
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Cell Dev. Biol. , 10 November 2022
Sec. Nuclear Organization and Dynamics
Volume 10 - 2022 | https://doi.org/10.3389/fcell.2022.1041938
This article is part of the Research Topic Nuclear Morphology in Development and Disease View all 8 articles
Nuclear import receptors ensure the recognition and transport of proteins across the nuclear envelope into the nucleus. In addition, as diverse processes as mitosis, post-translational modifications at mitotic exit, ciliogenesis, and phase separation, all share a common need for regulation by nuclear import receptors - particularly importin beta-1 and importin beta-2/transportin - independent on nuclear import. In particular, 1) nuclear import receptors regulate the mitotic spindle after nuclear envelope breakdown, 2) they shield cargoes from unscheduled ubiquitination, regulating their timely proteolysis; 3) they regulate ciliary factors, crucial to cell communications and tissue architecture during development; and 4) they prevent phase separation of toxic proteins aggregates in neurons. The balance of nuclear import receptors to cargoes is critical in all these processes, albeit in opposite directions: overexpression of import receptors, as often found in cancer, inhibits cargoes and impairs downstream processes, motivating the therapeutic design of specific inhibitors. On the contrary, elevated expression is beneficial in neuronal contexts, where nuclear import receptors are regarded as potential therapeutic tools in counteracting the formation of aggregates that may cause neurodegeneration. This paradox demonstrates the amplitude of nuclear import receptors-dependent functions in different contexts and adds complexity in considering their therapeutic implications.
Since the discovery that protein localisation depends on specific signal sequences (Blobel, 2000), nuclear transport receptors for such signals have been seen as global regulators of cellular organisation due to their ability to transport cargoes in and out of the nucleus, hence marking the identity of subcellular compartments.
Nuclear import receptors (NIRs) also play roles beyond transport. Here we will summarise our understanding of NIRs from nuclear import studies (Christie et al., 2016; Stewart, 2022; Wing et al., 2022), then examine how their mechanism(s) of action readapt to regulate processes independent on nuclear import, yet sensitive to regulation by NIRs: mitosis, mitotic exit, ciliogenesis, and phase separation.
Briefly, proteins to be imported within nuclei, carrying nuclear localisation signals (NLSs), are recognised by a receptor of the importin alpha family, which have subtle NLS sequence preference and tissue distribution (Kimura and Imamoto, 2014). NIRs of the importin beta family bind either the importin alpha/cargo dimer, or certain NLS proteins directly, and provide directionality to the complex. During transport, NIRs keep their interacting cargoes in a non-functional state, that will be reversed in the nucleus, where the GTPase RAN is activated by the chromatin-bound GTP exchange factor RCC1 (regulator of chromosomes condensation 1). Therein, RANGTP binding to the NIR dissociate the import complex and release functional NLS cargoes.
Nuclear import also relies on the interaction of NIRs with nuclear pore complexes (NPCs) that fenestrate the nuclear envelope (NE), providing gates to the nuclear periphery. About 30 nucleoporins (NUP) organise the NPC in a basket-shaped structure. Import complex “navigate” through the NPC central channel aided by the NUPs. Importin beta-1 interacts first with NUP358/RANBP2, the largest NUP, placed at the cytoplasmic base of the NPC. That interaction, directed by phenylalanine-glycine (FG)-rich regions of RANBP2 projecting from the NPC, helps orienting import complexes. Once attached to RANBP2, importin beta-1 interacts sequentially with FG-rich NUPs positioned along the NPC and forming a permeability barrier in nuclear pores (Bednenko et al., 2003). FG-rich domains are intrinsically disordered and can undergo phase separation, forming a dense meshwork that remains permeable to importin beta-type NIRs, but not to other macromolecules. The interaction of NIRs with FG domains enables the import complex passage through the NPC (Schmidt and Görlich, 2016). NUPs are therefore integral components of the nuclear transport machinery. The solubilisation of FG domains by NIRs indicates a widespread ability to disperse proteins in phase separations in different contexts (Yoshizawa and Guo, 2021). RANBP2 also has an additional function as a SUMO E3 ligase and SUMO-stabilising factor (Pichler et al., 2002). It interacts with the RAN GTPase-activating protein RANGAP1 and stabilises it in the SUMO-conjugated form, keeping SUMOylated RANGAP1 and the SUMO-conjugating enzyme UBC9 at the NPC base: this complex regulates the SUMOylation state of some transport cargoes (Ptak and Wozniak, 2017), and concomitantly determines a steep difference between the RANGTP-rich nucleus and the cytoplasm, which modulates entry and exit of transport complexes.
In proteomics studies, importin beta members have diversified partners and cargo preferences in different cell types, cell cycle phases and subcellular compartments (Roscioli et al., 2012; Kimura et al., 2013a, 2013b, 2017; Hugel et al., 2014; Mackmull et al., 2017; Baade et al., 2018; Di Francesco et al., 2018; Song et al., 2022). This indicates their potential to act in diverse cellular processes via their capacity to localise NLS-tagged cargoes and modulate protein interactions. The next sections summarise these processes and highlight how altered expression of NIRs can disrupt them, with pathogenetic consequences.
In mitosis, when nucleo-cytoplasmic transport ceases, NIRs localise a group of NLS factors, released from the nucleus at NE breakdown and regulating microtubule nucleation, interactions with kinetochores, and dynamic functions, collectively called spindle assembly factors (SAFs) (reviewed by Forbes et al., 2015; Cavazza and Vernos, 2016).
In interphase, the NE provides a physical barrier between subcellular compartments in which NIRs and RAN are differentially abundant. In mitosis, in the absence of any such barrier, spatial clues are critical in dictating the relative position of NIRs vs. RAN. Importin beta-1 interacts with the spindle microtubules and poles. RANGTP, generated by histone-bound RCC1, is abundant around chromosomes, including at kinetochores, where factors required for microtubule nucleation and kinetochore attachments are recruited. Centrosomes, the canonical microtubule-nucleation centers, also recruit a RAN fraction via the anchoring protein AKAP450. Thus, a topological map is established.
Regions of differential importin:RANGTP concentrations (high RANGTP around chromosomes, high NIRs at the mitotic apparatus) reorganise dynamically over varying distances during mitotic progression. During this time, the functional antagonism between NIRs and RANGTP continues to operate, and their relative abundance determines the functional state of SAFs at any given time and site in mitotic cells. Importin beta-1 keeps SAFs inactive while localising them, until chromosome- or centrosome-associated RANGTP binds to it and locally releases active SAFs (Roscioli et al., 2010). Importin beta-2 (transportin-1, TNPO1) also regulates SAFs via direct inhibition, with a binding mechanism that is partially RANGTP-reversible (Bernis et al., 2014). Thus, NIRs act as master regulators of the mitotic apparatus by governing the spatial programme of SAF activation.
After chromosome segregation has occurred, a coordinated dephosphorylation programme ensures chromatin decondensation and nuclear reassembly. The main actors at that stage are protein phosphatase 1 (PP1) and 2 (PP2A), representing the catalytic moiety of complexes that include regulatory or scaffolding subunits. Importin beta-1 plays roles in reconstituting the NE and resetting the interphase state and it interplays with both major phosphatases to achieve this programme (Table 1).
In late mitosis, importin beta-1 binds the scaffolding protein RepoMan in the N-terminal domain, which targets it around chromosomes, where NE reassembly initiates. Concomitantly, RepoMan binds PP1 via its C-terminal domain and also carries PP1 to the chromosome periphery to activate histone dephosphorylation (Vagnarelli et al., 2011), thus coupling chromatin remodelling and NE reorganisation. RepoMan inactivation impairs importin beta-1 recruitment at the nuclear rim, compromising the NE reformation. Importin beta-1 was also identified in a screening for mitotic exit regulators for co-purifying with PP2A/R1A/B55alpha complexes. These complexes have phosphatase activity over proteins previously phosphorylated by the mitotic Cdk1 kinase; their dephosphorylation is needed to complete NE reassembly and chromatin decondensation. Co-depletion of importin beta-1 and PP2A synergistically delays mitotic exit, demonstrating that they cooperate in post-mitotic reorganisation (Schmitz et al., 2010).
In summary, NIRs regulate the localisation:function relationship for factors acting in mitotic progression and exit. Overexpressed NIRs bypass modulation by RANGTP, preventing the release of active factors and disrupting the cell division steps in which they act, originating genetically unbalanced daughter cells that may initiate genomic instability, a cancer hallmark. Indeed, NIRs are overexpressed in many cancer types that display genetic instability and aneuploidy (Çağatay and Chook, 2018).
NIRs can modulate the accessibility of their interacting cargoes to external factors. As mitosis progresses towards completion, the anaphase-promoting complex (APC), the major mitotic ubiquitin ligase, acts in two waves: at metaphase completion, it ubiquitinates proteins that must be eliminated to enable chromosome segregation; at mitotic exit, it targets factors that would otherwise prevent interphase resetting. This temporal specificity is achieved by sequential binding of the APC/C to coactivators, CDC20 and CDH1, that have distinct windows of activity.
Two SAFS are identified as APC/C substrates (Table 1): Hepatoma Up-Regulated Protein (HURP), a mitotic microtubule stabiliser (Silljé et al., 2006), and Nucleolar and Spindle-Associated Protein 1 (NuSAP1), which promotes aster formation and fiber elongation, respectively regulated by Importin beta-1 and importin 7, also an importin beta family member (Ribbeck et al., 2006). HURP contains close binding sites for importin beta-1 and for APC/C. Importin beta-1 binding allosterically hides the APC/C site, preventing HURP recognition by APC/C. Importin beta-1 also competes with APC/C for NuSAP1 binding, protecting NuSAP1 from ubiquitination (Song and Rape, 2010). Thus, importin beta-1 shields both SAFs from premature ubiquitination. Microtubules also contribute to protect SAFs from ubiquitination. Simultaneously inhibiting importin beta-1 binding, and APC/C activity, impaired spindle function, indicating that importin beta-1 and APC/C cooperate in regulating the turnover of spindle regulators (Song et al., 2014). When importin beta-1 relocalises around the segregating chromosomes at anaphase, the SAFs become accessible to the APC/C and are conveyed towards degradation.
A similar mechanism operates in control of the mitotic checkpoint complex (MCC), which prevents premature chromosome segregation while microtubule attachments are ongoing. The MCC component BUB3, and its chaperone BuGZ, interact with either importin beta-1 or with the Ubr5 ubiquitin ligase. In prometaphase, when the checkpoint is active, importin beta-1 masks the accessibility of BuGZ and BUB3 to Ubr5. When all chromosomes are spindle-attached, highly concentrated RANGTP around chromosomes displaces importin beta-1, facilitating Ubr5 binding to BuGZ and BUB3: both proteins become ubiquitinated, which reduces MCC activity, triggering anaphase (Jiang et al., 2015).
Thus, importin beta-1 temporally regulates the disappearance of specific proteins by shielding them from premature ubiquitination-dependent proteolysis, modulating the timing of entry into the next phase.
NIRs localise factors to the primary cilium, a microtubule-based organelle required for the establishment of tissue architecture in almost every cell type and capable to integrate developmental and differentiation signals.
Cilia emerge from centrosomes in cells that exit the cell cycle. Centrosomes are composed of a “daughter” (newly duplicated) and a “mother” centriole, endowed with appendages that anchor it to the cell cortex. The mother centriole will form the cilium basal body in post-mitotic cells, originating a microtubule-based axoneme delimited by the ciliary membrane.
Proteomic and in situ studies of cilia have identified NIRs, RAN and RAN-binding proteins (Gupta et al., 2015; Arslanhan et al., 2020). Some cilium-associated proteins (Table 1) bear ciliary localisation signals (CLS) with which NIRs interact, sharing similarities with NLSs (Malicki and Avidor-Reiss, 2014). RANGTP accumulates at the basal body during ciliogenesis, where RAN-binding protein 1 (RANBP1), a negative regulator of nucleotide turnover on RAN, co-localises, suggesting a need for RANGTP modulation at cilia (Fan et al., 2011). NUPs also localise to the basal body, and global inhibition of NUP function prevents ciliary import of kinesin KIF17 (Kee et al., 2012). These data suggest that a size exclusion mechanism regulates protein ciliary entry (Kee and Verhey, 2013), and that RANGTP/GDP cycles control the ciliary release of proteins delivered by NIRs. A ciliary protein screening identified several CLS-tagged factors (Table 1), including retinitis pigmentosa 2 (RP2), as cargoes of importin beta-2/transportin-1 (TNPO1) (Madugula and Lu, 2016). RP2 harbours a nuclear transport signal typical of hnRNP proteins, termed M9, with which TNPO1 interacts. Most RP2 retinopathy-causing mutations fall within the M9 motif, preventing TNPO1 interaction and ciliary entry (Hurd et al., 2011).
Cilia concentrate developmental signals, including Hedgehog (Hh). The Gli2 transcriptional activator of Hh accumulates at cilia, dependent on a proline-tyrosine (PY)-NLS signal (Han et al., 2017), an NLS type recognised by TNPO1 (Lee et al., 2006). Gli2 mutations in the PY-NLS, or TNPO1 knockdown, block Gli2 targeting to cilia. Gli2 ciliary entry proved independent on the importin-alpha/beta-1 nuclear localisation pathway, yet still depends on the import machinery, as RANGTP overexpression, which destabilises import complexes reduces Gli2-expressing cilia (Torrado et al., 2016). Thus, TNPO1 regulates Hh signalling by delivering Gli factors to cilia. The TNPO1-encoding gene is a transcriptional target of Hh, defining a feedforward loop for Gli activation at cilia (Han et al., 2017).
Ciliogenesis is intertwined with intraflagellar transport (IFT) pathways that convey proteins into and along cilia and membrane channels to support cilia functions. Two major motor complexes mediate IFT: the heterotrimeric KIF3A/KIF3B/KAP3 complex, and the homodimeric KIF17 motor. NIRs regulate kinesins involved in IFT.
TNPO1 binds kinesin KIF17 via a CLS required for ciliary targeting. KIF17 is then released in the cilium. That suggests that ciliary entry requires a ciliary-cytoplasmic RAN gradient: TNPO1 cargo binding would take place in the presence of low cytoplasmic RANGTP, yet RANGTP would need be concentrated in cilia in order to disrupt the TNPO1/KIF17 complex and release KIF17. Indeed, expression of GTP-locked RAN in the cytoplasm disfavoured the TNPO1/KIF17 complex assembly and abolished KIF17 ciliary entry (Dishinger et al., 2010).
The other IFT pathway, utilising KIF3A, KIF3B, and KAP3, acts in anterograde transport (cilia base-to-tip). KAP3 has armadillo-repeats with which importin beta-1 interacts and required both for KAP3 nuclear import and ciliary targeting. Chlamydomonas, a model system with cilia regenerating capacity, proved informative to disentangle these processes, due to the evolutionary conservation of KAP3: in that system, assays combining importazole (an inhibitor of importin beta1), and cycloheximide (inhibiting ciliogenesis during regeneration), demonstrated the functional separation of ciliary transport and nuclear import of KAP3 (Huang et al., 2021).
In summary, NIRs control both cilia emergence and ciliary transport, with implications in developmental process, as exemplified by Hh and retinite pigmentosa. Ciliary defects yield complex, multiorgan developmental disorders (ciliopathies). Consequences are particularly severe in the central nervous system, as cilia regulate polarity in neuronal precursors, migration, organisation of layers, axon pathfinding and circuit formation during corticogenesis (Hasenpusch-Theil and Theil, 2021; Wilsch-Bräuninger and Huttner, 2021; Yang et al., 2021). Experimental depletion of NIRs disrupts cilia formation (Hurd et al., 2011; Han et al., 2017). NIR underexpression during early embryogenesis may therefore be unviable.
NIRs are global regulators of macromolecular transport in neurons, including retrograde transport, which this highly polarized cells use to transport signalling molecules from peripheral sites in axons into the nucleus of neurons; in that system, cytoplasmic pools of NIRs are poised to carry cargoes over a long distance (reviewed by Rishal and Fainzilber, 2014; Ferreira, 2019). NIRs exert additional roles in neurons, via their protein-dispersing ability and capacity to modulate phase separation of protein aggregates. We focus on NIR cargoes found to be mutated in amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) and summarise how NIRs mitigate the pathogenetic effects of these mutations. NIR functions on other cargoes also relevant to other neurodegenerative forms are listed in (Table 1).
Phase separation is a biophysical transition in which crucial activities concentrate at membraneless subcellular structures or organelles. It can be beneficial when concentrating activities for a specific biological scope, yet can be detrimental when deregulated.
The RNA-binding proteins (RBPs) FUS and TDP-43 harbour intrinsically disordered prion-like domain (PrLD) that can aggregate and undergo aberrant phase transition, associated with progressive neurodegeneration, as in ALS and FTD. These RBPs also contain NIR-interacting signal sequences: FUS bears a PY-NLS recognised by importin beta-2/TNPO1, and this binding not only enables FUS import in nuclei, but also disperses FUS cytoplasmic aggregates (Guo et al., 2018; Yoshizawa et al., 2018). Most ALS-causing FUS mutations fall in the PY-NLS, reducing PY-NLS affinity for TNPO1, with a consequent failure of nuclear import and the concomitant formation of toxic cytoplasmic aggregates (Zhang and Chook, 2012).
TDP-43 is also prone to form aggregates that can be solubilised by importin alpha/importin beta-1 binding to its bipartite NLS (Doll et al., 2022). TDP-43 mutations, also found in a proportion of ALS and FTD cases, yield cytoplasmic aggregates that mislocalise nucleo-cytoplasmic regulators, hence amplifying nuclear transport defects (Chou et al., 2018; Gasset-Rosa et al., 2019).
The C9ORF72 protein, when mutated, actually represents the most frequent cause of ALS and FTD, via many sophisticated mechanisms. The common theme in its pathogenetic mechanisms is the expansion of nucleotide repeats in an intron of the primary transcript, which accumulates in part in RNA foci, and in part is aberrantly translated (via non-AUG translation) into arginine-rich dipeptide repeat (R-DPRs) proteins. The R-DPRs present in mutant C9ORF72 generate aggregates that interfere with NIR binding to cargoes, and can also “plug” the NPC channel, hence blocking nuclear transport altogether, including wild-type TDP-43 which is found in the cytoplasm. Additionally, the repeats are transcribed in RNA that can bind to and sequester RAN regulators, which also disrupt nuclear import (Hayes et al., 2020). All C9ORF72 pathogenetic mechanisms are mitigated by overexpressing NIRs (Hutten et al., 2020), or NUPs (Freibaum et al., 2015), or by administering nuclear export inhibitors (Zhang et al., 2015).
In summary, NIRs binding to the NLS of proteins carrying aggregation-protein domains prevent aggregate formation, reversing phase separation properties and toxic effects. This capacity of NIRs is linked to both the recognition of NLSs, providing “entry points” for NIRs, and to their ability to engage regions of cargos via their large surfaces, with a mechanism distinct but conceptually reminiscent of that used during passage through the NPC and solubilisation of the hydrogels formed by FG-rich NUPs. These effects are independent on nuclear import. Actually, NLS-bearing RBPs can interact with multiple NIR members, suggesting they can utilise a network of NIRs as either chaperones in the cytoplasm or nuclear import vectors (Baade et al., 2021). The level of NIR expression is therefore crucial to prevent neurodegeneration (Moore et al., 2020; Springhower et al., 2020). Indeed, neurodegenerative disorders often display defective expression of NIRs (Pasha et al., 2021). These observations, together with the finding that increasing NIR levels dissolves protein aggregates and reverses their toxicity are raising interest for their potential to prevent or mitigate certain forms of neurodegeneration (Guo et al., 2019; Yoshizawa and Guo, 2021; Odeh et al., 2022).
Several processes of cellular reorganisation re-adapt NIR-dependent mechanisms operating in nuclear import. In mitosis, NIRs regulate spindle function via spatial control of their cargoes. They also regulate the accessibility of cargoes to modifying factors, shielding them from premature phosphorylation and ubiquitination. Roles are also emerging in ciliogenesis. Finally, NIRs can modulate phase separation of aberrant protein aggregates.
A challenging issue emerges: on the one hand, NIR overexpression in cycling cells inhibits spindle regulatory factors, causing faulty mitosis and genetic instability in daughter cells, predisposing them to cancer; on the other hand, NIR underexpression in neurons may impair ciliogenesis, causing ciliopathies, and fail to counteract toxic protein aggregates, causing neurodegeneration (Figure 1). Taking this into account, the therapeutic design of utilising the “beneficial” effects of NIRs should be carefully balanced against the risk represented by their intrinsic pro-oncogenic potential. Future molecular and structural developments will help mastering the challenges posed by these multifaceted regulators.
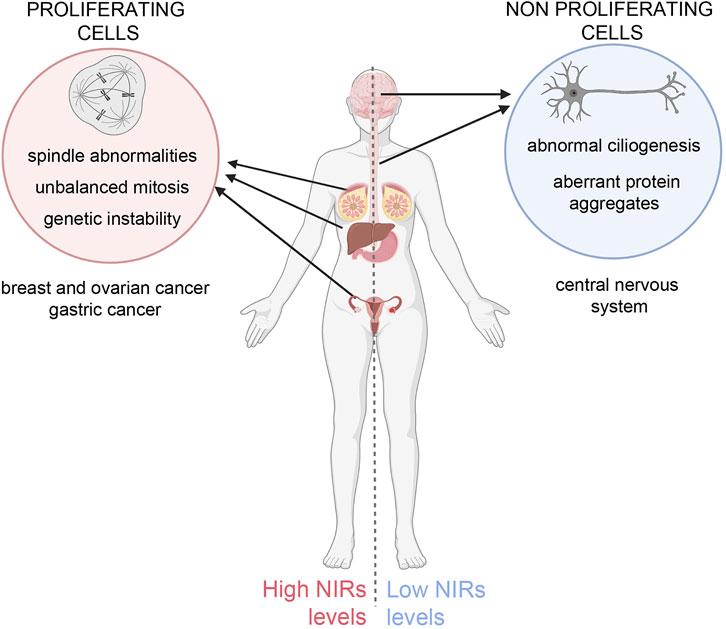
FIGURE 1. Unbalanced NIRs levels differentially affect cycling and non-proliferating cells. Left panel: overexpression of NIRs in proliferating cells causes mitotic abnormalities that can originate genetic instability, predisposing the cells to neoplastic growth. Indeed NIRs are found to be overexpressed in many tumour types, notably in breast, ovary, hepatocellular and gastric cancer (Çağatay and Chook, 2018; also see the TCGA database (https://www.cancer.gov/tcga, https://portal.gdc.cancer.gov/genes/ENSG00000108424). Right panel: in neurons, in which specific mutations in aggregation-prone proteins lead to the formation toxic aggregates that can undergo phase separation, NIRs can prevent or reverse the formation of such aggregates (see text for details) and mitigate their toxic effects, but fail to do so if expressed at insufficient ratios to the mutant proteins. Low levels of NIR can also impact on ciliary formation and function. The figure was created using BioRender.com.
PL conceived planned the manuscript, wrote sections and supervised its organisation, MD and LA wrote sections and prepared the Figures. All authors contributed to the article and approved the submitted version.
We apologise to many colleagues whose studies could not be cited for space reasons. Work in PL lab is supported by grants from MUR-PRIN 2017FNZRN3.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
ALS, amyotrophic lateral sclerosis; CLS, ciliary localisation signals; FG, phenylalanine-glycine; FTD, frontotemporal dementia; IFT, intraflagellar transport; NE, nuclear envelope; NIR, nuclear import receptor; NLS, nuclear localisation signal; NPC, nuclear pore complex; NUP, nucleoporin; PrLD, prion-like domain; PY-NLS, proline-tyrosine nuclear localisation signal; RBP, RNA-binding protein; R-DPR, arginine-rich dipeptide repeat; SAF, spindle assembly factor.
Arslanhan, M. D., Gulensoy, D., and Firat-Karalar, E. N. (2020). A proximity mapping journey into the Biology of the mammalian centrosome/cilium complex. Cells 9, 1–30. doi:10.3390/cells9061390
Baade, I., Hutten, S., Sternburg, E. L., Pörschke, M., Hofweber, M., Dormann, D., et al. (2021). The RNA-binding protein FUS is chaperoned and imported into the nucleus by a network of import receptors. J. Biol. Chem. 296, 100659. doi:10.1016/J.JBC.2021.100659
Baade, I., Spillner, C., Schmitt, K., Valerius, O., and Kehlenbach, R. H. (2018). Extensive identification and in-depth validation of importin 13 cargoes. Mol. Cell. Proteomics 17, 1337–1353. doi:10.1074/MCP.RA118.000623
Bednenko, J., Cingolani, G., and Gerace, L. (2003). Importin β contains a COOH-terminal nucleoporin binding region important for nuclear transport. J. Cell Biol. 162, 391–401. doi:10.1083/JCB.200303085
Bernis, C., Swift-Taylor, B., Nord, M., Carmona, S., Chook, Y. M., and Forbes, D. J. (2014). Transportin acts to regulate mitotic assembly events by target binding rather than Ran sequestration. Mol. Biol. Cell 25, 992–1009. doi:10.1091/MBC.E13-08-0506
Blobel, G. (2000). Protein targeting (Nobel lecture). Chembiochem 1, 86–102. doi:10.1002/1439-7633(20000818)1:2<86::AID-CBIC86>3.0.CO;2-A
Çağatay, T., and Chook, Y. M. (2018). Karyopherins in cancer. Curr. Opin. Cell Biol. 52, 30–42. doi:10.1016/J.CEB.2018.01.006
Cavazza, T., and Vernos, I. (2016). The RanGTP pathway: From nucleo-cytoplasmic transport to spindle assembly and beyond. Front. Cell Dev. Biol. 3, 82. doi:10.3389/FCELL.2015.00082
Chou, C. C., Zhang, Y., Umoh, M. E., Vaughan, S. W., Lorenzini, I., Liu, F., et al. (2018). TDP-43 pathology disrupts nuclear pore complexes and nucleocytoplasmic transport in ALS/FTD. Nat. Neurosci. 21, 228–239. doi:10.1038/S41593-017-0047-3
Christie, M., Chang, C. W., Róna, G., Smith, K. M., Stewart, A. G., Takeda, A. A. S., et al. (2016). Structural Biology and regulation of protein import into the nucleus. J. Mol. Biol. 428, 2060–2090. doi:10.1016/J.JMB.2015.10.023
Di Francesco, L., Verrico, A., Asteriti, I. A., Rovella, P., Cirigliano, P., Guarguaglini, G., et al. (2018). Visualization of human karyopherin beta-1/importin beta-1 interactions with protein partners in mitotic cells by co-immunoprecipitation and proximity ligation assays. Sci. Rep. 8, 1850. doi:10.1038/S41598-018-19351-9
Dishinger, J. F., Kee, H. L., Jenkins, P. M., Fan, S., Hurd, T. W., Hammond, J. W., et al. (2010). Ciliary entry of the kinesin-2 motor KIF17 is regulated by importin-beta2 and RanGTP. Nat. Cell Biol. 12, 703–710. doi:10.1038/ncb2073
Doll, S. G., Meshkin, H., Bryer, A. J., Li, F., Ko, Y.-H., Lokareddy, R. K., et al. (2022). Recognition of the TDP-43 nuclear localization signal by importin α1/β. Cell Rep. 39, 111007. doi:10.1016/J.CELREP.2022.111007
Dormann, D., Rodde, R., Edbauer, D., Bentmann, E., Fischer, I., Hruscha, A., et al. (2010). ALS-associated fused in sarcoma (FUS) mutations disrupt Transportin-mediated nuclear import. EMBO J. 29, 2841–2857. doi:10.1038/emboj.2010.143
Fan, S., Fogg, V., Wang, Q., Chen, X. W., Liu, C. J., and Margolis, B. (2007). A novel Crumbs3 isoform regulates cell division and ciliogenesis via importin beta interactions. J. Cell Biol. 178 (3), 387–398. doi:10.1083/jcb.200609096
Fan, S., Whiteman, E. L., Hurd, T. W., McIntyre, J. C., Dishinger, J. F., Liu, C. J., et al. (2011). Induction of ran GTP drives ciliogenesis. Mol. Biol. Cell 22, 4539–4548. doi:10.1091/mbc.E11-03-0267
Ferreira, P. A. (2019). The coming-of-age of nucleocytoplasmic transport in motor neuron disease and neurodegeneration. Cell. Mol. Life Sci. 76, 2247–2273. doi:10.1007/S00018-019-03029-0
Forbes, D. J., Travesa, A., Nord, M. S., and Bernis, C. (2015). Nuclear transport factors: Global regulation of mitosis. Curr. Opin. Cell Biol. 35, 78–90. doi:10.1016/j.ceb.2015.04.012
Freibaum, B. D., Lu, Y., Lopez-Gonzalez, R., Kim, N. C., Almeida, S., Lee, K. H., et al. (2015). GGGGCC repeat expansion in C9ORF72 compromises nucleocytoplasmic transport. Nature 525, 129–133. doi:10.1038/NATURE14974
Funabashi, T., Katoh, Y., Michisaka, S., Terada, M., Sugawa, M., and Nakayama, K. (2017). Ciliary entry of KIF17 is dependent on its binding to the IFT-B complex via IFT46-IFT56 as well as on its nuclear localization signal. Mol. Biol. Cell 28 (5), 624–633. doi:10.1091/mbc.E16-09-0648
Gasset-Rosa, F., Lu, S., Yu, H., Chen, C., Melamed, Z., Guo, L., et al. (2019). Cytoplasmic TDP-43 de-mixing independent of stress granules drives inhibition of nuclear import, loss of nuclear TDP-43, and cell death. Neuron 102, 339–357. e7. doi:10.1016/J.NEURON.2019.02.038
Guo, L., Fare, C. M., and Shorter, J. (2019). Therapeutic dissolution of aberrant phases by nuclear-import receptors. Trends Cell Biol. 29, 308–322. doi:10.1016/J.TCB.2018.12.004
Guo, L., Kim, H. J., Wang, H., Monaghan, J., Freyermuth, F., Sung, J. C., et al. (2018). Nuclear-import receptors reverse aberrant phase transitions of RNA-binding proteins with prion-like domains. Cell 173, 677–692. e20. doi:10.1016/J.CELL.2018.03.002
Gupta, G. D., Coyaud, É., Gonçalves, J., Mojarad, B. A., Liu, Y., Wu, Q., et al. (2015). A dynamic protein interaction landscape of the human centrosome-cilium interface. Cell 163, 1484–1499. doi:10.1016/J.CELL.2015.10.065
Han, Y., Xiong, Y., Shi, X., Wu, J., Zhao, Y., and Jiang, J. (2017). Regulation of Gli ciliary localization and Hedgehog signaling by the PY-NLS/karyopherin-β2 nuclear import system. PLoS Biol. 15, e2002063. doi:10.1371/journal.pbio.2002063
Hasenpusch-Theil, K., and Theil, T. (2021). The multifaceted roles of primary cilia in the development of the cerebral cortex. Front. Cell Dev. Biol. 9, 630161. doi:10.3389/fcell.2021.630161
Hayes, L. R., Duan, L., Bowen, K., Kalab, P., and Rothstein, J. D. (2020). C9orf72 arginine-rich dipeptide repeat proteins disrupt karyopherin-mediated nuclear import. Elife 9, e51685. doi:10.7554/ELIFE.51685
Huang, S., Dougherty, L. L., and Avasthi, P. (2021). Separable roles for RanGTP in nuclear and ciliary trafficking of a kinesin-2 subunit. J. Biol. Chem. 296, 100117. doi:10.1074/jbc.RA119.010936
Hugel, S., Depping, R., Dittmar, G., Rother, F., Cabot, R., Sury, M. D., et al. (2014). Identification of importin α 7 specific transport cargoes using a proteomic screening approach. Mol. Cell. Proteomics 13, 1286–1298. doi:10.1074/MCP.M112.026856
Hurd, T. W., Fan, S., and Margolis, B. L. (2011). Localization of retinitis pigmentosa 2 to cilia is regulated by Importin beta2. J. Cell Sci. 124, 718–726. doi:10.1242/jcs.070839
Hutten, S., Usluer, S., Bourgeois, B., Simonetti, F., Odeh, H. M., Fare, C. M., et al. (2020). Nuclear import receptors directly bind to arginine-rich dipeptide repeat proteins and suppress their pathological interactions. Cell Rep. 33, 108538. doi:10.1016/J.CELREP.2020.108538
Jiang, H., He, X., Feng, D., Zhu, X., and Zheng, Y. (2015). RanGTP aids anaphase entry through Ubr5-mediated protein turnover. J. Cell Biol. 211, 7–18. doi:10.1083/JCB.201503122
Kee, H. L., Dishinger, J. F., Lynne Blasius, T., Liu, C. J., Margolis, B., and Verhey, K. J. (2012). A size-exclusion permeability barrier and nucleoporins characterize a ciliary pore complex that regulates transport into cilia. Nat. Cell Biol. 14, 431–437. doi:10.1038/NCB2450
Kee, H. L., and Verhey, K. J. (2013). Molecular connections between nuclear and ciliary import processes. Cilia 2, 11. doi:10.1186/2046-2530-2-11
Kimura, M., and Imamoto, N. (2014). Biological significance of the importin-β family-dependent nucleocytoplasmic transport pathways. Traffic 15, 727–748. doi:10.1111/TRA.12174
Kimura, M., Kose, S., Okumura, N., Imai, K., Furuta, M., Sakiyama, N., et al. (2013a). Identification of cargo proteins specific for the nucleocytoplasmic transport carrier transportin by combination of an in vitro transport system and stable isotope labeling by amino acids in cell culture (SILAC)-based quantitative proteomics. Mol. Cell. Proteomics 12, 145–157. doi:10.1074/MCP.M112.019414
Kimura, M., Morinaka, Y., Imai, K., Kose, S., Horton, P., and Imamoto, N. (2017). Extensive cargo identification reveals distinct biological roles of the 12 importin pathways. Elife 6, e21184. doi:10.7554/ELIFE.21184
Kimura, M., Okumura, N., Kose, S., Takao, T., and Imamoto, N. (2013b). Identification of cargo proteins specific for importin-β with importin-α applying a stable isotope labeling by amino acids in cell culture (SILAC)-based in vitro transport system. J. Biol. Chem. 288, 24540–24549. doi:10.1074/JBC.M113.489286
Lee, B. J., Cansizoglu, A. E., Süel, K. E., Louis, T. H., Zhang, Z., and Chook, Y. M. (2006). Rules for nuclear localization sequence recognition by Karyopherinβ2. Cell 126, 543–558. doi:10.1016/j.cell.2006.05.049
Mackmull, M., Klaus, B., Heinze, I., Chokkalingam, M., Beyer, A., Russell, R. B., et al. (2017). Landscape of nuclear transport receptor cargo specificity. Mol. Syst. Biol. 13, 962. doi:10.15252/MSB.20177608
Madugula, V., and Lu, L. (2016). A ternary complex comprising transportin1, Rab8 and the ciliary targeting signal directs proteins to ciliary membranes. J. Cell Sci. 129, 3922–3934. doi:10.1242/jcs.194019
Malicki, J., and Avidor-Reiss, T. (2014). From the cytoplasm into the cilium: Bon voyage. Organogenesis 10, 138–157. doi:10.4161/org.29055
Moore, S., Rabichow, B. E., and Sattler, R. (2020). The hitchhiker’s guide to nucleocytoplasmic trafficking in neurodegeneration. Neurochem. Res. 45, 1306–1327. doi:10.1007/S11064-020-02989-1
Nanaura, H., Kawamukai, H., Fujiwara, A., Uehara, T., Aiba, Y., Nakanishi, M., et al. (2021). C9orf72-derived arginine-rich poly-dipeptides impede phase modifiers. Nat. Commun. 12, 5301. doi:10.1038/s41467-021-25560-0
Odeh, H. M., Fare, C. M., and Shorter, J. (2022). Nuclear-import receptors counter deleterious phase transitions in neurodegenerative disease. J. Mol. Biol. 434, 167220. doi:10.1016/J.JMB.2021.167220
Oostdyk, L. T., Wang, Z., Zang, C., Li, H., McConnell, M. J., and Paschal, B. M. (2020). An epilepsy-associated mutation in the nuclear import receptor KPNA7 reduces nuclear localization signal binding. Sci. Rep. 10 (1), 4844. doi:10.1038/s41598-020-61369-5
Pasha, T., Zatorska, A., Sharipov, D., Rogelj, B., Hortobágyi, T., and Hirth, F. (2021). Karyopherin abnormalities in neurodegenerative proteinopathies. Brain. 144, 2915–2932. doi:10.1093/BRAIN/AWAB201
Pichler, A., Gast, A., Seeler, J. S., Dejean, A., and Melchior, F. (2002). The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell 108, 109–120. doi:10.1016/S0092-8674(01)00633-X
Ptak, C., and Wozniak, R. W. (2017). SUMO and nucleocytoplasmic transport. Adv. Exp. Med. Biol. 963, 111–126. doi:10.1007/978-3-319-50044-7_7
Ribbeck, K., Groen, A. C., Santarella, R., Bohnsack, M. T., Raemaekers, T., Köcher, T., et al. (2006). NuSAP, a mitotic RanGTP target that stabilizes and cross-links microtubules. Mol. Biol. Cell 17, 2646–2660. doi:10.1091/MBC.E05-12-1178
Rishal, I., and Fainzilber, M. (2014). Axon-soma communication in neuronal injury. Nat. Rev. Neurosci. 15, 32–42. doi:10.1038/NRN3609
Roscioli, E., Bolognesi, A., Guarguaglini, G., and Lavia, P. (2010). Ran control of mitosis in human cells: Gradients and local signals. Biochem. Soc. Trans. 38, 1709–1714. doi:10.1042/BST0381709
Roscioli, E., Di Francesco, L., Bolognesi, A., Giubettini, M., Orlando, S., Harel, A., et al. (2012). Importin-β negatively regulates multiple aspects of mitosis including RANGAP1 recruitment to kinetochores. J. Cell Biol. 196, 435–450. doi:10.1083/jcb.201109104
Schmidt, H. B., and Görlich, D. (2016). Transport selectivity of nuclear pores, phase separation, and membraneless organelles. Trends biochem. Sci. 41, 46–61. doi:10.1016/J.TIBS.2015.11.001
Schmitz, M. H. A., Held, M., Janssens, V., Hutchins, J. R. A., Hudecz, O., Ivanova, E., et al. (2010). Live-cell imaging RNAi screen identifies PP2A-B55alpha and importin-beta1 as key mitotic exit regulators in human cells. Nat. Cell Biol. 12, 886–893. doi:10.1038/ncb2092
Silljé, H. H. W., Nagel, S., Körner, R., and Nigg, E. A. (2006). HURP is a Ran-importin beta-regulated protein that stabilizes kinetochore microtubules in the vicinity of chromosomes. Curr. Biol. 16, 731–742. doi:10.1016/J.CUB.2006.02.070
Song, D.-A., Alber, S., Doron-Mandel, E., Schmid, V., Albus, C. A., Leitner, O., et al. (2022). A new monoclonal antibody enables BAR analysis of subcellular importin β1 interactomes. Mol. Cell. Proteomics 2022, 100418. doi:10.1016/J.MCPRO.2022.100418
Song, L., Craney, A., and Rape, M. (2014). Microtubule-dependent regulation of mitotic protein degradation. Mol. Cell 53, 179–192. doi:10.1016/j.molcel.2013.12.022
Song, L., and Rape, M. (2010). Regulated degradation of spindle assembly factors by the anaphase-promoting complex. Mol. Cell 38, 369–382. doi:10.1016/j.molcel.2010.02.038
Springhower, C. E., Rosen, M. K., and Chook, Y. M. (2020). Karyopherins and condensates. Curr. Opin. Cell Biol. 64, 112–123. doi:10.1016/J.CEB.2020.04.003
Stewart, M. (2022). Function of the nuclear transport machinery in maintaining the distinctive compositions of the nucleus and cytoplasm. Int. J. Mol. Sci. 23, 2578. doi:10.3390/IJMS23052578
Torrado, B., Graña, M., Badano, J. L., and Irigoín, F. (2016). Ciliary entry of the Hedgehog transcriptional activator Gli2 is mediated by the nuclear import machinery but differs from nuclear transport in being Imp-α/β1-independent. PLoS One 11, e0162033. doi:10.1371/journal.pone.0162033
Vagnarelli, P., Ribeiro, S., Sennels, L., Sanchez-Pulido, L., de Lima Alves, F., Verheyen, T., et al. (2011). Repo-man coordinates chromosomal reorganization with nuclear envelope reassembly during mitotic exit. Dev. Cell 21, 328–342. doi:10.1016/j.devcel.2011.06.020
Wilsch-Bräuninger, M., and Huttner, W. B. (2021). Primary cilia and centrosomes in neocortex development. Front. Neurosci. 15, 755867. doi:10.3389/fnins.2021.755867
Wing, C. E., Fung, H. Y. J., and Chook, Y. M. (2022). Karyopherin-mediated nucleocytoplasmic transport. Nat. Rev. Mol. Cell Biol. 23, 307–328. doi:10.1038/S41580-021-00446-7
Yang, J., Hu, X., Ma, J., and Shi, S. H. (2021). Centrosome regulation and function in mammalian cortical neurogenesis. Curr. Opin. Neurobiol. 69, 256–266. doi:10.1016/j.conb.2021.06.003
Yoshizawa, T., Ali, R., Jiou, J., Fung, H. Y. J., Burke, K. A., Kim, S. J., et al. (2018). Nuclear import receptor inhibits phase separation of FUS through binding to multiple sites. Cell 173, 693–705. e22. doi:10.1016/J.CELL.2018.03.003
Yoshizawa, T., and Guo, L. (2021). Karyopherin-βs play a key role as a phase separation regulator. J. Biochem. 170, 15–23. doi:10.1093/JB/MVAB072
Zhang, K., Donnelly, C. J., Haeusler, A. R., Grima, J. C., Machamer, J. B., Steinwald, P., et al. (2015). The C9ORF72 repeat expansion disrupts nucleocytoplasmic transport. Nature 525, 56–61. doi:10.1038/NATURE14973
Keywords: nuclear transport, importin beta family, mitotic spindle, nucleoporins, ubiquitin/proteasome system, protein aggregates, neurodegenerative diseases, ciliogenesis
Citation: Damizia M, Altieri L and Lavia P (2022) Non-transport roles of nuclear import receptors: In need of the right balance. Front. Cell Dev. Biol. 10:1041938. doi: 10.3389/fcell.2022.1041938
Received: 11 September 2022; Accepted: 21 October 2022;
Published: 10 November 2022.
Edited by:
Daniel L. Levy, University of Wyoming, United StatesReviewed by:
Pavithra Chavali, Centre for Cellular and Molecular Biology (CCMB), IndiaCopyright © 2022 Damizia, Altieri and Lavia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Patrizia Lavia, cGF0cml6aWEubGF2aWFAdW5pcm9tYTEuaXQ=, cGF0cml6aWEubGF2aWFAY25yLml0
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.