- 1Department of Biochemistry and Chemistry, La Trobe Institute for Molecular Science, La Trobe University, Melbourne, VIC, Australia
- 2Research Centre for Molecular Cancer Prevention, La Trobe University, Melbourne, VIC, Australia
- 3Institute of Animal Ecology and Evolution, University of Veterinary Medicine Hannover, Foundation, Bünteweg, Hannover, Germany
- 4Department of Biochemistry and Pharmacology, University of Melbourne, Melbourne, VIC, Australia
- 5Department of Clinical Pathology, University of Melbourne, Melbourne, VIC, Australia
The innovation of multicellularity has driven the unparalleled evolution of animals (Metazoa). But how is a multicellular organism formed and how is its architecture maintained faithfully? The defining properties and rules required for the establishment of the architecture of multicellular organisms include the development of adhesive cell interactions, orientation of division axis, and the ability to reposition daughter cells over long distances. Central to all these properties is the ability to generate asymmetry (polarity), coordinated by a highly conserved set of proteins known as cell polarity regulators. The cell polarity complexes, Scribble, Par and Crumbs, are considered to be a metazoan innovation with apicobasal polarity and adherens junctions both believed to be present in all animals. A better understanding of the fundamental mechanisms regulating cell polarity and tissue architecture should provide key insights into the development and regeneration of all animals including humans. Here we review what is currently known about cell polarity and its control in the most basal metazoans, and how these first examples of multicellular life can inform us about the core mechanisms of tissue organisation and repair, and ultimately diseases of tissue organisation, such as cancer.
Introduction
Cell polarity refers to the intrinsic asymmetric distribution of macromolecules to distinct compartments of a cell to control directionality and coordinated polarisation. Cell polarity is associated with cell behaviours, such as migration and asymmetric cell division (Nelson 2003; Elsum et al., 2012; Butler and Wallingford 2017; Allam, Charnley, and Russell 2018; Stephens et al., 2018). The conservation through evolution of a vast majority of the cell polarity genes from basal metazoans to mammals highlights their significance and relevance in tissue architecture and cell behaviour (Goldstein and Macara 2007; Simons and Mlodzik 2008; Fahey and Degnan 2010; Elsum et al., 2012; Belahbib et al., 2018). Understanding cell polarity in basal metazoans may help unravel some of the mysteries of multicellularity and key processes that occurred during the transition from unicellular to multicellular organisms. The jump from unicellularity to multicellularity has occurred at least 25 times throughout evolution contributing to a complex tree of species, including: plants, fungi, amoeba and Bilateria to name a few (Grosberg and Strathmann 2007; Rokas 2008).
Advances in the understanding of the genetics of basal metazoans and unicellular organisms have provided opportunities to advance our understanding of signalling pathways that have been considered the main building blocks of multicellularity (Gerhart 1999). Here we extend this framework to include cell polarity signalling. Examination of multicellular events in unicellular organisms can shed light as to how the transition to multicellularity may have occurred and how early forms of cell polarity signalling may have enabled this. For example, Gram-negative bacteria P. aeruginosa demonstrates both kin selection (the progressive replication of a single cell to select for traits) (West et al., 2006) and cheating (the differential uptake of resources by certain cells, allowing for some cells to thrive at the cost of others) (Sandoz, Mitzimberg, and Schuster 2007; Dunny, Brickman, and Dworkin 2008). Another example is in the yeast species S. cerevisiae where cell polarity proteins Sro7 and Sro77, (homologues of D. melanogaster Lgl (Lethal 2) giant larvae)), regulate polarisation of the actin cytoskeleton and vesicle exocytosis (Lehman et al., 1999; Gangar et al., 2005; Hattendorf et al., 2007). There are different hypotheses as to how multicellularity occurred (King 2004; Knoll 2011; Richter and King 2013; Sogabe et al., 2019) of which all fundamentally agree on the significance of cells orientating spatio-temporally to allow for coordinated cell movement, migration, and adhesion.
A key concept in the exploration of multicellularity is co-option–the ability for a trait to switch and thus impact on function (McLennan 2008). Evolutionarily, this occurs in many different contexts and here can be demonstrated by the co-option of genes already present in the genome being redirected to polarising events to support multicellularity. Cell adhesion molecules are considered one of these key co-optive processes (Abedin and King 2010; Harden, Wang, and Krieger 2016), another example being the LAP family of adaptor genes containing leucin rich repeats (LRR) and PDZ domains giving rise to the Scribble cell polarity gene (Santoni et al., 2002). Of note, the choanoflagellate genome (the closest unicellular organism relating to animals) reveals a rich repertoire of adhesion, cell polarity and signalling genes (King 2004; Snell et al., 2006; King et al., 2008).
In this review we will highlight our current knowledge of cell polarity in basal metazoans to further understand the evolution and adaptation of cell polarity signalling. It should be noted that there is still vigorous debate as to the evolutionary order of these lower metazoan animals as to how they relate to the last known common ancestor of bilaterians. However, this extends outside the scope of this review and we direct the reader to other references that tackle this important issue (see. (Grosberg and Strathmann 2007; Nosenko et al., 2013; Srivastava 2015; Schierwater et al., 2021a).).
Transitioning to multicellularity: The first animals
Epithelial tissue is a key building block in the development of multicellularity due to the formation of epithelial sheets. ‘True’ epithelia is defined by 1) the presence of polarity between epithelial cells, 2) multiple junctions joining cells together, including: belt, septate, desmosome and tight junction, and 3) the presence of an extracellular matrix (Tyler 2003). The sheet formation acts as a barrier separating compartments of the organism, allowing for the regulation, diffusion and absorption of macromolecules (Tyler 2003; Fahey and Degnan 2012). To achieve such diverse functionality within an organism, epithelial cells need to be highly polarised, which is achieved by the asymmetric compartmentalising of cell polarity constituents (Rodriguez-Boulan and Nelson 1989; Elsum et al., 2012; Ebnet 2015; Wen and Zhang 2018). Basal metazoans are the first multicellular organisms and the ancient relatives to Bilateria, and more broadly the Eumetazoan subkingdom (Schierwater et al., 2021). They all contain examples of epithelial sheet formation, but only cnidarians have examples of true epithelia as explained above (Fahey and Degnan 2010; Rathbun, Everett, and Bergstralh 2022). The choanocytes in Poriferans (sponges) are considered epithelia-like (Simpson 1984) while the other epithelial cells lack key characteristics, like desmosomes and basal lamina (Fahey and Degnan 2010). Placozoans lack a basal lamina and key junctions associated with ‘true epithelia’. Although extracellular matrix (ECM) constituent genes such as collagen, integrin-β and laminin, are present and expressed in Placozoa. The absence of an actual ECM and basal lamina has been a peculiarity in the placozoans (Ringrose et al., 2013). These basal metazoans will be introduced briefly below.
Placozoa
Phylum Placozoa comprises flat sea-dwelling animals approximately 1–5 mm in diameter and 20 μm in height. They are morphologically considered to be one of the simplest animals with no distinguishable organs, nerve or muscle cells, basal lamina or extracellular matrix (Smith et al., 2014; DuBuc, Ryan, and Martindale 2019; Schierwater et al., 2021). The most well-known species of placozoans is Trichoplax adhaerens, although a number of other species have been described and studied (Schierwater et al., 2021; Neumann et al., 2022). Structurally, placozoans consist of six different cell types, with 80% of the animal comprised of epithelial cells (Smith et al., 2014). Most essential signalling pathways are present in placozoans, including Wnt, Notch, cell adhesion molecules, mitogen-activated protein kinase (MAPK) signalling, NFκB and TGF-β at both the transcriptome and proteome level (Srivastava et al., 2008; Ringrose et al., 2013; Belahbib et al., 2018). Placozoans show a very high regenerative potential including the ability to re-aggregate animals from single cells (A. Ruthmann and Terwelp 1979; Osigus et al., 2022). No placozoan has been identified as having cancer, even when exposed to high levels of radiation (Fortunato et al., 2021).
Porifera
Porifera, named due to their porous nature, encapsulates a diverse family of sponges. Their body plan consists of a labyrinth of small canals and chambers lined with choanocytes (cilia beating cells) that allows for the flow of water through the animal and the filtration of nutrients and microalgae (Soest et al., 2012). The well-studied marine Porifera Amphimedon queenslandica contains key regulatory, transcription, and signalling pathway genes including: Hox, Wnt, Hedgehog, TGF-ß, Notch, Jak/Stat, MAPK signalling pathway and cell adhesion molecules (Gerhart 1999; Nichols et al., 2006; Adamska et al., 2011; Wu et al., 2022). From a junctional perspective, Porifera have adherens junctions similar to those present in Bilateria, but there is no evidence of septate junctions or basal lamina (Srivastava et al., 2010). In the freshwater sponge E. muelleri, focal adhesion-like junctions and adherens junctions have been identified with highly conserved genes such as talin, integrin and focal adhesion kinase (Mitchell and Nichols 2019). Similar to other basal metazoans, sponges have the capacity to regenerate which has been reported to occur through the process of epithelial-to-mesenchymal transition (EMT) (Alexander et al., 2015; Lavrov et al., 2018; Ereskovsky et al., 2021; Wu et al., 2022).
Ctenophora
Ctenophores more commonly known as comb jellies, consist of over 200 species and differ from other basal metazoans in that they have a characteristic set of eight comb rows that run along their length (Pang and Mark 2008; Tamm 2014). Morphologically, ctenophores consist of an epithelial ectoderm and endoderm with a mesoglea layer containing collagen filaments (Freeman 1977; Harrison and Ruppert 1991; Simmons, Pang, and Martindale 2012). Gene analysis of the ctenophore M. leidyi reveals a canonical Wnt signalling pathway similar to bilaterians (J. F. Ryan et al., 2013). However, Ctenophores lack key signalling and polarity genes like Scribble and crumbs (Belahbib et al., 2018) and members required for non-canonical Wnt signalling pathway (J. F. Ryan et al., 2013). Similar to placozoans, many ctenophore species examined do not contain a basal lamina (Ringrose et al., 2013). In the ctenophore M. leidi, key binding domains such as the groove-binding motif and cytoplasmic binding domain of E-Cadherin showed a lack of conservation compared with Placozoa, Porifera and Bilateria (Belahbib et al., 2018). The analysis of Ctenophores show a lack of gene conservation and it has been suggested this is due to secondary loss (Belahbib et al., 2018). One example is the lack of the MAGUK protein Dlg in ctenophores, which is a highly conserved gene present well before basal metazoans e.g. choanoflagellates (Fahey and Degnan 2010; Belahbib et al., 2018; Schiller and Bergstralh 2021).
Cnidaria
Cnidarians encapsulate over 10,000 species that can be classified into two broad groups–sessile Anthozoa (e.g. the sea anemone Nematostella vectensis) and medusozoa (e.g. the freshwater Hydra vulgaris) (Technau and Steele 2011; Z.-Q. Zhang 2011). Similar to other basal metazoans, cnidarians have the capacity to regenerate lost or damaged body parts when both chemical or mechanical digestion occurs (P. M. Bode and Bode 1980; Layden, Rentzsch, and Röttinger 2016; Röttinger 2021). Studies of Hydra reveal the presence of ECM, cell-cell adhesion molecules, Wnt, hedgehog and notch signalling–which are all present and well conserved (Tucker and Adams 2014). A thorough review on the conservation of cell polarity signalling in Cnidaria has also recently been published (Rathbun, Everett, and Bergstralh 2022).
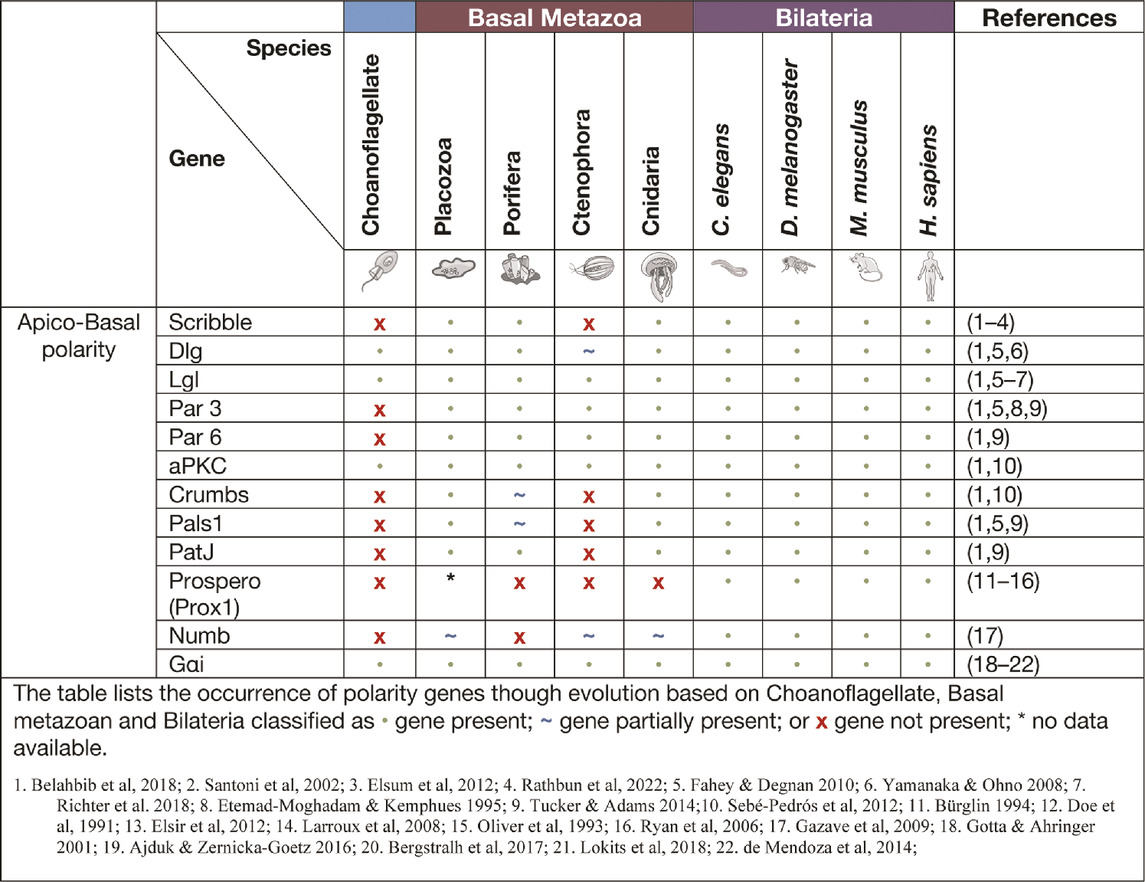
Core cell polarity signalling complexes in the basal metazoa. Several cell polarity signalling systems have developed through evolution, gaining complexity with evolving form and function of animal structures. In a few instances however, such as ctenophores or C. elegans, secondary loss of cell polarity genes have been observed (Belahbib et al., 2018). Analysis of genomic DNA sequences have identified the central cell polarity regulator complexes Scribble, Par and Crumbs in all basal metazoans (Srivastava et al., 2008; Fahey and Degnan 2010; Riesgo et al., 2014; Tucker and Adams 2014; Belahbib et al., 2018). These cell polarity signalling pathways remain fundamentally unexamined from a functional perspective in the basal metazoans. Here we seek to collate what is known of cell polarity signalling in basal metazoans, including the expression and function of cell polarity proteins, and to highlight the importance of these cell polarity mechanisms throughout evolution.
The par, crumbs, and scribble modules in apico-basal polarity regulation
Apico-basal polarity is largely specific to epithelial cells and involves the localisation of polarity modules to the apical and basolateral membranes (Figure 1A) (Bilder and Perrimon 2000; Nelson 2003; Margolis and Borg 2005). Apico-basal polarity is considered essential in the formation of epithelial sheet and barrier formation, a concept fundamental to metazoan development. The polarising events of apico-basal localisation within a cell allow for formation of junctions between cells. Notably zonula adherens and tight junctions in vertebrates, adherens and septate junctions in D. melanogaster and apical junctions in C. elegans (Alberts et al., 2002; Knust and Bossinger 2002; Guillot and Lecuit 2013). Apico-basal polarity is associated with three modules: Crumbs, Par and Scribble, that were first identified in the model organisms D. melanogaster and C. elegans (Bilder and Perrimon 2000; Nelson 2003; Margolis and Borg 2005). The spatial localisation of these modules, along with their mutually antagonistic relationship, allows for the establishment of tissue architecture (Figure 1A). Further, it allows for proper epithelial movement, junctional cell interaction, substrate secretion, cell proliferation and apoptosis, and regulation of cell signalling (Elsum et al., 2012; Margolis and Borg 2005; U. Tepass et al., 2001; Nelson 2003; Stephens et al., 2018). Disruptions to these polarity modules have been linked to a loss of tissue architecture, loss of junctional integrity, mis-localisation of other polarity proteins and aberrant cell signalling that can lead to increased cell proliferation and cancer (Bilder 2004; Elsum et al., 2012; Gödde et al., 2014; Stephens et al., 2018).
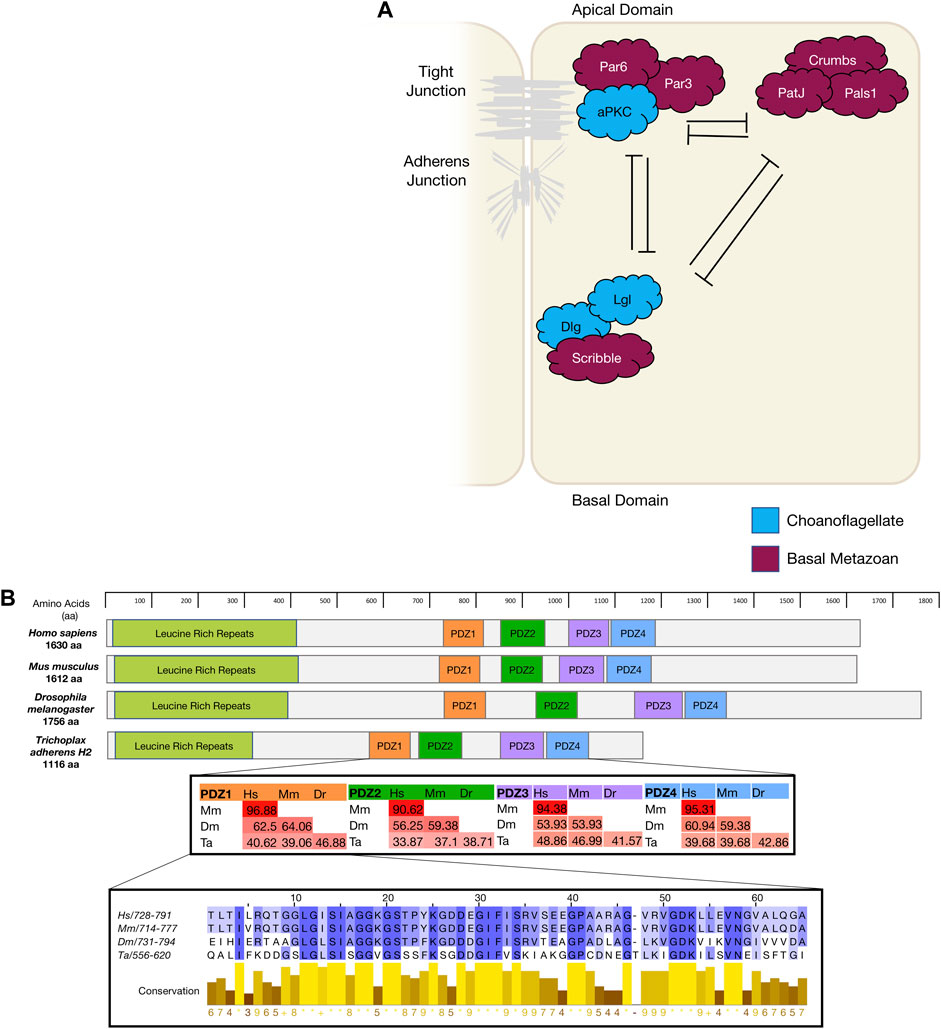
FIGURE 1. (A). The diagram depicts mammalian apico-basal polarity proteins and their interactions and localisation within an epithelial cell and the appearance of these genes in evolution. (B). Using Scribble as an example of gene conservation, a comparison has been made between the gene structure of four animals, a percentage map of key PDZ domains when compared between these four animals. PDZ1 has been expanded as an example of conservation and sequence similarity. Sequences were aligned in Clustal Omega and percentage conservation analysed in Jalview. Colour represented level of conservation. Sequences were sourced from uniport accessions: H. sapiens Q14160-1; M. musculus Q80U72-1; D. melanogaster Q7KRY7-1; and T. adherens A0A369S7Y8.
The par polarity complex first discovered in C Elegans (Kemphues et al., 1988) is considered to be a metazoan innovation (Fahey and Degnan 2010; Belahbib et al., 2018) and is responsible for the first asymmetric division in a zygote by establishing cortical polarity (Figure Figure1A and Figure 2) (Kemphues et al., 1988). The Par complex consists of scaffold proteins well known for their diverse roles in regulating cell polarity. In addition to asymmetric cell division, these proteins play an integral role in regulating many other polarity states including, apico-basal polarity, planar cell polarity and front-rear polarity (Petronczki and Knoblich 2001; Hurd et al., 2003; Goldstein and Macara 2007; Assemat et al., 2008; Etienne-Manneville 2008). The Par complex consists of three interacting proteins, Par3, Par6 and atypical protein kinase C (aPKC) that localise to junctional regions of epithelial cell. This allows for adherens junction and tight junction formation in vertebrates [Figure 1] (Matter and Balda 2003; St Johnston and Ahringer 2010; Wen and Zhang 2018).

FIGURE 2. The diagram represents protein localisation in the Drosophila neuroblast as it undergoes asymmetric cell division. Colours represent the evolutionary appearance of these genes. Green proteins are Drosophila neuroblast ACD specific.
Par complex genes have been identified in all the earliest basal metazoans and linked to a variety of polarity signalling contexts that co-evolved through evolution (Table 1) (Macara 2004; Magie and Martindale 2008; Belahbib et al., 2018). Indeed, this is illustrated by the strict evolutionary conservation of the interacting domains of Par proteins and the mechanisms regulating these interactions. For example, the lysine residue in PB1 (Phox and Bem1 binding module) domain of Par6 is responsible for the interaction between Par6 and aPKC, and the aPKC phosphorylation site (S/T) in Par3. This PBM domain remains highly conserved in all basal metazoans (Belahbib et al., 2018). Only a few functional experiments have been undertaken on the Par complex in basal metazoans. In the cnidarian N. vectensis functional investigation of the Par complex, a conserved role in maintaining cell-cell adhesion has been demonstrated. N. vectensis Par proteins (NvPar-3, NvPar-6, NvaPKC) were shown to localise within the cnidarian epithelium similarly to that seen in sheet epithelia of bilateria (Salinas-Saavedra et al., 2015; Salinas-Saavedra and Martindale, 2018). N. vectensis polyps expressing a dominant negative version of NvPar-3 showed leakage of fluorescent tracer dye demonstrating an ancestral role of the aPKC/Par complex in the maintenance of cell-cell adhesion and the paracellular boundary (SJs) of epithelial cells during animal development (Salinas-Saavedra and Martindale, 2018). Supporting this, knockout of Nvpar-6 and Nvpar-3 genes using CRISPR/Cas9 targeting resulted in loss of integrity of ectodermal epithelium including disruption of the cytoskeleton and adherens junctions (as visualised by ß-catenin localisation) (Salinas-Saavedra and Martindale, 2018). Clonal studies through single cell blastomere injections of CRISPR/Cas9 targeting Nvpar-3 showed that the resulting clones of NvPar-3 knockout epithelial cells also lost their structural integrity inducing in this case cell extrusion, thus demonstrating a cell-autonomous role for the Par Complex in regulation of epithelial cell polarity (Salinas-Saavedra and Martindale, 2018). Studies such as these reinforce the notion that these newly established polarity systems in the early metazoans played a critical role in the establishment of multicellularity.
The Crumbs polarity complex is well documented as a critical complex in the development and stabilisation of apical adherent and tight junctions (Dow and Humbert 2007; Bazellieres et al., 2009; Bivic 2013; Ebnet 2015). The Crumbs complex consists of two scaffold proteins, Pals1 (Protein associated Lin seven 1) and PatJ (Pals1-associated tight junction), and a transmembrane protein Crumbs (Tepass, 2012). The Crumbs complex proteins are all metazoan developments and first appear in basal metazoans (Belahbib et al., 2018). Crumbs was first discovered in D. melanogaster (U. Tepass et al., 1990) and is the central molecule that acts as a scaffold for PatJ and Pals1 (Figure 1A). Genomic analysis revealed that the placozoan T. adherens, the cnidarian N. vectensis, and the poriferan A. queenslandica have conserved domains of Crumbs. Whereas the ctenophore M. leidyi most strikingly had no crumbs or crumbs-like gene that has been identified (Table 1) (Belahbib et al., 2018). Furthermore, analysis of the genomic DNA sequence of A. queenslandica revealed multiple Crumbs-like coding regions that are either variants of the gene, pseudogenes or truncated forms (Fahey and Degnan 2010). However, there are some questions as to the functional capacity of Crumbs in A. queenslandica (Fahey and Degnan 2010; Srivastava et al., 2010; Belahbib et al., 2018). Structurally, Crumbs has extracellular epidermal growth factor (EGF) domains interspersed with laminin repeats and a cytoplasmic tail consisting of two motifs; the FERM-binding motif (FBM) and a Class II PDZ protein binding domain (PBM) essential for the function of Crumbs proteins (Knust, Tepaß, and Wodarz 1993; Bivic 2013). Of note, the FBM domain responsible for aPKC binding in higher order species is depleted of two phosphorylation sites in T. adherens and in studied sponges (Belahbib et al., 2018).
Pals1 is a member of the Membrane-Associated Guanylate Kinase (MAGUK) family. The MAGUK family includes cell polarity genes that cumulatively are responsible for the organisation of protein complexes within a cell or at a cell or synaptic junction. Their localisation governs the polarisation of cells and their cytoskeleton filament connections (Mendoza et al., 2010). MAGUK genes extend past the metazoan lineage and have been identified in the choanoflagellate M. brevicollis and protist C. owczarzaki (Mendoza et al., 2010). The other members of the Crumbs complex Pals1 and PatJ have not been identified in ctenophores and their presence in sponges is unclear. In the sponge A. queenslandica a relative of Pals1 gene MPP5/7, that is also a member of the MAGUK family, is present and may play a substitutional role in the Crumbs complex (Fahey and Degnan 2010; Bivic 2013).
The scribble polarity module consists of a triad of scaffold proteins, Scribble, Lgl (Lethal giant larvae) and Dlg (Discs large), that localise to the basolateral membrane of epithelial cells (Figure 1A). The module has an important role in the control of tissue architecture and morphogenesis, and in tumour suppression (Bilder et al., 2000; Humbert, Russell, and Richardson 2003). Proteins of the Scribble module are major regulators of epithelial apico-basal polarity with broader roles in other forms of cell polarity (Stephens et al., 2018). Dlg and Lgl have been identified in lower order unicellular species (choanoflagellates and fungi), whereas Scribble is considered a metazoan innovation (Srivastava 2015; Belahbib et al., 2018). Scribble is a member of the LAP (Leucine rich repeat and Post-synaptic density-95/Discs-Large/Zo-1) family. Structurally, Scribble contains 16 Leucine rich repeats (LRR, a highly conserved protein motif that forms an arc-like structure), a LAP-specific domain (a domain related to LRR) and four PSD-95, ZO-1 and Discs large (PDZ) domains that coordinate the majority of Scribble’s binding interactions (Figure 1B) (Bilder and Perrimon 2000; Humbert, Russell, and Richardson 2003; Stephens et al., 2018; Bonello and Peifer 2019). A Scribble or Scribble-like gene has not been identified in unicellular organisms and does not appear to be present in the ctenophore M. leidyi (Belahbib et al., 2018). In ctenophores, it is thought to be due a secondary loss of the gene, and while no functional studies have been completed, it is postulated that the absence of a Scribble gene may result in variations of polarity complex localisation (Belahbib et al., 2018). As noted above, ctenophores also appear to lack a Dlg gene (Schiller and Bergstralh 2021). Analysis of different porifera classes identified key polarity proteins, including Scribble, Lgl and Dlg, responsible for cell adhesion and epithelial development (Fahey and Degnan 2010; Riesgo et al., 2014). Dlg from a structural perspective, contains three PDZ domains, SH3 domain and a GUK domain. As a scaffold protein, Dlg is part of the Post-Synaptic Density (PSD) family. This family is responsible for the maintenance, anchorage and structural localisation of other PSD structures and proteins in relation to neurotransmitter receptors and signalling channels (Sakarya et al., 2007; Alié and Manuel 2010). In metazoan species that do not contain nerve structures, such as Placozoans and Porifera, it was found that these PSD proteins were present and contained near identical interacting domains when compared to their mammalian counterparts. Furthermore, it is suggested that Dlg and other PSD genes like Homer (scaffold protein involved in Ca2+ signalling and transport) may play significant roles in these metazoan species as Ca2+ receptors and signalling communicators (Alié and Manuel 2010). The significance of PSD proteins, specifically Dlg, is highlighted by the full conservation of their residues that interact with PDZ domains compared with their human orthologues (Sakarya et al., 2007). Of note, imaging of Placozoan epithelium using staining with a pan-human Dlg antibody show an identical basolateral cortical staining to that seen for Dlg in Bilateria epithelium suggesting that TaDlg may have a conserved function in the regulation of Trichoplax epithelium (Smith et al., 2014). Lgl is the most ancient gene with homologues found in yeast (Sro7 and Sro77) where it regulates polarised exocytosis (X. Zhang et al., 2005; Grosshans et al., 2006; Müsch et al., 2002). This has been similarly compared to mammalian Lgl in basolateral exocytosis (Müsch et al., 2002). High levels of conservation of polarity genes from the Scribble, Par and Crumbs complexes have been identified in cnidarians when compared to bilateria (Rathbun, Everett, and Bergstralh 2022).
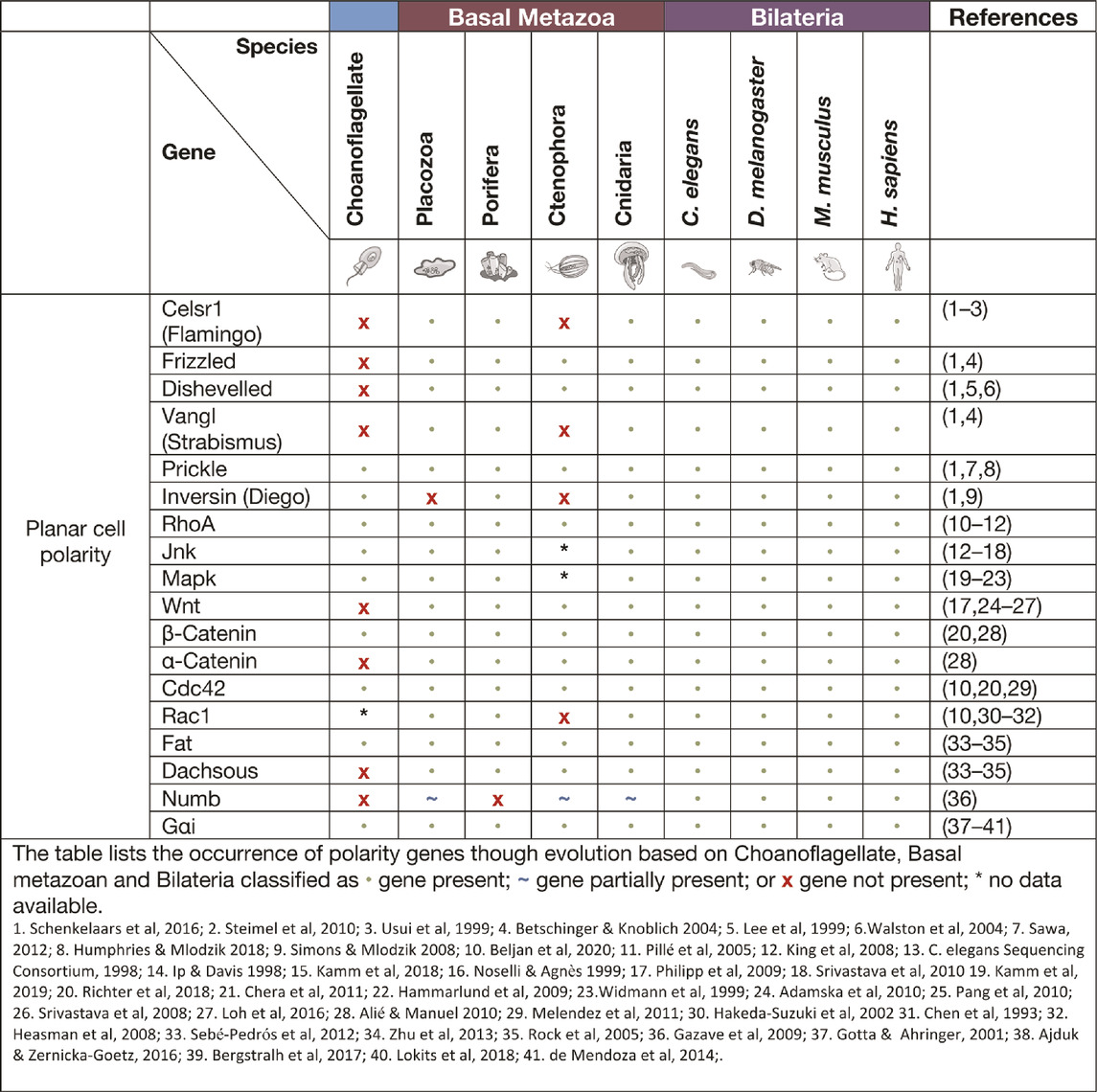
Planar cell polarity signalling
Planar cell polarity (PCP), also referred to as tissue polarity or the non-canonical Wnt signalling pathway, is the global organisation of cells along a x/y axis in a plane (Figure 3). PCP signalling is essential for normal tissue development, cell homeostasis, axis determination and tissue morphogenesis (Simons and Mlodzik 2008; Butler and Wallingford 2017). Junctional PCP genes were first identified and have been extensively studied in the fly D. melanogaster (Gubb and García-Bellido 1982; Axelrod 2001; Adler 2002; Hale and Strutt 2015). The organisation of six transmembrane proteins on opposing sides of a cell allow for communication and coordinated interactions, including polarising events. The polarising events allow for the asymmetric placement of cilia or hairs and the orientation of the mitotic spindle (Goodrich and David 2011; Schenkelaars et al., 2016; Butler and Wallingford 2017). Downstream from the core PCP signalling, the PCP protein Dishevelled interacts with Lgl, Cdc42, RhoA and Rac1. These interactions aid in cytoskeleton re-organisation, maintaining adherens junctions, and when interacting with Jnk, feeds into Wnt signalling pathway (Figure 3) (Milgrom-Hoffman and Humbert 2018; Wiese, Nusse, and van Amerongen 2018). On examination of PCP signalling, Frizzled, Dishevelled and Prickle have all been identified in the four basal metazoans, whereas Celsr1 (Flamingo) and Vangl (Strabismus) are not found in ctenophores, nor Inversin (Diego) in porifera (Table 2) (Adamska et al., 2010; Srivastava et al., 2008; Schenkelaars et al., 2016; Belahbib et al., 2018; J. F. Ryan et al., 2013; Momose, Kraus, and Houliston 2012). Phylogenetic analysis of prickle and prickle-like genes reveals an ancestor of the gene in choanoflagellates. In the basal metazoans placozoa and cnidaria, it diverges from one to two genes–prickle and testin (Schenkelaars et al., 2016).
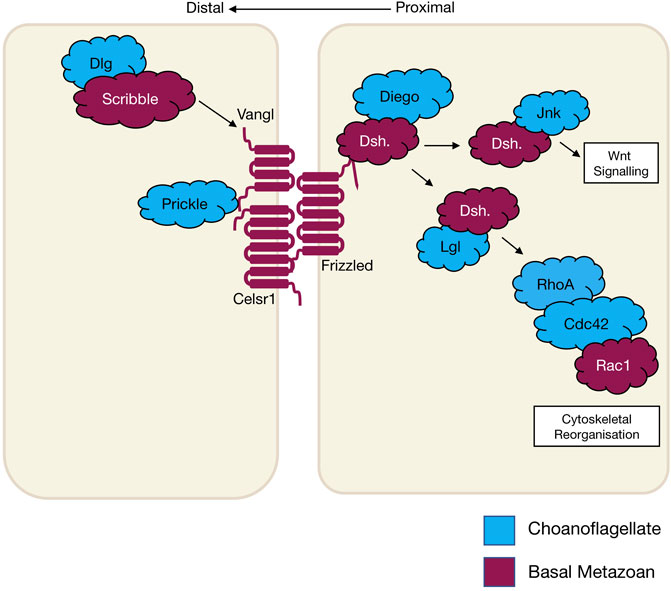
FIGURE 3. The schematic diagram shows core planar cell polarity pathways and their relationship to Wnt signalling and cytoskeletal reorganisation that occurs in wound healing. The different colours represent the evolutionary appearance of these genes.
Examination of the cnidarian C. hemisphaerica larva stages reveal established PCP characteristics of oral-arboreal polarity and the formation of directionally organised cilium in each epithelial cell similar to that described in bilaterians (Momose, Kraus, and Houliston 2012; Milgrom-Hoffman and Humbert 2018). Further, vangl mRNA expression levels were evident throughout embryogenesis, elongation and ciliogenesis with enrichment occurring to the axis of the developing hydrozoan (Momose, Kraus, and Houliston 2012). When vangl was knocked down in the cnidarian N. vectensis, the embryos failed to undergo gastrulation or primary invagination, however this did not impact ß-catenin nuclear localisation, which in bilaterians is tightly coupled. Thus cell fate specification of the endoderm may have developed separately to other PCP/Wnt signalling pathways (Kumburegama et al., 2011).
Non-canonical Wnt signalling investigations in the cnidarian Hydra revealed specific Wnt pathway genes (wnt5, wnt8, frizzled and dishevelled) are all required for correct evagination of the bud and tentacle of the Hydra. The upregulation of these genes can be correlated to the activation of Wnt/ß-catenin signalling during tissue morphogenesis and development of the Hydra pulp (Philipp et al., 2009). Planar cell polarity genes fat and fat-like genes are associated with cell directional migration and morphogenesis in asymmetric cell division in bilaterians (Matis and Axelrod 2013). The Hydra fat and dachsous genes localise to the body of the animals where continuous growth and migration of cells occur supporting the theory of a similar role to that of bilaterians (Brooun et al., 2019). Phylogenetic examination of frizzled in a variety of different poriferan, placozoan, cnidarian and ctenophore species show multiple orthologues of frizzled. In some porifera and cnidarians, up to four frizzled orthologues have been identified, with evidence that the vertebrate paralogue of frizzled is an amalgamation of ancestral frizzled genes (Schenkelaars et al., 2015). The genes flamingo, inversin and vangl are PCP genes considered to be secondarily lost from the ctenophore M. leidyi (Table. 2) (J. F. Ryan et al., 2013; Schenkelaars et al., 2016), whereas, dishevelled, frizzled and prickle are present in all metazoans (Srivastava et al., 2008; Schenkelaars et al., 2016). Functional studies relating to specific pathway significance between basal metazoan PCP signalling and its similarities or differences to higher order species are ongoing.
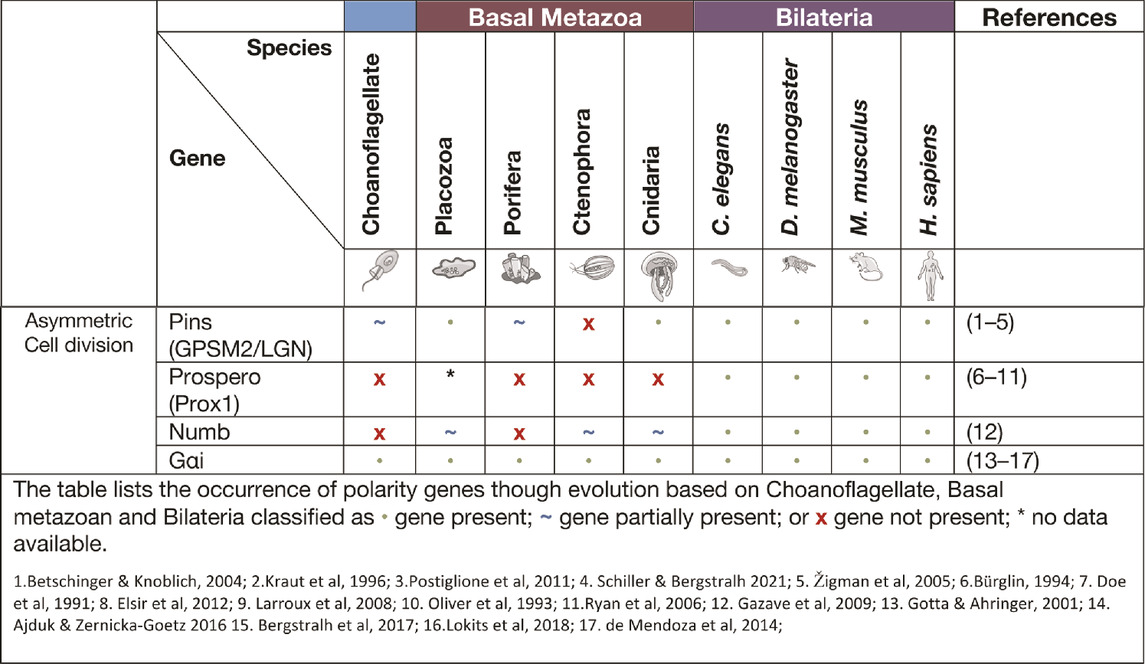
Asymmetric cell division signalling
Asymmetric cell division (ACD) refers to the specific localisation of cell fate determinants during cell division to establish two different cell characteristics (mother/daughter) (Knoblich 2001). In early cell division in the model organism D. melanogaster, asymmetric molecules Par3, Par6, aPKC, Inscuteable, Pins, Gαi and Mud localise to the apical cortex of the mitotic spindle, while cell fate factors Numb, Brat, Prospero, Pon and Miranda localise to the basal cortex (Figure 2) (Boyd et al., 1996; Tabuse et al., 1998; Hung and Kemphues 1999; Joberty et al., 2000; Kelsom and Lu 2012). Additionally, in Drosophila neuroblasts, the Scribble module proteins Scribble, Dlg and Lgl are important in ACD where they assist in mitotic spindle orientation (Elsum et al., 2012). The asymmetric localisation of these key polarity genes induces separation of the cells in an asymmetric fashion and therefore diversification of tissue types. A failure for polarity proteins to localise to the poles of the mitotic spindle is associated with defects in basal protein targeting, symmetric division, reduced spindle size or inverted neuroblast cell division (Bilder and Perrimon 2000; Albertson and Doe 2003; Neumüller and Knoblich 2009; Royer and Lu 2011). The diversification of ACD has been identified in prokaryote and eukaryotic organisms, basal metazoans and bilaterians (Table 3) (K. R. Ryan and Shapiro 2003; Knoblich 2001). It should be noted that cells at an early embryonic stage have the capacity to divide either asymmetrically, as described above, or symmetrically where two identical daughter cells are formed (Knoblich 2001; Schenkelaars et al., 2017). The selective differential distribution of protein and RNA into daughter cells is the foundation for the development of different tissue or cell types within an organism, referred to as cell fate (Jan and January 1998; Knoblich 2010).
Asymmetric cell division allows for the development of both germ cells and somatic cells that form different cell lineages and allow for plasticity of the cells in processes, such as reaggregation. In the cnidarian H. vulgaris, multipotent interstitial cells have the capacity to differentiate into gametes and almost all somatic cell lines (Bosch 2004; H. R. Bode 1996; Bosch and David 1987). The pliability of cnidarian cells and their capacity to adapt to their environment is remarkable, with the examples of an adult medusa metamorphosis into a polyp (Piraino et al., 1996). Another example is the cnidarian Podocoryne carnea that through the process of asymmetric cell division can differentiate medusae formed cells into an unrelated phenotype e.g. Muscle cells to nerve cells (Schmid and Alder 1984; Seipel, Yanze, and Schmid 2003). One of the proteins associated with ACD is Pins (also known as LGN or GPSM2). Pins has been shown to interact closely with Dlg in spindle orientation and this interaction is believed to have evolved in cnidarians (Schiller and Bergstralh 2021). The placozoan T. adhaerens and the sponge A. queenslandica do not contain the key linker regions required for GPSM2 to interact with Dlg, however they do contain other key motifs of GPSM2. It is postulated that these conserved regions may still be able to play a part in ACD, cell orientation and division (Schiller and Bergstralh 2021). In Porifera, during initial embryonic development, asymmetric division of macromeres to micromeres occur while later in embryonic development there is more evidence for higher levels of symmetric cell divisions. In the freshwater sponge E. fluviatilis the paralogue gene Musashi (a gene required for stem cell maintenance in Drosophila) has been identified as being specifically expressed in stem cells and regulates sustainable regeneration. This is the earliest occurrence of this gene in basal metazoans and of its role in ACD (Okamoto et al., 2012).
Cell junction complexes in the basal metazoa
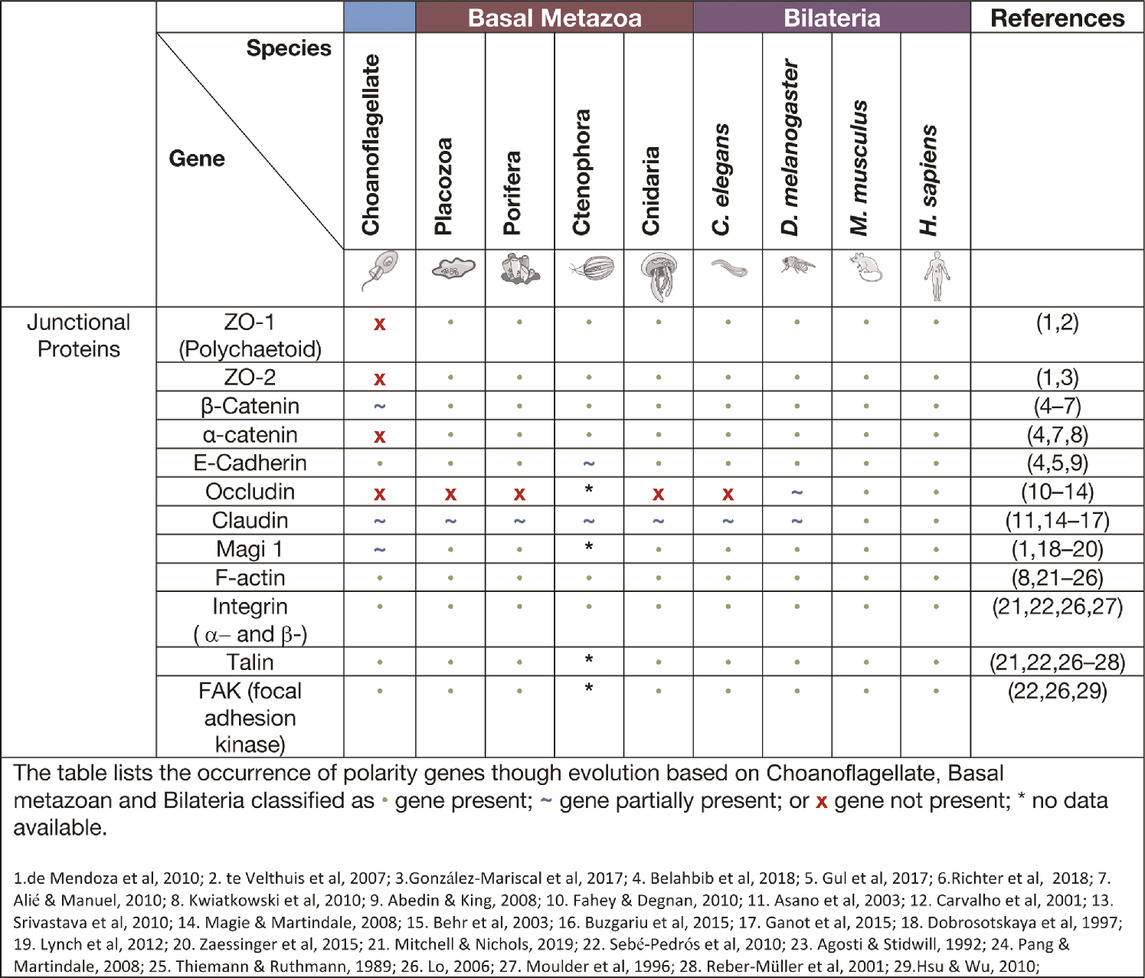
Adherens junctions
Adherens junctions, also known as Zonula Adherens, form belt-like junctions that act as a conduit between the apical and basal domains of epithelial cells (Figure 4). Adherens junctions are acknowledged as the most common junction in animal epithelia (Oda and Takeichi 2011; Hiroki 2012). Adherens junctions have been identified in placozoans, cnidarians and ctenophores with none so far identified in Porifera (table. 4) (T. J. C. Harris and Ulrich 2010; Salinas-Saavedra and Martindale, 2019). The presence of adherens junctions in placozoans appears crucial for their tissue integrity as no other junctions have been identified placozoans to date (Smith and Reese 2016).

FIGURE 4. Bilaterian representation of epithelial cell-cell and cell-ECM junctions and their emergence in evolution. It should be noted that for Claudin, whilst represented as a basal metazoan innovation, it has only been identified in Cnidaria. There are Occludin-like genes present in basal metazoans but it is not known if they have the same functional properties as in Bilateria.
Cadherin-catenin complexes
A major component of adherens junctions are cadherin-catenin complexes. Classical cadherins date back to the Urmetazoan (the hypothetical last common ancestor of all animals or metazoans) and are type I transmembrane proteins that consist of calcium-dependent transmembrane cell adhesion molecules (CAMs) that form adherens junctions associated with cell-cell adhesion, embryonic development, and cell morphogenesis (Hulpiau and van Roy 2011; T. J. C. Harris and Ulrich 2010; King, Hittinger, and Carroll 2003; Gul et al., 2017). The cadherins are a superfamily of proteins containing at least two cadherin repeats and can be classified into three families: Major cadherins, protocadherins and cadherin-related genes (Gul et al., 2017). Placozoans contain cadherin and cadherin-related genes, whereas cnidaria contain multiple genes of all three cadherin families (Gul et al., 2017; Hulpiau, Gul, and van Roy 2013; S. A. Nichols et al., 2012). When examining the current literature of cadherins in the basal metazoans it was found that placozoans, poriferans and cnidarians all have identifiable E-cadherin with necessary binding motifs (Table 4). M. leidyi (Ctenophora) has E-cadherin motifs but show high levels of divergence that raises doubt to its capacity to bind to known interacting genes such as ß-catenin and p120 (Belahbib et al., 2018; Hulpiau and van Roy 2011; S. A. Nichols et al., 2012; Ringrose et al., 2013; Srivastava et al., 2008).
Catenins that form part of adherens junctions can be placed into three sub-families: p120-, α- and β-, with examples of each subfamily identified in the basal metazoans, with β-catenin being identified in many unicellular organisms (Alié and Manuel 2010; Belahbib et al., 2018). Catenins, including α- and β-catenin bind filamentous actin (F-actin) within the cell and cadherins within the adherens junctions to form a semi-permeable barrier between anterior and posterior of the cell (Baum and Georgiou 2011; Gooding, Yap, and Ikura 2004; Tian et al., 2011; Nelson 2008; T. J. C. Harris and Ulrich 2010; Magie and Martindale 2008). A major contributor to adherens junction homeostasis is the presence of β-catenin in higher order metazoans and some basal metazoans. In the ctenophore M. leidyi, β-catenin does not localise to the cell junctions most likely due to the lack of a cytoplasmic domain essential for β-catenin binding. It was further concluded that this may indicate that the ancestral role of β-catenin was in cell-fate specification associated with Lef/Tcf co-factors that enter the nucleus to regulate canonical Wnt signalling rather than cell adhesion (Salinas-Saavedra and Martindale, 2019). The porifera A. queenslandica contain junctional proteins cadherin1 and α-catenin1-like gene, however within the middle of the gene a stretch sequence has been identified that is not otherwise seen in bilaterian counterparts (Fahey and Degnan 2010). The evidence is still lacking regarding other regulators of adherens junctions, except for the Par complex as discussed previously. Indeed, adherens junctions in the cnidarian N. vectensis ectodermal epithelial cells are responsible for the localisation of the Par complex and if disrupted a loss of integrity and loss of solute permeability has been observed (Salinas-Saavedra and Martindale, 2018).
Tight and septate junctions
Tight junctions are attributed to vertebrate species and act as a junctional barrier regulating the diffusion of macromolecules between and through cells (Matter and Balda 2003). Located at the apical region of cells, tight junctions consist of transmembrane signalling proteins, such as Claudin, Occludin, junctional adhesion molecules (JAMs), and adaptor proteins, such as ZO (Zonula Occludin) -1, -2, -3, polarity proteins Par -3, -6, Pals1, PatJ and Magi -1,-2 and -3 (Figure 4) (Fanning et al., 1998; Tsukita, Furuse, and Itoh 2001; Matter and Balda 2003; Niessen 2007; Steed, Balda, and Matter 2010; Hartmann et al., 2020). ZO-1 as a member of the MAGUK family is responsible for junctional organisation and regulation of proteins, such as ZO-2, Occludin and F-actin. These interactions allow linking and binding to the cortical actin cytoskeleton of the cell (Fanning et al., 1998; Itoh et al., 1999). The ZO proteins have been identified in all four basal metazoan lineages (Table 4) (Mendoza et al., 2010). Interestingly, electron microscopy studies have failed to reveal tight junctions in the placozoan T. adherens. This is a peculiarity as the genome contains Z O -1 and Claudins that are associated with tight junctions (Mendoza et al., 2010; González-Mariscal et al., 2017; Belahbib et al., 2018). Although tight junctions are ‘stricto-sensu’ vertebrate specific, genes associated with tight junctions have been identified in invertebrates, basal metazoans and choanoflagellates and hence referred to as ‘claudin-like’ (Ganot et al., 2015). For example, In the cnidarian Hydra, 14 claudin-like genes have been identified, with 10 of them specifically in the ectoderm and/or endoderm (Buzgariu et al., 2015) and claudin-like genes in Drosophila have been associated with septate junctions (Behr, Riedel, and Schuh 2003). Septate junctions are cell-cell junctions that appear ladder-like under electron microscope and aid in solute diffusion and structural support (Matter and Balda 2003). Septate junctions have been identified in Hydra, containing a ladder-like structure that in reaggregation studies forms within hours (Filshie and Flower 1977; Seybold, Salvenmoser, and Hobmayer 2016). A similar structure has also been noted in Trichoplax in the proximal cells of the animal that appear ‘ladder-like’ but are periodic in nature. No such junctions have been identified in porifera (Ruthmann et al., 1986; Ganot et al., 2015). Cnidarians display both the required genes and structure to form septate junctions similar to those found in Bilateria (Ganot et al., 2015; Rathbun, Everett, and Bergstralh 2022). This is not seen in ctenophores, where only claudin-like genes have been identified (Ganot et al., 2015).
Focal adhesions and integrin complexes
Focal adhesions are protein-rich structures where the integrin transmembrane proteins provide adhesion between cells and the extracellular matrix (ECM) (Figure 4). Like cadherins, integrins also represent signalling hubs (Michael and Parsons 2020). Focal adhesion genes surpass the age of the earliest metazoan lineages, believed to stretch back into the Cambrian time (S. A. Nichols et al., 2012). Whereas, components of the integrin machinery predate the metazoan lineage (Sebé-Pedrós et al., 2010). Integrin receptors that are composed of membrane-anchored heterodimer receptors have been reported in species of marine sponges (Müller 2003). More recently, the focal adhesion proteins, integrin, talin, and focal adhesion kinase (FAK), have been shown to form a complex that localises to the cell-cell junctions and extracellular matrix adhesions in the freshwater sponge E. mulleri (Mitchell and Nichols 2019). Of note, focal adhesion associated molecules integrin, vinculin, paxillin, talin and FAK are all found and expressed in Trichoplax (Srivastava et al., 2008), although a basement membrane structure does not appear present in these animals suggesting either a secondary loss in Placozoa or an independent gain in the other basal metazoans (Fidler et al., 2017).
Cell polarity in the basal metazoa and the origin of multicellularity
Here we have reviewed key signalling pathways regulating cell polarity and adhesion in the basal metazoan species and how this relates to the evolution of complex tissues such as epithelial structures. The examination of such pathways not only gives us knowledge into the ancient function of these genes and when they arose, but allows us to examine their role in the advent of multicellularity. A key challenge for a multicellular organism is to organise tissue architecture to drive cellular and organismic function. For the evolution of a multicellular animal to occur, a number of events are required including the development of cell differentiation and adhesive cell interactions within the epithelium, the orientation of division axis, and the ability to reposition daughter cells over long distances so as to establish and maintain a body plan. In addition, to obtain the division of labour that is linked with multicellularity, the process of differentiation that generates various cell types must be properly controlled. Asymmetry and cell polarity provides a universal tool for building multicellular tissue architecture. Although asymmetry can occur by stochastic means, extrinsic cues whether chemical or mechanical are more reliable and provide robustness to generate the asymmetry required for tissue architecture. Cell polarity and cell adhesion mechanisms relay these external cues internally to re-organise cell and tissue as well as provide a link with transcriptional programs required for tissue morphogenesis.
Apico-basal cell polarity mechanisms first appear in basal metazoans, and based on the simultaneous presentation of multicellularity and these cell polarity constituents, it is reasonable to propose that cell polarity mechanisms played a key role in this process. As discussed, other cell polarity genes are far more ancient and extend back into unicellular organisms. For example, β-catenin’s ancestral function appears related to TCF/LEF transcriptional regulation of Wnt signalling rather than junctional polarity and thus provides another example of co-option in cell polarity systems linking nuclear transcriptional programs to newly minted cell adhesion mechanisms (Salinas-Saavedra and Martindale, 2019). Interestingly, of the basal metazoans, ctenophores appear to be outliers at this point in terms of cell polarity mechanisms. The significant cell polarity associated gene loss in ctenophores raises interesting questions as to the alternative mechanisms by which ctenophores control various aspects of cell polarity, tissue organisation and its repair. In addition, the potential role of tight junction proteins in basal metazoans is an interesting enigma and could provide new insights into the evolution of these permeability barriers. As discussed, many basal metazoans produce tight-junction proteins (e.g. Zo-1) despite the absence of tight or septate junctions. This may indicate a more ancient divergent function for these genes that has been co-opted for the regulation of tight junctions. Interestingly, Polychaetoid, the Drosophila ZO-1 homologue, localises to adherens junctions and provides a link to actin regulation (Takahashi et al., 1998; Wei and Ellis 2001; Choi et al., 2011), pointing to a possible similar role for ZO-1 in basal organisms.
Research in the basal metazoans provides the opportunity to understand the fundamental building blocks of multicellularity and by extension its relationship to key events such as cancer. Indeed, examination of the evolutionary origin of cancer-related protein domains suggests two peaks, one at the time of the origin of the first cell and the other around the time of the evolution of the first multicellular organisms (Domazet-Lošo and Tautz 2010). Importantly, this second peak dubbed “gate-keeper” genes consist of oncogenes and tumour suppressors whose mutations promote tumour progression through altering cell proliferation, inhibiting differentiation or inhibiting cell death. This second peak also corresponds to the advent of the cell polarity signalling pathways in early basal metazoans described in this review. As many of these cell polarity regulators have been linked to tumour suppression in Bilateria (Stephens et al., 2018), examining the mechanisms of cell polarity and tissue architecture regulation in basal metazoans is likely to lead to fundamental insights into the origins of cancer. Almost all bilaterian animals have reported examples of cancer formation (Aktipis et al., 2015). Indeed, sponges and Hydra have reported cases of cancer that mimic that of higher order species, such as intrusive proliferation, loss of tissue architecture and a loss of specialised tissue (Hanahan and Weinberg 2000; Aktipis et al., 2015). The advent of multicellularity required new molecular mechanisms that allowed cellular cooperation and suppressed any cellular conflicts that enhance individual cell fitness to the detriment of the organism (Aktipis et al., 2015; Madan, Gogna, and Moreno 2018; Bowling, Lawlor, and Rodríguez 2019). From this point of view, cancer would represent a breakdown of this multicellular cooperation with over-competitive cells effectively “cheating”, leading to overall loss of fitness of the organism (Rainey 2007; Aktipis et al., 2015). Importantly, cell polarity and tissue architecture regulators play key roles in regulation of cell competition mechanism in Bilateria (Madan, Gogna, and Moreno 2018; Bowling, Lawlor, and Rodríguez 2019; Fahey-Lozano et al., 2019; Baker 2020). We therefore contend that cell competition mechanisms first appeared in basal metazoans and are mechanistically linked to the acquisition of the original cell polarity mechanisms required for the advent of multicellularity. The ability to generate tissue chimaeras in basal metazoans such as Trichoplax and Hydra (Klimovich, Wittlieb, and Bosch 2019; Schierwater et al., 2021) provides an attractive system to explore how cell competition mechanisms may have first appeared in basal metazoans to both control tissue architecture and enable cancer prevention.
The study of cell polarity and how it helped generate multicellularity in the basal metazoans represents a rich opportunity to identify the original mechanisms that establish and maintain the organisation of tissues. Because of the high conservation in gene function between basal metazoans and Bilateria, these studies are also likely to provide broader insights into regenerative medicine and human cancer. Furthermore, identification of any divergent cell polarity mechanisms between basal metazoan and bilaterians will inform us as to the diversity and evolution of these core cellular mechanisms.
Author contributions
BW: writing, editing, data collection and analysis; PH, MK, BS: editing.
Acknowledgments
The authors would like to thank Sarah Russell and Helena Richardson for their constructive and thorough reading of this manuscript. Many thanks to Yuliya Stepkina for assisting with the figure artwork. We would also like to thank the reviewers for their helpful comments that markedly helped improve this manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abedin, M., and King, N. (2008). The premetazoan ancestry of cadherins. Science 319 (5865), 946–948. doi:10.1126/science.1151084
AbedinKing, M. N. (2010). Diverse evolutionary paths to cell adhesion. Trends Cell. Biol. 20 (12), 734–742. doi:10.1016/j.tcb.2010.08.002
Adamska, M., Degnan, B. M., Green, K., and Zwafink, C. (2011). What sponges can tell us about the evolution of developmental processes. Zoology 114 (1), 1–10. doi:10.1016/j.zool.2010.10.003
Adamska, M., Larroux, C., Adamski, M., Green, K., Lovas, E., Koop, D., et al. (2010). Structure and expression of conserved wnt pathway components in the demosponge Amphimedon queenslandica. Evol. Dev. 12 (5), 494–518. doi:10.1111/j.1525-142X.2010.00435.x
Adler, P. N. (2002). Planar signaling and morphogenesis in Drosophila. Dev. Cell. 2 (5), 525–535. doi:10.1016/S1534-5807(02)00176-4
Agosti, C., and Stidwill, R. P. (1992). The contributions of microtubules and F-actin to the in vitro migratory mechanisms of Hydra nematocytes as determined by drug interference experiments. Exp. Cell. Res. 200 (1), 196–204. doi:10.1016/S0014-4827(05)80088-6
Ajduk, A., and Zernicka-Goetz, M. (2016). Polarity and cell division orientation in the cleavage embryo: From worm to human. Mol. Hum. Reprod. 22 (10), 691–703. doi:10.1093/molehr/gav068
Aktipis, C. M., Hibner, U., Hochberg, M. E., Maley, C. C., and Wilkinson, G. S. (2015). Cancer across the tree of life: Cooperation and cheating in multicellularity. Philos. Trans. R. Soc. Lond. B Biol. Sci. 370 (1673), 20140219. doi:10.1098/rstb.2014.0219
Alberts, B., Johnson, A., Lewis, J., Raff, M., Roberts, K., and Walter, P. (2002). Cell junctions. 1065–1126.
Albertson, R., and Doe, C. Q. (2003). Dlg, scrib and Lgl regulate neuroblast cell size and mitotic spindle asymmetry. Nat. Cell. Biol. 5 (2), 166–170. doi:10.1038/ncb922
Alexander, B. E., Achlatis, M., Osinga, R., Harm, G., Cleutjens, P. M., Schutte, B., et al. (2015). Cell kinetics during regeneration in the sponge halisarca caerulea: How local is the response to tissue damage? PeerJ 3, e820. doi:10.7717/peerj.820
Alié, A., and Manuel, M. (2010). The backbone of the post-synaptic density originated in a unicellular ancestor of choanoflagellates and metazoans. BMC Evol. Biol. 10, 34. doi:10.1186/1471-2148-10-34
Allam, A. H., Charnley, M., and Russell, S. M. (2018). Context-specific mechanisms of cell polarity regulation. J. Mol. Biol. 430 (19), 3457–3471. doi:10.1016/j.jmb.2018.06.003
Asano, A., Asano, K., Sasaki, H., Furuse, M., and Tsukita, S. (2003). Claudins in Caenorhabditis elegans: Their distribution and barrier function in the epithelium. Curr. Biol. 13 (12), 1042–1046. doi:10.1016/S0960-9822(03)00395-6
Assemat, E., Bazellieres, E., Pallesi-Pocachard, E., Le Bivic, A., and Massey-Harroche, D. (2008). Polarity complex proteins. Biochim. Biophys. Acta 1778 (3), 614–630. doi:10.1016/j.bbamem.2007.08.029
Axelrod, J. D., Werner, E. R., Leitner, S., Grobner, P., and Werner-Felmayer, G. (2001). Nitric oxide synthase is induced in sporulation of Physarum polycephalum. Genes. Dev. 15 (10), 1299–1309. doi:10.1101/gad.890501
Baker, N. E. (2020). Emerging mechanisms of cell competition. Nat. Rev. Genet. 21 (11), 683–697. doi:10.1038/s41576-020-0262-8
Baum, B., and Georgiou, M. (2011). Dynamics of adherens junctions in epithelial establishment, maintenance, and remodeling. J. Cell. Biol. 192 (6), 907–917. doi:10.1083/jcb.201009141
Bazellieres, E., Assemat, E., Arsanto, J. P., Le Bivic, A., and Massey-Harroche, D. (2009). Crumbs proteins in epithelial morphogenesis. Front. Biosci. 14, 2149–2169. doi:10.2741/3368
Behr, M., Riedel, D., and Schuh, R. (2003). The claudin-like megatrachea is essential in septate junctions for the epithelial barrier function in Drosophila. Dev. Cell. 5 (4), 611–620. doi:10.1016/S1534-5807(03)00275-2
Belahbib, H., Renard, E., Santini, S., Jourda, C., Borchiellini, C., Le Bivic, A., et al. (2018). New genomic data and analyses challenge the traditional vision of animal epithelium evolution. BMC Genomics 19 (1), 393. doi:10.1186/s12864-018-4715-9
Beljan, S., Herak Bosnar, M., and Ćetković, H. (2020). Rho family of ras-like GTPases in early-branching animals. Cells 9 (10), E2279. doi:10.3390/cells9102279
Bergstralh, D. T., Dawney, N. S., and Johnston., D. S. (2017). Spindle orientation: A question of complex positioning. Development 144 (7), 1137–1145. doi:10.1242/dev.140764
Betschinger, J., and Knoblich, J. A. (2004). Dare to Be different: Asymmetric cell division in Drosophila, C. Elegans and vertebrates. Curr. Biol. 14 (16), R674–R685. doi:10.1016/j.cub.2004.08.017
Bilder, D. (2004). Epithelial polarity and proliferation control: Links from the Drosophila neoplastic tumor suppressors. Genes. Dev. 18 (16), 1909–1925. doi:10.1101/gad.1211604
Bilder, D., Li, M., and Perrimon, N. (2000). Cooperative regulation of cell polarity and growth by Drosophila tumor suppressors. Science 289 (5476), 113–116. doi:10.1126/science.289.5476.113
Bilder, D., and Perrimon, N. (2000). Localization of apical epithelial determinants by the basolateral PDZ protein Scribble. Nature 403 (6770), 676–680. doi:10.1038/35001108
Bivic, A. L. (2013). Evolution and cell physiology. 4. Why invent yet another protein complex to build junctions in epithelial cells? Am. J. Physiol. Cell. Physiol. 305 (12), C1193–C1201. doi:10.1152/ajpcell.00272.2013
Bode, H. R. (1996). The interstitial cell lineage of Hydra: A stem cell system that arose early in evolution. J. Cell. Sci. 109 (6), 1155–1164. doi:10.1242/jcs.109.6.1155
Bode, P. M., and Bode, H. R. (1980). formation of pattern in regenerating tissue pieces of Hydra attenuata: I. Head-body proportion regulation. Dev. Biol. 78 (2), 484–496. doi:10.1016/0012-1606(80)90348-6
Bonello, T. T., and Peifer., M. (2019). Scribble: A master scaffold in polarity, adhesion, synaptogenesis, and proliferation. J. Cell. Biol. 218 (3), 742–756. doi:10.1083/jcb.201810103
Bosch, T. C. G. (2004). Control of asymmetric cell divisions: Will cnidarians provide an answer? BioEssays 26 (9), 929–931. doi:10.1002/bies.20108
Bosch, T. C. G., and David, C. N. (1987). Stem cells of Hydra magnipapillata can differentiate into somatic cells and germ line cells. Dev. Biol. 121 (1), 182–191. doi:10.1016/0012-1606(87)90151-5
Bowling, S., Lawlor, K., and Tristan, A. (2019). Cell competition: The winners and losers of fitness selection. Dev. Camb. Engl. 146 (13), dev167486. doi:10.1242/dev.167486
Boyd, L., Guo, S., Levitan, D., Stinchcomb, D. T., and Kemphues, K. J. (1996). PAR-2 is asymmetrically distributed and promotes association of P granules and PAR-1 with the cortex in C. Elegans embryos. Dev. Camb. Engl. 122 (10), 3075–3084. doi:10.1242/dev.122.10.3075
Brooun, M., Alexander, K., Bashkurov, M., Pearson, B. J., Steele, R. E., and McNeill, H. (2019). Ancestral role of fat-like cadherins in planar cell polarity. BioRxiv.
Bürglin, T. R. (1994). A Caenorhabditis elegans Prospero homologue defines a novel domain. Trends biochem. Sci. 19 (2), 70–71. doi:10.1016/0968-0004(94)90035-3
Butler, M. T., and Wallingford, J. B. (2017). Planar cell polarity in development and disease. Nat. Rev. Mol. Cell. Biol. 18 (6), 375–388. doi:10.1038/nrm.2017.11
Buzgariu, W., S Al Haddad, S. T., Wenger, Y., and Galliot, B. (2015). Multi-functionality and plasticity characterize epithelial cells in Hydra. Tissue Barriers 3 (4), e1068908. doi:10.1080/21688370.2015.1068908
Carvalho, A. B., Dobo, B. A., Vibranovski, M. D., and Clark, A. G. (2001). Identification of five new genes on the Y chromosome of Drosophila melanogaster. Proc. Natl. Acad. Sci. U. S. A. 98 (23), 13225–13230. doi:10.1073/pnas.231484998
C. elegans Sequencing Consortium (1998). Genome sequence of the nematode C. Elegans: A platform for investigating Biology. Sci. (New York, N.Y.) 282 (5396), 2012–2018. doi:10.1126/science.282.5396.2012
Chen, W., Lim, H. H., and Lim, L. (1993). A new member of the ras superfamily, the Rac1 homologue from Caenorhabditis elegans. Cloning and sequence analysis of CDNA, pattern of developmental expression, and biochemical characterization of the protein. J. Biol. Chem. 268 (1), 320–324. doi:10.1016/S0021-9258(18)54152-1
Chera, S., Ghila, L., Wenger, Y., and Galliot, B. (2011). Injury-Induced activation of the MAPK/CREB pathway triggers apoptosis-induced compensatory proliferation in Hydra head regeneration. Dev. Growth Differ. 53 (2), 186–201. doi:10.1111/j.1440-169X.2011.01250.x
Choi, W., Jung, K. C., Peifer, M., Fanning, A. S., and Beitel, G. J. (2011). The single Drosophila ZO-1 protein polychaetoid regulates embryonic morphogenesis in coordination with canoe/afadin and enabled. Mol. Biol. Cell. 22 (12), 2010–2030. doi:10.1091/mbc.E10-12-1014
Dobrosotskaya, I., Guy, R. K., and James, G. L. (1997). MAGI-1, a membrane-associated guanylate kinase with a unique arrangement of protein-protein interaction domains. J. Biol. Chem. 272 (50), 31589–31597. doi:10.1074/jbc.272.50.31589
Doe, C. Q., Chu-LaGraff, Q., Wright, D. M., and Scott, M. P. (1991). The Prospero gene specifies cell fates in the Drosophila central nervous system. Cell. 65 (3), 451–464. doi:10.1016/0092-8674(91)90463-9
Domazet-Lošo, T., and Tautz, D. (2010). Phylostratigraphic tracking of cancer genes suggests a link to the emergence of multicellularity in Metazoa. BMC Biol. 8 (1), 66. doi:10.1186/1741-7007-8-66
Dow, L. E., and Humbert, P. O. (2007). Polarity regulators and the control of epithelial architecture, cell migration, and tumorigenesis. Int. Rev. Cytol. 262, 253–302. doi:10.1016/S0074-7696(07)62006-3
DuBuc, T. Q., Ryan, J. F., and Martindale, M. Q. (2019). ‘Dorsal–Ventral’ genes are part of an ancient axial patterning system: Evidence from Trichoplax adhaerens (placozoa). Mol. Biol. Evol. 36 (5), 966–973. doi:10.1093/molbev/msz025
Dunny, G. M., Brickman, T. J., and Martin, D. (2008). Multicellular behavior in bacteria: Communication, cooperation, competition and cheating. BioEssays 30 (4), 296–298. doi:10.1002/bies.20740
Ebnet, K. (2015). Cell polarity 1: Biological role and basic mechanisms. Springer International Publishing. doi:10.1007/978-3-319-14463-4
Elsir, T., Smits, A., Lindström, M. S., and Nistér, M. (2012). Transcription factor PROX1: Its role in development and cancer. Cancer Metastasis Rev. 31 (3), 793–805. doi:10.1007/s10555-012-9390-8
Elsum, I., Yates, L., Humbert, P. O., and Richardson, H. E. (2012). The scribble–dlg–lgl polarity module in development and cancer: From flies to man. Essays Biochem. 53, 141–168. doi:10.1042/bse0530141
Ereskovsky, A., Borisenko, I. E., Bolshakov, F. V., and Lavrov, Andrey I. (2021). Whole-body regeneration in sponges: Diversity, fine mechanisms, and future prospects. Genes. 12 (4), 506. doi:10.3390/genes12040506
Etemad-Moghadam, B., Guo, S., and Kenneth, J. (1995). Asymmetrically distributed PAR-3 protein contributes to cell polarity and spindle alignment in early C. Elegans embryos. Cell. 83 (5), 743–752. doi:10.1016/0092-8674(95)90187-6
Etienne-Manneville, S. (2008). Polarity proteins in migration and invasion. Oncogene 27 (55), 6970–6980. doi:10.1038/onc.2008.347
Fahey, B., and Degnan, B. M. (2010). Origin of animal epithelia: Insights from the sponge genome. Evol. Dev. 12 (6), 601–617. doi:10.1111/j.1525-142X.2010.00445.x
Fahey, B., and Degnan, B. M. (2012). Origin and evolution of laminin gene family diversity. Mol. Biol. Evol. 29 (7), 1823–1836. doi:10.1093/molbev/mss060
Fahey-Lozano, N., Portela, M., Helena, E., and Richardson, H. E. (2019). Drosophila models of cell polarity and cell competition in tumourigenesis. Adv. Exp. Med. Biol. 1167, 37–64. doi:10.1007/978-3-030-23629-8_3
Fanning, A. S., Jameson, B. J., Jesaitis, L. A., and Anderson, J. M. (1998). The tight junction protein ZO-1 establishes a link between the transmembrane protein Occludin and the actin cytoskeleton. J. Biol. Chem. 273 (45), 29745–29753. doi:10.1074/jbc.273.45.29745
Fidler, A. L., Darris, C. E., Sergei, V., Vadim, K., Boudko, S. P., Kyle, L. B., et al. (2017). Collagen IV and basement membrane at the evolutionary dawn of metazoan tissues. ELife 6, e24176. doi:10.7554/eLife.24176
Filshie, B. K., and Flower, N. E. (1977). Junctional structures in Hydra. J. Cell. Sci. 23 (1), 151–172. doi:10.1242/jcs.23.1.151
Fortunato, A., Fleming, A., Aktipis, A., and Carlo, C. (2021). Upregulation of DNA repair genes and cell extrusion underpin the remarkable radiation resistance of Trichoplax adhaerens. PLoS Biol. 17 e3001471. doi:10.1101/2020.12.24.424349
Freeman, G. (1977). The establishment of the oral-aboral Axis in the ctenophore embryo. Development 24, 237–260. doi:10.1242/dev.42.1.237
Gangar, A., Rossi, G., Anna, A., Hales, R., and Brennwald, P. (2005). Structurally conserved interaction of Lgl family with SNAREs is critical to their cellular function. Curr. Biol. 15 (12), 1136–1142. doi:10.1016/j.cub.2005.05.046
Ganot, P., Zoccola, D., Tambutté, E., Voolstra, C. R., Aranda, M., Allemand, D., et al. (2015). Structural molecular components of septate junctions in Cnidarians point to the origin of epithelial junctions in eukaryotes. Mol. Biol. Evol. 32 (1), 44–62. doi:10.1093/molbev/msu265
Gazave, E., Pascal, L., Richards, G. S., Brunet, F., Ereskovsky, A. V., Degnan, B. M., et al. (2009). Origin and evolution of the notch signalling pathway: An overview from eukaryotic genomes. BMC Evol. Biol. 9 (1), 249. doi:10.1186/1471-2148-9-249
Gerhart, J. (1999). 1998 warkany lecture: Signaling pathways in development. Teratology 60 (4), 226–239. doi:10.1002/(SICI)1096-9926(199910)60:4<226::AID-TERA7>3.0.CO;2-W
Gödde, N. J., Pearson, H. B., Smith, L. K., and Humbert, P. O. (2014). Dissecting the role of polarity regulators in cancer through the use of mouse models. Exp. Cell. Res. 328 (2), 249–257. doi:10.1016/j.yexcr.2014.08.036
Goldstein, B., and Macara, I. G. (2007). The PAR proteins: Fundamental players in animal cell polarization. Dev. Cell. 13 (5), 609–622. doi:10.1016/j.devcel.2007.10.007
González-Mariscal, L., Miranda, J., Raya-Sandino, A., Domínguez-Calderón, A., and Cuellar-Perez, F. (2017). ZO-2, a tight junction protein involved in gene expression, proliferation, apoptosis, and cell size regulation: ZO-2, a protein with membrane and nuclear functions. Ann. N. Y. Acad. Sci. 1397 (1), 35–53. doi:10.1111/nyas.13334
Gooding, J. M., Yap, K. L., and Ikura, M. (2004). The cadherin–catenin complex as a focal point of cell adhesion and signalling: New insights from three-dimensional structures. BioEssays 26 (5), 497–511. doi:10.1002/bies.20033
Goodrich, L. V., and David, S. (2011). Principles of planar polarity in animal development. Development 138 (10), 1877–1892. doi:10.1242/dev.054080
Gotta, M., and Ahringer, J. (2001). Distinct roles for Galpha and Gbetagamma in regulating spindle position and orientation in Caenorhabditis elegans embryos. Nat. Cell. Biol. 3 (3), 297–300. doi:10.1038/35060092
Grosberg, R. K., and Strathmann, R. R. (2007). The evolution of multicellularity: A minor major transition? Annu. Rev. Ecol. Evol. Syst. 38, 621–654. doi:10.1146/annurev.ecolsys.36.102403.114735
Grosshans, B. L., Anna, A., Gangar, A., Niessen, S., Yates, J. R., Brennwald, P., et al. (2006). The yeast Lgl family member Sro7p is an effector of the secretory rab GTPase Sec4p. J. Cell. Biol. 172 (1), 55–66. doi:10.1083/jcb.200510016
Gubb, D., García-Bellido, A., and GarciA-Bellido, A. (1982). A genetic analysis of the determination of cuticular polarity during development in Drosophila melanogaster. Development 68, 37–57. doi:10.1242/dev.68.1.37
Guillot, C., and Lecuit, T. (2013). Mechanics of epithelial tissue homeostasis and morphogenesis. Science 340 (6137), 1185–1189. doi:10.1126/science.1235249
Gul, I. S., Hulpiau, P., Saeys, Y., and Frans van Roy, (2017). Evolution and diversity of cadherins and catenins. Exp. Cell. Res. 358 (1), 3–9. doi:10.1016/j.yexcr.2017.03.001
Hakeda-Suzuki, S., Ng, J., Julia, T., Dietzl, G., Sun, Y., Harms, M., et al. (2002). Rac function and regulation during Drosophila development. Nature 416 (6879), 438–442. doi:10.1038/416438a
Hale, R., and Strutt, D. (2015). Conservation of planar polarity pathway function across the animal kingdom. Annu. Rev. Genet. Vol. 49, 529–551. doi:10.1146/annurev-genet-112414-055224
Hammarlund, M., Nix, P., Hauth, L., Jorgensen, E. M., and Bastiani, M. (2009). Axon regeneration requires a conserved MAP kinase pathway. Science 323 (5915), 802–806. doi:10.1126/science.1165527
Hanahan, D., and Weinberg, R. A. (2000). The hallmarks of cancer. Cell. 100 (1), 57–70. doi:10.1016/S0092-8674(00)81683-9
Harden, N., Wang, S. H., and Krieger, C. (2016). Making the connection – shared molecular machinery and evolutionary links underlie the formation and plasticity of occluding junctions and synapses. J. Cell. Sci. 129 (16), 3067–3076. doi:10.1242/jcs.186627
Harris, T. J. C., and Ulrich, T. (2010). Adherens junctions: From molecules to morphogenesis. Nat. Rev. Mol. Cell. Biol. 11 (7), 502–514. doi:10.1038/nrm2927
Harrison, F. W., and Ruppert., E. E. (1991). Microscopic anatomy of invertebrates, placozoa, Porifera, Cnidaria, and ctenophora 2. New York: Wiley-Liss.
Hartmann, C., Otani, T., Furuse, M., and Ebnet, K. (2020). Physiological functions of junctional adhesion molecules (JAMs) in tight junctions. Biochim. Biophys. Acta. Biomembr. 1862 (9), 183299. doi:10.1016/j.bbamem.2020.183299
Hattendorf, D. A., Anna, A., Gangar, A., Brennwald, P. J., and Weis, W. I. (2007). Structure of the yeast polarity protein Sro7 reveals a SNARE regulatory mechanism. Nature 446 (7135), 567–571. doi:10.1038/nature05635
Heasman, S. J., and Ridley, A. J. (2008). Mammalian rho GTPases: New insights into their functions from in vivo studies. Nat. Rev. Mol. Cell. Biol. 9 (9), 690–701. doi:10.1038/nrm2476
Hiroki, O. (2012). “Evolution of the cadherin–catenin complex,” in Adherens junctions: From molecular mechanisms to tissue development and disease. Editor T. Harris (Dordrecht: Springer Netherlands). doi:10.1007/978-94-007-4186-7_2
Hsu, T-Y., and Wu, Y-C. (2010). Engulfment of apoptotic cells in C. elegans is mediated by integrin alpha/SRC signaling. Curr. Biol. 20 (6), 477–486. doi:10.1016/j.cub.2010.01.062
Hulpiau, P., Sahin Gul, I., and Frans van Roy, (2013). New insights into the evolution of metazoan cadherins and catenins. Prog. Mol. Biol. Transl. Sci. 116, 71–94. doi:10.1016/B978-0-12-394311-8.00004-2
Hulpiau, P., and van Roy, F. (2011). New insights into the evolution of metazoan cadherins. Mol. Biol. Evol. 28 (1), 647–657. doi:10.1093/molbev/msq233
Humbert, P., Russell, S., and Richardson, H. (2003). Dlg, Scribble and Lgl in cell polarity, cell proliferation and cancer. BioEssays 25 (6), 542–553. doi:10.1002/bies.10286
Humphries, A. C., and Mlodzik, M. (2018). From instruction to output: Wnt/PCP signaling in development and cancer. Curr. Opin. Cell. Biol. 51, 110–116. doi:10.1016/j.ceb.2017.12.005
Hung, T. J., and Kemphues, K. J. (1999). PAR-6 is a conserved PDZ domain-containing protein that colocalizes with PAR-3 in Caenorhabditis elegans embryos. Development 126 (1), 127–135. doi:10.1242/dev.126.1.127
Hurd, T. W., Gao, L., Roh, M. H., Macara, I. G., and Margolis, B. (2003). Direct interaction of two polarity complexes implicated in epithelial tight junction assembly. Nat. Cell. Biol. 5 (2), 137–142. doi:10.1038/ncb923
Ip, Y. T., and Davis, R. J. (1998). Signal transduction by the C-jun N-terminal kinase (JNK) — From inflammation to development. Curr. Opin. Cell. Biol. 10 (2), 205–219. doi:10.1016/S0955-0674(98)80143-9
Itoh, M., Furuse, M., Morita, K., Kubota, K., Saitou, M., and Tsukita, S. (1999). Direct binding of three tight junction-associated maguks, zo-1, zo-2, and zo-3, with the cooh termini of claudins. J. Cell. Biol. 147 (6), 1351–1363. doi:10.1083/jcb.147.6.1351
Jan, Y. N., and Jan, L. (1998). Asymmetric cell division. Nature 392 (6678), 775–778. doi:10.1038/33854
Joberty, G., Petersen, C., Gao, L., and Ian, G. (2000). The cell-polarity protein Par6 links Par3 and atypical protein kinase C to Cdc42. Nat. Cell. Biol. 2 (8), 531–539. doi:10.1038/35019573
Kamm, K., Osigus, H. J., Stadler, Pr F., DeSalle, R., and Schierwater, B. (2018). Trichoplax genomes reveal profound admixture and suggest stable wild populations without bisexual reproduction. Sci. Rep. 8 (1), 11168. doi:10.1038/s41598-018-29400-y
Kamm, K., Schierwater, B., and DeSalle, R. (2019). Innate immunity in the simplest animals–placozoans. BMC Genomics 20 (1), 5. doi:10.1186/s12864-018-5377-3
Kelsom, C., and Lu, W. (2012). Uncovering the link between malfunctions in Drosophila neuroblast asymmetric cell division and tumorigenesis. Cell. Biosci. 2 (1), 38. doi:10.1186/2045-3701-2-38
Kemphues, K. J., Priess, J. R., Morton, D. G., and Cheng, N. S. (1988). Identification of genes required for cytoplasmic localization in early C. Elegans embryos. Cell. 52 (3), 311–320. doi:10.1016/s0092-8674(88)80024-2
King, N., Hittinger, C. T., and Carroll, S. B. (2003). Evolution of key cell signaling and adhesion protein families predates animal origins. Science 301 (5631), 361–363. doi:10.1126/science.1083853
King, N. (2004). The unicellular ancestry of animal development. Dev. Cell. 7 (3), 313–325. doi:10.1016/j.devcel.2004.08.010
King, N., Westbrook, M. J., Young, S. L., Kuo, A., Abedin, M., Chapman, J., et al. (2008). The genome of the choanoflagellate monosiga brevicollis and the origin of metazoans. Nature 451 (7180), 783–788. doi:10.1038/nature06617
Klimovich, A., Wittlieb, J., and ThomasBosch, C. G. (2019). Transgenesis in Hydra to characterize gene function and visualize cell behavior. Nat. Protoc. 14 (7), 2069–2090. doi:10.1038/s41596-019-0173-3
Knoblich, J. A. (2001). Asymmetric cell division during animal development. Nat. Rev. Mol. Cell. Biol. 2 (1), 11–20. doi:10.1038/35048085
Knoblich, J. A. (2010). Asymmetric cell division: Recent developments and their implications for tumour Biology. Nat. Rev. Mol. Cell. Biol. 11 (12), 849–860. doi:10.1038/nrm3010
Knoll, A. H. (2011). The multiple origins of complex multicellularity. Annu. Rev. Earth Planet. Sci. 39 (1), 217–239. doi:10.1146/annurev.earth.031208.100209
Knust, E., and Bossinger, O. (2002). Composition and formation of intercellular junctions in epithelial cells. Sci. (New York, N.Y.) 298 (5600), 1955–1959. doi:10.1126/science.1072161
Knust, E., Ulrich, T., and Wodarz, A. (1993). Crumbs and stardust, two genes of Drosophila required for the development of epithelial cell polarity. Dev. Suppl. 119, 261–268. doi:10.1242/dev.119.Supplement.261
Kraut, R., Chia, W., Yeh Jan, L., Jan, Y. N., and Jürgen, A. (1996). Role of inscuteable in orienting asymmetric cell divisions in Drosophila. Nature 383 (6595), 50–55. doi:10.1038/383050a0
Kumburegama, S., Wijesena, N., Xu, R., and Athula, H. (2011). Strabismus-Mediated primary archenteron invagination is uncoupled from wnt/β-catenin-dependent endoderm cell fate specification in Nematostella vectensis (Anthozoa, Cnidaria): Implications for the evolution of gastrulation. EvoDevo 2 (1), 2. doi:10.1186/2041-9139-2-2
Kwiatkowski, V., Stephanie, L., Maiden, S. P., Choi, H. J., Benjamin, J. M., Lynch, A. M., et al. (2010). In vitro and in vivo reconstitution of the cadherin–catenin–actin complex from Caenorhabditis elegans. Proc. Natl. Acad. Sci. U. S. A. 107 (33), 14591–14596. doi:10.1073/pnas.1007349107
Larroux, C., Luke, G. N., Peter, K., Rokhsar, D. S., Shimeld, S. M., and Degnan, B. M. (2008). Genesis and expansion of metazoan transcription factor gene classes. Mol. Biol. Evol. 25 (5), 980–996. doi:10.1093/molbev/msn047
Lavrov, A. I., Bolshakov, F. V., Tokina, D. B., and Ereskovsky, A. V. (2018). Sewing up the wounds : The epithelial morphogenesis as a central mechanism of calcaronean sponge regeneration. J. Exp. Zool. B Mol. Dev. Evol. 330 (6–7), 351–371. doi:10.1002/jez.b.22830
Layden, M. J., Rentzsch, F., and Röttinger, E. (2016). The rise of the starlet sea anemone Nematostella vectensis as a model system to investigate development and regeneration. Wiley Interdiscip. Rev. Dev. Biol. 5 (4), 408–428. doi:10.1002/wdev.222
Lee, J-S., Ishimoto, A., and Yanagawa, S. (1999). Characterization of mouse dishevelled (dvl) proteins in wnt/wingless signaling pathway. J. Biol. Chem. 274 (30), 21464–21470. doi:10.1074/jbc.274.30.21464
Lehman, K., Rossi, G., Adamo, J. E., and Brennwald, P. (1999). Yeast homologues of tomosyn and lethal giant larvae function in exocytosis and are associated with the plasma membrane snare, Sec9. J. Cell. Biol. 146 (1), 125–140. doi:10.1083/jcb.146.1.125
Lo, S. H. (2006). Focal adhesions: What’s new inside. Dev. Biol. 294 (2), 280–291. doi:10.1016/j.ydbio.2006.03.029
Loh, K. M., van Amerongen, R., and Nusse, R. (2016). Generating cellular diversity and spatial form: Wnt signaling and the evolution of multicellular animals. Dev. Cell. 38 (6), 643–655. doi:10.1016/j.devcel.2016.08.011
Lokits, A. D., Indrischek, H., Meiler, J., Hamm, H. E., and Stadler, P. F. (2018). Tracing the evolution of the heterotrimeric G protein α subunit in Metazoa. BMC Evol. Biol. 18 (1), 51. doi:10.1186/s12862-018-1147-8
Lynch, A. M., Grana, T., Couthier, A., Cameron, M., et al. (2012). Theresa grana, elisabeth cox-paulson, annabelle couthier, michel cameron, ian chin-sang, jonathan pettitt, and jeff HardinA genome-wide functional screen shows MAGI-1 is an l1cam-dependent stabilizer of apical junctions in C. Elegans. Curr. Biol. 22 (20), 1891–1899. doi:10.1016/j.cub.2012.08.024
Macara, I. G. (2004). Parsing the polarity code. Nat. Rev. Mol. Cell. Biol. 5 (3), 220–231. doi:10.1038/nrm1332
Madan, E., Gogna, R., and Moreno, E. (2018). Cell competition in development: Information from flies and vertebrates. Curr. Opin. Cell. Biol. 55, 150–157. doi:10.1016/j.ceb.2018.08.002
Magie, C. R., and Martindale, M. Q. (2008). Cell-cell adhesion in the Cnidaria: Insights into the evolution of tissue morphogenesis. Biol. Bull. 214 (3), 218–232. doi:10.2307/25470665
Margolis, B., and Borg, J. P. (2005). Apicobasal polarity complexes. J. Cell. Sci. 118 (22), 5157–5159. doi:10.1242/jcs.02597
Matis, M., and Axelrod, J. D. (2013). Regulation of PCP by the fat signaling pathway. Genes. Dev. 27 (20), 2207–2220. doi:10.1101/gad.228098.113
Matter, K., and Balda, M. S. (2003). Signalling to and from tight junctions. Nat. Rev. Mol. Cell. Biol. 4 (3), 225–236. doi:10.1038/nrm1055
McLennan, D. A. (2008). The concept of Co-option: Why evolution often looks miraculous. Evo. Edu. Outreach 1 (3), 247–258. doi:10.1007/s12052-008-0053-8
Melendez, J., Grogg, M., and Zheng, Y. (2011). Signaling role of Cdc42 in regulating mammalian physiology. J. Biol. Chem. 286 (4), 2375–2381. doi:10.1074/jbc.R110.200329
Mendoza, A., Sebé-Pedrós, A., and Ruiz-Trillo, I. (2014). The evolution of the GPCR signaling system in eukaryotes: Modularity, conservation, and the transition to metazoan multicellularity. Genome Biol. Evol. 6 (3), 606–619. doi:10.1093/gbe/evu038
Mendoza, A., Suga, H., and Ruiz-Trillo, I. (2010). Evolution of the MAGUK protein gene family in premetazoan lineages. BMC Evol. Biol. 10 (1), 93. doi:10.1186/1471-2148-10-93
Michael, M., and Parsons, M. (2020). New perspectives on integrin-dependent adhesions. Curr. Opin. Cell. Biol. 63, 31–37. doi:10.1016/j.ceb.2019.12.008
Milgrom-Hoffman, M., and Humbert, P. O. (2018). Regulation of cellular and PCP signalling by the Scribble polarity module. Semin. Cell. Dev. Biol. 81, 33–45. doi:10.1016/j.semcdb.2017.11.021
Mitchell, J. M., and Nichols, S. A. (2019). Diverse cell junctions with unique molecular composition in tissues of a sponge (Porifera). EvoDevo 10 (1), 26. doi:10.1186/s13227-019-0139-0
Momose, T., Kraus, Y., and Houliston, E. (2012). A conserved function for strabismus in establishing planar cell polarity in the ciliated ectoderm during Cnidarian larval development. Development 139 (23), 4374–4382. doi:10.1242/dev.084251
Moulder, G. L., Huang, M. M., Waterston, R. H., and Barstead, R. J. (1996). Talin requires beta-integrin, but not vinculin, for its assembly into focal adhesion-like structures in the nematode Caenorhabditis elegans. Mol. Biol. Cell. 7 (8), 1181–1193. doi:10.1091/mbc.7.8.1181
Müller, W. E. G. (2003). The origin of metazoan complexity: Porifera as integrated animals. Integr. Comp. Biol. 43 (1), 3–10. doi:10.1093/icb/43.1.3
Müsch, A., Cohen, D., Yeaman, C., James Nelson, W., Rodriguez-Boulan, E., and Patrick, J. (2002). Mammalian homolog of Drosophila tumor suppressor lethal (2) giant larvae interacts with basolateral exocytic machinery in madin-darby canine kidney cells. Mol. Biol. Cell. 13 (1), 158–168. doi:10.1091/mbc.01-10-0496
Nelson, W. J. (2003). Adaptation of core mechanisms to generate cell polarity. Nature 422 (6933), 766–774. doi:10.1038/nature01602
Nelson, W. J. (2008). Regulation of cell–cell adhesion by the cadherin–catenin complex. Biochem. Soc. Trans. 36 (2), 149–155. doi:10.1042/BST0360149
Neumann, J., Tessler, M. S., Kamm, K., Osigus, H. J., Ershel, G., Narechania, A., et al. (2022). Phylogenomics and the first higher taxonomy of Placozoa, an ancient and enigmatic animal phylum. Front. Ecol. Evol. doi:10.3389/fevo.2022.1016357
Neumüller, Rh A., and Knoblich, J. A. (2009). Dividing cellular asymmetry: Asymmetric cell division and its implications for stem cells and cancer. Genes. Dev. 23 (23), 2675–2699. doi:10.1101/gad.1850809
Nichols, S. A., Dirks, W., Pearse, J. S., and King, N. (2006). Early evolution of animal cell signaling and adhesion genes. Proc. Natl. Acad. Sci. U. S. A. 103 (33), 12451–12456. doi:10.1073/pnas.0604065103
Nichols, S. A., Roberts, B. W., Richter, D. J., Fairclough, S. R., and King, N. (2012). Origin of metazoan cadherin diversity and the antiquity of the classical cadherin/-catenin complex. Proc. Natl. Acad. Sci. U. S. A. 109 (32), 13046–13051. doi:10.1073/pnas.1120685109
Niessen, C. M. (2007). Tight junctions/adherens junctions: Basic structure and function. J. Invest. Dermatol. 127 (11), 2525–2532. doi:10.1038/sj.jid.5700865
Noselli, S., and Agnès, F. (1999). Roles of the JNK signaling pathway in Drosophila morphogenesis. Curr. Opin. Genet. Dev. 9 (4), 466–472. doi:10.1016/S0959-437X(99)80071-9
Nosenko, T., Schreiber, F., Adamska, M., Adamski, M., Eitel, M., Hammel, J., et al. (2013). Deep metazoan phylogeny: When different genes tell different stories. Mol. Phylogenet. Evol. 67 (1), 223–233. doi:10.1016/j.ympev.2013.01.010
Oda, H., and Takeichi, M. (2011). Evolution: Structural and functional diversity of cadherin at the adherens junction. J. Cell. Biol. 193 (7), 1137–1146. doi:10.1083/jcb.201008173
Okamoto, K., Nakatsukasa, M., Alexandre, A., Masuda, Y., Agata, K., and Funayama, N. (2012). The active stem cell specific expression of sponge Musashi homolog EflMsiA suggests its involvement in maintaining the stem cell state. Mech. Dev. 129 (1), 24–37. doi:10.1016/j.mod.2012.03.001
Oliver, G., Sosa-Pineda, B., Geisendorf, S., Spana, E. P., Chris, Q., and Gruss, P. (1993). Prox 1, a prospero-related homeobox gene expressed during mouse development. Mech. Dev. 44 (1), 3–16. doi:10.1016/0925-4773(93)90012-M
Osigus, H. J., Eitel, M., Horn, K., Kamm, K., and Kosubek-Langer, J. (2022). “Studying PlacozoaPlacozoa WBRWhole-body regeneration (WBR)in the simplest metazoan animal, Trichoplax AdhaerensTrichoplax adhaerens,” in Whole-body regeneration: Methods and protocols, edited by simon blanchoud and brigitte galliot (New York, NY: Springer US). doi:10.1007/978-1-0716-2172-1_6
Pang, K., and Mark, Q. (2008). Ctenophores. Curr. Biol. 18 (24), R1119–R1120. doi:10.1016/j.cub.2008.10.004
Pang, K., Ryan, J. F., Mullikin, J. C., and Martindale, M. Q. (2010). Andreas D. Baxevanis, Mark Q. Martindale, and NISC comparative sequencing ProgramGenomic insights into wnt signaling in an early diverging metazoan, the ctenophore Mnemiopsis leidyi. EvoDevo 1 (1), 10. doi:10.1186/2041-9139-1-10
Petronczki, M., and Knoblich, J. A. (2001). DmPAR-6 directs epithelial polarity and asymmetric cell division of neuroblasts in Drosophila. Nat. Cell. Biol. 3 (1), 43–49. doi:10.1038/35050550
Philipp, I., Aufschnaiter, R., Ozbek, S., Pontasch, S., Jenewein, M., Watanabe, H., et al. (2009). Wnt/beta-catenin and noncanonical Wnt signaling interact in tissue evagination in the simple eumetazoan Hydra. Proc. Natl. Acad. Sci. U. S. A. 106 (11), 4290–4295. doi:10.1073/pnas.0812847106
Pillé, J. -Y., Denoyelle, C., Varet, J., Bertrand, J. -R., Soria, J., Opolon, P., et al. (2005). Anti-RhoA and anti-RhoC SiRNAs inhibit the proliferation and invasiveness of MDA-MB-231 breast cancer cells in vitro and in vivo. Mol. Ther. 11 (2), 267–274. doi:10.1016/j.ymthe.2004.08.029
Piraino, S., Boero, F., Aeschbach, B., and Schmid, V. (1996). Reversing the life cycle: Medusae transforming into polyps and cell transdifferentiation in turritopsis nutricula (Cnidaria, Hydrozoa). Biol. Bull. 190 (3), 302–312. doi:10.2307/1543022
Postiglione, M. P., Jüschke, C., Xie, Y., Haas, G. A., Charalambous, C., and Knoblich, J. A. (2011). Mouse inscuteable induces apical-basal spindle orientation to facilitate intermediate progenitor generation in the developing neocortex. Neuron 72 (2), 269–284. doi:10.1016/j.neuron.2011.09.022
Rathbun, L. I., Everett, C. A., and Bergstralh, D. T. (2022). Emerging Cnidarian models for the study of epithelial polarity. Front. Cell. Dev. Biol. 10, 854373. doi:10.3389/fcell.2022.854373
Reber-Müller, S., Studer, R., Müller, P., Yanze, N., and Schmid, V. (2001). Integrin and talin in the jellyfish podocoryne carnea. Cell. Biol. Int. 25 (8), 753–769. doi:10.1006/cbir.2000.0708
Richter, D. J., Fozouni, P., Eisen, M. B., and King, N. (2018). Gene family innovation, conservation and loss on the animal stem lineage. ELife 7, e34226. doi:10.7554/eLife.34226
Richter, D. J., and King., N. (2013). The genomic and cellular foundations of animal origins. Annu. Rev. Genet. 47 (1), 509–537. doi:10.1146/annurev-genet-111212-133456
Riesgo, A., Farrar, N., Windsor, P. J., Giribet, G., and Sally, P. (2014). The analysis of eight transcriptomes from all Poriferan classes reveals surprising genetic complexity in sponges. Mol. Biol. Evol. 31 (5), 1102–1120. doi:10.1093/molbev/msu057
Ringrose, J. H., van den Toorn, H. W. P., Eitel, M., Post, H., Neerincx, P., Schierwater, B., et al. (2013). Deep proteome profiling of Trichoplax adhaerens reveals remarkable features at the origin of metazoan multicellularity. Nat. Commun. 4, 1408. doi:10.1038/ncomms2424
Rock, R., Schrauth, S., and Gessler, M. (2005). Expression of mouse Dchs1, Fjx1, and fat-j suggests conservation of the planar cell polarity pathway identified in Drosophila. Dev. Dyn. 234 (3), 747–755. doi:10.1002/dvdy.20515
Rodriguez-Boulan, E., and Nelson, W. J. (1989). Morphogenesis of the polarized epithelial cell phenotype. Science 245 (4919), 718–725. doi:10.1126/science.2672330
Rokas, A. (2008). The origins of multicellularity and the early history of the genetic toolkit for animal development. Annu. Rev. Genet. 42, 235–251. doi:10.1146/annurev.genet.42.110807.091513
Röttinger, É. (2021). Nematostella vectensis, an emerging model for deciphering the molecular and cellular mechanisms underlying whole-body regeneration. Cells 10, 2692. doi:10.3390/cells10102692
Royer, C., and Lu, X. (2011). Epithelial cell polarity: A major gatekeeper against cancer? Cell. Death Differ. 18 (9), 1470–1477. doi:10.1038/cdd.2011.60
Ruthmann, A., Behrendt, G., and Wahl, R. (1986). The ventral epithelium of Trichoplax adhaerens (placozoa): Cytoskeletal structures, cell contacts and endocytosis. Zoomorphology 106 (2), 115–122. doi:10.1007/BF00312113
Ruthmann, A., and Terwelp, U. (1979). Disaggregation and reaggregation of cells of the primitive metazoon Trichoplax adhaerens. Differentiation 13 (3), 185–198. doi:10.1111/j.1432-0436.1979.tb01581.x
Ryan, J. F., Pang, K., Schnitzler, C. E., Moreland, R. T., Simmons, D. K., Koch, B. J., et al. (2013). The genome of the ctenophore Mnemiopsis leidyi and its implications for cell type evolution. Science 342 (6164), 1242592. doi:10.1126/science.1242592
Ryan, J. F., Patrick, M., Finnerty, J. R., Kwong, G. K., Mullikin, J. C., and Finnerty, J. R. (2006). The Cnidarian-bilaterian ancestor possessed at least 56 homeoboxes: Evidence from the starlet sea anemone, Nematostella vectensis. Genome Biol. 7 (7), R64. doi:10.1186/gb-2006-7-7-r64
Ryan, K. R., and Shapiro., L. (2003). Temporal and spatial regulation in prokaryotic cell cycle progression and development. Annu. Rev. Biochem. 72 (1), 367–394. doi:10.1146/annurev.biochem.72.121801.161824
Sakarya, O., Armstrong, K. A., Adamska, M., Adamski, M., Wang, I-F., Bruce, T., et al. (2007). A post-synaptic scaffold at the origin of the animal kingdom. PLoS ONE 2 (6), e506. doi:10.1371/journal.pone.0000506
Salinas-Saavedra, M., Dunn, C. W., and Martindale, M. Q. (2015). Par system components are asymmetrically localized in ectodermal epithelia, but not during early development in the sea anemone Nematostella vectensis. EvoDevo 6 (1), 20. doi:10.1186/s13227-015-0014-6
Salinas-Saavedra, M., and Martindale, M. Q. (2018). Germ layer-specific regulation of cell polarity and adhesion gives insight into the evolution of mesoderm. Elife 28, e36740. doi:10.7554/eLife.36740
Salinas-Saavedra, M., and Martindale, M. Q. (2019). β-Catenin has an ancestral role in cell fate specification but not cell adhesion. bioRxiv. doi:10.1101/520957
Sandoz, K. M., Mitzimberg, S. M., and Schuster., M. (2007). Social cheating in Pseudomonas aeruginosa quorum sensing. Proc. Natl. Acad. Sci. U. S. A. 104 (40), 15876–15881. doi:10.1073/pnas.0705653104
Santoni, M. J., Pontarotti, P., Birnbaum, D., and Borg, J. P. (2002). The LAP family: A phylogenetic point of view. Trends Genet. 18 (10), 494–497. doi:10.1016/s0168-9525(02)02738-5
Sawa, H. (2012). Control of cell polarity and asymmetric division in C. Elegans. Curr. Top. Dev. Biol. 101, 55–76. doi:10.1016/B978-0-12-394592-1.00003-X
Schenkelaars, Q., Fierro-Constain, L., Renard, E., and Borchiellini, C. (2016). Retracing the path of planar cell polarity. BMC Evol. Biol. 16 (1), 69. doi:10.1186/s12862-016-0641-0
Schenkelaars, Q., Fierro-Constain, L., Renard, E., Hill, A. L., and Borchiellini, C. (2015). Insights into frizzled evolution and new perspectives. Evol. Dev. 17 (2), 160–169. doi:10.1111/ede.12115
Schenkelaars, Q., Pratlong, M., Kodjabachian, L., Fierro-Constain, L., Vacelet, J., Bivic, A., et al. (2017). Animal multicellularity and polarity without wnt signaling. Sci. Rep. 7 (1), 1–11. doi:10.1038/s41598-017-15557-5
Schierwater, B., Osigus, H. J., Bergmann, T., Blackstone, N. W., Hadrys, H., Hauslage, J., et al. (2021). The enigmatic placozoa Part 1: Exploring evolutionary controversies and poor ecological knowledge. Bioessays. 43 (10), 2100080. doi:10.1002/bies.202100080
Schiller, E., and Bergstralh, D. T. (2021). Interaction between Discs large and Pins/LGN/GPSM2: A comparison across species. Biol. Open 10, bio058982. doi:10.1242/bio.058982
Schmid, V., and Alder, H. (1984). Isolated, mononucleated, striated muscle can undergo pluripotent transdifferentiation and form a complex regenerate. Cell. 38 (3), 801–809. doi:10.1016/0092-8674(84)90275-7
Sebé-Pedrós, A., Zheng, Y., Ruiz-Trillo, I., and Pan, D. (2012). Premetazoan origin of the hippo signaling pathway. Cell. Rep. 1 (1), 13–20. doi:10.1016/j.celrep.2011.11.004
Sebé-Pedrós, A., Arnau, A. J. R., Lang, F. B., King, N., and Ruiz-Trillo, I. (2010). Ancient origin of the integrin-mediated adhesion and signaling machinery. Proc. Natl. Acad. Sci. U. S. A. 107 (22), 10142–10147. doi:10.1073/pnas.1002257107
Seipel, K., Yanze, N., and Schmid, V. (2003). The germ line and somatic stem cell gene cniwi in the jellyfish Podocoryne carnea. Int. J. Dev. Biol. 48 (1), 1–7. doi:10.1387/ijdb.15005568
Seybold, A., Salvenmoser, W., and Hobmayer, B. (2016). Sequential development of apical-basal and planar polarities in aggregating epitheliomuscular cells of Hydra. Dev. Biol. 412 (1), 148–159. doi:10.1016/j.ydbio.2016.02.022
Simmons, D. K., Pang, K., and Martindale, M. Q. (2012). Lim homeobox genes in the ctenophore Mnemiopsis leidyi: The evolution of neural cell type specification. EvoDevo 3 (1), 2. doi:10.1186/2041-9139-3-2
Simons, M., and Mlodzik, M. (2008). Planar cell polarity signaling: From fly development to human disease. Annu. Rev. Genet. 42 (1), 517–540. doi:10.1146/annurev.genet.42.110807.091432
Smith, C. L., and Reese., T. S. (2016). Adherens junctions modulate diffusion between epithelial cells in Trichoplax adhaerens. Biol. Bull. 231 (3), 216–224. doi:10.1086/691069
Smith, C. L., Varoqueaux, F., Kittelmann, M., Azzam, R. N., Winters, C. A., Eitel, M., et al. (2014). Novel cell types, neurosecretory cells, and body plan of the early-diverging metazoan Trichoplax adhaerens. Curr. Biol. 24 (14), 1565–1572. doi:10.1016/j.cub.2014.05.046
Snell, E. A., M Brooke, N. W. R. T., Casane, D., Philippe, H., and Holland, P. W. H. (2006). An unusual choanoflagellate protein released by hedgehog autocatalytic processing. Proc. Biol. Sci. 273 (1585), 401–407. doi:10.1098/rspb.2005.3263
Soest, R., Boury-Esnault, N., Vacelet, J., Martin, D., Erpenbeck, D., NicoleSantodomingo, J. N., et al. (2012). Global diversity of sponges (Porifera). PLOS ONE 7 (4), e35105. doi:10.1371/journal.pone.0035105
Sogabe, S., Kocot, Kevin M., Say, T. E., Stoupin, Dl, Roper, K. E., Fernandez-Valverde, S. L., et al. (2019). Pluripotency and the origin of animal multicellularity. Nature 570 (7762), 519–522. doi:10.1038/s41586-019-1290-4
Srivastava, M., Begovic, E., Chapman, J., Putnam, N. H., Hellsten, U., Kawashima, T., et al. (2008). The Trichoplax genome and the nature of placozoans. Nature 454 (7207), 955–960. doi:10.1038/nature07191
Srivastava, M., Simakov, O., Chapman, J., Fahey, B., MarieGauthier, E. A., Mitros, T., et al. (2010). The Amphimedon queenslandica genome and the evolution of animal complexity. Nature 466 (7307), 720–726. doi:10.1038/nature09201
Srivastava, M. (2015). “A comparative genomics perspective on the origin of multicellularity and early animal evolution,” in Evolutionary transitions to multicellular life: Principles and mechanisms. Editors I. RuizTrillo, and A. M. Nedelcu (Dordrecht: Springer), 269–299.
St Johnston, D., and Ahringer, J. (2010). Cell polarity in eggs and epithelia: Parallels and diversity. Cell. 141 (5), 757–774. doi:10.1016/j.cell.2010.05.011
Steed, E., Balda, M. S., and Matter, K. (2010). Dynamics and functions of tight junctions. Trends Cell. Biol. 20 (3), 142–149. doi:10.1016/j.tcb.2009.12.002
Steimel, A., Wong, L., Ackley, B. D., Garriga, G., and Hutter, H. (2010). The flamingo ortholog FMI-1 controls pioneer-dependent navigation of follower axons in C. Elegans. Development 137 (21), 3663–3673. doi:10.1242/dev.054320
Stephens, R., Lim, K., Portela, M., Kvansakul, M., Humbert, P. O., and Richardson, H. E. (2018). The Scribble cell polarity module in the regulation of cell signaling in tissue development and tumorigenesis. J. Mol. Biol. 430 (19), 3585–3612. doi:10.1016/j.jmb.2018.01.011
Tabuse, Y., Izumi, Y., Piano, F., Kemphues, K. J., Miwa, J., and Ohno, S. (1998). Atypical protein kinase C cooperates with PAR-3 to establish embryonic polarity in Caenorhabditis elegans. Dev. Camb. Engl. 125 (18), 3607–3614. doi:10.1242/dev.125.18.3607
Takahashi, K., Matsuo, T., Katsube, T., Ueda, R., and Yamamoto, D. (1998). Direct binding between two PDZ domain proteins canoe and ZO-1 and their roles in regulation of the jun N-terminal kinase pathway in Drosophila morphogenesis. Mech. Dev. 78 (1–2), 97–111. doi:10.1016/s0925-4773(98)00151-8
Tamm, S. L. (2014). Cilia and the life of ctenophores. Invertebr. Biol. 133 (1), 1–46. doi:10.1111/ivb.12042
Technau, U., and Steele, R. E. (2011). Evolutionary crossroads in developmental Biology: Cnidaria. Development 138 (8), 1447–1458. doi:10.1242/dev.048959
Tepass, U. (2012). The apical polarity protein network in Drosophila epithelial cells: Regulation of polarity, junctions, morphogenesis, cell growth, and survival. Annu. Rev. Cell. Dev. Biol. 28 (1), 655–685. doi:10.1146/annurev-cellbio-092910-154033
Tepass, U., Tanentzapf, G., Ward, R., and Fehon, R. (2001). Epithelial cell polarity and cell junctions in Drosophila. Annu. Rev. Genet. 35, 747–784. doi:10.1146/annurev.genet.35.102401.091415
Tepass, U., Theres, C., and Knust, E. (1990). Crumbs encodes an EGF-like protein expressed on apical membranes of Drosophila epithelial cells and required for organization of epithelia. Cell. 61 (5), 787–799. doi:10.1016/0092-8674(90)90189-l
Thiemann, M., and Ruthmann, A. (1989). Microfilaments and microtubules in isolated fiber cells of trichoplax-adhaerens (placozoa). Zoomorphology 109 (2), 89–96. doi:10.1007/BF00312314
Tian, X., Liu, Z., Niu, B., Zhang, J., Tan, T. K., Lee, R., et al. (2011). E-Cadherin/β-Catenin complex and the epithelial barrier. J. Biomed. Biotechnol. 2011, e567305. doi:10.1155/2011/567305
Tsukita, S., Furuse, M., and Itoh, M. (2001). Multifunctional strands in tight junctions. Nat. Rev. Mol. Cell. Biol. 2 (4), 285–293. doi:10.1038/35067088
Tucker, R. P., and Adams., J. C. (2014). Adhesion networks of cnidarians: A postgenomic view. Int. Rev. Cell. Mol. Biol. 308, 323–377. doi:10.1016/B978-0-12-800097-7.00008-7
Tyler, S. (2003). Epithelium—the primary building block for metazoan complexity. Integr. Comp. Biol. 43 (1), 55–63. doi:10.1093/icb/43.1.55
Usui, T., Shima, Y., Shimada, Y., Hirano, S., Burgess, R. W., Schwarz, T. L., et al. (1999). Flamingo, a seven-pass transmembrane cadherin, regulates planar cell polarity under the control of frizzled. Cell. 98 (5), 585–595. doi:10.1016/S0092-8674(00)80046-X
Velthuis, A. J., Admiraal, J. F., and Bagowski, C. P. (2007). Molecular evolution of the MAGUK family in metazoan genomes. BMC Evol. Biol. 7 (1), 129. doi:10.1186/1471-2148-7-129
Walston, T., Tuskey, C., Edgar, L., Hawkins, N., Ellis, G., Bruce, B., et al. (2004). Multiple wnt signaling pathways converge to orient the mitotic spindle in early C. Elegans embryos. Dev. Cell. 7 (6), 831–841. doi:10.1016/j.devcel.2004.10.008
Wei, X., and Ellis, H. M. (2001). Localization of the Drosophila MAGUK protein polychaetoid is controlled by alternative splicing. Mech. Dev. 100 (2), 217–231. doi:10.1016/S0925-4773(00)00550-5
Wen, W., and Zhang, M. (2018). Protein complex assemblies in epithelial cell polarity and asymmetric cell division. J. Mol. Biol. 430 (19), 3504–3520. doi:10.1016/j.jmb.2017.09.013
West, S. A., Griffin, A. S., Gardner, A., and Diggle, S. P. (2006). Social evolution theory for microorganisms. Nat. Rev. Microbiol. 4 (8), 597–607. doi:10.1038/nrmicro1461
Widmann, C., Gibson, S., Jarpe, M. B., and Johnson, G. L. (1999). Mitogen-activated protein kinase: Conservation of a three-kinase module from yeast to human. Physiol. Rev. 79 (1), 143–180. doi:10.1152/physrev.1999.79.1.143
Wiese, K. E., Nusse, R., and van Amerongen, R. (2018). Wnt signalling: Conquering complexity. Development 145 (12), dev165902. doi:10.1242/dev.165902
Wu, Y. C., Franzenburg, S., Ribes, M., and Pita, L. (2022). Wounding response in Porifera (sponges) activates ancestral signaling cascades involved in animal healing, regeneration, and cancer. Sci. Rep. 12 (1), 1307. doi:10.1038/s41598-022-05230-x
Yamanaka, T., and Ohno, S. (2008). Role of lgl/dlg/scribble in the regulation of epithelial junction, polarity and growth. Front. Biosci. 13, 6693–6707. doi:10.2741/3182
Zaessinger, S., Zhou, Y., Bray, S. J., Tapon, N., and Alexandre, D. (2015). Drosophila MAGI interacts with RASSF8 to regulate E-cadherin-based adherens junctions in the developing eye. Development 142 (6), 1102–1112. doi:10.1242/dev.116277
Zhang, X., Wang, P., Gangar, A., Zhang, J., Brennwald, P., TerBush, D., et al. (2005). Lethal giant larvae proteins interact with the exocyst complex and are involved in polarized exocytosis. J. Cell. Biol. 170 (2), 273–283. doi:10.1083/jcb.200502055
Zhang, Z. Q. (2011). Animal biodiversity: An introduction to higher-level classification and taxonomic richness. Zootaxa 3148 (1), 7. doi:10.11646/zootaxa.3148.1.3
Zhu, H., Zhou, Z., Wang, D., Liu, W., and Zhu, H. (2013). Hippo pathway genes developed varied exon numbers and coevolved functional domains in metazoans for species specific growth control. BMC Evol. Biol. 13 (1), 76. doi:10.1186/1471-2148-13-76
Keywords: cell polarity, basal metazoa, multicellularity, asymmetry, signalling, tissue architecture
Citation: Wright BA, Kvansakul M, Schierwater B and Humbert PO (2022) Cell polarity signalling at the birth of multicellularity: What can we learn from the first animals. Front. Cell Dev. Biol. 10:1024489. doi: 10.3389/fcell.2022.1024489
Received: 21 August 2022; Accepted: 31 October 2022;
Published: 24 November 2022.
Edited by:
Alexander Ludwig, Nanyang Technological University, SingaporeReviewed by:
Pedro Martinez, University of Barcelona, SpainDan Bergstralh, University of Rochester, United States
Copyright © 2022 Wright, Kvansakul, Schierwater and Humbert. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Patrick O. Humbert, cC5odW1iZXJ0QGxhdHJvYmUuZWR1LmF1
 Bree A. Wright
Bree A. Wright Marc Kvansakul
Marc Kvansakul Bernd Schierwater
Bernd Schierwater Patrick O. Humbert
Patrick O. Humbert