- 1Department of Breast and Thyroid Surgery, Renmin Hospital of Wuhan University, Wuhan, China
- 2Cancer Center, Renmin Hospital of Wuhan University, Wuhan, China
- 3Department of Biochemistry and Molecular Medicine, The George Washington University School of Medicine and Health Sciences, Washington, DC, United States
- 4Department of Oncology, Georgetown Lombardi Comprehensive Cancer Center, Georgetown University Medical Center, Washington, DC, United States
Breast cancer susceptibility gene 1 (BRCA1) is a tumor suppressor gene, which is mainly involved in the repair of DNA damage, cell cycle regulation, maintenance of genome stability, and other important physiological processes. Mutations or defects in the BRCA1 gene significantly increase the risk of breast, ovarian, prostate, and other cancers in carriers. In this review, we summarized the molecular functions and regulation of BRCA1 and discussed recent insights into the detection and treatment of BRCA1 mutated breast cancer.
Introduction
Breast cancer (BC) is the most common malignancy all over the world, accounting for 11.7% of new cancer cases (Sung et al., 2021). Up to 7% of unselected BC patients have a definite germline genetic mutation called hereditary breast cancer (HBC) (Claus et al., 1996). Among them, breast cancer susceptibility gene 1 (BRCA1) is one of the most common tumor suppressor genes, which encodes a 220 kD nuclear protein and is detected in at least 5% of unselected patients with BC (Hall et al., 1990; Chen et al., 1996). BRCA1 plays an important role in DNA repair, replication fork protection, cell cycle regulation, and gene transcription regulation (Gudmundsdottir and Ashworth, 2006). When the BRCA1 gene is mutated or lost, the incidence of BC and ovarian cancer will increase significantly (Miki, et al., 1994). The cumulative risk of BC by 80 years of age in healthy female carriers of BRCA1 mutation is about 80% (Kuchenbaecker et al., 2017; King et al., 2003) while one in eight women will develop BC over the lifespan in the general population. Carriers of BRCA1 mutation are more likely to develop triple-negative breast cancer (TNBC), which suggests that BRCA1 mutation and the hormone receptor status are interlinked (Foulkes et al., 2004). BRCA1-mutated BC is associated with earlier onset, more aggressive disease, and a higher risk of relapse. Hence, it is important to investigate the function and dysregulation of BRCA1 in BC and treatment strategies for this population. In this article, we provide a comprehensive overview of BRCA1 in BC, including the BRCA1’s molecular function, its mutation detection, and the prevention and treatment of BC in mutated carriers.
BRCA1 Gene
Structure and Function of BRCA1 Gene
BRCA1 is an incomplete recessive gene on an autosome, located on chromosome 17q21 and encoded 220 kD protein-containing multi-function domains (Hall et al., 1990). There are 24 exons in BRCA1 whose exons 2–5 encode the RING domain and exons 15–23 encode the BRCA1 C-terminal (BRCT) domain (Figure 1) (Miki et al., 1994; Clark et al., 2012). The N-terminal RING domain has an E3 ligase activity, which interacts with its partner protein, the BRCA1-associated RING domain protein 1 (BARD1) to form a stable BRCA1-BARD1 heterodimer (Hashizume et al., 2001). The BRCT domain is associated with different phosphorylated interacting proteins. In addition to the N-terminal RING domain and C-terminal domain, there is a coiled-coil domain upstream of BRCT domains, which binds another coiled-coil domain at the N-terminus of PALB2. PALB2 also binds BRCA2 and serves as the molecular scaffold in the formation of the BRCA1-PALB2-BRCA2 complex (Shirley et al., 2009; Zhang et al., 2009). In mammalian cells, homologous recombination (HR) and non-homologous end-joining (NHEJ) are two major repair pathways to repair DNA double-strand breaks (DSBs) for genome integrity. Both the RING domain and BRCT domain of BRCA1 are essential for HR to maintain genome stability. Many clinically important mutations of BRCA1 gene frequently target these two domains.
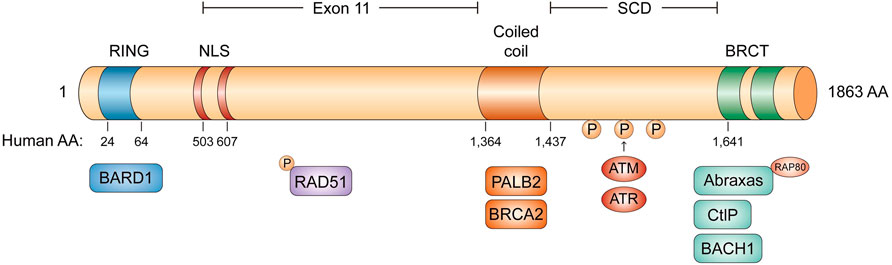
FIGURE 1. The domain structure of BRCA1. The RING domain in blue, the two NLS domain in red, the coiled coil domain in orange, and the two BRCT domains in green. BRCA1 can form four different complexes: BRCA1/RAP80/Abraxas complex, BRCA1/BACH1 complex, BRCA1/PALB2/BRCA2 complex and BRCA1/CtIP complex.
The BRCT domain is conserved in several DNA damage response (DDR) proteins and is responsible for BRCA1 to recognize a phospho-SPxF motif (S, serine; P, proline; x, varies; F, phenylalanine) (Wang, 2012; Chabanon et al., 2021). Figure 2 BRCA1 can form four different complexes in cells, through the association of different adaptor proteins with the BRCT domain, such as BRCA1/RAP80/Abraxas complex, BRCA1/BACH1(BRCA1 associated C-terminal helicase) complex, BRCA1/PALB2 (partner and localizer of BRCA2)/BRCA2 complex, and BRCA1/CtIP complex (Figure 1). BRCA1/RAP80/Abraxas complex is recruited to DNA DSBs through RAP80, a ubiquitin-binding protein. RAP80 could target this complex to MDC1-rH2AX-dependent K6 and K63-linked ubiquitin polymers at DSBs. BRCA1/RAP80/Abraxas complex prevents excessive end resection and potentially deleterious homology-directed DSB repair mechanisms (Huang and Zhou, 2020; Vohhodina et al., 2020). The helicase catalytic function of BRCA1/BACH1 is not only important for BRCA1-mediated DDR but also necessarily required to maintain DNA damage-induced G2/M checkpoint (Yu et al., 2003). As described previously, PALB2, the partner and localizer of BRCA2, could bind directly to BRCA1 to form BRCA1/PALB2/BRCA2 complex, which stimulates RAD51-mediated localization and repair at DNA breaks (Shirley et al., 2009; Ducy et al., 2019). Lastly, BRCA1/CtIP complex promotes the HR by DNA end resection.
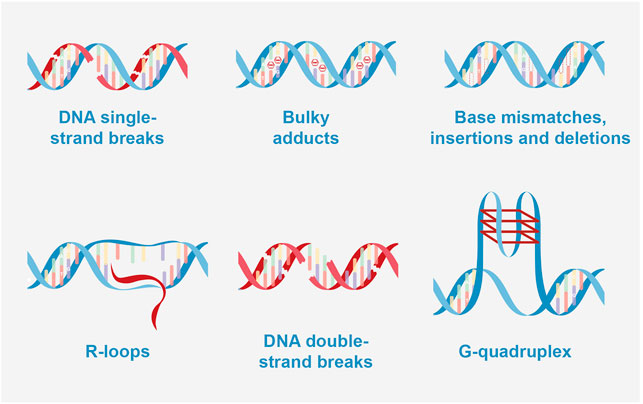
FIGURE 2. Types of DNA damage. DNA single-strand breaks: BER (base excision repair); bulky adducts: NER (nucleotide excision repair); base mismatches, insertions and deletions: MMR (Mismatch repair); R-loops caused double strand breaks (DSB): NHEJ (Non-homologous end joining) and HRR (homologous recombination repair); DNA double-strand breaks: NHEJ and HRR; G-quadruplex caused DSB: NHEJ, and HRR.
In addition to its critical roles in DSB repair, BRCA1 is also involved in the repair and restart of stalled and damaged DNA replication forks and in the protection from nucleolytic attack and degradation. BRCA1-mediated fork protection functions independently from its role in the HR-mediated repair of DNA DSBs (Schlacher et al., 2012; Long et al., 2014). Upon replication fork stress, BRCA1 protects nascent DNA strands from degradation by stabilizing RAD51 nucleofilaments that affect the exonuclease activity of MRE11 (Schlacher et al., 2012). RAD51 is also required for fork restart once halted forks are repaired in response to short replication blocks (Petermann et al., 2010). Moreover, BRCA1 also has important roles in gene transcription. Gerald M. Pao et al. have shown that the carboxyl terminus of BRCA1 transactivates the heterologous promoters (Pao et al., 2000). This BRCA1-mediated transactivation could be mediated by RNA polymerase II (Pol II) via RNA helicase A (RHA) and enhanced by transcriptional coactivators/acetyltransferases p300 and CBP (p300/CBP). Zhu et al. also showed that BRCA1 could bind to satellite DNA regions and ubiquitylates the histone H2A to maintain the heterochromatin structures (Zhu et al., 2011; Zhu et al., 2018); Bochar et al. (Bochar et al., 2000) reported that BRCA1 is a component of SWI/SNF chromatin remodeling complex and controls the transcription through the modulation of chromatin structure. RNA/DNA hybrid structures (R-loops) as normal transcriptional intermediates also affect transcription and genomic instability. BRCA1 is recruited to transcriptional pause sites and mediates the recruitment of senataxin (SETX) (Hatchi et al., 2015); SETX is involved in processing replication forks and resolves R-loops at transcriptional sites; thus, BRCA1/SETX addresses R-loop associated DNA damage arising at transcriptional pause sites. Recently, other studies showed that BRCA1 aberrantly retains at the transcription regions with increasing R-loop levels and decreases its distribution at DNA damage regions in Ewing sarcoma cells (Gorthi et al., 2018). As a result, these cells could not inhibit the transcription after DNA damage and showed a defect in HR repair. Furthermore, Steffi Herold et al. found more details about the relationship between BRCA1, R-loop, and transcriptional regulation in human neuroblastoma cells (Herold et al., 2019). In the MYCN-amplified cells, MYCN activation could increase the transcriptional elongation by inducing the escape of RNAPII from promoters. The recruitment of BRCA1 to the promoter-proximal regions could stabilize MYCN on the chromatin and prevent R-loop formation caused by the RNAPII stalling at the transcription-suspended sites.
The Mutation of BRCA1 and Breast Cancer
Since the first clone of the BRCA1 gene in 1994 (Miki et al., 1994), variable cut transcripts were found as a “naturally occurring” event in both tumor and normal tissues by many studies (Li et al., 2019). There are at least six alternative splicing transcripts of BRCA1 discovered, including BRCA1 exon 1a, exon 1b, exon 1c, BRCA1a (∆11q, ∆11), BRCA1b (∆9,10), and BRCA1-IRIS, which codes protein products with different molecular weights. Based on these observations, Walker et al. revised 77 published studies with 252 BRCA1 splicing analysis assays and found that some of the exon boundary variants may not perform as a loss of function, leading to a naturally occurring in-frame RNA isoform (Walker et al., 2013). Compared to the complete transcript of BRCA1, the variable-cut transcripts of BRCA1 can have similar or opposite functions and, in some cases, may have more unique functions (Braunschweig et al., 2013). Up to now, 1,800 mutations have been found in human BRCA1, including intron mutations, missense mutations, nonsense mutations, frameshift mutations, and other types. These mutations often occur in the RING and BRCT domains, which are the key domains in BRCA1 for genome integrity (Supplementary Data Sheet). Missense mutations in BRCA1 present a significant challenge for the prevention and treatment of patients. For example, BRCA1 c.5309G > T p. (Gly1770Val) has been shown to inhibit homologous recombination and could be considered as a disease-causing mutation (Tudini et al., 2018). As a benefit from bioinformatic analysis, more variants of BRCA1 can be found from public databases, such as the cBioPortal database (http://www.cbioportal.org/), ENIGMA (https://enigmaconsortium.org/), BRCA Exchange (https://brcaexchange.org), and ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/).
Patients with BRCA1 mutations have a higher risk for cancer. The estimated lifetime risk of BC is about 80%, and the lifetime risk of ovarian cancer is 40%–65% (King et al., 2003; Kuchenbaecker et al., 2017), which might alter according to the type and location of the mutations (Rebbeck, et al., 2015). BRCA1 gene deletion with or without p53 defect leads to a high incidence of basal-like BC and tends to form TNBC, which is the most aggressive type of BC (Tarsounas and Sung, 2020). Some studies show that the TNBC in BRCA1 mutation carriers originated from luminal progenitor cells, not basal stem cells (Lim et al., 2009; Molyneux et al., 2010). If BRCA1/p53 is perturbed in luminal progenitors, it could induce the abnormal alveolar differentiation premalignancy with pro-tumorigenic changes in the immune compartment. It belongs to cell autonomy and is caused by the dysregulation of transcription factors. This study explains how BRCA1 aberration impacts the state of nascent tumor cells and their microenvironment. Bach et al. found that breast cells with BRCA1 mutations undergo changes similar to those common changes in women during pregnancy (Bach et al., 2021). Based on the data, they proposed a model in which BRCA1/p53-driven transcriptional and epigenetic changes inadvertently promote innate differentiation programs in luminal progenitors accompanied by protumorigenic changes in the immune compartment, highlighting the decisive role of the origin cell and providing a potential explanation for the tissue specificity of BRCA1 tumors. Researchers have mapped early changes in seemingly healthy breast tissue before tumors appear, which may have great significance for the early diagnosis of BC (Bach et al., 2021).
In addition to familial BC, BRCA1 gene silencing due to promoter methylation can also lead to sporadic BC (Esteller et al., 2000). A study of tumor xenografts from TNBC patients (Ter Brugge et al., 2016) revealed a novel resistance mechanism in BRCA1-methylated PDX (patient-derived xenograft) tumors. Next-generation sequencing (NGS) data showed that the genome rearrangement places the BRCA1 gene under the transcriptional control of the heterologous promoter, which results in the re-expression of BRCA1 in a subset of BRCA1-mutated PDX tumors and leads to acquired resistance to PARP [poly (ADP-ribose) polymerase 1,2] inhibitor (PARPi) and cisplatin chemotherapy (Ter Brugge et al., 2016). This is a unique example of genomic plasticity that is caused by the treatment of BRCA1-deficient tumors, but it can lead to tumor regeneration.
Detection of BRCA1 Gene Mutations
The BRCA1 genetic test is designed to identify harmful changes in BRCA1 using a blood test. People who inherit mutation in BRCA1 gene are at an increased risk of developing BC and ovarian cancer than the general population. Therefore, the BRCA1 gene test has been widely used by physicians to develop risk-reducing strategies for those who are likely to have an inherited mutation based on personal or family history. Additionally, BRCA1 mutation is a prognostic and predictive biomarker for BC. Although studies provided conflicting interpretations of the prognostic value of BRCA1 mutation in BC patients (Lee et al., 2010; Copson et al., 2018), patients with BRCA1 mutation may be sensitive to platinum salts and PARP inhibitors, which could significantly prolong survival time. Hence, BRCA1 genetic testing is essential for making individualized therapy for selected BC patients. According to the consensus of experts and guidelines, the criteria for candidates to do BRCA1 genetic testing are (Daly et al., 2021): 1) BC patients before the age of 40 years; 2) BC patients before the age of 50 years old who had a second primary BC or a history of BC or pancreatic or prostate cancer in their relatives; 3) patients with TNBC before the age of 60 years old; 4) all male BC patients; 5) patients with bilateral BC; and 6) Patients with a relevant family history at any age, who want to be assessed for cancer risk.
The common gene detection methods for BRCA1 mutations include Sanger sequencing, NGS, multiplex ligation-dependent probe amplification (MLPA), massively parallel signature sequencing (MPSS), chromosomal microarray (CMA), and array comparative genomic hybridization (aCGH). (Toland et al., 2018). So far, no single technique can detect all mutations in the BRCA1 gene (Walsh et al., 2010).
In order to reduce the rate of missed detection, a combination of several methods is used to detect all mutations. Previously, BRCA1 gene detection is mainly performed by Sanger sequencing and MLPA, which can screen out single-nucleotide mutation, small fragment mutation, and large copy number mutation (CNVs). Nowadays, NGS is generally used for gene detection as a high-throughput gene sequencing technology (Plagnol et al., 2012). After the genomic DNA has been cut into small fragments, the end of the molecule is connected to the sequencing preparation library and the sequencing results are obtained after image collection and analysis (Gao and Smith, 2020). Compared with the previous gold standard Sanger sequencing, NGS is both cost and time effective, high throughput with simple operation, and operatable in clinical practice. The disadvantage is the increased error rate by introducing polymerase chain reaction in sequencing (Suryavanshi et al., 2017). For the detection of BRCA1 gene mutations in the real world, there are two major patterns. It is recommended to use NGS technology combined with large fragment deletion detection to detect all exons of BRCA gene and the junction region between exons and introns ±20 bp to explore BRCA1 gene mutation. If the mutation in the allele is identified from the proband in the family, it is appropriate to validate specific loci in the family using the Sanger sequencing method.
Most of the pathogenic mutations of BRCA1 are frameshift mutations and nonsense mutations caused by a single or several base changes. A large fragment rearrangement variant should be considered when no mutations are found by conventional gene sequencing in HBC/hereditary ovarian cancer families (Schmidt et al., 2017). MLPA is the most commonly used method to detect large fragment rearrangement in BRCA1 (Engert et al., 2008). When the polymorphic changes in the DNA of the binding site of the primers affect the binding force between the primers and the target fragment (allelic dropouts), it may lead to both false-positive and false-negative results. When a rearrangement variant is detected by an amplicon-based NGS panel, an additional MLPA assay may be considered for validation (Gomez et al., 2009). The targeting RNA-seq is used to analyze the naturally occurring splicing events of eight BC and/or ovarian cancer susceptibility genes (BRCA1, BRCA2, RAD51C, Rad51d, PTEN, STK11, CDH1, TP53). The results showed that the targeted RNA-seq could identify abnormal splicing events associated with BRCA1 genetic variation and successfully distinguish between complete and incomplete splicing events, which is of great significance in determining pathogenicity (Brandao et al., 2019).
Prevention Strategies in BRCA1 Mutation Carriers
Female carriers of a BRCA1 mutation face a higher lifetime risk to develop BC and ovarian cancer. In general, there are three risk-reducing strategies that have been recommended for these carriers: surveillance, risk-reducing surgery, and chemoprevention. Due to comprehensive considerations, the individual risk-reducing strategy should be discussed and made in terms of several factors including the risk from the specific mutation loci, age, general health status, and the life expectancy of the patient. This risk-reducing therapy should be discussed in a shared decision-making environment with a multidisciplinary team.
Until now, risk-reducing surgeries, a risk-reducing mastectomy (RRM) and risk-reducing salpingo-oophorectomy (RRSO), or a combination of both, have been considered to be most effective in preventing the onset of BC and ovarian cancer. RRM can reduce the risk of developing BC in BRCA1 carriers by more than 90% (Meijers-Heijboer et al., 2001; Heemskerk-Gerritsen et al., 2013; Li et al., 2016) and even reduce mortality from any cause (Honold and Camus, 2018). The psychosocial effect after RRM should not be ignored, with respect to negative impacts on body image and sexuality. Thus, the NCCN guidelines recommend that women with a BRCA1 mutation may undergo RRM with immediate bilateral breast reconstruction and multidisciplinary consultations before making treatment plans, and postoperative psychological counseling is necessary. The European Society of Medical Oncology (ESMO) (Cardoso et al., 2019) and NCCN (Esteller et al., 2000) of the United States stated that prophylactic RRSO may significantly reduce the risk of BC and ovarian cancer in women with BRCA1 gene mutation after the completion of reproductive needs. Several prospective clinical trials demonstrated the efficacy of selective estrogen receptor modulators (Cuzick et al., 2013) (i.e., tamoxifen, raloxifene) and aromatase inhibitors exemestane (Goss et al., 2011) and anastrozole (Cuzick et al., 2014) for preventing BC in unselected women. However, there is limited evidence supporting the efficacy of those risk-reducing endocrine therapy options for the carriers of BRCA1 mutations. Studies failed to show the efficacy of tamoxifen (King et al., 2001) or letrozole (Pujol et al., 2020) on decreasing BC incidence in women with germline BRCA1 mutations.
Treatment of Breast Cancer With BRCA1 Mutation: Current Practice and Future Directions
In general, the most common treatments for BC include a combination of surgery, radiation, chemotherapy, hormone therapy, targeted therapy, and immunotherapy. The commonly used surgical treatment methods include breast-conserving surgery (BCS), and mastectomy with the option of breast reconstruction. However, whether BCS is oncologically safe for BRCA mutation carriers has remained controversial (Cao et al., 2019; Davey et al., 2021). For BRCA1 mutation carriers, breast conservation, comprising of lumpectomy followed by whole breast radiation, was associated with higher local recurrence risk for BC patients with BRCA1 mutation; however, BRCA1 mutation was not associated with inferior survival outcomes. Since there is no prospective randomized controlled trial that directly compared BCS and ipsilateral mastectomy for BC patients with BRCA1 mutation, it should be careful to consider BCS (Cao et al., 2019; Davey et al., 2021). Regarding the significant increased risk of developing contralateral BC (Mavaddat et al., 2013), for BC patients with mutations in BRCA1, bilateral nipple-sparing mastectomy combined with reconstruction is a reasonable option for BRCA1 mutation carriers (Tung et al., 2020a).
PARP Inhibitor and Metastatic Breast Cancer With BRCA1 Mutations
PARP inhibitions could induce the death of BRCA1-deficient cells and tumors by interfering with DNA replication, playing a synthetic lethal effect (Farmer et al., 2005; Lord and Ashworth, 2017). Olaparib and talazoparib, as PARP inhibitors, could bring improvement in progression-free survival (PFS) and tumor response rates, and likely improve overall survival for metastatic germline BRCA1-mutated, HER2-negative BC patients based on the primary results from two phase III randomized controlled trials (OlympiAD (Robson et al., 2019) and EMBRACA (Litton et al., 2020)), respectively. Thus, they received approval from the US Food and Drug Administration (FDA) and the European Medicines Agency (EMA). More recently, olaparib monotherapy (Gelmon et al., 2021) and pamiparib (Sun et al., 2021), also showed a promising response in patients with advanced HER2-negative BC with a germline BRCA1 mutation. Additionally, veliparib (Somlo et al., 2017; Dieras et al., 2020; Xu et al., 2021), and niraparib (Turner et al., 2021) are also in investigation for BRCA1-mutated metastatic BC with modest benefit for patients. Those data revealed that the actions of different inhibitors as a PARPi are not the same. Therefore, based on the results above and studies currently under way, PARPi remain a very active area of investigation for BC with BRCA1 mutation.
Based on preliminary data, there is an ongoing trial to explore the efficacy and safety of PARP inhibitors combined with apatinib (NCT04296370). Ceralasertib, an ataxia telangiectasia and Rad3-related protein ATR inhibitor, targets DNA damage repair and cell cycle regulation and shows synergistic antitumor effects combined with olaparib in preclinical studies and a pilot clinical trial (Mahdi et al., 2021).
PARP Inhibitor and Early Breast Cancer With BRCA1 Mutations
In the neoadjuvant setting, a series of clinical trials explored the role of PARP inhibitors; however, the data cannot achieve a clear conclusion. In the BrightNess trial, with the patients with germline BRCA mutation, the pathological complete response (pCR) rate was 57% in the veliparib combined with the carboplatin/paclitaxel arm, 50% in the placebo-carboplatin/paclitaxel arm, and 41% in the control paclitaxel arm (Loibl et al., 2018a). The difference for adding veliparib was not significant, but the trial was not powered to detect it. In GeparOLA (Fasching et al., 2021), the olaparib-containing arm failed to reach its primary endpoint and had a similar pCR compared to the carboplatin-based arm (55.1% vs. 48.6%) in HER2-negative patients with germline BRCA1/2 mutation, or somatic BRCA1/2 mutation, or a high homologous recombination deficiency score. Of note, neoadjuvant single-agent talazoparib without chemotherapy showed promising antitumor activity with manageable toxicity (Litton et al., 2021) in BRCA1-mutated TNBC, which is close to standard chemotherapy in such patients based on previous studies.
In the multicentric, randomized, double-blinded, placebo-controlled phase III OlympiA trial (Tutt et al., 2021), adjuvant olaparib significantly reduced the risk of invasive disease recurrence or death by 42% in high-risk HER2-negative early BC with germline BRCA1/2 mutations. Subgroup analysis showed a consistent benefit in patients with BRCA1 mutations. Importantly, the rate of central nervous system (CNS) recurrence was lower with olaparib treatment than that of placebo arm, which suggests the action of olaparib. FDA-approved adjuvant olaparib for early HER2-negative BC patients with high-risk-carrying germline BRCA1/2 mutation. The definition of high risk in BC patients was defined in Table 1. OlympiA (Tutt et al., 2021) emphasizes the need to conduct the BRCA genetic test early to allow individualized treatment, which would maximize long-term outcomes for the BC patients with germline BRCA mutations.
Platinum Salts and Early Breast Cancer With BRCA1 Mutations
Retrospective clinical studies in the neoadjuvant setting have shown that early BC patients with BRCA1 mutations are more sensitive to platinum salts (Byrski et al., 2010; Arun et al., 2011; Saether et al., 2018; Holanek et al., 2019). However, the role of neoadjuvant platinum in patients with BRCA1 mutation is still unclear due to the conflicting data from several prospective clinical trials. GeparSixto (Hahnen et al., 2017; Loibl et al., 2018b) and CALGB 40603 (Sikov et al., 2015) trials demonstrated that early TNBC patients benefit from platinum-based neoadjuvant chemotherapy with higher pCR rates than that from platinum-free neoadjuvant chemotherapy. However, results of the post-hoc exploratory subgroup analyses from GeparSixto failed to demonstrate that BRCA1 mutation could predict higher pathological response. Furthermore, the addition of platinum could yield comparably high pCR for BRCA1 mutation and wild-type patients (Sharma et al., 2017). Nevertheless, pCR tended to be worse in the cisplatin- containing group than in the doxorubicin–cyclophosphamide group for BRCA carriers with early HER2-negative BC (Tung et al., 2020b). The BrightTNess trial demonstrated that the addition of carboplatin increased pCR (Loibl et al., 2018a) and improved event-free survival (Loibl, 2021) compared with paclitaxel alone in unselected TNBC patients. Therefore, for germline BRCA mutation carriers with BC treated with neoadjuvant therapy, the routine addition of platinum to anthracycline and taxane-based chemotherapy is not supported. Therefore, we need more evidence to explore the role of platinum salts in early BC with BRCA mutations. Several ongoing trials are conducted to investigate the efficacy and safety of neoadjuvant platinum-containing chemotherapy or combined with olaparib in neoadjuvant therapy for operable TNBC (NCT 02978495, NCT 04664972, NCT03150576).
Platinum Salts and Metastatic Breast Cancer With BRCA1 Mutations
Germline BRCA carriers could benefit from platinum agents in the treatment with metastatic BC (Kriege et al., 2009; Byrski et al., 2012). However, those results should be interpreted with caution due to small sample size. In a phase III trial TNT (Tutt et al., 2018), in the subset of patients with BRCA- mutated metastatic BC, patients benefit more from carboplatin versus docetaxel, who yielded a greater objective response rate (ORR) and longer PFS. However, no overall survival benefit was observed. In addition, TNT trial might also be underpowered due to a small sample size for BRCA1/2 mutation carriers (n = 55). In another small-sample-size (N = 11) single-arm trial, 6-mercaptopurine (6 MP) and methotrexate, which could selectively kill BRCA-defective cells in a xenograft model, failed to show anti-tumor activity for advanced BC with a BRCA1 mutation (Roberts et al., 2020).
Resistance to Platinum-Based Chemotherapy or PARP Inhibition
PARPi-based chemotherapy has shown great promise in clinics; however, not all patients with mutations in BRCA1 or genes associated with BRCAness will respond to PARPi, as different mutations may have differing effects on the DNA double-stand break repair function and sensitivities to PARP inhibition. There is very limited understanding of what factors may affect PARPi responses in the setting of BRCA1 mutations and other BRCAness genes. It is also likely that the therapeutic implications may differ in different cancer types, further reinforcing the importance of the context in which BRCA and other HR-related genes function in these malignancies.
In addition, lots of patients acquire PARPi resistance with prolonged PARPi treatment in clinics. Resistance to platinum-based chemotherapy is also a promising predictor for resistance to PARPi, suggesting that they may share a common mechanism. The main molecular mechanisms of PARPi resistance are the cellular availability of the inhibitor, reverse mutations, homologous recombination repair restoration, and restoration of replication fork protection. Firstly, in a murine model of BRCA1-deficient breast tumors, tumors with overexpressed drug-efflux transporter genes (Abcb1a and Abcb1b encoding for MDR1/P-gp and Abcg2) showed resistance to PARPi by influencing the cellular availability of the inhibitor. The coadministration of the MDR1 inhibitor could resensitize the tumors to the PARPi (Rottenberg, et al., 2008). Secondly, somatic reversion mutations were found in cfDNA (circulating cell-free) in BC patients who acquired resistance to platinum and/or PARP inhibitors. The BRCA1 reversion mutation could restore BRCA1 function(Weigelt, et al., 2017). Thirdly, the fork degradation of deprotected replication forks is mediated by at least three mechanisms, which, upon loss, leads to fork protection and thus to PARPi resistance. (Angelo et al., 2017). Lastly, HR reactivation is dependent on the ubiquitin E3 ligase RNF168 and the loss of 53BP1–RIF1–REV7–Shieldin axis in BRCA1-deficient and TP53BP1-deficient cells, leading to PARPi resistance (Nakada et al., 2012; Dev, et al., 2018). In addition, cancer stem cells (CSCs) in BRCA1 mutant TNBCs were resistant to PARP inhibition, and RAD51 protein levels and activity were elevated. ShRNA downregulated the expression of RAD51 and thereby made CSCs sensitive to PARPi. (Liu et al., 2016). Therefore, RAD51 is a functional biomarker to be used in the clinic to identify PARPi-sensitive cancer patients and select the population who may respond to PARPi therapy (C Cruz, et al., 2018). Moreover, epigenetic modification and restoration of ADPribosylation (PARylation) lead to PARPi resistance as well. Studies show that patients with high probability of resistance to PARPi may obtain a benefit from combinatorial treatment strategies. Since PALB2–BRCA2 recruitment to DNA breaks and RAD51 recruitment to stalled forks are both ATR dependent, the combination of PARPi with ATR inhibitors is expected to overcome PARPi resistance in tumors by restoring HR or restoring fork protection. (Stephanie A Yazinski et al., 2017). It is of great importance to clarify the clinical relevance of the different PARPi resistance mechanisms through more large patient cohorts. These studies will pave the way for patients in the clinic to improve diagnosis, therapy decisions, and outcome.
Hormone Therapy and Breast Cancer With BRCA1 Mutations
Carriers of BRCA1 mutation are more likely to develop TNBC (Atchley et al., 2008). Therefore, for those hormone receptor-positive BC patients with pathogenic BRCA1 mutations, hormone therapy is essential to delay the progression and prevent the onset of contralateral tumors. Adjuvant tamoxifen (Narod et al., 2000) and aromatase inhibitors (Gutierrez-Barrera et al., 2015) could significantly reduce the risk of contralateral tumors. The addition of contralateral breast irradiation was associated with a significant reduction of subsequent contralateral breast cancers and a delay in their onset (Evron et al., 2019).
Immunotherapy and Breast Cancer With BRCA1 Mutations
Immunotherapy is an important approach for cancer treatment, and DNA repair defects are an important factor in enhancing the anti-tumor immune response. The germline BRCA1 mutation is related to the high mutation burden of TNBC, and the combination of cisplatin and PD-1/CTLA-4 antibody has a more significant tumor inhibition effect than the use of cisplatin alone (Mouw et al., 2017). Therefore, the combination of immune checkpoint inhibitors and chemotherapy can effectively improve the efficacy of HR-deficient tumors. In addition, a number of clinical trials of PARP inhibitors combined with chemotherapy or immunotherapy are under way, and preliminary results show that PARP inhibitors combined with immunotherapy can enhance the killing effect of BRCA1 germline mutation BC (Mateo et al., 2019; Domchek et al., 2020).
Following the success of previous trials, there is a series of phases two clinical trials currently ongoing with immunotherapy or targeted therapy in BC patients with germline BRCA1 mutation (Table 2).
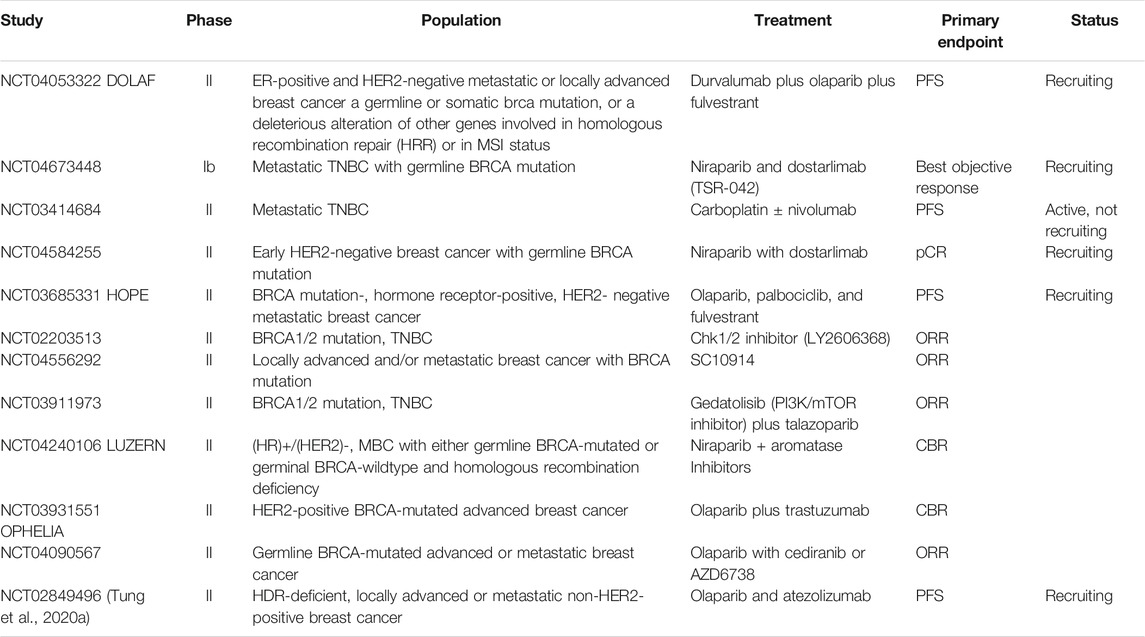
TABLE 2. Immunotherapy or targeted therapy-based treatment in metastatic BC with BRCA mutation: ongoing clinical trials.
Beyond BRCA1 Germline Mutation
In addition to BRCA1 and BRCA2 mutations, the BC patients with BARD1, PALB2, RAD51, FANCA, ATM, ATR, and CHK2 mutations also present an HRR defect and might also share sensitivity to platinum-based drugs and PARPi (Christopher J. Lord and Ashworth, 2016). The above HR genes share the same cancer genetic profile and are called BRCAness (Cai, 2020). All these genes’ mutations increase the sensitivity of tumor cells to PARP inhibitors, suggesting that PARP inhibitors may be effective against multiple tumors rather than specific tumors with BRCA1 mutations. TBCRC 048, a phase II study, emphasized that PARP inhibition is an effective treatment for patients with MBC and germline PALB2 or somatic BRCA1/2 mutations (Tung et al., 2020c). Furthermore, in treatment-naïve TNBCs, olaparib monotherapy yielded a high clinical response rate with BRCAness signature (Eikesdal et al., 2020), beyond germline HR mutations (NCT02849496, NCT03990896, NCT04892693). Many similar clinical trials are underway, which will greatly expand the usage of PARP inhibitors.
Conclusion
BRCA1 gene is a well-known tumor suppressor gene, and germline BRCA1 mutation is closely related to the occurrence, development, and treatment of BC. With the advancement and popularization of gene sequencing technology, BRCA testing has been widely used in clinical practice. For better outcomes of BC patients with germline BRCA1 mutation, it is necessary and critical to comprehensively consider the combination of the therapies, and a multidisciplinary consultant is required to make an appropriate individualized management plan. With further exploration on the BRCA1 gene, more and more patients with BRCA1 mutations will benefit.
Author Contributions
XF carried out the design and drafted the manuscript. WT and QS articipated in the draft of the manuscript. JL and HP conceived of the study, and participated in its design and coordination and helped to drafted the manuscript. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2022.813457/full#supplementary-material
Abbreviations
CBR, clinical benefit rate; HDR, homologous DNA repair; HER2, human epidermal growth factor receptor 2; HR, hormone receptor; ORR, overall response rate; PFS, progression-free survival; TNBC, triple-negative breast cancer.
References
Arun, B., Bayraktar, S., Liu, D. D., Gutierrez Barrera, A. M., Atchley, D., Pusztai, L., et al. (2011). Response to Neoadjuvant Systemic Therapy for Breast Cancer in BRCA Mutation Carriers and Noncarriers: a Single-Institution Experience. Jco 29, 3739–3746. doi:10.1200/JCO.2011.35.2682
Atchley, D. P., Albarracin, C. T., Lopez, A., Valero, V., Amos, C. I., Gonzalez-Angulo, A. M., et al. (2008). Clinical and Pathologic Characteristics of Patients with BRCA-Positive and BRCA-Negative Breast Cancer. Jco 26 (26), 4282–4288. doi:10.1200/jco.2008.16.6231
Bach, K., Pensa, S., Zarocsinceva, M., Kania, K., Stockis, J., Pinaud, S., et al. (2021). Time-resolved Single-Cell Analysis of Brca1 Associated Mammary Tumourigenesis Reveals Aberrant Differentiation of Luminal Progenitors. Nat. Commun. 12, 1502. doi:10.1038/s41467-021-21783-3
Bochar, D. A., Wang, L., Beniya, H., Kinev, A., Xue, Y., Lane, W. S., et al. (2000). BRCA1 Is Associated with a Human SWI/SNF-Related Complex. Cell 102, 257–265. doi:10.1016/s0092-8674(00)00030-1
Brandão, R. D., Mensaert, K., López‐Perolio, I., Tserpelis, D., Xenakis, M., Lattimore, V., et al. (2019). Targeted RNA‐seq Successfully Identifies normal and Pathogenic Splicing Events in Breast/ovarian Cancer Susceptibility and Lynch Syndrome Genes. Int. J. Cancer 145, 401–414. doi:10.1002/ijc.32114
Braunschweig, U., Gueroussov, S., Plocik, A. M., Graveley, B. R., and Blencowe, B. J. (2013). Dynamic Integration of Splicing within Gene Regulatory Pathways. Cell 152, 1252–1269. doi:10.1016/j.cell.2013.02.034
Byrski, T., Dent, R., Blecharz, P., Foszczynska-Kloda, M., Gronwald, J., Huzarski, T., et al. (2012). Results of a Phase II Open-Label, Non-randomized Trial of Cisplatin Chemotherapy in Patients with BRCA1-Positive Metastatic Breast Cancer. Breast Cancer Res. 14, R110. doi:10.1186/bcr3231
Byrski, T., Gronwald, J., Huzarski, T., Grzybowska, E., Budryk, M., Stawicka, M., et al. (2010). Pathologic Complete Response Rates in Young Women with BRCA1-Positive Breast Cancers after Neoadjuvant Chemotherapy. Jco 28, 375–379. doi:10.1200/JCO.2008.20.7019
Cai, C. (2020). A Novel Mechanism to Induce BRCAness in Cancer Cells. Cancer Res. 80, 2977–2978. doi:10.1158/0008-5472.CAN-20-1451
Cao, W., Xie, Y., He, Y., Li, J., Wang, T., Fan, Z., et al. (2019). Risk of Ipsilateral Breast Tumor Recurrence in Primary Invasive Breast Cancer Following Breast-Conserving Surgery with BRCA1 and BRCA2 Mutation in China. Breast Cancer Res. Treat. 175, 749–754. doi:10.1007/s10549-019-05199-8
Cardoso, F., Kyriakides, S., Ohno, S., Penault-Llorca, F., Poortmans, P., Rubio, I. T., et al. (2019). Early Breast Cancer: ESMO Clinical Practice Guidelines for Diagnosis, Treatment and Follow-Up. Ann. Oncol. 30, 1674. doi:10.1093/annonc/mdz189
Chabanon, F., Lord, J. L., Soria, J.-C., Pasero, P., and Postel-Vinay, S. (2021). Targeting the DNA Damage Response in Immuno-Oncology: Developments and Opportunities. Nat. Rev. Cancer 21 (11), 701–717.
Chen, Y., Farmer, A. A., Chen, C. F., Jones, D. C., Chen, P. L., and Lee, W. H. (1996). BRCA1 Is a 220-kDa Nuclear Phosphoprotein that Is Expressed and Phosphorylated in a Cell Cycle-dependent Manner. Cancer Res. 56 (14), 3168–3172.
Clark, S. L., Rodriguez, A. M., Snyder, R. R., Hankins, G. D. V., and Boehning, D. (2012). Structure-Function of the Tumor Suppressor BRCA1. Comput. Struct. Biotechnol. J. 1, e201204005. doi:10.5936/csbj.201204005
Claus, E. B., Schildkraut, J. M., Thompson, W. D., and Risch, N. J. (1996). The Genetic Attributable Risk of Breast and Ovarian Cancer. Cancer 77 (11), 2318–2324. doi:10.1002/(sici)1097-0142(19960601)77:11<2318:aid-cncr21>3.0.co;2-z
Copson, E. R., Maishman, T. C., Tapper, W. J., Cutress, R. I., Greville-Heygate, S., Altman, D. G., et al. (2018). Germline BRCA Mutation and Outcome in Young-Onset Breast Cancer (POSH): a Prospective Cohort Study. Lancet Oncol. 19 (2), 169–180. doi:10.1016/s1470-2045(17)30891-4
Cruz, C., Castroviejo-Bermejo, M., Gutiérrez-Enríquez, S., Llop-Guevara, A., Ibrahim, Y. H., Gris-Oliver, A., et al. (2018). RAD51 Foci as a Functional Biomarker of Homologous Recombination Repair and PARP Inhibitor Resistance in Germline BRCA-Mutated Breast Cancer. Ann. Oncol. 29 (5), 1203–1210. doi:10.1093/annonc/mdy099
Cuzick, J., Sestak, I., Bonanni, B., Costantino, J. P., Cummings, S., DeCensi, A., et al. (2013). Selective Oestrogen Receptor Modulators in Prevention of Breast Cancer: an Updated Meta-Analysis of Individual Participant Data. The Lancet 381, 1827–1834. doi:10.1016/S0140-6736(13)60140-3
Cuzick, J., Sestak, I., Forbes, J. F., Dowsett, M., Knox, J., Cawthorn, S., et al. (2014). Anastrozole for Prevention of Breast Cancer in High-Risk Postmenopausal Women (IBIS-II): an International, Double-Blind, Randomised Placebo-Controlled Trial. The Lancet 383, 1041–1048. doi:10.1016/S0140-6736(13)62292-8
Daly, M. B., Pal, T., Berry, M. P., Buys, S. S., Dickson, P., Domchek, S. M., et al. (2021). Genetic/Familial High-Risk Assessment: Breast, Ovarian, and Pancreatic, Version 2.2021, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Canc Netw. 19, 77–102. doi:10.6004/jnccn.2021.0001
Davey, M. G., Davey, C. M., Ryan, É. J., Lowery, A. J., and Kerin, M. J. (2021). Combined Breast Conservation Therapy versus Mastectomy for BRCA Mutation Carriers - A Systematic Review and Meta-Analysis. The Breast 56, 26–34. doi:10.1016/j.breast.2021.02.001
Dev, H., Chiang, T.-W. W., Lescale, C., de Krijger, I., Martin, A. G., Pilger, D., et al. (2018). Shieldin Complex Promotes DNA End-Joining and Counters Homologous Recombination in BRCA1-Null Cells. Nat. Cel Biol 20 (8), 954–965. doi:10.1038/s41556-018-0140-1
Diéras, V., Han, H. S., Kaufman, B., Wildiers, H., Friedlander, M., Ayoub, J.-P., et al. (2020). Veliparib with Carboplatin and Paclitaxel in BRCA-Mutated Advanced Breast Cancer (BROCADE3): a Randomised, Double-Blind, Placebo-Controlled, Phase 3 Trial. Lancet Oncol. 21, 1269–1282. doi:10.1016/S1470-2045(20)30447-2
Domchek, S. M., Postel-Vinay, S., Im, S.-A., Park, Y. H., Delord, J.-P., Italiano, A., et al. (2020). Olaparib and Durvalumab in Patients with Germline BRCA-Mutated Metastatic Breast Cancer (MEDIOLA): an Open-Label, Multicentre, Phase 1/2, Basket Study. Lancet Oncol. 21, 1155–1164. doi:10.1016/S1470-2045(20)30324-7
Ducy, M., Sesma-Sanz, L., Guitton-Sert, L., Lashgari, A., Gao, Y., Brahiti, N., et al. (2019). The Tumor Suppressor PALB2: Inside Out. Trends Biochem. Sci. 44 (3), 226–240. doi:10.1016/j.tibs.2018.10.008
Eikesdal, H. P., Yndestad, S., Elzawahry, A., Llop-Guevara, A., and Lnning, P. E. J. A. o. O. (2020). Olaparib Monotherapy as Primary Treatment in Unselected Triple Negative Breast Cancer. Clin. Trial 32, 240–249. doi:10.1016/j.annonc.2020.11.009
Engert, S., Wappenschmidt, B., Betz, B., Kast, K., Kutsche, M., Hellebrand, H., et al. (2008). MLPA Screening in theBRCA1gene from 1,506 German Hereditary Breast Cancer Cases: Novel Deletions, Frequent Involvement of Exon 17, and Occurrence in Single Early-Onset Cases. Hum. Mutat. 29, 948–958. doi:10.1002/humu.20723
Esteller, M., Silva, J. M., Dominguez, G., Bonilla, F., Matias-Guiu, X., Lerma, E., et al. (2000). Promoter Hypermethylation and BRCA1 Inactivation in Sporadic Breast and Ovarian Tumors. J. Natl. Cancer Inst. 92, 564–569. doi:10.1093/jnci/92.7.564
Evron, E., Ben-David, A. M., Goldberg, H., Fried, G., Kaufman, B., Catane, R., et al. (2019). Prophylactic Irradiation to the Contralateral Breast for BRCA Mutation Carriers with Early-Stage Breast Cancer. Ann. Oncol. 30, 412–417. doi:10.1093/annonc/mdy515
Farmer, H., McCabe, N., Lord, C. J., Tutt, A. N. J., Johnson, D. A., Richardson, T. B., et al. (2005). Targeting the DNA Repair Defect in BRCA Mutant Cells as a Therapeutic Strategy. Nature 434, 917–921. doi:10.1038/nature03445
Fasching, P. A., Link, T., Hauke, J., Seither, F., Jackisch, C., Klare, P., et al. (2021). Neoadjuvant Paclitaxel/olaparib in Comparison to Paclitaxel/carboplatinum in Patients with HER2-Negative Breast Cancer and Homologous Recombination Deficiency (GeparOLA Study). Ann. Oncol. 32, 49–57. doi:10.1016/j.annonc.2020.10.471
Foulkes, W. D., Metcalfe, K., Sun, P., Hanna, W. M., Lynch, H. T., Ghadirian, P., et al. (2004). Estrogen Receptor Status in BRCA1- and BRCA2-Related Breast Cancer. Clin. Cancer Res. 10 (6), 2029–2034. doi:10.1158/1078-0432.ccr-03-1061
Gao, G., and Smith, D. I. (2020). Clinical Massively Parallel Sequencing. Clin. Chem. 66, 77–88. doi:10.1373/clinchem.2019.303305
Gelmon, K. A., Fasching, P. A., Couch, F. J., Balmaña, J., Delaloge, S., Labidi-Galy, I., et al. (2021). Clinical Effectiveness of Olaparib Monotherapy in Germline BRCA-Mutated, HER2-Negative Metastatic Breast Cancer in a Real-World Setting: Phase IIIb LUCY Interim Analysis. Eur. J. Cancer 152, 68–77. doi:10.1016/j.ejca.2021.03.029
Gomez, L. C., Marzese, D. M., Adi, J., Bertani, D., Ibarra, J., Mol, B., et al. (2009). MLPA Mutation Detection in Argentine HNPCC and FAP Families. Fam. Cancer 8, 67–73. doi:10.1007/s10689-008-9200-1
Gorthi, A., Romero, J. C., Loranc, E., Cao, L., Lawrence, L. A., Goodale, E., et al. (2018). EWS-FLI1 Increases Transcription to Cause R-Loops and Block BRCA1 Repair in Ewing Sarcoma. Nature 555, 387–391. doi:10.1038/nature25748
Goss, P. E., Ingle, J. N., Alés-Martínez, J. E., Cheung, A. M., Chlebowski, R. T., Wactawski-Wende, J., et al. (2011). Exemestane for Breast-Cancer Prevention in Postmenopausal Women. N. Engl. J. Med. 364, 2381–2391. doi:10.1056/NEJMoa1103507
Gudmundsdottir, K., and Ashworth, A. (2006). The Roles of BRCA1 and BRCA2 and Associated Proteins in the Maintenance of Genomic Stability. Oncogene 25, 5864–5874. doi:10.1038/sj.onc.1209874
Gutierrez-Barrera, A. M., Lin, H. Y., and Arun, B. J. J. C. O. (2015). Aromatase Inhibitors and the Risk of Contralateral Breast Cancer in BRCA Mutation Carriers. J. Clin. Oncol., 28_Suppl. l 33.
Hahnen, E., Lederer, B., Hauke, J., Loibl, S., Kröber, S., Schneeweiss, A., et al. (2017). Germline Mutation Status, Pathological Complete Response, and Disease-free Survival in Triple-Negative Breast Cancer. JAMA Oncol. 3, 1378–1385. doi:10.1001/jamaoncol.2017.1007
Hall, J. M., Lee, M. K., Newman, B., Morrow, J. E., Anderson, L. A., Huey, B., et al. (1990). Linkage of Early-Onset Familial Breast Cancer to Chromosome 17q21. Science 250, 1684–1689. doi:10.1126/science.2270482
Hashizume, R., Fukuda, M., Maeda, I., Nishikawa, H., Oyake, D., Yabuki, Y., et al. (2001). The RING Heterodimer BRCA1-BARD1 Is a Ubiquitin Ligase Inactivated by a Breast Cancer-Derived Mutation. J. Biol. Chem. 276 (18), 14537–14540. doi:10.1074/jbc.c000881200
Hatchi, E., Skourti-Stathaki, K., Ventz, S., Pinello, L., Yen, A., Kamieniarz-Gdula, K., et al. (2015). BRCA1 Recruitment to Transcriptional Pause Sites Is Required for R-Loop-Driven DNA Damage Repair. Mol. Cel 57, 636–647. doi:10.1016/j.molcel.2015.01.011
Heemskerk-Gerritsen, B. A. M., Menke-Pluijmers, M. B. E., Jager, A., Tilanus-Linthorst, M. M. A., Koppert, L. B., Obdeijn, I. M. A., et al. (2013). Substantial Breast Cancer Risk Reduction and Potential Survival Benefit after Bilateral Mastectomy when Compared with Surveillance in Healthy BRCA1 and BRCA2 Mutation Carriers: a Prospective Analysis. Ann. Oncol. 24, 2029–2035. doi:10.1093/annonc/mdt134
Herold, S., Kalb, J., Büchel, G., Ade, C. P., Baluapuri, A., Xu, J., et al. (2019). Recruitment of BRCA1 Limits MYCN-Driven Accumulation of Stalled RNA Polymerase. Nature 567, 545–549. doi:10.1038/s41586-019-1030-9
Holánek, M., Bílek, O., Nenutil, R., Kazda, T., Selingerová, I., Zvaríková, M., et al. (2019). Effectiveness of Neoadjuvant Therapy with Platinum-Based Agents for Patients with BRCA1 and BRCA2 Germline Mutations - A Retrospective Analysis of Breast Cancer Patients Treated at MMCI Brno. Klin Onkol 32, 31–35. doi:10.14735/amko2019S31
Honold, F., and Camus, M. (2018). Prophylactic Mastectomy versus Surveillance for the Prevention of Breast Cancer in Women's BRCA Carriers. Medwave 18, e7160. doi:10.5867/medwave.2018.04.7160
Huang, R.-X., and Zhou, P.-K. (2020). DNA Damage Response Signaling Pathways and Targets for Radiotherapy Sensitization in Cancer. Sig Transduct Target. Ther. 5, 60. doi:10.1038/s41392-020-0150-x
King, M.-C., Marks, J. H., and Mandell, J. B. (2003). New York Breast Cancer StudyBreast and Ovarian Cancer Risks Due to Inherited Mutations in BRCA1 and BRCA2. Science 302, 643–646. doi:10.1126/science.1088759
King, M. C., Wieand, S., Hale, K., Lee, M., Walsh, T., Owens, K., et al. (2001). Tamoxifen and Breast Cancer Incidence Among Women with Inherited Mutations in BRCA1 and BRCA2. JAMA 286, 2251–2256. doi:10.1001/jama.286.18.2251
Kriege, M., Seynaeve, C., Meijers-Heijboer, H., Collee, J. M., Menke-Pluymers, M. B. E., Bartels, C. C. M., et al. (2009). Sensitivity to First-Line Chemotherapy for Metastatic Breast Cancer in BRCA1 and BRCA2 Mutation Carriers. Jco 27, 3764–3771. doi:10.1200/JCO.2008.19.9067
Kuchenbaecker, K. B., Hopper, J. L., Barnes, D. R., Phillips, K. A., Mooij, T. M., Roos-Blom, M. J., et al. (2017). Risks of Breast, Ovarian, and Contralateral Breast Cancer for BRCA1 and BRCA2 Mutation Carriers. JAMA 317, 2402–2416. doi:10.1001/jama.2017.7112
Lee, E.-H., Park, Sue. K., Park, S. K., Park, B., Kim, S.-W., Lee, M. H., et al. (2010). Effect of BRCA1/2 Mutation on Short-Term and Long-Term Breast Cancer Survival: a Systematic Review and Meta-Analysis. Breast Cancer Res. Treat. 122 (1), 11–25. doi:10.1007/s10549-010-0859-2
Li, D., Harlan-Williams, L. M., Kumaraswamy, E., and Jensen, R. A. (2019). BRCA1-No Matter How You Splice it. Cancer Res. 79, 2091–2098. doi:10.1158/0008-5472.CAN-18-3190
Li, X., You, R., Wang, X., Liu, C., Xu, Z., Zhou, J., et al. (2016). Effectiveness of Prophylactic Surgeries in BRCA1 or BRCA2 Mutation Carriers: A Meta-Analysis and Systematic Review. Clin. Cancer Res. 22, 3971–3981. doi:10.1158/1078-0432.CCR-15-1465
Lim, E., Vaillant, F., Vaillant, F., Wu, D., Forrest, N. C., Pal, B., et al. (2009). Aberrant Luminal Progenitors as the Candidate Target Population for Basal Tumor Development in BRCA1 Mutation Carriers. Nat. Med. 15, 907–913. doi:10.1038/nm.2000
Litton, J. K., Beck, J. T., Jones, J. M., Andersen, J., Blum, J. L., Mina, L. A., et al. (2021). Neoadjuvant Talazoparib in Patients with Germline BRCA1/2 (gBRCA1/2) Mutation-Positive, Early HER2-Negative Breast Cancer (BC): Results of a Phase 2 Study. Jco 39, 505. doi:10.1200/jco.2021.39.15_suppl.505
Litton, J. K., Hurvitz, S. A., Mina, L. A., Rugo, H. S., Lee, K.-H., Gonçalves, A., et al. (2020). Talazoparib versus Chemotherapy in Patients with Germline BRCA1/2-Mutated HER2-Negative Advanced Breast Cancer: Final Overall Survival Results from the EMBRACA Trial. Ann. Oncol. 31, 1526–1535. doi:10.1016/j.annonc.2020.08.2098
Liu, Y., Burness, M. L., Martin-Trevino, R., Guy, J., Bai, S., Harouaka, R., et al. (2016). RAD51 Mediates Resistance of Cancer Stem Cells to PARP Inhibition in Triple-Negative Breast Cancer. Clin. Cancer Res. 23 (2), 514–522. doi:10.1158/1078-0432.ccr-15-1348
Loibl, S. (2021). Event-free Survival (EFS), Overall Survival (OS), and Safety of Adding Veliparib (V) Plus Carboplatin (Cb) or Carboplatin Alone to Neoadjuvant Chemotherapy in Triple-Negative Breast Cancer (TNBC) after ≥4 Years of Follow-Up: BrighTNess, a Randomized Phase 3 Trial. ESMO Congress 2021, Abstract 119O.
Loibl, S., O'Shaughnessy, J., Untch, M., Sikov, W. M., Rugo, H. S., McKee, M. D., et al. (2018a). Addition of the PARP Inhibitor Veliparib Plus Carboplatin or Carboplatin Alone to Standard Neoadjuvant Chemotherapy in Triple-Negative Breast Cancer (BrighTNess): a Randomised, Phase 3 Trial. Lancet Oncol. 19, 497–509. doi:10.1016/S1470-2045(18)30111-6
Loibl, S., Weber, K. E., Timms, K. M., Elkin, E. P., Hahnen, E., Fasching, P. A., et al. (2018b). Survival Analysis of Carboplatin Added to an Anthracycline/taxane-Based Neoadjuvant Chemotherapy and HRD Score as Predictor of Response-Final Results from GeparSixto. Ann. Oncol. 29, 2341–2347. doi:10.1093/annonc/mdy460
Long, D. T., Joukov, V., Budzowska, M., and Walter, J. C. (2014). BRCA1 Promotes Unloading of the CMG Helicase from a Stalled DNA Replication fork. Mol. Cel 56, 174–185. doi:10.1016/j.molcel.2014.08.012
Lord, C. J., and Ashworth, A. (2016). BRCAness Revisited. Nat. Rev. Cancer 16 (2), 110–120. doi:10.1038/nrc.2015.21
Lord, C. J., and Ashworth, A. (2017). PARP Inhibitors: Synthetic Lethality in the Clinic. Science 355, 1152–1158. doi:10.1126/science.aam7344
Mahdi, H., Hafez, N., Doroshow, D., Sohal, D., Keedy, V., Do, K. T., et al. (2021). Ceralasertib-Mediated ATR Inhibition Combined With Olaparib in Advanced Cancers Harboring DNA Damage Response and Repair Alterations (Olaparib Combinations). JCO Precis. Oncol. 5 (5), 1432–1442. doi:10.1200/PO.20.00439
Mateo, J., Lord, C. J., Serra, V., Tutt, A., Balmaña, J., Castroviejo-Bermejo, M., et al. (2019). A Decade of Clinical Development of PARP Inhibitors in Perspective. Ann. Oncol. 30, 1437–1447. doi:10.1093/annonc/mdz192
Mavaddat, N., Peock, S., Frost, D., Ellis, S., Platte, R., Fineberg, E., et al. (2013). Cancer Risks for BRCA1 and BRCA2 Mutation Carriers: Results from Prospective Analysis of EMBRACE. J. Natl. Cancer Inst. 105, 812–822. doi:10.1093/jnci/djt095
Meijers-Heijboer, H., van Geel, B., van Putten, W. L. J., Henzen-Logmans, S. C., Seynaeve, C., Menke-Pluymers, M. B. E., et al. (2001). Breast Cancer after Prophylactic Bilateral Mastectomy in Women with aBRCA1orBRCA2Mutation. N. Engl. J. Med. 345, 159–164. doi:10.1056/NEJM200107193450301
Miki, Y., Swensen, J., Shattuck-Eidens, D., Futreal, P. A., Harshman, K., Tavtigian, S., et al. (1994). A strong Candidate for the Breast and Ovarian Cancer Susceptibility Gene BRCA1. Science 266, 66–71. doi:10.1126/science.7545954
Molyneux, G., Geyer, F. C., Magnay, F.-A., McCarthy, A., Kendrick, H., Natrajan, R., et al. (2010). BRCA1 Basal-like Breast Cancers Originate from Luminal Epithelial Progenitors and Not from Basal Stem Cells. Cell Stem Cell 7, 403–417. doi:10.1016/j.stem.2010.07.010
Mouw, K. W., Goldberg, M. S., Konstantinopoulos, P. A., and D'Andrea, A. D. (2017). DNA Damage and Repair Biomarkers of Immunotherapy Response. Cancer Discov. 7, 675–693. doi:10.1158/2159-8290.CD-17-0226
Nakada, S., Yonamine, R. M., and Matsuo, K. (2012). RNF8 Regulates Assembly of RAD51 at DNA Double-Strand Breaks in the Absence of BRCA1 and 53BP1. Cancer Res. 72 (19), 4974–4983. doi:10.1158/0008-5472.can-12-1057
Narod, S. A., Brunet, J.-S., Ghadirian, P., Robson, M., Heimdal, K., Neuhausen, S. L., et al. (2000). Tamoxifen and Risk of Contralateral Breast Cancer in BRCA1 and BRCA2 Mutation Carriers: a Case-Control Study. The Lancet 356, 1876–1881. doi:10.1016/s0140-6736(00)03258-x
Pao, G. M., Janknecht, R., Ruffner, H., Hunter, T., and Verma, I. M. (2000). CBP/p300 Interact with and Function as Transcriptional Coactivators of BRCA1. Proc. Natl. Acad. Sci. 97, 1020–1025. doi:10.1073/pnas.97.3.1020
Petermann, E., Orta, M. L., Issaeva, N., Schultz, N., and Helleday, T. (2010). Hydroxyurea-stalled Replication forks Become Progressively Inactivated and Require Two Different RAD51-Mediated Pathways for Restart and Repair. Mol. Cel 37, 492–502. doi:10.1016/j.molcel.2010.01.021
Plagnol, V., Curtis, J., Epstein, M., Mok, K. Y., Stebbings, E., Grigoriadou, S., et al. (2012). A Robust Model for Read Count Data in Exome Sequencing Experiments and Implications for Copy Number Variant Calling. Bioinformatics 28, 2747–2754. doi:10.1093/bioinformatics/bts526
Pujol, P., Roca, L., Lortholary, A., Lasset, C., Dugast, C., Berthet, P., et al. (2020). Five Year Letrozole versus Placebo in BRCA1/2 Germline Mutations Carriers: Final Results of LIBER, a Double-Blind Randomized Phase III Breast Cancer Prevention Trial. Jco 38, 1534. doi:10.1200/jco.2020.38.15_suppl.1534
Rebbeck, T. R., Mitra, N., Wan, F., Sinilnikova, O. M., Healey, S., McGuffog, L., et al. (2015). Association of Type and Location of BRCA1 and BRCA2 Mutations with Risk of Breast and Ovarian Cancer. JAMA 313, 1347–1361. doi:10.1001/jama.2014.5985
Roberts, C., Strauss, V. Y., Kopijasz, S., Gourley, C., Hall, M., Montes, A., et al. (2020). Results of a Phase II Clinical Trial of 6-mercaptopurine (6MP) and Methotrexate in Patients with BRCA-Defective Tumours. Br. J. Cancer 122, 483–490. doi:10.1038/s41416-019-0674-4
Robson, M. E., Tung, N., Conte, P., Im, S.-A., Senkus, E., Xu, B., et al. (2019). OlympiAD Final Overall Survival and Tolerability Results: Olaparib versus Chemotherapy Treatment of Physician's Choice in Patients with a Germline BRCA Mutation and HER2-Negative Metastatic Breast Cancer. Ann. Oncol. 30, 558–566. doi:10.1093/annonc/mdz012
Rottenberg, S., Jaspers, J. E., Kersbergen, A., van der Burg, E., Nygren, A. O., Zander, S. A., et al. (2008). High Sensitivity of BRCA1-Deficient Mammary Tumors to the PARP Inhibitor AZD2281 Alone and in Combination with Platinum Drugs. Proc. Natl. Acad. Sci. U S A. 105 (44), 17079–17084. doi:10.1073/pnas.0806092105
Sæther, N. H., Skuja, E., Irmejs, A., Maksimenko, J., Miklasevics, E., Purkalne, G., et al. (2018). Platinum-based Neoadjuvant Chemotherapy in BRCA1-Positive Breast Cancer: a Retrospective Cohort Analysis and Literature Review. Hered. Cancer Clin. Pract. 16. doi:10.1186/s13053-018-0092-2
Schlacher, K., Wu, H., and Jasin, M. (2012). A Distinct Replication fork protection Pathway Connects Fanconi Anemia Tumor Suppressors to RAD51-Brca1/2. Cancer Cell 22, 106–116. doi:10.1016/j.ccr.2012.05.015
Schmidt, A. Y., Hansen, T. v. O., Ahlborn, L. B., Jønson, L., Yde, C. W., and Nielsen, F. C. (2017). Next-Generation Sequencing-Based Detection of Germline Copy Number Variations in BRCA1/BRCA2. J. Mol. Diagn. 19, 809–816. doi:10.1016/j.jmoldx.2017.07.003
Sharma, P., López-Tarruella, S., García-Saenz, J. A., Ward, C., Connor, C. S., Gómez, H. L., et al. (2017). Efficacy of Neoadjuvant Carboplatin Plus Docetaxel in Triple-Negative Breast Cancer: Combined Analysis of Two Cohorts. Clin. Cancer Res. 23, 649–657. doi:10.1158/1078-0432.CCR-16-0162
Sikov, W. M., Berry, D. A., Perou, C. M., Singh, B., Cirrincione, C. T., Tolaney, S. M., et al. (2015). Impact of the Addition of Carboplatin And/or Bevacizumab to Neoadjuvant Once-Per-Week Paclitaxel Followed by Dose-Dense Doxorubicin and Cyclophosphamide on Pathologic Complete Response Rates in Stage II to III Triple-Negative Breast Cancer: CALGB 40603 (Alliance). Jco 33, 13–21. doi:10.1200/JCO.2014.57.0572
Somlo, G., Frankel, P. H., Arun, B. K., Ma, C. X., Garcia, A. A., Cigler, T., et al. (2017). Efficacy of the PARP Inhibitor Veliparib with Carboplatin or as a Single Agent in Patients with Germline BRCA1- or BRCA2-Associated Metastatic Breast Cancer: California Cancer Consortium Trial NCT01149083. Clin. Cancer Res. 23, 4066–4076. doi:10.1158/1078-0432.CCR-16-2714
Sun, T., Shi, Y., Cui, J., Yin, Y., Ouyang, Q., Liu, Q., et al. (2021). A Phase 2 Study of Pamiparib in the Treatment of Patients with Locally Advanced or Metastatic HER2-Negative Breast Cancer with Germline BRCA Mutation. Jco 39, 1087. doi:10.1200/jco.2021.39.15_suppl.1087
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A. Cancer J. Clin. 71, 209–249. doi:10.3322/caac.21660
Suryavanshi, M., Kumar, D., Panigrahi, M. K., Chowdhary, M., and Mehta, A. (2017). Detection of False Positive Mutations in BRCA Gene by Next Generation Sequencing. Fam. Cancer 16, 311–317. doi:10.1007/s10689-016-9955-8
Sy, S. M., Huen, M. S., and Chen, J. (2009). PALB2 Is an Integral Component of the BRCA Complex Required for Homologous Recombination Repair. Proc. Natl. Acad. Sci. U S A. 106 (17), 7155–7160. doi:10.1073/pnas.0811159106
Taglialatela, A., Alvarez, S., Leuzzi, G., Sannino, V., Ranjha, L., Huang, J. W., et al. (2017). Restoration of Replication Fork Stability in BRCA1- and BRCA2-Deficient Cells by Inactivation of SNF2-Family Fork Remodelers. Mol. Cel 68 (2), 414–e8. doi:10.1016/j.molcel.2017.09.036
Tarsounas, M., and Sung, P. (2020). The Antitumorigenic Roles of BRCA1-BARD1 in DNA Repair and Replication. Nat. Rev. Mol. Cel Biol 21, 284–299. doi:10.1038/s41580-020-0218-z
Ter Brugge, P., Kristel, P., van der Burg, E., Boon, U., de Maaker, M., Lips, E., et al. (2016). Mechanisms of Therapy Resistance in Patient-Derived Xenograft Models of BRCA1-Deficient Breast Cancer. JNCI J. Natl. Cancer Inst. 108, djw148. doi:10.1093/jnci/djw148
Toland, A. E., Forman, A., Forman, A., Couch, F. J., Culver, J. O., Eccles, D. M., et al. (2018). Clinical Testing of BRCA1 and BRCA2: a Worldwide Snapshot of Technological Practices. Npj Genomic Med. 3. doi:10.1038/s41525-018-0046-7
Tudini, E., Moghadasi, S., Parsons, M. T., van der Kolk, L., van den Ouweland, A. M. W., Niederacher, D., et al. (2018). Substantial Evidence for the Clinical Significance of Missense Variant BRCA1 c.5309G>T p.(Gly1770Val). Breast Cancer Res. Treat. 172, 497–503. doi:10.1007/s10549-018-4903-y
Tung, N., Arun, B., Hacker, M. R., Hofstatter, E., Toppmeyer, D. L., Isakoff, S. J., et al. (2020a). TBCRC 031: Randomized Phase II Study of Neoadjuvant Cisplatin versus Doxorubicin-Cyclophosphamide in Germline BRCA Carriers with HER2-Negative Breast Cancer (The INFORM Trial). Jco 38, 1539–1548. doi:10.1200/JCO.19.03292
Tung, N. M., Boughey, J. C., Pierce, L. J., Robson, M. E., Bedrosian, I., Dietz, J. R., et al. (2020b). Management of Hereditary Breast Cancer: American Society of Clinical Oncology, American Society for Radiation Oncology, and Society of Surgical Oncology Guideline. Jco 38, 2080–2106. doi:10.1200/JCO.20.00299
Tung, N. M., Robson, M. E., Ventz, S., Santa-Maria, C. A., Nanda, R., Marcom, P. K., et al. (2020c). TBCRC 048: Phase II Study of Olaparib for Metastatic Breast Cancer and Mutations in Homologous Recombination-Related Genes. Jco 38, 4274–4282. doi:10.1200/JCO.20.02151
Turner, N. C., Balmaña, J., Poncet, C., Goulioti, T., Tryfonidis, K., Honkoop, A. H., et al. (2021). Niraparib for Advanced Breast Cancer with Germline BRCA1 and BRCA2 Mutations: the EORTC 1307-BCG/BIG5-13/TESARO PR-30-50-10-C BRAVO Study. Clin. Cancer Res. 27, 5482–5491. doi:10.1158/1078-0432.CCR-21-0310
Tutt, A. N. J., Garber, J. E., Kaufman, B., Viale, G., Fumagalli, D., Rastogi, P., et al. (2021). Adjuvant Olaparib for Patients with BRCA1- or BRCA2-Mutated Breast Cancer. N. Engl. J. Med. 384, 2394–2405. doi:10.1056/NEJMoa2105215
Tutt, A., Tovey, H., Cheang, M. C. U., Kernaghan, S., Kilburn, L., Gazinska, P., et al. (2018). Carboplatin in BRCA1/2-Mutated and Triple-Negative Breast Cancer BRCAness Subgroups: the TNT Trial. Nat. Med. 24, 628–637. doi:10.1038/s41591-018-0009-7
Vohhodina, J., Toomire, K. J., Petit, S. A., Micevic, G., Kumari, G., Botchkarev, V. V., et al. (2020). RAP80 and BRCA1 PARsylation Protect Chromosome Integrity by Preventing Retention of BRCA1-B/C Complexes in DNA Repair Foci. Proc. Natl. Acad. Sci. USA 117, 2084–2091. doi:10.1073/pnas.1908003117
Walker, L. C., Whiley, P. J., Houdayer, C., Hansen, T. V. O., Vega, A., Santamarina, M., et al. (2013). Evaluation of a 5-tier Scheme Proposed for Classification of Sequence Variants Using Bioinformatic and Splicing Assay Data: Inter-reviewer Variability and Promotion of Minimum Reporting Guidelines. HUMAN MUTATION 34, 1424–1431. doi:10.1002/humu.22388
Walsh, T., Lee, M. K., Casadei, S., Thornton, A. M., Stray, S. M., Pennil, C., et al. (2010). Detection of Inherited Mutations for Breast and Ovarian Cancer Using Genomic Capture and Massively Parallel Sequencing. Proc. Natl. Acad. Sci. USA 107, 12629–12633. doi:10.1073/pnas.1007983107
Wang, B. (2012). BRCA1 Tumor Suppressor Network: Focusing on its Tail. Cell Biosci 2, 6. doi:10.1186/2045-3701-2-6
Weigelt, B., Comino-Méndez, I., de Bruijn, I., Tian, L., Meisel, J. L., García-Murillas, I., et al. (2017). Diverse BRCA1 and BRCA2 Reversion Mutations in Circulating Cell-free DNA of Therapy-Resistant Breast or Ovarian Cancer. Clin. Cancer Res. 23 (21), 6708–6720. doi:10.1158/1078-0432.ccr-17-0544
Xu, J., Keenan, T. E., Overmoyer, B., Tung, N. M., Gelman, R. S., Habin, K., et al. (2021). Phase II Trial of Veliparib and Temozolomide in Metastatic Breast Cancer Patients with and without BRCA1/2 Mutations. Breast Cancer Res. Treat. 189, 641–651. doi:10.1007/s10549-021-06292-7
Yazinski, S. A., Comaills, V., Buisson, R., Genois, M.-M., Nguyen, H. D., Ho, C. K., et al. (2017). ATR Inhibition Disrupts Rewired Homologous Recombination and fork protection Pathways in PARP Inhibitor-Resistant BRCA-Deficient Cancer Cells. Genes Dev. 31 (3), 318–332. doi:10.1101/gad.290957.116
Yu, X., Chini, C. C. S., He, M., Mer, G., and Chen, J. (2003). The BRCT Domain Is a Phospho-Protein Binding Domain. Science 302 (5645), 639–642. doi:10.1126/science.1088753
Zhang, F., Fan, Q., Ren, K., and Andreassen, P. R. (2009). PALB2 Functionally Connects the Breast Cancer Susceptibility Proteins BRCA1 and BRCA2. Mol. Cancer Res. 7, 1110–1118. doi:10.1158/1541-7786.mcr-09-0123
Zhu, Q., Hoong, N., Aslanian, A., Hara, T., Benner, C., Heinz, S., et al. (2018). Heterochromatin-Encoded Satellite RNAs Induce Breast Cancer. Mol. Cel 70, 842–853. doi:10.1016/j.molcel.2018.04.023
Keywords: BRCA1, gene mutation, breast cancer, detection, personalized and precision medicine (PPM)
Citation: Fu X, Tan W, Song Q, Pei H and Li J (2022) BRCA1 and Breast Cancer: Molecular Mechanisms and Therapeutic Strategies. Front. Cell Dev. Biol. 10:813457. doi: 10.3389/fcell.2022.813457
Received: 11 November 2021; Accepted: 18 January 2022;
Published: 01 March 2022.
Edited by:
Teng Ma, Capital Medical University, ChinaReviewed by:
Bing Xia, Rutgers Cancer Institute of New Jersey, United StatesJiadong Wang, Peking University Health Science Centre, China
Copyright © 2022 Fu, Tan, Song, Pei and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huadong Pei, aHVhZG9uZy5wZWlAZ2VvcmdldG93bi5lZHU=; Juanjuan Li, c25vd3kxMTUwMjE5QHNpbmEuY29t
 Xiaoyu Fu
Xiaoyu Fu Wei Tan3
Wei Tan3 Huadong Pei
Huadong Pei Juanjuan Li
Juanjuan Li