- Center for Reproductive Medicine, Center for Prenatal Genetics, First Hospital of Jilin University, Changchun, Jilin, China
Cervical cancer is one of the common tumors and often causes cancer-related death in women. Chemotherapy is a common cancer therapy, which displays a pivotal clinical benefit for cancer patients. However, chemoresistance becomes a big obstacle for failure of treatment in cancer patients. Recently, long noncoding RNAs (lncRNAs) have been identified to regulate drug resistance in human cancers, including cervical cancer. In this review, we describe the role of lncRNAs in regulation of chemotherapeutic resistance in cervical cancer. We also discuss the molecular mechanisms of lncRNA-mediated drug resistance in cervical cancer. Moreover, we describe that targeting lncRNAs could reverse drug resistance in cervical cancer. Therefore, lncRNAs could become effective therapeutic targets and chemotherapeutic sensitizers for cervical cancer patients.
Introduction
Cervical cancer is one of the common tumors and often causes cancer-related death in women (Sung et al., 2021; Siegel et al., 2022). Human papillomavirus (HPV) infection is the common cause for cervical oncogenesis (Okunade et al., 2022). The treatment for cervical cancer includes surgery, radiation, chemotherapy or combination therapies. Chemotherapy is a common cancer therapy for killing tumor cells (Miller et al., 2022). Chemotherapy has shown a pivotal clinical benefit for cancer patients. However, drug resistance becomes a key reason for failure of treatment in cancer patients. Many factors are associated with drug resistance in human cancers, including oncogenic action, inactivation of tumor suppressor genes, impaired DNA repair, cancer stem cells (CSCs), epithelial to mesenchymal transition (EMT) and exosomes (Friedmann Angeli et al., 2019; Marine et al., 2020; Bayik and Lathia, 2021; Passaro et al., 2021; Weiss et al., 2022). Multidrug resistance protein (MRP)-related ABC family includes ABCC11, ABCC12, MRP1-7 proteins. The G family member, ABCG2/BCRP, is also associated with drug resistance (Haimeur et al., 2004). EMT is a biological process: epithelial cells become mesenchymal cells after specific stimulations via loss of their cell polarity and adhesion and acquirement of invasive and migratory ability (Lambert and Weinberg, 2021). CSCs are specific cells with abilities of self-renewal, tumorigenicity and differentiation, which often have biomarkers of CSCs (Walcher et al., 2020). The tumor microenvironment (TME), which includes blood vessels, extracellular matrix, stromal cells and immune cells, is also involved in chemoresistance in cancer patients (Liu et al., 2022a; Barnestein et al., 2022; Balaji et al., 2021). Recently, noncoding RNAs (ncRNAs) have been identified to take part in drug resistance and immune resistance in human cancer (Jiang et al., 2020; Jiang et al., 2021; Xie et al., 2022; Zhou et al., 2022).
Long non-coding RNAs (lncRNAs) belong to non-coding RNA family and have larger than 200 nucleotides (Liu et al., 2021). Noncoding RNAs often have no functions for translation into proteins; however, noncoding RNAs play a pivotal role in epigenetic regulation (Statello et al., 2021; Zheng et al., 2022). Numerous studies suggested that lncRNAs are critically involved in carcinogenesis and tumor development in a variety of human malignancies (Liu et al., 2020a; Chen et al., 2022a; Liu and Shang, 2022). LncRNAs have been identified to participated in cervical carcinogenesis and progression via regulation of proliferation, autophagy, differentiation, apoptosis, invasion, migration, and metastasis (Cui et al., 2017; Aalijahan and Ghorbian, 2019; Caceres-Duran et al., 2020; Tornesello et al., 2020). LncRNAs could be prognostic and diagnostic biomarkers in cervical cancer patients (Chi et al., 2017). Moreover, circulating lncRNAs might be biomarkers for prediction of cervical cancer prognosis (Sun et al., 2018). In this review, we describe the role of lncRNAs in regulation of chemotherapeutic resistance and radio-resistance in cervical cancer. We also discuss the molecular mechanisms of lncRNA-mediated drug resistance in cervical cancer. Moreover, we describe that targeting lncRNAs could reverse drug resistance in cervical cancer. Therefore, lncRNAs could become effective therapeutic targets and chemotherapeutic sensitizers for cervical cancer patients.
LncRNAs regulate therapeutic resistance
Mounting evidence has demonstrated that numerous lncRNAs control the therapeutic resistance in human cancers, including chemoresistance, radio-resistance and immune-resistance. In the following sections, we describe how lncRNAs govern therapeutic resistance in cervical cancer.
LncRNA HOTAIR in cervical cancer
HOTAIR expression and clinical features of cervical cancer patients
Evidence revealed that lncRNA HOTAIR plays an essential role in tumorigenesis and progression in human cancers. Studies have showed that upregulation of HOTAIR was linked to tumor malignant progression (Hajjari and Salavaty, 2015; Kim et al., 2015; Sun et al., 2016; Zhou et al., 2020). One study measured the lncRNA HOTAIR expression levels in 218 cervical cancer samples and the matched adjacent normal samples by quantitative RT-PCR (Huang et al., 2014). This study found that HOTAIR expression was increased in cervical cancer tissues and correlated with age, FIGO stage, invasion and lymph node and tumor size (Huang et al., 2014). Moreover, high expression of HOTAIR was associated with worse OS and DFS in cervical cancer patients. Furthermore, HOTAIR expression level can act as an independent biomarker for OS in cervical cancer patients (Huang et al., 2014). Another study measured the circulating HOTAIR by QRT-PCR in the sera of 118 cervical cancer patients and 100 normal age-matched females (Li et al., 2015). The expression of HOTAIR in the sera of cancer patients was increased, which was correlated with invasion, lymphatic node metastasis, advanced tumor stages, tumor recurrence and short OS (Li et al., 2015).
HOTAIR regulates proliferation, migration and invasion
In vitro experiments showed that depletion of HOTAIR blocked proliferation, invasion and migration of cervical cancer cells (Kim et al., 2015). HOTAIR promoted cell motility and metastasis via modulation of MMP-9, VEGF and EMT-related genes in cervical cancer (Kim et al., 2015). HOTAIR overexpression increased cell growth and invasion via modulation of Notch signaling pathway in cervical cancer (Lee et al., 2016). Upregulation of HOTAIR regulated Notch-Wnt signaling pathway and EMT and caused induction of tumor growth in vitro and in vivo in cervical cancer (Lee et al., 2016). Another study revealed that HOTAIR upregulation stimulated cell viability, invasion, migration, and reduced apoptosis via modulation of HLA-G expression by sponging miR-148a in cervical cancer (Sun et al., 2016). Ji et al. (2018) reported that HOTAIR depletion reduced invasion, migration and proliferation via sponging miR-17-5p in cervical cancer. Liu et al. (2018) found that lncRNA HOTAIR facilitated tumor progression via regulation of BCL-2 expression by sponging miR-143-3p in cervical cancer cells. Zheng et al. (2018) observed that lncRNA HOTAIR elevated invasion and migration of cervical cancer cells via inhibition of miR-206 and modulation of megakaryoblastic leukemia 1 (MKL1) in HeLa cells. Moreover, silencing of HOTAIR retarded proliferation, invasion and migration of cervical cancer cells, and STAT3 promoted the activity of the HOTAIR. This work indicated that blockade of HOTAIR and STAT3 synergistically repressed invasion, migration and viability of cervical cancer cells (Zhang et al., 2018a). Again, lncRNA HOTAIR accelerated cell invasion, proliferation and migration via modulation of MAPK1 expression in DoTc2 cervical cancer cells (Liu et al., 2020b). Depletion of both HOTAIR and MAPK1 led to suppression of cell growth synergistically in DoTc2 cells (Liu et al., 2020b). Downregulation of HOTAIR inactivated Wnt/β-catenin signaling pathway and reduced the expression of several mRNAs, such as MAGI2, AJAP1, SOX17, PCDH10 and TET1 in cervical HeLa cells (Salmeron-Barcenas et al., 2019). Zhou et al. (2021) discovered that lncRNA HOTAIR inhibited apoptosis and enhanced proliferation via reduction of miR-214-3p expression and inducing Wnt/β-catenin signaling pathway in HPV16 positive cervical cancer cells.
HOTAIR is associated with HPV
LncRNAs have been known to associate with HPV infection in cervical cancer (Liu et al., 2022b; Chen et al., 2022c). HPV16 E7 oncoprotein can modulate the expression of HOTAIR in cervical cancer cells, suggesting that E7 oncoprotein was linked to lncRNA HORAIR expression in cervical cancer (Sharma et al., 2015). Prior study also revealed that genetic variation in the HOTAIR was correlated with HPV16 cervical cancer (Sharma Saha et al., 2016). This work found that rs2366152C was highly represented in HPV positive cervical cancer patients with lower expression of HOTAIR and higher expression of miR-22 (Sharma Saha et al., 2016). Moreover, miR-22 targeted rs2366152C of lncRNA HOTAIR, resulting in downregulation of lncRNA HOTAIR in cervical cancer cells (Sharma Saha et al., 2016). HPV16 E7 reduced the expression of HOTAIR, p53 and NRP2, but increased miR-33-3p, contributing to suppression of cell apoptosis and induction of cell proliferation in cervical cancer (Zhang et al., 2018b). LncRNA HOTAIR sponged miR-331-3p and elevated the expression of NRP2 in HPV positive cervical cancer, which established a negative feedback loop to govern the apoptosis of cervical cancer cells (Zhang et al., 2018b).
HOTAIR SNP in cervical cancer
Qiu et al. (2016) reported that HOTAIR single nucleotide polymorphism (SNP) rs920778 (C>T) was correlated with cervical cancer risk. Specifically, rs920778 genotype was analyzed in 215 cervical cancer patients and 430 normal controls and observed that TT genotype of rs920778 was associated with HOTAIR upregulation. TT genotype of rs920778 was correlated with higher risk of cervical cancer, advanced tumor stage, tumor grade, HPV infection and lymph node metastasis (Qiu et al., 2016). Cervical cancer patients with TT genotype exhibited resistance to the treatment of EBRT + ICBT + cisplatin, suggesting that rs920778 was linked to drug resistance in cervical cancer patients (Qiu et al., 2016). Guo et al. (2016) also revealed that SNP rs920778T was associated with cervical cancer predisposition via regulation of HOTAIR expression. TT genotype was correlated with TNM stage in cervical cancer patients, suggesting that TT genotype of rs920778 might be an indicator of cervical cancer progression (Guo et al., 2016). Similarly, Weng et al. (2018) uncovered that lncRNA HOTAIR SNP rs920778 was linked to worse OS and poorer cancer recurrence in cervical cancer patients. HOTAIR rs7958904 polymorphism has been found to be associated with increased cervical cancer risk (Jin et al., 2017). The rs7958904 CC genotype was linked to higher expression of HOTAIR and promotion of cell growth in cervical cancer (Jin et al., 2017).
HOTAIR regulates chemo-and radio-resistance
HOTAIR facilitated aggressive behaviors of cervical cancer and induced radio-resistance through inhibition of p21 (Jing et al., 2015). Upregulation of HOTAIR reduced apoptosis and enhanced proliferation, migration and invasion and promoted cell cycle in cervical cancer cells. Depletion of HOTAIR increased the expression of p21 and promoted the radio-sensitivity of C33A cervical cancer cells (Jing et al., 2015). In vivo data also showed that stable silencing of HOTAIR repressed tumor growth and increased the sensitivity of radiotherapy (Jing et al., 2015). Li et al. (2018) found that radiotherapy suppressed tumor growth in mice via downregulation of HOTAIR expression and HIF-1α in cervical cancer. Upregulation of HOTAIR abolished the efficacy of radiation on apoptosis and viability in C33A and HeLa cells through upregulation of HIF-1α (Li et al., 2018). Depletion of HIF-1α abrogated HOTAIR-induced viability of HeLa and C33A cells, revealing that radiotherapy suppress cell growth via regulation of HOTAIR/HIF-1α pathway in cervical cancer (Li et al., 2018). Cisplatin-resistant cells displayed stem cell features and increased migration and invasion, which had the upregulation of HOTAIR (Zhang et al., 2021). Downregulation of HOTAIR reduced the expression of stemness markers in cervical cancer. Notably, lncRNA HOTAIR facilitated cancer cell stemness via interaction with miR-203 and induction of ZEB1 expression in cervical cancer (Zhang et al., 2021). One direct evidence revealed that lncRNA HOTAIR enhanced chemoresistance via inducing EMT and modulating miR-29b/PTEN/PI3K pathway in cervical cancer (Zhang et al., 2022). HOTAIR increased the resistance of cisplatin, paclitaxel and docetaxel via sponging miR-29b in cervical cancer cells (Zhang et al., 2022).
LncRNA PVT1
LncRNA PVT1 has been known to play a critical role in oncogenesis and progression in a variety of human cancers (Lu et al., 2017; Pan et al., 2018; Wang et al., 2019; Bohosova et al., 2021). Evidence has dissected that lncRNA PVT1 could be chemotherapy and radiotherapy sensitizer in cancer (Yao et al., 2022). Iden et al. (2016) reported that high expression of lncRNA PVT1 was correlated with poor prognosis and cancer phenotype in cervical cancer. Knockdown of PVT1 reduced proliferation, invasion, migration, and increased cisplatin cytotoxicity. The expression of PVT1 was upregulated in cervical cancer cells after hypoxia and immune response stimulation (Iden et al., 2016). Studies have revealed that lncRNA PVT1 sponged miR-195 and regulated EMT in cervical cancer cells, resulting in chemoresistance (Shen et al., 2017). Depletion of HPV16 E7 reduced the expression of PVT1 and restored the expression of miR-195. LncRNA PVT1 can bind with EZH2 and miR-195, and increase the expression level of H3K27me3. Interestingly, overexpression of miR-195 also repressed the expression of PVT1 in cervical cancer cells (Shen et al., 2017). Importantly, PVT1 blocked paclitaxel-induced EMT and sensitized cervical cancer cells to paclitaxel treatment. This study implied that PVT1 modulated paclitaxel resistance of cervical cancer cells via regulating miR-195 and EMT (Shen et al., 2017).
LncRNA UCA1
LncRNA urothelial cancer associated 1 (UCA1) plays a necessary role in carcinogenesis and malignant development (Wang et al., 2017a; Yao et al., 2019a; Xuan et al., 2019). Wang et al. (2017b) reported that lncRNA UCA1 enhanced cisplatin resistance in cervical cancer. LncRNA UCA1 upregulation stimulated cell proliferation, reduced apoptosis and increased cisplatin resistance. Downregulation of UCA1 decreased cisplatin resistance in cervical cancer cells (Wang et al., 2017b). LncRNA UCA1 reduced the expression of caspase 3 and upregulated the expression of CDK2, leading to suppression of apoptosis of cervical cancer cells. UCA1 upregulated the expression of p21 and inhibited survivin expression level and caused enhancement of cell proliferation in cervical cancer cells (Wang et al., 2017b). Together, lncRNA UCA1 promoted cisplatin resistance in cervical cancer, suggesting that blockade of lncRNA UCA1 is a useful approach for cervical cancer therapy.
LncRNA GAS5
LncRNA growth arrest-specific transcript 5 (GAS5) acts as a pivotal tumor suppressor in various types of human cancers (Ma et al., 2016; Yang et al., 2020; Filippova et al., 2021; Kaur et al., 2022). LncRNA GAS5 has been verified to work as a tumor suppressor to control cisplatin resistance via targeting miR-21 in cervical cancer (Wen et al., 2017). LncRNA GAS5 overexpression repressed proliferation, invasion and migration of cervical cancer cells in vitro and in vivo. The low expression of GAS5 was detected in cervical cancer patients, which was associated with high expression of miR-21 (Wen et al., 2017). LncRNA GAS5 inhibited the expression of miR-21 in cervical cancer cells. Overexpression of lncRNA GAS5 promoted the sensitivity of cisplatin-resistant SiHa cells to cisplatin therapy. Mechanistically, GAS5 inhibited miR-21 expression and elevated the expression of PTEN and influenced the pAkt in cervical cancer cells, leading to suppression of cisplatin resistance (Wen et al., 2017). Yao et al. (2019b) found that lncRNA GAS5 attenuated cisplatin-mediated cell apoptosis through regulation of STAT3 pathway via targeting miR-21 in cervical cancer cells. Overexpression of GAS5 induced G0/G1 phase arrest, reduced colony formation and proliferation, attenuated migratory and invasive ability in cervical cancer cells. Moreover, overexpression of GAS5 reduced TIMP3 and PDCD4 expression and inactivated STAT3 and E2F3 in cervical cancer (Yao et al., 2019b). Similarly, Fang and coworkers found that low expression of lncRNA GAS5 could predict cisplatin resistance and worse survival in cervical cancer patients (Fang et al., 2020). Hence, lncRNA GAS5 might be a potential target for reversing cisplatin resistance in cervical cancer.
Other LncRNAs regulate drug resistance
The expression of lncRNA CASC2 was decreased in cervical cancer samples, which was related with poor prognosis and a shorter OS (Feng et al., 2017). Exogenous CASC2 amplified the repression of proliferation of cervical cancer cells induced by cisplatin treatment. Cisplatin-resistant cervical cancer patients had lower expression of lncRNA CASC2 (Feng et al., 2017). Upregulation of CASC2 increased the cisplatin sensitivity in cisplatin-resistant cervical cancer cells by suppression of miR-21 and upregulation of PTEN and inactivation of pAkt (Feng et al., 2017). LncRNA LINP1 caused DSBs repair via regulation of NHEJ pathway and reduced the sensitivity of cervical cancer cells to ionizing radiation (Wang et al., 2018a). Depletion of LINP1 elevated the expression levels of cleaved caspase 3 and PARP, contributing to cell apoptosis in cervical cancer cells after radiation (Wang et al., 2018a). LncRNA MALAT1 facilitated cisplatin resistance via modulating the PI3K/AKT pathway in cervical cancer (Wang et al., 2018b). Silencing of MALAT1 increased the expression of cleaved caspase-3, while upregulation of MALAT1 increased the mRNA level of BRWD1 and elevated the expression of p-PI3K and p-Akt in cervical cancer cells (Wang et al., 2018b). Upregulation of lncRNA ZFAS1 was observed in cervical cancer specimens and correlated with poor prognosis (Feng et al., 2019). ZFAS1 siRNA treatment led to suppression of migration, invasion and growth in cervical cancer cells. ZFAS1 siRNA also enhanced cisplatin sensitivity in cervical cancer cells and in nude mice (Feng et al., 2019).
LINC00511 downregulation blocked cell growth via reduction of MRP1, P-GP, MMP-2, MMP-9 and Bcl-2 and induction of cleaved caspase-3 and Bax expressions (Mao et al., 2019). Inhibition of LINC00511 increased cancer cell sensitivity to paclitaxel treatment in cervical cancer (Mao et al., 2019). LncRNA TUG1 expression was upregulated in cisplatin-resistant cervical cancer tissues and linked to a worse prognosis (Wei et al., 2019). TUG1 depletion inhibited the expression of RFX7 in cervical cancer cells and enhanced cisplatin sensitivity by activation of the MAPK pathway (Wei et al., 2019). Exosomal lncRNA HNF1A-AS1 enhanced cisplatin resistance via elevating the expression of TUFT1 by sponging microRNA-34b axis in cervical cancer cells (Luo et al., 2019). LncRNA miR503HG increased the cisplatin sensitivity via suppression of miR-155 and upregulating caspase-3 in recurrent cervical cancer (Zhao et al., 2020). LncRNA NNT-AS1 expression was upregulated in cisplatin-resistant cervical tumors. Silencing of NNT-AS1 attenuated cisplatin resistance via directly binding to miR-186 and subsequently increasing HMGB1 expression in cervical cancer (Liu et al., 2020c). Suppression of lncRNA NEAT1 increased the sensitivity of 5-FU via sponging miR-34a and elevating the expression of LDHA in cervical cancer cells (Shao et al., 2021). LDHA is a glycolysis key enzyme and regulates glycolysis rate, suggesting that lncRNA NEAT1 increases 5-FU resistance via regulation of glycolysis rate in cervical cancer cells (Shao et al., 2021). LncRNA DLG1-AS1 facilitated gemcitabine resistance via targeting miR-16-5p and modulating HDGF expression in cervical cancer cells (Zou et al., 2022). Repression of DLG1-AS1 inhibited proliferation of gemcitabine-resistant cervical tumor cells (Zou et al., 2022). Linc00958 was highly expressed in cisplatin-resistant SiHa cells. Moreover, linc00958 expression was correlated with a short survival in cervical cancer patients. Mechanistically, linc00958 regulated miR-185-5p and RSF-1 and modulated cisplatin resistance in cervical cancer cells via targeting AKT1/GSK3β/VEGFA pathway (Tian et al., 2022). Depletion of lncRNA NCK1-AS1 promoted cisplatin sensitivity to cisplatin via modulating miR-134-5p and restoring the expression of MSH2 in cervical cancer (Zhang et al., 2019). Altogether, lncRNAs regulate the chemotherapeutic resistance in cervical cancer treatment.
Conclusion
In summary, lncRNAs are critically taken part in therapeutic resistance in cervical cancer (Figure 1). Regulation of lncRNAs could overcome drug resistance to obtain good treatment benefit for cervical cancer patients. Chemotherapy is a standard approach for the treatment of cervical cancer. However, drug resistance is a huge challenge for treatment benefit. Hence, targeting lncRNAs is a good approach for reversing drug resistance. Propofol has been found to reduce tumor size and suppress cell proliferation as well as induce cell apoptosis via inhibition of HOTAIR and modulation of mTOR/p70S6K pathway in cervical cancer (Zhang et al., 2015). Artesunate inhibited the expression of HOTAIR and downregulated the COX-2 expression, leading to anti-metastatic ability of artesunate in cervical cancer (Zhang et al., 2016). HOTAIR interacted with COX-2 and increased COX-2 expression and catalytic activity, resulting in promotion of invasion and migration in cervical cancer (Zhang et al., 2016). Similarly, propofol repressed the cell viability, invasion, colony formation and migration in cervical cancer cells via targeting HOTAIR and modulating miR-129-5p/RPL14 axis (Sun et al., 2021). Further investigations are necessary to discover the compounds that regulate the expression of lncRNAs in cervical cancer.
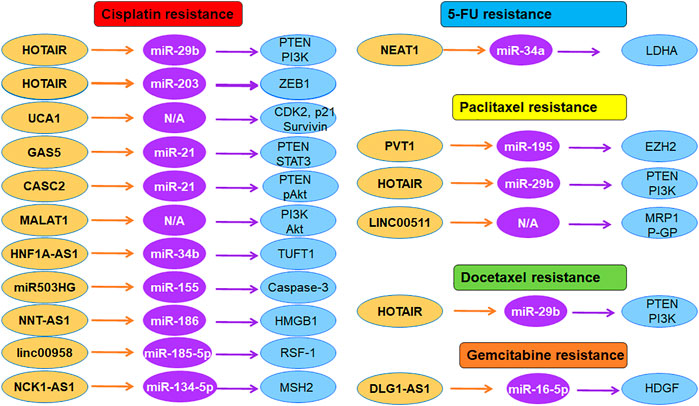
FIGURE 1. The role of lncRNAs in regulation of chemoresistance in cervical cancer. Multiple lncRNAs participate in cisplatin resistance, 5-FU resistance, paclitaxel resistance, and gemcitabine resistance in cervical cancer.
It is critical to mention several points. Besides miRNAs and lncRNAs, circRNAs also govern drug resistance in human cervical cancer (Wen et al., 2020). Hsa_circ_0023404 promoted chemoresistance via VEGFA and autophagy by targeting miR-5047 (Guo et al., 2019). Moreover, circMTO1 enhanced chemoresistance via sponging miR-6893 in cervical cancer (Chen et al., 2019). CircMYBL2 governed paclitaxel resistance via targeting miR-665/EGFR axis in cervical cancer (Dong et al., 2021). Hsa_circ_0074269 increased cisplatin resistance via mediating miR-485-5p and TUFT1 in cervical cancer (Chen et al., 2022b). CircEPSTI1 enhanced cisplatin resistance via upregulation of MSH2 in cervical cancer (Wu et al., 2022). Circ_ZFR regulated paclitaxel resistance via sponging miR-944 and upregulating IL-10 in cervical cancer (Long et al., 2022). Depletion of circ_CEP128 increased paclitaxel sensitivity via targeting miR-432-5p/MCL1 in cervical cancer (Zhao et al., 2022). In addition, one lncRNA has numerous downstream targets. If one lncRNA is modulated, how can we control the expression of its numerous targets? Altogether, targeting lncRNAs by natural compounds or their inhibitors is a promising approach for overcoming drug resistance in cervical cancer.
Author contributions
The manuscript was written by MZ and LL. JW made the figure. WL edited and revised the manuscript. All authors contributed to the manuscript and approved the final version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
CASC2, cancer susceptibility candidate 2; DFS, disease-free survival; DSBs, DNA double-strand breaks; EMT, epithelial to mesenchymal transition; HDGF, hepatoma-derived growth factor; HLA-G, human leucocyte antigen-G; HNF1A, human hepatocyte nuclear factor 1 homeobox A; HOTAIR, Hox transcript antisense intergenic RNA; HPV, human papillomavirus; LINP, lncRNA in non-homologous end joining pathway 1; MKL1, megakaryoblastic leukemia 1; MMP-9, matrix metalloproteinase-9; MRP, multidrug resistance protein; MSH2, Muts protein homolog 2; NEAT, nuclear-rich transcripts 1; NHEJ, non-homologous end joining; NNT-AS1, nicotinamide nucleotide transhydrogenase-antisense RNA 1; OS, overall survival; PVT1, plasmacytoma variant translocation 1; TME, tumor microenvironment; TNBC, triple-negative breast cancer; TUG1, Taurine-upregulated gene 1; VEGF, vascular endothelial growth factor; ZFAS1, zinc finger antisense 1.
References
Aalijahan, H., and Ghorbian, S. (2019). Long non-coding RNAs and cervical cancer. Exp. Mol. Pathol. 106, 7–16. doi:10.1016/j.yexmp.2018.11.010
Balaji, S., Kim, U., Muthukkaruppan, V., and Vanniarajan, A. (2021). Emerging role of tumor microenvironment derived exosomes in therapeutic resistance and metastasis through epithelial-to-mesenchymal transition. Life Sci. 280, 119750. doi:10.1016/j.lfs.2021.119750
Barnestein, R., Galland, L., Kalfeist, L., Ghiringhelli, F., Ladoire, S., and Limagne, E. (2022). Immunosuppressive tumor microenvironment modulation by chemotherapies and targeted therapies to enhance immunotherapy effectiveness. Oncoimmunology 11 (1), 2120676. doi:10.1080/2162402X.2022.2120676
Bayik, D., and Lathia, J. D. (2021). Cancer stem cell-immune cell crosstalk in tumour progression. Nat. Rev. Cancer 21 (8), 526–536. doi:10.1038/s41568-021-00366-w
Bohosova, J., Kubickova, A., and Slaby, O. (2021). lncRNA PVT1 in the pathogenesis and clinical management of renal cell carcinoma. Biomolecules 11 (5), 664. doi:10.3390/biom11050664
Caceres-Duran, M. A., Ribeiro-Dos-Santos, A., and Vidal, A. F. (2020). Roles and mechanisms of the long noncoding RNAs in cervical cancer. Int. J. Mol. Sci. 21 (24), E9742. doi:10.3390/ijms21249742
Chen, J., Wu, S., Wang, J., Sha, Y., and Ji, Y. (2022). Hsa_circ_0074269-mediated upregulation of TUFT1 through miR-485-5p increases cisplatin resistance in cervical cancer. Reprod. Sci. 29 (8), 2236–2250. doi:10.1007/s43032-022-00855-9
Chen, M., Ai, G., Zhou, J., Mao, W., Li, H., and Guo, J. (2019). circMTO1 promotes tumorigenesis and chemoresistance of cervical cancer via regulating miR-6893. Biomed. Pharmacother. 117, 109064. doi:10.1016/j.biopha.2019.109064
Chen, T., Liu, J., Zhang, H., Li, J., and Shang, G. (2022). Long intergenic noncoding RNA00265 enhances cell viability and metastasis via targeting miR-485-5p/USP22 Axis in osteosarcoma. Front. Oncol. 12, 907472. doi:10.3389/fonc.2022.907472
Chen, X., Liu, Y., Liu, H., Wang, Z. W., and Zhu, X. (2022). Unraveling diverse roles of noncoding RNAs in various human papillomavirus negative cancers. Pharmacol. Ther. 238, 108188. doi:10.1016/j.pharmthera.2022.108188
Chi, S., Shen, L., Hua, T., Liu, S., Zhuang, G., Wang, X., et al. (2017). Prognostic and diagnostic significance of lncRNAs expression in cervical cancer: A systematic review and meta-analysis. Oncotarget 8 (45), 79061–79072. doi:10.18632/oncotarget.18323
Cui, X., Jing, X., and Wu, X. (2017). The prognostic value of long non coding RNAs in cervical cancer: A meta-analysis. Oncotarget 8 (37), 62470–62477. doi:10.18632/oncotarget.17620
Dong, M., Li, P., Xie, Y., Wang, Z., and Wang, R. (2021). CircMYBL2 regulates the resistance of cervical cancer cells to paclitaxel via miR-665-dependent regulation of EGFR. Drug Dev. Res. 82 (8), 1193–1205. doi:10.1002/ddr.21834
Fang, X., Zhong, G., Wang, Y., Lin, Z., Lin, R., and Yao, T. (2020). Low GAS5 expression may predict poor survival and cisplatin resistance in cervical cancer. Cell. Death Dis. 11 (7), 531. doi:10.1038/s41419-020-2735-2
Feng, L. L., Shen, F. R., Zhou, J. H., and Chen, Y. G. (2019). Expression of the lncRNA ZFAS1 in cervical cancer and its correlation with prognosis and chemosensitivity. Gene 696, 105–112. doi:10.1016/j.gene.2019.01.025
Feng, Y., Zou, W., Hu, C., Li, G., Zhou, S., He, Y., et al. (2017). Modulation of CASC2/miR-21/PTEN pathway sensitizes cervical cancer to cisplatin. Arch. Biochem. Biophys. 623-624, 62320–62430. doi:10.1016/j.abb.2017.05.001
Filippova, E. A., Fridman, M. V., Burdennyy, A. M., Loginov, V. I., Pronina, I. V., Lukina, S. S., et al. (2021). Long noncoding RNA GAS5 in breast cancer: Epigenetic mechanisms and biological functions. Int. J. Mol. Sci. 22 (13), 6810. doi:10.3390/ijms22136810
Friedmann Angeli, J. P., Krysko, D. V., and Conrad, M. (2019). Ferroptosis at the crossroads of cancer-acquired drug resistance and immune evasion. Nat. Rev. Cancer 19 (7), 405–414. doi:10.1038/s41568-019-0149-1
Guo, J., Chen, M., Ai, G., Mao, W., Li, H., and Zhou, J. (2019). Hsa_circ_0023404 enhances cervical cancer metastasis and chemoresistance through VEGFA and autophagy signaling by sponging miR-5047. Biomed. Pharmacother. 115, 108957. doi:10.1016/j.biopha.2019.108957
Guo, L., Lu, X., Zheng, L., Liu, X., and Hu, M. (2016). Association of long non-coding RNA HOTAIR polymorphisms with cervical cancer risk in a Chinese population. PLoS One 11 (7), e0160039. doi:10.1371/journal.pone.0160039
Haimeur, A., Conseil, G., Deeley, R. G., and Cole, S. P. (2004). The MRP-related and BCRP/ABCG2 multidrug resistance proteins: Biology, substrate specificity and regulation. Curr. Drug Metab. 5 (1), 21–53. doi:10.2174/1389200043489199
Hajjari, M., and Salavaty, A. (2015). HOTAIR: An oncogenic long non-coding RNA in different cancers. Cancer Biol. Med. 12 (1), 1–9. doi:10.7497/j.issn.2095-3941.2015.0006
Huang, L., Liao, L. M., Liu, A. W., Wu, J. B., Cheng, X. L., Lin, J. X., et al. (2014). Overexpression of long noncoding RNA HOTAIR predicts a poor prognosis in patients with cervical cancer. Arch. Gynecol. Obstet. 290 (4), 717–723. doi:10.1007/s00404-014-3236-2
Iden, M., Fye, S., Li, K., Chowdhury, T., Ramchandran, R., and Rader, J. S. (2016). The lncRNA PVT1 contributes to the cervical cancer phenotype and associates with poor patient prognosis. PLoS One 11 (5), e0156274. doi:10.1371/journal.pone.0156274
Ji, F., Wuerkenbieke, D., He, Y., Ding, Y., and Du, R. (2018). Long noncoding RNA HOTAIR: An oncogene in human cervical cancer interacting with MicroRNA-17-5p. Oncol. Res. 26 (3), 353–361. doi:10.3727/096504017X15002869385155
Jiang, W., Pan, S., Chen, X., Wang, Z. W., and Zhu, X. (2021). The role of lncRNAs and circRNAs in the PD-1/PD-L1 pathway in cancer immunotherapy. Mol. Cancer 20 (1), 116. doi:10.1186/s12943-021-01406-7
Jiang, W., Xia, J., Xie, S., Zou, R., Pan, S., Wang, Z. W., et al. (2020). Long non-coding RNAs as a determinant of cancer drug resistance: Towards the overcoming of chemoresistance via modulation of lncRNAs. Drug resist. updat. 50, 100683. doi:10.1016/j.drup.2020.100683
Jin, H., Lu, X., Ni, J., Sun, J., Gu, B., Ding, B., et al. (2017). HOTAIR rs7958904 polymorphism is associated with increased cervical cancer risk in a Chinese population. Sci. Rep. 7 (1), 3144. doi:10.1038/s41598-017-03174-1
Jing, L., Yuan, W., Ruofan, D., Jinjin, Y., and Haifeng, Q. (2015). HOTAIR enhanced aggressive biological behaviors and induced radio-resistance via inhibiting p21 in cervical cancer. Tumour Biol. 36 (5), 3611–3619. doi:10.1007/s13277-014-2998-2
Kaur, J., Salehen, N., Norazit, A., Rahman, A. A., Murad, N. A. A., Rahman, N., et al. (2022). Tumor suppressive effects of GAS5 in cancer cells. Noncoding. RNA 8 (3), 39. doi:10.3390/ncrna8030039
Kim, H. J., Lee, D. W., Yim, G. W., Nam, E. J., Kim, S., Kim, S. W., et al. (2015). Long non-coding RNA HOTAIR is associated with human cervical cancer progression. Int. J. Oncol. 46 (2), 521–530. doi:10.3892/ijo.2014.2758
Lambert, A. W., and Weinberg, R. A. (2021). Linking EMT programmes to normal and neoplastic epithelial stem cells. Nat. Rev. Cancer 21 (5), 325–338. doi:10.1038/s41568-021-00332-6
Lee, M., Kim, H. J., Kim, S. W., Park, S. A., Chun, K. H., Cho, N. H., et al. (2016). The long non-coding RNA HOTAIR increases tumour growth and invasion in cervical cancer by targeting the Notch pathway. Oncotarget 7 (28), 44558–44571. doi:10.18632/oncotarget.10065
Li, J., Wang, Y., Yu, J., Dong, R., and Qiu, H. (2015). A high level of circulating HOTAIR is associated with progression and poor prognosis of cervical cancer. Tumour Biol. 36 (3), 1661–1665. doi:10.1007/s13277-014-2765-4
Li, N., Meng, D. D., Gao, L., Xu, Y., Liu, P. J., Tian, Y. W., et al. (2018). Overexpression of HOTAIR leads to radioresistance of human cervical cancer via promoting HIF-1α expression. Radiat. Oncol. 13 (1), 210. doi:10.1186/s13014-018-1153-4
Liu, F., Hu, L., Pei, Y., Zheng, K., Wang, W., Li, S., et al. (2020). Long non-coding RNA AFAP1-AS1 accelerates the progression of melanoma by targeting miR-653-5p/RAI14 axis. BMC Cancer 20 (1), 258. doi:10.1186/s12885-020-6665-2
Liu, H., Liu, J., and Zhao, G. (2020). Long non-coding RNA HOTAIR regulates proliferation, migration and invasion of human cervical cancer cells by modulating expression of MAPK1. Arch. Med. Sci. 16 (5), 1158–1165. doi:10.5114/aoms.2019.83512
Liu, J., Chen, T., Li, S., Liu, W., Wang, P., and Shang, G. (2022). Targeting matrix metalloproteinases by E3 ubiquitin ligases as a way to regulate the tumor microenvironment for cancer therapy. Semin. Cancer Biol. 86, 259–268. doi:10.1016/j.semcancer.2022.06.004
Liu, J., and Shang, G. (2022). The roles of noncoding RNAs in the development of osteosarcoma stem cells and potential therapeutic targets. Front. Cell. Dev. Biol. 10, 773038. doi:10.3389/fcell.2022.773038
Liu, M., Jia, J., Wang, X., Liu, Y., Wang, C., and Fan, R. (2018). Long non-coding RNA HOTAIR promotes cervical cancer progression through regulating BCL2 via targeting miR-143-3p. Cancer Biol. Ther. 19 (5), 391–399. doi:10.1080/15384047.2018.1423921
Liu, S. J., Dang, H. X., Lim, D. A., Feng, F. Y., and Maher, C. A. (2021). Long noncoding RNAs in cancer metastasis. Nat. Rev. Cancer 21 (7), 446–460. doi:10.1038/s41568-021-00353-1
Liu, Y., Guo, R., Qiao, Y., Han, L., and Liu, M. (2020). LncRNA NNT-AS1 contributes to the cisplatin resistance of cervical cancer through NNT-AS1/miR-186/HMGB1 axis. Cancer Cell. Int. 20, 190. doi:10.1186/s12935-020-01278-9
Liu, Y., Liu, H., Sheng, B., Pan, S., Wang, Z. W., and Zhu, X. (2022). The functions of lncRNAs in the HPV-negative cervical cancer compared with HPV-positive cervical cancer. Apoptosis. 27 (9-10), 685–696. doi:10.1007/s10495-022-01761-w
Long, X., Zheng, M., Yang, Y., Chen, Y., Zhang, X., and Zhang, H. (2022). circ_ZFR is linked to paclitaxel resistance in cervical cancer via miR-944 sponging and IL-10 upregulation. Anal. Cell. Pathol. 2022, 4807287. doi:10.1155/2022/4807287
Lu, D., Luo, P., Wang, Q., Ye, Y., and Wang, B. (2017). lncRNA PVT1 in cancer: A review and meta-analysis. Clin. Chim. Acta. 474, 1–7. doi:10.1016/j.cca.2017.08.038
Luo, X., Wei, J., Yang, F. L., Pang, X. X., Shi, F., Wei, Y. X., et al. (2019). Exosomal lncRNA HNF1A-AS1 affects cisplatin resistance in cervical cancer cells through regulating microRNA-34b/TUFT1 axis. Cancer Cell. Int. 19, 323. doi:10.1186/s12935-019-1042-4
Ma, C., Shi, X., Zhu, Q., Li, Q., Liu, Y., Yao, Y., et al. (2016). The growth arrest-specific transcript 5 (GAS5): A pivotal tumor suppressor long noncoding RNA in human cancers. Tumour Biol. 37 (2), 1437–1444. doi:10.1007/s13277-015-4521-9
Mao, B. D., Xu, P., Xu, P., Zhong, Y., Ding, W. W., and Meng, Q. Z. (2019). LINC00511 knockdown prevents cervical cancer cell proliferation and reduces resistance to paclitaxel. J. Biosci. 44 (2), 44. doi:10.1007/s12038-019-9851-0
Marine, J. C., Dawson, S. J., and Dawson, M. A. (2020). Non-genetic mechanisms of therapeutic resistance in cancer. Nat. Rev. Cancer 20 (12), 743–756. doi:10.1038/s41568-020-00302-4
Miller, K. D., Nogueira, L., Devasia, T., Mariotto, A. B., Yabroff, K. R., Jemal, A., et al. (2022). Cancer treatment and survivorship statistics. Ca. Cancer J. Clin. 72 (5), 409–436. doi:10.3322/caac.21731
Okunade, K. S., Adejimi, A. A., John-Olabode, S. O., Oshodi, Y. A., and Oluwole, A. A. (2022). An overview of HPV screening tests to improve access to cervical cancer screening amongst underserved populations: From development to implementation. Risk Manag. Healthc. Policy 15, 1823–1830. doi:10.2147/RMHP.S296914
Pan, X., Zheng, G., and Gao, C. (2018). LncRNA PVT1: A novel therapeutic target for cancers. Clin. Lab. 64 (5), 655–662. doi:10.7754/Clin.Lab.2018.171216
Passaro, A., Janne, P. A., Mok, T., and Peters, S. (2021). Overcoming therapy resistance in EGFR-mutant lung cancer. Nat. Cancer 2 (4), 377–391. doi:10.1038/s43018-021-00195-8
Qiu, H., Liu, Q., Li, J., Wang, X., Wang, Y., Yuan, Z., et al. (2016). Analysis of the association of HOTAIR single nucleotide polymorphism (rs920778) and risk of cervical cancer. APMIS 124 (7), 567–573. doi:10.1111/apm.12550
Salmeron-Barcenas, E. G., Illades-Aguiar, B., Del Moral-Hernandez, O., Ortega-Soto, A., and Hernandez-Sotelo, D. (2019). HOTAIR knockdown decreased the activity wnt/β-catenin signaling pathway and increased the mRNA levels of its negative regulators in hela cells. Cell. Physiol. biochem. 53 (6), 948–960. doi:10.33594/000000188
Shao, X., Zheng, X., Ma, D., Liu, Y., and Liu, G. (2021). Inhibition of lncRNA-NEAT1 sensitizes 5-Fu resistant cervical cancer cells through de-repressing the microRNA-34a/LDHA axis. Biosci. Rep. 41 (7), BSR20200533. doi:10.1042/BSR20200533
Sharma, S., Mandal, P., Sadhukhan, T., Roy Chowdhury, R., Ranjan Mondal, N., Chakravarty, B., et al. (2015). Bridging links between long noncoding RNA HOTAIR and HPV oncoprotein E7 in cervical cancer pathogenesis. Sci. Rep. 5, 11724. doi:10.1038/srep11724
Sharma Saha, S., Roy Chowdhury, R., Mondal, N. R., Chakravarty, B., Chatterjee, T., Roy, S., et al. (2016). Identification of genetic variation in the lncRNA HOTAIR associated with HPV16-related cervical cancer pathogenesis. Cell. Oncol. 39 (6), 559–572. doi:10.1007/s13402-016-0298-0
Shen, C. J., Cheng, Y. M., and Wang, C. L. (2017). LncRNA PVT1 epigenetically silences miR-195 and modulates EMT and chemoresistance in cervical cancer cells. J. Drug Target. 25 (7), 637–644. doi:10.1080/1061186X.2017.1307379
Siegel, R. L., Miller, K. D., Fuchs, H. E., and Jemal, A. (2022). Cancer statistics, 2022. CA Cancer J. Clin. 72 (1), 7–33. doi:10.3322/caac.21708
Statello, L., Guo, C. J., Chen, L. L., and Huarte, M. (2021). Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell. Biol. 22 (2), 96–118. doi:10.1038/s41580-020-00315-9
Sun, J., Chu, H., Ji, J., Huo, G., Song, Q., and Zhang, X. (2016). Long non-coding RNA HOTAIR modulates HLA-G expression by absorbing miR-148a in human cervical cancer. Int. J. Oncol. 49 (3), 943–952. doi:10.3892/ijo.2016.3589
Sun, N., Zhang, W., Liu, J., Yang, X., and Chu, Q. (2021). Propofol inhibits the progression of cervical cancer by regulating HOTAIR/miR-129-5p/RPL14 Axis. Onco. Targets. Ther. 14, 551–564. doi:10.2147/OTT.S279942
Sun, W., Wang, L., Zhao, D., Wang, P., Li, Y., and Wang, S. (2018). Four circulating long non-coding RNAs act as biomarkers for predicting cervical cancer. Gynecol. Obstet. Investig. 83 (6), 533–539. doi:10.1159/000487595
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71 (3), 209–249. doi:10.3322/caac.21660
Tian, J., Cheng, L., Kong, E., Gu, W., Jiang, Y., Hao, Q., et al. (2022). linc00958/miR-185-5p/RSF-1 modulates cisplatin resistance and angiogenesis through AKT1/GSK3β/VEGFA pathway in cervical cancer. Reprod. Biol. Endocrinol. 20 (1), 132. doi:10.1186/s12958-022-00995-2
Tornesello, M. L., Faraonio, R., Buonaguro, L., Annunziata, C., Starita, N., Cerasuolo, A., et al. (2020). The role of microRNAs, long non-coding RNAs, and circular RNAs in cervical cancer. Front. Oncol. 10, 150. doi:10.3389/fonc.2020.00150
Walcher, L., Kistenmacher, A. K., Suo, H., Kitte, R., Dluczek, S., Strauss, A., et al. (2020). Cancer stem cells-origins and biomarkers: Perspectives for targeted personalized therapies. Front. Immunol. 11, 1280. doi:10.3389/fimmu.2020.01280
Wang, B., Huang, Z., Gao, R., Zeng, Z., Yang, W., Sun, Y., et al. (2017). Expression of long noncoding RNA urothelial cancer associated 1 promotes cisplatin resistance in cervical cancer. Cancer biother. Radiopharm. 32 (3), 101–110. doi:10.1089/cbr.2016.2156
Wang, H., Guan, Z., He, K., Qian, J., Cao, J., and Teng, L. (2017). LncRNA UCA1 in anti-cancer drug resistance. Oncotarget 8 (38), 64638–64650. doi:10.18632/oncotarget.18344
Wang, N., Hou, M. S., Zhan, Y., Shen, X. B., and Xue, H. Y. (2018). MALAT1 promotes cisplatin resistance in cervical cancer by activating the PI3K/AKT pathway. Eur. Rev. Med. Pharmacol. Sci. 22 (22), 7653–7659. doi:10.26355/eurrev_201811_16382
Wang, W., Zhou, R., Wu, Y., Liu, Y., Su, W., Xiong, W., et al. (2019). PVT1 promotes cancer progression via MicroRNAs. Front. Oncol. 9, 609. doi:10.3389/fonc.2019.00609
Wang, X., Liu, H., Shi, L., Yu, X., Gu, Y., and Sun, X. (2018). LINP1 facilitates DNA damage repair through non-homologous end joining (NHEJ) pathway and subsequently decreases the sensitivity of cervical cancer cells to ionizing radiation. Cell. Cycle 17 (4), 439–447. doi:10.1080/15384101.2018.1442625
Wei, X., Zhou, Y., Qiu, J., Wang, X., Xia, Y., and Sui, L. (2019). Low expression of TUG1 promotes cisplatin sensitivity in cervical cancer by activating the MAPK pathway. J. BUON 24 (3), 1020–1026.
Weiss, F., Lauffenburger, D., and Friedl, P. (2022). Towards targeting of shared mechanisms of cancer metastasis and therapy resistance. Nat. Rev. Cancer 22 (3), 157–173. doi:10.1038/s41568-021-00427-0
Wen, Q., Liu, Y., Lyu, H., Xu, X., Wu, Q., Liu, N., et al. (2017). Long noncoding RNA GAS5, which acts as a tumor suppressor via microRNA 21, regulates cisplatin resistance expression in cervical cancer. Int. J. Gynecol. Cancer 27 (6), 1096–1108. doi:10.1097/IGC.0000000000001028
Wen, X., Liu, S., Sheng, J., and Cui, M. (2020). Recent advances in the contribution of noncoding RNAs to cisplatin resistance in cervical cancer. PeerJ 8, e9234. doi:10.7717/peerj.9234
Weng, S. L., Wu, W. J., Hsiao, Y. H., Yang, S. F., Hsu, C. F., and Wang, P. H. (2018). Significant association of long non-coding RNAs HOTAIR genetic polymorphisms with cancer recurrence and patient survival in patients with uterine cervical cancer. Int. J. Med. Sci. 15 (12), 1312–1319. doi:10.7150/ijms.27505
Wu, P., Qin, J., Liu, L., Tan, W., Lei, L., and Zhu, J. (2022). circEPSTI1 promotes tumor progression and cisplatin resistance via upregulating MSH2 in cervical cancer. Aging (Albany NY) 14 (13), 5406–5416. doi:10.18632/aging.204152
Xie, W., Chu, M., Song, G., Zuo, Z., Han, Z., Chen, C., et al. (2022). Emerging roles of long noncoding RNAs in chemoresistance of pancreatic cancer. Semin. Cancer Biol. 83, 303–318. doi:10.1016/j.semcancer.2020.11.004
Xuan, W., Yu, H., Zhang, X., and Song, D. (2019). Crosstalk between the lncRNA UCA1 and microRNAs in cancer. FEBS Lett. 593 (15), 1901–1914. doi:10.1002/1873-3468.13470
Yang, X., Xie, Z., Lei, X., and Gan, R. (2020). Long non-coding RNA GAS5 in human cancer. Oncol. Lett. 20 (3), 2587–2594. doi:10.3892/ol.2020.11809
Yao, F., Wang, Q., and Wu, Q. (2019). The prognostic value and mechanisms of lncRNA UCA1 in human cancer. Cancer Manag. Res. 11, 7685–7696. doi:10.2147/CMAR.S200436
Yao, T., Lu, R., Zhang, J., Fang, X., Fan, L., Huang, C., et al. (2019). Growth arrest-specific 5 attenuates cisplatin-induced apoptosis in cervical cancer by regulating STAT3 signaling via miR-21. J. Cell. Physiol. 234 (6), 9605–9615. doi:10.1002/jcp.27647
Yao, W., Li, S., Liu, R., Jiang, M., Gao, L., Lu, Y., et al. (2022). Long non-coding RNA PVT1: A promising chemotherapy and radiotherapy sensitizer. Front. Oncol. 12, 959208. doi:10.3389/fonc.2022.959208
Zhang, D., Zhou, X. H., Zhang, J., Zhou, Y. X., Ying, J., Wu, G. Q., et al. (2015). Propofol promotes cell apoptosis via inhibiting HOTAIR mediated mTOR pathway in cervical cancer. Biochem. Biophys. Res. Commun. 468 (4), 561–567. doi:10.1016/j.bbrc.2015.10.129
Zhang, L., Qian, H., Sha, M., Luan, Z., Lin, M., Yuan, D., et al. (2016). Downregulation of HOTAIR expression mediated anti-metastatic effect of artesunate on cervical cancer by inhibiting COX-2 expression. PLoS One 11 (10), e0164838. doi:10.1371/journal.pone.0164838
Zhang, M., Song, Y., and Zhai, F. (2018). ARFHPV E7 oncogene, lncRNA HOTAIR, miR-331-3p and its target, NRP2, form a negative feedback loop to regulate the apoptosis in the tumorigenesis in HPV positive cervical cancer. J. Cell. Biochem. 119 (6), 4397–4407. doi:10.1002/jcb.26503
Zhang, W., Liu, J., Wu, Q., Liu, Y., and Ma, C. (2021). HOTAIR contributes to stemness acquisition of cervical cancer through regulating miR-203 interaction with ZEB1 on epithelial-mesenchymal transition. J. Oncol. 2021, 4190764. doi:10.1155/2021/4190764
Zhang, W., Wu, Q., Liu, Y., Wang, X., Ma, C., and Zhu, W. (2022). LncRNA HOTAIR promotes chemoresistance by facilitating epithelial to mesenchymal transition through miR-29b/PTEN/PI3K signaling in cervical cancer. Cells Tissues Organs 211 (1), 16–29. doi:10.1159/000519844
Zhang, W. Y., Liu, Y. J., He, Y., and Chen, P. (2019). Suppression of long noncoding RNA NCK1-AS1 increases chemosensitivity to cisplatin in cervical cancer. J. Cell. Physiol. 234 (4), 4302–4313. doi:10.1002/jcp.27198
Zhang, Y., Cheng, X., Liang, H., and Jin, Z. (2018). Long non-coding RNA HOTAIR and STAT3 synergistically regulate the cervical cancer cell migration and invasion. Chem. Biol. Interact. 286, 106–110. doi:10.1016/j.cbi.2018.03.010
Zhao, S., Yu, M., and Wang, L. (2020). LncRNA miR503HG regulates the drug resistance of recurrent cervical squamous cell carcinoma cells by regulating miR-155/caspase-3. Cancer Manag. Res. 12, 1579–1585. doi:10.2147/CMAR.S225489
Zhao, Y., Lan, Y., Chi, Y., Yang, B., and Ren, C. (2022). Downregulation of circ-CEP128 enhances the paclitaxel sensitivity of cervical cancer through regulating miR-432-5p/MCL1. Biochem. Genet. doi:10.1007/s10528-022-10201-y
Zheng, C., Chu, M., Chen, Q., Chen, C., Wang, Z. W., and Chen, X. (2022). The role of lncRNA OIP5-AS1 in cancer development and progression. Apoptosis. 27 (5-6), 311–321. doi:10.1007/s10495-022-01722-3
Zheng, P., Yin, Z., Wu, Y., Xu, Y., Luo, Y., and Zhang, T. C. (2018). LncRNA HOTAIR promotes cell migration and invasion by regulating MKL1 via inhibition miR206 expression in HeLa cells. Cell. Commun. Signal. 16 (1), 5. doi:10.1186/s12964-018-0216-3
Zhou, X., Ao, X., Jia, Z., Li, Y., Kuang, S., Du, C., et al. (2022). Non-coding RNA in cancer drug resistance: Underlying mechanisms and clinical applications. Front. Oncol. 12, 951864. doi:10.3389/fonc.2022.951864
Zhou, Y., Wang, Y., Lin, M., Wu, D., and Zhao, M. (2021). LncRNA HOTAIR promotes proliferation and inhibits apoptosis by sponging miR-214-3p in HPV16 positive cervical cancer cells. Cancer Cell. Int. 21 (1), 400. doi:10.1186/s12935-021-02103-7
Zhou, Y. H., Cui, Y. H., Wang, T., and Luo, Y. (2020). Long non-coding RNA HOTAIR in cervical cancer: Molecular marker, mechanistic insight, and therapeutic target. Adv. Clin. Chem. 97, 117–140. doi:10.1016/bs.acc.2019.12.004
Keywords: lncRNAs, cervical cancer, resistance, mRNA, miRNAs, therapy
Citation: Zhou M, Liu L, Wang J and Liu W (2022) The role of long noncoding RNAs in therapeutic resistance in cervical cancer. Front. Cell Dev. Biol. 10:1060909. doi: 10.3389/fcell.2022.1060909
Received: 03 October 2022; Accepted: 20 October 2022;
Published: 09 November 2022.
Edited by:
Zichuan Liu, Tianjin University, ChinaCopyright © 2022 Zhou, Liu, Wang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wanru Liu, bGl1d3JAamx1LmVkdS5jbg==
 Meimei Zhou
Meimei Zhou Jing Wang
Jing Wang Wanru Liu
Wanru Liu