- 1Department of Oncology, Weifang Medical University, Weifang, China
- 2BGI Genomics, BGI-Shenzhen, Shenzhen, China
Epitranscriptomics has emerged as another level of epigenetic regulation similar to DNA and histone modifications. N6-methyladenosine (m6A) is one of the most prevalent and abundant posttranscriptional modifications, widely distributed in many biological species. The level of N6-methyladenosine RNA methylation is dynamically and reversibly regulated by distinct effectors including methyltransferases, demethylases, histone modification and metabolites. In addition, N6-methyladenosine RNA methylation is involved in multiple RNA metabolism pathways, such as splicing, localization, translation efficiency, stability and degradation, ultimately affecting various pathological processes, especially the oncogenic and tumor-suppressing activities. Recent studies also reveal that N6-methyladenosine modification exerts the function in immune cells and tumor immunity. In this review, we mainly focus on the regulatory mechanisms of N6-methyladenosine RNA methylation, the techniques for detecting N6-methyladenosine methylation, the role of N6-methyladenosine modification in cancer and other diseases, and the potential clinical applications.
1 Introduction
With the development of epigenetics, epitranscriptomics has emerged as another level of epigenetic regulation and has recently become a research hotspot. The epitranscriptome refers to the relevant functional changes of the transcriptome without any alteration of the RNA sequence. Conceptually, the epitranscriptome covers all the chemical modifications of RNA dynamically regulated by the removal and addition of various chemical groups in cells (Saletore et al., 2012). To date, over 170 RNA chemical modifications have been identified, including N6-methyladenosine (m6A), N1-methyladenosine (m1A), 5-hydroxymethylcytosine (hm5C), 5-methylcytidine (m5C), ribose 2′-O-methylation (Nm), 1-methylguanine (m1G), 6-methylguanine (m6G), 7-methylguanine (m7G), N4-acetylcytidine (ac4C) and pseudouridine (w) (Wiener and Schwartz, 2021), but most of their functions are largely unknown. Among them, 72 variants of methyl group modifications are conjugated at distinct positions in RNA bases. Since the first discovery of m6A RNA methylation in 1974, it has been identified as one of the most prevalent and abundant posttranscriptional modifications, widely distributed in many biological species, such as mammals (Desrosiers et al., 1974; Liu et al., 2022), plants (Yue et al., 2019; Yu Q. et al., 2021), zebrafish (Zhao et al., 2017), insects (Yang et al., 2021), yeast (Yadav and Rajasekharan, 2018), bacteria (Deng et al., 2015) and viruses (Bayoumi et al., 2020), accounting for approximately 50% of total methylated ribonucleotides in total RNA content (Wei et al., 1975). It is estimated that more than 7,000 mRNAs with m6A modification are distributed in mammalian cells, with a frequency of 0.1–0.6% of adenosines (Ke et al., 2015). In addition, m6A deposition also exists in other types of RNA, including rRNA, tRNA, small nuclear RNA (snRNA), small nucleolar RNA (snoRNA), long noncoding RNA (lncRNA), microRNA (miRNA), and circular RNA (circRNA) (Pendleton et al., 2017; van Tran et al., 2019; Dai et al., 2020).
In 2012, several decades after the first discovery of m6A RNA methylation, utilizing m6A -specific antibodies, two groups independently conducted fragmented RNA immunoprecipitation and subsequent high-throughput RNA deep sequencing (termed “MeRIP-seq” or “m6A-seq”) to map m6A throughout the transcriptome in humans and mice (Dominissini et al., 2012; Meyer et al., 2012). The results first revealed that m6A modification was widely distributed in mRNA, additionally, m6A modification mainly occurred in the common motif RRACH (R = G or A, H = A, C or U), but only 1–5% of these sites were methylated in cellular RNA. Notably, most m6A peaks were evolutionarily conserved between the human and mouse transcriptomes. More surprisingly, m6A modification on the RRACH motif was preferentially enriched in 3′-untranslated regions (3′-UTRs) and near stop codons of coding sequences (CDS) (Meyer et al., 2012), indicating that the RRACH motif is not sufficient for the determination of m6A modification. The m6A levels vary in distinct cell contexts and are involved in multiple RNA metabolism pathways, such as splicing, localization, translation efficiency, stability and degradation (Kasowitz et al., 2018; Liu et al., 2020; He and He, 2021), ultimately affecting various physiological and pathological processes.
m6A RNA methylation is dynamic and reversible and is also tightly regulated by three types of proteins, methyltransferases (“writers”), demethylases (“erasers”) and m6A binding proteins (“readers”). The m6A methylase complex was first purified in the 1990s (Bokar et al., 1994). METTL3 is identified as a predominant component and contains a catalytically active subunit. Another methyltransferase, METTL14, is essential for structural stability to facilitate the catalysis of m6A methylation. The larger methyltransferase holocomplex is composed of WTAP, HAKAI, RBM15, RBM15B, VIRMA and ZC3H13 and is approximately 1,000 kDa in size (Oerum et al., 2021). FTO was the first m6A demethylase identified in 2011, followed by ALKBH5 (Niu et al., 2013). These two enzymes can remove m6A methylation from RNA, posing a novel research field of regulation for epitranscriptomics. The functions of m6A RNA methylation are mediated by different m6A “readers” that selectively recognize m6A in a direct or indirect manner and conduct distinct functions (Shi et al., 2019). The m6A binding proteins include the YTH family, the heterogeneous nuclear ribonuclease (HNRNP) family and FMRP (Figure 1).
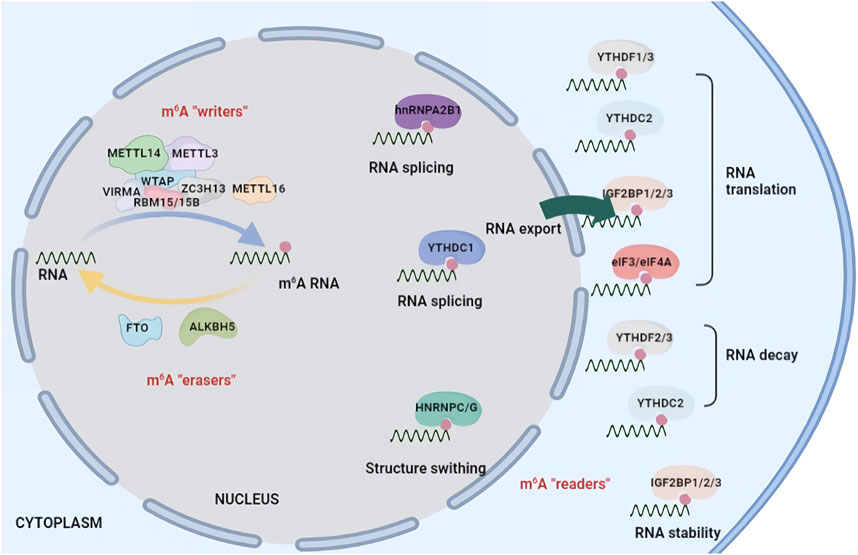
FIGURE 1. Chemical basis and molecular functions of m6A machinery. m6A methylation is catalysed by a multicomponent m6A methyltransferase complex (“m6A writers”) that is composed of two predominant proteins, METTL3 and METTL14, as well as their cofactors WTAP, HAKAI, RBM15, RBM15B, VIRMA and ZC3H13. m6A methylation can be removed by demethylases (“m6A erasers”) including FTO and ALKBH5. m6A modification affects RNA fate by recruiting m6A-binding proteins (“m6A readers”) such as YTHDF1/2/3, YTHDC1/2, IGF2BP1/2/3 and HNRNPC/A2B1. m6A methylation is involved in RNA splicing, localization, translation efficiency, stability and degradation.
In this review, we will address the regulatory mechanisms of m6A RNA methylation, the techniques for detecting m6A methylation, the role of m6A modification in cancer as well as the potential clinical applications.
2 Regulation of N6-methyladenosine RNA modification
m6A RNA methylation is functionally important and tightly modulated by several molecular mechanisms in eukaryotes. As described above, catalytic enzymes, methyltransferases and demethylases dynamically and reversibly direct the addition and removal of m6A RNA methylation, and are termed “m6A writers” and “m6A erasers”, respectively. Some binding proteins (“m6A readers”) recognize and function by decoding m6A methylation as well as recruiting downstream functional protein complexes to mediate biological activities. Histone modification also guides m6A deposition in stop codons of CDSs and 3′-UTRs. The dynamics of m6A regulation can be achieved by some transcription factors that recruit the m6A methyltransferase complex to specific RNA loci in distinct cellular contexts. In addition, nutrition and metabolites can reverse and modulate m6A methylation patterns (Figure 2).
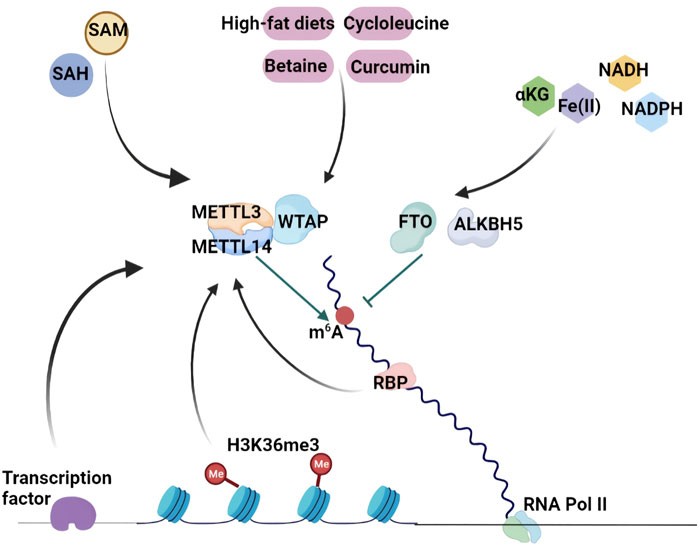
FIGURE 2. The biogenesis and regulatory mechanisms of RNA m6A Methylation. Methyltransferases and demethylases dynamically and reversibly direct the addition and removal of m6A RNA methylation. H3K36me3 guides m6A deposition in stop codons of CDSs and 3′-UTRs. The dynamics of m6A regulation can be achieved by some transcription factors that recruit the m6A methyltransferase complex to specific RNA loci in distinct cellular contexts. In addition, m6A methylation patterns are modulated by nutrition and metabolites including SAM, SAH, cycloleucine, betaine, curcumin, high-fat diets, αKG, NADH and NADPH.
2.1 RNA N6-methyladenosine machinery
m6A methylation is catalysed by a multicomponent m6A methyltransferase complex that is composed of two predominant proteins, methyltransferase-like 3 (METTL3) and methyltransferase-like 14 (METTL14), and their cofactors WTAP, HAKAI, RBM15, RBM15B, VIRMA and ZC3H13(Knuckles et al., 2018; Yue et al., 2018; Bawankar et al., 2021). Although both METTL3 and METTL14 have methyltransferase domains, only METTL3 contains a catalytically active subunit, which requires S-adenosylmethionine (SAM) as a substrate to mediate catalytic activity. The SAM binding pocket is distributed on one side of the central ß-sheet and is enclosed by the catalytic site loop (Wang et al., 2016). METTL14 is associated with the stabilization of the conformation between METTL3 and the RNA substrate (Zhou H. et al., 2021). The METTL3-METTL14 complex is formed in the cytoplasm and is located in the nucleus, and it induces m6A methylation (Scholler et al., 2018). Both in vitro methylation assays and CLIP combined with photoactivatable ribonucleoside-enhanced crosslinking (PAR-CLIP) suggested that the METTL3-METTL14 complex efficiently catalyses m6A methylation on the GGACU or GGAC motifs of RNAs, consistent with the RRACH motif of a previous study (Liu et al., 2014). The depletion of Mettl3 and/or Mettl14 could greatly reduce the peak numbers and the enrichment of m6A in the global transcriptome (Vu et al., 2017; Weng et al., 2018). Except for these two modulators, other m6A writers lack methyltransferase activity. WTAP guides METTL3 and METTL14 into nuclear speckles to efficiently methylate target RNAs (Ping et al., 2014). RBM15 and RBM15B have been confirmed to interact with WTAP by coimmunoprecipitation and bind to specific RNA regions that are adjacent to the DRACH sequence, suggesting that RBM15 and RBM15B can recruit the METTL3-WTAP complex and direct these methyltransferases to DRACH consensus sequence sites for m6A modification (Patil et al., 2016; Shi et al., 2019). RBM15 and RBM15B targets induce X-chromosome inactivation and gene silencing by binding to lncRNA XIST, and VIRMA prefers to mediate alternative polyadenylation and mRNA methylation near the 3′-UTRs and stop codon regions (Yue et al., 2018; Zhu et al., 2021). ZC3H13 complexes with WTAP or other cofactors to regulate nuclear m6A RNA methylation (Wen et al., 2018). Recently, METTL16, as an independent RNA methyltransferase, was shown to catalyse the m6A methylation of U6 spliceosomal RNA (snRNA) and U6-like hairpins of Mat2a mRNA (Shima et al., 2017). ZCCHC4, a new m6A methyltransferase, was found to specifically recognize the AAC motif associated with rRNA methylation (Ma et al., 2019).
After deposition, m6A methylation is reversible and can be removed by demethylases (“m6A erasers”). FTO belongs to the nonheme Fe(II)- and α-KG-dependent dioxygenase AlkB family. FTO was the first enzyme reported to modulate m6A demethylation in 2011. In addition, m6Am RNA, m1A RNA, m3T single-stranded DNA and m3U single-stranded RNA modifications can be demethylated by FTO (Wei et al., 2018). Mauer et al. found that the catalytic activity of FTO towards m6Am was approximately 10 times greater than that towards m6A (Mauer et al., 2017). It has been reported that snRNA and snoRNA are targets of FTO (Mauer et al., 2019). Another m6A demethylase, ALKBH5, is a member of the ALKB family and seems to be specific for m6A RNA methylation (Yu F. et al., 2021).
m6A modification affects RNA fate by recruiting m6A-binding proteins (m6A ‘readers’) such as YTH domain-containing proteins, insulin-like growth factor 2 mRNA-binding proteins IGF2BP1-3 and the heterogeneous nuclear ribonuclease (HNRNP) family (Zhao et al., 2020). In mammals, YTH domain-containing proteins contain five members: YTHDC1, YTHDC2, YTHDF1, YTHDF2, and YTHDF3. YTHDC1 play roles in alternative splicing events, nuclear export of RNAs into the cytoplasm and mRNA decay (Xiao et al., 2016). YTHDC2 promotes target mRNA translation (Mao et al., 2019). YTHDF1 interacts with the translation initiation factors eIF3 and eIF4A3 to enhance the translation efficiency of m6A-modified mRNAs (Liu et al., 2020; Cai et al., 2022). YTHDF2 and YTHDF3 are involved in the degradation of target mRNAs associated with p-bodies, and the depletion of Ythdf2 and Ythdf3 causes a considerable increase in m6A mRNA abundance in cells. hnRNPA2B1, hnRNPC and hnRNPG are related to mRNA splicing (Bi et al., 2019). In contrast, IGF2BP1/2/3, FMRP, and PRRC2A are essential for the stabilization of m6A-modified transcripts in a m6A-dependent manner (Huang et al., 2018; Edens et al., 2019; Wu et al., 2019).
2.2 Histone modification guides N6-methyladenosine deposition
Histone modification has an effect on m6A deposition. H3 lysine 36 trimethylation (H3K36me3) is a classical transcription activator that shows a similar m6A distribution. H3K36me3 chromatin immunoprecipitation (ChIP)-seq analysis indicated that approximately 70% of H3K36me3 sites overlapped with m6A peaks, suggesting a close connection between H3K36me3 and m6A modification (Huang et al., 2019) (Figure 2). SETD2 and KDM4A are H3K36me3 methyltransferase and H3K36me3 demethylase, respectively (Mar et al., 2017). More surprisingly, changes in H3K36me3 levels by utilizing dCas9-SETD2 or dCas9-KDM4A can significantly alter m6A abundance in human and mouse transcriptomes, revealing that H3K36me3 is a modulator of m6A deposition. Of note, most H3K36me3-dependent m6A sites are targeted by METTL3, METTL14 and WTAP, demonstrating the association between H3K36me3 and m6A modification. In addition, METTL14 can directly bind to H3K36me3, which leads to the recruitment of other m6A methyltransferases to activate RNA Pol II and controls m6A methylation on mRNA (Huang et al., 2019; Zhou X. L. et al., 2021). Therefore, the relationship between H3K36me3 and m6A provides a new way to enrich multiple aspects of gene expression regulation.
2.3 Transcription factors affect N6-methyladenosine deposition
Transcription factors also recruit m6A methyltransferases to regulate RNA modification in specific cell contexts. Zinc-finger protein 217 (ZFP217) is a transcriptional activator of some key pluripotency genes that are essential for maintaining self-renewal in mESCs. METTL3 can be bound and sequestered by ZFP217, preventing the formation of the m6A methyltransferase complex and m6A methylation on ZFP217 target transcripts (Aguilo et al., 2015). In contrast, the transcription factors SMAD2 and SMAD3 preferentially recruit the METTL3-METTL14-WTAP methyltransferase complex to their target transcripts and increase the m6A modification of target transcripts (Bertero et al., 2018). Different functions of m6A deposition on the target transcripts determine the distinct roles of ZFP217 and SMAD2/3 in ESCs. Another study reported that METTL3 could interact with the CAATT-box binding protein CEBPZ on target transcripts and mediate m6A modification to promote the translation of target mRNAs that maintain the leukaemic state in acute myeloid leukaemia (AML) cells (Barbieri et al., 2017). Frequently, these transcription factors manipulate m6A deposition on a subset of target transcripts in specific cellular contexts to implement dynamic regulation of gene expression (Figure 2).
2.4 Nutritional metabolism and metabolites regulate N6-methyladenosine deposition
Evidence has shown that nutritional challenge and metabolites play crucial roles in the manipulation of m6A deposition. Cycloleucine is a competitive inhibitor of methionine adenosyltransferase that decreases m6A RNA methylation levels by reducing SAM concentrations (Kang et al., 2018). Betaine, as a methyl donor for SAM synthesis, prompts m6A methylation by suppressing FTO expression in the adipose tissues of high-fat diet-fed mice, which increases the expression of the mitochondrial protein PGC-1α to improve metabolic disorder (Zhou et al., 2015). It has been reported that curcumin enhances m6A modification by decreasing ALKBH5 and increasing METTL3 and METTL14 expression in the livers of piglets (Ding et al., 2016). Undoubtedly, high-fat diets affect m6A modification in various models and tissues (Tung et al., 2010; Wu et al., 2020) (Figure 2).
SAM, a common methyl donor, is involved in most cellular methylation processes. The change in cellular SAM concentration affects DNA and histone methylation as well as RNA methylation (Duncan et al., 2013). METTL3 requires SAM as a substrate to mediate catalytic activity and m6A writing. Interestingly, the SAM binding affinity of METTL3 is regulated by substrate RNA availability. S-adenosyl homocysteine (SAH) is the metabolite of SAM during the methylation reaction that can strongly inhibit METTL3 methyltransferase activity (Li F. et al., 2016; Selberg et al., 2019). Demethylases, FTO and ALKBH5 are 2-oxoglutarate (αKG)- and Fe(II) dependent. The FTO and ALKBH5 mutants of the αKG-Fe(II) oxygenase domain lost the catalytic activities of m6A demethylation (Feng et al., 2014; Zhang X. et al., 2019). Recently, NADH and NADPH were identified as the direct binding partners of FTO by using a florescence quenching assay. Both NADH and NADPH could enhance FTO demethylase activity, indicating that reducing NADPH and NADH may attenuate demethylation reactions. Conversely, the induction of NADPH by glucose injection or a high-fat diet suppressed m6A modification (Figure 2). In contrast, the depletion of G6P dehydrogenase (G6PD) or NAD kinase (NADK) enhanced cellular m6A abundance, which was reversed by NADPH supplementation (Wang L. et al., 2020).
3 Approaches for detecting N6-methyladenosine RNA methylation
Several techniques have been developed for detecting m6A RNA methylation (Table 1). Although immuno-northern blot and m6A dot blot facilitate easier and faster observation of global m6A levels, the disadvantages are obvious with lower sensitivity and are semiquantitative accuracy (Nagarajan et al., 2019). High-performance liquid chromatography‒mass spectrometry (HPLC‒MS/MS) is used for quantifying m6A levels with high sensitivity; however, this approach cannot provide details about RNA sequence and localization information (Thuring et al., 2017). Site-specific cleavage and radioactive labelling followed by ligation-assisted extraction and thin-layer chromatography (SCARLET) is suitable for stoichiometric quantification, but it is very tedious and is only used to validate the known m6A changes at a given site (Liu and Pan, 2016). Above all, these techniques are not appropriate for widespread identification and localization of modified sites; subsequently, high-throughput sequencing methods have emerged and have been rapidly developed. m6A-specific antibody-based high-throughput sequencing strategies are widely used for the identification of m6A, which include m6A-Seq, MeRIPSeq, PA-m6A-Seq, m6A-CLIP and m6A individual-nucleotide resolution cross-linking and immunoprecipitation (miCLIP) (Dominissini et al., 2012; Meyer et al., 2012; Chen et al., 2015; Ke et al., 2015; Linder et al., 2015). MeRIP-seq was first identified in 2012, allowing for m6A analysis with 100- to 200-nucleotide resolution. In terms of MeRIP-seq, mRNA was fragmented into 100-nucleotide lengths and immunoprecipitated by a m6A-specific antibody with the combination of high-throughput deep sequencing. miCLIP enables the detection of m6A residues at precise positions with single-nucleotide resolution. However, the number of identified m6A peaks is limited due to the low cross-linking efficiency of this method. m6A-specific antibody-based sequencing approaches have some obvious drawbacks. m6A antibodies are not strikingly specific for m6A, may also bind to m6Am sites and other non-m6A-specific sequences. Distinct commercial m6A antibodies show differences in affinity for m6A (Haussmann et al., 2016). Endoribonuclease-based techniques include antibody-free m6A sequencing methods such as m6A-REF-Seq and MAZTER-Seq and rely on the endoribonuclease activity of MazF (Zhang Z. et al., 2019; Garcia-Campos et al., 2019). Therefore, the motif preference of endoribonuclease determines the limitation, and these methods detect a portion of the m6A sites. Recently, chemical labelling strategies have been reported, including m6A-SEAL, m6A-label-seq and m6A-SAC-seq (Wang Y. et al., 2020; Shu et al., 2020; Hu et al., 2022). Nevertheless, improvements in the chemical labelling efficiency are needed. A specific method for m6A detection in the global transcriptome should be validated by other techniques to obtain a more accurate m6A landscape.
4 Role of N6-methyladenosine RNA methylation in human diseases
m6A RNA methylation is involved in multiple RNA metabolism pathways, such as splicing, localization, translation efficiency, stability and degradation, ultimately affecting various physiological and pathological processes. For instance, m6A methylation has been demonstrated to regulate the haematopoietic system, the central nervous system, immunity stemness, mammalian spermatogenesis and brain development. The dysregulation of m6A methylation is associated with various diseases, especially cancer (Figure 3).
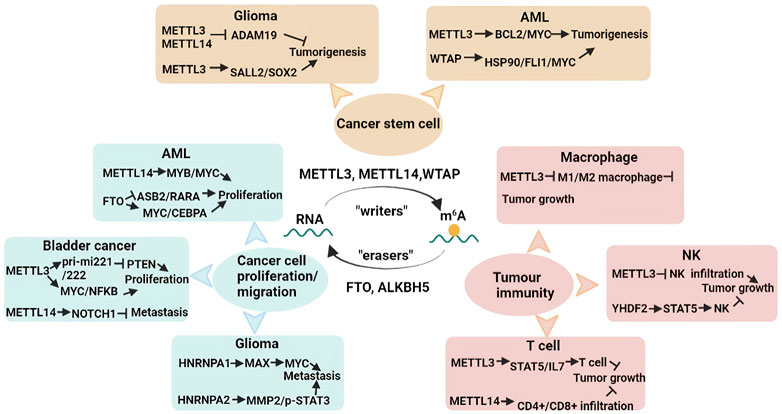
FIGURE 3. Diverse functions of m6A RNA methylation in cancers. m6A RNA methylation plays a crucial role in tumorigenesis and cancer progression. m6A RNA methylation is reversible and is also tightly regulated by “writers” and “readers”. m6A RNA methylation contributes to mediating CSC pluripotency, cancer proliferation, tumour metastasis, and tumor immunity. More details are shown in a graphical form.
4.1 N6-methyladenosine and cancer
4.1.1 N6-methyladenosine in cancer stem cells
Cancer stem cells (CSCs) are a type of cells possessing a stem cell-like capacity to self-renew, differentiate and survive and give rise to many types of cancers (Nassar and Blanpain, 2016). CSCs lead to the tolerance of standard therapeutics and CSCs, tumour recurrence, and distant metastasis (Walcher et al., 2020). Cui et al. found that m6A methylation played a role in the tumorigenesis of glioma stem cells (GSCs). Mettl3/14 knockdown prominently promoted GSC self-renewal and tumorigenesis by decreasing m6A levels, whereas METTL3 overexpression exerted negative effects (Cui et al., 2017; Zhang et al., 2017). MeRIP-seq revealed that silencing Mettl3/14 altered m6A enrichment and that m6A RNA methylation of ADAM19 regulated GSC self-renewal. Similarly, Visvanathan et al. confirmed that METTL3 was essential for glioma cell differentiation and GSC maintenance. Furthermore, METTL3 is also involved in radiosensitivity and DNA repair through the SOX2 axis in GSCs (Visvanathan et al., 2018).
Wang et al. provided the first evidence of the interplay between m6A methylation and osteosarcoma stem cells (OSCs). Compared with non-OSCs, METTL14 and FTO were significantly reduced in OSCs. Meanwhile, MeRIP-seq and RNA-seq analyses of OSCs and non-OSCs revealed that differentially expressed genes containing differentially methylated m6A peaks were associated with the Wnt pathway and the pluripotency of stem cells (Wang et al., 2019).
Ly P Vu and others revealed that METTL3 expression was elevated in leukaemia cells compared with normal haematopoietic cells. miCLIP analysis showed that METTL3 enhanced the m6A methylation of target genes such as Bcl2, Myc and Pten in the human acute myeloid leukaemia (AML) MOLM-13 cell line, which promoted the mRNA translation of these genes, thereby retaining pluripotency properties and inhibiting cell differentiation. Notably, silencing of Mettl3 in human myeloid leukemia cell lines promoted cell differentiation and cell apoptosis (Vu et al., 2017). WTAP is associated with haematopoietic stem cell (HSC) homeostasis and haematopoietic regeneration (Wang H. et al., 2018; Li et al., 2018). Recent research conducted by Bansal et al. suggested a role for m6A modification in myeloid leukaemia. WTAP expression was increased in AML cells derived from patients with AML, while knockdown of Wtap led to the repression of cell proliferation, the activation of cell differentiation and apoptosis in a leukaemia cell line (Bansal et al., 2014) (Figure 3).
4.1.2 N6-methyladenosine in cancer cell proliferation and migration
Numerous reports have elucidated that m6A modification is involved in cancer cell proliferation and tumour metastasis in different types of cancers. Recent findings revealed that METTL14 exerted an oncogenic function by increasing the expression levels of targets such as MYB and MYC in AML, while SPI downregulated the expression level of MEETL14 (Weng et al., 2018). FTO demethylase is elevated and plays an oncogenic role in AML. It has been demonstrated that a high level of FTO prompts cell proliferation and viability, whereas it reduces cell apoptosis and global m6A methylation by repressing the expression of ASB2 and RARA (Li Z. et al., 2017). Su et al. observed that R-2-hydroxyglutarate (R-2HG) attenuated FTO activity and augmented global m6A modification in R-2HG-sensitive AML cells, which decreased the stability of Cebpa/Myc mRNA and the activities of relevant cell signalling pathways (Su et al., 2018).
In bladder cancer, METTL3 interacts with DGCR8 to facilitate pri-miR221/222 maturation, which leads to a decrease in PTEN and ultimately promotes cell proliferation (Han et al., 2019). Similarly, another study showed that METTL3 was significantly elevated in bladder cancer, and METTL3 knockdown dramatically suppressed cancer cell proliferation, cell invasion, and tumour formation through the AFF4/MYC/NF-kB axis cell signalling pathway (Cheng et al., 2019). Xie et al. revealed that METTL3 also binds to YTHDF2, which induces the degradation of target tumour suppressor mRNAs, including Klf4 and Setd7, regulating the progression of bladder cancer (Xie et al., 2020). Nevertheless, METTL14 has been confirmed to be decreased in bladder cancer, and depletion of Mettl14 accelerated cell proliferation, tumour metastasis and self-renewal by decreasing the stability of m6A-modified Notch1 transcripts (Gu et al., 2019).
Evidence indicates that the hnRNPA1 expression level is elevated by EGFRvIII, leading to increased glycolytic gene expression in gliomas. Meanwhile, hnRNPA1 promotes Max mRNA splicing and then induces the generation of Delta Max, which promotes Myc-dependent cell transformation (Babic et al., 2013). Another study showed that silencing hnRNPA2 represses cancerous cell viability, cell invasion, tumour metastasis and chemoresistance by decreasing the expression of MMP-2 and phospho-STAT3. Notably, hnRNPA2 has been regarded as an oncogenic driver in gliomas (Deng et al., 2016) (Figure 3). Numerous studies have described how m6A methylation contributes to cell proliferation, cell invasion, and tumour metastasis in other cancers, including breast cancer, ovarian cancer, cervical cancer, prostate cancer, lung cancer, hepatocellular carcinoma, gastric carcinoma, pancreatic cancer and colorectal cancer (Hu et al., 2019; Yang et al., 2019; Ma et al., 2020; Yang et al., 2020; Guo et al., 2021; Zhang and Xu, Forthcoming 2022).
4.1.3 N6-methyladenosine in tumour immunity
Recent studies have revealed that m6A RNA methylation induces the activation and infiltration of various immune cells into the tumour microenvironment (TME), influencing the efficacy of cancer immunotherapy. Macrophages are closely associated with tumour initiation and progression. Yin et al. elucidated that METTL3 in macrophages regulates tumour development. Silencing of Mettl3 in macrophages facilitated tumour growth and lung metastasis. The TME was reshaped by inducing regulatory T (Treg) cells into tumour sites and promoting the infiltration of M1-and M2-like tumour-associated macrophages (Yin et al., 2021). Mettl14 knockdown in macrophages suppressed the antitumour activity of CD8+ T cells and improved tumour growth (Dong et al., 2021).
Natural killer (NK) cells play an important role in cancer immune surveillance and can directly recognize and kill cancer cells. YTHDF2 was critical for modulating NK-cell maturation, NK-cell homeostasis, IL-15-mediated survival, and antitumor activity due to the regulation of downstream target genes such as Stat5, Eomes and Tardbp (Ma et al., 2021). Song et al. found that METTL3 expression was decreased in tumour-infiltrating NK cells of cancer patients. In mice, they observed that depletion of Mettl3 enhanced NK-cell responsiveness to IL-15 and promoted tumour progression and metastasis by targeting SHP-2 (Song et al., 2021).
Silencing of Mettl3 in CD4+ T cells destroyed T-cell differentiation and homeostasis by repressing the activation of IL-7-mediated STAT5/suppressor of cytokine signalling (Li H. B. et al., 2017). Yao et al. revealed that conditional depletion of Mettl3 in CD4+ T cells inhibited T follicular helper differentiation and maturation, thereby preventing the antibody response of B cells by promoting the degradation of m6A-modified Tcf7 mRNA (Yao et al., 2021). In breast cancer, the expression levels of METTL14 have a positive correlation with the infiltration of CD4+ T cells, CD8+ T cells, dendritic cells, macrophages and neutrophils, but they negatively correlated with Treg cells in breast cancer (Gong et al., 2020) (Figure 3).
4.2 N6-methyladenosine and other human diseases
Emerging evidence has demonstrated that m6A RNA methylation is closely related to other human diseases, including cardiovascular disease, metabolic syndrome, psychiatric disorders and autoinflammatory disorders. Dorn et al. (Dorn et al., 2019) showed that the global m6A level of cardiomyocytes was significantly elevated in response to hypertrophic stimulation and that METTL3 played a vital role in cardiomyocyte hypertrophy. Notably, upregulated m6A modification resulted in compensated cardiac hypertrophy, whereas downregulated m6A levels led to eccentric cardiomyocyte remodelling and dysfunction. Overexpression of Mettl3 increased the expression levels of mitogen-activated protein (MAP)3K6, MAP4K5, and MAPK14 in cardiomyocytes, which was positively correlated with cardiomyocyte size, revealing that METTL3 is sufficient to drive cardiomyocyte hypertrophy. However, no histopathologic changes were observed in Mettl3-overexpressing mice.
Zhou et al. identified that YTHDC2 was significantly repressed in nonalcoholic fatty liver disease (NAFLD) patients, and Ythdc2-depleted hepatocytes led to the accumulation of excessive triglycerides (TGs) by reducing the expression levels of lipogenic genes, including fatty acid synthase, sterol regulatory element-binding protein 1c, and acetyl-CoA carboxylase 1 (Zhou B. et al., 2021). Furthermore, m6A sequencing was performed in human type 2 diabetes islets, and sequencing analysis showed that multiple hypomethylated transcripts were associated with insulin secretion, the insulin/IGF1 signalling pathway and cell cycle progression. Mettl14 knockout in mouse ß-cells caused a reduction in global m6A levels, giving rise to a similar islet phenotype in human T2D (De Jesus et al., 2019).
In Alzheimer’s disease mouse models, the global m6A level was increased in the hippocampus and the cortex compared to C57BL/6 mice, and the interaction of FTO and APOE contributed to the increase in Alzheimer’s disease risk. The overall m6A level was elevated in the cortex and the hippocampus of APP/PS1 (Alzheimer’s disease) mice compared to C57BL/6 control mice, and FTO was found to interact with APOE, which was associated with Alzheimer’s disease risk in a prospective cohort study (Keller et al., 2011; Han et al., 2020). Huang et al. found that ALKBH5/FAAH enhanced the expression of circSTAG1, which attenuated astrocyte dysfunction and depressive-like behaviours in vitro and in vivo (Huang et al., 2020).
m6A methylation also has a contributory effect on autoinflammatory disorders. Luo et al. identified that the expression levels of METTL14, ALKBH5 and YTHDF2 were downregulated in peripheral blood mononuclear cells of systemic lupus erythematosus (SLE) patients (Luo et al., 2020). In addition, multivariate logistic regression analysis showed that repression of ALKBH5 and YTHDF2 was considered a risk factor for SLE. However, direct mechanistic data should be provided for the function of m6A modification in SLE progression. Another study found that Mettl14 knockdown inhibited the activation of Treg cells, which impaired the balance between Th17 and Treg cells, leading to the development of spontaneous colitis (Lu et al., 2020).
5 Potential clinical applications of N6-methyladenosine RNA modification
Due to the major role of m6A RNA modification in tumour and other disease progression, m6A-associated proteins can be developed as potential therapeutic targets for tumours and other diseases (Table 2). METTL3 attenuates the sensitivity of colon cancer cells to chemotherapy of L-OHP and CPT-11 by upregulating the expression level of CBX8 in a m6A-dependent manner (Zhang Y. et al., 2019). Meanwhile, Ythdf1-depleted colon cancer cells are more sensitive to 5-FU and L-OHP (Nishizawa et al., 2018). Another study identified that gemcitabine drove the apoptosis of pancreatic cancer cells with low METTL3 expression (Taketo et al., 2018). The combination of m6A methylation and chemotherapeutic drugs contributes to resolving drug resistance in tumours. Cas13-directed methyltransferase has been used for cancer treatment, targeting m6A of specific RNA loci (Wilson et al., 2020; Lo et al., 2022).
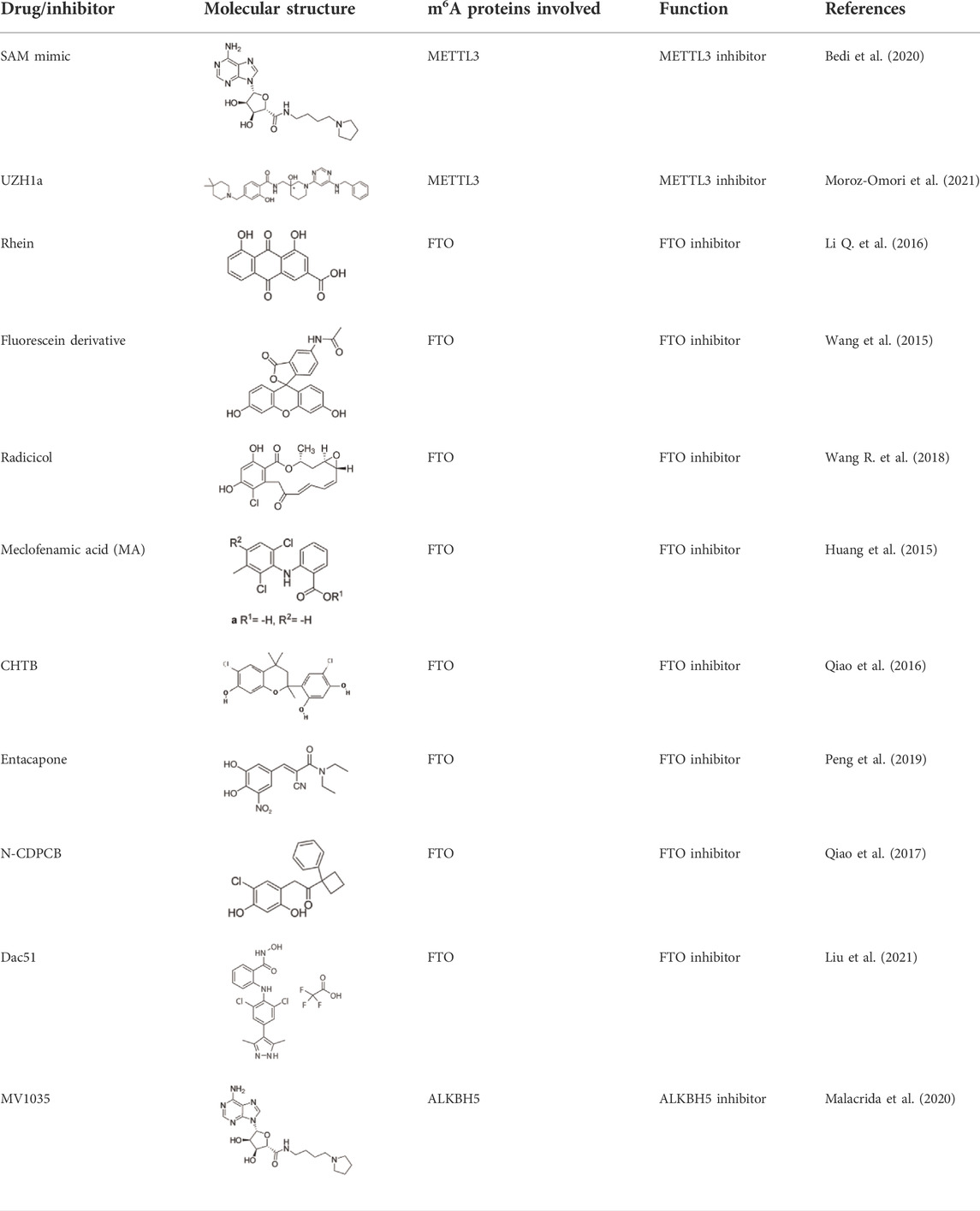
TABLE 2. Overview of the small-molecule drugs and m6A-related factor inhibitors described in the text.
Specific inhibitors based on m6A-related enzymes have been studied. Bedi et al. (Bedi et al., 2020) performed a virtual screening assay to identify potential METTL3 inhibitors from 4,000 adenosine derivatives. One compound, a SAM mimic, was found to be the first inhibitor of METTL3. However, the therapeutic value of this compound is still somewhat limited due to cell penetration issues and nonspecific targets. They also identified another METTL3 inhibitor, UZH1a, that decreased the m6A/A ratios in three different cell lines, MOLM-13, HEK293T and U2OS, but the specificity of UZH1a needs further improvement (Moroz-Omori et al., 2021).
Currently, more papers are focused on FTO inhibitors. Rhein, a competitive inhibitor of FTO, binds to the catalytic domain and blocks the catalytic activity of FTO (Li Q. et al., 2016). Fluorescein derivatives, radicicol, IOX3 and MA (a nonsteroidal anti-inflammatory drug) were subsequently identified to repress FTO expression (Huang et al., 2015; Wang et al., 2015; Wang R. et al., 2018). Similar to MA, MA2, an isomer of MA, interacts with FTO to increase the m6A level but possesses better cell penetration ability. In addition, CHTB, entacapone and N-CDPCB also repress the demethylase activity of FTO (Qiao et al., 2016; Qiao et al., 2017; Peng et al., 2019). Dac51 inhibits the activity of FTO, promotes T cell response and enhances the anti-PD-1 therapy (Liu et al., 2021). Small-molecule FTO inhibitors have been developed as potential drugs not only for tumour therapy but also for the treatment of neurological diseases, cardiovascular disease, metabolic syndrome and autoinflammatory disorders. To date, little is known about specific ALKBH5 inhibitors. However, the imidazobenzoxazin-5-thione MV1035 was found to be a potential candidate ALKBH5 inhibitor (Malacrida et al., 2020). ALK-04 as an inhibitor of ALKBH5 reduces the infiltration of myeloid-derived suppressor cells and Treg cells and suppresses tumour growth by enhancing the therapeutic effect of anti-PD-1 treatment (Li et al., 2020). (Table 2).
6 Conclusions and future prospects
Despite the initial discovery of m6A in 1974, m6A modification has received much less attention than histone and DNA epigenetic modifications. Since the establishment of MeRIP-Seq for mapping m6A deposition at a transcriptome-wide level in 2012, interest in studying m6A methylation has grown rapidly. The level of m6A is dynamically and reversibly regulated by various effectors termed “writers”, “erasers” and “readers”. The METTL3-14-WTAP methyltransferase complex is the core constituent of most m6A writers, whereas FTO and ALKBH5, as m6A demethylases, catalyse the removal of m6A. Currently, emerging evidence has shown that transcription factor and histone modification signatures together shape m6A deposition, suggesting that the contribution of transcription factor histone modification contributes to the modulation of m6A. The interplay between histone H3K36me3 and m6A modification provides a novel layer of gene expression regulation. Further crosstalk between m6A RNA modification and other epigenetic modifications should be carefully elucidated in the future. Nutritional metabolism and metabolites also influence the regulation of m6A. It is possible that tissue-specific m6A levels partially correlate with the metabolic activities of specific organs. Overall, the precise regulatory mechanisms of m6A are still in their infancy and need to be further investigated. Although high-throughput sequencing-based techniques have greatly promoted the research field of m6A methylation, none of the available approaches have simultaneously reached the achievements with single-base resolution, quantification of m6A disposition, and low input of RNA. A future method for the direct sequencing of m6A RNA will be better for illustrating its functions and dynamics in vivo.
As mentioned above, abnormal m6A methylation is closely related to various diseases, including cancer, cardiovascular disease, metabolic syndrome, psychiatric disorders and autoinflammatory disorders. Accordingly, m6A methylation is a double-edged sword for tumour: a lack of m6A modification on specific genes accelerates tumour development, while over-m6A modification of other genes also induces tumour progression. In fact, many major challenges remain in elucidating the relationships between m6A and cancer. First, whether the multiple roles proposed in cancer actually rely on m6A modification should be considered. Second, we should not ignore the notion that m6A-related effectors may mediate tumour development and the progression in a m6A-independent manner. Third, it is worth noting that we elucidate the regulatory association of noncoding RNAs and m6A methylation in tumour. Fourth, the effects of m6A regulators on tumor cells and immune cells are complicated and need to be carefully concerned. Fifth, over 170 RNA modifications have been identified, it is worth evaluating whether other RNA modifications in the same RNA transcripts affects the role of m6A methylation in human diseases. Finally, further studies should be carried out to assess the clinical value of m6A in diseases.
Many studies have demonstrated that m6A regulatory factors are suitable as therapeutic targets, and some inhibitors of m6A-related factors, especially FTO, have been discovered. Nevertheless, no clinical trials using m6A inhibitors for the treatment of cancer and other diseases have been reported yet. We also provide several reasonable strategies for driving m6A-based therapy: 1) Nanoparticles can specifically deliver m6A modification molecules to target immune cells for tumour immunotherapeutic treatment. 2) A programmable m6A gene-editing system by dCas13 or dRCas9 provides a potential tool for the treatment of diseases. 3) Chimeric antigen receptor (CAR) immune cells with lentivirus-mediated gene delivery of m6A effectors are beneficial to cancer immunotherapy. 4) It is feasible to treat cancer and other diseases by using a m6A inhibitor combined with other therapies. Further studies are urgently required for the understanding of RNA m6A modifications and clinical applications.
Author contributions
JL and JS made substantial contributions to conception and design. PL involved in drafting and revising the manuscript, YW revised the manuscript and draw the figures, YS drafted the tables.
Funding
This work was supported by the National Natural Science Foundation of China (NSFC 82104289), Weifang Science and Technology Bureau Plan Project (2021YX081), Science and Technology Innovation Plan from Weifang Medical College (041004), Yuandu Scholar Grant of Weifang City to JL and Postdoctoral Research Grant of Shenzhen to LPP (2237PT).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aguilo, F., Zhang, F., Sancho, A., Fidalgo, M., Di Cecilia, S., Vashisht, A., et al. (2015). Coordination of m(6)A mRNA methylation and gene transcription by ZFP217 regulates pluripotency and reprogramming. Cell Stem Cell 17 (6), 689–704. doi:10.1016/j.stem.2015.09.005
Babic, I., Anderson, E. S., Tanaka, K., Guo, D., Masui, K., Li, B., et al. (2013). EGFR mutation-induced alternative splicing of Max contributes to growth of glycolytic tumors in brain cancer. Cell Metab. 17 (6), 1000–1008. doi:10.1016/j.cmet.2013.04.013
Bansal, H., Yihua, Q., Iyer, S. P., Ganapathy, S., Proia, D. A., Penalva, L. O., et al. (2014). WTAP is a novel oncogenic protein in acute myeloid leukemia. Leukemia 28 (5), 1171–1174. doi:10.1038/leu.2014.16
Barbieri, I., Tzelepis, K., Pandolfini, L., Shi, J., Millan-Zambrano, G., Robson, S. C., et al. (2017). Promoter-bound METTL3 maintains myeloid leukaemia by m(6)A-dependent translation control. Nature 552 (7683), 126–131. doi:10.1038/nature24678
Bawankar, P., Lence, T., Paolantoni, C., Haussmann, I. U., Kazlauskiene, M., Jacob, D., et al. (2021). Hakai is required for stabilization of core components of the m(6)A mRNA methylation machinery. Nat. Commun. 12 (1), 3778. doi:10.1038/s41467-021-23892-5
Bayoumi, M., Rohaim, M. A., and Munir, M. (2020). Structural and virus regulatory insights into avian N6-methyladenosine (m6A) machinery. Front. Cell Dev. Biol. 8, 543. doi:10.3389/fcell.2020.00543
Bedi, R. K., Huang, D., Eberle, S. A., Wiedmer, L., Sledz, P., and Caflisch, A. (2020). Small-molecule inhibitors of METTL3, the major human epitranscriptomic writer. ChemMedChem 15 (9), 744–748. doi:10.1002/cmdc.202000011
Bertero, A., Brown, S., Madrigal, P., Osnato, A., Ortmann, D., Yiangou, L., et al. (2018). The SMAD2/3 interactome reveals that TGFβ controls m6A mRNA methylation in pluripotency. Nature 555 (7695), 256–259. doi:10.1038/nature25784
Bi, Z., Liu, Y., Zhao, Y., Yao, Y., Wu, R., Liu, Q., et al. (2019). A dynamic reversible RNA N(6) -methyladenosine modification: Current status and perspectives. J. Cell. Physiol. 234 (6), 7948–7956. doi:10.1002/jcp.28014
Bokar, J. A., Rath-Shambaugh, M. E., Ludwiczak, R., Narayan, P., and Rottman, F. (1994). Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei. Internal mRNA methylation requires a multisubunit complex. J. Biol. Chem. 269 (26), 17697–17704. doi:10.1016/s0021-9258(17)32497-3
Cai, J., Chen, Z., Zhang, Y., Wang, J., Zhang, Z., Wu, J., et al. (2022). CircRHBDD1 augments metabolic rewiring and restricts immunotherapy efficacy via m(6)A modification in hepatocellular carcinoma. Mol. Ther. Oncolytics 24, 755–771. doi:10.1016/j.omto.2022.02.021
Chen, K., Luo, G. Z., and He, C. (2015). High-resolution mapping of N⁶-Methyladenosine in transcriptome and genome using a photo-crosslinking-assisted strategy. Methods Enzymol. 560, 161–185. doi:10.1016/bs.mie.2015.03.012
Cheng, M., Sheng, L., Gao, Q., Xiong, Q., Zhang, H., Wu, M., et al. (2019). The m6A methyltransferase METTL3 promotes bladder cancer progression via AFF4/NF-κB/MYC signaling network. Oncogene 38 (19), 3667–3680. doi:10.1038/s41388-019-0683-z
Cui, Q., Shi, H., Ye, P., Li, L., Qu, Q., Sun, G., et al. (2017). m(6)A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 18 (11), 2622–2634. doi:10.1016/j.celrep.2017.02.059
Dai, F., Wu, Y., Lu, Y., An, C., Zheng, X., Dai, L., et al. (2020). Crosstalk between RNA m(6)A modification and non-coding RNA contributes to cancer growth and progression. Mol. Ther. Nucleic Acids 22, 62–71. doi:10.1016/j.omtn.2020.08.004
De Jesus, D. F., Zhang, Z., Kahraman, S., Brown, N. K., Chen, M., Hu, J., et al. (2019). m(6)A mRNA methylation regulates human beta-cell Biology in physiological states and in type 2 diabetes. Nat. Metab. 1 (8), 765–774. doi:10.1038/s42255-019-0089-9
Deng, J., Chen, S., Wang, F., Zhao, H., Xie, Z., Xu, Z., et al. (2016). Effects of hnRNP A2/B1 knockdown on inhibition of glioblastoma cell invasion, growth and survival. Mol. Neurobiol. 53 (2), 1132–1144. doi:10.1007/s12035-014-9080-3
Deng, X., Chen, K., Luo, G. Z., Weng, X., Ji, Q., Zhou, T., et al. (2015). Widespread occurrence of N6-methyladenosine in bacterial mRNA. Nucleic Acids Res. 43 (13), 6557–6567. doi:10.1093/nar/gkv596
Desrosiers, R., Friderici, K., and Rottman, F. (1974). Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. U. S. A. 71 (10), 3971–3975. doi:10.1073/pnas.71.10.3971
Ding, L., Li, J., Song, B., Xiao, X., Zhang, B., Qi, M., et al. (2016). Curcumin rescues high fat diet-induced obesity and insulin sensitivity in mice through regulating SREBP pathway. Toxicol. Appl. Pharmacol. 304, 99–109. doi:10.1016/j.taap.2016.05.011
Dominissini, D., Moshitch-Moshkovitz, S., Schwartz, S., Salmon-Divon, M., Ungar, L., Osenberg, S., et al. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485 (7397), 201–206. doi:10.1038/nature11112
Dong, L., Chen, C., Zhang, Y., Guo, P., Wang, Z., Li, J., et al. (2021). The loss of RNA N(6)-adenosine methyltransferase Mettl14 in tumor-associated macrophages promotes CD8(+) T cell dysfunction and tumor growth. Cancer Cell 39 (7), 945–957. e910. doi:10.1016/j.ccell.2021.04.016
Dorn, L. E., Lasman, L., Chen, J., Xu, X., Hund, T. J., Medvedovic, M., et al. (2019). The N(6)-methyladenosine mRNA methylase METTL3 controls cardiac homeostasis and hypertrophy. Circulation 139 (4), 533–545. doi:10.1161/CIRCULATIONAHA.118.036146
Duncan, T. M., Reed, M. C., and Nijhout, H. F. (2013). The relationship between intracellular and plasma levels of folate and metabolites in the methionine cycle: A model. Mol. Nutr. Food Res. 57 (4), 628–636. doi:10.1002/mnfr.201200125
Edens, B. M., Vissers, C., Su, J., Arumugam, S., Xu, Z., Shi, H., et al. (2019). FMRP modulates neural differentiation through m(6)a-dependent mRNA nuclear export. Cell Rep. 28 (4), 845–854. doi:10.1016/j.celrep.2019.06.072
Feng, C., Liu, Y., Wang, G., Deng, Z., Zhang, Q., Wu, W., et al. (2014). Crystal structures of the human RNA demethylase Alkbh5 reveal basis for substrate recognition. J. Biol. Chem. 289 (17), 11571–11583. doi:10.1074/jbc.M113.546168
Garcia-Campos, M. A., Edelheit, S., Toth, U., Safra, M., Shachar, R., Viukov, S., et al. (2019). Deciphering the "m(6)A code" via antibody-independent quantitative profiling. Cell 178 (3), 731–747. e716. doi:10.1016/j.cell.2019.06.013
Gong, P. J., Shao, Y. C., Yang, Y., Song, W. J., He, X., Zeng, Y. F., et al. (2020). Analysis of N6-methyladenosine methyltransferase reveals METTL14 and ZC3H13 as tumor suppressor genes in breast cancer. Front. Oncol. 10, 578963. doi:10.3389/fonc.2020.578963
Gu, C., Wang, Z., Zhou, N., Li, G., Kou, Y., Luo, Y., et al. (2019). Mettl14 inhibits bladder TIC self-renewal and bladder tumorigenesis through N(6)-methyladenosine of Notch1. Mol. Cancer 18 (1), 168. doi:10.1186/s12943-019-1084-1
Guo, J., Zheng, J., Zhang, H., and Tong, J. (2021). RNA m6A methylation regulators in ovarian cancer. Cancer Cell Int. 21 (1), 609. doi:10.1186/s12935-021-02318-8
Han, J., Wang, J. Z., Yang, X., Yu, H., Zhou, R., Lu, H. C., et al. (2019). METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol. Cancer 18 (1), 110. doi:10.1186/s12943-019-1036-9
Han, M., Liu, Z., Xu, Y., Liu, X., Wang, D., Li, F., et al. (2020). Abnormality of m6A mRNA methylation is involved in alzheimer's disease. Front. Neurosci. 14, 98. doi:10.3389/fnins.2020.00098
Haussmann, I. U., Bodi, Z., Sanchez-Moran, E., Mongan, N. P., Archer, N., Fray, R. G., et al. (2016). m(6)A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature 540 (7632), 301–304. doi:10.1038/nature20577
He, P. C., and He, C. (2021). m(6) A RNA methylation: from mechanisms to therapeutic potential. EMBO J. 40 (3), e105977. doi:10.15252/embj.2020105977
Hu, B. B., Wang, X. Y., Gu, X. Y., Zou, C., Gao, Z. J., Zhang, H., et al. (2019). N(6)-methyladenosine (m(6)A) RNA modification in gastrointestinal tract cancers: Roles, mechanisms, and applications. Mol. Cancer 18 (1), 178. doi:10.1186/s12943-019-1099-7
Hu, L., Liu, S., Peng, Y., Ge, R., Su, R., Senevirathne, C., et al. (2022). m(6)A RNA modifications are measured at single-base resolution across the mammalian transcriptome. Nat. Biotechnol. 40, 1210–1219. doi:10.1038/s41587-022-01243-z
Huang, H., Weng, H., Sun, W., Qin, X., Shi, H., Wu, H., et al. (2018). Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 20 (3), 285–295. doi:10.1038/s41556-018-0045-z
Huang, H., Weng, H., Zhou, K., Wu, T., Zhao, B. S., Sun, M., et al. (2019). Histone H3 trimethylation at lysine 36 guides m(6)A RNA modification co-transcriptionally. Nature 567 (7748), 414–419. doi:10.1038/s41586-019-1016-7
Huang, R., Zhang, Y., Bai, Y., Han, B., Ju, M., Chen, B., et al. (2020). N(6)-Methyladenosine modification of fatty acid amide hydrolase messenger RNA in circular RNA STAG1-regulated astrocyte dysfunction and depressive-like behaviors. Biol. Psychiatry 88 (5), 392–404. doi:10.1016/j.biopsych.2020.02.018
Huang, Y., Yan, J., Li, Q., Li, J., Gong, S., Zhou, H., et al. (2015). Meclofenamic acid selectively inhibits FTO demethylation of m6A over ALKBH5. Nucleic Acids Res. 43 (1), 373–384. doi:10.1093/nar/gku1276
Kang, H., Zhang, Z., Yu, L., Li, Y., Liang, M., and Zhou, L. (2018). FTO reduces mitochondria and promotes hepatic fat accumulation through RNA demethylation. J. Cell. Biochem. 119 (7), 5676–5685. doi:10.1002/jcb.26746
Kasowitz, S. D., Ma, J., Anderson, S. J., Leu, N. A., Xu, Y., Gregory, B. D., et al. (2018). Nuclear m6A reader YTHDC1 regulates alternative polyadenylation and splicing during mouse oocyte development. PLoS Genet. 14 (5), e1007412. doi:10.1371/journal.pgen.1007412
Ke, S., Alemu, E. A., Mertens, C., Gantman, E. C., Fak, J. J., Mele, A., et al. (2015). A majority of m6A residues are in the last exons, allowing the potential for 3' UTR regulation. Genes Dev. 29 (19), 2037–2053. doi:10.1101/gad.269415.115
Keller, L., Xu, W., Wang, H. X., Winblad, B., Fratiglioni, L., and Graff, C. (2011). The obesity related gene, FTO, interacts with APOE, and is associated with alzheimer's disease risk: A prospective cohort study. J. Alzheimers Dis. 23 (3), 461–469. doi:10.3233/JAD-2010-101068
Knuckles, P., Lence, T., Haussmann, I. U., Jacob, D., Kreim, N., Carl, S. H., et al. (2018). Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m(6)A machinery component Wtap/Fl(2)d. Genes Dev. 32 (5-6), 415–429. doi:10.1101/gad.309146.117
Li, F., Kennedy, S., Hajian, T., Gibson, E., Seitova, A., Xu, C., et al. (2016a). A radioactivity-based assay for screening human m6A-RNA methyltransferase, METTL3-METTL14 complex, and demethylase ALKBH5. J. Biomol. Screen. 21 (3), 290–297. doi:10.1177/1087057115623264
Li, H. B., Tong, J., Zhu, S., Batista, P. J., Duffy, E. E., Zhao, J., et al. (2017a). m(6)A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature 548 (7667), 338–342. doi:10.1038/nature23450
Li, N., Kang, Y., Wang, L., Huff, S., Tang, R., Hui, H., et al. (2020). ALKBH5 regulates anti-PD-1 therapy response by modulating lactate and suppressive immune cell accumulation in tumor microenvironment. Proc. Natl. Acad. Sci. U. S. A. 117 (33), 20159–20170. doi:10.1073/pnas.1918986117
Li, Q., Huang, Y., Liu, X., Gan, J., Chen, H., and Yang, C. G. (2016b). Rhein inhibits AlkB repair enzymes and sensitizes cells to methylated DNA damage. J. Biol. Chem. 291 (21), 11083–11093. doi:10.1074/jbc.M115.711895
Li, Z., Qian, P., Shao, W., Shi, H., He, X. C., Gogol, M., et al. (2018). Suppression of m(6)A reader Ythdf2 promotes hematopoietic stem cell expansion. Cell Res. 28 (9), 904–917. doi:10.1038/s41422-018-0072-0
Li, Z., Weng, H., Su, R., Weng, X., Zuo, Z., Li, C., et al. (2017b). FTO plays an oncogenic role in acute myeloid leukemia as a N(6)-methyladenosine RNA demethylase. Cancer Cell 31 (1), 127–141. doi:10.1016/j.ccell.2016.11.017
Linder, B., Grozhik, A. V., Olarerin-George, A. O., Meydan, C., Mason, C. E., and Jaffrey, S. R. (2015). Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 12 (8), 767–772. doi:10.1038/nmeth.3453
Liu, J., Yue, Y., Han, D., Wang, X., Fu, Y., Zhang, L., et al. (2014). A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10 (2), 93–95. doi:10.1038/nchembio.1432
Liu, L., Li, H., Hu, D., Wang, Y., Shao, W., Zhong, J., et al. (2022). Insights into N6-methyladenosine and programmed cell death in cancer. Mol. Cancer 21 (1), 32. doi:10.1186/s12943-022-01508-w
Liu, N., and Pan, T. (2016). Probing N⁶-methyladenosine (m⁶A) RNA modification in total RNA with SCARLET. Methods Mol. Biol. 1358, 285–292. doi:10.1007/978-1-4939-3067-8_17
Liu, T., Wei, Q., Jin, J., Luo, Q., Liu, Y., Yang, Y., et al. (2020). The m6A reader YTHDF1 promotes ovarian cancer progression via augmenting EIF3C translation. Nucleic Acids Res. 48 (7), 3816–3831. doi:10.1093/nar/gkaa048
Liu, Y., Liang, G., Xu, H., Dong, W., Dong, Z., Qiu, Z., et al. (2021). Tumors exploit FTO-mediated regulation of glycolytic metabolism to evade immune surveillance. Cell Metab. 33 (6), 1221–1233. e1211. doi:10.1016/j.cmet.2021.04.001
Lo, N., Xu, X., Soares, F., and He, H. H. (2022). The basis and promise of programmable RNA editing and modification. Front. Genet. 13, 834413. doi:10.3389/fgene.2022.834413
Lu, T. X., Zheng, Z., Zhang, L., Sun, H. L., Bissonnette, M., Huang, H., et al. (2020). A new model of spontaneous colitis in mice induced by deletion of an RNA m(6)A methyltransferase component METTL14 in T cells. Cell. Mol. Gastroenterol. Hepatol. 10 (4), 747–761. doi:10.1016/j.jcmgh.2020.07.001
Luo, Q., Fu, B., Zhang, L., Guo, Y., Huang, Z., and Li, J. (2020). Decreased peripheral blood ALKBH5 correlates with markers of autoimmune response in systemic lupus erythematosus. Dis. Markers 2020, 8193895. doi:10.1155/2020/8193895
Ma, H., Wang, X., Cai, J., Dai, Q., Natchiar, S. K., Lv, R., et al. (2019). N(6-)Methyladenosine methyltransferase ZCCHC4 mediates ribosomal RNA methylation. Nat. Chem. Biol. 15 (1), 88–94. doi:10.1038/s41589-018-0184-3
Ma, S., Yan, J., Barr, T., Zhang, J., Chen, Z., Wang, L. S., et al. (2021). The RNA m6A reader YTHDF2 controls NK cell antitumor and antiviral immunity. J. Exp. Med. 218 (8), e20210279. doi:10.1084/jem.20210279
Ma, X. X., Cao, Z. G., and Zhao, S. L. (2020). m6A methyltransferase METTL3 promotes the progression of prostate cancer via m6A-modified LEF1. Eur. Rev. Med. Pharmacol. Sci. 24 (7), 3565–3571. doi:10.26355/eurrev_202004_20817
Malacrida, A., Rivara, M., Di Domizio, A., Cislaghi, G., Miloso, M., Zuliani, V., et al. (2020). 3D proteome-wide scale screening and activity evaluation of a new ALKBH5 inhibitor in U87 glioblastoma cell line. Bioorg. Med. Chem. 28 (4), 115300. doi:10.1016/j.bmc.2019.115300
Mao, Y., Dong, L., Liu, X. M., Guo, J., Ma, H., Shen, B., et al. (2019). m(6)A in mRNA coding regions promotes translation via the RNA helicase-containing YTHDC2. Nat. Commun. 10 (1), 5332. doi:10.1038/s41467-019-13317-9
Mar, B. G., Chu, S. H., Kahn, J. D., Krivtsov, A. V., Koche, R., Castellano, C. A., et al. (2017). SETD2 alterations impair DNA damage recognition and lead to resistance to chemotherapy in leukemia. Blood 130 (24), 2631–2641. doi:10.1182/blood-2017-03-775569
Mauer, J., Luo, X., Blanjoie, A., Jiao, X., Grozhik, A. V., Patil, D. P., et al. (2017). Reversible methylation of m(6)Am in the 5' cap controls mRNA stability. Nature 541 (7637), 371–375. doi:10.1038/nature21022
Mauer, J., Sindelar, M., Despic, V., Guez, T., Hawley, B. R., Vasseur, J. J., et al. (2019). FTO controls reversible m(6)Am RNA methylation during snRNA biogenesis. Nat. Chem. Biol. 15 (4), 340–347. doi:10.1038/s41589-019-0231-8
Meyer, K. D., Saletore, Y., Zumbo, P., Elemento, O., Mason, C. E., and Jaffrey, S. R. (2012). Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell 149 (7), 1635–1646. doi:10.1016/j.cell.2012.05.003
Moroz-Omori, E. V., Huang, D., Kumar Bedi, R., Cheriyamkunnel, S. J., Bochenkova, E., Dolbois, A., et al. (2021). METTL3 inhibitors for epitranscriptomic modulation of cellular processes. ChemMedChem 16 (19), 3035–3043. doi:10.1002/cmdc.202100291
Nagarajan, A., Janostiak, R., and Wajapeyee, N. (2019). Dot blot analysis for measuring global N(6)-methyladenosine modification of RNA. Methods Mol. Biol. 1870, 263–271. doi:10.1007/978-1-4939-8808-2_20
Nassar, D., and Blanpain, C. (2016). Cancer stem cells: Basic concepts and therapeutic implications. Annu. Rev. Pathol. 11, 47–76. doi:10.1146/annurev-pathol-012615-044438
Nishizawa, Y., Konno, M., Asai, A., Koseki, J., Kawamoto, K., Miyoshi, N., et al. (2018). Oncogene c-Myc promotes epitranscriptome m(6)A reader YTHDF1 expression in colorectal cancer. Oncotarget 9 (7), 7476–7486. doi:10.18632/oncotarget.23554
Niu, Y., Zhao, X., Wu, Y. S., Li, M. M., Wang, X. J., and Yang, Y. G. (2013). N6-methyl-adenosine (m6A) in RNA: An old modification with a novel epigenetic function. Genomics Proteomics Bioinforma. 11 (1), 8–17. doi:10.1016/j.gpb.2012.12.002
Oerum, S., Meynier, V., Catala, M., and Tisne, C. (2021). A comprehensive review of m6A/m6Am RNA methyltransferase structures. Nucleic Acids Res. 49 (13), 7239–7255. doi:10.1093/nar/gkab378
Patil, D. P., Chen, C. K., Pickering, B. F., Chow, A., Jackson, C., Guttman, M., et al. (2016). m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537 (7620), 369–373. doi:10.1038/nature19342
Pendleton, K. E., Chen, B., Liu, K., Hunter, O. V., Xie, Y., Tu, B. P., et al. (2017). The U6 snRNA m(6)A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell 169 (5), 824–835. e814. doi:10.1016/j.cell.2017.05.003
Peng, S., Xiao, W., Ju, D., Sun, B., Hou, N., Liu, Q., et al. (2019). Identification of entacapone as a chemical inhibitor of FTO mediating metabolic regulation through FOXO1. Sci. Transl. Med. 11 (488), eaau7116. doi:10.1126/scitranslmed.aau7116
Ping, X. L., Sun, B. F., Wang, L., Xiao, W., Yang, X., Wang, W. J., et al. (2014). Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 24 (2), 177–189. doi:10.1038/cr.2014.3
Qiao, Y., Yang, Q., Song, C., and Chang, J. (2017). Computational insights into the origin of decrease/increase in potency of N-CDPCB analogues toward FTO. J. Biomol. Struct. Dyn. 35 (8), 1758–1765. doi:10.1080/07391102.2016.1193445
Qiao, Y., Zhou, B., Zhang, M., Liu, W., Han, Z., Song, C., et al. (2016). A novel inhibitor of the obesity-related protein FTO. Biochemistry 55 (10), 1516–1522. doi:10.1021/acs.biochem.6b00023
Saletore, Y., Meyer, K., Korlach, J., Vilfan, I. D., Jaffrey, S., and Mason, C. E. (2012). The birth of the epitranscriptome: Deciphering the function of RNA modifications. Genome Biol. 13 (10), 175. doi:10.1186/gb-2012-13-10-175
Scholler, E., Weichmann, F., Treiber, T., Ringle, S., Treiber, N., Flatley, A., et al. (2018). Interactions, localization, and phosphorylation of the m(6)A generating METTL3-METTL14-WTAP complex. RNA 24 (4), 499–512. doi:10.1261/rna.064063.117
Selberg, S., Blokhina, D., Aatonen, M., Koivisto, P., Siltanen, A., Mervaala, E., et al. (2019). Discovery of small molecules that activate RNA methylation through cooperative binding to the METTL3-14-WTAP complex active site. Cell Rep. 26 (13), 3762–3771. e3765. doi:10.1016/j.celrep.2019.02.100
Shi, H., Wei, J., and He, C. (2019). Where, when, and how: Context-dependent functions of RNA methylation writers, readers, and erasers. Mol. Cell 74 (4), 640–650. doi:10.1016/j.molcel.2019.04.025
Shima, H., Matsumoto, M., Ishigami, Y., Ebina, M., Muto, A., Sato, Y., et al. (2017). S-adenosylmethionine synthesis is regulated by selective N(6)-adenosine methylation and mRNA degradation involving METTL16 and YTHDC1. Cell Rep. 21 (12), 3354–3363. doi:10.1016/j.celrep.2017.11.092
Shu, X., Cao, J., Cheng, M., Xiang, S., Gao, M., Li, T., et al. (2020). A metabolic labeling method detects m(6)A transcriptome-wide at single base resolution. Nat. Chem. Biol. 16 (8), 887–895. doi:10.1038/s41589-020-0526-9
Song, H., Song, J., Cheng, M., Zheng, M., Wang, T., Tian, S., et al. (2021). METTL3-mediated m(6)A RNA methylation promotes the anti-tumour immunity of natural killer cells. Nat. Commun. 12 (1), 5522. doi:10.1038/s41467-021-25803-0
Su, R., Dong, L., Li, C., Nachtergaele, S., Wunderlich, M., Qing, Y., et al. (2018). R-2HG exhibits anti-tumor activity by targeting FTO/m(6)A/MYC/CEBPA signaling. Cell 172 (1-2), 90–105. doi:10.1016/j.cell.2017.11.031
Taketo, K., Konno, M., Asai, A., Koseki, J., Toratani, M., Satoh, T., et al. (2018). The epitranscriptome m6A writer METTL3 promotes chemo- and radioresistance in pancreatic cancer cells. Int. J. Oncol. 52 (2), 621–629. doi:10.3892/ijo.2017.4219
Thuring, K., Schmid, K., Keller, P., and Helm, M. (2017). LC-MS analysis of methylated RNA. Methods Mol. Biol. 1562, 3–18. doi:10.1007/978-1-4939-6807-7_1
Tung, Y. C., Ayuso, E., Shan, X., Bosch, F., O'Rahilly, S., Coll, A. P., et al. (2010). Hypothalamic-specific manipulation of Fto, the ortholog of the human obesity gene FTO, affects food intake in rats. PLoS One 5 (1), e8771. doi:10.1371/journal.pone.0008771
van Tran, N., Ernst, F. G. M., Hawley, B. R., Zorbas, C., Ulryck, N., Hackert, P., et al. (2019). The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Res. 47 (15), 7719–7733. doi:10.1093/nar/gkz619
Visvanathan, A., Patil, V., Arora, A., Hegde, A. S., Arivazhagan, A., Santosh, V., et al. (2018). Essential role of METTL3-mediated m(6)A modification in glioma stem-like cells maintenance and radioresistance. Oncogene 37 (4), 522–533. doi:10.1038/onc.2017.351
Vu, L. P., Pickering, B. F., Cheng, Y., Zaccara, S., Nguyen, D., Minuesa, G., et al. (2017). The N(6)-methyladenosine (m(6)A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat. Med. 23 (11), 1369–1376. doi:10.1038/nm.4416
Walcher, L., Kistenmacher, A. K., Suo, H., Kitte, R., Dluczek, S., Strauss, A., et al. (2020). Cancer stem cells-origins and biomarkers: Perspectives for targeted personalized therapies. Front. Immunol. 11, 1280. doi:10.3389/fimmu.2020.01280
Wang, H., Zuo, H., Liu, J., Wen, F., Gao, Y., Zhu, X., et al. (2018a). Loss of YTHDF2-mediated m(6)A-dependent mRNA clearance facilitates hematopoietic stem cell regeneration. Cell Res. 28 (10), 1035–1038. doi:10.1038/s41422-018-0082-y
Wang, L., Song, C., Wang, N., Li, S., Liu, Q., Sun, Z., et al. (2020a). NADP modulates RNA m(6)A methylation and adipogenesis via enhancing FTO activity. Nat. Chem. Biol. 16 (12), 1394–1402. doi:10.1038/s41589-020-0601-2
Wang, P., Doxtader, K. A., and Nam, Y. (2016). Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol. Cell 63 (2), 306–317. doi:10.1016/j.molcel.2016.05.041
Wang, R., Han, Z., Liu, B., Zhou, B., Wang, N., Jiang, Q., et al. (2018b). Identification of natural compound radicicol as a potent FTO inhibitor. Mol. Pharm. 15 (9), 4092–4098. doi:10.1021/acs.molpharmaceut.8b00522
Wang, T., Hong, T., Huang, Y., Su, H., Wu, F., Chen, Y., et al. (2015). Fluorescein derivatives as bifunctional molecules for the simultaneous inhibiting and labeling of FTO protein. J. Am. Chem. Soc. 137 (43), 13736–13739. doi:10.1021/jacs.5b06690
Wang, Y., Xiao, Y., Dong, S., Yu, Q., and Jia, G. (2020b). Antibody-free enzyme-assisted chemical approach for detection of N(6)-methyladenosine. Nat. Chem. Biol. 16 (8), 896–903. doi:10.1038/s41589-020-0525-x
Wang, Y., Zeng, L., Liang, C., Zan, R., Ji, W., Zhang, Z., et al. (2019). Integrated analysis of transcriptome-wide m(6)A methylome of osteosarcoma stem cells enriched by chemotherapy. Epigenomics 11 (15), 1693–1715. doi:10.2217/epi-2019-0262
Wei, C. M., Gershowitz, A., and Moss, B. (1975). Methylated nucleotides block 5' terminus of HeLa cell messenger RNA. Cell 4 (4), 379–386. doi:10.1016/0092-8674(75)90158-0
Wei, J., Liu, F., Lu, Z., Fei, Q., Ai, Y., He, P. C., et al. (2018). Differential m(6)A, m(6)Am, and m(1)A demethylation mediated by FTO in the cell nucleus and cytoplasm. Mol. Cell 71 (6), 973–985. e975. doi:10.1016/j.molcel.2018.08.011
Wen, J., Lv, R., Ma, H., Shen, H., He, C., Wang, J., et al. (2018). Zc3h13 regulates nuclear RNA m(6)A methylation and mouse embryonic stem cell self-renewal. Mol. Cell 69 (6), 1028–1038. e1026. doi:10.1016/j.molcel.2018.02.015
Weng, H., Huang, H., Wu, H., Qin, X., Zhao, B. S., Dong, L., et al. (2018). METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m(6)A modification. Cell Stem Cell 22 (2), 191–205. e199. doi:10.1016/j.stem.2017.11.016
Wiener, D., and Schwartz, S. (2021). The epitranscriptome beyond m(6)A. Nat. Rev. Genet. 22 (2), 119–131. doi:10.1038/s41576-020-00295-8
Wilson, C., Chen, P. J., Miao, Z., and Liu, D. R. (2020). Programmable m(6)A modification of cellular RNAs with a Cas13-directed methyltransferase. Nat. Biotechnol. 38 (12), 1431–1440. doi:10.1038/s41587-020-0572-6
Wu, J., Li, Y., Yu, J., Gan, Z., Wei, W., Wang, C., et al. (2020). Resveratrol attenuates high-fat diet induced hepatic lipid homeostasis disorder and decreases m(6)A RNA methylation. Front. Pharmacol. 11, 568006. doi:10.3389/fphar.2020.568006
Wu, R., Li, A., Sun, B., Sun, J. G., Zhang, J., Zhang, T., et al. (2019). A novel m(6)A reader Prrc2a controls oligodendroglial specification and myelination. Cell Res. 29 (1), 23–41. doi:10.1038/s41422-018-0113-8
Xiao, W., Adhikari, S., Dahal, U., Chen, Y. S., Hao, Y. J., Sun, B. F., et al. (2016). Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Mol. Cell 61 (4), 507–519. doi:10.1016/j.molcel.2016.01.012
Xie, H., Li, J., Ying, Y., Yan, H., Jin, K., Ma, X., et al. (2020). METTL3/YTHDF2 m(6) A axis promotes tumorigenesis by degrading SETD7 and KLF4 mRNAs in bladder cancer. J. Cell. Mol. Med. 24 (7), 4092–4104. doi:10.1111/jcmm.15063
Yadav, P. K., and Rajasekharan, R. (2018). The m(6)A methyltransferase Ime4 and mitochondrial functions in yeast. Curr. Genet. 64 (2), 353–357. doi:10.1007/s00294-017-0758-8
Yang, F., Jin, H., Que, B., Chao, Y., Zhang, H., Ying, X., et al. (2019). Dynamic m(6)A mRNA methylation reveals the role of METTL3-m(6)A-CDCP1 signaling axis in chemical carcinogenesis. Oncogene 38 (24), 4755–4772. doi:10.1038/s41388-019-0755-0
Yang, X., Wei, X., Yang, J., Du, T., Yin, C., Fu, B., et al. (2021). An unexpected role for Dicer as a reader of the unacetylated DNA binding domain of p53 in transcriptional regulation. Sci. Adv. 7 (19), eabi6684. doi:10.1126/sciadv.abi6684
Yang, Y., Wei, Q., Tang, Y., Yuanyuan, W., Luo, Q., Zhao, H., et al. (2020). Loss of hnRNPA2B1 inhibits malignant capability and promotes apoptosis via down-regulating Lin28B expression in ovarian cancer. Cancer Lett. 475, 43–52. doi:10.1016/j.canlet.2020.01.029
Yao, Y., Yang, Y., Guo, W., Xu, L., You, M., Zhang, Y. C., et al. (2021). METTL3-dependent m(6)A modification programs T follicular helper cell differentiation. Nat. Commun. 12 (1), 1333. doi:10.1038/s41467-021-21594-6
Yin, H., Zhang, X., Yang, P., Zhang, X., Peng, Y., Li, D., et al. (2021). RNA m6A methylation orchestrates cancer growth and metastasis via macrophage reprogramming. Nat. Commun. 12 (1), 1394. doi:10.1038/s41467-021-21514-8
Yu, F., Wei, J., Cui, X., Yu, C., Ni, W., Bungert, J., et al. (2021a). Post-translational modification of RNA m6A demethylase ALKBH5 regulates ROS-induced DNA damage response. Nucleic Acids Res. 49 (10), 5779–5797. doi:10.1093/nar/gkab415
Yu, Q., Liu, S., Yu, L., Xiao, Y., Zhang, S., Wang, X., et al. (2021b). RNA demethylation increases the yield and biomass of rice and potato plants in field trials. Nat. Biotechnol. 39 (12), 1581–1588. doi:10.1038/s41587-021-00982-9
Yue, H., Nie, X., Yan, Z., and Weining, S. (2019). N6-methyladenosine regulatory machinery in plants: Composition, function and evolution. Plant Biotechnol. J. 17 (7), 1194–1208. doi:10.1111/pbi.13149
Yue, Y., Liu, J., Cui, X., Cao, J., Luo, G., Zhang, Z., et al. (2018). VIRMA mediates preferential m(6)A mRNA methylation in 3'UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 4, 10. doi:10.1038/s41421-018-0019-0
Zhang, Q., and Xu, K. (Forthcoming 2022). The role of regulators of RNA m6A methylation in lung cancer. Genes & Dis. doi:10.1016/j.gendis.2021.12.017
Zhang, S., Zhao, B. S., Zhou, A., Lin, K., Zheng, S., Lu, Z., et al. (2017). m(6)A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell 31 (4), 591–606. e596. doi:10.1016/j.ccell.2017.02.013
Zhang, X., Wei, L. H., Wang, Y., Xiao, Y., Liu, J., Zhang, W., et al. (2019a). Structural insights into FTO's catalytic mechanism for the demethylation of multiple RNA substrates. Proc. Natl. Acad. Sci. U. S. A. 116 (8), 2919–2924. doi:10.1073/pnas.1820574116
Zhang, Y., Kang, M., Zhang, B., Meng, F., Song, J., Kaneko, H., et al. (2019b). m(6)A modification-mediated CBX8 induction regulates stemness and chemosensitivity of colon cancer via upregulation of LGR5. Mol. Cancer 18 (1), 185. doi:10.1186/s12943-019-1116-x
Zhang, Z., Chen, L. Q., Zhao, Y. L., Yang, C. G., Roundtree, I. A., Zhang, Z., et al. (2019c). Single-base mapping of m(6)A by an antibody-independent method. Sci. Adv. 5 (7), eaax0250. doi:10.1126/sciadv.aax0250
Zhao, B. S., Wang, X., Beadell, A. V., Lu, Z., Shi, H., Kuuspalu, A., et al. (2017). m(6)A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. Nature 542 (7642), 475–478. doi:10.1038/nature21355
Zhao, Y., Shi, Y., Shen, H., and Xie, W. (2020). m(6)A-binding proteins: the emerging crucial performers in epigenetics. J. Hematol. Oncol. 13 (1), 35. doi:10.1186/s13045-020-00872-8
Zhou, B., Liu, C., Xu, L., Yuan, Y., Zhao, J., Zhao, W., et al. (2021a). N(6) -methyladenosine reader protein YT521-B homology domain-containing 2 suppresses liver steatosis by regulation of mRNA stability of lipogenic genes. Hepatology 73 (1), 91–103. doi:10.1002/hep.31220
Zhou, H., Yin, K., Zhang, Y., Tian, J., and Wang, S. (2021b). The RNA m6A writer METTL14 in cancers: Roles, structures, and applications. Biochim. Biophys. Acta. Rev. Cancer 1876 (2), 188609. doi:10.1016/j.bbcan.2021.188609
Zhou, X., Chen, J., Chen, J., Wu, W., Wang, X., and Wang, Y. (2015). The beneficial effects of betaine on dysfunctional adipose tissue and N6-methyladenosine mRNA methylation requires the AMP-activated protein kinase α1 subunit. J. Nutr. Biochem. 26 (12), 1678–1684. doi:10.1016/j.jnutbio.2015.08.014
Zhou, X. L., Huang, F. J., Li, Y., Huang, H., and Wu, Q. C. (2021c). SEDT2/METTL14-mediated m6A methylation awakening contributes to hypoxia-induced pulmonary arterial hypertension in mice. Aging (Albany NY) 13 (5), 7538–7548. doi:10.18632/aging.202616
Keywords: RNA methylation, N6-methyladenosine, regulatory mechanisms, cancer, tumor therapy
Citation: Li P, Wang Y, Sun Y, Jiang S and Li J (2022) N6-methyladenosine RNA methylation: From regulatory mechanisms to potential clinical applications. Front. Cell Dev. Biol. 10:1055808. doi: 10.3389/fcell.2022.1055808
Received: 28 September 2022; Accepted: 24 October 2022;
Published: 03 November 2022.
Edited by:
Haitao Wang, Center for Cancer Research, National Cancer Institute (NIH), United StatesReviewed by:
Dawei Zhou, The Ohio State University, United StatesBo Zhu, University of Texas MD Anderson Cancer Center, United States
Copyright © 2022 Li, Wang, Sun, Jiang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sanjie Jiang, amlhbmdzYW5qaWVAYmdpLmNvbQ==; Jingjing Li, amluZ2ppbmdsaS5tZC5waGRAZ21haWwuY29t
†These authors have contributed equally to this work
 Peipei Li
Peipei Li Yuntao Wang
Yuntao Wang Yiwen Sun2
Yiwen Sun2