
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell Dev. Biol. , 02 February 2022
Sec. Developmental Epigenetics
Volume 10 - 2022 | https://doi.org/10.3389/fcell.2022.784128
 Zhen Xu1†
Zhen Xu1† Jiajia Shi1†
Jiajia Shi1† Yu Zhang2†
Yu Zhang2† Yuhan Liu1
Yuhan Liu1 Junzheng Zhao1
Junzheng Zhao1 Qian Chen1
Qian Chen1 Chenglin Song1
Chenglin Song1 Shuhui Geng1
Shuhui Geng1 Wei Xie2
Wei Xie2 Feizhen Wu3
Feizhen Wu3 Yun Bai1
Yun Bai1 Yang Yang1
Yang Yang1 Xiajun Li1*
Xiajun Li1*Zfp57 has both maternal and zygotic functions in mouse. It maintains genomic imprinting at most known imprinted regions and controls allelic expression of the target imprinted genes in mouse embryos. The DNA methylation imprint at many imprinting control regions (ICRs) is lost when both maternal and zygotic Zfp57 are absent in Zfp57 maternal–zygotic mutant mouse embryos. Interestingly, we found that DNA methylation at a few ICRs was partially lost without maternal Zfp57 in Zfp57 heterozygous mouse embryos derived from Zfp57 homozygous female mice. This suggests that maternal Zfp57 is essential for the maintenance of DNA methylation at a small subset of imprinted regions in mouse embryos. This maternal effect of Zfp57 was applied to allelic expression switch as well as expression levels of the corresponding imprinted genes. It is rather surprising that DNA methylation imprint was affected differently at Rasgrf1 and AK008011 imprinted regions in the female or male Zfp57 maternal–zygotic mutant embryos, with more significant loss of DNA methylation observed in the male mutant embryos. Loss of ZFP57 resulted in gender-specific differences in allelic expression switch and expression level changes of some imprinted genes in female or male mutant embryos. These results indicate maternal and sexually dimorphic effects of ZFP57 on genomic imprinting in mouse.
Genomic imprinting is a kind of parental effect on the progeny that is established in the female or male germline (Bartolomei and Ferguson-Smith, 2011; Li, 2013; Tucci et al., 2019). It is essential for mammalian embryonic growth and development. Most of approximately 150 known imprinted genes are clustered in over 20 known imprinted regions with each harboring a few imprinted genes (Monk et al., 2019). They are co-regulated by a cis-acting imprinting control region (ICR) containing germline-derived differential DNA methylation (Barlow and Bartolomei, 2014; Zeng and Chen, 2019). Based on definition, imprinted genes exhibit parent-of-origin–dependent monoallelic expression, although some are preferentially expressed from one parental allele, and others may be imprinted only in some tissues or organs (Williamson et al., 2004; Marcho et al., 2015; Plasschaert and Bartolomei, 2015; Freschi et al., 2018; Hsiao et al., 2019; Monk et al., 2019; Jiang et al., 2021; Prickett et al., 2021). Imprinting shares some similarities to other monoallelic gene expression phenomena in mammals (Lee and Bartolomei, 2013; Chess, 2016; Khamlichi and Feil, 2018; Bar and Benvenisty, 2019).
ZFP57 and ZFP445 are KRAB zinc finger proteins that play important roles in maintaining genomic imprinting (Hirasawa and Feil, 2008; Juan and Bartolomei, 2019; Tucci et al., 2019; Hanna and Kelsey, 2021). They are partially redundant in the maintenance of genomic imprinting, with ZFP57 being more dominant in mouse embryos (Takahashi et al., 2015; Takahashi et al., 2019). Human ZFP57 has similar functions in genomic imprinting, and mutations in human ZFP57 result in a number of human diseases, including transient neonatal diabetes (Mackay et al., 2008; Takikawa et al., 2013b; Monteagudo-Sanchez et al., 2020). Mouse Zfp57 is a maternal–zygotic effect gene (Li et al., 2008; Shamis et al., 2015). It has both maternal and zygotic functions. Loss of both maternal and zygotic Zfp57 in the maternal–zygotic mutant (M−Z-) embryos results in loss of DNA methylation imprinting at most ICRs and deregulation of target-imprinted genes at these imprinted regions, whereas loss of just zygotic Zfp57 in the zygotic mutant (M+Z−) embryos cause partial loss of DNA methylation imprint at these ICRs (Jiang et al., 2021). ZFP57 binds to almost all known ICRs, with higher binding affinity for the methylated DNA (Li et al., 2008; Quenneville et al., 2011; Liu et al., 2012; Strogantsev et al., 2015; Riso et al., 2016; Takahashi et al., 2019). Allelic expression switch occurs at some target imprinted genes when ZFP57 is lost in M−Z− embryos (Jiang et al., 2021).
Sexually dimorphic effect has been reported in many studies. There are gender-dependent phenotypes in cardiovascular diseases (Deegan et al., 2021; Walker et al., 2021). Males and female behave differently in neural behavior and brain disorders (Chen et al., 2019; Pfaff and Barbas, 2019; Simchovitz-Gesher and Soreq, 2020; Serrano-Saiz and Isogai, 2021). Gender-specific differences have been observed in gene expression and immune response (Gal-Oz et al., 2019). Even for COVID-19, males appear to be more susceptible to the viral infection and disease severity (Li and Li, 2020; Takahashi et al., 2020).
Sexual dimorphism has also been observed in genomic imprinting. Loss of the Peg3 imprinted gene causes more severe defects in the male placentas than the female ones (Tunster et al., 2018). There is an increased risk of type 2 diabetes (T2D) upon reduced expression of the KLF14 imprinted gene at the PEG1 imprinted region in females (Small et al., 2018). Cognition in childhood is impacted by the methylation at the PEG1/MEST imprinted region, with stronger effect observed in the males (Lorgen-Ritchie et al., 2019). Many miRNAs at the DLK1-DIO3 imprinted region were reported to show increased expression in male patients with multiple sclerosis (MS) but not in the female MS patients although their expression appeared to be lower in the healthy males compared with the healthy females (Baulina et al., 2019). Gender has an effect on the monoallelic expression of ATP10A that is more preferentially maternally expressed in the female human brains (Hogart et al., 2008). There are a few other studies indicating the gender-specific effects on DNA methylation at the imprinted regions or expression of the imprinted genes (Agba et al., 2017; Wang et al., 2020a; Wang et al., 2020b).
We noticed that the DNA methylation level was much higher at the Rasgrf1 ICR in the two Zfp57 maternal–zygotic mutant (M−Z-) embryos than at the Rasgrfl ICR in the other two M−Z− embryos in our previous study (Jiang et al., 2021). Interestingly, we found that DNA methylation imprint was more susceptible to loss of ZFP57 at the Rasgrf1 and AK008011 ICRs in the male M−Z- embryos than in the female M−Z− embryos. Furthermore, loss of ZFP57 caused sexually dimorphic effects on allelic expression switch or expression levels of some imprinted genes in mouse embryos. This is the first study to show sexually dimorphic effects of ZFP57 on genomic imprinting in mouse embryos. Zfp57 exhibits maternal–zygotic effect in genomic imprinting and embryonic lethality (Li et al., 2008; Takahashi et al., 2015; Takahashi et al., 2019; Jiang et al., 2021). In this study, we also found that loss of just maternal Zfp57 caused loss of DNA methylation at a subset of ICRs and loss of parent-of-origin–dependent expression of some imprinted genes in mouse embryos, indicating crucial maternal effect of Zfp57 on a small subset of imprinted regions.
Whole-genome bisulfite sequencing (WGBS) was performed to examine DNA methylation in the 129/DBA hybrid Zfp57+/− (M+Z+), Zfp57−/+ (M−Z+), and Zfp57−/− (M−Z−) embryos derived from timed mating with Zfp57+/− or Zfp57−/− 129 female mice being crossed with Zfp57+/− (DBA*) male mice mainly on the DBA/2J genetic background as reported in the previous study (Jiang et al., 2021). We found that the two M−Z- mutant embryos had much higher levels of DNA methylation at the Rasgrf1 imprinted region than two other M−Z- mutant embryos (Jiang et al., 2021). We tested these embryo samples to find out what may cause the differences of DNA methylation at this imprinted region when ZFP57 was absent.
We wonder if gender might contribute to the effects of ZFP57 on DNA methylation at the Rasgrf1 ICR as well as other ICRs. This may have resulted in the substantial differences of DNA methylation observed at the Rasgrf1 ICR among four M−Z− embryos in the previous WGBS study. There was no significant difference in DNA methylation at all 24 known ICRs comparing female M+Z+ embryos with the male M+Z+ embryos, except that DNA methylation was slightly significantly increased at the Nap1I5 ICR but decreased at the Grb10 ICR in the male M+Z+ embryos compared with the female M+Z+ embryos (Supplementary Figure S1A). DNA methylation was similar at all ICRs except for the slight increase of DNA methylation at the Peg3 ICR in the comparison of male M−Z+ embryos with the female ones (Supplementary Figure S1B). DNA methylation was significantly reduced at the AK008011 ICR in the comparison of male M−Z− embryos with the female ones (Supplementary Figure S1C). It was dramatically reduced at the Rasgrf1 ICR in the male M−Z- embryos compared with the female ones, although it was not statistically significant, which will be discussed later (Supplementary Figure S1C). Therefore, we examined DNA methylation at the ICRs in individual embryos subjected to WGBS in our previous study (Jiang et al., 2021). As expected, DNA methylation was similarly lost at most ICRs in the female or male M−Z− mutant embryos (Figures 1A, 1A’). It was largely retained at five imprinted regions (Peg10, Kcnq1ot1, Gpr1, Slc38a4, and H19) in both female or male M−Z- mutant embryos (Figures 1B, 1B’). Intriguingly, DNA methylation appeared to be more severely lost at the Peg13, Rasgrf1, and AK008011 ICRs in the male M−Z- mutant embryos than in the female M−Z- mutant embryos (Figures 1C, 1C’). This finding can be easily visualized on the methylation IGV plots for three ICRs in these M+Z+, M−Z+, and M−Z- embryos (Figure 2). One of two male M−Z− embryos displayed more severe loss of methylation at the Peg13 ICR, whereas DNA methylation was more severely lost at the Rasgrf1 and AK008011 ICRs in both male M−Z− embryos (Figures 1, 2). We also found that one female M−Z− mutant embryo had reduced DNA methylation at the Rasgrf1 ICR, whereas DNA methylation did not appear to be lost at the Rasgrf1 ICR in the other female M−Z− mutant embryos (Figures 1C, 2B).
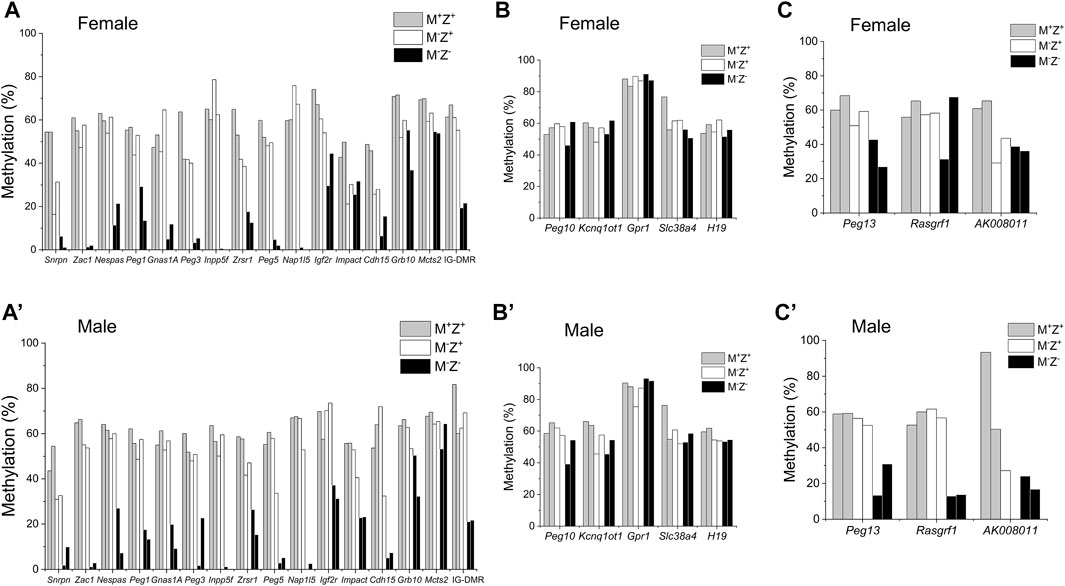
FIGURE 1. DNA methylation at three imprinted regions appeared to be more severely lost in male Zfp57 maternal–zygotic mutant embryos than in the female ones. Genomic DNA samples were isolated from the Zfp57+/− (M+Z+), Zfp57−/+ (M−Z+), and Zfp57−/− (M−Z-) hybrid E13.5 embryos from the timed mating between Zfp57+/− (or Zfp57−/−) 129 female mice and Zfp57+/− (DBA*) male mice mainly on the DBA/2J genetic background as reported in the previous study (Jiang et al., 2021). Two female embryos of each genotype and two male embryos of each genotype were used in this study. Whole-genome bisulfite sequencing (WGBS) was carried out to examine DNA methylation at previously known 21 maternally methylated and three paternally methylated ICRs of the imprinted regions. (A–C) ICR methylation in the female embryos. (A’–C’) ICR methylation in the male embryos. (A,A’) DNA methylation imprint appeared to be similarly affected at 16 ICRs in the female or male maternal–zygotic mutant (M−Z-) embryos. These include the Snrpn, Zac1, Nespas, Peg1, Gnas1A, Peg3, Inpp5f, Zrsr1, Peg5, Nap1l5, Igf2r, Impact, Cdh15, Grb10, and Mcts2 ICRs as well as the IG-DMR. (B,B’) DNA methylation imprint was mostly retained at the Peg10, Kcnq1ot1, Gpr1, Slc38A4, and H19 ICRs in the female or male maternal–zygotic mutant (M−Z-) embryos. (C,C’) DNA methylation imprint appeared to be more severely lost at the Peg13, Rasgrf1, and AK008011 ICRs in the male maternal–zygotic mutant (M−Z-) embryos compared with the female maternal–zygotic mutant (M−Z-) embryos. There were no sequence reads at the AK008011 ICR in the second male M−Z+ embryo in (C’).
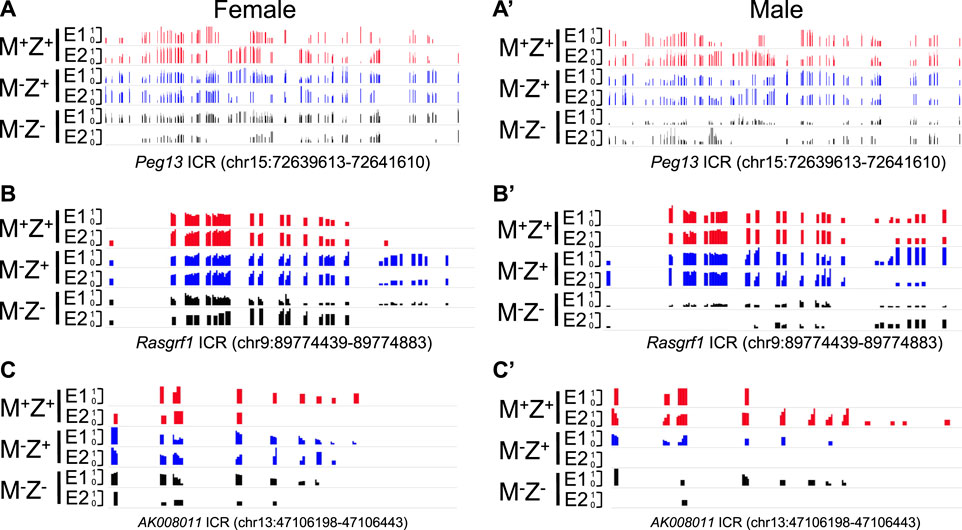
FIGURE 2. Methylation level was lower at the Peg13, Rasgrf1, and AK008011 ICRs in male Zfp57 maternal–zygotic mutant embryos than that in the female ones on the methylation IGV plot. Genomic DNA samples were isolated from Zfp57+/+ (M+Z+), Zfp57−/+ (M−Z+), and Zfp57−/− (M−Z-) hybrid 129/DBA embryos that were generated from the timed mating as reported in the previous study (Jiang et al., 2021). They were subjected to whole-genome bisulfite sequencing (WGBS) (Jiang et al., 2021). The methylation IGV plot with the scale of 0–1 is shown for the ICRs of the Peg13, Rasgrf1, and AK008011 imprinted regions in two female embryos (E1 and E2) or two male embryos (E1 and E2) of the M+Z+, M−Z+, and M−Z- genotypes. (A,A’) Methylation IGV plot of Peg13 ICR (mm9, chr15:72,639,613–72,641,610) in the female (A) or male (A’) embryos. (B,B’) Methylation IGV plot of Rasgrf1 ICR (mm9, chr9:89,774,439–89,774,883) in the female (B) or male (B’) embryos. (C,C’) Methylation IGV plot of AK008011 ICR (mm9, chr13:47,106,198–47,106,443) in the female (C) or male (C’) embryos. There were no sequence reads at the AK008011 ICR in the E2 sample of male M−Z+ in (C’). There were only six CpG sites with at least three unique reads in the E2 sample of female M−Z- in (C) and just two CpG sites with at least three unique reads in the E2 sample of male M−Z- in (C’).
DNA methylation was significantly reduced at the Peg13 ICR in the male M−Z− embryos compared with male M+Z+ and M-Z+ embryos, whereas it was close to being significantly reduced at the Peg13 ICR in the female M−Z− embryos compared with female M+Z+ embryos (Supplementary Figures S1D, S1D’). Indeed, DNA methylation was significantly lost at the Rasgrf1 ICR in the male M−Z− embryos compared with male M+Z+ and M−Z+ embryos, whereas it was not much different at the Rasgrf1 ICR comparing female M−Z− embryos with female M+Z+ or M−Z+ embryos (Supplementary Figures S1D, S1D’). DNA methylation at the AK008011 ICR was significantly reduced in both female M−Z+ and M−Z− embryos compared with the female M+Z+ embryos (Supplementary Figure S1D). It was close to being significantly reduced in both male M−Z+ and M−Z− embryos compared with the male M+Z+ embryos (Supplementary Figure S1D’). It was even more significantly lost at the AK008011 ICR in the male M−Z− embryos than in the female M−Z− embryos (Supplementary Figure S1C). We performed similar statistical comparisons for DNA methylation at the Snrpn, Impact, and Cdh15 ICRs that will be discussed for the maternal effect of Zfp57 on their DNA methylation below (Supplementary Figure S1E, S1E’).
To confirm if DNA methylation was, indeed, more severely lost at the Rasgrf1 ICR in the male M−Z− mutant embryos than in the female M−Z− mutant embryos, we carried out bacterial colony bisulfite sequencing analysis of this Rasgrf1 ICR with another set of M−Z+ and M−Z− embryos that consisted of 3–4 female and male embryos for each genotype (Figure 3). Since DNA methylation levels were similar at the Rasgrf1 ICR in the M+Z+ and M−Z+ embryos of the same gender based on WGBS (Figures 1C, 1C’), we think it should be sufficient to just perform bacterial colony bisulfite sequencing analysis of Rasgrf1 ICR to compare M−Z+ embryos with M−Z− embryos in this study. Much higher levels of DNA methylation were obtained from female and male M−Z+ embryos than with their M−Z− counterparts (Figures 3A, 3A’, 3B, 3B’). DNA methylation at the Rasgrf1 ICR was lower in four female M−Z− embryos, but it was lowest in three male M−Z− embryos (Figures 3B, 3B’). Indeed, DNA methylation was significantly reduced at the Rasgrf1 ICR in both female and male M−Z− embryos compared with their M−Z+ counterparts (Figures 3C, 3C’). It was similar in the female and male M−Z+ embryos (Figure 3D). However, DNA methylation was significantly reduced at the Rasgrf1 ICR in male M−Z− embryos compared with that of female M−Z− embryos (Figure 3E). Taken together, DNA methylation was more severely lost at the Rasgrf1 ICR in male M−Z− embryos than in female M−Z- embryos (Figure 3, Supplementary Figure S1).
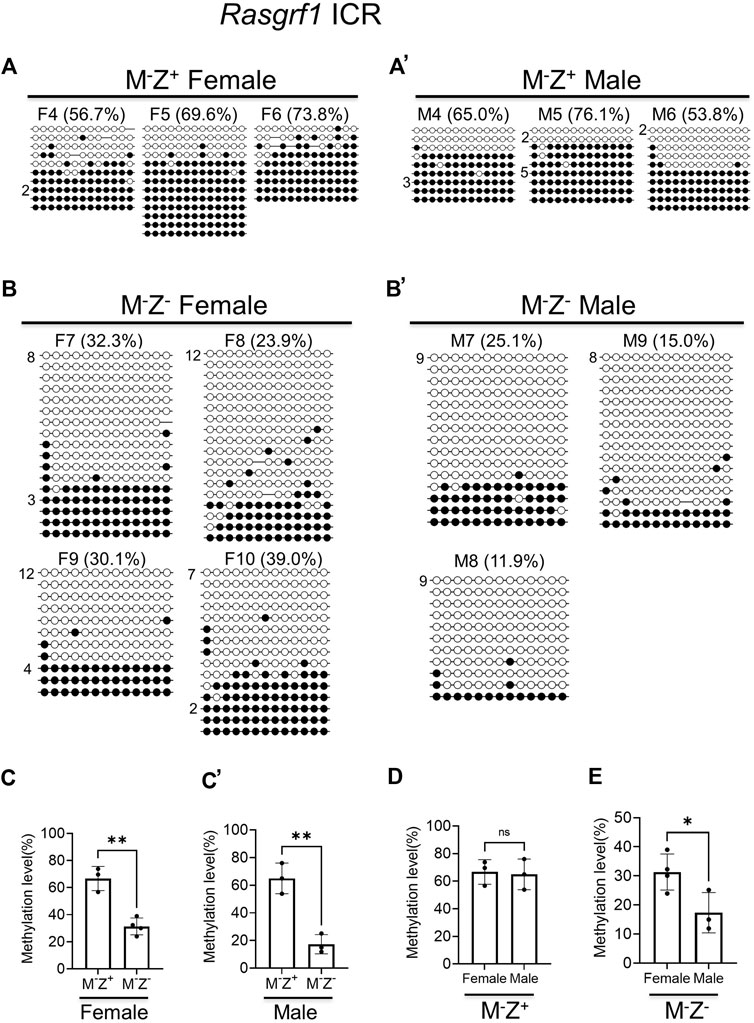
FIGURE 3. DNA methylation at the Rasgrf1 ICR was more severely lost in the male maternal–zygotic mutant embryos than in the female ones based on bacterial colony bisulfite sequencing analysis. Genomic DNA samples were isolated from at least three female or male Zfp57−/+ (M−Z+) and Zfp57−/− (M−Z-) E13.5 embryos derived from the timed mating between Zfp57 homozygous mutant female mice and heterozygous male mice. They were subjected to bisulfite mutagenesis followed by bacterial colony sequencing of the 321-bp bisulfite PCR product of the Rasgrf1 ICR region. The unconverted cytosine (C) residues were used to determine the unique clones for the bisulfite colony sequencing. Each row represents a unique clone, with the filled black circles for methylated CpG sites and unfilled white circles for unmethylated CpG sites. The number in front of a unique clone shows the number of sequenced non-unique clones containing the same sequence that cannot be distinguished by unconverted (C) residues. The percentage of DNA methylation in (A,B’) was calculated based on the number of methylated CpG sites divided by the total number of CpG sites for the sequenced unique clones of the bisulfite PCR product. Two-tailed Student’s t test was used for statistical analysis of DNA methylation level differences between two different genotypes of the same gender (C–C’) or within the same genotype between males and females (D,E). Statistical significance: *, p < 0.05; **, p < 0.01; and ***, p < 0.001. (A) Three M−Z+ female embryos (F4–F6). (A’) Three M−Z+ male embryos (M4–M6). (B) Four M−Z- female embryos (F7–F10). (B’) Three M−Z- male embryos (M7–M9). (C) DNA methylation at the Rasgrf1 ICR was significantly reduced in four M−Z- female embryos compared with three M−Z+ female embryos. (C’), DNA methylation at the Rasgrf1 ICR was significantly reduced in the three M−Z- male embryos compared with three M−Z+ male embryos. (D) DNA methylation at the Rasgrf1 ICR was similar comparing three M−Z+ female embryos with three M−Z- male embryos. (E) DNA methylation at the Rasgrf1 ICR was significantly reduced in the three M−Z- male embryos compared with four M−Z- female embryos.
Similarly, we performed bacterial colony bisulfite sequencing analysis of the AK008011 ICR in a set of M+Z+, M−Z+, and M−Z− embryos that consisted of 3–4 female and male embryos for each genotype (Figure 4). DNA methylation was significantly reduced at the AK008011 ICR in the male M−Z− embryos compared with male M+Z+ or M−Z+ embryos, whereas it was close to being significantly reduced in the female M−Z+ or M−Z− embryos compared with female M+Z+ embryos (Figures 4A, 4A’, 4B, 4B’, 4C, 4C’, 4D, 4D’). It was also significantly decreased in male M−Z+ embryos in comparison to male M+Z+ embryos (Figure 4D’). There was no significant difference in DNA methylation at the AK008011 ICR comparing female M+Z+ or M−Z+ embryos with their male counterparts, respectively (Figures 4A, 4A’, 4B, 4B’, 4E). Interestingly, DNA methylation was more significantly reduced at the AK008011 ICR in male M−Z− embryos than in female M−Z− embryos (Figure 4E). These results suggest that DNA methylation at the AK008011 ICR was more significantly lost in the male M−Z− embryos than in female M−Z− embryos (Supplementary Figure S2). Therefore, Zfp57 exhibited sexually dimorphic effect on DNA methylation at the AK008011 ICR in M−Z− embryos lacking both maternal and zygotic Zfp57.
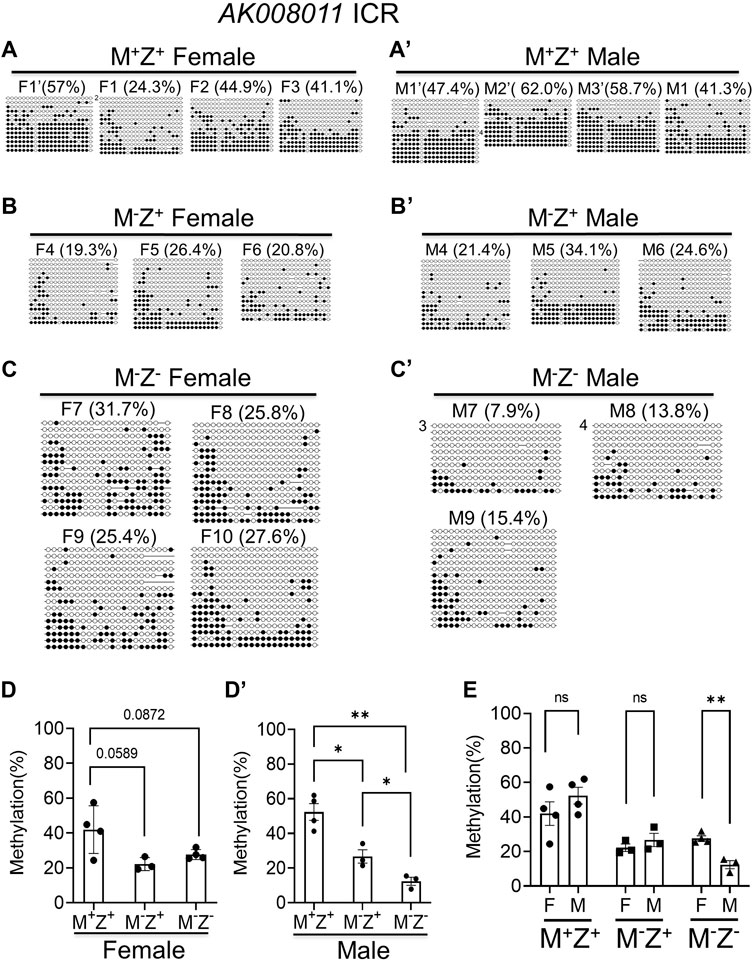
FIGURE 4. DNA methylation at the AK008011 ICR was more significantly reduced in the male M-Z- embryos compared with the female ones based on bacterial colony bisulfite sequencing analysis. Genomic DNA samples were isolated from at least three M+Z+, M−Z+, or M−Z- E13.5 embryos of each gender that had been used for RNA-seq analyses in this study too. They were subjected to bisulfite mutagenesis followed by bacterial colony sequencing of the 605-bp bisulfite PCR product of the AK008011 ICR region. The unconverted cytosine (C) residues were used to determine the unique clones for the bisulfite colony sequencing. Each row represents a unique clone, with the filled black circles for methylated CpG sites and unfilled white circles for unmethylated CpG sites. The number in front of a unique clone shows the number of sequenced non-unique clones containing the same sequence that cannot be distinguished by unconverted (C) residues. The percentage of DNA methylation in (A,C’) was calculated based on the number of methylated CpG sites divided by the total number of CpG sites for the sequenced unique clones of the bisulfite PCR product. Two-tailed Student’s t test was used for statistical analysis of DNA methylation level differences between two different genotypes of the same gender (D,D’) or within the same genotype between males and females (E). Statistical significance: *, p < 0.05; **, p < 0.01; ns, not statistically significant. (A’) Four M+Z+ female embryos (F1’ and F1–F3). (A’) Four M+Z+ male embryos (M1’–M3’ and M1). (B) Three M−Z+ female embryos (F4–F6). (B’) Three M−Z+ male embryos (M4–M6). (C) Four M−Z- female embryos (F7–F10). (C’) Three M−Z- male embryos (M7–M9). (D) DNA methylation at the AK008011 ICR was significantly reduced in four M−Z- or three M−Z+ female embryos compared with four M+Z+ female embryos. (D’) DNA methylation at the AK008011 ICR was significantly reduced in three M−Z- male embryos compared with four M+Z+ or three M−Z+ male embryos. It was also reduced in three M−Z+ male embryos compared with four M+Z+ male embryos. (E) DNA methylation at the AK008011 ICR was similar comparing M+Z+ or M−Z+ female embryos with their male counterparts. It was more significantly reduced in three M−Z- male embryos than in four M−Z- female embryos.
We examined DNA methylation at few other known ICRs. As expected, DNA methylation was similarly lost at the ICRs of Inpp5f, Zac1, and IG-DMR in the female and male M−Z- mutant embryos based on the methylation IGV plots (Supplementary Figure S2). Therefore, it seemed that loss of ZFP57 caused more severe loss of DNA methylation imprint at three ICRs in the male M−Z− embryos than in the female M−Z- embryos, with statistical significance observed at the Rasgrf1 and AK008011 ICRs.
DNA methylation appeared to be partially lost at a few ICRs in female M−Z+ or male M−Z+ embryos (Supplementary Figures 5A, 5A’). This caused partial allelic expression switch of many imprinted genes at the Snrpn imprinted region as well as Impact in male M−Z+ embryos, which is described in more detail below (Figures 5B, 5B’). Indeed, DNA methylation was partially lost at the Snrpn and Cdh15 ICRs in female M−Z+ embryos, although it was almost completely lost at these two ICRs in female M−Z− embryos (Figure 5A). One female M−Z+ embryo had more severe loss of DNA methylation at the Snrpn ICR than the other female M−Z+ embryo. DNA methylation was similarly partially lost at the Impact and AK008011 ICRs in female M−Z+ embryos as well as in female M−Z− embryos (Figure 5A). Similar results were obtained in bacterial colony bisulfite sequencing analysis for the AK008011 ICR (Figures 4A–4C). Indeed, these observations are confirmed by statistical analyses (Figure 4D, Supplementary Figures S1D–S1E). DNA methylation was significantly reduced at these four ICRs in the female M−Z+ embryos compared with those of the female M+Z+ embryos (Supplementary Figures S1D–S1E). Furthermore, it was more significantly or close to significantly reduced at the Snrpn and Cdh15 ICRs, but not at the Impact and AK008011 ICRs, while comparing the female M−Z− embryos with the female M−Z+ embryos (Supplementary Figures S1D–S1E). These results suggest that both maternal and zygotic Zfp57 are necessary but partially redundant for the maintenance of DNA methylation imprint at the Snrpn and Cdh15 ICRs in female embryos. However, only maternal Zfp57, but not zygotic Zfp57, is required for maintaining DNA methylation imprint at the Impact and AK008011 ICRs in female embryos.
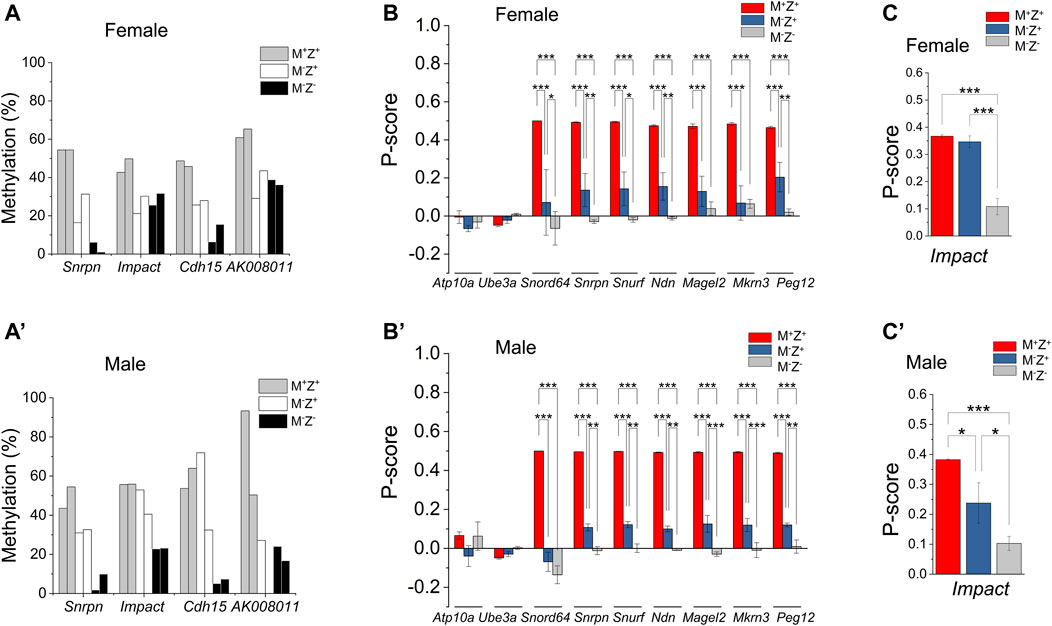
FIGURE 5. Maternal effect of Zfp57 was observed on DNA methylation imprint in a few imprinted regions as well as on the expression of a few imprinted genes. Genomic DNA samples were isolated from the Zfp57+/− (M+Z+), Zfp57−/+ (M−Z+), and Zfp57−/− (M−Z-) hybrid 129/DBA E13.5 embryos and subjected to whole-genome bisulfite sequencing (WGBS) analysis (Jiang et al., 2021). Two female embryos of each genotype and two male embryos of each genotype were used in this study for examination of DNA methylation imprint at the imprinted regions. For RNA-seq analysis, at least three RNA samples were used for each genotype (M+Z+, M−Z+, and M−Z-) of the female or male embryos. (A,A’) DNA methylation at the Snrpn, Impact, Cdh15, and AK008011 ICRs in the female embryos (A) or male embryos (A’). (B,B’) P-score was calculated to measure the expression of the paternal alleles of the imprinted genes at the Snrpn imprinted region in the female embryos (B) or male embryos (B’). These include the Atp10a, Ube3a, Snord64, Snrpn, Ndn, Magel2, Mkrn3, and Peg12 imprinted genes at the Snrpn imprinted region. Two-way ANOVA was used for statistical analysis of the P-score differences of each imprinted gene in the female M+Z+, M−Z+, and M−Z- or male M+Z+, M−Z+, and M−Z- embryos. Statistical significance: *, p < 0.05; **, p < 0.01; and ***, p < 0.001. (C,C’) P-score was calculated to measure the paternal allele expression of Impact in the female embryos (C) or male embryos (C’). One-way ANOVA was used for statistical analysis of the P-score differences of Impact in the female or male embryos. Statistical significance: * p < 0.05; ** p < 0.01; and ***, p < 0.001. Please refer to Materials and Methods for the definition of P-score and its calculation equation.
DNA methylation was also partially lost at the Snrpn ICR in both male M−Z+ embryos (Figure 5A’). It was partially lost at the Cdh15 ICR in one but not in the other male M−Z+ embryo (Figure 5A’). DNA methylation was partially lost at the Impact ICR in male M−Z− embryos, but it was largely intact at the Impact ICR in male M−Z+ embryos (Figure 5A’). DNA methylation was partially lost at the AK008011 ICR in one male M−Z+ embryo, which was similar to loss of DNA methylation at the AK008011 ICR in both male M−Z− embryos (Figure 5A’). Unfortunately, there were no sequence reads at the AK008011 ICR in the other male M−Z+ embryo and, therefore, we could not determine if DNA methylation was similarly lost at the AK008011 ICR in that male M−Z+ embryo based on WGBS. These results were also confirmed by statistical analyses (Supplementary Figures S1D’–S1E’). DNA methylation was significantly reduced at the AK008011 ICR in male M−Z+ embryos compared with male M+Z+ embryos in bacterial colony bisulfite sequencing analysis, and it was even more significantly reduced in male M−Z− embryos than in male M−Z+ embryos (Figures 4A’–D’). DNA methylation was close to significantly reduced at the Snrpn ICR in the male M−Z+ embryos compared with the male M+Z+ embryos, but even more significantly reduced in the male M−Z− embryos than in the male M+Z+ or M−Z+ embryos (Supplementary Figure S1E’). DNA methylation was only significantly reduced at the Cdh15 ICR in the male M−Z- embryos versus male M+Z+ embryos, whereas it was significantly reduced at the Impact ICR in the male M−Z− embryos compared with that in the male M+Z+ or M−Z+ embryos (Supplementary Figure S1E’).
Actually, DNA methylation was more severely lost at the AK008011 ICR in male M−Z− embryos compared with that in female M−Z− embryos (Figures 4E, 5A, 5A’, Supplementary Figure S1C). It seems that maternal Zfp57 is required for maintaining DNA methylation imprint at the AK008011 ICR in both female and male embryos, whereas zygotic Zfp57 seemed to contribute to maintenance of DNA methylation at the AK008011 ICR in male but not female embryos. Both maternal and zygotic Zfp57 play partially redundant roles in maintaining DNA methylation at the Impact ICR in male embryos, although only maternal but not zygotic Zfp57 is involved in maintaining DNA methylation at the Impact ICR in female embryos (Figure 5A, 5A’, Supplementary Figure S1C). These are also caused by the gender-specific effect of Zfp57 on ICR DNA methylation.
Loss of DNA methylation at these ICRs in female M−Z+ and M−Z− embryos or male M−Z+ and M−Z− embryos is also clearly seen on the methylation IGV plots (Figures 2, 6). DNA methylation was similarly partially lost at the AK008011 ICR in female and male M−Z+ embryos as well as in female M−Z− embryos, with more severe loss observed in the male M−Z− embryos than in the female ones (Figures 2C, 2C’). Partial loss of DNA methylation was observed at the Snrpn ICR in female M−Z+ and male M−Z+ embryos on the methylation IGV plots, although DNA methylation was almost completely missing at the Snrpn ICR in female M−Z− and male M−Z− embryos (Figures 6A, 6A’). Partial loss of DNA methylation was observed at the Impact ICR in male M−Z−, but not male M−Z+ embryos, whereas DNA methylation was partially lost at the Impact ICR in female M−Z+ as well as in female M−Z− embryos on the methylation IGV plots (Figure 6B, 6B’). Germline-derived DNA methylation imprint at the Snrpn and Impact ICRs was present on the maternal chromosomes in M+Z+ embryos as shown in the allelic methylation IGV plots (Supplementary Figure S3, Supplementary Table S1, S2). Indeed, loss of DNA methylation imprint occurred at the maternal Snrpn ICR in M−Z+ and M−Z− embryos based on the allelic methylation IGV plots (Supplementary Figure S3A-S3A′). DNA methylation was also lost at the maternal Impact ICR in the female M−Z+ and M−Z− embryos and in male M−Z− embryos based on the allelic methylation IGV plots (Supplementary Figure S3B-S3B′). The allelic methylation results further confirm what has been observed on the methylation IGV plots of the Snrpn and Impact ICRs (Figure 6, Supplementary Figure S3A). Partial loss of DNA methylation was observed at the Cdh15 ICR in both female M−Z+ embryos and one male M−Z+ embryo (Figures 6C, 6C’). DNA methylation was almost completely lost at the Cdh15 ICR in female M−Z− and male M−Z− embryos compared with M+Z+ embryos on the methylation IGV plots (Figures 6C, 6C’).
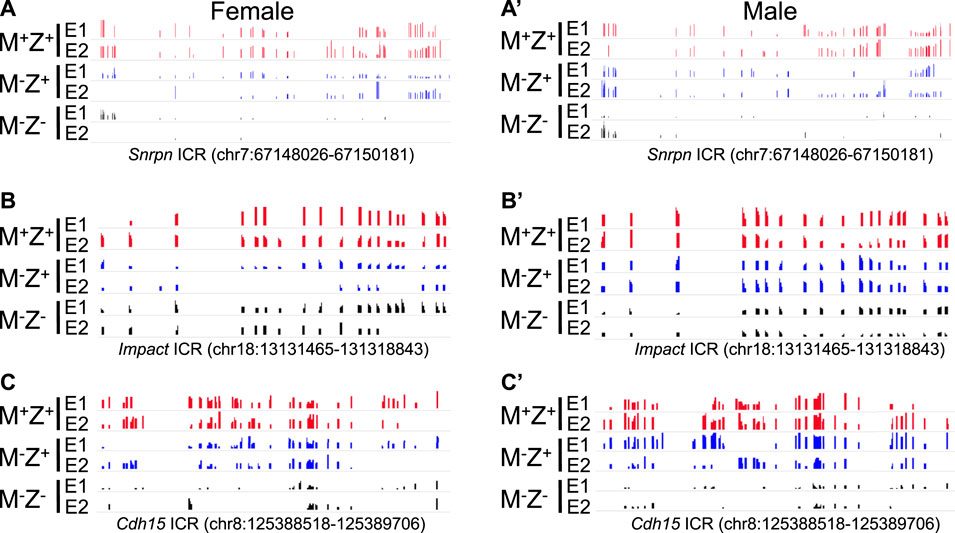
FIGURE 6. DNA methylation was partially lost at the Snrpn, Impact, and Cdh15 ICRs in the female or male embryos lacking just maternal Zfp57 on the methylation IGV plot. After WGBS analysis of the genomic DNA samples isolated from Zfp57+/+ (M+Z+), Zfp57−/+ (M−Z+), and Zfp57−/− (M−Z-) hybrid 129/DBA embryos, the methylation IGV plots with the scale of 0–1 are shown for the Snrpn, Impact, and Cdh15 ICRs in two female embryos (E1 and E2) or two male embryos (E1 and E2) for each of the M+Z+, M−Z+, and M−Z- genotypes. DNA methylation imprint was partially lost at these three imprinted regions in the M−Z+ embryos, although more severe loss of DNA methylation was observed in the M−Z- embryos. (A,A’) Methylation IGV plot of the Snrpn ICR (mm9, chr7:67,148,026–67,150,181) in the female (A) or male (A’) embryos. (B,B’) Methylation IGV plot of the Impact ICR (mm9, chr18: 13,131,465–1,31,318,843) in the female (B) or male (B’) embryos. (C,C’) Methylation IGV plot of the Cdh15 ICR (mm9, chr8:125,388,518–125,389,706) in the female (C) or male (C’) embryos.
Since DNA methylation imprint was partially lost at a few ICRs in M−Z+ embryos, we wonder if this may have any effect on the expression of the corresponding imprinted genes at these ICRs. RNA-seq analyses were carried out with another set of mouse embryos to examine expression of imprinted genes in the female M+Z+, M−Z+, and M−Z− 129/DBA hybrid E13.5 embryos and male M+Z+, M−Z+, and M−Z- 129/DBA hybrid E13.5 embryos, with at least three embryos in each group. These mouse embryos were different from the ones used for WGBS in the previous study. We used P-score for measuring the proportion of the transcripts expressed from the paternal allele of each imprinted gene (Figure 7, Supplementary Table S3–S4, Materials and Methods). Since there are no exonic SNPs on many imprinted genes, we could not analyze their P-score differences in these embryos. Ftx and Jpx were only two out of 48 known imprinted genes with an exonic SNP that showed significant P-score difference in statistical analysis (∆P-Score≥ 0.1, p < 0.05) comparing six female M+Z+ embryos with four male ones (Figure 7A). As expected, X-linked Ftx and Jpx, which are involved in X chromosome inactivation in the female embryos, along with Xist and Tsix, exhibited almost biallelic expression in the female M+Z+ embryos, whereas they were exclusively expressed from the maternal allele on the X chromosome in the male M+Z+ embryos (Figure 7A) (Collombet et al., 2020; Patrat et al., 2020). Thus, Ftx and Jpx serve as good internal positive controls for our data analyses. They were also the only two imprinted genes that displayed significant differences in the expression levels comparing six female M+Z+ embryos with four male ones that will be discussed further below (Figure 7B).
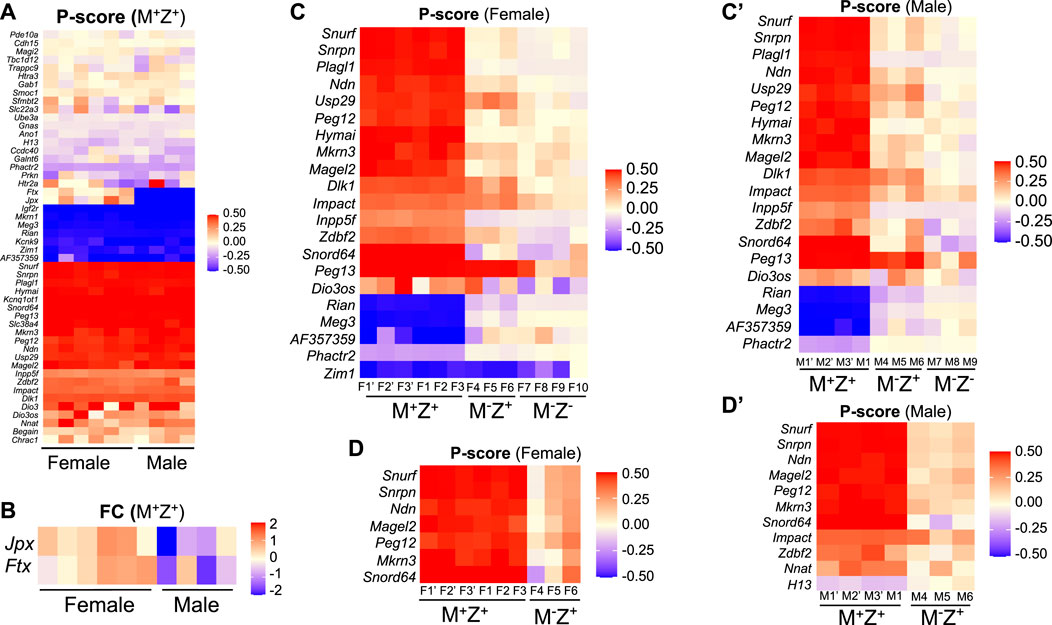
FIGURE 7. Allelic expression of some imprinted genes was primarily regulated by maternal Zfp57, and it may be affected differently in the female or male embryos lacking Zfp57. RNA-seq analysis was performed for 3–4 female 129/DBA hybrid embryos of the M+Z+, M−Z+, and M−Z- genotypes and three male 129/DBA hybrid embryos of the M+Z+, M−Z+, and M−Z- genotypes. P-score was calculated for assessing the proportion of the transcripts derived from the paternal allele of each imprinted gene carrying an SNP, and the intensity difference heatmaps were plotted accordingly ranging from 0.5 (red, paternal allele only) to −0.5 (blue, maternal allele only) Panel (A,C,C’,D,D’). Please refer to Materials and Methods for the P-score calculation and generation of intensity difference heatmaps. The Kruskal–Wallis test was used for statistical significance comparisons of the P-score differences of each imprinted gene across three different genotypes of the same gender, and then intensity difference heatmaps were generated for the imprinted genes containing the P-score difference above the threshold (∆P-Score≥ 0.1, p < 0.05) between M−Z- and M+Z+ embryos (C,C’) or between M−Z+ and M+Z+ embryos (D,D’). (A) P-score was generated and compared for the transcripts of 48 known imprinted genes with an exonic SNP between six female M+Z+ and four male M+Z+ embryos on an intensity difference heatmap. Red, paternal allele expression only with a P-score of 0.5; blue, maternal allele expression only with a P-score of −0.5. Most imprinted genes on the top of the panel are singleton ones with a P-score close to zero indicating biallelic expression. Jpx and Ftx located on the X chromosome exhibited almost biallelic expression in the female M+Z+ embryos and exclusively maternal allele expression in the male M+Z+ embryos as expected. They are good internal positive controls for our data analyses. (B) DESeq2 was used to analyze the RNA-seq data to sort out 105 imprinted genes with the TPM values above the threshold (TPM>1) in at least one embryo, out of 148 known mouse imprinted genes listed on the geneimprint website (www.geneimprint.com). Among 105 imprinted genes, only Jpx and Ftx showed significant differences, with |log2FC |> 0.3 (p < 0.05) as the threshold of significant difference, in their expression level comparison of six female M+Z+ embryos with four male M+Z+ embryos. The intensity difference heatmaps were generated for these two imprinted genes afterward. (C) Intensity difference heatmaps were shown for 21 imprinted genes in six M+Z+ (F1’–F3’, F1–F3), three M−Z+ (F4–F6), and four M−Z- (F7–F10) female embryos with a statistically different P-score (∆P-Score≥ 0.1, p < 0.05) comparing M−Z- with M+Z+ embryos. (C’) Intensity difference heatmaps were shown for 20 imprinted genes in four M+Z+(M1’–M3’, M1), three M−Z+ (M4–M6), and three M−Z- (M7–M9) male embryos with a statistically different P-score (∆P-Score≥ 0.1, p < 0.05) comparing M−Z- with M+Z+ embryos. (D) Allelic expression was significantly different (∆P-Score≥ 0.1, p < 0.05) for seven imprinted genes in the comparison of three female M−Z+ (F4–F6) with six female M+Z+ (F1’–F3’, F1–F3) embryos. (D’) Eleven imprinted genes exhibited statistically significantly different P-scores (∆P-Score≥ 0.1, p < 0.05) in the comparison of three male M−Z+ (M4–M6) with four male M+Z+ (M1’–M3’, M1) embryos.
The intensity difference heatmaps were generated for 21 imprinted genes with statistically different P-score (∆P-Score≥ 0.1, p < 0.05) by comparing the female M−Z− embryos with the female M+Z+ embryos and for 20 imprinted genes (∆P-Score≥ 0.1, p < 0.05) when the male M−Z− embryos were compared with the male M+Z+ embryos (Table 1, Figure 7C, 7C’). Zim1 at the Peg3 imprinted region was maternally expressed in female and male M+Z+ embryos or male M−Z− embryos, but it became partially biallelic in female M−Z− embryos (Supplementary Figure S4A).
Similarly, the intensity difference heatmaps (∆P-Score≥ 0.1, p < 0.05) were generated for seven imprinted genes by comparing three female M−Z+ embryos with six female M+Z+ embryos and for 11 imprinted genes when three male M−Z+ embryos were compared with four male M+Z+ embryos (Figures 7D, 7D’). Indeed, allelic expression of the imprinted genes at the Snrpn imprinted region was mostly affected in female or male M−Z+ embryos as well as in female or male M−Z− embryos compared with female or male M+Z+ embryos of the same gender (Figures 5B, 5B’, 7, Supplementary Table S3–S4). Snurf, Snrpn, Ndn, Magel2, Peg12, Mkrn3, and Snord64 at the Snrpn imprinted region were all paternally expressed in female and male M+Z+ embryos. They became biallelic in female or male M−Z− embryos, except that Snord64 was slightly preferentially maternally expressed in male M−Z− embryos (Figures 5B, 5B’, 7, Supplementary Table S3–S4). They were largely biallelic in female or male M−Z+ embryos at variable levels. Consistent with their tissue-specific or species-specific imprinting phenomena, as well as our previous results in mouse embryos, Atp10a and Ube3a at the Snrpn imprinted region were almost biallelic in the female M+Z+, M−Z+, and M−Z− embryos or male M+Z+, M−Z+, and M−Z− embryos (Figures 5B, 5B’, Supplementary Table S3-S4) (DuBose et al., 2010; Hsiao et al., 2019; Jiang et al., 2021).
Impact was preferentially paternally expressed in female and male M+Z+ embryos (Figure 5C-5C’, Figure 7C-7C’, Supplementary Table S3–S4). It became almost biallelic in female or male M−Z− embryos and partially biallelic in male M−Z+ embryos, whereas it remained preferentially paternally expressed in female M−Z+ embryos (Figure 5C-5C’, Figure 7C-7C’, 7D’). Zdbf2 and Nnat (Peg5) were preferentially paternally expressed in female or male M+Z+ embryos (Supplementary Figure S4, Supplementary Table S3-S4). Zdbf2 was partially biallelic in female M−Z+ embryos and almost completely biallelic in male M−Z+ embryos (Supplementary Figure S4B-S4B'). Nnat (Peg5) remained preferentially paternally expressed in female M−Z+ embryos, but it was almost completely biallelic in male M−Z+ embryos (Supplementary Figure S4C-S4C'). Slightly preferential maternal expression was observed for H13 in female and male M+Z+ embryos or female M−Z+ embryos (Supplementary Figure S4D-S4D'). H13 was biallelic in male M−Z+ embryos (Supplementary Figure S4D'). Taken together, loss of maternal Zfp57 caused variable allelic expression switch of Impact, Zdbf2, Nnat (Peg5), and H13 in the male M−Z+ embryos, whereas it only had a minor effect on allelic expression of Zdbf2 in female M−Z+ embryos. The gender effects on allelic expression of these imprinted genes will be further discussed below.
We also analyzed expression levels of some imprinted genes based on RNA-seq results. Among 105 analyzed imprinted genes with the TPM values above the threshold (see Materials and Methods), only two imprinted genes (Jpx and Ftx) had statistically significant differences (|log2FC |> 0.3, p < 0.05) in gene expression levels comparing the female M+Z+ embryos with the male ones (Figure 7B). Since they are involved in X chromosome inactivation only in the females (Furlan et al., 2018; Collombet et al., 2020; Patrat et al., 2020), it is expected that the expression of Jpx and Ftx was higher in the female M+Z+ embryos than in the male ones (Figure 7B).
The intensity difference heatmaps were generated for 39 imprinted genes with statistically significant changes (|log2FC |> 0.5, p < 0.05) in their expression levels when the female M−Z− embryos were compared with the female M+Z+ embryos and for 44 imprinted genes when the male M−Z− embryos were compared with the male M+Z+ embryos (Table 2; Figure 8A, 8A’). Similarly, the intensity difference heatmaps were generated for eight imprinted genes in the comparison of the female M−Z+ embryos with the female M+Z+ embryos and for nine imprinted genes when the male M−Z+ embryos were compared with the male M+Z+ embryos (Table 3, Figure 8B, 8B’). Eight imprinted genes at the Snrpn imprinted region were differentially expressed either in the comparison of the female M−Z+ embryos with the female M+Z+ embryos or in the comparison of the male M−Z+ embryos with the male M+Z+ embryos (Figure 8B, 8B’).
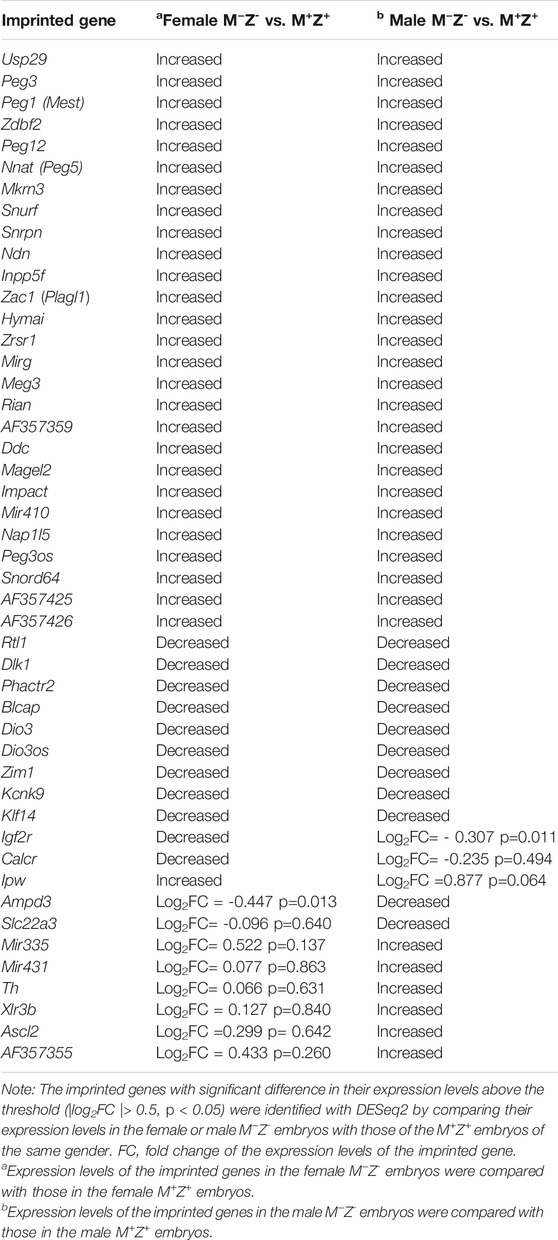
TABLE 2. Expression levels of these imprinted genes were significantly different in either the female or male M−Z- embryos compared with the M+Z+ embryos of the same gender.
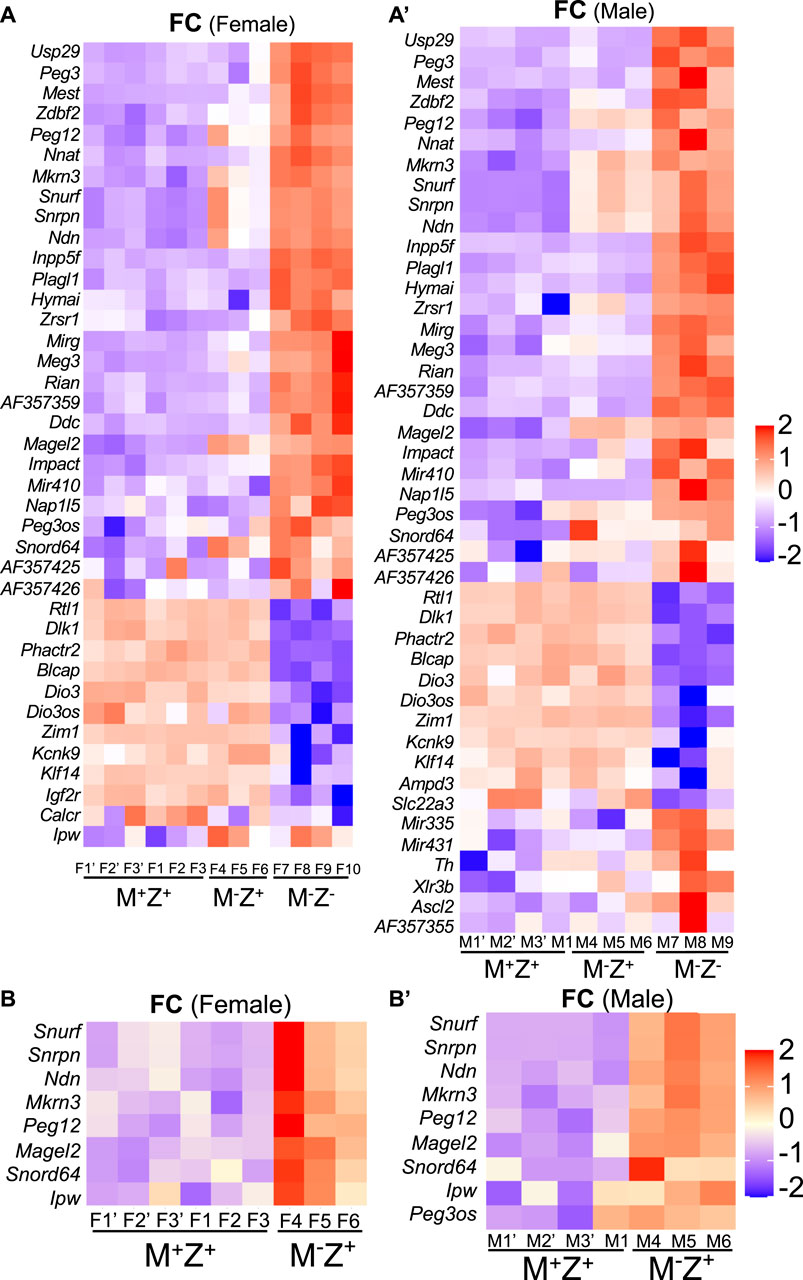
FIGURE 8. Some imprinted genes displayed sexually dimorphic effects in their expression in mouse embryos upon loss of Zfp57 or just maternal Zfp57. RNA-seq analysis was performed for female and male 129/DBA hybrid embryos of the M+Z+, M−Z+, and M−Z- genotypes. The log2FC values were used for quantification of the fold change (FC) of gene expression based on the ratios of TPM values for the imprinted gene transcripts in the M+Z+, M−Z+, and M−Z- embryos of the same gender. The imprinted genes with significant difference in their expression levels above the threshold (|log2FC |> 0.5, p < 0.05) were first identified with DESeq2 by comparing the RNA-seq data of female or male M−Z- embryos with those of the M+Z+ embryos of the same gender. Then, these imprinted genes were used for making the intensity difference heatmaps in the female M+Z+, M−Z+, and M−Z- embryos or male M+Z+, M−Z+, and M−Z- embryos. (A) Intensity difference heatmaps were generated for 39 imprinted genes in six M+Z+(F1’–F3’ and F1–F3), three M−Z+ (F4–F6) and four M−Z- (F7–F10) female embryos that displayed significantly different expression levels between the female M−Z- and female M+Z+ embryos. (A’) Intensity difference heatmaps were generated for 44 imprinted genes in four M+Z+(M1’–M3’ and M1), three M−Z+ (M4–M6), and three M−Z- (M7–M9) male embryos that displayed significantly different expression levels between the male M−Z- and male M+Z+ embryos. (B) Intensity difference heatmaps were shown for eight imprinted genes with significantly different expression levels in the comparison of three female M−Z+ (F4–F6) with six female M+Z+ (F1’–F3’ and F1–F3) embryos. (B’) Intensity difference heatmaps were shown for nine imprinted genes with significantly different expression levels in the comparison of three male M−Z+ (M4–M6) with four male M+Z+ (M1’–M3’ and M1) embryos.

TABLE 3. These imprinted genes showed significant difference in their expression levels in the female or male M−Z+ embryos compared with those in the female or male M+Z+ embryos of the same gender.
Peg3os was significantly increased in male M−Z+ embryos compared with male M+Z+ embryos (Figure 8B’). Impact was significantly increased in female M−Z+ embryos compared with female M+Z+ embryos (Supplementary Figure S5B-S6B', Supplementary Table S5-S6). There was not much change in the expression levels of Cdh15 in female M−Z+ and M−Z- embryos or male M−Z+ and M−Z− embryos compared with M+Z+ embryos (Supplementary Figure S5C-S6C', Supplementary Table S5-S6). Zrsr1 was also significantly increased in male M−Z+ embryos compared with male M+Z+ embryos (Supplementary Figure S8A-S8A', see below).
Taken together, loss of maternal Zfp57 caused increased expression of eight imprinted genes at the Snrpn imprinted region in both female and male M−Z+ embryos. It also resulted in increased expression of Peg3os and Zrsr1 in male M−Z+ embryos, as well as increased expression of Impact in female M−Z+ embryos.
Allelic expression switch occurs to some imprinted genes in mouse embryos when Zfp57 is lost (Jiang et al., 2021). Since DNA methylation imprint might be different at a few ICRs in the male M−Z− embryos compared with the female ones (Figure 9A), we wonder if allelic expression of the imprinted genes could also be differentially affected in the male M−Z− embryos compared with the female M−Z− embryos. Unfortunately, no exonic SNP was present in Rasgrf1 in the hybrid 129/DBA embryos, and its expression level was lower than the threshold of our RNA-seq TPM expression analysis (see Materials and Methods). There is no known imprinted gene located at the AK008011 imprinted region. Therefore, we could not determine in this study if there is any gender-specific effect on allelic expression switch and expression level differences for the corresponding imprinted genes at the Rasgrf1 and AK008011 imprinted regions in female or male M−Z− mutant embryos compared with the control M+Z+ embryos. This will be tested in a future study.
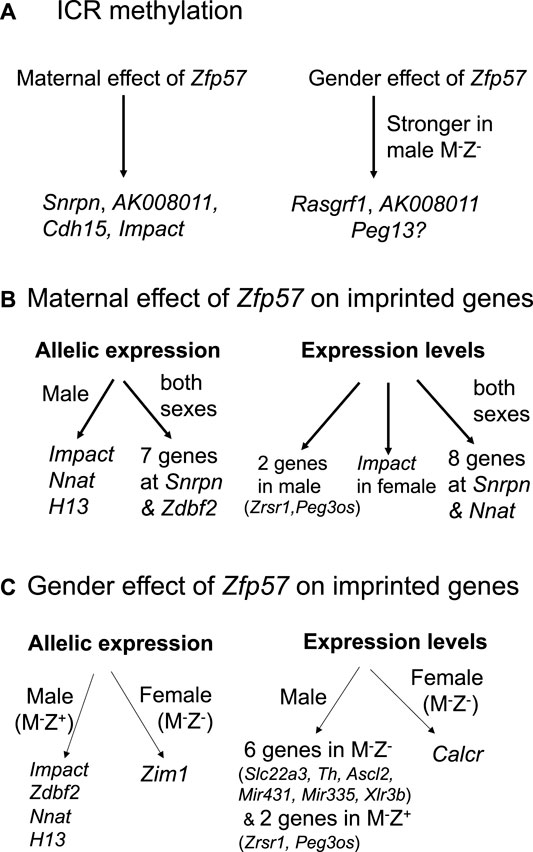
FIGURE 9. Zfp57 exhibits maternal effect and gender-specific effect in the maintenance of DNA methylation imprint and expression of the imprinted genes. The diagrams are shown for the maternal effect and gender-specific effect of Zfp57 in genomic imprinting. (A) Maternal Zfp57 is required for maintaining DNA methylation at the Snrpn, AK008011, Cdh15, and Impact ICRs, whereas there is gender effect of Zfp57 which is stronger in the male M−Z- embryos than female M−Z- embryos in the maintenance of DNA methylation at the Rasgrf1 and AK008011 ICRs, and possibly at the Peg13 ICR as well. (B) Maternal effect of Zfp57 is manifested in the allelic expression and expression levels of a few imprinted genes. Maternal Zfp57 is essential for the allelic expression of seven imprinted genes at the Snrpn imprinted region and Zdbf2 in both female and male embryos. Maternal Zfp57 is also required for the allelic expression of Impact, Nnat (Peg5), and H13 in male embryos. The expression levels of eight imprinted genes at the Snrpn imprinted region are regulated by maternal Zfp57 in both female and male embryos, whereas maternal Zfp57 regulates the expression levels of Impact in the female embryos and Zrsr1 and Peg3os in the male embryos, respectively. (C) Gender effect of Zfp57 is observed in allelic expression switch and change of expression levels of some imprinted genes in M−Z+ or M−Z- embryos. Allelic expression switch occurs to Zim1 in the female M−Z- embryos, whereas allelic expression of Impact, Zdbf2, Nnat (Peg5), and H13 is compromised in the male M−Z+ embryos. Expression of Calcr is only significantly decreased in female M−Z- embryos compared with female M+Z+ embryos. In contrast, the expression levels of six imprinted genes (Slc22a3, Th, Ascl2, Mir43, Mir335, and Xlr3b) were significantly deregulated in the male M−Z- embryos. The expression levels of Zrsr1 and Peg3os were only deregulated in the male M−Z+ embryos.
Peg13 was paternally expressed in female M+Z+ and M−Z+ embryos or male M+Z+ and M−Z+ embryos (Supplementary Figure S6A-S6A', Supplementary Table S3-S4). It became partially biallelic in female M−Z− and male M−Z− embryos (Supplementary Figure S6A-S6A', Supplementary Table S3-S4). By contrast, Kcnk9 at the Peg13 imprinted region was maternally expressed and Trappc9 at the Peg13 imprinted region was biallelic in female M+Z+, M−Z+, and M−Z− embryos or male M+Z+, M−Z+, and M−Z− embryos (Supplementary Figure S6A-S6A', Supplementary Table S3-S4). Thus, partial loss of DNA methylation at Peg13 ICR caused Peg13 to be partially biallelic in female M−Z- or male M−Z− embryos. However, it had no apparent effect on allelic expression of Kcnk9 and Trappc9 at the Peg13 imprinted region in female M−Z− or male M−Z− embryos. The residual DNA methylation at the Peg13 ICR may be sufficient to maintain allelic expression of Kcnk9 at the Peg13 imprinted region in female M−Z− or male M−Z− embryos. Despite this, expression of Kcnk9 was decreased in M−Z− embryos compared with M+Z+ and M−Z+ embryos, and it was more reduced in male M−Z− embryos than in female M−Z− embryos, although it was not statistically significant (Supplementary Figure S6B-S6B', Supplementary Figure S6C, Table S5-S6). This is consistent with DNA methylation at the Peg13 ICR in these embryos (Figure 1, Supplementary Figure S1).
Usp29 at the Peg3 imprinted region was paternally expressed in female M+Z+ and M−Z+ or male M+Z+ and M−Z+ embryos, but it was biallelic in female M−Z− or male M−Z− embryos (Supplementary Figure S4A, Supplementary Figure S4A', Supplementary Table S3-S4). Zim1 at the Peg3 imprinted region was maternally expressed in female M+Z+ and M−Z+ or male M+Z+ and M−Z+ embryos (Supplementary Figure S4A, Supplementary Figure S4A', Supplementary Table S3-S4). Although it was still maternally expressed in male M−Z− embryos, Zim1 was partially biallelic in female M−Z− embryos (Table 1, Supplementary Figure S4A, Supplementary Figure S4A', Supplementary Table S3-S4). Thus, Zim1 appeared to be affected differently in female M−Z− or male M−Z− embryos lacking ZFP57. Consistent with this, Zim1 is the only imprinted gene that displayed gender-specific effect of ZFP57 with significant P-score difference (∆P-Score≥ 0.1) in the comparisons of the female and male M−Z− embryos with their counterpart M+Z+ embryos (Figures 7C–7C’).
Impact was partially biallelic in male M−Z+ embryos, although it remained preferentially paternally expressed in female M−Z+ embryos (Figures 5C–5C’). Zdbf2 was biallelic in male M−Z+ embryos and partially biallelic in female M−Z+ embryos (Supplementary Figure S4B-S4B'). Nnat (Peg5) became almost biallelic in male M−Z+ embryos, whereas it was preferentially paternally expressed in female M−Z+ embryos (Supplementary Figure S4C-S4C'). H13 was slightly preferentially maternally expressed in female M−Z+ embryos, but it was biallelic in male M−Z+ embryos (Supplementary Figure S4D-S4D'). These four imprinted genes displayed gender-specific differences in allelic expression switch upon loss of maternal Zfp57.
We also examined allelic expression of the imprinted genes at the Inpp5f, Zac1, and Dlk1–Dio3 imprinted regions (Supplementary Figure S7, Supplementary Table S3-S4). For the tested imprinted genes at these three imprinted regions, they all became biallelic in female or male M−Z− embryos. Their allelic expression did not change upon loss of maternal Zfp57 in either female or male M−Z+ embryos. Therefore, allelic expression was only affected in M−Z- embryos for the imprinted genes at these three examined imprinted regions, and there was no difference in their allelic expression comparing male embryos with female embryos with or without ZFP57.
Loss of maternal Zfp57 resulted in mis-expression of eight imprinted genes at the Snrpn imprinted region in both female M−Z+ embryos and male M−Z+ embryos (Figures 8B, 8B’). Two other imprinted genes (Zrsr1 and Peg3os) were deregulated in the male M−Z+ embryos compared with male M+Z+ embryos, whereas the Impact expression level was significantly affected in the female M−Z+ embryos compared with female M+Z+ embryos (Figures 8B–8B’, 9B, Supplementary Figure S5B-S5B', Supplementary Figure S8A-S8A', Supplementary Figure S10A-10A').
We also observed some gender-specific differences in the expression levels of some imprinted genes in the female or male M−Z- embryos compared with their counterpart female or male M+Z+ embryos (Figure 9C, Supplementary Figure S8). Expression levels of Slc22a3, Ascl2, and Th were significantly affected in male M−Z− embryos compared with male M+Z+ embryos but not in female M−Z− embryos compared with female M+Z+ embryos (Figures 8A, 8A’, Supplementary Figure S8B-S8B', Supplementary Figure S8C-S8C'). By contrast, Calcr was only significantly reduced in female M−Z− embryos compared with female M+Z+ embryos (Figures 8A, 8A’, Supplementary Figure S8D-S8D').
Many imprinted genes showed similarly significant differences in their expression in both female and male M−Z− embryos compared with the M+Z+ embryos of the same gender (Table 2, Figure 8A–8A’). Indeed, most imprinted genes at the Dlk1–Dio3 imprinted region appeared to be similarly affected upon loss of ZFP57 comparing M−Z− embryos with M+Z+ embryos of the same gender (Table 2, Figure 8A–8A’, Supplementary Figure S9, Supplementary Table S5-S6). Begain, Dio3, Dio3os, Dlk1, and Rtl1 were reduced in the female M−Z− embryos compared with female M+Z+ embryos, whereas Rian, Meg3, Mirg, AF357359, AF357355, AF357425, and Mir410 displayed increased expression in the female M−Z− embryos compared with female M+Z+ embryos (Table 2, Supplementary Figure S9A). These genes behaved similarly in the male M−Z- embryos compared with male M+Z+ embryos (Table 2, Supplementary Figure S9A'). Expression of Mir431 and Mir335 was increased in male but not in female M−Z− embryos compared with M+Z+ embryos of the same gender (Table 2, Figures 8A, 8A’, Supplementary Figure S9). Therefore, Mir431 and Mir335 were more affected in the male embryos upon loss of Zfp57. Xlr3b was significantly increased in male M−Z− embryos compared with male M+Z+ embryos but not in the similar comparison of the female embryos (see Supplementary Figure S11B-S11B').
We also examined expression of the imprinted genes at the Peg3, Inpp5f, Zac1, and Peg5 imprinted regions in the female or male embryos (Table 2, Figures 8A, 8A’, Supplementary Figure S10, Supplementary Table S5-S6). For the genes at the Peg3 imprinted region, decreased expression of Zim1 and increased expression of Usp29, Peg3, and Peg3os were similarly observed comparing female M−Z− embryos with female M+Z+ embryos or comparing male M−Z− embryos with male M+Z+ embryos (Table 2, Supplementary Figure S10A-S10A'). Expression of Inpp5f was increased in M−Z− embryos compared with M+Z+ embryos, regardless of the gender (Table 2, Supplementary Figure S10B-S10B'). We observed increased expression of Zac1 (Plagl1) and Hymai and decreased expression of Phactr2 at the Zac1 imprinted region in M−Z− embryos compared with M+Z+ embryos, regardless of whether they are females or males (Table 2, Supplementary Figure S10C-S10C'). Increased Nnat (Peg5) expression and decreased Blcap expression were similarly observed at the Peg5 imprinted region in M−Z− embryos compared with M+Z+ embryos of the same gender (Table 2, Supplementary Figures S10D–S10D’). Decreased expression of Ampd3 was also similarly observed in both female and male M−Z− embryos compared with female and male M+Z+ embryos (Supplementary Figure S11A-S11A'). Therefore, no gender-specific effect was observed for the expression levels of these imprinted genes when Zfp57 was absent in the female or male embryos.
ZFP57 maintained DNA methylation imprint at most known ICRs in both male and female embryos. Intriguingly, more loss of DNA methylation imprint was observed at the ICRs of three imprinted regions upon loss of ZFP57 in the male M−Z− mutant embryos than in female M−Z− mutant embryos (Figures 1C–1C’, 3, 9A, Supplementary Figure S1). DNA methylation was unusually high at the Rasgrf1 ICR in one of two female M−Z− embryos based on WGBS in our previous study (Figure 1C) (Jiang et al., 2021). This was probably due to relatively low number of mapped sequence reads of the Rasgrf1 ICR in that female M−Z− embryo (Jiang et al., 2021). Accordingly, statistical significance could not be achieved when DNA methylation at the Rasgrf1 ICR was compared between two female M−Z− embryos and two male M−Z− embryos, despite that there was large difference in the DNA methylation level for the female M−Z− embryos compared with male M−Z− embryos (Supplementary Figure S1C). Nevertheless, DNA methylation was confirmed to be more severely reduced at the Rasgrf1 and AK008011 ICRs in male M−Z− embryos than in the female M−Z− embryos based on bisulfite bacterial colony sequencing (Figures 3E, 4E). It will be interesting to find out in the future if female hormones and their target genes may be involved in maintaining DNA methylation at these ICRs. It is also possible that some factors may be missing at these ICRs for the maintenance of DNA methylation in male M−Z- mutant embryos.
Although there was no allelic expression switch for Kcnk9 at the Peg13 imprinted region in female M−Z− and male M−Z− mutant embryos compared with M+Z+ embryos based on RNA-seq analysis of another set of mouse embryos that were different from the ones used in the previous WGBS experiments, the expression of Kcnk9 appeared to be more severely affected in male mutant embryos than in female mutant embryos (Supplementary Figure S6). There was partial biallelic expression of Peg13 in female and male M−Z− embryos compared with M+Z+ embryos (Supplementary Figure S6). This indicates that the residual DNA methylation at the Peg13 imprinted region was sufficient to maintain parent-of-origin–dependent monoallelic expression of Kcnk9, but not Peg13, in female and male M−Z− embryos, although two different sets of mouse embryos were used in RNA-seq and WGBS. Unfortunately, there is no SNP for the Rasgrf1 transcript and no other known transcripts at the Rasgrf1 and AK008011 imprinted regions in the hybrid 129/DBA embryos. Rasgrf1 transcripts in all embryo samples were below the TPM threshold (TPM>1) in our RNA-seq expression analysis. Therefore, we could not determine in this study if there was any gender effect on allelic expression switch and expression levels of the corresponding imprinted genes at these imprinted regions in M−Z− mutant embryos compared with the control M+Z+ embryos. These will be tested in the future research.
Zfp57 has both maternal and zygotic functions that are partially redundant in maintaining genomic imprinting at most ICRs (Jiang et al., 2021). In general, more severe loss of genomic imprinting is observed in Zfp57 maternal–zygotic mutant (M−Z−) embryos (Jiang et al., 2021). Indeed, DNA methylation imprint was almost completely lost at most known ICRs in M−Z− mutant embryos (Jiang et al., 2021). Interestingly, partial loss of DNA methylation imprint was observed at Snrpn, Cdh15, Impact, and AK008011 ICRs in the female or male M−Z+ embryos (Figures 4D, 4D’, 5A, 5A’, 9A). Furthermore, only maternal Zfp57 appeared to be necessary for maintaining DNA methylation at the AK008011 and Impact ICRs in female embryos. This suggests that zygotic Zfp57 is dispensable for the maintenance of DNA methylation at the AK008011 and Impact ICRs in female embryos. It also implies that the maintenance mechanisms for DNA methylation at these two ICRs in female embryos may be different from those at other ICRs in mouse embryos. Maternal Zfp57 is essential for the maintenance of DNA methylation imprint at these two ICRs in female embryos.
Maternal Zfp57 is also required for maintaining parent-of-origin–dependent monoallelic expression of some imprinted genes (Figure 9B). Upon partial loss of DNA methylation at the Snrpn ICR in the female or male M−Z+ embryos, loss of maternal Zfp57 caused partial allelic expression switch of seven imprinted genes at the Snrpn imprinted region as expected, although two different sets of mouse embryos were used in RNA-seq and WGBS (Figures 5B, 5B’, 9B). Complete loss of DNA methylation at the Snrpn ICR in the M−Z− embryos resulted in biallelic expression of almost all imprinted genes at the Snrpn imprinted region, that is, complete allelic expression switch of the corresponding imprinted genes (Figure 5B, 5B’). The expression levels of these imprinted genes changed accordingly, with almost 2-fold differences for most of them in the M−Z− embryos (Supplementary Figure S5A, S5A'). There were variable differences in the expression of these imprinted genes ranging from less than 2-fold–2-fold in the M−Z+ embryos (Figure 9B, Supplementary Figure S5A, S5A'). These are also consistent with the allelic switch models for the target imprinted genes proposed in our recent article, with partial or complete allelic switch in the M−Z+ or M−Z− embryos, respectively (Jiang et al., 2021).
In contrast with that in the female M−Z− embryos, partial loss of DNA methylation at the Impact ICR in female M−Z+ embryos did not cause allelic expression switch of the Impact imprinted gene according to the results obtained with two different sets of mouse embryos used in RNA-seq and WGBS (Figure 5). Its expression level was significantly increased in female M−Z+ embryos, albeit not as much as in female M−Z− embryos, compared with female M+Z+ embryos (Supplementary Figure S5B-S5B'). Intriguingly, partial allelic expression switch occurred to Impact in the female M−Z- or male M−Z− embryos even though there was still only partial loss of DNA methylation at the Impact ICR, with similar loss of DNA methylation to female M−Z+ embryos (Figure 5). Furthermore, partial allelic expression switch also occurred to Impact in the male M−Z+ embryos without significant loss of DNA methylation at its ICR (Figure 5). We suspect that there might be some other ZFP57-dependent factors besides DNA methylation at the known Impact ICR that is required for maintaining parent-of-origin–dependent expression of the Impact imprinted gene.
Since there is no known imprinted gene at the AK008011 imprinted region, we could not test the maternal effect of Zfp57 on expression of any imprinted gene in response to partial loss of DNA methylation at the AK008011 ICR in the M−Z+ embryos. Loss of maternal Zfp57 caused allelic expression switch of Zdbf2 in both female and male M−Z+ embryos, whereas it resulted in allelic expression switch of the three imprinted genes only in the male M−Z+ embryos (Figure 9B). Nnat (Peg5) expression level was affected in both female and male M−Z+ embryos, whereas loss of maternal Zfp57 caused deregulation of the expression levels of Zrsr1 and Peg3os in the male M−Z+ embryos and Impact in the female M−Z+ embryos, respectively. These will be further discussed below, together with the gender effect of ZFP57 on their expression. Taken together, the maternal effect of Zfp57 is manifested in the maintenance of DNA methylation imprint at a few imprinted regions, in particular Snrpn, as well as the parent-of-origin–dependent expression of some corresponding imprinted genes in mouse embryos. It may also result in deregulation of a few other imprinted genes without concomitant loss of DNA methylation imprint.
DNA methylation imprint was similarly lost at most known imprinted regions in M−Z- embryos compared with M+Z+ embryos of the same gender (Figure 1A, 1A’). Consequently, loss of ZFP57 caused similar allelic expression switch and expression level differences of most imprinted genes in M−Z- embryos compared with M+Z+ embryos of the same gender that we examined, although two different sets of mouse embryos were used in RNA-seq and WGBS (Tables 1, 2). Interestingly, we observed a sexually dimorphic effect of ZFP57 on the expression of some imprinted genes as discussed below (Figure 9C).
There was not much gender-specific difference in the allelic expression of 48 imprinted genes with an exonic SNP in the wild-type female and male mouse embryos that we examined in this study, except for two X-linked Jpx and Ftx genes (Figure 7A). Expression levels were mostly similar for 105 known imprinted genes in the wild-type female and male embryos, with only Jpx and Ftx showing significant differences in their expression levels (Figure 7B).
In this relatively comprehensive study, gender effect has been observed on allelic expression as well as expression levels of a few mouse imprinted genes in response to loss of Zfp57 or loss of just maternal Zfp57. Zim1 was switched from maternal allele–specific expression to become partially biallelic only in the female M−Z− embryos (Table 1, Figure 7C, 7C’). Zdbf2 was partially biallelic in the female M−Z+ embryos and biallelic in male M−Z+ embryos. Allelic expression of Impact, Nnat (Peg5), and H13 was also compromised in the male M−Z+ embryos, but not in the female ones (Figure 9B).
The sexually dimorphic effect of ZFP57 occurred to the expression levels of a few imprinted genes upon loss of Zfp57 (Figure 9C). Calcr was only significantly decreased in female M−Z- embryos compared with female M+Z+ embryos (Supplementary Figure S8). Expression levels of Slc22a3, Ascl2, and Th were significantly affected in male M−Z− embryos compared with male M+Z+ embryos, but not in the similar comparisons of female embryos (Supplementary Figure S8). Mir431 and Mir335 at the Dlk1–Dio3 imprinted region were also more severely affected in male M−Z− embryos (Table 2, Figures 8A, 8A’, Supplementary Figure S9). Xlr3b was only significantly increased in male M−Z− embryos compared with male M+Z+ embryos (Supplementary Figure S11B-S11B'). Furthermore, the expression levels of Zrsr1 and Peg3os were only deregulated in the male (but not female) M−Z+ embryos compared with male M+Z+ embryos. These results suggest that ZFP57 exerts a sexually dimorphic effect in the expression of some imprinted genes. It also means that we may need to analyze gender effect on some phenotypes caused by the loss of Zfp57 in the future.
In our current study, no imprinted genes exhibit gender-specific difference in their expression levels in the wild-type female and male embryos, except for two X-linked genes as expected. Zfp57 displays maternal–zygotic effect in maintaining genomic imprinting at most imprinted regions in mouse embryos. It also exerts maternal and sexual dimorphic effects on DNA methylation at a subset of imprinted regions. These effects of Zfp57 are manifested in allelic expression switch and expression level changes of a number of known imprinted genes.
The mice carrying the Zfp57 deleted mutant allele on the 129S6/SvEvTac genetic background were generated in the original study, and they were called the 129 mice in this study (Li et al., 2008). We also performed 12 backcrosses for the 129 mice carrying the Zfp57 deleted mutant allele with the wild-type DBA/2J mice to obtain Zfp57 heterozygous mice mainly on the DBA/2J background that was named Zfp57+/− (DBA*) in a recent study (Jiang et al., 2021). Then, timed pregnancy mating was set up between Zfp57−/− homozygous 129 female mice and Zfp57+/− (DBA*) male mice to obtain 129/DBA hybrid E13.5 embryos. The mouse vaginal plug was checked daily. The female mice were presumed to be potentially pregnant for 0.5 days when their vaginal plug was found around the noon of the day. The pregnant female mice were dissected for 129/DBA hybrid E13.5 embryos used for RNA-seq analysis as well as whole-genome bisulfite sequencing (WGBS) analysis.
To determine the gender of these 129/DBA hybrid E13.5 embryos, SRY genotyping was carried out with the primers SRY-F1 and SRY-R1 for a PCR product of 266 bp present in the male embryos but absent in the female embryos. The sequence for SRY-F1 is 5’- CCACTCCTCTGTGACACT, whereas the sequence for SRY-R1 is 5’- GAGAGCATGGAGGGCCAT.
Total RNA samples were obtained from the 129/DBA hybrid E13.5 embryos from the timed pregnancy mating. At least three female embryos of each genotype of M+Z+, M−Z+, and M−Z- were used for RNA purification and sent out for RNA-seq analysis. Similarly, total RNA samples were purified from at least three male embryos of each genotype of M+Z+, M−Z+, and M−Z- and subjected to RNA-seq analysis. The data analyses were performed as described below.
RNA-seq analysis was performed for the RNA samples obtained from the mouse embryos that were different from the ones used in WGBS of the previous study. At least three embryos were analyzed for each genotype of female M+Z+, M−Z+, and M−Z− 129/DBA hybrid E13.5 embryos and male M+Z+, M−Z+, and M−Z− 129/DBA hybrid E13.5 embryos.
First, the quality of the sequence reads obtained from RNA-seq was assessed by FastQC (v0.11.9) downloaded from the website (www.bioinformatics.babraham.ac.uk/projects/fastqc/). Then, the adapter sequences were trimmed from the sequence reads by using Trim_Galore (v0.6.6) (https://github.com/FelixKrueger/TrimGalore). Only good quality sequence reads were aligned to the mouse reference genome (mm9) on the UCSC website using STAR (v2.7.8a), with the setting of “–outFilterMultimapNmax 1 –alignEndsType EndToEnd–outSAMattributes NH HI NM MD” plus other default parameters (Dobin et al., 2013). The sequence reads were mapped to the annotated genes (UCSC mm9. gtf) with the featureCounts function (-O–s 2 option) of Subread (v.2.0.1) (Liao et al., 2014). Out of 148 mouse imprinted genes listed on the website of the geneimprint database: www.geneimprint.com, 139 had been mapped to the mouse reference genome (mm9) and then used for expression analyses in this study.
Transcripts per million (TPM) values were calculated for 139 mapped imprinted genes with more than 10 sequence reads. 105 imprinted genes with the TPM value of more than 1.0 in at least one embryo sample were selected for further analysis of their expression differences across different genotypes or genders. The log2FC values were used to quantify the fold change (FC) of gene expression based on the ratios of TPM for this imprinted gene in three genotypes of M+Z+, M−Z+, and M−Z−. The differentially expressed genes in the RNA-seq data of different genotypes were first identified by DESeq2 before they were used for constructing the intensity difference heatmaps (Love et al., 2014). The cutoff threshold was set for |log2FC |> 0.3 (p < 0.05) for the intensity difference heatmap when the expression levels of the imprinted genes were compared between six female M+Z+ embryos and four male M+Z+ embryos (Figure 7B). It was set for |log2FC |> 0.5 (p < 0.05) in other intensity difference heatmaps comparing their expression levels in the M+Z+, M−Z+, and M−Z− embryos of the same gender (Figure 8).
The SNPs present in the 129/DBA hybrid embryos were determined by using SNP calling with the Genome Analysis Toolkit (GATK) software package (McKenna et al., 2010; Van der Auwera et al., 2013). These SNPs were used for creating N-masked genomes from the reference genome (mm9) using BEDTools with incorporation of “N” at the position of an SNP (Quinlan and Hall, 2010). Mapping of the sequence reads to the N-masked genomes was performed using STAR (v2.5.4a), with the setting of “–outFilterMultimapNmax 1 –alignEndsType EndToEnd–outSAMattributes NH HI NM MD” and other default parameters (Dobin et al., 2013). The mapped sequence reads were assigned to the maternal (129) or paternal (DBA) genome using SNPsplit (v0.4.0), and then a file was generated with the SNPs (Krueger and Andrews, 2016). The allele-specific sequence reads were assigned to the maternal and paternal alleles of the imprinted genes using featureCounts (Liao et al., 2014). Further analyses in this study were limited to the genes with more than 10 counts based on the sum of the mapped reads of maternal and paternal alleles.
P-score was calculated for assessing the proportion of the transcripts derived from the paternal allele of each imprinted gene based on the following equation: P-score = P/(M + P)-0.5. P stands for the number of the paternal (P) allele transcript reads of an imprinted gene, whereas M stands for the number of the maternal (M) allele transcript reads of an imprinted gene. This was adapted from the calculation method for allelic expression of X-linked genes in a recent study (Yu et al., 2021). The imprinted genes with statistically significant allelic expression difference (∆P-Score≥ 0.1, p < 0.05) used for intensity difference heatmap analyses were identified by comparing their P-score values among different genotypes by using the Kruskal–Wallis test (Figure 7). Similar results were obtained by using the binomial test used in a recent study (Yu et al., 2021).
The WGBS data were based on the previous study (Jiang et al., 2021). The gender of these female or male embryos used for WGBS was determined by PCR genotyping of the Sry gene as described above. Please refer to the previous article for the WGBS sequence depth and genome coverage (Jiang et al., 2021). The WGBS sequence reads of these embryo samples were mapped to the abovementioned N-masked reference genome. The CpG sites present in at least three unique reads were used for quantification of DNA methylation of an ICR in the subsequent analyses. The methylated and unmethylated C residues were identified at each CpG site of an ICR. Then, the DNA methylation level was quantified for each CpG site of this ICR based on the number of methylated C residues divided by the total number of C residues for this CpG site. The percentage (%) of DNA methylation for the ICR was the average of DNA methylation levels of all CpG sites at this ICR.
SNPsplit (v0.4.0) was used to separate the allelic DNA methylation reads of the ICRs with the default parameters (Krueger and Andrews, 2016). Only CpG sites covered by at least one unique sequence read were subject to measurement of DNA methylation levels of the maternal or paternal ICR. Allelic DNA methylation was determined for only a few ICRs with SNPs.
Genomic DNA samples purified from the mouse embryos were subjected to bisulfite mutagenesis with the EZ DNA Methylation-Gold™ Kit (Zymo Research). Then, bisulfite PCR was carried out for a Rasgrf1 ICR region with two rounds of nested PCR reactions (Zuo et al., 2012; Takikawa et al., 2013a). The primers used for the first round of bisulfite PCR were Ras-Bis-OF with the sequence of 5’- GTTATTATTATGTGTTATGTGTAGTAAG and Ras-Bis-OR1 with the sequence of 5’- TAATACAACAACAACAATAACAATC. The primers used for the second round of nested bisulfite PCR were Ras-Bis-IF with the sequence of 5’- GGTGTAGAATATGGGGTTG and Ras-Bis-IR with the sequence of 5’- ATACAACAACAACAATAAC. Two specific PCR products of 321 bp and 408 bp were obtained after the second round of nested bisulfite PCR, and they had completely overlapping 321-bp long sequences at one end. Both specific PCR products were cloned into the pUCm-T vector (Sangon, cat# B522213), and the resultant bacterial colonies were sequenced to determine the methylation status of the CpG sites within the 321-bp region of the Rasgrf1 ICR. The obtained sequences were analyzed with the web-based program called QUMA (http://quma.cdb.riken.jp/). Then, DNA methylation status was determined for the CpG sites in the Rasgrf1 ICR as shown in Figure 3.
Bacterial colony bisulfite sequencing was similarly carried out for the AK008011 ICR with the genomic DNA samples derived from 3–4 M+Z+, M−Z+, or M−Z− E13.5 embryos of each gender that had also been used for RNA-seq analyses in this study (Figures 4, 7). Two rounds of nested PCR reactions were performed for the bisulfite genomic DNA product. The primers used for the first round of bisulfite PCR were AK-Bis-OFn1 with the sequence of 5’- GGTTTAGTTAGGGAAAGGGT and AK-Bis-ORn1 with the sequence of 5’- CACACACCTAAATCCTAACACT, with the PCR product of 765 bp. The primers used for the second round of nested bisulfite PCR were AK-Bis-OFn2 with the sequence of 5’- GTGGTTATATATTGTAGGGTAGG and AK-Bis-ORn2 with the sequence of 5’- CCTACATAATTAAAACCTACCTC. The obtained PCR product of 605 bp was cloned into the pUCm-T vector (Sangon, cat# B522213), and the resultant bacterial colonies were sequenced to determine the methylation status of the CpG sites of the AK008011 ICR. The obtained sequences were analyzed with the web-based program called QUMA (http://quma.cdb.riken.jp/). Then, DNA methylation status was determined for the CpG sites in the AK008011 ICR as shown in Figure 4.
Except for statistical analyses used to generate the intensity difference heatmaps, ANOVA (Fisher LSD) was used for statistical analysis of imprinted gene expression (FC value and P-score) differences among the RNA-seq samples of different genotypes of the same gender. And, two-tailed Student’s t-test was used for analyzing the statistical significance when expression of the imprinted genes was compared between the female samples and the male samples within the same genotype.
ANOVA (Fisher LSD) was similarly utilized in analyzing the statistical significance of DNA methylation at the ICRs of different genotypes of the same gender based on the WGBS data. And, two-tailed Student’s t-test was used to compare DNA methylation at the ICRs of the same genotype between the female samples and male samples. Calculation of the p-value for all statistical analyses was carried out by using the open source R software (https://www.R-project.org).
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/geo/, GSE165079; https://www.ncbi.nlm.nih.gov/geo/, GSE189761.
The animal study was reviewed and approved by the ShanghaiTech University.
XL designed the experiments. ZX, JS, YZ, YL, JZ, QC, CS, and SG performed the experiments. ZX, YZ, and XL carried out the data analyses. ZX, YZ, WX, FW, YB, YY, and XL wrote the manuscript.
The study in the authors’ laboratories has been supported by the grants from the Ministry of Science and Technology of the People’s Republic of China (Grant # 2018YFC1005004 to XL and YY), the Science and Technology Commission of Shanghai Municipality (Grant # 18PJ1407700 to XL), and the National Natural Science Foundation of China (Grant # 31670756 to YB).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We appreciate the help with mouse maintenance and breeding by the Animal Facility of Protein Center of Shanghai Zhangjiang Laboratory, with special thanks to Chaohua Zheng, Haojie Chen, and Hao Feng. We also would like to thank the Molecular and Cell Biology Core Facility (MCBCF) and Multi-Omics Core Facility (MOCF) at the School of Life Science and Technology in ShanghaiTech University for providing technical support. The RNA-seq data analyses in this study were performed on the HPC Platform of ShanghaiTech University.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2022.784128/full#supplementary-material
Agba, O. B., Lausser, L., Huse, K., Bergmeier, C., Jahn, N., Groth, M., et al. (2017). Tissue-, Sex-, and Age-specific DNA Methylation of Rat Glucocorticoid Receptor Gene Promoter and Insulin-like Growth Factor 2 Imprinting Control Region. Physiol. Genomics 49, 690–702. doi:10.1152/physiolgenomics.00009.2017
Bar, S., and Benvenisty, N. (2019). Epigenetic Aberrations in Human Pluripotent Stem Cells. EMBO J. 38. doi:10.15252/embj.2018101033
Barlow, D. P., and Bartolomei, M. S. (2014). Genomic Imprinting in Mammals. Cold Spring Harb Perspect. Biol. 6. doi:10.1101/cshperspect.a018382
Bartolomei, M. S., and Ferguson-Smith, A. C. (2011). Mammalian Genomic Imprinting. Cold Spring Harb Perspect. Biol. 3. doi:10.1101/cshperspect.a002592
Baulina, N., Osmak, G., Kiselev, I., Popova, E., Boyko, A., Kulakova, O., et al. (2019). MiRNAs from DLK1-DIO3 Imprinted Locus at 14q32 Are Associated with Multiple Sclerosis: Gender-specific Expression and Regulation of Receptor Tyrosine Kinases Signaling. Cells 8. doi:10.3390/cells8020133
Chen, P. B., Hu, R. K., Wu, Y. E., Pan, L., Huang, S., Micevych, P. E., et al. (2019). Sexually Dimorphic Control of Parenting Behavior by the Medial Amygdala. Cell 176, 1206–1221. doi:10.1016/j.cell.2019.01.024
Chess, A. (2016). Monoallelic Gene Expression in Mammals. Annu. Rev. Genet. 50, 317–327. doi:10.1146/annurev-genet-120215-035120
Collombet, S., Ranisavljevic, N., Nagano, T., Varnai, C., Shisode, T., Leung, W., et al. (2020). Parental-to-embryo Switch of Chromosome Organization in Early Embryogenesis. Nature 580, 142–146. doi:10.1038/s41586-020-2125-z
Deegan, D. F., Nigam, P., and Engel, N. (2021). Sexual Dimorphism of the Heart: Genetics, Epigenetics, and Development. Front. Cardiovasc. Med. 8, 668252. doi:10.3389/fcvm.2021.668252
Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., et al. (2013). STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 29, 15–21. doi:10.1093/bioinformatics/bts635
DuBose, A. J., Johnstone, K. A., Smith, E. Y., Hallett, R. A. E., and Resnick, J. L. (2010). Atp10a, a Gene Adjacent to the PWS/AS Gene Cluster, Is Not Imprinted in Mouse and Is Insensitive to the PWS-IC. Neurogenetics 11, 145–151. doi:10.1007/s10048-009-0226-9
Freschi, A., Hur, S. K., Valente, F. M., Ideraabdullah, F. Y., Sparago, A., Gentile, M. T., et al. (2018). Tissue-specific and Mosaic Imprinting Defects Underlie Opposite Congenital Growth Disorders in Mice. Plos Genet. 14, e1007243. doi:10.1371/journal.pgen.1007243
Furlan, G., Gutierrez Hernandez, N., Huret, C., Galupa, R., van Bemmel, J. G., Romito, A., et al. (2018). The Ftx Noncoding Locus Controls X Chromosome Inactivation Independently of its RNA Products. Mol. Cel 70, 462–472. doi:10.1016/j.molcel.2018.03.024
Gal-Oz, S. T., Maier, B., Yoshida, H., Seddu, K., Elbaz, N., Czysz, C., et al. (2019). ImmGen Report: Sexual Dimorphism in the Immune System Transcriptome. Nat. Commun. 10, 4295. doi:10.1038/s41467-019-12348-6
Hanna, C. W., and Kelsey, G. (2021). Features and Mechanisms of Canonical and Noncanonical Genomic Imprinting. Genes Dev. 35, 821–834. doi:10.1101/gad.348422.121
Hirasawa, R., and Feil, R. (2008). A KRAB Domain Zinc finger Protein in Imprinting and Disease. Developmental Cel. 15, 487–488. doi:10.1016/j.devcel.2008.09.006
Hogart, A., Patzel, K. A., and LaSalle, J. M. (2008). Gender Influences Monoallelic Expression of ATP10A in Human Brain. Hum. Genet. 124, 235–242. doi:10.1007/s00439-008-0546-0
Hsiao, J. S., Germain, N. D., Wilderman, A., Stoddard, C., Wojenski, L. A., Villafano, G. J., et al. (2019). A Bipartite Boundary Element restrictsUBE3Aimprinting to Mature Neurons. Proc. Natl. Acad. Sci. USA 116, 2181–2186. doi:10.1073/pnas.1815279116
Jiang, W., Shi, J., Zhao, J., Wang, Q., Cong, D., Chen, F., et al. (2021). ZFP57 Dictates Allelic Expression Switch of Target Imprinted Genes. Proc. Natl. Acad. Sci. U S A. 118. doi:10.1073/pnas.2005377118
Juan, A. M., and Bartolomei, M. S. (2019). Evolving Imprinting Control Regions: KRAB Zinc Fingers Hold the Key. Genes Dev. 33, 1–3. doi:10.1101/gad.322990.118
Khamlichi, A. A., and Feil, R. (2018). Parallels between Mammalian Mechanisms of Monoallelic Gene Expression. Trends Genet. 34, 954–971. doi:10.1016/j.tig.2018.08.005
Krueger, F., and Andrews, S. R. (2016). SNPsplit: Allele-specific Splitting of Alignments between Genomes with Known SNP Genotypes. F1000Res 5, 1479. doi:10.12688/f1000research.9037.2
Lee, J. T., and Bartolomei, M. S. (2013). X-inactivation, Imprinting, and Long Noncoding RNAs in Health and Disease. Cell 152, 1308–1323. doi:10.1016/j.cell.2013.02.016
Li, A. J., and Li, X. (2020). Sex-dependent Immune Response and Lethality of COVID-19. Stem Cel Res 50, 102116. doi:10.1016/j.scr.2020.102116
Li, X. (2013). Genomic Imprinting Is a Parental Effect Established in Mammalian Germ Cells. Curr. Top. Dev. Biol. 102, 35–59. doi:10.1016/b978-0-12-416024-8.00002-7
Li, X., Ito, M., Zhou, F., Youngson, N., Zuo, X., Leder, P., et al. (2008). A Maternal-Zygotic Effect Gene, Zfp57, Maintains Both Maternal and Paternal Imprints. Developmental Cel. 15, 547–557. doi:10.1016/j.devcel.2008.08.014
Liao, Y., Smyth, G. K., and Shi, W. (2014). featureCounts: an Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinformatics 30, 923–930. doi:10.1093/bioinformatics/btt656
Liu, Y., Toh, H., Sasaki, H., Zhang, X., and Cheng, X. (2012). An Atomic Model of Zfp57 Recognition of CpG Methylation within a Specific DNA Sequence. Genes Dev. 26, 2374–2379. doi:10.1101/gad.202200.112
Lorgen-Ritchie, M., Murray, A. D., Ferguson-Smith, A. C., Richards, M., Horgan, G. W., Phillips, L. H., et al. (2019). Imprinting Methylation in SNRPN and MEST1 in Adult Blood Predicts Cognitive Ability. PLoS One 14, e0211799. doi:10.1371/journal.pone.0211799
Love, M. I., Huber, W., and Anders, S. (2014). Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 15, 550. doi:10.1186/s13059-014-0550-8
Mackay, D. J. G., Callaway, J. L. A., Marks, S. M., White, H. E., Acerini, C. L., Boonen, S. E., et al. (2008). Hypomethylation of Multiple Imprinted Loci in Individuals with Transient Neonatal Diabetes Is Associated with Mutations in ZFP57. Nat. Genet. 40, 949–951. doi:10.1038/ng.187
Marcho, C., Bevilacqua, A., Tremblay, K. D., and Mager, J. (2015). Tissue-specific Regulation of Igf2r/Airn Imprinting during Gastrulation. Epigenetics & Chromatin 8, 10. doi:10.1186/s13072-015-0003-y
McKenna, A., Hanna, M., Banks, E., Sivachenko, A., Cibulskis, K., Kernytsky, A., et al. (2010). The Genome Analysis Toolkit: a MapReduce Framework for Analyzing Next-Generation DNA Sequencing Data. Genome Res. 20, 1297–1303. doi:10.1101/gr.107524.110
Monk, D., Mackay, D. J. G., Eggermann, T., Maher, E. R., and Riccio, A. (2019). Genomic Imprinting Disorders: Lessons on How Genome, Epigenome and Environment Interact. Nat. Rev. Genet. 20, 235–248. doi:10.1038/s41576-018-0092-0
Monteagudo-Sánchez, A., Hernandez Mora, J. R., Simon, C., Burton, A., Tenorio, J., Lapunzina, P., et al. (2020). The Role of ZFP57 and Additional KRAB-Zinc finger Proteins in the Maintenance of Human Imprinted Methylation and Multi-Locus Imprinting Disturbances. Nucleic Acids Res. 48, 11394–11407. doi:10.1093/nar/gkaa837
Patrat, C., Ouimette, J. F., and Rougeulle, C. (2020). X Chromosome Inactivation in Human Development. Development 147. doi:10.1242/dev.183095
Pfaff, D., and Barbas, H. (2019). Mechanisms for the Approach/Avoidance Decision Applied to Autism. Trends Neurosciences 42, 448–457. doi:10.1016/j.tins.2019.05.002
Plasschaert, R. N., and Bartolomei, M. S. (2015). Tissue-specific Regulation and Function of Grb10 during Growth and Neuronal Commitment. Proc. Natl. Acad. Sci. USA 112, 6841–6847. doi:10.1073/pnas.1411254111
Prickett, A. R., Montibus, B., Barkas, N., Amante, S. M., Franco, M. M., Cowley, M., et al. (2021). Imprinted Gene Expression and Function of the Dopa Decarboxylase Gene in the Developing Heart. Front. Cel Dev. Biol. 9, 676543. doi:10.3389/fcell.2021.676543
Quenneville, S., Verde, G., Corsinotti, A., Kapopoulou, A., Jakobsson, J., Offner, S., et al. (2011). In Embryonic Stem Cells, ZFP57/KAP1 Recognize a Methylated Hexanucleotide to Affect Chromatin and DNA Methylation of Imprinting Control Regions. Mol. Cel 44, 361–372. doi:10.1016/j.molcel.2011.08.032
Quinlan, A. R., and Hall, I. M. (2010). BEDTools: a Flexible Suite of Utilities for Comparing Genomic Features. Bioinformatics 26, 841–842. doi:10.1093/bioinformatics/btq033
Riso, V., Cammisa, M., Kukreja, H., Anvar, Z., Verde, G., Sparago, A., et al. (2016). ZFP57 Maintains the Parent-of-origin-specific Expression of the Imprinted Genes and Differentially Affects Non-imprinted Targets in Mouse Embryonic Stem Cells. Nucleic Acids Res. 44, 8165–8178. doi:10.1093/nar/gkw505
Serrano-Saiz, E., and Isogai, Y. (2021). Single-cell Molecular and Developmental Perspectives of Sexually Dimorphic Circuits Underlying Innate Social Behaviors. Curr. Opin. Neurobiol. 68, 159–166. doi:10.1016/j.conb.2021.03.010
Shamis, Y., Cullen, D. E., Liu, L., Yang, G., Ng, S.-F., Xiao, L., et al. (2015). Maternal and Zygotic Zfp57 Modulate NOTCH Signaling in Cardiac Development. Proc. Natl. Acad. Sci. USA 112, E2020–E2029. doi:10.1073/pnas.1415541112
Simchovitz-Gesher, A., and Soreq, H. (2020). Pharmaceutical Implications of Sex-Related RNA Divergence in Psychiatric Disorders. Trends Pharmacol. Sci. 41, 840–850. doi:10.1016/j.tips.2020.09.003
Small, K. S., Todorčević, M., Civelek, M., El-Sayed Moustafa, J. S., Wang, X., Simon, M. M., et al. (2018). Regulatory Variants at KLF14 Influence Type 2 Diabetes Risk via a Female-specific Effect on Adipocyte Size and Body Composition. Nat. Genet. 50, 572–580. doi:10.1038/s41588-018-0088-x
Strogantsev, R., Krueger, F., Yamazawa, K., Shi, H., Gould, P., Goldman-Roberts, M., et al. (2015). Allele-specific Binding of ZFP57 in the Epigenetic Regulation of Imprinted and Non-imprinted Monoallelic Expression. Genome Biol. 16, 112. doi:10.1186/s13059-015-0672-7
Takahashi, N., Coluccio, A., Thorball, C. W., Planet, E., Shi, H., Offner, S., et al. (2019). ZNF445 Is a Primary Regulator of Genomic Imprinting. Genes Dev. 33, 49–54. doi:10.1101/gad.320069.118
Takahashi, N., Gray, D., Strogantsev, R., Noon, A., Delahaye, C., Skarnes, W. C., et al. (2015). ZFP57 and the Targeted Maintenance of Postfertilization Genomic Imprints. Cold Spring Harb Symp. Quant Biol. 80, 177–187. doi:10.1101/sqb.2015.80.027466
Takahashi, T., Ellingson, M. K., Wong, P., Israelow, B., Lucas, C., Klein, J., et al. (2020). Sex Differences in Immune Responses that Underlie COVID-19 Disease Outcomes. Nature 588, 315–320. doi:10.1038/s41586-020-2700-3
Takikawa, S., Wang, X., Ray, C., Vakulenko, M., Bell, F. T., and Li, X. (2013b). Human and Mouse ZFP57 Proteins Are Functionally Interchangeable in Maintaining Genomic Imprinting at Multiple Imprinted Regions in Mouse ES Cells. Epigenetics 8, 1268–1279. doi:10.4161/epi.26544
Takikawa, S., Ray, C., Wang, X., Shamis, Y., Wu, T.-Y., and Li, X. (2013a). Genomic Imprinting Is Variably Lost during Reprogramming of Mouse iPS Cells. Stem Cel Res. 11, 861–873. doi:10.1016/j.scr.2013.05.011
Tucci, V., Isles, A. R., Kelsey, G., Ferguson-Smith, A. C., Tucci, V., Bartolomei, M. S., et al. (2019). Genomic Imprinting and Physiological Processes in Mammals. Cell 176, 952–965. doi:10.1016/j.cell.2019.01.043
Tunster, S. J., Boqué-Sastre, R., McNamara, G. I., Hunter, S. M., Creeth, H. D. J., and John, R. M. (2018). Peg3 Deficiency Results in Sexually Dimorphic Losses and Gains in the Normal Repertoire of Placental Hormones. Front. Cel Dev. Biol. 6, 123. doi:10.3389/fcell.2018.00123
Van der Auwera, G. A., Carneiro, M. O., Hartl, C., Poplin, R., Del Angel, G., Levy-Moonshine, A., et al. (2013). From FastQ Data to High Confidence Variant Calls: the Genome Analysis Toolkit Best Practices Pipeline. Curr. Protoc. Bioinformatics 43 (11 10 11), 11–33. doi:10.1002/0471250953.bi1110s43
Walker, C. J., Schroeder, M. E., Aguado, B. A., Anseth, K. S., and Leinwand, L. A. (2021). Matters of the Heart: Cellular Sex Differences. J. Mol. Cell Cardiol. 160, 42–55. doi:10.1016/j.yjmcc.2021.04.010
Wang, C., Plusquin, M., Ghantous, A., Herceg, Z., Alfano, R., Cox, B., et al. (2020a). DNA Methylation of Insulin-like Growth Factor 2 and H19 Cluster in Cord Blood and Prenatal Air Pollution Exposure to fine Particulate Matter. Environ. Health 19, 129. doi:10.1186/s12940-020-00677-9
Wang, K., Liu, S., Svoboda, L. K., Rygiel, C. A., Neier, K., Jones, T. R., et al. (2020b). Tissue- and Sex-specific DNA Methylation Changes in Mice Perinatally Exposed to Lead (Pb). Front. Genet. 11, 840. doi:10.3389/fgene.2020.00840
Williamson, C. M., Ball, S. T., Nottingham, W. T., Skinner, J. A., Plagge, A., Turner, M. D., et al. (2004). A Cis-Acting Control Region Is Required Exclusively for the Tissue-specific Imprinting of Gnas. Nat. Genet. 36, 894–899. doi:10.1038/ng1398
Yu, B., Qi, Y., Li, R., Shi, Q., Satpathy, A. T., and Chang, H. Y. (2021). B Cell-specific XIST Complex Enforces X-Inactivation and Restrains Atypical B Cells. Cell 184, 1790–1803. doi:10.1016/j.cell.2021.02.015
Zeng, Y., and Chen, T. (2019). DNA Methylation Reprogramming during Mammalian Development. Genes (Basel) 10. doi:10.3390/genes10040257
Zuo, X., Sheng, J., Lau, H.-T., McDonald, C. M., Andrade, M., Cullen, D. E., et al. (2012). Zinc finger Protein ZFP57 Requires its Co-factor to Recruit DNA Methyltransferases and Maintains DNA Methylation Imprint in Embryonic Stem Cells via its Transcriptional Repression Domain. J. Biol. Chem. 287, 2107–2118. doi:10.1074/jbc.m111.322644
Keywords: genomic imprinting, imprinting control region (ICR), imprinted genes, maternal effect, gender, sexually dimorphic, allelic expression, RNA-seq, WGBS, bisulfite sequencing
Citation: Xu Z, Shi J, Zhang Y, Liu Y, Zhao J, Chen Q, Song C, Geng S, Xie W, Wu F, Bai Y, Yang Y and Li X (2022) Zfp57 Exerts Maternal and Sexually Dimorphic Effects on Genomic Imprinting. Front. Cell Dev. Biol. 10:784128. doi: 10.3389/fcell.2022.784128
Received: 27 September 2021; Accepted: 04 January 2022;
Published: 02 February 2022.
Edited by:
Robert Feil, UMR5535 Institut de Génétique Moléculaire de Montpellier (IGMM), FranceReviewed by:
Andrea Riccio, University of Campania Luigi Vanvitelli, ItalyCopyright © 2022 Xu, Shi, Zhang, Liu, Zhao, Chen, Song, Geng, Xie, Wu, Bai, Yang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiajun Li, bGl4ajFAc2hhbmdoYWl0ZWNoLmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.