- 1Key Laboratory of Resource Biology and Biotechnology in Western China, Ministry of Education, Northwest University, Xi’an, China
- 2Provincial Key Laboratory of Biotechnology of Shaanxi Province, Northwest University, Xi’an, China
- 3State Key Laboratory of Cancer Biology and National Clinical Research Center for Digestive Diseases, Xijing Hospital of Digestive Diseases, The Fourth Military Medical University, Xi’an, China
Colorectal cancer (CRC) is one of the most common types of cancer and one of the leading causes of mortality worldwide. MicroRNAs (miRNAs) play central roles in normal cell maintenance, development, and other physiological processes. Growing evidence has illustrated that dysregulated miRNAs can participate in the initiation, progression, metastasis, and therapeutic resistance that confer miRNAs to serve as clinical biomarkers and therapeutic targets for CRC. Through binding to the 3′-untranslated region (3′-UTR) of target genes, miRNAs can lead to target mRNA degradation or inhibition at a post-transcriptional level. During the last decade, studies have found numerous miRNAs and their potential targets, but the complex network of miRNA/Targets in CRC remains unclear. In this review, we sought to summarize the complicated roles of the miRNA-target regulation network (Wnt, TGF-β, PI3K-AKT, MAPK, and EMT related pathways) in CRC with up-to-date, high-quality published data. In particular, we aimed to discuss the downstream miRNAs of specific pathways. We hope these data can be a potent supplement for the canonical miRNA-target regulation network.
Introduction
Colorectal cancer (CRC) remains one of the leading causes of related death worldwide. It is estimated that over 1.9 million new colorectal cancer (including anus) cases and 935,000 deaths occurred in 2020, representing about 1 in 10 cancer cases and deaths (Sung et al., 2021). Overall, colorectal cancer ranks third in terms of incidence and second in terms of mortality. Three main carcinogenic pathways determine CRC molecular profile: chromosomal instability (CIN), microsatellite instability (MSI), and CpG island methylator phenotype (CIMP) pathways (Pino and Chung, 2010). The accumulation of various genetic and epigenetic changes in colonic epithelial cells is a crucial factor driving the initiation, progression, and metastasis of CRC (Okugawa et al., 2015). In addition to canonical APC, KRAS and TP53 mutations in CRC, epigenetic changes also play a central role in the pathogenesis of CRC.
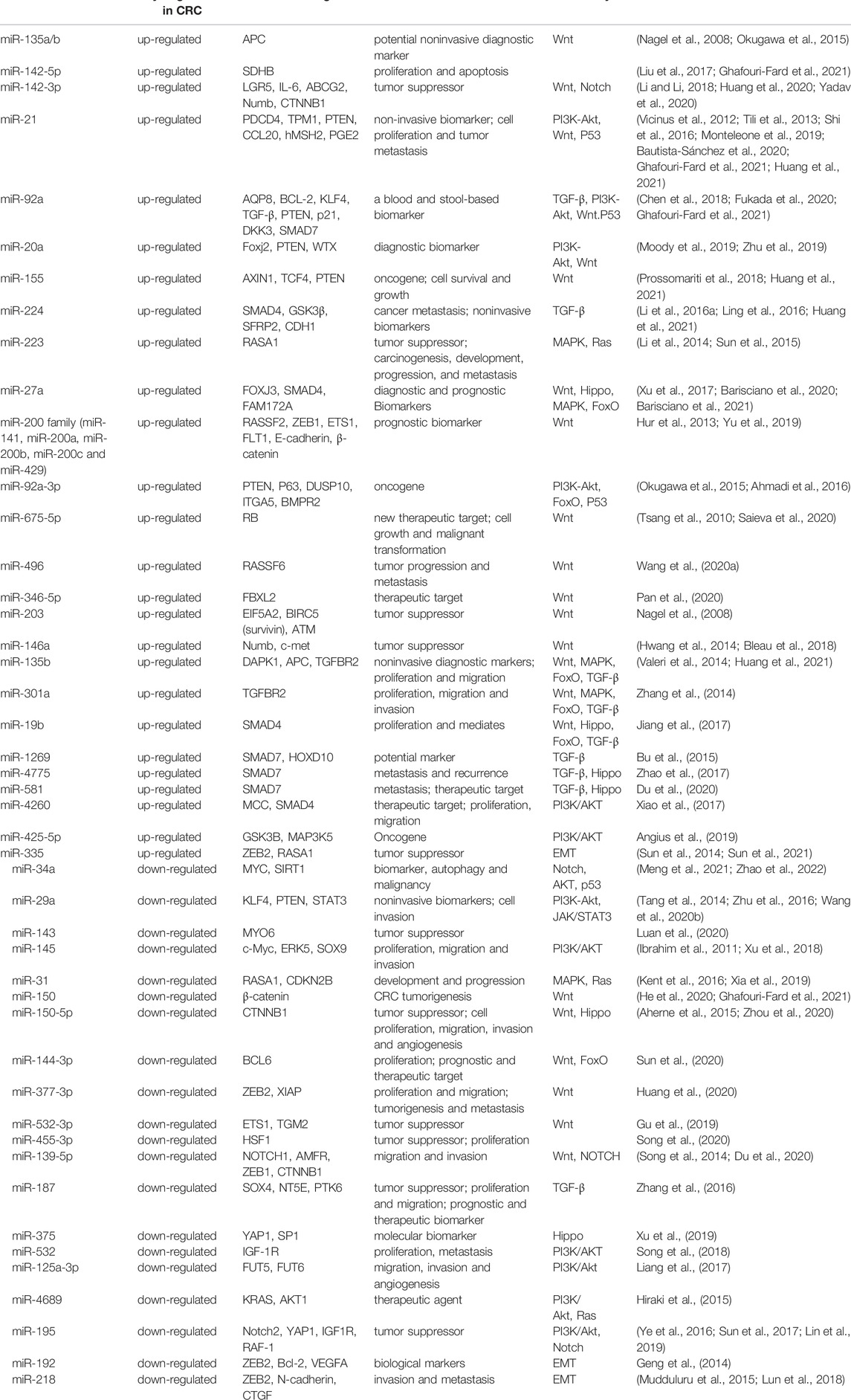
MicroRNAs (miRNAs) are non-coding RNAs that suppress mRNA expression through complementary base pairing. In most instances, miRNAs are transcribed by RNA polymerase II to form primary miRNAs (pri-miRNAs), and subsequently cleaved into precursor miRNAs (pre-miRNAs) processed by DROSHA and DCGR8. EXPO5 mediates the export of these pre-miRNAs to the cytoplasm for further processing by DICER, which cleaves pre-miRNAs to form mature miRNAs of approximately 20 nucleotides in length. One of the strands of mature miRNA (guide strand) gets incorporated into RISC involving DICER and AGO2 to bind to target genes (Rani and Sengar, 2022; Du and Palczewski, 2022). During the last decades, miRNAs have been shown to play an essential role in cancer progression (Rupaimoole and Slack, 2017). At all stages of CRC development, miRNAs can influence cancer-related pathways by suppressing corresponding targets at the post-transcriptional level. Single miRNA alone or panel miRNAs have been shown as a potential class of valuable biomarkers (Jung et al., 2020) (Table 1). Among these miRNAs, miR-21, and miR-92a are two well-characterized oncogenic miRNAs that are identified as the most promising biomarkers for CRC early detection and prognosis. The regulation and function studies of these miRNAs were subsequently investigated, which expanded our understanding of the sophisticated mechanism of CRC pathogenesis (Pu et al., 2019). MiRNAs exert their function (proliferation, invasion, or epithelial-mesenchymal transition) by targeting specific genes involved in cancer-related signaling pathways, including the Wnt/β-catenin, PI3K/AKT, TGF-β/Smads, and EGFR pathways (Niu et al., 2021a). However, since a single miRNA can target multiple genes, and one target gene could harbour tens of hundreds of binding miRNAs (Table 2), the complex miRNA-mRNA regulatory network remains to be fully illustrated (Okugawa et al., 2015).
In this review, we provide an overview of dysregulated miRNAs in CRC and highlight the promising miRNAs biomarkers involved in the CRC-related signaling pathways. We present a systematic search of up-to-date studies with high-quality profiling of the miRNAs involved in the progression or metastasis of CRC, mainly published from January 2019 to January 2022. In particular, we focus on the downstream miRNAs of specific pathways. We hope these data can be a potent supplement for the canonical miRNA-target regulation network.
Wnt Signaling Pathway
In different stages of development and particular cellular conditions, the canonical Wnt pathway is triggered by attaching the Wnt ligands to specific receptors. Stimulation of the canonical Wnt signaling leads to the inhibition of the GSK-3β, thus triggering the accumulation of stabilized β-catenin in the cytoplasm. β-catenin translocates into the nucleus and interacts with T-cell factor/lymphocyte enhancer factor (TCF/LEF) and subsequently loads Brg-1 and p300/CBP proteins onto promoters, thereby regulating the transcription of multiple target genes involved in cell proliferation, differentiation, and apoptosis (Azbazdar et al., 2021). Studies reported more than 80% of CRC samples have been detected with Wnt pathway activation, mainly caused by adenomatous polyposis coli (APC) or β-catenin (CTNNB1) coding gene mutations. Evidence has illustrated that the aberrant expression of microRNAs upstream of the Wnt cascade could be involved in CRC initiation and progression (Figure 1). As key oncogenes in the Wnt pathway, miR-135a/-135b directly target the 3′-UTR of APC to suppress its expression and induce downstream Wnt pathway activation (Nagel et al., 2008). Stool miR-135b expression is dramatically up-regulated in CRC patients. It targets the mRNA of ZNRF3, a zinc and ring finger protein that acts as a tumor suppressor in the intestinal stem cell zone by inhibiting the Wnt signaling pathway. MiR-135b may be a promising non-invasive biomarker for diagnosing CRC patients with TNM stage-III/IV (Li et al., 2020). Decreased levels of miR-142 and miR-150 were detected in CRC patients. In vitro function assays suggest that these two miRNAs could directly bind to CTNNB1 and thus inhibit CRC tumorigenesis (He et al., 2020; Liu et al., 2020). MiR-34a/b/c can directly act and inhibit the effector molecules of the Wnt signaling pathway, including WNT1, WNT3, LRP6, and CTNNB1 (Kim et al., 2011; Jiang and Hermeking, 2017), while inhibiting the WNT signaling pathway. MiR-224/GSK3β axis, identified by bioinformatics and luciferase function studies, can lead to the activation of Wnt/β-catenin signaling and the nuclear translocation of β-catenin (Li et al., 2016a). Saieva et al also reported that inhibition of the hypoxia-induced miR-675-5p could hamper the nuclear localization of β-catenin by regulating GSK-3β activity (Saieva et al., 2020). The ectopic expression of miR-92a-3p up-regulates cell cycle- and mitosis-related gene expression while down-regulates adhesion-related gene expression in endothelial cells by targeting Wnt regulatory protein DKK3 and Claudin-11 (Yamada et al., 2019). In addition, recent studies have also reported several miRNAs indirectly involved in Wnt pathway-mediated tumor progression and metastasis, such as elevated miR-496 (Wang et al., 2020a), miR-346-5p (Pan et al., 2020), and miR-203 (Peng et al., 2020), and down-regulated miR-144-3p (Sun et al., 2020), miR-377-3p (Huang et al., 2020), miR-532-3p (Gu et al., 2019), and miR-200c (Cong et al., 2021).
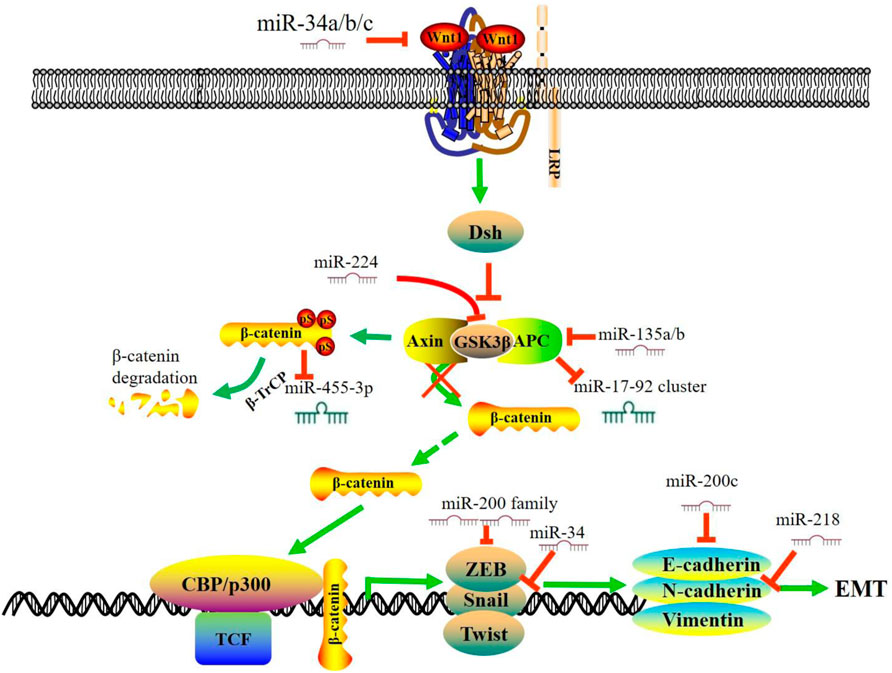
FIGURE 1. MiRNAs regulate Wnt/β-catenin signaling pathway and EMT in CRC. In the absence of Wnt, β-catenin forms a complex with APC, GSK3 and Axin. The β-catenin is phosphorylated by GSK3β and targeted for ubiquitin-dependent proteasomal degradation. This figure summarizes the interaction between miRNAs and the Wnt/β-catenin signaling pathway in CRC and a subset of downstream miRNAs that are involved in the EMT process. During the process of EMT, the expression of mesenchymal markers, including N-cadherin and vimentin, is up-regulated, and the expression of the epithelial markers, including E-cadherin and ZO-1, is down-regulated. miR, microRNA; EMT, epithelial-to-mesenchymal transition; APC, Adenomatous Polyposis Coli; TCF, T-cell factor.
Emerging studies have revealed a subset of miRNAs downstream of the Wnt signaling pathway. Activation of Wnt pathway alters the expression of these miRNAs and triggers the malignant phenotype. Heat shock transcription factor 1 (HSF1) is overexpressed in CRC and is vital for β-catenin to promote CRC development. β-catenin overexpression suppresses miR-455-3p generation to stimulate m6A modification. The translation of HSF1 mRNA is impaired after β-catenin depletion, accompanied by the up-regulation of miR-455-3p (Song et al., 2020). In KRAS-mutant cells, aberrantly activated Wnt signaling represses miR-139-5p through Transcription Factor 4 (TCF4), which forms a miR-139-5p/Wnt signaling feedback loop (Du et al., 2020). The APC and β-catenin protein could regulate miR-17–92 cluster, and the loss of APC can lead to the activation of β-catenin, thus activating the miR-17–92 promoter (Li et al., 2016b). Snail, a downstream effector and target of Wnt, induces the expression of miR-146a through the β-catenin-TCF4 complex. Meanwhile, miR-146a can target Numb to stabilize β-catenin, forming a feedback loop to maintain Wnt activity (Hwang et al., 2014).
TGF-β Signaling Pathway
The transforming growth factor—beta (TGF-β) signaling depends on Smad family proteins. Active TGF-β1 at the cell surface mediates signaling through the TGF-β type I and type II receptors (TβRI and TβRII) which are active serine/threonine kinases. The TGF-β/receptor complex subsequently phosphorylates receptor-activated Smads (R-Smads). R-Smads translocate to the nucleus and regulate the transcription of target genes (Tecalco-Cruz et al., 2018). TGF-β signaling pathway plays an essential role not only in normal colonic tissue homoeostasis but in CRC progression (Farooqi et al., 2019). This canonical pathway exerts a dual role in the carcinogenesis of various cancer types. In the early stages, the TGF-β pathway acts as a tumor suppressor, while in the advanced stages, and it enhances the malignant phenotype (Luo et al., 2019; Niu et al., 2021b; Vanpouille-Box and Formenti, 2018). Studies reported that more than 90% of microsatellite instability (MSI) tumors are affected by frameshift mutations in the TGFBR2 gene, leading to impaired receptor expression, and abrogated downstream signaling (Fricke et al., 2019). Due to the relatively long 3′-UTR of TGFBR2 (∼9.2 k), studies reported that many onco-miRNAs are involved in TGFBR2 regulation. MiR-21, miR-135b, and miR-301a exert their tumor-promoting role in proliferation, migration, and invasion in CRC cells by targeting TGFBR2 (Figure 2) (Yu et al., 2012; Zhang et al., 2014).
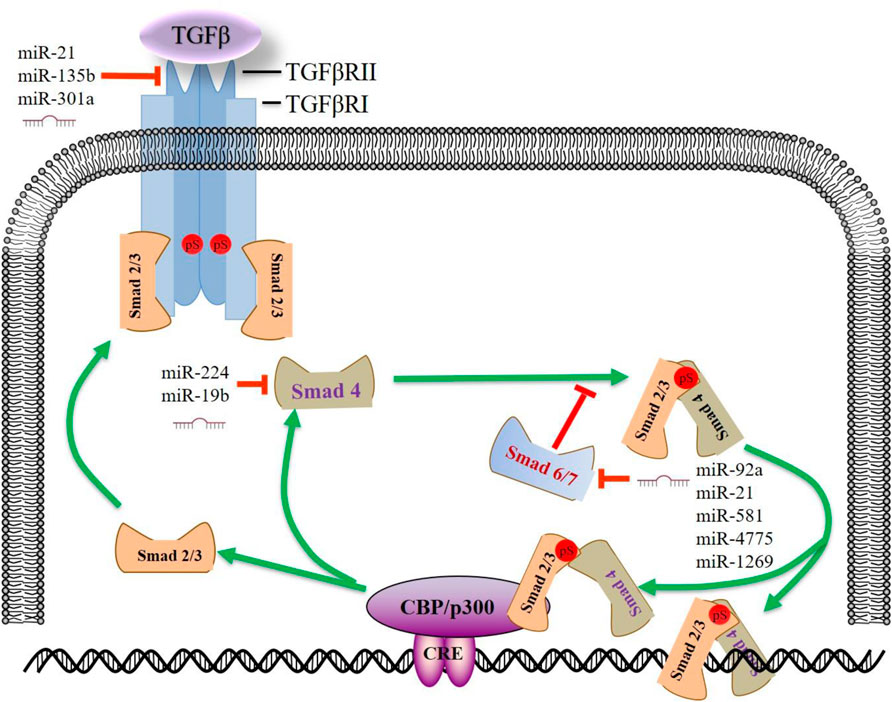
FIGURE 2. MiRNAs regulate the TGF-β signaling pathway in CRC. Upon TGF-β stimulation, activated TGFBR2 binds to and phosphorylates TGFBR1. Activated TGFBR1 phosphorylates SMAD2/SMAD3, enabling SMAD2/SMAD3 to bind SMAD4 and subsequent activation or repression of TGF-β responsive genes. Some miRNAs enhance the TGF-β signaling by targeting SMAD7, forming a positive feedback loop. This figure summarizes some prominent miRNAs implicated in CRC progression by targeting TGF-β signaling pathway. miR, microRNA; CRE, cAMP response element; TGFβ, transforming growth factor β; SMAD, SMAD Family Member; TGFβR, transforming growth factor β receptor.
In the SMAD-dependent pathway, phosphorylated Receptor-regulated-SMADs (R-SMADs, SMAD2 or 3) form functionally active complexes with joint partner SMAD4 and move into the nucleus to regulate a myriad of target genes (Itatani et al., 2019). A recent study showed that miR-224 could promote CRC metastasis, at least in part, through the regulation of tumor suppressor SMAD4. MiR-224 expression in primary CRC, alone or combined with SMAD4, may have prognostic value for the survival of patients with CRC (Ling et al., 2016). An elevated level of miR-19b was an independent prognostic factor associated with CRC patients’ survival. MiR-19b/SMAD4 promotes cell proliferation and mediates resistance to oxaliplatin-based chemotherapy (Jiang et al., 2017).
SMAD7, a negative regulator of the TGF-β/SMADs pathway, binds to TβR1 and competes with SMAD2/3, thus preventing the phosphorylation of R-SMADs (Stolfi et al., 2020). Meanwhile, SMAD7 can enhance the ubiquitination and proteasome-mediated degradation of TβR1 by SMURF1/2. In the last 5 years, the role of miRNA/SMAD7/TGFβ axis in CRC metastasis has been broadly discussed. MiR-21 (Jiao et al., 2017), miR-1269 (Bu et al., 2015), miR-4775 (Zhao et al., 2017), and miR-581 (Zhao et al., 2020) were uniquely up-regulated and negatively correlated with SMAD7 expression in metastatic CRC samples. Function assays confirmed that these miRNAs exert oncogenic roles only in regulating migration and invasion. Similarly, these miRNAs enhance TGF-β signaling by targeting SMAD7, forming a positive feedback loop. Based on the binding capacity and correlation analysis, bioinformatics also revealed that SMAD7 is among the top target of miR-92a, the blood and stool-based biomarker of CRC (Chen et al., 2018) (Table 1).
On the other hand, activation of the TGFβ pathway can trigger the alteration of some downstream miRNAs. MiR-187, a downstream effector of the TGFβ pathway, suppresses Smad-mediated epithelial-mesenchymal transition in CRC (Zhang et al., 2016). MiR-187 directly targets SOX4, NT5E, and PTK6, which are essential upstream effectors of the Smad pathway, resulting in a sustained TGFβ activation in cancer progression. Furthermore, TGFBR2 deficiency in HCT 116 cells leads to the down-regulation of miR-381-3p, which is expected to be of functional relevance and may contribute to MSI-specific tumor characteristics (Fricke et al., 2019). In addition, we analyzed the GEO dataset GSE53338 (TGF-β knock-down cells) and found that CRC biomarkers miR-31, miR-145, and miR-20a were down-regulated in the TGF-β knock-down cells. These results suggest that activation of TGF-β/SMADs could also influence Smad-dependent downstream miRNAs, thus contributing to CRC progression.
PI3K-AKT Pathway
The PI3K/Akt is a pathway that can promote cell survival and growth. Lipid kinase PI3K phosphorylates phosphatidylinositol 4,5-bisphosphate (PIP2) to generate phosphatidylinositol 3,4,5-trisphosphate (PIP3), which in turn phosphorylates protein kinase B (AKT) to cause a cascade of responses of cellular function. PIP3 can be dephosphorylated back to PIP2 by the lipid phosphatase PTEN (Hoxhaj and Manning, 2020). Activation of the PI3K/AKT/mTOR signaling is a prominent feature of human cancers (Hoxhaj and Manning, 2020; Fruman et al., 2017). PI3K/AKT signaling regulates cell survival and proliferation (Cantley, 2002; Ediriweera et al., 2019). Consequently, molecules in this pathway (p110, p85, AKT, mTOR, and PTEN, etc.) provide multiple novel targets for therapy (Mayer and Arteaga, 2016). MiRNAs could directly target the key molecules in the PI3K signaling pathway to promote the progression of CRC (Figure 3). MiR-21 expression is up-regulated in CRC and functions as an oncomir in CRC through PTEN (Xiong et al., 2013; Liu et al., 2019). Beyond that, miR-92a directly targets 3′-UTR of PTEN. The up-regulation of miR-92a inhibits the expression of PTEN, thus activating the PI3K/AKT signaling pathway and promoting CRC cell proliferation and lymph node metastasis (Ke et al., 2015; Chen et al., 2018). In a meta-analysis of cancer genome sequencing results, PIK3CA and PTEN are highly mutated genes in most human cancers (Lawrence et al., 2014). Herein, miR-375 represses CRC tumorigenesis by targeting PIK3CA by inactivating the PI3K/AKT signaling pathway. Moreover, miR-125a-3p directly inhibits FUT5 (fucosyltransferase) and FUT6, downstream targets of the PI3K/AKT signaling pathway in CRC. (Liang et al., 2017). This evidence indicates that the up-regulation of the PI3K/AKT signaling pathway is an essential aspect of miRNAs in CRC pathogenesis.
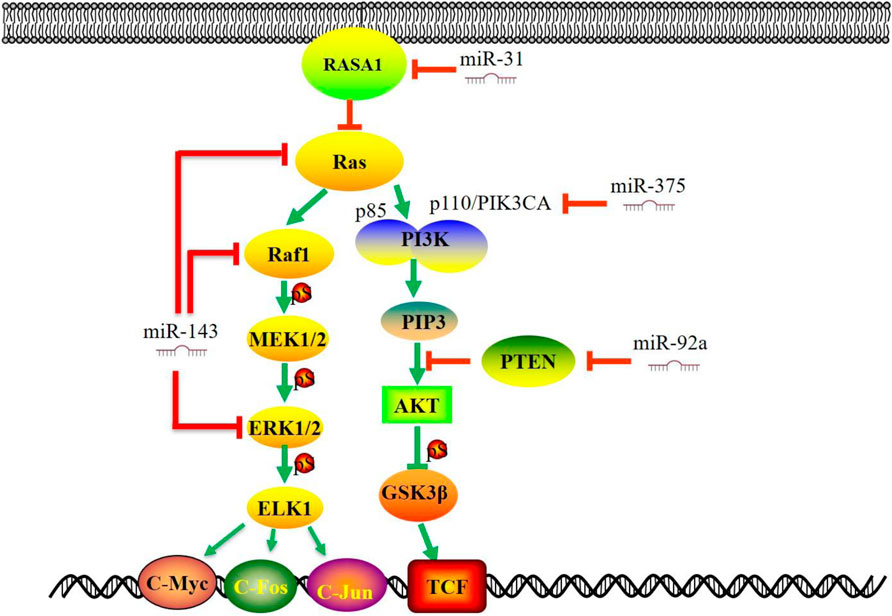
FIGURE 3. MiRNAs regulate Ras-Raf-MAPK and PI3K/AKT signaling pathways in CRC. Several studies have demonstrated that miRNAs target the key molecules in the MAPK signaling pathway (RAS, RAF, ERK, MEK5, ELK1 etc.). The diversity of alterations in the PI3K/AKT signaling pathway (p110, p85, AKT, PTEN, etc.) provides multiple molecular targets for therapy and raises the challenge of CRC. This figure summarizes that some significant miRNAs are involved in CRC progression by targeting critical molecules in MAPK and PI3K/AKT signaling pathway. miR, microRNA; PI3K, phosphatidylinositol 3-kinase; PTEN, phosphatase and tensin homolog; GSK3β, Glycogen Synthase Kinase 3 Beta; RASA1, RAS P21 Protein Activator 1; TCF, T-cell factor.
Ras-Raf-MAPK Signaling Pathway
The Ras-Raf-MAPK signaling pathway plays a central role in the survival and development of normal and tumor cells. (Downward, 2003; Collisson et al., 2014). Extracellular ligand (such as growth factor) binds to receptor tyrosine kinase (RTKs), which results in the recruitment of small G-proteins (RAS) to the cell membrane. In turn, RAS-GTP directly binds to RAF protein, thus phosphorylating MEK and ERK, followed by the activation of the mitogen-activated protein kinase (MAPK) pathway. (Figure 3). The MAPK pathway is a downstream effector of many growth-factor receptors, including the epidermal growth factor receptor (EGFR) (Fang and Richardson, 2005). Recent studies have shown that down-regulation of miR-143 and, or the up-regulation of miR-31 could trigger the RAS-RAF–MAPK pathway to enhance proliferation and modulating treatment responses (Sun et al., 2013; Akao et al., 2018). Furthermore, the expression of miR-143 was decreased in CRC specimens, and KRAS was a target gene of miR-143 (Luo et al., 2020). MiR-31 directly targets RAS p21 Protein Activator 1 (RASA1), a negative regulator of the RAS pathway, and modulates invasion behavior through interaction with p190RhoGAP and its downstream proteins, which plays essential roles in cell proliferation and differentiation (Buday and Downward, 2008; Sun et al., 2013). MiR-425-5p might participate in the pathogenesis of KRAS-mutated CRC and contribute to this subcategory of CRC (Angius et al., 2019). As a negative regulator, miR-4689 potently suppresses the oncogenic KRAS-driven EGFR signaling pathways through the direct inhibition of KRAS and AKT1. The loss of miR-195 lessens its binding and inhibitory effect to RAF-1, thus promoting colorectal cancer cell proliferation and cell survival (Ye et al., 2016).
The activation or mutation of BRAF and, or KRAS can trigger the abnormal expression of downstream miRNAs. Tumors with BRAF and KRAS mutations showed significantly elevated levels of miR-31 expression (Lundberg et al., 2018). MiR-193a, one of the candidate miRNAs in BRAF-mutated cells, acts as a tumor suppressor by regulating multiple cancer-related genes and affects cellular sensitivity to MAPK-related pathway inhibitors, such as BRAF inhibitors, MEK inhibitors, and, or anti-EGFR antibodies (Hiraide et al., 2021). MiR-139-5p could serve as a novel regulator of the crosstalk between the Ras and Wnt signaling pathways. Aberrantly activated Wnt signaling in KRAS-mutant cells represses miR-139-5p through TCF4, forming a double negative feedback loop of mir-139-5p/Wnt signal (Du et al., 2020). Mutant KRAS functions as a broad regulator of the EGFR signaling cascade by inhibiting miR-4689, negatively regulating both the RAS/MAPK and PI3K/AKT pathways (Hiraki et al., 2015). Therefore, targeting miRNAs that play a critical role in the Ras-Raf-MAPK signaling pathway might be an alternative therapy in CRC patients with aberrant Ras-Raf-MAPK signaling.
CRC Metastasis-Related Pathways
Epithelial-to-mesenchymal transition (EMT) is a process which cells lose their epithelial characteristics while gain mesenchymal properties, which has a critical role in tumor initiation, progression, and metastasis (Tsai and Yang, 2013; Dong et al., 2020; Taki et al., 2021). Growing evidence from preclinical studies has shown that some EMT biomarkers could serve as potential therapeutic targets (Zhang et al., 2021). The mesenchymal markers, including N-cadherin and Vimentin, are up-regulated, while the epithelial markers (E-cadherin, EpCAM, and ZO-1) are down-regulated during the EMT process (Loh et al., 2019; Satelli and Li, 2011). In addition, the transcription factors, such as Twists, Snails, Slug and ZEBs, also participate in this essential program. (Diepenbruck and Christofori, 2016; Mittal, 2018). Accordingly, miRNAs can regulate EMT by inhibiting the genes involved in EMT in CRC (Figure 1). The miR-200 family is a group of microRNAs with five members: miR-141, miR-200a, miR-200b, miR-200c, and miR-429, which target the EMT signaling pathway (O'Brien et al., 2018). In CRC patients, decreased expression of miR-429, miR-200a, and miR-200c is associated with worse survival. Gollavilli et al. reported that miR-200b/c low expressing cells could form a significantly lower number of liver metastasis in nude mice, which is in line with the partial EMT phenotype. In addition, a Seahorse metabolic analysis showed a drastic reduction of OXPHOS ability which further explain the loss of growth observed in miR-200b/c low-expressing cells. MiR-205-5p functions as a tumor suppressor and enhanced expression of miR-205-5p can significantly raise the levels of E-cadherin by targeting ZEB1 (Gulei et al., 2018). Furthermore, miRs-192, -218, and -335 (Figure 1) target ZEB2, and miR-218 also targets N-cadherin to promote EMT (Geng et al., 2014; Mudduluru et al., 2015). Transcription factors, including Snails and ZEBs, also regulate downstream miRNAs to affect the CRC malignant phenotype. In HT-29 cells, Snail overexpression repressed the level of miR-192 and miR-194 and increased the levels of miR-205 and let-7i (Przygodzka et al., 2019). Wang et al. confirmed that the chimeric [SNAIL/miR-34] [ZEB/miR-200] system might be the core axis for the EMT process (Wang et al., 2018).
Epidemiological and experimental studies have revealed that inflammatory signaling pathways are also involved in CRC metastasis (Świerczyński et al., 2021). NF-κB and STAT3 pathways are two critical regulators of inflammation. MiR-21 is also involved in the regulation of NF-κB and MyD88. MiR-21 reduces the tumor burden and decreases the expression of pro-inflammatory cytokines. Meanwhile, by depleting miR-21 in tumors, the PDCD4 expression increased, while the expression of STAT3, and BCL2 decreased (Bakirtzi et al., 2011; Shi et al., 2016). MiR-221 and miR-222 can also activate NF-κB and STAT3 by indirectly modulating their protein stability through miR-221/222-mediated positive feedback loops (Liu et al., 2014). IL-6/STAT3/MMPs signaling induces EMT and metastasis in colorectal cancer. An active IL-6R/STAT3/miR-34a loop is necessary for this program in CRC cells and this could affect nodal and distant metastasis in CRC patients (Rokavec et al., 2014). Yang and colleagues found that miR-1301 is significantly down-regulated in CRC samples. Functionally, miR-1301 regulates CRC cell migration and invasion capacity through the STAT3/MMPs axis (Yang et al., 2021). MiR-146 can function as a negative regulator of colonic inflammation. In myeloid cells, miR-146a prevents intestinal inflammation by limiting myeloid cell-mediated inflammatory IL-17 production or inhibiting tumorigenic IL-17R signaling in intestinal epithelial cells (Garo et al., 2021).
Summary and Future Perspective
Colorectal cancer usually requires the surgical removal of tumor tissue, chemotherapy and targeted therapy. At present, new targeted drugs are mainly aimed at the tumor angiogenesis endothelial growth factor (vascular endothelial growth factor, VEGF) and receptor (vascular endothelial growth factor receptor, VEGFR), or the epidermal growth factor receptor (EGFR) (Rio-Vilariño et al., 2021). However, more than 30% of cases are resistant to EGFR inhibitors, so it is crucial to find new therapeutic targets (Mangiapane et al., 2022). Anti-tumor immunotherapy has become another essential means of treating colorectal cancer after surgery, chemotherapy, radiotherapy, and targeted therapy. In this field, negative regulators of the immune system, which are called immune checkpoints, play a vital role in limiting anti-tumor immune responses. On this account, immune checkpoint-inhibiting agents, like those directed against cytotoxic T-lymphocyte antigen 4 (CTLA-4), programmed death-1 receptor (PD1), and its ligand PD-L1, have been developed as anti-tumor drugs, producing exciting results in preclinical and clinical studies (Ganesh et al., 2019).
Given the multiple studies on ncRNAs, miRNAs undoubtedly hold great potential as future biomarkers for the diagnosis, prognostication, and response prediction. Inhibitors targeting onco-miRNAs might be a fundamental treatment approach, at least at the hypothetical level. Despite the preclinical evidence, no clinical trials currently evaluate miRNA mimics specifically in CRC. A systematic understanding of the miRNAs’ regulatory mechanisms will explore their future clinical applications as biomarkers or their potential as therapeutic targets in CRC. In recent years, long noncoding RNAs (lncRNAs) have been demonstrated to be critical factors in colon cancer development and progression. LncRNAs can regulate gene transcription by binding miRNAs and acting as a sponge. LncRNAs, miRNAs and mRNA all together contribute to the biological processes in CRC, including metastasis, EMT, or inflammation formation. The in-depth studies in this field will provide new ideas for the diagnosis and treatment of CRC. However, the complex mechanism of LncRNA/miRNA/mRNA axis and assessing their values in CRC early detection or treatment would be a great challenge (Toiyama et al., 2018; Marcuello et al., 2019).
Taken together, this review summarized the up-to-date studies in the field of CRC-related miRNAs and the complex regulating network. We focused on miRNAs that could serve as important indicators for CRC and highlighted the dysregulated miRNAs downstream of specific pathways. This study would be helpful for further understanding of the non-coding RNA mechanism in CRC and provide new outlooks of these molecules for their utilization as targets for the treatment of CRC in the future.
Author Contributions
All authors participated in the writing of this manuscript. Conceptualization and original draft preparation: EC and FY. Table editing: YC, CW, and GX. Figure editing: FY, LC, and MZ. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by Grants from the National Natural Scientific Foundation of China (No. 82002038 and No. 82003115), the Subject Booster Program of Xijing Hospital (XJZT19MJ30), and the Natural Science Basic Research Plan in Shaanxi Province of China (2017JM3033).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to give our sincere appreciation to the reviewers for the helpful comments on this paper.
References
Aherne, S. T., Madden, S. F., Hughes, D. J., Pardini, B., Naccarati, A., Levy, M., et al. (2015). Circulating miRNAs miR-34a and miR-150 Associated with Colorectal Cancer Progression. BMC Cancer 15, 329. doi:10.1186/s12885-015-1327-5
Ahmadi, S., Sharifi, M., and Salehi, R. (2016). Locked Nucleic Acid Inhibits miR-92a-3p in Human Colorectal Cancer, Induces Apoptosis and Inhibits Cell Proliferation. Cancer Gene Ther. 23, 199–205. doi:10.1038/cgt.2016.10
Akao, Y., Kumazaki, M., Shinohara, H., Sugito, N., Kuranaga, Y., Tsujino, T., et al. (2018). Impairment of K‐Ras Signaling Networks and Increased Efficacy of Epidermal Growth Factor Receptor Inhibitors by a Novel Synthetic miR‐143. Cancer Sci. 109, 1455–1467. doi:10.1111/cas.13559
Angius, A., Pira, G., Scanu, A. M., Uva, P., Sotgiu, G., Saderi, L., et al. (2019). MicroRNA-425-5p Expression Affects BRAF/RAS/MAPK Pathways in Colorectal Cancers. Int. J. Med. Sci. 16, 1480–1491. doi:10.7150/ijms.35269
Azbazdar, Y., Karabicici, M., Erdal, E., and Ozhan, G. (2021). Regulation of Wnt Signaling Pathways at the Plasma Membrane and Their Misregulation in Cancer. Front. Cel Dev. Biol. 9, 631623. doi:10.3389/fcell.2021.631623
Bakirtzi, K., Hatziapostolou, M., Karagiannides, I., Polytarchou, C., Jaeger, S., Iliopoulos, D., et al. (2011). Neurotensin Signaling Activates microRNAs-21 and -155 and Akt, Promotes Tumor Growth in Mice, and Is Increased in Human colon Tumors. Gastroenterology 141, 1749–1761. doi:10.1053/j.gastro.2011.07.038
Barisciano, G., Colangelo, T., Rosato, V., Muccillo, L., Taddei, M. L., Ippolito, L., et al. (2020). miR-27a Is a Master Regulator of Metabolic Reprogramming and Chemoresistance in Colorectal Cancer. Br. J. Cancer 122, 1354–1366. doi:10.1038/s41416-020-0773-2
Barisciano, G., Leo, M., Muccillo, L., Pranzini, E., Parri, M., Colantuoni, V., et al. (2021). The miR-27a/FOXJ3 Axis Dysregulates Mitochondrial Homeostasis in Colorectal Cancer Cells. Cancers 13 (19), 4994. doi:10.3390/cancers13194994
Bautista-Sánchez, D., Arriaga-Canon, C., Pedroza-Torres, A., De La Rosa-Velázquez, I. A., González-Barrios, R., Contreras-Espinosa, L., et al. (2020). The Promising Role of miR-21 as a Cancer Biomarker and its Importance in RNA-Based Therapeutics. Mol. Ther. - Nucleic Acids 20, 409–420. doi:10.1016/j.omtn.2020.03.003
Bleau, A.-M., Redrado, M., Nistal-Villan, E., Villalba, M., Exposito, F., Redin, E., et al. (2018). miR-146a Targets C-Met and Abolishes Colorectal Cancer Liver Metastasis. Cancer Lett. 414, 257–267. doi:10.1016/j.canlet.2017.11.008
Bu, P., Wang, L., Chen, K.-Y., Rakhilin, N., Sun, J., Closa, A., et al. (2015). miR-1269 Promotes Metastasis and Forms a Positive Feedback Loop with TGF-β. Nat. Commun. 6, 6879. doi:10.1038/ncomms7879
Buday, L., and Downward, J. (2008). Many Faces of Ras Activation. Biochim. Biophys. Acta (Bba) - Rev. Cancer 1786, 178–187. doi:10.1016/j.bbcan.2008.05.001
Cantley, L. C. (2002). The Phosphoinositide 3-kinase Pathway. Science 296, 1655–1657. doi:10.1126/science.296.5573.1655
Caramés, C., Cristobal, I., Moreno, V., Marín, J. P., González-Alonso, P., Torrejón, B., et al. (2016). MicroRNA-31 Emerges as a Predictive Biomarker of Pathological Response and Outcome in Locally Advanced Rectal Cancer. Int. J. Mol. Sci. 17 (6), 878. doi:10.3390/ijms17060878
Chang, P.-Y., Chen, C.-C., Chang, Y.-S., Tsai, W.-S., You, J.-F., Lin, G.-P., et al. (2016). MicroRNA-223 and microRNA-92a in Stool and Plasma Samples Act as Complementary Biomarkers to Increase Colorectal Cancer Detection. Oncotarget 7, 10663–10675. doi:10.18632/oncotarget.7119
Chen, E., Li, Q., Wang, H., Yang, F., Min, L., and Yang, J. (2018). MiR-92a Promotes Tumorigenesis of Colorectal Cancer, a Transcriptomic and Functional Based Study. Biomed. Pharmacother. 106, 1370–1377. doi:10.1016/j.biopha.2018.07.098
Chen, J., Wang, W., Zhang, Y., Chen, Y., and Hu, T. (2014). Predicting Distant Metastasis and Chemoresistance Using Plasma miRNAs. Med. Oncol. 31, 799. doi:10.1007/s12032-013-0799-x
Collisson, E. A., Campbell, J. D., Brooks, A. N., Berger, A. H., Lee, W., Chmielecki, J., et al. (2014). Comprehensive Molecular Profiling of Lung Adenocarcinoma. Nature 511 (7511) 543–550. doi:10.1038/nature13385
Cong, J., Gong, J., Yang, C., Xia, Z., and Zhang, H. (2021). MiR-200c/FUT4 axis Prevents the Proliferation of colon Cancer Cells by Downregulating the Wnt/β-Catenin Pathway. BMC Cancer 21, 2. doi:10.1186/s12885-020-07670-y
Diepenbruck, M., and Christofori, G. (2016). Epithelial-mesenchymal Transition (EMT) and Metastasis: Yes, No, Maybe? Curr. Opin. Cel Biol. 43, 7–13. doi:10.1016/j.ceb.2016.06.002
Dong, B., Qiu, Z., and Wu, Y. (2020). Tackle Epithelial-Mesenchymal Transition with Epigenetic Drugs in Cancer. Front. Pharmacol. 11, 596239. doi:10.3389/fphar.2020.596239
Downward, J. (2003). Targeting RAS Signalling Pathways in Cancer Therapy. Nat. Rev. Cancer 3, 11–22. doi:10.1038/nrc969
Drebber, U., Lay, M., Wedemeyer, I., Vallböhmer, D., Bollschweiler, E., Brabender, J., et al. (2011). Altered Levels of the Onco-microRNA 21 and the Tumor-Supressor microRNAs 143 and 145 in Advanced Rectal Cancer Indicate Successful Neoadjuvant Chemoradiotherapy. Int. J. Oncol. 39, 409–415. doi:10.3892/ijo.2011.1036
Du, F., Cao, T., Xie, H., Li, T., Sun, L., Liu, H., et al. (2020). KRAS Mutation-Responsive miR-139-5p Inhibits Colorectal Cancer Progression and Is Repressed by Wnt Signaling. Theranostics 10, 7335–7350. doi:10.7150/thno.45971
Du, M., Liu, S., Gu, D., Wang, Q., Zhu, L., Kang, M., et al. (2014). Clinical Potential Role of Circulating microRNAs in Early Diagnosis of Colorectal Cancer Patients. Carcinogenesis 35, 2723–2730. doi:10.1093/carcin/bgu189
Du, S. W., and Palczewski, K. (2022). MicroRNA Regulation of Critical Retinal Pigment Epithelial Functions. Trends Neurosciences 45, 78–90. doi:10.1016/j.tins.2021.10.008
Duran-Sanchon, S., Moreno, L., Gómez-Matas, J., Augé, J. M., Serra-Burriel, M., Cuatrecasas, M., et al. (2021). Fecal MicroRNA-Based Algorithm Increases Effectiveness of Fecal Immunochemical Test-Based Screening for Colorectal Cancer. Clin. Gastroenterol. Hepatol. 19, 323–330. doi:10.1016/j.cgh.2020.02.043
Ediriweera, M. K., Tennekoon, K. H., and Samarakoon, S. R. (2019). Role of the PI3K/AKT/mTOR Signaling Pathway in Ovarian Cancer: Biological and Therapeutic Significance. Semin. Cancer Biol. 59, 147–160. doi:10.1016/j.semcancer.2019.05.012
Fang, J. Y., and Richardson, B. C. (2005). The MAPK Signalling Pathways and Colorectal Cancer. Lancet Oncol. 6, 322–327. doi:10.1016/s1470-2045(05)70168-6
Farooqi, A. A., de la Roche, M., Djamgoz, M. B. A., and Siddik, Z. H. (2019). Overview of the Oncogenic Signaling Pathways in Colorectal Cancer: Mechanistic Insights. Semin. Cancer Biol. 58, 65–79. doi:10.1016/j.semcancer.2019.01.001
Fricke, F., Mussack, V., Buschmann, D., Hausser, I., Pfaffl, M. W., Kopitz, J., et al. (2019). TGFBR2-dependent A-lterations of microRNA P-rofiles in E-xtracellular V-esicles and P-arental C-olorectal C-ancer C-ells. Int. J. Oncol. 55, 925–937. doi:10.3892/ijo.2019.4859
Fruman, D. A., Chiu, H., Hopkins, B. D., Bagrodia, S., Cantley, L. C., and Abraham, R. T. (2017). The PI3K Pathway in Human Disease. Cell 170, 605–635. doi:10.1016/j.cell.2017.07.029
Fukada, M., Matsuhashi, N., Takahashi, T., Sugito, N., Heishima, K., Akao, Y., et al. (2020). Tumor Tissue MIR92a and Plasma MIRs21 and 29a as Predictive Biomarkers Associated with Clinicopathological Features and Surgical Resection in a Prospective Study on Colorectal Cancer Patients. J. Clin. Med. 9 (8), 2509. doi:10.3390/jcm9082509
Ganesh, K., Stadler, Z. K., Cercek, A., Mendelsohn, R. B., Shia, J., Segal, N. H., et al. (2019). Immunotherapy in Colorectal Cancer: Rationale, Challenges and Potential. Nat. Rev. Gastroenterol. Hepatol. 16, 361–375. doi:10.1038/s41575-019-0126-x
Gao, J., Li, N., Dong, Y., Li, S., Xu, L., Li, X., et al. (2015). miR-34a-5p Suppresses Colorectal Cancer Metastasis and Predicts Recurrence in Patients with Stage II/III Colorectal Cancer. Oncogene 34, 4142–4152. doi:10.1038/onc.2014.348
Garo, L. P., Ajay, A. K., Fujiwara, M., Gabriely, G., Raheja, R., Kuhn, C., et al. (2021). MicroRNA-146a Limits Tumorigenic Inflammation in Colorectal Cancer. Nat. Commun. 12, 2419. doi:10.1038/s41467-021-22641-y
Geng, L., Chaudhuri, A., Talmon, G., Wisecarver, J. L., Are, C., Brattain, M., et al. (2014). MicroRNA-192 Suppresses Liver Metastasis of colon Cancer. Oncogene 33, 5332–5340. doi:10.1038/onc.2013.478
Ghafouri-Fard, S., Hussen, B. M., Badrlou, E., Abak, A., and Taheri, M. (2021). MicroRNAs as Important Contributors in the Pathogenesis of Colorectal Cancer. Biomed. Pharmacother. 140, 111759. doi:10.1016/j.biopha.2021.111759
Gu, C., Cai, J., Xu, Z., Zhou, S., Ye, L., Yan, Q., et al. (2019). MiR-532-3p Suppresses Colorectal Cancer Progression by Disrupting the ETS1/TGM2 axis-mediated Wnt/β-Catenin Signaling. Cell Death Dis 10, 739. doi:10.1038/s41419-019-1962-x
Gulei, D., Magdo, L., Jurj, A., Raduly, L., Cojocneanu-Petric, R., Moldovan, A., et al. (2018). The Silent Healer: miR-205-5p Up-Regulation Inhibits Epithelial to Mesenchymal Transition in colon Cancer Cells by Indirectly Up-Regulating E-Cadherin Expression. Cel Death Dis 9, 66. doi:10.1038/s41419-017-0102-8
He, Z., Dang, J., Song, A., Cui, X., Ma, Z., and Zhang, Y. (2020). The Involvement of miR-150/β-Catenin axis in Colorectal Cancer Progression. Biomed. Pharmacother. 121, 109495. doi:10.1016/j.biopha.2019.109495
Hiraide, S., Takahashi, M., Yoshida, Y., Yamada, H., Komine, K., and Ishioka, C. (2021). Tumor Suppressor miR‐193a‐3p Enhances Efficacy of BRAF/MEK Inhibitors in BRAF ‐mutated Colorectal Cancer. Cancer Sci. 112, 3856–3870. doi:10.1111/cas.15075
Hiraki, M., Nishimura, J., Takahashi, H., Wu, X., Takahashi, Y., Miyo, M., et al. (2015). Concurrent Targeting of KRAS and AKT by MiR-4689 Is a Novel Treatment against Mutant KRAS Colorectal Cancer. Mol. Ther. - Nucleic Acids 4, e231. doi:10.1038/mtna.2015.5
Hoxhaj, G., and Manning, B. D. (2020). The PI3K-AKT Network at the Interface of Oncogenic Signalling and Cancer Metabolism. Nat. Rev. Cancer 20, 74–88. doi:10.1038/s41568-019-0216-7
Huang, L., Liu, Z., Hu, J., Luo, Z., Zhang, C., Wang, L., et al. (2020). MiR-377-3p Suppresses Colorectal Cancer through Negative Regulation on Wnt/β-Catenin Signaling by Targeting XIAP and ZEB2. Pharmacol. Res. 156, 104774. doi:10.1016/j.phrs.2020.104774
Huang, X., Zhu, X., Yu, Y., Zhu, W., Jin, L., Zhang, X., et al. (2021). Dissecting miRNA Signature in Colorectal Cancer Progression and Metastasis. Cancer Lett. 501, 66–82. doi:10.1016/j.canlet.2020.12.025
Huang, Z., Huang, D., Ni, S., Peng, Z., Sheng, W., and Du, X. (2010). Plasma microRNAs Are Promising Novel Biomarkers for Early Detection of Colorectal Cancer. Int. J. Cancer 127, 118–126. doi:10.1002/ijc.25007
Hur, K., Toiyama, Y., Takahashi, M., Balaguer, F., Nagasaka, T., Koike, J., et al. (2013). MicroRNA-200c Modulates Epithelial-To-Mesenchymal Transition (EMT) in Human Colorectal Cancer Metastasis. Gut 62, 1315–1326. doi:10.1136/gutjnl-2011-301846
Hwang, W.-L., Jiang, J.-K., Yang, S.-H., Huang, T.-S., Lan, H.-Y., Teng, H.-W., et al. (2014). MicroRNA-146a Directs the Symmetric Division of Snail-Dominant Colorectal Cancer Stem Cells. Nat. Cel Biol 16, 268–280. doi:10.1038/ncb2910
Ibrahim, A. F., Weirauch, U., Thomas, M., Grünweller, A., Hartmann, R. K., and Aigner, A. (2011). MicroRNA Replacement Therapy for miR-145 and miR-33a Is Efficacious in a Model of colon Carcinoma. Cancer Res. 71, 5214–5224. doi:10.1158/0008-5472.can-10-4645
Igarashi, H., Kurihara, H., Mitsuhashi, K., Ito, M., Okuda, H., Kanno, S., et al. (2015). Association of MicroRNA-31-5p with Clinical Efficacy of Anti-EGFR Therapy in Patients with Metastatic Colorectal Cancer. Ann. Surg. Oncol. 22, 2640–2648. doi:10.1245/s10434-014-4264-7
Itatani, Y., Kawada, K., and Sakai, Y. (2019). Transforming Growth Factor-β Signaling Pathway in Colorectal Cancer and its Tumor Microenvironment. Int. J. Mol. Sci. 20 (23), 5822. doi:10.3390/ijms20235822
Jiang, L., and Hermeking, H. (2017). miR-34a and miR-34b/c Suppress Intestinal Tumorigenesis. Cancer Res. 77, 2746–2758. doi:10.1158/0008-5472.can-16-2183
Jiang, T., Ye, L., Han, Z., Liu, Y., Yang, Y., Peng, Z., et al. (2017). miR-19b-3p Promotes colon Cancer Proliferation and Oxaliplatin-Based Chemoresistance by Targeting SMAD4: Validation by Bioinformatics and Experimental Analyses. J. Exp. Clin. Cancer Res. 36, 131. doi:10.1186/s13046-017-0602-5
Jiao, W., Leng, X., Zhou, Q., Wu, Y., Sun, L., Tan, Y., et al. (2017). Different miR-21-3p Isoforms and Their Different Features in Colorectal Cancer. Int. J. Cancer 141, 2103–2111. doi:10.1002/ijc.30902
Jung, G., Hernández-Illán, E., Moreira, L., Balaguer, F., and Goel, A. (2020). Epigenetics of Colorectal Cancer: Biomarker and Therapeutic Potential. Nat. Rev. Gastroenterol. Hepatol. 17, 111–130. doi:10.1038/s41575-019-0230-y
Ke, T.-W., Wei, P.-L., Yeh, K.-T., Chen, W. T.-L., and Cheng, Y.-W. (2015). MiR-92a Promotes Cell Metastasis of Colorectal Cancer through PTEN-Mediated PI3K/AKT Pathway. Ann. Surg. Oncol. 22, 2649–2655. doi:10.1245/s10434-014-4305-2
Kent, O. A., Mendell, J. T., and Rottapel, R. (2016). Transcriptional Regulation of miR-31 by Oncogenic KRAS Mediates Metastatic Phenotypes by Repressing RASA1. Mol. Cancer Res. 14, 267–277. doi:10.1158/1541-7786.mcr-15-0456
Kim, N. H., Kim, H. S., Kim, N. G., Lee, I., Choi, H. S., Li, X. Y., et al. (2011). p53 and microRNA-34 Are Suppressors of Canonical Wnt Signaling. Sci. Signal. 4, ra71. doi:10.1126/scisignal.2001744
Kjersem, J. B., Ikdahl, T., Lingjaerde, O. C., Guren, T., Tveit, K. M., and Kure, E. H. (2014). Plasma microRNAs Predicting Clinical Outcome in Metastatic Colorectal Cancer Patients Receiving First-Line Oxaliplatin-Based Treatment. Mol. Oncol. 8, 59–67. doi:10.1016/j.molonc.2013.09.001
Koga, Y., Yasunaga, M., Takahashi, A., Kuroda, J., Moriya, Y., Akasu, T., et al. (2010). MicroRNA Expression Profiling of Exfoliated Colonocytes Isolated from Feces for Colorectal Cancer Screening. Cancer Prev. Res. 3, 1435–1442. doi:10.1158/1940-6207.capr-10-0036
Lawrence, M. S., Stojanov, P., Mermel, C. H., Robinson, J. T., Garraway, L. A., Golub, T. R., et al. (2014). Discovery and Saturation Analysis of Cancer Genes across 21 Tumour Types. Nature 505, 495–501. doi:10.1038/nature12912
Li, H., and Li, F. (2018). Exosomes from BM-MSCs Increase the Population of CSCs via Transfer of miR-142-3p. Br. J. Cancer 119, 744–755. doi:10.1038/s41416-018-0254-z
Li, L., Wang, A., Cai, M., Tong, M., Chen, F., and Huang, L. (2020). Identification of Stool miR-135b-5p as a Non-invasive Diaognostic Biomarker in Later Tumor Stage of Colorectal Cancer. Life Sci. 260, 118417. doi:10.1016/j.lfs.2020.118417
Li, T., Lai, Q., Wang, S., Cai, J., Xiao, Z., Deng, D., et al. (2016). MicroRNA-224 Sustains Wnt/β-Catenin Signaling and Promotes Aggressive Phenotype of Colorectal Cancer. J. Exp. Clin. Cancer Res. 35, 21. doi:10.1186/s13046-016-0287-1
Li, Y., Lauriola, M., Kim, D., Francesconi, M., D’Uva, G., Shibata, D., et al. (2016). Adenomatous Polyposis Coli (APC) Regulates miR17-92 Cluster through β-catenin Pathway in Colorectal Cancer. Oncogene 35, 4558–4568. doi:10.1038/onc.2015.522
Li, Z.-w., Yang, Y.-m., Du, L.-t., Dong, Z., Wang, L.-l., Zhang, X., et al. (2014). Overexpression of miR-223 Correlates with Tumor Metastasis and Poor Prognosis in Patients with Colorectal Cancer. Med. Oncol. 31, 256. doi:10.1007/s12032-014-0256-5
Liang, L., Gao, C., Li, Y., Sun, M., Xu, J., Li, H., et al. (2017). miR-125a-3p/FUT5-FUT6 axis Mediates Colorectal Cancer Cell Proliferation, Migration, Invasion and Pathological Angiogenesis via PI3K-Akt Pathway. Cel Death Dis 8, e2968. doi:10.1038/cddis.2017.352
Lin, X., Wang, S., Sun, M., Zhang, C., Wei, C., Yang, C., et al. (2019). miR-195-5p/NOTCH2-mediated EMT Modulates IL-4 Secretion in Colorectal Cancer to Affect M2-like TAM Polarization. J. Hematol. Oncol. 12, 20. doi:10.1186/s13045-019-0708-7
Ling, H., Pickard, K., Ivan, C., Isella, C., Ikuo, M., Mitter, R., et al. (2016). The Clinical and Biological Significance of MIR-224 Expression in Colorectal Cancer Metastasis. Gut 65, 977–989. doi:10.1136/gutjnl-2015-309372
Liu, H., Wang, J., Tao, Y., Li, X., Qin, J., Bai, Z., et al. (2019). Curcumol Inhibits Colorectal Cancer Proliferation by Targeting miR-21 and Modulated PTEN/PI3K/Akt Pathways. Life Sci. 221, 354–361. doi:10.1016/j.lfs.2019.02.049
Liu, P., Cao, F., Sui, J., Hong, Y., Liu, Q., Gao, X., et al. (2020). MicroRNA-142-3p Inhibits Tumorigenesis of Colorectal Cancer via Suppressing the Activation of Wnt Signaling by Directly Targeting to β-Catenin. Front. Oncol. 10, 552944. doi:10.3389/fonc.2020.552944
Liu, S., Sun, X., Wang, M., Hou, Y., Zhan, Y., Jiang, Y., et al. (2014). A microRNA 221- and 222-Mediated Feedback Loop Maintains Constitutive Activation of NFκB and STAT3 in Colorectal Cancer Cells. Gastroenterology 147, 847–859. e11. doi:10.1053/j.gastro.2014.06.006
Liu, S., Xiao, Z., Ai, F., Liu, F., Chen, X., Cao, K., et al. (2017). miR-142-5p Promotes Development of Colorectal Cancer through Targeting SDHB and Facilitating Generation of Aerobic Glycolysis. Biomed. Pharmacother. 92, 1119–1127. doi:10.1016/j.biopha.2017.05.134
Loh, C. Y., Chai, J. Y., Tang, T. F., Wong, W. F., Sethi, G., Shanmugam, M. K., et al. (2019). The E-Cadherin and N-Cadherin Switch in Epithelial-To-Mesenchymal Transition: Signaling, Therapeutic Implications, and Challenges. Cells 8 (10), 1118. doi:10.3390/cells8101118
Lopes-Ramos, C. M., Habr-Gama, A., Quevedo, B. d. S., Felício, N. M., Bettoni, F., Koyama, F. C., et al. (2014). Overexpression of miR-21-5p as a Predictive Marker for Complete Tumor Regression to Neoadjuvant Chemoradiotherapy in Rectal Cancer Patients. BMC Med. Genomics 7, 68. doi:10.1186/s12920-014-0068-7
Luan, Y., Li, X., Luan, Y., Zhao, R., Li, Y., Liu, L., et al. (2020). Circulating lncRNA UCA1 Promotes Malignancy of Colorectal Cancer via the miR-143/MYO6 Axis. Mol. Ther. - Nucleic Acids 19, 790–803. doi:10.1016/j.omtn.2019.12.009
Lun, W., Wu, X., Deng, Q., and Zhi, F. (2018). MiR-218 Regulates Epithelial-Mesenchymal Transition and Angiogenesis in Colorectal Cancer via Targeting CTGF. Cancer Cel Int 18, 83. doi:10.1186/s12935-018-0575-2
Lundberg, I. V., Wikberg, M. L., Ljuslinder, I., Li, X., Myte, R., Zingmark, C., et al. (2018). MicroRNA Expression in KRAS- and BRAF-Mutated Colorectal Cancers. Anticancer Res. 38, 677–683. doi:10.21873/anticanres.12272
Luo, J., Chen, X.-Q., and Li, P. (2019). The Role of TGF-β and its Receptors in Gastrointestinal Cancers. Translational Oncol. 12, 475–484. doi:10.1016/j.tranon.2018.11.010
Luo, Q., Song, H., Deng, X., Li, J., Jian, W., Zhao, J., et al. (2020). A Triple-Regulated Oncolytic Adenovirus Carrying MicroRNA-143 Exhibits Potent Antitumor Efficacy in Colorectal Cancer. Mol. Ther. - Oncolytics 16, 219–229. doi:10.1016/j.omto.2020.01.005
Maierthaler, M., Benner, A., Hoffmeister, M., Surowy, H., Jansen, L., Knebel, P., et al. (2017). Plasma miR-122 and miR-200 Family Are Prognostic Markers in Colorectal Cancer. Int. J. Cancer 140, 176–187. doi:10.1002/ijc.30433
Mangiapane, L. R., Nicotra, A., Turdo, A., Gaggianesi, M., Bianca, P., Di Franco, S., et al. (2022). PI3K-driven HER2 Expression Is a Potential Therapeutic Target in Colorectal Cancer Stem Cells. Gut 71, 119–128. doi:10.1136/gutjnl-2020-323553
Marcuello, M., Vymetalkova, V., Neves, R. P. L., Duran-Sanchon, S., Vedeld, H. M., Tham, E., et al. (2019). Circulating Biomarkers for Early Detection and Clinical Management of Colorectal Cancer. Mol. Aspects Med. 69, 107–122. doi:10.1016/j.mam.2019.06.002
Mayer, I. A., and Arteaga, C. L. (2016). The PI3K/AKT Pathway as a Target for Cancer Treatment. Annu. Rev. Med. 67, 11–28. doi:10.1146/annurev-med-062913-051343
Meng, F., Yang, M., Chen, Y., Chen, W., and Wang, W. (2021). miR-34a Induces Immunosuppression in Colorectal Carcinoma through Modulating a SIRT1/NF-Κb/b7-H3/tnf-α axis. Cancer Immunol. Immunother. 70, 2247–2259. doi:10.1007/s00262-021-02862-2
Mima, K., Nishihara, R., Yang, J., Dou, R., Masugi, Y., Shi, Y., et al. (2016). MicroRNA MIR21 (miR-21) and PTGS2 Expression in Colorectal Cancer and Patient Survival. Clin. Cancer Res. 22, 3841–3848. doi:10.1158/1078-0432.ccr-15-2173
Mittal, V. (2018). Epithelial Mesenchymal Transition in Tumor Metastasis. Annu. Rev. Pathol. Mech. Dis. 13, 395–412. doi:10.1146/annurev-pathol-020117-043854
Monteleone, N. J., Moore, A. E., Iacona, J. R., Lutz, C. S., and Dixon, D. A. (2019). miR-21-mediated Regulation of 15-hydroxyprostaglandin Dehydrogenase in colon Cancer. Sci. Rep. 9, 5405. doi:10.1038/s41598-019-41862-2
Moody, L., Dvoretskiy, S., An, R., Mantha, S., and Pan, Y. X. (2019). The Efficacy of miR-20a as a Diagnostic and Prognostic Biomarker for Colorectal Cancer: A Systematic Review and Meta-Analysis. Cancers (Basel) 11. doi:10.3390/cancers11081111
Mudduluru, G., Abba, M., Batliner, J., Patil, N., Scharp, M., Lunavat, T. R., et al. (2015). A Systematic Approach to Defining the microRNA Landscape in Metastasis. Cancer Res. 75, 3010–3019. doi:10.1158/0008-5472.can-15-0997
Nagel, R., le Sage, C., Diosdado, B., van der Waal, M., Oude Vrielink, J. A. F., Bolijn, A., et al. (2008). Regulation of the Adenomatous Polyposis Coli Gene by the miR-135 Family in Colorectal Cancer. Cancer Res. 68, 5795–5802. doi:10.1158/0008-5472.can-08-0951
Niu, B., Liu, J., Lv, B., Lin, J., Li, X., Wu, C., et al. (2021). Interplay between Transforming Growth Factor-β and Nur77 in Dual Regulations of Inhibitor of Differentiation 1 for Colonic Tumorigenesis. Nat. Commun. 12, 2809. doi:10.1038/s41467-021-23048-5
Niu, L., Yang, W., Duan, L., Wang, X., Li, Y., Xu, C., et al. (2021). Biological Implications and Clinical Potential of Metastasis-Related miRNA in Colorectal Cancer. Mol. Ther. - Nucleic Acids 23, 42–54. doi:10.1016/j.omtn.2020.10.030
O'Brien, S. J., Carter, J. V., Burton, J. F., Oxford, B. G., Schmidt, M. N., Hallion, J. C., et al. (2018). The Role of the miR-200 Family in Epithelial-Mesenchymal Transition in Colorectal Cancer: a Systematic Review. Int. J. Cancer 142, 2501–2511. doi:10.1002/ijc.31282
Okugawa, Y., Grady, W. M., and Goel, A. (2015). Epigenetic Alterations in Colorectal Cancer: Emerging Biomarkers. Gastroenterology 149, 1204–1225. e12. doi:10.1053/j.gastro.2015.07.011
Pan, S., Wu, W., Ren, F., Li, L., Li, Y., Li, W., et al. (2020). MiR-346-5p Promotes Colorectal Cancer Cell Proliferation In Vitro and In Vivo by Targeting FBXL2 and Activating the β-catenin Signaling Pathway. Life Sci. 244, 117300. doi:10.1016/j.lfs.2020.117300
Peng, Q., Shen, Y., Zhao, P., Cai, S., Feng, Z., Cheng, M., et al. (2020). Biomarker Exploration of microRNA-203 as a Promising Substrate for Predicting Poor Survival Outcome in Colorectal Cancer. BMC Cancer 20, 1003. doi:10.1186/s12885-020-07512-x
Pino, M. S., and Chung, D. C. (2010). The Chromosomal Instability Pathway in colon Cancer. Gastroenterology 138, 2059–2072. doi:10.1053/j.gastro.2009.12.065
Prossomariti, A., Piazzi, G., D'Angelo, L., Miccoli, S., Turchetti, D., Alquati, C., et al. (2018). miR-155 Is Downregulated in Familial Adenomatous Polyposis and Modulates WNT Signaling by Targeting AXIN1 and TCF4. Mol. Cancer Res. 16, 1965–1976. doi:10.1158/1541-7786.mcr-18-0115
Przygodzka, P., Papiewska-Pająk, I., Bogusz-Koziarska, H., Sochacka, E., Boncela, J., and Kowalska, M. A. (2019). Regulation of miRNAs by Snail during Epithelial-To-Mesenchymal Transition in HT29 colon Cancer Cells. Sci. Rep. 9, 2165. doi:10.1038/s41598-019-39200-7
Pu, M., Chen, J., Tao, Z., Miao, L., Qi, X., Wang, Y., et al. (2019). Regulatory Network of miRNA on its Target: Coordination between Transcriptional and post-transcriptional Regulation of Gene Expression. Cell. Mol. Life Sci. 76, 441–451. doi:10.1007/s00018-018-2940-7
Rani, V., and Sengar, R. S. (2022). Biogenesis and Mechanisms of microRNA‐mediated Gene Regulation. Biotech. Bioeng. 119, 685–692. doi:10.1002/bit.28029
Rashid, H., Hossain, B., Siddiqua, T., Kabir, M., Noor, Z., Ahmed, M., et al. (2020). Fecal MicroRNAs as Potential Biomarkers for Screening and Diagnosis of Intestinal Diseases. Front. Mol. Biosci. 7, 181. doi:10.3389/fmolb.2020.00181
Rio-Vilariño, A., Del Puerto-Nevado, L., García-Foncillas, J., and Cebrián, A. (2021). Ras Family of Small GTPases in CRC: New Perspectives for Overcoming Drug Resistance. Cancers 13 (15), 3757. doi:10.3390/cancers13153757
Rokavec, M., Öner, M. G., Li, H., Jackstadt, R., Jiang, L., Lodygin, D., et al. (2014). IL-6R/STAT3/miR-34a Feedback Loop Promotes EMT-Mediated Colorectal Cancer Invasion and Metastasis. J. Clin. Invest. 124, 1853–1867. doi:10.1172/jci73531
Rupaimoole, R., and Slack, F. J. (2017). MicroRNA Therapeutics: towards a new era for the Management of Cancer and Other Diseases. Nat. Rev. Drug Discov. 16, 203–222. doi:10.1038/nrd.2016.246
Saieva, L., Barreca, M. M., Zichittella, C., Prado, M. G., Tripodi, M., Alessandro, R., et al. (2020). Hypoxia-Induced miR-675-5p Supports β-Catenin Nuclear Localization by Regulating GSK3-β Activity in Colorectal Cancer Cell Lines. Int. J. Mol. Sci. 21 (11), 3832. doi:10.3390/ijms21113832
Satelli, A., and Li, S. (2011). Vimentin in Cancer and its Potential as a Molecular Target for Cancer Therapy. Cel. Mol. Life Sci. 68, 3033–3046. doi:10.1007/s00018-011-0735-1
Schetter, A. J., Leung, S. Y., Sohn, J. J., Zanetti, K. A., Bowman, E. D., Yanaihara, N., et al. (2008). MicroRNA Expression Profiles Associated with Prognosis and Therapeutic Outcome in colon Adenocarcinoma. Jama 299, 425–436. doi:10.1001/jama.299.4.425
Schou, J. V., Rossi, S., Jensen, B. V., Nielsen, D. L., Pfeiffer, P., Høgdall, E., et al. (2014). miR-345 in Metastatic Colorectal Cancer: a Non-invasive Biomarker for Clinical Outcome in Non-KRAS Mutant Patients Treated with 3rd Line Cetuximab and Irinotecan. PloS one 9, e99886. doi:10.1371/journal.pone.0099886
Shi, C., Yang, Y., Xia, Y., Okugawa, Y., Yang, J., Liang, Y., et al. (2016). Novel Evidence for an Oncogenic Role of microRNA-21 in Colitis-Associated Colorectal Cancer. Gut 65, 1470–1481. doi:10.1136/gutjnl-2014-308455
Siemens, H., Neumann, J., Jackstadt, R., Mansmann, U., Horst, D., Kirchner, T., et al. (2013). Detection of miR-34a Promoter Methylation in Combination with Elevated Expression of C-Met and β-Catenin Predicts Distant Metastasis of Colon Cancer. Clin. Cancer Res. 19, 710–720. doi:10.1158/1078-0432.ccr-12-1703
Slattery, M. L., Herrick, J. S., Mullany, L. E., Valeri, N., Stevens, J., Caan, B. J., et al. (2015). An Evaluation and Replication of miRNAs with Disease Stage and Colorectal Cancer-specific Mortality. Int. J. Cancer 137, 428–438. doi:10.1002/ijc.29384
Song, M., Yin, Y., Zhang, J., Zhang, B., Bian, Z., Quan, C., et al. (2014). MiR-139-5p Inhibits Migration and Invasion of Colorectal Cancer by Downregulating AMFR and NOTCH1. Protein Cell 5, 851–861. doi:10.1007/s13238-014-0093-5
Song, P., Feng, L., Li, J., Dai, D., Zhu, L., Wang, C., et al. (2020). β-Catenin Represses miR455-3p to Stimulate m6A Modification of HSF1 mRNA and Promote its Translation in Colorectal Cancer. Mol. Cancer 19, 129. doi:10.1186/s12943-020-01244-z
Song, Y., Zhao, Y., Ding, X., and Wang, X. (2018). microRNA-532 Suppresses the PI3K/Akt Signaling Pathway to Inhibit Colorectal Cancer Progression by Directly Targeting IGF-1R. Am. J. Cancer Res. 8, 435–449.
Stolfi, C., Troncone, E., Marafini, I., and Monteleone, G. (2020). Role of TGF-Beta and Smad7 in Gut Inflammation, Fibrosis and Cancer. Biomolecules 11 (1), 17. doi:10.3390/biom11010017
Sun, D., Wang, C., Long, S., Ma, Y., Guo, Y., Huang, Z., et al. (2015). C/EBP-β-activated microRNA-223 Promotes Tumour Growth through Targeting RASA1 in Human Colorectal Cancer. Br. J. Cancer 112, 1491–1500. doi:10.1038/bjc.2015.107
Sun, D., Yu, F., Ma, Y., Zhao, R., Chen, X., Zhu, J., et al. (2013). MicroRNA-31 Activates the RAS Pathway and Functions as an Oncogenic MicroRNA in Human Colorectal Cancer by Repressing RAS P21 GTPase Activating Protein 1 (RASA1). J. Biol. Chem. 288, 9508–9518. doi:10.1074/jbc.m112.367763
Sun, M., Song, H., Wang, S., Zhang, C., Zheng, L., Chen, F., et al. (2017). Integrated Analysis Identifies microRNA-195 as a Suppressor of Hippo-YAP Pathway in Colorectal Cancer. J. Hematol. Oncol. 10, 79. doi:10.1186/s13045-017-0445-8
Sun, N., Zhang, L., Zhang, C., and Yuan, Y. (2020). miR-144-3p Inhibits Cell Proliferation of Colorectal Cancer Cells by Targeting BCL6 via Inhibition of Wnt/β-Catenin Signaling. Cell Mol Biol Lett 25, 19. doi:10.1186/s11658-020-00210-3
Sun, X., Lin, F., Sun, W., Zhu, W., Fang, D., Luo, L., et al. (2021). Exosome-transmitted miRNA-335-5p Promotes Colorectal Cancer Invasion and Metastasis by Facilitating EMT via Targeting RASA1. Mol. Ther. - Nucleic Acids 24, 164–174. doi:10.1016/j.omtn.2021.02.022
Sun, Z., Zhang, Z., Liu, Z., Qiu, B., Liu, K., and Dong, G. (2014). MicroRNA-335 Inhibits Invasion and Metastasis of Colorectal Cancer by Targeting ZEB2. Med. Oncol. 31, 982. doi:10.1007/s12032-014-0982-8
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A. Cancer J. Clin. 71, 209–249. doi:10.3322/caac.21660
Świerczyński, M., Szymaszkiewicz, A., Fichna, J., and Zielińska, M. (2021). New Insights into Molecular Pathways in Colorectal Cancer: Adiponectin, Interleukin-6 and Opioid Signaling. Biochim. Biophys. Acta Rev. Cancer 1875, 188460. doi:10.1016/j.bbcan.2020.188460
Taki, M., Abiko, K., Ukita, M., Murakami, R., Yamanoi, K., Yamaguchi, K., et al. (2021). Tumor Immune Microenvironment during Epithelial-Mesenchymal Transition. Clin. Cancer Res. : official J. Am. Assoc. Cancer Res. 27, 4669–4679. doi:10.1158/1078-0432.ccr-20-4459
Tang, W., Zhu, Y., Gao, J., Fu, J., Liu, C., Liu, Y., et al. (2014). MicroRNA-29a Promotes Colorectal Cancer Metastasis by Regulating Matrix Metalloproteinase 2 and E-Cadherin via KLF4. Br. J. Cancer 110, 450–458. doi:10.1038/bjc.2013.724
Tecalco-Cruz, A. C., Ríos-López, D. G., Vázquez-Victorio, G., Rosales-Alvarez, R. E., and Macías-Silva, M. (2018). Transcriptional Cofactors Ski and SnoN Are Major Regulators of the TGF-β/Smad Signaling Pathway in Health and Disease. Sig Transduct Target. Ther. 3, 15. doi:10.1038/s41392-018-0015-8
Tili, E., Michaille, J.-J., and Croce, C. M. (2013). MicroRNAs Play a central Role in Molecular Dysfunctions Linking Inflammation with Cancer. Immunol. Rev. 253, 167–184. doi:10.1111/imr.12050
Toiyama, Y., Okugawa, Y., Fleshman, J., Richard Boland, C., and Goel, A. (2018). MicroRNAs as Potential Liquid Biopsy Biomarkers in Colorectal Cancer: A Systematic Review. Biochim. Biophys. Acta (Bba) - Rev. Cancer 1870, 274–282. doi:10.1016/j.bbcan.2018.05.006
Toiyama, Y., Takahashi, M., Hur, K., Nagasaka, T., Tanaka, K., Inoue, Y., et al. (2013). Serum miR-21 as a Diagnostic and Prognostic Biomarker in Colorectal Cancer. J. Natl. Cancer Inst. 105, 849–859. doi:10.1093/jnci/djt101
Tsai, J. H., and Yang, J. (2013). Epithelial-mesenchymal Plasticity in Carcinoma Metastasis. Genes Dev. 27, 2192–2206. doi:10.1101/gad.225334.113
Tsang, W. P., Ng, E. K. O., Ng, S. S. M., Jin, H., Yu, J., Sung, J. J. Y., et al. (2010). Oncofetal H19-Derived miR-675 Regulates Tumor Suppressor RB in Human Colorectal Cancer. Carcinogenesis 31, 350–358. doi:10.1093/carcin/bgp181
Valeri, N., Braconi, C., Gasparini, P., Murgia, C., Lampis, A., Paulus-Hock, V., et al. (2014). MicroRNA-135b Promotes Cancer Progression by Acting as a Downstream Effector of Oncogenic Pathways in colon Cancer. Cancer Cell 25, 469–483. doi:10.1016/j.ccr.2014.03.006
Vanpouille-Box, C., and Formenti, S. C. (2018). Dual Transforming Growth Factor-β and Programmed Death-1 Blockade: A Strategy for Immune-Excluded Tumors? Trends Immunol. 39, 435–437. doi:10.1016/j.it.2018.03.002
Vicinus, B., Rubie, C., Faust, S. K., Frick, V. O., Ghadjar, P., Wagner, M., et al. (2012). miR-21 Functionally Interacts with the 3′UTR of Chemokine CCL20 and Down-Regulates CCL20 Expression in miR-21 Transfected Colorectal Cancer Cells. Cancer Lett. 316, 105–112. doi:10.1016/j.canlet.2011.10.031
Wang, A., Deng, S., Chen, X., Yu, C., Du, Q., Wu, Y., et al. (2020). miR-29a-5p/STAT3 Positive Feedback Loop Regulates TETs in Colitis-Associated Colorectal Cancer. Inflamm. Bowel Dis. 26, 524–533. doi:10.1093/ibd/izz281
Wang, H., Yan, B., Zhang, P., Liu, S., Li, Q., Yang, J., et al. (2020). MiR‐496 Promotes Migration and Epithelial‐mesenchymal Transition by Targeting RASSF6 in Colorectal Cancer. J. Cell Physiol. 235, 1469–1479. doi:10.1002/jcp.29066
Wang, Y., Wu, Z., and Hu, L. (2018). The Regulatory Effects of Metformin on the [SNAIL/miR-34]:[ZEB/miR-200] System in the Epithelial-Mesenchymal Transition(EMT) for Colorectal Cancer(CRC). Eur. J. Pharmacol. 834, 45–53. doi:10.1016/j.ejphar.2018.07.006
Wu, C. W., Ng, S. S. M., Dong, Y. J., Ng, S. C., Leung, W. W., Lee, C. W., et al. (2012). Detection of miR-92a and miR-21 in Stool Samples as Potential Screening Biomarkers for Colorectal Cancer and Polyps. Gut 61, 739–745. doi:10.1136/gut.2011.239236
Xia, X., Wang, X., Zhang, S., Zheng, Y., Wang, L., Xu, Y., et al. (2019). miR‐31 Shuttled by Halofuginone‐induced Exosomes Suppresses MFC‐7 Cell Proliferation by Modulating the HDAC2/cell Cycle Signaling axis. J. Cell Physiol. 234, 18970–18984. doi:10.1002/jcp.28537
Xiao, J., Lv, D., Zhou, J., Bei, Y., Chen, T., Hu, M., et al. (2017). Therapeutic Inhibition of miR-4260 Suppresses Colorectal Cancer via Targeting MCC and SMAD4. Theranostics 7, 1901–1913. doi:10.7150/thno.19168
Xiong, B., Cheng, Y., Ma, L., and Zhang, C. (2013). MiR-21 Regulates Biological Behavior through the PTEN/PI-3 K/Akt Signaling Pathway in Human Colorectal Cancer Cells. Int. J. Oncol. 42, 219–228. doi:10.3892/ijo.2012.1707
Xu, Q., Tong, J.-L., Zhang, C.-P., Xiao, Q., Lin, X.-L., and Xiao, X.-Y. (2017). miR-27a Induced by colon Cancer Cells in HLECs Promotes Lymphangiogenesis by Targeting SMAD4. PloS one 12, e0186718. doi:10.1371/journal.pone.0186718
Xu, X., Chen, X., Xu, M., Liu, X., Pan, B., Qin, J., et al. (2019). miR-375-3p Suppresses Tumorigenesis and Partially Reverses Chemoresistance by Targeting YAP1 and SP1 in Colorectal Cancer Cells. Aging 11, 7357–7385. doi:10.18632/aging.102214
Xu, Y., Zhang, X., Hu, X., Zhou, W., Zhang, P., Zhang, J., et al. (2018). The Effects of lncRNA MALAT1 on Proliferation, Invasion and Migration in Colorectal Cancer through Regulating SOX9. Mol. Med. 24, 52. doi:10.1186/s10020-018-0050-5
Yadav, V. K., Huang, Y. J., George, T. A., Wei, P. L., Sumitra, M. R., Ho, C. L., et al. (2020). Preclinical Evaluation of the Novel Small-Molecule MSI-N1014 for Treating Drug-Resistant Colon Cancer via the LGR5/β-catenin/miR-142-3p Network and Reducing Cancer-Associated Fibroblast Transformation. Cancers (Basel) 12 (6), 1590. doi:10.3390/cancers12061590
Yamada, A., Horimatsu, T., Okugawa, Y., Nishida, N., Honjo, H., Ida, H., et al. (2015). Serum miR-21, miR-29a, and miR-125b Are Promising Biomarkers for the Early Detection of Colorectal Neoplasia. Clin. Cancer Res. 21, 4234–4242. doi:10.1158/1078-0432.ccr-14-2793
Yamada, N. O., Heishima, K., Akao, Y., and Senda, T. (2019). Extracellular Vesicles Containing MicroRNA-92a-3p Facilitate Partial Endothelial-Mesenchymal Transition and Angiogenesis in Endothelial Cells. Int. J. Mol. Sci. 20 (18), 4406. doi:10.3390/ijms20184406
Yang, F., Wang, H., Yan, B., Li, T., Min, L., Chen, E., et al. (2021). Decreased Level of miR-1301 Promotes Colorectal Cancer Progression via Activation of STAT3 Pathway. Biol. Chem. 402, 805–813. doi:10.1515/hsz-2020-0301
Ye, S., Song, W., Xu, X., Zhao, X., and Yang, L. (2016). IGF2BP2 Promotes Colorectal Cancer Cell Proliferation and Survival through Interfering withRAF-1degradation by miR-195. FEBS Lett. 590, 1641–1650. doi:10.1002/1873-3468.12205
Yu, C., Wan, H., Shan, R., Wen, W., Li, J., Luo, D., et al. (2019). The Prognostic Value of the MiR-200 Family in Colorectal Cancer: A Meta-Analysis with 1882 Patients. J. Cancer 10, 4009–4016. doi:10.7150/jca.27529
Yu, Y., Kanwar, S. S., Patel, B. B., Oh, P.-S., Nautiyal, J., Sarkar, F. H., et al. (2012). MicroRNA-21 Induces Stemness by Downregulating Transforming Growth Factor Beta Receptor 2 (TGF R2) in colon Cancer Cells. Carcinogenesis 33, 68–76. doi:10.1093/carcin/bgr246
Yuan, Z., Baker, K., Redman, M. W., Wang, L., Adams, S. V., Yu, M., et al. (2017). Dynamic Plasma microRNAs Are Biomarkers for Prognosis and Early Detection of Recurrence in Colorectal Cancer. Br. J. Cancer 117, 1202–1210. doi:10.1038/bjc.2017.266
Zhang, F., Luo, Y., Shao, Z., Xu, L., Liu, X., Niu, Y., et al. (2016). MicroRNA-187, a Downstream Effector of TGFβ Pathway, Suppresses Smad-Mediated Epithelial-Mesenchymal Transition in Colorectal Cancer. Cancer Lett. 373, 203–213. doi:10.1016/j.canlet.2016.01.037
Zhang, J.-X., Song, W., Chen, Z.-H., Wei, J.-H., Liao, Y.-J., Lei, J., et al. (2013). Prognostic and Predictive Value of a microRNA Signature in Stage II colon Cancer: a microRNA Expression Analysis. Lancet Oncol. 14, 1295–1306. doi:10.1016/s1470-2045(13)70491-1
Zhang, N., Ng, A. S., Cai, S., Li, Q., Yang, L., and Kerr, D. (2021). Novel Therapeutic Strategies: Targeting Epithelial-Mesenchymal Transition in Colorectal Cancer. Lancet Oncol. 22, e358–e368. doi:10.1016/s1470-2045(21)00343-0
Zhang, W., Zhang, T., Jin, R., Zhao, H., Hu, J., Feng, B., et al. (2014). MicroRNA-301a Promotes Migration and Invasion by Targeting TGFBR2 in Human Colorectal Cancer. J. Exp. Clin. Cancer Res. 33, 113. doi:10.1186/s13046-014-0113-6
Zhao, J., Lin, H., and Huang, K. (2022). Mesenchymal Stem Cell-Derived Extracellular Vesicles Transmitting MicroRNA-34a-5p Suppress Tumorigenesis of Colorectal Cancer through C-MYC/DNMT3a/PTEN Axis. Mol. Neurobiol. 59, 47–60. doi:10.1007/s12035-021-02431-9
Zhao, S., Sun, H., Jiang, W., Mi, Y., Zhang, D., Wen, Y., et al. (2017). miR-4775 Promotes Colorectal Cancer Invasion and Metastasis via the Smad7/TGFβ-Mediated Epithelial to Mesenchymal Transition. Mol. Cancer 16, 12. doi:10.1186/s12943-017-0585-z
Zhao, X., Liu, S., Yan, B., Yang, J., and Chen, E. (2020). MiR-581/SMAD7 Axis Contributes to Colorectal Cancer Metastasis: A Bioinformatic and Experimental Validation-Based Study. Int. J. Mol. Sci. 21 (18), 6499. doi:10.3390/ijms21186499
Zheng, G., Du, L., Yang, X., Zhang, X., Wang, L., Yang, Y., et al. (2014). Serum microRNA Panel as Biomarkers for Early Diagnosis of Colorectal Adenocarcinoma. Br. J. Cancer 111, 1985–1992. doi:10.1038/bjc.2014.489
Zhou, T., Wu, L., Ma, N., Tang, F., Zong, Z., and Chen, S. (2020). LncRNA PART1 Regulates Colorectal Cancer via Targeting miR-150-5p/miR-520h/CTNNB1 and Activating Wnt/β-Catenin Pathway. Int. J. Biochem. Cel Biol. 118, 105637. doi:10.1016/j.biocel.2019.105637
Zhu, G.-f., Xu, Y.-w., Li, J., Niu, H.-l., Ma, W.-x., Xu, J., et al. (2019). Mir20a/106a-WTX axis Regulates RhoGDIa/CDC42 Signaling and colon Cancer Progression. Nat. Commun. 10, 112. doi:10.1038/s41467-018-07998-x
Keywords: colorectal cancer, miRNA, biomarker, gene regulation, signaling pathway
Citation: Yang F, Xuan G, Chen Y, Cao L, Zhao M, Wang C and Chen E (2022) MicroRNAs Are Key Molecules Involved in the Gene Regulation Network of Colorectal Cancer. Front. Cell Dev. Biol. 10:828128. doi: 10.3389/fcell.2022.828128
Received: 03 December 2021; Accepted: 07 March 2022;
Published: 08 April 2022.
Edited by:
Karel Soucek, Academy of Sciences of the Czech Republic, CzechiaReviewed by:
Enikö Kallay, Medical University of Vienna, AustriaVeronika Holubekova, Comenius University, Slovakia
Copyright © 2022 Yang, Xuan, Chen, Cao, Zhao, Wang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Erfei Chen, Y2hlbmVyZmVpQG53dS5lZHUuY24=
 Fangfang Yang1,2
Fangfang Yang1,2 Guoyun Xuan
Guoyun Xuan Lichao Cao
Lichao Cao Erfei Chen
Erfei Chen