- 1Department of Ecology and Evolutionary Biology, University of Colorado, Boulder, CO, United States
- 2Department of Zoology, Comenius University in Bratislava, Bratislava, Slovakia
Vertebrates have distinct tissues which are not present in invertebrate chordates nor other metazoans. The rise of these tissues also coincided with at least one round of whole-genome duplication as well as a suite of lineage-specific segmental duplications. Understanding whether novel genes lead to the origin and diversification of novel cell types, therefore, is of great importance in vertebrate evolution. Here we were particularly interested in the evolution of the vertebrate musculoskeletal system, the muscles and connective tissues that support a diversity of body plans. A major component of the musculoskeletal extracellular matrix (ECM) is fibrillar collagens, a gene family which has been greatly expanded upon in vertebrates. We thus asked whether the repertoire of fibrillar collagens in vertebrates reflects differences in the musculoskeletal system. To test this, we explored the diversity of fibrillar collagens in lamprey, a jawless vertebrate which diverged from jawed vertebrates (gnathostomes) more than five hundred million years ago and has undergone its own gene duplications. Some of the principal components of vertebrate hyaline cartilage are the fibrillar collagens type II and XI, but their presence in cartilage development across all vertebrate taxa has been disputed. We particularly emphasized the characterization of genes in the lamprey hyaline cartilage, testing if its collagen repertoire was similar to that in gnathostomes. Overall, we discovered thirteen fibrillar collagens from all known gene subfamilies in lamprey and were able to identify several lineage-specific duplications. We found that, while the collagen loci have undergone rearrangement, the Clade A genes have remained linked with the hox clusters, a phenomenon also seen in gnathostomes. While the lamprey muscular tissue was largely similar to that seen in gnathostomes, we saw considerable differences in the larval lamprey skeletal tissue, with distinct collagen combinations pertaining to different cartilage types. Our gene expression analyses were unable to identify type II collagen in the sea lamprey hyaline cartilage nor any other fibrillar collagen during chondrogenesis at the stages observed, meaning that sea lamprey likely no longer require these genes during early cartilage development. Our findings suggest that fibrillar collagens were multifunctional across the musculoskeletal system in the last common ancestor of vertebrates and have been largely conserved, but these genes alone cannot explain the origin of novel cell types.
Introduction
The musculoskeletal system is a complex network of tissues which support the body and allow a broad range of motions. Composed of muscles and various connective tissues like skeleton, tendons, and ligaments, it has diversified across metazoans, permitting new morphologies and body plans. The vertebrate lineage has expanded considerably upon this musculoskeletal system, incorporating cytoskeletal and extracellular matrix (ECM) proteins which have diversified these tissues and their mechanical properties. These populations of musculoskeletal tissue have distinct functions that influence the vertebrate trunk, viscera, and head in different ways (Nathan et al., 2008; Harel et al., 2009; Sambasivan et al., 2009; Witten et al., 2010). Understanding how these tissues have diversified from those seen in invertebrate chordates has significant value in vertebrate evolution.
Vertebrates are thought to have undergone at least one whole-genome duplication, and gene duplication is an important mechanism through which they may have elaborated this musculoskeletal morphology (Van de Peer et al., 2009; Ohno, 2013; Martik et al., 2019; Simakov et al., 2020; Square et al., 2020). Although the metazoan muscular system’s origins are thought to be shared and innovated upon an ancestral contractile machinery (Seipel and Schmid, 2005; Steinmetz et al., 2012), vertebrates have duplicated actin and myosin heavy chain genes which are differentially expressed across smooth and striated muscle populations (Vandekerckhove and Weber, 1984; Mugue and Ozernyuk, 2006; Warkman et al., 2012; Witjes et al., 2019), forming the vertebrate-unique cardiac muscle and multi-layered skeletal muscles (Zoghbi et al., 2008; Shaffer and Gillis, 2010; Luther and Squire, 2014). Likewise, cartilage may be a connective tissue shared by bilaterians (Tarazona et al., 2016), but vertebrates have formed unique ECM-rich hyaline cartilages characterized by lectican proteins, all of which are likely vertebrate-specific duplications (Root et al., 2021a). Although tendons connect muscle and skeleton in both invertebrates and vertebrates, they are structurally different in both groups and have different developmental origins (Schweitzer et al., 2010), vertebrate tendons are often heterogeneous in morphology, but share their expression of the vertebrate-specific small interstitial leucine-rich repeat proteoglycans (SLRP’s) like fibromodulin, biglycan, decorin, and lumican (Yoon and Halper, 2005). All of these genes have been found across vertebrates but not invertebrate chordates like amphioxus, supporting the idea that these tendon-associated gene families arose from vertebrate-specific duplications (Matsushima et al., 2000). While aspects of vertebrate evolution are thought to have been predicated on gene duplication, it is difficult to find a single gene family that vertebrates have duplicated which specifically encompasses the musculoskeletal system and its various components.
It is because of this disparity that we were interested in investigating fibrillar collagens. They are the most abundant proteins in vertebrate bodies and are found all across the vertebrate musculoskeletal system (Bailey, 1968). These collagens confer biomechanical properties like load bearing, tensile strength, and torsional stiffness, and these proteins can even act as ligands or receptors for various signaling pathways (Di Lullo et al., 2002; Boraschi-Diaz et al., 2017; Huang et al., 2019). While fibrillar collagens have been found across all metazoans, this gene family has undergone multiple rounds of duplication in vertebrates (Exposito et al., 2010). Modern jawed vertebrates have eleven or twelve fibrillar collagen genes normally, and these are divided into three clades: Clade A (col1a1, col1a2 col2a1, col3a1 and col5a2), Clade B (col5a1, col5a3, col11a1, and col11a2), and Clade C (col24a1 and col27a1). These collagens are dispersed throughout the musculoskeletal system, and are major components of connective tissues. In gnathostomes, col2a1 is the predominant collagen in cartilage and col1a1 in bone, but the other genes have important if understudied roles. Furthermore, col2a1 and col1a1 have more pleiotropic expression than cartilage and bone exclusively. Previous work has identified col1a1 in developing tendons, dermis, and perichondrium (Fisher et al., 2003; Grimaldi et al., 2004; Leéjard et al., 2007; Enault et al., 2015; Gistelinck et al., 2016) while col2a1 has been reported in the perichondrium, notochord, and even in teleost osteocytes, although this bone expression is considerably lower than in cartilage (Eames et al., 2012; Enault et al., 2015). col1a2 is an alternate alpha chain of type I collagen that shares similar expression to its counterpart in bone, perichondrium, dermis, and tendons, and is also found in developing cartilage and skeletal muscle (Le Guellec et al., 2003; Shukunami et al., 2008; Gaspera et al., 2009; Eames et al., 2012; Gelse et al., 2012; Chen and Galloway, 2014; Enault et al., 2015; Sharma et al., 2019; Fan et al., 2020). col3a1 is found primarily in developing dermis and tendons, but is also expressed in skeletal muscle, bone, and cartilage, albeit in much lower quantities (Leéjard et al., 2007; Duance et al., 1977; Kuo et al., 2008; D’hondt et al., 2018; Owens et al., 2016; Session et al., 2016; Guerquin et al., 2013; Liu et al., 1997; Wang et al., 2020). Lastly, col5a2 is a minor component in almost all the aforementioned tissues, with lower levels of expression in cartilage, dermis, bone, and muscle (Rauch et al., 2003; Thisse, 2005; Session et al., 2016). Clade B genes share similar patterns of expression to their Clade A counterparts. col11a1 is a complementary collagen to Type II in gnathostome cartilage (Li et al., 1995; Yoshioka et al., 1995; Baas et al., 2009), but is also localized in tendons, bone, perichondrium, notochord, and dermis (Yoshioka et al., 1995; Imamura et al., 2000; Thisse, 2005; Baas et al., 2009; Fang et al., 2010; Mukhi et al., 2010; Wenstrup et al., 2011; Suzuki et al., 2012; Sun et al., 2020). col5a1 is another major Clade B collagen, with expression in the aforementioned tissues as well as developing muscle (Lincoln et al., 2006; Roulet et al., 2007; Bandyopadhyay et al., 2008; Gaspera et al., 2009; Fang et al., 2010; Hoffman et al., 2010; Wenstrup et al., 2011; Session et al., 2016; Sharma et al., 2019). col11a2 and col5a3 are poorly documented genes, as one or the other is often absent from gnathostome genomes. Their patterns of expression, however, closely resemble other Clade B genes, with col11a2 identified in developing cartilage, bone, perichondrium, notochord, and tendons (Tsumaki and Kimura, 1995; Iwai et al., 2008; Fang et al., 2010; Eames et al., 2012; Duran et al., 2015; Cao et al., 2018; Lawrence et al., 2018; Sun et al., 2020) while col5a3 is expressed in the aforementioned tissues as well as the dermis and skeletal muscle (Imamura et al., 2000; Yamaguchi et al., 2005; Fang et al., 2010; Hoffman et al., 2010; Kessels et al., 2014). The Clade C genes are a more recent discovery in this gene family (Koch et al., 2003; Pace et al., 2003), and their expression is poorly understood. What few studies exist, however, have found col24a1 musculoskeletal expression in the developing bone (Koch et al., 2003; Wang et al., 2012; Duran et al., 2015) whereas col27a1 was identified in cartilage, notochord, and some populations of muscle (Thisse et al., 2001; Pace et al., 2003; Jenkins et al., 2005; Christiansen et al., 2009; Mangos et al., 2010; Lien et al., 2013; Duran et al., 2015). While most vertebrates have twelve fibrillar collagen genes with diversified expression, only four have been found in invertebrate chordates, with each gene having largely overlapping expression in the oral ciri, notochord, muscles, and the nerve cord (Wada et al., 2006; Zhang and Cohn, 2006; Jandzik et al., 2015). If gene duplication played anportant role in the early evolution of vertebrate morphology, then structural proteins like fibrillar collagens may have co-diversified in the musculoskeletal system during this process.
The evolution of vertebrates from invertebrate chordates coincided with genomic expansion, but we know little as to how these genes acquired new roles or sub-functionalized previous ones. It is commonly accepted that at least one whole-genome duplication happened at the stem of the vertebrate lineage (Panopoulou and Poustka, 2005; Smith and Keinath, 2015; Simakov et al., 2020), but subsequent duplications may have occurred after extant vertebrate taxa diverged, meaning that some fibrillar collagens may be unique homologs with distinct expression domains. It has been recently suggested that jawed vertebrates (gnathostomes) and extant jawless vertebrates like lamprey and hagfish (cyclostomes) diverged before the second round of whole genome duplication (Simakov et al., 2020), meaning that many fibrillar collagens would be specific to each lineage. Gnathostomes and cyclostomes are thought to have diverged more than 500 million years ago (Kuraku and Kuratani, 2006), providing us an excellent opportunity to explore lineage-specific duplication and functionalization of collagens at one of the earliest nodes in vertebrate evolution. Both lamprey and hagfish lack epaxial-hypaxial divisions in somitic skeletal muscle like that seen in gnathostomes (Peters and Mackay, 1961; Kusakabe and Kuratani, 2007), and they additionally lack appendicular musculature from the lateral plate mesoderm (Nakao, 1977). Furthermore, cyclostomes have independently duplicated muscle actins and myosin chains which are differentially expressed in skeletal muscle, contrasting the somitic and head musculature from each other (Kusakabe et al., 2004). The head mesoderm of lampreys undergoes a split as its dorsal and ventral parts migrate around the eye as supraoptic and infraoptic streams; it has been previously shown that these muscles differentially express the myogenic markers pax3-7 and lbx-A (Kusakabe and Kuratani, 2005; Kusakabe et al., 2011). The tendons of the myosepta also differ greatly between both groups, as hagfish are thought to lack myoseptal tendons entirely while lamprey have only some of this structure, specifically the dorsal and ventral myorhabdoid tendons (Gemballa and Vogel, 2002). The gnathostome and cyclostome skeleton differ as well in that not only does the latter lacks bone and traditional hypertrophic cartilage, but it is also largely heterogeneous throughout the body and has acquired novel proteins in its extracellular matrix such as lamprin, myxinin, and pharymprin (Wright et al., 1983; Wright et al., 1984; Robson et al., 1993; McBurney et al., 1996; Yokoyama et al., 2019). Cyclostome cartilages are also distinct in that some species may no longer require type II collagen during early development. In the arctic lamprey Lethenteron camtschaticum, col2a1b transcripts have been identified in pharyngeal cartilage during early chondrogenesis (Ohtani et al., 2008). Conversely, type II collagen was reported in P. marinus in one study of the larval and adult hyaline cartilages (Zhang et al., 2006), but additional histological analyses were unable to corroborate any col2a1a or col2a1b expression during early development (McCauley, 2008; Cattell et al., 2011). Likewise, studies in adult hagfish have not found type II collagen in “hard” cartilage, thought to be homologous to gnathostome hyaline cartilage (Zhang and Cohn, 2006; Ota and Kuratani, 2010). These suggest that the sea lamprey have either deviated considerably from the ancestral vertebrate skeleton or have retained features of its diverse cartilages that were later lost in gnathostomes, a phenomenon previously noted with mineralized tissues in stem vertebrates (Eames et al., 2020). Overall, there are notable differences between the vertebrate and invertebrate chordate musculoskeletal system and even between that of cyclostomes and gnathostomes, and lineage-specific gene duplications may have facilitated an important role in these processes by permitting.
In this study, we used genomic and transcriptomic data from the sea lamprey Petromyzon marinus and the inshore hagfish Eptatretus burgeri to gain insight into the evolutionary history of fibrillar collagen genes and their activity in the musculoskeletal system, particularly in the branchial hyaline cartilage. We identified several lineage-specific duplications and deletions in cyclostomes, with linkage to other important gene families. We find mosaic expression of these genes across the larval lamprey skeleton, corresponding to previously documented subpopulations of cartilage, but we did not identify type II collagen in the developing pharyngeal chondrocytes. We also find a distribution of collagens across the early muscular system, dispersed throughout the dermomyotome as well as differential expression between the anterior and posterior myomeres.
Results
Identification of Fibrillar Collagens in Cyclostomes
We first searched for fibrillar collagens through transcriptomic reads of P. marinus and E. burgeri and verified their position on distinct genomic scaffolds. We identified 13 fibrillar collagens in lamprey: seven Clade A’s, five Clade B’s, and one Clade C. For hagfish, we identified 13 genes: six Clade A’s, five Clade B’s, and two Clade C’s. Before analyzing orthology, all fibrillar collagens were named according to their clade and the order in which they were identified in our transcriptome (colA1, colA2, colB1, etc).
All cyclostome fibrillar collagens display a similar protein architecture to their respective families. Most Clade A genes have Von Willebrand Factor Type C (VWC) domain at their N-terminus, almost all Clade B and C genes have a Thrombospondin-1 domain (TSPN) at their N-terminus, and all genes have the archetypal fibrillar collagen COLF C-terminal domain. The main exceptions to this domain architecture were the hagfish colC2 gene, which did not have the TSPN domain, and the lamprey colA4 and colA7 genes, which do not possess the VWC module. These lamprey features are shared with the gnathostome col1a2 gene, which has also been reported to not have the VWC domain in humans (Aouacheria et al., 2004). Upon reviewing the domain architecture of multiple col1a2 orthologs, our findings support this observation across gnathostomes. The lamprey colA4, hagfish colA3, and hagfish colA4 genes also lack the minor helix domain in the N-terminus which has been conserved across all other Clade A and Clade B genes but is notably absent from vertebrate Clade C genes. The COLF domain has been highly conserved across gnathostome and contains a recognition sequence with eight interspersed cysteine residues which are thought to mediate collagen trimer formation (Lees et al., 1997), and all lamprey and hagfish genes have maintained six of the eight residues (positions 1,4,5,6,7,8). Residue two is modified in the lamprey colA2, colB4, and colB5, and is changed in the hagfish colA2 and colA4 genes. This particular cysteine residue is known to be modified across gnathostome genes, specifically col1a2 and various Clade B collagens across different species (Lees et al., 1997; Exposito et al., 2010). Residue three is only modified in mammalian col5a2 orthologs, but this change can be found in the lamprey colA1, colA2, and colA4 genes, and is also distinct in the hagfish colA4 and colB2 genes. Overall, we find significant differences in the structure of cyclostome fibrillar collagen genes compared to other vertebrates on the amino acid level. How these structural differences influence their function, however, is beyond the scope of this study, but will be of future interest.
Phylogenetic and Syntenic Analysis of Cyclostome Fibrillar Collagens
We next performed phylogenetic analyses on the fibrillar collagens identified, with separate analyses for each clade. We reasoned that, if orthologs could be determined between lamprey and gnathostome genes, we could posit a better/deeper evolutionary scenario as to their acquisition and sub-functionalization over vertebrate evolution. Because fibrillar collagens can be found across bilaterians, we additionally tested the relationship of these genes between gnathostomes, cyclostomes, and invertebrates, using amphioxus (B. floridae), tunicate (C. intestinalis), purple urchin (S. purpuratus) and leech (H. robusta) as representatives for invertebrate chordates, non-chordate deuterostomes, and protostomes respectively. We then inspected the genomic locations of the genes to identify conserved synteny blocks across vertebrates in further support of our phylogenies.
We were interested in identifying potential pro-ortholog families between invertebrate fibrillar collagens and their vertebrate counterparts, so we first made phylogenetic trees to test the relationship between bilaterian Clade A (Supplementary Figure S1) and Clade B (Supplementary Figure S2) genes. We did not find strong evidence for any pro-ortholog families between invertebrate and vertebrate genes, and these invertebrate sequences weakened likelihood scores across earlier-diverging nodes. This is likely due to long branch attraction occurring with the protostome and invertebrate deuterostome sequences. We thus directed our phylogenetic analyses primarily to vertebrates and the relationships between gnathostome and cyclostome genes.
After adjusting our analyses to focus specifically on vertebrates, we were able to find support for the orthology of all hagfish Clade A genes as well as six of seven lamprey Clade A genes. Both lamprey and hagfish genes correspond to three gnathostome genes: col1a2, col2a1, and col3a1 (Figure 1). Previous work has identified col2a1 homologs in both lamprey and hagfish (Zhang and Cohn, 2006; Zhang et al., 2006; McCauley, 2008; Ohtani et al., 2008; Ota and Kuratani, 2010; Cattell et al., 2011), and our results support these findings as well. Likewise, previous work found an additional fibrillar collagen gene in lamprey but was unable to conclusively identify its orthology (Ohtani et al., 2008), but our phylogenies support its placement as a col1a2 ortholog. We will henceforth refer to this aforementioned gene, colA1 in our study, as col1a2a. Our results likewise reveal a topology for each gene that places each lamprey and hagfish gene closer to each other than to their paralogs. These findings suggest that the number of Clade A collagens in these lineages is due to cyclostome-specific duplications. We were unable to predict the orthology of the lamprey colA7 gene, as its sequence has deviated considerably from other Clade A genes. Gene topologies like this are common in lamprey due in part to a variety of factors such as GC bias in lamprey protein-coding genes (Smith et al., 2013), long branch attraction in divergent genes (Kuraku, 2008), or these genes could even reflect a diversity of homologs no longer present in extant gnathostomes (Kuraku, 2013). Because we were unable to identify a similar gene in hagfish, colA7 may either be a lamprey-specific duplicate or a highly divergent ortholog or paralog to an extant gnathostome gene. Our phylogenetic analyses present a topology of Clade A genes in vertebrates that suggests the loss of these genes in cyclostomes rather than their later acquisition in gnathostomes. Overall, our findings show that the last common ancestor of cyclostomes likely had five Clade A fibrillar collagens, each corresponding to the gnathostome col1a1, col1a2, col2a1, col3a1, and col5a2 genes. The repertoire of genes found in lamprey, therefore, is the result of cyclostome-specific gene duplications and at least one gene loss, as it remains possible that colA7 is a highly divergent ortholog of either col1a1 or col5a2. We therefore designated the lamprey colA1, colA2, colA3, colA4, colA5, and colA6 genes as col1a2α, col1a2β, col2a1α, col3a1β, col2a1β, and col3a1α, respectively. We likewise designated the hagfish Clade A genes with their respective lamprey ortholog.
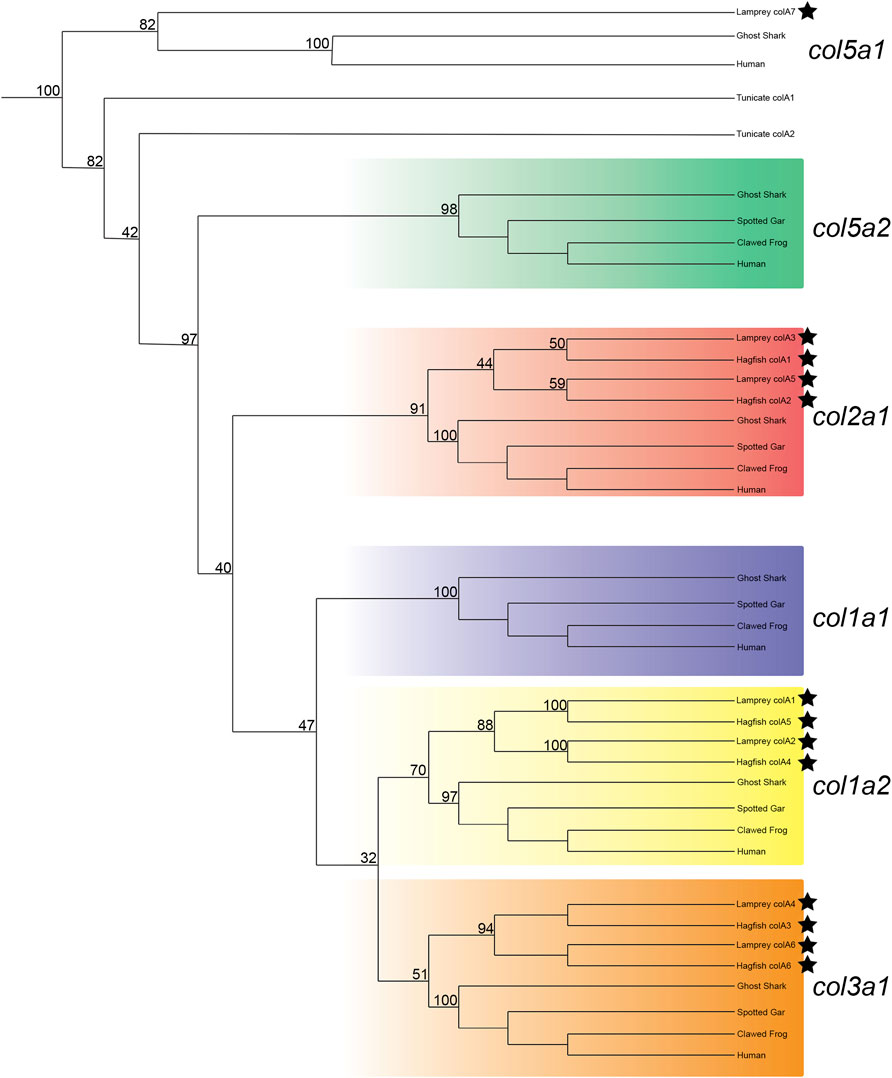
FIGURE 1. Recreated phylogenetic tree built from vertebrate and tunicate Clade A fibrillar collagen sequences. Maximum likelihood analysis scores are shown at the respective node. The human and ghost shark col5a1 genes were chosen as the most relevant outgroup. Cyclostome genes are labelled with a black star. Original tree can be found in Supplementary Figure S3. Accession numbers for all sequences can be found in Supplementary Table S3.
To support our phylogenetic analyses for Clade A genes, we analyzed the synteny of the lamprey collagen homologs. We were able to identify hox clusters syntenic to each of the six major Clade A genes, a phenomenon which has been previously observed in gnathostomes (Bailey et al., 1997) (Supplementary Figure S4). Despite this, we found that the adjacent genomic locations of the lamprey collagens did not share a higher number of linked genes with their gnathostome ortholog (Figure 2A). Of the six lamprey Clade A homologs, only colA6 shared more linked genes with its gnathostome ortholog (col3a1) than of the other gnathostome genes. When furthering analyzing the lamprey loci, we observed a large number of linked genes with other Clade A-containing lamprey scaffolds. These linkage clusters, however, did not correspond to those genes found near their paralog nor any particular collagen homolog. These data together suggest that the immediate genomic location of the Clade A collagens has undergone considerable rearrangement in lamprey and that the microsyntenic data for these genes alone cannot support the relationship between each lamprey and gnathostome fibrillar collagen.
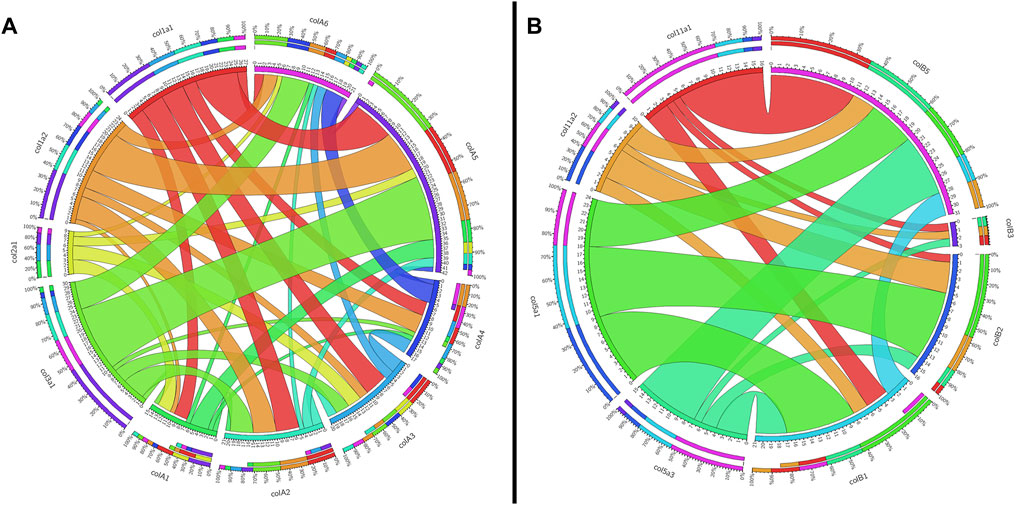
FIGURE 2. Circos plots showing the synteny between gnathostome and lamprey Clade (A) and Clade (B) fibrillar collagens. Inner circles are colored according to that gene, and they indicate the total number of linked genes and how many are syntenic with other fibrillar collagens. Outer circles show percentages of linked genes. Synteny between gnathostome genes is not shown. (A) Shared linkage between gnathostome and lamprey Clade A genes. All genes of interest in this plot as well as the grid used to generate it can be found in Supplementary Table S4. Because col3a1 and col5a2 are linked together, the latter is not shown on the plot. (B) Shared linkage between gnathostome and lamprey genes. All genes of interest in this plot as well as the grid used to generate it can be found in Supplementary Table S5.
We next deduced phylogenetic trees for the Clade B genes, but were unable to conclusively predict the orthology of all genes based on phylogenetics alone. We did find strong likelihood scores between complete lamprey and hagfish sequences, however, confirming that the diversity of genes found in cyclostomes was likely present in the last common ancestor. Our phylogenetic analyses provide moderate support for colB3 as a col5a3 ortholog, and we find some evidence for orthology of colB4 and colB5 within the col11a1/col5a3 gene family (Figure 3). Their topology suggests that they are either related to a related gene lost in gnathostomes or are highly divergent orthologs of col11a1. Overall, the lamprey colB3 gene is the only lamprey Clade B homolog which grouped with a gnathostome gene with moderate support.
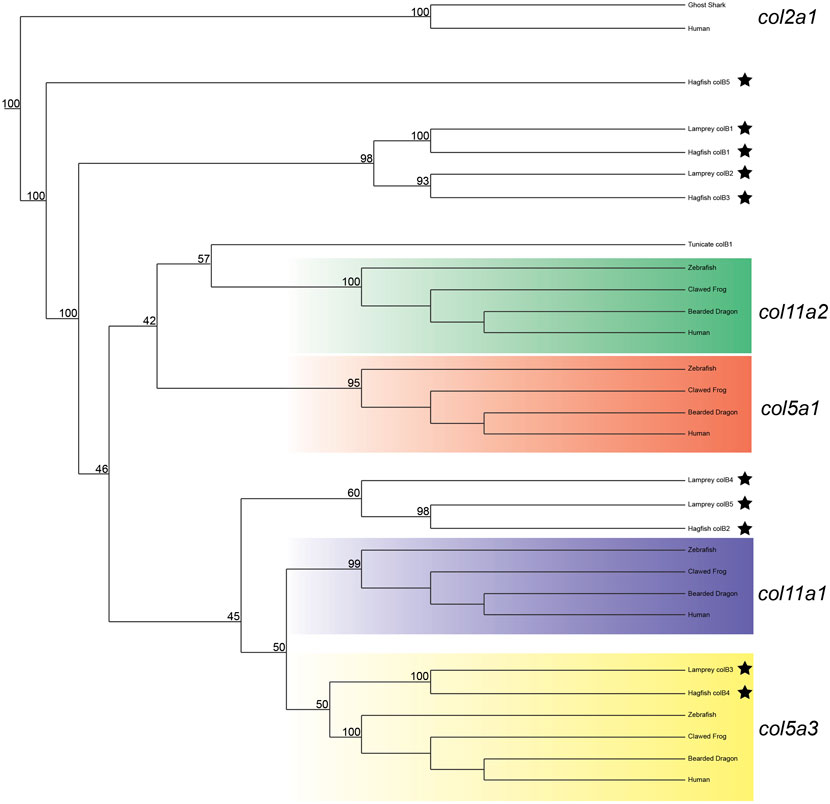
FIGURE 3. Recreated phylogenetic tree built from vertebrate and tunicate Clade B fibrillar collagen sequences. Maximum likelihood analysis scores are shown at the respective node. The human and ghost shark col2a1 genes were chosen as the most relevant outgroup. Cyclostome genes are labelled with a black star. Original tree can be found in Supplementary Figure S5. Accession numbers for all sequences can be found in Supplementary Table S6.
Given the degree of rearrangement seen in Clade A fibrillar collagens, we were interested to see if the genomic landscape of the Clade B lamprey genes would be similar. On the contrary, we saw very little evidence of rearrangement between the homologs (Figure 2B). We additionally observed lamprey genes with a higher degree of linkage with particular gnathostome collagens. Both colB1 and colB2 showed considerably more linkage with col5a1 than other genes. We found that the hagfish colB3, most likely the direct ortholog of lamprey ColB2, also showed a similar degree of linkage with col5a1. These together, paired with the paucity of rearrangement seen in these genes, indicate that lamprey colB1 and colB2 are likely col5a1 orthologs, and they have been designated col5a1α and col5a1β respectively. While the synteny of colB3 and colB4 provide little insight into their relationship with gnathostome genes, the genomic arrangement of colB5 suggests a relationship with col11a1 and col5a3, as it contains a higher proportion of conserved genes with these homologs than with the other Clade B collagens. Additionally, lamprey colB5 is distantly linked with the colC1 gene, a similar situation seen in gnathostomes between the col11a1 and col24a1 genes (Exposito and Lethias, 2013). These together indicate that colB5 is likely a col11a1 ortholog. Combined with phylogenetic data that suggests a paralogy between colB4 and colB5, we have thus designated these genes as col11a1α and col11a1β respectively. Overall, we were able to identify orthology for all lamprey Clade B genes by using a combination of phylogenetic and syntenic analyses, designating the colB1, colB2, colB3, colB4, and colB5 genes as col5a1α, col5a1β, col5a3, col11a1α, and col11a1β respectively. We further designated the hagfish colB1, colB2, colB3, and colB4 with their respective lamprey ortholog. Because the hagfish colB5 is only a partial sequence, we avoided assigning it an ortholog.
We lastly built a phylogenetic tree to test the relationship of lamprey colC1 to the gnathostome Clade C genes. The gene placed with very high certainty within the col24a1 gene family (Figure 4). Interestingly, the second hagfish Clade C homolog placed strongly outside of both gnathostome genes. Whether the second hagfish gene represents a lineage-specific duplicate, a highly divergent col27a1 gene, or an ancient homolog that both gnathostomes and lamprey lost, however, is unclear. Due to the very high affinity of the lamprey colC1 as a col24a1 ortholog and its linkage with the putative col11a1 ortholog, however, it is likely that the last common ancestor of cyclostomes and gnathostomes had two Clade C collagens, corresponding to the col24a1 and col27a1 genes.
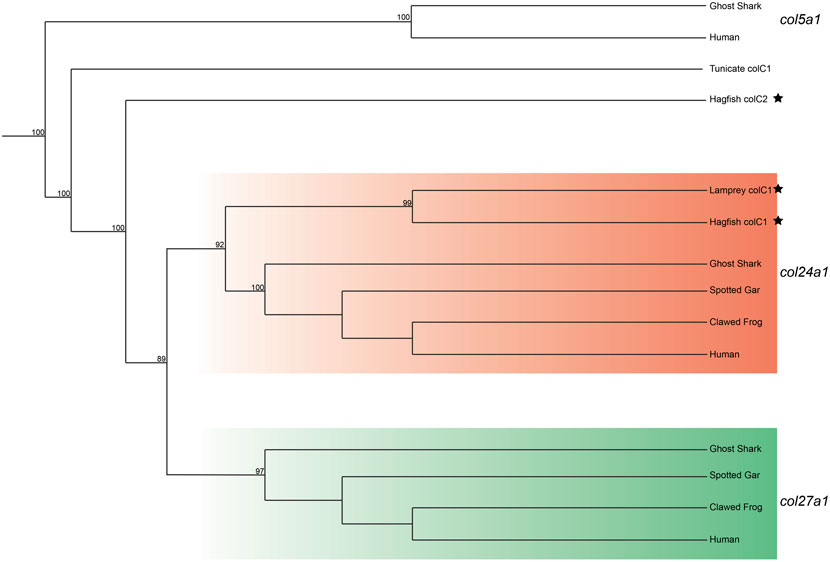
FIGURE 4. Recreated phylogenetic tree built from vertebrate and tunicate Clade C fibrillar collagen sequences. Maximum likelihood analysis scores are shown at the respective node. The human and ghost shark col5a1 genes were chosen as the most relevant outgroup. Cyclostome genes are labelled with a black star. Original tree can be found in Supplementary Figure S6. Accession numbers for all sequences can be found in Supplementary Table S7.
Expression of Fibrillar Collagens in Lamprey
We next characterized the expression of these fibrillar collagens in P. marinus embryos through developmental stages relevant during musculoskeletal histogenesis. Given the extent of lamprey-specific innovations, we opted to focus on stages which would be most relevant for comparison with gnathostome expression. We chose Tahara stages 26 through 28 (Yutaka, 1988), corresponding to mesenchymal condensations in the oropharyngeal skeleton and the differentiation of somitic components. To confirm our observations of expression in the musculoskeletal system, we compared our results to previous reports of the lamprey musculoskeletal system, such as the developing lamprey musculature (Kusakabe et al., 2004; Kusakabe and Kuratani, 2005; Kusakabe et al., 2011), hyaline cartilage (Root et al., 2021a) [Figure S7], and mucocartilage (Cerny et al., 2010; Root et al., 2021a; Root et al., 2021b).
Expression of Clade A Collagens
Previous work on the lamprey col1a2 ortholog did not analyze expression at later stages (Ohtani et al., 2008), so much of our results are new for this gene. We observed a dramatic difference in expression between lamprey col1a2 paralogs. We detected transcripts of colA1/col1a2α at stage T26 in the cranial ganglia, the ventral mesenchyme surrounding the endostyle, and in the somites, presumably the dermomyotome (Figure 5A). By stage T27, colA1/col1a2α transcripts were identified in the ectomesenchyme of the lower lip, ventral pharynx, oral hood, as well as the non-skeletogenic neural crest cells of the pharynx (Figures 5B,C,C′,C”). We also observed colA1/col1a2α expression in the mesenchyme of the developing fin fold as well as the dermomyotome, myomeres, and myosepta, likely presaging the myotendinous junctions (Figures 5B,D). By stage T28, this expression was limited to the mesenchyme of the oral hood and fin, the developing hypobranchial muscles, as well as the dermomyotome of the somites and the presumptive growth zone of the myomeres (Figures 5D,E,E’,E”). Conversely, we observed expression of colA2/col1a2β in the mesenchyme of the upper lip and lower lip at stage T27, albeit at low levels (Figures 5G,H,H”). We also observed weak expression in the developing myomeres at this stage (Figure 5G). By stage T28, expression was similar in all of the aforementioned structures as well as the fin mesenchyme.
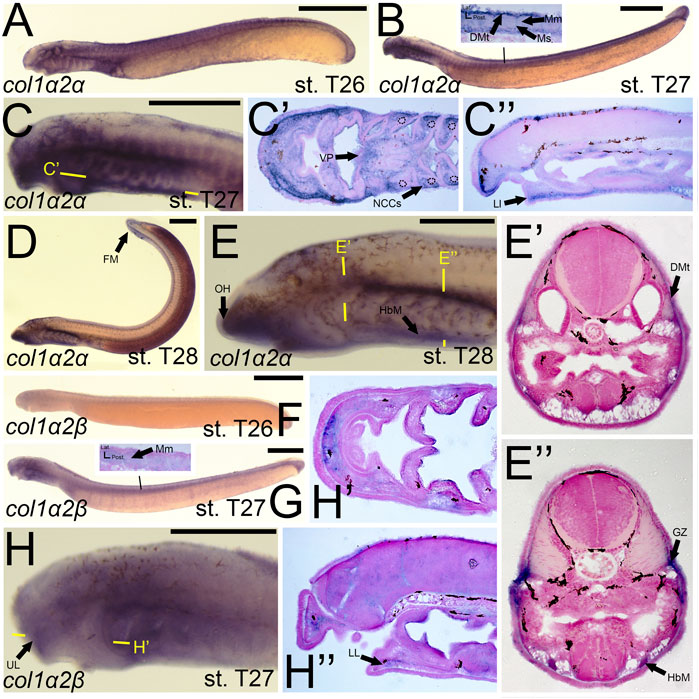
FIGURE 5. Expression of colA1/col1a2α and colA2/col1a2β in larval lamprey. All scale bars are approximately 500 μm. (A) col1a2α expression was in the cranial ganglia and throughout the head mesenchyme. (B,C) col1a2α expression was in the mesenchyme of the fin fold, lower lip, and ventral pharynx, the non-skeletogenic neural crest cells in the pharynx, the myosepta, myomeres, and the dermomyotome. Dotted circles indicate presumptive branchial arch skeleton. (D,E) col1a2α expression was in the mesenchyme of the fin fold, lower lip, and oral hood, as well as the hypobranchial musculature, the myomere growth zone, and the dermomyotome. (F) col1a2β expression was minimal at stage T26. (G,H) col1a2β expression was in the mesenchyme of the upper and lower lip, and was weak albeit present in the myomeres. Keywords: DMt: dermomyotome; FM: fin mesenchyme; GZ: growth zone; HbM: hypobranchial muscle; LL: lower lip; Mm: myomeres; Ms: myosepta; NCCs: neural crest cells; UL: upper lip; VP: ventral pharynx.
The expression of col2a1α and col2a1β has been documented previously, and our results largely support these findings. We detected transcripts of colA3/col2a1α in the facial ectoderm and the mesenchyme of the lower lip (Figure 6A). At stage T27, we observed colA3/col2a1α expression in the epidermis and myosepta, pharyngeal muscle, as well as the mesenchyme of the lower lip, oral hood, ventral pharynx, and of the fin fold (Figures 6B,D,D′,D”). We did not, however, see expression in the hyaline cartilage of the pharynx (Figure 6D”). By stage T28, colA3/col2a1α expression in the mesenchyme and epidermis was largely maintained (Figures 6C,E,E’). We likewise noticed additional expression in the hypobranchial muscles as well as the growth zone of the myomeres (Figure 6E”). Similar to its paralog, expression of colA5/col2a1β matched much of what has previously been identified. We detected transcripts of colA5/col2a1β at stage T26 in the cranial ganglia, the mesenchyme of the upper and lower lip, and the notochord (Figures 6F,G) By T27, it was additionally found in the fin fold mesenchyme (Figure 6H,H′,I). Along the trunk, we observed a transient expression of colA5/col2a1β in the developing myosepta, epidermis, and myomeres (Figure 6I). At stage T28, colA5/col2a1β was detected in the hypochord and lower lip mesenchyme (Figure 6K,K’). Similar to the other Clade A’s, we detected expression in the myomeres and hypobranchial muscles as well as the mesenchyme of the ventral pharynx and fin fold, but we also observed distinct expression in the parachordals (Figure 6K”).
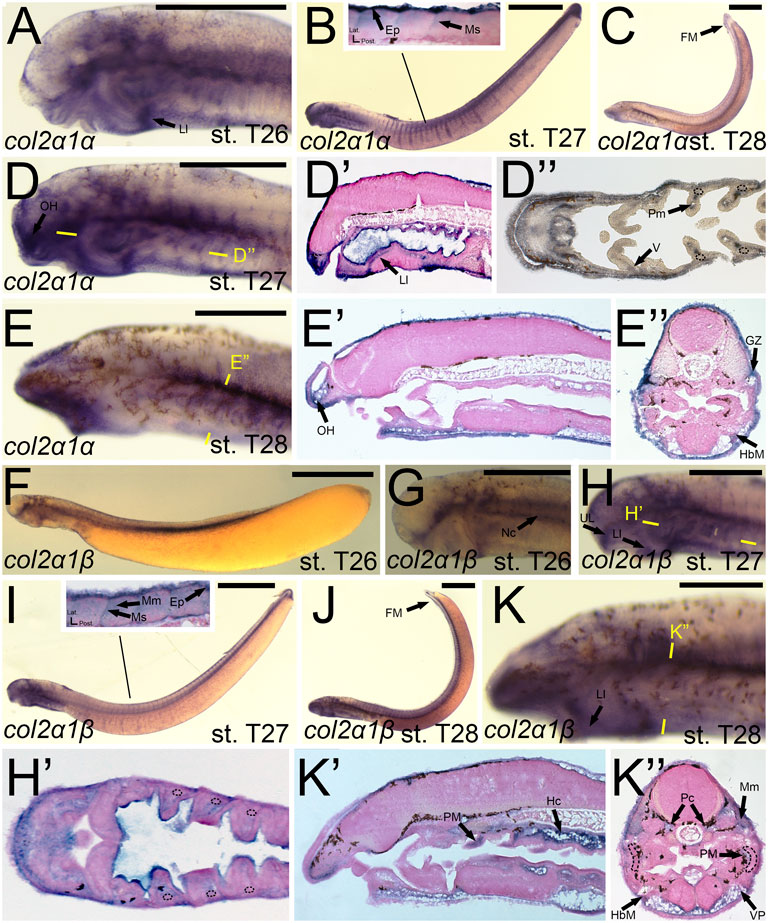
FIGURE 6. Expression of colA3/col2a1α and colA5/col2a1β in larval lamprey. All scale bars are approximately 500 μm. (A) col2a1α expression was in the facial ectoderm and the mesenchyme of the lower lip. (B,D) col2a1α expression was additionally found in the mesenchyme of the oral hood and lower lip as well as the lateral velum, pharyngeal muscle, epidermis, and myosepa. Dotted circles indicate presumptive branchial arch skeleton. (C,E) col2a1α expression was in the mesenchyme of the fin fold and oral hood, and was also in the hypobranchial musculature and growth zone. (F,G) col2a1β expression was in the notochord, cranial ganglia, and the mesenchyme of the upper and lower lip. (H,I) col2a1β expression was in the mesenchyme of the upper and lower lip, the pharyngeal muscle, the myomeres, epidermis, and the myosepta. (J,K) col2a1β expression was in the mesenchyme of the fin fold, ventral pharynx, and lower lip, the pharyngeal muscle, the myomeres, hypobranchial musculature, posterior hypochord, as well as the cartilages of the parachordals. Dotted circles indicate presumptive branchial arch skeleton. Keywords: Ep: epidermis; FM: fin mesenchyme; HbM: hypobranchial musculature; GZ: growth zone; Hc: hypochord; LL: lower lip; Mm: myomeres; Ms: myosepta; Nc: notochord; OH: oral hood; Pc: parachordals; PM: pharyngeal muscule; UL: upper lip; V: velum; VP: ventral pharynx.
We noticed a difference in expression between the col3a1 paralogs similar to that for the col1a2 genes. Expression of colA6/col3a1α at stage T26 was first seen in the developing ganglia and mesenchyme of the upper lip (Figures 7A,D). We detected expression of colA6/col3a1α at T27 in the mesenchyme of the lower lip, oral hood, and lateral velum (Figures 7B,E,E′,E”). We also observed expression of colA6/col3a1α at this stage in the non-skeletogenic neural crests of the pharynx (Figure 7E’). By stage T28, expression in the cranial mesenchyme was specific to the lower lip, oral hood, and ventral pharynx, but we additionally observed colA6/col3a1α activity in the fin fold mesenchyme starting at this stage (Figures 7C,F,F’). Expression in the trunk continued in the hypobranchial muscles as well as the dermomyotome of the somites and the myomeres (Figure 7F’). Transcripts of colA4/col3a1β, conversely, were detected in the mesenchyme of only the lower lip at stage T27, but we also find expression in the developing otic vesicle and the presumptive hypophysis (Figure 7H). No muscle expression was observed at this stage. At T28, we continued to see colA4/col3a1β activity in the supraoptic and infraoptic muscles (Figures 7I,J). We also observed new activity in the myomeres and hypobranchial muscles (Figure 7J’).
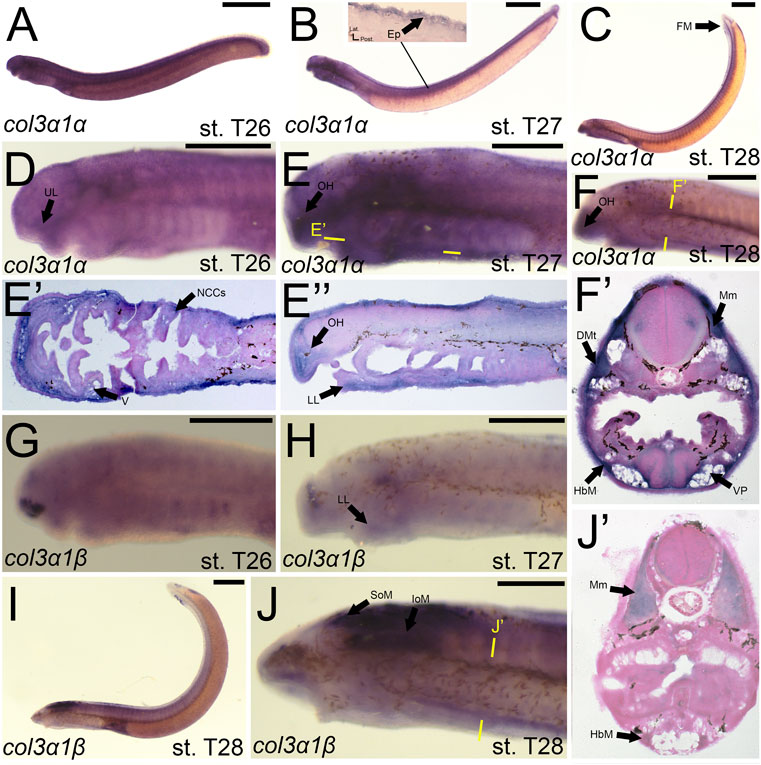
FIGURE 7. Expression of colA6/col3a1α and colA4/col3a1β in larval lamprey. All scale bars are approximately 500 μm. (A,D) col3a1α expression was in the cranial ganglia as well as the mesenchyme of the upper lip. (B,E) col3a1α expression was in the mesenchyme of the oral hood, lower lip, ventral pharynx, and velum, the non-skeletogenic neural crest cells of the pharynx, as well as the epidermis. (C,F) col3a1α expression was in the mesenchyme of the oral hood, lower lip, and ventral pharynx, the myomeres, hypobranchial musculature, and the dermomyotome. (G) col3a1β expression was in the rostral head in an unknown tissue. (H) col3a1β expression was in the forebrain and the mesenchyme of the lower lip. (I,J) col3a1β expression was in the infraoptic and supraoptic muscle populations as well as the hypobranchial musculature and myomeres. Keywords: DMt: dermomyotome Ep: epidermis; FM: fin mesenchyme; HbM: hypobranchial musculature; IoM: infraoptic muscles; LL: lower lip; Mm: myomeres; NCCs: neural crest cells; OH: oral hood; SoM: supraoptic muscles; UL: upper lip; V: velum; VP; ventral pharynx.
Although we were unable to predict the orthology of colA7, we were able to design a riboprobe to characterize its expression. We detected colA7 activity at stage T26 in the skin of the pharynx, albeit at low levels (Figure 8A). By T27, this activity expanded into the mesenchyme of the velum as well as the dermomyotome (Figure 8B,B′,D). Expression in the skin continued at this stage. colA7 expression in the velum and pharynx abated by T28, but we detected new signal in the mesenchyme of the oral hood and fin fold as well as the growth zone of the myomeres and the hypobranchial muscles (Figure 8C,C’,E).
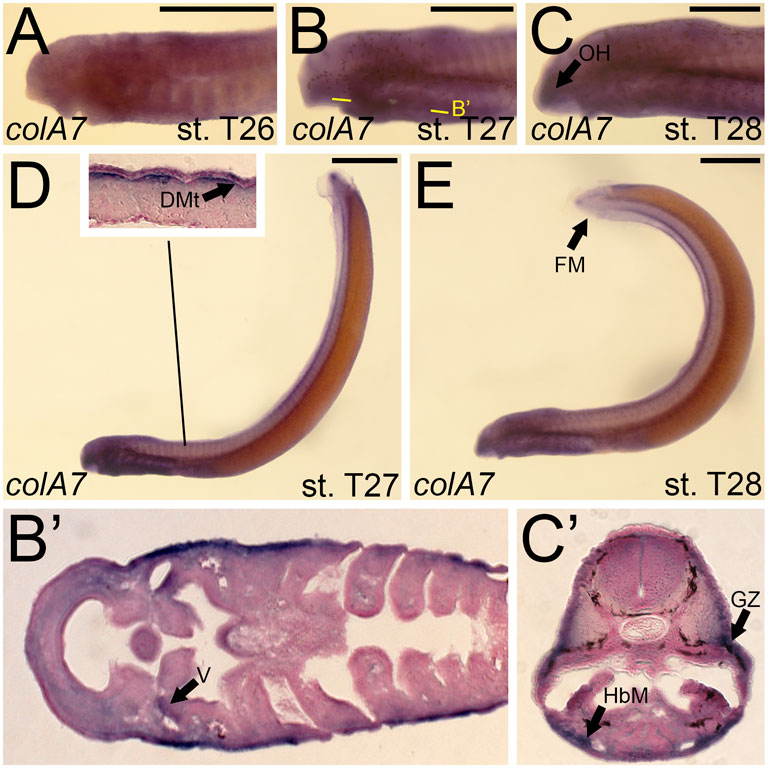
FIGURE 8. Expression of colA7 in larval lamprey. All scale bars are approximately 500 μm. (A) colA7 expression was weakly detected throughout the skin. (B,D) colA7 expression was in the medial velum as well as the dermomyotome. (C,E) colA7 expression was in the mesenchyme of the fin fold and oral hood, the growth zone, and the hypobranchial musculature. Keywords: DMt: dermomyotome; HbM: hypobranchial musculature; GZ: growth zone; OH: oral hood; V: velum.
Expression of Clade B and Clade C Collagens
We identified expression of colB1/col5a1α at T27 in the upper lip muscle as well as that of the pharynx and velum (Figures 9B,C). We additionally observed colB1/col5a1α expression in the developing myosepta and dermomyotome (Figure 9B). By stage 28, we continued to see expression in the aforementioned regions, with additional activity noted in the mesenchyme of the oral hood, the growth zone of the myomeres, and the hypobranchial muscles (Figure 9D,D′,D”). colB2/col5a1β expression was considerably different from that of colB1/col5a1α, being identified more uniformly across the developing pharynx at stage T27 (Figure 9G,G’). At this stage, we detected colB2/col5a1β activity in the mesodermal core, the ectoderm of the pharynx and lower mouth, and the presumptive vascular components of pharyngeal NCC derivatives (Figure 9G’). This expression largely abates by T28, being expressed primarily in the hypobranchial musculature and the myomeres of the somites (Figure 9I,I”.
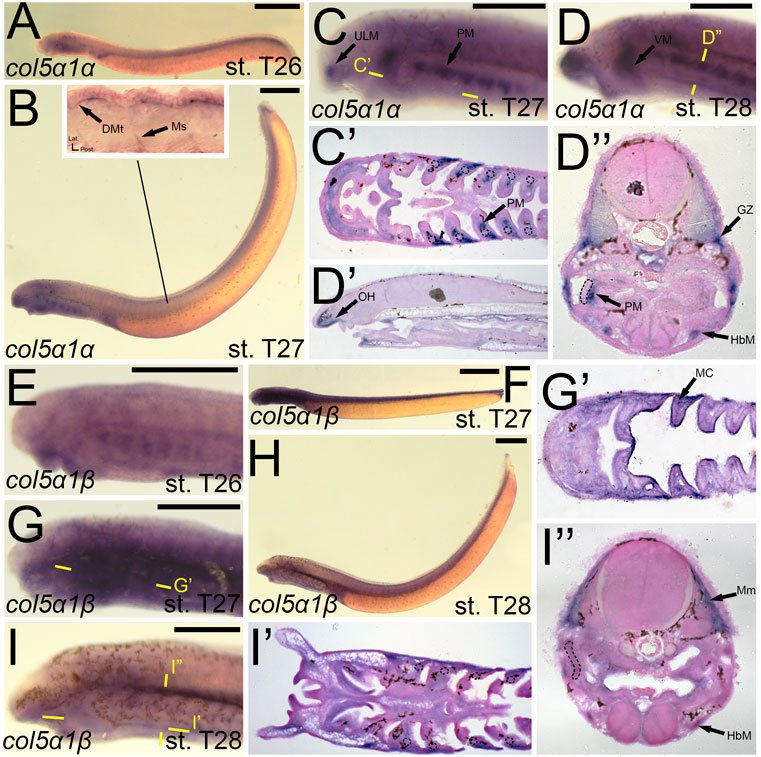
FIGURE 9. Expression of colB1/col5a1α and colB2/col5a1β in larval lamprey. All scale bars are approximately 500 μm. (A) col5a1α expression was primarily in the developing heart. (B,C) col5a1α expression was in the musculature of the pharynx, velum, and upper lip, and was also in the dermomyotome and myosepta. (D) col5a1α expression was in the mesenchyme of the oral hood, the growth zone, dermomyotome, hypobranchial musculature, and the pharyngeal muscles. Dotted circles indicate presumptive branchial arch skeleton. (E) col5a1β expression was in the oral ectoderm and throughout the pharynx albeit at low levels (F,G) col5a1β expression was in the mesodermal core of the pharynx and throughout the pharynx albeit at low levels. (H,I) col5a1β expression was in the hypobranchial musculature as well as the myomeres. Dotted circles indicate presumptive branchial arch skeleton. Keywords: DMt: dermomyotome; GZ: growth zone; HbM: hypobranchial musculature; MC: mesodermal core; Mm: myomeres; Ms: myosepta; OH: oral hood; PM: pharyngeal muscle; ULM: upper lip muscle; VM: velar muscle.
The expression of colB4/col11a1α largely mirrored that seen in colA1/col1a2a at stage 26, being expressed in similar populations of cranial ganglia, the mesenchyme of the upper lip and pharynx, and the somites (Figures 10A,B). At T27, colB4/col11a1α was identified in the mesenchyme of the oral hood, lower lip, and the non-skeletogenic neural crest of the pharynx (Figures 10C,D,D′,D”). We also observed expression along the trunk and tail, visualizing colB4/col11a1α transcripts in the presumptive sclerotome, myosepta, and in the base of the developing fin folds (Figure 10C). At stage 28, we saw additional expression in the pharyngeal muscles, hypobranchial muscles, the myomeres and their respective growth zone, and we continued to see expression in the mesenchyme of the oral hood, ventral pharynx and fin fold (Figure 10F,F′,F″,F‴,F””). colB5/col11a1β was exclusively detected in the epidermis at stage 26 (Figure 10G). At T27, this expression in the epidermis is largely the same, but additional transcription was identified in the mesenchyme of the inner velum (Figure 10H,H’). colB5/col11a1β was additionally identified in the velar epithelium as well as that of the oral hood. By stage 28, we detected expression only in the epidermis again, with weaker activity in and around the developing gill pores (Figure 10I).
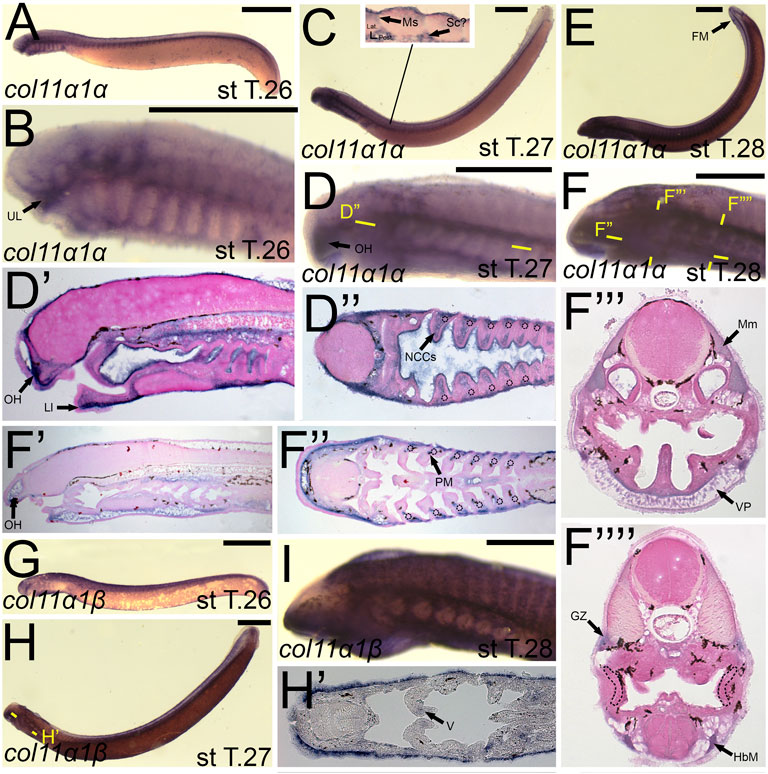
FIGURE 10. Expression of colB4/col11a1α and colB5/col11a1β in larval lamprey. All scale bars are approximately 500 μm. (A,B) col11a1α expression was in the cranial ganglia and the mesenchyme of the pharynx and upper and lower lip. (C,D) col11a1α expression was in the mesenchyme of the oral hood, lower lip, and fin fold, in the myosepta and presumptive sclerotome, and also in the non-skeletogenic neural crest cells in the pharynx. (E,F) col11a1α expression was in the mesenchyme of the fin fold, oral hood, lower lip, and ventral pharynx, the myomeres, hypobranchial musculature, pharyngeal muscles, and myomeres. Dotted circles indicate presumptive branchial arch skeleton. (G,I) col11a1β expression was superficial in the epidermis. (H) col11a1β expression was in the epidermis and in the medial velum. Keywords: Ep: epidermis; FM: fin mesenchyme; GZ: growth zone; HbM: hypobranchial musculature; LL: lower lip; Mm: myomeres; Ms: myosepta OH: oral hood; NCCs: neural crest cells; PM: pharyngeal muscle; Sc: sclerotome; UL: upper lip; VP: ventral pharynx.
We first found colB3/col5a3 activity at stage T26 in the developing ganglia and mesenchyme of the lower lip (Figures 11A,D). colB3/col5a3 was identified at T27 in the various pre-skeletogenic populations of mesenchyme, specifically the oral hood, lower lip, ventral pharynx, fin fold, as well as the lateral velum (Figures 11B,E,E′,E”). We also observed expression at this stage in the presumptive sclerotome and epidermis (Figure 11B,E’). At stage 28, colB3/col5a3 skeletal activity in the sclerotome was absent, but we did observe new expression in the hypochord and epibranchial bars and hypobranchial bars that are dorsal and ventral to the hyaline cartilage of the pharynx (Figure 11F,F′,F″,F‴). Expression in the oral hood, lower lip, fin fold, and ventral pharynx are similar to that seen at T27 (Figure 11F’,F‴). We additionally noted colB3/col5a3 in the hypobranchial musculature as well as the anterior myomeres and growth zone (Figure 11F”’).
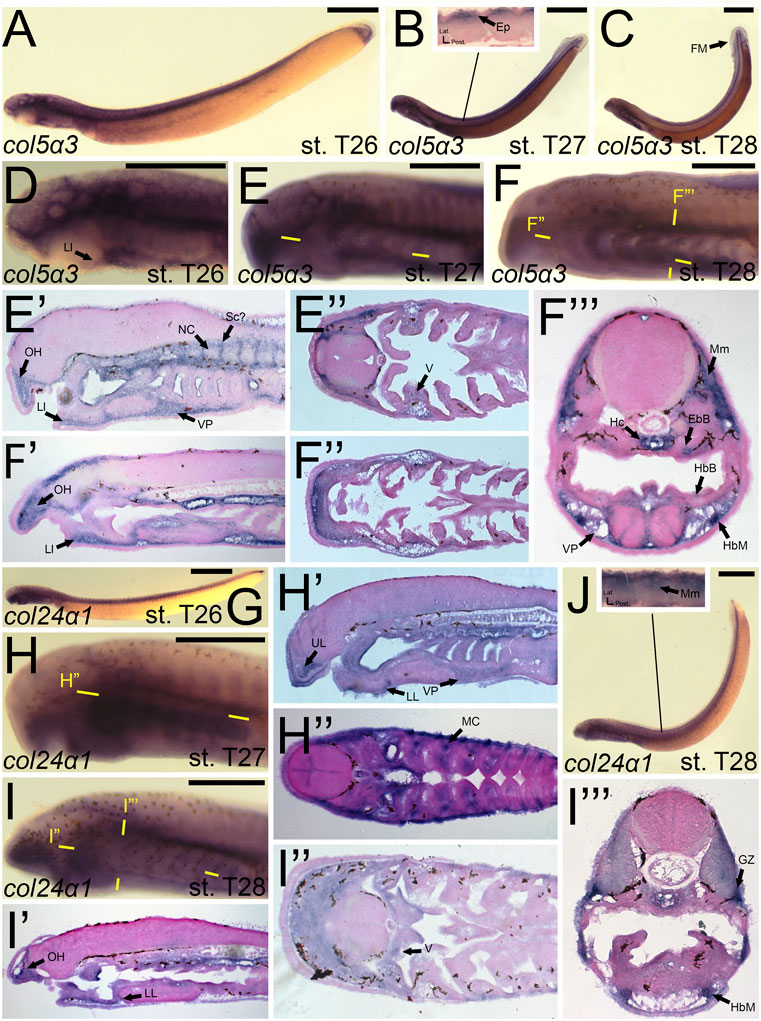
FIGURE 11. Expression of colB3/col5a3 and colC1/col24a1 in larval lamprey. All scale bars are approximately 500 μm. (A,D) col5a3 expression was in the cranial ganglia and the mesenchyme of the upper and lower lip and ventral pharynx. (B,E) col5a3 expression was in the mesenchyme of the oral hood, lower lip, ventral pharynx, lateral velum, fin fold, and presumptive sclerotome, as well as the notochord and epidermis. (C,F) col5a3 expression was in the mesenchyme of the aforementioned areas as well as the hypobranchial musculature, myomeres, hypochord, and the cartilages of the epibranchial bars and hypobranchial bars. Dotted circles indicate presumptive branchial arch skeleton. (G,H) col24a1 expression is in the mesodermal core of the pharyngeal arches as well as the mesenchyme of the upper and lower lip as well as the ventral pharynx. (I,J) col24a1 expression is in the mesenchyme of the oral hood, lower lip, ventral pharynx, and velum, and is also in the hypobranchial musculature, growth zone, and myomeres. Keywords: Ep: epidermis; EpB: epibranchial bars; FM: fin mesenchyme; GZ: growth zone; HbB: hypobranchial bars; HbM: hypobranchial musculature; Hc: hypochord; LL: lower lip; NC: notochord; OH: oral hood; MC: mesodermal core; Mm: myomeres; Sc: sclerotome; UL: upper lip; V: velum; VP: ventral pharynx.
colC1/col24a1 was expressed at both T26 and T27 in the mesenchyme of the upper and lower lip (Figures 11G,H,H′,H”). We also observed colC1/col24a1 activity in the mesodermal core of the pharyngeal arches, the otic vesicle, and the pharyngeal pouches (Figure 11H”). By stage 28, transcripts were further identified in the mesenchyme of the oral hood, ventral pharynx. and throughout the velum as well as the hypobranchial musculature, myomeres, the dermomyotome, and the growth zone (Figure 11I”).
Discussion
This study represents, to the best of our knowledge, the most comprehensive single study of fibrillar collagens and their gene expression in a vertebrate. Additionally, it is the first analysis of major and minor fibrillar collagens in a cyclostome. Future studies with collagens in lamprey connective tissues should use histochemical stains like picrosirius red to corroborate results, although this method still needs to be refined in lampreys (Supplementary Figure S8). Despite this, we used in situ hybridizations and sectioning to find multiple instances of fibrillar collagen co-expression across the musculoskeletal system, most dramatically in the hypobranchial musculature and the skeletogenic mesenchyme of the oral hood and fin fold. Our gene expression analyses support previous work for each gene which appear to be exclusive to specific groups. Overall, our findings are an important step in our understanding of fibrillar collagens and their evolution across vertebrates.
Gnathostomes and Lamprey Have Largely Conserved the Ancestral Collagen Repertoire
Our phylogenetic and syntenic analyses revealed distinct orthologs between the gene families in cyclostomes and gnathostomes as well as lineage-specific paralogs. Structural comparisons of the genes between these lineages, however, is unlikely to provide any new insights. Paired with macrosyntenic data, we can hypothesize the evolution of these gene families and its deviation in both cyclostomes and gnathostomes. The presence or absence of certain fibrillar collagens could provide insight into larger evolutionary trends.
Our phylogenetic analyses suggest that the last common ancestor of cyclostomes and gnathostomes had five Clade A fibrillar collagens. While gnathostomes have maintained all five genes, hagfish have likely lost both col1a1 and col5a2 whereas lamprey have lost at least one of these genes, the uncertainty stemming from whether colA7 is a highly divergent ortholog of one of these genes or a lamprey-specific duplicate of a different gene. Cyclostomes overall have more Clade A genes in part to three sets of paralogs, corresponding to col1a2, col2a1, and col3a1. There are considerable differences in expression between each set of paralogs. Interestingly, the developing skeleton of the fin expresses six of the seven Clade A genes. The loss of either col1a1 or col5a2 has interesting implications for the evolution of cyclostomes. There are no expression domains for col5a2 which are gene-specific, meaning that its function may be largely redundant. As seen with the loss of col5a3 and col11a2 in certain gnathostome taxa, it is possible that the gene was lost before the divergence of cyclostomes, its function being compensated by other genes. Conversely, the loss of cola1 may be more direct with a loss of morphology, being associated with the absence of mineralization in extant cyclostomes. Material studies have demonstrated the importance of type I collagen, composed of col1a1 proteins, in the formation of hydroxyapatite crystals during mineralization (Tomoaia and Pasca, 2015). Recent paleontological evidence now suggests a possible relationship between lamprey and hagfish and with conodonts, an ancient jawless vertebrate with mineralized teeth (Goudemand et al., 2011; Terrill et al., 2018). If conodonts were stem cyclostomes, then the loss of mineralization in modern lampreys and hagfishes may have coincided with the loss of col1a1 and other genes involved in the mineralization process. Both scenarios, however, depend on whether colA7 is orthologous to col5a2a or col1a1 or if it is a lamprey-specific duplicate instead.
Our results suggest that gnathostomes have maintained much of the ancestral repertoire of Clade B genes, but losses in this family are more common than in Clade A. Lamprey and hagfish no longer have the col11a2 gene but have duplicated the col5a1 gene and possibly col11a1 as well. Compared to Clade A, we see more variation in Clade B genes across gnathostomes, with several lineages having lost either col5a3 or col11a2. Similar to Clade A genes, we see considerable sub-functionalization between these paralogs, but their expression domains do not deviate beyond what we have seen in fibrillar collagens. These genes are a minor component of the collagenous ECM, so it is possible that their functions are similar enough that they can be compensatory for one another. Overall, the function of Clade B collagens has been largely maintained across extant vertebrates, being involved in most components of the musculoskeletal system, but they have drifted in function across lineages.
Our phylogenetic analyses suggest that the last common ancestor of cyclostomes and gnathostomes had two genes in the Clade C family, and the col27a1 gene was likely lost in lamprey. The extent of expression found in col24a1 is therefore interesting in contrast to gnathostomes, which is limited mostly to developing bone in the musculoskeletal system (Koch et al., 2003; Wang et al., 2012; Duran et al., 2015). The difference in expression between the gnathostome Clade C genes and their lamprey counterpart could be due in part to the sub-functionalization of these genes in the gnathostome lineage. Therefore, the lamprey col24a1 gene may reflect an ancestral domain of the gene in connective tissues, somewhat mirroring the range of expression seen in col1a2 or col1a1.
Fibrillar Collagens in Skeletal Muscle Are Largely Conserved Across Vertebrates
While fibrillar collagens are a less abundant component of the muscular ECM, their importance in these connective tissues remains an important area of study (Rowe, 1981; Eghbali and Weber, 1990; Rescan, 2019; Sleboda et al., 2020). Similar to expression patterns seen in gnathostomes, lamprey deploy fibrillar collagens in this connective tissue. An interesting pattern to note, however, is the contrast in lamprey expression between the medial myotome and lateral dermomyotome as well as between the anterior and posterior myotomes. Furthermore, the number of fibrillar collagen genes expressed in the musculature is also different between lamprey and gnathostomes. Jawed vertebrates only utilize type V collagen (col5a1 and col5a3 genes) in muscular connective tissue whereas lamprey use most of their Clade B genes to some extent. The abundance of lamprey fibrillar collagen genes in musculature is exemplified in the developing hypobranchial muscles, where col1a2a, col2a1a, col2a1b, col3a1a, col3a1b, col5a1a, col5a1b, col5a3, col11a1a, and col24a1 are all co-expressed.
The exact role of fibrillar collagens during muscle development in vertebrates is unclear. It is thought that the lateral and dorsoventral ends of the dermomyotome are a source of muscle fibers for secondary myogenesis, also known as stratified hyperplasia, and that these muscle fibers will have a distinct composition compared to the primary myofibers (Barresi et al., 2001; Georgiou et al., 2016; Chu et al., 2018). The presence of specific collagens in the medial or lateral domains of the dermomyotome indicate that, by these developmental stages, they are likely beginning to differentiate between the dermis and these secondary muscle fibers. These secondary muscle fibers are thought to contribute differentially to fast and slow-twitch muscle fibers, each having distinct collagen compositions and thus different mechanical properties (Kovanen et al., 1984; Nakamura et al., 2003). It is also possible, however, that these collagens are associated with the immature myoblasts of the dermomyotome, which control the rate at which differentiation and hyperplasia occur (Denetclaw et al., 2001; Ordahl et al., 2001; Steinbacher et al., 2006; Rescan et al., 2013). Further work must be done, therefore, to elucidate the role of fibrillar collagens in myogenesis and muscle homeostasis in lamprey.
The contrast in fibrillar collagens in the anterior and posterior myomeres may be attributed to the unique patterns of muscle migration in the lamprey head which have been previously described (Kusakabe and Kuratani, 2005; Kusakabe et al., 2011), but not all migratory muscles have similar collagen expression. Of particular interest is the infraoptic muscle, which did not express any collagens visualized in this study. It is thought that this muscle does not have a clear homolog in gnathostomes (Kusakabe et al., 2011), and its functions and mechanics remain unknown. Additionally, we observed a difference in fibrillar collagens between the anterior myomeres and the hypobranchial musculature, both of which are migratory muscle populations. This would suggest that there are some mechanical differences between the lamprey migratory muscles.
Taken together, our findings suggest that vertebrate fibrillar collagens have a conserved role during early myogenesis, with type V collagen being predominantly myogenic in distribution. They are expressed in both the early myotome and dermomyotome, and likely contribute to different myogenic populations. The main differences between lamprey and gnathostome fibrillar collagens in the developing muscles are the presence of type II, type XI, and type XXIV collagen in lamprey, where they are thoroughly co-expressed in the hypobranchial muscles and the myomeres and their respective growth zone. Whether this is due to a possible role in maintaining myoblasts or the development of secondary fibers, however, is uncertain. The function of collagens in the muscular system remains poorly understood, and further work is necessary to elaborate on the activity of these genes in the tissues. Overall, fibrillar collagens have a minor but conserved role in vertebrate musculature.
The Lamprey Collagenous Skeleton Differs Greatly From Gnathostomes
It has been noted for nearly two centuries that the lamprey larval and adult skeletons were very different from those seen in gnathostomes (Müller, 1834; Schneider, 1879; Parker, 1883; Johnels, 1948), but only in more recent years has a more descriptive genetic and molecular approach been possible. Among these observations were the identification of novel lamprey proteins like lamprin and pharymprin which are differentially expressed in the larval and adult skeletons (Wright et al., 1983; Robson et al., 1993; McBurney et al., 1996; Yokoyama et al., 2019). Additionally, we have learned more about the differences in gene expression and histology between the gnathostome-like hyaline cartilage of the larval trabeculae and branchial skeleton and that of mucocartilage, the avascular connective tissue scattered throughout the anterior head skeleton that has several characteristics of articular cartilage (Johnels, 1948; Cerny et al., 2010; Cattell et al., 2011; Yao et al., 2011; Root et al., 2021b). Recent work has shown that the lamprey larval skeleton expresses multiple Lectican homologs in distinct skeletal populations (Root et al., 2021a), an interesting feature which supports previous hypotheses that the lamprey larval skeleton may have as many as six cartilage types (Cattell et al., 2011).
Our results show that lamprey mucocartilage is rather homogenous for fibrillar collagens, particularly so in the oral region and fin fold, with each co-expressing five shared fibrillar collagens (col1a2a, col2a1a, col3a1a, col5a3, col11a1a). Additionally, the oral hood expresses col5a1a and col24a1 whereas the fin fold expresses col1a2b and col2a1b.
A rather intriguing find was the absence of fibrillar collagens in the branchial arch cartilages throughout all developmental stages noted. Previous studies have identified type II collagen in these arches at later developmental stages as well as the adult skeleton (Zhang et al., 2006), but it was unclear whether col2a1 orthologs were present in the prechondrogenic condensations or early chondrocytes of these arches in P. marinus. Our findings suggest that neither major nor minor fibrillar collagens are present in the early differentiation and condensation of sea lamprey branchial cartilage. This contrasts with collagen expression data across all metazoan cartilages, where Clade A collagens are present (Tarazona et al., 2016). Furthermore, type II collagen has been documented in the branchial cartilages of the arctic lamprey Lethenteron camtschaticum during early development (Ohtani et al., 2008), meaning that it is likely that P. marinus have lost type II collagen in these cartilages during these developmental stages. This sea lamprey deviation is an important insight into vertebrate chondrogenesis, as col2a1 is thought to be essential for normal cartilage development.
Although col2a1 is the most well-attested fibrillar collagen in the gnathostome cartilaginous skeleton and col1a1 in the bony skeleton, several other collagens are known to have transient roles in the development of these structures. It is therefore not surprising that the larval sea lamprey skeleton expresses a variety of fibrillar collagens during development (Figure 12). What is important to note, however, is the spatial differences in these collagens throughout the lamprey skeleton during these stages of development. Lamprey have sub-functionalized cartilage types in different regions of the skeleton, and we have found distinct collagen deployments throughout the head and fin skeleton that support these observations. Overall, these findings suggest that the ancestral vertebrate cartilage extracellular matrix utilized multiple fibrillar collagens, but sea lamprey have innovated upon this program to create distinct skeletal types and no longer require them for initial hyaline development.
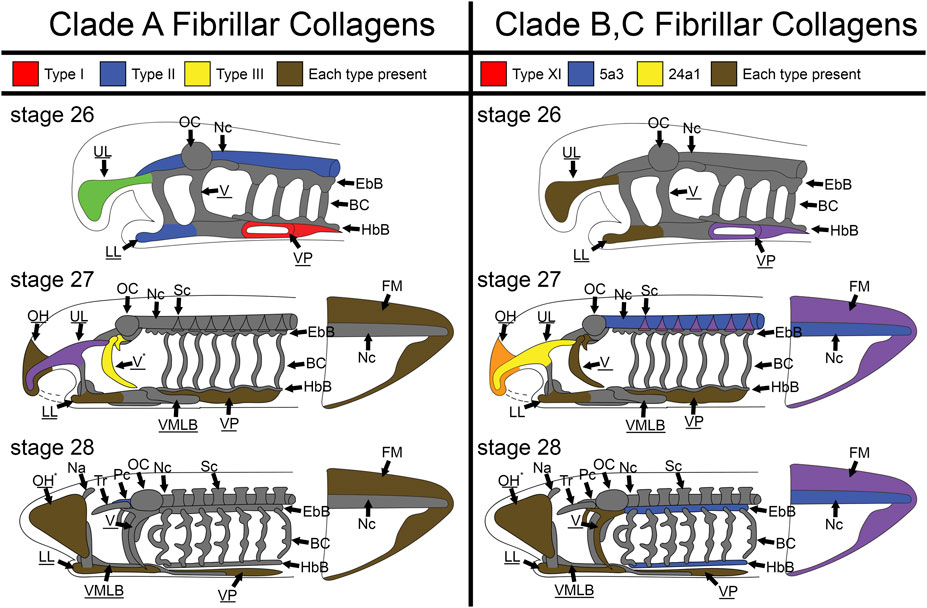
FIGURE 12. Skeletal heterogeneity of fibrillar collagens in larval lamprey during early development. For stage T26, only the head skeleton is shown, whereas both head and fin skeleton are shown for T27 and T28. Colors indicate specific subfamilies for each Clade. Minor components of the skeleton for each clade (colA7 and col5a1α for Clade A and Clade B respectively) are indicated by an asterisk near the tissue where they are expressed instead of a color. Non-primary colors signify a combination of collagen subfamilies, and gray indicates no fibrillar collagens. Underline keywords indicate skeletal populations which are considered mucocartilage. Although fibrillar collagens are largely ubiquitous the oral region and ventral pharynx later in development, they are entirely absent from the branchial cartilages, otic capsule, nasal capsule, and trabeculae throughout the stages observed, and are also particularly lacking in similar cartilages like the parachordals, epibranchial bars, and hypobranchial bars. Fibrillar collagens are also heterogeneous throughout the posterior mucocartilage. Keywords: BC: branchial cartilages; EbB: epibranchial bars; FM: fin mesenchyme; HbBL: hypobranchial bars; LL: lower lip; Na: nasal capsule; Nc: notochord; OC: otic capsule; OH: oral hood; Pc: parachordals; Sc: sclerotome; Tr: trabeculae; UL: upper lip; V: velum; VMLB: ventromedial longitudinal bar; VP: ventral pharynx.
Fibrillar Collagens Were Multifunctional in the Ancestral Vertebrate Musculoskeletal System
Fibrillar collagens have been previously documented in invertebrate chordates like tunicates and amphioxus, with most genes being expressed in the oral cirri, notochord, the tail bud, and the musculature (Wada et al., 2006; Zhang and Cohn, 2006; Jandzik et al., 2015). While the expansion of this gene family would have likely contributed to sub-functionalization amongst cell types in vertebrates, our results in lamprey, paired with gnathostome literature, suggest the contrary. We posit that the general function of the fibrillar collagen family has remained relatively unchanged over the course of chordate evolution, with a conserved role in the development and maintenance of connective tissues.
It remains of great interest why so many fibrillar collagens have been maintained in vertebrates if the majority of them are co-expressed across cell types to some extent. Our current understanding of gene duplication posits that redundant genes are eventually purged from the genome, but there may also be long-term benefits to this redundancy (Wapinski et al., 2007; Kuzmin et al., 2020). We see differences in specificity between orthologs in both lineages, with type II and type XI collagen being predominantly expressed in skeleton in gnathostomes and type V collagen being almost entirely expressed in muscle in lamprey. These together would suggest that has been some change of function for these genes. The extracellular matrix is a complicated meshwork of dozens of different types of proteins, and it is therefore possible that each collagen provides distinct structural support or even modulations to cell signaling. However, it is also possible that co-expression of fibrillar collagens helps overcome rate-limited transcription for such abundantly-needed ECM proteins, meaning that these collagens are redundant and interchangeable to some extent, at least in the musculoskeletal system. Whether these differences in co-expression are due to qualitative or quantitative differences is an important aspect of fibrillar collagen evolution that may help us further understand their roles in development.
The evolution of the vertebrate lineage coincided with multiple events of genomic expansion, meaning that most gene families are larger than their invertebrate counterparts. Understanding how these genes sub-functionalized and supported the rise of new tissue types is important for our understanding of vertebrate musculoskeletal evolution. Our results suggest that the function of fibrillar collagens was largely established before the divergence of extant vertebrates, having important roles throughout the developing skeleton and musculature. The overlap we see in these genes across the cell types is likely due to their piecemeal contribution to the mechanics and signaling of connective tissues, regardless of the progeny. The total diversity of fibrillar collagen genes present in vertebrates may be due to either redundancy or subtle sub-functionalization, with differences in their distribution in cell types being attributed to differences in signaling or mechanics. We therefore suggest that the main functions of fibrillar collagens were likely established before the divergence of modern vertebrates and that the evolution of the vertebrate musculoskeletal system involved upstream regulatory differences to the deployment of fibrillar collagens, allowing ancestral connective tissues to diversify in form and function.
Methods
Isolation of Lamprey and Hagfish Collagen Homologues
Lamprey collagen sequences were tiled from transcriptomic reads of Tahara st. 26.5 embryos and adult oral disc tissue that were previously gathered and submitted to GenBank (Wheeler et al., 2007). Hagfish collagen sequences were tiled from transcriptomic reads of Bashford Dean st 28–30 embryos generously provided by Juan Pascual Anaya at the Universidad de Malaga and were submitted to GenBank (Wheeler et al., 2007). To identify lamprey and hagfish sequences, we used zebrafish fibrillar collagen protein sequences to query P. marinus and E. burgeri transcriptomes using tBLASTN, collecting the top ten reads for each to ensure that zebrafish-specific biases were minimized. These reads were translated and searched against gnathostome genomes (D. rerio, C. milii, H. sapiens) to confirm whether they were Clade A, Clade B, or Clade C genes. To confirm that our lamprey transcriptomic sequences represented all possible fibrillar collagen genes, we used UCSC’s genome browser and the BLAT tool (Kent, 2002) to identify the locations in the lamprey genome where potential fibrillar collagens could be located. These sequences were additionally tested using BLASTp to identify which fibrillar collagen clade they likely corresponded to, letting us organize the primary sequences and remove partial sequences before our analyses.
Full-length sequences from these files were used for our phylogenetic and syntenic analyses. For in situ hybridizations for colA3/col2a1α and colA5/col2a1β, primers were designed from lamprey genomic sequence to amplify conserved exon sequences, which were cloned into the pJet1.2 vector. For the remainder of the lamprey genes, 500-550bp regions from transcriptomic sequences were selected and ordered as fragments in pUC57-amp vector from Synbio Tech©.
Phylogenetic Analyses
For our phylogenetic analyses, full-length gnathostome fibrillar collagens were chosen to best represent the breadth of gnathostome genomic and morphological diversity as well as select invertebrates where both Clade A and Clade B genes could be identified, and these sequences would be compared against lamprey and hagfish fibrillar collagen. Full-length human fibrillar collagen protein sequences were obtained from NCBI (Wheeler et al., 2007) and used to identify additional gnathostome and invertebrate collagen protein sequences using BLASTP (Altschul et al., 1990). GenBank accession numbers used are available in Tab. S3-S7. In the case of sturgeon (Acipenser oxyrhinchus) sequences, zebrafish collagen protein sequences were used to find transcripts using tBLASTN. These sequences were then translated and searched against multiple gnathostome genomes to confirm their identity. We then aligned all sequences for each analysis using the PROBALIGN (Roshan and Livesay, 2006) program on CIPRES (Miller et al., 2011) servers. We used a gap open penalty of 20 and a gap extension penalty of 1. To determine the optimal substitution model for our phylogenetic analysis, we used ProtTest v3.4.2 (Abascal et al., 2005) In the ProtTest settings, we allowed for invariant sites, empirical frequencies, and BIONJ-defined tree topology to determine our ideal model. For our main Clade A test, a JTT + I + G model was calculated with a log likelihood score of -111,200.671 under Akaike Information Criterion (AIC). For our main Clade B test, a JTT + I + G model was calculated with a log likelihood score of -97422.354 under AIC. For our Clade C test, a JTT + I + G model was calculated with a log likelihood score of -76554.941 under AIC. For our phylogenetic analysis, we used maximum likelihood analyses using RAxML-HPC2 Workflow on CIPRES servers. Using the aforementioned parameters provided by ProtTest, our likelihood scores were bootstrapped with 1,000 trees for each test to derive a consensus tree. For our Clade A and Clade C analyses, we used an outgroup of col5a1 homologues from human and ghost shark. For our Clade B analysis, we used an outgroup of col2a1 homologues from human and ghost shark. Our consensus trees were lastly visualized using FigTree v1. 4.4 (Rambaut, 2014).
Synteny Analyses
For our linkage analyses, we used peptide sequences of gnathostome fibrillar collagen genes from our phylogenetic analyses and found their respective genomic location using UCSC’s Genome Browser and the BLAT tool (Kent, 2002). For our estimate of gnathostome linkage groups, we looked at the loci of dog (C. lupus familiaris), chicken (G. gallus), zebrafish (D. rerio), and clawed frog (X. tropicalis), as these species have well-assembled genomes with chromosomes/scaffolds long enough to detect distant linkage. For our macrosynteny analyses, we gathered peptide sequences of gnathostome fibrillar collagens on NCBI and found their respective genomic location using UCSC’s Genome Browser and the BLAT tool (Kent, 2002) as well as ENSEMBL (Yates et al., 2020). We used sequences of human (Homo sapiens), clawed frog (Xenopus tropicalis or Xenopus laevis), spotted gar (Lepisosteus oculatus), zebrafish (Danio rerio), and elephant shark (Callorhinchus milii) to reconstruct the ancestral arrangement of genes around the fibrillar collagen loci. Because the lamprey genome has undergone considerable rearrangement, we did not limit our search for linked genes based on distance alone, instead identifying ≈27–30 genes immediately 5′ and 3′ of each collagen locus when possible. We annotated each gene according to the genome browser’s prediction. We then color-coded the lamprey genes based on which gnathostome fibrillar collagen it was mostly linked to. Due to the strong GC bias of lamprey protein coding sequences and the possibility of distinct duplicates, we included all homologs of a gene family at a particular locus. We then attempted to find similar linked genes around the hagfish collagen loci using the aforementioned methods. Due to the current condition of the hagfish genome assembly, we were unable to find any scaffolds that were long enough to effectively search. To better visualize our synteny results for the manuscript, we generated a Circos plot using the online version of Circos Table Viewer (Krzywinski et al., 2009), creating a non-reciprocal plot to show linked genes across gnathostome and lamprey fibrillar collagens.
Embryo Collection and Staging
Embryos for in situ hybridization were obtained from adult spawning-phase sea lampreys (Petromyzon marinus) collected from Lake Huron, MI, and kept in chilled holding tanks as previously described (Nikitina et al., 2009). Embryos were staged according to the method of Tahara (Yutaka, 1988), fixed in MEMFA (Mops buffer, EGTA, MgSO4, and formaldehyde), rinsed in Mops buffer, dehydrated into methanol, and stored at –20 °C.
In Situ Hybridization
Riboprobes were made for anti-sense fragments using SP6 RNA polymerase. Sequences for probes and genes are available upon request. In our experience, full-length P. marinus riboprobes, or riboprobes generated against untranslated regions of P. marinus transcripts, give higher background than short riboprobes against coding sequences. We believe that this is because lamprey noncoding sequences, especially 3′ UTRs, often have an excessive GC-repeat content, causing corresponding riboprobes to hybridize nonspecifically to off-targets. To mitigate this, we made short 550-bp riboprobes against coding regions and used a high-stringency hybridization protocol (Cerny et al., 2010; Square et al., 2016). Key parameters of this protocol include post-hybridization washes at 70 °C and the use of a low-salt, low-pH hybridization buffer (50% formamide; 1.3× SSC, pH 5.0; 5 mM EDTA, pH 8.0; 50 μg/ml tRNA; 0.2% Tween-20; 0.5% CHAPS; and 100 μg/ml heparin).
Histology, Histochemistry, and Sectioning.
After in situ hybridization, embryos were postfixed in 4% paraformaldehyde/PBS (4°C, overnight), rinsed in PBS, cryo-protected with 15% sucrose/PBS, embedded in 15% sucrose, 20% gelatin/PBS (37°C, overnight), and 20% gelatin/PBS (37°C overnight), frozen in liquid nitrogen, and mounted in OCT compound (Miles). Cryo-sections of 14 μm were collected on Super Frost Plus slides (Fisher Scientific), counterstained using Nuclear Fast Red (Vector Laboratories), and dehydrated and mounted in DPX (Fluka) (Jandzik et al., 2014). For lamprey colA3/col2a1α and colB5/col11a1β stage T27 sections, in situ hybridizations were performed on 10 μm paraffin sections which were deparaffinized in Histoclear immediately beforehand. For these sections, proteinase K, hybridization, and color development times were shortened. Picrosirius staining was performed on 14 μm paraffin sections which were deparaffinized in Histoclear and rehydrated, stained in 1% picrosirius red for 30 min, and differentiated twice in 1% acetic acid for 5 min each before being dehydrated and mounted.
Imaging
Whole-mount in situ hybridized P. marinus embryos and larvae were photographed using a Carl Zeiss Axiocam MRc5, Carl ZeissDiscovery V8 dissecting microscope, and Axiovision 4.9.1 software. Sections were photographed using a Carl Zeiss Imager A2 compound microscope.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The animal study was reviewed and approved by the IACUC Protocol 2392.
Author Contributions
ZR, CA, CG, MB, and DJ did the experiments. ZR wrote the manuscript and created the figures. Edits were done by ZR, DJ, and DM. Final manuscript was approved by ZR, DJ, and DM.
Funding
ZR and DM were supported by National Science Foundation grants IOS 1656843 and IOS 1257040 to DM. ZR, CA, CG, and MB were also supported by the Beverly Sears, Biological Sciences Initiative, and EBIO grants through the University of Colorado Boulder (Grants 13414815, 11060912, and 13410919). DJ was supported by the Scientific Grant Agency of the Slovak Republic VEGA grant No. 1/0450/21.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank Scott Miehls at the USGS Hammond Bay Biological Station for providing adult sea lampreys. They also thank Jeremiah J. Smith at the University of Kentucky, Juan Pascual Anaya at the Universidad de Malaga, and Shigeru Kuratani at the RIKEN Institute in Kobe, Japan for supplemental transcriptomic data.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2022.809979/full#supplementary-material
References
Abascal, F., Zardoya, R., and Posada, D. (2005). ProtTest: Selection of Best-Fit Models of Protein Evolution. Bioinformatics 21 (9), 2104–2105. doi:10.1093/bioinformatics/bti263
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic Local Alignment Search Tool. J. Mol. Biol. 215 (3), 403–410. doi:10.1016/s0022-2836(05)80360-2
Aouacheria, A., Cluzel, C., Lethias, C., Gouy, M., Garrone, R., and Exposito, J.-Y. (2004). Invertebrate Data Predict an Early Emergence of Vertebrate Fibrillar Collagen Clades and an Anti-incest Model. J. Biol. Chem. 279 (46), 47711–47719. doi:10.1074/jbc.m408950200
Baas, D., Malbouyres, M., Haftek-Terreau, Z., Le Guellec, D., and Ruggiero, F. (2009). Craniofacial Cartilage Morphogenesis Requires Zebrafish Col11a1 Activity. Matrix Biol. 28 (8), 490–502. doi:10.1016/j.matbio.2009.07.004
Bailey, W. J., Kim, J., Wagner, G. P., and Ruddle, F. H. (1997). Phylogenetic Reconstruction of Vertebrate Hox Cluster Duplications. Mol. Biol. Evol. 14 (8), 843–853. doi:10.1093/oxfordjournals.molbev.a025825
Bandyopadhyay, A., Kubilus, J. K., Crochiere, M. L., Linsenmayer, T. F., and Tabin, C. J. (2008). Identification of Unique Molecular Subdomains in the Perichondrium and Periosteum and Their Role in Regulating Gene Expression in the Underlying Chondrocytes. Develop. Biol. 321 (1), 162–174. doi:10.1016/j.ydbio.2008.06.012
Barresi, M. J. F., D'Angelo, J. A., Hernández, L. P., and Devoto, S. H. (2001). Distinct Mechanisms Regulate Slow-Muscle Development. Curr. Biol. 11 (18), 1432–1438. doi:10.1016/s0960-9822(01)00428-6
Boraschi-Diaz, I., Wang, J., Mort, J. S., and Komarova, S. V. (2017). Collagen Type I as a Ligand for Receptor-Mediated Signaling. Front. Phys. 5, 12. doi:10.3389/fphy.2017.00012
Cao, L., Li, L., Li, Y., Zhuang, J., Chen, Y., Wang, Y., et al. (2018). Loss of the Nodal Modulator Nomo Results in Chondrodysplasia in Zebrafish. Curr. Mol. Med. 18 (7), 448–458. doi:10.2174/1566524019666181212095307
Cattell, M., Lai, S., Cerny, R., and Medeiros, D. M. (2011). A New Mechanistic Scenario for the Origin and Evolution of Vertebrate Cartilage. PloS one 6 (7), e22474. doi:10.1371/journal.pone.0022474
Cerny, R., Cattell, M., Sauka-Spengler, T., Bronner-Fraser, M., Yu, F., and Medeiros, D. M. (2010). Evidence for the Prepattern/cooption Model of Vertebrate Jaw Evolution. Proc. Natl. Acad. Sci. 107 (40), 17262–17267. doi:10.1073/pnas.1009304107
Chen, J. W., and Galloway, J. L. (2014). The Development of Zebrafish Tendon and Ligament Progenitors. Development 141 (10), 2035–2045. doi:10.1242/dev.104067
Christiansen, H. E., Lang, M. R., Pace, J. M., and Parichy, D. M. (2009). Critical Early Roles for Col27a1a and Col27a1b in Zebrafish Notochord Morphogenesis, Vertebral Mineralization and post-embryonic Axial Growth. PLoS One 4 (12), e8481. doi:10.1371/journal.pone.0008481
Chu, W., Chen, D., Li, Y., Wu, P., Zhang, J., and Liu, L. (2018). Muscle Fiber Differentiation and Growth Patterns during Hyperplasia and Hypertrophy in the Ricefield Eel Regulated by Myogenic Regulatory Factors. North. Am. J. Aquacult. 80 (2), 180–186. doi:10.1002/naaq.10025
D'hondt, S., Guillemyn, B., Syx, D., Symoens, S., De Rycke, R., Vanhoutte, L., et al. (2018). Type III Collagen Affects Dermal and Vascular Collagen Fibrillogenesis and Tissue Integrity in a Mutant Col3a1 Transgenic Mouse Model. Matrix Biol. 70, 72–83. doi:10.1016/j.matbio.2018.03.008
Denetclaw, W. F., Berdougo, E., Venters, S. J., and Ordahl, C. P. (2001). Morphogenetic Cell Movements in the Middle Region of the Dermomyotome Dorsomedial Lip Associated with Patterning and Growth of the Primary Epaxial Myotome. Development 128 (10), 1745–1755. doi:10.1242/dev.128.10.1745
Di Lullo, G. A., Sweeney, S. M., Körkkö, J., Ala-Kokko, L., and San Antonio, J. D. (2002). Mapping the Ligand-Binding Sites and Disease-Associated Mutations on the Most Abundant Protein in the Human, Type I Collagen. J. Biol. Chem. 277 (6), 4223–4231. doi:10.1074/jbc.m110709200
Duance, V. C., Restall, D. J., Beard, H., Bourne, F. J., and Bailey, A. J. (1977). The Location of Three Collagen Types in Skeletal Muscle. FEBS Lett. 79 (2), 248–252. doi:10.1016/0014-5793(77)80797-7
Duran, I., Csukasi, F., Taylor, S. P., Krakow, D., Becerra, J., Bombarely, A., et al. (2015). Collagen Duplicate Genes of Bone and Cartilage Participate during Regeneration of Zebrafish Fin Skeleton. Gene Expr. Patterns 19 (1-2), 60–69. doi:10.1016/j.gep.2015.07.004
Eames, B. F., Amores, A., Yan, Y. L., and Postlethwait, J. H. (2012). Evolution of the Osteoblast: Skeletogenesis in Gar and Zebrafish. BMC Evol. Biol. 12 (1), 27–13. doi:10.1186/1471-2148-12-27
Eames, B. F., Medeiros, D. M., and Adameyko, I. (2020). On the Evolution of Skeletal Cells Befire Abd after Neural Crest" Evolving Neural Crest Cells. Boca Raton: CRC Press.
Eghbali, M., and Weber, K. T. (1990). Collagen and the Myocardium: Fibrillar Structure, Biosynthesis and Degradation in Relation to Hypertrophy and its Regression. Mol. Cel Biochem 96 (1), 1–14. doi:10.1007/BF00228448
Enault, S., Muñoz, D. N., Silva, W. T. A. F., Borday-Birraux, V., Bonade, M., Oulion, S., et al. (2015). Molecular Footprinting of Skeletal Tissues in the Catshark Scyliorhinus canicula and the Clawed Frog Xenopus Tropicalis Identifies Conserved and Derived Features of Vertebrate Calcification. Front. Genet. 6, 283. doi:10.3389/fgene.2015.00283
Exposito, J.-Y., and Lethias, C. (2013). “Invertebrate and Vertebrate Collagens,” in Evolution of Extracellular Matrix (Berlin: Springer), 39–72.
Exposito, J.-Y., Valcourt, U., Cluzel, C., and Lethias, C. (2010). The Fibrillar Collagen Family. Ijms 11 (2), 407–426. doi:10.3390/ijms11020407
Fan, Y., Liang, Y., Deng, K., Zhang, Z., Zhang, G., Zhang, Y., et al. (2020). Analysis of DNA Methylation Profiles during Sheep Skeletal Muscle Development Using Whole-Genome Bisulfite Sequencing. BMC genomics 21 (1), 327–415. doi:10.1186/s12864-020-6751-5
Fang, M., Adams, J. S., McMahan, B. L., Brown, R. J., and Oxford, J. T. (2010). The Expression Patterns of Minor Fibrillar Collagens during Development in Zebrafish. Gene Expr. Patterns 10 (7-8), 315–322. doi:10.1016/j.gep.2010.07.002
Fisher, S., Jagadeeswaran, P., and Halpern, M. E. (2003). Radiographic Analysis of Zebrafish Skeletal Defects. Develop. Biol. 264 (1), 64–76. doi:10.1016/s0012-1606(03)00399-3
Gaspera, B. d., Armand, A.-S., Sequeira, I., Lecolle, S., Gallien, C. L., Charbonnier, F., et al. (2009). The Xenopus MEF2 Gene Family: Evidence of a Role for XMEF2C in Larval Tendon Development. Develop. Biol. 328 (2), 392–402. doi:10.1016/j.ydbio.2009.01.039
Gelse, K., Ekici, A. B., Cipa, F., Swoboda, B., Carl, H. D., Olk, A., et al. (2012). Molecular Differentiation between Osteophytic and Articular Cartilage - Clues for a Transient and Permanent Chondrocyte Phenotype. Osteoarthritis and cartilage 20 (2), 162–171. doi:10.1016/j.joca.2011.12.004
Gemballa, S., and Vogel, F. (2002). Spatial Arrangement of white Muscle Fibers and Myoseptal Tendons in Fishes. Comp. Biochem. Physiol. A: Mol. Integr. Physiol. 133 (4), 1013–1037. doi:10.1016/s1095-6433(02)00186-1
Georgiou, S., Alami-Durante, H., Power, D. M., Sarropoulou, E., Mamuris, Z., and Moutou, K. A. (2016). Transient up- and Down-Regulation of Expression of Myosin Light Chain 2 and Myostatin mRNA Mark the Changes from Stratified Hyperplasia to Muscle Fiber Hypertrophy in Larvae of Gilthead Sea Bream (Sparus aurata L.). Cell Tissue Res 363 (2), 541–554. doi:10.1007/s00441-015-2254-0
Gistelinck, C., Gioia, R., Gagliardi, A., Tonelli, F., Marchese, L., Bianchi, L., et al. (2016). Zebrafish Collagen Type I: Molecular and Biochemical Characterization of the Major Structural Protein in Bone and Skin. Sci. Rep. 6 (1), 21540–21614. doi:10.1038/srep21540
Goudemand, N., Orchard, M. J., Urdy, S., Bucher, H., and Tafforeau, P. (2011). Synchrotron-aided Reconstruction of the Conodont Feeding Apparatus and Implications for the Mouth of the First Vertebrates. Proc. Natl. Acad. Sci. 108 (21), 8720–8724. doi:10.1073/pnas.1101754108
Grimaldi, A., Tettamanti, G., Martin, B. L., Gaffield, W., Pownall, M. E., Hughes, S. M., et al. (2004). Hedgehog Regulation of Superficial Slow Muscle Fibres in Xenopus and the Evolution of Tetrapod Trunk Myogenesis. Development 131 (14), 3249–3262. doi:10.1242/dev.01194
Guerquin, M. J., Charvet, B., Nourissat, G., Havis, E., Ronsin, O., Bonnin, M. A., et al. (2013). Transcription Factor EGR1 Directs Tendon Differentiation and Promotes Tendon Repair. J. Clin. Invest. 123 (8), 3564–3576. doi:10.1172/JCI67521
Harel, I., Nathan, E., Tirosh-Finkel, L., Zigdon, H., Guimarães-Camboa, N., Evans, S. M., et al. (2009). Distinct Origins and Genetic Programs of Head Muscle Satellite Cells. Develop. Cel. 16 (6), 822–832. doi:10.1016/j.devcel.2009.05.007
Hoffman, G. G., Branam, A. M., Huang, G., Pelegri, F., Cole, W. G., Wenstrup, R. M., et al. (2010). Characterization of the Six Zebrafish Clade B Fibrillar Procollagen Genes, with Evidence for Evolutionarily Conserved Alternative Splicing within the pro-α1(V) C-Propeptide. Matrix Biol. 29 (4), 261–275. doi:10.1016/j.matbio.2010.01.006
Huang, H., Wright, S., Zhang, J., and Brekken, R. A. (2019). Getting a Grip on Adhesion: Cadherin Switching and Collagen Signaling. Biochim. Biophys. Acta (Bba) - Mol. Cel Res. 1866 (11), 118472. doi:10.1016/j.bbamcr.2019.04.002
Imamura, Y., Scott, I. C., and Greenspan, D. S. (2000). The Pro-α3(V) Collagen Chain. J. Biol. Chem. 275 (12), 8749–8759. doi:10.1074/jbc.275.12.8749
Iwai, T., Murai, J., Yoshikawa, H., and Tsumaki, N. (2008). Smad7 Inhibits Chondrocyte Differentiation at Multiple Steps during Endochondral Bone Formation and Down-Regulates P38 MAPK Pathways. J. Biol. Chem. 283 (40), 27154–27164. doi:10.1074/jbc.m801175200
Jandzik, D., Garnett, A. T., Square, T. A., Cattell, M. V., Yu, J.-K., and Medeiros, D. M. (2015). Evolution of the New Vertebrate Head by Co-option of an Ancient Chordate Skeletal Tissue. Nature 518 (7540), 534–537. doi:10.1038/nature14000
Jandzik, D., Hawkins, M. B., Cattell, M. V., Cerny, R., Square, T. A., and Medeiros, D. M. (2014). Roles for FGF in Lamprey Pharyngeal Pouch Formation and Skeletogenesis Highlight Ancestral Functions in the Vertebrate Head. Development 141 (3), 629–638. doi:10.1242/dev.097261
Jenkins, E., Moss, J., Pace, J., and Bridgewater, L. (2005). The New Collagen Gene Contains SOX9-Responsive Enhancer Elements. Matrix Biol. 24 (3), 177–184. doi:10.1016/j.matbio.2005.02.004
Johnels, A. G. (1948). On the Development and Morphology of the Skeleton of the Head of Petromyzon. Acta zoologica 29 (1), 139–279. doi:10.1111/j.1463-6395.1948.tb00030.x
Kent, W. J. (2002). The Human Genome Browser at UCSC. Genome Res. 12 (6), 996–1006. doi:10.1101/gr.229102
Kessels, M. Y., Huitema, L. F. A., Boeren, S., Kranenbarg, S., Schulte-Merker, S., van Leeuwen, J. L., et al. (2014). Proteomics Analysis of the Zebrafish Skeletal Extracellular Matrix. PloS one 9 (3), e90568. doi:10.1371/journal.pone.0090568
Koch, M., Laub, F., Zhou, P., Hahn, R. A., Tanaka, S., Burgeson, R. E., et al. (2003). Collagen XXIV, a Vertebrate Fibrillar Collagen with Structural Features of Invertebrate Collagens. J. Biol. Chem. 278 (44), 43236–43244. doi:10.1074/jbc.m302112200
Kovanen, V., Suominen, H., and Heikkinen, E. (1984). Collagen of Slow Twitch and Fast Twitch Muscle Fibres in Different Types of Rat Skeletal Muscle. Europ. J. Appl. Physiol. 52 (2), 235–242. doi:10.1007/bf00433399
Krzywinski, M., Schein, J., Birol, İ., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an Information Aesthetic for Comparative Genomics. Genome Res. 19 (9), 1639–1645. doi:10.1101/gr.092759.109
Kuo, C. K., Petersen, B. C., and Tuan, R. S. (2008). Spatiotemporal Protein Distribution of TGF-Βs, Their Receptors, and Extracellular Matrix Molecules during Embryonic Tendon Development. Dev. Dyn. 237 (5), 1477–1489. doi:10.1002/dvdy.21547
Kuraku, S. (2013). Impact of Asymmetric Gene Repertoire between Cyclostomes and Gnathostomes. Semin. Cel. Dev. Biol. 24, 119–127. doi:10.1016/j.semcdb.2012.12.009
Kuraku, S. (2008). Insights into Cyclostome Phylogenomics: pre-2R or post-2R. Zoolog. Sci. 25 (10), 960–968. doi:10.2108/zsj.25.960
Kuraku, S., and Kuratani, S. (2006). Time Scale for Cyclostome Evolution Inferred with a Phylogenetic Diagnosis of Hagfish and Lamprey cDNA Sequences. Zoolog. Sci. 23 (12), 1053–1064. doi:10.2108/zsj.23.1053
Kusakabe, R., Kuraku, S., and Kuratani, S. (2011). Expression and Interaction of Muscle-Related Genes in the Lamprey Imply the Evolutionary Scenario for Vertebrate Skeletal Muscle, in Association with the Acquisition of the Neck and Fins. Develop. Biol. 350 (1), 217–227. doi:10.1016/j.ydbio.2010.10.029
Kusakabe, R., and Kuratani, S. (2005). Evolution and Developmental Patterning of the Vertebrate Skeletal Muscles: Perspectives from the Lamprey. Dev. Dyn. 234 (4), 824–834. doi:10.1002/dvdy.20587
Kusakabe, R., and Kuratani, S. (2007). Evolutionary Perspectives from Development of Mesodermal Components in the Lamprey. Dev. Dyn. 236 (9), 2410–2420. doi:10.1002/dvdy.21177
Kusakabe, R., Takechi, M., Tochinai, S., and Kuratani, S. (2004). Lamprey Contractile Protein Genes Mark Different Populations of Skeletal Muscles during Development. J. Exp. Zool. 302B (2), 121–133. doi:10.1002/jez.b.20009
Kuzmin, E., VanderSluis, B., Nguyen Ba, A. N., Wang, W., Koch, E. N., Usaj, M., et al. (2020). Exploring Whole-Genome Duplicate Gene Retention with Complex Genetic Interaction Analysis. Science 368 (6498), 368. doi:10.1126/science.aaz5667
Lawrence, E. A., Kague, E., Aggleton, J. A., Harniman, R. L., Roddy, K. A., and Hammond, C. L. (2018). The Mechanical Impact of Col11a2 Loss on Joints; Col11a2 Mutant Zebrafish Show Changes to Joint Development and Function, Which Leads to Early-Onset Osteoarthritis. Phil. Trans. R. Soc. B 373 (1759), 20170335. doi:10.1098/rstb.2017.0335
Le Guellec, D., Morvan-Dubois, G., and Sire, J. Y. (2003). Skin Development in Bony Fish with Particular Emphasis on Collagen Deposition in the Dermis of the Zebrafish (Danio rerio). Int. J. Dev. Biol. 48 (2-3), 217–231. doi:10.1387/ijdb.15272388
Leéjard, V., Brideau, G., Blais, F., Salingcarnboriboon, R., Wagner, G., Roehrl, M. H. A., et al. (2007). Scleraxis and NFATc Regulate the Expression of the Pro-α1 (I) Collagen Gene in Tendon Fibroblasts. J. Biol. Chem. 282 (24), 17665–17675. doi:10.1074/jbc.M610113200
Lees, J. F., Tasab, M., and Bulleid, N. J. (1997). Identification of the Molecular Recognition Sequence Which Determines the Type-specific Assembly of Procollagen. EMBO J. 16 (5), 908–916. doi:10.1093/emboj/16.5.908
Li, Y., Lacerda, D. A., Warman, M. L., Beier, D. R., Yoshioka, H., Ninomiya, Y., et al. (1995). A Fibrillar Collagen Gene, Col11a1, Is Essential for Skeletal Morphogenesis. Cell 80 (3), 423–430. doi:10.1016/0092-8674(95)90492-1
Lien, H.-W., Yang, C.-H., Cheng, C.-H., Hung, C.-C., Liao, W.-H., Hwang, P.-P., et al. (2013). A Novel Zinc finger Protein 219-like (ZNF219L) Is Involved in the Regulation of Collagen Type 2 Alpha 1a (Col2a1a) Gene Expression in Zebrafish Notochord. Int. J. Biol. Sci. 9 (9), 872–886. doi:10.7150/ijbs.7126
Lincoln, J., Florer, J. B., Deutsch, G. H., Wenstrup, R. J., and Yutzey, K. E. (2006). ColVa1 and ColXIa1 Are Required for Myocardial Morphogenesis and Heart Valve Development. Dev. Dyn. 235 (12), 3295–3305. doi:10.1002/dvdy.20980
Liu, X., Wu, H., Byrne, M., Krane, S., and Jaenisch, R. (1997). Type III Collagen Is Crucial for Collagen I Fibrillogenesis and for normal Cardiovascular Development. Proc. Natl. Acad. Sci. 94 (5), 1852–1856. doi:10.1073/pnas.94.5.1852
Luther, P., and Squire, J. (2014). The Intriguing Dual Lattices of the Myosin Filaments in Vertebrate Striated Muscles: Evolution and Advantage. Biology 3 (4), 846–865. doi:10.3390/biology3040846
Mangos, S., Lam, P. Y., Zhao, A., Liu, Y., Mudumana, S., Vasilyev, A., et al. (2010). The ADPKD Genes Pkd1a/b and Pkd2 Regulate Extracellular Matrix Formation. Dis. Model. Mech. 3 (5-6), 354–365. doi:10.1242/dmm.003194
Martik, M. L., Gandhi, S., Uy, B. R., Gillis, J. A., Green, S. A., Simoes-Costa, M., et al. (2019). Evolution of the New Head by Gradual Acquisition of Neural Crest Regulatory Circuits. Nature 574 (7780), 675–678. doi:10.1038/s41586-019-1691-4
Matsushima, N., Ohyanagi, T., Tanaka, T., and Kretsinger, R. H. (2000). Super-motifs and Evolution of Tandem Leucine-Rich Repeats within the Small Proteoglycans?biglycan, Decorin, Lumican, Fibromodulin, PRELP, Keratocan, Osteoadherin, Epiphycan, and Osteoglycin. Proteins 38 (2), 210–225. doi:10.1002/(sici)1097-0134(20000201)38:2<210:aid-prot9>3.0.co;2-1
McBurney, K. M., Keeley, F. W., Kibenge, F. S., and Wright, G. M. (1996). Spatial and Temporal Distribution of Lamprin mRNA during Chondrogenesis of Trabecular Cartilage in the Sea Lamprey. Anat. Embryol. (Berl) 193 (5), 419–426. doi:10.1007/BF00185873
McCauley, D. W. (2008). SoxE, Type II Collagen, and Evolution of the Chondrogenic Neural Crest. Zoolog. Sci. 25 (10), 982–989. doi:10.2108/zsj.25.982
Miller, M. A., Pfeiffer, W., and Schwartz, T. (2011). “The CIPRES Science Gateway: a Community Resource for Phylogenetic Analyses,” in Proceedings of the 2011 TeraGrid Conference: Extreme Digital Discovery, Salt Lake City, Utah, USA, July 18-21, 2011.
Mugue, N. S., and Ozernyuk, N. D. (2006). Comparative Structural Analysis of Myosin Light Chains and Gene Duplication in Fish. Biol. Bull. Russ. Acad. Sci. 33 (1), 30–34. doi:10.1134/s1062359006010043
Mukhi, S., Cai, L., and Brown, D. D. (2010). Gene Switching at Xenopus laevis Metamorphosis. Develop. Biol. 338 (2), 117–126. doi:10.1016/j.ydbio.2009.10.041
Nakamura, Y.-N., Iwamoto, H., Tabata, S., and Ono, Y. (2003). Comparison of Collagen Fibre Architecture between Slow-Twitch Cranial and Fast-Twitch Caudal Parts of Broiler M. Latissimus Dorsi. Br. Poult. Sci. 44 (3), 374–379. doi:10.1080/00071660310001598346
Nakao, T. (1977). Electron Microscopic Studies on the Myotomes of Larval Lamprey,Lampetra Japonica. Anat. Rec. 187 (3), 383–403. doi:10.1002/ar.1091870309
Nathan, E., Monovich, A., Tirosh-Finkel, L., Harrelson, Z., Rousso, T., Rinon, A., et al. (2008). The Contribution of Islet1-Expressing Splanchnic Mesoderm Cells to Distinct Branchiomeric Muscles Reveals Significant Heterogeneity in Head Muscle Development. Development 135 (4), 647–657. doi:10.1242/dev.007989
Nikitina, N., Bronner-Fraser, M., and Sauka-Spengler, T. (2009). Microinjection of RNA and Morpholino Oligos into Lamprey Embryos. Cold Spring Harb Protoc. 2009 (1), pdb. doi:10.1101/pdb.prot5123
Ohtani, K., Yao, T., Kobayashi, M., Kusakabe, R., Kuratani, S., and Wada, H. (2008). Expression of Sox and Fibrillar Collagen Genes in Lamprey Larval Chondrogenesis with Implications for the Evolution of Vertebrate Cartilage. J. Exp. Zool. 310B (7), 596–607. doi:10.1002/jez.b.21231
Ordahl, C. P., Berdougo, E., Venters, S. J., and Denetclaw, W. F. (2001). The Dermomyotome Dorsomedial Lip Drives Growth and Morphogenesis of Both the Primary Myotome and Dermomyotome Epithelium. Development 128 (10), 1731–1744. doi:10.1242/dev.128.10.1731
Ota, K. G., and Kuratani, S. (2010). Expression Pattern of Two Collagen Type 2 Alpha1 Genes in the Japanese Inshore Hagfish (Eptatretus burgeri) with Special Reference to the Evolution of Cartilaginous Tissue. J. Exp. Zool B Mol. Dev. Evol. 314 (2), 157–165. doi:10.1002/jez.b.21322
Owens, N. D. L., Blitz, I. L., Lane, M. A., Patrushev, I., Overton, J. D., Gilchrist, M. J., et al. (2016). Measuring Absolute RNA Copy Numbers at High Temporal Resolution Reveals Transcriptome Kinetics in Development. Cel Rep. 14 (3), 632–647. doi:10.1016/j.celrep.2015.12.050
Pace, J. M., Corrado, M., Missero, C., and Byers, P. H. (2003). Identification, Characterization and Expression Analysis of a New Fibrillar Collagen Gene, COL27A1. Matrix Biol. 22 (1), 3–14. doi:10.1016/s0945-053x(03)00007-6
Panopoulou, G., and Poustka, A. (2005). Timing and Mechanism of Ancient Vertebrate Genome Duplications - the Adventure of a Hypothesis. Trends. Genet. 21 (10), 559–567. doi:10.1016/j.tig.2005.08.004
Parker, W. K. (1883). On the Skeleton of the Marsipobranch Fishes. Part II. Petromyzon. Philos. Trans. R. Soc. Lond. 174, 411–457.
Peters, A., and Mackay, B. (1961). The Structure and Innervation of the Myotomes of the Lamprey. J. Anat. 95 (Pt 4), 575–585.
Rambaut, A., FigTree V1. 4.2, a Graphical Viewer of Phylogenetic Trees. Available from left angle bracket http://tree. bio. ed. ac. uk/software/figtree/right angle bracket (Accessed 30.8.2019), 2014.
Rauch, G. J., Lyons, D. A., Middendorf, I., Friedlander, B., Arana, N., Reyes, T., et al. (2003). Submission and Curation of Gene Expression Data(http://zfin.org).
Rescan, P.-Y. (2019). Development of Myofibres and Associated Connective Tissues in Fish Axial Muscle: Recent Insights and Future Perspectives. Differentiation 106, 35–41. doi:10.1016/j.diff.2019.02.007
Rescan, P. Y., Montfort, J., Fautrel, A., Rallière, C., and Lebret, V. (2013). Gene Expression Profiling of the Hyperplastic Growth Zones of the Late trout Embryo Myotome Using Laser Capture Microdissection and Microarray Analysis. BMC genomics 14 (1), 173–211. doi:10.1186/1471-2164-14-173
Robson, P., Wright, G. M., Sitarz, E., Maiti, A., Rawat, M., Youson, J. H., et al. (1993). Characterization of Lamprin, an Unusual Matrix Protein from Lamprey Cartilage. Implications for Evolution, Structure, and Assembly of Elastin and Other Fibrillar Proteins. J. Biol. Chem. 268 (2), 1440–1447. doi:10.1016/s0021-9258(18)54095-3
Root, Z. D., Gould, C., Brewer, M., Jandzik, D., and Medeiros, D. M. (2021). Comparative Approaches in Vertebrate Cartilage Histogenesis and Regulation: Insights from Lampreys and Hagfishes. Diversity 13 (9), 435. doi:10.3390/d13090435
Root, Z. D., Jandzik, D., Allen, C., Brewer, M., Romášek, M., Square, T., et al. (2021). Lamprey Lecticans Link New Vertebrate Genes to the Origin and Elaboration of Vertebrate Tissues. Develop. Biol. 476, 282–293. doi:10.1016/j.ydbio.2021.03.020
Roshan, U., and Livesay, D. R. (2006). Probalign: Multiple Sequence Alignment Using Partition Function Posterior Probabilities. Bioinformatics 22 (22), 2715–2721. doi:10.1093/bioinformatics/btl472
Roulet, M., Ruggiero, F., Karsenty, G., and LeGuellec, D. (2007). A Comprehensive Study of the Spatial and Temporal Expression of the Col5a1 Gene in Mouse Embryos: a Clue for Understanding Collagen V Function in Developing Connective Tissues. Cel Tissue Res 327 (2), 323–332. doi:10.1007/s00441-006-0294-1
Rowe, R. W. D. (1981). Morphology of Perimysial and Endomysial Connective Tissue in Skeletal Muscle. Tissue and Cell 13 (4), 681–690. doi:10.1016/s0040-8166(81)80005-5
Sambasivan, R., Gayraud-Morel, B., Dumas, G., Cimper, C., Paisant, S., Kelly, R. G., et al. (2009). Distinct Regulatory Cascades Govern Extraocular and Pharyngeal Arch Muscle Progenitor Cell Fates. Develop. Cel. 16 (6), 810–821. doi:10.1016/j.devcel.2009.05.008
Schneider, A. (1879). Anatomie und entwickelungsgeschichte von Petromyzon und ammocoetes. Beitrage zur vergleichenden. Anatomie und Entwickelungsgeschichte der Wirbeltiere. Berlin: Reimer., 85–92.
Schweitzer, R., Zelzer, E., and Volk, T. (2010). Connecting Muscles to Tendons: Tendons and Musculoskeletal Development in Flies and Vertebrates. Development 137 (17), 2807–2817. doi:10.1242/dev.047498
Seipel, K., and Schmid, V. (2005). Evolution of Striated Muscle: Jellyfish and the Origin of Triploblasty. Develop. Biol. 282 (1), 14–26. doi:10.1016/j.ydbio.2005.03.032
Session, A. M., Uno, Y., Kwon, T., Chapman, J. A., Toyoda, A., Takahashi, S., et al. (2016). Genome Evolution in the Allotetraploid Frog Xenopus laevis. Nature 538 (7625), 336–343. doi:10.1038/nature19840
Shaffer, J. F., and Gillis, T. E. (2010). Evolution of the Regulatory Control of Vertebrate Striated Muscle: the Roles of Troponin I and Myosin Binding Protein-C. Physiol. genomics 42 (3), 406–419. doi:10.1152/physiolgenomics.00055.2010
Sharma, P., Ruel, T. D., Kocha, K. M., Liao, S., and Huang, P. (2019). Single Cell Dynamics of Embryonic Muscle Progenitor Cells in Zebrafish. Development 146 (14), dev178400. doi:10.1242/dev.178400
Shukunami, C., Takimoto, A., Miura, S., Nishizaki, Y., and Hiraki, Y. (2008). Chondromodulin-I and Tenomodulin Are Differentially Expressed in the Avascular Mesenchyme during Mouse and Chick Development. Cel Tissue Res 332 (1), 111–122. doi:10.1007/s00441-007-0570-8
Simakov, O., Marlétaz, F., Yue, J.-X., O’Connell, B., Jenkins, J., Brandt, A., et al. (2020). Deeply Conserved Synteny Resolves Early Events in Vertebrate Evolution. Nat. Ecol. Evol. 4 (6), 820–830. doi:10.1038/s41559-020-1156-z
Sleboda, D. A., Stover, K. K., and Roberts, T. J. (2020). Diversity of Extracellular Matrix Morphology in Vertebrate Skeletal Muscle. J. Morphol. 281 (2), 160–169. doi:10.1002/jmor.21088
Smith, J. J., Kuraku, S., Holt, C., Sauka-Spengler, T., Jiang, N., Campbell, M. S., et al. (2013). Sequencing of the Sea Lamprey (Petromyzon marinus) Genome Provides Insights into Vertebrate Evolution. Nat. Genet. 45 (4), 415–422. doi:10.1038/ng.2568
Smith, J. J., and Keinath, M. C. (2015). The Sea Lamprey Meiotic Map Improves Resolution of Ancient Vertebrate Genome Duplications. Genome Res. 25 (8), 1081–1090. doi:10.1101/gr.184135.114
Square, T., Jandzik, D., Cattell, M., Hansen, A., and Medeiros, D. M. (2016). Embryonic Expression of Endothelins and Their Receptors in Lamprey and Frog Reveals Stem Vertebrate Origins of Complex Endothelin Signaling. Sci. Rep. 6 (1), 34282–34313. doi:10.1038/srep34282
Square, T. A., Jandzik, D., Massey, J. L., Romášek, M., Stein, H. P., Hansen, A. W., et al. (2020). Evolution of the Endothelin Pathway Drove Neural Crest Cell Diversification. Nature 585 (7826), 563–568. doi:10.1038/s41586-020-2720-z
Steinbacher, P., Haslett, J. R., Six, M., Gollmann, H. P., Sänger, A. M., and Stoiber, W. (2006). Phases of Myogenic Cell Activation and Possible Role of Dermomyotome Cells in Teleost Muscle Formation. Dev. Dyn. 235 (11), 3132–3143. doi:10.1002/dvdy.20950
Steinmetz, P. R. H., Kraus, J. E. M., Larroux, C., Hammel, J. U., Amon-Hassenzahl, A., Houliston, E., et al. (2012). Independent Evolution of Striated Muscles in Cnidarians and Bilaterians. Nature 487 (7406), 231–234. doi:10.1038/nature11180
Sun, M., Luo, E. Y., Adams, S. M., Adams, T., Ye, Y., Shetye, S. S., et al. (2020). Collagen XI Regulates the Acquisition of Collagen Fibril Structure, Organization and Functional Properties in Tendon. Matrix Biol. 94, 77–94. doi:10.1016/j.matbio.2020.09.001
Suzuki, T., Kusakabe, M., Nakayama, K., and Nishida, E. (2012). The Protein Kinase MLTK Regulates Chondrogenesis by Inducing the Transcription Factor Sox6. Development 139 (16), 2988–2998. doi:10.1242/dev.078675
Tarazona, O. A., Slota, L. A., Lopez, D. H., Zhang, G., and Cohn, M. J. (2016). The Genetic Program for Cartilage Development Has Deep Homology within Bilateria. Nature 533 (7601), 86–89. doi:10.1038/nature17398
Terrill, D. F., Henderson, C. M., and Anderson, J. S. (2018). New Applications of Spectroscopy Techniques Reveal Phylogenetically Significant Soft Tissue Residue in Paleozoic Conodonts. J. Anal. Spectrom. 33 (6), 992–1002. doi:10.1039/c7ja00386b
Thisse, B., Pflumio, S., Fürthauer, M., Loppin, B., Heyer, V., Degrave, A., et al. (2001). Expression of the Zebrafish Genome during Embryogenesis. ZFIN direct data submission.
Thisse, C. (2005). High Throughput Expression Analysis of ZF-Models Consortium Clones. ZFIN Direct Data Submission.
Tomoaia, G., and Pasca, R.-D. (2015). On the Collagen Mineralization. A Review. Med. Pharm. Rep. 88 (1), 15–22. doi:10.15386/cjmed-359
Tsumaki, N., and Kimura, T. (1995). Differential Expression of an Acidic Domain in the Amino-Terminal Propeptide of Mouse Pro- 2(XI) Collagen by Complex Alternative Splicing. J. Biol. Chem. 270 (5), 2372–2378. doi:10.1074/jbc.270.5.2372
Van de Peer, Y., Maere, S., and Meyer, A. (2009). The Evolutionary Significance of Ancient Genome Duplications. Nat. Rev. Genet. 10 (10), 725–732. doi:10.1038/nrg2600
Vandekerckhove, J., and Weber, K. (1984). Chordate Muscle Actins Differ Distinctly from Invertebrate Muscle Actins. J. Mol. Biol. 179 (3), 391–413. doi:10.1016/0022-2836(84)90072-x
Wada, H., Okuyama, M., Satoh, N., and Zhang, S. (2006). Molecular Evolution of Fibrillar Collagen in Chordates, with Implications for the Evolution of Vertebrate Skeletons and Chordate Phylogeny. Evol. Dev. 8 (4), 370–377. doi:10.1111/j.1525-142x.2006.00109.x
Wang, C., Brisson, B. K., Terajima, M., Li, Q., Hoxha, K. h., Han, B., et al. (2020). Type III Collagen Is a Key Regulator of the Collagen Fibrillar Structure and Biomechanics of Articular Cartilage and Meniscus. Matrix Biol. 85-86, 47–67. doi:10.1016/j.matbio.2019.10.001
Wang, W., Olson, D., Liang, G., Franceschi, R. T., Li, C., Wang, B., et al. (2012). Collagen XXIV (Col24α1) Promotes Osteoblastic Differentiation and Mineralization through TGF-β/Smads Signaling Pathway. Int. J. Biol. Sci. 8 (10), 1310–1322. doi:10.7150/ijbs.5136
Wapinski, I., Pfeffer, A., Friedman, N., and Regev, A. (2007). Natural History and Evolutionary Principles of Gene Duplication in Fungi. Nature 449 (7158), 54–61. doi:10.1038/nature06107
Warkman, A. S., Whitman, S. A., Miller, M. K., Garriock, R. J., Schwach, C. M., Gregorio, C. C., et al. (2012). Developmental Expression and Cardiac Transcriptional Regulation of Myh7b, a Third Myosin Heavy Chain in the Vertebrate Heart. Cytoskeleton 69 (5), 324–335. doi:10.1002/cm.21029
Wenstrup, R. J., Smith, S. M., Florer, J. B., Zhang, G., Beason, D. P., Seegmiller, R. E., et al. (2011). Regulation of Collagen Fibril Nucleation and Initial Fibril Assembly Involves Coordinate Interactions with Collagens V and XI in Developing Tendon. J. Biol. Chem. 286 (23), 20455–20465. doi:10.1074/jbc.m111.223693
Wheeler, D. L., Barrett, T., Benson, D. A., Bryant, S. H., Canese, K., Chetvernin, V., et al. (2007). Database Resources of the National center for Biotechnology Information. Nucleic Acids Res. 35 (Suppl. l_1), D5–D12. doi:10.1093/nar/gkl1031
Witjes, L., Van Troys, M., Vandekerckhove, J., Vandepoele, K., and Ampe, C. (2019). A New Evolutionary Model for the Vertebrate Actin Family Including Two Novel Groups. Mol. Phylogenet. Evol. 141, 106632. doi:10.1016/j.ympev.2019.106632
Witten, P. E., Huysseune, A., and Hall, B. K. (2010). A Practical Approach for the Identification of the many Cartilaginous Tissues in Teleost Fish. J. Appl. Ichthyology 26 (2), 257–262. doi:10.1111/j.1439-0426.2010.01416.x
Wright, G. M., Keeley, F. W., Youson, J. H., and Babineau, D. L. (1984). Cartilage in the atlantic Hagfish,Myxine glutinosa. Am. J. Anat. 169 (4), 407–424. doi:10.1002/aja.1001690404
Wright, G. M., Keeley, F. W., and Youson, J. H. (1983). Lamprin: a New Vertebrate Protein Comprising the Major Structural Protein of Adult Lamprey Cartilage. Experientia 39 (5), 495–497. doi:10.1007/bf01965172
Yamaguchi, K., Matsuo, N., Sumiyoshi, H., Fujimoto, N., Iyama, K., Yanagisawa, S., et al. (2005). Pro-α3(V) Collagen Chain Is Expressed in Bone and its Basic N-Terminal Peptide Adheres to Osteosarcoma Cells. Matrix Biol. 24 (4), 283–294. doi:10.1016/j.matbio.2005.03.006
Yao, T., Ohtani, K., Kuratani, S., and Wada, H. (2011). Development of Lamprey Mucocartilage and its Dorsal-Ventral Patterning by Endothelin Signaling, with Insight into Vertebrate Jaw Evolution. J. Exp. Zool. 316B (5), 339–346. doi:10.1002/jez.b.21406
Yates, A. D., Achuthan, P., Akanni, W., Allen, J., Allen, J., Alvarez-Jarreta, J., et al. (2020). Ensembl 2020. Nucleic Acids Res. 48 (D1), D682–D688. doi:10.1093/nar/gkz966
Yokoyama, H., Morino, Y., and Wada, H. (2019). Identification of a Unique Lamprey Gene with Tandemly Repeated Sequences and Pharyngeal Chondrocyte-specific Expression. Gene 701, 9–14. doi:10.1016/j.gene.2019.02.094
Yoon, J. H., and Halper, J. (2005). Tendon Proteoglycans: Biochemistry and Function. J. Musculoskelet. Neuronal Interact 5 (1), 22–34.
Yoshioka, H., Iyama, K.-I., Inoguchi, K., Khaleduzzaman, M., Ninomiya, Y., and Ramirez, F. (1995). Developmental Pattern of Expression of the Mouse α1(XI) Collagen Gene (Col11a1). Dev. Dyn. 204 (1), 41–47. doi:10.1002/aja.1002040106
Yutaka, T. (1988). Normal Stages of Development in the Lamprey, Lampetra Reissued (Dybowski). Zoolog. Sci. 5 (1), 109–118.
Zhang, G., and Cohn, M. J. (2006). Hagfish and Lancelet Fibrillar Collagens Reveal that Type II Collagen-Based Cartilage Evolved in Stem Vertebrates. Proc. Natl. Acad. Sci. 103 (45), 16829–16833. doi:10.1073/pnas.0605630103
Zhang, G., Miyamoto, M. M., and Cohn, M. J. (2006). Lamprey Type II Collagen and Sox9 Reveal an Ancient Origin of the Vertebrate Collagenous Skeleton. Proc. Natl. Acad. Sci. 103 (9), 3180–3185. doi:10.1073/pnas.0508313103
Keywords: lamprey, fibrillar collagen, evolution, cartilage, development
Citation: Root ZD, Allen C, Gould C, Brewer M, Jandzik D and Medeiros DM (2022) A Comprehensive Analysis of Fibrillar Collagens in Lamprey Suggests a Conserved Role in Vertebrate Musculoskeletal Evolution. Front. Cell Dev. Biol. 10:809979. doi: 10.3389/fcell.2022.809979
Received: 05 November 2021; Accepted: 18 January 2022;
Published: 15 February 2022.
Edited by:
Brian Frank Eames, University of Saskatchewan, CanadaReviewed by:
Christoph Winkler, National University of Singapore, SingaporePatrick Tschopp, University of Basel, Switzerland
Copyright © 2022 Root, Allen, Gould, Brewer, Jandzik and Medeiros. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zachary D. Root, emFjaGFyeS5yb290QGNvbG9yYWRvLmVkdQ==; Daniel M. Medeiros, ZGFuaWVsLm1lZGVpcm9zQGNvbG9yYWRvLmVkdQ==
 Zachary D. Root
Zachary D. Root Cara Allen1
Cara Allen1 David Jandzik
David Jandzik Daniel M. Medeiros
Daniel M. Medeiros