
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell Dev. Biol., 26 January 2022
Sec. Molecular and Cellular Oncology
Volume 10 - 2022 | https://doi.org/10.3389/fcell.2022.805507
This article is part of the Research TopicAngiogenesis and Tumor MetastasisView all 16 articles
 Xin-yu Li1,2†
Xin-yu Li1,2† Wei-Ning Ma3†
Wei-Ning Ma3† Li-xin Su1†
Li-xin Su1† Yuchen Shen1†
Yuchen Shen1† Liming Zhang1
Liming Zhang1 Yuhao Shao1
Yuhao Shao1 Deming Wang1
Deming Wang1 Zhenfeng Wang1
Zhenfeng Wang1 Ming-Zhe Wen1
Ming-Zhe Wen1 Xi-tao Yang1*
Xi-tao Yang1*Background: Several new blood vessels are formed during the process of tumor development. These new blood vessels provide nutrients and water for tumour growth, while spreading tumour cells to distant areas and forming new metastases in different parts of the body. The available evidence suggests that tumour angiogenesis is closely associated with the tumour microenvironment and is regulated by a variety of pro-angiogenic factors and/or angiogenic inhibitors.
Methods: In the present study, a comprehensive characterization of angiogenesis genes expression was performed in a pan-cancer analysis across the 33 human cancer types. Further, genetic data from several public databases were also used in the current study. An angiogenesis score was assigned to The Cancer Genome Atlas (TCGA) pan-cancer data, with one angiogenesis score as per sample for each tumour.
Results: It was found that angiogenesis genes vary across cancer types, and are associated with a number of genomic and immunological features. Further, it was noted that macrophages and iTreg infiltration were generally higher in tumours with high angiogenesis scores, whereas lymphocytes and B cells showed the opposite trend. Notably, NK cells showed significantly different correlations among cancer types. Furthermore, results of the present study showed that a high angiogenesis score was associated with poor survival and aggressive types of cancer in most of the cancer types.
Conclusion: In conclusion, the current study evidently showed that the expression of angiogenesis genes is a key feature of tumour biology that has a major impact on prognosis of patient with cancers.
Given that malignant tumours need supplies of oxygen and nutrients to survive and thrive, they require adequate vascularization to access the blood circulation system (Lugano et al., 2020). Previous studies have shown that rapid growth of tumours requires a large supply of nutrients from the blood as compared with dormant tumours. Therefore, it should be noted that initiation of tumour angiogenesis is a major factor for tumor development (Azoitei et al., 2010). Results of a previous clinical trial showed that anti-angiogenic therapy can be successfully used to treat cancer (Hurwitz et al., 2004). Recently, anti-tumour angiogenesis research has evolved from the early non-specific embolisation and severance of tumour vessels to a new level of specific and targeted blockade of tumour vessels (Hida et al., 2008; Mitsuhashi et al., 2015). However, it was found that the anti-angiogenesis therapy only provided a short-term relief and inhibition of tumour growth before resistance is developed (Lugano et al., 2020). Further, emerging evidence shows that angiogenesis and immunosuppression frequently occur simultaneously in response to this crosstalk. Accordingly, strategies combining both anti-angiogenic therapy and immunotherapy have potential to tip the balance of the tumor microenvironment and improve treatment response (Solimando et al., 2020).
The present study provides a comprehensive assessment of the genomic and clinical characteristics of angiogenesis genes in 33 solid tumours. In addition, the angiogenesis score of each patients with cancers was also assessed in the current study. Results of this study found that angiogenesis is correlated with distinct genomic and immunologic tumour characteristics. Moreover, it was found that expression of angiogenesis genes has prognostic and predictive value in outcomes of both patients and their response to immunotherapy. Therefore, the analyses reported in the current study provide the first comprehensive survey of angiogenesis genes expression across 33 cancer types.
The data of 33 tumors in The Cancer Genome Atlas (TCGA) including the mRNA data, mutation data, and clinical data were collected from UCSC Xena. The data on expression of gene for different tissues were retrieved from GTEx, whereas the angiogenesis relevant data was downloaded from the hallmark gene sets of the msigdb database. The flow of this articles is as presented in Figure 1.
The present study analyzed differential expression of angiogenesis genes in tumour samples (from TCGA database) as compared with that of the normal samples (from TCGA and GTEx database). The “limma” R package was used to identify differentially expressed genes (DEGs), with FDR <0.05, and |log2FC| ≥ 1 as the cut-offs. Next, “ggplot” and “reshape” R packages were used to generate a heatmap to visualize the results obtained.
The ‘Survival’ R package was used to conduct univariate Cox regression analysis for angiogenesis genes. Further, the p-value and HR-values were then extracted for heatmap presentation. The number of genes was then counted for which Cox analysis was significant in all tumours. Notably, a risk factor was considered if the score was increased by 1, whereas a protective factor was considered if the score was decreased by 1 and the final score was used as the risk score.
In the present study, Pearson’s r was used to explore correlations between different angiogenesis genes and the obtained results were presented using a heatmap. Genetic variation data was downloaded from the cbioportal database including mutations, fusions, amplifications, deletions, and multiple variants. Angiogenesis genes variants were then calculated for each tumour. Finally, mutation of angiogenesis genes in different cancers was explored using Gene Set Cancer Analysis (GSCA) (http://bioinfo.life.hust.edu.cn/GSCA).
Previous studies have suggested that DNA hypomethylation promote increased expression of many oncogenes, whereas DNA hypermethylation has also been shown to silence tumour suppressor genes (Wang et al., 2020). In the present study, the mRNA expression and methylation data for the angiogenesis genes were merged using the GSCA, followed by the analysis of the differential methylation. Further, a Student’s t test was performed to determine the methylation difference between tumor and normal samples, and the p-value was adjusted using the FDR. Notably, FDR ≤0.05 was considered statistically significant. Pearson’s or Spearman’s correlation was then performed to explore the association between methylation and expression levels. Finally, HR for prognostic value of gene methylation status was calculated using Cox regression analysis. The information of copy number variations (CNVs) of the 33 types of tumors from the TCGA database and determined using GISTIC 2.0 software were also totally collected. In addition, the percentage of CNVs and CNVs correlation with mRNA were calculated by Spearman correlation analyses by corrplot package. The CNVs were classified into 2 categories (homozygous and heterozygous), including amplification and deletion, which represents the presence of CNVs on only one chromosome or two chromosomes. It is worth noting that only genes with CNV >5% in cancer were discussed in the present study. The associations between paired mRNA expression and paired CNV percentage samples were explored based on the Person’s product moment correlation coefficient and t-distribution (Schlattl et al., 2011). The statistical significant threshold was set as FDR p-value ≤0.05.
The angiogenesis score for each sample in the present study was calculated using the “GSVA” R package (Kim et al., 2020). The size of the angiogenesis score in each tumour and explored the differences in angiogenesis score across clinical stages. Data visualization was done using “ggplot” R package (Shatalova et al., 2020). Next, the relationship between the angiogenesis score and prognoses in patients with cancer was explored. Survival estimates were calculated using Kaplan-Meier (KM) and Cox regression models. The “GSVA” R package was used to perform GSVA analysis, with t value >2 and FDR <0.05 as the cut-offs. Notably, the gene set used in this study was MsigDB dataset, HALLMARK pathway of the database. Single-sample gene set enrichment analysis (ssGSEA) was also employed to evaluate the enrichment scores for each sample. Therefore, the present study also evaluated the correlation between angiogenesis score and pathways. Finally, the correlation was visualized using “ggplot” R package (Shatalova et al., 2020).
Immune and stromal scores were calculated using the ESTIMATE algorithm with the help of “limma” and “estimate” R packages. The relationship between the angiogenesis score and tumor microenvironment in accordance with a previous study was also assessed to validate the results obtained in the current study (Zeng et al., 2019). Finally, the data was plotted using the “pheatmap” R package. Considering the association of immune infiltration level with survival and prognosis in cancers, the correlation between angiogenesis score and immune infiltration level was hence explored. On the other hand, the CIBERSORT algorithm was used to estimate data on tumor-infiltrating immune cells (Thorsson et al., 2018; Huang et al., 2021). Immune Cell Abundance Identifier and TIMER2 were used to evaluate the correlation of angiogenesis score with immune infiltration across all tumors in the TCGA database. Plots were then performed using “ggplot” R package (Shatalova et al., 2020). The use of immunotherapy in cancer treatment has evolved rapidly and several therapeutic antibodies have reached the clinical practices in recent years (Hultqvist et al., 2017; Wu and Shih, 2018). Therefore, the present study used immunotherapy data to explore the impact of high or low angiogenesis score on the prognosis of immunotherapy patients. Specifically, immunotherapy data was used from IMvigor210 dataset, GSE78220, GSE135222, and GSCA.
The correlation between immune genes and angiogenesis score was also analyzed in the present study. The analyzed target genes included MHC genes, chemokines, chemokine receptors, and immunosuppressive genes. Recently, tumor mutation burden (TMB) has emerged as a predictive indicator for tumor immunotherapy, which aids in prognostic prediction of immunotherapy in some tumors such as lung cancer, malignant melanoma, and colon cancer (Kelley et al., 2020; Saeed and Salem, 2020). Microsatellite instability (MSI) is a genetic change that has been shown to be closely associated with tumor prognosis (De Palma et al., 2019; Li et al., 2020a). In the present study, the TMB score was calculated using R software and corrected by dividing the total length of the exon. In addition, the MSI scores for all samples were obtained from the somatic mutation data which was downloaded from TCGA database. Finally, Spearman correlation analysis was performed to assess the correlation of angiogenesis score with TMB and MSI scores.
Using TCGA and GTEx datasets, we evaluated the differential expressions of angiogenesis-related genes between cancers and normal tissues. The angiogenesis genes were aberrantly expressed in 31 tumor types (p < 0.05, Figure 2A). We found that angiogenesis genes tended to be significantly downregulated in BRCA (Breast invasive carcinoma) and UCES(Uterine Corpus Endometrial Carcinoma), but upregulated in PAAD (Pancreatic adenocarcinoma), GBM (Glioblastoma multiforme). These findings are consistent with several previous studies (Ardito et al., 2008; Liu et al., 2019). For example, SPP1 was upregulated in most tumor types. SPP1 enhances tumor development and its overexpression is associated with poor prognostic outcomes for melanoma, while its silencing suppresses melanoma cell proliferation, migration, as well as invasion (Komatsu et al., 2012; Deng et al., 2020). These findings imply that dysregulated expressions of angiogenesis-associated genes are involved in cancer initiation and development. Since angiogenesis-related genes play critical roles in cancer metastasis (Firestone and Sundar, 2009), we evaluated the associations between their expression levels and survival outcomes. In at least one cancer type, all angiogenesis-related genes were associated with overall survival outcomes (Figure 2B). In many cancer types, patients with elevated angiogenesis-associated gene levels have significantly poor survival outcomes, compared to those with suppressed levels. Risk scores revealed that most genes were unfavourable for patient prognosis. These findings imply that angiogenesis-related gene levels are associated with prognostic outcomes in many human cancer types, with suppressed levels exhibiting protective effects (Figure 2C).
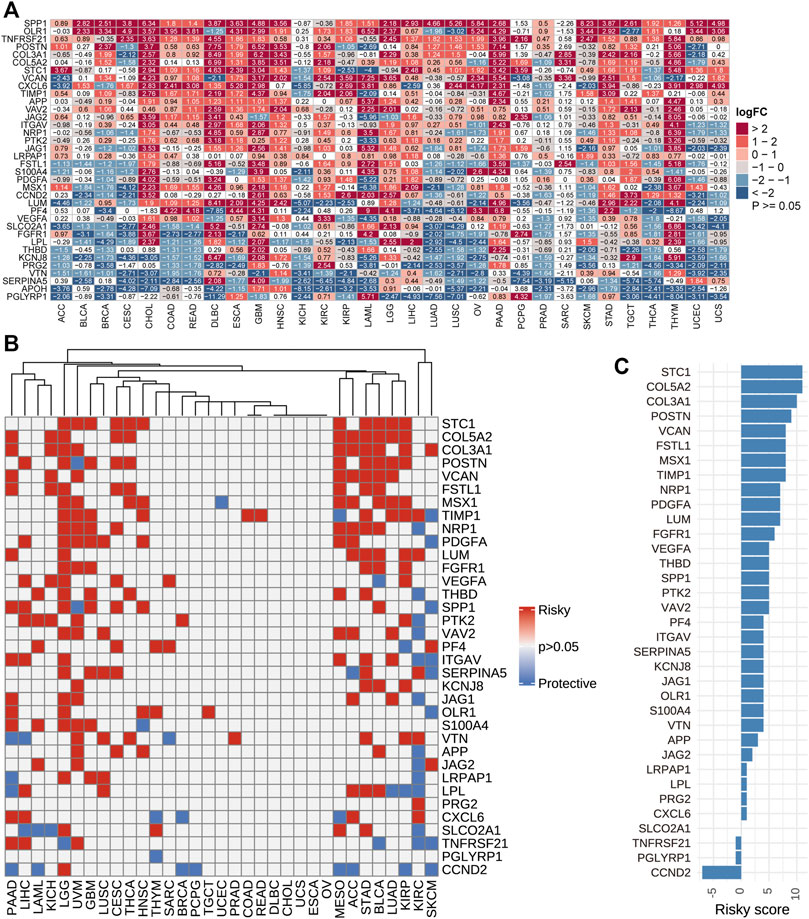
FIGURE 2. (A): Differential expressions of angiogenesis-related genes. We found the angiogenesis genes were aberrantly expressed in 31 tumor types. (B). Univariate Cox regression analysis of angiogenesis-associated genes in pan-cancer. Grey denotes p > 0.05, red represents p < 0.05 HR > 1while blue denotes HR < 1 (C). The number of genes with significant cox analysis in all tumors was counted. As a risk factor +1, as a protective factor -1, the final score as risky score.
The relationship between angiogenesis genes was established to be positively significant (Supplementary Figure S1A), suggesting that these genes could function individually or cooperatively. Notably, BRCA samples exhibited the highest number of variant genes. Genetic variabilities of STC1 and LPL genes were highest, and they were dominated by deletions while gene fusions were dominant among the other gene variants (Supplementary Figure S1B). The development of many tumors has been associated with point mutations and deletions (Agirre et al., 2006). The single nucleotide variation (SNV) was dominated by C > T (50%) (Supplementary Figures S2A-S2B). Supplementary Figure S2C show the frequencies of deleterious mutations in pan-caner. For instance, elevated VCAN levels in gastric cancer independently predict poor prognostic outcomes, and patients with VCAN mutations have a lower tumor grade, compared to those without (Li et al., 2020b; Li et al., 2020c). Moreover, Supplementary Figure S2D shows survival differences between mutant (deleterious) and wild type in the selected cancers. Aberrant methylation status are correlated with the development of various diseases, including cancer (Shao et al., 2017). Evaluation of the methylation status of angiogenesis-associated genes in this study was aimed at identifying epigenetic regulation. Supplementary Figure S2E shows the differences between tumor and normal samples with regards to gene methylation. In different tumors, the methylations of angiogenesis-associated genes were highly heterogeneous. Relative to hypomethylated genes, there were more hypermethylated genes in BRCA, prostate adenocarcinoma (PRAD), uterine corpus endometrial carcinoma (UCEC), and colon adenocarcinoma (COAD). In addition, in most cancers, PGLYRP1, KCNJ8, and LPL were hypermethylated while OLR1, VEGFA, and SPP1 were hypomethylated (Supplementary Figure S2E). Correlation analysis revealed that mRNA levels of angiogenesis-associated genes were negatively correlated with their methylation levels. However, in PRAD, skin cutaneous melanoma (SKCM), BRCA, liver hepatocellular carcinoma (LIHC), and uveal melanoma (UVM), PTK1 exhibited positive correlations between methylation and mRNA expression levels (Supplementary Figure S2F). Survival analysis showed that in most cancers, hypermethylations of PGLYRP1, PTK2, THBD, and CCND2 as well as hypomethylations of VAV2, OLR1, and ITGAV were associated with poor survival outcomes (Supplementary Figure S3). Hypermethylation of CCND2 was associated with female lung cancer and lung adenocarcinoma (Hung et al., 2018). Hypermethylation of CCND2 in lung and breast cancer is a potential biomarker and drug target (Hung et al., 2018).
CNV has been confirmed to have diagnostic, prognostic or therapeutic significances in various types of cancer (Su et al., 2018). In the present study, heterozygous amplifications and deletions were the main CNV types (Supplementary Figure S4A). For instance, CNV percentage analysis revealed that homozygous amplifications of PTK2 in OV, ESCA, BRCA LIHC, and UVM as well as CCND2, KCNJ8, and OLR1 in TGCT were all greater than 35% (Supplementary Figure S4B). Homozygous deletions of LPL and STC1 in PRAD were all greater than 35% (Supplementary Figure S4B). Heterozygous analysis showed that the amplified gene (PTGFA) in TGCT, READ, GBM, and KIRP was greater than 35% in all cases, while STC1 deletion in OV, LUSC, and LUAD was greater than 35% in all cases (Supplementary Figure S4C). Supplementary Figure S5 shows the differences in survival outcomes between CNV and wild types for selected cancers. These results imply that the CNV of angiogenesis-associated genes mediate their abnormal expressions, suggesting that it plays a significant role in cancer development.
For the 33 tumors, the highest angiogenesis scores were in PAAD while the lowest were in LAML (Supplementary Figure S6A). The scores in ACC, BLCA, BRCA, COAD, ESCA, LUSC, SKCM, STAD, and UVM varied across clinical stages. As clinical staging increased, so did the angiogenesis scores (Supplementary Figures S6B-J). Supplementary Figures S7A-D shows the associations between angiogenesis scores and pan-cancer overall survival (OS), disease free interval (DFI), progression-free interval (PFI), as well as disease-specific survival (DSS). Notably, HR was evaluated in the Cox proportional hazard models. In most cancers, high angiogenesis scores were associated with poor survival outcomes (Supplementary Figure S8). These results suggest a close association between angiogenesis scores and patient outcomes.
The related pathways network indicated that angiogenesis scores were significantly involved in cancer-related signaling pathways, including Kras signaling, TGF-beta signaling, apoptosis, IL-2 STAT5 signaling, TNF-a signaling via NFK-b, inflammatory responses, IL-6 JAK STAT3 signaling, notch signaling, and the P53 pathway (Figure 3A). However, these associations differed among tumors. For instance, in all cancer types, the IL-2 STAT5 signaling pathway was positively correlated with angiogenesis scores, while the DNA repair pathway exhibited the opposite trend. Besides, some pathways, such as those in pancreatic beta cells, exhibited different associations in different tumors. Using breast cancer as an example, enrichment analysis revealed a predominant enrichment in the PI3K-Akt signaling pathway, proteoglycans in cancer, human papillomavirus infections, and the relaxin signaling pathway (Figure 3B).
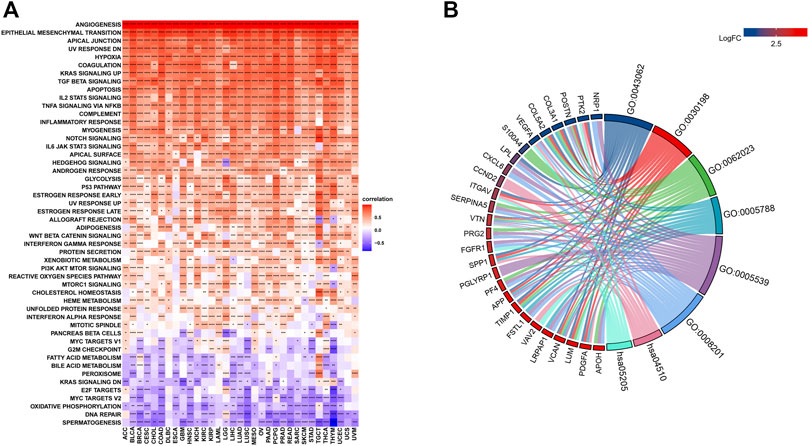
FIGURE 3. (A). Gene Set Variant Analysis (Red represents positive correlations while blue represents negative correlations. Angiogenesis scores were significantly involved in cancer-related signaling pathways, including Kras signaling, TGF-beta signaling, apoptosis, IL-2 STAT5 signaling, TNF-a signaling via NFK-b, inflammatory responses, IL-6 JAK STAT3 signaling, notch signaling, and the P53 pathway. *** denotes p < 0.001, ** denotes p < 0.01, while * denotes p < 0.05). (B). Enrichment analysis A predominant enrichment in the PI3K-Akt signaling pathway, proteoglycans in cancer, human papillomavirus infections, and the relaxin signaling pathway in BRCA.
The tumor microenvironment (TME) modulates tumor progression and treatment efficacy (Blanco‐Fernandez et al., 2021). Therefore, we evaluated the relationship between the angiogenesis score and the TME. Angiogenesis was significantly correlated with the TME (immune and stromal) (Figure 4A) while angiogenesis scores were strongly correlated with immune and stromal/metastasis-related pathways (Figure 4B). Next, the association between angiogenesis scores and infiltration levels of different immune cell types in 33 cancer types were evaluated. Most of the cancer types showed significant positive associations between angiogenesis scores and infiltration levels of macrophages, NK cells, as well as CD4+ T cells (Figures 5A,B), implying that angiogenesis scores are associated with a hot TME. For instance, THYM revealed a highly significant positive correlation between angiogenesis scores and infiltration levels of NK cells. In contrast, NK cells exhibited significant negative correlations with angiogenesis scores in TGCT, and weak or no correlations in ACC (Figures 5A–C).

FIGURE 4. Analysis of the tumor microenvironment. (*** denotes p < 0.001, ** denotes p < 0.01, while * denotes p < 0.05) (A). Correlations between angiogenesis scores and ImmuneScores, Stromalcores, ESTIMATEScores and TumorPurity. (B). Relationship between angiogenesis scores and pathways (Immune-related pathways, Stromal/transfer-related pathways and DNA damage repair-related pathways).
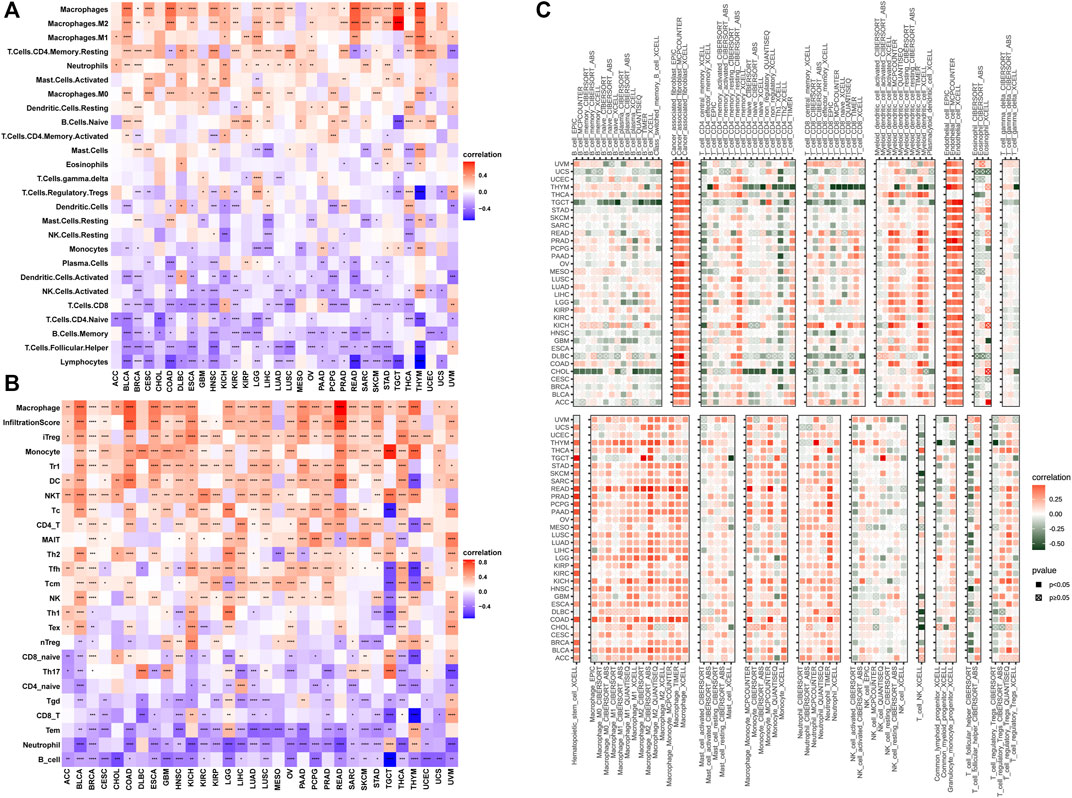
FIGURE 5. Angiogenesis and immune cell infiltration analysis (A–C): Most of the cancer types showed significant positive associations between angiogenesis scores and infiltration levels of macrophages, NK cells, as well as CD4+ T cells).
The core function of the immune system is to recognize self and eliminate non-self antigens to maintain normal physiological activities and fight disease (Bai et al., 2020; Forster and Radpour, 2020). This function is largely mediated by the major histocompatibility complex (MHC). The significance of MHC in tumor diagnosis and treatment is being evaluated in immunotherapy (Baba et al., 2007; Marcu et al., 2021). Chemokines are also involved in responses to cancer therapies, therefore, they are potential targets for immunotherapy and chemokine-targeted therapy (Samaniego et al., 2018). We found strong associations between angiogenesis scores and the above related genes (Supplementary Figures S9A-C). Then, we investigated the correlation between angiogenesis scores and expressions of immune checkpoints. In 33 cancer types, the angiogenesis score was positively correlated with expression levels of most of the immune checkpoints (Supplementary Figure S9D). A previous study revealed that TMB is associated with immunotherapeutic responses (Abou Khouzam et al., 2020). We found that angiogenesis scores were significantly correlated with TMB in STAD, HNSC, LUAD, KIRP, LIHC, CHOL, THYM, LAM, and LGG (Supplementary Figure S9E). MSI are repeated DNA sequences (Page and Graham, 2008). Impaired mismatch repair-associated MSI may be a mechanism in gastric cancer development, therefore, its significance is being evaluated by various studies. Our results showed a strong correlation between angiogenesis scores and MSI in BRCA, LUAD, LUSC, STAD, HNSC, and TGCT (Supplementary Figure S9F).
During the evaluation of angiogenesis-associated TME characteristics, we observed strong positive correlations between immune checkpoint levels and angiogenesis genes. Due to advances in cancer immunotherapy, we evaluated the expressions of angiogenesis-associated genes in immunotherapy-treated patients. We collected three datasets containing pre-treatment samples and immunotherapeutic data. These datasets had various patient information, including clinical manifestations (complete responses (CR) or partial responses (PR)) or no clinical benefits (progressive disease (PD) or stable disease (SD)). Compared to patients with low angiogenesis scores, patients with high angiogenesis scores exhibited poorer prognostic outcomes after immunotherapy. Higher scores were observed in progressive phase patients (Figure 6).
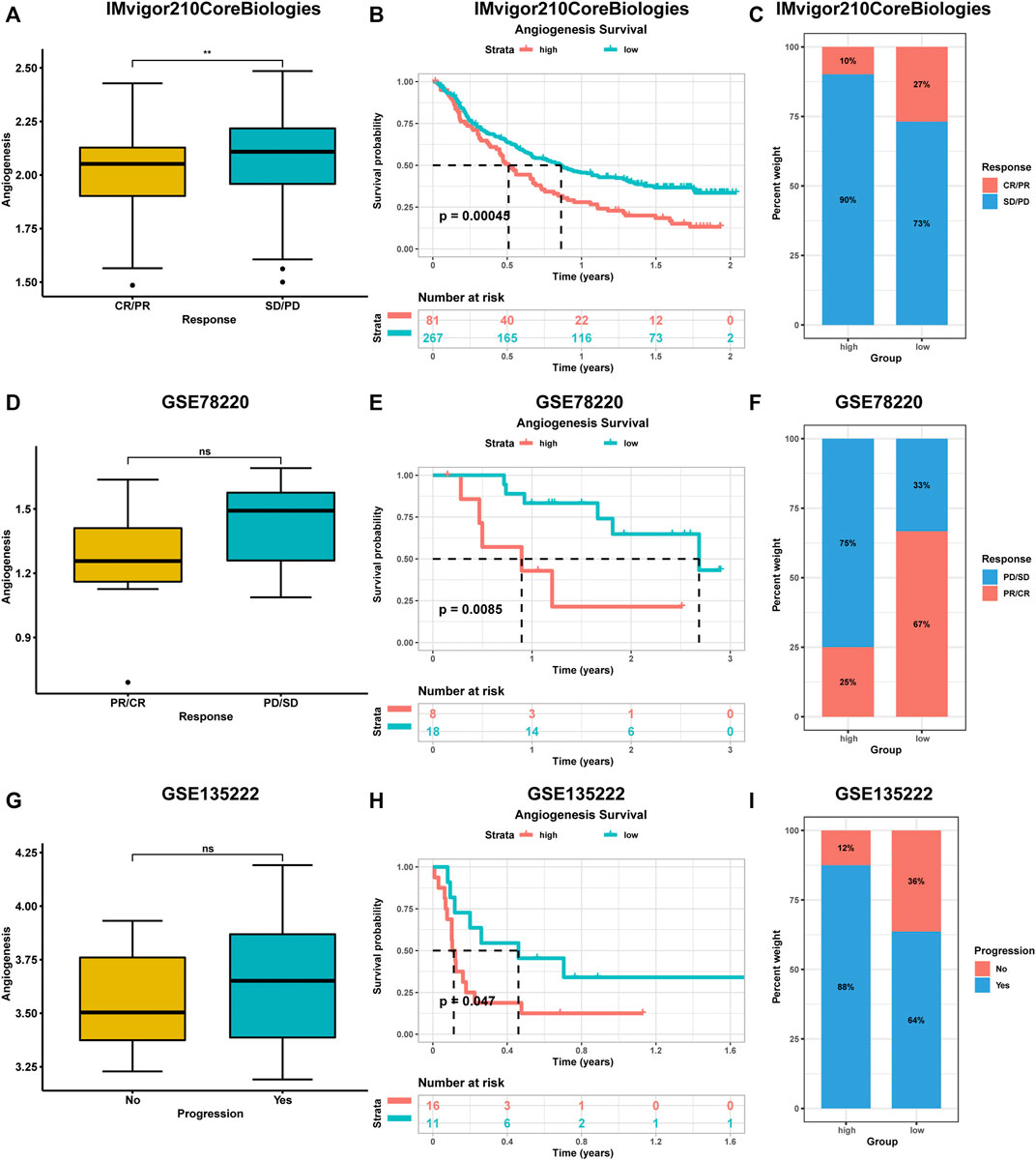
FIGURE 6. The impact of angiogenesis scores on the prognostic outcome of immunotherapy patients was analysed using multiple datasets (A-I). Patients with high angiogenesis scores exhibited poorer prognostic outcomes after immunotherapy. complete responses (CR), partial responses (PR), progressive disease (PD), stable disease (SD).
Currently, surgery and chemotherapy are the main therapeutic strategies for cancer, however, resistance to chemotherapeutic drugs and molecularly targeted therapies is a major barrier to cancer treatment (Wu et al., 2020). Spearman’s correlation analysis showed that drug sensitivity towards YM155, ZG-10, GW843682X, and S−Trityl−L−cysteine correlated with APP levels (positive correlation with IC50). However, resistance towards EHT 1864 and lisitinib correlated with expressions of APOH (negative correlation with IC50) (Supplementary Figure S10). In conclusion, dysregulated expressions of angiogenesis-associated genes may be involved in resistance to cancer therapies.
Non-coding RNAs (ncRNAs) are involved gene expression regulation. To ascertain whether angiogenesis genes are modulated by some ncRNAs, first, we predicted upstream miRNAs that have the potential to bind angiogenesis-associated genes. Network visualization was achieved using the Cytoscape software (Supplementary Figure S12). Hsa-miR-106a-5p was regulated and was central to most angiogenesis-associated genes. Therefore, in pan-cancer, hsa-miR-106a-5p might be a potential regulatory miRNA of angiogenesis-associated genes. Next, upstream lncRNAs of hsa-miR-106a-5p were predicted using the starbase database. Sixty eight potential lncRNAs were identified. Relationships visualization was performed using the Cytoscape software (Supplementary Figure S12). These results indicate that ncRNAs regulation of angiogenesis-associated genes might be involved in cancer progression.
Angiogenesis, the formation of new blood vessels, is important for tumor progression (Lin et al., 2010). In 1971, Folkman proposed that tumor growth is dependent on angiogenesis and that inhibition of angiogenesis is a potential therapeutic paradigm for solid tumors (Folkman, 1971). To trigger angiogenesis, tumors overexpress various angiogenic factors and cytokines, as well as endogenous angiogenesis enhancers (Folkman, 1996). Various angiogenic factors have been identified, their corresponding inhibitors developed, and their efficacies in cancer treatment demonstrated (Cao, 2010; Cao and Langer, 2010; Cao et al., 2011; Cao, 2014). Therefore, to understand tumorigenesis, and to investigate potential targets for clinical treatment, elucidation of angiogenesis in cancer is necessary.
In this study, we reveal multiple potential mechanisms of angiogenesis in cancer, including common angiogenesis-associated cancer pathways. According to the results of this study, we found a high frequency of CNVs of angiogenesis-associated genes. Moreover, the CNVs were positively correlated with angiogenesis-associated gene expressions, indicating that copy number variations may affect angiogenesis-associated gene expressions, contributing to tumorigenesis. For example, S100A4 was frequently amplified in PAAD and was associated with poor patient survival outcomes, consistent with findings from previous studies (Matsubara et al., 2005; Li and Bresnick, 2006). Hypermethylated CCND2 was associated with poor survival rates of KICH, implying that hypermethylated CCND2 may be a driver gene for KICH progression. This is in tandem with findings from previous studies that concluded that hypermethylation of CCND2 is associated with the progression of various cancers (Hung et al., 2018; Ding et al., 2019). However, in this study, there were inconsistencies between methylation levels and prognostic outcomes, therefore, we postulated that in certain contexts, genetic and epigenetic alterations of angiogenesis-associated genes might result in angiogenesis dysfunctions and promote tumorigenesis. Therefore, studies should be conducted to assess this postulate.
The TME is hypoxic and acidic in nature, and in this environment, tumor cells recruit a number of innate immune cells, the most representative of which are tumor-associated macrophages (TAMs), neutrophils, myeloid-derived suppressor cells (MDSC) and natural killer cells (NK) (Hamaguchi et al., 2020). TAMs are involved in every step of tumor angiogenesis (Komohara and Takeya, 2017). In early stages of tumor development, neutrophils play a key role in promoting angiogenesis. Neutrophils can release MMP-9 to activate endothelial cell growth signals and to promote angiogenesis (Eyileten et al., 2016). They also secrete myeloperoxidase (MPO), which is important for macrophage recruitment and platelet activation (Heslop et al., 2010). In addition, reduction in neutrophil counts significantly inhibits the link between VEGF and its receptors (Li et al., 2012).
Despite its significance in tumor therapy, immunotherapeutic responses are suboptimal, which may be attributed to immunosuppressive tumor microenvironments (Chan et al., 2020). The tumor vasculature carries essential nutrients and oxygen to the tumor tissue and plays an important role in the growth as well as progression of malignant tumors. Abnormal blood vessels can form a physical barrier that limits immunotherapeutic efficacies (Jain, 2005). Abnormal tumor vasculature may inhibit immunotherapeutic effectiveness through hypoxia and development of an acidic microenvironment, which enhances immunosuppression (Jain, 2005). In addition, it can induce a decrease in micro-environmental pH, affecting immune cell functions, therefore, anti-tumor immune cells, such as T lymphocytes and NK cells can become unresponsive in acidic environments and subsequently, undergo apoptosis (Huber et al., 2017). In contrast, immunosuppressive components (such as myeloid and Treg cells) enhance tumor growth in acidic environments (Huber et al., 2017). The combination of anti-angiogenic therapy and immunotherapy has improved prognostic outcomes for patients with different cancer types, including liver, kidney and breast cancers (Rini et al., 2019; Finn et al., 2020; Liu et al., 2020), resulting in improved PFS and OS. This combination provides additional clinical options for patients with liver metastases and positive driver genes (Finn et al., 2020). In addition, this combination has not increased toxicity but has an increased efficacy, while the overall safety profile is manageable.
Drug sensitivity analyses were performed to identify potential drugs that can modulate angiogenesis-associated genes. For instance, a positive correlation was found between APP and YM155, ZG-10, GW843682X, and S−Trityl−L−cysteine, suggesting that patients with elevated APP gene expressions may be resistant to these drugs. Thus, we postulate that targeting angiogenesis-associated genes may be an effective anticancer treatment approach. Our findings, which revealed variations in angiogenesis at all regulation levels, elucidate on regulation of angiogenesis-associated genes in tumors. These variations may in turn lead to differences in drug efficacies, treatment responses and patient survival outcomes. However, more studies should comprehensively investigate cancer heterogeneity and individual-based treatment approaches.
This study provides the first comprehensive description of angiogenesis-associated gene expressions in various tumor types. Moreover, we systematically investigated the impact of changes in expressions of all angiogenesis-associated genes on clinical outcomes of various cancer types. Angiogenesis-associated gene expressions were correlated with different genomic and immunological tumor characteristics, implying that they have prognostic values in both immunotherapeutic and standard settings.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
X-tY designed the experiments; W-NM, X-yL, YS, LZ, ZW, DW, L-xS, M-ZW, and YS performed the experiments and wrote the manuscript while X-tY reviewed the manuscript. The corresponding author of this article is X-tY. X-yL, W-NM, L-xS, and YS contributed equally to this work.
This study received Fundamental research program funding of Ninth People’s Hospital affiliated to Shanghai Jiao Tong University School of Medicine (No. JYZZ076), Clinical Research Program of Ninth People’s Hospital, Shanghai Jiao Tong University School of Medicine (No. JYLJ201801, JYLJ201911), the China Postdoctoral Science Foundation (No. 2017M611585) and the National Natural Science Foundation of China (No. 81871458).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Home for Researchers editorial team (www.home-for-researchers.com).
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2022.805507/full#supplementary-material
TCGA, The Cancer Genome Atlas; DEGs, Differentially expressed genes; OS, Overall survival; FDR, False discovery rate; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; HR, Hazard ratio; CI: Confidence interval; See.
Abou Khouzam, R., Goutham, H. V., Zaarour, R. F., Chamseddine, A. N., Francis, A., Buart, S., et al. (2020). Integrating Tumor Hypoxic Stress in Novel and More Adaptable Strategies for Cancer Immunotherapy. Semin. Cancer Biol. 65, 140–154. doi:10.1016/j.semcancer.2020.01.003
Agirre, X., Román-Gómez, J., Vázquez, I., Jiménez-Velasco, A., Garate, L., Montiel-Duarte, C., et al. (2006). Abnormal Methylation of the commonPARK2andPACRGpromoter Is Associated with Downregulation of Gene Expression in Acute Lymphoblastic Leukemia and Chronic Myeloid Leukemia. Int. J. Cancer 118 (8), 1945–1953. doi:10.1002/ijc.21584
Ardito, C. M., Briggs, C. D., and Crawford, H. C. (2008). Targeting of Extracellular Proteases Required for the Progression of Pancreatic Cancer. Expert Opin. Ther. Targets 12 (5), 605–619. doi:10.1517/14728222.12.5.605
Azoitei, N., Pusapati, G. V., Kleger, A., Moller, P., Kufer, R., Genze, F., et al. (2010). Protein Kinase D2 Is a Crucial Regulator of Tumour Cell-Endothelial Cell Communication in Gastrointestinal Tumours. Gut 59 (10), 1316–1330. doi:10.1136/gut.2009.206813
Baba, T., Hanagiri, T., Ichiki, Y., Kuroda, K., Shigematsu, Y., Mizukami, M., et al. (2007). Lack and Restoration of Sensitivity of Lung Cancer Cells to Cellular Attack with Special Reference to Expression of Human Leukocyte Antigen Class I And/or Major Histocompatibility Complex Class I Chain Related Molecules A/B. Cancer Sci. 98 (11), 1795–1802. doi:10.1111/j.1349-7006.2007.00586.x
Bai, B., Yang, Y., Wang, Q., Li, M., Tian, C., Liu, Y., et al. (2020). NLRP3 Inflammasome in Endothelial Dysfunction. Cell Death Dis 11 (9), 776. doi:10.1038/s41419-020-02985-x
Blanco‐Fernandez, B., Gaspar, V. M., Engel, E., and Mano, J. F. (2021). Proteinaceous Hydrogels for Bioengineering Advanced 3D Tumor Models. Adv. Sci. 8 (4), 2003129. doi:10.1002/advs.202003129
Cao, Y., Arbiser, J., D’Amato, R. J., D’Amore, P. A., Ingber, D. E., Kerbel, R., et al. (2011). Forty-Year Journey of Angiogenesis Translational Research. Sci. Transl. Med. 3 (114), 113r–114r. doi:10.1126/scitranslmed.3003149
Cao, Y., and Langer, R. (2010). Optimizing the Delivery of Cancer Drugs that Block Angiogenesis. Sci. Transl. Med. 2 (15), 13p–15p. doi:10.1126/scitranslmed.3000399
Cao, Y. (2010). Off-tumor Target-Beneficial Site for Antiangiogenic Cancer Therapy? Nat. Rev. Clin. Oncol. 7 (10), 604–608. doi:10.1038/nrclinonc.2010.118
Cao, Y. (2014). VEGF-targeted Cancer Therapeutics-Paradoxical Effects in Endocrine Organs. Nat. Rev. Endocrinol. 10 (9), 530–539. doi:10.1038/nrendo.2014.114
Chan, J. Y., Lim, J. Q., Yeong, J., Ravi, V., Guan, P., Boot, A., et al. (2020). Multiomic Analysis and Immunoprofiling Reveal Distinct Subtypes of Human Angiosarcoma. [J]. J. Clin. Invest. 130 (11), 5833–5846. doi:10.1172/JCI139080
De Palma, F., D’Argenio, V., Pol, J., Kroemer, G., Maiuri, M., and Salvatore, F. (2019). The Molecular Hallmarks of the Serrated Pathway in Colorectal Cancer. Cancers 11 (7), 1017. doi:10.3390/cancers11071017
Deng, G., Zeng, F., Su, J., Zhao, S., Hu, R., Zhu, W., et al. (2020). BET Inhibitor Suppresses Melanoma Progression via the Noncanonical NF-Κb/spp1 Pathway. Theranostics 10 (25), 11428–11443. doi:10.7150/thno.47432
Ding, Z. y., Li, R., Zhang, Q. j., Wang, Y., Jiang, Y., Meng, Q. y., et al. (2019). Prognostic Role of Cyclin D2/D3 in Multiple Human Malignant Neoplasms: A Systematic Review and Meta‐analysis. Cancer Med. 8 (6), 2717–2729. doi:10.1002/cam4.2152
Eyileten, C., Majchrzak, K., Pilch, Z., Tonecka, K., Mucha, J., Taciak, B., et al. (2016). Immune Cells in Cancer Therapy and Drug Delivery. Mediators Inflamm. 2016, 1–13. doi:10.1155/2016/5230219
Finn, R. S., Qin, S., Ikeda, M., Galle, P. R., Ducreux, M., Kim, T.-Y., et al. (2020). Atezolizumab Plus Bevacizumab in Unresectable Hepatocellular Carcinoma. N. Engl. J. Med. 382 (20), 1894–1905. doi:10.1056/NEJMoa1915745
Firestone, G. L., and Sundar, S. N. (2009). Anticancer Activities of Artemisinin and its Bioactive Derivatives. Expert Rev. Mol. Med. 11, e32. doi:10.1017/S1462399409001239
Folkman, J. (1971). Tumor Angiogenesis: Therapeutic Implications. N. Engl. J. Med. 285 (21), 1182–1186. doi:10.1056/NEJM197111182852108
Folkman, J. (1996). Tumor Angiogenesis and Tissue Factor. Nat. Med. 2 (2), 167–168. doi:10.1038/nm0296-167
Forster, S., and Radpour, R. (2020). Molecular Immunotherapy: Promising Approach to Treat Metastatic Colorectal Cancer by Targeting Resistant Cancer Cells or Cancer Stem Cells. Front. Oncol. 10, 569017. doi:10.3389/fonc.2020.569017
Hamaguchi, R., Narui, R., and Wada, H. (2020). Effects of Alkalization Therapy on Chemotherapy Outcomes in Metastatic or Recurrent Pancreatic Cancer. Anticancer Res. 40 (2), 873–880. doi:10.21873/anticanres.14020
Heslop, C. L., Frohlich, J. J., and Hill, J. S. (2010). Myeloperoxidase and C-Reactive Protein Have Combined Utility for Long-Term Prediction of Cardiovascular Mortality after Coronary Angiography. J. Am. Coll. Cardiol. 55 (11), 1102–1109. doi:10.1016/j.jacc.2009.11.050
Hida, K., Hida, Y., and Shindoh, M. (2008). Understanding Tumor Endothelial Cell Abnormalities to Develop Ideal Anti-angiogenic Therapies. Cancer Sci. 99 (3), 459–466. doi:10.1111/j.1349-7006.2007.00704.x
Huang, C., Huang, R., Chen, H., Ni, Z., Huang, Q., Huang, Z., et al. (2021). Chromatin Accessibility Regulates Gene Expression and Correlates with Tumor-Infiltrating Immune Cells in Gastric Adenocarcinoma. Front. Oncol. 10, 609940. doi:10.3389/fonc.2020.609940
Huber, V., Camisaschi, C., Berzi, A., Ferro, S., Lugini, L., Triulzi, T., et al. (2017). Cancer Acidity: An Ultimate Frontier of Tumor Immune Escape and a Novel Target of Immunomodulation. Semin. Cancer Biol. 43, 74–89. doi:10.1016/j.semcancer.2017.03.001
Hultqvist, G., Syvänen, S., Fang, X. T., Lannfelt, L., and Sehlin, D. (2017). Bivalent Brain Shuttle Increases Antibody Uptake by Monovalent Binding to the Transferrin Receptor. Theranostics 7 (2), 308–318. doi:10.7150/thno.17155
Hung, C.-S., Wang, S.-C., Yen, Y.-T., Lee, T.-H., Wen, W.-C., and Lin, R.-K. (2018). Hypermethylation of CCND2 in Lung and Breast Cancer Is a Potential Biomarker and Drug Target. Ijms 19 (10), 3096. doi:10.3390/ijms19103096
Hurwitz, H., Fehrenbacher, L., Novotny, W., Cartwright, T., Hainsworth, J., Heim, W., et al. (2004). Bevacizumab Plus Irinotecan, Fluorouracil, and Leucovorin for Metastatic Colorectal Cancer. N. Engl. J. Med. 350 (23), 2335–2342. doi:10.1056/nejmoa032691
Jain, R. K. (2005). Normalization of Tumor Vasculature: an Emerging Concept in Antiangiogenic Therapy. Science 307 (5706), 58–62. doi:10.1126/science.1104819
Kelley, R. K., Bridgewater, J., Gores, G. J., and Zhu, A. X. (2020). Systemic Therapies for Intrahepatic Cholangiocarcinoma. J. Hepatol. 72 (2), 353–363. doi:10.1016/j.jhep.2019.10.009
Kim, Y. J., Lee, S. C., Kim, S. E., Kim, S. H., Kim, S. K., and Lee, C. S. (2020). YAP Activity Is Not Associated with Survival of Uveal Melanoma Patients and Cell Lines. Sci. Rep. 10 (1), 6209. doi:10.1038/s41598-020-63391-z
Komatsu, Y., Waku, T., Iwasaki, N., Ono, W., Yamaguchi, C., and Yanagisawa, J. (2012). Global Analysis of DNA Methylation in Early-Stage Liver Fibrosis. BMC Med. Genomics 5, 5. doi:10.1186/1755-8794-5-5
Komohara, Y., and Takeya, M. (2017). CAFs and TAMs: Maestros of the Tumour Microenvironment. J. Pathol. 241 (3), 313–315. doi:10.1002/path.4824
Li, K., Luo, H., Huang, L., Luo, H., and Zhu, X. (2020). Microsatellite Instability: a Review of what the Oncologist Should Know. Cancer Cell Int 20, 16. doi:10.1186/s12935-019-1091-8
Li, W., Han, F., Fu, M., and Wang, Z. (2020). High Expression of VCAN Is an Independent Predictor of Poor Prognosis in Gastric Cancer. J. Int. Med. Res. 48 (1), 030006051989127. doi:10.1177/0300060519891271
Li, Y., Wang, J.-S., Zhang, T., Wang, H.-C., and Li, L.-P. (2020). Identification of New Therapeutic Targets for Gastric Cancer with Bioinformatics. Front. Genet. 11, 865. doi:10.3389/fgene.2020.00865
Li, Z.-H., and Bresnick, A. R. (2006). The S100A4 Metastasis Factor Regulates Cellular Motility via a Direct Interaction with Myosin-IIA. Cancer Res. 66 (10), 5173–5180. doi:10.1158/0008-5472.can-05-3087
Li, Z.-J., Zhu, H., Ma, B.-Y., Zhao, F., Mao, S.-H., Liu, T.-G., et al. (2012). Inhibitory Effect of Bifidobacterium Infantis-Mediated sKDR Prokaryotic Expression System on Angiogenesis and Growth of Lewis Lung Cancer in Mice. BMC cancer 12, 155. doi:10.1186/1471-2407-12-155
Lin, Y., Bai, L., Chen, W., and Xu, S. (2010). The NF-Κb Activation Pathways, Emerging Molecular Targets for Cancer Prevention and Therapy. Expert Opin. Ther. Targets 14 (1), 45–55. doi:10.1517/14728220903431069
Liu, J. J., Liu, Q., Li, Y., Li, Q., Su, F., Yao, H., et al. (2020). Efficacy and Safety of Camrelizumab Combined with Apatinib in Advanced Triple-Negative Breast Cancer: an Open-Label Phase II Trial]. J. Immunother. Cancer 8 (1), e000696. doi:10.1136/jitc-2020-000696
Liu, W., Zha, Z., and Wang, H. (2019). Upregulation of microRNA‐27a Inhibits Synovial Angiogenesis and Chondrocyte Apoptosis in Knee Osteoarthritis Rats through the Inhibition of PLK2. J. Cell Physiol 234 (12), 22972–22984. doi:10.1002/jcp.28858
Lugano, R., Ramachandran, M., and Dimberg, A. (2020). Tumor Angiogenesis: Causes, Consequences, Challenges and Opportunities. Cell. Mol. Life Sci. 77 (9), 1745–1770. doi:10.1007/s00018-019-03351-7
Marcu, A., Bichmann, L., Kuchenbecker, L., Kowalewski, D. J., Freudenmann, L. K., Backert, L., et al. (2021). HLA Ligand Atlas: a Benign Reference of HLA-Presented Peptides to Improve T-Cell-Based Cancer Immunotherapy. J. Immunother. Cancer 9 (4), e002071. doi:10.1136/jitc-2020-002071
Matsubara, D., Niki, T., Ishikawa, S., Goto, A., Ohara, E., Yokomizo, T., et al. (2005). Differential Expression of S100A2 and S100A4 in Lung Adenocarcinomas: Clinicopathological Significance, Relationship to P53 and Identification of Their Target Genes. Cancer Sci. 96 (12), 844–857. doi:10.1111/j.1349-7006.2005.00121.x
Mitsuhashi, A., Goto, H., Saijo, A., Trung, V. T., Aono, Y., Ogino, H., et al. (2015). Fibrocyte-like Cells Mediate Acquired Resistance to Anti-angiogenic Therapy with Bevacizumab. Nat. Commun. 6, 8792. doi:10.1038/ncomms9792
Page, K., and Graham, E. A. M. (2008). Cancer and Forensic Microsatellites. Forensic Sci. Med. Pathol. 4 (1), 60–66. doi:10.1007/s12024-008-9027-y
Rini, B. I., Powles, T., Atkins, M. B., Escudier, B., McDermott, D. F., Suarez, C., et al. (2019). Atezolizumab Plus Bevacizumab versus Sunitinib in Patients with Previously Untreated Metastatic Renal Cell Carcinoma (IMmotion151): a Multicentre, Open-Label, Phase 3, Randomised Controlled Trial. The Lancet 393 (10189), 2404–2415. doi:10.1016/S0140-6736(19)30723-8
Saeed, A., and Salem, M. E. (2020). Prognostic Value of Tumor Mutation burden (TMB) and INDEL burden (IDB) in Cancer: Current View and Clinical Applications. Ann. Transl Med. 8 (9), 575. doi:10.21037/atm-2020-75
Samaniego, R., Gutiérrez-González, A., Gutiérrez-Seijo, A., Sánchez-Gregorio, S., García-Giménez, J., Mercader, E., et al. (2018). CCL20 Expression by Tumor-Associated Macrophages Predicts Progression of Human Primary Cutaneous Melanoma. Cancer Immunol. Res. 6 (3), 267–275. doi:10.1158/2326-6066.CIR-17-0198
Schlattl, A., Anders, S., Waszak, S. M., Huber, W., and Korbel, J. O. (2011). Relating CNVs to Transcriptome Data at fine Resolution: Assessment of the Effect of Variant Size, Type, and Overlap with Functional Regions. Genome Res. 21 (12), 2004–2013. doi:10.1101/gr.122614.111
Shao, Z., Xu, P., Xu, W., Li, L., Liu, S., Zhang, R., et al. (2017). Discovery of Novel DNA Methyltransferase 3A Inhibitors via Structure-Based Virtual Screening and Biological Assays. Bioorg. Med. Chem. Lett. 27 (2), 342–346. doi:10.1016/j.bmcl.2016.11.023
Shatalova, A., Shatalov, I., and Lebedin, Y. (2020). X6: A Novel Antibody for Potential Use in Gluten Quantification. Molecules 25 (14), 3107. doi:10.3390/molecules25143107
Solimando, A. G., Summa, S. D., Vacca, A., and Ribatti, D. (2020). Cancer-Associated Angiogenesis: The Endothelial Cell as a Checkpoint for Immunological Patrolling. Cancers 12 (11), 3380. doi:10.3390/cancers12113380
Su, C., Li, D., Li, N., Du, Y., Yang, C., Bai, Y., et al. (2018). Studying the Mechanism of PLAGL2 Overexpression and its Carcinogenic Characteristics Based on 3'-untranslated Region in Colorectal Cancer. Int. J. Oncol. 52 (5), 1479–1490. doi:10.3892/ijo.2018.4305
Thorsson, V., Gibbs, D. L., Brown, S. D., Wolf, D., Bortone, D. S., Ou Yang, T. H., et al. (2018). The Immune Landscape of Cancer. Immunity 48 (4), 812–e14. doi:10.1016/j.immuni.2018.03.023
Wang, R., Zhao, A., Cao, N., Li, Z., Zhang, G., and Liu, F. (2020). The Value of Circulation Tumor DNA in Predicting Postoperative Recurrence of Colorectal Cancer: a Meta-Analysis. Int. J. Colorectal Dis. 35 (8), 1463–1475. doi:10.1007/s00384-020-03667-y
Wu, S.-G., and Shih, J.-Y. (2018). Management of Acquired Resistance to EGFR TKI-Targeted Therapy in Advanced Non-small Cell Lung Cancer. Mol. Cancer 17 (1), 38. doi:10.1186/s12943-018-0777-1
Wu, Z. X., Yang, Y., Wang, G., Wang, J. Q., Teng, Q. X., Sun, L., et al. (2020). Dual TTK/CLK2 Inhibitor, CC‐671, Selectively Antagonizes ABCG2‐mediated Multidrug Resistance in Lung Cancer Cells. Cancer Sci. 111 (8), 2872–2882. doi:10.1111/cas.14505
Keywords: angiogenesis, pan-cancer, prognosis, methylation, gene expression
Citation: Li X-y, Ma W-N, Su L-x, Shen Y, Zhang L, Shao Y, Wang D, Wang Z, Wen M-Z and Yang X-t (2022) Association of Angiogenesis Gene Expression With Cancer Prognosis and Immunotherapy Efficacy. Front. Cell Dev. Biol. 10:805507. doi: 10.3389/fcell.2022.805507
Received: 30 October 2021; Accepted: 03 January 2022;
Published: 26 January 2022.
Edited by:
Qiangzhe Zhang, Nankai University, ChinaReviewed by:
Domenico Ribatti, University of Bari Aldo Moro, ItalyCopyright © 2022 Li, Ma, Su, Shen, Zhang, Shao, Wang, Wang, Wen and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xi-tao Yang, eGl0YW8xMjM0NTZAMTI2LmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.