oligodendrocytes in the Zebrafish Hindbrain. J. Neurosci. 29 (8), 2322–2333. doi:10.1523/jneurosci.3755-08.2009
Keywords: dorsoventral patterning, hindbrain, Rnf220, serotonergic neuron, oligodendrocyte
Citation: Wang Y-B, Song N-N, Zhang L, Ma P, Chen J-Y, Huang Y, Hu L, Mao B and Ding Y-Q (2022) Rnf220 is Implicated in the Dorsoventral Patterning of the Hindbrain Neural Tube in Mice. Front. Cell Dev. Biol. 10:831365. doi: 10.3389/fcell.2022.831365
Received: 08 December 2021; Accepted: 08 February 2022;
Published: 24 March 2022.
Edited by:
Kunimasa Ohta, Kyushu University, JapanReviewed by:
Andreas H. Kottmann, City University of New York, United StatesCaitlin Winkler, University of Colorado, United States
Copyright © 2022 Wang, Song, Zhang, Ma, Chen, Huang, Hu, Mao and Ding. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bingyu Mao, bWFvQG1haWwua2l6LmFjLmNu; Yu-Qiang Ding, ZGluZ3l1cWlhbmdAdmlwLjE2My5jb20=
 Yu-Bing Wang
Yu-Bing Wang Ning-Ning Song
Ning-Ning Song Lei Zhang
Lei Zhang Pengcheng Ma
Pengcheng Ma Jia-Yin Chen
Jia-Yin Chen Ying Huang
Ying Huang Ling Hu
Ling Hu Bingyu Mao
Bingyu Mao Yu-Qiang Ding
Yu-Qiang Ding