
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol. , 12 December 2022
Sec. Signaling
Volume 10 - 2022 | https://doi.org/10.3389/fcell.2022.1059715
This article is part of the Research Topic Deregulated Signaling Pathways in Inflammation and Cancer View all 8 articles
Multiple myeloma (MM) remains a lethal hematologic cancer characterized by the expansion of transformed plasma cells within the permissive bone marrow (BM) milieu. The emergence of relapsed and/or refractory MM (RRMM) is provoked through clonal evolution of malignant plasma cells that harbor genomic, metabolic and proteomic perturbations. For most patients, relapsed disease remains a major cause of overall mortality. Transforming growth factors (TGFs) have pleiotropic effects that regulate myelomagenesis as well as the emergence of drug resistance. Moreover, TGF-β modulates numerous cell types present with the tumor microenvironment, including many immune cell types. While numerous agents have been FDA-approved over the past 2 decades and significantly expanded the treatment options available for MM patients, the molecular mechanisms responsible for drug resistance remain elusive. Multiple myeloma is uniformly preceded by a premalignant state, monoclonal gammopathy of unknown significance, and both conditions are associated with progressive deregulation in host immunity characterized by reduced T cell, natural killer (NK) cell and antigen-presenting dendritic cell (DC) activity. TGF-β promotes myelomagenesis as well as intrinsic drug resistance by repressing anti-myeloma immunity to promote tolerance, drug resistance and disease progression. Hence, repression of TGF-β signaling is a prerequisite to enhance the efficacy of current and future immunotherapeutics. Novel strategies that incorporate T cells that have been modified to express chimeric antigen receptor (CARs), T cell receptors (TCRs) and bispecific T cell engagers (BiTEs) offer promise to block TGF-β signaling, overcome chemoresistance and enhance anti-myeloma immunity. Here, we describe the effects of TGF-β signaling on immune cell effectors in the bone marrow and emerging strategies to overcome TGF-β-mediated myeloma growth, drug resistance and survival.
The transforming growth factor-β (TGF-β) pathway governs cellular growth, differentiation, apoptosis, invasion, and immunity (Massagué, 2008; Moses et al., 2016). A better understanding of the mechanisms that regulate the TGF-β pathway and downstream signaling steps has produced a coherent roadmap of the signaling cascade, as well as crosstalk with other pathways (Moustakas and Heldin, 2009; Massagué, 2012; Batlle and Massagué, 2019). Obstacles still exist in determining the precise role of TGF-β that originates from the context-dependent nature on various cell types. TGF-β1, 2 and 3 are evolutionarily conserved, secreted, isoforms that control cellular behavior under physiologic and pathologic conditions (Heldin and Moustakas, 2016; Malek et al., 2019; Tzavlaki and Moustakas, 2020; Kim et al., 2021). Activated TGF-β binds type I and type II TGF-β receptor kinase receptors (TGF-βRI and TGF-βRII), respectively. TGF-β binding promotes TGF-βRII heterotetramerization and phosphorylation to activate TGF-βRI, followed by Smad2/3 phosphorylation with subsequent activation of downstream signaling pathways. TGF-β has been shown to elicit both tumor suppressive and tumor promoting activities (Chen W. J et al., 2003; Fantini et al., 2004; Heldin and Moustakas, 2016; Dahmani and Delisle, 2018). In healthy cells, TGF-β functions as a potent growth inhibitor while in developed tumors, which produce excess TGF-β, it functions as a growth promoter. However, the homeostatic action of TGF-β is thwarted in oncogenically-activated cells. Within the tumor microenvironment (TME), TGF-β promotes cancer progression and metastasis (Swamydas et al., 2022). At later stages of disease, TGF-β targets vascular, fibroblastic, and immune cells (Blobe et al., 2000; Bhowmick et al., 2004; Massagué, 2012). SMAD is a canonical TGF-β signaling effector that controls metastasis and immune regulation (Chen et al., 2005; Laouar et al., 2005; Yu et al., 2006; Takimoto et al., 2010). TGF-β also modulates the epithelial to mesenchymal transition (EMT) in which epithelial cells convert to a mesenchymal phenotype that increases tumor growth (Blobe et al., 2000; Kim et al., 2020). Although the expression of TGF-β and TGF-βR1/II is widespread, TGF-β activation is localized to sites where the cytokine is released from latency (Shi et al., 2011). TGF-β is stored as a latent complex with the prodomain intact within the ECM. TGF-β1 activation requires the binding of αv integrin to an RGD sequence in the prodomain while TGF-β resides within the ECM with latent TGF-β (LTGFB) binding proteins. LTGFB is activated by several signals, e.g., integrins, thrombospondin and proteases. The ECM1 protein inhibits LTGFB activation and depletion of ECM1 leads to a massive R-Smads activation in the liver (Li et al., 2021). In healthy tissues, TGF-β maintains homeostasis to prevent tumor formation (Bhowmick et al., 2004; Mukherjee et al., 2010; Macfarlane et al., 2017).
Cancer cells develop resistance to the suppressive effects of TGF-β through mutation or epigenetic modification (Ikushima and Miyazono, 2010). For example, TGF-β1 signaling is hyperactivated in breast cancer cells and promotes tumorigenesis (Tang et al., 2018). SMAD7 is an antagonist of TGF-β1 signaling and complexes with the TGF-βRI to inhibit downstream signaling and pro-tumorigenic transcriptional activation. Proteins that regulate SMAD7 activity act to attenuate TGF-β1-mediated aggressive phenotypes in breast cancer (Hu et al., 2021). In colon cancer, SMAD7 deletion predisposes to a favorable prognosis, while SMAD7 amplification is linked to poor outcomes (Boulay et al., 2003). While TGF-β signaling activates Smad2/3, BMP (bone morphogenetic protein) signaling cascade activates Smad1/5/8. Both cascades share Smad4 but are inhibited by Smad6/7, while at the transcriptional level, TGF-β Smad4 complexes with Smad2/3, and BMP Smad4 complexes with Smad 1/5/8 (Dituri et al., 2019). BMP-7 deficiency accelerates tumorigenesis and negatively correlates with patient outcome (Tang et al., 2018). Prostate carcinoma demonstrates reduced BMP7 transcript levels relative to normal prostate gland. Recombinant BMP7 reduced tumor burden and metastasis in mice. Since the TGF-β signaling pathway represents an attractive target for cancer therapy, numerous modalities have been designed or are in development as anti-cancer strategies (Figure 1).
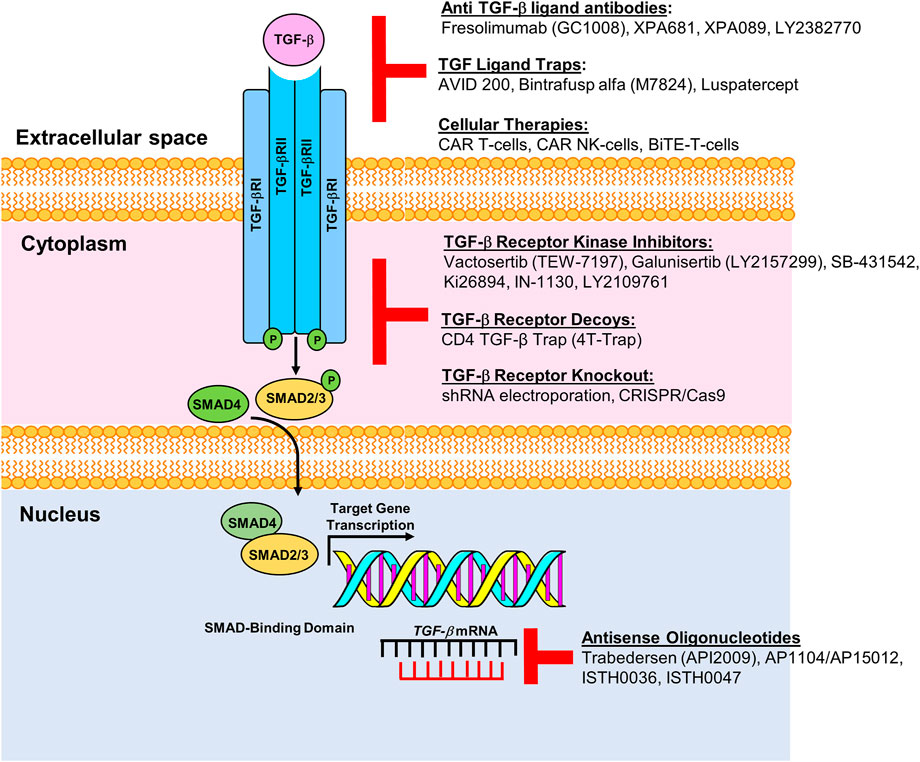
FIGURE 1. Pharmacologic and immunologic blockade of the TGF-β pathway. Schematic representation of multiple modalities developed or in development to target components of the TGF-β signaling pathway. Current roadblocks due to TGF-β mediated immune suppression can be overcome by targeting the TGF-β receptor with ligand antibodies, ligand traps or by inhibiting TGF-β receptors using specific kinase inhibitors, receptor decoys or by knocking out the TGF receptor gene. Antisense oligonucleotides, e.g., AP11014 and AP15012, can silence or suppress TGF-β at the transcriptional level. Figure is adapted from Kim et al., 2021.
TGF-β1 not only contributes to malignant progression but also promotes the development of drug resistance (Coşkun et al., 2006; Dong and Blobe, 2006). Elevated levels of TGF-β1 in MM patient serum correlates with the emergence of drug resistance, tumor progression and a poor prognosis (Coşkun et al., 2006). The proteasome is an evolutionarily conserved cylindrical structure of high molecular weight that demonstrates multiple endopeptidase activities (Driscoll, 1994; Driscoll et al., 2012). Plasma cells are protein-synthesizing factories that secrete copious amounts of antibodies and therefore rely heavily on protein homeostatic mechanisms. The ubiquitin-proteasome system degrades the vast majority of proteins in eukaryotic cells. Malignant plasma cells are exquisitely sensitive to agents that disrupt proteostasis. Proteasome inhibitors have been FDA-approved and are now frontline treatment for newly diagnosed and relapsed and/or refractory multiple myeloma (RRMM) as well as for Mantle Cell Lymphoma (Driscoll and Woodle, 2012). Moreover, the proteasome inhibitors bortezomib and carfilzomib have shown significant promise in targeting plasma cells that secrete HLA-antibodies and hinder organ transplantation (Tremblay et al., 2020; Woodle et al., 2020). TGF-β/Smad signaling has been shown to regulate expression of the proteasome catalytic subunit PSMB5 (Lai et al., 2018) and a number of studies suggest that PSMB5 contributes to proteasome inhibitor resistance (Oerlemans et al., 2008; Ignatz-Hoover et al., 2022). Studies in bortezomib-resistant myelomonocytic cells revealed PSMB5 overexpression, mutations within the highly conserved bortezomib binding pocket in PSMB5 and that PSMB5 knockdown restored drug sensitivity. RPMI 8226 cells grown in escalating doses of bortezomib generated drug resistant cells that overexpressed the proteasome β5 catalytic subunit PSMB5 at the messenger RNA (mRNA) and protein levels (Balsas et al., 2012). Bortezomib resistant lymphoma and leukemia cell lines also exhibit PSMB5 overexpression (Lü et al., 2008). Increased PSMB5 expression has been associated with a less favorable outcome in triple negative breast cancer (TNBC), while silencing PSMB5 sensitized TNBC cells to chemotherapy (Wei et al., 2018).
TGF-β regulates immune cell activity and suppresses immunosurveillance (Gorelik and Flavell, 2002; Swamydas et al., 2022) (Figure 2). TGF-β is secreted by malignant plasma cells that are characteristic of MM. TGF-β triggers interleukin (IL)-6 and vascular endothelial growth factor (VEGF) secretion in BM stromal cells (BMSCs), to increase paracrine IL-6 and VEGF-related tumor growth (Urashima et al., 1996; Hayashi et al., 2004). Within the BM, TGF-β regulates cytotoxic T cell and regulatory T cell (Tregs) (Wan and Flavell, 2008), natural killer (NK) cells (Regis et al., 2020), and macrophage activity (Zhang et al., 2016). A primary role of TGF-β is the generation of Tregs which suppress anti-myeloma immunity (Hadjiaggelidou and Katodritou, 2021). Antibodies and pharmacologics that block TGF-β signaling promote CD8+ T cell and NK-cell driven anti-tumor responses (Gunderson et al., 2020). Other agents elevate neutrophil-attracting chemokines to promote their recruitment. TGF-β silences IL-2 expression and TGF-β signaling blocks CD8+ T cells functional activity (Batlle and Massagué, 2019).
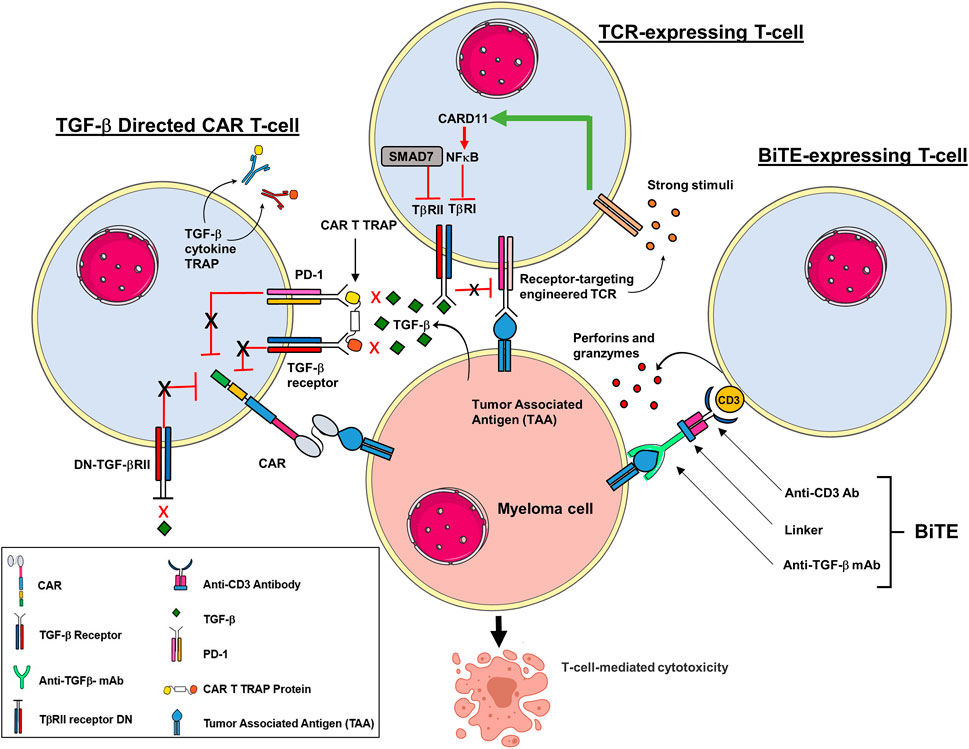
FIGURE 2. Adoptive T cell strategies to prevent immune suppression by TGF-β in TME. Multiple strategies are in development to modify T cells to target components of the TGB-β signaling pathway. T cells can be engineered to work against the immunosuppressive role of TGF-β, activated to induce a more efficient T cell cytotoxicity and prevent exhaustion. CAR T cells have been engineered to block the inhibitory effects of TGF-β by co-expressing a BCMA construct that expresses a dominant negative (DN) TGF-βRII armor. The DN TGF-βII lacks a functional receptor kinase domain which prevents TGF-β suppression despite prolonged exposure. CAR T cells have also been modified to express a decoy or a cytokine receptor that functionally converts inhibitory signals within the TME into pro-inflammatory signals. In addition, CAR T cells have been engineered to secrete a bispecific antibody that co-targets both PD-1 and TGF-βRII to improve anti-tumor immunity. T cells that express a TCR that engages the TGF-βRII to activate caspase recruitment domain-containing protein 11 (CARD11) and nuclear factor κ-B (NF-κB). Moreover, the inhibitory effects of SMAD7 on TGF-βRI promotes T cell mediated cytotoxicity. BiTE-expressing T cells simultaneous binding to CD3 in the TCR complex and the TGF-β to promote lytic enzymes that mediate CTL activity.
TGF-β stimulates Treg differentiation and expansion to promote immune evasion. TGF-β and IL-2 drive immune suppression by inducing Foxp3 expression in naïve CD4+ T cells (Chen C. H et al., 2003; Fantini et al., 2004). Foxp3 is a master transcription factor that regulates the Treg program and is induced by TGF-β (Takimoto et al., 2010). However, pro-inflammatory cytokines within the TME counteract TGF-β-driven Treg induction and drive CD4+ T cell conversion towards an effector phenotype (Wei et al., 2007; Battaglia et al., 2013). TGF-β reduces the production and activity of NK cells by suppressing IFN-γ as well as expression of the transcription factor T-bet (Batlle and Massagué, 2019). TGF-β also reduces expression of the surface receptor proteins NKG2D and NKp30 to impair recognition of stressed and transformed cells (Castriconi et al., 2003). Dendritic cells (DCs) are antigen-presenting cells (APCs) that have been shown to regulate Th1 and Treg-driven responses. TGF-β inhibits antigen presentation by DCs in vitro by suppressing MHC class II levels. Tumor cells induce the release of TGF-β by DCs which promotes the conversion of naïve CD4+ T cells into Tregs. Macrophages are polarized towards an M2 phenotype with anti-inflammatory and immune suppressive functions (Boutilier and Elsawa, 2021). Tumor-associated macrophages (TAMs) produce TGF-β and macrophage subsets mobilize TGF-β through integrin αv-β8 and MMP1(Kim et al., 2021). TGF-β acts as a chemoattractant that draws monocytes to inflammatory sites and upregulates adhesion molecules that facilitate monocyte attachment to the ECM. The TGF-β-rich TME further contributes to immune suppression by squelching macrophage activity (Ignatz-Hoover and Driscoll, 2022).
TGF-β1-mediated differentiation of myeloid-derived suppressor cells (MDSCs) leads to a potent anti-tumor phenotype suitable for adoptive T cell therapy (ACT) (Jayaraman et al., 2018). TGF-β controls MDSC differentiation and immunoregulatory activity while MDSCs enhance T cell anti-tumor effects. TGF-β increases monocytic MDSC (Mo-MDSC) expansion, the expression of immunosuppressive molecules, and suppression of CD4⁺ T cell proliferation (Lee et al., 2018).
TGF-β1 has been shown to be a potent enhancer of mRNA expression of BM stroma thrombopoietin (TPO), a commitment of lineage specificity. TPO induces TGF-β receptors I and II on megakaryoblasts at the mid-megakaryopoietic stage; at this stage, TGF-β1 was able to arrest the maturation of megakaryocyte colony-forming units (CFU-Meg) (Sakamaki et al., 1999). TGF-β1 released from destroyed platelets or megakaryocytes stimulates TPO synthesis in BMSC which then stimulates CD34+ stem cells to commit to the megakaryocyte lineage and to express TGF-β1 receptors that render them susceptible to suppression by TGF-β1 as part of a feedback regulatory loop. Megakaryocytes are an important component of the TME and directly supporting MM growth. Therapeutic interventions that interfere with MM-megakaryocyte interaction may help control disease (Yaccoby et al., 2005).
A hallmark of senescence is an accumulation of mutations in the machinery that controls cellular replication. TGF-β exercises its effects on cellular aging in a variety of ways owing to its vast array of pleiotropic effects (Oh and Li, 2013). TGF-β interferes with cell cycle regulators, and by suppressing proliferation factors (Zhang et al., 2017). TGF-β accelerates aging in fibroblasts, bronchial epithelial and cancer cells (Debacq-Chainiaux et al., 2005; Miyazono et al., 2018). Reactive oxygen species (ROS) is a hallmark of senescence and TGF-β increases ROS production within mitochondria of several cells, e.g., hepatocytes, lung epithelia (Albright et al., 2003; Yoon et al., 2005). TGF-β also exerts effects on expression of the telomerase reverse transcriptase (TERT) in fibroblasts and breast cancer cells (Hu et al., 2006; Li H. et al., 2006). The effects of TGF-β are also observed in non-protein entities, e.g., non-coding RNAs and microRNAs (Suzuki and Miyazono, 2011; Suzuki, 2018). TGF-β induces inhibitory Smads (I-Smads), Smad6 and Smad7 expression which contributes to negative feedback that contributes to senescence. TGF-β can be secreted in either an autocrine or paracrine manner.
During thymic development, self-reactive T cells undergo a goldilocks process where they are eliminated to prevent auto-reactive effects (Dzhagalov et al., 2013; Li M. O et al., 2006; Marie et al., 2006). There is evidence that TGF-β can dampen auto-reactive T cells that have avoided depletion during development (Ouyang et al., 2010). This phenomenon has implications for cancer biology, since TGF- β promotes the generation of immunosuppressive Tregs (Josefowicz et al., 2012). TGF-β induces Foxp3 in peripheral Tregs with concomitant expression of IL-2 (Chen W. J. et al., 2003; Davidson et al., 2007; Zheng et al., 2007). TGF-β1 is the predominant isoform secreted by immune cells and its primary role is to maintain a self-tolerant and functional T cell repertoire. However, TGF-β signaling has redundant effects and doubts persist on how Smad-dependent transcriptional activation controls T cell programs (Takimoto et al., 2010; Gu et al., 2012).
Importantly, TGF-β impacts T cell activation by inhibiting TCR signaling (Chen, C. H. et al., 2003) and downregulates expression of lineage-defining transcription factors, e.g., T-bet and GATA-3, which are critical for Th1 and Th2 CD4+ T-cell differentiation, respectively (Gorelik et al., 2000; Neurath et al., 2002). In contrast to inhibiting certain programs in effector T cells, TGF-β also enhances Th17 lineage T cell differentiation (Veldhoen et al., 2006). Another subset of CD4+ T cells affected by TGF-β are the Th9 cells which secret pro-inflammatory IL-9 when T cells are exposed to TGF-β and IL-4 (Jabeen and Kaplan, 2012).
Exhausted T cells display reduced proliferative capacity and impaired function activity, similar to that seen in viral infections (Gallimore et al., 1998; Urbani et al., 2006). Several studies have implicated TGF- β as having a role to induce T cell exhaustion that ultimately leads to T cell death (Tinoco et al., 2009). However, the specifics on this phenomenon have not been yet elucidated. Interestingly, B cell malignancies produce IL-10 and TGF-β (Gupta et al., 2012; Huai et al., 2021), while non-Hodgkin’s lymphoma cells upregulate CD70 expression on T cells through TGF-β (Yang et al., 2014). However, CD70 expression on MM cells is non-uniform (Zheng et al., 2013; Timmers et al., 2019). TGF-β also elevates the inhibitory markers CD39 and CD73 expressed on CD8+ T cells, limiting T cell activity (Li et al., 2017). TGF-β secreted from TAMs inhibits INF-γ and granzyme B release. In contrast, anti-TGF-β antibodies or a dominant negative (DN) TGF-β receptor have been shown to increase T cell activity (Wang et al., 2019).
CAR T cells have been shown high response rates for RRMM patients (Berdeja et al., 2021; Munshi et al., 2021) (Fig. 2). However, while a promising new anti-cancer strategy, CAR T cells present unique challenges (Maus et al., 2014; Rana et al., 2022). The TME is highly immunosuppressive and is comprised of many different cells that directly or indirectly impede the cytotoxic activity of T cells. TGF-β is secreted by many cells within the TME, e.g., stromal cells (Turley et al., 2015), and suppresses cytotoxic CD8+ T cells through transcriptional downregulation of perforins, granzymes and a plethora of cytotoxins (Thomas and Massagué, 2005; Trapani, 2005). TGF-β also suppresses the cytotoxic activity of Th1 cells and skews T cell differentiation toward a Th2 phenotype (Turley et al., 2015). TGF-β also inhibits TCR-CD28 signaling to promote hyporesponsive memory T cells (Broderick and Bankert, 2006). CD28 is a costimulatory receptor expressed on naïve T cells that is needed for naïve T cells activation following antigen recognition. Moreover, a CD28 module is also included as a costimulatory component of recombinant engineered CARs. The impact of TGF-β on CD28 signaling in naïve and CAR- expressing T cells remains to be determined. Additionally, TGF-β is known to convert CD4+ T- cells into Tregs to suppress anti-tumor immunity (Nakamura et al., 2001; Trapani, 2005).
Due to its vast array of negative effects, TGF-β can impair on T cells, antibodies against it have been developed as well as T cell modifications in order to limit its negative effects on the cytotoxic activity of CAR T cells. For example, an antibody against TGF-β has been shown to positively regulate T cells (Pu et al., 2018) as well as a TGF-β inhibitor (Sow et al., 2019). A CAR that responds to TGF-β enables the genetic modification of T cells that are stimulated by this cytokine (Hou et al., 2018). Genetic ablation of the TGF-β receptor II (TGFBR2) in CAR T cells reduces the conversion to Tregs and to reduce T cell exhaustion. TGFBR2-modified CAR T cells demonstrate improved in vivo tumor killing using either cell line–derived or patient-derived xenografts models (Tang et al., 2020). Knocking out TGFBRII improved the in vivo function of CAR T cells in TGF-β–rich TME. Expression of a dominant negative TGF-βRII receptor (DN-TGF-βRII), inhibits downstream signaling which is required CAR T cell activity, lymphocyte proliferation, cytokine secretion, resistance to exhaustion, and increased tumor killing as shown in cancer models and a phase 1 clinical trial (Narayan et al., 2022).
Alabanza et al. (2022) created an armored BCMA CAR-T cells that expressed resistance towards. TGF-β by co-expressing a BCMA-targeting CAR with DN-TGF-βIIR armor that confers resistance towards TGF-β suppression despite its prolonged exposure. This type of armored CAR T cell design could promote durable responses in MM patients. CD28-ζ CAR T cells retain more potency in resisting TGF-β repression and improving anti-tumor efficacy (Koehler et al., 2007). It was shown that CD28DLCK-ζ CAR T cells with a hybrid IL-7Ra/IL-2Rb receptor and transgenic IL-7 exhibit superior tumor killing (Golumba-Nagy et al., 2018).
While ACT that incorporate CARs and engineered TCRs for cancer treatment are promising, efficacy and sustained response remain outstanding issues. Chimeric switch receptors (CSRs) have been developed to reverse the actions of their original signaling pathways in order to yield immune cells with the ability to overcome the immunosuppressive TME (Tay et al., 2017). Activating CSRs exploit inhibitory molecules that are expressed by cancer cells to stimulate tumor antigen-specific T cells. On the contrary, inhibitory CSRs block the effects of tumor-reactive T cells on their targets. Noh et al. developed a TGF-β/IL-7 CSR encoding the cytokine-binding portion of the TGF-β receptor extracellular domain linked to the immunostimulatory I7R signaling endodomain (tTRII-I7R). CD19 CAR-tTRII-I7R T cells displayed a reduced level of phosphorylated SMAD2 and a higher level of target-specific cell killing in the presence of recombinant human TGF-β1. In B cell lymphoma, overall and recurrence-free survival were extended in mice that received CD19 CAR-tTRII-I7R T cells compared to mice that received control (Noh et al., 2021).
TCR-based ACT utilizes lymphocytes that have been genetically-engineered to target tumor-specific antigens or markers (Tsimberidou et al., 2021). The clinical use of TCR-based therapies entails a structured, multi-step process that involves patient screening and selection, generation of a transduced TCR product, lymphodepletion, and infusion of the TCR-based ACT (Zhao and Cao, 2019). Current CAR technology utilizes modularly-designed receptors introduced into immune cells. TCR-engineered immune cells use naturally occurring, or minimally modified, TCRs to develop ACTs (Figure 2). TCR-based ACT overcome the inhibitory effects of tumor-associated factors by secreting copious amounts of activated lymphocytes with known selectivity and potency. TCRs are heterodimers composed of α- and β- chains, covalently linked via a disulfide bridge between each chain (Huang et al., 2020). TCRs lack intrinsic signaling capacity and are activated by accessory signaling molecules (Birnbaum et al., 2014; Huang et al., 2020).
T cell activation is determined by affinity of TCR for the antigen-MHC complex. Autologous T cells have been harvested from tumors or PBMCs, expanded ex vivo and used as therapeutics. This approach is directed towards obtaining T cells that reflect the naturally occurring antigenic repertoire of TCRs. A recently developed approach features the genetically modified anti-tumor lymphocytes through insertion of genes that encode TCRs with defined specificity and affinity (Rosenberg and Restifo, 2015). Target antigens should be selectively expressed on tumor cells and not on normal cells (Chandran and Klebanoff, 2019).
Naïve T cells demonstrate active TGF-β signaling and TCR engagement that reduces TGF-β signaling by downregulating TGF-β type 1 receptor (TβRI) (Tu et al., 2018) (Figure 2). Moreover, SMAD7 inhibits TGF-β signaling during T cell activation (Hayashi et al., 1997; Fantini et al., 2009) (Figure 2). TCR-T cell therapies are have yielded sustained responses in patients with relapsed and/or refractory disease (Neelapu et al., 2017; Maude et al., 2018; Wang et al., 2020). Moreover, promising responses have been reported in metastatic melanoma (Rosenberg et al., 2011; Besser et al., 2013; Andersen et al., 2016; Forget et al., 2018), cervical carcinoma (Stevanović et al., 2015) and nasopharyngeal cancer (Comoli et al., 2005).
Bispecific T cell engagers (BiTEs) are recombinant proteins that are comprised of two single chain variable fragments (scFv) derived from one antibody that targets a tumor-specific antigen and the other that targets CD3 on the effector T cell. Consequently, T cells are then recruited to the tumor site to target cancer cells (Goebeler and Bargou, 2020). Glioblastoma (GBM) is the most common and most lethal malignant brain tumor. Current treatment for GBM includes surgical resection, radiation and temozolomide chemotherapy, which provide only incremental benefit and are limited by systemic toxicity that damages normal brain tissue (Choi et al., 2019). Choi et al. (2019) developed a bicistronic construct to drive expression of an EGFRvIII-specific CAR and a BiTE against EGFR, an antigen frequently overexpressed in glioblastoma but also expressed in normal tissues. CART. BiTE cells secreted EGFR-specific BiTEs to redirect CAR T cells and recruit untransduced bystander T cells against wild-type EGFR. EGFRvIII-specific CAR-T cells were unable to completely treat tumors with heterogeneous EGFRvIII expression, leading to outgrowth of EGFRvIII-negative, EGFR-positive glioblastoma.
The programmed death (PD)-1 pathway is implicated in immune suppression by reducing BiTE efficacy in patients. A recent case report described a 32-year-old male patient with refractory B-precursor acute lymphoblastic leukemia (ALL) resistant to treatment with blinatumomab (Köhnke et al., 2015). BM immunohistochemistry revealed T cell infiltrates and increased PD-ligand 1 (PD-L1)-positive ALL cells as a potential immune escape mechanism. The authors recapitulated the clinical observation in vitro by showing that blinatumomab was not able to mediate cytotoxicity of CD19+ ALL cells using autologous T cells. In contrast, the addition of healthy donor T cells led to lysis of ALL cells. These results strongly support further systematic evaluation of checkpoint molecules for blinatumomab treatment failure.
TGF-β impedes the anti-PD-1/PD-L1 directed agents and promotes the emergence of drug resistance. Hence, a therapeutic strategy that prevents the effects of TGF- β on the PD-1/PD-L1 axis would be valuable to relieve or prevent drug resistance. The bispecific antibody YM101 inhibits both the PD-1/PD-L1 axis and TGF-β in murine models (Yi et al., 2021).
Cancer cells, stromal fibroblasts and many other cell types that reside with the TME secrete TGF-β to promote myeloma growth and suppress anti-myeloma immunity. Consequently, the TGF-β signaling pathway impacts the anti-tumor efficacy of IMiDs, monoclonal antibodies, and T cells that have been genetically-engineered to express CARs, TCRs and BiTEs. CAR T cells offer the advantage of a single infusion with shorter treatment time, while disadvantages include cost, production, distribution, patient selection, equivocal persistence, and the generation of host antibodies against CAR antibodies. TCR T cells are generated by lentiviral transduction and circumvent the cumbersome in vitro production of specific immune cells. TCR T cell therapy also obviates the limitation of surface antigen expression on target cells. Barriers that limit the efficacy of CAR T cells and TCR T cells as viable, widespread options for solid tumor treatment remain significant. BiTEs combine antigen specificity with the ability to induce cytotoxicity through bystander T cells by linking an scFv that recognizes a tumor antigen and a second that recognizes CD3 on T cells. BiTE and CAR-based strategies are independent of the endogenous TCR specificity and independent of the MHC molecule on tumor surface. Cancer immunotherapy will continue to evolve and further revolutionize therapeutic strategies.
JD, PR, DS, and JK developed the concept, wrote, edited, made substantial contributions and approved the final version of the manuscript.
This research was supported by the NIH R01 (5R01AI139141 to JD), University Hospitals Cleveland Medical Center/Seidman Cancer Center, and the Case Comprehensive Cancer Center.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Alabanza, L. M., Xiong, Y., Vu, B., Webster, B., Wu, D., Hu, P., et al. (2022). Armored BCMA CAR T cells eliminate multiple myeloma and are resistant to the suppressive effects of TGF-β. Front. Immunol. 13, 832645. doi:10.3389/fimmu.2022.832645
Albright, C. D., Salganik, R. I., Craciunescu, C. N., Mar, M. H., and Zeisel, S. H. (2003). Mitochondrial and microsomal derived reactive oxygen species mediate apoptosis induced by transforming growth factor-beta1 in immortalized rat hepatocytes. J. Cell. Biochem. 89, 254–261. doi:10.1002/jcb.10498
Andersen, R., Donia, M., Ellebaek, E., Borch, T. H., Kongsted, P., Iversen, T. Z., et al. (2016). Long-Lasting complete responses in patients with metastatic melanoma after adoptive cell therapy with tumor-infiltrating lymphocytes and an attenuated il2 regimen. Clin. Cancer Res. 22, 3734–3745. doi:10.1158/1078-0432.CCR-15-1879
Balsas, P., Galán-Malo, P., Marzo, I., and Naval, J. (2012). Bortezomib resistance in a myeloma cell line is associated to PSMβ5 overexpression and polyploidy. Leuk. Res. 36, 212–218. doi:10.1016/J.LEUKRES.2011.09.011
Batlle, E., and Massagué, J. (2019). Transforming growth factor-β signaling in immunity and cancer. Immunity 50, 924–940. doi:10.1016/J.IMMUNI.2019.03.024
Battaglia, A., Buzzonetti, A., Baranello, C., Fanelli, M., Fossati, M., Catzola, V., et al. (2013). Interleukin-21 (IL-21) synergizes with IL-2 to enhance T-cell receptor-induced human T-cell proliferation and counteracts IL-2/transforming growth factor-β-induced regulatory T-cell development. Immunology 139, 109–120. doi:10.1111/IMM.12061
Berdeja, J. G., Madduri, D., Usmani, S. Z., Jakubowiak, A., Agha, M., Cohen, A. D., et al. (2021). Ciltacabtagene autoleucel, a B-cell maturation antigen-directed chimeric antigen receptor T-cell therapy in patients with relapsed or refractory multiple myeloma (CARTITUDE-1): A phase 1b/2 open-label study. Lancet 398, 314–324. doi:10.1016/S0140-6736(21)00933-8
Besser, M. J., Shapira-Frommer, R., Itzhaki, O., Treves, A. J., Zippel, D. B., Levy, D., et al. (2013). Adoptive transfer of tumor-infiltrating lymphocytes in patients with metastatic melanoma: Intent-to-treat analysis and efficacy after failure to prior immunotherapies. Clin. Cancer Res. 19, 4792–4800. doi:10.1158/1078-0432.CCR-13-0380
Bhowmick, N. A., Chytil, A., Plieth, D., Gorska, A. E., Dumont, N., Shappell, S., et al. (2004). TGF-beta signaling in fibroblasts modulates the oncogenic potential of adjacent epithelia. Science 303, 848–851. doi:10.1126/SCIENCE.1090922
Bierie, B., and Moses, H. L. (2006). Tumour microenvironment: TGFbeta: The molecular jekyll and hyde of cancer. Nat. Rev. Cancer 6, 506–520. doi:10.1038/nrc1926
Birnbaum, M. E., Berry, R., Hsiao, Y. S., Chen, Z., Shingu-Vazquez, M. A., Yu, X., et al. (2014). Molecular architecture of the αβ T cell receptor-CD3 complex. Proc. Natl. Acad. Sci. U. S. A. 111, 17576–17581. doi:10.1073/pnas.1420936111
Blobe, G. C., Schiemann, W. P., and Lodish, H. F. (2000). Role of transforming growth factor beta in human disease. N. Engl. J. Med. 342, 1350–1358. doi:10.1056/NEJM200005043421807
Boulay, J. L., Mild, G., Lowy, A., Reuter, J., Lagrange, M., Terracciano, L., et al. (2003). SMAD7 is a prognostic marker in patients with colorectal cancer. Int. J. Cancer 104, 446–449. doi:10.1002/IJC.10908
Boutilier, A. J., and Elsawa, S. F. (2021). Macrophage polarization states in the tumor microenvironment. Int. J. Mol. Sci. 22, 6995. doi:10.3390/IJMS22136995
Broderick, L., and Bankert, R. B. (2006). Membrane-associated TGF-beta1 inhibits human memory T cell signaling in malignant and nonmalignant inflammatory microenvironments. J. Immunol. 177, 3082–3088. doi:10.4049/jimmunol.177.5.3082
Castriconi, R., Cantoni, C., della Chiesa, M., Vitale, M., Marcenaro, E., Conte, R., et al. (2003). Transforming growth factor β1 inhibits expression of NKp30 and NKG2D receptors: Consequences for the NK-mediated killing of dendritic cells. Proc. Natl. Acad. Sci. U. S. A. 100, 4120–4125. doi:10.1073/PNAS.0730640100
Chandran, S. S., and Klebanoff, C. A. (2019). T cell receptor-based cancer immunotherapy: Emerging efficacy and pathways of resistance. Immunol. Rev. 290, 127–147. doi:10.1111/imr.12772
Chen, C. H., Seguin-Devaux, C., Burke, N. A., Oriss, T. B., Watkins, S. C., Clipstone, N., et al. (2003). Transforming growth factor β blocks Tec kinase phosphorylation, Ca2+ influx, and NFATc translocation causing inhibition of T cell differentiation. J. Exp. Med. 197, 1689–1699. doi:10.1084/jem.20021170
Chen, M. L., Pittet, M. J., Gorelik, L., Flavell, R. A., Weissleder, R., von Boehmer, H., et al. (2005). Regulatory T cells suppress tumor-specific CD8 T cell cytotoxicity through TGF-β signals in vivo. Proc. Natl. Acad. Sci. U. S. A. 102, 419–424. doi:10.1073/pnas.0408197102
Chen, W. J., Jin, W., Hardegen, N., Lei, K. J., Li, L., Marinos, N., et al. (2003). Conversion of peripheral CD4+CD25- naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J. Exp. Med. 198, 1875–1886. doi:10.1084/JEM.20030152
Choi, B. D., Yu, X., Castano, A. P., Bouffard, A. A., Schmidts, A., Larson, R. C., et al. (2019). CAR-T cells secreting BiTEs circumvent antigen escape without detectable toxicity. Nat. Biotechnol. 37, 1049–1058. doi:10.1038/S41587-019-0192-1
Comoli, P., Pedrazzoli, P., Maccario, R., Basso, S., Carminati, O., Labirio, M., et al. (2005). Cell therapy of stage IV nasopharyngeal carcinoma with autologous Epstein-Barr virus-targeted cytotoxic T lymphocytes. J. Clin. Oncol. 23, 8942–8949. doi:10.1200/JCO.2005.02.6195
Cook, G., Campbell, J. D. M., Carr, C. E., Boyd, K. S., and Franklin, I. M. (1999). Transforming growth factor β from multiple myeloma cells inhibits proliferation and IL-2 responsiveness in T lymphocytes. J. Leukoc. Biol. 66, 981–988. doi:10.1002/jlb.66.6.981
Coşkun, H. Ş., İlhan, O., Özcan, M., Ayaz, S., Dalva, K., Üstün, C., et al. (2006). Serum transforming growth factor beta 1 levels in multiple myeloma patients. Turk. J. Haematol. 23, 47–52.
Dahmani, A., and Delisle, J. S. (2018). TGF-β in T Cell biology: Implications for cancer immunotherapy. Cancers (Basel) 10, 194. doi:10.3390/CANCERS10060194
Davidson, T. S., DiPaolo, R. J., Andersson, J., and Shevach, E. M. (2007). Cutting edge: IL-2 is essential for TGF-β-mediated induction of Foxp3 + T regulatory cells. J. Immunol. 178, 4022–4026. doi:10.4049/jimmunol.178.7.4022
Debacq-Chainiaux, F., Borlon, C., Pascal, T., Royer, V., Eliaers, F., Ninane, N., et al. (2005). Repeated exposure of human skin fibroblasts to UVB at subcytotoxic level triggers premature senescence through the TGF-beta1 signaling pathway. J. Cell Sci. 118, 743–758. doi:10.1242/jcs.01651
Dituri, F., Cossu, C., Mancarella, S., and Giannelli, G. (2019). The interactivity between TGFβ and BMP signaling in organogenesis, fibrosis, and cancer. Cells 8, 1130. doi:10.3390/CELLS8101130
Dong, M., and Blobe, G. C. (2006). Role of transforming growth factor-β in hematologic malignancies. Blood 107, 4589–4596. doi:10.1182/blood-2005-10-4169
Driscoll, J. J., and Woodle, E. S. (2012). Targeting the Ubiquitin+Proteasome system in solid tumors. Semin. Hematol. 49, 277–283. doi:10.1053/J.SEMINHEMATOL.2012.04.002
Driscoll, J., Minter, A., Driscoll, A., and Burris, D., K. J. (2012). The Ubiquitin+Proteasome protein degradation pathway as a therapeutic strategy in the treatment of solid tumor malignancies. Anticancer. Agents Med. Chem. 11, 242–246. doi:10.2174/187152011795255948
Driscoll, J. (1994). The role of the proteasome in cellular protein degradation. Histol. Histopathol. 9, 197–202.
Dzhagalov, I. L., Chen, K. G., Herzmark, P., and Robey, E. A. (2013). Elimination of self-reactive T cells in the thymus: A timeline for negative selection. PLoS Biol. 11, e1001566. doi:10.1371/journal.pbio.1001566
Fantini, M. C., Becker, C., Monteleone, G., Pallone, F., Galle, P. R., and Neurath, M. F. (2004). Cutting edge: TGF-beta induces a regulatory phenotype in CD4+CD25- T cells through Foxp3 induction and down-regulation of Smad7. J. Immunol. 172, 5149–5153. doi:10.4049/JIMMUNOL.172.9.5149
Fantini, M. C., Rizzo, A., Fina, D., Caruso, R., Sarra, M., Stolfi, C., et al. (2009). Smad7 controls resistance of colitogenic T cells to regulatory T cell-mediated suppression. Gastroenterology 136, 1308–1316. doi:10.1053/j.gastro.2008.12.053
Forget, M. A., Haymaker, C., Hess, K. R., Meng, Y. J., Creasy, C., Karpinets, T., et al. (2018). Prospective analysis of adoptive TIL therapy in patients with metastatic melanoma: Response, impact of anti-CTLA4, and biomarkers to predict clinical outcome. Clin. Cancer Res. 24, 4416–4428. doi:10.1158/1078-0432.CCR-17-3649
Gallimore, A., Glithero, A., Godkin, A., Tissot, A. C., Plückthun, A., Elliott, T., et al. (1998). Induction and exhaustion of lymphocytic choriomeningitis virus-specific cytotoxic T lymphocytes visualized using soluble tetrameric major histocompatibility complex class I-peptide complexes. J. Exp. Med. 187, 1383–1393. doi:10.1084/jem.187.9.1383
Goebeler, M. E., and Bargou, R. C. (2020). T cell-engaging therapies - BiTEs and beyond. Nat. Rev. Clin. Oncol. 17, 418–434. doi:10.1038/S41571-020-0347-5
Golumba-Nagy, V., Kuehle, J., Hombach, A. A., and Abken, H. (2018). CD28-ζ CAR T cells resist TGF-β repression through IL-2 signaling, which can Be mimicked by an engineered IL-7 autocrine loop. Mol. Ther. 26, 2218–2230. doi:10.1016/j.ymthe.2018.07.005
Gorelik, L., Fields, P. E., and Flavell, R. A. (2000). Cutting edge: TGF-β inhibits Th type 2 development through inhibition of GATA-3 expression. J. Immunol. 165, 4773–4777. doi:10.4049/jimmunol.165.9.4773
Gorelik, L., and Flavell, R. A. (2002). Transforming growth factor-beta in T-cell biology. Nat. Rev. Immunol. 2, 46–53. doi:10.1038/NRI704
Gu, A., Wang, Y., Lin, L., Zhang, S. S., and Wan, Y. Y. (2012). Requirements of transcription factor Smad-dependent and -independent TGF-β signaling to control discrete T-cell functions. Proc. Natl. Acad. Sci. U. S. A. 109, 905–910. doi:10.1073/pnas.1108352109
Gunderson, A. J., Yamazaki, T., McCarty, K., Fox, N., Phillips, M., Alice, A., et al. (2020). TGFβ suppresses CD8+ T cell expression of CXCR3 and tumor trafficking. Nat. Commun. 11, 1749. doi:10.1038/S41467-020-15404-8
Gupta, M., Han, J. J., Stenson, M., Maurer, M., Wellik, L., Hu, G., et al. (2012). Elevated serum IL-10 levels in diffuse large B-cell lymphoma: A mechanism of aberrant JAK2 activation. Blood 119, 2844–2853. doi:10.1182/blood-2011-10-388538
Hadjiaggelidou, C., and Katodritou, E. (2021). Regulatory T-cells and multiple myeloma: Implications in tumor immune biology and treatment. J. Clin. Med. 10, 4588. doi:10.3390/JCM10194588
Hayashi, H., Abdollah, S., Qiu, Y., Cai, J., Xu, Y. Y., Grinnell, B. W., et al. (1997). The MAD-related protein Smad7 associates with the TGFbeta receptor and functions as an antagonist of TGFbeta signaling. Cell 89, 1165–1173. doi:10.1016/S0092-8674(00)80303-7
Hayashi, T., Hideshima, T., Nguyen, A. N., Munoz, O., Podar, K., Hamasaki, M., et al. (2004). Transforming growth factor β receptor I kinase inhibitor down-regulates cytokine secretion and multiple myeloma cell growth in the bone marrow microenvironment. Clin. Cancer Res. 10, 7540–7546. doi:10.1158/1078-0432.CCR-04-0632
Heldin, C. H., and Moustakas, A. (2016). Signaling receptors for TGF-β family members. Cold Spring Harb. Perspect. Biol. 8, a022053. doi:10.1101/CSHPERSPECT.A022053
Hou, A. J., Chang, Z. L., Lorenzini, M. H., Zah, E., and Chen, Y. Y. (2018). TGF-β-responsive CAR-T cells promote anti-tumor immune function. Bioeng. Transl. Med. 3, 75–86. doi:10.1002/btm2.10097
Hu, B., Tack, D. C., Liu, T., Wu, Z., Ullenbruch, M. R., and Phan, S. H. (2006). Role of Smad3 in the regulation of rat telomerase reverse transcriptase by TGFbeta. Oncogene 25, 1030–1041. doi:10.1038/sj.onc.1209140
Hu, Y., He, J., He, L., Xu, B., and Wang, Q. (2021). Expression and function of Smad7 in autoimmune and inflammatory diseases. J. Mol. Med. 99, 1209–1220. doi:10.1007/S00109-021-02083-1
Huai, G., Markmann, J. F., Deng, S., and Rickert, C. G. (2021). TGF-β-secreting regulatory B cells: Unsung players in immune regulation. Clin. Transl. Immunol. 10, e1270. doi:10.1002/cti2.1270
Huang, D., Miller, M., Ashok, B., Jain, S., and Peppas, N. A. (2020). CRISPR/Cas systems to overcome challenges in developing the next generation of T cells for cancer therapy. Adv. Drug Deliv. Rev. 158, 17–35. doi:10.1016/j.addr.2020.07.015
Ignatz-Hoover, J. J., and Driscoll, J. J. (2022). Therapeutics to harness the immune microenvironment in multiple myeloma. Cancer Drug resist. 5, 647–661. doi:10.20517/CDR.2022.23
Ignatz-Hoover, J. J., Murphy, E. v., and Driscoll, J. J. (2022). Targeting proteasomes in cancer and infectious disease: A parallel strategy to treat malignancies and microbes. Front. Cell. Infect. Microbiol. 12, 881. doi:10.3389/fcimb.2022.925804
Ikushima, H., and Miyazono, K. (2010). TGFbeta signalling: A complex web in cancer progression. Nat. Rev. Cancer 10, 415–424. doi:10.1038/NRC2853
Jabeen, R., and Kaplan, M. H. (2012). The symphony of the ninth: The development and function of Th9 cells. Curr. Opin. Immunol. 24, 303–307. doi:10.1016/j.coi.2012.02.001
Jayaraman, P., Parikh, F., Newton, J. M., Hanoteau, A., Rivas, C., Krupar, R., et al. (2018). TGF-β1 programmed myeloid-derived suppressor cells (MDSC) acquire immune-stimulating and tumor killing activity capable of rejecting established tumors in combination with radiotherapy. Oncoimmunology 7, e1490853. doi:10.1080/2162402X.2018.1490853
Josefowicz, S. Z., Lu, L. F., and Rudensky, A. Y. (2012). Regulatory T cells: Mechanisms of differentiation and function. Annu. Rev. Immunol. 30, 531–564. doi:10.1146/annurev.immunol.25.022106.141623
Kim, B. G., Malek, E., Choi, S. H., Ignatz-Hoover, J. J., and Driscoll, J. J. (2021). Novel therapies emerging in oncology to target the TGF-β pathway. J. Hematol. Oncol. 14, 55. doi:10.1186/S13045-021-01053-X
Kim, B. N., Ahn, D. H., Kang, N., Yeo, C. D., Kim, Y. K., Lee, K. Y., et al. (2020). TGF-β induced EMT and stemness characteristics are associated with epigenetic regulation in lung cancer. Sci. Rep. 10, 10597–10611. doi:10.1038/s41598-020-67325-7
Koehler, H., Kofler, D., Hombach, A., and Abken, H. (2007). CD28 costimulation overcomes transforming growth factor-β-mediated repression of proliferation of redirected human CD4+ and CD8 + T cells in an antitumor cell attack. Cancer Res. 67, 2265–2273. doi:10.1158/0008-5472.CAN-06-2098
Köhnke, T., Krupka, C., Tischer, J., Knösel, T., and Subklewe, M. (2015). Increase of PD-L1 expressing B-precursor ALL cells in a patient resistant to the CD19/CD3-bispecific T cell engager antibody blinatumomab. J. Hematol. Oncol. 8, 111. doi:10.1186/s13045-015-0213-6
Lai, G., Sun, R., Wu, J., Zhang, B., and Zhao, Y. (2018). 20-HETE regulated PSMB5 expression via TGF-β/Smad signaling pathway. Prostagl. Other Lipid Mediat. 134, 123–130. doi:10.1016/j.prostaglandins.2017.08.005
Laouar, Y., Sutterwala, F. S., Gorelik, L., and Flavell, R. A. (2005). Transforming growth factor-beta controls T helper type 1 cell development through regulation of natural killer cell interferon-gamma. Nat. Immunol. 6, 600–607. doi:10.1038/ni1197
Lee, C. R., Lee, W., Cho, S. K., and Park, S. G. (2018). Characterization of multiple cytokine combinations and TGF-β on differentiation and functions of myeloid-derived suppressor cells. Int. J. Mol. Sci. 19, 869. doi:10.3390/IJMS19030869
Li, H., Xu, D., Li, J., Berndt, M. C., and Liu, J. P. (2006). Transforming growth factor β suppresses human telomerase reverse transcriptase (hTERT) by Smad3 interactions with c-Myc and the hTERT gene. J. Biol. Chem. 281, 25588–25600. doi:10.1074/jbc.M602381200
Li, J., Wang, L., Chen, X., Li, L., Li, Y., Ping, Y., et al. (2017). CD39/CD73 upregulation on myeloid-derived suppressor cells via TGF-β-mTOR-HIF-1 signaling in patients with non-small cell lung cancer. Oncoimmunology 6, e1320011. doi:10.1080/2162402X.2017.1320011
Li, M. O., Wan, Y. Y., Sanjabi, S., Robertson, A. K. L., and Flavell, R. A. (2006). Transforming growth factor-β regulation of immune responses. Annu. Rev. Immunol. 24, 99–146. doi:10.1146/annurev.immunol.24.021605.090737
Li, Y., Fan, W., Link, F., Wang, S., and Dooley, S. (2021). Transforming growth factor β latency: A mechanism of cytokine storage and signalling regulation in liver homeostasis and disease. JHEP Rep. 4, 100397. doi:10.1016/J.JHEPR.2021.100397
Lü, S., Chen, Z., Yang, J., Chen, L., Gong, S., Zhou, H., et al. (2008). Overexpression of the PSMB5 gene contributes to bortezomib resistance in T-lymphoblastic lymphoma/leukemia cells derived from Jurkat line. Exp. Hematol. 36, 1278–1284. doi:10.1016/J.EXPHEM.2008.04.013
Macfarlane, E. G., Haupt, J., Dietz, H. C., and Shore, E. M. (2017). TGF-Β family signaling in connective tissue and skeletal diseases. Cold Spring Harb. Perspect. Biol. 9, a022269. doi:10.1101/CSHPERSPECT.A022269
Malek, E., Hwang, S., de Lima, M., Caimi, P., Gallogly, M. M., Metheny, L., et al. (2019). Preclinical studies and phase I trial of vactosertib in combination with pomalidomide in relapsed multiple myeloma: A corticosteroid-free approach by targeting TGF-β signaling pathway. Blood 134, 3232. doi:10.1182/BLOOD-2019-126728
Massagué, J. (2012). TGFβ signalling in context. Nat. Rev. Mol. Cell Biol. 13, 616–630. doi:10.1038/NRM3434
Maude, S. L., Laetsch, T. W., Buechner, J., Rives, S., Boyer, M., Bittencourt, H., et al. (2018). Tisagenlecleucel in children and young adults with B-cell lymphoblastic leukemia. N. Engl. J. Med. 378, 439–448. doi:10.1056/nejmoa1709866
Maus, M. v., Grupp, S. A., Porter, D. L., and June, C. H. (2014). Antibody-modified T cells: CARs take the front seat for hematologic malignancies. Blood 123, 2625–2635. doi:10.1182/BLOOD-2013-11-492231
Miyazono, K., Katsuno, Y., Koinuma, D., Ehata, S., and Morikawa, M. (2018). Intracellular and extracellular TGF-β signaling in cancer: Some recent topics. Front. Med. 12, 387–411. doi:10.1007/s11684-018-0646-8
Moses, H. L., Roberts, A. B., and Derynck, R. (2016). The discovery and early days of TGF-β: A historical perspective. Cold Spring Harb. Perspect. Biol. 8, a021865. doi:10.1101/CSHPERSPECT.A021865
Moustakas, A., and Heldin, C. H. (2009). The regulation of TGFbeta signal transduction. Development 136, 3699–3714. doi:10.1242/DEV.030338
Mukherjee, P., Winter, S. L., and Alexandrow, M. G. (2010). Cell cycle arrest by transforming growth factor beta1 near G1/S is mediated by acute abrogation of prereplication complex activation involving an Rb-MCM interaction. Mol. Cell. Biol. 30, 845–856. doi:10.1128/MCB.01152-09
Munshi, N. C., Anderson, L. D., Shah, N., Madduri, D., Berdeja, J., Lonial, S., et al. (2021). Idecabtagene vicleucel in relapsed and refractory multiple myeloma. N. Engl. J. Med. 384, 705–716. doi:10.1056/NEJMOA2024850
Nakamura, K., Kitani, A., and Strober, W. (2001). Cell contact-dependent immunosuppression by CD4(+)CD25(+) regulatory T cells is mediated by cell surface-bound transforming growth factor beta. J. Exp. Med. 194, 629–644. doi:10.1084/jem.194.5.629
Narayan, V., Barber-Rotenberg, J. S., Jung, I. Y., Lacey, S. F., Rech, A. J., Davis, M. M., et al. (2022). PSMA-Targeting tgfβ-insensitive armored CAR T cells in metastatic castration-resistant prostate cancer: A phase 1 trial. Nat. Med. 28 (4 28), 724–734. doi:10.1038/s41591-022-01726-1
Neelapu, S. S., Locke, F. L., Bartlett, N. L., Lekakis, L. J., Miklos, D. B., Jacobson, C. A., et al. (2017). Axicabtagene ciloleucel CAR T-cell therapy in refractory large B-cell lymphoma. N. Engl. J. Med. 377, 2531–2544. doi:10.1056/nejmoa1707447
Neurath, M. F., Weigmann, B., Finotto, S., Glickman, J., Nieuwenhuis, E., Iijima, H., et al. (2002). The transcription factor T-bet regulates mucosal T cell activation in experimental colitis and Crohn’s disease. J. Exp. Med. 195, 1129–1143. doi:10.1084/jem.20011956
Noh, K.-E., Lee, J.-H., Choi, S.-Y., Jung, N.-C., Nam, J.-H., Oh, J.-S., et al. (2021). TGF-β/IL-7 chimeric switch receptor-expressing CAR-T cells inhibit recurrence of CD19-positive B cell lymphoma. Int. J. Mol. Sci. 22, 8706. doi:10.3390/ijms22168706
Oerlemans, R., Franke, N. E., Assaraf, Y. G., Cloos, J., van Zantwijk, I., Berkers, C. R., et al. (2008). Molecular basis of bortezomib resistance: Proteasome subunit beta5 (PSMB5) gene mutation and overexpression of PSMB5 protein. Blood 112, 2489–2499. doi:10.1182/BLOOD-2007-08-104950
Oh, S. A., and Li, M. O. (2013). TGF-Β: guardian of T cell function. J. Immunol. 191, 3973–3979. doi:10.4049/JIMMUNOL.1301843
Ouyang, W., Beckett, O., Ma, Q., and Li, M. O. (2010). Transforming growth factor-β signaling curbs thymic negative selection promoting regulatory T cell development. Immunity 32, 642–653. doi:10.1016/j.immuni.2010.04.012
Pu, N., Zhao, G., Gao, S., Cui, Y., Xu, Y., Lv, Y., et al. (2018). Neutralizing TGF-β promotes anti-tumor immunity of dendritic cells against pancreatic cancer by regulating T lymphocytes. Cent. Eur. J. Immunol. 43, 123–131. doi:10.5114/ceji.2018.77381
Rana, P. S., Murphy, E. v., Kort, J., and Driscoll, J. J. (2022). Road testing new CAR design strategies in multiple myeloma. Front. Immunol. 13, 957157. doi:10.3389/FIMMU.2022.957157
Regis, S., Dondero, A., Caliendo, F., Bottino, C., and Castriconi, R. (2020). NK cell function regulation by TGF-β-induced epigenetic mechanisms. Front. Immunol. 11, 311. doi:10.3389/FIMMU.2020.00311
Rosenberg, S. A., and Restifo, N. P. (2015). Adoptive cell transfer as personalized immunotherapy for human cancer. Science 348, 62–68. doi:10.1126/science.aaa4967
Rosenberg, S. A., Yang, J. C., Sherry, R. M., Kammula, U. S., Hughes, M. S., Phan, G. Q., et al. (2011). Durable complete responses in heavily pretreated patients with metastatic melanoma using T-cell transfer immunotherapy. Clin. Cancer Res. 17, 4550–4557. doi:10.1158/1078-0432.CCR-11-0116
Sakamaki, S., Hirayama, Y., Matsunaga, T., Kuroda, H., Kusakabe, T., Akiyama, T., et al. (1999). Transforming growth factor-β1 (TGF-β1) induces thrombopoietin from bone marrow stromal cells, which stimulates the expression of TGF-β receptor on megakaryocytes and, in turn, renders them susceptible to suppression by TGF-β itself with high specificity. Blood 94, 1961–1970. doi:10.1182/blood.v94.6.1961.418k17_1961_1970
Shi, M., Zhu, J., Wang, R., Lizhi, M., Walz, T., Springer, T. A., et al. (2011). Latent TGF-β structure and activation. Nature 474, 343–349. doi:10.1038/nature10152
Sow, H. S., Ren, J., Camps, M., Ossendorp, F., and ten Dijke, P. (2019). Combined inhibition of TGF-β signaling and the PD-L1 immune checkpoint is differentially effective in tumor models. Cells 8, 320. doi:10.3390/cells8040320
Stevanović, S., Draper, L. M., Langhan, M. M., Campbell, T. E., Kwong, M. L., Wunderlich, J. R., et al. (2015). Complete regression of metastatic cervical cancer after treatment with human papillomavirus-targeted tumor-infiltrating T cells. J. Clin. Oncol. 33, 1543–1550. doi:10.1200/JCO.2014.58.9093
Suzuki, H. I. (2018). MicroRNA control of TGF-β signaling. Int. J. Mol. Sci. 19, 1901. doi:10.3390/ijms19071901
Suzuki, H. I., and Miyazono, K. (2011). Emerging complexity of microRNA generation cascades. J. Biochem. 149, 15–25. doi:10.1093/jb/mvq113
Swamydas, M., Murphy, E. v., Ignatz-Hoover, J. J., Malek, E., and Driscoll, J. J. (2022). Deciphering mechanisms of immune escape to inform immunotherapeutic strategies in multiple myeloma. J. Hematol. Oncol. 15, 17. doi:10.1186/s13045-022-01234-2
Takimoto, T., Wakabayashi, Y., Sekiya, T., Inoue, N., Morita, R., Ichiyama, K., et al. (2010). Smad2 and Smad3 are redundantly essential for the TGF-β–mediated regulation of regulatory T plasticity and Th1 development. J. Immunol. 185, 842–855. doi:10.4049/jimmunol.0904100
Tang, J., Gifford, C. C., Samarakoon, R., and Higgins, P. J. (2018). Deregulation of negative controls on TGF-β1 signaling in tumor progression. Cancers (Basel) 10, 159. doi:10.3390/CANCERS10060159
Tang, N., Cheng, C., Zhang, X., Qiao, M., Li, N., Mu, W., et al. (2020). TGF-β inhibition via CRISPR promotes the long-term efficacy of CAR T cells against solid tumors. JCI Insight 5, e133977. doi:10.1172/JCI.INSIGHT.133977
Tay, J., Levesque, J. P., and Winkler, I. G. (2017). Cellular players of hematopoietic stem cell mobilization in the bone marrow niche. Int. J. Hematol. 105, 129–140. doi:10.1007/S12185-016-2162-4
Thomas, D. A., and Massagué, J. (2005). TGF-β directly targets cytotoxic T cell functions during tumor evasion of immune surveillance. Cancer Cell 8, 369–380. doi:10.1016/j.ccr.2005.10.012
Timmers, M., Roex, G., Wang, Y., Campillo-Davo, D., van Tendeloo, V. F. I., Chu, Y., et al. (2019). Chimeric antigen receptor-modified T cell therapy in multiple myeloma: Beyond B cell maturation antigen. Front. Immunol. 10, 1613. doi:10.3389/fimmu.2019.01613
Tinoco, R., Alcalde, V., Yang, Y., Sauer, K., and Zuniga, E. I. (2009). Cell-intrinsic transforming growth factor-β signaling mediates virus-specific CD8+ T cell deletion and viral persistence in vivo. Immunity 31, 145–157. doi:10.1016/j.immuni.2009.06.015
Trapani, J. A. (2005). The dual adverse effects of TGF-β secretion on tumor progression. Cancer Cell 8, 349–350. doi:10.1016/j.ccr.2005.10.018
Tremblay, S., Driscoll, J. J., Rike-Shields, A., Hildeman, D. A., Alloway, R. R., Girnita, A. L., et al. (2020). A prospective, iterative, adaptive trial of carfilzomib-based desensitization. Am. J. Transpl. 20, 411–421. doi:10.1111/AJT.15613
Tsimberidou, A. M., van Morris, K., Vo, H. H., Eck, S., Lin, Y. F., Rivas, J. M., et al. (2021). T-Cell receptor-based therapy: An innovative therapeutic approach for solid tumors. J. Hematol. Oncol. 14 (1 14), 102–122. doi:10.1186/S13045-021-01115-0
Tu, E., Chia, C. P. Z., Chen, W., Zhang, D., Park, S. A., Jin, W., et al. (2018). T cell receptor-regulated TGF-β type I receptor expression determines T cell quiescence and activation. Immunity 48, 745–759. doi:10.1016/j.immuni.2018.03.025
Turley, S. J., Cremasco, V., and Astarita, J. L. (2015). Immunological hallmarks of stromal cells in the tumour microenvironment. Nat. Rev. Immunol. 15, 669–682. doi:10.1038/nri3902
Tzavlaki, K., and Moustakas, A. (2020). TGF-Β signaling. Biomolecules 10, 487. doi:10.3390/BIOM10030487
Urashima, M., Ogata, A., Chauhan, D., Hatziyanni, M., Vidriales, M., Dedera, D., et al. (1996). Transforming growth factor-beta1: Differential effects on multiple myeloma versus normal B cells. Blood 87, 1928–1938. doi:10.1182/BLOOD.V87.5.1928.1928
Urbani, S., Amadei, B., Tola, D., Massari, M., Schivazappa, S., Missale, G., et al. (2006). PD-1 expression in acute hepatitis C virus (HCV) infection is associated with HCV-specific CD8 exhaustion. J. Virol. 80, 11398–11403. doi:10.1128/jvi.01177-06
Veldhoen, M., Hocking, R. J., Flavell, R. A., and Stockinger, B. (2006). Signals mediated by transforming growth factor-β initiate autoimmune encephalomyelitis, but chronic inflammation is needed to sustain disease. Nat. Immunol. 7, 1151–1156. doi:10.1038/ni1391
Wan, Y. Y., and Flavell, R. A. (2008). TGF-beta and regulatory T cell in immunity and autoimmunity. J. Clin. Immunol. 28, 647–659. doi:10.1007/S10875-008-9251-Y
Wang, D., Yang, L., Yue, D., Cao, L., Li, L., Wang, D., et al. (2019). Macrophage-derived CCL22 promotes an immunosuppressive tumor microenvironment via IL-8 in malignant pleural effusion. Cancer Lett. 452, 244–253. doi:10.1016/j.canlet.2019.03.040
Wang, M., Munoz, J., Goy, A., Locke, F. L., Jacobson, C. A., Hill, B. T., et al. (2020). KTE-X19 CAR T-cell therapy in relapsed or refractory mantle-cell lymphoma. N. Engl. J. Med. 382, 1331–1342. doi:10.1056/nejmoa1914347
Wei, J., Duramad, O., Perng, O. A., Reiner, S. L., Liu, Y. J., and Qin, F. X. F. (2007). Antagonistic nature of T helper 1/2 developmental programs in opposing peripheral induction of Foxp3+ regulatory T cells. Proc. Natl. Acad. Sci. U. S. A. 104, 18169–18174. doi:10.1073/PNAS.0703642104
Wei, W., Zou, Y., Jiang, Q., Zhou, Z., Ding, H., Yan, L., et al. (2018). PSMB5 is associated with proliferation and drug resistance in triple-negative breast cancer. Int. J. Biol. Markers 33, 102–108. doi:10.5301/IJBM.5000283
Woodle, E. S., Tremblay, S., Rossi, A., Rojas, C. C., Alloway, R., Roskin, K., et al. (2020). Plasma cell targeting to prevent antibody-mediated rejection. Am. J. Transpl. 20, 33–41. doi:10.1111/AJT.15889
Wu, X., Wang, Y., Xu, J., Luo, T., Deng, J., and Hu, Y. (2017). MM-BMSCs induce naïve CD4+ T lymphocytes dysfunction through fibroblast activation protein α. Oncotarget 8, 52614–52628. doi:10.18632/oncotarget.17538
Yaccoby, S., Saha, R., Kozlowska, E., Ge, Y., Ling, W., Cottler-Fox, M., et al. (2005). Myeloma cells cultivate megakaryocytes to promote lytic bone disease. Blood 106, 2495. doi:10.1182/BLOOD.V106.11.2495.2495
Yang, Z. Z., Grote, D. M., Xiu, B., Ziesmer, S. C., Price-Troska, T. L., Hodge, L. S., et al. (2014). TGF-β upregulates CD70 expression and induces exhaustion of effector memory T cells in B-cell non-Hodgkin’s lymphoma. Leukemia 28, 1872–1884. doi:10.1038/leu.2014.84
Yi, M., Zhang, J., Li, A., Niu, M., Yan, Y., Jiao, Y., et al. (2021). The construction, expression, and enhanced anti-tumor activity of YM101: A bispecific antibody simultaneously targeting TGF-β and PD-L1. J. Hematol. Oncol. 14, 27. doi:10.1186/s13045-021-01045-x
Yoon, Y. S., Lee, J. H., Hwang, S. C., Choi, K. S., and Yoon, G. (2005). TGF beta1 induces prolonged mitochondrial ROS generation through decreased complex IV activity with senescent arrest in Mv1Lu cells. Oncogene 24, 1895–1903. doi:10.1038/sj.onc.1208262
Yu, J., Wei, M., Becknell, B., Trotta, R., Liu, S., Boyd, Z., et al. (2006). Pro- and antiinflammatory cytokine signaling: Reciprocal antagonism regulates interferon-gamma production by human natural killer cells. Immunity 24, 575–590. doi:10.1016/j.immuni.2006.03.016
Zhang, F., Wang, H., Wang, X., Jiang, G., Liu, H., Zhang, G., et al. (2016). TGF-β induces M2-like macrophage polarization via SNAIL-mediated suppression of a pro-inflammatory phenotype. Oncotarget 7, 52294–52306. doi:10.18632/ONCOTARGET.10561
Zhang, Y., Alexander, P. B., and Wang, X. F. (2017). TGF-β family signaling in the control of cell proliferation and survival. Cold Spring Harb. Perspect. Biol. 9, a022145. doi:10.1101/cshperspect.a022145
Zhao, L., and Cao, Y. J. (2019). Engineered T cell therapy for cancer in the clinic. Front. Immunol. 10, 2250. doi:10.3389/fimmu.2019.02250
Zheng, S. G., Wang, J., Wang, P., Gray, J. D., and Horwitz, D. A. (2007). IL-2 is essential for TGF-β to convert naive CD4 + CD25 − cells to CD25 + Foxp3 + regulatory T cells and for expansion of these cells. J. Immunol. 178, 2018–2027. doi:10.4049/jimmunol.178.4.2018
Zheng, W., Liu, D., Fan, X., Powers, L., Goswami, M., Hu, Y., et al. (2013). Potential therapeutic biomarkers in plasma cell myeloma: A flow cytometry study. Cytom. B Clin. Cytom. 84, 222–228. doi:10.1002/cyto.b.21083
ACT Adoptive cell-based therapy
BiTEs Bispecific T cell engagers
BCMA B cell maturation antigen
BM Bone marrow
BMSCs bone marrow stromal cells
BMP bone morphogenic protein
CARD11 Caspase recruitment domain-containing protein 11
CAR-T Chimeric antigen receptor T cells
CRISPR/Cas9 Clustered Regularly Interspaced Short Palindromic Repeats And CRISPR-Associated Protein 9
CSRs Chimeric switch receptors
CTL Cytotoxic T lymphocyte
CTLA-4 Cytotoxic T lymphocyte antigen-4
DC Dendritic cell
ECM extracellular matrix
EMT Epithelial to mesenchymal transition
Fc Fragment crystallizable
FDA Food and drug administration
Fox-P3 forkhead box P3
IFN Interferon
IL interleukin
IMiDs immunomodulatory drugs
ITGB3 integrin β3
MDSC Myeloid-derived suppressor cell
MHC Major histocompatibility complex
MM Multiple myeloma
MMP Matrix metalloproteinase
Mo-MDSC monocytic MDSC
mRNA messenger ribonucleic acid
NK Natural Killer
NKG2D Natural Killer Group 2D
OS Overall Survival
OTUD1 OTU domain-containing protein 1
PBMC Peripheral blood mononuclear cell
PD-1 Programmed death 1
PD-L1 programmed death ligand 1
PFS Progression free survival
PSMB5 Proteasome subunit beta type-5
ROS Reactive oxygenated species
RRMM Relapsed and/or refractory MM
SASP Senescence-associated secretory phenotype
scFv Single chain variable fragments
SMAD Drosophila mothers against decapentaplegic
TAMs Tumor-associated macrophages
TCR T cell receptor
Teff effector T cells
TERT Telomerase reverse transcriptase
TGF-β Transforming Growth Factor-beta
TGF-βR Transforming Growth Factor-beta receptor
Th T-helper
TNBC triple negative breast cancer
Treg Regulatory T cell
TME Tumor microenvironment
VEGF Vascular endothelial growth factor
ZEB Zinc finger E-box-binding homeobox
Keywords: transforming growth factor-β, multiple myeloma, T-cell, drug resistance, CAR (chimeric antigen receptor) T-cells, bispecific T-cell engager (BiTE) therapy
Citation: Rana PS, Soler DC, Kort J and Driscoll JJ (2022) Targeting TGF-β signaling in the multiple myeloma microenvironment: Steering CARs and T cells in the right direction. Front. Cell Dev. Biol. 10:1059715. doi: 10.3389/fcell.2022.1059715
Received: 01 October 2022; Accepted: 29 November 2022;
Published: 12 December 2022.
Edited by:
Javier Vaquero, Institut d'Investigacio Biomedica de Bellvitge (IDIBELL), SpainReviewed by:
Stefanie Maurer, Icahn School of Medicine at Mount Sinai, United StatesCopyright © 2022 Rana, Soler, Kort and Driscoll. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: James J. Driscoll, SmFtZXMuZHJpc2NvbGxAVUhob3NwaXRhbHMub3Jn
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.