
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol. , 07 October 2022
Sec. Nuclear Organization and Dynamics
Volume 10 - 2022 | https://doi.org/10.3389/fcell.2022.1030950
This article is part of the Research Topic Is Aberrant Genome Organization a Cause or Consequence of Specific Diseases? View all 10 articles
Cardiomyopathy is a myocardial disorder, in which the heart muscle is structurally and functionally abnormal, often leading to heart failure. Dilated cardiomyopathy is characterized by a compromised left ventricular function and contributes significantly to the heart failure epidemic, which represents a staggering clinical and public health problem worldwide. Gene mutations have been identified in 35% of patients with dilated cardiomyopathy. Pathogenic variants in LMNA, encoding nuclear A-type lamins, are one of the major causative causes of dilated cardiomyopathy (i.e. CardioLaminopathy). A-type lamins are type V intermediate filament proteins, which are the main components of the nuclear lamina. The nuclear lamina is connected to the cytoskeleton on one side, and to the chromatin on the other side. Among the models proposed to explain how CardioLaminopathy arises, the “chromatin model” posits an effect of mutated A-type lamins on the 3D genome organization and thus on the transcription activity of tissue-specific genes. Chromatin contacts with the nuclear lamina via specific genomic regions called lamina-associated domains lamina-associated domains. These LADs play a role in the chromatin organization and gene expression regulation. This review focuses on the identification of LADs and chromatin remodeling in cardiac muscle cells expressing mutated A-type lamins and discusses the methods and relevance of these findings in disease.
The global prevalence of heart failure is approximately 26 million patients and the economic load related to this condition is approximately US$120 billion (Groenewegen et al., 2020). The large burden of heart failure implies that developing compelling management strategies is paramount. Cardiomyopathy is a condition associated with contractile dysfunction of the heart, often leading to heart failure. Cardiomyopathies are a clinically heterogeneous group of cardiac muscle disorders, which can be either genetic (Figure 1) or systemic (Schultheiss et al., 2019; Hershberger et al., 2021). Dilated cardiomyopathy, the most common form, has an estimated prevalence of >0.4% in the general population. Dilated cardiomyopathy is a condition where the heart muscle becomes enlarged and weakened, resulting in poor left ventricular function defined as a left ventricular ejection fraction <45% (Ziaeian and Fonarow, 2016). Despite current strategies to aggressively manage dilated cardiomyopathy, it remains a common cause of heart failure and a reference for cardiac transplantation. Notwithstanding progresses in reducing heart failure-related mortality, hospitalizations for heart failure remain very frequent and rates of readmissions continue to rise. Presumably, it is important and necessary to increase our knowledge on the pathophysiology of cardiomyopathies to unveil novel molecular/cellular mechanisms for future therapeutic approaches (Leviner et al., 2015).
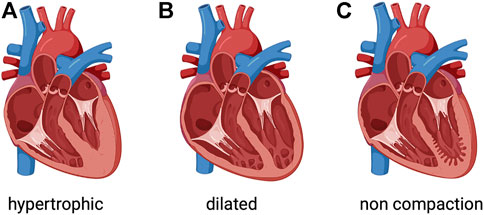
FIGURE 1. Phenotypic groups of inherited cardiomyopathies. (A) Hypertrophic cardiomyopathy involves thickened myocardium, which affects the septum. (B) Dilated cardiomyopathy is characterized by a dilated left ventricle with or without right ventricular involvement. (C) Left ventricular noncompaction is characterized by a noncompacted endocardial layer surrounded by a compacted epicardial layer with marked trabeculation. Created with BioRender.com.
Among the most common genes implicated in dilated cardiomyopathy, it has been estimated that mutations in LMNA gene accounts for about a 10th of familial dilated cardiomyopathy, thus representing one of the major causative genes (Taylor et al., 2003). Affected patients often exhibit early conduction defects before left ventricle dysfunction and dilatation occur. CardioLaminopathy usually presents in early to mid-adulthood with symptomatic conduction system disease or arrhythmias, or with dilated cardiomyopathy. CardioLaminopathy has an intrusive clinical progression with higher rates of aggressive arrhythmias and faster course towards heart failure than most other cardiomyopathies (Taylor et al., 2003). Given the increased awareness among physicians, cardiologists now use defibrillators in order to avoid sudden death from aggressive ventricular arrhythmias, and pharmacological interventions to improve heart failure symptoms (Meune et al., 2006). Once dilated cardiomyopathy is clinically detected, the management for CardioLaminopathy follows the standard of care for heart failure. It is unclear whether early use of these therapeutic drugs prior detectable cardiac dysfunction can modify the aggressive nature of CardioLaminopathy. There is no definitive treatment for the progressive cardiac dilatation and loss of contractility in CardioLaminopathy, short of heart transplantation.
LMNA encodes nuclear A-type lamins. Lamins are class V intermediate filament proteins forming the main component of the nuclear lamins, a fibrous meshwork underlining the inner nuclear membrane of most eukaryotic cells (Wong et al., 2022). In mammalian somatic cells, four major lamin isoforms are found, encoded by two different genes. LMNB1 and LMNB2 genes encode lamins B1 and B2 respectively, which are ubiquitously expressed in cells during development. LMNA gene encodes A-type lamins, lamins A and C, obtained via an alternative pre-mRNA splicing (Figure 2A), which are mainly expressed in most differentiated cells (Dittmer and Misteli, 2011). A-type lamins interact both with the cytoskeleton in the cytoplasm through the LINC complex (Linker of nucleoskeleton and cytoskeleton), and with the chromatin in the nucleoplasm (Méjat, 2010) (Figure 2B). The LINC complex is a large protein complex composed of SUN and KASH domain proteins, present at the inner and outer nuclear membrane respectively (Haque et al., 2006; McGee et al., 2006). SUN proteins interact with several components of the nucleoskeleton, whereas KASH proteins interact with cytoskeleton through their large cytoplasmic protein domains (Jahed et al., 2021). The LINC complex has been shown to be involved in several biological processes including meiosis, DNA damage repair and gene expression (Wong et al., 2021b). By its location and protein interactions, the LINC complex provides a physical continuum between cytoskeleton and nuclear proteins, allows to withstand and transfer forces across the nucleus (Lombardi et al., 2011).
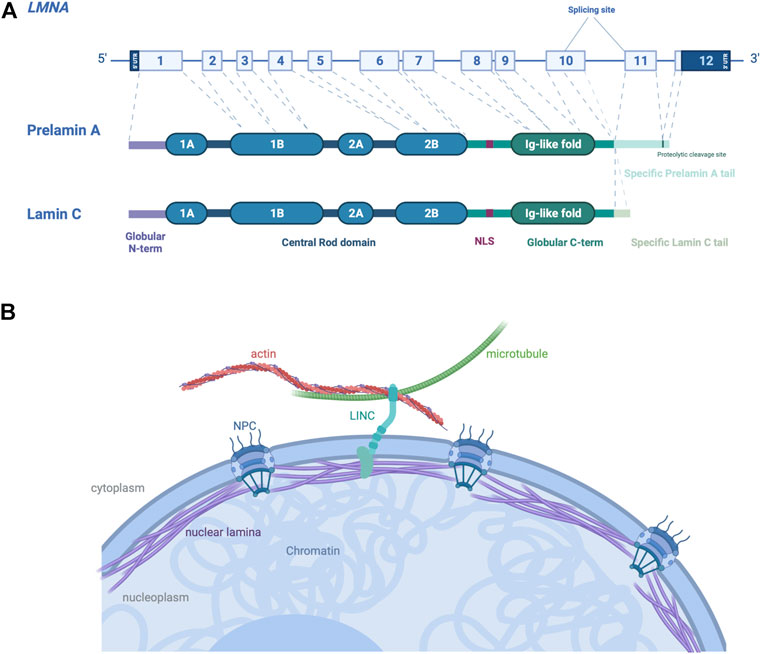
FIGURE 2. Schematic representation of A-type A/C and their localization in cells. (A) LMNA gene encodes the A-type lamins, with prelamin A and lamin C generated by alternative RNA splicing being the major somatic cell isoforms. (B) Schematic view of A-type lamins, proteins of the nuclear lamina on the inner aspect of the inner nuclear membrane, cause autosomal dominant EDMD. The interactions between A-type lamins with SUN and KASH proteins form the LINC complex, connecting the nucleus to cytoskeleton in the cytoplasm of somatic cells. Created with BioRender.com.
Given their interaction with LINC complex and chromatin, A-type lamins are involved in a plethora of biophysical and biochemical processes from the extracellular environment to the nuclear interior. A-type lamins participate to the signal mechanotransduction and regulation of the nucleus stiffness and shape (Broers et al., 2004; Lammerding et al., 2004), as well as in chromatin organization, gene regulation, DNA replication, RNA splicing and genome protection from mechanical cues (Dechat et al., 2008; Shimi et al., 2008; Bertero et al., 2019a; Cho et al., 2019).
Cardiomyocytes, by their intrinsic properties, are constantly subject to mechanical stress. Hence, the nuclei must resist these mechanical cues and correctly transmit the mechanical signal inside the nuclei to convert into a biochemical signal and modulate gene expression. In cardiomyocytes, the level of A-type lamins is finely regulated in response to a mechanical stimulus, to prevent nuclear rupture and protect the genome integrity (Cho et al., 2019). Mutations in A-type lamins could alter the signal mechanotransduction and thus contribute to the pathogenesis of CardioLaminopathy (Jaalouk and Lammerding, 2009). The mammalian nucleus is a highly specialized organelle, which maintains the genome integrity. The nuclear lamina plays an essential role of mechanotransducer, mechanosensor and participate to the chromatin organization. However, the specific mechanistic roles of A-type lamins in this last process in a cardiomyocyte remain obscure (Carmosino et al., 2014). In this review, we discuss recent findings supporting the role of A-type lamins in the organization and regulation of chromatin in Cardiolaminopathy.
A-type lamins have been shown to be involved in genomic organization (Guelen et al., 2008; Paulsen et al., 2018; Guerreiro and Kind, 2019), recruitment of epigenetic regulators (Auguste et al., 2020) and gene expression (van Steensel and Belmont, 2017). In mammalian nucleus, the genome is highly organized in a tissue-dependent manner. Each chromosome is located in a distinct region called “chromosome territories” (Figure 3) (Cremer and Cremer, 2010), containing two different chromosomal compartments, named A and B (Lieberman-Aiden et al., 2009). A-compartments are gene-rich chromatin regions, enriched in active chromatin marks (H3K36me3, H3K79me2, H3K27ac and H3K4me3) and are preferentially embedded in the center of the nucleus. B-compartments are poor-gene and repressive chromatin marks enriched at the nuclear periphery (H3K9me2/3, H3K27me3) (Bertero and Rosa-Garrido, 2021). The only genes present in B-compartments are silenced or weakly transcribed (Guelen et al., 2008). In B-compartments, large chromatin domains bound to nuclear lamins are referred as lamina-associated domains (LADs) (Figure 3) (Guelen et al., 2008; Kind et al., 2013; Amendola and Steensel, 2015; Leemans et al., 2019).
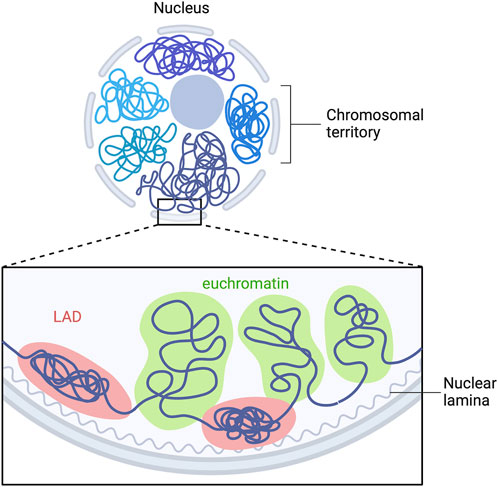
FIGURE 3. LADs organization in mammalian cells. Model depicting the interaction of multiple LADs with the nuclear lamina. Repressed LADs are located at the nuclear periphery and are composed of “gene-desert” regions, which are enriched in heterochromatin marks. Euchromatin is located within the nucleoplasm and is transcriptional active. Adapted from “Chromosome Organization in Nucleus: TADs”, by BioRender.com. Retrieved from https://app.biorender.com/biorender-templates.
In mammals, A-type lamins interact with hundreds of LADs, which are regions ranging from 0,1–10 megabases in size detected on all chromosomes (Briand and Collas, 2020). LADs were first mapped by DNA adenine methyltransferase (DamID) identification in Drosophila (Steensel and Henikoff, 2000; Pickersgill et al., 2006) and later by chromatin immunoprecipitation sequencing (ChIP-seq) (Lund et al., 2014, 2015; Gesson et al., 2016; Cheedipudi et al., 2019; Shah et al., 2021). LADs are predominantly located at the nuclear periphery and are composed of “gene-desert” regions enriched in heterochromatin marks (Kind et al., 2013; Harr et al., 2015).
During mitosis (Wong et al., 2021a) or disease (Cheedipudi et al., 2019), LADs can be reorganized leading to a compartment change and aberrant gene expression (Zheng et al., 2018; Kim et al., 2019) (Figure 2). A-type lamins have been traditionally seen as associated with heterochromatin and transcriptional silencing. However, recent genome-wide approaches show that A-type lamins could be associated with promoters and enhancers outside the LADs, and regulate transcription and chromatin topology of key differentiation gene programs (Leemans et al., 2019).
A comprehensive knowledge of the structural features and dynamics of chromatin is essential to understand cellular mechanisms involving DNA. Though, little is known about the dynamics of chromatin arrangement despite that the scientific community has gained important insights into the higher-order spatial organization of eukaryotic genomes. Several methods have enabled the discovery of higher-order chromatin structure, and notably to study and visualize the LADs organization within the nucleus.
A large variety of proteins bind to specific parts of the genome to regulate gene expression and chromatin structure. In ChIP experiment (Figure 4A), the interacting part of DNA with a protein of interest (lamin A/C for example) are chemically cross-linked together, and DNA is sheared into small fragments. Next, an antibody specific to the protein of interest is used to extract the DNA-protein complex by immunoprecipitation and the extracted DNA sequences are identified by sequencing (ChIP-seq) approaches (Park, 2009).
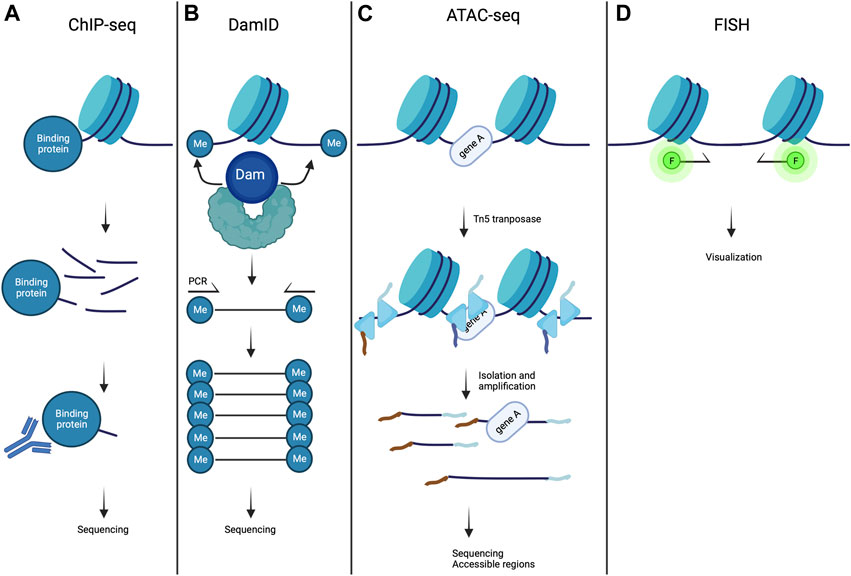
FIGURE 4. Methods to study chromatin organization. (A) In ChIP-seq experiments, chromatin is fragmented and magnetic beads conjugated to antibody specific to the target protein are used to precipitate chromatin fragments bound to the protein of interest. This is then followed by sequencing and mapped onto the genome assembly. (B) In DamID, a fusion protein is created consisting of dam methyltransferase and a protein of interest, resulting in local methylation of adenines in GATC sequences on the chromatin. The genomic methylation pattern can be mapped using sequencing. (C) ATAC- seq is a methodology used for identifying open chromatin regions. Chromatin is incubated with Tn5 transposase to simultaneously fragment and index the exposed DNA fragments. This is then followed by sequencing. (D) In FISH methods, DNA loci of interest are labeled with immunofluorescent DNA sequence specific or chromosome paint probes and are visualized by fluorescent microscopy. Created with BioRender.com.
DamID is a powerful method used to map the genomic interaction sites of these proteins (Figure 4B) (Greil et al., 2006). It is based on fusing a protein of interest to Escherichia coli DNA adenine methyltransferase (Dam). Expression of this fusion protein in vivo leads to the addition of a methylation group to the adenine in GATC sequences (adenine-6-methylation (m6A)). Because adenine methylation does not occur endogenously in eukaryotic cells, it provides a unique tag to mark protein interaction sites. The adenine-methylated DNA fragments are next isolated using a specific endonuclease, and are sequenced after PCR amplification. This method allows the identification of LADs in diseases caused by mutations in A-type lamins (Guelen et al., 2008; Peric-Hupkes et al., 2010; Zullo et al., 2012; Perovanovic et al., 2016; Harr et al., 2020). In combination with the DamID technology, m6A-Tracer method turns out to be a powerful tool to track LADs in living cells (Kind et al., 2013). ChIP-seq and DamID methods are both genomic DNA-binding profiling methods. However, DamID and ChIP-seq approaches each have their strengths and weaknesses. ChIP-seq method provides a snapshot of protein occupancy at a single location on DNA, whereas DamID method relies on DNA adenine methylation, which occurs over a period of several hours. Also, DamID method could give information of chromatin-binding events in vivo, whereas ChIP-seq method is only performed in vitro (Aughey et al., 2019). For a review of an elusive comparison between DamID and ChIP-seq approaches, please refer to (Aughey and Southall, 2016).
The arrangement of chromatin being intimately linked to the gene expression, it is of relevance to study the open state of the chromatin. This has been made possible by the assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq) (Figure 4C). This approach allows to map the chromatin accessibility at the genome-wide scale with an in vitro transposition of sequencing adaptors into native chromatin (Buenrostro et al., 2013; Grandi et al., 2022). Prokaryotic Tn5 transposase is integrated into accessible chromatin regions. The combination of sequencing adaptors and Tn5 transposase integration allows to simultaneously fragment and sequence the open chromatin regions (Buenrostro et al., 2015). ATAC-seq method can be used in combination with ChIP-seq and DamID to correlate LADs spatial organization and gene expression with a chromatin region opening (Lee et al., 2019). A recent methodology development, ATAC-see, allowed to visualize open chromatin regions, by replacing the sequencing by fluorescent adaptors (Chen et al., 2016).
It is also essential to visualize regions of the genome, in order to reveal their individual relationships with nuclear structures in single cells. This is achieved by DNA fluorescence in situ hybridization (FISH) and more recently, by 3D-FISH (FISH on 3D-preserved nuclei with immunofluorescence) in cells (Clements et al., 2016). 3D-FISH allows to study individual locus or discrete number of loci within the nucleus (Szabo et al., 2021). In 3D-FISH methods, (Figure 4D) DNA loci of interest are labeled with immunofluorescent DNA sequence specific or chromosome paint probes and visualized by fluorescent microscopy (Jensen, 2014). This approach is efficient to study the nuclear organization at the single-cell level. More recently, a quantitative high-resolution imaging approach, which combines FISH, array tomography imaging, and multiplexed immunostaining, has been implemented for investigating chromatin organization in complex tissues (Linhoff et al., 2015). 3D-FISH is thus a method of choice to study and visualize LADs and its location in Cardiolaminopathy (Kind et al., 2013; Paulsen et al., 2017; Bertero et al., 2019b; Salvarani et al., 2019).
Specific cellular and molecular mechanisms of pathogenesis and progression of Cardiolaminopathy remain unclear and are still under investigation. Several hypotheses have been proposed attempting to link the pathophysiology of this disease to known or emerging functions of A-type lamins, among which the “chromatin hypothesis”. This model is based on the interaction between A-type lamins and chromatin via LADs. This hypothesis suggests that a cell-type specific chromatin reorganization caused by a disruption of A-type lamins protein expression in Cardiolaminopathy, leads to an abnormal gene expression and thus participates to the pathogenesis.
The “chromatin model” is supported by different studies. It emerges from recent works suggesting that CardioLaminopathy results from dysregulated gene expression as a consequence of LADs reorganization in cardiomyocytes. In fact, these LADs play a role for chromatin organization and gene expression regulation (Guelen et al., 2008; Shimi et al., 2008; Zullo et al., 2012; Solovei et al., 2013). It has been shown that the organization of the LADs is altered in CardioLaminopathy (Perovanovic et al., 2016; Shah et al., 2021). Due to the interaction between A-type lamins and chromatin, dysregulated gene expression has emerged as a plausible mechanism to partially explain the pathogenesis of CardioLaminopathy. One study focused on the organization of LADs in CardioLaminopathy using ChIP-Seq approach and RNA-sequencing in explanted hearts from patients (Cheedipudi et al., 2019). The authors highlighted the role of LADs in the regulation of gene expression, and identified several transcription factors involved in biological processes, such as cell death/survival, cell cycle and metabolism (Cheedipudi et al., 2019). These findings demonstrated that a reorganization of LADs associated with alteration of gene profile expression are occurring in CardioLaminopathy. This was recently strongly supported by a study from another group (Shah et al., 2021). This study was based on the differentiation of cardiomyocytes, adipocytes and hepatocytes derived from iPSCs from patients with cardiolaminopathy. The authors showed that mutations in A-type lamins result in abnormal gene regulation from peripheral chromatin regions only in cardiomyocytes cell types. With this study, Shah and collaborators provided evidence not only of the LADs reorganization in Cardiolaminopathy but also of the cell-type specific organization of several LADs and the link between LADs-chromatin interaction with cell identity. This study thus supports the tissue-specificity of the phenotypes observed in Cardiolaminopathy. Bertero and others studied chromosome conformation in cardiomyocytes derived from iPSCs from patient with CardioLaminopathy carrying the LMNA p. R225X mutation (Bertero et al., 2019b). The authors reported a switch from A- and B-compartments restricted at only ∼1% of the genome, resulting in transcriptional activation of genes shifting from nuclear periphery to the nuclear interior. The limited chromatin compartment change thus challenges the “chromatin model”, as chromosomal compartmentation may not be the primary pathogenic mechanism in CardioLaminopathy. The distribution of open chromatin was biased towards the nuclear periphery in cardiomyocytes derived from iPSCs from patient with CardioLaminopathy carrying the LMNA p. K117fs mutation, compared with isogenic control cells (Lee et al., 2019). Using a combination of ChIP-seq and ATAC-seq approaches (see section ‘methods to identify and visualize LADs’), Lee and collaborators have shown that the abnormal conformation of open chromatin is accompanied by an increase of open chromatin marks of the LADs gene promoters and a decrease of LADs association with A-type lamins. These results suggest that LMNA p. K117f mutation leads to local changes in LADs leading to transcriptional activation. Another study using cardiomyocytes derived from iPSCs from patient with CardioLaminopathy carrying the LMNA p. K219T mutation showed that conduction abnormalities are caused by repression of SCN5A, a gene encoding a sodium channel, due to epigenetics changes (Salvarani et al., 2019). SCN5A may be included in a LAD that shifts from the nuclear interior towards the nuclear periphery in CardioLaminopathy (Lund et al., 2013). Hence, epigenetic state of tissue-specific genes could lead to transcriptional silencing in LADs (Leemans et al., 2019). These data showed that even a slightly change in chromatin organization in mutant cells can still participates to phenotype observed in Cardiolaminopathy.
A-type lamins are mostly located at the nuclear periphery and bind LADs but a pool of these proteins can be associated with the open chromatin (euchromatin) (Gesson et al., 2014). Recently, two studies have shown an alteration of A-type lamins interaction with euchromatin in dilated cardiomyopathy (Zhang et al., 2021; Feng et al., 2022). Collectively, all these studies participate to understand the underlying mechanisms of the disease pathogenesis.
The mammalian genome is organized in different three-dimensional levels, which are finely regulated during normal and pathological development (Zheng and Xie, 2019). In this review, we focused on the LADs organization and reorganization in cardiac disease context but other levels of three-dimensional genome organization can be disrupted in cardiac diseases (Meaburn et al., 2007; Mewborn et al., 2010; Rosa-Garrido et al., 2017; Chapski et al., 2021). A better knowledge of the regulation at different three-dimensional scales of genome organization could then help to better understand the development of cardiac diseases.
This review targets the genome organization specifically in cardiomyocytes. However, studying other cardiac cell type present in the heart environment (e.g., cardiac fibroblasts and endothelial cells) could participate to further understand the pathogenesis of Cardiolaminopathy (Mewborn et al., 2010; Perovanovic et al., 2016; Ramirez-Martinez et al., 2021).
Recent advances to study chromatin organization shed exciting new light on the pathogenesis of CardioLaminopathy. Here, we have outlined recent findings and methods that uncovered the role of chromatin compartment dysregulation in both mice and humans, giving novel insights into the nature of cardiomyocyte dysfunction during disease progression. Nevertheless, there is still a long list of open questions that needs to be answered. While recent studies have elegantly identified the role of chromatin regulation during the pathogenesis of CardioLaminopathy, the distinct roles of chromosomal compartmentation remain elusive. Application of selective tools targeting only the context-dependent A-type lamins gene expression by Cre recombinase-mediated gene targeting or defined drugs might provide new insights into the nature of chromatin organization in the heart. To date, little is known about the temporal regulation of chromatin organization and the transcriptional level during development of CardioLaminopathy. Analyzing chromatin organization and gene regulation in heart along the progression of the disease will illuminate another aspect of genome regulation and help to identify specialized functions for the pathogenesis of age-related disease. In addition, whether such chromatin phenotypes are detrimental or beneficial for disease progression still remain unclear. Answering such critical questions, together with profiling heterogeneous subclusters of chromatin organization in healthy and diseased situations, can open new avenues for the development of therapeutic targeting of cardiomyocytes in CardioLaminopathy. These studies open up new perspectives in an attempt to explain the pathophysiology of Cardiolaminopathies and pinpoint possible new A-type lamins functions still unexplored.
AMu and MK performed data collection (literature reviewing) and prepared the original draft of the manuscript. AMu, AMe and MK revised and wrote the final version of the manuscript. All authors revised and edited the manuscript for critically important intellectual content.
This work was supported by the Association Française contre les Myopathies, Institut National de la Santé et de la Recherche Médicale and Sorbonne Université. MK received a funding from Région Occitanie.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Amendola, M., and Steensel, B. (2015). Nuclear lamins are not required for lamina‐associated domain organization in mouse embryonic stem cells. EMBO Rep. 16, 610–617. doi:10.15252/embr.201439789
Aughey, G. N., Cheetham, S. W., and Southall, T. D. (2019). DamID as a versatile tool for understanding gene regulation. Development 146, dev173666. doi:10.1242/dev.173666
Aughey, G. N., and Southall, T. D. (2016). Dam it’s good! DamID profiling of protein‐DNA interactions. Wiley Interdiscip. Rev. Dev. Biol. 5, 25–37. doi:10.1002/wdev.205
Auguste, G., Rouhi, L., Matkovich, S. J., Coarfa, C., Robertson, M. J., Czernuszewicz, G., et al. (2020). BET bromodomain inhibition attenuates cardiac phenotype in myocyte-specific lamin A/C–deficient mice. J. Clin. Invest. 130, 4740–4758. doi:10.1172/JCI135922
Bertero, A., Fields, P. A., Ramani, V., Bonora, G., Yardimci, G. G., Reinecke, H., et al. (2019a). Dynamics of genome reorganization during human cardiogenesis reveal an RBM20-dependent splicing factory. Nat. Commun. 10, 1538. doi:10.1038/s41467-019-09483-5
Bertero, A., Fields, P. A., Smith, A. S. T., Leonard, A., Beussman, K., Sniadecki, N. J., et al. (2019b). Chromatin compartment dynamics in a haploinsufficient model of cardiac laminopathy. J. Cell Biol. 218, 2919–2944. doi:10.1083/jcb.201902117
Bertero, A., and Rosa-Garrido, M. (2021). Three-dimensional chromatin organization in cardiac development and disease. J. Mol. Cell. Cardiol. 151, 89–105. doi:10.1016/j.yjmcc.2020.11.008
Briand, N., and Collas, P. (2020). Lamina-associated domains: Peripheral matters and internal affairs. Genome Biol. 21, 85. doi:10.1186/s13059-020-02003-5
Broers, J. L. V., Peeters, E. A. G., Kuijpers, H. J. H., Endert, J., Bouten, C. V. C., Oomens, C. W. J., et al. (2004). Decreased mechanical stiffness in LMNA−/− cells is caused by defective nucleo-cytoskeletal integrity: Implications for the development of laminopathies. Hum. Mol. Genet. 13, 2567–2580. doi:10.1093/hmg/ddh295
Buenrostro, J. D., Giresi, P. G., Zaba, L. C., Chang, H. Y., and Greenleaf, W. J. (2013). Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218. doi:10.1038/nmeth.2688
Buenrostro, J. D., Wu, B., Chang, H. Y., and Greenleaf, W. J. (2015). ATAC‐seq: A method for assaying chromatin accessibility genome‐wide. Curr. Protoc. Mol. Biol. 109, 1–21. doi:10.1002/0471142727.mb2129s109
Carmosino, M., Torretta, S., Procino, G., Gerbino, A., Forleo, C., Favale, S., et al. (2014). Role of nuclear Lamin A/C in cardiomyocyte functions: Lamin A/C in cardiomyocytes physiology. Biol. Cell 106, 346–358. doi:10.1111/boc.201400033
Chapski, D. J., Cabaj, M., Morselli, M., Mason, R. J., Soehalim, E., Ren, S., et al. (2021). Early adaptive chromatin remodeling events precede pathologic phenotypes and are reinforced in the failing heart. J. Mol. Cell. Cardiol. 160, 73–86. doi:10.1016/j.yjmcc.2021.07.002
Cheedipudi, S. M., Matkovich, S. J., Coarfa, C., Hu, X., Robertson, M. J., Sweet, M., et al. (2019). Genomic reorganization of lamin-associated domains in cardiac myocytes is associated with differential gene expression and DNA methylation in human dilated cardiomyopathy. Circ. Res. 124, 1198–1213. doi:10.1161/CIRCRESAHA.118.314177
Chen, X., Shen, Y., Draper, W., Buenrostro, J. D., Litzenburger, U., Cho, S. W., et al. (2016). ATAC-see reveals the accessible genome by transposase-mediated imaging and sequencing. Nat. Methods 13, 1013–1020. doi:10.1038/nmeth.4031
Cho, S., Vashisth, M., Abbas, A., Majkut, S., Vogel, K., Xia, Y., et al. (2019). Mechanosensing by the lamina protects against nuclear rupture, DNA damage, and cell-cycle arrest. Dev. Cell 49, 920–935. e5. doi:10.1016/j.devcel.2019.04.020
Clements, C. S., Bikkul, U., Ahmed, M. H., Foster, H. A., Godwin, L. S., and Bridger, J. M. (2016). “Visualizing the spatial relationship of the genome with the nuclear envelope using fluorescence in situ hybridization,” in The nuclear envelope methods in molecular Biology. Editors S. Shackleton, P. Collas, and E. C. Schirmer (New York, NY: Springer New York), 387–406. doi:10.1007/978-1-4939-3530-7_24
Cremer, T., and Cremer, M. (2010). Chromosome territories. Cold Spring Harb. Perspect. Biol. 2, a003889. doi:10.1101/cshperspect.a003889
Dechat, T., Pfleghaar, K., Sengupta, K., Shimi, T., Shumaker, D. K., Solimando, L., et al. (2008). Nuclear lamins: Major factors in the structural organization and function of the nucleus and chromatin. Genes Dev. 22, 832–853. doi:10.1101/gad.1652708
Dittmer, T. A., and Misteli, T. (2011). The lamin protein family. Genome Biol. 12, 222. doi:10.1186/gb-2011-12-5-222
Feng, Y., Cai, L., Hong, W., Zhang, C., Tan, N., Wang, M., et al. (2022). Rewiring of 3D chromatin topology orchestrates transcriptional reprogramming and the development of human dilated cardiomyopathy. Circulation 145, 1663–1683. doi:10.1161/CIRCULATIONAHA.121.055781
Gesson, K., Rescheneder, P., Skoruppa, M. P., von Haeseler, A., Dechat, T., and Foisner, R. (2016). A-type lamins bind both hetero- and euchromatin, the latter being regulated by lamina-associated polypeptide 2 alpha. Genome Res. 26, 462–473. doi:10.1101/gr.196220.115
Gesson, K., Vidak, S., and Foisner, R. (2014). Lamina-associated polypeptide (LAP)2α and nucleoplasmic lamins in adult stem cell regulation and disease. Semin. Cell Dev. Biol. 29, 116–124. doi:10.1016/j.semcdb.2013.12.009
Grandi, F. C., Modi, H., Kampman, L., and Corces, M. R. (2022). Chromatin accessibility profiling by ATAC-seq. Nat. Protoc. 17, 1518–1552. doi:10.1038/s41596-022-00692-9
Greil, F., Moorman, C., and van Steensel, B. (2006). “[16] DamID: Mapping of in vivo protein–genome interactions using tethered DNA adenine methyltransferase,” in Methods in enzymology (Germany: Elsevier), 342–359. doi:10.1016/S0076-6879(06)10016-6
Groenewegen, A., Rutten, F. H., Mosterd, A., and Hoes, A. W. (2020). Epidemiology of heart failure. Eur. J. Heart Fail. 22, 1342–1356. doi:10.1002/ejhf.1858
Guelen, L., Pagie, L., Brasset, E., Meuleman, W., Faza, M. B., Talhout, W., et al. (2008). Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453, 948–951. doi:10.1038/nature06947
Guerreiro, I., and Kind, J. (2019). Spatial chromatin organization and gene regulation at the nuclear lamina. Curr. Opin. Genet. Dev. 55, 19–25. doi:10.1016/j.gde.2019.04.008
Haque, F., Lloyd, D. J., Smallwood, D. T., Dent, C. L., Shanahan, C. M., Fry, A. M., et al. (2006). SUN1 interacts with nuclear lamin A and cytoplasmic nesprins to provide a physical connection between the nuclear lamina and the cytoskeleton. Mol. Cell. Biol. 26, 3738–3751. doi:10.1128/MCB.26.10.3738-3751.2006
Harr, J. C., Luperchio, T. R., Wong, X., Cohen, E., Wheelan, S. J., and Reddy, K. L. (2015). Directed targeting of chromatin to the nuclear lamina is mediated by chromatin state and A-type lamins. J. Cell Biol. 208, 33–52. doi:10.1083/jcb.201405110
Harr, J. C., Schmid, C. D., Muñoz-Jiménez, C., Romero-Bueno, R., Kalck, V., Gonzalez-Sandoval, A., et al. (2020). Loss of an H3K9me anchor rescues laminopathy-linked changes in nuclear organization and muscle function in an Emery-Dreifuss muscular dystrophy model. Genes Dev. 34, 560–579. doi:10.1101/gad.332213.119
Hershberger, R. E., Cowan, J., Jordan, E., and Kinnamon, D. D. (2021). The complex and diverse genetic architecture of dilated cardiomyopathy. Circ. Res. 128, 1514–1532. doi:10.1161/CIRCRESAHA.121.318157
Jaalouk, D. E., and Lammerding, J. (2009). Mechanotransduction gone awry. Nat. Rev. Mol. Cell Biol. 10, 63–73. doi:10.1038/nrm2597
Jahed, Z., Domkam, N., Ornowski, J., Yerima, G., and Mofrad, M. R. K. (2021). Molecular models of LINC complex assembly at the nuclear envelope. J. Cell Sci. 134, jcs258194. doi:10.1242/jcs.258194
Jensen, E. (2014). Technical review: In situ hybridization: AR insights. Anat. Rec. 297, 1349–1353. doi:10.1002/ar.22944
Kim, Y., Zheng, X., and Zheng, Y. (2019). Role of lamins in 3D genome organization and global gene expression. Nucleus 10, 33–41. doi:10.1080/19491034.2019.1578601
Kind, J., Pagie, L., Ortabozkoyun, H., Boyle, S., de Vries, S. S., Janssen, H., et al. (2013). Single-cell dynamics of genome-nuclear lamina interactions. Cell 153, 178–192. doi:10.1016/j.cell.2013.02.028
Lammerding, J., Schulze, P. C., Takahashi, T., Kozlov, S., Sullivan, T., Kamm, R. D., et al. (2004). Lamin A/C deficiency causes defective nuclear mechanics and mechanotransduction. J. Clin. Invest. 113, 370–378. doi:10.1172/JCI19670
Lee, J., Termglinchan, V., Diecke, S., Itzhaki, I., Lam, C. K., Garg, P., et al. (2019). Activation of PDGF pathway links LMNA mutation to dilated cardiomyopathy. Nature 572, 335–340. doi:10.1038/s41586-019-1406-x
Leemans, C., van der Zwalm, M. C. H., Brueckner, L., Comoglio, F., van Schaik, T., Pagie, L., et al. (2019). Promoter-intrinsic and local chromatin features determine gene repression in LADs. Cell 177, 852–864. e14. doi:10.1016/j.cell.2019.03.009
Leviner, D. B., Hochhauser, E., and Arad, M. (2015). Inherited cardiomyopathies—novel therapies. Pharmacol. Ther. 155, 36–48. doi:10.1016/j.pharmthera.2015.08.003
Lieberman-Aiden, E., van Berkum, N. L., Williams, L., Imakaev, M., Ragoczy, T., Telling, A., et al. (2009). Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293. doi:10.1126/science.1181369
Linhoff, M. W., Garg, S. K., and Mandel, G. (2015). A high-resolution imaging approach to investigate chromatin architecture in complex tissues. Cell 163, 246–255. doi:10.1016/j.cell.2015.09.002
Lombardi, M. L., Jaalouk, D. E., Shanahan, C. M., Burke, B., Roux, K. J., and Lammerding, J. (2011). The interaction between nesprins and sun proteins at the nuclear envelope is critical for force transmission between the nucleus and cytoskeleton. J. Biol. Chem. 286, 26743–26753. doi:10.1074/jbc.M111.233700
Lund, E. G., Duband-Goulet, I., Oldenburg, A., Buendia, B., and Collas, P. (2015). Distinct features of lamin A-interacting chromatin domains mapped by ChIP-sequencing from sonicated or micrococcal nuclease-digested chromatin. Nucleus 6, 30–39. doi:10.4161/19491034.2014.990855
Lund, E., Oldenburg, A. R., and Collas, P. (2014). Enriched domain detector: A program for detection of wide genomic enrichment domains robust against local variations. Nucleic Acids Res. 42, e92. doi:10.1093/nar/gku324
Lund, E., Oldenburg, A. R., Delbarre, E., Freberg, C. T., Duband-Goulet, I., Eskeland, R., et al. (2013). Lamin A/C-promoter interactions specify chromatin state–dependent transcription outcomes. Genome Res. 23, 1580–1589. doi:10.1101/gr.159400.113
McGee, M. D., Rillo, R., Anderson, A. S., and Starr, D. A. (2006). UNC-83 is a KASH protein required for nuclear migration and is recruited to the outer nuclear membrane by a physical interaction with the SUN protein UNC-84. Mol. Biol. Cell 17, 1790–1801. doi:10.1091/mbc.e05-09-0894
Meaburn, K. J., Cabuy, E., Bonne, G., Levy, N., Morris, G. E., Novelli, G., et al. (2007). Primary laminopathy fibroblasts display altered genome organization and apoptosis. Aging Cell 6, 139–153. doi:10.1111/j.1474-9726.2007.00270.x
Méjat, A., and Misteli, T. (2010). LINC complexes in health and disease. Nucleus 1, 40–52. doi:10.4161/nucl.1.1.10530
Meune, C., Van Berlo, J. H., Anselme, F., Bonne, G., Pinto, Y. M., and Duboc, D. (2006). Primary prevention of sudden death in patients with lamin A/C gene mutations. N. Engl. J. Med. 354, 209–210. doi:10.1056/NEJMc052632
Mewborn, S. K., Puckelwartz, M. J., Abuisneineh, F., Fahrenbach, J. P., Zhang, Y., MacLeod, H., et al. (2010). Altered chromosomal positioning, compaction, and gene expression with a lamin A/C gene mutation. PLoS ONE 5, e14342. doi:10.1371/journal.pone.0014342
Park, P. J. (2009). ChIP–seq: Advantages and challenges of a maturing technology. Nat. Rev. Genet. 10, 669–680. doi:10.1038/nrg2641
Paulsen, J., Liyakat Ali, T. M., and Collas, P. (2018). Computational 3D genome modeling using Chrom3D. Nat. Protoc. 13, 1137–1152. doi:10.1038/nprot.2018.009
Paulsen, J., Sekelja, M., Oldenburg, A. R., Barateau, A., Briand, N., Delbarre, E., et al. (2017). Chrom3D: Three-dimensional genome modeling from hi-C and nuclear lamin-genome contacts. Genome Biol. 18, 21. doi:10.1186/s13059-016-1146-2
Peric-Hupkes, D., Meuleman, W., Pagie, L., Bruggeman, S. W. M., Solovei, I., Brugman, W., et al. (2010). Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol. Cell 38, 603–613. doi:10.1016/j.molcel.2010.03.016
Perovanovic, J., Dell’Orso, S., Gnochi, V. F., Jaiswal, J. K., Sartorelli, V., Vigouroux, C., et al. (2016). Laminopathies disrupt epigenomic developmental programs and cell fate. Sci. Transl. Med. 8, 335ra58. doi:10.1126/scitranslmed.aad4991
Pickersgill, H., Kalverda, B., de Wit, E., Talhout, W., Fornerod, M., and van Steensel, B. (2006). Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat. Genet. 38, 1005–1014. doi:10.1038/ng1852
Ramirez-Martinez, A., Zhang, Y., Chen, K., Kim, J., Cenik, B. K., McAnally, J. R., et al. (2021). The nuclear envelope protein Net39 is essential for muscle nuclear integrity and chromatin organization. Nat. Commun. 12, 690. doi:10.1038/s41467-021-20987-x
Rosa-Garrido, M., Chapski, D. J., Schmitt, A. D., Kimball, T. H., Karbassi, E., Monte, E., et al. (2017). High-resolution mapping of chromatin conformation in cardiac myocytes reveals structural remodeling of the epigenome in heart failure. Circulation 136, 1613–1625. doi:10.1161/CIRCULATIONAHA.117.029430
Salvarani, N., Crasto, S., Miragoli, M., Bertero, A., Paulis, M., Kunderfranco, P., et al. (2019). The K219T-Lamin mutation induces conduction defects through epigenetic inhibition of SCN5A in human cardiac laminopathy. Nat. Commun. 10, 2267. doi:10.1038/s41467-019-09929-w
Schultheiss, H. P., Fairweather, D., Caforio, A. L. P., Escher, F., Hershberger, R. E., Lipshultz, S. E., et al. (2019). Dilated cardiomyopathy. Nat. Rev. Dis. Primer 5, 32. doi:10.1038/s41572-019-0084-1
Shah, P. P., Lv, W., Rhoades, J. H., Poleshko, A., Abbey, D., Caporizzo, M. A., et al. (2021). Pathogenic LMNA variants disrupt cardiac lamina-chromatin interactions and de-repress alternative fate genes. Cell Stem Cell 28, 938–954.e9. e9. doi:10.1016/j.stem.2020.12.016
Shimi, T., Pfleghaar, K., Kojima, S., Pack, C.-G., Solovei, I., Goldman, A. E., et al. (2008). The A- and B-type nuclear lamin networks: Microdomains involved in chromatin organization and transcription. Genes Dev. 22, 3409–3421. doi:10.1101/gad.1735208
Solovei, I., Wang, A. S., Thanisch, K., Schmidt, C. S., Krebs, S., Zwerger, M., et al. (2013). LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell 152, 584–598. doi:10.1016/j.cell.2013.01.009
Steensel, B., and Henikoff, S. (2000). Identification of in vivo DNA targets of chromatin proteins using tethered Dam methyltransferase. Nat. Biotechnol. 18, 424–428. doi:10.1038/74487
Szabo, Q., Cavalli, G., and Bantignies, F. (2021). “Higher-order chromatin organization using 3D DNA fluorescent in situ hybridization,” in Capturing chromosome conformation methods in molecular Biology. Editors B. Bodega, and C. Lanzuolo (New York, NY: Springer US), 221–237. doi:10.1007/978-1-0716-0664-3_13
Taylor, M. R. G., Fain, P. R., Sinagra, G., Robinson, M. L., Robertson, A. D., Carniel, E., et al. (2003). Natural history of dilated cardiomyopathy due to lamin A/C gene mutations. J. Am. Coll. Cardiol. 41, 771–780. doi:10.1016/S0735-1097(02)02954-6
van Steensel, B., and Belmont, A. S. (2017). Lamina-associated domains: Links with chromosome architecture, heterochromatin, and gene repression. Cell 169, 780–791. doi:10.1016/j.cell.2017.04.022
Wong, X., Hoskins, V. E., Melendez-Perez, A. J., Harr, J. C., Gordon, M., and Reddy, K. L. (2021a). Lamin C is required to establish genome organization after mitosis. Genome Biol. 22, 305. doi:10.1186/s13059-021-02516-7
Wong, X., Loo, T.-H., and Stewart, C. L. (2021b). LINC complex regulation of genome organization and function. Curr. Opin. Genet. Dev. 67, 130–141. doi:10.1016/j.gde.2020.12.007
Wong, X., Melendez-Perez, A. J., and Reddy, K. L. (2022). The nuclear lamina. Cold Spring Harb. Perspect. Biol. 14, a040113. doi:10.1101/cshperspect.a040113
Zhang, X., Shao, X., Zhang, R., Zhu, R., and Feng, R. (2021). Integrated analysis reveals the alterations that LMNA interacts with euchromatin in LMNA mutation-associated dilated cardiomyopathy. Clin. Epigenetics 13, 3. doi:10.1186/s13148-020-00996-1
Zheng, H., and Xie, W. (2019). The role of 3D genome organization in development and cell differentiation. Nat. Rev. Mol. Cell Biol. 20, 535–550. doi:10.1038/s41580-019-0132-4
Zheng, X., Hu, J., Yue, S., Kristiani, L., Kim, M., Sauria, M., et al. (2018). Lamins organize the global three-dimensional genome from the nuclear periphery. Mol. Cell 71, 802–815. e7. doi:10.1016/j.molcel.2018.05.017
Ziaeian, B., and Fonarow, G. C. (2016). Epidemiology and aetiology of heart failure. Nat. Rev. Cardiol. 13, 368–378. doi:10.1038/nrcardio.2016.25
Keywords: LMNA, nuclear lamins, cardiomyopathy, genome organization, LADs, lamina-associated domains
Citation: Kervella M, Jahier M, Meli AC and Muchir A (2022) Genome organization in cardiomyocytes expressing mutated A-type lamins. Front. Cell Dev. Biol. 10:1030950. doi: 10.3389/fcell.2022.1030950
Received: 29 August 2022; Accepted: 26 September 2022;
Published: 07 October 2022.
Edited by:
Eric C Schirmer, University of Edinburgh, United KingdomReviewed by:
James Holaska, Cooper Medical School of Rowan University, United StatesCopyright © 2022 Kervella, Jahier, Meli and Muchir. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Antoine Muchir, YS5tdWNoaXJAaW5zdGl0dXQtbXlvbG9naWUub3Jn
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.