
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell Dev. Biol. , 15 December 2022
Sec. Epigenomics and Epigenetics
Volume 10 - 2022 | https://doi.org/10.3389/fcell.2022.1035033
This article is part of the Research Topic Protein Modifications in Epigenetic Dysfunctional Diseases: Mechanisms and Potential Therapeutic Strategies View all 15 articles
Objective: This study aimed to identify ROS1 fusion partners in Chinese patients with solid tumors.
Methods: Next-generation sequencing (NGS) analysis was used to detect ROS1 rearrangement in 45,438 Chinese patients with solid tumors between 2015 and 2020, and the clinical characteristics and genetic features of gene fusion were evaluated. H&E staining of the excised tumor tissues was conducted. Samples with a tumor cell content ≥ 20% were included for subsequent DNA extraction and sequencing analysis.
Results: A total of 92 patients with ROS1 rearrangements were identified using next-generation sequencing, and the most common histological type lung cancer. From the 92 ROS1 fusion cases, 24 ROS1 fusion partners had been identified, including 14 novel partners and 10 reported partners. Of these, CD74, EZR, SDC4, and TPM3 were the four most frequently occurring partners. Fourteen novel ROS1 fusion partners were detected in 16 patients, including DCBLD1-ROS1, FRK-ROS1, and VGLL2-ROS1. In many patients, the ROS1 breakpoint was located between exons 32 and 34.
Conclusion: This study describes 14 novel ROS1 fusion partners based on the largest ROS1 fusion cohort, and the ROS1 breakpoint was mostly located between exons 32 and 34. Additionally, next-generation sequencing is an optional method for identifying novel ROS1 fusions.
1. This study detected ROS1 fusion partners and the ROS1 fusion breakpoint in solid tumors of Chinese patients in the largest ROS1 fusion cohort to date.
2. A total of 14 novel ROS1 fusions in solid tumor of Chinese patients were identified.
3. The majority of patients had a ROS1 breakpoint between exons 32 and 34.
Several gene-targeting therapies have been developed to treat human malignancies. With an in-depth study of the biological mechanism of tumor cells, the status of target genes in gene targeting therapy, such as EGFR, MET, and ROS1, has gradually become prominent (Cao et al., 2020). ROS1 is a receptor tyrosine kinase whose activation is reported to be linked to the growth and proliferation of malignant tumors. Previous studies have reported that ROS1 undergoes gene rearrangement in many malignant tumors, such as lung cancer and liver cancer. In addition, ROS1 rearrangements rarely overlap with alterations in EGFR, KRAS, or other targeted oncogenes (Zhu et al., 2018). ROS1 was originally identified in 1986 as a viral proto-oncogene with unique oncogenic effects in the UR2 avian sarcoma virus. Hybridization analysis revealed that ROS1 was located in the human chromosome region 6ql6–q22 and was further positioned on chromosome 6q22.1 (Birchmeier et al., 1986; Matsushime et al., 1986).
ROS1 gene fusion expression can drive cell proliferation and induce malignant transformation, which is common in many tumor cells, such as malignant gliomas (Sievers et al., 2021). ROS1 fusions occur in 1%–2% of NSCLC, and the prevalence is higher among patients in Asian countries (including China) than in Western countries (Cai and Su, 2013; Davies and Doebele, 2013; Melosky et al., 2021). A synthetic study has introduced the role of ROS1 in various cancers. It has been reported that 26 genes fuse with ROS1. With the advances in sequencing technology, many new genes have been reported to fuse with ROS1. Natural ROS1 rearrangement was first found in the human brain glioblastoma cell line U118MG (Birchmeier et al., 1987; Charest et al., 2003). The deletion of chromosome six led to the fusion of the ROS1 gene into the FIG gene, which was observed in samples from patients with hepatobiliary carcinoma and ovarian cancer (Birch et al., 2011; Suehara et al., 2012). Numerous studies have shown that crizotinib achieves good results in NSCLC patients with positive ROS1 rearrangement (Shaw et al., 2011; Camidge et al., 2012). AP26113, a specific inhibitor of ROS1, exhibits additional inhibitory activity against oncogenic ROS1 fusions that are involved in the clinical treatment of patients with advanced solid tumors (Anjum et al., 2012). Thus, it is important to detect and evaluate ROS1 rearrangements and gene fusion in patients with malignancies.
Currently, there are many methods for detecting ROS1 fusion genes, including qRT-PCR, FISH, IHC, and NGS (Sakai et al., 2019). These methods have advantages and drawbacks. For example, owing to the continuous increase in ROS1 gene fusion partners, some positive cases may be missed by RT-PCR (Shaw et al., 2011). Fluorescence in situ hybridization (FISH) requires advanced technology, which limits its use. Immunohistochemistry (IHC) is suitable for screening existing fusion expression, whereas only NGS can detect novel ROS1 fusion partners (Zhang et al., 2019). More than 30 ROS1 fusion gene partners have been reported in lung cancer, glioma, and hepatic angiosarcoma, containing CD74, SLC34A2, GOPC (Zhu et al., 2019). NGS is regarded as a powerful tool for detecting ROS1 rearrangements because of its accuracy, sensitivity, and specificity.
Although some studies have reported fusion partners of ROS1, more novel fusions are still being reported. In addition, the distribution of ROS1 fusions varies among different types of cancer. Exploring ROS1 fusions in different cancers may help identify more precise therapies. Therefore, this study aimed to detect the ROS1 partners of Chinese patients with solid tumors using NGS. This study may offer a novel understanding of the treatment of solid tumors based on the ROS1 fusion profile.
A total of 45,438 Chinese patients with solid tumor were treated in the 3Dmed Lab (Shanghai, China) between 2015 and 2020. ROS1 gene fusion was confirmed by NGS. This study was approved by Shanghai East Hospital, Tongji University School of Medicine (No. 2020-093).
Pathological results were used to screen the samples for subsequent analysis. Hematoxylin and eosin (H&E) staining of the removed tumor tissues was conducted. Samples with tumor cell content ≥ 20% were included for subsequent analysis. Genomic DNA (gDNA) was extracted using the ReliaPrep™ FFPE gDNA Miniprep System (Promega) and quantified using a Qubit™ dsDNA HS Assay Kit (Thermo Fisher Scientific). The cfDNA in plasma was extracted using the QIAamp Circulating Nucleic Acid Kit (Qiagen), and the gDNA in white blood cells was extracted using the QIAamp DNA Mini Kit (Qiagen).
Sequencing libraries were established as described previously (Shu et al., 2017). Probe-based hybridization was carried out on the index library with a custom NGS panel, in which the probe bait was a single compound 5 biotinylated 120 bp DNA oligonucleotide. All these contain introns of ROS1 for fusion detection. Repetitive elements were screened and removed from the baits of introns, as previously described (Karolchik et al., 2004).
The extracted DNA was analyzed using a NovaSeq 6,000 platform (Illumina) to screen for targeted gene rearrangements (Xia et al., 2020). The detection approach for variants was based on a binomial test, and an R package was developed to analyze these variants through the analysis of unique supporting read depth, strand bias, and base quality (Su et al., 2017). The variants were analyzed using an automatic false-positive filtering pipeline to ensure specificity and sensitivity when the allele frequency was ≥ 5%. ANNOVAR was performed against dbSNP (v138), 1000Genome, and ESP6500 to annotate the SNPs, insertions, and deletions. Only missense, stop-gain, frameshift, and non-frameshift indel mutations were retained for gene rearrangement analysis.
A total of 45,438 solid tumor patients from the 3Dmed lab were evaluated, of which 92 patients were identified to have ROS1 rearrangements in the blood or tumor tissues (Figure 1). The basic features of the 92 patients were analyzed. Their average age was 57 years (range, 16–82 years) and 63.0% (58/92) of the patients were female (Table 1). As shown in Supplementary Table S1, there were 82 cases of lung cancer, two cases of gastric cancer, two cases of retroperitoneal neoplasm, and one case of liver cancer, epithelioid hemangioendothelioma, liposarcoma, schwannoma, colorectal cancer, and squamous cell carcinoma. Lung adenocarcinoma was diagnosed in 78.9% (68/92) of the patients (Table 1).
From 92 ROS1 fusion cases, 24 ROS1 fusion partners were identified, including 14 novel partners and 10 reported partners. Cluster of differentiation 74 (CD74), ezrin (EZR), syndecan 4 (SDC4), and tropomyosin 3 (TPM3) were the four most frequently occurring fusion partners. The reported ROS1 fusion partners were identified in 75 cases (81.5%, 75/92) and their distribution was as follows: CD74-ROS1 (38.0%, 35/92), EZR-ROS1 (19.6%, 18/92), SDC4-ROS1 (12.0%, 11/92), GOPC-ROS1 (4.3%, 4/92), SLC34A2-ROS1 (3.2%, 3/92), TPM3-ROS1 (3.2%, 3/92), CAPRIN1-ROS1 (1.1%, 1/92), CCDC6-ROS1(1.1%, 1/92), LRIG3-ROS1(1.1%, 1/92), and TPR-ROS1 (1.1%, 1/92) (Figure 2A).
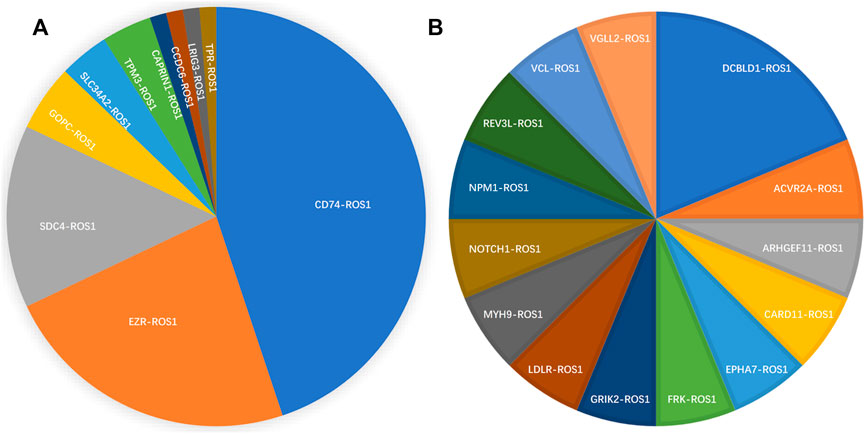
FIGURE 2. Spectrum of ROS1 fusion partners. (A) 10 reported ROS1 fusion partners. (B) 14 novel ROS1 fusion partners.
Fourteen novel ROS1 fusion partners were identified in the 16 patients. Of these, novel DCBLD1-ROS1 was observed three times in three patients with lung cancer, and other novel fusion partners were observed only once in six lung cancer cases. Moreover, seven novel partners were identified in two retroperitoneal neoplasm patients (FRK-ROS1 and VGLL2-ROS1), one gastric cancer patient (ARHGEF11-ROS1), one liver cancer patient (REV3L-ROS1), one liposarcoma patient (EPHA7-ROS1), one patient with epithelioid hemangioendothelioma (NOTCH1-ROS1), and one squamous cell carcinoma patient (CARD11-ROS1). In addition, two patients with lung cancer harbored two ROS1 fusions (Figure 2B). One patient had CD74-ROS1 and SLC34A2-ROS1 fusions, and the other patient had novel DCBLD1-ROS1 and GOPC-ROS1 fusions.
The distribution of ROS1 fusion partners and ROS1 breakpoints was investigated. The results showed that the ROS1 breakpoint was mostly located between exons 32 and 34, which was more apparently in reported ROS1 fusion types (Figures 3A–C), without affecting the transmembrane and tyrosine kinase domains of the ROS1 protein. A breakpoint at exon 35 was found in 15 patients, which disrupted the transmembrane domain (Tables 2, 3).
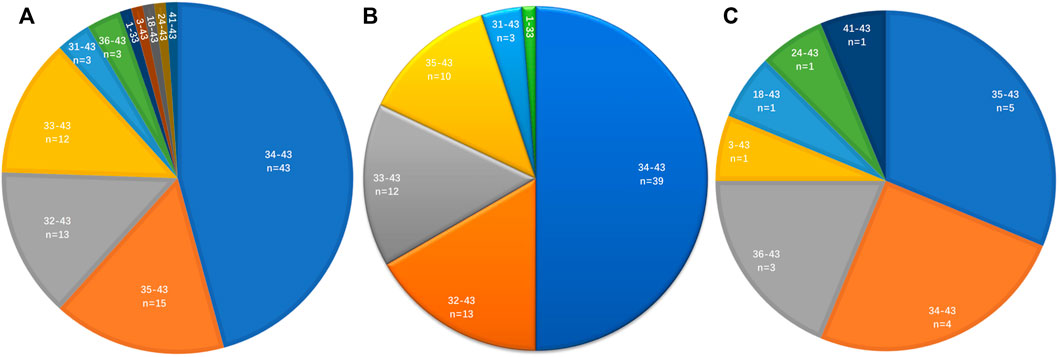
FIGURE 3. Distribution of ROS1 breakpoints. (A) ROS1 breakpoints of ROS1 fusion patients. (B) ROS1 breakpoints of reported ROS1 fusion types. (C) ROS1 breakpoints of novel ROS1 fusion types in patients.
Gene fusion is an ideal target for cancer therapy; therefore, reliable detection of gene fusion is necessary. Many methods have been developed for detecting gene fusion, such as FISH, IHC, qRT-PCR, and NGS, which have different characteristics and applications. In clinical practice, FISH is considered the gold standard for detecting ROS1 fusions (Chiari et al., 2020; Shen et al., 2020). Since FISH is a confirmation test, it may be applied to detect existing ROS1 fusions with high accuracy. However, it is challenging to identify novel ROS1 partners using FISH and simultaneous detection of ROS1 fusion is limited. Due to this facile manipulation, IHC seems to be user-friendly, cost-effective, and highly sensitive. However, it is mostly a complementary tool to other methods, since the results should be observed and analyzed by skilled personnel. Similar to FISH and IHC, qRT-PCR can be used to detect reported fusions, and it exhibits better throughput, sensitivity, and specificity. NGS is an emerging tool for screening ROS1 fusions. A unique advantage of NGS is that it is an innovative and optimal assay for identifying novel fusions. Woo et al. detected glioblastomas harboring gene fusions using NGS in 356 diffuse gliomas. They identified 53 patients harboring various oncogenic gene fusions, including MET, EGFR, and FGFR. They also identified two patients with novel CCDC6-RET fusions (Woo et al., 2020). NGS is a promising approach for obtaining the distribution information of fusions. Thus, our study also used NGS fusion assays to detect the profiles of ROS1 rearrangement in 45,438 patients with malignancies in China, which is the largest ROS1 fusion cohort screened to date, which led to reliable results. It is important to screen for new ROS1 fusions, which can be further confirmed and applied as therapeutic targets.
Another advantage of NGS for detecting ROS1 fusion is that blood samples can be used, including plasma and white blood cells. In this study, positive ROS1 fusion was observed in the blood samples of patients with lung cancer (n = 15) and other cancers (n = 5). This indicated that ROS1 fusions could also be detected in plasma samples, even for the detection of novel fusion types, which was the same as that in tissue samples.
ROS1 fusions were identified in 92 patients. A total of 10 reported and 14 novel ROS1 fusions were found in the solid tumors of Chinese patients with various malignancies. CD74, EZR, SDC4, and TPM3 were 4 most frequently occurring fusion partners. Matsuura et al. (2013) detected common fusion genes in 114 NSCLCs using RT-PCR. They found that the CD74-ROS1 fusion was involved in the carcinogenesis of a subpopulation of NSCLC, which may assist in clarifying the features of tumors and guiding treatment. The CD74-ROS1 fusion gene was reported for the first time in an inflammatory breast cancer patient by NGS (Hu et al., 2021). Cui et al. found that CD74-ROS1 was the most frequently occurring fusion protein in NSCLC (Cui et al., 2020), which is in line with the results of this study. Tatjana et al. (2022) first reported the EZR-ROS1 fusion identified in renal cell carcinoma by molecular sequencing. This new fusion was significant, as crizotinib may be effective in future treatment. The existing ROS1 fusions can be detected in samples from patients, and the positive results may provide information for subsequent therapies, such as the use of kinase inhibitors.
Different ROS1 fusions were observed in one case. Our results indicated that two patients with lung cancer harbored two ROS1 fusions. One patient had CD74-ROS1 and SLC34A2-ROS1 fusions, and the other patient had novel DCBLD1-ROS1 and GOPC-ROS1 fusions. This finding has also been reported in previous studies. Using NGS, Xu et al. found two ROS1 fusions [SDC4-ROS1 (EX2:EX32) and ROS1-GK (EX31:EX13)] in a patient with lung adenocarcinoma. These results indicated that the patient might be sensitive to ROS1 inhibitors (Xu et al., 2021). Patients harboring ROS1 fusions may benefit from crizotinib if their tumors are metastatic (Cai et al., 2019).
With the large number of patients included in our study, we revealed that 16 patients harbored 14 novel ROS1 fusion partners, all of whom had lung cancer. Of these, a novel DCBLD1-ROS1 fusion was observed in three cases, while other fusions occurred once in six cases, and seven novel partners occurred in two retroperitoneal neoplasm patients (FRK-ROS1, VGLL2-ROS1), one gastric cancer patient (ARHGEF11-ROS1), one liver cancer patient (REV3L-ROS1), one liposarcoma patient (EPHA7-ROS1), one patient with epithelioid hemangioendothelioma (NOTCH1-ROS1), and one squamous cell carcinoma patient (CARD11-ROS1). These novel ROS1 fusion partners should be further explored and may also be linked to promising therapies. For example, FPK is a Fyn-related kinase that functions as a tumor suppressor. It has been reported to inhibit glioma progression by suppressing ITGB1/FAK signaling. The effects of specific kinase inhibitors in FRK-ROS1 fusion-positive cases should be explored further.
Variable genomic breakpoints have been identified in Chinese patients through GNS, most location was between exons 32 and 34, and exon 35was also a common. Most of canonical ROS1 fusions were sensitive to crizotinib, especially CD74-ROS1 fusion. Many novel uncommon ROS1 fusions have been found using NGS, most of which were reported to be sensitive to matched targeted therapy, similar to the canonical fusions (Hung et al., 2022). Clinical significance of some genomic breakpoints remained unclear. Simultaneously, more in-depth studies should be conducted to confirm and explore the mechanism underlying these fusions.
This study had numerous limitations. First, the response of the novel ROS1 fusion types to the ROS1 inhibitors was not clear, and even the same ROS1 fusion types with different fusion breakpoints of the partners or the ROS1 gene should be collected, which will be beneficial for clinical therapy. In addition, the specific biological function of gene fusion has not been experimentally determined, and technical errors in NGS analyses cannot be completely excluded. Finally, although NGS is an established and powerful tool, there are barriers to its extensive application. It is expensive for patients and requires complicated equipment and skilled personnel to perform sequencing and subsequent bioinformatics analysis.
In summary, this study was performed to detect ROS1 fusion partners and ROS1 fusion breakpoints in solid tumors of Chinese patients in the largest ROS1 fusion cohort to date. Fourteen novel ROS1 fusions were identified in the solid tumors of Chinese patients, and the ROS1 breakpoint was located between exons 32 and 34 in many patients. Moreover, this study showed that NGS fusion assays can be used on plasma and tissue samples. NGS is a potent tool for reliably identifying novel ROS1 fusions and for detecting molecular alterations.
The data presented in the study are deposited in the Genome Sequence Archive for human (GSA-human) repository, accession number HRA003505, under the BioProject PRJCA013094.
The study was approved by the Institutional Review Boards of Shanghai East Hospital (Approved Number: 2020-093). Each patient included in this study has signed the informed consent form.
NL and MH contributed to the data collection. NL and MQ contributed to paper assessment. DZ, MH, and NL contributed to manuscript writing. FJ and MQ contributed to manuscript revising. All authors contributed to read and approved the final version.
This work was supported by grants 81972280 (to MQ) from National Natural Science Foundation of China, and by Shanghai “Rising Stars of Medical Talent” Youth Development Program (to MQ), and by the Top-level Clinical Discipline Project of Shanghai Pudong (PWYgf 2021-07).
Authors DZ and MH were employed by the company 3D Medicines Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2022.1035033/full#supplementary-material
Anjum, R., Zhang, S., Squillace, R., Clackson, T., Garner, A. P., and Rivera, V. M. (2012). 164 the dual ALK/EGFR inhibitor AP26113 also potently inhibits. Eur. J. Cancer 48, 50. doi:10.1016/s0959-8049(12)71962-7
Birch, A. H., Arcand, S. L., Oros, K. K., Rahimi, K., Watters, A. K., Provencher, D., et al. (2011). Chromosome 3 anomalies investigated by genome wide SNP analysis of benign, low malignant potential and low grade ovarian serous tumours. PloS one 6 (12), e28250. doi:10.1371/journal.pone.0028250
Birchmeier, C., Birnbaum, D., Waitches, G., Fasano, O., and Wigler, M. (1986). Characterization of an activated human ros gene. Mol. Cell. Biol. 6 (9), 3109–3116. doi:10.1128/mcb.6.9.3109
Birchmeier, C., Sharma, S., and Wigler, M. (1987). Expression and rearrangement of the ROS1 gene in human glioblastoma cells. Proc. Natl. Acad. Sci. U. S. A. 84 (24), 9270–9274. doi:10.1073/pnas.84.24.9270
Cai, C. L., Zhang, M. Q., Guo, J., Wang, L. W., Zhao, J. Q., Guo, W. J., et al. (2019). Response to crizotinib in a patient with metastatic lung spindle cell carcinoma harboring TPM3-ROS1 fusion. Chin. Med. J. 132 (24), 3003–3005. doi:10.1097/CM9.0000000000000556
Cai, W. J., and Su, B. (2013). A new target in non-small cell lung cancer: ROS1 fusion gene. Zhonghua Zhong Liu Za Zhi 35 (1), 1–4. doi:10.3760/cma.j.issn.0253-3766.2013.01.001
Camidge, D. R., Bang, Y. J., Kwak, E. L., Iafrate, A. J., Varella-Garcia, M., Fox, S. B., et al. (2012). Activity and safety of crizotinib in patients with ALK-positive non-small-cell lung cancer: Updated results from a phase 1 study. Lancet. Oncol. 13 (10), 1011–1019. doi:10.1016/S1470-2045(12)70344-3
Cao, Z., Wu, W., Zhang, W., Li, Z., Gao, C., Huang, Y., et al. (2020). ALK and ROS1 rearrangement tested by ARMS-PCR in non-small-cell lung cancer patients via cytology specimens: The experience of Shanghai Pulmonary Hospital. Diagn. Cytopathol. 48 (6), 524–530. doi:10.1002/dc.24404
Charest, A., Lane, K., McMahon, K., Park, J., Preisinger, E., Conroy, H., et al. (2003). Fusion of FIG to the receptor tyrosine kinase ROS in a glioblastoma with an interstitial del(6)(q21q21). Genes Chromosom. Cancer 37 (1), 58–71. doi:10.1002/gcc.10207
Chiari, R., Ricciuti, B., Landi, L., Morelli, A. M., Delmonte, A., Spitaleri, G., et al. (2020). ROS1-rearranged non-small-cell lung cancer is associated with a high rate of venous thromboembolism: Analysis from a phase II, prospective, multicenter, two-arms trial (METROS). Clin. Lung Cancer 21 (1), 15–20. doi:10.1016/j.cllc.2019.06.012
Cui, M., Han, Y., Li, P., Zhang, J., Ou, Q., Tong, X., et al. (2020). Molecular and clinicopathological characteristics of ROS1-rearranged non-small-cell lung cancers identified by next-generation sequencing. Mol. Oncol. 14 (11), 2787–2795. doi:10.1002/1878-0261.12789
Davies, K. D., and Doebele, R. C. (2013). Molecular pathways: ROS1 fusion proteins in cancer. Clin. Cancer Res. 19 (15), 4040–4045. doi:10.1158/1078-0432.CCR-12-2851
Hu, H., Ding, N., Zhou, H., Wang, S., Tang, L., Xiao, Z., et al. (2021). Association of subjective cognitive decline with risk of cognitive impairment and dementia: A systematic review and meta-analysis of prospective longitudinal studies. J. Prev. Alzheimers Dis. 15 (1), 277–285. doi:10.14283/jpad.2021.27
Hung, M. S., Lin, Y. C., Chen, F. F., Jiang, Y. Y., Fang, Y. H., Lu, M. S., et al. (2022). The potential and limitation of targeted chromosomal breakpoint sequencing for the ROS1 fusion gene identification in lung cancer. Am. J. Cancer Res. 12 (5), 2376–2386. eCollection 2022.
Karolchik, D., Hinrichs, A. S., Furey, T. S., Roskin, K. M., Sugnet, C. W., Haussler, D., et al. (2004). The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 32, D493–D496. doi:10.1093/nar/gkh103
Matsushime, H., Wang, L. H., and Shibuya, M. (1986). Human c-ros-1 gene homologous to the v-ros sequence of UR2 sarcoma virus encodes for a transmembrane receptorlike molecule. Mol. Cell. Biol. 6 (8), 3000–3004. doi:10.1128/mcb.6.8.3000
Matsuura, S., Shinmura, K., Kamo, T., Igarashi, H., Maruyama, K., Tajima, M., et al. (2013). CD74-ROS1 fusion transcripts in resected non-small cell lung carcinoma. Oncol. Rep. 30 (4), 1675–1680. doi:10.3892/or.2013.2630
Melosky, B., Wheatley-Price, P., Juergens, R. A., Sacher, A., Leighl, N. B., Tsao, M. S., et al. (2021). The rapidly evolving landscape of novel targeted therapies in advanced non-small cell lung cancer. Lung cancer (Amsterdam, Neth. 160, 136–151. doi:10.1016/j.lungcan.2021.06.002
Sakai, K., Ohira, T., Matsubayashi, J., Yoneshige, A., Ito, A., Mitsudomi, T., et al. (2019). Performance of Oncomine Fusion Transcript kit for formalin-fixed, paraffin-embedded lung cancer specimens. Cancer Sci. 110 (6), 2044–2049. doi:10.1111/cas.14016
Shaw, A. T., Yeap, B. Y., Solomon, B. J., Riely, G. J., Gainor, J., Engelman, J. A., et al. (2011). Effect of crizotinib on overall survival in patients with advanced non-small-cell lung cancer harbouring ALK gene rearrangement: A retrospective analysis. Lancet. Oncol. 12 (11), 1004–1012. doi:10.1016/S1470-2045(11)70232-7
Shen, L., Qiang, T., Li, Z., Ding, D., Yu, Y., and Lu, S. (2020). First-line crizotinib versus platinum-pemetrexed chemotherapy in patients with advanced ROS1-rearranged non-small-cell lung cancer. Cancer Med. 9 (10), 3310–3318. doi:10.1002/cam4.2972
Shu, Y., Wu, X., Tong, X., Wang, X., Chang, Z., Mao, Y., et al. (2017). Circulating tumor DNA mutation profiling by targeted next generation sequencing provides guidance for personalized treatments in multiple cancer types. Sci. Rep. 7 (1), 583. doi:10.1038/s41598-017-00520-1
Sievers, P., Stichel, D., Sill, M., Schrimpf, D., Sturm, D., Selt, F., et al. (2021). GOPC:ROS1 and other ROS1 fusions represent a rare but recurrent drug target in a variety of glioma types. Acta Neuropathol. 142 (6), 1065–1069. doi:10.1007/s00401-021-02369-1
Su, D., Zhang, D., Chen, K., Lu, J., Wu, J., Cao, X., et al. (2017). High performance of targeted next generation sequencing on variance detection in clinical tumor specimens in comparison with current conventional methods. J. Exp. Clin. Cancer Res. 36 (1), 121. doi:10.1186/s13046-017-0591-4
Suehara, Y., Arcila, M., Wang, L., Hasanovic, A., Ang, D., Ito, T., et al. (2012). Identification of KIF5B-RET and GOPC-ROS1 fusions in lung adenocarcinomas through a comprehensive mRNA-based screen for tyrosine kinase fusions. Clin. Cancer Res. 18 (24), 6599–6608. doi:10.1158/1078-0432.CCR-12-0838
Tatjana, A., Tjota, M. Y., O'Donnell, P. H., Eggener, S. E., Agarwal, P. K., Haridas, R., et al. (2022). EZR-ROS1 fusion renal cell carcinoma mimicking urothelial carcinoma: Report of a previously undescribed gene fusion in renal cell carcinoma. Virchows Arch. 480 (2), 487–492. doi:10.1007/s00428-021-03138-x
Woo, H. Y., Na, K., Yoo, J., Chang, J. H., Park, Y. N., Shim, H. S., et al. (2020). Glioblastomas harboring gene fusions detected by next-generation sequencing. Brain Tumor Pathol. 37 (4), 136–144. doi:10.1007/s10014-020-00377-9
Xia, H., Xue, X., Ding, H., Ou, Q., Wu, X., Nagasaka, M., et al. (2020). Evidence of NTRK1 fusion as resistance mechanism to EGFR TKI in EGFR+ NSCLC: Results from a large-scale survey of NTRK1 fusions in Chinese patients with lung cancer. Clin. Lung Cancer 21 (3), 247–254. doi:10.1016/j.cllc.2019.09.004
Xu, L., Chen, X., Huo, H., Liu, Y., Yang, X., Gu, D., et al. (2021). Case report: Detection of double ROS1 translocations, SDC4-ROS1 and ROS1-GK, in a lung adenocarcinoma patient and response to crizotinib. Front. Med. 8, 649177. doi:10.3389/fmed.2021.649177
Zhang, L., Wang, Y., Zhao, C., Shi, J., Zhao, S., Liu, X., et al. (2019). High feasibility of cytological specimens for detection of ROS1 fusion by reverse transcriptase PCR in Chinese patients with advanced non-small-cell lung cancer. Onco. Targets. Ther. 12, 3305–3311. doi:10.2147/OTT.S198827
Zhu, Y. C., Lin, X. P., Li, X. F., Wu, L. X., Chen, H. F., Wang, W. X., et al. (2018). Concurrent ROS1 gene rearrangement and KRAS mutation in lung adenocarcinoma: A case report and literature review. Thorac. Cancer 9 (1), 159–163. doi:10.1111/1759-7714.12518
Keywords: solid tumor, next-generation sequencing, ROS1 fusions partners, ROS1 breakpoint, lung cancer
Citation: Li N, Chen Z, Huang M, Zhang D, Hu M, Jiao F and Quan M (2022) Detection of ROS1 gene fusions using next-generation sequencing for patients with malignancy in China. Front. Cell Dev. Biol. 10:1035033. doi: 10.3389/fcell.2022.1035033
Received: 02 September 2022; Accepted: 01 December 2022;
Published: 15 December 2022.
Edited by:
Bin Liu, Jiangsu Ocean Universiity, ChinaReviewed by:
Xuguang Yang, Shanghai University of Traditional Chinese Medicine, ChinaCopyright © 2022 Li, Chen, Huang, Zhang, Hu, Jiao and Quan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Feng Jiao, amlhb2ZlbmdAcmVuamkuY29t; Ming Quan, bXF1YW5AdG9uZ2ppLmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.