
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell Dev. Biol. , 27 April 2022
Sec. Cellular Biochemistry
Volume 10 - 2022 | https://doi.org/10.3389/fcell.2022.813701
This article is part of the Research Topic Exosomal Biomarkers: Roles in Diagnostics and Therapeutics View all 9 articles
Background: Hepatocellular carcinoma (HCC) is one of the fifty most common cancers globally, having a high mortality rate being the second most common cause of cancer-related deaths. However, little attention has been paid to the involvement of exosomes and ceRNA in HCC.
Method: The study aimed to explore exosome data from exoRBase database and a free online database to estimate possible binding miRNA from mRNA, lncRNA, and circRNA and discover useful exosome biomarkers for HCC therapy.
Results: The results indicated that a total of 159 mRNAs, 60 lncRNAs, and 13 circRNAs were differentially expressed, with HIST2H3C exhibiting the highest log2FC change, CTD-2031P19 exhibiting the most relevant lncRNA, and CTD-2031P19 exhibiting the most relevant lncRNA. MARCH8, SH3PXD2A, has-circ-0014088, hsa-miR-186-5p, and hsa-miR-613 were identified as hub biomarkers used by Cytoscape. According to the KEGG pathway analysis results, the differentially expressed proteins were primarily enriched in the MAPK signaling network, central carbon metabolism in cancer, the glucagon signaling pathway, glutamatergic synapse, and spliceosome. Furthermore, immunohistochemical images from the Human Protein Atlas (HPA) online tool were used to directly evaluate the protein expression of SMARCA5, CDC42, and UBC between normal and cancer tissues, and the results showed that these three gene expressions were significantly higher in tumor tissues.
Conclusion: This study discovered atypical signature exosomes for HCC prognostic prediction based on an online database. The signals could mimic exosome microenvironmental disorders providing potential biomarkers for exosome treatment.
Hepatocellular carcinoma (HCC) is one of the fifty most common cancers globally, having a high mortality rate being the second most common cause of cancer-related deaths, and is diagnosed at a rate of 500,000 patients each year globally (Torre et al., 2015). The most prevalent causes of cirrhosis are viral hepatitis and nonalcoholic steatohepatitis, with roughly 80% of cases progressing to HCC (Tang et al., 2016; Coskun, 2017). Due to the recurrence of HCC, the prognosis of HCC remains bleak, with a 5-year overall survival rate of approximately 34–50% (Lang et al., 2007). Despite significant advancements in medical technology, there are no useful curative therapies for HCC patients (Jiao et al., 2018). Radical treatment can enhance the survival probability of individuals diagnosed with HCC early on and offer possible long-term treatments (De Lope et al., 2012). Various serum biomarkers such as alpha-fetoprotein (AFP) and alkaline phosphatase (AKP or ALP) are well known in clinical practice, but they have the demerit of low sensitivity and specificity (Shen et al., 2017). As a result, developing efficient biomarkers for HCC diagnosis and treatment is critical.
Exosomes are small membrane vesicles rich in lipids, proteins, RNA, and DNA secreted by most cell types due to the fusion of multivesicular late lysosomes/endosomes with the plasma membrane, thereby influencing the biological behavior of nearby or distant cells (Denzer et al., 2000). Exosomes are involved in the initiation and progression of cancer. Many studies reported that the cancer cells release more exosomes than normal cells, affecting tumor cell biology [such as proliferation (Richards et al., 2017), metastasis (Costa -Silva et al., 2015; Soung et al., 2016), drug resistance (Yousafzai et al., 2018), and broad biological processes associated with immunity (Han et al., 2019)], with microRNA (miRNA) playing a significant role. The HCC-derived exosomes have been shown to reduce the cytotoxicity of T cells and NK cells, which are important mediators of the host antitumor immune response and tumor cell immune escape (Chen et al., 2018). Exosomes from highly metastatic MHCC97H cells are connected with low metastatic HCC cells, boosting their migration, chemotaxis, and invasion (Zhang et al., 2020). Exosomal circUHRF1 induces NK cell dysfunction, which causes immunosuppression and may lead to resistance to anti-PD1 therapy in HCC (Greening et al., 2015). Exosomes have been studied for their predictive significance in HCC. However, the true role and mechanism remain unknown. According to a recent study, BAK1 expression is driven by exosome circ-0051443 via competitive interaction with miR-331-3p and acts as a predictor of HCC and a possible therapeutic target (Chen et al., 2020a). Similarly, Huang et al. (2020) discovered that exosomes circRNA-100338 influence cell proliferation, angiogenesis, permeability, angiogenesis mimics, and tumor metastasis. Furthermore, Lu et al. (2020) discovered three lncRNA compositions associated with AFP, implying that they serve as a fingerprint in predicting HCC metastasis. There is little doubt that exosome research provides a novel viewpoint on tumor mechanisms; nevertheless, no systematic examination of exosome biomarkers in HCC patients is reported. The co-expression and competing endogenous RNA (ceRNA) networks may hamper immunotherapy efficacy in HCC via PD-L1+ exosome activity (Wei et al., 2021). Furthermore, the lncRNA FAL1 was overexpressed in serum exosomes and acted as a ceRNA pathway to promote cell proliferation and metastasis in HCC (Li et al., 2018a). Moreover, exosomal SENP3-EIF4A1 was reported with the ability to suppress tumor growth in vivo via the ceRNA pathway (Wang et al., 2020). However, little attention has been paid to the involvement of exosomes and ceRNA in HCC. Since high-throughput expression data are now available, it is feasible to use global gene expression data to investigate the relationship between exosome analysis and clinical outcomes in HCC patients. The study aimed to explore exosome data from exoRBase database and a free online database to estimate possible binding miRNA from mRNA, lncRNA, and circRNA and discover useful exosome biomarkers for HCC therapy.
The exoRBase (http://www.exoRBase.org) (Li et al., 2018b) is an online database containing circular RNA (circRNA), long noncoding RNA (lncRNA), and messenger RNA (mRNA) extracted from RNA-seq investigations of human blood exosomes. ExoRBase’s initial version comprises 58,330 circular RNAs, 15,501 long noncoding RNAs, and 18,333 mRNAs. The exoRBase also contains 32 normal and 21 HCC samples from which differential mRNA, lncRNA, and circRNA expression between normal and HCC was detected.
By evaluating the expression profiles of the 32 cancer types included in the TCGA study, the ENCORI pan-cancer analysis platform seeks to decode the pan-cancer network of lncRNA, miRNA, pseudogenes, snoRNA, RNA binding protein (RBP), and all protein-coding genes. TargetScan identifies miRNA biological targets by searching for conserved 8mer, 7mer, and 6mer sequences that match the seed region of each miRNA (Lewis et al., 2005). The miRanda is the first miRNA target gene prediction software developed and is not species-specific (Liu et al., 2019), while the miRcode is based on an exhaustive transcriptome prediction of human microRNA targets, including 10,000 long noncoding RNA genes (Jeggari et al., 2012). In addition, the StarBase was designed to decode precancerous and interacting lncRNA, miRNA, competitive endogenous RNA (ceRNA), RBP, and mRNA networks from large-scale CLIP-Seq data and tumor samples (Li et al., 2014). Thus, TargetScan and miRanda databases were used in this study in conjunction with the ENCORI platform to predict possible miRNA binding to mRNA, with only those satisfying both databases being deemed prospective miRNA. The miRcode was used to estimate the potential binding miRNA of lncRNA, and StarBase was utilized to predict the potential binding miRNA of circRNA via the ENCORI platform.
Cytoscape (version 3.7.1) is a bioinformatics software used for constructing and visualizing networks of molecular interactions (Shannon et al., 2003). Cytoscape was also used to create ceRNA networks by constricting the link between mRNAs, lncRNAs, and circRNAs. Furthermore, the DAVID online database (Huang et al., 2009) was also accessed to investigate the network’s mRNAs’ functional annotations and signaling pathways. The Benjamini–Hochberg method was used for converting p-values to FDR values. The R (version 3.5.3) and R Bioconductor software packages analyzed all data, while Perl was used for creating data matrixes and performing all the data processing on p < 0.05.
After standardizing the microarray results, primarily, the mRNAs, lncRNAs, and circRNAs were selected, which are differentially expressed between normal and HCC samples. The results indicated that a total of 159 mRNAs, 60 lncRNAs, and 13 circRNAs were differentially expressed, with HIST2H3C exhibiting the highest log2FC change and CTD-2031P19 exhibiting the most relevant lncRNA. Supplementary Tables S1–S3 represented that the most significant circRNA was hsa circ 0001380. Meanwhile, Figure 1 depicts the heatmap of the top 20 upregulated and downregulated mRNAs, lncRNAs, and circRNAs based on the log2FC value.
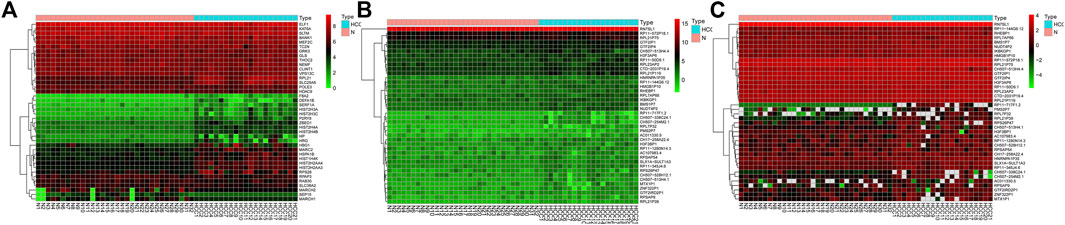
FIGURE 1. The heatmap of top 20 upregulated and downregulated mRNAs, lncRNAs, and circRNAs. (A) mRNAs; (B) lncRNAs; (C) circRNAs.
The online database tools were used to determine the possible binding miRNAs of mRNAs, lncRNAs, and circRNAs between the normal and HCC samples. Supplementary Tables S4–S6 contain specific information on each putative miRNA binding partner. Cytoscape was then utilized to create ceRNA networks by constricting the links between mRNAs, lncRNAs, and circRNAs. Cytoscape plug-in Molecular Complex Detection (MCODE) was used to identify densely linked regions in ceRNA networks. Cytoscape enabled the construction of the ceRNA networks, while MCODE helped to select the most significant module within the ceRNA networks. Finally, MARCH8, SH3PXD2A, has-circ-0014088, hsa-miR-186-5p, and hsa-miR-613 were identified as hub biomarkers with degrees ≥10. The OS analysis revealed no statistically significant difference in survival for MARCH8 and SH3PXD2A, where a high expression of HIST2H3C indicated poor survival (Figure 5A). Figure 2 depicts the detailed ceRNA networks and the most significant module. Meanwhile, the mRNA from the ceRNA network was extracted for the purpose of building a protein–protein network, identifying SMARCA5, CDC42, and UBC as the hub genes in the network (Figure 3). Furthermore, immunohistochemical images from the Human Protein Atlas (HPA) online tool were used to directly evaluate the protein expression of these three genes between normal and cancer tissues, and the results showed that the SMARCA5, CDC42, and UBC expression were significantly higher in tumor tissues (Figure 4). Using cBioPortal to investigate genetic modifications in SMARCA5, CDC42, and UBC, it has been reported that the genes were altered in 13 (4%) of 366 HCC patients (Figure 5). The overall survival analysis of SMARCA5, CDC42, and UBC in HCC was also investigated using the Gene Expression Profiling Interactive Analysis (GEPIA) survival analysis module. The statistical difference between the curves was assessed using the log-rank test (Li et al., 2021). The findings revealed that high levels of SMARCA5, CDC42, and UBC expression are associated with poor survival (p < 0.05) (Figures 5B–D). The genes discovered in this study may have a role in HCC carcinogenesis.
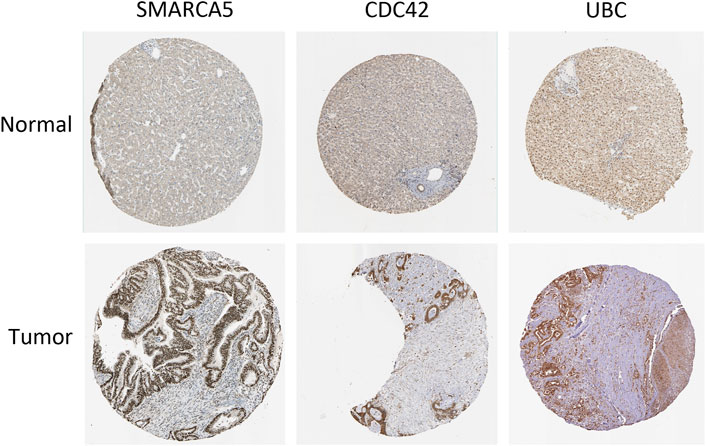
FIGURE 4. Human Protein Atlas (HPA) online tool directly compares the protein expression of these three genes among normal and cancer tissues.
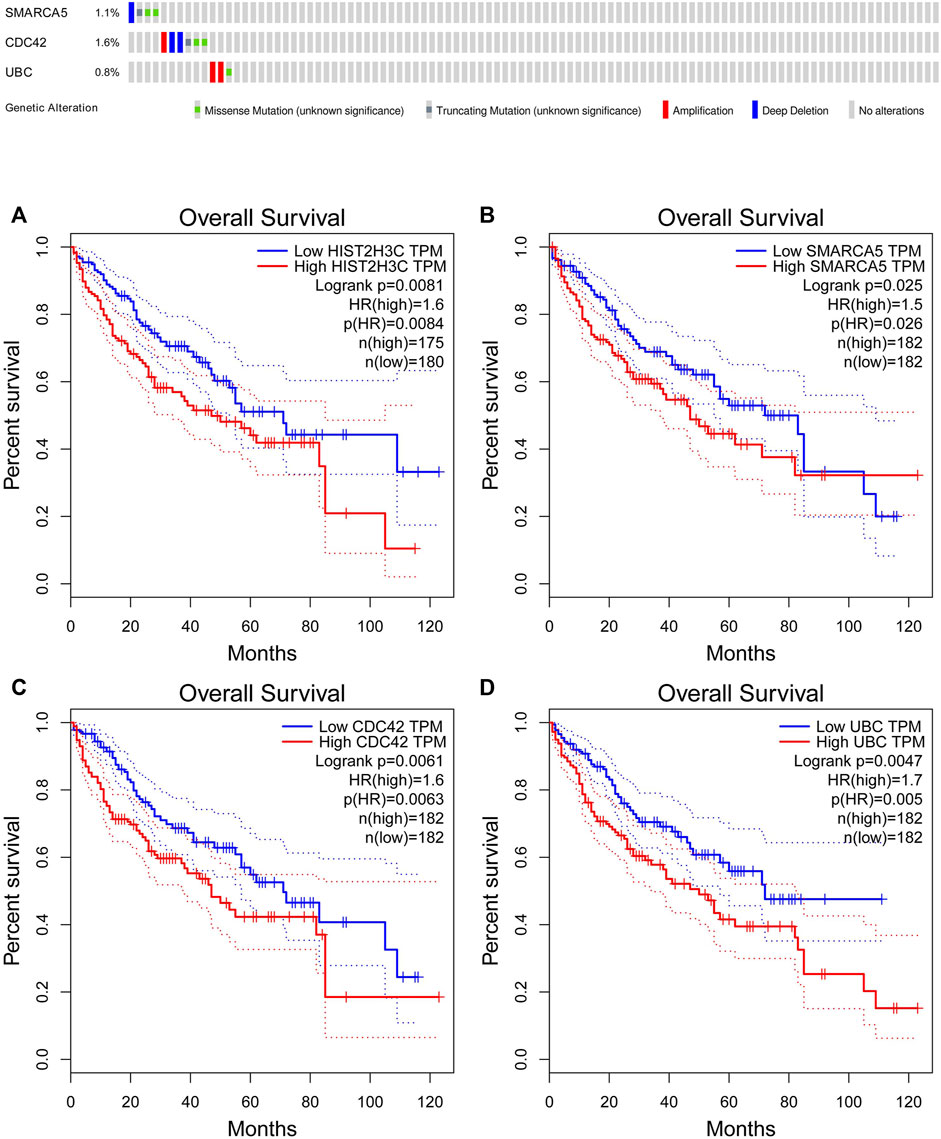
FIGURE 5. cBioPortal genetic alterations and GEPIA overall survival results. (A) HIST2H3C; (B) SMARCA5; (C) CDC42; (D) UBC.
The DAVID online database was used to investigate the network’s mRNAs’ functional annotation information and signaling pathways. The results of the GO analysis revealed that changes in biological processes (BP) of differentially expressed proteins were enriched in protein phosphorylation, osteoblast differentiation, positive transcription regulation, DNA-templated, and nucleosome assembly. Histone binding and protein kinase activity were the most affected by changes in molecular function (MF). The most abundant cell components (CC) were the cytoplasm, nucleoplasm, and cytosol. According to the KEGG pathway analysis results, the differentially expressed proteins were primarily enriched in the MAPK signaling network, central carbon metabolism in cancer, the glucagon signaling pathway, glutamatergic synapse, and spliceosome (Supplementary Table S7).
Tumor-related exosomes or tumor-derived exosomes are considered critical for controlling carcinogenesis and its development. Identifying and analyzing tumor exosomes can help with early diagnosis, efficacy evaluation, and prognosis analysis of cancers, which are envisaged to significantly advance the oncology research, cancer clinical diagnosis, and introduce novel opportunities for cancer treatment. Some novel and effective exosome biomarkers were identified in this study based on online databases, indicating that the signature has great predictive value. The findings of this study may also represent the exosome status of individuals with liver cancer and prospective biomarkers and therapeutic targets in exosome signaling pathways.
This study used a complete exosome analysis using the exoRBase database on normal and HCC samples; 159 differentially expressed mRNAs, 60 differentially expressed lncRNAs, and 13 differentially expressed circRNAs were identified. The most meaningful mRNAs, lncRNAs, and circRNAs were RGPD6, CTD-2031P19.4, and has-circ-0001380, respectively. Microduplication of RGPD6 was earlier reported in a family with liver cirrhosis and other diseases (Chen et al., 2017). Targets of hsa-miR-363-5p were found in RGPD6, and reduced expression of hsa-miR-363-5p was linked to improved overall survival in HCC patients (Zhang et al., 2017). There was no systematic examination of the involvement of RGPD6, CTD-2031P19.4, and has-circ-0001380 in HCC yet; hence, the findings of this study may be useful for future research. The ceRNA networks were then built, and MARCH8, SH3PXD2A, has-circ-0014088, hsa-miR-186-5p, and hsa-miR-613 were identified as hub biomarkers. Membrane-related RING-CH 8 (MARCH8) is one of the 11 members of the MARCH family of RING-finger E3 ubiquitin ligase. Research reports claim that MARCH8 is an effective antiviral protein that targets viral envelope glycoproteins and reduces their incorporation into the viral particles (Tada et al., 2015). The immunomodulatory role of MARCH8 in the regulation of intestinal epithelial cells (IEC), MHC II expression, and graft-versus-host disease severity was confirmed (Lineburg et al., 2018). According to a study, the lncRNA SH3PXD2A-AS1 knockdown decreased colorectal cancer cell proliferation, migration, and invasion in vitro and suppressed carcinogenesis in vivo (Ma et al., 2018). SH3PXD2A, a gene encoding Tks5, plays a crucial role in invasion, body formation and function, cell migration, and matrix destruction (Pilar et al., 2014). The SH3PXD2A-AS1/miR-330-5p/UBA2 network may influence colorectal cancer growth via the Wnt/-catenin pathway (Guo et al., 2021). According to another research, miR-186-5p levels in hepatocellular carcinoma tissues were significantly lower than those in surrounding normal tissues (Lan et al., 2017). The HOXD-AS1/miR-186-5p/PIK3R3 is a novel route in epithelial ovarian cancer that promotes cell migration, invasion, and epithelial-mesenchymal transition (EMT) (Dong et al., 2019); hence, circ-PRKCI may also boost the survival, invasion, and migration of HCC cells by sponging miR-186-5p to increase FOXK1 expression levels (Chen et al., 2021). The miR-613 ectopic expression decreased the proliferation and invasion of Hep3B and SMMC-7721 HCC cells (Wang et al., 2016), whereas the lower miR-613 expression was linked to tumor growth, vascular invasion, and a worse prognosis in HCC patients (Jiang et al., 2018). The RMRP regulates HCC carcinogenesis by functioning as a ceRNA for miR-613 and decreases the miR-613 expression, which is adversely linked with NEAT1 expression in the HCC tissues (Wang et al., 2017; Zhou et al., 2019). There have previously been no similar studies investigating the role of circ-0014088 in malignancies. Therefore, this study may provide potentially useful information for further research. HIST2H3C is a histone H3 gene, and the K27M mutation in HIST2H3C has been reported to cause higher metastatic recurrences in glioma (Castel et al., 2015). The histone H3 variations can control gene expression by altering chromatin organization during cellular processes, contributing to cancer pathogenesis (Rashid et al., 2021). This study discovered some promising prospective exosome indicators in HCC, but further investigation is needed.
The functional annotation information and signaling pathways of the mRNAs in the network were then investigated. According to the KEGG pathway analysis results, the differentially expressed mRNAs were primarily abundant in the MAPK signaling pathway, central carbon metabolism in cancer, the glucagon signaling pathway, glutamatergic synapse, and spliceosome. The Ras/MAPK pathway is activated in 50–100% of human HCC; thus, it is linked to a poor prognosis (Delire and Stärkel, 2015). miR-370 has also been shown in studies to downregulate BEX2 gene and suppress the activation of the MAPK/JNK signaling pathway, thereby preventing the development of HCC (Wang et al., 2019). Similarly, suppressed BMP2 in HepG2 cells inhibits tumor development, progression, and angiogenesis in HCC via inactivating the MAPK/p38 signaling pathway (Feng et al., 2019). As a result, it is anticipated that the differentially expressed mRNAs identified in this study may play an essential role in the MAPK signaling pathway, but further research is required. The TCA cycle is required to handle glucose and glutamine-derived carbon in biosynthetic pathways, which are crucial to tumor growth. According to research, the central carbon metabolic pathway provides energy and produces biosynthetic precursors to aid T-cell homeostasis, proliferation, and immune function (Chen et al., 2020b). The enzymes involved in central carbon metabolism and energy production are essential mediators of bacterial physiology, persistence, and pathogenicity, making them naturally appealing for drug discovery (Hards et al., 2020). This research discovered that these mRNAs were primarily enriched in metabolism-related pathways, and exosomes may be associated with central carbon metabolism and the respiratory chain of Mycobacterium tuberculosis. However, the signatures predict that exosomes must be validated in multiple independent cohorts. Though this study provided more information, however, there were some limitations, where the findings have not been validated in clinical samples due to lack of clinicopathologic data, the function of clinical variables in exosomes not being investigated, and the number of genes investigated being insufficient to generalize the importance of exosome transcript upregulation and downregulation in HCC.
This study discovered atypical signature exosomes for HCC prognostic prediction based on an online database. The signals could mimic exosome microenvironmental disorders providing potential biomarkers for exosome treatment. Future research work is certain to undertake large-scale related research, which will eventually be translated into developing new methods for accurate cancer medicine.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Z-HW designed and analyzed the research study. Z-HW and RL wrote and revised the manuscript. Z-HW, CL, and Y-JZ collected the data. All authors read and approved the manuscript.
This work was supported by the National Natural Science Foundation of China (81974068).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2022.813701/full#supplementary-material
Supplementary Table S1 | | 159 differential mRNA expression and the most meaningful mRNA was RGPD6
Supplementary Table S2 | | 60 differential lncRNA expression and the most meaningful lncRNA was CTD-2031P19.4.
Supplementary Table S3 | | 13 differential circRNA expression and the most meaningful circRNA were has-circ-0001380.
Supplementary Table S4 | | Detailed information of mRNA potential binding miRNAs.
Supplementary Table S5 | | Detailed information of lncRNA potential binding miRNAs.
Supplementary Table S6 | | Detailed information of circRNA potential binding miRNAs.
Supplementary Table S7 | | Results of the DAVID online database explored the functional annotation information and signaling pathways of mRNAs.
HCC, hepatocellular carcinoma; AFP, alpha-fetoprotein; miRNA, microRNA; ceRNA, endogenous RNA.
Castel, D., Philippe, C., Calmon, R., Le Dret, L., Truffaux, N., Boddaert, N., et al. (2015). Histone H3F3A and HIST1H3B K27M Mutations Define Two Subgroups of Diffuse Intrinsic Pontine Gliomas with Different Prognosis and Phenotypes. Acta Neuropathol. 130 (6), 815–827. doi:10.1007/s00401-015-1478-0
Chen, C.-P., Lin, S.-P., Lee, C.-L., Chern, S.-R., Wu, P.-S., Chen, Y.-N., et al. (2017). Recurrent 2q13 Microduplication Encompassing MALL , NPHP1 , RGPD6 , and BUB1 Associated with Autism Spectrum Disorder, Intellectual Disability, and Liver Disorder. Taiwanese J. Obstet. Gynecol. 56 (1), 98–101. doi:10.1016/j.tjog.2016.12.003
Chen, L., Guo, P., He, Y., Chen, Z., Chen, L., Luo, Y., et al. (2018). HCC-derived Exosomes Elicit HCC Progression and Recurrence by Epithelial-Mesenchymal Transition through MAPK/ERK Signalling Pathway. Cell Death Dis. 9 (5), 513. doi:10.1038/s41419-018-0534-9
Chen, W., Li, Y., Zhong, J., and Wen, G. (2021). Circ-PRKCI Targets miR-1294 and miR-186-5p by Downregulating FOXK1 Expression to Suppress Glycolysis in Hepatocellular Carcinoma. Mol. Med. Rep. 23 (6), 464. doi:10.3892/mmr.2021.12103
Chen, W., Quan, Y., Fan, S., Wang, H., Liang, J., Huang, L., et al. (2020). Exosome-transmitted Circular RNA Hsa_circ_0051443 Suppresses Hepatocellular Carcinoma Progression. Cancer Lett. 475, 119–128. doi:10.1016/j.canlet.2020.01.022
Chen, X., Sherman, J. W., and Wang, R. (2020). Radioisotope-Based Protocol for Determination of Central Carbon Metabolism in T Cells. Methods Mol. Biol. 2111, 257–265. doi:10.1007/978-1-0716-0266-9_20
Coskun, M. (2017). Hepatocellular Carcinoma in the Cirrhotic Liver: Evaluation Using Computed Tomography and Magnetic Resonance Imaging. Exp. Clin. Transpl. 15 (Suppl. 2), 36–44. doi:10.6002/ect.TOND16.L10
Costa -Silva, B., Aiello, N. M., Ocean, A. J., Singh, S., Zhange, H., Becker, A., et al. (2015). Pancreatic Cancer Exosomes Initiate Pre-metastatic Niche Formation in the Liver. Nat. Cel Biol. 17, 816–826. doi:10.1038/ncb3169
De Lope, C. R., Tremosini, S., Forner, A., Reig, M., and Bruix, J. (2012). Management of HCC. J. Hepatol. 56 (Suppl. 1), S75–S87. doi:10.1016/s0168-8278(12)60009-9
Delire, B., and Stärkel, P. (2015). The Ras/MAPK Pathway and Hepatocarcinoma: Pathogenesis and Therapeutic Implications. Eur. J. Clin. Invest. 45 (6), 609–623. doi:10.1111/eci.12441
Denzer, K., Kleijmeer, M. J., Heijnen, H. F., Stoorvogel, W., and Geuze, H. J. (2000). Exosome: from Internal Vesicle of the Multivesicular Body to Intercellular Signaling Device. J. Cel Sci 113, 3365–3374. doi:10.1242/jcs.113.19.3365
Dong, S., Wang, R., Wang, H., Ding, Q., Zhou, X., Wang, J., et al. (2019). HOXD-AS1 Promotes the Epithelial to Mesenchymal Transition of Ovarian Cancer Cells by Regulating miR-186-5p and PIK3R3. J. Exp. Clin. Cancer Res. 38 (1), 110. doi:10.1186/s13046-019-1103-5
Feng, P.-C., Ke, X.-F., Kuang, H.-L., Pan, L.-L., Ye, Q., and Wu, J.-B. (2019). BMP2 Secretion from Hepatocellular Carcinoma Cell HepG2 Enhances Angiogenesis and Tumor Growth in Endothelial Cells via Activation of the MAPK/p38 Signaling Pathway. Stem Cel Res Ther 10 (1), 237. doi:10.1186/s13287-019-1301-2
Greening, D. W., Gopal, S. K., Xu, R., Chen, W., and Simpson, R. (2015). Exosomes and Their Roles in Immune Regulation and Cancer. Semin. Cel Dev Biol. 40, 72–81. doi:10.1016/j.semcdb.2015.02.009
Guo, S., Zhu, K. x., Yu, W. h., Wang, T., Li, S., Wang, Y. x., et al. (2021). SH3PXD2A‐AS1/miR ‐330‐5p/UBA2 ceRNA Network Mediates the Progression of Colorectal Cancer through Regulating the Activity of the Wnt/β‐catenin Signaling Pathway. Environ. Toxicol. 36 (10), 1969–1980. doi:10.1002/tox.23038
Han, Q., Zhao, H., Jiang, Y., Yin, C., and Zhang, J. (2019). HCC-derived Exosomes: Critical Player and Target for Cancer Immune Escape. Cells 8 (6), 558. doi:10.3390/cells8060558
Hards, K., Adolph, C., Harold, L. K., McNeil, M. B., Cheung, C.-Y., Jinich, A., et al. (2020). Two for the price of One: Attacking the Energetic-Metabolic Hub of Mycobacteria to Produce New Chemotherapeutic Agents. Prog. Biophys. Mol. ABiol. 152, 35–44. doi:10.1016/j.pbiomolbio.2019.11.003
Huang, D. W., Sherman, B. T., and Lempicki, R. A. (2009). Systematic and Integrative Analysis of Large Gene Lists Using DAVID Bioinformatics Resources. Nat. Protoc. 4 (1), 44–57. doi:10.1038/nprot.2008.211
Huang, X.-Y., Huang, Z.-L., Huang, J., Xu, B., Huang, X.-Y., Xu, Y.-H., et al. (2020). Exosomal circRNA-100338 Promotes Hepatocellular Carcinoma Metastasis via Enhancing Invasiveness and Angiogenesis. J. Exp. Clin. Cancer Res. 39 (1), 20. doi:10.1186/s13046-020-1529-9
Jeggari, A., Marks, D. S., and Larsson, E. (2012). miRcode: a Map of Putative microRNA Target Sites in the Long Non-coding Transcriptome. Bioinformatics 28 (15), 2062–2063. doi:10.1093/bioinformatics/bts344
Jiang, X., Wu, J., Zhang, Y., Wang, S., Yu, X., Li, R., et al. (2018). MiR-613 Functions as Tumor Suppressor in Hepatocellular Carcinoma by Targeting YWHAZ. Gene 659, 168–174. doi:10.1016/j.gene.2018.03.036
Jiao, Y., Fu, Z., Li, Y., Meng, L., and Liu, Y. (2018). High EIF2B5 mRNA Expression and its Prognostic Significance in Liver Cancer: a Study Based on the TCGA and GEO Database. Cmar 10, 6003–6014. doi:10.2147/cmar.s185459
Lan, T., Yan, X., Li, Z., Xu, X., Mao, Q., Ma, W., et al. (2017). Long Non-coding RNA PVT1 Serves as a Competing Endogenous RNA for miR-186-5p to Promote the Tumorigenesis and Metastasis of Hepatocellular Carcinoma. Tumour Biol. 39 (6), 101042831770533. doi:10.1177/1010428317705338
Lang, H., Sotiropoulos, G. C., Brokalaki, E. I., Schmitz, K. J., Bertona, C., Meyer, G., et al. (2007). Survival and Recurrence Rates after Resection for Hepatocellular Carcinoma in Noncirrhotic Livers. J. Am. Coll. Surgeons 205 (1), 27–36. doi:10.1016/j.jamcollsurg.2007.03.002
Lewis, B. P., Burge, C. B., and Bartel, D. P. (2005). Conserved Seed Pairing, Often Flanked by Adenosines, Indicates that Thousands of Human Genes Are microRNA Targets. Cell 120 (1), 15–20. doi:10.1016/j.cell.2004.12.035
Li, B., Mao, R., Liu, C., Zhang, W., Tang, Y., and Guo, Z. (2018). LncRNA FAL1 Promotes Cell Proliferation and Migration by Acting as a CeRNA of miR-1236 in Hepatocellular Carcinoma Cells. Life Sci. 197, 122–129. doi:10.1016/j.lfs.2018.02.006
Li, C., Tang, Z., Zhang, W., Ye, Z., and Liu, F. (2021). GEPIA2021: Integrating Multiple Deconvolution-Based Analysis into GEPIA. Nucleic Acids Res. 49 (W1), W242–W246. doi:10.1093/nar/gkab418
Li, J. H., Liu, S., Zhou, H., Qu, L. H., and Yang, J. H. (2014). starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and Protein-RNA Interaction Networks from Large-Scale CLIP-Seq Data. Nucleic Acids Res. 42 (Database issue), D92–D97. doi:10.1093/nar/gkt1248
Li, S., Li, Y., Chen, B., Zhao, J., Yu, S., Tang, Y., et al. (2018). exoRBase: a Database of circRNA, lncRNA and mRNA in Human Blood Exosomes. Nucleic Acids Res. 46 (D1), D106–D112. doi:10.1093/nar/gkx891
Lineburg, K. E., Le Texier, L., Melino, M., Wang, R., Clouston, A., Blazar, B. R., et al. (2018). Ubiquitin Ligase MARCH8 Attenuates Graft versus Host Disease via Regulation of Gut Epithelial Cell Surface MHC II Expression. Transplantation 102, S300. doi:10.1097/01.tp.0000543013.84163.3a
Liu, S., Xie, X., Lei, H., Zou, B., and Xie, L. (2019). Identification of Key circRNAs/lncRNAs/miRNAs/mRNAs and Pathways in Preeclampsia Using Bioinformatics Analysis. Med. Sci. Monit. 25, 1679–1693. doi:10.12659/msm.912801
Lu, Y., Duan, Y., Xu, Q., Zhang, L., Chen, W., Qu, Z., et al. (2020). Circulating Exosome‐derived Bona Fide Long Non‐coding RNAs Predicting the Occurrence and Metastasis of Hepatocellular Carcinoma. J. Cel Mol Med 24 (2), 1311–1318. doi:10.1111/jcmm.14783
Ma, Z., Peng, P., Zhou, J., Hui, B., Ji, H., Wang, J., et al. (2018). Long Non-coding RNA SH3PXD2A-AS1 Promotes Cell Progression Partly through Epigenetic Silencing P57 and KLF2 in Colorectal Cancer. Cell Physiol Biochem 46 (6), 2197–2214. doi:10.1159/000489589
Pilar, C. M., Angela, Y., Nicole, V., Lock, P., Courtneidge, S., Diaz, B., et al. (2014). Genetic Disruption of the Sh3pxd2a Gene Reveals an Essential Role in Mouse Development and the Existence of a Novel Isoform of Tks5. PLoS ONE 9 (9), e107674. doi:10.1371/journal.pone.0107674
Rashid, M., Shah, S. G., Verma, T., Chaudhary, N., Rauniyar, S., Patel, V. B., et al. (2021). Tumor-specific Overexpression of Histone Gene, H3C14 in Gastric Cancer Is Mediated through EGFR-FOXC1 axis. Biochim. Biophys. Acta Gene Regul. Mech. 1864 (4-5), 194703. doi:10.1016/j.bbagrm.2021.194703
Richards, K. E., Zeleniak, A. E., and Fishel, M. L. (2017). Cancer -associated Fibroblast Exosomes Regulate Survival and Proliferation of Pancreatic Cancer Cells. Oncogene 36 (13), 1770–1778.
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 13, 2498–2504. doi:10.1101/gr.1239303
Shen, Y., Bu, L., Li, R., Chen, Z., Tian, F., Lu, N., et al. (2017). Screening Effective Differential Expression Genes for Hepatic Carcinoma with Metastasis in the Peripheral Blood Mononuclear Cells by RNA-Seq. Oncotarget 8 (17), 27976–27989. doi:10.18632/oncotarget.15855
Soung, Y. H., Nguyen, T., Cao, H., Lee, J., and Chung, J. (2016). Emerging Roles of Exosomes in Cancer Invasion and Metastasis[J]. BMB Rep. 49 (1), 18–25. doi:10.5483/BMBRep.2016.49.1.239
Tada, T., Zhang, Y., Koyama, T., Tobiume, M., Tsunetsugu-Yokota, Y., Yamaoka, S., et al. (2015). MARCH8 Inhibits HIV-1 Infection by Reducing Virion Incorporation of Envelope Glycoproteins. Nat. Med. 21 (12), 1502–1507. doi:10.1038/nm.3956
Tang, Y., Wang, H., Ma, L., Zhang, X., Yu, G., Li, J., et al. (2016). Diffusion-weighted Imaging of Hepatocellular Carcinomas: a Retrospective Analysis of Correlation between Apparent Diffusion Coefficients and Histological Grade. Abdom. Radiol. 41 (8), 1539–1545. doi:10.1007/s00261-016-0715-x
Torre, L. A., Bray, F., Siegel, R. L., Ferlay, J., Lortet-Tieulent, J., and Jemal, A. (2015). Global Cancer Statistics, 2012. CA Cancer J. Clin. 65 (2), 87–108. doi:10.3322/caac.21262
Wang, J., Pu, J., Zhang, Y., Yao, T., Luo, Z., Li, W., et al. (2020). Exosome-transmitted Long Non-coding RNA SENP3-Eif4a1 Suppresses the Progression of Hepatocellular Carcinoma. Aging 12 (12), 11550–11567. doi:10.18632/aging.103302
Wang, W., Zhang, H., Wang, L., Zhang, S., and Tang, M. (2016). miR-613 Inhibits the Growth and Invasiveness of Human Hepatocellular Carcinoma via Targeting DCLK1. Biochem. Biophysical Res. Commun. 473 (4), 987–992. doi:10.1016/j.bbrc.2016.04.003
Wang, X., Zhu, W., Xu, C., Wang, F., Zhu, X., Sun, Y., et al. (2019). MicroRNA-370 Functions as a Tumor Suppressor in Hepatocellular Carcinoma via Inhibition of the MAPK/JNK Signaling Pathway by Targeting BEX2. J. Hum. Genet. 64 (12), 1203–1217. doi:10.1038/s10038-019-0653-x
Wang, Z., Zou, Q., Song, M., and Chen, J. (2017). NEAT1 Promotes Cell Proliferation and Invasion in Hepatocellular Carcinoma by Negative Regulating miR-613 Expression. Biomed. Pharmacother. 94, 612–618. doi:10.1016/j.biopha.2017.07.111
Wei, Y., Tang, X., Ren, Y., Yang, Y., Song, F., Fu, J., et al. (2021). An RNA-RNA Crosstalk Network Involving HMGB1 and RICTOR Facilitates Hepatocellular Carcinoma Tumorigenesis by Promoting Glutamine Metabolism and Impedes Immunotherapy by PD-L1+ Exosomes Activity. Sig Transduct Target. Ther. 6 (1), 421. doi:10.1038/s41392-021-00801-2
Yousafzai, N. A., Wang, H., Wang, Z., Zhu, Y., Zhu, L., and Jin, H., (2018). Exosome Mediated Multidrug Resistance in Cancer[J]. Am. J. Cancer Res. 8 (11), 2210–2226.
Zhang, J., Fan, J., Zhou, C., and Qi, Y. (2017). miR-363-5p as Potential Prognostic Marker for Hepatocellular Carcinoma Indicated by Weighted Co-expression Network Analysis of miRNAs and mRNA. BMC Gastroenterol. 17 (1), 81. doi:10.1186/s12876-017-0637-2
Zhang, P.-F., Gao, C., Huang, X.-Y., Lu, J.-C., Guo, X.-J., Shi, G.-M., et al. (2020). Cancer Cell-Derived Exosomal circUHRF1 Induces Natural Killer Cell Exhaustion and May Cause Resistance to Anti-PD1 Therapy in Hepatocellular Carcinoma. Mol. Cancer 19 (1), 110. doi:10.1186/s12943-020-01222-5
Keywords: hepatocellular carcinoma, exosomes, data mining, exoRBase, circRNA
Citation: Wu Z-H, Li C, Zhang Y-J and Lin R (2022) Bioinformatics Study Revealed Significance of Exosome Transcriptome in Hepatocellular Carcinoma Diagnosis. Front. Cell Dev. Biol. 10:813701. doi: 10.3389/fcell.2022.813701
Received: 12 November 2021; Accepted: 23 March 2022;
Published: 27 April 2022.
Edited by:
Dwijendra K. Gupta, Jai Prakash Vishwavidyalaya, IndiaReviewed by:
Manikandan Murugesan, Bharathidasan University, IndiaCopyright © 2022 Wu, Li, Zhang and Lin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rong Lin, c2VsaW5hbGluMzVAaG90bWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.